Abstract
The PrrBA two-component activation system of Rhodobacter sphaeroides plays a major role in the induction of photosynthesis gene expression under oxygen-limiting or anaerobic conditions. The PrrB histidine kinase is composed of two structurally identifiable regions, the conserved C-terminal kinase/phosphatase domain and the N-terminal membrane-spanning domain with six transmembrane helices framing three periplasmic and two cytoplasmic loops. Using a set of PrrB mutants with lesions in the transmembrane domain, we demonstrate that the central portion of the PrrB transmembrane domain including the second periplasmic loop plays an important role in both sensing and signal transduction. Signal transduction via the transmembrane domain is ultimately manifested by controlling the activity of the C-terminal kinase/phosphatase domain. The extent of signal transduction is determined by the ability of the transmembrane domain to sense the strength of the inhibitory signal received from the cbb3 terminal oxidase (J.-I Oh, and S. Kaplan, EMBO J. 19:4237–4247, 2000). Therefore, the intrinsic (“default”) state of PrrB is in the kinase-dominant mode. It is also demonstrated that the extent of prrB gene expression is subject to the negative autoregulation of the PrrBA system.
In prokaryotes, two-component signal transduction systems composed of a histidine kinase and a cognate response regulator play major roles in cellular adaptation to various environmental conditions (4, 12). Rhodobacter sphaeroides 2.4.1 possesses extensive physiological versatility, i.e., it can grow chemoheterotrophically, chemolithotrophically, photoheterotrophically, or photolithotrophically. When oxygen tensions fall below ∼3%, the intracytoplasmic membrane (ICM) housing the photosynthetic apparatus is synthesized through invaginations of the cytoplasmic membrane. Thus, oxygen is the primary signal determining ICM formation. Anaerobic induction of the photosynthetic apparatus is mediated by at least three major regulatory systems, the PrrBA two-component activation system, the AppA-PpsR antirepressor-repressor system, and FnrL (23, 33).
The PrrBA two-component system (RegBA in Rhodobacter capsulatus) is involved in the positive regulation of photosynthesis (PS) gene expression as well as the expression of genes responsible for CO2 and N2 fixation (8, 9, 14, 26, 28). PrrB is a membrane-localized histidine kinase consisting of 462 amino acid residues of which the C-terminal cytoplasmic kinase/phosphatase domain (amino acids 183 to 462) contains highly conserved regions (H, N, G1, F, and G2 boxes) characteristic of histidine kinases (8, 12). From sequence analyses it was predicted that autophosphorylation occurs at the conserved histidine residue (His-221) in the H box (8). PrrA, the cognate response regulator, is phosphorylated at Asp-63 by PrrB (J. M. Eraso and S. Kaplan, personal communication). It has been proposed that RegB (PrrB) has both kinase and phosphatase activities (1, 6, 10). Phosphorylated PrrA is thought to be the active form, capable of activating the transcription of those target genes belonging to the PrrBA regulon. In some cases, it has been shown that PrrA (RegA) functions as a repressor, as exemplified by the regulation of regB, regCA, and hupSLC in R. capsulatus (3, 5) as well as the ccoNOQP operon encoding the cbb3 cytochrome c oxidase in R. sphaeroides (J. M. Eraso and S. Kaplan, unpublished data). Phosphorylated RegA was shown to be stable, unlike CheY, as judged by its decaying kinetics (1). Therefore, the phosphatase activity of PrrB (RegB) appears to play a major role in controlling the ratio between phosphorylated and unphosphorylated PrrA (RegA) in the cell in response to changes in O2 tensions.
We have previously provided evidence that the cbb3 cytochrome c oxidase forms a signal transduction pathway together with the PrrBA two-component system and that the former serves as an oxygen sensor, i.e., the volume of electron flow through the cbb3 oxidase serves as the signal which is perceived and transduced to the PrrBA two-component system through the membrane-localized PrrC protein (7, 23, 24). The greater the volume of electron flow through the cbb3 oxidase, the stronger the inhibitory signal, which shifts the equilibrium of PrrB activity from the kinase mode to the phosphatase mode, resulting in the repression of PS gene expression.
Since the cbb3 oxidase, PrrC, and PrrB are localized in the cytoplasmic membrane, the transmembrane domain (amino acids 1 to 182) of PrrB is likely to be involved in sensing and transducing the signal derived from an upstream component of the cbb3-PrrBA signal transduction pathway in order to control the relative activity of the PrrB kinase/phosphatase. As a first effort to address this question, we have characterized a set of PrrB mutants with lesions in the transmembrane domain and suggest that the central portion of the PrrB transmembrane domain is important for the sensing function of PrrB and that the “unsignaled” or “default” state of PrrB is kinase dominant.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. R. sphaeroides and Escherichia coli strains were grown as described previously (22).
TABLE 1.
Bacterial strains and plasmids used in this work
| Strain or plasmid | Relevant phenotype or genotype | Source or reference |
|---|---|---|
| R. sphaeroides | ||
| 2.4.1 | Wild type | 31 |
| PRRB1 | 2.4.1 derivative, Δ prrB::ΩSpr/Str | 8 |
| PRRA2 | 2.4.1 derivative, Δ prrA::ΩSpr/Str | 8 |
| E. coli | ||
| DH5α | (Φ80dlacZΔM15)ΔlacU169 recA1 endA1 hsdR17 supE44 thil gyrA96 relA1 | 13 |
| S17-1 | Pro− Res− Mob+recA; integrated plasmid RP4-Tc::Mu-Km::Tn7 | 29 |
| Plasmids | ||
| pUC19 | Apr; lacPOZ′ | 32 |
| pBSIIKS+ | Apr; lacPOZ′ | Stratagene |
| pT7-7 | Apr; T7 promoter, RBS, and translation start codon overlapping with NdeI site | 30 |
| pRK415 | Tcr; Mob+lacZα IncP | 15 |
| pU1655 | pBSIIKS+::0.47-kb StyI fragment of pufBA | J. K. Lee |
| pUCP6.37 | pUC19::3.2-kb PvuII fragment containing the rrnB operon | S. C. Dryden |
| pUI1643 | pBSIIKS+::4.0-kb BamHI-HindIII fragment containing prrB and prrA | 8 |
| pUI1649 | pRK415/BamHI-Asp718, 1.9-kb SmaI-ClaI fragment containing prrB; prrB is in divergent orientation to lacZ | 8 |
| pUI1663 | Spr Str Kmr; IncQ; puf::lacZYA′ | 6 |
| pCF1010 | Spr Str Tcr; IncQ | 18 |
| pPRRB1-1 | pBSIIKS+::2.0-kb ApaI fragment containing prrB | This study |
| pPRRB2 | pBSIIKS+::429-bp PCR product containing six His codons at the 3′ end of prrB; prrB is in divergent orientation to lacZ | This study |
| pPRRB3 | pPRRB1-1::400-bp PflMI-KpnI from pPRRB2 | This study |
| pPRRB4 | pRK415::1.65-kb PstI-KpnI fragment containing prrB from pPRRB3; prrB is in colinear orientation to lacZ | This study |
| pPRRB5 | pUC19::1.65-kb PstI-KpnI fragment containing prrB from pPRRB3 | This study |
| pBLP1,3,5 | pBSIIKS+::1.65-kb PstI-KpnI fragment containing prrB with five-Ala codon insertion mutation and six His codons at the 3′ end of prrB | This study |
| pLP1-pLP5 | pPRRB4 in which five Ala codons are inserted in prrB | This study |
| pTM1Δ | pPRRB4 with a deletion mutation in prrB | This study |
| pOYH1 | pT7-7::1.42-kb NdeI-PstI fragment containing prrB | This study |
| pOYH2 | pBSIIKS+::1.43-kb XbaI-PstI fragment from pOYH1 | This study |
| pA-9 | pRK415::1.45-kb XbaI-KpnI fragment from pOYH2; prrB is in colinear orientation to lacZ | This study |
| pPRRBLAC | pCF1010::0.6-kb PstI-XbaI fragment of prrB promoter region | This study |
DNA manipulations and conjugation techniques.
