Abstract
Infections caused by the bacterium Staphylococcus aureus continue to pose threats to human health and put a financial burden on the healthcare system. The overuse of antibiotics has contributed to mutations leading to the emergence of methicillin-resistant S. aureus, and there is a critical need for the discovery and development of new antibiotics to evade drug-resistant bacteria. Medicinal plants have shown promise as sources of new small-molecule therapeutics with potential uses against pathogenic infections. The principal Rhode Island secondary metabolite (PRISM) library is a botanical extract library generated from specimens in the URI Youngken Medicinal Garden by upper-division undergraduate students. PRISM extracts were screened for activity against strains of methicillin-susceptible S. aureus (MSSA). An extract generated from the tulip tree (Liriodendron tulipifera) demonstrated growth inhibition against MSSA, and a bioassay-guided approach identified a sesquiterpene lactone, laurenobiolide, as the active constituent. Intriguingly, its isomers, tulipinolide and epi-tulipinolide, lacked potent activity against MSSA. Laurenobiolide also proved to be more potent against MSSA than the structurally similar sesquiterpene lactones, costunolide and dehydrocostus lactone. Laurenobiolide was the most abundant in the twig bark of the tulip tree, supporting the twig bark’s historical and cultural usage in poultices and teas.
Introduction
Infections caused by pathogenic bacteria claim millions of lives yearly and accrue billions of dollars in healthcare costs.1 The discovery and implementation of antibiotics to combat pathogens have saved millions of lives worldwide since the discovery of penicillin by Sir Alexander Fleming in 1928.2,3 Apart from inhibitors of cell wall biosynthesis like the β-lactam antibiotics, other classes of antibiotic molecules with distinct mechanisms of action such as the inhibition of protein biosynthesis, the inhibition of DNA synthesis, or the inhibition of folic acid metabolism are now routinely utilized in the clinic.4 The pharmacophores for the majority of these drugs were initially identified from natural products, primarily from microbial sources.5 However, the emergence and spread of antibiotic drug resistance among bacterial pathogens now represent an existential threat to our healthcare systems.6
Staphylococcus aureus causes severe health and economic problems associated with morbidity, mortality, and extended hospital stays due to invasive infections.7 While methicillin-resistant S. aureus (MRSA) is of major concern in hospital settings, methicillin-susceptible S. aureus (MSSA) can be the primary cause of invasive S. aureus infections in the hospital.8 Examining an infant cohort with invasive S. aureus infections from 1997–2012 showed that infant mortality after infection was similar for MRSA and MSSA, and MSSA actually caused more infant infections and more deaths than MRSA.9 Methicillin-susceptible S. aureus (MSSA) can infect multiple human organs, including the skin which is the most prevalent. Untreated MSSA can lead to invasive infections in the bloodstream that lead to life threatening conditions such as sepsis.10
Plant metabolites offer promise in providing new molecular scaffolds to use as antibiotics either in isolation or in combination with currently used treatments to increase efficacy.11,12 The same specialized metabolites that a plant may use to combat microbes in the environment can potentially be harnessed to combat human pathogens. The tulip tree (Liriodendron tulipifera) has been of interest to medicinal chemists for its purported biological properties and cultural and historical use. L. tulipifera is a large flowering deciduous tree endemic across the eastern United States. Indigenous Americans valued the tulip tree for its timber. Medicinally it was utilized as a tonic, antipyretic, and anti-malarial agent.13−15 During the Civil War, Samuel Preston Moore, Surgeon General of the Confederacy, responded to the blockade of medicines by the Union Army by building new field hospitals and commissioning a study of medicinal plants of the Southeast to treat soldiers. Francis Porcher led the search for alternative plant medicines, conducting a deep study of the Southeast principally based on the cultural legacy of Cherokee healers. He ultimately compiled a book containing plant remedies native to the Southern States to help soldiers deal with injuries.16 One of the plants he highlighted was the tulip tree, which was used for rheumatism, gout, laxative, headache, malaria, and other purposes.16−18 The described bioactivities were later validated by the discovery of anti-plasmodial aporphine alkaloids and sesquiterpene lactones from the tree.19 While antibacterial activity has been demonstrated historically from plant preparations and recently from extracts,15 no studies have yet identified the antibacterial constituents of the tulip tree.
The Youngken Medicinal Garden at the University of Rhode Island is a botanical collection of nearly 300 medicinal plant species. In 2019, we developed an extract library of aqueous and organic extracts of the aerial and root portions of specimens found in the garden.20 This resource, the principal Rhode Island secondary metabolite (PRISM) library, was screened for antibiotic activity against MSSA. The library had 110 extracts from 26 plants at the time this work was carried out.
Herein, we identified the organic (methanol) extract of the tulip tree leaves (Liriodendron tulipifera) as having growth inhibitory properties against S. aureus DMS 1104, an MSSA strain. Using bioassay-guided isolation, the bioactive constituent was identified as the sesquiterpene lactone laurenobiolide. We further investigated the relative abundance of this compound in various parts of the tulip tree demonstrating that the compound was present in higher concentrations in the twig bark when compared to other plant parts. Additionally, we examined other Magnoliaceae specimens to determine if they produced laurenobiolide and other related sesquiterpene lactones. Finally, we evaluated a panel of structurally similar sesquiterpene lactones (Figure 1) for their ability to inhibit MSSA.
Figure 1.
Primary active compound against MSSA was determined to be the sesquiterpene lactone laurenobiolide in this study. Other sesquiterpene lactones identified from Liriodendron tulipifera and magnolias were tulipinolide, epi-tulipinolide, costunolide, and dehydrocostus lactone.
Results and Discussion
Screening the PRISM library for anti-MSSA activity revealed five active extracts from the 110 tested. These were the methanol extracts of Amorpha fruticosa flowers and leaves, a combined leaf and flower methanol extract from Isatis tinctora, a methanol extract from the aerial parts of a Datura sp., and a methanol extract derived from L. tulipifera leaves that provided reproducible activity against MSSA (Figure S1). This L. tulipifera extract was subsequently prioritized for further studies due to its documented historical uses as an antimicrobial treatment. High-performance liquid chromatography (HPLC) (Figure S2) was used to visualize the complex metabolite profile leading to our strategy for bioassay-guided fractionation. The tulip tree extract was first fractionated on a C18 column using a stepwise gradient of CH3OH in H2O. Five fractions were generated, and the most robust activity against MSSA was observed from constituents that eluted with 60 and 80% CH3OH (Figure S3). A more polar fraction also initially displayed antibacterial activity, but upon retesting individually isolated components, it seemed to be an additive effect of multiple compounds rather than a single potent entity responsible for the activity. Upon visual analysis of the HPLC chromatograms for each fraction, the most active fractions contained two large peaks that eluted at 30 min from the column (Figure S3). We speculated that a compound in one of these peaks could be responsible for the observed activity.
