Abstract
The type III secretion pathway is broadly distributed across many parasitic bacterial genera and serves as a mechanism for delivering effector proteins to eukaryotic cell surface and cytosolic targets. While the effectors, as well as the host responses elicited, differ among type III systems, they all utilize a conserved set of 9 to 11 proteins that together form a bacterial envelope-associated secretory organelle or needle complex. The general structure of the needle complex consists of a transenvelope base containing at least three ring-forming proteins (MxiD, MxiJ, and MxiG in Shigella) that is connected to a hollow needle-like extension that projects away from the cell surface. Several studies have shown that the initial steps in needle complex assembly require interactions among the base proteins, although specific details of this process remain unknown. Here we identify a role for another base element in Shigella, MxiM, in interactions with the major outer-membrane-associated ring-forming protein, MxiD. MxiM affects several features of MxiD, including its stability, envelope association, and assembly into homomultimeric structures. Interestingly, many of the effects were also elicited by the inner-membrane-associated base element, MxiJ. We confirmed that MxiM-MxiD and MxiJ-MxiD interactions occur in vivo in the cell envelope, and we present evidence that together these base elements can form a transmembrane structure which is likely an important intermediary in the process of needle complex assembly.
The gram-negative cell envelope presents a formidable hydrophobic barrier against protein secretion to the microbial cell surface or into the extracellular matrix. In gram-negative bacteria there are five conserved pathways or mechanisms (designated types I to V) that collectively mediate the processes required for recognition of secretion substrates at the cytoplasmic face of the inner membrane (IM) and active transport of the substrates across the outer membrane (OM) (reviewed in reference 29). Different sets of components, consisting of 1 to more than 40 elements, distinguish each pathway.
The type III secretion pathway is a largely virulence-specialized pathway that has demonstrated importance in parasitic interactions between a diverse set of bacterial pathogens and their mammalian or plant host targets (14, 24). Virulence protein secretion via the type III pathway is induced by close bacterium-host cell apposition and is specifically directed to the host cell membrane or cytosol. The components of each type III system, usually encoded in pathogenicity islands, are grouped into several functional classes, including (i) transcriptional regulatory proteins, which mediate type III gene expression in response to environmental cues (for example, growth at 37°C with Shigella); (ii) secreted substrates, consisting of translocators (pore-forming proteins delivered to host membranes) and effectors (delivered through translocator pores to intracytosolic targets); (iii) cytoplasmic chaperones, which influence secreted substrate synthesis at the level of mRNA translation or protein stability; and (iv) envelope-associated structural subunits, which consist of approximately 20 proteins that assemble into hollow transmembrane secretory channels or needle complexes. Conservation of the protein sequence that defines the type III pathway is largely restricted to structural subunits, suggesting that needle complex substructure and function are shared by different type III systems.
Type III needle complexes were recently purified from Salmonella and Shigella and were visualized at high resolution by transmission electron microscopy (6, 19). The supramolecular structure of each system consists of an envelope-spanning pair of stacked rings joined by a central rod (together called the base structure), which is connected to an axial needle-like extension that projects into the extracellular environment. When they looked at needle structures in membranes of osmotically shocked shigellae, Blocker et al. (6) also observed a bulb-like projection that extended from the base into the cytoplasm. The base elements probably anchor needle complexes in the envelope and provide the bulk of a transenvelope channel; the cytoplasmic bulb may consist of export proteins that recognize substrates and energize their translocation through the base and needle extension, toward eukaryotic cell surface and cytosolic targets. Three major constituents of the Shigella base structure have been defined previously: MxiD, MxiJ, and MxiG (5, 6, 28). MxiD is a member of a family of secretory proteins called secretins, which multimerize into stacked OM rings. A large periplasmic extension of MxiD may project through the peptidoglycan layer to the IM (22, 23). Salmonella PrgH and PrgK, homologs of MxiG and MxiJ, form stacked rings that correspond to IM sections of a base substructure (16). MxiG and MxiJ are integral IM proteins (1, 2) that likely form similar membrane complexes. MxiD, MxiG, and MxiJ all have cleavable N-terminal sec-dependent export signals and are processed and translocated into the envelope in the absence of other type III proteins. After export, the MxiG and MxiJ rings in the IM presumably interact with the periplasmic extension of MxiD to form the basic framework of a transmembrane structure. The export proteins of the cytoplasmic bulb and subunits of the needle extension may then nucleate within and around the envelope-spanning base and allow completion of the needle complex.
While MxiD, MxiG, and MxiJ are the only base elements identified thus far in the Shigella secreton, at least one additional protein may be required. Like MxiD, MxiG, and MxiJ, MxiM of Shigella has a cleavable sec-dependent signal sequence and is required for type III secretion (25). MxiM is anchored to the inner face of the OM via a lipid moiety, where it could interact with the OM ring structure formed by the MxiD secretin. MxiM may, in fact, represent a class of secretory proteins called pilots (9, 13, 17, 26), which are OM-linked lipoproteins that stabilize secretins during assembly processes and promote secretin insertion into the OM. In addition to this periplasmic chaperone-like function, pilots can also be structural elements attached to the periphery of secretin rings in the OM (22). Since pilots like InvH of Salmonella enterica serovar Typhimurium can be lost during purification and visualization of secretion structures (7, 9), a similar process may explain why MxiM has not been detected previously in Shigella needle complexes (5, 28). MxiM could, therefore, be an overlooked element of the Mxi-Spa system that is required for interactions among base proteins, like MxiD, which assemble transmembrane structures during Mxi-Spa synthesis.
In this study, we sought to identify MxiM interactions and functions with respect to the MxiD secretin. MxiM interacts with MxiD in the cell envelope, influencing both MxiD stability and multimerization. Our findings are consistent with MxiM being the MxiD pilot and a structural element. Thus, MxiM is an important base element. Interestingly, we also found that MxiD interacts with the IM protein MxiJ and that this interaction affects MxiD stability and multimerization in a manner similar to the manner in which MxiM affects MxiD stability and multimerization. We found that MxiM-MxiD-MxiJ interactions alone could form a complex linking the IM and the OM.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The following Shigella flexneri strains were used in this study: 2457T, a wild-type serotype 2a strain (11); BS103, a virulence plasmid-cured derivative of 2457T (21); BS612, a mxiD mutant (3); and BS547 (25), a mxiM mutant. Escherichia coli DH5α (Gibco BRL) was used for standard genetic manipulations. Strains were grown at 37°C either in Luria broth (LB) with aeration or on tryptic soy broth plates with 1.5% agar and 0.025% Congo red (Sigma), unless indicated otherwise. Antibiotics were used at the following concentrations: ampicillin, 100 μg ml−1; tetracycline, 15 μg ml−1; kanamycin, 50 μg ml−1; and chloramphenicol, 10 μg ml−1. To induce PBAD expression, growth media were supplemented with 0.2% arabinose.
Plasmid construction.
