Abstract
The SARS-CoV-2 virus has complex and divergent immune alterations in differing hosts and over disease evolution. Much of the nuanced COVID-19 disease immune dysregulation was originally dominated by innate cytokine changes, which has since been replaced with a more complex picture of innate and adaptive changes characterized by simultaneous hyperinflammatory and immunosuppressive phenomena in effector cells. These intricacies are summarized in this review as well as potential relevance from acute infection to a multisystem inflammatory syndrome commonly seen in children. Additional consideration is made for the influence of variant to variant host cellular changes and the impact of potential vaccination upon these phenotypes. Finally, therapeutic benefit for immune alterations are discussed.
Keywords: COVID-19, SARS-CoV-2, Immunology, Host immune response, Innate, Adaptive
1. Introduction
Immune dysregulation in the context of the SARS-CoV-2 virus (COVID-19 disease) is complex. Early research in the COVID-19 disease pandemic identified a pathophysiology of cytokine storms leading to damaging hyperinflammation while additional research demonstrated profound long-term immune suppression [1,2]. Although COVID-19 patients may exhibit elevated levels of inflammatory cytokines compared to non-critically-ill patients, a study comparing the immune profiles of COVID-19 and influenza noted that while a 3–4% subset of COVID-19 patients exhibited hyperinflammation characteristic of a cytokine storm, they more commonly demonstrated immunosuppression [3]. COVID-19 associated lymphopenia is predictive of poor outcomes and is a risk factor for secondary hospital-acquired infections, accounting for 50% of estimated mortality secondary to COVID-19 [4,5]. Given its severity and complexity, we performed a narrative literature review to evaluate the mechanisms of immune dysregulation that is specific to cell type, disease, and individual immune systems to unlock targeted therapies for COVID-19 disease and identify future paths for immunologic research.
2. Innate immune system
Both the innate and adaptive immune systems experience dysregulation in COVID-19. Responsible for the initial antiviral activity, the innate system functions as a single defense mechanism, crucial for host response and illness protection. Pattern recognition receptors (PRRs) on innate immune cells detect pathogen-associated molecular patterns (PAMPs) from invading microbes and damage-associated molecular patterns (DAMPs) released from dying host cells [6]. In COVID-19, possible PAMPs include proteins from the SARS-CoV-2 viral envelope, spikes, and nucleoproteins (N) as well as single stranded RNA [6]. DAMPs may include S100A8/A9 and nucleic acids from dead cells [6]. This activation leads to NF-κB and AP-1 transcription of antiviral and inflammatory cytokines designed to induce apoptosis and inhibit viral replication [6].
Paradoxically, in COVID-19 pneumonia, the innate immune system fails to mount an effective antiviral response while also inducing potentially damaging inflammation. Severe cases are especially marked by decreased early production of type I and type III interferons (IFN) allowing for SARS-CoV-2 to replicate and cause severe cellular damage in the lungs [3,[6], [7], [8]]. Not only is this antiviral response of IFN delayed and reduced, but also accompanied by an unusually early and strong production of cytokines [9]. This overexaggerated and unregulated inflammatory response includes interleukin-6 (IL-6), interleukin-8 (IL-8), interleukin-10 (IL-10), and tumor necrosis factor-α (TNF-α), all of which predict disease severity and mortality [[9], [10], [11]]. Importantly, the ensuing hyperinflammation is believed to induce edema, fibrosis, and thrombosis in the lungs leading to hypoxia, acute respiratory distress syndrome (ARDS), and death [6]. However, hyperstimulation by inflammatory cytokines of cell surface receptors on adaptive immune cells such as lymphocytes and monocytes may cause a paradoxical immunosuppressive effect, a concept that will be explored in more detail in subsequent sections.
In addition to PRR pathways in COVID-19, dysregulation occurs in other sensors of cellular stress including the transcription factors, nuclear factor, erythroid 2-related factor (Nrf2) and hypoxia-inducible factor 1a (HIF-1a). Nrfr2 activation inhibits SARS-CoV-2 replication through type 1 IFN signaling and inhibits inflammatory cytokine release [6,12,13]. However, lungs of COVID-19 patients exhibit Nrf2 suppression, possibly contributing to decreased antiviral action and increased cytokine levels [13]. Conversely, HIF-1α activation is increased in severe COVID-19 [6]. HIF-1α is correlated with mortality in elderly COVID-19 patients and may contribute to the induction of inflammatory organ damage [6,14].
Hyperinflammatory damage with additionally immune paralyzing effects to host innate responses provide a complex sequence of dysregulation creating specific cell type chaos that leads to altering degrees of disease severity, and varies during the course of the disease.
3. Monocytes and macrophages
The immune profile of monocytes and macrophages in COVID-19 is multifaceted. Several single-cell profiling studies of bronchoalveolar lavages have identified an increase in hyperinflammatory monocytes in critical COVID-19 with an accompanying decrease in anti-inflammatory alveolar macrophages leading to cell death and fibrosis of the lungs [6,8,15]. In addition, a transcriptomic study found peripheral monocytes to have significantly increased S100A8/A9, a marker for inflammation, believed to contribute to a possible cytokine storm in severe COVID-19 [16].
