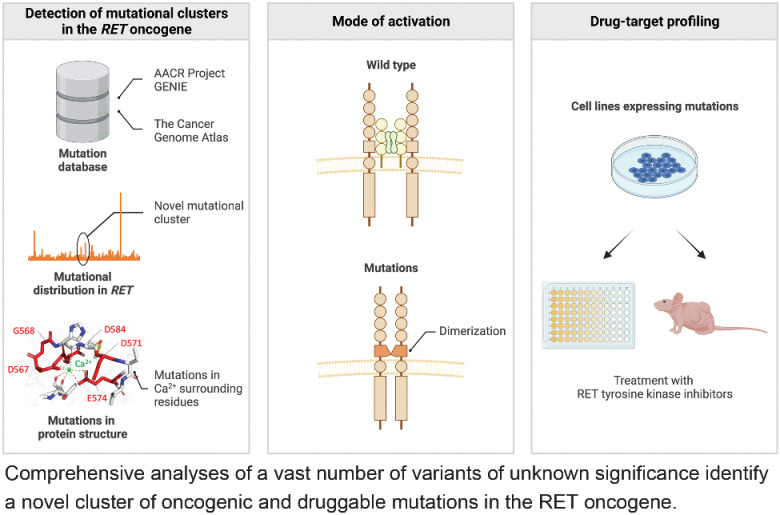
Comprehensive proteogenomic and in silico analyses of a vast number of VUSs identify a novel set of oncogenic and druggable mutations in the well-characterized RET oncogene.
Abstract
Distinguishing oncogenic mutations from variants of unknown significance (VUS) is critical for precision cancer medicine. Here, computational modeling of 71,756 RET variants for positive selection together with functional assays of 110 representative variants identified a three-dimensional cluster of VUSs carried by multiple human cancers that cause amino acid substitutions in the calmodulin-like motif (CaLM) of RET. Molecular dynamics simulations indicated that CaLM mutations decrease interactions between Ca2+ and its surrounding residues and induce conformational distortion of the RET cysteine-rich domain containing the CaLM. RET-CaLM mutations caused ligand-independent constitutive activation of RET kinase by homodimerization mediated by illegitimate disulfide bond formation. RET-CaLM mutants possessed oncogenic and tumorigenic activities that could be suppressed by tyrosine kinase inhibitors targeting RET. This study identifies calcium-binding ablating mutations as a novel type of oncogenic mutation of RET and indicates that in silico–driven annotation of VUSs of druggable oncogenes is a promising strategy to identify targetable driver mutations.
Significance:
Comprehensive proteogenomic and in silico analyses of a vast number of VUSs identify a novel set of oncogenic and druggable mutations in the well-characterized RET oncogene.
Graphical Abstract
Introduction
The identification of oncogenic mutations that drive tumorigenesis is important because these mutations are therapeutic targets for inhibitory drugs. Specific protein kinase inhibitors have improved the prognosis of patients with activating mutations in oncogenic kinase genes (1). Hotspot mutations affecting a single or a few amino acid residues associated with human cancers have been intensively studied as oncogenic and druggable mutations because of their cell-transforming activity and drug sensitivity. These include mutations in the BRAF (V600E; ref. 2), KRAS (G12C; ref. 3), EGFR (L858R; ref. 4), and RET (M918T; refs. 5, 6) genes. However, next-generation sequencing-based genomic analysis of tens of thousands of cancer cases shows that many cancer genomes also carry “apparently non-clustered (scattered)” mutations in well-known oncogenes (7, 8). Such mutations are called variants of unknown significance (VUS), and their role in tumorigenesis and drug sensitivity has not been studied extensively because of their low frequency of occurrence and thus low therapeutic potential. There is an urgent need to predict the oncogenic effects of such VUSs in precision cancer medicine because this could help to select cases who would benefit from molecular targeted therapies using existing inhibitors. The existence of vast numbers of VUSs led us to speculate that their functional annotation can be rapidly performed using in silico methods, such as AlphaFold2 analysis, which predicts protein structures from amino acid sequences (9). However, the gap between structural prediction and functional annotation hampers us from distinguishing the few mutations with functional significance (“drivers”) from biologically neutral mutations (“passengers”; ref. 10).
In the present study, we sought to identify oncogenic and druggable mutations among the large number of VUSs of a therapeutic target oncogene by conducting an in silico–driven functional (in vitro) analysis. A well-known traditional oncogene, RET, was chosen as a model case for several reasons. First, hotspot mutations of RET in thyroid cancers are used as drug targets (11, 12). Recently developed RET-specific tyrosine kinase inhibitors (TKI) have shown remarkable therapeutic effects in cancer patients with such oncogenic RET mutations (5, 6). Second, the molecular mechanisms underlying the oncogenicity conferred by hotspot mutations have been elucidated. RET hotspot mutations result in the substitution of amino acid residues in the kinase domain or in cysteine residues that form intramolecular disulfide bonds under physiologic conditions. These cysteine residues are included in the core cysteine motif (CCM), which spans the extracellular cysteine-rich domain (CRD) and the transmembrane domain (Fig. 1A). Kinase domain mutations increase kinase activity by enhancing enzymatic activity and/or substrate presentation for autophosphorylation (12). CCM mutations generate unpaired cysteine residues that form intermolecular disulfide bonds, leading to constitutive RET dimerization and constitutive enzyme activation (11, 13). Therefore, identifying additional mutations with oncogenic/therapeutic potential will increase the population of cancer patients who benefit from RET-TKI therapy.
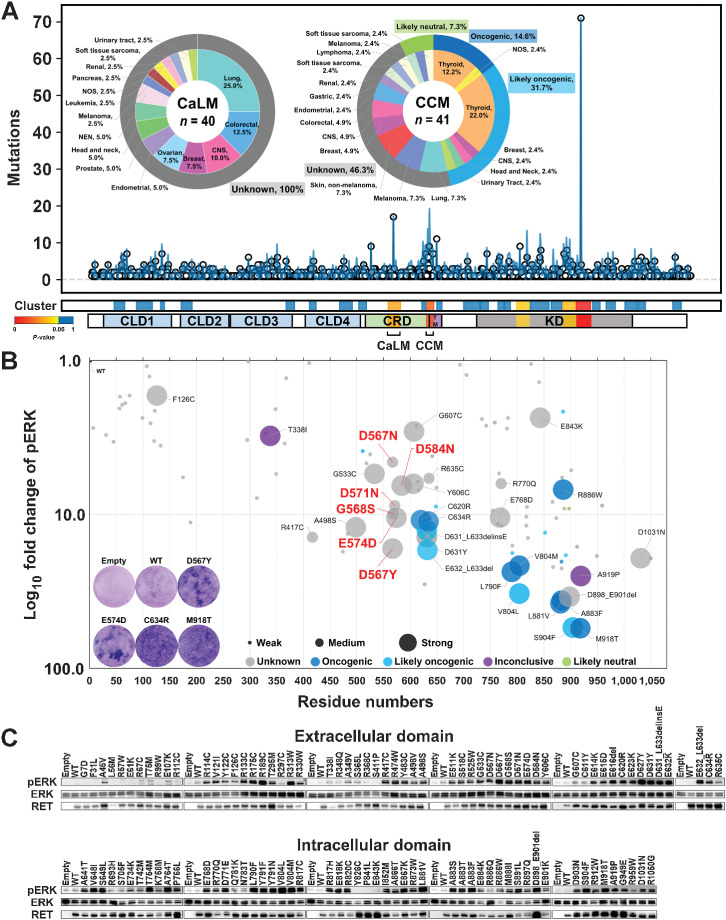
Figure 1.
Identification of the RET-CaLM mutation cluster. A, Mutational modeling for positive selection. The top graph shows the incidence of individual 1,290 RET mutations (circles) with a smoothed view of distribution (blue lines). The bottom bars show mutational clusters determined by OncodriveCLUSTL with P values and distribution of domains/motifs. Pie charts show the proportion of cancer types and OncoKB annotations (37) for the CaLM and CCM mutation clusters. The numbers in the center of the pie charts refer to the total number of mutations in the clusters. The percentages in the pie charts refer to the fractions of mutations in each cancer type among total mutations. CLD1, cadherin-like domain 1, residues 29–154; CLD2, cadherin-like domain 2, residues 172–261; CLD3, cadherin-like domain 3, residues 265–379; CLD4, cadherin-like domain 4, residues 404–506; CRD, cysteine-rich domain, residues 516–634; TM, transmembrane domain, residues 635–657; KD, kinase domain, residues 723–1012; CaLM, calmodulin-like motif, residues 557–580; CCM, core cysteine motif, residues 629–643. B, Bubble plot showing NIH3T3 cell–transforming ability and ERK phosphorylation activity of RET mutations. The horizontal axis indicates the amino acid residue numbers, and the vertical axis shows the log10-fold increase in ERK phosphorylation compared with WT RET. The size of the plot for each mutation indicates the magnitude of focus formation in three grades. The color of each plot indicates the significance grades of OncoKB annotation (37). Blue, oncogenic; light blue, likely oncogenic; gray, unknown significance; purple, inconclusive; green, likely neutral. Representative images of Giemsa-stained NIH3T3 cells are shown in the graph area. C, ERK phosphorylation assay. HEK293H cells transfected with empty vector or WT RET, 110 RET mutants, and kinase-dead mutant (K758M)-expressing plasmids were subjected to immunoblot analysis of phosphorylated ERK (pERK), ERK, and RET. The relative pERK levels compared with those in the WT are plotted in B.
