Abstract
Purpose
To elucidate the differences in ocular biometric parameters by generation and gender and to identify axial length (AL)-associated genetic variants in Japanese individuals, we analyzed Tohoku Medical Megabank Organization (ToMMo) Eye Study data.
Design
We designed the ToMMo Eye Study, examined AL variations, and conducted genome-wide association studies (GWASs).
Participants
In total, 33 483 participants aged > 18 years who were recruited into the community-based cohort (CommCohort) and the birth and three-generation cohort (BirThree Cohort) of the ToMMo Eye Study were examined.
Methods
Each participant was screened with an interview, ophthalmic examinations, and a microarray analysis. The GWASs were performed in 22 379 participants in the CommCohort (discovery stage) and 11 104 participants in the BirThree Cohort (replication stage). We evaluated the associations of single nucleotide polymorphisms (SNPs) with AL using a genome-wide significance threshold (5 × 10-8) in each stage of the study and in the subsequent meta-analysis.
Main Outcome Measures
We identified the association of SNPs with AL and distributions of AL in right and left eyes and individuals of different sexes and ages.
Results
In the discovery stage, the mean AL of the right eye (23.99 mm) was significantly greater than that of the left eye (23.95 mm). This difference was reproducible across sexes and ages. The GWASs revealed 703 and 215 AL-associated SNPs with genome-wide significance in the discovery and validation stages, respectively, and many of the SNPs in the discovery stage were replicated in the validation stage. Validated SNPs and their associated loci were meta-analyzed for statistical significance (P < 5 × 10-8). This study identified 1478 SNPs spread over 31 loci. Of the 31 loci, 5 are known AL loci, 15 are known refractive-error loci, 4 are known corneal-curvature loci, and 7 loci are newly identified loci that are not known to be associated with AL. Of note, some of them shared functional relationships with previously identified loci.
Conclusions
Our large-scale GWASs exploiting ToMMo Eye Study data identified 31 loci linked to variations in AL, 7 of which are newly reported in this article. The results revealed genetic heterogeneity and similarity in SNPs related to ethnic variations in AL.
Keywords: Axial length, Genome-wide association study, Population-based cohort study
Abbreviations and Acronyms: AL, axial length; BirThree, birth and three-generation cohort; CommCohort, community-based cohort; GWAS, genome-wide association study; MAF, minor allele frequency; PCA, principal component analysis; TMM, Tohoku Medical Megabank; ToMMo, Tohoku Medical Megabank Organization
Myopia, a type of refractive error, is one of the most common eye disorders, and its most severe form eventually causes blindness.1 Visual impairments due to severe myopia are often associated with structural changes in the eye, such as degeneration of the retina and choroid.2 Refraction is mainly determined by axial length (AL), corneal curvature, and anterior chamber depth. Of the 3 factors, AL is recognized as the most basic anatomic parameter associated with refraction.1 As an ocular parameter, AL is the major variable influencing the optical quality of the image on the retina and is an important predictor for ocular diseases. The increasing prevalence of myopia in modern society implies that the prevalence of related complications, such as myopic maculopathy and neuropathy, retinal detachment, cataracts, and various types of glaucoma, is also increasing.3, 4, 5
Many population-based and cross-sectional studies have revealed a significantly increased prevalence of myopia in Europe, North America, Australia, and Asia.6,7 Of these regions, Asia exhibits a significantly higher prevalence of myopia than Europe,8, 9, 10 as indicated in several previous studies, including the Beijing Eye Study,11 the Tanjong Pagar Survey,12 and the Tajimi Study.13 Although the precise ethnic differences in myopia prevalence remain to be clarified, myopia is known to be a multifactorial disease under the influence of both genetic and environmental factors.14
Elucidation of the causative factors and complexities of myopia is an important step that will provide eye clinicians and scientists with new insights into the genetics of ocular phenotypes. Many common diseases are multifactorial and complex, failing to exhibit typical Mendelian inheritance attributable to a single gene.15,16 Via exploitation of catalogs of millions of common single nucleotide polymorphisms (SNPs), genome-wide association studies (GWASs) have enabled the identification of many susceptibility loci for ophthalmic phenotypes; for instance, SNPs related to myopia have been identified across populations of several ethnicities.17,18
We previously established the Tohoku Medical Megabank (TMM) project, which is strategically conducting 2 cohort studies and is also constructing an integrated biobank.19 Tohoku University and Iwate Medical University have been cooperating on the TMM study, and Tohoku University is responsible for the Tohoku Medical Megabank Organization (ToMMo) Study. More than 157 000 individuals voluntarily participated in the TMM study by March 2016, and approximately 33 000 individuals received detailed physiological examinations, including ophthalmic examination.19 We also conducted microarray analysis by using a Japonica array, an ethnicity-specific microarray developed in the ToMMo Study,20 with genotype imputation.
Whereas several studies have examined the AL in population-based and cross-sectional studies,11,12,21, 22, 23, 24, 25 only a limited number of studies have challenged the identification of AL-specific SNPs and associated loci by means of GWASs.26, 27, 28 By contrast, regarding refractive error, several large studies, such as the International Consortium for Refractive Error and Myopia17 and the Consortium for Refractive Error and Myopia combined with the UK Biobank Eye and Vision Consortium,29,30 have reported comprehensive results. Additionally, data from these studies have been meta-analyzed together with 23andMe and have been reported.18,31
Although these studies identified many novel loci and pathways involved in refractive error, genetic factors leading to the increase in myopia prevalence remain largely elusive. Therefore, to understand the genetic and environmental influences on AL, we established the ToMMo Eye Study and collected data on AL and related factors from 2 cohorts strategically designed for the Japanese population. We examined basic characteristics related to eye functions and conducted GWASs of AL. The results identified 1478 SNPs related to AL that were spread over 31 loci, including many new loci associated with AL and myopia.
Methods
Cohort Design and Study Population
The TMM is conducting a population-based genome cohort study and constructing an integrated biobank that includes the ophthalmic examination data of the participants.19 Two prospective cohort studies have been established in TMM: the community-based cohort (CommCohort) study, which recruited 84 073 participants,32 and the birth and three-generation cohort (BirThree Cohort) study, which recruited 73 529 participants.33 Within TMM, Tohoku University is conducting the ToMMo Study in Miyagi Prefecture. The ToMMo CommCohort Study recruited participants at specific health checkup sites in 28 municipalities and collected basic information and samples after recruitment. Approximately 65% of the residents we asked were enrolled with informed consent. The BirThree Cohort Study recruited pregnant participants at the obstetric hospital, and the enrollment rate was similar. More than 33 000 participants in ToMMo cohort studies received additional detailed examinations at 7 community support centers to assess many variables related to health conditions.19
We conducted the same set of ophthalmic examinations in all community support centers. The ophthalmic examinations include assessments of AL and intraocular pressure and examination of the fundus. In this project, we also obtained the following information from the participants: answers to a wide range of questions regarding family history, residential area, education, and various environmental factors; results of physiological examinations; and results of multi-omics analyses of participant biospecimens. Participants who had ocular disease, for example, retinal detachment, central serous chorioretinopathy, macular edema due to diabetic retinopathy, or age-related macular degeneration, as well as those who had undergone ophthalmic surgery, including cataract surgery, were excluded from the study. Information on ocular disease and family history was obtained from questionnaires.
For the discovery-stage GWAS on AL, we used the CommCohort Study with 22 635 genotyped subjects with AL measurements. For the validation-stage GWAS on AL, we used the BirThree Cohort Study with 11 188 genotyped subjects with AL measurements. For the meta-analysis stage, we analyzed 33 483 genotyped subjects of both cohort studies after removing the subjects who met the exclusion criteria.
Our genome medical research coordinators informed eligible individuals of the aims and protocols of the TMM study and obtained their informed consent. At baseline, we explained the protocols of the cohort studies, biobanking, and the general research methods for genomic analyses and omics analyses. Institutional Review Board approval was obtained from the Ethics Committee of the ToMMo (2012-4-617, 2013-4-103, 2020-4-155, 2020-4-156). This study was conducted in accordance with the Declaration of Helsinki, the Ethical Guidelines for Human Genome/Gene Analysis Research, and other appropriate guidelines.
Eye Examination Procedures
Comprehensive eye examinations included measurements of AL and intraocular pressure. Axial length was measured with an OA-1000 (Tomey), which uses the same partial coherence interferometry technique as the IOL Master instrument (Carl Zeiss AG) to measure signals from the tear film and retinal pigment epithelium. Ten valid AL readings were taken and averaged. Intraocular pressure was measured 3 times for each eye and averaged using a TX-20P Full Auto Tonometer (Canon).
Genotyping, Imputation, and Quality Control
To prepare direct and imputed genotype datasets, 99 564 samples, including samples from the CommCohort Study (for the discovery study) and the BirThree Cohort Study (for the validation study), were genotyped with an Affymetrix Axiom Japonica Array (v2) separately in 21 batches. For quality control, we excluded plates with an average call rate < 0.95 and removed samples with a DishQC metric < 0.82 or a step 1 call rate < 0.97 before batch genotyping. We excluded samples with a call rate < 0.95 or with unusually high identical by descent values compared with most of the other samples. We applied an SNP quality control step to each batch to exclude variants with a Hardy-Weinberg equilibrium test P value < 1.00 × 10-5, a minor allele frequency (MAF) < 0.01, or a missing rate > 0.01. We merged the imputed genotype datasets for the 21 batches using QCTOOL (v2.0.4) (https://www.well.ox.ac.uk∼gavqctool). Thus, we obtained an imputed genotype dataset in the Oxford BGEN format for 99 564 Japanese individuals with 54 034 112 variants. We also obtained a direct genotype dataset in the PLINK BED format for 659 328 variants in these 99 564 individuals by merging the genotype datasets before imputation for the 21 batches.