In contrast to the inflammatory nature of focal myeloid lineage cells in infected lung tissue, functional analysis of peripheral blood mononuclear cells by ex vivo lipopolysaccharide stimulation and quantification of TNF-a producing cells demonstrated a 3-fold decrease in TNF-a-producing peripheral monocytes in COVID-19 patients compared to other critically ill patients revealing significant immune suppression [2].
An analysis of 54 patients, 28 of whom were diagnosed with COVID-19-associated severe respiratory failure, demonstrated that these two opposite phenotypes may be patient-dependent or not mutually exclusive [17]. Among the patients with severe respiratory failure, one-fourth were characterized as having macrophage-activation syndrome [17]. However, the majority demonstrated immune paralysis detailed by significant decreases in HLA-DR expression of CD14 monocytes similar to that seen in sepsis [17] and confirmed in other studies [3,18]. Importantly, macrophages have been identified as producing high levels of IL-6 in COVID-19, a cytokine with varying effects on many immune cells including neutrophils and lymphocytes [19]. These signaling pathways will be explored throughout this article. Together, these findings demonstrate the heterogeneity of immune dysregulation in severe COVID-19 and need for treatment that is individualized to a patient's immune profile.
4. Neutrophils
Neutrophils exhibit hyperinflammatory activity in COVID-19 and may account for significant cellular damage. Numerous studies have identified neutrophilia as characteristic of critical COVID-19 and is associated with disease severity, ARDS, and death [17,20,21]. Neutrophils are recruited to the lungs by increased levels of IL-8 and induce inflammation through the formation of neutrophil extracellular traps (NETs), induction of epithelial cell apoptosis, and release of DAMPs [6,20]. Elevated levels of IL-6 have been shown to induce this inflammatory action of neutrophils through STAT3 signaling [22,23]. However, these IL-6-stimulated neutrophils can suppress T lymphocyte activity through program death (PD) via program death ligand 1 (PDL1) signaling [23]. Current anti-IL-6 treatments (tocilizumab and sarulimab) seem to be effective at different times in bacterial and viral infections, however there is speculation whether IL-6 represents the extent of inflammatory response or the cause of organ injury [24]. Together, this research demonstrates how neutrophils may be responsible for significant inflammatory organ damage in COVID-19 while simultaneously contributing to a systemic picture of immune suppression.
5. Complement cascade
Critical COVID-19 is marked by early and stark complement activation that may result in inflammatory damage in the lungs. Circulating levels of complement factors including C3, C4d, and C5a are significant elevated in patients with severe critical illness due to COVID-19 [6,[25], [26], [27]]. While elevated complement levels were not associated with decreased viral load, they were correlated with the development of ARDS [25,28]. The proposed mechanism for activation and inflammation involves indirect viral N antigen interaction with mannan-binding lectin serine protease 2 (MASP-2) and activation of the lectin pathway leading to cellular deposition of the membrane attack complex (MAC) [6,[25], [26], [27], [28]]. This MAC deposition occurs in vascular walls, bronchial epithelial cells, macrophages, and lymphocytes, and the ensuing cellular damage and intravascular coagulation is believed to contribute to the pathogenesis of ARDS [[28], [29], [30]].
6. Epithelial cells
SARS-CoV-2 is able to infect both types I and II alveolar epithelial cells [6] by binding to the angiotensin converting enzyme 2 receptor (ACE2) [31]. Multiple studies examining postmortem lung samples from COVID-19 patients identified significant apoptosis and necroptosis of these alveolar cell types through caspase-8 activation and inflammatory infiltration leading to necrosis and fibrosis [32,33]. The apoptotic pathways and inflammatory destruction of epithelial cells are important in the pathogenesis of ARDS in COVID-19 and demonstrate potential targets for treatment of this grave complication [6].
7. Eosinophils
The role of cells associated with allergic reactions such as basophils and eosinophils in COVID-19 has not been investigated to the same extent as other cell types [6]. However, at least two studies have identified peripheral eosinopenia at presentation as reliably and independently correlated with severity in COVID-19 pneumonia [28,34]. Interestingly, severe cases were also marked by extended local infiltration and inflammatory activity of eosinophils in the lungs possibly pointing to a type I hypersensitivity reaction involving Th2 lymphocyte activity [28].
8. Dendritic cells
Plasmacytoid dendritic cells (pDCs) are essential for the innate immune response to COVID-19. pDCs recognize viral RNA through the endosomal TLR7 and produce type I and type III IFNs as an antiviral response [6,35]. Early in COVID-19 onset, a significant decrease in circulating pDCs is observed. This depletion indicates severity and is accompanied by failure to mount a strong initial antiviral response through type I IFN production [35,36]. Apoptotic signaling of pDCs is increased in severe disease, and antigen-presenting cDCs also appear to be inhibited as demonstrated by a decrease in major histocompatibility complex (MHC) class II genes [37]. Activation of the STAT3 pathway has been demonstrated to inhibit DC function in cancer and may explain a mechanism for the observed decrease in immune response and antigen presentation in high levels of IL-6 associated with severe COVID-19 [38].
