Abstract
Cancer is one of the leading causes of death in the world. Efforts to find and develop cancer drugs from natural products continue with the exploration of trisindoline, a substance that is isolated from marine sponges Hyrtios altum. Trisindoline is an indole trimer alkaloid compound that has been successfully synthesized into trisindoline 1, 3 and 4. Trisindoline is cytotoxic in cell lines and in this study, trisindoline was able to induce apoptosis in the in silico and in vitro tests that were carried out. The in silico test was carried out through molecular docking using the Autodock Vina method and the Molecular Dynamics (MD) Simulation QM / MM AMBER. The target proteins used were protein p53 and caspase −9 which played a role in the apoptotic pathway and cyclin D1 which played a role in cell proliferation. Meanwhile, cytotoxicity analysis was carried out using the MTT method (3- (4,5-dimethyltiazol −2-yl) −2,5 -dipenyl tetrazolium bromide). Nevertheless, the ability of trisindoline to induce phagocytosis is still unrevealed. The phagocytosis assay was carried out by assessing the macrophage capacity and phagocytic index using the latex-beads model. The in silico results showed that the binding affinity values between the target protein Cdk-2 and the trisindoline 1, trisindoline 3 and trisindoline 4 ligands were −7.3 kcal / mol, −7.7 kcal / mol and −6.6 kcal / mol respectively. The binding affinity values between the target protein p53 and the trisindoline 1, trisindoline 3 and trisindoline 4 ligands were −7.5 kcal / mol, −7.4 kcal / mol and −7.5 kcal / mol respectively. The binding affinity values between the target protein caspase-9 and the trisindoline 1, trisindoline 3 and trisindoline 4 ligands were −7.5 kcal / mol, −7.1 kcal / mol and −7.2 kcal / mol respectively. The results of RMSD (Root Mean Square Deviation), RMSF (Root Mean Square Fluctuation), and hydrogen bonds in the MD (Molecular Dynamics) Simulation showed that Cdk-2 formed a protein complex with trisindoline 3, protein p53 with trisindoline 1 and caspase-9 with trisindoline 1. The cytotoxicity assay was carried out in the MCF-7 cell line and the IC50 value obtained for trisindoline 1 was 2.059 μM, for trisindoline 3 was 3.9759 μM, for trisindoline 4 was 15.46 μM and for doxorubicin was 9.88 μM. Furthermore, the phagocytosis test was carried out using trisindoline 1, 3 and 4. Our results showed that 6.25 µg mL−1 of trisindoline 1 and trisindoline 3 were able to induce the phagocytosis capacity of macrophage cells of 38.34; whereas trisindoline 4 at a concentration of 50 µg mL−1 induces a phagocytosis capacity of 32.89. Trisindoline 1, 3 and 4 showed potentials of immunostimulants at low concentrations but showed potentials of immunosuppressants at high concentrations. The overall results demonstrated that trisindoline 1 and 3 are potential anti-cancer candidates capable of activating the apoptotic pathway.
Keywords: Trisindoline, P53, Caspase 9, In Silico, MCF-7
1. Introduction
The development of anti-cancer drugs can be done by utilizing both the intrinsic and extrinsic pathways of apoptosis. Cancer drugs generally function to inhibit cell proliferation without killing normal cells and are multidrug-resistant (MDR) or resistant to various cancer drugs (Mansoori et al., 2017). For this reason, research to find anti-cancer drug candidates is needed. One of the anti-cancer drug candidates being developed is trisindoline. Trisindoline was first isolated from Vibrio sp., which is a bacteria symbiont of sea sponge Hyrtios altum. (Kobayashi et al., 1994). The extrinsic pathway in apoptosis occurs through the induction of genes encoding transmembrane proteins, namely: Tumor Necrosis Factor Receptor (TNF) and Fas. This protein activates the transducer signal which in turn activates the caspase, p-53 induces the Fas protein through binding elements on the promoter and introns and activates TNF in response to DNA damage and through the caspase complex will trigger cell death. Meanwhile, the intrinsic pathway occurs through pro-apoptotic proteins, one of which is cyclin-dependent kinase 2 (CDK2) which triggers the release of cytochrome C from the mitochondria and activates caspase 9 which acts as an apoptosis inhibitor (Khan et al., 2014).
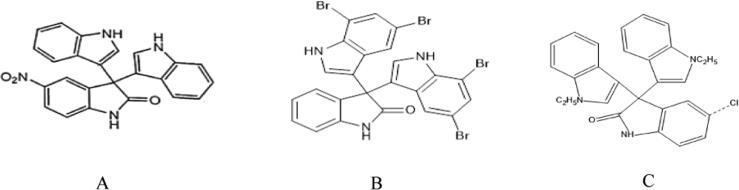
The trisindoline group has been extensively developed in recent anti-cancer research due to the success of the synthesis method and its high potential for cytotoxicity. Santoso & Mursyidah (2010) succeeded in synthesizing and modifying trisindoline into 4 compounds namely trisindoline 1: 5′-nitro- [3,3 ': 3′, 3”-terindoline] −2′-one (B), which is the result of the synthesis of trisindoline with the addition of the nitro group shown in Fig. 1.A, trisindoline 3: 5,5 “, 7,7″ -tetrabromo- [3,3 ': 3′, 3 ”-terindoline] −2′-one, which is the trisindoline compound with the.
Fig. 1.
Chemical Structure of Trisindoline (A) Trisindoline 1 (B) Trisindoline 3 and (C) Trisindoline 4.
addition of the bromo group shown in Fig. 1.B and trisindoline 4: 5′-chloro-1,1 “-diethyl-1H, 1″ H- [3,3 ': 3′, 3 ”-indol] − 2 '(1′H) -one, which is a trisindoline synthesis compound with the addition of a chloro group shown in Fig. 1.C. The three compounds have not yet been tested for cytotoxicity. The success of the synthesis method and the potential for trisindoline derivative (1a) can be further developed as a new anti-cancer group that is cytotoxic to cancer cells but safe for normal cells. Trisindoline compounds have cytotoxic activity against several cell lines. The trisindoline group has been widely developed in the latest anti-cancer research due to the success of the synthesis method and its high cytotoxicity potential. In particular, shikonin has been shown to exert anti-cancer properties such as inducing cellular apoptosis through mitochondria-mediated pathway in some cancer cells.
Trisindoline showed activity against HepG2 liver cancer cells with an IC50 of 20.3 μM, A549 lung cancer cells with an IC50 of 8.6 μM, SK-N-SH brain cancer cells with an IC50 of 11.3 μM, MCF-7 breast cancer cells with an IC50 of 49. 8 μM, prostate cancer cells DU-145 with an IC50 of 8.7 μM (Kamal et al., 2010), uterine cancer cells MES-SA / DX5 with an IC50 of 3.51 μM and HCT15 colon cancer cells with an IC50 of 6.63 μM (Yoo et al., 2008). Results from previous studies showed that trisindoline 1 had higher cytotoxic activity on HepG2 cells compared to Vero cells with an IC50 value of 2.837 μg / ml and induced apoptosis during the 24-hour incubation period by activating p53. Based on those reports of IC50 value that range 1–100 μM, Trisindoline is being categorized as possessing a high cytotoxicity. However, further investigation is required to determine whether a high cytotoxicity possess a necrotic or apoptosis pathway. In addition, anticancer drugs are investigated recently, whether their pathways are works through 1) Stimulates apoptosis, 2) Regulates the cell cycle and controls checkpoints and 3) Inhibits growth factors and the signal transduction.
Molecular docking is a computational biology and bioinformatic methods for supporting in vivo and in vitro assessment of natural products as an anti-cancer. This method is used to model the interaction between natural compounds and the target proteins (Jensen, 2007, Lyskov and Gray, 2008, Sri et al., 2011). The binding site area between amino acid residues plays an important role in binding with drugs or ligands (Arwansyah et al., 2014). Molecular docking integrates the computational and experimental strategies and has been of great assistance in the identification and development of promising and novel compounds. Broadly used in modern drug design, molecular docking methods explore the ligand conformations adopted within the binding sites of macromolecular targets. This approach also estimates the ligand-receptor binding free energy by evaluating critical phenomena involved in the intermolecular recognition process (Ferreira et al., 2015).
Previous research showed that the binding affinity of p53 with trisindoline 1, 3 and 4 were −5.5; −8.3; and −8.1 kcal mol−1 respectively and is in the form of static molecules (Joerger et al., 2015). Therefore, Molecular Dynamics (MD) Simulation which describes the conditions of temperature, pressure, and solvents in the biological system of the human body is required. One method of MD is the Quantum Mechanics / Molecular Mechanics (QM / MM) using the Assisted Model Building with Energy Refinement (AMBER) software. The hybrid quantum mechanics/molecular mechanics (QM / MM) is a molecular simulation method that combines the QM (accuracy) and MM (velocity) approaches, making it possible to study chemical processes in a chemical reaction of a system. The conventional (“energy mixing”) QM / MM method (convQM / MM) defines the total energy function for an entire system consisting of three components namely, a) the energy of the QM model applied to the atoms in an area, b) the energy of the MM model applied on atoms that have velocity, and c) the energy interaction between the two systems (Xu et al, 2013). For this reason, investigation on whether Trisindoline works as a cancer adjuvant on apoptotic pathway or not is required. Furthermore, we employ molecular docking of trisindoline 1, trisindoline 3, trisindoline 4 on cdk-2 gen that regulates cell cycle and growth factor, p53 and caspase −9 genes that regulate apoptosis. At the same time, we analyse phagocytosis activity against the induction of phagocytosis in macrophage cells of mice (M. musculus).
2. Experimental
2.1. Research design
The docking process were conducted at the Department of Biology and the Department of Chemistry, Institut Teknologi Sepuluh Nopember, Surabaya, Indonesia and at the Department of Biochemistry, Chulalongkorn University, Bangkok, Thailand. Cytotoxicity assay and phagocytosis test were carried out at the Laboratory of Parasitology and Pharmacology, Faculty of Medicine, Gadjah Mada University, Yogyakarta, Indonesia. This research was conducted from December 2019 to June 2020.
2.2. Material and methods
2.2.1. In silico analysis
2.2.1.1. Protein preparation
Protein structures were taken from the RCSB Protein Data Bank database (Burley et al., 2019) with PDB ID: 2UZE for cdk-2 (Richardson et al., 2007), 1TUP for p53 (Cho et al., 1994), and 1NW9 for caspase-9 (Shiozaki et al., 2003). The ligands, ions, and carried water were cleaned using VMD and saved in.pdb file format.
2.2.1.2. Ligand preparation
Structures of the three trisindoline ligands were drawn using MarvinSketch (https://chemaxon.com/products/marvin) to obtain 3D molecules from 2D structures and saved in.pdb file format. The software supported the valence checking, atom and bond query, stereochemistry, and user-defined templates.
2.2.1.3. Molecular docking
The blind docking method was used in this study to demonstrate ligands attachment to all active sites of the target proteins. The Autodock Vina software (https://vina.scripps.edu/) from the Laboratory of Molecular Graphics, The Scripps Research Institute was used for the docking process. The PDBQT molecular structure file format was used in AutoDock (Trott and Olson, 2010). The experiment was repeated 100 times and the protein–ligand complex that had the best binding affinity (Kcal/mol) was selected.
