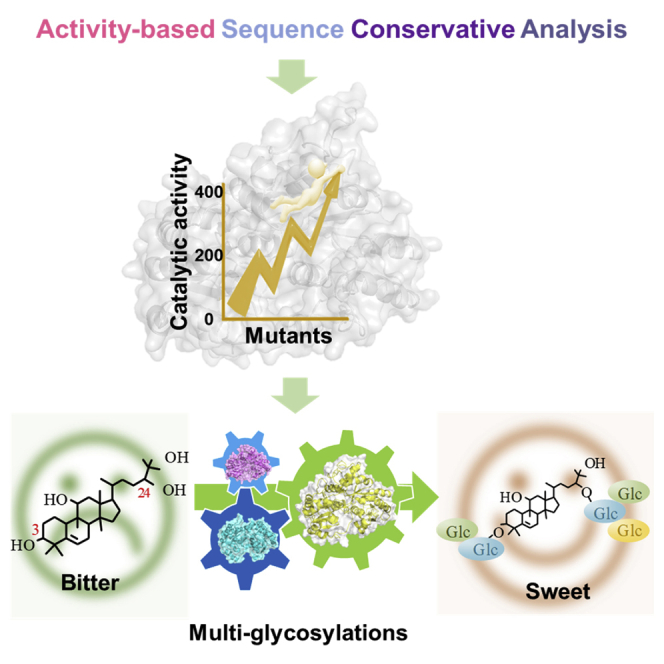
Summary
Mogrosides are widely served as natural zero-calorie sweeteners. To date, the biosynthesis of high-intensity sweetness mogrosides V from mogrol has not been achieved because of inefficient and uncontrollable multi-glycosylation process. To address this challenge, we reported three UDP-glycosyltransferases (UGTs) catalyzing the primary and branched glycosylation of mogrosides and increased the catalytic efficiency by 74–400-folds toward branched glycosylation using an activity-based sequence conservative analysis engineering strategy. The computational studies provided insights into the origin of improved catalytic activity. By virtue of UGT mutants, we provided regio- and bond-controllable multi-glycosylation routes, successfully facilitating sequential glycosylation of mogrol to three kinds of mogroside V in excellent yield of 91–99%. Meanwhile, the feasibility of the routes was confirmed in engineered yeasts. It suggested that the multi-glycosylation routes would be combined with mogrol synthetic pathway to de novo produce mogrosides from glucose by aid of metabolic engineering and synthetic biology strategies in the future.
Subject areas: Chemistry, Biosynthesis, Biotechnology
Graphical abstract
Highlights
-
•
Three UGTs sequentially converting mogrol to mogrosides were characterized
-
•
Enzyme engineering of one UGT improved its catalytic efficiency by 74–400-folds
-
•
Development of multi-glycosylation route facilitated biosynthesis of mogroside V
-
•
Engineered yeast successfully converts bitter mogrosides to sweet mogrosides
Chemistry; Biosynthesis; Biotechnology
Introduction
Nowadays, excessive consumption of sucrose and high-fructose corn syrup leads to the disease risk of dental caries, diabetes mellitus, and obesity. Consumers came to focus on replacing sucrose in food with natural low-calorie sweeteners, such as mogrosides, steviosides, glycyrrhizin and so on (Kinghorn and Soejarto, 2002; Pawar et al., 2013). Mogrosides are usually extracted from Siraitia grosvenorii, a traditional Chinese medicine and tea in China (Gong et al., 2019); they exhibit high sweetness and various pharmacological activities and have been approved as “generally recognized as safe” substances by the US Food and Drug Administration. To date, the manufacture of mogrosides mainly depends on the extraction from S. grosvenorii, but the low content limits their large-scale commercial application. In addition, such extraction processes suffer from low yield, high production cost, and loss and destruction of some value components. A great interest focuses on the de novo biosynthesis of mogrosides via metabolic engineering and synthetic biology strategies. Although several attempts have been made to synthesize mogrol using such approaches, the multi-glycosylation of mogrol to high-intensity sweetener mogrosides has never been achieved.
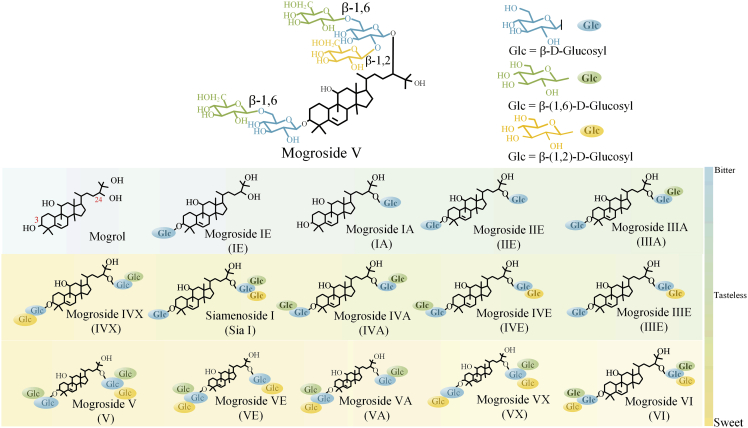
Mogrosides are a group of cucurbitane-type triterpenoid glycosides (Li et al., 2014) and possess different numbers of sugar moieties attached to mogrol (as aglycone) with β-bond linkage (Chiu et al., 2013). The number and locations of sugar moieties and type of glycosidic bond can influence the sweetness of mogrosides (Figure 1) (Matsumoto et al., 1990). For example, mogroside V (V), Siamenoside I (Sia I) and mogroside IV (IV) owing more than four glucose moieties at different location are over 300–500-times sweeter than sucrose (Matsumoto et al., 1990; Wang et al., 2019), whereas mogrosides with less than three glucose moieties, such as mogroside III and IIE, are tasteless or bitter (Li et al., 2007). The glycosylation location at 11-hydroxy of mogroside IIIE results in a bitter taste compared with the substrate (Li et al., 2014). The β-glycosidic bond may contribute to the enhancement of the sweet taste experience, but the α-glycosidic bond does not have this capability in all cases (Wang et al., 2014). Therefore, toward biosynthesis of natural and high-density sweetness mogrosides V, how to precisely control the glycosyl numbers, types, location, and bond type is indeed a major challenge.
Figure 1.
The chemical structure of mogroside. In the cartoon representation, the aglycone is represented by a black skeleton
The glucose residues are marked by individually colored spheres. Glucose residues showing β (1–6) and β (1–2) glycolic bonds are marked in green and yellow. And glucose directly attached to aglycone are labeled in blue.
In plant, multi-glycosylation process mainly relies on uridine diphosphate-dependent glycosyltransferases (UGTs; EC 2.4.x.y), which catalyze the transfer of glucose residues from UDP-glucose to acceptor (Itkin et al., 2016; Cantarel et al., 2009). Several candidate UGTs, including UGT74AC1, UGT74-345-2, UGT75-281-2, UGT-720-269-1, UGT720-269-4, UGT94-289-3, 289-2, and 289-1, were functionally characterized to show activity on mogrol and/or mogrosides (Dai et al., 2015; Itkin et al., 2016). UGT-720-269-1 and UGT94-289-3 have been predicted involved in the proposed pathway of mogrosides biosynthesis pathway in plant (Itkin et al., 2016). However, to date, the feasibility of the direct conversion of mogrol to mogroside V either in vitro or in vivo has not been confirmed. In addition, nowadays, because of deficient information of natural product synthesis pathway, multi-glycosylation of many flavonoid and triterpene aglycones were uneasy to achieved in vitro. Therefore, the development of UGTs for efficiently catalyzing primary and branched glycosylation is in urgence for manufacturing glycosides possessing multi-glycosyl units.
To date, enzyme engineering of UGTs for improved catalytic properties have been achieved. For example, we have engineered a UGT74AC1 from S. grosvenorii and delivered a mutant with 104-fold improvement in catalytic efficiency by means of directed evolution and sequence/structure based engineering strategies (Li et al., 2020). Another UGT94-289-2 from S. grosvenorii was engineered to enhance its activity toward particular substrate IIIE (Xu et al., 2022a, 2022b). The engineering of UGTs for improved regiospecificity and increased substrate spectrum of sugar donor have also been mentioned (Zhang et al., 2021a, 2021b; Xu et al., 2022a, 2022b). The success of UGT engineering encourages us to improve primary and branched glycosylation activity of mogrol with aid of structure/sequence-based enzyme engineering methods.
In this study, we presented the artificial multi-glycosylation routes for manufacturing sweeter mogroside V from mogrol in a controllable procedure as follows: (1) Primary glycosylation, in which sugar moieties were transferred from active UDP-sugar to mogrol at 3-OH and 24-OH sites; (2) branched glycosylation, in which the glycosylation process was carried out to elongate the sugar chain with β (1–6) bond and β (1–2) bond types. Three UGTs catalyzing the primary and branched glycosylation reactions were confirmed, and their catalytic efficiency was significantly improved through protein engineering. We successfully synthesized several natural and unnatural sweet mogroside V from aglycone mogrol with high conversion yield through in vitro cascade reactions. And the feasibility of designed multi-glycosylation routes was further confirmed in vivo through strain engineering.
