Abstract
As one of the promising next-generation probiotics (NGPs), Akkermansia muciniphila, a well-known mucin-degrading bacterium, has been proven to be closely related to the metabolic diseases of its human host. However, the role of A. muciniphila in the host’s intestinal health remains ambiguous. Here, we comprehensively summarize and discuss the characteristics, the distribution, and the colonization of A. muciniphila in the human gastrointestinal tract (GIT). We propose that the application of A. muciniphila as a biomarker for longevity, for diagnostics and prognostics of intestinal diseases, or for intestinal health should be cautiously considered. Precise dietary regulation can mediate the treatment of intestinal diseases by altering the abundance of A. muciniphila. Although the beneficial role of A. muciniphila and its component in intestinal inflammation has been discovered, in gnotobiotic mice with specific gut microbiota, certain genotype, and colorectal cancer, or in animal models infected with a specific pathogen, A. muciniphila may be related to the occurrence and development of intestinal diseases. Genomic analysis, emphasizing the strain-level phylogenetic differences of A. muciniphila, indicates that a clear description and discussion of each strain is critical before its practical application. Our review provides much needed insight for the precise application of A. muciniphila.
Subject terms: Applied microbiology, Symbiosis
Introduction
As a mucin utilizing specialist1, Akkermansia muciniphila has been highly considered as one of the next-generation probiotics (NGPs) and is regarded to play an important role in the maintenance of the intestinal epithelial barrier. A typical cycle of intestinal inflammation is driven by abnormal interactions among genetic risk factors, environmental triggers (microbiota), modifiers, and the host’s immune system2. Akkermansia muciniphila widely exists in the GIT of multiple animals including humans, mice3, cattle4, guinea pigs5, swine6, rabbits7, ostriches8 and chickens9. In infants and healthy adults, A. muciniphila can account for 1~3% of total fecal cells10, during which the excessive degradation of mucin allows pathogens to invade the sloughed intestinal mucosa11. In such cases, supplement with adequate numbers of A. muciniphila, or heat-killed A. muciniphila may safely improve the intestinal barrier in obese humans12 and mice fed high-fat diets13,14. However, an excessive enrichment of A. muciniphila in mice with a specific intestinal environment may lead to the aggravation of intestinal inflammation caused by epithelial barrier damage15–17. Although the effect of A. muciniphila on intestinal inflammation has been gradually studied, how it works is still unclear. Meanwhile, factors including host, intestinal segmentation, age, intestinal disease, and diet, affecting the distribution of A. muciniphila in the GIT and how A. muciniphila interacts with the host to maintain intestinal health is mainly unknown. In this review, we bring together the latest research to comprehensively discuss the potential of A. muciniphila as a NGP to intervene in the intestinal homeostasis in humans and animals.
The characteristics and safety of A. muciniphila in the GIT
Belonging to the phylum Verrucomicrobia, A. muciniphila has been described as an oval-shaped, non-mobile, Gram-negative, non-spore forming, and strictly anaerobic bacterium. However, more than 90% number of A. muciniphila ATCC BAA-835 can survive in 95% oxygen and 5% CO2 for 1 h18. Different strains and phylogroups of A. muciniphila differ in their sensitivity to oxygen19, and most of the known A. muciniphila strains can utilize mucin as the sole carbon and nitrogen sources. The bacterium can grow on the Brain Heart Infusion (BHI) and Columbia medium, and mucin-derived monosaccharides, such as fucose, galactose, and N-acetylglucosamine, can also be used by A. muciniphila as growth substrates20.
The complete genome of type strain, A. muciniphila ATCC BAA-835, is 2,664,102 bp long, and has 2,176 predicted protein-coding genes, which suggest it can metabolize different kinds of carbohydrates and mucin21. Phylogenetic analysis of A. muciniphila classified it into three22 or four23 species-level phylogroups. Akkermansia muciniphila MucT strain is resistant to several antibiotics, such as chloramphenicol, clindamycin, streptomycin, erythromycin, vancomycin, and metronidazole24,25. The MucT strain is also abundantly colonized in the GIT of individuals treated with broad-spectrum antibiotics25, which may be due to the fact that A. muciniphila is an open-pangenome microorganism that can continually acquire genes from other bacteria via lateral gene transfer22.
Nowadays, A. muciniphila is widely studied as a promising probiotic to improve metabolic syndrome and obesity. However, its safety and toxicity are a growing concern. Long-term oral high-dosage of A. muciniphila, or pasteurized A. muciniphila (1010 bacteria per day), are safe and well tolerated in overweight and obese individuals12. The bacterial reverse mutation, in vitro mammalian cell micronucleus test, and a subchronic toxicity test (lasting 90 days in rat), show that pasteurized A. muciniphila has no-adverse effects26. Recently, pasteurized A. muciniphila has been recognized as a new food by the European Union27. Based on these findings and policies, the utilization of A. muciniphila in metabolic syndrome and in healthy individuals may be safe. However, whether A. muciniphila treatment is safe, when intestinal diseases occur, still needs to be confirmed.
The colonization and abundance of A. muciniphila in GIT
Location-dependent colonization of A. muciniphila in the GIT
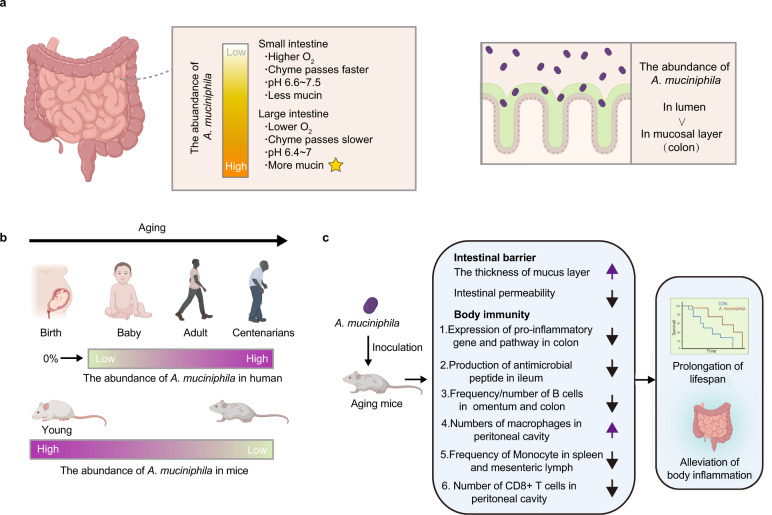
The abundance of A. muciniphila in the GIT seems to be location-dependent. Bacteria, belonging to the phylum Verrucomicrobia, not specified to Akkermansia, can be detected in human duodenal biopsies (0.0688%) and mucus (0.0387%)28. Akkermansia species, with an average relative abundance of 0.01%, are also found in the jejunal content of humans29, but the abundance of Verrucomicrobia-related bacteria can make up 5% of the bacteria density in the distal ileum of humans, and as much as 6% and 9% in the ascending colon and rectum mucosal biopsies, respectively30. Compared to the small intestine, the passage time of chyme is much longer (9–46 h)31 and the mucosal layer is thicker in the large intestine, which is presumed to provide multiple substrates for A. muciniphila32. As a mucin-degrading bacterium, A. muciniphila is abundantly found in the mucin-rich intra-intestinal location33, for which it is positively correlated with the concentration of mucin34. For instance, inoculated A. muciniphila is found to efficiently colonize (13.08% of total microbes) in the caecum of chickens9. In humans, there are approximate 1.45 × 104 cells of A. muciniphila per gram of ascending, or sigmoid colonic mucosal biopsies35. Moreover, the proportion of Akkermanisa in the lumen (0.57%) is found higher than that in colonic mucosa of healthy individuals (0.21%)36. The pH value may be another factor affecting the distribution of A. muciniphila in different intestinal segments. The pH value of the small and large intestine is 6.6~7.5 and 6.4~7.0, respectively37 (Fig. 1a). Using a model of the human digestive system, Simulator of the Human Intestinal Microbial Ecosystem (SHIME), when the pH value of the distal colon is 6.6~6.9, the abundance of A. muciniphila is at its highest38.
