Abstract
The molecular heterogeneity of feline mammary carcinomas (FMCs) represents a prognostic and therapeutic challenge. RNA-Seq-based comparative transcriptomic profiling serves to identify recurrent and exclusive differentially expressed genes (DEGs) across sample types and molecular subtypes. Using mass-parallel RNA-Seq, we identified DEGs and performed comparative function-based analysis across 15 tumours (four basal-like triple-negative [TN], eight normal-like TN, and three luminal B fHER2 negative [LB fHER2−]), two cell lines (CL, TiHo-0906, and TiHo-1403) isolated from the primary tumours (LB fHER2−) of two cats included in this study, and 13 healthy mammary tissue controls. DEGs in tumours were predominantly upregulated; dysregulation of CLs transcriptome was more extensive, including mostly downregulated genes. Cell-cycle and metabolic-related DEGs were upregulated in both tumours and CLs, including therapeutically-targetable cell cycle regulators (e.g. CCNB1, CCNB2, CDK1, CDK4, GTSE1, MCM4, and MCM5), metabolic-related genes (e.g. FADS2 and SLC16A3), heat-shock proteins (e.g. HSPH1, HSP90B1, and HSPA5), genes controlling centrosome disjunction (e.g. RACGAP1 and NEK2), and collagen molecules (e.g. COL2A1). DEGs specifically upregulated in basal-like TN tumours were involved in antigen processing and presentation, in normal-like TN tumours encoded G protein-coupled receptors (GPCRs), and in LB fHER2− tumours were associated with lysosomes, phagosomes, and endosomes formation. Downregulated DEGs in CLs were associated with structural and signalling cell surface components. Hence, our results suggest that upregulation of genes enhancing proliferation and metabolism is a common feature among FMCs and derived CLs. In contrast, the dissimilarities observed in dysregulation of membrane components highlight CLs’ disconnection with the tumour microenvironment. Furthermore, recurrent and exclusive DEGs associated with dysregulated pathways might be useful for the development of prognostically and therapeutically-relevant targeted panels.
Subject terms: Cancer, Molecular biology, Biomarkers, Oncology
Introduction
Mammary carcinomas in cats are predominantly triple-negative (Oestrogen Receptor negative [ER-], Progesterone Receptor negative [PR-], and feline orthologous of the human epidermal growth factor receptor 2 negative [fHER2−]) resembling aggressive human breast cancer (HBC) subtypes1–5. As observed in human breast oncology6–9, feline mammary carcinomas (FMCs) represent a heterogeneous group of diseases with distinctive molecular traits influencing therapeutic response and survival intervals2,5,10. Accordingly, a detailed characterisation of FMCs molecular subtypes and derived in vitro model’s gene dysregulation is pivotal for the development and testing of molecularly-targeted therapies11.
The heterogeneity, biological behaviour, and prognostic differences among FMCs have been realised through histopathology, immunohistochemical profiling and more recently by the emergence of in-depth nucleic acid analyses2,5,10,12–15. Molecular subtypes described include luminal A (LA), luminal B fHER2 negative (LB fHER2−), LB fHER2 positive (LB fHER2+), fHER2 positive [fHER2+], basal-like triple-negative (TN), and normal-like TN. Moreover, FMCs and HBCs molecular subtypes are intrinsically heterogeneous6–9,16–18. Thus, characterising gene dysregulation through deeper molecular analyses is imperative to identify common and specific molecular markers and therapeutic targets across molecular subtypes.
Immortalised CLs are useful to study specific features of the neoplastic process and to test novel treatment modalities under controlled conditions. Despite establishment and characterisation of FMC-derived CLs19–26, their actual potential to model distinctive aspects of the neoplastic transformation and suitability for the testing of specific therapeutic alternatives is yet to be elucidated.
High-resolution transcriptome sequencing serves to detect up-, or downregulated DEGs in neoplastic lesions and derived CLs in comparison to healthy tissues. Furthermore, investigating DEGs using available online resources for function-based analysis (e.g. Gene Ontology and DAVID functional analysis) serves to provide biological insight into molecular mechanisms behind cancer, as DEGs are categorised into gene sets based on cellular location, shared biological function, and/or involvement in molecular pathways. Comparing results of function-based DEGs analysis across different molecular subtypes has the potential of revealing specific features characterising the neoplastic dysregulation and possible prognostic and therapeutic implications, on the other hand, a comparison across CLs and tumour samples, helps to reveal changes associated with cell seeding and subculturing favouring clonal selection.
Thus, this study aimed to identify recurrent and exclusive DEGs across 15 tumours representing three immunohistochemically defined molecular subtypes (basal-like TN, normal-like TN, and LB fHER2−) and two CLs derived from LB fHER2− tumours in comparison with healthy mammary tissues including control specimens derived from the same individuals. To disclose possible similarities and differences among primary lesions and established cellular models, we analysed recurrent and exclusive DEG sets for enrichment in specific cellular components, biological processes, molecular functions, and signalling pathways.
Results
Animals and samples characterisation
We generated and analysed RNA-seq libraries of 15 selected primary tumours (eight formalin-fixed paraffin-embedded [FFPE] and seven fresh-frozen tissue [FT] samples), three samples representing two cell lines (CLs) at different passages (TiHo-1403 passage 10, and TiHo-090626 passage 7 and 77), and 13 healthy mammary tissue controls (FFPE = 12 and FT = 1). Seven of the primary tumour samples had a healthy mammary tissue control counterpart obtained from the same animal; CLs were isolated from the primary tumours of two animals (Fig. S1). Characteristics of patients and tumours included in this study are summarised in Table S1. After immunohistochemical evaluation of ER, PR, fHER2, CK5/6, and Ki-67 and validation of fHER2 amplification using mass-parallel sequencing10, tumours were classified following the St. Gallen guidelines as basal-like TN (n = 4, 26.67%), normal-like TN (n = 8, 53.33%), and LB fHER2− (n = 3, 20%). CL samples were derived from LB fHER2− tumours. TiHo-1403 displayed 26 h doubling time at passage 10 and was able to proliferate until passage 33. TiHo-0906 cellular morphology and proliferation rate during prolonged subculturing have been previously documented26.
Animals included were followed up for two years. After censoring and survival analysis, cats with basal-like TN tumours displayed the poorest survival (n = 4; disease-free interval [DFI]: 3.35 ± 1.71 months, and cancer-specific overall survival [CSS]: 3.73 ± 1.67 months), followed by normal-like TN (n = 7; DFI: 7.22 ± 4.87 months, and CSS: 9.34 ± 4.28 months), and LB fHER2− (n = 2; DFI: 12 ± 5.9, and CSS: 14.3 ± 4.2 months), as previously reported2,5,10. Additional outcome information of included cases is provided in Table S2.
The transcriptome of CLs is more dysregulated and includes more downregulated genes compared to tumours
Importantly, gene expression profiles were more similar among samples of the same type (i.e., CL, and FT and FFPE tumours) than to molecular subtype (i.e., LA, LB fHER2−, normal-like and basal-like TN, Fig. S1). Based on their expression profiles, CLs and FT samples formed a cluster that was clearly separated from the cluster formed by FFPE samples. Both clusters included a mixture of controls and neoplastic samples. Paired samples (control/neoplastic and cell line/original tumour) were separated into different subclusters.
Independent differential gene expression analysis between each neoplastic sample group (i.e., CLs, LB fHER2− tumours, normal-like TN tumours, and basal-like TN tumours) and control samples identified a total of 7,454 DEGs out of 18,504 detected protein-coding genes (adjusted P value < 0.05 and log2 fold-changes ≥ 2 or ≤ −2; Methods). Among all sample groups studied, CLs displayed the highest number of DEGs (n = 6210), with 62% of them being exclusively differentially expressed in this sample group. Among primary neoplastic samples, normal-like TN tumours had the highest number of DEGs (n = 2406), followed by LB fHER2− (n = 1310), and basal-like TN (n = 1279). DEGs exclusive to tumour samples represented 10.39–32.87% of DEGs, depending on the subtype (Fig. 1a). While most DEGs in tumours were upregulated, DEGs in CLs were predominantly downregulated (Fig. 1a). Furthermore, CLs featured the highest number of extremely dysregulated genes (log2 fold-changes ≥ 5 and ≤ −5), representing 12.25% of DEGs in this sample group. Indeed, most DEGs (97.25–99.42%, depending on the subtype) in tumours were only moderately dysregulated (log2 fold-changes ≥ 2 but ≤ 5, and ≤ -2 but ≥ −5). Only 349 DEGs (~ 5% of all DEGs, Fig. 1b) were recurrently dysregulated across all sample groups. Specifically, 203 DEGs were consistently upregulated and 137 were downregulated across all sample groups (Table S3), while the additional nine genes were dysregulated in different directions depending on the sample group.
Figure 1.
