Significance
Trophoblast cell–guided restructuring of the uterus is an essential event in the establishment of the hemochorial placenta. Establishment of a suitable animal model for investigating regulatory mechanisms in this critical developmental process is a key to better understanding the etiology of diseases of placentation, such as early pregnancy loss, preeclampsia, intrauterine growth restriction, and preterm birth. The rat exhibits deep trophoblast cell invasion, as seen in human hemochorial placentation. Similarities in the transcriptomes of rat and human invasive trophoblast cells led to the discovery of conserved candidate regulators of the invasive trophoblast cell lineage. This creates opportunities to test hypotheses underlying the pathophysiologic basis of trophoblast cell–guided uterine transformation and provides insights into the etiology of diseases of placentation.
Keywords: trophoblast, placentation, single cell genomic analysis, rat
Abstract
The hemochorial placentation site is characterized by a dynamic interplay between trophoblast cells and maternal cells. These cells cooperate to establish an interface required for nutrient delivery to promote fetal growth. In the human, trophoblast cells penetrate deep into the uterus. This is not a consistent feature of hemochorial placentation and has hindered the establishment of suitable animal models. The rat represents an intriguing model for investigating hemochorial placentation with deep trophoblast cell invasion. In this study, we used single-cell RNA sequencing to characterize the transcriptome of the invasive trophoblast cell lineage, as well as other cell populations within the rat uterine–placental interface during early (gestation day [gd] 15.5) and late (gd 19.5) stages of intrauterine trophoblast cell invasion. We identified a robust set of transcripts that define invasive trophoblast cells, as well as transcripts that distinguished endothelial, smooth muscle, natural killer, and macrophage cells. Invasive trophoblast, immune, and endothelial cell populations exhibited distinct spatial relationships within the uterine–placental interface. Furthermore, the maturation stage of invasive trophoblast cell development could be determined by assessing gestation stage–dependent changes in transcript expression. Finally, and most importantly, expression of a prominent subset of rat invasive trophoblast cell transcripts is conserved in the invasive extravillous trophoblast cell lineage of the human placenta. These findings provide foundational data to identify and interrogate key conserved regulatory mechanisms essential for the development and function of an important compartment within the hemochorial placentation site that is essential for a healthy pregnancy.
The uterine–placental interface is a dynamic site where trophoblast and uterine cells cooperate to support growth and maturation of the fetus. These tasks are accomplished by specialized trophoblast cells, which arise from a multilineage cell differentiation pathway (1). Among specialized trophoblast cells are those that acquire invasive properties and enter the uterine parenchyma where they facilitate the transformation of the uterine environment (2–5). In some primates and rodents, trophoblast cells penetrate the uterine vasculature and are directly bathed by maternal blood, a process referred to as hemochorial placentation (6, 7). Trophoblast cells entering the uterus are generically termed invasive trophoblast cells. In the human placenta, they are called extravillous trophoblast (EVT) cells. Invasive trophoblast cells migrate inside uterine blood vessels where they replace the endothelium (endovascular) and external to the vasculature where they move among uterine stromal cells (interstitial). Impairments in trophoblast-directed uterine transformation are linked to pregnancy-related diseases (2, 3, 8). However, there is a limited understanding of the regulatory mechanisms controlling the function of invasive trophoblast cells.
Interactions between invasive trophoblast cells and uterine cells are critical for the establishment of pregnancy and may be best investigated in vivo. Depth and extent of intrauterine trophoblast cell invasion show prominent species differences (9), affecting selection of suitable in vivo models. Human hemochorial placentation is characterized by deep intrauterine trophoblast cell invasion, a feature not shared with the mouse (6). In contrast to the mouse, the rat uterine–placental interface is characterized by deep intrauterine trophoblast cell invasion (9–11). This structural feature of the rat uterine–placental interface, and the availability of single-cell RNA sequencing (scRNA-seq) technology, make the capture of invasive trophoblast cells remarkably straightforward.
In this report, scRNA-seq was performed on the rat uterine–placental interface to profile cell populations within the structure and to identify conserved candidate regulators of invasive trophoblast cell lineage development.
Results
Identification of Cell Clusters within the Uterine–Placental Interface.
Intrauterine trophoblast cell invasion in the rat is first detected at midgestation in the form of endovascular invasive trophoblast cells lining arterioles that penetrate the mesometrial decidua (10). After gestation day (gd) 13.5, trophoblast cells exit the junctional zone of the placenta, penetrating beyond the decidua and deep into the mesometrial uterine parenchyma (10, 12). Both endovascular and interstitial invasive trophoblast cells are evident beginning on gd 14.5 (10, 13). The uterine–placental interface is the nodule of uterine tissue juxtaposed to the decidua and placenta and the destination of trophoblast cell invasion. This structure is retained in the uterus following removal of the placenta and adherent decidua and is easily dissected for further analysis (Fig. 1A) (14). The uterine–placental interface consists of an assortment of cell types, including invasive trophoblast, endothelial, immune, stromal, and smooth muscle cells. We performed scRNA-seq of the uterine–placental interface on gd 15.5 and 19.5, which reflect early and mature stages of intrauterine trophoblast cell invasion, respectively.
Fig. 1.
Single cell interrogation of the rat uterine–placental interface. (A) Schematic showing isolation of the rat uterine–placental interface from gd 15.5 or 19.5. Uterine tissue at the site of trophoblast invasion was peeled away from placental tissue and associated decidua. (B and C) Visualization of cell clusters from gd 15.5 and 19.5, respectively, plotted using UMAP. (D and E) Dot plots for gd 15.5 and 19.5 showing marker transcripts used to identify cell clusters. Dot size represents the average percentage of cells expressing the transcript. Dot colors correspond to cell types. For transcript expression level, see SI Appendix, Fig. S2 and Dataset S1. Abbreviations: UPI, uterine–placental interface; JZ, junctional zone; LZ, labyrinth zone.
