Abstract
Three transposon Tn5367 mutagenesis vectors (phAE94, pPR28, and pPR29) were used to create a collection of insertion mutants of Mycobacterium bovis strain BCG. A strategy to select for transposon-generated mutants that cannot make coenzyme F420 was developed using the nitroimidazopyran-based antituberculosis drug PA-824. One-third of 134 PA-824-resistant mutants were defective in F420 accumulation. Two mutants that could not make F420-5,6 but which made the biosynthesis intermediate FO were examined more closely. These mutants contained transposons inserted in two adjacent homologues of Mycobacterium tuberculosis genes, which we have named fbiA and fbiB for F420 biosynthesis. Homologues of fbiA were found in all seven microorganisms that have been fully sequenced and annotated and that are known to make F420. fbiB homologues were found in all but one such organism. Complementation of the fbiA mutant with fbiAB and complementation of the fbiB mutant with fbiB both restored the F420-5,6 phenotype. Complementation of the fbiA mutant with fbiA or fbiB alone did not restore the F420-5,6 phenotype, but the fbiA mutant complemented with fbiA produced F420-2,3,4 at levels similar to F420-5,6 made by the wild-type strain, but produced much less F420-5. These data demonstrate that both genes are essential for normal F420-5,6 production and suggest that the fbiA mutation has a partial polar effect on fbiB. Reverse transcription-PCR data demonstrated that fbiA and fbiB constitute an operon. However, very low levels of fbiB mRNA are produced by the fbiA mutant, suggesting that a low-level alternative start site is located upstream of fbiB. The specific reactions catalyzed by FbiA and FbiB are unknown, but both function between FO and F420-5,6, since FO is made by both mutants.
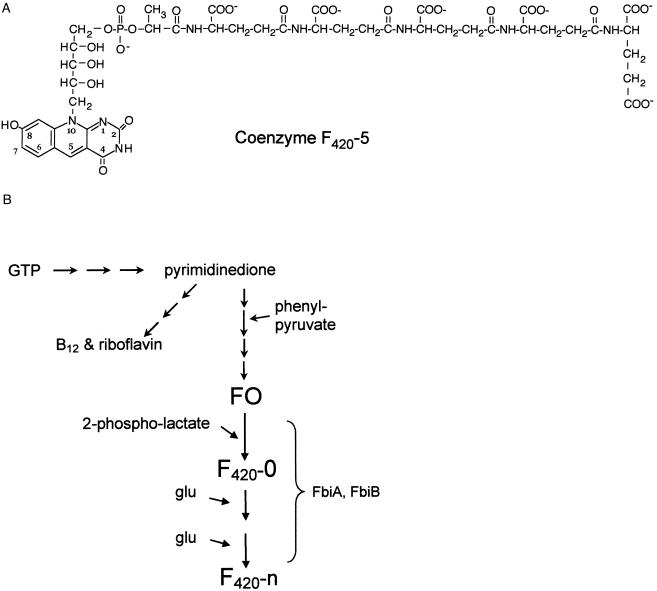
F420 is a 7,8-didemethyl-8-hydroxy 5-deazaflavin electron transfer coenzyme that is present in few microorganisms. It was first described in detail in methanogens (11, 12). Methanobacterium F420 contains two glutamates (12), while Mycobacterium contains primarily five- and six-glutamate forms (F420-5 and F420-6). Figure 1A shows the structure of F420-5 (4). FO is a biosynthetic intermediate that contains the deazaflavin ring and ribityl, but lacks phosphate, lactyl, and glutamate moieties. In archaea, F420 is essential for methylenetetrahydromethanopterin reductase, some hydrogenases, formate and methylenetetrahydromethanopterin dehydrogenases, some alcohol dehydrogenases, and quinone oxidoreductase activities (18, 21, 23, 26, 27, 47). F420 is used by Streptomyces for tetracycline and lincomycin biosynthesis (8, 31, 39) and perhaps in mitomycin C biosynthesis (29). In Mycobacterium and Nocardia, F420 is used by F420-dependent glucose-6-phosphate dehydrogenase (35, 36) and is required for activation of the experimental antituberculosis drug PA-824 by Mycobacterium tuberculosis and Mycobacterium bovis strain BCG (45). The green alga Scenedesmus, the cyanobacterium Synechocystis, and the archaeon Halobacterium use F420 in their photolyase (14, 15, 32).
FIG. 1.
(A) Structure of coenzyme F420-5 from Mycobacterium spp.(4). (B) Overview of the hypothesized pathway for F420 biosynthesis, which is based on our modifications of the reactions proposed by Bacher and Thauer and their colleagues (13, 22, 38), and including the role of 2-phospholactate proposed by Graupner and White (17).
The pathway by which F420 is made has been explored with labeling approaches in methanogens (13, 22, 38). Also, recent enzymatic work with methanogens has shown that 2-phospholactate serves as a precursor of the lactyl and phosphate groups of F420 (17).
A simplified overview of the current hypothetical pathway is shown in Fig. 1B. The genes used for F420 biosynthesis have never been described. Because of our interest in F420 biosynthesis and in the metabolism of M. tuberculosis, we have undertaken a study to determine the genes involved in F420 biosynthesis in Mycobacterium spp. Identification of these genes will enable us to create knockout mutants of M. tuberculosis to study in animal models in order to determine if F420 is important for virulence. We believe that a role for F420 in virulence is possible because preliminary studies have indicated that M. bovis mutants not producing F420 are more sensitive than the wild-type strain to some oxidative stress agents (K.-P. Choi, unpublished observations).
We anticipated that mycobacteria would be more amenable than most other organisms for a study of F420 biosynthesis for three reasons. First, since Streptomyces mutants lacking F420 biosynthetic capabilities can grow without F420 supplementation (8, 31, 39), we expected that F420 would not be required for in vitro growth of the closely related mycobacteria, whereas in several archaea F420 is essential for growth. Thus, with mycobacteria, no supplementation with F420 should be required in the screening medium (F420 is not commercially available and is expensive to produce, e.g., compared to the purchase of NADP.). Second, these organisms are considerably easier to grow than methanogens or Archaeoglobus. Third, several good systems exist for creating Tn5367 insertion mutants in Mycobacterium species (5, 30, 34).
We describe here our development of a strategy to select transposon insertion mutants of M. bovis which cannot make F420. We then demonstrate the utility of this method to identify the M. bovis homologues of the Rv3261 and Rv3262 genes from M. tuberculosis as essential for a step or steps in the pathway between FO and F420-5,6, after the deazaflavin ring is formed.
MATERIALS AND METHODS
Bacteria, plasmids, and culture conditions.
The bacteria and plasmids used in this study are given in Table 1. M. bovis was typically grown in Middlebrook 7H9 liquid medium or on 7H10 agar plates supplemented with 0.05% Tween 80 (liquid medium only), 0.2% glycerol, and Bacto Middlebrook ADC enrichment. Escherichia coli JM109 and pGEM were used as recombinant host and vector, respectively. E. coli was grown at 37°C on Luria-Bertani medium; ampicillin (100 μg/ml), isopropyl-β-d-thiogalactopyranoside (IPTG; 0.5 mM), and 3-indolyl-β-d-galactopyranoside (X-Gal; 80 μg/ml) were incorporated to select for and identify recombinants.
TABLE 1.