The two prominent peaks were isolated using semipreparative HPLC and the latter eluting peak exhibited the largest zone of inhibition in the MSSA assay (Figure S4A). The peak purity was checked using a phenyl-hexyl column paired with HPLC–diode-array detector (DAD) analysis. We determined that the earlier eluting peak (peak 1) was a single pure compound while the later eluting peak was two separate analytes (Figure S4B). Upon repurification and retesting of the three compounds, it was determined that only peak 2.1 had antimicrobial activity, while peaks 1 and 2.2 did not show any observable activity in the disk diffusion assay (Figure S4C). Analysis and dereplication via 1D and 2D NMR (Figures S5–S9 and S11) and HRESIMS (Figure S10) revealed that these three metabolites were isomers and the active peak 2.1 was determined to be laurenobiolide. HRESIMS of 2.1 gave an m/z of 313.1418 ([M + Na]+, calcd: 313.1416 for C17H22O4Na). The specific rotation measured for peak 2.1: [α]D23 16.8 (c 1.0, EtOH) closely matched the literature value of [α]D 17.1 (EtOH) for laurenobiolide.21 Additionally, examination of the 1H NMR spectra of peak 2.1 showed an envelope of olefinic protons from δ 4.70 to 5.50 consistent with the original characterization of this metabolite by Tada and Takeda in 1971,22 and the 1H NMR spectrum clearly showed the presence of conformational isomers, a phenomenon previously described for laurenobiolide during NMR analysis (Figure S6).23,24 Examination of correlations from COSY, HSQC, and HMBC NMR data confirmed the identification (Figures S7–S9). Examination of 1H NMR spectra identified peak 1 as epi-tulipinolide (Figure S5), and peak 2.2 as tulipinolide (Figure S11). Evidence included a multiplet resonance at δ 5.72 in peak 1 corresponding to H-8, which was in a cluster of resonances between δ 4.76 and 5.03 in peak 2.2. Furthermore, the acetyl methyl groups at δ 2.09 and 2.06 were consistent with those of tulipinolide and epi-tulipinolide, respectively, as detailed in the original isolation and elucidation work, as were the olefinic methyl resonances.25 Interestingly, among these three sesquiterpene lactones that are extremely similar in structure (Figure 1), only laurenobiolide displayed antibiotic activity toward S. aureus DMS 1104. The minimum inhibitory concentration (MIC) for laurenobiolide and a panel of sesquiterpene lactones was determined via a broth dilution assay (Table 1). Laurenobiolide showed the strongest potency with an MIC of 7.8 μg/mL. Costunolide, one of the terpenoids devoid of an acetyl group, had an MIC value of 31 μg/mL. Epi-tulipinolide, tulipinolide, and dehydrocostus lactone had MIC values of 62, 250, and 62 μg/mL, respectively.
Table 1. MIC Values for Sesquiterpene Lactones Tested against S. aureus DSM 1104.
| compound | MIC (μg/mL) |
|---|---|
| laurenobiolide | 7.8 |
| epi-tulipinolide | 62 |
| tulipinolide | 250 |
| costunolide | 31 |
| dehydrocostus lactone | 62 |
| honokiol | 7.8 |
| gentamicin sulfate (positive control) | 6.3 |
Laurenobiolide was evaluated for its cytotoxic effects to HaCaT cells, a commonly studied human keratinocyte cell line. The compound demonstrated cytotoxic effects at 2.9 μg/mL showing 13% cell viability as compared to the vehicle control (Figure S12). Epi-tulipinolide and tulipinolide showed less cytotoxic activity against the HaCaT cells at 29 μg/mL with cell viability of 66 and 84%, respectively (Figure S12). Doses at 0.29 μg/mL and lower did not show cytotoxicity. These observations are consistent with those of Dettweiler and co-workers, who showed that branch bark extracts of L. tulipifera possessed some mammalian toxicity when tested against cells.15
Next, we investigated which parts of the tulip tree contained the highest concentrations of laurenobiolide. HPLC–DAD and liquid chromatography–mass spectrometry (LC–MS/MS) analyses were performed to evaluate the presence of this constituent in the trunk bark, leaves, and twigs of the tulip tree. Laurenobiolide was most abundant in the twigs, and low amounts were detected in the leaves, but no laurenobiolide was detected in the trunk bark (Figure S13). These findings could be an artifact of the sampling approach, where outer bark pieces were removed non-invasively so as to not damage the tree. Further analysis showed the presence of laurenobiolide in the twig bark, but not the inner twig tissue (Figure S14). These observations are consistent with some of the documented traditional Cherokee uses of the twig bark for medicine,26 and these data will now inform future investigations of plant parts while provoking intriguing questions about the ecological role of laurenobiolide. Additionally, the twig itself showed zones of inhibition when tested against MSSA, while other plant parts tested did not show activity (Figure 2). A poultice-like preparation made with twigs and jojoba oil showed a zone of inhibition, while a portion of “stripped twig” with the outer covering removed did not (Figure S15). This helps connect the historic use of the twig bark poultices with the data on laurenobiolide discovered in this report.
Figure 2.
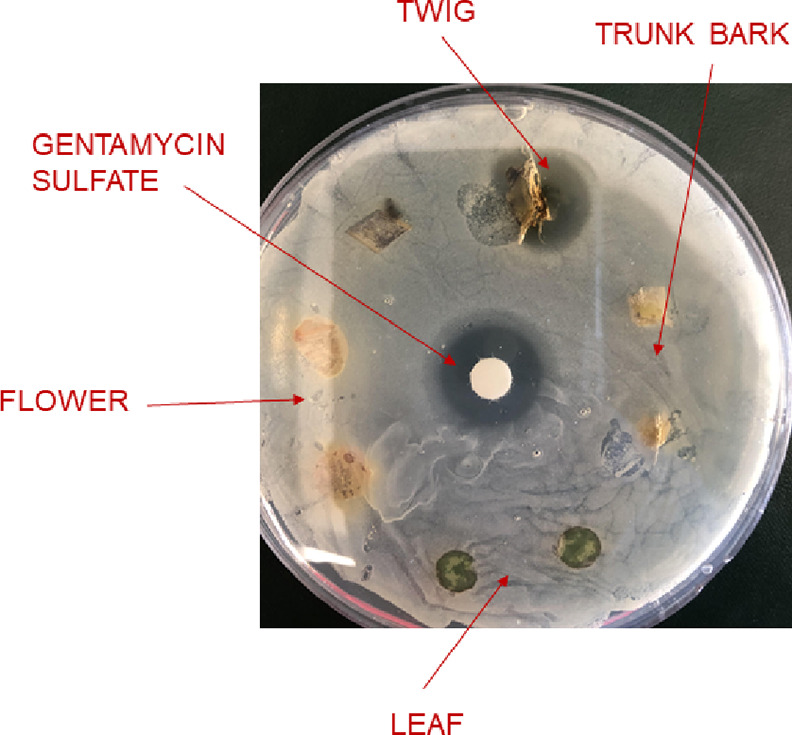
MSSA inhibition assay with L. tulipifera plant parts. Twig, trunk bark, flower, and leaf were directly placed onto MSSA plates. Plant parts were surface sterilized with methanol and dried prior to evaluation. The zone of inhibition can be seen for the twig specimen. Gentamicin sulfate was used as the positive control.