Standard protocols were used for DNA manipulation and for S. flexneri and E. coli transformations. PCR amplification procedures for cloning and plasmid screening were performed by using the Pfu (Stratagene) and Taq (Qiagen, Inc.) DNA polymerases, respectively. PCR fidelity was confirmed in several cases by DNA sequencing using an ABI Prism dye terminator cycle sequencing core kit and an ABI Prism 377 DNA sequencer.
The following vectors were used: pBluescript SK+ (Stratagene), a PLAC expression vector (ColE1 origin, Ampr); pBAD18 (12), a PBAD expression vector (ColE1 origin, Ampr); pBAD24 (12), a PBAD expression vector (ColE1 origin, Ampr); pBAD33 (12), a PBAD expression vector (p15A origin, Cmr); pWSK129 (30), a PLAC expression vector (pSC101 origin, Kmr); pDBLeu (Gibco BRL), a GAL4 DNA binding domain fusion vector (Kanr); and pPC86 (Gibco BRL), a GAL4 activation domain fusion vector (Ampr). All inserts were generated by PCR and cloned into the vectors by using primer-encoded restriction sites. The following mxiD alleles were used: mxiD, encoding wild-type MxiD (566 residues); mxiDHIS, encoding MxiD fused at its C terminus to six histidine residues; mxiD2, encoding processed MxiD lacking the 21-residue N-terminal signal sequence; mxiD2HIS, encoding mature MxiD that was C terminally His tagged; and mxiD46, encoding only 46 C-terminal residues. The following mxiM alleles were used: mxiM, encoding wild-type MxiM (142 residues); mxiM2, a mutant derivative bearing the G23R substitution (25); and mxiM3, encoding mature MxiM which lacked a 23-residue N-terminal signal sequence required for membrane insertion (3). The following mxiJ alleles were used: mxiJ, encoding wild-type MxiJ (241 residues); mxiJFLAG, encoding MxiJ fused at its C terminus to a Flag epitope (MDYKDDDDK); mxiJ2, encoding mature MxiJ lacking the 17-residue N-terminal signal sequence; and mxiJ2FLAG, encoding the mature form of Flag-tagged MxiJ. For constructions involving pBAD18, pBAD24, and pBAD33 and for constructions involving pBluescript SK+ and pWSK129, inserts were expressed from PBAD and PLAC promoters, respectively. For constructions in pDBLeu and constructions in pPC86, inserts were fused at the C termini of GAL4 DNA binding and activation domains, respectively.
Analysis of MxiDHIS in whole-cell protein extracts.
To examine MxiDHIS stability and multimerization, BS103 derivatives were grown overnight in LB and diluted the following day in LB containing arabinose. At an optical density at 600 nm (OD600) of ∼0.6, culture aliquots were removed and washed with phosphate-buffered saline. A portion of each sample was plated and used for enumeration, and the remainder was suspended in a loading buffer containing 0.5% sodium dodecyl sulfate (SDS) and 3% β-mercaptoethanol. For analyses of protein stability, samples were boiled for 10 min (this allowed all multimeric MxiD to be converted into monomers). For analyses of multimerization, duplicate samples were incubated at 37°C for 5 min or in boiling water for 3 min. Proteins were then separated by SDS-polyacrylamide gel electrophoresis (PAGE) and analyzed by immunoblotting with anti-His antibodies.
SDS-PAGE and immunoblotting.
Samples were boiled in Laemmli buffer (20) for 10 min unless otherwise indicated. Proteins were separated in 12.5% SDS- polyacrylamide minigels, transferred to polyvinylidene difluoride membranes (Schleicher & Schuell, Inc.), and treated with a blocking agent (1% casein hydrolysate in Tris-buffered saline). Immunodetection was performed by using anti-penthistidine (Qiagen, Inc.), anti-FLAG M2 (Stratagene), anti-BlaM (5′-3′, Inc.), and anti-MxiM (25) antisera. The activity of an alkaline phosphatase-labeled secondary antibody was visualized by using the chemiluminescent substrate CDP-Star (Roche).
Yeast two-hybrid analysis.
The ProQuest system of Gibco BRL was used for yeast two-hybrid analysis. Culturing of yeasts, transformation, and screening were performed as described by the manufacturer. The pDBLeu and pPC86 expression vectors, which encode GAL4 DNA binding and activation domains, respectively, were provided with the kit. Constructs were generated by fusing the GAL4 C terminus to the N terminus of an Mxi protein. The reporter Saccharomyces cerevisiae strain used was MaV203 (MATα leu2-3,112 trp1-901 his3Δ200 ade2-101 gal4Δ gal80Δ SPAL10::URA3 GAL1::lacZ HIS3UAS GAL1::HIS3@LYS2 can1R cyh2R). Plasmid pairs were cotransformed into MaV203 and plated on minimal yeast synthetic complete medium lacking tryptophan and leucine (SC-Leu-Trp). Up to 25 transformants were replica plated onto SC-Leu-Trp-His supplemented with 25 mM 3-aminotriazole to examine activation of an HIS3 reporter. The lacZ reporter was evaluated with a liquid assay that measured β-galactosidase activity in yeast cultures exposed to chlorophenol red-β-d-galactopyranoside (CPRG). The reporter activation data were compared to the data for five control strains provided with the two-hybrid kit, which displayed no, weak, moderately strong, strong, and very strong interactions.
Protein cross-linking.
The strains were grown in LB to an OD600 of 0.8 (∼1 × 109 cells/ml), washed with cross-linking buffer (20 mM NaPO4, 150 mM NaCl; pH 7.2), and concentrated 10-fold in the same buffer. Samples were incubated in the presence of 1 mM dithiobis succinimidyl propionate (DSP) for 30 min at 37°C and quenched with 20 mM Tris for an additional 15 min at 37°C.
Purification of His-tagged complexes.
The DSP-treated cultures were harvested and suspended in an ice-cold sucrose solution (0.75 M sucrose, 10 mM Tris [pH 7.8], 1 mM phenylmethylsulfonyl fluoride, 0.2 mg of lysozyme per ml) for 15 min at 4°C and then for 15 min at 37°C. The resulting spheroplasts were lysed with 0.1% Triton X-100–10 mM MgSO4 and mild sonication. Debris was removed by centrifugation (20,000 × g for 20 min at 4°C), NaCl was added to each cleared lysate to a concentration of 0.3 M, and the preparation was incubated at 4°C for 30 min. The membrane fraction was pelleted by centrifugation (110,000 × g for 30 min at 4°C) and resuspended in 1.0 ml of urea buffer (8 M urea, 10 mM Tris, 100 mM Na2HPO4, 1% Triton X-100, 0.2% Sarkosyl; pH 8.0), and proteins were solublized overnight at 4°C with gentle agitation. Insoluble material was pelleted by centrifugation (11,000 × g for 30 min at 4°C), and the resulting supernatant was mixed with 100 μl of washed 50% nickel-nitrilotriacetic acid (Ni-NTA) agarose (Qiagen). After overnight incubation at 4°C, the agarose was recovered and washed five times (10 min each) with 8 M urea–10 mM Tris–100 mM Na2HPO4–1% Triton X-100 (pH 6.3) and 0.5 M NaCl–20 mM Tris–5 mM imidazole–0.1% SDS (pH 8.0). Bound proteins were eluted in 8 M urea–50 mM Tris–2% SDS–0.4 M imidazole (pH 6.8), boiled for 5 min in Laemmli buffer containing 5% β-mercaptoethanol, and examined by SDS-PAGE and immunoblotting.
