Abstract
Estrogen and estrogen receptor alpha (ERα)-induced gene transcription is tightly associated with ERα-positive breast carcinogenesis. ERα-occupied enhancers, particularly super-enhancers, have been suggested to play a vital role in regulating such transcriptional events. However, the landscape of ERα-occupied super-enhancers (ERSEs) as well as key ERα-induced target genes associated with ERSEs remain to be fully characterized. Here, we defined the landscape of ERSEs in ERα-positive breast cancer cell lines, and demonstrated that bromodomain protein BRD4 is a master regulator of the transcriptional activation of ERSEs and cognate ERα target genes. RET, a member of the tyrosine protein kinase family of proteins, was identified to be a key ERα target gene of BRD4-regulated ERSEs, which, in turn, is vital for ERα-induced gene transcriptional activation and malignant phenotypes through activating the RAS/RAF/MEK2/ERK/p90RSK/ERα phosphorylation cascade. Combination therapy with BRD4 and RET inhibitors exhibited additive effects on suppressing ERα-positive breast cancer both in vitro and in vivo, comparable with that of standard endocrine therapy tamoxifen. Furthermore, combination therapy re-sensitized a tamoxifen-resistant ERα-positive breast cancer cell line to tamoxifen treatment. Taken together, our data uncovered the critical role of a super-enhancer-associated positive feedback loop constituting BRD4/ERα–RET–ERα in ERα-positive breast cancer, and suggested that targeting components in this loop would provide a new therapeutic avenue for treating ERα-positive breast cancer in the clinic.
INTRODUCTION
Breast cancer has surpassed lung cancer as the most commonly diagnosed cancer, with an estimated 2.3 million new cases in 2020 (1). Breast cancer is classified into five subtypes, namely luminal A, luminal B, HER2-positive, basal-like and normal-like. Luminal A and B subtypes together belong to the so-called estrogen receptor (ER)-positive group, which has the most heterogeneous copy number changes, gene expression patterns, mutation spectra and patient outcomes (2). First-line treatment for ER-positive breast cancer is endocrine therapy, including selective ER modulators (SERMs) (e.g. tamoxifen), aromatase inhibitors (e.g. letrozole) and selective ER down-regulators (SERDs) (e.g. fulvestrant) (3). However, there are myriad and prevalent side effects associated with endocrine therapies, such as hot flushes, arthralgias, night sweats and effects on bone and sexual health (3). Resistance towards endocrine therapy inevitably occurs in ER+ metastatic breast cancer (4). Searching for more effective treatments to overcome endocrine therapy resistance remains a challenging topic in the breast cancer field. ERα-dependent transcription plays a pivotal role in ER-positive breast tumorigenesis, which requires highly coordinated and complex interplay between various transcription factors, epigenetic enzymes, epigenetic readers and chromatin remodelers. These ERα cofactors represent a class of promising therapeutic targets in ERα-positive breast cancer.
Super-enhancers (SEs) are large clusters of enhancers that drive the expression of genes with implications in cell identity and diseases (5,6). SEs and SE-associated genes have been shown to be essential for cancer development (5). BRD4, a member of the bromodomain and extra-terminal (BET) family of proteins, has been shown to play critical roles in human diseases, such as cancer (7,8), cardiovascular disease (9,10), inflammatory disease (11) and central neural system (CNS) disorders (12). As part of the general transcription machinery, BRD4 is enriched at transcription start sites (TSSs), typical enhancer and SE regions, where it acts as an epigenetic reader to recognize acetylated lysine on histone tails and regulate transcription by recruiting the positive transcription elongation factor b (P-TEFb). BRD4-occupied SEs have been characterized in different types of cancers, which drive the expression of a large cohort of oncogenic genes to promote cancer development (13–15). A large number of small-molecule inhibitors targeting BRD4 have since been developed. JQ1 is the first reported and the most studied BET family inhibitor that can bind competitively to acetyl-lysine recognition motifs or bromodomains (16). Other BRD4 inhibitors including CPI203, MS417 (17) and OTX015 (18) were also reported. Dual BET-kinase inhibitors are efficacious against JAK2-driven cell lines and the neoplastic growth of hematopoietic progenitor cells from myeloproliferative neoplasm (MPN) patients (19). A phase I study of the phosphatidylinositol 3-kinase/BRD4 inhibitor SF1126 for the treatment of hepatocellular carcinoma and neuroblastoma was approved by the FDA. Despite the fact that BET inhibitors are promising therapeutic agents for the treatment of cancers, the rapid emergence of side effects and acquired resistance necessitates the investigation of combination therapies. For example, BRD4 inhibitors were combined with HDAC inhibitors to inhibit growth and induce apoptosis in a number of tumor models, including breast cancer (20), acute myeloid leukemia (21,22), pancreatic ductal adenocarcinoma (23), neuroblastoma (24) and cutaneous T-cell lymphoma (25). THe BET inhibitor OTX015 and the proteasome inhibitor carfilzomib synergistically induced apoptosis in TERT-rearranged neuroblastoma (26). Strong synergy was observed in small cell lung cancer (SCLC) when co-treated with the BET inhibitor ABBV-075 and BCL2 inhibitors (27). Co-treatment with JQ1 and the CDK7 inhibitor THZ1 impaired cell proliferation, and induced apoptosis and senescence in head and neck squamous cell carcinoma (28). We demonstrated that JQ1 overcame CAR-T cell therapy-induced immune resistance in glioblastoma (GBM) (29). Similarly, combination therapy with inhibitors targeting BRD4 and BRD4 downstream target genes would also represent an effective way for treating cancers.
RET is a transmembrane receptor and a member of the tyrosine protein kinase family of proteins. In general, a RET complex contains a dimerized RET receptor, a glial cell-derived neurotrophic factor (GDNF) family ligand and two GDNF family receptor alpha (GFRα) co-receptors (30). When ligands bind to GFRα, RET undergoes dimerization and subsequent phosphorylation, leading to the activation of different intracellular signaling cascades (31). RET plays critical roles in renal organogenesis and enteric neurogenesis (32). RET overexpression, fusion and mutations have been characterized as oncogenic drivers in various cancer types, showing great potential as a therapeutic target (33–42). RET expression was induced by estrogens, and RET signaling enhanced estrogen-driven proliferation in breast cancer (33,43–47). Recently, two RET-specific, mutant-effective inhibitors, BLU-667 (Pralsetinib) and LOXO-292 (Selpercatinib), were approved by the FDA for treating non-small cell lung cancer (NSCLC) and thyroid cancers.
In the current study, we defined the landscape of ERα-occupied SEs (ERSEs) in multiple ERα-positive breast cancer cell lines, and demonstrated that BRD4 is critical for the transcriptional activation of ERSEs and cognate ERα target genes. Furthermore, RET was identified to be a key downstream target gene of BRD4, promoting the malignant phenotypes of ERα-positive breast cancer cells. Combination therapy with BRD4 and RET inhibitors was effective in suppressing ERα-positive breast cancer cell growth both in vitro and in vivo.
MATERIALS AND METHODS
Cell culture
MCF7, T47D and HEK293T cells were obtained from the American Type Culture Collection (ATCC). MCF7, tamoxifen-resistant MCF7 and HEK293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; 01-052-1ACS, Biological Industries) supplemented with 10% fetal bovine serum (FBS; 04-001-1ACS, Biological Industries), and T47D cells were cultured in RPMI 1640 medium (01-100-1ACS, Biological Industries) supplemented with 10% FBS. Tamoxifen-resistant MCF7 cells were developed by culturing MCF7 cells in the presence of 2 μM tamoxifen (HY-13757A, MedChemExpress) for >12 months. Tamoxifen-resistant MCF7 cells were then maintained in the presence of 1 μM tamoxifen. All cells were cultured in a humidified incubator with 5% CO2 at 37°C. If estrogen (E2, E8875, Sigma) was added, cells were maintained in stripping medium (phenol red free) plus 5% charcoal-depleted FBS for 72 h before treatment.
Cloning procedures
Full-length human RET (NM_001355216.1) cDNA was cloned into the lentiviral vector pCDH-CMV-MCS-EF1-Puro (VT1480, Youbio). Short hairpin RNAs (shRNAs) targeting RET (shRET#1, CCTGTTTGTGAATGACACCAA; shRET#2, GCTGCATGAGAACAAC TGGAT) or BRD4 (shBRD4#1, AAGCTGAGAAAGTTGATGTGA; shBRD4#2, AAGACACTATGGAAACACCAG) were cloned into the lentiviral vector pLKO.1.
Lentivirus packaging and infection
HEK293T cells were transfected with the mixture of the lentiviral and packaging vectors, psPAX2 (12260, Addgene) and pMD2.G (12259, Addgene), using polyethylenimine (PEI, 24765-2, Polysciences) following the manufacturer's instructions. Lentivirus-containing supernatants were harvested 60 h post-transfection. Viruses were collected, filtered and added in the presence of 10 μg/ml polybrene (Sigma, H9268), followed by centrifugation for 30 min at 1500 g at 37°C. The medium was replaced 24 h later.
SiRNA transfection, RNA isolation and RT–qPCR
Small interfering RNA (siRNA) transfections were performed using Lipofectamine™ 2000 Transfection Reagent (11668500, Invitrogen) according to the manufacturer's protocol. MCF7 cells were transfected with siRNAs targeting RET (siRET#1, GCATCAACGTCCAGTACAA; siRET#2, GGACGGTTGTCACTTATGAAG) or BRD4 (siBRD4#1, GCGTTTCCACGGTACCAAA; siBRD4#2, GGAAACCTCAAG CTGAGAA). Total RNA was isolated using the Eastep Super Total RNA Extraction Kit (LS1040, Promega) following the manufacturer's protocol. First-strand cDNA synthesis from total RNA was performed using the GoScript Reverse Transcription System (A5001, Promega), followed by quantitative polymerase chain reaction (qPCR) using an AriaMx Real-Time PCR machine (Agilent Technologies). Sequence information for all primers used to check gene expression is presented in Supplementary Table S3.
Cell proliferation assay
Cell viability was measured by using a CellTiter 96 AQueous one solution cell proliferation assay kit (G3582, Promega) following the manufacturer's protocol. Briefly, MCF7 cells were maintained in stripping medium (phenol red free) for 2 days before treating or not with estrogen (E2, 10−7 M) for different times followed by cell proliferation assay. When BRD4 or RET knockdown cells were subjected to cell proliferation assay, cells were maintained in stripping medium for 2 days and then re-seeded at the same density before treating or not with estrogen (E2, 10−7 M) for different time followed by cell proliferation assay. To measure cell viability, 20 μl of CellTiter 96 AQueous one solution reagent was added per 100 μl of culture medium, and the culture plates were incubated for 1 h at 37°C in a humidified, 5% CO2 atmosphere incubator. The reaction was stopped by adding 25 μl of 10% sodium dodecylsulfate (SDS). Data were recorded at a wavelength of 490 nm using a Thermo Multiskan MK3 Microplate Reader.
Colony formation assay
BRD4 or RET knockdown cells were maintained in stripping medium for 2 days and re-seeded at the same density in a 6-well plate before treating or not with estrogen (E2, 10−7 M). Colonies were examined 20 days later. Briefly, colonies were fixed with methanol/acid solution (3:1) for 5 min and stained with 0.1% crystal violet for 15 min. For quantification, the crystal violet dye was released into 10% acetic acid and data were recorded at a wavelength of 590 nm.
Wound healing assay
BRD4 or RET knockdown cells were re-seeded and grown to 90% confluence in 6-well culture plates, and linear scratch wounds were created by a sterile 200 μl pipet tip in the cell monolayer. The wells were washed by phosphate-buffered saline (PBS) three times to remove detached cells. An inverted microscope (Carl Zeiss) was used to capture the images to calculate the rate of wound closure at 0 and 48 h after wounding.
Transwell assay
Cell invasion was investigated by a transwell chamber containing 50 ng/ml Matrigel (Corning) according to the manufacturer's instructions. Briefly, BRD4 or RET knockdown cells were re-seeded on the top compartment of trans-well Boyden chambers (8 μm, Corning) in serum-free medium, and then allowed to migrate to the lower compartment containing complete medium with 10% FBS in a humidified, 5% CO2 atmosphere incubator at 37°C for 36 h. Cells that did not migrate into the lower compartment were wiped away with a cotton swab. The inserts were fixed with methanol/acid solution (3:1) for 5 min and stained with 0.1% crystal violet for 20 min. After washing with PBS extensively, invaded cells were photographed and quantified using Image J.
Fluorescence-activated cell sorting (FACS) analysis
MCF7 cells were maintained in stripping medium for 2 days before treating or not with estrogen (E2, 10−7 M) followed by FACS analysis. Specifically, cells were trypsinized, washed with cold PBS and fixed with cold 75% ethanol at –20°C overnight. Cells were then washed with PBS and stained with propidium iodide (PI)/Triton X-100 staining solution [0.1% (v/v) Triton X-100, 0.2 mg/ml DNase-free RNase A (Sigma) and 0.02 mg/ml PI (Roche)] at 37°C for 15 min. DNA content was then measured and ∼105 events were analyzed for each sample. Data were analyzed using ModFit LT (Verity Software House).