Bacterial strains, transposon, plasmids, and DNA primers used in this study
| Strain, transposon, plasmid, or primer | Description or DNA sequence | Source or reference |
|---|---|---|
| Strains | ||
| M. bovis BCG | Montreal M. bovis strain BCG | C. Kendall Stover, PathoGenesis Corp. |
| M. smegmatis mc2 155 | Efficient plasmid transformant mutant (ept-1) | William R. Jacobs Jr., Albert Einstein College of Medicine (44) |
| E. coli JM109 | RecA−, pGEM host strain | Promega, Madison Wis. |
| M. bovis fbiA mutant | M. bovis strain with fbiA::Tn5367 | This study |
| M. bovis fbiB mutant | M. bovis strain with fbiB::Tn5367 | This study |
| Transposon Tn5367 | IS 1096 with kanamycin resistance cassette | Brigitte Gicquel, Institut Pasteur (5, 30, 34) |
| Plasmids and phasmid | ||
| pGEM | T/A PCR cloning vector | Promega, Madison, Wis. |
| pFbiAB | pGEM containing fbiA and fbiB | This study |
| pPR28 | sacB (Ts) delivery plasmid pPR27 with Tn5367 insert | Brigitte Gicquel, Institut Pasteur (34) |
| pPR29 | sacB(Ts) delivery plasmid pPR27 with Tn5367 insert | Brigitte Gicquel, Institut Pasteur (34) |
| phAE94 | Temperature-sensitive shuttle plasmid with a Tn5367 insert; contains oriM(-Ts); no replication at 37°C | William R. Jacobs Jr., Albert Einstein College of Medicine (5) |
| pSMT3 | Mycobacterium complementation vector | Larry Schlesinger (33) |
| p3261 | pSMT3 containing fbiA as 1,150-bp insert | This study |
| p3262 | pSMT3 containing fbiB as 1,454-bp insert | This study |
| p61-62 | pSMT3 containing fbiA and fbiB as 2,425-bp insert | This study |
| Sequencing primers (5′ → 3′) | ||
| T7 | TAATACGACTCACTATAGGG | Sigma-Genosys |
| SP6 | CATACGATTTAGGTGACACTATAG | Sigma-Genosys |
| Primers for inverse and semirandom PCR and RT-PCR (5′ → 3′) | ||
| Tnp2 | GTGGACGTTGTTGCCATGGA | This study |
| Tnp3 | CGCGTTGAGGATTCTTCACC | This study |
| Tnp4 | CCATTAGCTTCTGCAGCAAC | This study |
| HOPS1 | GGCGTAGGAACCTCCATCATC | Bardarov et al. (5) |
| HOPS2 | CTTGCTCTTCCGCTTCTTCTCC | Bardarov et al. (5) |
| YMB1 | GTACGTCACCGTCATCATCG | This study |
| YMB2 | GAGTCACGCTGTCACGTACA | This study |
| Semi-rand 2-1 | GGCCACGCGTCGACTAGTAC-(N)10-GCAGC | This study |
| Semi-rand 4 | (CUA)-GGCCACGCGTCGACTAGTAC | Chun et al. (7) |
| F1 | GAAACTTGGCACGCCATG | This study |
| R2 | CTCGCATTTGGACACCACCTTG | This study |
| F3 | CGATACGTGCCTTTCGGTTATC | This study |
| R4 | CATGGTGTCGGTGATGAC | This study |
| F5 | GGCTGCTAGATCGGATGA | This study |
| R6 | TGTGCAGCCAGGTAAAAAGG | This study |
| 3260-2 | GAATTGATCCGTCGGGAGGTTCT | This study |
| 3260-3 | CTCCGCCTCCTCACTCGATAGA | This study |
| 3260-9 | GTGGACGGATCCAACGTC | This study |
| 3261-1 | GCAATGAGTTGGTGGTCACC | This study |
| 3262-2 | CAAAGATCGTCGAGCCGATC | This study |
| 3260C-1 | CTGGATATCCGATGTGAAGGTCACCGTTCTG | This study |
| 3260C-2 | GTGAAGCTTGAATTGATCCGTCGGGAGGTTG | This study |
| 3261C-2 | CACAAGCTTACCTTGCTGGTAACCACCAC | This study |
| 3262C-1 | CATGATATCGGTGCGACCTTGCGGGAGTGGTA | This study |
| Primers for checking if fgd contained an insert | ||
| FGD-left | TTCTCGCTG(G/T) C(C/G) TGGATGA | This study |
| FGD-right | CGGTTCGCGACCTAGCA | This study |
HPLC.
F420 and FO were separated by high-pressure liquid chromatography (HPLC) using a Beckman System Gold Nouveau 126 HPLC with a Shimadzu RF-10Axl fluorescence detector (excitation at 400 nm, emission at 470 nm). HPLC was performed using a method similar to that of Gorris (16), with modifications. An α-Bondpack C-18 (Supelco) column (3.9 mm by 300 mm) was eluted at 1 ml/min. Buffer A was 27.5 mM sodium acetate (pH 4.7) containing 2% acetonitrile; buffer B was 100% acetonitrile. The portion of buffer B in the elution buffer was varied as follows: 0 to 2 min, 0%; 2 to 6 min, 0 to 2% (linear gradient); 6 to 15 min, 2 to 10%; 15 to 22 min, 28%; 22 to 27 min, 28 to 0%. Extracts of cultures to be examined were prepared by heating approximately equal volumes of a wet cell pellet and buffer A at >90°C for 15 min, followed by centrifugation to remove the pellet.
Molecular biology techniques.
Chromosomal DNA was purified according to Husson et al. (19). Access reverse transcription (RT)-PCR System and Wizard minicolumns for plasmid purification were from Promega (Madison, Wis.). RNA was purified according to Mangan et al. (28) and processed with an RNeasy Kit (Qiagen). Standard E. coli cloning protocols were used (40). Primers are described in Table 1. Sequencing was done at the University of Iowa DNA Facility using an Applied Biosystems 373A sequencer. PA-824 (a nitroimidazopyran used to select F420-minus mutants) was a gift of the PathoGenesis Corporation.
Insertion mutants of M. bovis were created with the phage phAE94 using techniques described by Bardarov et al. (5), but with the incorporation of PA-824 selection. Phage prepared from Mycobacterium smegmatis grown at 30°C was incubated with the target M. bovis. The infected culture was then grown at 37°C on 7H9-ADC agar medium containing 20 μg of kanamycin/ml, which did not allow growth of the phage due to a temperature-sensitive mutation. Colonies from these plates were combined and spread on plates of 7H9-ADC agar with kanamycin (20 μg/ml) and PA-824 (10 μg/ml). Colonies that grew on these plates were examined by PCR for the presence of uninterrupted fgd (fgd codes for F420-dependent glucose-6-phosphate dehydrogenase [35, 36]). Insertion mutants with an intact fgd that survived this selection were further propagated and analyzed by HPLC for the presence of 5-deazaflavins.
Transposon insertions in F420-minus mutants created with phAE94 were identified using a semirandom two-step PCR procedure modified from the original method (7). The primers that corresponded to the known transposon sequence, HOPS1 and HOPS2, were taken from Bardarov et al. (5). The random primer Semi-rand 2–1 was independently developed, but Semi-rand 4 was the same as primer 4 of Chun et al. (7). Semirandom PCR was carried out as described previously (7), but with the #3 buffer from the Boehringer Mannheim Expand Long Template PCR kit, and Platinum Taq polymerase (Gibco).