We phased the genotype data of each batch using SHAPEIT2 (v2. r837).34 Genotype imputation was performed for the phased genotype data of each batch with IMPUTE2 (ver. 2.3.2)35 using a phased reference panel of 3552 Japanese individuals from cohort studies of the TMM project.36 We then merged the imputed genotype datasets using QCTOOL (v2.0.4) (https://www.well.ox.ac.uk∼gavqctool). Ultimately, we obtained an imputed genotype dataset in Oxford BGEN format and a direct genotype dataset in PLINK BED format.
We applied an SNP quality control step to the imputed and direct genotype datasets for the discovery and validation stages to exclude variants with Hardy-Weinberg equilibrium test P values < 1.00 × 10-5, MAF values < 0.01, or missing rates > 0.05. The number of remaining variants in each dataset for GWAS was 8 585 409 in the discovery stage and 8 590 037 in the validation stage. Then, we extracted 22 635 individuals who were from the CommCohort Study along with their AL phenotypes and covariate values for the discovery stage, and we extracted 11 188 individuals who were from the BirThree Cohort Study along with their AL phenotypes and covariate values for the validation stage. After exclusion of subjects who showed gender/sex inconsistency, identical twins, extreme outlier individuals as detected by principal component analysis (PCA), and subjects with a history of ocular disease as determined by questionnaires, we selected 22 379 subjects for the discovery stage and 11 104 subjects for the validation stage.
GWASs and Statistical Analysis
We used BOLT-LMM v2.3.237 for linear mixed model analysis to test the additive genetic effects of SNPs on AL. In our GWAS analysis, biases from the population stratification as well as familial and cryptic relatedness were controlled by considering the genetic correlation matrix in the linear mixed model.38 To reduce the skewness and kurtosis of the distribution of AL, Box-Cox transformation was applied to these data by using an R package (car ver. 2.1.5). The following variables were used as covariates for the AL adjustment: age and sex. In the GWAS for each phenotype, we inputted the direct genotype dataset and the imputed genotype dataset into BOLT-LMM. We merged the results for the direct genotype dataset and the results for the imputed genotype dataset by overwriting the latter results with the former results for variants existing in both datasets.
We examined associated SNPs in the discovery GWAS in the Community-Based Cohort using the BirThree Cohort as the validation cohort. For meta-analysis genome-wide association scans, we used METAL software.39 Regional association plots for the target regions were generated using LocusZoom.40
Results
Study Populations and Baseline Characteristics of the ToMMo Eye Study
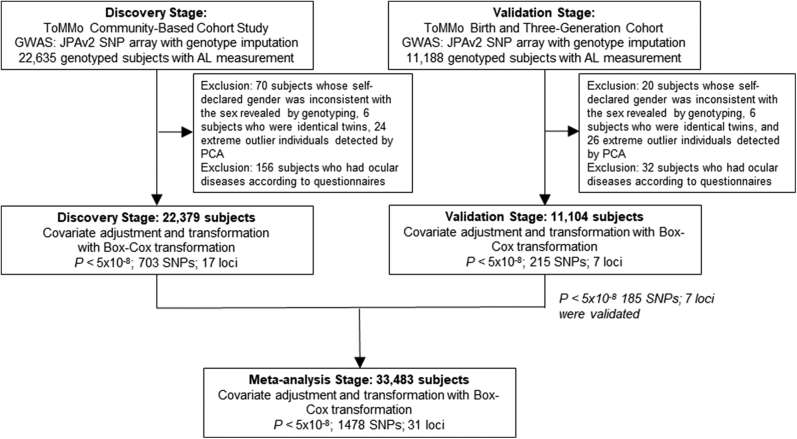
In the ToMMo Eye Study, ophthalmic examination data from the baseline analyses of 2 ToMMo population-based genome cohort studies were analyzed. We analyzed data from 22 635 participants in the CommCohort Study with imputed genotype and AL information for the discovery-stage GWAS (Fig 1). We excluded 70 participants because their self-reported gender was inconsistent with the sex indicated by genotyping, 6 participants who had identical twins, 24 extreme outlier participants who were detected by PCA, and 156 participants who had ocular diseases based on their questionnaire responses.
Figure 1.
Genome-wide association study (GWAS) design for investigating the genetic correlates of axial length (AL) in 2 cohorts. The inclusion criteria of the 2 population cohorts and a scheme of the GWASs for the discovery and validation stages are shown. For the discovery-stage GWAS, we used the CommCohort Study, which included 22 379 genotyped subjects with AL measurements. For the validation-stage GWAS, we used the BirThree Cohort Study, which included 11 104 genotyped subjects with AL measurements after the exclusion criteria were applied. We evaluated the associations of single nucleotide polymorphisms (SNPs) with AL with a genome-wide significance threshold (5 × 10-8) in each stage and in the meta-analysis with 33 483 subjects. PCA = principal component analysis.
We also examined data from 11 188 participants in the BirThree Cohort with imputed genotype and AL information for the validation-stage GWAS (Fig 1). We excluded 20 participants because of gender/sex inconsistency, 6 participants because they had identical twins, 26 extreme outlier participants who were detected by PCA, and 32 participants who had ocular diseases based on their questionnaire responses.
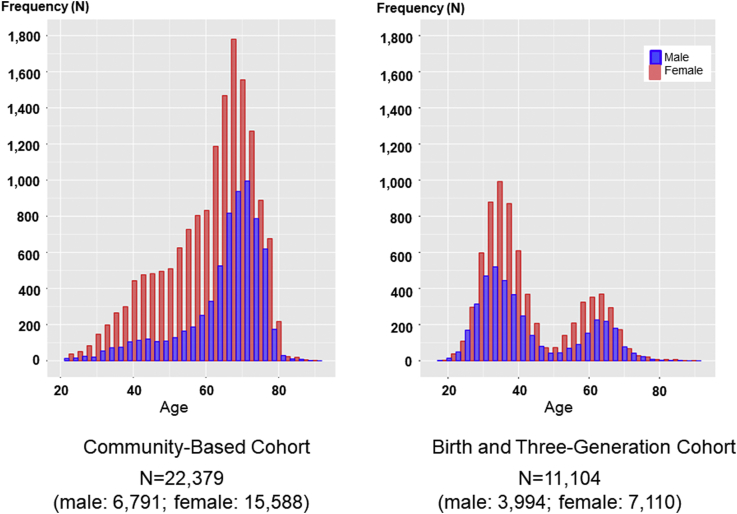
As shown in Table 1, we obtained AL data from 22 379 participants in the TMM CommCohort Study, which included 6791 men and 15 588 women with a mean age of 61.82 ± 12.38 years. We also obtained AL data from the 11 104 participants in the BirThree Cohort, which included 3994 men and 7110 women with a mean age of 42.67 ± 13.64 years. We used the former as a discovery study and the latter as a validation study. Because of the nature of the prospective cohort studies, female participants outnumbered male participants in both studies.
Table 1.
Baseline Characteristics of the Subjects in the Tohoku Medical Megabank Organization Eye Study
| Community-Based Cohort | Birth and Three-Generation Cohort | |
|---|---|---|
| No., total | 22 379 | 11 104 |
| No., male (%) | 6791 (30.35) | 3994 (35.97) |
| Mean age, yrs (SD) | 61.82 (12.38) | 42.67 (13.64) |
| Mean intraocular pressure, mmHg (SD) | 13.99 (2.97) | 14.13 (3.04) |
| Intraocular pressure right, mmHg (SD) | 13.96 (3.11) | 14.17 (3.16) |
| Intraocular pressure left, mmHg (SD) | 14.02 (3.21) | 14.09 (3.27) |
| Height, cm (SD) | 158.36 (7.91) | 162.42 (8.51) |
| Higher educational background (SD) | 2.43 (0.85) | 2.81 (0.94) |
| Family history father, number | 1951 | 1628 |
| Family history mother, number | 1763 | 1630 |
| AL > 26.0 mm in both eyes, number/percentage | 1558/6.96% | 1464/13.18% |
AL = axial length; SD = standard deviation.
The AL examinations of these 2 studies were conducted at the same facility and with the same machines and protocol. Nonetheless, there are several differences between these studies, and 2 of them seem to be important for interpretation of the results. One is that the age distribution of the participants differed substantially between these 2 genome cohort studies. As shown in Figure 2, the discovery study participants were primarily aged in their 60s to 70s, whereas the validation study participants were primarily in their 30s and 60s. The other is that 6.96% of participants in the discovery study had high myopia with ALs greater than 26.0 mm in both eyes, and 13.18% of the participants in the validation study had these characteristics (Table 1). Thus, although the 2 cohort studies in the current analysis together covered wide-ranging populations, these differences should be noted with caution.
Figure 2.
Age distribution of the participants. Axial length examination was performed in the 7 Community Support Centers. A total of 22 379 participants (mean age, 61.82 ± 12.38 years; 6791 men and 15 588 women) were recruited for the discovery stage, and a total of 11 104 participants (mean age, 42.67 ± 13.64 years; 3994 men and 7110 women) were recruited for the validation stage. Participants aged in their 60s to 70s made up the majority of the population in the discovery stage, whereas there were 2 age peaks (i.e., 30s and 60s) in the validation stage.
Axial Length Differences between the Right and Left Eyes and between Sexes
Among the 22 379 participants in the discovery cohort, the mean AL was 23.99 mm in the right eye and 23.95 mm in the left eye (Table 2). Among men, the mean AL was 24.31 mm in the right eye and 24.27 mm in the left eye, whereas among women, it was 23.85 mm in the right eye and 23.81 mm in the left eye. Among the 11 104 participants in the validation cohort, the mean AL was 24.54 mm in the right eye and 24.50 mm in the left eye. Among men, the mean AL was 24.83 mm in the right eye and 24.79 mm in the left eye, whereas among women, it was 24.38 mm in the right eye and 24.34 mm in the left eye.
Table 2.