Standard protocols (27) or manufacturer's instructions were followed for recombinant DNA manipulations. Mobilization of plasmids from E. coli strains into R. sphaeroides strains was performed as described elsewhere (2).
Construction of plasmids. (i) pPRRB4.
A 2.0-kb ApaI fragment containing prrB from pUI1643 was cloned into pBSIIKS+ to give pPRRB1-1. A 0.43-kb fragment containing the 3′ portion of prrB with a tail of six histidine codons immediately upstream of its stop codon was generated by PCR using primers 6HIS+ (5′-GGCGCCGGATCAGTGGTGGTGATGGTGGTGGGTCTGGATCAGGACGTTCTC-3′) and 6HIS− (5′-CAACCTCATTCAGAATGCCGTCGA-3′) and pPRRB1-1 as a template. The PCR product was cloned into pBSIIKS+ digested with HincII to give plasmid pPRRB2. A 0.4-kb PflMI-KpnI fragment from pPRRB2 was cloned into pPRRB1-1 restricted with the same enzymes, resulting in plasmid pPRRB3. Finally a 1.6-kb PstI-KpnI fragment containing the whole prrB with six histidine codons was cloned into pRK415, yielding pPRRB4.
(ii) pA-9.
To construct pA-9, prrB was amplified using primers 5′-TTGCCCCATATGATACTCGGTCCCGAC-3′ (NdeI site is underlined) and 5′-ACTCTGCAGTCAGGTCTGGATCAGGAC-3′ (PstI site is underlined) to generate a 1.4-kb product containing prrB with NdeI and PstI sites at both ends. The PCR product was restricted with NdeI and PstI and cloned into pT7-7 in order to provide the optimal ribosome binding site (RBS) to the prrB gene, yielding pOYH1. A 1.4-kb XbaI-PstI fragment from pOYH1 was cloned into pBSIIKS+, resulting in pOYH2. Finally the 1.4-kb XbaI-KpnI fragment from pOYH2 was cloned into pRK415 to give pA-9.
(iii) pPRRBLAC.
To construct the prrB::lacZ transcriptional fusion, the prrB promoter region was amplified with primers 5′-GAAGAACTGCAGCTGCTGTTCGGGCGTGTC-3′ (PstI site is underlined) and 5′-CGGACCTCTAGATACCAGTCGGTCACG-3′ (XbaI site is underlined) and pUI1643 as the template to generate a 605-bp product. The PCR product was digested with PstI and XbaI and cloned into promoterless lacZ vector pCF1010 digested with the same enzymes, yielding plasmid pPRRBLAC.
All PCRs were carried out employing Pfu Turbo polymerase (Stratagene, La Jolla, Calif.).
Site-directed mutagenesis.
Plasmids (pLP1 to pLP5) carrying a set of insertion mutations in those regions of prrB encoding the PrrB transmembrane region were constructed as follows. A 1.65-kb PstI-KpnI fragment from pPRRB3 was cloned into pUC19 to give pPRRB5. A string of five alanine codons was inserted by recombinant PCR using the primers listed in Table 2. Two rounds of the PCR were carried out using Pfu Turbo polymerase. With pPRRB5 as the template, two primary PCRs were performed with primers LP#+ and OUT− and with LP#− and OUT+ to generate two DNA fragments each containing a 37-bp overlapping region (# indicates a number from 1 to 5). The two primary PCR products were used as templates for the secondary PCR, which was performed using primers OUT+ and OUT−. In constructing pLP1, pLP3, and pLP5, 1.0-kb secondary PCR products were restricted with PstI and EcoRI and each 0.77-kb PstI-EcoRI fragment was cloned into pPRRB3 digested with PstI and EcoRI to yield pBLP1, pBLP3, and pBLP5. Following the verification of these insertion mutations by DNA sequencing, 1.65-kb PstI-KpnI fragments from pBLP1, pBLP3, and pBLP5 were cloned into pRK415 to give pLP1, pLP3, and pLP5, respectively. To construct pLP2 and pLP4, 1.0-kb secondary PCR products were digested with NotI and PstI. A 0.9-kb NotI-PstI fragment from each PCR product was cloned into pPRRB5 digested with the same enzymes, and a 1.65-kb PstI-KpnI fragment from pPRRB5 was finally cloned into pRK415, resulting in pLP2 and pLP4.
TABLE 2.
