Abstract
The lymphoma of the mucosa-associated lymphoid tissue (MALT) of the stomach has been linked to Helicobacter pylori infection, but the mechanisms involved in B-cell proliferation remain elusive. In a search for putative H. pylori-specific monoclonal immunoglobulin production, an H. pylori strain was isolated from 10 patients with MALT lymphoma and used to detect the specific serum antibody response to the homologous strain by immunoblotting. Moreover, the antigenicity of the different strains was compared by using each of the 10 sera. We found that the different strains induced highly variable patterns of systemic immunoglobulin G antibody response, although several bacterial antigens, such as the 60-kDa urease B, were often recognized by the different sera. The cagA marker was detected in the strains by PCR with specific primers and by dot blot analysis, and the CagA protein was found in the sera of 4 of the 10 patients by immunoblotting. In conclusion, MALT lymphoma patients, like other patients with H. pylori gastritis, exhibit a polymorphic systemic antibody response, despite an apparently similar antigenic profile. The CagA marker of pathogenicity is not associated with this disease.
Since the discovery of Helicobacter pylori and its pathogenic role in gastric and duodenal ulceration, it has also been associated with gastric adenocarcinomas. Recently, an association between the presence of H. pylori and the development of mucosa-associated lymphoid tissue (MALT) B-cell gastric lymphoma has been documented (12). H. pylori infection was found in 85 to 92% of patients with this malignancy (17, 24). Carlson et al. observed the progression of H. pylori gastritis with polyclonal lymphoid hyperplasia to a MALT lymphoma with a monoclonal lymphoid population (4). Moreover, among a series of six patients with low-grade MALT lymphoma, five patients displayed complete regression of their lymphomas upon eradication of H. pylori by antibiotherapy. Therefore, MALT lymphoma appears in the stomach in response to colonization by the bacterium and regresses after treatment of H. pylori infection (1, 2, 4, 13–15, 19, 23, 24). Gastric MALT lymphoma has a low incidence of occurrence (seven cases per 1 million people per year in the United States), but it is the most common type of extranodal lymphoma (8). It seems to occur more frequently in certain parts of Europe, such as northeastern Italy (9).
The mechanisms by which this bacterial infection leads to the development of MALT lymphoma have not yet been elucidated. MALT is not found in normal gastric mucosa but is assumed to develop after infection. It is possible that the pattern of evolution of low-grade MALT lymphomas is dependent on a local immune response to a specific antigen. In the case of gastric lymphomas, an abnormal immune response to H. pylori in the gastric mucosa and gastric lymph nodes may be associated with proliferation of neoplastic B cells. There are few cases where the strains and the patients’ sera are available. Therefore, the immune response of the patient to his or her homologous strain has not been previously studied.
The aim of this study was to analyze, by immunoblotting, the serum antibody response to H. pylori strains from 10 patients with MALT lymphoma, in order to define a typical pattern for this pathology. In addition, because the cag pathogenicity island has been associated with severe diseases due to H. pylori, the immune response to the CagA marker was studied. Strains were also analyzed by PCR and dot blot analysis to detect the presence of the cagA gene.
Patients.
Ten patients (four females and six males) bearing B-lymphocytic low-grade gastric MALT lymphomas (stage IE or IIE) were studied. For each patient, two gastric biopsy specimens were collected, one at the site of the lesion and one at a distance from it. Biopsies were transported to the laboratory by using Portagerm pylori medium (bioMérieux, Marcy l’Etoile, France) and processed as follows: they were ground in brucella broth and inoculated onto nonselective Wilkins-Chalgren medium and GC agar base supplemented with 5% human blood. After 12 days of incubation at 37°C under microaerobic conditions, growing colonies were identified as H. pylori by morphology and positive reactions for urease, catalase, and oxidase. At the time of the sampling, blood was drawn, and serum was collected, aliquoted, and kept frozen at −20°C until use. Eight of these patients have subsequently received an omeprazole-clarithromycin-amoxicillin therapy which was successful, and seven of them are still in remission.
ELISA and immunoblot analysis.
