Abstract
The Saccharomyces cerevisiae zinc finger protein Rim101p is activated by cleavage of its C-terminal region, which resembles PEST regions that confer susceptibility to proteolysis. Here we report that Rim20p, a member of the broadly conserved PalA/AIP1/Alix family, is required for Rim101p cleavage. Two-hybrid and coimmunoprecipitation assays indicate that Rim20p binds to Rim101p, and a two-hybrid assay shows that the Rim101p PEST-like region is sufficient for Rim20p binding. Rim101p-Rim20p interaction is conserved in Candida albicans, supporting the idea that interaction is functionally significant. Analysis of Rim20p mutant proteins indicates that some of its broadly conserved regions are required for processing of Rim101p and for stability of Rim20p itself but are not required for interaction with Rim101p. A recent genome-wide two-hybrid study (T. Ito, T. Chiba, R. Ozawa, M. Yoshida, M. Hattori, and Y. Sakaki, Proc. Natl. Acad. Sci. USA 98:4569–4574, 2000) indicates that Rim20p interacts with Snf7p and that Snf7p interacts with Rim13p, a cysteine protease required for Rim101p proteolysis. We suggest that Rim20p may serve as part of a scaffold that places Rim101p and Rim13p in close proximity.
Intracellular proteolytic systems can either inactivate or activate target proteins. Inactivation generally occurs through degradation of a polypeptide; activation generally occurs through site-specific cleavage (11, 39, 49). Proteolysis substrates are diverse, and proteolytic systems have pivotal roles in cell cycle progression, developmental pathways, stress responses, and apoptosis.
There are two major classes of cytoplasmic proteolysis systems in eukaryotes, the proteasome and cysteine proteases. The proteasome is a large multiprotein complex (2, 4). Protein substrates are often targeted for degradation or site-specific cleavage by ubiquitination, which occurs through binding of an E3 enzyme (ubiquitin-protein ligase) to the substrate (12). Cells express many different E3 proteins, permitting recognition of a variety of substrate proteins. Cysteine proteases include the calpains (30, 33) and caspases (44). These proteases can promote either extensive degradation or site-specific cleavage of their substrates (33, 39, 41, 42). It is not clear how cysteine proteases recognize their substrates in general, but in vitro experiments indicate that μ-calpain may bind directly to the substrate IκB (41).
Several protein motifs can channel a substrate into a proteolytic system (2). Among these is the PEST sequence, a segment rich in proline, aspartate, glutamate, serine, and threonine (36). The PEST regions of several proteins confer susceptibility to ubiquitination and proteasomal degradation (13, 38, 53). However, the IκB C-terminal PEST sequence promotes its degradation by μ-calpain in vitro (41). Therefore, PEST sequences may serve as recognition motifs for both proteasome and cysteine protease systems.
A C-terminal PEST-like region plays a central role in the activity of Saccharomyces cerevisiae transcription factor Rim101p. Newly synthesized full-length Rim101p is inactive; proteolytic removal of the C-terminal ∼100-residue PEST-like region yields active Rim101p, a positive regulator of invasive growth and sporulation (23). The Rim101p C-terminal region lacks proline residues, distinguishing it from true PEST sequences (36). Rim101p is homologous to the Aspergillus nidulans pH response regulator PacC, which undergoes a cleavage reaction that removes ∼420 C-terminal residues (28). The cleavage requirements for PacC are well defined genetically and include PalB, a cysteine protease (6), and PalA, a protein of uncertain biochemical function (31). This cleavage pathway is conserved, because Rim101p cleavage depends on the PalB homolog Rim13p/Cpl1p (22) and, as we report here, the PalA homolog Rim20p.
Among PacC/Rim101p cleavage pathway members, PalA/Rim20p is very broadly conserved; homologs have been found in every sequenced metazoan genome, including at least two in Homo sapiens. Prior studies suggest that metazoan PalA/Rim20p family members may govern cysteine protease activity; the mouse homolog AIP1/Alix interacts with ALG-2 (29, 48), a putative cysteine protease regulatory subunit (24) that is required for apoptosis (20, 47). (We note that the name AIP1 also refers to an unrelated gene product that interacts with the actin cytoskeleton [14, 37].) Our findings here argue that Rim20p has a fairly direct role in the Rim101p cleavage reaction and yield testable predictions for extrapolation to other PalA/Rim20p homologs. Our findings support a model for Rim101p cleavage that is distinct from a recent proposal for PacC cleavage (10); this distinction highlights differences in the respective cleavage reactions that we reconcile in the Discussion.
MATERIALS AND METHODS
Strains and media.
S. cerevisiae strains were derived from the SK-1 background (18) except for the two-hybrid strains Y187 and Y190 (8). The epitope-tagged RIM101-HA2 allele has been described (23). The rim101Δ::His3MX6, rim20Δ::His3MX6, and bro1Δ::His3MX6 mutations remove each entire open reading frame (ORF) and were introduced by PCR product-directed gene disruption, using plasmid pFA6a-His3MX6 (25) as the His3MX6 template. PCR with outside primers confirmed genotypes; primer sequences are available by request. Strain construction involved standard methods, including mating, meiotic crosses, and transformations (17). For the experiments in Fig. 1 and 5A to C, all strains are derived from WXY169 (MATα RIM101-HA2 ura3 leu2 trp1 his3 lys2 gal80::LEU2 rme1Δ5::LEU2 ho::LYS2). For the experiments in Fig. 2, all strains are derived from KB395 (MATa ura3 leu2 trp1 his3 lys2 gal80::LEU2 ho::LYS2). In this report, we use the prefix Ca for Candida albicans genes and gene products to distinguish them from their S. cerevisiae homologs. Medium composition followed standard recipes (17).
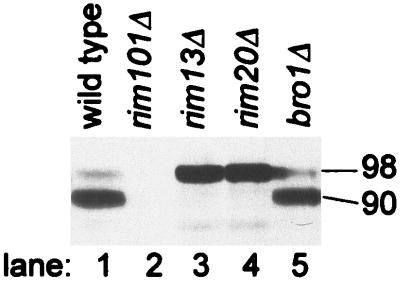
FIG. 1.
Processing of epitope-tagged Rim101-HA2p in wild-type and mutant strains. Extracts from yeast strains grown in YPD medium were analyzed on an anti-HA Western blot to visualize Rim101-HA2p. The masses of Rim101-HA2p forms, indicated along the right margin (in kilodaltons), were estimated by comparison with size standards. The strains included WXY169 (RIM101-HA2; lane 1), WXY220 (rim101Δ; lane 2), WXY185 (RIM101-HA2 rim13Δ; lane 3), WXY219 (RIM101-HA2 rim20Δ; lane 4), and WXY225 (RIM101-HA2 bro1Δ; lane 5).
