Abstract
The surface protein P65 is a constituent of the Mycoplasma pneumoniae cytoskeleton and is present at reduced levels in mutants lacking the cytadherence accessory protein HMW2. Pulse-chase studies demonstrated that P65 is subject to accelerated turnover in the absence of HMW2. P65 was also less abundant in noncytadhering mutants lacking HMW1 or P30 but was present at wild-type levels in mutants lacking proteins A, B, C, and P1. P65 exhibited a polar localization like that in wild-type M. pneumoniae in all mutants having normal levels of HMW1 and HMW2. Partial or complete loss of these proteins, however, correlated with severe reduction in the P65 level and the inability to localize P65 properly.
Mycoplasma pneumoniae is a major cause of bronchitis and pneumonia in humans. Adherence of this cell wall-less bacterium to host respiratory epithelium (cytadherence) is pivotal to successful colonization and ensuing pathogenesis (6) and is mediated largely by a differentiated terminal structure, the attachment organelle, which is also believed to function in gliding motility and cell division (reviewed in reference 13). Protein P1 is a major adhesin (12) and localizes primarily to the attachment organelle in wild-type M. pneumoniae cells. Loss of HMW2, whether by frameshift mutation or transposon insertion, results in the inability to cytadhere and reduced levels of the cytadherence-associated proteins HMW1, HMW3, and P65 (8, 11, 15). These proteins are components of a Triton X-100 (TX)-insoluble network that comprises the mycoplasma cytoskeleton, or Triton shell, and are collectively required for the development of a fully functional attachment organelle, including the proper localization of the adhesin P1 to this structure (2, 9, 13, 14, 24).
Proteins HMW1 and HMW3 are largely dissimilar but have in common a central acidic and proline-rich (APR) domain which is defined by its amino acid composition but not its sequence (1, 7, 20). The genes for HMW1 and HMW3 are coexpressed as part of a large transcriptional unit, the hmw operon (7, 30). Both proteins exhibit a distinctive subcellular distribution (25–27); HMW3 is a major component of the attachment organelle, while HMW1 localizes to the filamentous extensions of the mycoplasma cell, including the attachment organelle. The loss of HMW1 and HMW3 in hmw2 mutants occurs posttranslationally, probably a consequence of accelerated turnover by housekeeping protease activity (21). Newly synthesized HMW1 in the cytoplasmic pool associates with the mycoplasma cytoskeleton and is translocated to the mycoplasma surface as a peripheral membrane protein. Translocation and stabilization of HMW1 are much less efficient in the absence of HMW2, resulting in its removal (1).
Like HMW1 and HMW3, protein P65 is a component of the M. pneumoniae Triton shell (23) and is found at significantly reduced levels in hmw2 mutants (15). Furthermore, like HMW1, P65 is peripherally associated with the mycoplasma membrane (22; M. F. Balish and D. C. Krause, unpublished data). P65 contains an APR domain (23), which, unlike that of HMW1 and HMW3, is found at its N terminus. The APR domain of P65 has limited sequence similarity to that of HMW3. The gene encoding P65 is the first gene in the operon of the same name and immediately precedes the gene for HMW2 (15); thus, P65 has not only structural similarity with HMW1 and HMW3 but also a functional association with HMW1, HMW2, and HMW3.
Seto et al. recently localized P65 to the attachment organelle in wild-type M. pneumoniae by using immunofluorescence microscopy (25). However, P65 was not detected by this technique with several cytadherence mutants despite its reported presence at normal levels in those mutants by Western immunoblotting (25). In the current study we evaluated protein P65 further, demonstrating reduced levels of P65 in some but not all cytadherence mutants tested by both Western immunoblotting and immunofluorescence microscopy and establishing that P65 is subject to accelerated turnover in hmw2 mutants, as are HMW1 and HMW3.