Insertion mutants were created in M. bovis with plasmid pPR28 or pPR29 using techniques similar to those described by Pelicic et al. (34), but with incorporation of PA-824 selection. pPR28 (1 μg) produced in E. coli JM109 was electroporated into 0.1 ml of electrocompetent M. bovis cells. Electroporated cells were supplemented with 5 ml of 7H9-ADC liquid medium and incubated at 32°C for 1 day. Transformants were selected by growth on 7H9-ADC agar medium containing gentamicin (5 μg/ml) and kanamycin (20 μg/ml) for 4 to 6 weeks at 32°C. One colony was used to inoculate 100 ml of 7H9-ADC-kanamycin medium. After 3 weeks of incubation at 32°C with shaking at 170 rpm, 100 μl of this culture was spread on 7H9-ADC plates containing kanamycin (20 μg/ml), PA-824 (5 μg/ml), and sucrose (2%, wt/vol) and then incubated at 39°C for 3 weeks. Colonies were picked and grown in 7H9-ADC liquid medium.
pPR29 (1 μg) produced in E. coli JM109 was electroporated into 0.4 ml of electrocompetent M. bovis cells. Electroporated cells were supplemented with 4 ml of 7H9-ADC and incubated at 32°C for 1 day. Transformants were selected by growth in 50 ml of 7H9-ADC liquid medium containing gentamicin (5 μg/ml) and kanamycin (20 μg/ml) for 4 to 6 weeks at 32°C. Cultures were centrifuged, the pellet (≈0.3 ml of packed cells) was diluted with 0.1 ml of 10% glycerol in water, and 0.2-ml fractions were spread-plated onto 7H10-ADC agar plates containing kanamycin (20 μg/ml), PA-824 (5 μg/ml), and sucrose (2%, wt/vol). These plates were grown at 39°C for 4 to 6 weeks, and the resultant colonies were used to inoculate 7H10-ADC agar medium or 7H9-ADC liquid medium supplemented with sucrose, kanamycin, and PA-824. Strains mutated by pPR28 or pPR29 that survived this selection were further propagated and analyzed by HPLC for 5-deazaflavin. Dry weights were estimated by measuring the postboiling pellet volume (milliliters) and dividing by 8 ml/g (a conversion factor determined experimentally with M. bovis).
Transposon insertion sites created with pPR28 and pPR29 were found by an inverse PCR technique similar to that described by David Bowtell (http://www.pmci.unimelb.edu.au/manual/molbiol). With inverse PCR, chromosomal DNA from mutants was digested with EagI. Two EagI sites in the transposon of pPR28 and pPR29 allowed a short internal section of the transposon to be excised, and at some distance upstream and downstream of the insertion other EagI sites resulted in chromosomal DNA cleavage. This resulted in two independent segments with EagI-cut ends, with DNA adjacent to the insertion site and either the left or right portion of the transposon. These segments were ligated to create circular DNA. To identify the pPR28 sites, inverse PCR was conducted with primers YMB1 and YMB2. To identify the pPR29 sites, inverse PCR was conducted using TNP3 and TNP4 (for molecules containing the left transposon component) or HOPS1 and TNP2 (for molecules containing the right transposon component). Fragments of approximately 500 to 1,500 bp were usually obtained and were then gel purified and subcloned into pGEM, and one round of single-strand sequencing was conducted.
M. bovis homologues of both Rv3261 and Rv3262 were amplified by Pfu DNA polymerase PCR using primers 3260–2 and 3260–3. A terminal A overhang was introduced onto both ends of this product by Taq polymerase, and the final product was cloned into pGEM to make pFbiAB. Using this clone as a template, PCR products were constructed with an EcoRV site on the 5′ end and a HindIII site on the 3′ end. The insert containing both fbiA and fbiB that was used to create p61–62 was constructed using primers 3260C-1 and 3260C-2. The insert containing fbiA used to create p3261 was constructed using primers 3260C-1 and 3261C-2. The insert containing fbiB used to create p3262 was constructed using primers 3262C-1 and 3260C-2. These inserts were subcloned in frame into pSMT3 after digesting this plasmid with both restriction enzymes. pSMT3 was used as an expression vector to complement insertion mutants (33), using hygromycin-supplemented liquid or agar medium (50 μg/ml for M. bovis).
Electroduction/electroporation to confirm that pSMT3 containing an insert was present in complemented cells was performed by a modification of the method described by O Gaora (33). One M. bovis colony was transferred to 100 μl of 10% glycerol in water along with ≈0.1 ml of 1-mm glass beads in a 1.5-ml screw-cap plastic tube, and the tube was shaken for 30 s in a Mini Beadbeater (Biospec Products, Bartlesville, Okla.) at 5,000 rpm to disrupt clumps but not break all cells. After the beads settled, 50 μl of the supernatant was used for electroporation of E. coli, which were then plated on LB-hygromycin (250 μg/ml) to select transformants. Plasmids prepared from the E. coli were then examined by agarose gel electrophoresis. The remaining supernatant from the Beadbeater was spread on 7H10-ADC medium supplemented with sucrose and kanamycin, and after growth, cells were examined to confirm F420 content.
RT-PCR was used to determine if genes were coexpressed in an operon. The reverse transcriptase reaction was performed with Access RTase at 48°C (45 min) according to instructions provided by the manufacturer. PCR was performed with primers pairs F1 and R2, F3 and R4, and F5 and R6 (Table 1), and the products were analyzed by agarose gel electrophoresis. RT-PCR to determine if the fbiB transcript was made by fbiA::Tn5367 was conducted with primer pairs 3260–9 and 3262–2, and 3261–1 and 3262–2.
Computer analysis of sequences.
Comparison of derived amino acid sequences to the NCBI protein database was performed by the NCBI BlastP (1) and ψ-Blast (2) programs (http://www.ncbi.nlm.nih.gov). Comparison of M. tuberculosis sequences to M. bovis sequences not in the NCBI database was made by the TBLASTN program at the Sanger Centre (http://www.sanger.ac.uk). Genes in the Mycobacterium leprae genome were examined at the Institut Pasteur Leproma site (http://genolist.pasteur.fr). Sequences were aligned using ClustalW (46) or PIMA (43) multiple alignment programs with default settings available at the Baylor College of Medicine Search Launcher (http://dot.imgen.bcm.tmc.edu). Alignments were prepared for examination by shading with the Boxshade program at the Swiss Institute for Experimental Cancer Research (http://www.isrec.isb-sib.ch). Relationships of protein sequences were examined by the NCBI COGs program.
A computer program was written in C++ to examine any genome for sites with high, medium, and low probability for Tn5367 insertion [4(A/T), 3(A/T), and 2(A/T), respectively] (3). This program was used to examine the M. tuberculosis nucleotide sequence (9) at the Sanger Centre web site to identify the frequency and locations of all such sequences in the M. tuberculosis genome and to calculate maximum distances and average distances between high-, medium-, and low-probability sites. To characterize Tn5367 insertion sites, the duplicated 8-bp sequences of all 50 available sequences were analyzed by the WebLogo program (41) (available at http://www.bio.cam.ac.uk/cgi-bin/seqlogo/logo.cgi). Chi square analysis was used to examine the significance of the AT percentages.
RESULTS AND DISCUSSION
Transposon mutagenesis and selection for PA-824-resistant F420-minus mutants of M. bovis.