Results
Characterization of glycosyltransferases for primary glycosylation
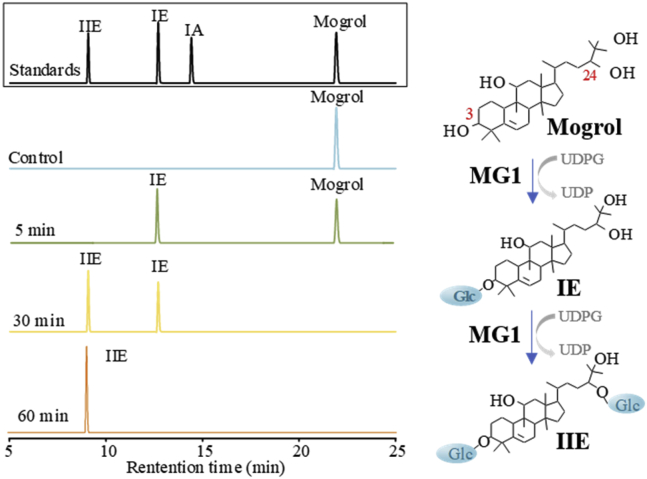
The saponin mogrol has four potential glycosylated hydroxyl groups at C3, C11, C24, and C25. To proceed with the primary glycosylation at C3 and C24, desirable UGTs with high regioselectivity and activity were needed. The mutant UGT74AC1T79Y/L48M/R28H/L109I/S15A/M76L/H47R (UGTMG1) mentioned in our previous study exhibited high catalytic efficiency in glycosylation at the 3-OH of mogrol (Li et al., 2020). Here, we discovered that this enzyme could further convert the generated IE to IIE, which having two glucose moieties at C3 and C24 sites of mogrol. (Figures 2 and S2). The results indicated that enzyme UGTMG1 can be used for primary glycosylation in our designed multi-glycosylation routes. The two glucose moieties of IIE denoted as glucose 1-C3 (glucose attached to the C3 end of aglycone) and glucose 1-C24 (glucose attached to the C24 end of the aglycone) will provide acceptor sites for all subsequent glucose additions in branched glycosylation process.
Figure 2.
HPLC analysis of UGTMG1 enzymatic reaction products using mogrol and UDPG as substrates
Identification of glycosyltransferases for branched glycosylation
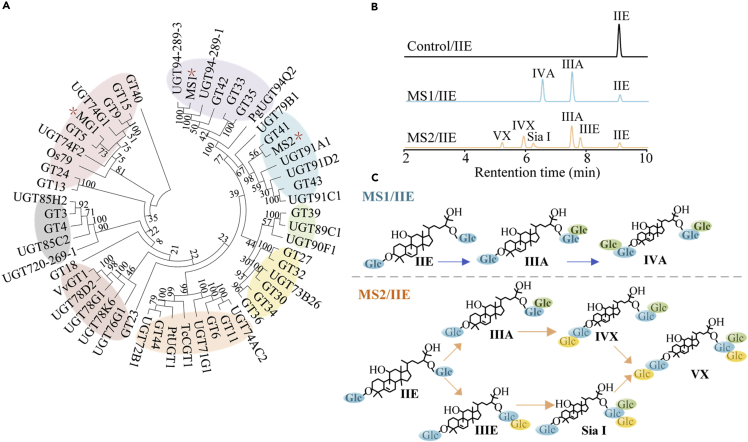
Enzyme UGT94-289-3 has been demonstrated with the branched glycosylation to mogrosides (Itkin et al., 2016). We initially cloned its gene from genome S. grosvenorii and expressed in Escherichia coli BL21 (DE3). The results showed that the enzyme activity of UGT94-289-3 to mogroside IIE were extremely low, and only trace products were detected by high-performance liquid chromatography (HPLC) (Figure S3). To obtain UGTs with a high enzymatic capability in catalyzing branched glycosylation, we performed transcriptomic analysis of the sample of S. grosvenorii under different growth phases (data no shown), and ∼40 candidate SgUGT genes were discovered. The candidate UGTs were clustered into UGT74-94 family which involved in the UGTs from plant (Carbohydrate Active Enzyme database, http://www.cazy.org/). Sequence alignment showed that MS1 is clustered with UGT94-289-3, UGT94-289-1, and PgUGT94Q2 into UGT94 families (Figure 3A), indicating that they may possess similar activity in the glycosylation. MS2 was clustered into UGT91 family.
Figure 3.
Identification of UGTs for branched glycosylation of mogrosides
(A) Phylogenetic tree of UGTs studied in this research with previously characterized flavonoid/triterpenoid UGTs. Different UGT families were shown in different background color. MEGA6 software was used to align amino acid sequences of UGTs and draw the Neighbor–Joining tree. Accession numbers are shown in the Materials and Methods.
(B and C) Enzymatic products using mogroside IIE as substrates catalyzed by UGTMS1 and UGTMS2.
The catalytic capability of these UGTs was tested with mogroside IIE as sugar acceptors and UDP-glucose (UDPG) as the sugar donor. Among all candidate UGTs from S. grosvenorii, only enzyme UGTMS1 could convert IIE to IIIA by forming a β (1–6) glycosidic bond with the 6-hydroxyl of glucose 1-C24 of IIE. Compared with UGT94-289-3 (0.021 mU/mg), this enzyme misses 21 amino acids in N-terminal, but exhibited higher catalytic ability to IIE (12.42 mU/mg). When IIE was used as substrate for UGTMS1, the generated intermediate product IIIA was subsequently glycosylated at glucose 1-C3 to form IVA with β (1–6) glycosidic bond (Figures 3B and 3C). The conversion rate of this reaction was 82%. Of interest, this enzyme also presented catalytic ability in branched glycosylation of other substrates, such as mogroside III (IIIE and IIIA) and mogroside IV (IVA, IVE), with a conversion rate of 51–82% (Figures S4 and S5). Typically, it converted IIIE to sweetness and pleasant taste Sia I and IVE, the latter further could be glycosylated to mogroside V. According to the bond type and glycosylation site, this enzyme has application potential in elongating the sugar chain at glucose 1-C3 and-C24 of mogrosides by solely forming β (1–6) glycosidic bond.
Given that natural mogrosides have both β (1–6) glycolic and β (1–2) glycolic bonds (Figure 1), the UGTs catalyzing the branched glycosylation by forming β (1–2) glycolic bond was also required. Enzyme UGTMS2 from Oryza sativa, namely OsUGT91C1, undergo β (1–2) glycosylation of the C2 of the 13-O-glucose of a steviol glycoside in vivo and has been applied to producing Reb E and Reb D (Zhang et al., 2021a, 2021b). Here, we measured its catalytic capability with mogroside IIE as sugar acceptor and UDPG as the sugar donor and discovered that this enzyme converted IIE to several mogrosides including two mogroside IIIs (IIIE and IIIA), two mogroside IVs (Sia I and IVX), and one mogroside VX with the total conversion rate of 85% (Figures 3B and 3C). The obtained IIIA bonded a new glucose at 6-hydroxyl of glucose 1-C24 by forming β (1–6) glycosidic bond; whereas with β (1–2) glycosidic bond for product IIIE at the same site. Meanwhile, this enzyme exhibited branched glycosylation to other mogrosides such as IIIE, IIIA, IVA, IVE, Sia I, and V (Figures S4 and S5). Surprisingly, it converted mogroside V to VI possessing three glucoses by β (1–6) and β (1–2) bonds at each C3 and C24 sites of mogrol saponin. These findings demonstrated that UGTMS2 was not only capable of catalyzing β (1–6) glycosylation but also β (1–2) glycosylation of mogrosides.
We then measured the enzyme kinetic value of UGTMS1 and UGTMS2 and discovered that the Km value of UGTMS1 was 0.034 mM, indicating its high affinity to substrate (Table S1). However, the catalytic efficiencies of UGTMS1 and UGTMS2 were extremely low, which influenced the branched glycosylation reaction rate. Based on the performance concerning affinity for branched glycosylation, UGTMS1 was selected for further engineering.