9. Natural killer (NK) cells
Multiple studies have demonstrated the cytotoxic activity of NK cells on SARS-CoV-2-infected cells and identified an inverse relationship between viral load and NK cell activity [39,40]. However, severe COVID-19 is marked by NK cell cytopenia [17,36]. Compared to milder cases, NK cells in patients with severe COVID-19 produce less IFN-γ and TNF-α [21,40]. One study demonstrated increased NK cell expression of NKG2A, an inhibitory receptor leading to cell exhaustion in viral illnesses, in COVID-19 and identified it as a possible target for immune therapy [21]. An in vitro cell culture model and in vivo xenograft model using peripheral blood and tissue samples in esophageal squamous cell carcinoma demonstrated the ability of both IL-6 and IL-8 to inhibit NK cells function through the STAT3 pathway [41]. This further contributes to the picture of immune dysregulation from an increase in these cytokines and an inability to generate an adequate antiviral response in COVID-19.
10. Adaptive immune system
Cellular and humoral activity of the adaptive immune system is critical for developing a balanced and efficient host response to invading pathogens while also conferring immunologic memory for future infections with similar coronaviruses. Additionally, the activation of this process demonstrates particular importance in terms of vaccination efficacy and longevity [42,43]. Conversion from the innate immune response to the adaptive arm of the immune system is a critical function in acute COVID-19, and therefore, quantitative analysis of this response could maintain prognostic value. Early antibody production is associated with disease severity, and T cell production of IFN- ɣ is correlated with disease moderation [44]. Unfortunately, the adaptive immune system demonstrates significant dysregulation in COVID-19 as well.
11. B cells
Many COVID-19 patients demonstrate a robust memory B cell and antibody-secreting B cell plasmablast response early in infection [3,45,46]. However, research on B cell levels over the course of illness is conflicting with some studies identifying an increase of plasmablasts [3,46] and another identifying a decrease in B cell frequency [45]. Additionally, an imbalance of IL-6 and IL-10 production by B cells has been observed in COVID-19 [47]. However, most B cell alterations experienced in acute COVID-19 are recovered in convalescent patients [47].
12. T cells
CD4+ helper T cells and CD8+ cytotoxic T cells have been identified as crucial in the immunologic response to SARS-CoV-2 infection. CD4+ T cells are responsive to the virus's spike protein, and the presence of CD8+ T cell expansion in bronchoalveolar lavage is correlated with illness moderation [15,48].
However, one of the most remarkable characteristics of immune dysregulation in COVID-19 is an immense depletion of CD4+ and CD8+ T cells associated with disease severity [2,11,17,21,22,[48], [49], [50]]. While lymphopenia is observed in other respiratory viral illnesses such as influenza A H3N2 viral infection, COVID-19 induced lymphocytic depletion is distinctive for its magnitude and longevity [3,48,51]. Additionally, CD8+ T cells, crucial for their cytotoxic activity against virally infected cells, may experience the more stark reduction [48]. While CD8+ T cell have some cellular markers of activity indicating specificity to SARS-CoV-2, markers of exhaustion indicate dysregulated function [48]. This lymphopenia has been identified as an important risk factor in the development of secondary infections in hospitalized COVID-19 patients [5].
The lack of intense lymphocytic infiltration found in the lungs of critical COVID-19 patients demonstrates that the peripherally observed lymphopenia may be occurring through a mechanism beyond simply recruitment to the infection site [15,48]. Using an enzyme-linked immune absorbent spot (ELISpot) assay as a tool to quantitate peripheral immune cell function in patients with COVID-19, researchers not only confirmed T cell lymphopenia, but also identified functional suppression of T cells as measured by IFN-ɣ [2]. Decreased production of IFN-ɣ by CD4+ T cells correlates with COVID-19 severity [11]. T cell counts in COVID-19 patients are inversely proportional to IL-6, IL-10, and TNF-α concentrations and shown to express elevated levels of PD-1. These cytokines and signaling pathways have been identified as possible mechanisms for the observed immune exhaustion and dysregulation in COVID-19 [[48], [49], [50]].
13. Proposed mechanism
The mechanism of simultaneous hyperinflammatory damage in context of systemic immune suppression is undoubtedly complex. Yet, the production and function of IL-6, IL-8, and IL-10 provides a possible explanation concerning dysregulation of innate immunity and direct and indirect dysregulation of adaptive immunity [48].
Macrophages demonstrated elevated production of IL-6 in COVID-19 accompanied by increased levels of IL-8 [3,9,10,19]. Neutrophils recruited to the site of infection by IL-8 are activated by IL-6 to induce inflammatory activity and epithelial cell damage [20,22,23]. However, these IL-6-activated neutrophils suppress T lymphocytes through programmed death ligand (PD1/PDL1) signaling [23]. PD-1 expression of T cells has been identified as significantly upregulated in COVID-19 patients, resulting in the apoptosis that may account for the severe lymphopenia seen in critical COVID-19 [[48], [49], [50]].