Purification of FLAG-tagged complexes.
Membrane proteins were prepared as described above for His-tagged complexes, except that no cross-linker was used and overnight membrane protein solublization at 4°C was performed in 10 mM Tris (pH 7.5)–10 mM EDTA–2% Triton X-100. After membrane protein solublization, samples were centrifuged (11,000 × g for 30 min at 4°C), and the resulting supernatant was mixed with 150 mM NaCl and 75 μl of washed anti-FLAG M2 affinity gel (Sigma). The preparation was incubated overnight at 4°C with gentle agitation. Resin was then recovered and washed five times (15 min each) with 0.5 M Tris (pH 7.4)–1.5 M NaCl. The samples were boiled for 5 min in Laemmli buffer containing 5% β-mercaptoethanol and examined by SDS-PAGE and immunoblotting. The secondary antibody used to detect the anti-FLAG antibody was anti-mouse immunoglobulin G (γ-chain specific, alkaline phosphatase conjugated).
Protease accessibility of BlaM.
Cultures were grown to an OD600 of ∼0.8, and standardized culture volumes were removed, washed with proteinase K buffer (5 mM CaCl2, 50 mM Tris-HCl; pH 7.5), and resuspended in proteinase K buffer supplemented with tetracycline. Equivalent aliquots were then incubated in the presence or absence of Congo red (40 μM) and/or proteinase K (100 μg ml−1) for 20 min at 37°C without agitation. After one wash in proteinase K buffer, samples were titrated, boiled in Laemmli buffer, and analyzed by SDS-PAGE and immunoblotting.
RESULTS
Periplasmic MxiM stabilizes MxiD.
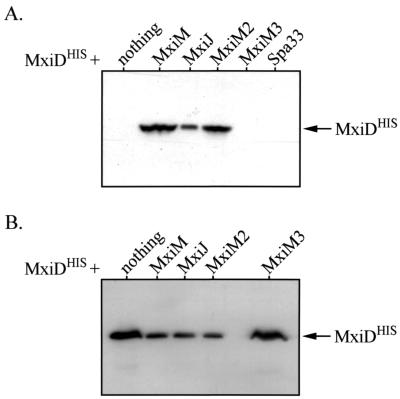
The effect of MxiM on MxiD stability was tested in a virulence plasmid-cured Shigella background (BS103) expressing MxiDHIS (from pBAD33, a low-copy-number vector) in the presence or absence of MxiM (from pBluescript, a high-copy-number vector). Since the type III Mxi-Spa system of Shigella is plasmid encoded (21), no other type III secretory proteins were present in the backgrounds used here. In the absence of MxiM, MxiDHIS was not detected by immunoblotting in the whole-cell protein extract of 1 × 108 bacteria (Fig. 1A). Coexpression with MxiM, however, resulted in high MxiDHIS levels, which suggested that MxiM can stabilize MxiD. To examine the influence of MxiM localization, MxiDHIS was coexpressed with either MxiM2 or MxiM3. MxiM2 is a periplasmic form that is neither lipidated nor OM anchored (25). Even without OM anchoring, periplasmic MxiM still stabilized MxiDHIS (Fig. 1A). MxiM3 lacks an N-terminal signal sequence required for membrane insertion (which is normally found within MxiM [3]) and is not able to insert into the bacterial envelope (data not shown). The resulting absence of MxiM from the envelope ablates its protective effect on MxiDHIS (Fig. 1A). MxiM, therefore, can probably stabilize MxiD within the envelope, perhaps via a direct interaction in the periplasm.
FIG. 1.
Analysis of MxiDHIS stability in the presence or absence of other Mxi-Spa proteins. Whole-cell protein extracts of various BS103 derivatives were separated by SDS-PAGE and analyzed by immunoblotting with anti-His antibodies. The position of MxiDHIS (∼62 kDa) is indicated by the arrows. (A) Induction of MxiDHIS (from low copy-number-vector pBAD33) in either the absence or presence of different Mxi-Spa proteins (expressed from pBluescript SK+). Protein from 1 × 108 bacteria was examined in each case. (B) Induction of MxiDHIS (from high-copy-number vector pBAD18) in the absence or presence of different Mxi-Spa proteins (expressed from pBAD33). For lanes in which MxiDHIS was expressed with nothing or MxiM3, whole-cell extracts from 1 × 106 bacteria were used. For the remaining lanes, protein from 1 × 107 bacteria was used.
Periplasmic MxiM can also destabilize MxiD.
To extend our studies, we reversed the expression vectors, so that MxiDHIS was expressed from a high-copy-number vector (pBAD18) and MxiM was expressed from a low-copy-number vector (pBAD33). When these conditions were used, we surprisingly observed results opposite those described above. First, when MxiDHIS was expressed alone, it was detected at high levels, in the extract of only 1 × 106 bacteria (Fig. 1B). Expression of MxiDHIS from a high-copy-number vector bypassed the need for the stabilizing function of MxiM. Second, coinduction with MxiM yielded much lower MxiDHIS levels (levels that were more than 10-fold lower), suggesting that MxiM has a destabilizing effect. Since coexpression of MxiDHIS with the periplasmic (but not OM-anchored) MxiM2 protein had a destabilizing effect, while coexpression with the nonperiplasmic MxiM3 protein did not, the effect observed revealed that MxiM must be located in the periplasm. Together, our results show that the effects of MxiM change (stabilization versus destabilization) depending on the relative levels of MxiDHIS. The contradictory nature of our findings may be explained as follows: (i) a periplasmic protease which can degrade MxiD when it is expressed from a low-copy-number vector in the absence of MxiM may be overwhelmed when MxiD is expressed from a high-copy-number vector; and (ii) MxiM is part of a mechanism that stabilizes or destabilizes MxiD in the periplasm depending on the level of MxiD.
Periplasmic MxiJ affects MxiD stabilization and destabilization.