Following sequencing and data quality control (SI Appendix, Fig. S1), 65,842 cells were present in gd 15.5 samples and 33,617 cells in gd 19.5 (SI Appendix, Fig. S2). Cell clusters were then defined based on their transcript profiles (Materials and Methods, Fig. 1 B and C, and Dataset S1). Endothelial, macrophage, natural killer (NK), invasive trophoblast, and smooth muscle cell clusters were identified using established biomarkers for the cell types (Fig. 1 D and E and SI Appendix, Fig. S2). Additional cell clusters were detected in the analysis, but their identities were not precisely determined (Dataset S1). These cell clusters broadly resembled fibroblasts, stromal cells, mesothelial cells, smooth muscle cells, or leukocytes. Invasive trophoblast cells were conspicuous in their epithelial character (e.g., expression of cytokeratins: Krt7, Krt8, and Krt18) and expression of members of the expanded prolactin (PRL) gene family (Prl7b1, Prl5a1, Prl4a1, Prl2a1, Prl5a2, Prl2c1, and Prl6a1) (Dataset S1). To date, expression of cytokeratin and a subset of PRL gene family members have formed the basis of tracking invasive trophoblast cells within the rat uterine–placental interface (10, 16–21).
Spatial Relationships of Invasive Trophoblast Cells and Uterine Cell Populations.
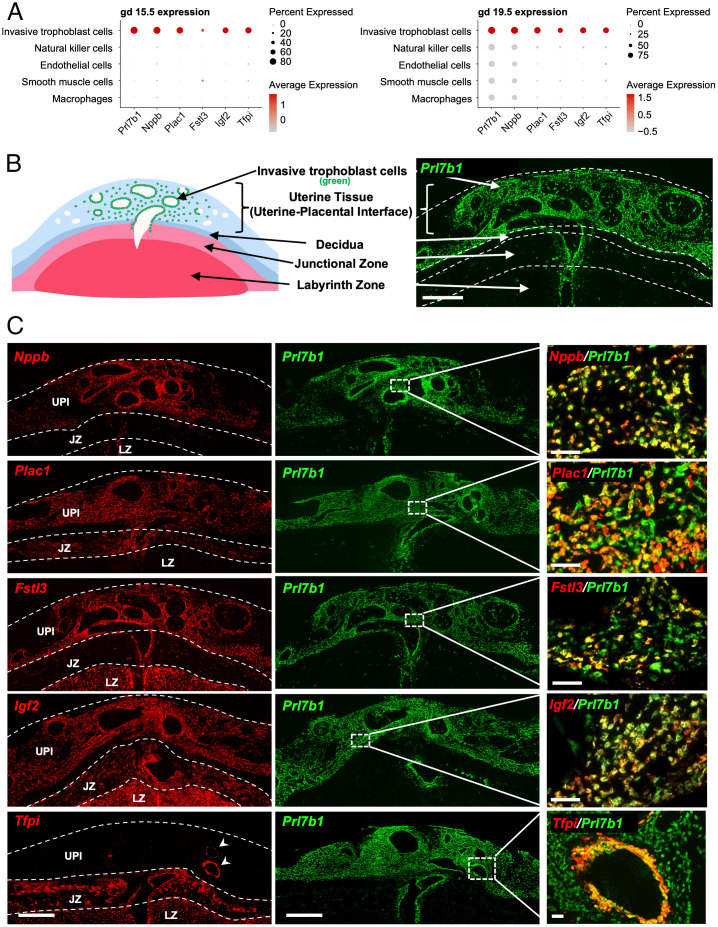
In situ hybridization was used to investigate spatial relationships of the uterine–placental interface cellular constituents at gd 15.5 and 19.5. Probes for identifying the cell types were selected based on the scRNA-seq cluster profiles where the selected genes had a high expression level in a large percentage of invasive trophoblast cells (Fig. 2A). Invasive trophoblast cells were readily identified by the expression of the epithelial cell marker, Krt8, and by expression of the invasive trophoblast cell marker, Prl7b1 (10, 16), which were colocalized to both endovascular and interstitial trophoblast cells within the gd 19.5 uterine–placental interface (Fig. 2B and SI Appendix, Fig S3). Distributions of other transcripts, enriched in the invasive trophoblast cell cluster, within the gd 19.5 rat placentation site were tracked by colocalization with Prl7b1 (Fig. 2C). Although we were not able to resolve subpopulations of invasive trophoblast cells through scRNA-seq analysis, we were able to define distinct expression profiles for specific transcripts enriched in the invasive trophoblast cell cluster, which included expression patterns within the placentation site and among invasive trophoblast cells.
Fig. 2.
Distribution of transcripts enriched in invasive trophoblast cells at the rat UPI. (A) Dot plots for gd 15.5 (Left) and 19.5 (Right) showing expression levels of invasive trophoblast cell enriched transcripts. Dot size and color represents the average percentage of cells expressing the transcript and the average level of expression, respectively. (B) Schematic depicting a gd 19.5 placentation site and a section through the central region of a gd 19.5 placentation site. Distribution of invasive trophoblast cells was determined by in situ hybridization of Prl7b1 transcripts. (Scale bar: 1,000 μm.) (C) Localization of invasive trophoblast cell enriched transcripts [Nppb, Plac1 (LOC102550080), Fstl3, Igf2, Tfpi; red] within the gd 19.5 placentation site. Transcripts were detected using in situ hybridization and colocalized to the distribution of Prl7b1 (green). (Scale bars Left and Middle: 1,000 μm; Right: 100 μm.) For abbreviations, see Fig. 1 legend.
In situ hybridization was performed for several transcripts identified in the invasive trophoblast cell cluster. A range of placentation site–associated patterns were observed, which are highlighted in Fig. 2. Prl7b1 and Nppb were notable in their enrichment in only invasive trophoblast cells (Fig. 2C). Plac1 (LOC102550080) was expressed in both invasive trophoblast cells and the junctional zone, whereas Fstl3 was enriched in invasive trophoblast cells and the labyrinth zone (Fig. 2C). Igf2 and Tfpi exhibited expression throughout the placentation site (Fig. 2C). Among invasive trophoblast cell enriched transcripts, Prl7b1, Igf2, and Nppb showed similar distributions in both endovascular and interstitial trophoblast cells (Fig. 2C). Tfpi expression was enriched in endovascular trophoblast cells (Fig. 2C), whereas Plac1 was prominently expressed in interstitial trophoblast cells and a subset of endovascular trophoblast cells (Fig. 2C). Fstl3 was localized to subsets of endovascular and interstitial trophoblast cells (Fig. 2C). Plac1 and Fstl3 showed some overlap in expression mainly in interstitial trophoblast cells (SI Appendix, Fig. S4). The expression patterns are intriguing and imply that there are subpopulations of invasive trophoblast cells, which were not resolved by scRNA-seq.
