Abstract
The nodZ gene, which is present in various rhizobial species, is involved in the addition of a fucose residue in an α1-6 linkage to the reducing N-acetylglucosamine residue of lipo-chitin oligosaccharide signal molecules, the so-called Nod factors. Fucosylation of Nod factors is known to affect nodulation efficiency and host specificity. Despite a lack of overall sequence identity, NodZ proteins share conserved peptide motifs with mammalian and plant fucosyltransferases that participate in the biosynthesis of complex glycans and polysaccharides. These peptide motifs are thought to play important roles in catalysis. NodZ was expressed as an active and soluble form in Escherichia coli and was subjected to site-directed mutagenesis to investigate the role of the most conserved residues. Enzyme assays demonstrate that the replacement of the invariant Arg-182 by either alanine, lysine, or aspartate results in products with no detectable activity. A similar result is obtained with the replacement of the conserved acidic position (Asp-275) into its corresponding amide form. The residues His-183 and Asn-185 appear to fulfill functions that are more specific to the NodZ subfamily. Secondary structure predictions and threading analyses suggest the presence of a “Rossmann-type” nucleotide binding domain in the half C-terminal part of the catalytic domain of fucosyltransferases. Site-directed mutagenesis combined with theoretical approaches have shed light on the possible nucleotide donor recognition mode for NodZ and related fucosyltransferases.
The symbiosis between rhizobial species and leguminous plants, resulting in the formation of nitrogen-fixing root nodules, is a species-specific process that is mediated by signal molecules from both the plant and the bacterium. During the initial phases of nodulation (root hair curling and bacterial entry), flavonoids secreted by the host plant induce an activation of nod genes that are involved in the synthesis of lipo-chitin oligosaccharides, called Nod factors. Once Nod factors have opened the root hair door to the invading rhizobia, additional bacterial signals appear necessary for continued development of the infection (for recent reviews, see references 9, 41, and 51). The basic Nod factors comprise a chitin backbone formed by the assembly of a few β1,4-linked N-acetyl-d-glucosamine (GlcNAc) residues and an acyl chain attached at the nonreducing end (16). The nature of the fatty acyl chain, the number of core GlcNAc residues, and the presence or absence of extra substituents together determine the host specificity and nodulation efficiency of the bacterium. Among the Nod factor substituents, fucose is frequently observed on C-6 of the reducing GlcNAc residue, where it may play subtle roles in nodulation, perhaps by permitting interaction with certain plants or by protecting Nod factors from degradation (3, 17, 37, 41).
The NodA, NodB, and NodC proteins are required for the synthesis of the acylated core of Nod factors (32, 47). Fucose addition to Nod factors is encoded by nodZ in many rhizobia (31, 45, 46, 52). Indeed, recombinant NodZ catalyzes the α1,6-fucosylation of the chitin oligosaccharide backbone (45, 46). Interestingly, this enzyme seems to have an activity comparable to eukaryotic enzymes that fucosylate the chitobiosyl core of complex-type N-linked oligosaccharide (46).
Fucosyltransferases (FucTs) are enzymes catalyzing the transfer of fucose from GDP-fucose to various oligosaccharide acceptor substrates. This class of enzymes is involved in the synthesis of many oligosaccharides of biological importance in eukaryotes (13). Many fucosyltransferase genes have been cloned in humans and in various animal species (13, 36), as have a few genes from invertebrates (15, 29, 56) and plants (19, 28, 42). In prokaryotes, in addition to the cloned bacterial nodZ genes from Rhizobium species (45, 46), several studies have reported the cloning and expression of fucosyltransferase genes from the pathogenic bacterium Helicobacter pylori (23, 30, 61). All fucosyltransferases that have been cloned to date transfer fucose either in a α1,2-linkage to a galactose residue, or in α1,3-, α1,4-, or α1,6-linkage to a GlcNAc residue.
The eukaryotic fucosyltransferases are type II membrane proteins sharing the same general topology as other Golgi-resident glycosyltransferases, i.e., a short amino-terminal cytoplasmic tail, a transmembrane domain, a stem region, and a large globular catalytic domain facing the luminal side (38). In contrast, the bacterial enzymes appear to lack such transmembrane segment. Despite a lack of overall sequence identity, a highly conserved peptide motif has been previously identified in the catalytic domain of all α1,2-FucTs and α1,6-FucTs of prokaryotic or eukaryotic origin (7). The α1,3-FucTs appear to form a distinct family, since they lack the consensus peptide, but some regions might display structural similarities to the α1,2- and α1,6-FucTs (6, 7, 36). These observations suggest that fucosyltransferases share common structural and catalytic features, at least in the conserved regions.
For many years, our knowledge of the mechanism of action of glycosyltransferases was hampered by the lack of three-dimensional (3D) structures. To date, eight crystallographic structures, from prokaryotes and eukaryotes, have been determined (11, 21, 22, 24, 39, 43, 58, 60). These structures have provided a wealth of information on the basis of substrate binding, specificity, and catalysis. They have also shed light on the role played by short conserved peptide motifs, such as the aspartate-any residue-aspartate (DXD) motif. This motif has been identified in many different glycosyltransferase families (4, 5, 62) and was shown, in several crystal structures, to interact mainly with the phosphate groups of nucleotide donor through the coordination of a metal cation (11, 21, 39, 43, 58). Indeed, enzymes sharing this motif have an absolute requirement for divalent cations for activity. Structural information has also been obtained for glycosyltransferases that are not metal dependent (24, 60). In the latter cases, basic residues were shown to make direct contacts with the pyrophosphate moiety of the nucleotide donor. From these structural data it appears that the recognition of donor substrate is mediated by residues that are found in the most conserved regions as demonstrated for the large α3- and β4-galactosyltransferase families (21, 22). In striking contrast, site-directed mutagenesis studies performed on α1,3-FucTs indicate that the residues involved in acceptor binding are mostly located in more variable regions (18, 27, 34, 63).
At the present time, there is no structural information for fucosyltransferases and the detailed mechanism of action remains unclear. We failed to identify a conserved canonical DXD motif in fucosyltransferases. Instead, the consensus peptide motif shared by all of the known α1,2- and α1,6-FucTs comprises conserved basic residues (7, 36). In addition, the mammalian α6-FucTs (59, 64) were shown to be insensitive to EDTA. Taken together, these data suggest that for NodZ and related proteins the interaction with the nucleotide donor could be mediated by basic residues. To further our understanding of the NodZ reaction mechanism and of its functional similarities with the eukaryotic α1,2-/α1,6-FucTs, we investigated the role of the most conserved residues identified in the catalytic domain of this large enzyme family. Threading analyses strongly suggest the presence of a Rossmann fold in the C-terminal part of the NodZ catalytic domain. Extensive protein sequence analysis coupled to mutagenesis allowed the prediction of a role for some of the most conserved residues present in this large protein family.
MATERIALS AND METHODS
Materials.
GDP-fucose, ATP, and l-fucose were obtained from Sigma. Dowex AG 1x8 resin (200 to 400 mesh; formate− form) was from Bio-Rad. GDP-[14C]fucose (310 mCi/mmol) was obtained from Amersham. The plasmid pUCNZ, harboring the nodZ gene from Azorhizobium caulinodans, type strain ORS571, was kindly provided by M. Holsters (Universiteit Gent, Ghent, Belgium). The chitooligosaccharide (GlcNAc)5 was a gift from E. Samain (CERMAV, Grenoble, France). Oligonucleotides for PCR were purchased from Cybergene (St-Malo, France).
