Significance
Sexual reproduction has not been observed in unicellular red algae and Glaucophyceae, early branching groups in Archaeplastida, in which red algae and Viridiplantae independently evolved multicellular sexual life cycles. The finding of sexual reproduction in the unicellular red alga Galdieria provides information on the missing link of life cycle evolution in Archaeplastida. In addition, the metabolic plasticity, the polyextremophilic features, a relatively small genome, transcriptome data for the diploid and haploid, and the genetic modification tools developed here provide a useful platform for understanding the evolution of Archaeplastida, photosynthesis, metabolism, and environmental adaptation. For biotechnological use of the information and tools of Galdieria, the newly found cell wall–less haploid makes cell disruption less energy/cost intensive than the cell-walled diploid.
Keywords: Galdieria, sexual reproduction, homeobox, actin, microalgae
Abstract
Sexual reproduction is widespread in eukaryotes; however, only asexual reproduction has been observed in unicellular red algae, including Galdieria, which branched early in Archaeplastida. Galdieria possesses a small genome; it is polyextremophile, grows either photoautotrophically, mixotrophically, or heterotrophically, and is being developed as an industrial source of vitamins and pigments because of its high biomass productivity. Here, we show that Galdieria exhibits a sexual life cycle, alternating between cell-walled diploid and cell wall–less haploid, and that both phases can proliferate asexually. The haploid can move over surfaces and undergo self-diploidization or generate heterozygous diploids through mating. Further, we prepared the whole genome and a comparative transcriptome dataset between the diploid and haploid and developed genetic tools for the stable gene expression, gene disruption, and selectable marker recycling system using the cell wall–less haploid. The BELL/KNOX and MADS-box transcription factors, which function in haploid-to-diploid transition and development in plants, are specifically expressed in the haploid and diploid, respectively, and are involved in the haploid-to-diploid transition in Galdieria, providing information on the missing link of the sexual life cycle evolution in Archaeplastida. Four actin genes are differently involved in motility of the haploid and cytokinesis in the diploid, both of which are myosin independent and likely reflect ancestral roles of actin. We have also generated photosynthesis-deficient mutants, such as blue-colored cells, which were depleted in chlorophyll and carotenoids, for industrial pigment production. These features of Galdieria facilitate the understanding of the evolution of algae and plants and the industrial use of microalgae.
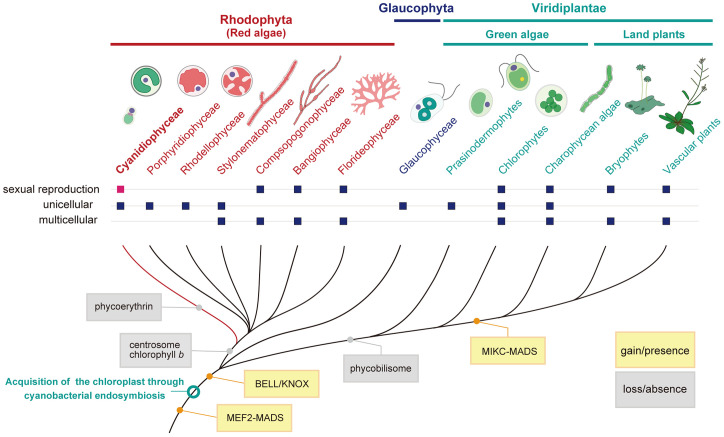
Cyanidiophyceae are unicellular red algae that exhibit a blue–green color because of a lack of phycoerythrin, a red protein pigment complex found in other red algae, and thrive in a thermo-acidic environment (pH of 0.05 to 5.0, <56 °C) worldwide (1). This group is estimated to have branched off from other red algae during early eukaryotic evolution (1.3 to 1.4 billion y ago; Fig. 1) (2). Red algae belong to Archaeplastida, a major eukaryotic group, which evolved from a unicellular eukaryotic ancestor that acquired the chloroplast through a cyanobacterial endosymbiotic event more than 1 billion y ago. Within Archaeplastida, Glaucophyceae, a group consisting of a limited number of unicellular algae, and red algae branched earlier than diversification of Viridiplantae (Fig. 1, green algae and land plants) (3, 4). In addition, many other eukaryotic lineages acquired their chloroplasts through secondary and tertiary endosymbiotic events with unicellular red algae (3). Therefore, cyanidialean unicellular red algae are important for elucidating the evolutionary history of all algae and plants (Fig. 1).
Fig. 1.
Evolutionary position of the red algae Cyanidiophyceae to which the genus Galdieria belongs. Cladogram showing gain and loss of some traits and genes. Also shown is whether respective groups are unicellular or multicellular organisms and whether sexual reproduction has been observed. The topologies in red algae (Rhodophyta) (66, 67) and Viridiplantae (4) are based on previous studies. The order of branching of red algae and Glaucophyta remains unclear, although a recent study based on large-scale genomic data suggests that red algae branched first in Archaeplastida (4).
Based on their evolutionary position, genomic analyses of cyanidialean unicellular red algae have contributed to the elucidation of the evolutionary process of eukaryotes (5–7). However, sexual reproduction has not been observed in unicellular red algae and Glaucophyceae, early branching groups in Archaeplastida (8), unlike some unicellular green algae (Fig. 1) (9). Thus, it is still not known how the sexual life cycle in Archaeplastida originated nor how red algae and Viridiplantae independently evolved multicellular sexual life cycles from their unicellular ancestors. Because the genome of Cyanidiophyceae encodes genes for meiosis (8), they may represent a missing evolutionary link in the Archaeplastida.
Cyanidiophyceae, and especially the genus Galdieria, are attracting attention in studies of photosynthesis, metabolic plasticity, and microbial environmental adaptation (1, 7, 10). Galdieria shows remarkable metabolic capabilities and grows photoautotrophically, mixotrophically, and heterotrophically by using more than 50 different carbon sources (1, 11), including organic wastes (12, 13), unlike other obligately photoautotrophic genera, such as Cyanidium, Cyanidiococcus, and Cyanidioschyzon (14). Moreover, Galdieria is a polyextremophile, tolerating higher salt (up to 1.5 M NaCl) and heavy metal concentrations than the other cyanidialean genera (1). Despite these additional characteristics, Galdieria spp. have a very small genome size, comparable to those of the other genera (12 to 18 megabases [Mb]) (6, 7).
Galdieria is also considered an emerging system for biotechnology applications because it grows to a relatively high density (∼100 g dried algae/L) under mixotrophic and heterotrophic conditions and is rich in proteins and vitamins like the other Cyanidiophyceae (15–17). Its acidic culture conditions reduce the risk of contamination by undesirable microorganisms in outdoor cultivation (18). Based on these features, Galdieria is being developed for wastewater treatment (12), production of food ingredients (19), and pigments (13). However, Galdieria cells are relatively small (3 to 10 µm) and are surrounded by a thick and rigid cell wall (1, 17, 20), requiring energy-intensive physical processing to extract the cellular contents and preventing the introduction of exogenous DNA and, thus, genetic modification. These difficulties in developing molecular genetic tools in Galdieria and a lack of information on a clear life cycle of Cyanidiophyceae have limited the use of these microalgae for basic and applied research.
Here, we show that the known cell-walled form of Galdieria, which dominates in their natural habitat, is a diploid that generates a cell wall–less haploid and undergoes isogamous sexual reproduction. We also sequenced the whole genome, compared transcriptome datasets of diploid and haploid cells, and developed molecular genetic tools using the cell wall–less haploid. These tools and information will facilitate understanding evolution of Archaeplastida, photosynthesis, metabolic plasticity, microbial environmental adaptation, and industrial use of microalgae. We demonstrate the role of transcription factors conserved in Archaeplastida in the haploid-to-diploid transition, myosin-independent roles of actin, which likely reflect ancestral roles of actin, and generation of a blue-colored algal cell for industrial production of pigments.
Results and Discussion
The Known Cell-Walled Form of Galdieria Is Diploid and Generates Cell Wall–Less Haploids, Both of Which Can Proliferate Asexually.