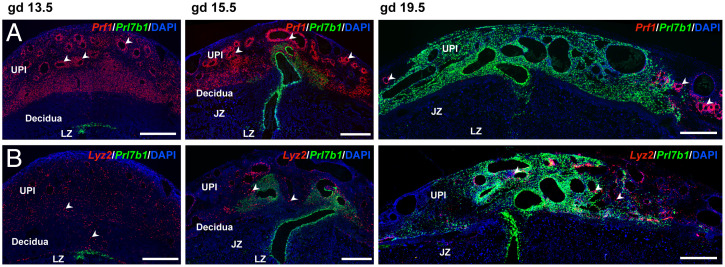
Differences were also identified in the spatial relationships of invasive trophoblast cells with immune cells. Invasive trophoblast cells (Prl7b1 positive) and NK cells (Prf1 positive) exhibited a reciprocal relationship and showed little overlap in their spatial distribution (Fig. 3A and SI Appendix, Fig. S5A), while macrophage populations (Lyz2 positive) were interspersed among invasive trophoblast cells within the uterine–placental interface (Fig. 3B and SI Appendix, Fig. S5B). In situ hybridization of endothelial cell cluster transcripts indicated the existence of subsets of endothelial cells, including the presence of peripherally located blood vessels in late gestation placentation sites that were not restructured by endovascular invasive trophoblast cells (SI Appendix, Fig. S6). Adgrl4 and Cdh5 showed some evidence for dual expression in endothelial cells and endovascular trophoblast cells, while Cdh5 also exhibited expression in interstitial invasive trophoblast cells (SI Appendix, Figs. S6 and S7). Potential cell–cell communication in the form of ligand–receptor expression profiles for invasive trophoblast cells with endothelial cells, macrophages, or NK cells was determined using CellPhoneDB (SI Appendix, Fig. S8 and Dataset S2).
Fig. 3.
Distribution of NK cells and macrophages within the rat uterine–placental interface. (A) NK cell and invasive trophoblast cells were monitored in gd 13.5, 15.5, and 19.5 placentation sites using in situ hybridization for Prf1 (red) and Prl7b1 (green), respectively. (B) Macrophages and invasive trophoblast cells were monitored in gd 13.5, 15.5, and 19.5 placentation sites using in situ hybridization for Lyz2 (red) and Prl7b1 (green), respectively. (Scale bar: 1,000 μm.) Arrowheads indicate examples of the distribution of NK cells (Top, Prf1 positive) and macrophages (Bottom, Lyz2 positive).
Gestation Stage–Dependent Invasive Trophoblast Cell Gene Expression.
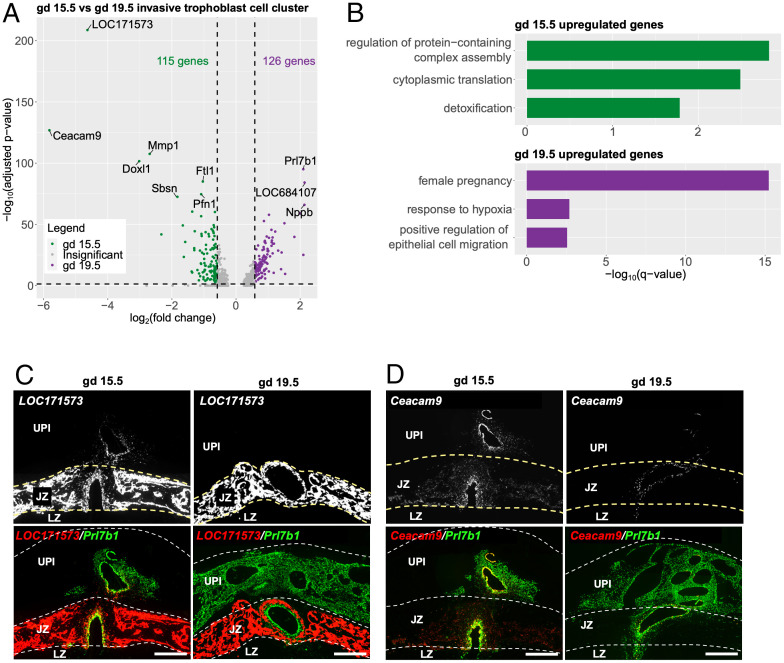
Differential patterns of transcript expression were observed for invasive trophoblast cells during the initial phase of intrauterine invasion (gd 15.5: 115 enriched transcripts) versus late-stage invasion (gd 19.5: 126 transcripts) (Fig. 4A and Dataset S3). To explore the biological functions associated with the differentially expressed transcripts, we carried out gene ontology (GO) analysis. GO terms related to protein translation [“cytoplasmic translation,” −log10(q-value) = 2.49] and detoxification [“detoxification,” −log10(q-value) = 1.78] were significantly enriched with transcripts up-regulated at gd 15.5, whereas transcripts up-regulated at gd 19.5 were significantly linked to female pregnancy [“female pregnancy,” −log10(q-value) = 15.23] and epithelial cell migration [“positive regulation of epithelial cell migration,” −log10(q-value) = 2.54] (Fig. 4B and Dataset S3). Some of the top transcripts differentially expressed at gd 15.5 and gd 19.5 by invasive trophoblast cells were unique to rodents (gd 15.5: LOC171573, Ceacam9, and Doxl1; gd 19.5: Prl7b1 and LOC684107) (Fig. 4A). LOC171573 and Ceacam9 were conspicuous in the initial wave of invasive trophoblast cells penetrating the uterus (Fig. 4 C and D). The biology of LOC171573, also called spleen protein 1 precursor, and CEACAM9 in placentation is not well understood (22, 23). LOC171573 has previously been localized to rat endovascular invasive trophoblast cells lining decidual arterioles at gd 11.5, and Ceacam9 has been shown to be expressed in differentiating rat trophoblast giant cells (23). CEACAM9 is a member of the larger carcinoembryonic antigen family, which has been implicated in cell adhesion, epithelial barrier function, and inflammatory responses (24). An implication from these observations may be that invasive trophoblast cells first entering deep into the potentially hostile uterus activate species-specific protective mechanisms, which are not required later in gestation as the uterine parenchyma becomes conditioned by infiltrating invasive trophoblast cells.
Fig. 4.