Subcloning of NodZ into an E. coli expression vector.
The 987-bp open reading frame (ORF) of NodZ was amplified by PCR from the plasmid pUCNZ with the forward primer (5′-ACTGGGATCCTATAACTCTGCCTGTCCTG) and the reverse primer (5′-ACTCAAGCTTAACCGCTCGATGCGCC) to create BamHI and HindIII restriction sites at each end of the gene. The PCR product was obtained by using the Taq polymerase (Promega) in 30 cycles, with each cycle comprising 45 s at 94°C, 1 min of annealing at 50°C, and 2 min of elongation at 72°C, and then was ligated to the T cloning vector pCRII (Invitrogen). The resulting recombinant plasmid pCRNZ was used to transform Escherichia coli XL1-Blue cells (Stratagene). Recombinant clones were identified by restriction analysis and subsequently verified by DNA sequencing. A 992-bp fragment was excised from a suitable clone with BamHI and HindIII and cloned into the BamHI/HindIII sites of the expression vector pET29a to give the plasmid pET-NodZ. By using this expression system, the recombinant enzymes are produced as an N-terminal fusion with the S-Tag.
Site-directed mutagenesis.
Mutant forms of NodZ were prepared by using the plasmid pET-NodZ as the template. PCR-based mutagenesis (QuikChange site-directed mutagenesis kit; Stratagene) was used for all mutations. The primers used to create mutants of NodZ are described in Table 1. All of the recombinant plasmids were propagated into XL1-Blue cells. The entire gene was sequenced to confirm the desired mutation and to check PCR fidelity.
TABLE 1.
Primers used in PCR to obtain each of the desired mutations
| Enzyme mutation | Primera | Positionb |
|---|---|---|
| Arg-182→Ala | 5′-GATCGGCGTCCATATAGCGCACGGTAATGGCG-3′ (sense) | 528–559 |
| 5′-CGCCATTACCGTGCGCTATATGGACGCCGATC-3′ (antisense) | 559–528 | |
| Arg-182→Asp | 5′-CGTGATCGGCGTCCATATAGACCACGGTAATGGCGAAG-3′ (sense) | 525–562 |
| 5′-CTTCGCCATTACCGTGGTCTATATGGACGCCGATCACG-3′ (antisense) | 562–525 | |
| Arg-182→Lys | 5′-GATCGGCGTCCATATAAAGCACGGTAATGGCGAAG-3′ (sense) | 528–562 |
| 5′-CTTCGCCATTACCGTGCTTTATATGGACGCCGATC-3′ (antisense) | 562–528 | |
| His-183→Ala | 5′-CGGCGTCCATATAAGGGCCGGTAATGGCGAAG-3′ (sense) | 531–562 |
| 5′-CTTCGCCATTACCGGCCCTTATATGGACGCCG-3′ (antisense) | 562–531 | |
| His-183→Arg | 5′-GCGTCCATATAAGGCGCGGTAATGGCGAAG-3′ (sense) | 533–562 |
| 5′-CTTCGCCATTACCGCGCCTTATATGGACGC-3′ (antisense) | 562–533 | |
| Asn-185→Ala | 5′-GTCCATATAAGGCACGGTGCTGGCGAAGATATATTGG-3′ (sense) | 535–571 |
| 5′-CCAATATATCTTCGCCAGCACCGTGCCTTATATGGAC-3′ (antisense) | 535–571 | |
| Asn-185→Asp | 5′-CCATATAAGGCACGGTGATGGCGAAGATATATTGG-3′ (sense) | 537–571 |
| 5′-CCAATATATCTTCGCCATCACCGTGCCTTATATGG-3′ (antisense) | 571–537 | |
| Asp-275→Ala | 5′-GCTGCGCTGGTCGCTATGCAACTGCTTTC-3′ (sense) | 811–839 |
| 5′-GAAAGCAGTTGCATAGCGACCAGCGCAGC-3′ (antisense) | 839–811 | |
| Asp-275→Asn | 5′-CGTTGCTGCGCTGGTCAATATGCAACTGCTTTC-3′ (sense) | 807–839 |
| 5′-GAAAGCAGTTGCATATTGACCAGCGCAGCAACG-3′ (antisense) | 839–807 |
Primer bases in boldface indicate mutations.
Positions of primers are given in comparison with adenine residue of the initiation codon assigned as base one.
Expression of recombinant enzymes.
E. coli BL21(DE3)/pLysS cells were used for expression of recombinant native and mutated NodZ. Cells transformed with pET29a were used as control. A total of 50 ml of fresh Luria-Bertani medium, containing 30 μg of kanamycin/ml and 34 μg of chloramphenicol/ml, was inoculated with 0.5 ml of an overnight preculture and incubated at 37°C. Optimal protein production was achieved by induction with 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) when the cultures had reached an optical density at 600 nm of 0.8, and incubation was continued overnight at 30°C. Cells were harvested by centrifugation, washed once with phosphate-buffered saline, resuspended in 5 ml of chilled 20 mM HEPES (pH 7), and broken by three passages through the French press (18,000 lb/in2). Cell extracts were cleared by centrifugation (50,000 × g, 30 min, 4°C), and supernatants were divided into aliquots and stored at −80°C until they were used.
Western blot analysis.
Recombinant enzymes were detected by Western blot analysis. Proteins were separated on sodium dodecyl sulfate–10% polyacrylamide gels and electrotransferred to a polyvinylidene difluoride membrane (Pall Gelman Laboratory). The presence of the S-Tag epitope was probed by using the S-Tag Western blotting kit with alkaline phosphatase S-protein conjugate (Novagen) according to the manufacturer's instructions.
Fucosyltransferase assays.
The fucosyltransferase activity of NodZ and mutants was assayed by a modification (8) of the method of Prieels et al. (44). Standard reactions were conducted at 27°C for 15 min in a final volume of 50 μl in the presence of 50 μM GDP-fucose, 110,000 cpm of GDP-[14C]fucose, 5 mM acceptor (GlcNAc)5, 10 mM MgCl2, 1 mM ATP, and 10 mM l-fucose in 20 mM HEPES-NaOH (pH 7). The reaction was initiated by addition of cell extract and stopped by the addition of 1 ml of a mixed bed resin slurry AG 1-X8 (1:4 [wt/vol] in water). Samples were vortexed, centrifuged, and washed once more with 600 μl of water. Supernatants (2 × 600 μl) were pooled, and the radioactivity was measured by scintillation counting. Parallel control reactions were performed in the absence of acceptor. An apparent Km value for GDP-fucose was obtained by using 8 to 200 μM GDP-fucose with 5 mM acceptor and, for the acceptor, by using 0.1 to 10 mM (GlcNAc)5 with 100 μM GDP-fucose. The metal cation requirement of recombinant NodZ was assessed by performing reactions in the presence of 10 mM concentrations of various divalent cations or EDTA. Product of the enzyme reaction was identified in preliminary experiments by thin-layer chromatography as described elsewhere (46).