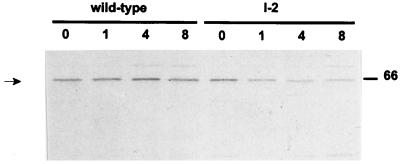
The steady-state level of P65 was previously shown to be substantially lower in hmw2 mutants than in wild-type M. pneumoniae (15). Pulse-chase analysis of protein synthesis and turnover in the hmw2 mutants I-2 and A3 demonstrates that HMW1 and HMW3 are synthesized at wild-type levels but are subject to accelerated turnover (21). We conducted pulse-chase/radioimmunoprecipitation analysis to determine if the same were true for P65. Mycoplasmas were pulse labeled with [35S]methionine, incubated for up to 8 h in fresh Hayflick medium with excess unlabeled methionine, and processed for radioimmunoprecipitation as described previously (21). Samples were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography using X-Omat AR film (Kodak, Rochester, N.Y.) with exposure for 3 to 7 days. The level of P65 in the wild-type M. pneumoniae profiles remained stable throughout the 8-h chase (Fig. 1). Furthermore, the amounts of P65 in the wild-type and mutant I-2 profiles were comparable at the 0-h time point. However, the level of P65 in the mutant decreased during the chase period, as was seen previously for HMW1 and HMW3 (21). A similar pattern (data not shown) was observed with transposon insertion mutant A3 (11). Analysis of the spent medium for wild-type and mutant cultures yielded no evidence of P65 release (data not shown). Thus, like HMW1 and HMW3, in the absence of HMW2, P65 is subject to accelerated turnover, probably by housekeeping protease activity.
FIG. 1.
Pulse-chase/radioimmunoprecipitation analysis of P65 in wild-type and mutant I-2 M. pneumoniae. The chase time (in hours) is shown above the lanes. The position of the 66-kDa protein size standard is indicated on the right. P65 levels (arrow) remained constant in wild-type M. pneumoniae throughout the 8-h time course. The level of P65 in mutant I-2 was comparable to that in wild-type M. pneumoniae at the 0-h time point but decreased subsequently.
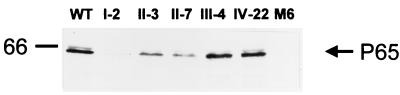
We evaluated P65 steady-state levels in other noncytadhering M. pneumoniae mutants by Western immunoblotting (Fig. 2). Mutant III-4 (14) lacks proteins A, B, and C; the identity and corresponding gene for protein A are not known, but proteins B and C are the 90- and 40-kDa products, respectively, of M. pneumoniae gene MPN142 (5; previously designated E07_orf1218 or orf6 of the P1 operon) and are associated with the attachment organelle (18). Mutant IV-22 lacks protein P1 in addition to A, B, and C (14). Mutants II-7 and II-3 have a truncated cytadherence-associated protein P30 and no P30, respectively (3, 4, 14), while mutant M6 lacks HMW1 and has a truncated P30 (17) but also exhibits reduced levels of HMW2 (M. J. Willby, M. F. Balish, and D. C. Krause, unpublished data). Substantially lower steady-state levels of P65 were associated not only with the loss of HMW2, but also with the loss or truncation of P30 and the loss of HMW1, but not the loss of A, B, and C or A, B, C, and P1 (Fig. 2). For control purposes we also examined the level of FtsH in parallel immunoblots of identical samples. No differences were observed in the levels of FtsH in the wild-type and noncytadhering mutant profiles (data not shown).
FIG. 2.
Western immunoblotting analysis of P65 in wild-type (WT) M. pneumoniae and noncytadhering mutants I-2, II-3, II-7, III-4, IV-22, and M6, as indicated (50 μg of protein per lane). Anti-P65 serum was used at a dilution of 1:7,500. The position of the 66-kDa protein size standard is shown on the left.
