Significance
Actin plays essential roles in cellular processes such as muscle contraction, cell motility, and cytokinesis. Humans express six noninterchangeable actin isoforms. Mutations in all the isoforms have been linked to devastating human diseases. Despite its significance, there is no reliable method to produce fully native recombinant actin, featuring native posttranslational modifications and free of contamination from endogenous actin of expressing cells. We report a method to obtain high yields of recombinant actin in human cells that addresses the shortcomings of previous methods. This method opens the way to studies that address how differences among actin isoforms dictate their specialized cellular functions and will help define the molecular bases of disease-causing mutations, which should accelerate the development of targeted therapies.
Keywords: recombinant actin, human cell expression, posttranslational modification, actin isoforms
Abstract
Actin is the most abundant protein in the cytoplasm of eukaryotic cells and interacts with hundreds of proteins to perform essential functions, including cell motility and cytokinesis. Numerous diseases are caused by mutations in actin, but studying the biochemistry of actin mutants is difficult without a reliable method to obtain recombinant actin. Moreover, biochemical studies have typically used tissue-purified α-actin, whereas humans express six isoforms that are nearly identical but perform specialized functions and are difficult to obtain in isolation from natural sources. Here, we describe a solution to the problem of actin expression and purification. We obtain high yields of actin isoforms in human Expi293F cells. Experiments along the multistep purification protocol demonstrate the removal of endogenous actin and the functional integrity of recombinant actin isoforms. Proteomics analysis of endogenous vs. recombinant actin isoforms confirms the presence of native posttranslational modifications, including N-terminal acetylation achieved after affinity-tag removal using the actin-specific enzyme Naa80. The method described facilitates studies of actin under fully native conditions to determine differences among isoforms and the effects of disease-causing mutations that occur in all six isoforms.
As a major component of the cytoskeleton, actin plays crucial roles in countless cellular functions, including organelle and cell motility, muscle contraction, and cytokinesis (1). Actin also participates in more protein–protein interactions than most known proteins, including the interaction with itself to form the actin filament (2). The evolutionary pressure to account for so many interactions has made actin one of the most highly conserved proteins in nature, on par only with histones. Actin is also exceptionally abundant, with a cellular concentration of 150 to 200 μM (3), approximately evenly distributed between its monomeric (G-actin) and filamentous (F-actin) forms. Owing to these unique features, eukaryotic cells have evolved specialized pathways for actin folding and posttranslational processing. Actin is an obligate substrate of the hetero-oligomeric eukaryotic chaperonin TRiC, which promotes a stepwise conformational progression of nascent actin to the native state (4). Actin’s N terminus also undergoes co- and posttranslational processing unique among eukaryotic proteins, including the removal of the acetylated initiator methionine and reacetylation by a dedicated N-terminal acetyl transferase, Naa80 (5, 6). Another enzyme, SETD3, is specifically responsible for actin methylation at residue H73, a posttranslational modification (PTM) conserved in animals and plants that is essential for function (7). The unique folding and posttranslational processing requirements of actin may explain why we still do not have a reliable method to obtain recombinant actin in its native form.
Most biochemical studies have used skeletal α-actin purified from muscle tissues (8). While valuable, this method cannot be used to study the many actin mutations now implicated in human diseases (9). Furthermore, while the six actin isoforms expressed in humans share high sequence identity (93 to 99%) (10), they have distinct functions and tissue specificity (11). Early observations showed that cytoplasmic and muscle actin isoforms were not functionally interchangeable (12–14), and more recent data show differences in the localization and activity of cytoplasmic β- and γ-actin (15–21). Biochemical data are also beginning to reveal differences in the polymerization activities of different actin isoforms, both alone and driven by actin assembly factors (20, 22, 23). Reliable isoform comparisons, however, are not available due to the inability to obtain fully native and pure preparations of individual actin isoforms.
Several actin expression methods have been reported, including expression in bacteria (24, 25), yeast (26), insect cells (27–29), and mammalian cells (30) (SI Appendix, Table S1). These methods suffer from at least one of three major limitations. First, they fail to demonstrate complete removal of endogenous actin isoforms from the expressing cells. This is particularly concerning when studying the effects of disease-causing mutations, since small amounts of contaminating endogenous actin can alter the biochemical analyses of mutants. Second, they fail to demonstrate the presence of native PTMs that play essential roles in actin function (31, 32). Insect cells in particular cannot fully account for PTMs occurring in human cells (33, 34). Third, they fail to demonstrate the native-like functionality of the expressed actin. For instance, we show here that a commonly used method of chymotryptic digestion to remove a C-terminal affinity tag produces a variable amount of internally cleaved actin that reduces the polymerization activity. Here, we describe an actin expression method that resolves these issues and is applicable to any actin isoform or mutant. We obtain high yields of actin isoforms in Expi293F cells, a human cell line that grows in suspension to high density and contains the native PTM machinery. The purification protocol uses an N-terminal affinity tag that is specifically removed by cleavage with tobacco etch virus (TEV) protease, followed by N-terminal acetylation (Nt-acetylation) with Naa80. Proteomics and biochemical analyses demonstrate the presence of native PTMs and the functional equivalence of recombinant and tissue-purified actin isoforms. This method facilitates reliable studies of functional differences among actin isoforms and disease-causing actin mutations.
Results
Actin Expression in Expi293F Cells and Separation from Endogenous Actin.
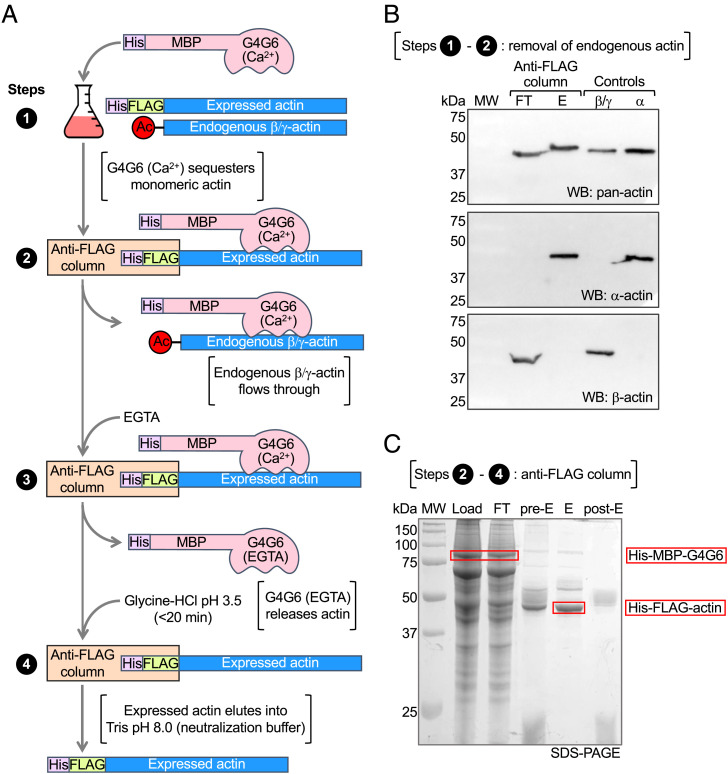
The actin expression and purification method described here uses human Expi293F cells and an N-terminal His-FLAG dual-affinity tag (Materials and Methods). Expi293F cells are derived from human embryonic kidney (HEK) cells and adapted to grow in suspension to high density. These cells can be transiently transfected, express high levels of recombinant protein, and have the endogenous protein folding and PTM machineries necessary for native actin expression. For removal of the affinity tag, a TEV protease cleavage site was added immediately N-terminal to the first amino acid of mature, postprocessed actin, which depending on the actin isoform can be either D or E (10). Cleavage with TEV protease removes the N-terminal tags, leaving no extra amino acids on actin. In nonmuscle cells, such as Expi293F, the most abundant actin isoforms are cytoplasmic β- and γ-actin (35), which must be removed during purification. Endogenous β/γ-actin is separated from recombinant actin by incubating cell lysates overnight with a large amount (1 mg per g of wet cell pellet) of Escherichia coli–expressed His-MBP-G4G6 (Ca2+) (Fig. 1 A, step 1). G4G6 consists of human gelsolin subdomains 4 to 6 (amino acids 434 to 782) and binds monomeric actin in a Ca2+-dependent manner. The use of G4G6 for actin purification was reported previously (36) and adapted here by adding N-terminal dual affinity His and maltose-binding protein (MBP) tags, which were used both for purification and to permit clear separation from actin by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (His-G4G6 and actin run similarly by SDS-PAGE). His-MBP-G4G6 (Ca2+) binds both endogenous and recombinant actin in the monomeric state, which allows for complete separation of the two pools of actin on an anti-FLAG affinity column; the complex of His-FLAG-actin with His-MBP-G4G6 (Ca2+) remains on the column while the complex of endogenous actin with His-MBP-G4G6 (Ca2+) flows through (Fig. 1 A, step 2). After extensive washing (150× column volumes), EGTA is added to dissociate His-MBP-G4G6 from His-FLAG-actin, which is then eluted with glycine-HCl, pH 3.5 (Fig. 1 A, steps 3 and 4). To neutralize the low pH buffer to which the actin is exposed only briefly (<20 min), His-FLAG-actin is eluted directly into Tris pH 8.0 buffer. The protocol was first optimized for expression of skeletal α-actin for which highly specific monoclonal antibodies are available. The evolution of the purification protocol on the anti-FLAG affinity column and separation from endogenous β/γ-actin were verified by Western blot and SDS-PAGE analyses (Fig. 1 B and C).