Sequence analysis.
Protein sequences were retrieved from GenBank and protein data banks and analyzed by using BLAST programs (1), LALIGN (25) and CLUSTALW (55). The accession numbers (from GenBank/EBI Databank) for the selected peptide sequences displayed in the multiple sequence alignment are as follows: M35531 (human, H), U17894 (human, Se), U80026_2 (Caenorhabditis elegans, CE2FT-1), U46859_3 (Yersinia enterocolitica, WbcH), AF076779 (H. pylori, FucT2), AF154111_1 (Arabidopsis thaliana, AtFT1), D89289 (human, FucT-VIII), AF022968_5 (C. elegans, D), and L18897_8 (Azorhizobium caulinodans, NodZ). Secondary structure predictions were obtained with programs available at the NPS@ server (12) and the JPred server (14), starting from either a single peptide sequence or a multiple sequence alignment. The sensitive hydrophobic cluster analysis (HCA) method was used to compare protein sequences with very low level of sequence identity (10). HCA is a graphical method based on the detection and comparison of hydrophobic clusters that are presumed to correspond to the regular secondary structure elements constituting the architecture of globular proteins. Plots were obtained from the Drawhca server (http://www.lmcp.jussieu.fr/∼soyer/www-hca/hca-form.html). Fold recognition analyses were performed with the program ProFIT that is based on an energy-function potential (ProCeryon package, King's Beech Biosoftware Solution) (20, 50). All runs were carried out with the default settings and by scanning the database of 3D structures provided with the program as well as a homemade 3D database that comprised all of the known carbohydrate- and nucleotide-interacting proteins (∼150 protein structures). Protein sequences selected for the fold recognition analysis are those displayed in the multialignment in Fig. 1A (see above). Eight α3-FucT sequences were also included: the human FucT-III (X53578), FucT-IV (M58596), and FucT-VII (X78031); mouse FucT-IX (AB015426); H. pylori FucTA (AF194963); C. elegans CEFT-1 (Z466497_3); Schistosoma mansoni FucT-A (AF183577); and mung bean FucT-C3 (Y18529). The transmembrane and stem regions of eukaryotic sequences were deleted. For each sequence, the 10 first best scores were taken into consideration, and they were given a value ranging from 10 (first score) to 1 (tenth score). The selected folds were those giving the highest score upon addition of individual values obtained for the most representative protein members of each FucT group. Information about protein fold type was extracted from the SCOP database (33).
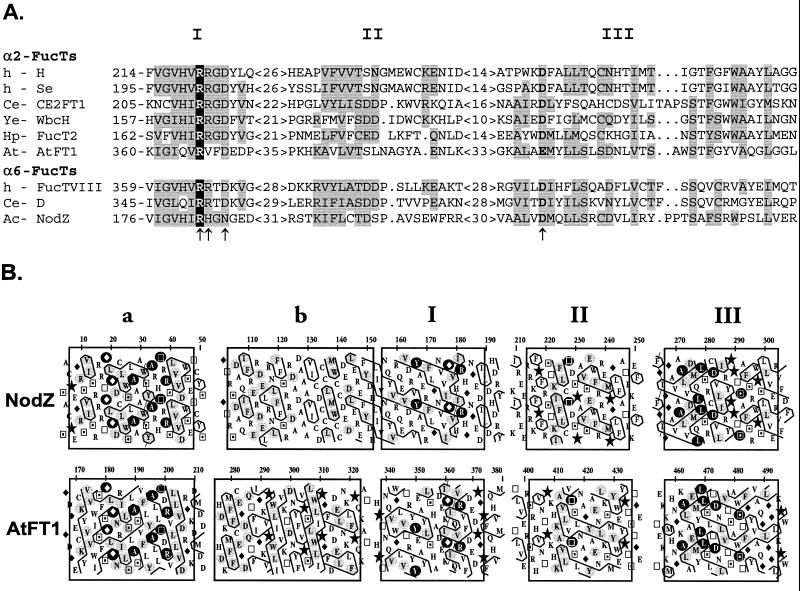
FIG. 1.
Protein sequence analysis of α1,2- and α1,6-fucosyltransferase sequences. (A) Sequence alignment of the three most conserved regions (I, II, and III) present in all α1,2- and α1,6-FucTs. The only invariant arginine residue is indicated in white on a black background. Similar amino acids significantly present in both fucosyltransferase groups are shaded in gray (groups of similar residues are defined as follows: AILMV, FWY, DENQ, RHK, and CST). Residues of NodZ that have been mutated in the present study are indicated by arrows. Accession numbers for the selected peptide sequences are given in Materials and Methods. Abbreviations: Ac, Azorhizobium caulinodans; At, Arabidopsis thaliana; Ce, C. elegans; h, human; Hp, H. pylori; Ye, Y. enterocolitica. (B) HCA comparison of NodZ and AtFT1. The protein sequences are written on a duplicated α-helical net, and the contour of clusters of hydrophobic residues is drawn. The one-letter code for amino acids is used except for Gly, Pro, Ser, and Thr, which are represented by diamonds, stars, squares with solid dots, and open squares, respectively. The five best-conserved regions have been boxed. Residues that are strictly invariant in all NodZ proteins and plant α2-FucTs are indicated in white on a black background, and those that are highly conserved are shaded in gray.
RESULTS AND DISCUSSION
Identification of conserved residues in NodZ.
NodZ is a 358-amino-acid protein with no predicted transmembrane domain. Despite a lack of overall sequence identity, we have previously demonstrated that NodZ displays similarities with other α1,2- and α1,6-FucTs from prokaryotic and eukaryotic origin, suggesting a common origin (7, 36). The recent cloning of plant α1,2-FucT genes introduced further divergence that led us to refine our sequence analysis. Since BLAST searches with known α1,2- and α1,6-FucT protein sequences yielded ca. 100 entries (of these entries, ∼90% encode known or putative α1,2-FucTs), we selected the most representative protein members of each group, namely, six α1,2-FucTs and three α1,6-FucTs, to generate the multiple sequence alignment displayed in Fig. 1A. The criterion for selection was to include the more divergent sequences originating from animal, bacteria, and plants. Of particular interest is the presence of the highly conserved peptide motif present in the central part of the catalytic domain of these enzymes (motif I [Fig. 1]). Although less conserved, a second region of similarity was also identified (motif II). Reexamination of all of these sequences allowed the identification of a third conserved region at the C terminus that is shared by all of the α1,2- and α1,6-FucTs (motif III). All of these motifs display significant similarities that are clearly evidenced by the bidimensional HCA method. This unconventional method has proven to be particularly sensitive for detecting similarities in protein sequences sharing low levels of sequence identity (10). Interestingly, the newly cloned plant α1,2-FucTs, although catalyzing a similar reaction as the mammalian α1,2-FucTs, display much more similarities with the bacterial α1,6-FucTs (NodZ), as judged from the HCA plot comparison (Fig. 1B). This observation further supports the hypothesis of a common genetic origin for all of these enzymes. It is striking to note that the similarities cover almost all of the catalytic domain of NodZ. In addition to the three above-mentioned motifs, NodZ proteins display two other regions of similarity with the plant α1,2-FucT sequences (motifs a and b), which are located at the N-terminal side of the catalytic domain. Motif a, which is particularly well conserved, is also partially present in a putative α1,2-FucT, WbcH, from Y. enterocolitica (data not shown). However, we failed to detect motif a in other protein members of the large α1,2/α1,6-FucT family. From the revised sequence alignment presented in Fig. 1, a few residues were considered as good candidates for site-directed mutagnesis experiments. Extensive sequence analysis (including all of the known sequences) revealed the presence of a unique invariant arginine residue located in motif I (R182 in NodZ) and a conserved acidic position in motif III. We have therefore postulated a role for this basic residue in the transfer reaction, more precisely in donor substrate binding, since the common feature of all of these enzymes is the use of the same nucleotide sugar. Two residues in motif I, which correspond to amino acid positions specific to NodZ proteins (H183 and N185), and the conserved acidic position in motif III (D275) were also selected. Conservative and nonconservative mutations were done to evaluate the functional importance of the selected residues.
