Abstract
Estrogen and estrogen receptor alpha (ERα) have been implicated in systemic lupus erythematosus (SLE) pathogenesis. ERα signaling influences dendritic cell (DC) development and function, as well as inflammation and downstream immune responses. We previously reported that ERα modulates multiple TLR-stimulated pathways in both conventional and plasmacytoid DCs in lupus-prone mice. For example, CD11chiMHCII+ cell numbers are reduced in mice with global ERα deficiency or when expressing a short variant of ERα. Herein, RNA-seq analysis of CD11chi cells from bone marrow of NZM2410 mice expressing WT ERα vs. ERα short vs. ERα null revealed differentially expressed complement genes, interferon-related genes and cytokine signaling (e.g., IL-17 and Th17 pathways). To better understand the role of ERα in CD11c+ cells, lupus prone NZM2410 mice with selective deletion of the Esr1 gene in CD11c+ cells were generated. Phenotype and survival of these mice were similar with the exception of Cre positive (CrePos) female mice. CrePos females, but not males, all died unexpectedly prior to 35 weeks. DC subsets were not significantly different between groups. Since ERα is necessary for robust development of DCs, this result suggests that DC fate was determined prior to CD11c expression and subsequent ERα deletion (i.e. proximally in DC ontogeny). Overall, findings point to a clear functional role for ERα in regulating cytokine signaling and inflammation, suggesting that further study into ERα-mediated regulatory mechanisms in DCs and other immune cell types is warranted.
Keywords: Systemic lupus erythematosus (SLE), estrogen receptor alpha (ERα), cytokines, Toll-like receptors, dendritic cells
Introduction
A variety of risk factors including genetic predisposition, environmental exposures and hormonal status lead to the development of SLE. Being female is a risk factor for developing autoimmune diseases (SLE, Sjogren’s syndrome, systemic sclerosis, among others)1, 2. In SLE, greater than 80% of those diagnosed are women of reproductive age, when most hormonally active1, 3, suggesting sex hormones and their receptors are important to examine for underlying mechanisms of disease risk in females. Estrogen, and estrogen receptor alpha (ERα) in particular, have been implicated in SLE disease pathogenesis4–7; however, the data at times have been contradictory and precise mechanisms have yet to be elucidated.
Estrogen receptors are expressed in a variety of innate and adaptive immune cells, including dendritic cells (DCs), macrophages, monocytes, natural killer cells, as well as B and T cells8, 9 and have variable influences on immune cell development and function. While the effects of some sex hormones can be described as either generally anti-inflammatory (androgen and progesterone)10, 11 or proinflammatory (prolactin)12, estrogens produce highly pleiotropic immune effects, depending on concentration, timing, cell/tissue type, and disease state, among other variables13. The mechanisms by which high and low physiological doses of estradiol differentially alter ER activity to modulate immune responses are not well-defined8. One possibility is that distinct ER-containing transcriptional complexes (with coactivators/corepressors) are formed in response to varying hormone levels leading to differential expression of genes that promote or dampen inflammation. ERα variant expression is a another contributor to the diversity of responses. Binding partners and transcriptional machinery are likely also cell-type specific. Overall, ER expression and function are dependent on cellular milieu, and responses are heterogeneous.
Adding to the complexity, timing of estrogen signaling differentially impacts immune responses (ex. impacts on genes are different at early vs. late time-points throughout cellular development and function). For example, SLE patient B cells have distinct epigenetic profiles, influenced by early growth response (EGR) transcription factors that impact both early and late estrogen response gene sets19. In innate immune cells, ERα signaling stimulates the early development of both conventional and plasmacytoid dendritic cells16. CD11b+ DC differentiation is driven by ERα signaling, inducing IRF4 in the early stages of myeloid cell development20. Conversely, ERα signaling in mature DCs (late effects) inhibits certain DC functions. DC treatment with estrogen/SERMs impairs their ability to upregulate co-stimulatory molecules (ex. CD80/86 and CD40). They are less effective antigen presenting cells compared to control DCs21. Multiple other studies have shown the impact of ERα signaling on proinflammatory cytokine expression in DCs8, 18, 22.
Our previous studies demonstrated an important role for ERα and ERα variants in lupus disease expression in lupus prone mice (NZM2410 and MRL/lpr)23, 24. While ERβ knockout had no effect on disease, female mice with a complete knockout of ERα were protected from disease, but only if they were not ovariectomized (likely due to high testosterone levels from hypergonadism of ovary-intact females)25. Of note, an important feature of all global ERα knockouts is aberrant sex hormone levels due to loss of endocrine feedback loops that negatively regulate pituitary gonadotropin secretion. These mice have elevated levels of both estrogen and testosterone (females have levels akin to males) as well as low prolactin26–28. All of these sex hormones have significant immune modulating capacity (and ERα-mediated consequences) and must be controlled for experimentally. We recently demonstrated that female mice expressing an N-terminally truncated ERα (structurally similar to a naturally ocurring short ERα variant) exhibited significantly less lupus renal disease and increased survival of female mice. Estrogen was required for this protective phenotype29. These data revealed that ERα deficiency by itself was not protective, but that, in the presence of estrogen, an ERα short variant provided protection. Multiple Toll-like receptor (TLR)-stimulated immune responses were significantly altered in B cells, mesangial cells and DCs depending on the presence or absence of the full-length ERα26, 30.
In this study, RNA-seq experiments were undertaken to further examine the role of ERα in DCs (comparing DCs expressing WT ERα vs. ERα short vs. ERα null). In addition, to better understand the effects of ERα on DCs in lupus disease expression, a tissue-specific knockout of ERα in CD11c+ cells in NZM2410 mice was created. Results of this study revealed differences in immune-related genes and pathways, including complement genes, cytokines and genes that impact interferon, all of which are modulated by ERα. These results suggest that differential expression of ERα and its variants in DCs likely impact autoimmune disease risk in females. Further mechanistic studies are needed to understand the complex nature of ERα effects and the differential expression of ERα variants.
Materials and Methods
Mice
Mice were maintained at the Ralph H. Johnson VAMC Animal Facility (Charleston, SC). Animal protocols followed the principles outlined in the Guide for the Care and Use of Laboratory Animals, and were approved by the VA’s IACUC. Mice were maintained on a 12-hr. light/dark cycle with access to food and water ad libitum. The NZM2410 mouse strain was originally purchased from Jackson Laboratory (Bar Harbor, ME). Mice carrying the Esr1tm4.2Ksk allele (Stock No. 026176, The Jackson Laboratory) are ERα null, and have no tissue responses to estrogen or estrogen receptor alpha activity27. ERαKO mice (ERα short) on the C57BL/6 background were a kind gift of Dr. Ken Korach (NIEHS) and are also available commercially (Stock No. 004744, The Jackson Laboratory). In contrast to the ERα null strain, mice carrying the Esr1tm1Ksk allele express a truncated form of ERα (ERα short) and have residual estrogen responsivity. Mice were backcrossed onto the NZM2410 background for 10 generations and have different lupus disease phenotypes29. Mice with global disruption of ERα were ovariectomized (OVX) pre-disease at 4–8 weeks of age (peri-puberty) in order to combat hypergonadism (high testosterone, high estrogen) that results from loss of normal endocrine feedback to the pituitary27. These groups subsequently received 0.25 mg, 90-day sustained release 17β-estradiol pellet (Innovative Research of America, Sarasota, FL, USA) implanted sub-dermally to replete physiologic E2 levels31. For RNA-seq, experimental mice (n=12) were female and littermates when possible. Mice were sacrificed at 10–13 weeks and bone marrow-derived dendritic cells (BM-DCs) generated as outlined below.
The floxed-estrogen receptor alpha (ERα) mice and Cre-CD11c mice were generous gifts from Dr. Ken Korach (NIEHS, Raleigh, NC) and Dr. Zihai Li (The Ohio State University, Columbus, OH), respectively. Floxed-ERα (ERαflox/flox) mice and Cre-CD11c mice were each backcrossed onto NZM2410 lupus-prone background for 10 generations. ERα expression was specifically disrupted in CD11chi cells using this Cre/LoxP system, creating the experimental CrePos (conditional ERα−/−) and CreNeg animals. A survival study contained both male and female experimental mice (cohort 1, n=26). For the phenotypic study (cohort 2, n=29), only female mice were used. Littermates were used when possible. Mice were sacrificed when they reached sacrifice requirements: >10% loss of weight, >500mg urine protein as assessed by dipstick, upon recommendation by the animal facility, or >52 weeks for the survival experiment, and 9–26 weeks for the phenotypic study: pre-disease (9–13 weeks) and disease (23–26 weeks).
Genotyping
The following primer pairs were utilized to confirm the genotypes of the mice: floxed-ERα null mice - N6delcKF (5’ GACTCGCTACTGTGCCGTGTGC 3’) and N6del3R (5’ CTTCCCTGGCATTACCACTTCTCCT 3’) and Cre-CD11c - CD11c-Cre Forward (5’ ACTTGGCAGCTGTCTCCAAG 3’) and CD11c-Cre Reverse (5’ GCGAACATCTTCAGGTTCTG 3’). DNA was isolated from tail snips of 4 week-old mice using DirectPCR (Tail) Lysis Reagent and Proteinase K solution (Viagen, Los Angeles, CA). DNA samples were incubated at 56°C overnight with a final digestion at 95°C for ten minutes. PCR conditions for the Floxed-ERα null (−/−) mice are as follows: 94°C for 1 minute, with 30 cycles of 94°C for 30 seconds, 60°C for 30 seconds, 72°C for 30 seconds, and a final cycle of 72°C for 5 minutes. A 275bp fragment indicates the presence of the WT allele and the Flox-ERα band is detected at 475bp. PCR conditions for genotyping the Cre-CD11c mice were: 95°C for 3 minutes, 35 cycles of 95°C for 30 seconds, 60°C for 1 minute, 72°C for 1 minute, and one cycle of 72°C for 5 minutes. The Cre-CD11c transgene was observed at approximately 313bp. All experimental mice were confirmed to have the Floxed-ERα null (−/−) genotype and were categorized based on their being positive or negative for Cre-CD11c. Conditional ERα−/− mice were validated using quantitative Real-Time PCR. CD11chi BM-DCs were sorted by FACS. RNA was isolated using the Qiagen RNeasy kit (Qiagen, Germantown, MD) and cDNA synthesized with the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA). Real-Time PCR was performed using the iQ SYBR Green Supermix (Bio-Rad, Hercules, CA), and Esr1 and GAPDH primers (Qiagen, Germantown, MD). PCR was carried out using the CFX connect Real-time PCR Detection System (Bio-Rad) and relative expression was determined using the delta CT method.
