Significance
Many droplet-like cellular structures lack membranes and are referred to as biomolecular condensates. One fundamental process associated with condensates is gene expression, in which the protein transcription machinery generates RNA de novo from a DNA template. We used mitochondrial transcription as a model system to ask how the structure of a condensate is influenced by the transcription that occurs within it. We were able to reconstitute mitochondrial transcription under condensate-forming conditions and show that the presence of the condensate reduces the efficiency of transcription. In turn, we also found that the production of RNA alters the structure of the condensate. These results reveal that structure and function are inherently coupled in transcriptional condensates.
Keywords: biomolecular condensates, phase separation, transcription, mitochondrial genome, vesicles
Abstract
In live cells, phase separation is thought to organize macromolecules into membraneless structures known as biomolecular condensates. Here, we reconstituted transcription in condensates from purified mitochondrial components using optimized in vitro reaction conditions to probe the structure–function relationships of biomolecular condensates. We find that the core components of the mt-transcription machinery form multiphasic, viscoelastic condensates in vitro. Strikingly, the rates of condensate-mediated transcription are substantially lower than in solution. The condensate-mediated decrease in transcriptional rates is associated with the formation of vesicle-like structures that are driven by the production and accumulation of RNA during transcription. The generation of RNA alters the global phase behavior and organization of transcription components within condensates. Coarse-grained simulations of mesoscale structures at equilibrium show that the components stably assemble into multiphasic condensates and that the vesicles formed in vitro are the result of dynamical arrest. Overall, our findings illustrate the complex phase behavior of transcribing, multicomponent condensates, and they highlight the intimate, bidirectional interplay of structure and function in transcriptional condensates.
Proteins and nucleic acids form diverse biomolecular condensates that arise via macromolecular phase separation (1–4). Condensates emerge via demixing of protein, RNA, and DNA components from their cellular surroundings, leading to the formation of distinct, non-membrane-bound cellular structures. The formation of condensates via macromolecular phase separation is mediated by multivalent homotypic and heterotypic interactions among proteins and nucleic acids (1). Prominent biomolecular condensates include P granules and stress granules in the cytoplasm (5, 6) as well as the nucleolus and RNA splicing factor speckles in the nucleus (7, 8).
Condensates are thought to contribute to many cellular functions, including genome organization and transcription (9–11). Major architectural chromatin proteins such as the linker histone H1 (12) and the heterochromatin protein HP1α form condensates in vitro and in vivo (13, 14). Phase separation, in different manifestations, has been suggested to contribute to higher-order organization of genomes into domains and compartments (15–17). In particular, various components of the transcription machinery spontaneously concentrate into condensed phases in the mammalian nucleus, including prominently at sites of superenhancers (18). This behavior has been attributed to the intrinsically disordered regions (IDRs) found in many transcription factors and chromatin proteins (19). IDRs are thought to mediate an array of multivalent protein–protein interactions that give rise to dynamic, nonstoichiometric condensed assemblies (20, 21).
Condensates are enriched in functional components, such as transcription factors and RNA processing factors. It has been proposed that increased concentrations of bioactive macromolecules within condensates enhances reaction rates and increase the overall efficiencies of key biochemical reactions within condensates (22). However, this hypothesis rests on the assumption that condensates are well-mixed reactors, without considerations of complexities of molecular transport within condensates. It thus remains largely unclear how condensate structure relates to function (2). A major hurdle in elucidating relationships between mesoscale structures and functions in vivo condensates has been the difficulty of reconstituting functionally active condensates in vitro with all the biochemically relevant components.
The mitochondrial genome (mtDNA) and its own dedicated gene expression machinery are also organized via phase separation (23). Human mitochondria contain hundreds of copies of their own 16 kb, circular genome (24) that assemble into mitochondrial (mt-) nucleoids, which are membraneless, nucleoprotein complexes of ∼100 nm in diameter containing mtDNA and associated proteins (25, 26). In support of phase separation as a driver of mt-nucleoid organization, the major mt-genome architectural protein TFAM phase separates in vitro and in vivo, and, combined with mtDNA, forms condensates that recapitulate the behavior of mt-nucleoids in cells (23). The mt-nucleoids also serve as sites of transcription of long, polycistronic mt-RNA, which becomes further processed in adjacent mt-RNA granules that are also thought to form via phase separation (27).
The relative simplicity of mt-transcription, involving only a small number of essential components, makes for a unique and tractable model system to probe relationships between mesoscale structures formed via phase separation and the functional outputs of a biologically relevant transcriptional condensate. Mt-transcription can be reconstituted under soluble conditions with only four components: mtDNA, the single-subunit mtRNA polymerase POLRMT, and two transcription factors, TFAM and TFB2M (28, 29). Here, we study mt-transcription under condensate-forming conditions in vitro. We demonstrate that the mt-transcription machinery forms multiphasic condensates in vitro, leading to dynamically arrested mesoscale structures with dampened transcriptional kinetics compared with equivalent reactions in bulk solutions. Importantly, we find that the production of nascent RNA during transcription alters the structure of the condensate. Our results demonstrate a close interplay between the physical behavior and functional activity of an archetypal biomolecular condensate.
Results
Individual Components of the Mitochondrial Transcription Machinery Undergo Phase Separation In Vitro.
The mt-transcription has previously been reconstituted with three proteins and a DNA template containing a mitochondrial promoter under dilute conditions in solution (29). Given the organization of mt-nucleoids into condensates within the crowded matrix in vivo (9), we sought to establish in vitro conditions for mt-transcription within condensates.
Taking a bottom-up approach, we first established the individual phase behavior of the minimal components required for mt-transcription. The transcription factors TFAM and TFB2M combined with the polymerase POLRMT are the minimal components of the human mt-transcription machinery. Structural studies (30) and bioinformatics analysis show that these proteins contain a combination of ordered domains and IDRs (Fig. 1A). Computational predictions for disordered proteins suggest that unbound TFAM is the most disordered of the three proteins, with a flexible linker that bridges two DNA binding domains (high-mobility groups A and B) and a disordered C terminus. These features are consistent with conformational heterogeneity that has been reported for TFAM molecules in solution (31). TFB2M and POLRMT contain well-folded, functional domains (28, 32). However, both proteins also contain disordered regions at their N termini.
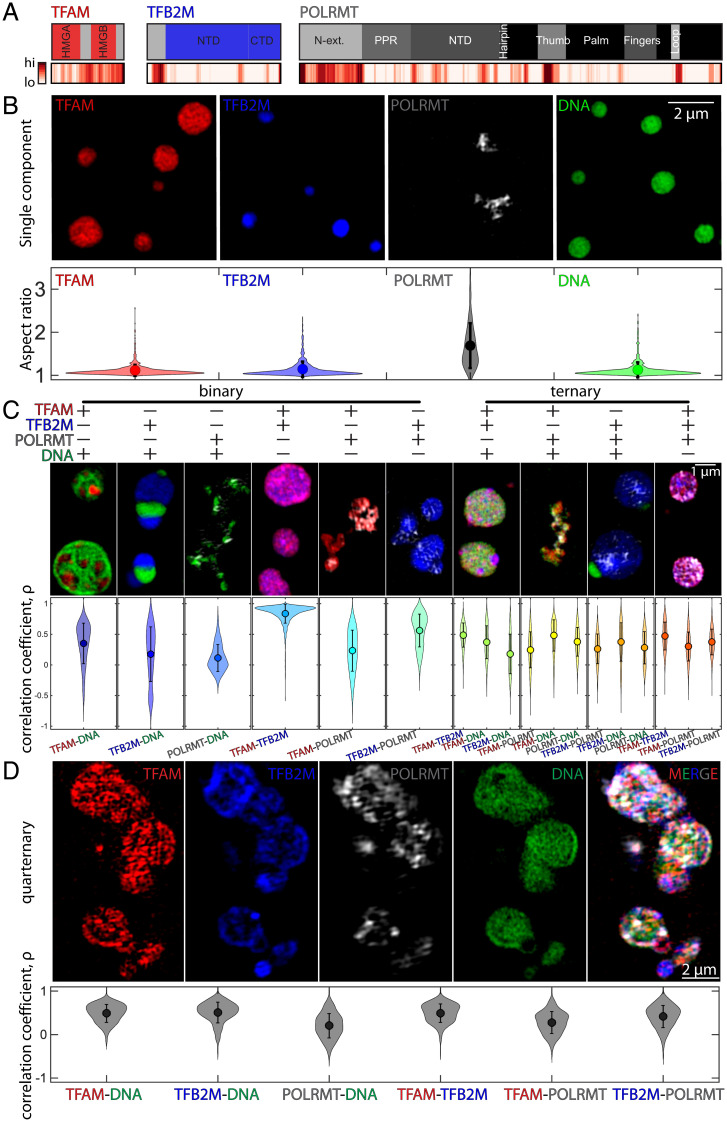
Fig. 1.
