Abstract
Purpose
To select individuals and families with a low genetic burden for age-related macular degeneration (AMD), to inform the clinical diagnosis of macular disorders, and to find novel genetic variants associated with maculopathies.
Design
Genetic association study based on targeted and whole-exome sequencing.
Participants
A total of 758 subjects (481 individuals with maculopathy and 277 controls), including 316 individuals in 72 families.
Methods
We focused on 150 genes involved in the complement, coagulation, and inflammatory pathways. Single-variant tests were performed on 7755 variants shared among ≥ 5 subjects using logistic regression. Gene-based tests were used to evaluate aggregate effects from rare and low-frequency variants (at minor allele frequency [MAF] ≤ 5% or ≤ 1%) in a gene using burden tests. For families whose affected members had a low burden of genetic risk based on known common and rare variants related to AMD, we searched for rare variants (MAF < 0.001) whose risk alleles occurred in ≥ 80% of affected individuals but not in controls. Immunohistochemistry was performed to determine the protein expression of a novel gene (coagulation factor II thrombin receptor-like 2 [F2RL2]) in retinal tissues.
Main Outcome Measures
Genotypes and phenotypes of macular degeneration.
Results
We confirmed the association of a synonymous variant in complement factor H (Ala473, rs2274700, proxy to intronic rs1410996, r2 = 1) with maculopathy (odds ratio, 0.64; P = 4.5 × 10–4). Higher AMD polygenic risk scores (PRSs) were associated with intermediate and advanced AMD. Among families with low PRSs and no known rare variants for maculopathy, we identified 2 novel, highly penetrant missense rare variants in ADAM15, A disintegrin and metalloprotease, metallopeptidase domain 15 (p.Arg288Cys) and F2RL2 (p.Leu289Arg). Immunohistochemistry analyses revealed F2RL2 protein expression in cone photoreceptor outer segments and Müller glia cells of human and pig retinas. Coagulation factor II thrombin receptor-like 2 expression appeared increased in fibrotic areas in advanced AMD samples with neovascularization, suggesting that F2RL2 may play a role in the progression to advanced macular disease.
Conclusions
New missense rare variants in the genes ADAM15 and F2RL2 were associated with maculopathies. Results suggest that novel genes related to the coagulation and immune pathways may be involved in the pathogenesis of macular diseases.
Keywords: Coagulation pathway, Immune pathways; Macular degeneration; Maculopathy; Targeted sequencing; Whole-exome sequencing
Abbreviations and Acronyms: AMD, age-related macular degeneration; ATP, adenosine triphosphate; CADD, Combined Annotation Dependent Depletion; CFH, complement factor H; CFI, complement factor I; C3, complement component 3; C9, complement component 9; ENG, endoglin; F2RL2, coagulation factor II thrombin receptor-like 2; FANTOM5, functional annotation of the mammalian genome; GS, glutamine synthetase; GWAS, genome-wide association studies; MAF, minor allele frequency; PECAM1, Platelet Endothelial Cell Adhesion Molecule 1; PRS, polygenic risk score; SKAT, sequence kernel association testing; SNP, single nucleotide polymorphism; TPM, tags per million; WES, whole-exome sequencing
Age-related macular degeneration (AMD) is a multifactorial neurodegenerative disease determined by human aging and genetic and environmental factors.1 Age-related macular degeneration is the leading cause of irreversible central vision impairment and blindness worldwide. In the United States alone, AMD affects approximately 11 million individuals. Some macular dystrophies and macular degenerations can resemble the phenotypes of AMD, especially in the late stages, including Stargardt disease and some of the pattern dystrophies.2 Both AMD and macular dystrophy phenotypes can also occur in the same families. In cases without Mendelian familial transmission or in the absence of classic signs of dystrophies versus AMD, the etiology might only be confirmed by genetic analyses.
Genome-wide association studies (GWAS) have revealed common and rare variants related to AMD.3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14 About half of those loci are in genes related to immune function, the alternative complement system, and inflammatory pathways, including complement factor H (CFH), complement factor I (CFI), complement component 3 (C3), complement component 9 (C9), complement factor B, and others. Functional studies have expanded knowledge about the essential roles of these genes involved in those immune pathways in AMD.15, 16, 17, 18, 19, 20 Despite the success of GWAS, the contribution of rare variants to AMD is more difficult to explore because of the low occurrence of rare variants in the general population. Compared with large-scale GWAS in unrelated individuals, family-based designs provide unique opportunities to detect pathogenic variants that are extremely rare in the general population but have high penetrance in affected families.
In 2010, we reported a method to find novel genetic associations in families with AMD.21 Our approach was based on the hypothesis that if an affected family had less than the expected genetic load because of known common or rare variants associated with AMD, it would suggest that there were likely other unknown rare, more penetrant variants in the family to explain the burden of disease. We calculated an AMD polygenic risk score (PRS) for each family member as initially described22 and later expanded23, 24, 25, 26 and then selected those families with low scores. These families suspected to be at higher risk of harboring new variants were good candidates for finding novel variants by performing DNA sequencing. The application of this method has yielded many new variants in family-based analyses in the past decade. One of the most striking findings is the association our team discovered between AMD and the rare variant, CFH R1210C (rs121913059) with a minor allele frequency (MAF) of 0.025% in the European population, which has a 20-fold increased risk of AMD for individuals carrying the risk allele.8 We also found that this variant was associated with earlier onset of AMD,8,27 a phenotype characterized by a high burden of macular and extramacular drusen,27 as well as earlier progression to advanced disease.23, 24, 25,28 We detected other highly penetrant rare variants in CFH in other families with AMD, including R53C, D90G, R175P, and C192F, as well as rare variants in the complement pathway genes C3, CFI, and C9, using whole-exome sequencing (WES) and targeted sequencing approaches.9,11,12,16
In this study, we focused on genes involved in the inflammation, coagulation, and complement pathways. Several connections among these cascades have been described and are involved at various steps in the development of clinical inflammatory and thrombotic events.29 Targeted sequencing and WES were performed to identify novel risk variants and their role in the pathogenesis of macular degeneration. We selected families with low PRS based on genes known to be related to AMD with no known pathogenic rare variants associated with AMD or other forms of maculopathy. We identified novel rare protein-coding variants that were highly penetrant in affected subjects in those families. The functional impact of a novel gene (coagulation factor II thrombin receptor-like 2 [F2RL2]) was assessed by immunohistochemistry to identify its expression in human and pig retinas and in retinas with advanced AMD pathologies.
Methods
Study Samples and Phenotypes
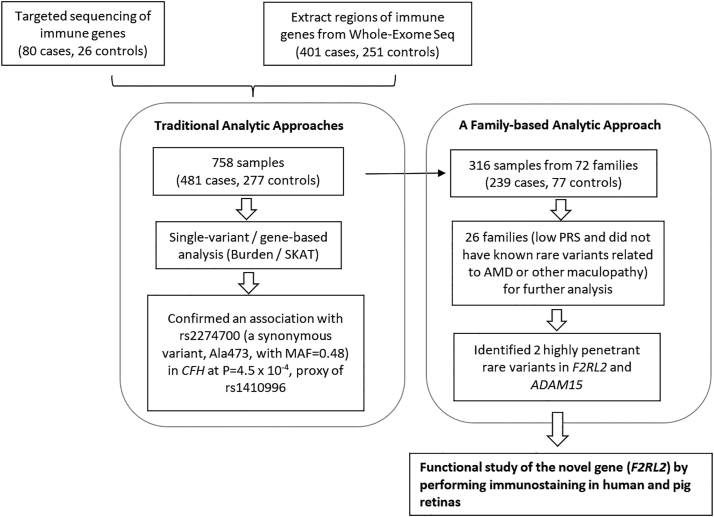
The overall design of this study is depicted in Figure 1. We performed targeted sequencing of exonic regions of 150 genes involved in the inflammation, coagulation, or complement pathways for 106 samples and WES for an additional 652 samples. We focused on the overlapping genomic regions measured by targeted sequencing and WES. We selected patients with a relatively low burden of known genetic risk of AMD or younger age of onset of macular disease, as well as macular dystrophies not typical for AMD. There were 758 participants of European ancestry, including 481 cases with signs of advanced macular degeneration (mean age, 79 years; 59% women) and 277 unaffected subjects (mean age, 80 years; 37% women). Among those participants, there were 316 participants from 72 families. All participants were previously enrolled in ongoing genetic and epidemiologic studies.1,25 All affected and unaffected individuals were evaluated by board-certified ophthalmologists, and ocular records, fundus photographs, autofluorescence, and OCT images were reviewed to determine grades according to the Clinical Age-Related Maculopathy Staging system.30 Briefly, the grades were defined as follows: (1) no AMD; (2) early AMD; (3) intermediate AMD; (4) advanced dry AMD with central or noncentral geographic atrophy; (5) neovascular AMD; and (6) maculopathy other than AMD. Cases with advanced disease had drusenoid retinal pigment epithelial detachment, central or noncentral macular atrophy, or neovascular disease. Unaffected status for this analysis was defined as no sign of macular degeneration or only a few small drusen. Approval for this research was obtained from the institutional review board, and informed consent was obtained from all participants.