Fig. 1. The abundance and role of A. muciniphila with spatial and temporal change in the GIT.
a The distribution of A. muciniphila along the GIT (small and large intestine) and in the lumen and mucosal layer. b The schematic diagram of A. muciniphila abundance changing with age in the human and mouse GIT. c The mechanisms of A. muciniphila ameliorating aging in mice. All figures are created with Biorender.com.
Aging-dependent colonization of A. muciniphila in GIT
Tracking the fecal bacterial composition in 98 infants from birth to 12 months old shows a gradual increase (0% to 0.57%) in the relative abundance of A. muciniphila39. Similar increase (0.14% to 4.25%) is found in children aged from 1 to 4 years40. In addition, the abundance of A. muciniphila is high in long-living Chinese people (≥90 years old)41. Moreover, the abundance of A. muciniphila is especially higher in the gut of older populations aged from 105 to 109 years old, compared to other age groups42 (Fig. 1b). These findings43–46 (Table 1) give rise to the consideration of exogenous A. muciniphila inoculation to alleviate the negative effects of aging47,48 (Fig. 1c). However, opposite results are found in studies using rodents. The abundance of Akkermansia appears to be decreaed in aged mice or rats3,49,50. Although A. muciniphila may be a potential biomarker of longevity in humans, mice may not be a natural research model to study this relationship in humans.
Table 1.
The abundance of A. mucinihlia varies with age.
| Author/Year | Volunteers | Geographic area | Method | Main Findings |
|---|---|---|---|---|
| Elena Biagi et al. 201642 | 22–48 years: n = 15 65–75 years: n = 15 99–104 years: n = 15 105–109 years: n = 24 | Emilia Romagna and surrounding area, Italy | 16S rRNA gene sequencing | The relative abundance of A. muciniphila is increased in 105–109 years old humans. |
| Fanli Kong et al. 201641 | 24–64 years: n = 47 65–83 years: n = 54 90–102 years: n = 67 | Dujiangyan and Ya’an, Sichuan, China | 16S rRNA gene sequencing | Relative abundance of Akkermansia OTUs in 90–102 years old humans is higher than that in younger people. |
| Simone Rampelli et al. 202046 | 22–48 years: n = 11 65–75 years: n = 13 99–104 years: n = 15 105–109 years: n = 23 | Emilia Romagna, Italy | Shotgun sequencing | Compared with younger individuals, long-lived humans show a significantly increase of A. muciniphila. |
| Nuria Salazar et al. 201944 | <50 years: n = 49 50–65 years: n = 58 66–80 years: n = 19 >80 years: n = 27 | The central area of the Asturias Region, northern Spain | Real-time PCR | The counts of Akkermansia in older humans (>80 years) is higher than that in younger population. |
| Bong-Soo Kim et al. 201945 | 26–43 years: 9 67–79 years: 17 95–108 years: 30 | The neighboring counties of Gurye, Gokseong, Sunchang, and Damyang, located in the southwestern part of Korea | 16S rRNA gene sequencing | Centenarians have higher levels of Akkermansia in their gut. |
Factors influencing the colonization and abundance of A. muciniphila in the GIT
The abundance of A. muciniphila related to different intestinal diseases
Inflammatory bowel disease (IBD), including Crohn’s disease (CD) and ulcerative colitis (UC), is a known risk factor for the development of colorectal cancer (CRC), like colitis-associated colorectal cancer (CAC)51, the third leading cause of cancer-related death in humans52. The number of A. muciniphila in healthy individuals is higher than that in IBD patients53,54 (Supplementary Table 1), especially in the hindgut55. The relative abundance of A. muciniphila can be as high as 2.9% in healthy populations, but is found to sharply decline in noninflamed UC (0.03%), inflamed UC (0.02%), noninflamed CD (0.62%), and inflamed CD (0.20%) patients56. Moreover, A. muciniphila are more abundant in CD patients than in UC patients54,56.
However, the higher abundance of A. muciniphila may not be negatively correlated with IBD. A surprising result shows in both CRC patients and CRC mice, the abundance of A. muciniphila is higher than that in healthy people57,58 (Supplementary Table 1). Moreover, A. muciniphila is enriched in the early stage of CRC59. The abundance of A. muciniphila may also be increased by pathogenic infection60,61 (Supplementary Table 1).
Diet and lifestyle can regulate the abundance of A. muciniphila
Diet is an important factor that cannot be ignored to shape the gut microbiota62,63. We summarized previous studies and focused on the relationship between the abundance of A. muciniphila and dietary ingredients, which are associated with host health and intestinal diseases. The high-concentration of cellulose in the diet can relieve the inflammation of dextran sodium sulfate (DSS)-induced mice, while increasing the abundance of A. muciniphila64. A diet enriched with rye bran and wheat aleurone is reported to increase the relative abundance of Akkermansia in C57BL/6 J mice, accompanied by changes in glycine betaine metabolism65. Both sugarcane bagasse, a water-soluble fiber, and xylo-oligosaccharide can also increase the abundance of Akkermansia in Fischer 344 rats66. Milk and its products, for example, breast milk can promote the growth of A. muciniphila in mice transplanted with microbiota from infant67, which may be triggered by galacto-N-biose68. Another study revealed that the consumption of cheese is negatively associated with the abundance of A. muciniphila69. The increase of A. muciniphila by dietary supplement of polyphenol containing grape proanthocyanidin, chlorogenic acid, and resveratrol is accompanied by the improvement of metabolic profile and anti-inflammatory activities of host, especially in mice with DSS-induced colitis70–72. Interestingly, grape proanthocyanidin may indirectly induce the intestinal bloom of A. muciniphila, in vivo, in mice, but shows no effect on the quantity of A. muciniphila in vitro70. Probiotics, such as Lactobacillus fermentum and Bacillus subtilis, can alleviate DSS-induced colits in mice and increase the abundance of Akkermansia73,74. In contrast, other probiotics, such as Bifidobacteria adolescentis, is found to inhibit the excessive growth of A. muciniphila during the therapy of DSS-induced chronic colitis75. Similarly, Pediococcus pentosaceus and Lactobacillus coryniformis can ameliorate CRC in mice via regulating gut microbiota, including increasing the abundance of A. muciniphila76,77. Particular dietary patterns, such as low-calorie diet, ketogenic diet, and fasting, are reported to increase the abundance of A. muciniphila in healthy individuals, or IBD patients78–81. It is worth noting that gut microbial composition can be influenced by many factors, especially stool consistency and fecal transit time, which are closely connected with the abundance of A. muciniphila82,83. To summarize, A. muciniphila may participate in the effect of diet on IBD, but whether the change of A. muciniphila abundance is the cause, or result, remains to be determined.