We also examined several magnolia species for the presence of laurenobiolide and other sesquiterpene lactones. Laurenobiolide, epi-tulipinolide, and tulipinolide were only detected in L. tulipifera (Figure 3A,B). However, costunolide was detected in L. tulipifera, Magnolia virginiana, and Magnolia acuminata (Figures 3C and S16), and dehydrocostus lactone was detected in Magnolia macrophylla and M. virginiana (Figure S17). This was confirmed by comparison of the extracts to authentic standards of dehydrocostus lactone and costunolide. Additionally, costunolide and dehydrocostus lactone were identified as library hits on the GNPS social molecular networking website following library analysis (Figures S16 and S17).
Figure 3.
Sesquiterpene lactones in Liriodendron tulipifera and three other Magnolia spp. (A) Epi-tulipinolide (E), laurenobiolide (L), and tulipinolide (T) were only detected in L. tulipifera extracts. (B) Extracted ion chromatogram (m/z 231) shows the base peak of the three acetylated sesquiterpene lactones in L. tulipifera, and an unidentified metabolite in M. virginiana and M. acuminata. (C) Extracted ion chromatogram (m/z 233) shows the presence of costunolide in L. tulipifera, M. virginiana and M. acuminata. (D) Among the magnolias tested, only the M. macrophylla extract showed anti-MSSA activity. Gentamicin sulfate, an aminoglycoside antibiotic, was used as the positive control (20 μg/disk; upper left quadrant on each plate). The test concentration of the extracts was 200 μg/disk, and each extract was tested in duplicate. Negative controls are in the bottom left quadrant of each plate.
Interestingly, other than L. tulipifera, only the M. macrophylla extract showed inhibition of MSSA among the magnolia extracts tested (Figure 3D). To determine the active components in the M. macrophylla extract, we conducted bioassay-guided isolation. Following additional inhibition tests, we characterized honokiol as the major antibacterial component in the M. macrophylla extract (MIC = 7.8 μg/mL against MSSA). Honokiol has reported potent anti-MSSA activity with MIC values as low as 2.5 μg/mL.27 The Magnolia denudata extract did not show the presence of any of the known sesquiterpene lactones on which we focused (Figure S18) and its extract was inactive against MSSA (Figure 2D). Even though M. virginiana and M. acuminata extracts contained costunolide, which showed modest inhibitory activity against MSSA, the concentration was likely too low to show antibacterial activity.
While Magnolia and Liriodendron are both genera in the magnolia family, Liriodendron is the only genus in the Liriodendroidae subfamily, which consists of two species: L. tulipifera and Liriodendron chinense.28 Single nucleotide polymorphism analysis from DNA sequencing results of the specimen used in the current report clearly showed that the specimen was a Liriodendron and specifically L. tulipifera (Figure S19). It will be intriguing to examine authentic specimens of L. chinense to compare the metabolite profile and sesquiterpene lactone composition between the two Liriodendron species.
We searched for biosynthetic genes that could putatively generate sesquiterpene lactones, such as laurenobiolide in publicly available L. tulipifera RNAseq data. The search was based on the pathway identified in Helianthus annuus.29 This pathway synthesizes a sesquiterpene lactone from inunolide or eupatolide using germacrene-A synthase/hydroxylase, and a cytochrome P450 enzyme. This pathway is predicted to occur widely in Viridiplantae,30 and germacrene-A has been identified in the closely related genus Magnolia.31 We recovered L. tulipifera transcripts for farnesyl pyrophosphate synthase, cytochrome P450 (E-class, group IV), acetyl-CoA acetyltransferase (cytosolic), and multiple terpene cyclases including germacrene D synthase (Table S1). Although we found seven distinct terpene cyclases, we did not find a transcript with sufficient homology to indicate the presence of germacrene A synthase or germacrene-A hydroxylase in the L. tulipifera transcriptome. We would predict that laurenobiolide would derive from germacrene A acid. Terpene cyclases can have wide substrate affinity,32 and it is possible that laurenobiolide is being synthesized via a novel biosynthetic pathway. However, due to the lack of a reference genome (this analysis utilized RNAseq data), it is also possible these germacrene A genes were simply not expressed in the tissue at the time of collection. Further genomic and transcriptomic investigations are needed to discern whether this compound is being synthesized via an undescribed biosynthetic pathway, or with germacrene-A, as in H. annuus.
There has been a consistent effort to develop drugs with unique chemical scaffolds and new mechanisms of action to combat pathogen resistance.33 While relatively few antibacterial compounds have been identified in the terpene class, terpenes and terpenoids are an incredibly diverse group of phytochemicals that possess potential as potent antimicrobials.34 The exact mechanism of action of many terpene antibiotics is unknown, but Griffin and co-workers identified that certain terpene metabolites inhibit two essential processes, which include oxygen uptake and oxidative phosphorylation.35
Conclusions
The tulip tree extract has documented antimicrobial activity,15 yet the constituents responsible for the activity had not been identified until now. In this report, we have determined for the first time that the metabolite laurenobiolide isolated from L. tulipifera possesses potent anti-MSSA activity and is the likely component responsible for the antiseptic activity ascribed to the tulip tree. This work sheds light on the intriguing story of the cultural and historical use of tulip tree preparations. The structural features that allow laurenobiolide to inhibit MSSA growth, while very closely related structures do not inhibit growth, remains an intriguing observation that we plan to explore in future studies. The most likely anti-MSSA structural features would appear to be the position of double bonds and/or the absolute configuration about the ring juncture. Furthermore, we have demonstrated the utility of the PRISM library as a source of molecular potential in therapeutic areas, encouraging its future expansion and use in screening efforts. Future research will focus on the mechanism of action of laurenobiolide and its ecological role for the tree. Additionally, we will continue to search for antibacterial components from magnolias and other botanical specimens.
Materials and Methods
General Experimental Procedures
Optical rotations were measured using a Jasco P-2000 polarimeter. NMR spectra were recorded using an Agilent NMR 500 MHz spectrometer with (CD3)2SO (referenced to residual DMSO at δH 2.50 and δC 39.5) or CDCl3 (referenced to residual CHCl3 at δH 7.26 and δC 77.2) at 25 °C. HRESIMS analysis was performed using a SCIEX TripleTOF 4600 mass spectrometer with Analyst software. MS/MS data were recorded on this same instrument using the product ion function. LC–MS/MS was performed using a Thermo Scientific LTQ XL mass spectrometer with an electrospray ionization (ESI) source, and certain mass spectrometry experiments (standard analysis) were completed using a Thermo Scientific ISQ mass spectrometer. Both the LTQ and ISQ were coupled to Dionex Ultimate 3000 HPLC systems equipped with a microvacuum degasser, an autosampler, and DAD. Semipreparative and analytical HPLC was carried out using a Dionex UltiMate 3000 HPLC system equipped with a microvacuum degasser, an autosampler, and a DAD. Standard compounds, costunolide and dehydrocostus lactone, were purchased from Sigma-Aldrich.