An enzyme-linked immunosorbent assay (ELISA) for H. pylori was performed with the experimental Pylori Check enzyme immunoassay kit (Hoffmann-La Roche, Basel, Switzerland). Immunoblot analysis was performed with the Helico-Blot 2.0 kit (Genelabs Diagnostics, Geneva, Switzerland). The strain of H. pylori used in the Helico-Blot 2.0 was a clinical isolate (ATCC 43256) from an ulcer. These two assays were conducted following the manufacturer’s recommendations.
An in-house immunoblot was also used. The antigens used were made from H. pylori strains isolated from the patients’ biopsies. Colonies from two semiconfluent plates were harvested, washed twice in phosphate-buffered saline (PBS), resuspended in 0.3 ml of PBS, and sonicated for 3 min with a Vibra cell apparatus (Sonics and Materials Inc., Danbury, Conn.). The sonicates were centrifuged to discard debris, and the supernatants were retained. After determination of the protein concentration with a protein assay (Bio-Rad, Ivry sur Seine, France), the sonicates were adjusted to 1 mg of protein per ml, aliquoted, and frozen at −20°C until use. Immediately before use, sonicates were diluted in sample loading buffer (0.5 M Tris-HCl [pH 6.8], 0.5% bromophenol blue, 8% glycerol, 4% sodium dodecyl sulfate [SDS], 4% 2-mercaptoethanol) and the mixture was heated at 95°C for 5 min. After cooling, 5 μg of H. pylori proteins was loaded on each lane of SDS–10% polyacrylamide gels, under reducing conditions, and proteins were separated by electrophoresis.
Molecular weight standards (Bethesda Research Laboratories, Bethesda, Md.), which included proteins ranging from 14.3 to 200 kDa, were treated similarly. Proteins were then transferred to nitrocellulose sheets (Bio-Rad, Richmond, Calif.) by using a semidry electroblotter (Biolyon, Dardilly, France) at 12 V for 10 min, followed immediately by 24 V for 1 h. Nonspecific binding sites on the nitrocellulose membrane were then saturated with Tris-buffered saline containing 0.5% Tween 20 and 3% fat-free milk for 1 h at room temperature. After 3 washes with Tris-buffered saline–Tween buffer, the nitrocellulose tracks were incubated with the different sera diluted 1/50 and 1/100 in washing buffer, under gentle agitation, for 2 h at room temperature. Membranes were washed three times and incubated with horseradish peroxidase-labelled goat anti-human immunoglobulin G (IgG) conjugate (Nordic, Tilburg, The Netherlands) for 1 h diluted 1/500 in washing buffer. After another three washes, immunoblots were developed by using 4-chloro-naphthol (Sigma, L’Isle-d’Abeau-Chêne, France) as a substrate.
The immunoblot patterns were analyzed with Gelcompar software (Applied Maths, Kortrijk, Belgium): molecular sizes of the stained bands were determined by comparing them with protein markers ranging from 14.3 to 200 kDa, and the intensity of the staining was calculated following densitometric integration of the nitrocellulose tracks. Faint bands (less than 3% staining for any given track) were not considered for data interpretation.
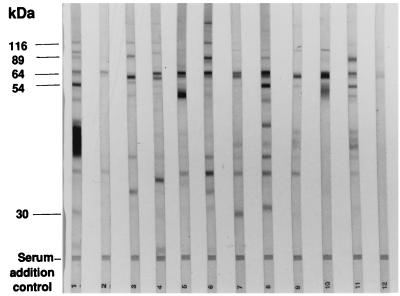
The sera were tested by using a Pylori Check enzyme immunoassay and immunoblot (Helico-Blot 2.0), and found to contain IgG antibodies against H. pylori in both assays (Table 1). The intensity of the ELISA was usually positively correlated with the number and the intensity of the bands seen in the immunoblot (Fig. 1). However, some discrepancies were noted between these two assays (Fig. 1, lane 2). Major differences appeared when the band patterns in the immunoblot were compared for these 10 patients. Several bands, such as those corresponding to the 19- to 20-, 26.5-, 35-, and 116-kDa antigens, were recognized by very few patients.
TABLE 1.