Characteristics of Axial Length in the Study Population According to Age in the Community-Based Cohort (Discovery Cohort)
| Variable | Community-Based Cohort, Whole Population (N = 22 379) Range: 20.0–90.1 Years Old | Total | P Value∗ | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Age (yrs) | ∼29 | 30–39 | 40–49 | 50–59 | 60–69 | 70–79 | 80 ∼ | all | ||
| N | 330 | 1374 | 2384 | 3550 | 8196 | 6411 | 134 | 22 379 | ||
| Right AL (mm) | Mean | 24.59 | 24.61 | 24.52 | 24.11 | 23.85 | 23.74 | 23.54 | 23.99 | <0.001 |
| 95% CI | [24.45–24.73] | [24.54–24.68] | [24.46–24.57] | [24.07–24.16] | [23.82–23.88] | [23.71–23.78] | [23.33–23.75] | [23.97–24.01] | ||
| Left AL (mm) | Mean | 24.57 | 24.56 | 24.48 | 24.08 | 23.82 | 23.70 | 23.48 | 23.95 | <0.001 |
| 95% CI | [24.43–24.72] | [24.49–24.62] | [24.42–24.53] | [24.03–24.12] | [23.79–23.85] | [23.67–23.73] | [23.28–23.67] | [23.93–23.97] | ||
| Male (N = 6791) | ||||||||||
| N | 65 | 276 | 444 | 686 | 2352 | 2906 | 62 | 6791 | ||
| Right AL (mm) | Mean | 24.99 | 25.03 | 24.76 | 24.63 | 24.32 | 24.08 | 23.76 | 24.31 | <0.001 |
| 95% CI | [24.64–25.34] | [24.87–25.19] | [24.64–24.87] | [24.52–24.74] | [24.27–24.38] | [24.03–24.12] | [23.49–24.04] | [24.27–24.34] | ||
| Left AL (mm) | Mean | 24.98 | 25.01 | 24.71 | 24.55 | 24.29 | 24.05 | 23.82 | 24.27 | <0.001 |
| 95% CI | [24.62–25.33] | [24.85–25.16] | [24.58–24.83] | [24.45–24.66] | [24.24–24.34] | [24.00–24.09] | [23.54–24.09] | [24.24–24.30] | ||
| Female (N = 15 588) | ||||||||||
| N | 265 | 1098 | 1940 | 2864 | 5844 | 3505 | 72 | 15 588 | ||
| Right AL (mm) | Mean | 24.50 | 24.50 | 24.46 | 23.99 | 23.66 | 23.47 | 23.35 | 23.85 | <0.001 |
| 95% CI | [24.35–24.65] | [24.42–24.58] | [24.40–24.52] | [23.94–24.04] | [23.63–23.69] | [23.42–23.51] | [23.04–23.66] | [23.83–23.87] | ||
| Left AL (mm) | Mean | 24.48 | 24.44 | 24.43 | 23.96 | 23.63 | 23.41 | 23.19 | 23.81 | <0.001 |
| 95% CI | [24.33–24.63] | [24.37–24.52] | [24.37–24.49] | [23.91–24.01] | [23.60–23.66] | [23.36–23.45] | [22.93–23.45] | [23.79–23.83] | ||
|
Birth and Three-Generation Cohort (Validation Cohort) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Variable | Birth and Three-Generation Cohort, Whole Population (N = 11 104) Range: 17.8–88.6 years old | Total | P Value∗ | |||||||
| Age (yrs) | ∼ 29 | 30–39 | 40–49 | 50–59 | 60–69 | 70–79 | 80 ∼ | all | ||
| N | 1512 | 5001 | 1407 | 1110 | 1819 | 232 | 23 | 11 104 | ||
| Right AL (mm) | Mean | 24.68 | 24.80 | 24.76 | 24.03 | 23.95 | 23.81 | 23.03 | 24.54 | <0.001 |
| 95% CI | [24.61–24.75] | [24.76–24.84] | [24.68–24.83] | [23.95–24.11] | [23.89–24.01] | [23.65–23.97] | [22.70–23.36] | [24.51–24.57] | ||
| Left AL (mm) | Mean | 24.64 | 24.76 | 24.72 | 23.99 | 23.92 | 23.82 | 23.05 | 24.50 | <0.001 |
| 95% CI | [24.57–24.70] | [24.72–24.80] | [24.64–24.79] | [23.91–24.07] | [23.86–23.98] | [23.66–23.98] | [22.72–23.38] | [24.47–24.53] | ||
| Male (N = 3994) | ||||||||||
| N | 519 | 1736 | 588 | 279 | 736 | 129 | 7 | 3994 | ||
| Right AL (mm) | Mean | 24.97 | 25.06 | 24.99 | 24.45 | 24.34 | 24.06 | 23.02 | 24.83 | |
| 95% CI | [24.85–25.08] | [25.00–25.12] | [24.87–25.10] | [24.29–24.61] | [24.25–24.43] | [23.86–24.26] | [22.25–23.78] | [24.78–24.87] | ||
| Left AL (mm) | Mean | 24.92 | 25.02 | 24.96 | 24.4 | 24.33 | 24.06 | 23.05 | 24.79 | |
| 95% CI | [24.80–25.03] | [24.96–25.08] | [24.85–25.07] | [24.25–24.55] | [24.24–24.42] | [23.87–24.25] | [22.31–23.80] | [24.75–24.84] | ||
| Female (N = 7110) | ||||||||||
| N | 993 | 3,265 | 819 | 831 | 1083 | 103 | 16 | 7110 | ||
| Right AL (mm) | Mean | 24.53 | 24.66 | 24.59 | 23.89 | 23.69 | 23.50 | 23.04 | 24.38 | <0.001 |
| 95% CI | [24.45–24.61] | [24.62–24.71] | [24.50–24.69] | [23.80–23.98] | [23.61–23.76] | [23.26–23.74] | [22.63–23.45] | [24.34–24.41] | ||
| Left AL (mm) | Mean | 24.49 | 24.62 | 24.55 | 23.85 | 23.64 | 23.52 | 23.05 | 24.34 | <0.001 |
| 95% CI | [24.41–24.57] | [24.57–24.67] | [24.45–24.64] | [23.76–23.94] | [23.57–23.72] | [23.26–23.78] | [22.64–23.45] | [24.30–24.37] | ||
AL = axial length; CI = confidence interval.
Kruskal–Wallis rank-sum test.
These results revealed that the right eye AL was significantly longer than the left eye AL in both women and men in the discovery study. This AL difference between the right and left eyes (P < 0.01) was unequivocally replicated in the validation study (P < 0.05). The difference was 0.04 mm and consistent in both the discovery and validation studies and in both male and female participants.
A significant sex-related difference in the AL also existed; the mean AL in men was significantly longer than in women for all generations. The differences were 0.45 to 0.46 mm, approximately 10 times larger than the difference between the right and left eyes. This result shows very good agreement with that of a previous study.41
Axial Length Distribution in Age Groups
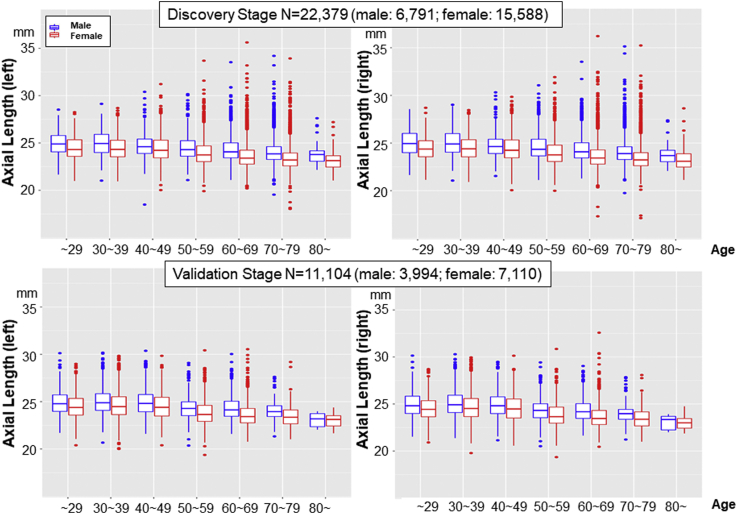
We then examined the relation of age and right eye/left eye differences with AL and found that the ALs of both the right and left eyes changed significantly with age in the discovery study (Fig 3). The AL of the right eye was 0.87 mm longer in participants aged 30 to 39 years than in participants aged 70 to 79 years, whereas the AL of the left eye was 0.86 mm longer in participants aged 30 to 39 years than in participants aged 70 to 79 years; these differences were consistent with those revealed when male and female eyes were assessed separately (Table 2).
Figure 3.
Axial length (AL) distribution by age group. The mean AL was 23.99 ± 1.4 mm in the right eye and 23.95 ± 1.4 mm in the left eye in the discovery stage and 24.54 ± 1.4 mm in the right eye and 24.50 ± 1.4 mm in the left eye in the validation stage. Note that the ALs of both the right and left eyes changed significantly with age in both female and male participants.
These differences were well replicated in the analyses of the validation cohort. Regarding the distribution of AL by age group, the ALs of the right and left eyes changed significantly with age (Fig 3). The mean differences in AL in 30- to 39-year-old participants versus the 70- to 79-year-old participants in the validation cohort were 0.99 mm in men and 0.94 mm in women, and differences for male and female eyes together were again consistent with those revealed for male and female eyes separately (Table 2).
These data revealed that the AL was longer in younger age groups and that the right/left difference in the AL was consistent in these age groups. Because this study was not a longitudinal assessment, it remains to be clarified whether this difference reflects dynamic changes in populations and society, including height and the education system.
Factors Associated with Axial Length
We next conducted multiple regression analyses to determine the factors associated with AL. In the CommCohort, height, higher educational background, family history, and intraocular pressure were positively associated with AL, but age was negatively associated with AL (Table 3, top). These associations were reproduced clearly in the BirThree Cohort (Table 3, bottom).