Animal experiments
To examine the effects of RET knockdown on tumor growth, four groups (five mice per group) of female nude mice (BALB/C, 20–25 g, 4–6 weeks old) (Shanghai SLAC Laboratory Animal Center) were subcutaneously implanted with 5 × 106 control or RET knockdown MCF7 cells suspended in 100 μl of PBS. Each nude mouse was brushed with estrogen (E2, 10−2 M) on the neck every 3 days for the duration of the experiments to induce tumor formation. Mice were sacrificed before the tumor burden exceeded the limit (∼1500 mm3). Tumors were then excised, photographed and weighed.
To examine the effects of combination treatment with JQ1 and BLU-667, MCF7 cells stably expressing firefly luciferase (MCF7-fluc) were suspended in PBS (5.0 × 106 cells/mouse) and inoculated subcutaneously into female nude mice. Each nude mouse was brushed with estrogen (E2, 10−2 M) on the neck every 3 days for the duration of the experiments to induce tumor formation. Tumors were allowed to grow for 14 days until the mean flux reached ∼1 × 108 p/s/cm2/sr before treatment (day 0). Mice were then randomly assigned to four groups (five mice/group): CTL (PBS, intraperitoneal injection), JQ1 (50 mg/kg, intraperitoneal injection, every other day), BLU-667 (20 mg/kg, gavage injection, every other day) or JQ1 (25 mg/kg, intraperitoneal injection) combined with BLU-667 (10 mg/kg, gavage injection). Tumor progression was monitored by bioluminescence using the Xenogen IVIS Lumina imaging system (Caliper Life Sciences). Each mouse was injected intraperitoneally with beetle luciferin (1.5 mg, E1605, Promega) and then imaged 6–8 min later with an exposure time of 3 min. Luminescence images were analyzed using the Living Image software (Caliper Life Sciences). Tumor volume (V) was calculated using the formula: V = 1/2 (length × width2). The survival of mice was monitored daily. Mice were treated for 32 days and observed for 120 days.
To compare the effects of combination treatment with the standard therapy tamoxifen, five groups (five mice per group) of female nude mice were subcutaneously implanted with 5 × 106 MCF7 cells suspended in PBS. Estrogen was brushed on the neck every 3 days for the duration of the experiments. After 2 weeks, mice were randomized for treatment with tamoxifen (5 mg/kg, gavage injection, every other day), JQ1 (50 mg/kg, intraperitoneal injection, every other day), BLU-667 (20 mg/kg, gavage injection, every other day) or JQ1 (25 mg/kg, intraperitoneal injection) combined with BLU-667 (10 mg/kg, gavage injection) for another 32 days. Mice were sacrificed before the tumor burden exceeded the limit (∼1500 mm3). Tumors were then excised, photographed and weighed.
To assess the effects of combination treatment in tamoxifen-resistant breast tumor model, five groups (five mice per group) of female nude mice were subcutaneously implanted with 5 × 106 tamoxifen-resistant MCF7 cells suspended in PBS. After 2 weeks, mice were randomized for treatment with tamoxifen (5 mg/kg, gavage injection, every other day), JQ1 (25 mg/kg, intraperitoneal injection) combined with BLU-667 (10 mg/kg, gavage injection) or JQ1 (25 mg/kg, intraperitoneal injection) and BLU-667 (10 mg/kg, gavage injection) combined with tamoxifen (5 mg/kg, gavage injection, every other day) for another 32 days. Mice were sacrificed before the tumor burden exceeded the limit (∼1500 mm3). Tumors were then excised, photographed and weighed.
All animals were maintained in an animal room with 12 h light/12 h dark cycles and cared for with free access to standard rodent chow and water in accordance with institutional guidelines. All animal experiments were conducted in accordance with a protocol approved by the Animal Care and Use Committee of Xiamen University.
RNA sequencing (RNA-seq)
The Eastep Super Total RNA Extraction Kit (LS1040, Promega) was used for RNA isolation. DNase I was included in the column digestion to ensure RNA quality. RNA library preparation was performed using the NEBNext® Ultra™ Directional RNA Library Prep Kit for Illumina (E7420L, Illumina). Paired-end sequencing was performed using an Illumina HiSeq 3000 system. Sequencing reads were aligned to the hg19 RefSeq database using Tophat (http://ccb.jhu.edu/software/tophat/index.shtml). Cuffdiff was used to quantify the expression of RefSeq annotated genes with the option -M (reads aligned to repetitive regions were masked) and -u (multiple aligned reads were corrected using the ‘rescue method’). Coding genes with FPKM (fragments per kilobase per million mapped reads) >0.5 in at least one condition were included in our analysis. Up- or down-regulated genes were determined by the fold change (FC) of the gene’s FPKM. The FPKM of a gene was calculated as mapped reads on exons divided by exonic length and the total number of mapped reads. Box plots and heat maps were generated using R software, and statistical significance was determined using Student's t-test.
Chromatin immunoprecipitation coupled with high-throughput sequencing (ChIP-seq)
MCF7 cells were maintained in stripping medium for 3 days before treating or not with estrogen (E2, 10−7 M) for 1 h. Cells were then fixed with disuccinimidyl glutarate (DSG) (2 mM) (20593, Thermo Fisher Scientific) for 45 min at room temperature, washed twice with PBS and double-fixed with 1% formaldehyde (689316, Sigma) for another 10 min at room temperature. Fixation was stopped by adding glycine (1610718, Bio-Rad; 0.125 M) for 5 min at room temperature, followed by washing with PBS twice. Chromatin DNA was sheared to an average size of 300–500 bp by sonication. The resultant protein was immunoprecipitated with anti-BRD4 antibody (2 μg, A301-985A100, Bethyl Laboratories) overnight at 4°C, followed by incubation with protein G magnetic beads (0.4 mg/ml, 1614023, Bio-Rad) for an additional 4 h. After washing, the protein–DNA complex was reversed by heating at 65°C overnight. Immunoprecipitated DNA was purified using the QIAquick PCR Purification Kit (28104, Qiagen) and subjected to high-throughput sequencing.
Super-enhancer definition
ERα ChIP-Seq was aligned using Bowtie2 (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml) with default parameters to the human reference genome hg19. Both uniquely aligned reads and reads that align to repetitive regions were kept for downstream analysis (if a read aligned to multiple genomic locations, only one location with the best score was chosen). Clonal amplification was circumvented by allowing a maximum of one tag for each unique genomic position.
The identification of ChIP-seq peaks was performed using HOMER (http://homer.ucsd.edu/homer/). ChIP-Seq reads aligning to the region were extended by 200 bp. The threshold for the number of tags that determined a valid peak was selected at a false discovery rate (FDR) of 0.001. Four-fold more tags relative to the local background region (10 kb) were also required to avoid identifying regions with genomic duplications or non-localized binding. Genomic distribution was done by using the default parameters from HOMER with minor modifications, in which promoter peaks were defined as those with their peak center falling between 200 bp downstream and 5000 bp upstream of a TSS. Enhancers were defined as peaks which locate at non-promoter regions. In order to accurately identify dense clusters of enhancers, we stitched enhancers within 15 kb together. The tag density of clusters of enhancers was the sum of tag density at each enhancer involved.
To identify SEs, we first ranked all enhancer clusters by ERα ChIP-seq tag density, and then plotted them in order. The x-axis point at which a line with a slope of 1 was tangent to the curve was then identified. SEs were defined as enhancers above this point, and typical enhancers as enhancers below this point.
To define ERSE-associated genes, we first looked for genes inside an SE or the closest (both upstream and downstream) to an SE; if these genes are induced by estrogen, we continued to look for genes right next to them in both directions until we found genes that are not induced by estrogen. We counted these estrogen-induced genes as genes in the vicinity, nearby or associated with ERSE.
Global run-on sequencing (Gro-seq)
MCF7 cells were washed three times with cold PBS buffer and then incubated in swelling buffer [10 mM Tris–HCl (pH 7.5), 2 mM MgCl2, 3 mM CaCl2] for 5 min on ice and harvested. Cells were first re-suspended and lysed in lysis buffer (swelling buffer with 0.5% IGEPAL and 10% glycerol). The resultant nuclei were washed once again with 10 ml of lysis buffer and then re-suspended in 100 μl of freezing buffer [50 mM Tris–HCl (pH 8.3), 40% glycerol, 5 mM MgCl2, 0.1 mM EDTA]. For run-on assay, re-suspended nuclei were mixed with an equal volume of reaction buffer [10 mM Tris–HCl (pH 8.0), 5 mM MgCl2, 1 mM dithiothreitol (DTT), 300 mM KCl, 20 U of SUPERase In, 1% sarkosyl, 500 μM ATP, GTP and Br-UTP, and 2 μM CTP) and incubated for 5 min at 30°C. The nuclear run-on RNA (NRO-RNA) was then extracted with TRIzol LS reagent (Invitrogen) following the manufacturer's instructions. NRO-RNA was then subjected to base hydrolysis on ice for 40 min followed by treatment with DNase I and antarctic phosphatase. To purify the Br-UTP-labeled nascent RNA, the NRO-RNA was immunoprecipitated with anti-bromodeoxyuridine (BrdU) agarose beads (Santa Cruz Biotech) in binding buffer (0.5× SSPE, 1 mM EDTA, 0.05% Tween-20) for 1 h at 4°C while rotating. To repair the end, the immunoprecipitated BrU-RNA was re-suspended in a 50 μl reaction [43 μl of DEPC water, 5.2 μl of T4 PNK buffer, 1 μl of SUPERase In and 1 μl of T4 PNK (New England BioLabs)] and incubated at 37°C for 1 h. RNA was then extracted, precipitated using acidic phenol–chloroform (Ambion) and subjected to poly(A) tailing reaction by using poly(A) polymerase (New England BioLabs) for 30 min at 37°C. Subsequently, reverse transcription was performed using oNTI223 primers (5′pGATCGTCGGACTGTAGAACTCT; CAAGCAGAAGACGGCATACGATTTTTTTTTTTTTTTTTTTTVN-3′) where the p indicates 5′ phosphorylation, ‘;’ indicates the abasic dSpacer furan and VN indicates degenerate nucleotides. Specifically, tailed RNA (8.0 μl) was subjected to reverse transcription using Superscript III (Invitrogen), after which the cDNA products were separated on a 10% polyacrylamide TBE–urea gel. The extended first-strand product (100–500 bp) was excised and recovered by gel extraction. After that, the first-strand cDNA was circularized by CircLigase (Epicenter) and relinearized by ApeI (New England BioLabs). Relinearized single-stranded cDNA (sscDNA) was separated in a 10% polyacrylamide TBE gel as described above and the product of the needed size was excised (∼170–400 bp) for gel extraction. Finally, sscDNA template was amplified by PCR using the Phusion High-Fidelity enzyme (NEB) according to the manufacturer's instructions. Primers oNTI200 (5′-CAAGCAGAAGACGGCATA-3′) and oNTI201 (5′-AATGATACGGCGACCACCGACAGGTTCAGAGTTCTACAGTCCGACG-3′) were used to generate DNAs for deep sequencing. Sequencing was performed on the Illumina Genome Analyzer II (GA II) according to the manufacturer's instructions with small RNA sequencing primer 5′-CGACAGGTTCAGAGTTCTACAGTCCGACGATC-3′.
Traveling ratio calculation
The traveling ratio (TR) was defined as the relative ratio of Gro-seq tag density in the promoter-proximal region and the gene body. The TR was defined as the ratio of Gro-seq read density at the promoter-proximal bin (from 50 bp upstream to 300 bp downstream of the TSS) and that at the gene body bin [from 300 bp downstream of the TSS to the transcription termination site (TTS) or 30 kb downstream of the TSS if the distance from 300 bp downstream of the TSS to the TTS is >30 kb]. The significance of the change of the TR between control and knockdown samples was displayed using a box plot and determined using Student's t-test.
Immunoblotting analysis
Immunoblotting was performed as described previously (48). The following primary antibodies were used: anti-BRD4 antibody (A301-985A100, Bethyl Laboratories), anti-RET antibody (ab134100, Abcam), anti-RET (phospho Y1015) antibody (ab74154, Abcam), anti-Raf1 antibody (A19638, Abclonal), anti-phospho-Raf1-S259 antibody (AP1012, Abclonal), anti-MEK1/MEK2 antibody (A4868, Abclonal), anti-phospho-MAP2K1-S217/MAP2K2-S221 antibody (AP0209, Abclonal), anti-ERK1/2 antibody (A4782, Abclonal), anti-phospho-ERK1-T202/Y204 + ERK2-T185/Y187 antibody (AP0472, Abclonal), anti-p90Rsk antibody (A4695, Abclonal), anti-phospho-P90RSK-S380 antibody (AP0562, Abclonal), anti-ERα antibody (ab32063, Abcam), anti-phospho-ERα (Ser167) antibody (64508s, Cell Signaling Technology), anti-phospho-ERα (Ser118) antibody (ab32396, Abcam), anti-CCND1 antibody (ab40754, Abcam), anti-c-MYC antibody (SC-40, Santa Cruz Biotech) and anti-ACTIN antibody (8432, Cell Signaling Technology).