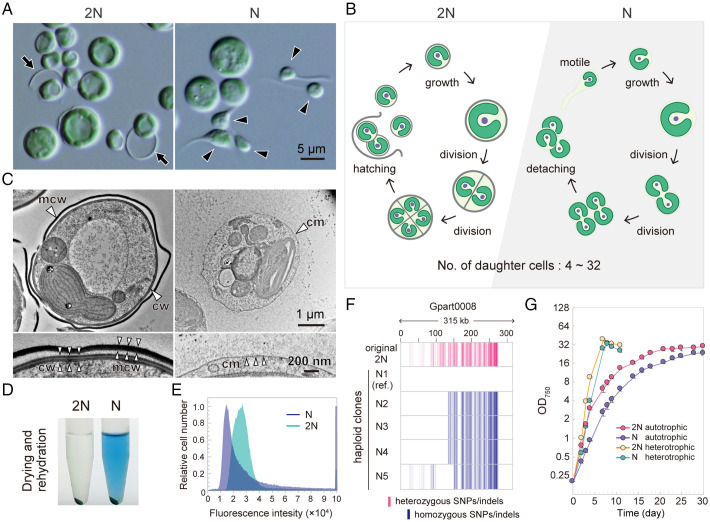
As observed in Galdieria spp. (G. sulphuraria, G. daedala, G. partita, and G. phlegrea) since its first description of G. sulphuraria in 1899 (1), G. partita NBRC102759 cells cultured at pH 2.0, known as the optimum pH (21), are spherical and nonmotile, proliferating by forming 4 to 32 daughter cells in a mother cell by successive cell divisions before hatching out of the mother cell wall (Fig. 2 A and B, 2N, and Movies S1 and S2). In addition to the known form, we discovered tadpole-shaped cells, which moved over surfaces in static cultures when G. partita was cultured at pH 1.0 (Fig. 2 A, N, and Movie S3). The tail of the tadpole-shaped cell is not a cilium because red algae lost centrosomes and cilia, accompanied by the loss of related genes, in their common ancestor (22, 23). When a single tadpole-shaped cell was isolated and cultured at pH 1.0, the clonal culture contained both tadpole-shaped and spherical cells (Fig. 2 A, N). As shown by electron microscopy and genome and transcriptome analyses below, both tadpole-shaped and spherical G. partita cells in the culture at pH 1.0 are haploids. The same phenomenon was also observed in two strains of G. sulphuraria (NIES-550 and SAG108.79; SI Appendix, Fig. S1 A and B). Electron microscopy showed that both the tadpole-shaped and spherical cells in the newly obtained population of G. partita did not possess any cell wall (Fig. 2C and SI Appendix, Fig. S1C), in contrast to the cells of the original culture, which have a thick cell wall (70 nm thick), as also observed in several strains of G. sulphuraria (1, 20) (Fig. 2C and SI Appendix, Fig. S1C). However, after cell division, daughter cells were surrounded and connected by an extracellular matrix (SI Appendix, Fig. S1C). Consistent with this observation, the cellular content was easily extracted by drying and rehydrating the newly generated cell wall–less population (Fig. 2 D, N; the blue in the supernatant is a photosynthetic pigment, phycocyanin extracted from the chloroplast) in contrast to the original form (Fig. 2 D, 2N). Electron microscopy also showed that the tail of the tadpole-shaped cell contained filamentous structures (SI Appendix, Fig. S1C). Nuclear DNA staining with SYTOX Green (SI Appendix, Fig. S1D) and subsequent flow cytometric analysis showed that the nuclear DNA content of the newly generated cell wall–less form is about half of that of the original cell-walled form (Fig. 2E). These results indicate that the cell-walled and cell wall–less forms are diploid and haploid, respectively, and that both diploid and haploid can proliferate asexually.
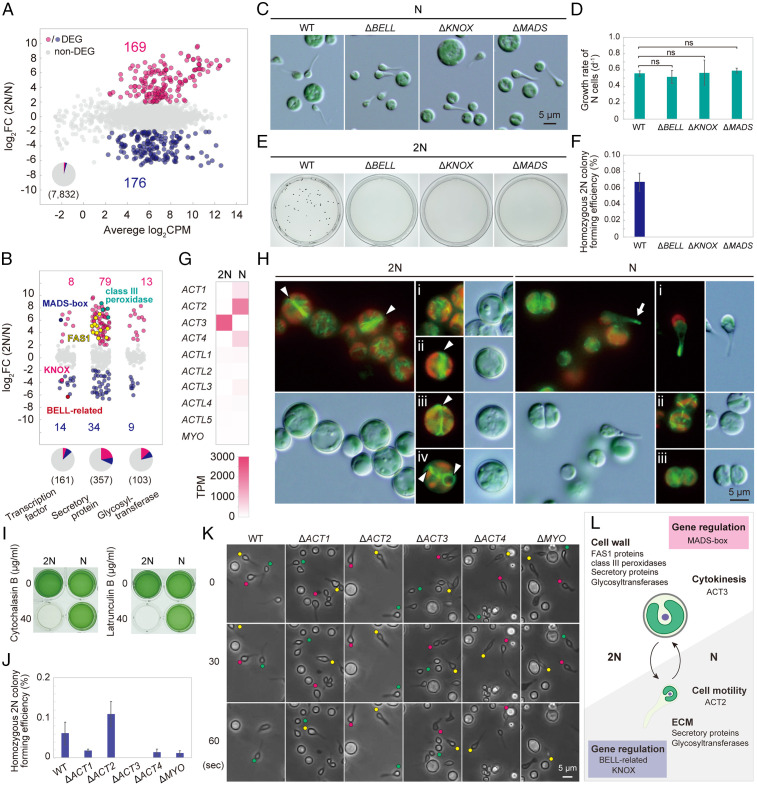
Fig. 2.
The generation of the cell wall–less haploids from cell-walled diploids of G. partita. (A) Micrographs of the original diploid (2N) clone of G. partita obtained from the Biological Resource Center, NITE, and haploid (N; clone N1) cells. The black arrow indicates the mother cell wall released upon hatching of daughter cells, and the black arrowhead indicates tadpole-shaped cells. See also SI Appendix, Fig. S1 A and B, for other Galdieria spp. (B) Proliferation mode of 2N (Movies S1 and S2) and N (Movies S2–S6) cells based on time-lapse observations. (C) Electron micrographs of 2N and N cells; cm, cell membrane; cw, daughter cell wall; mcw, mother cell wall. More details are indicated in SI Appendix, Fig. S1C. Fluorescent visualization of membranous organelles in 2N and N cells is shown in SI Appendix, Fig. S3. (D) The 2N and N cell pellets were dried, rehydrated with phosphate-buffered saline, and centrifuged. (E) Flow cytometric analysis of 2N and N cells stained with SYTOX Green. Images by fluorescence microscopy are shown in SI Appendix, Fig. S1D. (F) Mapping of SNP/indel positions in the original 2N and five distinct N clones. Sites that match the reference genome sequence (haploid clone N1) are shown in white, heterozygous SNPs/indels are shown in red, and homozygous SNPs/indels are shown in blue. (G) Growth curves of 2N and N cells in rotating suspension cultures under photoautotrophic (in an inorganic medium in the light) and heterotrophic (in a medium supplemented with glucose in the dark) conditions. Data represent the mean and SD of three independent cultures. Growth rates of 2N and N cells at various pH values are shown in SI Appendix, Fig. S1E.
To further advance the analysis at the whole-genome level, we determined the nuclear (∼17.8 Mb and 7,832 protein-coding genes), chloroplast, and mitochondrial genome sequences of a haploid clone (clone N1; SI Appendix, Table S1 and Fig. S2 A and B). The nuclear genome contained numerous duplicated regions (SI Appendix, Fig. S2C). Numerous heterozygous single nucleotide polymorphisms (SNPs)/insertions/deletions (indels) were detected when genomic reads of the original diploid form were mapped to the genome of the haploid clone N1, indicating that the original clone was a heterozygous diploid (Fig. 2F). Different SNP/indel patterns were also detected depending on the clone when genomic reads of other haploid clones (N2 through N5) were mapped to the haploid clone N1 genome, indicating segregation of recombined chromosomes into haploid cells (Fig. 2F).
A motile tadpole-shaped cell transformed into a nonmotile spherical cell (Movie S4), which grew and multiplied by successive cell divisions during haploid asexual reproduction (Movie S5). Eventually, the spherical daughter cells detached and again transformed into motile tadpole-shaped cells (Fig. 2B and Movie S6). No obvious differences in the number and morphology of intracellular organelles were observed between the diploid and haploid cells other than the presence or absence of a cell wall by fluorescence (SI Appendix, Fig. S3) and electron microscopy (SI Appendix, Fig. S1C). The absence of a cell wall in the haploid form probably enables the haploid gamete to move and undergo mating (cell fusion), as observed in several lineages of eukaryotes (24).
We then examined the physiological differences between diploid and haploid cells. Both forms proliferated by asexual reproduction, photoautotrophically in an inorganic medium and heterotrophically in a medium supplemented with glucose (Fig. 2G). The optimal pH for the growth of diploid and haploid cells was between 0.5 and 3.0 and between 0.5 and 1.0, respectively (SI Appendix, Fig. S1E). Thus, the diploid cells observed in the natural habitat exhibit a broader environmental fitness.
Development of Procedures for Genetic Modification in the Cell Wall–Less Haploid of G. partita.
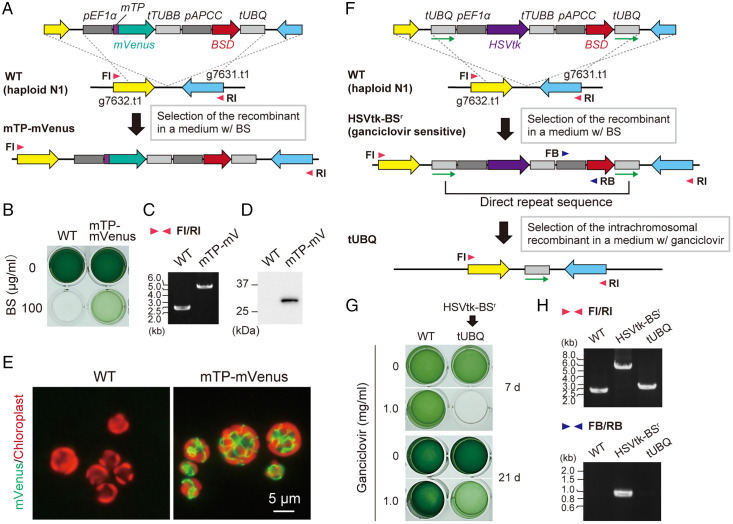
By using the cell wall–less haploid cells, we succeeded in genetic modification of G. partita by introducing exogenous DNA using a polyethylene glycol (PEG)–mediated method previously developed in Cyanidioschyzon merolae 10D, a cell wall–less member of the class Cyanidiophyceae (25), as follows. Linear DNA encoding a mitochondria-targeted mVenus (mTP-mVenus) and a blasticidin S (BS) deaminase selectable marker (BSD) were introduced into the haploid clone N1 and integrated into a chromosomal intergenic region by homologous recombination (Fig. 3A). In the transformant selected in an inorganic medium with BS, mitochondrial transit peptide (mTP)–mVenus was stably expressed (Fig. 3 B–E). We also generated several other transformants in which mVenus was localized in distinct membranous organelles (SI Appendix, Fig. S3). Transgenes have been expressed without any silencing activity, which often obstructs genetic modification in eukaryotic algae (26).