Enzyme engineering to improve the catalytic efficiency of UGTMS1
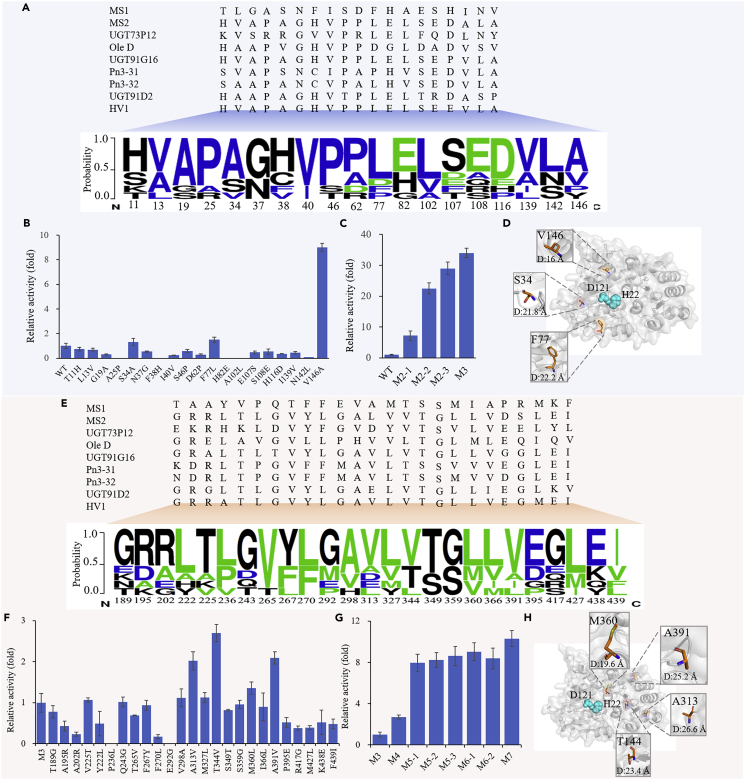
With the development of genome sequence and bioinformatic tools, more enzymatic functions were predicted and characterized by experimental data. Enzymes presenting catalytic activity to one class of substrates usually have the conservative amino acid sites or catalytic region (Liu et al., 2016). However, the catalytic efficiency of the same class enzymes from different organisms varies greatly, resulting from the amino acid sequence difference within or surrounding the catalytic region. It demonstrated that the catalytic capability can be evolved based on consensus sequence analysis (Porebski and Buckle, 2016). Activity-based sequence conservative analysis (ASCA) strategy has showed potential in the improvement of the enzyme activity (Li et al., 2020). Here, to improve accuracy of ASCA strategy and screen UGTMS1 mutants with improved activity to IIE, eight UGT sequences (UGT73P12 from Licorice (Nomura et al., 2019), OleD from Streptomyces antibioticus (Zhang et al., 2020), Pn3-31 and 32 from Ginseng (Wang et al., 2020a, 2020b), UGT91G16 from Oat (Orme et al., 2019), HV1 from Human (Matsumoto et al., 2011), UGT91D2 from Stevia rebaudiana (Zhang et al., 2021a, 2021b), and UGTMS2 from O. sativa (Wang et al., 2020a, 2020b)) exhibiting the branched glycosylation activity toward triterpenes were collected and aligned with UGTMS1 (Figures 4 and S6). Considering that the N-terminal domain of UGTs interacts with the sugar-acceptor, conservative degrees of amino acid from 1 to 233 of UGTMS1 were analyzed, and 19 mutants were identified based on ASCA method and verified by experiment (Figure 4B). The results showed that the mutation of S34A, F77L, and V146A increased the enzyme activity by 1.3-, 1.5-, and 9.0-folds relative to that of wild type (WT), respectively (Table 1, Figures S7 and S8). Then, the three mutations were sequentially combined, and the resulting mutant MS1V146A/S34A/F77L (M3) increased the catalytic efficiency (kcat/Km) to 14.03mM−1s−1, which was 34-folds higher than that of WT and meanwhile maintained the similar substrate affinity compared with WT (Figure 4C and Table 1).
Figure 4.
Enzyme engineering for improvement of catalytic efficiency of UGTMS1
The sequence conservation analysis of N-terminal (A) and C-terminal (E) of based on 9 known UGTs showing branched glycosylation activity to triterpenes.
(B and C) The effect of single and combinational mutation in N-terminal domain on relative activities.
(F and G) The effect of single and combinational mutation in C-terminal domain on relative activities.
(D and H) The location of the activity-enhanced amino acid residues in structure of UGTMS1. M2-1:S34A/F77L; M2-2:S34A/V146A; M2-3:F77L/V146A; M3: S34A/F77L/V146A; M4: S34A/F77L/V146A/T344V; M5-1: S34A/F77L/V146A/T344V/A313V; M5-2: S34A/F77L/V146A/T344V/M360L; M5-3: S34A/F77L/V146A/T344V/A391V; M6-1: S34A/F77L/V146A/T344V/A313V/A391V; M6-2: S34A/F77L/V146A/T344V/M360L/A391V; M7: S34A/F77L/V146A/T344V/A313V/M360L/A391V.
Table 1.
Kinetic parameters of UGTMS1 and its mutants toward mogroside IIE
| Enzyme | Km (μM) | kcat (s−1) | kcat/Km (mM−1·s−1) | Folda |
|---|---|---|---|---|
| WT | 34.31 ± 0.5 | 0.014 ± 0.002 | 0.41 ± 0.01 | 1 |
| M1 | 34.19 ± 0.8 | 0.13 ± 0.01 | 3.83 ± 0.12 | 9 |
| M2 | 35.20 ± 0.7 | 0.42 ± 0.02 | 12.11 ± 0.26 | 29 |
| M3 | 38.76 ± 1.1 | 0.54 ± 0.02 | 14.03 ± 0.18 | 34 |
| M4 | 30.85 ± 0.4 | 1.17 ± 0.06 | 37.94 ± 0.47 | 92 |
| M5 | 15.82 ± 0.3 | 1.90 ± 0.09 | 118.75 ± 2.45 | 293 |
| M6 | 14.70 ± 0.5 | 1.85 ± 0.05 | 132.14 ± 3.26 | 307 |
| M7 | 13.08 ± 0.2 | 1.87 ± 0.06 | 143.85 ± 4.02 | 351 |
WT: wild type; M1: V146A; M2: F77L/V146A; M3: S34A/F77L/V146A.
M4: S34A/F77L/V146A/T344V; M5: S34A/F77L/V146A/T344V/A391V.
M6: S34A/F77L/V146A/A313V/T344V/A391V.
M7: S34A/F77L/V146A/A313V/T344V/M360L/A391V.
Fold change over catalytic efficiency (kcat/Km) of WT.
The C-terminal domain of UGTs generally possesses the highly conserved motif which determines the recognition and binding of the UDP-sugar donor. Considering the high effectiveness of ASCA method in the identification of key amino acid sites in N-terminal domain related to catalytic efficiency, we analyzed the C-terminal domain using the similar method. A total of 24 sites were selected for site mutagenesis with mutant M3 as the template (Figures 4E and S6). Among them, four variants, namely, A313V, T344V, M360L, and A391V, exhibited 2.0-, 2.7-, 1.4-, and 2.1-fold increase in activity compared with that of mutant M3, respectively (Figure 4F). The kcat/Km value of resulting mutant MS1S34A/F77L/V146A/T344V (M4) increased to 37.94mM−1s−1, which was 92-fold higher than that of WT (Table 1 and Figure S8). Then, three positive mutations (A313V, M360L, and A391V) were combined with M4 (Figure 4G), resulting in mutants MS1S34A/F77L/V146A/T344V/A313V (M5-1), MS1S34A/F77L/V146A/T344V/M360L (M5-2), and MS1S34A/F77L/V146A/T344V/A391V (M5-3), respectively. The catalytic efficiency of mutant M5-3 further increased, and its Km value decreased compared with that of WT, indicating the improved substrate affinity. When mutation A313V and M360L were further introduced into mutant M5-3, the resulting mutant MS1S34A/F77L/V146A/T344V/A313V/M360L/A391V (M7) presented a 351-fold increase in the catalytic efficiency compared with WT, and still maintained a higher substrate affinity relative to M5-3.
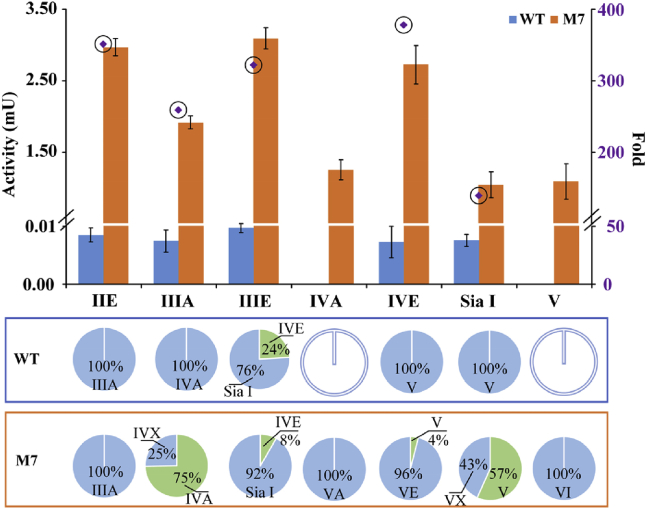
We further measured the catalytic properties of mutant M7 to other mogrosides, such as IIIA, IIIE, IVA, IVE, Sia I, and V. To our surprise, mutant M7 enzyme activity displayed 73- to 400-fold increase in the measured substrates (Figures 5, S9, S10, S11, S12, S13, S14, S15, and Table S2). In addition, mutant M7 transferred glucose to glucose 1-C3 of mogroside IIIA and Sia I by forming β (1–2) glycolic bonds, which had not been achieved by WT. Mutant M7 also displayed catalytic activity toward IVA and V, which were not glycosylated by WT. The results showed that mutant M7 was more promiscuous than the WT in the branched glycosylation of mogrosides.
Figure 5.
The comparison of enzymatic activity and product composition for MS1 and mutant M7 under different mogroside substrates
To detect the product types and composition glycosylated by MS1 and its mutants, the glycosylation processes were controlled, in which one glucose residue was transferred to substrates. Error bars represent the standard deviation from three repeats.
