Summary
In microgravity, cells undergo profound changes in their properties. However, how human cardiac progenitors respond to space microgravity is unknown. In this study, we evaluated the effect of space microgravity on differentiation of human induced pluripotent stem cell (hiPSC)-derived cardiac progenitors compared with 1G cultures on the International Space Station (ISS). Cryopreserved 3D cardiac progenitors were cultured for 3 weeks on the ISS. Compared with 1G cultures, the microgravity cultures had 3-fold larger sphere sizes, 20-fold higher counts of nuclei, and increased expression of proliferation markers. Highly enriched cardiomyocytes generated in space microgravity showed improved Ca2+ handling and increased expression of contraction-associated genes. Short-term exposure (3 days) of cardiac progenitors to space microgravity upregulated genes involved in cell proliferation, survival, cardiac differentiation, and contraction, consistent with improved microgravity cultures at the late stage. These results indicate that space microgravity increased proliferation of hiPSC-cardiomyocytes, which had appropriate structure and function.
Keywords: cardiomyocytes, differentiation, function, human induced pluripotent stem cells, microgravity, proliferation, cardiac progenitors, calcium handling, gene experssison, spaceflight
Graphical abstract
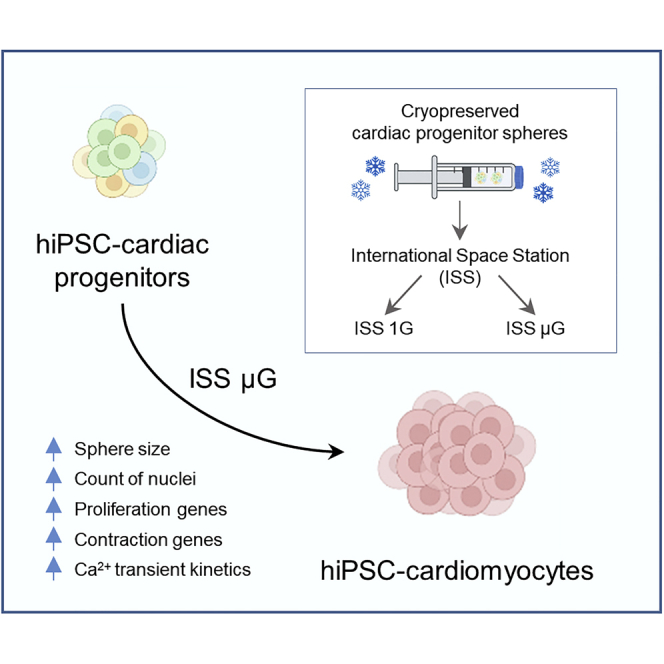
Highlights
-
•
Cryopreserved 3D hiPSC-cardiac progenitors differentiated efficiently in space
-
•
Microgravity cultures had increased sphere sizes and cellular proliferation
-
•
Beating cardiomyocytes in microgravity cultures had improved Ca2+ handling
-
•
Microgravity cultures had upregulated genes in cardiac contraction
In this article, Xu and colleagues show that cryopreserved 3D cardiac progenitors differentiated into highly enriched cardiomyocytes on the International Space Station (ISS). Compared with ISS 1G cells, ISS microgravity cells had increased proliferation and improved Ca2+ transients. Short-term microgravity also increased the expression of genes associated with proliferation and cardiac differentiation and contraction.
Introduction
A leading candidate cell source for regenerative cardiac therapy is the cardiomyocytes derived from human induced pluripotent stem cells (hiPSC-CMs) (Laflamme and Murry, 2005; Chong et al., 2014). Targeting the intermediate steps from human induced pluripotent stem cells (hiPSCs) to cardiomyocytes could help improve the efficiency of hiPSC-CM production. Exposure of cardiac progenitors to microgravity presents a novel method to achieve cardiomyocyte differentiation at high efficiency and high yield, as microgravity can profoundly modulate cell properties (Barzegari and Saei, 2012; Becker and Souza, 2013; Ingber, 1999), including proliferation of stem cells (Chen et al., 2006; Kawahara et al., 2009; Li et al., 2009). For example, under simulated microgravity generated with a random positioning machine, microscale three-dimensional (3D) cardiac progenitors from hiPSCs showed increased proliferation and survival compared with parallel cultures under standard gravity (Jha et al., 2016), resulting in the production of enriched cardiomyocytes at high cell yield. These cardiomyocytes also had improved structural and functional maturation features (Jha et al., 2016), which are highly desirable for improving the safety of cell therapy since transplantation of immature cardiomyocytes increases the risk of graft-induced arrhythmias (Chong et al., 2014).
The International Space Station (ISS)-US National Laboratory provides an extraordinary environment to study the effect of space microgravity on cell properties that is not achievable elsewhere. While the random positioning machine and similar devices can simulate some aspects of microgravity and weightless environment during spaceflight, they only provide a good approximation to microgravity environment on Earth (Grimm et al., 2014; Zhang et al., 2013). Gravitational forces are still present under simulated microgravity, affecting cell properties. Research on the ISS has shown that space microgravity can indeed modulate cell properties (Unsworth and Lelkes, 1998) and provide beneficial effects on the cells for possible therapeutic use on Earth (Freed and Vunjak-Novakovic, 2002; Sharma et al., 2022; Yuge et al., 2006).
Cell biology studies on the ISS are usually conducted using live, non-cryopreserved cell cultures maintained in modules that required CO2 (Wnorowski et al., 2019). To facilitate the study of space microgravity on the culture and differentiation of 3D cardiac progenitors, we have recently developed methods to cryopreserve the 3D cardiac progenitors and culture them in a CO2-independent medium (Rampoldi et al., 2021). In this study, we conducted a spaceflight experiment (MVP-CELL-03 project) with cryopreserved hiPSC-derived cardiac progenitors sent to the ISS through the SpaceX-20 mission. The astronauts successfully cultured the cells for 3 weeks and returned live, beating cardiomyocytes back to us. We then comprehensively assessed the cellular, molecular, and functional characteristics of the cells. We also assessed the molecular effect of a short-term (3 days) exposure of cardiac progenitors to space microgravity. Here, we report that space microgravity increased cell proliferation and that the cardiomyocytes generated in space microgravity cultures showed appropriate structural and functional features.
Results
Recovery of live cells following cell culture of cryopreserved 3D cardiac progenitors without CO2 on the ISS
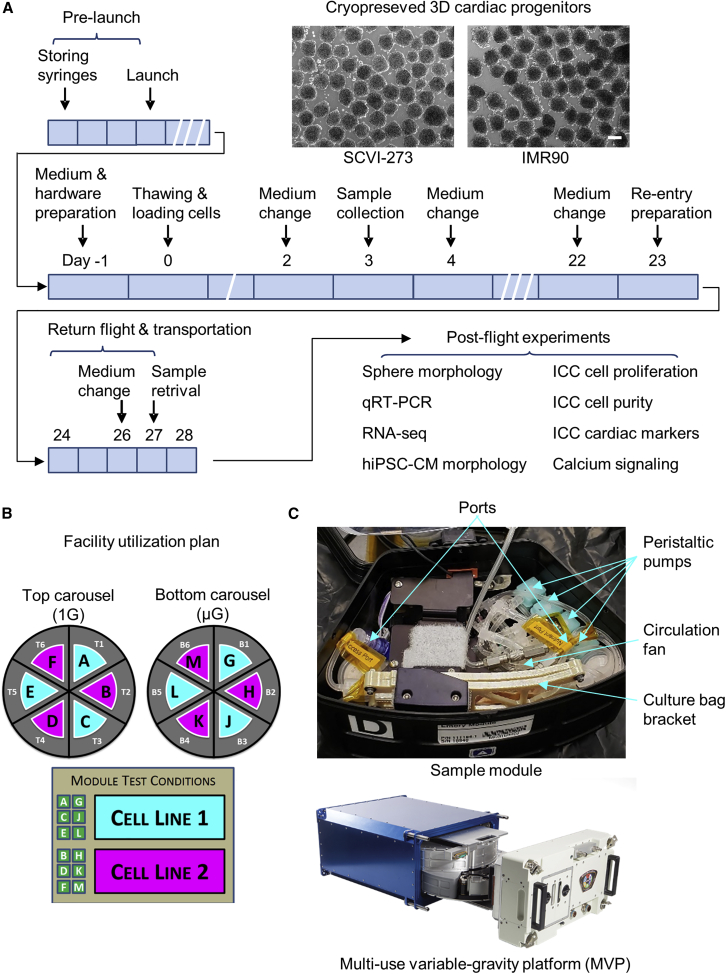
For the spaceflight experiment (MVP-CELL-03 project), we prepared cryopreserved cardiac progenitor spheres from two hiPSC lines: SCVI-273 and IMR90 (Figure S1 and supplemental information). The cryopreserved cardiac progenitors were flown to the ISS through the SpaceX-20 mission. As illustrated in Figure 1A, the astronauts on board the ISS thawed the cardiac progenitor spheres into the CO2-independent medium with a ROCK (Rho-associated coiled-coil containing protein kinase) inhibitor. The cells were cultured at 37°C using the Multi-specimen Variable-gravity Platform (MVP) configured to load multiple cultures under both ISS microgravity (ISS μG) and ISS 1G conditions; the 1G condition on the ISS was achieved by centrifugation of one carousel within the same MVP system (Figures 1B and 1C). Specifically, we designed our experiments to examine the growth and differentiation of 3D cardiac progenitors from two hiPSC cell lines under ISS μG and ISS 1G conditions with each condition run in triplicate. A total of 12 MVP experiment modules (six for each cell line) were used.
