Abstract
The triple-helix structure at the 3′ end of metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), a long non-coding RNA, has been considered to be a target for modulating the oncogenic functions of MALAT1. This study examines the binding of quercetin—a known triplex binding molecule—to the MALAT1 triplex. By employing UV-visible spectroscopy, circular dichroism spectroscopy, and isothermal titration calorimetry, we observed that quercetin binds to the MALAT1 triplex with a stoichiometry of 1:1 and Kd of 495 ± 61 nM, along with a negative change in free energy, indicating a spontaneous interaction. Employing real-time PCR measurements, we observed around 50% downregulation of MALAT1 transcript levels in MCF7 cells, and fluorescence in situ hybridization (FISH) experiments showed concomitantly reduced levels of MALAT1 in nuclear speckles. This interaction is likely a result of a direct interaction between the molecule and the RNA, as indicated by a transcription-stop experiment. Further, transcriptome-wide analysis of alternative splicing changes induced by quercetin revealed modulation of MALAT1 downstream genes. Collectively, our study shows that quercetin strongly binds to the MALAT1 triplex and modulates its functions. It can thus be used as a scaffold for further development of therapeutics or as a chemical tool to understand MALAT1 functions.
Keywords: MT: Oligonucleotides: Therapies and Applications, MALAT1, triple helix, quercetin, small-molecule binding, isothermal titration calorimetry, in silico docking, RNA FISH
Graphical abstract

The small molecule quercetin and MALAT1 RNA 3′ triplex structure interaction indicates major groove binding. A physical interaction is predicted by in silico docking, biophysics, and a transcription-stop assay. The interaction decreases MALAT1 levels in cellulo, as seen by real-time PCR and RNA FISH. Decreased MALAT1 affects alternative splicing, as demonstrated by RNA sequencing.
Introduction
Less than 2% of the human genome codes for proteins.1 However, we now know that transcription is pervasive across the genome and leads to a wide array of functional coding and non-coding transcripts regulated by cellular states.2 In the class of non-protein-coding RNA transcripts, long non-coding RNAs (lncRNAs) comprise the largest subset and are characterized by having a length of 200 nucleotides or more. Although there are close to 100,000 lncRNA transcripts encoded by the human genome, only a few have been functionally characterized.3 lncRNAs are generally poorly conserved among species at the sequence level, are expressed at low levels in cells, and are heterogeneous in function and expression pattern.4 They may be encoded in gene bodies or gene deserts and be transcribed from intergenic locations such as introns, exons, or enhancer regions.5, 6, 7 This variability and lack of sequence conservation seen among lncRNAs suggests a role for lncRNAs that may be sequence independent.4 From the ones already annotated, many lncRNAs are reported to undergo mRNA-like processing, in that they are transcribed by RNA polymerase II (Pol II), undergo 5′ end capping, are polyadenylated, and are even spliced. Several lncRNAs are known to play important roles in functions like regulation of RNA and proteins, transcriptional modulation, modification of chromatin structure, epigenetic regulation, organization of nuclear domains, gene imprinting, dosage compensation, and splicing regulation.8, 9, 10, 11, 12, 13, 14 However, despite the large number of lncRNAs that have been reported in the literature, very little is known so far about their structure-function relationships and their effects in modulating cellular states. lncRNAs interact with proteins not just through their primary sequence, but also through secondary and tertiary structural motifs. An increasing number of studies point toward the importance of RNA structure as a critical regulator of function, and such structures might be formed both locally (within the RNA) and with other intracellular molecules.15, 16, 17, 18
One such lncRNA, MALAT1 (metastasis-associated lung adenocarcinoma transcript 1), has an expression that is dysregulated in many cancers.19, 20, 21 High MALAT1 levels in patients also confer resistance to chemotherapy and radiotherapy.22,23 Among certain cancers, such as human breast cancer, it is upregulated and its expression leads to poor relapse-free survival.24 A role for MALAT1 has also been shown in pancreatic cancer, where high MALAT1 levels are found in metastasized cells compared with localized tumors. Similarly, high MALAT1 levels in human pancreatic ductal adenocarcinoma tissues are correlated with low patient survival.25 In gall bladder cancer, high levels of MALAT1 are shown to be correlated with larger tumor size, lymphatic metastasis, and poor patient survival rate.26 Similar associations have been reported for MALAT1 in renal cell carcinomas, bladder cancer, ovarian cancer, and a number of other cancer types.27, 28, 29 Furthermore, it has also been shown that targeting MALAT1 using antisense oligonucleotides (ASOs) leads to reduced metastasis in mouse lung cancer and the development of fewer tumor nodules after primary tumor implantation. This indicates that the targeting of MALAT1 can have potential therapeutic aspects.30 Among its other functions, MALAT1 in nuclear speckles alters the phosphorylation of SR (serine/ arginine-rich) proteins and modulates the cellular distribution and activity of SR splicing factors, thereby influencing alternative splicing of pre-mRNAs.31
MALAT1 is not polyadenylated, yet is transcribed at very high levels inside the nucleus by RNA Pol II.1 MALAT1 owes its nuclear stability to the triple-helix motif at its 3′ end, which sequesters the end from recognition and degradation by 3′–5′ exonucleases, which in turn leads to a higher half-life of the lncRNA.32,33 A U-rich hairpin motif termed element for nuclear expression (ENE) (which was later deemed a misnomer), was reported to modulate the stability of the lncRNA. The ENE, in reality, sequesters a genomically encoded A-rich tract (from the 3′ end of the lncRNA) to form the triple helix, which in turn protects the lncRNA from degradation.34,35 This triple helix is flanked on both ends by stem-loop structures named the P1 helix and the P2 apical helix.
Targeting lncRNAs and modulating their function with small molecules has not been well studied, primarily because of limited understanding of lncRNA structures. Interestingly, for MALAT1, the blunt-ended triplex structure formed at its 3′ end is well characterized. Brown et al. deciphered the structure of the MALAT1 triplex region of the MALAT1 3′ end at a 3.1 Å resolution and implicated it in counteracting the rapid phase of RNA decay.36 The structure was described as an ENE (U-rich hairpin) structure, with an A-rich tract sequestered within (ENE + A). The MALAT1 transcript is not polyadenylated; rather, its A-rich tract is genomically encoded. A bipartite triple helix (that is, two shorter U⋅A-U triple helices) is formed with U⋅A-U triplets separated by an intervening C+⋅G-C triplet and a C-G doublet. The C-G doublet maintains the alignment between the Hoogsteen and the Watson-Crick strand. Similar triple-helical regions have also been observed in some viral RNAs, but with a lower number of triples and an absence of the intervening doublet in their triple-helical region. Despite these differences, there also exist structural similarities in the triplex between viral and vertebrate RNA. Up to six consecutive base triplets have been observed in nature, in contrast to 11 base triplets when chemically synthesized.37 The MALAT1 triplex has recently been characterized using several biophysical techniques.38,39 It is proposed that, since the triplex structure formed in MALAT1 provides stability to the RNA, selective inhibition of this triple helix may lead to a reduction in MALAT1 levels for better prognosis in cancer. A small molecule binding stably to a triplex may also preclude binding of other factors that stabilize the triple helix, leading to its opening up and affecting the degradation of the lncRNA.
The triple helix is a structural motif that is susceptible to targeting with small molecules. A recent report published by Abulwerdi et al. shows the use of a small-molecule microarray of more than 26,000 molecules to identify two molecules that cause a decrease in MALAT1 RNA levels.40 The identified molecules were shown to physically bind with the MALAT1 triplex with unique modes of interaction. Two reports by Donlic et al. also demonstrated the screening of small molecules based on the diphenylfuran scaffold by using molecular docking to determine selectivity toward the triple helix.41,42 These studies revealed that rod-shaped ligands have the maximum specificity for the triplex structure. Using RNaseR exonuclease assays, they also showed that these molecules protect MALAT1 from exonucleolytic degradation.41
However, detailed understanding of the interaction of small molecules with the MALAT1 triplex has been lacking, particularly with respect to the complete biophysical characterization of such interactions. In this study, we have characterized a known triplex-binding small molecule, quercetin, for its ability to bind to the MALAT1 triple-helix structure. Quercetin is a flavonoid (subgroup flavonol) that occurs naturally in plants, many of which form a part of our diet. It is reported to be an antioxidant,43 with anti-inflammatory and anticarcinogenic properties.44 It has a generally recognized as safe (GRAS) status and is safe for animal and human consumption in low doses when taken in purified form.45 Further, quercetin has been reported to bind the RNA U-A-U triplex.46 However, there is no report that describes the binding of quercetin to a naturally existing triplex that is biologically relevant, like that of MALAT1. We have studied the interaction between quercetin and the MALAT1 triplex using different biophysical experiments. We have also checked the levels of MALAT1 upon treatment with quercetin in cells and in nuclear speckles by quantitative real-time PCR and small-molecule fluorescence in situ hybridization (FISH) techniques, respectively. Further, we have studied the effect on the splicing of genes upon MALAT1 perturbation by quercetin. Taken together, our study reveals that quercetin binds the MALAT1 triplex selectively and modulates its biological functions.
Results
CD spectroscopy and UV melting studies on the MALAT1 triplex motif
We employed circular dichroism (CD) to assess the structural aspect of triplex formation using the full-length 94-nt wild-type sequence. We also used a truncated 73-nt sequence as a control in our study, where 21 nt of the A-rich strand at the 3′ end of the sequence have been omitted so that the canonical triplex structure cannot form (Figures 1A and 1B). CD spectra of both oligonucleotides were measured at 25°C at pH 7. The 94-nt as well as the control 73-nt structures exhibit the typical spectra of a nucleic acid duplex in the “A” conformation; that is, the positive band at around ∼260 nm has a magnitude similar to that of the negative band at ∼210 nm. In addition to these peaks, a weak positive peak at around 219 nm and a negative peak at ∼243 nm were seen for the 94-nt sequence, whereas a weak negative peak at ∼243 nm was observed for the 73-nt sequence. The 94-nt sequence showed a larger magnitude band at 210 nm compared with the 73-nt, which may be characteristic of triple-helix formation (Figure 2A).