Statistical analysis
Comparison of two groups or data points was performed using two-tailed t-test. Statistical analyses of tumor growth data between multiple groups were performed using one-way analysis of variance (ANOVA) with Holm–Šídák's multiple comparisons test or two-way ANOVA with Dunnett's multiple comparisons test as needed. Survival curves were constructed according to the Kaplan–Meier method and compared using the log-rank (Mantel–Cox) test. Statistical significance was set at P <0.05.
RESULTS
ERSEs are highly active in ERα-positive breast cancer cells
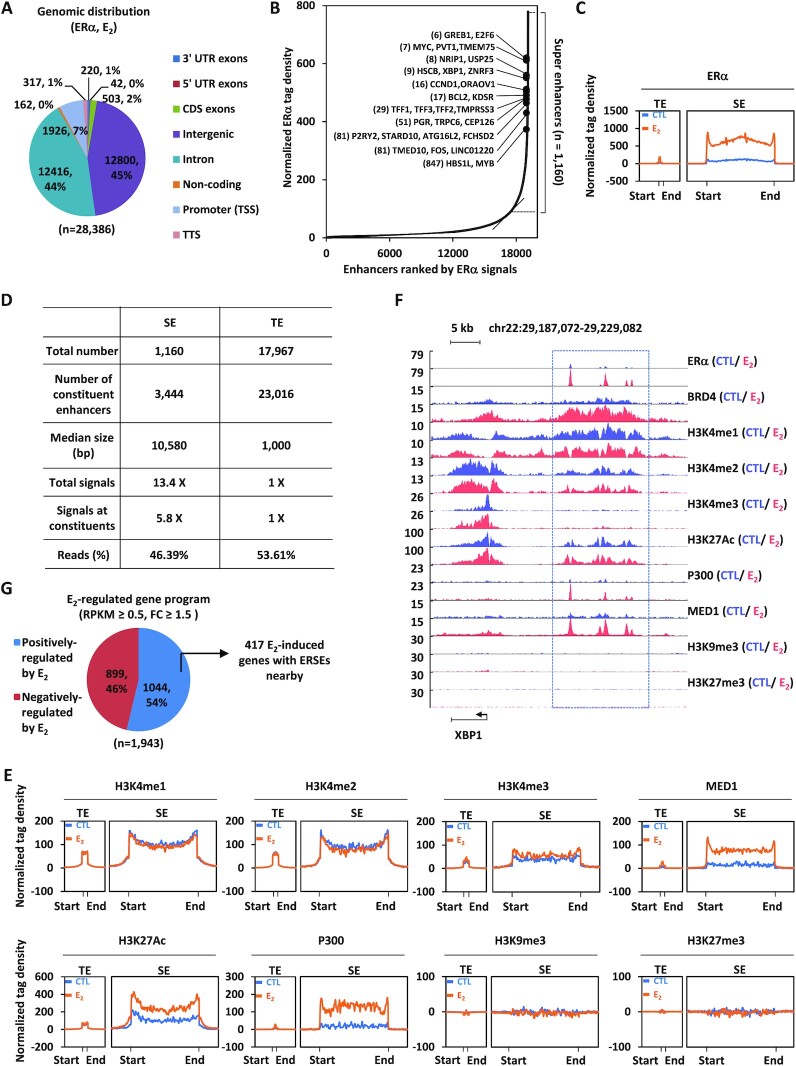
To better understand the molecular mechanisms underlying transcriptional activation of ERα-induced genes, we sought to define the landscape of ERSEs in MCF7 cells, a model cell line of ERα-positive breast cancer. ERα ChIP-seq analysis in MCF7 cells treated or not with estrogen revealed that, upon estrogen treatment, a large number of ERα-binding sites (n = 28 386) were found to be significantly induced (Figure 1A). Nearly 93% of these estrogen-induced ERα-binding sites were localized at distal regions (non-promoter regions) (Figure 1A). SE analysis results revealed that 1160 SEs could be defined based on these estrogen-induced ERα-binding sites (Figure 1B; Supplementary Figure S1A; Supplementary Table S1). Many of the well-known estrogen target genes, such as GREB1, MYC, NRIP1, XBP1, CCND1, BCL2, TFF1, PGR, P2RY2, FOS and MYB, were found to be in the vicinity of SEs (Figure 1B). These 1160 SEs differed greatly from typical enhancers in metagene representation of ERα occupancy (Figure 1C), number of constituent enhancers, median size, total ERα signals, ERα signals on constituent enhancers and number of total reads (Figure 1D). Specifically, there were ∼3 constituent enhancers on average for each SE (Figure 1D). The SEs had a median size of 10.58 kb, whereas that for typical enhancers was 1.0 kb (Figure 1D). We also defined the SEs based on ERα intensity without aggregation. If we compared the top 1160 enhancers defined based on intensity with those 1160 ERSEs defined above, there are 1003 in common, indicating that strong enhancers are often part of SEs (Supplementary Figure S1B). The ERSEs in two other ERα-positive breast cancer cell lines, T47D and ZR-75-1, were also analyzed. The results showed that there were 636 and 1344 ERSEs in T47D and ZR-75-1, respectively (Supplementary Figure S1C–F; Supplementary Table S1). Similar to what was observed in MCF7 cells, these SE differed greatly from typical enhancers in metagene representation of ERα occupancy (Supplementary Figure S1G, H), number of constituent enhancers, median size, total ERα signals, ERα signals on constituent enhancers and number of total reads (Supplementary Figure S1I, J). There were 214 ERSEs shared by all three cell lines, MCF7, T47D and ZR-75-1 (Supplementary Figure S1K; Supplementary Table S1). In particular, ERSEs were found in the vicinity of oncogenic estrogen target genes, such as MYC, CCND1, FOS and MYB (Supplementary Table S1).
Figure 1.
ERSEs are highly active in ERα-positive breast cancer. (A) MCF7 cells treated with estrogen (E2, 10−7 M, 1 h) were subjected to ChIP-seq with anti-ERα-specific antibody. Genomic distribution of ERα-binding sites is shown by a pie chart. (B) Normalized ERα ChIP-seq tag density in enhancer regions after clustering is shown. Estrogen-induced genes in the vicinity of representative ERSEs are shown. The number in parentheses indicates the rank of ChIP-seq tag density. (C) Metagene representation of ERα occupancy at typical enhancers (TEs) and super-enhancers (SEs) is shown. The x-axis shows the start and end of the TE (left) or SE (right) regions flanked by a 3 kb sequence. The y-axis shows the normalized tag density. (D) The characteristics of TEs and SEs defined by ERα are shown. (E) Metagene representation of H3K4me1, H3K4me2, H3K4me3, MED1, H3K27Ac, P300, H3K9me3 and H3K27me3 occupancy at TEs and SEs is shown. The x-axis shows the start and end of the TE (left) or SE (right) regions flanked by ± 3 kb sequence. The y-axis shows the normalized tag density. (F) UCSC Genome browser views of ERα, BRD4, H3K4me1, H3K4me2, H3K4me3, H3K27Ac, P300, MED1, H3K9me3 and H3K27me3 ChIP-seq in the presence or absence of estrogen on the SE region in the vicinity of the estrogen-induced gene, XBP1. The boxed region indicates SEs. (G) MCF7 cells treated or not with estrogen (E2, 10−7 M, 1 h) were subjected to Gro-seq. Genes positively and negatively regulated by estrogen based on Gro-seq, and those with an SE nearby are shown.
We then characterized the ERSEs in MCF7 cells in detail. Characteristics of enhancers, namely highly enriched H3K4me1/2, but low levels of H3K4me3, were evident from tag density distribution and a heat map for both ERα-occupied typical enhancers and SEs, supporting that they are bona fide enhancers (Figure 1E; Supplementary Figure S1L). Furthermore, these enhancer sites were enriched with activation markers including H3K27Ac, P300 and MED1, but devoid of repressive markers including H3K9me3 and H3K27me3, indicating that they are active enhancers (Figure 1E; Supplementary Figure S1L). SEs differed greatly from typical enhancers in metagene representation of H3K4me1/2, H3K27Ac, P300 and MED1 (Figure 1E; Supplementary Figure S1L). Representative ERSEs are shown, such as those in the vicinity of estrogen target genes XBP1, BCL2 and ITPK1 (Figure 1 F; Supplementary Figure S1M, N). To examine whether ERSEs themselves and genes in the vicinity are transcriptionally active, Gro-seq was performed in MCF7 cells treated or not with estrogen. Enhancer RNAs (eRNAs), the sign of enhancer activation, were strongly induced on ERSEs upon estrogen treatment (see below). Furthermore, 1044 genes were induced by estrogen, out of which 417 were found to be right near to ERSEs [reads per kilobase of transcript per million mapped reads (RPKM) ≥0.5, FC ≥1.5] (Figure 1G). Taken together, ERSEs are associated with active chromatin signatures and estrogen-induced transcriptional activation.
BRD4 is a master regulator of the transcriptional activation of ERSEs as well as estrogen target genes in their vicinity
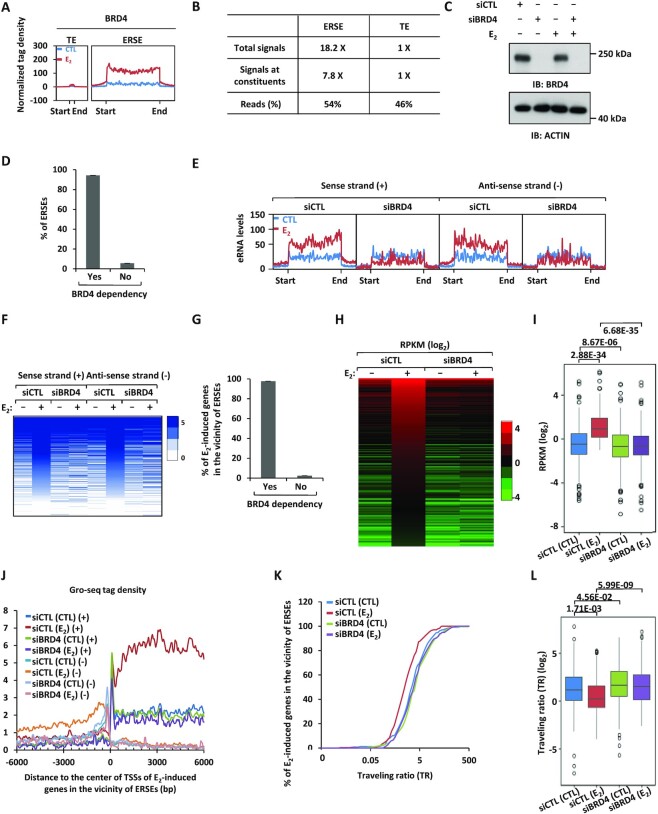
BRD4 has been shown to be associated with SEs in multiple cancer types (49–51). We directly tested whether BRD4 binds to ERSEs in ERα-positive breast cancer cells. To this end, BRD4 ChIP-seq was performed in MCF7 cells treated or not with estrogen. Upon estrogen treatment, BRD4 was highly and significantly induced on ERSEs (Figure 2A, B). The metagene representation of BRD4 occupancy, total BRD4 signals, BRD4 signals on constituent enhancers and number of total reads were much higher for BRD4-occupied ERSEs than typical enhancers (Figure 2A, B). We next asked whether BRD4 is required for the transcriptional activation of ERSEs as well as estrogen-induced genes in the vicinity. Gro-seq analysis results revealed that the activation of 94% of ERSEs, represented by eRNA levels on both the sense and antisense strand, was dependent on BRD4, as shown by the bar graph, tag density plot and heat map (Figure 2C–F). Accordingly, nearly all estrogen-induced genes (98%) in the vicinity of ERSEs were dependent on BRD4 for expression, as shown by the bar graph (Figure 2G), heat map (Figure 2H) and box plot (Figure 2I). The effects of BRD4 on the activation of estrogen-induced genes in the vicinity of ERSEs including PGR, GREB1, TFF1, SMAD7, SIAH2, CCND1 and MYC were demonstrated by RT–qPCR analysis (Supplementary Figure S2A, B; Supplementary Table S2), which were further validated by JQ1 treatment (Supplementary Figure S2C; Supplementary Table S2). The requirement of BRD4 for the activation of estrogen-induced genes was independently confirmed in another ERα-positive breast cancer cell line, T47D, through RT–qPCR analysis (Supplementary Figure S2D; Supplementary Table S2).
Figure 2.
