Abstract
The nascent light-emitting organ of newly hatched juveniles of the Hawaiian sepiolid squid Euprymna scolopes is specifically colonized by cells of Vibrio fischeri that are obtained from the ambient seawater. The mechanisms that promote this specific, cooperative colonization are likely to require a number of bacterial and host-derived factors and activities, only some of which have been described to date. A characteristic of many host-pathogen associations is the presence of bacterial mechanisms that allow attachment to specific tissues. These mechanisms have been well characterized and often involve bacterial fimbriae or outer membrane proteins (OMPs) that act as adhesins, the expression of which has been linked to virulence regulators such as ToxR in Vibrio cholerae. Analogous or even homologous mechanisms are probably operative in the initiation and persistence of cooperative bacterial associations, although considerably less is known about them. We report the presence in V. fischeri of ompU, a gene encoding a 32.5-kDa protein homolog of two other OMPs, OmpU of V. cholerae (50.8% amino acid sequence identity) and OmpL of Photobacterium profundum (45.5% identity). A null mutation introduced into the V. fischeri ompU resulted in the loss of an OMP with an estimated molecular mass of about 34 kDa; genetic complementation of the mutant strain with a DNA fragment containing only the ompU gene restored the production of this protein. The expression of the V. fischeri OmpU was not significantly affected by either (i) iron or phosphate limitation or (ii) a mutation that renders V. fischeri defective in the synthesis of a homolog of the OMP-regulatory protein ToxR. The ompU mutant grew normally in complex nutrient media but was more susceptible to growth inhibition in the presence of either anionic detergents or the antimicrobial peptide protamine sulfate. Interestingly, colonization experiments showed that the ompU null mutant initiated a symbiotic association with juvenile light organ tissue with only about 60% of the effectiveness of the parent strain. When colonization did occur, it proceeded more slowly and resulted in an approximately fourfold-smaller bacterial population. Surprisingly, there was no evidence that in a mixed infection with its parent, the ompU-defective strain had a competitive disadvantage, suggesting that the presence of the parent strain provided a shared compensatory activity. Thus, the OmpU protein appears to play a role in the normal process by which V. fischeri initiates its colonization of the nascent light organ of juvenile squids.
Essentially all multicellular eukaryotes normally exist in association with a suite of cooperative microorganisms, many of which have been recognized to play essential, benign roles in the development and health of their hosts (5). Such interactions can be either consortial, being composed of many microbial species, as in the enteric tracts of all animals (6), or monospecific, as in the root nodules of plants (42) and the light-emitting organs of marine squids and fishes (34). In most of these associations, microorganisms are transmitted horizontally to each new host generation when bacteria present in the ambient environment come in contact with and colonize a particular tissue in their animal and plant hosts. Species specificity is important in these relationships, yet remarkably little is known about how each association is established and persists over the lifetime of the host. One common mechanism of cell-cell recognition is specific attachment structures, which provide the bacterium with the ability to first identify and then remain in close contact with the appropriate target tissue.
Bacteria utilize a number of external structures to attach to specific host tissues and, in some instances, to bring about changes in the biochemical and cellular activity of the host cells to which they adhere (12). Among the best studied of these adhesive structures are pili (fimbriae) and outer membrane proteins (OMPs, or porins), both of which project into the bacterium's environment. The regulation, structure, and specificity of fimbriae and OMPs have been best described in certain pathogenic bacterial species (23, 29, 35), including the human intestinal pathogen Vibrio cholerae (2). In contrast, less is known about the mechanisms by which benign bacteria initiate specific, cooperative, and often obligate associations that can persist throughout the life of the host.
The marine bacterium Vibrio fischeri is the specific symbiont of the light-emitting organ of the sepiolid squid Euprymna scolopes (20). The nascent light organ of a newly hatched E. scolopes juvenile is axenic, but cells of V. fischeri present in the surrounding seawater serve as an inoculum that passes through pores on the surface of the organ and proliferates within epithelium-lined internal crypt spaces (27). The colonization process requires that the bacteria migrate past several different host cell types on their way into the organ (26) and then become securely associated with the microvillar surface of the crypt cells (14). Periodic expulsion of over 95% of the symbiotic bacterial population every morning (33) may further select for closely adhering V. fischeri cells, which become increasingly invested in the microvilli during the first few days after colonization (14). In addition, the presence of the bacteria induces both reversible and irreversible stages in the program of normal light organ development (22, 44), suggesting that signaling is occurring between the bacteria and their host.
The mechanisms by which V. fischeri cells attach to and colonize the light organ tissues of juvenile hosts are just beginning to be described in detail. For example, aggregation of V. fischeri cells in a host-derived mucus-like matrix is an early event that is required for these cells to find and enter the pores that lead to the nascent light organ crypts (27). In addition, evidence exists that mannose residues present on the cells lining the crypts may function as receptors for the colonizing bacteria (21) and that bacterial fimbriae are involved in this process (B. Feliciano and E. G. Ruby, Abstr. 99th Annu. Meet. Am. Soc. Microbiol., abstr. 462, 1999). Reports that the OmpU protein of some strains of V. cholerae might serve in attachment of this pathogen to host tissue (38) suggested to us that an examination of the OMPs of V. fischeri might lead to a better understanding of the role of extracellular structures in the symbiotic colonization of the squid light organ.
MATERIALS AND METHODS
Bacterial strains and media.
V. fischeri strain ESR1, a rifampin-resistant derivative of wild-type strain ES114 (8), was used as the parent strain for all mutant constructions (Table 1). Escherichia coli strain DH5α (4) was the recipient for most cloning experiments, and plasmids were passaged through a dam mutant E. coli strain prior to introduction into V. fischeri by electroporation (45).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Characteristicsa | Reference or source |
|---|---|---|
| V. fischeri | ||
| ES114 | Wild-type isolate | 1 |
| ESR1 | ES114 derivative, spontaneous Rfr | 9 |
| OM3 | ESR1 with a portion of ompU replaced with the Cmr gene | This study |
| KR-tox1 | ESR1 toxR null mutant, Cmr | K. Reich |
| Plasmids | ||
| pBluescriptII | Commercial cloning vector | Stratagene Inc. |
| pCR2.1 | Commercial cloning vector | Invitrogen Inc. |
| pVO8 | V. fischeri cloning vector, mob site, Cmr Emr | 46 |
| pKV36 | Derivative of pUC19, ColE1 ori, Apr Cmr | K. Visick |
| pEVS20 | Derivative of pCR2.1, ColE1 ori, F1 ori, Apr Knr Tpr | E. Stabb |
| pOV2 | 11-kbp BglII fragment of ES114 DNA containing ompU in pVO8 | This study |
| pFA3 | 3.3-kbp HindIII fragment of pOV2 containing ompU in pVO8 | This study |
| pFA5 | pFA3 with ompU PstI/HincII fragment replaced with Cmr gene | This study |
| pFA8 | pFA5 with Tpr gene from pEVS20 inserted in NotI site | This study |
| pFA9 | 2.7-kbp SacI fragment containing intact ES114 ompU in pVO8 | This study |
Rf, rifampin; Em, erythromycin; Kn, kanamycin; Ap, ampicillin.