Bone marrow-derived dendritic cells
Bone marrow (BM) was flushed from the femurs and tibias of mice, dissociated through a 70μm strainer, and depleted of red blood cells using red blood cell lysis buffer (144mM NH4Cl and 17mM Tris, pH 7.6). Cells are spun immediately (3min low speed spin) and resuspended in cold PBS. To generate BM-DCs for RNA-seq, total BM was plated at a concentration of 1.5×106 cells/ml in RPMI 1640 media containing 10% FBS, 1% penicillin/streptomycin/ampicillin, and L-glutamine with 20 ng/ml GM-CSF and 10 ng/ml IL-4. Media and cytokines were replenished on day 3 and BM-DCs were harvested on day 7. For flow cytometry experiments, BM-DCs were generated using 10% supernatant from a Flt3L-producing cell line (a kind gift of Dr. Stephania Gallucci, Temple University, Philadelphia, PA) in complete RPMI 1640 media. BM-DCs were harvested on day 7, and washed with PBS prior to staining.
RNA-sequencing and Heatmap visualization
CD11chi BM-DCs were generated from n=3 pooled mice per group and sorted by FACS. Experimental triplicates were treated with vehicle or loxoribine (200 μmol/ml), a TLR7 agonist, and CpG (1 μg/ml), a TLR9 agonist (Invivogen, San Diego, CA) for 24 hours. Total RNA (100–200 ng) was used to prepare RNA-Seq libraries using the TruSeq RNA Sample Prep Kit (Illumina, CA, USA), following the protocol described by the manufacturer. Paired-end RNA sequencing was performed using an Illumina HiSeq2500 in the MUSC Center for Genomic Medicine (CGM), with each sample sequenced to a minimum depth of ~50 million reads. Illumina Casava1.8 software used for base calling.
Two types of heatmaps were generated. First, three sets of comparisons were analyzed, using only TLR-stimulated BM-DCs from NZM2410 mice, including 1) WT vs. ERα short; 2) ERα short OVX vs. ERα null OVX; and 3) ERα short OVX vs. ERα short. The comparison criteria were utilized to control for the fact that not all of the animals were OVX’ed for the sequencing experiments. The resulting intersection of all three comparisons should control for hormonal changes due to OVX vs non-OVX’ed animals. To accomplish this, genes with absolute log2 fold change greater than 1 for the first two comparisons were selected and then those with absolute log fold change less than 1 for the third comparison were included. The genes that intersected all three lists were included in Fig. 1A. The heatmap was generated using log10-transformed Fragments Per Kilobase of transcript per Million mapped reads (FPKM) using the pheatmap() function in the R package pheatmap. For the second set of heatmaps in Fig. 1B, WT vs. ERα null OVX TLR was exclusively compared using the absolute degree of expression, arranged in descending order by absolute fold change between the two strains. The heatmap was generated as described above.
Figure 1 – Cytokines and cytokine signaling altered in CD11chi lupus prone NZM2410 mice expressing different forms of ERα.
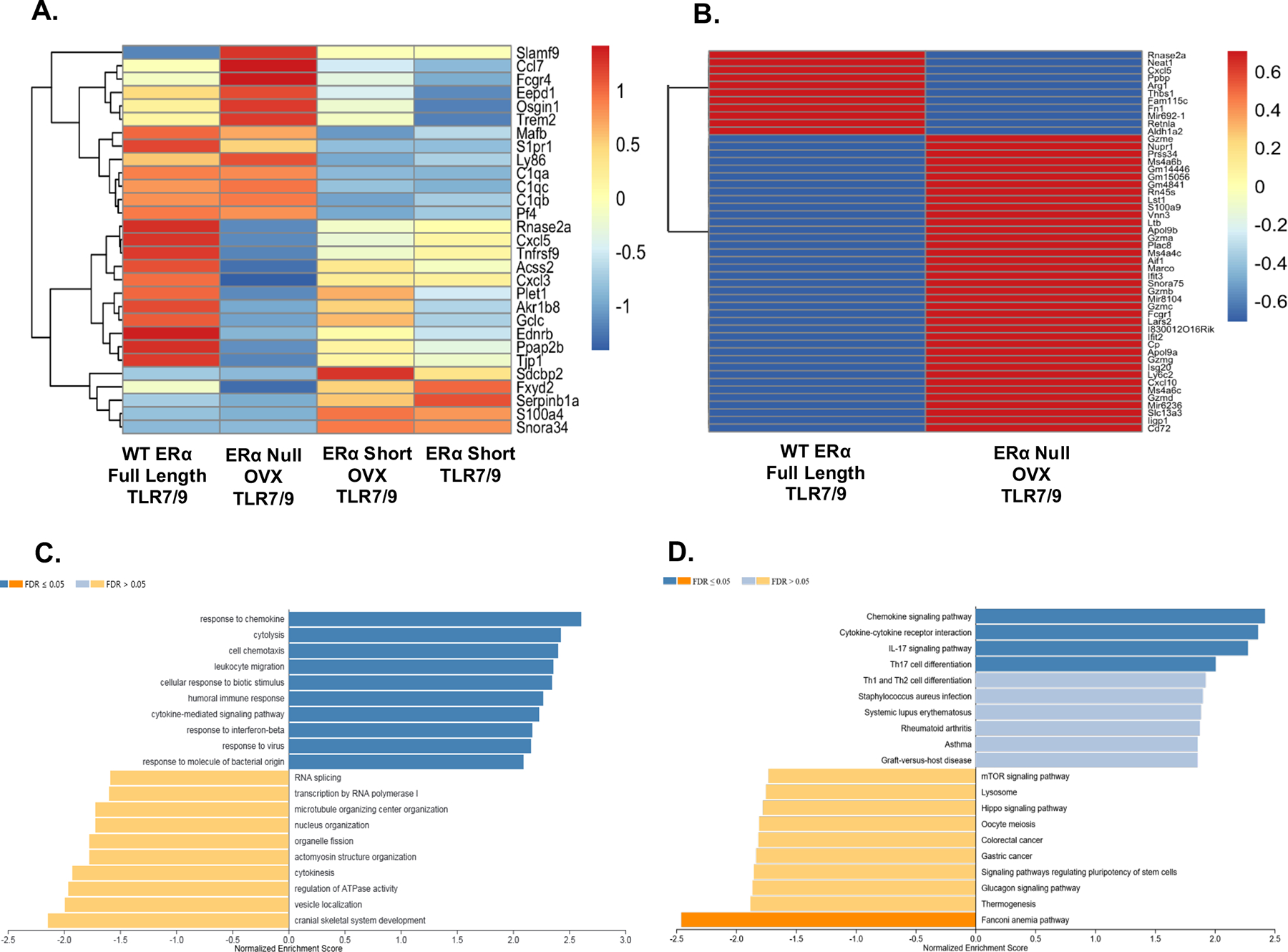
Heatmaps revealing differentially expressed genes (DEGs) in CD11c hi cells isolated from lupus mouse strains with different forms of ERα. CD11chi cells were derived from murine bone marrow cultures with GM-CSF and IL-4. Panel A depicts DEGs across all four strains of mice (with and without ovariectomy). Values are in log10-based FPKM. All cells were stimulated with TLR7/9 and hormonal differences were accounted for in the analysis. Panel B shows the DEGs across WT and ERα null only. The same parameters used in Panel A apply to Panel B. The bar plot in Panel C depicts the GO analysis results for genes between WT and ERα null that have an absolute fold change over 1 and included a total of 1912 genes. Panel D represents the results of the KEGG pathway analysis using the same set of genes as in Panel C. For Panels C and D the WT group was used as the reference gene set.
Gene set enrichment analysis
Gene set enrichment analysis was completed for the WT TLR vs. ERα null OVX TLR comparison using the WebGestalt (WEB-based Gene SeT AnaLysis Toolkit: http://www.webgestalt.org/#). Specifically, we implemented the module Gene Set Enrichment Analysis (GSEA) based on Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotations, using the genes with absolute log2 fold change greater than 1 (Figure 1C and D). In addition, we also implemented GSEA GO analysis using all the genes, where absolute log2 fold changes were used as scores (Supplementary Figure 1).
Cytokine Multiplexing
Magnet bead-based Luminex multiplex assays with 14 analytes were purchased from R&D Systems (Minneapolis, MN). Analytes assessed were GM-CSF, TNFα, IL-12p70, IL-1β, IL-4, IL-6, IL-17a, IFNγ, G-CSF, IL-23p19, M-CSF, IL-1α, BAFF, and IL-9. Serum samples were taken from WT NZM2410 mice (n=11), NZM ERα short mice (n=12), NZM ERα short mice +E2 (n=8), and NZM ERα null mice +E2 (n=6) at the time of death (32 weeks or when renal failure occurred). The assay was completed according to manufacturer’s protocol and perfomed on a FlexMap 3D instrument (Luminex, Austin, TX).
Staining and flow cytometry
BM-DCs and spleen cells were resuspended (4×106 per sample) in staining buffer (0.5% BSA, 0.02% sodium azide in 1X PBS) and stained with one of two panels. Panel 1: CD11c-Brilliant violet 605 (1:100), CD8a-Brilliant violet 421 (1:100), CD11b-PE (1:400), MHCII-APC (1:200), Siglec H-PerCP/Cy5.5 (1:100), B220-PE/Cy7 (1:100), F4/80-BV510 (1:200) CD3/CD19-FITC (1:200; both stained with FITC in order to remove populations for analysis) and Live/Dead-APC/Cy7. Panel 2: F4/80-Brilliant violet 421 (1:100), CD19-PerCP/Cy5.5 (1:100), CD3-Brilliant violet 605 (1:100), CD49b-PE (1:400), pDCA1-APC (1:200), and Live/Dead-APC/Cy7. All antibodies were purchased from Biolegend (San Diego, CA). Cells were incubated with antibodies in the dark for 30 minutes on ice. Viability was assessed using LIVE/DEAD Fixable Dead Cell stain (Life Technologies, Carlsbad, CA) at a concentration of 1μl/million cells. Cells were washed twice with staining buffer and resuspended in 300μL of staining buffer for flow cytometry. Cells were acquired on an LSRFortessa cell analyzer (BD Biosciences, San Jose, CA) and analysis was performed using FlowJo software (FlowJo LLC, Ashland, Oregon). For real-time PCR assays, BM-DCs were sorted on CD11chi cells via a BDFacs Aria sorter (BD Biosciences, San Jose, CA).