Anti-H. pylori antibody response in serum from 10 patients with MALT lymphoma.
| Patient | ELISA optical density | No. of bands in Helico-Blot 2.0 | Result
fora:
|
|||
|---|---|---|---|---|---|---|
| PCR
|
Dot blot (cagA) | Antibodies against CagA | ||||
| cagA1-cagA2 and cagA3-cagA4 | cagA5-cagA6 | |||||
| 1 | 2.521 | 16 | + | + | + | + |
| 2 | 2.411 | 11 | − | − | − | − |
| 3 | 2.303 | 12 | + | + | + | + |
| 4 | 1.900 | 11 | + | + | + | + |
| 5 | 1.853 | 4 | − | − | − | − |
| 6 | 1.800 | 9 | + | + | + | + |
| 7 | 1.577 | 10 | − | − | − | − |
| 8 | 1.503 | 7 | − | − | − | − |
| 9 | 1.010 | NDb | − | − | − | − |
| 10 | 0.801 | 16 | − | − | − | |
The Cag region was also amplified by PCR of the cagA gene marker in gastric biopsies with primer pairs cagA1-cagA2, cagA3-cagA4, and cagA5-cagA6 and was detected by dot blot analysis (probe cagA5-cagA6).
ND, Not determined.
FIG. 1.
Anti-H. pylori serum antibody response assessed by the Helico-Blot 2.0 immunoblot in nine patients with MALT lymphoma. Patterns displayed are for patients 1, 5, 6, 3, 8, 4, 7, 2, and 10 (lanes 1 to 9, respectively) and for positive and negative controls (lanes 10 and 11, respectively).
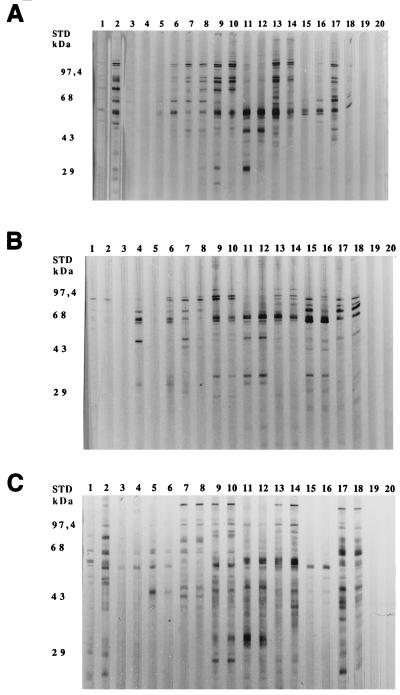
The results obtained with the Helico-Blot 2.0 assay prompted us to analyze the immunoblot patterns with the homologous strain, i.e., the strain infecting each patient. As an example, Fig. 2 displays the immunoblot patterns for the strains isolated from patients 1, 5, and 6, tested with the serum from the corresponding patient (lanes 1 and 2, 3 and 4, and 7 and 8, respectively). The antibody response against the antigens of the homologous strain (1/50 and 1/100 dilutions in odd- and even-numbered lanes, respectively) was generally greater than that against the antigens of the strain used in the Helico-Blot 2.0, in terms of number of bands. The same set of bands were recognized by serum antibodies, including proteins migrating at 20, 26 to 28, 30, 37, 40 to 42, 50, 54, 60, 67, 70, and 89 kDa. However, not all the sera reacted with all of these antigens. The sera with a mild response against the antigens used in the Helico-Blot 2.0 stained very few bands when tested against the homologous strain. A more precise comparison of this response did not single out any specific antigen(s) present in the strain isolated from patients with lymphomas and absent in the reference strain.
FIG. 2.
Anti-H. pylori serum antibody response assessed by immunoblot by using homologous and heterologous strains. Lysates from the strains isolated from patients 1, 5, and 6 (A, B, and C, respectively) were used as antigens to detect anti-H. pylori antibodies in the serum from seven patients with MALT lymphoma. The sera tested were from patients 1 (lanes 1 and 2), 5 (lanes 3 and 4), 9 (lanes 5 and 6), 6 (lanes 7 and 8), 3 (lanes 9 and 10), 8 (lanes 11 and 12), 4 (lanes 13 and 14), and 7 (lanes 15 and 16). Positive and negative control sera (lanes 17 and 18 and lanes 19 and 20, respectively) were included. Two dilutions were tested: 1/50 (odd-numbered lanes) and 1/100 (even-numbered lanes).