2.2.1.4. Molecular dynamic simulation
Trisindoline ligands were optimized using AMBER Tools 18 (https://ambermd.org/GetAmber.php) by removing the hydrogen molecules using GaussView and saved in.gjf file format. Next, hydrogen was added and optimized using the AMBER Tools software (Case et al., 2018). Protein-ligand complexes was saved in the.gjf file format. MD simulations were run using AMBER for 20 ns (Rungnim et al., 2015).
2.2.1.5. Quantification and Visualization
Visualization of the hydrogen bond distances was conducted using Discovery Studio 2019 and VMD. The obtained calculations (RMSD, RMSF, inter and intramolecular hydrogen bonds) from previous simulations were displayed in the form of graphs using OriginLab.
Root mean square deviation (RMSD) can be defined for two structures containing identical numbers and types of atoms. If the two atomic coordinates become and then.
where and refer to subsets of the entire molecule, such as Cα atoms, backbone atoms or heavy atoms. The RMSD value depends on the exact definition of (Kovács and Péter, 2016).
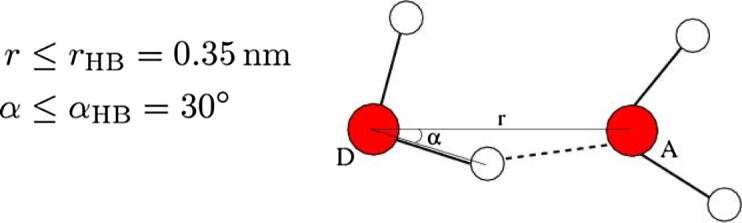
The root mean square fluctuation (RMSF) is the deviation between the position of particle i and several reference positions. If the two atomic coordinates become and , subsequently, to determine whether there is an H-bond between donor D and acceptor A, then the geometric criterion is used:
The time of occurrence of hydrogen bonds can be calculated from the autocorrelation function, using the following equation:
where s(t) = {0,1} is a function of the presence of the H-bond at time t. The integral of C(τ) gives the approximate time of occurrence of the hydogen bond. MD shows the location of the intra and intermolecular hydrogen bonds in the COX bond complex with IBP (Menéndez et al., 2016).
2.2.2. Cytotoxicity assay
The cytotoxicity test of the three trisindoline compounds against the MCF-7 cell line (breast cancer cells) was performed using the MTT (3-(4,5-dimethyltiazol-2-yl)-2,5-diphenyl tetrazolium bromide) method. Cell viability was measured using a microplate reader at a wavelength of 595 nm. The cytotoxicity test was carried out as an initial process to analyse the potential toxicity of the four compounds on the growth of the MCF-7 cell line (breast cancer cells).
2.2.3. Phagocytosis assay
2.2.3.1. Macrophage culture
The macrophages used were isolated from 8 weeks old Swiss strain M. musculus having a weight of 25–30 g. Isolation and culture of macrophages was conducted in accordance with Davies et al., 2001, Rios et al., 2017. The macrophage cells density used in this study was 2.5x106 /mL. Cells were incubated for 24 h in a 5% CO2 incubator at a temperature of 37 °C.
2.2.3.2. Phagocytosis activity
The phagocytic activity test of macrophage cells was conducted in accordance to modified protocols from Davies et al., 2001, Rios et al., 2017. The concentration of trisindolines were varied at 6.25; 12.5; 25 and 50 μg/mL. A 200 µL latex suspension was added to each well and incubated for 60 min. The observation of macrophages was done under a light microscope with a magnification of 400 x. The observations of macrophages and latex were carried out in 10 fields of view (a total of 300 macrophages). The macrophage capacity is the number of macrophage phagocytic latex particles in 300 macrophages observed. The phagocytic index was calculated using the following formula: phagocytic index = (total number of ingested cells/total number of counted macrophages) × (number of macrophages containing ingested cells/total number of counted macrophages) × 100.
2.2.3.3. Data analysis and design experiment
Phagocytosis index and macrophage capacity studies were designed using Completely Randomized Design (CRD). The data obtained from the phagocytosis index and macrophage capacity were then tested with One Way ANOVA in the SPSS software to determine the effect of trisindoline compounds on macrophage phagocytosis with a 95% confidence level.
3. Results and discussion
3.1. In silico analysis
3.1.1. Molecular docking visualization
The potential of a compound can be predicted by comparing their docking scores to other compounds. The molecule with the lowest score (minus value) shows good affinity (Purnomo, 2011). RMSD is a two-pose measurement that compares the atomic position between the experimental structure and the docking or predicted structure (Irina and Ruben, 2012). MSD values < 2.0 Å are usually used as a criterion for the success of the docking method (Huang and Zou, 2006, Santos et al., 2020). As the RMSD value approaches zero, the pose of the original ligand and the copy ligand becomes similar. The docking scores of trisindolines against cdk2, p53, and caspace 9 are listed in Table 1, Table 2 and Table 3 respectively.
Table 1.
Molecular docking scores against Cdk2.
| Model | Tris 1 |
Tris 3 |
Tris 4 |
|||
|---|---|---|---|---|---|---|
| aff | dist | aff | dist | aff | dist | |
| 1 | −7.4 | 0.00 | −8.0 | 0.00 | −6.9 | 0.00 |
| 2 | −7.3 | 29.54 | −7.7 | 31.04 | −6.6 | 30.31 |
| 3 | −7.2 | 2.66 | −7.5 | 5.73 | −6.5 | 8.79 |
| 4 | −7.2 | 2.39 | −7.4 | 22.34 | −6.4 | 15.07 |
| 5 | −7.2 | 2.25 | −7.3 | 4.70 | −6.3 | 1.04 |
Note: Tris: Trisindoline; aff: Binding affinity (kcal/mol); dist:Distribution of RMSD.
Table 2.
Molecular docking scores against p53.
| Model | Tris 1 |
Tris 3 |
Tris 4 |
|||
|---|---|---|---|---|---|---|
| aff | dist | Aff | dist | aff | dist | |
| 1 | −8.3 | 0.00 | −8.1 | 0.00 | −7.5 | 0.00 |
| 2 | −7.8 | 2.76 | −7.9 | 23.96 | −7.4 | 19.12 |
| 3 | −7.5 | 2.37 | −7.8 | 25.21 | −7.4 | 2.50 |
| 4 | −7.5 | 1.50 | −7.8 | 3.38 | −7.4 | 23.75 |
| 5 | −7.5 | 2.10 | −7.8 | 22.79 | −7.3 | 1.07 |
Note: Tris: Trisindoline; aff: Binding Affinity (kcal/mol); dist: Distribution of RMSD.
Table 3.
Molecular docking scores against caspase 9.
| Model | Tris 1 |
Tris 3 |
Tris 4 |
|||
|---|---|---|---|---|---|---|
| aff | dist | aff | dist | aff | dist | |
| 1 | −7.3 | 0.00 | −7.9 | 0.00 | −7.4 | 0.00 |
| 2 | −7.3 | 36.58 | −7.5 | 2.35 | −7.1 | 1.95 |
| 3 | −7.2 | 8.05 | −7.5 | 10.70 | −7.1 | 1.26 |
| 4 | −7.2 | 39.66 | −7.4 | 10.55 | −6.9 | 35.55 |
| 5 | −7.2 | 40.11 | −7.4 | 34.58 | −6.9 | 1.11 |
Note: Tris: Trisindoline; aff: Binding Affinity (kcal/mol); dist: distribution of RMSD.
In the validation evaluation, the parameters that were observed were RMSD and visual pose.It was observed that the developed protocol was acceptable and can be further developed for virtual screening in an effort to discover new compounds (de Sousa et al., 2020). Visualization of the molecular docking is needed to determine which amino acids play a role in maintaining the stability of these compounds at their receptors (Purnomo, 2011). Visualization of the validation pose between the original ligand and the copy ligand shows that each atom within the structures of the two molecules have similar positions and angles. At RMSD > 2.0 Å, it can be seen that the two molecules have significantly different positions and angles, even though they have the same number of atoms. The protein complex obtained from the molecular docking process is then used in the MD simulation.
3.1.1.1. Molecular docking Visualization of trisindoline with Cdk-2
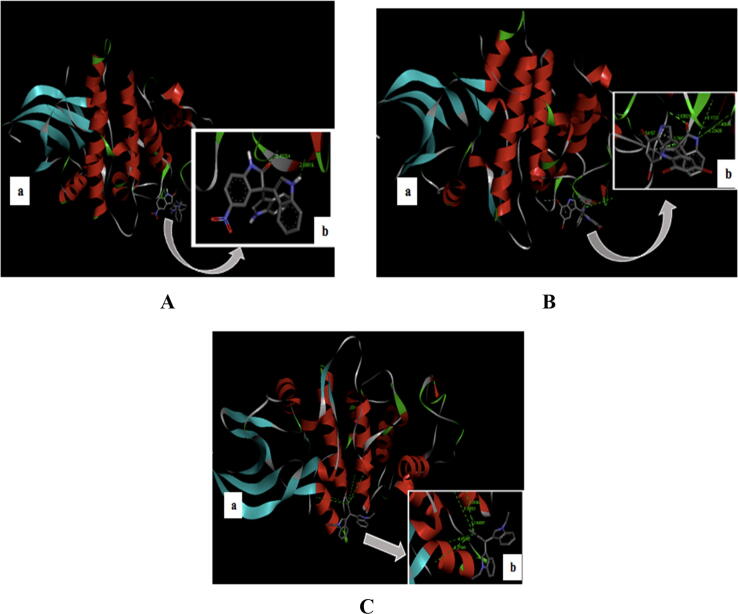
The highest binding affinity value from the molecular docking between the Cdk-2 target protein and the trisindoline 1, trisindoline 3, and trisindoline 4 ligands were −7.3 kcal/mol, −7.7 kcal/mol and −6.6 kcal/mol respectively. The results of the protein complex formed in the molecular docking are marked in bold in Table 1 and shown in Fig. 2.
Fig. 2.
(A) Visualization of the molecular docking results of Cdk-2 with Trisindoline 1 (a) and the hydrogen bonds formed (b); (B) Visualization of the molecular docking results of Cdk-2 with Trisindoline 3 (a) and the hydrogen bonds formed (b) and (C) Visualization of the molecular docking results of Cdk-2 with Trisindoline 4 (a) and the hydrogen bonds formed (b).
3.1.1.2. Molecular docking Visualization of trisindoline with p53
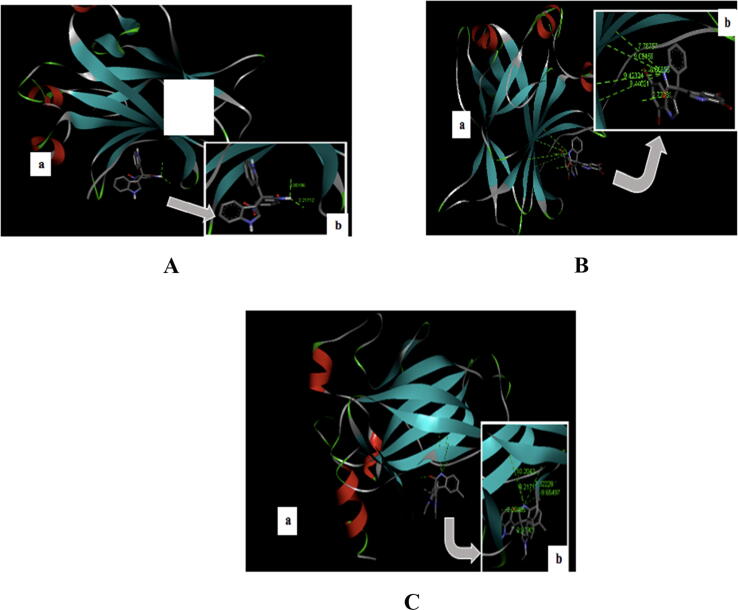
The highest binding affinity value from the molecular docking between the target protein p53 and the trisindoline 1, trisindoline 3, and trisindoline 4 ligands were −7.5 kcal / mol, −7.4 kcal / mol and −7.5 kcal / mol respectively. The results of the protein complex formed in the molecular docking are marked in bold in Table 2 and shown in Fig. 3.