Unless otherwise noted, V. fischeri strains were grown at 28°C with shaking in one of two nutrient-rich media: SWT (1), which contains 0.5% (wt/vol) tryptone-peptone (Difco, Sparks, Md.), 0.3% (wt/vol) yeast extract, and 0.3% (vol/vol) glycerol in 70% seawater, or LBS (7), which contains 1% (wt/vol) tryptone-peptone, 0.5% (wt/vol) yeast extract, 2% NaCl, and 0.3% (vol/vol) glycerol in 50 mM Tris-HCl (pH 7.5). E. coli strains were grown in Luria-Bertani (LB) broth (4). Agar was added to a concentration of 1.5% to solidify media. Antibiotics were added to media when appropriate to achieve the following final concentrations: ampicillin, 100 μg/ml; chloramphenicol, 25 μg/ml for E. coli and 2.5 μg/ml for V. fischeri; erythromycin, 150 μg/ml for E. coli and 5 μg/ml for V. fischeri; trimethoprim, 10 μg/ml for E. coli and 2 μg/ml for V. fischeri.
In certain cases, cultures of V. fischeri cells were grown in media that were designed to be growth limiting for either phosphate, nitrogen, or iron. Phosphate-limited growth was achieved in an artificial-seawater-based minimal medium containing 300 mM NaCl, 50 mM MgSO4 · 7H2O, 10 mM CaCl2 · 2H2O, 10 mM KCl, 10 mM NH4Cl, 0.01 mM FeSO4, 50 mM Tris-HCl (pH 7.5), and 20 mM ribose or other carbon source (1) to which no phosphate was added. A nitrogen-limiting variation of this medium had the same composition except that the NH4Cl was omitted and K2HPO4 was added to a final concentration of 0.33 mM. Iron limitation was achieved in a medium containing both NH4Cl and K2HPO4 by the addition of the iron chelator ethylenediamine-di-o-hydroxyphenylacetic acid (EDDHA) to a concentration of 30 μM (10).
Growth yields were also determined in LBS medium to which sodium dodecyl sulfate (SDS), the antimicrobial peptide protamine sulfate, sodium deoxycholate, or bovine bile (Sigma Chemical Co., St. Louis, Mo.) was added (31). Medium containing bile was sterilized by passage through a 0.45-μm-pore-size membrane filter. Because this medium had a dark brown color that made it impossible to estimate bacterial concentrations by optical density, cell yields were obtained by determining the CFU per milliliter present in the culture after 24 h of growth on SWT agar medium. In the other media, final growth yields were determined by measuring the optical density of the culture at a wavelength of 600 nm (OD600).
Protein analyses.
Cellular protein extracts were obtained from cultures of V. fischeri cells grown to mid-exponential phase (OD of approximately 0.2) in SWT medium. Cells were harvested by centrifugation and washed with seawater. Total soluble proteins were extracted from washed cell pellets that were resuspended in a cold lysing buffer containing 50 mM Tris-HCl (pH 7.9), 50 mM EDTA, 15% (wt/vol) sucrose, and lysozyme (final concentration, 0.5 mg/ml). This suspension was incubated for 30 min on ice and centrifuged for 5 min at 12,000 × g. OMP-enriched fractions were obtained from the resulting pellet using a 1% N-lauroylsarcosine detergent extraction as previously described (19). Soluble proteins were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and visualized by staining with Coomassie brilliant blue. Western blot analyses using a Photobacterium profundum OmpL antibody were performed as described by Welch and Bartlett (47).
Cloning procedures.
V. fischeri DNA was manipulated using previously described methods (10, 39, 46). DNA fragments obtained by restriction endonuclease digestion were separated by agarose gel electrophoresis, and the desired fragments were extracted from gel slices using GeneClean (Bio 101, Inc., Vista, Calif.). T4 DNA ligase was used to join two fragments together, the ligated fragments were transformed into E. coli DH5α cells made competent by CaCl2 treatment (4), and plasmid-carrying strains were isolated on selective antibiotic-containing media. Enzymes were obtained from Promega, Inc. (Madison, Wis.) or New England Biolabs (Beverly, Mass.).
Squid colonization assays.
Juveniles of E. scolopes were inoculated within 4 h of hatching with V. fischeri strains as described previously (32). Briefly, individual squids in vials containing 4 ml of seawater were exposed for 3 h to an inoculum of either the ompU mutant or its parent strain. After this inoculation, the animals were transferred to symbiont-free seawater and maintained for up to 3 days. Measurements of the luminescence of the juvenile squid were performed using a Turner 20/20 luminometer (Turner, Sunnyvale, Calif.) and used as an indication of successful colonization (33). At specific times following inoculation, the juveniles were homogenized, and dilutions of the homogenates were spread on SWT agar medium to determine the number of CFU in the light organ (1). To examine whether the ompU mutant had a competitive disadvantage compared to the parent strain, juvenile squids were inoculated with a 1:1 mixture of the mutant and the parent, and the proportion of the resulting symbiotic population was determined by plating light organ homogenates on antibiotic-containing media that differentiated between the two strains (46).
In some experiments, V. fischeri cells were exposed to either preimmune serum or antiserum directed against the P. profundum OmpL protein prior to inoculation (47). The bacterial inoculum was then added to seawater containing the juvenile squids, and the progress of the colonization was monitored as described above.
Nucleotide sequence accession number.
The GenBank nucleotide accession number for the V. fischeri ompU gene sequence is AY050511.
RESULTS
OMPs of V. fischeri.