Table 3.
Associations between Background Characteristics: Community-Based Cohort (Discovery Stage)
| Sex | Age | Height | Higher Educational Background | Family History Father | Family History Mother | Intraocular Pressure (R) | Intraocular Pressure (L) | Axial Length (R) | |
|---|---|---|---|---|---|---|---|---|---|
| Age | −0.20 | ||||||||
| Height | −0.66 | −0.20 | |||||||
| Higher educational background | −0.03 | −0.24 | 0.16 | ||||||
| Family history father | 0.04 | −0.17 | 0.04 | 0.15 | |||||
| Family history mother | 0.04 | −0.20 | 0.04 | 0.13 | 0.24 | ||||
| Intraocular pressure (R) | 0.00 | −0.14 | 0.02 | 0.04 | 0.04 | 0.02 | |||
| Intraocular pressure (L) | −0.01 | −0.10 | 0.02 | 0.04 | 0.03 | 0.02 | 0.77 | ||
| AL (R) | −0.15 | −0.21 | 0.27 | 0.18 | 0.21 | 0.19 | 0.07 | 0.05 | |
| AL (L) | −0.16 | −0.22 | 0.27 | 0.18 | 0.21 | 0.19 | 0.06 | 0.06 | 0.95 |
|
Birth and Three-Generation Cohort (Validation Stage) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Sex | Age | Height | Higher Educational Background | Family History Father | Family History Mother | Intraocular Pressure (R) | Intraocular Pressure (L) | Axial Length (R) | |
| Age | −0.04 | ||||||||
| Height | −0.73 | −0.20 | |||||||
| Higher educational background | −0.05 | −0.20 | 0.14 | ||||||
| Family history father | 0.02 | −0.11 | 0.02 | 0.19 | |||||
| Family history mother | 0.03 | −0.15 | 0.02 | 0.15 | 0.33 | ||||
| Intraocular pressure (R) | −0.09 | −0.02 | 0.05 | 0.01 | 0.03 | 0.02 | |||
| Intraocular pressure (L) | −0.10 | 0.02 | 0.05 | 0.01 | 0.03 | 0.00 | 0.79 | ||
| AL (R) | −0.15 | −0.24 | 0.25 | 0.23 | 0.24 | 0.22 | 0.03 | 0.02 | |
| AL (L) | −0.16 | −0.24 | 0.26 | 0.23 | 0.24 | 0.22 | 0.03 | 0.02 | 0.97 |
AL = axial length.
Each column shows Pearson correlation coefficients (r).
We extended the survey of the association of AL with educational background. To this end, we categorized the educational backgrounds of the participants into 5 categories (Table 4). Inspection of the categories revealed that in the CommCohort, 83% and 64% of women and men, respectively, had received up to a high school, college, or technical college education, whereas 8.9% and 23% of women and men, respectively, had received advanced education (i.e., >16 education years). Likewise, in the BirThree Cohort, 75% and 58% of women and men, respectively, had received up to a high school, college, or technical college education, whereas 21% and 35% of women and men, respectively, had received advanced education. Thus, the participants in the BirThree Cohort represented much younger generations than those of the CommCohort but included participants with more advanced education.
Table 4.
Distribution of Axial Length by Educational Background
| Variable |
Community-Based Cohort (N = 22 379) Discovery Stage |
P Value∗ | P for Trend† | Kendall's Rank Correlation Tau (P Value) | ||||
|---|---|---|---|---|---|---|---|---|
| Educational Background | Elementary School, Junior High School | High School | Vocational School, College, Technical College | University | Graduate School | |||
| Education duration (yrs) | 6–9 | 12 | 14 | 16 | 18–21 | |||
| N | 2165 | 11 533 | 5743 | 2791 | 147 | |||
| (Male/female) | (895/1270) | (3494/8039) | (846/4897) | (1473/1318) | (83/64) | |||
| Mean age (yrs) | 69.02 | 63.02 | 58.45 | 58.63 | 53.34 | |||
| SD | 9.27 | 11.35 | 12.61 | 14.36 | 15.91 | |||
| Right AL (mm)‡ | 23.80 | 23.95 | 23.99 | 24.24 | 24.39 | <0.0001 | 0.0001 | 0.17 (P < 0.0001) |
| 95% CI | [23.78–23.81] | [23.95–23.96] | [23.98–24.01] | [24.22–24.25] | [24.31–24.47] | |||
| Left AL (mm)‡ | 23.76 | 23.92 | 23.96 | 24.20 | 24.35 | <0.0001 | 0.0001 | 0.17 (P < 0.0001) |
| 95% CI | [23.74–23.78] | [23.91–23.92] | [23.95–23.97] | [24.19–24.22] | [24.27–24.43] | |||
| Variable |
Birth and Three-Generation Cohort (N =11 104) Validation Stage |
P Value∗ | P for Trend† | Kendall's Rank Correlation Tau (P Value) | ||||
|---|---|---|---|---|---|---|---|---|
| Educational Background | Elementary School, Junior High School | High School | Vocational School, College, Technical College | University | Graduate School | |||
| Education duration (yrs) | 6–9 | 12 | 14 | 16 | 18–21 | |||
| N | 541 | 4293 | 3321 | 2676 | 273 | |||
| (Male/female) | (251/290) | (1605/2688) | (711/2610) | (1252/1424) | (175/98) | |||
| Mean age, yrs | 50.58 | 45.05 | 41.11 | 39.59 | 39.03 | |||
| SD | 17.29 | 14.67 | 12.21 | 11.74 | 9.16 | |||
| Right AL (mm)‡ | 24.31 | 24.48 | 24.51 | 24.68 | 24.79 | <0.0001 | 0.0001 | 0.15 (P < 0.0001) |
| 95% CI | [24.26–24.36] | [24.47–24.50] | [24.50–24.53] | [24.67–24.70] | [24.75–24.83] | |||
| Left AL (mm)‡ | 24.28 | 24.45 | 24.47 | 24.64 | 24.75 | <0.0001 | 0.0001 | 0.15 (P < 0.0001) |
| 95% CI | [24.23–24.32] | [24.43–24.46] | [24.46–24.49] | [24.63–24.66] | [24.71–24.80] | |||
Elementary School, Junior High School: <9 years of school; High School: 12 years of school; Vocational School, College, Technical College: 16 years of school; University: 16 years of school; Graduate School: ≥18 years of school.
AL = axial length; CI = confidence interval; SD = standard deviation.
One-way analysis of variance was performed after adjusting for age and sex.
Jonckheere-Terpstra test (number of permutations for the reference distribution = 10 000).
Adjusted for sex, age, and height.
Notably, AL differed by education level in the CommCohort (P < 0.0001) (Table 4 and Fig S1, available at www.ophthalmologyscience.org). The ALs of the right and left eyes incrementally increased with the education level from 23.80 mm and 23.76 mm in individuals with an elementary school/junior high school education to 24.39 mm and 24.35 mm in individuals with a graduate school education, respectively. Despite the differences in the distributions of age and education levels of the participants, these relationships were well recapitulated in the BirThree Cohort participants. Notably, although there were significant differences in mean age among the levels, the difference due to the education level was significant after adjustment of the AL for age and sex in both the discovery and validation stages. These results support the hypothesis that more years of education may contribute to an increase in AL.
GWAS for the ToMMo Eye Study
We next conducted a GWAS on AL using the AL data and microarray data for the post-quality control samples from the CommCohort Study (discovery stage) and the BirThree Cohort Study (validation stage). After stringent quality control, a total of 659 328 SNPs and imputed SNPs of 8 585 409 in the discovery stage and 8 590 037 in the validation stage, respectively, were included in the analyses. In the quantile-quantile plots, the genomic control inflation factor (λ) for the discovery stage and validation stage showed evidence of inflation (mean λ GC = 1.146 and 1096, respectively; Fig S2, available at www.ophthalmologyscience.org).
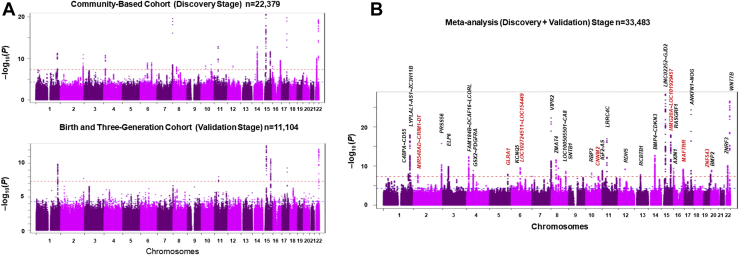
We identified 703 SNPs that cleared the genome-wide significance threshold of P < 5 × 10-8 in the discovery-stage screening. These SNPs were clustered into 17 distinct genomic regions (Fig 4A). These 17 loci in the discovery stage included 4 loci known as AL loci, 10 loci associated with refractive error, and 2 loci associated with corneal curvature. Additionally, we identified a new locus: MAFTRR (Table 5). The 3 most significant associations were found in the LINC02252;GJD2 locus (OMIM: 607058; rs16959560; P = 2.3 × 10-21) on Chr 15q14, ANKFN1∼NOG locus (OMIM: 602991; rs151278468; P =1.7 × 10-20) on Chr 17q22, and VIPR2 locus (vasoactive intestinal peptide receptor 2; OMIM: 601970; rs141313179; P = 2.5 × 10-20) on Chr 7q36.3.
Figure 4.
A, Genome-wide association study of axial length (AL) in the discovery stage and validation stage. B, Genome-wide association study of AL in the meta-analysis. The data for both directly genotyped and imputed single nucleotide polymorphisms (SNPs) are presented in the Manhattan plot. The y axis represents –log10(P) for the association with AL, and the x axis represents the chromosome and base-pair position based on human genome build 37. The horizontal red dotted lines indicate the genome-wide significance level of P < 5.0 × 10-8, and the blue dotted lines indicate the nominal genome-wide significance level of P < 5.0 × 10-5. Genes (locus) reported are shown in black, and genes (locus) newly identified in this study are shown in red (B).