Serum anti-dsDNA and proteinuria measurements
Serum was collected at 25- and 35-week time points and at time of sacrifice. Serum anti-dsDNA was measured by an in-house ELISA assay as described previously (23). Mice were housed in metabolic cages for 24-hour urine collection at 24–25 week and 35-week time points, when possible. To prevent bacterial growth, antibiotics (ampicillin 25ug/mL, gentamicin 50 ug/mL, chloramphenicol 200 ug/mL) were added to the collection tube. After 24 hours, urine quantity was determined, and samples were frozen at −20°C for future analysis via mouse albumin ELISA with known standards.
Statistics
See above for statistics related to RNA-seq analysis. Log rank (Mantel-Cox) analysis was used to compare trends in animal survival, except for median time of death (two-tailed t test). For other experiments, depending on whether the normality assumption is violated or not, either Kruskal-Wallis test with post-hoc Dunn’s multiple comparison test, or one-way analysis of variance (ANOVA) with post-hoc Tukey’s test for multiple comparisons, were utilized to test for significance. Standard error of the means was reported where applicable. P values < 0.05 were considered significant.
Results
The impact of ERα on gene expression in CD11chi cells of lupus prone NZM2410 mice
We and others have demonstrated a critical role for ERα in murine DC development and function. We examined the impact of ERα on gene expression in CD11chi BM-DCs to obtain a broader understanding of ERα impact on inflammatory-type DCs (generated with GM-CSF and IL-4). RNA-seq was performed on BM-DCs from two sets of female lupus prone NZM2410 mice expressing full length ERα vs. an ERα short mutant (missing the AF-1 activation domain) vs. a global knockout of ERα (ERα null). The first set included TLR7/9-treated BM-DCs from NZM WT and NZM ERα short mice. To control for the hormonal impact of ERα disruption in mice (e.g., hypergonadism resulting in supra-physiologic levels of estrogen and testosterone), a second set of female ERα short and ERα null mice were ovariectomized (OVX’ed) and E2-repleted. As in the first set, BM-DCs were cultured and TLR7/9-treated prior to RNA-seq. Thus, ERα expression was either present, present in the truncated form, or deleted completely from the CD11chi BM-DCs. As described in the methods section, comparisons were made across the four sets of female mice under a strict set of guidelines to control for known hormonal impacts.
The heatmap shown in Fig. 1A depicts the results of a comparison of sorted CD11chi cells across all four sets of mice and shows a total of the 29 most highly differentially expressed genes (DEG)s. Several genes that impact both the innate and adaptive immune system differed in expression across all four sets of mice, including Slamf9, Fcgr4, complement factors C1qa, C1qb and C1qc, as well as cytokines Ccl7, Cxcl5 and Cxcl3. Distinct differences were observed between ERα short compared with both WT ERα and ERα null mice, supporting the theory that a truncated version (AF-1 mutant) of ERα differentially and significantly impacts the immune system. Fig. 1B shows a direct comparison of DEGs between WT and ERα null mice, to further illustrate the impact of a complete deletion of ERα on DCs. Multiple genes important to the function of the innate and adaptive immune system were differentially expressed between WT and ERα null mice. Cytokines (Cxcl5 and Cxcl10), several granzymes, Arg1 which impacts immune defense and immune cell regulation, and interferon related genes Ifit2, Ifit3, and Apol9, were all differentially expressed between WT and ERα null. GO and KEGG pathway analyses were performed on the WT vs. ERα null comparisons, with WT being the reference set, and are shown in Fig. 1C and 1D. Gene sets involved with several immune processes including response to chemokine, cytolysis, cytokine-mediated signaling pathway and response to interferon beta were enriched, whereas gene sets involved in more basic biological processes, such as regulation of ATPase and vesicle localization were down regulated (Fig. 1C). KEGG pathway analysis revealed similar results, with immunological pathways associated with cytokine signaling enriched and biological processes (ex. glucagon signaling) down regulated (Fig. 1D). Of note, upregulated pathways included those associated with autoimmune disease and down regulated pathways were associated with cancer, diseases that can sometimes have rival immune effects. Interestingly, the mTOR pathway, which is known to be activated in SLE was also down regulated in ERα null mice by KEGG analysis (Fig. 1D).
Cytokine protein levels vary in mice expressing different forms of ERα
Based on the number of differentially expressed cytokines and the prevalence of cytokine gene sets and pathways observed by RNA-seq, we examined the protein levels for a select number of cytokines in these lupus mouse strains using multiplex technology. Serum taken from all female OVX’ed mice expressing intact ERα, ERα short or ERα null with E2 repletion, was utilized to determine cytokine expression levels. Cytokines were expressed at different levels across ERα genotypes, also reflecting diverse roles for full length (WT) ERα vs. ERα short (Fig. 2). Consistent with our RNA-seq data, IL-17a expression and production was greatly impacted by the absence of ERα. However, the results were opposite for mRNA levels vs. protein levels for IL-17. The IL-17 inflammatory pathway was upregulated in in ERα null mice in the RNAseq experiment, but IL-17 protein was undetectable in these mice. Message levels of IL-17 may be induced in response to lack of negative feedback (since the IL-17 cytokine is missing in these lupus-prone mice, despite being in an inflamed disease state). Thus, these results suggest that ERα is critical for regulating the IL-17 pathway and will be further investigated in future studies.
Figure 2 – Cytokine protein expression levels in NZM2410 mice with variable forms of ERα expression ± E2.
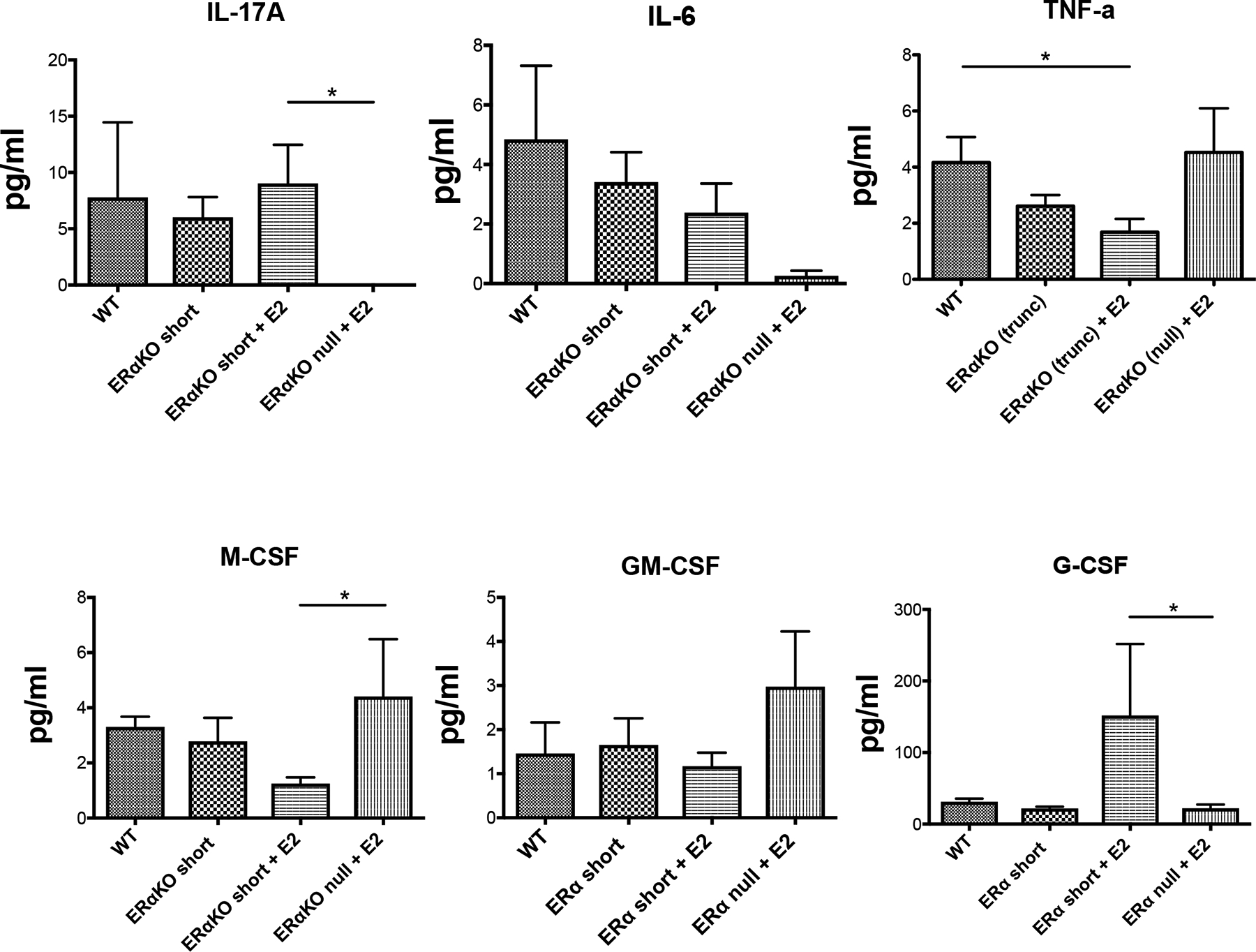
Cytokine protein expression levels were determined using a multiplex array (FlexMap). Observed results in pg/ml for IL-17A (Panel A), IL-6 (Panel B), TNFα (Panel C), M-CSF (Panel D), GM-CSF (Panel E), and G-CSF (Panel F) are displayed. All mice were ovariectomized (pre-pubertal), and serum samples were taken at sacrifice. Significance is based on a t-test, with the exception of Panel C, whose results were significant based on ANOVA with post-hoc Tukey’s test for multiple comparisons.