Figure 1.
MALAT1 triplex sequence and control sequence along with the chemical structure of quercetin
(A) Structure of the last 94 bp at the 3′ end of the MALAT1 RNA (complete triplex region). Nucleotide base numbers are marked. (B) The 73-nt control sequence with missing second strand, which does not form the triple helix. (C) The chemical structure of quercetin.
Figure 2.
CD signatures between MALAT1 triplex and control non-triplex-containing sequences are different
(A) CD spectra of MALAT1 triplex (blue) and control duplex (red), both at 2 μM, shows a pronounced triplex-forming signature in the case of the 94-nt triplex. (B) Thermal melting profiles of MALAT1 triplex (blue) and control duplex (red) show characteristic melting temperatures of the MALAT1 triplex. (C) First derivative curves of the raw absorbance signal of MALAT1 triplex (blue) and control duplex (red) along with fitted curves depict different melting temperatures of the respective structural components. Experiments were performed three times in 10 mM sodium cacodylate buffer (pH 7) in the presence of 100 mM NaCl and 0.5 mM MgCl2. The plotted curves are representative of the same.
UV melting experiments were conducted at pH 7.0 and were used to characterize the helix-coil transition of the structured RNAs under these conditions. Figure 2B shows typical melting curves for both oligonucleotides. Melting curves for both follow the characteristic sigmoidal behavior for the unfolding of a nucleic acid helix. Reversible melting and refolding profiles were obtained for both oligonucleotides without any hysteresis. UV melting profiles for both oligonucleotides show two transitions, a broad transition at lower temperature and a sharp transition at higher temperature. Plots of the first derivative of the absorbance at 260 nm with respect to temperature are presented in Figure 2C. The derivative curve for the 94-nt oligonucleotide indicated that there were two resolved transitions with peaks at 58°C and a high-temperature transition at 78°C (Figure 2C). The derivative curve for the 73-nt sequence showed similar behavior, with two resolved transitions with peaks at 45°C and a high-temperature transition at 76°C. The high melting temperatures for the 94-nt as well as the 73-nt sequence (76°C and 78°C, respectively) correspond to the denaturation of the Watson-Crick base-paired duplex from the P2 region into single strands. This is consistent with the observation made earlier under similar conditions.33 The transition at 58°C for the 94-nt oligonucleotide structure is due to the displacement of the Hoogsteen base-paired strand, as was reported earlier.33 Transition at 45°C for the 73-nt oligonucleotide is due to the 5-bp P1 helix. Ageeli et al.38 have reported similar melting behavior in that melting of the P1 helix occurs together with the triplex. Taken together, from these observations it may be concluded that the 94-nt sequence forms a stable triplex (hereafter termed the MALAT1 triplex) flanked by two duplexes, P1 and P2, and the 73-nt oligonucleotide, which lacks the A-rich tail, forms a stable duplex (hereafter termed the control duplex) under the conditions used in this study.
Quercetin binds the MALAT1 triplex motif in vitro
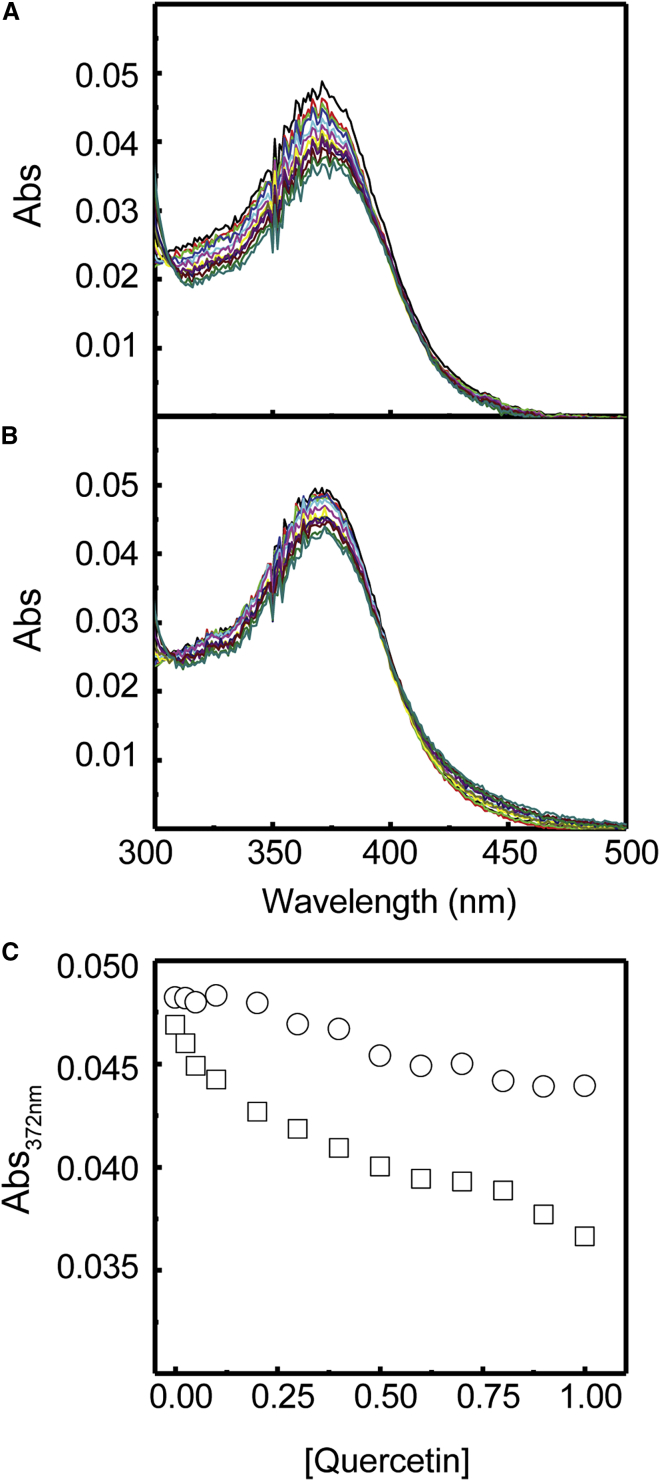
UV-visible (UV-vis) absorption spectroscopy is routinely used to gain insight into the interaction between a ligand and a nucleic acid structure. To understand the binding behavior between quercetin (Figure 1C) and the MALAT1 triplex, UV spectra of this ligand in the absence and presence of the MALAT1 triplex and the control duplex were obtained at 25°C. Figure 3 shows the changes in the absorption spectrum of 2.5 M quercetin when titrated against increasing amounts of triplex up to 1 M. Quercetin shows an absorbance spectrum with a positive peak at 369 nm. Upon addition of the MALAT1 triple-helix structure, a 23% hypochromic shift (Figure 3C) was observed along with a red shift of 9 nm from 369 to 378 nm (Figure 3A). In contrast, only a 10% hypochromic shift (Figure 3C) along with a red shift of 3 nm from 369 to 372 nm (Figure 3B) was observed when the control duplex was added. A higher change in hypochromicity and red shift in the absorption spectrum of quercetin in the presence of the MALAT1 triplex, in comparison with the changes in the presence of the control duplex, indicate that quercetin binds to the MALAT1 triplex stronger than to the control duplex. Hence, quercetin shows binding to the MALAT1 triplex, demonstrated by the appearance of isosbestic points in the UV spectra. Similar observations in the binding of flavonoids to an RNA triplex were also made by others.46,47
Figure 3.
UV titration of quercetin with triplex and control sequences
Absorbance spectra of 0.5 μM quercetin with successive additions of (A) MALAT1 triplex and (B) control duplex in 10 mM sodium cacodylate (pH 7), 0.5 mM MgCl2, and 150 mM NaCl at 25°C. (C) Variation in absorbance at 372 nm as a function of quercetin concentration for the triplex (□) and the control (○) structures shows a sharper rate of decline in absorbance for the triplex-forming sequence compared with the 73-nt control sequence, indicating a stronger interaction of quercetin with the 94-nt triplex sequence.
Quercetin binds the MALAT1 triplex motif with favorable enthalpy and entropy
Advances in the sensitivity of isothermal titration calorimetry allow for more accurate determination of thermodynamic parameters such as the binding enthalpy change (ΔH), Gibbs free energy change (ΔG), entropy change (ΔS), and number of binding sites (n). Therefore, this technique was used to characterize the thermodynamic profiles of the MALAT1 triplex and quercetin complex formation. Preformed triple helix and control duplex were titrated with quercetin at 25°C (Figure 4), and an isothermal titration calorimetry (ITC) binding isotherm was generated. ITC was also performed to determine heat of dilution of quercetin, where identical amounts of quercetin were injected into the buffer alone (Figure 4A).
Figure 4.

Binding parameters of quercetin to the MALAT1 triplex indicate clear binding to the MALAT1 triplex sequence
Thermograms for the calorimetric titration of 100 μM quercetin in 10 mM sodium cacodylate (pH 7), 0.5 mM MgCl2, and 150 mM NaCl into (A) buffer or (B) 6 μM 94-nt complete triplex or (C) 73-nt control structure at 25°C indicate binding of quercetin with the triplex. (D) Plot of integrated heats versus quercetin to triplex (□) molar ratio and duplex (○) molar ratio indicates no binding to quercetin in the case of control duplex and clear binding in the case of triplex. The first data point was eliminated in the data fit.