Fig. 3.
(A) Visualization of the molecular docking results of P53 with Trisindoline 1 (a) and the hydrogen bonds formed (b); (B) Visualization of the molecular docking results of P53 with Trisindoline 3 (a) and the hydrogen bonds formed (b) and (C) Visualization of the molecular docking results of P53 with Trisindoline 4 (a) and thehydrogen bonds formed (b).
Based on the visualization and validation of the molecular docking results of p53 with the three trisindoline compounds, trisindoline 1 exhibited the best potential to form protein complexes with p53 with a binding affinity value of −7.5 kcal / mol with a bond distance of 2 to 3 Å. The protein complex formed between p53 protein and trisindoline 1 has an RMSD value between 1.50 Å and 2 Å. Therefore, it can be used as one of the criteria for the success of the docking method (Jensen, 2007).
3.1.1.3. Molecular docking Visualization of trisindoline with Caspase-9
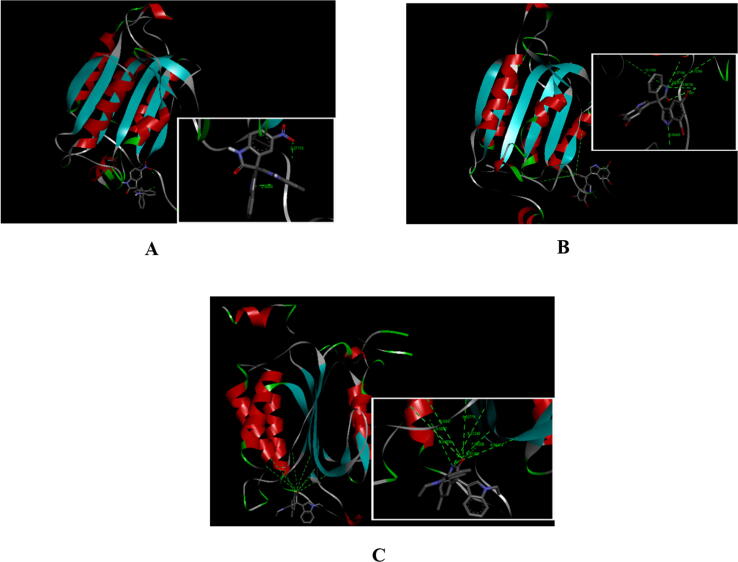
The highest binding affinity value from the molecular docking between the target protein caspase-9 and the trisindoline 1, trisindoline 3 and trisindoline ligands were −7.5 kcal / mol, −7.1 kcal / mol and −7.2 kcal / mol respectively. The results of the protein complex formed in the molecular docking are marked in bold in Table 3 and shown in Fig. 4.
Fig. 4.
(A) Visualization of the molecular docking results of Caspase-9 with Trisindoline 1 (a) and the hydrogen bonds formed (b); (B) Visualization of the molecular docking results of Caspase-9 with Trisindoline 3 (a) and the hydrogen bonds formed (b) and (C) Visualization of the molecular docking results of Caspase-9 with Trisindoline 4 (a) and the hydrogen bonds formed (b).
Based on the visualization and validation of the molecular docking results of caspase-9 with the three trisindoline compounds, trisindoline 1 has the best potential to form protein complexes with caspase-9 with a binding affinity value of −7.5 kcal / mol. Trisindoline 1 also has the highest number of bonds. The bonds distance is between 3 Å and 10 Å. The protein complex formed between caspase-9 and trisindoline 4 has an RMSD value between 1.90 Å and 2 Å. Therefore, it can be used as one of the criteria for the success of the docking method (Jensen, 2007).
3.1.2. Molecular dynamics simulation
Molecular docking provides an overview of the interaction, bonding, and affinity of a ligand (drug) with its receptor, as well as an enzyme with its substrate or inhibitor. Interaction between ligands and proteins in molecular docking occurs when there is a match in shape and volume between the ligand molecule and the protein binding site (Motiejunas and Wade, 2006). The results obtained from molecular docking are still in the form of static molecules, so it is followed by the MD Simulation method which describes the conditions in which temperature, pressure and solvents correspond to the biological system in the human body. MD Simulation is a computer simulation method used to analyse the physical motion of atoms and molecules. Atoms and molecules that interact over a period of time, provides a picture of the changes in the dynamic system. The trajectories of atoms and molecules are determined by Newton's equations of motion for the system of interacting particles, where the force between the particles and the potential energy is calculated using the interatomic potential or molecular mechanical force field (Cruz et al., 2006).
3.1.2.1. Molecular dynamics simulation complex protein Cdk-2
The concentration of CDKs is relatively constant during the cell cycle. CDKs in the free state (unbonded) is inactive because of the catalytic site, where ATP and the substrate bind are blocked by the C-terminal ends of the CKIs. Cyclin can remove this blockage. CDKs become active when they bind and form complexes with cyclin proteins (Manson et al., 2006).
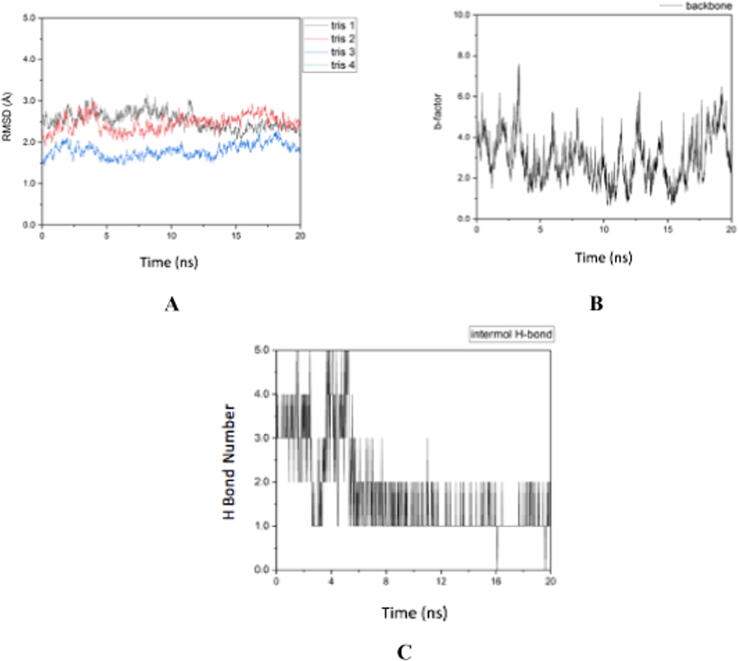
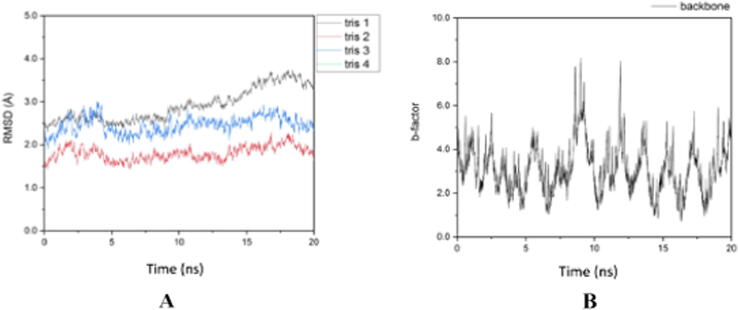
Based on the level of fluctuation of the RMSD results shown in Fig. 5A, it can be seen that the Cdk-2 protein complex is able to bind more stably with trisindoline 3 (blue), indicated by the low peaks and valleys (peaks and valleys) compared to trisindoline 1 (black). While the RMSF in Fig. 5B shows that the level of flexibility of Cdk-2 is quite high (Roe and Cheatham, 2013). When Cdk-2 forms a protein complex with trisindoline 3, a hydrogen bond is formed which activates Cdk-2.
Fig. 5.
(A) RMSD of the Cdk-2 protein complex (B) RMSF of the Cdk-2 protein complex (C) Intermolecular hydrogen bonds of the Cdk-2 protein complex with trisindoline 3.
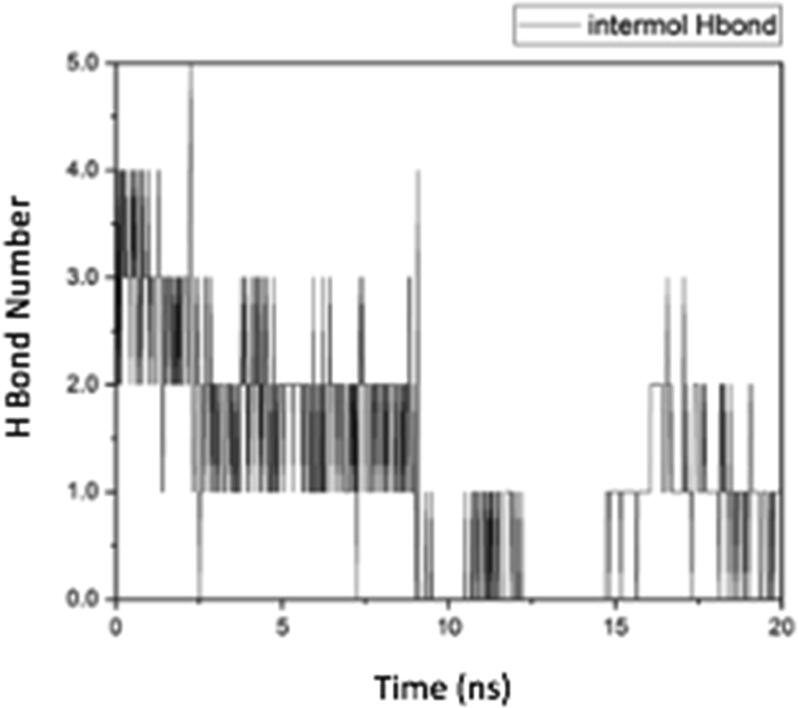
Hydrogen bonds are bonds of hydrogen atoms that bridge two atoms that have large electronegativity. The hydrogen bonds formed before the MD simulation of the Cdk-2 protein complex with trisindoline 3 were SER_231 @ H13 (Serine231 with Hydrogen13), SER_180 @ O1 (Serine 180 with Oxygen1), SER_180 @ H13 (Serine180 with Hydrogen13) and ASN_271 @ H1 (Asparagin271 with Hydrogen1). After 20 ns there was only 1 bond, namely TYR_178 @ H13 (Tyrosine178 with Hydrogen13). The change in the formed hydrogen bond can be seen in Fig. 1 (Roe and Cheatham, 2013). When Cdk-2 is active, it will stimulate the downstream process by phosphorylating specific proteins. The activated Cdk-2 stimulates the cell cycle continuously by phosphorylating and therefore activating specific proteins in the cell that are necessary for the transition to the next stage. For example, at the beginning of mitotic prophase, the breakdown of the nuclear membrane is initiated by the phosphorylation of lamins, which forms part of the nuclear framework.