Characterization of native and mutant forms of NodZ expressed in E. coli.
The entire NodZ ORF was cloned into the expression vector pET29a to create pET-NodZ. In order to facilitate the detection and further purification of the recombinant protein, NodZ was produced as an N-terminal fusion with the S-Tag epitope. Cell fractionation studies demonstrated that with the E. coli BL21(DE3)pLysS cells, the recombinant protein was expressed for the most part (∼90%) as an active and soluble form in the cytoplasm, whereas the remaining fraction that is recovered either as inclusion bodies or membrane associated is totally inactive (data not shown).
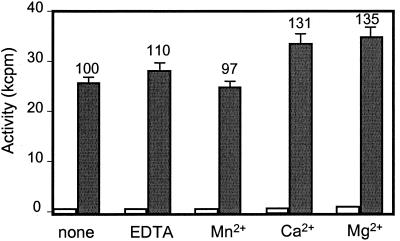
Many glycosyltransferases exhibit an absolute requirement for divalent metal cation to be active. However, the mammalian α1,6-fucosyltransferases were shown to be insensitive to metal cations (59, 64). Therefore, the effects of EDTA and various cations on NodZ activity were investigated. The NodZ activity was assessed by using as an acceptor a penta-chitin-oligosaccharide substrate since it was previously found to be more effective than the lipo-chitin oligosaccharides (46). The results given in Fig. 2 clearly show that NodZ is fully active in the presence of 10 mM EDTA, and only a slight stimulation of enzyme activity was observed for certain divalent cations such as Mg2+ and Ca2+. The characteristics of NodZ are therefore very similar to those of the mammalian α1,6-FucTs and are indicative of metal-independent donor substrate binding.
FIG. 2.
Effects of EDTA and divalent cations on NodZ activity. Fucose transfer activity of NodZ was determined in the presence (gray bars) or in the absence (white bars) of the (GlcNAc)5 acceptor as described in Materials and Methods. EDTA or the different divalent cations were added at a final concentration of 10 mM. Each value above a bar indicates the percent enzyme activity relative to the control with no additive and is the average of at least three independent determinations.
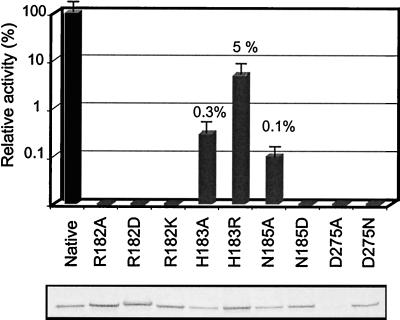
As seen in Fig. 3, eight of the nine mutants were produced at levels equivalent to the native enzyme in the cytoplasm of E. coli. However, one mutant (D275A) was poorly expressed. In region I, the substitution of the invariant basic residue R182 by either alanine (A), lysine (K), or aspartic acid (D) resulted in a complete loss of enzyme activity. These results indicate that the invariant R182 is crucial for enzyme function and that both the positive charge and the size of the side chain are necessary for activity. The conserved acidic position in motif III (D275) is also crucial for enzyme activity since its replacement into the corresponding amide form (D275N) completely abolished enzyme activity. Mutants H183A and H183R exhibited very low but significant levels of activity relative to the native enzyme (0.3 and 5%, respectively). Interestingly, if the mutation N185A resulted in a dramatic decrease in NodZ activity, with only 0.1% of the level of the wild-type enzyme, the mutation N185D yielded a completely inactive enzyme. Our data suggest that these two amino acids (H183 and N185), which are observed only in NodZ proteins, cannot be replaced by residues that are present in the mammalian α1,2- and α1,6-FucTs. A possible explanation is that these amino acids have evolved to fulfill functions that are more specific to the NodZ subfamily.
FIG. 3.
Enzyme activity and expression levels of native NodZ and mutants. NodZ was mutated at positions indicated in the legend to Fig. 1. The yield of recombinant protein was determined by Western blotting after sodium dodecyl sulfate-polyacrylamide gel electrophoresis (lower panel). Cell extracts were adjusted to give equivalent amounts of native and mutant enzymes for enzyme assays. Fucosyltransferase activity of mutants (gray bars) is expressed relative to the native enzyme (black bar). Values are the averages of at least three determinations.
Kinetic analysis of the native and mutant enzymes.
Kinetic parameters for the donor and acceptor were measured for the native enzyme and for the only mutant that retained enough enzyme activity (H183R). The apparent Kms for GDP-fucose and the acceptor (GlcNAc)5 of native NodZ are 30 μM and 1.54 mM, respectively (Table 2). Rather similar apparent Km values have been obtained with the mutant H183R. Therefore, the decrease in activity can be attributed only to a Vmax effect. These data indicate that a basic residue in this position in NodZ is required for enzyme activity, with a His residue providing higher intrinsic activity. Therefore, His-183 does not directly participate in donor or acceptor substrate binding, but presumably it has a more complex function either by participating in catalysis or by maintaining an active enzyme conformation.
TABLE 2.
Kinetic parameters for the native NodZ and H183R mutant
| NodZ | GDP-Fuc
|
(GlcNAc)5
|
||
|---|---|---|---|---|
| Km (μM) | Vmax (nmol/min/ml) | Km (mM) | Vmax (nmol/min/ml) | |
| Native | 30 | 20.02 | 1.54 | 1.01 |
| H183R | 26 | 0.73 | 2.32 | 0.065 |
Recently, the roles of some of the most conserved residues in motif I of human α1,6-FucT have been explored by using a similar approach (54), and it was shown that the replacement of the invariant Arg (R365 in the human enzyme) by Ala or Lys led to a complete loss of enzyme activity. Also, substitution of the neighboring R366 (corresponding to H183 in NodZ) by Ala and Lys resulted in a decrease in activity to ca. 3% of the level of the wild type. These results are consistent with those obtained in the present study with NodZ. However, in striking contrast, the kinetic data of Takahashi et al. (54) showed that the replacement of R366 by Ala or Lys significantly alters the apparent Km values for both the donor and acceptor and also the Kcat,app determined by varying the GDP-Fuc concentration. The data obtained with the human enzyme are in favor of a direct participation of R366 in sugar donor binding. These observations suggest that subtle differences probably occur in the active sites of these two enzymes in a way to accommodate the donor substrate.
Prediction of the fold of NodZ and related enzymes.