Here, the in silico genome-wide modeling of 71,756 variants in different human cancers for positive selection identified a novel mutational cluster in the RET gene that undergoes mutagenesis beyond the local background level of tri- or pentanucleotide substitutions (14). The mutations resulted in a three-dimensional (3D) cluster of amino acid substitutions in the calmodulin-like motif (CaLM), which was recently identified by cryo-electron microscopy (EM) analysis of the RET extracellular domain (ECD; ref. 15). Molecular dynamics (MD) simulation using the high-performance supercomputer Fugaku (16) indicated that mutations in CaLM destabilize binding of the RET–Ca2+ ion complex. Gene-wide functional analyses of 110 representative RET variants, including in vitro cell transformation and signal transduction assays, validated the CaLM mutations as a novel cluster of activating mutations, whereas other variants resulting in amino acid substitutions in the ECD were mostly neutral. The CaLM mutations caused ligand-independent homodimerization of RET kinase through illegitimate disulfide bond formation, which occurs in wild-type RET only under artificial Ca2+-free conditions. These results indicate that annotating VUSs scattered in oncogenes is a promising strategy for precision oncology by identifying novel oncogenic mutations in a traditional oncogene.
Materials and Methods
RET mutation selection
For the discovery cohort, mutational data of 71,756 variants from 79,720 tumors excluding gross alterations, such as fusion and copy-number alterations, were downloaded from the American Association for Cancer Research Project GENIE (version 7.0-public) website (https://genie.cbioportal.org/; Supplementary Fig. S1A). Variant allele frequencies within a sample were calculated through the relative abundance of a variant over total sequence read counts obtained from the GENIE data set. Mutations were analyzed using OncodriveCLUSTL (14), a nucleotide sequence-based clustering algorithm that detects cancer driver genes based on a local background model derived from the trinucleotide context mutational probabilities of the cohort under study (i.e., GENIE; Supplementary Fig. S1B, left). The GENIE mutational data set was used as the mutational profile data for processing. The background data set (training cohort) consisted of somatic mutation data in the Mutation Annotation Format of whole-exome sequencing compiled by the MC3 Working Group from The Cancer Genome Atlas (TCGA; version 0.2.8; ref. 17) because it contains similar fractions of different mutation types to those of the GENIE data set (Supplementary Fig. S1B, right). Statistical significance was examined with a smoothing and clustering window of 9 bp and a simulation window of 35 bp to increase sensitivity at a single gene level. A total of 110 RET mutants including six CaLM, 35 known (5, 6, 11, 12, 18, 19), and 69 other mutants recurrently observed in three or more GENIE cases were subjected to gene-wide cell-based assays.
Cell lines and reagents
HEK293H cells (catalog no. 11631017, RRID:CVCL_6643), Flp-in T-REx 293 cells (catalog no. R78007, RRID:CVCL_U427), and Sf21 cells (catalog no. 12682019, RRID:CVCL_0518) were purchased from Thermo Fisher Scientific. Ba/F3 cells were provided by Dr. Yasuda Hiroyuki of Keio University School of Medicine. NIH3T3 cells were provided by Dr. Hiroyuki Mano of the National Cancer Center Research Institute. 293FT cells (catalog no. R70007, RRID:CVCL_6911) were obtained from Invitrogen in 2013. WEHI-3B cells (catalog no. RCB2853, RRID:CVCL_2239) were obtained from the RIKEN BioResource Center in 2016. U-2 OS cells (catalog no. HTB-96, RRID:CVCL_0042) were obtained from the ATCC. The 293H and NIH-3T3 cells were tested for Mycoplasma contamination with a MycoAlert detection kit (Lonza) prior to the study and the in vivo experiments, respectively, whereas no testing was performed for the other cell lines during the experiments. The 293H, Flp-in T-REx 293, and 293FT cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS) purchased from Thermo Fisher Scientific. WEHI-3B cells were cultured in an RPMI medium containing 10% FBS. Ba/F3 cells were cultured in the RPMI medium containing 10% FBS and 10% WEHI-3B–conditioned medium (a source of IL3). All cells were incubated at 37°C in 5% CO2. Primary antibodies against RET (catalog no. 14556, RRID:AB_2798509), phospho-RET (Tyr905; catalog no. 3221, RRID:AB_2179887), p44/42 MAPK (ERK1/2; catalog no. 9102, RRID:AB_330744), phospho-p44/42 MAPK (ERK1/2; Thr202/Tyr204; catalog no. 4370, RRID:AB_2315112), SHC (catalog no. 2432, RRID:AB_2254631), and phospho-SHC (Tyr239/240; catalog no. 2434, RRID:AB_10841301), and secondary antibodies against rabbit IgG (catalog no. 7074, RRID:AB_2099233) and mouse IgG (catalog no. 7076, RRID:AB_330924) were purchased from Cell Signaling Technology. A primary antibody against β-actin (catalog no. ab8227, RRID:AB_2305186) was purchased from Abcam. A primary antibody against FLAG (catalog no. F1804, RRID:AB_262044) was purchased from Sigma-Aldrich. A primary antibody against GST (catalog no. 27457701, RRID:AB_771432) was purchased from Cytiva. Recombinant human glial cell-derived neurotrophic factor (GDNF; catalog no. 212-GD) and GFRα1 (catalog no. 714-GR) were purchased from R&D Systems. Selpercatinib (catalog no. S8781) and pralsetinib (catalog no. S8716) were purchased from Selleck. These compounds were dissolved in DMSO to yield 10 mmol/L stocks and stored at −20°C.
Construction of lentiviral vectors expressing wild-type and mutant RET cDNAs
Full-length wild-type and mutant RET-51 cDNAs (NM_020975) were synthesized by GeneArt, a part of Thermo Fisher Scientific, and ligated into pLenti6.3/V5-DEST plasmids (catalog no. V53306, RRID:Addgene_56658; Invitrogen). The final constructs were verified by Sanger sequencing. Expression of cDNA products was confirmed by immunoblotting of transiently transfected 293H cells. Lentiviruses were generated in 293FT cells (6 × 106 cells per 10 cm plate) transfected with pLenti6.3/V5-DEST plasmid and ViraPower packaging mix (Invitrogen) using Lipofectamine 3000 (Invitrogen). Viral supernatants were collected 24 to 48 hours after the medium change and used for infection.
Phosphorylation assay
HEK293H cells seeded in 6-well plates were transiently transfected with 0.25 μg of plasmid DNA using Lipofectamine 3000. The cells were cultured under serum-starvation conditions for 48 hours after transfection and lysed in RIPA buffer [20 mmol/L Tris-HCl (pH 7.5), 150 mmol/L NaCl, 1 mmol/L Na2EDTA, 1 mmol/L EGTA, 1% NP-40, 1% sodium deoxycholate, 2.5 mmol/L sodium pyrophosphate, 1 mmol/L β-glycerophosphate, 1 mmol/L Na3VO4, and 1 μg/mL leupeptin] containing a protease/phosphatase inhibitor cocktail (Cell Signaling Technology). Cell lysates were centrifuged at 14,000 rpm for 15 minutes, and the supernatants were collected. The supernatants were subjected to SDS-PAGE, followed by immunoblotting onto polyvinylidene difluoride membranes. The membranes were blocked for 1 hours with TBS containing 0.1% Tween 20 (TBST) and 1.0% bovine serum albumin, and then probed with the following primary antibodies: anti-RET (CST #14556), anti-p44/42 MAPK (ERK1/2; CST #4695), and anti–phospho-p44/42 MAPK (ERK1/2; Thr202/Tyr204; CST #4370). After washing with TBST, the membranes were incubated with a horseradish peroxidase–conjugated secondary antibody against rabbit IgG (CST #7074) and then visualized with an enhanced chemiluminescence reagent (PerkinElmer). The intensity of signals was quantified using a LAS3000 imaging system (Quansys Biosciences) and Multi-Gauge software (Fujifilm). For conditional experiments in a medium with or without calcium ions, cells were cultured in calcium-containing DMEM (catalog no. 08459-35, Nacalai Tesque) or calcium-free DMEM (catalog no. 16972-45, Nacalai Tesque) for 48 hours after transfection under serum starvation. The phosphorylation of RET and ERK was confirmed using an antibody against phospho-RET (Tyr905; CST #3221) and phospho-p44/42 MAPK (ERK1/2; Thr202/Tyr204; CST #4370). For the phosphorylation assay, GDNF and GFRα1 were added at 48 hours after transfection and incubated for an additional 6 hours. The indicated drugs were added 48 hours after transfection and incubated for an additional 6 hours. Assays were independently repeated three times.