Plant Harvesting and Preparation of Plant Extracts
Specimens of the tulip tree (L. tulipifera L., Magnoliaceae), the yulan magnolia (M. denudata Desr., Magnoliaceae, Kiri-01292), the sweetbay magnolia (Magnolia virginiana L., Magnoliaceae, Kiri-01276), the cucumber tree (Magnolia acuminata L., Magnoliaceae, Kiri-01293), and the bigleaf magnolia (M. macrophylla Michx., Magnoliaceae, Kiri-01288) were harvested from the University of Rhode Island campus, and voucher specimens of the magnolias can be found in the KIRI herbarium. Biological material from the L. tulipifera specimen is held in the lab of the corresponding author (voucher 8038051126). Plant extracts in the PRISM library were prepared as previously described from specimens in the Youngken Medicinal Garden.20 Briefly, plants were separated into aerial and underground portions and then further divided (twigs, leaves, etc.) for extraction in both aqueous and organic (methanol) solvents. Each solvent was vacuum filtered to remove particulates, then either freeze-dried, or evaporated under reduced pressure to remove the remaining solvent. The dried product was reconstituted at 10 mg/mL in DMSO or CH3OH depending on solubility.
Methicillin-Susceptible S. aureus Inhibition Assays
S. aureus strain DSM 1104 was cryopreserved and stored at −80 °C. The strain was grown in Luria Bertani (LB) broth (Difco, Fisher scientific, Waltham, MA) at 37 °C, 150 rpm for 20 h. Plant extracts were prepared as described above and 20 μL were loaded onto sterile antimicrobial susceptibility test disks (Oxoid, Thermo Fisher, Franklin, MA). The disks were air dried for 30 min. Control disks were prepared with 20 μL MeOH and similarly dried. The disks were then added to Petri dishes (100 × 15 mm) containing 15 mL of LB agar (Alpha biosciences, Baltimore, MD) inoculated with DSM 1104. Plant parts that were directly tested against DSM 1104 were first sterilized by wiping with CH3OH and then dried before testing. Gentamicin sulfate (20 μg/disk) was the positive control for MSSA. Negative vehicle controls were DMSO or CH3OH (20 μL). Plates were incubated at 37 °C for 18 h, after which the diameters of the zones of growth inhibition were measured to the nearest mm.
MIC of Sesquiterpene Lactones
MICs were determined in duplicate by broth microdilution in a LB medium as previously described36 and in accordance with Clinical Laboratory Standards Institute (CLSI) standards.37,38S. aureus DSM 1104 was obtained from the American Type Culture Collection (ATCC 25923). Test samples were prepared at 10 mg/mL in DMSO. Gentamicin sulfate (2 mg/mL) in H2O was the positive control and DMSO was the negative control.
Bioassay Guided Fractionation of Crude Plant Extracts
We used 6.97 g of dried leaves for initial extraction in CH3OH, which resulted in 140.5 mg of extract. The tulip tree leaf organic extract was further fractionated using a C18 SPE cartridge to purify samples before additional testing. Five fractions were produced from the extract, beginning with the most polar (80% H2O: 20% CH3OH) and ending with the least polar (100% CH3OH). Following solvent removal, the chromatography fractions were reconstituted at 10 mg/mL in the mobile phase followed by HPLC-DAD analysis using a Thermo Scientific Acclaim 3 μm C18 column (150 × 3 mm). A 70 min gradient method was utilized beginning with 15% acetonitrile (CH3CN) for 10 min, then increasing the concentration of CH3CN until 45 min, with a 20 min hold at 100% CH3CN, followed by a return to starting conditions for 5 min. Chromatograms were visually evaluated in parallel with repeated antimicrobial assays to identify potentially active compounds.
Isolation of Sesquiterpene Lactones and Honokiol from L. tulipifera and Magnolia macrophylla
Having identified the potential active components in the tulip tree chromatography fractions following initial chromatogram analysis, fractions 3 (60% H2O/40% CH3OH) and 4 (80% H2O/20% CH3OH) were combined and subjected to reversed-phase semipreparative HPLC using a Kinetex 5 μm C18 column (250 × 10 mm) and a gradient method. The mobile phase was 60% CH3CN/40% H2O with 0.05% formic acid (FA) added to each solvent with a flow rate of 3.0 mL/min. Initial conditions were held for 10.5 min followed by a linear gradient to 12.0 min reaching 100% CH3CN and holding at this concentration for 4.0 min, ultimately returning to initial conditions from min 17–20. Two chromatographic peaks were collected at tR 9.5 (peak 1, epi-tulipinolide, 3.3 mg) and 10.0 min (peak 2), respectively. Peak 2 was further purified using a Luna phenyl-hexyl 5 μm column (250 × 10 mm) and an isocratic method. The mobile phase was 55% CH3CN/45% H2O with 0.05% FA added to each solvent and a flow rate of 3 mL/min. Peaks 2.1 (laurenobiolide, 1.7 mg; tR 13.5 min) and 2.2 (tulipinolide, 1.5 mg; tR 14.25 min) were isolated. The CH3OH branch extract of M. macrophylla was subjected to RP-HPLC using the same isocratic method as described above. The mobile phase was 55% CH3CN/45% H2O with 0.05% FA added to each solvent and a flow rate of 3 mL/min. A single peak eluting at 18.25 min was collected and identified as honokiol (4 mg).
Laurenobiolide (Major Conformer)
Laurenobiolide (major conformer): colorless oil; [α]D23 +16.8 (c 1.0, EtOH) 1H NMR (DMSO-d6, 500 MHz): δ 6.14 (1H, br s, H-13a), 5.86 (1H, br s, H-13b), 5.45 (1H, t, J = 9.9 Hz, H-6), 5.15 (1H, m, H-1), 4.72 (1H, t, J = 10.8 Hz, H-5), 4.24 (1H, dd, J = 9.5, 6.5 Hz, H-8), 3.27 (1H, m, H-7), 2.32 (1H, m, H-2a), 2.14 (1H, m, H-3a), 2.11 (2H, m, H-9), 2.10 (1H, m, H-2b), 1.98 (1H, m, H-3b), 2.03 (3H, s, H-17), 1.59 (3H, s, H-15), 1.58 (3H, s, H-14); 13C NMR (DMSO-d6, 125 MHz): δ 169.7 (C, C-12), 169.6 (C, C-16), 139.3 (C, C-4), 138.1 (C, C-10), 130.3 (C, C-11), 128.9 (CH2, C-13), 126.4 (CH, C-5), 125.4 (CH, C-1), 79.8 (CH, C-8), 71.8 (CH, C-6), 50.2 (CH, C-7). 38.4 (CH2, C-3), 35.9 (CH2, C-9), 24.7 (CH2, C-2), 23.1 (CH3, C-17), 21.3 (CH3, C-15), 21.2 (CH3, C-14); HRESIMS m/z 313.1418 ([M + Na]+) (calcd for C17H22O4Na, 313.1416).