Figure 1.
Overall study design.
In the discovery phase, single-variant and gene-based association analyses were performed for 758 samples. We also searched for highly penetrant variants in families with ≥ 2 affected members, low PRS among affected family members, and no known complement rare variants (26 families). Two rare variants displayed high penetrance in >80% of affected members in 2 families. Immunochemistry was performed to determine the expression of a new gene F2RL2 in pig and human retinas. AMD = age-related macular degeneration; CFH = complement factor H; F2RL2 = coagulation factor II thrombin receptor-like 2; MAF = minor allele frequency; PRS = polygenic risk score; SKAT = sequence kernel association testing.
Targeted Sequencing and Quality Control
Blood specimens were collected, and DNA extraction and quantitation were performed using standard procedures. We selected 150 genes in the complement, coagulation, and inflammatory pathways and sequenced their coding exons, splice junctions, nonrepetitive 3’ and 5’ untranslated regions, and ± 50 bp of these regions. The libraries were pooled together in 24 sets of 5 or 6 samples for target capture using custom SeqCap EZ Probes. The HyperCap Workflow from Roche (version 2.3) was used for library capture. Samples and pools were quantified with Agilent Bioanalyzer and Qubit Fluorometric quantification. The 24 sets of enriched libraries were combined into 2 equimolar pools for sequencing. Each pool was loaded onto 2 lanes of an Illumina HiSeq 3000 flow cell and sequenced using paired-end 150 cycle reads. The reads from the samples were demultiplexed using Illumina’s bcl2fastq utility based on the single indexes of the library fragments.
Sequence reads were mapped to the human reference build hg19 using NovoAlign. Sentieon software was used to remove duplicate reads, realign reads around indels, recalibrate quality scores, and call variants. Variants were called in the target region using the Sentieon DNAscope variant caller. Variant numbers across the sample were plotted to determine the normal distribution of variants. Samples with sex inconsistencies and with more variants than expected based on the normal distribution were removed. Additionally, all samples were cross-compared with each other based on sequence variants to identify related samples. There were 106 samples, including 80 cases and 26 unaffected subjects in the targeted sequencing.
WES and Quality Control
The samples for WES were sequenced at the Perkin Elmer Center for Genome Innovation at the University of Connecticut.11,12 Genomic DNA extracted from blood samples was used to construct Illumina libraries with indexing barcodes to uniquely identify each sequenced individual. Samples were combined from the indexed libraries at an equimolar ratio and then hybridized to the Nimblegen 3.0 human exome reagent, designed to capture a total of 60 Mb consisting of the coding exons and splice junctions, nonrepetitive 3ʹ and 5ʹ untranslated regions, and selected noncoding RNA sequences. We isolated the hybridized library fragments, quantitated with quantitative polymerase chain reaction, produced 2 × 100 bp reads, and then sequenced them with the Illumina HiSeq2000 TM platform. After the deconvolution of barcodes from each lane, individual reads were aligned to the human reference genome hg19 using the Burrows–Wheeler Aligner resulting in BAM files. Variant calling was performed using the best practices recommendations of the tools in the Genome Analysis Toolkit, version 3.1 (www.broadinstitute.org/gatk/guide/best-practices).
We removed variants that did not pass variant quality score recalibration using the Genome Analysis Toolkit best practices thresholds and a tranche sensitivity threshold of 99.0% or that were present in < 95% of samples. Samples were removed based on the following criteria: (1) samples with < 20× depth for > 80% or < 10× depth for > 90% of the targeted regions in the WES; (2) samples showing sex inconsistency using the PLINK sex-check tool; (3) samples with more exome-wide heterozygosity than expected; (4) samples showing unexpected relatedness or unrelatedness with other samples in the datasets; (5) samples that were self-reported White but did not cluster closely with other members of the dataset; and (6) samples with < 95% of the variants genotyped. There were 652 subjects, including 401 cases and 251 controls, included in the WES. We extracted the genomic regions of the 150 targeted sequenced genes from the WES ∗.bam files.
Single-Variant Association Analysis
Genotypes of 14 180 biallelic variants (after removing duplicated sites) for 758 samples were combined. Single-variant association analysis required the minor allele to occur in at least 5 sequenced samples. Genetic variant effects were modeled additively. Logistic regression was used to model the association between each variant (single nucleotide polymorphism [SNP], as the independent variable) and case status (0 or 1, as the outcome), implemented in the glmr R package. Covariates included age, sex, ever smoking, 10 principal components computed from genotyping data, and pedigrees.
Gene-Based Burden and Sequence Kernel Association Testing Analysis
Gene-level analysis was used to evaluate aggregate effects from rare and low-frequency variants at MAF ≤ 5% (T5 test) or MAF ≤ 1% (T1 test) in a gene using the standard burden and sequence kernel association testing (SKAT) analysis, implemented in the SKAT R package. Variants residing within the sequenced region of each gene were grouped for tests. Covariates included age, sex, ever smoking, 10 principal components, and pedigrees. Burden tests assumed that all variants in a gene either increased or decreased the disease risk. Sequence kernel association testing allows variants with opposite directions of effect to reside in the same gene. Odds ratio estimates of the burden were derived by logistic regression using the Wald test on the collapsed burden.
Identifying Highly Penetrant Variants in Families
We used the family-based analysis method developed previously to search for rare variants that have a high penetrance in maculopathy families, suggesting a dominant inheritance pattern.21 We selected families for analysis based on the following criteria: (1) there were at least 2 family members affected with signs of advanced macular disease; (2) most of the affected family members were diagnosed before age 75; (3) the affected family members had a lower PRS for AMD, that is, the mean value of the PRS of the affected subjects in the family was < 1. Data from our previous targeted sequencing and whole-genome genotyping studies were used to calculate the PRS and to identify carriers of known AMD variants in 5581 subjects in our longitudinal cohort study (Fig 1).1,8,9,11,14,25 Polygenic risk score was calculated using the weighted sum of the number of disease risk alleles in each subject using the model previously described22,26 (an online application of most predictive variants is available at www.seddonamdriskscore.org)25; and (4) family members that did not carry known deleterious rare mutations associated with AMD in C3 (K155Q), CFI (59 rare variants),9 CFH (R1210C, D90G, R53C, R175P, C192F, and P503A), and C9 (P167S).
Because we searched for highly penetrant variants, our assumption was that such variants would have strong functional effects. We only focused on variants that were predicted to alter protein-coding sequences (missense, nonsense, read-through, or splice variants) by SnpEff/SnpSift (https://pcingola.github.io/SnpEff/)31 and were also very rare in the population (at MAF < 0.1% in 1000 Genomes and gnomAD databases). A highly penetrant variant was defined as following: (1) the minor allele was carried by ≥ 80% of the affected family members when there are > 4 affected members in the family; or (2) only 1 affected member did not carry the variant in a family when there were 3 to 4 affected members in the family; and (3) there were no carriers among all sequenced unaffected subjects. We used Combined Annotation Dependent Depletion (CADD)32 and PolyPhen233 to predict the functions of identified variants. A scaled CADD score of 20 means that a variant is among the top 1% of deleterious variants in the human genome. The PolyPhen2 scores ranged from 0 to 1. Variants with scores closer to 1 in PolyPhen2 suggested the variant to be deleterious.
We further confirmed that members of the 2 families (pedigrees A and B) did not carry pathogenic or likely pathogenic rare variants in other maculopathies, including rare variants in adenosine triphosphate (ATP) binding cassette subfamily A member 4, bestrophin 1, catenin alpha-1, interphotoreceptor matrix proteoglycan 1, interphotoreceptor matrix proteoglycan 2, peripherin 2, and epidermal growth factor (EGF) containing fibulin extracellular matrix protein 1. The rare variants related to maculopathies were downloaded from the Leiden Open Variation Database (www.lovd.nl), ClinVar (www.ncbi.nih.gov/clinvar), and the Human Gene Mutation Database (www.hgmd.cf.ac.uk).
Immunohistochemistry of F2RL2 in Human and Pig Retinas
The human retinal tissues were obtained from patient donors with AMD who participated in our eye donation program.34,35 The clinical diagnosis and severity of AMD were determined by reviewing and grading ocular examination records and multimodal imaging of eye donors in the Seddon Longitudinal Cohort by J.M.S.. The collection of human donor tissue was performed according to the protocol approved by the Institutional Review Board of the University of Massachusetts. We performed immunohistochemistry and immunofluorescence analyses on retinal cross sections of 6 human donor eyes from 6 donors (mean age, 88 years; 4 women and 2 men). Three eyes had no clinical evidence of AMD, and 3 eyes had neovascular AMD. The time from death to enucleation ranged from 2 to 9 hours.