The study by Seto et al. (25) included noncytadhering mutants M5, M6, and M7; mutant M6 was the same in both studies, while M5 appears to be similar but not necessarily identical to mutant III-4 from this study. Mutants M7 and II-7 are similar in that each produces a truncated P30 but differ in the extent of truncation. The study by Seto et al. reported no difference by Western immunoblot analysis in P65 steady-state levels in wild-type M. pneumoniae and mutants M5, M6, or M7 (25). For mutant M5, this is consistent with our observation for mutant III-4 (Fig. 2), but otherwise our findings differed from what they reported. The reason for this discrepancy is unclear, particularly for mutant M6, which was presumably identical in both studies.
We analyzed wild-type and mutant M. pneumoniae by immunofluorescence microscopy in an attempt to reconcile the results from the two studies. Frozen stocks of mycoplasmas stored at −80°C in Hayflick broth were thawed, passed through a 25-gauge needle five times to disperse aggregates, inoculated into 500 μl of Hayflick medium in 24-well dishes containing glass cover slips which had been treated with poly-l-lysine, and incubated for 2 h at 37°C. Pretreatment with poly-l-lysine was included to enhance attachment of the noncytadhering mutants to the glass surface. Cells were fixed with 3.4% paraformaldehyde (wt/vol) in Hayflick medium for 15 min at room temperature and 45 min at 4°C. The cover slips were removed and rinsed four times for 5 min each with phosphate-buffered saline (PBS), then treated for 5 min at room temperature in PBS–0.1% Triton X-100 (vol/vol) for permeabilization, as described by Seto et al. (25). Mycoplasmas were again washed four times for 5 min each in PBS, then incubated for 1 h in a blocking solution of PBS–5% (wt/vol) bovine serum albumin (BSA) at room temperature to limit nonspecific labeling, and probed with either rabbit anti-P65 or rabbit anti-FtsH (1) serum diluted 1:100 in PBS–1% BSA overnight at 4°C in a moisture chamber. After rinsing in PBS four times for 5 min each, the cover slips were incubated for 1 h at room temperature with indocarbocyanine (Cy3)-conjugated donkey anti-rabbit immunoglobulin G antibody (Jackson ImmunoResearch Laboratories, West Grove, Pa.) diluted 1:75 in PBS–1% BSA, then rinsed four times for 5 min each in PBS and once in distilled H2O and mounted on microscope slides using Prolong Antifade (Molecular Probes, Eugene, Oreg.).
Cells were examined using a Nikon TE300 epifluorescence microscope with a tetramethyl rhodamine isothiocyanate filter cube (528 to 552 nm) and equipped with phase-contrast optics. Samples were viewed using a 100× objective, and images were digitized using a Micromax charge-coupled device camera (Princeton Scientific Instruments, Monmouth Junction, N.J.). The charge-coupled device exposure time was set at 0.6 s for fluorescence. Digital images were false colorized and superimposed using Adobe Photoshop version 5.0 (Adobe Systems, Mountain View, Calif.). Phase-contrast images were placed in the green channel, while the corresponding fluorescence images were placed in the red channel. The brightness and contrast of the blue channel were adjusted to give a black image.