IL-6 levels were also decreased in ERα null mice, although this did not reach significance. Few differences were observed in the cytokine protein levels between WT and ERα short if no ligand was available indicating an active role for ERα short in modulating cytokine release (rather than being a result of deficiency of full length ERα). With the addition of E2, TNFα levels were significantly decreased in NZM ERα short compared to WT, but unaffected in ERα null mice (Fig. 2C). A statistically significant decrease in M-CSF protein expression was also observed in E2-replete ERα short-expressing mice compared to ERα null mice, and G-CSF was coordinately significantly decreased in ERα null mice (Fig. 2D and F). GM-CSF levels were increased in the ERα null animals but did not rise to significance (Fig. 2E). In most cases, the effect of ERα null resulted in cytokine levels that were opposite of those observed for ERα short. Overall, these results illustrate the stark differences between the presence and absence of ERα in immune responses in females. They also illustrate differences mediated by the short ERα variant, whose role may be to modulate the pro-inflammatory actions of full length ERα.
Generation and survival of DC-specific ERα−/− NZM2410 lupus prone mice
In order to better understand the function of ERα in DCs of lupus prone mice, experimental CrePos (Cre-CD11c × floxed-ERα) and CreNeg lupus prone mice were created to study selective disruption of the Esr1 gene in NZM2410 cDCs. Disruption of the Esr1 gene was determined by measuring mRNA expression in the CrePos and CreNeg animals. Both male and female mice were studied. Figure 3A demonstrates a large and statistically significant reduction in expression of the Esr1 gene in CD11chi bone marrow-derived DCs from the CrePos animals compared to CreNeg animals. Expression of the Esr1 gene was more than 90% reduced in CrePos BM-DCs that were isolated by fluorescence-activated cell sorting (FACS), gated on CD11chi cells. There was no statistically significant difference in the overall survival rate between CrePos and CreNeg (floxed-ERα) mice (Fig. 3B, C) when considering both males and females. However, CrePos female mice died significantly earlier than the CreNeg female mice (Fig. 3B, C). None of the CrePos female mice survived past 35 weeks, while nearly half of CreNeg females were alive at 52 weeks (cohort 1).
Figure 3 -. Selective disruption of ERα expression in dendritic cells (CD11c-Cre/ERαflox/flox) of NZM2410 lupus-prone mice.
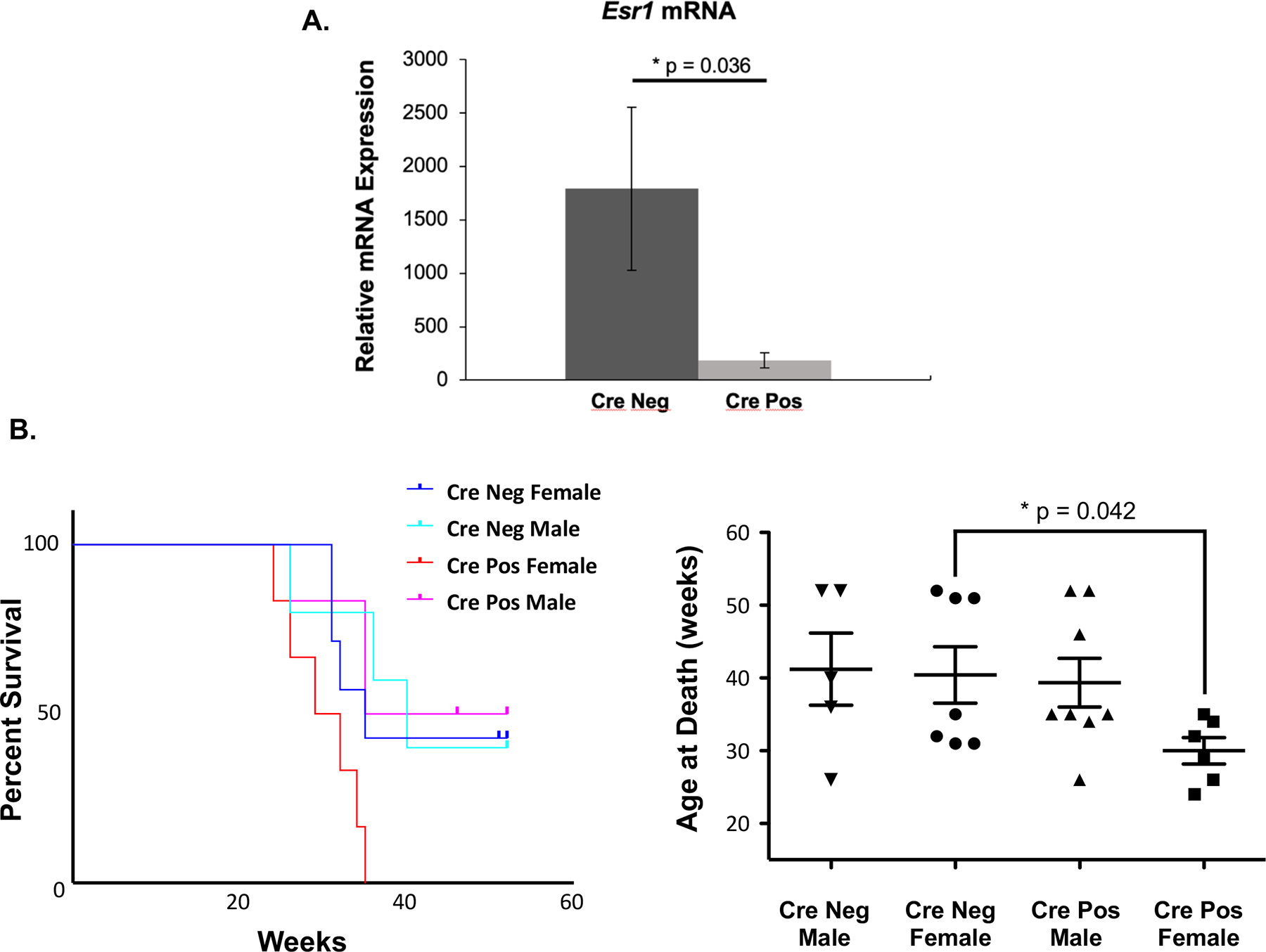
(A) Relative mRNA expression of the Esr1 gene in CD11chi cells isolated from CrePos (CD11c-Cre/ERαflox/flox) and CreNeg (CD11c-Cre/WTloxP-ERα) mice. Total RNA was isolated from flow sorted BM-DCs from female mice 20–27 weeks of age (n=3 for each group). Real time PCR was performed in triplicate on transcribed cDNA with primers to the Esr1 gene and normalized to Gapdh housekeeping gene. Survival of CrePos and CreNeg mice in Cohort 1 by sex (n=26) illustrated by Kapalan-Meier curve (B) and scatter-plot of individual mouse groups (C). Pre-determined sacrifice at 52 weeks.
Characterization of the disease phenotype in NZM2410 lupus-prone CD11c-Cre/ERαflox/flox mice
Body and spleen weight, autoantibody production and 24h proteinuria were measured in male and female NZM CD11c-Cre/ERαflox/flox mice, both in the survivors of the first cohort and in a second cohort with a pre-determined sacrifice date. Despite an accelerated pattern of death in female CrePos mice, no differences were observed in body or spleen weight between CrePos and CreNeg female mice (Fig. 4A). Additionally, no significant differences were observed in serum autoantibody levels between CrePos and CreNeg mice (female and male mice). Interestingly, male CrePos mice had significantly increased autoantibody production compared to female CrePos mice (Fig. 4B). Of note, autoantibody levels did not parallel disease activity in these mice. For example, autoantibody levels were similar between CrePos and CreNeg female mice (Fig. 4B), while survival between those groups was significantly different (Fig. 3). This result again suggests an uncoupling of autoantibody development (autoimmunity/loss of tolerance) and lupus disease expression (ex. nephritis) in the setting of altered sex hormone signaling, as we have seen in our other studies23, 25, 29. With regard to proteinuria as a reflection of nephritis, a trend towards more severe proteinuria was observed in the CrePos mice, which was driven by a few mice and the results were not statistically significant (Fig. 4C). Since most of the CrePos female mice died unexpectedly prior to 35 weeks, we were unable to obtain 24 urine albumin data to help determine renal impact on these mice (mice either died in cage or were not healthy enough to undergo 24h metabolic cage collection).
Figure 4 – Characterization of disease phenotype in NZM2410 lupus-prone CD11c-Cre/ERαflox/flox mice.