In the absence of a crystal structure of any related protein, protein sequence analysis and mutagenesis studies constitute alternative approaches to provide insights into the structure-function relationships of fucosyltransferases. From this work and previous studies (7, 19, 36, 54), it is now clear that all α1,2- and α1,6-FucTs constitute a large protein family sharing structural similarities on a large part of the catalytic domain. The α3-FucTs constitute a homogeneous and distinct group showing no sequence similarity with the α2- and α6-FucTs. Comparison of all of the known α1,3-FucT sequences revealed the presence of a highly conserved motif located in the C-terminal half of the catalytic domain (7, 30, 36). This motif comprises a few invariant residues, namely, one Lys, two Glu, and two aromatic residues. However, little is known regarding the role of these conserved residues. It was recently demonstrated for a human enzyme that the invariant Lys residue in the conserved α1,3-FucT motif is probably involved in GDP-fucose binding (49). In a previous study (6), Breton et al. proposed, based on HCA and threading analyses, that all fucosyltransferases share, at least partially, a similar topology and postulated the presence of a “Rossmann-type” nucleotide binding domain within the catalytic domain, similar to the one present in a β-glucosyltransferase (BGT) from phage T4 (60). It must be noted that the BGT was at that time the only available glycosyltransferase crystal structure. In the past 2 years, seven new crystal structures of glycosyltransferases, which use various UDP-sugars as a donor, have been determined (11, 21, 22, 24, 39, 43, 58). Interestingly, comparison of crystal structures reveals that glycosyltransferases are probably comprised of an unexpected small number of protein folds. Although they belong to different glycosyltransferase families showing no primary sequence identity, the various 3D structures reported so far share, partially, a common structural feature: a similar class of fold consisting in a three-layer α/β/α sandwich that resembles the “Rossmann fold” (5, 57).
In the light of these new crystallographic data, fucosyltransferase sequences were reexamined by using fold recognition methods. The program ProFIT (ProCeryon package) was used throughout this study (20, 50). Representative protein members of each FucT group (six α2-FucTs, three α6-FucTs, and eight α3-FucTs) were selected for the fold recognition analysis. Selection was made to include the more divergent protein sequences originating from mammals, invertebrates, bacteria, and sometimes plants. Statistically, the protein folds that gave the best scores by using the various FucT sequences are given in Table 3. Results were obtained by using a homemade database which contained ca. 150 protein folds, all of which are carbohydrate and nucleotide interaction proteins. These data strongly indicated that all fucosyltransferases share an α/β fold (also confirmed by using various secondary structure prediction methods) that most probably consists of the standard three-layer α/β/α sandwich. This type of fold occurs in many nucleotide binding proteins as well as in the known glycosyltransferase structures. The three-layer α/β/α sandwich was also given as a highly probable fold for most FucTs when the basic PDB-derived database supplied with the ProCeryon package was used. It is striking that among the selected folds, three correspond to glycosyltransferases (BGT, MurG, and SpsA). From these results, the BGT fold (PDB code 1C3J) still appears to be the most probable one since it was given the highest score for the α1,2- and α1,6-FucT peptide sequences. Interestingly, it is also given as the most probable fold for the α1,3-FucT group.
TABLE 3.
Fold prediction results for the different fucosyltransferase families
| PDB code | Scorea
|
Protein | Classb | Descriptionb | Reference | |
|---|---|---|---|---|---|---|
| α1,3-FucT | α1,2/1,6 FucT | |||||
| 1C3J | 35 | 30 | BGT (β-GlcT) | α/β | Three-layer α/β/α sandwich (Rossmann fold; β-sheet) | 60 |
| 1CTN | – | 14 | Chitinase A | α/β | TIM β/α barrel | 40 |
| 1DEA | 19 | 15 | Glucosamine-6-phosphate deaminase | α/β | Three-layer α/β/α sandwich (Rossmann-like; β-sheet) | 35 |
| 1DPG | 8 | 16 | Glucose-6-phosphate dehydrogenase | α/β α+β | Domain 1: Rossmann fold Domain 2: two-layer sandwich (mixed β-sheet) | 48 |
| 1ETU | 9 | 23 | Elongation factor TU | α/β | Three-layer α/β/α sandwich P-loop containing NTP hydrolases (mixed β-sheet) | 26 |
| 1FOK | 21 | 10 | MurG (β-GlcNAcT) | α/β | Three-layer α/β/α sandwich (Rossmann fold) | 24 |
| 1GKY | 19 | 6 | Guanylate kinase | α/β | Three-layer α/β/α sandwich P-loop containing NTP hydrolases | 53 |
| 1QG8 | 23 | 7 | SpsA (hypothetical glycosyltransferase) | α/β | Three-layer α/β/α sandwich (mixed β-sheet) | 11 |
| 1UAG | 13 | 7 | MurD (UDP-N-acetylmuramoyl-l-alanine:d-glutamate ligase) | α/β | Three-layer α/β/α sandwich (mixed β-sheet) | 2 |
Statistical analysis was performed as described in Materials and Methods. The best significant scores for each FucT family are in boldface. –, not predicted.
Class and description of the fold are extracted from the SCOP database (33). TIM, triosephosphate isomerase; NTP, nucleoside triphosphate.
If we assume that fucosyltransferases share a Rossmann fold in their catalytic domain, the next step consists in predicting its location. The three best-conserved regions that were identified in all α1,2- and α1,6-FucT sequences probably constitute an essential part of the active site. In addition, since enzymes of this large family have only in common the use of the same sugar donor, a role for the invariant Arg residue in GDP-fucose binding can reasonably be postulated. Threading methods have become more reliable for the detection of remote evolutionary relationships and particularly for recognizing the correct fold. However, if high-scoring sequences may be correctly aligned with their corresponding target profiles, most often the threading alignments provided are of poor quality. As for NodZ, because the sequence-structure alignment does not seem satisfactory, we used a combination of HCA and secondary structure predictions in order to produce the best sequence alignment between NodZ and the target protein model (BGT). When all of the information (conserved motifs, site-directed mutagenesis, secondary structure elements, etc.) is managed effectively, we can expect the prediction to be more accurate.
As shown in Fig. 4A, the region in NodZ sequence that is predicted to correspond to the nucleotide binding domain covers a large part of the C-terminal half of the catalytic domain of NodZ and comprises the three best-conserved regions (I, II, and III). The structurally conserved regions corresponding to the secondary structure elements that form the protein core of this domain have been determined by using the above-mentioned methods. In the proposed model, the nucleotide binding domain in NodZ is composed of a six-stranded parallel β-sheet (order 321456) flanked by at least three α-helices (Fig. 4B). By analogy to the BGT and from the present mutagenesis results, two residues of NodZ are proposed to interact with GDP-fucose: (i) the invariant R182, which makes direct contacts with the phosphate groups, and (ii) D275, which possibly interacts with the ribose ring. Since acidic residues are the prime candidates to act as catalytic residues and also because its replacement by alanine or asparagine led to a totally inactive enzyme, a direct role in the catalysis of the residue D275 is conceivable.
FIG. 4.