Other type III secretory proteins were tested to determine their effects on MxiD stability in BS103. Structural elements that lack sec-dependent signals (such as Spa33) and do not insert into the BS103 envelope did not influence MxiDHIS stability (Fig. 1A). MxiJ, a base element with an sec-dependent signal, altered the MxiDHIS stability profile in a manner identical to the manner observed with MxiM. Depending on the MxiJ/MxiDHIS ratio, MxiJ had either a stabilizing effect (Fig. 1A) or a destabilizing effect (Fig. 1B). The effects observed also required a periplasmic form of MxiJ, as the MxiJ derivative lacking a signal sequence, MxiJ2, did not influence MxiDHIS stability (data not shown). A direct interaction between MxiJ and MxiD in the envelope may have an effect on MxiD stability
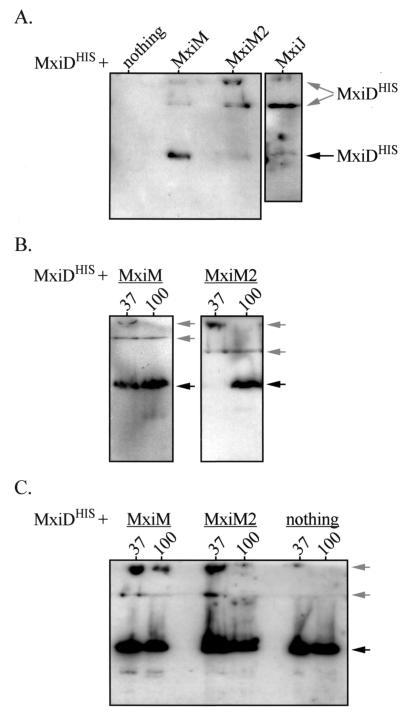
MxiM and MxiJ affect MxiD multimerization.
Secretin family members, including PulD, OutD, and YscC, form high-molecular-weight homomultimers that are detectable in stacking gels after SDS-PAGE (13, 17, 26). These homomultimers represent OM ring structures formed by secretins. When MxiDHIS alone was induced from a high-copy-number vector, high-molecular-weight multimers were detected at only very low levels. The ∼62-kDa monomer was the primary MxiDHIS species detected in whole-cell protein extracts of 1 × 106 bacteria loaded, unboiled in protein sample buffer (37°C sample), into SDS-PAGE gels (Fig. 2C). Similarly, only low-level MxiDHIS multimerization was observed upon coinduction with the nonperiplasmic MxiM3 and MxiJ2 proteins (data not shown). When MxiDHIS was coinduced with MxiM, MxiM2, or MxiJ (each from pBAD33), we detected two prominent multimeric species in the stacking gel (Fig. 2C and data not shown). Therefore, MxiDHIS oligomerizes much more efficiently when it is coexpressed with its putative interacting partners. We also observed that only the multimers formed in the presence of MxiM (and not the multimers formed in the presence of MxiM2 or MxiJ) displayed heat resistance (Fig. 2C and data not shown). The MxiDHIS complex is, therefore, most stable in the presence of OM-anchored MxiM.
FIG. 2.
Formation of MxiDHIS homomultimers in the presence or absence of other Mxi-Spa proteins. Whole-cell protein extracts of various BS103 derivatives were separated by SDS-PAGE. The stacking and separating gels were transferred to polyvinylidene difluoride membranes and analyzed by immunoblotting with anti-His antibodies. The positions of MxiDHIS monomers (∼62 kDa) are indicated by black arrows, while the positions of the two MxiDHIS multimers (>200 kDa) are indicated by gray arrows. The lower multimer is at the interface between the stacking and separating gels. (A) Induction of MxiDHIS (from low-copy-number vector pBAD33) in either the presence or the absence of mxiM, mxiM2, or mxiJ (expressed from pBluescript SK+). In each case, protein from 2.5 × 108 bacteria was boiled for 3 min and examined. (B) Resistance of MxiDHIS multimers to heat. MxiDHIS was induced (from pBAD33) in the presence of either MxiM or MxiM2 (expressed from pBluescript SK+), and samples were incubated at either 37°C (lanes 37) or 100°C (lanes 100). Protein from 3 × 108 bacteria was examined in each case. (C) Induction of MxiDHIS (from high-copy-number vector pBAD18) in either the presence or the absence of MxiM or MxiM2 (expressed from pBAD33). Samples were incubated at either 37°C (lanes 37) or 100°C (lanes 100). Protein from either 3 × 107 bacteria (for MxiM- and MxiM2-expressing strains) or 1 × 106 bacteria (for the strain expressing only MxiDHIS) was used.
We also examined the oligomerization of MxiDHIS expressed from a low-copy-number number vector (pBAD33) in the presence of MxiM, MxiM2, or MxiJ. In these backgrounds (in which MxiM, MxiM2, and MxiJ stabilize MxiDHIS), we detected both monomeric and oligomeric MxiDHIS (Fig. 2A). Whereas the major form of MxiDHIS coexpressed with MxiM was clearly the monomer, there was a pronounced shift toward oligomers upon coinduction with either MxiM2 or MxiJ. Thus, OM-anchored MxiM not only enhances the stability of MxiDHIS multimers, but it can also restrict the extent of multimerization. This effect on multimer formation was also observed in the experiment whose results are shown in Fig. 2B, as was the sensitivity of the multimers to heat.
Two-hybrid analysis of MxiD-MxiM and MxiD-MxiJ interactions.
The effects of MxiM and MxiJ on MxiD stability and multimerization likely reflect direct interactions between these proteins. To study this possibility, we utilized the yeast two-hybrid system. Derivatives of plasmids pDBLeu (GAL4 DNA binding domain vector) and pPC86 (GAL4 activation domain vector) were introduced into yeast strain MaV203, and pairwise tests for HIS3 and lacZ reporter activation were performed (Table 1). The mature forms of MxiM, MxiD, and MxiJ were used in our study because they lack N-terminal signal sequences which may impair nuclear translocation in the yeast reporter system and which are probably absent during in vivo interactions in Shigella cells. A strong interaction was detected between MxiM and either mature MxiD (His tagged or not His tagged) or a C-terminal 46-residue fragment of MxiD. This fragment corresponded to the pilot interaction domain for the secretin family (8, 9, 26), indicating that MxiM is the MxiD pilot. Other interactions, albeit weaker, were detected between both the wild-type and tagged forms of MxiD and MxiJ. No interaction between MxiM and MxiJ was observed.
TABLE 1.
Results of two-hybrid analysesa
| pDBLeu insert or control | pPC86 insert | His+ (3ATR)b | β-Galactosidase activity (U)c |
|---|---|---|---|
| None | None | No | 0.0005 |
| No-interaction control | No | 0.001 | |
| Weak-interaction control | Yes | 0.18 | |
| Moderate-interaction control | Yes | 1.5 | |
| Moderately-strong-interaction control | Yes | 70.0 | |
| mxiD | mxiM | Yes | 19.5 |
| mxiM | mxiD | Yes | 14.8 |
| mxiM | mxiDHIS | Yes | 16.9 |
| mxiD46 | mxiM | Yes | 26.5 |
| mxiM | mxiD46 | Yes | 24.0 |
| mxiJ | mxiD | Yes | 2.1 |
| mxiD | mxiJ | Yes | 1.9 |
| mxiDHIS | mxiJFLAG | Yes | 2.4 |
| mxiM | mxiJ | No | 0.003 |
Control interactions were examined, but the results are not shown. For each of the pDBLeu and pPC86 inserts we analyzed control strains in which each of the test constructs was cotransformed with either a pDBLeu vector or a pPC86 vector lacking the insert. For each insert, the N-terminal signal sequence was not present. The values obtained with the self-activation controls were routinely <0.001 U of β-galactosidase activity, and these controls did not grow on minimal medium in the absence of histidine.
