Abstract
Blood lipids are heritable modifiable causal factors for coronary artery disease. Despite well-described monogenic and polygenic bases of dyslipidemia, limitations remain in discovery of lipid-associated alleles using whole genome sequencing (WGS), partly due to limited sample sizes, ancestral diversity, and interpretation of clinical significance. Among 66,329 ancestrally diverse (56% non-European) participants, we associate 428M variants from deep-coverage WGS with lipid levels; ~400M variants were not assessed in prior lipids genetic analyses. We find multiple lipid-related genes strongly associated with blood lipids through analysis of common and rare coding variants. We discover several associated rare non-coding variants, largely at Mendelian lipid genes. Notably, we observe rare LDLR intronic variants associated with markedly increased LDL-C, similar to rare LDLR exonic variants. In conclusion, we conducted a systematic whole genome scan for blood lipids expanding the alleles linked to lipids for multiple ancestries and characterize a clinically-relevant rare non-coding variant model for lipids.
Subject terms: Genome-wide association studies, Cardiovascular genetics, Genetic markers
Although the common genetic variants contributing to blood lipid levels have been studied, the contribution of rare variants is less understood. Here, the authors perform a rare coding and noncoding variant association study of blood lipid levels using whole genome sequencing data.
Introduction
The discovery of rare alleles linked to plasma lipids (i.e., low-density lipoprotein cholesterol [LDL-C], high-density lipoprotein cholesterol [HDL-C], total cholesterol [TC], and triglycerides [TG]) continue to yield important translational insights toward coronary artery disease (CAD), including PCSK9 and ANGPTL3 inhibitors now available in clinical practice1–5. The monogenic and polygenic bases of plasma lipids are well-suited to population-based discovery analyses and confer broader insights for genetic analyses of complex traits. We now evaluate numerous newly catalogued, largely rare, alleles never previously systematically analyzed with lipids.
Analyses of imputed array-derived genome-wide genotypes and whole exome sequences in hundreds of thousands of increasingly diverse individuals continue to uncover low-frequency protein-coding variants linked to lipids. Due to purifying selection, causal variants conferring large effects tend to occur relatively more recently, and are thus rare and often specific to families or communities6. Most discovery analyses for large-effect rare alleles have focused on the analysis of disruptive protein-coding variants given (1) well-recognized constraint in coding regions, (2) incomplete genotyping of rare non-coding sequence given relative sparsity of deep-coverage (i.e., >30X) whole genome sequencing (WGS), and (3) better prediction of coding versus non-coding sequence variation consequence1,7–12. We recently described a statistical framework incorporating multi-dimensional reference datasets paired with genomic data to improve rare coding and non-coding variant analyses for WGS analysis of lipids and other complex traits13,14. Furthermore, including individuals of non-European ancestry facilitates the discovery of both novel alleles at established loci as well as novel loci14–16.
Here, we examine the full allelic spectrum with plasma lipids using whole genome sequences and harmonized lipids from the National Heart, Lung, and Blood Institute (NHLBI) Trans-Omics for Precision Medicine (TOPMed) program17,18. We studied 66,329 participants and 428 million variants across multiple ancestry groups—44.48% European, 25.60% Black, 21.02% Hispanic, 7.11% Asian, and 1.78% Samoan. We identified robust allelic heterogeneity at known loci with several novel variants at these loci; we additionally identified novel loci and pursued replication in independent cohorts. We then explored the association of genome-wide rare variants with lipids, with detailed explorations of rare coding and non-coding variant models at known Mendelian dyslipidemia genes. Our systemic effort yields new insights for plasma lipids and provides a framework for population-based WGS analysis of complex traits.
Results
Overview
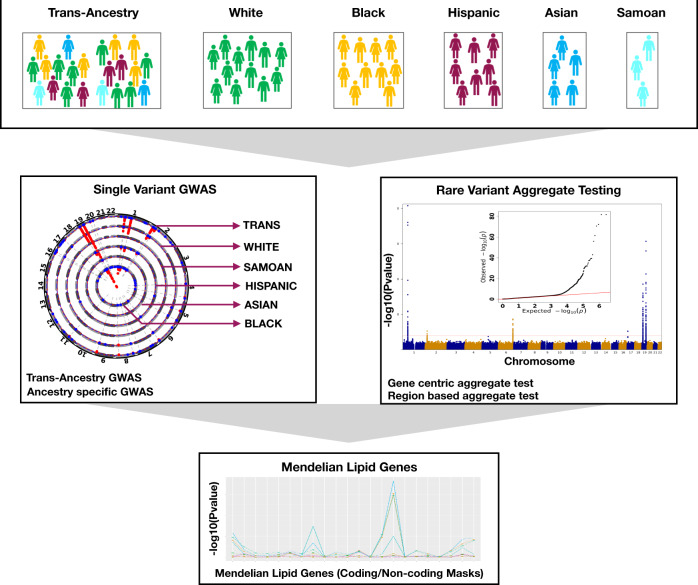
We studied the TOPMed Freeze8 dataset of 66,329 samples from 21 studies and performed genome-wide association studies (GWAS) separately for the four plasma lipid phenotypes (i.e., LDL-C, HDL-C, TC, and TG) using 28 M individual autosomal variants (minor allele count [MAC] >20) and aggregated rare autosomal variant (minor allele frequency [MAF] <1%) association testing for 417 M variants (Fig. 1, Supplementary Fig. 1). Secondarily, we associated individual variants with minor allele frequencies (MAF) >0.01% within each ancestry group to detect ancestry-specific lipid-associated alleles. We intersected our results with currently published array-based GWAS results15 to identify novel associations with lipids. We performed replication analyses for the putative novel associations identified, in up to ~45,000 independent samples with array-based genotyping imputed to TOPMed and 400 K samples from UK Biobank (UKB) imputed genotypes. Finally, we conducted rare variant association studies as multiple aggregate tests across the genome to identify gene-specific functional categories and non-coding genomic regions influencing plasma lipid concentrations. We replicated the significant rare variant aggregates in ~130 K whole genomes from UKB.
Fig. 1. Overall study schematic.
The analyses were conducted using the multi-ancestral TOPMed freeze8 data to associate whole genome sequence variation with lipid phenotypes (i.e., LDL-C, HDL-C, TC, and TG). A total of 66,329 samples with lipids quantified data from five ancestry groups were analyzed. Single variant GWAS were carried out using SAIGE on the Encore platform using SNPs with MAC >20. Both trans-ancestry and ancestry-specific GWAS were conducted. Genome-wide rare variant (MAF <1%) gene-centric and region-based aggregate tests were grouped and analyzed using STAARpipeline. Finally, single variant and rare variant associations at Mendelian dyslipidemia genes were investigated in further detail. TOPMed Trans-Omics for Precision Medicine, HDL-C high-density lipoprotein cholesterol, LDL-C low-density lipoprotein cholesterol, TC total cholesterol, TG triglycerides, GWAS genome wide association study, SAIGE Scalable and Accurate Implementation of GEneralized mixed model, MAC minor allele count, MAF minor allele frequency, SNPs single nucleotide polymorphisms.
TOPMed baseline characteristics
The TOPMed Informatics Research Center (IRC) and TOPMed Data Coordinating Center (DCC) performed quality control, variant calling, and calculated the relatedness of population structures of Freeze 8 data17. We studied 66,329 samples across 21 cohorts, and 41,182 (62%) were female. The ancestry distribution was 29,502 (44.46%) White, 16,983 (25.60%) Black, 13,943 (21.02%) Hispanic, 4719 (7.11%) Asian, and 1182 (1.78%) Samoan (Supplementary Data 1). The mean (standard deviation [SD]) age of the full cohort was 53 (15.00) years which varied by cohort from 25 (3.56) years for Coronary Artery Risk Development in Young Adults (CARDIA) to 73 (5.38) years for Cardiovascular Health Study (CHS). The Amish cohort had a higher-than-average concentration of LDL-C (140 [SD 43] mg/dL) and HDL-C (56 [SD 16] mg/dL) as well as lower TG (median 63 [IQR 50] mg/dL) consistent with the known founder mutations in APOB and APOC37,8,14. In the Women’s Health Initiative (WHI) cohort, the TC (230 [SD 41] mg/dL) and TG (median 129 [IQR 87] mg/dL) concentrations were higher than for other cohorts as previously described12. We accounted for lipid-lowering medications and fasting status and inverse rank normalized the phenotypes as before12,14 which are further detailed in the Methods. The adjusted normalized lipid concentrations for the four lipids were similar across the cohorts.
A total of 428 M variants passed the quality criteria with an average depth >30X in 22 autosomes. 202 M variants were singletons, 417 M were rare variants (MAF <1%), and 11 M were common or low-frequency variants (MAF >1%) with differences by cohort (Supplementary Data 2).
Individual variant associations with lipids
We performed single variant analysis of ~28 M variants with a MAC > 20 for four lipid phenotypes. We identified significant genomic risk loci for each lipid level (Supplementary Data 3) and considered a p-value <5 × 10−9 to claim significance as previously recommended for whole genome sequencing common variant association studies14,19. The total numbers of variants that met our significance threshold were 2214, 2314, 2697, and 2442 for LDL-C, HDL-C, TC and TG, respectively, and after clumping20 the numbers of variants were 357, 338, 324, and 289, respectively. Of these variants, 99% were previously demonstrated to be associated with plasma lipids either at the variant- or locus-level15 (Supplementary Data 4, Supplementary Fig. 2).
To identify putative novel variant associations, we compared our results to a recent multi-ethnic lipid GWAS among 312,571 participants of the Million Veteran Program (MVP)15 as well as the GWAS Catalog (All associations(v1.0) file dated 06/04/2020) (Fig. 2). We clumped (window 250 kb, r2 0.5) significant variants using Plink20 and queried these in the GWAS Catalog and MVP. Among genome-wide significant variants, we tabulated ‘known-position’ (variant previously associated), ‘known-loci’ (variants not previously significantly associated with the corresponding lipid phenotype but within 500 kb of a known locus, thereby representing additional allelic heterogeneity), and ‘novel’ variants (variants not in a known lipid locus) (Supplementary Data 4).
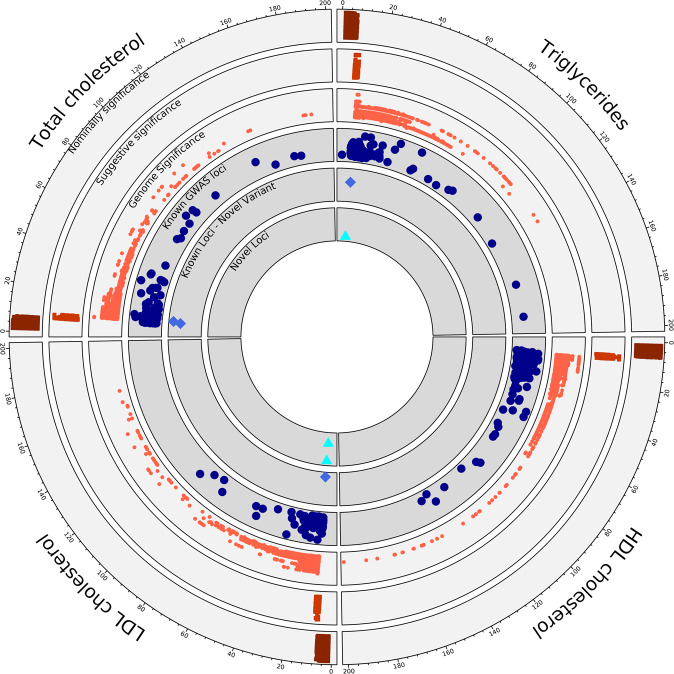
Fig. 2. Summary of single variant genome-wide association.
Representation of the single variant GWAS results from TOPMed Freeze 8 whole genome sequenced data of 66,329 samples. Each quarter represents a different lipid phenotype, and dots extending in clock-wise fashion represent variants with increasing evidence of association as noted by −log10(p-value), which was truncated at 200. The outer three circles show the GWAS data from TOPMed freeze8 where variants binned to nominally significant (p-value 0.05–5 × 10−07), suggestive significant (p-value 5 × 10−07–5 × 10−09) and genome wide significant (p-value < 5 × 10−09). The inner three circles compare our TOPMed results with known significantly associated lipid loci and variants from the MVP summary statistics and GWAS catalog to the identified novel variants and loci that are genome-wide significant from the current study, respectively. The figure represents the outputs from two-sided genetic association testing preformed using SAIGE-QT model, where the model was adjusted for all the covariates; see Methods. TOPMed Trans-Omics for Precision Medicine, GWAS genome wide association study, MVP million veteran program.