BRD4 is a master regulator of the transcriptional activation of ERSEs as well as of estrogen target genes in the vicinity. (A) MCF7 cells treated or not with estrogen (E2, 10−7 M, 1 h) were subjected to ChIP-seq with anti-BRD4-specific antibody. Metagene representation of BRD4 occupancy at typical enhancers (TEs) and ERSEs is shown. The x-axis shows the start and end of the TE (left) or SE (right) regions flanked by ± 3 kb sequence. The y-axis shows the normalized tag density. (B) Statistics of BRD4 ChIP-seq on TEs and ERSEs. (C) MCF7 cells transfected with control siRNA (siCTL) or siRNAs specifically targeting BRD4 (siBRD4) were treated or not with estrogen (E2, 10−7 M, 1 h), followed by immunoblotting (IB) analysis using antibodies as indicated. Molecular weight is indicated on the right (in kDa). (D) MCF7 cells as described in (C) were subjected to Gro-seq analysis. The levels of eRNA were calculated, and the percentage of BRD4-dependent SEs is shown. (E, F) eRNA levels on both sense and antisense strands on ERSEs as detected by Gro-seq as described in (D) are shown by tag density plot (E) and heat map (F). (G) The percentage of ERSE-associated and estrogen-induced genes that are BRD4 dependent is shown. (H, I) The expression of ERSE-associated and estrogen-induced genes that are BRD4 dependent is shown by heat map (H) and box plot (I) (unpaired Student's t-test, two-tailed). (J) Gro-seq tag density, both sense (+) and antisense (−), centered on the TSSs (± 6000 bp) of ERSE-associated and estrogen-induced genes that are BRD4 dependent is shown. (K) Traveling ratio (TR) distribution calculated based on Gro-seq tag density for ERSE-associated and estrogen-induced genes that are BRD4 dependent. (L) The change of the TR as shown in (K) is shown by box plot (unpaired Student's t-test, two-tailed).
One of the major functions of BRD4 is to recruit the P-TEFb complex to induce transcriptional pausing release and subsequent transcriptional elongation (52,53). Close examination of the Gro-seq tag distribution surrounding TSSs (6 kb upstream and downstream of TSSs) revealed that there was a decrease in tag density at the promoter-proximal region, but an increase along the gene body upon estrogen treatment for estrogen-induced genes in the vicinity of ERSEs, resembling a typical pause release (Figure 2J). Knockdown of BRD4 attenuated the estrogen effects on pausing release (Figure 2J). We then calculated the TR based on the Gro-seq tag density in the promoter-proximal region and the gene body. Indeed, the vast majority of estrogen-induced genes in the vicinity of ERSEs experience promoter-proximal pausing, which was released upon estrogen treatment (i.e. the TR was decreased upon estrogen treatment) (Figure 2K). Importantly, knockdown of BRD4 abolished estrogen-induced pausing release (Figure 2K). The change of the TR caused by estrogen treatment and the impact of BRD4 on such a change was visualized by box plot analysis (Figure 2L). It should be noted that there are also a large number of BRD4-occupied enhancers that are not responsive to estrogen treatment. For instance, BRD4 binding on the enhancers of RTEL1 and GAPDH genes is not responsive to estrogen treatment (Supplementary Figure S2E, G). Knockdown of BRD4 exhibits no significant impact on the expression of these genes (Supplementary Figure S2F, H). Taken together, our data suggest that BRD4 is a master regulator of the transcriptional activation of ERSEs as well as estrogen-induced genes in the vicinity.
BRD4 promotes the malignant behaviors of ERα-positive breast cancer cells
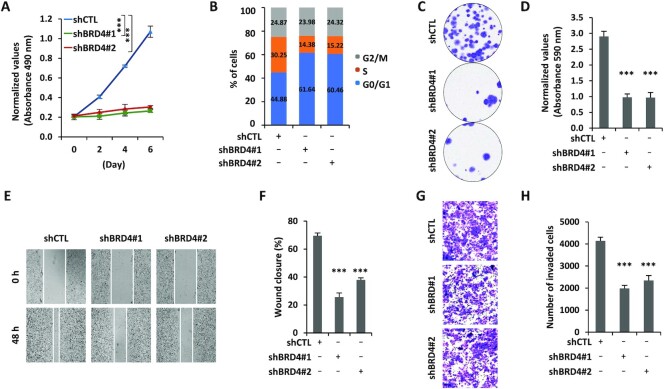
We next tested whether BRD4 promotes the malignant behaviors of ERα-positive breast cancer cells due to the critical role of estrogen-induced transcriptional events in ERα-positive breast cancer development. Knockdown of BRD4 significantly inhibited the growth of MCF7 cells (Figure 3A). FACS analysis results revealed that cells were arrested in G1 phase upon BRD4 knockdown (Figure 3B). BRD4’s effects on MCF7 cell growth were further demonstrated by colony formation assay (Figure 3C, D). Furthermore, the migration and invasion of MCF7 cells were inhibited when BRD4 was knocked down (Figure 3E–H). The effects of BRD4 on cell proliferation, cell cycle progression, colony formation, migration and invasion were independently confirmed by siRNA-mediated BRD4 knockdown (Supplementary Figure S3A–H). Furthermore, the BET inhibitor JQ1 exhibited similar effects (Supplementary Figure S3I–P). The requirement for BRD4 in cell proliferation and cell cycle progression was independently confirmed in T47D cells (Supplementary Figure S3Q, R). To strengthen the importance of BRD4, it is expressed at a significantly higher level in ER-positive breast tumor tissues compared with normal tissues (Supplementary Figure S3S). Taken together, our data suggest that BRD4 promotes the malignant phenotypes of ERα-positive breast cancer cells.
Figure 3.
BRD4 promotes the malignant behaviors of ERα-positive breast cancer cells. (A–C, E, G) MCF7 cells infected with lentiviral control shRNA (shCTL) or two independent shRNAs specifically targeting BRD4 (shBRD4#1 and shBRD4#2) were maintained in the presence of estrogen (E2, 10−7 M) followed by cell proliferation assay (A), FACS analysis (B), colony formation assay (C), wound healing (E) and transwell assay (G). (D, F, H) Quantification of the crystal violet dye as shown in (C) (D), wound closure in (E) (F) and the number of invaded cells in (G) (H) (± SEM, n = 3, ***P <0.001). Representative images are shown.
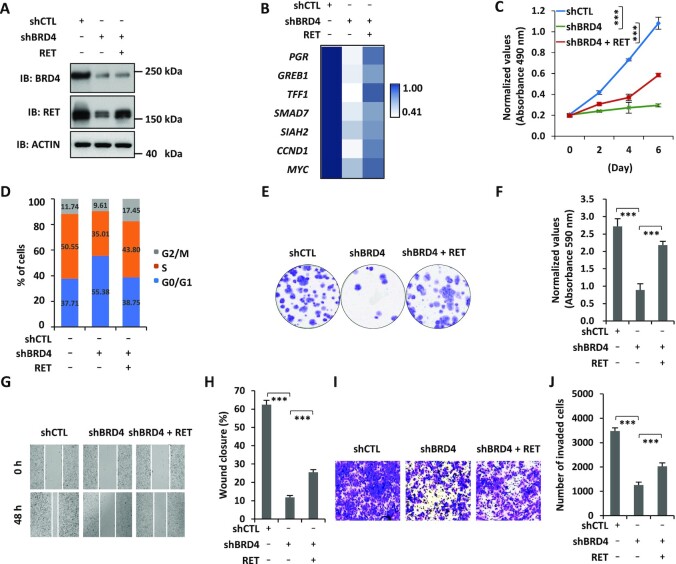
RET is associated with BRD4-regulated ERSE, and promotes the malignant behaviors of ERα-positive breast cancer cells
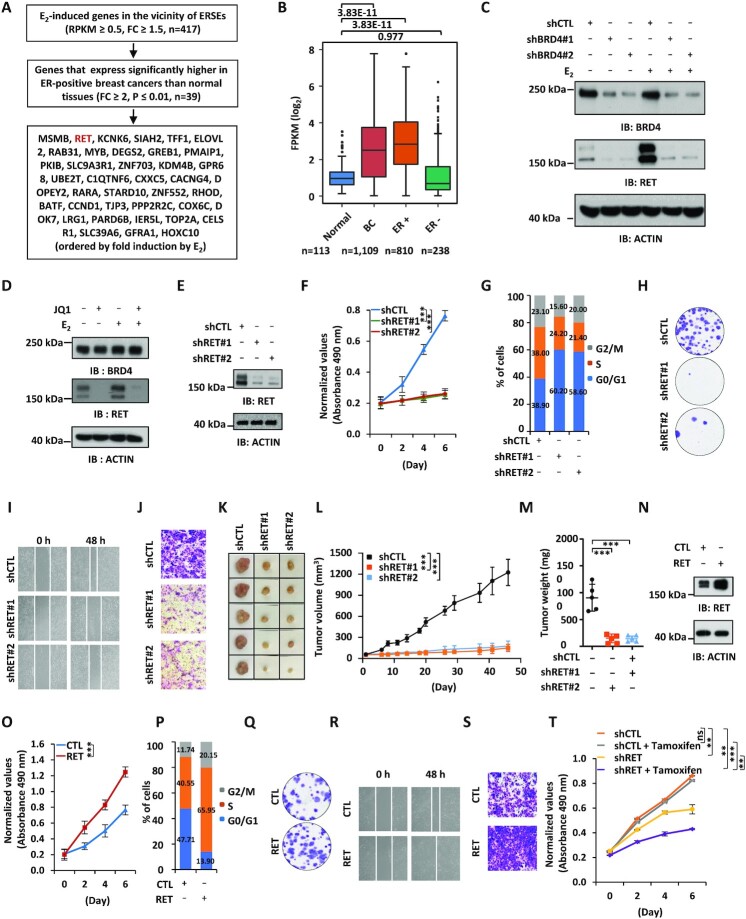
SEs are believed to control the expression of key oncogenes to drive tumorigenesis (5). To identify the key estrogen-induced genes in the vicinity of BRD4-regulated ERSEs, we first searched for genes that are clinically relevant. This led to the identification of 39 genes that are highly and significantly expressed in ER-positive breast tumor tissues compared with normal tissues in the clinic (FC ≥2, P ≤0.01). RET, a member of the tyrosine protein kinase family of proteins, was associated with ERSE and among the top genes most induced by estrogen (Figure 4A, B). Two of the constituent enhancers inside the ERSE we defined for the RET gene were previously reported to be critical for the transcriptional activation of RET (46) (Supplementary Figure S4A) (29). We first confirmed that RET could be significantly induced by estrogen, which was attenuated when BRD4 was knocked down (Figure 4C). JQ1 also attenuated estrogen-induced RET expression (Figure 4D). To examine whether ERSE-associated RET promotes the malignant phenotypes of ERα-positive breast cancer cells, we infected MCF7 cells with lentiviruses expressing RET shRNAs. Cell proliferation, cell cycle progression, colony formation, migration and invasion of MCF7 cells were significantly affected when RET was knocked down, which was similar to what was observed for BRD4 (Figure 4F–J; Supplementary Figure S4B–D). The effects of RET were independently confirmed by siRNA-mediated RET knockdown (Supplementary Figure S4E–M). Similar results were obtained with RET inhibitors including BLU-667, LOXO-292 and AST-487 (Supplementary Figure S4N–U). The requirement of RET for cell proliferation and cell cycle progression was independently confirmed in T47D cells (Supplementary Figure S4V, W). To test whether RET affects tumor growth in vivo, we injected nude mice subcutaneously with control MCF7 cells or MCF7 cells with RET knockdown. It was found that RET knockdown led to a significant reduction of tumor growth rate, tumor volume and tumor weight (Figure 4K–M).
Figure 4.
RET is associated with BRD4-regulated ERSEs, and promotes the malignant behaviors of ERα-positive breast cancer cells. (A) Flowchart of identifying ERSE-associated estrogen target genes that are clinically relevant. (B) The expression of RET in normal, breast cancer (BC), ER-positive (ER+) breast cancer and ER-negative (ER−) breast cancer samples from the TCGA database is shown by box plot (unpaired Student's t-test, two-tailed). (C) MCF7 cells infected with lentiviral control shRNA (shCTL) or two independent shRNAs specifically targeting BRD4 (shBRD4#1 and shBRD4#2) were maintained in the presence or absence of estrogen (E2, 10−7 M, 6 h) followed by immunoblotting (IB) analysis using antibodies as indicated. Molecular weight is indicated on the right (in kDa). (D) MCF7 cells pre-treated or not with JQ1 (250 nM, 30 min) were then treated or not with estrogen (E2, 10−7 M, 6 h) followed by IB analysis using antibodies as indicated. Molecular weight is indicated on the right (in kDa). (E–J) MCF7 cells infected with lentiviral control shRNA (shCTL) or two independent shRNAs specifically targeting RET (shRET#1 and shRET#2) were maintained in the presence of estrogen (E2, 10−7 M) followed by IB analysis (E), cell proliferation assay (F), FACS analysis (G), colony formation assay (H), wound healing assay (I) and transwell assay (J). (K) Xenograft experiments were performed by injecting shRNA or shRET lentivirus-infected MCF7 cells into female BALB/C nude mice (five mice per group). Tumor growth was monitored daily, and tumors were then excised, photographed and weighed at the end of the experiment. (L) The growth curve of tumors as in (K) is shown (± SD, ***P <0.001). (M) The weight of tumors as in (K) is shown (± SD, ***P <0.001). (N–S) MCF7 cells infected with lentiviral control vector (CTL) or vector expressing RET were maintained in the presence of estrogen (E2, 10−7 M) followed by IB analysis (N), cell proliferation assay (O), FACS analysis (P), colony formation assay (Q), wound healing assay (R) and transwell assay (S). (T) Tamoxifen-resistant MCF7 cells infected with shCTL or shRET were treated or not with tamoxifen (2 μM), followed by cell proliferation assay (± SEM, **P <0.01; ***P <0.001; ns, non-significant).