The representative raw ITC profiles resulting from the titrations of triplex and control duplex with quercetin are shown in Figures 4B and 4C, respectively. Each of the injections of quercetin into MALAT1 triplex solution produced a large exothermic heat signal (Figure 4B), which continued to decrease until the 12th injection and then became similar to the titration of quercetin into buffer (Figure 4A). In contrast, when quercetin was injected into the control duplex solution, exothermic heat signals for each and every injection were more or less the same and also were similar to the heat of dilution (Figure 4C). The resulting corrected injection heats plotted against the respective molar ratios are shown in Figure 4D. In Figure 4D, the data points reflect the experimental injection heats, while the solid line reflects the calculated fit of data using a single site binding model. We observed that quercetin binds to MALAT1 triplex with a stoichiometry of 1:1 with the Kd in the range of 495 ± 61.6 nM, accompanied by negative changes in free energy (−8.61 kcal mol−1), indicating that the interaction was spontaneous. Further investigation showed that the negative free energy results from a net balance of favorable entropy (+7.08 cal mol−1 K−1) and favorable enthalpy contributions (−6.5 ± 0.184 kcal mol−1). A favorable binding enthalpy is associated with the presence of strong hydrogen bonds and van der Waals interactions between quercetin and triplex. A favorable binding entropy is usually associated with the release of bound water molecules upon complex formation.
Quercetin binds the MALAT1 triplex motif through the major groove
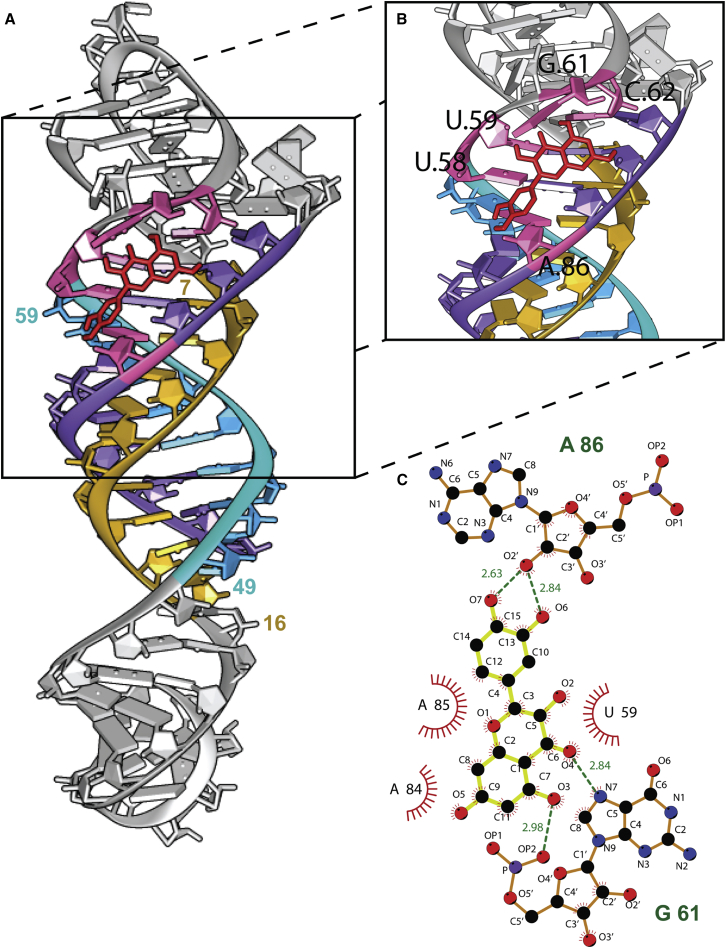
The interaction of the quercetin molecule with the core ENE hairpin and A-rich tract from MALAT1 (PDB: 4PLX) was studied using in silico docking. The search-space encompassing the complete structure revealed that the ligand predominantly binds to major grooves (Figure 5A). The most efficient binding pose of quercetin was found to interact with the binding energy of −5.98 kcal mol-1 (Figure 5B). Atomistic detail of the docked conformation revealed six non-covalent electrostatic interactions between the ligand and the nucleotides of the MALAT1 triplex helix crystal structure (Figure 5C), where the O5 and O7 atoms of quercetin participated in two electrostatic bonds each, while atoms O1 and O6 participated in one bond each. The six non-covalent bonds can be further categorized as five of them being “moderate, mostly electrostatic,” while one is “weak, electrostatic.”48 The nucleotides involved are U58, U59, G61, C62, and A86 (Figure 5B). The nucleotides U59 and A84 and A85 participated also in the hydrophobic interaction, as depicted in Figure 5C (with red sun-flared semicircle). There were four oxygen-oxygen interacting pairs and two oxygen-nitrogen non-covalent interaction pairs found in the quercetin-MALAT1 triplex complex.
Figure 5.
In silico docking of quercetin against the MALAT1 triplex
(A) Docking of quercetin to the MALAT1 triple-helix crystal structure. The flanking strands are colored in sea green (49–59) and golden yellow (7–16), while the middle strand is colored in purple (83–93). The interacting nucleotides are colored in pink. (B) The lowest-energy docking pose of the ligand (represented in red). (C) Atomistic representation of the ligand bound to the MALAT1 triplex. The ligand is depicted in yellow bonds, while the bonds of the lncRNA are shown in brown. Carbon, oxygen, nitrogen, and phosphorus atoms are depicted in black, red, blue, and purple, respectively. The hydrogen bonds between the ligand and the receptor are shown as a green dashed line with the distance marked in angstroms. The nucleotides involved in the hydrophobic interaction are shown with a red sun-flare semicircle. (The nucleotide residue numbers in all images correspond to the numbering of the 94-nt MALAT1 triplex as shown in Figure 1.)
As shown in Figure S1, there was a mismatch between the 76-nt sequence of the crystal structure (4PLX, used for docking) and the 94-nt MALAT1 triplex sequence. The alignment shown highlights the missing regions in the PDB structure. All the nucleotide positions mentioned above for docking reflect the numbering of the 94-nt MALAT1 triplex sequence. To assess the effect of quercetin binding on the MALAT1 triplex, we used temperature-scanning UV experiments to measure the melting temperatures of the MALAT1 triplex bound to quercetin. As is shown in Figure S2, quercetin binding does not appreciably influence the melting temperature of the MALAT1 triplex.
Quercetin binding to the MALAT1 triplex motif causes a decrease in MALAT1 levels
Recently, utilizing a novel small-molecule microarray strategy, Abulwerdi et al. identified two small molecules that bind very selectively to MALAT1 triplex.40 They observed reduced MALAT1 expression in HEK293 T cells on treatment with these compounds. As observed from our study, quercetin also binds strongly to the MALAT1 triplex, so we investigated the MALAT1 expression level in MCF7 cells upon treatment with quercetin. After treatment of cells with quercetin at concentrations ranging from 10 nM to 16 μM for 24 h, the total RNA was isolated (TRIzol, Invitrogen) and qRT-PCR was carried out for MALAT1 (Figure 6A). The U6 snRNA level was used to normalize the expression levels of MALAT1. Indeed, a significant and sustained decrease in MALAT1 levels was observed in cells treated with quercetin compared with the cells treated with 1% DMSO that served as controls. A 50% decrease in MALAT1 RNA levels was observed when cells were treated with 1 μM quercetin. As a comparison, we tested for the levels of NEAT1, which is another lncRNA and is encoded by the same chromosome as MALAT1 and also forms a triple helix similar to the one found at the 3′ end of MALAT1. Using the same protocol of qRT-PCR (and normalization), we observed no change in the levels of NEAT1 lncRNA, indicating that the effect of quercetin on MALAT1 is specific (Figure 6B). Further, to query whether the difference in MALAT1 was the result of a physical interaction between the small molecule and the RNA or a result of transcription level alteration, a transcription-stop assay was performed using α-amanitin. After 12 h of treatment with the transcription inhibitor, when MCF7 cells were exposed to quercetin, a chase experiment for MALAT1 levels using RT-PCR revealed that at 20 min post-quercetin treatment, MALAT1 RNA levels were brought down to half of the initial amount (Figure 6C). This indicated that the reduction in MALAT1 level was not a result of an effect on transcription; rather, it was due to a physical interaction between the molecule and the RNA.
Figure 6.
Change in transcript levels upon quercetin treatment
Quantitative real-time PCR showing (A) MALAT1 expression levels in MCF7 cells upon treatment with the indicated concentrations of the small molecule quercetin for 24 h. (B) Fold change in NEAT1 levels relative to control upon 1 μM quercetin treatment for 24 h in MCF7 cells. (C) Transcription-stop assay using α-amanitin. Bars depict fold change in levels of MALAT1 lncRNA after treatment with quercetin. The x axis shows time following quercetin treatment. All results presented are an average of three biological replicates (n = 3). Error bars represent SEM; asterisks denote Student’s two-tailed t test p values (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001). n.s. refers to non-significant.
Quercetin suppresses MALAT1 in nuclear speckles
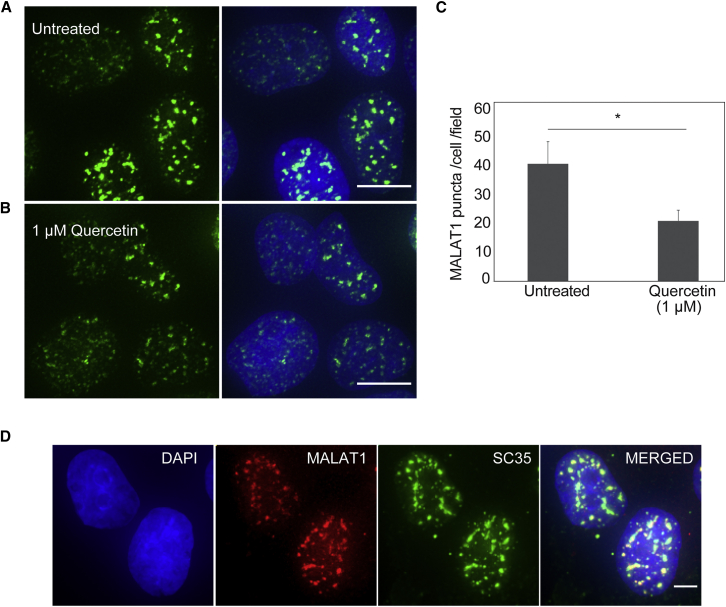
Subcellular localization determination via qRT-PCR and FISH demonstrated that MALAT1 localizes to both the nucleus and the cytoplasm. However, it is more abundant in the nucleus and forms a characteristic speckled pattern.49 From the qRT-PCR studies, we observed that MALAT1 levels in cells are suppressed by quercetin treatment. However, it was not indicated whether MALAT1 suppression by quercetin affects MALAT1 located in both the nucleus and the cytoplasm or whether suppression occurs only in the nucleus or in the cytoplasm. As MALAT1 is mostly expressed in the nucleus, quercetin requires entry into the nucleus to suppress MALAT1 expression effectively. However, in drug-resistant cancer cells, only a small percentage of drugs finally reach the nucleus, as cancer cells develop intracellular resistance mechanisms to limit the access of cytosolic drugs to the nucleus.50 To determine if quercetin is able to reach the nucleus and suppress MALAT1 effectively, we examined the abundance of MALAT1 using single-molecule RNA FISH, along with co-immunostaining using an antibody specific for the SR splicing factor SC35. In line with the previous reports, our FISH experiments along with co-immunostaining showed that MALAT1 is located in the nuclear speckles (Figure 7D). However, upon treatment with quercetin, the FISH signals decrease, indicating a decrease in MALAT1 concentration in the nuclear speckles (Figures 7A and 7B). These experiments clearly demonstrate that quercetin is able to localize in the nucleus and target MALAT1, thus reducing the MALAT1 concentration in speckles. The number of MALAT1 puncta reduces by ∼50% on treatment of cells with 1 μM quercetin (Figure 7C), which is in line with what was observed by qRT-PCR.