Primers used to construct the PrrB mutants
| Plasmid | Primer | Nucleotide sequence (5′–3′)a | Insertion or deletion site in PrrB |
|---|---|---|---|
| pLP1 | LP1+ | TGGGGGTCCGCGCGGCGGCCGCAGCGCTGCCGATGGGGCTCTGCTTC | Insertion of five Ala residues between R48 and L49 |
| LP1− | CCCATCGGCAGCGCTGCGGCCGCCGCGCGGACCCCCAGATACCAGTC | ||
| pLP2 | LP2+ | TCTTCCCGCAGGCGGCGGCAGCCGCGAACCGCCGCCTGACCGAGTTC | Insertion of five Ala residues between Q74 and N75 |
| LP2− | AGGCGGCGGTTCGCGGCTGCCGCCGCCTGCGGGAAGACGAAGGTCGC | ||
| pLP3 | LP3+ | TCCTGCTGTTCGCGGCGGCCGCAGCGCTGACCGGGGGGCTCACCAAC | Insertion of five Ala residues between F99 and L100 |
| LP3− | CCCCCGGTCAGCGCTGCGGCCGCCGCGAACAGCAGGAACGACAGCTG | ||
| pLP4 | LP4+ | TCGAGCTGCGCGCGGCGGCAGCCGCGACGACCGTCATTCTCGGGGCC | Insertion of five Ala residues between R126 and T127 |
| LP4− | ATGACGGTCGTCGCGGCTGCCGCCGCGCGCAGCTCGAGCGCGAGCGC | ||
| pLP5 | LP5+ | ACGGGTCGAGCGCGGCGGCCGCAGCGCTGTCCGTCCCGCGCATGTTC | Insertion of five Ala residues between S156 and L157 |
| LP5− | GGGACGGACAGCGCTGCGGCCGCCGCGCTCGACCCGTCGGCGAGGAT | ||
| pTM1Δ | TM1+ | GACTGCGCACCCTGATCCTCACCGAGTTCCAGGCGCTGAT | Deletion from L26 to L78 |
| TM1− | ATCAGCGCCTGGAACTCGGTGAGGATCAGGGTGCGCAGTC | ||
| Outer primer | OUT+ | ACCATGATTACGCCAAGCTTG | |
| OUT− | TTGATCGTGGCCAGCGGCGTG |
Inserted nucleotides are shown in boldface.
For the construction of pTM1Δ, recombinant PCR was also carried out using pPRRB5 as the template. The secondary PCR product was restricted with PstI and NotI and cloned into pPRRB5. After verification of the construction by sequencing, the PstI-KpnI fragment containing prrB from pPRRB5 was cloned into pRK415 to yield pTM1Δ.
The resulting plasmids, pLP1 to pLP5 and pTM1Δ, were introduced into R. sphaeroides PrrB1, a PrrB null mutant strain.
RNA isolation and analysis.
Total RNA was isolated from R. sphaeroides strains as described by Oelmuller et al. (20). Northern hybridization experiments were performed using the AlkPhos DIRECT system (American Pharmacia Biotech, Piscataway, N.J.) as instructed by the manufacturer. Quantitation of signals was performed with National Institutes of Health Imager, version 1.62. The signal levels were normalized by those of processed 23S rRNA (14S).
Preparation of solubilized membrane proteins.
The harvested cells were resuspended in buffer A (20 mM Tris-HCl, pH 7.5) and disrupted by passage through a French pressure cell. Crude cell extracts were obtained following centrifugation at 27,000 × g for 20 min at 4°C to remove unbroken cells and cell debris. Membrane fractions were isolated by ultracentrifugation of crude extracts at 150,000 × g for 1 h at 4°C. After membrane fractions (pellets) were washed twice with buffer A, the membranes were solubilized in buffer A containing 0.5% (wt/vol) n-dodecyl maltoside by pipetting and then centrifuged at 20,000 × g for 10 min at 4°C in a benchtop minicentrifuge. The supernatant was taken as solubilized membrane proteins.
Immunoblotting analysis.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting using an anti-His4 antibody (Qiagen Inc., Santa Clarita, Calif.) were performed as described elsewhere (17, 19).
Quantitative analysis of spectral complexes.
The B800-850 and B875 complex levels were determined spectrophotometrically as described previously (22).
β-Galactosidase activity assay and protein determination.
Preparation of crude cell extracts and determination of β-galactosidase activities were performed as described previously (22). Protein concentration was determined by the bicinchoninic acid protein assay (Pierce, Rockford, Ill.) using bovine serum albumin as the standard protein.
RESULTS
Effect of mutations in the transmembrane domain of PrrB on PS gene expression.
The transmembrane domain of PrrB (amino acids 1 to 182) has been assumed to be the signal-sensing domain of PrrB (25). To investigate which portion(s) of the transmembrane domain of PrrB is important to PrrB function, i.e., effectively controlling the on or off state of PS genes belonging to the PrrBA regulon, a variety of insertion and deletion mutations were constructed. These were introduced into those regions of prrB encoding the PrrB transmembrane region as shown in Fig. 1A. This study was based on the topology of PrrB in the cell membrane as determined by Ouchane and Kaplan (25).
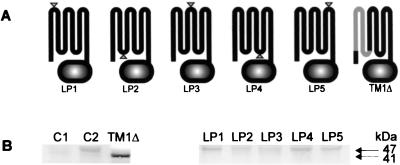
FIG. 1.
Schematic diagram depicting the mutant sites in PrrB (A) and immunoblot analysis of PrrB from the set of PrrB mutants (B). Triangles, insertion sites of a string of five alanines. The membrane portion (gray) indicates the deleted region. Identical amounts of membrane fractions (70 μg of protein) were run on an sodium dodecyl sulfate–12.5% polyacrylamide gel, and PrrB was detected with the anti-His4 antibody. C1 and C2, negative control PrrB1 with pRK415 and positive control PrrB1 with pPRRB4, respectively. Strains were grown anaerobically with 0.5% (vol/vol) DMSO as the terminal electron acceptor by continuous sparging with a mixture of 95% N2–5% CO2 to an optical density at 600 nm of 0.45 to 0.55. See Materials and Methods for additional details about preparation of the membrane fraction.
As described in Materials and Methods, mutagenesis was performed on the plasmid-borne prrB gene with a string of six histidine codons immediately upstream of its stop codon. The addition of a six-histidine tag at the C terminus of PrrB was designed in order to determine whether or not the mutant PrrB is incorporated into the membrane by virtue of immunoassay using the anti-His4 antibody. The plasmids carrying the mutated forms of prrB (pLP1 to pLP5 and pTM1Δ) as well as the parental plasmid pPRRB4 were introduced into PrrB null mutant strain PrrB1 by conjugation. When PrrB1 was mated with E. coli S17-1 carrying pLP3, we repeatedly failed to isolate exconjugants on Sistrom's medium A (SIS) plates incubated under aerobic conditions. However, exconjugants from this mating could be isolated under anaerobic photosynthetic conditions at medium light intensity, where a PrrB null mutant cannot grow. In contrast, the other exconjugants harboring pLP1, pLP2, pLP4, pLP5, or pTM1Δ were isolated under aerobic conditions. When streaked onto SIS plates, the LP3 mutant, obtained under anaerobic conditions, was unstable under aerobic conditions, i.e., it gave rise to colonies with variegated colorization. This suggested that the LP3 mutant able to grow under aerobic conditions appeared to acquire a secondary mutation(s). For this reason we were unable to further characterize this mutant under aerobic conditions. The LP2 and LP4 mutants were also slightly unstable under aerobic conditions, whereas the LP1, LP5, and TM1Δ mutants were stable under the same conditions.
(i) Integration of the altered PrrB into the membrane.