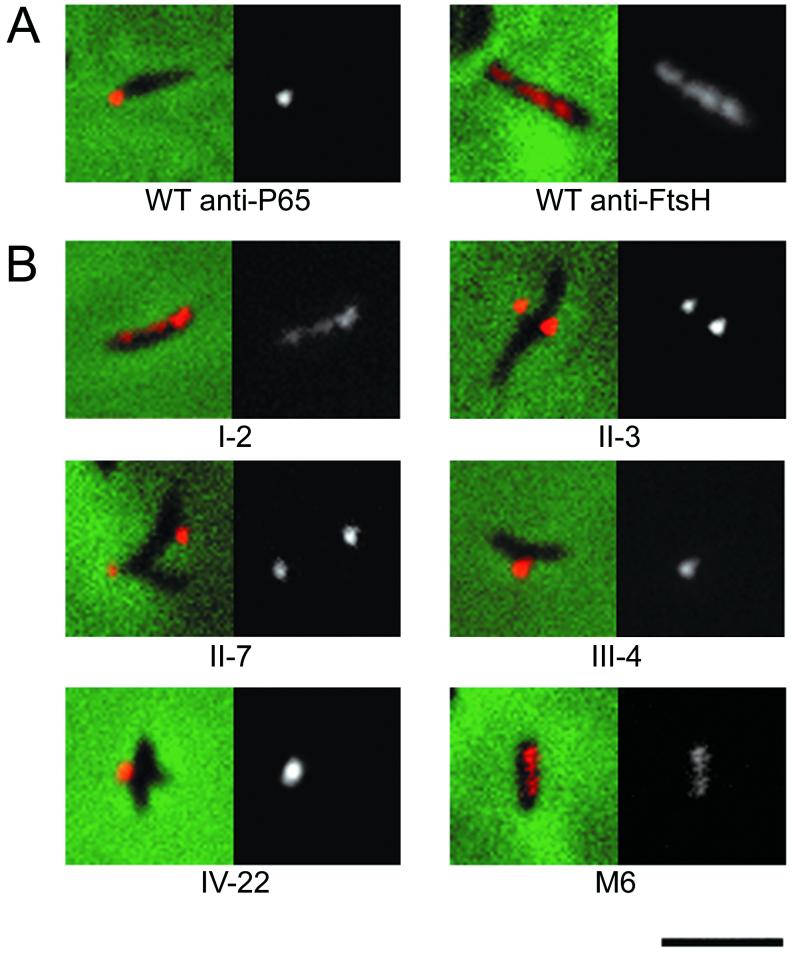
Immunofluorescence microscopy revealed a polar distribution for P65 in wild-type M. pneumoniae cells, with individual cells generally labeled at only one pole (Fig. 3), consistent with results reported by Seto et al. (25). The first panel for each cell type is a merged image of a representative individual cell from fluorescence and phase-contrast microscopy. The second panel is the corresponding fluorescent image alone. We examined mutants I-2, II-3, II-7, III-4, IV-22, and M6 for P65 localization in the same manner (Fig. 3), observing that fluorescence intensity varied among the mutants. The intensity and distribution of fluorescence for mutants II-3, II-7, III-4, and IV-22 were comparable to those of wild-type cells, although more cell branching was observed with the mutants, as has been described previously (24, 25). In marked contrast, mutants I-2 and M6 exhibited substantially lower fluorescence, consistent with the P65 levels detected by Western blots, and with a widespread, patchy distribution.
FIG. 3.
(A) Analysis of P65 and FtsH distribution in wild-type M. pneumoniae. (B) P65 localization in noncytadhering mutants I-2, II-3, II-7, III-4, IV-22, and M6. The first panel for each is the merged fluorescent and phase-contrast images of individual cells. The second panel is the fluorescent image from each composite. Bar, 2.0 μm (same for panels A and B).
As a control for diffuse distribution of a membrane protein, we examined FtsH in wild-type M. pneumoniae, observing a widespread distribution similar to that in Escherichia coli (28). It is noteworthy that evaluation by fluorescence microscopy distinguished major decreases in P65 levels, (for example, with mutants M6 and I-2), but not more subtle decreases (mutants II-3 and II-7, for example). Thus, significant reductions in the level of P65 correlated with partial or total loss of HMW1 or HMW2 and with an inability to localize properly in the mycoplasma cell.
Interestingly, a generally widespread and patchy distribution for P65 was observed with wild-type M. pneumoniae cells not permeabilized with 0.1% TX (data not shown). Furthermore, a similar pattern was observed when cells were analyzed as described elsewhere (27) by immunoelectron microscopy (data not shown). However, a strictly polar labeling pattern was observed when unpermeabilized wild-type cells were probed with affinity-purified anti-P65 antibodies (data not shown), suggesting that this apparent difference in P65 distribution with and without permeabilization reflects antibody reactivity in the anti-P65 serum to a surface component extractable with TX. The lack of a P65− mutant or P65-specific monoclonal antibodies prevents a more definitive determination.