Figure 7.
MALAT1 RNA puncta inside cells are reduced upon quercetin treatment
Representative images showing (A) untreated vs. (B) 1 μM quercetin-treated cells. The left shows MALAT1 puncta (green) alone and the right shows MALAT1 puncta along with DAPI (blue). Scale bars, 10 μm. (C) Quantification of MALAT1 puncta (of size 1–1,000 pixels) per cell per field, counted for at least 100 cells per treatment; Error bar represents SEM from n=10 replicate fields; asterisk denotes Student’s two-tailed t test p value (∗p < 0.05). (D) Representative images showing the colocalization of MALAT1 and SC35 in MCF7 cells. DNA is stained with DAPI and represented in blue. Scale bar, 5 μm.
Quercetin treatment causes global splicing alteration
One of the major roles of MALAT1 in regulating cell fate has been through its modulation of RNA splicing through SR protein phosphorylation.31 The control of alternative splicing has been studied extensively, and large ongoing efforts have been made to develop synthetic modulators of alternative splicing that can control cellular function, resulting in potential novel therapeutics. Since the treatment with quercetin reduced MALAT1 expression levels, we asked whether the altered expression of MALAT1 in the presence of quercetin may affect the alternative splicing scenario that is under the control of MALAT1 coordination.
In previous studies, MALAT1 perturbation has been performed using sequence-specific knockdown approaches leading to discovery of diverse molecular pathways regulated by MALAT1. To explore the effect of small-molecule-induced MALAT1 changes on the transcriptome, we treated MCF7 cells with 1 μM quercetin and performed whole-RNA sequencing on two biological replicates. We were able to obtain only 168 transcripts (Table S1) that were significantly differentially regulated between 1% DMSO (control)- and quercetin-treated samples (Figure 8A). Surprisingly, we found that MALAT1 perturbation through quercetin, too, recapitulated major RNA regulatory pathways (ribosome biogenesis, RNA transport, etc.) at the level of transcript expression (Figure S3).51 This indicates that the pattern of gene expression in quercetin-treated MALAT1 knockdown cells is similar to that in cells where MALAT1 is specifically downregulated using siRNA.
Figure 8.
Differentially expressed transcripts and distribution of splicing events between DMSO-treated (control) and quercetin-treated MCF7 cells
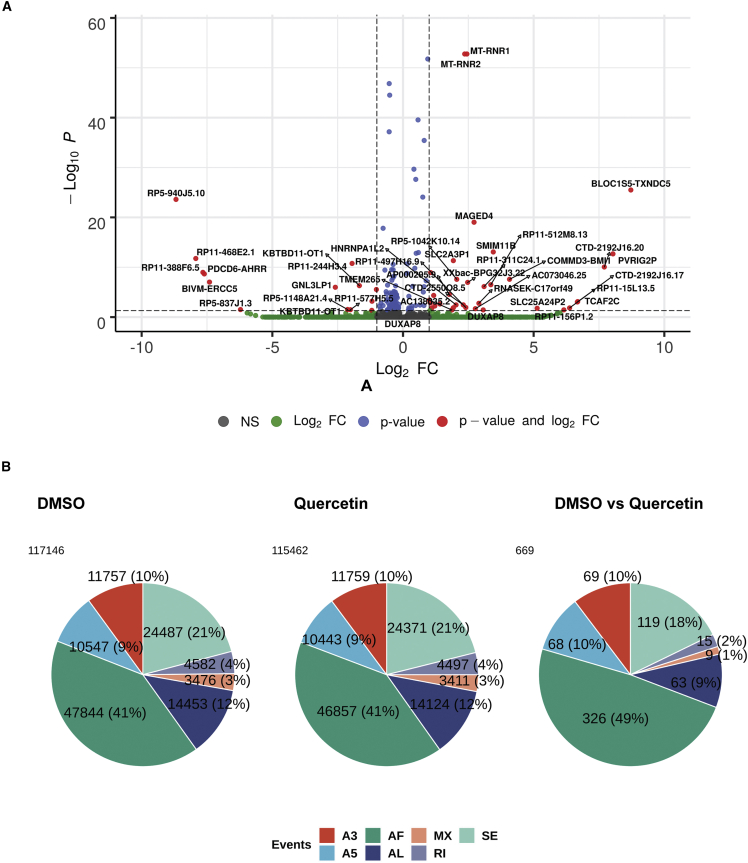
(A) Volcano plot to show differentially expressed transcripts. Dashed lines represent –log10 P and log2 FC cutoff of ±1. (B) Pie chart to show differentially spliced events. Events are denoted as A3, alternative 3′ usage; A5, alternative 5′ end of transcript; AF, alternative first exon; AL, alternative last exon; MX, mutually exclusive usage; RI, intron retention event; SE, exon skipping event.
Alternative splicing being a significant outcome of MALAT1-mediated SR protein phosphorylation, we investigated if quercetin treatment recapitulated a similar regulation at the transcript level. We observed more than 100,000 alternative splicing local events each in the control and the quercetin-treated samples (Figure 8B), which were classified as events on mapping to Gencode data. This high number of events come up due to the specific annotation used for exon, intron boundaries, etc., in Gencode. However, 669 events were observed when we looked at only the significantly differential alternative splicing events between control and quercetin-treated cells. These may be classified into six different categories based on the type of event (Figure 8B) (Table S2). Pathway analysis for this set of alternatively spliced genes showed a prominent association with RNA-regulatory pathways (spliceosome, spliceosomal complex, mRNA splicing, etc.) and mitochondrial pathways (Figures S4A and S4B). This indicates that quercetin treatment-led MALAT1 downregulation affects the splicing of genes involved in RNA regulation as well as mitochondrial processes. Sashimi plots of a few obtained transcripts showing their differently spliced events are shown in Figure S5.
There were some splicing events obtained that appeared exclusively in the control condition, and not in the treatment, and vice versa. We understood these events to appear as a result of quercetin-induced MALAT1 downregulation. Closer inspection of these mutually exclusive splicing events between the two conditions revealed an enrichment of exon-skipping events, alternative first exon usage, and alternative 5′ exon events in the quercetin-treated samples (Figure S6). These lead to a decrease in alternative 3′ exon usage, intron retention, and alternative last exon usage. Overall, we can conclude that quercetin treatment in MCF7 led to gross changes in alternative splicing, possibly acting through the MALAT1 splicing axis.
In vivo quercetin effect is MALAT1 specific
We asked whether quercetin treatment causes any global change in the lncRNA transcriptome. Our RNA-sequencing data analysis identified 168 transcripts (Table S1) that were significantly differentially regulated (Figure 8A), of which only 6 were lncRNAs (RP5-837J1.3, MIR181A1HG, FAM225B, AC138035.2, RP11-497H16.9, RP11-15L13.5). We checked the sequences of these lncRNAs for putative U-A-U triplex-forming motifs and found that no such motifs were present in any of these six lncRNA transcripts, suggesting that these lncRNAs (unlike MALAT1) are probably not targets of quercetin-triplex interaction and may be an outcome of other non-specific effects.
Discussion
lncRNAs have recently been well studied and have emerged as important regulators of gene expression. They can fold into complex structures and interact with proteins, thus modulating biological activity inside a cell. Generally, lncRNAs are structurally conserved, and these structural motifs are crucial for maintaining their function. MALAT1 is an extensively characterized multifunctional lncRNA that is involved in transcriptional regulation, alternative splicing, microRNA sponging, and many more functions. Recent studies indicate that elevated MALAT1 expression levels promote cell proliferation and are correlated with poor overall survival in various cancer types. It has also been shown that an elevated MALAT1 level correlates with larger tumor size, advanced tumor stage, and overall poor prognosis. Thus, selective inhibition of MALAT1 can be a strategy to halt its oncogenic activity from a therapeutic perspective. The 3.1-Å-resolution crystal structural study has recently identified a triple helix at the 3′ end of MALAT1 containing stacks of five and four U⋅A-U triples separated by a C+⋅G-C triplet and C-G doublet, extended by two A-minor interactions and a 3′-terminal A-rich tract. Similar triple-helix-forming structural elements were also observed in other non-coding and genomic RNAs of diverse viruses.52 Targeting these intramolecular triple-helical structures with small molecules could thus provide valuable insights into the roles of these structures in lncRNA functions and will consequently help to establish these structures as small-molecule-druggable targets. It also offers opportunities to therapeutically modulate numerous cellular processes.