Prior to characterization of the PrrB mutants, they were examined by Western blotting using the anti-His4 antibody to determine whether or not the mutant forms of PrrB were integrated into the membrane. The PrrB mutant strains, as well as the control strains (PrrB1 with pRK415 and PrrB1 with pPRRB4) were grown under dark anaerobic conditions with dimethyl sulfoxide (DMSO) as the terminal electron acceptor. The isolated membrane fractions were employed for the immunoassay.
As shown in Fig. 1B, the altered PrrB polypeptides of proper size were detected in the membrane. For mutants LP1 to LP5 and control strain PrrB1 with pPRRB4 these were predicted to be approximately 47 kDa, and for mutant TM1Δ they were predicted to be approximately 41 kDa. For mutants LP2 and LP3, the levels of the altered PrrB were lower than those observed for the other strains.
(ii) Spectral complex formation in the PrrB mutants.
An initial insight into PS gene expression involving the PrrB mutant strains was obtained following an examination of spectral complex levels in cells grown under both aerobic and anaerobic conditions (Table 3). As expected, the wild-type 2.4.1 with blank vector pRK415 did not produce spectral complexes under aerobic conditions. A marginal increase in the B875 complex in the PrrB1 mutant was observed with pRK415 grown under aerobic conditions, which can be accounted for by the nonspecific phosphorylation of PrrA (10) and absence of PrrB phosphatase activity in the PrrB1 mutant. The PrrB1 mutant with pPRRB4, from which the prrB gene is transcribed from both its own promoter and the promoter of the Tc resistance gene, synthesized even more spectral complexes under aerobic conditions than the PrrB1 mutant with pRK415. The aerobic formation of the spectral complexes in this strain appears to be due to the overexpression effect of prrB. When grown under aerobic conditions, all of the PrrB mutants tested produced four- to fivefold-increased and four- to sixfold-increased levels of B875 and B800-850 complexes, respectively, compared to the control strain (PrrB1 with pPRRB4) grown under the same conditions. The levels of spectral complexes synthesized in the mutant strains are comparable to those observed for a Cco mutant strain (24). Due to the relative instability of LP2 and LP4 under aerobic conditions, it is possible that the levels of spectral complexes determined in these mutants could be underestimated, although they are strongly elevated in the presence of oxygen. Although we do not know the precise reason for the inability of the LP3 mutant strain to grow under aerobic conditions, we previously observed that mutant strains such as a PpsR null mutant and PrrBL78P, in which PS gene expression is highly derepressed under aerobic conditions, form variegated colonies (from deep red to colorless colonies) under aerobic conditions and that their growth is retarded under these same conditions (8, 11). Therefore, it is reasonable to suggest that PS gene expression in this mutant may be very high under aerobic conditions relative to that observed for the other mutant strains. Taken together, the results indicate that, when the transmembrane domain of PrrB is altered by constructing either insertion or deletion mutations, the activity of the mutant form of PrrB is in the kinase-dominant mode, even under aerobic conditions, i.e., the altered PrrB is unable to sense the inhibitory signal originating from the cbb3 oxidase, which normally shifts the equilibrium of PrrB activity toward the phosphatase mode and away from the kinase mode under aerobic conditions.
TABLE 3.
Levels of spectral complexes in R. sphaeroides strains grown under anaerobic and aerobic conditionsa
| Strain | Plasmid | Level of indicated complex. (nmol/mg of protein) in:
|
|||
|---|---|---|---|---|---|
| Dark-DMSO
|
30% O2
|
||||
| B800–850 | B875 | B800–850 | B875 | ||
| 2.4.1 | pRK415 | 20.23 ± 1.72 | 5.74 ± 0.21 | 0.05 ± 0.00 | 0.02 ± 0.01 |
| PrrB1 | pRK415 | 1.36 ± 0.14 | 5.52 ± 0.27 | 0.06 ± 0.02 | 0.36 ± 0.12 |
| PrrB1 | pPRRB4 | 21.13 ± 0.26 | 7.73 ± 0.11 | 0.11 ± 0.01 | 0.74 ± 0.01 |
| PrrB1 | pLP1 | 24.83 ± 1.86 | 8.67 ± 0.61 | 0.40 ± 0.08 | 3.66 ± 0.09 |
| PrrB1 | pLP2 | 19.12 ± 0.27 | 8.42 ± 0.21 | 0.53 ± 0.09 | 2.95 ± 0.11 |
| PrrB1 | pLP3 | 9.39 ± 0.11 | 6.57 ± 0.06 | n.d. | n.d. |
| PrrB1 | pLP4 | 19.15 ± 1.68 | 8.99 ± 0.95 | 0.67 ± 0.09 | 3.60 ± 0.42 |
| PrrB1 | pLP5 | 25.55 ± 0.03 | 9.37 ± 0.26 | 0.42 ± 0.07 | 3.52 ± 0.24 |
| PrrB1 | pTM1Δ | 21.67 ± 0.27 | 10.04 ± 0.14 | 0.55 ± 0.06 | 3.76 ± 0.11 |
Strains were grown anaerobically in the dark with DMSO (Dark-DMSO) or aerobically by sparging with 30% O2–69% N2–1% CO2 to an optical density at 600 nm of 0.4 to 0.5. All values are the averages of two independent determinations. n.d., not determined.
Under anaerobic conditions spectral-complex formation in negative-control strain PrrB1 with pRK415 was severely affected. The levels of the B875 complex in this strain were similar to those observed for the wild type with pRK415, which can be explained by nonspecific phosphorylation of PrrA by another histidine kinase(s), such as HupT (10), coupled with the hierarchical formation of spectral complexes in the order of the reaction center, B875, and B800-850 complexes (33). Positive-control strain PrrB1 with pPRRB4 as well as LP1, LP2, LP4, LP5, and TM1Δ produced comparable, high levels of the spectral complexes under anaerobic conditions. The LP3 mutant was, in part, impaired in spectral-complex formation under anaerobic conditions. However, the levels of the spectral complexes synthesized in this mutant strain were still well above those observed for the PrrB1 mutant with pRK415, indicating that this mutant form of PrrB, although impaired, is still functional with regard to the anaerobic formation of the spectral complexes. The fact that all mutant strains synthesized much more spectral complexes than negative-control strain PrrB1 (pRK415) indicates that the suspected dimerization of the mutant forms of PrrB appears not to be affected, since the prerequisite for kinase activity of histidine kinases is the formation of the homodimer quaternary structure (4).
(iii) Expression of the puf operon in the PrrB mutants.
