Abstract
DNA microarrays were constructed by using 271 open reading frame (ORFs) from the genome of the archaeon Pyrococcus furiosus. They were used to investigate the effects of elemental sulfur (S°) on the levels of gene expression in cells grown at 95°C with maltose as the carbon source. The ORFs included those that are proposed to encode proteins mainly involved in the pathways of sugar and peptide catabolism, in the metabolism of metals, and in the biosynthesis of various cofactors, amino acids, and nucleotides. The expression of 21 ORFs decreased by more than fivefold when cells were grown with S° and, of these, 18 encode subunits associated with three different hydrogenase systems. The remaining three ORFs encode homologs of ornithine carbamoyltransferase and HypF, both of which appear to be involved in hydrogenase biosynthesis, as well as a conserved hypothetical protein. The expression of two previously uncharacterized ORFs increased by more than 25-fold when cells were grown with S°. Their products, termed SipA and SipB (for sulfur-induced proteins), are proposed to be part of a novel S°-reducing, membrane-associated, iron-sulfur cluster-containing complex. Two other previously uncharacterized ORFs encoding a putative flavoprotein and a second FeS protein were upregulated more than sixfold in S°-grown cells, and these are also thought be involved in S° reduction. Four ORFs that encode homologs of proteins involved in amino acid metabolism were similarly upregulated in S°-grown cells, a finding consistent with the fact that growth on peptides is a S°-dependent process. An ORF encoding a homolog of the eukaryotic rRNA processing protein, fibrillarin, was also upregulated sixfold in the presence of S°, although the reason for this is as yet unknown. Of the 20 S°-independent ORFs that are the most highly expressed (at more than 20 times the detection limit), 12 of them represent enzymes purified from P. furiosus, but none of the products of the 34 S°-independent ORFs that are not expressed above the detection limit have been characterized. These results represent the first derived from the application of DNA microarrays to either an archaeon or a hyperthermophile.
Hyperthermophiles are microorganisms that grow optimally at temperatures of 80°C and higher, and most are classified as archaea (65). They are a rather diverse group with respect to their metabolic capabilities, but many of them utilize peptides as a carbon source and reduce elemental sulfur (S°) to H2S (66). Of these, the majority are obligately proteolytic and show little if any growth unless S° is added to the growth medium. The exceptions include some species of Pyrococcus that metabolize poly- and oligosaccharides, as well as peptides (3, 4, 15). For example, Pyrococcus furiosus grows on a disaccharide (maltose) to high cell densities in the absence of S° and produces H2 as an end product rather than H2S. On the other hand, P. furiosus requires S° in the growth medium when it utilizes peptides as a carbon source (1). A comparison of the activities of some key metabolic enzymes in cells grown either on peptides (with S°) or on maltose (with or without S°) provided the first evidence for a highly regulated fermentative-based metabolism in P. furiosus, with S° or its metabolites playing a key regulatory role (1). The molecular mechanism by which S° achieves this control is not known.
The pathways by which P. furiosus metabolizes peptides and carbohydrates are reasonably well established. The organism contains a modified Embden-Meyerhof pathway in which hexokinase and phosphofructokinase are ADP rather than ATP dependent (29, 68). In addition, the expected glyceraldehyde-3-phosphate dehydrogenase and phosphoglycerate kinase are replaced by a single phosphate-independent enzyme, glyceraldehyde-3-phosphate ferredoxin oxidoreductase (GAPOR [42]), which is regulated at the transcriptional level (69). The pyruvate produced by this pathway is converted to the end product acetate in two steps involving pyruvate ferredoxin oxidoreductase (POR [5]) and acetyl-coenzyme A (CoA) synthetases I and II (40, 61), with the concomitant production of ATP from ADP and phosphate. The oxidative steps in the pathway of acetate production from glucose therefore involve two ferredoxin-dependent enzymes (GAPOR and POR), and a similar mechanism is present during peptide catabolism by P. furiosus. The organism contains, in addition to POR, three other, ferredoxin-dependent, 2-keto acid oxidoreductases, and these convert transaminated amino acids into their corresponding CoA derivatives (5, 19, 39), which are then transformed to their corresponding organic acids by the two acetyl-CoA synthetases with concomitant ATP synthesis (40).
How P. furiosus and related species couple the oxidation of the reduced ferredoxin generated by the catabolic pathways to the reduction of S° is not known. The organism contains three different hydrogenases: two cytoplasmic, NAD(P)H-dependent enzymes, and one as part of a membrane-bound complex (9, 38, 59). The cytoplasmic hydrogenases were thought to play a role in S° reduction since they can reduce S° in vitro (37), but this appears not to be the case as the total hydrogenase activity of S°-grown cells is dramatically lower than that of cells grown without S° (1). The mechanism by which S° affects hydrogenase activity is not understood. Moreover, the nature of the enzyme system that reduces S° remains unknown. In mesophilic organisms, the only S°-reducing enzyme that has been well-characterized is the trimeric, membrane-bound, molybdenum (Mo)-containing, polysulfide reductase of the mesophilic bacterium, Wolinella succinogenes (32), but the genome of P. furiosus (54) does not contain ORFs that would encode a homolog of the mesophilic enzyme. Moreover, P. furiosus is not known to utilize Mo but contains at least three enzymes that utilize the analogous element tungsten (W [27]), perhaps suggesting a role for W in S° reduction.
The ability of P. furiosus to grow well both with and without S°, together with the availability of its complete genome sequence, provide an opportunity to investigate S° metabolism by expression analysis by using DNA microarrays. In fact, the genus Pyrococcus is unique in that complete genome sequences are available from three species; P. horikoshii, P. abyssi, and P. furiosus (28, 54; D. Prieur, P. Forterre, J.-C. Thierry, and J. Dietrich [http://www.genoscope.cns.fr/Pab/]). While DNA microarrays (60) have revolutionized functional genomics in eukaryotic systems (e.g., references 14, 56, and 63), there have been far fewer applications with prokaryotes. Studies initially focused on pathogens (11, 17, 58, 67, 74) and more recently have included Escherichia coli (2, 30, 53, 73, 79), Bacillus subtilus (76, 78) and a cyanobacterium (21). As yet there have been no reports of using this technique with either a hyperthermophile or an archaeon. The genome of P. furiosus contains ca. 2,200 open reading frames (ORFs) with >50% of unknown function. As a prelude to a complete genomic analysis, we focus here on 271 ORFs that encode proteins that are proposed to be involved in the primary metabolic pathways, energy conservation, and metal metabolism. The results indicate that S° or its metabolites play a major regulatory role at the transciptional level and that S° reduction appears to be accomplished by a new type of enzyme system involving the products of previously uncharacterized ORFs.
MATERIALS AND METHODS
Primer design and PCR.
Gene sequences were obtained from the P. furiosus genome site (http://comb5-156.umbi.umd.edu/) (54). Of the 275 ORFs that were examined in this study, 87 of them are listed in Tables 1 to 4. A complete list of the 275 ORFs is available at http://adams.bmb.uga.edu/pubs/sup228.pdf. Primers were designed for the ORFs by using the Primer 3 program (Whitehead Institute, MIT) and were purchased from Stanford University and from MWG Biotech (High Point, N.C.). For the initial round of PCR the primer pairs were designed to give products corresponding to the complete ORFs, and this yielded products for 249 of them as verified by gel electrophoresis. The remaining 42 primer pairs were designed to yield a product of ca. 1 kb (unless the target ORF was smaller). Of these only four ORFs did not yield suitable PCR products, and these were not pursued further. PCR products were purified with a Strataprep 96 PCR purification kit (Stratagene, La Jolla, Calif.) and eluted in 33% (vol/vol) dimethyl sulfoxide. They were spotted in duplicate onto aminosilane coated slides (Sigma, St. Louis, Mo.) in four subarrays by using a robotic slide printer (Omnigrid; Genemachines, San Carlos, Calif.). The slides were processed as previously described (8).
TABLE 1.