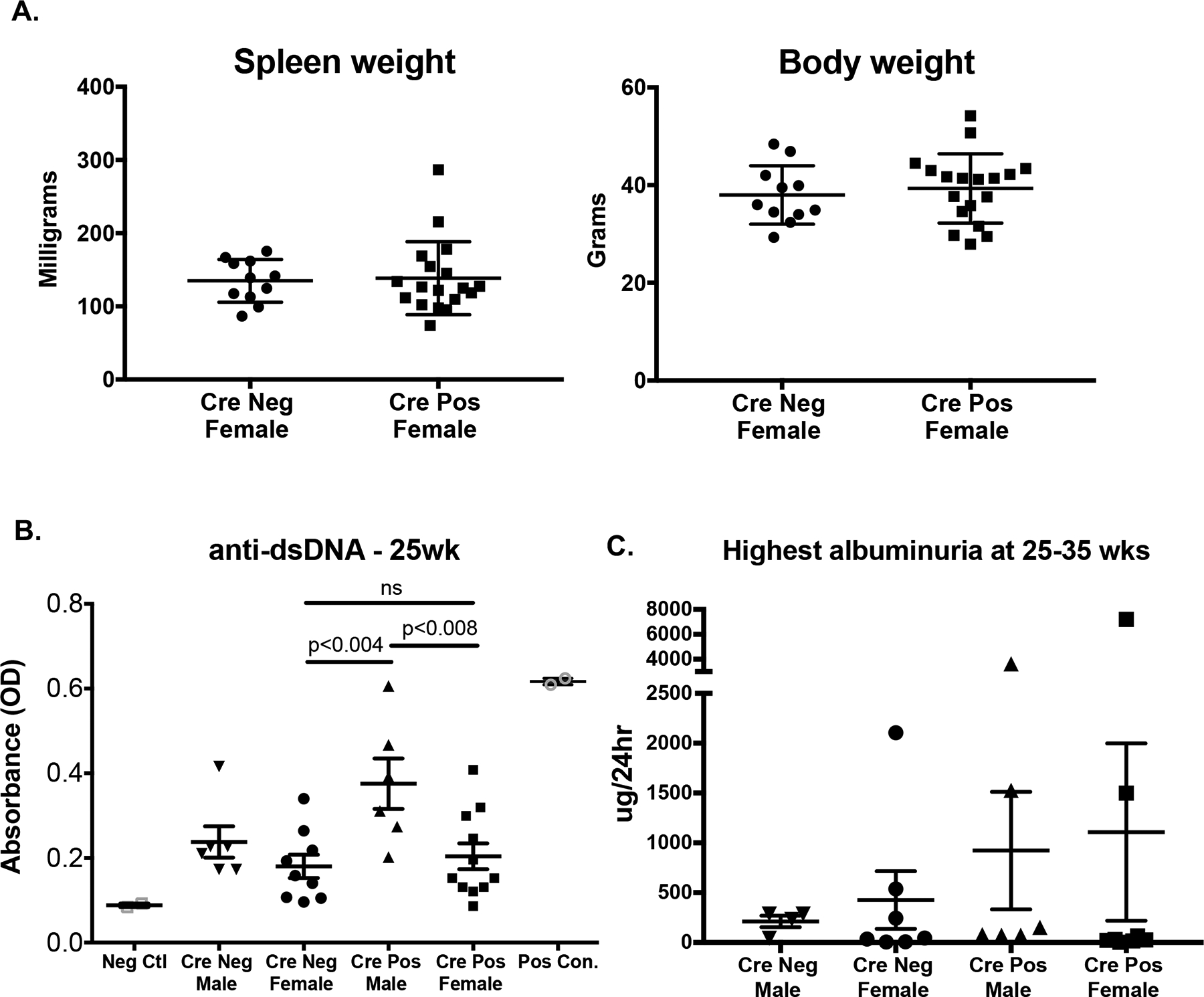
Data was collected from Cohort 1 (male and female, as above) plus a second all-female cohort sacrificed at 25 weeks. (A) Body and spleen weight for CrePos and CreNeg female mice. (B) serum anti-dsDNA measured by ELISA for all mice at the 25 week time point. Positive controls from diseased MRL/lpr mice (open grey circles) and negative controls from C57BL/6J mice (open grey square) are included. Panel C demonstrates albuminuria in mice ages 25–35 weeks old (highest recorded) as measured by 24hr urine collection on metabolic cages.
Determination of spleen cell immunophenotype in female NZM2410 lupus-prone CD11c-Cre/ERαflox/flox mice
Given the significant difference in female survival rate (Fig. 3B) despite the lack of other observable phenotypic differences, we investigated the impact of ERα deletion on the immunophenotype of spleen cells in female CrePos and CreNeg mice (cohort 2). The gating strategy utilized to evaluate the effects of ERα on classical DCs (cDCs) in the spleen is outlined in Fig. 5A and further described in Supplementary Fig. 2. No differences were observed in the percent splenic plasmacytoid DCs (using SiglecH+B220+ markers), although there was a trend towards increased percent pDCs and increased percent MHCII+ cells in CrePos females (Fig. 5B). Percent live cDCs or total number cDCs (data not shown) were also not found to be different. These results suggest that the impact of ERα on DC development may be minimal in the DC-specific ERα−/− on this NZM2410 background (interferon-rich), or that DC fate was determined prior to CD11c expression and subsequent ERα deletion.
Figure 5 – Immunophenotype of spleen cells from NZM2410 lupus-prone Cre-CD11c-Floxed-ERα mice.
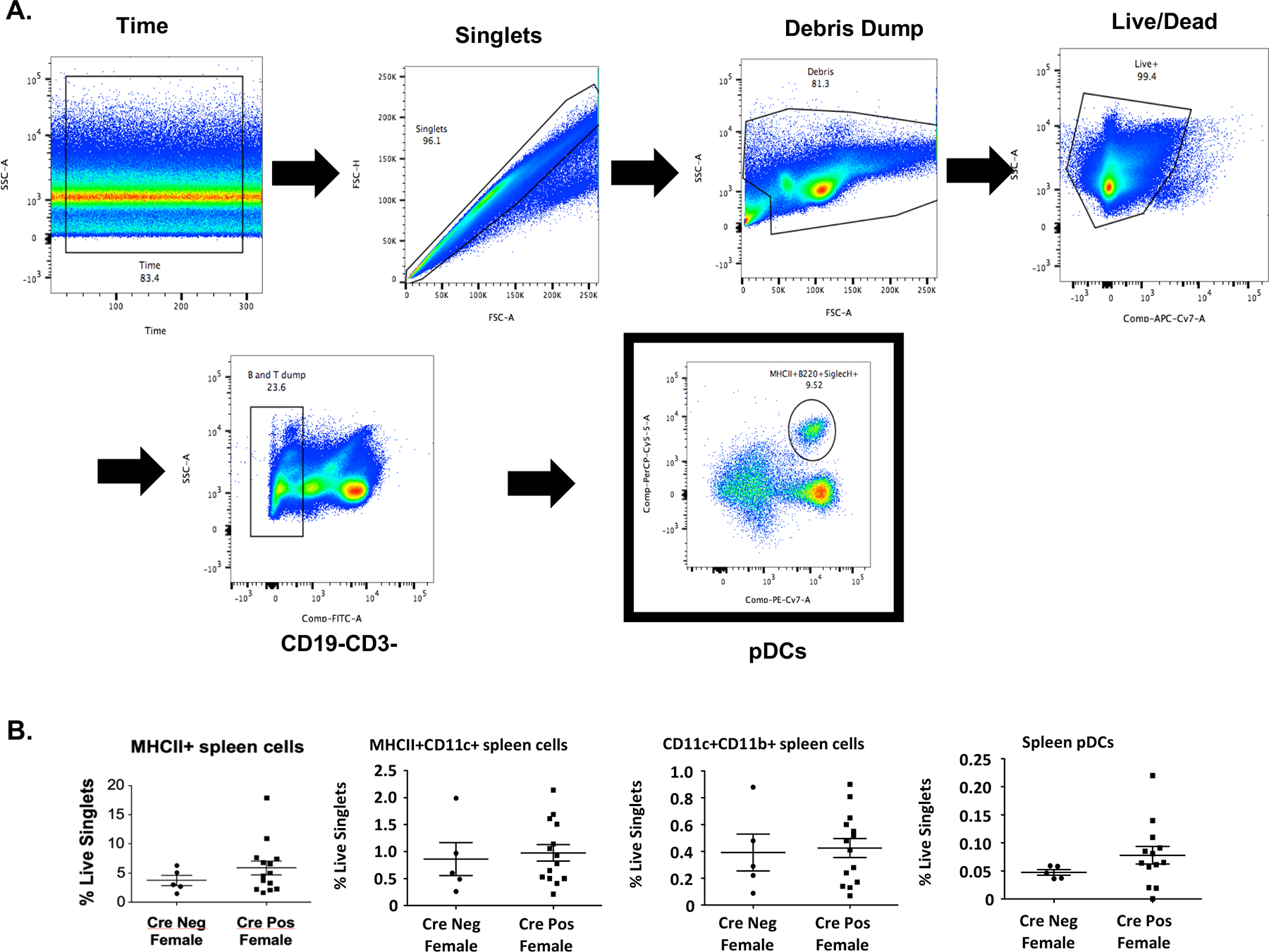
Panel A shows the preliminary gating strategy for isolating immune cells (Time, singlets, debris dump, live/dead staining). Subsequent gating is as follows: pDCs (Lin-, F4/80−, MHCII+, B220+/SiglecH+); cDCs (Lin−, F4/80−, MHCII+, CD11c++, CD11blo); B cells (CD19). All flow experiments utilized FMOs, found in Supplementary Figure 2. Panel B depicts differences in the % live of cDCs and pDCs.
Discussion
Compared with other immune cells, more is known about the requirement for estrogen and ERα for normal/robust cDC and pDC early development and differentiation18, 21, 22, 29, 32. Other sex hormones play a role similar to estrogen signaling on DC maturation and function (i.e. late responses, such as progesterone modulation of TLR3/4-induced cytokine production)33. While a complete knockout of ERα in lupus prone mice did not significantly affect DC numbers, bone marrow-derived DCs had distinct differences in subsets with an increase in cDC1s and a concomitant decrease in cDC2 subsets29. In ERα short mice treated with estrogen, we observed decreased numbers of cDCs as well as decreased plasmacytoid DCs that had reduced IL-6 and IFNα production compared to mice expressing a full length ERα22. Our previous work using ERα variants and mutants, combined with that of others, exemplifies the complex role of ERα in immune cells, and profound impact on DCs in particular.
This study examines the role of ERα in myeloid cells, mainly CD11chi dendritic cells, focusing on the immune response in lupus prone NZM2410 mice. RNA-seq experiments examining the role of ERα in DCs (comparing DCs expressing WT ERα vs. ERα short vs. ERα null) revealed differences in immune related genes and pathways, including complement genes, cytokines and genes that impact interferon. It identifies differentially regulated genes involved in both the innate and adaptive immune response across TLR7 and TLR9-stimulated cells from mice expressing full-length ERα, a truncated version of ERα, or no ERα. Multiple chemokine and cytokine signaling pathways are affected by deletion of ERα, indicating its importance in immunomodulation. Selective deletion of the Esr1 gene in CD11c cells of lupus prone mice led to significantly decreased female (CrePos) survival, although the mechanism behind this has not yet been elucidated. Due to unexpectedly early deaths in this group, proteinuria and renal histology were not fully captured, and thus we were not powered to observe significant differences in those lupus disease endpoints. We did observe significant differences in anti-dsDNA antibody levels. However, similar to our previous studies in ERα mutant mice, there was no correlation between autoantibody levels and disease severity consistent with multiple distinct and sometimes opposing roles for sex hormones and ERα in lupus23, 25, 29. CrePos male mice had significantly higher anti-dsDNA levels compared to CrePos female mice, and no change in survival. In addition, there were surprisingly few differences detected in spleen DC populations in CrePos mice lacking ERα, suggesting that lineage commitments had already been made prior to CD11c expression, since ERα is necessary for normal DC expansion and function in mice8, 18, 34, 35.