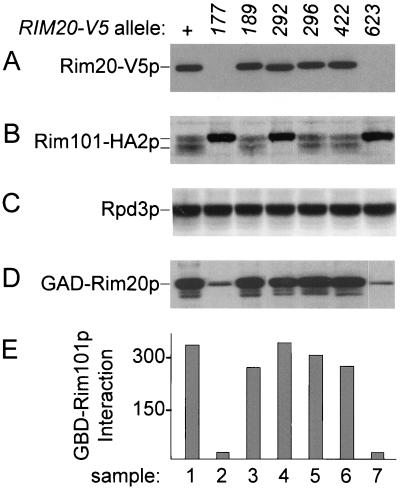
FIG. 5.
Functional analysis of Rim20p mutant derivatives. Strain WXY219 (rim20Δ::His3MX6 RIM101-HA2) was transformed with plasmids specifying Rim20-V5p, Rim20-V5–177p, Rim20-V5–189p, Rim20-V5–292p, Rim20-V5–296p, Rim20-V5–422p, or Rim20-V5–623p (samples 1 to 7, respectively). Extracts of transformants were used for Western analysis with anti-V5 (panel A), anti-HA (panel B), or anti-Rpd3p (panel C). Strain Y187 was transformed with plasmids specifying wild-type GAD-Rim20p and each respective mutant derivative. Extracts were used for Western analysis with anti-HA (panel D), and strains were used for two-hybrid interaction assays with GBD-Rim101p (panel E). In panel E, the y-axis shows the β-galactosidase activity (in Miller units).
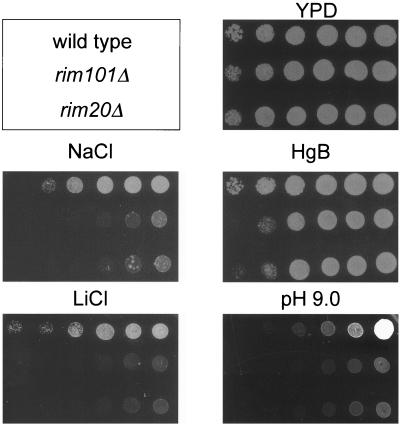
FIG. 2.
Comparison of rim20Δ and rim101Δ mutant phenotypes. Yeast strains were grown to saturation in YPD, and then growth of serially diluted culture samples was compared on YPD (control), YPD containing hygromycin B (HgB), NaCl, or LiCl, or YPD titrated to pH 9. Cell density of the inoculum decreases from right to left in each row. Strains were KB395 (top row; wild type) and its otherwise isogenic derivatives WXY265 (middle row; rim101Δ) and WXY263 (bottom row; rim20Δ).
Growth assays.
Wild-type strain KB395 and otherwise isogenic strains WXY265 (rim101Δ::His3MX6) and WXY263 (rim20Δ::His3MX6) were grown in YPD to approximately the same cell density. Each culture was diluted 1:5, 1:25, 1:125, 1:625, and 1:3,125 with fresh YPD medium, and 3 μl of each of the dilutions was spotted onto a YPD plate, a YPD plate buffered with 100 mM HEPES at pH 9.0, or YPD plates supplemented with 50 μg of hygromycin B per ml, 0.4 M NaCl, or 20 mM LiCl. The plates were incubated at 30°C for 1 to 4 days before the growth was scored.
GBD and GAD fusion plasmids.
To generate fusions of the Gal4p DNA-binding domain (GBD) to Rim101p (residues 1 to 628, 99 to 628, 297 to 628, 400 to 628, 501 to 628, 547 to 628, 1 to 546, 99 to 546, and 297 to 546) or CaRim101p (residues 408 to 604), an NcoI site was engineered in forward primers (pRIM101-F/300F/900F/1200F/1500F/1650F and pCaRIM101 to 1200F), and a BamHI site in reverse primers (pRIM101-R2/1600R and pCaRIM101-R). (Primer sequences are available by request.) The ATG within the NcoI site is in frame with the coding sequences. Genomic DNA from S. cerevisiae strain AMP108 (MATα ura3 leu2 trp1 lys2 ho::LYS2) or C. albicans strain BWP17 (ura3/ura3 his1/his1 arg4/arg4 [3]) was used as the PCR template. PCR products were first cloned into pGEM-Easy vectors (Promega). Inserts released by NcoI-BamHI double digestions were moved into NcoI-BamHI-digested vector pAS1-CYH2. Gal4p activation domain (GAD)-Rim101p fusions were generated by a similar strategy.
To generate fusions of GAD to Rim20p (residues 1 to 661 and 353 to 661) or CaRim20p (404 to 613), an NcoI site was engineered in forward primers (pRIM20-F/1050F and pCaRIM20–1200F) and a BamHI site was engineered in reverse primers (pRIM20-R and pCaRIM20-R). PCR products were first cloned into pGEM-Easy vectors (Promega). Inserts released by NcoI-BamHI double digestions were moved into NcoI-BamHI-digested vector pACTII. GBD-Rim20p fusions were generated by a similar strategy.
Construction of RIM20 substitution mutants and V5 epitope-tagged RIM20.
RIM20 alleles with different point mutations were created by PCR-directed sequence alterations. To create rim20–177, first-round PCR products were generated with S. cerevisiae strain AMP108 genomic DNA as the template and oligonucleotide pRIM20-F3 paired with pRIM20-AQAQE-R and pRIM20-AQAQE-F paired with pRIM20-R. Two PCR products were purified and used as the template in the second-round PCR with oligonucleotide pRIM20-F3 paired with pRIM20-R. The resulting 2.1-kb DNA fragment was purified and cloned into vector pGEM-Easy to generate plasmid pWX73. Plasmids pWX74, -75, -76, -77, and -78, which carry the other five RIM20 mutant alleles, were created by the same strategy. The nucleotide sequence of each entire ORF was verified by direct determination. Inserts released from plasmids pWX73 to -78 by NcoI-BamHI double digestions were moved into NcoI-BamHI-digested vector pACTII to create plasmids pWX121 to -126.