The novel variants, tabulated in Table 1, are divided into two subsets—‘novel variants’ or variants at established lipid loci for another lipid phenotype, and ‘novel loci,’ representing new loci associations for any lipid phenotype. For example, the CETP locus is well-known for its link to HDL-C, but we now found that rs183130 (16:56957451:C:T, MAF 28.3%) at the locus is associated with LDL-C. Similarly, the variants rs7140110 (13:113841051:T:C, MAF 27.8%) GAS6 and rs73729083 (7:137875053:T:C, MAF 4.5%) CREB3L2 are newly associated with TC, while previous studies showed that rs73729083 associates with LDL-C21 and rs7140110 associates with LDL-C22 and TG23. Index variants at novel loci were typically low-frequency variants often observed in non-European ancestries, so we also conducted ancestry-specific association analyses for these alleles (Supplementary Data 5). For example, 12q23.1 (12:97352354:T:C, MAF 0.3%) and 4q34.2 (4:176382171:C:T, MAF 0.2%) associations with LDL-C are specific to Hispanic (MAF 1.3%) and Black (MAF 0.6%) populations, respectively and among Asians (MAF 1.5%) alone, 11q13.3 (11:69219641:C:T, MAF 0.2%) was associated with TG. One variant initially passing the novel locus filter for HDL-C (RNF111 - rs112147665, beta = 8.664, p-value = 6.51 × 10−10), was in LD (r = 0.7) with LIPC p.Thr405Met (rs113298164) which is known to be associated with HDL-C. The lead variant from MVP was 604 kb away from the RNF111 variant but the rare LIPC missense variant p.Thr405Met was 421 kb away. Conditional analysis accounting for LIPC p.Thr405Met rendered the non-coding variant near RNF111 variant non-significant (beta = 4.351, p-value = 2.47 × 10−02), therefore we reclassified RNF111 variant as a known-position variant. Ancestry-specific GWAS did not yield additional novel loci beyond our larger trans-ancestry GWAS. The majority of genome-significant single variants were captured by previous lipid GWAS15, but ancestry-specific novel-hits are unique to WGS TOPMed data.
Table 1.
Putative novel variants identified in TOPMed and evidence for replication
| Associated lipid phenotype | Novel variant class | Variants (Gene) | Discovery Cohort TOPMed Freeze8 (N = 66,329) | Replication Cohort Meta Analysis (METASOFT) MGB Biobank (N = 25,137); Penn Medicine Biobank (N = 20,079); UK Biobank (N = 424,955) | ||||
|---|---|---|---|---|---|---|---|---|
| Effect estimate | p-value | MAF | Beta | p-value | Std.Err | |||
| LDL-C | Novel locus | 12:97352354:T:C | −12.439 | 4.88 × 10−09 | 0.003 | 3.316 | 3.62 x 10−01 | 3.634 |
| LDL-C | Novel variant | 16:56957451:C:T (CETP) | −1.568 | 2.88 × 10−09 | 0.283 | −1.459 | 8.74 x 10−84 | 0.075 |
| LDL-C | Novel locus | 4:176382171:C:T | −16.086 | 2.82 × 10−09 | 0.002 | −0.980 | 7.80 x 10−01 | 3.514 |
| TC | Novel variant | 13:113841051:T:C (GAS6) | 1.731 | 1.12 × 10−09 | 0.278 | 1.262 | 1.29 x 10−38 | 0.097 |
| TC | Novel variant | 7:137875053:T:C (CREB3L2) | −4.106 | 7.54 × 10−11 | 0.045 | −3.538 | 7.70 x 10−07 | 0.716 |
| TG | Novel locus | 11:69219641:C:T | 0.232 | 1.98 × 10−09 | 0.002 | −0.030 | 6.04 x 10−01 | 0.059 |
| TG | Novel variant | 13:107551611:C:T (FAM155A) | 0.052 | 6.78 × 10−10 | 0.045 | 0.015 | 2.20 x 10−02 | 0.006 |
Variants identified as novel after comparing with the GWAS catalog and MVP summary statistics for associations with lipid phenotypes, including LDL-C, TC, and TG. All effect estimates are in mg/dL units, except for TG which was log-transformed in analysis thereby representing fractional change. Variants are categorized as novel loci or novel variant (i.e., known locus associated with another lipid phenotype) and the genes assigned to the variants per TOPMed whole genome sequence annotations (WGSA) are listed. Data is provided for the discovery (TOPMed freeze8) and replication cohorts (Imputed datasets from MGB Biobank, Penn Medicine Biobank and UK Biobank). Meta-analysis with the replication cohorts was carried out and the corresponding beta, p-values and standard-errors are provided. All the effect-estimates and p-values are reported from two-sided association testing with all independent samples from each cohort (Discovery-TOPMed: 66,329; Replication-MGB Biobank: 25,137; UK Biobank: 424,955; Penn Biobank: 20,079).
GWAS genome wide association study, MVP million veteran program, LDL-C low-density lipoprotein cholesterol, TC total cholesterol, TG triglycerides, TOPMed trans-omics for precision medicine, WGSA whole genome sequence annotations.
For the single variant GWAS, we pursued replication with two genome-wide array-based genotyped datasets imputed to TOPMed WGS17,24: Mass General Brigham (MGB) Biobank (N = 25,137) and Penn Medicine Biobank (N = 20,079)25,26, these replication cohorts had diverse ancestry distribution, where non-European samples accounted for 15.77% in MGB Biobank and 51.20% in Penn Medicine Biobank. We also conducted replication using UKB imputed data which accounted for 16.10% of non-European samples (Supplementary Data 6). We brought seven putative novel variants with p-values < 5 × 10−9 forward for replication. The three common variants, rs183130 (CETP), rs7140110 (GAS6), and rs73729083 (CREB3L2), that were associated with both LDL-C and TC in TOPMed replicated in MGB and UKB along with rs77687061 for TG and two of these (rs183130, rs73729083) replicated in Penn Biobank at an alpha level of 0.05 and consistent direction of effect (Supplementary Data 5). The two variants that were associated in all three replication studies were most significantly associated among African Americans in TOPMed (rs183130: beta = −2.762 mg/dL, p-value = 5.71 × 10−07; rs73729083: beta = −3.725 mg/dL, p-value = 5.25 × 10−07). We meta-analyzed the single variant replication from the three cohorts and identified three common variants with suggestive p-value (5 × 10−5) (Table 1). Low-frequency variants from specific ancestry groups associated with lipids in TOPMed were not replicated but we cannot rule out the possibility of reduced power due to the general underrepresentation of non-white ancestry groups in the replication data. In exploratory analyses, we extended the same approach for variants discovered to have 5 × 10−9 < p-value < 5 × 10−7 but did not observe replication (Supplementary Data 7).
In-silico analysis to gain mechanic insights from single variant GWAS results
Prioritization and functional enrichment analysis
We first mapped the variants to genes and to functional regions using ANNOVAR. Second, we determined gene tissue specificity, relating tissue-specific gene expression with disease-gene associations, using MAGMA. Significantly associated variants were enriched in intronic and intergenic regions (Supplementary Fig. 3). Using GTEx, tissue-specific gene expression was enriched among liver, stomach, and pancreatic tissues (Supplementary Fig. 4) with top tissue-gene sets tabulated in Supplementary Data 8. Using the STRING protein-protein interaction database examining liver-specific genes, we highlight that the HDL-C protein network uniquely harbored metal-ions related genes (MT1A, MT1B, MF1F, MT1G, MT1H) and anticipated LCAT-CETP interactions (Supplementary Fig. 5). Enriched pathways from Reactome, GeneOntology and other curated and canonical pathways (Supplementary Data 9) with a p-value < 2.5 × 10−06 were observed including response to metal ions, lipoprotein assembly, and chylomicron remodeling.
The enrichment analysis was carried out with the full single variant summary statistics, where we identified that most of the prioritized loci/genes were previously documented for lipid associations. Next, we specifically investigated the novel variants that we identified from this study. Out of the seven variants documented in Table 1, four were low frequency variants, 12:97352354:T:C (rs189010847) closest to NEDD1, 4:176382171:C:T (rs115489644) closest to SPCS3, 11:69219641:C:T (rs74791751) near to SMIM38, are all intergenic variants and 13:107551611:C:T (rs77687061) is an intronic variant in FAM155A. We did not find any information for these variants in the Open Target Genetics database27. Finally, two of the common novel-loci variants (rs183130 and rs7140110) were present in eQTL and sQTL databases28, therefore, we performed analysis to determine the correlation among effects and the importance of these variants more in detail.
CETP locus, HDL-C, and LDL-C
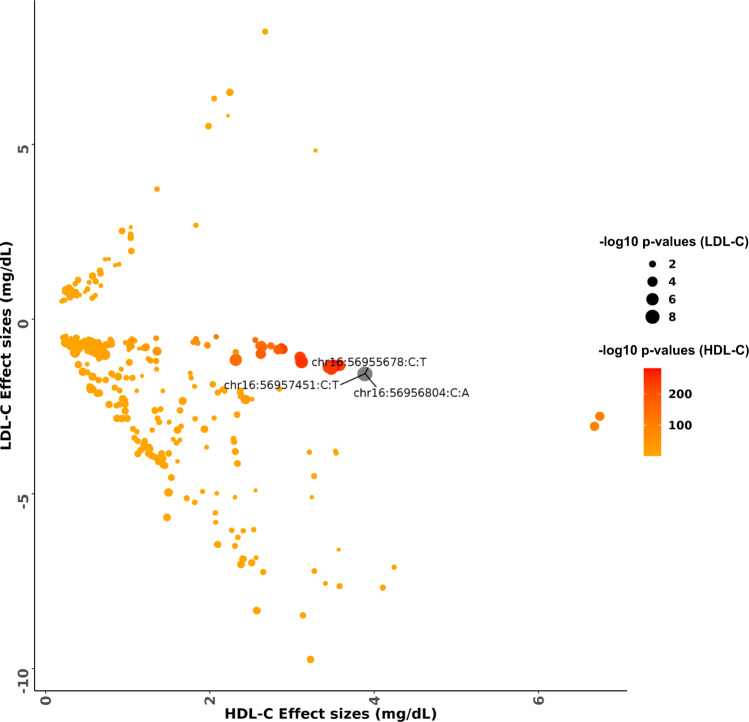
CETP is a well-recognized Mendelian HDL-C gene and the locus was previously known to be significantly associated with HDL-C, TC, and TG at genome-wide significance15. Pharmacologic CETP inhibitors have shown strong associations with increased HDL-C but mixed effects for LDL-C reduction in clinical trials29–32. We found that the CETP locus variant rs183130 (chr16:56957451:C:T, MAF 28.3%, intergenic variant) was associated with reduced LDL-C concentration (beta = −1.568 mg/dL, SE = 0.264, p-value = 2.88 × 10−09). The lead HDL-C-associated variant at the locus, rs3764261 (chr16:56959412:C:A, MAF 30.3%), was associated with 3.5 mg/dL increased HDL-C (p-value = 8.03 × 10−283), and rs183130 was associated with 3.9 mg/dL increased HDL-C (p-value < 1 × 10−284) as well. Among the ancestry groups analyzed, rs183130 was most significantly associated with LDL-C among those of African ancestry (beta = −2.762 mg/dL, p-value = 5.71 × 10−07) (Supplementary Data 10). We next investigated variants by their HDL-C and LDL-C effects within this locus (+/−500 kb of rs183130 and rs3764261) (Fig. 3). We identified five variants showing at least suggestive (p-value < 5 × 10−07) association with both HDL-C and LDL-C. Though variants with strong LD (linkage disequilibrium) existed, ancestry-specific analyses showed that the stronger LDL-C effects were among those of African ancestry.
Fig. 3. Comparison of effects estimates for HDL-C and LDL-C among variants in the CETP locus.
The color scale of the data points was based on −log10 p-values from HDL-C association and the size of each data point was based on −log10 p-values of LDL-C association. Variants which are genome wide significant with LDL-C are represented as chromosome:position:reference allele:alternate allele. The effect estimates and p-values were calculated from two-sided genetic association testing preformed using SAIGE-QT model, where the model was adjusted for all the covariates; see Methods. HDL-C high-density lipoprotein cholesterol, LDL-C low-density lipoprotein cholesterol.
To better understand the mechanisms for HDL-C and LDL-C effects at the CETP locus, we pursued colocalization with eQTLs from three tissues (Liver, Adipose Subcutaneous and Adipose Visceral [Omentum]) from GTEx28. We analyzed 5 LDL-C and 441 HDL-C associated (p-values <5 × 10−07) variants. We correlated eQTL effect estimates for genes at the locus with lipid outcome effect estimates. Indeed, CETP gene expression effects were strongly negatively correlated with HDL-C effects (Liver: ρ −0.933, p-value 4.01 × 10−17; Adipose Subcutaneous: ρ −0.762, p-value 8.87 × 10−12; Adipose Visceral: ρ −0.739, p-value 5.52 × 10−10) (Supplementary Fig. 6). However, CETP expression effects were not significantly correlated with LDL-C (Liver: ρ 0.007, p-value 0.99; Adipose Subcutaneous: ρ 0.344, p-value 0.57; Adipose Visceral: ρ −0.59, p-value 0.29). Given the possibility that the observed lack of correlation for LDL-C could be due to reduced power from a limited number of variants attaining a suggestive p-value (<5 × 10−07), we repeated the analysis with a subset of 122 nominally significant (p-value < 0.05) LDL-C associated variants in this locus. Indeed, CETP gene expression effects were strongly positively correlated with LDL-C effects (Liver: ρ 0.957, p-value 2.28 × 10−08; Adipose Subcutaneous: ρ 0.922, p-value 1.34 × 10−15; Adipose Visceral: ρ 0.868, p-value 6.09 × 10−11).