3.1.2.2. Molecular dynamics simulation of the p-53 protein complex
One of the roles of p53 is to monitor cellular stress and induce apoptosis when DNA lesions are irreversible. Apoptosis is a highly regulated multilevel process characterized by cell shrinkage, chromatin condensation and cell and nuclear fragmentation. In its development, apoptosis is also often referred to as programmed cell death, which continues throughout the life process with the aim of maintaining tissue homeostasis, namely the balance between proliferation and cell death (Bai and Zhu, 2006, Miettinen, 2009).
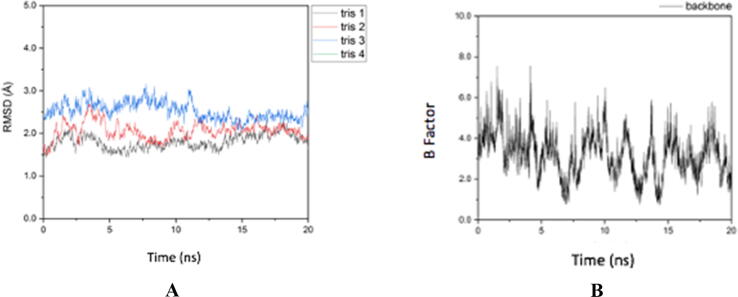
The RMSD results in Fig. 3 shows that trisindoline 1 has the lowest level of fluctuation with the p53 protein complex. Meanwhile, trisindoline 3 experienced high fluctuations at the beginning of the simulation. For trisindoline 4, there were no results because the ionic charge of the formed protein complex could not be neutralized (charge 0). Fig. 3 shows that the RMSF of the p53 protein has high flexibility and fluctuation (b-factor) (Roe and Cheatham, 2013). Based on the results of the fluctuations in the RMSD shown in Fig. 4 and the RMSF shown in Fig. 3, p53 forms a stable protein complex with trisindoline 1.
The hydrogen bonds formed before the MD simulation of the p53 protein complex with trisindoline 1 were ASN_115 @ H13 (Asparagine115 with Hydrogen13), SER_1 @ N1 (Serine1 with Nitrogen1), ARG_114 @ H1 (Arginine114 with Hydrogen1), ARG_63 @ N2 (Arginine63 with Nitrogen). After 20 ns, there was only 1 bond, namely ASP_113 @ H13 (Aspartic Acid113 with Hydrogen13). Changes in the hydrogen bonds formed can be seen in Fig. 7 (Roe and Cheatham, 2013).
Fig. 7.
Intermolecular hydrogen bonds of the p53 protein complex with trisindoline 3.
When p53, a tumor suppressor protein, acts as a master regulator then the various signaling pathways are activated. The role of the p53 protein as a tumor suppressor includes the ability to hold the cell cycle, DNA repair, and make apoptosis occur. The activity of the p53 protein can be stabilized by inhibiting the interaction between p53 and the ligands (Collavin et al., 2010, Rivlin et al., 2011).
3.1.2.3. Molecular dynamics simulation complex protein Caspase-9
Caspase is a family of protease enzymes that play a key role in cellular processes such as apoptosis and inflammation (Mace et al., 2014). Caspase, known as the apoptotic executor, is a selective cysteine protease that controls all stages of apoptosis. Procaspase-9 is an inactive caspase-9, and when it activates it becomes caspase-9 (initiator caspase) (Wong, 2011).
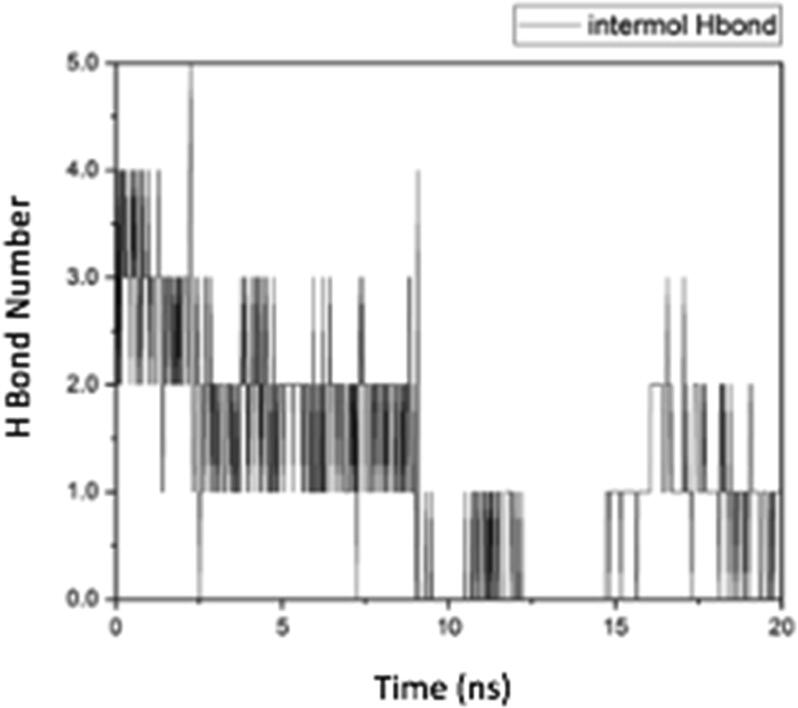
Based on the level of fluctuation of the RMSD results shown in Fig. 8 (A) caspase-9 forms hydrogen bonds and a stable protein complex with trisindoline 1 (black color) which can be seen in Fig. 8 (A). The RMSF shown in Fig. 8 (B) demonstrated the high degree of fluctuation and flexibility of the caspase-9 protein (see Fig. 9).
Fig. 8.
(A) RMSD Caspase-9 protein complex (B) RMSF Caspase-9 protein complex.
Fig. 9.
Intermolecular hydrogen bonds of the Caspase-9 protein complex with trisindoline 3.
The hydrogen bonds formed before the MD simulation of the caspase-9 protein complex with trisindoline 1 were THR_337 @ O2 (Threonine337 with Oxygen2), PRO_336 @ H1 (Proline336 with Hydrogen1). After 20 ns, there was only 1 bond, namely PRO_336 @ H1 (Proline336 with Hydrogen1) (Roe and Cheatham, 2013).
In the MD simulation it was concluded that Cdk-2 protein formed a stable bond with trisindoline 3, p53 formed a stable bond with trisindoline 1 and caspase-9 formed a stable bond with trisindoline 1.
After the MD simulation was carried out on the three trisindolines, it was found that the trisindoline 1 and trisindoline 3 ligands had the potential to become anti-cancer drugs. Both trisindoline 1 and trisindoline 3 were able to trigger the apotosis process. Apoptosis was triggered by multiple signaling pathways and regulated by complex extrinsic and intrinsic ligands.
When Cdk-2 forms a complex with trisindoline 3, it induces apoptosis via the intrinsic pathway. Furthermore, it will induce the activity of BH-3 and continue to activate BAX and the mitochondrial outer membrane (MOMP). MOMP will then induce the release of cytochrome c from the intermembrane mitochondrial space (IMS) into the cytosol which in turn activates caspase and apoptosis (Dewson and Kluck, 2009, Sáez and Villunger, 2016).
Meanwhile, when trisindoline 1 forms a protein complex with p53, it induces apoptosis via the extrinsic pathway, where the active p53 induces genes encoding 3 transmembrane proteins, namely: Tumor Necrosis Factor receptor 1 (TNFR1), Fas, and Death Receptor-5 (DR- 5). This protein activates the transducer signal which in turn activates the caspase. p53 induces Fas protein through binding elements on the promoter and introns. p53 also activates DR-5 and TNFR1 in response to DNA damage and through caspase-9, triggers cell death (Elmore, 2007).
Trisindoline 1 was also able to form bonds with caspase-9. The bonds are formed in the catalytic domain that converts the procaspase-9 into caspase-9. The activated caspase-9 induces the procaspase-3 stream. The caspase-3 protein breaks down various types of substrates, including DNA repair enzymes such as poly-ADP Ribose Polymerase (PARP) and DNA protein kinases such as cellular and nuclear structural proteins, including the core mitotic apparatus, nuclear laminae and actin and endonucleases such as the Caspase-Activated Deoxyribonuclease (ICAD) inhibitor and other cellular constituents (Wong, 2011). Caspase-3 is an effector caspase that carries out the apoptosis process (Wong, 2011).
3.2. Cytotoxicity assay
The results of the cytotoxicity test and the IC50 values obtained for the three trisindoline compounds against the MCF-7 breast cancer cell line are shown in Table 4.
Table 4.
IC50 Value of Four Test Compounds against Cell Line.
| No | Compund | IC50 |
|---|---|---|
| 1 | Trisindoline 1 | 2.059 μM |
| 2 | Trisindoline 3 | 3.9759 μM |
| 3 | Trisindoline 4 | 15.46 μM |
| 4 | Doxorubicin | 9.88 Μm |
Based on Table 4, trisindoline 1 shows an IC50 value in the range of compounds that have high potential. Trisindoline 3 has a higher cytotoxicity than trisindoline 4 (Fortes et al., 2016). The best activity was exhibited by trisindoline 1 with an IC50 value of 2.059 μM. Based on the results of the cytotoxicity test, trisindoline 1 and trisindoline 3 possess a good cytotoxicity effect and are more effective at inducing apoptosis in MCF-7 breast cancer cell line compared to the chemotherapy agent Doxorubicin. Whilst trisindoline 4 is considered potential but less effective as a chemotherapy agent when compared to Doxorubicin, a compound commonly used in the treatment of breast cancer. The potency of Doxorubicin is well known. It is used in the treatment of breast cancer through two mechanisms, the first is the intercalation of doxorubicin into DNA and the disruption of topoisomerase-II-mediated DNA repair and the second is the generation of free radicals and their damage to cellular membranes, DNA and proteins (de Campos-Nebel et al., 2010, Thorn et al., 2011). However, according to National Cancer International, the three compounds, namely trisindoline 1, trisindoline 3 and trisindoline 4 are classified as compounds that have the potential to be developed as anti-cancer agents due to the fact that the three compounds has an IC50 value of<20 µg/mL(Munandar et al., 2015).
The following is a brief comparison between the capabilities of the trisindoline and doxorubicin mechanisms. Doxorubicin is oxidized into semiquinone, an unstable metabolite, which is converted back to doxorubicin in a process that releases reactive oxygen species (Zhang et al., 2009). Reactive oxygen species leads to lipid peroxidation and membrane damage, DNA damage, oxidative stress, and triggers apoptotic pathways of cell death (Quinzii et al., 2010, Su et al., 2019) whilst trisindoline in oxidureductase in metabolic pathways results in hydrolysis of the third C of trisindoline. This hydrolysis produces 2 indole molecules and 1 isatin molecule. Isatin and indole condensation forms the indirubin structure (Yoo et al., 2008). Indirubin induces cytotoxicity by inhibiting cyclin-dependent kinase (CDK) 2, by attaching its molecule to the phosphorylated Thr-160 CDK2 site to form an inactive CDK-2 structure, shown in Appendix 6 (Davies et al., 2001). This results in inhibited cell synthesis and cell death.
