Significance
Genomic DNA is being continuously damaged and repaired. DNA damage that evades repair can block the replicative DNA polymerase, a critical enzyme responsible for copying genetic material. Translesion synthesis (TLS) alleviates this blockade by using specialized TLS polymerases to copy damaged DNA. Because TLS polymerases are mutagenic, their access to parental DNA must be restricted. How TLS polymerases are selectively enabled only when replication is blocked remains unclear. In this study, we describe a molecular mechanism that selectively enriches TLS polymerases near stalled sites of replication in Escherichia coli. Furthermore, we show that it may be a general mechanism for enzymes that respond to replication stress.
Keywords: DNA replication, translesion synthesis, DNA damage response, single-molecule imaging, superresolution microscopy
Abstract
Translesion synthesis (TLS) polymerases bypass DNA lesions that block replicative polymerases, allowing cells to tolerate DNA damage encountered during replication. It is well known that most bacterial TLS polymerases must interact with the sliding-clamp processivity factor to carry out TLS, but recent work in Escherichia coli has revealed that single-stranded DNA-binding protein (SSB) plays a key role in enriching the TLS polymerase Pol IV at stalled replication forks in the presence of DNA damage. It remains unclear how this interaction with SSB enriches Pol IV in a stalling-dependent manner given that SSB is always present at the replication fork. In this study, we use single-molecule imaging in live E. coli cells to investigate this SSB-dependent enrichment of Pol IV. We find that Pol IV is enriched through its interaction with SSB in response to a range of different replication stresses and that changes in SSB dynamics at stalled forks may explain this conditional Pol IV enrichment. Finally, we show that other SSB-interacting proteins are likewise selectively enriched in response to replication perturbations, suggesting that this mechanism is likely a general one for enrichment of repair factors near stalled replication forks.
Translesion synthesis (TLS) is a DNA damage-tolerance pathway, conserved across all domains of life, that can rescue replication forks stalled at sites of DNA damage (1, 2). In this process, a TLS polymerase exchanges with the replicative polymerase and continues DNA synthesis past the lesion on the templating strand. By allowing processive replication to continue downstream of the lesion, TLS helps cells avoid the deleterious consequences of a stalled replication fork, including replication-fork collapse and toxic double-strand DNA breaks. Proper regulation of TLS is critical, however, because most TLS polymerases are error prone and can introduce harmful mutations if their access to the DNA template is not restricted (3).
Previously we used live-cell single-molecule imaging to gain insight into the regulation of the most abundant TLS polymerase in Escherichia coli, Pol IV (4). We found that Pol IV is only modestly enriched near sites of replication under normal growth conditions but becomes strongly colocalized with the replisome when cells are treated with the DNA-damaging agent methyl methanesulfonate (MMS). This lack of enrichment of Pol IV near replication forks in the absence of DNA damage likely limits the access of Pol IV to the DNA template during normal replication in order to minimize mutagenesis.
It is well established that Pol IV and other TLS polymerases interact with the replication processivity factor, or the sliding clamp, and that this interaction is critical for TLS (5, 6). In addition to TLS polymerases, clamps bind a number of other proteins involved in DNA replication and repair (7–9). The E. coli β clamp is a dimer with two equivalent binding cleft sites for clamp-interacting proteins (10). The replicative polymerase Pol III binds to both sites through its α and ε subunits (11, 12), which can occlude Pol IV access (13). Less is known, however, about the importance of other protein–protein interactions in TLS. Previous studies have reported that Pol IV also interacts with single-stranded DNA-binding protein (SSB) (14) and both the RecA recombinase and the UmuD subunit of the TLS polymerase Pol V (15, 16). Like the clamp, E. coli SSB is known to interact with a wide range of DNA replication and repair proteins, which bind to the conserved C-terminal peptide (17).
Recently, our laboratory showed that the Pol IV–SSB interaction enriches Pol IV near replication forks stalled by DNA damage (18). Locally concentrating Pol IV enables it to outcompete Pol III and gain access to the β clamp, which is critical for TLS on both the leading and lagging strands. The interaction with SSB might also help Pol IV outcompete other clamp-binding proteins that do not interact with SSB. The clustering of the SSB C-terminal tail appears to be a key feature required for Pol IV localization, likely due to the relatively weak (low micromolar) binding affinity between Pol IV and a single SSB C-terminal tail.
The role of the SSB interaction in enriching Pol IV near replication forks raises several new questions. First, we initially observed this enrichment when cells were treated with MMS, which generates DNA lesions that Pol IV is capable of bypassing (4). Is Pol IV only enriched near replication forks in the presence of such “cognate” DNA lesions, or is enrichment a general consequence of replication stalling independent of the cause? Second, SSB binds transiently exposed single-stranded DNA (ssDNA) on the lagging strand during replication; thus, it is a constitutive component of normal replication forks. Yet, we observed little Pol IV enrichment near replication forks in the absence of DNA damage (4). What features differentiate SSB at stalled forks from SSB at moving forks such that Pol IV is selectively enriched upon stalling? Finally, E. coli SSB interacts with at least 17 different SSB interacting proteins (SIPs) (17, 19). Are other SIPs selectively enriched near stalled replication forks upon replication stalling, or is this mechanism unique to Pol IV?
In this study, we use single-molecule fluorescence imaging in live E. coli cells to address these questions. We find that Pol IV is enriched near replication forks in the presence of “noncognate” forms of DNA damage, meaning lesions that it cannot bypass, as well as upon nucleotide depletion, suggesting that Pol IV enrichment is a general consequence of replication stalling. Furthermore, we show that this enrichment requires interactions with the β clamp and SSB, with SSB playing the major role. By imaging single SSB molecules, we find that SSB dynamics change upon replication stress, with an increase in the relative population of statically bound SSB molecules and a concomitant decrease in the mobile population. In addition to a modest increase in SSB copy number at replication forks, we observe a substantial increase in the SSB binding lifetime upon replication perturbation, providing a possible mechanism for selective Pol IV enrichment. Finally, we show that two other SIPs, PriA and RecG, are also enriched near sites of replication only upon DNA damage, suggesting that changes in SSB dynamics may provide a general mechanism for enriching factors that can respond to stalled replication forks.
Results
Pol IV Is Strongly Enriched at Replication Forks in the Presence of Cognate DNA Damage.
Previously, we constructed and validated a functional C-terminal fusion of Pol IV to the photoactivatable fluorescent protein PAmCherry (20) at the endogenous dinB locus, where dinB is the gene encoding Pol IV (4). To prevent large changes in copy number in response to DNA damage, we introduced this fusion in the lexA51-strain background, in which the SOS DNA damage response is constitutively activated due to a truncation of the LexA repressor (21, 22); the SOS response increases Pol IV levels by approximately 10-fold (5, 23). To visualize sites of DNA replication in the cell, we introduced an orthogonal C-terminal fusion of SSB to the yellow fluorescent protein variant mYPet (24). Because replacement of all SSB copies with a C-terminal fusion has been reported to be lethal (25) or to impair protein–protein interactions (19), we introduced this SSB fusion as a second copy at the isopropyl β-d-1thiogalactopyranoside (IPTG)-inducible lacZ locus.
To determine whether and under what conditions Pol IV is colocalized with sites of replication, we imaged Pol IV–PamCherry in live E. coli cells in a custom-built, single-molecule fluorescence microscope using particle-tracking photoactivation localization microscopy (PALM) (26, 27). In brief, a 405-nm near-ultraviolet laser was used to photoactivate PamCherry molecules from a dark (nonfluorescent) state to a bright (fluorescent) state that was excited using 561-nm laser excitation (SI Appendix, Fig. S1 A and C). The motion of each individual Pol IV–PAmCherry molecule was tracked until the molecule photobleached, converting it irreversibly to a dark state. In the same cells, we used 514-nm laser excitation to excite SSB–mYPet, which formed foci at replication forks (SI Appendix, Fig. S1 B and D), as previously reported (28). Previously, we observed two populations of Pol IV in normally growing E. coli cells: a fast-moving diffusive population and an immobile statically bound population (4). This immobile population should include molecules specifically bound to the β clamp or to SSB, although it may also include molecules that are transiently bound to DNA in a nonspecific manner (29). To resolve immobile molecules selectively, we used a long exposure time of 250 ms, which blurs out fast-moving molecules, and identified static molecules based on the width of their point spread function (PSF) (4, 27).