Differential invasive trophoblast cell gene expression on gd 15.5 versus gd 19.5. (A) Volcano plot showing differentially expressed genes (DEGs) in gd 15.5 and 19.5 invasive trophoblast cell clusters. Top 10 most significantly changed genes are labeled. (B) Bar plots showing selected significantly enriched gene ontology terms for transcripts up-regulated at gd 15.5 (Top) and transcripts up-regulated at gd 19.5 (Bottom). For full list of enriched terms, see Dataset S2). (C and D) Differential expression of LOC171573 and Cecam9 transcripts within placentation sites at gd 15.5 and gd 19.5. Transcripts were detected using in situ hybridization and colocalized to the distribution of Prl7b1 (green). Yellow arrows indicate the intrauterine invasive cells expressing LOC171573 and Ceacam9 at gd 15.5. (Scale bar: 1,000 μm.).
Conservation of Rat and Human Invasive Trophoblast Cell–Specific Transcripts.
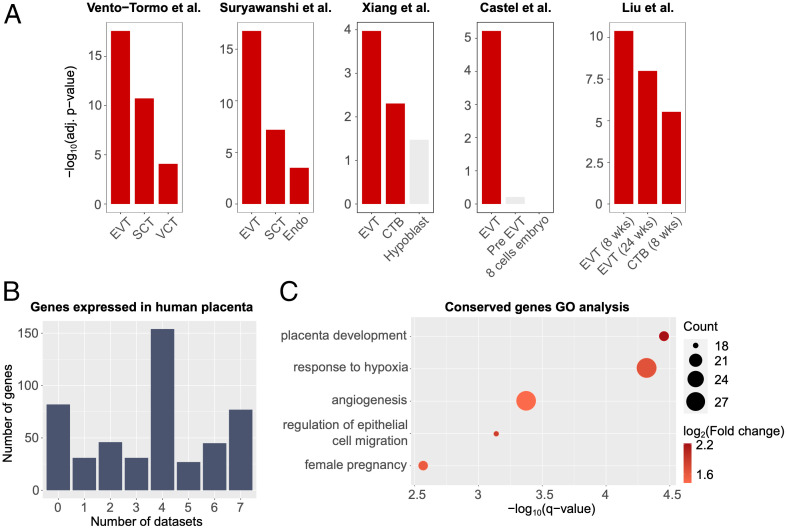
To explore invasive trophoblast cell transcripts conserved in rats and humans, we started by using the PlacentaCellEnrich webtool (25). Briefly, PlacentaCellEnrich groups genes that have cell-type–specific expression according to scRNA-seq data generated in human placenta and then calculates cell-type–specific enrichment for a given set of input genes (e.g., invasive trophoblast cell cluster marker genes). PlacentaCellEnrich employs the definitions of cell-type–specific gene groups from the Human Protein Atlas (26), in which a gene can be cell-type specific if it is highly expressed in a group of cell types compared to the rest of the cell types. Therefore, a single gene can be classified as having cell-type–specific expression in multiple trophoblast cell types. Interestingly, we observed that transcripts marking the invasive trophoblast clusters at gd 15.5 and gd 19.5 were most significantly enriched for EVT cell–specific genes when using cell-type–specific groups defined through five independent single cell human placenta or early embryo culture datasets (Fig. 5A) (27–31).
Fig. 5.
Conserved transcript expression in rat invasive trophoblast cells and human EVT cells. (A) Bar plots showing that transcripts highly expressed in the invasive trophoblast cell clusters at gd 15.5 and 19.5 in rats share similar profiles to EVT cells in human placenta. Enrichment analyses were carried out using human placenta single cell data as references: Vento-Tormo et al. (29), Suryawanshi et al. (28), Xiang et al. (30), Castel et al. (31), and Liu et al. (27). A significant enrichment has an adjusted P value ≤0.05, fold change ≥1.5, and number of observed genes ≥5. Red, significant enrichment; gray, insignificant enrichment. (B) Bar plot showing the number of rat invasive trophoblast cell genes (y axis) expressed in different human EVT cell datasets. Seven EVT cell datasets were analyzed (Dataset S4) and the x axis shows the total number of EVT cell datasets in which the rat invasive trophoblast gene was either an EVT cell marker or expressed in EVT cells. (C) Dot plots showing GO enrichment for rat invasive trophoblast cell genes that are also expressed in human placenta. Dot size represents the number of observed genes related to a term; dot colors correspond to log2(fold change) of the terms. Selected gene ontology terms are shown. For a full list of enriched terms, see SI Appendix, Fig. S9. Abbreviations: SCT, syncytiotrophoblast; VCT, villous cytotrophoblast; Endo, vascular endothelial cells (villi); CTB, cytotrophoblast; EVT (8 wk), EVT from villi at 8 wk of pregnancy; EVT (24 wk), EVT from decidua at 24 wk of pregnancy; CTB (8 wk), CTB from villi at 8 wk of pregnancy.
Motivated by the results from PlacentaCellEnrich, we further annotated all 493 of the invasive trophoblast cell cluster transcripts in rat at either gd 15.5 or gd 19.5 that have expressed human orthologs (Materials and Methods) in not only single cell data but also bulk RNA-seq data obtained from EVT cells differentiated from human trophoblast stem cells (32) and EVT cells isolated from first trimester human placenta (33). The rat invasive trophoblast cell cluster transcripts showed some evidence of species specificity as there were 82 (16.63%) transcripts that did not have human orthologs or were not expressed in human (Fig. 5B and Dataset S4). However, the vast majority of transcripts (411, 83.37%) from the rat invasive trophoblast scRNA-seq cell clusters were also expressed in at least one of the human EVT datasets (Fig. 5B and Dataset S4). Next, we used GO analysis on invasive trophoblast cell genes expressed in at least one dataset and observed that the genes are enriched for processes related to low oxygen conditions [“response to hypoxia,” −log10(q-value) = 4.32], pregnancy and placenta development [“female pregnancy,” −log10(q-value) = 2.57; “placenta development,” −log10(q-value) = 4.45], cell migration [“regulation of epithelial cell migration,” −log10(q-value) = 3.14], and vasculature development [“angiogenesis,” −log10(q-value) = 3.37] (Fig. 5C and SI Appendix, Fig. S9 and Dataset S5). Collectively, these observations reinforce the merits of modeling trophoblast-guided uterine transformation in the rat.