Differentially expressed genes (DEGs) relative to control samples for the four sample groups considered in this study. (a) Number of up- and down-regulated DEGs. (b) UpSet plot27 showing disjoint intersections of DEGs in different comparisons. LB fHER2-, luminal B fHER2 negative tumours; and TN triple-negative tumours. A total of 349 DEGs were recurrently dysregulated across all four sample groups.
For each sample group, we then identified ten “top DEGs” (Table S4; Methods). RACGAP1 was among the top DEGs in all sample groups. NEK2 was among the top DEGs in triple-negative subtypes and was upregulated across all samples. Furthermore, top upregulated genes across different tumour subtypes and CLs included several proliferation-related and metabolic-related therapeutic targets in cancer. On the other hand, top downregulated genes included ATP-binding cassette (ABC) transporters, chemotherapy resistance-associated genes, metabolic-related genes, cell adhesion molecules, and membrane receptors (Table S4).
Recurrently upregulated genes across all sample groups were associated with cell cycle and metabolic-related pathways
DEGs that were recurrently upregulated across all sample groups (n = 203) were associated with pathways related to cell cycle and metabolic networks (Table S5 Methods). Consistently, similar pathways were also enriched among the DEGs detected in each sample group (Tables S6–9). Besides the upregulation of cell cycle and metabolic-related pathways observed for all sample groups, several other pathways supporting replication, transcription, translation, and protein processing were specifically associated with the DEGs upregulated in CLs (Table S9).
In agreement with the pathway enrichment analysis, gene ontology (GO) analysis revealed that DEGs that were recurrently upregulated across all sample groups represented protein complexes relevant during DNA replication and cellular division (e.g., DNA highly conserved mini-chromosome maintenance [MCM], and Ndc80 complexes), or were involved in movement elicitation (e.g. kinesin complexes) and cellular parts pivotal during cytokinesis (e.g. kinetochore, microtubule cytoskeleton, mitotic spindle, and midbody). Accordingly, GO terms representing molecular functions and biological processes were mostly related to cell cycle, motor activity, and cytoskeleton reorganisation during mitosis (Fig. S2).
Upregulated genes through all tumour subtypes and cell lines were associated with three cell cycle-related pathways
DEGs that were recurrently upregulated across all sample groups (n = 137) were associated with three interconnected pathways governing key events during cell cycle progression and control: DNA replication, cell cycle, and p53 signalling (Table S5). These pathways were also enriched among the DEGs detected in each sample group (Tables S6–9). Recurrently upregulated DEGs across all sample groups associated with enhanced proliferation included core cell cycle regulators (e.g., CCNB1, CCNB2, CDK1, and CDK4) and genes promoting DNA synthesis, and/or repair (e.g., MCM4 and MCM5). In addition to these genes, the different sample groups featured their own specific cell cycle regulators. For example, the histone deacetylase gene HDAC1 was upregulated across all sample groups except for LB fHER2− tumours, and the histone deacetylase gene HDAC2 was only upregulated in CLs (Tables S6–9).
The concurrent up-regulation of TP53 and its downstream effectors related to proliferation suggests a loss of p53 tumour suppressor activity in tumours and derived cell lines
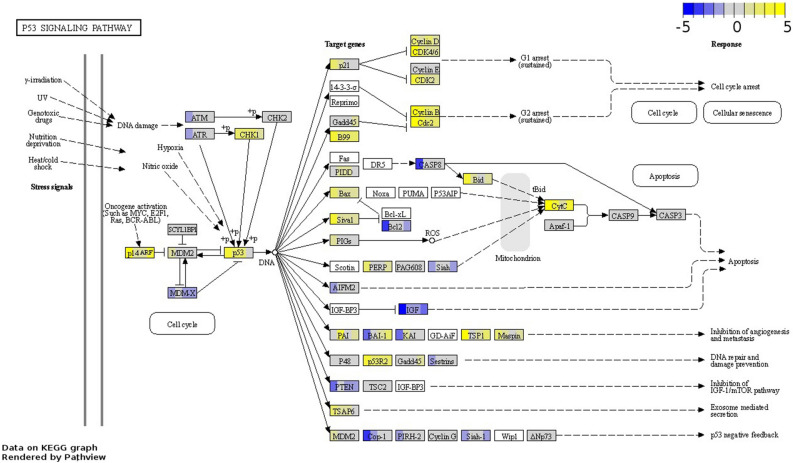
The p53 pathway is implicated in the cellular homeostatic mechanisms controlling cell proliferation in response to a variety of intrinsic and extrinsic stress signals and is commonly disrupted in cancer28–30. Interestingly, in all sample groups, TP53 and various p53 target genes were upregulated. Typically, p53 targets (e.g., CDKN1A, GTSE1, GADD45A) interact with cyclins and cyclin-dependent kinases involved in cell cycle progression to inhibit their expression31. But surprisingly, the cyclins and cyclin-dependent kinases CCNB1, CCNB2, CDK1, and CDK4 were upregulated in all sample groups (Table S3). Moreover, also the p53 activation mediator CHEK1, the p53 downstream effector RRM2—involved in DNA repair—, and GTSE1—which represses p53’s ability to induce apoptosis (Table S5 and Fig. 2)32–35—were upregulated in all sample groups. On the other hand, kinases favouring p53 activation in response to stress cues, such as ATM and ATR —which signals are integrated by CHEK1— were only downregulated in CLs. Overall, a stronger dysregulation of the p53 pathway was observed in CLs when compared to tumours (Tables S6–9). Notably, mechanisms underlying oncogene-mediated activation of p53 appear to be more active in CLs and basal-like tumours, as evidenced by the upregulation in both groups of CDKN2A—which indirectly activates p53 by repressing MDM236,37. However, the main negative feedback of TP53 (MDM2) was upregulated in CLs, while MDM4 which inhibits p53 by binding its transcriptional activation domain29, was downregulated in this group. Finally, despite upregulation of TP53 and p53 target genes, downstream related proliferation signals remained active across all sample groups.
Figure 2.
Differentially expressed genes associated with the p53 signalling pathway (fca04115) across all sample groups. The boxes illustrating DEGs are uniformly divided by the number of sample groups; representing from left to right: CLs, LB fHER2− tumours, normal-like TN tumours, and basal-like TN tumours. Gene log2 fold-change relative to controls is indicated in yellow (upregulated) or blue (downregulated), white boxes represent genes not detected. Data on KEGG graph rendered by “Pathview” (https://pathview.uncc.edu/).
PI3K/Akt/mTOR pathway activation was downregulated in CLs while AKT-related cell proliferation and survival remained active across all sample groups except for basal-like tumours
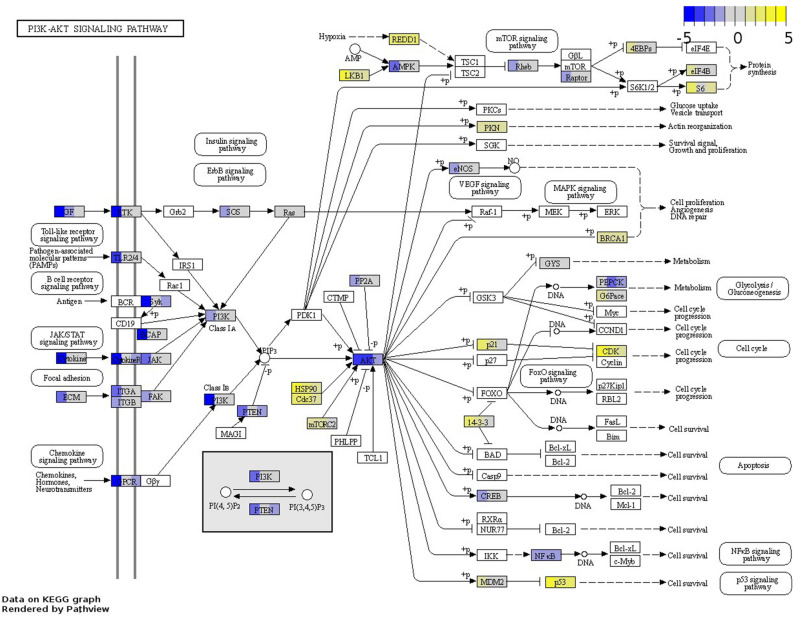
The PI3K/Akt/mTOR pathway is implicated in the regulation of cell growth, proliferation, metabolism, and motility38, and several of its components are commonly activated in cancer39. And indeed, the DEGs in each of the sample groups except for basal-like tumours were significantly associated with this pathway. Specifically, CLs exhibited downregulation of many membrane receptors and ECM molecules participating in the activation of the PI3K/Akt/mTOR pathway, while their expression in tumours varied (Tables S6–9). Furthermore, PI3Ks—whose phosphorylation activates AKT—were downregulated in CLs, but not dysregulated in tumours. Heat-shock proteins (HSPs) —also phosphorylating AKT—were upregulated in CLs and normal-like TN tumours; some of them—HSP90AA1 and HSP90B1—were also upregulated in basal-like TN tumours, and one—HSP90B1— was upregulated across all sample groups. Finally, AKT1 was upregulated in CLs, while AKT3 was downregulated across all sample groups except for basal-like TN tumours, while AKT antagonising factors such as PTEN were only downregulated in CLs and normal-like TN tumours. Interestingly, despite differences in pathway activation between tumours and CLs, AKT downstream effectors related to cell cycle, cell survival regulation, and p53 signalling activation were predominantly upregulated across all sample groups but basal-like tumours (Fig. 3). In contrast, members of the mTOR signalling pathway exhibited no dominant dysregulation pattern except DDIT4, which was upregulated across all sample groups (Table S6–9).