ORFs whose expression is dramatically (>5-fold) downregulated by S°
| ORFa | Descriptionb | Mean intensity ratio (log2 ± SD)c | Change in expression (fold)c |
|---|---|---|---|
| 577932 | [Hydrogenase expression and/or formation regulatory protein, hypF] | 5.28 ± 0.89 | 39.0 |
| 1337916 | Membrane-bound hydrogenase ORF1, mbh1d | 4.14 ± 0.80 | 17.6 |
| 1251888 | Hydrogenase II gamma, hyhG2 (38) | 3.78 ± 1.47 | 13.7 |
| 1338167 | Membrane-bound hydrogenase ORF2, mbh2d | 3.77 ± 1.29 | 13.6 |
| 1339081 | Membrane-bound hydrogenase ORF5, mbh5d | 3.68 ± 1.30 | 12.8 |
| 1339520 | Membrane-bound hydrogenase ORF6, mbh6d | 3.65 ± 0.77 | 12.6 |
| 1253842 | Hydrogenase II alpha, hyhL2 (38) | 3.57 ± 1.45 | 11.8 |
| 1341399 | Membrane-bound hydrogenase ORF8 (like cooM and mbh8)d | 3.49 ± 0.79 | 11.2 |
| 1338538 | Membrane-bound hydrogenase ORF3, mbh3d | 3.31 ± 1.66 | 9.9 |
| 1252601 | Hydrogenase II delta, hyhS2 (38) | 3.25 ± 1.43 | 9.5 |
| 1342770 | Membrane-bound hydrogenase ORF11, mbh11d | 3.06 ± 1.22 | 8.3 |
| 866528 | Hydrogenase I delta, hyhS1 (37) | 2.95 ± 0.66 | 7.7 |
| 1345018 | Membrane-bound hydrogenase ORF13 (like hydC, cooK, echB, and mbh13)d | 2.77 ± 0.85 | 6.8 |
| 1345434 | Membrane-bound hydrogenase ORF14 (like hycF, echF, cooX, and mbh14)d | 2.73 ± 1.40 | 6.6 |
| 1344050 | Membrane-bound hydrogenase ORF12, catalytic NiFe subunit, mbh12 (59) | 2.68 ± 0.59 | 6.4 |
| 864857 | Hydrogenase I beta, hyhB1 (37) | 2.66 ± 1.08 | 6.3 |
| 1251025 | Hydrogenase II beta, hyhB2 (38) | 2.65 ± 1.04 | 6.3 |
| 1342256 | Membrane-bound hydrogenase ORF10, small subunit homolog, mbh10d | 2.59 ± 0.87 | 6.0 |
| 51760 | [Conserved hypothetical protein] | 2.59 ± 1.14 | 6.0 |
| 1338785 | Membrane-bound hydrogenase ORF 4, mbh4d | 2.54 ± 2.16 | 5.8 |
| 615154 | OTCase, argF (36) | 2.43 ± 0.49 | 5.4 |
ORF designation is the end nucleotide number (54).
The ORF description is derived either from annotation by homology (given within brackets) or from the indicated reference where there is experimental data to support the ORF assignment specifically in P. furiosus (given without brackets).
The intensity ratio is expressed as a log2 value so that the standard deviation can be given. For case of comparison between ORFs, the apparent change in the expression level of a given ORF is also indicated.
This work (see text for details).
TABLE 4.
Poorly expressed S°-independent ORFsa
| ORF | Descriptionb |
|---|---|
| 53135 | [Conserved hypothetical protein] |
| 349245 | [Flagellum-related protein D, putative] |
| 350457 | [Conserved hypothetical protein] |
| 350944 | [Flagellin B2 precursor] |
| 351748 | [Flagellin B2 precursor] |
| 373060 | [ABC transporter, OppBC family] |
| 379218 | [β-Galactosidase precursor] |
| 488057 | [DNA mismatch repair protein, MutS] |
| 562899 | [Molybdopterin converting factor, subunit 1, moaD] |
| 575520 | [NADH oxidase, noxA-4/nitrite reductase] |
| 637268 | [Molybdopterin-guanine dinucleotide biosynthesis protein, mobA] |
| 637303 | [Hydrogenase maturation protease, hycI] |
| 722839 | [Conserved hypothetical protein] |
| 738742 | [Conserved hypothetical protein] |
| 743892 | [Putative proline depeptidase] |
| 834030 | [Conserved hypothetical protein] |
| 838710 | [Conserved hypothetical protein] |
| 880926 | [Ferric enterobactin transport ATP-binding protein homolog] |
| 881669 | [Iron(III) ABC transporter, permease protein, hemU-1] |
| 882732 | [Iron(III) ABC transporter ATP-binding protein, hemV-2] |
| 962748 | [Alkaline phosphatase IV precursor] |
| 1012695 | [Phosphoglycerate kinase] |
| 1138558 | [Transcriptional regulator (FurR family)] |
| 1158892 | [Dissimilatory sulfate adenylyl transferase] |
| 1165967 | [4-Aminobutyrate aminotransferase] |
| 1197174 | [Putative nucleolar protein II, Nol1-Nop2-sun family] |
| 1208389 | [Conserved hypothetical protein] |
| 1417217 | [Sugar-binding transport ATP-binding protein] |
| 1647980 | [2-keto-acid:ferredoxin oxidoreductase subunit alpha] |
| 1668574 | [Sarcosine oxidase, alpha subunit, SoxA] |
| 1669077 | [Putative polyferredoxin, muhB] |
| 1711295 | [Molybdenum cofactor biosynthesis protein, moaC] |
| 1873595 | [Nitrogen reductase, N terminus] |
| 1873914 | [Ferredoxin-family protein] |
Growth of P. furiosus and RNA preparation.
P. furiosus was grown in batch mode in a 20-liter custom fermentor at 95°C with maltose as the primary carbon source and was harvested in mid-log phase (1). The only variable was the presence or absence of S°. The cells that were used to prepare RNA for the microarray analyses were from experiments that have been described previously (1). In that case, activity assays were determined for more than 20 enzymes involved in the primary metabolic pathways by using cytoplasmic and membrane fractions, and these results are referred to below. Samples (2,000 ml) of the same cultures were cooled on ice, and total RNA was extracted by using acid-phenol extraction (71). No significant contamination of the RNA with genomic DNA could be detected by Northern blot hybridization or preliminary microarray experiments (data not shown), and so a DNase treatment was not used.
Preparation of cDNA and hybridization conditions.
Fluorescently labeled cDNA was prepared with the ARES DNA Labeling Kit (Molecular Probes, Eugene, Oreg.). In brief, 15 μg of total RNA was reversed transcribed in a total volume of 20 μl by using Stratascript RT (Stratagene, La Jolla, Calif.) in the presence of 1 mM dATP, 1 mM CTP, 1 mM GTP, 0.3 mM dTTP, 0.5 mM aminoallyl dUTP, and 1 μg of random 9-mers (Stratagene) according to the manufacturer's instructions. After a 1.5 h of incubation at 42°C, RNA was destroyed by the addition of 0.1 N NaOH, followed by a 10-min incubation at 70°C. After neutralization with 0.1 N HCl, amine-modified cDNA was purified by using a QIAquick PCR Purification Kit (Qiagen, Valencia, Calif.), except that the wash buffer was replaced with 75% (vol/vol) ethanol, and the cDNA was eluted with 45 μl of distilled water and dried under vacuum. The amine modified cDNA was labeled with Alexa 488 or Alexa 594 dyes (Molecular Probes) according to the manufacturer's instructions. Alexa 546 was used as third dye for the triple-labeling experiments. The labeled cDNA was purified with the Qiagen kit as described above and was dried under vacuum. Differentially labeled (Alexa 488 and Alexa 594) cDNA derived from P. furiosus cells grown in the presence or absence of S° was pooled and hybridized to the microarrays. The labeled cDNA was dissolved in 20 μl of hybridization buffer (Sigma), and hybridization was performed under a coverslip at 65°C in a humidity chamber (Arrayit, Sunnyville, Calif.) for between 10 and 15 h. The slides were then washed twice for 5 min in each of 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% sodium dodecyl sulfate (SDS), and 0.2× SSC–0.1% SDS and then rinsed in distilled water and blown dry with compressed air. The intensities of the Alexa 488 and Alexa 594 dyes were measured by using a Scan Array 5000 spectrometer (Packard, Meriden, Conn.) with the appropriate laser and filter settings and analyzed by using Quantarray (Packard).
Data analysis.
To compute signal intensities for each ORF, the areas surrounding each spot were taken as the background, and 17 control spots, where only spotting buffer was printed, were included on the 384-spot array. The control spots were corrected for local background, and the average was used to correct the signals from each ORF after their background correction. The relative amounts of the transcripts (with or without S°) are presented in a linear fashion by converting all ratios to a log2 function. For the two different growth conditions tested, a negative value of less than −2.3 represents a >5-fold downregulation of expression from a given ORF by S°, and this corresponds to a green color in a false color overlay. Conversely, a positive value graeater than 2.3 represents a >5-fold upregulation and corresponds to a red color in a false color overlay. ORFs that display log2 values between approximately −1 and 1 are minimally affected by S° and show a yellow color on the overlay. The detection limit of fluorescent signals was set arbitrary to 1,000 intensity units (see Fig. 1) and such spots are not visible on the false overlay. Only ORFs that display intensities more than twice the detection limit are considered valid. Conversion of intensity ratios enables the data to be presented as a log2 value with a standard deviation (SD). This represents an average of six hybridization experiments with cDNA derived from five different cultures of P. furiosus: two grown with S° and three grown without it. In preliminary experiments we did not observe any significance difference in the quality of the data between different Alexa dyes (Molecular Probes) nor when the Alexa dyes were replaced with CY3 and CY5 (Amersham, Piscataway, N.J.), dyes that are more usually used as fluorescent labels (12). The advantage of the Alexa dyes is that labeling with three dyes is possible with the lasers and filters available in the Scanarray 5000 (Packard). Statistical significance of the observed fluorescence signal ratios was obtained by a paired t test analysis by using the statistical software JMP4 (SAS Institute, Cary, N.C.).
FIG. 1.