Assays of Ba/F3 cells
Ba/F3 cells (4.0 × 105) were infected with lentivirus in the presence of 10 μg/mL polybrene (Sigma-Aldrich) by centrifugation at 3,000 × g for 150 minutes at 32°C. Following overnight incubation at 37°C in 5% CO2, the cells were seeded into 24-well plates and selected in medium containing IL3 and 8 μg/mL blasticidin (Invitrogen) for 10 days. The blasticidin-resistant cells were grown in IL3-free medium for 2 weeks. The expression of exogenous RET proteins was confirmed by immunoblotting. For the drug treatment, 5 × 103 Ba/F3 cells were plated in quadruplicate in 96-well plates and serially diluted inhibitors were added to the wells. Cell viability was measured at 72 hours after drug treatment using the CellTiter-Glo luminescent cell viability reagent (Promega) with EnVision (PerkinElmer). Cell viability was calculated as the cell count in drug-treated samples relative to that in untreated samples (n = 6). The data were displayed graphically using GraphPad Prism version 9.0 (GraphPad Software Inc., RRID:SCR_002798).
NIH3T3 transformation assay
NIH3T3 cells were infected with lentiviruses in the presence of 8 μg/mL polybrene (Sigma-Aldrich) for 24 hours and cultured in DMEM-F12 supplemented with 5% calf serum for up to 2 weeks as previously described (20). Focus formation assays for infected NIH3T3 cells were performed in the absence of GDNF/GFRα1 or any other ligands for screening, and functional studies for alanine substitution mutations were performed in the presence of 5% calf serum for 14 days. When evaluating the addition of ligand and coreceptor, infected NIH3T3 cells were cultured with 0, 1, or 2.5 ng/mL GDNF and GFRα1 in the presence of 5% calf serum for 14 days. Cell transformation was assessed by staining with Giemsa solution. Reproducibility was confirmed by performing the same experiment three times. The focus area was quantified using ImageJ software (NIH, RRID:SCR_003070).
RET dimerization assay
T-REx 293 cells expressing full-length wild-type, D567N, D567Y, and C634R RET cDNAs and an empty control were established using the Flp-in T-REx-293 system according to the manufacturer's protocol. The cloned cells were cultured in normal (calcium-containing) or calcium-free DMEM supplemented with 0.1 μg/mL doxycycline (Fujifilm) for 6 hours. In this assay, the protein extracts were prepared using RIPA buffer with or without 40 μmol/L dithiothreitol (Cell Signaling Technology). An antibody against FLAG (catalog no. F1804, Sigma-Aldrich) was used as the primary antibody in the immunoblot analysis.
For the Sf21 cell assay, pFastBac (Thermo Fisher Scientific) baculoviral plasmids containing cDNAs encoding the CLD4-CRD (amino acids 382–635) region of RET were transfected into Sf21 cells using Cellfectin II Reagent (Thermo Fisher Scientific). Transfected cells were lysed in extraction buffer (50 mmol/L HEPES, 0.3 M NaCl, and 0.2% NP-40, pH 7.5) with or without 5 mmol/L EDTA. Following centrifugation (10,000 × g), GST-Sepharose beads were added to the supernatant and mixed by rotation at 4°C for 30 minutes. The beads were washed thoroughly with extraction buffer and mixed in loading buffer (50 mmol/L Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, and 0.1% bromophenol blue) with or without 1% 2-mercaptoethanol (2-ME) and incubated at 95°C for 5 minutes. The samples were electrophoresed, and dimer formation was detected by immunoblotting using an anti-GST antibody as the primary antibody or by Coomassie brilliant blue (CBB) staining.
NanoBiT assay
U-2 OS cells were transfected with plasmids expressing the Small BiT (SmBit; N197 pBiT2.1-C [TK/SmBiT] Vector) or Large BiT (LgBit; N196 pBiT1.1-C [TK/LgBiT] Vector) luciferase subunits (Promega) fused to the C-terminus of the RET-ECD domain (encoding amino acids 1–662) of wild-type (WT), C634R, D567N, and D567Y RET via a peptide linker. Transfected cells were then seeded onto 96-well polylysine-coated plates. Next, cells (1.13 × 104 per well) were transfected with 5 ng of a mixture of SmBit and LgBiT plasmid DNA at a mass ratio of 1:1. After 24 hours, cells were serum-starved with Opti-MEM for 5 hours. Fifteen minutes after the addition of the NanoGlo live-cell luciferase substrate, GDNF and GFRα1 were added to the cells to a final concentration of 7.4 μg/mL, and the kinetics of luminescence was measured for 45 minutes using a Synergy HTX plate reader (BioTek).
In vivo tumorigenesis
NIH3T3 cells expressing the RET C634R, D567N, and D567Y mutants were selected by exposure to 8 μg/mL blasticidin for 20 days after lentiviral infection. Cells were counted and resuspended in a 1:1 mixture of 100 μL culture medium and 100 μL Matrigel (BD Biosciences) on ice. Cells (5 × 106 cells/mouse) were injected subcutaneously into the flanks of 6-week-old female BALB/c-nu: CAnN.Cg-Foxn1nu/CrlCrlj mice (Charles River; RRID:IMSR_CRL:194). Mice were divided into three groups for each mutant, and treatments began when tumors measured between approximately 100 and 200 mm3. For all mutant experiments, mice were dosed twice daily with vehicle or 3 mg/kg selpercatinib or pralsetinib. Tumor growth was measured at regular intervals using calipers, and tumor volume was calculated using the following formula: tumor volume (mm3) = 0.5 × length × width × width. The subcutaneous tumor weight was measured at the end of the experiment, and the mice were sacrificed according to standard protocols. The experiment was approved by the National Cancer Center Animal Ethical Committee (approval number: A277bM1-21).
MD simulation of monomers
The initial cryo-EM structural data of the RET-CRD were obtained from the Protein Data Bank (PDB code: 6Q2N). Monomer models for the MD simulation were constructed using residues 508–622 of the E chain, and the other residues were deleted. Bond order assignment and hydrogenation were performed using the Protein Preparation module in Maestro 2020-2 (Schrödinger, Inc.). The structures of disordered loops (residues 508–514 and 547–548) were modeled using Prime (21, 22). The N- and C-termini of all protein models were capped with acetyl and N-methyl groups, respectively. RET mutation models were created using “Sequence viewer” in Maestro (Schrödinger, Inc.) and were based on the 6Q2N structure using the same protocol. H-bond optimization and energy minimization were performed using PROPKA (23) and the OPLS3e force field (24).
MD simulations for interaction analysis and evaluation of binding free energy were performed using Desmond (Schrödinger, Inc.) The Molecular Dynamics System Setup module in Maestro was used for preparing hydration models. All energy-minimized models were placed in an orthorhombic box with a buffer distance of 10 Å to create a hydration model. The SPC water model (25) was used for the hydration model. NaCl (0.15 M) was included as the counter ion to neutralize the system. The cutoff radii for Van der Waals interactions was 9 Å, and the time step, initial temperature, and pressure of the system were set to 2.0 fs, 300 K, and 1.01325 bars, respectively. The sampling interval of the trajectory frame was set to 100 ps. The final production run was performed for 1 μs using the number of molecules, pressure, and temperature (NPT) ensemble.
All trajectories from the MD simulation were aligned to the initial frame with protein Cα. The root-mean-square deviation (RMSD) and distance analyses were performed using the Simulation Interactions Diagram module in Maestro and based on the coordinates of the first frame. The MM/PB(GB)SA (26) method was used to predict the binding free energy of Ca ions to the CRD. Binding free energy analysis was performed at 100 frame intervals for each trajectory, and Prime (21, 22) was used for the analysis.
MD simulation of the RET–GDNF–GFRA1 extracellular complex
The initial structural data of the RET/GFRα1/GDNF extracellular complex were obtained from the PDB (code: 6Q2N). The structures of disordered loops were modeled using the Structure Preparation module in the MOE program (Chemical Computing Group Inc.). The N- and C-termini of all protein models were capped with acetyl and N-methyl groups, respectively. The dominant protonation state at pH 7.0 was assigned for titratable residues. A D567Y or D567N mutation was introduced into the structure of WT RET using MOE.
All MD simulations were performed using the GROMACS 2021 program (27). The Amber ff99SB-ILDN force field (28) was used for protein and ions, and TIP3P (29) was used to model water molecules. Approximately 280,000 water molecules were placed around each complex model with an encompassing distance of 10 Å. Sodium and chloride ions (150 mmol/L each) were introduced into the simulation box to neutralize the system. Electrostatic interactions were calculated using the particle mesh Ewald method (30) with a cutoff radius of 10 Å. The cutoff for Van der Waals interactions was set at 10 Å. Virtual sites for hydrogen atoms were used to allow for a time step of 4 fs (31). The P-LINCS algorithm was used to constrain all bond lengths at their equilibrium value (32). After energy minimization, each system was equilibrated for 100 ps in a constant number of molecules, volume, and temperature ensemble and run for 100 ps in a constant NPT ensemble, with positional restraints applied on protein heavy atoms. The temperature was maintained at 310 K by stochastic velocity rescaling (33), and a Parrinello–Rahman barostat was used to maintain the pressure at 1 bar (34). The temperature and pressure time constants were set to 0.1 and 2 ps, respectively. Three independent 1 μs production runs were performed with different velocities for each WT RET and its D567Y and D567N mutants. The contact area between the CRD and GDNF/GFRα1 was calculated with a probe radius of 1.4 Å using High-Throughput Molecular Dynamics (HTMD) environment 1.14.0 (35).
