Abstract
Schizophrenia is a highly heritable mental disorder characterized by functional dysconnectivity across the brain. However, the relationships between polygenic risk factors and connectome-wide neural mechanisms are unclear. Here, combining genetic and multiparadigm fMRI data of 623 healthy Caucasian adults drawn from the Human Connectome Project, we found that higher schizophrenia polygenic risk scores were significantly correlated with lower functional connectivity in a large-scale brain network primarily encompassing the visual system, default-mode system, and frontoparietal system. Such correlation was robustly observed across multiple fMRI paradigms, suggesting a brain-state-independent neural phenotype underlying individual genetic liability to schizophrenia. Moreover, using an independent clinical dataset acquired from the Consortium for Neuropsychiatric Phenomics, we further demonstrated that the connectivity of the identified network was reduced in patients with schizophrenia and significantly correlated with general cognitive ability. These findings provide the first evidence for connectome-wide associations of schizophrenia polygenic risk at the systems level and suggest that disrupted integration of sensori–cognitive information may be a hallmark of genetic effects on the brain that contributes to the pathogenesis of schizophrenia.
Introduction
The high heritability of schizophrenia renders the understanding of its genetic mechanisms critically important. Since genetic effects on human behaviors are mediated by the functions of neural circuits, the investigation of functional network changes associated with genetic risk for schizophrenia is a pivotal strategy to reach this goal. To date, such investigations have primarily focused on the associations of a typical schizophrenia risk gene (e.g., DRD2, ZNF804A, and CACNA1C) with neuroimaging measures [1, 2] or probed for an “imaging intermediate phenotype” based on first-degree relative or twin data [3, 4]. These two strategies have complementary limitations: the former due to extremely small effects of single genetic markers on disease risk and the latter due to the inability to scale individual differences in the degree of genetic risk and to isolate genetic risk from shared nongenetic familial risk.
One approach to assessing genetic risk at the individual level is to compute a “polygenic risk score (PRS)”, which essentially represents an overall additive genetic vulnerability one has for developing schizophrenia [5, 6]. By this, each individual can be proxied by a single PRS index, facilitating the evaluation of neurobiological phenotypes related to variations in individual genomic profiles. Combining PRS and functional neuroimaging, previous studies have shown that variations in schizophrenia PRS can at least partly explain individual differences in the function of multiple brain regions, such as the prefrontal cortical activity during working memory [7–9], hippocampal activity during episodic memory [10], and the activity of the ventral striatum during reward anticipation [11]. While these studies have provided important evidence for brain functional correlates of schizophrenia genetic risk, they nevertheless have only been focused on a priori regions based upon specific imaging paradigms. A critical but unsolved question is whether and how PRS would influence functional connectivity both at the whole-brain level and in a paradigm-independent way.
The investigation of this question is of particular importance to advance the understanding of the pathogenesis of schizophrenia. For one, schizophrenia is characterized by neural dysconnectivity that is not confined to a certain area but widely distributed across the whole brain [12]. As a result, connectivity changes related to genetic risk may also involve a broad range of regions and their connections [13]. The identification of these regions and connections would help us illuminate which parts of the brain are particularly vulnerable to genetic risk factors and thus may be relevant to the pathophysiology of schizophrenia. For another, since the PRS per se reflects an individual trait, it should theoretically also relate to certain neural “traits” at the brain level that are independent of any functional “state” in which the brain is involved. While results from previous studies may only reflect a state-dependent relationship, searching for state-independent neurobiological associations of PRS may offer us a more fundamental understanding of the genetic mechanisms of schizophrenia.
In this study, we investigated connectome-wide associations of individual genetic risk using multiparadigm functional magnetic resonance imaging (fMRI) data acquired from the Human Connectome Project (HCP [14]). Following the procedures of our prior publications [15, 16], we constructed paradigm-independent whole-brain networks for each individual in a sample of 623 healthy participants who completed a total of eight fMRI paradigms, and performed a network-based statistical analysis to link these individual connectivity patterns to their PRSs. For the observed connectomic associations, we further examined clinical and behavioral correlates of the finding in a completely independent sample drawn from the Consortium for Neuropsychiatric Phenomics (CNP [17]). Specifically, as a proof of concept, we investigated whether the connectivity of the PRS-associated network would be altered in patients with schizophrenia, and would be correlated with cognition and clinical symptoms in patients. We expected to see a significant paradigm-independent association between brain connectivity and individual PRSs, with evidence that the detected imaging phenotype is present and related to behavioral phenotypes in schizophrenia.
Materials and methods
Subjects
This study included two single-site samples. The discovery sample drawn from HCP consisted of 623 healthy subjects (age 28.86 ± 3.63 years, 302 males). The subjects were selected according to the following criteria: (1) Caucasian race; (2) Subjects had fMRI data available for all eight paradigms used in the project (resting state, working memory, emotional processing, gambling, motor, social cognition, relational processing, and language processing); and (3) Subjects had imputed genetic data available. The protocol of the HCP project was approved by the Institutional Review Board (IRB) at Washington University in St. Louis, and our use of genetic and imaging data from the HCP project was approved by the IRB at Yale University. All subjects provided written informed consent for the project.
The CNP sample was used to examine the clinical associations of the observed findings (Table S1), including 44 patients with schizophrenia (SZ, age 35.80 ± 8.94 years, 34 males), 43 patients with bipolar disorders (BD, age 35.21 ± 8.87 years, 25 males), 34 patients with attention deficit hyperactivity disorder (ADHD, age 31.09 ± 9.85 years, 18 males), and 77 healthy controls (HCs, age 30.70 ± 8.54 years, 43 males). All subjects completed a battery of seven paradigms employed in the cohort (resting state, risk taking, working memory, episodic memory encoding, episodic memory retrieval, stop signal, and task switching), and provided written informed consent following procedures approved by the IRBs at UCLA and the Los Angeles County Department of Mental Health. See Supplementary Materials for sample details and data acquisition.