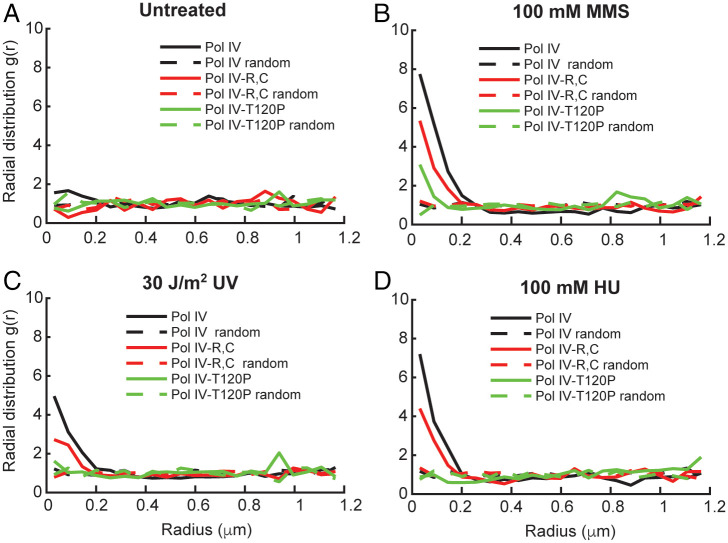
To measure the degree of colocalization between these static Pol IV molecules and sites of DNA replication, we used radial distribution function analysis (Methods) (4, 30, 31). The magnitude of the radial distribution function, g(r), at each value of r reflects the fold enrichment of Pol IV at that distance from SSB relative to the random distribution. Consistent with our previous report, we observed little enrichment of Pol IV near replication forks in untreated E. coli cells (Fig. 1A and SI Appendix, Table S1) (4). However, Pol IV was strongly enriched at sites of replication, with g(r) ∼ 8 at short Pol IV–SSB distances (Fig. 1B), in cells treated with a 100-mM concentration of the alkylating agent MMS for 20 min (Methods). Previous work has shown that cells can recover from a short duration of exposure to this concentration of MMS (27) and that Pol IV can efficiently bypass the lesions generated by this drug, particularly the replication-blocking N3-methyladenine lesion (18, 32). Importantly, this effect was dose dependent; when cells were treated with a lower MMS concentration of 10 mM, Pol IV was still enriched at sites of replication, but less strongly [maximum g(r) ∼ 4] (SI Appendix, Fig. S3A). An intermediate dose of 25 mM MMS was estimated to generate ∼400 DNA lesions/min in E. coli (27).
Fig. 1.
Effect of MMS, UV, and HU treatment on single-cell colocalization of Pol IV–PAmCherry and SSB–mYPet. Graphs of the radial distribution function g(r) between each static Pol IV–PAmCherry track and the nearest SSB–mYPet focus for Pol IVWT (black), Pol IVR,C (red), and Pol IVT120P (green) in (A) untreated cells (n = 1,482, 1,090, and 878, respectively) and cells treated with (B) 100 mM MMS (n = 2,754, 2,894, and 2,331, respectively), (C) 30 J/m2 UV light (n = 2,681, 3,261, and 1,142, respectively), and (D) 100 mM HU (n = 1,539, 1,479, and 1,207, respectively).
This behavior was not unique to constitutively SOS-induced cells, as we observed a similar conditional Pol IV–SSB enrichment in an isogenic lexA+ strain, with little to no colocalization in untreated cells but strong enrichment upon 100 mM MMS treatment [maximum g(r) ∼ 9] (SI Appendix, Fig. S3B). Furthermore, we did not observe any colocalization in untreated cells after fixation (SI Appendix, Fig. S3C), indicating that the enrichment was not an artifact of biasing the Pol IV population toward immobile molecules. The colocalization was, however, sensitive to protein–protein interactions known to be important for TLS. Mutating the two clamp-binding sites on Pol IV (Pol IVR,C), which contact the rim and cleft of the β clamp, led to a reduction in the maximum g(r) value to ∼5 in cells treated with 100 mM MMS (Fig. 1B) (4). Likewise, the T120P mutation, which abrogates Pol IV binding to SSB (Pol IVT120P), produced a larger reduction in the maximum g(r) to ∼3 (Fig. 1B) (18). From these results, we previously concluded that SSB plays the primary role in enrichment of Pol IV at stalled replication forks, whereas the interaction with the β clamp stabilizes Pol IV at the primer-template junction and is essential for synthesis past the lesion. Neither mutant was enriched near sites of replication in untreated cells, with a maximum g(r) ∼ 1 for both (Fig. 1A).
Pol IV Is Enriched at Replication Forks Even in the Presence of Noncognate DNA Damage.
In contrast to MMS, there are DNA-damaging agents that generate lesions that Pol IV cannot efficiently bypass. Next, we asked whether Pol IV would be enriched at replication forks in the presence of such noncognate DNA lesions or whether localization requires Pol IV recognition of a cognate lesion. We treated cells with 30 J/m2 254 nm ultraviolet (UV) light, which generates strongly blocking DNA lesions including cyclobutane pyrimidine dimers and 6–4 photoproducts (33). A similar dose was estimated to generate approximately one cyclobutane–pyrimidine dimer per 9 kb of DNA and found to inhibit DNA replication without having a major impact on survival of wild-type (WT) E. coli cells (34–36). We observed moderate enrichment of Pol IV at sites of replication upon UV treatment [maximum g(r) ∼ 5] (1C), consistent with a prior report (23). Importantly, this enrichment was dependent on the same protein–protein interactions as in MMS-treated cells. The maximum g(r) value was reduced by nearly twofold [g(r) ∼ 3] for the Pol IVR,C mutant and by ∼2.5-fold for the Pol IVT120P mutant [g(r) ∼ 2] (Fig. 1C). Although the maximum value of g(r) for a given treatment condition depends on factors including the lesion density and the dynamics of Pol IV binding, this colocalization analysis indicates that Pol IV is enriched at replication forks through interactions with the β clamp and SSB in the presence of noncognate forms of DNA damage. While photochemical cyclobutane pyrimidine dimers and 6–4 photoproducts are expected to be the primary lesions generated by short-wavelength, 254-nm UVC irradiation, we note that UV exposure can also produce oxidative lesions like 8-oxoguanine or thymine glycol (37–39), which Pol IV might be able to bypass more efficiently. Nevertheless, these results suggest that Pol IV is enriched at stalled forks even when it is unable to carry out TLS. They also support our view that the static population of Pol IV includes both molecules actively synthesizing DNA as well as molecules that are bound at the replication fork but not carrying out TLS.
Pol IV Is Enriched at Replication Forks Upon DNA Damage-Independent Replication Stalling.
Since both cognate DNA damage and noncognate DNA damage enrich Pol IV at the replication fork in an SSB-dependent manner, our results suggest that any obstacles to processive replication may act as a molecular signal for the recruitment of Pol IV. To test this possibility, we treated cells with the ribonucleotide reductase (RNR) inhibitor hydroxyurea (HU) (40). RNR inhibition by HU depletes cellular deoxynucleoside triphosphate pools, ultimately leading to replication stalling and cell death (41). Cells lacking Pol IV are not sensitized to HU relative to WT cells (SI Appendix, Fig. S2B), indicating that Pol IV does not contribute to survival upon HU treatment.
We found that Pol IV was strongly enriched at replication forks when cells were treated with 100 mM HU [maximum g(r) ∼ 7] (Fig. 1D), with a slightly lower level of enrichment upon treatment with 20 mM HU (SI Appendix, Fig. S3D). To test whether Pol IV enrichment was specific to HU treatment or resulted generally from nucleotide depletion, we tested a different RNR inhibitor, guanazole (40, 42). Pol IV enrichment was comparable for cells treated with a 100 mM concentration of guanazole and with the same concentration of HU (SI Appendix, Fig. S3D), suggesting that the enrichment is a general result of nucleotide depletion and not specific to HU. Next, we asked whether the same molecular interactions were required for Pol IV enrichment upon this DNA damage–independent stalling. We measured a reduction of almost twofold in the maximum g(r) value for the β-binding deficient Pol IVR,C mutant in comparison to Pol IVWT in cells treated with 100 mM HU. This reduction is comparable to the loss of enrichment observed for this mutant relative to Pol IVWT upon MMS treatment [g(r) ∼ 4] (Fig. 1D). Furthermore, the SSB-binding deficient Pol IVT120P mutant was not enriched at all relative to random colocalization upon HU treatment (Fig. 1D). These results support a model where the Pol IV–SSB interaction plays the dominant role in Pol IV recruitment upon general replication stalling and not just in response to DNA damage–induced stalling.
SSB Dynamics Change Upon Perturbations to Replication.
Taken together, our data suggest that interactions with SSB enrich Pol IV at stalled replication forks, independent of whether Pol IV is capable of resolving the stall. SSB is present at the replication fork during normal replication, where it transiently binds ssDNA exposed on the lagging strand. Thus, our results raise the question of why Pol IV is not strongly enriched at the replication fork through interactions with SSB during normal replication but only upon replication stalling. We reasoned that one possible mechanism could be changes in SSB behavior upon perturbations to replication.