Fig. 1.
Actin expression in Expi293F cells and separation from endogenous actin. (A) Diagram depicting steps 1 to 4 of the purification method. His-MBP-G4G6 (Ca2+) is added to the lysate of Expi293F cells expressing His-FLAG-actin. His-FLAG-actin with bound His-MBP-G4G6 remains on the anti-FLAG column, whereas endogenous β/γ-actin flows through. His-MBP-G4G6 is eluted with EGTA, which disrupts its Ca2+-dependent interaction with actin. His-FLAG-actin is eluted with low-pH glycine buffer and collected directly into Tris pH 8 neutralization buffer. (B) Western blot analysis using pan-actin and α-actin and β-actin specific antibodies showing the separation of recombinant α-actin from the endogenous actin of Expi293F cells. Blots show the flowthrough (FT) and elution (E) from the anti-FLAG column, demonstrating that only recombinant α-actin is present in E, whereas endogenous β-actin is in FT. Cytoplasmic β/γ-actin purified from untransfected Expi293F cells and tissue-purified α-actin serve as controls and are recognized by their respective isoform-specific antibodies and the pan-actin antibody. (C) SDS-PAGE analysis of the purification steps on the anti-FLAG column showing the load, flowthrough (FT), preelution resin (pre-E), elution (E), and postelution resin (post-E).
Affinity Tag Removal and Nt-Acetylation.
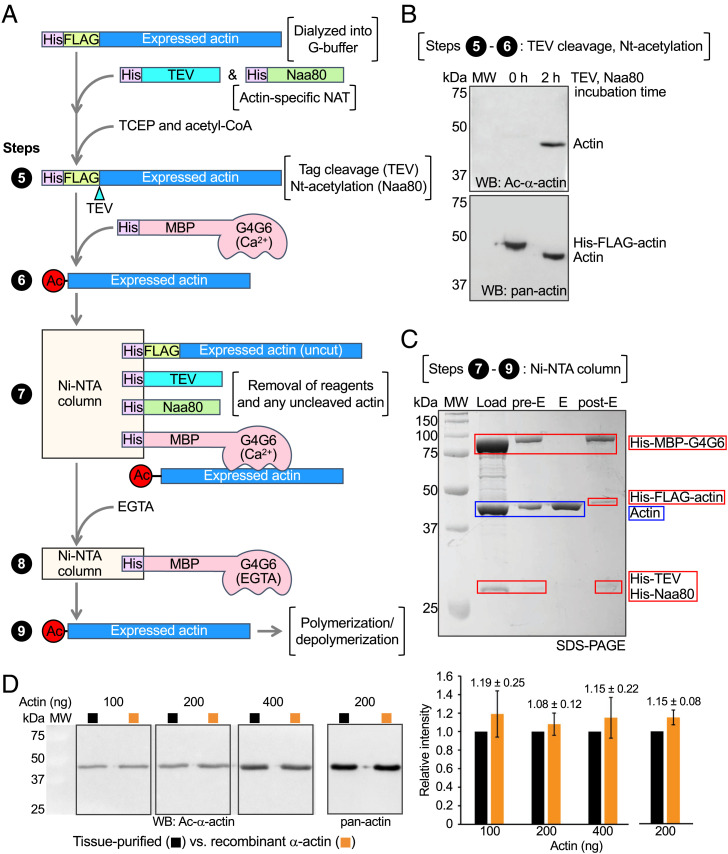
After elution from the anti-FLAG affinity column, the N-terminal affinity tag must be precisely removed, and the N-terminal amino acid of actin (D or E, depending on isoform) must be acetylated. To accomplish this, His-FLAG-actin is incubated at 37 °C for 2 h with two highly efficient enzymes, His-TEV protease (1:10 mass ratio with actin) and His-Naa80 (1:50 molar ratio with actin) (Fig. 2 A, steps 5 and 6). Tris(2-carboxyethyl)phosphine (TCEP, 0.5 mM) and acetyl-CoA (∼10 times the actin concentration) are added as cofactors of TEV protease and Naa80, respectively. The Naa80 construct used here corresponds to the N-terminally His-tagged fragment 78 to 308, called Naa80ΔN in a recent study where we showed that the N-terminal region is not necessary for actin Nt-acetylation in vitro (6). His-MBP-G4G6 (Ca2+) is then added at a 1.2:1 molar ratio with actin to sequester the Nt-acetylated actin [His-MBP-G4G6 does not bind to Naa80-bound actin (6)] and the mixture is loaded onto a Ni-NTA affinity column (Fig. 2 A, step 7). Pure recombinant actin is eluted from the column with the addition of EGTA (Fig. 2 A, step 8), whereas all the His-tagged components, including His-TEV, His-Naa80, His-MBP-G4G6, and any uncleaved His-FLAG-actin remain on the column. Western blot analysis using an Nt-acetylation-specific anti–α-actin antibody verifies tag removal and Nt-acetylation (Fig. 2B), and SDS-PAGE analysis verifies the efficient removal of purification reagents and any remaining tagged actin (Fig. 2C). Western blot–based quantification reveals similar levels of Nt-acetylation in tissue-purified and recombinant α-actin (Fig. 2D and SI Appendix, Fig. S2). Final yields of pure, Nt-acetylated α-actin are ∼3 mg/L of Expi293F cell culture.
Fig. 2.
Affinity tag removal and Nt-acetylation. (A) Diagram depicting steps 5 to 9 of the purification method. His-FLAG-actin in G-buffer is incubated with His-TEV (TCEP) and His-Naa80 (acetyl-CoA). TEV protease cleaves the affinity tag precisely before the first amino acid of mature postprocessed α-actin (10) (aspartic acid number 1, according to the actin numbering convention). Actin is then Nt-acetylated by Naa80. His-MBP-G4G6 (Ca2+) is added to bind Ac-α-actin. The complex is loaded onto a Ni-NTA affinity column. His-TEV, His-Naa80, uncleaved His-FLAG-actin, and the complex of His-MBP-G4G6 (Ca2+) with recombinant Ac-α-actin bind to the resin. Ac-α-actin is eluted with EGTA. (B) Western blot analysis of tag removal and Nt-acetylation using α-actin– and Nt-acetylation–specific (Top) and pan-actin (Bottom) antibodies before (0 h) and after (2 h) incubation with TEV protease and Naa80. The shift of the actin band reflects the removal of the purification/cleavage tag (3.2 kDa). (C) SDS-PAGE analysis of the purification step on the Ni-NTA column showing the load, preelution resin (Pre-E), elution (E), and postelution resin (post-E). (D) Western blot analysis of different amounts (100 to 400 ng) of tissue-purified and recombinant α-actin using α-actin– and Nt-acetylation–specific (Left) and pan-actin (Middle) antibodies. (Right) Quantification of the band intensity of recombinant α-actin normalized to the corresponding band intensity of tissue-purified α-actin within each blot. The recombinant α-actin intensity corresponds to the mean ± SD from six similar Westerns (see also SI Appendix, Fig. S2).