GAS6 locus, LDL-C/TG, and TC
Variants at GAS6 were previously associated with LDL-C and TG22,23, but in our analysis, rs7140110 was now significantly associated with TC. We performed colocalization analysis of the variants+/−500 Kb from rs7140110 in liver and adipose tissues from GTEx. Across the three lipid-related tissues (liver, adipose subcutaneous, and adipose visceral), strong colocalization was observed in liver for all three lipid phenotypes (TG 46.6%; LDL-C 33.3%; TC 28%). The TG and LDL-C-associated variants were eQTLs for the GAS6 gene only. However, the TC-associated eQTLs at this locus influenced the cis expression of multiple genes, including GAS6, antisense genes of GAS6 (AS1, AS2) as well as other genes (i.e., TFDP1, CHAMP1, LINC00565, ADPRHL1, RASA3, UPF3A, GRTP1, AL442125.1, C13orf46, DCUN1D2, CDC16, TMEM255B, GRTP1-AS1, ATP4B, TMCO3). In addition to GAS6, the TC-associated rs7140110 is an sQTL for TMEM255B in adipose subcutaneous tissue (p-value 5.6 × 10−08), with further support from TC colocalization analysis and was not significant for other lipid levels.
Phenome-wide association with complex traits
We conducted a phenome-wide association (PheWAS) of 1572 binary complex traits using UK Biobank for the three replicated common variants (16:56957451:C:T (CETP); 13:113841051:T:C (GAS6); 7:137875053:T:C (CREB3L2)) adjusting for PC1–10, age, age2, sex, and race. We claimed significance at FDR of 0.05 and identified various complex traits significant, including ischemic heart disease for the CETP variant and heart failure/atherosclerosis, hypercholesterolemia traits for GAS6 variant. The summary statistics from PheWAS analysis for the significant complex traits are tabulated in Supplementary Data 11.
Rare variant aggregates associated with lipids
Gene-Centric associations
We next evaluated the association of aggregated rare (MAF < 1%) variants, linked to protein-coding genes (‘gene-centric’). We employed a Bonferroni-corrected significance threshold of 0.05/20,000 = 2.5 × 10−06 for coding and non-coding gene-centric rare variant analyses (Supplementary Fig. 7). We identified 102 coding and 160 non-coding gene-centric rare variant aggregates significantly associated with at least one of the four plasma lipid phenotypes in nonconditional analysis (Supplementary Data 12, 13). We secondarily conditioned our significant aggregate sets on variants individually associated with lipid levels from the GWAS catalog, MVP summary statistics and the TOPMed data. We identified 74 coding and 25 non-coding rare variants aggregates associated with at least one lipid level after conditional analyses (Supplementary Data 14, 15).
Most of the coding gene-centric sets remained significant after secondary conditioning, while a minority of non-coding gene-centric sets remained significant after conditioning. Significant genes identified from coding rare variant analyses included multiple known Mendelian lipid genes including LCAT, LDLR, and APOB (Supplementary Data 13). RFC2 putative loss-of-function mutations (combined allele frequency < 0.002%) were significantly associated with triglycerides (p-value 2 × 10−06) representing a putative novel association for triglycerides. The RFC2 aggregate set (plof) was associated with reduced TG (beta = −0.89 for log[TG]). The persistently significant regions identified from non-coding rare variant analyses linked to genes included the UTR (untranslated region) for CETP and promoter-CAGE (CAGE—Cap Analysis of Gene Expression sites) around APOA1 for HDL-C, and APOE promoter-CAGE, APOE enhancer-DHS (DHS—DNase hypersensitivity sites), and EHD3 promoter-DHS for total cholesterol (Supplementary Data 15). Most of the coding aggregates had larger effects compared to non-coding aggregates, and among the non-coding aggregates SPC24 non-coding aggregate (enhancer-CAGE) at the LDLR locus had the strongest effect for LDL-C (beta = 2.320 mg/dL; p-value = 1.75 × 10−05).
We analyzed the UK Biobank whole genome sequences among ~130 K participants to provide evidence of replication for the significant coding and non-coding aggregate sets. We used a Bonferroni-corrected significance threshold based on the number of genes tested in each type of aggregate-based test. For gene centric-coding aggregates, we conducted replication of 21 genes (p-value < 0.05/21 = 2.38 × 10−03) and for non-coding aggregates we replicated the findings from 13 genes (p-value < 0.05/13 = 3.85 × 10−03). At Bonferroni significance, 71% and 62% of genes replicated for at least one coding and non-coding aggregate set, respectively (Supplementary Data 14, 15). We observed that most of the Mendelian lipid genes replicated for coding aggregates including ABCA1, ABCG5, LCAT, APOB, LDLR, PCSK9, and LPL. For the non-coding aggregate set, the most significant replications were observed for the APOB, LDLR (SPC24), and PCSK9 loci, further corroborating the observation that both coding and noncoding rare variant signals contribute to variation in lipid levels at these loci.
Region-based associations
We also performed unbiased region-based rare variant association analyses tiled across the genome with both static and dynamic window sizes. We first evaluated 2.6 M regions statically at 2 kb size and 1 kb window overlap by the sliding window approach. Statistical significance was assigned at 0.05/(2.6 × 1−06)=1.88 × 10−08. We identified 28 significantly associated windows with at least one lipid phenotype. After conditioning on variants individually associated with the corresponding lipid phenotype, we identified two regions at LDLR still significantly associated with both total cholesterol and LDL-C, although these regions included both intronic and exonic variants (Supplementary Data 16). LDLR intron 1, which encodes LDLR-AS1 (LDLR antisense RNA 1) on the minus strand, had suggestive evidence for association with TC (p-value 3.17 × 10−6) with −2.76 mg/dL reduction in TC. A prior study identified that a common variant (rs6511720, MAF 0.11) in LDLR intron 1 is associated with increased LDLR expression in a luciferase assay and reduction in LDL-C33. When adjusting for rs6511720, the significance improved (p-value 1.43 × 10−8) with −3.35 mg/dL reduction in TC.
For dynamic window scanning of the genome, we implemented the SCANG method34. The SCANG procedure accounts for multiple testing by controlling the genome-wide error rate (GWER) at 0.134. In the dynamic window-based workflow, STAAR-O detected 51 regions significantly associated with at least one lipid phenotype after conditioning on known variants (Supplementary Data 17). Most of the regions mapped to known Mendelian lipid genes, including LCAT (8.7 × 10−13) for HDL-C, and LDLR (2.4 × 10−28, 7.3 × 10−26) and PCSK9 (2.9 × 10−12, 5.5 × 10−12) for LDL-C and TC, respectively. Exon 4 aggregates of LDLR were specifically associated with 20 mg/dL increase in LDL-C. PCSK9 Exon2-Intron2 region spanning chr1:55043782–55045960 had significantly reduced LDL-C by 6 mg/dL (p-value = 3 × 10−13), and the effect persisted even with only Intron 2 rare variants of PCSK9 (−5 mg/dL, p-value = 2 × 10−8). Strikingly, in secondary analyses, we found evidence for very large effects for rare variants in LDLR Introns 2 and 3 (+21 mg/dL, p-value = 7 × 10−4) and LDLR Introns 16 and 17 (+17 mg/dL, p-value = 0.02), similar to rare coding LDLR mutations. While 32 of the significant dynamic windows also included exonic regions, there were also several dynamic windows significantly independently associated with lipids not containing exonic regions. For example, four non-coding windows (two overlapping) at 2p24.1, which harbors the Mendelian APOB gene, were significantly associated with LDL-C. Intronic non-coding regions were associated with both LDL-C and TC -associated windows at LPAL2-LPA-SLC22A3; for example, LPAL2 Intron 3 was associated with a 3.7 mg/dL increase in TC. Non-coding TC-associated significant dynamic windows were near TOMM40/APOE. One rare variant signal observed was at TOMM40 Intron 6, where the ‘poly-T’ variant in this region is on the APOE4 haplotype and influences expressivity for Alzheimer’s disease age-of-onset35,36. For HDL-C, we identified significant non-coding windows at an intergenic region near LPL and CD36 Intron 4. In the generation of the spontaneously hypertensive rat model, the deletion of intron 4 in CD36 with resultant CD36 deficiency has been mapped to defective fatty acid metabolism in this model37. Several regions significant in SCANG were not even nominally significant in burden association analyses indicating the likelihood of causal variants with bidirectional effects.
We replicated 28 sliding and 51 dynamic window aggregate sets using UKB whole genomes, at a Bonferroni-corrected alpha threshold of 0.05/no. of regions for each approach separately. At Bonferroni significance, 61% of the regions from each of the sliding window (p-value < 0.05/28 = 1.79 × 10−03) and dynamic window (p-value < 0.05/51 = 9.80 × 10−04) approaches significantly replicated (Supplementary Data 16, 17). Multiple regions linked to LDLR, PCSK9, CETP, APOC3, and ABCA1 were highly significant.
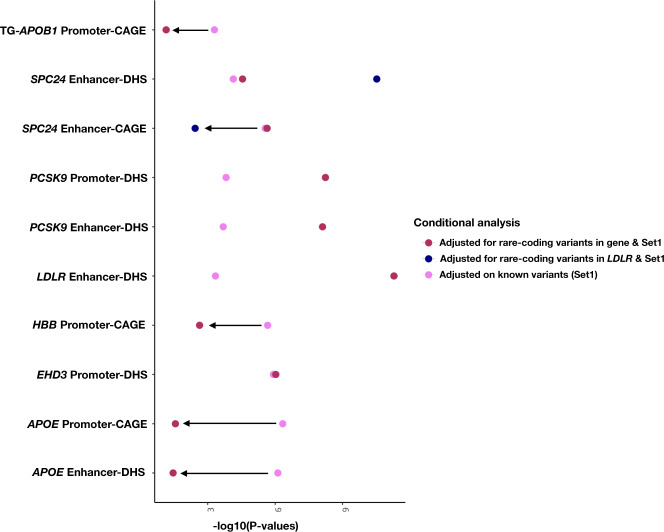
Several gene-centric non-coding aggregates associated with lipids near known monogenic lipid genes but mapped to another gene at the locus via annotations. Therefore, we performed downstream conditional analyses adjusting the gene-centric non-coding results for rare coding variants (MAF < 1%) within known lipid monogenic genes (Supplementary Data 18). When accounting for both common and rare coding variants at the nearby familial hypercholesterolemia LDLR gene, SPC24-enhancer DHS was significantly associated with total cholesterol (p-value = 3.01 × 10−11) and with suggestive evidence for LDL-C (p-value = 1.57 × 10−06). In a similarly adjusted model, LDLR-enhancer-DHS showed a strong association with TC (p-value 5.18 × 10−12). When adjusting for known common variants as well as rare coding variants in PCSK9, both PCSK9-enhancer DHS and PCSK9-promoter DHS were significantly associated with total cholesterol (Fig. 4, Supplementary Fig. 8). Through this procedure, CETP UTR retained significance for its independent association with HDL-C as well as the putatively novel gene EHD3-promoter DHS association with TC. However, the non-coding gene-centric APOC3 and APOE associations were rendered non-significant for HDL-C and TC, respectively.
Fig. 4. Conditional analysis of coding rare-variants from the same gene and a near-by gene.
Non-coding rare variant sets significantly associated with TC and TG after the conditional analysis on known variants are shown with additional adjustment on rare-coding variants. The additional adjustment for rare-coding variants were carried out for the same gene of the aggregate set and for certain gene aggregates (SPC24) the conditional analysis was carried out with a nearby Mendelian gene. After adjusting for rare-coding variants and known variants, EHD3 signal drops minimally, whereas signal from PCSK9 (promoter-DHS, enhancer-DHS), LDLR-loci (enhancer-DHS, SPC24 enhancer-DHS) enhances significantly. APOB1, SPC24 (enhancer-CAGE), HBB and APOE signal drops after the conditional analysis on rare-coding variants. The different colored dots on the plot represents the conditional STAAR-O p-values when adjusting for known variants (Set1) and rare-coding variants of the same or near-by gene. The p-values were calculated from two-sided aggregate testing preformed using STAAR gene-centric model, where the model was adjusted for all the covariates; see Methods. STAAR variant-Set Test for Association using annotation information, TC total cholesterol, TG triglycerides, CAGE cap analysis of gene expression, DHS DNase hypersensitivity.