The laurenobiolide 1H NMR data have been deposited at NP-MRD (np-mrd.org/natural_products) (NP0048592).
LC–MS/MS Analysis of L. tulipifera Plant Parts and Magnolia Specimens and Database Searching
Samples of tulip tree trunk, twig, and leaf were taken from the L. tulipifera specimen on the URI campus and methanol extracts were made using 1.0 g portions of each L. tulipifera plant part. Additionally, twig extracts were generated from 1 g portions of M. denudata, M. virginiana, M. acuminata, and M. macrophylla. Each extract was subjected separately to HPLC–DAD and LC–MS/MS analyses with a specific scan event recording MS/MS spectra in the data-dependent acquisition mode. A Kinetex 5 μm C18 column (150 × 4.6 mm) was used for separation of analytes in the extracts. The LC method consisted of a linear gradient from 15 to 100% CH3CN in H2O + 0.1% FA over 20 min, followed by an isocratic period at 100% CH3CN of 5 min. The flow rate was held constant at 0.4 mL/min. The MS spray voltage was 3.5 kV with a capillary temperature of 325 °C. For the MS/MS component, the CID isolation width was 1.0 and the collision energy was 35.0 eV. Following acquisition, the raw data files were converted to the .mgf format using MSConvert from the ProteoWizard suite (http://proteowizard.sourceforge.net/tools.shtml). Library searching was performed using the online platform at the Global Natural Products Social Molecular Networking website (gnps.ucsd.edu).39 Both raw LC–MS/MS files and converted .mzXML files are available in a MassIVE data set—accession MSV000089948.
Detection of Sesquiterpene Lactones in L. tulipifera Plant Parts and Magnolias
L. tulipifera twig, leaf, and trunk bark CH3OH extracts (10 mg/mL) were subjected to HPLC–DAD analysis to evaluate the presence of laurenobiolide. Branch CH3OH extracts from L. tulipifera, M. denudata, M. virginiana, M. acuminata, and M. macrophylla were subjected to HPLC–DAD and LC–MS/MS analyses to specifically investigate the presence of the sesquiterpene lactones epi-tulipinolide, laurenobiolide, and tulipinolide. The chromatography parameters for HPLC–DAD and LC–MS/MS were identical. The Luna phenyl-hexyl 5 μm column (250 × 4.6 mm) was used for separation of analytes. The mobile phase was 55% CH3CN/45% H2O with 0.05% formic acid added to each solvent and a flow rate of 0.6 mL/min. Following the HPLC–DAD and LC–MS/MS analyses, the presence of costunolide and dehydrocostus lactone was confirmed by chromatographic comparison to authentic standards using the methods described above. These raw LC–MS/MS files and converted .mzXML files are also available in the same MassIVE data set—accession MSV000089948.
Culturing HaCaT
Human keratinocytes (HaCaT) were a kind gift from the laboratory of Dr. Navindra Seeram at URI and originally purchased from American Type Cell Culture Collection. The culture was maintained in Dulbecco’s modified Eagle’s medium (DMEM) and the DMEM was supplemented with a 10% fetal bovine serum. An incubator was kept at 37 °C with 5% CO2 at constant humidity. All test samples were dissolved in sterile DMSO for viability assessments.
Cytotoxic Evaluation of Keratinocytes (HaCaT) in the Presence of L. tulipera Compounds
The viability of HaCaT cells in the presence of laurenobiolide, tulipinolide, and epi-tulipinolide was evaluated using cell titer glo (CTG 2.0). HaCaT cells were seeded in a sterile 96-well plate at 5 × 103 cells per well and incubated for 12 h. Postincubation, the media was removed, and compounds were added along with new media, followed by a 24 h incubation. The CTG 2.0 reagent was added to each well and shaken at 200 rpm for 2 min using an orbital shaker. The plate was allowed to equilibrate at room temperature for 10 min, and then the luminescence was recorded using a Spectramax M2 plate reader. Viability was determined by comparing luminescence values of treated wells to those of the vehicle control (DMSO) (n = 4 per treatment). Cannabidiol (31 μg/mL) was used as a positive control.40
Identifying Putatively Useful Nuclear Markers from WGS Data
We used the SISRS bioinformatics package41 to develop nuclear genetic markers for the (1) delineation of Liriodendron spp. from other Magnoliaceae and (2) to delineate Liriodendron chinese and L. tulipifera. Whole-genome sequence (WGS) data for 6 L. tulipifera, 6 L. chinese, and 83 specimens of Magnolia spp. were downloaded from the NCBI SRA database (https://www.ncbi.nlm.nih.gov/sra). All read data were trimmed using the bbmap suite (https://sourceforge.net/projects/bbmap/) using a sliding-window with a Q10 cutoff and removing reads with a final average quality less than Q15 and/or length less than 50 bp. To enrich each data set for nuclear data, we then used bbmap to remove any reads that were mapped against a pooled set of 253 chloroplast assemblies and 5 mitochondrial assemblies downloaded from NCBI. Based on a nuclear genome size estimate for the group of 2.5 Gb, we subset reads from each species such that the final combined read depth was ∼25 Gb (10X coverage), with reads sampled evenly across the 69 species and specimens therein. These subset reads were then pooled and used as an input for a genome assembly using Ray with default parameters,42 with the resulting assembly representing loci that are conserved enough among species to be assembled and compared easily among taxa.
Identifying Fixed Sites for Use in Species and Genus Delineation Assays
To identify genomic sites that would be useful for species delineation between Liriodendron species, we mapped the trimmed reads from all 12 Liriodendron specimens against the SISRS composite genome using bbmap and identified all sites that were (1) fixed and identical within species for all six L. tulipifera or L. chinese specimens and (2) variable between species. A total of 164,710 species-specific SNPs were identified.
To identify SNPs that can delineate Liriodendron from Magnolia, we first identified all sites in the composite genome that were fixed and identical among all 12 Liriodendron specimens. Next, after setting aside eight Magnolia specimens for downstream testing, we removed any sites where those fixed Liriodendron alleles were found in any of the remaining 73 Magnolia samples. Magnolia samples had a variable read coverage and not finding a Liriodendron specific allele could be due to either (1) the allele not being present or (2) the allele not being sampled. To make a more conservative estimate of robust genus-specific SNPs, we only considered SNPs where there was a read coverage in at least half (n = 38) of the Magnolia specimens. A total of 630,741 Liriodendron-specific SNPs were identified.