To further strengthen the critical role of RET in promoting the malignant phenotypes of ERα-positive breast cancer cells, we overexpressed RET in MCF7 cells followed by cell proliferation assay, FACS analysis, colony formation assay, wound healing assay and transwell assay. Overexpression of RET was found to promote cell growth, G1 to S progression, colony formation, migration and invasion of MCF7 cells (Figure 4N–S; Supplementary Figure S4X–Z). Furthermore, RET knockdown re-sensitized tamoxifen-resistant MCF7 cells to tamoxifen treatment (Figure 4T). Taken together, our data suggest that RET, a gene in the vicinity of BRD4-regulated ERSEs, is essential for the malignant phenotypes of ERα-positive breast cancer cells.
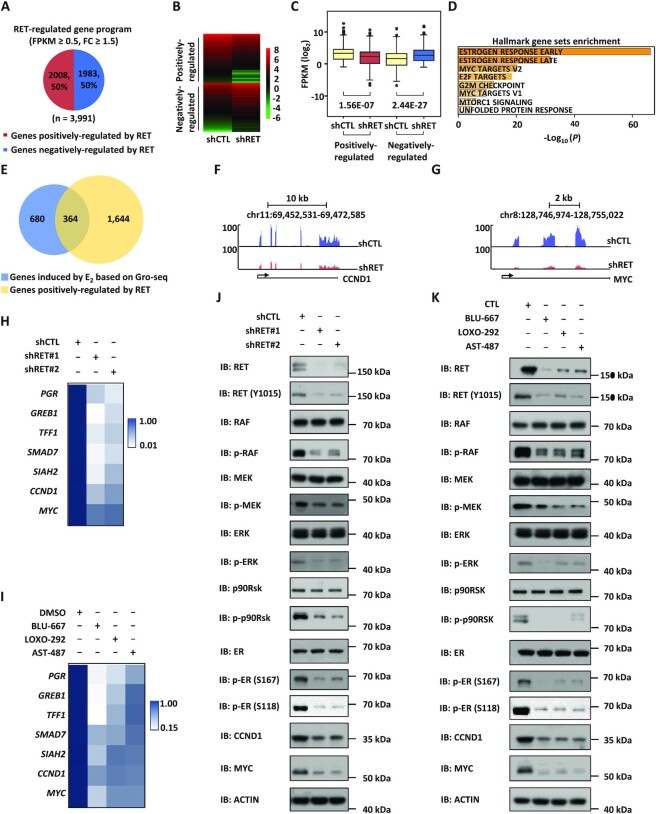
RET is a downstream target of BRD4-regulated ERSEs
To further demonstrate that RET is a key target gene of BRD4-regulated ERSEs, we performed rescue experiments in which RET was introduced when BRD4 was knocked down (Figure 5A). The expression of BRD4-regulated, ERSE-associated estrogen target genes, such as PGR, GREB1, TFF1, SMAD7, SIAH2, CCND1 and MYC, was significantly down-regulated when BRD4 was knocked down, which was largely rescued by introduction of RET (Figure 5B; Supplementary Table S2). Accordingly, RET partially rescued the defects of cell proliferation, cell cycle progression, colony formation, migration and invasion caused by BRD4 knockdown (Figure 5C–J). Our data therefore suggest that RET is a key downstream target gene of BRD4-regulated ERSEs.
Figure 5.
RET is a downstream target of BRD4-regulated ERSEs. (A–E, G, I) MCF7 cells infected with control lentiviral vector or lentiviral vector expressing shBRD4 in the presence or absence of lentiviral vector expressing RET were subjected to immunoblotting (IB) analysis (A), RT–qPCR analysis to examine the expression of selected estrogen target genes (B), cell proliferation assay (C), FACS analysis (D), colony formation assay (E), wound healing assay (G) and transwell assay (I). Molecular weight is indicated on the right. Statistical significance for (B) is shown in Supplementary Table S2 (± SEM, *P <0.05; **P <0.01; ***P <0.001). (F, H, J) Quantification of the crystal violet dye as shown in (E) (F), wound closure in (G) (H) and the number of invaded cells in (I) (J) (± SEM, n = 3, ***P <0.001). Representative images are shown.
RET activates the RAS/RAF/MEK/ERK/p90RSK/ERα phosphorylation cascade to promote the expression of estrogen target genes
To understand the molecular mechanisms through which RET regulates the malignant behaviors of ERα-positive breast cancer cells, we knocked down RET and then performed RNA-seq analysis in MCF7 cells. RNA-seq analysis results revealed that RET regulated the expression of a large cohort of genes, with 2008 and 1983 genes being positively and negatively regulated by RET, respectively (Figure 6A–C). In particular, estrogen-responsive genes are the most enriched hallmarks for genes positively regulated by RET (Figure 6D). Indeed, nearly 35% of estrogen-induced genes were dependent on RET for expression (Figure 6E). RNA-seq views for estrogen-induced genes from the UCSC genome browser track, such as CCND1 and MYC, are shown (Figure 6F, G). The dependency on RET for estrogen target genes, such as PGR, GREB1, TFF1, SMAD7, SIAH2, CCND1 and MYC, was confirmed by RT–qPCR analysis in both MCF7 and T47D cells (Figure 6H; Supplementary Figure S5A; Supplementary Table S2). Furthermore, the expression of these genes was similarly inhibited by the RET inhibitors BLU-667, LOXO-292 and AST-487 (Figure 6I; Supplementary Table S2). Overexpression of RET was shown to activate the RET/RAS/RAF/MEK/ERK/p90RSK signaling cascade, and ERK and p90RSK can both phosphorylate ERα at Ser118 and Ser167, respectively, to promote the expression of ERα target genes (54–56). Indeed, the phosphorylation of RAF, MEK, ERK, p90RSK and ERα was significantly attenuated when RET was knocked down (Figure 6J) or inhibited (Figure 6K). As expected, similar observations were made when BRD4 was knocked down (Supplementary Figure S5B). RET knockdown also led to a significant decrease of ER phosphorylation in tamoxifen-resistant MCF7 cells (Supplementary Figure S5C). Taken together, our data suggest that RET promotes the phosphorylation of ERα via the RAS/RAF/MEK/ERK/p90RSK signaling pathway to activate estrogen target genes and promote the malignant phenotypes of ERα-positive breast cancer cells.
Figure 6.
RET activates the RAS/RAF/MEK/ERK/p90RSK/ERα phosphorylation cascades to promote the expression of estrogen target genes. (A) MCF7 cells were infected with lentiviral control shRNA (shCTL) or shRNA specifically targeting RET (shRET) followed by RNA-seq analysis. Genes positively and negatively regulated by RET are shown by a pie chart. (B, C) Heat map (B) and box blot (C) representation of the expression (FPKM, log2) for genes regulated by RET (unpaired Student's t-test, two-tailed). (D) Hallmark gene set enrichment analysis for genes positively regulated by RET. (E) The overlap of genes induced by E2 and genes positively regulated by RET is shown by a Venn diagram. (F, G) UCSC Genome browser views of the expression of CCND1 and MYC from RNA-seq are shown. Blue, shCTL; red, shRET. (H) MCF7 cells infected with control shRNA (shCTL) or two independent shRNAs specifically targeting RET (shRET#1 and shRET#2) were maintained in the presence of estrogen (E2, 10−7 M, 6 h) followed by RT–qPCR analysis to examine selected estrogen-induced genes. Statistical significance is shown in Supplementary Table S2 (± SEM, *P <0.05; **P <0.01; ***P <0.001). (I) MCF7 cells pre-treated with LOXO-292 (1 μM), BLU-667 (1 μM) or AST487 (1 μM) were then treated with estrogen (E2, 10−7 M, 6 h) followed by RT–qPCR analysis to examine selected estrogen-induced genes. Statistical significance is shown in Supplementary Table S2 (± SEM, *P <0.05; **P <0.01; ***P <0.001). (J, K) MCF7 cells as described in (H) or (I) were subjected to immunoblotting (IB) analysis using antibodies as indicated. Molecular weight is indicated on the right (in kDa).
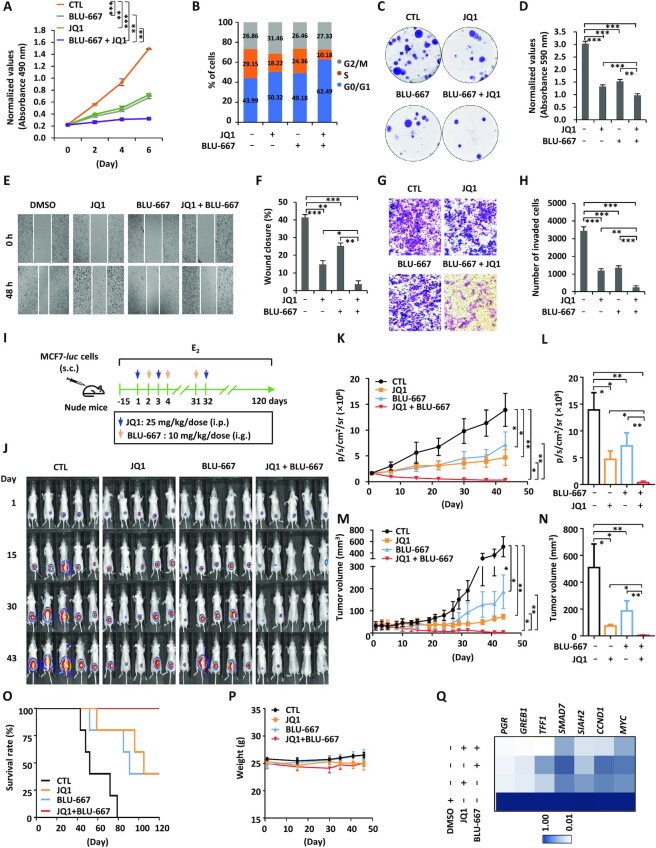
Combination treatment with BRD4 and RET inhibitors suppresses ERα-positive breast cancer both in vitro and in vivo
Our data above suggest that BRD4 is a master regulator of ERSEs and estrogen-induced genes in the vicinity of ERSEs as well as ERα-positive breast cancer cell growth. RET serves as a critical downstream target of BRD4. This prompted us to propose that simultaneously blocking BRD4 and RET might be an effective way to suppress ERα-positive breast cancer. To this end, MCF7 cells were treated with JQ1 or BLU-667 alone or in combination followed by cell proliferation assay. Co-treatment with both JQ1 and BLU-667 exhibited additive effects on cell proliferation compared with treatment with either JQ1 or BLU-667 alone (Figure 7A). The additive effects of BRD4 and RET inhibitor were also seen in cell cycle progression, colony formation, cell migration and cell invasion (Figure 7B–H). Similar observations were made in T47D cells (Supplementary Figure S6A, B).
Figure 7.
Combination treatment with BRD4 and RET inhibitors suppresses ERα-positive breast cancer cell growth both in vitro and in vivo. (A–C, E, G) MCF7 cells treated or not with JQ1 (250 nM) in the presence or absence of BLU-667 (1 μM) were subjected to cell proliferation assay (A), FACS analysis (B), colony formation assay (C), wound healing assay (E) and transwell assay (G). (D, F, H) Quantification of the crystal violet dye as shown in (C) (D), wound closure in (E) (F) and the number of invaded cells in (G) (H) (± SEM, n = 3, **P <0.01; ***P <0.001). Representative images are shown. (I) Six-week-old female nude mice were injected subcutaneously (s.c.) with MCF7 cells stably expressing a luciferase reporter (MCF7-luc, 5.0 × 106 cells/mouse), and randomized for treatment 15 days later (day 1, five mice/group). Mice were treated with CTL [PBS, intraperitoneal (i.p.) injection], JQ1 (50 mg/kg, i.p. injection), BLU-667 [20 mg/kg, gavage (i.g.) injection] or JQ1 (25 mg/kg) combined with BLU-667 (10 mg/kg) following the protocol as depicted. Mice were brushed with estrogen (E2, 10−2 M) on the neck every 3 days for the duration of the experiments to induce tumor formation. (J) Tumor growth was monitored by bioluminescence imaging. (K, M) The tumor growth curves based on bioluminescence (K) and tumor volumes (M) are shown (± SEM, n = 5, *P <0.05; **P <0.01; ***P <0.001, two-way ANOVA). (L, N) The average of bioluminescence (L) and tumor volume (N) based on (K) and (M), respectively, is shown (± SEM, *P <0.05; **P <0.01; ***P <0.001). (O) The survival curves of mice as shown in (J). (P) The average of mice weight as shown in (J). (Q) Tumors as shown in (J) were excised and subjected to RNA extraction followed by RT–qPCR analysis to examine the expression of genes as indicated. Statistical significance is shown in Supplementary Table S2 (± SEM, *P <0.05; **P <0.01; ***P <0.001).