Figure 1.
Spaceflight experimental design
(A) Schematic of spaceflight operational plan. Scale bar, 200 μm.
(B) Schematic of Multi-specimen Variable-gravity Platform (MVP) sample collection plan.
(C) Schematic of MVP system. ICC, immunocytochemistry. See also Figure S1.
For short-term exposure of microgravity, after 3 days of cell culture on the ISS, an aliquot of samples was harvested from the experimental modules, fixed using RNAProtect, and stored at −20°C. For long-term exposure of space microgravity, live cultures were returned to ground via warm storage after having been cultured for 22 days on the ISS for further analysis. The cardiac spheres from post-flight ISS cultures recovered beating activity 2–3 days after they were transferred to an incubator, with beat rates of 10–15 beats/min in ISS 1G cultures and 9–17 beats/min in ISS μG cultures.
Space microgravity increases the size of hiPSC-CM spheres and hiPSC-CM proliferation
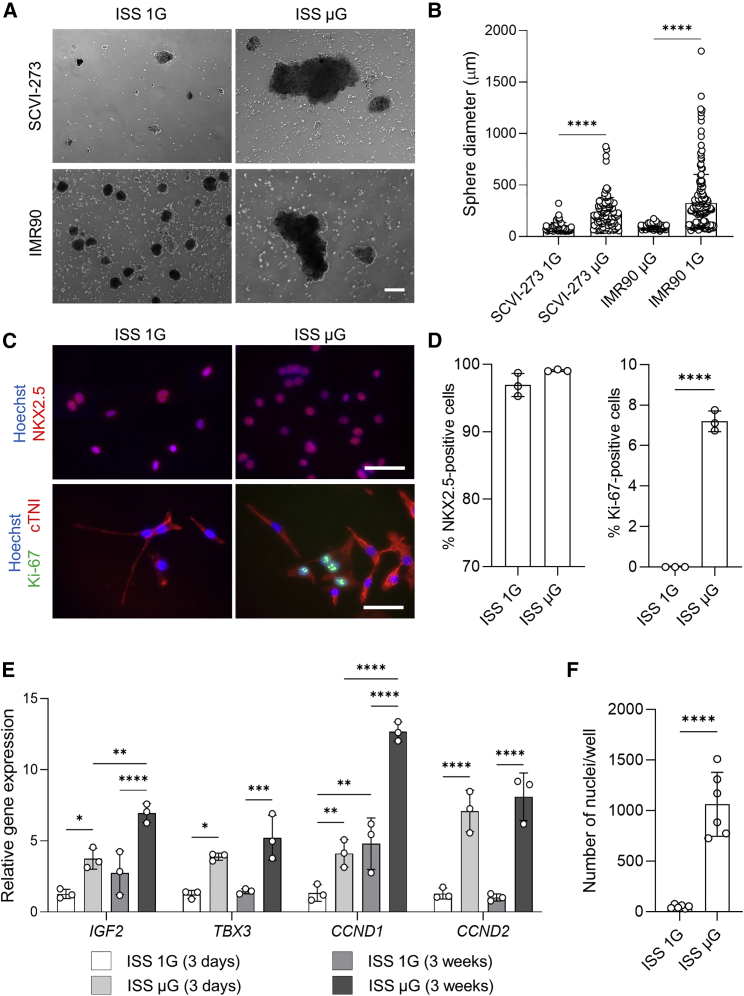
Cardiac sphere morphology was examined under a phase-contrast microscope after the samples were returned from the ISS. Phase-contrast photos were taken (Figure 2A) and sphere diameters were measured by software ImageJ. In the IMR90 ISS μG cultures, cardiac spheres were on average three times bigger compared with ISS 1G cardiac spheres (Figure 2B). Similar results were observed in the SCVI-273 ISS cultures (Figures 2A and 2B).
Figure 2.
Space microgravity increases cardiac sphere size and improves proliferation of enriched hiPSC-CMs
(A) Cell morphology of cardiac spheres derived from SCVI-273 and IMR90 hiPSCs from International Space Station (ISS) cultures post flight (scale bar, 200 μm).
(B) Diameters of cardiac spheres derived from SCVI-273 and IMR90 hiPSCs (n = 72–180 spheres).
(C) Immunocytochemistry analysis of IMR90 cultures to detect cardiomyocyte purity by cardiac marker NKX2.5 (red) and proliferation by Ki-67 (green) and cardiac marker cTNI (red) (scale bar, 50μm).
(D) Percentage of NKX2.5-positive cells and Ki-67-positive cells in IMR90 cultures. Aliquots of ISS cultures post flight were dissociated, replated, and subjected to immunocytochemistry (n = 3 cultures).
(E) Expression of genes associated with proliferation in IMR90 cultures (3 weeks on the ISS) and SCVI-273 cultures (3 days on the ISS) (n = 3 cultures).
(F) Counts of nuclei in samples from IMR90 cultures (n = 6 wells) after the cells were dissociated, replated, and stained. Statistical analyses were performed with unpaired, two-tailed Student’s t test for (B), (D), and (F), and one-way ANOVA for (E). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001. Data are presented as mean ± SD. ISS 1G, the 1G condition on the ISS; ISS μG, the microgravity condition on the ISS.
Immunocytochemical analysis was performed to examine cardiomyocyte purity and cardiomyocyte proliferation of the ISS cultures. As shown in Figures 2C and 2D, the majority of the cells were positive for NKX2.5 (NK2 homeobox 5) (>95%) in both ISS 1G and μG conditions. These results were similar to the pre-testing results done before the samples were flown to the ISS (Figures S1C and S1D). In addition, approximately 7% of cells were positive for Ki-67 in ISS μG cultures, while no Ki-67-positive cells were detected in ISS 1G cultures (Figures 2C and 2D). As expected, almost all cells were positive for cardiac marker cTNI (cardiac troponin I) in both ISS μG and ISS 1G cultures. Therefore, ISS cultures of the cryopreserved cardiac progenitor spheres differentiated into highly enriched cardiomyocytes, and ISS μG cultures contained more cells at the active phase of cell cycle than did ISS 1G cultures.
We examined the expression of genes related to cell cycle and cell proliferation in both short-term (3 days) and long-term (3 weeks) ISS cultures by qRT-PCR analysis (Figure 2E). The expression of cyclin D2 (CCND2), cyclin D1 (CCND1), T-box transcription factor 3 (TBX3), and insulin like growth factor 2 (IGF2) was significantly upregulated in ISS μG cells compared with ISS 1G cells from both short-term and long-term cultures. Compared with the short-term ISS μG cultures, the long-term ISS μG cultures had increased expression of CCND1 and IGF2. In addition, the nuclei counts of live cells (after the long-term culture cells were replated) were ∼20 times higher in cells from ISS μG cultures compared with ISS 1G cultures (Figure 2F). These results were consistent with the observations on increased proliferation capacity in ISS μG cells.
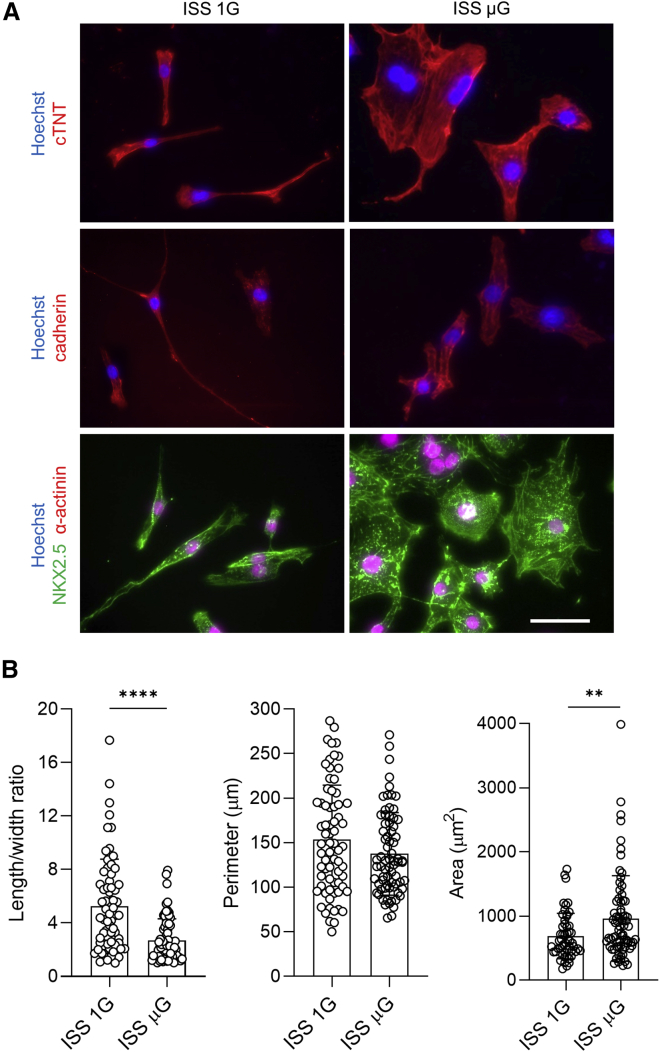
Space microgravity improves cellular and structural parameters in hiPSC-CMs
Immunocytochemical analysis was performed in ISS μG and ISS 1G cultures for other cardiac structural markers including α-actinin, cardiac troponin T (cTNT), and pan-cadherin (Figure 3A). Almost all cells from ISS μG and ISS 1G cultures were positive for these cardiac structural markers. ISS μG cells had larger cell area but similar perimeter compared with ISS 1G control. In addition, ISS 1G cells were more elongated, while ISS μG cells expanded in both length and width (Figure 3B).
