Summary
Human expansion in space is hampered by the physiological risks of spaceflight. The muscle and the liver are among the most affected tissues during spaceflight and their relationships in response to space exposure have never been studied. We compared the transcriptome response of liver and quadriceps from mice on NASA RR1 mission, after 37 days of exposure to spaceflight using GSEA, ORA, and sparse partial least square-differential analysis. We found that lipid metabolism is the most affected biological process between the two organs. A specific gene cluster expression pattern in the liver strongly correlated with glucose sparing and an energy-saving response affecting high energy demand process gene expression such as DNA repair, autophagy, and translation in the muscle. Our results show that impaired lipid metabolism gene expression in the liver and muscle atrophy gene expression are two paired events during spaceflight, for which dietary changes represent a possible countermeasure.
Subject areas: Space medicine, Omics, Space sciences, Astronautics
Highlights
-
•
Lipid metabolic genes are the most affected among mice muscle and liver in spaceflight
-
•
Glucose metabolic genes are the most DEG on mice quadriceps in spaceflight
-
•
Muscle atrophy gene expression correlates with a liver lipid gene cluster expression
-
•
Hepatokines are likely the effector of this organ communication
Space medicine; Omics; Space sciences; Astronautics.
Introduction
The presence of humans in space has considerably increased since the beginning of space conquest and the space population is expected to continue to grow with tourism and private research activities. The NASA Artemis program plans to take humans back to the Moon for 2024, and a walk on Mars soil is estimated in the course of the 2030s (Bukley, 2020). Meanwhile, space research is driving more and more astronauts in low Earth orbit as an invaluable tool used to deepen humanity’s understanding of the universe and our place within it. But not only do humans want to visit and explore space, humans want to live in space. Outer space settlement has become a new goal of space agencies and the private sector. The Artemis program, SpaceX, the Moon Village Association, and the Mars Society are examples of the driving force of human performance in space. Recently, the USA Air Force Space Command reported eight most plausible scenarios to the year 2060, all of which anticipating a permanent occupation of the Moon surface ranging from a small-sized crew to colony of thousands of people (Air Force Space Command, 2019). Over fifty years after the first foot imprint on the lunar surface, humans could very soon become an interplanetary species.
This endeavor is hampered by space stressors such as space radiation, microgravity, confinement, isolation, hostile conditions, and distance from Earth, that all induce physiological changes (Afshinnekoo et al., 2020). These changes can be seen as physiological adaptation to the space environment, but they can become maladaptive and deleterious over time and upon gravity re-exposure during a planetary mission such as to Mars. Nine major spaceflight systemic and physiological health risks have been identified: cardiovascular dysregulation, central nervous system impairment, increased cancer risk, muscle degeneration, bone loss, liver dysfunction and lipid dysregulation, circadian rhythm dysregulation, immune dysfunction, and space-associated neuro-ocular syndrome (SANS) (Afshinnekoo et al., 2020). Muscle atrophy is a major risk for these missions as it impairs mobility, thus compromising critical mission operations such as post-landing vehicle egress. Muscle atrophy is characterized by unbalanced protein degradation and the loss of muscle mass. However, protein supplementation fails to counteract muscle loss in sarcopenia, cachexia, and spaceflight-induced muscle atrophy (Gao and Chilibeck, 2020). Many studies have documented spaceflight-induced muscle atrophy as a consequence of weightlessness and decreased muscle solicitation (Fitts et al., 2000). Daily exercises as countermeasures succeed in slowing down the process but the muscle still shrinks during missions. Despite the important advances in human physiology in space, the molecular mechanisms underlying these physiological changes are poorly understood. Therefore, molecular analyses are needed to enable a new countermeasure approach to monitor astronaut health and ensure safety of future habited space missions.
The liver and the muscle are among the most affected tissues during spaceflight. The liver and the muscle are two master organs of the metabolism, with the muscle tissue representing up to 50% of the total body mass. Consistently, astronauts and living organisms exposed to space environment display many features of dysregulated metabolism.
Rats on the SpaceLab-2 mission displayed increased serum cholesterol levels (Popova et al., 1999). In mice, activated lipotoxic pathways are associated with abnormal liver lipid accumulation and increased lipid metabolism and lipid localization gene expression (Jonscher et al., 2016; Beheshti et al., 2019). Beheshti et al. recently showed using a multi-omic analysis that lipid dysregulation in mice liver and lipotoxic pathways was a specific response to space stressors alone (Beheshti et al., 2019). Dysregulated lipid metabolism, especially lipid accumulation, is strongly associated with non-alcoholic fatty acid liver diseases (NAFLD), liver dysfunction, insulin resistance (IR), and diabetes (Kovacs and Stumvoll, 2005; Perry et al., 2014; Zhang et al., 2010). Of note, mice showed early signs of liver injury on a short space flight (STS-135), and subclinical diabetogenic changes have been shown to occur during spaceflight and bedrest studies (Jonscher et al., 2016; Tobin et al., 2002). In mice flown aboard the BION-M1 biosatellite, the transcriptome response of the longissimus dorsi was mainly linked to metabolism, especially insulin signaling and sensitivity, highlighting the impact of spaceflight on glucose metabolism in skeletal muscle (Gambara et al., 2017a). Glucagon and insulin, two hormones of glucose metabolism, were predicted to be the common upstream regulators of spaceflight metabolic shift (Beheshti et al., 2019). In the Twin Study, increased levels of lactic acid were found in the urine of the flight astronaut, consistent with metabolomics data showing an increased lactic acid/pyruvic acid ratio suggesting a metabolic shift (Garrett-Bakelman et al., 2019). Interestingly, insulin resistance and liver diseases are associated with muscle atrophy and a potential reciprocal influence of insulin resistance and muscle disuse atrophy (Chakravarthy et al., 2020; Meyer et al., 2020; Rudrappa et al., 2016; Samuel and Shulman, 2012). As such, metabolic stress is a priority research for human space exploration (Bergouignan et al., 2016). The roles of the liver and the muscle are often interlinked, and muscle-liver crosstalk are described in metabolic diseases NAFLD, NASH, IR, and diabetes (Chakravarthy et al., 2020; Soeters and Soeters, 2012). However, the relationship of the two organs in the response to space environment has not been yet elucidated.
In this study, we describe for the first time the relationship of the response of both the liver and the muscle of mice during spaceflight using the R software package mixOmics widely used in the omic field. The mixOmics package enables datasets comparison, correlations, and associations visualization in various research areas (Duruflé et al., 2021; González et al., 2012).
Results
Lipid metabolic processes are the most common biological processes affected in mice quadriceps and liver during spaceflight
Previous studies showed that the muscle and the liver are differentially affected by metabolic changes during spaceflight (da Silveira et al., 2020). Here, we first investigated the commonalities between the two organs. We chose to focus on the quadriceps, which is the most important in term of size and as such potentially in terms of effects.
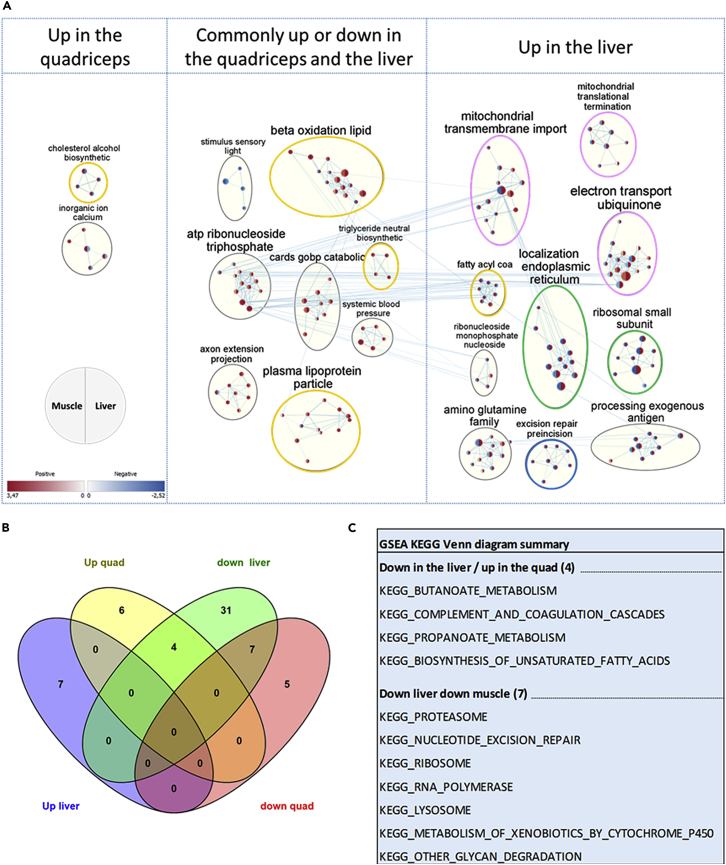
Transcriptomic data from C57BL/6J mouse liver and quadriceps from the NASA Rodent Research 1 (RR1) protocol were analyzed by the Gene Set Enrichment Analysis (GSEA) method and visualized with Cytoscape software (Figure 1A). In the quadriceps, “fatty coA process” and “axon extension negative” were the most enriched processes. In the liver, most enriched processes were related to mitochondria (“proton triphosphate gsea”, “electron transport cytochrome”, and “mitochondrial translational elongation”) followed by lipid metabolism-related processes (“oxidation fatty acyl”, “plasma lipoprotein particle”, and “positive lipid process”) (Figure S1A, Related to Figure 1). Similarly, lipid metabolism-related pathways were also the most enriched reported by KEGG pathways over representation analysis in the liver (Figure S1B, Related to Figure 1).