Fig. 3.
Genetic manipulation of G. partita haploids and selectable marker removal. (A) DNA encoding an mTP was fused to mVenus orf. The fusion orf was connected with EF1a promotor (pEF1 a) and β-TUBULIN terminator (tTUBB). BSD orf was connected with an APCC promotor (pAPCC) and UBIQUITIN terminator (tUBQ). These gene cassettes were integrated into a chromosomal intergenic region between g7632.t1 and g7631.t1 loci by homologous recombination in haploid (N) clone N1. (B) The transformed cells (mTP–mVenus) were selected in an inorganic medium with BS in the light. (C) The targeted integration of the transgenes into the chromosome was confirmed by PCR using primers FI and RI. (D and E) Expression of mTP–mVenus was confirmed by immunoblotting with an anti-green fluorescent protein antibody (D) and fluorescence microscopy (E); green, mVenus fluorescence; red, chloroplast fluorescence. The wild type (WT) served as a negative control. (F) A schematic diagram of the targeted integration and subsequent removal of the BSD selectable marker. The HSVtk suicide marker, connected with pEF1a and tTUBB, and the BSD selectable marker, connected using pAPCC, were sandwiched between a directly repeated tUBQ (indicated by green arrows). This construct was integrated into the intergenic region between g7632.t1 and g7631.t1 loci of wild-type haploid clone N1 by homologous recombination. After selecting the transformant (HSVtk-BSr) in the presence of BS, the selectable marker was removed through homologous recombination between two copies of tUBQ and selection with ganciclovir, which is converted to a toxic product by HSVtk. (G) The HSVtk-BSr cells were cultured photoautotrophically for 21 d in the presence or absence of ganciclovir. Wild-type clone N1 served as a negative control. (H) Confirmation of the recombination events by PCR using the primers indicated by arrowheads in F (FI, RI, FB, and RB).
Furthermore, we developed a procedure to remove the BSD selectable marker from a transformant by a combination of the herpes simplex virus thymidine kinase (HSVtk) suicide marker (27), which converts ganciclovir into a toxic product, and intrachromosomal recombination (Fig. 3 F–H). Using this system, multiple chromosomal loci can be edited in a stepwise manner (shown later), and, for industrial use, genetically modified lines can, in some cases, be converted to self-cloning lines that do not contain any heterologous DNA sequence.
Generation of Homozygous Diploids by Self-Diploidization and Heterozygous Diploids through Isogamous Mating.
Haploid cells can be converted into diploid cells with cell walls by self-diploidization upon acetic acid stress or by mating between different haploid clones. Diploid cells generated through either case were stably maintained in medium at pH 2.0 once formed.
When a single haploid clone was exposed to acetic acid stress, it transformed into a homozygous diploid cell (SI Appendix, Fig. S4), although it is unclear how the stress induces diploidization at this point. The homozygous diploids lack interallelic polymorphism, which facilitates a comparison between the diploid and haploid, as shown later.
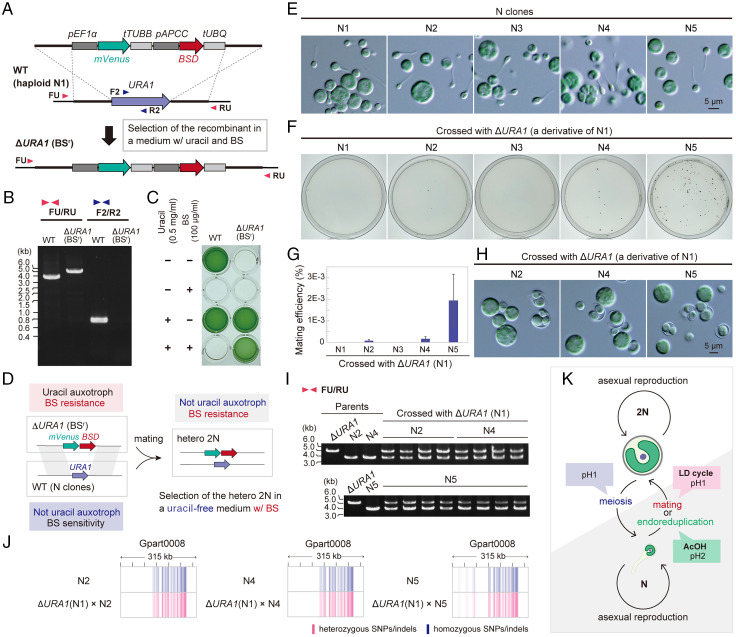
Then, to investigate whether mating between different haploid clones generates heterozygous diploids, we constructed a uracil-auxotrophic haploid N1 by replacing the chromosomal URA1 (dihydroorotate oxidase) locus with mVenus-BSD cassettes (Fig. 4 A–C). This way, we were able to select heterozygous diploids that possessed both BSD and URA1 (thus BS resistant but not uracil auxotrophic) after haploid N1 (ΔURA1) and other haploid clones (wild type) mated (Fig. 4D). No colonies appeared when haploid N1 (ΔURA1) and N1 or N3 (wild type) were cocultured under a 12-h light/12-h dark cycle for 7 d in a medium supplemented with uracil for mating and subsequently spread on BS plate medium without uracil to select mated diploids (Fig. 4 E–G). However, crosses between haploid N1 (ΔURA1) and haploid N2, N4, or N5 (wild type) generated diploid colonies with cell walls (Fig. 4 E–H). The heterozygosity of the obtained diploid clones was confirmed by PCR amplifying the URA1 locus (Fig. 4I) and by whole-genome resequencing (Fig. 4J). The results suggest that G. partita possesses two complementary mating types (type 1, N1 and N3; type 2, N2, N4, and N5) of the same cell size and morphology (Fig. 4E) and undergoes isogamous sexual reproduction (Fig. 4K).
Fig. 4.
Generation of homozygous and heterozygous diploids of G. partita. See also SI Appendix, Fig. S4, for the generation of homozygous diploids. (A) To select heterozygous diploids, a uracil-auxotrophic BS-resistant haploid was generated. To knock out URA1 and provide BS resistance to G. partita haploid clone N1, mVENUS orf, connected with pEF1a and tTUBB, and BSD selectable marker, connected with pAPCC and tUBQ, were integrated into the chromosomal URA1 locus of haploid clone N1 by homologous recombination. (B) Replacement of the chromosomal URA1 orf with the mVENUS and BSD markers in the resultant ΔURA1 (BSr) clone was confirmed by PCR with the primers FU, F2, RU, and R2, indicated by the arrowheads in A. The wild-type haploid clone N1 (WT) served as a control. (C) The uracil auxotroph and BS resistance of the ΔURA1 (BSr) clone was confirmed through photoautotrophic cultivation (for 7 d) of the cells in the presence or absence of uracil and BS. Wild type served as a control. (D) A scheme showing the selection of heterozygous diploids, which possess both BSD marker and the URA1 gene, after mating ΔURA1 (BSr) haploid clone with a wild-type haploid clone. (E) Micrographs of the haploid clones N1 through N5 obtained from the original diploid clone. (F) ΔURA1 (BSr) N clone N1 was crossed with wild-type N clones, and heterozygous diploids (2N) were selected on gellan gum–solidified medium with BS. (G) Mating efficiency of respective combinations of N clones. Data represent the mean and SD of three independent experiments. (H) Micrographs of the cells in a colony of respective combinations. (I) Heterozygosity was confirmed by PCR using primers FU and RU (primer positions are indicated in A). (J) Confirmation of heterozygosity by mapping SNP/indel positions of heterozygous 2N clones. Sites that match the reference genome sequence (N clone N1) are shown in white, heterozygous SNPs/indels are shown in red, and homozygous SNPs/indels are shown in blue. (K) Schematic representation of the generation of homozygous and heterozygous 2N.
Regarding the self-diploidization (generation of homozygous diploids) described above, there are two known ways in other eukaryotes: homothallism (mating type switching) (28) and endoreduplication (nucleus replicates its DNA without division) (29). For example, the homothallic unicellular green alga Chlamydomonas monoica undergoes intrastrain differentiation into opposite mating types, which then mate (28). The generation of homozygous diploids also occasionally happens in cultures of the budding yeast by mating type switching. However, in yeast, endoreduplication also occasionally occurs, generating homozygous diploids, and the frequency of endoreduplication is higher than mating type switching (29). In G. partita, all five haploid clones (N1 through N5) generated homozygous diploids in their respective clonal cultures upon acetic acid stress (SI Appendix, Fig. S4), but N1 (ΔURA1) and N1 (wild type) did not produce a hybrid diploid (Fig. 4 F and G). These results suggest that the self-diploidization observed in G. partita probably occurred through endoreduplication rather than the mating of two clonal cells by mating type switching (Fig. 4K).
Difference in the Transcriptomes between the Diploid and Haploid Cells of G. partita.
The transcriptomes of photoautotrophically grown cells were compared between haploid N1 and homozygous diploid cells derived from N1 (Fig. 5A and Dataset S1) to gain insights into the genetic basis of the phenotypic difference between diploid and haploid cells. The same analysis was conducted between haploid N2 and the homozygous diploid derived from N2, which showed similar results (SI Appendix, Fig. S5A and Dataset S1). As a result, 345 (4.4%) differentially expressed genes (DEGs; false discovery rate [FDR] < 0.01, log counts per million [CPM] > 2, log fold change [FC] > 2 or log FC < −2) were identified (Fig. 5A and Dataset S1).