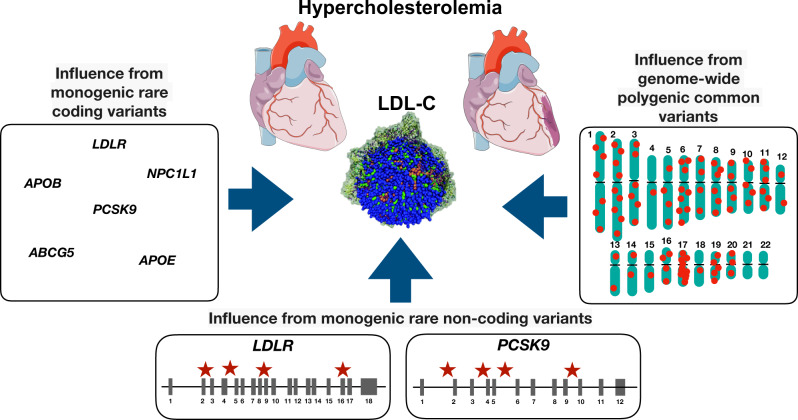
Since we cannot rule out the possibility of reduced power for genome-wide rare variant analyses, we leveraged current knowledge of 22 Mendelian lipid genes for more focused exploratory analyses14. We validated most genes in rare variant coding analyses. The genes with the strongest coding signals typically had at least nominal evidence of gene-centric non-coding rare variant associations (Supplementary Data 19, Supplementary Fig. 9). When rare coding variants were introduced into the model, the evidence for non-coding rare variant associations were largely unchanged. Our findings expanding the currently described genetic basis for hypercholesterolemia to include rare non-coding variation at LDLR and PCSK9 (Fig. 5).
Fig. 5. Influence of common and rare variants with hypercholesterolemia.
In addition to monogenic contributions from rare variants in Mendelian hypercholesterolemia genes, multiple genome-wide significant LDL-C-associated common variants also yield a polygenic basis for hypercholesterolemia. In the present work, we now identify rare non-coding variants in proximity of Mendelian hypercholesterolemia genes, specifically LDLR and PCSK9, that also contribute to the genetic basis of hypercholesterolemia. Parts of the figure were generated using pictures from Servier Medical Art. Servier Medical Art by Servier is licensed under a Creative Commons Attribution 3.0 Unported License (https://creativecommons.org/licenses/by/3.0/). LDL-C low-density lipoprotein cholesterol.
Heritability contributions from rare variants
To understand the contribution of rare variants towards lipid trait heritability, we examined heritability of lipids by variant allele frequency across three ancestral samples (White, Black, and Hispanic) in TOPMed. We calculated trait heritability using Greml-LDMS38 following the steps as implemented by Wainschtein et al.39. Using the TOPMed WGS, we grouped the variants into 4 MAF bins for the three ancestral samples. In each MAF bin, we grouped variants based on the LD scores into four quartiles and calculated variance contributed by the SNPs (h2) for each of the lipids using unrelated individuals from each ancestral group (Supplementary Fig. 10) and set negative estimate to zero. We observed that rare variants from the lower MAF bins contributed to trait heritability but have large standard errors (Supplementary Data 20). We observed an increase in h2 values including WGS variants relative to estimates obtained from array-genotypes as reported by Cadby et al.40 for the European samples. We also compared the h2 estimates from all the variants from WGS TOPMed cohort against array-genotypes captured in MGB Biobank to understand the differences contributed by these two sequencing methods. As expected, the h2 estimates from array-genotypes were reduced corresponding to missing heritability from the lower MAF bins captured by WGS. The heritability estimates from array-genotypes were markedly higher for European samples relative to African and Hispanic sample sets indicating that WGS better captured heritability for the latter groups.
Discussion
Conducting one of the largest population-based WGS association analyses, we now simultaneously interrogate and establish a common, rare coding, and rare non-coding variant model for a complex trait. Utilizing 66,329 diverse individuals with deep-coverage WGS, we interrogated 428 M variants with plasma lipids expanding the allelic series to rare non-coding variants, often within introns, of Mendelian lipid genes with prior robust rare coding variant support. Our observations have important implications for plasma lipids as well as the genetic basis of complex traits more broadly.
WGS of diverse ancestries enables both allelic and locus heterogeneity for complex traits. Population genetic analyses have largely been enriched for individuals of European descent41. Genetic association of plasma lipids using arrays or whole exome sequencing among Europeans have yielded several important insights regarding plasma lipids and the causal determinants of CAD4,5,42–44. Similar increasingly larger studies among non-Europeans have often yielded new genetic loci and sometimes new genes, such as PCSK91,15,16,45,46. Such differences have also led to concerns about the use of polygenic risk scores gleaned from much larger European GWAS of complex traits for non-Europeans47. Aided by the availability of WGS data, we identify new putative loci associated with lipids in non-Europeans. Furthermore, our study enabled the discovery of several novel alleles at known loci, with richly distinct allelic heterogeneity across ancestry groups. For example, HDL-C-raising CETP locus variants linked to CETP gene expression were only associated with LDL-C reduction among those of African ancestry. While all pharmacologic CETP inhibitors increase HDL-C, only those that decrease LDL-C also reduce cardiovascular disease risk29–32. Given the contribution of genetic differences, clinical trials with more diverse samples would show insights.
Our study now provides increasingly robust evidence for a rare non-coding variant model for complex traits. Our rare non-coding variant associations in both gene-centric and sliding window models were largely restricted to the introns of Mendelian lipid genes with prior robust rare coding variant support consistent with biologic plausibility48. Rare intronic variants, often impacting splicing, have been previously implicated in afflicted Mendelian families or small exceptional case series, often through candidate gene approaches49–52. We discovered one example of a rare non-coding signal without prior rare coding support—i.e., EHD3 which also nominally replicated in the independent UKB WGS cohort. We obtained estimates of phenotypic effect using burden tests. For most regions, even nominal significance was not detected using burden testing indicating the likelihood of variants with bidirectional effects further complicating clinical interpretation. When burden signals were detected, observed effects were typically larger than common non-coding variants and less than rare coding variants, with the exception of LDLR, consistent with whole genome mutational constraint models53–55.
The detection of independent rare non-coding variant signals has remained elusive largely due to limited sample sizes with requisite WGS and limitations in the interpretation of rare non-coding variation functional consequence. Previously, we used annotated functional non-coding sequence in 16,324 TOPMed participants, and found that rare non-coding gene regions associated with lipid levels, but they were not independent of individually associated single variants14. Using STAAR, we observed putative rare non-coding variant associations for lipids independent of individual variants associated with lipids in TOPMed.
WGS can improve diagnostic yield beyond the current standard of next-generation gene panel sequencing for dyslipidemias. A very small fraction with severe hypercholesterolemia and features consistent with strong genetic predisposition have a familial hypercholesterolemia variant in LDLR, APOB, or PCSK956,57. The presence of familial hypercholesterolemia variants is independently prognostic for CAD, beyond lipids, and merits the consideration of more costly lipid-lowering medications56–59. We now observe that rare LDLR variants in Introns 2, 3, 16, and 17 lead to ~0.5 standard deviation increase in LDL-C, approximating effects observed with clinically reported exonic familial hypercholesterolemia variants in LDLR59. Small studies have indicated the possibility of rare intronic LDLR variants causing familial hypercholesterolemia due to altered splicing, which we now observe in our unbiased population-based WGS study60,61. A WGS approach to lipid disorders, particularly for familial hypercholesterolemia, will markedly improve the diagnostic yield beyond existing limited approaches.
Our dynamic window approach may also improve the clinical curation of exonic variants. Among the data used to curate exonic variants is the use of in silico functional prediction tools62. Although evolutionary constraint measures are typically employed, such tools are largely agnostic to functional domain. As it relates to lipids, disruptive APOB and PCSK9 exonic variants can lead to strikingly opposing directions with large effects for LDL-C depending on locations1,8,63,64. Using SCANG34, we detect a significant association with large effect for LDLR Exon 4 itself. This observation supports the pathogenicity of LDLR Exon 4 disruptive variants among patients with severe hypercholesterolemia. The majority of familial hypercholesterolemia variants worldwide occur in Exon 4 of LDLR65–68. Conventional rare coding variant analyses aggregate all exonic variants for a transcript. Here, we demonstrate an opportunity for exon-level rare variant association testing.
Our discovery analyses with replication as well as heritability assessment are consistent with the notion that both rare coding and non-coding alleles, not well-captured by genome-wide arrays. Furthermore, we observe that heritability gains relative to genome-wide genotyping arrays are more significant for individuals of European-ancestry likely indicative of Eurocentric array designs. A tradeoff for WGS, however, is the greater cost. However, as costs continue to decrease as well as cheaper WGS implementations via reduced coverage, cost may no longer be a downside.
Our study has important limitations. First, while our study is large for a WGS study by contemporary standards, it is dwarfed by existing GWAS datasets limiting power for novel discovery. Nevertheless, by using WGS in diverse ancestries, we can study hundreds of millions new variants. Second, prediction of rare non-coding variation consequence to prioritize causal variants remains a challenge thereby limiting power69. The striking difference for most STAAR and burden results also highlights bidirectional effects for rare non-coding variants within the same region and further challenges for clinical utility. Third, given the paucity of multi-ancestral WGS datasets with lipids, our analyses are largely restricted to TOPMed and replication to European rich UK Biobank WGS data. For single variant associations, we pursued TOPMed-imputed GWAS datasets but were limited by the lack of ancestral diversity. As TOPMed is a consortium of multiple different cohorts, we demonstrate consistencies by cohort. Furthermore, rare variant non-coding signals were largely restricted to regions with rare variant coding signals supporting biological plausibility.
In conclusion, using WGS and lipids among 66,329 ancestrally diverse individuals we expand the catalog of alleles associated with lipids, including allelic heterogeneity at known loci and locus heterogeneity by ancestry. We characterize the common, rare coding, and rare non-coding variant model for lipids and replicated the results. Lastly, we now demonstrate a monogenic-equivalent model for rare LDLR intronic variants predisposing to marked alterations in LDL-C, currently not recognized in current population or clinical models for LDL-C.
Methods
Dataset
Contributing studies
The discovery cohort includes the whole genome sequenced (WGS) data of 66,329 samples from 21 studies of the Trans-Omics for Precision Medicine (TOPMed) program with blood lipids available17. The overall goal of TOPMed is to generate and use trans-omics, including whole genome sequencing, of large numbers of individuals from diverse ancestral backgrounds with rich phenotypic data to gain novel insights into heart, lung, blood, and sleep disorders. The Freeze 8 data includes 140,306 samples out of which 66,329 samples qualified with lipid phenotype. Freeze 8 dataset passed the central quality control protocol implemented by the TOPMed Informatics Research Core (described below) and was deposited in the dbGaP TOPMed Exchange Area.
The studies included in the current dataset, along with their abbreviations and sample sizes, contains the Old Order Amish (Amish, n = 1083), Atherosclerosis Risk in Communities study (ARIC, n = 8016), Mt Sinai BioMe Biobank (BioMe, n = 9848), Coronary Artery Risk Development in Young Adults (CARDIA, n = 3,056), Cleveland Family Study (CFS, n = 579), Cardiovascular Health Study (CHS, n = 3,456), Diabetes Heart Study (DHS, n = 365), Framingham Heart Study (FHS, n = 3992), Genetic Studies of Atherosclerosis Risk (GeneSTAR, n = 1757), Genetic Epidemiology Network of Arteriopathy (GENOA, n = 1046), Genetic Epidemiology Network of Salt Sensitivity (GenSalt, n = 1772), Genetics of Lipid-Lowering Drugs and Diet Network (GOLDN, n = 926), Hispanic Community Health Study - Study of Latinos (HCHS_SOL, n = 7714), Hypertension Genetic Epidemiology Network and Genetic Epidemiology Network of Arteriopathy (HyperGEN, n = 1853), Jackson Heart Study (JHS, n = 2847), Multi-Ethnic Study of Atherosclerosis (MESA, n = 5290), Massachusetts General Hospital Atrial Fibrillation Study (MGH_AF, n = 683), San Antonio Family Study (SAFS, n = 619), Samoan Adiposity Study (SAS, n = 1182), Taiwan Study of Hypertension using Rare Variants (THRV, n = 1982) and Women’s Health Initiative (WHI, n = 8263) (Please see Supplementary Note 1 for additional details). The multi-ancestral data set included individuals from White (44%), Black (26%), Hispanic (21%), Asian (7%), and Samoan (2%) ancestries. Study participants granted consent per each study’s Institutional Review Board (IRB) approved protocol. Secondarily, these data were analyzed through a protocol approved by the Massachusetts General Hospital IRB. Supplementary Data 1 details the number of samples across different studies and ancestral group.