To probe changes in the behavior of single SSB molecules upon replication stalling, we created an SSB–PAmCherry fusion, introduced as a second copy at the lacZ locus in the same manner as the mYPet fusion. PALM imaging allowed us to visualize individual SSB–PAmCherry molecules as bright spots in the cell (SI Appendix, Fig. S1C). As a replisome marker, we used a previously characterized fusion of mYPet to the proofreading exonuclease subunit ε (encoded by the dnaQ gene) of the replicative polymerase, Pol III (4). This ε–mYPet fusion forms distinct foci at sites of replication (SI Appendix, Fig. S1D). Cells bearing the SSB–PAmCherry fusion were not sensitized to MMS treatment relative to WT cells (SI Appendix, Fig. S2A), indicating that the SSB fusion does not appear to impair TLS or other DNA damage–response pathways.
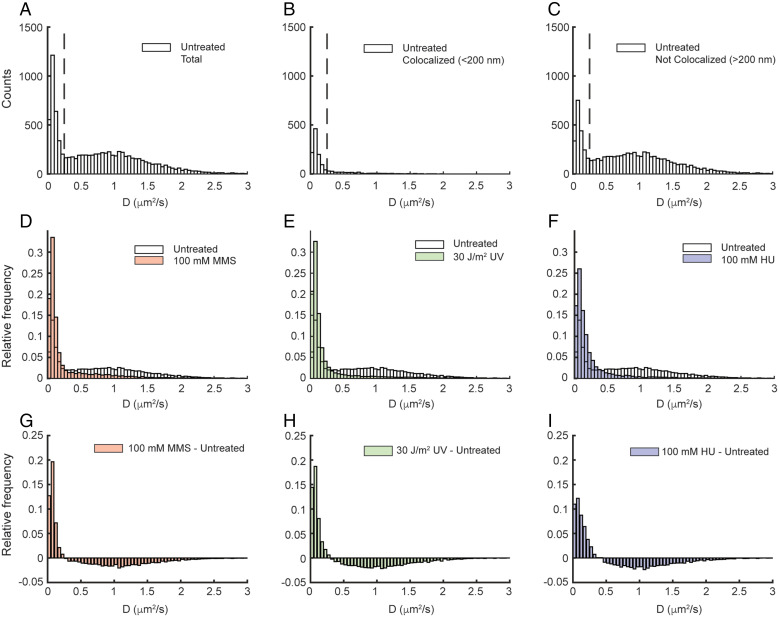
To characterize SSB mobility, we imaged untreated E. coli cells containing this SSB–PAmCherry fusion using a short 13.3-ms exposure time. Two broad populations of SSB molecules were observed in cells: a static fraction, with an apparent diffusion coefficient D* ∼ 0.08 µm2/s and a mobile fraction with D* ∼ 1 µm2/s (Fig. 2A); D* for the static molecules is slightly larger than 0, due to the localization precision of ∼17 nm, which leads to some apparent motion of immobile molecules. As for Pol IV and other DNA-binding proteins, this static fraction comprises DNA-bound molecules, whereas the mobile fraction represents free SSB molecules (4, 27). The static population of SSB molecules is expected to be disproportionately localized near sites of replication, where SSB is bound to Okazaki fragments. Consistent with this prediction, SSB molecules localized within 200 nm of the nearest ε–mYPet focus are almost entirely static (Fig. 2B), whereas both static and mobile populations are represented outside that radius (Fig. 2C). There are several possible explanations for the presence of static SSB molecules that do not appear to colocalize with replication forks, including the presence of ssDNA gaps on incomplete Okazaki fragments or at sites of DNA repair, SSB molecules that are transiently static but are not actually DNA bound, or missed detection of ε–mYPet foci.
Fig. 2.
Effect of MMS, UV, and HU treatment on the apparent diffusion coefficient (D*) of SSB–PAmCherry. D* distributions of SSB–PAmCherry in untreated cells for (A) all molecules (n = 8,722), (B) molecules <200 nm from Pol III ε–mYPet foci (n = 1,276), and (C) molecules >200 nm from Pol III ε–mYPet foci (n = 7,446). The dashed lines indicate the threshold D* value for bound molecules (D* < 0.25 µm2/s). D* distributions of SSB–PAmCherry for all molecules in untreated cells and cells treated with (D) 100 mM MMS (red; n = 11,797), (E) 30 J/m2 UV light (green; n = 16,542), and (F) 100 mM HU (blue; n = 5,690). (G–I) The difference in D* distributions of SSB–PAmCherry between untreated cells and MMS-, UV-, and HU-treated cells, respectively.
Upon treatment of cells with 100 mM MMS, 30 J/m2 UV, or 100 mM HU, there was a dramatic reduction in the mobile fraction of SSB and a corresponding increase in the static fraction (Fig. 2 D–I and SI Appendix, Tables S2 and S3). To quantify these changes, the apparent diffusion coefficient distributions were fit to a three-species model (SI Appendix, Fig. S4 and Methods). The three populations had mean apparent diffusion coefficients in the ranges of 0.06 to 0.08 µm2/s, 0.15 to 0.25 µm2/s, and 1.0 to 1.2 µm2/s, with the exact values depending on the treatment condition (SI Appendix, Table S4). The first and third populations represent stably bound and mobile SSB, respectively, whereas the intermediate population may represent SSB molecules interacting transiently with DNA. In untreated cells, static molecules represented 24% of the total SSB population; this fraction doubled in cells treated with MMS, UV, or HU (to 47%, 50%, and 45%, respectively). Thus, there is a dramatic change in the mobility of SSB molecules upon replication perturbation, which could act as a molecular signal to enrich Pol IV in a replication stalling–specific manner.
The Binding Lifetime of SSB at the Replication Fork Increases Upon Perturbations to Replication.
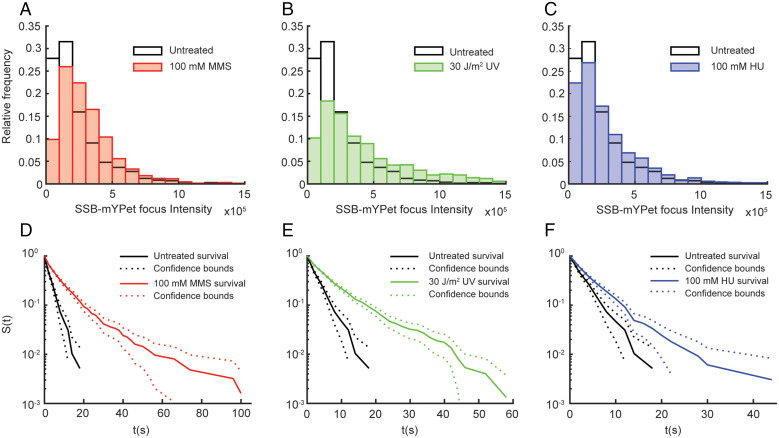
The increase in the static fraction of SSB molecules upon replication stalling suggests an increase in the average SSB copy number at the replication fork, but it may also partially reflect an increase in the average SSB-binding lifetime at the replication fork. Either of these changes could contribute to a larger fraction of static SSB molecules in the distribution of apparent diffusion coefficients. To test the first of these possible changes, we carried out an analysis of SSB–mYPet foci in untreated cells and cells treated with 100 mM MMS, 30 J/m2 UV, and 100 mM HU. In brief, SSB–mYPet foci were fit to symmetric two-dimensional Gaussian functions and the integrated area was determined (see Methods for further details). For the 100 mM MMS and 100 mM HU conditions, increases of ∼50% and 25% in the median integrated intensity of the SSB foci were observed relative to untreated cells. For 30 J/m2 UV, the increase in the median was more than twofold for the integrated SSB–mYPet intensity (Fig. 3 A–C and SI Appendix, Table S5). Under all treatment conditions, there was a minimal change in the mean number of SSB–mYPet foci per cell (SI Appendix, Table S6).
Fig. 3.
Effect of MMS, UV, and HU treatment on SSB–mYPet focus intensity and survival time of static SSB–PAmCherry tracks. Distributions of integrated SSB–mYPet focus intensity in untreated cells (white; n = 2,138) and (A) cells treated with 100 mM MMS (red; n = 1,309), (B) 30 J/m2 UV light (green; n = 2,015), and (C) 100 mM HU (blue; n = 1,207). Raw survival curves, given by the survivor function (S(t)), of static SSB–PAmCherry tracks in untreated (black; n = 545) cells and (D) MMS-treated (red; n = 1,171), (E) UV-treated (green; n = 1,755), and (F) HU-treated (blue; n = 956) cells measured with a 2-s stroboscopic interval.