To construct the plasmid that expresses V5 epitope-tagged RIM20 from its native promoter, RIM20 gene 5′-flanking sequence (−1000 to +60) and 3′-flanking sequence (from 60 bp upstream of the stop codon to 1 kb downstream of the stop codon) were first cloned into vector pRS314 to create plasmid pWX179. pWX179 was then linearized by BamHI and purified. PCR products generated with plasmid pWX60 (pGEM-RIM20) as the template and oligonucleotide pRIM20-F3 paired with pRIM20-R were cotransformed with linearized pWX179 into yeast strain WXY219 for in vivo recombination in order to generate plasmid pWX192 (pRS314-RIM20). pWX192 was linearized by SacI and purified. PCR products generated with pYES2/GS (Invitrogen) as the template and oligonucleotide pRIM20C-V5-F paired with pRS314-V5-R2 were cotransformed with linearized pWX192 into yeast strain WXY219 for in vivo recombination in order to generate plasmid pWX272 (pRS314-RIM20-V5). The plasmids carrying mutant RIM20 alleles, pWX273 to -278, were created by the same strategy.
In vitro transcription template.
A PCR product including RIM101 codons 297 to 628, amplified from genomic DNA of strain AMP108, was cloned into vector pGEM-Easy. The T7 promoter on the vector was used to direct in vitro transcription, and the ATG contained within the SphI site on the vector served as the start codon.
Two-hybrid interaction assays.
Strain Y190, carrying various GBD fusion plasmids, was mated with strain Y187, carrying various GAD fusion plasmids, and diploids were selected on plates of SC medium lacking Trp and Leu (SC−Trp−Leu plates). For assays, 24-h cultures in SC−Trp−Leu were diluted 1:25 into SC−Trp−Leu medium and harvested after three doublings. β-Galactosidase assays were conducted on permeabilized cells as previously described (45).
Western blot assays.
Cells from 10 ml of mid-log-phase culture in YPD were pelleted, resuspended in 100 μl of 3× Laemmli buffer, and boiled for 5 min. After centrifugation, 5 to 20 μl of the supernatants was fractionated on a sodium dodecyl sulfate (SDS)–9% polyacrylamide gel and transferred to nitrocellulose. For hemagglutinin (HA) epitope detection, the filter was probed with anti-HA monoclonal antibody 12CA5 (Babco; 10−4 dilution) and goat anti-mouse immunoglobulin G (IgG) conjugated to peroxidase (Boehringer; 10−4 dilution) and developed with enhanced chemiluminescence (ECL) detection reagents (Amersham). For V5 epitope detection, the filter was probed with anti-V5-horseradish peroxidase antibody (Invitrogen; 1:5,000 dilution in TBST [Tris-buffered saline plus Tween]) and developed with ECL detection reagents (Amersham). The control protein Rpd3p was detected as described previously (21).
In vitro binding assays.
35S-labeled Rim101p(297–628) and the luciferase control were produced using the TnT Quick Coupled transcription/translation kit (Promega), following the manufacturer's instructions. Whole-cell extracts were prepared (27) from mid-log-phase SC−Leu cultures of strain Y187 carrying GAD or GAD-Rim20p plasmids. For assays, 100 μl of extract (10 μg of total protein/μl), 5 μl of in vitro translation mix containing Rim101p(297–628), 5 μl of in vitro translation mix containing the luciferase control, 100 μl of bovine serum albumin (New England Biolabs; 10 μg/μl), and 100 μl of extraction buffer (50 mM Tris-Cl [pH 7.4], 100 mM NaCl, 50 mM KCl, 2 mM EDTA, 5% glycerol, 0.1% β-mercaptoethanol, plus 0.1 mg of phenylmethylsulfonyl fluoride and 1 μg each of leupeptin, aprotinin, and pepstatin per ml) were mixed in a 1.5-ml tube. Then 3 μl of anti-HA monoclonal antibody 12CA5 (Babco) or H2O was added to the experimental and control tubes, respectively. After 10 min at room temperature, 100 μl of a 50% protein A-Sepharose bead suspension (Sigma) was added to each tube. Tubes were incubated at 4°C for 45 min. The beads were washed three times in extraction buffer and boiled for 5 min in 3× Laemmli buffer. After centrifugation, supernatants were fractionated on an SDS–9% polyacrylamide gel and transferred to nitrocellulose. The filter was first exposed to a phosphor screen (Molecular Dynamics) for 36 h to detect 35S-labeled Rim101p(297–628) and luciferase and then probed with anti-HA monoclonal antibody to detect GAD and GAD-Rim20p.
RESULTS
Identification of S. cerevisiae RIM20
The S. cerevisiae genes YOR275C (henceforth RIM20) and BRO1 (32) are homologs of A. nidulans PalA. To determine whether their gene products are required for Rim101p processing, we examined the electrophoretic mobility of epitope-tagged Rim101-HA2p in rim20Δ and bro1Δ null mutants (Fig. 1). Rim101-HA2p was detected primarily as a 90-kDa processed form in a wild-type control extract (lane 1) and as a 98-kDa unprocessed form in a rim13Δ control extract (lane 3). The anti-HA reaction was specific for Rim101-HA2p, because no 90- or 98-kDa protein was detected in a rim101Δ extract (lane 2). We found only the 98-kDa form in a rim20Δ extract (lane 4), and failure to produce 90-kDa processed Rim101-HA2p cosegregated with an independently constructed rim20Δ mutation in two meiotic tetrads (data not shown). The bro1Δ mutation did not affect Rim101-HA2p processing (Fig. 1, lane 5). These results indicate that Rim20p is required for Rim101p proteolytic processing.
Mutations that block Rim101p processing cause the same set of phenotypes as rim101Δ mutations (22, 23). Thus, we expected that rim20Δ and rim101Δ mutants should have similar phenotypes. We found that both mutations caused sensitivity to NaCl, LiCl, and hygromycin B and caused weak growth in medium titrated to pH 9 (Fig. 2). Also, the rim20Δ mutation caused weak sporulation and invasive growth (data not shown), as does a rim101Δ mutation (23). These observations are consistent with the finding that Rim20p is required for Rim101p processing.
Interaction of Rim20p and Rim101p.
To gain mechanistic insight into the role of Rim20p in Rim101p processing, we used two-hybrid assays to determine whether Rim101p and Rim20p interact with each other. We created plasmids that express GAD and GBD fusions to full-length Rim20p and Rim101p. Expression of a gal1-lacZ reporter in strains carrying pairs of fusions was then used as a measure of interaction in two-hybrid assays. These assays revealed detectable Rim20p-Rim20p interaction (data not shown) and Rim101p-Rim20p interaction.