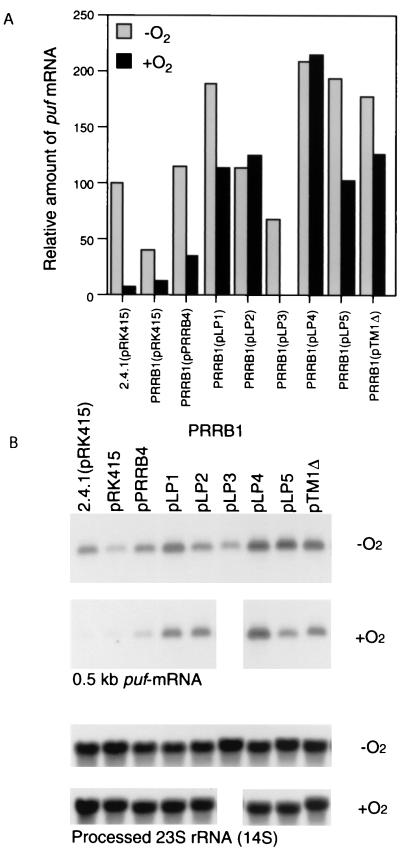
Since the puf operon encoding the apoproteins of the B875 complex and the L and M polypeptides of the reaction center appears to be regulated exclusively by the PrrBA two-component system, the level of puf mRNA was determined in order to assess the activity of the PrrBA system in the PrrB mutant strains grown under aerobic or anaerobic dark conditions in the presence of DMSO (Fig. 2).
FIG. 2.
Analysis of puf mRNA in R. sphaeroides strains grown anaerobically with DMSO by continuous sparging with a mixture of 95% N2–5% CO2 to an optical density at 600 nm of 0.45 to 0.55 or aerobically by sparging with 30% O2–69% N2–1% CO2 to an OD600 of 0.4 to 0.5. Approximately 20 μg of total RNA was loaded in each lane. The Northern blots were probed with a labeled 0.47-kb StyI fragment from pUI655 or a 0.5-kb HindIII-PstI fragment from pUCP6.37, which is specific for puf mRNA or processed 23S rRNA (14S), respectively (B). After background correction the values were normalized to the levels of processed 23S rRNA (14S). The normalized values are plotted above the Northern blots. The transcript level in the wild type (2.4.1) with pRK415 grown under anaerobic condition was set at 100 (A). PRRB1, the prrB-negative strain PrrB1. Electrophoresis and Northern blotting were performed at the same time using the same agarose gel and nylon membrane.
Under aerobic conditions the expression of the puf operon was minimal in the strains containing the wild-type form of PrrB (2.4.1 with pRK415 and PrrB1 with pPRRB4). In contrast, the puf operon was derepressed in the LP1, LP2, LP4, LP5, and TM1Δ mutant strains grown under aerobic conditions, which is consistent with the oxygen-insensitive formation of the spectral complexes observed in these mutants, as presented in Table 3.
Mutants LP1, LP5, and TM1Δ were shown to retain the ability to induce puf operon expression in the absence of O2 by a comparison of the transcript levels of the puf operon in these mutants grown under aerobic and anaerobic conditions. In contrast, the constitutive expression levels of the puf operon were virtually the same in the LP2 and LP4 mutant strains regardless of the presence or absence of O2. These results suggested that PrrB function with regard to the anaerobic induction and aerobic repression of PS genes, as determined from the analysis of puf operon expression, in response to changes in O2 tensions is lost in these mutant strains. The transcript level of the puf operon in the LP3 mutant grown under anaerobic conditions was significantly decreased compared with that in control strain PrrB1 (pPRRB4) grown under the same conditions, indicating a partial impairment of its ability to induce PS genes in response to reduced oxygen tensions. As described above, PS gene expression in the LP3 mutant strain under aerobic conditions was suggested to be high, as inferred from its instability under aerobic growth conditions. If this is true, the function of this mutant form of PrrB most strikingly deviates from the normal function of PrrB with regard to the signaling state in response to O2 availability.
Multiple-copy effect of the prrB gene on PS gene expression.
To examine whether the overexpression of prrB affects PS gene expression, the prrB gene was introduced in trans into PrrB null mutant strain PrrB1 and levels of the light-harvesting complexes (B800-850 and B875) as well as the promoter activity of the puf operon were determined in these strains, grown under 30% O2 conditions (Table 4). As a control, wild-type strain 2.4.1 carrying the vector alone was included in this experiment. As anticipated, the wild type with pRK415 synthesized virtually no spectral complexes under 30% O2 conditions. In contrast, the PrrB1 mutant with pA-9 in trans, on which the transcription of prrB is driven from the plasmid-borne Tc resistance promoter (the prrB promoter is completely removed) and the RBS upstream of prrB is optimized, produced significant levels of the spectral complexes under the same conditions. However, when the prrB gene is transcribed from its own promoter, pUI1649, the multiple-copy effect of prrB was not observed.
TABLE 4.
Levels of spectral complexes and β-galactosidase activities in R. sphaeroides strains grown under aerobic conditionsa
| Strain | Plasmid | Level (nmol/mg of protein) of:
|
Activity (nmol/min/mg of protein) of strain carrying puf::lacZ | |
|---|---|---|---|---|
| B800–850 | B875 | |||
| 2.4.1 | pRK415 | 0.05 ± 0.02 | 0.00 ± 0.00 | 72 ± 9 |
| PrrB1 | pA-9 | 0.13 ± 0.01 | 1.36 ± 0.01 | 251 ± 5 |
| PrrB1 | pUI1649 | 0.02 ± 0.01 | 0.11 ± 0.01 | 53 ± 3 |
Strains carrying the corresponding plasmids were grown aerobically by sparging with 30% O2–69% N2–1% CO2 to an optical density of 600 nm of 0.4 to 0.5. For determination of promoter activities of the puf operon, strains carrying puf::lacZ transcriptional fusion plasmid pUI1663 were grown aerobically by sparging with 30% O2–69% N2–1% CO2 to an optical density of 600 nm of 0.3 to 0.35. All values are the averages of two independent determinations.
Consistent with these data, the promoter activity of the puf operon was increased in the PrrB1 mutant with pA-9 by 3.5-fold compared to that for control strain 2.4.1 with pRK415. puf expression remained uninduced in the PrrB1 mutant with pUI1649, as observed for the control strain.
Taken together, these data clearly revealed that overexpression of prrB leads to increased PS gene expression under aerobic conditions, resulting in O2-insensitive spectral-complex formation. There are several possible reasons why the PrrB1 mutant carrying pUI1649 does not produce spectral complexes under aerobic conditions, although multiple copies of prrB are present. The promoter of prrB may be too weak to bring about O2-insensitive formation of the spectral complexes. Alternatively, the PrrBA two-component system may negatively regulate prrB expression and such regulation may be inoperative when a heterologous promoter is used to express prrB.
Expression of the prrB gene.