A. muciniphila and intestinal homeostasis of host
A. muciniphila and the intestinal physical barrier of host
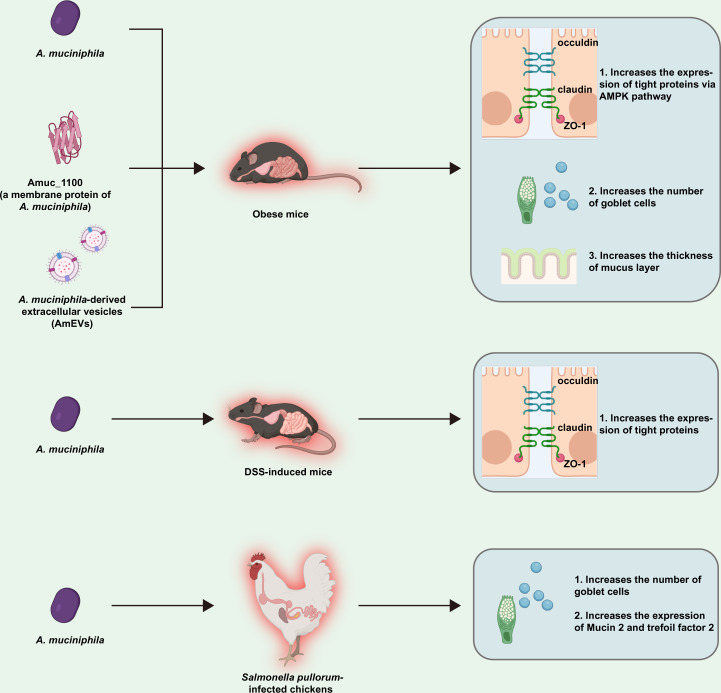
Live A. muciniphila bacteria have been repeatedly confirmed to be related to the improvement of the intestinal barrier. Oral gavage with live A. muciniphila can increase the expression of tight junction proteins (TJs), such as zonula occludens (ZO-1) and occludin, in DSS-induced mice84. In vitro, active A. muciniphila bacteria are also found to increase the transepithelial electrical resistance (TER), a recognized parameter to reflect the cell integrity of the cell membrane85 of cocultured Caco-2 cells after 24 or 48 h18,86. Particularly, some cellular components of A. muciniphila have also been shown to improve the intestinal permeability. One of them is extracellular vesicles (AmEVs), the lipid bilayer secreted by A. muciniphila. Compared to obese mice induced by high-fat diet, or lipopolysaccharide (LPS)-induced Caco-2 cell, the expression of occludin, ZO-1, and claudin-5 is enhanced (in vivo and in vitro) by activating the adenosine monophosphate (AMP)-activated protein kinase (AMPK) pathway in a dose-dependent manner with oral administration of 10 μg AmEVs87. Moreover, after pasteurization88, a stable outer membrane of A. muciniphila, Amuc_1100, has been shown to increase the TER in vitro86 and the expression of TJ genes in the small intestine of obese mice induced by high-fat diet in vivo14. Amuc-1100 belongs to a gene cluster related to the formation of pilus86 and was recently used in mice with metabolic and intestinal diseases14,88.
As a mucin-specialist, the abundance of A. muciniphila is closely related to the thickness of the intestinal mucosa. A similar result is found in Apoe−/− mice fed western-diet89. Goblet cells, a specialized epithelial cell that secretes mucins, have attracted much attention because of their important role in maintaining the integrity of the inner mucus layer90. A gavage with 1.0 × 108 CFU/day of A. muciniphila (DSM 22959) can increase the density of goblet cells in the ileum of mice with a long-term feeding of high-fat diet91. Similarly, A. muciniphila bacteria are believed to increase the number of goblet cells and up-regulate the expression of Mucin 2 (MUC2) and trefoil factor 2 (Tff2) in Salmonella pullorum-infected chickens92. A genome-wide association study (GWAS) based on 288 pigs revealed a correlation between the relative abundance of A. muciniphila and a gene encoding carbohydrate sulfotransferase 1293, a required gene for the biosynthesis of glycosaminoglycan and the formation of mucin94,95. It should be highlighted that the genome of A. muciniphila (ATCC BAA-835) lacks mucus-binding domains21, which is verified by an in vitro study that A. muciniphila can barely adhere to the mucus18. These results describe the protective effect of A. muciniphila on intestinal mucosa, which may be related to the increase of goblet cells (Fig. 2).
Fig. 2. The possible mechanisms of A. muciniphila regulating intestinal barrier summarized according to existing references.
All figures are created with Biorender.com.
A. muciniphila and the intestinal immunity of the host
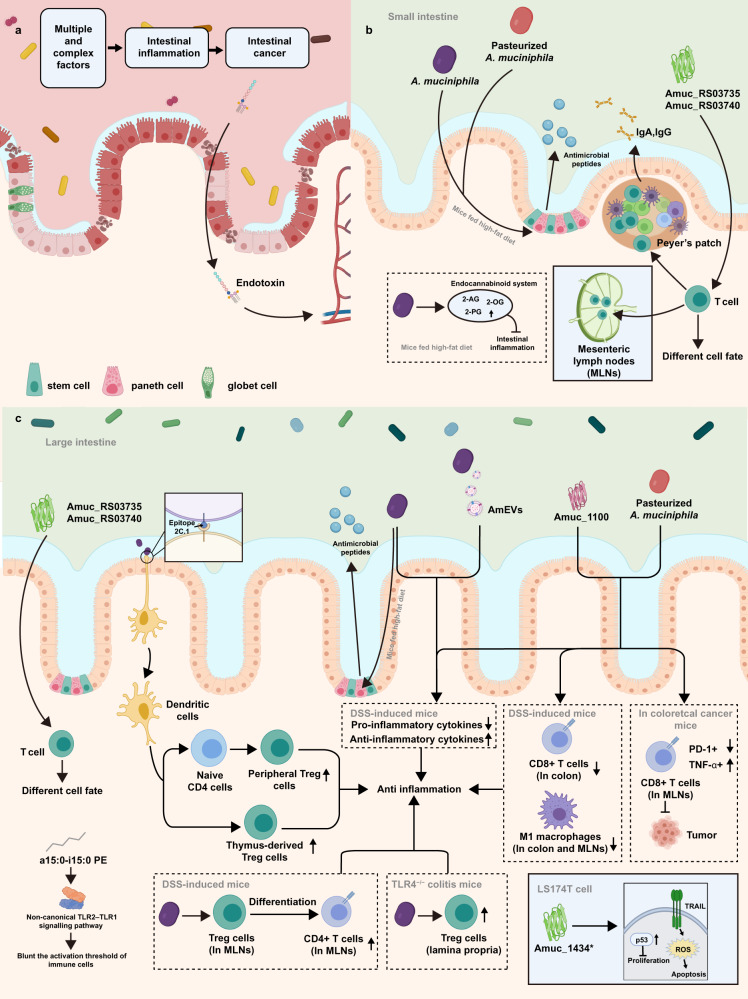
The intestinal inflammation involves the complex interaction of host genes, host immunity, microbiota and environmental factors (Fig. 3a). As a mucin-degrader in the gut, A. muciniphila can easily induce the immune response of the host due to its frequent communication with intestinal epithelial cells (IECs) (Fig. 3b and c). For instance, A. muciniphila increases the expression of genes encoding 2-oleoylglycerol, 2-arachidonoylglycerol and 2-palmitoylglycerol in the ileum of mice13, which are associated with the endocannabinoid system involving intestinal homeostasis and improved intestinal barriers96. When A. muciniphila is present in the intestine of specified pathogen free (SPF) mice, T cells response to A. muciniphila are localized to the Peyer’s patches (PPs), large intestine, small intestine lamina propria and mesenteric lymph nodes (mLNs), which is regulated by the outer membrane proteins Amuc_RS03735 and Amuc_RS0374097. In mice with oral treatment of live A. muciniphila, the differentiation of peripheral regulatory T cells (pTregs), the proliferation of residual thymus-derived Tregs (tTregs) in the colon (which reprogramed by epitope 2 C.1 from A. muciniphila98), and the differentiation of Foxp3+ Treg from CD4+ T cells in MLNs are found to be promoted99. Akkermansia muciniphila is also found to be positively correlated with TLR4 receptor and against TLR4−/− induced colitis in mice by increasing the proportion of RORγt+ Treg cells that enhances the immune response100. Whereas, in altered Schaedler flora (ASF) mice, the treatment of A. muciniphila specifically impacted the number of T follicular helper (TFH) cells only in the Peyer’s patches (PPs)97. As the TFH cells are important for the secretion of immunoglobulins (e.g. IgA), the variation in the quantity of these cells may help to slow down the advanced-stage intestinal inflammation101. Besides the proliferation, the development of immune cells is also involved in the abundance of A. muciniphila. In addition, both pasteurised A. muciniphila and Amuc_1100 can