Testing Genus- and Species-Specific Genetic Markers
To assess the ability of the genetic SNP markers to identify samples to the genus and species levels, we mapped read data from eight Magnolia specimens, two L. tulipifera samples, and one L. chinese sample against the SISRS composite genome. Alleles at sites identified above were queried for all samples, and the proportion of sites matching either (1) the Liriodendron genus-specific allele or (2) the L. tulipifera or L. chinese specific allele was tabulated.
For genus-level screening, individual samples had a coverage for 83,908–594,416 genus SNPs per specimen. The three Liriodendron samples had the matching Liriodendron allele at 99.2–99.9% of surveyed sites, while no more than 0.44% of sites had the corresponding allele across the eight Magnolia samples. For species-level screening, the three Liriodendron samples had coverage for 22,094–152,985 sites per specimen. The L. chinese-specific allele was detected at over 99% of sites in the L. chinese sample, while fewer than 2% of sites contained the allele in either L. tulipifera sample. In parallel, 97.5–99.4% of sites in the two L. tulipifera samples had the L. tulipifera-specific allele, while it was found in only 0.21% of sites for the L. chinese sample.
Evaluation of Potential Biosynthetic Genes
Publicly available L. tulipifera RNAseq data originally collected from apical shoots were retrieved from SRA (SRR16546781, SRR16546782, SRR16546783, SRR16546784, SRR16546785, SRR16546803, and SRR16546804), trimmed with Trimmomatic (v0.39),43 and assembled using rnaSPAdes (v3.15.3).44 This resulted in 332,056 transcripts, with an average length of 1,011 bp. Transdecoder (https://github.com/TransDecoder/TransDecoder) was then run to predict reading frames of the resulting transcripts, resulting in 189,683 predicted proteins. These proteins were annotated with KEGG,45 Interpro,46 and a custom BLAST47 database comprised biosynthetic genes that produced sesquiterpenes such as costunolide in other plant species.
Acknowledgments
We would like to recognize that the historical knowledge of Liriodendron tulipifera was the result of the efforts of indigenous people and their use of the tulip tree in traditional medicines. The University of Rhode Island itself occupies the traditional stomping ground of the Narragansett Nation and the Niantic People. We honor and respect the enduring and continuing relationship between the Indigenous people and this land by teaching and learning more about their history and present-day communities and by becoming stewards of the land we, now too, inhabit. We thank Dr. Navindra Seeram at URI and his coworkers for the HaCaT cells. In this study, the acquisition of high-resolution mass spectrometry, polarimetry, UV–vis data, and NMR data in this publication was made possible by the use of spectrometric and spectroscopic equipment and services available through the RIINBRE Centralized Research Core Facility, which is supported by the Institutional Development Award (IDeA) Network for Biomedical Research Excellence from the National Institute of General Medical Sciences of the National Institutes of Health under Grant P20GM103430. We additionally gratefully acknowledge research support for N.O. from the American Society of Pharmacognosy Summer Research Fellowship and the URI MARC U*STAR grant from the National Institute of General Medical Sciences of the National Institutes of Health under grant number T34GM131948.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.2c03539.
HPLC chromatograms of tulip tree extracts and chromatography fractions and bioassay-guided isolation information, NMR and MS data for epi-tulipinolide, laurenobiolide, tulipinolide, and honokiol, cytotoxicity data for sesquiterpene lactones to HaCaT cells, and chromatography data on distribution of metabolite in plant parts and magnolia specimens, sequence analysis, and putative biosynthetic enzymes (PDF)
Author Contributions
Conceptualization, M.J.B., R.D.K., and E.L.; methodology, R.D.K., M.E.R, N.O., T.M.J., M.A.C., C.W., A.M.K., and M.J.B.; formal analysis, R.D.K., M.E.R., E.S.H., R.L., S.M.H., D.C.R., and M.J.B.; writing—original draft preparation, R.D.K., M.J.B.; writing—review and editing, R.D.K., M.E.R., T.M.J., E.L., D.C.R., and M.J.B. All authors have read and agreed to the published version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Dadgostar P. Antimicrobial Resistance: Implications and Costs. Infect. Drug Rests. 2019, 12, 3903–3910. 10.2147/idr.s234610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S.; Chattopadhyay M. K.; Grossart H. P. The multifaceted roles of antibiotics and antibiotic resistance in nature. Front. Microbiol. 2013, 4, 47. 10.3389/fmicb.2013.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventola C. L. The antibiotic resistance crisis: part 1: causes and threats. Pharm. Therapeut. 2015, 40, 277–283. [PMC free article] [PubMed] [Google Scholar]
- Kapoor G.; Saigal S.; Elongavan A. Action and resistance mechanisms of antibiotics: a guide for clinicians. J. Anaesthesiol. Clin. Pharmacol. 2017, 33, 300–305. 10.4103/joacp.joacp_349_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman D. J.; Cragg G. M. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. 10.1021/acs.jnatprod.9b01285. [DOI] [PubMed] [Google Scholar]
- Martens E.; Demain A. L. The antibiotic resistance crisis, with a focus on the United States. J. Antibiot. 2017, 70, 520–526. 10.1038/ja.2017.30. [DOI] [PubMed] [Google Scholar]