Genetic data processing
The overall data processing pipeline is present in Fig. 1. The imputed genetic data were downloaded from https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001364.v1.p1. The downloaded data underwent standard quality control using PLINK 1.9 [18] before the computation of PRS. Specifically, single-nucleotide polymorphisms (SNPs) with genotype certainty <0.9, minor allele frequency <0.05, genotype call rate <0.95, and Hardy–Weinberg equilibrium probability <10−6, as well as individuals with a call rate <0.95 were excluded from the analysis. In addition, due to the high linkage disequilibrium (LD) in the major histocompatibility complex region, all SNPs on chromosome 6 between 26,000 and 33,000 kb were excluded from further analysis. The final sample included a total of 5,732,973 SNPs on 623 individuals in the HCP data.
Fig. 1. Flowchart of the data processing pipeline used in the study.
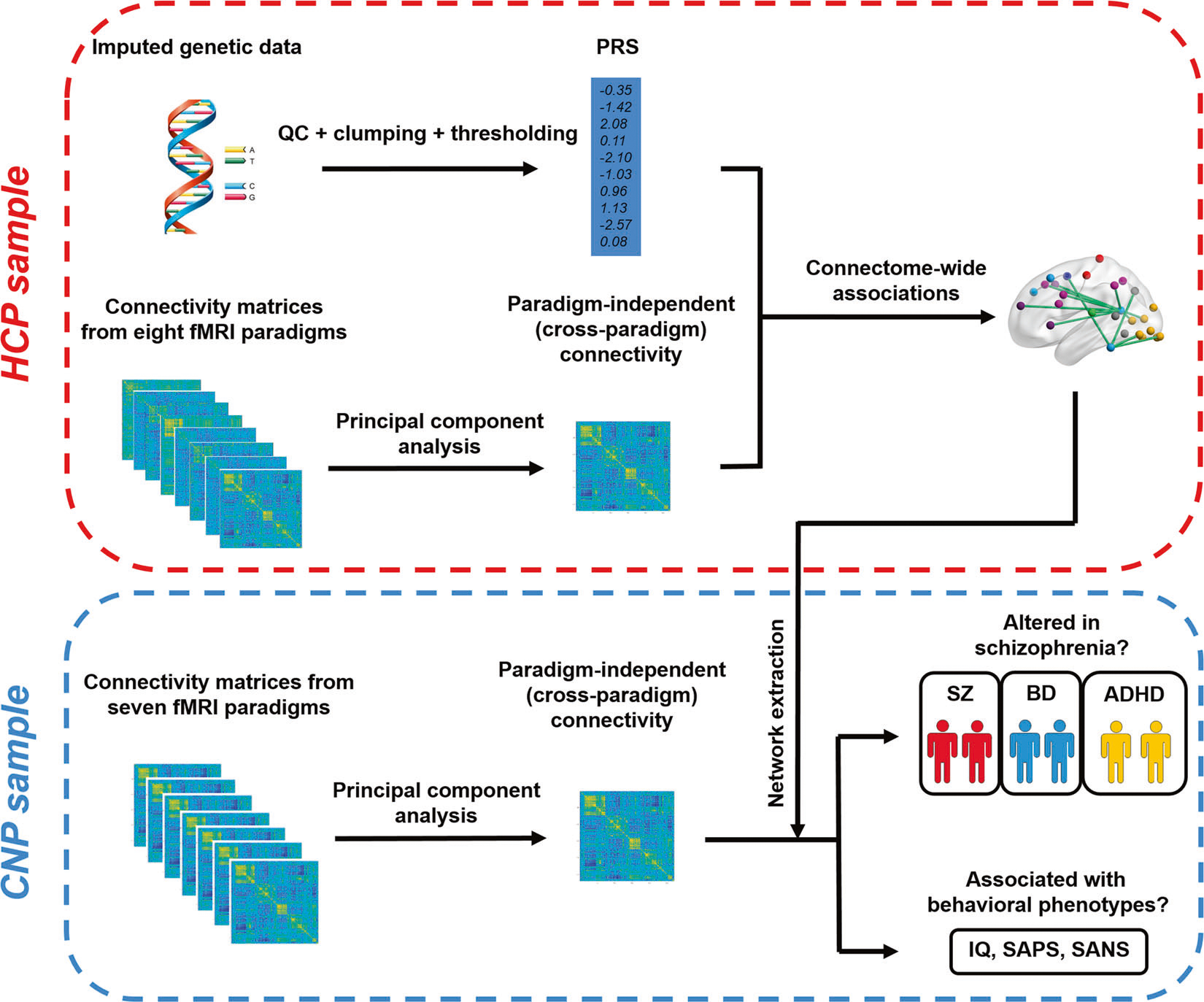
The connectomic associations were identified in the HCP sample and further investigated for clinical associations in the CNP sample.
The PRS analysis was performed using the PRSice toolbox [19] based on the genome-wide association study (GWAS) results from the Psychiatric Genomics Consortium [20] (downloaded from https://www.med.unc.edu/pgc/results-and-downloads). All matched SNPs between the base and target datasets were clumped based on the LD threshold of R2 < 0.2 within a 500 kb window. The scores were calculated as the sum of genome-wide risk alleles for each individual, weighted by the corresponding odds ratios to schizophrenia. We report our main findings based on the GWAS-significant P-value threshold (5 × 10−8). In addition, to test the robustness of our findings, we also calculated PRSs with a set of other thresholds ranging from 5 × 10−7 to 5 × 10−2.