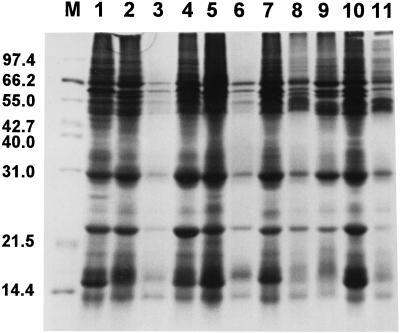
The protein profile of the strains was performed by SDS-polyacrylamide gel electrophoresis (PAGE). The cells were washed twice in 1 ml of PBS solution and centrifuged at 6,000 × g for 5 min. The pellets were resuspended in 200 μl of PBS and boiled at 100°C for 5 min. Proteins (15 μl) were separated on an SDS-polyacrylamide gel (29% acrylamide, 1% N,N′-methylenebisacrylamide) through a stacking gel (5%) for 1 h at 100 V and through a resolving gel (10%) for 4 h at 135 V (20). Polypeptides separated by SDS-PAGE were stained with Coomassie brilliant blue for 1 h and destained in a methanol-acetyl acid solution (45% [vol/vol] of each reagent) overnight. The profiles of the different strains were indeed similar (Fig. 3).
FIG. 3.
SDS-PAGE of the proteins of the 10 strains of H. pylori. The proteins were from strains isolated from patients 5, 9, 6, 3, 8, 4, 7, 10, 2, and 1 (lanes 1 to 10, respectively) and from strain 26695 (lane 11). M, molecular mass standard proteins (Mid-Range proteins; Promega). The numbers on the left indicate the molecular weights (in thousands) of the proteins.
Each of the 10 sera was tested against the nine heterologous strains isolated from the nine other patients. Representative patterns are displayed in Fig. 2 for strains isolated from three patients, against which seven serum samples from seven patients were tested (1/50 dilution, odd-numbered lanes, and 1/100 dilution, even-numbered lanes). The immune response against H. pylori also appeared very different from one patient to another, as already suggested by the Helico-Blot 2.0 assay. Several antigens elicited a strong antibody response with the heterologous sera, such as the strain from patient 1 with serum from patient 4 or 8 (Fig. 2B, lanes 11 to 14), with a major band at 60 kDa, as judged from the staining with a heterologous strain, although they were mildly recognizable with the homologous strain (Fig. 2A, lane 2). This same antigen was strongly recognized with serum from patient 7, incubated with its homologous strain (not shown) as well as with the heterologous strains from patients 1 and 5 (Fig. 2A and B, lanes 15 and 16, respectively). When the antibody response to the homologous strain was weak, in terms of number of bands, it reacted similarly toward the heterologous strains. This was the case for serum from patient 5 with strains from patients 1, 5, and 6 (Fig. 2A, B, and C, lanes 3 and 4, respectively). Globally, the patterns were very different for any given strain and the 10 sera tested with it.
Analysis of the cagA gene of H. pylori.
The presence of the cagA gene was analyzed by PCR and dot blot analysis. The DNA of H. pylori cultures was extracted by the standard phenol-chloroform procedure. Briefly, the cells were harvested in 2 ml of brucella broth from two 48-h-old culture plates, and they were centrifuged at 4,000 × g for 15 min. The pellets of bacteria were resuspended in 2 ml of a buffer containing 10 mM Tris-HCl (pH 8.0) and 1 mM EDTA. They were then treated successively with SDS (1%) and proteinase K (0.1 g/liter). Proteins were eliminated by solvent extraction (twice with phenol and once with chloroform). DNA was precipitated with 95% ethanol at −20°C in the presence of 0.3 M sodium (pH 4.8). The DNA pellet was washed with 75% ethanol, dried, and resuspended in water.