To determine if the SSB-binding lifetime also increased upon treatment, we imaged single SSB–PAmCherry molecules using a long 250-ms integration time with a stroboscopic illumination pattern of one excitation frame every 2 s; this stroboscopic illumination allowed us to extend the time until PAmCherry photobleaching and thus probe longer binding events. The resulting distributions of SSB-binding lifetimes for untreated cells and cells treated with 100 mM MMS, 30 J/m2 UV, and 100 mM HU (SI Appendix, Fig. S5 and Table S7) were converted to survival curves (SI Appendix, Fig. S6 and Table S8, and Methods). The resulting survival curves (Fig. 3 D–F) revealed an increase in the SSB-binding lifetime upon perturbations to replication, which was apparent as an increase in the population of molecules with longer binding times and a concomitant decrease in the population with shorter binding times. To quantify these results, each survival curve was fit to a double exponential function and corrected for PAmCherry photobleaching (see SI Appendix, Figs. S6 and S7 and Table S9, and Methods). The lifetime increased from 4.5 s in untreated cells to 32.5 s, 56.7 s, and 8.9 s in cells treated with MMS, UV, and HU, respectively, indicating a substantial increase in the SSB-binding lifetime upon replication perturbation. Directly measured lifetime distributions give qualitatively similar results (SI Appendix, Figs. S5 and S6 and Table S7). The lifetime in untreated cells is consistent with findings of a previous study that measured an SSB exchange timescale of ∼2.5 s during processive replication in E. coli cells using fluorescence recovery after photobleaching (43).
In addition to this increase in the binding lifetime of SSB molecules, we observed an increase in the number of static SSB molecules using these imaging conditions (Methods, SI Appendix, Fig. S8 and Table S10). The average number of SSB-binding events increased by approximately twofold for MMS, threefold for UV treatment, and ∼20% for HU treatment in the 2-s stroboscopic condition (SI Appendix, Fig. S8 and Table S10). Although these measurements do not report directly on the copy number of SSB at the replication fork, because they do not include molecules that are bound only briefly, the behavior is consistent with an increase in the static, or more stable, population of SSB.
Other SSB-Interacting Proteins Are Enriched at Replication Forks in the Presence of DNA Damage.
Taken together, our results show that Pol IV is enriched at replication forks when replication is perturbed, primarily through interactions with SSB, and that this enrichment is likely a consequence of changes in SSB copy number and binding lifetime upon replication stalling. Importantly, this enrichment occurs even when Pol IV is unable to resolve the stall by performing TLS. In addition to Pol IV, the C-terminal tail of E. coli SSB is known to interact with at least 17 other SIPs (17, 19), raising the possibility that selective enrichment to stalled replication forks is general rather than specific to Pol IV. To test this possibility, we created C-terminal fusions of PAmCherry to two other SIPs, PriA and RecG, at their native loci. PriA is a helicase and a member of the primosome that plays a role in replication initiation and in the restart of stalled replication forks (44–47). RecG is a helicase that likewise plays a role in replication restart at stalled forks (47–49).
The functionality of the PriA and RecG fusions was tested by assaying their sensitivity to MMS. The survival of the PriA–PAmCherry fusion strain was essentially identical to the untagged PriA strain, both in the MG1655 WT background and in the lexA51 imaging-strain background (SI Appendix, Fig. S2C). The RecG–PAmCherry fusion showed a modest reduction in survival at higher MMS concentrations compared with untagged RecG. Cells bearing a ΔrecG knockout, however, were unable to tolerate even the lowest MMS concentration tested, indicating that the RecG–PamCherry fusion still retains substantial activity (SI Appendix, Fig. S2D).
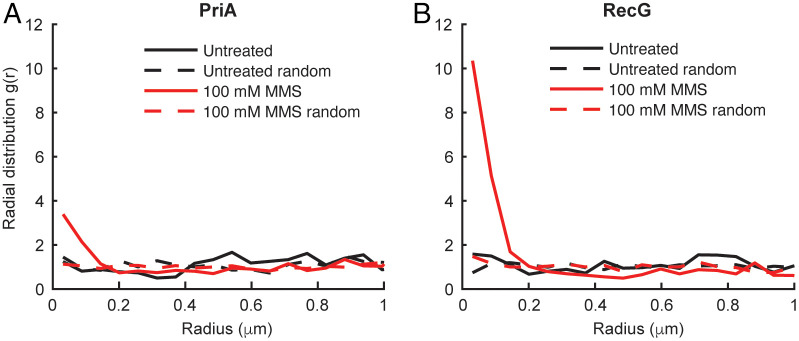
Unperturbed E. coli cells bearing a PriA–PAmCherry or RecG–PAmCherry fusion and an SSB–mYPet replisome marker were imaged using 250 ms integration times to resolve static molecules. There was no enrichment of either PriA or RecG near sites of replication in the absence of damage (Fig. 4A and B), as observed for Pol IV. Upon treatment with 100 mM MMS, however, both proteins were strongly enriched at replication forks, with an enrichment of approximately fourfold for PriA and 10-fold for RecG. These results suggest that the replication stalling–dependent enrichment of SIPs may be a general mechanism triggered by changes in SSB-binding dynamics, which acts to recruit DNA repair and DNA damage tolerance proteins when replication is challenged.
Fig. 4.
Effect of MMS treatment on the single-cell colocalization of SSB-interacting proteins PriA and RecG with SSB. (A) Radial distribution function g(r) between static PriA–PAmCherry molecules and SSB–mYPet foci in untreated cells (black; n = 781) and cells treated with 100 mM MMS (red; n = 2,412). (B) Radial distribution function g(r) between static RecG–PAmCherry molecules and SSB–mYPet foci in untreated (black; n = 1,189) and MMS-treated (red; n = 1,694) cells.
Discussion
DNA damage acts to block the progress of the replisome. Numerous polymerases, helicases, and other enzymes not believed to be constitutive members of the replisome are employed to resolve these blockades. Previously, we showed that the TLS polymerase Pol IV is enriched near replication forks upon treatment with the cognate DNA–damaging agent MMS through interactions with the β clamp and SSB (4, 18). Although MMS-induced lesions are a strong block for the replicative polymerase Pol III, Pol IV is able to bypass them efficiently. (32, 18) In this study, we found that Pol IV is likewise selectively enriched when replication is perturbed by a noncognate form of DNA damage, UV lesions, or by nucleotide depletion caused by HU or guanazole treatment. Thus, stable enrichment of Pol IV is not contingent on its ability to resolve the stall. Furthermore, this enrichment requires the same β clamp and SSB interactions as in the case of MMS, with the SSB interaction playing the major role. These results are consistent with a model where Pol IV is rapidly enriched near the fork in response to general replication perturbations through the same interactions that mediate its response to cognate lesions.
Although interactions between Pol IV and the β clamp are essential for TLS, the interaction with SSB is critical for Pol IV to gain access to the clamp (18). While clamps may reside on DNA behind the replication fork (50), there is presumably only one clamp near a lesion site. Thus, binding sites on the clamp near a DNA lesion are limited. The β clamp is a dimer, with an equivalent binding cleft site on each protomer (5). The Pol III α subunit binds tightly to one of these clefts, while the Pol III ε subunit binds weakly to the other cleft (11, 12). Although this interaction has a lower binding affinity, ε is present at high effective local concentration due to α binding and acts as a molecular gate to regulate other factors from binding the clamp (13). In addition, there are a number of other clamp-binding proteins that may compete with Pol IV for a cleft site. Thus, SSB acts as a platform that locally concentrates Pol IV near the clamp, allowing it to outcompete other clamp-binding proteins that do not bind SSB (18).
SSB is a constitutive component of replication forks, where it binds ssDNA on the lagging strand. A previous imaging study revealed an average of ∼8 SSB tetramers present at a single replication fork, a value consistent with typical Okazaki fragment sizes (650 to 2,000 bp depending on temperature) (51, 52) and SSB binding footprint (35 or 65 bp). (53, 17) Thus, some mechanism must prevent Pol IV enrichment through the interaction with SSB during processive replication. In this study, we looked for changes in SSB behavior upon replication perturbation that might explain this selective Pol IV enrichment. By directly imaging single SSB molecules and characterizing their diffusion, we identified both static (DNA-bound) and mobile populations. As expected, static SSB molecules were preferentially enriched near replication forks. Upon different forms of replication perturbation, we observed a significant shift in SSB dynamics, with the static fraction roughly doubling from 25% to 50% of the total SSB population. We propose that this change in SSB dynamics could represent a damage-dependent signal for Pol IV enrichment, explaining why Pol IV is not enriched near replication forks in the absence of replication stress. Although the SSB diffusion measurements are indicative of an increase in the total number of statically bound SSB molecules, they are convoluted with a second possible effect, a change in SSB-binding lifetime upon replication perturbation. We explored both of these possible mechanisms in this study.