decrease the colonic infiltration of CD8+ cytotoxic T lymphocytes (CTLs), which aggravates colitis by mediating the production of cytokines102,103, and can suppress the proliferation of proinflammatory CD16/32+ macrophages in the MLNs and decrease the mRNA level of pro-inflammatory cytokines in mice with DSS-induced colitis88. In a mice model with CRC, pasteurised A. muciniphila and Amuc_1100 increased the activation of CTLs in the MLN and the proportion of tumor necrosis factor-alpha (TNF-α)+ CTLs to promote the apoptosis of tumor cells. Meanwhile, the proportion of PD-1+ CTLs in MLN can be decreased to suppress the growth of tumor88. Another protein of A. muciniphila, Amuc_1434, an aspartic protease can degrade MUC2 in vitro104, can inhibit the proliferation of LS174T cells and block the G0/G1 phase of cell cycle of LS174T cells by increasing the expression of tumor protein 53 (p53) in vitro105. Further, Amuc_1434* treatment promotes the apoptosis of LS174T cells and increases the level of mitochondrial reactive oxygen species (ROS) by upregulating tumor-necrosis-factor-related apoptosis-inducing ligand (TRAIL)105. The concentration of inflammatory cytokines can be used as an important indicator to assess the severity of intestinal inflammation. The pretreatment of A. muciniphila was found to suppress the expression of pro-inflammatory cytokines, such as interferon gamma (IFN-γ), interleukin-17 (IL-17), TNF-α, interleukin-1beta (IL-1β) and nitric oxide synthase 2 (NOS2), in the colon of mice with DSS-induced colitis106. Similarly, the mRNA level of pro-inflammatory cytokines, TNF-α, IFN-γ, IL-1β, IL-6, IL-18 and IL-33, in the colon of mice with DSS-induced colitis can be also decreased by the treatment of pasteurised A. muciniphila (1.5×108 CFU) or 3 µg of Amuc_110088. In vitro, the level of IL-6 in colonic epithelial cells (CT26), challenged by E. coli-derived extracellular vesicle, can be reduced by the pre-treatment of AmEVs in a dose-dependent manner107. Adiacyl phosphatidylethanolamine, with two branched chains (a15:0-i15:0 PE), isolated from A. muciniphila can cause the release of specific inflammatory cytokines by acting on the non-classical TLR2-TLR1 heterodimer, and at low doses, can blunt the activation threshold of immune cells108.
Fig. 3. The possible mechanisms of A. muciniphila regulating intestinal immunity in host with intestinal inflammation and colon cancer.
All figures are created with Biorender.com.
The production of antigen-specific T cell-dependent IgA and IgG1 in the serum of ASF mice is reported to be induced by acquiring A. muciniphila vertically from mothers97. Live A. muciniphila bacteria markedly increases the expression of regenerating islet derived 3-gamma (Reg3g)13, a lectin protecting the intestinal mucosa against the invasion of pathogens109, in the colon of mice fed high-fat diet. In contrast, both live and pasteurized A. muciniphila improved the expression of lysozyme C-1 (Lyz1) in the small intestine of obese mice induced by high-fat diet14 (Fig. 3b and c).
The interaction between A. muciniphila and the intestinal epithelium
A few studies suggest a direct effect of A. muciniphila on IECs. A linear discriminate analysis clearly shows the enrichment of A. muciniphila in the early regenerative mucosa of mice. The intrarectal administration of active A. muciniphila remarkably facilitate the closure of injured mucosa (from 43.7% to 74.14%) in mice by promoting the proliferation and migration of intestinal stem cells (ISCs) and accelerating the regeneration of the wound in SK-CO15 monolayers in vitro110. This requires the participation of formyl peptide receptor 1 (FPR1) and neutrophilic NADPH oxidase (NOX1) to increase ROS in the wound edge and the phosphorylation of extracellular-signal-regulated kinase (ERK) in colonic epithelial cells. In addition, the gavage of AmEVs isolated from A. muciniphila can alleviate dysplasia in C57BL/6 mice induced by 2% DSS107. Amuc_1100 (3 μg) can also relieve the shortening of colon and the histological injuries in the proximal colon in mice with DSS-induced colitis88, indicating an alleviation or even the repair of injured intestinal epithelium by A. muciniphila, or its derivatives.
The steady renewal of the IECs is fueled by ISCs lying at the basilar part of crypts111, which is particularly important in case of disrupted intestinal homeostasis. The colonization of A. muciniphila in the chicken colon is found to regulate the proliferation of ISCs though the classical Wnt/β-catenin signaling pathway92. In addition, A. muciniphila can closely bind to laminin18, one of the important components of extracellular matrix which can regulate the migration, differentiation and anti-inflammatory responses of IECs112–114. A GWAS based study showed a strong connection between laminin β1 chain encoding gene and the susceptibility of UC115, and showed the laminin γ1 chain encoding gene as a susceptible locus of IBD116. However, the interaction between A. muciniphila and laminin is still poorly understood. Therefore, as a bacterium that is directly communicated with intestinal mucosa, A. muciniphila displays an intervention in the proliferation and/or differentiation of IECs and ISCs, which represents a very complex cross-talk to be further discussed.