Growth was assessed 2 days after replica plating onto Sc-Leu-Trp-His plates supplemented with 10 mM 3-aminotriazole (3AT). yes, growth; no, no growth.
Quantitative analysis of β-galactosidase activity in lysed yeast suspensions when CPRG was used as a substrate. One unit was the amount of enzyme activity that hydrolyzed 1.0 μmol of CPRG to chloramphenicol red and d-galactose per min.
MxiD-MxiM and MxiD-MxiJ interactions in the envelope of BS103.
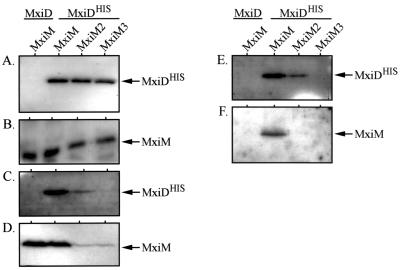
To confirm our two-hybrid results, we examined whether MxiM could be coprecipitated with MxiDHIS in vivo. His-tagged MxiD was used, and Ni-NTA resin, which binds the His tag, was used to precipitate the resulting complexes. In BS103 expressing MxiDHIS (from the high-copy-number vector pBAD18) and MxiM (from the low-copy-number vector pBAD33), high levels of both proteins were observed in whole-cell protein extracts (Fig. 3A and B) and in the cross-linked, solublized membrane proteins (Fig. 3C and D). When Ni-NTA resin was used, complexes containing both MxiDHIS and MxiM were precipitated from the membrane fraction (Fig. 3E and F). No coprecipitation was observed when, as a control, wild-type MxiD was used instead of MxiDHIS. In the presence of either MxiM2 or MxiM3, MxiDHIS was barely detectable, if it was detectable at all, in the membrane fractions (Fig. 3C and D) or the precipitated proteins (Fig. 3E and F). These results indicate that there is a direct MxiM-MxiDHIS interaction within the envelope. Additionally, MxiM must be OM anchored to promote the stable envelope insertion or association of MxiDHIS.
FIG. 3.
In vivo interaction between MxiM and MxiDHIS. BS103 derivatives expressing either MxiD or MxiDHIS (from pBAD18) in the presence of MxiM, MxiM2, or MxiM3 (expressed from pBAD33) were examined in coprecipitation analyses. The presence of MxiDHIS and MxiM (and its derivatives) was monitored in cultures prior to cross-linking with DSP (panels A and B, respectively), in membrane fractions from cross-linked cells (panels C and D, respectively), and after purification from membrane fractions using Ni-NTA beads (panels E and F, respectively). Cross-linked proteins were released prior to analysis by reducing DSP with 5% β-mercaptoethanol. The samples were then resolved on SDS–10% polyacrylamide gels and immunodetected with either anti-His or anti-MxiM antibodies. The arrows indicate the positions of MxiM (∼13 kDa) and MxiDHIS (∼62 kDa).
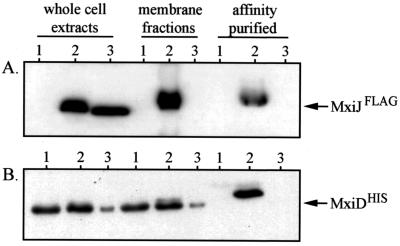
We next examined whether MxiDHIS coprecipitated with MxiJFLAG when an anti-FLAG affinity gel was used. Coinduction of MxiDHIS (from the low-copy-number vector pBAD33) and MxiJFLAG (from the high-copy-number vector pBAD24) yielded detectable levels of both proteins in whole-cell and solublized-membrane-protein extracts of BS103 (Fig. 4A and B). The anti-FLAG resin recovered MxiJFLAG-MxiDHIS complexes from the solublized fraction, suggesting that there is an in vivo interaction between these proteins in the envelope. The specificity of the interaction was confirmed by the lack of MxiDHIS precipitation in strains expressing either wild-type MxiJ (which lacks the FLAG epitope) or the MxiJ2 derivative (which does not associate with the envelope). The MxiJFLAG-MxiDHIS interaction differed from the MxiM-MxiDHIS interaction, as it was detected in either the presence (data not shown) or the absence (Fig. 4) of cross-linker. Why an OM complex is disruptable during solublization (in the absence of cross-linker) while the OM-IM complex is not disruptable is not known.
FIG. 4.
In vivo interaction between MxiJFLAG and MxiDHIS. BS103 derivatives expressing either MxiJ (lanes 1), MxiJFLAG (lanes 2), or MxiJ2FLAG (lanes 3) from pBAD24 in the presence of MxiDHIS (expressed from low-copy-number vector pBAD33) were examined in coprecipitation analyses. The presence of MxiJFLAG and MxiDHIS was monitored in whole-cell protein extracts of each strain, in the resulting membrane fractions of lysed cells, and in proteins affinity purified from the membranes by using an anti-FLAG affinity gel. No cross-linker was used in this experiment. The samples were resolved on SDS–10% polyacrylamide gels and immunodetected with either anti-FLAG or anti-MxiM antibodies. The arrows indicate the positions of MxiJFLAG (27 kDa) and MxiDHIS (∼62 kDa).
Method to study MxiM-MxiD-MxiJ complex formation.
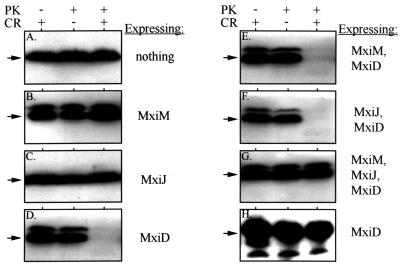
Together, the MxiM-MxiD and MxiJ-MxiD interactions could span the OM and IM and form an important intermediary in Mxi-Spa assembly. To detect an MxiM-MxiD-MxiJ structure, we exploited our observation that MxiD expression in BS103 made periplasmic β-lactamase (BlaM) susceptible to degradation by extracellular proteinase K when the dye Congo red was present (Fig. 5D). In similarly treated cells, a cytoplasmic marker, H-NS, remained stable (Fig. 5H). Additionally, BlaM expressed in the absence of MxiD or in the presence of either MxiM or MxiJ was stable in the presence of Congo red (Fig. 5A, B, and C). The loss of BlaM depended on expression of MxiD and the presence of Congo red. Congo red is a dye that interacts with Mxi-Spa surface elements and triggers the opening of a secretory pore (4). Our results suggest that a MxiD pore at the bacterial surface (represented by the low level of multimers in Fig. 2C) could bind Congo red, allowing the protease access to periplasmic proteins through the open pore. When MxiD was expressed with either MxiM or MxiJ (in strains which express high levels of MxiDHIS multimers), the Congo red treatment still destabilized the BlaM pool (Fig. 5E and F), suggesting that interactions with either of these proteins cannot block the loss of BlaM. Only when all three elements (MxiM, MxiJ, and MxiD) were coexpressed did we observe restoration of periplasmic integrity and the subsequent protease resistance of BlaM. Presumably, expression of all three proteins results in the formation of a stable complex in which the MxiD pore no longer provides the protease with access to periplasmic components (Fig. 6). It is likely that the protease enters the pore (and not that BlaM leaks out), since the BlaM pool is always stable in the presence of Congo red alone. The involvement of OM (MxiM and MxiD) and IM (MxiJ) components suggests that access to the periplasm is blocked by connection of the OM components to the component in the IM. Since we did not observe subsequent proteolysis of cytoplasmic components (Fig. 5H), additional Mxi-Spa elements must be necessary to form a continuous channel from the OM to the cytoplasm.
