Abstract
Inflammatory bowel diseases (IBDs) are characterized by inflammation in the gastrointestinal tract and include Ulcerative Colitis and Crohn’s Disease. These diseases are costly to health services, substantially reduce patients’ quality of life, and can lead to complications such as cancer and even death. Symptoms include abdominal pain, stool bleeding, diarrhea, and weight loss. The treatment of these diseases is symptomatic, seeking disease remission. The intestine is colonized by several microorganisms, such as fungi, viruses, and bacteria, which constitute the intestinal microbiota (IM). IM bacteria promotes dietary fibers fermentation and produces short-chain fatty acids (SCFAs) that exert several beneficial effects on intestinal health. SCFAs can bind to G protein-coupled receptors, such as GPR41 and GPR43, promoting improvements in the intestinal barrier, anti-inflammatory, and antioxidant effects. Thus, SCFAs could be a therapeutic tool for IBDs. However, the mechanisms involved in these beneficial effects of SCFAs remain poorly understood. Therefore, this paper aims to provide a review addressing the main aspects of IBDs, and a more detailed sight of SCFAs, focusing on the main effects on different aspects of the intestine with an emphasis on IBDs.
Keywords: Ulcerative colitis, Crohn’s disease, Short-chain fatty acid, Butyrate, Inflammatory bowel diseases, Free fatty acid receptor
Core Tip: This review addresses the main epidemiological, and pathophysiological aspects of inflammatory bowel diseases (IBDs), characterizes the intestinal microbiota, and describes, in more details, the production, metabolism, and main effects of short-chain fatty acids (SCFAs) on various aspects of intestinal health, elucidating potential therapeutic effects of SCFAs in IBDs. In addition, this review addresses aspects of the modulation of SCFA production and encourages further studies on the subject seeking clinical application.
INTRODUCTION
Inflammatory bowel diseases (IBDs) comprise ulcerative colitis (UC) and Crohn’s disease (CD) and are characterized by inflammation in the gastrointestinal tract (GIT)[1]. They are expensive diseases to health services, and their prevalence is increasing every year[2]. Abdominal pain, stool bleeding, diarrhea, and weight loss are some of the most frequent symptoms of IBDs[3].
The pathogenesis of IBDs remains unclear, but there is evidence of a relationship between genetic factors associated with the dysregulation of the immune system and intestinal microbiota (IM)[4]. The treatment of these diseases aims to relieve the initial symptoms and seek to keep the patient in remission. Anti-inflammatory drugs, immunosuppressants, analgesics, and monoclonal antibodies are used for this[5].
Under physiological conditions, the intestine presents a heterogeneous and symbiotic community of microorganisms such as fungi, viruses, and bacteria[6]. These microorganisms that colonize the intestine constitute the IM and provide beneficial effects to the host as protection against pathogens, strengthening of intestinal barrier integrity, and production of short-chain fatty acids (SCFAs)[7].
SCFAs are produced from dietary fibers fermentation by IM. Acetate, Propionate, and Butyrate are the most abundant SCFAs and exert several beneficial effects on the body[8]. Among the three most abundant types of SCFAs, Butyrate has been widely studied due to its anti-inflammatory and antioxidant effects[9]. SCFAs can bind to G-protein-coupled receptors such as GPR41 and GPR43 and exert several intracellular effects that benefit intestinal health[10-12]. In this context, SCFAs may represent an alternative to conventional therapy for IBDs. Therefore, this review aims to provide the main aspects of IBDs and a more detailed sight of SCFAs, elucidating their main effects, possible mechanisms involved, and possible applications in IBDs.
INFLAMMATORY BOWEL DISEASES
IBDs are characterized by inflammation in GIT and comprise CD and UC[13]. These diseases are a serious health problem because they substantially reduce patients’ quality of life, are expensive to health services, and can lead to complications such as cancer and even death[14].
Between 1990 and 2017, the overall number of people with IBDs increased from 3.7 million to more than 6.8 million and affected more women (57%) than men (43%)[2]. Countries with a high sociodemographic index had higher rates of age-standardized prevalence, with Western Europe and North America presenting the highest age-standardized mortality rates in 2017[2].
The diagnosis of IBDs requires endoscopic/colonoscopic, imaging, and laboratory exams associated with biopsy of the affected areas, and despite having similar symptoms, CD and UC have considerable pathophysiological differences[1]. CD can affect, usually discontinuously, any segment of the GIT. Microscopically, it is possible to observe transmural inflammation, penetrating ulcers, and the presence of granulomas[15,16]. UC, in turn, is characterized by a limited inflammation of the colon, usually continuous, which begins in the rectum and can extend to the colon at varying distances[3]. Microscopic findings of UC include reduction of goblet cells, superficial and mucosal-limited inflammation, and presence of ulcers and abscesses[16-18].
The pathogenesis of IBDs is still not fully understood[19-21], but several studies indicate that it tends to be caused by an association between genetic susceptibility and environmental alterations in the gut microbiota, along with an intestinal barrier dysfunction, causing a dysregulation of the immune system[22,23]. Genome-wide association studies identified 201 IBD-related loci, 41 of which were specific for CD and 30 for UC[23,24]. Mutations in the NOD2 gene, as well as mutations in the interleukin (IL) 10 receptor gene region and polymorphisms in the 16-like 1 gene are examples of genetic alterations related to the development of IBDs[25-28]. In addition, many of the IBD-related loci are also associated with autoimmune and immunodeficiency diseases, indicating the important relationship of IBDs to immune system disorders[23,29].
The host-microbiota interaction also plays an important role in the pathogenesis of CD and UC and involves gene regions that regulate microbial defense and intestinal inflammation[4,30-32]. Changes in the gut microbiota are frequent among individuals with IBDs[33,34]. However, it is not known whether the alteration of the microbiota is a cause or a consequence of these diseases[35,36]. Furthermore, eating habits are also related to IBDs[37]. A diet rich in processed foods, with high amounts of saturated fat and filled with protein is often associated with a higher chance of developing IBDs[38,39], while a high fiber diet reduces the chances of developing these disorders[40,41].
Treatment for IBDs seeks to control initial symptoms, normalizes evacuations frequency, promotes histological recovery of the mucosa, and prevents recurrences[5,42]. Therapeutic strategies include the use of anti-inflammatory drugs, analgesics, immunosuppressants, monoclonal antibodies, and, in more severe cases, surgeries[43-46]. In addition to conventional treatments for IBDs, several studies have investigated the modulation of the IM as one of the possible therapeutic/adjuvant approaches in the treatment of IBDs[47-49].
INTESTINAL MICROBIOTA
GIT harbors a complex and heterogeneous population of bacteria, fungi, viruses, and other microorganisms[6]. These microorganisms constitute the IM and establish a symbiotic relationship with the organism[50]. About 70% of all human microbiota is found in the intestine[51] and it is estimated that there are about 1011 to 1012 bacterial cells per gram of colonic content[52] and more than 1000 bacterial species in the human intestine[53].
In the colon, of all microorganisms that constitute the IM, anaerobic bacteria are seen in greater quantity, with a predominance of the phyla Firmicutes and Bacteroidetes, followed by Actinobacteria and Proteobacteria, although this composition may vary between individuals and among the different structures of the GIT[54,55]. The profile of these bacteria is influenced by diet and by the use of medications, particularly antibiotics[56-58].
IM can interact with the immune system, causing tolerance to symbiotic microorganisms and an effector response to pathogenic microorganisms[50,59]. In addition to being involved in this mechanism of communication with the immune system[60,61], the IM contributes to the protection against pathogens[62] through the production of bactericidal and/or bacteriostatic molecules, competition for nutrients, and stimulation of mucus secretion[63-65].
In addition, a healthy IM, under symbiosis conditions, promotes structural benefits to the host such as strengthening the integrity and modulation of the intestinal barrier[7] and metabolic benefits through the fermentation of complex carbohydrates, SCFAs production, energy uptake, increasing of intestinal hormones such as the Peptide YY (PYY), glucagon-like peptide 1 (GLP1) and vitamin synthesis[66].
Many of these beneficial effects of IM have been related to the production of SCFAs, which, in addition to being one of the main energy sources for colonocytes, can interfere in cellular mechanism, modifying the expression of some genes and promoting changes, including, to extra-intestinal levels such as in the Central Nervous System (CNS)[67-69]. In this context, SCFAs has been the target of studies by the scientific community due to its beneficial effects in GIT and to the possible therapeutic potential in some diseases, including IBDs[70,71].
When a variation in the composition and diversity of the microorganisms that make up the IM happens, it generates an imbalance between symbiotic and pathogenic microorganisms, promoting an instability in the IM-host relationship[20,33]. This harmful alteration of the IM is called dysbiosis[72]. Intestinal dysbiosis leads to a pro-inflammatory response with the activation of immune system cells and, in addition, causes a reduction in the synthesis of vitamins as well as in the carbohydrate metabolism, consequently, the beneficial effects of a healthy IM are suppressed, and individuals become more susceptible to the development of diseases[73,74]. Dysbiosis is associated with the pathogenesis of several diseases, including IBDs[34,75-77].
SHORT-CHAIN FATTY ACIDS
Production and metabolism
SCFAs are produced by IM bacteria through the fermentation of dietary fibers[67]. SCFAs are saturated carboxylic acids, with a chain of one to six carbons, and among these, Acetate, Propionate, and Butyrate are the most abundant SCFAs in the colon[78,79] in a molar ratio of 60:20:20, respectively, although this proportion is different along the large intestine[78,80]. The production of SCFAs is influenced by the composition of IM, the anatomical site of the GIT and by the diet, differing in the type and quantity produced according to different substrates[79,81].
The fermentation of dietary fibers and the production of SCFAs is promoted by specific enzymes of IM microorganisms[82]. Under certain physiological conditions, the main SCFAs, namely the acetate, propionate and butyrate are produced by intestinal bacteria such as, for example, the Bacteroides spp., the Bifidobacterium spp. and the Faecalibacterium prausnitzii[83-85]. Acetate is produced from pyruvate via acetyl-CoA[83]. Propionate is produced via the succinate, acrylate, and propanediol pathways[84,86]. In turn, butyrate is produced from two molecules of acetyl-CoA, which are transformed into butyrate via phosphotransbutyrylase and butyrate kinase[85]. The butyryl-CoA:acetate CoA-transferase also leads to the production of butyrate, and some bacteria use acetate as a substrate for butyrate production[82].
In a dysbiosis condition, the composition and diversity of SCFA-producing microorganisms are altered[33,34]. A particular reduction in butyrate-producing bacteria such as Faecalibacterium prausnitzii is seen in patients with IBDs[87], as well as reduced levels of propionate acetate and butyrate in the feces of these patients [88,89].
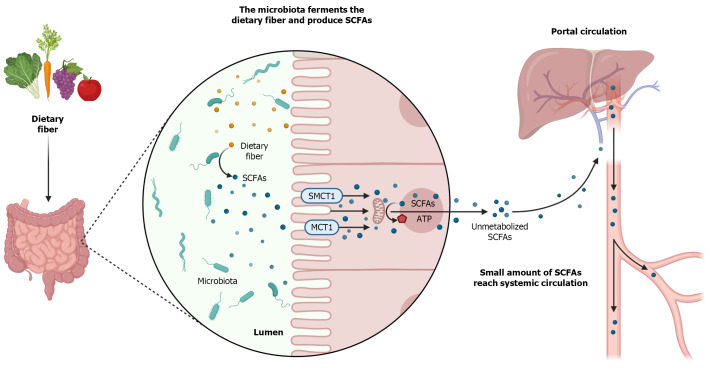
Once produced, most SCFAs are absorbed by the colonocytes. This absorption can occur by passive diffusion, through an exchange with bicarbonate, and mainly through monocarboxylate transporters (MCTs), by active transport (Figure 1)[67]. There are several types of MCTs, with different affinities for different types of SCFAs, which transport the SCFAs to the intracellular environment, including in the CNS, liver, kidneys, heart, lungs, skeletal muscle, and defense cells[90-94].
Figure 1.
Production and metabolism of shortchain fatty acids. Short-chain fatty acids (SCFAs) are produced by the intestinal microbiota from the fermentation of dietary fibers. SCFAs are absorbed by colonocytes mainly through monocarboxylate transporter 1 (MCT1), sodium-dependent monocarboxylate transporter 1 (SMCT1) and passive diffusion. At intracellular levels, SCFAs are metabolized and converted into ATP that will serve as energy source for these cells. Unmetabolized SCFAs cross the basolateral membrane and reach the portal circulation. In the liver, SCFAs are used in the synthesis of cholesterol and glucose. Only a small amount of SCFAs not used by the liver reach the systemic circulation[221]. The authors have obtained the permission for figure using from the BioRender.com (Supplementary material).
Among the MCTs, MCT1 stands out, due to its ubiquitous feature, and the sodium- dependent MCT1 (SMCT1), due to the greater amount in the large intestine. MCT1 has an affinity for Acetate, Propionate, and Butyrate is the main carrier responsible for the absorption of Butyrate, and transports SCFAs in a dependent way of H+ ions[67,92-94]. In turn, SMCT1 also has an affinity for Acetate, Propionate, and Butyrate, especially for Butyrate, and performs the transport in a sodium-dependent manner[95-97]. After being absorbed by the colonocytes at mitochondrial levels, SCFAs are converted into adenosine triphosphate (ATP), serving as an energy source for these cells (Figure 1)[69].