Table 5.
Loci and Lead SNPs Associated with Axial Length (31 Loci)
| Chr | Lead SNP∗ | Position† | Nearest Gene | RA | EA | IMP | EA Freq | Discovery Stage |
Validation Stage |
Meta-Analysis |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B | SE | P | B | SE | P | Het/sq | Het P Value | P | ||||||||
| 15 | rs16959560 | 35006600 | LINC02252∼GJD2 | A | G | 1 | 0.535 | 0.084 | 0.009 | 2.3E-21 | 0.091 | 0.013 | 3.5E-13 | 0 | 0.658 | 4.47E-29 |
| 17 | rs151278468 | 54634412 | ANKFN1∼NOG | G | A | 1 | 0.012 | −0.397 | 0.044 | 1.7E-20 | −0.378 | 0.060 | 5.0E-10 | 0 | 0.812 | 4.66E-25 |
| 7 | rs141313179 | 158906675 | VIPR2 | A | G | 1 | 0.010 | 0.364 | 0.040 | 2.5E-20 | 0.303 | 0.057 | 2.1E-7 | 0 | 0.408 | 5.35E-23 |
| 22 | rs10453459 | 46366127 | WNT7B | G | C | 1 | 0.668 | −0.085 | 0.009 | 1.2E-19 | −0.095 | 0.013 | 1.4E-12 | 0 | 0.558 | 2.69E-27 |
| 11 | rs7936359 | 40148976 | LRRC4C | T | A | 1 | 0.204 | 0.078 | 0.011 | 1.4E-13 | 0.091 | 0.016 | 1.0E-8 | 0 | 0.510 | 1.14E-17 |
| 15 | rs13380109 | 79378775 | RASGRF1 | G | A | 1 | 0.513 | 0.060 | 0.009 | 3.3E-12 | 0.082 | 0.013 | 1.1E-10 | 48.1 | 0.165 | 1.16E-18 |
| 1 | NA | 219743871 | LYPLAL1-AS1∼ ZC3H11B | AT | ATTT | 1 | 0.521 | 0.060 | 0.009 | 5.8E-12 | 0.082 | 0.013 | 1.3E-10 | 46.8 | 0.170 | 1.10E-18 |
| 2 | rs77311538 | 233386148 | PRSS56 | C | G | 1 | 0.093 | −0.103 | 0.015 | 1.3E-11 | −0.129 | 0.023 | 1.8E-8 | 0 | 0.372 | 1.83E-16 |
| 4 | NA | 17883603 | FAM184B∼DCAF16 ∼LCORL | GA | GAA | 1 | 0.231 | −0.072 | 0.011 | 1.9E-11 | −0.056 | 0.015 | 1.4E-4 | 0 | 0.430 | 4.52E-13 |
| 22 | rs4823003 | 29410232 | ZNRF3 | A | C | 1 | 0.595 | −0.058 | 0.009 | 1.4E-10 | −0.037 | 0.013 | 3.7E-3 | 37.8 | 0.205 | 7.90E-11 |
| 14 | rs10459508 | 54485490 | BMP4∼CDKN3 | A | G | 1 | 0.521 | −0.056 | 0.009 | 3.2E-10 | −0.057 | 0.013 | 2.0E-5 | 0 | 0.966 | 1.97E-13 |
| 16 | rs4889024 | 79796196 | MAFTRR | C | A | 1 | 0.716 | −0.062 | 0.010 | 5.1E-10 | −0.035 | 0.014 | 1.4E-2 | 54.7 | 0.137 | 6.88E-10 |
| 6 | rs7744813 | 73643289 | KCNQ5 | C | A | 0 | 0.763 | 0.061 | 0.010 | 2.8E-9 | 0.048 | 0.015 | 1.1E-3 | 0 | 0.509 | 2.95E-10 |
| 12 | rs3138142 | 56115585 | RDH5 | C | T | 1 | 0.049 | −0.117 | 0.021 | 6.4E-9 | −0.104 | 0.031 | 8.2E-4 | 0 | 0.736 | 6.30E-10 |
| 10 | rs11204213 | 48388228 | RBP3 | C | T | 0 | 0.018 | 0.188 | 0.033 | 7.9E-9 | 0.126 | 0.048 | 6.3E-3 | 3.2 | 0.309 | 6.20E-9 |
| 8 | rs16890057 | 40726582 | ZMAT4 | G | A | 0 | 0.068 | −0.100 | 0.017 | 1.2E-8 | −0.124 | 0.026 | 1.7E-6 | 0 | 0.461 | 2.72E-12 |
| 16 | rs3848363 | 370484 | AXIN1 | T | C | 1 | 0.405 | −0.050 | 0.009 | 2.6E-8 | −0.047 | 0.013 | 1.9E-4 | 0 | 0.870 | 3.91E-10 |
| 8 | rs36005291 | 60179048 | LOC100505501∼CA8 | CA | C | 1 | 0.442 | −0.048 | 0.009 | 6.6E-8 | −0.049 | 0.013 | 1.1E-4 | 0 | 0.923 | 3.32E-10 |
| 3 | rs7651194 | 47540917 | ELP6 | G | A | 1 | 0.551 | −0.047 | 0.009 | 9.9E-8 | −0.054 | 0.013 | 1.6E-5 | 0 | 0.668 | 1.33E-10 |
| 20 | rs146314970 | 6757519 | BMP2 | T | TA | 1 | 0.185 | 0.059 | 0.011 | 3.0E-7 | 0.061 | 0.016 | 1.4E-4 | 0 | 0.928 | 1.26E-9 |
| 1 | NA | 207410874 | C4BPA∼CD55 | TA | TAAA | 1 | 0.514 | −0.042 | 0.009 | 7.7E-7 | −0.047 | 0.013 | 2.0E-4 | 0 | 0.769 | 1.15E-8 |
| 19 | rs11325378 | 57836180 | ZNF543 | TA | T | 1 | 0.541 | 0.044 | 0.009 | 1.2E-6 | 0.044 | 0.013 | 4.2E-4 | 0 | 0.971 | 1.19E-8 |
| 6 | rs58353542 | 170491975 | LOC102724511∼ LOC154449 | T | G | 1 | 0.128 | −0.062 | 0.013 | 1.2E-6 | −0.065 | 0.019 | 4.4E-4 | 0 | 0.932 | 4.51E-8 |
| 13 | NA | 50134748 | RCBTB1 | AT | ATT | 1 | 0.124 | −0.063 | 0.013 | 2.6E-6 | −0.076 | 0.020 | 1.2E-4 | 0 | 0.619 | 1.66E-8 |
| 11 | rs3741210 | 2169540 | IGF2-AS | A | G | 1 | 0.454 | −0.042 | 0.009 | 4.9E-6 | −0.056 | 0.013 | 7.5E-6 | 0 | 0.391 | 1.48E-9 |
| 4 | rs147732642 | 55089323 | GSX2∼PDGFRA | C | CT | 1 | 0.152 | −0.055 | 0.012 | 5.2E-6 | −0.084 | 0.018 | 1.0E-6 | 39.3 | 0.199 | 1.54E-9 |
| 15 | rs965480 | 77781926 | HMG20A∼ LOC101929457 | A | G | 1 | 0.407 | 0.040 | 0.009 | 7.9E-6 | 0.047 | 0.013 | 2.0E-4 | 0 | 0.642 | 4.70E-8 |
| 8 | rs72609833 | 121669010 | SNTB1 | C | A | 1 | 0.375 | 0.039 | 0.009 | 8.1E-6 | 0.063 | 0.013 | 2.0E-6 | 49.2 | 0.161 | 2.20E-9 |
| 10 | rs7902218 | 104740210 | CNNM2 | A | G | 1 | 0.285 | −0.043 | 0.010 | 1.5E-5 | −0.052 | 0.014 | 1.6E-4 | 0 | 0.591 | 4.59E-8 |
| 5 | rs35305813 | 151225351 | GLRA1 | T | TA | 1 | 0.104 | 0.061 | 0.014 | 1.9E-5 | 0.090 | 0.020 | 9.2E-6 | 21.2 | 0.260 | 1.27E-8 |
| 2 | rs60806750 | 36577213 | MIR548AD∼ CRIM1-DT | T | C | 1 | 0.330 | 0.036 | 0.009 | 2.2E-4 | 0.067 | 0.014 | 8.9E-7 | 68 | 0.077 | 1.80E-8 |
Chr = chromosome; EA = effect allele; EA freq = effect allele frequency in tommo_4772; HetIsq = heterogeneity I squared; Het P value: heterogeneity P value; IMP = imputed; NA = not applicable; RA = reference allele; SE = standard error; SNP = single nucleotide polymorphisms.
Bold characters represent a statistically significant P value.
The lead SNPs of each locus identified in the discovery analysis are presented.
The position is based on NCBI human genome build 37.
We also identified 215 SNPs that cleared the genome-wide significance threshold of P < 5 × 10-8 in the validation-stage screening (Fig 4A). These SNPs were clustered into 7 distinct genomic regions: LINC02252∼GJD2, WNT7B, RASGRF1, LYPLAL1-AS1∼ZC3H11B, ANKFN1∼NOG, LRRC4C, and PRSS56. These 7 loci corresponded to the top hit loci of the discovery-stage GWAS (Table 5). When we compared the SNP P values of the other SNPs with those of the discovery stages, we found that although they did not reach standard statistical significance, they were relatively well correlated, indicating that the discovery-stage SNP P values were generally well replicated in the validation stage.
Of the 488 SNPs that were not replicated in the validation stage, some were rare variants with MAFs < 0.02. For example, VIPR2 rs141313179 and RBP3 showed MAFs of 0.012 and 0.018, respectively, in the ToMMo 4.7K allele frequency panel.36 By contrast, common SNPs were well replicated in the validation stage. We surmise that the former SNPs did not replicate well because of the low MAFs.