Pig eyes were obtained from animal medicine, and all procedures involving animals were in compliance with the Association for Research in Vision and Ophthalmology Statement for the Use of Animals in Ophthalmic and Vision Research. Eyes were processed by fixing the superior quadrants of each eye in 4% paraformaldehyde, embedding in optimal cutting media, and then processing for cryo-sectioning. Dissection, fixation, and embedding of human and pig tissue were performed and processed for immunohistochemistry as previously described.36 In short, retinas were sectioned at 20-μm thickness. Samples were permeabilized in phosphate-buffered saline with 0.3% Triton X-100 and 0.01% sodium dodecyl sulfate for 24 hours at room temperature. The following primary antibodies were used: (1) rabbit anti-F2RL2 (1:300, MyBioSource, Cat no: MBS2523405); (2) mouse anti-glutamine synthetase (GS) (1:300, Millipore, Cat no: MAB302); and (3) mouse anti-Platelet Endothelial Cell Adhesion Molecule 1 (PECAM1; aka: CD31) (1:300, Invitrogen (JC/70A), Cat no: MA5-13188). The following reagent had a chromophore conjugate: fluorescein peanut agglutinin lectin (1:500, Vector Laboratories, Cat no: FL1071). All primary and secondary antibodies were diluted in phosphate-buffered saline with 0.3% Triton X-100 and 5% bovine serum albumin (Cell Signaling Technology). Incubation of primary antibody was performed overnight at 4° C, whereas the incubation of secondary antibody was performed at room temperature for 2 hours. Secondary antibody controls were performed in parallel, using the same incubation times, while omitting the primary antibody. All fluorescent secondary antibodies were purchased from Jackson ImmunoResearch and were purified F(ab)2 fragments that displayed minimal cross-reactivity with other species. Nuclei were counterstained with 4',6-diamidino-2-phenylindole (Sigma-Aldrich; Cat no. 9542) for the immunofluorescence staining. For the immunohistochemistry staining, a horseradish peroxidase-conjugated anti-rabbit antibody (1:500, Cell Signaling; Cat no: 711-036-152) was used with the ImmPACT VIP kit (Vector Laboratories, Cat no: SK-4605). All images were visualized using a Leica DM6 Thunder microscope with a 12-bit color camera and a 16-bit monochrome camera for fluorescent images.
Results
Single-Variant and Gene-Based Association Analyses
Single-variant association tests were performed for the 758 samples on 7755 variants occurring in at least 5 sequenced samples. The genomic control lambda was 1.04. The top result of single-variant analysis was rs2274700 (a synonymous variant, Ala473, with MAF = 0.48) in CFH (odds ratio, 0.64; P = 4.5 × 10–4). This variant, rs2274700, is a perfect proxy of the previously discovered intronic SNP rs1410996 in CFH (linkage disequilibrium, r2 = 1) associated with advanced AMD4 and is the most significant common variant related to AMD.
Gene-based burden and SKAT test results revealed a burden of 8 low-frequency variants with MAF < 0.05 (T5 test) in endoglin (ENG) associated with disease status at the suggestive significant threshold (PBurden = 2.7 × 10–4 and PSKAT = 2.2 × 10–3). The 8 low-frequency variants in ENG, including 4 synonymous variants, 2 missense variants, and 2 intronic variants, were found more often in patients than in controls (2.7-fold). Endoglin has been reported to be involved in retinal neovascularization.37
PRS and Identification of Highly Penetrant Variants
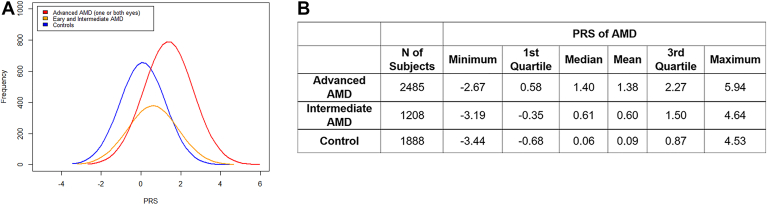
Figure 2 shows the distribution of the PRS for AMD in 5581 samples in our cohort, including 2485 advanced subjects with AMD, 1208 with early and intermediate AMD, and 1888 controls, whose genotypes were measured in previous studies.1,4, 5, 6, 7, 8, 9,14,22, 23, 24, 25, 26 The median PRS values for advanced AMD, early and intermediate AMD, and controls were 1.4, 0.61, and 0.06, respectively (P values for advanced group vs. control group, and for intermediate group vs. control group were < 1e-16, by 2-sided t test). These results demonstrate a definite relationship between higher genetic risk score and stage of AMD, with higher scores for more advanced disease. They also underscored the importance of common and rare genetic risk for risk stratification of both onset and progression to advanced AMD subtypes as in previous models.22, 23, 24, 25,28
Figure 2.
Polygenic risk score (PRS) distribution in 5581 samples.
A, Polygenic risk score distribution in advanced AMD (n = 2485), early and intermediate AMD (n = 1208), and controls (n = 1888). B, Summary of PRS values in advanced AMD (median PRS = 1.4), early and intermediate disease (median PRS = 0.61), and controls (median PRS = 0.06). Genotypes previously measured by HumanCoreExome arrays were used to calculate the PRS, using 26 genotyped variants known to be associated with AMD.14,22, 23, 24, 25, 26 AMD = age-related macular degeneration; PRS = polygenic risk score based on known variants associated with age-related macular degeneration.
Among the 72 families in this study, there were 26 families that had affected members with relatively low PRS (the average PRS of affected members was < 1), and there were 6 families with PRS < 0. Each family had ≥ 2 family members who were diagnosed with signs of advanced macular degeneration before age < 75 years. We extended our analysis in these 26 families to search for highly penetrant variants that may confer disease risk in some of those families. We hypothesized, a priori, that there were unknown variants carried by affected members of those families, which may be very rare in the general population and could not be identified in previous GWAS. We focused on 95 rare variants with MAF < 0.1% (based on 1000 Genomes and gnomAD cohorts), which were also predicted in protein-altering sequence. We identified 2 rare variants showing penetrance in > 80% of affected members of 2 families including rs757672473 (p.Arg288Cys) in ADAM15 and rs147969213 (p.Leu289Arg) in F2RL2 (Table 1). Both rs757672473 and rs147969213 affect highly conserved amino acid residues (Fig S1, available at www.ophthalmologyscience.org.) and are predicted to be deleterious with PolyPhen2 score = 1 and CADD score of > 20.
Table 1.
Potential Pathogenic Protein-coding Rare Variants Identified in 2 Families
| Gene | dbSNP ID | Chr | Position (hg19) | Function | CADD | PolyPhen2 (Score) | Minor/Major | MAF (gnomAD) | Family | Mean PRS of Affected Members | Number of Affected Members Carrying the Risk Allele (Total Number of Affected Members in the Family) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ADAM15 | rs757672473 | 1 | 155028673 | Missense, p.Arg288Cys | 26.1 | Probably damaging (1) | T/C | 0.0003 | A | 0.47 | 6 (7) |
| F2RL2 | rs147969213 | 5 | 75913666 | Missense, p.Leu289Arg | 24.7 | Probably damaging (1) | G/A | 0.0001 | B | –0.6 | 5 (5) |
ADAM15 = a disintegrin and metalloprotease, metallopeptidase domain 15; CADD = Combined Annotation Dependent Depletion; dbSNP ID = single nucleotide polymorphism database identifier; Chr = chromosome; F2RL2 = coagulation factor II thrombin receptor-like 2; MAF = minor allele frequency; PRS = polygenic risk score based on known variants associated with age-related macular degeneration.
Phenotypic Characterization of ADAM15 and F2RL2 Rare Variants
Table 2 and Figures 3 and 4 display the phenotypic characteristics of the 2 families, each harboring a rare loss-of-function variant in ADAM15 (pedigree A) or F2RL2 (pedigree B) in association with their macular degeneration. In pedigree A, there were 7 affected family members. The mean PRS of the 7 affected family members was 0.47. Six of the 7 individuals carried the ADAM15 rare variant (rs757672473), except for subject III:2. Three of the 6 subjects with atrophy also had neovascularization. Four of the 7 affected members of pedigree A (II:2, III:2, III:3, and III:6), including one without the ADAM15 variant (III:2), carried a rare nonpathogenic variant rs148919174 in fatty acid elongase 4 (ELOVL4) (MAF = 0.0012), which has been reported in 2 Turkish patients with Stargardt disease but was labeled as a nonpathogenic variant in that study.38 Of note, 2 of these family members also have Fuchs' corneal dystrophy. Another member of this family recently examined and not displayed in the table, the daughter of III:1, has advanced AMD with drusen and atrophy diagnosed at age 50, and she also has Fuchs' corneal dystrophy (not yet genotyped).
Table 2.