Molecular basis of enhanced branched glycosylation activity
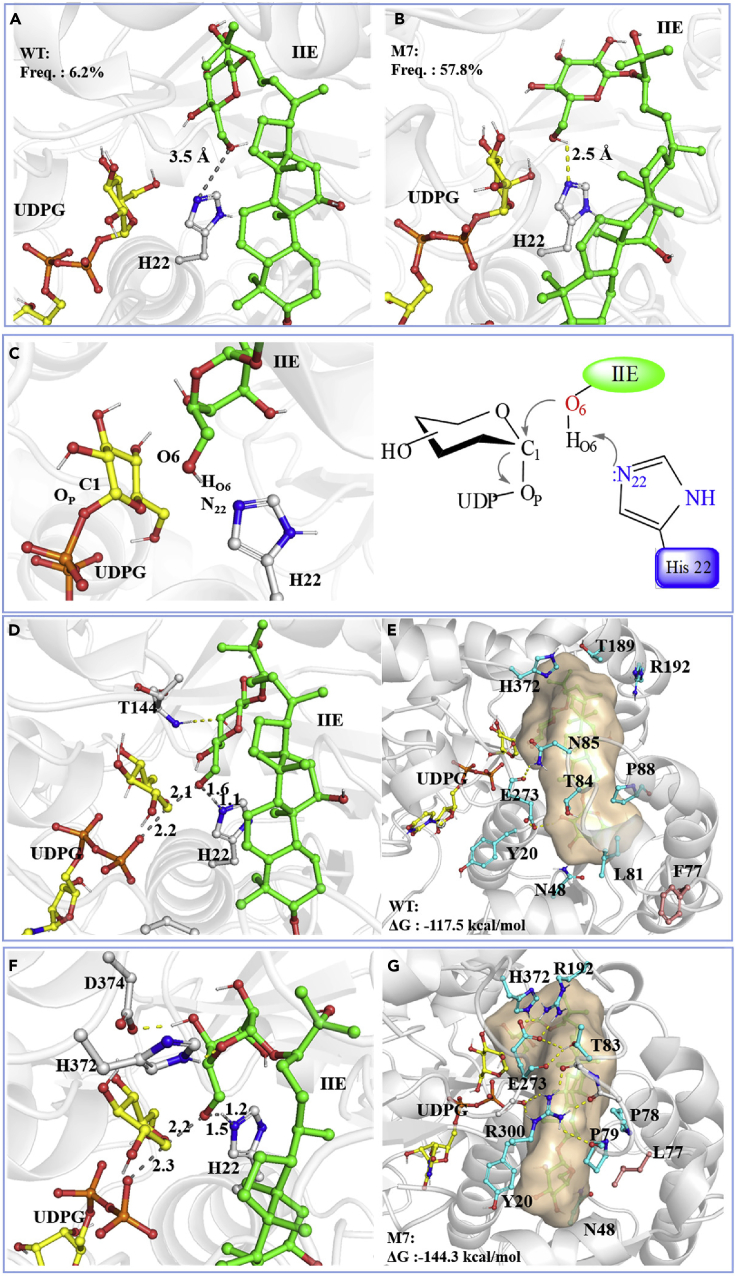
To gain insights into the source of increased catalytic efficiency of M7 to IIE, we in silico built homology models for M7 and WT by means of Schrödinger with multiple known crystal structure as templates and refined them by molecular dynamics (MD) simulations. The representative structures of complexes M7-UDPG-IIE and WT-UDPG-IIE were generated from 3∗5000 docking trajectories. According to the UGT reaction mechanism, glycosylation occurred when the distance between the O6 atom of glucose 1-C24 and the N22 atom of catalytic residue His22 was less than 3.6 Å, and the angle of HO6-O6-N22 was larger than 135° (Li et al., 2020). The catalytic conformation result showed that the distance in the structure complex M7-UDPG-IIE was 2.5 Å (Figure 6B), which was less than that in complex WT-UDPG-IIE (3.5 Å) (Figure 6A), indicating that proton transfer between HO6 and N22 occurred more easily. The emerging frequency of catalytic conformations for each complex was calculated by analyzing the MD trajectories. The frequency value in complex M7-UDPG-IIE was 57.8%, approximately nine times than that observed in WT-UDPG-IIE (6.2%) (Figures 6A, 6B, and S16). This finding revealed that IIE was bonded in a favorable orientation for reaction in M7-UDPG-IIE compared with WT-UDPG-IIE.
Figure 6.
Structural comparison between MS1 and its mutant
The representative structures of complex WT-UDPG-IIE (A) and M7-UDPG-IIE (B).
(C) The catalytic mechanism of UGTMS1. His22 deprotonates the 6-hydroxyl of IIE and makes it a nucleophile to attack the sugar donor UDPG. Geometrical representation of the TS-like structure for WT-UDPG-IIE (D) and M7-UDPG-IIE (F) obtained from the QM/MM simulations and their corresponding binding pocket (E) and (G). Distances are depicted by gray dot lines. Hydrogen bond interactions are shown in yellow dot lines. Notes: Freq.: The emerging frequency of catalytic conformations for complex WT-UDPG-IIE and M7-UDPG-IIE. ▵G: The binding affinity of catalytic conformations for each the TS-like complex structure.
To better understand the substrate-enzyme interaction of high-energy intermediate in WT and M7, the proton and glycosyl moiety transfer process were calculated for M7-UDPG-IIE and WT-UDPG-IIE by means of quantum mechanical/molecular mechanical (QM/MM) simulations. The proton HO6 of glucose 1-C24 was transferred to N22 and generated nucleophile attacks on the anomeric C1 carbon of glucose in UDPG using SN2-like mechanism (Figure 6C) (Li et al., 2020). The transition state like (TS-like) structure of M7-UDPG-IIE showed that the C24 sugar of IIE was tightly fixed by His372 and Asp374 with hydrogen bonds (Figure 6F). However, only one hydrogen bond formed with Thr144 in WT-UDPG-IIE (Figure 6D). In addition, the substitutions of phenylalanine to leucine at residue 77 triggered a large loop movement and enabled the side chain of IIE to extend into the active pocket (Figures 6E and 6G). The hydrogen bond network around the acceptor binding pocket in M7-UDPG-IIE contributed to a stronger pocket compactness than WT-UDPG-IIE, leading to tight bonding to acceptor IIE. Furthermore, the binding free energy was calculated using the QM molecular mechanics generalized born surface area (QM/MMGBSA) method. The binding affinity of TS-like structure in M7-UDPG-IIE was 26.8 kcal/mol lower than that in WT-UDPG-IIE. The finding further confirmed that the high-energy intermediate in the TS-like structure of M7-UDPG-IIE was effectively stabilized, leading to a high catalytic efficiency. Overall, although the mutation sites in M7 were far from the active center (Figures 4D and 4H), they contributed to position IIE in favorable orientation for glycosylation reaction, and then increased the frequency of catalytic conformations and stabilized the TS-like configuration during the reaction process, which endowed M7 with a high catalytic efficiency.
In vitro multi-glycosylation of mogrol to high-intensity sweetener mogrosides
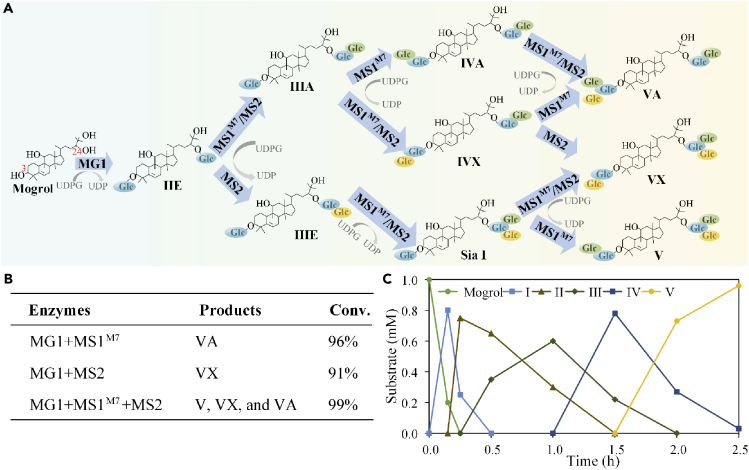
In this study, we attempted to perform the multi-glycosylation of mogrol to mogroside V in an oriented manner as follows: Enzyme UGTMG1 catalyzed the subsequent glycosylation of mogrol at 3-OH and 24-OH sites resulting in IIE. Then, the glucose 1-C3 and 1-C24 of IIE served as the glucose acceptor sites for branched glycosylation catalyzed by mutants UGTMS1-M7 and UGTMS2 along with the formation of β (1–6) bond and β (1–2) bond types. In the designed routes, mogroside VA can be synthesized using enzyme UGTMG1 and UGTMS1-M7; mogroside VX was produced using UGTMG1 and UGTMS2. If UGTMG1, UGTMS1-M7, and UGTMS2 were combined together, three kinds of mogroside Vs would be obtained (Figure 7A).
Figure 7.
In vitro multi-glycosylation of mogrol to mogrosides
(A) The designed multi-glycosylation routes for conversion of mogrol to mogrosides through combination of MG1, MS1-M7, and MS2; (B) The glycosylated products obtained from different combination of three UGTs; (C) Time-course of one-pot cascade reaction for mogrosides production using MG1, MS1-M7, and MS2 as substrates.
To verify the feasibility of this designed multi-glycosylation route, we initially measured the optimal reaction conditions for UGTMG1, UGTMS1-M7, and UGTMS2. The optimal pH for UGTMG1, UGTMS1-M7, and UGTMS2 were 7.0, 8.0, and 8.0, respectively, and the optimal temperatures were 50°C, 45°C, and 45°C, respectively. Moreover, the addition of Mg2+ contributed to increase enzymatic activity of all three enzymes (Figure S17). Then, the one-pot cascade reaction based on UGTMG1, UGTMS1-M7, and UGTMS2 was performed in vitro under the reaction pH and temperature of 8.0 and 45°C, respectively. The results showed that substrate mogrol was rapidly depleted in the initial 10 min, and I (monoglycoside, IE) and II (diglycoside, IIE) accumulated accordingly (Figure 7C). By virtue of the enhanced catalytic activity of UGTMS1-M7, intermediate products III (including IIIE and IIIA) reached highest conversion within 0.5 h, and then were converted into IV and V eventually. After the reaction for 2 h, three mogrosides V (VA, VX, and V) were synthesized with a total conversion rate of >99%. The molar ratio of VA, VX, and V was 2:1:1.5. When enzymes UGTMG1 and UGTMS2 were used, a single mogroside VX was obtained with the total conversion rate of 91%; the combination of UGTMG1 and UGTMS1-M7 led to mogroside VA with a conversion rate of 96% (Figure 7B). To our best knowledge, our results presented the first report for in vitro biosynthesis of sweet mogrosides V directly from mogrol.