Suitability of the Method for Other Actin Isoforms.
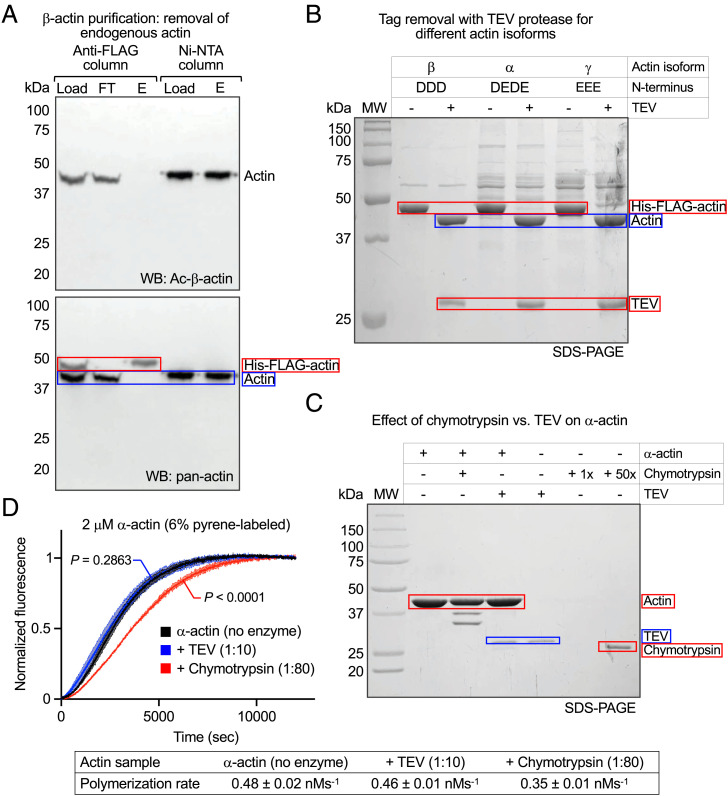
To test whether this method works for isoforms other than α-actin, we expressed and purified cytoplasmic β-actin and smooth muscle γ-actin. Western blot analysis using Nt-acetylation–specific anti–β-actin and pan-actin antibodies showed that endogenous Ac-β-actin flowed through in the anti-FLAG column, whereas His-FLAG-β-actin was retained during washing and then eluted with glycine-HCl pH 3.5 (Figs. 1A and 3A). The elution fraction only contained His-FLAG-β-actin (higher band), recognized by the pan-actin antibody, whereas endogenous Ac-β-actin, recognized by both antibodies, was absent. Eluted His-FLAG-β-actin was then incubated with His-TEV and His-Naa80, complexed with His-MBP-G4G6 (Ca2+), loaded onto the Ni-NTA column, and eluted with EGTA (Fig. 3A). The fractions loaded and eluted from the Ni-NTA column were recognized by both antibodies, confirming Nt-acetylation of recombinant β-actin.
Fig. 3.
Suitability of the method for other actin isoforms. (A) Western blot analysis of endogenous actin removal, tag cleavage, and Nt-acetylation during recombinant β-actin purification using β-actin– and Nt-acetylation–specific and pan-actin antibodies. The blot shows the load, flowthrough (FT), and elution (E) from the anti-FLAG column and the load and elution (E) from the Ni-NTA column. The same blot was first stained with Ac-β-actin antibody (Top) and then restained with pan-actin (Bottom). Endogenous Ac-β-actin is detected in load and FT, but not in E from the anti-FLAG column, whereas His-FLAG-β-actin is detected both in load and E, demonstrating the removal of endogenous actin. Prior to loading onto the Ni-NTA column, the N-terminal His-FLAG tag is removed and the recombinant β-actin is Nt-acetylated by Naa80 and bound to His-MBP-G4G6. The recombinant actin is detected by the Ac-β-actin antibody in both load and E from the Ni-NTA column. (B) SDS-PAGE analysis shows that TEV protease efficiently removes the His-FLAG tag from actin isoforms β, α, and γ with different N termini, as reflected by a 3.2-kDa shift in the actin band. (C) SDS-PAGE analysis shows that chymotrypsin (1:80 wt/wt with actin, 15 min, room temperature) cleaves tissue-purified α-actin at several locations, whereas TEV (1:10 wt/wt with actin, 2 h, 37 °C) does not. Protease cleavage reactions were stopped with 1 mM PMSF. (D) Polymerization time course of tissue-purified α-actin (2 μM, 6% pyrene-labeled) before and after treatment with proteases (as in C) shows a decrease in polymerization for chymotrypsin-treated but not TEV-treated α-actin. Data are shown as the average curve from three independent experiments with SD bars in lighter shade. Polymerization rates are reported at the bottom of the figure as the mean ± SD. P values for polymerization rate comparisons of enzyme-treated vs. untreated actin were calculated using one-way ANOVA with Dunnett’s test for multiple comparisons.
TEV protease removed the affinity tag from three isoforms (α-, β-, and γ-actin) that represent the spectrum of amino acids found at the mature N termini of the six human actin isoforms (Fig. 3B). This result and our previous finding that Naa80 acetylates the N termini of all six actin isoforms (5) suggest that the method described here is applicable to all isoforms.
The most popular actin expression/purification methods to date use chymotrypsin to remove a C-terminal affinity tag (37–39), since chymotrypsin cleaves precisely after the last natural amino acid of actin (F375). However, actin’s susceptibility to internal chymotrypsin cleavage has been known since the 1970s (40, 41). We considered other proteases for tag removal but, except TEV and enterokinase, they all leave extra (nonnative) amino acids at the N or C terminus. TEV was chosen because it is easy to produce and use. To further verify TEV’s suitability for tag removal, tissue-purified α-actin was treated with TEV protease under the conditions used here (1:10 mass ratio with actin, 2 h, 37 °C) or chymotrypsin under published conditions (1:80 mass ratio with actin, 15 min, room temperature) (38). SDS-PAGE analysis revealed the appearance of several chymotryptic fragments and lack of internal cleavage by TEV (Fig. 3C). When protein gels are shown, lower-molecular-weight degradation bands are observed in published work that used chymotrypsin for tag removal (37–39). While this can be alleviated through additional purification steps (30), yields are compromised as a result and degradation subproducts typically persist and are difficult to control for. In our laboratory, we have also found it difficult to obtain actin uncontaminated by additional chymotrypsin cleavage, which the method described here overcomes. In actin polymerization assays, the chymotryptic digestion of actin led to a decrease in activity compared to control, whereas TEV-treated actin polymerized normally (Fig. 3D).
Posttranslational Modification of Recombinant Actin.