Fluorescence intensities of DNA microarrays. (A) cDNA versus cDNA derived from the same cultures of cells grown on maltose (no S°). (B) cDNA versus cDNA derived from cells grown on maltose with or without S°. The upper and lower diagonal lines indicate fivefold changes in the signal intensities. See the text for details.
RESULTS AND DISCUSSION
Selection of ORFs.
Of the ca. 2,200 ORFs in the P. furiosus genome (54), 271 were selected for microarray analysis. The targets were putative genes involved in the primary metabolic pathways of carbon, nitrogen, and energy metabolism, and those that are, or might be, related to the metabolism of S° and H2. They included ORFs that are proposed to encode various enzymes and proteins involved in the pathways of sugar and peptide catabolism; the utilization of metals (such as Fe, Ni, W, and Mo); and the biosynthesis of cofactors, amino acids, lipids, polyamines, ribosomes, and nucleotides, together with putative chaperonins, ATPases, and transcriptional regulators. Unless otherwise indicated, each ORF is referred to by its end nucleotide number and by its current annotation, which is based on homology searches (depicted in brackets [see Table 1 and reference 54]). It is specifically noted in Tables 1 to 4 where there are experimental data derived by using P. furiosus to support the ORF assignment (given without brackets and with a reference, see Table 1).
Experimental protocols and data analysis.
The efficacy of the microarray experiment with mRNA derived from P. furiosus cells is shown in Fig. 1. Figure 1A shows data with differentially labeled cDNA samples of RNA prepared from the same batch of cells, grown with maltose in the absence of S°, and hybridized to the same microarray. The intensity of the fluorescence signals varied over a range of more than 103, a finding consistent with what has been reported with other organisms (26, 34, 46, 78). Those ORFs with intensities of below ca. 2,000 (Fig. 1A) are considered below the detection limit, and it cannot be concluded that these ORFs are expressed to a significant degree. From Fig. 1A it is clear that ORFs that give rise to low-intensity signals show a high deviation because of background fluorescence, whereas the data derived from ORFs that give high signal intensities lie near or on the diagonal.
As shown in Fig. 1B, S° and/or its metabolites clearly have a dramatic effect on the expression of many of the 271 ORFs that were examined. It was previously shown that the presence of S° in the growth medium affects the activities of several metabolic enzymes (1). The microarray data indicate that this regulation is primarily at the transcriptional level. We focus below on ORFs whose expression appears to be strongly regulated by S°, where the signal intensity changes by at least fivefold (shown by the upper and lower diagonal lines in Fig. 1B). The standard deviations (log2 intensity ratios) for almost all of these ORFs are <2, and they are differentially regulated with at least a 95% confidence level as determined by using a paired t test analysis (P > 0.95). However, there are three exceptions. These are ORFs 1419794 (formate dehydrogenase α-chain), 156299 (3-isopropyl malate dehydrogenase), and 438413 (phosphoribosylglycinamide formyltransferase) whose log2 intensities showed deviations of >2 (P < 0.90). For these ORFs a large variation in the ratios was obtained by using the same growth condition (maltose without S°). This was verified when cDNA samples from three independently grown cultures were separately labeled with the Alexa 488, Alexa 546, or Alexa 594 dyes, and all three cDNAs were hybridized to the same slide. Only these three (of 271) ORFs showed distinct false colors, indicating differential expression in one of the three samples. The reasons for this are not clear, but these ORFs appear not to be regulated by S° and are not considered further.
ORFs strongly downregulated by S°.
The expression of 21 of the 271 ORFs examined decreased by more than a factor of 5 when RNA was derived from cells grown with S° (Fig. 1B), and these are listed in Table 1. Of these, 18 ORFs encode subunits associated with the three different hydrogenase systems that have been characterized from P. furiosus. These include those encoding six of the eight subunits of the two cytoplasmic hydrogenases (I and II [38, 50]), whose expression decreases between 6- and 14-fold in the presence of S° (Table 1). Expression of the other two ORFs (encoding the α- and γ-subunits of hydrogenase I) decrease by 2.2- to 4.8-fold. These data are in accord with the report that the presence of S° in the medium decreased the total hydrogenase activity of the cytoplasmic fraction by ca. 16-fold (1). Since the specific activity of hydrogenase I is ca. 10-fold higher that of hydrogenase II in in vitro assays, it was not possible to determine from the activity analyses if hydrogenase II activity was regulated by S°. However, the microarray data clearly show that both hydrogenases are strongly regulated. Moreover, S° regulates the expression of the genes encoding the cytoplasmic hydrogenases in a negative fashion, rather than S° (or its metabolites) having some inhibitory effect on activity. Obviously, these hydrogenases are unlikely to play a role in S° reduction, as originally suggested (37).
It was previously shown that the activity of the membrane-bound hydrogenase of P. furiosus decreases by almost 30-fold when cells are grown in the presence of S° (1). After solubilization and purification from cells grown in the absence of S°, the enzyme contained two subunits (Mbh11 and Mbh12) and, based on their N-terminal sequences, the genes encoding them were proposed to be part of a 14-ORF operon (Mbh1 to -14 [59]). All but one (Mbh7) of the 14 ORFs were included on the DNA microarray. As shown in Table 1, the expression of all but one of them decreased by more than 5-fold in the presence of S° (the remainder, Mbh9, decreased by 4.2-fold). These data confirm that the 14 ORFs do indeed constitute an operon and that its products are involved in the metabolism of H2. The average fluorescence intensities associated with the 13 ORFs of the membrane-bound hydrogenase operon are shown in Fig. 2, along with the values for the eight subunits that encode the two cytoplasmic hydrogenases. The ORFs are depicted according to their order in their respective operons. All show a similar pattern in that they are downregulated to close to the detection limit (see Fig. 1B) in the presence of S°, a finding consistent with barely detectable hydrogenase activities in cell extracts (1). The apparent exception is the ORF encoding the α-subunit of hydrogenase I. Although this ORF is expressed at about five times the detection limit, the results show that it is regulated by S° as determined by the paired t test analysis (P > 0.98). However, the 3′ end of this ORF (ORF 867811) overlaps by two nucleotides with the 3′ end of an ORF (ORF 867808) on the opposing strand. If the untranslated 3′ end of ORF 867811 mRNA extends far enough into the complementary strand of hyhA, this could give rise to a “false signal” due to cross-hybridization. ORF 867811 encodes a putative ABC transporter (ATP binding protein) and was not included on the microarray. The interference of overlapping mRNAs in microarray analyses has been reported (30), although it has not been systematically analyzed with any genome. With the 18 hydrogenase-related ORFs (Fig. 2), there is considerable variation in the absolute signal intensities, but this appears to arise to some extent from the differences in ORF sizes, several of which are <500 bp.
FIG. 2.
Expression of ORFs encoding hydrogenase-related subunits with or without S°. The ORFs encoding the subunits of the cytoplasmic hydrogenase I (A), cytoplasmic hydrogenase II (B), and the membrane-bound hydrogenase (C) are arranged according to their positions in their respective operons. These are plotted against signal intensities for cDNA obtained from cells grown with (solid bars) or without (shaded bars) S°. The results with all subunits have a confidence level of at least 96% (P > 0.98 for the cytoplasmic hydrogenases and P > 0.96 for the membrane-bound enzyme).
Hence, of the 21 ORFs (from the total 271 ORFs) that are strongly downregulated (>5-fold) in the presence of S°, 18 represent structural genes for the three hydrogenases. Of the other three S°-responsive ORFs, one (ORF 577932) showed an almost 40-fold decrease in expression, the largest of any of the downregulated ORFs (Table 1). ORF 577932 is annotated as hypF and shows 37% sequence identity to its closest homolog, HypF, from the aerobic bacterium Ralstonia eutrophicus (75). HypF is thought to be involved in biosynthesis of the metal site of [NiFe]-hydrogenases. There is only one gene of this type in P. furiosus so it appears that, as in E. coli (25), one HypF protein is involved in the maturation of three different hydrogenases (two cytoplasmic and one membrane bound). One would expect HypFA, as a processing enzyme, to be present at low intracellular concentrations, and the reason why its gene appears to be so strongly regulated by S° is not clear at this point. P. furiosus contains homologs of several other genes that are involved in hydrogenase maturation and metal insertion in mesophilic bacteria. These include hycI (ORF 637303) and hypACDE (ORFs 636092, 566869, 567973, and 626147; a version of hypB appears not to be present in the P. furiosus genome). These ORFs were also included on the microarray. The expression of the hyp genes, which are all expressed at significant levels (1.5 to 9.6 times the detection limit; see Fig. 1B) decreased by two- to threefold when S° was added, but the expression of hycI was below the detection limit under both conditions.