Additionally, IL-6 and IL-10 may act on T cells and monocytes more directly. A study using mass spectrometry to identify upregulated signaling pathways in COVID-19 patients found significantly increased levels of STAT3 phosphorylation in myeloid and lymphocytic cells [18]. STAT3 is a transcription factor downstream of IL-6 and IL-10 signaling [52]. This upregulation of STAT3 was accompanied by decreased myeloid cell activation in addition to CD4+ and CD8+ T cell IFN-ɣ production, suggesting STAT3 signaling as a potentially unifying pathway for the dysregulation observed in COVID-19 [18].
While STAT3 is associated with some levels of inflammatory activity, it has also been shown to induce an anti-inflammatory phenotype when overexpressed. Its activation has been associated with proviral function in viruses such as HBV, HCV, HSV-1, VZV, human CMV, and measles virus [53,54]. STAT3 activation in tumor cells has also been demonstrated to inhibit the function of dendritic cells which may contribute to failure of successful antigen presentation in COVID-19 [38].
A proposed mechanism for IL-6/STAT3 signaling to induce the inactive phenotypes of NK cells and CD8+ T cells in COVID-19 is by increasing expression of NKG2A, an inhibitory receptor [21,22]. In the context of cancer, both IL-6 and IL-8 have been shown to impair NK function through the STAT3 pathway [41]. A failure for the cytotoxic cells to be activated sufficiently against SARS-CoV-2 infected cells would impair viral clearance and possibly worsen disease outcomes.
14. Multisystem inflammatory syndrome in children (MIS-C)
One severe presentation of dysregulated immune function in COVID-19 is multisystem inflammatory syndrome in children (MIS-C), defined as systemic inflammation with multiple organ involvement in children four to six weeks after exposure to SARS-CoV-2 [55]. MIS-C, characterized as a post-infectious syndrome, has characteristics that are similar to Kawasaki disease and toxic shock syndrome [[55], [56], [57]]. Speculation on the connection between SARS-CoV-2 and MIS-C is of great interest to researchers as providers are increasingly called to take action on these diagnoses in clinical practice.
The SARS-CoV-2 spike protein has been identified as a possible superantigen capable of T cell activation and interaction with MHC II [55,56]. Evidence also suggests MIS-C patients experience an activation of innate immune system with dysregulated IFN-ɣ signaling and a cytokine producing phenotype [55]. While inflammatory monocytes are also potentially responsible for hyperinflammation, anti-inflammatory components on monocytes and neutrophils in a cohort of patients are prototypical of the immune dysregulation discussed in this review [55].
Similar to COVID-19, studies have identified lymphopenia as a hallmark of disease in MIS-C [55]. Additionally, elevated levels of PD-1 in MIS-C patients suggest prolonged CD4+ and CD8+ T cell activation and exhaustion [55]. In summary, while MIS-C is characterized by a massive proinflammatory cytokine production caused by the innate immune system, there seems to be a simultaneous dysregulation of T and B cells in the adaptive immune system [55]. By studying the immune dysregulation in MIS-C, the role of a SARS-CoV-2 superantigen can be further explored and applied to that of COVID-19 and its treatments.
15. T cells, vaccination, and variants of concern
Understanding and addressing immune dysregulation remains paramount towards vaccination efficacy against SARS-CoV-2. As new variants of concern (VOCs) arise, mutations may allow viral particle to evade vaccination-induced antibodies directed toward spike proteins [58]. One study on the success of vaccinations in mice identified that memory CD4+ and CD8+ T cells were crucial for limiting disease severity during exposure to the beta variant [58]. Memory T cells are believed to be key in responding to VOCs because they recognize more viral epitopes that may be conserved compared to antibodies that only recognize spike proteins [42]. This evidence supports that establishing a functional adaptive immune response to COVID-19 is not only important in controlling a patient's initial infection but also in terms of vaccination efficacy and reinfection.
16. Treatment
The immune dysregulation observed in COVID-19 has strong implications for its treatment. Due to the widely-held belief that COVID-19 is characterized predominantly by a hyperinflammatory cytokine storm, many medications aiming to suppress the immune system have been considered, including anti-IL-6 receptor antibodies, IL-1 receptor antagonists, and JAK/STAT inhibitors [2]. However, in the context of significant immunosuppression explored in this review, anti-inflammatory therapies may be harmful and further diminish innate and adaptive immune responses necessary for viral control and illness moderation [24]. Instead, treatments that suppress the immune system may only be appropriate in the smaller subset of patients who are experiencing cytokine storms [3]. A randomized control trial found that the administration of dexamethasone reduced mortality in COVID, but only for critical patients needing respiratory support [59]. In theory, such treatments may be even more effective when targeted exclusively to patients with hyperinflammatory phenotypes.
Personalized immune profiling is already used in cancer immunotherapy and presents a promising possibility in COVID-19 [24]. One study identified ELISpot assays as potential tools in this phenotypic identification. Using peripheral blood samples from COVID-19 patients and ex vivo stimulation, researchers were able to quantitate lymphocyte and monocyte function through IFN-ɣ and TNF-α production respectively and identify personalized immune profiles for individual patients [2]. And as most patients demonstrated signs of immune suppression, proinflammatory therapies may result in better outcomes.