Biosynthesis of high-intensity sweetener mogrosides using engineered yeast
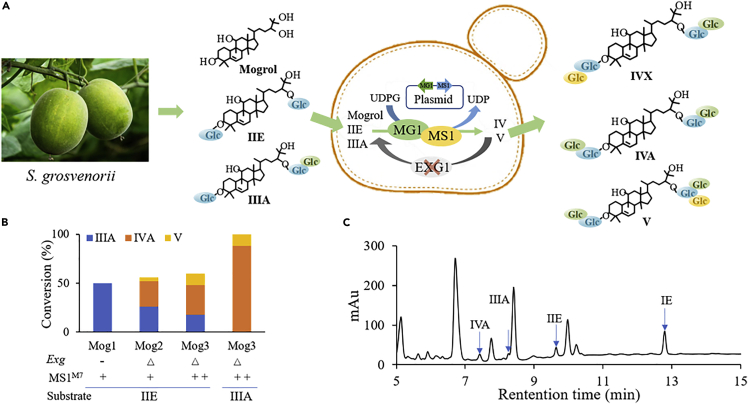
Although mogrol can be effectively converted to mogrosides V by in vitro cascade reaction in our study, the high cost of UDPG limits its large-scale application. Here, the feasibility of our designed multi-glycosylation routes in vivo was further measured (Figure 8A). We initially introduced gene of MS1M7 into Saccharomyces cerevisiae, which has been widely used as a host for biosynthesis of natural products such as triterpene saponins (Sun et al., 2019). The resulting strain Mog1 converted IIE to IIIA with a conversion rate of 50% but did not generate IV and V (Figure 8B). Given that glucosidase EXG1 in S. cerevisiae hydrolyses glycosyl units from mogroside V (Wang et al., 2015), the corresponding gene EXG1 was further deleted in strain Mog1. The resulting strain Mog2 successfully produced IV and V from IIE. To further enhance the gene expression of UGTMS1-M7, we introduced double copies of UGTMS1-M7 into glucosidase EXG1 inactive strain resulting strain Mog3. It led to two-fold higher amount of IV and V from IIE compared with strain Mog2. The total amount of sweetness mogrosides IV and V reached to 49.21 mg/L, with a conversion yield of 62%. When IIIA was used as the substrate, strain Mog3 produced IV and V with total amount of 114.45 mg/L and a conversion rate of >99%.
Figure 8.
Biosynthesis of sweet mogrosides using engineered S. cerevisiae
(A) A schematic diagram for the sweet mogrosides producing yeast.
(B) Summary of mogrosides production by five engineered yeast strains (Mog1: S. cerevisiae with one copy of UGTMS1-M7, Mog2: S. cerevisiae with one copy of UGTMS1-M7 and knockout of gene Exg, Mog3: S. cerevisiae with two copies of UGTMS1-M7 and knockout of gene Exg) using IIE or IIIA as substrates.
(C) HPLC analysis of mogrosides production by engineered strain Mog-4 (S. cerevisiae with UGTMG1 and UGTMS1-M7 and knockout of gene Exg) with mogrol as substrate.
To measure the efficiency of our designed routes in multi-glycosylation of mogrol, we introduced both genes of UGTMG1 and UGTMS1-M7 into glucosidase EXG1 inactive S. cerevisiae resulting in strain Mog4 and then cultured with mogrol and glucose. After fermentation for 94 h, mogrol was completely consumed, and product mogrosides IE, IIE, IIIA, and IVA (total 79.30 mg/L) were detected (Figure 8C). The results confirmed the feasibility of multi-glycosylation of mogrol in vivo.
Discussion
De novo biosynthesis of mogrosides via metabolic engineering and synthetic biology strategies has been gaining great interest. Although several attempts have been made to synthesize mogrol using such approaches, the multi-glycosylation of mogrol to high-intensity sweetener mogrosides has never been achieved (Wang et al., 2022). A key obstacle was the inefficient and uncontrollable multi-glycosylation process, particularly for deficiency of UGTs in catalyzing branched glycosylation of two primary glycosyl moieties (Itkin et al., 2016). In this work, we presented three UGTs (MG1, MS1-M7, and MS2) which exhibited primary and branched glycosylation activity to several mogrosides by forming desired β (1–6) and β (1–2) bonds. For the primary glycosylation of mogrol to mogrosides IIE, UGT720-269-1 has been confirmed with such catalytic ability (Itkin et al., 2016), but suffered low enzymatic activity (Figure S3). Compared to the UGT720-269-1, the enzyme component UGTMG1 in this work was more effective (Table S3).
For branched glycosylation of IIE to mogrosides V, we found that the WT UGTMS1 has 21 amino acids deficiency in N-terminal compared with the reported UGT94-289-3. UGTMS1 can sequentially converted IIE to IVA with a conversion yield of 80%; however, the catalytic ability of UGT94-289-3 to mogroside IIE was scarcely detected under the same reaction conditions. The 21 amino acids protein tag N-terminal may influence the UGT enzyme acitvity. Furthermore, we engineered UGTMS1 using activity-based sequence conservative analysis engineering strategy (ASCA) (Li et al., 2020), and obtained mutant UGTMS1-M7 with remarkable increase (74–400-folds) in catalytic efficiency, broad substrate promiscuity, and new glycosylic bond formation compared with WT. The origins of improved catalytic activity of these UGT mutants were revealed using MD and QM/MM simulations method. Our results showed that the sequence-based ASCA enzyme engineering strategy proved to be an effective approach for engineering other UGTs to obtain high catalytic efficiency. Enzyme UGTMS2, previously reported as OsUGT91C1, has been characterized with the catalytic ability in branched glycosylation of steviol glycosides (Zhang et al., 2021a, 2021b); however, its activity to mogrosides has never been reported. In this study, we discovered that this enzyme displayed catalytic ability to various glycosides from II to V by forming β (1–6) and β (1–2) bonds. The combination of UGTMS1-M7 and UGTMS2 would create a library of structural diversity mogrosides available for food and pharmaceutical area.
By virtue of efficient enzyme components UGTMG1, UGTMS1-M7, and UGTMS2, we designed an artificial multi-glycosylation pathway in an oriented manner and successfully converted mogrol to three kinds of mogroside V in excellent yield under in vitro processes, which has not been achieved before. Although natural mogroside biosynthesis pathway in the fruit of S. grosvenorii has been proposed, its feasibility has not been confirmed in studies (Itkin et al., 2016). We performed the one-pot cascade reaction based on the UGT720-269-1 and UGT94-289-3 predicted in natural mogroside biosynthesis pathway. However, mogrosides IV and V have not been detected probably because of the low catalytic property of UGT720-269-1 in the sequential glycosylation of mogrol to IIE (Figure S3). By contrast, our designed multi-glycosylation pathway provided a highly efficient method for manufacturing mogrosides V. In addition, this unnatural multi-glycosylation process would be combined with the in situ UDPG recycling methods (Dai et al., 2018) to provide a low-cost way for manufacturing high-intensity sweetness mogrosides.
Here, the feasibility of artificial multi-glycosylation pathway was also confirmed in yeast. Typically, the engineered yeast converted mogrosides IIE and IIIA to IV and V in high yield. Given that immature S. grosvenorii fruits contain high amounts of bitter mogrosides IIIA and IIE (Li et al., 2004), which influence the commercial value of natural mogroside extracts, our engineered strain demonstrated the high application potential in improving the sweetness property of immature S. grosvenorii fruits extracts. We also tested the synthetic efficiency of the engineered strain with mogrol as substrate of which the conversion rate was nearly 100% and successfully obtained sweetness IVA with a concentration of 19.26 mg/L. However, the concentration ratio of IVA and V was low, which was probably because of the limited amount of intracellular UDPG in yeast (Zhuang et al., 2017). In this work, we only provided the elementary strain for multi-glycosylation. In future work, the synthesis of intracellular UDPG can be enhanced (Wang et al., 2020a, 2020b) and the fermentation conditions can be optimized to increase the amount ratio of mogroside V. In addition, the multi-glycosylation routes would be combined with mogrol synthetic pathway to de novo produce series of high-density sweetness mogrosides IVA and V from glucose by aid of metabolic engineering and synthetic biology strategies in future. Our study suggests that the multi-glycosylation approach based on UGT engineering and artificial synthetic pathway construction allow access to glycosylation of more flavonoid and triterpene aglycones for manufacturing valuable glycosides.
Limitations of the study
Our study provided efficient multi-glycosylation pathways to biosynthesis different kinds of mogroside V. One of the key weaknesses for the in vitro multi-glycosylation is the utilization of expensive UDPG. Therefore, an economic in situ UDPG recycling system should be constructed and developed by coupling UGTs with sucrose synthase. On the other hand, although we tested and confirmed the feasibility of multi-glycosylation pathway in yeast, the titer of mogrosides IV and V was low. In future work, more complicated and detailed metabolic regulations should be executed to improve the yield of mogrosides IV and V.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| Escherichia coli DH5α | Tsingke | TSC-C01 |
| Escherichia coli BL21 | AngYuBio | G6030 |
| Recombinant DNA | ||
| pET32-MS1 | This paper | N/A |
| pET32-MS2 | This paper | GenBank: XP_015629141 |
| Software and algorithms | ||
| WebLogo 3 | N/A | http://weblogo.threeplusone.com/create.cgi |
| Schrödinger | N/A | https://www.schrodinger.com/ |
| Amber 16 | N/A | http://ambermd.org/index.php |
| Rosetta | N/A | https://www.rosettacommons.org/software |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Professor Yuanxia Sun (sun_yx@tib.cas.cn).