Demographic, Clinical, and Genetic Characteristics of Pedigrees A and B
| ID | Sex, Ageat LatestRecord | RareGeneticVariant | AMD PRS | Age atDiagnosis orEarliest ClinicalRecord of Disease | CARMS InitialGrade | CARMS LastKnown Gradeand Follow-upTime (yrs) | Baseline Visual Acuity (OD; OS) | Last Known Visual Acuity (OD; OS) | Ocular Phenotypic Description |
|---|---|---|---|---|---|---|---|---|---|
| -- Pedigree A | |||||||||
| II:1 | F, 95 yrs | ADAM15 | 0.90 | 55 yrs | 4, 5B | 4, 5B (16) | 20/80; 20/400 | CF; CF | Macular atrophy with temporal and peripheral drusen OU; area of fibrosis, consistent with NV OS; peripheral patches of atrophy OU |
| II:2 | M, 97 yrs | ADAM15 | 0.87 | 84 yrs | 4, 4 | 5B, 5B (13) | 20/200; 20/80 | CF; CF | Atrophy OU (OD > OS) progressed to NV with scar OU. Nasal and peripheral drusen |
| III:1 | M, 88 yrs | ADAM15 | –0.42 | 58 yrs | 2A, 1 | 4, 5B (29) | 20/15; 20/15 | 20/40; 20/50 | Progression from few drusen to large drusen to drusenoid RPED, then atrophy OD; progression from no/few drusen to large drusen, drusenoid RPED, then NV OS; reticular drusen and peripheral reticular pigment changes |
| III:2 | M, 76 yrs | No | –0.17 | 55 yrs | 3A, 3A | 4, 5B (23) | 20/20; 20/20 | CF; CF | Progression from large drusen to atrophy OD and NV OS. Large drusen around atrophy; peripheral reticular pigment; Fuchs' corneal dystrophy |
| III:3 | M, 64 yrs | ADAM15 | 0.81 | 60 yrs | 3A, 3A | 3B, 3A (3) | 20/20; 20/20 | 20/80; 20/25 | Large drusen OU with progression to drusenoid RPED OD |
| III:4 | F, 82 yrs | ADAM15 | 1.75 | 53 yrs | 3B, 3B | 4, 4 (31) | 20/20; 20/40 | 20/200; 20/400 | Drusenoid RPED OU evolved to atrophy OU; peripheral drusen and reticular pigment changes |
| III:5 | F, 79 yrs | No | –0.25 | 65 yrs | 1, 1 | 2C, 2C (12) | 20/20; 20/20 | 20/30; 20/30 | No AMD to early AMD OU |
| III:6 | M, 83 yrs | ADAM15 | –1.11 | 70 yrs | 2A, 2A | 4, 3A (17) | 20/20; 20/20 | 20/30; 20/25 | Progressed from early AMD to large drusen OU, then atrophy OD; extramacular drusen; Fuchs' corneal dystrophy |
| -- Pedigree B | |||||||||
| II:1 | F, 94 yrs | F2RL2 | –1.07 | 74 yrs | 2A, 2C | 6, 6 (19) | 20/40; 20/50 | CF; CF | Progression from few drusen and minimal pigment irregularities to irregular shaped, circular pattern of atrophy in macula OU |
| II:2 | F, 83 yrs | F2RL2 | –0.27 | 78 yrs | 4, 5B | – | CF; CF | – | OD - central atrophy; OS - atrophy, fibrosis, and pigment mottling. Laser treatment for NV OS |
| II:3 | M, 85 yrs | No | 0.34 | 75 yrs | 2A, 2C | 3A, 3A (10) | 20/20; 20/25 | 20/30; 20/200 | OD - drusen; OS - pigment irregularity and drusen |
| II:5 | F, 83 yrs | No | 0.50 | 83 yrs | 3A, 3A | – | 20/30; 20/25 | 20/30; 20/25 | Drusen and pigment irregularities OU based on medical records |
| II:2-spouse | M, 75 yrs | No | –1.95 | – | 1, 1 | – | 20/40; 20/40 | – | No sign of AMD, no drusen |
| III:1 | F, 72 yrs | F2RL2 | –1.61 | 58 yrs | 3A, 3A | 5B, 3B (9) | 20/20; 20/20 | 20/40; 20/30 | Large drusen with small drusenoid RPED OU, progressed to larger RPED OU and NV OD. Anti-VEGF injections OD |
| III:2 | F, 71 yrs | F2RL2 | –0.98 | 66 yrs | 3A, 4 | 3A, 4 (4) | 20/25; 20/25 | 20/30; 20/30 | Few small drusen OD and patches of atrophy OS |
| III:4 | M, 83 yrs | F2RL2 | –1.10 | 58 yrs | 6, 6 | 6, 6 (20) | 20/30; 20/25 | CF; CF | Drusen and central atrophy with foveal sparing OU |
ADAM15 = a disintegrin and metalloprotease, metallopeptidase domain 15; AMD = age-related macular degeneration; CARMS = Clinical Age-Related Maculopathy Staging30; CF = count fingers visual acuity; F2RL2 = coagulation factor II thrombin receptor-like 2; NV = neovascularization; OD = oculus dexter, right eye; OS = oculus sinister, left eye; OU = oculus uterque, both eyes; PRS = polygenic risk score; RPED = retinal pigment epithelial detachment.
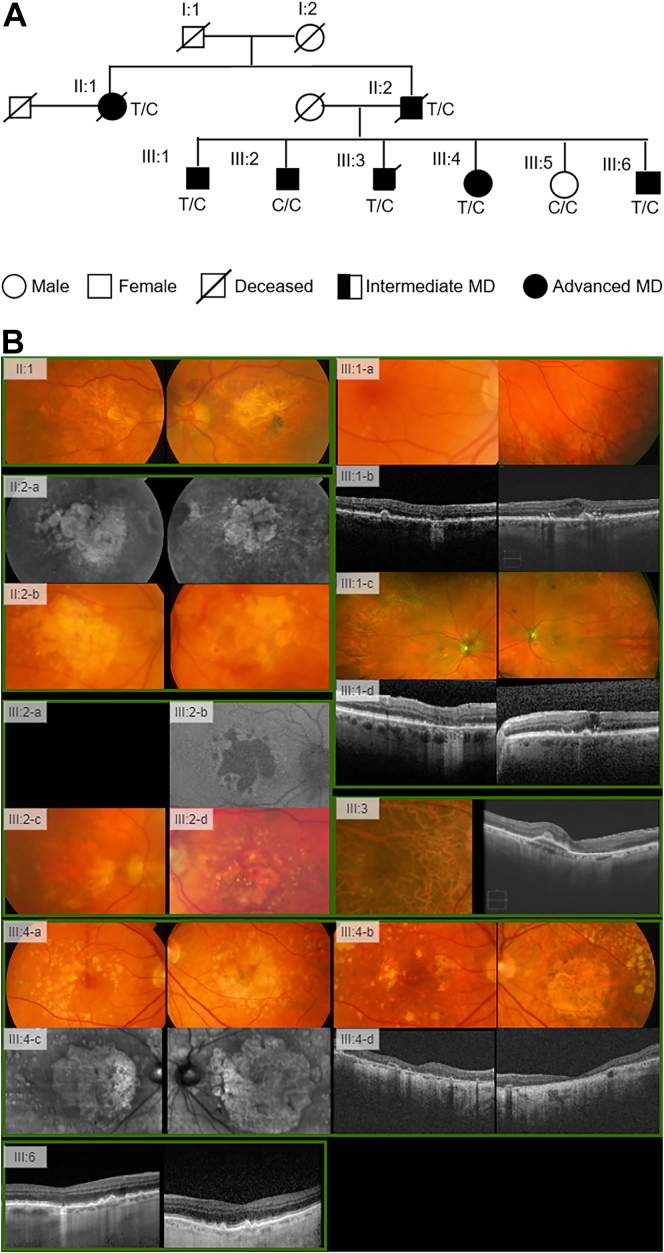
Figure 3.
Pedigree A, carrying the ADAM15 rare variant rs757672473 (p.Arg288Cys).
A, The pedigree diagram. B, Multimodal retinal imaging of subjects in pedigree A.
II:1 had macular atrophy with temporal and peripheral drusen in both eyes (OU) and fibrosis consistent with neovascular age-related macular degeneration (AMD) in the left eye (OS). II:2, brother of II:1, had atrophy OU seen on infrared imaging OU (II:2-a) with progression to neovascular disease (NV) (II:2-b). III:1, son of II:2, had few drusen OU (III:1-a), which progressed to geographic atrophy (GA) OD and NV OS (III:1-b-d). The son, III:2, a noncarrier, had large drusen that progressed to GA in the right eye (OD) and NV OS (III:2-a-d). The son III:3 had large drusen OU with progression to drusenoid retinal pigment epithelial detachment OD. III:4, daughter of II:2, had drusenoid retinal pigment epithelial detachment OU that progressed to GA OU (III:4-a-d). Individual III:6, also son of II:2, progressed from early AMD to large drusen OU and GA OD.