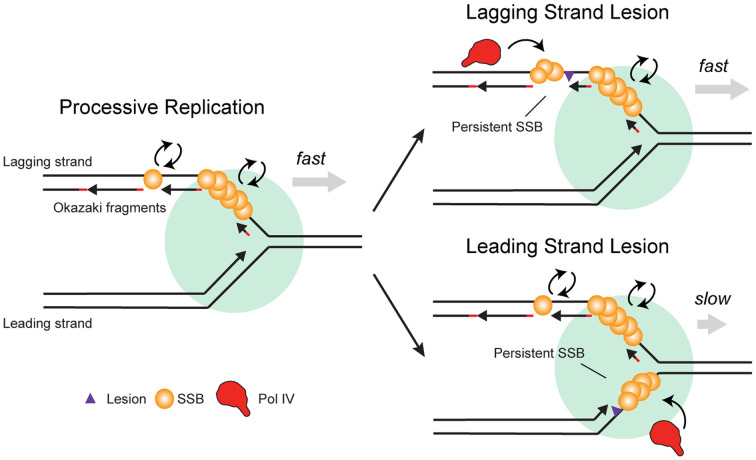
The effect of replication perturbations on the amount of exposed ssDNA, and thus on the copy number of bound SSB, likely depends on the nature of the perturbation, and the effects may be different on the leading and lagging strand. During normal replication, there is no significant SSB binding on the leading strand, due to the continuous nature of leading-strand synthesis (Fig. 5, Left). Stalling of the leading-strand replicative polymerase at a lesion, however, leads to uncoupling of the polymerase and helicase, exposing ssDNA on the leading strand (Fig. 5, Bottom Right) (54). Alternatively, the presence of lesions on either the leading or lagging strand may result in repriming, in which case an ssDNA gap remains behind the moving fork (13, 55, 56). We would expect UV irradiation, which produces highly blocking DNA lesions, to lead to repriming and ssDNA gaps behind the fork. (55, 57, 54) Nucleotide depletion by HU could lead to polymerase–helicase uncoupling and the accumulation of ssDNA on the leading strand (58, 59). By measuring the intensity of SSB–mYPet foci, we found a twofold increase upon UV treatment but smaller increases of 50% and 25% for MMS and HU treatment, respectively. The relatively modest increase in focus intensity upon MMS treatment in comparison to UV may reflect the increased propensity for Pol IV-mediated TLS at the replication fork versus resolution by repriming. Despite the relatively modest increase in bound SSB for HU treatment, there was still a significant enrichment of Pol IV under this condition, suggesting that SSB copy number alone is not likely to be the sole determinant of Pol IV enrichment and that other factors, like the stability of Pol IV at the primer-template junction, may contribute. Because SSB clustering is needed to overcome the weak affinity of a single Pol IV–SSB C terminal-tail interaction (18), an increase in the copy number of bound SSB may be required for Pol IV enrichment upon replication stalling.
Fig. 5.
Model of the selective recruitment of Pol IV and other SIPs to stalled replication forks by SSB. During processive replication (Left), SSB turns over rapidly on the lagging strand. In the case of a lagging strand lesion (Top Right), SSB bound at persistent lagging-strand ssDNA gaps enriches Pol IV or other SIPs. In the case of a leading-strand lesion (Bottom Right), continued helicase translocation after Pol III stalling creates a persistent leading-strand ssDNA gap. SSB binding at this gap enriches Pol IV or other SIPs.
Like SSB copy number, the average binding lifetime of SSB may also change upon replication perturbation. During processive replication, the lifetime of an SSB molecule on the lagging strand is likely limited by eviction due to the synthesizing polymerase (Fig. 5, Left). Indeed, we measured an SSB lifetime of ∼4 s (SI Appendix, Table S9), consistent with estimated replication speeds of 500 to 1,000 base pairs/s and Okazaki fragments of up to 2,000 base pairs in length. Recycling of SSB from one Okazaki fragment to the next would increase the measured binding lifetime (43). However, this is probably not the predominant mechanism in cells, given that high in vivo SSB concentrations (300 to 600 nM) favor free SSB binding from solution (43). Stalling at lesions on the lagging strand template is believed to be resolved by rapid repriming, resulting in persistent gaps and SSB stabilization (Fig. 5, Top Right) (60). Likewise, SSB bound in gaps downstream of a stalled leading-strand polymerase should remain bound longer than lagging-strand SSB during processive replication (Fig. 5, Bottom Right). Consistent with our expectations, perturbing replication resulted in increases in SSB binding lifetime from twofold to greater than 10-fold. Thus, this reduction in turnover of bound SSB could lead to a corresponding reduction in Pol IV turnover and, as a result, an increase in average Pol IV enrichment.
We provide evidence that changes in SSB copy number and dynamics alone may trigger selective recruitment of Pol IV upon replication-fork stalling. Other mechanisms are possible, however, either independent of the changes we observe or downstream of them. For example, posttranslational modifications of SSB triggered by perturbations to replication could increase its affinity for Pol IV; in eukaryotes, posttranslational modifications to the ssDNA-binding protein replication protein A have been shown to play a role in the DNA damage response (61). Although SSB phosphorylation has been reported in E. coli, no direct connection to the DNA-damage response has been elucidated (62). Alternatively, a change in SSB binding conformation in response to stalling could result in selective enrichment. DNA binding is known to release the C-terminal tail of SSB, which can interact with the oligonucleotide/oligosaccharide–binding (OB) fold domain in free SSB, making it available for interaction with SIPs (63). Further conformational changes in SSB upon replication-fork stalling are not known, however, and proposals that SIP binding remodels the SSB filament (64, 65) do not explain how SSB selectively enriches factors at stalled forks. Recent observations show that SSB readily forms phase-separated condensates in vitro (66, 67). These condensates may preferentially form at clusters of persistent SSB, thus leading to selective enrichment of SIPs at stalled replication forks. However, further work is needed to determine if SSB condensates are physiologically relevant and whether their formation is required for SIP recruitment in cells. Finally, we note that our experiments were performed in the constitutively SOS-activated lexA51 background; physiological changes due to SOS induction are an important component of TLS polymerase regulation in vivo and may also play a role in regulating Pol IV enrichment.
In E. coli, there are at least 17 other SIPs that are involved in various genome maintenance processes. Among these are RecG and PriA, which help resolve stalled replication forks. Prior studies have not found strong evidence for the formation of replisome-associated foci of RecG (68–70) or PriA (68–71) at native expression levels during processive replication, nor have they investigated whether these SIPs are selectively enriched near replication forks upon replication perturbation. To address these questions, we imaged PriA and RecG with single-molecule resolution and quantified their colocalization with sites of replication. We found that both PriA and RecG are strongly enriched near replication forks upon MMS treatment, but not in unperturbed cells, similar to Pol IV. Although the SSB-binding interfaces are not known for these proteins, and thus we cannot make mutations to eliminate binding, it is likely that this enrichment is mediated through interactions with SSB. Previously, we showed that a chimeric protein composed of the C-terminal Pol IV little-finger domain and the RecQ SSB-binding winged-helix domain was selectively enriched upon DNA damage and that this enrichment was lost when the SSB-binding residues of RecQ were mutated (18). As for Pol IV, this selective enrichment in response to replication stress likely helps PriA and RecG gain access to the DNA when their activity may be needed. Our results suggest that selective enrichment of SIPs in response to replication perturbations is a general effect, but future work is needed to elucidate whether other interactions help to establish a hierarchical response of different SIPs upon replication stalling and to determine the functional consequences for cell survival upon replication perturbation.
Methods
Bacterial Strain Construction.
Bacterial strains containing fluorescent protein fusions were constructed using Lambda Red recombineering (72, 73), P1vir transduction, and Flp–FRT recombination (74), as described previously (4) and in the SI Appendix, Methods and Tables S11–S13.
Culture Conditions and Sample Preparation for Microscopy.
Cell culture and sample preparation methods were as previously described (4). In brief, cells were streaked from glycerol stocks onto Luria broth (LB) agar plates containing the appropriate antibiotics and incubated overnight at 37 °C. Single colonies were picked and used to inoculate “overday” starter cultures in 3 mL of LB, and these were incubated for ∼8 h on a roller drum at 37 °C. Overnight cultures in 3 mL of supplemented M9 glucose medium (0.4% glucose, 1 mM thiamine hydrochloride, 0.2% casamino acids, 2 mM MgSO4, and 0.1 mM CaCl2) were inoculated with a 1:1,000 dilution of the overday cultures and then grown in a roller drum at 37 °C overnight. The next day, imaging cultures were prepared in 50 mL of supplemented M9 medium, inoculated with a 1:200 dilution of the corresponding overnight cultures, and incubated at 37 °C, shaking at 225 rpm. Antibiotics were not included in liquid cultures, but 0.5 mM IPTG was included in the M9 overnight and imaging cultures to induce expression of fusions inserted at the lacZ locus.
When imaging cultures reached early exponential phase (optical density at 600 nm [OD600nm] ∼ 0.15), a 1-mL aliquot was removed and centrifuged at 8,609g. The supernatant was removed, the cell pellet was resuspended in a few microliters of the remaining liquid, and <1 µL was deposited on an agarose pad (3% concentration of NuSieve GTG agarose) prepared with supplemented M9 glucose medium (not including thiamine hydrochloride, casamino acids, or IPTG). The agarose pad was sandwiched between two glass coverslips. Coverslips were cleaned by 30-min cycles of sonication in ethanol and then 1 M KOH (two cycles of each, alternating) and rinsed thoroughly with deionized water.
Sensitivity Assays and Treatment Conditions for Microscopy.