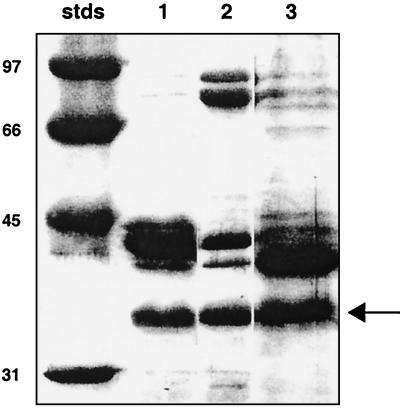
A characterization of the presence and regulation of OMPs in V. fischeri cells was begun by growing the bacteria in media limited for either iron or phosphate, two nutrients that often require transport through specific porins. SDS-PAGE analysis of OMPs present in cell extracts of V. fischeri strain ES114 (or its derivative, ESR1) grown in SWT medium revealed three major protein bands with molecular masses of approximately 34, 40, and 41 kDa (Fig. 1). Two additional minor proteins (approximately 74 and 88 kDa) were also present and were significantly induced in cells cultured in medium containing the iron chelator EDDHA, suggesting that they might be in the class of iron siderophore porins described for other Vibrio species (17). Similarly, growth in a phosphate-limited medium resulted in enhancement of the 40-kDa OMP band relative to the 41-kDa band (Fig. 1). There was no indication that growth in any of these media resulted in a significant change in the intensity of the 34-kDa band.
FIG. 1.
SDS-polyacrylamide gel of OMPs isolated from cells of V. fischeri strain ESR1 grown under different culture conditions. Lane 1, cells grown in nutrient medium SWT (7.2 μg of protein loaded); lane 2, cells grown in SWT under iron limitation (6.5 μg of protein loaded); lane 3, cells grown in a minimal medium under phosphate limitation (8.0 μg of protein loaded). Molecular mass standards (stds) are indicated. The arrow indicates the 34-kDa band.
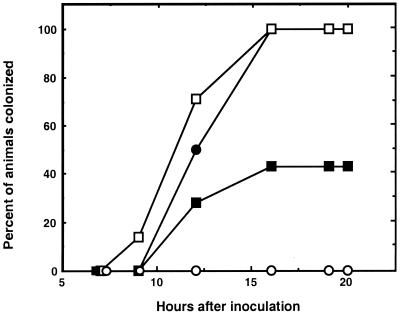
Western blot analysis of V. fischeri OMP gels revealed that a protein with an apparent molecular mass of 34 kDa had epitopes that reacted with antiserum raised against the OmpL protein of P. profundum (data not shown) (48). Experiments were performed to determine whether treatment of V. fischeri cells with this OmpL antiserum prior to using them to inoculate juvenile E. scolopes squids would affect their efficiency of colonization. Analysis of several trials showed that pretreatment with the OmpL antiserum but not preimmune serum (or an antiserum made to another protein) resulted in an extent of colonization that was approximately 60% less efficient (Fig. 2). Together these data suggested that (i) V. fischeri cells had an OMP that was related to P. profundum OmpL, a homolog of the OmpU of V. cholerae (38) and (ii) this V. fischeri OMP might play a role in symbiotic colonization.
FIG. 2.
Inhibition of light organ colonization by V. fischeri cells treated with an antiserum directed against P. profundum OmpL, a homolog of V. fischeri OmpU. Groups of between 7 and 10 newly hatched, uncolonized juveniles of E. scolopes were individually inoculated by placing them for 3 h in 5 ml of seawater containing approximately 104 cells of V. fischeri ESR1 that were either untreated (●) or treated with OmpL antiserum (▪) or preimmune serum (□). Control animals (○) were incubated in seawater without added V. fischeri cells. The data shown are representative of those obtained in four separate experiments.
Isolation of V. fischeri ompU homolog.
To identify the presence of a V. cholerae ompU homolog in the genome of V. fischeri, we first aligned the V. cholerae gene (37) and the homologous ompL gene from P. profundum (48). Based on a conserved sequence at the C terminus, two oligonucleotide PCR primers, OMP3 (5′-GACGCTACTTACTACTTC-3′) and OMP4 (5′-AAGTCGTAACGTACACC-3′) that would amplify a predicted 134-bp fragment corresponding to nucleotide positions 928 to 1061 in the V. cholerae ompU gene were designed. PCR amplification using these primers and V. fischeri chromosomal DNA as the target resulted in a 137-bp fragment that, when sequenced, aligned with 80% identity to the expected corresponding region of the V. cholerae ompU sequence.
This PCR product was used to probe a Southern blot of a gel of V. fischeri chromosomal DNA digested with one of a number of restriction enzymes, and a BglII band at about 12 kb was detected. A size-fractionated library of BglII fragments of V. fischeri DNA cloned into the vector pVO8 was constructed, and clones of these plasmids carried in E. coli were subsequently probed. Two positive clones were obtained, and both produced a 137-bp PCR fragment with the expected DNA sequence. The 11.6-kbp BglII fragment in plasmid pOV2 was further subcloned in pBluescriptII, and a 3.3-kbp HindIII fragment (carried in a plasmid designated pFA3) was found that contained the 137-bp ompU-like sequence. Analysis of the sequence of this fragment revealed the presence of one complete open reading frame (ORF1) and a second, partial one (ORF2). Analysis of the region upstream of ORF1 revealed putative −10 and −35 promoter site sequences, as well as a possible ribosome-binding site (data not shown). Interestingly, repeated attempts to subclone a larger, 8.6-kbp ClaI fragment carrying the ompU-like sequence and a larger amount of flanking DNA were unsuccessful, perhaps because this fragment encoded a gene that was lethal to E. coli when carried in the multicopy pBluescriptII vector.
Alignment of V. fischeri ompU gene homolog with other OMP genes.
As expected, the complete ORF1 aligned well with the V. cholerae ompU and P. profundum ompL genes. There was 50.8 and 45.5% identity between the deduced amino acid sequence of the V. fischeri ORF1 and those of V. cholerae OmpU and P. profundum OmpL, respectively, leading us to propose that ORF1 should be designated the V. fischeri ompU gene. The N terminus of the deduced V. fischeri OmpU protein (Fig. 3) contains a 21-amino-acid leader sequence that is 52% identical to the processed 21-amino-acid signal peptide of P. profundum OmpL (48). Based on the assumption that this peptide sequence is also processed in V. fischeri, the mature OmpU protein of V. fischeri would be 300 residues long, with a deduced molecular mass of 32.5 kDa. This molecular mass is consistent with the OMP SDS-polyacrylamide gel band at approximately 34 kDa (Fig. 1). Twenty-eight base pairs downstream of the stop codon of ompU is a putative rho-independent terminator, beyond which is ORF2, which encodes a partial protein sequence with highest identity to the E. coli penicillin-binding protein 4 gene, called dacB. The V. cholerae ompU gene is also located directly upstream of a dacB gene homolog (11).