SCFAs that are not metabolized by the colonocytes, cross the basolateral membrane, and reach the portal circulation to the liver. In the liver, these SCFAs will serve as an energy source for hepatocytes and will participate in the synthesis of cholesterol and glucose[98,99]. A small amount of SCFAs, not metabolized by colonocytes and hepatocytes, actually reach the systemic circulation and have access to other organs (Figure 1)[67,100].
Shortchain fatty acid receptors
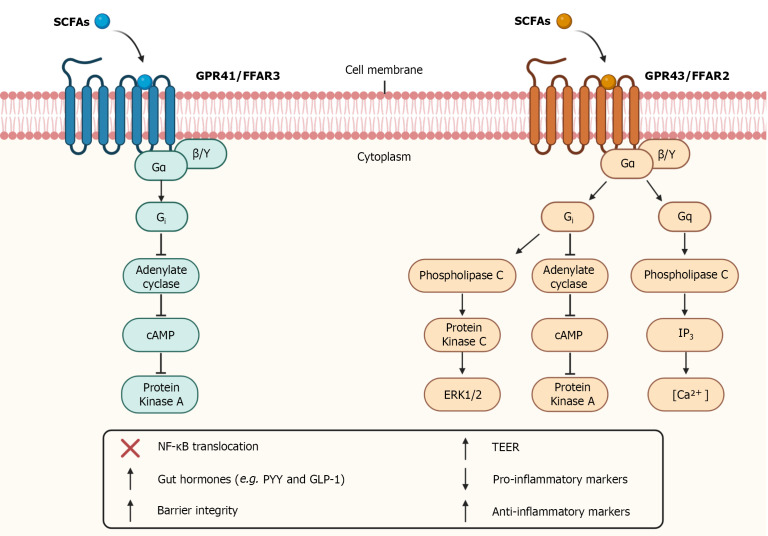
In addition to entering the intracellular environment, SCFAs can also bind to G-protein-coupled receptors. The main SCFAs receptors studied are GPR43 and GPR41, also known as free fatty acid receptor-2 (FFAR2) and free fatty acid receptor-3 (FFAR3), respectively[67,101]. They are transmembrane receptors, which recognize SCFAs, with different affinities for the different types of SCFAs, and promote signaling cascades mediated by G Protein (Figure 2)[12].
Figure 2.
Short-chain fatty receptors and intracellular signaling. Short-chain fatty acids (SCFAs) binds to G-protein coupled receptors such as GPR41 and GPR43 receptors also known as free fatty acid receptors (FFAR) 3 and FFAR2, respectively. The activation of GPR41 and GPR43 receptors through Gi subunit promotes inhibition of adenylate cyclase which inhibit cyclic adenosine monophosphate (cAMP) that inhibit protein kinase A. GPR43 receptor activation through Gq subunit promotes activation of phospholipase C which activate inositol 1,4,5-trisphosphate (IP3) leading to the elevation of intracellular calcium levels. GPR43 receptor activation through Gq subunit also promotes activation of phospholipase C which activate protein kinase C leading to activation of extracellular signal-regulated kinase 1/2 (ERK1/2) cascade. The activation of these receptors inhibits translocation of nuclear factor kappa B (NF- ΚB), altering the expression of certain proteins, promotes an increase in the release of gut hormones such as Peptide YY (PYY) and Glucagon-like peptide 1 (GLP-1), increase intestinal barrier integrity and Transepithelial Electrical Resistance (TEER), reduce pro-inflammatory markers levels, and increase anti-inflammatory markers levels. The authors have obtained the permission for figure using from the BioRender.com (Supplementary material).
Both the GPR41 receptor and the GPR43 receptor are coupled to the G-protein subunit Gi, however only the GPR43 receptor is coupled to the Gq subunit[102,103]. Activation of GPR41 and GPR43 receptors at Gi level promotes inhibition of cyclic adenosine monophosphate (cAMP) production and activation of extracellular signal-regulated kinases (ERK) cascade. In turn, GPR43 receptor activation through Gq promotes the elevation of calcium levels and activation of the mitogen-activated protein kinases (MAPKs) cascade[102-104]. The activation of these receptors is also capable of inhibiting the nuclear translocation of nuclear factor kappa B (NF-ΚB), altering the expression of certain proteins (Figure 2)[105].
GPR43 is expressed mainly in the colonic epithelium, enteroendocrine L-cells, and immune system cells[11,106]. In turn, GPR41 is expressed in the colon, kidneys, blood vessels, peripheral nervous system, and enteric nervous system (ENS)[107-109]. The activation of these receptors contributes to the maintenance of intestinal homeostasis and plays important roles in pathological conditions such as IBDs.
Effects of SCFAs on the ntestinal
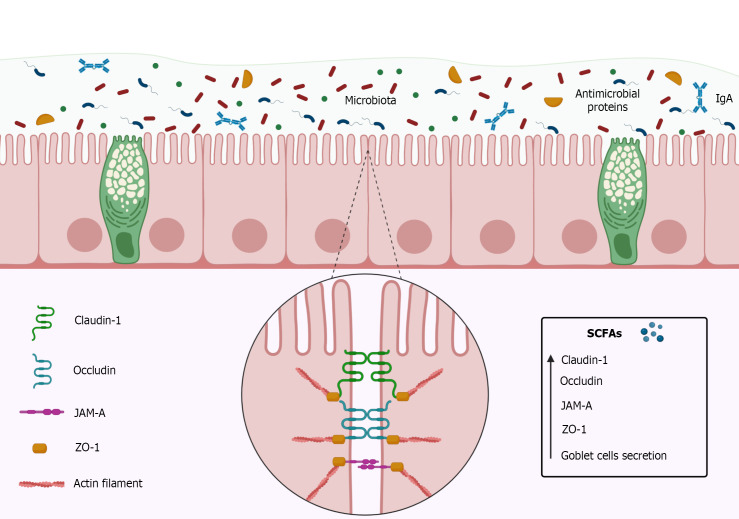
The intestinal barrier consists of simple cylindrical epithelial cells, mucus, immunoglobulins (Ig), and intercellular binding proteins such as tight junction proteins (Figure 3)[110,111]. The function of the intestinal barrier is to prevent the passage of pathogens, toxins, and other undesirable substances from the intestinal lumen into the paracellular space[110]. Tight junctions play an important role in the effectiveness of the intestinal barrier[112] by keeping epithelial cells well adhered to each other, and are composed of transmembrane proteins such as occludin, claudins, junctional adhesion molecules (JAM), and accessory cytoplasmic proteins such as zonulas occludens (ZOs)[113-115].
Figure 3.
Intestinal barrier constitution and short-chain fatty acids effects. The intestinal barrier is composed by simple cylindrical epithelial cells, mucus, microbiota, immunoglobulins A (IgA), antimicrobial proteins, and intercellular binding proteins such as tight junction proteins. Tight junctions play an important role in the integrity of the intestinal barrier by keeping epithelial cells well adhered to each other, and are composed of transmembrane proteins such as Claudins, Occludin, Junctional adhesion molecules (JAM), and accessory cytoplasmic proteins such as Zonulas Occludens (ZOs). Claudin-1 pairs with other Claudin-1 Loops of the adjacent cell, decreasing paracellular permeability. Like Claudin-1, Occludin pairs with other Occludin loops of the adjacent cell forming a barrier mainly to macromolecules. JAM-A belongs to immunoglobulins superfamily and can form homophilic interactions adjacent to tight junctions and interacting with integrins or other members of the JAM family. ZO-1 are located on the cytoplasm and perform the anchorage of proteins joining them to the cytoskeleton through actin filaments. Short-chain fatty acids (SCFAs), mainly Butyrate, are capable of increase Claudin-1, Occludin, JAM, ZO proteins and increase mucus secretion by goblet cells. The authors have obtained the permission for figure using from the BioRender.com (Supplementary material).
The occludins form a barrier mainly to macromolecules[116,117], while the claudins, particularly claudin-1, can maintain the intestinal barrier functionality even in the absence of other tight junctions[113,118-120]. The JAM, a protein belonging to the Immunoglobulins (Ig) superfamily[121-123], is capable of forming homophilic interactions adjacent to tight junctions as well as interacting with integrins or other members of the JAM family[122,123], playing an important role in the constitution of the intestinal barrier[115,124,125]. The ZOs proteins, such as ZO-1, ZO-2, and ZO-3 are located in the cytoplasm[126,127] acting as an anchor for proteins by joining them to the cytoskeleton and, therefore, are key to maintaining the integrity of the intestinal barrier, mainly due to its relationship with the claudins and the occludins[114,128].
A deregulated intestinal barrier is associated with IBDs. It is not known for sure if the corrupted intestinal barrier is the cause or consequence of these diseases, but there are reports that an increase in intestinal permeability is an antecedent to recurrence of CD[129,130], which provides evidence that the functionality of the intestinal barrier is linked to the development/manifestation of IBDs. Architectural and functional disorganizations of the intestinal barrier, mucus reduction, and changes in the expression of tight junctions are associated with IBDs[131-133]. In addition, the loss of intestinal barrier integrity exposes the mucosa to luminal microbial antigens, which trigger inflammatory responses, sometimes hyper-responsive, causing extra-intestinal manifestations[132,134].
In this context, Butyrate, more than others SCFAs, has shown an interesting increase in intestinal barrier function[135] and could represent a possible ally in the treatment of IBDs, regarding the integrity of the intestinal barrier. In vitro and in vivo studies using Butyrate as intervention reported increased Transepithelial Electrical Resistance, rapid structuring of the intestinal barrier, and increase of tight junctions proteins (Figure 3)[136,137].
Although most of the beneficial effects of SCFAs at the intestinal barrier are attributed to the butyrate, some studies have shown recently that the propionate is also able to improve the functionality of such barrier[138]. In an experimental model of UC, propionate was able to attenuate the decrease in the expression of tight junctions such as ZO-1 and occludin in the large intestine[139]. These SCFAs were also able to attenuate the intestinal barrier dysfunction induced in Caco-2 Cell monolayers, however, the amount of propionate needed in order to observe its effects on the intestinal barrier is greater than that of butyrate[140]. In addition, propionate also appears to influence the differentiation and proliferation of the intestinal epithelium[141].
The molecular mechanisms behind these effects are unclear, but there are reports that Butyrate is capable of promoting positive regulation in the expression of tight junctions proteins by activating AMP-activated protein kinase (AMPK), mediated by activation of receptors such as GPR41 and GPR43, reduction of the phosphorylation of the myosin light chain 2 (MLC2), increased phosphorylation of the protein kinase C β2 (PKCβ2) or by negative regulation of channel formers proteins, such as claudin-2[137,142,143].
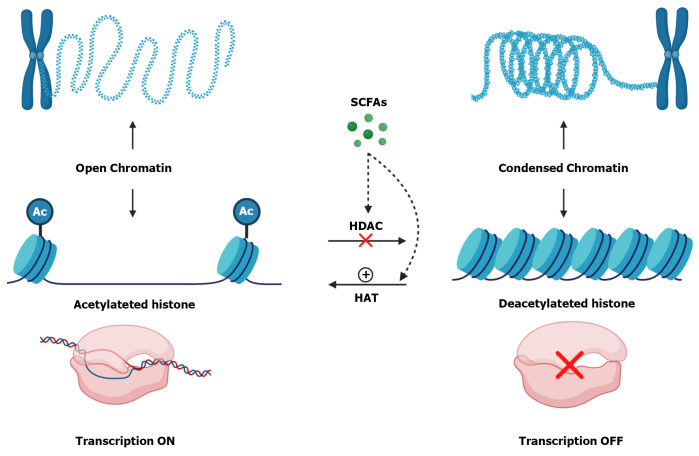
Another possible mechanism involved in the regulation of junction proteins by Butyrate may be associated with its ability to inhibit histone deacetylases[144,145]. When histones are deacetylated, that is, without acetyl groups, their conformation is more tangled which makes chromatin more condensed and promotes gene silencing[146,147]. On the other hand, when these histones are acetylated, there is a repulsion between these groups, so histone becomes more distant from each other, causing chromatin to become decondensed and, consequently, activate gene transcription (Figure 4)[148,149].
Figure 4.
Histone modifications and short-chain fatty acids influence. Histones are proteins that interacts with DNA and play an important role in organizing the double strand. When histones contain acetyl (Ac) groups they become acetylated and there is a repulsion between these groups, so histone becomes more distant from each other, causing chromatin to become decondensed and, consequently, more accessible which activate transcription. Otherwise, when histones are deacetylated, without Ac groups, DNA becomes more tangled which makes chromatin more condensed and promotes gene silencing. Histone Deacetylases (HDAC) are enzymes that remove Ac groups from Histones making them deacetylated. Otherwise, Histone Acetylases (HAT) are enzymes that insert Ac groups in Histones making them acetylated. Short-chain fatty acids (SCFAs), mainly Butyrate, inhibit HDAC and increase HAT activity. The authors have obtained the permission for figure using from the BioRender.com (Supplementary material).