Meta-analysis of Two GWASs for the ToMMo Eye Study
As we concluded that the validation cohort did not retain sufficient power of resolution and could not obtain statistically significant replications, we decided to challenge a meta-analysis of these discovery-stage and validation-stage GWASs. This meta-analysis using METAL software provides intriguing results in which we identified 31 loci across 19 chromosomes without chromosomes 9, 18, and 21 (Fig 4B). These loci included 5 AL loci, 15 loci associated with refractive error, and 4 loci associated with corneal curvature. Additionally, we identified 7 new loci: MAFTRR, ZNF543, GLRA1, MIR548AD∼CRIM1-DT, LOC102724511∼LOC154449, CNNM2, and HMG20A∼LOC101929457. These loci had not been identified previously as having any association with AL.
The 5 loci that are reported to be associated with the AL phenotype are GJD2, WNT7B, ZC3H11B, ZNRF3, and BMP2.27,28 GJD2, WNT7B, and ZC3H11B were in the top hit group (P < 1.0 × 10-17). By contrast, ANKFN1-NOG, VIPR2, LRRC4C, RASGRF1, PRSS56, BMP4, KCNQ5, RDH5, ZMAT4, AXIN1, LOC100505501∼CA8, ELP6, C4BPA∼CD55, RCBTB1, and SNTB1 are associated with spherical equivalent or refractive error,17,18,31,42, 43, 44 and PRSS56 is also associated with angle-closure glaucoma.45 ANKFN1∼NOG and VIPR2 showed particularly low P values. Four loci—LCORL, RBP3, IGF2-AS, and GSX2∼PDGFRA—were related to corneal curvature.29,46,47
Figure S3 (available at www.ophthalmologyscience.org) shows regional plots of the 8 top hit loci in the meta-analysis stage with direct genotyping and genotype imputation data. We found differences in the number and accumulation pattern of SNPs at each locus; nonetheless, each of the 8 loci harbored a cluster of SNPs surrounding the SNP with the most significant association, denoted with a purple diamond. These results validated the identified AL-associated loci. Table S1 (available at www.ophthalmologyscience.org) assesses the effect of the top hit 8 loci between this study and a previous European study.31 We compared the β value, which is the coefficient of linear regression and indicates the effect sizes. The absolute β value was associated with 3 types. This comparison showed that the degree of Japanese β values was higher (ANKFN1∼NOG, VIPR2), almost similar (WNT7B), or lower (LINC02252∼GJD2, RASGRF1, LRRC4C, PRSS56) than that of European β values.18
Discussion
Alterations in eye AL are common optical aberrations and have a considerable effect on refractive error. Although genetic background influences AL and the onset of myopia, environmental factors also play important roles.48 In this study, we conducted extensive analyses of the associations of AL with age and sex and compared the ALs of the right and left eyes. We found that AL was longer in younger generations and that men had longer ALs than women. The AL of the right eye was longer than that of the left eye across ages and sexes, and the AL increased with the level of education. In this study, we also performed an extensive GWAS on AL and identified 1478 SNPs located over 31 genetic loci. Because the National Human Genome Research Institute–European Bioinformatics Institute GWAS Catalog (https://www.ebi.ac.ukgwas) lists 28 candidate loci for AL from 5 studies,49 this GWAS adds much to the current body of knowledge on genetic factors related to AL. Thus, our results on environmental and genetic factors provide new insights into the factors influencing AL in the general Japanese population.
Axial length measurement has been conducted in several population-based cohort studies in Australia and Asia.11,22,25 Intriguingly, the mean AL in our discovery study (23.99 mm, right eye) was considerably longer than that in the Blue Mountains Eye Study of elderly Australians (23.44 mm, right eye),22 suggesting the presence of ethnic differences in AL. This notion is supported by the results of the Nagahama cohort study in Japan, in which the mean AL of the right eye was 24.12 mm.25 In contrast, the presence of interocular asymmetry in AL appears to be transethnic. In this study, we found that the mean ocular AL of the left eye was markedly shorter than that of the right eye. Of note, this right eye/left eye asymmetry has been reported in several eye cohort studies,8,41,50, 51, 52 and the asymmetry may be related more to eye dominance than right-left laterality.41 Investigating the link between accommodation and eye dominance is warranted.
There was a difference that 6.96% of participants in the discovery study had high myopia with ALs > 26.0 mm in both eyes, whereas 13.18% of the participants in the validation study had these characteristics (Table 1). This may be due to the difference in the number of younger-generation participants between the studies. Although 2 cohort studies in the current analysis together covered wide-ranging populations, their differences should be considered carefully. We surmise that the longer AL observed in younger generations may be due to environmental factors. One plausible explanation for the observation may be socioeconomic, for example, poorer nutrition in older generations. In this study, we also found an apparent elongation in AL in populations with higher levels of education. More advanced education may be associated with more time spent on close-up activities and a lack of outdoor light exposure, which are currently proposed environmental risk factors for myopia.7,48,53, 54, 55 Analysis of the association between education and myopia using a 5-level classification system revealed that AL increased with increasing education level, supporting the notion that increased durations of desk work and reduced outdoor light exposure are related to myopia. In this regard, a preceding study revealed stronger correlations between the AL and education levels than this study.11 Likewise, a significant correlation was also shown between genetic variations and AL, and many genetic loci influencing AL have been identified.56 However, the contribution of the individual refractive error associated locus to the phenotypic variance is relatively small,17,18,27,31 and many more loci must be identified to explain the genetic architecture. We believe that our current GWAS may contribute in this regard.
We designed a 2-stage GWAS and meta-analysis based on our statistical evaluations. A significant association of the gap junction protein delta-2 (GJD2) locus with AL was found in our discovery stage, validation stage, and meta-analysis. One of the first GWASs for refractive error in European populations identified a GJD2 locus.57 The association of this locus with myopia has been replicated by multiple independent studies in various ethnicities.17,18,27,31 Loss of GJD2 gene orthologs was found to cause refractive error in zebrafish,58 and it was hypothesized that the uncoupling of retinal gap junctions inhibits ocular growth, constituting functional evidence for a link between GJD2 and refractive error, including AL elongation. A second significant association with the WNT7B locus was found in our meta-analysis, showing very good agreement with the results of previous Japanese studies.28,59 WNT7B is localized to retinal ganglion cells, and its expression is significantly upregulated in experimental myopic eyes.28 We also found a concomitant association of ZNRF3, which was identified independently as an AL-associated locus in another GWAS.27 ZNRF3 is a membrane-bound protein that acts as a negative regulator of the Wnt signaling pathway.60 Overexpression of a dominant-negative variant of human ZNRF3 in zebrafish embryos induces small eye development or the absence of eyes.60 Given these reports, WNT7B and ZNRF3 seem to form a significant pathway involved in the regulation of AL.
A third significant association with the ANKFN1∼NOG locus was found in our meta-analysis, which showed very good agreement with the results of previous meta-analyses.18,31,43 The secreted polypeptide encoded by NOG binds and inactivates members of the transforming growth factor-beta superfamily signaling proteins, including bone morphogenetic protein, and NOG is required during skeletogenesis. Molecular genetics revealed that NOG mutation is associated with autosomal dominant disorder characterized by the premature onset of joint fusions, that is, multiple synostoses syndrome.61
A fourth significant association was found with the VIPR2 locus in our meta-analysis. Although we identified a statistically significant rare variant SNP, VIPR2 rs141313179 (MAF 0.010) in the discovery stage, this SNP did not clear the significance threshold in the validation stage (P = 8.7 × 10-8). For rare variants, this critical P value is too stringent to detect certain associations and is useful only when studies have adequate statistical power.62 Available lines of evidence suggest that VIPR2 is related to novel pathways for anterior-segment morphology, a susceptibility locus for quantitative trait refractive errors and age of diagnosis of myopia.18
Refraction depends on a balance between these features, which are sensitive to genetic and environmental conditions. Previous studies have revealed strong correlations between AL and both education background and body height,11 and significant genetic correlations between them.63, 64, 65 The changes in AL and height suggest that they shift concomitantly with age, and greater changes are always observed in younger children.38,64 On comparing the effect, it should be considered that our cohort comprises an adult population. Bivariate genetic analysis reported that the genetic correlation between AL and height was moderate (0.42), whereas the genetic correlation between AL and corneal curvature was high (0.85) in emmetropic eyes.65 Correlation analysis of background characteristics revealed that the correlation between AL and height was statistically significant (P < 0.01). However, Pearson correlation coefficients (r), 0.25 (validation stage) to 0.27 (discovery stage), were lower than those reported (Table 3). We speculate that the correlation between AL and height is lower in the Japanese population because of the action of other environmental and socioeconomic factors. However, exposure to more years of education likely contributes to the rising prevalence of myopia in Europeans.56 Notably, AL differed by education level in the discovery and validation stages in this study (Table 4; P < 0.0001), supporting the hypothesis that more years of education contribute to an increase in AL and a causal determinant of myopia.
The main ocular structural features related to refractive error are AL and corneal curvature. Although a small number of genetic variants may influence AL but not corneal curvature, and vice versa, AL and corneal curvature loci are highly linked and related to refractive error. Of 31 AL-related loci identified in this study, 15 were refractive error–related loci and 4 were corneal curvature–related loci. By contrast, 7 loci were newly identified to be associated with AL. These 7 loci included transcription factors and genes encoding some zinc-finger family of proteins, chromatin-associated protein, transmembrane protein, and ligand-gated chloride channels, almost all of which were expressed in the retina. Our genetic observations were consistent with the current notion that refractive errors are caused by a retina-to-sclera signaling cascade that induces scleral remodeling in response to light stimuli.18 Absolute β values in the assessment of effect sizes of top hit loci (Table S1, available at www.ophthalmologyscience.org) shows that the degrees of Japanese β values were higher, almost similar, or lower than those of European β values. Comparing the β value between AL and refractive error indicates the nonlinearity between Japanese and European populations. Genetic overlap is plausible, but a difference should exist in the per-allele effect size as previously suggested.17,18
The severity of myopia is often categorized as mild, moderate, or high. Mild myopia usually does not increase the risk of eye disease, but high myopia is significantly associated with vision-threatening eye problems and is called “degenerative myopia.” In one GWAS of subjects with high myopia (spherical equivalent refraction < −9.0 diopters in at least 1 eye), 9 loci were identified with genome-wide significance.59 Of the 9 loci, we identified only 3 in this study: ZC3H11B, GJD2, and RASGRF1. Because our study included the general population and we used only the AL, differences may exist in the pathogeneses of degenerative myopia and simple high myopia.