FIG. 3.
Alignment of deduced amino acid sequence encoded by the V. fischeri (VFIS) ompU gene with those of V. cholerae (VCHO) ompU and P. profundum (PPRO) ompL genes. Residues that are identical to those in the V. fischeri sequence are indicated by an asterisk (*), and gaps that were inserted to optimize alignment are indicated by periods.
Expression of OmpU in V. fischeri.
Experiments were conducted to determine whether OmpU was differentially expressed during growth in either minimal medium or the tryptone-based SWT medium. Cells of V. fischeri strain ESR1 were harvested at the early, middle, and late phases of exponential growth, and SDS-polyacrylamide protein gels were run to examine the relative intensity of the OmpU 34-kDa band. While generally produced to the same level in all media tested, this band appeared somewhat more prominent in cells grown to late exponential phase in SWT medium (data not shown). Culturing the cells under conditions that were limited for iron, phosphate (Fig. 1), or oxygen (i.e., anaerobic culture) had no significant effect on the intensity of the 34-kDa band.
Construction of V. fischeri ompU null mutant.
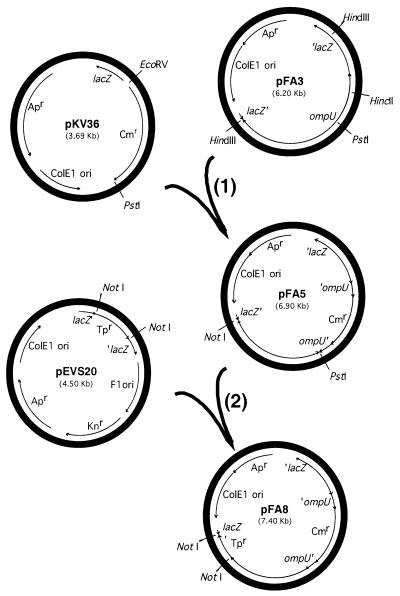
To determine the possible function(s) of OmpU in V. fischeri, we constructed a mutation in the ompU gene by marker exchange. Briefly, a 1.1-kbp PstI/EcoRV fragment from pKV36 (provided by K. Visick), which contains a chloramphenicol resistance (Cmr) gene, was used to replace a 560-bp PstI/HincII fragment within the ompU coding region in pFA3 to form pFA5 (Fig. 4, step 1). A trimethoprim resistance (Tpr) gene was obtained as a NotI fragment from pEVS20 (provided by E. Stabb) and inserted into the NotI site in pFA5, creating pFA8 (Fig. 4, step 2). pFA8 was electroporated (45) into V. fischeri ESR1, and Cmr clones were selected. Because the ColE1 origin does not replicate well in V. fischeri, plasmid pFA8 is lost over time in the absence of selection. Clones were passaged several times on medium without antibiotic to select cells that had integrated the plasmid by homologous recombination. Such clones were then serially passaged on chloramphenicol-containing medium and periodically patched onto trimethoprim-containing medium to screen for a second recombinational event that had led to excision of the plasmid (resulting in the loss of Tpr) and replacement of the wild-type copy of ompU with the insertionally inactivated one. One of a few such Cmr and Tps clones obtained was designated V. fischeri strain OM3. Gene replacement in OM3 was confirmed by PCR amplification of the ompU locus from chromosomal DNA. Only a single band was produced, and it had the size expected for the disrupted ompU gene rather than the full-length wild-type gene.
FIG. 4.
Two-step scheme for constructing an insertional mutation in the V. fischeri ompU gene. In step 1, a Cmr cassette is used to replace an internal portion of the ompU gene in pFA3, resulting in pFA5. In step 2, to facilitate screening of recombinants, a Tpr cassette is also added to pFA5, yielding pFA8. This plasmid was then introduced into V. fischeri to produce an ompU mutation by double recombination. Abbreviations: lacZ, β-galactosidase gene; ColE1 ori, ColE1 origin of replication; F1 ori, F1 origin of replication; Apr, ampicillin resistance gene; Cmr, chloramphenicol resistance gene; Knr, kanamycin resistance gene; Tpr, trimethoprim resistance gene.
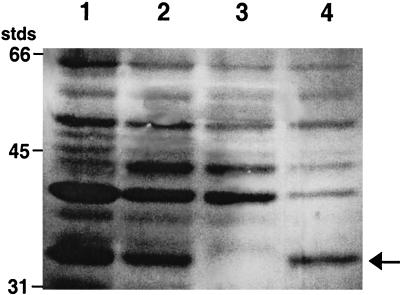
When proteins extracted from strain OM3 were separated by SDS-PAGE, the normally present 34-kDa band was missing (Fig. 5). As predicted, this defect could be complemented in trans by introducing pFA9 into strain OM3, which restored a wild-type copy of ompU to the mutant strain and resulted in the reappearance of the 34-kDa band (Fig. 5).
FIG. 5.
SDS-polyacrylamide gel of proteins isolated from different strains of V. fischeri. Cultures were grown at room temperature in SWT medium for 14 to 18 h with shaking. Lane 1, KR-tox1 (toxR omp+); lane 2, ESR1 (parent strain, ompU+); lane 3, OM3 (ompU gene replacement mutant); lane 4, OM3 pFA9 (OM3 complemented with pFA9, ompU+). Positions of molecular mass standards (stds) are indicated. The arrow indicates the 34-kDa OmpU protein.
Unlike in V. cholerae (32), expression of ompU in V. fischeri appears normal in a toxR null mutant strain (Fig. 5). However, the V. fischeri toxR mutant does seem to have reduced expression of its 41-kDa OMP relative to the 40-kDa one. Interestingly, this response is similar to that observed in wild-type cells that are grown under conditions of phosphate limitation (Fig. 1).
Growth characteristics of V. fischeri ompU mutant in culture.