A spot dilution assay was used to determine cell sensitivity to MMS and HU. In brief, 50 mL LB cultures of 50 mL each were inoculated with a 1:1,000 dilution of the corresponding overnight cultures and incubated shaking at 37 °C until reaching OD600nm ∼ 1.0. Aliquots were removed and serially diluted in 0.9% NaCl. A dilution series was stamped or pipetted on LB agar plates containing MMS (2, 4, or 6 mM), HU (4, 7, and 10 mM), or without any drug. All plates included 0.5 mM IPTG to induce SSB expression. Plates were photographed after a 16-h incubation at 37 °C.
As described previously (4, 27), cells were treated with MMS included at 10 mM or 100 mM concentration (in most experiments) in the standard agarose pad. Cells were deposited on the pad as normal, then incubated at room temperature in a humidified chamber for 20 min before imaging. Cells were treated with 20 mM HU or 100 mM HU or 100 mM guanazole for 20 min, following the same approach. UV treatment was performed using 254-nm light (General Electric G15T8 15W bulb) at an irradiance of 2 W/m2 measured with a UVP UVX Radiometer (no. 97–0015-02) and UVX-25 sensor. Cells were deposited on an agarose pad as normal but without the coverslip in place, and then exposed to a fluence of 30 J/m2. After treatment, the coverslip was placed on the agarose pad and the sample was imaged immediately.
Cells were fixed by treatment with formaldehyde, as described previously (4, 27). In brief, cells were harvested, concentrated by centrifugation at 7,197g, and resuspended in a 2.5% solution of formaldehyde in phosphate-buffered saline. After a 45-min incubation at room temperature, cells were resuspended and washed with fresh supplemented M9 glucose medium before imaging.
Microscopy.
The fluorescence microscope used in this study was described previously (4). In brief, a Nikon TE2000 inverted microscope was equipped with a 514-nm laser (Coherent Sapphire, 150 mW) for mYPet excitation, a 405-nm laser (Coherent OBIS, 100 mW) for PAmCherry activation, and a 561-nm laser (Coherent Sapphire, 200 mW) for PAmCherry excitation. The lasers were passed through excitation filters (Chroma ZET405/20X, ZET514/10X, and ZET561/10X), and combined with dichroic filters (Chroma ZT405rdc, Chroma ZT514rdc, and a mirror). Imaging was performed using highly inclined thin illumination (75), or near-total internal reflection fluorescence, in which a 400-mm focal length lens was used to focus the beams to the back focal plane of a Nikon CFI Apo 100×/1.49 NA total internal reflection fluorescence (TIRF) objective. Two-color imaging was performed using a Chroma 91032 Laser TIRF Cube containing a ZT405/514/561rpc dichroic filter, ZET442/514/561m emission filter, and ET525lp longpass filter. Images were collected on a Hamamatsu ImageEM C9100-13 EMCCD camera. A brightfield image was recorded for each field of view using white-light transillumination.
Movies were recorded with an integration time of 13.3 ms for short-exposure imaging and 250 ms for long-exposure imaging, as described previously (4). Stroboscopic imaging was implemented using computer-controlled shutters (Uniblitz VS14) and custom-written LabVIEW (National Instruments) scripts. All short- and long-exposure PALM movies were initiated with a 561-nm prebleaching period, followed by 514-nm excitation to image mYPet fusions, followed by simultaneous, low-power 405-nm excitation and 561-nm excitation for photoactivation and imaging of PAmCherry fusions. The 405-nm power was increased twice during the course of the PALM movie. Laser power densities at the sample were as follows: ∼120 W/cm2 (short exposure) or ∼12.5 W/cm2 (long exposure) for 561-nm excitation; ∼0.23 W/cm2 (SSB–mYPet for long-exposure focus-intensity analysis to avoid focus saturation), ∼0.4 W/cm2 (SSB–mYPet in other long-exposure imaging), 1.4 W/cm2 (ε–mYPet long exposure), or 16.6 W/cm2 (ε–mYPet short exposure) for 514-nm excitation; and ∼2.5 to 17.5 mW/cm2 405-nm excitation.
Image Analysis.
Automated image analysis was carried out in MATLAB using the approach and parameters described in detail previously (4). In brief, the MicrobeTracker (76) and u-track (77, 78) packages were used for cell segmentation of brightfield images and fluorescence spot detection and tracking, respectively. Fluorescent spots were fit to symmetrical two-dimensional (2D) Gaussian PSFs. Static molecules in long-exposure PALM imaging were identified by comparing the average PSF width over all localizations in the track with the mean PSF width measured in fixed cells (4). SSB–mYPet and ε–mYPet foci were fit to 2D Gaussian PSFs after averaging the first five frames of 514-nm excitation; to avoid spurious detection of broad and weak fluorescence spots, foci with background values below the camera offset level were discarded. A very small number of cells containing saturated foci were likewise removed from analysis. For PALM analysis, a few cells were removed from analysis if they met any of the following criteria: 1) maximum pixel intensities >15,000 counts in the prebleaching period; 2) localizations in the first frame of the PALM excitation (to avoid possible crosstalk from 514-nm foci); or 3) >50 trajectories.
Data Analysis.
Data analysis methods were as described in detail previously (4).
Diffusion-coefficient analysis.
Apparent 2D diffusion coefficients (D*) were calculated for short-exposure PALM imaging. First, the mean-squared displacement was determined for tracks with least five localizations as follows:
where x and y are the track coordinates. Then D* was calculated as
where Δt is the time interval between localizations in the track.
Diffusion-coefficient distribution fitting.
Probability distributions of apparent distribution coefficients were fit to an analytical expression for three diffusing species where trajectories contain exactly four steps (79):
with the constraint
Here, An represents the fraction of molecules corresponding to the nth species and Dn represents the diffusion coefficient of the nth species. For this analysis, trajectories longer than four steps were truncated and the apparent diffusion calculated as above.
Radial distribution function analysis.
Colocalization was quantified using radial distribution function analysis (30, 31), as described previously (4). In brief, a distribution was generated of the mean distance between each Pol IV–PAmCherry trajectory within a cell and the nearest SSB–mYPet focus. Then a random Pol IV–SSB distance distribution was generated by taking the same cell outline and SSB focus position(s) and simulating the same number of Pol IV localizations randomly across the cell. The same procedure was repeated for all cells in the dataset to yield aggregated experimental and simulated Pol IV–SSB distance distributions. Finally, the radial distribution function g(r) was calculated by normalizing the experimental distance distribution by the simulated one. A g(r) value of 1 indicates no enrichment relative to random chance, whereas values greater than 1 indicate enrichment. As described previously, 100 different, simulated random distributions were generated to account for variability, giving 100 g(r) curves. The final g(r) curve was taken as the mean of these 100 curves. Independently, another random distance distribution was simulated and normalized by the same 100 simulated random distributions to give a mean random g(r) curve; deviations in this random g(r) curve from 1 may arise due to the finite sample size. This procedure was repeated for colocalization analysis of other PAmCherry-labeled proteins and mYPet-labeled replisome markers. As another measure of variability, we calculated the SEM for the average value of g(r) at the smallest r value for the 100 g(r) replicates (SI Appendix, Table S1). In all cases, the SEM was very small relative to differences in this g(r) value between different proteins and treatment conditions.
Survival curve analysis.
The empirical cumulative distribution function, F(t), with 95% confidence bounds, was tabulated for each set (untreated, MMS-damaged, UV-damaged, and HU-treated) of SSB–PAmCherry molecule lifetimes using the MATLAB ecdf function. The survivor function, the complement, was then tabulated using the relationship S(t) = 1 – F(t). The result was fit to a normalized double exponential, as follows:
with the requirement that A + B = 1. The quantity A × T1 + B × T2 represents the weighted estimated survival time T of SSB–PAmCherry. For pre-exponential factors A and B, there is a constant time-correction factor t0 = 250 ms, such that A = A′ × e0.25/T1 and B = Bʹ × e0.25/T2. Thus, A + B = 1 at t0, or 250 ms; this factor is included because 250 ms is the minimum event time detected in our analysis.
Photobleaching analysis.
The survival analysis described above was generated for continuous imaging as well as 1-s and 2-s stroboscopic imaging intervals. As described previously (80, 81), this allows us to plot the measured off-rate of any given SSB–PAmCherry molecule as a function of total stroboscopic interval. Here, we assume that while the off-rate of the molecule from the replisome is a constant, the off-rate contribution from photobleaching is weighted by the total laser exposure within that time interval, according to the following expression:
In this expression, knet is the inverse of T, or the measured weighted survival time of the SSB–PAmCherry molecule. For this analysis, we included all events and fit the data with a double-exponential function (SI Appendix, Fig. S6). We used a net survival time T as a sum of the two survival times weighted by their pre-exponential factors as the treatment condition–dependent value. The net observed off-rate is the sum of the rate of photobleaching and the off-rate of the particle itself, but the photobleaching rate is weighted by tlight/ttot, where tlight is the exposure time of the light frame and ttot is the duration of the total stroboscopic interval (including light and dark frames). Thus, when plotting knetttot versus ttot, the intercept is kbleachtlight, or the expected number of photobleaching events per light frame, and the slope is koff, an estimated value for the off-rate of the fluorescent molecule corrected for the impact of photobleaching (SI Appendix, Fig. S7).