Using recently reported transposon mutagenesis methods (5, 34), we were confident that we could create insertion mutants of Mycobacterium spp. that would not produce F420. However, it was not obvious how to identify mutants which did not make F420 except to screen thousands of colonies by HPLC. As a rapid alternative to HPLC, we tried direct spectrofluorometric analysis of cell extracts, but the low levels of F420 present compared to the amounts produced by methanogens and the presence of other fluorescent material made this unsuitable for mutant screening (E. Schoenberger, unpublished).
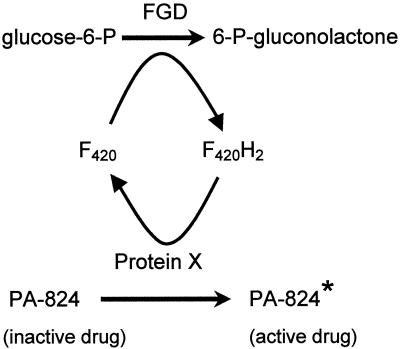
We then encountered an approach to enrich the mutant population for F420-minus mutants using an experimental antituberculosis drug (a nitroimidazopyran, PA-824) (45). F420-dependent glucose-6-phosphate dehydrogenase (FGD) (35, 36) is required to activate PA-824, which can then kill members of the M. tuberculosis complex; other mycobacteria (e.g., M. smegmatis) are not inhibited (45). We developed the hypothesis (Fig. 2) that PA-824 is activated by a reductive reaction that is dependent on reduced F420 (F420H2). Thus, some M. tuberculosis or M. bovis mutants that are resistant to this drug would be defective in FGD activity or unable to produce F420. This allowed us to substantially reduce the number of mutants to be examined by HPLC.
FIG. 2.
Hypothesis for the role of F420 in PA-824 activation. Protein X is a hypothetical enzyme that transfers electrons from F420H2 to PA-824.
The transposon vectors phAE94, pPR28, and pPR29 all successfully transformed M. bovis, giving rise to colonies on plates supplemented with kanamycin and PA-824. While the phAE94 method was initially easier and less susceptible to generating sibling mutants, the plasmid method proved superior due to its ability to routinely generate large numbers of mutants. We examined cell extracts from 134 cultures derived from these colonies by HPLC and compared F420 and FO levels to those in the wild-type strain. FO levels were considerably lower than F420 levels, and in some cases it was difficult to determine conclusively if FO was absent. About one-third (n = 48) of the PA-824-resistant mutants were defective in F420 accumulation.
Identification of genes interrupted in F420-minus mutants.
With either inverse PCR or semirandom two-step PCR, we located transposon insertion sites in PA-824-resistant mutants. Among 48 PA-824-resistant F420-minus mutants, we identified 31 distinct genes with a transposon insertion. Several genes were found more than once due to independent insertions or siblings. Insertions in 19 genes were found in mutants which had an F420-minus and FO-minus phenotype, and interruptions in 12 genes were found in mutants with an F420-minus and FO-plus phenotype.
Characteristics of Tn5367 insertion sites.
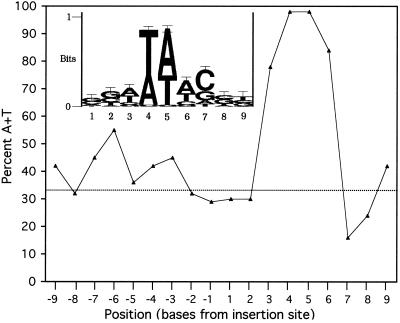
As shown in Fig. 3, examination of our transposon insertion sites and sites reported by others (5, 30, 34) showed that in the 8-bp duplicated region, positions 4 and 5 are nearly 100% A or T and positions 3 and 6 are very high in A+T; this is a significant deviation from the 34% A+T genome content (α < 0.001). Position 7 is G+C rich (16% A+T; α < 0.005). The inset sequence logo graph portrays the approximately equal amounts of A and T at positions 3 to 6 and the predominance of C at position 7. This agrees with the general observations (that the duplicated region was AT-rich) by others who had each examined a smaller number of insertions (5, 30, 34).
FIG. 3.
Sequence characteristics of the Tn5367 transposon insertion sites. In the large graph, percent A+T at each position to the right (positive values) and to the left (negative values) of the insertion site is given for nine bases on both sides of the 31 gene insertion sites observed in our experiments and the 8 duplicated bases (positive values 1 to 8) for an additional 19 insertion sites reported by others (5, 30, 34). The horizontal dotted line indicates the A+T content of the M. tuberculosis genome (34.4%). The inset graph provides a sequence logo plot to graphically emphasize the incidence of individual base types, if they predominate at positions 1 to 9.
The sequence on the other side of the insertion site (the unduplicated region corresponding to positions −1 through −9), for which we have data for only our mutants, was not conserved; it was not AT-rich (Fig. 3), nor did any bases predominate (data not shown). We cannot confidently conclude if position −6 is GC rich (α < 0.025).
We wondered if this transposon would insert into most mycobacterial genes, which have a G+C content of 65.6%. We examined the M. tuberculosis H37Rv genome with a computer program to determine how many genes had sites that Tn5367 would insert into with high probability (four adjacent A's or T's) or with a moderate probability (three adjacent A's or T's) (3). Of the 3,924 genes in the genome, 103 (2.6%) do not contain at least one high-probability site, but all have a moderate-probability site. All of our 31 transposon-inserted genes had a high-probability site. We conclude that, although this transposon is not truly random for insertion in mycobacteria, insertion is very likely in >97% of M. tuberculosis and M. bovis genes, and ultimately all genes will be interrupted if a large number of mutants are examined.
M. bovis mutants with an insertion in Rv3261 or Rv3262 homolog.
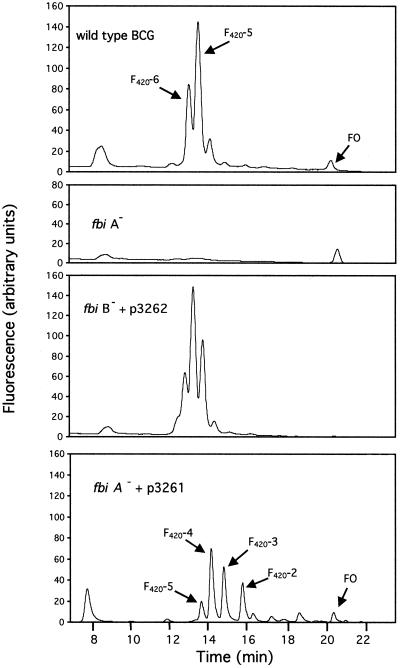
Of 18 genes interrupted by pPR29 which gave an F420-minus phenotype, one each had an independent insertion in the adjacent M. bovis homolog of M. tuberculosis H37Rv sequences Rv3261 (790 bp downstream from the start codon) and Rv3262 (801 bp downstream from the start codon), as shown in Fig. 4. We have named these genes fbiA and fbiB, respectively, for F420 biosynthesis. These mutants made no F420 but did make FO. An example of the HPLC profile of the fbiA mutant is given in Fig. 5, in comparison with the wild-type strain. The fbiB mutant had about the same HPLC profile as the fbiA mutant at the scale shown in Fig. 5 (data not shown). Since FO is produced by both mutants, the products of these genes were expected to be involved in steps between FO and F420-5,6 (Fig. 1B).