Phase behavior of individual, binary, ternary, and quaternary condensates comprising mt-transcriptional components. (A) Protein domain analysis for core mt-transcription proteins TFAM, TFB2M, and POLRMT; Top illustrates the known protein domains (red, blue, and dark gray, respectively), and the unfolded, intrinsically disordered domains are indicated in light gray. Bottom shows probability of intrinsically disordered sequences as predicted by several models using D2P2, where high and low likelihoods for disorder are indicated in red (high) and white (low) (SI Appendix). (B) Top shows SIM (Structured Illumination Microscopy) images of condensates formed for individual components at room temperature on pluronic-treated coverslips: 10 µM TFAM in 10% PEG (red), 10 µM TFB2M in 10% PEG (blue), 1.5 µM POLRMT in 5% PEG (gray), and 500 nM DNA in 10% PEG (green). Bottom contains violin plots of the aspect ratio for all condensates analyzed; n = 4 experimental replicates, average (dot) values are indicated, and error bar = SD. (Scale bar, 2 µm.) (C) Binary and ternary compound droplets. Violin plots of correlation coefficient measured for each pair of channels; correlation coefficient = 1 denotes complete colocalization; n = 4 experimental replicates, average (dot) values are indicated, and error bar = SD (see SI Appendix for concentrations). (Scale bar, 1 µm.) (D) Quaternary droplets at room temperature. Top includes individual channels of ∼6 µM TFAM (red), ∼6 µM TFB2M (blue), ∼6 µM POLRMT (gray), and ∼500 nM DNA (green), and the merged image in 5% PEG. (Scale bar, 2 µm.) Bottom contains violin plots of the correlation coefficient for all pairs of channels; n = 3 experimental replicates, average (dot) values are indicated, and error bar = SD. Buffer for all conditions was 20 mM Tris⋅HCl, pH ∼8.0, 20 mM BME, 10 mM MgCl2, and ∼100 mM NaCl at room temperature.
We find that TFAM, TFB2M, and POLRMT as well as DNA individually phase separate to form dense phases in the presence of the macromolecular crowder, polyethylene glycol (PEG, molecular weight of ∼3 kDa). Conditions that promote phase separation of individual components include 10 µM TFAM or TFB2M in 10% PEG; 1.5 µM to 10 µM POLRMT in 5 to 10% PEG; and 500 nM DNA in 10% PEG (Fig. 1B and SI Appendix, Fig. S1 A–C). In the presence of crowder, condensates formed by TFAM, TFB2M, or DNA are highly spherical, with an aspect ratio of approximately one (Fig. 1B and SI Appendix, Fig. S1 A–C). In contrast, POLRMT assembles into highly irregular structures, with an aspect ratio of greater than one (Fig. 1B and SI Appendix, Fig. S1 A–C). The structures formed by POLRMT show limited recoveries after photobleaching (SI Appendix, Fig. S1 D and E) and fit the description of being dynamically arrested phases (33–35). Overall, our results show that all components of the minimal mt-transcription machinery can undergo phase separation via macromolecular crowder-mediated homotypic interactions. However, each of the condensates has distinct dynamics and/or overall morphologies.
Multicomponent Systems Form Inhomogeneously Organized and Dynamically Arrested Condensates In Vitro.
Next, we documented the joint phase behaviors of multiple components by characterizing the structures formed in binary and ternary mixtures of components of the minimal mt-transcription machinery (Fig. 1C). Using the correlation coefficient as a metric of their colocalization, we find that TFAM and DNA form multiphase condensates, containing micrometer-sized subdomains that are either TFAM-rich or DNA-rich (Fig. 1C). In contrast, TFB2M and POLRMT show lower degrees of colocalization with DNA (Fig. 1C). When the proteins were mixed in pairs without DNA, TFAM and TFB2M colocalize with one another, although they do not colocalize as well as with POLRMT (Fig. 1C). This, again, is a likely consequence of POLRMT driving the formation of dynamically arrested phases.
For ternary combinations of pairs of proteins with DNA, the organization of condensates remained inhomogeneous for all combinations, with micrometer-sized domains forming for TFAM–TFB2M–DNA and TFB2M–POLRMT–DNA (Fig. 1C). However, for the ternary mixture of all proteins without DNA, the condensates became more well mixed, implying that heterotypic interactions of proteins with DNA contribute to the emergence of spatially distinct coexisting phases of the multicomponent condensates (Fig. 1C). These results demonstrate differences in phase behaviors of the various components of the mt-transcription machinery.
To begin to build transcriptionally competent condensates, all four biomolecular components (DNA, POLRMT, TFAM, and TFB2M) were combined in equimolar protein ratios (∼6 µM TFAM, TFB2M, and POLRMT with 500 nM DNA) in the presence of 5% PEG (Fig. 1D). In this mixture, all biomolecules collectively partition into condensates (Fig. 1D). However, these condensates do not fully fuse with neighboring ones. Instead, they come into contact and form dynamically arrested, higher-order structures (Fig. 1D). Moreover, condensate features appear to be dependent on the stoichiometries of components (SI Appendix, Fig. S1F). For example, increasing levels of TFAM relative other components lead to significant increases in condensate size (SI Appendix, Fig. S1F). Alternatively, relatively low amounts of POLRMT lead to more round shapes (SI Appendix, Fig. S1F). These results suggest that differences in concentration and stoichiometry of components can influence emergent phase behavior.
We also found that localization of individual components within the multiphasic structures remained inhomogeneous: POLRMT showed the lowest level of colocalization with all other components, while TFB2M tended to accumulate more peripherally (Fig. 1D). Importantly, the colocalization correlation coefficients for specific pairs tended to be higher in the quaternary condensates than those found in the binary or ternary condensates (Fig. 1 C and D). This behavior is consistent with the dual nature of TFAM, which has affinity for DNA via its N-terminal DNA-binding domain , but also for other proteins, such as POLRMT, via its disordered C terminus (23). Together, these results show that components of the minimal mt-transcription machinery assemble into multiphasic condensates in vitro.
Dampening of Transcription Rates under Condensate-Forming Conditions.
Next, we sought to reconstitute mt-transcription in vitro under condensate-forming conditions. We first characterized the effect of increasing concentrations of reaction components starting with standard, soluble in vitro mt-transcription reactions (29, 36). We added a full set of nucleotides (nucleoside 5′-triphosphates [NTPs]) to the mixture of 0.6 µM TFAM, 0.6 µM TFB2M, 0.6 µM POLRMT, and 50 nM DNA (1×) in the absence of any crowder (−PEG). After 30 min of incubation at ∼35 °C (SI Appendix), transcriptional activity was measured by detection of a ∼300-nt RNA product using a PCR-amplified template containing the LSP promoter and radioactively labeled nucleotides, as previously described (36) (Fig. 2A). Using these conditions (1×, −PEG) as a starting point, we increased the concentration of all components proportionally over a 10-fold range. Transcriptional activity increased roughly linearly over an approximately seven-fold range of initial concentrations, but then dropped off at higher concentrations of the macromolecular components (Fig. 2 A and B). Furthermore, by individually titrating various components, we found that high levels of TFAM relative to TFB2M, POLRMT, and DNA led to a reduction of the transcription reaction, while lower concentrations of POLRMT relative to TFAM, TFB2M, and DNA also reduced RNA production (SI Appendix, Fig. S2 A and B). By using 1:1:1 stoichiometries of proteins at approximately 10 times the molar concentration of template DNA, we were able to produce significant amounts of RNA (Fig. 2 and SI Appendix, Fig. S2A).