Relationship between A. muciniphila and other intestinal bacteria during intestinal inflammation
Although A. muciniphila is found to negatively correlate with total mucin-degrading bacteria, its decreased number may result in the proliferation of mucin-associated bacteria when intestinal inflammation occurs56. This can reduce the degradation of mucus and maintain a relatively stable intestinal barrier56. Several studies provide direct evidence for such interaction between A. muciniphila and other mucosa-associated bacteria. When cocultured with mucolytic bacteria like Bacteroides vulgatus, Ruminococcus gnavus, or Ruminococcus torques, in a defined medium with MUC2 as sole carbon source, the growth of A. muciniphila is inhibited while the growth of other bacteria is promoted56,117. On the other hand, A. muciniphila may influence the intestinal microbiota by regulating the intestinal immunity of the host13,118. Akkermansia muciniphila treatment accelerates the normalization of the microbial community in mice with colitis, and reverses the decreased ratio of Firmicutes/Bacteroidetes bacteria in the cecum caused by high-fat diet119. A correlation between the abundance of A. muciniphila and Faecalibacterium prausnitzii is also confirmed in the feces of CD patients120. Moreover, six genera (Prevotella, Sutterella, Klebsiella, Dorea, Parabacteroides, and Akkermansia) are found to flourish in CD patients with remission121. Furthermore, both A. muciniphila-F. prausnitzii, and A. muciniphila-Bacteroides thetaiotaomicron, in IBD patients, are lower than in healthy individuals54, suggesting a relationship between mutualistic symbiosis of mucolytic bacteria and IBD.
The negative effect of A. muciniphila in specific GIT environment
In several cases, A. muciniphila may have a negative impact on intestinal health (Table 2). Specifically, in a gnotobiotic C3H mouse model with eight bacterial species normally found in humans, the infection of Salmonella typhimurium with the pro-colonization of A. muciniphila makes the former a dominant bacterium in this limited microbiota accompanied by more severe intestinal inflammation16. Another study shows that A. muciniphila is able to induce colitis in specific-pathogen-free and germ-free Il10−/− mice and its colonization is mediated by Nod-like receptor 617. Low-fiber diet promotes expansion of A. muciniphila and other mucus-degrading bacteria in mice colonizing with a synthetic human gut microbiota, which promotes the degradation of the mucus layer and increases the colitis caused by Citrobacter rodentium infection15. In CRC mice transplanted with the fecal microbiota from CRC patients, Akkermansia bacteria are positively correlated with increased tumor burden122. In addition, gavage of A. muciniphila into intestine-specific Apc mutant mice (FabplCre; Apc15lox/+) aggravates the development of colorectal cancer by increasing the number of tumors123. In conclusion, A. muciniphila may be at risk of exacerbating pathogenic infections and inflammation of intestine, which is a common problem to be considered in mucin-degrading bacteria124.
Table 2.
The negative effects of A. muciniphila on intestinal disease in some special cases.
| Author/Year | Object | Model | Experimental design | Negative effect |
|---|---|---|---|---|
| Mahesh S. Desai et al.15 | Mouse | Low-fiber diet and pathogen infection | Gnotobiotic mice are constructed with a synthetic gut microbiota from fully sequenced human gut bacteria, fed a fiber-deprivation diet (chronic or intermittent) and used Citrobacter rodentium to infect mice with two diet models to investigate the mechanistic connections between dietary fiber deficiency and microbiota composition, as well as the resulting effects on the mucus barrier. | Low-fiber diet promotes expansion and activity of mucus-degrading bacteria, such as A. muciniphila, which alleviates the degradation of the mucus layer and increases the susceptibility of pathogen-associated colitis. |
| Sergey S. Seregin et al.17 | Mouse | Immune deficiency disorders associated with IBD | 16 S rRNA sequencing is used to analyze the change of gut microbiota in Il10−/− mice with spontaneous colitis and innate immune receptor NLRP6 deficiency, and oral gavage of screened strains is performed to investigate its effects in these mice. |
1. The relative abundance of A. muciniphila is significantly increased in Il10−/− Nlrp6−/− mice. 2. A. muciniphila promotes colitis represented by the decreasing of body weight, as well as the increase of the colonic histological scores, weight of spleen, inflammation indication of colon, level of fecal Lcn-2, bacterial translocation to MLNs and pro-inflammatory mediators in the colons of both SPF Il10−/− mice and germ-free Il10−/− mice. |
| Héctor Argüello et al.131 | Pig | S. typhimurium infection | 16 S rRNA sequencing is used to analyze the composition of mucosa microbiome in the ileum of 28 days old pigs with S. typhimurium infection. |
1. Genus Akkermansia increases within the mucosa of the S. typhimurium infected pigs. 2. Epithelial damage is positively correlated to taxa belonging to the phyla Verrucomicrobia such as A. muciniphila. |
| Bhanu Priya Ganesh et al.16 | Mouse | S. typhimurium infection | Oral gavage of A. muciniphila followed by subsequently infection of S. typhimurium in gnotobiotic C3H mouse model with a background microbiota of eight bacterial species to research the impact of A. muciniphila on inflammatory and infectious symptoms. |
1. After 5 days infection, S. typhimurium become the predominant species representing 94.03% of total bacteria in the cecum of mice co-colonized by A. muciniphila and S. typhimurium. 2. Co-colonization of A. muciniphila and S. typhimurium causes significantly higher histological scores and elevates the mRNA levels of pro-inflammatory cytokines, especially IFN-γ, IP-10, TNF-α, IL-12, IL-6, IL-17 in the cecum and colon of the infected mice. 3. The number of mucin-filled goblet cells, the thickness of mucus and mucus sulphation are significantly decreased by the co-colonization of A. muciniphila and S. typhimurium. 4. The existence of A. muciniphila may induce the deeper colonization of S. typhimurium in cecal tissue and encourages the recruitment of macrophages into the cecal lamella propria and submucosa. |
| Nielson T Baxter et al.122 | Mouse | CRC | The fecal microbiota from three CRC patients and three healthy individuals are transplanted into germ-free mice, respectively. then, these mice are chemically induced to CRC resulting in different levels of tumorigenesis. The change of gut microbiome is investigated using 16 S rRNA sequencing and metagenomic analysis. | The taxa most strongly positively correlate with increased tumor burden are several Gram-negative species including Akkermansia. |