Figure 3.
Space microgravity cultures generated enriched cardiomyocytes with increased cell size
(A) Immunocytochemical analysis of cardiac proteins in IMR90 cells from ISS cultures post flight, including cTNT (red), pan-cadherin (red), α-actinin (green), and NKX2.5 (red) (scale bar, 50 μm).
(B) Comparison of cellular parameters of IMR90 hiPSC-CMs. Statistical analyses were performed with unpaired, two-tailed Student’s t test. ∗∗p < 0.01, and ∗∗∗∗p < 0.0001. Data are presented as mean ± SD (ISS 1G, n = 53 cells; ISS μG, n = 73 cells).
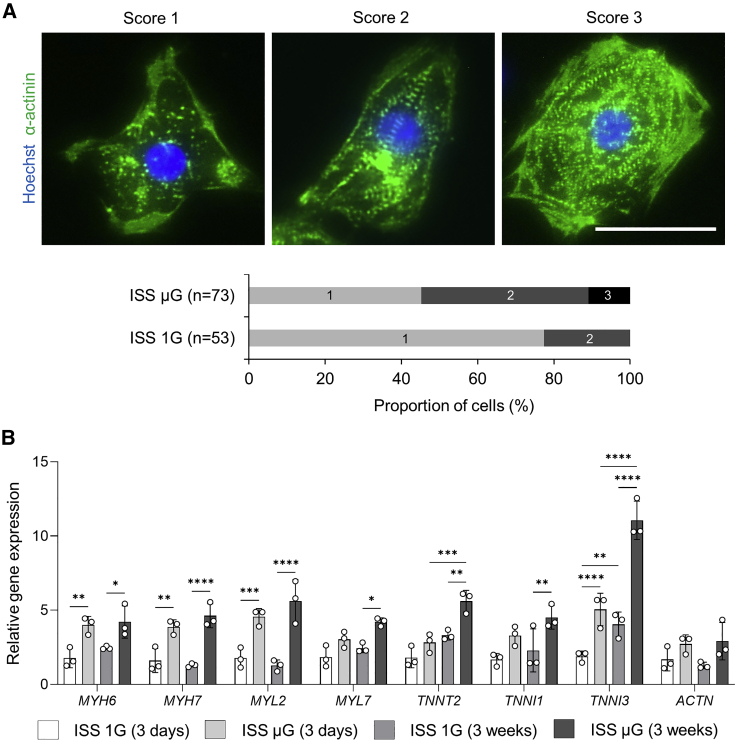
We evaluated cells for their overall appearance of sarcomeric striations based on the levels of the organization of the Z-line protein α-actinin and categorized them into three different levels as described previously (Jha et al., 2016; Nguyen et al., 2014; Ribeiro et al., 2015). As shown in Figure 4A, cells with score 1 were α-actinin-positive cells but without clear sarcomeric striations, cells with score 2 were cells with a diffuse and punctate staining pattern of α-actinin staining, and cell with score 3 were cells with more organized myofibrillar structure with distinct paralleled bands of z-discs. Compared with ISS 1G cardiomyocytes, cardiac spheres exposed to space microgravity had more cells with a higher score of defined myofibrillar structure, indicating an improved structural development (Figure 4A).
Figure 4.
Space microgravity improves cardiac structure of hiPSC-CMs
(A) Structural analysis of IMR90 hiPSC-CMs from ISS cultures post flight. Cells were dissociated, replated, and stained for sarcomeric α-actinin (green) and Hoechst (blue). Overall appearance of myofibrillar structure was categorized into three different levels and percentage of the cells by the scores was generated by counting n = 53 cells from ISS 1G and n = 73 cells from ISS μG cultures.
(B) qRT-PCR panel showing relative mRNA expression levels of gene associated with cardiac structure in cells derived from IMR90 hiPSCs (3 weeks on the ISS) and SCVI-273 hiPSCs (3 days on the ISS) (n = 3 cultures). Statistical analyses was performed with one-way ANOVA. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001. Data are presented as mean ± SD (n = 3 cultures).
In addition, we examined the expression of genes related to cardiac structural proteins in intact cardiac spheres. Among the genes examined, myosin light chain 2V (MYL2), cardiac troponin I3 (TNNI3), myosin heavy chain 6 (MYH6), and myosin heavy chain 7 (MYH7) were upregulated in ISS μG cells compared with ISS 1G cells from both short-term (3 days) and long-term (3 weeks) ISS cultures (Figure 4B); MYL2, TNNI3, and MYH7 are isoforms in more mature cardiomyocytes. In long-term ISS cultures, myosin light chain 2A (MYL7) and cardiac troponin T2 (TNNT2) were also upregulated in ISS μG cells compared with ISS 1G cells. In addition, compared with the short-term ISS μG cells, the long-term ISS μG cells had increased expression of TNNT2 and TNNI3. Such increased cardiac structural proteins are major parameters of cardiomyocyte maturation (Guo and Pu, 2020).
Space microgravity improves Ca2+ signaling in hiPSC-CMs
After the cardiac spheres were transferred to low-adhesion dishes, ISS μG cardiac spheres recovered beating activity quite fast, between 16 and 36 h. In ISS 1G cultures, spheres showed beating activity 48 h after being transferred. To assess the function of hiPSC-CMs from the ISS cultures at single-cell level, we performed Ca2+ signaling analysis after the cardiac spheres were dissociated and replated. The cells recovered beating activity after they were maintained in RPMI 1640 with 2% B27 supplement for 72 h, and were then loaded with calcium-sensitive dye Fluo-4 for Ca2+ imaging.
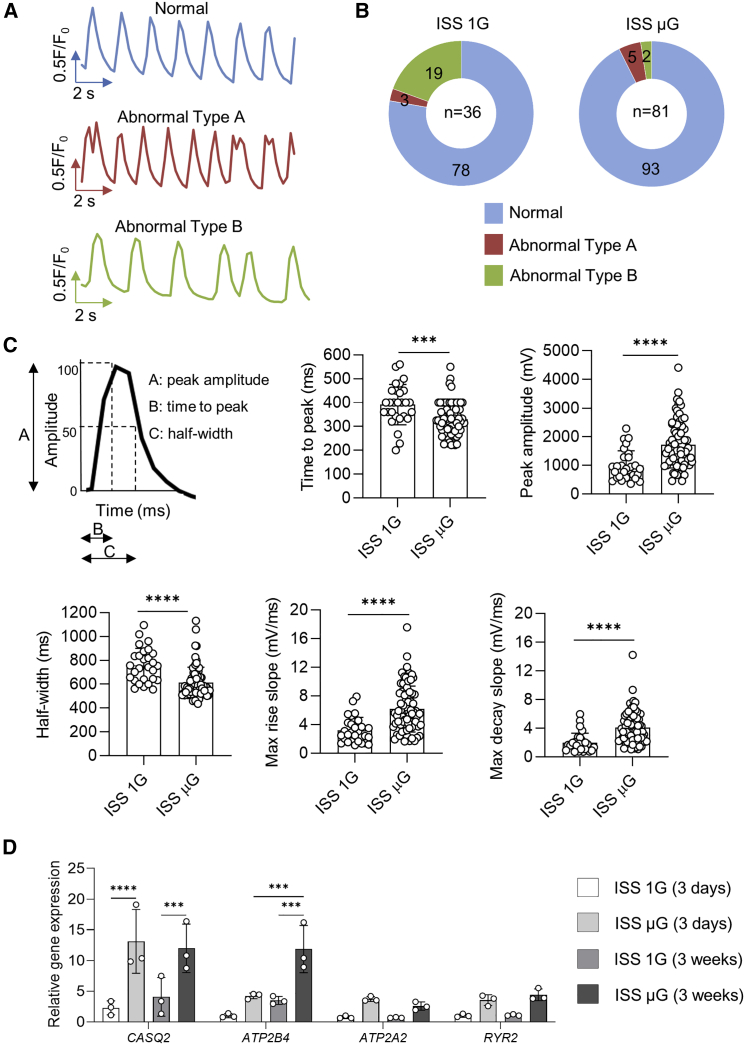
Among these beating cells, we observed three types of Ca2+ transients: (1) normal Ca2+ transients, (2) abnormal Ca2+ transients with spontaneous Ca2+ leak showing a single notch of diastolic Ca2+ signal, and (3) abnormal Ca2+ transients with inconsistent beating period (Figure 5A). Transients were categorized as normal if they had mostly consistent amplitudes and beat periods with typical cardiac transient morphology of upstroke and decay kinetics, while transients were categorized as abnormal if they exhibited spontaneous Ca2+ release between transients: oscillations of diastolic cytosolic Ca2+ and inconsistent beating (Preininger et al., 2016). ISS μG samples had more cells with normal Ca2+ transients compared with ISS 1G samples (93% in ISS μG cells versus 78% in ISS 1G cells) (Figure 5B). The proportion of the cells with abnormal Ca2+ transients and the types of abnormal Ca2+ transients in ISS 1G cultures were comparable with typical hiPSC-CM cultures in ground-based studies (Forghani et al., 2021; Lan et al., 2013; Liu et al., 2020a; Saraf et al., 2021). Most of these abnormal cells had minor abnormality with inconsistent beating period (type B) and a few cells had transients with a single notch of additional Ca2+ spike (diastolic Ca2+ signal) before the following Ca2+ transient had initiated (type A) (Figures 5A and 5B). These minor abnormal types of Ca2+ transients are likely due to immature nature of hiPSC-CMs. In both ISS μG and ISS 1G samples, we did not observe cells with other types of abnormal Ca2+ transients associated with arrhythmias such as tachycardia-like events or transients with multiple notches of additional Ca2+ spikes as in hiPSC-CMs from patients with heart disease or hiPSC-CMs treated with drugs.