UV melting studies showed that the MALAT1 triplex melts in two sequential well-resolved steps. The high-temperature transition is due to the melting of the apical P2 stem and the low-temperature transition to the thermally induced release of the third strand from the triplex to form the core duplex and the free single strand. These observations are consistent with the recent data reported by Ageeli et al.38 The U-rich third strand separation from the triplex occurs at 59°C ± 1.0°C, while the duplex strand denaturation occurs at 78°C ± 1.0°C in 10 mM sodium cacodylate (pH 7.0), 150 mM Na+, and 0.5 mM MgCl2. We also observed two melting transitions for the control duplex. The high-temperature transition for the control duplex occurred within the same temperature range as observed in the case of the MALAT1 triplex, confirming that the transition corresponds to apical P2 stem melting. However, low-temperature transition for the control duplex happened at 45°C ± 1.0°C, which is 14°C lower than the triplex melting. The amplitude of this transition is around 40% lower than the triplex transition, confirming that the transition is due to melting of the short P1 helix. Melting of the P1 helix at lower temperature was also noticed by Ageeli et al.38
There are a considerable number of small molecules known in the literature that specifically bind to triplex structures. We chose quercetin to determine biophysical insights into the interaction between quercetin and MALAT1 triplex because quercetin is known to bind specifically to RNA triplexes and is known for its potential pharmacological relevance.46,53 The results presented above clearly indicate that quercetin binds to the MALAT1 triplex. The binding of quercetin to the MALAT1 triplex resulted in a modest red shift (9 nm) as well as a modest hypochromicity (23%) at λ366nm. In general, small molecules binding to nucleic acid structures through an intercalation mode are generally accompanied by large red shifts and high hypochromicities.54 On the other hand, in the case of minor groove binding, a hypochromic effect can be observed, but the λmax absorption band virtually does not change.55, 56, 57 In the present study, since we observed modest shifts in the UV spectra of quercetin upon binding with the MALAT1 triplex, it can be argued that the binding mode is different from intercalation or minor groove binding. Currently, there are no high-resolution structures of a triple helix in complex with the aforementioned compounds, although various binding modes are predicted, such as intercalation and minor groove binding. The perturbation in the UV spectrum with hypochromic effect and red shifts upon increase in triplex concentration can be interpreted as an indication that the bound ligand is in a less polar environment. Our computational study supports the observation that the ligand predominantly binds to major grooves. This was further confirmed by the UV melting study, where we observed that the Tm of the triplex motif does not change in the presence of quercetin. Although not many small molecules are known that bind to RNA tertiary structures through a major groove, there are many natural products, such as pluramycins, aflatoxins, azinomycins, leinamycins, amino sugars, and neocarzinostatins, that are known to bind to DNA tertiary structures through the major groove.58 It has been shown that major groove binders do not stabilize nucleic acid tertiary structures, corroborating our observation.59 ITC experiments showed that, while quercetin binds to the MALAT1 triplex with a Kd of 495 ± 61.6 nM, it does not bind to the control duplex, hence confirming the selectivity of quercetin toward the MALAT1 triplex. Quercetin binding to MALAT1 triplex at 25°C yielded a negative enthalpy change (ΔH) of −6.5 ± 0.184 kcal mol−1 and an entropy change (ΔS) of +7.08 cal mol−1 with an overall favorable free energy (ΔGobs) of −8.61 kcal mol−1. The favorable binding of quercetin comes from a combination of enthalpy and entropy terms. The negative values of ΔH and positive values of ΔS are consistent with the characteristics of a combination of van der Waals, hydrophobic, and electrostatic interactions in the binding process. However, a large amount of entropy gain (TΔS = 2.11 kcal mol−1) was observed. Upon analysis of large scale data from existing experimental observations on thermodynamic features of drug-DNA interactions, Chaires et al. in previous reports have found that intercalation is primarily enthalpy driven, whereas groove binding is entropy driven.60 Correlating with this observation, the large entropy gain in the binding process indicates that quercetin interacts with MALAT1 triplex through the major groove binding mode. We observed an overall favorable free energy (ΔGobs) of −8.61 kcal mol−1 upon interaction, which is also more favorable than that obtained in the case of intercalation. For example, binding free energies (ΔGobs) of −6.7 and −7.49 kcal mol−1 were determined for ethidium and propidium, respectively, at 25°C in a buffer containing 0.2 M NaCl.61,62 Therefore, an additional contribution to the favorable binding free energy could indicate the formation of H bonds, in addition to the hydrophobic and van der Waals interactions, which generally happen in cases of interaction binding for ethidium and propidium. All these findings are also largely consistent with our computational observation. We found that a large network of hydrophobic and electrostatic interactions stabilize the quercetin-MALAT1 complex.
Our real-time qPCR data clearly indicate that quercetin is capable of targeting the triplex and bringing pharmacological perturbation to MALAT1 expression levels. We observed almost 50% downregulation in the MALAT1 levels in MCF7 cells upon treatment of 1 μM or higher quercetin without affecting the expression of NEAT1, which contains a structurally similar ENE triplex compared with MALAT1, thereby confirming the structural selectivity of quercetin toward the MALAT1 triplex. Further, by transcription-stop assay, a physical interaction between the small molecule and the RNA is indicated (over a perturbation in transcription) as a probable cause of MALAT1 RNA level decrease. Decreased levels of MALAT1 were further supported by the observation of depletion of MALAT1 in nuclear speckles in MCF7 cells upon treatment with quercetin. As MALAT1’s role in alternative splicing has been discussed,31 any perturbation in the levels of MALAT1 by such targeting of the triplex by small molecules could be expected to manifest into visible effects in alternative splicing. Indeed, we observed successful splicing perturbation events in more than 600 cases due to quercetin treatment (leading to MALAT1 level downregulation). Moreover, quercetin exhibits minimal effects on the lncRNA transcriptome.
The role of small-molecule-mediated targeting of triple helixes is gaining momentum with the recent focus on identification of highly specific lead molecules.41,42 A few comprehensive studies have reported the development of novel tools for targeting the triple helix in the lncRNA MALAT1.40 These studies have largely relied upon unbiased screening of large libraries of compounds using the MALAT1 triple helix as a target. Although these lead molecules are expected to bind to the MALAT1 triple helix with high specificity, detailed studies under complex intracellular conditions will further ascertain the utility of these compounds, but these are going to be restricted due to their lack of commercial availability. Quercetin binds to the MALAT1 triplex with dissociation constants in the nanomolar range and is as strong as the newly discovered MALAT1 triplex binders. Commercial availability of quercetin may help to overcome such restrictions.
To sum up, this study entails an integrative approach wherein we combine both biophysical methods and molecular assays to demonstrate quercetin as a scaffold to target a triplex that is present at the 3′ end of the MALAT1 lncRNA. The spectroscopic and calorimetric studies have unanimously shown efficient and selective binding of quercetin to MALAT1 triplex. Upon binding to the triplex of MALAT1, quercetin decreases MALAT1 levels, which in turn modulates alternative splicing for certain genes. Overall, our study helps to establish the triplex that is present in the 3′ end of MALAT1 as a druggable target.
Materials and methods
Procurement of oligos and quercetin
Quercetin (3,3′,4′,5,7-pentahydroxyflavone, 2-(3,4-dihydroxyphenyl)-3,5,7-trihydroxy-4H-1-benzopyran-4-one, C15H10O7) was obtained from Sigma (Q4951, purity ≥95%, solid) and used without any further purification. All other reagents were of analytical grade. MilliQ water was used throughout all the experiments. The concentration of quercetin was determined by measuring the absorbance at 370 nm using the molar extinction coefficient of 1.4 × 104 M−1 cm−1. The lyophilized and HPLC-purified 94-nt (sequence: 5′-GGAAGGUUUUUCUUUUCCUGAGAAAACAACACGUAUUGUUUUCUCAGGUUUUGCUUUUUGGCCUUUUUCUAGCUUAAAAAAAAAAAAAGCAAAA-3′) and 73-nt control ENE (sequence: 5′-GGAAGGUUUUUCUUUUCCUGAGAAAACAACACGUAUUGUUUUCUCAGGUUUUGCUUUUUGGCCUUUUUCUAGC-3′) unmodified RNA oligonucleotides were obtained from Genscript (Biotech Desk, India) and dissolved in nuclease-free water. The solution concentrations of each of the oligonucleotides were determined optically at 260 nm and 25°C using the molar extinction coefficients (M−1 cm−1 of strands) of 955,700 for the 94-nt oligonucleotide and 713,000 for the 73-nt oligonucleotide. The folded triplex was obtained by heating the solutions to 100°C for 5 min and then keeping on ice for 5 min, followed by a slow return to room temperature. All biophysical experiments were performed in a 10 mM sodium cacodylate buffer (pH 7.0) containing 150 mM NaCl and 0.5 mM MgCl2 at 25°C, unless otherwise specified. All experiments were replicated three times.
UV titration spectroscopy
Absorption spectra were measured on a Cary 100 UV-vis double-beam spectrophotometer with a 1-cm path length quartz cell. UV-vis absorption titrations were carried out by the stepwise addition of triplex and control duplex solutions to a quartz cuvette of 1-cm path length containing 700 μL of 0.5 μM quercetin. The triplex was titrated into the cuvette from an initial concentration of 0.025 μM up to a concentration of 1.0 μM. The absorbance spectra were recorded from 300 to 500 nm at 25°C. Experiments were replicated thrice.
Temperature-dependent UV spectroscopy (UV melting)
Absorbance vs. temperature profiles (melting curves) for triplex (1 μM) and control duplex (1 μM) were measured at 260 nm with a thermoelectrically controlled Cary 100 (Varian) spectrophotometer. A temperature range of 25°C–95°C was used to monitor the absorbance at 260 nm at a heating/cooling rate of 0.25°C min−1. First derivatives of the melting profiles were then computed using Origin 7.5 software (OriginLab). The melting temperatures, Tms, were determined by fitting the derivative plots using multiple peak Gaussian fitting algorithms within the software. All reported Tms are the average of at least three independent measurements and within ±1°C. Experiments were replicated thrice.