The replication cohorts for single variant GWAS include TOPMed-imputed genome-wide array data from the Mass General Brigham (MGB), Penn Medicine Biobanks and UK Biobank (UKB) imputed data which consist of 25,137, 20,079, and 424,955 samples respectively25,26,70. The replication cohort for rare variant aggregates test include UKB whole genome sequenced data which consists of a subset of 133,360 UKB participants, where we removed unconsented and related individuals. We curated the MGB Biobank and Penn Medicine Biobank phenotype data from the corresponding electronic health record databases in accordance with corresponding institutional IRB approvals. The UKB data included volunteer residents of the UK aged 40–69 and were recruited between 2006 and 2010. Consent was previously obtained from each participant regarding storage of biological specimens, genetic sequencing, access to all available electronic health record (EHR) data, and permission to recontact for future studies. All UKB participants gave written informed consent per UKB primary protocol. The MGB Biobank consists of 54%, Penn Medicine Biobank consist of 52% and UK Biobank imputed data consist of 54% of female samples and average ages were 55.89, 58.35 and 56.55 years, respectively (Supplementary Data 6).
Phenotypes
The primary outcomes in this study included LDL cholesterol (LDL-C), HDL cholesterol (HDL-C), total cholesterol (TC), and triglycerides (TG) phenotypes. LDL-C was either directly measured or calculated by the Friedewald equation when triglycerides were <400 mg/dL. Given the average effect of lipid lowering-medicines, when lipid-lowering medicines were present, we adjusted the total cholesterol by dividing by 0.8 and LDL-C by dividing by 0.7, as previously done14. Triglycerides remained natural log transformed for analysis. Fasting status was accounted for with an indicator variable.
We harmonized the phenotypes across each cohort18 and inverse rank normalization of the residuals of each race within each cohort scaled by the standard deviation of the trait and adjusted for covariates12. We included covariates such as age, age2, sex, PC1–11, study-groups as well as Mendelian founder lipid variants APOB p.R3527Q and APOC3 p.R19X for the Amish cohort7,8,71. Supplementary Data 1 provides the distributions of each of the four lipid phenotypes by cohort, ancestral groups, and gender. For the UK Biobank, we curated the first instance of the four lipids (data field numbers: HDL-C-30760; LDL-C-30780; TC-30690; TG-30870). The lipid measurements from mmol/L were converted to mg/dL by multiplying TG measurements by 88.57 and for other lipids by multiplying by 38.67. We executed similar steps of phenotype harmonization and normalization for the replication cohorts. In addition, we adjusted the MGB Biobank for study-center and array-type, and Penn Medicine Biobank for ancestry and BMI in addition to the other common covariates.
Genotypes
Whole genome sequencing of goal >30X coverage was performed at seven centers (Broad Institute of MIT and Harvard, Northwest Genomics Center, New York Genome Center, Illumina Genomic Services, PSOMAGEN [formerly Macrogen], Baylor College of Medicine Human Genome Sequencing Center, and McDonnell Genome Institute [MGI] at Washington University). In most cases, all samples for a given study within a given Phase were sequenced at the same center (Supplementary Note 1). The reads were aligned to human genome build GRCh38 using a common pipeline across all centers (BWA-MEM).
The TOPMed Informatics Research Core at the University of Michigan performed joint genotype calling on all samples in Freeze 8. The variant calling “GotCloud” pipeline (https://github.com/statgen/topmed_variant_calling) is under continuous development and details on each step can be accessed through TOPMed website for Freeze817. The resulting BCF files were split by study and consent group for distribution to approved dbGaP users. Quality control was performed centrally by the TOPMed IRC and the TOPMed Data Coordinating Center (DCC) as previously described17. Briefly, the two sequence quality criteria used in freeze 8 are: estimated DNA sample contamination below 10%, and 95% or more of the genome covered to 10× or greater. The variant filtering in TOPMed Freeze 8 is performed by (1) first calculating Mendelian consistency scores using known familial relatedness and duplicates, and (2) training a Support Vector Machine (SVM) classifier between known variant sites (positive labels) and Mendelian inconsistent variants. A small number of sex mismatches were detected as annotated females with low X and high Y chromosome depth or annotated males with high X and low Y chromosome depth. These samples were either excluded from the sample set to be released on dbGaP or their sample identities were resolved using information from prior array genotype comparisons and/or pedigree checks. Details regarding WGS data acquisition, processing and quality control vary among the TOPMed data freezes. Freeze-specific methods are described on the TOPMed website (https://www.nhlbiwgs.org/data-sets) and in documents included in each TOPMed accession released on dbGaP. The VCF/BCF files were converted to GDS (Genomic Data Structure) format by the DCC and were deposited into the dbGap TOPMed Exchange Area.
The genetic relationship matrix (GRM) is an N*N matrix of relatedness information of the samples included in the study and was computed centrally using ‘PC-relate’ R package (version: 1.24.0)72. Using the ‘Genesis’ R package (version:2.20.1)73 we generated subsetted GRM for the samples with plasma lipid profiles. The GDS files with the variants were annotated internally by curating data from multiple database sources using Functional Annotation of Variant–Online Resource (FAVOR (http://favor.genohub.org)13. This study used the resultant aGDS (annotation GDS) files.
The MGB Biobank replication cohort was genotyped using three different arrays (Multiethnic Exome Global (Meg), Human multi-ethnic array (Mega), and Expanded multi-ethnic genotyping array (Megex)), and we separately imputed the data using TOPMed imputation server with default parameters74,75. This study applied the Version-r2 of the imputation panel, it includes 97,256 reference samples and ~300 M genetic variants. The Illumina Global Screening array was used to genotype the Penn Medicine Biobank. Penn Medicine Biobank TOPMed imputation was performed using EAGLE75 and Minimac76 software. For this study, we downloaded variants that passed a min R2 threshold of 0.3. The TOPMed imputation panel is robust, built from 97,256 deeply sequenced human genomes and contains 308,107,085 genetic variants from multi-ethnic samples. Imputation was performed in independent non-overlapping samples agnostic to phenotypes. The UKB imputed data was derived using merged UK10K77, 1000 Genomes phase2 reference panels and was combined to the Haplotype reference Consortium78 (HRC) using IMPUTE 4 program (https://jmarchini.org/software/). The UKB WGS data consist of whole genomes of 150,119 UKB participants with an average coverage of 32.5X. We used joint called VCFs from GraphTyper, which consist of 710,913,648 variants79. We used VCFs provided on the UK Biobank and conducted all the analysis in UKB Research Analysis Platform (UKB RAP).
Single variant association
We performed genome-wide single variant association analyses for autosomal variants with minor allele frequency (MAF) >0.1% across the dataset with each of the four lipid phenotypes. We implemented the SAIGE-QT80 method, which employs fast linear mixed models with kinship adjustment, in Encore (https://encore.sph.umich.edu/) for single variant association analyses. We additionally adjusted the model for covariates (PC1-PC11, age, sex, age2, and study-groups [cohort-race subgrouping]).
We conducted single variant association replications for putative novel variants. After comparing the results with published lipid GWAS summary statistics, we filtered putative novel GWAS variants based on a stringent whole genome-wide significant threshold (alpha = 5 × 10−9)81. Replication was performed in the MGB, Penn Medicine Biobanks and UK Biobank where linear regression models were fitted and adjusted for covariates as indicated above. In addition, we adjusted the MGB Biobank for study recruitment center and array and Penn Medicine Biobank for ancestry and BMI. In the MGB Biobank, we selected lipid concentrations closest to the sample acquisition time point and adjusted for statins if prescribed within one year prior to sample acquisition. In the Penn Biobank, we utilized each participant’s median lipid concentration for replication; statins prescribed prior to lipid concentration used were adjusted in the models. In addition, we carried out meta-analysis using fixed effects model based on inverse-variance-weighted effect size for the two replication cohorts using METASOFT82.
Rare variant association test
We performed rare variant association (RVA) using the Variant-Set Test for Association using Annotation infoRmation (STAAR) pipeline13,83. STAARpipeline is a regression-based framework that permits adjustment of covariates, population structure, and relatedness by fitting linear and logistic mixed models for quantitative and dichotomous traits83–85. We chose STAAR to leverage the annotation information and associated scores that were available for TOPMed Freeze 8 data to incorporate the analysis of rare non-coding variants from whole genome sequencing. The method implements genome-wide scanning of rare variants (MAF <0.01) in gene-centric and region-based workflows. For each variant set, STAARpipeline calculates a set-based p-value using the STAAR method, which increases the analysis power by incorporating multiple in silico variant functional annotation scores capturing diverse genomic features and biochemical readouts13. We aggregated rare variants into multiple groups for coding and non-coding analyses. For the coding region, we defined five different aggregate masks of rare variants 1) plof (putative loss-of-function), plof-Ds (putative loss-of-function or disruptive missense), missense, disruptive-missense, and synonymous. For the non-coding regions, we used seven rare variant masks: (1) promoter-CAGE (promoter variants within Cap Analysis of Gene Expression [CAGE] sites86), (2) promoter-DHS (promoter variants within DNase hypersensitivity [DHS] sites87), (3) enhancer-CAGE (enhancer within CAGE sites88,89), (4) enhancer-DHS (enhancer variants within DHS sites87,89), (5) UTR (rare variants in 3′ untranslated region [UTR] and 5′ UTR untranslated region), (6) upstream, and (7) downstream. Detailed explanations of the regions defined based on these masks is discussed within STAARpipeline13,83.
In the gene-centric workflows, for both coding (within exonic boundaries) and non-coding (promoter: +/-3 kb window of transcription starting site (TSS), enhancer: GeneHancer predicted regions, UTR (both 5′ and 3′ UTR regions)/upstream/downstream: GENCODE Variant Effect Predictor (VEP) categories) regions, we considered only genes with at least two rare variants (i.e., 18,445 genes in all 22 autosomes). In the region-based workflows, we implemented two protocols: (1) a ‘sliding window’ approach, where we aggregated rare variants within 2-kb sliding windows and with 1-kb overlap length, and (2) a ‘dynamic window’ approach, where we executed SCANG34 method and aggregated dynamically variant-sets between 40–300 variants per set, where the method systematically scans the whole genome with overlapping windows of varying sizes. The STAARpipeline R-package implements multiple rare-variants aggregate tests including SKAT90, Burden91 and ACAT92 and integrates them as STAAR-O13,83. We performed gene-centric and region-based rare variant tests using annotated GDS files of TOPMed.
We completed aggregate tests as three-step process. In the first step, we fitted a null model using glmmkin() function. The null model was fitted for each of the four lipid phenotypes adjusted for all covariates and relatedness except the genotype of interest. In the second step, we ran genome-wide gene-centric and region-based rare-variant aggregate tests. The third step directed conditional analyses, where the results were adjusted for previously known significantly lipid-associated (i.e., p < 5 × 10−8 in external datasets) individual variants from GWAS Catalog93 and Million Veterans Program (MVP)15 GWAS summary statistics. To obtain effect estimates of significant aggregate sets, we associated the cumulative genotypes (binary scores) based on the variants forming the aggregates and used Glmm.Wald test from GMMAT R package83(version 1.3.1). For significantly associated window-based rare variant aggregations, we trimmed the exonic variants and estimated the effects with only non-coding variants.
For the rare variant replication in UKB WGS data, we curated the rare variant aggregate sets in UKB RAP for the gene-centric coding/non-coding and region-based significant sets and applied STAAR workflow as demonstrated by the STAARpipeline (https://github.com/xihaoli/STAARpipeline) and describe above.
Computational mining of single variant GWAS
Gene-set enrichment using FUMA
We performed enrichment analysis with single variant GWAS summary stats from the four lipids using FUMA94 (version 1.3.7) with default parameters and significance at 5 × 10−9. FUMA is an integrated platform which efficiently facilitates functional mapping and enrichment of GWAS-associated genes using multiple useful resources. The method uses 18 different biological data repositories and tools to process GWAS data. We additionally used MAGMA95 (version 1.08) gene-based analysis enrichment workflow within FUMA with the complete GWAS summary data for eQTL based tissue enrichment. The functionally prioritized genes were visualized based on their protein-protein interaction networks using the STRING database96.
CETP and GAS6 gene expression and lipid trait colocalization
We studied the correlation of LDL-C and HDL-C effects with eQTL effects at chromosome 16q13, which includes CETP and correlation of LDL-C and TC with eQTLs at rs7140110 of GAS6. We downloaded GTEx eQTL build 38 (version8) data for liver, adipose subcutaneous, and adipose visceral (omentum) tissues from GTEx on 16/APR/202097. For the CETP variant analysis, we selected eQTLs with nominal significance (p-value < 0.05) and utilized the eQTL-gene pairs with the most significant p-values. Genes with at least 5 eQTLs were selected for the colocalization analysis. We selected variants with a suggestive significance (p-value <5 × 10−7) for LDL-C or HDL-C effects within 500 kb of the lead locus variant. For the GAS6 variant analysis, we curated all the GWAS variants within 500 kb of the lead variant with nominal significance (p-value < 0.05) and matched them to eQTL data where the transcription starting site of the corresponding gene is within +/−500 kb. We conducted colocalization analysis using the coloc.abf() function98 and identified nominally significant (PP.H4 > 1 × 10−03) genes-eQTL pairs. The coloc methodology implements an efficient statistical framework to identify shared variants from two association signals through posteriors probabilities. Finally, we used the colocalized signals and compared the significant genes using STRING96, a protein-protein interaction database. All the correlation tests were conducted in R, where we calculated Pearson correlations between the lipid effect estimates and gene expression effects (slope) from GTEx.