Materials availability
This study did not generate new reagents.
Experimental model and subject detail
Expression and purification of glycosyltransferases
The genes of UGTs from S. grosvenorii were amplified from the cDNA of S. grosvenorii using a specific primer pair (Table S4). The gene of MS2 (EUGT11, GenBank accession no. AK121682.1) was synthesized by GenScript (Nanjing) Co., Ltd and inserted into the expression plasmid pET32. Then, the recombinant plasmids were transformed into Escherichia coli BL21 (DE3), respectively. The E. coli BL21 harboring UGTs plasmid was cultivated in LB medium containing 100 μg/mL ampicillin at 37°C and 200 rpm. When OD600 reached 0.6–0.8, the recombinants were induced with 0.4 mM isopropyl-β-D-thiogalactopyranoside (IPTG, final concentration) at 16°C and 200 rpm for 20 h. The cell pellets were harvested by centrifugation at 6,000 rpm for 10 min at 4°C and suspended in lysis buffer (50 mM Tris-HCl, 50 mM NaCl, and 25 mM imidazole, pH 7.0). After sonication, the crude UGT enzymes were obtained by centrifugation at 20,000 rpm and 4°C. To obtain pure recombinant proteins, we applied the supernatant to Ni-NTA agarose affinity column as before (Dai et al., 2015). Enzyme purity and concentration were assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and a NanoDrop 2000 (Thermo Scientific, USA).
Method details
Sequence alignment and phylogenetic analysis
A neighbor-joining tree was constructed using the MEGA 6 software. The following UGTs were used in this analysis: UGT85C2 (GenBank: AAR06916.1), UGT720-269-1, UGT94-MS1, UGT94-289-1, PgUGT94Q2 (GenBank: AGR44632.1), UGT79B1 (GenBank: OAO90958.1), UGT91A1 (GenBank: RDX99130.1), UGT91C1 (GenBank: AAO63454.1), UGT91D2 (GenBank: ACE87855.1), UGT78D2 (GenBank: OAO89857.1), UGT89C1 (GenBank: AHL38967.1), UGT74F2 (GenBank: OAP07463.1), UGT72B1 (GenBank: KAF5519386.1), Os79 (GenBank: CAE01609.2), UGT78K6 (GenBank: BAF49297.1), VvGT1 (GenBank: AAB81682.1), UGT78G1 (GenBank: ABI94025.1), UGT85H2 (GenBank: ABI94024.1), TcCGT1 (GenBank: QCZ42162.1), PtUGT1 (GenBank: AUO17147.1), and MS2 (GenBank: XP_015629141).
Site-directed mutagenesis of glycosyltransferases
Sequence conservative analysis was performed to identify candidate mutation residues. Eight UGTs, which showed branched glycosylation activity toward triterpenoids, were searched and obtained from the National Center for Biotechnology Information database (NCBI), and sequential conservative analysis was conducted using WebLogo 3 (http://weblogo.threeplusone.com/create.cgi). Multiple sequence alignments of MS1, MS2, and the above characterized UGTs were performed by using the Clustal W program in MEGA 6 (Li et al., 2020). The plasmids pET32-MS2 and pET32-MS1 were used as templates to perform site-directed mutagenesis. Table S4 provides all the primers used.
Enzyme characterization
To preliminarily verify the activity of candidate UGTs towards mogrol and mogroside IIE, 300 μL reactions, including 0.2 mM mogrol or IIE, 1 mM UDPG, 10 mM MgCl2 and 300 μL crude UGT enzymes, were incubated at 40°C overnight. To detect the primary glycosylation of MG1 towards mogrol, the UGT activity assay was conducted in 300 μL of the reaction buffer (0.2 mM mogrol, 10 mM UDPG, 10 mM MgCl2, 50 mM Tris−HCl (pH 8.0), and 300 μg of the purified enzyme) at 40°C for 1 h, and the sample was detected at 10, 30, and 60 min. To measure the enzyme activity of UGTs towards different mogroside substrates, reactions (300 μL) containing 50 mM Tris−HCl (pH 8.0), 0.2 mM mogrosides, 1 mM UDPG, 10 mM MgCl2 and 100 μg of MS2, or 100 μg MS1 enzyme were performed at 40°C. Substrates were dissolved by dimethyl sulfoxide for mogrol and mogrosides I to III, and mogrosides IV and V were dissolved in ddH2O. Then, the reactions were analyzed by HPLC with a reverse-phase Ultimate C18 column (4.6 × 250 mm2, 5 μm particle, Welch, Shanghai, China) and an ultraviolet detector at 210 nm. Solvent A (H2O with 0.1% formic acid) and solvent B (acetonitrile with 0.1% formic acid) were utilized as the mobile phase at a flow rate of 1 mL/min using the following gradients:0–30 min, 25%–80% B.
The kinetic parameters of UGT and their variants were determined in 300 μL reaction buffer (including 1 mM UDPG, 10 mM MgCl2, 50 mM Tris−HCl, pH 8.0, and appropriate of the purified enzyme) at 40°C for 30 min, with the concentration of mogrol or mogroside IIE varied from 0.01 to 0.4 mM. The reaction was stopped by adding 300 μL of methanol. The substrate consumptions were then measured using HPLC method. To identify the effects of pH and temperature on the activity of UGTs, we carried out reactions under various pH (from pH 5.0 to pH 10.0; CH3COOH-CH3COONa: 5.0 and 6.0; Tris-HCl: 6.0, 7.0, 8.0 and 9.0; Glycine-NaOH: 9.0 and 10.0) and temperatures (from 20 to 55°C). Furthermore, the effect of metal ions (MnCl2, ZnCl2, CaCl2, CoCl2, CuCl2, MgCl2, and EDTA) on enzyme activity was tested.
To detect the product types and composition glycosylated by MS1 and its mutants, the glycosylation processes were controlled, in which one glucose residue was transferred to substrates. The UGT activity assay was as follow: 1 mM UDPG, 10 mM MgCl2, 50 mM Tris−HCl (pH 8.0), 0.2 mM mogrosides and 10–100 μg of the purified enzyme at 40°C for 15–30 min (depending on the activity of mutants).
Cascade reaction for multi-glycosylation of mogrol
The in vitro cascade reaction was conducted in the reaction medium (10 mL) containing 1 mM mogrol, 10 mM UDPG, 10 mM MgCl2, 50 mM Tris−HCl (pH 8.0), 600 μg/mL MG1, 600 μg/mL MS1-M7 and/or MS2 at 40°C. Samples at different reaction times were collected and analyzed by HPLC.
Biosynthesis of sweet mogrosides by engineered S. cerevisiae
The gene sequences of MG1 and MS1-M7 were synthesized by GenScript (Nanjing, China) with codon optimization based on that of S. cerevisiae (Figure S24). Then, the gene of MS1-M7 was constructed in plasmid pYZ290 with GPD1 as the promoter and ADH1 as the terminator, resulting in plasmid pYZ291-MS1. The gene of MG1 was inserted into plasmid pYZ291 with TEF1 as the promoter and ACT1 as the terminator, resulting in plasmid pYZ291-MG1. To construct the recombinant plasmid containing the genes of MG1 and MS1-M7, we inserted the expression cassette of PTEF1-MS1-TACT1 amplified from plasmid pYZ291-MG1 into plasmid pYZ291-MS1, resulting in plasmid pYZ291-MS1-MG1. Similarly, the cassette PTEF1-MS1-TACT1 gene was further cloned into plasmid pYZ291-MS1, resulting in plasmid pYZ291-MS1-MS1 with two copies of MS1-M7 gene. To delete the EXG1 gene encoding glucosidase in yeast, we integrated gene G418 into the EXG1 gene sites. Then, the recombinant plasmid pYZ-MS1-1 was introduced into S. cerevisiae resulting in strains Mog1. The recombinant plasmids pYZ291-MS1, pYZ291-MS1-MS1, pYZ291-MS1-MG1 were introduced into EXG1 knockout strain Saccharomyces cerevisiae▵EXG1, respectively, resulting in strains Mog2, Mog3, and Mog4. (Table S4). The engineered yeast strains were cultivated in SD-URA media (100 mL medium in 250 mL flasks) at 30°C and 220 rpm. Then, the cells were harvested after incubation for 60 h and resuspended in 5mL PB buffer (20 mM, pH 7.0, with 5% glucose) with a final OD600 100. Then, 0.1 mM mogrol or mogroside IIE/IIIA was added to the medium. The cell transformation was performed at 40°C and 150 rpm for 72 h, and the samples were collected and analyzed by HPLC.
NMR spectroscopic characterization and structural analysis of products
The reactions were performed in 100 mL reaction buffer (50 mM Tris-HCl pH 8.0), containing 50 mg acceptor, 200 mg UDPG, 1 mM MgCl2, concentrates of 2 L cell lysates of MS2, at 40°C for 10 h and the reaction was stopped by adding same volume of Methanol. Then, the reaction mixture was evaporated to 5 mL. To identify the structure of products, glycosides were purified by a preparative HPLC system using reverse-phase Ultimate C18 column2 and re-dissolved in dimethylsulfoxide-d6. And finally, purified glycosylated products were subjected to AVANCE III 600MHz spectrometer and characterized by 1H NMR, 13C NMR, COSY, TOCSY, HSQC and HMBC.