Our findings by fluorescence microscopy for P65 in wild-type M. pneumoniae agree with those of Seto et al. (25). Unlike that study, however, our results from analysis of noncytadhering mutants by immunofluorescence microscopy were consistent with the steady-state levels of P65 detected by Western immunoblotting. While the discrepancy between the two studies might be attributed to strain differences for mutants M5 and M7 compared to III-4 and II-7, respectively, this would not seem to be the case for M6, the only mutant identical in both studies. Differences in the sensitivity of the optical systems might account for the conflicting immunomicroscopy results for mutant M6, but the reason for discrepancy in the Western immunoblot data is not known.
The studies presented here establish clearly that P65, like HMW1 and HMW3, is unstable in the absence of HMW2. It is not known if P65 turnover requires C-terminal sequences previously suggested to be targeted in HMW1 and HMW3. Specifically, the motif P-X-R-X(0–5)-S-S was implicated in the HMW1 C-terminal domain, which is necessary for accelerated turnover in hmw2 mutants (21). This motif does not occur in P65; hence, P65 might be lost indirectly as a secondary consequence of HMW1/HMW3 turnover, perhaps because of failure to anchor properly with the cytoskeleton or at the mycoplasma surface. Alternatively, additional features in the primary or secondary structure of HMW1, HMW3, and P65 may be targeted in their accelerated turnover. HMW1 in mutant I-2 cells fails to associate in a stable manner with the cytoskeleton and is exported less efficiently to the mycoplasma surface, resulting in targeting of the TX-soluble population of HMW1 for accelerated turnover (1). Pulse-chase data presented here, combined with cell fractionation studies published previously (22, 23), suggest that, like HMW1, accelerated turnover of P65 is a consequence of failure to associate efficiently with the mycoplasma cytoskeleton and cell surface. Significantly, the greatest reduction in P65 levels corresponded with partial and/or complete loss of HMW1 or HMW2. Furthermore, the patchy distribution of P65 in these mutants suggests that the loss of P65 in mutants I-2 and M6 might also reflect improper localization and therefore a requirement for HMW1, HMW2, or both in that process.
The observation that the loss of P30 results in decreased levels of P65 suggests that HMW1, HMW2, and HMW3 are not sufficient to stabilize P65 regardless of the mechanism of its loss and despite its proper localization in those mutants. Perhaps significantly, P30 is also not required for localization of P1 to the attachment organelle (24). However, P65 levels were unaffected by the loss of proteins A, B, C, and P1 (Fig. 2); thus, these proteins are not required to stabilize P65. We favor a scenario in which HMW1-HMW3, P30, and P65 are all elements in the assembly pathway of the attachment organelle (13). Failure to incorporate each protein at the appropriate time appears to have downstream consequences. The reduced levels of P65 in some mutants suggest that P65 may be a late addition in the assembly process, consistent with previous predictions (1, 25). However, the role of P65 in the functions of the terminal organelle, including cytadherence, gliding motility, and cell division, remains unclear and requires additional investigation.
Acknowledgments
We thank Richard Herrmann for his generous donation of P65-specific antiserum.
This work was supported by Public Health Service Research grant AI23362 to D.C.K.