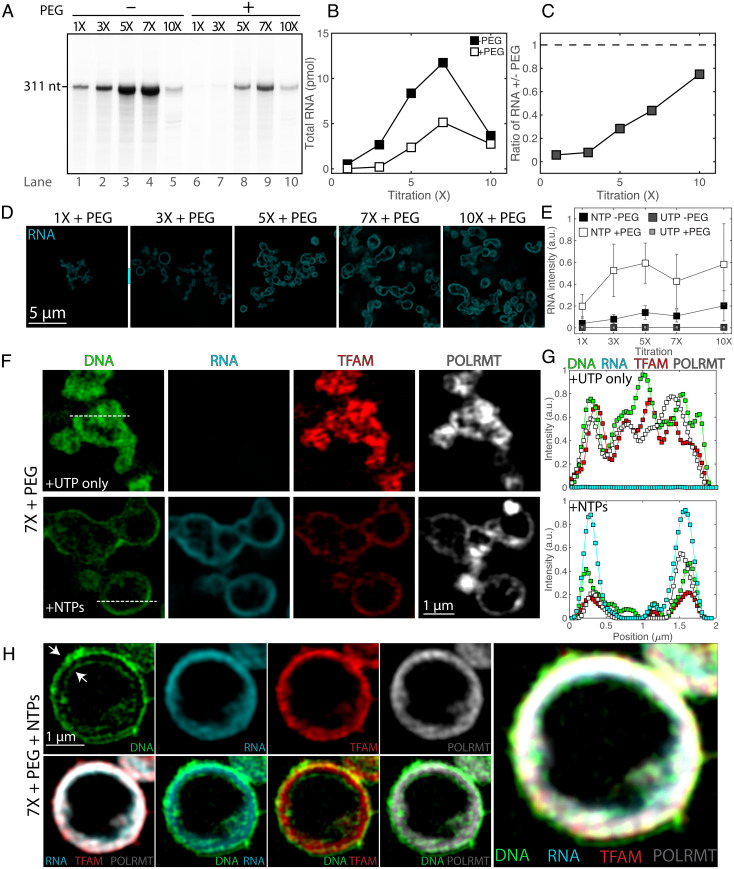
Fig. 2.
In vitro transcription under soluble and condensed conditions leads to changes in condensate organization. (A) RNA production rates at soluble and condensed states. The transcription run-off assay was performed using increasing concentrations (1× to 10×) of components of the transcription machinery (TFAM, TFB2M, and POLRMT) and DNA in the absence (lanes 1 to 5) or presence (lanes 6 to 10) of 5% PEG. (B) Quantification of RNA product from A as a function of reactant titration, where 0% PEG corresponds to filled squares, and 5% PEG corresponds to open squares. (C) Ratio of RNA production of crowded (5% PEG) to soluble (0% PEG) states as a function of reactant titration. Values in A–C illustrate a representative experiment. (D) SIM images of reactions after fixation and RNA fluorescence in situ hybridization (FISH), following 1 h of reactions under the same concentrations of transcription machinery and DNA as well as buffer composition as A with 2 mM NTPs. (Scale bar, 5 µm.) All images are at the same contrast settings. (E) Quantification of RNA FISH intensity under soluble (0% PEG) or condensed (5% PEG) conditions for the reactions (NTPs) and of the negative control (UTP); n = 3 experimental replicates, values represent averages, and error bars = SD. (F) SIM images of core mt-transcription components in condensates at 7× and 5% PEG for negative control (UTP only) and for reactions (NTPs), where DNA is in green, RNA FISH is in cyan, TFAM is in red, and POLRMT is in gray scale. (Scale bar, 1 µm.) Dashed lines indicate line profile. (G) Smoothed line profile for all components from F. (H) Images of a large vesicle after 1 h of transcription (7×, 5% PEG, NTPs). Panels include single channel of DNA, RNA, TFAM, and POLRMT (Top Left), overlays of RNA/TFAM/POLRMT, DNA/RNA, DNA/TFAM, and DNA/POLRMT (Bottom Left), and a four-channel overlay (Right). Arrows indicate the outer and inner lining of DNA. (Scale bar, 1 µm.)
To reconstitute transcriptionally active condensates, we proportionally increased protein and DNA concentrations in the presence of a crowder (+PEG) and nucleotides (Fig. 2A). Condensates were transcriptionally active, but the transcriptional output of condensates was 1.3-fold to 20-fold lower than under the corresponding solution conditions where condensates do not form (Fig. 2 B and C). Decreased rates of transcriptional output in condensates occurred most significantly at equimolar ratios of macromolecular components and approached unity with increasing reactant concentration (Fig. 2 A–C).
To confirm the formation of condensates under conditions that support transcription and to relate mesoscale structures of condensates to function, we compared the morphologies of condensates under different conditions (Fig. 2 D–H and SI Appendix, Figs. S2C and S3). In the absence of crowder, corresponding to dilute conditions (1×, −PEG), we did not observe condensates (SI Appendix, Fig. S3 C–F). Under transcriptionally competent, condensate-forming conditions (1× to 10×, +PEG), RNA and all transcriptional components localized to the periphery of condensates, forming vesicle-like morphologies within 60 min (Fig. 2 D–H, SI Appendix, Figs. S2C and S3 B–F, and ref. 37). In these vesicles, DNA tended to associate with the outermost and innermost layers, whereas RNA and proteins colocalized in the middle layer (Fig. 2H and SI Appendix, Fig. S3I). Visualization of fluorescently labeled PEG suggests that the crowder is localized to the aqueous phase of the lumen within these structures, supporting the vesicular nature of the mt-transcriptional condensates (SI Appendix, Fig. S3G). The peripheral localization of the transcription machinery appears to be a consequence of active transcription, as identical, but transcription-incompetent, condensates generated in the presence of only uridine 5′-triphosphate (UTP) nucleotides tended to retain their filled, nonvesicular structures (Fig. 2 E–H and SI Appendix, Figs. S2C and S3; see “Newly Synthesized RNA Transcripts Shape Condensate Structure”).
Similarly, we examined the morphologies of the mt-transcriptional condensates under several stoichiometries (SI Appendix, Fig. S2C). Increasing levels of TFAM led to more-pronounced vesicular morphologies, suggesting that TFAM supports vesicle formation. In contrast, lower levels of POLRMT—associated with reduced RNA production (SI Appendix, Fig. S2B)—drastically reduced the amount of the condensed phase, and the few remaining condensates tended to retain their droplet-like morphologies. We note that these different stoichiometries exhibit different phase behaviors under non-reacting conditions (SI Appendix, Fig. S1F), and are associated with different transcriptional efficiencies, or, effectively, RNA levels (SI Appendix, Fig. S2 A and B). Thus, the phase behaviors that we observe upon the reaction are functions of not only the protein and DNA components but also of the amount of de novo RNA generated.
Overall, our results demonstrate transcriptional activity and changes to phase behavior in reconstituted mt-condensates, and that transcription is dampened in condensates compared with the bulk solution.
Newly Synthesized RNA Transcripts Shape Condensate Structure.
Newly synthesized RNA transcripts tended to localize to the periphery of mt-transcription condensates (Fig. 2 D–H). To determine whether nascent RNA is exclusively produced at the edge of the condensates or is generated internally and accumulates over time at the periphery, we performed time course experiments (Fig. 3 A and B). RNA can be detected as early as 5 min after the start of the reaction in the condensate interior (Fig. 3 A and B). At early time points of 5 and 10 min, all components of the transcription machinery localize throughout the interior of the condensate (Fig. 3 A and B). In contrast, at later time points of 20, 40, and 60 min, we detected significant changes i organization, whereby condensates with pronounced vacuoles start to appear (Fig. 3 A and B, Movies S1 and S2, and SI Appendix, Fig. S4 H–J). All the components become peripherally located with increasing reaction time, and these morphological changes were concomitant with RNA production (Fig. 3 A and B). Quantification of component intensities within the condensates over time shows that RNA and protein levels accumulate over the course of ∼20 min and then plateau, suggesting an arrest of transcription (SI Appendix, Fig. S4A). However, quantification of DNA, as measured by DAPI, shows a decrease in intensity, corresponding to, potentially, either strand separation resulting from the transcription reaction or an efflux of DNA to accommodate the accumulation of negatively charged RNA (SI Appendix, Fig. S4A).