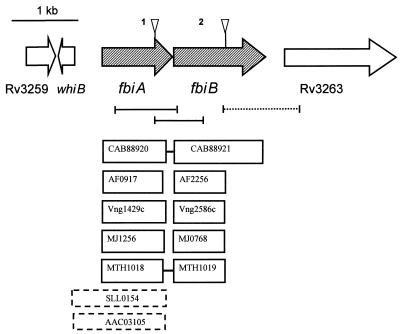
FIG. 4.
Gene arrangement of the fbiAB cluster in M. bovis BCG, corresponding transposon insertion sites, and alignment of these genes with homologs from other microorganisms. Triangles indicate transposon insertion sites. CAB, Streptomyces coelicolor; MTH, Methanobacterium thermoautotrophicum; AF, Archaeoglobus fulgidus; Vng, Halobacterium sp; SLL, Synechocystis sp.; AAC, Nostoc sp. Numbers refer to gene designations in the corresponding genome sequences. Rv3259 and Rv3263 are used to indicate the M. bovis homologs of these M. tuberculosis H37Rv genes, since the M. bovis sequence is not yet annotated, and thus M. bovis genes have not been assigned unique designations. The lines below the M. bovis genes indicate the sizes and locations of RT-PCR products expected; a solid line indicates that a product was seen, and a dotted line indicates that no product was observed. The primer pairs used were (from left to right) F1-R2, F3-R4, and F5-R6. Boxes with solid lines indicate genes identified with default NCBI Blast settings. Boxes with dashed lines indicate genes identified with ψ-Blast iterations. Boxes connected by bars indicate the genes are adjacent, and those not connected are not adjacent. Figure is approximately to scale, with boxes indicating the sizes of all putative homologous genes.
FIG. 5.
HPLC elution profiles of extracts made from M. bovis BCG wild-type, the fbiA mutant (Rv3261 homolog interrupted), the fbiB mutant (Rv3262 homolog interrupted) that was complemented with p3262, and the fbiA mutant that was complemented with p3261.
We cloned and sequenced a 2,718-bp region from M. bovis containing fbiA and fbiB, creating pFbiAB. Our sequences for both genes precisely matched the Rv3261 and Rv3262 sequences and the Sanger database M. bovis homologs of these genes. As shown in Table 2, Blast analysis of the deduced sequence of FbiA against the NCBI database revealed that this gene has homologs in all fully sequenced microorganisms which make F420, including M. tuberculosis (9), M. leprae (10), Archaeoglobus fulgidis (25), Halobacterium sp. (32), Methanococcus jannaschii (6), Methanobacterium thermoautotrophicum (42), and Synechocystis sp. (24). However, the Synechocystis homolog was not identified by default BlastP settings, but was identified by ψ-Blast (2).
TABLE 2.
Blast analysis results when M. bovis BCG FbiA and FbiB were compared to homologs from other F420 producersa
| FbiA
|
FbiB
|
||||||
|---|---|---|---|---|---|---|---|
| Homolog (organism) | Score | E value | % identity | Homolog (organism) | Score | E value | % identity |
| Rv3261 (M. tuberculosis) | 575 | 1 × 10−163 | 89 | Rv3262 (M. tuberculosis) | 655 | 0 | 81 |
| ML0759 (M. leprae) | 450 | 1 × 10−125 | 70 | ML0758 (M. leprae) | 531 | 1 × 10−150 | 65 |
| CAB88920 (Streptomyces) | 315 | 2 × 10−85 | 52 | CAB88921 (Streptomyces) | 251 | 9 × 10−66 | 39 |
| AF0917 (Archaeoglobus) | 126 | 3 × 10−28 | 30 | AF2256 (Archaeoglobus) | 84 | 2 × 10−15 | 30 |
| MJ1256 (Methanococcus) | 112 | 5 × 10−24 | 25 | MJ0768 (Methanococcus) | 78 | 1 × 10−13 | 26 |
| Vng1429c (Halobacterium) | 96 | 5 × 10−19 | 27 | Vng2586c (Halobacterium) | 73 | 5 × 10−12 | 31 |
| MTH1018 (Methanobacterium) | 83 | 4 × 10−15 | 26 | MTH1019 (Methanobacterium) | 65 | 1 × 10−9 | 27 |
| SLL0154 (Synechocystis) | ψ272 | ψ3 × 10−72 | 17 | ||||
| AAC03105 (Anabaena) | ψ268 | ψ6 × 0−71 | 17 | ||||
Default BlastP analysis of FbiA and FbiB sequences versus NCBI database, using a low-complexity filter, except that ψ-BlastP iteration was used for identification of Synechocystis and Anabaena genes. The low-complexity filter results in a conservative identity score, since sequences of low complexity are counted as not identical. Without the filtering, identity is usually higher; e.g., the M. tuberculosis and M. bovis sequences are 100% identical but are given as 89 and 81% identical in the BlastP results. Original papers describing genome or partial sequences: M. tuberculosis (9), M. leprae (10), S. coelicolor (37), A. fulgidis (25), Halobacterium sp. (32), Methanococcus jannaschii (6), Methanobacterium thermoautotrophicum (42), Synechocystis sp. (24), and Nostoc sp. (also known as Anabaena; NCBI database; data submitted by H.-S. Yoon and J. W. Golden).
When MTH1018 was analyzed by BlastP against Synechocystis alone, a weak hit was obtained with SLL0154 (score = 44; E = 10−5). The ψ-Blast analysis also revealed a homolog from a Nostoc sp. (also known as Anabaena), which may contain F420 (since it is a cyanobacterium), although the genome of this organism has not been sequenced. We are cautiously optimistic that these weak hits in cyanobacteria have identified true functional homologs of FbiA, but will wait for further supportive data (e.g., complementation of our fbiA mutant with Synechococcus fbiA and the M. bovis fbiB on the same plasmid) before reaching a final decision.
Strong FbiA hits were also obtained with Streptomyces coelicolor (37), another F420 producer for which the genome sequence is largely completed but not yet annotated. Figure 4 provides an overview of the arrangement of these homologs (the M. leprae homolog, ML0759, which was the same size as Rv3261, is listed in Table 2 but was omitted from Fig. 4 to conserve space). Homologs for FbiB were found in all sequenced F420-producing organisms except Synechocystis (Table 2 and Fig. 4). The Mycobacterium, Streptomyces, and Methanobacterium fbiA and fbiB homologs were adjacent, while in the other organisms these two genes were distant from each other. M. tuberculosis and M. bovis have exactly the same gene arrangement in the region shown in Fig. 4. M. leprae has a very similar arrangement, except that another open reading frame (ORF) (ML0756) was identified between fbiB and the Rv3263 homolog.
Transcription in the fbiAB cluster.
As shown in Fig. 4, the whiB gene upstream of fbiA is transcribed in the opposite direction. Downstream of fbiB there is a 296- or 371-bp gap (depending on the start codon chosen) before the start of Rv3263. Downstream of Rv3263, rmlA2 (not shown in Fig. 4) is transcribed in the opposite direction. This arrangement left open the possibility that fbiA, fbiB, and Rv3263 were transcribed together or that some or all were transcribed independently. Thus, we used RT-PCR to examine transcription from these three genes.
As shown in Fig. 4, mRNA species existed which coded for significant sections of both fbiA and fbiB. The sizes of the amplified sections within these two genes were as predicted. However, no product was obtained when primers were used to see if mRNA arising from both fbiB and Rv3263 was present (Fig. 4). This supports the concept that fbiA and fbiB are expressed as an operon which does not include Rv3263. We also used RT-PCR to examine mRNA production by the fbiA mutant, since it was possible that significant fbiB transcripts could result from a promoter upstream of fbiB. Using mRNA at the same concentration as above, we saw no fbiB transcript, but when the mRNA level was increased 10-fold, a weak but significant band of the correct size was observed. This suggests that although most fbiB transcription occurs from a promoter upstream of fbiA, a small but possibly significant amount of monocistronic fbiB transcription may occur from an alternative site.