Furthermore, the innate immune system demonstrates potential for targeted therapy. As previously discussed, there is a failure to mount an early antiviral response to severe COVID-19. Therapies such as exogenous IFN-α may be able to counter this phenomenon and inhibit viral replication early in disease [6]. Additionally, other dysregulated transcription factors such as Nrf2 or HIF-1α, associated with disease severity, could be targets for modification. One group suggested the possibility of using anti-NKG2A monoclonal antibodies to block the inhibition of NK cells, a treatment that could affect the adaptive immune system by increasing CD8+ T cell function [22].
Finally, there are ways to reverse the adaptive immune suppression observed in COVID-19. IL-7, a pluripotent inflammatory cytokine, has been shown to ameliorate the activity of T cells in COVID-19 and has been proposed as a potential treatment [2,24]. Additionally, as T cell expression of PD-1 is upregulated in severe disease, the use of PD-1 inhibitors including nivolumab and pembrolizumab should be considered [24].
17. Conclusion
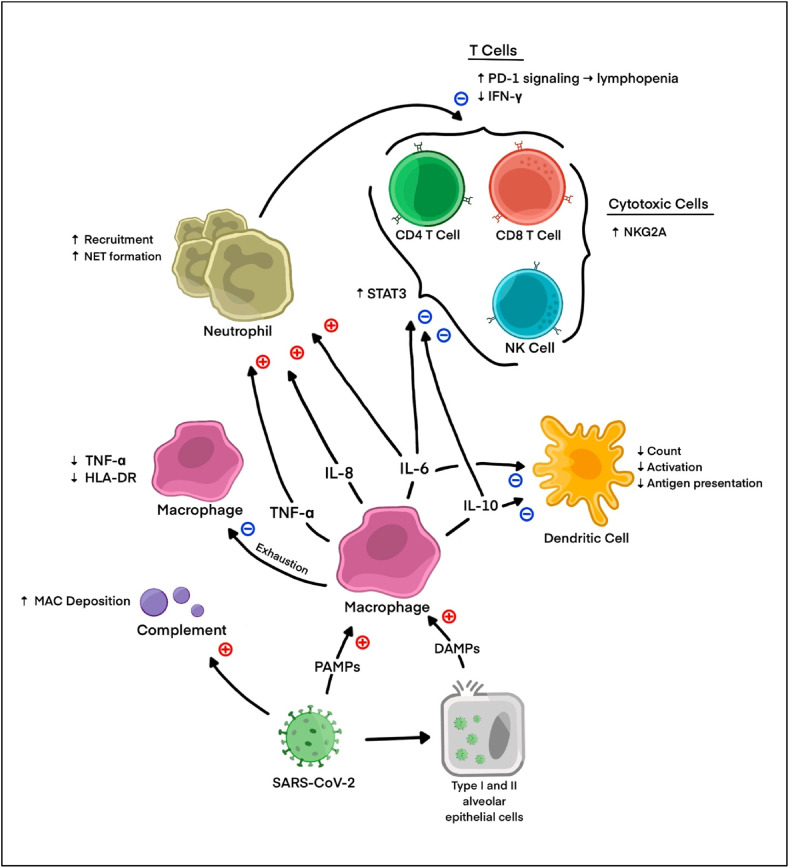
While COVID-19 was originally characterized as hyperinflammatory in its pathophysiology, emerging evidence demonstrates the possibility of a strongly immunosuppressive phenotype in more critical disease states. While immune activation from neutrophils and complement may contribute to inflammatory damage in the lungs, decreased antiviral responses, dysregulated macrophages and dendritic cells, and severe lymphopenia contribute to a suppressed state in which viral replication and secondary infections are prone. IL-6/STAT3 signaling may be an important contributor to this immune dysregulation and deserves further investigation. Ultimately, when considering the future of immune therapy in COVID-19, it will be essential to understand each patient's immune profile and adjust treatments accordingly (see Fig. 1 ).
Fig. 1.
Proposed pathways for inflammation and immune suppression in COVID-19 immune dysregulation.
Five key advances needed to guide COVID-19 therapies
-
•
Need a rapid test that can indicate the status of cellular immunity to SARS-CoV-2, i.e., does the individual or patient have a potent T cell response against SARS-CoV-2 spike and/or matrix antigens as a result of either primary infection or vaccines that would provide protect against infection.
-
•
Need a rapid test that can indicate the status of antibody immunity to SARS-CoV-2, i.e., does the individual or patient have potent effective antibodies against spike and/or matrix proteins of the SARS-CoV-2 virus due either to vaccination or primary infection
-
•
Ability to determine the relative importance of T cell versus B cell immunity against SAR-CoV-2 infection.
-
•
Ability to determine the functional status of the uninfected individual innate and adaptive immunity. Is the patient going to be able to mount an effective immune response against viral infection or vaccination or, is the patient immune suppressed due to immunosenescence, biologic agents, corticosteroids, or cancer and not able to develop potent immunity?