Imaging data processing
The preprocessed imaging data were downloaded from https://db.humanconnectome.org/. The paradigm-independent networks for each individual were computed using principal component analysis (PCA) following the procedures described in our prior work [15, 16]. Specifically, the mean time series for each of the 270 nodes in the expanded Power brain atlas [15, 21, 22] were extracted from the preprocessed images. These time series were corrected for scanner and physiological noises and head motion, and were used to generate a 270 × 270 pairwise connectivity matrix for each subject during each paradigm using Pearson correlation. See Supplementary Methods for details.
The computed connectivity matrices for eight paradigms in each subject were then vectorized, concatenated, and decomposed into a set of principal components (PCs) using singular value decomposition. By definition, the first PC scores generated from the PCA analysis extracted the majority of shared variance across all paradigms in a subject and thus were reflective of individualized state-independent network architecture (Fig. S1). These individual-specific first PC matrices were termed as “cross-paradigm connectivity (CPC) matrices” [15] and were used for further analysis.
Network-based statistic in the HCP sample
The network-based statistic (NBS) analysis [23] was performed to investigate connectome-wide associations of PRSs. The procedures followed our previous publications [4, 15, 24]. In brief, an initial linear regression model was applied on each of the N(N − 1)/2 = 36,315 (N = 270) edges in the CPC matrices, with PRS, age, sex, mean frame-wise displacement (FD) across all paradigms as regressors. This step generated a P-value matrix representing the probability of accepting the null hypothesis on the PRS effect for each edge. All edges with a P value < 0.001 were then thresholded into a set of suprathreshold links, and connected clusters within this set were identified using breadth first search. The family-wise error (FWE) rate of the identified clusters was controlled by permutation testing (10,000 permutations). Specifically, the PRSs for each subject were randomized and the maximal size of the identified clusters was recalculated at the same threshold during each permutation. The corrected P value for a given cluster was determined by the proportion of the derived cluster sizes in the permutation distribution that were larger than the observed PRS effect. A P value < 0.05 indicated FWE-controlled significance of the identified cluster.
Clinical relevance of findings in the CNP sample
We used the CNP sample to examine (1) whether the identified connectomic finding could be detected in patients with schizophrenia, and (2) whether the finding could be related to behavioral deficits in patients. Here, the CPC analyses followed exactly the same procedure as described above. After acquiring the paradigm-independent CPC matrices for each individual, the identified network indicating the polygenic risk of schizophrenia was extracted from these CPC matrices in the CNP sample, and was further averaged to generate a subject-specific metric. To examine whether altered connectivity in this network was present in patients with schizophrenia, an analysis of covariance model was performed to compare these metrics between groups, covarying for age and sex. For significant main effects, post hoc analyses were performed to compare the differences between each pair of groups. Significance was set at P < 0.05 after Bonferroni correction. To examine the associations of the identified network with behavioral phenotypes, partial Pearson correlations were performed to test the effects on IQ and on scores for the Scale for the Assessment of Positive Symptoms (SAPS [25]) and the Scale for the Assessment of Negative Symptoms (SANS [25]), controlling for age and sex.
Results
Connectome-wide associations of PRS
In the HCP sample, the CPC matrices explained on average 68% of total variance in connectivity matrices across all paradigms (Fig. S1). The NBS analysis revealed a significant association (PFWE < 0.05) between schizophrenia PRSs and a large-scale network involving a total of 69 edges connecting between 54 nodes (Fig. 2a). These nodes were predominantly distributed in the brain’s visual system (including e.g., calcarine sulcus, superior, middle, and inferior occipital gyrus, lingual gyrus, and fusiform gyrus), default-mode system (including e.g., medial frontal cortex, middle and inferior temporal gyrus, and precuneus), and frontoparietal system (including e.g., superior and inferior frontal gyrus, and inferior parietal cortex). In addition to these three major systems, this network also involved several other systems such as the sensorimotor, auditory, salience, attention, and subcortex, although with fewer nodes (see Table S2 for details of the identified nodes and edges in the network). Here, higher PRSs were associated with lower connectivity for all connections in the identified network. The correlation coefficient between the PRSs and the mean CPC of the network was R = −0.37 (Fig. 2b), suggesting that ~14% (R2) of total variance in the detected network can be explained by the polygenic risk of schizophrenia.
Fig. 2. Connectome-wide associations of PRS in the HCP data.