We examined the characteristics and genetics of AL using the ToMMo Eye Study, which comprises 2 strategically designed cohorts with a respectable sample size because of its use of ToMMo Eye Study data (N = 33 483 participants), suggesting genetic heterogeneity between populations. Because our study inherently harbors a selection bias in recruiting participants, we examined the validation stage to overcome this limitation.
In conclusion, the ToMMo Eye Study identifies multiple new and previously reported loci linked to AL variations of Japanese individuals, and comparison of the results with those of international studies suggests the presence of genetic heterogeneity between populations. Ethnicity-specific genetic variations provide new insights into the genetic factors underlying the phenotype of AL in the general Japanese population. We plan to pursue longitudinal follow-up studies of these 2 cohorts because such studies should be informative and will form a strong and indispensable basis for new insights into the genetic background underlying AL. Such investigation will, in turn, contribute to the design of strategies for the prevention of common eye diseases.
Acknowledgments
The authors acknowledge the Iwate Tohoku Medical Megabank for discussion and support in performing the cohort study. We thank the members of the TMM, including the Genome Medical Research Coordinators and the office and administrative personnel, for their assistance with the eye study in this project. https://www.megabank.tohoku.ac.jp/english/a210901/.
Manuscript no. D-21-00095
Footnotes
Supplemental material available atwww.ophthalmologyscience.org.
Disclosure(s):
All authors have completed and submitted the ICMJE disclosures form.
The author(s) have made the following disclosure(s): The TMM is supported by grants from the Reconstruction Agency; the Ministry of Education, Culture, Sports, Science and Technology; the Japan Agency for Medical Research and Development (JP 20km0105001, JP 20km0105002 and 20km0405001); and the Agency for Medical Research and Development Advanced Genome Research and Bioinformatics Study to Facilitate Medical Innovation project (JP20km0405203). The funding organization participated in the design of the TMM study. This work was also supported by Ministry of Education, Culture, Sports, Science andTechnologyJapan Society for the Promotion of Science KAKENHI19K09927.
HUMAN SUBJECTS: Human subjects were included in this study. The human ethics committees at the Tohoku University Graduate School of Medicine approved the study. All research adhered to the tenets of the Declaration of Helsinki. All participants provided informed consent.
No animal subjects were used in this study.
Author Contributions:
Conception and design: Fuse, Sakurai, Motoike, Takai-Igarashi, Nakaya, Obara, Miyazawa, Homma, Hozawa, Kuriyama, Yaegashi, Kure, Kinoshita, Yamamoto
Data collection: Fuse, Sakurai, Motoike, Kojima, Takai-Igarashi, Nakaya, Tsuchiya, Nakamura, Ishikuro, Obara, Miyazawa, Homma, Ido, Taira, Kobayashi, Shimizu, Uruno, Kodama, Suzuki, Hamanaka, Tomita, Sugawara, Suzuki, Nagami, Ogishima, Katsuoka, Minegishi, Hozawa, Kuriyama, Yaegashi, Kure, Kinoshita, Yamamoto
Analysis and interpretation: Fuse, Sakurai, Motoike, Kojima, Takai-Igarashi, Nakaya, Tsuchiya, Nakamura, Ishikuro, Obara, Miyazawa, Homma, Ido, Hozawa, Kuriyama, Kinoshita, Yamamoto
Obtained funding: N/A
Overall responsibility: Fuse, Sakurai, Motoike, Kojima, Takai-Igarashi, Nakaya, Tsuchiya, Ido, Taira, Katsuoka, Hozawa, Kuriyama, Kinoshita, Yamamoto
Supplementary Data
References
- 1.Tideman J.W., Snabel M.C., Tedja M.S., et al. Association of axial length with risk of uncorrectable visual impairment for Europeans with myopia. JAMA Ophthalmol. 2016;134:1355–1363. doi: 10.1001/jamaophthalmol.2016.4009. [DOI] [PubMed] [Google Scholar]
- 2.Ohsugi H., Ikuno Y., Oshima K., et al. Morphologic characteristics of macular complications of a dome-shaped macula determined by swept-source optical coherence tomography. Am J Ophthalmol. 2014;158:162–170 e161. doi: 10.1016/j.ajo.2014.02.054. [DOI] [PubMed] [Google Scholar]
- 3.Saw S.M., Gazzard G., Shih-Yen E.C., et al. Myopia and associated pathological complications. Ophthalmic Physiol Opt. 2005;25:381–391. doi: 10.1111/j.1475-1313.2005.00298.x. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki Y., Iwase A., Araie M., et al. Risk factors for open-angle glaucoma in a Japanese population: the Tajimi Study. Ophthalmology. 2006;113:1613–1617. doi: 10.1016/j.ophtha.2006.03.059. [DOI] [PubMed] [Google Scholar]
- 5.Flitcroft D.I. The complex interactions of retinal, optical and environmental factors in myopia aetiology. Prog Retin Eye Res. 2012;31:622–660. doi: 10.1016/j.preteyeres.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 6.Morgan I.G., Ohno-Matsui K., Saw S.M. Myopia. Lancet. 2012;379:1739–1748. doi: 10.1016/S0140-6736(12)60272-4. [DOI] [PubMed] [Google Scholar]
- 7.Pan C.W., Ramamurthy D., Saw S.M. Worldwide prevalence and risk factors for myopia. Ophthalmic Physiol Opt. 2012;32:3–16. doi: 10.1111/j.1475-1313.2011.00884.x. [DOI] [PubMed] [Google Scholar]
- 8.Wang Q., Klein B.E., Klein R., et al. Refractive status in the Beaver Dam Eye Study. Invest Ophthalmol Vis Sci. 1994;35:4344–4347. [PubMed] [Google Scholar]
- 9.Attebo K., Ivers R.Q., Mitchell P. Refractive errors in an older population: the Blue Mountains Eye Study. Ophthalmology. 1999;106:1066–1072. doi: 10.1016/S0161-6420(99)90251-8. [DOI] [PubMed] [Google Scholar]
- 10.Kempen J.H., Mitchell P., Lee K.E., et al. The prevalence of refractive errors among adults in the United States, Western Europe, and Australia. Arch Ophthalmol. 2004;122:495–505. doi: 10.1001/archopht.122.4.495. [DOI] [PubMed] [Google Scholar]
- 11.Yin G., Wang Y.X., Zheng Z.Y., et al. Ocular axial length and its associations in Chinese: the Beijing Eye Study. PLoS One. 2012;7 doi: 10.1371/journal.pone.0043172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong T.Y., Foster P.J., Ng T.P., et al. Variations in ocular biometry in an adult Chinese population in Singapore: the Tanjong Pagar Survey. Invest Ophthalmol Vis Sci. 2001;42:73–80. [PubMed] [Google Scholar]
- 13.Sawada A., Tomidokoro A., Araie M., et al. Refractive errors in an elderly Japanese population: the Tajimi study. Ophthalmology. 2008;115:363–370 e363. doi: 10.1016/j.ophtha.2007.03.075. [DOI] [PubMed] [Google Scholar]
- 14.Young T.L., Metlapally R., Shay A.E. Complex trait genetics of refractive error. Arch Ophthalmol. 2007;125:38–48. doi: 10.1001/archopht.125.1.38. [DOI] [PubMed] [Google Scholar]
- 15.McCarthy M.I., Abecasis G.R., Cardon L.R., et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 16.Manolio T.A., Collins F.S., Cox N.J., et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verhoeven V.J., Hysi P.G., Wojciechowski R., et al. Genome-wide meta-analyses of multiancestry cohorts identify multiple new susceptibility loci for refractive error and myopia. Nat Genet. 2013;45:314–318. doi: 10.1038/ng.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tedja M.S., Wojciechowski R., Hysi P.G., et al. Genome-wide association meta-analysis highlights light-induced signaling as a driver for refractive error. Nat Genet. 2018;50:834–848. doi: 10.1038/s41588-018-0127-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuriyama S., Yaegashi N., Nagami F., et al. The Tohoku Medical Megabank Project: design and mission. J Epidemiol. 2016;26:493–511. doi: 10.2188/jea.JE20150268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawai Y., Mimori T., Kojima K., et al. Japonica array: improved genotype imputation by designing a population-specific SNP array with 1070 Japanese individuals. J Hum Genet. 2015;60:581–587. doi: 10.1038/jhg.2015.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fledelius H.C., Goldschmidt E. Oculometry findings in high myopia at adult age: considerations based on oculometric follow-up data over 28 years in a cohort-based Danish high-myopia series. Acta Ophthalmol. 2010;88:472–478. doi: 10.1111/j.1755-3768.2008.01472.x. [DOI] [PubMed] [Google Scholar]
- 22.Fotedar R., Wang J.J., Burlutsky G., et al. Distribution of axial length and ocular biometry measured using partial coherence laser interferometry (IOL Master) in an older white population. Ophthalmology. 2010;117:417–423. doi: 10.1016/j.ophtha.2009.07.028. [DOI] [PubMed] [Google Scholar]
- 23.Asakuma T., Yasuda M., Ninomiya T., et al. Prevalence and risk factors for myopic retinopathy in a Japanese population: the Hisayama Study. Ophthalmology. 2012;119:1760–1765. doi: 10.1016/j.ophtha.2012.02.034. [DOI] [PubMed] [Google Scholar]
- 24.Fu T., Song Y.W., Chen Z.Q., et al. Ocular biometry in the adult population in rural central China: a population-based, cross-sectional study. Int J Ophthalmol. 2015;8:812–817. doi: 10.3980/j.issn.2222-3959.2015.04.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshikawa M., Yamashiro K., Miyake M., et al. Comprehensive replication of the relationship between myopia-related genes and refractive errors in a large Japanese cohort. Invest Ophthalmol Vis Sci. 2014;55:7343–7354. doi: 10.1167/iovs.14-15105. [DOI] [PubMed] [Google Scholar]