One of the two remaining ORFs that are strongly downregulated by S° (Table 1) may also be related to hydrogenase biosynthesis. This is ORF 615154, which encodes ornithine carbamoyltransferase (OTCase). This enzyme has been purified from P. furiosus, its gene has been cloned and expressed, and its crystal structure is available (36, 70). OTCase catalyzes the transfer of a carbamoyl group from carbamoyl phosphate to ornithine, generating citrulline in the arginine biosynthetic pathway. The potential link between OTCase and the hydrogenases stems from the recent proposal that the source of the C1 (CO and CN) ligands to the metals at the active site of hydrogenases is the carbamoyl group of carbamoyl phosphate (48). In view of the regulation of both OTCase and hydrogenase expression by S° in P. furiosus, we speculate that citrulline is the precursor of the carbamoyl group for the synthesis of the active sites of the three hydrogenases in this organism. Citrulline may well be preferred to carbamoyl phosphate because of the high thermal lability of the latter compound (35). One would expect citrulline also to be needed for arginine biosynthesis even in the presence of S°, and this appears to be the case, since OTCase is expressed at close to three times the detection level in S°-grown cells.
The remaining ORF that is strongly downregulated by S° is ORF 51760, and this is annotated as a conserved hypothetical protein (Table 1). It is adjacent to the gene (ORF 49183) encoding PEP synthetase, a gluconeogenic enzyme recently purified from P. furiosus (24; see also below), and an ORF (ORF 53135) encoding a conserved hypothetical protein, neither of which showed significant (<2-fold) changes in expression according to the microarray data. The function of the S°-responsive ORF 51760 is therefore unclear at present.
ORFs strongly upregulated by S°.
Of the 271 ORFs examined, the expression of 12 of them was strongly upregulated (>5-fold) when S° was present in the growth medium (Table 2). In contrast to the ORFs that were downregulated by S°, all of the upregulated ORFs represent putative proteins since none of their products have been characterized from P. furiosus. Moreover, the two most strongly regulated ORFs, 1871822 and 1872873, which are upregulated more than 25-fold, are next to each other on the genome. The products of these two ORFs show no sequence similarity to any known protein. Nevertheless, they will be referred to as SipA (1871822) and SipB (1872873) for “sulfur-induced protein” since expression of their genes clearly responds to S° or its metabolites. Both genes are also very tightly regulated, since there appears to be little or no expression from them when cells are grown in the absence of S° (their signal intensities are less than the detection limit).
TABLE 2.
ORFs whose expression is dramatically (>5-fold) upregulated by S°
| ORFa | Descriptiona | Mean intensity ratio (log2 ± SD)a | Change in expression (fold)a |
|---|---|---|---|
| 1871822 | [Conserved hypothetical protein, sipA]b | 5.94 ± 1.43 | 61.4 |
| 1872873 | [Putative polyferredoxin, sipB]b | 4.65 ± 1.90 | 25.1 |
| 1487371 | [Tryptophan synthase, subunit beta, trpB-1] | 2.98 ± 0.69 | 7.9 |
| 1805557 | [Conserved hypothetical protein] | 2.93 ± 1.29 | 7.6 |
| 1008251 | [Aspartokinase II alpha subunit] | 2.92 ± 0.89 | 7.6 |
| 1131551 | [NADH oxidase, noxA-2] | 2.88 ± 0.39 | 7.4 |
| 1825269 | [Thermosome, single subunit] | 2.83 ± 1.69 | 7.1 |
| 900019 | [Acetolacetate synthase] | 2.78 ± 1.02 | 6.9 |
| 65527 | [Fibrillarin-like pre-rRNA processing protein] | 2.70 ± 0.80 | 6.5 |
| 204761 | [Oligopeptide transport system permease protein] | 2.68 ± 0.45 | 6.4 |
| 102519 | [Glutaredoxin-like protein] | 2.61 ± 1.72 | 6.1 |
| 1352206 | [NADH dehydrogenase subunit] | 2.59 ± 0.59 | 6.0 |
See Table 1 for details.
See text for details.
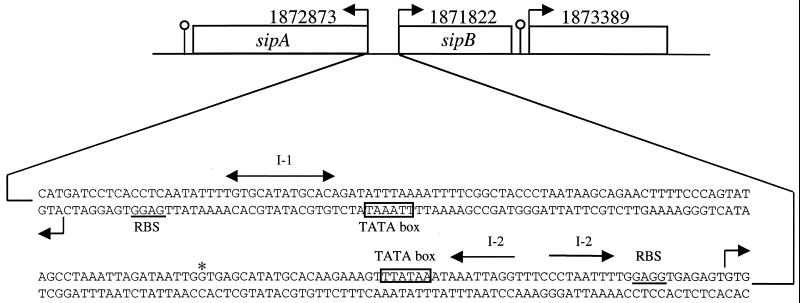
What is now termed sipA was included in the list of ORFs for microarray analysis because its product had been previously identified (from the N-terminal sequence) after gel electrophoresis of P. furiosus extracts. The protein was present in washed membrane preparations of cells grown with S° but was not detected in cells grown without S° (23). SipA is a 19-kDa protein that contains a single Cys residue. Although it is strongly associated with the membrane, sequence analyses do not reveal any transmembrane helix motifs (23). What we now term sipB is currently annotated as a “putative polyferredoxin,” and it contains Cys motifs that could coordinate two [4Fe-4S] clusters. However, the closest homolog (25% sequence identity) to this putative 15-kDa protein is actually the δ-subunit of the P. furiosus enzyme 2-ketoisovalerate ferredoxin oxidoreductase (VOR; see below), a protein that also contains two [4Fe-4S] centers (see reference 41). Interestingly, although sipA and sipB are side by side, they are transcribed in opposite directions. Figure 3 shows a detailed analysis of putative transcriptional and ribosomal binding sites for sipA and sipB, and these strongly suggest that both genes are expressed from a shared promoter domain, as previously shown for the genes encoding β-glucosidase and alcohol dehydrogenase of P. furiosus (72). Homologs of sipA and sipB are present in the genomes of the S°-reducing Pyrococcus species, P. abyssi (PAB1692 and PAB0578), and P. horikoshii (PH1227 and PH0982), but their genes are not arranged back to back such since they are in P. furiosus and SipB is much larger (by ∼15 kDa) in these other two species. In P. furiosus, this SipB 15-kDa extension is encoded by a separate ORF (1873389) that is adjacent to sipB (see Fig. 3). The product of ORF 1873389 shows no significant sequence similarity to any characterized protein. This ORF was included in the microarray analyses, but it is not expressed above the detection limit when cells were grown with or without S° (Table 4). This difference in genome arrangement among the Pyrococcus species might be related to the ability of P. furiosus, but not P. abyssi and P. horikoshii, to grow in the absence of S°.
FIG. 3.
Genome organization of sipA and sipB. The back to back ORFs coding for SipA (179 amino acids) and SipB (115 amino acids) are shown with the indicated directions for transcription. Putative transcription stop sites are indicated with a “ball on a stick,” and putative translational start sites are indicated with right-angled arrows. The putative start sites are preceded with possible ribosomal binding sites (underlined) and TATA boxes (boxed). The start site for sipB shown here is not the same as that given in the genome annotation (which is indicated with an asterisk [54]). Both sipA and sipB contain an inverted repeat (I-1 and I-2) directly next to the proposed TATA boxes. The position of the ORF (1873389) encoding the SipB extension (142 amino acids) is indicated.
Considering the tight regulation of sipA and sipB by S° and the likely electron transfer ability of SipB, it seems reasonable to suggest that SipA and SipB form part of a membrane-associated complex that directly reduces S° to H2S. Two of the other ORFs listed in Table 2 that are strongly upregulated by S°, ORFs 1131551 and 102519, may also be directly involved in S° reduction since both show sequence similarity to the disulfide oxidoreductase family of enzymes. ORF 1131551 is annotated as NADH oxidase (noxA-2) and would encode a 49-kDa cytoplasmic protein that shows significant sequence similarity to NADH peroxidase, a flavoprotein with an active site cysteine (64). ORF 102519 is predicted to encode a 26-kDa cytoplasmic protein that contains four Cys residues (as two -CxxC- motifs). Both ORFs are expressed at near the detection limit in the absence of S° but increase to the “highly expressed” category (>10-fold the detection limit; see below) in S°-grown cells, and neither appears to be part of an operon. Homologs of both ORFs are also present in P. abyssi (PAB0936 and PAB2245) and P. horikoshii (PH0572 and PH0178).
Two of the other S°-induced proteins that might be related to S° metabolism are ORF 1805557, which is annotated as a conserved hypothetical protein, and ORF 1352727, which is annotated as an NADH dehydrogenase (Table 2). The expression of ORF 1805557 changes from below the detection limit to twice the detection limit when cells are grown with S°. This ORF would encode a 27-kDa cytoplasmic protein where the N-terminal part has high sequence similarity to ATP/GTP-binding proteins (13), while the C-terminal half contains six Cys residues, including a CxxCxxxC motif. ORF 1352727 is part of a putative operon containing 13 ORFs, most of which have homology to membrane-bound NADH dehydrogenases. Hagen and coworkers suggested that this operon encodes a (fourth) hydrogenase system in P. furiosus (62), but this seems unlikely in view of the upregulation of ORF 1352727 by S° and the corresponding dramatic decrease in membrane-associated hydrogenase activity (1). Moreover, two other ORFs (1350388 and 1359534) in this putative operon were included in the microarray analysis, and these are also upregulated by S° (three- and fourfold [data not shown]). It is tempting to postulate that ORFs 1805557 and 1352727 (and possibly other ORFs in the putative operon) are involved in nucleotide binding and electron transfer during S° reduction, although further evidence is obviously required to substantiate this.