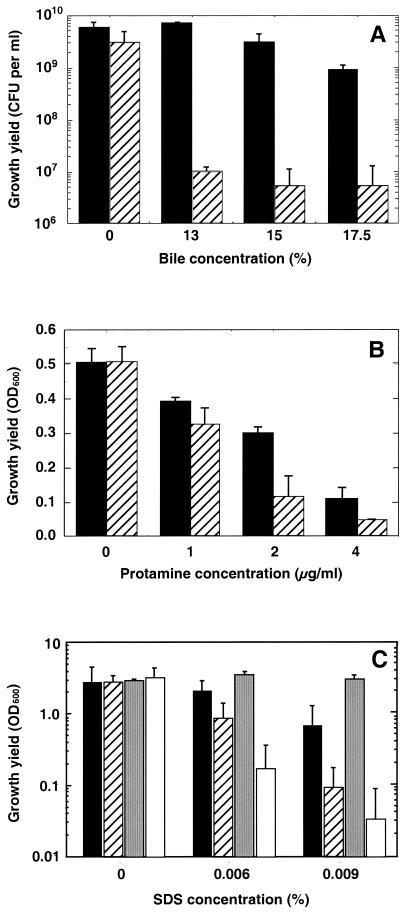
When grown in either SWT or LBS, which are rich nutrient media containing tryptic peptides and yeast extract, OM3 and its parent ESR1 grew at the same rate and to about the same cell yield (Fig. 6 and data not shown), suggesting that the absence of OmpU creates no significant defect in general bacterial metabolism. However, growth of strain OM3 is restricted by a significantly lower concentration of bile salts, the antimicrobial peptide protamine sulfate, or SDS compared to the ompU+ parent strain (Fig. 6). A similar growth defect was observed with cells of OM3 grown in the presence of deoxycholate (data not shown). This defect in OM3 could be complemented by pFA9, which carries an intact ompU gene (Table 1), but not by the vector control, pVO8 (Fig. 6C). Perhaps the apparently increased SDS resistance of the complemented OM3 strain reflected the increased gene dosage of ompU resulting from carriage of the multicopy pFA9 plasmid. Taken together, these data suggest that OmpU is required to maintain normal cell integrity.
FIG. 6.
Effects of bile, protamine, and SDS on the growth of strains OM3 and ESR1. Cultures of the ompU mutant strain OM3 (hatched bars) or its parent, ESR1 (solid bars), were inoculated into LBS medium containing different concentrations of bovine bile, protamine sulfate, or SDS. (A) After 24 h of growth at 28°C, dilutions of the bile-containing cultures were spread on LBS agar medium, and the numbers of CFU in the cultures were calculated. The value of each bar is the average of two separate experiments. Similarly, LBS cultures containing either protamine sulfate (B) or SDS (C) were incubated for 24 h at 28°C, and the resulting relative cell density was estimated spectrophotometrically. Cultures of strain OM3 carrying either a wild-type copy of ompU on pFA9 (gray bars) or the vector control, pVO8 (open bars), were also tested on SDS-containing LBS medium. Error bars indicate standard errors of the means.
Colonization characteristics of V. fischeri ompU mutant.
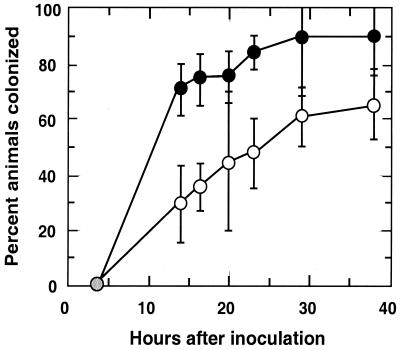
Experiments were conducted to determine whether the mutation in ompU resulted in a symbiotic defect in V. fischeri. A comparison of the light organ colonization efficiency of strain OM3 relative to its parent, ESR1, indicated that when presented at a concentration of between 1,000 and 2,000 CFU/ml of seawater, the mutant strain was less effective at initiating the symbiosis (Fig. 7). Specifically, while about 70% of the animals exposed to strain ESR1 were colonized at 14 h postinoculation, fewer than 30% of the animals exposed to the ompU mutant were infected. Similarly, by 38 h about 90% of the ESR1-exposed animals were colonized, while only 65% of the mutant-exposed animals were.
FIG. 7.
Colonization efficiency of V. fischeri ompU mutant strain. Groups of newly hatched, uncolonized juvenile squids were placed into seawater containing between 700 and 1,500 cells of either the ompU mutant, OM3, or its parent strain, ESR1, per ml for 3 h. In each experiment the OM3 inoculum was determined to have at least as many cells as the ESR1 inoculum. Starting 14 h after inoculation and continuing at different intervals, the animals were assayed for the production of luminescence, an indication of successful colonization. The percentage of animals in the OM3-inoculated group (○) and the ESR1-inoculated group (●) that produced luminescence was determined. Data points are the means of three separate experiments, and error bars indicate the standard errors of the means.
In another set of experiments, the extent of colonization was determined in the animals that had become symbiotically infected with either strain ESR1 or strain OM3 (Table 2). In all cases, the ompU mutant strain was able to maintain only about one-quarter as many bacteria in the light organ as ESR1. Interestingly, experiments in which animals were exposed to an inoculum containing equal numbers of both ESR1 and OM3 did not support the notion that the mutant was at a competitive disadvantage. Specifically, in four trials in which animals were examined at 24 and 48 h postinoculation, the average proportion of mutant cells in the symbiotic colonization was not significantly different from the proportion in the inoculum (Table 3).
TABLE 2.
Comparison between light organ colonization levels achieved by V. fischeri ompU mutant OM3 and its parent ESR1 at different times after inoculationa
| Time postinoculation (h) | 105 CFU/light organ
|
Ratio of colonization effectiveness (CFU of ESR1/CFU of OM3) | |
|---|---|---|---|
| ESR1 | OM3 | ||
| 24 | 5.1 | 0.9 | 5.7 |
| 39 | 4.2 | 2.4 | 1.8 |
| 41 | 4.2 | 0.4 | 9.3 |
| 60 | 2.0 | 1.0 | 2.0 |
| 72 | 7.8 | 3.5 | 2.2 |
| Mean | 4.2 | ||
In each of the five separate experiments, the number of CFU present in the inoculum of strain OM3 was between 1.2 and 1.6 times greater than that of strain ESR1. The total concentration of V. fischeri cells (OM3 or ESR1) in the inoculum of each experiment was between 2,000 and 4,700 CFU/ml of seawater.
TABLE 3.
Colonization effectiveness of ompU mutant strain in competition with the wild typea
| Expt | % Strain OM3 (ompU)
|
Ratiob | |
|---|---|---|---|
| In inoculum | In colonization | ||
| 1 | 63 | 67 | 0.94 |
| 2 | 54 | 41 | 1.23 |
| 3 | 65 | 80 | 0.73 |
| 4 | 47 | 52 | 0.90 |
| Mean ± SD | 57 ± 8 | 60 ± 17 | 0.95 ± 0.21 |
In each of four separate colonization experiments, an inoculum was prepared by mixing cells of strain OM3 and its wild-type parent in a proportion of approximately 1:1. The percentage of OM3 cells in this inoculum and in the resulting symbiotic population 24 to 48 h after inoculation was determined as described in the text.