Figure 1.
Lipid metabolic processes are the most commonly biological processes affected in mice quadriceps and liver during spaceflight
(A) Cytoscape network of most common biological processes shared by the liver and the quadriceps reported by Gene Ontology biological Pathway (GOBP) Gene Set Enrichment Analysis (GSEA) in mice during spaceflight (p-value 0.05, FDR 0.05). Yellow circles indicate lipid metabolism related processes, pink circles indicate mitochondrial function related processes, green circles indicates protein post-translational processes, and the blue circle indicates a DNA Repair-related process.
(B) Venn diagram of common KEGG GSEA pathways between the liver and the quadriceps in mice during spaceflight.
(C) Summary of the common KEGG GSEA pathways between mice liver and quadriceps. FDR: False Discovery Rate, GOBP: Gene Ontology Biological Process, GSEA: Gene Set Enrichment Analysis, KEGG: Kyoto Encyclopedia of Genes and Genomes.
Among the 24,961 transcripts identified in the liver samples and the 22,657 transcripts in the identified quadriceps sample, the most enriched gene ontology biological processes altered by spaceflight assessed by GSEA commonly shared were related to lipid metabolism (Figure 1A). “β-oxidation lipid”, “plasma lipoprotein particle”, and “triglycerides neutral biosynthetic” genesets were upregulated in both organs, while “cholesterol alcohol biosynthetic” and “fatty acyl coa” displayed opposed expression patterns between the two organs. Consistent with previous studies, mitochondria-related process genesets were also among the most shared processes and were upregulated in the liver, while downregulated in the quadriceps. Endoplasmic reticulum and ribosome-related process genesets were also among the most shared process genesets, potentially suggesting impaired protein metabolism.
Both positively (upregulated) and negatively (downregulated) enriched KEGG pathways obtained by GSEA for each organ were compared together in a Venn diagram (Figures 1B and 1C). Among the 70 pathways analyzed, 11 pathways were common to the two organs. Again, lipid metabolism-related pathways were the most represented among the common ones (Figure 1B), with “KEGG_BIOSYNTHESIS_OF_UNSATURED_FATTY_ACIDs” and the two short chain fatty acid metabolism pathways “KEGG_BUTANOATE_METABOLISM” and “KEGG_PROPANOATE_METABOLISM” (Figure 1C). Protein metabolism (“KEGG_RIBOSOME”, “KEGG_PROTEASOME”, “KEGG_RNA_POLYMERASE”) and DNA repair-related pathways were also among the common downregulated pathways between the two organs. Thus, spaceflights induce a strong transcriptional response of the metabolism, especially for lipid metabolism in the liver and in the quadriceps.
Hypoglycemic signaling pathways genes are the most significantly downregulated in mice quadriceps during spaceflight
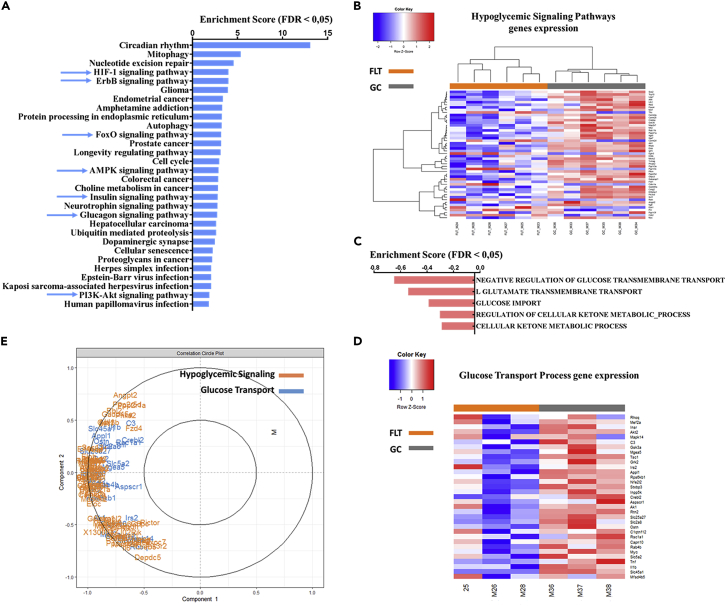
In the quadriceps, 972 genes were significantly differentially expressed, among which 562 were downregulated and 410 were upregulated. KEGG pathways over representation analysis (ORA) in the online software Webgesalt reported 30 most enriched pathways (Figure 2A). Pathways involved in metabolism are particularly enriched, especially those regulating glycemia: HIF1, ErbB, FoxO, AMPK, Insulin, and PI3K-Akt signaling pathways. Hypoglycemic signaling pathway gene expression was decreased in flight mice compared to ground control mice (Figure 2B). Interestingly, glucose transport processes were among the most negatively enriched in GSEA reports, and glucose transport process gene expression was decreased in flight mice compared to ground control mice (Figures 2C and 2D).
Figure 2.
Hypoglycemic signaling pathways genes are the most significantly downregulated in mice quadriceps during spaceflight
(A) Webgesalt ORA enriched KEGG pathways of DEG in mice quadriceps during spaceflight. Blue arrow indicates hypoglycemic pathways.
(B) Heatmap showing level expression of hypoglycemic genes in inflight mice (FLT) versus control (GC) mice. Color Key shows row z-score level.
(C) GOBP GSEA of the whole transcriptome of mice quadriceps RR1 (p-value 0.05, FDR 0.05).
(D) Heatmap showing level expression of glucose transport process genes expression in inflight mice (FLT) versus control (GC) mice (p-value 0.05, FDR 0.05). Color key shows row z-score level.
(E) Correlation circle plot between muscle hyperglycemic pathways and glucose transport process gene expression from sPLS-DA of mice quadriceps transcriptome during spaceflight. DEG: Differentially Expressed Genes, FDR: False Discovery Rate, GOBP: Gene Ontology Biological Process, GSEA: Gene Set Enrichment Analysis, KEGG: Kyoto Encyclopedia of Genes and Genomes, ORA: Over Representation Analysis, sPLS-DA: sparse Partial Least Square- Differential Analysis.
We investigated the relationships between the downregulation of glucose transport process genes and the downregulation of hypoglycemic signaling pathway genes using the supervised method sPLS-DA (sparse partial least square-differential analysis) with the R-mixOmics package. We compared the reported genes by the sPLS-DA performed at 100% (22657), 75% (16992), 50% (11328), and 25% (5664) (according to the process suggested in Duruflé et al., 2021) transcript input in a Venn diagram, and we found that 3463 genes were commonly reported with the strongest importance for discrimination (“discriminating genes”) (Figure S2A, Related to Figure 2). KEGG pathways ORA on these specific genes reported several enriched pathways among which were the previously mentioned hypoglycemic signaling pathways. Moreover, 491 significant differentially expressed genes (DEG) were still present among the 3463 sPLS-DA common genes, and KEGG pathway ORA on these genes demonstrated fewer enriched pathways than on the original analysis of the 972 DEG (Figure 2A) and on the 3463 sPLS-DA common genes (Figure S2A, Related to Figure 2), but the hypoglycemic ErbB, FoxO, AMPK, Insulin, and HIF-1 signaling pathways were still among the most enriched (Figure S2B, Related to Figure 2). Principal component analysis (PCA) showed that inflight mice and ground control mice groups were clearly discriminated by gene variability (Figure S2C, Related to Figure 2), despite PCA being an unsupervised method. Finally, we found that glucose transport process and hypoglycemic signaling pathway gene expression correlate (Figure 2E), and this correlation is strong as indicated by the correlation intensity level shown on the correlation heatmap (Figure S2D, Related to Figure 2).