Figure 5.
Space microgravity improves intracellular Ca2+ handling in hiPSC-CMs
(A) Representative traces of normal Ca2+ transients or abnormal Ca2+ transients of IMR90 hiPSC-CMs from ISS cultures post flight.
(B) Pie chart showing the percentage of cells exhibiting normal Ca2+ transients or abnormal Ca2+ transients (ISS 1G, n = 36 cells; ISS μG, n = 81 cells).
(C) Ca2+ transient analyses with parameters presented as mean ± SD (ISS 1G, n = 28 cells; ISS μG, n = 75 cells).
(D) qRT-PCR panel showing relative mRNA expression levels of genes associated with Ca2+ handling in cells derived from IMR90 hiPSCs (3 weeks on the ISS) and SCVI-273 hiPSCs (3 days on the ISS) (n = 3 cultures). Statistical analyses were performed with unpaired, two-tailed Student’s t test for (C) and one-way ANOVA for (D). ∗∗∗p < 0.001; and ∗∗∗∗p < 0.0001.
Among the cells with normal Ca2+ transients, ISS μG cells had reduced time to peak, increased peak amplitude, reduced half-width, and increased maximum rise slope and maximum decay slope compared with ISS 1G cells (Figure 5C), indicating faster Ca2+-transient kinetics, a functional feature of more mature cardiomyocytes.
In addition, the expression of calcium-handling proteins/ion channels (calsequestrin 2 [CASQ2] and ATPase plasma membrane Ca2+ transporting 4 [ATP2B4]) was significantly upregulated in ISS μG cardiac spheres compared with ISS 1G cardiac spheres from both short-term (3 days) and long-term (3 weeks) ISS cultures (Figure 5D).
These results suggest that space microgravity reduced abnormal Ca2+ signaling and increased Ca2+-handling kinetics, which was consistent with increased expression of Ca2+-handling proteins in ISS μG cells.
RNA-seq analysis reveals increased proliferation and differentiation during short-term exposure to space microgravity
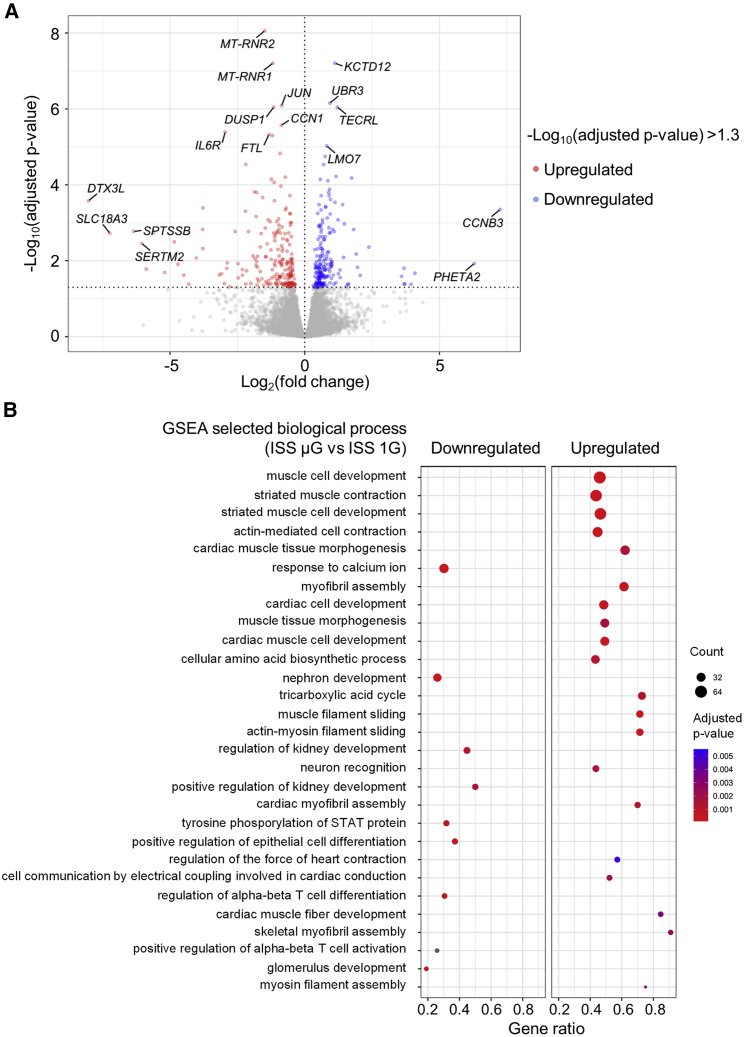
We next examined how a short-term exposure of cardiac progenitors to space microgravity affected the expression of genes associated with expansion and differentiation of these cells. Using cardiac spheres collected 3 days after thawing onboard the ISS, we performed RNA sequencing (RNA-seq) analysis to compare global gene expression profiles of SCVI-273 hiPSC-CMs in ISS μG versus ISS 1G conditions. As detected by RNA-seq, 195 genes were significantly upregulated, and 207 downregulated in ISS μG cells compared with ISS 1G cells. Among the significantly upregulated genes (Figure 6A; Table S1), several are involved in cell cycle, proliferation, survival, and regeneration. They include cyclin B3 (CCNB3), which promotes metaphase-anaphase transition in cell cycle (Li et al., 2019); reelin (RELN), which promotes cardiac regeneration and repair by improving cell survival after heart injury (Liu et al., 2020b); and ubiquitin protein ligase E3 component N-recognin 3 (UBR3), which regulates apurinic/apyrimidinic endodeoxyribonuclease 1 (APE1), a protein involved in DNA damage repair, cell survival, and regulation of transcription, to reduce genome instability (Meisenberg et al., 2012).
Figure 6.
Differentially expressed genes and GO terms identified by RNA sequencing analysis of hiPSC-CMs exposed to space microgravity
(A) Volcano plot illustrating differentially expressed genes between ISS μG and ISS 1G samples (n = 3 cultures) collected from SCVI-273 cultures of 3 days on the ISS (short-term exposure to microgravity).
(B) Dot plot showing up- and downregulated GO terms of biological processes. GSEA, Gene Set Enrichment Analysis. See also Figures S2–S7.
Several upregulated genes are also involved in heart development. They include Lim-domain only 7 (LMO7), a transcriptional regulator of emerin involved in beta-catenin signaling (Holaska et al., 2006); collagen type XIV alpha 1 (COL14A1), encoding type XIV collagen, which is important for growth and structural integrity of the myocardium (Tao et al., 2012); and heparan sulfate proteoglycan 2 (HSPG2), encoding protein perlecan (a major structural components of the basement membrane surrounding cells in the myocardium; Martinez et al., 2018). Several other genes are also involved in modulating cardiomyocyte contractility. They include C-X-C motif chemokine receptor 4 (CXCR4) (Pyo et al., 2006) and gap junction protein alpha 5 (GJA5) (Chaldoupi et al., 2009), which are responsible for contraction and the electrical coupling of cardiomyocytes. Space microgravity also increased the expression of genes involved in fatty acid metabolism, including trans-2,3-enoyl-CoA reductase like TECRL), whose mutations are linked to catecholaminergic polymorphic ventricular tachycardia (Moscu-Gregor et al., 2020).
Among significantly downregulated genes (Figure 6A & Table S1), mitochondrially encoded 12S RRNA (MT-RNR1) and mitochondrially encoded 16S RRNA (MT-RNR2) are functional ncRNAs that protect cells from mitochondrial apoptosis (Bitar et al., 2017); MT-RNR1 was upregulated in hiPSC-CMs subjected to ionizing radiation during differentiation (Baljinnyam et al., 2017). Another downregulated gene, cytokine angiopoietin-2 (ANGPT2), is a promising predictor of heart disease (Pöss et al., 2015) and possesses proinflammatory and apoptosis-promoting abilities (Scholz et al., 2015). Other downregulated genes are involved in the Jak-Stat- and mitogen-activated protein kinase (MAPK)-pathways. They include dual specificity phosphatase 1 (DUSP1) and dual specificity phosphatase 2 (DUSP2), encoding a subclass of tyrosine phosphatases that regulate the activity of MAPK, mediating stress responses, inflammation, and apoptosis (Lang and Raffi, 2019); and serpin family E member 1 (SERPINE1), a serine protease inhibitor of plasminogen activator and an activator of the JAK/STAT pathway associated with cellular stress (Simone et al., 2014) and cardiac diseases (Basisty et al., 2020).