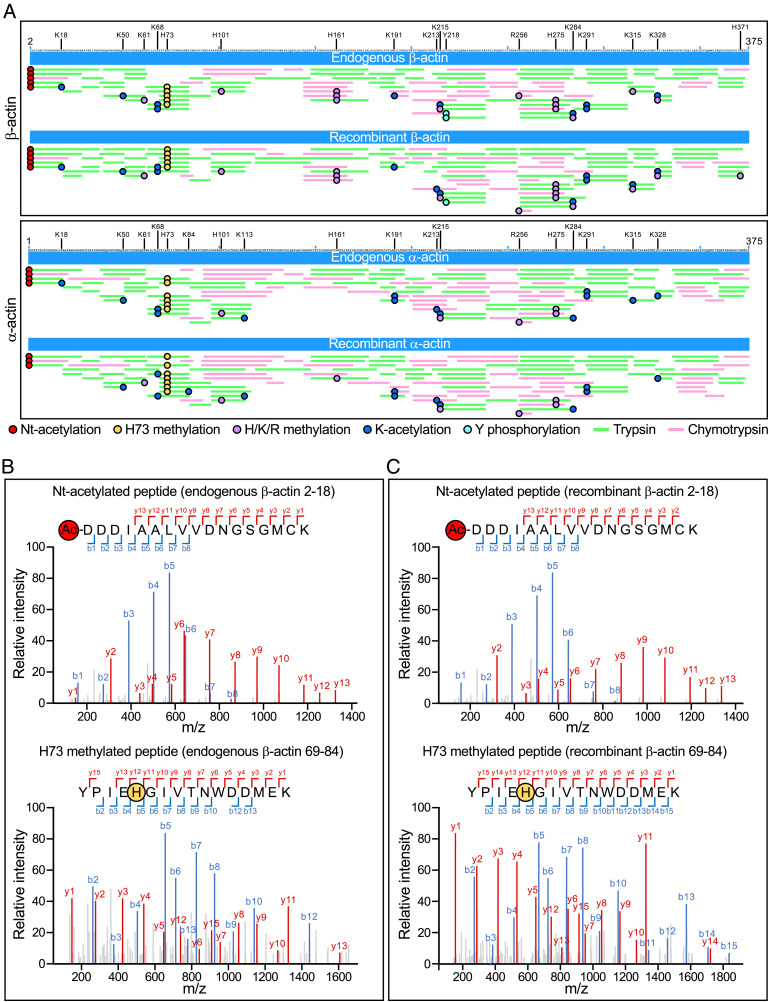
An advantage of human cells for actin expression is that they intrinsically contain the necessary components for actin folding and posttranslational processing. We performed mass spectrometry (MS)-based proteomics analysis to check whether the PTMs present in recombinant α- and β-actin matched those present in endogenous α- and β-actin. For comparisons, skeletal α-actin was purified from muscle tissue (8) and a mixture of β/γ-actin was purified from untransfected Expi293F cells using G4G6, as we previously described (5). To ensure full peptide coverage, each actin sample was digested with two enzymes, trypsin and chymotrypsin. The identity of peptides in the MS spectra was determined by comparing their masses to those of peptides in the α-, β-, and γ-actin sequences, considering potential PTMs and using a false discovery rate (FDR) threshold of <1%. Full peptide coverage was obtained for all the actin samples (Fig. 4A). We observed several PTMs, including tyrosine phosphorylation, histidine/lysine/arginine methylation, and lysine acetylation (Datasets S1 and S2). Table 1 and Fig. 4A show a subset of PTMs observed with the highest confidence, based on the presence of b (forward) and y (backward) fragment ions encompassing the PTM in the MS2 spectra (Fig. 4 B and C and SI Appendix, Figs. S3 and S4) and occupancy >5% in at least one of the four actin samples. For each PTM-containing peptide, we define the PTM occupancy as the ratio of the number of peptides containing the PTM to the total number of observed peptides. Notably, Nt-acetylation and H73 methylation, two modifications essential for actin function that implicate specialized enzymes (5, 7), were observed in 92 to 100% of the observed peptides in both endogenous and recombinant β-actin (Fig. 4 B and C) and α-actin (SI Appendix, Fig. S4). This result confirmed the successful and precise removal of the N-terminal affinity tag by TEV protease and Nt-acetylation by Naa80. In contrast, only one phosphorylation site (Y218) was observed above the 5% occupancy cutoff in both endogenous and recombinant β-actin. This is likely due to the lack of phosphatase inhibitors during purification (42); the large-scale use of phosphatase inhibitors is cost-prohibitive, and we chose to use the same protein preparations in PTM and biochemical analyses (see below). γ-Actin differs from β-actin only at four positions, all located near the N terminus. Consistently, N-terminal peptides corresponding to the γ-actin sequence were observed in the endogenous but not the recombinant β-actin sample, further confirming the complete removal of endogenous actin by the purification method (SI Appendix, Fig. S5). Overall, the location and occupancy of PTMs are similar between recombinant and endogenous samples of the same actin isoform and, to a notable extent, between the two actin isoforms analyzed here (Fig. 4A and Table 1).
Fig. 4.
PTM analysis of recombinant and endogenous β- and α-actin. (A) MS peptide coverage maps of endogenous and recombinant β-actin (Top) and α-actin (Bottom) relative to the protein sequence. Green and magenta lines indicate trypsin and chymotrypsin digestion peptides, respectively, whose masses were identified by MS with FDR <1% (see also Datasets S1 and S2). Red, orange, purple, blue, and cyan circles indicate Nt-acetylation, H73 methylation, H/K/R methylation, K acetylation, and Y phosphorylation, respectively. Only PTMs observed with the highest confidence are shown, i.e., those with b (forward) and y (backward) fragment ions encompassing the PTM in MS2 spectra and occupancy >5% (see also Table 1). MS2 spectra of Nt-acetylated and H73 methylated peptides are shown for endogenous (B) and recombinant (C) β-actin. The spectra show the b (backward, blue) and y (forward, red) fragment ions observed by MS for each peptide. SI Appendix, Figs. S3 and S4 show all the MS2 spectra of PTM-containing β- and α-actin peptides observed with highest confidence.
Table 1.
List of PTMs observed with highest confidence in endogenous and recombinant β- and α-actin, based on the presence of b (forward) and y (backward) fragment ions encompassing the PTM in MS2 spectra and occupancy >5% in at least one of the actin samples
| β-Actin | α-Actin | |||
|---|---|---|---|---|
| PTM | Endogenous | Recombinant | Endogenous | Recombinant |
| Nt-acetylation | 100 (61/61) | 98 (96/98) | 99 (200/203) | 100 (258/259) |
| K18 acetylation | 11 (7/61) | 11 (11/98) | 5 (10/203) | |
| K50 acetylation | 7 (16/217) | 7 (16/231) | 12 (42/337) | 15 (53/353) |
| K61 methylation | 6 (8/134) | 8 (12/146) | 5 (11/214) | |
| K68 acetylation | 19 (10/52) | 16 (8/49) | 17 (9/54) | 9 (4/46) |
| H73 methylation | 94 (246/263) | 93 (286/308) | 96 (412/430) | 92 (314/341) |
| K84 acetylation | 11 (40/361) | |||
| H101 methylation | 10 (25/240) | 10 (26/264) | 6 (19/300) | 5 (16/336) |
| K113 acetylation | 5 (16/300) | 5 (16/336) | ||
| H161 methylation | 15 (57/372) | 17 (68/410) | 7 (33/501) | |
| K191 acetylation | 9 (21/224) | 11 (31/284) | 15 (74/493) | 18 (125/706) |
| K213 acetylation | 17 (5/30) | 8 (5/65) | 12 (18/146) | |
| K215 acetylation | 13 (6/47) | 13 (7/52) | 20 (24/119) | 8 (14/176) |
| K215 methylation | 13 (6/47) | 12 (6/52) | 5 (6/119) | 5 (8/176) |
| Y218 phosphorylation | 18 (32/179) | 11 (22/201) | ||
| R256 methylation | 7 (1/15) | 8 (2/24) | 7 (16/242) | 6 (29/451) |
| H275 methylation | 28 (13/47) | 32 (26/82) | 8 (52/664) | 5 (45/965) |
| K284 acetylation | 14 (8/59) | 13 (11/82) | 11 (46/419) | 15 (63/420) |
| K284 methylation | 14 (8/59) | 9 (7/82) | ||
| K291 acetylation | 17 (46/268) | 20 (60/298) | 21 (75/358) | 21 (85/400) |
| K315 acetylation | 7 (1/14) | 30 (3/10) | ||
| K315 methylation | 25 (1/4) | 21 (3/14) | ||
| K328 acetylation | 20 (2/10) | 35 (6/17) | 64 (14/22) | 40 (6/15) |
| K328 methylation | 20 (2/10) | 12 (2/17) | ||
| H371 methylation | 5 (5/110) | |||
For each PTM, the percentile occupancy is defined as 100 × the ratio of the number of peptides containing the PTM to the total number of observed peptides (shown in parentheses). See also Fig. 4 and SI Appendix, Figs. S3 and S4.
Native-Like Polymerization Activity of Recombinant Actin.