Muscle glucose transport gene expression correlates with decreased muscle DNA repair, autophagy, and translation processes gene expression
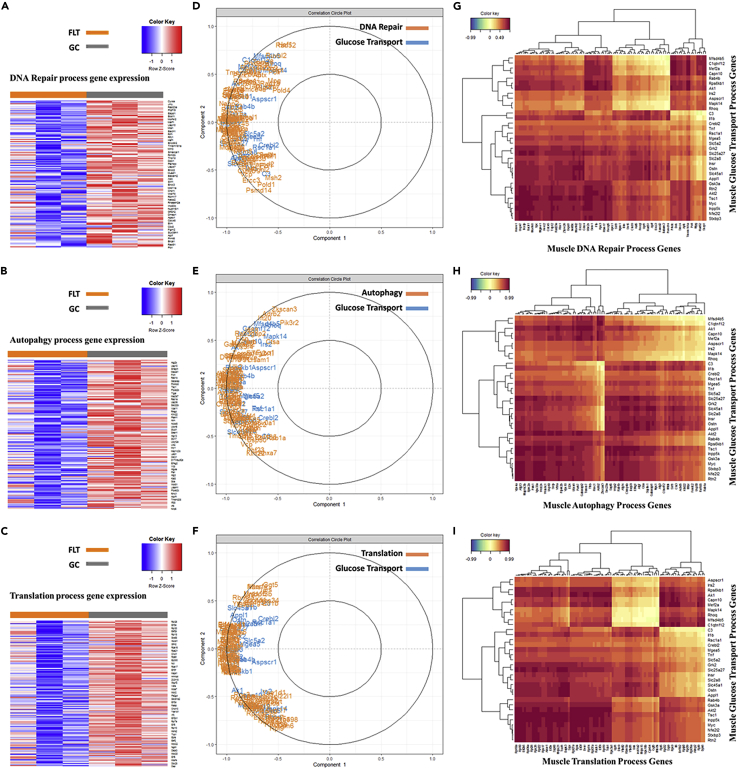
In the quadriceps, DNA repair, autophagy, and translation are among the most enriched processes and pathways reported by GSEA and ORA. DNA repair, autophagy, and translation process genes are decreased in inflight mice compared to ground control mice (Figures 3A–3C). Muscle glucose transport gene expression strongly correlates with decreased muscle DNA repair, autophagy, and translation process gene expression as shown by the correlation circle plot (Figures 3D–3F) and the correlation heatmap (Figures 3G–3I). These processes are both high energy demand processes. Thus, decreased energy intake gene expression correlated with an energy-saving transcriptome in mice quadriceps during spaceflight.
Figure 3.
Muscle glucose transport genes expression correlate with decreased muscle DNA repair, autophagy, and translation processes gene expression
(A–C) Heatmap showing the expression level of DNA repair (A), autophagy (B), and translation (C) process genes in the muscle in inflight mice (FLT) versus control (GC) mice. Color key shows row z-score level (p-value 0.05, FDR 0.05).
(D–F) Correlation circle plots between muscle glucose transport and DNA repair (D), autophagy (E), and translation (F) gene expression from sPLS-DA of mice quadriceps transcriptome during spaceflight.
(G–I) Correlation heatmap between muscle glucose transport and DNA repair (G), autophagy (H), and translation (I) gene expression from sPLS of mice quadriceps transcriptome during spaceflight. Color key shows correlation intensity. FDR: False Discovery Rate, sPLS-DA: sparse Partial Least Square- Differential Analysis.
Lipid metabolic processes gene expression in the liver correlates with gene expression changes in the muscle and muscle atrophy phenotype
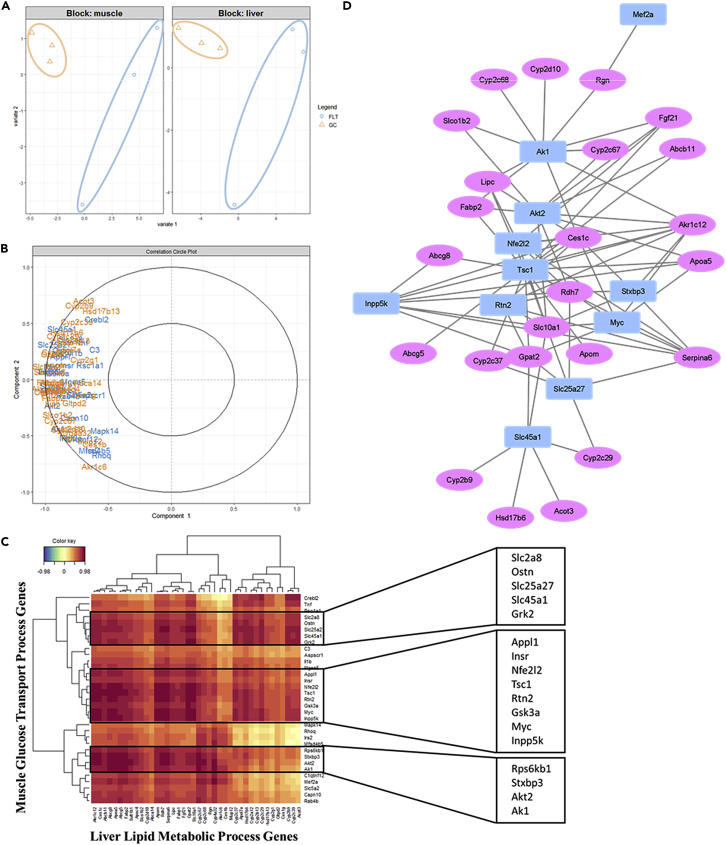
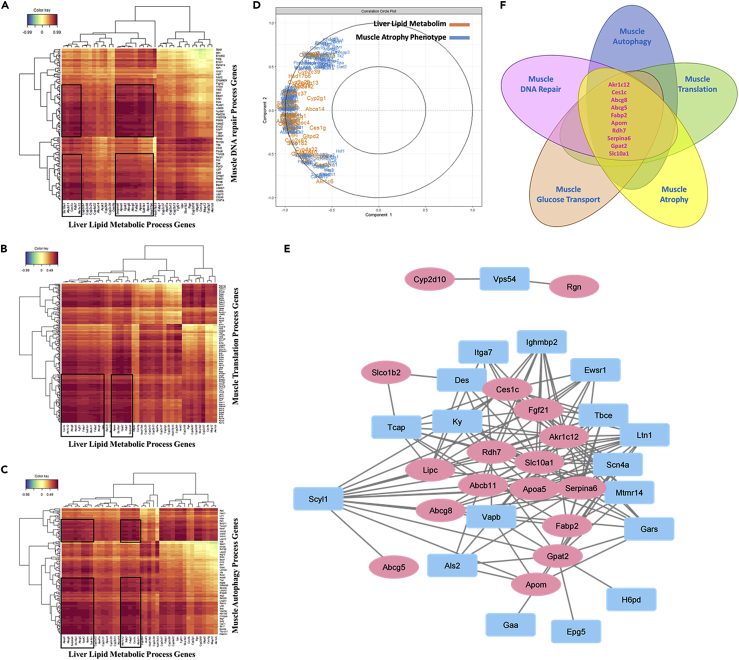
Lipid metabolism pathways are among the most reported by GSEA and ORA (Figures 1 and S1). In addition, discriminating genes after 100%, 75%, 50%, and 25% sPLS-DA and post DEG 100%, 75%, 50%, and 25% sPLS-DA still maintain this pattern as shown by KEGG ORA (Figures S3A and S3B, Related to Figure 4). Of note, the two short chain fatty acid “butanoate” and ‘propanoate” were always among the most enriched KEGG pathways (Figure S3C, Related to Figure 4). Since fatty liver diseases are associated with IR and diabetes, we investigated whether altered glucose transport gene expression was linked to liver lipid metabolism. We found that glucose transport process gene expression strongly correlated with liver lipid metabolism gene expression during spaceflight (Figures 4B and 4C). Consistently, liver lipid metabolism gene expression also correlated with muscle autophagy, DNA repair, and translation process gene expression (Figures 5A–5C). Furthermore, liver lipid metabolism gene expression strongly correlated with muscle atrophy phenotype gene expression (Figure 5D). Indeed, liver lipid metabolism and muscle atrophy genes formed a gene correlation network (Figure 5E). Akr1c12, Ces1c, Abcg8, Abcg5, Fabp2, Apom, Rdh7, Serpina6, Gpat2, and Slc10a1 formed a gene cluster strongly correlating with muscle gene expression (Figure 5F).