According to gene set enrichment analysis (GSEA), space microgravity upregulated the gene ontology (GO) terms of biological processes associated with cardiac muscle cell development, muscle activity, or cell contractions (Figure 6B; supplemental information) and the GO terms of cellular components and molecular functions associated with structural constituent of muscle, sarcomeric structure, and voltage-gated calcium channel activity involved in cardiac muscle cell action potential and sodium channel activity (Figure S2; supplemental information). In addition, space microgravity downregulated GO terms associated with processes, functions, or components of non-cardiac cells, such as nephron development and neuronal cell body (Figures 6B and S2; supplemental information). These results were consistent with efficient differentiation and maturation of ISS μG cells into cardiomyocytes.
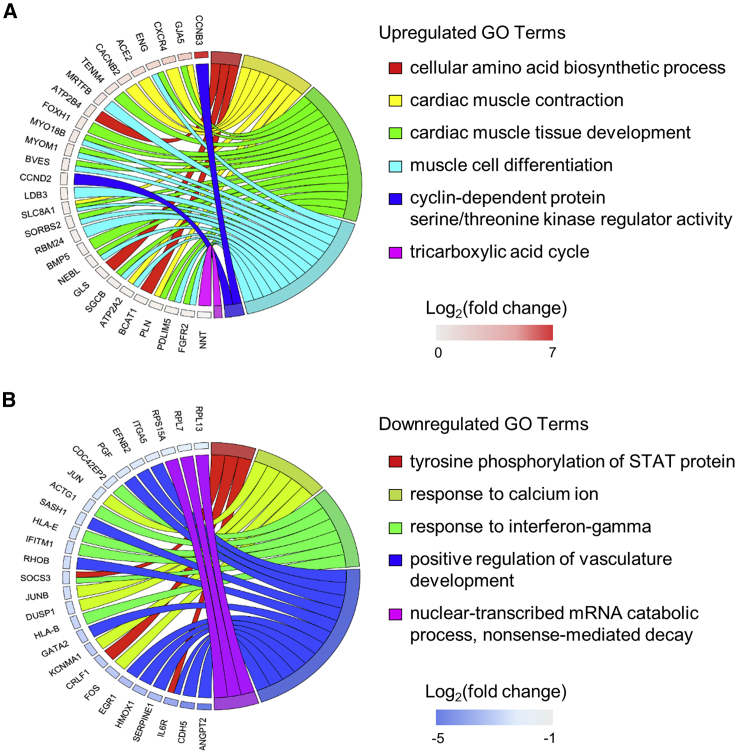
We next examined the link between selected GO terms of biological processes and specific differentially expressed genes (Figure 7; supplemental information). Notably, ISS μG cells had upregulated GO term of cyclin-dependent protein serine/threonine kinase regulator activity that was linked to upregulated genes of CCNB3 and CCND2. Among upregulated genes, CCNB3 had the highest increase in ISS μG cultures (Log2[fold change] = 7.25) (Table S1). CCNB3 is known to regulate the G2/M transition of mitotic cells (Li et al., 2019), whereas CCND2 regulates the G1/S and its overexpression is associated with increased survival and regeneration potency in hiPSC-CMs (Zhu et al., 2018). In addition, cellular amino acid biosynthetic process and the tricarboxylic acid cycle were also upregulated, indicating that ISS μG cells were metabolically more activate than ISS 1G cells.
Figure 7.
Chord diagrams showing the relationship between GO terms and differentially expressed genes in hiPSC-CMs exposed to space microgravity
(A) Chord diagram of selected upregulated GO terms and genes in ISS μG versus ISS 1G of SCVI-273 cultures of 3 days on the ISS.
(B) Chord diagram of selected downregulated GO terms and genes. GO terms are presented on the right, genes on the left, and colored squares on the left indicate Log2(fold change) value from highest to lowest (n = 3 cultures).
Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis showed that several pathways were upregulated by space microgravity, including calcium signaling pathway, cardiac muscle contraction, and adrenergic signaling in cardiomyocytes, which is tightly connected to calcium signaling, cell contraction, and cardiomyocyte maturation. Regulations of the specific genes associated with these pathways and cell cycle are shown in Figures S3–S5. The MAPK signaling pathway and ribosomal subunits proteins were downregulated in ISS μG cells (Figures S6 and S7). Since cell differentiation is associated with the downregulation of rRNA transcription (Hayashi et al., 2014), these results suggest that ISS μG cells actively differentiated into cardiomyocytes.
Together, both the GO term and KEGG enrichment analyses indicate that ISS μG cells were in a state of increased cell growth and cardiac differentiation compared with ISS 1G cells.
Discussion
Despite recent advances in understanding cell behavior under extracellular forces, few studies have investigated changes in proliferation and differentiation of cells under space microgravity (Baio et al., 2018b; Huang et al., 2020; Wnorowski et al., 2019). Our analysis shows that the expansion of hiPSC-derived cardiac progenitors under space microgravity resulted in increased cell proliferation and efficient generation of highly enriched cardiomyocytes with appropriate features. Specifically, the beating cardiac spheres were detected in the cultures, containing >90% cardiomyocytes. Compared with cells from ISS 1G cultures, cardiac spheres from ISS μG cultures were bigger in size and had appropriate molecular and functional properties. At single-cell level, cells from ISS μG cultures had increased cell size and clearer sarcomere structure. The cells also had an increased peak amplitude and faster kinetics of Ca2+ transients. RNA-seq analysis showed upregulation of genes associated with cardiac development, cell cycle, proliferation, survival, and cardiac functions, and downregulation of genes related to extracellular matrix and apoptosis in ISS μG cultures compared with ISS 1G cultures.
An innovative aspect of this study is the direct comparison of cells cultured under both microgravity condition and 1G condition in the MVP system on the ISS. Unlike typical ISS experiments where microgravity cells are compared with the ground control, the MVP system consisted of both ISS μG and ISS 1G modules and thus allowed us to better characterize the impact of space microgravity alone on physiology, structure, and gene expression of hiPSC-CMs and examine whether space microgravity altered the growth and differentiation of cardiac progenitors. Therefore, we could focus on the effect of space microgravity without background noise of space environment, including space radiation that could potentially alter or mask microgravity effects on cellular features and gene expression. The MVP system had an automatic imaging device for each cell culture module but did not provide clear images for us to monitor the presence of beating cells. Further improvement of the flight hardware with more advanced and automatic imaging with higher resolutions would be desirable.
Multiple assessments indicate that long-term exposure of cardiac progenitors to space microgravity generated enriched cardiomyocytes with improved proliferation. The size of the spheres under ISS μG was three times bigger on average than that of ISS 1G controls. Compared with ISS 1G cultures, ISS μG cultures also had increased features of proliferation, including increased expression of proliferation marker Ki-67: 7% of ISS μG cells were positive for Ki-67, while Ki-67 was not detected in ISS 1G cultures. These results were further confirmed by upregulation of selected genes in ISS μG cultures, including TBX3 (Ribeiro et al., 2007), IGF2 (Shen et al., 2020), CCND1 (Gan et al., 2019), and CCND2 (Zhu et al., 2018), which have specific roles in cell cycle, cell proliferation, and heart regeneration. Furthermore, the counts of live cell nuclei in ISS μG cultures were 20-fold higher than those in ISS 1G cultures.
Both structural and functional assessments indicate that cardiomyocytes from ISS μG cultures had improved features. Cells from ISS μG cultures had increased cell size and contained more cells with better-developed and clearer sarcomeres compared with cells from ISS 1G cultures and therefore were structurally improved. The improved cardiomyocyte structure could contribute to the increased stability of Ca2+ signaling we observed in ISS μG cells: the proportions of the cells with normal Ca2+ transients were higher in ISS μG cells than in ISS 1G cells. ISS μG cells also had an increased peak amplitude and faster kinetics of Ca2+ transients. Consistently, ISS μG cells had increased expression of genes encoding cardiac structural and Ca2+-handling proteins, including MYL2, MYL7, TNNI3, TNNT2, MYH6, MYH7, CASQ2, and ATP2B4, indicating that space microgravity could have a beneficial effect on the structure and function of cardiac cells.