Circular dichroism
CD spectra were recorded in a JASCO 815 spectropolarimeter equipped with a thermoelectrically controlled cell holder and a cuvette with a path length of 1 cm. CD spectra for the triplex and control duplex (both 2 μM) were recorded between 220 and 350 nm at 25°C, and the spectrum obtained was the average of three scans.
Isothermal titration calorimetry
ITC measurements were performed using a Microcal PEAQ-ITC (Malvern Panalytical) instrument. The triplex (6 μM) was kept in the sample cell, and quercetin (100 μM) in the same buffer (with 2% final DMSO concentration) was placed in a syringe of volume 40 μL. Quercetin was added sequentially in 2-μL aliquots (for a total of 16 injections, 4 s duration) into the cell with 150 s of spacing time. All experiments were performed at 25°C. The injections were done from a stirring syringe rotating at 750 rpm, with detection on high feedback. The heat of dilution was determined in parallel experiments by titrating a quercetin solution of the same concentration into the same buffer in the cell. The generated heat burst curve (microwatts per second) was integrated with respect to time to give total heat per injection. The data were fit according to one set of site parameters using MicroCal PEAQ ITC Analysis software.
In silico docking against MALAT1 triplex
Virtual docking of quercetin was performed against the core ENE hairpin and A-rich tract from MALAT1 (PDB: 4PLX) using Autodock 4.63 The ligand was drawn using Marvin Sketch (https://www.chemaxon.com), a computational tool for drawing three- and two-dimensional chemical structures. The ligand’s structure was randomized and minimized prior to docking. A blind docking study was performed for the ligand wherein the complete receptor lncRNA was considered for search-space. Two hundred GA (genetic algorithm) runs were performed and the conformations with the 2.0 cluster tolerance from the 200 independent runs were selected for the cluster analysis. The best-docked position was further minimized using GROMACS 5.14 with 50,000 minimization steps and 0.01 step size.64 The analysis was performed using Autodock ADT and UCSF Chimera and LigPlot+.65, 66, 67
Maintenance of cells in culture and treatment with small molecules
In cellulo experiments were performed in the MCF7 breast cancer cell line (procured from the European Collection of Authenticated Cell Cultures [ECACC]). MCF7 was maintained in DMEM (GIBCO) with 10% FBS (GIBCO) at 37°C with 5% CO2. Quercetin in solid (powder) form was procured from SIGMA (Q4951; HPLC purified with >95% purity) and reconstituted in 100% DMSO. All experiments had volume-equivalent vehicle (DMSO)-treated cells as controls. The final concentration of DMSO was kept at 1% or below in all cell culture experiments (referred to as 1% DMSO). Fresh quercetin solution was prepared for each experiment.
RNA isolation and quantitative polymerase chain reaction
Total RNA was isolated from approximately 500,000 cells using Ambion TRIzol (cat. no. 15596018) reagent. RNA was quantified using the Nanodrop spectrophotometer. Only RNA with ideal ratios (A260nm/A280nm > 1.8 and A260nm/A230nm > 2.0) were processed for qPCR. DNase digestion and cDNA preparation were performed using the Qiagen Quantitect reverse transcription kit for 1 μg of RNA in a 20-μL reaction volume. Gene-specific reverse primers were used instead of the random reverse primers provided in the kit. RNA was diluted two times in nuclease-free water (AMBION) before use (to get 25 ng per reaction) in qRT-PCR. cDNA was amplified using the SYBR Green master mix (TaKaRa Tli RNase H PLus) in a Roche LightCycler 480 instrument with compatible reaction plates. Cycling conditions for qRT-PCR were as follows: 95°C for 3 min followed by 40 cycles of 95°C for 10 s, 60°C for 30 s, and 72°C for 30 s. Three technical replicates were set on the qPCR plate. Amplification of MALAT1 lncRNA (NR_002819.4) was done to produce a 217-bp amplicon from the 3′ end of MALAT1, and U6 snRNA (NR_004394.1) was amplified to give a 93-bp amplicon under the same conditions, as a control. A no-RT control was used to check for any residual DNA contamination, which may contribute toward amplification. A high Ct value (different by >10 Ct) was obtained each time to indicate no residual DNA in the sample. A PCR negative control (with no cDNA) was also used to check for any contamination of reagents or during setting up the reaction. A single melting curve was observed at the end of 40 cycles, which indicated the generation of a single species of product. Fold change for the analysis was performed using the 2−ΔΔCt method. Three biological replicates were used to calculate average fold change.
α-amanitin transcription-stop assay
Sixty percent confluent MCF7 cells (maintained as described above) were treated with 10 μg/mL α-amanitin (A2263, Sigma Aldrich) for 12 h in a 12-well plate format. Following this, the cells were treated with 1 μM quercetin or volume-equivalent DMSO (as vehicle control). At fixed time points post-treatment, quercetin-treated as well as DMSO-treated cells were washed with PBS and harvested in Ambion TRIzol for RNA isolation, followed by qPCR using protocols as described above. Results presented are an average of three biological replicates.
RNA sequencing
Total RNA was isolated from MCF7 cells using Ambion TRIzol reagent. RNA was quantified using the Qubit RNA BR Assay. RNA purity was checked using QIAxpert, and RNA integrity was assessed on TapeStation using RNA ScreenTapes. The NEB Ultra II Directional RNA-Seq Library Prep kit protocol was used to prepare the libraries for total RNA sequencing (NEB cat. no. E7760L). Ribosomal RNA (which constitutes approximately 95% of the total RNA population) was first removed from 200 ng of total RNA by using biotinylated, target-specific oligos combined with Ribo-Cop rRNA removal beads. Following purification, the ribo-depleted RNA was fragmented using divalent cations under elevated temperatures. The cleaved RNA fragments were copied into first-strand cDNA using reverse transcriptase. Second-strand cDNA synthesis was performed using DNA Pol I and RNase H enzyme. The cDNA fragments were then subjected to a series of enzymatic steps that repair the ends and tail the 3′ end with a single A base, followed by ligation of the adapters. The adapter-ligated products were then purified and enriched using the following thermal conditions: initial denaturation at 98°C for 30 s; 13 cycles of denaturation at 98°C for 10 s, annealing at 65°C for 75 s; final extension at 65°C for 5 min. PCR products were then purified and checked for fragment size distribution on TapeStation using the High Sensitivity D1000 ScreenTape assay (Agilent, cat. no. 5067-5584). Prepared libraries were quantified using the Qubit High Sensitivity Assay (Invitrogen, cat. no. Q32854) and were pooled and diluted to the final optimal loading concentration before cluster amplification. The clustered flow cell was loaded on the Illumina NovaSeq 6000 instrument to generate 150-bp paired-end reads.
Immuno-FISH for MALAT1 and SC35
MCF7 cells were seeded onto coverslips introduced into a six-well plate. One day post-seeding, the cells were treated with quercetin. Twenty-four hours after treatment, the cells were washed with PBS and then fixed and permeabilized for immunofluorescence staining with SC35 antibody (Abcam Ab11826) in a humid chamber overnight. The next day, the cells were washed and incubated with a secondary antibody labeled with Alexa Fluor having an excitation wavelength of 488 nm (Thermo Fisher A11011). To probe for MALAT1, these cells were further fixed and permeabilized according to the Stellaris FISH protocol (LGC Bioresearch Technologies) and then hybridized with the stellaris MALAT1 probe, which was conjugated with Quasar 570 dye (cat. no. VSMF-2210-5), in a humidified chamber overnight. The next day, the coverslip was washed and then briefly stained with DAPI before fixing onto a slide for visualization. The images were captured at 60× in three channels using the z-stacks option in the DeltaVision microscope. Images were projected for maximum intensity and then analyzed in ImageJ. The far-red channel (for MALAT1 intensity) was processed by converting to 8 bits and then subjected to auto-thresholding. Particles in the size range of 0–1,000 pixels were counted and compared between treated and untreated samples. The results presented are averaged over 100 cells per sample.
Gene expression analysis
Libraries were generated using the TruSeq Stranded Total RNA Sample Prep Kit (Illumina) followed by sequencing on the Illumina NovaSeq 6000 platform (>45 million reads per condition).
Raw reads were checked for quality using FastQC (v.0.11.5).68 Trimmomatic (v.0.39) was used to trim adaptor sequences and filter low-quality reads.69 High-quality reads were mapped to the Human Gencode (GRCh38.p13)80 reference genome and transcriptome using STAR (v.2.7.9a) aligner with RSEM (v.1.3.1) to estimate gene expression.70,71 Gene read counts from RSEM were normalized using TMM (weighted trimmed mean of M values). This was followed by differential expression analysis using quasi-likelihood methods in edgeR (v.3.30.3).72,73 Genes showing differential expression compared with the control condition were plotted using EnhancedVolcano (v.1.6.0).74 Gene enrichment and pathway analysis were done using the R interface of Enrichr.75, 76, 77
Bioinformatics analysis of alternatively spliced transcripts
Reads were aligned against the Human Gencode80 (GRCh38.p13) transcriptome by Salmon (v.1.4.0)78 in quasi-mapping-based mode. Following this, SUPPA2 was used to generate, calculate, and compare the alternative splicing events (SE, A3, A5, MX, RI, AF, AL) between the quercetin-treated and the DMSO control samples.79,81 Splicing events with delta percentage or proportion spliced-in (ΔPSI) ≥ |0.10| and p < 0.05 were considered significant differential events. Pie charts were generated using the ggpubr and ggplot2 R package.
Statistical analysis
Data were expressed as the mean ± SEM for control and experiment cases for three biological replicates. Statistical analysis was performed by two-tailed Student’s t test using MS Excel. Experimental results leading to p < 0.05 were considered statistically significant. Asterisks denote p values in the figures as ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001, respectively. Biological replicates were similar to those employed in the field.
Primer sequences used in this study were as follows:
MALAT1 forward primer, CTTCCTGTGGCAGGAGAGAC;
MALAT1 reverse primer, CGCTTGAGATTTGGGCTTTA;
NEAT1 forward primer, TCTCCTGGCTATTCCAGGCT;
NEAT1 reverse primer, TAGCCACACAGTGGCAGAAG;
U6 forward primer, CTCGCTTCGGCAGCACATATACT;
U6 reverse primer, ACGCTTCACGAATTTGCGTGTC.