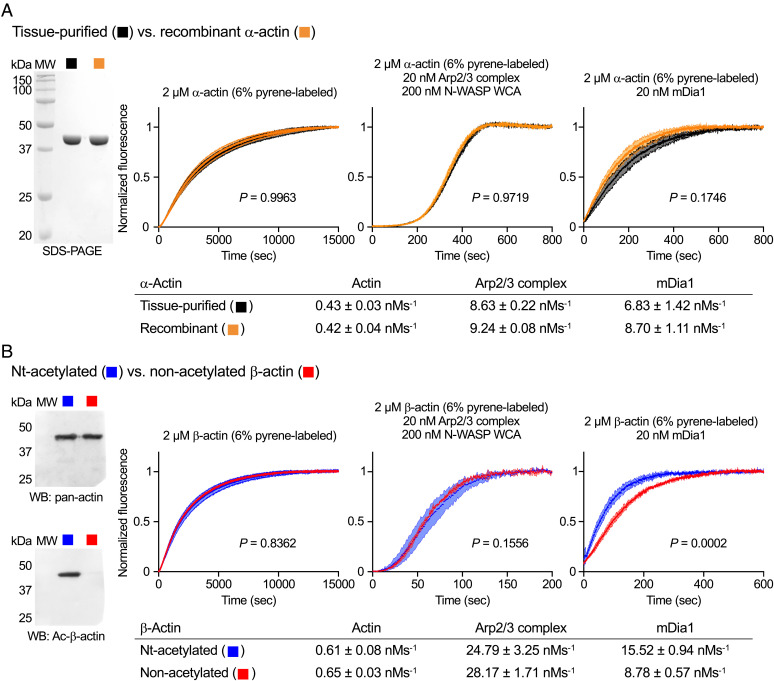
Muscle tissue–purified actin consists mostly of a single isoform, α-actin (8), allowing for direct activity comparison with recombinant α-actin. Both α-actin preparations were supplemented with 6% pyrene-labeled α-actin (tissue-purified), and their polymerization kinetics were assessed using the pyrene-actin polymerization assay (43). Time courses of polymerization were recorded for α-actin alone and catalyzed by two common actin-assembly factors, Arp2/3 complex and the formin mDia1. Under all conditions, the two α-actin preparations displayed similar polymerization rates (Fig. 5A).
Fig. 5.
Native-like polymerization activity of recombinant actin. (A, Left) SDS-PAGE analysis of tissue-purified and recombinant α-actin. (A, Right) Time courses of α-actin polymerization alone and with Arp2/3 complex or mDia1 (experimental conditions shown in the figure). (B, Left) Western blot analysis of recombinant β-actin before and after Nt-acetylation using pan-actin (Top) and isoform- and Nt-acetylation–specific (Bottom) antibodies. (B, Right) Time courses of β-actin polymerization alone and with Arp2/3 complex or mDia1 (experimental conditions shown in the figure). Data are shown as the average curve from three independent experiments with SD bars in lighter shade. Polymerization rates are reported below the graphs as the mean ± SD. P values for polymerization rate comparisons were calculated using one-way ANOVA with Sidak’s test for multiple comparisons. P values for comparisons between acetylated recombinant α- and β-actin are 0.0043, < 0.0001, and 0.0001, respectively for actin alone, with Arp2/3 complex, or with mDia1.
For many applications the presence of Nt-acetylation may not be crucial, particularly when comparing wild-type/mutant combinations of the same actin isoform. Indeed, most biochemical studies use E. coli–expressed proteins lacking most PTMs. As part of our previous characterization of Naa80 (5), we had established that Nt-acetylation affected mostly filament elongation by formins, which mediate the processive incorporation of actin subunits at the barbed end of the filament. In contrast, the polymerization of actin alone or by the Arp2/3 complex was unaffected. However, that study used a mixture of β/γ-actin purified from cells. Here, we directly compared the polymerization activity of recombinant β-actin ± Nt-acetylation. The consistency among protein preparations was high, and notable differences in polymerization were not observed as a function of the presence/absence of Nt-acetylation for β-actin alone or with Arp2/3 complex. However, consistent with our previous findings (5), the rate of polymerization with mDia1, which drives both nucleation and elongation, was reduced nearly twofold for nonacetylated β-actin (Fig. 5B). Therefore, for most applications, it may be acceptable to bypass the Naa80-mediated Nt-acetylation step.
Discussion
More than 20 studies have been devoted to developing novel actin expression methods or improvements upon existing ones. Yet, the methods published thus far suffer from at least one of several shortcomings including deficient folding, low yields, absence of PTMs critical for actin activity, contamination with endogenous actin from the expressing cells, lack of evidence of native-like activity, and deficient affinity-tag removal (SI Appendix, Table S1 summarizes published methods and their shortcomings). The specific requirements of actin folding (4) and posttranslational processing (5, 7, 10, 31, 32) are more evidently unmet in methods that have used E. coli for expression (24, 25). Saccharomyces cerevisiae has a single actin gene that if substituted by that of mammalian muscle actin affects viability (44). Thus, a compromising solution has been to introduce a limited number of mammalian actin amino acids into the yeast actin gene to study their function (45). More generally, however, lower eukaryotes, including yeast, Dictyostelium, and even insect Sf9 cells that adequately fulfill actin’s folding requirements, perform posttranslational modifications that differ in important ways from those of mammalian cells. Yeast, for instance, lacks SETD3 and Naa80, responsible for H73 methylation (7) and Nt-acetylation (5), respectively. A recent study attempts to address this issue by coexpressing SETD3 and Naa80 along with actin in Pichia pastoris (39). While H73 methylation and Nt-acetylation were achieved by this method (albeit possibly partial), the yields were low (0.5 mg/L) and chymotrypsin was used for affinity tag removal which, as shown above, leads to actin degradation that is difficult to control (Fig. 3C). While Sf9 cells more closely resemble human cells, studies have shown PTM differences in proteins expressed in insect cells compared to their human counterparts (33, 34).
Both the N- and C-terminal ends of actin are essential for interactions with actin-binding proteins and with actin itself within the filament (2). Even the absence of the small acetyl group at the N terminus of actin in Naa80 knockout cells leads to increased F- to G-actin ratios, augmented filopodia and lamellipodia formation, and accelerated cell motility (5). N- and C-terminal affinity tags must therefore be precisely removed, such that no extra amino acids or deletions are present at either end. Despite early evidence of actin’s susceptibility to chymotrypsin digestion (40, 41), this enzyme is still used for affinity tag removal by most expression methods. The chymotrypsin approach was first used to cleave a thymosin-β4-His affinity tag added at the C terminus of actin expressed in Dictyostelium (37). The novelty of this approach was the incorporation of thymosin-β4 as part of the affinity tag. Thymosin-β4 is a 44-amino-acid actin monomer sequestering protein, which helps to keep the recombinant actin mostly monomeric and segregated from the endogenous Dictyostelium actin, albeit the latter has not been formally demonstrated. The thymosin-β4-His tag is then cleaved with chymotrypsin, which has a high-affinity site after the last native amino acid of actin (F375). Subsequently, this method was adopted for actin expression using baculovirus-Sf9 (38), P. pastoris (39), and mammalian cells (30), carrying over concerns with the chymotryptic digestion of actin at other sites (Fig. 3C).
Another issue afflicting most existing actin expression methods is contamination of recombinant actin with endogenous actin of the expressing cells, including yeast, Dictyostelium, or Sf9 cells. Some studies have simply overlooked this problem, whereas others have incompletely addressed it, and not a single study has conclusively demonstrated full removal of endogenous actin. In fact, when specifically investigated, contamination with endogenous actin has been invariably found (22, 46). Contamination with endogenous actin precludes an accurate analysis of polymerization differences among actin isoforms or mutants. The presence of even small amounts of endogenous actin also prevents the analysis of PTMs using highly sensitive MS-based proteomics. For instance, we observed here acetylated N-terminal peptides of both β- and γ-actin in the endogenous actin sample but only β-actin peptides were observed in the recombinant sample (Fig. 4, SI Appendix, Fig. S5, and Dataset S1). This analysis would not have been possible if the recombinant β-actin had been contaminated with endogenous actin.