Following the sampling of SCVI-273 cultures at day 3 for RNA-seq analysis, only a limited amount of the cells was left for late-stage characterization. Because of this limitation, our characterization of the late-stage cells was focused on the IMR90 cultures but not the SCVI-273 cultures. However, the cell morphologies of both the cell lines were similar at the late stage. In addition, molecular profiling of the short-term cultures of SCVI-273 cells reveals gene regulations that are consistent with the improved cell proliferation observed in the long-term cultures of IMR90 cells. Our RNA-seq analysis of the cardiac spheres that had short-term exposure of microgravity revealed upregulation of key genes and pathways involved in cardiomyocytes differentiation, cardiac structural maturation, contractility, and cell proliferation in ISS μG cells compared with ISS 1G cells. We also detected downregulation of genes and pathways associated with apoptosis, inflammation, and cellular stress in ISS μG cells compared with ISS 1G cells. These results indicate that the short-term exposure of cardiac progenitors to space microgravity was able to implement significant changes in their transcriptomic profiles.
Previous studies indicate that cells can undergo profound changes at the morphological, molecular, and functional levels in response to microgravity and spaceflight (Freed and Vunjak-Novakovic, 2002; Unsworth and Lelkes, 1998). For example, neonatal cardiovascular progenitors (CPCs) had enhanced cell proliferation and changes in cytoskeletal organization and migration after the cells were cultured aboard the ISS for 30 days (Baio et al., 2018b). These CPCs also exhibited elevated expression of Ca2+-handling and signaling genes, which corresponded to the activation of protein kinase C alpha, a calcium-dependent protein kinase (Baio et al., 2018a). In another spaceflight study, human mesenchymal stem cells cultured on the ISS for 7 and 14 days had more potent immunosuppressive capacity than did the ground control (Huang et al., 2020). In addition, simulated microgravity potentiated the proliferation of bone marrow-derived human mesenchymal stem cells (Yuge et al., 2006) and adipose-derived stem cells (Zhang et al., 2015).
Our RNA-seq results provide insights into genes and molecular pathways linked to cardiomyocyte survival and differentiation. For example, ISS μG cells had upregulated expression of genes that support cell proliferation, survival, and cardiac development, including CCNB3, CCND2, T-box transcription factor 1 (TBX1), and T-box transcription factor 2 (TBX2). Among them, CCNB3 was the most upregulated gene in ISS μG cells. The role of CCNB3 in cardiomyocyte proliferation, survival, and differentiation has not been reported, although another cyclin gene, CCND2, is known to be able to improve cardiomyocyte proliferation and cardiac regeneration (Zhu et al., 2018). Further study on CCNB3 and other genes identified in our transcriptomic analysis is likely to be fruitful given significant challenge in graft survival of hiPSC-CMs for regenerative medicine; in nonhuman primate model studies, even with the prosurvival pretreatment, ∼90% of the transplanted cells died post injection (Chong and Murry, 2014).
Our RNA-seq results also highlight that the enhanced cardiomyocyte differentiation in ISS μG cultures was associated with decreased expression of genes associated with differentiation of non-cardiac lineages. For example, the expression of genes related to the GO term of positive regulation of vasculature development and the GO terms associated with processes, functions, or components of neurons and nephrons were downregulated in ISS μG cells. Suppression of differentiation of other cell types would be expected during efficient cardiomyocyte differentiation as, for example, endothelial and cardiac cells are derived from the same progenitors. The results of the molecular profiling could be exploited to facilitate efficient production of cardiomyocytes under standard gravity. Modulating gene expression during early-stage cardiomyocyte differentiation could significantly affect the efficiency of differentiation. For example, downregulation of a Wnt-signaling gene ( leucine rich repeat containing G protein-coupled receptor 5 [LGR5]) inhibited cardiomyocyte differentiation but potentiated endothelial differentiation, while a typical differentiation cultures without suppression of LGR5 resulted in higher levels of cardiomyocytes but very few endothelial cells (Jha et al., 2017).
Therapeutic application of hiPSC-CMs requires not only large amounts of the cells with improved ability for engraftment but also cells with high quality including improved maturation and function in order to improve the safety of cell therapy. Our transcriptomic analysis showed an upregulation of genes and pathways that support cell contractility and calcium signaling. In addition to increased expression of genes associated with Ca2+ handling, structure, and contractility in late-stage ISS μG cells, ISS μG cells at early stage had reduced expression of several genes related to potassium channel activity, including potassium two pore domain channel subfamily K member 5 (KCNK5) and potassium calcium-activated channel subfamily M alpha 1 (KCNMA1). Cardiac potassium channels regulate the shape and duration of the cardiac action potential, and limit the depolarization duration of the cell membrane and the time course of the contractions and the refractory periods (Tamargo et al., 2004). The upregulation of genes involved in calcium channel activity related to contraction and downregulation of genes related to potassium channel activity may be in part contributing to the reduced abnormal intracellular Ca2+ transients observed in ISS μG cells.
In conclusion, we have demonstrated that culture of cryopreserved 3D cardiac progenitors under space microgravity resulted in efficient differentiation of cardiomyocytes. The combination of microgravity and 3D culture employed in this study provides a novel method to increase the proliferation and differentiation of cardiac progenitors. This method also results in cardiomyocytes with improved proliferation, structure, and cardiac function, which are highly desirable for future application of hiPSC-CMs in regenerative medicine. In addition, we have identified potential genes and pathways involved in cardiomyocyte proliferation, survival, and differentiation. Targeting these genes and pathways may provide alternative strategies used on Earth to mimic the effect of space microgravity on improved proliferation, survival, and differentiation of hiPSC-CMs.
Experimental procedures
Cell culture and cardiomyocyte differentiation
SCVI-273 and IMR90 hiPSCs were cultured in a feeder-free condition and subjected to cardiomyocyte differentiation by small molecule (Lian et al., 2012) and growth factors, respectively (Jha et al., 2015; Laflamme et al., 2007).
Formation and cryopreservation of cardiac progenitor spheres
Cardiac progenitor spheres were generated from differentiation day 6 cultures that were dissociated using 0.25% trypsin-EDTA (Thermo Fisher Scientific). The dissociated cells were seeded into the Aggrewell 400 plates at 1.8 × 106 cells/well (1,500 cells/microwell) and cultured in RPMI/B27 medium (RPMI 1640 with 2% B27 supplement with insulin) with 10 μM ROCK inhibitor Y-27632. After 24 h, cardiac progenitor spheres were collected and resuspended in cryopreservation medium (90% fetal bovine serum and 10% dimethyl sulfoxide with 10 μM ROCK inhibitor) and transferred into cryosyringes at 0.5 mL/cryosyringe. The cryosyringes were cooled at 4°C for 25 min and then stored at −80°C in a cooling box (Rampoldi et al., 2021).
ISS cell culture facility and spaceflight operation
The cryopreserved cardiac progenitor spheres were pre-tested and sent to the ISS through the SpaceX-20 mission, a mission launched by the aerospace company SpaceX on March 6, 2020 (https://www.issnationallab.org/launches/spacex-crs-20/). On the ISS, the astronauts thawed the cryopreserved cardiac progenitor spheres, and cultured the cells using the MVP system from Techshot. The MVP system allows loading of multiple cultures/modules under both ISS μG and ISS 1G conditions (the 1G condition on the ISS was achieved by centrifugation). Each condition was run in triplicate for each cell line.
For thawing cells, cryosyringes containing cardiac progenitor spheres were placed in a thermoblock at 37°C for 5 min. The cells were then injected into cell culture chambers of the MVP modules containing the CO2-independent medium with 10 μM ROCK inhibitor (Table S2) (Rampoldi et al., 2021). The MVP modules were re-installed into the MVP facility, which started with a medium flush cycle to replace the medium with new culture medium (20 mL per chamber; ∼2× chamber volume), to flush out the DMSO in the cryopreserved cell solution (0.5 mL/cryosyringe). The cells were cultured at 37°C in the MVP system with medium exchange every other day.
For short-term exposure of space microgravity, after 3 days of cell culture on the ISS, the following samples were harvested, fixed using RNAProtect (Qiagen), and stored at −20°C: three samples from SCVI-273 hiPSCs at ISS μG and three samples from SCVI-273 hiPSCs at ISS 1G (one sample each from the three cell culture modules, and 9 mL for each sample). The remaining cells were returned to the MVP for the duration of the experiment. For long-term exposure of space microgravity, live cultures were returned to ground via warm storage after 22 days of culture on the ISS.
Upon arrival at Emory University, cardiac spheres were transferred immediately into an incubator and allowed to recover overnight. The following day cardiac spheres were transferred from the collection bags into low-adhesion dishes in RPMI 1640 medium with 2% B27 supplement, and were then maintained overnight. Spheres were imaged using an inverted microscope (Axio Vert.A1) and analyzed by ImageJ software.
Quantification and statistical analysis
Data were analyzed in GraphPad Prism 7.04. Comparisons were conducted via an unpaired, two-tailed Student’s t test and one-way ANOVA test with significant differences defined by ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001. Data are presented as mean ± SD.
Supplemental experimental procedures include cell culture, immunocytochemical analysis, high-content imaging analysis, structural analysis of hiPSC-CMs, RNA-seq analyses, real-time qRT-PCR, and calcium imaging.
Author contributions
A.R., K.M., and C.X. designed experiments. A.R., P.F., D.L., H.H., L.C.A., and J.M. performed experiments and analyzed data. J.F. and G.B. contributed to the MVP hardware design and testing. A.R. and C.X. wrote the manuscript. All authors reviewed and approved the manuscript.