Four of the remaining six ORFs that are upregulated more than fivefold by S° (Table 2) appear to encode proteins that are involved in amino acid metabolism. These are annotated as an oligopeptide permease (ORF 204761), acetolactate synthase (which is involved in the biosynthesis of branched chain amino acids [ORF 900019]), and as subunits of aspartokinase (ORF 1008251) and tryptophan synthase (ORF 1487371). Although all are putative proteins at present, these data support the relationship between S° and amino acid metabolism that was recently established by growth studies of P. furiosus, in which it was shown that peptides can only serve as the primary carbon source if S° is present (1). All four of these ORFs are expressed at significant levels in the absence of S° (more than twice the detection limit), but all four fall into the highly expressed category (>10-fold the detection limit [see below]) in S°-grown cells. Of the other two upregulated ORFs in Table 2, one is annotated as the thermosome (ORF 1825269). This protein is highly conserved in archaea and is a type II chaperonin, a class that also includes the eukaryotic TCP-1(CCT) proteins (7). The thermosome is expressed at about twice the detection limit in the absence of S°, although why its expression should increase ∼7-fold in the presence of S° is not clear.
The other S°-responsive ORF in Table 2 is annotated as a fibrillarin-like protein (ORF 62257). It shows more than 50% sequence identity to a key eukaryotic nucleolar protein that associates with small nucleolar RNAs (snoRNAs) directing 2′-O-ribose methylation of the rRNA. Homologs of fibrillarin and some of its associated proteins, such as Nop56/58, as well as snoRNAs, have only recently been identified in archaea, although their function in these organisms is not clear (16, 33, 44, 45, 47). In P. furiosus, the ORF encoding the fibrillarin homolog is directly adjacent to a homolog of Nop56/58 (ORF 66216). The microarray data indicate that these ORFs are expressed at twice and six times the detection limit, respectively, in the absence of S°, but their expression increases a further six- and threefold, respectively, when S° is present. Indeed, the microarray experiment included two copies of the ORF encoding the fibrillarin homolog, and both gave remarkably similar results with RNA from cells grown with (an intensity of 22,237 ± 1,074, see Fig. 1B) and without S° (3,590 ± 185). P. furiosus also contains five homologs of a family of RNA m5C methyl transferases (52), and four were included in the microarray analysis (ORFs 1191037, 674979, 179810, and 1197174). All of them are annotated as a nucleolar NOL1-NOP2-sun family protein, but none appear to be regulated by S°. Thus, it is not obvious why S° should cause the genes encoding homologs of eukaryotic nucleolar rRNA processing proteins to be expressed at such high levels. Fibrillarin is also found in archaea that do not metabolize S°, such as methanogens (20), so this protein must have a more general role, albeit one that is associated with S° metabolism, at least in P. furiosus.
Sulfur-independent ORFs.
In the preceding discussion we have considered all 33 of the 271 ORFs that show a >5-fold change in expression when the cells were grown in the presence of S°. In addition to these 33, there are 84 ORFs that showed between a two- to fivefold response to S° (data not shown). At this point further comment on the nature and function of these ORFs seems premature since there are no obvious trends and corroborating experimental data are required. That leaves 154 of the 271 ORFs remaining that show little if any response to S° (<2-fold), and these can be separated into three groups: (i) 20 ORFs that appear to be the most highly expressed, where the transcripts are at >10 times the detection level (> 20,000 intensity units [see Fig. 1B]); (ii) 34 ORFs that appear to be poorly expressed, if at all, where the transcripts are below the detection level (<2,000 intensity units); and (iii) 100 ORFs that appear to be moderately expressed (2,000 to 20,000 intensity units). It should be noted that the relative signal intensities do not necessarily correlate with the degrees of expression of different ORFs because intensity depends on many factors, including PCR product size and efficiencies of labeling and hybridization. However, the results and discussion below show that there may be some merit in this general assumption.
As discussed above, it was anticipated that S° reduction by P. furiosus would involve Mo- or W-containing proteins, analogous to the situation in mesophilic S° reducers. For example, the polysulfide (sulfur) reductase of the mesophilic bacterium, W. succinogenes, is a molybdoenzyme (32). Consequently, the microarray for P. furiosus included numerous ORFs involved in the metabolism of these metals, as well as putative molybdo- and tungstoenyzmes. In fact, the genome contains two ORFs (1175771 and 1419794) that show sequence similarity to members of the ubiquitous molybdenum-containing family of enzymes (22). However, both ORFs are expressed either below or close to the detection limit and show no response to the presence of S° (data not shown). In addition, P. furiosus appears to contain five distinct tungstoenzymes. Three of them have been purified and characterized (abbreviated AOR, GAPOR, and FOR and encoded by ORFs 356987, 478142, and 1145403, respectively), and two are putative (WOR4 and WOR5, encoded by ORFs 1812948 and 1385199, respectively [see reference 57]). FOR, GAPOR, and AOR are in the highly expressed category (>10-fold detection limit [see Table 3]), while WOR4 and WOR5 are moderately expressed and are only slightly upregulated in the presence of S° (data not shown). Similarly, P. furiosus contains several ORFs that would encode homologs of proteins involved in the biosynthesis of pterin (designated moa, mob, and moe [see reference 51]), the cofactor that binds the Mo and W in molybdo- and tungstoenzymes. None of these ORFs showed any significant changes in expression when cells were grown with S°, with the exception of ORF 1805557 (discussed above). In light of these data, it is concluded that S° reduction in P. furiosus does not involve Mo-, W-, or pterin-containing proteins—or at least those that can be identified by sequence similarity to known proteins of this type.
TABLE 3.
Highly expressed S°-independent ORFs
| ORFa | Descriptionb |
|---|---|
| 49183 | Phosphoenolpyruvate synthetase, ppsA (24) |
| 143318 | [Conserved hypothetical protein] |
| 232621 | Enolase (2-phosphoglycerate dehydratase) (49) |
| 236793 | [Hexulose-6-phosphate synthase] |
| 358419 | Aldehyde ferredoxin oxidoreductase, aor (43) |
| 478142 | GAPOR, gor (42) |
| 683389 | [Methylmalonyl-CoA decarboxylase, subunit alpha, mmdA] |
| 720985 | [Alkyl hydroperoxide reductase subunit C] |
| 925374 | POR beta, porB (6) |
| 926380 | POR alpha, porA (6) |
| 927947 | VOR subunit beta, vorB (19) |
| 928888 | VOR subunit alpha, vorA (19) |
| 1145403 | Formaldehyde ferredoxin oxidoreductase, for (57) |
| 1208774 | [LSU ribosomal protein L10] |
| 1210188 | Superoxide reductase, sor (77) |
| 1210814 | [Rubrerythrin] |
| 1425928 | [Ethylene-inducible protein homolog] |
| 1493675 | Glutamate dehydrogenase, gdh (55) |
| 1599202 | Intracellular protease, pfpI (18) |
| 1619038 | [Putative trehalose synthase] |
Some comment is also necessary on the nature of the proteins encoded by the ORFs that are designated as either poorly or highly expressed. For example, the ORF with the highest intensity on the microarray (∼23 times the detection limit) encodes phosphoenolpyruvate synthase (ppsA). This enzyme was recently purified from P. furiosus and was shown to be present at extremely high cellular concentrations (24), a finding consistent with the highly expressed connotation. In fact, of the 20 ORFs that show the highest intensities on the microarray (>10 times the detection limit; see Table 3), 12 of them encode proteins that have been purified from P. furiosus. The biochemical data therefore supports the notion that, in general, these ORFs encode proteins that are indeed present at relatively high cellular concentrations and that signal intensity is a reasonable measure of this. Further evidence for this qualitative relationship comes from the 34 ORFs that are in the poorly expressed category, since none of the proteins that they are proposed to encode have been characterized from P. furiosus, see Table 4 (although one, encoding a putative phosphoglycerate kinase, has been cloned and expressed from the related organism, P. woesei [10]). Of the 100 (of 271) ORFs that are in the moderately expressed category, the products of 12 of them have been characterized (data not shown).