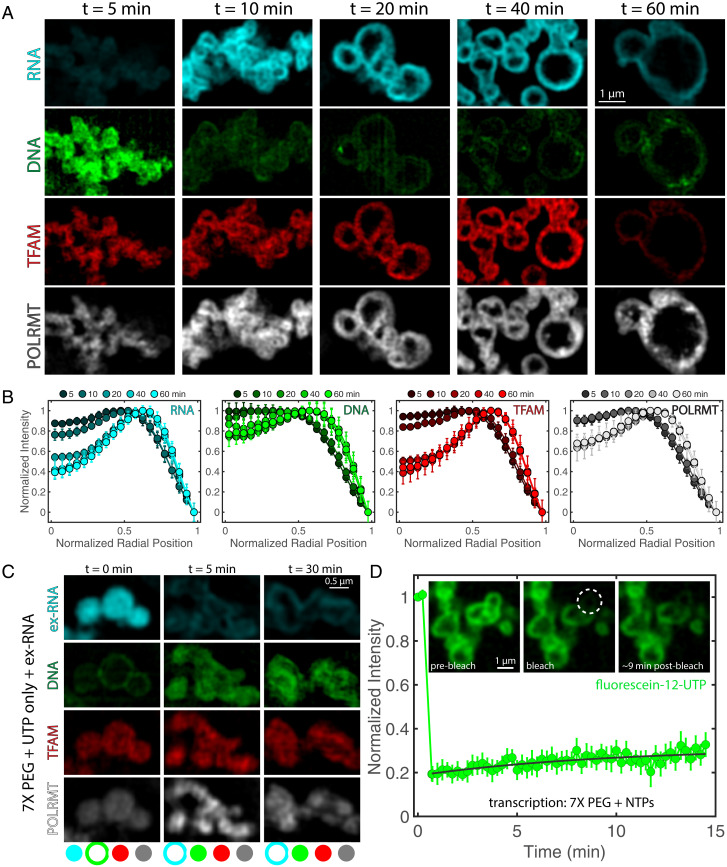
Fig. 3.
Dynamics of transcriptional condensate organization. (A) Time course of condensate morphology under reaction conditions (7× PEG, 5% PEG). Condensates were fixed and imaged after t = 5, 10, 20, 40, or 60 min at 35 °C. Single channels for RNA (cyan), DNA (green), TFAM (red), and POLRMT (grayscale) are shown; n = 4 experimental replicates. (Scale bar, 1 µm.) Intensity of green channel was adjusted for visibility due to decrease in DAPI signal with time; all other channels are set at the same contrast settings (see SI Appendix, Fig. S4A for intensity quantification). (B) Quantification of normalized intensity profiles of each component in the condensate, where r = 0 is the center of the condensate, and r = 1 is the normalized perimeter, for each channel in A. Shading indicates the time point corresponding to the average line profile, where darker colors are early time points, and lighter colors are late time points; n = 4 experimental replicates, and error bars = SEM. (C) (Top) Organization of condensates after addition of ex-RNA (SI Appendix) to nonreacting droplets at t = 0 (RNA added before all other proteins/DNA), t = 5 min (RNA added after condensates assembled for 5 min at 35 °C), or t = 30 min (RNA added after condensates assembled for 30 min at 35 °C). Buffer was the same as that used in the negative control (8 mM UTP). Condensates were fixed onto coverslips after 1 h of incubation at 35 °C; n = 3 experimental replicates. (Scale bar, 0.5 µm.) Bottom contains schematic illustration indicating localization (peripheral or internal) of each component: RNA (cyan), DNA (green), TFAM (red), and POLRMT (grayscale). (D) FRAP recovery for transcribing droplets (NTPs, each 2 mM) for 7× and 5% PEG conditions. Inset shows condensates prebleach, bleach, and 9 min postbleach. Dashed circle represents region that was bleached; n = 9 droplets, and error bars = SEM. (Scale bar, 1 µm.)
The observed reorganization of transcription components during the reaction suggests that the presence of RNA, and its increase in concentration over time, leads to structural changes of condensates. We performed an order-of-addition experiment to test the role of the newly synthesized RNA in determining condensate structure: We added exogenous RNA (ex-RNA)—comparable in sequence and length—to the mt-components at various time points in the presence of only UTP. The goal was to mimic interactions that arise from the presence of RNA despite the absence of transcriptional activity. Addition of ex-RNA at the beginning of mixing resulted in condensates with a layered structure, where ex-RNA, TFAM and POLRMT were in the interior, surrounded by a shell of DNA (Fig. 3C). In contrast, addition of ex-RNA after the DNA and protein components had mixed and had time to form condensates (t = 5, 30 min) resulted in condensates that had the reverse layered organization: DNA, TFAM, and POLRMT were in the interior, surrounded by a peripheral shell of ex-RNA (Fig. 3C). The peripheral localization of ex-RNA suggests that it coats pre-formed protein–DNA-rich condensates.
We found that, across the conditions we investigated, the proteins TFAM and POLRMT tended to partition with ex-RNA, suggesting that protein–RNA interactions are energetically favorable. However, in all cases, there was little colocalization of DNA with RNA, implying that DNA and RNA repel each other and that DNA–RNA interactions are energetically unfavorable. Indeed, combining only DNA and RNA yielded condensates with layered organization, where DNA was internal and surrounded by a shell of RNA, suggesting inherent immiscibility between DNA and RNA (SI Appendix, Fig. S4 E–G). Moreover, RNA is prone to forming irregular structures on its own, suggesting that RNA tends to self-associate in arrested structures, as has been demonstrated previously (34). The ability of proteins to alter the organization of nucleic acids within condensates supports the idea that proteins can help mitigate the immiscibility between DNA and RNA in transcriptional condensates (SI Appendix, Fig. S4 E–G).
Our observations, showing that localization of components within condensates depends on the order in which different components are added, suggest that active transcription contributes to shaping condensate structure. The distinct morphologies of condensates depend on when RNA is added, thereby indicating that active mt-transcriptional vesicular condensates represent nonequilibrium structures.
Next, we tested whether these structures were dynamically arrested by performing fluorescence recovery after photobleaching (FRAP). We used FRAP to measure the mobility of actively transcribed RNA molecules by detection of fluorescently labeled nucleotides (fluorescein-12-UTP). After ∼30 min of reaction, minimal recovery occurred over the course of ∼15 min (Fig. 3D). As a control, free nucleotides in a transcription-incompetent condensate rapidly exchanged, resulting in only limited bleach depth (SI Appendix, Fig. S4 B–D). Taken together with how organization depends on the order of addition of macromolecular components, the slow internal dynamics of newly synthesized RNA molecules led us to conclude that vesicular structures, previously shown to be non-equilibrium structures (38), are, in fact, dynamically arrested condensates.
Organization of Mitochondrial Nucleoids Is Altered upon Depletion of Core Transcription Components In Vivo.
To further probe relationships between mesoscale structures and functions of mt-condensates in vivo, we depleted key mt transcription components and assessed their effects on mt-nucleoid organization.
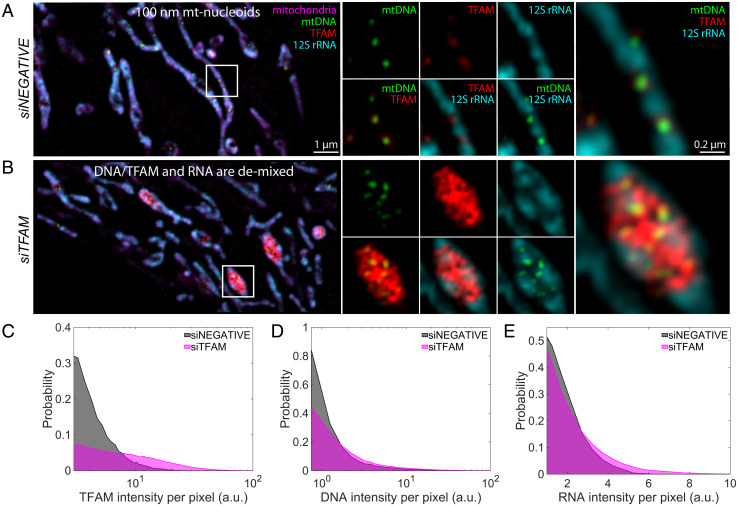
Using conditions of partial knockdown by RNA interference, we find that reduced levels of TFAM lead to a significant reorganization of mt-nucleoids (SI Appendix, Figs. S4B, S5, and S6), as previously observed in TFAM heterozygous knockout mice (39). After small interfering RNA (siRNA) treatment, we found that the number of mt-nucleoids per cell, based on staining for TFAM, was reduced significantly (SI Appendix, Fig. S5). In line with prior observations (39), dramatically enlarged mt-nucleoids were observed in HeLa cells that were partially depleted of TFAM (Fig. 4B). Interestingly, these remaining mt-nucleoids resembled the heterogeneous condensates observed in vitro (23). These clustered mt-nucleoids allowed us to better resolve the spatial organization of the mitochondrial (ribo)nucleoprotein complexes (Fig. 4B): Mitochondrial 12S rRNA localized peripherally, demixed from TFAM and mtDNA (Fig. 4B and SI Appendix, Fig. S6), similar to in vitro condensates that were formed when ex-RNA was added at later time points (Fig. 3C), and supporting the conclusion that mt-nucleoids and mt-RNA exist as spatially distinct phases in live mitochondria (23, 27).