Complementation of fbiA and fbiB mutants.
Using complementation, we confirmed that interruption of these genes was responsible for the F420-minus phenotype. PCR of the cloned M. bovis fbi genes yielded subclones containing each gene separately (p3261 and p3262) and one clone containing both genes (p61–62). The strong mycobacterial promoter in pSMT3 located upstream of the cloned gene(s) ensured adequate expression. Electroduction confirmed that pSMT3 in complemented cells contained an insert of the correct size.
Figure 5 shows examples of the HPLC profiles of extracts from the wild-type strain, the fbiA mutant, the fbiB mutant complemented with p3262, and the fbiA mutant complemented with p3261. Transformation of the fbiA mutant with p61–62 (HPLC data not shown) and of the fbiB mutant with p3262 (HPLC data shown in Fig. 5) resulted in full restoration of F420-5,6 production, as shown quantitatively in Table 3 for both complemented mutants. The fbiA mutant complemented with p3262 did not make F420-5,6 (Table 3); FO was the only significant 5-deazaflavin peak observed.
TABLE 3.
F420 production by wild-type M. bovis BCG, by fbiA and fbiB mutants, and by mutants complemented with these genesa
| Complementation status | F420-5,6 level (pmol/culture)
|
F420-2,3,4 level (pmol/culture)
|
||||
|---|---|---|---|---|---|---|
| Wild type | fbiA mutant | fbiB mutant | Wild type | fbiA mutant | fbiB mutant | |
| Uncomplemented | 448 | 6 | 5 | 83 | 4 | 15 |
| Control with pSMT3 | NDb | 22 | 8 | ND | 15 | 7 |
| Complemented (p3261) | ND | 62 | 6 | ND | 438 | 6 |
| Complemented (p3262) | ND | 16 | 363 | ND | 12 | 201 |
| Complemented (p61-62) | ND | 464 | ND | ND | 136 | ND |
Cultures were of approximately the same packed cell volume (about 200 mg [wet weight] of cells) taken from a 100-ml liquid culture or the surface of one petri plate. M. bovis wild-type F420 production in this experiment was 0.02 nmol per mg of dry cell weight.
ND, not determined.
However, as shown in Table 3, complementation of the fbiA mutant with p3261 led to a slight but significant production of F420-5,6 (comprised only of F420-5). As shown in Fig. 5, this complementation also led to the accumulation of compounds that are almost certainly F420-2,3,4 (F420-2, F420-3, and F420-4), since coinjection of standard F420-2 and F420-5,6 showed precisely coincident elution with putative F420-2 and F420-5, and the fluorescent properties of these peaks are consistent with their being F420 species. Prior experience with F420-4 also agrees with this conclusion (4). It is possible that smaller peaks at 17 to 20 min include F420-0 and F420-1. F420-2,3,4 peaks of this magnitude were not observed in the uncomplemented mutants or in mutants complemented with p3262 or p61–62, but the wild-type strain and some of the mutants often showed small peaks in this region that were about one-tenth (or less) of the size seen in the fbiA mutant complemented with p3261. The appearance of several F420 species with fewer than five or six glutamates also suggests that glutamates are not added exclusively as an already assembled five- or six-glutamate piece, but are instead added individually, at least after F420-2 is formed.
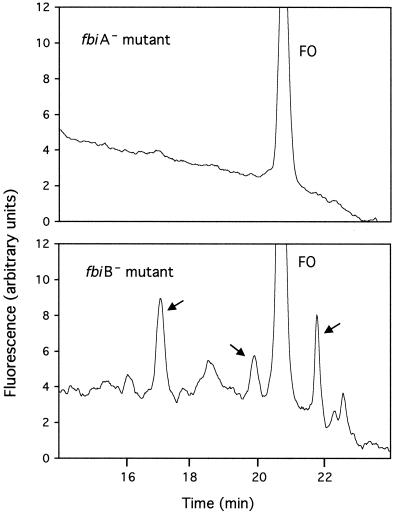
The discovery of intermediates in the p3261-complemented fbiA mutant led us to more carefully examine the HPLC profiles of the fbiA and fbiB mutants. As shown in Fig. 6, aside from FO, the uncomplemented fbiB mutant accumulated material showing up as three fluorescent HPLC peaks (indicated by arrows) that eluted between 16 and 22 min. The peaks at 18.5 and 22.5 min were not always seen in the fbiB mutant. Note that the scale in Fig. 6 is considerably magnified compared to Fig. 5, allowing these peaks to be seen. The three fbiB mutant peaks were not seen with the fbiA mutant. Peaks eluting in this region are expected to contain no glutamate, and thus these peaks could arise from F420-0 or closely related structures. This would be consistent with a role for FbiA in making F420-0 (F0-phosphate-lactyl), and with the inability of the fbiB mutant to add glutamates to F420-0.
FIG. 6.
HPLC elution profiles of extracts made from the fbiA mutant and the fbiB mutant. Compare the scale to the HPLC data in Fig. 5; the scale here is enhanced by 13.3-fold.
The complementation data clearly show that FbiB is essential, since replacing the inactive gene with an active one fully restores the original phenotype. The complication in interpreting these data is the behavior of the fbiA mutant which has been complemented with p3261. If the genes are monocistronic, we would expect to see full restoration of F420-5,6 production when the fbiA mutant was complemented with p3261, but instead, although a little F420-5 was made, mostly the smaller species were made. If the two genes were entirely transcribed as an operon from a promoter or promoters upstream of fbiA, then we should see no F420-2,3,4,5 made when the fbiA mutant is complemented with p3261, since no FbiB could be produced. A reasonable explanation of these data, in light of the transcriptional data, is that although the fbiA mutant makes no FbiA, it can make a small amount of FbiB due to a leaky polar effect, i.e., a small amount of fbiB transcript is made from an alternative start site just upstream of fbiB.
Consideration of the roles of FbiA and FbiB.
Since the mutated strains make FO, FbiA and FbiB must play roles in the later portion of the pathway shown in Fig. 1B, between FO and F420-n. This portion of the pathway involves the construction, by mostly unknown mechanisms, of a side group that is composed of a phosphate, a lactyl, and five or six glutamyl groups. Although it would be logical to initially add a phosphate to FO by an FO kinase (in analogy to flavin mononucleotide biosynthesis), our repeated efforts to demonstrate such a reaction using ATP or GTP have been unsuccessful (20). Recent work with methanogens has demonstrated that 2-phospholactate is a precursor of the phosphate and lactyl groups of F420 (17), suggesting that FO kinase may not be present in mycobacteria. It also makes sense that an enzyme transfers individual glutamyl groups to F420-0 (since we see intermediates with two, three, four, five, and six glutamyl groups), but we have not sought such an enzyme yet.