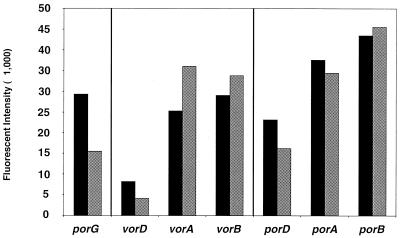
Four of the highly expressed ORFs listed in Table 3 encode the two α-subunits and the two β-subunits of two closely related, heterotetrameric enzymes, POR (6) and VOR (19). Both of these enzymes have been purified from P. furiosus. The genes encoding them form three adjacent operons, in which the third is a common gene encoding their γ subunits (31). The gene arrangement and their transcript intensities in cells with or without S° are shown in Fig. 4. Although the twofold difference in expression of porG is statistically significant (P > 0.95), S° does not dramatically affect the expression of POR and, accordingly, the specific activity of POR is comparable in the two cell types (1). Moreover, while the apparent amounts of the mRNA species encoding the two α-subunits and the two β-subunits are consistent with the high cellular concentrations of the enzymes (1, 6, 19), this is not true for the other subunits and particular for the ORF encoding the δ-subunit of VOR, which is barely above the detection limit. How these three operons are regulated such that they produce two functional enzymes at high cellular concentrations and with a common subunit is not clear from the transcript levels measured by using the DNA microarray. Thus, while there may be some overall relationship between transcript intensities on the microarray slide and cellular concentration, that encoding the δ-subunit of VOR is a notable exception. On the other hand, the biochemical and electrophoretic data strongly support the dramatic downregulation of the expression of the three hydrogenases by S°, and the dramatic upregulation of at least one of the “conserved hypothetical” ORFs (sipA) that are now proposed to form a new type of S°-responsive complex. Further analyses of this type are now required to determine the nature of this complex, whether it is directly involved in S° reduction, and the nature of the effector molecule that mediates the S° response.
FIG. 4.
Expression of the ORFs encoding POR and VOR with or without S°. The genes (A, B, and D) encoding the α-, β-, and δ-subunits of POR and of VOR are arranged according to their positions in their respective operons, along with the gene (G) encoding the γ-subunit that is shared by the two enzymes (31). For each ORF, the signal intensities are indicated for cDNA obtained from cells grown with (solid bars) or without (shaded bars) S°.
ACKNOWLEDGMENTS
This research was funded by grants to M.W.W.A. from the National Institutes of Health (GM 60329), the National Science Foundation (MCB 9904624, MCB 9809060, and BES-0004257), and the Department of Energy (FG05-95ER20175 and contract 992732401 with Argonne National Laboratory) and to J.Z. from the Department of Energy under the Microbial Genome Program and the Natural and Accelerated Bioremediation Research Program of the Office of Biological and Environmental Research. Oak Ridge National Laboratory is managed by the University of Tennessee-Battelle LLC for the Department of Energy under contract DE-AC05-00OR22725.
We thank Gary Li, Scott Lee, and Marc Sudman for their help in preparing the microarray slides; F. Robb and R. Weiss for making the P. furiosus sequence available prior to publication; and Frank E. Jenney, Jr., Angeli Lal Menon, James F. Holden, Eleanor Green, and Rajat Sapra for many helpful discussions.
REFERENCES
- 1.Adams M W W, Holden J F, Menon A L, Schut G J, Grunden A M, Hou C, Hutchins A M, Jenney F E, Jr, Kim C, Ma K, Pan G, Roy R, Sapra R, Story S V, Verhagen M F. Key role for sulfur in peptide metabolism and in regulation of three hydrogenases in the hyperthermophilic archaeon Pyrococcus furiosus. J Bacteriol. 2001;183:716–724. doi: 10.1128/JB.183.2.716-724.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arfin S M, Long A D, Ito E T, Tolleri L, Riehle M M, Paegle E S, Hatfield G W. Global gene expression profiling in Escherichia coli K12. The effects of integration host factor. J Biol Chem. 2000;275:29672–29684. doi: 10.1074/jbc.M002247200. [DOI] [PubMed] [Google Scholar]
- 3.Barbier G, Godfroy A, Meunier J R, Querellou J, Cambon M A, Lesongeur F, Grimont P A, Raguenes G. Pyrococcus glycovorans sp. nov., a hyperthermophilic archaeon isolated from the East Pacific Rise. Int J Syst Bacteriol. 1999;49(Pt. 4):1829–1837. doi: 10.1099/00207713-49-4-1829. [DOI] [PubMed] [Google Scholar]
- 4.Blamey J, Chiong M, Lopez C, Smith E. Optimization of the growth conditions of the extremely thermophilic microorganisms Thermococcus celer and Pyrococcus woesei. J Microbiol Methods. 1999;38:169–175. doi: 10.1016/s0167-7012(99)00092-5. [DOI] [PubMed] [Google Scholar]
- 5.Blamey J M, Adams M W W. Characterization of an ancestral type of pyruvate ferredoxin oxidoreductase from the hyperthermophilic bacterium Thermotoga maritima. Biochemistry. 1994;33:1000–1007. doi: 10.1021/bi00170a019. [DOI] [PubMed] [Google Scholar]
- 6.Blamey J M, Adams M W W. Purification and characterization of pyruvate ferredoxin oxidoreductase from the hyperthermophilic archaeon Pyrococcus furiosus. Biochim Biophys Acta. 1993;1161:19–27. doi: 10.1016/0167-4838(93)90190-3. [DOI] [PubMed] [Google Scholar]
- 7.Bosch G, Baumeister W, Essen L O. Crystal structure of the beta-apical domain of the thermosome reveals structural plasticity in the protrusion region. J Mol Biol. 2000;301:19–25. doi: 10.1006/jmbi.2000.3955. [DOI] [PubMed] [Google Scholar]
- 8.Brown P O, Botstein D. Exploring the new world of the genome with DNA microarrays. Nat Genet. 1999;21:33–37. doi: 10.1038/4462. [DOI] [PubMed] [Google Scholar]
- 9.Bryant F O, Adams M W W. Characterization of hydrogenase from the hyperthermophilic archaebacterium Pyrococcus furiosus. J Biol Chem. 1989;264:5070–5079. [PubMed] [Google Scholar]
- 10.Crowhurst G, McHarg J, Littlechild J A. Phosphoglycerate kinases from bacteria and archaea. Methods Enzymol. 2001;331:90–104. doi: 10.1016/s0076-6879(01)31049-2. [DOI] [PubMed] [Google Scholar]
- 11.de Saizieu A, Certa U, Warrington J, Gray C, Keck W, Mous J. Bacterial transcript imaging by hybridization of total RNA to oligonucleotide arrays. Nat Biotechnol. 1998;16:45–48. doi: 10.1038/nbt0198-45. [DOI] [PubMed] [Google Scholar]
- 12.Duggan D J, Bittner M, Chen Y, Meltzer P, Trent J M. Expression profiling using cDNA microarrays. Nat Genet. 1999;21:10–14. doi: 10.1038/4434. [DOI] [PubMed] [Google Scholar]
- 13.Eaves D J, Palmer T, Boxer D H. The product of the molybdenum cofactor gene mobB of Escherichia coli is a GTP-binding protein. Eur J Biochem. 1997;246:690–697. doi: 10.1111/j.1432-1033.1997.t01-1-00690.x. [DOI] [PubMed] [Google Scholar]
- 14.Edman C F, Mehta P, Press R, Spargo C A, Walker G T, Nerenberg M. Pathogen analysis and genetic predisposition testing using microelectronic arrays and isothermal amplification. J Investig Med. 2000;48:93–101. [PubMed] [Google Scholar]
- 15.Fiala G, Stetter K O. Pyrococcus furiosus sp. nov. represents a novel genus of marine heterotrophic archaebacteria growing optimally at 100°C. Arch Microbiol. 1986;145:56–61. [Google Scholar]
- 16.Gaspin C, Cavaille J, Erauso G, Bachellerie J P. Archaeal homologs of eukaryotic methylation guide small nucleolar RNAs: lessons from the Pyrococcus genomes. J Mol Biol. 2000;297:895–906. doi: 10.1006/jmbi.2000.3593. [DOI] [PubMed] [Google Scholar]
- 17.Gingeras T R, Ghandour G, Wang E, Berno A, Small P M, Drobniewski F, Alland D, Desmond E, Holodniy M, Drenkow J. Simultaneous genotyping and species identification using hybridization pattern recognition analysis of generic Mycobacterium DNA arrays. Genome Res. 1998;8:435–448. doi: 10.1101/gr.8.5.435. [DOI] [PubMed] [Google Scholar]