Number of static SSB-binding events analysis.
The number of static SSB-binding events was measured as the number of detected static SSB–PAmCherry trajectories per cell over the course of a movie. These values do not reflect the total number of SSB molecules present at the replication fork, because not all SSB–PAmCherry molecules are photoactivated under our imaging conditions. These values decrease as the stroboscopic interval increases (see SI Appendix, Table S10), because the movie duration was held constant across imaging conditions and thus the total number of illuminated frames was smaller for longer stroboscopic intervals.
Statistical analysis.
The two-sided Wilcoxon rank-sum test (MATLAB function ranksum) was used to compare different distributions, with statistically significant differences determined as P < 0.05.
Imaging Dataset.
SI Appendix, Table S14 contains the number of imaging days, imaging replicates (defined as independent imaging cultures), cells, and tracks or foci for all imaging data.
Supplementary Material
Acknowledgments
We thank members of the J.J.L. laboratory for helpful feedback. We also thank Talley Lambert and the Nikon Imaging Center at Harvard Medical School for help with the collection of preliminary data. This work was supported by NIH grants R01 GM114065 (to J.J.L.), F32 GM113516 (to E.S.T.), and T32 GM008313 (to S.C.P.), as well as a Landry Cancer Biology Research Fellowship (to S.C.P.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2208875119/-/DCSupplemental.
Data, Materials, and Software Availability
The data and custom MATLAB analysis code from this study are archived in Zenodo (DOI: 10.5281/zenodo.7007581) (82).
References
- 1.Fuchs R. P., Fujii S., Translesion DNA synthesis and mutagenesis in prokaryotes. Cold Spring Harb. Perspect. Biol. 5, a012682 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goodman M. F., Woodgate R., Translesion D. N. A., Translesion DNA polymerases. Cold Spring Harb. Perspect. Biol. 5, a010363 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sale J. E., Lehmann A. R., Woodgate R., Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat. Rev. Mol. Cell Biol. 13, 141–152 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thrall E. S., Kath J. E., Chang S., Loparo J. J., Single-molecule imaging reveals multiple pathways for the recruitment of translesion polymerases after DNA damage. Nat. Commun. 8, 2170 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sutton M. D., Coordinating DNA polymerase traffic during high and low fidelity synthesis. Biochim. Biophys. Acta 1804, 1167–1179 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pagès V., Fuchs R. P., How DNA lesions are turned into mutations within cells? Oncogene 21, 8957–8966 (2002). [DOI] [PubMed] [Google Scholar]
- 7.López de Saro F. J., O’Donnell M., Interaction of the beta sliding clamp with MutS, ligase, and DNA polymerase I. Proc. Natl. Acad. Sci. U.S.A. 98, 8376–8380 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kurz M., Dalrymple B., Wijffels G., Kongsuwan K., Interaction of the sliding clamp β-subunit and Hda, a DnaA-related protein. J. Bacteriol. 186, 3508–3515 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ozaki S., et al. , A replicase clamp-binding dynamin-like protein promotes colocalization of nascent DNA strands and equipartitioning of chromosomes in E. coli. Cell Rep. 4, 985–995 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Kong X.-P., Onrust R., O’Donnell M., Kuriyan J., Three-dimensional structure of the β subunit of E. coli DNA polymerase III holoenzyme: A sliding DNA clamp. Cell 69, 425–437 (1992). [DOI] [PubMed] [Google Scholar]
- 11.Jergic S., et al. , A direct proofreader-clamp interaction stabilizes the Pol III replicase in the polymerization mode. EMBO J. 32, 1322–1333 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toste Rêgo A., Holding A. N., Kent H., Lamers M. H., Architecture of the Pol III-clamp-exonuclease complex reveals key roles of the exonuclease subunit in processive DNA synthesis and repair. EMBO J. 32, 1334–1343 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang S., et al. , A gatekeeping function of the replicative polymerase controls pathway choice in the resolution of lesion-stalled replisomes. Proc. Natl. Acad. Sci. U.S.A. 116, 25591–25601 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Furukohri A., Nishikawa Y., Akiyama M. T., Maki H., Interaction between Escherichia coli DNA polymerase IV and single-stranded DNA-binding protein is required for DNA synthesis on SSB-coated DNA. Nucleic Acids Res. 40, 6039–6048 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Godoy V. G., et al. , UmuD and RecA directly modulate the mutagenic potential of the Y family DNA polymerase DinB. Mol. Cell 28, 1058–1070 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cafarelli T. M., et al. , A single residue unique to DinB-like proteins limits formation of the polymerase IV multiprotein complex in Escherichia coli. J. Bacteriol. 195, 1179–1193 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shereda R. D., Kozlov A. G., Lohman T. M., Cox M. M., Keck J. L., SSB as an organizer/mobilizer of genome maintenance complexes. Crit. Rev. Biochem. Mol. Biol. 43, 289–318 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang S., et al. , Compartmentalization of the replication fork by single-stranded DNA binding protein regulates translesion synthesis. Nat. Struct. Mol. Biol https://doi.org/10.1038/s41594-022-00827-2 (2022). [DOI] [PMC free article] [PubMed]
- 19.Shinn M. K., Kozlov A. G., Nguyen B., Bujalowski W. M., Lohman T. M., Are the intrinsically disordered linkers involved in SSB binding to accessory proteins? Nucleic Acids Res. 47, 8581–8594 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Subach F. V., et al. , Photoactivatable mCherry for high-resolution two-color fluorescence microscopy. Nat. Methods 6, 153–159 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ennis D. G., Little J. W., Mount D. W., Novel mechanism for UV sensitivity and apparent UV nonmutability of recA432 mutants: Persistent LexA cleavage following SOS induction. J. Bacteriol. 175, 7373–7382 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernández De Henestrosa A. R., et al. , Identification of additional genes belonging to the LexA regulon in Escherichia coli. Mol. Microbiol. 35, 1560–1572 (2000). [DOI] [PubMed] [Google Scholar]
- 23.Henrikus S. S., et al. , DNA polymerase IV primarily operates outside of DNA replication forks in Escherichia coli. PLoS Genet. 14, e1007161 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nguyen A. W., Daugherty P. S., Evolutionary optimization of fluorescent proteins for intracellular FRET. Nat. Biotechnol. 23, 355–360 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Antony E., et al. , Multiple C-terminal tails within a single E. coli SSB homotetramer coordinate DNA replication and repair. J. Mol. Biol. 425, 4802–4819 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manley S., et al. , High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat. Methods 5, 155–157 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Uphoff S., Reyes-Lamothe R., Garza de Leon F., Sherratt D. J., Kapanidis A. N., Single-molecule DNA repair in live bacteria. Proc. Natl. Acad. Sci. U.S.A. 110, 8063–8068 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reyes-Lamothe R., Possoz C., Danilova O., Sherratt D. J., Independent positioning and action of Escherichia coli replisomes in live cells. Cell 133, 90–102 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stracy M., et al. , Transient non-specific DNA binding dominates the target search of bacterial DNA-binding proteins. Mol. Cell 81, 1499–1514.e6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zawadzki P., et al. , The localization and action of topoisomerase IV in Escherichia coli chromosome segregation is coordinated by the SMC complex, MukBEF. Cell Rep. 13, 2587–2596 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garza de Leon F., Sellars L., Stracy M., Busby S. J. W., Kapanidis A. N., Tracking low-copy transcription factors in living bacteria: The case of the lac repressor. Biophys. J. 112, 1316–1327 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bjedov I., et al. , Involvement of Escherichia coli DNA polymerase IV in tolerance of cytotoxic alkylating DNA lesions in vivo. Genetics 176, 1431–1440 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sinha R. P., Häder D.-P., UV-induced DNA damage and repair: A review. Photochem. Photobiol. Sci. 1, 225–236 (2002). [DOI] [PubMed] [Google Scholar]
- 34.Courcelle J., Donaldson J. R., Chow K.-H., Courcelle C. T., Damage-Induced Replication Fork Regression D. N. A., DNA damage-induced replication fork regression and processing in Escherichia coli. Science 299, 1064–1067 (2003). [DOI] [PubMed] [Google Scholar]
- 35.Courcelle C. T., Chow K.-H., Casey A., Courcelle J., Nascent DNA processing by RecJ favors lesion repair over translesion synthesis at arrested replication forks in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 103, 9154–9159 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soubry N., Wang A., Reyes-Lamothe R., Replisome activity slowdown after exposure to ultraviolet light in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 116, 11747–11753 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leadon S. A., Production of thymine glycols in DNA by radiation and chemical carcinogens as detected by a monoclonal antibody. Br. J. Cancer Suppl. 8, 113–117 (1987). [PMC free article] [PubMed] [Google Scholar]