RNA-seq experiments revealed distinct differences in DCs between WT and ERα null mice (Fig. 1 A and B), with a majority of upregulated genes from immune processes impacting cytokine signaling, cytolysis, and immune defense. Several interferon inducible genes were upregulated in DCs from mice deficient in ERα, and thus may have protective effects (if down-regulated in the presence of ERα). One example is the murine orthologue of Apol1, which is induced by interferon, plays a role in autophagy, and has been linked to the time of progression to end stage renal disease in African Americans36, 37. It is well established that Type I IFNs promote plasmacytoid DC development, and interferon signature genes are increased in most lupus patients and patients with lupus nephritis, serving as a disease biomarker in a large subset of patients38–41. Arg1, which has ties to IL-4, nitric oxide synthase, and Th1/2 responses42 was also upregulated in DCs from ERα null mice (Fig. 1B). This is interesting as the Th1/Th2 pathway also appears to be differentially regulated in DCs from WT vs. ERα null lupus prone mice (Fig. 1D). Arg-1 activity and expression is increased in SLE patients and correlated with disease activity43. Further, the Th17 response and cytokine production in myeloid-derived suppressor cells from SLE patients was found to be dependent on Arg-143.
Other pathways significantly impacted by deletion of ERα include the IL-17 signaling pathway, Th17 cell differentiation and the mTOR signaling pathway, all of which have importance in SLE. Serum IL-17 and Th17 cell frequency are enhanced in SLE patients and have a positive correlation with SLEDAI scores44 and increased levels of mTOR have been detected in SLE T cells45. That ERα affects these pathways is consistent with our previous observations indicating that estradiol and ERα impact IL-23 secretion and promote the Th17 response in murine DCs46. Results of the cytokine multiplex analysis in murine serum were generally consistent with the RNA-seq analysis. While not in the top 50 DEGs from the RNAseq experiment, the trends we observed in serum cytokine protein expression between WT and ERα null mice mirrored those observed in the RNA-seq data (e.g. IL-6 was downregulated in ERα null compared to WT in both sets of experiments). This was true for all genes except IL-17A, which showed different levels of protein production in mice expressing different forms of ERα (Fig. 2). IL-17A protein was undetectable in ERα null mice, suggesting a role for ERα in IL-17 induction and an impact on Th17 cells that occurs after RNA processing. ERα has multiple other inflammatory cytokine targets and our previous work demonstrates the impact of ERα short on IL-6, IL-1β and CCL2 expression8, 20, 22, 30, 47–49. Serum TNFα was significantly reduced in E2-treated ERα short mice (vs. WT NZM) and these are the mice that were protected from severe lupus-related renal disease24. We did not expect to see a reduction in TNFα in ERα null mice since these mice had similar survival to WT NZM. We were surprised to see a disconnect between disease state and IL-17 and IL-6 (both undetectable in ERα null mice), suggesting ERα is singularly required to regulate their induction even in an already inflamed disease state; this will be further investigated.
Colony stimulating factors, M-CSF, GM-CSF and G-CSF are associated with autoimmune disease50–54 and GM-CSF promotes DC development50, 55, 56. GM-CSF and G-CSF are partially regulated by Fli-1, another transcription factor implicated in SLE pathogenesis53, 57 and DC differentiation from hematopoietic progenitors is stimulated by estradiol/ERα signaling20. In this study, we show that there were clear opposing effects on M-CSF, GM-CSF and G-CSF expression in ERα null mice vs. mice expressing ERα short in the presence of estradiol compared with WT mice (Fig. 2). These results suggest that the previously observed protective effect of ERα short in lupus prone mice may be in part associated with regulation of CSF expression impacting DC and other myeloid cell development, and thus downstream impacts on antigen presenting cell function and inflammatory potential.
To gain a better understanding of the role of ERα in DCs, the Esr1 gene was selectively deleted from DCs in NZM2410 lupus prone mice (CrePos). While the overall survival rates between CrePos and CreNeg mice did not differ, CrePos female mice died rapidly, with 0% of the first cohort surviving to the predetermined sacrifice date of 52 weeks, while survival of CrePos male mice was not different from CreNeg lupus prone mice (Fig. 3). Renal disease and survival of unmanipulated NZM2410J mice is bimodal, and is routinely observed in these mice; we do not observe a spectrum of pathologic renal disease25, 29. In this study, CrePos mice lacking Esr1 in DCs were unable to tolerate any amount of renal disease, which could be attributed to an anti-inflammatory effect of Esr1 that is missing (perhaps lack of an ERα variant)29. Mice had similar overall body and spleen weights (Fig. 4). Autoantibody levels between female CrePos and CreNeg mice also did not differ, suggesting that while they may be necessary for lupus disease onset, they were not sufficient to drive disease in these mice. Interestingly, CrePos male mice had significantly increased anti-dsDNA antibodies compared to CrePos females, but again, this did not impact proteinuria or mortality. Unfortunately, we were not able to capture proteinuria or renal histology on most female CrePos mice since they died unexpectedly early. The proteinuria data we do have is biased towards the healthier mice. Thus, it is not possible to say with certainty that the female CrePos NZM2410 mice died as a result of kidney disease. When a second cohort of mice was sacrificed early (25 weeks) a large percent did not yet have significant proteinuria, adding speculation that the female CrePos mice dying early may have had an alternative cause of death than renal disease. Limitations of this study include a lack of functional studies and an inability to study additional mice to investigate the mechanisms of accelerated death in female CrePos mice. Given the breeding difficulties and the challenges of the times, there was an inability to maintain the colony lines which have been lost.
Analysis of spleen DCs with ERα deleted revealed no significant differences in cDCs or overall MHCII positivity, but there was a trend towards increased pDCs (Fig. 5B), suggesting that selective deletion of ERα in CD11c+ cells mainly occurred subsequent to cDC and pDC developmental decisions. These results are consistent with previous findings suggesting that while some level of ERα is required for cDC development58, abundant (or even full-length) ERα may not be required25. Further supporting evidence from our laboratory demonstrated that an ERα short variant does play a role in specific DC subset development and is consistent with findings that the AF-1 domain of ERα may affect early progenitor DC development through IRF-4 upregulation16, 18, 59. Despite the total number of DCs being largely unchanged, it is possible that DC function may have been altered. Functional studies to determine if ERα deletion affected cellular responses to antigen presentation or TLR stimulation may be interesting. Unfortunately, due to the rapid deaths in cohort 1, along with the ultimate loss of the line, these studies were not performed.
This study defines the effect and immunophenotype of DC-specific ERα deletion in lupus prone mice, which ultimately affects female survival. Complete deletion of ERα in CD11c+ cells did not confer protection in lupus-prone mice. In contrast, it caused female mice to die significantly earlier than their female counterparts that retained ERα expression in CD11c+ cells. These studies have generated many questions regarding the complex and diverse role of ERα in the development and function of myeloid cells like DCs. The data reported in this study continues to provide evidence that deletion of full length ERα (whether global or in specific immune cell subsets) does not provide protection from lupus disease expression despite having some anti-inflammatory effects (ex. reduced IL-17 expression). These current data also further support the need for more study of ERα variants in immunity and autoimmunity. The fact that a variant of ERα, but not loss of ERα in CD11chi cells affects DC development highlights the complexity behind ERα expression and signaling. DC RNA-seq and protein expression data indicate that multiple cytokine signaling pathways, but not all, are impacted by ERα deletion. Importantly, the distinct differences in gene expression observed between mice expressing full length ERα and the short variant of ERα provide further support for the fact that ERα variants differentially impact the immune response.
Supplementary Material
Supplementary Figure 1 – GO gene set analysis for all gene comparisons between WT and ERα null. Bar chart revealing the results of GO analysis on all DEGs between WT and ERα null. The list supplied to GSEA includes a total of 24,000 genes, where absolute log2 fold change between the two strains were used as scores.
Supplementary Figure 2 – Flow cytometry analysis gating strategy. Gating strategy for flow cytometry analysis used in Figure 4. Includes florescence minus one (FMO)s for time, singlets, live, Lin−, F4/80−, MHCII+, CD11chi, and CD11b+.
Acknowledgements:
The authors wish to thank our long-time collaborator Ken Korach at NIEHS for both ERαKO strains of mice as well as his insight through the years. We would also like to acknowledge Ivan Molano for help with experimental troubleshooting and colony management. We would like to express our thanks to Cell Evaluation & Therapy Shared Resource at the Hollings Cancer Center for help with flow cytometry set-up and helping us to produce high quality flow data for reproducibility.
Funding and Disclosure Statement:
The authors report no conflict of interest. This research was supported by the National Institute of Arthritis and musculoskeletal and Skin Diseases grant AR068471; the South Carolina Clinical and Translational Research (SCTR) Institute under grant NIH UL1 TR000062, KL2 TR000060; and supported in part by the Cell Evaluation & Therapy Shared Resource, Hollings Cancer Center, Medical University of South Carolina (P30 CA138313).