Data availability
The data generated in this study are available within the article and its Supplementary data files. The data and materials used in this study are available upon request from the corresponding authors.
Results
CaLM as a hidden cluster of oncogenic mutations
To discover hidden clusters of oncogenic mutations among the large number of VUSs, we performed genome-wide trinucleotide enrichment analysis using the OncodriveCLUSTL program (14). A total of 71,756 variants obtained from GENIE version 7.0 cohorts (version 7; 79,720 tumors; ref. 36), which are real-world targeted captured sequencing data containing diagnosis codes for 686 tumor types from 32 tissues from 19 of the leading cancer centers in the world, were tested against a background mutation rate model based on the mutational trinucleotide context generated from 9,104 samples from TCGA (Supplementary Fig. S1A and S1B). For VUS investigation, we focused on RET as a druggable oncogene because it possesses many types (860 kinds of genetic alterations) of missense and in-frame indel mutations in 1,290 mutation tumors (1.6%), most of which are annotated as “unknown significance” by OncoKB (Supplementary Fig. S1A, S1C, and S1D; ref. 37). The CRD of RET kinase has two mutational cluster motifs, a known CCM cluster containing a C634R hotspot mutation and a novel cluster at residues 557–580 that only contains VUSs (Fig. 1A). Identification of three other clusters in the intracellular domain (ICD), each containing the hotspot mutations V804M, R886W, and M918T (12), demonstrated the authenticity of the mutational modeling. Residues 557–580 correspond to the CaLM of the ECD, which was recently identified by cryo-EM analysis (15). The CaLM mutations were observed in various tumor types, whereas CCM variants were enriched for thyroid cancers (Fig. 1A). The variant allele frequencies of the CaLM mutations were similar to those of CCM (Supplementary Fig. S1E; Supplementary Table S1). They were mostly observed as independent, but not as compound, mutations similar to those in the CCM (Supplementary Fig. S1F). These data were consistent with the positively selected characters.
We next performed gene-wide cell-based assays using 110 RET mutants, selected by the following three criteria: (i) mutations occurring in CaLM: six, (ii) known mutations annotated as oncogenic in OncoKB or previously reported: 35 (5, 6, 11, 12, 18, 19), and (iii) mutations recurrently observed in three or more samples in the same residue in the GENIE cohort: 69 (Supplementary Tables S1 and S2). The effect of the introduction of RET mutant cDNAs on transforming activity in NIH3T3 cells in the absence of GDNF/GFRα1 or any other ligands and downstream ERK phosphorylation in HEK293H cells was examined (Fig. 1B and C). The results confirmed CaLM as a cluster of oncogenic variants with similar magnitude as CCM and ICD hotspot mutants, whereas most variants, especially in the ECD, were functionally neutral except for R417C, D631Y, A498S, G533C, Y606C, and R635C, which were annotated as “unknown significance” (Fig. 1B). The biological significance of CaLM mutations was verified by the increased phosphorylation of RET, SHC, which is a direct substrate of RET, and downstream ERK upon expression of the representative D567N and D567Y CaLM mutants (Supplementary Fig. S1G).
Structural destabilizations of the CRD caused by CaLM mutations
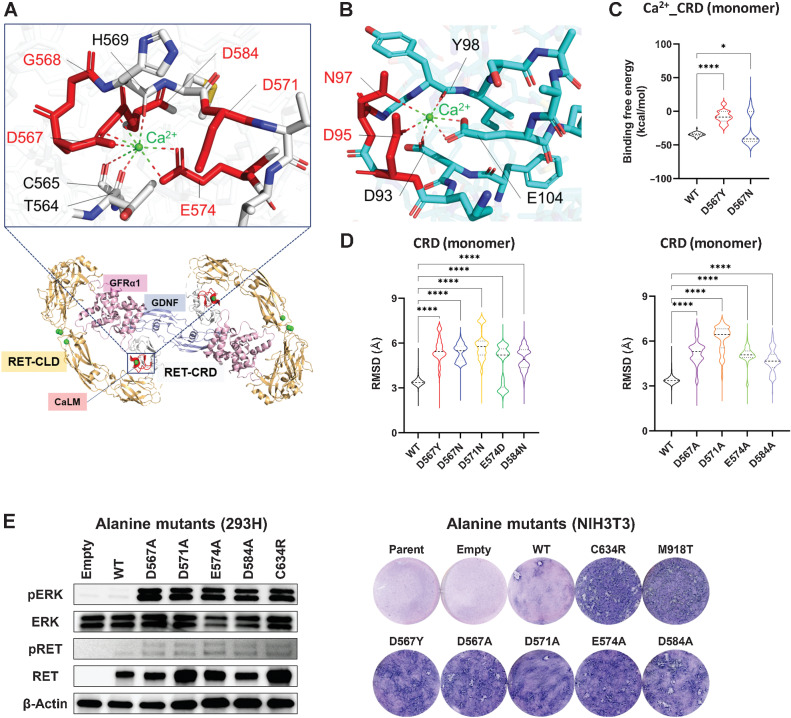
The CaLM of RET captures the Ca2+ ion by forming coordinate bonds with negatively charged side chains or the backbone carbonyl of contributing amino acids (Fig. 2A; ref. 15), similar to calmodulin (Fig. 2B). RET-CaLM mutations result in the formation of a 3D cluster with substitutions in amino acids involved in Ca2+ ion-binding (i.e., D567Y and D567N, G568S, D571N, and E574D). Because three-dimensional mapping of D584N outside CaLM to the protein structure shows a residue directly binding to the Ca2+ ion, the mutation is integrated into CaLM mutations in subsequent analyses. This is similar to the germline mutations at D95 and N97 residues in the CALM1 gene (encoding calmodulin) causing long-QT syndrome. Because these CALM1 mutations are associated with destabilized Ca2+ ion holding (38, 39), we performed MD simulation to predict the impact of CaLM mutations on the conformational integrity of the RET–Ca2+ ion complex.
Figure 2.
Three-dimensional fluctuation of CRD caused by CaLM mutation. A, 3D view of CaLM mutations. The cartoon shows an overhead view of the extracellular portion of the RET–GDNF–GFRα1 hexamer (PDB: 6Q2N) with an expanded view of CaLM. The expanded image shows a stick and cartoon representation showing the locations of Ca2+ ions (green sphere) and amino acid residues contributing to Ca2+ ion holding. Mutated residues are highlighted in red. Bottom, CLD, orange; cysteine-rich domain (CRD), gray; glial cell-derived neurotrophic factor (GDNF), light blue; GDNF family receptor alpha1 (GFRα1), pink. B, 3D view of calmodulin mutations. The stick and cartoon model represents the Ca2+-binding motif of calmodulin (PDB code: 2BE6). Mutated residues in long-QT syndrome patients are highlighted in red. C, Violin plot of binding-free energy between the Ca2+ ion and a RET-CRD monomer at 10 ns intervals. The width of each strip represents the fraction of time points showing a binding-free energy value. The black dashed line represents the median value. Each dotted line represents the interval between the end of the first quartile and the beginning of the fourth quartile. *, P < 0.05; ****, P < 0.0001 by a one-way ANOVA with the Tukey multiple comparisons test. D, Violin plots of RMSD of backbone Cα atoms of CRD monomers. The RMSDs of heavy atoms of each amino acid at 100 ps intervals were calculated with respect to the initial position and plotted. Left, RMSD of WT and native CaLM mutants. Right, artificial alanine mutants. E, Oncogenic properties of artificial CaLM mutants. Left, ERK phosphorylation assay. HEK293H cells transfected with empty vector or WT RET, 110 RET mutants, and kinase-dead mutant (K758M)-expressing plasmids were subjected to immunoblot analysis of pERK, ERK, pRET, RET, and β-actin. Right, NIH3T3 focus formation. The transforming ability of lentiviruses expressing artificial CaLM mutants was examined using parental cells (uninfected), empty virus infection, and lentiviruses to express D567Y (native CaLM mutant), C634R (CCM mutant), and M918T (kinase domain mutant) proteins.