FIG. 5.
Susceptibility of periplasmic BlaM and cytoplasmic H-NS to extracellular protease. BS103 derivatives expressing either MxiM, MxiJ, or MxiD or various combinations of these proteins (indicated to the right of each panel) were incubated with proteinase K (PK) and/or Congo red (CR) for 20 min. Whole-cell protein extracts of each strain were resolved on SDS–10% polyacrylamide gels and examined by immunoblotting with either anti-BlaM (A to G) or anti-H-NS (H) antibodies. The arrow to the left of each panel indicates the position of BlaM (∼29 kDa) or H-NS (∼15 kDa). In these backgrounds, MxiD was expressed from the high-copy-number vector pBAD18, while MxiJ was expressed from pBAD33. MxiM was expressed from either pBAD33 (when it was used alone or with MxiD) or from pWSK129. Protein from 5 × 108 bacteria was used in each lane of panels A to G; protein from 5 × 107 bacteria was used in each lane of panel H. The minor higher-Mr band present in some panels probably corresponds to unprocessesed β-lactamase.
FIG. 6.
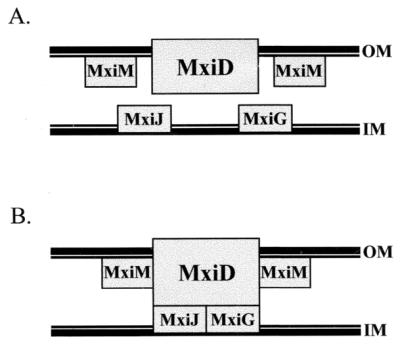
Interactions and localizations of Mxi-Spa base elements in the S. flexneri envelope. (A) Positions of MxiM, MxiD, and MxiJ. MxiM is anchored via a lipid moiety to the inner face of the OM (25), while the MxiD secretin is an integral OM protein (3). MxiJ is predicted to be anchored to the outer face of the IM (2, 14). (B) Interactions among MxiM, MxiD, and MxiJ predicted on the basis of our study. Interactions are indicated by rectangles that touch each other. MxiM and MxiD probably interact in the OM, while the MxiD-MxiJ interaction spans the periplasmic space.
DISCUSSION
Many structural components from different type III systems are highly conserved, explaining why needle complexes of Shigella and Salmonella (and probably those of other type III systems) are similar in appearance. Steps in the assembly of these structures have been studied in biochemical and electron microscope analyses of membrane complexes of type III mutants or strains expressing only one or two secretory proteins (5, 6, 16, 18, 27, 28). Findings obtained in these studies imply that the membrane-spanning base forms first, creating a structure that nucleates extracellular needle and cytoplasmic bulb proteins. The base itself assembles from at least three proteins, an OM ring-forming secretin (MxiD in Shigella) and a pair of IM ring-forming proteins (MxiG and MxiJ in Shigella). These all have sec-dependent export signals and can insert into the envelope in the absence of other type III proteins. The actual interactions among base elements and whether other proteins have roles in base assembly are unclear.
Several features of MxiM suggest that this protein plays a role in base assembly. MxiM is a 142-residue lipoprotein of Shigella which has an sec-dependent export signal, is anchored to the periplasmic face of the OM, and is required for type III secretion. MxiM is 18.6% identical to InvH of Salmonella and, like InvH, is encoded two genes upstream of a secretin open reading frame. InvH is required for proper needle complex formation in Salmonella (27) and belongs to a group of proteins called secretin pilots (7, 9) that includes YscW of the Yersinia type III system (17), PulS and OutS of the Klebsiella oxytoca (13) and Erwinia chrysanthemi (26) type II secretion systems, respectively, and PilP of the Neisseria gonorrhoeae pilus system (10). While the levels of sequence homology among pilots can be low (∼18% identity), the pilots (i) are small (120- to 150-residue) OM lipoproteins; (ii) are encoded two to four genes upstream of a secretin; (iii) are chaperone-like proteins that protect secretins from proteolysis in the periplasm and promote OM insertion of secretins; and (iv) are capable of binding a ∼50-residue C-terminal region in their respective secretins. Nouwen et al. (22) also showed that PulS forms peripheral spokes around a cylindrical PulD pore in the OM. Therefore, pilots can act like both periplasmic chaperones and structural elements, interacting with secretins in the process.
In this study we examined the role of MxiM in base assembly and in particular looked at whether this protein is a pilot for the MxiD secretin. For this study, we used virulence plasmid-cured Shigella strains (i.e., strains with no type III system) expressing mxiM and/or mxiDHIS in trans from various expression vectors. Each allele, regardless of the vector used, fully complemented its mutant background (data not shown), thus demonstrating that each protein is functional and is expressed at levels which support type III secretion. In many of the previous studies of pilot-secretin interactions the workers used such reconstituted backgrounds, expressing the pilot and/or secretin in trans in the absence of other secretory elements (and using tagged proteins as well).
We show that MxiM influences MxiD stability. As observed in several pilot-secretin studies, MxiD may not be detected in the absence of MxiM. We found that whether this protective effect is observed depends on the relative amounts of MxiM and MxiD. When MxiD is expressed at low levels compared to the level of MxiM, MxiM acts like a chaperone and stabilizes MxiD (Fig. 1A). When MxiD is expressed at higher levels, it can be detected in the absence of MxiM (Fig. 1B); not only is the chaperone-like function obviated, but coinduction with MxiM also greatly reduces the MxiD pool. The stabilizing and destabilizing functions described above require that MxiM be periplasmic, although not OM anchored. Presumably, MxiM-MxiD interactions in the periplasm either prevent MxiD proteolysis (by directly or indirectly blocking a protease-sensitive site) or promote a proteolytic activity. MxiM itself does not appear to be a protease, based on a lack of sequence similarity to known proteases. As the relative amounts of MxiM and MxiD determine the effect seen, there may be a system to ensure proper stoichiometry of these secretory components during assembly.