In silico model generation and preparations
To obtain the three-dimensional structure models of MS1-WT and MS1-M7, we used three protein structure homology-modeling and prediction servers, including Phyre2 sever (Kelley et al., 2015), trRosetta server (Du et al., 2021), SWISS-MODEL server (Waterhouse et al., 2018), prime module of Schrödinger (Friesner et al., 2004) and Amber 16 molecular dynamics package (Salomon-Ferrer et al., 2013), to find suitable templates for model generation. Three reported crystal structure PDB 5V2K, 6LZX, and 3HBF were obtained to build a chimera model. The pose of cofactor UDPG in PDB 2ACW was superimposed into this chimera model to obtain the conformation of the protein–cofactor complex. Then, the protein–cofactor complexes for MS1-WT and MS1-M7 were refined by Amber 16 MD program for side-chain refinement and overall structure relaxation. Each system was solvated in TIP3P water box with a buffer distance of 10 Å and neutralized by adding Na+ counter-ions. Total 2000 steps of steepest descent and 1000 steps of conjugate gradient minimizations were carried out to remove poor contacts in the system. The system was slowly heated up to 300 K in 25,000 steps and underwent 50 ps density simulation with 50 kcal/mol restraint force on the backbone. Afterward, 2 ns constant pressure equilibration MD run was performed at 10 kcal/mol restraint force on the backbone. Finally, 3∗50 ns unconstrained production MD was performed for each system at 300 K and 1 atm with 2 fs integration time step. The frames in the last 30 ns were aligned and clustered based on the backbone atoms of the protein using average linkage algorithm. The most representative conformation from each trajectory was selected for further use.
Molecule docking and MD simulations
Three parallel MD simulations led to three different representative conformations for each system. The dockings of MS1-WT and MS1-M7 with ligand IIE were performed using the RosettaLigand protocol in the Rosetta software suite (Meiler and Baker, 2006). The preparations of ligand structures were carried out in Schrödinger Maestro software, and the ligand conformer library generation was performed using MacroModel module. In this study, we used transform mover to perform a Monte Carlo search of the ligand binding site. The maximum rotation angle per step in the Monte Carlo search was set to 20 degrees and the maximum translation per step was set to 0.25 angstroms. A total of 500 steps of a Monte Carlo simulation was performed. Total 5000 docking trajectories were computed for each conformation to generate a comprehensive ensemble of complex conformations. To determine the possible substrate binding mode, we first filtered the final docked conformations based on the distance of N atom of catalytic residue His and O atoms of the substrate in accordance with the reaction mechanism. All configurations in the 3∗5000 docking trajectories with a distance shorter than 4 Å were selected and clustered based on the backbone atoms of the substrate with RMS threshold of 3 Å. Then, the representative conformations for each complex were obtained. To further evaluate the conformation bias for MS1-WT and MS1-M7, their representative conformation was submitted to 3∗50 ns unconstrained MD simulation as described above, and the emerging frequency of catalytic conformations was calculated for each system.
QM/MM simulations
QM/MM simulations were applied to describe the reaction process of glycosyl moiety transfer from UDPG to the O6 group of IIE for MS1-WT and MS1-M7. The initial structures came from the representative conformation of ensemble docking. Co-factor UDPG, the catalytic residue His22, and substrate IIE were treated by QM and the rest of the system was treated by MM. At first, 10000 steps (0.001 ps/step) of QM/MM equilibration were performed with the distance between the O6 atom of IIE and N22 atom of His22, and the O6 atom of IIE and C1 atom of UDPG restrained. Then, we selected the last structure from the ensemble and performed 50,000 steps (0.001 ps/step) of steered molecular dynamics (SMD) simulation. Two adaptively biased MD (ABMD) collective variables (CV, also referred to as reaction coordinates) were used to describe the reaction. The linear combination of r(N22…HO6) and r(HO6 …O6 ) [RC1 = r(N22 … HO6)-r(HO6 …O6 )] was used to describe the proton transfer process, and the linear combination of r(O6 … C1) and r(C1 … Op) [RC2 = r(O6 … C1)-r(C1 … Op)] was used to depict the glycosyl moiety transfer process. To study the binding mode of transition state like (TS-like) structures, we selected PM3 semi-empirical quantum chemical Hamiltonian for QM atoms and performed the two-dimensional reaction process search to find the structure close to the TS (TS-like structure). To evaluate the corresponding binding free energies of the TS-like structures, we performed the QM/MMGBSA calculation using the self-consistence charge density functional tight binding (SCC-DFTB) Hamiltonian (Seabra et al., 2007). All simulations were carried out using AMBER package. TIP3P water model was employed for the solvent. The non-bonded cutoff was set as 8 Å for QM and MM regions, and the harmonic restraint force constant used to force the steered CV along its defined path was 1000kcalmol−1 Å−2.
Acknowledgments
This work was supported by National Key R&D Program of China (2020YFA0908000), National Natural Science Foundation of China (No. 32101885), as well as Tianjin Synthetic Biotechnology Innovation Capacity Improvement Project (No. TSBICIP-CXRC-023). Youth Promotion Association of Chinese Academy of Sciences (2021176). We appreciate Yi Cai helping on NMR analysis.
Author contributions
‡J.L. and S.M. contributed equally to this work. They performed the protein engineering experiments, analyzed the data, and prepared the manuscript. C.L. performed the MD simulation and analyzed the data. Y.Z. and Y.C. purified the products and analyzed their structure. P.C. and Y.Z. performed the biotransformation of yeast. Y.S. and J. Y. designed the experiment and revised the manuscript.
Declaration of interests
The engineered mutants and strains described in this paper are covered by patents CN202210272559.4 and CN202210273343. X. J. L., S. M., J.Y., Y. Z., and Y.S. are listed as co-inventors of the patents. The rest of the authors declare no competing interests.
Published: October 21, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105222.
Contributor Information
Jiangang Yang, Email: yang_jg1@tib.cas.cn.
Yuanxia Sun, Email: sun_yx@tib.cas.cn.
Supplemental information
Data and code availability
All data reported in this article are available within the paper and the Supplementary Information files. The UGT protein sequences used in this work were downloaded from NCBI database (www.ncbi.nlm.nih.gov/). Multiple sequence alignments of MS1, MS2 with seven characterized UGTs was performed using the Clustal W program in MEGA 6 and the conserved motif is presented using WebLogo 3 (http://weblogo.threeplusone.com/create.cgi). Three crystal structures of UGTs (PDB ID: 5V2K, 6LZX, and 3HBF) downloaded from PDB database (www.rcsb.org) was used as a template for modeling of MS1. Data will be shared by the lead contact on request.
This article does not report original code.
Any additional information required to reanalyze the data reported in this article is available from the lead contact on request.