The impact on the local steric integrity was first investigated using a 1 μs simulation of RET-CRD monomers with and without CaLM mutations. The binding free energy (ΔGbind) of the Ca2+ ion to the CRD of D567Y and D567N mutants was higher than that of the WT (Fig. 2C; Supplementary Fig. S2A), indicating destabilization of the CRD–Ca2+ complex due to CaLM mutations. In the simulation, the locations of the Ca2+ ion fluctuated toward the opposite side of the mutated residues at position 567 (Supplementary Fig. S2B and S2C; Supplementary Movies S1–3). The instability of the Ca2+ ion location caused by CaLM mutations was suggested by the increased RMSD of the Ca2+ ion from the initial location (Supplementary Fig. S2D). In parallel with this, the structural fluctuation of the CRD associated with the D567 mutation was indicated by the RMSD of backbone Cα atoms (Fig. 2D, left; Supplementary Fig. S2E and S2F; Supplementary Movies S1–3). Similar results were obtained for other CaLM mutations (Fig. 2D, left), strongly indicating that destabilization of the CaLM-Ca2+ complex underlies the oncogenic properties. To validate the results of the simulation, we examined the properties of artificial mutants generated by removing the effects of side chains contributing to Ca2+ ion holding (i.e., mutants with substitution to alanine; Fig. 2D, right). The alanine mutants showed high RMSD values similar to those of native CaLM mutants. The oncogenic properties of the alanine mutants were verified by downstream ERK activation as well as NIH3T3 cell transformation (Fig. 2E).
Illegitimate intermolecular disulfide bond formation induced by CaLM mutations
Under physiologic conditions, the RET protein homodimerizes by binding with GDNF ligands and coreceptors, such as GDNF family receptor alpha1 (GFRα1), resulting in a hexamer complex (Fig. 2A; ref. 15). To address the effects of CaLM mutations on the structure of RET and its interaction with ligands/coreceptors, we performed microsecond-timescale MD simulations starting from a cryo-EM structure (PDB: 6Q2N) consisting of a dimerized extracellular RET, two GFRα1 molecules, and two GDNF molecules. Compared with the WT RET complex, the CaLM mutants exhibited structural distortion of the CRD concomitant with loss of interactions between the Ca2+ ion and its surrounding residues (Fig. 3A, left, middle, and Supplementary Fig. S3A). The distorted CRD conformations of the CaLM mutants increased the solvent-accessible surface area (SASA) of cysteine residues C565, C570, and C585 (Fig. 3A, right), which are physiologically buried inside the CRD domain and form intra-domain disulfide bonds in the WT RET structure (15). These simulation results prompted us to investigate whether intermolecular disulfide bond formation may be induced by CaLM mutations.
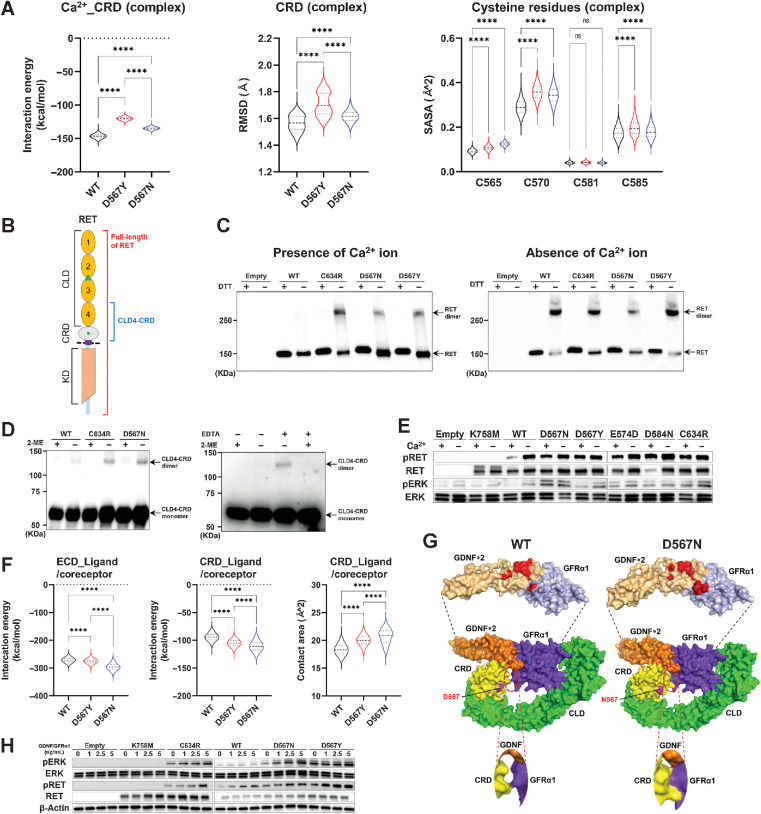
Figure 3.
Illegitimate intermolecular disulfide bond formation induced by CaLM mutations. A, Violin plots showing the interaction energy between the Ca2+ ion and the CRD (left), the RMSD of the CRD (middle), and the SASA of cysteine residues (C565, C570, C581, and C585) in the CRD (right) estimated from MD simulation of the RET–GDNF–GFRα1 extracellular complex. MD simulation was performed in triplicate for each WT and RET mutant, and structures extracted from the 0.5–1 μs trajectories every 20 ps were subjected to calculation. The black dashed line represents the median values. Each dotted line represents the interval between the end of the first quartile and the beginning of the fourth quartile. B, Schematic diagram of the RET protein. CLD, cadherin-like domain; CRD, cysteine-rich domain; KD, kinase domain; green circle, Ca2+ ion-binding sites; dashed line, cell membrane. C, Intermolecular RET dimer formation. Flp-in T-REx 293 cells with or without doxycycline-inducible FLAG-tagged full-length RET cDNAs were cultured for 48 hours with doxycycline in calcium-containing (left) or calcium-free (right) medium. Whole-cell lysates were prepared under reducing or nonreducing conditions, i.e., presence or absence of dithiothreitol, and resolved by SDS-PAGE, followed by immunoblot analysis using an anti-FLAG antibody. D, Intermolecular dimer formation of CLD4-CRD. GST-tagged RET-CRD-CLD4 polypeptides expressed in Sf21 cells were purified under reducing or nonreducing conditions, i.e., presence or absence of 2-ME, and resolved by SDS-PAGE, followed by immunoblot analysis using an anti-GST antibody. Left, intermolecular dimer formation of WT, C634R, and D567N mutant polypeptides. Right, intermolecular dimer formation of WT RET polypeptides purified with or without calcium chelation, i.e., presence or absence of EDTA. E, RET autophosphorylation and ERK phosphorylation induced by calcium depletion. HEK293H cells transfected with WT or mutant full-length RET cDNAs were cultured for 48 hours in calcium-containing or calcium-free medium. RET-CaLM mutants were examined by detecting K758M (kinase-dead) and C634R (CCM) mutants. Protein lysates were subjected to immunoblot analysis of pRET, RET, pERK, and ERK. F, Violin plots of the interaction energy between RET-ECD and GDNF/GFRα1 (left) or RET-CRD and GDNF/GFRα1 (middle) and contact area between the CRD and GDNF/GFRα1 (right) estimated from the MD simulation of the RET–GDNF–GFRα1 extracellular complex. The black dashed line represents the median values. Each dotted line represents the interval between the end of the first quartile and the beginning of the fourth quartile. G, D567N mutation–induced increase in contact areas between RET-CRD and ligand/coreceptor. The mean structure of a WT (left) or D567N (right) RET subunit bound to two GDNFs and a GFRα1 molecule was calculated using three 0.5–1 μs trajectories obtained from the MD simulation of the RET extracellular complex and is represented by the surface model. Top, the CRD binding interfaces of two GDNFs and a GFRα1 molecule whose amino acid residues located within 5 Å from the CRD domain are highlighted in red. An enlarged view of the CRD–GFRα1 binding interface is shown in the bottom panels. H, ERK phosphorylation assay. HEK293H cells transfected with empty vector or WT RET, K758M (kinase dead), C634R (CCM), D567N, and D567Y RET mutant–expressing plasmids and treated with GDNF/GFRα1 as described were subjected to immunoblot analysis of pERK, ERK, pRET, RET, and β-actin. n.s., not significant; ****, P < 0.0001 by a one-way ANOVA with the Tukey multiple comparisons test.
Exogenous expression of full-length RET cDNA in HEK293 cells caused CaLM mutants to dimerize through illegitimate intermolecular disulfide bonds similar to the C634R CCM mutant (Fig. 3B and C, left). Depletion of the Ca2+ ion from the culture medium, which mimics the ablation of Ca2+ ion holding in the CaLM, also caused dimerization of WT RET through the formation of intermolecular disulfide bonds (Fig. 3C, right), indicating that CaLM suppresses illegitimate RET dimerization. RET has three other Ca2+ ion-binding sites spanning cadherin-like domains (CLD) 2 and 3 (Fig. 3B; ref. 15). Dimerization assays using purified CLD4-CRD proteins verified the suppressive role of CaLM on dimerization; CaLM mutations increased disulfide bond–mediated dimerization in the presence of Ca2+ ions, whereas Ca2+ ion depletion increased disulfide bond–mediated dimerization of WT RET (Fig. 3D; Supplementary Fig. S3B and S3C). Consistently, after Ca2+ ion depletion, RET and downstream ERK phosphorylation were more evident in WT RET than in CaLM mutants (Fig. 3E).