Practice points
-
•
Severe lymphopenia should be considered as potential immune paralysis in T cells. Further immunosuppressive therapies such as corticosteroids should be considered with caution.
-
•
Directed targeted therapies (barcitinib) to key pathways such as JAK/STAT must be provided early in disease course for benefit.
-
•
Further evaluation of targeted drug therapies should include functional immune assays that evaluate cellular function.
Declaration of competing interest
There are no conflicts to report. Dr. Remy is funded by the National Institutes of Health, National Institute of General Medical Sciences 7K08GM129763. Dr. Hotchkiss is funded by the NIH, NIGMS 5R35GM126928 and 5R01GM139046.
References
- 1.Moore J.B., June C.H. Cytokine release syndrome in severe COVID-19. Science. 2020;368(6490):473–474. doi: 10.1126/science.abb8925. [DOI] [PubMed] [Google Scholar]
- 2.Remy K.E., Mazer M., Striker D.A., Ellebedy A.H., Walton A.H., Unsinger J., et al. Severe immunosuppression and not a cytokine storm characterizes COVID-19 infections. JCI insight. 2020;5(17) doi: 10.1172/jci.insight.140329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mudd P.A., Crawford J.C., Turner J.S., Souquette A., Reynolds D., Bender D., et al. Distinct inflammatory profiles distinguish COVID-19 from influenza with limited contributions from cytokine storm. Sci Adv. 2020;6(50) doi: 10.1126/sciadv.abe3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ripa M., Galli L., Poli A., Oltolini C., Spagnuolo V., Mastrangelo A., et al. Secondary infections in patients hospitalized with COVID-19: incidence and predictive factors. Clin Microbiol Infect. 2021;27(3):451–457. doi: 10.1016/j.cmi.2020.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paludan S.R., Mogensen T.H. Innate immunological pathways in COVID-19 pathogenesis. Sci Immunol. 2022;7(67) doi: 10.1126/sciimmunol.abm5505. eabm5505. [DOI] [PubMed] [Google Scholar]
- 7.Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369(6504):718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wauters E., Van Mol P., Garg A.D., Jansen S., Van Herck Y., Vanderbeke L., et al. Discriminating mild from critical COVID-19 by innate and adaptive immune single-cell profiling of bronchoalveolar lavages. Cell Res. 2021;31(3):272–290. doi: 10.1038/s41422-020-00455-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galani I.-E., Rovina N., Lampropoulou V., Triantafyllia V., Manioudaki M., Pavlos E., et al. Untuned antiviral immunity in COVID-19 revealed by temporal type I/III interferon patterns and flu comparison. Nat Immunol. 2021;22(1):32–40. doi: 10.1038/s41590-020-00840-x. [DOI] [PubMed] [Google Scholar]
- 10.Del Valle D.M., Kim-Schulze S., Huang H.-H., Beckmann N.D., Nirenberg S., Wang B., et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat Med. 2020;26(10):1636–1643. doi: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H., et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest. 2020;130(5):2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muri J., Kopf M. Redox regulation of immunometabolism. Nat Rev Immunol. 2021;21(6):363–381. doi: 10.1038/s41577-020-00478-8. [DOI] [PubMed] [Google Scholar]
- 13.Olagnier D., Farahani E., Thyrsted J., Blay-Cadanet J., Herengt A., Idorn M., et al. SARS-CoV2-mediated suppression of NRF2-signaling reveals potent antiviral and anti-inflammatory activity of 4-octyl-itaconate and dimethyl fumarate. Nat Commun. 2020;11(1):1–12. doi: 10.1038/s41467-020-18764-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tian M., Liu W., Li X., Zhao P., Shereen M.A., Zhu C., et al. HIF-1α promotes SARS-CoV-2 infection and aggravates inflammatory responses to COVID-19. Signal Transduct Targeted Ther. 2021;6(1):1–13. doi: 10.1038/s41392-021-00726-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med. 2020;26(6):842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 16.Ren X., Wen W., Fan X., Hou W., Su B., Cai P., et al. COVID-19 immune features revealed by a large-scale single-cell transcriptome atlas. Cell. 2021;184(7):1895–1913. doi: 10.1016/j.cell.2021.01.053. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giamarellos-Bourboulis E.J., Netea M.G., Rovina N., Akinosoglou K., Antoniadou A., Antonakos N., et al. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe. 2020;27(6):992–1000. doi: 10.1016/j.chom.2020.04.009. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turnbull I.R., Fuchs A., Remy K.E., Kelly M.P., Frazier E.P., Ghosh S., et al. Dysregulation of the leukocyte signaling landscape during acute COVID-19. PLoS One. 2022;17(4) doi: 10.1371/journal.pone.0264979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paces J., Strizova Z., Daniel S., Cerny J. COVID-19 and the immune system. Physiol Res. 2020;69(3):379. doi: 10.33549/physiolres.934492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gustine J.N., Jones D. Immunopathology of hyperinflammation in COVID-19. Am J Pathol. 2021;191(1):4–17. doi: 10.1016/j.ajpath.2020.