References
- 1.Libert C, Dejager L, Pinheiro I. The X chromosome in immune functions: when a chromosome makes the difference. Nat Rev Immunol. 2010;10(8):594–604. Epub 2010/07/24. doi: 10.1038/nri2815. [DOI] [PubMed] [Google Scholar]
- 2.Whitacre CC. Sex differences in autoimmune disease. Nature immunology. 2001;2(9):777–80. doi: 10.1038/ni0901-777. [DOI] [PubMed] [Google Scholar]
- 3.Pons-Estel GJ, Alarcon GS, Scofield L, Reinlib L, Cooper GS. Understanding the epidemiology and progression of systemic lupus erythematosus. Semin Arthritis Rheum. 2010;39(4):257–68. Epub 2009/01/13. doi: 10.1016/j.semarthrit.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buyon JP, Petri MA, Kim MY, Kalunian KC, Grossman J, Hahn BH, Merrill JT, Sammaritano L, Lockshin M, Alarcon GS, Manzi S, Belmont HM, Askanase AD, Sigler L, Dooley MA, Von Feldt J, McCune WJ, Friedman A, Wachs J, Cronin M, Hearth-Holmes M, Tan M, Licciardi F. The effect of combined estrogen and progesterone hormone replacement therapy on disease activity in systemic lupus erythematosus: a randomized trial. Ann Intern Med. 2005;142(12 Pt 1):953–62. Epub 2005/06/22. [DOI] [PubMed] [Google Scholar]
- 5.Cooper GS, Dooley MA, Treadwell EL, St Clair EW, Gilkeson GS. Risk factors for development of systemic lupus erythematosus: allergies, infections, and family history. Journal of clinical epidemiology. 2002;55(10):982–9. [DOI] [PubMed] [Google Scholar]
- 6.Roubinian J, Talal N, Siiteri PK, Sadakian JA. Sex hormone modulation of autoimmunity in NZB/NZW mice. Arthritis Rheum. 1979;22(11):1162–9. Epub 1979/11/01. [DOI] [PubMed] [Google Scholar]
- 7.Talal N, Ahmed SA. Sex hormones, CD5+ (Lyl+) B-cells, and autoimmune diseases. Isr J Med Sci. 1988;24(12):725–8. Epub 1988/12/01. [PubMed] [Google Scholar]
- 8.Kovats S Estrogen receptors regulate innate immune cells and signaling pathways. Cell Immunol. 2015;294(2):63–9. Epub 2015/02/16. doi: 10.1016/j.cellimm.2015.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cunningham M, Gilkeson G. Estrogen receptors in immunity and autoimmunity. Clin Rev Allergy Immunol. 2011;40(1):66–73. Epub 2010/03/31. doi: 10.1007/s12016-010-8203-5. [DOI] [PubMed] [Google Scholar]
- 10.Gubbels Bupp MR, Jorgensen TN. Androgen-Induced Immunosuppression. Front Immunol. 2018;9:794. Epub 2018/05/15. doi: 10.3389/fimmu.2018.00794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taneja V Sex Hormones Determine Immune Response. Front Immunol. 2018;9:1931. Epub 2018/09/14. doi: 10.3389/fimmu.2018.01931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borba VV, Zandman-Goddard G, Shoenfeld Y. Prolactin and Autoimmunity. Front Immunol. 2018;9:73. Epub 2018/02/28. doi: 10.3389/fimmu.2018.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cutolo M, Straub RH. Sex steroids and autoimmune rheumatic diseases: state of the art. Nat Rev Rheumatol. 2020;16(11):628–44. Epub 2020/10/04. doi: 10.1038/s41584-020-0503-4. [DOI] [PubMed] [Google Scholar]
- 14.Calippe B, Douin-Echinard V, Laffargue M, Laurell H, Rana-Poussine V, Pipy B, Guery JC, Bayard F, Arnal JF, Gourdy P. Chronic estradiol administration in vivo promotes the proinflammatory response of macrophages to TLR4 activation: involvement of the phosphatidylinositol 3-kinase pathway. J Immunol. 2008;180(12):7980–8. Epub 2008/06/05. doi: 10.4049/jimmunol.180.12.7980. [DOI] [PubMed] [Google Scholar]
- 15.Campbell L, Emmerson E, Williams H, Saville CR, Krust A, Chambon P, Mace KA, Hardman MJ. Estrogen receptor-alpha promotes alternative macrophage activation during cutaneous repair. J Invest Dermatol. 2014;134(9):2447–57. Epub 2014/04/29. doi: 10.1038/jid.2014.175. [DOI] [PubMed] [Google Scholar]
- 16.Laffont S, Seillet C, Guery JC. Estrogen Receptor-Dependent Regulation of Dendritic Cell Development and Function. Front Immunol. 2017;8:108. Epub 2017/02/28. doi: 10.3389/fimmu.2017.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Douin-Echinard V, Laffont S, Seillet C, Delpy L, Krust A, Chambon P, Gourdy P, Arnal JF, Guery JC. Estrogen receptor alpha, but not beta, is required for optimal dendritic cell differentiation and [corrected] CD40-induced cytokine production. J Immunol. 2008;180(6):3661–9. [DOI] [PubMed] [Google Scholar]
- 18.Seillet C, Rouquie N, Foulon E, Douin-Echinard V, Krust A, Chambon P, Arnal JF, Guery JC, Laffont S. Estradiol promotes functional responses in inflammatory and steady-state dendritic cells through differential requirement for activation function-1 of estrogen receptor alpha. J Immunol. 2013;190(11):5459–70. Epub 2013/04/30. doi: 10.4049/jimmunol.1203312. [DOI] [PubMed] [Google Scholar]
- 19.Scharer CD, Blalock EL, Mi T, Barwick BG, Jenks SA, Deguchi T, Cashman KS, Neary BE, Patterson DG, Hicks SL, Khosroshahi A, Eun-Hyung Lee F, Wei C, Sanz I, Boss JM. Epigenetic programming underpins B cell dysfunction in human SLE. Nat Immunol. 2019;20(8):1071–82. Epub 2019/07/03. doi: 10.1038/s41590-019-0419-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kovats S Estrogen receptors regulate an inflammatory pathway of dendritic cell differentiation: mechanisms and implications for immunity. Horm Behav. 2012;62(3):254–62. Epub 2012/05/09. doi: 10.1016/j.yhbeh.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nalbandian G, Paharkova-Vatchkova V, Mao A, Nale S, Kovats S. The selective estrogen receptor modulators, tamoxifen and raloxifene, impair dendritic cell differentiation and activation. J Immunol. 2005;175(4):2666–75. Epub 2005/08/06. doi: 10.4049/jimmunol.175.4.2666. [DOI] [PubMed] [Google Scholar]
- 22.Cunningham MA, Naga OS, Eudaly JG, Scott JL, Gilkeson GS. Estrogen receptor alpha modulates Toll-like receptor signaling in murine lupus. Clin Immunol. 2012;144(1):1–12. Epub 2012/06/05. doi: 10.1016/j.clim.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Svenson JL, EuDaly J, Ruiz P, Korach KS, Gilkeson GS. Impact of estrogen receptor deficiency on disease expression in the NZM2410 lupus prone mouse. Clin Immunol. 2008;128(2):259–68. Epub 2008/06/03. doi: 10.1016/j.clim.2008.03.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cunningham MA, Richard ML, Wirth JR, Scott JL, Eudaly J, Ruiz P, Gilkeson GS. Novel mechanism for estrogen receptor alpha modulation of murine lupus. Journal of autoimmunity. 2018. Epub 2018/11/13. doi: 10.1016/j.jaut.2018.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scott JL, Wirth JR, Eudaly J, Ruiz P, Cunningham MA. Complete knockout of estrogen receptor alpha is not directly protective in murine lupus. Clinical immunology (Orlando, Fla. 2017;183:132–41. Epub 2017/08/22. doi: 10.1016/j.clim.2017.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kos M, Denger S, Reid G, Korach KS, Gannon F. Down but not out? A novel protein isoform of the estrogen receptor alpha is expressed in the estrogen receptor alpha knockout mouse. J Mol Endocrinol. 2002;29(3):281–6. [DOI] [PubMed] [Google Scholar]
- 27.Hewitt SC, Kissling GE, Fieselman KE, Jayes FL, Gerrish KE, Korach KS. Biological and biochemical consequences of global deletion of exon 3 from the ER alpha gene. FASEB J. 2010;24(12):4660–7. doi: 10.1096/fj.10-163428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.KJ H, Y A, KS K. Estrogen hormone physiology: reproductive findings from estrogen receptor mutant mice. Reproductive biology. 2014;14(1). doi: 10.1016/j.repbio.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cunningham MA, Richard ML, Wirth JR, Scott JL, Eudaly J, Ruiz P, Gilkeson GS. Novel mechanism for estrogen receptor alpha modulation of murine lupus. J Autoimmun. 2019;97:59–69. Epub 2018/11/13. doi: 10.1016/j.jaut.2018.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Svenson J, Cunningham M, Dasgupta S, Gilkeson GS. Estrogen receptor alpha modulates mesangial cell responses to toll-like receptor ligands. Am J Med Sci. 2014;348(6):492–500. Epub 2014/10/25. doi: 10.1097/MAJ.0000000000000339. [DOI] [PubMed] [Google Scholar]
- 31.Cunningham MA, Wirth JR, Scott JL, Eudaly J, Collins EL, Gilkeson GS. Early Ovariectomy Results in Reduced Numbers of CD11c+/CD11b+ Spleen Cells and Impacts Disease Expression in Murine Lupus. Front Immunol. 2016;7:31. doi: 10.3389/fimmu.2016.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nalbandian G, Kovats S. Understanding sex biases in immunity: effects of estrogen on the differentiation and function of antigen-presenting cells. Immunol Res. 2005;31(2):91–106. Epub 2005/03/22. doi: 10.1385/IR:31:2:091. [DOI] [PubMed] [Google Scholar]
- 33.Jones LA, Kreem S, Shweash M, Paul A, Alexander J, Roberts CW. Differential Modulation of TLR3- and TLR4-Mediated Dendritic Cell Maturation and Function by Progesterone2010. doi: 10.4049/jimmunol.0901155. [DOI] [PubMed] [Google Scholar]