The removal of acetyl groups is done by the histone deacetylases enzymes[146,150]. Inhibition of these enzymes can occur directly, through intracellular SCFAs, which entered through MCTs or SMCTs, or indirectly, through the activation of SCFAs receptors[52]. On the other hand, Butyrate also increases the activity of histone acetylase enzymes, which promote the acetylation of histone[60].
Another effect of SCFAs, mainly Butyrate and Acetate, on the intestinal barrier is related to increased mucus production, which acts as a lubricant and physical barrier against microorganisms, toxins, and acidity resulting from the digestion process[111,151]. Studies with mice have reported that Butyrate is capable of stimulating the production of mucin and affected the expression of the mucus-producing gene, MUC-2[152,153]. In this sense, molecules, such as Butyrate, that can strengthen the intestinal barrier or recover its functionality in IBDs seem to be an interesting therapeutic strategy to avoid the development/recurrence of these diseases.
Effects of SCFAs on Intestinal Inflammation
SCFAs, especially Butyrate, also play anti-inflammatory roles[154,155]. Studies have reported that Butyrate is capable of promoting a reduction in pro-inflammatory cytokines such as interleukin (IL)-8, IL-6, IL-12 and tumor necrosis factor (TNF)-α and an increase in the production of anti-inflammatory cytokines such as IL-10[60] and IL-18, which is important for the maintenance of intestinal barrier epithelium[156]. It is believed that the anti-inflammatory effects of Butyrate are related due to its ability to inhibit the signaling pathway of NF-κβ and Histone Deacetylases[157-159], reducing macrophage activation, oxidative stress, recruitment of inflammatory cells, and consequently intestinal inflammation[145,160,161].
In addition, SCFAs also influences the differentiation, maturation, and activation of immune system cells such as macrophages, dendritic cells, and lymphocytes[60,162], and this influence can be mediated by activation of GPR43 receptors that are present in immune system cells[67,104,106]. In synergy with its anti-inflammatory effects, Butyrate also has antioxidant effects indicated by the increase of antioxidant enzymes such as catalase and superoxide dismutase-2 and by the reduction of uric acid, glutathione, and IL-β levels[163,164].
In animal models, Butyrate, by inhibiting Histone Deacetylase-1, was able to maintain a balance between Th17 lymphocytes (auxiliary T lymphocytes) and Treg lymphocytes (regulatory T lymphocytes) and to exert anti-inflammatory effects by inhibiting IL-6, signal transducer, and activator of transcription 3 (STAT-3) and IL-17[165]. In addition, in a double-blind, placebo-controlled study with patients with IBDs, Butyrate considerably reduced fecal calprotectin levels in patients with UC[166].
In experimental Colitis induced by 2,4,6-trinitrobenzene sulfonic acid (TNBS), treatment with Sodium Butyrate was able to improve intestinal inflammation, reduce histological injury scores, promote significant recovery of animal weight, and increase IL-10 levels[167]. In dextran sulfate sodium (DSS)-induced colitis, pretreatment with Sodium Butyrate was able to prevent weight loss of animals, recover colon shortening caused by colitis, reduce intestinal mucosa lesions, attenuate the production of pro-inflammatory cytokines such as IL-6 and TNF-α and increase IL-10 levels[160].
Still in a DSS-induced colitis model, the propionate was able to inhibit the expression of pro-inflammatory markers such as IL-6, IL-1β and TNF-α in the colon[139]. Moreover, the propionate is also capable of modulating the activation of immune system cells and reducing the levels of reactive oxygen species in the tissues[168,169].
Although many anti-inflammatory and antioxidant effects of SCFAs, most notably Butyrate, have been described in the literature, the mechanisms involved in these effects remain unclear. However, these properties have attracted the attention of the scientific community, as these metabolites could be configured as therapeutic tools in the prevention of recurrences and/or as adjuvants in the treatment of active IBDs.
Effects of SCFAs on the Enteric Nervous System
The GIT has an extensive intrinsic nervous system called the ENS, which consists of an interconnected network of neurons, axons, and enteric glial cells[170]. The ENS can even isolated from the CNS, control several functions[171] such as intestinal motility, regulation of blood flow, regulation of fluids through the mucosa, secretion of digestive substances and intestinal hormones, and communication with the immune system[170,172].
The ENS has two plexus: the myenteric plexus (Auerbach’s plexus) and the submucosal plexus (Meissner plexus)[170,173]. The myenteric plexus is located, throughout the digestive system, between the outer longitudinal muscle layer and the circular muscle layer and is mainly responsible for intestinal motility[170,173]. The submucosal plexus is located between the circular muscle layer and the muscularis mucosae layer of the small and large intestines and its function is related to the control of intestinal secretions and blood flow of the mucosa[170].
In IBDs, even in mild cases, promotes changes in neurons and nerve endings of the ENS, as well as in the patterns of motility and secretion of the intestine[172]. These changes may persist even after the resolution of inflammation in the intestine, as it generates prolonged hyperexcitability of enteric neurons, persisting symptoms such as cramps, abdominal pain, and diarrhea[174].
Studies with experimental models have shown that intestinal inflammation causes changes in the number and size of enteric ganglia, glia, and neurons in addition to degeneration, necrosis, and neuronal apoptosis[174-177] In addition to morphological changes, colitis also promotes changes in the expression of neurotransmitters and their receptors, consequently altering the chemical code of neurons[178,179].
It is known that enteric neurons have GPR41 receptors and are responsive to SCFAs[180]. Studies have reported that Butyrate also has effects on ENS[181] and is capable of causing increased excitatory motor neurons, increased intestinal motility and contractile response induced by electrical stimulus[182]. However, the way the interaction of SCFAs with ENS occurs in the face of intestinal inflammation and how it interferes in the different neuronal classes, enteric glial cells, in the expression of SCFAs receptors, and the neurochemical dynamics is not yet clarified[183].
The results so far available in the literature encourage further investigations on the effects of SCFAs on enteric neurons, once ENS exerts a remarkable influence on homeostasis and intestinal functionality and the beneficial effects of SCFAs could recover/prevent changes in ENS due to IBDs and other intestinal diseases.
MODULATION OF SCFAs PRODUCTION AND FUTURE PROSPECTS
The production of SCFAs depends mainly on two major variables: the characteristic of IM and the supply of substrates used by microorganisms. For SCFAs production, there must be a healthy IM rich in fermenting microorganisms[184]. The logic behind the modulation of SCFAs production involves two pathways: increase of living beneficial microorganisms (probiotics) and increase of substrates used by microorganisms (prebiotics), giving the host health benefits[185,186].
In this context, the production of SCFAS, such as Butyrate, for example, can be increased through the use of probiotics with butyrogenic microorganisms, that is, capable of producing Butyrate or by the use of prebiotics, such as fibers and complex starches, which acts as energetic substrates for microorganisms, increasing the proliferation of IM microorganisms and the production of SCFAs[82,187,188].
In other words, a person’s diet exerts a strong influence on the composition of their IM and, consequently, on the modulation of the SCFAs production[187,189]. A plant-based diet, such as the Mediterranean diet, tends to increase the source of dietary fiber and, consequently, the production of SCFAs[190-192]. On the other hand, a diet low in vegetables and rich in meats, sugars or saturated fats, such as the Western diet, not only alters the IM profile, but reduces the availability of fiber and, consequently, the production of SCFAs[193,194]. In this sense, the association between the use of probiotics, prebiotics and a Mediterranean-like diet optimizes the production of SCFAs in the intestine[184,190,195,196].
Many animal models of IBDs seek to identify a possible effectiveness of the use of probiotics as a treatment for this disease[197]. In the TNBS-induced UC models, the microorganisms Bifidobacterium infantis and Bifidobacterium bifidum demonstrated beneficial effects in animals by attenuating the clinical manifestations of UC, promoting a greater preservation of the mucus layer and a reduction of pro-inflammatory cytokines such as the IL-10 and the IL-1β in the intestine[198,199]. In the DSS-induced UC model, the microorganisms Bifidobacterium longum subsp. infantis BB-02 and the Bifidobacterium animalis subsp. lactis BB12 also demonstrated to have a positive influence on the clinical, histological and inflammatory manifestations of this disorder[200,201]. Ultimately, a reduction of potentially pathogenic microorganisms was observed after the administration of Bifidobacterium lactis[202]. The improvement of the results observed with the administration of these probiotics may be related to an increase in the production of the SCFAs[203].
As for the clinical studies, the effectiveness of using probiotics in patients with IBDs is controversial[204]. The Bifidobacterium bifidum, Lactobacillus acidophilus, Escherichia coli Nissle1917 and Bifidobacterium breve appear to exert favorable effects on UC remission [205-207]. In addition, the use of the referred probiotics caused an increase of butyrate and propionate levels in the subject’s fecal matter[207]. The combination of some microorganisms has also been used in clinical studies. This is the case of VSL#3, consisting essentially of 8 strains of microorganisms: Lactobacillus paracasei, Lactobacillus plantarum, Lactobacillus acidophilus, Lactobacillus delbrueckii subsp. bulgaricus, Bifidobacterium longum, Bifidobacterium breve, Bifidobacterium infantis and Streptococcus thermophilus[208]. Patients with UC who used VSL#3 showed a clinical improvement, with a reduction in fecal bleeding as well as a decrease in the frequency of stool evacuation[209,210]. These findings do not appear to be applicable to CD[211]. It is important to mention that VSL#3 microorganisms are producers of SCFAs and this may be related to the improvements observed with the use of this probiotic for UC[203]. Pathophysiological differences between CD and UC, the great abundance of probiotics studied and the difficulty in obtaining a standardized sample population are some of the limitations of studies involving the use of probiotics and prebiotics in IBDs[48].
Another approach studied for the modulation of SCFAs production and for the treatment of IBDs is the Fecal Microbiota Transplantation (FMT)[212]. FMT is able to change the composition of the recipient’s IM, in order to reduce the proliferation of potentially pathogenic microorganisms and increase the production of SCFAs, especially butyrate[213-215]. Furthermore, fecal levels of Eubacterium and Lactobacillus spp. increased after the FMT, which are producers of Butyrate[213,216,217]. Although FMT is classically studied for the treatment of Clostridium difficile infection, several studies have observed that FMT has proven to be an effective, safe, and promising alternative to the treatment of IBDs, including UC and CD[212,218-220].
CONCLUSION
This review approached the main points about SCFAs, its main characteristics and contributions for gut health and potential benefits related to IBDs. In this context, this review encourages further studies on SCFAs, reinforces the importance of IM and healthy eating habits for the maintenance of intestinal health.
ACKNOWLEDGEMENTS
The authors would like to thank the Research Support Foundation of the State of São Paulo (FAPESP, Brazil) and Higher Education Personnel Improvement Coordination (CAPES, Brazil) for financial support.
Footnotes
Conflict-of-interest statement: Authors declare no conflict of interests for this article.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Corresponding Author’s Membership in Professional Societies: American Gastroenterological Association.
Peer-review started: June 19, 2022
First decision: July 11, 2022
Article in press: August 25, 2022
Specialty type: Anatomy and morphology
Country/Territory of origin: Brazil
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): E
P-Reviewer: Nikolić M, Croatia; Sitkin S, Russia; Zhao G, China S-Editor: Chen YL L-Editor: A P-Editor: Chen YL
Contributor Information
Marcos Antônio Ferreira Caetano, Department of Anatomy, Institute of Biomedical Sciences, University of São Paulo, São Paulo 05508900, SP, Brazil.
Patricia Castelucci, Department of Anatomy, Institute of Biomedical Sciences, University of São Paulo, São Paulo 05508900, SP, Brazil. pcastel@usp.br.