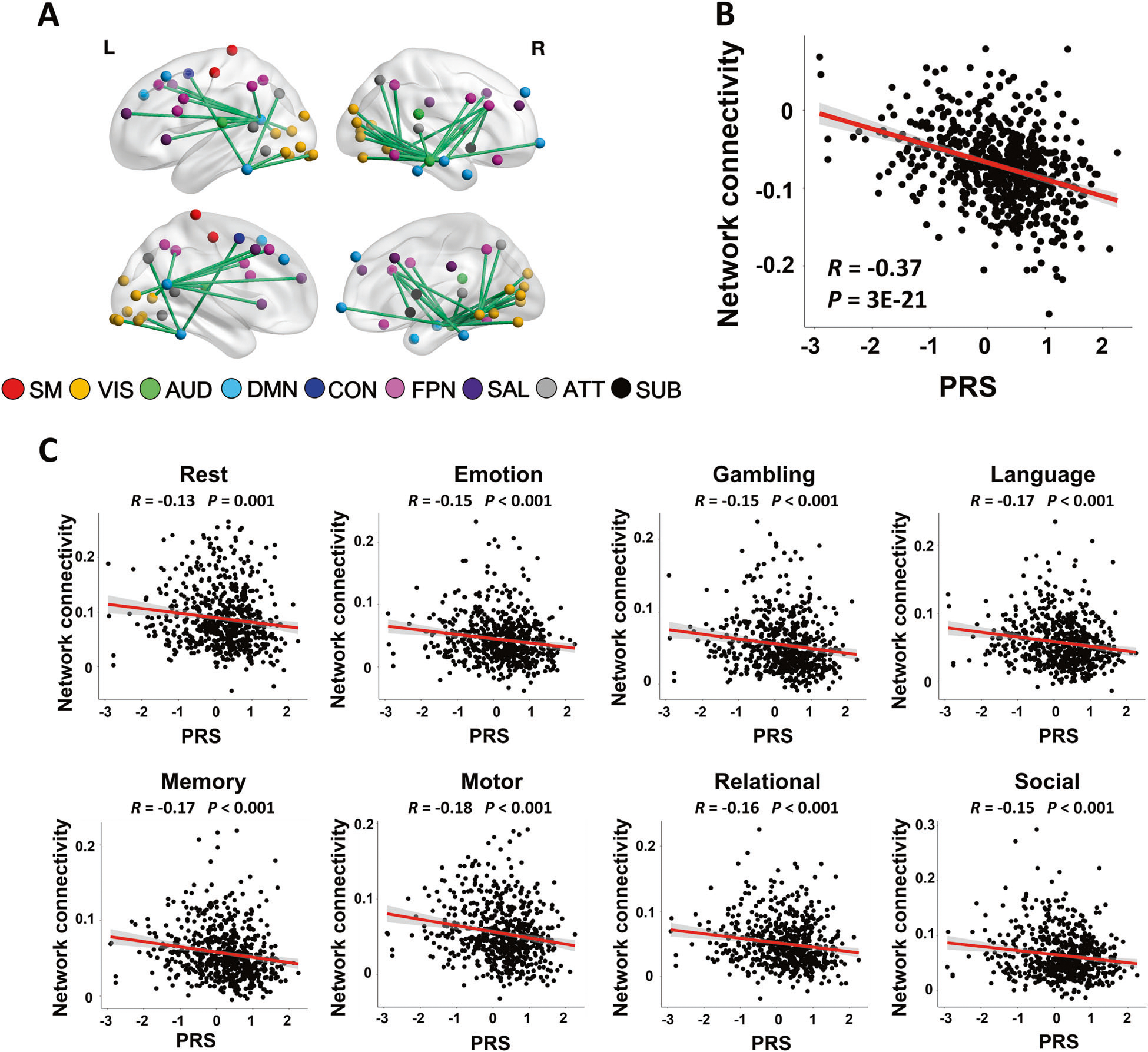
a The identified network consisted of 54 nodes and 69 edges predominantly located in the visual system, default-mode system, and frontoparietal system (SM sensorimotor, VIS visual, AUD auditory, DMN default-mode, CON cingulo-opercular, FPN frontoparietal, SAL salience, ATT attention, SUB subcortex). b The scatter plot showed significant correlation between PRSs and the mean cross-paradigm connectivity of the identified network. Note that the cross-paradigm connectivity values were defined at the PCA space and were rescaled to be mean centered at zero. c The functional connectivity strengths of the identified network were significantly correlated with PRSs in each of the eight studied paradigms, confirming that the observed association is brain-state-independent.
To further confirm that the detected associations were paradigm-independent and not driven by any particular fMRI tasks, we extracted the functional connectivity of the identified network during each of the eight paradigms and examined the correlations of these measures with PRSs. As expected, significant correlations were shown for all eight paradigms (R < −0.13, P < 0.001, Fig. 2c), indicating that the detected connectomic associations indeed reflect a trait-like neural phenotype that is independent of fMRI paradigm.
To examine the robustness of the connectomic finding, we further calculated PRSs for a range of GWAS P-value thresholds from 5 × 10−7 to 5 × 10−2 and correlated the connectivity of the detected network with these PRSs. Significant correlations were present at all of these thresholds (R < −0.10, P < 0.02, Fig. S2), suggesting that the detected connectomic association finding is not limited to the preselected threshold (i.e., GWAS significant) for PRS calculation.
To ensure that the observed result was not driven by head motion, we examined the association between the connectivity of the detected network and mean FD across all paradigms. No significant correlation was found for this analysis (R = 0.06, P = 0.14), suggesting that the detected connectomic associations are unlikely to be driven by head motion.
Presence of the observed network in patients
In the CNP sample, the connectivity of the detected network differed significantly between groups (P = 0.025, Fig. 3a, K–S test for normality P = 0.2, Levene’s test for variance equality P = 0.59). The post hoc analysis showed that the difference was driven by significantly decreased connectivity in patients with schizophrenia compared with that in controls (PBonferroni = 0.03). In contrast, no significant differences were shown between other pairs of groups (PBonferroni > 0.23), though the trend was for a gradient decline of the connectivity in the identified network with increasing genetic proximity to schizophrenia across groups (i.e., SZ < BD < ADHD < HC).
Fig. 3. Clinical and cognitive relevance of the PRS-related network in the CNP data.