The combination treatment was further tested in an MCF7 cell-derived xenograft model. Nude mice were subcutaneously implanted with MCF7 cells stably expressing a luciferase gene, and treated with JQ1 and/or BLU-667 as indicated (Figure 7I). Tumor growth was monitored via bioluminescence imaging and caliper-based sizing. Consistent with what was observed in cultured cells, co-treatment with both JQ1 and BLU-667 exhibited additive effects on both bioluminescence intensity and tumor volume compared with treatment with either JQ1 or BLU-667 alone (Figure 7J–N). All mice in the control group died within 80 days (Figure 7O). In the JQ1- or BLU-667-treated group, only two-fifths of the mice survived and they were with tumors (Figure 7O). All mice were alive at the end of observation (120 days) in the co-treated group, and two-fifths were tumor-free (Figure 7O). All treatments exhibited no apparent side effects on body weight (Figure 7P). JQ1 and BLU-667 co-treatment exhibited additive effects on the expression of estrogen target genes including PGR, GREB1, TFF1, SMAD7, SIAH2, CCND1 and MYC in the tumor tissues isolated (Figure 7Q; Supplementary Table S2).
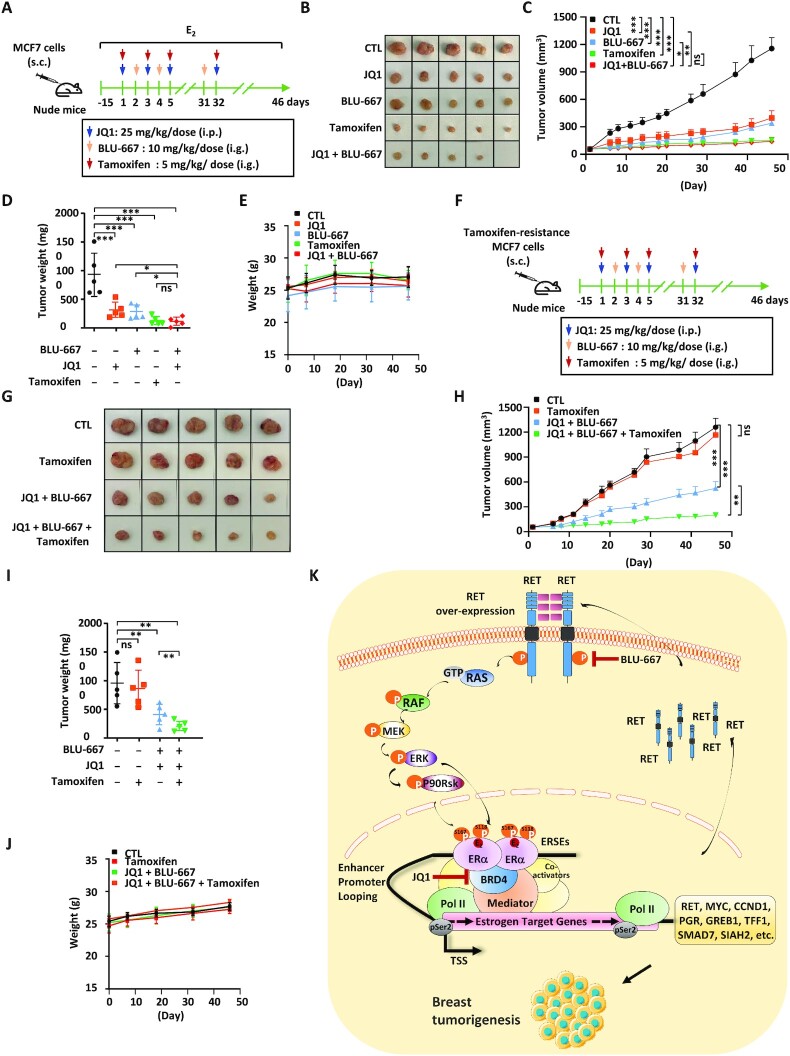
In order to explore the clinical application potential, we compared the therapeutic effects of combination therapy with JQ1 and BLU-667 with that of one of the standard therapies for ER-positive breast cancer, tamoxifen. Nude mice were subcutaneously implanted with MCF7 cells, and treated with tamoxifen, JQ1 and/or BLU-667 (Figure 8A). Tumor growth was monitored via caliper-based sizing. The tumor-suppressive effect of JQ1 or BLU-667 treatment alone was less effective than tamoxifen. However, co-treatment with JQ1 and BLU-667 was as effective as tamoxifen treatment (Figure 8B–D). All treatments exhibited no apparent side effects on body weight (Figure 8E). Furthermore, we tested whether combination treatment with JQ1 and BLU-667 could re-sensitize tamoxifen-resistant MCF7 cells to tamoxifen treatment in a xenograft mouse model. Nude mice were subcutaneously implanted with tamoxifen-resistant MCF7 cells, and treated with tamoxifen, JQ1 and/or BLU-667 (Figure 8F). Tumor growth was monitored via caliper-based sizing. As expected, tamoxifen-resistant MCF7 cell-derived tumors exhibited no responses to tamoxifen treatment. However, treatment with JQ1 and BLU-667 re-sensitized tamoxifen-resistant tumors to tamoxifen treatment, as co-treatment with JQ1, BLU-66, and tamoxifen exhibited the most dramatic effects on tumor suppression (Figure 8G–I). All treatments exhibited no apparent side effects on body weight (Figure 8J). Taken together, our data suggest that simultaneously blocking BRD4 and RET function by JQ1 and BLU-667, respectively, is an effective way to suppress ERα-positive breast cancer as well as those that have developed resistance to endocrine therapy.
Figure 8.
Combination treatment with BRD4 and RET inhibitors is comparable with tamoxifen treatment, and can re-sensitize tamoxifen-resistant cells. (A) Six-week-old female nude mice were injected subcutaneously (s.c.) with MCF7 cells (5.0 × 106 cells/mouse), and randomized for treatment 15 days later (day 1, five mice/group). Mice were treated with CTL [corn oil, intraperitoneal (i.p.) injection], JQ1 (25 mg/kg, i.p. injection), BLU-667 [10 mg/kg, gavage (i.g.) injection], tamoxifen (5 mg/kg, i.g. injection) or JQ1 (25 mg/kg) combined with BLU-667 (10 mg/kg) following the protocol as depicted. Mice were brushed with estrogen (E2, 10−2 M) on the neck every 3 days for the duration of the experiments to induce tumor formation. Tumor growth was monitored daily, and tumors were then excised, photographed and weighed. (B) Tumors as described in (A) were excised and photographed. (C) The tumor growth curve for tumors as described in (A) is shown (± SEM, n = 5, *P <0.05; **P <0.01; ***P <0.001, two-way ANOVA). (D) The weight of tumors as described in (A) is shown (± SD, ***P <0.001). (E) The body weight of mice as described in (A) is shown (± SD). (F) Six-week-old female nude mice were injected s.c. with tamoxifen-resistant MCF7 cells (5.0 × 106 cells/mouse), and randomized for treatment 15 days later (day 1, five mice/group). Mice were treated with CTL (corn oil), tamoxifen (5 mg/kg), JQ1 (25 mg/kg) combined with BLU-667 (10 mg/kg) or tamoxifen (5 mg/kg) combined with JQ1 (25 mg/kg) and BLU-667 (10 mg/kg) following the protocol as depicted. (G) Tumors as described in (F) were excised and photographed. (H) The tumor growth curve for tumors as described in (F) is shown (± SEM, n = 5, **P <0.01; ***P <0.001; ns, non-significant, two-way ANOVA). (I) The weight of tumors as described in (G) is shown (± SD, **P <0.01; ns, non-significant). (J) The body weight of mice as described in (F) is shown (± SD). (K) A proposed working model for an SE-controlled positive feedback loop constituting BRD4/ERα–RET–ERα in promoting ERα-positive breast cancer. Estrogen-induced BRD4 binding on ERSEs triggers the expression of a large cohort of oncogenic estrogen targets including RET, which activates the RAS/RAF/MEK/ERK/p90RSK signaling cascades to induce ERα phosphorylation and the expression of estrogen target genes. The amplification of oncogenic signaling from this positive feedback loop constituting BRD4/ERα–RET–ERα contributes to ERα-positive breast carcinogenesis. Simultaneously targeting two critical components in this loop, BRD4 and RET, is effective in suppressing ERα-positive breast cancer both in vitro and in vivo.
DISCUSSION
It is well recognized that cancer cells rely on aberrant transcription propelled by SEs, and such dependence offers valuable targets for cancer therapy (57–59). For instance, proteins governing the activation of SEs as well as protein products of genes downstream of SEs are appealing drug targets. In our study, we defined ERSEs in ERα-positive breast cancer cells, and demonstrated that BRD4 is a master regulator of the transcriptional activation of ERSEs and estrogen target genes in their vicinity. One of these ERSE-associated genes, RET, was found to enhance ERα phosphorylation to promote estrogen-induced gene transcription, forming a positive feedback loop to drive ERα-positive breast cancer development. Targeting BRD4 and RET in this loop was effective in suppressing breast cancer cell and tumor growth (Figure 8K).
We set out to define the landscape of ERSEs based on ERα-binding sites in multiple ERα-positive breast cancer cells, and a large cohort of ERSEs were shared. There was 67.35% overlap between the SEs identified by Bojcsuk et al. and those defined by us here in MCF7 cells (60). It has been well documented that chromatin loops are formed among ERSEs, promoters and/or gene bodies of estrogen-induced target genes such as TFF1, GREB1, SIAH2, PGR, MYC, CCND1, BCL2, ITPK1 and RET, providing the molecular basis for transcriptional activation induced by estrogen (61–64). Importantly, well-known oncogenes, such as MYC, CCND1, FOS and MYB, were found to be associated with ERSEs, supporting the functionality of ERSEs. SEs are particularly sensitive to drug intervention and therefore proteins that govern the activation of SEs show great promise as therapeutic targets (57). Due to the critical role of BRD4 in SE regulation and disease development (49–51,65), we tested directly whether BRD4 similarly regulates the activation of ERSEs and estrogen target genes in the vicinity as well as ERα-positive breast cancer. Strikingly, nearly all ERSEs and ERSE-associated, estrogen target genes are dependent on BRD4 for activation, strengthening the therapeutic potential of BRD4 inhibitors in treating ERα-positive breast cancer.
To date, many small-molecule inhibitors targeting BRD4 have been discovered, and a number of them are in clinical trials for treating diseases including cancer, inflammatory diseases (11), cardiovascular diseases (9) and CNS disorders (12). However, limited potency and acquired resistance from BRD4 inhibitors represent the main challenges for clinical use. Therefore, combination therapy is urgently needed to improve the use of these inhibitors. We focus on searching for key downstream target genes of BRD4-occupied ERSEs, and propose that combination therapy with BRD4 inhibitors and inhibitors targeting the protein products of these key genes would be effective in treating ERα-positive breast cancer. There are several criteria to be a key target gene for BRD4-occupied ERSEs: this gene is associated with ERSEs; this gene is regulated by BRD4; this gene is highly expressed in ER-positive breast cancer samples; and this gene can rescue the defects in ERSE-induced gene transcription and ER-positive breast cancer cell growth when BRD4 is knocked down. This led to the discovery of 39 genes, of which we focused on RET due to its importance in breast cancer reported previously as well as its therapeutic value. It should be emphasized that, though they are not highly expressed in ER-positive breast cancer, oncogenes such as CCND1 and c-Myc should also, at least partially, be responsible for BRD4’s oncogenic role. Two RET-specific inhibitors, BLU-667 and LOXO-292, were recently approved for treating NSCLC and thyroid cancer (MTC) by the FDA. As we proposed, co-treatment with the BRD4 inhibitor JQ1 and the RET inhibitor BLU-667 was much more effective than treatment with either inhibitor alone. The tumor-suppressive effects of combination treatment are comparable with that of tamoxifen, a standard therapy for ERα-positive breast cancer in the clinic, in MCF7 cell-derived xenografts. In addition, combination therapy re-sensitized tamoxifen-resistant cells to tamoxifen treatment. Despite additive effects on tumor suppression seen from combination treatment, potential cumulative side effects should also be taken into consideration in the clinic. It should be noted that there are many other downstream target genes of BRD4-occupied ERSEs, such as KDM4B and TOP2A, and testing the effects of combination therapy with BRD4 inhibitors and inhibitors targeting these proteins is worthy of future investigation.
As a member of the tyrosine protein kinase family, RET can be activated upon GDNF binding with its co-receptor GFRα, undergoing dimerization and subsequent phosphorylation. Alternatively, RET overexpression can lead to self-activation. RET can activate several downstream pathways, including RAS/MAPK, PI3K/AKT, JAK/STAT, PKA and PKC pathways (54–56,66–71). Based on our RNA-seq and immunoblotting analysis, RET was found to regulate the estrogen-induced program through RAS/RAF/MEK/ERK/p90RSK-mediated ERα phosphorylation. Therefore, a positive feedback loop in estrogen-induced gene activation was revealed, such that BRD4/ERα-occupied ERSE drives the expression of RET, which in turn activates ERα to stimulate the expression of itself as well as other estrogen target genes, leading to uncontrolled cell growth.
In summary, we found that targeting SE and SE-driven transcriptional events is an effective way to suppress ERα-positive breast cancer. This strategy may also be effective in treating other cancers. Future investigation is required to determine whether this therapeutic strategy can be translated into clinical settings.