The difference in value from the cytotoxicity test results of the three trisindoline compounds was due to the addition of the different groups in each compound. Trisindoline 1, which received the addition of a nitro group, showed an increase in activity compared to the other trisindoline compounds that was not added with the nitro group. In theory, this is in accordance with Elomri et al. (1999) which stated that the reduction of nitro compounds to amino compounds increases the toxicity of acronisine compounds against L1210 leukemia cells.
However, the addition of the nitro group can have a different effect, depending on the type of cell being treated. Nepali et al., 2019, Olender et al., 2018 stated that the aromatic nitro compound, has a better toxicity than aromatic amino against P388 leukemia cells. Aromatic amino compounds are known to be the result of reduction from nitro compounds.
The nitro group affects the reduction of the cyclin E1 protein expression as an activator of the cyclin kinase-dependent enzyme (CDK) 2 enzyme by inhibiting the expression of the CCNE1 gene (Chandarlapaty and Razavi, 2019). If E1 protein expression is inhibited, CDK-2 as a regulatory enzyme at the check point from phase G1 to phase S becomes inactive. This causes cell growth to stop (Bertoli et al., 2013, Mazumder et al., 2004). Trisindoline 1, which has the best IC50 value compared to the other three test compounds proves that the inhibition mechanism of the E1 expression is the best mechanism of cytotoxicity. This is because MCF-7 cells overexpress E1 protein (Bertoli et al., 2013, Mazumder et al., 2004).
In addition, trisindoline 3, which has an IC50 value of 3.9759 μM, was also classified as a compound with good activity. This value is thought to be obtained due to the addition of the bromo group. When compared to the results of the trisindoline compound toxicity test against MCF-7 Cell Line, trisindoline 3 is considered to have better cytotoxicity activity, because trisindoline compounds have a higher IC50 value, namely 49.8 μM. This result is inconsistent with the research conducted by Fortes et al. (2016) which states that the addition of a donor electron group (MeO), one of which is 5-bromo, to 3-thiocyanato-1H-indole reduces the level of toxicity of a compound. The Bromo group itself is one of the most commonly used anti-cancer compounds, namely cisplatin (Granchi et al., 2014). The 5-Bromo-deoxyuridin compound in cisplatin, is a pyruvate halogenization compound that works by inhibiting hexokinase, a catalytic enzyme in the initial glycolysis process, which transfers one phosphate from ATP to glucose molecules and produces 6-phosphate, according to Fig. 8 (Granchi et al., 2014). In addition, 5-Bromo-deoxyuridin also works by inhibiting glyceraldehyde-3-phosphate dehydrogenase (GAPDH) through its reaction with the nucleophile –SH site of the enzyme (Granchi et al., 2014).
Furthermore, the results of the cytotoxicity test for trisindoline 4 yielded an IC50 value of 15.46 μM. This value is still categorized as a good value of IC50. These results are in accordance with the theory put forward by Fessenden & Fessenden (1986) which states that members of the halogen group, one of which is the Chloro (Cl) compound, is more reactive when bonded to alkyl carbon, for example ethyl, methyl and butyl. Trisindoline 4, namely trisindoline with the addition of a dimethyl-nitro group, is an example of a bond between members of the halogen group and activated carbon. Fortes et al. (2016) also stated that the chloro derivative of the 3-thiocyanato-1H-indole compound showed good cytotoxicity potential against MCF-7 breast cancer cells as well as other types of cancer cells.
In addition, the two chloride ligands also become the leaving group of cisplatin which supports its toxicity mechanism by cross-linking with purine bases in DNA (Dasari and Tchounwou, 2014).
Based on the results of the cytotoxicity test, it can be concluded that the best addition of groups to the trisindoline compound is the addition of the nitro and bromo groups. The addition of a chloro group may cause an increase in the cytotoxicity activity of trisindoline, but it was less significant..
3.3. Phagocytosis assay
3.3.1. Macrophage morphology of Mus musculus
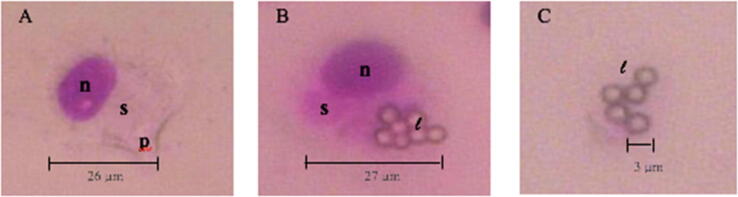
In vitro phagocytosis activity test of macrophage cells is an initial method for screening active ingredients that affect immune responses. In this study, an immunomodulatory response is an increase of the immune system. Increased ability of macrophage is measured by their activity in phagocytic infectious agents, such as latex beads or latex particles. Phagocytic activity of macrophage is expressed by the phagocytosis index and macrophage capacity (Karavitis and Kovacs, 2011, Tatarczuch et al., 2002). Macrophages were isolated from the peritoneal cavity (PM). The average macrophage cell has a size of 18–28 μm. Analysis of macrophage cell morphology was carried out using a light microscope at 400x magnification using Optilab assistance and is shown in Fig. 10.
Fig. 10.
Morphology of macrophage cells in mice (Mus musculus) with a magnification of 400x. A) Normal macrophages, B) macrophages that phagocytes latex particles, C) Latex particles free. Note: n = nucleus, s = cytoplasm, p = pseudopodia, l = latex.
Normal macrophage cells that have not been infected by antigens appear in a state that has not been activated (Fig. 10A). The morphology of large round cells with eccentric nuclei does not indicate the presence of pseudopodia macrophages (Nakada et al., 2014). Histologically the macrophages are round, about 10–30 µm in diameter, and the surface of the cell is uneven and has finger-like bumps (Zhang et al., 2008). Trisindoline 1, 3 and 4 in macrophage cells activates the recognition receptors of macrophage cells towards latex beads thus increasing the ability of macrophage cells to phagocyte latex beads (Fig. 10B). These active macrophage cells are characterized by the cytoplasm having a small / solid vesicles and an oval / eccentric nuclei (Zhang et al., 2008). Fig. 10A shows macrophage cells that do not phagocyte normal latex particles. Fig. 10B shows a macrophage cell that phagocytes latex particles. The rounded cell shape has pseudopods which phagocytes latex. (Fig. 10A and 10B). Active macrophages have large cell sizes (Baranov et al., 2021, Fröhlich, 2015, Murray and Wynn, 2011). Round cell forms with sizes of around 15–40 μm, have pseudopods and the p53 nuclei tend to be large. This study used 2–3 μm polystyrene latex particles as antigens. According to Hirota and Terada (2012) have the ability to phagocyte particles with sizes between 1 and 10 μm thus the latex used is considered to be responded well by macrophages.
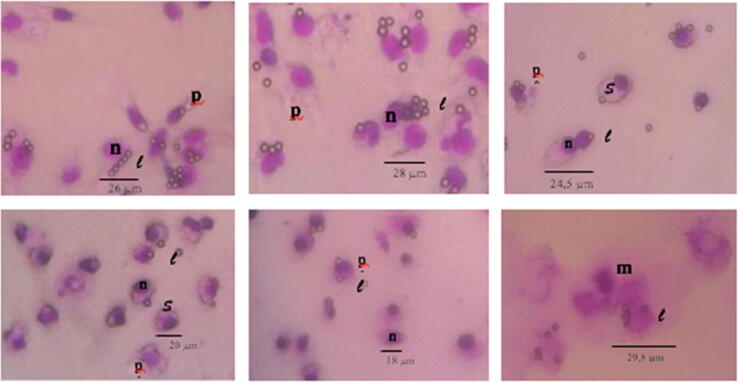
Normal macrophage cells have not been infected by antigens in the treatment with a concentration of 0 µg / mL (Fig. 11a). In large macrophage cells with homogeneous pseudopodia, the cell nucleus is conspicuous and purple. The macrophages treated with trisindoline causes activation (Fig. 11b). Activation of this macrophage occurs at a low concentration of 6.25 μg / mL, where the cell size is larger (Fig. 11b), whereas at concentrations of 50 µg / mL appeared with a larger cell size, a shorter pseudopodia and a smaller nuclei (Fig. 11e). Activated macrophages have a larger cell size and a widened pseudopodia (Hirota and Terada, 2012). Large macrophages with a widened pseudopodia are the characteristics of activated macrophages (Glanz et al., 2019, Sheikh et al., 2015). The activation of macrophages occurs due to the receptor bonds on their ligands. After the receptors recognizes the ligands, the subsequent initiation of the rearrangement of cytoskeleton proteins causes membrane protrusion and remodeling of actin filaments and as a result the cytoskeleton become wider (Fletcher and Mullins, 2010, Tang and Gerlach, 2017). This bond initiates changes in the composition of proteins in the cytoskeleton and causes changes in the shape of the cytoskeleton. In this study the ligands were trisindoline and latex particles. In the treatment with doxorubicin, macrophage cells were damaged by morphological changes. The shape of the cell becomes irregular and some cells of the macrophage are fragmented into separate smaller parts (Fig. 11f). This is presumably because doxorubicin rapidly enters cells and strongly binds to base pairs in DNA. The bond between doxorubicin and DNA causes damage to the double strand of DNA and triggers cell apoptosis (Gregory and Devitt, 2004, Tacar et al., 2012, Yang et al., 2014).
Fig. 11.
The morphology of macrophage cells of mice treated with trisindoline with concentrations of (a) 0 μg/mL, (b) 6.25 μg/mL, (c) 12.5 μg/mL, (d) 25 μg/mL, (e) 50 μg/mL, and with doxorubicin (f) with a concentration of 12.5 μg/mL. A magnification of 400x was used. Note: m = macrophages n = nucleus, s = cytoplasm, p = pseudopodia, l = latex.
3.3.2. Phagocytosis capacity of macrophage
ANOVA test on compound treatment showed significant results (p < 0.05) and showed significant differences. The subsequent Tukey test showed that trisindoline 1, 3 and 4 exhibited significant differences. Trisindoline 1, 3 and 4 were able to increase the phagocytosis capacity of the macrophage cells of mice (Table 5). Significant increase of the phagocytic capacity in trisindoline 1 and 3 were in a concentration of 6.25 µg/mL. This is in accordance to the research conducted by (Hubbard et al., 2015) that used alkaloid compounds and found that the optimal concentration to increase macrophage activity was 10 µg/mL, which is also a low concentration (see Table 6).
Table 5.
Phagocytosis capacity of macrophage cells of mice induced by trisindoline 1, trisindoline 3, trisindoline 4 and doxorubicin.
| Compound concentration | Phagocytosis capacity |
|||
|---|---|---|---|---|
| trisindoline 1 | trisindoline 3 | trisindoline 4 | doxorubicin | |
| 0 μg/mL | 31.83 ± 4.01c | 33.17 ± 1.18b | 21.33 ± 5.24a | 44.17 ± 1.18c |
| 6.25 μg/mL | 58.33 ± 2.35c | 59.00 ± 0.95b | 23.44 ± 7.16a | 9.11 ± 2.79ab |
| 12.5 μg/mL | 21.16 ± 1.18b | 16.50 ± 1.65a | 21.89 ± 1.68a | 15.64 ± 7.89ab |
| 25 μg/mL | 9.00 ± 0.94a | 13.00 ± 2.36a | 17.33 ± 2.90a | 17.71 ± 3.44b |
| 50 μg/mL | 11.50 ± 0.24a | 10.67 ± 1.41a | 32.89 ± 1.49a | 3.53 ± 0.91a |
Note: Different superscripts in the same column showed significant differences (p < 0.05).