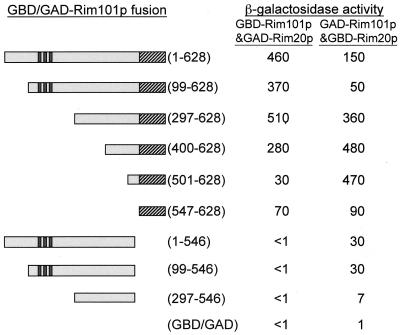
In assays of GAD-Rim20p with GBD-Rim101p derivatives (Fig. 3), we observed interaction with the Rim101p segments 1 to 628, 99 to 628, 297 to 628, and 400 to 628 and slightly weaker interaction with Rim101p segments 501 to 628 and 547 to 628. No interaction was detected with Rim101p segments 1 to 546, 99 to 546, and 297 to 546 or with the GBD domain alone. Largely similar results were obtained with two-hybrid assays of GBD-Rim20p with GAD-Rim101p derivatives (Fig. 3), but these assays revealed interaction between Rim20p and two nonoverlapping segments of Rim101p. N-terminal segments 1 to 546, 99 to 546, and 297 to 546 were capable of weak interaction, and the C-terminal region 547 to 628 was capable of moderate interaction. GAD-Rim101p derivatives were detectable through Western analysis, whereas GBD-Rim101p derivatives were not (data not shown). Therefore, we have greater confidence in the results obtained with GAD-Rim101p derivatives. Together, these results indicate that Rim20p interacts with two regions of Rim101p and that the Rim101p C-terminal PEST-like region is sufficient for Rim20p interaction.
FIG. 3.
Two-hybrid assays of Rim101p-Rim20p interaction. Expression of a gal1-lacZ reporter gene (β-galactosidase activity in Miller units) was measured in strains expressing either GBD-Rim101p derivatives along with GAD-Rim20p or GAD-Rim101p derivatives along with GBD-Rim20p. Features of Rim101p diagrammed include three zinc fingers (dark stripes) and a C-terminal PEST-like region (hatched box). The GBD/GAD-Rim101p fusions included the residues indicated in parentheses; the bottom entry is a vector control. β-Galactosidase activity is the mean for three independent transformants, and the range of values was less than 20% of the mean value for each pair of fusions. All GBD-Rim101p and GAD-Rim101p derivatives yielded <1 U of activity when coexpressed with GAD or GBD (lacking Rim20p sequences).
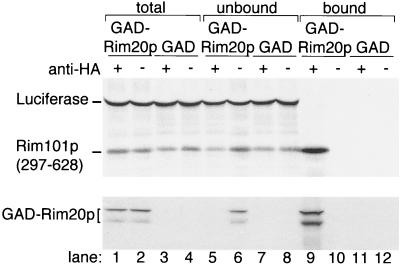
An in vitro binding assay provided biochemical confirmation of Rim101p-Rim20p interaction. Here we measured binding of an in vitro-synthesized Rim101p C-terminal segment (residues 297 to 628) to GAD-Rim20p in yeast crude extracts. Rim101p(297–628) and a control protein, firefly luciferase, were synthesized and 35S labeled through in vitro transcription and translation reactions (Fig. 4, lane 1). Aliquots of the reaction were incubated with extracts of yeast expressing GAD-Rim20p or GAD alone to permit binding; the GAD segment includes an HA epitope tag. Then, monoclonal anti-HA antibody was added, immune complexes were bound to protein A-Sepharose and pelleted, and both supernatant and pellet proteins were visualized after SDS-polyacrylamide gel electrophoresis (PAGE) (Fig. 4, lanes 5 to 8 and 9 to 12, respectively). Rim101p(297–628) was recovered along with GAD-Rim20p (lane 9). Interaction was specific, because luciferase was not recovered in anti-HA immune complexes containing GAD-Rim20p. Also, Rim101p(297–628) was not recovered from immune complexes containing GAD in place of GAD-Rim20p (lane 11) or in pellets that lacked anti-HA antibody (lanes 10 and 12). These results verify that Rim20p interacts with the C-terminal region of Rim101p.
FIG. 4.
Binding of the Rim101p C-terminal region to GAD-Rim20p. Radiolabeled Rim101p C-terminal region (residues 297 to 628) and firefly luciferase were synthesized in vitro and added to extracts of yeast strain Y187 containing GAD-Rim20p (lanes 1, 2, 5, 6, 9, and 10) or GAD (lanes 3, 4, 7, 8, 11, and 12). The mixtures were incubated with (+, odd lanes) or without (−, even lanes) anti-HA IgG, and immune complexes were collected with protein A-Sepharose. After SDS-PAGE, Rim101p and luciferase were visualized with a PhosphorImager (upper panel) and GAD-Rim20p was visualized by Western analysis (lower panel). The samples include the complete binding mixtures (lanes 1 to 4; 1/20 of total reaction), the supernatants after removal of protein A-Sepharose (lanes 5 to 8; 1/20 of total reaction), and the protein A-Sepharose pellets (lanes 9 to 12; 1/2 of entire reaction). We note that GAD-Rim20p is recovered in two forms that are resolved by electrophoresis (lane 1 compared to lane 3). We believe that the smaller form is an artifactual in vitro cleavage product.
If Rim101p-Rim20p interaction has functional significance, then it should be conserved in other species. We used C. albicans CaRim101p and CaRim20p to test this prediction. CaRIM101 and CaRIM20 were identified recently through homology searches (3, 35, 51), and genetic studies indicate that CaRim20p acts upstream of CaRim101p in a signal transduction pathway (3). CaRIM101 and CaRIM20 have two and five CUG codons, respectively, which are subject to nonstandard translation in C. albicans (40). To minimize mistranslation in the S. cerevisiae two-hybrid host, we used GBD-CaRim101p and GAD-CaRim20p fusions that included only C-terminal segments, with zero and two CUG codons, respectively (Table 1). Two-hybrid assays revealed that the CaRim101p C-terminal region interacts with the CaRim20p C-terminal region (Table 1). In parallel studies with S. cerevisiae protein segments, we determined that Rim101p also interacts with the Rim20p C-terminal region (Table 1). Therefore, Rim101p-Rim20p interaction is conserved in two fungi. This finding argues that Rim101p-Rim20p interaction has functional significance.
TABLE 1.