Acknowledgments
The authors are thankful to Dr. Kausik Chakraborty for guidance and help with RNA FISH analysis and Dr. Mary Krishna Ekka for proofreading the manuscript. We are grateful to Dr. Beena Pillai for suggestions and guidance on the RNA sequencing portion of the article. Dr. Souvik Maiti’s lab members are acknowledged for helpful advice and discussions. This study is funded by the Council for Scientific and Industrial Research (CSIR), Government of India, project MLP2104, “Role of Unusual Secondary Structures in lncRNA Functions,” and project MLP0139, “Targeting RNA Driven Processes: Novel Chemical Biology Approaches to Identify New Classes of RNA Modulators.”
Author contributions
S.M. conceptualized the work and wrote the manuscript, I.R. performed experiments and wrote the manuscript. A.H.A. and D.J. performed the RNA-sequencing analysis and A.R. did docking experiments.
Declaration of interests
The authors declare there are no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2022.09.016.
Supplemental information
Data availability
Sequencing data relevant to the study have been deposited in SRA (Sequence Read Archive, NCBI) under BioProject ID PRJNA772554 and SRA accessions SRR16477292, SRR16477293, SRR16477294, and SRR16477295.
References
- 1.Djebali S., Davis C.A., Merkel A., Dobin A., Lassmann T., Mortazavi A., Tanzer A., Lagarde J., Lin W., Schlesinger F., et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kapranov P., Cheng J., Dike S., Nix D.A., Duttagupta R., Willingham A.T., Stadler P.F., Hertel J., Hackermüller J., Hofacker I.L., et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 3.Zhao Y., Li H., Fang S., Kang Y., Wu W., Hao Y., Li Z., Bu D., Sun N., Zhang M.Q., Chen R. NONCODE 2016: an informative and valuable data source of long non-coding RNAs. Nucleic Acids Res. 2016;44:D203–D208. doi: 10.1093/nar/gkv1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., Guernec G., Martin D., Merkel A., Knowles D.G., et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacob F., Monod J. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 1961;3:318–356. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- 6.Kapranov P., Willingham A.T., Gingeras T.R. Genome-wide transcription and the implications for genomic organization. Nat. Rev. Genet. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 7.Kim T.K., Hemberg M., Gray J.M., Costa A.M., Bear D.M., Wu J., Harmin D.A., Laptewicz M., Barbara-Haley K., Kuersten S., et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kopp F., Mendell J.T. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hung T., Wang Y., Lin M.F., Koegel A.K., Kotake Y., Grant G.D., Horlings H.M., Shah N., Umbricht C., Wang P., et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat. Genet. 2011;43:621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Plath K., Fang J., Mlynarczyk-Evans S.K., Cao R., Worringer K.A., Wang H., De la Cruz C.C., Otte A.P., Panning B., Zhang Y. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 11.Leeper T.C., Varani G. The structure of an enzyme-activating fragment of human telomerase RNA. RNA. 2005;11:394–403. doi: 10.1261/rna.7222505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forrest M.E., Khalil A.M. Review: regulation of the cancer epigenome by long non-coding RNAs. Cancer Lett. 2017;407:106–112. doi: 10.1016/j.canlet.2017.03.040. [DOI] [PubMed] [Google Scholar]
- 13.Gupta R.A., Shah N., Wang K.C., Kim J., Horlings H.M., Wong D.J., Tsai M.C., Hung T., Argani P., Rinn J.L., et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghosh A., Pandey S.P., Ansari A.H., Sundar J.S., Singh P., Khan Y., Ekka M.K., Chakraborty D., Maiti S. Alternative splicing modulation mediated by G-quadruplex structures in MALAT1 lncRNA. Nucleic Acids Res. 2021;1 doi: 10.1093/nar/gkab1066. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Hacisuleyman E., Goff L.A., Trapnell C., Williams A., Henao-Mejia J., Sun L., McClanahan P., Hendrickson D.G., Sauvageau M., Kelley D.R., et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat. Struct. Mol. Biol. 2014;21:198–206. doi: 10.1038/nsmb.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quinn J.J., Ilik I.A., Qu K., Georgiev P., Chu C., Akhtar A., Chang H.Y. Revealing long noncoding RNA architecture and functions using domain-specific chromatin isolation by RNA purification. Nat. Biotechnol. 2014;32:933–940. doi: 10.1038/nbt.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brimacombe R., Stiege W. Structure and function of ribosomal RNA. Biochem. J. 1985;229:1–17. doi: 10.1042/bj2290001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conrad N.K. The emerging role of triple helices in RNA biology. Wiley Interdiscip. Rev. RNA. 2014;5:15–29. doi: 10.1002/wrna.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ji P., Diederichs S., Wang W., Böing S., Metzger R., Schneider P.M., Tidow N., Brandt B., Buerger H., Bulk E., et al. MALAT-1, a novel noncoding RNA, and thymosin β4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 20.Sun Y., Ma L. New insights into long non-coding rna MALAT1 in cancer and metastasis. Cancers. 2019;11 doi: 10.3390/cancers11020216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li P., Zhang X., Wang H., Wang L., Liu T., Du L., Yang Y., Wang C. MALAT1 is associated with poor response to oxaliplatin-based chemotherapy in colorectal cancer patients and promotes chemoresistance through EZH2. Mol. Cancer Therapeut. 2017;16:739–751. doi: 10.1158/1535-7163.MCT-16-0591. [DOI] [PubMed] [Google Scholar]
- 22.Jin C., Yan B., Lu Q., Lin Y., Ma L. The role of MALAT1/miR-1/slug axis on radioresistance in nasopharyngeal carcinoma. Tumour Biol. 2016;37:4025–4033. doi: 10.1007/s13277-015-4227-z. [DOI] [PubMed] [Google Scholar]
- 23.YiRen H., YingCong Y., Sunwu Y., Keqin L., Xiaochun T., Senrui C., Ende C., XiZhou L., Yanfan C. Long noncoding RNA MALAT1 regulates autophagy associated chemoresistance via miR-23b-3p sequestration in gastric cancer. Mol. Cancer. 2017;16:174. doi: 10.1186/s12943-017-0743-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z., Katsaros D., Biglia N., Shen Y., Fu Y., Loo L.W.M., Jia W., Obata Y., Yu H. High expression of long non-coding RNA MALAT1 in breast cancer is associated with poor relapse-free survival. Breast Cancer Res. Treat. 2018;171:261–271. doi: 10.1007/s10549-018-4839-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gong R., Jiang Y. Non-coding RNAs in pancreatic ductal adenocarcinoma. Front. Oncol. 2020;10:309. doi: 10.3389/fonc.2020.00309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang S.H., Zhang W.J., Wu X.C., Zhang M.D., Weng M.Z., Zhou D., Wang J.D., Quan Z.W. Long non-coding RNA MALAT1 promotes gallbladder cancer development by acting as a molecular sponge to regulate miR-206. Oncotarget. 2016;7:37857–37867. doi: 10.18632/oncotarget.9347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang H.m., Yang F.Q., Chen S.J., Che J., Zheng J.H. Upregulation of long non-coding RNA MALAT1 correlates with tumor progression and poor prognosis in clear cell renal cell carcinoma. Tumour Biol. 2015;36:2947–2955. doi: 10.1007/s13277-014-2925-6. [DOI] [PubMed] [Google Scholar]
- 28.Ying L., Chen Q., Wang Y., Zhou Z., Huang Y., Qiu F. Upregulated MALAT-1 contributes to bladder cancer cell migration by inducing epithelial-to-mesenchymal transition. Mol. Biosyst. 2012;8:2289–2294. doi: 10.1039/c2mb25070e. [DOI] [PubMed] [Google Scholar]
- 29.Zhou R.P., Lin S.J., Wan W.B., Zuo H.L., Yao F.F., Ruan H.B., Xu J., Song W., Zhou Y.C., Wen S.Y., et al. The long noncoding RNA MALAT-1 is highly expressed in ovarian cancer and induces cell growth and migration. PLoS One. 2016;11:e0166751. doi: 10.1371/journal.pone.0155250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gutschner T., Hämmerle M., Eissmann M., Hsu J., Kim Y., Hung G., Revenko A., Arun G., Stentrup M., Gross M., et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tripathi V., Ellis J.D., Shen Z., Song D.Y., Pan Q., Watt A.T., Freier S.M., Bennett C.F., Sharma A., Bubulya P.A., et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilusz J.E., JnBaptiste C.K., Lu L.Y., Kuhn C.D., Joshua-Tor L., Sharp P.A. A triple helix stabilizes the 3' ends of long noncoding RNAs that lack poly(A) tails. Genes Dev. 2012;26:2392–2407. doi: 10.1101/gad.204438.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown J.A., Valenstein M.L., Yario T.A., Tycowski K.T., Steitz J.A. Formation of triple-helical structures by the 3′-end sequences of MALAT1 and MENβ noncoding RNAs. Proc. Natl. Acad. Sci. USA. 2012;109:19202–19207. doi: 10.1073/pnas.1217338109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitton-Fry R.M., DeGregorio S.J., Wang J., Steitz T.A., Steitz J.A. Poly(A) tail recognition by a viral RNA element through assembly of a triple helix. Science. 2010;330:1244–1247. doi: 10.1126/science.1195858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown J.A., Kinzig C.G., Degregorio S.J., Steitz J.A. Hoogsteen-position pyrimidines promote the stability and function of the MALAT1 RNA triple helix. RNA. 2016;22:743–749. doi: 10.1261/rna.055707.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brown J.A., Bulkley D., Wang J., Valenstein M.L., Yario T.A., Steitz T.A., Steitz J.A. Structural insights into the stabilization of MALAT1 noncoding RNA by a bipartite triple helix. Nat. Struct. Mol. Biol. 2014;21:633–640. doi: 10.1038/nsmb.