References
- 1.Kaistha A, Levine J. Inflammatory bowel disease: the classic gastrointestinal autoimmune disease. Curr Probl Pediatr Adolesc Health Care. 2014;44:328–334. doi: 10.1016/j.cppeds.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 2.GBD 2017 Inflammatory Bowel Disease Collaborators. The global, regional, and national burden of inflammatory bowel disease in 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol Hepatol. 2020;5:17–30. doi: 10.1016/S2468-1253(19)30333-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fumery M, Singh S, Dulai PS, Gower-Rousseau C, Peyrin-Biroulet L, Sandborn WJ. Natural History of Adult Ulcerative Colitis in Population-based Cohorts: A Systematic Review. Clin Gastroenterol Hepatol. 2018;16:343–356.e3. doi: 10.1016/j.cgh.2017.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu S, Kurilshikov A, Zhernakova A, Weersma R. IBD Genetics and the Gut Microbiome. In: Molecular Genetics of Inflammatory Bowel Disease. Springer International Publishing, 2019: 231-248. [Google Scholar]
- 5.Taylor K, Gibson PR. Conventional therapy of ulcerative colitis: Corticosteroids. In: Crohn’s Disease and Ulcerative Colitis: From Epidemiology and Immunobiology to a Rational Diagnostic and Therapeutic Approach: Second Edition. Springer International Publishing, 2017: 399-412. [Google Scholar]
- 6.Milani C, Duranti S, Bottacini F, Casey E, Turroni F, Mahony J, Belzer C, Delgado Palacio S, Arboleya Montes S, Mancabelli L, Lugli GA, Rodriguez JM, Bode L, de Vos W, Gueimonde M, Margolles A, van Sinderen D, Ventura M. The First Microbial Colonizers of the Human Gut: Composition, Activities, and Health Implications of the Infant Gut Microbiota. Microbiol Mol Biol Rev. 2017;81 doi: 10.1128/MMBR.00036-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reunanen J, Kainulainen V, Huuskonen L, Ottman N, Belzer C, Huhtinen H, de Vos WM, Satokari R. Akkermansia muciniphila Adheres to Enterocytes and Strengthens the Integrity of the Epithelial Cell Layer. Appl Environ Microbiol. 2015;81:3655–3662. doi: 10.1128/AEM.04050-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartoszek A, Moo EV, Binienda A, Fabisiak A, Krajewska JB, Mosińska P, Niewinna K, Tarasiuk A, Martemyanov K, Salaga M, Fichna J. Free Fatty Acid Receptors as new potential therapeutic target in inflammatory bowel diseases. Pharmacol Res. 2020;152:104604. doi: 10.1016/j.phrs.2019.104604. [DOI] [PubMed] [Google Scholar]
- 9.Chen G, Ran X, Li B, Li Y, He D, Huang B, Fu S, Liu J, Wang W. Sodium Butyrate Inhibits Inflammation and Maintains Epithelium Barrier Integrity in a TNBS-induced Inflammatory Bowel Disease Mice Model. EBioMedicine. 2018;30:317–325. doi: 10.1016/j.ebiom.2018.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stilling RM, van de Wouw M, Clarke G, Stanton C, Dinan TG, Cryan JF. The neuropharmacology of butyrate: The bread and butter of the microbiota-gut-brain axis? Neurochem Int. 2016;99:110–132. doi: 10.1016/j.neuint.2016.06.011. [DOI] [PubMed] [Google Scholar]
- 11.Karaki S, Mitsui R, Hayashi H, Kato I, Sugiya H, Iwanaga T, Furness JB, Kuwahara A. Short-chain fatty acid receptor, GPR43, is expressed by enteroendocrine cells and mucosal mast cells in rat intestine. Cell Tissue Res. 2006;324:353–360. doi: 10.1007/s00441-005-0140-x. [DOI] [PubMed] [Google Scholar]
- 12.Kimura I, Ichimura A, Ohue-Kitano R, Igarashi M. Free Fatty Acid Receptors in Health and Disease. Physiol Rev. 2020;100:171–210. doi: 10.1152/physrev.00041.2018. [DOI] [PubMed] [Google Scholar]
- 13.Sairenji T, Collins KL, Evans DV. An Update on Inflammatory Bowel Disease. Prim Care. 2017;44:673–692. doi: 10.1016/j.pop.2017.07.010. [DOI] [PubMed] [Google Scholar]
- 14.Malik TA. Inflammatory Bowel Disease: Historical Perspective, Epidemiology, and Risk Factors. Surg Clin North Am. 2015;95:1105–1122, v. doi: 10.1016/j.suc.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 15.Wilkins T, Jarvis K, Patel J. Diagnosis and management of Crohn's disease. Am Fam Physician. 2011;84:1365–1375. [PubMed] [Google Scholar]
- 16.Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 17.Sartor RB. Mechanisms of disease: pathogenesis of Crohn's disease and ulcerative colitis. Nat Clin Pract Gastroenterol Hepatol. 2006;3:390–407. doi: 10.1038/ncpgasthep0528. [DOI] [PubMed] [Google Scholar]
- 18.Graham DB, Xavier RJ. Pathway paradigms revealed from the genetics of inflammatory bowel disease. Nature. 2020;578:527–539. doi: 10.1038/s41586-020-2025-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abraham C, Cho JH. Inflammatory bowel disease. N Engl J Med. 2009;361:2066–2078. doi: 10.1056/NEJMra0804647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manichanh C, Borruel N, Casellas F, Guarner F. The gut microbiota in IBD. Nat Rev Gastroenterol Hepatol. 2012;9:599–608. doi: 10.1038/nrgastro.2012.152. [DOI] [PubMed] [Google Scholar]
- 21.Stange EF, Schroeder BO. Microbiota and mucosal defense in IBD: an update. Expert Rev Gastroenterol Hepatol. 2019;13:963–976. doi: 10.1080/17474124.2019.1671822. [DOI] [PubMed] [Google Scholar]
- 22.Shouval DS, Rufo PA. The Role of Environmental Factors in the Pathogenesis of Inflammatory Bowel Diseases: A Review. JAMA Pediatr. 2017;171:999–1005. doi: 10.1001/jamapediatrics.2017.2571. [DOI] [PubMed] [Google Scholar]
- 23.Ramos GP, Papadakis KA. Mechanisms of Disease: Inflammatory Bowel Diseases. Mayo Clin Proc. 2019;94:155–165. doi: 10.1016/j.mayocp.2018.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGovern DP, Kugathasan S, Cho JH. Genetics of Inflammatory Bowel Diseases. Gastroenterology. 2015;149:1163–1176.e2. doi: 10.1053/j.gastro.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glocker EO, Kotlarz D, Boztug K, Gertz EM, Schäffer AA, Noyan F, Perro M, Diestelhorst J, Allroth A, Murugan D, Hätscher N, Pfeifer D, Sykora KW, Sauer M, Kreipe H, Lacher M, Nustede R, Woellner C, Baumann U, Salzer U, Koletzko S, Shah N, Segal AW, Sauerbrey A, Buderus S, Snapper SB, Grimbacher B, Klein C. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N Engl J Med. 2009;361:2033–2045. doi: 10.1056/NEJMoa0907206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cooney R, Baker J, Brain O, Danis B, Pichulik T, Allan P, Ferguson DJ, Campbell BJ, Jewell D, Simmons A. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med. 2010;16:90–97. doi: 10.1038/nm.2069. [DOI] [PubMed] [Google Scholar]
- 27.Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, Magalhães JG, Yuan L, Soares F, Chea E, Le Bourhis L, Boneca IG, Allaoui A, Jones NL, Nuñez G, Girardin SE, Philpott DJ. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- 28.Kuballa P, Huett A, Rioux JD, Daly MJ, Xavier RJ. Impaired autophagy of an intracellular pathogen induced by a Crohn's disease associated ATG16L1 variant. PLoS One. 2008;3:e3391. doi: 10.1371/journal.pone.0003391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, Essers J, Mitrovic M, Ning K, Cleynen I, Theatre E, Spain SL, Raychaudhuri S, Goyette P, Wei Z, Abraham C, Achkar JP, Ahmad T, Amininejad L, Ananthakrishnan AN, Andersen V, Andrews JM, Baidoo L, Balschun T, Bampton PA, Bitton A, Boucher G, Brand S, Büning C, Cohain A, Cichon S, D'Amato M, De Jong D, Devaney KL, Dubinsky M, Edwards C, Ellinghaus D, Ferguson LR, Franchimont D, Fransen K, Gearry R, Georges M, Gieger C, Glas J, Haritunians T, Hart A, Hawkey C, Hedl M, Hu X, Karlsen TH, Kupcinskas L, Kugathasan S, Latiano A, Laukens D, Lawrance IC, Lees CW, Louis E, Mahy G, Mansfield J, Morgan AR, Mowat C, Newman W, Palmieri O, Ponsioen CY, Potocnik U, Prescott NJ, Regueiro M, Rotter JI, Russell RK, Sanderson JD, Sans M, Satsangi J, Schreiber S, Simms LA, Sventoraityte J, Targan SR, Taylor KD, Tremelling M, Verspaget HW, De Vos M, Wijmenga C, Wilson DC, Winkelmann J, Xavier RJ, Zeissig S, Zhang B, Zhang CK, Zhao H International IBD Genetics Consortium (IIBDGC), Silverberg MS, Annese V, Hakonarson H, Brant SR, Radford-Smith G, Mathew CG, Rioux JD, Schadt EE, Daly MJ, Franke A, Parkes M, Vermeire S, Barrett JC, Cho JH. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mangan PR, Harrington LE, O'Quinn DB, Helms WS, Bullard DC, Elson CO, Hatton RD, Wahl SM, Schoeb TR, Weaver CT. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 2006;441:231–234. doi: 10.1038/nature04754. [DOI] [PubMed] [Google Scholar]
- 31.Eckburg PB, Relman DA. The role of microbes in Crohn's disease. Clin Infect Dis. 2007;44:256–262. doi: 10.1086/510385. [DOI] [PubMed] [Google Scholar]
- 32.Lobionda S, Sittipo P, Kwon HY, Lee YK. The Role of Gut Microbiota in Intestinal Inflammation with Respect to Diet and Extrinsic Stressors. Microorganisms. 2019;7 doi: 10.3390/microorganisms7080271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Santana PT, Rosas SLB, Ribeiro BE, Marinho Y, de Souza HSP. Dysbiosis in Inflammatory Bowel Disease: Pathogenic Role and Potential Therapeutic Targets. Int J Mol Sci. 2022;23 doi: 10.3390/ijms23073464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pisani A, Rausch P, Bang C, Ellul S, Tabone T, Marantidis Cordina C, Zahra G, Franke A, Ellul P. Dysbiosis in the Gut Microbiota in Patients with Inflammatory Bowel Disease during Remission. Microbiol Spectr. 2022;10:e0061622. doi: 10.1128/spectrum.00616-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sartor RB, Wu GD. Roles for Intestinal Bacteria, Viruses, and Fungi in Pathogenesis of Inflammatory Bowel Diseases and Therapeutic Approaches. Gastroenterology. 2017;152:327–339.e4. doi: 10.1053/j.gastro.2016.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shan Y, Lee M, Chang EB. The Gut Microbiome and Inflammatory Bowel Diseases. Annu Rev Med. 2022;73:455–468. doi: 10.1146/annurev-med-042320-021020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Serrano-Moreno C, Brox-Torrecilla N, Arhip L, Romero I, Morales Á, Carrascal ML, Cuerda C, Motilla M, Camblor M, Velasco C, Bretón I. Diets for inflammatory bowel disease: What do we know so far? Eur J Clin Nutr. 2022 doi: 10.1038/s41430-021-01051-9. [DOI] [PubMed] [Google Scholar]
- 38.Roncoroni L, Gori R, Elli L, Tontini GE, Doneda L, Norsa L, Cuomo M, Lombardo V, Scricciolo A, Caprioli F, Costantino A, Scaramella L, Vecchi M. Nutrition in Patients with Inflammatory Bowel Diseases: A Narrative Review. Nutrients. 2022;14 doi: 10.3390/nu14040751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crooks B, McLaughlin J, Matsuoka K, Kobayashi T, Yamazaki H, Limdi JK. The dietary practices and beliefs of people living with inactive ulcerative colitis. Eur J Gastroenterol Hepatol. 2021;33:372–379. doi: 10.1097/MEG.0000000000001911. [DOI] [PubMed] [Google Scholar]
- 40.Yusuf K, Saha S, Umar S. Health Benefits of Dietary Fiber for the Management of Inflammatory Bowel Disease. Biomedicines. 2022;10 doi: 10.3390/biomedicines10061242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ananthakrishnan AN, Khalili H, Konijeti GG, Higuchi LM, de Silva P, Korzenik JR, Fuchs CS, Willett WC, Richter JM, Chan AT. A prospective study of long-term intake of dietary fiber and risk of Crohn's disease and ulcerative colitis. Gastroenterology. 2013;145:970–977. doi: 10.1053/j.gastro.2013.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Colombel JF, Rutgeerts P, Reinisch W, Esser D, Wang Y, Lang Y, Marano CW, Strauss R, Oddens BJ, Feagan BG, Hanauer SB, Lichtenstein GR, Present D, Sands BE, Sandborn WJ. Early mucosal healing with infliximab is associated with improved long-term clinical outcomes in ulcerative colitis. Gastroenterology. 2011;141:1194–1201. doi: 10.1053/j.gastro.2011.06.054. [DOI] [PubMed] [Google Scholar]
- 43.Ordás I, Feagan BG, Sandborn WJ. Early use of immunosuppressives or TNF antagonists for the treatment of Crohn's disease: time for a change. Gut. 2011;60:1754–1763. doi: 10.1136/gutjnl-2011-300934. [DOI] [PubMed] [Google Scholar]