DATA AVAILABILITY
The authors confirm that the data supporting the findings of this study are available within the article and/or its supplementary data, or can be made available upon reasonable request. ERα ChIP-seq in MCF7 was from GSE45822; ERα ChIP-seq in T47D was from GSE72249 and GSE80367; ERα ChIP-seq in ZR-75–1 was from GSE72249 and GSE141582; H3K27Ac and P300 ChIP-seq were from GSE62229; MED1 ChIP-seq was from GSE60272; H3K4me3, H3K9me3 and H3K27me3 ChIP-seq were from GSE23701. All other sequencing data were deposited in the Gene Expression Omnibus database under accession GSE186646.
Supplementary Material
ACKNOWLEDGEMENTS
We thank all members of the Liu laboratory for helpful discussions during the development of this project.
Contributor Information
Zao-zao Zheng, State Key Laboratory of Cellular Stress Biology, School of Pharmaceutical Sciences, Faculty of Medicine and Life Sciences, Xiamen University, Xiang’an South Road, Xiamen, Fujian 361102, China; Fujian Provincial Key Laboratory of Innovative Drug Target Research, School of Pharmaceutical Sciences, Faculty of Medicine and Life Sciences, Xiamen University, Xiang’an South Road, Xiamen, Fujian 361102, China.
Lin Xia, State Key Laboratory of Cellular Stress Biology, School of Pharmaceutical Sciences, Faculty of Medicine and Life Sciences, Xiamen University, Xiang’an South Road, Xiamen, Fujian 361102, China; Fujian Provincial Key Laboratory of Innovative Drug Target Research, School of Pharmaceutical Sciences, Faculty of Medicine and Life Sciences, Xiamen University, Xiang’an South Road, Xiamen, Fujian 361102, China.
Guo-sheng Hu, State Key Laboratory of Cellular Stress Biology, School of Pharmaceutical Sciences, Faculty of Medicine and Life Sciences, Xiamen University, Xiang’an South Road, Xiamen, Fujian 361102, China; Fujian Provincial Key Laboratory of Innovative Drug Target Research, School of Pharmaceutical Sciences, Faculty of Medicine and Life Sciences, Xiamen University, Xiang’an South Road, Xiamen, Fujian 361102, China.
Jun-yi Liu, State Key Laboratory of Molecular Vaccinology and Molecular Diagnostics, National Institute of Diagnostics and Vaccine Development in Infectious Diseases, School of Public Health, Faculty of Medicine and Life Sciences, Xiamen University, Xiang’an South Road, Xiamen, Fujian 361102, China.
Ya-hong Hu, State Key Laboratory of Cellular Stress Biology, School of Pharmaceutical Sciences, Faculty of Medicine and Life Sciences, Xiamen University, Xiang’an South Road, Xiamen, Fujian 361102, China; Fujian Provincial Key Laboratory of Innovative Drug Target Research, School of Pharmaceutical Sciences, Faculty of Medicine and Life Sciences, Xiamen University, Xiang’an South Road, Xiamen, Fujian 361102, China.
Yu-jie Chen, State Key Laboratory of Cellular Stress Biology, School of Pharmaceutical Sciences, Faculty of Medicine and Life Sciences, Xiamen University, Xiang’an South Road, Xiamen, Fujian 361102, China; Fujian Provincial Key Laboratory of Innovative Drug Target Research, School of Pharmaceutical Sciences, Faculty of Medicine and Life Sciences, Xiamen University, Xiang’an South Road, Xiamen, Fujian 361102, China.
Jia-yin Peng, State Key Laboratory of Cellular Stress Biology, School of Pharmaceutical Sciences, Faculty of Medicine and Life Sciences, Xiamen University, Xiang’an South Road, Xiamen, Fujian 361102, China; Fujian Provincial Key Laboratory of Innovative Drug Target Research, School of Pharmaceutical Sciences, Faculty of Medicine and Life Sciences, Xiamen University, Xiang’an South Road, Xiamen, Fujian 361102, China.
Wen-juan Zhang, Department of Laboratory Medicine, First Affiliated Hospital of Gannan Medical University, No. 23, Qingnian Road, Ganzhou, Jiangxi 341000, China.
Wen Liu, State Key Laboratory of Cellular Stress Biology, School of Pharmaceutical Sciences, Faculty of Medicine and Life Sciences, Xiamen University, Xiang’an South Road, Xiamen, Fujian 361102, China; Fujian Provincial Key Laboratory of Innovative Drug Target Research, School of Pharmaceutical Sciences, Faculty of Medicine and Life Sciences, Xiamen University, Xiang’an South Road, Xiamen, Fujian 361102, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by the Ministry of Science and Technology of China [2020YFA0112300 and 2020YFA0803600]; National Natural Science Foundation of China [82125028, 31871319, 91953114, 81761128015 and 81861130370]; Natural Science Foundation of Fujian Province of China [2020J02004]; and the Fundamental Research Funds for the Central University [20720190145 and 20720220003 to W.L.]
Conflict of interest statement. None declared.
REFERENCES
- 1. Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F.. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021; 71:209–249. [DOI] [PubMed] [Google Scholar]
- 2. Koboldt D.C., Fulton R.S., McLellan M.D., Schmidt H., Kalicki-Veizer J., McMichael J.F., Fulton L.L., Dooling D.J., Ding L., Mardis E.R.et al.. Comprehensive molecular portraits of human breast tumours. Nature. 2012; 490:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Burstein H.J. Systemic therapy for estrogen receptor-positive, HER2-negative breast cancer. Reply. N. Engl. J. Med. 2021; 384:1176–1177. [DOI] [PubMed] [Google Scholar]
- 4. Hanker A.B., Sudhan D.R., Arteaga C.L.. Overcoming endocrine resistance in breast cancer. Cancer Cell. 2020; 37:496–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hnisz D., Abraham B.J., Lee T.I., Lau A., Saint-Andre V., Sigova A.A., Hoke H.A., Young R.A.. Super-enhancers in the control of cell identity and disease. Cell. 2013; 155:934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Whyte W.A., Orlando D.A., Hnisz D., Abraham B.J., Lin C.Y., Kagey M.H., Rahl P.B., Lee T.I., Young R.A.. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013; 153:307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Qin Z.Y., Wang T., Su S., Shen L.T., Zhu G.X., Liu Q., Zhang L., Liu K.W., Zhang Y., Zhou Z.H.et al.. BRD4 promotes gastric cancer progression and metastasis through acetylation-dependent stabilization of Snail. Cancer Res. 2019; 79:4869–4881. [DOI] [PubMed] [Google Scholar]
- 8. Shi C.Z., Ye Z., Han J., Ye X.Q., Lu W.C., Ji C.X., Li Z.Z., Ma Z.Y., Zhang Q.L., Zhang Y.C.et al.. BRD4 as a therapeutic target for nonfunctioning and growth hormone pituitary adenoma. Neuro Oncol. 2020; 22:1114–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Anand P., Brown J.D., Lin C.Y., Qi J., Zhang R., Artero P.C., Alaiti M.A., Bullard J., Alazem K., Margulies K.B.et al.. BET bromodomains mediate transcriptional pause release in heart failure. Cell. 2013; 154:569–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim S.Y., Zhang X., Schiattarella G.G., Altamirano F., Ramos T.A.R., French K.M., Jiang N., Szweda P.A., Evers B.M., May H.I.et al.. Epigenetic reader BRD4 (Bromodomain-Containing protein 4) governs nucleus-encoded mitochondrial transcriptome to regulate cardiac function. Circulation. 2020; 142:2356–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brown J.D., Lin C.Y., Duan Q., Griffin G., Federation A., Paranal R.M., Bair S., Newton G., Lichtman A., Kung A.et al.. NF-kappaB directs dynamic super enhancer formation in inflammation and atherogenesis. Mol. Cell. 2014; 56:219–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liang D., Yu Y., Ma Z.. Novel strategies targeting bromodomain-containing protein 4 (BRD4) for cancer drug discovery. Eur. J. Med. Chem. 2020; 200:112426. [DOI] [PubMed] [Google Scholar]
- 13. Delmore J.E., Issa G.C., Lemieux M.E., Rahl P.B., Shi J.W., Jacobs H.M., Kastritis E., Gilpatrick T., Paranal R.M., Qi J.et al.. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011; 146:903–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nagarajan S., Hossan T., Alawi M., Najafova Z., Indenbirken D., Bedi U., Taipaleenmaki H., Ben-Batalla I., Scheller M., Loges S.et al.. Bromodomain protein BRD4 is required for estrogen receptor-dependent enhancer activation and gene transcription. Cell Rep. 2014; 8:460–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Feng Q., Zhang Z., Shea M.J., Creighton C.J., Coarfa C., Hilsenbeck S.G., Lanz R., He B., Wang L., Fu X.et al.. An epigenomic approach to therapy for tamoxifen-resistant breast cancer. Cell Res. 2014; 24:809–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Filippakopoulos P., Qi J., Picaud S., Shen Y., Smith W.B., Fedorov O., Morse E.M., Keates T., Hickman T.T., Felletar I.et al.. Selective inhibition of BET bromodomains. Nature. 2010; 468:1067–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li Z., Guo Z., Lan R., Cai S., Lin Z., Li J., Wang J., Li Z., Liu P.. The poly(ADP-ribosyl)ation of BRD4 mediated by PARP1 promoted pathological cardiac hypertrophy. Acta Pharm. Sin. B. 2021; 11:1286–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Filippakopoulos P., Picaud S., Mangos M., Keates T., Lambert J.P., Barsyte-Lovejoy D., Felletar I., Volkmer R., Muller S., Pawson T.et al.. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012; 149:214–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ember S.W., Lambert Q.T., Berndt N., Gunawan S., Ayaz M., Tauro M., Zhu J.Y., Cranfill P.J., Greninger P., Lynch C.C.et al.. Potent dual BET bromodomain-kinase inhibitors as value-added multitargeted chemical probes and cancer therapeutics. Mol. Cancer Ther. 2017; 16:1054–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Borbely G., Haldosen L.A., Dahlman-Wright K., Zhao C.. Induction of USP17 by combining BET and HDAC inhibitors in breast cancer cells. Oncotarget. 2015; 6:33623–33635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fiskus W., Sharma S., Qi J., Valenta J.A., Schaub L.J., Shah B., Peth K., Portier B.P., Rodriguez M., Devaraj S.G.et al.. Highly active combination of BRD4 antagonist and histone deacetylase inhibitor against human acute myelogenous leukemia cells. Mol. Cancer Ther. 2014; 13:1142–1154. [DOI] [PubMed] [Google Scholar]
- 22. Latif A.L., Newcombe A., Li S., Gilroy K., Robertson N.A., Lei X., Stewart H.J.S., Cole J., Terradas M.T., Rishi L.et al.. BRD4-mediated repression of p53 is a target for combination therapy in AML. Nat. Commun. 2021; 12:241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mazur P.K., Herner A., Mello S.S., Wirth M., Hausmann S., Sanchez-Rivera F.J., Lofgren S.M., Kuschma T., Hahn S.A., Vangala D.et al.. Combined inhibition of BET family proteins and histone deacetylases as a potential epigenetics-based therapy for pancreatic ductal adenocarcinoma. Nat. Med. 2015; 21:1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shahbazi J., Liu P.Y., Atmadibrata B., Bradner J.E., Marshall G.M., Lock R.B., Liu T.. The bromodomain inhibitor JQ1 and the histone deacetylase inhibitor panobinostat synergistically reduce N-Myc expression and induce anticancer effects. Clin. Cancer Res. 2016; 22:2534–2544. [DOI] [PubMed] [Google Scholar]
- 25. Zhao L., Okhovat J.P., Hong E.K., Kim Y.H., Wood G.S.. Preclinical studies support combined inhibition of BET family proteins and histone deacetylases as epigenetic therapy for cutaneous T-cell lymphoma. Neoplasia. 2019; 21:82–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen J.W., Nelson C., Wong M., Tee A.E., Liu P.Y., La T., Fletcher J.I., Kamili A., Mayoh C., Bartenhagen C.et al.. Targeted therapy of TERT-rearranged neuroblastoma with BET bromodomain inhibitor and proteasome inhibitor combination therapy. Clin. Cancer Res. 2021; 27:1438–1451. [DOI] [PubMed] [Google Scholar]
- 27. Lam L.T., Lin X., Faivre E.J., Yang Z., Huang X., Wilcox D.M., Bellin R.J., Jin S., Tahir S.K., Mitten M.et al.. Vulnerability of small-cell lung cancer to apoptosis induced by the combination of BET bromodomain proteins and BCL2 inhibitors. Mol. Cancer Ther. 2017; 16:1511–1520. [DOI] [PubMed] [Google Scholar]
- 28. Zhang W., Ge H., Jiang Y., Huang R., Wu Y.P., Wang D.M., Guo S.S., Li S., Wang Y.L., Jiang H.B.et al.. Combinational therapeutic targeting of BRD4 and CDK7 synergistically induces anticancer effects in head and neck squamous cell carcinoma. Cancer Lett. 2020; 469:510–523. [DOI] [PubMed] [Google Scholar]