Fig. 4.
Depletion of mt-transcription components in vivo. (A and B) SIM images of mt components after 72 h of siRNA treatment: siNEGATIVE (A) and siTFAM (B). Left are the merged images of the zoomed-out version of the mitochondrial network, where mitochondria are in magenta (MitoTracker Red), mtDNA is in green (anti-DNA), TFAM is in red (anti-TFAM), and 12S rRNA is in cyan (RNA FISH). (Scale bar, 1 µm.) White box indicates region of interest (ROI). Middle are signal or two-channel overlays of the ROI. Right are the three-channel overlays for the ROI. (Scale bar, 0.2 µm.) Intensities across each channel are matched across siRNA conditions; n = 4 independent experimental replicates. (C) Probability distribution of TFAM pixel intensity within a segmented mt-nucleoid, where gray denotes siNEGATIVE, and magenta denotes siTFAM. (D) Probability distribution of DNA pixel intensity within a segmented nucleoid. (E) RNA pixel intensity surrounding mt-nucleoids. Data represent n = 4 independent experimental replicates that were pooled together.
We further noticed altered phase behavior upon perturbation of other mt-nucleoid components involved in transcription (SI Appendix, Fig. S5). The mt-nucleoid associated protein MTERF2 (mt-transcription termination factor 2) is an abundant mt-nucleoid protein, present at ∼60:1 copies relative to mtDNA in vivo (40). Partial knockdown of MTERF2 also led to significantly reduced cell number and altered mt-nucleoid number (SI Appendix, Fig. S5), associated with a pronounced population of swollen mitochondria. These mitochondria corresponded to accumulation of RNA puncta alongside the swollen membrane (SI Appendix, Fig. S6A). TFAM accumulated in these mitochondria and appeared to wet the inner surface of 12S rRNA foci. However, mtDNA remained organized as ∼100-nm puncta that were surrounded by TFAM and frequently positioned adjacent to bright puncta of 12S rRNA (SI Appendix, Fig. S6), which further supports the idea of coexisting (ribo)nucleoprotein phases. Additionally, depletion of mtDNA achieved using a mitochondrially targeted endonuclease (41) led to complete dissolution of mt-nucleoids, including TFAM, and reduction of 12S rRNA signal (SI Appendix, Fig. S6). The dissolution of DNA- and RNA-rich condensates in the mt-matrix upon mtDNA depletion suggests that mtDNA is involved in nucleating mt-transcriptional condensates in live cells.
Computational Simulation of Transcription-Mediated Reorganization.
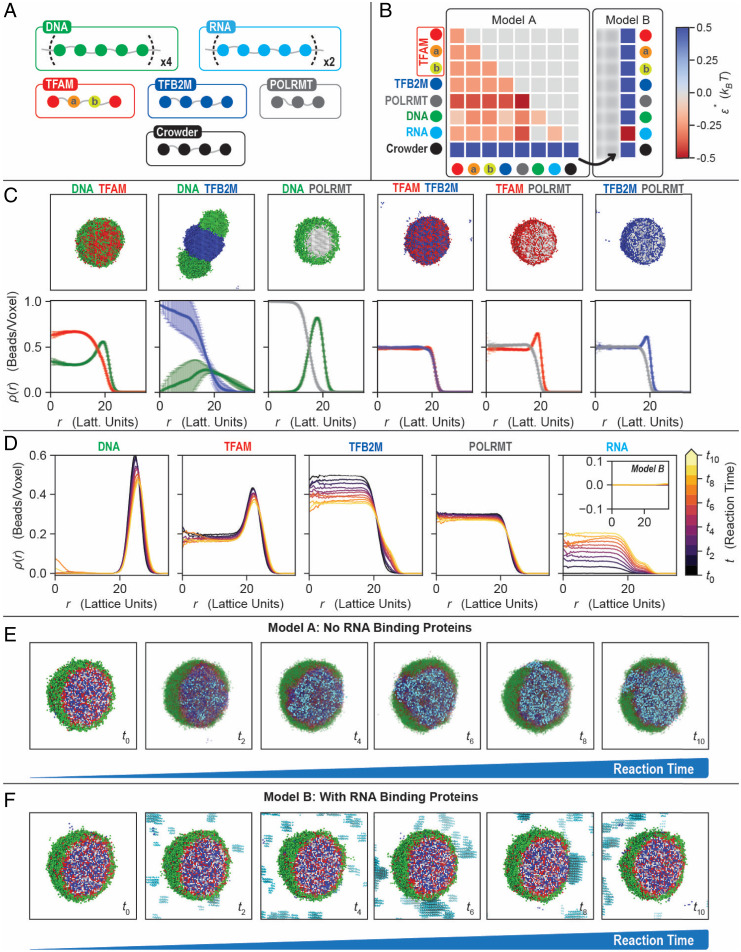
To interpret the disparate organizations found in vitro and in vivo, we used computational modeling to recapitulate the effect of RNA production on phase behavior of mt-transcriptional condensates. Using the simulation engine Lattice Simulation Engine for Sticker and Spacer Interactions (LaSSI) (42), we performed Monte Carlo simulations of coarse-grained (CG) models of binary and higher-order mixtures to probe the effects of RNA production on equilibrium condensates (Fig. 5). To preserve the overall length scales and interaction hierarchies of the macromolecules involved in mt-transcriptional condensates in the simulations, DNA molecules were modeled as chains of 20 beads, RNA as chains of 10 beads, TFAM and TFB2M as chains of 4 beads, POLRMT as chains of 3 beads, and crowders as chains of 4 beads (Fig. 5A).
Fig. 5.
Computational modeling of mt-transcriptional condensates. (A) CG model for the mt-transcriptional components. All beads are connected by implicit linkers of two-lattice sites. DNA and RNA are modeled as chains of 20 and 10 beads, respectively. TFAM is modeled as four beads (‘X–a–b–X’), where the central two beads, a and b, interact more favorably with DNA. To account for TFAM’s weak dimerization upon DNA binding, bead b has an additional interaction energy (−2 kBT) for another b bead, yielding a local anisotropic interaction. TFB2M, POLRMT, and crowder are chains with four, three, and four beads, respectively. (B) Interaction matrices for the two models considered. Model A lacks RNA-binding proteins and captures the organization of the condensates seen in vitro. The crowder has repulsive interactions with every species, including itself. The DNA and RNA have no interactions, while the rest of the molecules have favorable interactions. Model B mimics the effective inclusion of RNA-binding proteins by making the RNA–crowder interactions favorable, modeling an effective RNA binding protein as occurs in vivo. (C) Representative snapshots and density profiles for the binary mixtures shown in Fig. 1C. For clarity, the crowder is not shown. (D) Density profiles of each component with different RNA amounts for model A. Inset corresponds to RNA profiles for model B and shows no RNA accumulation inside the condensate when RNA binding proteins are present. (E and F) Representative snapshots of the condensates with increasing RNA for models A (E) and B (F). A higher amount of RNA in the system corresponds to a later time in the transcription reactions. For clarity, the crowder is not shown, and non-RNA components are made transparent. The RNA is evenly distributed inside the condensates and continues to be accumulated as the reaction continues (E). With a suitable RNA binding protein, the RNA can be prevented from going inside the condensates, as observed in vivo (F).
We first parameterized the contact energies between pairs of molecules by reproducing experimentally observed morphologies of single and binary mixtures (Fig. 5C; compare with Fig. 1 B and C) (23). The experimentally measured colocalization and condensate homogeneity were used as proxies for stronger heterotypic interactions, while spatial inhomogeneities within the condensates were used as proxies for stronger homotypic interactions. We included an additional anisotropic interaction for TFAM to account for its weak dimerization upon DNA binding (Fig. 5B) (43). Simulations of the binary mixtures of transcriptional condensate components generated morphologies (Fig. 5C and SI Appendix, Fig. S7A) with spatial organizations, quantified in terms of radial density profiles, that recapitulate the experimental results (Fig. 1C). For example, TFAM–DNA and POLRMT–DNA formed multiphase droplets in the simulations, while TFB2M and DNA behaved as distinct coexisting phases, and pairs of proteins tended to form well-mixed droplets, as observed in vitro (Figs. 1C and 5C).