Phenome wide association analysis
The complex trait information was curated from UK Biobank resource, where we curated multiple disease phenotypes for UKB samples into International Classification of Diseases (ICD)-based phecodes based on phecode map (https://phewascatalog.org) using the PheWAS R package (version PheWAS_0.99.5-4). We conducted a phenome-wide association analysis (PheWAS) using a logistic regression model glm() in R. We adjusted the models for PC1–10, age, age2, sex, and race.
Calculation of heritability estimates from TOPMed WGS data
We calculated heritabilities estimated for the four lipids using TOPMed WGS data using Greml-LDMS approach39, where we binned the variants into four MAF bins based on minor allele frequency and grouped the variants to four LD quartiles based on LD score calculated by GCTA method99. The four MAF bins used in this study includes >=0.05, >=0.01 to <0.05, >=0.001 to <0.01 and >=0.0001 to <0.001. We excluded any variant with MAF < 0.0001 from this analysis. The hereditary estimation was calculated for three ancestral groups (African, European, Hispanic) where only unrelated samples (kinship score < 0.025) were included in the analysis. We excluded the other two ancestral groups (i.e., Asian and Samoan) from this analysis due to insufficient sample sizes. In total we included 9640, 21568 and 10631 in African, European and Hispanic ancestries respectively. For each MAF bin, we implemented certain quality control (QC) measures using PLINK software20, which includes; genotype missingness (--geno 0.05), sample missingness (--mind 0.05), Hardy-Weinberg equilibrium (--hwe 10−6) and LD pruned variants (--indep-pairwise 50 5 0.1) as implemented by Wainschtein et al.39. Next, we implemented Greml-LDMS with LD score region as 200 and GRM cut-off as 0.05 for the four lipid phenotypes. We calculated 20 principal components from the QC passed variants in each MAF bin and implemented GCTA workflow with --reml-no-constrain, --reml-no-lrt and --reml-maxit 10,000 parameters to avoid the no-convergence issues and negative h2 estimates. For comparing the h2 estimates between variants from WGS data and array-genotypes, first, we used QC passed WGS variants as mentioned above, second, we curated the variants from MGB Biobank array data and intersected them with WGS variants from TOPMed. Next, we calculated heritability estimates for array-genotype variants and compared with h2 estimates from WGS variants for the three ancestral groups.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
Whole genome sequencing (WGS) for the Trans-Omics in Precision Medicine (TOPMed) program was supported by the National Heart, Lung and Blood Institute (NHLBI). P.N. is supported by grants from the National Heart, Lung, and Blood Institute (R01HL142711, R01HL148050, R01HL151283, R01HL148565, R01HL135242, R01HL151152), Fondation Leducq (TNE-18CVD04), and Massachusetts General Hospital (Paul and Phyllis Fireman Endowed Chair in Vascular Medicine). G.M.P. is supported by NIH grants R01HL142711 and R01HL127564. X.Lin is supported by grants R35-CA197449, U19-CA203654, R01-HL113338, and U01-HG009088. Prior to his employment at Novartis and during this work S.A.L. was supported by NIH grants R01HL139731, R01HL157635, and American Heart Association 18SFRN34250007. We like to acknowledge all the grants that supported this study, R01 HL121007, U01 HL072515, R01 AG18728, X01HL134588, HL 046389, HL113338, and 1R35HL135818, K01 HL135405, R03 HL154284, U01HL072507, R01HL087263, R01HL090682, P01HL045522, R01MH078143, R01MH078111, R01MH083824, U01DK085524, R01HL113323, R01HL093093, R01HL140570, R01HL142711, R01HL127564, R01HL148050, R01HL148565, HL105756, and Leducq TNE-18CVD04. The views expressed in this manuscript are those of the authors and do not necessarily represent the views of the National Heart, Lung, and Blood Institute; the National Institutes of Health; or the U.S.Department of Health and Human Services. Detailed acknowledgements provided in Supplementary Note 2.
Author contributions
M.S.S., G.M.P., and P.N. designed the study. M.S.S. carried out all the primary analysis with critical inputs from G.M.P. and P.N. M.S.S., Xih.L, Z.L., A.P., D.Y.Z., J.P., S.A., J.C.B., J.A.B., B.E.C., L.M.C., R.H.C., J.E.C., L.F., P.S.V., R.D., B.I.F., M.G., X.G., N.H.C., B.H., C.M.H., M.R.I., T.N.K., B.G.K., L.L., Xia.L, M.L., S.A.L., A.W.M., P.M., M.E.M., A.C.M., T.N., J.R.O.C., N.D.P., P.A.P., M.S.R., J.A.S., X.S., K.D.T., R.P.T., M.Y.T., Z.W., Y.W., B.W., J.T.W., L.R.Y., W.Z., D.K.A., J. Blangero, E.B., D.W.B., Y.I.C., A.C., L.A.C., S.K.D., P.T.E., M.F., S. Gabriel, S. Germer, R.G., J.H., R.C.K., S.L.R.K., R. Kim, C.K., R.J.F.L., K.M., R.A.M., S.T.M., B.D.M., D.N., K.E.N., B.M.P., S. Redline, A.P.R., R.S.V., S.S.R., C.W., J.I.R., D.J.R., X.Lin., G.M.P., and P.N. acquired, analyzed or interpreted data. M.S.S., G.M.P. and P.N. wrote the first draft of the manuscript and all others provided intellectual revisions. G.M.P. and P.N. and NHLBI TOPMed Lipids Working Group provided administrative, technical, or material support.
Peer review
Peer review information
Nature Communications thanks David Meyre and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Data availability
Individual whole-genome sequence data for TOPMed and harmonized lipids at individual sample level are available through restricted access via the TOPMed dbGaP Exchange area. Summary level genotype data from TOPMed are available through the BRAVO browser (https://bravo.sph.umich.edu/). The UK Biobank (UKB) whole-genome sequence data can be accessed through UKB Research Analysis Platform (RAP), through the UKB approval system (https://www.ukbiobank.ac.uk). The Mass General Brigham Biobank (MGBB) individual-level data are available from https://personalizedmedicine.partners.org/Biobank/Default.aspx, where the data is available through institutional review board (IRB) approval, therefore not publicly available. Individual-level data from Penn Medicine BioBank is not publicly available due to research participants privacy concerns. The summary data captured using whole exome sequencing can be accessed through PMBB Genome Browser (https://pmbb.med.upenn.edu/allele-frequency/). The dbGaP accessions for TOPMed cohorts are as follows: Old Order Amish (Amish) phs000956 and phs00039; Atherosclerosis Risk in Communities study (ARIC) phs001211 and phs000280; Mt Sinai BioMe Biobank (BioMe) phs001644 and phs000925; Coronary Artery Risk Development in Young Adults (CARDIA) phs001612 and phs000285; Cleveland Family Study (CFS) phs000954 and phs000284; Cardiovascular Health Study (CHS) phs001368 and phs000287; Diabetes Heart Study (DHS) phs001412 and phs001012; Framingham Heart Study (FHS) phs000974 and phs000007; Genetic Studies of Atherosclerosis Risk (GeneSTAR) phs001218 and phs000375; Genetic Epidemiology Network of Arteriopathy (GENOA) phs001345 and phs001238; Genetic Epidemiology Network of Salt Sensitivity (GenSalt) phs001217 and phs000784; Genetics of Lipid-Lowering Drugs and Diet Network (GOLDN) phs001359 and phs000741; Hispanic Community Health Study - Study of Latinos (HCHS_SOL) phs001395 and phs000810; Hypertension Genetic Epidemiology Network and Genetic Epidemiology Network of Arteriopathy (HyperGEN) phs001293 and phs001293; Jackson Heart Study (JHS) phs000964 and phs000286; Multi-Ethnic Study of Atherosclerosis (MESA) phs001416 and phs000209; Massachusetts General Hospital Atrial Fibrillation Study (MGH_AF) phs001062 and phs001001; San Antonio Family Study (SAFS) phs001215 and phs000462; Samoan Adiposity Study (SAS) phs000972 and phs000914; Taiwan Study of Hypertension using Rare Variants (THRV) phs001387 and phs001387; Women’s Health Initiative (WHI) phs001237 and phs000200.
Code availability
Codes used to implement STAAR workflows are available at https://github.com/xihaoli/STAAR and https://github.com/xihaoli/STAARpipeline. Workflow implemented for whole genome heritability calculations are available at https://github.com/CNSGenomics/Heritability_WGS.
Competing interests
P.N. reports investigator-initiated grant support from Amgen, Apple, AstraZeneca, and Boston Scientific, personal fees from Apple, AstraZeneca, Blackstone Life Sciences, Foresite Labs, Genentech, TenSixteen Bio, and Novartis, scientific advisory board membership of geneXwell and TenSixteen Bio, and spousal employment at Vertex, all unrelated to the present work. B.P. serves on the Steering Committee of the Yale Open Data Access Project funded by Johnson & Johnson. M.E.M. receives funding from Regeneron Pharmaceutical Inc. unrelated to this work. S.A. has employment and equity in 23andMe, Inc. The spouse of C.J.W. works at Regeneron. S.A.L. is a full-time employee of Novartis as of July 18, 2022. S.A.L. has received sponsored research support from Bristol Myers Squibb, Pfizer, Boehringer Ingelheim, Fitbit, Medtronic, Premier, and IBM, and has consulted for Bristol Myers Squibb, Pfizer, Blackstone Life Sciences, and Invitae. X. Lin is a consultant of AbbVie Pharmaceuticals and Verily Life Sciences. The remaining authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors jointly supervised this work: Gina M. Peloso, Pradeep Natarajan.
A list of authors and their affiliations appears at the end of the paper.
Contributor Information
Gina M. Peloso, Email: gpeloso@bu.edu
Pradeep Natarajan, Email: pnatarajan@mgh.harvard.edu.
NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium:
Namiko Abe, Gonçalo Abecasis, Francois Aguet, Christine Albert, Laura Almasy, Alvaro Alonso, Seth Ament, Peter Anderson, Pramod Anugu, Deborah Applebaum-Bowden, Kristin Ardlie, Dan Arking, Allison Ashley-Koch, Tim Assimes, Paul Auer, Dimitrios Avramopoulos, Najib Ayas, Adithya Balasubramanian, John Barnard, Kathleen Barnes, R. Graham Barr, Emily Barron-Casella, Lucas Barwick, Terri Beaty, Gerald Beck, Diane Becker, Lewis Becker, Rebecca Beer, Amber Beitelshees, Emelia Benjamin, Takis Benos, Marcos Bezerra, Larry Bielak, Thomas Blackwell, Russell Bowler, Ulrich Broeckel, Jai Broome, Deborah Brown, Karen Bunting, Esteban Burchard, Carlos Bustamante, Erin Buth, Jonathan Cardwell, Vincent Carey, Julie Carrier, Cara Carty, Richard Casaburi, Juan P. Casas Romero, James Casella, Peter Castaldi, Mark Chaffin, Christy Chang, Yi-Cheng Chang, Daniel Chasman, Sameer Chavan, Bo-Juen Chen, Wei-Min Chen, Yii-Der Ida Chen, Michael Cho, Seung Hoan Choi, Mina Chung, Clary Clish, Suzy Comhair, Matthew Conomos, Elaine Cornell, Carolyn Crandall, James Crapo, L. Adrienne Cupples, Jeffrey Curtis, Brian Custer, Coleen Damcott, Dawood Darbar, Sean David, Colleen Davis, Michelle Daya, Mariza de Andrade, Michael DeBaun, Ranjan Deka, Dawn DeMeo, Scott Devine, Huyen Dinh, Harsha Doddapaneni, Qing Duan, Shannon Dugan-Perez, Ravi Duggirala, Jon Peter Durda, Charles Eaton, Lynette Ekunwe, Adel El Boueiz, Leslie Emery, Serpil Erzurum, Charles Farber, Jesse Farek, Tasha Fingerlin, Matthew Flickinger, Nora Franceschini, Chris Frazar, Mao Fu, Stephanie M. Fullerton, Lucinda Fulton, Weiniu Gan, Shanshan Gao, Yan Gao, Margery Gass, Heather Geiger, Bruce Gelb, Mark Geraci, Robert Gerszten, Auyon Ghosh, Chris Gignoux, Mark Gladwin, David Glahn, Stephanie Gogarten, Da-Wei Gong, Harald Goring, Sharon Graw, Kathryn J. Gray, Daniel Grine, Colin Gross, C. Charles Gu, Yue Guan, Namrata Gupta, David M. Haas, Jeff Haessler, Michael Hall, Yi Han, Patrick Hanly, Daniel Harris, Nicola L. Hawley, Ben Heavner, Susan Heckbert, Ryan Hernandez, David Herrington, Craig Hersh, Bertha Hidalgo, James Hixson, Brian Hobbs, John Hokanson, Elliott Hong, Karin Hoth, Chao Agnes Hsiung, Jianhong Hu, Yi-Jen Hung, Haley Huston, Chii Min Hwu, Rebecca Jackson, Deepti Jain, Cashell Jaquish, Jill Johnsen, Andrew Johnson, Craig Johnson, Rich Johnston, Kimberly Jones, Hyun Min Kang, Shannon Kelly, Eimear Kenny, Michael Kessler, Alyna Khan, Ziad Khan, Wonji Kim, John Kimoff, Greg Kinney, Barbara Konkle, Holly Kramer, Christoph Lange, Ethan Lange, Cathy Laurie, Cecelia Laurie, Meryl LeBoff, Jiwon Lee, Sandra Lee, Wen-Jane Lee, Jonathon LeFaive, David Levine, Dan Levy, Joshua Lewis, Yun Li, Henry Lin, Honghuang Lin, Simin Liu, Yongmei Liu, Yu Liu, Kathryn Lunetta, James Luo, Ulysses Magalang, Michael Mahaney, Barry Make, Alisa Manning, JoAnn Manson, Lisa Martin, Melissa Marton, Susan Mathai, Susanne May, Patrick McArdle, Merry-Lynn McDonald, Sean McFarland, Daniel McGoldrick, Caitlin McHugh, Becky McNeil, Hao Mei, James Meigs, Vipin Menon, Luisa Mestroni, Ginger Metcalf, Deborah A. Meyers, Emmanuel Mignot, Julie Mikulla, Nancy Min, Mollie Minear, Ryan L. Minster, Matt Moll, Zeineen Momin, Courtney Montgomery, Donna Muzny, Josyf C. Mychaleckyj, Girish Nadkarni, Rakhi Naik, Sergei Nekhai, Sarah C. Nelson, Bonnie Neltner, Caitlin Nessner, Osuji Nkechinyere, Jeff O’Connell, Tim O’Connor, Heather Ochs-Balcom, Geoffrey Okwuonu, Allan Pack, David T. Paik, James Pankow, George Papanicolaou, Cora Parker, Juan Manuel Peralta, Marco Perez, James Perry, Ulrike Peters, Lawrence S. Phillips, Jacob Pleiness, Toni Pollin, Wendy Post, Julia Powers Becker, Meher Preethi Boorgula, Michael Preuss, Pankaj Qasba, Dandi Qiao, Zhaohui Qin, Nicholas Rafaels, Laura Raffield, Mahitha Rajendran, Ramachandran S. Vasan, D. C. Rao, Laura Rasmussen-Torvik, Aakrosh Ratan, Robert Reed, Catherine Reeves, Elizabeth Regan, Alex Reiner, Ken Rice, Rebecca Robillard, Nicolas Robine, Dan Roden, Carolina Roselli, Ingo Ruczinski, Alexi Runnels, Pamela Russell, Sarah Ruuska, Kathleen Ryan, Ester Cerdeira Sabino, Danish Saleheen, Shabnam Salimi, Sejal Salvi, Steven Salzberg, Kevin Sandow, Vijay G. Sankaran, Jireh Santibanez, Karen Schwander, David Schwartz, Frank Sciurba, Christine Seidman, Jonathan Seidman, Frédéric Sériès, Vivien Sheehan, Stephanie L. Sherman, Amol Shetty, Aniket Shetty, Wayne Hui-Heng Sheu, M. Benjamin Shoemaker, Brian Silver, Edwin Silverman, Robert Skomro, Albert Vernon Smith, Josh Smith, Nicholas Smith, Tanja Smith, Sylvia Smoller, Beverly Snively, Michael Snyder, Tamar Sofer, Nona Sotoodehnia, Adrienne M. Stilp, Garrett Storm, Elizabeth Streeten, Jessica Lasky Su, Yun Ju Sung, Jody Sylvia, Adam Szpiro, Daniel Taliun, Hua Tang, Margaret Taub, Matthew Taylor, Simeon Taylor, Marilyn Telen, Timothy A. Thornton, Machiko Threlkeld, Lesley Tinker, David Tirschwell, Sarah Tishkoff, Hemant Tiwari, Catherine Tong, Dhananjay Vaidya, David Van Den Berg, Peter VandeHaar, Scott Vrieze, Tarik Walker, Robert Wallace, Avram Walts, Fei Fei Wang, Heming Wang, Jiongming Wang, Karol Watson, Jennifer Watt, Daniel E. Weeks, Joshua Weinstock, Bruce Weir, Scott T. Weiss, Lu-Chen Weng, Jennifer Wessel, Kayleen Williams, L. Keoki Williams, Carla Wilson, James Wilson, Lara Winterkorn, Quenna Wong, Joseph Wu, Huichun Xu, Ivana Yang, Ketian Yu, Seyedeh Maryam Zekavat, Yingze Zhang, Snow Xueyan Zhao, Wei Zhao, Xiaofeng Zhu, Michael Zody, and Sebastian Zoellner
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-022-33510-7.
References
- 1.Cohen JC, Boerwinkle E, Mosley TH, Hobbs HH. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N. Engl. J. Med. 2006;354:1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 2.Cohen J, et al. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat. Genet. 2005;37:161–165. doi: 10.1038/ng1509. [DOI] [PubMed] [Google Scholar]
- 3.Musunuru K, et al. Exome sequencing, ANGPTL3 mutations, and familial combined hypolipidemia. N. Engl. J. Med. 2010;363:2220–2227. doi: 10.1056/NEJMoa1002926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stitziel NO, et al. ANGPTL3 deficiency and protection against coronary artery disease. J. Am. Coll. Cardiol. 2017;69:2054–2063. doi: 10.1016/j.jacc.2017.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dewey FE, et al. Genetic and pharmacologic inactivation of ANGPTL3 and cardiovascular disease. N. Engl. J. Med. 2017;377:211–221. doi: 10.1056/NEJMoa1612790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pollin TI, et al. A null mutation in human APOC3 confers a favorable plasma lipid profile and apparent cardioprotection. Science. 2008;322:1702–1705. doi: 10.1126/science.1161524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen H, et al. Familial defective apolipoprotein B-100 and increased low-density lipoprotein cholesterol and coronary artery calcification in the old order amish. Arch. Intern. Med. 2010;170:1850–1855. doi: 10.1001/archinternmed.2010.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saleheen D, et al. Human knockouts and phenotypic analysis in a cohort with a high rate of consanguinity. Nature. 2017;544:235–239. doi: 10.1038/nature22034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Exome Aggregation Consortium. et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Samocha KE, et al. A framework for the interpretation of de novo mutation in human disease. Nat. Genet. 2014;46:944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Natarajan P, et al. Chromosome Xq23 is associated with lower atherogenic lipid concentrations and favorable cardiometabolic indices. Nat. Commun. 2021;12:2182. doi: 10.1038/s41467-021-22339-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X, et al. Dynamic incorporation of multiple in silico functional annotations empowers rare variant association analysis of large whole-genome sequencing studies at scale. Nat. Genet. 2020;52:969–983. doi: 10.1038/s41588-020-0676-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Natarajan P, et al. Deep-coverage whole genome sequences and blood lipids among 16,324 individuals. Nat. Commun. 2018;9:3391. doi: 10.1038/s41467-018-05747-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klarin D, et al. Genetics of blood lipids among ~300,000 multi-ethnic participants of the Million Veteran Program. Nat. Genet. 2018;50:1514–1523. doi: 10.1038/s41588-018-0222-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu Y, et al. Minority-centric meta-analyses of blood lipid levels identify novel loci in the Population Architecture using Genomics and Epidemiology (PAGE) study. PLoS Genet. 2020;16:e1008684. doi: 10.1371/journal.pgen.1008684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium. et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature. 2021;590:290–299. doi: 10.1038/s41586-021-03205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stilp, A. M. et al. A System for Phenotype Harmonization in the NHLBI Trans-Omics for Precision Medicine (TOPMed) Program. Am. J. Epidemiol. 10.1093/aje/kwab115 (2021). [DOI] [PMC free article] [PubMed]
- 19.Fadista J, Manning AK, Florez JC, Groop L. The (in)famous GWAS P-value threshold revisited and updated for low-frequency variants. Eur. J. Hum. Genet. 2016;24:1202–1205. doi: 10.1038/ejhg.2015.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bentley AR, et al. Multi-ancestry genome-wide gene-smoking interaction study of 387,272 individuals identifies new loci associated with serum lipids. Nat. Genet. 2019;51:636–648. doi: 10.1038/s41588-019-0378-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ripatti P, et al. Polygenic hyperlipidemias and coronary artery disease risk. Circ. Genom. Precis. Med. 2020;13:e002725. doi: 10.1161/CIRCGEN.119.002725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Leeuwen EM, et al. Meta-analysis of 49 549 individuals imputed with the 1000 Genomes Project reveals an exonic damaging variant in ANGPTL4 determining fasting TG levels. J. Med. Genet. 2016;53:441–449. doi: 10.1136/jmedgenet-2015-103439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nielsen JB, et al. Loss-of-function genomic variants highlight potential therapeutic targets for cardiovascular disease. Nat. Commun. 2020;11:6417. doi: 10.1038/s41467-020-20086-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aragam KG, et al. Limitations of contemporary guidelines for managing patients at high genetic risk of coronary artery disease. J. Am. Coll. Cardiol. 2020;75:2769–2780. doi: 10.1016/j.jacc.2020.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park J, et al. Exome-wide evaluation of rare coding variants using electronic health records identifies new gene-phenotype associations. Nat. Med. 2021;27:66–72. doi: 10.1038/s41591-020-1133-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mountjoy E, et al. An open approach to systematically prioritize causal variants and genes at all published human GWAS trait-associated loci. Nat. Genet. 2021;53:1527–1533. doi: 10.1038/s41588-021-00945-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lonsdale J, et al. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barter PJ, et al. Effects of torcetrapib in patients at high risk for coronary events. N. Engl. J. Med. 2007;357:2109–2122. doi: 10.1056/NEJMoa0706628. [DOI] [PubMed] [Google Scholar]
- 30.Schwartz GG, et al. Effects of dalcetrapib in patients with a recent acute coronary syndrome. N. Engl. J. Med. 2012;367:2089–2099. doi: 10.1056/NEJMoa1206797. [DOI] [PubMed] [Google Scholar]
- 31.The HPS3/TIMI55–REVEAL Collaborative Group. Effects of anacetrapib in patients with atherosclerotic vascular disease. N. Engl. J. Med. 2017;377:1217–1227. doi: 10.1056/NEJMoa1706444. [DOI] [PubMed] [Google Scholar]
- 32.Lincoff AM, et al. Evacetrapib and cardiovascular outcomes in high-risk vascular disease. N. Engl. J. Med. 2017;376:1933–1942. doi: 10.1056/NEJMoa1609581. [DOI] [PubMed] [Google Scholar]
- 33.Fairoozy RH, White J, Palmen J, Kalea AZ, Humphries SE. Identification of the functional variant(s) that explain the low-density lipoprotein receptor (LDLR) GWAS SNP rs6511720 association with lower LDL-C and risk of CHD. PLoS ONE. 2016;11:e0167676. doi: 10.1371/journal.pone.0167676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Z, et al. Dynamic scan procedure for detecting rare-variant association regions in whole-genome sequencing studies. Am. J. Hum. Genet. 2019;104:802–814. doi: 10.1016/j.ajhg.2019.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roses AD, et al. A TOMM40 variable-length polymorphism predicts the age of late-onset Alzheimer’s disease. Pharmacogenomics J. 2010;10:375–384. doi: 10.1038/tpj.2009.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li G, et al. TOMM40 intron 6 poly-T length, age at onset, and neuropathology of AD in individuals with APOE ε3/ε3. Alzheimers Dement. J. Alzheimers Assoc. 2013;9:554–561. doi: 10.1016/j.jalz.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glazier AM, Scott J, Aitman TJ. Molecular basis of the Cd36 chromosomal deletion underlying SHR defects in insulin action and fatty acid metabolism. Mamm. Genome . J. Int. Mamm. Genome Soc. 2002;13:108–113. doi: 10.1007/s00335-001-2132-9. [DOI] [PubMed] [Google Scholar]