Acknowledgments
We thank Astronaut Jessica Meir and Astronaut Andrew Morgan for performing cell culture experiments aboard the ISS and Gregory Tharp and Kathryn Pellegrini at Emory University for RNA-seq analysis. We also thank Dr. Bill McLamb and Dr. Marc Giulianotti of the CASIS for guidance and discussions. The graphical abstract was created with BioRender.com. This study was supported in part by the Center for Advancement of Science in Space (GA-2017-266), the National Institutes of Health (R01HL136345 and R01AA028527), and the National Science Foundation and the Center for Advancement of Science in Space (CBET 1926387).
Conflicts of interest
J.F. and G.B. were employees of Techshot. All other authors declare no competing interests.
Published: September 8, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2022.08.007.
Supplemental information
Data and code availability
The accession number for RNA-seq data reported in this paper is GEO: GSE188793.
References
- Baio J., Martinez A.F., Bailey L., Hasaniya N., Pecaut M.J., Kearns-Jonker M. Spaceflight activates protein kinase C alpha signaling and modifies the developmental stage of human neonatal cardiovascular progenitor cells. Stem Cells Dev. 2018;27:805–818. doi: 10.1089/scd.2017.0263. [DOI] [PubMed] [Google Scholar]
- Baio J., Martinez A.F., Silva I., Hoehn C.V., Countryman S., Bailey L., Hasaniya N., Pecaut M.J., Kearns-Jonker M. Cardiovascular progenitor cells cultured aboard the International Space Station exhibit altered developmental and functional properties. NPJ Microgravity. 2018;4:13. doi: 10.1038/s41526-018-0048-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baljinnyam E., Venkatesh S., Gordan R., Mareedu S., Zhang J., Xie L.H., Azzam E.I., Suzuki C.K., Fraidenraich D. Effect of densely ionizing radiation on cardiomyocyte differentiation from human-induced pluripotent stem cells. Physiol. Rep. 2017;5:e13308. doi: 10.14814/phy2.13308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barzegari A., Saei A.A. An update to space biomedical research: tissue engineering in microgravity bioreactors. Bioimpacts. 2012;2:23–32. doi: 10.5681/bi.2012.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basisty N., Kale A., Jeon O.H., Kuehnemann C., Payne T., Rao C., Holtz A., Shah S., Sharma V., Ferrucci L., et al. A proteomic atlas of senescence-associated secretomes for aging biomarker development. PLoS Biol. 2020;18:e3000599. doi: 10.1371/journal.pbio.3000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker J.L., Souza G.R. Using space-based investigations to inform cancer research on Earth. Nat. Rev. Cancer. 2013;13:315–327. doi: 10.1038/nrc3507. [DOI] [PubMed] [Google Scholar]
- Bitar M., Kuiper S., O'Brien E., Barry G. Using human iPSC-derived neurons to uncover activity-dependent non-coding RNAs. Genes. 2017;8:E401. doi: 10.3390/genes8120401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaldoupi S.M., Loh P., Hauer R.N.W., de Bakker J.M.T., van Rijen H.V.M. The role of connexin40 in atrial fibrillation. Cardiovasc. Res. 2009;84:15–23. doi: 10.1093/cvr/cvp203. [DOI] [PubMed] [Google Scholar]
- Chen X., Xu H., Wan C., McCaigue M., Li G. Bioreactor expansion of human adult bone marrow-derived mesenchymal stem cells. Stem Cell. 2006;24:2052–2059. doi: 10.1634/stemcells.2005-0591. [DOI] [PubMed] [Google Scholar]
- Chong J.J.H., Murry C.E. Cardiac regeneration using pluripotent stem cells-Progression to large animal models. Stem Cell Res. 2014;13:654–665. doi: 10.1016/j.scr.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J.J.H., Yang X., Don C.W., Minami E., Liu Y.W., Weyers J.J., Mahoney W.M., Van Biber B., Cook S.M., Palpant N.J., et al. Human embryonic-stem-cell-derived cardiomyocytes regenerate non-human primate hearts. Nature. 2014;510:273–277. doi: 10.1038/nature13233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forghani P., Rashid A., Sun F., Liu R., Li D., Lee M.R., Hwang H., Maxwell J.T., Mandawat A., Wu R., et al. Carfilzomib treatment causes molecular and functional alterations of human induced pluripotent stem cell-derived cardiomyocytes. J. Am. Heart Assoc. 2021;10:e022247. doi: 10.1161/JAHA.121.022247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed L.E., Vunjak-Novakovic G. Spaceflight bioreactor studies of cells and tissues. Adv. Space Biol. Med. 2002;8:177–195. doi: 10.1016/s1569-2574(02)08019-x. [DOI] [PubMed] [Google Scholar]
- Gan J., Tang F.M.K., Su X., Lu G., Xu J., Lee H.S.S., Lee K.K.H. microRNA-1 inhibits cardiomyocyte proliferation in mouse neonatal hearts by repressing CCND1 expression. Ann. Transl. Med. 2019;7:455. doi: 10.21037/atm.2019.08.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D., Wehland M., Pietsch J., Aleshcheva G., Wise P., van Loon J., Ulbrich C., Magnusson N.E., Infanger M., Bauer J. Growing tissues in real and simulated microgravity: new methods for tissue engineering. Tissue Eng. Part B Rev. 2014;20:555–566. doi: 10.1089/ten.TEB.2013.0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Pu W.T. Cardiomyocyte maturation: new phase in development. Circ. Res. 2020;126:1086–1106. doi: 10.1161/CIRCRESAHA.119.315862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y., Kuroda T., Kishimoto H., Wang C., Iwama A., Kimura K. Downregulation of rRNA transcription triggers cell differentiation. PLoS One. 2014;9:e98586. doi: 10.1371/journal.pone.0098586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holaska J.M., Rais-Bahrami S., Wilson K.L. Lmo7 is an emerin-binding protein that regulates the transcription of emerin and many other muscle-relevant genes. Hum. Mol. Genet. 2006;15:3459–3472. doi: 10.1093/hmg/ddl423. [DOI] [PubMed] [Google Scholar]
- Huang P., Russell A.L., Lefavor R., Durand N.C., James E., Harvey L., Zhang C., Countryman S., Stodieck L., Zubair A.C. Feasibility, potency, and safety of growing human mesenchymal stem cells in space for clinical application. NPJ Microgravity. 2020;6:16. doi: 10.1038/s41526-020-0106-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingber D. How cells (might) sense microgravity. FASEB J. 1999;13(Suppl):S3–S15. doi: 10.1096/fasebj.13.9001.s3. [DOI] [PubMed] [Google Scholar]
- Jha R., Singh M., Wu Q., Gentillon C., Preininger M.K., Xu C. Downregulation of LGR5 expression inhibits cardiomyocyte differentiation and potentiates endothelial differentiation from human pluripotent stem cells. Stem Cell Rep. 2017;9:513–527. doi: 10.1016/j.stemcr.2017.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha R., Wile B., Wu Q., Morris A.H., Maher K.O., Wagner M.B., Bao G., Xu C. Molecular beacon-based detection and isolation of working-type cardiomyocytes derived from human pluripotent stem cells. Biomaterials. 2015;50:176–185. doi: 10.1016/j.biomaterials.2015.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha R., Wu Q., Singh M., Preininger M.K., Han P., Ding G., Cho H.C., Jo H., Maher K.O., Wagner M.B., Xu C. Simulated microgravity and 3D culture enhance induction, viability, proliferation and differentiation of cardiac progenitors from human pluripotent stem cells. Sci. Rep. 2016;6:30956. doi: 10.1038/srep30956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y., Manabe T., Matsumoto M., Kajiume T., Matsumoto M., Yuge L. LIF-free embryonic stem cell culture in simulated microgravity. PLoS One. 2009;4:e6343. doi: 10.1371/journal.pone.0006343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laflamme M.A., Chen K.Y., Naumova A.V., Muskheli V., Fugate J.A., Dupras S.K., Reinecke H., Xu C., Hassanipour M., Police S., et al. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat. Biotechnol. 2007;25:1015–1024. doi: 10.1038/nbt1327. [DOI] [PubMed] [Google Scholar]
- Laflamme M.A., Murry C.E. Regenerating the heart. Nat. Biotechnol. 2005;23:845–856. doi: 10.1038/nbt1117. [DOI] [PubMed] [Google Scholar]
- Lan F., Lee A.S., Liang P., Sanchez-Freire V., Nguyen P.K., Wang L., Han L., Yen M., Wang Y., Sun N., et al. Abnormal calcium handling properties underlie familial hypertrophic cardiomyopathy pathology in patient-specific induced pluripotent stem cells. Cell Stem Cell. 2013;12:101–113. doi: 10.1016/j.stem.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang R., Raffi F.A.M. Dual-specificity phosphatases in immunity and infection: an update. Int. J. Mol. Sci. 2019;20:E2710. doi: 10.3390/ijms20112710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Ma Z., Niu Z., Qian H., Xuan D., Hou R., Ni L. NASA-approved rotary bioreactor enhances proliferation and osteogenesis of human periodontal ligament stem cells. Stem Cells Dev. 2009;18:1273–1282. doi: 10.1089/scd.2008.0371. [DOI] [PubMed] [Google Scholar]