Extending this modeling approach to the quaternary condensates, we find the formation of heterogenous droplets that are consistent with our experimental results. This is best illustrated by comparing results summarized in Fig. 5E, t = 0, with results shown in Fig. 1D. Simulations show the formation of layered droplets with DNA being localized almost exclusively to the periphery (Fig. 5D and SI Appendix, Fig. S7B). The three proteins tend to be enriched in the interior of the droplet, enveloped by a shell of DNA. This multiphase organization is consistent with the heterogeneity and colocalization of components in the four-component dynamically arrested droplets observed in vitro (Fig. 1D).
Next, we explored how transcription affects the morphologies of condensates in silico. We did not explicitly model the active process of creating RNA transcripts; instead, we performed a series of independent equilibrium simulations, each with a distinct amount of RNA. These simulations thus allowed us to query the equilibrium morphologies that would be achieved using our model for different amounts of RNA, corresponding to different time points of the reaction. We then compared these morphologies with those observed in vitro. The model we used is based on pairwise interaction energies derived from reconstitutions with fewer components. As summarized next, the equilibrium morphologies we observed in the simulations are not the vesicular structures that were observed in vitro.
The details are as follows: In each simulation, we query the arrangement of the system for fixed amounts of RNA, corresponding to a particular time point of the reaction. For example, at the initial time point t0, all reactants are fully mixed, and no RNA is present, implying that , while, at later times, ti > 0, the system contains increasing amounts of RNA, implying that . Here, represents the total concentration or amount of RNA in the system at a given time, . With increasing amounts of RNA in our simulations, or our proxy for time, we observe three features: First, the fixed amounts of RNA that are included in each simulation are incorporated exclusively in the interior of existing droplets (the fifth panel in Fig. 5D and SI Appendix, Fig. S7B); second, as shown in the third panel of Fig. 5D, there is an appreciable reduction of TFB2M in the interior, and a concomitant increase of TFB2M near the periphery; and third, DNA persists at the periphery, while TFAM and POLRMT remain in the interior, largely unaffected by different RNA levels (Fig. 5 D and E and SI Appendix, Fig. S7B).
The computational results summarized in Fig. 5 D and E do not show vesicular structures observed in vitro. However, they capture the poor mixing of DNA and RNA. Taken together with the observations in vitro that RNA localization depends on the order of addition of RNA into the system (Figs. 2 and 3C), we take the discrepancy between the equilibrium morphologies observed in the simulations and the vesicular structures observed in vitro as evidence that the vesicles represent dynamically arrested structures.
Importantly, vesicular organization is not observed in vivo either. This is likely because newly synthesized mt-RNA molecules are bound by RNA-binding proteins and processed in mt-RNA granules, which are phase-separated structures that are often located adjacent to mt-nucleoids (27, 44). To test for this possibility, we incorporated an additional favorable interaction between the crowder and RNA in our simulations, to mimic the presence of association of RNA-binding proteins to the newly synthesized RNA under steady-state conditions in vivo (Fig. 5B). Under these conditions, RNA no longer associates with the DNA- and protein-rich droplet but condenses separately in the bulk (Fig. 5 D, Inset and F and SI Appendix, Fig. S7C). The removal or exclusion of RNA from the mt-transcriptional condensate by an effective RNA binding protein in our simulations thus mirrors their distribution in vivo (Fig. 4 and SI Appendix, Figs. S5 and S6).
Discussion
We report here the in vitro reconstitution of transcriptionally active, multiphasic condensates using the human mt-transcription machinery as a model system. We show that, when compared with bulk reactions in solution, the transcriptional rate is reduced under condensate-forming conditions. We also observe that the production of RNA alters the spatial organization of condensates, thus providing direct evidence for a dynamic interplay between the structure of condensates and the functional activities they harbor.
A key observation in our study is the finding of reduced transcriptional output when phase separation occurs, compared with bulk solution conditions. Decreased rates of transcription within condensates are likely due to the slower internal dynamics associated with arrested phases, which represent metastable, nonequilibrium structures, wherein one or more macromolecules are immobile because they are a part of highly cross-linked networks. The slow dynamics associated with an arrested environment suggest that mt-transcriptional condensates experience transport-limited kinetics. In support, our rough estimates suggest that less than one round of transcription occurs under condensate-forming conditions, indicating that not all DNA templates are actively transcribed. This behavior is in line with the situation in live cells, where only a minority of <5% of the mt-nucleoids are actively transcribing at any given time (45, 46).
Comparisons of in vitro and in vivo mt-transcription and phase behavior are complicated by differences in the stoichiometry and DNA templates required to reconstitute efficient mt-transcription in vitro. For example, in vivo TFAM is present in roughly 1,000 copies per copy of mtDNA (25), or, equivalently, 0.06 molecules TFAM per base pair of DNA and in significant stoichiometric excess of TFB2M and POLRMT (47), whereas our in vitro reconstitution system requires use lower TFAM/DNA ratios (∼0.02 molecules TFAM per base pair of DNA; SI Appendix) and roughly equimolar ratios of proteins to generate detectable RNA product (28, 48). We find that increasing TFAM levels closer to physiological levels significantly reduced the reaction in vitro (SI Appendix, Fig. S2) (29). Moreover, the mt-genome is circular and 16 kb in size; conversely, in vitro transcription assays require short linear DNA templates of ∼0.5 kb, since longer transcripts cannot be efficiently generated in vitro without the presence of other protein factors (30). The RNA molecules synthesized in vivo are long transcripts that are quickly bound and modified by RNA-binding proteins (49), while our in vitro system generates short (∼300 nt) RNAs in the absence of any such RNA-modifying proteins.
Importantly, we find that the mesoscale structures of mt-transcriptional condensates are affected by their activity. Production of RNA in the otherwise DNA- and protein-rich mt-transcriptional condensate leads to nonequilibrium changes in condensate organization, reflected by the emergence of vesicle-like morphologies. Intriguingly, similar vesicle morphologies have been observed for simple in vitro RNA–protein systems (37), in vivo liquid spherical shells of the DNA- and RNA-binding protein TDP-43 (50), and in vitro liquid spherical shells of DNA and poly-L-Lysine (51). RNA has also been shown to form a corona on the surface of engineered condensates, directly impeding their coarsening (52).
The vesicles observed due to RNA generation in mt-transcriptional condensates in vitro differ from that of the canonical mt-nucleoid organization in mitochondria in vivo (25, 26, 53), where mt-nucleoids remain as condensed, ∼100-nm droplet-like structures, and mt-RNA localizes to separate mt-RNA–processing granules (27). Our simulations, which are based on experimentally determined interaction parameters, suggest that, during transcription, mt-RNA is tethered to the mt-nucleoid as it is being transcribed by POLRMT and is effectively handed off from the mt-nucleoids to the more energetically favorable mt-RNA–processing granules, which are themselves condensates (27). This scenario is supported by our computational simulations which demonstrate that the presence of RNA-binding activities outside the condensate is sufficient to remove the accumulating RNA from the condensate, restoring morphologies concordant with the maintenance of local equilibrium. This feature, whereby the newly transcribed RNA molecules are sequestered in RNA granules juxtaposed to non-vesicular mt-nucleoids, appears to help with maintaining local thermodynamic equilibrium, a phenomenon recently observed for P granules (54). It is worth emphasizing that our simulations do not directly model active transcription. Instead, we perform a series of separate simulations, each with a predetermined amount of RNA. This approach allows us to model the mesoscale structures of condensates that form at equilibrium by minimizing the global free energy in the presence of specific amounts of RNA (Fig. 5). Other approaches have been introduced in the literature to model phase equilibria in the presence of chemical reactions of the phase-separating components (55, 56).
There are growing numbers of reports of transcription occurring within condensates in vivo. RNA Pol I transcribes ribosomal RNA (rRNA) in the multiphase nucleolus (7, 57), and RNA Pol II has been reported to produce messenger RNA in transcriptional condensates (58, 59). By extension, POLRMT generates long, polycistronic mt-RNA in the droplet-like mt-nucleoid (23, 27, 30). While condensate formation is not an absolute requirement for transcription, since soluble conditions can support in vitro transcription of mt-RNA (60), there is growing evidence that condensate formation may offer advantages for regulation of transcription in vivo. First, the condensed phase enriches for specific reactants, which may contribute to mass action effects (22, 61). In addition, increased dwell times of proteins associated with the condensate microenvironment may also be conducive for assembly of reacting complexes (62). In support of this hypothesis, an early FRAP study showed that the kinetics of multiple RNA Pol I components could only be explained by inclusion of a slow kinetic component prior to binding of the polymerase subunits to the promoter, possibly reflecting slowed diffusion in a nucleolar condensate (63).