Two pairs of primers previously described (21) were used for detection of two internal regions of the cagA gene (GenBank accession no. U60176). First, 5′-GAT AAC AGG CAA CGT TTT CAG GGA-3′ (positions 19119 to 19142) and 5′-CCG AAC GGA TCA AAA ATT CAT GG-3′ (19512 to 19490), named cagA1 and cagA2, were used to amplify a 394-bp section of the cagA region (21). Second, 5′-ATG GGG AGT CAT GAT GGC ATA GAA CC-3′ (19872 to 19897) and 5′-GGT GGC TGT CTT TAA TTT GCC TAA T-3′ (20588 to 20564), named cagA3 and cagA4, were used to amplify a 717-bp fragment (21). PCR conditions were optimized with the reference strain CCUG 17864 by using a 480 thermal cycler (Perkin-Elmer Corp., Norwalk, Conn.). PCR amplification was performed in a final volume of 50 μl containing 1 ng of H. pylori DNA with 67 mM Tris-HCl (pH 8.8), 16 mM (NH4)2SO4, 0.01% Tween 20, 1.5 mM MgCl2, 50 pmol of each primer, 1 U of Gold Start Red Taq DNA polymerase (Eurogentec Bel. A., Seraing, Belgium), and a 0.2 mM concentration of deoxynucleoside triphosphate mixture (Eurobio). The amplicons were analyzed on a 1% agarose gel.
The amplification cycles consisted of an initial denaturation of the target DNA at 94°C for 5 min and then denaturation at 94°C for 1 min, primer annealing at 62°C for 1 min, and extension at 72°C for 1 min (2 min for the second set of primers), repeated in 35 consecutive cycles. The final cycle included an extension step at 72°C for 5 min to ensure full extension of the product. Furthermore, two other primers were used for amplification of the complete cagA gene (cagA5 and cagA6). The primers, chosen by comparing the sequences of the cagA gene in strains J99, 26695, and CCUG 17874 (GenBank accession no. U60176), are as follows: cagA5, 5′-TAG GTA TAG TAA GGA GAA ACA ATG ACT AAC G-3′ (positions 18942 to 18972), and cagA6, 5′-CCT TAA TCC TTT AAG ATT TTT GGA AAC CAC C-3′ (22416 to 22386). All the primers were synthesized by Eurogentec. The last PCR was performed with a 9600 thermal cycler (Perkin-Elmer). The amplification cycles consisted of an initial denaturation at 94°C for 1 min, primer annealing at 60°C for 2 min, and extension at 72°C for 5 min; the final cycle included an extension step at 72°C for 7 min. All PCR products were run on 1% agarose gels and stained with ethidium bromide.
The amplified products cagA5 and cagA6 of the reference strain obtained by PCR were used as probes in the hybridization experiments. The amplicons were electrophoresed on a 1% agarose gel, cut out of the gel, and purified with a QIAEX II gel extraction kit (Qiagen, Courtaboeuf, France). DNA probes were labelled with an enhanced chemiluminescence kit according to the conditions (hybridization at 42°C and washing steps at 55°C) specified by the supplier (Amersham, Little Chalfont, Buckinghamshire, United Kingdom). For the dot blot analysis, 300 ng of undigested denatured total DNA was transferred to a blot membrane by means of a Bio-Rad slot blot apparatus.
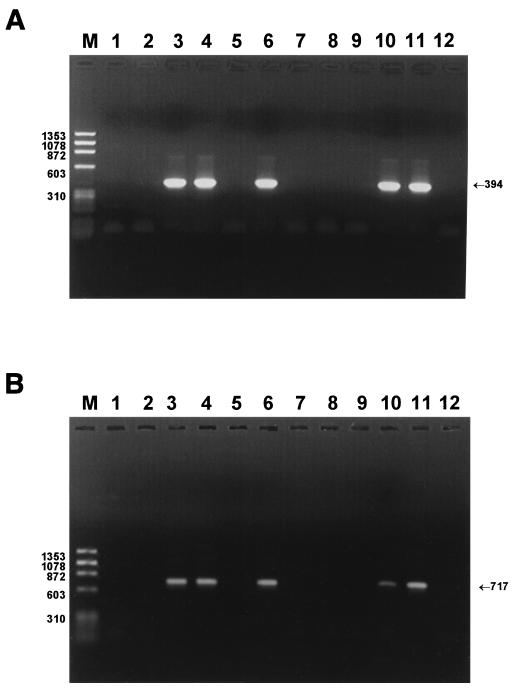
Four of the 10 strains (from patients 1, 3, 4, and 6) had a positive result, with primer pairs cagA1-cagA2 and cagA3-cagA4 detecting the cagA gene (Fig. 4). In the same strains, the complete cagA gene sequence was also detected with a pair of primers from the extremity of the cagA gene, allowing the amplification of 3,474 bp. For six strains, the cagA gene was absent and false-negative results were ruled out by amplification of the 16S rRNA gene of H. pylori. Dot blot analysis was then performed on all strains with the cagA5-cagA6 probe, and it confirmed the PCR results. Immunoblot analysis with the 10 sera against the strain isolated from PCR-positive patient 6 (Fig. 2C) showed the presence of a 120- to 140-kDa band, corresponding in size to the CagA protein, in the four patients for whom the cagA gene was present.