Fig. 5.
Comparison of transcriptomes and differences in the function of actin genes between diploids and haploids in G. partita. (A) MA plot showing 169 genes up-regulated in diploids (homozygous 2N derived from N1) and 176 genes up-regulated in haploids (N; clone N1) among 7,832 nucleus-encoded genes (DEGs, FDR < 0.01, log CPM > 2, and log FC > 2 or log FC < −2 in three independent cultures). The result of clone N2 is shown in SI Appendix, Fig. S5A. (B) Ratio of mRNA abundance of genes encoding transcription factors, secretory proteins, and glycosyltransferases between 2N and N cells. (C) Micrographs of the wild-type (WT), ΔBELL, ΔKNOX, and ΔMADS N cells. (D) Photoautotrophic growth rate of wild-type, ΔBELL, ΔKNOX, and ΔMADS N cells. Data represent the mean and SD of three independent cultures. (E and F) Efficiency of generation of homozygous 2N colonies on gellan gum plates supplemented with acetate of wild-type, ΔBELL, ΔKNOX, and ΔMADS cells. (G) Comparison of mRNA abundance of actin (ACT), actin-like (ACTL), and MYO genes between 2N and N cells. The phylogenetic relationship of ACT1, ACT2, ACT3, and ACT4 proteins is shown in SI Appendix, Fig. S6A. (H) Actin filaments visualized using Lifeact-mVenus in 2N and N cells; green, Lifeact fluorescence indicating actin filaments; red, chloroplast fluorescence. (I) Comparison of cytochalasin/latrunculin sensitivity between 2N and N cells. (J) Efficiency of generation of homozygous 2N colonies on gellan gum plates supplemented with acetate of the wild type and actin and myosin knockouts. Data represent the mean and SD of three independent cultures. Photographs of the plates and cells are shown in SI Appendix, Fig. S6B. (K) Time-lapse observation of WT, ΔACT, and ΔMYO N cells in static liquid cultures. Tadpole-shaped cells are indicated by yellow, red, and green circles. (L) Schematic summary of the difference between 2N and N cells.
Consistent with the presence of a cell wall in diploid cells, 11 genes encoding fasciclin (FAS1) domain proteins and four class III peroxidase genes were expressed in the diploid but not the haploid cells (Fig. 5B). The FAS1 domain protein is a membrane-anchored glycoprotein involved in building cell wall structures in plants (30). The class III peroxidase is involved in cell wall loosening/hardening in plants and is a major and acid-tolerant protein in the cell wall of G. sulphuraria 074G (a derivative of 074W) (31). Many other genes encoding secretory and glycosyltransferase proteins, many of which are involved in extracellular matrix (32) and cell wall (33) synthesis in algae and plants, were also enriched in the DEGs in diploid or haploid cells, respectively (χ2 test, P < 0.05; Fig. 5B). These secretory proteins and polysaccharides, synthesized by the glycosyltransferase proteins, likely constitute the diploid cell wall and the haploid extracellular matrix, both resistant to a highly acidic environment.
In addition, the DEGs included genes encoding BAR-domain proteins (Dataset S1). The BAR-domain proteins are expected to be components of eisosomes, which are trough-shaped invaginations of the cell membrane whose functions are unclear (20). Among microalgae, eisosomes have been observed only in cell-walled species, including G. sulphuraria CCMEE 5587.1 (20). The G. partita genome consists of ten genes encoding BAR-domain proteins, of which seven genes are expressed in both diploid and haploid cells and three are predominantly expressed in haploid cells. Because BAR genes are expressed in haploid cells that possess an extracellular matrix, they are likely related to both the cell wall in the diploid (seven genes) and the extracellular matrix in the haploid (ten genes).
The Haploid-Specific BELL and KNOX TALE-homeodomain and the Diploid-Specific MADS-box Genes Are Required for the Haploid-to-Diploid Transition in G. partita.
In the DEGs, transcription factors were enriched (χ2 test, P < 0.05), including a haploid-specific BELL-related gene (only a single BELL-related gene encoded in the genome), a KNOX gene (KNOX-Red1 described below, one of the two KNOX genes encoded in the genome), and a diploid-specific MEF2-type MADS-box gene (only a single MADS-box gene encoded in the genome) (Fig. 5B).
BELL-related and the KNOX TALE-homeodomain family of transcription factors regulate haploid-to-diploid transitions in green algae and organ differentiation in land plants (34). Recent research showed that the mRNA level of the KNOX gene changes according to life cycle transitions in the multicellular red alga Pyropia yezoensis (Bangiophyceae in Fig. 1) (35). In the unicellular green alga Chlamydomonas reinhardtii, the BELL-related (GSP1) or KNOX (GSM1) gene is expressed only in mating-type–plus or mating-type–minus gametes, respectively, and the two proteins heteromerize, trigger nuclear and other organellar fusions between the two mating types (36), and activate diploid gene expression after mating (37). As previously reported in G. sulphuraria 074W (38), the G. partita genome encodes two KNOX proteins, KNOX-Red1, which is closely related to the KNOX (GSM1) gene of Viridiplantae, KNOX-Red2, and one BELL protein. In contrast to the green alga C. reinhardtii, both BELL and KNOX (KNOX-Red1) genes were expressed in the same mating type in G. partita (clones N1 and N2; Fig. 5B and SI Appendix, Fig. S5B and Dataset S1). In plants, MIKC-type MADS-box genes also regulate development, including male and female gametophyte development (39). MIKC-MADS genes are conserved in charophycean algae and land plants and evolved from MEF2-type MADS-box genes, which exist in green and red algae, by acquiring additional domains (I region and K domain) (39). When the haploid-specific BELL or KNOX gene or the diploid-specific MADS gene was disrupted (SI Appendix, Fig. S5D), ΔBELL, ΔKNOX, and ΔMADS haploid cells exhibited little difference in morphology and growth rate from the wild type (Fig. 5 C and D). However, these mutant haploids could not undergo self-diploidization (Fig. 5 E and F), suggesting that BELL, KNOX, and MADS genes are required for haploid-to-diploid transition in G. partita. These expression patterns and functions of BELL, KNOX, and MADS genes in G. partita likely reflect their ancestral roles in Archaeplastida.
Actin-Dependent but Myosin-Independent Cytokinesis of the Diploid and Motility of the Haploid.
Actin and myosin are involved in cellular motility and contraction of cells during cell division in eukaryotes (40). The G. partita genome encodes four actin genes, ACT1, ACT2, ACT3, and ACT4. The phylogenetic analysis suggested that ACT1 and ACT2 proteins branched in the ancestor of Galdieria and are closely related to proteins from other red algal lineages (SI Appendix, Fig. S6A), whereas ACT3 and ACT4 are closely related to each other and are specific to Cyanidiophyceae (SI Appendix, Fig. S6A). The transcriptome analysis showed that ACT1, ACT2, and ACT4 are specifically expressed in haploid cells, and ACT3 is specifically expressed in diploid cells (Fig. 5G and SI Appendix, Fig. S5C and Dataset S1). In contrast to multiple actin genes, the genome encodes only one myosin gene (MYO) belonging to a previously uncharacterized member of the myosin family found only in Galdieria spp. and a cryptophyte alga (41). MYO was expressed both in the diploid and haploid cells, although the mRNA level was very low (Fig. 5G and SI Appendix, Fig. S5C and Dataset S1).
Labeling F-actin with Lifeact-mVenus (which labels all four types of actin proteins) (42) showed that actin rings form at the cell division plane in diploid cells (Fig. 5 H, 2N, stages ii to iv) but not in haploid cells (Fig. 5 H, N, stages ii to iii). In the haploid, F-actin was enriched in the tips of protrusion of the tadpole-shaped cells (Fig. 5 H, N, stage i). Consistent with this observation, haploid, but not diploid, cells could proliferate in the presence of the actin polymerization inhibitors cytochalasin B and latrunculin B (Fig. 5I), suggesting that cytokinesis of diploid, but not haploid, cells depends on F-actin.
To correlate the difference in expression of actin genes and the phenotypes between the diploid and haploid cells, ACT and MYO genes, respectively, were disrupted in the haploid clone N1 (SI Appendix, Fig. S5D) and further converted into diploids by self-diploidization (SI Appendix, Fig. S6 B and C). ΔACT3 could not generate diploid cells (Fig. 5J and SI Appendix, Fig. S6 B and C), and ΔACT2 lost the motility of the tadpole-shaped haploid cells (Fig. 5K and Movie S7), consistent with the specific expression of ACT3 and ACT2 in diploid and haploid cells, respectively. By contrast, other ACT and MYO mutants did not exhibit obvious phenotypes (Fig. 5 J and K and SI Appendix, Fig. S6 B and C and Movie S7). These results suggest that cell division in the diploid cells and cell motility in the haploid cells require ACT3 and ACT2, respectively (Fig. 5L). Furthermore, it is also suggested that actin-based cell division and cell motility do not require myosin in G. partita. These results agree with the hypothesis that cytokinesis of the earliest eukaryotes and some lineages of extant eukaryotes might somehow involve actin but not myosin, unlike that performed by the contractile actomyosin (actin and type II myosin) ring (43). Because the actin ring is formed in the cell-walled diploid cells but not cell wall–less haploid cells, the role of the cytokinetic actin ring in the ancestor of Archaeplastida is likely related to cell wall ingression at the cleavage furrow. This assumption is also consistent with the observation that the actin ring forms during cytokinesis in the cell-walled Cyanidium but not in the cell wall–less C. merolae in Cyanidiophyceae (17, 44).