Conservation of two-hybrid interaction between Rim101p and Rim20p C-terminal regionsa
| Rim20p/Rim101p source | GAD fusion | GBD fusion | β-Galactosidase activity |
|---|---|---|---|
| C. albicans | CaRim20p (404-613) | CaRim101p (408-604) | 34.6 |
| CaRim20p (404-613) | Vector | <1 | |
| Vector | CaRim101p (408-604) | <1 | |
| S. cerevisiae | Rim20p (353-661) | Rim101p (1-628) | 160 |
| Rim20p (353-661) | Vector | <1 | |
| Vector | Rim101p (1-628) | <1 |
RIM20 and RIM101 ORF segments from the species indicated were cloned into GAD and GBD fusion vectors and transformed into the two-hybrid host Y187 × Y190. Numbers in parentheses indicate the ORF segments, in codon numbers, that were fused to GAD or GBD. Vector refers to plasmids with no insert. Transformants were grown in selective medium, and interaction was measured through activation of gal1-lacZ expression. β-Galactosidase activity, in Miller units (17), is the average for three independent transformants. The range of values was less than 20% of the mean value for each pair of fusions.
Role of conserved Rim20p regions in Rim101p processing.
Rim20p is a member of a broadly conserved protein family. Some family members presumably have the same function as Rim20p, such as A. nidulans PalA and C. albicans Rim20p, which act in the respective organisms' PacC/Rim101p processing pathway. Other family members may have distinct functions, such as S. cerevisiae Bro1p and mammalian Alix. We carried out a mutational analysis of conserved Rim20p residues to establish a basis for structural comparison. Clusters of two or three highly conserved residues were replaced with alanine residues (Table 2), and accumulation and function of the six resulting mutant Rim20p derivatives were assessed (Fig. 5).
TABLE 2.
Amino acid substitutions in conserved regions of Rim20pa
| Rim20p region | Mutant protein
|
||
|---|---|---|---|
| Designation | Accumulation | Function | |
| 176-AQAQE-180 | Rim20-177p | − | (−) |
| 189-DKHKD-193 | Rim20-189p | + | + |
| 289-AQRDN-293 | Rim20-292p | + | − |
| 293-NEFIY-297 | Rim20-296p | + | + |
| 420-LNEEA-424 | Rim20-422p | + | + |
| 621-TAFYE-625 | Rim20-623p | − | (−) |
These amino acid regions are conserved among S. cerevisiae Rim20p and homologs C. albicans Rim20p, A. nidulans PalA, S. cerevisiae Bro1p, Drosophila melanogaster CG12896, Caenorhabditis elegans Ynk1, Xenopus laevis Xp95, and Mus musculus Alix-AIP1. Residues printed in boldface are identical in at least six proteins of the group. The residues indicated by underlining were replaced with alanine to create mutant proteins. Accumulation of each mutant protein is summarized from the data in Fig. 5. The function of each mutant protein is summarized from the Rim101p processing assays in Fig. 5. (−), absence of detectable function for mutant proteins with accumulation defects.
Accumulation of wild-type and mutant derivatives was compared through use of RIM20-V5, which specifies Rim20p with a C-terminal V5 epitope (43). DNA sequences specifying the epitope and a CYC1 transcriptional terminator were introduced at the 3′ end of the RIM20 coding region on a low-copy-number plasmid that also includes RIM20 promoter sequences. Rim20-V5p was identified on an anti-V5 immunoblot as an ∼85-kDa protein (Fig. 5A, lane 1) present only in strains carrying the RIM20-V5 plasmid (data not shown). Mutant derivatives Rim20-V5–189p, Rim20-V5–292p, Rim20-V5–296p, and Rim20-V5–422p accumulated at levels comparable to that of wild-type Rim20-V5p (Fig. 5A, lanes 3 to 6 compared to lane 1), but Rim20-V5–177p and Rim20-V5–623p were not detectable (lanes 2 and 7). All of the samples had comparable amounts of extract protein, as verified by probing for control protein Rpd3p (Fig. 5C). We found similar relative accumulation levels of each overexpressed GAD-Rim20p derivative (Fig. 5D). Therefore, the substitutions in Rim20-V5–189p, Rim20-V5–292p, Rim20-V5–296p, and Rim20-V5–422p do not cause obvious defects in stability.
Functional activity of each Rim20-V5p derivative was determined through assessment of Rim101-HA2p processing in rim20Δ RIM101-HA2 strains carrying each RIM20-V5 plasmid (Fig. 5B). The wild-type RIM20-V5 allele was functional, as indicated by the presence of the 90-kDa Rim101-HA2p form (Fig. 5B, lane 1). Mutants RIM20-V5–177, -292, and -623 were nonfunctional (lanes 2, 4, and 7); mutants RIM20-V5–189, -296, and -422 were functional (lanes 3, 5, and 6). We infer that the V5 epitope does not alter mutant protein activity substantially, because we obtained comparable results with Rim20p derivatives lacking the V5 epitope tag (data not shown). Therefore, among mutant proteins that are expressed, only Rim20-V5–292p has a severe defect in activity.
A defect in Rim20p functional activity may reflect a defect in interaction with Rim101p. We used two-hybrid assays of GBD-Rim101p and each GAD-Rim20p derivative to measure this interaction (Fig. 5E). GAD-Rim20–177p and GAD-Rim20–623p had apparent interaction defects, as expected from their accumulation defects (Fig. 5D, lanes 2 and 7). All other GAD-Rim20p derivatives were fully capable of interaction with Rim101p (Fig. 5E). Therefore, this set of highly conserved Rim20p residues, as represented by the stable mutant derivatives, is not required for Rim101p-Rim20p interaction. In addition, we conclude that the functional defect of Rim20-V5–292p does not arise from an inability to interact with Rim101p.
DISCUSSION
Rim20p represents a family of eukaryotic proteins whose molecular functions are largely unknown. Its closest homologs include PalA and C. albicans Rim20p, which act in conserved PacC-Rim101p processing pathways, and here we confirmed the requirement for Rim20p in S. cerevisiae Rim101p processing. Our analysis indicates that Rim20p interacts with the Rim101p C-terminal PEST-like region in both S. cerevisiae and C. albicans. The conservation of this interaction argues that it is functionally important, and the interaction between a processing pathway member and the cleavage target suggests that Rim20p has a relatively direct role in the processing reaction, rather than an indirect role in the pH-sensing signal transduction pathway.