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruszkowska A., Ruszkowski M., Hulewicz J.P., Dauter Z., Brown J.A. Molecular structure of a U⋅A-U-rich RNA triple helix with 11 consecutive base triples. Nucleic Acids Res. 2020;48:3304–3314. doi: 10.1093/nar/gkz1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ageeli A.A., McGovern-Gooch K.R., Kaminska M.M., Baird N.J. Finely tuned conformational dynamics regulate the protective function of the lncRNA MALAT1 triple helix. Nucleic Acids Res. 2019;47:1468–1481. doi: 10.1093/nar/gky1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yonkunas M.J., Baird N.J. A highly ordered, nonprotective MALAT1 ENE structure is adopted prior to triplex formation. RNA. 2019;25:975–984. doi: 10.1261/rna.069906.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abulwerdi F.A., Xu W., Ageeli A.A., Yonkunas M.J., Arun G., Nam H., Schneekloth J.S., Dayie T.K., Spector D., Baird N., Le Grice S.F.J. Selective small-molecule targeting of a triple helix encoded by the long noncoding RNA, MALAT1. ACS Chem. Biol. 2019;14:223–235. doi: 10.1021/acschembio.8b00807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Donlic A., Zafferani M., Padroni G., Puri M., Hargrove A.E. Regulation of MALAT1 triple helix stability and in vitro degradation by diphenylfurans. Nucleic Acids Res. 2020;48:7653–7664. doi: 10.1093/nar/gkaa585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Donlic A., Morgan B.S., Xu J.L., Liu A., Roble C., Hargrove A.E. Discovery of small molecule ligands for MALAT1 by tuning an RNA-binding scaffold. Angew Chem. Int. Ed. Engl. 2018;57:13242–13247. doi: 10.1002/anie.201808823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu D., Hu M.J., Wang Y.Q., Cui Y.L. Antioxidant activities of quercetin and its complexes for medicinal application. Molecules. 2019;24:1123. doi: 10.3390/molecules24061123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harwood M., Danielewska-Nikiel B., Borzelleca J.F., Flamm G.W., Williams G.M., Lines T.C. A critical review of the data related to the safety of quercetin and lack of evidence of in vivo toxicity, including lack of genotoxic/carcinogenic properties. Food Chem. Toxicol. 2007;45:2179–2205. doi: 10.1016/j.fct.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 45.Bansal M., Singh N., Pal S., Dev I., Ansari K.M. Chemopreventive role of dietary phytochemicals in colorectal cancer. Adv. Mol. Toxicol. 2018;12:69–121. [Google Scholar]
- 46.Pradhan A.B., Bhuiya S., Haque L., Das S. Role of hydroxyl groups in the B-ring of flavonoids in stabilization of the Hoogsteen paired third strand of Poly(U).Poly(A)∗Poly(U) triplex. Arch. Biochem. Biophys. 2018;637:9–20. doi: 10.1016/j.abb.2017.11.008. [DOI] [PubMed] [Google Scholar]
- 47.Tiwari R., Haque L., Bhuiya S., Das S. Third strand stabilization of poly(U)·poly(A)∗ poly(U) triplex by the naturally occurring flavone luteolin: a multi-spectroscopic approach. Int. J. Biol. Macromol. 2017;103:692–700. doi: 10.1016/j.ijbiomac.2017.05.115. [DOI] [PubMed] [Google Scholar]
- 48.An introduction to hydrogen bonding. Choice Rev. Online. 1998;35 [Google Scholar]
- 49.Cabili M.N., Dunagin M.C., McClanahan P.D., Biaesch A., Padovan-Merhar O., Regev A., Rinn J.L., Raj A. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol. 2015;16:20. doi: 10.1186/s13059-015-0586-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Duvvuri M., Krise J.P. Intracellular drug sequestration events associated with the emergence of multidrug resistance: a mechanistic review. Front. Biosci. 2005;10:1499–1509. doi: 10.2741/1634. [DOI] [PubMed] [Google Scholar]
- 51.Chen R., Liu Y., Zhuang H., Yang B., Hei K., Xiao M., Hou C., Gao H., Zhang X., Jia C., et al. Quantitative proteomics reveals that long non-coding RNA MALAT1 interacts with DBC1 to regulate p53 acetylation. Nucleic Acids Res. 2017;45:9947–9959. doi: 10.1093/nar/gkx600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tycowski K.T., Shu M.D., Borah S., Shi M., Steitz J.A. Conservation of a triple-helix-forming RNA stability element in noncoding and genomic rnas of diverse viruses. Cell Rep. 2012;2:26–32. doi: 10.1016/j.celrep.2012.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kashyap D., Garg V.K., Tuli H.S., Yerer M.B., Sak K., Sharma A.K., Kumar M., Aggarwal V., Sandhu S.S. Fisetin and quercetin: promising flavonoids with chemopreventive potential. Biomolecules. 2019;9 doi: 10.3390/biom9050174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ghazaryan A.A., Dalyan Y.B., Haroutiunian S.G., Tikhomirova A., Taulier N., Wells J.W., Chalikian T.V. Thermodynamics of interactions of water-soluble porphyrins with RNA duplexes. J. Am. Chem. Soc. 2006;128:1914–1921. doi: 10.1021/ja054070n. [DOI] [PubMed] [Google Scholar]
- 55.Shah A., Nosheen E., Munir S., Badshah A., Qureshi R., Rehman Z.U., Muhammad N., Hussain H. Characterization and DNA binding studies of unexplored imidazolidines by electronic absorption spectroscopy and cyclic voltammetry. J. Photochem. Photobiol. B. 2013;120:90–97. doi: 10.1016/j.jphotobiol.2012.12.015. [DOI] [PubMed] [Google Scholar]
- 56.Shi J.H., Chen J., Wang J., Zhu Y.Y. Binding interaction between sorafenib and calf thymus DNA: spectroscopic methodology, viscosity measurement and molecular docking. Spectrochim. Acta Mol. Biomol. Spectrosc. 2015;136 Pt B:443–450. doi: 10.1016/j.saa.2014.09.056. [DOI] [PubMed] [Google Scholar]
- 57.Sarwar T., Rehman S.U., Husain M.A., Ishqi H.M., Tabish M. Interaction of coumarin with calf thymus DNA: deciphering the mode of binding by in vitro studies. Int. J. Biol. Macromol. 2015;73:9–16. doi: 10.1016/j.ijbiomac.2014.10.017. [DOI] [PubMed] [Google Scholar]
- 58.Hamilton P.L., Arya D.P. Natural product DNA major groove binders. Nat. Prod. Rep. 2012;29:134–143. doi: 10.1039/c1np00054c. [DOI] [PubMed] [Google Scholar]
- 59.Tartakoff S.S., Finan J.M., Curtis E.J., Anchukaitis H.M., Couture D.J., Glazier S. Investigations into the DNA-binding mode of doxorubicinone. Org. Biomol. Chem. 2019;17:1992–1998. doi: 10.1039/c8ob02344a. [DOI] [PubMed] [Google Scholar]
- 60.Chaires J.B. A thermodynamic signature for drug-DNA binding mode. Arch. Biochem. Biophys. 2006;453:26–31. doi: 10.1016/j.abb.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 61.Chaires J.B. Energetics of drug-DNA interactions. Biopolymers. 1997;44:201–215. doi: 10.1002/(SICI)1097-0282(1997)44:3<201::AID-BIP2>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 62.Chaires J.B. Dissecting the free energy of drug binding to DNA. Anti Cancer Drug Des. 1996;11:569–580. [PubMed] [Google Scholar]
- 63.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. Software news and updates AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., Lindahl E. High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. [Google Scholar]
- 65.Huey R., Morris G.M., Olson A.J., Goodsell D.S. A semiempirical free energy force field with charge-based desolvation. J. Comput. Chem. 2007;28:1145–1152. doi: 10.1002/jcc.20634. [DOI] [PubMed] [Google Scholar]
- 66.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera - a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 67.Laskowski R.A., Swindells M.B. Multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011;51:2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- 68.Babraham S.A. Bioinformatics - FastQC A quality control tool for high throughput sequence data. Babraham Inst. 2020;1 [Google Scholar]
- 69.Bolger A.M., Lohse M., Usadel B. A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li B., Dewey C.N. Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Robinson M.D., McCarthy D.J., Smyth G.K. A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McCarthy D.J., Chen Y., Smyth G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Blighe K., Rana S., Lewis M. EnhancedVolcano: publication-ready volcano plots with enhanced colouring and labeling. R-Package. 2019 [Google Scholar]
- 75.Xie Z., Bailey A., Kuleshov M.V., Clarke D.J.B., Evangelista J.E., Jenkins S.L., Lachmann A., Wojciechowicz M.L., Kropiwnicki E., Jagodnik K.M., et al. Gene set knowledge discovery with Enrichr. Curr. Protoc. 2021;1:e90. doi: 10.1002/cpz1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma'ayan A. Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Patro R., Duggal G., Love M.,I., Irizarry R.,A., Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Trincado J.L., Entizne J.C., Hysenaj G., Singh B., Skalic M., Elliott D.J., Eyras E. Fast, accurate, and uncertainty-aware differential splicing analysis across multiple conditions. Genome Biol. 2018;19:40. doi: 10.1186/s13059-018-1417-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Frankish A., Diekhans M., Jungreis I., Lagarde J., Loveland J.E., Mudge J.M., et al. GENCODE 2021. Nucleic Acids Res. 2021;49:D916–D923. doi: 10.1093/nar/gkaa1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Alamancos G.P., Pagès A., Trincado J.L., Bellora N., Eyras E. Leveraging transcript quantification for fast computation of alternative splicing profiles. RNA. 2015;21:1521–1531. doi: 10.1261/rna.051557.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data relevant to the study have been deposited in SRA (Sequence Read Archive, NCBI) under BioProject ID PRJNA772554 and SRA accessions SRR16477292, SRR16477293, SRR16477294, and SRR16477295.