Figure 3.
Differentially expressed genes enriching the PI3K-AKT signalling pathway (fca04151) in CLs, LB fHER2-, and normal-like TN tumours. The boxes illustrating DEGs are uniformly divided by the number of sample groups; representing from left to right: CLs, LB fHER2- tumours, and Normal-like TN tumours. Gene log2 fold change relative to controls is indicated in yellow (upregulated) or blue (downregulated), white boxes represent genes not detected. Data on KEGG graph rendered by “Pathview” (https://pathview.uncc.edu/).
Upregulated DEGs in basal-like TN tumours were associated with the antigen processing and presentation pathway
Antigen processing and presentation is pivotal for immune evasion during carcinogenesis and its modulation might be promising for the development of targeted immunotherapies40. Upregulated genes basal-like TN tumours were associated with the antigen processing and presentation pathway (fca04612) including genes participating in the MHC I and MHC II pathways—mediating the presentation of endogen cytoplasmic and exogenous (intravesicular) antigens to T and/or NK cells (Fig. 4, and Table S6). Upregulated genes included CALR (calreticulin), and several HSPs implicated in tumour immune evasion and critical for maintaining cellular protein homeostasis41–43.
Figure 4.
Upregulated genes enriching the Antigen Processing and Presentation pathway (fca04612) in basal-like triple-negative tumours. Gene log2 fold change relative to controls is indicated in yellow (upregulated) or blue (downregulated), white boxes represent genes not detected. Data on KEGG graph rendered by “Pathview” (https://pathview.uncc.edu/).
Upregulated GPCRs associated with the neuroactive ligand-receptor interaction pathway are promising oncotarget candidates in normal-like TN tumours
DEGs in normal-like TN tumours were associated with the neuroactive ligand-receptor interaction pathway. Upregulated DEGs associated with this pathway included several neurotransmitters and neuropeptides receptors (Table S7) triggering cell growth and metastasis through downstream signalling in cancer44. Among them, neuropeptide G protein-coupled receptors (GPCRs) —which represent the first gate through which outside signals are transmitted into the cell—have been postulated as possible therapeutic targets44,45. Upregulated GPCRs that have been proposed as therapeutic targets in cancer included the adenosine receptors ADORA1 and ADORA3, and important GPCRs for autocrine growth factors in cancer, such as bombesin receptor NMBR, and neurotensin receptor NTSR145,46. Additional GPCRs reported to promote proliferation and migration of cancer cells included secretin receptor SCTR, and lysophosphatidic acid (LPA) receptor LPAR247,48. Finally, several upregulated hormone receptors were associated with the neuroactive receptor interaction pathway, including those for melanin-concentrating hormone, thyroid-stimulating hormone, calcitonin, corticotrophin-releasing hormone, follicle-stimulating hormone, luteinizing hormone beta, and glucagon-like peptide (Fig. 5). In contrast to normal-like TN tumours, neurotransmitters receptors were predominantly downregulated in CLs and not dysregulated in other tumour subtypes.
Figure 5.
DEGs enriching the Neuroactive Ligand-Receptor Interaction pathway (fca04080) in normal-like triple-negative tumours. Gene log2 fold change relative to controls is indicated in yellow (upregulated) or blue (downregulated), white boxes represent genes not detected. Data on KEGG graph rendered by “Pathview” (https://pathview.uncc.edu/).
Components of lysosomes, phagosomes and exosomes might be promising targets in LB fHER2− tumours
Lysosomes and phagosomes increase energy supply through the recycling of intracellular substances and autophagy and have been reported to enhance cell communication, cancer progression, and metastasis through exosome formation49–53. An up-regulation of specific exosome components and the lysosome and phagosome pathways were exclusively detected in LB fHER2− tumours (Fig. 6 and Table S8). Accordingly, up-regulated DEGs in this group were annotated with GO terms representing extracellular exosomes, lysosomes, vacuoles, and microtubule cytoskeleton (Fig. S3 and Table S8).
Figure 6.
Upregulated genes enriching the (a) Lysosome (fca04142), and (b) Phagosome (fca04145) pathways in LB fHER2- tumours. Gene log2 fold change relative to controls is indicated in yellow (upregulated) or blue (downregulated), white boxes represent genes not detected. Data on KEGG graph rendered by “Pathview” (https://pathview.uncc.edu/).
Upregulated metabolic genes in LB fHER2− tumours and derived CLs were associated with the glycolysis/gluconeogenesis pathway
Notably, genes associated with the glycolysis/gluconeogenesis pathway including LDHA —whose overexpression in cancer is associated with proliferation and malignancy54–56— and several glycolytic genes (e.g. HK1, VDAC1, and VDAC2) were only upregulated in LB fHER2− tumours and derived CLs (Tables S8 and 9). However, members of this pathway that are possible therapeutic targets such as SIX1 and some of its targets (e.g., ENO1, PFKL, and PGK1)57 were upregulated in all sample types were also upregulated across all sample types. Moreover, several members of the solute carrier family-16—also known as monocarboxylate transporter (MCT) family— catalysing the transport of lactic acid and pyruvate across the plasma membranes were upregulated across different sample groups (Table S6–9).
Dysregulation of cell lines transcriptome highlights loss of interaction with the tumour microenvironment
We observed differences in the regulation of pathways related to reception and processing of environmental cues between tumours and CLs. For instance, DEGs associated with the ECM-receptor interaction pathway (also involved in PI3K/Akt/mTOR activation) were predominantly upregulated (~ 80%) in LB fHER2− tumours, and represented a combination of approximately half up- and downregulated DEGs in TN tumours (Tables S6–8). In contrast, DEGs in this pathway were predominantly downregulated (~ 70%) in CLs. (Table S9). Notoriously, recurrently downregulated DEGs in this pathway represented membrane-, and/or extracellular matrix-associated components shared between the ECM-receptor interaction and the focal adhesion pathways. Additional cell surface components (e.g. laminins and collagens) were also predominantly downregulated in CLs. Similarly, laminins were predominantly downregulated across tumour sample groups among which LAMA2 and LAMA3 were downregulated across all sample groups. On the other hand, collagen molecules were up or downregulated across different tumour subtypes and only COL2A1 was upregulated across all sample groups (Table S10). Additional pathways behind cell adhesion and actin cytoskeleton remodelling were also associated with downregulated DEGs in CLs (e.g. Gap junction, Rap1 signalling, and cell adhesion molecules [CAMs]), as well as, several immune system and endocrine system-related pathways (Table S9). To sum up, cell surface components mediating direct cellular interaction and the transduction of multiple environmental signals including nervous and sensory systems, as well as, endocrine- and immune system-related pathways were significantly associated with downregulated DEGs in CLs highlighting the lack of interaction with structural and non-structural components of the tumour microenvironment (Table S9).
Discussion
In this study, we comparatively analysed the transcriptome of primary neoplastic samples representing three molecular subtypes of FMCs characterised by poor survival (basal-like TN, DFI: 3.35 ± 1.71 months, CSS: 3.73 ± 1.67 months], normal-like TN [DFI: 7.22 ± 4.87 months, and CSS: 9.34 ± 4.28 months], and LB fHER2− [DFI: 12 ± 5.9, and CSS: 14.3 ± 4.2 months]) aiming to identify common and exclusive therapeutic targets, prognostic markers and clinically relevant gene expression changes eliciting cancer progression. Furthermore, we analysed two LB fHER2− derived CLs to uncover similarities and differences that might impact their ability to model distinct aspects of the tumour dysregulation. Limitations of this study include the small sample size, the lack of representation of molecular subtypes with better prognosis, and the poor quality of the RNA isolated from FFPE samples, reflected by the higher similarity observed between FT and CL samples. Furthermore, only one FT sample was included in the control group. Additionally, tumour tissues include a certain amount of normal cells, and usually display clonal heterogeneity, which in combination with patient-specific effects might impact the number of detectable dysregulated genes. Therefore, as RNA was indistinctively isolated from all cellular subpopulations included in the tumour sample, a contamination with healthy mammary tissue is possible.