- 29. Xia L., Liu J.Y., Zheng Z.Z., Chen Y.J., Ding J.C., Hu Y.H., Hu G.S., Xia N.S., Liu W.. BRD4 inhibition boosts the therapeutic effects of epidermal growth factor receptor-targeted chimeric antigen receptor T cells in glioblastoma. Mol. Ther. (2021); 29:3011–3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Airaksinen M.S., Saarma M.. The GDNF family: signalling, biological functions and therapeutic value. Nat. Rev. Neurosci. 2002; 3:383–394. [DOI] [PubMed] [Google Scholar]
- 31. Ichihara M., Murakumo Y., Takahashi M.. RET and neuroendocrine tumors. Cancer Lett. 2004; 204:197–211. [DOI] [PubMed] [Google Scholar]
- 32. Schuchardt A., D’Agati V., Larsson-Blomberg L., Costantini F., Pachnis V.. Defects in the kidney and enteric nervous system of mice lacking the tyrosine kinase receptor RET. Nature. 1994; 367:380–383. [DOI] [PubMed] [Google Scholar]
- 33. Boulay A., Breuleux M., Stephan C., Fux C., Brisken C., Fiche M., Wartmann M., Stumm M., Lane H.A., Hynes N.E.. The RET receptor tyrosine kinase pathway functionally interacts with the ERalpha pathway in breast cancer. Cancer Res. 2008; 68:3743–3751. [DOI] [PubMed] [Google Scholar]
- 34. Mechera R., Soysal S.D., Piscuoglio S., Ng C.K.Y., Zeindler J., Mujagic E., Daster S., Glauser P., Hoffmann H., Kilic E.et al.. Expression of RET is associated with oestrogen receptor expression but lacks prognostic significance in breast cancer. BMC Cancer. 2019; 19:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Subbiah V., Cote G.J.. Advances in targeting RET-dependent cancers. Cancer Discov. 2020; 10:498–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Komminoth P., Roth J., Muletta-Feurer S., Saremaslani P., Seelentag W.K., Heitz P.U.. RET proto-oncogene point mutations in sporadic neuroendocrine tumors. J. Clin. Endocrinol. Metab. 1996; 81:2041–2046. [DOI] [PubMed] [Google Scholar]
- 37. Lanzi C., Borrello M.G., Bongarzone I., Migliazza A., Fusco A., Grieco M., Santoro M., Gambetta R.A., Zunino F., Della Porta G.et al.. Identification of the product of two oncogenic rearranged forms of the RET proto-oncogene in papillary thyroid carcinomas. Oncogene. 1992; 7:2189–2194. [PubMed] [Google Scholar]
- 38. Santoro M., Rosati R., Grieco M., Berlingieri M.T., Damato G.L., Defranciscis V., Fusco A.. The ret protooncogene is consistently expressed in human pheochromocytomas and thyroid medullary carcinomas. Oncogene. 1990; 5:1595–1598. [PubMed] [Google Scholar]
- 39. Xing S., Smanik P.A., Oglesbee M.J., Trosko J.E., Mazzaferri E.L., Jhiang S.M.. Characterization of ret oncogenic activation in MEN2 inherited cancer syndromes. Endocrinology. 1996; 137:1512–1519. [DOI] [PubMed] [Google Scholar]
- 40. Carlson K.M., Dou S., Chi D., Scavarda N., Toshima K., Jackson C.E., Wells S.A. Jr, Goodfellow P.J., Donis-Keller H. Single missense mutation in the tyrosine kinase catalytic domain of the RET protooncogene is associated with multiple endocrine neoplasia type 2B. Proc. Natl Acad. Sci. USA. 1994; 91:1579–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dawson D.M., Lawrence E.G., MacLennan G.T., Amini S.B., Kung H.J., Robinson D., Resnick M.I., Kursh E.D., Pretlow T.P., Pretlow T.G.. Altered expression of RET proto-oncogene product in prostatic intraepithelial neoplasia and prostate cancer. J. Natl Cancer Inst. 1998; 90:519–523. [DOI] [PubMed] [Google Scholar]
- 42. Ito Y., Okada Y., Sato M., Sawai H., Funahashi H., Murase T., Hayakawa T., Manabe T.. Expression of glial cell line-derived neurotrophic factor family members and their receptors in pancreatic cancers. Surgery. 2005; 138:788–794. [DOI] [PubMed] [Google Scholar]
- 43. Gattelli A., Nalvarte I., Boulay A., Roloff T.C., Schreiber M., Carragher N., Macleod K.K., Schlederer M., Lienhard S., Kenner L.et al.. Ret inhibition decreases growth and metastatic potential of estrogen receptor positive breast cancer cells. EMBO Mol. Med. 2013; 5:1335–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gattelli A., Garcia Sola M.E., Roloff T.C., Cardiff R.D., Kordon E.C., Chodosh L.A., Hynes N.E.. Chronic expression of wild-type Ret receptor in the mammary gland induces luminal tumors that are sensitive to Ret inhibition. Oncogene. 2018; 37:4046–4054. [DOI] [PubMed] [Google Scholar]
- 45. Gattelli A., Hynes N.E., Schor I.E., Vallone S.A.. Ret receptor has distinct alterations and functions in breast cancer. J. Mammary Gland Biol. Neoplasia. 2020; 25:13–26. [DOI] [PubMed] [Google Scholar]
- 46. Stine Z.E., McGaughey D.M., Bessling S.L., Li S., McCallion A.S.. Steroid hormone modulation of RET through two estrogen responsive enhancers in breast cancer. Hum. Mol. Genet. 2011; 20:3746–3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tan S.K., Lin Z.H., Chang C.W., Varang V., Chng K.R., Pan Y.F., Yong E.L., Sung W.K., Cheung E.. AP-2 gamma regulates oestrogen receptor-mediated long-range chromatin interaction and gene transcription. EMBO J. 2011; 30:2750–2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Xia L., Zheng Z.Z., Liu J.Y., Chen Y.J., Ding J.C., Xia N.S., Luo W.X., Liu W.. EGFR-targeted CAR-T cells are potent and specific in suppressing triple-negative breast cancer both in vitro and in vivo. Clin. Transl. Immunology. 2020; 9:e01135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liu W., Ma Q., Wong K., Li W., Ohgi K., Zhang J., Aggarwal A., Rosenfeld M.G.. Brd4 and JMJD6-associated anti-pause enhancers in regulation of transcriptional pause release. Cell. 2013; 155:1581–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chapuy B., McKeown M.R., Lin C.Y., Monti S., Roemer M.G.M., Qi J., Rahl P.B., Sun H.H., Yeda K.T., Doench J.G.et al.. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell. 2013; 24:777–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gryder B.E., Yohe M.E., Chou H.C., Zhang X.H., Marques J., Wachtel M., Schaefer B., Sen N., Song Y., Gualtieri A.et al.. PAX3-FOXO1 establishes myogenic super enhancers and confers BET bromodomain vulnerability. Cancer Discov. 2017; 7:884–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jang M.K., Mochizuki K., Zhou M.S., Jeong H.S., Brady J.N., Ozato K.. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell. 2005; 19:523–534. [DOI] [PubMed] [Google Scholar]
- 53. Yang Z.Y., Yik J.H.N., Chen R.C., He N.H., Jang M.K., Ozato K., Zhou Q.. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell. 2005; 19:535–545. [DOI] [PubMed] [Google Scholar]
- 54. Plaza-Menacho I., Morandi A., Robertson D., Pancholi S., Drury S., Dowsett M., Martin L.A., Isacke C.M.. Targeting the receptor tyrosine kinase RET sensitizes breast cancer cells to tamoxifen treatment and reveals a role for RET in endocrine resistance. Oncogene. 2010; 29:4648–4657. [DOI] [PubMed] [Google Scholar]
- 55. Morandi A., Plaza-Menacho I., Isacke C.M.. RET in breast cancer: functional and therapeutic implications. Trends Mol. Med. 2011; 17:149–157. [DOI] [PubMed] [Google Scholar]
- 56. Lo Nigro C., Rusmini M., Ceccherini I.. RET in breast cancer: pathogenic implications and mechanisms of drug resistance. Cancer Drug Resist. 2019; 2:1136–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Loven J., Hoke H.A., Lin C.Y., Lau A., Orlando D.A., Vakoc C.R., Bradner J.E., Lee T.I., Young R.A.. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013; 153:320–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chipumuro E., Marco E., Christensen C.L., Kwiatkowski N., Zhang T., Hatheway C.M., Abraham B.J., Sharma B., Yeung C., Altabef A.et al.. CDK7 inhibition suppresses super-enhancer-linked oncogenic transcription in MYCN-driven cancer. Cell. 2014; 159:1126–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Christensen C.L., Kwiatkowski N., Abraham B.J., Carretero J., Al-Shahrour F., Zhang T., Chipumuro E., Herter-Sprie G.S., Akbay E.A., Altabef A.et al.. Targeting transcriptional addictions in small cell lung cancer with a covalent CDK7 inhibitor. Cancer Cell. 2014; 26:909–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bojcsuk D., Nagy G., Balint B.L.. Inducible super-enhancers are organized based on canonical signal-specific transcription factor binding elements. Nucleic Acids Res. 2017; 45:3693–3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Le Dily F., Bau D., Pohl A., Vicent G.P., Serra F., Soronellas D., Castellano G., Wright R.H., Ballare C., Filion G.et al.. Distinct structural transitions of chromatin topological domains correlate with coordinated hormone-induced gene regulation. Genes Dev. 2014; 28:2151–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Panigrahi A.K., Foulds C.E., Lanz R.B., Hamilton R.A., Yi P., Lonard D.M., Tsai M.J., Tsai S.Y., O’Malley B.W. SRC-3 coactivator governs dynamic estrogen-induced chromatin looping interactions during transcription. Mol. Cell. 2018; 70:679–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Achinger-Kawecka J., Valdes-Mora F., Luu P.L., Giles K.A., Caldon C.E., Qu W.J., Nair S., Soto S., Locke W.J., Yeo-Teh N.S.et al.. Epigenetic reprogramming at estrogen-receptor binding sites alters 3D chromatin landscape in endocrine-resistant breast cancer. Nat. Commun. 2020; 11:320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rodriguez J., Ren G., Day C.R., Zhao K., Chow C.C., Larson D.R.. Intrinsic dynamics of a human gene reveal the basis of expression heterogeneity. Cell. 2019; 176:213–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sabari B.R., Dall’Agnese A., Boija A., Klein I.A., Coffey E.L., Shrinivas K., Abraham B.J., Hannett N.M., Zamudio A.V., Manteiga J.C.et al.. Coactivator condensation at super-enhancers links phase separation and gene control. Science. 2018; 361:eaar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Besset V., Scott R.P., Ibanez C.F.. Signaling complexes and protein–protein interactions involved in the activation of the ras and phosphatidylinositol 3-kinase pathways by the c-Ret receptor tyrosine kinase. J. Biol. Chem. 2000; 275:39159–39166. [DOI] [PubMed] [Google Scholar]
- 67. Hayashi H., Ichihara M., Iwashita T., Murakami H., Shimono Y., Kawai K., Kurokawa K., Murakumo Y., Imai T., Funahashi H.et al.. Characterization of intracellular signals via tyrosine 1062 in RET activated by glial cell line-derived neurotrophic factor. Oncogene. 2000; 19:4469–4475. [DOI] [PubMed] [Google Scholar]
- 68. Qian Y.Y., Chai S.J., Liang Z.Y., Wang Y.F., Zhou Y., Xu X., Zhang C.C., Zhang M., Si J.X., Huang F.T.et al.. KIF5B–RET fusion kinase promotes cell growth by multilevel activation of STAT3 in lung cancer. Mol. Cancer. 2014; 13:176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Asai N., Fukuda T., Wu Z., Enomoto A., Pachnis V., Takahashi M., Costantini F.. Targeted mutation of serine 697 in the ret tyrosine kinase causes migration defect of enteric neural crest cells. Development. 2006; 133:4507–4516. [DOI] [PubMed] [Google Scholar]
- 70. Fukuda T., Kiuchi K., Takahashi M.. Novel mechanism of regulation of rac activity and lamellipodia formation by RET tyrosine kinase. J. Biol. Chem. 2002; 277:19114–19121. [DOI] [PubMed] [Google Scholar]
- 71. Li X.Z., Yan J., Huang S.H., Zhao L., Wang J., Chen Z.Y.. Identification of a key motif that determines the differential surface levels of RET and TrkB tyrosine kinase receptors and controls depolarization enhanced RET surface insertion. J. Biol. Chem. 2012; 287:1932–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and/or its supplementary data, or can be made available upon reasonable request. ERα ChIP-seq in MCF7 was from GSE45822; ERα ChIP-seq in T47D was from GSE72249 and GSE80367; ERα ChIP-seq in ZR-75–1 was from GSE72249 and GSE141582; H3K27Ac and P300 ChIP-seq were from GSE62229; MED1 ChIP-seq was from GSE60272; H3K4me3, H3K9me3 and H3K27me3 ChIP-seq were from GSE23701. All other sequencing data were deposited in the Gene Expression Omnibus database under accession GSE186646.