- 44.Hazel K, O'Connor A. Emerging treatments for inflammatory bowel disease. Ther Adv Chronic Dis. 2020;11:2040622319899297. doi: 10.1177/2040622319899297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shen B, Kochhar G, Navaneethan U, Farraye FA, Schwartz DA, Iacucci M, Bernstein CN, Dryden G, Cross R, Bruining DH, Kobayashi T, Lukas M, Shergill A, Bortlik M, Lan N, Tang SJ, Kotze PG, Kiran RP, Dulai PS, El-Hachem S, Coelho-Prabhu N, Thakkar S, Mao R, Chen G, Zhang S, Suárez BG, Lama YG, Silverberg MS, Sandborn WJ. Practical guidelines on endoscopic treatment for Crohn's disease strictures: a consensus statement from the Global Interventional Inflammatory Bowel Disease Group. Lancet Gastroenterol Hepatol. 2020;5:393–405. doi: 10.1016/S2468-1253(19)30366-8. [DOI] [PubMed] [Google Scholar]
- 46.Magro F, Cordeiro G, Dias AM, Estevinho MM. Inflammatory Bowel Disease - Non-biological treatment. Pharmacol Res. 2020;160:105075. doi: 10.1016/j.phrs.2020.105075. [DOI] [PubMed] [Google Scholar]
- 47.Tan P, Li X, Shen J, Feng Q. Fecal Microbiota Transplantation for the Treatment of Inflammatory Bowel Disease: An Update. Front Pharmacol. 2020;11:574533. doi: 10.3389/fphar.2020.574533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Akutko K, Stawarski A. Probiotics, Prebiotics and Synbiotics in Inflammatory Bowel Diseases. J Clin Med. 2021;10 doi: 10.3390/jcm10112466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang XF, Guan XX, Tang YJ, Sun JF, Wang XK, Wang WD, Fan JM. Clinical effects and gut microbiota changes of using probiotics, prebiotics or synbiotics in inflammatory bowel disease: a systematic review and meta-analysis. Eur J Nutr. 2021;60:2855–2875. doi: 10.1007/s00394-021-02503-5. [DOI] [PubMed] [Google Scholar]
- 50.Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol. 2016;16:341–352. doi: 10.1038/nri.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ho JT, Chan GC, Li JC. Systemic effects of gut microbiota and its relationship with disease and modulation. BMC Immunol. 2015;16:21. doi: 10.1186/s12865-015-0083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dave M, Higgins PD, Middha S, Rioux KP. The human gut microbiome: current knowledge, challenges, and future directions. Transl Res. 2012;160:246–257. doi: 10.1016/j.trsl.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 53.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Jian M, Zhou Y, Li Y, Zhang X, Qin N, Yang H, Wang J, Brunak S, Doré J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J MetaHIT Consortium, Bork P, Ehrlich SD, Wang J. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jandhyala SM, Talukdar R, Subramanyam C, Vuyyuru H, Sasikala M, Nageshwar Reddy D. Role of the normal gut microbiota. World J Gastroenterol. 2015;21:8787–8803. doi: 10.3748/wjg.v21.i29.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Louis P, Flint HJ. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett. 2009;294:1–8. doi: 10.1111/j.1574-6968.2009.01514.x. [DOI] [PubMed] [Google Scholar]
- 56.Wu S, Bhat ZF, Gounder RS, Mohamed Ahmed IA, Al-Juhaimi FY, Ding Y, Bekhit AEA. Effect of Dietary Protein and Processing on Gut Microbiota-A Systematic Review. Nutrients. 2022;14 doi: 10.3390/nu14030453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mamieva Z, Poluektova E, Svistushkin V, Sobolev V, Shifrin O, Guarner F, Ivashkin V. Antibiotics, gut microbiota, and irritable bowel syndrome: What are the relations? World J Gastroenterol. 2022;28:1204–1219. doi: 10.3748/wjg.v28.i12.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Patangia DV, Anthony Ryan C, Dempsey E, Paul Ross R, Stanton C. Impact of antibiotics on the human microbiome and consequences for host health. Microbiologyopen. 2022;11:e1260. doi: 10.1002/mbo3.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takiishi T, Fenero CIM, Câmara NOS. Intestinal barrier and gut microbiota: Shaping our immune responses throughout life. Tissue Barriers. 2017;5:e1373208. doi: 10.1080/21688370.2017.1373208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zuo T, Ng SC. The Gut Microbiota in the Pathogenesis and Therapeutics of Inflammatory Bowel Disease. Front Microbiol. 2018;9:2247. doi: 10.3389/fmicb.2018.02247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Corrêa-Oliveira R, Fachi JL, Vieira A, Sato FT, Vinolo MA. Regulation of immune cell function by short-chain fatty acids. Clin Transl Immunology. 2016;5:e73. doi: 10.1038/cti.2016.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bäumler AJ, Sperandio V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature. 2016;535:85–93. doi: 10.1038/nature18849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kommineni S, Bretl DJ, Lam V, Chakraborty R, Hayward M, Simpson P, Cao Y, Bousounis P, Kristich CJ, Salzman NH. Bacteriocin production augments niche competition by enterococci in the mammalian gastrointestinal tract. Nature. 2015;526:719–722. doi: 10.1038/nature15524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Johansson ME, Jakobsson HE, Holmén-Larsson J, Schütte A, Ermund A, Rodríguez-Piñeiro AM, Arike L, Wising C, Svensson F, Bäckhed F, Hansson GC. Normalization of Host Intestinal Mucus Layers Requires Long-Term Microbial Colonization. Cell Host Microbe. 2015;18:582–592. doi: 10.1016/j.chom.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ubeda C, Djukovic A, Isaac S. Roles of the intestinal microbiota in pathogen protection. Clin Transl Immunology. 2017;6:e128. doi: 10.1038/cti.2017.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.LeBlanc JG, Milani C, de Giori GS, Sesma F, van Sinderen D, Ventura M. Bacteria as vitamin suppliers to their host: a gut microbiota perspective. Curr Opin Biotechnol. 2013;24:160–168. doi: 10.1016/j.copbio.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 67.Dalile B, Van Oudenhove L, Vervliet B, Verbeke K. The role of short-chain fatty acids in microbiota-gut-brain communication. Nat Rev Gastroenterol Hepatol. 2019;16:461–478. doi: 10.1038/s41575-019-0157-3. [DOI] [PubMed] [Google Scholar]
- 68.De Vadder F, Grasset E, Mannerås Holm L, Karsenty G, Macpherson AJ, Olofsson LE, Bäckhed F. Gut microbiota regulates maturation of the adult enteric nervous system via enteric serotonin networks. Proc Natl Acad Sci U S A. 2018;115:6458–6463. doi: 10.1073/pnas.1720017115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schönfeld P, Wojtczak L. Short- and medium-chain fatty acids in energy metabolism: the cellular perspective. J Lipid Res. 2016;57:943–954. doi: 10.1194/jlr.R067629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Carretta MD, Quiroga J, López R, Hidalgo MA, Burgos RA. Participation of Short-Chain Fatty Acids and Their Receptors in Gut Inflammation and Colon Cancer. Front Physiol. 2021;12:662739. doi: 10.3389/fphys.2021.662739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wong C, Harris PJ, Ferguson LR. Potential Benefits of Dietary Fibre Intervention in Inflammatory Bowel Disease. Int J Mol Sci. 2016;17 doi: 10.3390/ijms17060919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Petersen C, Round JL. Defining dysbiosis and its influence on host immunity and disease. Cell Microbiol. 2014;16:1024–1033. doi: 10.1111/cmi.12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sanders DJ, Inniss S, Sebepos-Rogers G, Rahman FZ, Smith AM. The role of the microbiome in gastrointestinal inflammation. Biosci Rep. 2021;41 doi: 10.1042/BSR20203850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Thursby E, Juge N. Introduction to the human gut microbiota. Biochem J. 2017;474:1823–1836. doi: 10.1042/BCJ20160510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vijay A, Valdes AM. Role of the gut microbiome in chronic diseases: a narrative review. Eur J Clin Nutr. 2022;76:489–501. doi: 10.1038/s41430-021-00991-6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 76.Lloyd-Price J, Arze C, Ananthakrishnan AN, Schirmer M, Avila-Pacheco J, Poon TW, Andrews E, Ajami NJ, Bonham KS, Brislawn CJ, Casero D, Courtney H, Gonzalez A, Graeber TG, Hall AB, Lake K, Landers CJ, Mallick H, Plichta DR, Prasad M, Rahnavard G, Sauk J, Shungin D, Vázquez-Baeza Y, White RA 3rd IBDMDB Investigators, Braun J, Denson LA, Jansson JK, Knight R, Kugathasan S, McGovern DPB, Petrosino JF, Stappenbeck TS, Winter HS, Clish CB, Franzosa EA, Vlamakis H, Xavier RJ, Huttenhower C. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569:655–662. doi: 10.1038/s41586-019-1237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Miyoshi J, Chang EB. The gut microbiota and inflammatory bowel diseases. Transl Res. 2017;179:38–48. doi: 10.1016/j.trsl.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cummings JH, Pomare EW, Branch WJ, Naylor CP, Macfarlane GT. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut. 1987;28:1221–1227. doi: 10.1136/gut.28.10.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Macfarlane S, Macfarlane GT. Regulation of short-chain fatty acid production. Proc Nutr Soc. 2003;62:67–72. doi: 10.1079/PNS2002207. [DOI] [PubMed] [Google Scholar]
- 80.den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud DJ, Bakker BM. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res. 2013;54:2325–2340. doi: 10.1194/jlr.R036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Baxter NT, Schmidt AW, Venkataraman A, Kim KS, Waldron C, Schmidt TM. Dynamics of Human Gut Microbiota and Short-Chain Fatty Acids in Response to Dietary Interventions with Three Fermentable Fibers. mBio. 2019;10 doi: 10.1128/mBio.02566-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Koh A, De Vadder F, Kovatcheva-Datchary P, Bäckhed F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell. 2016;165:1332–1345. doi: 10.1016/j.cell.2016.05.041. [DOI] [PubMed] [Google Scholar]
- 83.Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol. 2014;12:661–672. doi: 10.1038/nrmicro3344. [DOI] [PubMed] [Google Scholar]
- 84.Scott KP, Martin JC, Campbell G, Mayer CD, Flint HJ. Whole-genome transcription profiling reveals genes up-regulated by growth on fucose in the human gut bacterium "Roseburia inulinivorans". J Bacteriol. 2006;188:4340–4349. doi: 10.1128/JB.00137-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Duncan SH, Barcenilla A, Stewart CS, Pryde SE, Flint HJ. Acetate utilization and butyryl coenzyme A (CoA):acetate-CoA transferase in butyrate-producing bacteria from the human large intestine. Appl Environ Microbiol. 2002;68:5186–5190. doi: 10.1128/AEM.68.10.5186-5190.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hetzel M, Brock M, Selmer T, Pierik AJ, Golding BT, Buckel W. Acryloyl-CoA reductase from Clostridium propionicum. An enzyme complex of propionyl-CoA dehydrogenase and electron-transferring flavoprotein. Eur J Biochem. 2003;270:902–910. doi: 10.1046/j.1432-1033.2003.03450.x. [DOI] [PubMed] [Google Scholar]
- 87.Kostic AD, Xavier RJ, Gevers D. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology. 2014;146:1489–1499. doi: 10.1053/j.gastro.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Imhann F, Vich Vila A, Bonder MJ, Fu J, Gevers D, Visschedijk MC, Spekhorst LM, Alberts R, Franke L, van Dullemen HM, Ter Steege RWF, Huttenhower C, Dijkstra G, Xavier RJ, Festen EAM, Wijmenga C, Zhernakova A, Weersma RK. Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease. Gut. 2018;67:108–119. doi: 10.1136/gutjnl-2016-312135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Marchesi JR, Holmes E, Khan F, Kochhar S, Scanlan P, Shanahan F, Wilson ID, Wang Y. Rapid and noninvasive metabonomic characterization of inflammatory bowel disease. J Proteome Res. 2007;6:546–551. doi: 10.1021/pr060470d. [DOI] [PubMed] [Google Scholar]