Coreceptors/ligands contribute to bringing RET molecules together for dimerization (40). A temporal bimolecular interaction assay of the RET-ECD domain using a NanoBiT system (41) demonstrated that not only WT RET but also CaLM (D567N and D567Y) and CCM (C634R) mutant dimerization depend on the addition of ligands/coreceptors (Supplementary Fig. S3D, left). These responses were mediated by active dimerization including ligands/coreceptors, and not by inactive dimerization without these molecules, as indicated by the introduction of R77E and R144E double mutations that suppress inactive dimerization (15), which did not abolish the response (Supplementary Fig. S3D, right). The present MD simulation suggests that the D567N RET mutant forms a more stable RET–GDNF–GFRα1 hexamer complex than WT RET and the D567Y mutant via enhanced RET–ligand/coreceptor contacts (Fig. 3F and G). Consistently, the addition of GDNF and GFRα increased the NIH3T3 cell–transforming and ERK activation abilities of the D567N mutant more evidently than those of the D567Y and C634R mutants (Fig. 3H; Supplementary Fig. S3E). These data suggest that the oncogenic properties of the D567N mutant increase in the presence of ligands/coreceptors, consistent with their enhanced interaction observed in the MD simulation.
RET-CaLM mutants as a target for RET-TKIs
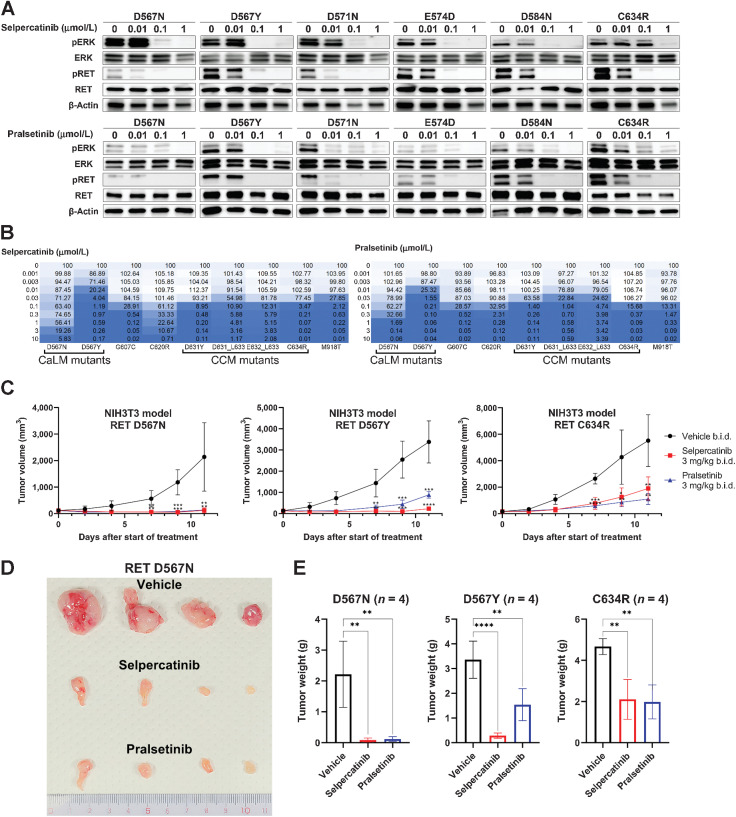
Finally, we examined whether RET-CaLM mutants are targetable by existing RET-TKIs. Enhanced ERK phosphorylation caused by exogenously expressed CaLM mutants in HEK293H cells was suppressed by selpercatinib and pralsetinib, two RET-specific TKIs approved by the FDA (Fig. 4A; ref. 18). Stable expression of RET-CaLM mutants enabled Ba/F3 cells to grow in an IL3-independent manner similar to C634R and M918T hotspot mutants, thus confirming the oncogenic activity of CaLM mutants. The growth of Ba/F3 cells expressing CaLM mutants was suppressed by selpercatinib and pralsetinib at similar concentrations as those that were effective against C634R and M918T mutants (Fig. 4B; Supplementary Fig. S4A). NIH3T3 cells with stable expression of D567N and D567Y formed tumors in nude mice similar to those expressing the C634R mutant. Both selpercatinib and pralsetinib suppressed the growth of NIH3T3 cells expressing D567N and D567Y similar to their effect on NIH3T3 cells expressing C634R mutants (Fig. 4C–E; Supplementary Fig. S4B). These results indicate that RET-CaLM mutants are targetable by existing RET-TKIs.
Figure 4.
RET-CaLM mutants as a target for RET-TKIs. A, Suppression of signal transduction by RET inhibitors. HEK293H cells expressing RET-CaLM and C634R (CCM) mutants were treated with selpercatinib (top) or pralsetinib (bottom) for 6 hours. Cell lysates were subjected to immunoblot analysis of pERK, ERK, pRET, RET, and β-actin. B, Suppression of RET-dependent cell growth by RET inhibitors. IL3-independent growth of Ba/F3 cells expressing RET-CaLM, CCM, and M918T kinase domain mutants was examined in the presence of the indicated concentrations of selpercatinib (left) or pralsetinib (right) for 72 hours. Cell viability was measured using the CellTiter-Glo assay. The relative viability (%) is shown in a color-coded manner. C, Suppression of allograft growth of NIH3T3 cells expressing RET mutants by RET inhibitors. Drug treatment was started when the subcutaneous tumors grew to a volume of approximately 100 to 200 mm3. All mutants were treated with vehicle or 3 mg/kg selpercatinib or pralsetinib twice daily. Four mice were included in each treatment group. Data are expressed as the mean ± SD. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; one-way ANOVA with a Dunnett multiple comparisons test against the vehicle group. b.i.d., twice daily. D, Tumors of D567N mutant-expressing NIH3T3 cells after treatment with vehicle (top), selpercatinib (middle), or pralsetinib (bottom). After treatment completion, tumors were dissected from subjects. E, Tumor weights at the endpoint. Data represent the mean ± SD. **, P < 0.01; ****, P < 0.0001; one-way ANOVA with the Tukey multiple comparisons test against the vehicle group.
Discussion
The present in silico analysis–guided study identified a novel set of oncogenic and targetable mutations among a large number of VUSs of the RET gene. These were Ca2+-binding ablating mutations of CaLM, which cause constitutive RET kinase activation by inducing illegitimate dimerization. The mutants were identified using two strategies: a genome-wide in silico mutation modeling to identify positively selected mutations, and gene-wide cell-based screening of representative mutants. CaLM was defined as the location of a novel 3D cluster of oncogenic mutations in RET. The CaLM mutations are not found in thyroid cancer and are distinct from the CCM mutations enriched in thyroid cancer. In thyroid cancer, including medullary thyroid carcinoma, most mutations have been studied extensively, e.g., M918T and C634R/W. Mutations other than the established hotspots are unlikely to occur. This mutational pattern is inconsistent with that of other cancer lineages, in which non-hotspot mutations are likely to occur. This suggests that thyroid cancer has a stronger selection pressure for the acquisition of major oncogenic mutations such as M918T or C634R during the oncogenic process than other cancer lineages. This speculation will be confirmed by collecting additional genomic data and performing positive enrichment mutation analysis across cancer lineages. The functional effects of the CaLM mutations were predicted using MD simulations of both CRD monomers and the RET/ligand/coreceptor complex. Cell- and protein-based assays supported the prediction and demonstrated the oncogenic and targetable properties of RET-CaLM mutations. The present study strongly indicates that cancer genomes harbor unidentified oncogenic/targetable mutations in well-known oncogenes. Notably, not only D567N, which was recurrently observed in >10 cases, but also other mutations observed only in one or two cases, such as D567Y, were included in the 3D cluster of CaLM mutations. The EGFR kinase activation loop shows a highly recurrent L858R hotspot mutation, although it also contains other infrequent but similarly oncogenic mutations such as L861Q and L861R (20). Therefore, comprehensive annotation of apparent non-hotspot mutations by incorporating a 3D perspective that focuses on protein motifs could be a powerful method to expand the list of targetable mutations in known oncogenes.
The results suggest that RET-CaLM mutations cause homodimerization of RET proteins through illegitimate disulfide bond formation. Depletion of Ca2+ ions from the culture medium caused dimerization of the WT RET protein by inducing intermolecular disulfide bond formation, indicating that CaLM negatively regulates intermolecule dimerization by preserving its proper conformation for holding Ca2+ ions. However, CaLM mutations caused intermolecular disulfide bond formation even in the presence of Ca2+ ions, suggesting that altered Ca2+ ion holding in CaLM mutants triggers illegitimate disulfide bond formation similar to that in the Ca2+ ion-depleted condition. MD simulation of the RET-ECD–ligand–coreceptor complex suggested that RET-CaLM mutations cause the exposure of cysteine residues that form intramolecular disulfide bonds under physiologic conditions. Thus, this structural change might increase the chances of intermolecular interaction between cysteine residues, which may lead to illegitimate disulfide bond formation between two RET molecules. The present study elucidated the mechanisms underlying the oncogenic properties of CaLM mutations in human cancer and shed light on the physiologic role of RET-CaLM, which was poorly understood (15). In this mechanism, the Ca2+ ion is the key to protein structural stabilization, and the destabilization induced by the CaLM mutations is a novel critical hallmark of RET oncogenic conversion. Another receptor tyrosine kinase (RTK) gene with a couple of Ca2+ ion-binding motifs and their surrounding mutations has not been found. However, future structural studies may identify an RTK gene with a similar mechanism of activation.