| Joseph P. Zackular et al.132 | Mouse | CRC | The development of microbiome during the tumorigenesis in a mouse model with inflammation-driven colon cancer is investigated using 16 S rRNA sequencing. |
1. Tumor-bearing mice show an enrichment in OTUs affiliated with members of Akkermansia. 2. The tumorigenesis in the colon of germ-free mice transplanted with the fecal microbiota from mice with tumor is increased. |
| Celia Dingemanse et al.123 | Mouse | CRC | Shotgun metagenomic sequencing plus quantitative PCR is used to analyze the gut microbiota in intestine-specific conditional Apc mutant mice (FabplCre; Apc15lox/+) with large intestine tumor. Then, the FabplCre; Apc15lox/+ mice are treated with the identified specific bacteria by orally gavage to investigate their impact on the development of tumor. |
1. Metagenomic sequencing shows that the genus Akkermansia is responsible for the overrepresentation in the conventional samples with more intestinal tumors. 2. The oral gavage of A. muciniphila to antibiotic-pretreated FabplCre; Apc15lox/+ mice significantly increases the number of intestinal tumors. 3. A. muciniphila significantly increases the thickness of intestinal mucus layer and the goblet cell ratio in FabplCre; Apc15lox/+ mice which may aggravate adenomatous in tumor-susceptive mice. |
The inspiration of precise application: strain-specific role of A. muciniphila on host intestinal health associated with its genetic and phenotypic properties
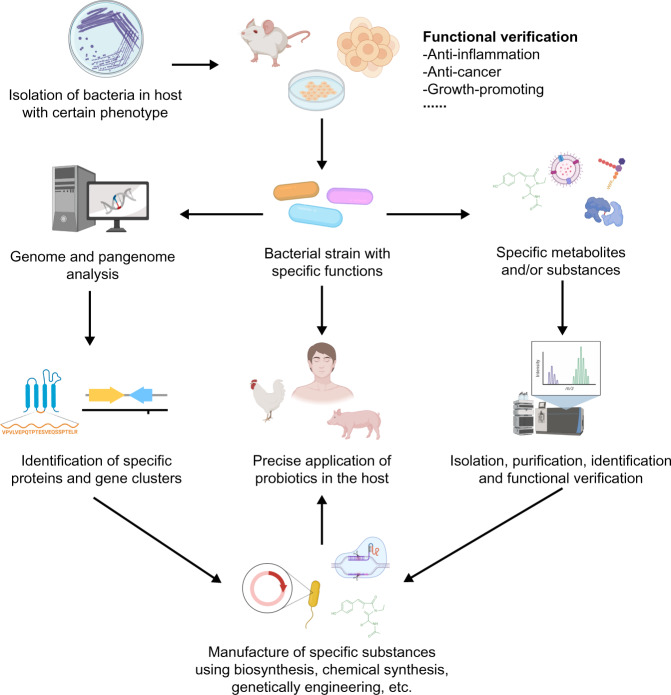
The role of probiotics largely depends on the bacterial strains used, which is essential for their clinical effects125. Different bacterial strains have distinct genomic homology leading to discrepant function126,127, which makes it reasonable to consider the practical application of different strains. A total of 106 A. muciniphila metagenome-assembled genomes (MAGs) have been reconstructed based on the available metagenomic datasets of human, mouse and pig, which revealed three phylogroups of A. muciniphila, AmI, AmII and AmIII with different relative abundance22. Based on the whole-genome shotgun sequencing of 39 isolates of A. muciniphila, from human and mouse feces, three A. muciniphila phylogroups (AmI, AmII and AmIII) are identified and the functional annotation shows their distinct metabolic and functional features22. The comparative genomic analysis based on 35 metagenome-assembled genomes (MAGs) and 40 publicly available genomes further reveals at least four phylogroups of A. muciniphila (AmI to AmIV) and some strains in specific phylogroup have the genes and ability to vitamin B12 biosynthesis23. A study including genomic analysis and phenotypic test shows distinct characteristics of these phylogroups, including oxygen tolerance, cell adherence, the activation of toll-like receptor 2, sulfur acquisition and the colonization of the bacterium in GIT19. A large-scale metagenomic-based genomic analysis further confirms that the genomic difference may diversify the effect of A. muciniphila strains on host health128,129, and results of in vivo and in vitro studies support this hypothesis. In mice with chronic colitis, A. muciniphila strain ATCC 835 presents better anti-inflammatory properties than strain 13999. Of 11 human-derived A. muciniphila strains, only the supernatant from a culture of the AK32 strain can increase the size of small intestine-derived organoids in vitro130. It can be assumed that the function of different A. muciniphila strains may be various, possibly due to the diversity in their cellular components and metabolites, although most related studies focus on A. muciniphila ATCC 835. Moreover, function-specific component of different A. muciniphila strains, or their metabolites may be mass produced or recombined to investigate and reveal the effects and mechanism of A. muciniphila targeting diseases (Fig. 4). Based on the understanding of functional characterization of A. muciniphila strains, studies on the phenotypes of A. muciniphila in vitro and its effect on the host are required for the precise application of A. muciniphila in disease treatment.
Fig. 4. A schematic diagram of workflow on the precise application of NGP.
All figures are created with Biorender.com.
In summary, regardless of host animal species, A. muciniphila is found to be more abundant in the hindgut. The abundance of A. muciniphila in the human GIT increases with age, which is contrary to that in mice. Types of intestinal diseases, dietary supplements, as well as other mucus-associated microbes can influence the abundance of A. muciniphila, but cautious consideration should be given to A. muciniphila as a biomarker for indicating an intestinal health risk. Akkermansia muciniphila may safety be administered in healthy individuals or those with metabolic syndrome (excess fat around the waist, high blood sugar, increased blood pressure, and abnormal cholesterol levels). Akkermansia muciniphila may also be beneficial to the maintenance of intestinal homeostasis of the host. However, in some cases, such as the lack of dietary fiber, pathogenic infection, or specific host genotypes, the accumulation of A. muciniphila in the GIT may exacerbate the damage of the intestinal epithelium, indicating that A. muciniphila may have a double-edged effect on the intestinal health of the host. In view of the strain-specific genome and phenotype of A. muciniphila, a clear description and discussion of each strain is critical before its practical application.
Supplementary information
Acknowledgements
This work is supported by the National Natural Science Foundation of China (grant numbers 31872369 and 32072743). The authors thank Yifan Bao (Department of Physiological Chemistry, University of Vienna) to help draw figures.
Author contributions
Y.L., C.L., H.L. and Q.O. collected the references, conceived and wrote the manuscript and share equal contribution; F.K. and A.W. helped to carry out pan genomic analysis. Z.R., G.T. J.C., and A.D.W. helped to collect references and revise the manuscript; B.Y. and J.H. helped to prepare and organize the tables and figures. All authors have read the completed version of the manuscript and agreed to its publication.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yuheng Luo, Cong Lan, Hua Li, Qingyuan Ouyang.
Supplementary information
The online version contains supplementary material available at 10.1038/s41522-022-00338-4.