Top up- and downregulated DEGs were similarly dysregulated across all sample groups. However, only RACGAP1 was among the top upregulated DEGs across all sample groups, and NEK2 across TN subtypes. RACGAP1 and NEK2 have a pivotal role during cytokinesis and are known prognostic markers in HBC: high expression levels of these genes are correlated with high proliferation and poor prognosis58–61. Importantly, metabolic-related genes including proposed therapeutic targets in different human cancers42,62–66 were among the top upregulated DEGs (e.g. FADS2, HSPH1, VLDLR, TF, and ASNS) across different sample groups, emphasizing the importance of new therapeutic strategies targeting metabolic reprogramming. In line with the picture emerging from the top upregulated DEGs, recurrently upregulated genes across all sample groups were also associated with cell cycle- and metabolic-related pathways. Among them, we identified prognostic and therapeutic targets (e.g. CCNB1, CCNB2, CDK1, CDK4, GTSE1, MCM4, and MCM5) implicated in DNA replication, cell cycle, and signalling pathways controlling proliferation (e.g. p53, and PI3K-AKT).
The main recurrent dysregulated pathways in this study were the p53 and PI3K-AKT, whose interactions play a significant role in the determination of cell death/survival and have been previously reported as important in FMCs67–72. The observed concurrent up-regulation of TP53 and downstream effectors CCNB1, CCNB2, CDK1, and CDK4—normally inhibited by genes whose expression is promoted by p53—suggests as previously documented on FMCs the presence of TP53 mutations or post-transcriptional modifications resulting in the loss of p53 tumour suppressor activity70–72. Furthermore, in tumours retaining wild-type TP53, p53 function might still be impaired, with MDM2 and MDM4 being the most important regulators of its activity29. However, in this study MDM2 and MDM4 were only dysregulated in CL samples. Accordingly, future research should focus on characterising mutations, post-transcriptional modifications, and possibly targeting the mutant p53 protein and/or restoring wild-type p53 activity30. The more extensive dysregulation of p53 signalling pathway observed in CLs might facilitate cell survival, replication, and adaptation to culture conditions, as gene dysregulation behind p53 signalling presumably mediates different responses aiming to protect neoplastic cells against different transformation-stress stimuli28. As in the p53 pathway, AKT downstream effectors related to proliferation and survival remained upregulated across all sample groups. Moreover, in contrast to previous reports in HBC73–75, PIK3CA and other PI3Ks activating AKT as well as AKT1 and AKT3 were downregulated or not differentially expressed in tumours, suggesting that subtypes evaluated here are not likely to respond to therapies targeting PI3Ks and AKT inhibitors.
Upregulated DEGs included several proposed prognostic markers and therapeutic targets in cancer-relevant for DNA replication, cell cycle- and gene expression regulation. For instance, MCM4 and MCM5 are essential for the initiation of genome replication76,77, and were recurrently upregulated across all sample groups. Additional members of the MCM complex (i.e. MCM2, MCM3, MCM6, and MCM7) were upregulated in all sample groups but basal-like TN tumours. These results are in line with previous studies on HBC in which independent or simultaneous upregulation/overexpression of MCM members was correlated with poor outcomes78–80, suggesting a role of MCM members as biomarkers of poor prognosis and possible therapeutic targets in FMCs. Other genes involved in regulating the cell cycle that were recurrently upregulated across all sample groups included GTSE1, whose upregulation has been associated with poor prognosis in several human malignancies33–35, and is believed to enhance metastasis in human TNBC81 and to regulate cancer progression by affecting p53 function32,34. GTSE1 knockdown reduces cell proliferation and increases sensitivity to radio- and chemotherapy in in vitro models by blocking the expression of LHDA82–85. Hence, it is a potential therapeutic target in FMCs. Furthermore, we identified upregulated DEGs in TN tumours including drug targets histone deacetylase enzymes (e.g. HDAC1, HDAC2, and HDAC11) implicated in the epigenetic regulation of gene expression86,87. Approved HDAC inhibitors (HDACis) for human cancer include vorinostat, romidepsin, panobinostat, and belinostat-like. Furthermore, several HDACis have been tested in vitro in FMCs88, and in vitro and in vivo different canine tumours, documenting antineoplastic activity, good tolerability, and clinical response in some cases89,90.
Considering the identification of recurrent DEGs across all sample groups associated with several metabolic pathways, FMCs might be susceptible to therapies targeting cancer metabolic rewiring91,92. Among possible metabolic targets, HSP90B1 was upregulated across all sample groups. HSP90B1 is reported to enhance apoptosis evasion and its overexpression is correlated with metastasis and poor prognosis in HBC93–95. Furthermore, we detected an upregulation in CLs and across different tumour subtypes of heat-shock proteins (e.g. HSP90AA1, HSP90AB1, HSP90B1, HSPA5, HSPA8, HSPD1, and HSPH1) associated with malignancy and implicated in the stabilization of important cancer-related proteins (e.g. KIT, MET, BRAF and AKT), the formation of exosomes, and the presentation of tumour antigens43,96–98. The upregulation of HSP90 family members is targetable with small molecule inhibitors (e.g., ganetespib, 17-AAG, and STA-1474). Moreover, STA-1474 has been tested in dogs with spontaneous tumours demonstrating tolerability, and biological activity against several neoplasms99,100. Besides HSP90 family members, molecules targeting HSPA5 (upregulated across all sample groups) master regulator of endoplasmic reticulum homeostasis have also been reported as a therapeutic strategy in human melanoma101,102. Finally, several monocarboxylate transporters (MCTs) implicated in the export of lactic acid and pyruvate across the plasma membrane were upregulated in different sample groups. Among them, the Solute Carrier Family 16 Member 3 SLC16A3 (MCT4) was upregulated across all sample groups. MCT4 supports tumour proliferation, and has been associated with poor prognosis and proposed as therapeutic target in different human cancers103–105.
Cell–cell and cell-matrix interactions eliciting cell proliferation and migration are determined by the tumour microenvironment characteristics, which are to great extent affected by dysregulation of ECM components106,107. In this study, ECM molecules in tumours included both up- and downregulated genes—some of them being exclusively dysregulated in specific molecular subtypes. Dysregulation of specific ECM components might enhance local invasion and metastasis. For instance, the collagen network reorganization might facilitate cell migration, which has been reported as a predictive factor of worst outcome in FMCs108. Among all collagen molecules, only collagen type II alpha 1 chain (COL2A1) was found to be upregulated across all sample groups. This in agreement with a previous report documenting a positive association between COL2A1 protein levels and canine mammary tumours development109. On the other hand, COL2A1 overexpression is associated with poor clinical response and cell culture resistance to combinatorial HER2 and PI3K inhibition in HBC110. Furthermore, COL2A1 mutations are associated with chondrosarcoma development in humans111–113, and it was recently reported as a biomarker of melanoma repopulating cells exerting cancer stem-like cell properties114. Considering the emerging roles of COL2A1 in cancer dysregulation, findings of this study support the evaluation of COL2A1 role on FMCs progression, as well as, possible prognostic and therapeutic implications.
To identify DEGs that might be relevant to the development of targeted panels or to the design of subtype-specific targeted therapies, we analysed the function of the DEGs in each tumour subtype. The studied subtypes displayed distinctive groups of DEGs that might contribute to molecular events characterising carcinogenesis. For instance, only LB fHER2− tumours exhibited upregulated genes in the lysosome and phagosome pathways. Hence, it is plausible that this subtype might be especially susceptible to blocking the formation of autophagolysosomes, as well as, other autophagy-targeted therapies53. Furthermore, LB fHER2− sample groups (tumours and CLs) displayed specific upregulation of several glycolytic genes including HK1, VDAC1, and VDAC2. Similarly, LDHA—which converts pyruvate to lactic acid—was only upregulated in LB fHER2− sample groups, highlighting a particular susceptibility to glycolytic inhibitors, and the potential of the CLs described here for testing this therapeutic strategy. On the other hand, normal-like TN tumours exhibited upregulated genes in the Neuroactive ligand-receptor interaction pathway, including major components of the superfamily of rhodopsin-like G protein-coupled receptors (GPCRs). GPCRs function as autocrine growth factors in cancer and have been suggested as suitable therapeutic targets45,47,48. Finally, basal-like TN tumours showed an exclusive upregulation of several HCM class I related heat-shock proteins (HSPs) and genes associated with the Antigen Processing and Presentation pathway, suggesting a more important role of immune surveillance evasion and specific sensibility to inhibitors of the proteasome, HSPAs, and cancer immunotherapy40,42,115.
To sum up, our results suggest that upregulation of genes related to cell cycle control and metabolic reprogramming remain similar after culture establishment and subculturing. In agreement with previous studies116–118, genes that would normally mediate cellular adhesion and prevent migration were predominantly downregulated in both tumours and CLs. However, downregulated genes in CLs included signalling molecules and membrane receptors such as protein kinases and phosphatases, their substrates, and various adapter proteins. In particular, the extensive downregulation of cell surface components and signalling pathways mediating cell–cell and cell–matrix cross-talks observed in CLs highlights cells disconnection with the cellular and non-cellular components of the tumour microenvironment and points out limitations of immortalised cells to model complex interactions between the tumour and the hosting organism during cancer development and progression. As previously described117,119,120, CLs evaluated in this study might fail to address the role of the immune, endocrine, and neurologic systems stimuli during cancer progression. The higher number of DEGs—particularly, downregulated genes— detected in CLs in comparison to tumours most likely evidences adaptation to culturing conditions, but may also partially reflect clonal selection and poor tissue sample quality.