Several genetic alterations that cause constitutive RET kinase activation are effective targets for cancer therapy using RET-TKIs (42). Especially, genetic alterations that cause ligand-independent dimerization of RET kinase in lung and thyroid cancers, such as RET fusion and hotspot cysteine mutations (11, 13, 43, 44), are strongly associated with a good therapeutic response to inhibitory drugs specifically targeting RET kinase (5, 45). These RET alterations are present in several cancers other than lung and thyroid cancers (19, 46), and the therapeutic effects of RET kinase inhibitors have been observed in a tumor agnostic manner (47–49). The RET-CaLM mutations identified in this study are present in multiple types of cancers, such as lung, colorectal, central nervous system, and blood cancers. As an example of the importance of RET-CaLM mutations in clinical specimens, the available transcript data from colon adenocarcinoma with D571N mutations, a member of CaLM mutations, listed in the TCGA cohort demonstrated the mRNA expression of the D571N-mutated allele (Supplementary Fig. S5A). In addition, a comparison of expression values by z-score obtained from cBioPortal (https://www.cbioportal.org/) showed that the expression of the RET D571N mutant is similar to that of other oncogenic gene aberrations including gene fusions and M918T and C634R mutations, and that the expression of GDNF and GFRA1 is correlated with the expression of RET (Supplementary Fig. S5B), supporting the significance of RET-CaLM mutations in clinical samples. Further case analysis is required to confirm the results. One obstacle limiting the delivery of precision medicine to patients with these mutations is that the CaLM DNA segment is not covered by the present companion diagnostics for detecting RET mutations. The present study indicated that tumors with CaLM mutations respond to RET inhibitory therapy in vivo. Therefore, CaLM mutations should be added to the list of RET alterations that increase the benefit from RET inhibitory therapy and should be analyzed to expand the population of cancer patients that may benefit from RET-TKI therapy.
Lastly, the present study used MD simulation to predict the functional effects of mutations. MD simulation has exclusively been used in clinical oncology to analyze drug resistance/sensitivity according to secondary mutations in target kinases in response to kinase inhibitory therapy (50–54). Thus, the present study expands the utility of MD simulation in oncology, i.e., for predicting the functional effects of VUSs based on conformational fluctuations. Native and artificial CaLM mutants caused increased fluctuation of the CRD as well as decreased Ca2+ ion interactions. Furthermore, a long simulation of a protein hexamer consisting of a dimerized extracellular RET, two GFRα1, and two GDNF molecules was performed using the high-performance supercomputer “Fugaku” (16). The simulation provided insight into the function of CaLM mutants, such as intermolecular disulfide bond formation triggered by exposure of cysteine residues from the CRD and increased oncogenicity of the D567N mutant induced by ligand/coreceptor molecules. Oncogenic proteins ubiquitously express their properties by forming complexes with signaling molecules (55). MD simulation of protein complexes could guide functional annotation of the vast number of VUSs in human cancer.
Supplementary Material
Acknowledgments
This work was supported in part by grants-in-aid from the Japan Agency for Medical Research and Development (AMED; JP21cm0106701, JP21ck0106522, and JP22ck0106721 to T. Nakaoku and 22ym0126804j0001/21-A-08 to T. Kohno), the National Cancer Center Research and Development Fund (2021-A-10 to T. Kohno), JSPS KAKENHI (grant number 20H00545 to T. Kohno and JP21K06510 to M. Araki), MEXT as the “Program for Promoting Researches on the Supercomputer Fugaku (Application of Molecular Dynamics Simulation to Precision Medicine Using Big Data Integration System for Drug Discovery)” (to Y. Okuno), the Foundation for Computational Science (FOCUS) Establishing Supercomputing Center of Excellence (to Y. Okuno), the Takeda Science Foundation (to T. Nakaoku and A. Ui), the Naito Foundation (to A. Ui), and the Uehara Memorial Foundation (to T. Nakaoku). This research used computational resources of the supercomputer Fugaku provided by the RIKEN Center for Computational Science through the HPCI System Research Project (Project ID: hp200129 and hp210172). This research was partially supported by the Platform Project for Supporting Drug Discovery and Life Science Research [Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS)] from AMED under grant number JP21am0101112 (support number 1198). The authors thank K. Shiraishi for performing genome data processing.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Disclosures
J. Tabata reports grants from the Foundation for Promotion of Cancer Research in Japan during the conduct of the study. M. Sekijima reports grants from the Japan Agency for Medical Research and Development during the conduct of the study and grants from the Japanese Society for the Promotion of Science (JSPS) outside the submitted work. Y. Okuno reports other support from MEXT and grants from Foundation for Computational Science during the conduct of the study. T. Kohno reports grants from AMED, National Cancer Center, and JSPS during the conduct of the study; personal fees from Eli Lilly and grants from Sysmex and Chugai outside the submitted work; in addition, T. Kohno has a patent 9216172 issued to Method for determining effectiveness of cancer treatment by assessing the presence of a KIF5B-RET chimeric gene. No disclosures were reported by the other authors.
Authors' Contributions
J. Tabata: Data curation, formal analysis, investigation, visualization, methodology, writing–original draft. T. Nakaoku: Conceptualization, resources, data curation, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. M. Araki: Data curation, software, formal analysis, funding acquisition, investigation, visualization, methodology, writing–original draft, writing–review and editing. R. Yoshino: Data curation, software, formal analysis, funding acquisition, investigation, visualization, methodology, writing–original draft, writing–review and editing. S. Kohsaka: Investigation, writing–review and editing. A. Otsuka: Investigation. M. Ikegami: Investigation, writing–review and editing. A. Ui: Funding acquisition, investigation, methodology, writing–review and editing. S.-i. Kanno: Investigation, methodology, writing–review and editing. K. Miyoshi: Investigation. S. Matsumoto: Software, investigation, writing–review and editing. Y. Sagae: Software, investigation, writing–review and editing. A. Yasui: writing–review and editing. M. Sekijima: Writing–review and editing. H. Mano: Writing–review and editing. Y. Okuno: Funding acquisition, writing–review and editing. A. Okamoto: writing–review and editing. T. Kohno: Conceptualization, resources, supervision, funding acquisition, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Howlader N, Forjaz G, Mooradian MJ, Meza R, Kong CY, Cronin KA, et al. The effect of advances in lung-cancer treatment on population mortality. N Engl J Med 2020;383:640–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med 2011;364:2507–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Skoulidis F, Li BT, Dy GK, Price TJ, Falchook GS, Wolf J, et al. Sotorasib for lung cancers with KRAS p.G12C mutation. N Engl J Med 2021;384:2371–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 2010;362:2380–8. [DOI] [PubMed] [Google Scholar]
- 5. Wirth LJ, Sherman E, Robinson B, Solomon B, Kang H, Lorch J, et al. Efficacy of selpercatinib in RET-altered thyroid cancers. N Engl J Med 2020;383:825–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Subbiah V, Hu MI, Wirth LJ, Schuler M, Mansfield AS, Curigliano G, et al. Pralsetinib for patients with advanced or metastatic RET-altered thyroid cancer (ARROW): a multi-cohort, open-label, registrational, phase 1/2 study. Lancet Diabetes Endocrinol 2021;9:491–501. [DOI] [PubMed] [Google Scholar]
- 7. Ikemura S, Yasuda H, Matsumoto S, Kamada M, Hamamoto J, Masuzawa K, et al. Molecular dynamics simulation-guided drug sensitivity prediction for lung cancer with rare EGFR mutations. Proc Nat Acad Sci U S A 2019;116:10025–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Foley SB, Rios JJ, Mgbemena VE, Robinson LS, Hampel HL, Toland AE, et al. Use of whole genome sequencing for diagnosis and discovery in the cancer genetics clinic. EBioMedicine 2015;2:74–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021;596:583–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Buel GR, Walters KJ. Can AlphaFold2 predict the impact of missense mutations on structure? Nat Struct Mol Biol 2022;29:1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mulligan LM. RET revisited: expanding the oncogenic portfolio. Nat Rev Cancer 2014;14:173–86. [DOI] [PubMed] [Google Scholar]
- 12. Plaza-Menacho I, Barnouin K, Goodman K, Martinez-Torres RJ, Borg A, Murray-Rust J, et al. Oncogenic RET kinase domain mutations perturb the autophosphorylation trajectory by enhancing substrate presentation in trans. Mol Cell 2014;53:738–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Romei C, Ciampi R, Elisei R. A comprehensive overview of the role of the RET proto-oncogene in thyroid carcinoma. Nat Rev Endocrinol 2016;12:192–202. [DOI] [PubMed] [Google Scholar]