- 26.Fan Q., Barathi V.A., Cheng C.Y., et al. Genetic variants on chromosome 1q41 influence ocular axial length and high myopia. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1002753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng C.Y., Schache M., Ikram M.K., et al. Nine loci for ocular axial length identified through genome-wide association studies, including shared loci with refractive error. Am J Hum Genet. 2013;93:264–277. doi: 10.1016/j.ajhg.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyake M., Yamashiro K., Tabara Y., et al. Identification of myopia-associated WNT7B polymorphisms provides insights into the mechanism underlying the development of myopia. Nat Commun. 2015;6:6689. doi: 10.1038/ncomms7689. [DOI] [PubMed] [Google Scholar]
- 29.Fan Q., Pozarickij A., Tan N.Y.Q., et al. Genome-wide association meta-analysis of corneal curvature identifies novel loci and shared genetic influences across axial length and refractive error. Commun Biol. 2020;3:133. doi: 10.1038/s42003-020-0802-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tideman J.W.L., Parssinen O., Haarman A.E.G., et al. Evaluation of shared genetic susceptibility to high and low myopia and hyperopia. JAMA Ophthalmol. 2021;139:601–609. doi: 10.1001/jamaophthalmol.2021.0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hysi P.G., Choquet H., Khawaja A.P., et al. Meta-analysis of 542,934 subjects of European ancestry identifies new genes and mechanisms predisposing to refractive error and myopia. Nat Genet. 2020;52:401–407. doi: 10.1038/s41588-020-0599-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hozawa A., Tanno K., Nakaya N., et al. Study profile of The Tohoku Medical Megabank Community-Based Cohort Study. J Epidemiol. 2020;31:65–76. doi: 10.2188/jea.JE20190271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuriyama S., Metoki H., Kikuya M., et al. Cohort Profile: Tohoku Medical Megabank Project Birth and Three-Generation Cohort Study (TMM BirThree Cohort Study): rationale, progress and perspective. Int J Epidemiol. 2020;49:18–19m. doi: 10.1093/ije/dyz169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delaneau O., Marchini J., Zagury J.F. A linear complexity phasing method for thousands of genomes. Nat Methods. 2011;9:179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 35.Howie B.N., Donnelly P., Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tadaka S., Katsuoka F., Ueki M., et al. 3.5KJPNv2: an allele frequency panel of 3552 Japanese individuals including the X chromosome. Hum Genome Var. 2019;6:28. doi: 10.1038/s41439-019-0059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loh P.R., Tucker G., Bulik-Sullivan B.K., et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat Genet. 2015;47:284–290. doi: 10.1038/ng.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Z., Ersoz E., Lai C.Q., et al. Mixed linear model approach adapted for genome-wide association studies. Nat Genet. 2010;42:355–360. doi: 10.1038/ng.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pruim R.J., Welch R.P., Sanna S., et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mahroo O.A., Williams C., Hysi P.G., et al. Interocular asymmetries in axial length and refractive error in 4 cohorts. Ophthalmology. 2015;122:648–649. doi: 10.1016/j.ophtha.2014.10.021. [DOI] [PubMed] [Google Scholar]
- 42.Pickrell J.K., Berisa T., Liu J.Z., et al. Detection and interpretation of shared genetic influences on 42 human traits. Nat Genet. 2016;48:709–717. doi: 10.1038/ng.3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fan Q., Verhoeven V.J., Wojciechowski R., et al. Meta-analysis of gene-environment-wide association scans accounting for education level identifies additional loci for refractive error. Nat Commun. 2016;7:11008. doi: 10.1038/ncomms11008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han X., Ong J.S., An J., et al. Association of myopia and intraocular pressure with retinal detachment in European descent participants of the UK Biobank Cohort: A Mendelian Randomization Study. JAMA Ophthalmol. 2020;138:671–678. doi: 10.1001/jamaophthalmol.2020.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nair K.S., Hmani-Aifa M., Ali Z., et al. Alteration of the serine protease PRSS56 causes angle-closure glaucoma in mice and posterior microphthalmia in humans and mice. Nat Genet. 2011;43:579–584. doi: 10.1038/ng.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Han S., Chen P., Fan Q., et al. Association of variants in FRAP1 and PDGFRA with corneal curvature in Asian populations from Singapore. Hum Mol Genet. 2011;20:3693–3698. doi: 10.1093/hmg/ddr269. [DOI] [PubMed] [Google Scholar]
- 47.Chen P., Miyake M., Fan Q., et al. CMPK1 and RBP3 are associated with corneal curvature in Asian populations. Hum Mol Genet. 2014;23:6129–6136. doi: 10.1093/hmg/ddu322. [DOI] [PubMed] [Google Scholar]
- 48.Morgan I., Rose K. How genetic is school myopia? Prog Retin Eye Res. 2005;24:1–38. doi: 10.1016/j.preteyeres.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 49.MacArthur J., Bowler E., Cerezo M., et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog) Nucleic Acids Res. 2017;45:D896–D901. doi: 10.1093/nar/gkw1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Katz J., Tielsch J.M., Sommer A. Prevalence and risk factors for refractive errors in an adult inner city population. Invest Ophthalmol Vis Sci. 1997;38:334–340. [PubMed] [Google Scholar]
- 51.Wensor M., McCarty C.A., Taylor H.R. Prevalence and risk factors of myopia in Victoria, Australia. Arch Ophthalmol. 1999;117:658–663. doi: 10.1001/archopht.117.5.658. [DOI] [PubMed] [Google Scholar]
- 52.Wolfram C., Hohn R., Kottler U., et al. Prevalence of refractive errors in the European adult population: the Gutenberg Health Study (GHS) Br J Ophthalmol. 2014;98:857–861. doi: 10.1136/bjophthalmol-2013-304228. [DOI] [PubMed] [Google Scholar]
- 53.Dirani M., Shekar S.N., Baird P.N. The role of educational attainment in refraction: the Genes in Myopia (GEM) twin study. Invest Ophthalmol Vis Sci. 2008;49:534–538. doi: 10.1167/iovs.07-1123. [DOI] [PubMed] [Google Scholar]
- 54.Mirshahi A., Ponto K.A., Hoehn R., et al. Myopia and level of education: results from the Gutenberg Health Study. Ophthalmology. 2014;121:2047–2052. doi: 10.1016/j.ophtha.2014.04.017. [DOI] [PubMed] [Google Scholar]
- 55.Williams K.M., Bertelsen G., Cumberland P., et al. Increasing prevalence of myopia in Europe and the impact of education. Ophthalmology. 2015;122:1489–1497. doi: 10.1016/j.ophtha.2015.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mountjoy E., Davies N.M., Plotnikov D., et al. Education and myopia: assessing the direction of causality by Mendelian randomisation. BMJ. 2018;361 doi: 10.1136/bmj.k2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Solouki A.M., Verhoeven V.J., van Duijn C.M., et al. A genome-wide association study identifies a susceptibility locus for refractive errors and myopia at 15q14. Nat Genet. 2010;42:897–901. doi: 10.1038/ng.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Quint W.H., Tadema K.C.D., de Vrieze E., et al. Loss of Gap Junction Delta-2 (GJD2) gene orthologs leads to refractive error in zebrafish. Commun Biol. 2021;4:676. doi: 10.1038/s42003-021-02185-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meguro A., Yamane T., Takeuchi M., et al. Genome-wide association study in Asians identifies novel loci for high myopia and highlights a nervous system role in its pathogenesis. Ophthalmology. 2020;127:1612–1624. doi: 10.1016/j.ophtha.2020.05.014. [DOI] [PubMed] [Google Scholar]
- 60.Hao H.X., Xie Y., Zhang Y., et al. ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature. 2012;485:195–200. doi: 10.1038/nature11019. [DOI] [PubMed] [Google Scholar]
- 61.Gong Y., Krakow D., Marcelino J., et al. Heterozygous mutations in the gene encoding noggin affect human joint morphogenesis. Nat Genet. 1999;21:302–304. doi: 10.1038/6821. [DOI] [PubMed] [Google Scholar]
- 62.Sham P.C., Purcell S.M. Statistical power and significance testing in large-scale genetic studies. Nat Rev Genet. 2014;15:335–346. doi: 10.1038/nrg3706. [DOI] [PubMed] [Google Scholar]
- 63.Zhang J., Hur Y.M., Huang W., et al. Shared genetic determinants of axial length and height in children: the Guangzhou twin eye study. Arch Ophthalmol. 2011;129:63–68. doi: 10.1001/archophthalmol.2010.323. [DOI] [PubMed] [Google Scholar]
- 64.Wang D., Ding X., Liu B., et al. Longitudinal changes of axial length and height are associated and concomitant in children. Invest Ophthalmol Vis Sci. 2011;52:7949–7953. doi: 10.1167/iovs.11-7684. [DOI] [PubMed] [Google Scholar]
- 65.Guggenheim J.A., Zhou X., Evans D.M., et al. Coordinated genetic scaling of the human eye: shared determination of axial eye length and corneal curvature. Invest Ophthalmol Vis Sci. 2013;54:1715–1721. doi: 10.1167/iovs.12-10560. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.