Generation of Photosynthesis-Deficient Mutants and Blue-Colored Cells for Pigment Production.
Chloroplasts descended from a cyanobacterial ancestor; however, they were gradually remodeled during algal and plant evolution (45, 46). Red algae and cyanobacteria possess phycobilisomes as light-harvesting antennas associated with photosystem II, unlike in Viridiplantae, which lost them during evolution. Like Viridiplantae, red algae possess the light-harvesting complex associated with photosystem I, whereas cyanobacteria do not. Thus, the red algal photosynthetic apparatus exhibits an intermediate character between that of cyanobacteria and Viridiplantae (45, 46).
The cyanidialean unicellular red alga C. merolae is genetically tractable; however, this species is an (ecologically) obligate photoautotroph. Recent studies showed that C. merolae could grow heterotrophically in the dark for a limited period (∼six generations) in medium supplemented with a high concentration (≥200 mM) of glycerol. However, the growth rate under artificial heterotrophic conditions is slower than under photoautotrophic conditions, and a daily light pulse is required for continuous heterotrophic growth (47). Another study showed that transgenic C. merolae strains, in which a plasma membrane sugar transporter of G. sulphuraria 074G is expressed, could grow heterotrophically in the dark in a medium supplemented with glucose. However, the transformants could grow in the presence of a photosynthetic inhibitor but required light for efficient heterotrophic growth (48). Thus, studies generating/using photosynthesis-deficient mutants are still not feasible.
By contrast, the generation of such mutants is probably feasible in Galdieria because the cells can grow heterotrophically, as are photosynthesis-deficient mutants of the green alga C. reinhardtii in medium supplemented with acetate as an organic carbon source (49). In addition, the phycocyanin of Cyanidiophyceae is stable at a lower pH and higher temperature (50, 51) and is thus likely tolerant to pasteurization compared with the cyanobacterium Spirulina (Arthrospira platensis), which has been used as a natural blue colorant in certain food products (16). By developing a blue-colored, cell wall–less, photosynthesis-deficient Galdieria mutant, where photosynthetic pigments other than phycocyanin are depleted, the costs of extraction and purification of phycocyanin would be reduced. The blue-colored mutant would be depleted in chlorophyll (a green pigment harvesting light energy for photosynthesis) and carotenoids (yellow and orange pigments involved in photoprotection by excess energy dissipation and radical quenching in the photosynthetic apparatus) (45).
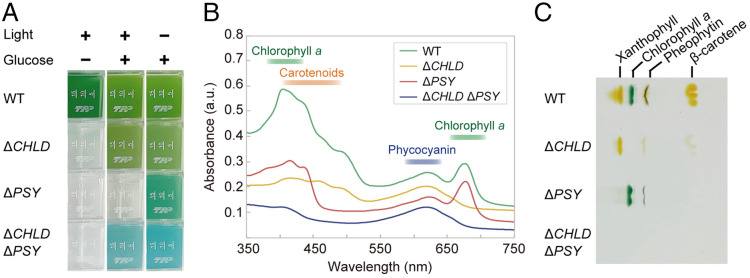
To test these possibilities, we generated single and double knockouts of the CHLD (magnesium chelatase subunit D) and PSY (phytoene synthase) genes, which are key enzymes in the chlorophyll (52) and carotenoid (53) synthetic pathways, respectively (Fig. 6 and SI Appendix, Fig. S7 A–C; ΔCHLD ΔPSY was generated by disrupting these genes one by one by the procedure shown in Fig. 3 F–H). All three mutants grew heterotrophically in the dark in a medium supplemented with glucose (Fig. 6A and SI Appendix, Fig. S7D). As expected, ΔPSY exhibited a more bluish-green color than the wild type, and ΔCHLD ΔPSY exhibited a blue color similar to that of phycocyanin (Fig. 6A), the level of which was comparable to the wild type (SI Appendix, Fig. S7E). Consistent with this observation, absorption spectrometry and thin-layer chromatography (TLC) analyses showed absence of chlorophyll, carotenoids, and both in ΔCHLD, ΔPSY, and ΔCHLD ΔPSY, respectively (Fig. 6 B and C and SI Appendix, Fig. S7E).
Fig. 6.
The generation of photosynthesis-deficient mutants and blue algal culture by genetic modification of G. partita. (A) Liquid cultures of the wild-type (WT), ΔCHLD, ΔPSY, and ΔCHLD ΔPSY haploid (N) in an inorganic medium in the light and in an organic (supplemented with 100 mM glucose) medium in the light or dark. The growth rate is shown in SI Appendix, Fig. S7D. (B and C) Absorption spectrum (B) and TLC (C) analysis of wild type, ΔCHLD, ΔPSY, and ΔCHLD ΔPSY N cells that are cultured heterotrophically (in a medium supplemented with glucose in the dark). The chlorophyll, carotenoid, and phycocyanin contents are shown in SI Appendix, Fig. S7E.
Although the cell contained chlorophyll and phycocyanin, in addition to ΔCHLD and ΔCHLD ΔPSY, ΔPSY did not grow photoautotrophically (Fig. 6A and SI Appendix, Fig. S7D). ΔCHLD and ΔCHLD ΔPSY, but not ΔPSY, grew under light circumstances in a medium supplemented with glucose (Fig. 6A and SI Appendix, Fig. S7D). These results are consistent with the result of a PSY mutant of the green alga C. reinhardtii (54) and indicate that the presence of chlorophyll, but not carotenoids, under light circumstances causes cell death also in red algae, although the structure of the photosynthetic apparatus is different between green and red algae.
The Life Cycle and Functional Genomics of Galdieria as a Platform for Elucidating Evolution of Archaeplastida and Industrial Use of Microalgae.
Cyanidiophyceae branched at an early point in the evolution and diversification of Archaeplastida. In this study, we have elucidated the life cycle of Galdieria, prepared genomic and transcriptomic information, and developed procedures for genetic modification in G. partita, which serves as a powerful platform for studying the evolution of photosynthetic eukaryotes based on the following features. 1) Sexual reproduction, the transition between haploid and diploid, and both the autotrophic and heterotrophic growth are operated by the relatively simple genome (7,832 genes). In addition, cellular architecture is also relatively simple. 2) Both the diploid and haploid can proliferate asexually. 3) In the genetic modification, transgenes are stably expressed without any silencing activity. 4) The heterotrophic growth capacity allows the generation and analysis of photosynthesis-deficient mutants.
Galdieria haploids and diploids could proliferate asexually, and we could obtain homozygous diploids by endoreduplication of haploid clones in addition to heterozygous diploids by the mating of different haploid clones under laboratory conditions. However, only diploids have been observed in natural habitats, and the original G. partita strain isolated from the natural habitat was a heterozygous diploid. Thus, in the natural habitat, the diploid is formed by the mating and dominant phase, and the haploids are most likely to emerge in a limited environment and time.
Regarding other members of Cyanidiophyceae, C. merolae is the best studied and the only member that lacks a cell wall. Among the haploid- or diploid-specific genes/proteins of G. partita discussed above, the C. merolae genome encodes a single copy of FAS1 domain protein (diploid specific, likely involved in cell wall synthesis in G. partita; CMI147C in C. merolae), BELL, KNOX1 (haploid specific, required for haploid-to-diploid transition in G. partita; CMR176C and CMR153C, respectively, in C. merolae), and MADS (diploid specific, required for haploid-to-diploid transition in G. partita; CMA095C in C. merolae). Among them, however, FAS1 and MADS are not expressed in C. merolae based on available genome/transcriptome datasets (5). In addition, the C. merolae genome encodes an unexpressed actin gene (CMM237C); in G. partita, ACT3 is expressed specifically in the diploid. Based on these observations, the cell wall–less C. merolae might represent a haploid stage of an as-yet-identified cell-walled diploid, although further studies are required to test this assumption.
Red algae evolved a multicellular system of development and reproduction independently from Viridiplantae (Fig. 1) (55, 56). Sexuality and transition between haploid and diploid have been observed in unicellular green algae, which share a common ancestor with land plants (Fig. 1) (3, 4). Due to lack of information on sexual reproduction of unicellular red algae so far, studies on the evolution of the sexual life cycle in photosynthetic eukaryotes have been limited to Viridiplantae (34, 39). In this regard, the haploid-specific expression of BELL and KNOX genes, the diploid-specific expression of the MADS gene, and the involvement of these genes in the haploid-to-diploid transition in G. partita suggest that the life cycle based on these genes was already developed in a unicellular common ancestor of red algae and Viridiplantae.
In terms of industrial application, Galdieria grows to a very high density in acidic medium, reducing the risk of microbial contamination. However, the diploid cells studied and developed thus far possess a rigid cell wall that requires mechanical disruption to release the cellular contents (57). By contrast, the haploid cells lack a cell wall, thus making cell disruption less energy/cost intensive. In addition, the cell wall–less haploid is genetically tractable, and genetic modification by self-cloning is also feasible. Thus, the procedures to control the life cycle and genetic modification in Galdieria developed here also provide a useful platform for the biotechnological use of microalgae.
Materials and Methods
Algal Culture.
Generally, algal cells were cultured photoautotrophically in MA medium (an inorganic medium) (58) in the presence of light unless otherwise indicated (e.g., cultivation of photosynthesis-deficient mutants under heterotrophic or mixotrophic conditions in MA medium supplemented with glucose). The diploid cells (i.e., known cell-walled form of Galdieria spp.) were cultivated at pH 2.0 (the pH is indicated by Biological Resource Center, NITE [NBRC]). By contrast, the haploid cells (cell wall–less form found in this study) were cultivated at pH 1.0 to prevent self-diploidization unless otherwise indicated (e.g., the diploid and haploid growth rates were compared at pH 1.0). The pH of the medium was adjusted with sulfuric acid.