Ratio = percent in inoculum/percent in colonization.
DISCUSSION
Bacterial OMPs constitute a class of cell envelope proteins whose members are involved in an array of diverse surface-mediated phenomena that include attachment, nutrient acquisition, and host-cell signal transduction (13, 35). In the genus Vibrio, OMPs function as porins in iron, phosphate, and sugar accumulation (15, 19, 41) and in attachment to inanimate and animate surfaces (37, 40). Considerable emphasis has been placed on understanding the role of Vibrio OMPs in pathogenesis, yet important issues have remained unresolved. For instance, in V. cholerae strain O1, OmpU has been reported to be involved in bacterial adhesion to host tissue (37), while other evidence suggests it is not required for adherence to the intestinal epithelium of rabbits (24). We report here the presence in V. fischeri of an ompU homolog that encodes a 32.5-kDa OMP. A V. fischeri ompU null mutant grew normally in both complex and minimal nutrient media; however, it was more susceptible to growth inhibition in the presence of either anionic detergents or the cationic antimicrobial peptide protamine. In addition, the mutant had a decreased ability to colonize the light organ of juvenile E. scolopes squid except when it was presented together with its ompU+ parent. This work constitutes the first report that ties the presence of a specific bacterial OMP to the initiation of a successful cooperative association with an animal host.
The V. fischeri ompU gene has a number of characteristics in common with its homologs in V. cholerae and P. profundum, including a putative N-terminal processing site and similar adjacent genetic loci. However, there are several differences as well. First, while these two other bacterial species express a second OMP whose abundance is regulated in an inverse manner (e.g., when V. cholerae OmpU expression is depressed, OmpT is increased, and vice versa) (30, 48), no analogous protein was detected in V. fischeri (Fig. 5). A second difference is that the regulation of expression of the V. fischeri OmpU was distinct from that reported for either OmpU in V. cholerae or OmpL in P. profundum. For instance, unlike these two OMPs (30), expression of the V. fischeri OmpU protein is not significantly affected by a null mutation in the porin regulatory protein ToxR (Fig. 5). This difference may not be too surprising, since other Vibrio species have also shown variations in OMP numbers and regulation (16, 17, 19, 31). Similarly, during the evolution of some but not all Vibrio species, a porin regulon has even been recruited to control the expression of horizontally transferred virulence determinants (30). Taken together, these findings suggest that there has been considerable functional and adaptive divergence in the biology of OMPs within the Vibrionaceae.
The fact that the expression of certain Vibrio OMPs responds to physiological changes in the environment has long been recognized (3, 24) and has indicated a role for these proteins in (i) the transport of specific nutrients and (ii) protecting the bacterium from disruptive chemical agents. An examination of the growth of the V. fischeri ompU mutant in media of different compositions did not identify any nutrients (e.g., peptides, inorganic iron, or phosphate) whose transport required OmpU. In contrast, the V. fischeri ompU mutants expressed an increased sensitivity to bile and other detergents (Fig. 6) that was similar to what has been reported for a V. cholerae ompU mutant (31). The absence of an obvious chemical or structural similarity between the anionic detergents and the cationic protamine further suggests that the effects of these agents may not be due to a change in the mutant of a specific structure in the outer membrane. Thus, while the functional basis for the V. fischeri ompU mutant's increased sensitivity to cell membrane-disrupting agents remains unclear, it may simply indicate that this protein plays a significant role in the integrity of the outer membrane. The fact that 30 to 60% of the OMPs in V. cholerae are OmpU (2) indicates the potential importance of this protein and provides support for this hypothesis.
Perhaps the most intriguing phenotype of the V. fischeri ompU mutant is its inability to colonize the squid light organ normally. The process by which juvenile E. scolopes become infected is typically a well-programmed and predictable one (22). If they are present in the ambient seawater, V. fischeri cells become aggregated on mucous strands emanating from the nascent light organ of newly hatched squid (27). After a period of 2 or 3 h, these cells begin to move out of the aggregates and towards the pores that lead to epithelium-lined crypts inside the nascent light organ. Only V. fischeri cells are able to survive and complete this process, and within 5 h they have begun to proliferate into a symbiotic population of several hundred thousand (44). The progression of this process is remarkably consistent and is reflected in the onset and level of luminescence emitted by the squid (33). The ompU mutant, while able to colonize the light organ, is defective in at least two aspects of the process. First, it initiates the colonization significantly more slowly and with less than 70% of the effectiveness of its ompU+ parent (Fig. 7). Interestingly, the delay they exhibit is similar to that seen when wild-type cells are treated with an antibody that reacts with OmpU (Fig. 2), suggesting that the inability of the cell to present this protein on its surface is responsible for this initiation defect.
The second defect expressed by the mutant is that, on average, the population level it can achieve in the light organ is only 20 to 25% of that of the parent strain (Table 2). The reason for this reduced colonization effectiveness is unknown, but it may indicate either that the cells have a diminished capacity to survive some condition within the crypts or, alternatively, that the host reacts to the mutant by providing fewer nutrients to support the proliferation of the bacterial population (9).
A method that has proven useful for examining the basis of symbiotic defects in V. fischeri mutants has been to determine their ability to compete with wild-type cells in mixed-colonization experiments. Previous work has suggested that such competition assays reveal subtle defects in mutants that, as a monoculture, would otherwise colonize to normal levels (43, 46). Thus, it was surprising to see no evidence of a competitive disadvantage in the ompU mutant. Instead, when it was coinoculated with its parent, there were twice as many cells of the mutant present in the light organ (average = 3.4 × 105) than when it was the sole strain in the inoculum (average = 1.6 × 105; Table 2). This result suggests that the presence of the parent strain provided an activity that complemented the ompU defect in the mutant, i.e., both the mutant and parent shared the benefit resulting from the activity. The basis for this effect is as yet unknown, but we hypothesize that one way in which OmpU may function in the symbiosis is to attach to a receptor on the host epithelium. This attachment may initiate a host response, such as provision of nutrients, that is required to support a normal level of symbiotic colonization. Support for this hypothesis comes from reports of bacterial OMPs that bind to host tissue and specifically modify its activities. Examples of such OMPs include Opa and Opc of Neisseria spp. (18) and OmpA-like proteins in Acinetobacter spp. (28) and members of the family Enterobacteriaceae (36). Future investigations will be focused on testing the hypothesis in V. fischeri and identifying the mechanism(s) underlying the role of OmpU in promoting benign colonization by V. fischeri.