References
- 1.Derrien M, Vaughan EE, Plugge CM, de Vos WM. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int. J. Syst. Evolut. Microbiol. 2004;54:1469–1476. doi: 10.1099/ijs.0.02873-0. [DOI] [PubMed] [Google Scholar]
- 2.Sheehan D, Shanahan F. The Gut Microbiota in Inflammatory Bowel Disease. Gastroenterol. Clin. North Am. 2017;46:143–154. doi: 10.1016/j.gtc.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 3.Alam MS, Gangiredla J, Hasan NA, Barnaba T, Tartera C. Aging-Induced Dysbiosis of Gut Microbiota as a Risk Factor for Increased Listeria monocytogenes Infection. Front. Immunol. 2021;12:672353. doi: 10.3389/fimmu.2021.672353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dowd, S. E. et al. Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP. 8, 125 (2008). BMC Microbiol. 8, 125 (2008). [DOI] [PMC free article] [PubMed]
- 5.Hildebrand F, et al. A comparative analysis of the intestinal metagenomes present in guinea pigs (Cavia porcellus) and humans (Homo sapiens). BMC Genomics. 2012;13:514. doi: 10.1186/1471-2164-13-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCormack UM, et al. Exploring a Possible Link between the Intestinal Microbiota and Feed Efficiency in Pigs. Appl. Environ. Microbiol. 2017;83:e00380–17. doi: 10.1128/AEM.00380-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fang S, et al. Faecal microbiota and functional capacity associated with weaning weight in meat rabbits. Microb. Biotechnol. 2019;12:1441–1452. doi: 10.1111/1751-7915.13485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Videvall E, et al. Major shifts in gut microbiota during development and its relationship to growth in ostriches. Mol. Ecol. 2019;28:2653–2667. doi: 10.1111/mec.15087. [DOI] [PubMed] [Google Scholar]
- 9.Kubasova T, et al. Gut Anaerobes Capable of Chicken Caecum Colonisation. Microorganisms. 2019;7:597. doi: 10.3390/microorganisms7120597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Derrien M, Collado MC, Ben-Amor K, Salminen S, de Vos WM. The Mucin degrader Akkermansia muciniphila is an abundant resident of the human intestinal tract. Appl. Environ. Microbiol. 2008;74:1646–1648. doi: 10.1128/AEM.01226-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindén SK, Florin THJ, McGuckin MA. Mucin dynamics in intestinal bacterial infection. PloS one. 2008;3:e3952. doi: 10.1371/journal.pone.0003952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Depommier C, et al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: a proof-of-concept exploratory study. Nat. Med. 2019;25:1096–1103. doi: 10.1038/s41591-019-0495-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Everard A, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. USA. 2013;110:9066–9071. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plovier H, et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat. Med. 2017;23:107–113. doi: 10.1038/nm.4236. [DOI] [PubMed] [Google Scholar]
- 15.Desai MS, et al. A Dietary Fiber-Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility. Cell. 2016;167:1339–1353. doi: 10.1016/j.cell.2016.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ganesh BP, Klopfleisch R, Loh G, Blaut M. Commensal Akkermansia muciniphila exacerbates gut inflammation in Salmonella Typhimurium-infected gnotobiotic mice. PloS one. 2013;8:e74963. doi: 10.1371/journal.pone.0074963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seregin SS, et al. NLRP6 Protects Il10−/− Mice from Colitis by Limiting Colonization of Akkermansia muciniphila. Cell Rep. 2017;19:733–745. doi: 10.1016/j.celrep.2017.03.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reunanen J, et al. Akkermansia muciniphila Adheres to Enterocytes and Strengthens the Integrity of the Epithelial Cell Layer. Appl. Environ. Microbiol. 2015;81:3655–3662. doi: 10.1128/AEM.04050-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Becken B, et al. Genotypic and Phenotypic Diversity among Human Isolates of Akkermansia muciniphila. mBio. 2021;12:e00478–21. doi: 10.1128/mBio.00478-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ottman N, et al. Genome-Scale Model and Omics Analysis of Metabolic Capacities of Akkermansia muciniphila Reveal a Preferential Mucin-Degrading Lifestyle. Appl. Environ. Microbiol. 2017;83:e01014–e01017. doi: 10.1128/AEM.01014-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Passel MWJ, et al. The genome of Akkermansia muciniphila, a dedicated intestinal mucin degrader, and its use in exploring intestinal metagenomes. PloS one. 2011;6:e16876. doi: 10.1371/journal.pone.0016876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo X, et al. Genome sequencing of 39 Akkermansia muciniphila isolates reveals its population structure, genomic and functional diverisity, and global distribution in mammalian gut microbiotas. BMC genomics. 2017;18:800. doi: 10.1186/s12864-017-4195-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirmiz N, et al. Comparative Genomics Guides Elucidation of Vitamin B12 Biosynthesis in Novel Human-Associated Akkermansia Strains. Appl. Environ. Microbiol. 2020;86:e02117–e02119. doi: 10.1128/AEM.02117-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cozzolino A, et al. Preliminary Evaluation of the Safety and Probiotic Potential of Akkermansia muciniphila DSM 22959 in Comparison with Lactobacillus rhamnosus GG. Microorganisms. 2020;8:189. doi: 10.3390/microorganisms8020189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dubourg G, et al. High-level colonisation of the human gut by Verrucomicrobia following broad-spectrum antibiotic treatment. Int. J. antimicrobial agents. 2013;41:149–155. doi: 10.1016/j.ijantimicag.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 26.Druart C, et al. Toxicological safety evaluation of pasteurized Akkermansia muciniphila. J. Appl. Toxicol.: JAT. 2021;41:276–290. doi: 10.1002/jat.4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turck D, et al. Safety of pasteurised Akkermansia muciniphila as a novel food pursuant to Regulation (EU) 2015/2283. EFSA J. Eur. Food Saf. Auth. 2021;19:e06780. doi: 10.2903/j.efsa.2021.6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li G, et al. Diversity of Duodenal and Rectal Microbiota in Biopsy Tissues and Luminal Contents in Healthy Volunteers. J. Microbiol. Biotechnol. 2015;25:1136–1145. doi: 10.4014/jmb.1412.12047. [DOI] [PubMed] [Google Scholar]
- 29.Rogers MB, et al. Disturbances of the Perioperative Microbiome Across Multiple Body Sites in Patients Undergoing Pancreaticoduodenectomy. Pancreas. 2017;46:260–267. doi: 10.1097/MPA.0000000000000726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang M, Ahrné S, Jeppsson B, Molin G. Comparison of bacterial diversity along the human intestinal tract by direct cloning and sequencing of 16S rRNA genes. FEMS Microbiol. Ecol. 2005;54:219–231. doi: 10.1016/j.femsec.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 31.Madsen JL. Effects of gender, age, and body mass index on gastrointestinal transit times. Digestive Dis. Sci. 1992;37:1548–1553. doi: 10.1007/BF01296501. [DOI] [PubMed] [Google Scholar]
- 32.Johansson MEV, et al. Composition and functional role of the mucus layers in the intestine. Cell. Mol. life Sci.: CMLS. 2011;68:3635–3641. doi: 10.1007/s00018-011-0822-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Derrien M, et al. Modulation of Mucosal Immune Response, Tolerance, and Proliferation in Mice Colonized by the Mucin-Degrader Akkermansia muciniphila. Front. Microbiol. 2011;2:166. doi: 10.3389/fmicb.2011.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van den Abbeele P, et al. Arabinoxylans and inulin differentially modulate the mucosal and luminal gut microbiota and mucin-degradation in humanized rats. Environ. Microbiol. 2011;13:2667–2680. doi: 10.1111/j.1462-2920.2011.02533.x. [DOI] [PubMed] [Google Scholar]