Discussion
Trophoblast cell–guided transformation of the uterus is a defining feature of the hemochorial placenta (6, 7). A specialized lineage of trophoblast cells referred to as invasive trophoblast or EVT cells executes these vital functions (5, 6). This fundamental process includes restructuring uterine spiral arteries to optimize blood flow into the placenta (3, 5). In human pregnancies, failures in trophoblast cell–guided uterine vasculature remodeling underly disease states such as preeclampsia, intrauterine growth restriction, and preterm birth (8). There is a need for modeling this key trophoblast cell–uterine interaction in vivo; however, arguments have been levied that the process as it occurs in human placentation is unique (34). Unfortunately, this opinion has stifled progress in understanding regulatory mechanisms controlling deep placentation. In this report, compelling evidence is provided supporting the utility of the rat as a model for investigating the uterine–placental interface. Two important characteristics of the rat placentation site are key features demonstrating its efficacy as a model: 1) deep intrauterine trophoblast cell invasion (9, 11, 35) and 2) physical separation of intrauterine invasive trophoblast cells from the placenta (36). Cellular constituents of the rat uterine–placental interface were defined and are consistent with previous scRNA-seq analyses of the first trimester human uterine–placental interface (27–29). Furthermore, core conservation in the transcriptomes of rat and human invasive trophoblast/EVT cells was revealed.
There are two main populations of invasive trophoblast cells: 1) interstitial and 2) endovascular. These two invasive trophoblast cell types exhibit positional and functional differences and are characteristic of both rat and human placentation sites (3, 5, 9, 11, 35). Invasive trophoblast cells and EVT cells did not resolve into clusters reflecting the two invasive trophoblast cell types in the present analysis or prior scRNA-seq efforts of the first trimester human placentation site (27–29). Spatial transcriptomic differences of interstitial versus endovascular trophoblast cells were resolved by in situ hybridization. This is especially noteworthy for Tfpi and Mmp12 transcripts, which reside within the singular invasive trophoblast cell cluster, but based on in situ hybridization are enriched in endovascular trophoblast cells (19, 36). The reason for this discrepancy is not understood. It may relate to unequal recovery of interstitial versus endovascular trophoblast cells during the single cell isolation process. Alternatively, it may reflect the plasticity of the isolated invasive trophoblast cells and the requirement of positional cues for displaying phenotypic differences, which disappear once the cells are dissociated. Application of genome-wide spatial transcriptomic analyses (37) could further define phenotypic differences between interstitial and endovascular trophoblast cells.
Dynamic relationships exist for immune cell populations and invasive trophoblast cells within the uterine–placental interface. For example, NK cells were predominant early and subsequently disappeared as invasive trophoblast cells infiltrated the uterine–placental interface. A similar reciprocal relationship of NK cells and invasive trophoblast cells has been reported (10, 14). NK cell deficiencies lead to accelerated endovascular trophoblast cell invasion (17, 38), whereas deficits in invasive trophoblast cells result in a retention of NK cells (36). These results infer that cell–cell communication is engineering the cellular composition of the uterine–placental interface. Signaling may be direct between NK cells and invasive trophoblast cells or indirect through any of the other cellular constituents of the uterine–placental interface. Cellular transcriptomes defined in this report possess clues for understanding the gestation stage–dependent reciprocal distributions of NK cells and invasive trophoblast cells.
Conservation in gene regulatory networks controlling rat and human invasive trophoblast and EVT cell lineages has been previously demonstrated and include Mmp12, Ascl2, and Tfpi (19, 21, 36). scRNA-seq analysis has expanded the list of candidate conserved regulators. The top functional categories for conserved candidates most notably included biological activities required for cell migration and invasion. Among the conserved candidates are transcripts encoding transcriptional regulators (e.g., ASCL2, ARID3A, CITED2, ETS2, GATA3, JUN, NCOR2, NR2F6, NRIP1, PPARG, SIN3B, SNAI1, TFAP2C, and WWTR1); ligands, binding proteins, and protein processing enzymes (e.g., C1QTNF6, DPP3, FSTL3, FURIN, IGF2, IGFBP2, NPPB, PGF, and SUB1); regulatory hubs in signaling pathways (e.g., CDKN1C, DUSP9, DYRK2, GNG12, GRB2, RGS16, and SEMA4C); proteins contributing to cell–cell and cell matrix interactions (DSP, FBLIM1, GPC1, ITGA5, and PXN); extracellular matrix proteins and modulators (e.g., AGRN, COL4A1, CRISPLD2, FIBLN1, LAMA4, MMP12, MMP15, NID1, SERPINE1, and SERPINH1); and proteins known to directly affect cell movement (e.g., ANXA2, DAAM1, and EZR). Strategies exist in the rat for testing in vivo roles of candidates using global and conditional genome editing (39) and trophoblast cell–specific lentiviral-mediated gene manipulation (40, 41). Dysregulation of candidate genes may directly affect the development and function of the invasive trophoblast cell lineage and indirectly affect uterine cell dynamics at the uterine–placental interface. These approaches can be further leveraged through comparative analysis in vitro using rat and human trophoblast stem cells (32, 42). In addition to conservation at the uterine–placental interface there are also elements of species specificity, which is exemplified by the hormone producing capacity of invasive trophoblast and EVT cells (e.g., rat: prolactin family; human: chorionic gonadotropin and pregnancy-specific glycoproteins).
In summary, conservation exists within the invasive trophoblast cell lineages of the rat and human providing a rationale for using the rat as an experimental model to elucidate regulatory mechanisms controlling uterine–placental cellular dynamics.
Materials and Methods
Animals and Tissue Collection.
Holtzman Sprague–Dawley rats were obtained from Envigo. Rats were housed on a 14-h light/10-h dark photoperiod with free access to food and water. Timed pregnancies were obtained by mating adult males (>10 wk) and females (8 to 12 wk). Pregnancies were confirmed with a sperm positive vaginal lavage, defined as gd 0.5. Uterine–placental interface tissues of the rat placentation site (also termed the metrial gland) from gd 15.5 and 19.5 were dissected as previously described (36) and used for scRNA-seq. Uteroplacental/fetal sites were also frozen in dry-ice–cooled heptane and used for in situ hybridization localization of transcripts. Protocols for research with animals were approved by the University of Kansas Medical Center (KUMC) Animal Care and Use Committee.
Cell Isolation and scRNA-Seq.