Secondary structure predictions and topology diagram of the nucleotide binding domain of NodZ. (A) Schematic representation of NodZ showing the location of the three best-conserved regions (I to III). Structurally conserved regions (β-strands and α-helices) between BGT and NodZ were determined by combining results from secondary structure predictions and HCA analysis; they are shaded in gray in the sequence alignment. Residues in boldface are proposed to interact with the donor sugar GDP. (B) Topology diagram (Rossmann fold) deduced from the C-terminal domain of the BGT crystal structure (60), showing the locations of the two residues of NodZ (R182 and D275 in bold in Fig. 4A). β-Strands are represented as broad arrows, and α-helices are represented as cylinders.
In the absence of an experimental 3D structure, site-directed mutagenesis combined with theoretical approaches (HCA and fold recognition) have shed light on the possible nucleotide donor recognition mode for fucosyltransferases. It is postulated that for NodZ and related proteins, the interaction with the nucleotide donor is mediated by basic and acidic residues, in a way similar to what is observed in the crystal structure of the BGT in complex with UDP. The proposed model may contribute to a better understanding of the molecular mechanisms underlying substrate specificity of this class of glycosyltransferases.
ACKNOWLEDGMENTS
We thank Rafael Oriol for careful reading of the manuscript.
This work was completed as part of the French network “GT-rec,” which is partially supported by MENRT grant ACC SV 9514111 and CNRS Program PCV. C.B. is a full-time researcher at the Institut National de la Recherche Agronomique (INRA).
Footnotes
Dedicated to the memory of André Verbert.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bertrand J A, Auger G, Fanchon E, Martin L, Blanot D, van Heijenoort J, Dideberg O. Crystal structure of UDP-N-acetylmuramoyl-l-alanine:d-glutamate ligase from Escherichia coli. EMBO J. 1997;16:3416–3425. doi: 10.1093/emboj/16.12.3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bras C P, Jorda M A, Wijfjes A H, Harteveld M, Stuurman N, Thomas-Oates J E, Spaink H P. A Lotus japonicus nodulation system based on heterologous expression of the fucosyl transferase NodZ and the acetyl transferase NoIL in Rhizobium leguminosarum. Mol Plant-Microbe Interact. 2000;13:475–479. doi: 10.1094/MPMI.2000.13.4.475. [DOI] [PubMed] [Google Scholar]
- 4.Breton C, Bettler E, Joziasse D H, Geremia R A, Imberty A. Sequence-function relationships of prokaryotic and eukaryotic galactosyltransferases. J Biochem. 1998;123:1000–1009. doi: 10.1093/oxfordjournals.jbchem.a022035. [DOI] [PubMed] [Google Scholar]
- 5.Breton C, Imberty A. Structure/function studies of glycosyltransferases. Curr Opin Struct Biol. 1999;9:563–571. doi: 10.1016/s0959-440x(99)00006-8. [DOI] [PubMed] [Google Scholar]
- 6.Breton C, Oriol R, Imberty A. Sequence alignment and fold recognition of fucosyltransferases. Glycobiology. 1996;6:vii–xii. doi: 10.1093/glycob/6.7.647-a. [DOI] [PubMed] [Google Scholar]
- 7.Breton C, Oriol R, Imberty A. Conserved structural features in eukaryotic and prokaryotic fucosyltransferases. Glycobiology. 1998;8:87–94. doi: 10.1093/glycob/8.1.87. [DOI] [PubMed] [Google Scholar]
- 8.Britten C J, Bird M I. Chemical modification of an α3-fucosyltransferase; definition of amino acid residues essential for enzyme activity. Biochim Biophys Acta. 1997;1334:57–64. doi: 10.1016/s0304-4165(96)00076-1. [DOI] [PubMed] [Google Scholar]
- 9.Broughton W J, Jabbouri S, Perret X. Keys to symbiotic harmony. J Bacteriol. 2000;182:5641–5652. doi: 10.1128/jb.182.20.5641-5652.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Callebaut I, Labesse G, Durand P, Poupon A, Canard L, Chomilier J, Henrissat B, Mornon J-P. Deciphering protein sequence information through hydrophobic cluster analysis (HCA): current status and perspectives. Cell Mol Life Sci. 1997;53:621–645. doi: 10.1007/s000180050082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charnock S J, Davies G J. Structure of the nucleotide-diphospho-sugar transferase, SpsA from Bacillus subtilis, in native and nucleotide-complexed forms. Biochemistry. 1999;38:6380–6385. doi: 10.1021/bi990270y. [DOI] [PubMed] [Google Scholar]
- 12.Combet C, Blanchet C, Geourjon C, Deleage G. NPS@: network protein sequence analysis. Trends Biochem Sci. 2000;25:147–150. doi: 10.1016/s0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- 13.Costache M, Cailleau A, Fernandez-Mateos P, Oriol R, Mollicone R. Advances in molecular genetics of α-2- and α-3/4-fucosyltransferases. Transf Clin Biol. 1997;4:367–382. doi: 10.1016/s1246-7820(97)80042-0. [DOI] [PubMed] [Google Scholar]
- 14.Cuff J A, Clamp M E, Siddiqui A S, Finlay M, Barton G J. JPred: a consensus secondary structure prediction server. Bioinformatics. 1998;14:892–893. doi: 10.1093/bioinformatics/14.10.892. [DOI] [PubMed] [Google Scholar]
- 15.DeBose-Boyd R A, Nyame A K, Cummings R D. Molecular cloning and characterization of an α1,3 fucosyltransferase, CEFT-1, from Caenorhabditis elegans. Glycobiology. 1998;8:905–917. doi: 10.1093/glycob/8.9.905. [DOI] [PubMed] [Google Scholar]
- 16.Dénarié J, Debelle F, Promé J-C. Rhizobium lipo-chitooligosaccharide nodulation factors: signaling molecules mediating recognition and morphogenesis. Annu Rev Biochem. 1996;65:503–535. doi: 10.1146/annurev.bi.65.070196.002443. [DOI] [PubMed] [Google Scholar]
- 17.D'Haeze W, Mergaert P, Promé J-C, Holsters M. Nod factor requirements for efficient stem and root nodulation of the tropical legume Sesbania rostrata. J Biol Chem. 2000;275:15676–15684. doi: 10.1074/jbc.275.21.15676. [DOI] [PubMed] [Google Scholar]
- 18.Dupuy F, Petit J-M, Mollicone R, Oriol R, Julien R, Maftah A. A single amino acid in the hypervariable stem domain of vertebrate α1,3/1,4-fucosyltransferases determines the type 1/type 2 transfer. Characterization of acceptor substrate specificity of the Lewis enzyme by site-directed mutagenesis. J Biol Chem. 1999;274:12257–12262. doi: 10.1074/jbc.274.18.12257. [DOI] [PubMed] [Google Scholar]