- 18.Halio S B, Blumentals I I, Short S A, Merrill B M, Kelly R M. Sequence, expression in Escherichia coli, and analysis of the gene encoding a novel intracellular protease (PfpI) from the hyperthermophilic archaeon Pyrococcus furiosus. J Bacteriol. 1996;178:2605–2612. doi: 10.1128/jb.178.9.2605-2612.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heider J, Mai X, Adams M W W. Characterization of 2-ketoisovalerate ferredoxin oxidoreductase, a new and reversible coenzyme A-dependent enzyme involved in peptide fermentation by hyperthermophilic archaea. J Bacteriol. 1996;178:780–787. doi: 10.1128/jb.178.3.780-787.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hickey A J, Macario A J, Conway de Macario E. Identification of genes in the genome of the archaeon Methanosarcina mazeii that code for homologs of nuclear eukaryotic molecules involved in RNA processing. Gene. 2000;253:77–85. doi: 10.1016/s0378-1119(00)00235-3. [DOI] [PubMed] [Google Scholar]
- 21.Hihara Y, Kamei A, Kanehisa M, Kaplan A, Ikeuchi M. DNA microarray analysis of cyanobacterial gene expression during acclimation to high light. Plant Cell. 2001;13:793–806. doi: 10.1105/tpc.13.4.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hille R. The mononuclear molybdenum enzymes. Chem Rev. 1996;96:2757–2816. doi: 10.1021/cr950061t. [DOI] [PubMed] [Google Scholar]
- 23.Holden J F, Poole F L, Tollaksen S L, Giometti C S, Lim H, Yates J R, Adams M W W. Identification of membrane proteins in the hyperthermophilic archaeon Pyrococcus furiosus using proteomics and prediction programs. Comp Funct Genomics. 2001;2:275–288. doi: 10.1002/cfg.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hutchins A M, Holden J F, Adams M W. Phosphoenolpyruvate synthetase from the hyperthermophilic archaeon Pyrococcus furiosus. J Bacteriol. 2001;183:709–715. doi: 10.1128/JB.183.2.709-715.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobi A, Rossmann R, Bock A. The hyp operon gene products are required for the maturation of catalytically active hydrogenase isoenzymes in Escherichia coli. Arch Microbiol. 1992;158:444–451. doi: 10.1007/BF00276307. [DOI] [PubMed] [Google Scholar]
- 26.Jia M H, Larossa R A, Lee J M, Rafalski A, Derose E, Gonye G, Xue Z. Global expression profiling of yeast treated with an inhibitor of amino acid biosynthesis, sulfometuron methyl. Physiol Genomics. 2000;3:83–92. doi: 10.1152/physiolgenomics.2000.3.2.83. [DOI] [PubMed] [Google Scholar]
- 27.Johnson M K, Rees D C, Adams M W W. Tungstoenzymes. Chem Rev. 1996;96:2817–2839. doi: 10.1021/cr950063d. [DOI] [PubMed] [Google Scholar]
- 28.Kawarabayasi Y, Sawada M, Horikawa H, Haikawa Y, Hino Y, Yamamoto S, Sekine M, Baba S, Kosugi H, Hosoyama A, Nagai Y, Sakai M, Ogura K, Otsuka R, Nakazawa H, Takamiya M, Ohfuku Y, Funahashi T, Tanaka T, Kudoh Y, Yamazaki J, Kushida N, Oguchi A, Aoki K, Kikuchi H. Complete sequence and gene organization of the genome of a hyper-thermophilic archaebacterium, Pyrococcus horikoshii OT3 (supplement) DNA Res. 1998;5:147–155. doi: 10.1093/dnares/5.2.147. [DOI] [PubMed] [Google Scholar]
- 29.Kengen S W, Tuininga J E, de Bok F A, Stams A J, de Vos W M. Purification and characterization of a novel ADP-dependent glucokinase from the hyperthermophilic archaeon Pyrococcus furiosus. J Biol Chem. 1995;270:30453–30457. doi: 10.1074/jbc.270.51.30453. [DOI] [PubMed] [Google Scholar]
- 30.Khodursky A B, Peter B J, Cozzarelli N R, Botstein D, Brown P O, Yanofsky C. DNA microarray analysis of gene expression in response to physiological and genetic changes that affect tryptophan metabolism in Escherichia coli. Proc Natl Acad Sci USA. 2000;97:12170–12175. doi: 10.1073/pnas.220414297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kletzin A, Adams M W W. Molecular and phylogenetic characterization of pyruvate and 2-ketoisovalerate ferredoxin oxidoreductases from Pyrococcus furiosus and pyruvate ferredoxin oxidoreductase from Thermotoga maritima. J Bacteriol. 1996;178:248–257. doi: 10.1128/jb.178.1.248-257.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krafft T, Gross R, Kroger A. The function of Wolinella succinogenes psr genes in electron transport with polysulphide as the terminal electron acceptor. Eur J Biochem. 1995;230:601–606. doi: 10.1111/j.1432-1033.1995.0601h.x. [DOI] [PubMed] [Google Scholar]
- 33.Lafontaine D L, Tollervey D. Birth of the snoRNPs: the evolution of the modification-guide snoRNAs. Trends Biochem Sci. 1998;23:383–388. doi: 10.1016/s0968-0004(98)01260-2. [DOI] [PubMed] [Google Scholar]
- 34.Lee H, Greeley G H, Englander E W. Age-associated changes in gene expression patterns in the duodenum and colon of rats. Mech Ageing Dev. 2001;122:355–371. doi: 10.1016/s0047-6374(00)00254-2. [DOI] [PubMed] [Google Scholar]
- 35.Legrain C, Stalon V, Noullez J P, Mercenier A, Simon J P, Broman K, Wiame J M. Structure and function of ornithine carbamoyltransferases. Eur J Biochem. 1977;80:401–409. doi: 10.1111/j.1432-1033.1977.tb11895.x. [DOI] [PubMed] [Google Scholar]
- 36.Legrain C, Villeret V, Roovers M, Gigot D, Dideberg O, Pierard A, Glansdorff N. Biochemical characterisation of ornithine carbamoyltransferase from Pyrococcus furiosus. Eur J Biochem. 1997;247:1046–1055. doi: 10.1111/j.1432-1033.1997.01046.x. [DOI] [PubMed] [Google Scholar]
- 37.Ma K, Schicho R N, Kelly R M, Adams M W W. Hydrogenase of the hyperthermophile Pyrococcus furiosus is an elemental sulfur reductase or sulfhydrogenase: evidence for a sulfur-reducing hydrogenase ancestor. Proc Natl Acad Sci USA. 1993;90:5341–5344. doi: 10.1073/pnas.90.11.5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma K, Weiss R, Adams M W W. Characterization of hydrogenase II from the hyperthermophilic archaeon Pyrococcus furiosus and assessment of its role in sulfur reduction. J Bacteriol. 2000;182:1864–1871. doi: 10.1128/jb.182.7.1864-1871.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mai X, Adams M W W. Indolepyruvate ferredoxin oxidoreductase from the hyperthermophilic archaeon Pyrococcus furiosus. A new enzyme involved in peptide fermentation. J Biol Chem. 1994;269:16726–16732. [PubMed] [Google Scholar]
- 40.Mai X H, Adams M W W. Purification and characterization of two reversible and ADP-dependent acetyl coenzyme-A synthetases from the hyperthermophilic archaeon Pyrococcus furiosus. J Bacteriol. 1996;178:5897–5903. doi: 10.1128/jb.178.20.5897-5903.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Menon A L, Hendrix H, Hutchins A, Verhagen M F J M, Adams M W W. The delta-subunit of pyruvate ferredoxin oxidoreductase from Pyrococcus furiosus is a redox-active, iron-sulfur protein: evidence for an ancestral relationship with 8Fe-type ferredoxins. Biochemistry. 1998;37:12838–12846. doi: 10.1021/bi980979p. [DOI] [PubMed] [Google Scholar]
- 42.Mukund S, Adams M W W. Glyceraldehyde-3-phosphate ferredoxin oxidoreductase, a novel tungsten-containing enzyme with a potential glycolytic role in the hyperthermophilic archaeon Pyrococcus furiosus. J Biol Chem. 1995;270:8389–8392. doi: 10.1074/jbc.270.15.8389. [DOI] [PubMed] [Google Scholar]
- 43.Mukund S, Adams M W W. The novel tungsten-iron-sulfur protein of the hyperthermophilic archaebacterium, Pyrococcus furiosus, is an aldehyde ferredoxin oxidoreductase. Evidence for its participation in a unique glycolytic pathway. J Biol Chem. 1991;266:14208–14216. [PubMed] [Google Scholar]
- 44.Newman D R, Kuhn J F, Shanab G M, Maxwell E S. Box C/D snoRNA-associated proteins: two pairs of evolutionarily ancient proteins and possible links to replication and transcription. RNA. 2000;6:861–879. doi: 10.1017/s1355838200992446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Noon K R, Bruenger E, McCloskey J A. Posttranscriptional modifications in 16S and 23S rRNAs of the archaeal hyperthermophile Sulfolobus solfataricus. J Bacteriol. 1998;180:2883–2888. doi: 10.1128/jb.180.11.2883-2888.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oh M K, Liao J C. Gene expression profiling by DNA microarrays and metabolic fluxes in Escherichia coli. Biotechnol Prog. 2000;16:278–286. doi: 10.1021/bp000002n. [DOI] [PubMed] [Google Scholar]
- 47.Omer A D, Lowe T M, Russell A G, Ebhardt H, Eddy S R, Dennis P P. Homologs of small nucleolar RNAs in Archaea. Science. 2000;288:517–522. doi: 10.1126/science.288.5465.517. [DOI] [PubMed] [Google Scholar]
- 48.Paschos A, Glass R S, Bock A. Carbamoyl phosphate requirement for synthesis of the active center of [NiFe]-hydrogenases. FEBS Lett. 2001;488:9–12. doi: 10.1016/s0014-5793(00)02408-x. [DOI] [PubMed] [Google Scholar]