Uterine–placental interface tissues were harvested from gd 15.5 (n = 3 pregnancies/gd) and 19.5 (n = 4 pregnancies/gd) rat placentation sites and placed in ice-cold Hank’s balanced salt solution (HBSS). Tissues were minced into small pieces with a razor blade and digested with Dispase II (1.25 units/mL, D4693, Sigma-Aldrich), 0.4 mg/mL collagenase IV (C5138, Sigma-Aldrich), and DNase I (80 units/mL, D4513, Sigma-Aldrich) in HBSS for 30 min. Red blood cells were lysed using ammonium-chloride-potassium lysis buffer (A10492-01, Thermo Fisher) with rotation at room temperature for 5 min. Intact cells were washed with HBSS supplemented with 2% fetal bovine serum (Thermo Fisher), and DNase1 (Sigma-Aldrich) and passed through a 100-µm cell strainer (100ICS, Midwest Scientific). Following enzymatic digestion, cellular debris was removed using MACS Debris Removal Solution (130-109-398, Miltenyi Biotec). Cells were then filtered through a 40-µm cell strainer (40ICS, Midwest Scientific) and cell viability was assessed, which ranged from 90 to 93%. Cells were used for the preparation of single cell libraries using the 10× Genomics Chromium system (10× Genomics) and sequenced with an Illumina NovaSEq. 6000 (Illumina) by the KUMC Genome Sequencing Facility.
scRNA-Seq Data Analysis.
scRNA-seq preprocessing.
Raw data were aligned to the rat genome (Rnor 6.0, Ensembl 98) (43) and quantified using Cell Ranger Software (version 4.0.0). The R package Seurat (version 4.1.0) (44) was used for quality control and downstream analysis. Hereinafter, if not mentioned, all parameter settings are as default. We retained the cells with the number of unique genes between 500 and 3,500 and less than 20% of mitochondrial genes (SI Appendix, Fig. S1). Next, we merged the replicates of gd 15.5 together using Seurat function Merge(). Since replicates of gd 19.5 were generated in two different batches, Seurat functions FindIntegrationAnchors() and IntegrateData() were used to perform batch correction and integrate gd 19.5 samples (SI Appendix, Fig. S1). All mitochondrial genes were then removed from the resulting Seurat objects. To mitigate technical noise, we normalized and selected the top 2,000 most variable transcripts in each time point, and then scaled the data. Principal component analysis (PCA) was then carried out with the top 2,000 variable transcripts to reduce dimensions of the data. We accessed the significance of the top 100 principal components (PCs) in each time point with Seurat functions JackStraw() and ElbowPlot(). The analyses showed that PC 72 was the last significant component in the gd 15.5 sample (P value ≤0.05); hence, we retained the first 72 PCs in gd 15.5 for downstream analyses. Similarly, the first 77 PCs were kept for the gd 19.5 sample (SI Appendix, Fig. S2).
scRNA-seq clustering and cluster annotation.
With the retained significant PCs, we utilized K-nearest neighbor (KNN) graphs and the original Louvain algorithm (45), which are implemented through the Seurat functions FindNeighbors() and FindClusters(), to identify cell clusters in each time point (resolution = 0.8). The cell clusters were then visualized with uniform manifold approximation and projection (UMAP). To identify marker transcripts for each cell cluster at each gestation day, we used the Seurat function FindAllMarker() with the Wilcoxon rank sum test, which compares gene expression in each cell cluster to the expression across all of the other cell clusters. Original assays of individual replicates were inputs for the FindAllMarker() differential expression tests. A transcript was considered a marker of one cell cluster if it was expressed in ≥10% of the cells, its adjusted P value was ≤0.05, and its average fold change was ≥log2(1.5). We annotated the cell clusters using the following cell type markers: Prl5a1, Prl7b1, Krt7, Krt8, and Krt18 (invasive trophoblast cells); Nkg7, Prf1, Gzmm, and Gzmbl2 (NK cells); Cdh5, Plvap, Adgrl4, and Egfl7 (endothelial cells); C1qa, Lyz2, Aif1, and Cybb (macrophages); and Acta2, Myl9, Tagln, and Myh11 (smooth muscle cells). A cell cluster was annotated to a cell identity if it had ≥75% of the previously identified cell type markers. Insights into the identities of other cell clusters were obtained by running cluster-associated transcripts through the Enrichr web tool using default parameters and the “Cell Types” results (46).
Identification of ligand–receptor interactions between cell types.
To predict potential ligand–receptor interactions of invasive trophoblast cells with endothelial cells, macrophages, or NK cells, we used CellPhoneDB (47) with default settings. Normalized gene expression level, cell identities, and all expressed genes (genes with mean expression level >0 in 10% of cells) were used as inputs for the command “cellphonedb method statistical_analysis.” Rat genes were mapped to human genes using Ensembl version 103. (48) The resulting P values were then corrected using the Benjamini–Hochberg procedure (49). A pair was considered significant if its adjusted P value was ≤0.05.
scRNA-seq comparison between timepoints.
To compare the expression profiles of trophoblast clusters between the two gestation days, we carried out differential expression analysis. We used the Seurat function FindMarkers() with the original assays of the samples as inputs for the differential tests. A differentially expressed gene at a gestation day was one with adjusted P value ≤0.05 and average fold change ≥1.5.
Gene ontology enrichment analysis.
To explore gene set functions, we carried out GO enrichment analysis using the R package clusterProfiler (version 3.16.1) (50). Gene annotation was obtained from the R package org.Rn.eg.db (version 3.11.4) (51). A fold change for each term was calculated as GeneRatio/BgRatio. Nonredundant terms were then obtained using the simplify() function in clusterProfiler. A GO term was considered enriched if it was nonredundant, its q-value was ≤0.05, fold change was ≥2, and the number of observed genes was ≥5.
Identification of conserved trophoblast genes between human and rat.