The purpose of this research was to unravel biologically relevant gene expression differences and similarities across three molecular subtypes (basal-like TN, normal-like TN, and LB fHER2−) of feline mammary tumours, and among primary lesions and established cell lines. Pathways and DEGs characterising cell cycle control and metabolic reprogramming are recurrent across different subtypes and remain similar in derived cellular models. However, a higher number of downregulated genes specifically enriching cell components and pathways involved in cellular communication characterised the dysregulation of CLs and might impact in vitro modelling. Importantly, we identified recurrent and subtype-specific dysregulated genes susceptible to different targeted therapies including histone deacetylases, heat-shock proteins, metabolic-related genes, and genes controlling centrosome disjunction and cell cycle. Moreover, molecular subtype-specific DEG sets associated with lysosomes, phagosomes and exosomes formation in LB fHER2− tumours; GPCRs in normal-like TN tumours; and antigen processing and presentation in basal-like TN tumours highlight the importance of interaction with the tumour microenvironment during FMCs progression and provide an avenue for the development of molecular subtype targeted panels and possible subtype-tailored therapeutic approaches.
Methods
Animals and samples
Primary neoplastic samples (CL, FF, and FT) were collected from female cats diagnosed with FMCs, surgically treated and followed-up for at least two years at the Small Animal Clinic, University of Veterinary Medicine Hannover. Patients included in this study are part of a larger cohort in which mass-parallel genomic sequencing was used to characterise copy-number variations10. Moreover, copy-number variations affecting one of the CLs included in this study (TiHo-0906) were also characterised using mass-parallel genomic sequencing in a previous publication from our group26. Patients included in this study were followed up during a two-year postoperative period, animals without recurrence or alive at the end of the study were excluded from DFI and cancer-specific survival analysis, respectively. All samples included in this study were obtained from tissue specimens surgically retrieved during medically necessary treatment and diagnosis after owner’s written approval. Therefore, in accordance with the German Animal Welfare Act this study was not considered an animal experiment and ethical approval was not required.
Cell lines culture and RNA extraction
TiHo-1403 and TiHo-0906 were isolated from the primary tumours of two animals included in this study (Fig. S1) and were established and cultured as previously described26.Cultures with growth medium (medium 199 with 10% foetal bovine serum [FBS, Biochrom], and 2% penicillin–streptomycin) were incubated in a humidified atmosphere (5% CO2) at 37 °C. To obtain pellets for RNA isolation, cells were detached using a dissociating agent (TrypLETM, Gibco). After centrifugation (1000 g for 10 min) of cell suspensions, the supernatant was discarded to obtain cell pellets. Cell pellets were resuspended in 1 mL freezing medium (medium 199 with 20% FBS, 2% penicillin–streptomycin and 10% DMSO [AppliChem]) and transferred into vials for cryogenic storage. Vials were preserved in a − 80 °C freezer during 24 h, and afterward, stored in a − 150 °C freezer until RNA extraction. For nucleic acids extraction, samples containing 4 × 106 cells (TiHo-0906 p7 and p77, and TiHo-1403 p10) were homogenized using QIAshredderTM columns (QIAGEN). For all samples types (FFPE, FT and CL), RNA extraction was performed using the AllPrep DNA/RNA Mini Kit (QIAGEN) according to manufacturer’s instructions. RNA yields and purity (260/280 ratio) were measured with the Synergy 2 plate-reader (BioTek).
RNA-seq
Sequencing libraries were prepared using the NEBNext Ultra Library Prep Kit for Illumina. Sequencing was conducted on an Illumina NextSeq500, single-reads with 75 bp were generated. Reads were mapped to the feline genome (FelCat5) using BWA121.
Counts from sequencing libraries corresponding to different passages of the TiHo-0906 cell line were added together.
Differential expression analysis
Differential expression analysis was conducted with the DESeq2 R package (version 1.24)122. Only protein-coding genes were considered for the analysis. Samples were divided into five sample groups: normal-like TN primary neoplastic samples, LB fHER2− primary neoplastic samples, basal-like TN primary neoplastic samples, LB fHER2 negative-derived cell lines, and healthy mammary tissue samples. Expression was compared between each of the primary neoplastic or cell line sample groups and the healthy mammary tissue sample group. Genes that had an adjusted P-value (false discovery rate, FDR) smaller than or equal to 0.05 and exhibited a fold-change greater than or equal to 2 (or less than or equal to 0.5) were considered differentially expressed.
The “top differentially expressed genes” (“top DEGs”) were defined for each sample group as the set consisting of the 5 upregulated and 5 downregulated DEGs with the smallest FDR.
Expression values
Expression values were obtained by applying the regularized logarithm transformation implemented in the rlog() function of the DESeq2 R package122 to the raw read counts; no experimental design (“design = ~ 1”) was used for this purpose.
Functional and pathway analysis
Functional analysis of DEGs was performed with DAVID (Database for annotation, visualization and integrated discovery tool)123,124. Specifically, the following ontologies and databases were considered: GOTERM_MF_ALL, GOTERM_BP_ALL, GOTERM_CC_ALL, GOTERM_MF_DIRECT, GOTERM_MF_FAT, GOTERM_CC_FAT, and KEGG pathways. The DAVID default list of corresponding genome-wide genes for the species was used as background. Terms with a false discovery rate (FDR) equal to or less than 5% were considered significant.
PATHVIEW was used to visualise enriched KEGG pathways (https://pathview.uncc.edu)125,126. Furthermore, all analyses were performed for total DEGs (up- and downregulated genes together), and up-, and downregulated genes separately. REVIGO was used to summarize and visualise the enriched GO terms127.
Statistics
Descriptive statistics, and survival analysis were performed using the statistical software SPSS (IBM SPSS Statistics for Windows, Version 23.0. Armonk, NY, USA). DFI and CSS were calculated as months from surgery to tumour recurrence (local or distant), and to death, respectively. Cat deaths without recurrence were censored from DFI analysis, cats alive at the end of the study period (24 months) and animals that died due to non-tumour related causes were censored from cancer-specific OS analysis. All data is displayed as mean ± standard deviation unless indicated otherwise. For all statistical analyses, a p ≤ 0.05 was considered significant.
Supplementary Information
Acknowledgements
The authors wish to thank Prof. Dr. H.-H. Kreipe (Department of Pathology, Hannover Medical School) who kindly provided some of the positive controls used for immunohistochemistry. Some of the experiments reported here also feature in the doctoral thesis of José Luis Granados-Soler (https://d-nb.info/1191752739/34).
Author contributions
H.M.E., I.N., J.L.G.-S., and M.H.-T. performed the primary study design; L.T., I.N., H.M.E., J.J., V.N., M.H.-T., and F.F. performed the manuscript revision and final approval; K.B.-K., J.B., E.S., and B.B. performed the RNA sequencing; and L.T. performed the bioinformatics assessment of sequencing data. J.L.G.-S. retrieved the clinical data, performed the RNA isolation, analysed the data and wrote the manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Hugo Murua Escobar and Ingo Nolte.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-20874-5.
References
- 1.Adega F, Borges A, Chaves R. Cat mammary tumors: Genetic models for the human counterpart. Vet. Sci. 2016;3(3):17–17. doi: 10.3390/vetsci3030017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soares M, et al. Molecular based subtyping of feline mammary carcinomas and clinicopathological characterization. Breast. 2016;27:44–51. doi: 10.1016/j.breast.2016.02.016. [DOI] [PubMed] [Google Scholar]
- 3.Wiese DA, et al. Feline mammary basal-like adenocarcinomas: A potential model for human triple-negative breast cancer (TNBC) with basal-like subtype. BMC Cancer. 2013;13(1):403. doi: 10.1186/1471-2407-13-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caliari D, et al. Triple-negative vimentin-positive heterogeneous feline mammary carcinomas as a potential comparative model for breast cancer. BMC Vet. Res. 2014;10(1):1–12. doi: 10.1186/s12917-014-0185-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soares M, et al. St Gallen molecular subtypes in feline mammary carcinoma and paired metastases-disease progression and clinical implications from a 3-year follow-up study. Tumour Biol. 2016;37(3):4053–4064. doi: 10.1007/s13277-015-4251-z. [DOI] [PubMed] [Google Scholar]
- 6.Burstein MD, et al. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin. Cancer Res. 2015;21(7):1688–1698. doi: 10.1158/1078-0432.CCR-14-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prat A, Pascual T, Adamo B. Intrinsic molecular subtypes of HER2+ breast cancer. Oncotarget. 2017;8(43):73362–73363. doi: 10.18632/oncotarget.20629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cejalvo JM, et al. Intrinsic subtypes and gene expression profiles in primary and metastatic breast cancer. Cancer Res. 2017;77(9):2213–2221. doi: 10.1158/0008-5472.CAN-16-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poudel P, et al. Heterocellular gene signatures reveal luminal-A breast cancer heterogeneity and differential therapeutic responses. NPJ Breast Cancer. 2019;5:21. doi: 10.1038/s41523-019-0116-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Granados-Soler JL, et al. Analysis of copy-number variations and feline mammary carcinoma survival. Sci. Rep. 2020;10(1):1003. doi: 10.1038/s41598-020-57942-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Londhe P, Gutwillig M, London C. Targeted therapies in veterinary oncology. Vet. Clin. North Am. Small Anim. Pract. 2019;49(5):917–931. doi: 10.1016/j.cvsm.2019.04.005. [DOI] [PubMed] [Google Scholar]
- 12.Misdorp, W., et al.Histological classification of mammary tumors of the dog and cat. 2nd ed. 1999, Washington DC. 1–59.