The eukaryotic RNA Pol II transcription machinery has long been known to be organized into transcription factories in human nuclei (64), where nonspecific interactions stabilize the clusters (65)—many of which exhibit condensate-like properties (18, 58). Initially, Pol II and its transcription machinery assemble into dynamic, droplet-like structures, particularly around superenhancers, and, as RNA is generated, it demixes from the DNA-rich phase (66, 67), reminiscent of a microphase separation process (68). Interestingly, for RNA Pol II condensates, rapid, local RNA production has been shown to result in complete dissolution of the condensate, underscoring a feedback mechanism between phase behavior and RNA production, as also observed here for mt-transcription (69). Conversely, transcriptional condensates are not always associated with increased activity: During embryonic development, such hubs may act as repressors (70–73). Similar condensation events of transcription have also been seen in the bacterial nucleoid, where RNAP clusters with transcription factors at specific sites, particularly rDNA, in the bacterial genome (74, 75). The commonality that begins to emerge from these observations is that RNA is not retained in DNA-rich phases, pointing to an intrinsic energetic barrier for their mixing.
Our in vitro studies describe the formation of arrested phases of mt-transcriptional components, suggesting that equilibrium morphologies observed in silico can become dynamically inaccessible because of energetic barriers. These barriers appear to be negotiated to alleviate arrest and metastability in vivo by achieving local thermodynamic equilibria upon transcription through the presence of RNA-associating proteins—a feature that is readily reproduced in simulations. The resulting multiway interplay of phase separation, active transcription, and sinks for new transcripts likely gives rise to local thermodynamic equilibrium (54), that is, the maintenance of local detailed balance. It is attractive to postulate that local equilibria are being actively maintained in other cellular processes, where there is coupling of phase separation and biochemical reactions.
Materials and Methods
All details and protocols are described in full in SI Appendix. Briefly, core components of the human mt-transcription machinery (TFAM, TFB2M, and POLRMT) were purified using bacterial expression, and short template DNA (∼0.5 kb) containing the mitochondrial light-strand promoter was amplified using PCR. Phase separation assays were imaged using Structured Illumination Microscopy or Laser-Scanning Confocal Microscopy. In vitro transcription reactions were performed in the absence and presence of the crowder PEG to capture soluble and condensed states, respectively. In vivo organization of mt-transcription components was observed in HeLa cells upon siRNA and mtDNA depletion. Simulations on the phase behavior of mt-transcriptional condensates were performed using a LaSSI simulation engine.
Supplementary Material
Acknowledgments
We are grateful for discussions with T.M. group members; we thank J. Jones and M. Taylor for protein expression and purification of TFAM at the NIH/National Cancer Institute (NCI)/Center for Cancer Research (CCR) Protein Production Core; and we thank T. Karpova and D. Ball for assistance with Structured Illumination Microscopy and laser-scanning confocal microscopy as part of the NIH/NCI/CCR Optical Imaging Core. F.D. and R.V.P. are also grateful to K. M. Ruff for many helpful and clarifying discussions. Research in the T.M. laboratory is supported by funding from the Intramural Research Program of the NIH, NCI, and CCR (project 1-ZIA-BC010309); M.F. was supported by a Postdoctoral Research Associate Training fellowship from the National Institute of General Medical Sciences (grant 1Fi2GM128585-01). D.T. is supported by a grant from NIH ( R35 GM131832). This work in the R.V.P. laboratory was supported by grants from the NIH (R01NS121114 and 5R01NS056114), the Air Force Office of Scientific Research (FA9550-20-1-0241), and the St. Jude Collaborative Research Consortium on the Biology of Membraneless Organelles.
Footnotes
Competing interest statement: R.V.P. is a member of the Scientific Advisory Board of Dewpoint Therapeutics Inc. The work here was not supported or influenced by this affiliation.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2207303119/-/DCSupplemental.
Data, Materials, and Software Availability
All data supporting the findings are available in the main text and supporting information. Images and image analysis code can be found in the GitHub repository: https://github.com/fericm/mito-transcriptional-condensates (76).
References
- 1.Banani S. F., Lee H. O., Hyman A. A., Rosen M. K., Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shin Y., Brangwynne C. P., Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Sabari B. R., Dall’Agnese A., Young R. A., Biomolecular condensates in the nucleus. Trends Biochem. Sci. 45, 961–977 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boeynaems S., et al. , Protein phase separation: A new phase in cell biology. Trends Cell Biol. 28, 420–435 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brangwynne C. P., et al. , Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324, 1729–1732 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Sanders D. W., et al. , Competing protein-RNA interaction networks control multiphase intracellular organization. Cell 181, 306–324.e328 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feric M., et al. , Coexisting liquid phases underlie nucleolar subcompartments. Cell 165, 1686–1697 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greig J. A., et al. , Arginine-enriched mixed-charge domains provide cohesion for nuclear speckle condensation. Mol. Cell 77, 1237–1250.e1234 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hnisz D., Shrinivas K., Young R. A., Chakraborty A. K., Sharp P. A., A phase separation model for transcriptional control. Cell 169, 13–23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feric M., Misteli T., Phase separation in genome organization across evolution. Trends Cell Biol. 31, 671–685 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H., Han M., Qi L. S., Engineering 3D genome organization. Nat. Rev. Genet. 22, 343–360 (2021). [DOI] [PubMed] [Google Scholar]
- 12.Gibson B. A., et al. , Organization of chromatin by intrinsic and regulated phase separation. Cell 179, 470–484.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strom A. R., et al. , Phase separation drives heterochromatin domain formation. Nature 547, 241–245 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larson A. G., et al. , Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 547, 236–240 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Falk M., et al. , Heterochromatin drives compartmentalization of inverted and conventional nuclei. Nature 570, 395–399 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erdel F., Rippe K., Formation of chromatin subcompartments by phase separation. Biophys. J. 114, 2262–2270 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strickfaden H., et al. , Condensed chromatin behaves like a solid on the mesoscale in vitro and in living cells. Cell 183, 1772–1784.e1713 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Sabari B. R., et al. , Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boija A., et al. , Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell 175, 1842–1855.e1816 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chong S., et al. , Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science 361, eaar2555 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bremer A., et al. , Deciphering how naturally occurring sequence features impact the phase behaviours of disordered prion-like domains. Nat. Chem. 14, 196–207 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lyon A. S., Peeples W. B., Rosen M. K., A framework for understanding the functions of biomolecular condensates across scales. Nat. Rev. Mol. Cell Biol. 22, 215–235 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feric M., et al. , Self-assembly of multi-component mitochondrial nucleoids via phase separation. EMBO J. 40, e107165 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen X. J., Butow R. A., The organization and inheritance of the mitochondrial genome. Nat. Rev. Genet. 6, 815–825 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Kukat C., et al. , Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA. Proc. Natl. Acad. Sci. U.S.A. 108, 13534–13539 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garrido N., et al. , Composition and dynamics of human mitochondrial nucleoids. Mol. Biol. Cell 14, 1583–1596 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rey T., et al. , Mitochondrial RNA granules are fluid condensates positioned by membrane dynamics. Nat. Cell Biol. 22, 1180–1186 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hillen H. S., Morozov Y. I., Sarfallah A., Temiakov D., Cramer P., Structural basis of mitochondrial transcription initiation. Cell 171, 1072–1081.e10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Litonin D., et al. , Human mitochondrial transcription revisited: Only TFAM and TFB2M are required for transcription of the mitochondrial genes in vitro. J. Biol. Chem. 285, 18129–18133 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hillen H. S., Temiakov D., Cramer P., Structural basis of mitochondrial transcription. Nat. Struct. Mol. Biol. 25, 754–765 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rubio-Cosials A., et al. , Human mitochondrial transcription factor A induces a U-turn structure in the light strand promoter. Nat. Struct. Mol. Biol. 18, 1281–1289 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Ringel R., et al. , Structure of human mitochondrial RNA polymerase. Nature 478, 269–273 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Sciortino F., Bansil R., Stanley H. E., Alstrom P., Interference of phase separation and gelation: A zeroth-order kinetic model. Phys. Rev. E Stat. Phys. Plasmas Fluids Relat. Interdiscip. Topics 47, 4615–4618 (1993). [DOI] [PubMed] [Google Scholar]