We carried out cell enrichment analysis using the PlacentaCellEnrich webtool (25). Cell-type–specific genes were identified based on human placenta or embryo culture single cell data (27–31). The datasets from Suryawanshi et al. (28) and Vento-Tormo et al. (29) were used as supplied in PlacentaCellEnrich (25). The datasets from Xiang et al. (30) and Castel et al. (31) were obtained from Seetharam et al. (52). The data from Liu et al. (27) was reanalyzed to build a reference dataset with transcripts per million (TPM)-like expression values following the PlacentaCellEnrich procedure (25). An enrichment was considered significant if its adjusted P value was ≤0.05, fold change was ≥1.5, and the number of observed genes was ≥5. Code to carry out PlacentaCellEnrich analysis was adapted from the TissueEnrich R package (53). A gene was considered expressed in human EVT cells in Liu et al. (27), Suryawanshi et al. (28), Vento-Tormo et al. (29), Xiang et al. (30), and Castel et al. (31) datasets if its TPM-like expression was ≥1; expressed in Morey et al. human EVT cells (33) if its TPM was ≥1, and expressed in Okae et al. human EVT cells (32) if its FPKM (fragments per kilobase of TPM mapped reads) was ≥1.
In Situ Hybridization.
Localization of transcript expression within placentation sites was performed on cryosections (20 µm) prepared from gd 15.5 and 19.5 rat placentation sites. RNAScope Multiplex Fluorescent V2 assay (Advanced Cell Diagnostics) was used to detect transcripts within the uterine–placental interface according to protocols provided by the manufacturer. Probes were prepared to detect invasive trophoblast cells (Krt8: NM_199370.1, 873041-C2, target region: 134 to 1,472; Prl7b1: NM_153738.1, 860181, 860181-C2, target region: 28 to 900; Igf2: NM_031511.2, 444561, target region: 1,475 to 2,634; Fstl3: NM_053629.3, 862961, target region: 2 to 766; Nppb: NM_031545.1, 583051, target region: 3 to 531; Plac1: NM_001024894.1, 860141, target region: 3 to 944; Tfpi: NM_017200.1, 878371, target region: 2 to 1,138; LOC171573: NM_138537.2, 1104761, target region: 2 to 576; Ceacam9: NM_053919.2, 1166771, target region: 130 to 847); endothelial cell (Adgrl4: NM_022294.1, 878411-C2, target region: 886 to 1,871; Plvap: NM_020086.1, 878401, target region: 61 to 1,225), macrophage (Lyz2: NM_012771.3, 888811-C2, target region: 82 to 1,181); natural killer cell (Prf1: NM_017330.2, 871601-C2, target region: 451 to 1,452). A Nikon 80i upright microscope (Nikon) and Photometrics CoolSNAP-ES monochrome was used to capture fluorescence images.
Supplementary Material
Acknowledgments
We thank Brandi Miller and Stacy Oxley for their assistance. This work was supported by the NIH National Research Service Award Predoctoral Fellowship, F31HD104495 (R.L.S.); by NIH grants: HD020676 (M.J.S.), ES029280 (M.J.S.), HD099638 (M.J.S.), HD104033 (M.J.S. and G.T.), HD105734 (M.J.S.); and by the Sosland Foundation (M.J.S.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2210633119/-/DCSupplemental.
Data, Materials, and Software Availability
scRNA-seq datasets are available at the Gene Expression Omnibus website [https://www.ncbi.nlm.nih.gov/geo/; accession no. GSE206086 (54)]. All data generated and analyzed during this study are included in the published article and the supporting information. All code used for the analyses are available at https://github.com/Tuteja-Lab/MetrialGland-scRNA-seq (55).
References
- 1.Gardner R. L., Beddington R. S., Multi-lineage ‘stem’ cells in the mammalian embryo. J. Cell Sci. Suppl. 10, 11–27 (1988). [DOI] [PubMed] [Google Scholar]
- 2.Kaufmann P., Black S., Huppertz B., Endovascular trophoblast invasion: Implications for the pathogenesis of intrauterine growth retardation and preeclampsia. Biol. Reprod. 69, 1–7 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Pijnenborg R., Vercruysse L., Hanssens M., The uterine spiral arteries in human pregnancy: Facts and controversies. Placenta 27, 939–958 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Soares M. J., Varberg K. M., Iqbal K., Hemochorial placentation: Development, function, and adaptations. Biol. Reprod. 99, 196–211 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knöfler M., et al. , Human placenta and trophoblast development: Key molecular mechanisms and model systems. Cell. Mol. Life Sci. 76, 3479–3496 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pijnenborg R., Robertson W. B., Brosens I., Dixon G., Review article: Trophoblast invasion and the establishment of haemochorial placentation in man and laboratory animals. Placenta 2, 71–91 (1981). [DOI] [PubMed] [Google Scholar]
- 7.Roberts R. M., Green J. A., Schulz L. C., The evolution of the placenta. Reproduction 152, R179–R189 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brosens I., Pijnenborg R., Vercruysse L., Romero R., The “Great Obstetrical Syndromes” are associated with disorders of deep placentation. Am. J. Obstet. Gynecol. 204, 193–201 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pijnenborg R., Vercruysse L., “Animal models of deep trophoblast invasion” in Placental Bed Disorders: Basic Science and its Translation to Obstetrics, Pijnenborg R., Brosens I., Romero R., Eds. (Cambridge University Press, 2010), pp. 127–139. [Google Scholar]
- 10.Ain R., Canham L. N., Soares M. J., Gestation stage-dependent intrauterine trophoblast cell invasion in the rat and mouse: Novel endocrine phenotype and regulation. Dev. Biol. 260, 176–190 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Soares M. J., Chakraborty D., Karim Rumi M. A., Konno T., Renaud S. J., Rat placentation: An experimental model for investigating the hemochorial maternal-fetal interface. Placenta 33, 233–243 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosario G. X., Konno T., Soares M. J., Maternal hypoxia activates endovascular trophoblast cell invasion. Dev. Biol. 314, 362–375 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Konno T., Rempel L. A., Arroyo J. A., Soares M. J., Pregnancy in the brown Norway rat: A model for investigating the genetics of placentation. Biol. Reprod. 76, 709–718 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Ain R., Konno T., Canham L. N., Soares M. J., Phenotypic analysis of the rat placenta. Methods Mol. Med. 121, 295–313 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Wiemers D. O., Ain R., Ohboshi S., Soares M. J., Migratory trophoblast cells express a newly identified member of the prolactin gene family. J. Endocrinol. 