Table 6.
Phagocytosis capacity of macrophage cells of mice induced by trisindoline 1, trisindoline 3, trisindoline 4 and doxorubicin.
| Compound concentration | Phagocytosis capacity |
|||
|---|---|---|---|---|
| trisindoline 1 | trisindoline 3 | trisindoline 4 | doxorubicin | |
| 0 μg/mL | 1.94 ± 0.01abc | 2.31 ± 0.17b | 1.80 ± 0.30b | 2.31 ± 0.17b |
| 6.25 μg/mL | 2.73 ± 0.23c | 2.96 ± 0.59b | 1.95 ± 0.18a | 0.41 ± 0.40a |
| 12.5 μg/mL | 2.52 ± 0.46bc | 1.43 ± 0.13a | 1.99 ± 0,18a | 0.92 ± 0.65a |
| 25 μg/mL | 1.59 ± 0.24ab | 1.40 ± 0.18a | 2.21 ± 0.39a | 0.37 ± 0.11a |
| 50 μg/mL | 1.27 ± 0.02a | 1.22 ± 0.57a | 1.92 ± 0.21a | 0.07 ± 0.01a |
Note: Different superscripts in the same column showed significant differences (p < 0.05).
The optimal trisindoline 4 needed to increase the value of the phagocytic capacity is 50 μg/mL. In the doxorubicin treatment, the value of the phagocytic capacity increased at concentrations of 6.75, 12.5, and 25 μg/mL, then decreased at concentrations of 50 and 100 μg/mL. This is related to the toxicity of trisindoline at high concentrations.
3.3.3. Phagocytocic index of Macrophage
The phagocytosis index of macrophage cells is calculated based on percentage of latex particles ingested by active macrophage cells (Seyrantepe et al., 2010, Wang et al., 2017). The optimal phagocytosis index of macrophage cells was found at a concentration of 6.25 μg/mL.
The lowest index value was at a concentration of 50 μg/mL. The trisindoline 3 treatment showed the highest index value of 2.96 at a concentration of 6.25 μg / mL, followed by an index value of 2.31 at the control of 0 μg / mL, 1.43 at a concentration of 12.5 μg/mL, 1.40 at a concentration of 25 μg/mL, and the lowest index value was 1.22 at a concentration of 50 μg/mL group. There were no significant differences for trisindoline 4 at concentrations of 6.25, 12.5, 25 and 50 µg/mL, and was only significantly different at the control of 0 µg/mL. In the doxorubicin treatment there was no difference between treatments, even the control showed a high phagocytic index value. This shows that doxorubicin was not able to induce the phagocytosis ability of macrophage cells.
Trisindoline 1 is a trisindoline aromatic compound with the addition of a Nitro (NO2) group or commonly called aromatic nitration (Fig. 4). The nitro aromatic group in cells has a high reactivity and as a result easily binds to receptors on macrophages and causes an increase in NFĸB receptor activity21. NFĸB in macrophage cells is a transcription factor iNOS which plays a role in increasing the response of macrophages to antigens. In trisindoline 3 (Fig. 5) there are additional elements of Br, which belongs to the halogen group, and it is known that halogenated aromatic ligands increases affinity or strengthens interactions between ligands and receptors making the activation more optimal.
Trisindoline 3 can better activate macrophages through the AhR receptor but only at low concentrations of 6.25 μg / mL. Trisindoline 4 is a member of the halogen group, one of which is Kloro (Cl) compound, and is more reactive if it is bound to alkyl carbon, for example ethyl, methyl and butyl. Trisindoline 4 (Fig. 6) with the addition of the dimethyl-nitro group, is an example of the bond between members of the halogen group and activated carbon22. Furthermore, the two chloride ligands are the leaving group of cisplatin which supports its toxicity through the formation of crosslinks with purine bases in DNA (Tacar et al., 2012).
Fig. 6.
(A) RMSD of the Cdk-2 protein complex (B) RMSF of the Cdk-2 protein complex.
The capacity of macrophages is the number of macrophages that ingests latex particles in the observed 300 macrophages18. A concentration of 6.25 μg/mL is the optimum trisindoline concentration to induce macrophage phagocytic capacity. This is related to the addition of trisindoline which causes the innate immune system performance to increase when given indole derivative compounds (Hirayama et al., 2018, Wang et al., 2016). Indole compounds such as indirubin and methyl indole isomers are able to modulate immune cell activity in the gastrointestinal tract. Indol compounds activates Arylhydrocarbon Receptor (AhR) in macrophages and subsequently through secretion of cytokines, increases the immune system activity (Naga and Rajeshwari, 2014). Active macrophages experience changes in their morphology and function thus causing active macrophages to have a higher ability to do phagocytosis compared to inactive macrophages (Hirayama et al., 2018). The trisindoline compound itself is an indole compound included in the class of alkaloid compounds (Netz and Opatz, 2015) which metabolized through oxygenase pathways, namely oxidation into two indole groups and one isatin group. The presence of oxygen condenses indole with isatin and produces indirubin. Indirubin and indole are known to have the ability to activate AhR in macrophages (Benson and Shepherd, 2011, Hubbard et al., 2015). AhR is a transcriptional factor activated by ligands (Zhang et al., 2015) such as natural materials that include compounds in food ingredients such as flavonoids and indoles, tryptophan amino acid derivative metabolites and other compounds with a basic framework in the form of indole structures (Thelen et al., 2010). Although the mechanism is not yet clearly known, the activation of AhR can control the secretion of cytokines, induce anti-inflammatory factors in macrophages and cause modulated immune cell activity (Zhang et al., 2015).
Higher concentrations of trisindoline may reduce the capacity of macrophages due to the fact that trisindoline has a fairly high cytotoxic activity as the concentration of compounds increases (Munandar et al., 2015) and it is thought to interfere with the performance of immune cells. For this reason, trisindoline 3 compounds are not suitable to be used as immunostimulant compounds. In the treatment with doxorubicin, macrophages were still able to ingest latex particles, but the value of their capacity was far below the control. This is because doxorubicin, when used as a chemotherapy drug, has side effects in the form of immunosuppression and as a result, the components of the immune system may not work normally. In addition, macrophage cells also experienced apoptosis because doxorubicin binds enzymes associated with DNA, disturbing the base pairs of DNA causing DNA damage which leads to cell death (Berraondo et al., 2019, Larionova et al., 2019, Zhao et al., 2021).
Latex is a protein antigen derived from rubber latex, with a total BM ± 500 (Nucera et al., 2020). Latex beads are polystyrene micro particles consisting of water, polymers (polysaccharides / proteins), surfactants and inorganic salts (Hoshino et al., 2014).
Trisindoline 1, 3 and 4 in macrophage cells activates recognition receptors against latex bead and as a result the ability of macrophage cells to ingest latex beads increases (Zhang et al., 2015). The phagocytosis process of latex beads by macrophages begins with the introduction of latex particles by the MARCO receptor located on the surface of macrophage cells. The next process is the ingestion of latex particles by macrophages. The process of bacterial ingestion occurs because phagocytes form pseudopodia protrusions on the plasma membrane (Munandar et al., 2015). Then phagosomes enter the cytoplasm, they will fuse with lysosomes and form phagolisosomes.
Nonopsonized latex can be recognized by the MARCO receptor on macrophages, namely one member of the Scavenger Receptor (SR) - AI / II (Arredouani et al., 2006) which is a family of ligand recognition receptors, including lipoprotein and protein, apolipoprotein AI and E, collagen, asbestos, silica, fukoidan, carrageenan, lipopolysaccharide (LPS), apoptotic cells, etc (Zhang et al., 2015). Phagocytic particles via the MARCO receptor caused the expression of immunoregulatory cytokines such as IL-10 and TGF-β. Both cytokines are known to cause polarized macrophages to become anti-inflammatory macrophages (M2) which have particle elimination activities that impacts the increasing in phagocytic activity (Hirayama et al., 2018, Murray and Wynn, 2011).
4. Conclusion
Based on research on the anti-cancer potency of trisindoline compounds 1, 3 and 4 against the anti-cancer mechanism through the apoptotic pathway, it can be concluded that the best RMSD for the Cdk-2 protein complex was with trisindoline 3, the best RMSD for the p53 protein complex was with trisindoline 1 and the best RMSD for the caspase-9 protein complex was with trisindoline. The ligands that have the potential to become anti-cancer drug candidates are trisindoline 1 and trisindoline 3. Trisindoline 1,3 and 4 are immunostimulants at low concentrations but are immunosuppressants at high concentrations. The RMSD of the Cdk-2 protein complex with trisindoline 3 was the best, indicated by the lowest level of fluctuation. The RMSF showed that Cdk-2 has a high degree of flexibility. The number of intramolecular hydrogen bonds ranged from 1 to 5. The RMSD of the p53 protein complex with trisindoline 1 was the best. The RMSF showed that p53 has a high degree of flexibility. The number of intramolecular hydrogen bonds ranged from 1 to 5. The results of the cytotoxicity test that was carried out showed the potential for cytotoxicity against MCF-7 breast cancer cell line of all three trisindolines. Trisindoline 1 had an IC50 value of 2.059 μM, trisindoline 3 had an IC50 value of 3,9759 μM and trisindoline 4 had an IC50 value of 15.46 μM. The trisindoline 1 and trisindoline 3 compounds at a concentration of 6.25 µg/mL were able to induce phagocytosis capacity of macrophage cells by 38.34; whereas trisindoline 4 was able to induce phagocytosis capacity of 32.89 at a concentration of 50 µg/mL. Trisindoline 1, 3 and 4 are immunostimulant at low concentrations and are immunosuppressant at higher concentration.
Funding
This research was funded by The Ministry of Education, Culture, Research and Technology, Republic of Indonesia (Grant number : 8/E1/KPT/2021 874/PKS/ITS/2021).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
The authors thank to Sukorini, Dyah, Rizqi, Dimas, Silvia, Novia, Andina, and Zulfina for supporting this research.
Footnotes
Peer review under responsibility of King Saud University.