- 13.Soares M, et al. Immunophenotyping of primary and metastatic lesions in feline mammary tumors - Are they equal? Microsc. Microanal. 2013;19(S4):19–20. doi: 10.1017/S1431927613000718. [DOI] [Google Scholar]
- 14.Brunetti B, et al. Molecular phenotype in mammary tumours of queens: Correlation between primary tumour and lymph node metastasis. J. Comp. Pathol. 2013;148(2–3):206–213. doi: 10.1016/j.jcpa.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 15.Govoni, V. M. et al. Genetic variants of BRCA1 and BRCA2 genes in cats with mammary gland carcinoma. Vet. Comp. Oncol.19(2), 404–408 (2021). [DOI] [PubMed]
- 16.Dagher E, et al. Bcl-2 expression and prognostic significance in feline invasive mammary carcinomas: A retrospective observational study. BMC Vet. Res. 2019;15:25. doi: 10.1186/s12917-018-1772-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dagher E, et al. Androgen receptor and FOXA1 coexpression define a "luminal-AR" subtype of feline mammary carcinomas, spontaneous models of breast cancer. BMC Cancer. 2019;19(1):1267. doi: 10.1186/s12885-019-6483-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Truchot Y, et al. Unfavorable prognostic effects of the stem cell pluripotency factor Sox2 in feline invasive mammary carcinomas. Front. Vet. Sci. 2020;7:622019. doi: 10.3389/fvets.2020.622019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Norval M, Maingay J, Else RW. Characteristics of a feline mammary carcinoma cell line. Res. Vet. Sci. 1985;39(2):157–164. doi: 10.1016/S0034-5288(18)31739-9. [DOI] [PubMed] [Google Scholar]
- 20.Minke JM, et al. Isolation of two distinct epithelial cell lines from a single feline mammary carcinoma with different tumorigenic potential in nude mice and expressing different levels of epidermal growth factor receptors. Can. Res. 1991;51(15):4028–4037. [PubMed] [Google Scholar]
- 21.Muleya JS, et al. Establishment and characterization of a new cell line derived from feline mammary tumor. The J. Vet. Med. Sci. / The Jpn. Soc. Vet. Sci. 1998;60(8):931–935. doi: 10.1292/jvms.60.931. [DOI] [PubMed] [Google Scholar]
- 22.Uyama R, et al. Establishment and characterization of eight feline mammary adenocarcinoma cell lines. J. Vet. Med. Sci. 2005;67(12):1273–1276. doi: 10.1292/jvms.67.1273. [DOI] [PubMed] [Google Scholar]
- 23.Barbieri F, et al. Isolation of stem-like cells from spontaneous feline mammary carcinomas: Phenotypic characterization and tumorigenic potential. Exp. Cell Res. 2012;318(7):847–860. doi: 10.1016/j.yexcr.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 24.Pang LY, et al. Feline mammary carcinoma stem cells are tumorigenic, radioresistant, chemoresistant and defective in activation of the ATM/p53 DNA damage pathway. Vet. J. 2013;196(3):414–423. doi: 10.1016/j.tvjl.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Borges A, Adega F, Chaves R. Establishment and characterization of a new feline mammary cancer cell line, FkMTp. Cytotechnology. 2016;68(4):1529–1543. doi: 10.1007/s10616-015-9912-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Granados-Soler JL, et al. TiHo-0906: A new feline mammary cancer cell line with molecular, morphological, and immunocytological characteristics of epithelial to mesenchymal transition. Sci. Rep. 2018;8(1):13231. doi: 10.1038/s41598-018-31682-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lex A, et al. UpSet: Visualization of intersecting sets. IEEE Trans. Vis. Comput. Graph. 2014;20(12):1983–1992. doi: 10.1109/TVCG.2014.2346248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Colman MS, Afshari CA, Barrett JC. Regulation of p53 stability and activity in response to genotoxic stress. Mutat. Res. 2000;462(2–3):179–188. doi: 10.1016/S1383-5742(00)00035-1. [DOI] [PubMed] [Google Scholar]
- 29.Jiang L, Zawacka-Pankau J. The p53/MDM2/MDMX-targeted therapies—a clinical synopsis. Cell Death Dis. 2020;11(4):237. doi: 10.1038/s41419-020-2445-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao D, et al. Molecularly targeted therapies for p53-mutant cancers. Cell. Mol. Life. Sci. 2017;74(22):4171–4187. doi: 10.1007/s00018-017-2575-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fischer M. Census and evaluation of p53 target genes. Oncogene. 2017;36(28):3943–3956. doi: 10.1038/onc.2016.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monte M, et al. The cell cycle-regulated protein human GTSE-1 controls DNA damage-induced apoptosis by affecting p53 function. J. Biol. Chem. 2003;278(32):30356–30364. doi: 10.1074/jbc.M302902200. [DOI] [PubMed] [Google Scholar]
- 33.Lai, W. et al. GTSE1 promotes prostate cancer cell proliferation via the SP1/FOXM1 signaling pathway. Lab Invest.101(5), 554–563 (2021). [DOI] [PubMed]
- 34.Lin F, et al. GTSE1 is involved in breast cancer progression in p53 mutation-dependent manner. J. Exp. Clin. Cancer Res. 2019;38(1):152. doi: 10.1186/s13046-019-1157-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu X, et al. GTSE1 promotes cell migration and invasion by regulating EMT in hepatocellular carcinoma and is associated with poor prognosis. Sci. Rep. 2017;7(1):5129. doi: 10.1038/s41598-017-05311-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Agrawal A, et al. Regulation of the p14ARF-Mdm2-p53 pathway: An overview in breast cancer. Exp. Mol. Pathol. 2006;81(2):115–122. doi: 10.1016/j.yexmp.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 37.Stott FJ, et al. The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 1998;17(17):5001–5014. doi: 10.1093/emboj/17.17.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang N, et al. Role of PI3K/AKT pathway in cancer: The framework of malignant behavior. Mol. Biol. Rep. 2020;47(6):4587–4629. doi: 10.1007/s11033-020-05435-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Porta C, Paglino C, Mosca A. Targeting PI3K/Akt/mTOR Signaling in Cancer. Front. Oncol. 2014;4:64. doi: 10.3389/fonc.2014.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mpakali A, Stratikos E. The role of antigen processing and presentation in cancer and the efficacy of immune checkpoint inhibitor immunotherapy. Cancers (Basel) 2021;13(1):134. doi: 10.3390/cancers13010134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu YC, Weng WC, Lee H. Functional roles of calreticulin in cancer biology. Biomed. Res. Int. 2015;2015:526524. doi: 10.1155/2015/526524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sojka DR, et al. Functional redundancy of HSPA1, HSPA2 and other HSPA proteins in non-small cell lung carcinoma (NSCLC); an implication for NSCLC treatment. Sci. Rep. 2019;9(1):14394. doi: 10.1038/s41598-019-50840-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murshid A, Gong J, Calderwood SK. The role of heat shock proteins in antigen cross presentation. Front. Immunol. 2012;3(63):63. doi: 10.3389/fimmu.2012.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pouya FD, Rasmi Y, Asl ER. Role of neurotransmitters and neuropeptides in breast cancer metastasis. Biochem. (Moscow), Suppl. Ser. A: Membr. Cell Biol. 2020;14(2):107–116. doi: 10.1134/S1990747820020142. [DOI] [Google Scholar]
- 45.Moody, T. W., Ramos-Alvarez, I. & Jensen, R. T. Neuropeptide G protein-coupled receptors as oncotargets. Front. Endocrinol.9, 345 (2018). [DOI] [PMC free article] [PubMed]
- 46.Gorain B, et al. Adenosine receptors as novel targets for the treatment of various cancers. Curr. Pharm. Des. 2019;25(26):2828–2841. doi: 10.2174/1381612825666190716102037. [DOI] [PubMed] [Google Scholar]
- 47.Kang S, et al. SCTR regulates cell cycle-related genes toward anti-proliferation in normal breast cells while having pro-proliferation activity in breast cancer cells. Int. J. Oncol. 2015;47(5):1923–1931. doi: 10.3892/ijo.2015.3164. [DOI] [PubMed] [Google Scholar]
- 48.Mills GB, Moolenaar WH. The emerging role of lysophosphatidic acid in cancer. Nat. Rev. Cancer. 2003;3(8):582–591. doi: 10.1038/nrc1143. [DOI] [PubMed] [Google Scholar]