- 38.Kielbassa C., Roza L., Epe B., Wavelength dependence of oxidative DNA damage induced by UV and visible light. Carcinogenesis 18, 811–816 (1997). [DOI] [PubMed] [Google Scholar]
- 39.Schuch A. P., da Silva Galhardo R., de Lima-Bessa K. M., Schuch N. J., Menck C. F. M., Development of a DNA-dosimeter system for monitoring the effects of solar-ultraviolet radiation. Photochem. Photobiol. Sci. 8, 111–120 (2009). [DOI] [PubMed] [Google Scholar]
- 40.Moore E. C., Hurlbert R. B., The inhibition of ribonucleoside diphosphate reductase by hydroxyurea, guanazole and pyrazoloimidazole (IMPY). Pharmacol. Ther. 27, 167–196 (1985). [DOI] [PubMed] [Google Scholar]
- 41.Sinha N. K., Snustad D. P., Mechanism of inhibition of deoxyribonucleic acid synthesis in Escherichia coli by hydroxyurea. J. Bacteriol. 112, 1321–1324 (1972). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Larsen I. K., Sjöberg B. M., Thelander L., Characterization of the active site of ribonucleotide reductase of Escherichia coli, bacteriophage T4 and mammalian cells by inhibition studies with hydroxyurea analogues. Eur. J. Biochem. 125, 75–81 (1982). [DOI] [PubMed] [Google Scholar]
- 43.Spenkelink L. M., et al. , Recycling of single-stranded DNA-binding protein by the bacterial replisome. Nucleic Acids Res. 47, 4111–4123 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sandler S. J., Marians K. J., Role of PriA in replication fork reactivation in Escherichia coli. J. Bacteriol. 182, 9–13 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cadman C. J., McGlynn P., PriA helicase and SSB interact physically and functionally. Nucleic Acids Res. 32, 6378–6387 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gabbai C. B., Marians K. J., Recruitment to stalled replication forks of the PriA DNA helicase and replisome-loading activities is essential for survival. DNA Repair (Amst.) 9, 202–209 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bianco P. R., DNA helicase-SSB interactions critical to the regression and restart of stalled DNA replication forks in Escherichia coli. Genes (Basel) 11, 471 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McGlynn P., Lloyd R. G., Genome stability and the processing of damaged replication forks by RecG. Trends Genet. 18, 413–419 (2002). [DOI] [PubMed] [Google Scholar]
- 49.Buss J. A., Kimura Y., Bianco P. R., RecG interacts directly with SSB: Implications for stalled replication fork regression. Nucleic Acids Res. 36, 7029–7042 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moolman M. C., et al. , Slow unloading leads to DNA-bound β2-sliding clamp accumulation in live Escherichia coli cells. Nat. Commun. 5, 5820 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zechner E. L., Wu C. A., Marians K. J., Coordinated leading- and lagging-strand synthesis at the Escherichia coli DNA replication fork. II. Frequency of primer synthesis and efficiency of primer utilization control Okazaki fragment size. J. Biol. Chem. 267, 4045–4053 (1992). [PubMed] [Google Scholar]
- 52.Yao N. Y., Georgescu R. E., Finkelstein J., O’Donnell M. E., Single-molecule analysis reveals that the lagging strand increases replisome processivity but slows replication fork progression. Proc. Natl. Acad. Sci. U.S.A. 106, 13236–13241 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lohman T. M., Ferrari M. E., Escherichia coli single-stranded DNA-binding protein: Multiple DNA-binding modes and cooperativities. Annu. Rev. Biochem. 63, 527–570 (1994). [DOI] [PubMed] [Google Scholar]
- 54.Marians K. J., Lesion bypass and the reactivation of stalled replication forks. Annu. Rev. Biochem. 87, 217–238 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yeeles J. T. P., Marians K. J., The Escherichia coli replisome is inherently DNA damage tolerant. Science 334, 235–238 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ikeda M., et al. , DNA polymerase IV mediates efficient and quick recovery of replication forks stalled at N2-dG adducts. Nucleic Acids Res. 42, 8461–8472 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yeeles J. T. P., Marians K. J., Dynamics of leading-strand lesion skipping by the replisome. Mol. Cell 52, 855–865 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sogo J. M., Lopes M., Foiani M., Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297, 599–602 (2002). [DOI] [PubMed] [Google Scholar]
- 59.Feng W., et al. , Genomic mapping of single-stranded DNA in hydroxyurea-challenged yeasts identifies origins of replication. Nat. Cell Biol. 8, 148–155 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McInerney P., O’Donnell M., Functional uncoupling of twin polymerases: Mechanism of polymerase dissociation from a lagging-strand block. J. Biol. Chem. 279, 21543–21551 (2004). [DOI] [PubMed] [Google Scholar]
- 61.Maréchal A., Zou L., RPA-coated single-stranded DNA as a platform for post-translational modifications in the DNA damage response. Cell Res. 25, 9–23 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mijakovic I., et al. , Bacterial single-stranded DNA-binding proteins are phosphorylated on tyrosine. Nucleic Acids Res. 34, 1588–1596 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kozlov A. G., Cox M. M., Lohman T. M., Regulation of single-stranded DNA binding by the C termini of Escherichia coli single-stranded DNA-binding (SSB) protein. J. Biol. Chem. 285, 17246–17252 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wessel S. R., et al. , PriC-mediated DNA replication restart requires PriC complex formation with the single-stranded DNA-binding protein. J. Biol. Chem. 288, 17569–17578 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bhattacharyya B., et al. , Structural mechanisms of PriA-mediated DNA replication restart. Proc. Natl. Acad. Sci. U.S.A. 111, 1373–1378 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Harami G. M., et al. , Phase separation by ssDNA binding protein controlled via protein-protein and protein-DNA interactions. Proc. Natl. Acad. Sci. U.S.A. 117, 26206–26217 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kozlov A. G., et al. , How glutamate promotes liquid-liquid phase separation and DNA binding cooperativity of E. coli SSB protein. bioRxiv [Preprint] (2022) 10.1101/2022.01.17.476650. [DOI] [PMC free article] [PubMed]
- 68.Upton A. L., et al. , Cellular location and activity of Escherichia coli RecG proteins shed light on the function of its structurally unresolved C-terminus. Nucleic Acids Res. 42, 5702–5714 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bentchikou E., et al. , Are the SSB-interacting proteins RecO, RecG, PriA and the DnaB-interacting protein rep bound to progressing replication forks in Escherichia coli? PLoS One 10, e0134892 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yu C., et al. , SSB binds to the RecG and PriA helicases in vivo in the absence of DNA. Genes Cells 21, 163–184 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lesterlin C., Ball G., Schermelleh L., Sherratt D. J., RecA bundles mediate homology pairing between distant sisters during DNA break repair. Nature 506, 249–253 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Datsenko K. A., Wanner B. L., One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sharan S. K., Thomason L. C., Kuznetsov S. G., Court D. L., Recombineering: A homologous recombination-based method of genetic engineering. Nat. Protoc. 4, 206–223 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cherepanov P. P., Wackernagel W., Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158, 9–14 (1995). [DOI] [PubMed] [Google Scholar]
- 75.Tokunaga M., Imamoto N., Sakata-Sogawa K., Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat. Methods 5, 159–161 (2008). [DOI] [PubMed] [Google Scholar]
- 76.Sliusarenko O., Heinritz J., Emonet T., Jacobs-Wagner C., High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics. Mol. Microbiol. 80, 612–627 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jaqaman K., et al. , Robust single-particle tracking in live-cell time-lapse sequences. Nat. Methods 5, 695–702 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Aguet F., Antonescu C. N., Mettlen M., Schmid S. L., Danuser G., Advances in analysis of low signal-to-noise images link dynamin and AP2 to the functions of an endocytic checkpoint. Dev. Cell 26, 279–291 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stracy M., et al. , Live-cell superresolution microscopy reveals the organization of RNA polymerase in the bacterial nucleoid. Proc. Natl. Acad. Sci. U.S.A. 112, E4390–E4399 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gebhardt J. C. M., et al. , Single-molecule imaging of transcription factor binding to DNA in live mammalian cells. Nat. Methods 10, 421–426 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ho H. N., Zalami D., Köhler J., van Oijen A. M., Ghodke H., Identification of multiple kinetic populations of DNA-binding proteins in live cells. Biophys. J. 117, 950–961 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.E.S. Thrall, S.C. Piatt, S. Chang, J.J. Loparo, Replication stalling activates SSB for recruitment of DNA damage tolerance factors. Zenodo. https://zenodo.org/record/7007581#.Yy2pBnZBy3A. Deposited 22 August 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data and custom MATLAB analysis code from this study are archived in Zenodo (DOI: 10.5281/zenodo.7007581) (82).