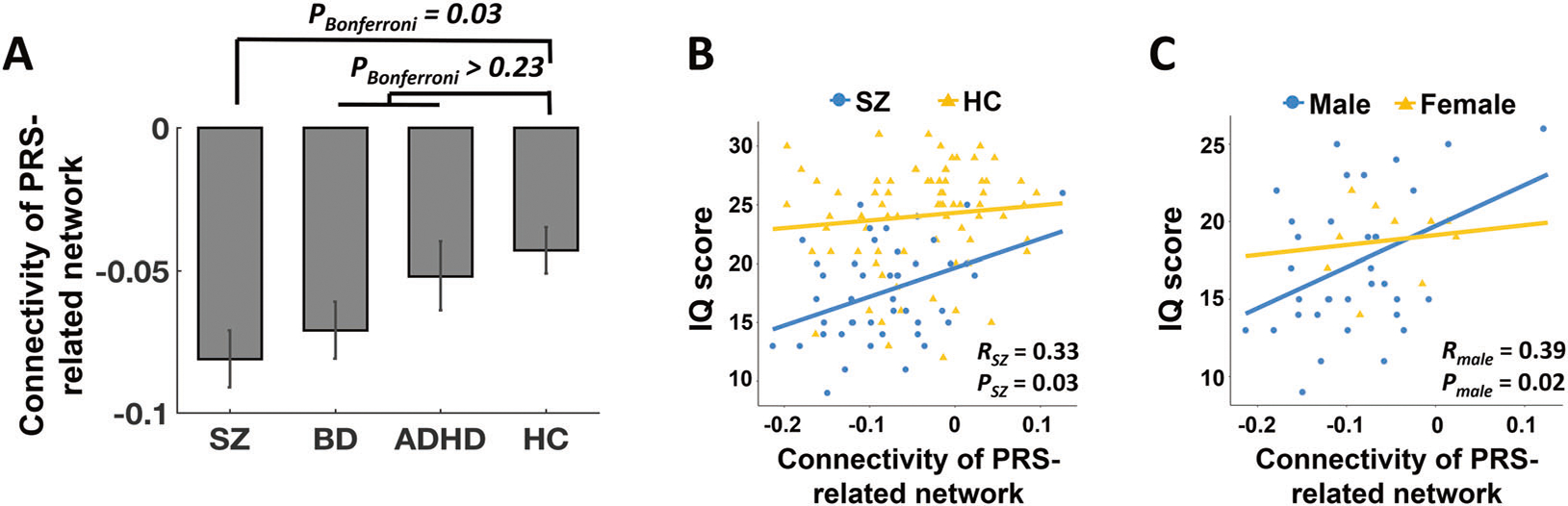
a Significant group differences were shown for the mean cross-paradigm connectivity of the identified network, which was particularly driven by the difference between schizophrenia and controls. b Lower connectivity of the identified network was significantly correlated with lower scores for IQ in patients with schizophrenia but not healthy controls. c In patients, significant correlation between network connectivity and IQ was only observed in males but not females. The error bars indicate standard errors.
Associations with cognition and clinical symptoms
There was a significant positive correlation between the connectivity of the identified network and IQ scores in patients with schizophrenia (R = 0.33, P = 0.03) but not in healthy subjects (R = 0.06, P = 0.64, Fig. 3b). The correlation remained highly significant in the combined sample (R = 0.29, P = 0.001), indicating that patients with lower connectivity in the PRS-related network have lower general cognitive ability. Considering that sex may play an important role in cognitive functioning among schizophrenia [26, 27], we further investigated such association separated by sex. We found that the detected correlation in patients was only significant in males (R = 0.39, P = 0.02) but not in females (R = 0.14, P = 0.71). This may suggest a potential interaction between polygenic risk and sex in contribution to cognitive deficits in schizophrenia.
The partial correlations did not reveal any significant associations between the connectivity of the identified network and clinical symptoms in patients with schizophrenia, nor in the combined sample of SZ and BD. Additional results see Supplementary Materials and Fig. S3.
Discussion
Combining genetic and multiparadigm fMRI data in the HCP dataset, we observed significant connectome-wide associations of schizophrenia polygenic risk in healthy Caucasian population. The identified connectomic associations primarily involved connections within and between the visual system, default-mode system, and frontoparietal system, with higher PRSs associated with lower connectivity in this network. Moreover, using data from an independent clinical consortium, we further showed that connectivity of the same network indexing genetic liability was reduced in patients with schizophrenia and was correlated with IQ scores. Overall, to the best of our knowledge, this study has provided the first evidence for schizophrenia PRS-related neural phenotypes across the whole brain.
Prior work based on group comparisons between high-risk relatives and controls has demonstrated a wealth of connectome-based intermediate phenotypes related to the genetic risk of schizophrenia [28, 29], which mainly include connectivity of the prefrontal cortex [30, 31], lateral and medial temporal cortex [4, 30, 32], parietal cortex [30, 32], and subcortex [4, 31]. In addition, the few studies to date focusing on the effects of PRS on brain connectivity have found significant negative correlations of risk scores with paradigm-dependent functional connectivity measures, including connectivity between prefrontal cortex and hippocampus during resting state [33, 34] and between visual cortex and limbic system during emotional face processing [35]. Highly consistent with these previous findings in both location and directionality, our current work revealed that individuals with higher PRSs had significantly lower connectivity in a large-scale network predominantly encompassing the visual, default-mode, and frontoparietal systems. Notably, our analysis was based on individual connectivity patterns that were independent of any particular fMRI paradigm, and our results were robustly detected across different fMRI paradigms, supporting that these findings are likely to reflect a state-independent neural phenotype influenced by individual genetic vulnerability for schizophrenia. As a proof of concept, the connectivity of the same network was decreased in patients with schizophrenia but not with other major mental disorders when compared with healthy subjects, suggesting particular relevance to schizophrenia. Together, these results point to an individualized imaging connectomic phenotype indexing schizophrenia risk for future genetic and clinical studies.