ACKNOWLEDGMENTS
Early stages of this work were performed with the help and guidance of T. Welch and D. Bartlett. We also thank D. Bartlett for providing P. profundum OmpL antiserum and J. Kaper for providing V. cholerae OmpU antiserum. K. Reich donated the toxR null mutant of V. fischeri ESR1, and J. Sanders provided technical assistance. Data on outer membrane protein patterns in V. fischeri were generously provided by S. Hensey and M. McFall-Ngai. D. Millikan, E. Stabb, and K. Visick provided insightful comments on both experimentation and the manuscript.
This work was supported in part by National Institutes of Health grant RR-12294 to E.G.R. and M. McFall-Ngai and by National Science Foundation grant IBN-9904601 to M. McFall-Ngai and E.G.R. F.A. was supported by a grant from the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Boettcher K J, Ruby E G. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J Bacteriol. 1990;172:3701–3706. doi: 10.1128/jb.172.7.3701-3706.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chakrabarti S R, Chaudhuri K, Sen K, Das J. Porins of Vibrio cholerae: purification and characterization of OmpU. J Bacteriol. 1996;178:524–530. doi: 10.1128/jb.178.2.524-530.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davey M L, Hancock R E, Mutharia L M. Influence of culture conditions on the expression of the 40-kilodalton porin protein of Vibrio anguillarum serotype O2. Appl Environ Microbiol. 1998;64:138–146. doi: 10.1128/aem.64.1.138-146.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davis R W, Botstein D, Roth J R. Advanced bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1980. [Google Scholar]
- 5.Douglas A E. Symbiotic interactions. Oxford, England: Oxford Science Publications; 1994. [Google Scholar]
- 6.Duncan H E, Edberg S C. Host-microbe interaction in the gastrointestinal tract. Crit Rev Microbiol. 1995;21:85–100. doi: 10.3109/10408419509113535. [DOI] [PubMed] [Google Scholar]
- 7.Dunlap P V. Regulation of luminescence by cyclic AMP in cya-like and crp-like mutants of Vibrio fischeri. J Bacteriol. 1989;171:1199–1202. doi: 10.1128/jb.171.2.1199-1202.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graf J, Dunlap P V, Ruby E G. Effect of transposon-induced motility mutations on colonization of the host light organ by Vibrio fischeri. J Bacteriol. 1994;176:6986–6991. doi: 10.1128/jb.176.22.6986-6991.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graf J, Ruby E G. Host-derived amino acids support the proliferation of symbiotic bacteria. Proc Natl Acad Sci USA. 1998;95:1818–1822. doi: 10.1073/pnas.95.4.1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graf J, Ruby E G. Novel effects of a transposon insertion in the Vibrio fischeri glnD gene: defects in iron uptake and symbiotic persistence in addition to nitrogen utilization. Mol Microbiol. 2000;37:168–179. doi: 10.1046/j.1365-2958.2000.01984.x. [DOI] [PubMed] [Google Scholar]
- 11.Heidelberg J F, Eisen J A, Nelson W C, Clayton R A, Gwinn M L, Dodson R J, Haft D H, Hickey E K, Peterson J D, Umayam L, Gill S R, Nelson K E, Read T D, Tettelin H, Richardson D, Ermolaeva M D, Vamathevan J, Bass S, Qin H, Dragol I, Sellers P, McDonald L, Utterback T, Fleishmann R D, Nierman W C, White O, Salzberg S L, Smith H O, Colwell R R, Mekalanos J J, Venter J C, Fraser C M. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henderson B, Wilson M, McNab R, Lax A J. Cellular microbiology: bacteria-host interactions in health and disease. New York, N.Y: John Wiley and Sons; 1999. [Google Scholar]
- 13.Koebnik R, Locher K P, Van Gelder P. Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol Microbiol. 2000;37:239–253. doi: 10.1046/j.1365-2958.2000.01983.x. [DOI] [PubMed] [Google Scholar]
- 14.Lamarcq L H, McFall-Ngai M J. Induction of a gradual, reversible morphogenesis of its host's epithelial brush border by Vibrio fischeri. Infect Immun. 1998;66:777–785. doi: 10.1128/iai.66.2.777-785.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lang H, Jonson G, Holmgren J, Palva E T. The maltose regulon of Vibrio cholerae affects production and secretion of virulence factors. Infect Immun. 1994;62:4781–4788. doi: 10.1128/iai.62.11.4781-4788.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li C C, Crawford J A, DiRita V J, Kaper J B. Molecular cloning and transcriptional regulation of ompT, a ToxR-repressed gene in Vibrio cholerae. Mol Microbiol. 2000;35:189–203. doi: 10.1046/j.1365-2958.2000.01699.x. [DOI] [PubMed] [Google Scholar]
- 17.Litwin C M, Byrne B L. Cloning and characterization of an outer membrane protein of Vibrio vulnificus required for heme utilization: regulation of expression and determination of the gene sequence. Infect Immun. 1998;66:3134–3141. doi: 10.1128/iai.66.7.3134-3141.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lorenzen D R, Gunther D, Pandit J, Rudel T, Brandt E, Meyer T. Neisseria gonorrhoeae porin modifies the oxidative burst of human professional phagocytes. Infect Immun. 2000;68:6215–6222. doi: 10.1128/iai.68.11.6215-6222.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCarter L L, Silverman M. Phosphate regulation of gene expression in Vibrio parahaemolyticus. J Bacteriol. 1987;169:3441–3449. doi: 10.1128/jb.169.8.3441-3449.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McFall-Ngai M J. Consequences of evolving with bacterial symbionts: insights from the squid-vibrio associations. Annu Rev Ecol Syst. 1999;30:235–256. [Google Scholar]
- 21.McFall-Ngai M J, Brennan C, Weiss V, Lamarcq L. Mannose adhesin-glycan interactions in the Euprymna scolopes-Vibrio fischeri symbiosis. In: Le Gal Y, Halvorson H, editors. New developments in marine biotechnology. New York, N.Y: Plenum Press; 1998. pp. 273–276. [Google Scholar]
- 22.McFall-Ngai M J, Ruby E G. Developmental biology in marine invertebrate symbioses. Curr Opin Microbiol. 2000;3:603–607. doi: 10.1016/s1369-5274(00)00147-8. [DOI] [PubMed] [Google Scholar]
- 23.Merz A J, So M. Interactions of pathogenic neisseriae with epithelial cell membranes. Annu Rev Cell Dev Biol. 2000;16:423–457. doi: 10.1146/annurev.cellbio.16.1.423. [DOI] [PubMed] [Google Scholar]