- Chatterjee A.; Rai S.; Guddattu V.; Mukhopadhyay C.; Saravu K. Is methicillin-resistant Staphylococcus Aureus infection associated with higher mortality and morbidity in hospitalized patients? A cohort study of 551 patients from South Western India. Risk Manag. Healthc. Pol. 2018, 11, 243–250. 10.2147/rmhp.s176517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crandall H.; Kapusta A.; Killpack J.; Heyrend C.; Nilsson K.; Dickey M.; Daly J. A.; Ampofo K.; Pavia A. T.; Mulvey M. A.; Yandell M.; Hulten K. G.; Blaschke A. J. Clinical and molecular epidemiology of invasive Staphylococcus aureus infection in Utah children; continued dominance of MSSA over MRSA. PLoS One 2020, 15, e0238991 10.1371/journal.pone.0238991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericson J. E.; Popoola V. O.; Smith P. B.; Benjamin D. K.; Fowler V. G.; Benjamin D. K. Jr.; Clark R. H.; Milstone A. M. Burden of InvasiveStaphylococcus aureusInfections in Hospitalized Infants. JAMA Pediatr. 2015, 169, 1105–1111. 10.1001/jamapediatrics.2015.2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hal S. J.; Jensen S. O.; Vaska V. L.; Espedido B. A.; Paterson D. L.; Gosbell I. B. Predictors of Mortality in Staphylococcus aureus Bacteremia. Clin. Microbiol. Rev. 2012, 25, 362–386. 10.1128/cmr.05022-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazly B. S.; Khameneh B.; Zahedian M. R.; Hosseinzadeh H. In vitro evaluation of antibacterial activity of verbascoside, lemon verbena extract and caffeine in combination with gentamicin against drug-resistant Staphylococcus aureus and Escherichia coli clinical isolates. Avicenna J. Phytomed. 2018, 8, 246–253. [PMC free article] [PubMed] [Google Scholar]
- Rossiter S. E.; Fletcher M. H.; Wuest W. M. Natural products as platforms to overcome antibiotic resistance. Chem. Rev. 2017, 117, 12415–12474. 10.1021/acs.chemrev.7b00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y. F.; Liu C. M.; Kao C. L.; Chen C. Y. Antioxidant and anticancer constituents from the leaves of Liriodendron tulipifera. Molecules 2014, 19, 4234–4245. 10.3390/molecules19044234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quassinti L.; Maggi F.; Ortolani F.; Lupidi G.; Petrelli D.; Vitali L. A.; Miano A.; Bramucci M. Exploring new applications of tulip tree (Liriodendron tulipifera L.): leaf essential oil as apoptotic agent for human glioblastoma. Environ. Sci. Pollut. Res. 2019, 26, 30485–30497. 10.1007/s11356-019-06217-4. [DOI] [PubMed] [Google Scholar]
- Dettweiler M.; Lyles J. T.; Nelson K.; Dale B.; Dale R. M.; Zurawski D. V.; Reddinger C.; Zurawski D. V.; Quave C. L. American Civil War plant medicines inhibit growth, biofilm formation, and quorum sensing by multidrug-resistant bacteria. Sci. Rep. 2019, 9, 7692. 10.1038/s41598-019-44242-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porcher F. P.Resources of the Southern Fields and Forests, Medical, Economical, and Agricultural: Being Also a Medical Botany of the Confederate States, with Practical Information on the Properties of the Trees, Plants, and Shrubs; Steam-power press of Evans and Cogswell: Charleston, SC, 1863. [Google Scholar]
- Spencer C. F.; Koniuszy F. R.; Rogers E. F.; Shavel J.; Easton N. R.; Kaczka E. A.; Kuehl F. A.; Phillips R. F.; Walti A.; Folkers K.; Malanga C.; Seeler A. O.. Survey of Plants for Antimalarial Activity; Lloydia, 1947; Vol. 10, pp 145–174. [Google Scholar]
- Thacher J.American Medical Biography; or, Memoirs of Eminent Physicians Who Have Flourished in America, to Which is Prefixed a Succinct History of Medical Science in the United States from the First Settlement of the Country; Milford House: New York, 1967. [Google Scholar]
- Graziose R.; Rathinasabapathy T.; Lategan C.; Poulev A.; Smith P. J.; Grace M.; Lila M. A.; Raskin I. Antiplasmodial activity of aporphine alkaloids and sesquiterpene lactones from Liriodendron tulipifera L. J. Ethnopharmacol. 2011, 133, 26–30. 10.1016/j.jep.2010.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk R. D.; Carro M. A.; Wu C.; Aldine M.; Wharton A. M.; Goldstein D. G.; Rosario M. E.; Gallucci G. M.; Zhao Y.; Leibovitz E.; Bertin M. J. Integrating natural product chemistry workflows into medicinal chemistry laboratory training: building the PRISM library and cultivating independent research. J. Chem. Educ. 2020, 98, 410–415. 10.1021/acs.jchemed.0c00396. [DOI] [Google Scholar]
- Doskotch R. W.; Wilton J. H.; Harraz F. M.; Fairchild E. H.; Huang C.-T.; El-Feraly F. S. Six additional sesquiterpene lactones from Liriodendron tulipifera. J. Nat. Prod. 1983, 46, 923–929. 10.1021/np50030a016. [DOI] [Google Scholar]
- Tada H.; Takeda K. Structure of the sesquiterpene lactone laurenobiolide. J. Chem. Soc. D 1971, 21, 1391–1392. 10.1039/c29710001391. [DOI] [Google Scholar]
- Tori K.; Horibe I.; Kuriyama K.; Tada H.; Takeda K. Conformational isomers of laurenobiolide, a new ten-membered-ring sesquiterpene lactone. J. Chem. Soc. D 1971, 21, 1393–1394. 10.1039/c29710001393. [DOI] [Google Scholar]
- Tori K.; Horibe I.; Tamura Y.; Kuriyama K.; Tada H.; Takeda K. Re-investigation of the conformation of laurenobiolide, a ten-membered ring sesquiterpene lactone by variable-temperature carbon-13 NMR spectroscopy. Evidence for the presence of four conformational isomers in solution. Tetrahedron Lett. 1976, 17, 387–390. 10.1016/s0040-4039(00)93739-0. [DOI] [Google Scholar]
- Doskotch R. W.; El-Feraly F. S. Antitumor agents. IV. Structure of tulipinolide and epitulipinolide. Cytotoxic sesquiterpenes from Liriodendron tulipifera L. J. Org. Chem. 1970, 35, 1928–1936. 10.1021/jo00831a046. [DOI] [PubMed] [Google Scholar]
- Moerman D. E.Native American Ethnobotany; Timber Press Inc.: Portland, OR, 1998. [Google Scholar]
- Choi E.-J.; Kim H.-I.; Kim J.-A.; Jun S. Y.; Kang S. H.; Park D. J.; Son S.-J.; Kim Y.; Shin O. S. The herbal-derived honokiol and magnolol enhances immune response to infection with methicillin-sensitive Staphylococcus aureus (MSSA) and methicillin-resistant S. aureus (MRSA). Appl. Microbiol. Biotechnol. 2015, 99, 4387–4396. 10.1007/s00253-015-6382-y. [DOI] [PubMed] [Google Scholar]
- Chen J.; Hao Z.; Guang X.; Zhao C.; Wang P.; Xue L.; Zhu Q.; Yang L.; Sheng Y.; Zhou Y.; Xu H.; Xie H.; Long X.; Zhang J.; Wang Z.; Shi M.; Lu Y.; Liu S.; Guan L.; Zhu Q.; Yang L.; Ge S.; Cheng T.; Laux T.; Gao Q.; Peng Y.; Liu N.; Yang S.; Shi J. Liriodendron genome sheds light on angiosperm phylogeny and species-pair differentiation. Nat. Plants 2019, 5, 18–25. 10.1038/s41477-018-0323-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey M.; Schmauder K.; Pateraki I.; Spring O. Biosynthesis of Eupatolide-A Metabolic Route for Sesquiterpene Lactone Formation Involving the P450 Enzyme CYP71DD6. ACS Chem. Biol. 2018, 13, 1536–1543. 10.1021/acschembio.8b00126. [DOI] [PubMed] [Google Scholar]