- 90.Vijay N, Morris ME. Role of monocarboxylate transporters in drug delivery to the brain. Curr Pharm Des. 2014;20:1487–1498. doi: 10.2174/13816128113199990462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gopal E, Umapathy NS, Martin PM, Ananth S, Gnana-Prakasam JP, Becker H, Wagner CA, Ganapathy V, Prasad PD. Cloning and functional characterization of human SMCT2 (SLC5A12) and expression pattern of the transporter in kidney. Biochim Biophys Acta. 2007;1768:2690–2697. doi: 10.1016/j.bbamem.2007.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pierre K, Pellerin L. Monocarboxylate transporters in the central nervous system: distribution, regulation and function. J Neurochem. 2005;94:1–14. doi: 10.1111/j.1471-4159.2005.03168.x. [DOI] [PubMed] [Google Scholar]
- 93.Halestrap AP, Meredith D. The SLC16 gene family-from monocarboxylate transporters (MCTs) to aromatic amino acid transporters and beyond. Pflugers Arch. 2004;447:619–628. doi: 10.1007/s00424-003-1067-2. [DOI] [PubMed] [Google Scholar]
- 94.Sepponen K, Ruusunen M, Pakkanen JA, Pösö AR. Expression of CD147 and monocarboxylate transporters MCT1, MCT2 and MCT4 in porcine small intestine and colon. Vet J. 2007;174:122–128. doi: 10.1016/j.tvjl.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 95.Ganapathy V, Thangaraju M, Gopal E, Martin PM, Itagaki S, Miyauchi S, Prasad PD. Sodium-coupled monocarboxylate transporters in normal tissues and in cancer. AAPS J. 2008;10:193–199. doi: 10.1208/s12248-008-9022-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Coady MJ, Chang MH, Charron FM, Plata C, Wallendorff B, Sah JF, Markowitz SD, Romero MF, Lapointe JY. The human tumour suppressor gene SLC5A8 expresses a Na+-monocarboxylate cotransporter. J Physiol. 2004;557:719–731. doi: 10.1113/jphysiol.2004.063859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Martin PM, Gopal E, Ananth S, Zhuang L, Itagaki S, Prasad BM, Smith SB, Prasad PD, Ganapathy V. Identity of SMCT1 (SLC5A8) as a neuron-specific Na+-coupled transporter for active uptake of L-lactate and ketone bodies in the brain. J Neurochem. 2006;98:279–288. doi: 10.1111/j.1471-4159.2006.03878.x. [DOI] [PubMed] [Google Scholar]
- 98.Bloemen JG, Venema K, van de Poll MC, Olde Damink SW, Buurman WA, Dejong CH. Short chain fatty acids exchange across the gut and liver in humans measured at surgery. Clin Nutr. 2009;28:657–661. doi: 10.1016/j.clnu.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 99.Boets E, Gomand SV, Deroover L, Preston T, Vermeulen K, De Preter V, Hamer HM, Van den Mooter G, De Vuyst L, Courtin CM, Annaert P, Delcour JA, Verbeke KA. Systemic availability and metabolism of colonic-derived short-chain fatty acids in healthy subjects: a stable isotope study. J Physiol. 2017;595:541–555. doi: 10.1113/JP272613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Boets E, Deroover L, Houben E, Vermeulen K, Gomand SV, Delcour JA, Verbeke K. Quantification of in Vivo Colonic Short Chain Fatty Acid Production from Inulin. Nutrients. 2015;7:8916–8929. doi: 10.3390/nu7115440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Brown AJ, Goldsworthy SM, Barnes AA, Eilert MM, Tcheang L, Daniels D, Muir AI, Wigglesworth MJ, Kinghorn I, Fraser NJ, Pike NB, Strum JC, Steplewski KM, Murdock PR, Holder JC, Marshall FH, Szekeres PG, Wilson S, Ignar DM, Foord SM, Wise A, Dowell SJ. The Orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J Biol Chem. 2003;278:11312–11319. doi: 10.1074/jbc.M211609200. [DOI] [PubMed] [Google Scholar]
- 102.Bolognini D, Tobin AB, Milligan G, Moss CE. The Pharmacology and Function of Receptors for Short-Chain Fatty Acids. Mol Pharmacol. 2016;89:388–398. doi: 10.1124/mol.115.102301. [DOI] [PubMed] [Google Scholar]
- 103.Hudson BD, Smith NJ, Milligan G. Experimental challenges to targeting poorly characterized GPCRs: uncovering the therapeutic potential for free fatty acid receptors. Adv Pharmacol. 2011;62:175–218. doi: 10.1016/B978-0-12-385952-5.00006-3. [DOI] [PubMed] [Google Scholar]
- 104.Le Poul E, Loison C, Struyf S, Springael JY, Lannoy V, Decobecq ME, Brezillon S, Dupriez V, Vassart G, Van Damme J, Parmentier M, Detheux M. Functional characterization of human receptors for short chain fatty acids and their role in polymorphonuclear cell activation. J Biol Chem. 2003;278:25481–25489. doi: 10.1074/jbc.M301403200. [DOI] [PubMed] [Google Scholar]
- 105.Lee SU, In HJ, Kwon MS, Park BO, Jo M, Kim MO, Cho S, Lee S, Lee HJ, Kwak YS, Kim S. β-Arrestin 2 mediates G protein-coupled receptor 43 signals to nuclear factor-κB. Biol Pharm Bull. 2013;36:1754–1759. doi: 10.1248/bpb.b13-00312. [DOI] [PubMed] [Google Scholar]
- 106.Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, Schilter HC, Rolph MS, Mackay F, Artis D, Xavier RJ, Teixeira MM, Mackay CR. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nøhr MK, Egerod KL, Christiansen SH, Gille A, Offermanns S, Schwartz TW, Møller M. Expression of the short chain fatty acid receptor GPR41/FFAR3 in autonomic and somatic sensory ganglia. Neuroscience. 2015;290:126–137. doi: 10.1016/j.neuroscience.2015.01.040. [DOI] [PubMed] [Google Scholar]
- 108.Pluznick J. A novel SCFA receptor, the microbiota, and blood pressure regulation. Gut Microbes. 2014;5:202–207. doi: 10.4161/gmic.27492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tazoe H, Otomo Y, Karaki S, Kato I, Fukami Y, Terasaki M, Kuwahara A. Expression of short-chain fatty acid receptor GPR41 in the human colon. Biomed Res. 2009;30:149–156. doi: 10.2220/biomedres.30.149. [DOI] [PubMed] [Google Scholar]
- 110.Baumgart DC, Dignass AU. Intestinal barrier function. Curr Opin Clin Nutr Metab Care. 2002;5:685–694. doi: 10.1097/00075197-200211000-00012. [DOI] [PubMed] [Google Scholar]
- 111.Capaldo CT, Powell DN, Kalman D. Layered defense: how mucus and tight junctions seal the intestinal barrier. J Mol Med (Berl) 2017;95:927–934. doi: 10.1007/s00109-017-1557-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Suzuki T. Regulation of the intestinal barrier by nutrients: The role of tight junctions. Anim Sci J. 2020;91:e13357. doi: 10.1111/asj.13357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tsukita S, Tanaka H, Tamura A. The Claudins: From Tight Junctions to Biological Systems. Trends Biochem Sci. 2019;44:141–152. doi: 10.1016/j.tibs.2018.09.008. [DOI] [PubMed] [Google Scholar]
- 114.Shen L, Weber CR, Raleigh DR, Yu D, Turner JR. Tight junction pore and leak pathways: a dynamic duo. Annu Rev Physiol. 2011;73:283–309. doi: 10.1146/annurev-physiol-012110-142150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Van Itallie CM, Anderson JM. Phosphorylation of tight junction transmembrane proteins: Many sites, much to do. Tissue Barriers. 2018;6:e1382671. doi: 10.1080/21688370.2017.1382671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Furuse M, Hirase T, Itoh M, Nagafuchi A, Yonemura S, Tsukita S. Occludin: a novel integral membrane protein localizing at tight junctions. J Cell Biol. 1993;123:1777–1788. doi: 10.1083/jcb.123.6.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Saitou M, Furuse M, Sasaki H, Schulzke JD, Fromm M, Takano H, Noda T, Tsukita S. Complex phenotype of mice lacking occludin, a component of tight junction strands. Mol Biol Cell. 2000;11:4131–4142. doi: 10.1091/mbc.11.12.4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Günzel D, Yu AS. Claudins and the modulation of tight junction permeability. Physiol Rev. 2013;93:525–569. doi: 10.1152/physrev.00019.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Slifer ZM, Blikslager AT. The Integral Role of Tight Junction Proteins in the Repair of Injured Intestinal Epithelium. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21030972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tamura A, Tsukita S. Paracellular barrier and channel functions of TJ claudins in organizing biological systems: advances in the field of barriology revealed in knockout mice. Semin Cell Dev Biol. 2014;36:177–185. doi: 10.1016/j.semcdb.2014.09.019. [DOI] [PubMed] [Google Scholar]
- 121.Matter K, Balda MS. Signalling to and from tight junctions. Nat Rev Mol Cell Biol. 2003;4:225–236. doi: 10.1038/nrm1055. [DOI] [PubMed] [Google Scholar]
- 122.Ebnet K, Suzuki A, Ohno S, Vestweber D. Junctional adhesion molecules (JAMs): more molecules with dual functions? J Cell Sci. 2004;117:19–29. doi: 10.1242/jcs.00930. [DOI] [PubMed] [Google Scholar]
- 123.Luissint AC, Nusrat A, Parkos CA. JAM-related proteins in mucosal homeostasis and inflammation. Semin Immunopathol. 2014;36:211–226. doi: 10.1007/s00281-014-0421-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Nomme J, Fanning AS, Caffrey M, Lye MF, Anderson JM, Lavie A. The Src homology 3 domain is required for junctional adhesion molecule binding to the third PDZ domain of the scaffolding protein ZO-1. J Biol Chem. 2011;286:43352–43360. doi: 10.1074/jbc.M111.304089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Garrido-Urbani S, Bradfield PF, Imhof BA. Tight junction dynamics: the role of junctional adhesion molecules (JAMs) Cell Tissue Res. 2014;355:701–715. doi: 10.1007/s00441-014-1820-1. [DOI] [PubMed] [Google Scholar]
- 126.Buckley A, Turner JR. Cell Biology of Tight Junction Barrier Regulation and Mucosal Disease. Cold Spring Harb Perspect Biol. 2018;10 doi: 10.1101/cshperspect.a029314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kim S, Kim GH. Roles of claudin-2, ZO-1 and occludin in leaky HK-2 cells. PLoS One. 2017;12:e0189221. doi: 10.1371/journal.pone.0189221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Shen L. Tight junctions on the move: molecular mechanisms for epithelial barrier regulation. Ann N Y Acad Sci. 2012;1258:9–18. doi: 10.1111/j.1749-6632.2012.06613.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.D'Incà R, Di Leo V, Corrao G, Martines D, D'Odorico A, Mestriner C, Venturi C, Longo G, Sturniolo GC. Intestinal permeability test as a predictor of clinical course in Crohn's disease. Am J Gastroenterol. 1999;94:2956–2960. doi: 10.1111/j.1572-0241.1999.01444.x. [DOI] [PubMed] [Google Scholar]
- 130.Arnott ID, Kingstone K, Ghosh S. Abnormal intestinal permeability predicts relapse in inactive Crohn disease. Scand J Gastroenterol. 2000;35:1163–1169. doi: 10.1080/003655200750056637. [DOI] [PubMed] [Google Scholar]
- 131.Wapenaar MC, Monsuur AJ, van Bodegraven AA, Weersma RK, Bevova MR, Linskens RK, Howdle P, Holmes G, Mulder CJ, Dijkstra G, van Heel DA, Wijmenga C. Associations with tight junction genes PARD3 and MAGI2 in Dutch patients point to a common barrier defect for coeliac disease and ulcerative colitis. Gut. 2008;57:463–467. doi: 10.1136/gut.2007.133132. [DOI] [PubMed] [Google Scholar]
- 132.Chelakkot C, Ghim J, Ryu SH. Mechanisms regulating intestinal barrier integrity and its pathological implications. Exp Mol Med. 2018;50:1–9. doi: 10.1038/s12276-018-0126-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Antoni L, Nuding S, Wehkamp J, Stange EF. Intestinal barrier in inflammatory bowel disease. World J Gastroenterol. 2014;20:1165–1179. doi: 10.3748/wjg.v20.i5.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Catalioto RM, Maggi CA, Giuliani S. Intestinal epithelial barrier dysfunction in disease and possible therapeutical interventions. Curr Med Chem. 2011;18:398–426. doi: 10.2174/092986711794839179. [DOI] [PubMed] [Google Scholar]
- 135.Lewis K, Lutgendorff F, Phan V, Söderholm JD, Sherman PM, McKay DM. Enhanced translocation of bacteria across metabolically stressed epithelia is reduced by butyrate. Inflamm Bowel Dis. 2010;16:1138–1148. doi: 10.1002/ibd.21177. [DOI] [PubMed] [Google Scholar]
- 136.Plöger S, Stumpff F, Penner GB, Schulzke JD, Gäbel G, Martens H, Shen Z, Günzel D, Aschenbach JR. Microbial butyrate and its role for barrier function in the gastrointestinal tract. Ann N Y Acad Sci. 2012;1258:52–59. doi: 10.1111/j.1749-6632.2012.06553.x. [DOI] [PubMed] [Google Scholar]