FIG. 4.
Detection of the cagA gene by PCR in strains obtained from MALT lymphoma patients. (A) PCR amplifications with primers cagA1 and cagA2 (394 bp). (B) PCR amplifications with primers cagA3 and cagA4 (717 bp). The amplicons are for strains isolated from patients 5, 9, 6, 3, 8, 4, 7, 10, 2, and 1 (lanes 1 to 10, respectively); reference strain 26695 (lane 11); and the negative control (lane 12). M, φX174 DNA/Hae III markers (Eurobio). The numbers on the left indicate the molecular sizes (in base pairs) of DNA.
Normal gastric mucosa does not contain lymphoid tissue, but lymphocytes are recruited upon colonization of the stomach by the bacterium. In vitro experiments have shown that H. pylori stimulates the proliferation of B-lymphoma cells in cultures of gastric biopsies (11, 12). This cellular proliferation is abolished when the T lymphocytes associated with the MALT are eliminated. Therefore, the occurrence of B-lymphoma cells is dependent on the presence of T cells.
The malignant proliferation is monoclonal, as shown by the study of the molecular rearrangement of the immunoglobulin heavy-chain genes (3, 16). It is not known whether the bacterium triggers or selects B lymphocytes specific for its own antigen; i.e., it has not been shown thus far whether the B-lymphoma cells are capable of producing an H. pylori-specific monoclonal antibody. Another unanswered question deals with the nature of the stimulating antigen, if it exists.
A comparative study by immunoblot of the serum antibody response against H. pylori in 10 patients with MALT lymphoma was undertaken. All the patients studied produced anti-H. pylori antibodies, as shown by ELISA and Western blot analysis. However, the immunoblot pattern was very different among the patients, suggesting great variability in individuals’ capacities to react to H. pylori antigens. In view of these differences, the antibody response against the homologous strains isolated from the gastric biopsies was studied. As with the commercial reagent, there was considerable variation among the individual patients’ responses toward their own strains. No band that was recognized by all the sera tested and was not known to be present in other H. pylori-associated diseases was detected.
A positive conclusion of this study is the variability in the systemic immune response observed which does not differentiate MALT lymphoma patients from other infected persons. The cause of this variability is believed to be linked to the immune response rather than to the antigenic composition of the strain. However, patients 1 and 5 displayed some specificity for their own strains. This fact could be explained by a broadening, over time, of the specificity of the immune response, but sera collected at different times in the natural course of the infection were not available to confirm this hypothesis. Alternatively, it is possible that, in fact, only a few clones of H. pylori with minor protein variation, even if undetectable by SDS-PAGE, do exist. Indeed, the results of the randomly amplified polymorphic DNA analysis performed on the strains showed different profiles (data not shown).
Moreover, a similar conclusion can be drawn from the comparative study of the response against the nine heterologous strains isolated from the nine other patients. The results strongly suggested that no typical pattern was associated with this particular disease state. For most patients and strains, no abnormally strong reaction against any antigen could be seen. This indicates that no monoclonal antibody against H. pylori was circulating in the serum of these patients, at least at a titer high enough to be detected. Alternatively, this putative monoclonal antibody may be confined to the gastric tissue and not released into the bloodstream. Another possibility is that it does not belong to the IgG isotype. We tried to detect an IgA response to H. pylori, but antibody titers were always very low or undetectable (data not shown). However, the systemic antibody response might not reflect very well the local gastric antibody response. To exclude a local monoclonal antibody response, immunoglobulin from the gastric juice should have been tested. Unfortunately, such samples were not available.
Another interesting antigen of H. pylori is the CagA protein. It is a high-molecular-mass protein, approximately 120 to 140 kDa, which is present in 80 to 100% of the strains responsible for duodenal ulcers and in 70% of the strains responsible for gastric ulcers, but only in 50 to 60% of the strains responsible for nonulcer dyspepsia (6) (percentages are based on strains isolated from Western countries). This immunodominant protein is encoded by the cagA gene and is a marker for the presence of the cag pathogenicity island (5). In the present study, 4 out of the 10 sera tested reacted against a protein of this size and possessed the cagA gene, as assessed by different tests.