Figure 4.
Lipid metabolic processes gene expression in the liver correlates with glucose transport processes gene expression in the muscle
(A) PCA in muscle glucose transport and liver lipid metabolism process.
(B) Correlation circle plot between muscle glucose transport and liver lipid metabolism process gene expression from sPLS-DA of mice quadriceps and liver transcriptomes during spaceflight.
(C) Correlation heatmap between muscle glucose transport and liver lipid metabolism process gene expression from sPLS of mice quadriceps and liver transcriptomes during spaceflight. Color key shows correlation intensity.
(D) Correlation network between muscle glucose transport and liver lipid metabolism process gene expression from sPLS-DA of mice quadriceps and liver transcriptomes during spaceflight. sPLS-DA: sparse Partial Least Square- Differential Analysis.
Figure 5.
Liver lipid metabolism gene expression correlates with muscle atrophy phenotype, and DNA repair, autophagy, and translation genes decreased expression
(A–C) Correlation heatmap between liver lipid metabolism and muscle DNA repair (A), translation (B), and autophagy (C) process gene expression from sPLS of mice quadriceps and liver transcriptomes during spaceflight. Color key shows correlation intensity.
(D) Correlation circle plot between muscle atrophy phenotype and liver lipid metabolism process gene expression from sPLS-DA of mice quadriceps and liver transcriptomes during spaceflight.
(E) Correlation network between muscle atrophy phenotype and liver lipid metabolism process gene expression from sPLS-DA of mice quadriceps and liver transcriptomes during spaceflight.
(F) Venn diagram of strongest correlating liver lipid metabolism process genes in common between muscle atrophy phenotype, autophagy, DNA repair, translation, and glucose transport process genes. sPLS-DA: sparse Partial Least Square- Differential Analysis.
Discussion
Despite the systemic and physiological health risk of spaceflight, it is a fact that humans have become a space-faring species. Muscle atrophy has been shown to occur in both weightlessness and bed rest studies. The decreased gravity load on the locomotive apparatus is often presented as the main driver of muscle atrophy and bone loss. Previous studies showed that mitochondrial function and lipid metabolism were impaired in both astronauts and mice during spaceflight. Da Silveira and colleagues previously showed that a mitochondrial stress is a central hub for spaceflight-induced physiological changes (da Silveira et al., 2020). Mitochondria are complex organelles inside the cell where fundamental biomolecules are metabolized through the tricarboxylic acid cycle (TCA) and coupled to energy production by the electron transport chain. Thus, mitochondria play a key role in metabolism and energy management. Here, we found that lipid metabolism is the most affected biological process between the quadriceps and the liver transcriptome in mice during spaceflight. Impaired lipid metabolism and fatty acid accumulation is associated with muscle atrophy. Of note, butanoate and propanoate metabolism were the most affected in the two organs analyzed, decreased in the liver while increased in the muscle. Butanoate and propanoate, also known as butyrate and propionate, are short chain fatty acids produced in the intestinal lumen by bacterial fermentation of undigested carbohydrates, especially resistant starch, dietary fiber, and to a lesser extent proteins. Resistant starch is found in some grains, beans, and legumes. The gut microbiome is disturbed during spaceflight; astronauts’ gut microbiome showed an alteration in the composition pattern of bacteria of the phylum Firmicutes, the main butyrate-producing bacteria in the human gut (Venegas et al., 2019), with the presence of different genus increasing or decreasing during spaceflight but also showing a consistent decrease in bacteria from the taxa Pseudobutyrivibrio and Akkermansia, also known to produce butyrate in the intestine (Garrett-Bakelman et al., 2019; Voorhies et al., 2019; Venegas et al., 2019). Microbiome analysis from RR1 mice showed an overall increase of bacteria of the phylum Firmicutes, but the authors linked that to similar alterations that occur with aging (Jiang et al., 2019). This Firmicute increase in older mice is seen as a sign of dysbiosis and this alteration was shown to be followed by a ∼70% decrease in butyrate levels (Spychala et al., 2018). The upregulation of butyrate and propionate pathways in the muscle can reflect a possible need for these fatty acids and derivatives. In this context, dietary changes could represent a feasible countermeasure to spaceflight-induced physiological changes.
The TCA cycle activity and the glycolysis/gluconeogenesis ratio are decreased in muscle during spaceflight suggesting disturbed energy production. Consistently, we found that spaceflight induced a hyperglycemic response in mouse muscle involving the downregulation of hypoglycemic signaling pathways downstream of the insulin receptor (HIF, ErbB, AMPK, FoxO, and PI3K-Akt). This signaling response mostly affected glucose import gene expression. In addition, high energy demand processes such as DNA repair, autophagy, and translation gene expression were decreased in inflight mice quadriceps, suggesting a shift toward an energy-saving mode in the muscle during spaceflight. The muscle is a high energy demand organ. Since the muscle represent up to 50% of the total body mass, its metabolism radically affects glucose availability in the body. Consequently, glucose sparing and energy saving by the muscle in detrimental conditions can compensate for the energy needs of other such as the brain that requires stable and continuous glucose supply. Oxidative stress and DNA damage overload, that can impair mitochondrial function and the metabolism, are examples of detrimental conditions occurring during spaceflight (Afshinnekoo et al., 2020; Garrett-Bakelman et al., 2019). Also, mitochondrial dysfunction has been linked to muscle atrophy (Abrigo et al., 2019). The HIF-1 signaling enriched pathways suggest that oxidative stress occurred in RR1 inflight mice. Oxidative stress and mitochondrial dysfunction promote insulin resistance and diabetes (Bashan et al., 2009; Di Meo et al., 2017; Sivitz and Yorek, 2010). Our results, in line with previous space studies, suggest an insulin resistance phenotype. Meanwhile, insulin resistance has been proposed as an evolutionary mechanism to spare glucose that can benefit survival in various states such as starvation, immune activation growth, and cancer (Soeters and Soeters, 2012).
Insulin resistance and diabetes are often associated with dysregulated lipid metabolism and liver diseases. Fatty acids inhibit the insulin signaling pathways by directly interfering in the signaling through diacylglycerol or by overloading the TCA with the precursor acetyl CoA (Kovacs and Stumvoll, 2005; Perry et al., 2014; Zhang et al., 2010). During spaceflight, liver lipid metabolic gene expression correlated with muscle glucose import gene expression in RR1 mice. Insulin resistance, NAFLD, non-alcoholic steatohepatitis (NASH), and cirrhosis are associated with muscle atrophy and their severity correlates positively (Chakravarthy et al., 2020; Meyer et al., 2020; Samuel and Shulman, 2012). During a stress starvation, fatty acids together with ketone bodies become the primary source of energy for cells while peripheral organs such as the muscle, but also the skin and bones, provide amino acids taken up by central organs such as the liver, the spleen, immune cells, and healing tissues, ultimately leading to protein loss (Soeters and Soeters, 2012). During a spaceflight, all cells are exposed to space stressors such as radiation, endocrine system perturbation induced by altered circadian rhythm or psychological issues, and microgravity, all promoting oxidative stress, DNA damage, and mitochondrial dysfunction. In addition, it has been shown that spaceflight induces a shift from slow aerobic and highly enriched mitochondria muscle fibers (“slow fiber”, “type I”) toward fast anaerobic and fast low-enriched mitochondria muscle fiber (“fast fiber”, “type II”) (Gambara et al., 2017b; Ulanova et al., 2015). Mitochondria are the major source of reactive oxygen species. As such, spaceflight-induced fiber shift and mitochondrial dysfunction can be seen as a biological mechanism to decrease mitochondrial activity and mitigate oxidative stress. If mitochondrial stress is a central biological hub during spaceflight, and if the mitochondria are the master organelle of metabolism regulation and energy management, we can then expect a general metabolic shift in the body toward a starvation-like phenotype that promotes decreased mitochondrial activity, decreased metabolism and protein loss in the muscle, and increased lipid metabolism in the liver, to the benefit of all body cells during spaceflight. Consistently, liver lipid metabolism gene expression strongly correlated with muscle DNA repair, autophagy, translation, and finally muscle atrophy phenotype gene expression. Butanoate and propanoate metabolism-related genes were still among the one with the strongest correlation. Interestingly, sodium butyrate and butyrate diet supplementation improve mitochondrial function, fat accumulation, insulin resistance, diabetes, and liver diseases (Gao et al., 2009; Khan and Jena, 2016; Mollica et al., 2017; Ye et al., 2018; Zhang et al., 2019; Zhou et al., 2018). Moreover, sodium butyrate and butyrate diet supplementation decreased muscle atrophy in various conditions (Chang et al., 2001; Tang et al., 2022; Walsh et al., 2015). Thus, butyrate supplementation or dietary modification to stimulate microbial production represents simple options to counteract muscle atrophy while preventing insulin resistance and lipid accumulation during spaceflight.