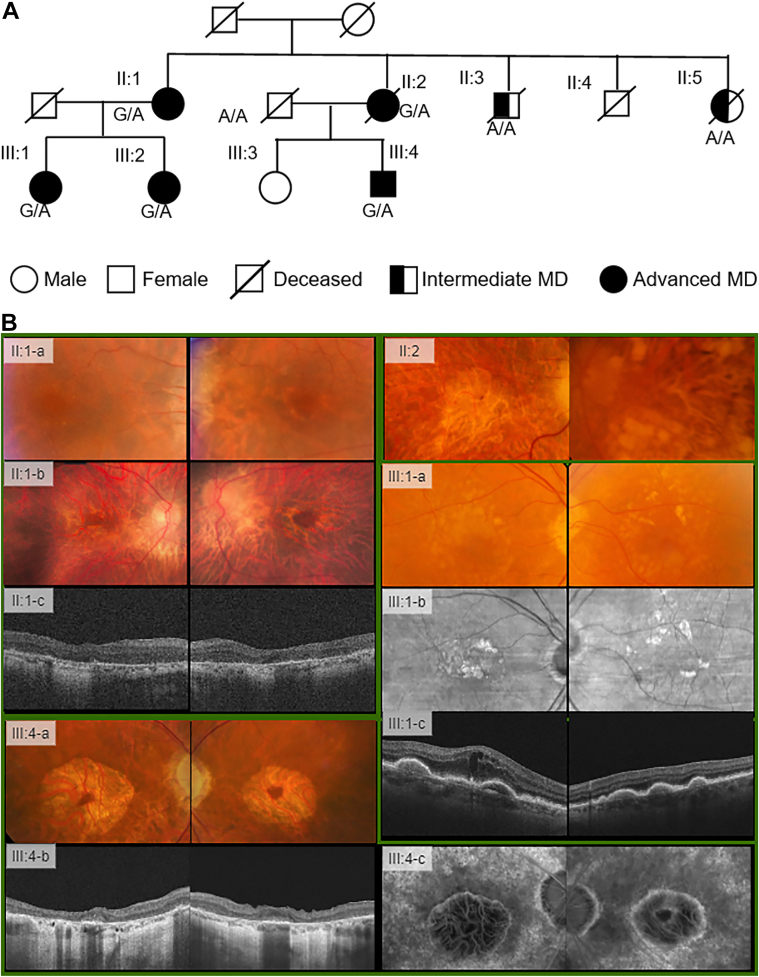
Figure 4.
Pedigree B, carrying the F2RL2 rare variant rs147969213 (p.Leu289Arg).
A, The pedigree diagram. B, Multimodal retinal imaging of subjects in pedigree B.
Individual II:1, a female aged 77 years (II:1-a), with progression to a circular pattern of atrophy shown in fundus color both eyes (OU) 16 years later (II:1-b) and atrophy seen on OCT at the same time (II:1-c). The sister, II:2, had atrophy right eye (OD) and atrophy left eye (OS) with fibrosis and pigment mottling with a history of laser treatment for neovascular disease (NV), seen on fundus color photograph OU at age 82. III:1, the daughter of II:1, had large drusen with drusenoid retinal pigment epithelial detachment seen on fundus color OU at age 65 years (III;1a), which progressed to larger retinal pigment epithelial detachment seen on infrafred imaging OU (III:1-b) and OCT OU, and 3 years later had neovascular disease (NV) OD and received anti-VEGF injections (III:1-c). III-4, the son of individual II:2, had drusen and central geographic atrophy (GA) with foveal sparing OU beginning at age 58 years, seen on fundus photo, OCT, and fluorescein angiography OU at age 82 (III:4-a-c). F2RL2 = coagulation factor II thrombin receptor-like 2.
In pedigree B, there were 5 family members with advanced macular disease. The mean PRS of the 5 affected family members was –0.6. All 5 affected members carried the F2RL2 rare variant (rs147969213), including 2 individuals with neovascular AMD. The other 3 family members with intermediate macular degeneration (n = 2) and no AMD (n = 1) did not have this rare variant. Two family members, II:1 and her nephew, III:4, had atrophic fundus features, which are atypical for AMD.
Localization of F2RL2 Protein in the Retina
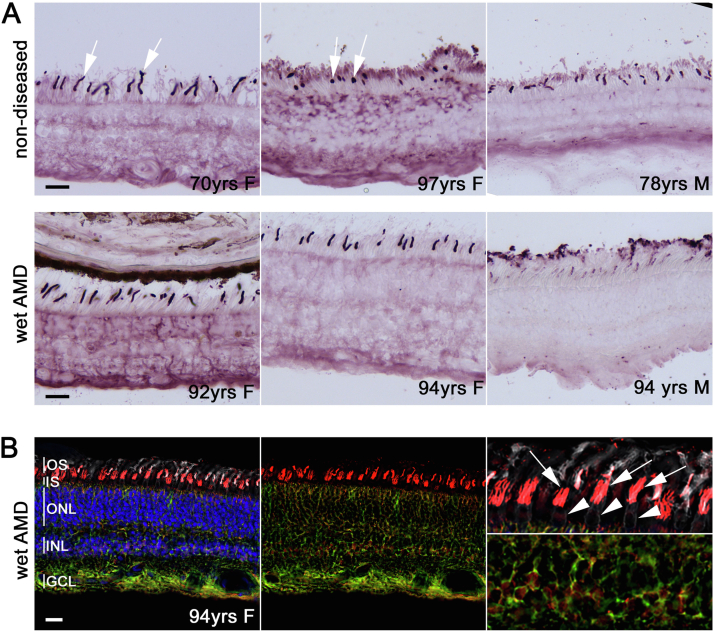
One previous study showed that ADAM15 might promote VEGF-induced ocular neovascularization in mice.39 F2RL2 has not yet been reported to be associated with any retinal disorders. Therefore, we further performed a functional analysis of the novel gene F2RL2. As shown in publicly available repositories of RNA-sequencing data measured in multiple tissues (https://www.proteinatlas.org/ENSG00000164220-F2RL2/tissue), the RNA expression level of F2RL2 is specific in the retina (the scaled tags per million [TPM] is 38.1 in the retina, which is 3 times higher than its expression in the other 59 tissues; Fig S2A, available at www.ophthalmologyscience.org.). Figure S2B shows the distribution of log transformed TPM values for 18 156 genes in 60 tissues from functional annotation of the mammalian genome (FANTOM5).40 The mean TPM value for all genes was 63.3. Therefore, F2RL2 showed a relatively low expression. Single-cell RNA-sequencing showed F2RL2 was expressed in rod photoreceptor cells in human retinas (TPM = 23, Fig S2C).41 To further elucidate F2RL2 function in pathology, we performed immunohistochemistry of F2RL2 in human retinas from 6 eyes from 6 donors (mean age, 88 years, 4 women and 2 men, Fig 5A). Three eyes had no clinical evidence of AMD, and 3 eyes had neovascular AMD. Coagulation factor II thrombin receptor-like 2 protein was enriched in cone photoreceptor outer segments, at the level of the ganglion cell layer where the first vascular plexus is located, and in a diffuse pattern throughout the retina, suggesting a possible expression in Müller glia cells. The expression in cone outer segments seemed to be higher than in other areas (Fig 5A). To confirm the expression in Müller glia cells, we performed fluorescent immunolabeling with an antibody against GS to identify Müller glia cells in addition to F2RL2 expression (Fig 5B). There was a strong colabeling of Müller glia cell processes and F2RL2 expression. The use of peanut agglutinin lectin also confirmed that the outer segment expression of F2RL2 was indeed in cone outer segments. To further confirm the expression, we performed immunofluorescence for F2RL2 and GS on pig retinas (Fig 6A–C). The staining confirmed the predominant expression of F2RL2 in cone outer segments and the relatively weaker expression in Müller glia. Overall, the expression of F2RL2 between diseased and nondiseased individuals did not reveal any disease-specific difference in regions without advanced pathology (Fig 5A).
Figure 5.
F2RL2 expression patterns in human retinas.
A, Superior-oriented peripheral retinas of 3 nondiseased (top row; left to right: 70-year-old woman, 97-year-old woman, and 78-year-old man) and 3 neovascular patients with AMD (bottom row; left to right: 92-year-old women, 94-year-old women, and 94-year-old man) stained for expression of F2RL2 by immunohistochemistry. Expression seems high in a subset of photoreceptor outer segments and intermediate where the first retinal vascular plexus and the ganglion cells are located. A diffuse pattern across the retina resembling Müller glia staining is seen as well. B, Immunofluorescence staining for F2RL2 expression (red signal) in retina of an eye with neovascular AMD (94-year-old woman as shown in panel A). Retina was also stained with an antibody against glutamine synthetase (green signal) to visualize Müller glia cells, peanut agglutinin lectin (white signal) to detect cones, and nuclear 4’,6-diamidino-2-phenylindole (blue signal) to visualize nuclei. The cone outer segment signal of F2RL2 (arrows) can be seen on top of cone inner segments (white arrowheads; top right panel). Yellow signal throughout the retina confirms the expression of F2RL2 in Müller glia cells. Higher magnification of region where Müller glia cell bodies reside is shown in lower right panel. Scale bars: 25 mm. AMD = age-related macular degeneration; F2RL2 = coagulation factor II thrombin receptor-like 2; GCL = ganglion cell layer; INL = inner nuclear layer; IS = inner segments; ONL = outer nuclear layer; OS = outer segments.