References
- Chiu C.H., Wang R., Lee C.C., Lo Y.C., Lu T.J. Biotransformation of mogrosides from Siraitia grosvenoriiswingle by Saccharomyces cerevisiae. J. Agric. Food Chem. 2013;61:7127–7134. doi: 10.1021/jf402058p. [DOI] [PubMed] [Google Scholar]
- Cantarel B.L., Coutinho P.M., Rancurel C., Bernard T., Lombard V., Henrissat B. The carbohydrate-active enzymes data-base (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L., Liu C., Zhu Y., Zhang J., Men Y., Zeng Y., Sun Y. Functional characterization of cucurbitadienol synthase and triterpene glycosyltransferase involved in biosynthesis of mogrosides from Siraitia grosvenorii. Plant Cell Physiol. 2015;56:1172–1182. doi: 10.1093/pcp/pcv043. [DOI] [PubMed] [Google Scholar]
- Dai L., Liu C., Li J., Dong C., Yang J., Dai Z., Zhang X., Sun Y. One-pot synthesis of ginsenoside Rh2 and bioactive unnatural ginsenoside by coupling promiscuous glycosyltransferase from Bacillus subtilis 168 to sucrose synthase. J. Agric. Food Chem. 2018;66:2830–2837. doi: 10.1021/acs.jafc.8b00597. [DOI] [PubMed] [Google Scholar]
- Du Z., Su H., Wang W., Ye L., Wei H., Peng Z., Anishchenko I., Baker D., Yang J. The trRosetta server for fast and accurate protein structure prediction. Nat. Protoc. 2021;16:5634–5651. doi: 10.1038/s41596-021-00628-9. [DOI] [PubMed] [Google Scholar]
- Friesner R.A., Banks J.L., Murphy R.B., Halgren T.A., Klicic J.J., Mainz D.T., Repasky M.P., Knoll E.H., Shelley M., Perry J.K., et al. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- Gong X., Chen N., Ren K., Jia J., Wei K., Zhang L., Lv Y., Wang J., Li M. The fruits of Siraitia grosvenorii: a review of a Chinese food-medicine. Front. Pharmacol. 2019;10:1627. doi: 10.3389/fphar.2019.01400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itkin M., Davidovich-Rikanati R., Cohen S., Portnoy V., Doron-Faigenboim A., Oren E., Freilich S., Tzuri G., Baranes N., Shen S., et al. The biosynthetic pathway of the nonsugar, high-intensity sweetener mogroside V from Siraitia grosvenorii. Proc. Natl. Acad. Sci. USA. 2016;113:E7619–E7628. doi: 10.1073/pnas.1604828113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinghorn A.D., Soejarto D.D. Discovery of terpenoid and phenolic sweeteners from plants. Pure Appl. Chem. 2002;74:1169–1179. [Google Scholar]
- Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Lin L.-M., Sui F., Wang Z.-M., Huo H.-R., Dai L., Jiang T.-L. Chemistry and pharmacology of Siraitia grosvenorii: a review. Chin. J. Nat. Med. 2014;12:89–102. doi: 10.1016/S1875-5364(14)60015-7. [DOI] [PubMed] [Google Scholar]
- Li D., Ikeda T., Huang Y., Liu J., Nohara T., Sakamoto T., Nonaka G.-I. Seasonal variation of mogrosides in Lo Han Kuo (Siraitia grosvenori) fruits. J. Nat. Med. 2007;61:307–312. [Google Scholar]
- Li J., Yang J., Mu S., Shang N., Liu C., Zhu Y., Cai Y., Liu P., Lin J., Liu W., et al. Efficient O-glycosylation of triterpenes enabled by protein engineering of plant glycosyltransferase UGT74AC1. ACS Catal. 2020;10:3629–3639. [Google Scholar]
- Li D.P., Chen Y.Y., Pan Z.H., Zhang H.R. Study on variation of mogrol glycosides from fruits of Siraitia grosvenorii in different growing ages. Guihaia. 2004;24:546–549. [Google Scholar]
- Liu Y., Yan Z., Lu X., Xiao D., Jiang H. Improving the catalytic activity of isopentenyl phosphate kinase through protein coevolution analysis. Sci. Rep. 2016;6:24117. doi: 10.1038/srep24117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K., Kasai R., Ohtani K., Tanaka O. Minor cucurbitane-glycosides from fruits of Siraitia grosvenori (cucurbitaceae) Chem. Pharm. Bull. 1990;38:2030–2032. [Google Scholar]
- Matsumoto T., Tanaka T., Sakai H., Amano N., Kanamori H., Kurita K., Kikuta A., Kamiya K., Yamamoto M., Ikawa H., Fujii N., Hori K., Itoh T., Sato K. Comprehensive sequence analysis of 24,783 barley full-length cDNAs derived from 12 clone libraries. Plant physiol. 2011;156:20–28. doi: 10.1104/pp.110.171579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiler J., Baker D. ROSETTALIGAND: protein–small molecule docking with full side-chain flexibility. Proteins. 2006;65:538–548. doi: 10.1002/prot.21086. [DOI] [PubMed] [Google Scholar]
- Nomura Y., Seki H., Suzuki T., Ohyama K., Mizutani M., Kaku T., Tamura K., Ono E., Horikawa M., Sudo H., et al. Functional specialization of UDP-glycosyltransferase 73P12 in Licorice to produce a sweet triterpenoid saponin, glycyrrhizin. Plant J. 2019;99:1127–1143. doi: 10.1111/tpj.14409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orme A., Louveau T., Stephenson M.J., Appelhagen I., Melton R., Cheema J., Li Y., Zhao Q., Zhang L., Fan D., et al. A noncanonical vacuolar sugar transferase required for biosynthesis of antimicrobial defense compounds in Oat. Proc. Natl. Acad. Sci. USA. 2019;116:27105–27114. doi: 10.1073/pnas.1914652116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawar R.S., Krynitsky A.J., Rader J.I. Sweeteners from plants—with emphasis on Stevia rebaudiana (bertoni) and Siraitia grosvenorii (swingle) Anal. Bioanal. Chem. 2013;405:4397–4407. doi: 10.1007/s00216-012-6693-0. [DOI] [PubMed] [Google Scholar]
- Porebski B.T., Buckle A.M. Consensus protein design. Protein Eng. Des. Sel. 2016;29:245–251. doi: 10.1093/protein/gzw015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W., Qin L., Xue H., Yu Y., Ma Y., Wang Y., Li C. Novel trends for producing plant triterpenoids in yeast. Crit. Rev. Biotechnol. 2019;39:618–632. doi: 10.1080/07388551.2019.1608503. [DOI] [PubMed] [Google Scholar]
- Salomon-Ferrer R., Case D.A., Walker R.C. An overview of the Amber biomolecular simulation package. WIREs. Comput. Mol. Sci. 2013;3:198–210. [Google Scholar]
- Seabra G., Walker R.C., Elstner M., Case D.A., Roitberg A.E. Implementation of the SCC–DFTB method for hybrid QM/MM simulations within the Amber molecular dynamics package. J. Phys. Chem. A. 2007;111:5655–5664. doi: 10.1021/jp070071l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Yang Z., Lu F., Liu J., Song Y., Li D. Cucurbitane glycosides derived from mogroside IIE: structure-taste relationships, antioxidant activity, and acute toxicity. Molecules. 2014;19:12676–12689. doi: 10.3390/molecules190812676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Chen Y.C., Lai Y.J., Lu T.J., Huang S.T., Lo Y.C. Dekkera bruxellensis, a beer yeast that specifically bioconverts mogroside extracts into the intense natural sweetener Siamenoside I. Food Chem. 2019;276:43–49. doi: 10.1016/j.foodchem.2018.09.163. [DOI] [PubMed] [Google Scholar]
- Wang D., Wang J., Shi Y., Li R., Fan F., Huang Y., Li W., Chen N., Huang L., Dai Z., Zhang X. Elucidation of the complete biosynthetic pathway of the main triterpene glycosylation products of panax notoginseng using a synthetic biology platform. Metab. Eng. 2020;61:131–140. doi: 10.1016/j.ymben.2020.05.007. [DOI] [PubMed] [Google Scholar]
- Wang Z., Hong J., Ma S., Huang T., Ma Y., Liu W., Liu W., Liu Z., Song H. Heterologous expression of EUGT11 from Oryza sativa in Pichia pastoris for highly efficient one-pot production of rebaudioside D from rebaudioside A. Int. J. Biol. Macromol. 2020;163:1669–1676. doi: 10.1016/j.ijbiomac.2020.09.132. [DOI] [PubMed] [Google Scholar]
- Wang R., Lin P.-Y., Huang S.-T., Chiu C.-H., Lu T.J., Lo Y.-C. Hyperproduction of β-glucanase Exg1 promotes the bioconversion of mogrosides in Saccharomyces cerevisiae mutants defective in mannoprotein deposition. J. Agric. Food Chem. 2015;63:10271–10279. doi: 10.1021/acs.jafc.5b03909. [DOI] [PubMed] [Google Scholar]
- Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L., et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Xu X., Lv X., Liu Y., Li J., Du G., Liu L. Construction and Optimization of the de novo Biosynthesis Pathway of Mogrol in Saccharomyces cerevisiae. Front. Bioeng. Biotechnol. 2022;10:919526. doi: 10.3389/fbioe.2022.919526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Liu S., Bian L., Li Z., Luo C., Chen Y., Wu X. Engineering of a UDP-glycosyltransferase for the efficient whole-cell biosynthesis of Siamenoside I in Escherichia coli. J. Agric. Food Chem. 2022;70:1601–1609. doi: 10.1021/acs.jafc.1c07699. [DOI] [PubMed] [Google Scholar]
- Xu Z., Hong L.L., Liu C.S., Kong J.Q. Protein engineering of PhUGT, a donor promiscuous glycosyltransferase, for the improved enzymatic synthesis of antioxidant quercetin 3-O-N-acetylgalactosamine. J. Agric. Food Chem. 2022;70:4076–4085. doi: 10.1021/acs.jafc.2c01029. [DOI] [PubMed] [Google Scholar]
- Zhang J., Tang M., Chen Y., Ke D., Zhou J., Xu X., Yang W., He J., Dong H., Wei Y., et al. Catalytic flexibility of rice glycosyltransferase OsUGT91C1 for the production of palatable steviol glycosides. Nat. Commun. 2021;12:7030. doi: 10.1038/s41467-021-27144-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Xu S., Jin Y., Dai Y., Chen Y., Wu X. Efficient biocatalytic preparation of rebaudioside KA: highly selective glycosylation coupled with UDPG regeneration. Sci. Rep. 2020;10:6230. doi: 10.1038/s41598-020-63379-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Yang Y., Lyu C., Chen J., Li D., Liu Y., Zhang Z., Liu Y., Wu W. Identification of the key residues of the uridine diphosphate glycosyltransferase 91D2 and its effect on the accumulation of steviol glycosides in Stevia rebaudiana. J. Agric. Food Chem. 2021;69:1852–1863. doi: 10.1021/acs.jafc.0c07066. [DOI] [PubMed] [Google Scholar]
- Zhuang Y., Yang G., Chen X., Liu Q., Zhang X., Deng Z., Feng Y. Biosynthesis of plant-derived ginsenoside Rh2 in yeast via repurposing a key promiscuous microbial enzyme. Metab. Eng. 2017;42:25–32. doi: 10.1016/j.ymben.2017.04.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this article are available within the paper and the Supplementary Information files. The UGT protein sequences used in this work were downloaded from NCBI database (www.ncbi.nlm.nih.gov/). Multiple sequence alignments of MS1, MS2 with seven characterized UGTs was performed using the Clustal W program in MEGA 6 and the conserved motif is presented using WebLogo 3 (http://weblogo.threeplusone.com/create.cgi). Three crystal structures of UGTs (PDB ID: 5V2K, 6LZX, and 3HBF) downloaded from PDB database (www.rcsb.org) was used as a template for modeling of MS1. Data will be shared by the lead contact on request.
This article does not report original code.
Any additional information required to reanalyze the data reported in this article is available from the lead contact on request.