Gambara et al. previously identified insulin- and glucose metabolism-linked transcripts as main DEG in BION-M1 biosatellite flown mice longissimus dorsi. In the present study, 17 and 13 differentially expressed genes in the liver and the muscle, respectively, were common to the 89 previously identified (Gambara et al., 2017a). Acas1a, Cpt2, Agt, Lcn2, and Prkcd, which were among the most DEG in Gambara’s study were not found significantly differentially expressed neither in the liver nor the quadriceps in the present study. The present study is based on the analyses of discriminating genes identified by the spls-DA performed with the mixOmics script. This method is designed to identify variations that are correlated, here gene expression in the liver and gene expression in the quadriceps. Also, 31 over the 9753, among which Agt and Cpt2, and 24 over the 3463 discriminating genes in the liver and the quadriceps, respectively, were in common with the 89 of the Gambara’s study. These differences may reflect the variability of gene expression between the different muscle types. Nonetheless, in line with the literature, both converge to an altered insulin transcriptome, thus highlighting a general adaptive response in muscle during spaceflight despite different molecular pathways. Moreover, the present study highlights the importance of discriminating genes over DEG in deciphering omic data. We found that discriminating genes are mainly linked to lipid metabolism in the liver and insulin signaling and glucose metabolism in the quadriceps. This means that lipid metabolism gene expression in the liver correlates with insulin signaling and glucose metabolism gene expression in the quadriceps in mice during spaceflight. We identified a cluster of lipid metabolism processes-related genes in the liver, including Akr1c12, Ces1c, Abcg8, Abcg5, Fabp2, Apom, Rdh7, Serpina6, Gpat2, and Slc10a1, correlating the most with glucose transport gene expression and potentially insulin resistance in the muscle. Among these genes, some are already described for their insulin sensitivity. Fabp2, Abcg5, and Abcg8 genes polymorphisms are associated with insulin sensitivity while Apom might protect against insulin resistance (Gylling et al., 2004; Kurano et al., 2020; Weiss et al., 2002; Yu et al., 2020). Interestingly, Sestrin 1 (Sesn1), a well-known regulator of cell metabolism upregulated in longissimus dorsi of BION-M1 mice, was reported as a discriminating gene in both the liver and the quadriceps (Gambara et al., 2017a). Sestrin proteins are involved in the metabolism of reactive oxygen species, autophagy, and insulin signaling (Lee et al., 2013). Sesn1 together with the present lipid metabolism-related gene cluster may thus account in insulin resistance research.
The liver influence on quadriceps gene expression we identified needs to be carried out by organ communication mechanisms and we identified hepatokines as the most likely effector. Hepatokines are proteins secreted by the liver that can modulate signaling pathways associated with energy metabolism in the muscle and influence the development of chronic metabolic diseases (Seo et al., 2021). Important for our context, an increased blood concentration of FGF21, ANGPTL4, FST, and Andropin increases insulin sensitivity, muscle hypertrophy, and mitochondrial function and diminishes fat mass in the muscle (Seo et al., 2021). Interestingly, increased concentration of Fetuin-A showed to have the inverse effect of the previously described hepatokines (Jensen-Cody and Potthoff, 2021; Seo et al., 2021). The levels of Fetuin-A increase after 60 day of bed rest, a ground model for microgravity effects, and we found in the literature a positive correlation between the Fetuin-A levels and the gene expression of Serpina6 and Rdh7, two genes we identified in our gene cluster (Herrmann et al., 2020; Thakur et al., 2008; Ward et al., 2020). Interestingly, Fetuin-A, as well as FGF21 and ANGPTL4 genes are among the correlating genes in the liver of mice during spaceflight reported by the liver versus quadriceps sPLS-DA. We hypothesize that the gene expression correlation identified in our work likely involves an increased Fetuin-A production by the liver and a diminished production of FGF21, ANGPTL4, FST, and Andropin.
In conclusion, we described for the first time a strong correlation between liver lipid metabolism gene expression and the downregulation of hypoglycemic and energy demand process-related genes. In the light of current knowledge, our result suggests that a starvation-like phenotype could promote a metabolic shift toward an energy-saving mode, resource sparring, and lipid mobilization through a tight liver-muscle crosstalk contributing to muscle atrophy during spaceflight. Dietary changes based on starch supplementation and microbiome modulation represent an affordable and sustainable countermeasure to support our planetary goals and outer space settlement. Meanwhile, improving food habits and body energy monitoring is an overall human challenge both on Earth and beyond.
Limitations of the study
Sample size was only six mice to match sample availability in the quadriceps and the liver transcriptome. Future space studies should be designed to match individuals for any factors (organs, environmental parameters, psychological parameters, etc.).
Here, we studied biological processes and pathways at a transcriptional level. Transcription regulation, post-transcriptional stabilization, and post-translational modifications impact cell response from gene expression to the final protein activity. In addition, transcriptomics provides a static view of one single moment in the cell. However, spatial repartition also impacts molecular dynamics, and a transcriptional ecosystem theory has been defined (Silveira and Bilodeau, 2018). To further understand the biological significance of the transcriptomic response to spaceflight in mice quadriceps, both functional analysis and multi-omics studies are needed.
Additional factors account for inducing muscle atrophy during spaceflight. Circadian rhythm pathways genes are among those enriched in both the quadriceps and the liver in mice during spaceflight (Figure S1B, Related to Figures 1 and 2A), and circadian rhythm perturbation is associated with muscle atrophy (Choi et al., 2020). Astronauts experience continuous circadian rhythm stressors on board the ISS, with sixteen sunsets per day and sustained artificial light exposure as the only illumination source. However, assessing and deciphering circadian rhythm dynamics and correlation at molecular levels require proper protocol and experimental design (Zeitgeber time method) that should be implemented in future space biology research.
We used the method implemented in mixOmics between the same molecular variables. This method is a powerful tool to compare any types of datasets. In this context, multiple variables comparisons, e.g., temperature variation inside the spacecrafts, food intake levels, dietary type, cognitive performances, psychological parameters, and allowing multiscale comparisons, will considerably widen our understanding of spaceflight’s impact on living organisms. The list of possible datasets combinations is infinite, as much as the number of studies which can raise from.
STAR★Methods
Key resources table
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact Dr Willian Abraham da Silveira, willian.dasilveira@staffs.ac.uk.
Materials availability
This study did not generate new unique reagents.
Method details
Full experimental procedures are available at: https://genelab-data.ndc.nasa.gov/genelab/accession/GLDS-103/ and https://genelab-data.ndc.nasa.gov/genelab/accession/GLDS-168.
The omics data of mice liver and muscle were obtained from the NASA Rodent Research 1 (RR1) protocol accessible on the NASA’s GeneLab public omics repository. Also, all animal procedures, ethics, and tissues harvesting, disposal, and experiments have been previously conducted and performed by external experimenters. Thus, animal procedures were performed according to the relevant guide-lines at each institution and were approved by the Institutional Animal Care and Use Committee (IACUC). Full experimental procedures are available at: https://genelab-data.ndc.nasa.gov/genelab/accession/GLDS-103/ and https://genelab-data.ndc.nasa.gov/genelab/accession/GLDS-168.