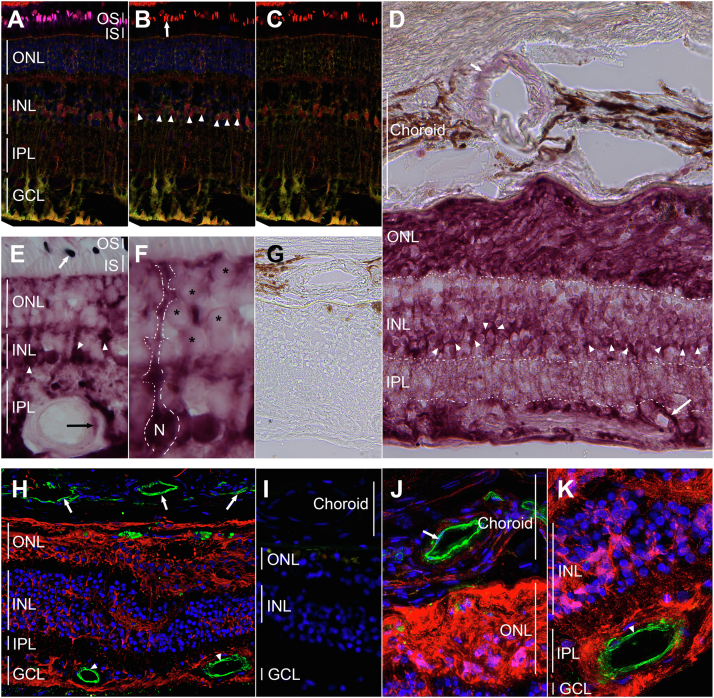
Figure 6.
F2RL2 expression in pig retinas and regions of vascular pathology.
A-C, Immunofluorescence for F2RL2 expression (red signal) in pig retinas. Like in humans, F2RL2 expression is primarily seen in cone outer segments (OS, arrow) and Müller glia cells (green signal). Müller glia cell bodies (arrowheads) show clear expression of F2RL2 (magenta: peanut agglutinin lectin marking cone segments; red: F2RL2; green: glutamine synthetase [GS] marking Müller glia cells; blue: nuclear 4’,6-diamidino-2-phenylindole [DAPI] marking nuclei). A, Shows all 4 colors. B, Shows F2RL2, GS, and DAPI. C, Shows F2RL2 and GS. D-K, Images of immunohistochemistry (D-G) and immunofluorescence (H-K) staining from regions of human retinas with neovascular pathology and fibrotic scars. D, Regions of fibrotic scar in the outer nuclear layer (ONL), where photoreceptors used to reside, show the strongest expression of F2RL2 protein. Expression in Müller glia cell bodies is seen as a band of cells (arrowheads) across the INL. Expression of F2RL2 is also seen around the choroidal and retinal vasculature (arrows). Boundaries of different retinal layers are demarcated by dotted lines or vertical lines (for choroid). E, F, F2RL2 expression outside region of vascular pathology on same retina as shown in (D). Müller glia (arrowheads) cell morphology is discernable by the strong expression of F2RL2. F, Shows higher magnification of a region shown in (E) with dotted line tracing the outlines of a Müller glia cell. Weak expression is also seen in photoreceptor cells (asterisks). Outer segment expression in cones is indicated by arrow. G, Absence of primary antibody control staining shows no signal. Incubation was performed in parallel, omitting the primary antibody. H, Immunofluorescence for F2RL2 (red signal) and PECAM1 (green signal) to determine if F2RL2 is also expressed in retinal (arrowhead) and/or choroidal (arrows) endothelial cells. No apparent overlap is seen between the red and green signal. Fibrotic scar tissue in ONL shows few photoreceptor nuclei (blue: DAPI) left. I, Omission of primary antibody as a control for the secondary antibodies used in (H). J, K, Higher magnification of same staining shown in (H), showing choroidal (J, arrow) and retinal (K, arrowhead) vasculature (green signal) surrounded by F2RL2 but not overlapping with F2RL2 (blue: nuclear DAPI). Human retina in (D-K) is from a 94-year-old woman (same as shown in Fig 5) with neovascular AMD pathology. AMD = age-related macular degeneration; F2RL2 = coagulation factor II thrombin receptor-like 2; GCL = ganglion cell layer; INL = inner nuclear layer; IPL = inner plexiform layer; IS = inner segments; ONL = outer nuclear layer; OS = outer segments; PECAM1 = Platelet Endothelial Cell Adhesion Molecule 1.
To investigate the role of the F2RL2 protein in advanced AMD, we analyzed regions of neovascular pathology (Fig 6D–K). Expression was high in the fibrotic scar tissue that formed around the remaining photoreceptor nuclei in eyes with neovascular AMD that had retinal fluid. Choroidal and retinal vasculatures also showed some F2RL2 positive signal by immunohistochemistry. To determine if F2RL2 was expressed in endothelial cells, we performed double immunofluorescence with an antibody against PECAM1 to identify endothelial cells and F2RL2. Although F2RL2 protein did surround vascular cells of choroidal and retinal origin, it did not overlap with PECAM1, suggesting that it is not expressed in endothelial cells, or its expression is below the detectable levels of the assay. Overall, expression of F2RL2 appeared to correlate with increased formation of fibrotic tissue, in particular in regions of neovascular pathology, suggesting a potential role of F2RL2 in the pathogenesis of advanced macular degeneration.
Discussion
The PRS was highly related to the stage of AMD, with the highest scores in individuals with advanced disease. Such scores have been used in other diseases to inform clinical diagnoses and decisions and for risk stratification. By selecting families with low PRS and performing genetic association analysis of genes involved in the inflammation, coagulation, or complement pathways, we confirmed a common synonymous variant in CFH in the overall cohort, and in families with low PRS and various forms of macular degeneration, we identified 2 new rare missense variants in ADAM15 and F2RL2. Our functional studies have further implicated the potential roles of the novel gene F2RL2 in the pathogenesis of macular degeneration.
A large GWAS of AMD confirmed the previously reported associations and identified some new variants in 52 independent SNPs (45 common and 7 rare) in 43 loci in association with AMD in 16 144 patients and 17 832 controls using the HumanCoreExome array platform (i.e., exome chip) and 1000 Genome-imputed genotypes.13 Other large GWAS also used this platform in 4332 cases and 25 268 controls and identified protective coding variants and new loci.14 The array-based platform is limited to discovery of variants that only reside in the platform. The imputed genotypes of rare variants (MAF < 1%) are uncertain. Extremely rare penetrant variants (MAF < 0.1%) can only be detected by sequencing. To overcome this limitation, some studies implemented a sequencing-based platform, such as WES. However, because of the limitation of small sample size, some WES studies limited their discovery only to known AMD-associated loci.11,12,42,43 Few published WES of AMD reported variants in new loci, beyond the known AMD loci.
Because the complement pathway is known to play a key role in AMD, we prioritized our discovery to function-altering regions in 150 targeted genes in related pathways including inflammation and coagulation. A unique advantage of our study is that we selected families whose affected members had a low burden of known AMD risk variants, which indicated that there could be undiscovered rare variants. In this analysis, we identified 2 highly penetrant variants in 2 families, rs757672473 (p.Arg288Cys) in ADAM15 (pedigree A) and rs147969213 (p.Leu289Arg) in F2RL2 (pedigree B). In each pedigree, > 80% of the sequenced affected subjects carried the risk allele. The variants were not found in any of the controls. We further confirmed that members of the 2 families did not carry known pathogenic or likely pathogenic rare variants in other maculopathies. In pedigree A, 4 of the 7 subjects (II:2, III:2, III:3, III:6) with advanced macular disease also carried the risk allele for variant rs148919174 (p.Glu272Gln in ELOVL4), which was reported in 2 Turkish patients with Stargardt disease but was reported as a nonpathogenic variant in the same research.38
Phenotypes of some members of pedigree B resemble both macular dystrophy with atrophy and advanced atrophic macular degeneration. Phenotypes of macular dystrophies can resemble those of AMD. Late-onset Stargardt disease, as well as central areolar choroidal dystrophy and other macular dystrophies, can appear as central atrophy similar to the atrophic stage of advanced AMD. For example, case III:4 in pedigree B resembles a macular dystrophy but did not have variants in ATP binding cassette subfamily A member 4, peripherin 2, or other macular dystrophy genes.