The regions and systems in the observed network are known to be implicated in schizophrenia. Specifically, dysfunction in the primary sensory systems such as the visual cortex may impair visual perception and visual information inputs to the higher-order cognitive systems, and thus has been considered to be related to symptoms such as hallucinations and emotional dysregulation [4, 24, 36, 37]. The frontoparietal system is one of the major cognitive control systems in humans whose dysfunction is a well-established phenotype linked to cognitive deficits in patients [38, 39]. The abnormal activity and connectivity of the default-mode system may lead to an imbalance between internally focused thoughts and externally goal-directed behaviors and thus contribute to the disturbances in attention and processing of salience information in patients [40, 41]. The overall negative associations of the connectivity between these three systems with PRSs may indicate that accumulating genetic liability to schizophrenia is likely to cause difficulties in integrating sensori–cognitive information across the cerebral cortex, which in turn renders the individuals more prone to developing a psychotic state.
Such interpretation is further corroborated by the observation that the connectivity of the same network was significantly correlated with IQ in patients with schizophrenia, suggesting that functional disturbances in the observed network indeed contribute to impairments in cognitive functioning. Notably, cognitive deficits are a well-known trait for schizophrenia and psychotic disorders, and have long been considered as a fundamental abnormality that underlies clinical symptoms and predicts disease development [42–44]. Genetic studies have also shown a dramatic overlap between genetic variants for schizophrenia and genetic variants for cognition [45–47], implying a strong association between pathogenesis of schizophrenia and cognitive dysfunctioning at the molecular level. Consistent with these previous findings, our results demonstrated a common connectomic phenotype linking PRSs and IQ at the systems level, suggesting that the genetic risk may affect general cognitive ability in patients via the modulation of sensori–cognitive information integration. Moreover, the observed correlation was only significant in male patients in the studied sample, suggesting a potential interaction between genetic risk and sex in contribution to cognitive impairments. This observation is in line with previous publications showing that cognitive functioning in schizophrenia differs between males and females, with more severe cognitive declines in males [26, 27, 48]. These findings together may suggest that males are more vulnerable to the polygenic risk. However, it should be noted that the present sample size is relatively small (especially for females), and thus the observed sex difference needs to be interpreted with caution and be further investigated in larger samples.
We would like to note some limitations of this study. First, our work only examined neural correlates of additive effects of common genetic variants for schizophrenia. However, the difference in estimated heritability between twin studies and SNP-based studies (0.8 vs. 0.2) [49, 50] suggests that nonadditive genetic effects actually contribute to the majority of genetic risk for schizophrenia. The neural mechanisms underlying these nonadditive genetic effects are largely unknown and should be investigated in the future. Second, the identified connectomic associations may only reflect part of the additive genetic effects for schizophrenia, depending on how much of these effects could be captured by PRS. While we reported our main findings based on the most stringent genome-wide threshold to maximize “real” genetic signals and to minimize irrelevant genetic noise, it may not be able to capture the entire additive genetic risk. However, the robustness of such finding across multiple thresholds further supports the idea that additive genetic risk would generally disrupt the connectivity between sensory and cognitive systems, and our results in this sense may reflect the lower bound of such effect. Third, our study lacks an independent sample to replicate the observed connectomic associations with PRS. Fourth, the CNP sample is relatively small, and therefore the observed clinical associations of the identified network merit further replication in larger samples. Fifth, the two samples in this study used different fMRI paradigms. However, such difference would actually strengthen the argument that the observed connectomic associations are state-independent.
To sum up, using genetic and multiparadigm fMRI data, our study demonstrates that higher schizophrenia PRSs are associated with lower connectivity in the visual, default-mode, and frontoparietal systems in healthy adults. Connectivity of this network is decreased in patients with schizophrenia and associated with general cognitive ability in patients. Our results provide the first evidence for connectome-wide associations of schizophrenia polygenic risk.
Code availability
All toolboxes used in this study are freely available. PLINK 1.9 is available at https://www.cog-genomics.org/plink2, PRSice-2 is available at https://choishingwan.github.io/PRSice/, and the NBS toolbox is available at https://www.nitrc.org/projects/nbs/.
Supplementary Material
Acknowledgements
This work was supported by the Brain and Behavior Research Foundation NARSAD Young Investigator Grant (No. 27068) to HC, by National Institute of Health (NIH) grants U01 MH081902 to TDC, and by gifts from the Staglin Music Festival for Mental Health and International Mental Health Research Organization to TDC. Authors would like to thank Kevin Anderson (Yale Psychology) for suggestions on genetic data analysis.
Footnotes
Conflict of interest TDC has served as a consultant for Boehringer-Ingelheim Pharmaceuticals and Lundbeck A/S. The other authors report no conflicts of interest.
Compliance with ethical standards
Supplementary information The online version of this article (https://doi.org/10.1038/s41380-020-0699-3) contains supplementary material, which is available to authorized users.