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng M., Gao Y., Wang G., Song G., Liu S., Sun D., et al. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell Mol Immunol. 2020;17(5):533–535. doi: 10.1038/s41423-020-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Antonioli L., Fornai M., Pellegrini C., Blandizzi C. NKG2A and COVID-19: another brick in the wall. Cell Mol Immunol. 2020;17(6):672–674. doi: 10.1038/s41423-020-0450-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng Y., Li H., Deng Y., Tai Y., Zeng K., Zhang Y., et al. Cancer-associated fibroblasts induce PDL1+ neutrophils through the IL6-STAT3 pathway that foster immune suppression in hepatocellular carcinoma. Cell Death Dis. 2018;9(4):1–11. doi: 10.1038/s41419-018-0458-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Remy K.E., Brakenridge S.C., Francois B., Daix T., Deutschman C.S., Monneret G., et al. Immunotherapies for COVID-19: lessons learned from sepsis. Lancet Respir Med. 2020;8(10):946–949. doi: 10.1016/S2213-2600(20)30217-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holter J.C., Pischke S.E., de Boer E., Lind A., Jenum S., Holten A.R., et al. Systemic complement activation is associated with respiratory failure in COVID-19 hospitalized patients. Proc Natl Acad Sci USA. 2020;117(40):25018–25025. doi: 10.1073/pnas.2010540117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ali Y.M., Ferrari M., Lynch N.J., Yaseen S., Dudler T., Gragerov S., et al. Lectin pathway mediates complement activation by SARS-CoV-2 proteins. Front Immunol. 2021;12:2645. doi: 10.3389/fimmu.2021.714511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perico L., Benigni A., Casiraghi F., Ng L.F., Renia L., Remuzzi G. Immunity, endothelial injury and complement-induced coagulopathy in COVID-19. Nat Rev Nephrol. 2021;17(1):46–64. doi: 10.1038/s41581-020-00357-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim D.-M., Kim Y., Seo J.-W., Lee J., Park U., Ha N.-Y., et al. Enhanced eosinophil-mediated inflammation associated with antibody and complement-dependent pneumonic insults in critical COVID-19. Cell Rep. 2021;37(1) doi: 10.1016/j.celrep.2021.109798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma L., Sahu S.K., Cano M., Kuppuswamy V., Bajwa J., McPhatter J.N., et al. Increased complement activation is a distinctive feature of severe SARS-CoV-2 infection. Sci Immunol. 2021;6(59) doi: 10.1126/sciimmunol.abh2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noris M., Benigni A., Remuzzi G. The case of complement activation in COVID-19 multiorgan impact. Kidney Int. 2020;98(2):314–322. doi: 10.1016/j.kint.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Varga Z., Flammer A.J., Steiger P., Haberecker M., Andermatt R., Zinkernagel A.S., et al. Endothelial cell infection and endotheliitis in COVID-19. Lancet. 2020;395(10234):1417–1418. doi: 10.1016/S0140-6736(20)30937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li S., Zhang Y., Guan Z., Li H., Ye M., Chen X., et al. SARS-CoV-2 triggers inflammatory responses and cell death through caspase-8 activation. Signal Transduct Targeted Ther. 2020;5(1):1–10. doi: 10.1038/s41392-020-00334-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y., Garron T.M., Chang Q., Su Z., Zhou C., Qiu Y., et al. Cell-type apoptosis in lung during SARS-CoV-2 infection. Pathogens. 2021;10(5):509. doi: 10.3390/pathogens10050509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cazzaniga M., Fumagalli L.A., D'angelo L., Cerino M., Bonfanti G., Fumagalli R.M., et al. Eosinopenia is a reliable marker of severe disease and unfavourable outcome in patients with COVID‐19 pneumonia. Int J Clin Pract. 2021;75(7) doi: 10.1111/ijcp.14047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van der Sluis R.M., Cham L.B., Gris‐Oliver A., Gammelgaard K.R., Pedersen J.G., Idorn M., et al. TLR2 and TLR7 mediate distinct immunopathological and antiviral plasmacytoid dendritic cell responses to SARS‐CoV‐2 infection. EMBO J. 2022;41(10) doi: 10.15252/embj.2021109622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laing A.G., Lorenc A., Del Molino Del Barrio I., Das A., Fish M., Monin L., et al. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat Med. 2020;26(10):1623–1635. doi: 10.1038/s41591-020-1038-6. [DOI] [PubMed] [Google Scholar]
- 37.Saichi M., Ladjemi M.Z., Korniotis S., Rousseau C., Ait Hamou Z., Massenet-Regad L., et al. Single-cell RNA sequencing of blood antigen-presenting cells in severe COVID-19 reveals multi-process defects in antiviral immunity. Nat Cell Biol. 2021;23(5):538–551. doi: 10.1038/s41556-021-00681-2. [DOI] [PubMed] [Google Scholar]
- 38.Wang T., Niu G., Kortylewski M., Burdelya L., Shain K., Zhang S., et al. Regulation of the innate and adaptive immune responses by Stat-3 signaling in tumor cells. Nat Med. 2004;10(1):48–54. doi: 10.1038/nm976. [DOI] [PubMed] [Google Scholar]