We have examined protein sequence databases with several search methods (COGS, BlastP, and ψ-Blast) to deduce possible functions for FbiA and FbiB and have conducted multiple alignments of FbiA and FbiB with homologs from other F420 producers. None of these methods provided firm support for assignment of specific functions to FbiA and FbiB, which is not surprising because these enzymes have never been studied. However, taken together, our data suggest three hypotheses. (i) FbiA is involved in making F420-0, and FbiB participates in the addition of glutamates to F420-0. (ii) FbiA and FbiB are subunits of one enzyme which adds glutamates to F420-0; neither protein is active in glutamyl transfer by itself. (iii) FbiB is involved in making F420-0, and FbiA adds glutamates to F420-0. Our data cannot confidently distinguish between the three hypotheses. Nonetheless, we are most attracted to hypothesis i, that FbiA participates in the conversion of FO to F420-0, and that FbiB plays a role in the addition of glutamates. This hypothesis is consistent with intermediate accumulation in the fbiB mutant, a lack of intermediate accumulation in the fbiA mutant, and a leaky polar effect seen with the fbiA-complemented fbiA mutant. Development of assays in Mycobacterium species and with overexpressed FbiA and FbiB for the 2-phospholactate and glutamyl transfer steps will be very useful for examination of these hypotheses.
ACKNOWLEDGMENTS
We thank C. Kendall Stover, Paul Warrener, David Sherman, and Ying Yuan of the PathoGenesis Corporation for the gift of PA-824, an expensive material that is not commercially available. We thank the Brigitte Gicquel-Vladimir Pelicic group at the Institut Pasteur, Paris, the William Jacobs Jr. HHMI group at the Albert Einstein College of Medicine for providing phAE94, pPR28, and pPR29, and Larry Schlesinger at Iowa for providing pSMT3. We are very appreciative of the excellent technical work contributed by Seong-Ae Kang. We thank Elena Schoenberger for screening for F420-minus mutants of M. smegmatis. We thank Robert White and Marion Graupner for making us aware of their work on the role of 2-phospholactate in F420 biosynthesis prior to publication. We appreciate advice from Frances Ufkes on statistical analysis of our insertion sites.
This work was supported by National Institutes of Health grant GM56177 and U.S. Department of Agriculture grant 4132008 to L.D.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bair T B. Methods to identify genes involved in F420 biosynthesis by Mycobacterium. Ph.D. thesis. Iowa City: University of Iowa; 2001. [Google Scholar]
- 4.Bair T B, Isabelle D W, Daniels L. Structures of coenzyme F420 in Mycobacterium species. Arch Microbiol. 2001;176:37–43. doi: 10.1007/s002030100290. [DOI] [PubMed] [Google Scholar]
- 5.Bardarov S, Kriakov J, Carriere C, Yu S, Vaamonde C, McAdam R A, Bloom B R, Hatfull G F, Jacobs W R., Jr Conditionally replicating mycobacteriophages: a system for transposon delivery to Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 1997;94:10961–10966. doi: 10.1073/pnas.94.20.10961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Weidman J F, Fuhrmann J L, Presley E A, Nguyen D, Utterback T R, Kelley J M, Peterson J D, Sadow P W, Hanna M C, Cotton M D, Hurst M A, Roberts K M, Kaine B P, Borodovsky M, Klenk H-P, Fraser C M, Smith H O, Woese C R, Venter J C. Complete genome sequence of the methanogenic archaeon Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 7.Chun K T, Edenberg H J, Kelley M R, Goebl M G. Rapid amplification of uncharacterized transposon-tagged DNA sequences from genomic DNA. Yeast. 1997;13:233–240. doi: 10.1002/(SICI)1097-0061(19970315)13:3<233::AID-YEA88>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 8.Coats J H, Li G P, Kuo M S, Yurek D A. Discovery, production, and biological assay of an unusual flavenoid cofactor involved in lincomycin biosynthesis. J Antibiot (Tokyo) 1989;42:472–474. doi: 10.7164/antibiotics.42.472. [DOI] [PubMed] [Google Scholar]
- 9.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry C E, 3rd, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail M A, Rajandream M A, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston J E, Taylor K, Whitehead S, Barrell B G. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 10.Cole S T, Eiglmeier K, Parkhill J, James K D, Thomson N R, Wheeler P R, Honore N, Garnier T, Churcher C, Harris D, Mungall K, Basham D, Brown D, Chillingworth T, Connor R, Davies R M, Devlin K, Duthoy S, Feltwell T, Fraser A, Hamlin N, Holroyd S, Hornsby T, Jagels K, Lacroix C, Maclean J, Moule S, Murphy L, Oliver K, Quail M A, Rajandream M A, Rutherford K M, Rutter S, Seeger K, Simon S, Simmonds M, Skelton J, Squares R, Squares S, Stevens K, Taylor K, Whitehead S, Woodward J R, Barrell B G. Massive gene decay in the leprosy bacillus. Nature. 2001;409:1007–1011. doi: 10.1038/35059006. [DOI] [PubMed] [Google Scholar]
- 11.Eirich L D, Vogels G D, Wolfe R S. Distribution of coenzyme F420 and properties of its hydrolytic fragments. J Bacteriol. 1979;140:20–27. doi: 10.1128/jb.140.1.20-27.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eirich L D, Vogels G D, Wolfe R S. Proposed structure for coenzyme F420 from Methanobacterium. Biochemistry. 1978;17:4583–4593. doi: 10.1021/bi00615a002. [DOI] [PubMed] [Google Scholar]
- 13.Eisenreich W, Schwarzkopf B, Bacher A. Biosynthesis of nucleotides, flavins, and deazaflavins in Methanobacterium thermoautotrophicum. J Biol Chem. 1991;266:9622–9631. [PubMed] [Google Scholar]
- 14.Eker A P, Hessels J K C, van de Velde J. Photoreactivating enzyme from the green alga Scenedesmus acutus. Evidence for the presence of two different flavin chromophores. Biochemistry. 1988;27:1758–1765. [Google Scholar]
- 15.Eker A P, Kooiman P, Hessels J K, Yasui A. DNA photoreactivating enzyme from the cyanobacterium Anacystis nidulans. J Biol Chem. 1990;265:8009–8015. [PubMed] [Google Scholar]
- 16.Gorris L G M, van der Drift C. Separation and quantitation of cofactors from methanogenic bacteria by high-performance liquid chromatography: optimum and routine analysis. J Microbiol Methods. 1988;8:175–190. [Google Scholar]
- 17.Graupner M, White R H. Biosynthesis of the phosphodiester bond in coenzyme F420 in the Methanoarchaea. 2001. Biochemistry, in press. [DOI] [PubMed] [Google Scholar]
- 18.Hartzell P L, Zvilius G, Escalante-Semerena J C, Donnelly M I. Coenzyme F420 dependence of the methylenetetrahydromethanopterin dehydrogenase of Methanobacterium thermoautotrophicum. Biochem Biophys Res Commun. 1985;133:884–890. doi: 10.1016/0006-291x(85)91218-5. [DOI] [PubMed] [Google Scholar]
- 19.Husson R N, James B E, Young R A. Gene replacement and expression of foreign DNA in mycobacteria. J Bacteriol. 1990;172:519–524. doi: 10.1128/jb.172.2.519-524.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Isabelle D W. Coenzyme F420 biosynthesis in Mycobacterium smegmatis. M.S. thesis. Iowa City: University of Iowa; 2001. [Google Scholar]