It is very challenging to validate these extremely rare variants in independent studies. In our cohort, the risk alleles for the 2 rare variants rs757672473 (ADAM15) and rs147969213 (F2RL2) did not appear in other families in our cohort. Further validation of these 2 rare variants in independent genetic studies is needed to confirm the findings. Given the limitations in validating those 2 extremely rare variants associated with AMD in independent genetic studies, we explored the functional correlations of ADAM15 and F2RL2 with macular degeneration. The protein coded by ADAM15 is a member of the ADAM (a disintegrin and metalloproteinase) protein family, which can mediate inflammation and has been found to be involved in the pathogenesis of many inflammatory diseases.44 Several members of the ADAM family, such as ADAM8, ADAM9, ADAM15, and ADAM17, have also been found to play critical and distinct roles in pathologic neovascularization. Absence of ADAM9, ADAM15, or ADAM17 reduces revascularization of the retinal capillary bed in mice.45 Xie et al39 reported that ADAM15 might promote VEGF-induced ocular neovascularization. It is possible that ADAM15 is involved in ischemia-induced retinal neovascularization that may be related to retinal pathology. The rare missense variant rs757672473 (p.Arg288Cys) resides in the reprolysin domain (Fig S3, available at www.ophthalmologyscience.org.) and was predicted as deleterious by CADD and PolyPhen2. The amino acid changes from the positively charged Arg to Cys within the metalloproteinase domain of ADAM15 may alter its peptide structure and its proteinase function.
In the ADAM15 rare variant family, 3 family members (1 carrier, 1 noncarrier, and 1 not yet genotyped) had Fuchs' dystrophy, which is an incurable and inherited condition affecting the cornea. Previous research has shown that Fuchs' dystrophy and AMD share some interesting similarities. Risk factors such as advanced age, female sex, and smoking are common in both diseases. One article explored the association between Fuchs' and drusen because, in Fuchs' there is deposition of extracellular matrix in the form of guttae, and in AMD, there is deposition of material in the form of drusen. However, no association was found between Fuchs' and drusen in that study of 213 cases and 181 controls with normal corneas based on OCT.46 The genetics of classic late-onset Fuchs' dystrophy is complex and multifactorial. Genetic variants in ZEB1, SLC4A11, and LOXHD1 have been reported in association with the late-onset form, but each variant has a very small effect. An early-onset form of Fuchs' dystrophy that occurs in the first decades of life is caused by rare mutations (MAF < 0.01) in the COL8A2 gene.47 Common variants (MAF > 0.05) in COL8A1 and COL4A3 are associated with AMD.7,13,14,26 COL8A1 and COL8A2 encode 2 subunits of collagen VIII, and COL4A3 encodes a subunit of collagen IV in the extracellular matrix pathway. There may be a shared dysregulation of the extracellular matrix pathway underlying the pathogenesis of AMD and Fuchs' dystrophy. Although Fuchs' dystrophy and AMD occurred in the same family as reported here, a common genetic origin of Fuchs' dystrophy and AMD has not been established, and further research is needed to explain the relationship between these 2 diseases.
Coagulation factor II thrombin receptor-like 2, also known as Proteinase-activated receptor 3, is a 7-transmembrane G-protein coupled receptor. The rare missense variant rs147969213 (p.Leu289Arg) resides in the cytoplasmic nontransmembrane domain, is highly conserved, and is predicted as probably deleterious (PolyPhen2 score = 1, CADD > 20). To our knowledge, there is no previous study showing the involvement of F2RL2 in a retinal disease. We performed immunohistochemistry in donor retinas from 3 eyes of patients with neovascular AMD and 3 eyes of controls and showed that F2RL2 expression is enriched in cone outer segments, Müller glia cells, and endothelial cells. The increased expression in astrocytes during gliosis and its enrichment in the inner lumen of large blood vessels in neovascular regions of the retina suggest a potential role of F2RL2 in the regulation of leakage from neovascular blood vessels. Data suggest that F2RL2 might play an important role in the retina and in retinal disease. It remains to be determined if its expression changes in the retinas of patients with diabetic retinopathy. Because the variants identified presumably cause a loss of function of F2RL2, its increased expression in pathologic blood vessels might play an important protective role during the pathogenesis of AMD.
The gene-based analysis suggested that ENG was associated with maculopathy in our study cohort. The ENG gene provides instructions for making a protein called ENG. Endoglin is a type I membrane glycoprotein and is part of the transforming growth factor-β receptor complex. This protein is found on the surface of cells, especially in the lining of developing arteries, and is involved in the development of blood vessels. In particular, this complex is involved in the specialization of new blood vessels into arteries or veins. Endoglin has been implicated as being involved in epithelial-mesenchymal transition or fibrosis and is expressed in endothelial cells in choroidal neovascular membranes.36 Based on previous findings, we speculate that ENG might be involved in the pathogenesis of macular degeneration by promoting new blood vessels, fibrosis, or other functions. Additional studies are needed to further explore associations between this gene and maculopathies.
In conclusion, we confirmed the value of a PRS to indicate the likelihood of AMD and its severity and to select families with lower PRS who may carry novel rare variants. We confirmed a known common CFH variant in the overall cohort, and in selected families, we detected 2 novel rare variants in ADAM15 and F2RL2 associated with maculopathy. We also showed that F2RL2 protein expression occurred in regions with neovascular pathology, suggesting a role for this F2RL2 variant in the immune and coagulation pathways in AMD pathogenesis. Our study also suggests that by performing genetic association analysis and DNA sequencing in maculopathy families with a low burden of known genetic variants, we may find new genes and variants associated with AMD or other forms of macular degeneration. Novel genes and rare variants can provide targets for new therapies.
Manuscript no. XOPS-D-22-00031R1.
Footnotes
Supplemental material available atwww.ophthalmologyscience.org.
Presented at the Association for Research in Vision & Ophthalmology 2021 Annual Meeting, May 2-6, 2021, San Francisco, California.
Disclosure(s):
All authors have completed and submitted the ICMJE disclosures form.
The authors made the following disclosures: J.M.S.: Stock options – Gemini Therapeutics and Apellis Pharmaceuticals; Consultant – Laboratoires Théa.
Supported by NIH R01-EY011309, R01-EY028602, R01-EY023570; Bethesda, MD; American Macular Degeneration Foundation, Northampton, MA; Macular Degeneration Research Fund, University of Massachusetts Medical School, Department of Ophthalmology and Visual Sciences, Worcester, MA. The sponsor or funding organization had no role in the design or conduct of this research.
HUMAN SUBJECTS: Human subjects were included in this study. Informed Consent was obtained from all participants. Institutional review board approval was obtained. All research adhered to the tenets of the Declaration of Helsinki. All information presented in this study is HIPAA-compliant.
Non-human animals were used in this study. Protocol was approved by the IACUC of the University of Massachusetts.