- Hawkins C.; Ginzburg D.; Zhao K.; Dwyer W.; Xue B.; Xu A.; Rice S.; Cole B.; Paley S.; Karp P.; Rhee S. Y. Plant Metabolic Network 15: A resource of genome-wide metabolism databases for 126 plants and algae. J. Integr. Plant Biol. 2021, 63, 1888–1905. 10.1111/jipb.13163. [DOI] [PubMed] [Google Scholar]
- Farag M. A.; El Din R. S.; Fahmy S. Headspace analysis of volatile compounds coupled to chemometrics in leaves from the Magnoliaceae family. Rec. Nat. Prod. 2015, 9, 153–158. [Google Scholar]
- Pazouki L.; Memari H. R.; Kännaste A.; Bichele R.; Niinemets Ü. Germacrene A synthase in yarrow (Achillea millefolium) is an enzyme with mixed substrate specificity: gene cloning, functional characterization and expression analysis. Front. Plant Sci. 2015, 6, 111. 10.3389/fpls.2015.00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coates A. R. M.; Halls G.; Hu Y.. Novel classes of antibiotics or more of the same?. Br. J. Pharmacol. 2011, 163, 184–194. 10.1111/j.1476-5381.2011.01250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahizan N. A.; Yang S.; Moo C.; Song A. A.; Chong C.; Chong C.; Abushelaibi A.; Lim S. E.; Lai K. Terpene Derivatives as a potential agent against antimicrobial resistance (AMR) Pathogens. Molecules 2019, 24, 2631. 10.3390/molecules24142631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin S. G.; Wyllie S. G.; Markham J. L.; Leach D. N. The role of structure and molecular properties of terpenoids in determining their antimicrobial activity. Flavour Fragrance J. 1999, 14, 322–332. . [DOI] [Google Scholar]
- Deering R. W.; Whalen K. E.; Alvarez I.; Daffinee K.; Beganovic M.; LaPlante K. L.; Kishore S.; Zhao S.; Cezairliyan B.; Yu S.; Rosario M.; Mincer T. J.; Rowley D. C. Identification of a bacteria-produced benzisoxazole with antibiotic activity against multi-drug resistant Acinetobacter baumannii. J. Antibiot. 2021, 74, 370–380. 10.1038/s41429-021-00412-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute . Methods for Dilution of Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically; Approved Standard–10th Edition; CLSI Document M07-A10; Clinical and Laboratory Standards Institute: Wayne, PA, 2015. [Google Scholar]
- Clinical and Laboratory Standards Institute . Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fifth Informational Supplement; CLSI Document M100-S25; Clinical and Laboratory Standards Institute: Wayne, PA, 2015. [Google Scholar]
- Wang M.; Carver J. J.; Phelan V. V.; Sanchez L. M.; Garg N.; Peng Y.; Nguyen D. D.; Watrous J.; Kapono C. A.; Luzzatto-Knaan T.; Porto C.; Bouslimani A.; Melnik A. V.; Meehan M. J.; Liu W.-T.; Crüsemann M.; Boudreau P. D.; Esquenazi E.; Sandoval-Calderón M.; Kersten R. D.; Pace L. A.; Quinn R. A.; Duncan K. R.; Hsu C.-C.; Floros D. J.; Gavilan R. G.; Kleigrewe K.; Northen T.; Dutton R. J.; Parrot D.; Carlson E. E.; Aigle B.; Michelsen C. F.; Jelsbak L.; Sohlenkamp C.; Pevzner P.; Edlund A.; McLean J.; Piel J.; Murphy B. T.; Gerwick L.; Liaw C.-C.; Yang Y.-L.; Humpf H.-U.; Maansson M.; Keyzers R. A.; Sims A. C.; Johnson A. R.; Sidebottom A. M.; Sedio B. E.; Klitgaard A.; Larson C. B.; Boya P C. A.; Torres-Mendoza D.; Gonzalez D. J.; Silva D. B.; Marques L. M.; Demarque D. P.; Pociute E.; O’Neill E. C.; Briand E.; Helfrich E. J. N.; Granatosky E. A.; Glukhov E.; Ryffel F.; Houson H.; Mohimani H.; Kharbush J. J.; Zeng Y.; Vorholt J. A.; Kurita K. L.; Charusanti P.; McPhail K. L.; Nielsen K. F.; Vuong L.; Elfeki M.; Traxler M. F.; Engene N.; Koyama N.; Vining O. B.; Baric R.; Silva R. R.; Mascuch S. J.; Tomasi S.; Jenkins S.; Macherla V.; Hoffman T.; Agarwal V.; Williams P. G.; Dai J.; Neupane R.; Gurr J.; Rodríguez A. M. C.; Lamsa A.; Zhang C.; Dorrestein K.; Duggan B. M.; Almaliti J.; Allard P.-M.; Phapale P.; Nothias L.-F.; Alexandrov T.; Litaudon M.; Wolfender J.-L.; Kyle J. E.; Metz T. O.; Peryea T.; Nguyen D.-T.; VanLeer D.; Shinn P.; Jadhav A.; Müller R.; Waters K. M.; Shi W.; Liu X.; Zhang L.; Knight R.; Jensen P. R.; Palsson B. Ø.; Pogliano K.; Linington R. G.; Gutiérrez M.; Lopes N. P.; Gerwick W. H.; Moore B. S.; Dorrestein P. C.; Bandeira N. Sharing and Community Curation of Mass Spectrometry Data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivas-Aguirre M.; Torres-López L.; Valle-Reyes J. S.; Hernández-Cruz A.; Pottosin I.; Dobrovinskaya O. Cannabidiol directly targets mitochondria and disturbs calcium homeostasis in acute lymphoblastic leukemia. Cell Death Dis. 2019, 10, 779. 10.1038/s41419-019-2024-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz R. S.; Harkins K. M.; Stone A. C.; Cartwright R. A. A composite genome approach to identify phylogenetically informative data from next-generation sequencing. BMC Bioinf. 2015, 16, 193. 10.1186/s12859-015-0632-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert S.; Laviolette F.; Corbeil J. Ray: Simultaneous assembly of reads from a mix of high-throughput sequencing technologies. J. Comp. Biol. 2010, 17, 1519–1533. 10.1089/cmb.2009.0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A. M.; Lohse M.; Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushmanova E.; Antipov D.; Lapidus A.; Prjibelski A. D. rnaSPADES: a de novo transcriptome assembler and its application to RNA-Seq data. GigaScience 2019, 8, giz100. 10.1093/gigascience/giz100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M.; Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum M.; Chang H.-Y.; Chuguransky S.; Grego T.; Kandasaamy S.; Mitchell A.; Nuka G.; Paysan-Lafosse T.; Qureshi M.; Raj S.; Richardson L.; Salazar G. A.; Williams L.; Bork P.; Bridge A.; Gough J.; Haft D. H.; Letunic I.; Marchler-Bauer A.; Mi H.; Natale D. A.; Necci M.; Orengo C. A.; Pandurangan A. P.; Rivoire C.; Sigrist C. J. A.; Sillitoe I.; Thanki N.; Thomas P. D.; Tosatto S. C. E.; Wu C. H.; Bateman A.; Finn R. D. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 2021, 49, D344–D354. 10.1093/nar/gkaa977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F.; Gish W.; Miller W.; Myers E. W.; Lipman D. J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. 10.1016/s0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.