The association of cagA+ strains with MALT lymphoma is controversial. The first studies did not show an association: de Jong et al. (7) and Witherell et al. (22) reported 12 and 32 cases, respectively. Later Eck et al. (10) claimed that 95% of MALT lymphomas occurred with cagA+ strains. Their series included 46 high-grade and 22 low-grade MALT lymphomas. Peng et al. (18) subsequently confirmed that cagA+ strains are more frequent in high-grade than in low-grade MALT lymphomas.
This study demonstrates that a large variety of H. pylori strains can be considered responsible for the induction of MALT lymphoma, as judged from the heterogeneity of their antigenic properties, and that the cagA marker does not seem to be associated with the disease. However, the local response as well as other markers remains to be studied.
REFERENCES
- 1.Bayerdörffer E, Neubauer A, Rudolph B, Thiede C, Lehn N, Eidt S, Stolte M. Regression of primary gastric lymphoma of mucosa-associated lymphoid tissue type after cure of Helicobacter pylori infection. Lancet. 1995;345:1591–1594. doi: 10.1016/s0140-6736(95)90113-2. [DOI] [PubMed] [Google Scholar]
- 2.Boot H, de Jong D, van Heerde P, Taal B. Role of Helicobacter pylori eradication in high-grade MALT lymphoma. Lancet. 1995;346:448–449. doi: 10.1016/s0140-6736(95)92823-5. [DOI] [PubMed] [Google Scholar]
- 3.Calvert R, Randerson P E, Evans P, Cawkwell L, Lewis F, Dixon M F, Morgan G J. Genetic abnormalities during transition from Helicobacter pylori associated gastritis to low-grade MALToma. Lancet. 1995;345:2626–2627. doi: 10.1016/s0140-6736(95)91154-5. [DOI] [PubMed] [Google Scholar]
- 4.Carlson S J, Yokoo H, Vanagunas A. Progression of gastritis to monoclonal B-cell lymphoma with resolution and recurrence following eradication of Helicobacter pylori. JAMA. 1996;275:937–939. [PubMed] [Google Scholar]
- 5.Censini S, Lange C, Xiang Z, Crabtree J E, Ghiara P, Borodovski M, Rappuoli R, Covacci A. Cag, a pathogenicity island of Helicobacter pylori, encodes type 1-specific and disease-associated virulence factors. Proc Natl Acad Sci USA. 1996;93:14648–14653. doi: 10.1073/pnas.93.25.14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Covacci A, Censini S, Bugnoli M, Petracca R, Burroni D, Macchia G, Massone A, Papini E, Xiang Z, Figura N, Rappuoli R. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc Natl Acad Sci USA. 1993;90:5791–5795. doi: 10.1073/pnas.90.12.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Jong D, van der Hulst R W, Pals G, van Dijk W C, van der Ende A, Tytgat G N, Taal B G, Boot H. Gastric non-Hodgkin lymphomas of mucosa-associated lymphoid tissue are not associated with more aggressive Helicobacter pylori strains as identified by CagA. Am J Clin Pathol. 1996;106:670–675. doi: 10.1093/ajcp/106.5.670. [DOI] [PubMed] [Google Scholar]
- 8.Delchier J C. Lymphomes gastriques. Aspects cliniques. In: Mégraud F, Lamouliatte H, editors. Helicobacter pylori, clinique, traitement. Vol. 2. Paris, France: Elsevier Option Bio; 1997. pp. 151–160. [Google Scholar]
- 9.Doglioni C, Wotherspoon A C, Moschini A, de Boni M, Isaacson P G. High incidence of primary gastric lymphoma in northeastern Italy. Lancet. 1992;339:834–835. doi: 10.1016/0140-6736(92)90280-g. [DOI] [PubMed] [Google Scholar]
- 10.Eck M, Schmausser B, Haas R, Greiner A, Czub S, Muller-Hermelink H K. MALT-type lymphoma of the stomach is associated with Helicobacter pylori strains expressing the CagA protein. Gastroenterology. 1997;112:1482–1486. doi: 10.1016/s0016-5085(97)70028-3. [DOI] [PubMed] [Google Scholar]