Author Contributions:
Conception and design: Huan, Daly, Seddon
Data collection: Huan, Cheng, Tian, Punzo, Seddon
Analysis and interpretation: Huan, Cheng, Punzo, Lin, Daly, Seddon
Obtained funding: Seddon
Overall responsibility: Huan, Punzo, Daly, Seddon
Supplementary Data
References
- 1.Seddon J.M. Macular degeneration epidemiology: nature-nurture, lifestyle factors, genetic risk, and gene-environment interactions–the Weisenfeld Award Lecture. Invest Ophthalmol Vis Sci. 2017;58:6513–6528. doi: 10.1167/iovs.17-23544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saksens N.T., Fleckenstein M., Schmitz-Valckenberg S., et al. Macular dystrophies mimicking age-related macular degeneration. Prog Retin Eye Res. 2014;39:23–57. doi: 10.1016/j.preteyeres.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Klein R.J., Zeiss C., Chew E.Y., et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maller J., George S., Purcell S., et al. Common variation in three genes, including a noncoding variant in CFH, strongly influences risk of age-related macular degeneration. Nat Genet. 2006;38:1055–1059. doi: 10.1038/ng1873. [DOI] [PubMed] [Google Scholar]
- 5.Maller J.B., Fagerness J.A., Reynolds R.C., et al. Variation in complement factor 3 is associated with risk of age-related macular degeneration. Nat Genet. 2007;39:1200–1201. doi: 10.1038/ng2131. [DOI] [PubMed] [Google Scholar]
- 6.Fagerness J.A., Maller J.B., Neale B.M., et al. Variation near complement factor I is associated with risk of advanced AMD. Eur J Hum Genet. 2009;17:100–104. doi: 10.1038/ejhg.2008.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neale B.M., Fagerness J., Reynolds R., et al. Genome-wide association study of advanced age-related macular degeneration identifies a role of the hepatic lipase gene (LIPC) Proc Natl Acad Sci U S A. 2010;107:7395–7400. doi: 10.1073/pnas.0912019107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raychaudhuri S., Iartchouk O., Chin K., et al. A rare penetrant mutation in CFH confers high risk of age-related macular degeneration. Nat Genet. 2011;43:1232–1236. doi: 10.1038/ng.976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seddon J.M., Yu Y., Miller E.C., et al. Rare variants in CFI, C3 and C9 are associated with high risk of advanced age-related macular degeneration. Nat Genet. 2013;45:1366–1370. doi: 10.1038/ng.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fritsche L.G., Chen W., Schu M., et al. Seven new loci associated with age-related macular degeneration. Nat Genet. 2013;45 doi: 10.1038/ng.2578. 433–439.e1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu Y., Triebwasser M.P., Wong E.K., et al. Whole-exome sequencing identifies rare, functional CFH variants in families with macular degeneration. Hum Mol Genet. 2014;23:5283–5293. doi: 10.1093/hmg/ddu226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wagner E.K., Raychaudhuri S., Villalonga M.B., et al. Mapping rare, deleterious mutations in factor H: association with early onset, drusen burden, and lower antigenic levels in familial AMD. Sci Rep. 2016;6 doi: 10.1038/srep31531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fritsche L.G., Igl W., Bailey J.N., et al. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat Genet. 2016;48:134–143. doi: 10.1038/ng.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu Y., Wagner E.K., Souied E.H., et al. Protective coding variants in CFH and PELI3 and a variant near CTRB1 are associated with age-related macular degeneration. Hum Mol Genet. 2016;25:5276–5285. doi: 10.1093/hmg/ddw336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ambati J., Atkinson J.P., Gelfand B.D. Immunology of age-related macular degeneration. Nat Rev Immunol. 2013;13:438–451. doi: 10.1038/nri3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Triebwasser M.P., Roberson E.D., Yu Y., et al. Rare variants in the functional domains of complement factor H are associated with age-related macular degeneration. Invest Ophthalmol Vis Sci. 2015;56:6873–6878. doi: 10.1167/iovs.15-17432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kavanagh D., Yu Y., Schramm E.C., et al. Rare genetic variants in the CFI gene are associated with advanced age-related macular degeneration and commonly result in reduced serum factor I levels. Hum Mol Genet. 2015;24:3861–3870. doi: 10.1093/hmg/ddv091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Java A., Baciu P., Widjajahakim R., et al. Functional analysis of rare genetic variants in complement factor I (CFI) using a serum-based assay in advanced age-related macular degeneration. Transl Vis Sci Technol. 2020;9 doi: 10.1167/tvst.9.9.37. 37–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McMahon O., Hallam T.M., Patel S., et al. The rare C9 P167S risk variant for age-related macular degeneration increases polymerization of the terminal component of the complement cascade. Hum Mol Genet. 2021;30:1188–1199. doi: 10.1093/hmg/ddab086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Java A., Nicola P., Schroeder M.C., et al. Functional analysis of rare genetic variants in complement factor I in advanced age-related macular degeneration. Hum Mol Genet. 2022 doi: 10.1093/hmg/ddac103. Advance access publication date: May 9, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sobrin L., Maller J.B., Neale B.M., et al. Genetic profile for five common variants associated with age-related macular degeneration in densely affected families: a novel analytic approach. Eur J Hum Genet. 2010;18:496–501. doi: 10.1038/ejhg.2009.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seddon J.M., Reynolds R., Maller J., et al. Prediction model for prevalence and incidence of advanced age-related macular degeneration based on genetic, demographic, and environmental variables. Invest Ophthalmol Vis Sci. 2009;50:2044–2053. doi: 10.1167/iovs.08-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seddon J.M., Reynolds R., Yu Y., Rosner B. Three new genetic loci (R1210C in CFH, variants in COL8A1 and RAD51B) are independently related to progression to advanced macular degeneration. PLoS One. 2014;9 doi: 10.1371/journal.pone.0087047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seddon J.M., Silver R.E., Kwong M., Rosner B. Risk prediction for progression of macular degeneration: 10 common and rare genetic variants, demographic, environmental, and macular covariates. Invest Ophthalmol Vis Sci. 2015;56:2192–2202. doi: 10.1167/iovs.14-15841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seddon J.M., Rosner B. Validated prediction models for macular degeneration progression and predictors of visual acuity loss identify high-risk individuals. Am J Ophthalmol. 2019;198:223–261. doi: 10.1016/j.ajo.2018.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu Y., Bhangale T.R., Fagerness J., et al. Common variants near FRK/COL10A1 and VEGFA are associated with advanced age-related macular degeneration. Hum Mol Genet. 2011;20:3699–3709. doi: 10.1093/hmg/ddr270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferrara D., Seddon J.M. Phenotypic characterization of complement factor H R1210C rare genetic variant in age-related macular degeneration. JAMA Ophthalmol. 2015;133:785–791. doi: 10.1001/jamaophthalmol.2015.0814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seddon J.M., Widjajahakim R., Rosner B. Rare and common genetic variants, smoking, and body mass index: progression and earlier age of developing advanced age-related macular degeneration. Invest Ophthalmol Vis Sci. 2020;61 doi: 10.1167/iovs.61.14.32. 32–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oikonomopoulou K., Ricklin D., Ward P.A., Lambris J.D. Interactions between coagulation and complement—their role in inflammation. Semin Immunopathol. 2012;34:151–165. doi: 10.1007/s00281-011-0280-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seddon J.M., Sharma S., Adelman R.A. Evaluation of the clinical age-related maculopathy staging system. Ophthalmology. 2006;113:260–266. doi: 10.1016/j.ophtha.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 31.Cingolani P., Platts A., Wang L.L., et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rentzsch P., Witten D., Cooper G.M., et al. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47:D886–D894. doi: 10.1093/nar/gky1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adzhubei I.A., Schmidt S., Peshkin L., et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seddon J.M., McLeod D.S., Bhutto I.A., et al. Histopathological insights into choroidal vascular loss in clinically documented cases of age-related macular degeneration. JAMA Ophthalmol. 2016;134:1272–1280. doi: 10.1001/jamaophthalmol.2016.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lutty G.A., McLeod D.S., Bhutto I.A., et al. Choriocapillaris dropout in early age-related macular degeneration. Exp Eye Res. 2020;192 doi: 10.1016/j.exer.2020.107939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng S.Y., Cipi J., Ma S., et al. Altered photoreceptor metabolism in mouse causes late stage age-related macular degeneration-like pathologies. Proc Natl Acad Sci U S A. 2020;117:13094–13104. doi: 10.1073/pnas.2000339117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grisanti S., Canbek S., Kaiserling E., et al. Expression of endoglin in choroidal neovascularization. Exp Eye Res. 2004;78:207–213. doi: 10.1016/j.exer.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 38.Bardak H., Gunay M., Ercalik Y., et al. Analysis of ELOVL4 and PRPH2 genes in Turkish Stargardt disease patients. Genet Mol Res. 2016;15 doi: 10.4238/gmr15048774. [DOI] [PubMed] [Google Scholar]
- 39.Xie B., Shen J., Dong A., et al. An Adam15 amplification loop promotes vascular endothelial growth factor-induced ocular neovascularization. FASEB J. 2008;22:2775–2783. doi: 10.1096/fj.07-099283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Noguchi S., Arakawa T., Fukuda S., et al. FANTOM5 CAGE profiles of human and mouse samples. Sci Data. 2017;4 doi: 10.1038/sdata.2017.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Menon M., Mohammadi S., Davila-Velderrain J., et al. Single-cell transcriptomic atlas of the human retina identifies cell types associated with age-related macular degeneration. Nat Commun. 2019;10:4902. doi: 10.1038/s41467-019-12780-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ratnapriya R., Acar İ.E., Geerlings M.J., et al. Family-based exome sequencing identifies rare coding variants in age-related macular degeneration. Hum Mol Genet. 2020;29:2022–2034. doi: 10.1093/hmg/ddaa057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Corominas J., Colijn J.M., Geerlings M.J., et al. Whole-exome sequencing in age-related macular degeneration identifies rare variants in COL8A1, a component of Bruch’s membrane. Ophthalmology. 2018;125:1433–1443. doi: 10.1016/j.ophtha.2018.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Charrier-Hisamuddin L., Laboisse C.L., Merlin D. ADAM-15: a metalloprotease that mediates inflammation. FASEB J. 2008;22:641–653. doi: 10.1096/fj.07-8876rev. [DOI] [PubMed] [Google Scholar]
- 45.Maretzky T., Blobel C.P., Guaiquil V. Characterization of oxygen-induced retinopathy in mice carrying an inactivating point mutation in the catalytic site of ADAM15. Invest Ophthalmol Vis Sci. 2014;55:6774–6782. doi: 10.1167/iovs.14-14472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matthaei M., Elsner E., Caramoy A., et al. Fuchs endothelial corneal dystrophy and macular drusen: evidence for coincidence? Eye (Lond) 2018;32:840–841. doi: 10.1038/eye.2017.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gottsch J.D., Sundin O.H., Liu S.H., et al. Inheritance of a novel COL8A2 mutation defines a distinct early-onset subtype of Fuchs corneal dystrophy. Invest Ophthalmol Vis Sci. 2005;46:1934–1939. doi: 10.1167/iovs.04-0937. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.