Using the same experimental conditions, we determined that β-actin polymerized faster than α-actin: ∼1.5-fold alone, ∼3-fold with Arp2/3 complex, and ∼2-fold with mDia1 (Fig. 5). These differences are surprisingly large, considering that the two actin isoforms share ∼94% sequence identity and most amino acids variations tend to be conservative. In the absence of actin degradation or contamination with endogenous actin, these differences between isoforms are likely reliable and warrant further investigation.
In summary, we described here a dependable method to obtain high yields of recombinant actin in its native form. Biochemical experiments demonstrated that this method overcomes deficiencies of prior actin expression methods, such as incomplete removal of endogenous actin, absence of key PTMs, and unproven native-like polymerization activity. Such deficiencies imply that the conclusions of some studies may not be fully reliable, especially when studying disease-causing mutations in actin. The method described here produces recombinant actin that closely matches actin in human cells, offering the opportunity to revisit prior conclusions. The ability to obtain pure recombinant actin free of endogenous actin contamination also enables studies to understand how minor differences in actin isoforms result in substantially different activities in cells and different biochemical properties in vitro.
Materials and Methods
Proteins.
The following complementary DNAs (cDNAs) were used in cloning: human skeletal α-actin (UniProt P68133), human cytoplasmic β-actin (UniProt P60709), human smooth muscle γ-actin (UniProt P63267), human gelsolin (UniProt P06396-1), human Naa80 (UniProt Q93015-2), mouse mDia1 (UniProt O08808), and mouse N-WASP (UniProt Q91YD9). The cDNAs were either available in the laboratory from previous work (N-WASP, mDia1, Naa80) (5) or purchased from DNASU (actin isoforms) or Open Biosystems (gelsolin). Recombinant proteins were obtained as follows.
Actin.
Construct His-FLAG-actin was obtained using overlap extension PCR to add the sequence encoding for a His-FLAG-TEV affinity/cleavage tag (MAHHHHHH-SG-DYKDDDDK-TS-ENLYFQ) N-terminally to the α-actin cDNA [lacking the first two N-terminal residues that are removed posttranslationally (10)]. The construct was inserted between the SalI and BamHI sites of vector pJCX4 (Addgene ID: 170756) (47, 48). A SpeI site was silently inserted into the TEV-encoding sequence, and other actin isoforms (β cytoplasmic and γ smooth) were inserted between the SpeI and BamHI sites. SI Appendix, Table S2 lists the primers used in this study. Proteins were expressed in Expi293F cells grown in suspension in Expi293 Expression Medium and 8% CO2 (Thermo Fisher Scientific). Cells, at a density of 2.5 million per mL, were transfected with 1 mg DNA per liter of culture. The transfection mixture was prepared in Opti-MEM Reduced Serum Media using polyethylenimine-MAX (PEI-MAX) for transfection at a 3:1 vol/wt ratio to DNA (3 mL PEI-MAX per 1 mg plasmid). Transfected cells were grown for 72 h at 37 °C, pelleted at 3,000 × g for 10 min, and stored at −80 °C. Cell pellets were thawed in a cool water bath and diluted in actin lysis buffer (10 mM Tris⋅HCl pH 8, 10 mM CaCl2, 2 mM MgCl2, 0.5 mM ATP) supplemented with 25 mM sucrose, 0.1% octyl-β-glucoside, 10 µg/mL aprotonin, 10 µg/mL leupeptin, 1:1,000 dilution of protease inhibitor mixture P8340 (Sigma-Aldrich) and 2 mM phenylmethanesulfonyl fluoride (PMSF). Unless otherwise specified, purification was performed at 4 °C. Cells were lysed using a Dounce homogenizer, followed by centrifugation at 50,000 × g for 20 min. To sequester monomeric actin, clarified lysates were incubated with 1 mg His-MBP-G4G6 per gram of wet cell pellet. The following day, lysates were centrifuged (50,000 × g for 20 min) to pellet any precipitate, mixed with anti-FLAG resin (equilibrated with actin lysis buffer, ∼2 mL of anti-FLAG resin per 10 g of cell pellet) and incubated for 2 h with gentle rocking. The resin was then transferred into a column and washed with 50 column volumes (CV) of FLAG-wash-buffer (20 mM Tris⋅HCl pH 8, 10 mM CaCl2, 1 mM MgCl2, 0.5 mM ATP) followed by 150 CV of FLAG-wash-buffer supplemented with 100 mM KCl. To dissociate His-MBP-G4G6 from His-FLAG-actin, the resin was first washed with 5 CV of 20 mM Tris⋅HCl pH 8, 1 mM MgCl2, 0.5 mM ATP and incubated for 10 min with 5 CV of the same buffer supplemented with 3 mM EGTA. The column was then washed with 10 CV of FLAG-wash-buffer to restore Ca2+. His-FLAG-actin was eluted from the anti-FLAG column in 1-mL fractions after incubation for ∼20 min with 100 mM glycine pH 3.5, 10 mM CaCl2, 1 mM MgCl2, 0.5 mM ATP. To restore the pH, actin fractions were collected directly into 340-µL aliquots of 100 mM Tris⋅HCl pH 8, 10 mM CaCl2, 1 mM MgCl2, 0.5 mM ATP and analyzed by SDS-PAGE. Actin-containing fractions were pooled and dialyzed overnight into G-buffer (5 mM Tris⋅HCl pH 8, 0.2 mM CaCl2, 0.2 mM ATP).
The actin concentration was estimated from the ultraviolet (UV) absorbance at 290 nm (ε = 26,600 M−1⋅cm−1). N-terminal tag removal and Nt-acetylation were performed in parallel by incubating His-FLAG-actin with His-TEV (1:10 mass ratio to actin, 0.5 mM TCEP) and His-Naa80 (1:50 molar ratio to actin, acetyl-CoA at ∼10 times the actin concentration) for 2 h at 37 °C. Actin was then incubated for 30 min with a 1.2 molar excess of His-MBP-G4G6, supplemented with 2 mM MgCl2 and 5 mM CaCl2 to promote G4G6 binding. The complex, supplemented with 10 mM imidazole pH 8, 100 mM KCl and 5 mM ATP, was loaded onto a Ni-NTA column equilibrated with 20 mM Tris⋅HCl pH 8, 10 mM imidazole, 100 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 0.5 mM ATP (∼2 mL Ni-NTA resin per 10 g wet cell pellet processed). The column was washed with 50 CV of equilibration buffer, followed by 10 CV of 10 mM Tris⋅HCl pH 8, 1 mM CaCl2, 1 mM MgCl2, 0.5 mM ATP. Actin was eluted by inducing its dissociation from G4G6 with 10 mM Tris⋅HCl pH 8, 3 mM EGTA, 1 mM MgCl2, 0.5 mM ATP for 30 min. Elution fractions (2 mL) were supplemented with 2 mM MgCl2 and analyzed by SDS-PAGE. Actin-containing fractions were pooled and dialyzed overnight into G-buffer. Purified actin was polymerized by addition of 100 mM KCl, 2 mM MgCl2, and 1 mM EGTA for 1 h at room temperature, and F-actin was pelleted by centrifugation at 100,000 × g for 30 min. Actin was depolymerized in 1 mL G-buffer and dialyzed against G-buffer for 3 d. The sample was centrifuged at 100,000 × g for 30 min and the actin concentration in the supernatant was measured by UV.
His-MBP-G4G6.
PCR products of His-MBP and G4G6 (human gelsolin residues 434 to 782) were ligated and inserted between the PstI and HindIII sites of vector pRSF1 (Novagen). The protein was expressed in ArcticExpress (DE3) RIL cells (Agilent Technologies) grown in Terrific Broth (TB) medium for 6 h at 37 °C to an optical density at 600 nm (OD600) of ∼1.5, followed by 24 h growth at 10 °C in the presence of 0.5 mM isopropyl-β-d-thiogalactoside (IPTG). Cells were harvested by centrifugation (50,000 × g for 20 min), resuspended in 20 mM Tris⋅HCl pH 8, 500 mM NaCl, 5 mM imidazole, and 1 mM PMSF, and lysed by Microfluidizer (Microfluidics). The protein was purified on a Ni-NTA affinity column, followed by ion exchange chromatography on a MonoQ column (GE Healthcare) in 50 mM Tris⋅HCl pH 8 and 50 to 500 mM NaCl gradient. The protein was dialyzed against 20 mM Hepes pH 8, 100 mM NaCl, concentrated to 15 mg/mL, and stored at −80 °C.