References
- Arredouani M.S., Yang Z., Imrich A., Ning Y., Qin G., Kobzik L. The macrophage scavenger receptor SR-AI/II and lung defense against pneumococci and particles. Am. J. Respir. Cell Mol. Biol. 2006;35(4):474–478. doi: 10.1165/rcmb.2006-0128OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arwansyah, Ambarsari L., Sumaryada T.I. Simulasi Docking Senyawa Kurkumin dan Analognya Sebagai Inhibitor Reseptor Androgen pada Kanker Prostat. Curr. Biochem. 2014;1:11–19. doi: 10.29244/cb.1.1.11-19. [DOI] [Google Scholar]
- Bai L., Zhu W.-G. p53: Structure, Function dan Therapeutic Applications. J. Cancer Mol. 2006;2:141–153. [Google Scholar]
- Baranov M.V., Kumar M., Sacanna S., Thutupalli S., van den Bogaart G. Modulation of Immune Responses by Particle Size and Shape. Front. Immunol. 2021;11:1–23. doi: 10.3389/fimmu.2020.607945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson J.M., Shepherd D.M. Dietary ligands of the aryl hydrocarbon receptor induce anti-inflammatory and immunoregulatory effects on murine dendritic cells. Toxicol. Sci. 2011;124:327–338. doi: 10.1093/toxsci/kfr249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berraondo P., Sanmamed M.F., Ochoa M.C., Etxeberria I., Aznar M.A., Pérez-Gracia J.L., Rodríguez-Ruiz M.E., Ponz-Sarvise M., Castañón E., Melero I. Cytokines in clinical cancer immunotherapy. Br. J. Cancer. 2019;120(1):6–15. doi: 10.1038/s41416-018-0328-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertoli C., Skotheim J.M., de Bruin R.A.M. Control of cell cycle transcription during G1 and S phases. Nat. Rev. Mol. Cell Biol. 2013;14(8):518–528. doi: 10.1038/nrm3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley S.K., Berman H.M., Bhikadiya C., Bi C., Chen L., Di Costanzo L., Christie C., Dalenberg K., Duarte J.M., Dutta S., Feng Z., Ghosh S., Goodsell D.S., Green R.K., Guranović V., Guzenko D., Hudson B.P., Kalro T., Liang Y., Lowe R., Namkoong H., Peisach E., Periskova I., Prlić A., Randle C., Rose A., Rose P., Sala R., Sekharan M., Shao C., Tan L., Tao Y.P., Valasatava Y., Voigt M., Westbrook J., Woo J., Yang H., Young J., Zhuravleva M., Zardecki C. RCSB Protein Data Bank: Biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Res. 2019;47:D464–D474. doi: 10.1093/nar/gky1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandarlapaty S., Razavi P. Cyclin e mRNA: Assessing cyclin-dependent kinase (CDK) activation state to elucidate breast cancer resistance to CDK4/6 Inhibitors. J. Clin. Oncol. 2019;37(14):1148–1150. doi: 10.1200/JCO.19.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y., Gorina S., Jeffrey P.D., Pavletich N.P. Crystal structure of a p53 tumor suppressor-DNA complex: Understanding tumorigenic mutations. Science (80-. 1994;265(5170):346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- Collavin L., Lunardi A., Del Sal G. P53-family proteins and their regulators: Hubs and spokes in tumor suppression. Cell Death Differ. 2010;17(6):901–911. doi: 10.1038/cdd.2010.35. [DOI] [PubMed] [Google Scholar]
- Cruz F.J.A.L., Lopes J.N.C., Calado J.C.G. Molecular dynamics simulations of molten calcium hydroxyapatite. Fluid Phase Equilib. 2006;241(1-2):51–58. doi: 10.1016/j.fluid.2005.12.021. [DOI] [Google Scholar]
- Dasari S., Tchounwou P.B. Cisplatin in cancer therapy: Molecular mechanisms of action. Eur. J. Pharmacol. 2014;740:364–378. doi: 10.1016/j.ejphar.2014.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies T.G., Tunnah P., Meijer L., Marko D., Eisenbrand G., Endicott J.A., Noble M.E.M. Inhibitor binding to active and inactive CDK2: The crystal structure of CDK2-cyclin A/indirubin-5-sulphonate. Structure. 2001;9(5):389–397. doi: 10.1016/S0969-2126(01)00598-6. [DOI] [PubMed] [Google Scholar]
- de Campos-Nebel M., Larripa I., González-Cid M., Harris R.S. Topoisomerase ii-mediated DNA damage is differently repaired during the cell cycle by non-homologous end joining and homologous recombination. PLoS ONE. 2010;5(9):e12541. doi: 10.1371/journal.pone.0012541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Sousa A.C.C., Combrinck J.M., Maepa K., Egan T.J. Virtual screening as a tool to discover new β-haematin inhibitors with activity against malaria parasites. Sci. Rep. 2020;10:3374. doi: 10.1038/s41598-020-60221-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewson G., Kluck R.M. Mechanisms by which Bak and Bax permeabilise mitochondria during apoptosis. J. Cell Sci. 2009;122:2801–2808. doi: 10.1242/jcs.038166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore S. Apoptosis: A Review of Programmed Cell Death. Toxicol. Pathol. 2007;35(4):495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elomri A., Michel S., Seguin E., Tillequin F., Koch M., Pierre A., Atassi G. Synthesis and Cytotoxic Activity of 11-Nitro and 11-Amino Derivatives of Acronycine and 6-Demethoxyacronicine. Chem. Pharm. Bull. 1999;47:1604–1606. doi: 10.1248/cpb.47.1604. [DOI] [PubMed] [Google Scholar]
- Ferreira L., dos Santos R., Oliva G., Andricopulo A. Molecular docking and structure-based drug design strategies. Molecules. 2015;20(7):13384–13421. doi: 10.3390/molecules200713384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fessenden, R., Fessenden, J.S., 1986. Organic Chemistry Third Edition. Brooks/Cole Publishing Company, Belmont California. 10.1007/s10750-007-9195-x. [DOI]
- Fletcher D.A., Mullins R.D. Cell mechanics and the cytoskeleton. Nature. 2010;463(7280):485–492. doi: 10.1038/nature08908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortes M.P., Da Silva P.B.N., Da Silva T.G., Kaufman T.S., Militão G.C.G., Silveira C.C. Synthesis and preliminary evaluation of 3-thiocyanato-1H-indoles as potential anticancer agents. Eur. J. Med. Chem. 2016;118:21–26. doi: 10.1016/j.ejmech.2016.04.039. [DOI] [PubMed] [Google Scholar]
- Fröhlich E. Value of phagocyte function screening for immunotoxicity of nanoparticles in vivo. Int. J. Nanomedicine. 2015;10:3761–3778. doi: 10.2147/IJN.S83068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glanz V., Myasoedova V.A., Sukhorukov V., Grechko A., Zhang D., Romaneneko E.B., Orekhova V.A., Orekhov A. Transcriptional Characteristics of Activated Macrophages. Curr. Pharm. Des. 2019;25(3):213–217. doi: 10.2174/1381612825666190319120132. [DOI] [PubMed] [Google Scholar]
- Granchi C., Fancelli D., Minutolo F. An update on therapeutic opportunities offered by cancer glycolytic metabolism. Bioorganic Med. Chem. Lett. 2014;24(21):4915–4925. doi: 10.1016/j.bmcl.2014.09.041. [DOI] [PubMed] [Google Scholar]
- Gregory C.D., Devitt A. The macrophage and the apoptotic cell: An innate immune interaction viewed simplistically? Immunology. 2004;113(1):1–14. doi: 10.1111/j.1365-2567.2004.01959.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama D., Iida T., Nakase H. The Phagocytic Function of Macrophage-Enforcing Innate Immunity and Tissue Homeostasis. Int. J. Mol. Sci. 2018;19:92. doi: 10.3390/ijms19010092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota, K., Terada, H., 2012. Endocytosis of Particle Formulations by Macrophages and Its Application to Clinical Treatment, in: Molecular Regulation of Endocytosis. InTech. 10.5772/4582. [DOI]
- Hoshino Y.u., Lee H., Miura Y. Interaction between synthetic particles and biomacromolecules: Fundamental study of nonspecific interaction and design of nanoparticles that recognize target molecules. Polym. J. 2014;46(9):537–545. doi: 10.1038/pj.2014.33. [DOI] [Google Scholar]
- Huang S.-Y., Zou X. Efficient molecular docking of NMR structures: Application to HIV-1 protease. Protein Sci. 2006;16:43–51. doi: 10.1110/ps.062501507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard T.D., Murray I.A., Bisson W.H., Lahoti T.S., Gowda K., Amin S.G., Patterson A.D., Perdew G.H. Adaptation of the human aryl hydrocarbon receptor to sense microbiota-derived indoles. Sci. Rep. 2015;5:1–13. doi: 10.1038/srep12689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irina K., Ruben A. Methods of protein structure comparison. Methods Mol. Biol. 2012:231–257. doi: 10.1007/978-1-61779-588-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen F. Second Edition. Wiley; Odense: 2007. Introduction to Computational Chemistry. [Google Scholar]
- Joerger A.C., Bauer M.R., Wilcken R., Baud M.G.J., Harbrecht H., Exner T.E., Boeckler F.M., Spencer J., Fersht A.R. Exploiting Transient Protein States for the Design of Small-Molecule Stabilizers of Mutant p53. Structure. 2015;23(12):2246–2255. doi: 10.1016/j.str.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamal A., Srikanth Y.V.V., Khan M.N.A., Shaik T.B., Ashraf M.d. Synthesis of 3,3-diindolyl oxyindoles efficiently catalysed by FeCl 3 and their in vitro evaluation for anticancer activity. Bioorganic Med. Chem. Lett. 2010;20(17):5229–5231. doi: 10.1016/j.bmcl.2010.06.152. [DOI] [PubMed] [Google Scholar]
- Karavitis J., Kovacs E.J. Macrophage phagocytosis: effects of environmental pollutants, alcohol, cigarette smoke, and other external factors. J. Leukoc. Biol. 2011;90(6):1065–1078. doi: 10.1189/jlb.0311114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan K.H., Blanco-Codesido M., Molife L.R. Cancer therapeutics: Targeting the apoptotic pathway. Crit. Rev. Oncol. Hematol. 2014;90(3):200–219. doi: 10.1016/j.critrevonc.2013.12.012. [DOI] [PubMed] [Google Scholar]
- Kobayashi M., Aoki S., Gato K., Matsunami K., Kurosu M., Kitagawa I. Marine Natural Products. XXXIV. Trisindoline, a New Antibiotic Indole Trimer, Produced by a Bacterium of Vibrio sp. Separated from the Marine Sponge Hyrtios altum. Chem. Pharm. Bull. 1994;42(12):2449–2451. doi: 10.1248/cpb.42.2449. [DOI] [PubMed] [Google Scholar]
- Kovács B., Péter P. Pázmány Péter Catholic University; Budapest: 2016. Computing RMSD dan fitting protein structures : how I do it dan how others do it. [Google Scholar]
- Larionova I., Cherdyntseva N., Liu T., Patysheva M., Rakina M., Kzhyshkowska J. Interaction of tumor-associated macrophages and cancer chemotherapy. Oncoimmunology. 2019;8:1–15. doi: 10.1080/2162402X.2019.1596004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyskov S., Gray J.J. The RosettaDock server for local protein-protein docking. Nucleic Acids Res. 2008;36:233–238. doi: 10.1093/nar/gkn216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mace, P.D., Riedl, S.J., Salvesen, G.S., 2014. Caspase enzymology and activation mechanisms. In: Methods in Enzymology. Elsevier Inc., pp. 161–178. 10.1016/B978-0-12-417158-9.00007-8. [DOI] [PubMed]
- Manson, A., Jones, E., Morris, A., 2006. The Molecular Basis of Genetics, in: Cell Biology Dan Genetics 2nd Edition. Mosby, London.