G. partita NBRC102759, G. sulphuraria NIES-550, and G. sulphuraria SAG108.79 were obtained from NBRC, the Microbial Culture Collection at the National Institute of Environmental Studies, and the Culture Collection of Algae at Goettingen University, respectively. A single cell of a given strain was isolated from the stock cultures under an inverted microscope and transferred into 1 mL of MA medium at pH 2.0. Each isolated cell was cultured in one well of a 24-well plate (92424; TPP Techno Plastic Products) statically in a 2% CO2 incubator at 42 °C in the light (50 μmol photons m−2 s−1) to generate a clonal heterozygous diploid population of respective strains (defined as original heterozygous diploid clones). The diploid clones of individual strains were maintained in MA medium at pH 2.0.
To generate haploid from diploid clones described above, the cells in MA medium at pH 2.0 were transferred to MA medium at pH 1.0 to give a concentration of 5 cells/mL and cultivated in a 24-well plate (1 mL/well) statically in a CO2 incubator in the light as described above for 1 wk. The resulting culture contained tadpole-shaped haploid cells, and a single tadpole-shaped cell was isolated and transferred into 1 mL of MA medium at pH 1.0. The isolated cell was cultured in one well of a 24-well plate statically in a CO2 incubator in the presence of light above to generate a clonal haploid population. The haploid clones of respective strains were maintained in MA medium at pH 1.0. For G. partita, cultures of five independent haploid clones (N1, N2, N3, N4, and N5) were generated from the original diploid clone, respectively.
To generate a homozygous diploid from a G. partita haploid clone, 1 × 105 cells of a growing haploid culture in MA medium at pH 1.0 were spread on a gellan gum-solidified MA medium at pH 2.0 (0.5% gellan gum, 9-cm Petri dish) supplemented with 1 mM sodium acetate. The cells were then cultured in a CO2 incubator in the light as above for 3 wk, and a single colony on the medium was isolated as a homozygous diploid clone. The diploid colony formation efficiency was determined as the number of colonies per number of cells spread on a gellan gum–solidified plate medium. The homozygous diploid clones were maintained in MA medium at pH 2.0.
To generate and select a heterozygous diploid clone by crossing two different G. partita haploid clones, growing cells of the haploid clone N1 ΔURA1 (BSr; SI Appendix, Material and Methods) and one of the wild-type haploid clones N1, N2, N3, N4, or N5 were inoculated into 1 mL of MA medium at pH 1.0 supplemented with 0.5 mg/mL uracil to give a concentration of optical density at 750 nm (OD750) of 0.1 for each clone (OD750 = 0.2). Cells of two different haploid clones were cocultured in one well of a 24-well plate enclosed in an AnaeroPack CO2 generator (Mitsubishi Gas Chemical) statically in an incubator at 42 °C under 12-h light/12-h dark conditions (100 μmol photons m−2 s−1) for 1 wk for haploid cell mating. The cells were then harvested by centrifugation at 1,500 × g for 5 min and resuspended in 1.05 mL of MA medium at pH 2.0; 50 µL was used for counting the cell numbers, and the remaining 1 mL was spread on a gellan gum–solidified MA medium at pH 2.0 (9-cm Petri dish, 0.5% gellan gum) supplemented with 100 μg/mL BS hydrochloride (FUJIFILM Wako Pure Chemical Corporation). The cells were then cultured in a CO2 incubator in the presence of light as described above to generate colonies of heterozygous diploid, which shows BS resistance but not uracil auxotrophy (Fig. 4F). Mating efficiency was determined as the number of colonies per number of cells spread on a gellan gum–solidified plate medium. PCR checked the heterozygosity of respective clones (primers are listed in Dataset S2). The heterozygous diploid clones were maintained in MA medium at pH 2.0.
To compare the growth rates between the diploid and haploid cells of G. partita, growing cells of the original heterozygous diploid and haploid clones N1, respectively, were inoculated into 20 mL of MA medium with or without 100 mM glucose at pH 1.0. The cells were cultured in 25-cm2 tissue culture flasks (90026, TPP Techno Plastic Products) in an incubator at 42 °C in the light (50 μmol photons m−2 s−1; CO2 incubator) or dark (normal incubator) on a rotary shaker (140 rpm).
To determine the optimal pH condition for diploid and haploid cells of G. partita, the original heterozygous diploid and haploid clones N1 and N2 grown in MA medium at pH 2.0 were inoculated into 1 mL of MA medium at six separate pH values (from pH 0.25 to pH 3.0) to give an OD750 of 0.2, respectively. Individual cells were cultured statically in one well of a 24-well plate in a CO2 incubator in the presence of light for 1 wk.
To compare cytochalasin B/latrunculin B sensitivity between diploid and haploid cells of G. partita, homozygous diploid obtained from N1 and haploid clone N1 grown in MA medium at pH 1.0 were inoculated into 1 mL of MA medium at pH 1.0 supplemented with 40 µg/mL cytochalasin B/latrunculin B to give an OD750 of 0.2, respectively. Respective cells were cultured in one well of a 24-well plate statically in a CO2 incubator in the presence of light for 1 wk.
Genetic Manipulation of G. partita.
PEG-mediated transformation of G. partita haploid clone N1 and its derivatives was performed according to the C. merolae transformation procedure (59) with the following modifications.
To prepare cells for transformation, except for the photosynthesis-deficient mutant (ΔCHLD), haploid cells grown in MA medium at pH 1.0 (OD750 = 1.5 to 3) were inoculated into 50 mL of MA medium at pH 1.0 to give an OD750 of 0.5. Then, the cells were cultured in a 100-mL test tube at 42 °C with aeration (0.3 L 2% CO2 min−1) under a 12-h light/12-h dark cycle (100 μmol photons m−2 s−1). At the end of the third light period, when the percentage of S-phase cells became the highest (60), Tween-20 was added to the culture to give a final concentration of 0.001%, and the cells were harvested by centrifugation at 2,000 × g for 5 min at room temperature. To knockout PSY in ΔCHLD, cells for transformation were prepared as described above, except that MA medium was supplemented with 100 mM glucose.
Cells were then resuspended in MA medium to give an OD750 of 500. To prepare ∼60% (wt/vol) PEG solution, 0.3 g of PEG4000 (Sigma-Aldrich) was dissolved in 225 μL of MA2 medium (an inorganic medium) (59, 61) at 95 °C for 5 min and then kept at 42 °C on a heat block until use. Then, 20 μg of linear DNA (SI Appendix, Material and Methods) was diluted into 45 μL of water. The 45 μL of DNA solution, 5 μL of 10× transformation solution (400 mM [NH4]2SO4, 40 mM MgSO4, 0.3% H2SO4), and 62.5 μL of PEG solution (total of 112.5 µL) were mixed by pipetting in a 1.5-mL tube. A 12.5-μL cell suspension was added to the 112.5 μL of transformation-DNA-PEG mixture (final concentration of PEG was ∼30% [wt/vol]), the tube was vigorously inverted ten times, and the content was immediately transferred to 10 mL of MA medium (for ΔCHLD, ΔPSY, and ΔCHLD ΔPSY supplemented with 100 mM glucose) at pH 1.0 in one well of a six-well culture plate (VTC-P6, VIOLAMO). The six-well culture plate was incubated statically in a CO2 incubator at 42 °C in the presence of light (50 μmol photons m−2 s−1; in the dark for ΔCHLD, ΔPSY, and ΔCHLD ΔPSY) for 3 d. Then, the cells were harvested by centrifugation at 2,000 × g for 5 min and resuspended in 1 mL of MA medium (for ΔCHLD, ΔPSY, and ΔCHLD ΔPSY supplemented with 100 mM glucose) at pH 1.0, and 100 μL of cell suspension was inoculated into 1 mL of MA medium (for ΔCHLD, ΔPSY, and ΔCHLD ΔPSY supplemented with 100 mM glucose) at pH 1.0 supplemented with 100 μg/mL BS. The cells were cultured in one well of a 24-well plate. The 24-well plate was incubated statically in a CO2 incubator at 38 °C in the light (50 μmol m−2 s−1; in the dark for ΔCHLD, ΔPSY, and ΔCHLD ΔPSY) for 3 wk to pick BS-resistant transformants. Single clones were obtained by limiting dilution of the culture in a 96-well plate (92696, TPP Techno Plastic Products).
To remove the HSVtk-BSD marker from a chromosome of a transformant through intrachromosomal homologous recombination, the cells in which the HSVtk-BSD marker was integrated were inoculated into 1 mL of MA medium at pH 1.0 with (for ΔCHLD [HSVtk-BSr] cells) or without (for HSVtk-BSr cells) 100 mM glucose in the presence of 1 mg/mL ganciclovir (TCI) to give an OD750 of 0.2. Then, the cells were cultured in one well of a 24-well plate statically in a CO2 incubator at 38 °C in the light (50 μmol photons m−2 s−1; for HSVtk-BSr line) or in the dark (for ΔCHLD [HSVtk-BSr] line) for 3 wk to select cells that had lost the HSVtk-BSD marker. Single clones were obtained by limiting dilution of the culture in a 96-well plate.
Microscopy.
For observation of G. partita cells using differential interference contrast and fluorescence microscopy, diploid and haploid cells were cultured statically in 20 mL of MA medium at pH 2.0 and 1.0, respectively, in 25-cm2 tissue culture flasks in a CO2 incubator at 42 °C in light (50 μmol photons m−2 s−1) unless otherwise indicated. Images of the cells were captured using a fluorescence microscope (BX51, Olympus) equipped with a three charge–coupled device camera system (DP71, Olympus). To detect mVenus and chloroplast fluorescence, filter sets NIBA (Olympus) and WIG (Olympus) were used, respectively.