Briefly, the study was conducted aboard the International Space Station (ISS) in 2014 on twelves female C57BL/6J mice, six in flight mice (M23, M24, M25, M26, M27, M28) referred to as the FLT group, and six ground controls mice (M33, M34, M35, M36, M37, M38), referred to as the GC group, for 37 days upon 12h day/night cycle, and fed with Nutrient Upgraded Rodent Food Bar (NuRFB)63. The research platform used for this experiment was NASA’s Rodent Research Hardware System also referred to as the Rodent Research Habitat System. Mice were sacrificed with Euthasol injection followed by cervical dislocation on orbit on the ISS for the FLT group, and on the ground at the same time for the GC group. The entire carcasses of 10 mice were stored at −80C until processing. Two FLT mice were dissected on-board the ISS, and livers were isolated on-orbit and stored in the MELFI. Frozen FLT mice carcasses were then returned on Earth by the SpaceX-4 spacecraft and delivered to the NASA Ames Research Center (ARC) for disposal. The ten frozen mice carcasses, GC and FLT, were dissected at room temperature, and harvested samples stored at −80C.
RNA extraction from 20–30mg mice liver and muscle were realized at 4C using a lysis buffer (Qiagen, Valenica, CA) and a beta-mercaptoethanol (1:100)/Buffer RLT solution, lysis buffer (Qiagen, Valenica, CA) and kept on ice. Samples were then homogenized for 20 s at 21,000 RPM using a Polytron PT1300D handheld homogenizer with a 5 mm standard dispersing aggregate (Kinematica, Bohemia, NY). Homogenates were centrifuged for 3 min at room temperature to remove tissue debris. RNA from homogenates were further purified using the Qiagen AllPrep DNA/RNA Mini Kit (Qiagen, Valencia, CA), and eluted in DNase/RNase-free water. RNA concentrations were measured using the NanoDrop 2000 UV-Vis Spectrophotometer (Thermo Fisher Scientific, Waltham, MA), and RNA quality was assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). RNA sequencing and library construction was performed on 1 μg of RNA having RIN values of 7 or above, using Illumina HiSeq 4000 kit and experimental protocol.
Quantification and statistical analysis
RNAseq data processing
Raw FASTQ files were proceed using the “GeneLab RNAseq data processing protocol” as reported at https://genelab-data.ndc.nasa.gov/genelab/accession/GLDS-103/ and https://genelab-data.ndc.nasa.gov/genelab/accession/GLDS-168. On this work we used the mentioned protocol up to the step “Quantification data was imported to R (version 3.6.0) with tximport”.
R
Differential Expression (DE) analysis has been performed using the R studio software Version 1.2.5001 and the DESeq2 package version 1.30.1 that uses shrinkage estimation for dispersion and fold change. RSEM expected counts were extracted and rounded up to the next integer and used as input for DE analysis. Statistical significance of differentially expressed genes (FDR ≤0.05) were assessed using Benjamini-Hochberg multiple testing adjustment procedure.
Biological process, pathways, networks and clusters analysis
For biological process, pathways, networks and clusters analysis, normalized count from RR1 mice quadriceps and liver were processed by the DEseq2 package and assessed for Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways Over Representation Analysis on the online Webgesalt platform, and for Gene Ontology Biological Process (GOBP) and KEGG pathways GeneSets Enrichment by the Gene Set Enrichment Analysis (GSEA) software. The Cytoscape software was used to build geneset networks from GSEA results. GSEAs were run with a ‘‘Flight vs Ground Control’’ comparison setting. The ranked list of genes was defined by the signal-to-noise metric, and the statistical significance were determined by 1000 permutations of the gene sets. Enrichment importance were sorted according to the enrichment score calculated by GSEA (Müller, 2019). Venn diagrams were generated on the online platform Venny available at: https://bioinfogp.cnb.csic.es/tools/venny/index2.0.2.html.
mixOmics
mixomics is a R package developed by Kim-anh Lê Cao, Sébastien Déjean and collaborators to visualize associations between paired omics datasets using correlation circles plots, heatmaps, and networks (Duruflé et al., 2021; González et al., 2012; Rohart et al., 2017). In the present study we used mixOmics package version 6.14.1. More information can be found on the online mixOmics platform available at: http://mixomics.org/.
sPLS and sPLS-DA
Transcriptome from 6 matched NASA RR1 mice, 3 ground controls (M36, M37, M38) and 3 inflight (M25, M26, M28), between the liver and the quadriceps were analyzed by sPLS (Partial Least Square) and sPLS-DA (sparse Partial Least Square-Differential Analysis). Correlation circle and correlation network were generated from sPLS-DA performed on 100%, 75%, 50%, and 25% of total normalized counts according to the process suggested in Duruflé et al. (2021). This method is an internal method to validate that the pattern of genes evaluated with the strongest importance for discrimination maintain across input reduction. These genes are referred to as to “discriminating genes” in this study. The correlation circle plot is a 1 sized radius were variable pools located between |0,5| and |1| have the strongest correlation. Correlation heatmaps are generated from sPLS analyses. Post 100%, 75%, 50%, and 25% sPLS-DA DEG are DEG still present in reported genes from 100%, 75%, 50%, and 25% sPLS-DA. R scripts used for this study are available at: http://mixomics.org/case-studies/spls-liver-toxicity-case-study/.
Acknowledgments
W.A.d.S. acknowledge this work was partially funded by the ESA grant/contract 4000131202/20/NL/PG/pt ‘‘Space Omics: Toward an integrated ESA/NASA – omics database for spaceflight and ground facilities experiments’’ awarded to Raul Herranz. V.W. acknowledges this work was partially funded by the ESA grant/contract 4000134990/21/UK/AL. This work was allowed by the free access online repository data resources NASA genelab. The Rodent Research 1 data collection is supervised by Jonathan Galazka, Project Scientist, NASA GeneLab and Ruth Globus, RR1 Project Scientist NASA ARC.
Author contributions
G.V. analyzed and interpreted data, generated graphical abstract, and wrote and edited the manuscript. R.F. and G.V. analyzed and interpreted data and edited the manuscript. A.B. and G.M. participated in the exchange of ideas, edited the manuscript. T.L., V.W., and W.A.d.S. designed the study and revised the manuscript. V.W. and W.A.d.S. secured funding, supervised the project, and wrote and edited the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: October 21, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105213.
Contributor Information
Virginia Wotring, Email: virginia.wotring@isunet.edu.
Willian Abraham da Silveira, Email: willian.dasilveira@staffs.ac.uk.
Supplemental information
Data and code availability
-
•
The original/source data for all GeneLab datasets (GLDS) in the paper is available on GeneLab (https://genelab.nasa.gov/) with the specific GLDS identifier numbers in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
-
•
This paper does not report original code.