- Li Y., Wang L., Zhang L., He Z., Feng G., Sun H., Wang J., Li Z., Liu C., Han J., et al. Cyclin B3 is required for metaphase to anaphase transition in oocyte meiosis I. J. Cell Biol. 2019;218:1553–1563. doi: 10.1083/jcb.201808088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian X., Hsiao C., Wilson G., Zhu K., Hazeltine L.B., Azarin S.M., Raval K.K., Zhang J., Kamp T.J., Palecek S.P. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc. Natl. Acad. Sci. USA. 2012;109:E1848–E1857. doi: 10.1073/pnas.1200250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R., Li D., Sun F., Rampoldi A., Maxwell J.T., Wu R., Fischbach P., Castellino S.M., Du Y., Fu H., et al. Melphalan induces cardiotoxicity through oxidative stress in cardiomyocytes derived from human induced pluripotent stem cells. Stem Cell Res. Ther. 2020;11:470. doi: 10.1186/s13287-020-01984-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., De la Cruz E., Gu X., Balint L., Oxendine-Burns M., Terrones T., Ma W., Kuo H.H., Lantz C., Bansal T., et al. Lymphoangiocrine signals promote cardiac growth and repair. Nature. 2020;588:705–711. doi: 10.1038/s41586-020-2998-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J.R., Dhawan A., Farach-Carson M.C. Modular proteoglycan perlecan/HSPG2: mutations, phenotypes, and functions. Genes. 2018;9:E556. doi: 10.3390/genes9110556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisenberg C., Tait P.S., Dianova I.I., Wright K., Edelmann M.J., Ternette N., Tasaki T., Kessler B.M., Parsons J.L., Kwon Y.T., Dianov G.L. Ubiquitin ligase UBR3 regulates cellular levels of the essential DNA repair protein APE1 and is required for genome stability. Nucleic Acids Res. 2012;40:701–711. doi: 10.1093/nar/gkr744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscu-Gregor A., Marschall C., Müntjes C., Schönecker A., Schuessler-Hahn F., Hohendanner F., Parwani A.S., Boldt L.H., Ott C.E., Bennewiz A., et al. Novel variants in TECRL cause recessive inherited CPVT type 3 with severe and variable clinical symptoms. J. Cardiovasc. Electrophysiol. 2020;31:1527–1535. doi: 10.1111/jce.14446. [DOI] [PubMed] [Google Scholar]
- Nguyen D.C., Hookway T.A., Wu Q., Jha R., Preininger M.K., Chen X., Easley C.A., Spearman P., Deshpande S.R., Maher K., et al. Microscale generation of cardiospheres promotes robust enrichment of cardiomyocytes derived from human pluripotent stem cells. Stem Cell Rep. 2014;3:260–268. doi: 10.1016/j.stemcr.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöss J., Ukena C., Kindermann I., Ehrlich P., Fuernau G., Ewen S., Mahfoud F., Kriechbaum S., Böhm M., Link A. Angiopoietin-2 and outcome in patients with acute decompensated heart failure. Clin. Res. Cardiol. 2015;104:380–387. doi: 10.1007/s00392-014-0787-y. [DOI] [PubMed] [Google Scholar]
- Preininger M.K., Jha R., Maxwell J.T., Wu Q., Singh M., Wang B., Dalal A., McEachin Z.T., Rossoll W., Hales C.M., et al. A human pluripotent stem cell model of catecholaminergic polymorphic ventricular tachycardia recapitulates patient-specific drug responses. Dis. Model. Mech. 2016;9:927–939. doi: 10.1242/dmm.026823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyo R.T., Sui J., Dhume A., Palomeque J., Blaxall B.C., Diaz G., Tunstead J., Logothetis D.E., Hajjar R.J., Schecter A.D. CXCR4 modulates contractility in adult cardiac myocytes. J. Mol. Cell. Cardiol. 2006;41:834–844. doi: 10.1016/j.yjmcc.2006.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampoldi A., Jha R., Fite J., Boland G., Xu C. Cryopreservation and CO2-independent culture of 3D cardiac progenitors for spaceflight experiments. Biomaterials. 2021;269:120673. doi: 10.1016/j.biomaterials.2021.120673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro I., Kawakami Y., Büscher D., Raya A., Rodríguez-León J., Morita M., Rodríguez Esteban C., Izpisúa Belmonte J.C. Tbx2 and Tbx3 regulate the dynamics of cell proliferation during heart remodeling. PLoS One. 2007;2:e398. doi: 10.1371/journal.pone.0000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro M., Tertoolen L., Guadix J., Bellin M., Kosmidis G., D'Aniello C., Monshouwer-Kloots J., Goumans M., Wang Y., Feinberg A., et al. Functional maturation of human pluripotent stem cell derived cardiomyocytes in vitro--correlation between contraction force and electrophysiology. Biomaterials. 2015;51 doi: 10.1016/j.biomaterials.2015.01.067. [DOI] [PubMed] [Google Scholar]
- Saraf A., Rampoldi A., Chao M., Li D., Armand L., Hwang H., Liu R., Jha R., Fu H., Maxwell J.T., Xu C. Functional and molecular effects of TNF-alpha on human iPSC-derived cardiomyocytes. Stem Cell Res. 2021;52:102218. doi: 10.1016/j.scr.2021.102218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz A., Plate K.H., Reiss Y. Angiopoietin-2: a multifaceted cytokine that functions in both angiogenesis and inflammation. Ann. N. Y. Acad. Sci. 2015;1347:45–51. doi: 10.1111/nyas.12726. [DOI] [PubMed] [Google Scholar]
- Sharma A., Clemens R.A., Garcia O., Taylor D.L., Wagner N.L., Shepard K.A., Gupta A., Malany S., Grodzinsky A.J., Kearns-Jonker M., et al. Biomanufacturing in low Earth orbit for regenerative medicine. Stem Cell Rep. 2022;17:1–13. doi: 10.1016/j.stemcr.2021.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., Gan P., Wang K., Darehzereshki A., Wang K., Kumar S.R., Lien C.L., Patterson M., Tao G., Sucov H.M. Mononuclear diploid cardiomyocytes support neonatal mouse heart regeneration in response to paracrine IGF2 signaling. Elife. 2020;9:e53071. doi: 10.7554/eLife.53071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simone T.M., Higgins C.E., Czekay R.P., Law B.K., Higgins S.P., Archambeault J., Kutz S.M., Higgins P.J. SERPINE1: a molecular switch in the proliferation-migration dichotomy in wound-"Activated" keratinocytes. Adv. Wound Care. 2014;3:281–290. doi: 10.1089/wound.2013.0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamargo J., Caballero R., Gómez R., Valenzuela C., Delpón E. Pharmacology of cardiac potassium channels. Cardiovasc. Res. 2004;62:9–33. doi: 10.1016/j.cardiores.2003.12.026. [DOI] [PubMed] [Google Scholar]
- Tao G., Levay A.K., Peacock J.D., Huk D.J., Both S.N., Purcell N.H., Pinto J.R., Galantowicz M.L., Koch M., Lucchesi P.A., et al. Collagen XIV is important for growth and structural integrity of the myocardium. J. Mol. Cell. Cardiol. 2012;53:626–638. doi: 10.1016/j.yjmcc.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unsworth B.R., Lelkes P.I. Growing tissues in microgravity. Nat. Med. 1998;4:901–907. doi: 10.1038/nm0898-901. [DOI] [PubMed] [Google Scholar]
- Wnorowski A., Sharma A., Chen H., Wu H., Shao N.Y., Sayed N., Liu C., Countryman S., Stodieck L.S., Rubins K.H., et al. Effects of spaceflight on human induced pluripotent stem cell-derived cardiomyocyte structure and function. Stem Cell Rep. 2019;13:960–969. doi: 10.1016/j.stemcr.2019.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuge L., Kajiume T., Tahara H., Kawahara Y., Umeda C., Yoshimoto R., Wu S.L., Yamaoka K., Asashima M., Kataoka K., Ide T. Microgravity potentiates stem cell proliferation while sustaining the capability of differentiation. Stem Cells Dev. 2006;15:921–929. doi: 10.1089/scd.2006.15.921. [DOI] [PubMed] [Google Scholar]
- Zhang S., Liu P., Chen L., Wang Y., Wang Z., Zhang B. The effects of spheroid formation of adipose-derived stem cells in a microgravity bioreactor on stemness properties and therapeutic potential. Biomaterials. 2015;41:15–25. doi: 10.1016/j.biomaterials.2014.11.019. [DOI] [PubMed] [Google Scholar]
- Zhang S., Zhang Y., Chen L., Liu T., Li Y., Wang Y., Geng Y. Efficient large-scale generation of functional hepatocytes from mouse embryonic stem cells grown in a rotating bioreactor with exogenous growth factors and hormones. Stem Cell Res. Ther. 2013;4:145. doi: 10.1186/scrt356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W., Zhao M., Mattapally S., Chen S., Zhang J. CCND2 overexpression enhances the regenerative potency of human induced pluripotent stem cell-derived cardiomyocytes: remuscularization of injured ventricle. Circ. Res. 2018;122:88–96. doi: 10.1161/CIRCRESAHA.117.311504. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for RNA-seq data reported in this paper is GEO: GSE188793.