- 49.Peak M J, Peak J G, Stevens F J, Blamey J, Mai X, Zhou Z H, Adams M W W. The hyperthermophilic glycolytic enzyme enolase in the archaeon Pyrococcus furiousus: comparison with mesophilic enolases. Arch Biochem Biophys. 1994;313:280–286. doi: 10.1006/abbi.1994.1389. [DOI] [PubMed] [Google Scholar]
- 50.Pedroni P, Della Volpe A, Galli G, Mura G M, Pratesi C, Grandi G. Characterization of the locus encoding the [Ni-Fe] sulfhydrogenase from the archaeon Pyrococcus furiosus: evidence for a relationship to bacterial sulfite reductases. Microbiology. 1995;141:449–458. doi: 10.1099/13500872-141-2-449. [DOI] [PubMed] [Google Scholar]
- 51.Rajagopalan K V. Biosynthesis and processing of the molybdenum cofactors. Biochem Soc Trans. 1997;25:757–761. doi: 10.1042/bst0250757. [DOI] [PubMed] [Google Scholar]
- 52.Reid R, Greene P J, Santi D V. Exposition of a family of RNA m(5)C methyltransferases from searching genomic and proteomic sequences. Nucleic Acids Res. 1999;27:3138–3145. doi: 10.1093/nar/27.15.3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Richmond C S, Glasner J D, Mau R, Jin H, Blattner F R. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 1999;27:3821–3835. doi: 10.1093/nar/27.19.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robb F T, Maeder D L, Brown J R, DiRuggiero J, Stump M D, Yeh R K, Weiss R B, Dunn D M. Genomic sequence of hyperthermophile Pyrococcus furiosus: implications for physiology and enzymology. Methods Enzymol. 2001;330:134–157. doi: 10.1016/s0076-6879(01)30372-5. [DOI] [PubMed] [Google Scholar]
- 55.Robb F T, Park J B, Adams M W W. Characterization of an extremely thermostable glutamate dehydrogenase: a key enzyme in the primary metabolism of the hyperthermophilic archaebacterium Pyrococcus furiosus. Biochim Biophys Acta. 1992;1120:267–272. doi: 10.1016/0167-4838(92)90247-b. [DOI] [PubMed] [Google Scholar]
- 56.Ross D T, Scherf U, Eisen M B, Perou C M, Rees C, Spellman P, Iyer V, Jeffrey S S, Van de Rijn M, Waltham M, Pergamenschikov A, Lee J C, Lashkari D, Shalon D, Myers T G, Weinstein J N, Botstein D, Brown P O. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet. 2000;24:227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- 57.Roy R, Menon A L, Adams M W. Aldehyde oxidoreductases from Pyrococcus furiosus. Methods Enzymol. 2001;331:132–144. doi: 10.1016/s0076-6879(01)31052-2. [DOI] [PubMed] [Google Scholar]
- 58.Salama N, Guillemin K, McDaniel T K, Sherlock G, Tompkins L, Falkow S. A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc Natl Acad Sci USA. 2000;97:14668–14673. doi: 10.1073/pnas.97.26.14668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sapra R, Verhagen M F, Adams M W W. Purification and characterization of a membrane-bound hydrogenase from the hyperthermophilic archaeon Pyrococcus furiosus. J Bacteriol. 2000;182:3423–3428. doi: 10.1128/jb.182.12.3423-3428.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schena M, Shalon D, Davis R W, Brown P O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 61.Schönheit P, Schäfer T. Metabolism of hyperthermophiles. World J Microbiol Biotech. 1995;11:26–57. doi: 10.1007/BF00339135. [DOI] [PubMed] [Google Scholar]
- 62.Silva P J, van den Ban E C, Wassink H, Haaker H, de Castro B, Robb F T, Hagen W R. Enzymes of hydrogen metabolism in Pyrococcus furiosus. Eur J Biochem. 2000;267:6541–6551. doi: 10.1046/j.1432-1327.2000.01745.x. [DOI] [PubMed] [Google Scholar]
- 63.Staudt L M, Brown P O. Genomic views of the immune system. Annu Rev Immunol. 2000;18:829–859. doi: 10.1146/annurev.immunol.18.1.829. [DOI] [PubMed] [Google Scholar]
- 64.Stehle T, Ahmed S A, Claiborne A, Schulz G E. Structure of NADH peroxidase from Streptococcus faecalis 10C1 refined at 2.16 Å resolution. J Mol Biol. 1991;221:1325–1344. [PubMed] [Google Scholar]
- 65.Stetter K O. Extremophiles and their adaptation to hot environments. FEBS Lett. 1999;452:22–25. doi: 10.1016/s0014-5793(99)00663-8. [DOI] [PubMed] [Google Scholar]
- 66.Stetter K O. Hyperthermophilic procaryotes. Fems Microbiol Rev. 1996;18:149–158. [Google Scholar]
- 67.Talaat A M, Hunter P, Johnston S A. Genome-directed primers for selective labeling of bacterial transcripts for DNA microarray analysis. Nat Biotechnol. 2000;18:679–682. doi: 10.1038/76543. [DOI] [PubMed] [Google Scholar]
- 68.Tuininga J E, Verhees C H, van der Oost J, Kengen S W, Stams A J, de Vos W M. Molecular and biochemical characterization of the ADP-dependent phosphofructokinase from the hyperthermophilic archaeon Pyrococcus furiosus. J Biol Chem. 1999;274:21023–21028. doi: 10.1074/jbc.274.30.21023. [DOI] [PubMed] [Google Scholar]
- 69.van der Oost J, Schut G, Kengen S W, Hagen W R, Thomm M, de Vos W M. The ferredoxin-dependent conversion of glyceraldehyde-3-phosphate in the hyperthermophilic archaeon Pyrococcus furiosus represents a novel site of glycolytic regulation. J Biol Chem. 1998;273:28149–28154. doi: 10.1074/jbc.273.43.28149. [DOI] [PubMed] [Google Scholar]
- 70.Villeret V, Clantin B, Tricot C, Legrain C, Roovers M, Stalon V, Glansdorff N, Van Beeumen J. The crystal structure of Pyrococcus furiosus ornithine carbamoyltransferase reveals a key role for oligomerization in enzyme stability at extremely high temperatures. Proc Natl Acad Sci USA. 1998;95:2801–2806. doi: 10.1073/pnas.95.6.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Voorhorst W G, Eggen R I, Luesink E J, de Vos W M. Characterization of the celB gene coding for β-glucosidase from the hyperthermophilic archaeon Pyrococcus furiosus and its expression and site-directed mutation in Escherichia coli. J Bacteriol. 1995;177:7105–7111. doi: 10.1128/jb.177.24.7105-7111.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Voorhorst W G, Gueguen Y, Geerling A C, Schut G, Dahlke I, Thomm M, van der Oost J, de Vos W M. Transcriptional regulation in the hyperthermophilic archaeon Pyrococcus furiosus: coordinated expression of divergently oriented genes in response to beta-linked glucose polymers. J Bacteriol. 1999;181:3777–3783. doi: 10.1128/jb.181.12.3777-3783.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wei Y, Lee J M, Richmond C, Blattner F R, Rafalski J A, LaRossa R A. High-density microarray-mediated gene expression profiling of Escherichia coli. J Bacteriol. 2001;183:545–556. doi: 10.1128/JB.183.2.545-556.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wilson M, DeRisi J, Kristensen H H, Imboden P, Rane S, Brown P O, Schoolnik G K. Exploring drug-induced alterations in gene expression in Mycobacterium tuberculosis by microarray hybridization. Proc Natl Acad Sci USA. 1999;96:12833–12838. doi: 10.1073/pnas.96.22.12833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wolf I, Buhrke T, Dernedde J, Pohlmann A, Friedrich B. Duplication of hyp genes involved in maturation of [NiFe] hydrogenases in Alcaligenes eutrophus H16. Arch Microbiol. 1998;170:451–459. doi: 10.1007/s002030050666. [DOI] [PubMed] [Google Scholar]
- 76.Ye R W, Tao W, Bedzyk L, Young T, Chen M, Li L. Global gene expression profiles of Bacillus subtilis grown under anaerobic conditions. J Bacteriol. 2000;182:4458–4465. doi: 10.1128/jb.182.16.4458-4465.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yeh A P, Hu Y, Jenney F E, Jr, Adams M W W, Rees D C. Structures of the superoxide reductase from Pyrococcus furiosus in the oxidized and reduced states. Biochemistry. 2000;39:2499–2508. doi: 10.1021/bi992428k. [DOI] [PubMed] [Google Scholar]
- 78.Yoshida K, Kobayashi K, Miwa Y, Kang C M, Matsunaga M, Yamaguchi H, Tojo S, Yamamoto M, Nishi R, Ogasawara N, Nakayama T, Fujita Y. Combined transcriptome and proteome analysis as a powerful approach to study genes under glucose repression in Bacillus subtilis. Nucleic Acids Res. 2001;29:683–692. doi: 10.1093/nar/29.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zimmer D P, Soupene E, Lee H L, Wendisch V F, Khodursky A B, Peter B J, Bender R A, Kustu S. Nitrogen regulatory protein C-controlled genes of Escherichia coli: scavenging as a defense against nitrogen limitation. Proc Natl Acad Sci USA. 2000;97:14674–14679. doi: 10.1073/pnas.97.26.14674. [DOI] [PMC free article] [PubMed] [Google Scholar]