References
- 1.Esslinger C, Walter H, Kirsch P, Erk S, Schnell K, Arnold C, et al. Neural mechanisms of a genome-wide supported psychosis variant. Science. 2009;324:605. [DOI] [PubMed] [Google Scholar]
- 2.Bigos KL, Mattay VS, Callicott JH, Straub RE, Vakkalanka R, Kolachana B, et al. Genetic variation in CACNA1C affects brain circuitries related to mental illness. Arch Gen Psychiatry. 2010;67:939–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cannon TD, Thompson PM, van Erp TGM, Toga AW, Poutanen VP, Huttunen M, et al. Cortex mapping reveals regionally specific patterns of genetic and disease-specific gray-matter deficits in twins discordant for schizophrenia. Proc Natl Acad Sci USA. 2002;99:3228–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao H, Bertolino A, Walter H, Schneider M, Schafer A, Taurisano P, et al. Altered functional subnetwork during emotional face processing: a potential intermediate phenotype for schizophrenia. JAMA Psychiatry. 2016;73:598–605. [DOI] [PubMed] [Google Scholar]
- 5.International Schizophrenia C, Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wray NR, Lee SH, Mehta D, Vinkhuyzen AAE, Dudbridge F, Middeldorp CM. Research review: polygenic methods and their application to psychiatric traits. J Child Psychol Psychiatry. 2014;55:1068–87. [DOI] [PubMed] [Google Scholar]
- 7.Kauppi K, Westlye LT, Tesli M, Bettella F, Brandt CL, Mattingsdal M, et al. Polygenic risk for schizophrenia associated with working memory-related prefrontal brain activation in patients with schizophrenia and healthy controls. Schizophr Bull. 2015;41:736–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller JA, Scult MA, Conley ED, Chen Q, Weinberger DR, Hariri AR. Effects of schizophrenia polygenic risk scores on brain activity and performance during working memory subprocesses in healthy young adults. Schizophr Bull. 2018;44:844–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walton E, Geisler D, Lee PH, Hass J, Turner JA, Liu J, et al. Prefrontal inefficiency is associated with polygenic risk for schizophrenia. Schizophr Bull. 2014;40:1263–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Q, Ursini G, Romer AL, Knodt AR, Mezeivtch K, Xiao E, et al. Schizophrenia polygenic risk score predicts mnemonic hippocampal activity. Brain. 2018;141:1218–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lancaster TM, Linden DE, Tansey KE. Polygenic risk of psychosis and ventral striatal activation during reward processing in healthy adolescentspg. JAMA Psychiatry. 2016;73:852–61. [DOI] [PubMed] [Google Scholar]
- 12.Fornito A, Zalesky A, Pantelis C, Bullmore ET. Schizophrenia, neuroimaging and connectomics. Neuroimage. 2012;62:2296–314. [DOI] [PubMed] [Google Scholar]
- 13.Dezhina Z, Ranlund S, Kyriakopoulos M, Williams SCR, Dima D. A systematic review of associations between functional MRI activity and polygenic risk for schizophrenia and bipolar disorder. Brain Imaging Behav. 2019;13:862–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Essen DC, Smith SM, Barch DM, Behrens TE, Yacoub E, Ugurbil K, et al. The WU-Minn human connectome project: an overview. Neuroimage. 2013;80:62–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cao H, Chen OY, Chung Y, Forsyth JK, McEwen SC, Gee DG, et al. Cerebello-thalamo-cortical hyperconnectivity as a state-independent functional neural signature for psychosis prediction and characterization. Nat Commun. 2018;9:3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cao H, Ingvar M, Hultman CM, Cannon T. Evidence for cerebello-thalamo-cortical hyperconnectivity as a heritable trait for schizophrenia. Transl Psychiatry. 2019;9:192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poldrack RA, Congdon E, Triplett W, Gorgolewski KJ, Karlsgodt KH, Mumford JA, et al. A phenome-wide examination of neural and cognitive function. Sci Data. 2016;3:160110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Euesden J, Lewis CM, O’Reilly PF. PRSice: polygenic risk score software. Bioinformatics. 2015;31:1466–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schizophrenia Working Group of the Psychiatric Genomics C. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Power JD, Cohen AL, Nelson SM, Wig GS, Barnes KA, Church JA, et al. Functional network organization of the human brain. Neuron. 2011;72:665–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cao H, McEwen SC, Forsyth JK, Gee DG, Bearden CE, Addington J, et al. Toward leveraging human connectomic data in large consortia: generalizability of fMRI-based brain graphs across sites, sessions, and paradigms. Cereb Cortex. 2019;29:1263–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zalesky A, Fornito A, Bullmore ET. Network-based statistic: identifying differences in brain networks. Neuroimage. 2010;53:1197–207. [DOI] [PubMed] [Google Scholar]
- 24.Cao H, Harneit A, Walter H, Erk S, Braun U, Moessnang C, et al. The 5-HTTLPR polymorphism affects network-based functional connectivity in the visual-limbic system in healthy adults. Neuropsychopharmacology. 2018;43:406–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andreasen NC. Methods for assessing positive and negative symptoms. Schizophrenia: positive and negative symptoms and syndromes. Karger: Basel, Switzerland, 1990, 73–88. [DOI] [PubMed] [Google Scholar]
- 26.Mendrek A, Mancini-Marie A. Sex/gender differences in the brain and cognition in schizophrenia. Neurosci Biobehav Rev. 2016;67:57–78. [DOI] [PubMed] [Google Scholar]