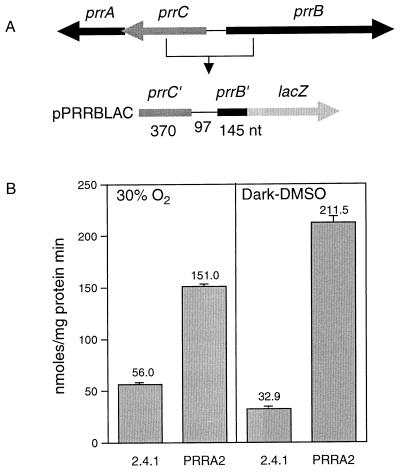
To ascertain whether the prrB gene is negatively regulated by the PrrBA two-component system, the transcriptional activity of prrB was determined in wild-type strain 2.4.1 and PrrA null mutant strain PrrA2 grown under 30% O2 conditions as well as under anaerobic, dark conditions in the presence of DMSO. The promoter activity of prrB was monitored by using prrB::lacZ transcriptional fusion plasmid pPRRBLAC (Fig. 3).
FIG. 3.
Promoter activities of the prrB gene in R. sphaeroides strains. (A) Schematic diagram of prrB::lacZ transcriptional fusion plasmid pPRRBLAC. (B) Wild-type (2.4.1) and prrA-negative (PRRA2) strains containing pPRRBLAC were grown aerobically by sparging with 30% O2–69% N2–1% CO2 to an optical density at 600 nm of 0.3 or anaerobically in the dark with 0.5% (vol/vol) DMSO. All values provided are the averages of two independent determinations.
Under both aerobic and anaerobic conditions the promoter activity of prrB was derepressed in the PrrA2 mutant strain by factors of 2.7 and 6.4, respectively, compared to that in the wild-type strain grown under these same conditions. This indicates that PrrA normally represses the expression of prrB under both aerobic and anaerobic conditions and that “activated” PrrA appears to affect prrB transcription more negatively than “nonactivated” PrrA. Furthermore, anaerobic repression of prrB was observed in the wild type, whereas the PrrA2 mutant strain did not show such a phenotype, confirming that phosphorylated PrrA represses the prrB gene more efficiently than unphosphorylated PrrA.
DISCUSSION
The PrrB (RegB) protein is a bifunctional enzyme which possesses both kinase and phosphatase activities (1, 6). The kinase-dominant state is favored in low-oxygen or anaerobic conditions, resulting in the induction of PS gene expression, whereas the relative propensity for the phosphatase-dominant state increases with increasing oxygen tensions in the environment.
We previously showed that the cbb3 cytochrome c oxidase is an oxygen sensor and that, when electron flow through the oxidase occurs under aerobic conditions, it generates a signal shifting the equilibrium of PrrB activity toward the phosphatase-dominant mode, leading to repression of PS gene expression (23, 24). The inhibitory nature of the signal emanating from the cbb3 oxidase implies that the default state of PrrB is in the kinase-dominant mode. One of the simplest approaches to examine the default state of PrrB is to alter the ratio of cbb3 oxidase to PrrB by increasing the cellular level of PrrB and to determine the expression levels of those genes belonging to the PrrBA regulon. It was reasoned that, when prrB is overexpressed and the cellular level of its product is in excess, some fraction of PrrB lies outside the cbb3-PrrBA signal transduction pathway and thus the “excess” PrrB activity is not susceptible to the inhibitory signal. The overexpression of prrB led to increased PS gene expression under aerobic conditions, indicating that the default state (epistatic to the signal transduction pathway) of PrrB is in the kinase-dominant mode.
The prrB gene is also subject to the negative autoregulation of the PrrBA system, as evidenced by the following findings. The prrB gene is significantly derepressed in the PrrA null mutant under both aerobic and anaerobic conditions in comparison with its expression in the wild type grown under the same conditions. The anaerobic repression of prrB in the wild type further implies that activated PrrA is more effective in the repression of prrB than unactivated PrrA. It has been shown that both phosphorylated and unphosphorylated forms of purified RegA (PrrA homologue) of R. capsulatus are able to bind to the promoter region of the puc operon, but phosphorylated RegB shows a higher DNA-binding affinity than unphosphorylated RegA (1). Therefore, the reduction in prrB expression in the wild type under anaerobic conditions versus aerobic conditions may result from a higher affinity of phosphorylated PrrA than unphosphorylated PrrA for binding to the regulatory region of prrB . The net effect of the negative autoregulation of prrB is to maintain low cellular levels of PrrB and to blunt the induction of PS gene expression, thereby preventing the excess formation of the spectral complexes. It has been reported that a similar autoregulation mechanism is operative in R. capsulatus (3).
PrrB comprises two topologically distinct regions, the conserved C-terminal kinase/phosphatase domain and the N-terminal membrane-spanning domain with six transmembrane helices framing three periplasmic and two cytoplasmic loops (25). The kinase/phosphatase domain of PrrB (amino acids 183 to 462) has the domain organization characteristic of a class I histidine kinase, of which EnvZ is a member, i.e., the DHp (dimerization and histidine phosphotransfer) region is directly linked to the CA (catalytic and ATP-binding) region (4, 8). This domain is directly involved in phosphorylation and dephosphorylation of cognate response regulator PrrA (1). On the other hand, the functional importance of the N-terminal transmembrane domain of PrrB has yet to be assessed systematically. As our initial effort, we constructed a series of insertion and deletion mutations which affect the transmembrane domain of PrrB. It is conceivable that, when a string of five alanines is inserted into a loop region of the PrrB transmembrane domain, the structure and conformation of the altered loop are severely affected. Furthermore, the steric strain imposed by such an insertion might also affect the conformation of the neighboring regions. To construct the PrrB deletion mutant used here, a pair of adjoining transmembrane helices were removed to ensure the correct orientation of the remaining transmembrane helices within the membrane.