- 34.Kadel S, Kovats S. Sex Hormones Regulate Innate Immune Cells and Promote Sex Differences in Respiratory Virus Infection. Front Immunol. 2018;9:1653. Epub 2018/08/07. doi: 10.3389/fimmu.2018.01653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Griesbeck M, Ziegler S, Laffont S, Smith N, Chauveau L, Tomezsko P, Sharei A, Kourjian G, Porichis F, Hart M, Palmer CD, Sirignano M, Beisel C, Hildebrandt H, Cenac C, Villani AC, Diefenbach TJ, Le Gall S, Schwartz O, Herbeuval JP, Autran B, Guery JC, Chang JJ, Altfeld M. Sex Differences in Plasmacytoid Dendritic Cell Levels of IRF5 Drive Higher IFN-alpha Production in Women. J Immunol. 2015;195(11):5327–36. Epub 2015/11/01. doi: 10.4049/jimmunol.1501684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Freedman BI, Langefeld CD, Andringa KK, Croker JA, Williams AH, Garner NE, Birmingham DJ, Hebert LA, Hicks PJ, Segal MS, Edberg JC, Brown EE, Alarcon GS, Costenbader KH, Comeau ME, Criswell LA, Harley JB, James JA, Kamen DL, Lim SS, Merrill JT, Sivils KL, Niewold TB, Patel NM, Petri M, Ramsey-Goldman R, Reveille JD, Salmon JE, Tsao BP, Gibson KL, Byers JR, Vinnikova AK, Lea JP, Julian BA, Kimberly RP, Lupus Nephritis-End-Stage Renal Disease C. End-stage renal disease in African Americans with lupus nephritis is associated with APOL1. Arthritis & rheumatology. 2014;66(2):390–6. Epub 2014/02/08. doi: 10.1002/art.38220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iwamoto T, Niewold TB. Genetics of human lupus nephritis. Clinical immunology. 2017;185:32–9. Epub 2016/10/04. doi: 10.1016/j.clim.2016.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Banchereau R, Hong S, Cantarel B, Baldwin N, Baisch J, Edens M, Cepika AM, Acs P, Turner J, Anguiano E, Vinod P, Kahn S, Obermoser G, Blankenship D, Wakeland E, Nassi L, Gotte A, Punaro M, Liu YJ, Banchereau J, Rossello-Urgell J, Wright T, Pascual V. Personalized Immunomonitoring Uncovers Molecular Networks that Stratify Lupus Patients. Cell. 2016;165(3):551–65. Epub 2016/04/05. doi: 10.1016/j.cell.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Der E, Ranabothu S, Suryawanshi H, Akat KM, Clancy R, Morozov P, Kustagi M, Czuppa M, Izmirly P, Belmont HM, Wang T, Jordan N, Bornkamp N, Nwaukoni J, Martinez J, Goilav B, Buyon JP, Tuschl T, Putterman C. Single cell RNA sequencing to dissect the molecular heterogeneity in lupus nephritis. JCI Insight. 2017;2(9). Epub 2017/05/05. doi: 10.1172/jci.insight.93009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Der E, Suryawanshi H, Morozov P, Kustagi M, Goilav B, Ranabothu S, Izmirly P, Clancy R, Belmont HM, Koenigsberg M, Mokrzycki M, Rominieki H, Graham JA, Rocca JP, Bornkamp N, Jordan N, Schulte E, Wu M, Pullman J, Slowikowski K, Raychaudhuri S, Guthridge J, James J, Buyon J, Tuschl T, Putterman C, Accelerating Medicines Partnership Rheumatoid A, Systemic Lupus Erythematosus C. Tubular cell and keratinocyte single-cell transcriptomics applied to lupus nephritis reveal type I IFN and fibrosis relevant pathways. Nat Immunol. 2019;20(7):915–27. Epub 2019/05/22. doi: 10.1038/s41590-019-0386-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deng Y, Tsao BP. Updates in Lupus Genetics. Curr Rheumatol Rep. 2017;19(11):68. Epub 2017/10/07. doi: 10.1007/s11926-017-0695-z. [DOI] [PubMed] [Google Scholar]
- 42.Hesse M, Modolell M, La Flamme AC, Schito M, Fuentes JM, Cheever AW, Pearce EJ, Wynn TA. Differential regulation of nitric oxide synthase-2 and arginase-1 by type 1/type 2 cytokines in vivo: granulomatous pathology is shaped by the pattern of L-arginine metabolism. J Immunol. 2001;167(11):6533–44. Epub 2001/11/21. doi: 10.4049/jimmunol.167.11.6533. [DOI] [PubMed] [Google Scholar]
- 43.Wu H, Zhen Y, Ma Z, Li H, Yu J, Xu ZG, Wang XY, Yi H, Yang YG. Arginase-1-dependent promotion of TH17 differentiation and disease progression by MDSCs in systemic lupus erythematosus. Sci Transl Med. 2016;8(331):331ra40. Epub 2016/03/25. doi: 10.1126/scitranslmed.aae0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shah K, Lee WW, Lee SH, Kim SH, Kang SW, Craft J, Kang I. Dysregulated balance of Th17 and Th1 cells in systemic lupus erythematosus. Arthritis Res Ther. 2010;12(2):R53. Epub 2010/03/26. doi: 10.1186/ar2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fernandez DR, Telarico T, Bonilla E, Li Q, Banerjee S, Middleton FA, Phillips PE, Crow MK, Oess S, Muller-Esterl W, Perl A. Activation of mammalian target of rapamycin controls the loss of TCRzeta in lupus T cells through HRES-1/Rab4-regulated lysosomal degradation. J Immunol. 2009;182(4):2063–73. Epub 2009/02/10. doi: 10.4049/jimmunol.0803600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Relloso M, Aragoneses-Fenoll L, Lasarte S, Bourgeois C, Romera G, Kuchler K, Corbi AL, Munoz-Fernandez MA, Nombela C, Rodriguez-Fernandez JL, Diez-Orejas R. Estradiol impairs the Th17 immune response against Candida albicans. J Leukoc Biol. 2012;91(1):159–65. Epub 2011/10/04. doi: 10.1189/jlb.1110645. [DOI] [PubMed] [Google Scholar]
- 47.Franco HL, Nagari A, Kraus WL. TNFalpha signaling exposes latent estrogen receptor binding sites to alter the breast cancer cell transcriptome. Mol Cell. 2015;58(1):21–34. Epub 2015/03/11. doi: 10.1016/j.molcel.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nwachukwu JC, Srinivasan S, Bruno NE, Parent AA, Hughes TS, Pollock JA, Gjyshi O, Cavett V, Nowak J, Garcia-Ordonez RD, Houtman R, Griffin PR, Kojetin DJ, Katzenellenbogen JA, Conkright MD, Nettles KW. Resveratrol modulates the inflammatory response via an estrogen receptor-signal integration network. Elife. 2014;3:e02057. Epub 2014/04/29. doi: 10.7554/eLife.02057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cunningham MA, Wirth JR, Naga O, Eudaly J, Gilkeson GS. Estrogen Receptor Alpha Binding to ERE is Required for Full Tlr7- and Tlr9-Induced Inflammation. SOJ Immunol. 2014;2(1). Epub 2014/07/26. doi: 10.15226/soji.2014.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hamilton JA. Colony-stimulating factors in inflammation and autoimmunity. Nat Rev Immunol. 2008;8(7):533–44. Epub 2008/06/14. doi: 10.1038/nri2356. [DOI] [PubMed] [Google Scholar]
- 51.Becher B, Tugues S, Greter M. GM-CSF: From Growth Factor to Central Mediator of Tissue Inflammation. Immunity. 2016;45(5):963–73. Epub 2016/11/17. doi: 10.1016/j.immuni.2016.10.026. [DOI] [PubMed] [Google Scholar]
- 52.Hirota K, Hashimoto M, Ito Y, Matsuura M, Ito H, Tanaka M, Watanabe H, Kondoh G, Tanaka A, Yasuda K, Kopf M, Potocnik AJ, Stockinger B, Sakaguchi N, Sakaguchi S. Autoimmune Th17 Cells Induced Synovial Stromal and Innate Lymphoid Cell Secretion of the Cytokine GM-CSF to Initiate and Augment Autoimmune Arthritis. Immunity. 2018;48(6):1220–32 e5. Epub 2018/05/29. doi: 10.1016/j.immuni.2018.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lennard Richard ML, Brandon D, Lou N, Sato S, Caldwell T, Nowling TK, Gilkeson G, Zhang XK. Acetylation impacts Fli-1-driven regulation of granulocyte colony stimulating factor. Eur J Immunol. 2016;46(10):2322–32. Epub 2016/07/20. doi: 10.1002/eji.201646315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Denny MF, Yalavarthi S, Zhao W, Thacker SG, Anderson M, Sandy AR, McCune WJ, Kaplan MJ. A distinct subset of proinflammatory neutrophils isolated from patients with systemic lupus erythematosus induces vascular damage and synthesizes type I IFNs. J Immunol. 2010;184(6):3284–97. doi: 10.4049/jimmunol.0902199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Naik SH, Metcalf D, van Nieuwenhuijze A, Wicks I, Wu L, O’Keeffe M, Shortman K. Intrasplenic steady-state dendritic cell precursors that are distinct from monocytes. Nat Immunol. 2006;7(6):663–71. Epub 2006/05/09. doi: 10.1038/ni1340. [DOI] [PubMed] [Google Scholar]
- 56.Schmid MA, Kingston D, Boddupalli S, Manz MG. Instructive cytokine signals in dendritic cell lineage commitment. Immunol Rev. 2010;234(1):32–44. Epub 2010/03/03. doi: 10.1111/j.0105-2896.2009.00877.x. [DOI] [PubMed] [Google Scholar]
- 57.Wang X, Lennard Richard M, Li P, Henry B, Schutt S, Yu XZ, Fan H, Zhang W, Gilkeson G, Zhang XK. Expression of GM-CSF Is Regulated by Fli-1 Transcription Factor, a Potential Drug Target. J Immunol. 2021;206(1):59–66. Epub 2020/12/04. doi: 10.4049/jimmunol.2000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Paharkova-Vatchkova V, Maldonado R, Kovats S. Estrogen preferentially promotes the differentiation of CD11c+ CD11b(intermediate) dendritic cells from bone marrow precursors. J Immunol. 2004;172(3):1426–36. Epub 2004/01/22. doi: 10.4049/jimmunol.172.3.1426. [DOI] [PubMed] [Google Scholar]
- 59.Kovats S, Carreras E. Regulation of dendritic cell differentiation and function by estrogen receptor ligands. Cell Immunol. 2008;252(1–2):81–90. Epub 2008/02/19. doi: 10.1016/j.cellimm.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1 – GO gene set analysis for all gene comparisons between WT and ERα null. Bar chart revealing the results of GO analysis on all DEGs between WT and ERα null. The list supplied to GSEA includes a total of 24,000 genes, where absolute log2 fold change between the two strains were used as scores.
Supplementary Figure 2 – Flow cytometry analysis gating strategy. Gating strategy for flow cytometry analysis used in Figure 4. Includes florescence minus one (FMO)s for time, singlets, live, Lin−, F4/80−, MHCII+, CD11chi, and CD11b+.