His-TEV.
The His-TEV expression plasmid was obtained from Addgene (ID: 92414). The protein was expressed in BL21(DE3) cells (Agilent Technologies) grown in TB media for 6 h at 37 °C to an OD600 of ∼0.8, followed by 12-h growth at 19 °C in the presence of 0.4 mM IPTG. Cells were harvested by centrifugation, resuspended in 20 mM Hepes pH 7.5, 300 mM NaCl, 10 mM imidazole, and 1 mM PMSF, and lysed by Microfluidizer. The protein was purified on a Ni-NTA affinity column, followed by gel filtration chromatography on a SD200HL 26/600 column (GE Healthcare) in 20 mM Hepes pH 7.5, 200 mM NaCl, 2 mM ethylenediaminetetraacetic acid (EDTA), 1 mM dithiothreitol (DTT). Fractions containing His-TEV were dialyzed overnight into 20 mM Hepes pH 7.5, 20 mM NaCl, 2 mM EDTA, 1 mM DTT, concentrated to 2.5 mg/mL, and stored at −80 °C.
mDia1, His-Naa80, Arp2/3 complex, N-WASP, endogenous β/γ-actin, and skeletal α-actin.
The cloning (pTYB12 vector), expression [ArcticExpress (DE3) RIL cells], and purification (chitin affinity, followed by intein self-cleavage) of mDia1 and Naa80 (amino acids 78 to 308) were described by us previously (5). One change here was to add an N-terminal His-tag to Naa80 by PCR extension. Naa80 was purified on a chitin affinity column, followed by intein self-cleavage. N-WASP (amino acids 426 to 501) was cloned into vector pTYB12, expressed in BL21(DE3) cells and purified on a chitin affinity column. Arp2/3 complex was purified from bovine brain (49). Endogenous β/γ-actin was purified from untransfected Expi293F cells using G4G6 (5), and α-actin was purified from rabbit skeletal muscle acetone powder (8).
Plasmid Deposition.
The plasmids developed here for the expression and purification of actin were deposited with Addgene. ID numbers are as follows: His-FLAG-TEV-α-actin, 188452; His-FLAG-TEV-β-actin, 188453; His-MBP-G4G6, 188454; His-Naa80ΔN, 188455.
Antibodies.
SI Appendix, Table S3 lists commercial antibodies used in this study.
Proteomics.
Liquid chromatography tandem MS analysis was performed by the Proteomics and Metabolomics Facility of The Wistar Institute using a Q Exactive Plus mass spectrometer (Thermo Fisher Scientific) coupled to a Nano-ACQUITY ultraperformance liquid chromatography (UPLC) system (Waters). Gel bands of purified proteins were excised and digested in-gel with trypsin or chymotrypsin and injected into the UPLC system. Peptides were separated by reversed-phase high-performance liquid chromatography on a BEH C18 nanocapillary analytical column (Waters) using a 90-min gradient of solvent-A (0.1% formic acid in water) and solvent-B (0.1% formic acid in acetonitrile). A 30-min blank gradient was run between sample injections. Eluted peptides were analyzed by the mass spectrometer, set to repetitively scan m/z 300 to 1,800 in positive ion mode. The full MS scan was collected at 70,000 resolution, followed by data-dependent MS/MS scans at 17,500 resolution on the 20 most abundant ions exceeding a minimum threshold of 10,000. Other settings included: preferred peptide match, exclude isotopes, and charge-state screening (to reject unassigned and more than five charged ions). The MS raw spectra were processed using MetaMorepheus version 0.0.316 (50). The calibrate and search tasks of the program were used with default parameters, including precursor mass tolerance of 5 ppm, enzyme specific cleavage, and up to two missed cleavages. The G-PTM-D task was used with common PTMs, including N-terminal acetylation, H/K/R methylation, S/T/Y phosphorylation, and K acetylation. We did not specifically search for uncommon PTMs such as D/E/K carboxylation, K/R dimethylation, K/R trimethylation, K/N/P hydroxylation, and C/Y nitrosylation. An FDR cutoff of <1% was employed at the peptide and protein levels. Raw proteomics data files were deposited with MassIVE (accession nos. MSV000089132 and MSV000089919).
Actin Polymerization Assay.
Actin polymerization was measured as the time course of the fluorescence increase resulting from the incorporation of pyrene-actin (excitation 365 nm, emission 407 nm) into filaments using a Cary Eclipse fluorescence spectrophotometer (Varian Medical Systems). Tissue-purified actin was additionally purified by gel filtration to remove impurities and polymerization seeds and ensure reproducibility in polymerization assays. The recombinant actin did not require this step, presumably because the purification protocol removes impurities and seeds. The polymerization of actin alone was carried out by mixing 4 μL of buffer alone with 196 μL of Mg-ATP-actin at 2 μM concentration (6% pyrene-labeled tissue-purified α-actin) in F-buffer (10 mM Tris⋅HCl pH 8, 1 mM MgCl2, 50 mM KCl, 1 mM EGTA, 0.2 mM ATP, 0.5 mM DTT, 0.1 mM NaN3). Polymerization by actin assembly factors was carried out similarly, by mixing Mg-ATP-actin with 20 nM mDia1 or Arp2/3 complex (with 200 nM N-WASP WCA). Experiments were conducted at 25 °C and were repeated at least three times. Data acquisition started 10 s after mixing. Polymerization rates are reported as the maximal slope of a polymerization curve (between 0.1 and 0.4 of the maximum fluorescence) normalized by the change in fluorescence.
Supplementary Material
Acknowledgments
This work was supported by NIH Grants R01 GM073791 to R.D., R01 DK128282 to R.O.H., and F31 DK127610 to R.H.C and a Blavatnik Family Foundation predoctoral fellowship to P.J.C. R.O.H. acknowledges support from the Irma and Norman Braman and the Suzi and Scott Lustgarten Center endowments.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2209150119/-/DCSupplemental.
Data, Materials, and Software Availability
Proteomics data have been deposited in the MassIVE Repository [accession nos. MSV000089132 (51) and MSV000089919 (52)].