- 14. Arnedo-Pac C, Mularoni L, Muinos F, Gonzalez-Perez A, Lopez-Bigas N. OncodriveCLUSTL: a sequence-based clustering method to identify cancer drivers. Bioinformatics 2019;35:4788–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li J, Shang G, Chen YJ, Brautigam CA, Liou J, Zhang X, et al. Cryo-EM analyses reveal the common mechanism and diversification in the activation of RET by different ligands. Elife 2019;8:e47650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jung J, Kobayashi C, Kasahara K, Tan C, Kuroda A, Minami K, et al. New parallel computing algorithm of molecular dynamics for extremely huge scale biological systems. J Comput Chem 2021;42:231–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ellrott K, Bailey MH, Saksena G, Covington KR, Kandoth C, Stewart C, et al. Scalable open science approach for mutation calling of tumor exomes using multiple genomic pipelines. Cell Syst 2018;6:271–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Thein KZ, Velcheti V, Mooers BHM, Wu J, Subbiah V. Precision therapy for RET-altered cancers with RET inhibitors. Trends Cancer 2021;7:1074–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kohno T, Tabata J, Nakaoku T. REToma: a cancer subtype with a shared driver oncogene. Carcinogenesis 2020;41:123–9. [DOI] [PubMed] [Google Scholar]
- 20. Kohsaka S, Nagano M, Ueno T, Suehara Y, Hayashi T, Shimada N, et al. A method of high-throughput functional evaluation of EGFR gene variants of unknown significance in cancer. Sci Transl Med 2017;9:eaan6566. [DOI] [PubMed] [Google Scholar]
- 21. Jacobson MP, Friesner RA, Xiang Z, Honig B. On the role of the crystal environment in determining protein side-chain conformations. J Mol Biol 2002;320:597–608. [DOI] [PubMed] [Google Scholar]
- 22. Jacobson MP, Pincus DL, Rapp CS, Day TJ, Honig B, Shaw DE, et al. A hierarchical approach to all-atom protein loop prediction. Proteins 2004;55:351–67. [DOI] [PubMed] [Google Scholar]
- 23. Li H, Robertson AD, Jensen JH. Very fast empirical prediction and rationalization of protein pKa values. Proteins 2005;61:704–21. [DOI] [PubMed] [Google Scholar]
- 24. Roos K, Wu C, Damm W, Reboul M, Stevenson JM, Lu C, et al. OPLS3e: extending force field coverage for drug-like small molecules. J Chem Theory Comput 2019;15:1863–74. [DOI] [PubMed] [Google Scholar]
- 25. Berendsen HJC, Grigera JR, Straatsma TP. The missing term in effective pair potentials. J Phys Chem 1987;91:6269–71. [Google Scholar]
- 26. Kollman PA, Massova I, Reyes C, Kuhn B, Huo S, Chong L, et al. Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc Chem Res 2000;33:889–97. [DOI] [PubMed] [Google Scholar]
- 27. Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, et al. GROMACS: high-performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015;1–2:19–25. [Google Scholar]
- 28. Lindorff-Larsen K, Piana S, Palmo K, Maragakis P, Klepeis JL, Dror RO, et al. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins 2010;78:1950–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys 1983;79:926–35. [Google Scholar]
- 30. Darden T, York D, Pedersen L. Particle mesh Ewald: an N⋅log(N) method for Ewald sums in large systems. J Chem Phys 1993;98:10089–92. [Google Scholar]
- 31. Feenstra KA, Hess B, Berendsen HJC. Improving efficiency of large time-scale molecular dynamics simulations of hydrogen-rich systems. J Comput Chem 1999;20:786–98. [DOI] [PubMed] [Google Scholar]
- 32. Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. LINCS: a linear constraint solver for molecular simulations. J Comput Chem 1997;18:1463–72. [Google Scholar]
- 33. Bussi G, Donadio D, Parrinello M. Canonical sampling through velocity rescaling. J Chem Phys 2007;126:014101. [DOI] [PubMed] [Google Scholar]
- 34. Parrinello M, Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J Appl Phys 1981;52:7182–90. [Google Scholar]
- 35. Doerr S, Harvey MJ, Noé F, De Fabritiis G. HTMD: high-throughput molecular dynamics for molecular discovery. J Chem Theory Comput 2016;12:1845–52. [DOI] [PubMed] [Google Scholar]
- 36. AACR Project GENIE Consortium. AACR project GENIE: powering precision medicine through an international consortium. Cancer Discov 2017;7:818–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol 2017;2017:PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang K, Holt C, Lu J, Brohus M, Larsen KT, Overgaard MT, et al. Arrhythmia mutations in calmodulin cause conformational changes that affect interactions with the cardiac voltage-gated calcium channel. Proc Natl Acad Sci U S A 2018;115:E10556–E65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Priori SG, Napolitano C, Memmi M, Colombi B, Drago F, Gasparini M, et al. Clinical and molecular characterization of patients with catecholaminergic polymorphic ventricular tachycardia. Circulation 2002;106:69–74. [DOI] [PubMed] [Google Scholar]
- 40. Bigalke JM, Aibara S, Roth R, Dahl G, Gordon E, Dorbeus S, et al. Cryo-EM structure of the activated RET signaling complex reveals the importance of its cysteine-rich domain. Sci Adv 2019;5:eaau4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dixon AS, Schwinn MK, Hall MP, Zimmerman K, Otto P, Lubben TH, et al. NanoLuc complementation reporter optimized for accurate measurement of protein interactions in cells. ACS Chem Biol 2016;11:400–8. [DOI] [PubMed] [Google Scholar]
- 42. Subbiah V, Yang D, Velcheti V, Drilon A, Meric-Bernstam F. State-of-the-art strategies for targeting RET-dependent cancers. J Clin Oncol 2020;38:1209–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kohno T, Ichikawa H, Totoki Y, Yasuda K, Hiramoto M, Nammo T, et al. KIF5B-RET fusions in lung adenocarcinoma. Nat Med 2012;18:375–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gainor JF, Curigliano G, Kim DW, Lee DH, Besse B, Baik CS, et al. Pralsetinib for RET fusion-positive non-small-cell lung cancer (ARROW): a multi-cohort, open-label, phase 1/2 study. Lancet Oncol 2021;22:959–69. [DOI] [PubMed] [Google Scholar]
- 45. Drilon A, Oxnard GR, Tan DSW, Loong HHF, Johnson M, Gainor J, et al. Efficacy of selpercatinib in RET fusion-positive non-small-cell lung cancer. N Engl J Med 2020;383:813–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rich TA, Reckamp KL, Chae YK, Doebele RC, Iams WT, Oh MS, et al. Analysis of cell-free DNA from 32,989 advanced cancers reveals novel co-occurring activating RET alterations and oncogenic signaling pathway aberrations. Clin Cancer Res 2019;25:5832–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Watanabe S, Takeda M, Otani T, Yoshida T, Sakai K, Olek E, et al. Complete response to selective RET inhibition with selpercatinib (LOXO-292) in a patient with RET fusion-positive breast cancer. JCO Precis Oncol 2021;5:PO.20.00282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tsui DCC, Kavanagh BD, Honce JM, Rossi C, Patil T, Camidge DR. Central nervous system response to selpercartinib in patient with RET-rearranged non-small cell lung cancer after developing leptomeningeal disease on pralsetinib. Clin Lung Cancer 2021;23:e5–e8. [DOI] [PubMed] [Google Scholar]
- 49. Ortiz MV, Gerdemann U, Raju SG, Henry D, Smith S, Rothenberg SM, et al. Activity of the highly specific RET inhibitor selpercatinib (LOXO-292) in pediatric patients with tumors harboring RET gene alterations. JCO Precis Oncol 2020;4:PO.19.00401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yoshizawa T, Uchibori K, Araki M, Matsumoto S, Ma B, Kanada R, et al. Microsecond-timescale MD simulation of EGFR minor mutation predicts the structural flexibility of EGFR kinase core that reflects EGFR inhibitor sensitivity. NPJ Precis Oncol 2021;5:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Assadollahi V, Rashidieh B, Alasvand M, Abdolahi A, Lopez JA. Interaction and molecular dynamics simulation study of Osimertinib (AstraZeneca 9291) anticancer drug with the EGFR kinase domain in native protein and mutated L844V and C797S. J Cell Biochem 2019;120:13046–55. [DOI] [PubMed] [Google Scholar]
- 52. Zabriskie MS, Eide CA, Tantravahi SK, Vellore NA, Estrada J, Nicolini FE, et al. BCR-ABL1 compound mutations combining key kinase domain positions confer clinical resistance to ponatinib in Ph chromosome-positive leukemia. Cancer Cell 2014;26:428–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Eide CA, Zabriskie MS, Savage Stevens SL, Antelope O, Vellore NA, Than H, et al. Combining the allosteric inhibitor asciminib with ponatinib suppresses emergence of and restores efficacy against highly resistant BCR-ABL1 mutants. Cancer Cell 2019;36:431–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nakaoku T, Kohno T, Araki M, Niho S, Chauhan R, Knowles PP, et al. A secondary RET mutation in the activation loop conferring resistance to vandetanib. Nat Commun 2018;9:625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lourenco C, Resetca D, Redel C, Lin P, MacDonald AS, Ciaccio R, et al. MYC protein interactors in gene transcription and cancer. Nat Rev Cancer 2021;21:579–91. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are available within the article and its Supplementary data files. The data and materials used in this study are available upon request from the corresponding authors.