- 19.Faik A, Bar-Peled M, DeRocher A E, Zeng W, Perrin R M, Wilkerson C, Raikhel N V, Keegstra K. Biochemical characterization and molecular cloning of an α-1,2-fucosyltransferase that catalyzes the last step of cell wall xyloglucan biosynthesis in pea. J Biol Chem. 2000;275:15082–15089. doi: 10.1074/jbc.M000677200. [DOI] [PubMed] [Google Scholar]
- 20.Flöckner H, Braxenthaler M, Lackner P, Jaritz M, Ortner M, Sippl M J. Progress in fold recognition. Proteins Struct Funct Genet. 1995;23:376–386. doi: 10.1002/prot.340230311. [DOI] [PubMed] [Google Scholar]
- 21.Gastinel L N, Bignon C, Misra A K, Hindsgaul O, Shaper J H, Joziasse D H. Bovine α1,3-galactosyltransferase catalytic domain structure and its relationship with ABO histo-blood group and glycosphingolipid glycosyltransferases. EMBO J. 2001;20:638–649. doi: 10.1093/emboj/20.4.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gastinel L N, Cambillau C, Bourne Y. Crystal structures of the bovine β4-galactosyltransferase catalytic domain and its complex with uridine diphosphogalactose. EMBO J. 1999;18:3546–3557. doi: 10.1093/emboj/18.13.3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ge Z, Chan N W C, Palcic M M, Taylor D E. Cloning and heterologous expression of an α1,3-fucosyltransferase gene from the gastric pathogen Helicobacter pylori. J Biol Chem. 1997;272:21357–21363. doi: 10.1074/jbc.272.34.21357. [DOI] [PubMed] [Google Scholar]
- 24.Ha S, Walker D, Shi Y, Walker S. The 1.9 Å crystal structure of Escherichia coli MurG, a membrane-associated glycosyltransferase involved in peptidoglycan biosynthesis. Prot Sci. 2000;9:1045–1052. doi: 10.1110/ps.9.6.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang X, Miller W. A time-efficient, linear-space local similarity algorithm. Adv Appl Math. 1991;12:337–357. [Google Scholar]
- 26.La Cour T F, Nyborg J, Thirup S, Clark B F. Structural details of the binding of guanosine diphosphate to elongation factor Tu from E. coli as studied by X-ray crystallography. EMBO J. 1985;4:2385–2388. doi: 10.1002/j.1460-2075.1985.tb03943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Legault D J, Kelly R J, Natsuka Y, Lowe J B. Human α(1,3/1,4)-fucosyltransferases discriminate between different oligosaccharide acceptor substrates through a discrete peptide fragment. J Biol Chem. 1995;270:20987–20996. doi: 10.1074/jbc.270.36.20987. [DOI] [PubMed] [Google Scholar]
- 28.Leiter H, Mucha J, Staudacher E, Grimm R, Glössl J, Altmann F. Purification, cDNA cloning, and expression of GDP-l-Fuc:Asn-linked GlcNAc α1,3-fucosyltransferase from mung beans. J Biol Chem. 1999;274:21830–21839. doi: 10.1074/jbc.274.31.21830. [DOI] [PubMed] [Google Scholar]
- 29.Marques E T A J, Weiss J B, Strand M. Molecular characterization of a fucosyltransferase encoded by Schistosoma mansoni. Mol Biochem Parasitol. 1998;93:237–250. doi: 10.1016/s0166-6851(98)00033-4. [DOI] [PubMed] [Google Scholar]
- 30.Martin S L, Edbrooke M R, Hodgman T C, van den Eijnden D H, Bird M I. Lewis X biosynthesis in Helicobacter pylori. Molecular cloning of an α(1,3)-fucosyltransferase gene. J Biol Chem. 1997;272:21349–21356. doi: 10.1074/jbc.272.34.21349. [DOI] [PubMed] [Google Scholar]
- 31.Mergaert P, D'Haeze W, Fernandez-Lopez M, Geelen D, Goethals K, Promé J-C, van Montagu M, Holsters M. Fucosylation and arabinosylation of Nod factors in Azorhizobium caulinodans: involvement of nolK, nodZ as well as noeC and/or downstream genes. Mol Microbiol. 1996;21:409–419. doi: 10.1046/j.1365-2958.1996.6451366.x. [DOI] [PubMed] [Google Scholar]
- 32.Mergaert P, van Montagu M, Holsters M. Molecular mechanisms of Nod factor diversity. Mol Microbiol. 1997;25:811–817. doi: 10.1111/j.1365-2958.1997.mmi526.x. [DOI] [PubMed] [Google Scholar]
- 33.Murzin A G, Brenner S E, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 34.Nguyen A T, Holmes E H, Whitaker J M, Ho S, Shetterly S, Macher B A. Human α1,3/4-fucosyltransferases. I. Identification of amino acids involved in acceptor substrate binding by site-directed mutagenesis. J Biol Chem. 1998;273:25244–25249. doi: 10.1074/jbc.273.39.25244. [DOI] [PubMed] [Google Scholar]
- 35.Oliva G, Fontes M R, Garratt R C, Altamirano M M, Calcagno M L, Horjales E. Structure and catalytic mechanism of glucosamine 6-phosphate deaminase from Escherichia coli at 2.1 Å resolution. Structure. 1995;3:1323–1332. doi: 10.1016/s0969-2126(01)00270-2. [DOI] [PubMed] [Google Scholar]
- 36.Oriol R, Mollicone R, Cailleau A, Balanzino L, Breton C. Divergent evolution of fucosyltransferase genes from vertebrates, invertebrates, and bacteria. Glycobiology. 1999;9:233–334. doi: 10.1093/glycob/9.4.323. [DOI] [PubMed] [Google Scholar]
- 37.Ovtsyna A O, Schultze M, Tikhonovich I A, Spaink H P, Kondorosi E, Kondorosi A, Staehelin C. Nod factors of Rhizobium leguminosarum bv. viciae and their fucosylated derivatives stimulate a Nod factor cleaving activity in pea roots and are hydrolyzed in vitro by plant chitinases at different rates. Mol Plant-Microbe Interact. 2000;13:799–807. doi: 10.1094/MPMI.2000.13.8.799. [DOI] [PubMed] [Google Scholar]
- 38.Paulson J C, Colley K J. Glycosyltransferases: structure, localization, and control of cell type-specific glycosylation. J Biol Chem. 1989;264:17614–17618. [PubMed] [Google Scholar]
- 39.Pedersen L C, Tsuchida K, Kitagawa H, Sugahara K, Darden T A, Negishi M. Heparan/chondroitin sulfate biosynthesis: structure and mechanism of human glucuronyltransferase I. J Biol Chem. 2000;275:34580–34585. doi: 10.1074/jbc.M007399200. [DOI] [PubMed] [Google Scholar]
- 40.Perrakis A, Tews I, Dauter Z, Oppenheim A B, Chet I, Wilson K S, Vorgias C E. Crystal structure of a bacterial chitinase at 2.3 Å resolution. Structure. 1994;2:1169–1180. doi: 10.1016/s0969-2126(94)00119-7. [DOI] [PubMed] [Google Scholar]
- 41.Perret X, Staehelin C, Broughton W J. Molecular basis of symbiotic promiscuity. Microbiol Mol Biol Rev. 2000;64:180–201. doi: 10.1128/mmbr.64.1.180-201.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perrin R M, DeRocher A E, Bar-Peled M, Zeng W, Norambuena L, Orellana A, Raikhel N V, Keegstra K. Xyloglucan fucosyltransferase, an enzyme involved in plant cell wall biosynthesis. Science. 1999;284:1976–1979. doi: 10.1126/science.284.5422.1976. [DOI] [PubMed] [Google Scholar]