- 34.Boeynaems S., et al. , Spontaneous driving forces give rise to protein-RNA condensates with coexisting phases and complex material properties. Proc. Natl. Acad. Sci. U.S.A. 116, 7889–7898 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seim I., et al. , Dilute phase oligomerization can oppose phase separation and modulate material properties of a ribonucleoprotein condensate. Proc. Natl. Acad. Sci. U.S.A. 119, e2120799119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sarfallah A., Temiakov D., “In vitro reconstitution of human mitochondrial transcription” inMitochondrial Gene Expression, Minczuk M., Rorbach J., Eds. (Springer, 2021), pp. 35–41. [DOI] [PubMed] [Google Scholar]
- 37.Alshareedah I., Moosa M. M., Raju M., Potoyan D. A., Banerjee P. R., Phase transition of RNA–protein complexes into ordered hollow condensates. Proc. Natl. Acad. Sci. U.S.A. 117, 15650–15658 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Banerjee P. R., Milin A. N., Moosa M. M., Onuchic P. L., Deniz A. A., Reentrant phase transition drives dynamic substructure formation in ribonucleoprotein droplets. Angew. Chem. Int. Ed. Engl. 56, 11354–11359 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.West A. P., et al. , Mitochondrial DNA stress primes the antiviral innate immune response. Nature 520, 553–557 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pellegrini M., et al. , MTERF2 is a nucleoid component in mammalian mitochondria. Biochim. Biophys. Acta Bioenergetics 1787, 296–302 (2009). [DOI] [PubMed] [Google Scholar]
- 41.Spadafora D., Kozhukhar N., Chouljenko V. N., Kousoulas K. G., Alexeyev M. F., Methods for efficient elimination of mitochondrial DNA from cultured cells. PLoS One 11, e0154684 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choi J. M., Dar F., Pappu R. V., LASSI: A lattice model for simulating phase transitions of multivalent proteins. PLOS Comput. Biol. 15, e1007028 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ngo H. B., Lovely G. A., Phillips R., Chan D. C., Distinct structural features of TFAM drive mitochondrial DNA packaging versus transcriptional activation. Nat. Commun. 5, 3077 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jourdain A. A., Boehm E., Maundrell K., Martinou J.-C., Mitochondrial RNA granules: Compartmentalizing mitochondrial gene expression. J. Cell Biol. 212, 611–614 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tu Y.-T., Barrientos A., The human mitochondrial DEAD-box protein DDX28 resides in RNA granules and functions in mitoribosome assembly. Cell Rep. 10, 854–864 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brüser C., Keller-Findeisen J., Jakobs S., The TFAM-to-mtDNA ratio defines inner-cellular nucleoid populations with distinct activity levels. Cell Rep. 37, 110000 (2021). [DOI] [PubMed] [Google Scholar]
- 47.Cotney J., Wang Z., Shadel G. S., Relative abundance of the human mitochondrial transcription system and distinct roles for h-mtTFB1 and h-mtTFB2 in mitochondrial biogenesis and gene expression. Nucleic Acids Res. 35, 4042–4054 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sologub M., Litonin D., Anikin M., Mustaev A., Temiakov D., TFB2 is a transient component of the catalytic site of the human mitochondrial RNA polymerase. Cell 139, 934–944 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xavier V. J., Martinou J. C., RNA granules in the mitochondria and their organization under mitochondrial stresses. Int. J. Mol. Sci. 22, 9502 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu H., et al. , HSP70 chaperones RNA-free TDP-43 into anisotropic intranuclear liquid spherical shells. Science 371, eabb4309 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shakya A., King J. T., DNA local-flexibility-dependent assembly of phase-separated liquid droplets. Biophys. J. 115, 1840–1847 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cochard A., et al. , RNA at the surface of phase-separated condensates impacts their size and number. Biophys. J. 121, 1675–1690 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Iborra F. J., Kimura H., Cook P. R., The functional organization of mitochondrial genomes in human cells. BMC Biol. 2, 9 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fritsch A. W., et al. , Local thermodynamics govern formation and dissolution of Caenorhabditis elegans P granule condensates. Proc. Natl. Acad. Sci. U.S.A. 118, e2102772118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zwicker D., Seyboldt R., Weber C. A., Hyman A. A., Jülicher F., Growth and division of active droplets provides a model for protocells. Nat. Phys. 13, 408–413 (2017). [Google Scholar]
- 56.Kirschbaum J., Zwicker D., Controlling biomolecular condensates via chemical reactions. J. R. Soc. Interface 18, 20210255 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lafontaine D. L. J., Riback J. A., Bascetin R., Brangwynne C. P., The nucleolus as a multiphase liquid condensate. Nat. Rev. Mol. Cell Biol. 22, 165–182 (2021). [DOI] [PubMed] [Google Scholar]
- 58.Cho W.-K., et al. , Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361, 412–415 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Henninger J. E., et al. , RNA-mediated feedback control of transcriptional condensates. Cell 184, 207–225.e24 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Beckert B., Masquida B., “Synthesis of RNA by in vitro transcription” in RNA: Methods and Protocols, Nielsen H., Ed. (Springer, 2011), pp. 29–41. [DOI] [PubMed] [Google Scholar]
- 61.Peeples W., Rosen M. K., Mechanistic dissection of increased enzymatic rate in a phase-separated compartment. Nat. Chem. Biol. 17, 693–702 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Case L. B., Zhang X., Ditlev J. A., Rosen M. K., Stoichiometry controls activity of phase-separated clusters of actin signaling proteins. Science 363, 1093–1097 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dundr M., et al. , A kinetic framework for a mammalian RNA polymerase in vivo. Science 298, 1623–1626 (2002). [DOI] [PubMed] [Google Scholar]
- 64.Iborra F. J., Pombo A., Jackson D. A., Cook P. R., Active RNA polymerases are localized within discrete transcription "factories’ in human nuclei. J. Cell Sci. 109, 1427–1436 (1996). [DOI] [PubMed] [Google Scholar]
- 65.Brackley C. A., Taylor S., Papantonis A., Cook P. R., Marenduzzo D., Nonspecific bridging-induced attraction drives clustering of DNA-binding proteins and genome organization. Proc. Natl. Acad. Sci. U.S.A. 110, E3605–E3611 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pancholi A., et al. , RNA polymerase II clusters form in line with surface condensation on regulatory chromatin. Mol. Syst. Biol. 17, e10272 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guo Y. E., et al. , Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature 572, 543–548 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hilbert L., et al. , Transcription organizes euchromatin via microphase separation. Nat. Commun. 12, 1360 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Henninger J. E., et al. , RNA-mediated feedback control of transcriptional condensates. Cell 184, 207–225.e224 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ghavi-Helm Y., et al. , Enhancer loops appear stable during development and are associated with paused polymerase. Nature 512, 96–100 (2014). [DOI] [PubMed] [Google Scholar]
- 71.Espinola S. M., et al. , Cis-regulatory chromatin loops arise before TADs and gene activation, and are independent of cell fate during early Drosophila development. Nat. Genet. 53, 477–486 (2021). [DOI] [PubMed] [Google Scholar]
- 72.Treen N., Shimobayashi S. F., Eeftens J., Brangwynne C. P., Levine M., Properties of repression condensates in living Ciona embryos. Nat. Commun. 12, 1561 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Trojanowski J., et al. , Transcription activation is enhanced by multivalent interactions independent of phase separation. Mol. Cell 82, 1878–1893.e1810 (2022). [DOI] [PubMed] [Google Scholar]
- 74.Ladouceur A.-M., et al. , Clusters of bacterial RNA polymerase are biomolecular condensates that assemble through liquid–liquid phase separation. Proc. Natl. Acad. Sci. U.S.A. 117, 18540–18549 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mata Martin C., Sun Z., Zhou Y. N., Jin D. J., Extrachromosomal nucleolus-like compartmentalization by a plasmid-borne ribosomal RNA operon and its role in nucleoid compaction. Front. Microbiol. 9, 1115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Feric M., et al. , Mesoscale structure–function relationships in mitochondrial transcriptional condensates. GitHub. https://github.com/fericm/mito-transcriptional-condensates. Deposited 27 July 2022. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings are available in the main text and supporting information. Images and image analysis code can be found in the GitHub repository: https://github.com/fericm/mito-transcriptional-condensates (76).