179, 335–346 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Caluwaerts S., Vercruysse L., Luyten C., Pijnenborg R., Endovascular trophoblast invasion and associated structural changes in uterine spiral arteries of the pregnant rat. Placenta 26, 574–584 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Chakraborty D., Rumi M. A. K., Konno T., Soares M. J., Natural killer cells direct hemochorial placentation by regulating hypoxia-inducible factor dependent trophoblast lineage decisions. Proc. Natl. Acad. Sci. U.S.A. 108, 16295–16300 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Konno T., et al. , Chromosome-substituted rat strains provide insights into the genetics of placentation. Physiol. Genomics 43, 930–941 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chakraborty D., et al. , HIF-KDM3A-MMP12 regulatory circuit ensures trophoblast plasticity and placental adaptations to hypoxia. Proc. Natl. Acad. Sci. U.S.A. 113, E7212–E7221 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kozai K., et al. , Protective role of IL33 signaling in negative pregnancy outcomes associated with lipopolysaccharide exposure. FASEB J. 35, e21272 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Varberg K. M., et al. , ASCL2 reciprocally controls key trophoblast lineage decisions during hemochorial placenta development. Proc. Natl. Acad. Sci. U.S.A. 118, e2016517118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Finkenzeller D., et al. , Carcinoembryonic antigen-related cell adhesion molecule 10 expressed specifically early in pregnancy in the decidua is dispensable for normal murine development. Mol. Cell. Biol. 23, 272–279 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kent L. N., Konno T., Soares M. J., Phosphatidylinositol 3 kinase modulation of trophoblast cell differentiation. BMC Dev. Biol. 10, 97 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuespert K., Pils S., Hauck C. R., CEACAMs: Their role in physiology and pathophysiology. Curr. Opin. Cell Biol. 18, 565–571 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jain A., Tuteja G., PlacentaCellEnrich: A tool to characterize gene sets using placenta cell-specific gene enrichment analysis. Placenta 103, 164–171 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uhlén M., et al. , Proteomics. Tissue-based map of the human proteome. Science 347, 1260419 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Liu Y., et al. , Single-cell RNA-seq reveals the diversity of trophoblast subtypes and patterns of differentiation in the human placenta. Cell Res. 28, 819–832 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suryawanshi H., et al. , A single-cell survey of the human first-trimester placenta and decidua. Sci. Adv. 4, eaau4788 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vento-Tormo R., et al. , Single-cell reconstruction of the early maternal-fetal interface in humans. Nature 563, 347–353 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiang L., et al. , A developmental landscape of 3D-cultured human pre-gastrulation embryos. Nature 577, 537–542 (2020). [DOI] [PubMed] [Google Scholar]
- 31.Castel G., et al. , Induction of human trophoblast stem cells from somatic cells and pluripotent stem cells. Cell Rep. 33, 108419 (2020). [DOI] [PubMed] [Google Scholar]
- 32.Okae H., et al. , Derivation of human trophoblast stem cells. Cell Stem Cell 22, 50–63.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Morey R., et al. , Transcriptomic drivers of differentiation, maturation, and polyploidy in human extravillous trophoblast. Front. Cell Dev. Biol. 9, 702046 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carter A. M., Unique aspects of human placentation. Int. J. Mol. Sci. 22, 8099 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shukla V., Soares M. J., Modeling trophoblast cell-guided uterine spiral artery transformation in the rat. Int. J. Mol. Sci. 23, 2947 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muto M., et al. , Intersection of regulatory pathways controlling hemostasis and hemochorial placentation. Proc. Natl. Acad. Sci. U.S.A. 118, e2111267118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rao A., Barkley D., França G. S., Yanai I., Exploring tissue architecture using spatial transcriptomics. Nature 596, 211–220 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Renaud S. J., Scott R. L., Chakraborty D., Rumi M. A. K., Soares M. J., Natural killer-cell deficiency alters placental development in rats. Biol. Reprod. 96, 145–158 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chenouard V., et al. , Advances in genome editing and application to the generation of genetically modified rat models. Front. Genet. 12, 615491 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee D. S., Rumi M. A., Konno T., Soares M. J., In vivo genetic manipulation of the rat trophoblast cell lineage using lentiviral vector delivery. Genesis 47, 433–439 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chakraborty D., Muto M., Soares M. J., Ex vivo trophoblast-specific genetic manipulation using lentiviral delivery. Bio Protoc. 7, e2652 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Asanoma K., et al. , FGF4-dependent stem cells derived from rat blastocysts differentiate along the trophoblast lineage. Dev. Biol. 351, 110–119 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cunningham F., et al. , Ensembl 2019. Nucleic Acids Res. 47, D745–D751 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R., Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blondel V. D., Guillaume J. L., Hendrickx J. M., de Kerchove C., Lambiotte R., Local leaders in random networks. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 77, 036114 (2008). [DOI] [PubMed] [Google Scholar]
- 46.Kuleshov M. V., et al. , Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Efremova M., Vento-Tormo M., Teichmann S. A., Vento-Tormo R., CellPhoneDB: Inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat. Protoc. 15, 1484–1506 (2020). [DOI] [PubMed] [Google Scholar]
- 48.Howe K. L., et al. , Ensembl 2021. Nucleic Acids Res. 49, D884–D891 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Benjamini Y., Hochberg Y., Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 (1995). [Google Scholar]
- 50.Yu G., Wang L.-G., Han Y., He Q.-Y., clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carlson M., org.RN.eg.db: Genome wide annotation for Rat. R package version 3.11.4 (2019). 10.18129/B9.bioc.org.Rn.eg.db. [DOI]
- 52.Seetharam A. S., et al. , The product of BMP-directed differentiation protocols for human primed pluripotent stem cells is placental trophoblast and not amnion. Stem Cell Reports 17, 1289–1302 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jain A., Tuteja G., TissueEnrich: Tissue-specific gene enrichment analysis. Bioinformatics 35, 1966–1967 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.R. Scott, K. Iqbal, M. J. Soares, G. Tuteja, Conservation at the uterine-placental interface. NCBI: GEO. http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE206086. Deposited 20 September 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ha T. H. Vu, MetrialGland-scRNA-seq. Github. https://github.com/Tuteja-Lab/MetrialGland-scRNA-seq. Deposited 5 August 2022. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
scRNA-seq datasets are available at the Gene Expression Omnibus website [https://www.ncbi.nlm.nih.gov/geo/; accession no. GSE206086 (54)]. All data generated and analyzed during this study are included in the published article and the supporting information. All code used for the analyses are available at https://github.com/Tuteja-Lab/MetrialGland-scRNA-seq (55).