- 43.Persson K, Ly H D, Dieckelmann M, Wakarchuk W W, Withers S G, Strynadka N C. Crystal structure of the retaining galactosyltransferase LgtC from Neisseria meningitidis in complex with donor and acceptor sugar analogs. Nat Struct Biol. 2001;8:166–175. doi: 10.1038/84168. [DOI] [PubMed] [Google Scholar]
- 44.Prieels J-P, Monnom D, Dolmans M, Beyer T A, Hill R L. Copurification of the Lewis blood group N-acetylglucosaminide α1 goes to 4 fucosyltransferase and an N-acetylglucosaminide α1 goes to 3 fucosyltransferase from human milk. J Biol Chem. 1981;256:10456–10463. [PubMed] [Google Scholar]
- 45.Quesada-Vincens D, Fellay R, Nasim T, Viprey V, Burger U, Promé J-C, Broughton W J, Jabbouri S. Rhizobium sp. strain NGR234 NodZ protein is a fucosyltransferase. J Bacteriol. 1997;179:5087–5093. doi: 10.1128/jb.179.16.5087-5093.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quinto C, Wijfjes A H M, Bloemberg G V, Blok-Tip L, López-Lara I M, Lugtenberg B J J, Thomas-Oates J E, Spaink H P. Bacterial nodulation protein NodZ is a chitin oligosaccharide fucosyltransferase which can also recognize related substrates of animal origin. Proc Natl Acad Sci USA. 1997;94:4336–4341. doi: 10.1073/pnas.94.9.4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roche P, Maillet F, Plazanet C, Debelle F, Ferro M, Truchet G, Promé J-C, Dénarié J. The common nodABC genes of Rhizobium meliloti are host-range determinants. Proc Natl Acad Sci USA. 1996;93:15305–15310. doi: 10.1073/pnas.93.26.15305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rowland P, Basak A K, Gover S, Levy H R, Adams M J. The three-dimensional structure of glucose 6-phosphate dehydrogenase from Leuconostoc mesenteroides refined at 2.0 Å resolution. Structure. 1994;2:1073–1087. doi: 10.1016/s0969-2126(94)00110-3. [DOI] [PubMed] [Google Scholar]
- 49.Sherwood A L, Davis W C, Ho S, Macher B A, Stroud M R, Upchurch D A, Holmes E H. A GDP-fucose-protected, pyridoxal-5′-phosphate/NaBH(4)-sensitive Lys residue common to human α1,3-fucosyltransferases corresponds to Lys(300) in FucT-IV. Biochem Biophys Res Commun. 2000;273:870–876. doi: 10.1006/bbrc.2000.3018. [DOI] [PubMed] [Google Scholar]
- 50.Sippl M J, Flöckner H. Threading thrills and threats. Structure. 1996;4:15–19. doi: 10.1016/s0969-2126(96)00005-6. [DOI] [PubMed] [Google Scholar]
- 51.Spaink H P. Root nodulation and infection factors produced by rhizobial bacteria. Annu Rev Microbiol. 2000;54:257–288. doi: 10.1146/annurev.micro.54.1.257. [DOI] [PubMed] [Google Scholar]
- 52.Stacey G, Luka S, Sanjuan J, Banfalvi Z, Nieuwkoop A J, Chun J Y, Forsberg L S, Carlson R. NodZ, a unique host-specific nodulation gene, is involved in the fucosylation of the lipooligosaccharide nodulation signal of Bradyrhizobium japonicum. J Bacteriol. 1994;176:620–633. doi: 10.1128/jb.176.3.620-633.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stehle T, Schulz G E. Refined structure of the complex between guanylate kinase and its substrate GMP at 2.0 Å resolution. J Mol Biol. 1992;224:1127–1141. doi: 10.1016/0022-2836(92)90474-x. [DOI] [PubMed] [Google Scholar]
- 54.Takahashi T, Ikeda Y, Tateishi A, Yamaguchi Y, Ishikawa M, Taniguchi N. A sequence motif involved in the donor substrate binding by α1,6-fucosyltransferase: the role of the conserved arginine residues. Glycobiology. 2000;10:503–510. doi: 10.1093/glycob/10.5.503. [DOI] [PubMed] [Google Scholar]
- 55.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trottein F, Mollicone R, Fontaine J, de Mendonca R, Piller F, Pierce R, Oriol R, Capron M. Molecular cloning of a putative α3-fucosyltransferase from Schistosoma mansoni. Mol Biochem Parasitol. 2000;107:279–287. doi: 10.1016/s0166-6851(00)00213-9. [DOI] [PubMed] [Google Scholar]
- 57.Ünligil U M, Rini J M. Glycosyltransferase structure and mechanism. Curr Opin Struct Biol. 2000;10:510–517. doi: 10.1016/s0959-440x(00)00124-x. [DOI] [PubMed] [Google Scholar]
- 58.Ünligil U M, Zhou S, Yuwaraj S, Sarkar M, Schachter H, Rini J M. X-ray crystal structure of rabbit N-acetylglucosaminyltransferase I: catalytic mechanism and a new protein superfamily. EMBO J. 2000;19:5269–5280. doi: 10.1093/emboj/19.20.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Uozumi N, Yanagidani S, Miyoshi E, Ihara Y, Sakuma T, Gao C X, Teshima T, Fujii S, Shiba T, Taniguchi N. Purification and cDNA cloning of porcine brain GDP-l-Fuc:N-acetyl-β-d-glucosaminide α1–6fucosyltransferase. J Biol Chem. 1996;271:27810–27817. doi: 10.1074/jbc.271.44.27810. [DOI] [PubMed] [Google Scholar]
- 60.Vrielink A, Ruger W, Driessen H P, Freemont P S. Crystal structure of the DNA modifying enzyme β-glucosyltransferase in the presence and absence of the substrate uridine diphosphoglucose. EMBO J. 1994;13:3413–3422. doi: 10.1002/j.1460-2075.1994.tb06646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang G, Boulton P G, Chan N W, Palcic M M, Taylor D E. Novel Helicobacter pylori α1,2-fucosyltransferase, a key enzyme in the synthesis of Lewis antigens. Microbiology. 1999;145:3245–3253. doi: 10.1099/00221287-145-11-3245. [DOI] [PubMed] [Google Scholar]
- 62.Wiggins C A R, Munro S. Activity of the yeast MNN1 α-1,3-mannosyltransferase requires a motif conserved in many other families of glycosyltransferases. Proc Natl Acad Sci USA. 1998;95:7945–7950. doi: 10.1073/pnas.95.14.7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu Z, Vo L, Macher B A. Structure-function analysis of human α1,3-fucosyltransferase. Amino acids involved in acceptor substrate specificity. J Biol Chem. 1996;271:8818–8823. doi: 10.1074/jbc.271.15.8818. [DOI] [PubMed] [Google Scholar]
- 64.Yanagidani S, Uozumi N, Ihara Y, Miyoshi E, Yamaguchi N, Taniguchi N. Purification and cDNA cloning of GDP-l-Fuc:N-acetyl-β-d-glucosaminide:α1–6 fucosyltransferase (α1–6 FucT) from human gastric cancer MKN45 cells. J Biochem. 1997;121:626–632. doi: 10.1093/oxfordjournals.jbchem.a021631. [DOI] [PubMed] [Google Scholar]