References
- Abrigo J., Simon F., Cabrera D., Vilos C., Cabello-Verrugio C. Mitochondrial dysfunction in skeletal muscle pathologies. Curr. Protein Pept. Sci. 2019;20:536–546. doi: 10.2174/1389203720666190402100902. [DOI] [PubMed] [Google Scholar]
- Air Force Space Command The future of space 2060 and implications for U.S. Strategy: report on the space futures, workshop Air force space Command. 2019. https://aerospace.csis.org/wp-content/uploads/2019/09/Future-of-Space-2060-v2-5-Sep.pdf
- Afshinnekoo E., Scott R.T., MacKay M.J., Pariset E., Cekanaviciute E., Barker R., Gilroy S., Hassane D., Smith S.M., Zwart S.R., et al. Fundamental biological features of spaceflight: advancing the field to enable deep-space exploration. Cell. 2020;183:1162–1184. doi: 10.1016/j.cell.2020.10.050. doi: 10.1016/j.cell.2020.10.050. Erratum Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashan N., Kovsan J., Kachko I., Ovadia H., Rudich A. Positive and negative regulation of insulin signaling by reactive oxygen and nitrogen species. Physiol. Rev. 2009;89:27–71. doi: 10.1152/physrev.00014.2008. [DOI] [PubMed] [Google Scholar]
- Beheshti A., Chakravarty K., Fogle H., Fazelinia H., Silveira W.A.d., Boyko V., Polo S.H.L., Saravia-Butler A.M., Hardiman G., Taylor D., et al. Multi-omics analysis of multiple missions to space reveal a theme of lipid dysregulation in mouse liver. Sci. Rep. 2019;9:1–13. doi: 10.1038/s41598-019-55869-2. doi: 10.1038/s41598-019-55869-2. Erratum Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergouignan A., Stein T.P., Habold C., Coxam V., O’gorman D., Blanc S. Towards human exploration of space: the THESEUS review series on nutrition and metabolism research priorities. npj Microgravity. 2016;2:16029. doi: 10.1038/npjmgrav.2016.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukley A.P. To the moon and beyond: challenges and opportunities for nasa’s artemis program. Cent. Space Pol. Strat. Space agenda. 2020;2021 https://csps.aerospace.org/sites/default/files/2021-08/Bukley_TheMoon_20201027.pdf [Google Scholar]
- Chakravarthy M.V., Siddiqui M.S., Forsgren M.F., Sanyal A.J. Harnessing muscle–liver crosstalk to treat nonalcoholic steatohepatitis. Front. Endocrinol. 2020;11:592373. doi: 10.3389/fendo.2020.592373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J.G., Hsieh-Li H.M., Jong Y.J., Wang N.M., Tsai C.H., Li H. Treatment of spinal muscular atrophy by sodium butyrate. Proc. Natl. Acad. Sci. USA. 2001;98:9808–9813. doi: 10.1073/pnas.171105098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y., Cho J., No M.H., Heo J.W., Cho E.J., Chang E., Park D.H., Kang J.H., Kwak H.B. Re-setting the circadian clock using exercise against sarcopenia. Int. J. Mol. Sci. 2020;21:E3106. doi: 10.3390/ijms21093106. https://pubmed.ncbi.nlm.nih.gov/32354038/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silveira W.A., Fazelinia H., Rosenthal S.B., Laiakis E.C., Kim M.S., Meydan C., Kidane Y., Rathi K.S., Smith S.M., Stear B., et al. Comprehensive multi-omics analysis reveals mitochondrial stress as a central biological hub for spaceflight impact. Cell. 2020;183:1185–1201.e20. doi: 10.1016/j.cell.2020.11.002. https://linkinghub.elsevier.com/retrieve/pii/S0092867420314616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Meo S., Iossa S., Venditti P. Skeletal muscle insulin resistance: role of mitochondria and other ROS sources. J. Endocrinol. 2017;233:R15–R42. doi: 10.1530/JOE-16-0598. [DOI] [PubMed] [Google Scholar]
- Duruflé H., Selmani M., Ranocha P., Jamet E., Dunand C., Déjean S. A powerful framework for an integrative study with heterogeneous omics data: from univariate statistics to multi-block analysis. Brief. Bioinform. 2021;22:bbaa166. doi: 10.1093/bib/bbaa166. [DOI] [PubMed] [Google Scholar]
- Fitts R.H., Riley D.R., Widrick J.J. Physiology of a microgravity environment invited review: microgravity and skeletal muscle. J. Appl. Physiol. 2000;89:823–839. doi: 10.1152/jappl.2000.89.2.823. [DOI] [PubMed] [Google Scholar]
- Gambara G., Salanova M., Ciciliot S., Furlan S., Gutsmann M., Schiffl G., Ungethuem U., Volpe P., Gunga H.C., Blottner D. Microgravity-induced transcriptome adaptation in mouse paraspinal longissimus dorsi muscle highlights insulin resistance-linked genes. Front. Physiol. 2017;8:279. doi: 10.3389/fphys.2017.00279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambara G., Salanova M., Ciciliot S., Furlan S., Gutsmann M., Schiffl G., Ungethuem U., Volpe P., Gunga H.C., Blottner D. Gene expression profiling in slow-Type calf soleus muscle of 30 days space-flown mice. PLoS One. 2017;12 doi: 10.1371/journal.pone.0169314. e0169314–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R., Chilibeck P.D. Nutritional interventions during bed rest and spaceflight: prevention of muscle mass and strength loss, bone resorption, glucose intolerance, and cardiovascular problems. Nutr. Res. 2020;82:11–24. doi: 10.1016/j.nutres.2020.07.001. [DOI] [PubMed] [Google Scholar]
- Gao Z., Yin J., Zhang J., Ward R.E., Martin R.J., Lefevre M., Cefalu W.T., Ye J. Butyrate improves insulin sensitivity and increases energy expenditure in mice. PMID: 19366864. Diabetes. 2009;58:1509–1517. doi: 10.2337/db08-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett-Bakelman F.E., Darshi M., Green S.J., Gur R.C., Lin L., Macias B.R., McKenna M.J., Meydan C., Mishra T., Nasrini J., et al. The NASA twins study: a multidimensional analysis of a year-long human spaceflight. Science. 2019;364:eaau8650. doi: 10.1126/science.aau8650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gylling H., Hallikainen M., Pihlajamäki J., Agren J., Laakso M., Rajaratnam R.A., Rauramaa R., Miettinen T.A. Polymorphisms in the ABCG5 and ABCG8 genes associate with cholesterol absorption and insulin sensitivity. J. Lipid Res. 2004;45:1660–1665. doi: 10.1194/jlr.M300522-JLR200. [DOI] [PubMed] [Google Scholar]
- González I., Cao K.A.L., Davis M.J., Déjean S. Visualising associations between paired ‘omics’ data sets. BioData Min. 2012;5:19–23. doi: 10.1186/1756-0381-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann M., Babler A., Moshkova I., Gremse F., Kiessling F., Kusebauch U., Nelea V., Kramann R., Moritz R.L., McKee M.D., Jahnen-Dechent W. Lumenal calcification and microvasculopathy in fetuin-A-deficient mice lead to multiple organ morbidity. PLoS One. 2020;15 doi: 10.1371/journal.pone.0228503. e0228503–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonscher K.R., Alfonso-Garcia A., Suhalim J.L., Orlicky D.J., Potma E.O., Ferguson V.L., Bouxsein M.L., Bateman T.A., Stodieck L.S., Levi M., et al. Correction: spaceflight activates lipotoxic pathways in mouse liver. PLoS One. 2016;11:e0155282. doi: 10.1371/journal.pone.0155282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen-Cody S.O., Potthoff M.J. Hepatokines and metabolism: deciphering communication from the liver. Mol. Metab. 2021;44:101138. doi: 10.1016/j.molmet.2020.101138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P., Green S.J., Chlipala G.E., Turek F.W., Vitaterna M.H. Reproducible changes in the gut microbiome suggest a shift in microbial and host metabolism during spaceflight. Microbiome. 2019;7:113–118. doi: 10.1186/s40168-019-0724-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S., Jena G. Sodium butyrate reduces insulin-resistance, fat accumulation and dyslipidemia in type-2 diabetic rat: a comparative study with metformin. Chem. Biol. Interact. 2016;254:124–134. doi: 10.1016/j.cbi.2016.06.007. [DOI] [PubMed] [Google Scholar]
- Kovacs P., Stumvoll M. Fatty acids and insulin resistance in muscle and liver. Best Pract. Res. Clin. Endocrinol. Metab. 2005;19:625–635. doi: 10.1016/j.beem.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Kurano M., Tsukamoto K., Shimizu T., Kassai H., Nakao K., Aiba A., Hara M., Yatomi Y. Protection against insulin resistance by apolipoprotein M/Sphingosine-1-Phosphate. Diabetes. 2020;69:867–881. doi: 10.2337/db19-0811. [DOI] [PubMed] [Google Scholar]