- 137.Miao W, Wu X, Wang K, Wang W, Wang Y, Li Z, Liu J, Li L, Peng L. Sodium Butyrate Promotes Reassembly of Tight Junctions in Caco-2 Monolayers Involving Inhibition of MLCK/MLC2 Pathway and Phosphorylation of PKCβ2. Int J Mol Sci. 2016;17 doi: 10.3390/ijms17101696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Suzuki T, Yoshida S, Hara H. Physiological concentrations of short-chain fatty acids immediately suppress colonic epithelial permeability. Br J Nutr. 2008;100:297–305. doi: 10.1017/S0007114508888733. [DOI] [PubMed] [Google Scholar]
- 139.Tong LC, Wang Y, Wang ZB, Liu WY, Sun S, Li L, Su DF, Zhang LC. Propionate Ameliorates Dextran Sodium Sulfate-Induced Colitis by Improving Intestinal Barrier Function and Reducing Inflammation and Oxidative Stress. Front Pharmacol. 2016;7:253. doi: 10.3389/fphar.2016.00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Elamin EE, Masclee AA, Dekker J, Pieters HJ, Jonkers DM. Short-chain fatty acids activate AMP-activated protein kinase and ameliorate ethanol-induced intestinal barrier dysfunction in Caco-2 cell monolayers. J Nutr. 2013;143:1872–1881. doi: 10.3945/jn.113.179549. [DOI] [PubMed] [Google Scholar]
- 141.Zhang Y, Chen H, Zhu W, Yu K. Cecal Infusion of Sodium Propionate Promotes Intestinal Development and Jejunal Barrier Function in Growing Pigs. Animals (Basel) 2019;9 doi: 10.3390/ani9060284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Peng L, Li ZR, Green RS, Holzman IR, Lin J. Butyrate enhances the intestinal barrier by facilitating tight junction assembly via activation of AMP-activated protein kinase in Caco-2 cell monolayers. J Nutr. 2009;139:1619–1625. doi: 10.3945/jn.109.104638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Daly K, Shirazi-Beechey SP. Microarray analysis of butyrate regulated genes in colonic epithelial cells. DNA Cell Biol. 2006;25:49–62. doi: 10.1089/dna.2006.25.49. [DOI] [PubMed] [Google Scholar]
- 144.Kazemi Sefat NA, Mohammadi MM, Hadjati J, Talebi S, Ajami M, Daneshvar H. Sodium Butyrate as a Histone Deacetylase Inhibitor Affects Toll-Like Receptor 4 Expression in Colorectal Cancer Cell Lines. Immunol Invest. 2019;48:759–769. doi: 10.1080/08820139.2019.1595643. [DOI] [PubMed] [Google Scholar]
- 145.Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci U S A. 2014;111:2247–2252. doi: 10.1073/pnas.1322269111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Huang B, Li G, Jiang XH. Fate determination in mesenchymal stem cells: a perspective from histone-modifying enzymes. Stem Cell Res Ther. 2015;6:35. doi: 10.1186/s13287-015-0018-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Marks PA, Richon VM, Miller T, Kelly WK. Histone deacetylase inhibitors. Adv Cancer Res. 2004;91:137–168. doi: 10.1016/S0065-230X(04)91004-4. [DOI] [PubMed] [Google Scholar]
- 148.Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- 150.Marmorstein R, Trievel RC. Histone modifying enzymes: structures, mechanisms, and specificities. Biochim Biophys Acta. 2009;1789:58–68. doi: 10.1016/j.bbagrm.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Johansson ME. Fast renewal of the distal colonic mucus layers by the surface goblet cells as measured by in vivo labeling of mucin glycoproteins. PLoS One. 2012;7:e41009. doi: 10.1371/journal.pone.0041009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gaudier E, Rival M, Buisine MP, Robineau I, Hoebler C. Butyrate enemas upregulate Muc genes expression but decrease adherent mucus thickness in mice colon. Physiol Res. 2009;58:111–119. doi: 10.33549/physiolres.931271. [DOI] [PubMed] [Google Scholar]
- 153.Barcelo A, Claustre J, Moro F, Chayvialle JA, Cuber JC, Plaisancié P. Mucin secretion is modulated by luminal factors in the isolated vascularly perfused rat colon. Gut. 2000;46:218–224. doi: 10.1136/gut.46.2.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Cox MA, Jackson J, Stanton M, Rojas-Triana A, Bober L, Laverty M, Yang X, Zhu F, Liu J, Wang S, Monsma F, Vassileva G, Maguire M, Gustafson E, Bayne M, Chou CC, Lundell D, Jenh CH. Short-chain fatty acids act as antiinflammatory mediators by regulating prostaglandin E(2) and cytokines. World J Gastroenterol. 2009;15:5549–5557. doi: 10.3748/wjg.15.5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Liu L, Li L, Min J, Wang J, Wu H, Zeng Y, Chen S, Chu Z. Butyrate interferes with the differentiation and function of human monocyte-derived dendritic cells. Cell Immunol. 2012;277:66–73. doi: 10.1016/j.cellimm.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 156.Macia L, Tan J, Vieira AT, Leach K, Stanley D, Luong S, Maruya M, Ian McKenzie C, Hijikata A, Wong C, Binge L, Thorburn AN, Chevalier N, Ang C, Marino E, Robert R, Offermanns S, Teixeira MM, Moore RJ, Flavell RA, Fagarasan S, Mackay CR. Metabolite-sensing receptors GPR43 and GPR109A facilitate dietary fibre-induced gut homeostasis through regulation of the inflammasome. Nat Commun. 2015;6:6734. doi: 10.1038/ncomms7734. [DOI] [PubMed] [Google Scholar]
- 157.Davie JR. Inhibition of histone deacetylase activity by butyrate. J Nutr. 2003;133:2485S–2493S. doi: 10.1093/jn/133.7.2485S. [DOI] [PubMed] [Google Scholar]
- 158.Lührs H, Gerke T, Müller JG, Melcher R, Schauber J, Boxberge F, Scheppach W, Menzel T. Butyrate inhibits NF-kappaB activation in lamina propria macrophages of patients with ulcerative colitis. Scand J Gastroenterol. 2002;37:458–466. doi: 10.1080/003655202317316105. [DOI] [PubMed] [Google Scholar]
- 159.Song M, Xia B, Li J. Effects of topical treatment of sodium butyrate and 5-aminosalicylic acid on expression of trefoil factor 3, interleukin 1beta, and nuclear factor kappaB in trinitrobenzene sulphonic acid induced colitis in rats. Postgrad Med J. 2006;82:130–135. doi: 10.1136/pgmj.2005.037945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Lee C, Kim BG, Kim JH, Chun J, Im JP, Kim JS. Sodium butyrate inhibits the NF-kappa B signaling pathway and histone deacetylation, and attenuates experimental colitis in an IL-10 independent manner. Int Immunopharmacol. 2017;51:47–56. doi: 10.1016/j.intimp.2017.07.023. [DOI] [PubMed] [Google Scholar]
- 161.Ji J, Shu D, Zheng M, Wang J, Luo C, Wang Y, Guo F, Zou X, Lv X, Li Y, Liu T, Qu H. Microbial metabolite butyrate facilitates M2 macrophage polarization and function. Sci Rep. 2016;6:24838. doi: 10.1038/srep24838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Rodrigues HG, Takeo Sato F, Curi R, Vinolo MAR. Fatty acids as modulators of neutrophil recruitment, function and survival. Eur J Pharmacol. 2016;785:50–58. doi: 10.1016/j.ejphar.2015.03.098. [DOI] [PubMed] [Google Scholar]
- 163.Nielsen DSG, Jensen BB, Theil PK, Nielsen TS, Knudsen KEB, Purup S. Effect of butyrate and fermentation products on epithelial integrity in a mucus-secreting human colon cell line. J Funct Foods. 2018;40:9–17. [Google Scholar]
- 164.Hamer HM, Jonkers DM, Bast A, Vanhoutvin SA, Fischer MA, Kodde A, Troost FJ, Venema K, Brummer RJ. Butyrate modulates oxidative stress in the colonic mucosa of healthy humans. Clin Nutr. 2009;28:88–93. doi: 10.1016/j.clnu.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 165.Zhou L, Zhang M, Wang Y, Dorfman RG, Liu H, Yu T, Chen X, Tang D, Xu L, Yin Y, Pan Y, Zhou Q, Zhou Y, Yu C. Faecalibacterium prausnitzii Produces Butyrate to Maintain Th17/Treg Balance and to Ameliorate Colorectal Colitis by Inhibiting Histone Deacetylase 1. Inflamm Bowel Dis. 2018;24:1926–1940. doi: 10.1093/ibd/izy182. [DOI] [PubMed] [Google Scholar]
- 166.Facchin S, Vitulo N, Calgaro M, Buda A, Romualdi C, Pohl D, Perini B, Lorenzon G, Marinelli C, D'Incà R, Sturniolo GC, Savarino EV. Microbiota changes induced by microencapsulated sodium butyrate in patients with inflammatory bowel disease. Neurogastroenterol Motil. 2020;32:e13914. doi: 10.1111/nmo.13914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Zhang M, Zhou Q, Dorfman RG, Huang X, Fan T, Zhang H, Zhang J, Yu C. Butyrate inhibits interleukin-17 and generates Tregs to ameliorate colorectal colitis in rats. BMC Gastroenterol. 2016;16:84. doi: 10.1186/s12876-016-0500-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Tedelind S, Westberg F, Kjerrulf M, Vidal A. Anti-inflammatory properties of the short-chain fatty acids acetate and propionate: a study with relevance to inflammatory bowel disease. World J Gastroenterol. 2007;13:2826–2832. doi: 10.3748/wjg.v13.i20.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Vinolo MA, Rodrigues HG, Hatanaka E, Sato FT, Sampaio SC, Curi R. Suppressive effect of short-chain fatty acids on production of proinflammatory mediators by neutrophils. J Nutr Biochem. 2011;22:849–855. doi: 10.1016/j.jnutbio.2010.07.009. [DOI] [PubMed] [Google Scholar]
- 170.Furness JB. The Enteric Nervous System. Oxford: Blackwell Publishing, 2006: 1-102. [Google Scholar]
- 171.Furness JB, Johnson PJ, Pompolo S, Bornstein JC. Evidence that enteric motility reflexes can be initiated through entirely intrinsic mechanisms in the guinea-pig small intestine. Neurogastroenterol Motil. 1995;7:89–96. doi: 10.1111/j.1365-2982.1995.tb00213.x. [DOI] [PubMed] [Google Scholar]
- 172.Furness JB. The enteric nervous system and neurogastroenterology. Nat Rev Gastroenterol Hepatol. 2012;9:286–294. doi: 10.1038/nrgastro.2012.32. [DOI] [PubMed] [Google Scholar]
- 173.Furness JB. Types of neurons in the enteric nervous system. J Auton Nerv Syst. 2000;81:87–96. doi: 10.1016/s0165-1838(00)00127-2. [DOI] [PubMed] [Google Scholar]
- 174.Lakhan SE, Kirchgessner A. Neuroinflammation in inflammatory bowel disease. J Neuroinflammation. 2010;7:37. doi: 10.1186/1742-2094-7-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.da Silva MV, Marosti AR, Mendes CE, Palombit K, Castelucci P. Submucosal neurons and enteric glial cells expressing the P2X7 receptor in rat experimental colitis. Acta Histochem. 2017;119:481–494. doi: 10.1016/j.acthis.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 176.Linden DR, Couvrette JM, Ciolino A, McQuoid C, Blaszyk H, Sharkey KA, Mawe GM. Indiscriminate loss of myenteric neurones in the TNBS-inflamed guinea-pig distal colon. Neurogastroenterol Motil. 2005;17:751–760. doi: 10.1111/j.1365-2982.2005.00703.x. [DOI] [PubMed] [Google Scholar]
- 177.Evangelinellis MM, Souza RF, Mendes CE, Castelucci P. Effects of a P2X7 receptor antagonist on myenteric neurons in the distal colon of an experimental rat model of ulcerative colitis. Histochem Cell Biol. 2022;157:65–81. doi: 10.1007/s00418-021-02039-z. [DOI] [PubMed] [Google Scholar]
- 178.de Fontgalland D, Brookes SJ, Gibbins I, Sia TC, Wattchow DA. The neurochemical changes in the innervation of human colonic mesenteric and submucosal blood vessels in ulcerative colitis and Crohn's disease. Neurogastroenterol Motil. 2014;26:731–744. doi: 10.1111/nmo.12327. [DOI] [PubMed] [Google Scholar]
- 179.Linden DR. Colitis is associated with a loss of intestinofugal neurons. Am J Physiol Gastrointest Liver Physiol. 2012;303:G1096–G1104. doi: 10.1152/ajpgi.00176.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Kaji I, Akiba Y, Furuyama T, Adelson DW, Iwamoto K, Watanabe M, Kuwahara A, Kaunitz JD. Free fatty acid receptor 3 activation suppresses neurogenic motility in rat proximal colon. Neurogastroenterol Motil. 2018;30 doi: 10.1111/nmo.13157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Leonel AJ, Alvarez-Leite JI. Butyrate: implications for intestinal function. Curr Opin Clin Nutr Metab Care. 2012;15:474–479. doi: 10.1097/MCO.0b013e32835665fa. [DOI] [PubMed] [Google Scholar]
- 182.Soret R, Chevalier J, De Coppet P, Poupeau G, Derkinderen P, Segain JP, Neunlist M. Short-chain fatty acids regulate the enteric neurons and control gastrointestinal motility in rats. Gastroenterology. 2010;138:1772–1782. doi: 10.1053/j.gastro.2010.01.053. [DOI] [PubMed] [Google Scholar]