For staining with quinacrine to fluorescently label vacuoles in G. partita, diploid and haploid cells cultured as above were harvested by centrifugation at 2,000 × g for 5 min and resuspended in 1 mL of MA medium at pH 2.0. The cells were again centrifuged and resuspended in 1 mL of MA medium at pH 2.0. Then, 0.1 mL of 1 M Tris-HCl (pH 8.0) was added to the sample to neutralize. Quinacrine dihydrochloride was added to give a concentration of 40 µg/mL and incubated for 15 min at room temperature. The stained cells were harvested using centrifugation and resuspended in MA medium at pH 2.0 and observed using fluorescence microcopy. For quinacrine fluorescence detection, the filter set NIBA (Olympus) was used.
For long-term time-lapse imaging of G. partita, original heterozygous diploid clone and haploid clone N1, the cells were cultured in 20 mL of MA medium supplemented with 100 mM glucose at pH 2.0 and 1.0, respectively, in 25-cm2 tissue culture flasks on a rotary shaker (120 rpm) in an incubator at 42 °C in the light (50 μmol photons m−2 s−1) and subjected to time-lapse observation for 5 to 20 h (images were taken every 2 s) according to the method previously developed for observing C. merolae (62).
For short-term time-lapse imaging of G. partita wild-type (clone N1), ΔACT1, ΔACT2, ΔACT3, ΔACT4, and ΔMYO haploid, the cells were cultured in 20 mL of MA medium at pH 1.0 statically in 25-cm2 tissue culture flasks in a CO2 incubator at 42 °C in light (50 μmol photons m−2 s−1). Then, 3 mL of respective cultures was transferred to a 35-mm glass-bottom dish (D11130H, Matsunami). The culture in the dish was kept at 42 °C on ThermoPlate (SX-100, Tokai Hit) and observed for 3 min (images were taken every 0.2 s) using an inverted microscope (CKX41, Olympus).
For observation of G. partita original heterozygous diploid clone and haploid clone N1 using transmission electron microscopy, cells were cultured in MA medium at pH 2.0 and 1.0, respectively, in 25-cm2 tissue culture flasks on a rotary shaker (120 rpm) in a 5% CO2 incubator at 42 °C under light conditions (50 μmol photons m−2 s−1). The cells were harvested using centrifugation and fixed using a high-pressure freezing method (Leica, EM PACT2 and EM AFS2) followed by OsO4 fixation and embedding in Spurr’s low-viscosity resin (63) with a minor modification. Thin sections (∼70-nm thick) were stained using uranyl acetate and lead citrate and examined using a JEM-1400 Plus electron microscope (JEOL).
Supplementary Material
Acknowledgments
We thank Drs. T. Kuroiwa, H. Kuroiwa, H. Nozaki, K. Tanaka, C. Saito, Y. Kashiyama, Y. Kanesaki, Y. Hirose, T. Torisawa, R. Ohbayashi, and Y. Kobayashi and members of the DIC Corporation for their advice and K. Hashimoto, R. Ujigawa, and U. Sugimoto for their technical support. This work was supported by the JST-MIRAI Program of the Japan Science and Technology Agency (grant no. JPMJMI22E1 to S.-y.M.), by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (20K06778 to S.H., 20H00477 to S.-y.M., and 26650016 to A.H.I), and by DIC Corporation (to S.-y.M.).
Footnotes
Competing interest statement: The Japan Science and Technology Agency has filed patent applications related to the generation and maintenance of Galdieria spp. haploid cells on behalf of S.H. and S.-y.M. The National Institute of Genetics and the DIC Corporation have filed patent applications related to the genetic modification on behalf of S.H., T.F., and S.-y.M. All other authors declare they have no competing interests.
This article is a PNAS Direct Submission.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2210665119/-/DCSupplemental.
Data, Materials, and Software Availability
The genome sequence data of G. partita have been deposited in the DNA Data Bank of Japan (DDBJ)/ European Molecular Biology Laboratory (EMBL)/ GenBank (BioProject accession no. PRJDB12742 (64); BioSample accession nos. SAMD00436389–SAMD00436398; Whole Genome Shotgun accession nos. BQMJ01000001–BQMJ01000080; chloroplast DNA accession no. AP025529; mitochondrial DNA accession no. AP025530; DDBJ Sequence Read Archive [DRA] accession no. DRA013290). The RNA-sequencing data of G. partita have been deposited in the DDBJ/EMBL/GenBank (BioProject accession no. PRJDB12743 (65); BioSample accession nos. SAMD00434483–SAMD00434494; DRA accession no. DRA013289). All other data are included in the manuscript and/or supporting information.
References
- 1.Seckbach J., Ed., Evolutionary Pathways and Enigmatic Algae: Cyanidium caldarium (Rhodophyta) and Related Cells (Springer Science+Business Media, Dordrecht, The Netherlands, 1981). [Google Scholar]
- 2.Yoon H. S., Hackett J. D., Ciniglia C., Pinto G., Bhattacharya D., A molecular timeline for the origin of photosynthetic eukaryotes. Mol. Biol. Evol. 21, 809–818 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Adl S. M., et al. , Revisions to the classification, nomenclature, and diversity of eukaryotes. J. Eukaryot. Microbiol. 66, 4–119 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.One Thousand Plant Transcriptomes Initiative, One thousand plant transcriptomes and the phylogenomics of green plants. Nature 574, 679–685 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matsuzaki M., et al. , Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 428, 653–657 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Schönknecht G., et al. , Gene transfer from bacteria and archaea facilitated evolution of an extremophilic eukaryote. Science 339, 1207–1210 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Rossoni A. W., et al. , The genomes of polyextremophilic cyanidiales contain 1% horizontally transferred genes with diverse adaptive functions. eLife 8, e45017 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Umen J., Coelho S., Algal sex determination and the evolution of anisogamy. Annu. Rev. Microbiol. 73, 267–291 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Goodenough U., Lee J.-H., Snell W. J., “The sexual cycle” in The Chlamydomonas Sourcebook, Goodenough U., Ed. (Academic Press, ed. 3, 2022), vol. 1, in press. [Google Scholar]
- 10.Curien G., et al. , Mixotrophic growth of the extremophile Galdieria sulphuraria reveals the flexibility of its carbon assimilation metabolism. New Phytol. 231, 326–338 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barbier G., et al. , Comparative genomics of two closely related unicellular thermo-acidophilic red algae, Galdieria sulphuraria and Cyanidioschyzon merolae, reveals the molecular basis of the metabolic flexibility of Galdieria sulphuraria and significant differences in carbohydrate metabolism of both algae. Plant Physiol. 137, 460–474 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henkanatte-Gederaa S. M., et al. , Removal of dissolved organic carbon and nutrients from urban wastewaters by Galdieria sulphuraria: Laboratory to field scale demonstration. Algal Res. 24, 450–456 (2017). [Google Scholar]
- 13.Sloth J. K., Jensen H. C., Pleissner D., Eriksen N. T., Growth and phycocyanin synthesis in the heterotrophic microalga Galdieria sulphuraria on substrates made of food waste from restaurants and bakeries. Bioresour. Technol. 238, 296–305 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Liu S. L., Chiang Y. R., Yoon H. S., Fu H. Y., Comparative genome analysis reveals Cyanidiococcus gen. nov., a new extremophilic red algal genus sister to Cyanidioschyzon (Cyanidioschyzonaceae, Rhodophyta). J. Phycol. 56, 1428–1442 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Schmidt R. A., Wiebe M. G., Eriksen N. T., Heterotrophic high cell-density fed-batch cultures of the phycocyanin-producing red alga Galdieria sulphuraria. Biotechnol. Bioeng. 90, 77–84 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Lang I., Bashir S., Lorenz M., Rader S., Weber G., Exploiting the potential of Cyanidiales as a valuable resource for biotechnological applications. Appl. Phycol., 10.1080/26388081.2020.1765702 (2020). [Google Scholar]
- 17.Miyagishima S. Y., Tanaka K., The unicellular red alga Cyanidioschyzon merolae-the simplest model of a photosynthetic eukaryote. Plant Cell Physiol. 62, 926–941 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirooka S., et al. , Efficient open cultivation of cyanidialean red algae in acidified seawater. Sci. Rep. 10, 13794 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graziani G., et al. , Microalgae as human food: Chemical and nutritional characteristics of the thermo-acidophilic microalga Galdieria sulphuraria. Food Funct. 4, 144–152 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Lee J. H., Heuser J. E., Roth R., Goodenough U., Eisosome ultrastructure and evolution in fungi, microalgae, and lichens. Eukaryot. Cell 14, 1017–1042 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seckbach J., “Overview on cyanidian biology” in Red Algae in the Genomic Age, Seckbach J., Chapman D. J., Eds. (Springer, The Netherlands, 2010), pp. 345–356. [Google Scholar]
- 22.Graham L. D., Wilcox L. W., Algae (Prentice Hall, Upper Saddle River, NJ, 2000), p. 640. [Google Scholar]
- 23.Qiu H., Price D. C., Yang E. C., Yoon H. S., Bhattacharya D., Evidence of ancient genome reduction in red algae (Rhodophyta). J. Phycol. 51, 624–636 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Goodenough U., Lee J.-H., “Cell walls” in The Chlamydomonas Sourcebook, Goodenough U., Ed. (Academic Press, ed. 3, 2022), vol. 1, in press. [Google Scholar]