A prominent feature of secretins is their formation of homomultimers, corresponding to OM rings of type III base elements. These multimers are very stable, displaying various degrees of resistance to SDS and boiling, depending on the secretin. We found that MxiD also forms high-molecular-weight multimers that are partially SDS and heat resistant (Fig. 2). High levels of MxiD multimers were observed only upon coexpression with MxiM or MxiM2 (periplasmic forms). Without MxiM in the envelope, MxiD was primarily (although not exclusively) detected as a monomer (Fig. 2C). Presumably, an MxiM-MxiD interaction begins in the periplasm and favors MxiD multimerization. The extent to which multimers form and their subsequent stability were influenced by the OM anchoring of MxiM. If MxiM is not in the OM (like MxiM2), a majority of the MxiD pool forms multimers that display little heat resistance (Fig. 2C). If MxiM does anchor to the OM, the extent of MxiD multimerization is restricted and heat resistance occurs (suggesting that a more stable complex forms). These findings support the hypothesis that MxiM has a structural role, controlling multimerization and stabilizing the MxiD complex by virtue of its OM anchoring.
It was surprising that high MxiD levels were induced from pBAD18, considering that coinduction with MxiM resulted in such low levels (Fig. 1B). The stability of MxiD when it is expressed alone (i.e., from pBAD18) may stem from the fact that it remains primarily a monomer (Fig. 2C). The multimerization that occurs in the presence of MxiM may be extensive enough to activate a periplasmic stress response system like the Cpx two-component pathway. The Cpx system of uropathogenic E. coli responds to inappropriate aggregation of pilin monomers in the periplasm by activating a periplasmic protease, DegP (15). A similar response may affect the destabilizing influence of MxiM. Excessive MxiD multimerization induces the Cpx pathway (or another stress response), which upregulates periplasmic proteases that degrade the multimers. This can be part of the above-mentioned system which ensures that the levels of MxiM and MxiD in the envelope are roughly equivalent.
The influence of MxiM on MxiD stability strongly suggests that these proteins interact within the envelope. Using two-hybrid and coprecipitation studies (with solublized envelope extracts), we confirmed that this interaction occurs. During this work, we also showed that unlike MxiM, MxiM2 supports only a weak membrane association for MxiD. This finding also supports the hypothesis that MxiM has a structural role, anchoring and stabilizing MxiD in the envelope via a direct interaction. Finally, we identified an MxiM binding domain in the C-terminal 46-residue region of MxiD. This domain corresponds to the pilot-binding site identified in several secretins. This interaction, combined with the stabilizing effect of MxiM, is certainly consistent with the hypothesis that MxiM is the Shigella pilot. Features of MxiM that have not previously been identified in pilot family members include its potential to destabilize MxiD and its ability to promote MxiD multimerization.
Our findings suggest that the MxiM requirement in type III secretion stems from interactions with MxiD during base assembly and in completed Mxi-Spa needle complexes. These interactions can have chaperone, anti-chaperone, and structural functions, controlling MxiD stability and promoting multimerization and stable association of MxiD with the envelope. MxiM, therefore, joins MxiD as a component required to establish and maintain a proper OM region of the base. As the base is predicted to consist of both OM and IM proteins interacting across the periplasm to form a transmembrane bridge (Fig. 6), we also tried to determine MxiM or MxiD interactions with the IM base proteins, MxiJ and MxiG. Vectors encoding MxiG were too unstable for interaction analyses (data not shown). We did, however, use the two-hybrid system and coprecipitation studies to show that interactions between MxiD and MxiJ occur. The large periplasmic domain predicted for MxiD can be envisioned as extending toward and interacting with an IM ring structure formed by MxiJ. Interestingly, in an analysis of MxiDHIS stability in the presence and absence of MxiJ, we noted an effect nearly identical to that observed with MxiM. We found that when there is excess MxiD, MxiJ destabilizes (Fig. 1B); however, when MxiD is expressed at low levels, MxiJ stabilizes (Fig. 1A). MxiJ also promotes the formation of MxiD multimers. The fact that MxiM and MxiJ influence MxiD in such similar ways suggests these effects are general effects of interactions between MxiD and structural proteins of the base (i.e., they are not due to a specific chaperone-like function). Our results, therefore, support the hypothesis that there is an MxiM-MxiD-MxiJ complex in the base, perhaps spanning the IM and OM (Fig. 6).
A method to identify the structure formed by MxiM, MxiD, and MxiJ was developed based on two findings: (i) colonies of BS103 expressing MxiD (or MxiDHIS) bound the dye Congo red, whereas BS103 alone did not; and (ii) periplasmic BlaM is digested by extracellular protease in BS103/pBAD18::mxiD (or mxiDHIS) treated with Congo red. Congo red normally binds to type III system surface elements, destabilizing the secretory pore and inducing secretion (4). With BS103/pBAD18::mxiD, Congo red presumably binds to and destabilizes MxiD pore structures at the OM, thereby exposing periplasmic BlaM (but not a cytoplasmic marker) to extracellular protease. When MxiD was expressed with either MxiM or MxiJ, periplasmic BlaM was still degraded. When MxiD was expressed with both MxiM and MxiJ, however, the BlaM pool was stable. In the presence of MxiM at the OM, MxiD may have a more stable interaction with MxiJ in the IM and physically block access to periplasmic BlaM via the MxiD pore. This experiment provided indirect evidence that the transmembrane MxiM-MxiD-MxiJ structure shown in Fig. 6 is formed. The fact that we did not see cytoplasmic leakage in this system (data not shown) suggests that additional type III accessory proteins are needed to complete a channel from the OM to the cytoplasm. Future uses for the system described here should include addition of other base elements and/or IM export proteins in order to allow cytoplasmic leakage or to provide the ability to distinguish substrates and secrete them. The benefit of this genetic method for studying type III system assembly is the ease with which secretory components can be added in a stepwise manner to elicit progressive formation of surface structures. Functional information obtained in this way could complement the structural information obtained by the electron microscope analyses currently used to probe intermediaries in type III assembly.
ACKNOWLEDGMENTS
This work was supported by grant AI24656 from the National Institute of Allergy and Infectious Diseases and by grant RO7385 from the Uniformed Services University of the Health Sciences.
We thank Mike Flora (Biomedical Instrumentation Center, Uniformed Services University of the Health Sciences) for sequencing and primer synthesis and William Day, Colleen Kane, and Rachel Binet for critical reading of the manuscript.