All mutations of the PrrB transmembrane domain confer a phenotype leading to the oxygen-insensitive formation of spectral complexes. This phenotype was previously observed for the PrrBL78P mutant, in which Leu-78 within the PrrB transmembrane domain is replaced by proline (8). Together with the assumption that the N-terminal transmembrane domain of PrrB is a sensor domain receiving and transducing a signal derived from upstream element(s) of the cbb3-PrrBA signal transduction pathway, this observation indicates that, when the ability of PrrB to sense is disrupted, its default state is similar to that of low-oxygen conditions, i.e., a kinase-dominant state. Thus, the “signal” appears to exist under high-oxygen conditions, as proposed previously (23, 24), and when PrrB senses this signal, it shifts from the kinase-dominant default state to that found under high-oxygen conditions. Most importantly, the LP2 and LP4 mutant strains have apparently lost their ability to respond to changes in oxygen tensions. They exhibit constitutive expression of the puf operon regardless of the presence or absence of O2. The fact that the LP2 and LP4 mutant strains differ in the ultimate levels of puf operon expression implies that the LP2 and LP4 forms of PrrB might be locked into a conformation which renders them different in their abilities to transduce the change of their default states. Alternatively, the difference in the level of PrrB incorporated into the cytoplasmic membrane in these mutants (Fig. 1B) might explain the various expression levels of the puf operon. In contrast, LP1, LP5, and TM1Δ are still capable of inducing the puf operon when oxygen tensions are reduced, although the puf operon is partially derepressed in the presence of O2. In other words, these forms of PrrB, although impaired, are still functional in terms of O2 sensing. On the basis of this observation, we can draw the following conclusions. (i) Transmembrane helices 1 and 2 (numbered from the N terminus of PrrB) as well as periplasmic loop 1 and cytoplasmic loop 1 are not essential for sensing the inhibitory signal, since the TM1Δ form of PrrB is still responsive to lowering O2 tensions. (ii) The closer the site of the alanine insertion is to the central portion of the transmembrane domain of PrrB (the second periplasmic loop), the more severely affected the function of PrrB with regard to responding to the inhibitory signal. This suggests that periplasmic loop 2 and flanking transmembrane helices 3 and 4 might be the most important segment of the transmembrane-spanning domain for the sensing function of PrrB. In agreement with this conclusion, we see that the region encompassing transmembrane helix 3, periplasmic loop 2, and transmembrane helix 4 is well conserved at the level of amino acid sequence, when the corresponding regions of the PrrB homologues from several photosynthetic bacteria are multiply aligned (Fig. 4). A conspicuous characteristic found within this conserved region is the presence of conserved leucine residues at positions 88, 94, 97, 98, 100, 104, 110, and 113 (numbering is based on PrrB sequence). This leucine-rich repeat is found in a variety of proteins and in many cases is involved in protein-protein interactions (16). This finding raises the possibility that PrrB receives the signal from an upstream component of the cbb3-PrrBA signal transduction pathway by means of a protein-protein interaction.
FIG. 4.
Multiple alignment of the N-terminal transmembrane domain of PrrB homologues. The highly conserved region in the central part is underlined, and the corresponding part is depicted in gray in the schematic model of PrrB. The identical or conservatively substituted residues are indicated by asterisks or colons, respectively. Abbreviations: R.c., R. capsulatus; R.d., Roseobacter denitrificans; PrrB-R.s., PrrB from R. sphaeroides; RegB-R.s., RegB from Rhodovulum sulfidophilum.
It is noteworthy that mutant LP3 does not grow properly and is very unstable under aerobic conditions. We know that when the cellular level of porphyrin intermediates is high in the presence of O2, the intermediates are toxic to the cell. The PrrBA two-component system is involved in the anaerobic induction of the tetrapyrrole biosynthetic pathway, at least at the level of hemA encoding 5-aminolevulinic acid synthase, hemN and hemZ encoding the isoenzymes of coproporphyrinogen III oxidase, and some of the genes encoding bacteriochlorophyll biosynthetic enzymes (21, 23). Taken together, these observations might imply that PS gene expression in the LP3 mutant strain may be so high that the strain's growth under aerobic conditions is severely compromised. In addition, mutant LP3 is impaired in spectral-complex formation and puf operon induction under anaerobic conditions. These interpretations and observations seem to present a paradox. If we assume that there is a high level of expression of PS genes in this mutant under aerobic conditions, then the LP3 form of PrrB responds to alteration in the inhibitory signal in a way opposite to that of the normal PrrB. In any event, the LP3 form of PrrB deviates from the normal function of PrrB in terms of anaerobic induction and aerobic repression of PS gene expression in the most striking way, corroborating our suggestion that periplasmic loop 2 and its neighboring transmembrane region(s) are critical for the sensing function of PrrB.
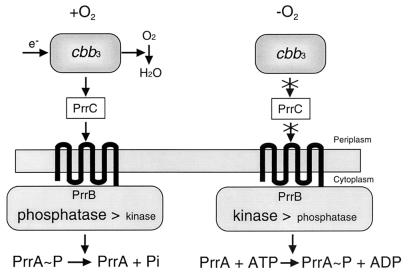
On the basis of the results presented here, as well as those reported previously (23), we present a model describing O2 sensing through the cbb3-PrrBA signal transduction pathway (Fig. 5). The intrinsic or default state of PrrB is in the kinase-dominant mode. The signal which controls PrrB activity in response to changes in O2 tensions exists under high-oxygen conditions. In the presence of O2, electron flow through the cbb3 oxidase generates the signal which shifts the equilibrium of PrrB activity from the kinase mode to the phosphatase mode, resulting in dephosphorylation of PrrA. Under these conditions, the genes belonging to the PrrBA regulon remain uninduced. This signal is likely transduced to PrrB via the PrrC membrane-spanning protein. We placed PrrC between the cbb3 oxidase and PrrB in the signal transduction pathway, since a PrrC null mutant containing normal cbb3 oxidase activity exhibited substantial increases in the level of PS complexes and PS gene expression under aerobic conditions, as observed for Cco mutant strains (7). Further, the prrC gene forms an operon together with prrA (7). The central portion of the PrrB transmembrane domain including periplasmic loop 2 likely plays important roles in both sensing and transmembrane signal transduction. When O2 tensions are reduced or under anaerobic conditions, the interruption of electron flow through the cbb3 oxidase abolishes or alleviates the inhibitory signal and PrrB returns to its default state, i.e., the kinase-dominant mode, to induce PS gene expression. Therefore, inactivation of any of the upstream component(s) of the cbb3-PrrBA signal transduction pathway such as the cbb3 oxidase or PrrC brings about the oxygen-insensitive formation of the spectral complexes.
FIG. 5.
Model for O2 signaling through the cbb3-PrrBA signal transduction pathway. The cbb3 cytochrome c oxidase (cbb3) and PrrC as well as PrrB are localized in the cytoplasmic membrane. PrrB is thought to exist as a homodimer.
ACKNOWLEDGMENTS
Jeong-Il Oh and In-Jeong Ko contributed equally to this work.
This work was supported by grant GM15590 to S.K.