- Lee J.H., Budanov A.V., Karin M. Sestrins orchestrate cellular metabolism to attenuate aging. Cell Metab. 2013;18:792–801. doi: 10.1016/j.cmet.2013.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Wang J., Jaehnig E.J., Shi Z., Zhang B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019;47:W199–W205. doi: 10.1093/nar/gkz401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer F., Bannert K., Wiese M., Esau S., Sautter L.F., Ehlers L., Aghdassi A.A., Metges C.C., Garbe L.A., Jaster R., et al. Molecular mechanism contributing to malnutrition and sarcopenia in patients with liver cirrhosis. Int. J. Mol. Sci. 2020;21:53577–E5419. doi: 10.3390/ijms21155357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollica M.P., Mattace Raso G., Cavaliere G., Trinchese G., De Filippo C., Aceto S., Prisco M., Pirozzi C., Di Guida F., Lama A., et al. Butyrate regulates liver mitochondrial function, efficiency, and dynamics in insulin-resistant obese mice. Diabetes. 2017;66:1405–1418. doi: 10.2337/db16-0924. [DOI] [PubMed] [Google Scholar]
- Müller A.C. Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat. Protoc. 2019;22:924–934. doi: 10.1038/s41596-018-0103-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry R.J., Samuel V.T., Petersen K.F., Shulman G.I. The role of hepatic lipids in hepatic insulin resistance and type 2 diabetes. Nature. 2014;510:84–91. doi: 10.1038/nature13478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popova I.A., Zabolotskaia I.V., Kurkina L.M. Vliianie kosmicheskogo poleta na lipidnyĭ sostav krovi, nadpochechnikov i pecheni krys [The effects of space flights on the lipid composition of blood, adrenal glands and liver in rats] Aviakosm. Ekolog. Med. 1999;33:47–51. Aerospace and environmental medicine. [PubMed] [Google Scholar]
- R Core Team . R Foundation for Statistical Computing; 2021. R: A language and environment for statistical computing.https://www.R-project.org/ [Google Scholar]
- Rohart F., Gautier B., Singh A., Lê Cao K.A. mixOmics: an R package for 'omics feature selection and multiple data integration. PLoS Comput. Biol. 2017;13:e1005752. doi: 10.1371/journal.pcbi.1005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudrappa S.S., Wilkinson D.J., Greenhaff P.L., Smith K., Idris I., Atherton P.J. Human skeletal muscle disuse atrophy: effects on muscle protein synthesis, breakdown, and insulin resistance-a qualitative review. Front. Physiol. 2016;7:361. doi: 10.3389/fphys.2016.00361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel V.T., Shulman G.I. Mechanisms for insulin resistance: common threads and missing links. Cell. 2012;148:852–871. doi: 10.1016/j.cell.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo D.Y., Park S.H., Marquez J., Kwak H.B., Kim T.N., Bae J.H., Koh J.H., Han J. Hepatokines as a molecular transducer of exercise. J. Clin. Med. 2021;10:385. doi: 10.3390/jcm10030385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergushichev A. An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation. bioRxiv. 2016 doi: 10.1101/060012. Preprint at. [DOI] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silveira M.A.D., Bilodeau S. Defining the transcriptional ecosystem. Mol. Cell. 2018;72:920–924. doi: 10.1016/j.molcel.2018.11.022. [DOI] [PubMed] [Google Scholar]
- Sivitz W.I., Yorek M.A. Mitochondrial dysfunction in diabetes: from molecular mechanisms to functional significance and therapeutic opportunities. Antioxid. Redox Signal. 2010;12:537–577. doi: 10.1089/ars.2009.2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeters M.R., Soeters P.B. The evolutionary benefit of insulin resistance. Clin. Nutr. 2012;31:1002–1007. doi: 10.1016/j.clnu.2012.05.011. [DOI] [PubMed] [Google Scholar]
- Spychala M.S., Venna V.R., Jandzinski M., Doran S.J., Durgan D.J., Ganesh B.P., Ajami N.J., Putluri N., Graf J., Bryan R.M., McCullough L.D. Age-related changes in the gut microbiota influence systemic inflammation and stroke outcome. Ann. Neurol. 2018;84:23–36. doi: 10.1002/ana.25250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G., Du Y., Guan H., Jia J., Zhu N., Shi Y., Rong S., Yuan W. Butyrate ameliorates skeletal muscle atrophy in diabetic nephropathy by enhancing gut barrier function and FFA2-mediated PI3K/Akt/mTOR signals. Br. J. Pharmacol. 2022;179:159–178. doi: 10.1111/bph.15693. [DOI] [PubMed] [Google Scholar]
- Thakur A., Bollig A., Wu J., Liao D.J. Gene expression profiles in primary pancreatic tumors and metastatic lesions of Ela-c-myc transgenic mice. Mol. Cancer. 2008;7:11–16. doi: 10.1186/1476-4598-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin B.W., Uchakin P.N., Leeper-Woodford S.K. Insulin secretion and sensitivity in space flight: diabetogenic effects. Nutrition. 2002;18:842–848. doi: 10.1016/s0899-9007(02)00940-1. [DOI] [PubMed] [Google Scholar]
- Ulanova A., Gritsyna Y., Vikhlyantsev I., Salmov N., Bobylev A., Abdusalamova Z., Rogachevsky V., Shenkman B., Podlubnaya Z. Isoform composition and gene expression of thick and thin filament proteins in striated muscles of mice after 30-day space flight. BioMed Res. Int. 2015;2015:104735. doi: 10.1155/2015/104735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parada Venegas D., De La Fuente M.K., Landskron G., González M.J., Quera R., Dijkstra G., Harmsen H.J.M., Faber K.N., Hermoso M.A. Short chain fatty acids (SCFAs)mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front. Immunol. 2019;10:277. doi: 10.3389/fimmu.2019.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhies A.A., Mark Ott C., Mehta S., Pierson D.L., Crucian B.E., Feiveson A., Oubre C.M., Torralba M., Moncera K., Zhang Y., et al. Study of the impact of long-duration space missions at the International Space Station on the astronaut microbiome. Sci. Rep. 2019;9:9911–9917. doi: 10.1038/s41598-019-46303-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh M.E., Bhattacharya A., Sataranatarajan K., Qaisar R., Sloane L., Rahman M.M., Kinter M., Van Remmen H. The histone deacetylase inhibitor butyrate improves metabolism and reduces muscle atrophy during aging. Aging Cell. 2015;14:957–970. doi: 10.1111/acel.12387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward K., Mulder E., Frings-Meuthen P., O’Gorman D.J., Cooper D. Fetuin-A as a potential biomarker of metabolic variability following 60 Days of bed rest. Front. Physiol. 2020;11:573581. doi: 10.3389/fphys.2020.573581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss E.P., Brown M.D., Shuldiner A.R., Hagberg J.M. Fatty acid binding protein-2 gene variants and insulin resistance: gene and gene-environment interaction effects. Physiol. Genom. 2002;10:145–157. doi: 10.1152/physiolgenomics.00070.2001. [DOI] [PubMed] [Google Scholar]
- Ye J., Lv L., Wu W., Li Y., Shi D., Fang D., Guo F., Jiang H., Yan R., Ye W., Li L. Butyrate protects mice against methionine-choline-deficient diet-induced non-alcoholic steatohepatitis by improving gut barrier function, attenuating inflammation and reducing endotoxin levels. Front. Microbiol. 2018;9:1967. doi: 10.3389/fmicb.2018.01967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Zhang J., Yao S., Pan L., Luo G., Xu N. Apolipoprotein M overexpression through adeno-associated virus gene transfer improves insulin secretion and insulin sensitivity in Goto-Kakizaki rats. J. Diabetes Investig. 2020;11:1150–1158. doi: 10.1111/jdi.13261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Keung W., Samokhvalov V., Wang W., Lopaschuk G.D. Role of fatty acid uptake and fatty acid β-oxidation in mediating insulin resistance in heart and skeletal muscle. Biochim. Biophys. Acta. 2010;1801:1–22. doi: 10.1016/j.bbalip.2009.09.014. [DOI] [PubMed] [Google Scholar]
- Zhang W.Q., Zhao T.T., Gui D.K., Gao C.L., Gu J.L., Gan W.J., Huang W., Xu Y., Zhou H., Chen W.N., et al. Sodium butyrate improves liver glycogen metabolism in type 2 diabetes mellitus. J. Agric. Food Chem. 2019;67:7694–7705. doi: 10.1021/acs.jafc.9b02083. [DOI] [PubMed] [Google Scholar]
- Zhou D., Chen Y.W., Zhao Z.H., Yang R.X., Xin F.Z., Liu X.L., Pan Q., Zhou H., Fan J.G. Sodium butyrate reduces high-fat diet-induced non-alcoholic steatohepatitis through upregulation of hepatic GLP-1R expression. Exp. Mol. Med. 2018;50:1–12. doi: 10.1038/s12276-018-0183-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The original/source data for all GeneLab datasets (GLDS) in the paper is available on GeneLab (https://genelab.nasa.gov/) with the specific GLDS identifier numbers in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
-
•
This paper does not report original code.