REFERENCES
- 1.Allaoui A, Sansonetti P J, Ménard R, Barzu S, Mounier J, Phalipon A, Parsot C. MxiG, a membrane protein required for secretion of Shigella spp. Ipa invasins: involvement in entry into epithelial cells and in intercellular dissemination. Mol Microbiol. 1995;17:461–470. doi: 10.1111/j.1365-2958.1995.mmi_17030461.x. [DOI] [PubMed] [Google Scholar]
- 2.Allaoui A, Sansonetti P J, Parsot C. MxiJ, a lipoprotein involved in secretion of Shigella Ipa invasins, is homologous to YscJ, a secretion factor of the Yersinia Yop proteins. J Bacteriol. 1992;174:7661–7669. doi: 10.1128/jb.174.23.7661-7669.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allaoui A, Sansonetti P J, Parsot C. MxiD: an outer membrane protein necessary for the secretion of the Shigella flexneri Ipa invasins. Mol Microbiol. 1993;7:59–68. doi: 10.1111/j.1365-2958.1993.tb01097.x. [DOI] [PubMed] [Google Scholar]
- 4.Bahrani F K, Sansonetti P J, Parsot C. Secretion of Ipa proteins by Shigella flexneri: inducer molecules and kinetics of activation. Infect Immun. 1997;65:4005–4010. doi: 10.1128/iai.65.10.4005-4010.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blocker A, Jouihri N, Larquet E, Gounon P, Ebel F, Parsot C, Sansonetti P, Allaoui A. Structure and composition of the Shigella flexneri ‘needle complex,’ a part of its type III secreton. Mol Microbiol. 2001;39:652–663. doi: 10.1046/j.1365-2958.2001.02200.x. [DOI] [PubMed] [Google Scholar]
- 6.Blocker A, Gounon P, Larquet E, Niebuhr K, Cabiaux V, Parsot C, Sansonetti P. The tripartite type III secreton of Shigella flexneri inserts IpaB and IpaC into host membranes. J Cell Biol. 1999;147:683–693. doi: 10.1083/jcb.147.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crago A M, Koronakis V. Salmonella InvG forms a ring-like multimer that requires the InvH lipoprotein for outer membrane localization. Mol Microbiol. 1998;30:47–56. doi: 10.1046/j.1365-2958.1998.01036.x. [DOI] [PubMed] [Google Scholar]
- 8.Daefler S, Guilvout I, Hardie K R, Pugsley A P, Russel M. The C-terminal domain of the secretin PulD contains the binding site for its cognate chaperone, PulS, and confers PulS dependence on pIVf1 function. Mol Microbiol. 1997;24:465–475. doi: 10.1046/j.1365-2958.1997.3531727.x. [DOI] [PubMed] [Google Scholar]
- 9.Daefler S, Russel M. The Salmonella typhimurium InvH protein is an outer membrane lipoprotein required for the proper localization of InvG. Mol Microbiol. 1998;28:1367–1380. doi: 10.1046/j.1365-2958.1998.00908.x. [DOI] [PubMed] [Google Scholar]
- 10.Drake S L, Sandstedt S A, Koomey M. PilP, a pilus biogenesis lipoprotein in Neisseria gonorrhoeae, affects expression of PilQ as a high-molecular-mass multimer. Mol Microbiol. 1997;23:657–668. doi: 10.1046/j.1365-2958.1997.2511618.x. [DOI] [PubMed] [Google Scholar]
- 11.Formal S B, Dammin G J, LaBrec E H, Schneider H. Experimental Shigella infections: characteristics of a fatal infection produced in guinea pigs. J Bacteriol. 1958;75:604–610. doi: 10.1128/jb.75.5.604-610.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guzman L-M, Belin D, Carson M J, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hardie K R, Seydel A, Guilvout I, Pugsley A P. The secretin-specific, chaperone-like protein of the general secretory pathway: separation of proteolytic protection and piloting functions. Mol Microbiol. 1996;22:967–976. doi: 10.1046/j.1365-2958.1996.01539.x. [DOI] [PubMed] [Google Scholar]
- 14.Hueck C J. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hung D L, Raivio T L, Jones C H, Silhavy T J, Hultgren S J. Cpx signaling pathway monitors biogenesis and affects assembly and expression of P pili. EMBO J. 2001;20:1508–1518. doi: 10.1093/emboj/20.7.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kimbrough T G, Miller S I. Contribution of Salmonella typhimurium type III secretion components to needle complex formation. Proc Natl Acad Sci USA. 2000;97:11008–11013. doi: 10.1073/pnas.200209497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koster M, Bitter W, de Cock H, Allaoui A, Cornelis G R, Tommassen J. The outer membrane component, YscC, of the Yop secretion machinery of Yersinia enterocolitica forms a ring-shaped multimeric complex. Mol Microbiol. 1997;26:789–797. doi: 10.1046/j.1365-2958.1997.6141981.x. [DOI] [PubMed] [Google Scholar]
- 18.Kubori T, Sukhan A, Aizawa S-I, Galan J E. Molecular characterization and assembly of the needle complex of the Salmonella typhimurium type III protein secretion system. Proc Natl Acad Sci USA. 2000;97:10225–10230. doi: 10.1073/pnas.170128997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kubori T, Matsushima Y, Nakamura D, Uralil J, Lara-Tejero M, Sukhan A, Galán J E, Aizawa S-I. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science. 1998;280:602–605. doi: 10.1126/science.280.5363.602. [DOI] [PubMed] [Google Scholar]
- 20.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.Maurelli A T, Blackmon B, Curtiss R., III Loss of pigmentation in Shigella flexneri 2a is correlated with loss of virulence and virulence-associated plasmid. Infect Immun. 1984;43:397–401. doi: 10.1128/iai.43.1.397-401.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nouwen N, Ranson N, Saibil H, Wolpensinger B, Engel A, Ghazi A, Pugsley A P. Secretin PulD: association with pilot PulS, structure, and ion-conducting channel formation. Proc Natl Acad Sci USA. 1999;96:8173–8177. doi: 10.1073/pnas.96.14.8173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nouwen N, Stahlberg H, Pugsley A P, Engel A. Domain structure of secretin PulD revealed by limited proteolysis and electron microscopy. EMBO J. 2000;19:2229–2236. doi: 10.1093/emboj/19.10.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Plano G V, Day J B, Ferracci F. Type III export: new uses for an old pathway. Mol Microbiol. 2001;40:284–293. doi: 10.1046/j.1365-2958.2001.02354.x. [DOI] [PubMed] [Google Scholar]
- 25.Schuch R, Maurelli A T. The Mxi-Spa type III secretory pathway of Shigella flexneri requires an outer membrane lipoprotein, MxiM, for invasin translocation. Infect Immun. 1999;67:1982–1991. doi: 10.1128/iai.67.4.1982-1991.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shevchik V E, Condemine G. Functional characterization of the Erwinia chrysanthemi OutS protein, an element of a type II secretion system. Microbiology. 1998;144:3219–3228. doi: 10.1099/00221287-144-11-3219. [DOI] [PubMed] [Google Scholar]
- 27.Sukhan A, Kubori T, Wilson J, Galan J E. Genetic analysis of assembly of the Salmonella enterica serovar Typhimurium type III secretion-associated needle complex. J Bacteriol. 2001;183:1159–1167. doi: 10.1128/JB.183.4.1159-1167.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamano K, Aizawa S-I, Katayama E, Nonaka T, Imajoh-Ohmi S, Kuwae A, Nagai S, Sasakawa C. Supramolecular structure of the Shigella type III secretion machinery: the needle part is changeable in length and essential for delivery of effectors. EMBO J. 2000;19:3876–3887. doi: 10.1093/emboj/19.15.3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thanassi D G, Hultgren S J. Multiple pathways allow protein secretion across the bacterial outer membrane. Curr Opin Cell Biol. 2000;12:420–430. doi: 10.1016/s0955-0674(00)00111-3. [DOI] [PubMed] [Google Scholar]
- 30.Wang R F, Kushner S R. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]