- 183.Obata Y, Pachnis V. The Effect of Microbiota and the Immune System on the Development and Organization of the Enteric Nervous System. Gastroenterology. 2016;151:836–844. doi: 10.1053/j.gastro.2016.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Verbeke KA, Boobis AR, Chiodini A, Edwards CA, Franck A, Kleerebezem M, Nauta A, Raes J, van Tol EA, Tuohy KM. Towards microbial fermentation metabolites as markers for health benefits of prebiotics. Nutr Res Rev. 2015;28:42–66. doi: 10.1017/S0954422415000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Gibson GR, Hutkins R, Sanders ME, Prescott SL, Reimer RA, Salminen SJ, Scott K, Stanton C, Swanson KS, Cani PD, Verbeke K, Reid G. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat Rev Gastroenterol Hepatol. 2017;14:491–502. doi: 10.1038/nrgastro.2017.75. [DOI] [PubMed] [Google Scholar]
- 186.Sanders ME. Probiotics: definition, sources, selection, and uses. Clin Infect Dis. 2008;46 Suppl 2:S58–61; discussion S144. doi: 10.1086/523341. [DOI] [PubMed] [Google Scholar]
- 187.Garcia-Mantrana I, Selma-Royo M, Alcantara C, Collado MC. Shifts on Gut Microbiota Associated to Mediterranean Diet Adherence and Specific Dietary Intakes on General Adult Population. Front Microbiol. 2018;9:890. doi: 10.3389/fmicb.2018.00890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Moens F, Van den Abbeele P, Basit AW, Dodoo C, Chatterjee R, Smith B, Gaisford S. A four-strain probiotic exerts positive immunomodulatory effects by enhancing colonic butyrate production in vitro. Int J Pharm. 2019;555:1–10. doi: 10.1016/j.ijpharm.2018.11.020. [DOI] [PubMed] [Google Scholar]
- 189.Trakman GL, Fehily S, Basnayake C, Hamilton AL, Russell E, Wilson-O'Brien A, Kamm MA. Diet and gut microbiome in gastrointestinal disease. J Gastroenterol Hepatol. 2022;37:237–245. doi: 10.1111/jgh.15728. [DOI] [PubMed] [Google Scholar]
- 190.Kasti A, Petsis K, Lambrinou S, Katsas K, Nikolaki M, Papanikolaou IS, Hatziagelaki E, Triantafyllou K. A Combination of Mediterranean and Low-FODMAP Diets for Managing IBS Symptoms? Microorganisms. 2022;10 doi: 10.3390/microorganisms10040751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Kimble R, Gouinguenet P, Ashor A, Stewart C, Deighton K, Matu J, Griffiths A, Malcomson FC, Joel A, Houghton D, Stevenson E, Minihane AM, Siervo M, Shannon OM, Mathers JC. Effects of a mediterranean diet on the gut microbiota and microbial metabolites: A systematic review of randomized controlled trials and observational studies. Crit Rev Food Sci Nutr. 2022:1–22. doi: 10.1080/10408398.2022.2057416. [DOI] [PubMed] [Google Scholar]
- 192.Maldonado-Contreras A, Noel SE, Ward DV, Velez M, Mangano KM. Associations between Diet, the Gut Microbiome, and Short-Chain Fatty Acid Production among Older Caribbean Latino Adults. J Acad Nutr Diet. 2020;120:2047–2060.e6. doi: 10.1016/j.jand.2020.04.018. [DOI] [PubMed] [Google Scholar]
- 193.Veettil SK, Wong TY, Loo YS, Playdon MC, Lai NM, Giovannucci EL, Chaiyakunapruk N. Role of Diet in Colorectal Cancer Incidence: Umbrella Review of Meta-analyses of Prospective Observational Studies. JAMA Netw Open. 2021;4:e2037341. doi: 10.1001/jamanetworkopen.2020.37341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Hou H, Chen D, Zhang K, Zhang W, Liu T, Wang S, Dai X, Wang B, Zhong W, Cao H. Gut microbiota-derived short-chain fatty acids and colorectal cancer: Ready for clinical translation? Cancer Lett. 2022;526:225–235. doi: 10.1016/j.canlet.2021.11.027. [DOI] [PubMed] [Google Scholar]
- 195.Derrien M, van Hylckama Vlieg JE. Fate, activity, and impact of ingested bacteria within the human gut microbiota. Trends Microbiol. 2015;23:354–366. doi: 10.1016/j.tim.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 196.De Filippis F, Pellegrini N, Vannini L, Jeffery IB, La Storia A, Laghi L, Serrazanetti DI, Di Cagno R, Ferrocino I, Lazzi C, Turroni S, Cocolin L, Brigidi P, Neviani E, Gobbetti M, O'Toole PW, Ercolini D. High-level adherence to a Mediterranean diet beneficially impacts the gut microbiota and associated metabolome. Gut. 2016;65:1812–1821. doi: 10.1136/gutjnl-2015-309957. [DOI] [PubMed] [Google Scholar]
- 197.Jakubczyk D, Leszczyńska K, Górska S. The Effectiveness of Probiotics in the Treatment of Inflammatory Bowel Disease (IBD)-A Critical Review. Nutrients. 2020;12 doi: 10.3390/nu12071973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Duranti S, Gaiani F, Mancabelli L, Milani C, Grandi A, Bolchi A, Santoni A, Lugli GA, Ferrario C, Mangifesta M, Viappiani A, Bertoni S, Vivo V, Serafini F, Barbaro MR, Fugazza A, Barbara G, Gioiosa L, Palanza P, Cantoni AM, de'Angelis GL, Barocelli E, de'Angelis N, van Sinderen D, Ventura M, Turroni F. Elucidating the gut microbiome of ulcerative colitis: bifidobacteria as novel microbial biomarkers. FEMS Microbiol Ecol. 2016;92 doi: 10.1093/femsec/fiw191. [DOI] [PubMed] [Google Scholar]
- 199.Satish Kumar CS, Kondal Reddy K, Boobalan G, Gopala Reddy A, Sudha Rani Chowdhary CH, Vinoth A, Jayakanth K, Srinivasa Rao G. Immunomodulatory effects of Bifidobacterium bifidum 231 on trinitrobenzenesulfonic acid-induced ulcerative colitis in rats. Res Vet Sci. 2017;110:40–46. doi: 10.1016/j.rvsc.2016.10.010. [DOI] [PubMed] [Google Scholar]
- 200.Chae JM, Heo W, Cho HT, Lee DH, Kim JH, Rhee MS, Park TS, Kim YK, Lee JH, Kim YJ. Erratum to: Orally-Administered Bifidobacterium animalis subsp. lactis Strain BB12 on Dextran Sodium Sulfate-Induced Colitis in Mice. J Microbiol Biotechnol. 2019;29:665. doi: 10.4014/jmb.2019.2904.665. [DOI] [PubMed] [Google Scholar]
- 201.Elian SD, Souza EL, Vieira AT, Teixeira MM, Arantes RM, Nicoli JR, Martins FS. Bifidobacterium longum subsp. infantis BB-02 attenuates acute murine experimental model of inflammatory bowel disease. Benef Microbes. 2015;6:277–286. doi: 10.3920/BM2014.0070. [DOI] [PubMed] [Google Scholar]
- 202.Veiga P, Gallini CA, Beal C, Michaud M, Delaney ML, DuBois A, Khlebnikov A, van Hylckama Vlieg JE, Punit S, Glickman JN, Onderdonk A, Glimcher LH, Garrett WS. Bifidobacterium animalis subsp. lactis fermented milk product reduces inflammation by altering a niche for colitogenic microbes. Proc Natl Acad Sci U S A. 2010;107:18132–18137. doi: 10.1073/pnas.1011737107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Cheng Y, Liu J, Ling Z. Short-chain fatty acids-producing probiotics: A novel source of psychobiotics. Crit Rev Food Sci Nutr. 2021:1–31. doi: 10.1080/10408398.2021.1920884. [DOI] [PubMed] [Google Scholar]
- 204.Abraham BP, Quigley EMM. Probiotics in Inflammatory Bowel Disease. Gastroenterol Clin North Am. 2017;46:769–782. doi: 10.1016/j.gtc.2017.08.003. [DOI] [PubMed] [Google Scholar]
- 205.Hegazy SK, El-Bedewy MM. Effect of probiotics on pro-inflammatory cytokines and NF-kappaB activation in ulcerative colitis. World J Gastroenterol. 2010;16:4145–4151. doi: 10.3748/wjg.v16.i33.4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Kruis W, Fric P, Pokrotnieks J, Lukás M, Fixa B, Kascák M, Kamm MA, Weismueller J, Beglinger C, Stolte M, Wolff C, Schulze J. Maintaining remission of ulcerative colitis with the probiotic Escherichia coli Nissle 1917 is as effective as with standard mesalazine. Gut. 2004;53:1617–1623. doi: 10.1136/gut.2003.037747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Kato K, Mizuno S, Umesaki Y, Ishii Y, Sugitani M, Imaoka A, Otsuka M, Hasunuma O, Kurihara R, Iwasaki A, Arakawa Y. Randomized placebo-controlled trial assessing the effect of bifidobacteria-fermented milk on active ulcerative colitis. Aliment Pharmacol Ther. 2004;20:1133–1141. doi: 10.1111/j.1365-2036.2004.02268.x. [DOI] [PubMed] [Google Scholar]
- 208.Chapman TM, Plosker GL, Figgitt DP. VSL#3 probiotic mixture: a review of its use in chronic inflammatory bowel diseases. Drugs. 2006;66:1371–1387. doi: 10.2165/00003495-200666100-00006. [DOI] [PubMed] [Google Scholar]
- 209.Sood A, Midha V, Makharia GK, Ahuja V, Singal D, Goswami P, Tandon RK. The probiotic preparation, VSL#3 induces remission in patients with mild-to-moderately active ulcerative colitis. Clin Gastroenterol Hepatol. 2009;7:1202–1209, 1209.e1. doi: 10.1016/j.cgh.2009.07.016. [DOI] [PubMed] [Google Scholar]
- 210.Derwa Y, Gracie DJ, Hamlin PJ, Ford AC. Systematic review with meta-analysis: the efficacy of probiotics in inflammatory bowel disease. Aliment Pharmacol Ther. 2017;46:389–400. doi: 10.1111/apt.14203. [DOI] [PubMed] [Google Scholar]
- 211.Fedorak RN, Feagan BG, Hotte N, Leddin D, Dieleman LA, Petrunia DM, Enns R, Bitton A, Chiba N, Paré P, Rostom A, Marshall J, Depew W, Bernstein CN, Panaccione R, Aumais G, Steinhart AH, Cockeram A, Bailey RJ, Gionchetti P, Wong C, Madsen K. The probiotic VSL#3 has anti-inflammatory effects and could reduce endoscopic recurrence after surgery for Crohn's disease. Clin Gastroenterol Hepatol. 2015;13:928–35.e2. doi: 10.1016/j.cgh.2014.10.031. [DOI] [PubMed] [Google Scholar]
- 212.Khan R, Roy N, Ali H, Naeem M. Fecal Microbiota Transplants for Inflammatory Bowel Disease Treatment: Synthetic- and Engineered Communities-Based Microbiota Transplants Are the Future. Gastroenterol Res Pract. 2022;2022:9999925. doi: 10.1155/2022/9999925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.El-Salhy M, Valeur J, Hausken T, Gunnar Hatlebakk J. Changes in fecal short-chain fatty acids following fecal microbiota transplantation in patients with irritable bowel syndrome. Neurogastroenterol Motil. 2021;33:e13983. doi: 10.1111/nmo.13983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Zhang J, Guo Y, Duan L. Features of Gut Microbiome Associated With Responses to Fecal Microbiota Transplantation for Inflammatory Bowel Disease: A Systematic Review. Front Med (Lausanne) 2022;9:773105. doi: 10.3389/fmed.2022.773105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Biliński J, Jasiński M, Basak GW. The Role of Fecal Microbiota Transplantation in the Treatment of Acute Graft-versus-Host Disease. Biomedicines. 2022;10 doi: 10.3390/biomedicines10040837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Cremon C, Guglielmetti S, Gargari G, Taverniti V, Castellazzi AM, Valsecchi C, Tagliacarne C, Fiore W, Bellini M, Bertani L, Gambaccini D, Cicala M, Germanà B, Vecchi M, Pagano I, Barbaro MR, Bellacosa L, Stanghellini V, Barbara G. Effect of Lactobacillus paracasei CNCM I-1572 on symptoms, gut microbiota, short chain fatty acids, and immune activation in patients with irritable bowel syndrome: A pilot randomized clinical trial. United European Gastroenterol J. 2018;6:604–613. doi: 10.1177/2050640617736478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.El-Salhy M, Hatlebakk JG, Gilja OH, Bråthen Kristoffersen A, Hausken T. Efficacy of faecal microbiota transplantation for patients with irritable bowel syndrome in a randomised, double-blind, placebo-controlled study. Gut. 2020;69:859–867. doi: 10.1136/gutjnl-2019-319630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Shi Y, Dong Y, Huang W, Zhu D, Mao H, Su P. Fecal Microbiota Transplantation for Ulcerative Colitis: A Systematic Review and Meta-Analysis. PLoS One. 2016;11:e0157259. doi: 10.1371/journal.pone.0157259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Tan XY, Xie YJ, Liu XL, Li XY, Jia B. A Systematic Review and Meta-Analysis of Randomized Controlled Trials of Fecal Microbiota Transplantation for the Treatment of Inflammatory Bowel Disease. Evid Based Complement Alternat Med. 2022;2022:8266793. doi: 10.1155/2022/8266793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Allegretti JR, Kelly CR, Grinspan A, Mullish BH, Hurtado J, Carrellas M, Marcus J, Marchesi JR, McDonald JAK, Gerardin Y, Silverstein M, Pechlivanis A, Barker GF, Miguens Blanco J, Alexander JL, Gallagher KI, Pettee W, Phelps E, Nemes S, Sagi SV, Bohm M, Kassam Z, Fischer M. Inflammatory Bowel Disease Outcomes Following Fecal Microbiota Transplantation for Recurrent C. difficile Infection. Inflamm Bowel Dis. 2021;27:1371–1378. doi: 10.1093/ibd/izaa283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.BioRender.com (2022) Metabolism of SCFAs. Available from https://app.biorender.com/biorender-templates .