REFERENCES
- 1.Balish M F, Hahn T-W, Popham P L, Krause D C. Stability of Mycoplasma pneumoniae cytadherence accessory protein HMW1 correlates with its association with the Triton shell. J Bacteriol. 2001;183:3680–3688. doi: 10.1128/JB.183.12.3680-3688.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baseman J B, Cole R M, Krause D C, Leith D K. Molecular basis for cytadsorption of Mycoplasma pneumoniae. J Bacteriol. 1982;151:1514–1522. doi: 10.1128/jb.151.3.1514-1522.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baseman J B, Morrison-Plummer J, Drouillard D, Puleo-Scheppke B, Tryon V V, Holt S C. Identification of a 32 kilodalton protein of Mycoplasma pneumoniae associated with hemadsorption. Isr J Med Sci. 1987;23:474–479. [PubMed] [Google Scholar]
- 4.Dallo S F, Lazzell A L, Chavoya A, Reddy S P, Baseman J B. Biofunctional domains of the Mycoplasma pneumoniae P30 adhesin. Infect Immun. 1996;64:2595–2601. doi: 10.1128/iai.64.7.2595-2601.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dandekar T, Huynen M, Regula J T, Ueberle B, Zimmermann C U, Andrade M A, Doerks T, Sánchez-Pulido L, Snel B, Suyama M, Yuan Y P, Herrmann R, Bork P. Re-annotating the Mycoplasma pneumoniae genome sequence: adding value, function and reading frames. Nucleic Acids Res. 2000;28:3278–3288. doi: 10.1093/nar/28.17.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denny F W, Clyde W A J, Glezen W P. Mycoplasma pneumoniae disease: clinical spectrum, pathophysiology, epidemiology, and control. J Infect Dis. 1971;123:74–92. doi: 10.1093/infdis/123.1.74. [DOI] [PubMed] [Google Scholar]
- 7.Dirksen L B, Proft T, Hilbert H, Plagens H, Herrmann R, Krause D C. Nucleotide sequence analysis and characterization of the hmw gene cluster of Mycoplasma pneumoniae. Gene. 1996;171:19–25. doi: 10.1016/0378-1119(96)00050-9. [DOI] [PubMed] [Google Scholar]
- 8.Fisseha M, Göhlmann H W H, Herrmann R, Krause D C. Identification and complementation of frameshift mutations associated with loss of cytadherence in Mycoplasma pneumoniae. J Bacteriol. 1999;181:4404–4410. doi: 10.1128/jb.181.14.4404-4410.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hahn T-W, Willby M J, Krause D C. HMW1 is required for cytadhesin P1 trafficking to the attachment organelle in Mycoplasma pneumoniae. J Bacteriol. 1998;180:1270–1276. doi: 10.1128/jb.180.5.1270-1276.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayflick L. Tissue cultures and mycoplasmas. Tex Rep Biol Med. 1965;23(Suppl. 1):285–303. [PubMed] [Google Scholar]
- 11.Hedreyda C T, Krause D C. Identification of a possible cytadherence regulatory locus in Mycoplasma pneumoniae. Infect Immun. 1995;63:3479–3483. doi: 10.1128/iai.63.9.3479-3483.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu P-C, Collier A, Baseman J B. Surface parasitism by Mycoplasma pneumoniae of respiratory epithelium. J Exp Med. 1977;145:1328–1343. doi: 10.1084/jem.145.5.1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krause D C, Balish M F. Structure, function, and assembly of the terminal organelle of Mycoplasma pneumoniae. FEMS Microbiol Lett. 2001;198:1–7. doi: 10.1111/j.1574-6968.2001.tb10610.x. [DOI] [PubMed] [Google Scholar]