- 21.Jacobson F S, Daniels L, Fox J A, Walsh C T, Orme-Johnson W H. Purification and properties of an 8-hydroxy-5-deazaflavin-reducing hydrogenase from Methanobacterium thermoautotrophicum. J Biol Chem. 1982;257:3385–3388. [PubMed] [Google Scholar]
- 22.Jaenchen R, Schonheit P, Thauer R K. Studies on the biosynthesis of coenzyme F420 in methanogenic archaea. Arch Microbiol. 1984;137:362–365. doi: 10.1007/BF00410735. [DOI] [PubMed] [Google Scholar]
- 23.Jones J B, Stadtman T C. Reconstitution of a formate-NADP+ oxidoreductase from formate dehydrogenase and a 5-deazaflavin-linked NADP+ reductase isolated from Methanococcus vannielii. J Biol Chem. 1980;255:1049–1053. [PubMed] [Google Scholar]
- 24.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 25.Klenk H P, Clayton R A, Tomb J F, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, Richardson D L, Kerlavage A R, Graham D E, Kyrpides N C, Fleischmann R D, Quackenbush J, Lee N H, Sutton G G, Gill S, Kirkness E F, Dougherty B A, McKenney K, Adams M D, Loftus B, Venter J C. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 26.Kunow J, Linder D, Stetter K O, Thauer R K. F420H2: quinone oxidoreductase from Archaeoglobus fulgidus: characterization of a membrane-bound multisubunit complex containing FAD and iron-sulfur clusters. Eur J Biochem. 1994;223:503–511. doi: 10.1111/j.1432-1033.1994.tb19019.x. [DOI] [PubMed] [Google Scholar]
- 27.Ma K, Thauer R K. Purification and properties of N5,N10-methylenetetrahydromethanopterin reductase from Methanobacterium thermoautotrophicum (strain Marburg) Eur J Biochem. 1990;191:187–193. doi: 10.1111/j.1432-1033.1990.tb19109.x. [DOI] [PubMed] [Google Scholar]
- 28.Mangan J A, Sole K M, Mitchison D A, Butcher P D. An effective method of RNA extraction from bacteria refractory to disruption, including mycobacteria. Nucleic Acids Res. 1997;25:675–676. doi: 10.1093/nar/25.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mao Y, Varoglu M, Sherman D H. Molecular characterization and analysis of the biosynthetic gene cluster for the antitumor antibiotic mitomycin C from Streptomyces lavendulae NRRL 2564. Chem Biol. 1999;6:251–263. doi: 10.1016/S1074-5521(99)80040-4. [DOI] [PubMed] [Google Scholar]
- 30.McAdam R A, Weisbrod T R, Martin J, Scuderi J D, Brown A M, Cirillo J D, Bloom B R, Jacobs W R., Jr In vivo growth characteristics of leucine and methionine auxotrophic mutants of Mycobacterium bovis BCG generated by transposon mutagenesis. Infect Immun. 1995;63:1004–1012. doi: 10.1128/iai.63.3.1004-1012.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCormick J R D, Morton G O. Identity of cosynthetic factor 1 of Streptomyces aureofaciens and fragment FO from coenzyme F420 of Methanobacterium sp. J Am Chem Soc. 1982;104:4014–4015. [Google Scholar]
- 32.Ng W V, Kennedy S P, Mahairas G G, Berquist B, Pan M, Shukla H D, Lasky S R, Baliga N S, Thorsson V, Sbrogna J, Swartzell S, Weir D, Hall J, Dahl T A, Welti R, Goo Y A, Leithauser B, Keller K, Cruz R, Danson M J, Hough D W, Maddocks D G, Jablonski P E, Krebs M P, Angevine C M, Dale H, Isenbarger T A, Peck R F, Pohlschroder M, Spudich J L, Jung K H, Alam M, Freitas T, Hou S, Daniels C J, Dennis P P, Omer A D, Ebhardt H, Lowe T M, Liang P, Riley M, Hood L, DasSarma S. Genome sequence of Halobacterium species NRC-1. Proc Natl Acad Sci USA. 2000;97:12176–12181. doi: 10.1073/pnas.190337797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O Gaora P. Expression of genes in mycobacteria. In: Parish T, Stoker N G, editors. Mycobacteria protocols. Vol. 101. Totowa, N.J: Humana Press; 1998. p. 472. [Google Scholar]
- 34.Pelicic V, Jackson M, Reyrat J M, Jacobs W R, Jr, Gicquel B, Guilhot C. Efficient allelic exchange and transposon mutagenesis in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 1997;94:10955–10960. doi: 10.1073/pnas.94.20.10955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Purwantini E, Daniels L. Purification of a novel coenzyme F420-dependent glucose-6-phosphate dehydrogenase from Mycobacterium smegmatis. J Bacteriol. 1996;178:2861–2866. doi: 10.1128/jb.178.10.2861-2866.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Purwantini E, Gillis T, Daniels L. Presence of F420-dependent glucose-6-phosphate dehydrogenase in Mycobacterium and Nocardia species, but absence in Streptomyces and Corynebacterium species and methanogenic Archaea. FEMS Microbiol Lett. 1997;146:129–134. doi: 10.1111/j.1574-6968.1997.tb10182.x. [DOI] [PubMed] [Google Scholar]
- 37.Redenbach M, Kieser H M, Denapaite D, Eichner A, Cullum J, Kinashi H, Hopwood D A. A set of ordered cosmids and a detailed genetic and physical map for the 8 Mb Streptomyces coelicolor A3(2) chromosome. Mol Microbiol. 1996;21:77–96. doi: 10.1046/j.1365-2958.1996.6191336.x. [DOI] [PubMed] [Google Scholar]
- 38.Reuke B, Korn S, Eisenreich W, Bacher A. Biosynthetic precursors of deazaflavins. J Bacteriol. 1992;174:4042–4049. doi: 10.1128/jb.174.12.4042-4049.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rhodes P M, Winskill N, Friend E J, Warren M. Biochemical and genetic comparison of Streptomyces rimosus mutants impaired in oxytetracycline biosynthesis. J Gen Microbiol. 1981;124:329–338. [Google Scholar]
- 40.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1989. [Google Scholar]
- 41.Schneider T D, Stephens R M. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, Harrison D, Hoang L, Keagle P, Lumm W, Pothier B, Qiu D, Spadafora R, Vicaire R, Wang Y, Wierzbowski J, Gibson R, Jiwani N, Caruso A, Bush D, Safer H, Patwell D, Prabhaker S, McDougall S, Shimer G, Goyal A, Pietrokovski S, Church G M, Daniels C J, Mao J-I, Rice P, Nolling J, Reeve J N. Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith R F, Smith T F. Pattern-induced multisequence alignment (PIMA) algorithm employing secondary structure-dependent gap penalties for use in comparative protein modeling. Protein Eng. 1992;5:35–41. doi: 10.1093/protein/5.1.35. [DOI] [PubMed] [Google Scholar]
- 44.Snapper S B, Melton R E, Mustafa S, Kieser T, Jacobs W R., Jr Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol. 1990;4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 45.Stover C K, Warrener P, VanDevanter D R, Sherman D R, Arain T M, Langhorne M H, Anderson S W, Towell J A, Yuan Y, McMurray D N, Kreiswirth B N, Barry C E, Baker W R. A small-molecule nitroimidazopyran drug candidate for the treatment of tuberculosis. Nature. 2000;405:962–966. doi: 10.1038/35016103. [DOI] [PubMed] [Google Scholar]
- 46.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Widdel F, Wolfe R S. Expression of secondary alcohol dehydrogenase in methanogenic bacteria and purification of the F420-specific enzyme from Methanogenium thermophilum strain TCI. Arch Microbiol. 1989;152:322–328. [Google Scholar]