All S. cerevisiae homologs of PacC processing pathway proteins are required for Rim101p processing (5, 22, 46; W. Xu and A. P. Mitchell, unpublished results), so the mechanisms of Rim101p and PacC processing are almost certainly similar. However, our results underscore a key difference between the processing reactions. Rim101p is activated by cleavage adjacent to the PEST-like region (near residue 530), whereas PacC is activated by cleavage far from the PEST-like region (near residue 253). Truncation mutations that remove only the PEST-like region of Rim101p or PacC yield activated proteins that can function independently of Rim20p/PalA and the protease Rim13p/PalB, but these truncations have very different molecular consequences. The Rim101p truncation product accumulates at the approximate size of the primary translation product, whereas PacC truncation products are cleaved, independently of PalA and PalB, to remove an additional ∼320-residue C-terminal segment. Thus, in wild-type strains, PacC cleavage may occur in two steps: an initial cleavage removes the ∼100-residue PEST-like region, and a second cleavage removes a further ∼320-residue C-terminal segment. The first cleavage would be analogous to Rim101p cleavage and dependent on PalA and PalB; the second cleavage would be dependent on the first cleavage but otherwise independent of PalA and PalB. This model explains why Rim101p and PacC both have C-terminal PEST-like regions and have homologous processing pathways yet exist as quite different mature products.
This model also reconciles the differences in sequence requirements for proteolytic activation of Rim101p and PacC. For Rim101p, the finding that Rim20p binds to the PEST-like region suggests that this region has a vital role in the proteolytic activation of Rim101p. However, for PacC, deletion derivatives that lack its PEST-like region undergo proteolytic cleavage, and an internal PacC region is required for PacC cleavage (10, 28). According to our model, the internal PacC region directs cleavage by gene products distinct from the conserved Pal-Rim gene products; the PacC PEST-like region directs cleavage by PalA, PalB, and other conserved Pal-Rim gene products.
How do our findings reflect on the functions of more distant Rim20p homologs? Sequence identity between Rim20p and mouse Alix is 26%, which points to significant common structural features. We found that alanine substitutions in two conserved regions cause protein accumulation defects, as expected if the mutant proteins were misfolded (50). A simple interpretation is that these two regions are critical for proper folding of Rim20p. Also, alanine substitutions for the highly conserved residues D292 and N293 abolished Rim20p function but did not affect its accumulation or interaction with Rim101p. This region may be required for a second critical interaction (such as with Snf7p, as discussed below), a conformational or oligomeric change, or a possible catalytic activity.
Our most surprising observation is that alanine substitutions in three highly conserved regions have no detectable effect on Rim20p function. These regions may make redundant contacts with other molecules, so that the defects are overcome by compensatory interactions elsewhere in Rim20p. These observations may guide the types of mutations and analyses used to characterize Rim20p homologs. Finally, we note that the Rim20p C-terminal half is sufficient for Rim101p interaction. The C-terminal half of Rim20p is less conserved than the N-terminal half, with 23 and 30% identity to Alix, respectively, for example. This observation raises the interesting possibility that the diversity of C-terminal sequences among Rim20p homologs reflects the diversity of possible protease substrates that they recognize.
Two general models for Rim20p function can account for our observations. One model is that Rim20p promotes interaction between Rim101p and the Rim13p protease. Rim20p may act as a scaffold that binds to both Rim101p and Rim13p, or it may act as a targeting subunit that directs Rim101p to the subcellular location of Rim13p. A second model is that Rim20p acts as a chaperone that ensures that the Rim101p cleavage site is exposed to solvent and thus accessible to Rim13p. These models are not mutually exclusive, in that Rim20p may have several roles in the cleavage reaction.
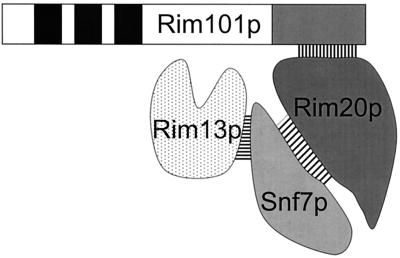
The genome-wide two-hybrid analysis of Ito et al. (16) lends support to the first model. Ito et al. detected interactions between Rim13p and Snf7p and between Snf7p and Yor275Cp, the gene product shown here to be Rim20p. A simple interpretation is that the Rim20p-Snf7p complex provides a scaffold that holds Rim13p and the Rim101p PEST-like region in close proximity, promoting Rim101p cleavage and activation (Fig. 6). In principle, the need for a scaffold might be bypassed by overexpression of Rim13p, so that mass action would promote Rim101p-Rim13p interaction. However, two different RIM13 overexpression plasmids fail to promote Rim101p processing in a rim20Δ mutant, though they do complement a rim13Δ mutant (W. Xu and A. P. Mitchell, unpublished results). Thus, it is possible that Rim20p has a more complex role than simply to promote interaction of protease and target. We note that the scaffold Ste5p is thought to act both to tether mitogen-activated protein kinase cascade components and to promote signal transmission through dimerization and membrane localization (9, 15, 26, 34, 52). Like Ste5p, Rim20p may have roles in both scaffolding and signaling.
FIG. 6.
Model for Rim101p cleavage complex. The Rim13p-Snf7p and Rim20p-Snf7p interactions were reported by Ito et al. (16), and here we report that Rim20p and Rim101p interact. We propose that the Rim20p-Snf7p complex acts as a scaffold to promote interaction between the Rim101p cleavage site and the Rim13p protease.
The finding that Rim20p is an Snf7p-interacting protein that promotes Rim101p cleavage has an interesting implication for recognition of the external signal, alkaline pH, that stimulates Rim101p cleavage (23). Snf7p is an endosome-associated protein that acts in the Vps4p complex to promote fusion of the late endosome with the vacuole (1, 19). This endosome includes surface receptors, so it is possible that a surface receptor governing Rim101p processing is active only after its endocytosis, as may be the case for dynamin-dependent receptors of higher eukaryotes (7). A second possibility is that the acidification of endocytosed fluid is used by the Rim101p processing pathway as a measure of external pH. Clearly, it will be vital to determine whether Rim101p processing depends on Snf7p and on endosome-vacuole fusion as a first test of these models.
ACKNOWLEDGMENTS
We thank Teresa Lamb for comments on the manuscript and all members of this lab for many helpful discussions.
This work was supported by research grant GM39531 from the NIH.