- 49.Tang T, et al. The role of lysosomes in cancer development and progression. Cell Biosci. 2020;10(1):131. doi: 10.1186/s13578-020-00489-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kalluri R. The biology and function of exosomes in cancer. J. Clin. Invest. 2016;126(4):1208–1215. doi: 10.1172/JCI81135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu, J., Camfield, R. & Gorski, S. M. The interplay between exosomes and autophagy - partners in crime. J. Cell. Sci.131(15), jcs215210 (2018). [DOI] [PubMed]
- 52.Radisavljevic Z. Lysosome activates AKT inducing cancer and metastasis. J. Cell. Biochem. 2019;120(8):12123–12127. doi: 10.1002/jcb.28752. [DOI] [PubMed] [Google Scholar]
- 53.Udristioiu A, Nica-Badea D. Autophagy dysfunctions associated with cancer cells and their therapeutic implications. Biomed. Pharmacother. 2019;115:108892. doi: 10.1016/j.biopha.2019.108892. [DOI] [PubMed] [Google Scholar]
- 54.Zhu W, et al. The molecular mechanism and clinical significance of LDHA in HER2-mediated progression of gastric cancer. Am. J. Transl. Res. 2018;10(7):2055–2067. [PMC free article] [PubMed] [Google Scholar]
- 55.Hussien R, Brooks GA. Mitochondrial and plasma membrane lactate transporter and lactate dehydrogenase isoform expression in breast cancer cell lines. Physiol. Genomics. 2011;43(5):255–264. doi: 10.1152/physiolgenomics.00177.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koukourakis MI, et al. Lactate dehydrogenase 5 isoenzyme overexpression defines resistance of prostate cancer to radiotherapy. Br. J. Cancer. 2014;110(9):2217–2223. doi: 10.1038/bjc.2014.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li L, et al. Transcriptional regulation of the Warburg effect in cancer by SIX1. Cancer Cell. 2018;33(3):368–385. doi: 10.1016/j.ccell.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 58.Sahin S, et al. Clinicopathological significance of the proliferation markers Ki67, RacGAP1, and topoisomerase 2 alpha in breast cancer. Int. J. Surg. Pathol. 2016;24(7):607–613. doi: 10.1177/1066896916653211. [DOI] [PubMed] [Google Scholar]
- 59.Milde-Langosch K, et al. Validity of the proliferation markers Ki67, TOP2A, and RacGAP1 in molecular subgroups of breast cancer. Breast Cancer Res. Treat. 2013;137(1):57–67. doi: 10.1007/s10549-012-2296-x. [DOI] [PubMed] [Google Scholar]
- 60.Pliarchopoulou K, et al. Prognostic significance of RACGAP1 mRNA expression in high-risk early breast cancer: A study in primary tumors of breast cancer patients participating in a randomized Hellenic Cooperative Oncology Group trial. Cancer Chemother. Pharmacol. 2013;71(1):245–255. doi: 10.1007/s00280-012-2002-z. [DOI] [PubMed] [Google Scholar]
- 61.Fang Y, Zhang X. Targeting NEK2 as a promising therapeutic approach for cancer treatment. Cell Cycle. 2016;15(7):895–907. doi: 10.1080/15384101.2016.1152430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu Q, et al. Knockdown of asparagine synthetase (ASNS) suppresses cell proliferation and inhibits tumor growth in gastric cancer cells. Scand. J. Gastroenterol. 2016;51(10):1220–1226. doi: 10.1080/00365521.2016.1190399. [DOI] [PubMed] [Google Scholar]
- 63.Shen Y, et al. Transferrin receptor 1 in cancer: A new sight for cancer therapy. Am. J. Cancer Res. 2018;8(6):916–931. [PMC free article] [PubMed] [Google Scholar]
- 64.Young RSE, et al. Apocryphal FADS2 activity promotes fatty acid diversification in cancer. Cell Rep. 2021;34(6):108738. doi: 10.1016/j.celrep.2021.108738. [DOI] [PubMed] [Google Scholar]
- 65.He L, et al. Up-regulated expression of type II very low density lipoprotein receptor correlates with cancer metastasis and has a potential link to beta-catenin in different cancers. BMC Cancer. 2010;10:601. doi: 10.1186/1471-2407-10-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zappasodi R, et al. HSPH1 inhibition downregulates Bcl-6 and c-Myc and hampers the growth of human aggressive B-cell non-Hodgkin lymphoma. Blood. 2015;125(11):1768–1771. doi: 10.1182/blood-2014-07-590034. [DOI] [PubMed] [Google Scholar]
- 67.Maniscalco L, et al. Activation of AKT in feline mammary carcinoma: A new prognostic factor for feline mammary tumours. Vet. J. (London, England: 1997) 2012;191(1):65–71. doi: 10.1016/j.tvjl.2010.12.016. [DOI] [PubMed] [Google Scholar]
- 68.Maniscalco L, et al. Activation of mammalian target of rapamycin (mTOR) in triple negative feline mammary carcinomas. BMC Vet. Res. 2013;9(1):80. doi: 10.1186/1746-6148-9-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ressel L, et al. Reduced PTEN protein expression and its prognostic implications in canine and feline mammary tumors. Vet. Pathol. 2009;46(5):860–868. doi: 10.1354/vp.08-VP-0273-P-FL. [DOI] [PubMed] [Google Scholar]
- 70.Mayr B, et al. Presence of p53 mutations in feline neoplasms. Res. Vet. Sci. 2000;68(1):63–70. doi: 10.1053/rvsc.1999.0339. [DOI] [PubMed] [Google Scholar]
- 71.Mayr B, Reifinger M, Loupal G. Polymorphisms in feline tumour suppressor gene p53. Mutations in an osteosarcoma and a mammary carcinoma. The Vet. J. 1998;155:103–106. doi: 10.1016/S1090-0233(98)80044-0. [DOI] [PubMed] [Google Scholar]
- 72.Mayr B, et al. Sequence of an exon of tumour suppressor p53 gene–a comparative study in domestic animals: Mutation in a feline solid mammary carcinoma. Br. Vet. J. 1995;151(3):325–329. doi: 10.1016/S0007-1935(95)80183-9. [DOI] [PubMed] [Google Scholar]
- 73.Samuels Y, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304(5670):554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 74.Lee JW, et al. PIK3CA gene is frequently mutated in breast carcinomas and hepatocellular carcinomas. Oncogene. 2005;24(8):1477–1480. doi: 10.1038/sj.onc.1208304. [DOI] [PubMed] [Google Scholar]
- 75.Levine DA, et al. Frequent mutation of the PIK3CA gene in ovarian and breast cancers. Clin. Cancer Res. 2005;11(8):2875–2878. doi: 10.1158/1078-0432.CCR-04-2142. [DOI] [PubMed] [Google Scholar]
- 76.Maiorano D, Lutzmann M, Mechali M. MCM proteins and DNA replication. Curr. Opin. Cell Biol. 2006;18(2):130–136. doi: 10.1016/j.ceb.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 77.Lei M. The MCM complex: Its role in DNA replication and implications for cancer therapy. Curr. Cancer Drug Targets. 2005;5(5):365–380. doi: 10.2174/1568009054629654. [DOI] [PubMed] [Google Scholar]
- 78.Issac MSM, et al. MCM2, MCM4, and MCM6 in breast cancer: Clinical utility in diagnosis and prognosis. Neoplasia. 2019;21(10):1015–1035. doi: 10.1016/j.neo.2019.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kwok HF, et al. Prognostic significance of minichromosome maintenance proteins in breast cancer. Am. J. Cancer Res. 2015;5(1):52–71. [PMC free article] [PubMed] [Google Scholar]
- 80.Jurikova M, et al. Ki67, PCNA, and MCM proteins: Markers of proliferation in the diagnosis of breast cancer. Acta Histochem. 2016;118(5):544–552. doi: 10.1016/j.acthis.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 81.Stelitano D, et al. GTSE1: A novel TEAD4-E2F1 target gene involved in cell protrusions formation in triple-negative breast cancer cell models. Oncotarget. 2017;8(40):67422–67438. doi: 10.18632/oncotarget.18691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Subhash VV, et al. GTSE1 expression represses apoptotic signaling and confers cisplatin resistance in gastric cancer cells. BMC Cancer. 2015;15:550. doi: 10.1186/s12885-015-1550-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Guo L, et al. Silencing GTSE-1 expression inhibits proliferation and invasion of hepatocellular carcinoma cells. Cell Biol. Toxicol. 2016;32(4):263–274. doi: 10.1007/s10565-016-9327-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lei X, et al. Knockdown GTSE1 enhances radiosensitivity in non-small-cell lung cancer through DNA damage repair pathway. J. Cell Mol. Med. 2020;24(9):5162–5167. doi: 10.1111/jcmm.15165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen L, et al. Downregulation of GTSE1 leads to the inhibition of proliferation, migration, and Warburg effect in cervical cancer by blocking LHDA expression. J. Obstet. Gynaecol. Res. 2021;47(11):3913–3922. doi: 10.1111/jog.15000. [DOI] [PubMed] [Google Scholar]