References
- 1.Pollard T. D., Actin and actin-binding proteins. Cold Spring Harb. Perspect. Biol. 8, a018226 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dominguez R., Holmes K. C., Actin structure and function. Annu. Rev. Biophys. 40, 169–186 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Funk J., et al. , Profilin and formin constitute a pacemaker system for robust actin filament growth. eLife 8, e50963 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balchin D., Milicic G., Strauss M., Hayer-Hartl M., Hartl F. U., Pathway of actin folding directed by the eukaryotic chaperonin TRiC. Cell 174, 1507–1521.e1516 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Drazic A., et al. , NAA80 is actin’s N-terminal acetyltransferase and regulates cytoskeleton assembly and cell motility. Proc. Natl. Acad. Sci. U.S.A. 115, 4399–4404 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rebowski G., et al. , Mechanism of actin N-terminal acetylation. Sci. Adv. 6, eaay8793 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilkinson A. W., et al. , SETD3 is an actin histidine methyltransferase that prevents primary dystocia. Nature 565, 372–376 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pardee J. D., Spudich J. A., Purification of muscle actin. Methods Enzymol. 85 Pt B, 164–181 (1982). [DOI] [PubMed] [Google Scholar]
- 9.Parker F., Baboolal T. G., Peckham M., Actin mutations and their role in disease. Int. J. Mol. Sci. 21, 3371 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arnesen T., Marmorstein R., Dominguez R., Actin’s N-terminal acetyltransferase uncovered. Cytoskeleton (Hoboken) 75, 318–322 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kashina A. S., Regulation of actin isoforms in cellular and developmental processes. Semin. Cell Dev. Biol. 102, 113–121 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaech S., Fischer M., Doll T., Matus A., Isoform specificity in the relationship of actin to dendritic spines. J. Neurosci. 17, 9565–9572 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mounier N., Perriard J. C., Gabbiani G., Chaponnier C., Transfected muscle and non-muscle actins are differentially sorted by cultured smooth muscle and non-muscle cells. J. Cell Sci. 110, 839–846 (1997). [DOI] [PubMed] [Google Scholar]
- 14.von Arx P., Bantle S., Soldati T., Perriard J. C., Dominant negative effect of cytoplasmic actin isoproteins on cardiomyocyte cytoarchitecture and function. J. Cell Biol. 131, 1759–1773 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perrin B. J., Ervasti J. M., The actin gene family: Function follows isoform. Cytoskeleton (Hoboken) 67, 630–634 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bunnell T. M., Burbach B. J., Shimizu Y., Ervasti J. M., β-Actin specifically controls cell growth, migration, and the G-actin pool. Mol. Biol. Cell 22, 4047–4058 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katz Z. B., et al. , β-Actin mRNA compartmentalization enhances focal adhesion stability and directs cell migration. Genes Dev. 26, 1885–1890 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baranwal S., et al. , Nonredundant roles of cytoplasmic β- and γ-actin isoforms in regulation of epithelial apical junctions. Mol. Biol. Cell 23, 3542–3553 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dugina V., et al. , Interaction of microtubules with the actin cytoskeleton via cross-talk of EB1-containing +TIPs and γ-actin in epithelial cells. Oncotarget 7, 72699–72715 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen A., Arora P. D., McCulloch C. A., Wilde A., Cytokinesis requires localized β-actin filament production by an actin isoform specific nucleator. Nat. Commun. 8, 1530 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vedula P., Kashina A., The makings of the ‘actin code’: Regulation of actin’s biological function at the amino acid and nucleotide level. J. Cell Sci. 131, jcs215509 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bergeron S. E., Zhu M., Thiem S. M., Friderici K. H., Rubenstein P. A., Ion-dependent polymerization differences between mammalian beta- and gamma-nonmuscle actin isoforms. J. Biol. Chem. 285, 16087–16095 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boiero Sanders M., et al. , Specialization of actin isoforms derived from the loss of key interactions with regulatory factors. EMBO J. 41, e107982 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frankel S., Sohn R., Leinwand L., The use of sarkosyl in generating soluble protein after bacterial expression. Proc. Natl. Acad. Sci. U.S.A. 88, 1192–1196 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tamura M., Ito K., Kunihiro S., Yamasaki C., Haragauchi M., Production of human β-actin and a mutant using a bacterial expression system with a cold shock vector. Protein Expr. Purif. 78, 1–5 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Aspenström P., Karlsson R., Interference with myosin subfragment-1 binding by site-directed mutagenesis of actin. Eur. J. Biochem. 200, 35–41 (1991). [DOI] [PubMed] [Google Scholar]
- 27.Anthony Akkari P., et al. , Production of human skeletal alpha-actin proteins by the baculovirus expression system. Biochem. Biophys. Res. Commun. 307, 74–79 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Joel P. B., Fagnant P. M., Trybus K. M., Expression of a nonpolymerizable actin mutant in Sf9 cells. Biochemistry 43, 11554–11559 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Miller B. M., Trybus K. M., Functional effects of nemaline myopathy mutations on human skeletal alpha-actin. J. Biol. Chem. 283, 19379–19388 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.A M., et al. , Regulation of INF2-mediated actin polymerization through site-specific lysine acetylation of actin itself. Proc. Natl. Acad. Sci. U.S.A. 117, 439–447 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Terman J. R., Kashina A., Post-translational modification and regulation of actin. Curr. Opin. Cell Biol. 25, 30–38 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Varland S., Vandekerckhove J., Drazic A., Actin post-translational modifications: The Cinderella of cytoskeletal control. Trends Biochem. Sci. 44, 502–516 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Amann T., Schmieder V., Faustrup Kildegaard H., Borth N., Andersen M. R., Genetic engineering approaches to improve posttranslational modification of biopharmaceuticals in different production platforms. Biotechnol. Bioeng. 116, 2778–2796 (2019). [DOI] [PubMed] [Google Scholar]
- 34.McKenzie E. A., Abbott W. M., Expression of recombinant proteins in insect and mammalian cells. Methods 147, 40–49 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Khaitlina S. Y., Functional specificity of actin isoforms. Int. Rev. Cytol. 202, 35–98 (2001). [DOI] [PubMed] [Google Scholar]
- 36.Ohki T., Ohno C., Oyama K., Mikhailenko S. V., Ishiwata S., Purification of cytoplasmic actin by affinity chromatography using the C-terminal half of gelsolin. Biochem. Biophys. Res. Commun. 383, 146–150 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Noguchi T. Q., Kanzaki N., Ueno H., Hirose K., Uyeda T. Q., A novel system for expressing toxic actin mutants in Dictyostelium and purification and characterization of a dominant lethal yeast actin mutant. J. Biol. Chem. 282, 27721–27727 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Lu H., Fagnant P. M., Bookwalter C. S., Joel P., Trybus K. M., Vascular disease-causing mutation R258C in ACTA2 disrupts actin dynamics and interaction with myosin. Proc. Natl. Acad. Sci. U.S.A. 112, E4168–E4177 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hatano T., Sivashanmugam L., Suchenko A., Hussain H., Balasubramanian M. K., Pick-ya actin—A method to purify actin isoforms with bespoke key post-translational modifications. J. Cell Sci. 133, jcs241406 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Collins J. H., Elzinga M., The primary structure of actin from rabbit skeletal muscle. Three cyanogen bromide peptides that are insoluble at neutral pH. J. Biol. Chem. 250, 5906–5914 (1975). [PubMed] [Google Scholar]
- 41.Jacobson G. R., Rosenbusch J. P., ATP binding to a protease-resistant core of actin. Proc. Natl. Acad. Sci. U.S.A. 73, 2742–2746 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van Hoof C., Goris J., Phosphatases in apoptosis: To be or not to be, PP2A is in the heart of the question. Biochim. Biophys. Acta 1640, 97–104 (2003). [DOI] [PubMed] [Google Scholar]
- 43.Doolittle L. K., Rosen M. K., Padrick S. B., Measurement and analysis of in vitro actin polymerization. Methods Mol. Biol. 1046, 273–293 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McKane M., et al. , A mammalian actin substitution in yeast actin (H372R) causes a suppressible mitochondria/vacuole phenotype. J. Biol. Chem. 280, 36494–36501 (2005). [DOI] [PubMed] [Google Scholar]
- 45.McKane M., Wen K. K., Meyer A., Rubenstein P. A., Effect of the substitution of muscle actin-specific subdomain 1 and 2 residues in yeast actin on actin function. J. Biol. Chem. 281, 29916–29928 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Bookwalter C. S., Trybus K. M., Functional consequences of a mutation in an expressed human alpha-cardiac actin at a site implicated in familial hypertrophic cardiomyopathy. J. Biol. Chem. 281, 16777–16784 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Carman P. J., Dominguez R., Novel protein production method combining native expression in human cells with an intein-based affinity purification and self-cleavable tag. Bio Protoc. 12, e4363 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carman P. J., Barrie K. R., Dominguez R., Novel human cell expression method reveals the role and prevalence of posttranslational modification in nonmuscle tropomyosins. J. Biol. Chem. 297, 101154 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fregoso F. E., et al. , Molecular mechanism of Arp2/3 complex inhibition by Arpin. Nat. Commun. 13, 628 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Solntsev S. K., Shortreed M. R., Frey B. L., Smith L. M., Enhanced global post-translational modification discovery with MetaMorpheus. J. Proteome Res. 17, 1844–1851 (2018). [DOI] [PubMed] [Google Scholar]
- 51.R. H. Ceron et al., Proteomics analysis of endogenous and recombinant β-actin. MassIVE. https://massive.ucsd.edu/ProteoSAFe/dataset.jsp?accession=MSV000089132. Deposited 26 March 2022. [Google Scholar]
- 52.R. H. Ceron et al., Proteomics analysis of endogenous and recombinant α-actin. MassIVE. https://massive.ucsd.edu/ProteoSAFe/dataset.jsp?accession=MSV000089919. Deposited 19 July 2022. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Proteomics data have been deposited in the MassIVE Repository [accession nos. MSV000089132 (51) and MSV000089919 (52)].