REFERENCES
- 1.Babst M, Wendland B, Estepa E J, Emr S D. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 1998;17:2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ciechanover A. The ubiquitin-proteasome pathway: on protein death and cell life. EMBO J. 1998;17:7151–7160. doi: 10.1093/emboj/17.24.7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis D, Wilson R B, Mitchell A P. RIM101-dependent and -independent pathways govern pH responses in Candida albicans. Mol Cell Biol. 2000;20:971–978. doi: 10.1128/mcb.20.3.971-978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeMartino G N, Slaughter C A. The proteasome, a novel protease regulated by multiple mechanisms. J Biol Chem. 1999;274:22123–22126. doi: 10.1074/jbc.274.32.22123. [DOI] [PubMed] [Google Scholar]
- 5.Denison S H, Negrete-Urtasun S, Mingot J M, Tilburn J, Mayer W A, Goel A, Espeso E A, Penalva M A, Arst H N., Jr Putative membrane components of signal transduction pathways for ambient pH regulation in Aspergillus and meiosis in saccharomyces are homologous. Mol Microbiol. 1998;30:259–264. doi: 10.1046/j.1365-2958.1998.01058.x. [DOI] [PubMed] [Google Scholar]
- 6.Denison S H, Orejas M, Arst H N., Jr Signaling of ambient pH in Aspergillus involves a cysteine protease. J Biol Chem. 1995;270:28519–28522. doi: 10.1074/jbc.270.48.28519. [DOI] [PubMed] [Google Scholar]
- 7.Di Fiore P P, De Camilli P. Endocytosis and signaling, an inseparable partnership. Cell. 2001;106:1–4. doi: 10.1016/s0092-8674(01)00428-7. [DOI] [PubMed] [Google Scholar]
- 8.Durfee T, Becherer K, Chen R, Yeh S H, Yang Y, Killburn A E, Lee W H, Elledge S J. The retinoblastoma protein associates with protein the phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 9.Elion E A. Pheromone response, mating and cell biology. Curr Opin Microbiol. 2000;3:573–581. doi: 10.1016/s1369-5274(00)00143-0. [DOI] [PubMed] [Google Scholar]
- 10.Espeso E A, Roncal T, Diez E, Rainbow L, Bignell E, Alvaro J, Suarez T, Denison S H, Tilburn J, Arst H N, Jr, Penalva M A. On how a transcription factor can avoid its proteolytic activation in the absence of signal transduction. EMBO J. 2000;19:719–728. doi: 10.1093/emboj/19.4.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hersh D, Monack D M, Smith M R, Ghori N, Falkow S, Zychlinsky A. The Salmonella invasin SipB induces macrophage apoptosis by binding to caspase-1. Proc Natl Acad Sci USA. 1999;96:2396–2401. doi: 10.1073/pnas.96.5.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoppe T, Rape M, Jentsch S. Membrane-bound transcription factors: regulated release by RIP or RUP. Curr Opin Cell Biol. 2001;13:344–348. doi: 10.1016/s0955-0674(00)00218-0. [DOI] [PubMed] [Google Scholar]
- 13.Huggins G S, Chin M T, Sibinga N E, Lee S L, Haber E, Lee M E. Characterization of the mUBC9-binding sites required for E2A protein degradation. J Biol Chem. 1999;274:28690–28696. doi: 10.1074/jbc.274.40.28690. [DOI] [PubMed] [Google Scholar]
- 14.Iida K, Yahara I. Cooperation of two actin-binding proteins, cofilin and Aip1, in Saccharomyces cerevisiae. Genes Cells. 1999;4:21–32. doi: 10.1046/j.1365-2443.1999.00235.x. [DOI] [PubMed] [Google Scholar]
- 15.Inouye C, Dhillon N, Thorner J. Ste5 RING-H2 domain: role in Ste4-promoted oligomerization for yeast pheromone signaling. Science. 1997;278:103–106. doi: 10.1126/science.278.5335.103. [DOI] [PubMed] [Google Scholar]
- 16.Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaiser C, Michaelis S, Mitchell A. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 18.Kane S, Roth R. Carbohydrate metabolism during ascospore development in yeast. J Bacteriol. 1974;118:8–14. doi: 10.1128/jb.118.1.8-14.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kranz A, Kinner A, Kolling R. A family of small coiled-coil-forming proteins functioning at the late endosome in yeast. Mol Biol Cell. 2001;12:711–723. doi: 10.1091/mbc.12.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krebs J, Klemenz R. The ALG-2/AIP-complex, a modulator at the interface between cell proliferation and cell death? A hypothesis. Biochim Biophys Acta. 2000;1498:153–161. doi: 10.1016/s0167-4889(00)00091-4. [DOI] [PubMed] [Google Scholar]
- 21.Lamb T M, Mitchell A P. Coupling of Saccharomyces cerevisiae early meiotic gene expression to DNA replication depends upon RPD3 and SIN3. Genetics. 2001;157:545–556. doi: 10.1093/genetics/157.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lamb T M, Xu W, Diamond A, Mitchell A P. Alkaline response genes of S. cerevisiae and their relationship to the RIM101 pathway. J Biol Chem. 2001;276:1850–1856. doi: 10.1074/jbc.M008381200. [DOI] [PubMed] [Google Scholar]
- 23.Li W, Mitchell A P. Proteolytic activation of Rim1p, a positive regulator of yeast sporulation and invasive growth. Genetics. 1997;145:63–73. doi: 10.1093/genetics/145.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lo K W, Zhang Q, Li M, Zhang M. Apoptosis-linked gene product ALG-2 is a new member of the calpain small subunit subfamily of Ca2+-binding proteins. Biochemistry. 1999;38:7498–7508. doi: 10.1021/bi990034n. [DOI] [PubMed] [Google Scholar]
- 25.Longtine M S, McKenzie A, 3rd, Demarini D J, Shah N G, Wach A, Brachat A, Philippsen P, Pringle J R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 26.Mahanty S K, Wang Y, Farley F W, Elion E A. Nuclear shuttling of yeast scaffold Ste5 is required for its recruitment to the plasma membrane and activation of the mating MAPK cascade. Cell. 1999;98:501–512. doi: 10.1016/s0092-8674(00)81978-9. [DOI] [PubMed] [Google Scholar]
- 27.Malathi K, Xiao Y, Mitchell A P. Interaction of yeast repressor-activator protein Ume6p with glycogen synthase kinase 3 homolog Rim11p. Mol Cell Biol. 1997;17:7230–7236. doi: 10.1128/mcb.17.12.7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mingot J M, Tilburn J, Diez E, Bignell E, Orejas M, Widdick D A, Sarkar S, Brown C V, Caddick M X, Espeso E A, Arst H N, Jr, Penalva M A. Specificity determinants of proteolytic processing of Aspergillus PacC transcription factor are remote from the processing site, and processing occurs in yeast if pH signaling is bypassed. Mol Cell Biol. 1999;19:1390–1400. doi: 10.1128/mcb.19.2.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Missotten M, Nichols A, Rieger K, Sadoul R. Alix, a novel mouse protein undergoing calcium-dependent interaction with the apoptosis-linked-gene 2 (ALG-2) protein. Cell Death Differ. 1999;6:124–129. doi: 10.1038/sj.cdd.4400456. [DOI] [PubMed] [Google Scholar]