- 86.Ropero S, Esteller M. The role of histone deacetylases (HDACs) in human cancer. Mol. Oncol. 2007;1(1):19–25. doi: 10.1016/j.molonc.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat. Rev. Cancer. 2004;4(2):143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 88.Almeida F, et al. Histone deacetylase inhibitors and microtubule inhibitors induce apoptosis in feline luminal mammary carcinoma cells. Animals. 2021;11(2):502. doi: 10.3390/ani11020502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cohen LA, et al. Treatment of canine haemangiosarcoma with suberoylanilide hydroxamic acid, a histone deacetylase inhibitor. Vet. Comp. Oncol. 2004;2(4):243–248. doi: 10.1111/j.1476-5810.2004.00057.x. [DOI] [PubMed] [Google Scholar]
- 90.Wittenburg LA, et al. The histone deacetylase inhibitor valproic acid sensitizes human and canine osteosarcoma to doxorubicin. Cancer Chemother. Pharmacol. 2011;67(1):83–92. doi: 10.1007/s00280-010-1287-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Harrelson JP, Lee MW. Expanding the view of breast cancer metabolism: Promising molecular targets and therapeutic opportunities. Pharmacol. Ther. 2016;167:60–73. doi: 10.1016/j.pharmthera.2016.07.014. [DOI] [PubMed] [Google Scholar]
- 92.Jiang B. Aerobic glycolysis and high level of lactate in cancer metabolism and microenvironment. Genes Dis. 2017;4(1):25–27. doi: 10.1016/j.gendis.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cawthorn TR, et al. Proteomic analyses reveal high expression of decorin and endoplasmin (HSP90B1) are associated with breast cancer metastasis and decreased survival. PLoS ONE. 2012;7(2):e30992. doi: 10.1371/journal.pone.0030992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liu S, et al. GRP94 overexpression as an indicator of unfavorable outcomes in breast cancer patients. Int. J. Clin. Exp. Pathol. 2018;11(6):3061–3067. [PMC free article] [PubMed] [Google Scholar]
- 95.Dejeans N, et al. Overexpression of GRP94 in breast cancer cells resistant to oxidative stress promotes high levels of cancer cell proliferation and migration: Implications for tumor recurrence. Free Radic. Biol. Med. 2012;52(6):993–1002. doi: 10.1016/j.freeradbiomed.2011.12.019. [DOI] [PubMed] [Google Scholar]
- 96.Choi HK, Lee K. Recent updates on the development of ganetespib as a Hsp90 inhibitor. Arch. Pharmacal. Res. 2012;35(11):1855–1859. doi: 10.1007/s12272-012-1101-z. [DOI] [PubMed] [Google Scholar]
- 97.Pacey S, et al. Hsp90 inhibitors in the clinic. Handb Exp. Pharmacol. 2006;172:331–358. doi: 10.1007/3-540-29717-0_14. [DOI] [PubMed] [Google Scholar]
- 98.Liu, H. et al. Plasma HSP90AA1 predicts the risk of breast cancer onset and distant metastasis. Front. Cell Dev. Biol.9, 639596 (2021). [DOI] [PMC free article] [PubMed]
- 99.London CA, et al. Consecutive day HSP90 inhibitor administration improves efficacy in murine models of KIT-driven malignancies and canine mast cell tumors. Clin. Cancer Res. 2018;24(24):6396–6407. doi: 10.1158/1078-0432.CCR-18-0703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.London CA, et al. Phase I evaluation of STA-1474, a prodrug of the novel HSP90 inhibitor ganetespib, in dogs with spontaneous cancer. PLoS ONE. 2011;6(11):e27018. doi: 10.1371/journal.pone.0027018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cerezo M, Rocchi S. New anti-cancer molecules targeting HSPA5/BIP to induce endoplasmic reticulum stress, autophagy and apoptosis. Autophagy. 2017;13(1):216–217. doi: 10.1080/15548627.2016.1246107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang J, et al. HSPA5 Gene encoding Hsp70 chaperone BiP in the endoplasmic reticulum. Gene. 2017;618:14–23. doi: 10.1016/j.gene.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Baek G, et al. MCT4 defines a glycolytic subtype of pancreatic cancer with poor prognosis and unique metabolic dependencies. Cell Rep. 2014;9(6):2233–2249. doi: 10.1016/j.celrep.2014.11.025. [DOI] [PubMed] [Google Scholar]
- 104.Martins SF, et al. Significance of glycolytic metabolism-related protein expression in colorectal cancer, lymph node and hepatic metastasis. BMC Cancer. 2016;16:535. doi: 10.1186/s12885-016-2566-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kuo TC, et al. Monocarboxylate transporter 4 is a therapeutic target in non-small cell lung cancer with aerobic glycolysis preference. Mol. Ther. Oncol. 2020;18:189–201. doi: 10.1016/j.omto.2020.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Brassart-Pasco S, et al. Tumor microenvironment: Extracellular matrix alterations influence tumor progression. Front. Oncol. 2020;10:397. doi: 10.3389/fonc.2020.00397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Henke E, Nandigama R, Ergun S. Extracellular matrix in the tumor microenvironment and its impact on cancer therapy. Front. Mol. Biosci. 2019;6:160. doi: 10.3389/fmolb.2019.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rosen S, et al. Intratumoral collagen signatures predict clinical outcomes in feline mammary carcinoma. PLoS ONE. 2020;15(8):e0236516. doi: 10.1371/journal.pone.0236516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wu CC, et al. Proteome analyses reveal positive association of COL2A1, MPO, TYMS, and IGFBP5 with canine mammary gland malignancy. Proteomics Clin. Appl. 2019;13(4):e1800151. doi: 10.1002/prca.201800151. [DOI] [PubMed] [Google Scholar]
- 110.Hanker AB, et al. Extracellular matrix/integrin signaling promotes resistance to combined inhibition of HER2 and PI3K in HER2(+) breast cancer. Cancer Res. 2017;77(12):3280–3292. doi: 10.1158/0008-5472.CAN-16-2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tarpey PS, et al. Frequent mutation of the major cartilage collagen gene COL2A1 in chondrosarcoma. Nat. Genet. 2013;45(8):923–926. doi: 10.1038/ng.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Totoki Y, et al. Unique mutation portraits and frequent COL2A1 gene alteration in chondrosarcoma. Genome Res. 2014;24(9):1411–1420. doi: 10.1101/gr.160598.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chow, W. A. Chondrosarcoma: Biology, genetics, and epigenetics. F1000Res. 7, F1000 Faculty Rev-1826 (2018). [DOI] [PMC free article] [PubMed]
- 114.Talluri, B. et al. COL2A1 is a novel biomarker of melanoma tumor repopulating cells. Biomedicines. 8(9), 360 (2020). [DOI] [PMC free article] [PubMed]
- 115.Seliger B, Kloor M, Ferrone S. HLA class II antigen-processing pathway in tumors: Molecular defects and clinical relevance. Oncoimmunology. 2017;6(2):e1171447. doi: 10.1080/2162402X.2016.1171447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ertel A, et al. Pathway-specific differences between tumor cell lines and normal and tumor tissue cells. Mol. Cancer. 2006;5:55. doi: 10.1186/1476-4598-5-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Katt ME, et al. In vitro tumor models: Advantages, disadvantages, variables, and selecting the right platform. Front. Bioeng. Biotechnol. 2016;4:12. doi: 10.3389/fbioe.2016.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Taher L, et al. Comparative high-resolution transcriptome sequencing of lymphoma cell lines and de novo lymphomas reveals cell-line-specific pathway dysregulation. Sci. Rep. 2018;8(1):6279. doi: 10.1038/s41598-018-23207-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Vincent KM, Postovit LM. Investigating the utility of human melanoma cell lines as tumour models. Oncotarget. 2017;8(6):10498–10509. doi: 10.18632/oncotarget.14443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yu K, et al. Comprehensive transcriptomic analysis of cell lines as models of primary tumors across 22 tumor types. Nat. Commun. 2019;10(1):3574. doi: 10.1038/s41467-019-11415-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.da Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucl. Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 125.Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019;28(11):1947–1951. doi: 10.1002/pro.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Luo W, Brouwer C. Pathview: An R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics. 2013;29(14):1830–1831. doi: 10.1093/bioinformatics/btt285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Supek F, et al. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE. 2011;6(7):e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.