- 38.The LifeLines Cohort Study. et al. Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nat. Genet. 2015;47:1114–1120. doi: 10.1038/ng.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wainschtein P, et al. Assessing the contribution of rare variants to complex trait heritability from whole-genome sequence data. Nat. Genet. 2022;54:263–273. doi: 10.1038/s41588-021-00997-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cadby G, et al. Heritability of 596 lipid species and genetic correlation with cardiovascular traits in the Busselton Family Heart Study. J. Lipid Res. 2020;61:537–545. doi: 10.1194/jlr.RA119000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Global Lipids Genetics Consortium. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 2013;45:1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Willer CJ, et al. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 2013;45:1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.ENGAGE Consortium. et al. The impact of low-frequency and rare variants on lipid levels. Nat. Genet. 2015;47:589–597. doi: 10.1038/ng.3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.The Myocardial Infarction Genetics Consortium Investigators. Inactivating mutations in NPC1L1 and protection from coronary heart disease. N. Engl. J. Med. 2014;371:2072–2082. doi: 10.1056/NEJMoa1405386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.GLGC Consortium. et al. Exome chip meta-analysis identifies novel loci and East Asian–specific coding variants that contribute to lipid levels and coronary artery disease. Nat. Genet. 2017;49:1722–1730. doi: 10.1038/ng.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoffmann TJ, et al. A large electronic-health-record-based genome-wide study of serum lipids. Nat. Genet. 2018;50:401–413. doi: 10.1038/s41588-018-0064-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martin AR, et al. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet. 2019;51:584–591. doi: 10.1038/s41588-019-0379-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peloso GM, Natarajan P. Insights from population-based analyses of plasma lipids across the allele frequency spectrum. Curr. Opin. Genet. Dev. 2018;50:1–6. doi: 10.1016/j.gde.2018.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kremer LS, et al. Genetic diagnosis of Mendelian disorders via RNA sequencing. Nat. Commun. 2017;8:15824. doi: 10.1038/ncomms15824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cummings BB, et al. Improving genetic diagnosis in Mendelian disease with transcriptome sequencing. Sci. Transl. Med. 2017;9:eaal5209. doi: 10.1126/scitranslmed.aal5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Genome Aggregation Database Production Team. et al. Transcript expression-aware annotation improves rare variant interpretation. Nature. 2020;581:452–458. doi: 10.1038/s41586-020-2329-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mendes de Almeida R, et al. Whole gene sequencing identifies deep-intronic variants with potential functional impact in patients with hypertrophic cardiomyopathy. PLoS ONE. 2017;12:e0182946. doi: 10.1371/journal.pone.0182946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vitsios D, Dhindsa RS, Middleton L, Gussow AB, Petrovski S. Prioritizing non-coding regions based on human genomic constraint and sequence context with deep learning. Nat. Commun. 2021;12:1504. doi: 10.1038/s41467-021-21790-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.di Iulio J, et al. The human noncoding genome defined by genetic diversity. Nat. Genet. 2018;50:333–337. doi: 10.1038/s41588-018-0062-7. [DOI] [PubMed] [Google Scholar]
- 55.Genome Aggregation Database Consortium. et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Khera AV, et al. Diagnostic yield and clinical utility of sequencing familial hypercholesterolemia genes in patients with severe hypercholesterolemia. J. Am. Coll. Cardiol. 2016;67:2578–2589. doi: 10.1016/j.jacc.2016.03.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Benn M, Watts GF, Tybjærg-Hansen A, Nordestgaard BG. Mutations causative of familial hypercholesterolaemia: screening of 98 098 individuals from the Copenhagen General Population Study estimated a prevalence of 1 in 217. Eur. Heart J. 2016;37:1384–1394. doi: 10.1093/eurheartj/ehw028. [DOI] [PubMed] [Google Scholar]
- 58.Grundy SM, et al. 2018 AHA/ACC/AACVPR/AAPA/ABC/ACPM/ADA/AGS/APhA/ASPC/NLA/PCNA Guideline on the Management of Blood Cholesterol: Executive Summary. J. Am. Coll. Cardiol. 2019;73:3168–3209. doi: 10.1016/j.jacc.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 59.Sturm AC, et al. Clinical genetic testing for familial hypercholesterolemia. J. Am. Coll. Cardiol. 2018;72:662–680. doi: 10.1016/j.jacc.2018.05.044. [DOI] [PubMed] [Google Scholar]
- 60.Reeskamp, L. F. et al. A Deep intronic variant in LDLR in familial hypercholesterolemia: time to widen the scope? Circ. Genomic Precis. Med. 11, e002385 (2018). [DOI] [PubMed]
- 61.Calandra S, Tarugi P, Bertolini S. Altered mRNA splicing in lipoprotein disorders. Curr. Opin. Lipidol. 2011;22:93–99. doi: 10.1097/MOL.0b013e3283426ebc. [DOI] [PubMed] [Google Scholar]
- 62.on behalf of the ACMG Laboratory Quality Assurance Committee. et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–423. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Peloso GM, et al. Rare protein-truncating variants in APOB, lower low-density lipoprotein cholesterol, and protection against coronary heart disease. Circ. Genom. Precis. Med. 2019;12:e002376. doi: 10.1161/CIRCGEN.118.002376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Abifadel M, et al. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat. Genet. 2003;34:154–156. doi: 10.1038/ng1161. [DOI] [PubMed] [Google Scholar]
- 65.Jiang L, et al. The distribution and characteristics of LDL receptor mutations in China: a systematic review. Sci. Rep. 2015;5:17272. doi: 10.1038/srep17272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Arráiz N, et al. Novel mutations identification in exon 4 of LDLR gene in patients with moderate hypercholesterolemia in a Venezuelan population. Am. J. Ther. 2010;17:325–329. doi: 10.1097/MJT.0b013e3181c1234d. [DOI] [PubMed] [Google Scholar]
- 67.Gudnason V, et al. Identification of recurrent and novel mutations in exon 4 of the LDL receptor gene in patients with familial hypercholesterolemia in the United Kingdom. Arterioscler. Thromb. J. Vasc. Biol. 1993;13:56–63. doi: 10.1161/01.ATV.13.1.56. [DOI] [PubMed] [Google Scholar]
- 68.Goldmann R, et al. Genomic characterization of large rearrangements of the LDLR gene in Czech patients with familial hypercholesterolemia. BMC Med. Genet. 2010;11:115. doi: 10.1186/1471-2350-11-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zuk O, et al. Searching for missing heritability: Designing rare variant association studies. Proc. Natl Acad. Sci. USA. 2014;111:E455–E464. doi: 10.1073/pnas.1322563111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bycroft C, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Soria LF, et al. Association between a specific apolipoprotein B mutation and familial defective apolipoprotein B-100. Proc. Natl Acad. Sci. USA. 1989;86:587–591. doi: 10.1073/pnas.86.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Conomos MP, Reiner AP, Weir BS, Thornton TA. Model-free estimation of recent genetic relatedness. Am. J. Hum. Genet. 2016;98:127–148. doi: 10.1016/j.ajhg.2015.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gogarten SM, et al. Genetic association testing using the GENESIS R/Bioconductor package. Bioinforma. Oxf. Engl. 2019;35:5346–5348. doi: 10.1093/bioinformatics/btz567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Das S, et al. Next-generation genotype imputation service and methods. Nat. Genet. 2016;48:1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Loh P-R, et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nat. Genet. 2016;48:1443–1448. doi: 10.1038/ng.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fuchsberger C, Abecasis GR, Hinds DA. minimac2: faster genotype imputation. Bioinformatics. 2015;31:782–784. doi: 10.1093/bioinformatics/btu704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.UK10K Consortium. et al. Improved imputation of low-frequency and rare variants using the UK10K haplotype reference panel. Nat. Commun. 2015;6:8111. doi: 10.1038/ncomms9111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.the Haplotype Reference Consortium. A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 2016;48:1279–1283. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Halldorsson, B. V. et al. The sequences of 150,119 genomes in the UK biobank. Nature607, 732–740 (2022). [DOI] [PMC free article] [PubMed]
- 80.Zhou W, et al. Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat. Genet. 2018;50:1335–1341. doi: 10.1038/s41588-018-0184-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pulit SL, de With SAJ, de Bakker PIW. Resetting the bar: statistical significance in whole-genome sequencing-based association studies of global populations. Genet. Epidemiol. 2017;41:145–151. doi: 10.1002/gepi.22032. [DOI] [PubMed] [Google Scholar]
- 82.Han B, Eskin E. Random-effects model aimed at discovering associations in meta-analysis of genome-wide association studies. Am. J. Hum. Genet. 2011;88:586–598. doi: 10.1016/j.ajhg.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li, Z. et al. A framework for detecting noncoding rare variant associations of large-scale whole-genome sequencing studies. 10.1101/2021.11.05.467531 (2021). [DOI] [PMC free article] [PubMed]
- 84.Chen H, et al. Control for population structure and relatedness for binary traits in genetic association studies via logistic mixed models. Am. J. Hum. Genet. 2016;98:653–666. doi: 10.1016/j.ajhg.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen H, et al. Efficient variant set mixed model association tests for continuous and binary traits in large-scale whole-genome sequencing studies. Am. J. Hum. Genet. 2019;104:260–274. doi: 10.1016/j.ajhg.2018.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.The FANTOM Consortium and the RIKEN PMI and CLST (DGT A promoter-level mammalian expression atlas. Nature. 2014;507:462–470. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.The FANTOM Consortium. et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fishilevich, S. et al. GeneHancer: genome-wide integration of enhancers and target genes in GeneCards. Database J. Biol. Databases Curation2017, (2017). [DOI] [PMC free article] [PubMed]
- 90.Wu MC, et al. Rare-variant association testing for sequencing data with the sequence kernel association test. Am. J. Hum. Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Madsen BE, Browning SR. A groupwise association test for rare mutations using a weighted sum statistic. PLoS Genet. 2009;5:e1000384. doi: 10.1371/journal.pgen.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liu Y, et al. ACAT: a fast and powerful p value combination method for rare-variant analysis in sequencing studies. Am. J. Hum. Genet. 2019;104:410–421. doi: 10.1016/j.ajhg.2019.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Buniello A, et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47:D1005–D1012. doi: 10.1093/nar/gky1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 2017;8:1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: Generalized Gene-Set Analysis of GWAS Data. PLoS Comput. Biol. 2015;11:e1004219. doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Szklarczyk D, et al. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.GTEx Consortium. Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Giambartolomei C, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10:e1004383. doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
Individual whole-genome sequence data for TOPMed and harmonized lipids at individual sample level are available through restricted access via the TOPMed dbGaP Exchange area. Summary level genotype data from TOPMed are available through the BRAVO browser (https://bravo.sph.umich.edu/). The UK Biobank (UKB) whole-genome sequence data can be accessed through UKB Research Analysis Platform (RAP), through the UKB approval system (https://www.ukbiobank.ac.uk). The Mass General Brigham Biobank (MGBB) individual-level data are available from https://personalizedmedicine.partners.org/Biobank/Default.aspx, where the data is available through institutional review board (IRB) approval, therefore not publicly available. Individual-level data from Penn Medicine BioBank is not publicly available due to research participants privacy concerns. The summary data captured using whole exome sequencing can be accessed through PMBB Genome Browser (https://pmbb.med.upenn.edu/allele-frequency/). The dbGaP accessions for TOPMed cohorts are as follows: Old Order Amish (Amish) phs000956 and phs00039; Atherosclerosis Risk in Communities study (ARIC) phs001211 and phs000280; Mt Sinai BioMe Biobank (BioMe) phs001644 and phs000925; Coronary Artery Risk Development in Young Adults (CARDIA) phs001612 and phs000285; Cleveland Family Study (CFS) phs000954 and phs000284; Cardiovascular Health Study (CHS) phs001368 and phs000287; Diabetes Heart Study (DHS) phs001412 and phs001012; Framingham Heart Study (FHS) phs000974 and phs000007; Genetic Studies of Atherosclerosis Risk (GeneSTAR) phs001218 and phs000375; Genetic Epidemiology Network of Arteriopathy (GENOA) phs001345 and phs001238; Genetic Epidemiology Network of Salt Sensitivity (GenSalt) phs001217 and phs000784; Genetics of Lipid-Lowering Drugs and Diet Network (GOLDN) phs001359 and phs000741; Hispanic Community Health Study - Study of Latinos (HCHS_SOL) phs001395 and phs000810; Hypertension Genetic Epidemiology Network and Genetic Epidemiology Network of Arteriopathy (HyperGEN) phs001293 and phs001293; Jackson Heart Study (JHS) phs000964 and phs000286; Multi-Ethnic Study of Atherosclerosis (MESA) phs001416 and phs000209; Massachusetts General Hospital Atrial Fibrillation Study (MGH_AF) phs001062 and phs001001; San Antonio Family Study (SAFS) phs001215 and phs000462; Samoan Adiposity Study (SAS) phs000972 and phs000914; Taiwan Study of Hypertension using Rare Variants (THRV) phs001387 and phs001387; Women’s Health Initiative (WHI) phs001237 and phs000200.
Codes used to implement STAAR workflows are available at https://github.com/xihaoli/STAAR and https://github.com/xihaoli/STAARpipeline. Workflow implemented for whole genome heritability calculations are available at https://github.com/CNSGenomics/Heritability_WGS.