REFERENCES
- 1.Bird T H, Du S, Bauer C E. Autophosphorylation, phosphotransfer, and DNA-binding properties of the RegB/RegA two-component regulatory system in Rhodobacter capsulatus. J Biol Chem. 1999;274:16343–16348. doi: 10.1074/jbc.274.23.16343. [DOI] [PubMed] [Google Scholar]
- 2.Davis J, Donohue T J, Kaplan S. Construction, characterization, and complementation of a Puf− mutant of Rhodobacter sphaeroides. J Bacteriol. 1988;170:320–329. doi: 10.1128/jb.170.1.320-329.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Du S, Kouadio J L, Bauer C E. Regulated expression of a highly conserved regulatory gene cluster is necessary for controlling photosynthesis gene expression in response to anaerobiosis in Rhodobacter capsulatus. J Bacteriol. 1999;181:4334–4341. doi: 10.1128/jb.181.14.4334-4341.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dutta R, Qin L, Inouye M. Histidine kinases: diversity of domain organization. Mol Microbiol. 1999;34:633–640. doi: 10.1046/j.1365-2958.1999.01646.x. [DOI] [PubMed] [Google Scholar]
- 5.Elsen S, Dischert W, Colbeau A, Bauer C E. Expression of uptake hydrogenase and molybdenum nitrogenase in Rhodobacter capsulatus is coregulated by the RegB-RegA two-component regulatory system. J Bacteriol. 2000;182:2831–2837. doi: 10.1128/jb.182.10.2831-2837.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eraso J M, Kaplan S. Complex regulatory activities associated with the histidine kinase PrrB in expression of photosynthesis genes in Rhodobacter sphaeroides 2.4.1. J Bacteriol. 1996;178:7037–7046. doi: 10.1128/jb.178.24.7037-7046.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eraso J M, Kaplan S. From redox flow to gene regulation: role of the PrrC protein of Rhodobacter sphaeroides 2.4.1. Biochemistry. 2000;39:2052–2062. doi: 10.1021/bi9923858. [DOI] [PubMed] [Google Scholar]
- 8.Eraso J M, Kaplan S. Oxygen-insensitive synthesis of the photosynthetic membranes of Rhodobacter sphaeroides: a mutant histidine kinase. J Bacteriol. 1995;177:2695–2706. doi: 10.1128/jb.177.10.2695-2706.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eraso J M, Kaplan S. prrA, a putative response regulator involved in oxygen regulation of photosynthesis gene expression in Rhodobacter sphaeroides. J Bacteriol. 1994;176:32–43. doi: 10.1128/jb.176.1.32-43.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gomelsky M, Kaplan S. Isolation of regulatory mutants in photosynthesis gene expression in Rhodobacter sphaeroides 2.4.1 and partial complementation of a PrrB mutant by the HupT histidine-kinase. Microbiology. 1995;141:1805–1819. doi: 10.1099/13500872-141-8-1805. [DOI] [PubMed] [Google Scholar]
- 11.Gomelsky M, Kaplan S. Molecular genetic analysis suggesting interactions between AppA and PpsR in regulation of photosynthesis gene expression in Rhodobacter sphaeroides 2.4.1. J Bacteriol. 1997;179:128–134. doi: 10.1128/jb.179.1.128-134.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grebe T W, Stock J B. The histidine protein kinase superfamily. Adv Microb Physiol. 1999;41:139–227. doi: 10.1016/s0065-2911(08)60167-8. [DOI] [PubMed] [Google Scholar]
- 13.Jessee J. New subcloning efficiency competent cells: >1 × 106 transformants/μg. Focus. 1986;8:9. [Google Scholar]
- 14.Joshi H M, Tabita F R. A global two component signal transduction system that integrates the control of photosynthesis, carbon dioxide assimilation, and nitrogen fixation. Proc Natl Acad Sci USA. 1996;93:14515–14520. doi: 10.1073/pnas.93.25.14515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 16.Kobe B, Deisenhofer J. The leucine-rich repeat: a versatile binding motif. Trends Biochem Sci. 1994;19:415–421. doi: 10.1016/0968-0004(94)90090-6. [DOI] [PubMed] [Google Scholar]
- 17.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 18.Lee J K, Kaplan S. Transcriptional regulation of puc operon expression in Rhodobacter sphaeroides. Analysis of the cis-acting downstream regulatory sequence. J Biol Chem. 1995;270:20453–20458. [PubMed] [Google Scholar]
- 19.Mouncey N J, Kaplan S. Redox-dependent gene regulation in Rhodobacter sphaeroides 2.4.1(T): effects on dimethyl sulfoxide reductase (dor) gene expression. J Bacteriol. 1998;180:5612–5618. doi: 10.1128/jb.180.21.5612-5618.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oelmuller U, Kruger N, Steinbuchel A, Friedrich C G. Isolation of procaryotic RNA and detection of specific mRNA with biotinylated probes. J Microbiol Methods. 1990;11:73–84. [Google Scholar]
- 21.Oh J-I, Eraso J M, Kaplan S. Interacting regulatory circuits involved in orderly control of photosynthesis gene expression in Rhodobacter sphaeroides 2.4.1. J Bacteriol. 2000;182:3081–3087. doi: 10.1128/jb.182.11.3081-3087.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oh J-I, Kaplan S. The cbb3 terminal oxidase of Rhodobacter sphaeroides 2.4.1: structural and functional implications for the regulation of spectral complex formation. Biochemistry. 1999;38:2688–2696. doi: 10.1021/bi9825100. [DOI] [PubMed] [Google Scholar]
- 23.Oh J-I, Kaplan S. Generalized approach to the regulation and integration of gene expression. Mol Microbiol. 2001;39:1116–1123. doi: 10.1111/j.1365-2958.2001.02299.x. [DOI] [PubMed] [Google Scholar]
- 24.Oh J-I, Kaplan S. Redox signaling: globalization of gene expression. EMBO J. 2000;19:4237–4247. doi: 10.1093/emboj/19.16.4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ouchane S, Kaplan S. Topological analysis of the membrane-localized redox-responsive sensor kinase PrrB from Rhodobacter sphaeroides 2.4.1. J Biol Chem. 1999;274:17290–17296. doi: 10.1074/jbc.274.24.17290. [DOI] [PubMed] [Google Scholar]
- 26.Qian Y, Tabita F R. A global signal transduction system regulates aerobic and anaerobic CO2 fixation in Rhodobacter sphaeroides. J Bacteriol. 1996;178:12–18. doi: 10.1128/jb.178.1.12-18.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 28.Sganga M W, Bauer C E. Regulatory factors controlling photosynthetic reaction center and light-harvesting gene expression in Rhodobacter capsulatus. Cell. 1992;68:945–954. doi: 10.1016/0092-8674(92)90037-d. [DOI] [PubMed] [Google Scholar]
- 29.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 30.Tabor S, Richardson C C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Neil C B. The culture, general physiology, morphology, and classification of the non-sulfur purple and brown bacteria. Bacteriol Rev. 1944;8:1–118. doi: 10.1128/br.8.1.1-118.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 33.Zeilstra-Ryalls J H, Gomelsky M, Eraso J M, Yeliseev A A, O'Gara J, Kaplan S. Control of photosystem formation in Rhodobacter sphaeroides. J Bacteriol. 1998;180:2801–2809. doi: 10.1128/jb.180.11.2801-2809.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]