- 30.Molinari M, Carafoli E. Calpain: a cytosolic proteinase active at the membranes. J Membr Biol. 1997;156:1–8. doi: 10.1007/s002329900181. [DOI] [PubMed] [Google Scholar]
- 31.Negrete-Urtasun S, Denison S H, Arst H N., Jr Characterization of the pH signal transduction pathway gene palA of Aspergillus nidulans and identification of possible homologs. J Bacteriol. 1997;179:1832–1835. doi: 10.1128/jb.179.5.1832-1835.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nickas M E, Yaffe M P. BRO1, a novel gene that interacts with components of the Pkc1p-mitogen-activated protein kinase pathway in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2585–2593. doi: 10.1128/mcb.16.6.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ono Y, Sorimachi H, Suzuki K. Structure and physiology of calpain, an enigmatic protease. Biochem Biophys Res Commun. 1998;245:289–294. doi: 10.1006/bbrc.1998.8085. [DOI] [PubMed] [Google Scholar]
- 34.Pryciak P M, Huntress F A. Membrane recruitment of the kinase cascade scaffold protein Ste5 by the Gbetagamma complex underlies activation of the yeast pheromone response pathway. Genes Dev. 1998;12:2684–2697. doi: 10.1101/gad.12.17.2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramon A M, Porta A, Fonzi W A. Effect of environmental pH on morphological development of Candida albicans is mediated via the PacC-related transcription factor encoded by PRR2. J Bacteriol. 1999;181:7524–7530. doi: 10.1128/jb.181.24.7524-7530.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rechsteiner M, Rogers S W. PEST sequences and regulation by proteolysis. Trends Biochem Sci. 1996;21:267–271. [PubMed] [Google Scholar]
- 37.Rodal A A, Tetreault J W, Lappalainen P, Drubin D G, Amberg D C. Aip1p interacts with cofilin to disassemble actin filaments. J Cell Biol. 1999;145:1251–1264. doi: 10.1083/jcb.145.6.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roth A F, Sullivan D M, Davis N G. A large PEST-like sequence directs the ubiquitination, endocytosis, and vacuolar degradation of the yeast a-factor receptor. J Cell Biol. 1998;142:949–961. doi: 10.1083/jcb.142.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sahara S, Aoto M, Eguchi Y, Imamoto N, Yoneda Y, Tsujimoto Y. Acinus is a caspase-3-activated protein required for apoptotic chromatin condensation. Nature. 1999;401:168–173. doi: 10.1038/43678. [DOI] [PubMed] [Google Scholar]
- 40.Santos M A, Tuite M F. The CUG codon is decoded in vivo as serine and not leucine in Candida albicans. Nucleic Acids Res. 1995;23:1481–1486. doi: 10.1093/nar/23.9.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shumway S D, Maki M, Miyamoto S. The PEST domain of IκBα is necessary and sufficient for in vitro degradation by mu-calpain. J Biol Chem. 1999;274:30874–30881. doi: 10.1074/jbc.274.43.30874. [DOI] [PubMed] [Google Scholar]
- 42.Smith G C, di Fagagna F, Lakin N D, Jackson S P. Cleavage and inactivation of ATM during apoptosis. Mol Cell Biol. 1999;19:6076–6084. doi: 10.1128/mcb.19.9.6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Southern J A, Young D F, Heaney F, Baumgartner W K, Randall R E. Identification of an epitope on the P and V proteins of simian virus 5 that distinguishes between two isolates with different biological characteristics. J Gen Virol. 1991;72:1551–1557. doi: 10.1099/0022-1317-72-7-1551. [DOI] [PubMed] [Google Scholar]
- 44.Stennicke H R, Salvesen G S. Properties of the caspases. Biochim Biophys Acta. 1998;1387:17–31. doi: 10.1016/s0167-4838(98)00133-2. [DOI] [PubMed] [Google Scholar]
- 45.Stern M, Jensen R, Herskowitz I. Five SWI genes are required for expression of the HO gene in yeast. J Mol Biol. 1984;178:853–868. doi: 10.1016/0022-2836(84)90315-2. [DOI] [PubMed] [Google Scholar]
- 46.Treton B, Blanchin-Roland S, Lambert M, Lepingle A, Gaillardin C. Ambient pH signaling in ascomycetous yeasts involves homologues of the Aspergillus nidulans genes palF and paIH. Mol Gen Genet. 2000;263:505–513. doi: 10.1007/s004380051195. [DOI] [PubMed] [Google Scholar]
- 47.Vito P, Lacana E, D'Adamio L. Interfering with apoptosis: Ca2+-binding protein ALG-2 and Alzheimer's disease gene ALG-3. Science. 1996;271:521–525. doi: 10.1126/science.271.5248.521. [DOI] [PubMed] [Google Scholar]
- 48.Vito P, Pellegrini L, Guiet C, D'Adamio L. Cloning of AIP1, a novel protein that associates with the apoptosis-linked gene ALG-2 in a Ca2+-dependent reaction. J Biol Chem. 1999;274:1533–1540. doi: 10.1074/jbc.274.3.1533. [DOI] [PubMed] [Google Scholar]
- 49.Wang G, Wang B, Jiang J. Protein kinase A antagonizes Hedgehog signaling by regulating both the activator and repressor forms of Cubitus interruptus. Genes Dev. 1999;13:2828–2837. doi: 10.1101/gad.13.21.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wickner S, Maurizi M R, Gottesman S. Posttranslational quality control: folding, refolding, and degrading proteins. Science. 1999;286:1888–1893. doi: 10.1126/science.286.5446.1888. [DOI] [PubMed] [Google Scholar]
- 51.Wilson R B, Davis D, Mitchell A P. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J Bacteriol. 1999;181:1868–1874. doi: 10.1128/jb.181.6.1868-1874.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yablonski D, Marbach I, Levitzki A. Dimerization of Ste5, a mitogen-activated protein kinase cascade scaffold protein, is required for signal transduction. Proc Natl Acad Sci USA. 1996;93:13864–13869. doi: 10.1073/pnas.93.24.13864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yaglom J, Linskens M H, Sadis S, Rubin D M, Futcher B, Finley D. p34Cdc28-mediated control of Cln3 cyclin degradation. Mol Cell Biol. 1995;15:731–741. doi: 10.1128/mcb.15.2.731. [DOI] [PMC free article] [PubMed] [Google Scholar]