- 27.Torniainen M, Suvisaari J, Partonen T, Castaneda AE, Kuha A, Perala J, et al. Sex differences in cognition among persons with schizophrenia and healthy first-degree relatives. Psychiatry Res. 2011;188:7–12. [DOI] [PubMed] [Google Scholar]
- 28.Cao H, Dixson L, Meyer-Lindenberg A, Tost H. Functional connectivity measures as schizophrenia intermediate phenotypes: advances, limitations, and future directions. Curr Opin Neurobiol. 2016;36:7–14. [DOI] [PubMed] [Google Scholar]
- 29.Fornito A, Bullmore ET. Connectomic intermediate phenotypes for psychiatric disorders. Front Psychiatry. 2012;3:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rasetti R, Sambataro F, Chen Q, Callicott JH, Mattay VS, Weinberger DR. Altered cortical network dynamics: a potential intermediate phenotype for schizophrenia and association with ZNF804A. Arch Gen Psychiatry. 2011;68:1207–17. [DOI] [PubMed] [Google Scholar]
- 31.Fornito A, Harrison BJ, Goodby E, Dean A, Ooi C, Nathan PJ, et al. Functional dysconnectivity of corticostriatal circuitry as a risk phenotype for psychosis. JAMA Psychiatry. 2013;70:1143–51. [DOI] [PubMed] [Google Scholar]
- 32.Rasetti R, Mattay VS, White MG, Sambataro F, Podell JE, Zoltick B, et al. Altered hippocampal-parahippocampal function during stimulus encoding: a potential indicator of genetic liability for schizophrenia. JAMA Psychiatry. 2014;71:236–47. [DOI] [PubMed] [Google Scholar]
- 33.Liu S, Li A, Liu Y, Yan H, Wang M, Sun Y, et al. Polygenic effects of schizophrenia on hippocampal grey matter volume and hippocampus-medial prefrontal cortex functional connectivity. Br J Psychiatry. 2019: 1–8. 10.1192/bjp.2019.127 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 34.Wang T, Zhang X, Li A, Zhu M, Liu S, Qin W, et al. Polygenic risk for five psychiatric disorders and cross-disorder and disorder-specific neural connectivity in two independent populations. Neuroimage Clin. 2017;14:441–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lieslehto J, Kiviniemi VJ, Nordstrom T, Barnett JH, Murray GK, Jones PB, et al. Polygenic risk score for schizophrenia and face-processing network in young adulthood. Schizophr Bull. 2019;45:835–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zmigrod L, Garrison JR, Carr J, Simons JS. The neural mechanisms of hallucinations: a quantitative meta-analysis of neuroimaging studies. Neurosci Biobehav Rev. 2016;69:113–23. [DOI] [PubMed] [Google Scholar]
- 37.Silbersweig DA, Stern E, Frith C, Cahill C, Holmes A, Grootoonk S, et al. A functional neuroanatomy of hallucinations in schizophrenia. Nature. 1995;378:176–9. [DOI] [PubMed] [Google Scholar]
- 38.Baker JT, Holmes AJ, Masters GA, Yeo BT, Krienen F, Buckner RL, et al. Disruption of cortical association networks in schizophrenia and psychotic bipolar disorder. JAMA Psychiatry. 2014;71:109–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baker JT, Dillon DG, Patrick LM, Roffman JL, Brady RO Jr., Pizzagalli DA, et al. Functional connectomics of affective and psychotic pathology. Proc Natl Acad Sci USA. 2019;116:9050–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whitfield-Gabrieli S, Ford JM. Default mode network activity and connectivity in psychopathology. Annu Rev Clin Psychol. 2012;8:49–76. [DOI] [PubMed] [Google Scholar]
- 41.Orliac F, Naveau M, Joliot M, Delcroix N, Razafimandimby A, Brazo P, et al. Links among resting-state default-mode network, salience network, and symptomatology in schizophrenia. Schizophr Res. 2013;148:74–80. [DOI] [PubMed] [Google Scholar]
- 42.Kahn RS, Keefe RS. Schizophrenia is a cognitive illness: time for a change in focus. JAMA Psychiatry. 2013;70:1107–12. [DOI] [PubMed] [Google Scholar]
- 43.Elvevag B, Goldberg TE. Cognitive impairment in schizophrenia is the core of the disorder. Crit Rev Neurobiol. 2000;14:1–21. [PubMed] [Google Scholar]
- 44.Seidman LJ, Shapiro DI, Stone WS, Woodberry KA, Ronzio A, Cornblatt BA, et al. Association of neurocognition with transition to psychosis: baseline functioning in the second phase of the North American Prodrome Longitudinal Study. JAMA Psychiatry. 2016;73:1239–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toulopoulou T, Picchioni M, Rijsdijk F, Hua-Hall M, Ettinger U, Sham P, et al. Substantial genetic overlap between neurocognition and schizophrenia—genetic modeling in twin samples. Arch Gen Psychiatry. 2007;64:1348–55. [DOI] [PubMed] [Google Scholar]
- 46.Lencz T, Knowles E, Davies G, Guha S, Liewald DC, Starr JM, et al. Molecular genetic evidence for overlap between general cognitive ability and risk for schizophrenia: a report from the Cognitive Genomics consorTium (COGENT). Mol Psychiatry. 2014;19:168–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hubbard L, Tansey KE, Rai D, Jones P, Ripke S, Chambert KD, et al. Evidence of common genetic overlap between schizophrenia and cognition. Schizophr Bull. 2016;42:832–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Halari R, Kumari V, Mehrotra R, Wheeler M, Hines M, Sharma T. The relationship of sex hormones and cortisol with cognitive functioning in Schizophrenia. J Psychopharmacol. 2004;18:366–74. [DOI] [PubMed] [Google Scholar]
- 49.Hilker R, Helenius D, Fagerlund B, Skytthe A, Christensen K, Werge TM, et al. Heritability of schizophrenia and schizophrenia spectrum based on the Nationwide Danish Twin Register. Biol Psychiatry. 2018;83:492–8. [DOI] [PubMed] [Google Scholar]
- 50.Lee SH, Ripke S, Neale BM, Faraone SV, Purcell SM, Perlis RH, et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nature Genetics. 2013;45:984–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