- Mansoori B., Mohammadi A., Davudian S., Shirjang S., Baradaran B. The different mechanisms of cancer drug resistance: A brief review. Adv. Pharm. Bull. 2017;7:339–348. doi: 10.15171/apb.2017.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder S., DuPree E., Almasan A. A Dual Role of Cyclin E in Cell Proliferation and Apotosis May Provide a Target for Cancer Therapy. Curr. Cancer Drug Targets. 2004;4:65–75. doi: 10.2174/1568009043481669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menéndez C.A., Accordino S.R., Gerbino D.C., Appignanesi G.A., Salsbury F. Hydrogen bond dynamic propensity studies for protein binding and drug design. PLoS ONE. 2016;11(10):e0165767. doi: 10.1371/journal.pone.0165767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miettinen S. University of Tampere; 2009. Targetting the Growth of Ovarian Cancer Cell. [Google Scholar]
- Motiejunas D., Wade R. In: Comprehensive Medicinal Chemistry II Volume 4. Taylor dan J.B., Triggle D.J., editors. Computer-Assisted Drug Design. Elsevier; Philadelphia: 2006. Structural, Energetics, dan Dynamics Aspect of Ligand-Receptor Interactions. [DOI] [Google Scholar]
- Munandar A., Mustopa A.Z., Tarman K., Nurhayati T. AKTIVITAS ANTIKANKER PROTEIN KAPANG Xylaria psidii KT30 (Anticancer Activity of Protein Fungus Xylaria psidii KT30) J. Perikan. dan Kelaut. 2015;5:65–70. [Google Scholar]
- Murray P.J., Wynn T.A. Protective and pathogenic functions of macrophage subsets. Nat. Rev. Immunol. 2011;11(11):723–737. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naga P.N., Rajeshwari P. An Overview on Immunomodulators. Int. J. Curr. Pharm. Clin. Res. 2014;4:108–114. [Google Scholar]
- Nakada K., Fujisawa K., Horiuchi H., Furusawa S. Studies on morphology and cytochemistry in blood cells of ayu Plecoglossus altivelis altivelis. J. Vet. Med. Sci. 2014;76:693–704. doi: 10.1292/jvms.13-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nepali K., Lee H.-Y., Liou J.-P. Nitro-Group-Containing Drugs. J. Med. Chem. 2019;62(6):2851–2893. doi: 10.1021/acs.jmedchem.8b00147. [DOI] [PubMed] [Google Scholar]
- Netz N., Opatz T. Marine Indole Alkaloids. Mar. Drugs. 2015;13:4814–4914. doi: 10.3390/md13084814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nucera E., Aruanno A., Rizzi A., Centrone M. Latex allergy: Current status and future perspectives. J. Asthma Allergy. 2020;13:385–398. doi: 10.2147/JAA.S242058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olender D., Żwawiak J., Zaprutko L. Multidirectional efficacy of biologically active nitro compounds included in medicines. Pharmaceuticals. 2018;11:1–29. doi: 10.3390/ph11020054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purnomo H. Pustaka Pelajar; Yogyakarta: 2011. Kimia Komputasi: Molecular Docking Plants Penambatan Molekul Plants [Protein-Ligand-Ant-System] (Ilmu Semut) [Google Scholar]
- Quinzii C.M., Lopez L.C., Gilkerson R.W., Dorado B., Coku J., Naini A.B., Lagier‐Tourenne C., Schuelke M., Salviati L., Carrozzo R., Santorelli F., Rahman S., Tazir M., Koenig M., Dimauro S., Hirano M. Reactive oxygen species, oxidative stress, and cell death correlate with level of CoQ10 deficiency. FASEB J. 2010;24(10):3733–3743. doi: 10.1096/fj.09-152728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson C.M., Nunns C.L., Williamson D.S., Parratt M.J., Dokurno P., Howes R., Borgognoni J., Drysdale M.J., Finch H., Hubbard R.E., Jackson P.S., Kierstan P., Lentzen G., Moore J.D., Murray J.B., Simmonite H., Surgenor A.E., Torrance C.J. Discovery of a potent CDK2 inhibitor with a novel binding mode, using virtual screening and initial, structure-guided lead scoping. Bioorganic Med. Chem. Lett. 2007;17(14):3880–3885. doi: 10.1016/j.bmcl.2007.04.110. [DOI] [PubMed] [Google Scholar]
- Rios F.J., Touyz R.M., Montezano A.C. Isolation and differentiation of human macrophages. Methods Mol. Biol. 2017;1527:311–320. doi: 10.1007/978-1-4939-6625-7_24. [DOI] [PubMed] [Google Scholar]
- Rivlin N., Brosh R., Oren M., Rotter V. Mutations in the p53 tumor suppressor gene: Important milestones at the various steps of tumorigenesis. Genes and Cancer. 2011;2(4):466–474. doi: 10.1177/1947601911408889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe D.R., Cheatham T.E. PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013;9(7):3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- Rungnim C., Phunpee S., Kunaseth M., Namuangruk S., Rungsardthong K., Rungrotmongkol T., Ruktanonchai U. Co-solvation effect on the binding mode of the α-mangostin/β-cyclodextrin inclusion complex. Beilstein J. Org. Chem. 2015;11:2306–2317. doi: 10.3762/bjoc.11.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sáez A.J.G., Villunger A. MOMP in the absence of BH3-only proteins. Genes Dev. 2016;30:878–880. doi: 10.1101/gad.281519.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos K.B., Guedes I.A., Karl A.L.M., Dardenne L.E. Highly Flexible Ligand Docking: Benchmarking of the DockThor Program on the LEADS-PEP Protein-Peptide Data Set. J. Chem. Inf. Model. 2020;60(2):667–683. doi: 10.1021/acs.jcim.9b00905. [DOI] [PubMed] [Google Scholar]
- Seyrantepe V., Iannello A., Liang F., Kanshin E., Jayanth P., Samarani S., Szewczuk M.R., Ahmad A., Pshezhetsky A.V. Regulation of phagocytosis in macrophages by neuraminidase 1. J. Biol. Chem. 2010;285(1):206–215. doi: 10.1074/jbc.M109.055475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh Z., Brooks P.J., Barzilay O., Fine N., Glogauer M. Macrophages, foreign body giant cells and their response to implantable biomaterials. Materials (Basel). 2015;8:5671–5701. doi: 10.3390/ma8095269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiozaki E.N., Chai J., Rigotti D.J., Riedl S.J., Li P., Srinivasula S.M., Alnemri E.S., Fairman R., Shi Y. Mechanism of XIAP-Mediated Inhibition of Caspase-9 fector caspases are produced in cells as catalytically. Mol. Cell. 2003;11(2):519–527. doi: 10.1016/s1097-2765(03)00054-6. [DOI] [PubMed] [Google Scholar]
- Sri R.T., Sathyanathan V., Praveen Kumar D., Manasa Chowdhari C. DOCKING STUDIES ON SYNTHESIZED QUINAZOLINE COMPOUNDS AGAINST ANDROGEN RECEPTOR Author for Correspondence. Int. J. Pharm. Ind. Res. 2011;1:266–269. [Google Scholar]
- Su L.J., Zhang J.H., Gomez H., Murugan R., Hong X., Xu D., Jiang F., Peng Z.Y. Reactive Oxygen Species-Induced Lipid Peroxidation in Apoptosis, Autophagy, and Ferroptosis. Oxid. Med. Cell. Longev. 2019;2019:1–13. doi: 10.1155/2019/5080843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacar O., Sriamornsak P., Dass C.R. Doxorubicin: an update on anticancer molecular action, toxicity and novel drug delivery systems. J. Pharm. Pharmacol. 2012;65:157–170. doi: 10.1111/j.2042-7158.2012.01567.x. [DOI] [PubMed] [Google Scholar]
- Tang D.D., Gerlach B.D. The roles and regulation of the actin cytoskeleton, intermediate filaments and microtubules in smooth muscle cell migration. Respir. Res. 2017;18:1–12. doi: 10.1186/s12931-017-0544-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatarczuch L., Bischof R.J., Philip C.J., Lee C.-S. Phagocytic capacity of leucocytes in sheep mammary secretions following weaning. J. Anat. 2002;201(5):351–361. doi: 10.1046/j.0021-8782.2002.00104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelen T., Hao Y., Medeiros A.I., Curtis J.L., Serezani C.H., Kobzik L., Harris L.H., Aronoff D.M. The Class A Scavenger Receptor, Macrophage Receptor with Collagenous Structure, Is the Major Phagocytic Receptor for Clostridium sordellii Expressed by Human Decidual Macrophages. J. Immunol. 2010;185(7):4328–4335. doi: 10.4049/jimmunol.1000989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorn C.F., Oshiro C., Marsh S., Hernandez-Boussard T., McLeod H., Klein T.E., Altman R.B. Doxorubicin pathways: Pharmacodynamics and Adverse effect. Pharmacogenet. Genomics. 2011;21:440–446. doi: 10.1097/FPC.0b013e32833ffb56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O., Olson A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Wang W., Cai H., Du B., Zhang L., Ma W., Hu Y., Feng S., Miao G. MACC1 facilitates chemoresistance and cancer stem cell-like properties of colon cancer cells through the PI3K/AKT signaling pathway. Mol. Med. Rep. 2017;16:8747–8754. doi: 10.3892/mmr.2017.7721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Wang Z., Yang P., Xia L., Zhou M., Wang S., Du J., Zhang J. PER1 prevents excessive innate immune response during endotoxin-induced liver injury through regulation of macrophage recruitment in mice. Cell Death Dis. 2016;7 doi: 10.1038/cddis.2016.9. e2176 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong R.S.Y. Apoptosis in cancer: From pathogenesis to treatment. J. Exp. Clin. Cancer Res. 2011;30:87. doi: 10.1186/1756-9966-30-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Mengang, -, et al. Induced fit docking, and the use of QM/MM methods in docking. Drug Discov Today Technol. 2013;10 doi: 10.1016/j.ddtec.2013.02.003. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Teves S.S., Kemp C.J., Henikoff S. Doxorubicin, DNA torsion, and chromatin dynamics. Biochim. Biophys. Acta - Rev. Cancer. 2014;1845(1):84–89. doi: 10.1016/j.bbcan.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo M., Choi S.-U., Choi K.Y., Yon G.H., Chae J.-C., Kim D., Zylstra G.J., Kim E. Trisindoline synthesis and anticancer activity. Biochem. Biophys. Res. Commun. 2008;376(1):96–99. doi: 10.1016/j.bbrc.2008.08.092. [DOI] [PubMed] [Google Scholar]
- Zhang X., Goncalves R., Mosser D.M. The isolation and characterization of murine macrophages. Curr. Protoc. Immunol. 2008;83(1) doi: 10.1002/0471142735.im1401s83. https://doi.org/10.1002/0471142735.2008.83.issue-110.1002/0471142735.im1401s83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.-H., Yu H.-L., Wang F.-J., Han Y.-L., Yang W.-L. Pim-2 modulates aerobic glycolysis and energy production during the development of colorectal tumors. Int. J. Med. Sci. 2015;12(6):487–493. doi: 10.7150/ijms.10982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.-W., Shi J., Li Y.-J., Wei L. Cardiomyocyte death in doxorubicin-induced cardiotoxicity. Arch. Immunol. Ther. Exp. (Warsz) 2009;57(6):435–445. doi: 10.1007/s00005-009-0051-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Wu L., Yan G., Chen Y., Zhou M., Wu Y., Li Y. Inflammation and tumor progression: signaling pathways and targeted intervention. Signal Transduct. Target. Ther. 2021;6:1–46. doi: 10.1038/s41392-021-00658-5. [DOI] [PMC free article] [PubMed] [Google Scholar]