- 11.Hussell T, Isaacson P G, Crabtree J E, Spencer J. The response of cells from low-grade B-cell gastric lymphomas of mucosa-associated lymphoid tissue to Helicobacter pylori. Lancet. 1993;342:571–574. doi: 10.1016/0140-6736(93)91408-e. [DOI] [PubMed] [Google Scholar]
- 12.Isaacson P G, Spencer J. The biology of low grade MALT lymphoma. J Clin Pathol. 1995;48:395–397. doi: 10.1136/jcp.48.5.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levine M S, Elmas N, Furth E E, Rubesin S E, Goldwein M I. Helicobacter pylori and gastric MALT lymphoma. Am J Roentgenol. 1996;166:85–86. doi: 10.2214/ajr.166.1.8571912. [DOI] [PubMed] [Google Scholar]
- 14.Montalban C, Boixeda D, Bellas C. Helicobacter pylori eradication in gastric mucosa-associated lymphoid tissue lymphomas. Ann Intern Med. 1996;124:275. doi: 10.7326/0003-4819-124-2-199601150-00023. [DOI] [PubMed] [Google Scholar]
- 15.Montalban C, Manzanal A, Boixeda D, Redondo C, Bellas C. Treatment of low-grade gastric MALT lymphoma with Helicobacter pylori eradication. Lancet. 1995;345:798–799. doi: 10.1016/s0140-6736(95)90679-7. [DOI] [PubMed] [Google Scholar]
- 16.Okazaki K, Morita M, Yamamoto Y. Gene rearrangements, Helicobacter pylori, and gastric MALT lymphoma. Lancet. 1994;343:1636. doi: 10.1016/s0140-6736(94)93087-2. [DOI] [PubMed] [Google Scholar]
- 17.Parsonnet J, Hansen S, Rodriguez L, Geld A B, Warnke R A, Jellum E, Orentreich N, Vogelman J H, Friedman G D. Helicobacter pylori infection and gastric lymphoma. N Engl J Med. 1994;330:1267–1270. doi: 10.1056/NEJM199405053301803. [DOI] [PubMed] [Google Scholar]
- 18.Peng H, Ranaldi R, Diss T C, Isaacson P G, Bearzi I, Pan L. High frequency of CagA+ Helicobacter pylori infection in high-grade gastric MALT B-cell lymphomas. J Pathol. 1998;185:409–412. doi: 10.1002/(SICI)1096-9896(199808)185:4<409::AID-PATH121>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 19.Roggero E, Zucca E, Pinotti G, Pascarella A, Capella C, Savio A, Pedrinis E, Paterlini A, Venco A, Cavalli F. Eradication of Helicobacter pylori infection in primary low-grade gastric lymphoma of mucosa-associated lymphoid tissue. Ann Intern Med. 1995;122:767–769. doi: 10.7326/0003-4819-122-10-199505150-00006. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 21.Tummuru M K, Cover T L, Blaser M J. Cloning and expression of a high-molecular-mass major antigen of Helicobacter pylori: evidence of linkage to cytotoxin production. Infect Immun. 1993;61:1799–1809. doi: 10.1128/iai.61.5.1799-1809.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Witherell H L, Hansen S, Jellum E, Orentreich N, Vogelman J H, Parsonnet J. Risk for gastric lymphoma in persons with CagA+ and CagA− Helicobacter pylori infection. J Infect Dis. 1997;176:1641–1644. doi: 10.1086/517346. [DOI] [PubMed] [Google Scholar]
- 23.Wotherspoon A C, Doglioni C, de Boni M, Spencer J, Isaacson P G. Antibiotic treatment for low-grade gastric MALT lymphoma. Lancet. 1994;343:1503. [PubMed] [Google Scholar]
- 24.Wotherspoon A C, Doglioni C, Diss T C, Pan L, Moschini A, de Boni M, Isaacson P G. Regression of primary low-grade B-cell gastric lymphoma of mucosa-associated lymphoid tissue type after eradication of Helicobacter pylori. Lancet. 1993;342:575–577. doi: 10.1016/0140-6736(93)91409-f. [DOI] [PubMed] [Google Scholar]