- 14.Krause D C, Leith D K, Wilson R M, Baseman J B. Identification of Mycoplasma pneumoniae proteins associated with hemadsorption and virulence. Infect Immun. 1982;35:809–817. doi: 10.1128/iai.35.3.809-817.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krause D C, Proft T, Hedreyda C T, Hilbert H, Plagens H, Herrmann R. Transposon mutagenesis reinforces the correlation between Mycoplasma pneumoniae cytoskeletal protein HMW2 and cytadherence. J Bacteriol. 1997;179:2668–2677. doi: 10.1128/jb.179.8.2668-2677.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 17.Layh-Schmitt G, Hilbert H, Pirkl E. A spontaneous hemadsorption-negative mutant of Mycoplasma pneumoniae exhibits a truncated adhesin-related 30-kilodalton protein and lacks the cytadherence accessory protein HMW1. J Bacteriol. 1995;177:843–846. doi: 10.1128/jb.177.3.843-846.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Layh-Schmitt G, Herrmann R. Spatial arrangement of gene products of the P1 operon in the membrane of Mycoplasma pneumoniae. Infect Immun. 1994;62:974–979. doi: 10.1128/iai.62.3.974-979.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lipman R P, Clyde W A., Jr The interrelationship of virulence, cytadsorption and peroxide formation in Mycoplasma pneumoniae. Proc Soc Exp Biol Med. 1969;131:1163–1167. doi: 10.3181/00379727-131-34061. [DOI] [PubMed] [Google Scholar]
- 20.Ogle K F, Lee K K, Krause D C. Nucleotide sequence analysis reveals novel features of the phase-variable cytadherence accessory protein HMW3 of Mycoplasma pneumoniae. Infect Immun. 1992;60:1633–1641. doi: 10.1128/iai.60.4.1633-1641.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Popham P L, Hahn T-W, Krebes K, Krause D C. Loss of HMW1 and HMW3 in noncytadhering mutants of Mycoplasma pneumoniae occurs post-translationally. Proc Natl Acad Sci USA. 1997;94:13979–13984. doi: 10.1073/pnas.94.25.13979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Proft T, Herrmann R. Identification and characterization of hitherto unknown Mycoplasma pneumoniae proteins. Mol Microbiol. 1994;13:337–348. doi: 10.1111/j.1365-2958.1994.tb00427.x. [DOI] [PubMed] [Google Scholar]
- 23.Proft T, Hilbert H, Layh-Schmitt G, Herrmann R. The proline-rich P65 protein of Mycoplasma pneumoniae is a component of the Triton X-100-insoluble fraction and exhibits size polymorphism in strains M129 and FH. J Bacteriol. 1995;177:3370–3378. doi: 10.1128/jb.177.12.3370-3378.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Romero-Arroyo C E, Jordan J, Peacock S J, Willby M J, Farmer M A, Krause D C. Mycoplasma pneumoniae protein P30 is required for cytadherence and associated with proper cell development. J Bacteriol. 1999;181:1079–1087. doi: 10.1128/jb.181.4.1079-1087.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seto S, Layh-Schmitt G, Kenri T, Miyata M. Visualization of the attachment organelle and cytadherence proteins of Mycoplasma pneumoniae by immunofluorescence microscopy. J Bacteriol. 2001;183:1621–1630. doi: 10.1128/JB.183.5.1621-1630.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stevens M K, Krause D C. Localization of the Mycoplasma pneumoniae cytadherence-associated proteins HMW1 and HMW4 in the cytoskeleton-like Triton shell. J Bacteriol. 1991;173:1041–1050. doi: 10.1128/jb.173.3.1041-1050.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stevens M K, Krause D C. Mycoplasma pneumoniae cytadherence phase-variable protein HMW3 is a component of the attachment organelle. J Bacteriol. 1992;174:4265–4274. doi: 10.1128/jb.174.13.4265-4274.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tomoyasu T, Yamanaka K, Murata K, Suzaki T, Bouloc P, Kato A, Niki H, Hiraga S, Ogura T. Topology and subcellular localization of FtsH protein in Escherichia coli. J Bacteriol. 1993;175:1352–1357. doi: 10.1128/jb.175.5.1352-1357.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waldo R H, Popham P L, Romero-Arroyo C E, Mothershed E A, Lee K K, Krause D C. Transcriptional analysis of the hmw gene cluster of Mycoplasma pneumoniae. J Bacteriol. 1999;181:4978–4985. doi: 10.1128/jb.181.16.4978-4985.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]