- 25.Fujiwara T., Hirooka S., Miyagishima S. Y., A cotransformation system of the unicellular red alga Cyanidioschyzon merolae with blasticidin S deaminase and chloramphenicol acetyltransferase selectable markers. BMC Plant Biol. 21, 573 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neupert J., et al. , An epigenetic gene silencing pathway selectively acting on transgenic DNA in the green alga Chlamydomonas. Nat. Commun. 11, 6269 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Capecchi M. R., Altering the genome by homologous recombination. Science 244, 1288–1292 (1989). [DOI] [PubMed] [Google Scholar]
- 28.Vanwinkle-Swift K. P., Hahn J. H., The search for mating-type-limited genes in the homothallic alga Chlamydomonas monoica. Genetics 113, 601–619 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harari Y., Ram Y., Rappoport N., Hadany L., Kupiec M., Spontaneous changes in ploidy are common in yeast. Curr. Biol. 28, 825–835.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Silva J., Ferraz R., Dupree P., Showalter A. M., Coimbra S., Three decades of advances in arabinogalactan-protein biosynthesis. Front Plant Sci 11, 610377 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oesterhelt C., Vogelbein S., Shrestha R. P., Stanke M., Weber A. P., The genome of the thermoacidophilic red microalga Galdieria sulphuraria encodes a small family of secreted class III peroxidases that might be involved in cell wall modification. Planta 227, 353–362 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Ulvskov P., Paiva D. S., Domozych D., Harholt J., Classification, naming and evolutionary history of glycosyltransferases from sequenced green and red algal genomes. PLoS One 8, e76511 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amos R. A., Mohnen D., Critical review of plant cell wall matrix polysaccharide glycosyltransferase activities verified by heterologous protein expression. Front Plant Sci 10, 915 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bowman J. L., Sakakibara K., Furumizu C., Dierschke T., Evolution in the cycles of life. Annu. Rev. Genet. 50, 133–154 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Mikami K., Li C., Irie R., Hama Y., A unique life cycle transition in the red seaweed Pyropia yezoensis depends on apospory. Commun. Biol. 2, 299 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kariyawasam T., et al. , TALE homeobox heterodimer GSM1/GSP1 is a molecular switch that prevents unwarranted genetic recombination in Chlamydomonas. Plant J. 100, 938–953 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Lee J. H., Lin H., Joo S., Goodenough U., Early sexual origins of homeoprotein heterodimerization and evolution of the plant KNOX/BELL family. Cell 133, 829–840 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Joo S., et al. , Common ancestry of heterodimerizing TALE homeobox transcription factors across Metazoa and Archaeplastida. BMC Biol. 16, 136 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thangavel G., Nayar S., A survey of MIKC type MADS-box genes in non-seed plants: Algae, bryophytes, lycophytes and ferns. Front Plant Sci 9, 510 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kollmar M., Mühlhausen S., Myosin repertoire expansion coincides with eukaryotic diversification in the Mesoproterozoic era. BMC Evol. Biol. 17, 211 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pasha S. N., Meenakshi I., Sowdhamini R., Revisiting myosin families through large-scale sequence searches leads to the discovery of new myosins. Evol. Bioinform. Online 12, 201–211 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Riedl J., et al. , Lifeact: A versatile marker to visualize F-actin. Nat. Methods 5, 605–607 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Onishi M., Umen J. G., Cross F. R., Pringle J. R., Cleavage-furrow formation without F-actin in Chlamydomonas. Proc. Natl. Acad. Sci. U.S.A. 117, 18511–18520 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suzuki K., et al. , Cytokinesis by a contractile ring in the primitive red alga Cyanidium caldarium RK-1. Eur. J. Cell Biol. 67, 170–178 (1995). [PubMed] [Google Scholar]
- 45.Blankenship R. E., Ed., Molecular Mechanisms of Photosynthesis (Wiley, West Sussex, ed. 3, 2002), pp. 1–328. [Google Scholar]
- 46.Allen J. F., de Paula W. B., Puthiyaveetil S., Nield J., A structural phylogenetic map for chloroplast photosynthesis. Trends Plant Sci. 16, 645–655 (2011). [DOI] [PubMed] [Google Scholar]
- 47.Moriyama T., Mori N., Nagata N., Sato N., Selective loss of photosystem I and formation of tubular thylakoids in heterotrophically grown red alga Cyanidioschyzon merolae. Photosynth. Res. 140, 275–287 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Fujiwara T., et al. , Integration of a Galdieria plasma membrane sugar transporter enables heterotrophic growth of the obligate photoautotrophic red alga Cynanidioschyzon merolae. Plant Direct 3, e00134 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li X., et al. , A genome-wide algal mutant library and functional screen identifies genes required for eukaryotic photosynthesis. Nat. Genet. 51, 627–635 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moon M., et al. , Isolation and characterization of thermostable phycocyanin from Galdieria sulphuraria. Korean J. Chem. Eng. 31, 490–495 (2014). [Google Scholar]
- 51.Athané A., et al. , The safety evaluation of phycocyanin-enriched Galdieria sulphuraria extract using 90-day toxicity study in rats and in vitro genotoxicity studies. Toxicology Res. Appl. 4, 1–15 (2020). [Google Scholar]
- 52.Tanaka R., Tanaka A., Tetrapyrrole biosynthesis in higher plants. Annu. Rev. Plant Biol. 58, 321–346 (2007). [DOI] [PubMed] [Google Scholar]
- 53.Ruiz-Sola M. A., Rodríguez-Concepción M., Carotenoid biosynthesis in Arabidopsis: A colorful pathway. Arabidopsis Book 10, e0158 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Santabarbara S., et al. , The requirement for carotenoids in the assembly and function of the photosynthetic complexes in Chlamydomonas reinhardtii. Plant Physiol. 161, 535–546 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Searles R. B., The strategy of the red algal life history. Am. Nat. 116, 113–120 (1980). [Google Scholar]
- 56.Grosberg R. K., Strathmann R., The evolution of multicellularity: A minor major transition? Annu. Rev. Ecol. Evol. Syst. 38, 621–654 (2007). [Google Scholar]
- 57.Eppink M. H. M., et al. , From current algae products to future biorefinery practices: A review. Adv. Biochem. Eng. Biotechnol. 166, 99–123 (2019). [DOI] [PubMed] [Google Scholar]
- 58.Minoda A., Sakagami R., Yagisawa F., Kuroiwa T., Tanaka K., Improvement of culture conditions and evidence for nuclear transformation by homologous recombination in a red alga, Cyanidioschyzon merolae 10D. Plant Cell Physiol. 45, 667–671 (2004). [DOI] [PubMed] [Google Scholar]
- 59.Fujiwara T., Ohnuma M., “Procedures for transformation and their applications in Cyanidioschyzon merolae” in Cyanidioschyzon Merolae: A New Model Eukaryote for Cell and Organelle Biology, Kuroiwa T., et al., Eds. (Springer Nature, Singapore, 2017), pp. 87–103. [Google Scholar]
- 60.Jong L. W., Fujiwara T., Hirooka S., Miyagishima S. Y., Cell size for commitment to cell division and number of successive cell divisions in cyanidialean red algae. Protoplasma 258, 1103–1118 (2021). [DOI] [PubMed] [Google Scholar]
- 61.Ohnuma M., Yokoyama T., Inouye T., Sekine Y., Tanaka K., Polyethylene glycol (PEG)-mediated transient gene expression in a red alga, Cyanidioschyzon merolae 10D. Plant Cell Physiol. 49, 117–120 (2008). [DOI] [PubMed] [Google Scholar]
- 62.Ichinose T. M., Iwane A. H., Long-term live cell cycle imaging of single Cyanidioschyzon merolae cells. Protoplasma 258, 651–660 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ichinose T. M., Iwane A. H., “Cytological analyses by advanced electron microscopy” in Cyanidioschyzon Merolae: A New Model Eukaryote for Cell and Organelle Biology, Kuroiwa T. et al., Eds. (Springer Nature, Singapore, 2017), pp. 129–151. [Google Scholar]
- 64.S. Hirooka et al., Genome sequencing of the unicellular red alga Galdieria partita. National Center for Biotechnology Information. https://www.ncbi.nlm.nih.gov/bioproject/PRJDB12742. Deposited 15 December 2021. [Google Scholar]
- 65.S. Hirooka et al., Transcriptome analysis between diploid and haploid in the unicellular red alga Galdieria partita. National Center for Biotechnology Information. https://www.ncbi.nlm.nih.gov/bioproject/PRJDB12743. Deposited 15 December 2021. [Google Scholar]
- 66.Yoon H. S., Müller K. M., Sheath R. G., Ott F. D., Bhattacharya D., Defining the major lineages of red algae (Rhodophyta). J. Phycol. 42, 482–492 (2006). [Google Scholar]
- 67.Muñoz-Gómez S. A., et al. , The new red algal subphylum Proteorhodophytina comprises the largest and most divergent plastid genomes known. Curr. Biol. 27, 1677–1684.e4 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome sequence data of G. partita have been deposited in the DNA Data Bank of Japan (DDBJ)/ European Molecular Biology Laboratory (EMBL)/ GenBank (BioProject accession no. PRJDB12742 (64); BioSample accession nos. SAMD00436389–SAMD00436398; Whole Genome Shotgun accession nos. BQMJ01000001–BQMJ01000080; chloroplast DNA accession no. AP025529; mitochondrial DNA accession no. AP025530; DDBJ Sequence Read Archive [DRA] accession no. DRA013290). The RNA-sequencing data of G. partita have been deposited in the DDBJ/EMBL/GenBank (BioProject accession no. PRJDB12743 (65); BioSample accession nos. SAMD00434483–SAMD00434494; DRA accession no. DRA013289). All other data are included in the manuscript and/or supporting information.