- 39.Krämer B., Knoll R., Bonaguro L., ToVinh M., Raabe J., Astaburuaga-García R., et al. Early IFN-α signatures and persistent dysfunction are distinguishing features of NK cells in severe COVID-19. Immunity. 2021;54(11):2650–2669. doi: 10.1016/j.immuni.2021.09.002. e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Witkowski M., Tizian C., Ferreira-Gomes M., Niemeyer D., Jones T.C., Heinrich F., et al. Untimely TGFβ responses in COVID-19 limit antiviral functions of NK cells. Nature. 2021;600(7888):295–301. doi: 10.1038/s41586-021-04142-6. [DOI] [PubMed] [Google Scholar]
- 41.Wu J., Gao F-x, Wang C., Qin M., Han F., Xu T., et al. IL-6 and IL-8 secreted by tumour cells impair the function of NK cells via the STAT3 pathway in oesophageal squamous cell carcinoma. J Exp Clin Cancer Res. 2019;38(1):1–15. doi: 10.1186/s13046-019-1310-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vardhana S., Baldo L., Morice W.G., Wherry E.J. Understanding T cell responses to COVID-19 is essential for informing public health strategies. Science immunology. 2022;7(71) doi: 10.1126/sciimmunol.abo1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sette A., Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021;184(4):861–880. doi: 10.1016/j.cell.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan A.T., Linster M., Tan C.W., Le Bert N., Chia W.N., Kunasegaran K., et al. Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Cell Rep. 2021;34(6) doi: 10.1016/j.celrep.2021.108728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tan M., Liu Y., Zhou R., Deng X., Li F., Liang K., et al. Immunopathological characteristics of coronavirus disease 2019 cases in Guangzhou, China. Immunology. 2020;160(3):261–268. doi: 10.1111/imm.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mathew D., Giles J.R., Baxter A.E., Oldridge D.A., Greenplate A.R., Wu J.E., et al. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020;369(6508) doi: 10.1126/science.abc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shuwa H.A., Shaw T.N., Knight S.B., Wemyss K., McClure F.A., Pearmain L., et al. Alterations in T and B cell function persist in convalescent COVID-19 patients. Med. 2021;2(6):720–735. doi: 10.1016/j.medj.2021.03.013. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen Z., John Wherry E. T cell responses in patients with COVID-19. Nat Rev Immunol. 2020;20(9):529–536. doi: 10.1038/s41577-020-0402-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Diao B., Wang C., Tan Y., Chen X., Liu Y., Ning L., et al. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19) Front Immunol. 2020:827. doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He L., Zhang Q., Zhang Y., Fan Y., Yuan F., Li S. Single-cell analysis reveals cell communication triggered by macrophages associated with the reduction and exhaustion of CD8+ T cells in COVID-19. Cell Commun Signal. 2021;19(1):1–18. doi: 10.1186/s12964-021-00754-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mudd P.A., Remy K.E. Prolonged adaptive immune activation in COVID-19: implications for maintenance of long-term immunity? J Clin Invest. 2021;131(1) doi: 10.1172/JCI143928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Murray P.J. Understanding and exploiting the endogenous interleukin-10/STAT3-mediated anti-inflammatory response. Curr Opin Pharmacol. 2006;6(4):379–386. doi: 10.1016/j.coph.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 53.Hillmer E.J., Zhang H., Li H.S., Watowich S.S. STAT3 signaling in immunity. Cytokine Growth Factor Rev. 2016;31:1–15. doi: 10.1016/j.cytogfr.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chang Z., Wang Y., Zhou X., Long J.-E. STAT3 roles in viral infection: antiviral or proviral? Future Virol. 2018;13(8):557–574. doi: 10.2217/fvl-2018-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mazer M.B., Bulut Y., Brodsky N.N., Lam F.W., Sturgill J.L., Miles S.M., et al. Multisystem inflammatory syndrome in children: host immunologic responses. Pediatr Crit Care Med. 2022;23(4):315–320. doi: 10.1097/PCC.0000000000002897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kouo T., Chaisawangwong W. SARS-CoV-2 as a superantigen in multisystem inflammatory syndrome in children. J Clin Investig. 2021;131(10) doi: 10.1172/JCI149327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Feldstein L.R., Tenforde M.W., Friedman K.G., Newhams M., Rose E.B., Dapul H., et al. Characteristics and outcomes of US children and adolescents with multisystem inflammatory syndrome in children (MIS-C) compared with severe acute COVID-19. JAMA. 2021;325(11):1074–1087. doi: 10.1001/jama.2021.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kingstad-Bakke B., Lee W., Chandrasekar S.S., Gasper D.J., Salas-Quinchucua C., Cleven T., et al. Vaccine-induced systemic and mucosal T cell immunity to SARS-CoV-2 viral variants. Proc Natl Acad Sci USA. 2022;119(20) doi: 10.1073/pnas.2118312119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Horby P.L.W., Emberson J.R., Mafham M., Bell J.L., Linsell L., et al. Dexamethasone in hospitalized patients with Covid-19. N Engl J Med. 2021;384(8):693–704. doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]