- 24.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakasone N, Iwanaga M. Characterization of outer membrane protein OmpU of Vibrio cholerae O1. Infect Immun. 1998;66:4726–4728. doi: 10.1128/iai.66.10.4726-4728.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nyholm S V, McFall-Ngai M J. Sampling the microenvironment of the Euprymna scolopes light organ: description of a population of host cells with the bacterial symbiont Vibrio fischeri. Biol Bull. 1998;195:89–97. doi: 10.2307/1542815. [DOI] [PubMed] [Google Scholar]
- 27.Nyholm S V, Stabb E V, Ruby E G, McFall-Ngai M J. Establishment of an animal-bacterial association: recruiting symbiotic vibrios from the environment. Proc Natl Acad Sci USA. 2000;97:10231–10235. doi: 10.1073/pnas.97.18.10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ofori-Darko E, Zavros Y, Rieder G, Tarle S A, Van Antwerp M, Merchant J L. An OmpA-like protein from Acinetobacter spp. stimulates gastrin and interleukin-8 promoters. Infect Immun. 2000;68:3657–3666. doi: 10.1128/iai.68.6.3657-3666.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Payne S H, Lawlor K M. Molecular studies on iron acquisition by non-Escherichia coli species. In: Iglewski B H, Clark V L, editors. Molecular basis of bacterial pathogenesis. New York, N.Y: Academic Press; 1990. pp. 225–248. [Google Scholar]
- 30.Provenzano D, Klose K E. Altered expression of the ToxR-regulated porins OmpU and OmpT diminishes Vibrio cholerae bile resistance, virulence factor expression, and intestinal colonization. Proc Natl Acad Sci USA. 2000;97:10220–10224. doi: 10.1073/pnas.170219997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Provenzano D, Schumacher D A, Barker J L, Klose K E. The virulence regulatory protein ToxR mediates enhanced bile resistance in Vibrio cholerae and other pathogenic Vibrio species. Infect Immun. 2000;68:1491–1497. doi: 10.1128/iai.68.3.1491-1497.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruby E G. Lessons from a cooperative, bacterial-animal association: the Vibrio fischeri-Euprymna scolopes light organ symbiosis. Annu Rev Microbiol. 1996;50:591–624. doi: 10.1146/annurev.micro.50.1.591. [DOI] [PubMed] [Google Scholar]
- 33.Ruby E G, Asato L M. Growth and flagellation of Vibrio fischeri during initiation of the sepiolid squid light organ symbiosis. Arch Microbiol. 1993;159:160–167. doi: 10.1007/BF00250277. [DOI] [PubMed] [Google Scholar]
- 34.Ruby E G, Lee K H. The Vibrio fischeri-Euprymna scolopes light organ association: current ecological paradigms. Appl Environ Microbiol. 1998;64:805–812. doi: 10.1128/aem.64.3.805-812.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soto G E, Hultgren S J. Bacterial adhesins: common themes and variations in architecture and assembly. J Bacteriol. 1999;181:1059–1071. doi: 10.1128/jb.181.4.1059-1071.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soulas C, Baussant T, Aubry J P, Delneste Y, Barillat N, Caron G, Renno T, Bonnefoy J Y, Jeannin P. Outer membrane protein A (OmpA) binds to and activates human macrophages. J Immunol. 2000;165:2335–2340. doi: 10.4049/jimmunol.165.5.2335. [DOI] [PubMed] [Google Scholar]
- 37.Sperandio V, Bailey C, Giron J A, DiRita V J, Silveira W D, Vettore A L, Kaper J B. Cloning and characterization of the gene encoding the OmpU outer membrane protein of Vibrio cholerae. Infect Immun. 1996;64:5406–5409. doi: 10.1128/iai.64.12.5406-5409.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sperandio V, Giron J, Silveira W D, Kaper J B. The OmpU outer membrane protein, a potential adherence factor of Vibrio cholerae. Infect Immun. 1995;63:4433–4438. doi: 10.1128/iai.63.11.4433-4438.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stabb E V, Reich K A, Ruby E G. Vibrio fischeri genes hvnA and hvnB encode secreted NAD+-glycohydrolases. J Bacteriol. 2001;183:309–317. doi: 10.1128/JB.183.1.309-317.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tarsi R, Pruzzo C. Role of surface proteins in Vibrio cholerae attachment to chitin. Appl Environ Microbiol. 1999;65:1348–1351. doi: 10.1128/aem.65.3.1348-1351.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tashima K T, Carroll P A, Rogers M B, Calderwood S B. Relative importance of three iron-regulated outer membrane proteins for in vivo growth of Vibrio cholerae. Infect Immun. 1996;64:1756–1761. doi: 10.1128/iai.64.5.1756-1761.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Rhijn P, Vanderleyden J. The Rhizobium-plant symbiosis. Microbiol Rev. 1995;59:124–142. doi: 10.1128/mr.59.1.124-142.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Visick K L, Foster J, Doino J, McFall-Ngai M, Ruby E G. Vibrio fischeri lux genes play an important role in colonization and development of the host light organ. J Bacteriol. 2000;182:4578–4586. doi: 10.1128/jb.182.16.4578-4586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Visick K L, McFall-Ngai M J. An exclusive contract: specificity in the Vibrio fischeri-Euprymna scolopes partnership. J Bacteriol. 2000;182:1779–1787. doi: 10.1128/jb.182.7.1779-1787.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Visick K L, Ruby E G. Construction and symbiotic competence of a luxA deletion mutant of Vibrio fischeri. Gene. 1996;175:89–94. doi: 10.1016/0378-1119(96)00129-1. [DOI] [PubMed] [Google Scholar]
- 46.Visick K L, Ruby E G. The periplasmic, group III catalase of Vibrio fischeri is required for normal symbiotic competence and is induced both by oxidative stress and by approach to stationary phase. J Bacteriol. 1998;180:2087–2092. doi: 10.1128/jb.180.8.2087-2092.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Welch T J, Bartlett D H. Identification of a regulatory protein required for pressure-responsive gene expression in the deep-sea bacterium Photobacterium species strain SS9. Mol Microbiol. 1998;27:977–985. doi: 10.1046/j.1365-2958.1998.00742.x. [DOI] [PubMed] [Google Scholar]
- 48.Welch T J, Bartlett D H. Isolation and characterization of the structural gene for OmpL, a pressure-regulated porin-like protein from the deep-sea bacterium Photobacterium sp. strain SS9. J Bacteriol. 1996;178:5027–5031. doi: 10.1128/jb.178.16.5027-5031.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]