- 35.Lyra A, et al. Comparison of bacterial quantities in left and right colon biopsies and faeces. World J. Gastroenterol. 2012;18:4404–4411. doi: 10.3748/wjg.v18.i32.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ringel Y, et al. High throughput sequencing reveals distinct microbial populations within the mucosal and luminal niches in healthy individuals. Gut microbes. 2015;6:173–181. doi: 10.1080/19490976.2015.1044711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Evans DF, et al. Measurement of gastrointestinal pH profiles in normal ambulant human subjects. Gut. 1988;29:1035–1041. doi: 10.1136/gut.29.8.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Herreweghen F, et al. In vitro colonisation of the distal colon by Akkermansia muciniphila is largely mucin and pH dependent. Beneficial microbes. 2017;8:81–96. doi: 10.3920/BM2016.0013. [DOI] [PubMed] [Google Scholar]
- 39.Bäckhed F, et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell host microbe. 2015;17:690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 40.Guo M, et al. Developmental differences in the intestinal microbiota of Chinese 1-year-old infants and 4-year-old children. Sci. Rep. 2020;10:19470. doi: 10.1038/s41598-020-76591-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kong F, et al. Gut microbiota signatures of longevity. Curr. Biol.: CB. 2016;26:R832–R833. doi: 10.1016/j.cub.2016.08.015. [DOI] [PubMed] [Google Scholar]
- 42.Biagi E, et al. Gut Microbiota and Extreme Longevity. Curr. Biol.: CB. 2016;26:1480–1485. doi: 10.1016/j.cub.2016.04.016. [DOI] [PubMed] [Google Scholar]
- 43.Bárcena C, et al. Healthspan and lifespan extension by fecal microbiota transplantation into progeroid mice. Nat. Med. 2019;25:1234–1242. doi: 10.1038/s41591-019-0504-5. [DOI] [PubMed] [Google Scholar]
- 44.Salazar N, et al. Age-Associated Changes in Gut Microbiota and Dietary Components Related with the Immune System in Adulthood and Old Age: A Cross-Sectional Study. Nutrients. 2019;11:1765. doi: 10.3390/nu11081765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim B-S, et al. Comparison of the Gut Microbiota of Centenarians in Longevity Villages of South Korea with Those of Other Age Groups. J. Microbiol. Biotechnol. 2019;29:429–440. doi: 10.4014/jmb.1811.11023. [DOI] [PubMed] [Google Scholar]
- 46.Rampelli S, et al. Shotgun Metagenomics of Gut Microbiota in Humans with up to Extreme Longevity and the Increasing Role of Xenobiotic Degradation. mSystems. 2020;5:e00124–20. doi: 10.1128/mSystems.00124-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van der Lugt B, et al. Akkermansia muciniphila ameliorates the age-related decline in colonic mucus thickness and attenuates immune activation in accelerated aging Ercc1-/Δ7 mice. Immun. Ageing.: I A. 2019;16:6. doi: 10.1186/s12979-019-0145-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bodogai M, et al. Commensal bacteria contribute to insulin resistance in aging by activating innate B1a cells. Sci. Transl. Med. 2018;10:aat4271. doi: 10.1126/scitranslmed.aat4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van der Lugt B, et al. Integrative analysis of gut microbiota composition, host colonic gene expression and intraluminal metabolites in aging C57BL/6J mice. Aging. 2018;10:930–950. doi: 10.18632/aging.101439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang X, et al. Age-related compositional changes and correlations of gut microbiome, serum metabolome, and immune factor in rats. GeroScience. 2021;43:709–725. doi: 10.1007/s11357-020-00188-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grivennikov SI. Inflammation and colorectal cancer: colitis-associated neoplasia. Semin. Immunopathol. 2013;35:229–244. doi: 10.1007/s00281-012-0352-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jemal A, et al. Cancer Statistics, 2007. CA: A Cancer J. Clinicians. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 53.Vigsnæs LK, Brynskov J, Steenholdt C, Wilcks A, Licht TR. Gram-negative bacteria account for main differences between faecal microbiota from patients with ulcerative colitis and healthy controls. Beneficial microbes. 2012;3:287–297. doi: 10.3920/BM2012.0018. [DOI] [PubMed] [Google Scholar]
- 54.Zhang T, et al. Alterations of Akkermansia muciniphila in the inflammatory bowel disease patients with washed microbiota transplantation. Appl. Microbiol. Biotechnol. 2020;104:10203–10215. doi: 10.1007/s00253-020-10948-7. [DOI] [PubMed] [Google Scholar]
- 55.Earley H, et al. The abundance of Akkermansia muciniphila and its relationship with sulphated colonic mucins in health and ulcerative colitis. Sci. Rep. 2019;9:15683. doi: 10.1038/s41598-019-51878-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Png CW, et al. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am. J. Gastroenterol. 2010;105:2420–2428. doi: 10.1038/ajg.2010.281. [DOI] [PubMed] [Google Scholar]
- 57.Song C-H, et al. Changes in Microbial Community Composition Related to Sex and Colon Cancer by Nrf2 Knockout. Front. Cell. Infect. Microbiol. 2021;11:636808. doi: 10.3389/fcimb.2021.636808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lang M, et al. Crypt residing bacteria and proximal colonic carcinogenesis in a mouse model of Lynch syndrome. Int. J. cancer. 2020;147:2316–2326. doi: 10.1002/ijc.33028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Han S, et al. Adequate Lymph Node Assessments and Investigation of Gut Microorganisms and Microbial Metabolites in Colorectal Cancer. OncoTargets Ther. 2020;13:1893–1906. doi: 10.2147/OTT.S242017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vakili B, Fateh A, Asadzadeh Aghdaei H, Sotoodehnejadnematalahi F, Siadat SD. Characterization of Gut Microbiota in Hospitalized Patients with Clostridioides difficile Infection. Curr. Microbiol. 2020;77:1673–1680. doi: 10.1007/s00284-020-01980-x. [DOI] [PubMed] [Google Scholar]
- 61.Borton MA, et al. Chemical and pathogen-induced inflammation disrupt the murine intestinal microbiome. Microbiome. 2017;5:47. doi: 10.1186/s40168-017-0264-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bolte LA, et al. Long-term dietary patterns are associated with pro-inflammatory and anti-inflammatory features of the gut microbiome. Gut. 2021;70:1287–1298. doi: 10.1136/gutjnl-2020-322670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.David, L. A. et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature505, 10.1038/nature12820 (2014). [DOI] [PMC free article] [PubMed]
- 64.Kim Y, et al. Dietary cellulose prevents gut inflammation by modulating lipid metabolism and gut microbiota. Gut microbes. 2020;11:944–961. doi: 10.1080/19490976.2020.1730149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Koistinen VM, et al. Contribution of gut microbiota to metabolism of dietary glycine betaine in mice and in vitro colonic fermentation. Microbiome. 2019;7:103. doi: 10.1186/s40168-019-0718-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pelpolage SW, et al. Colonic fermentation of water soluble fiber fraction extracted from sugarcane (Sacchurum officinarum L.) bagasse in murine models. Food Chem. 2019;292:336–345. doi: 10.1016/j.foodchem.2019.04.063. [DOI] [PubMed] [Google Scholar]
- 67.Li N, et al. Human milk and infant formula modulate the intestinal microbiota and immune systems of human microbiota-associated mice. Food Funct. 2021;12:2784–2798. doi: 10.1039/D0FO03004J. [DOI] [PubMed] [Google Scholar]
- 68.Rubio-Del-Campo A, et al. Infant gut microbiota modulation by human milk disaccharides in humanized microbiome mice. Gut microbes. 2021;13:1–20. doi: 10.1080/19490976.2021.1914377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Partula V, et al. Associations between usual diet and gut microbiota composition: results from the Milieu Intérieur cross-sectional study. Am. J. Clin. Nutr. 2019;109:1472–1483. doi: 10.1093/ajcn/nqz029. [DOI] [PubMed] [Google Scholar]
- 70.Zhang L, et al. Grape proanthocyanidin-induced intestinal bloom of Akkermansia muciniphila is dependent on its baseline abundance and precedes activation of host genes related to metabolic health. J. nutritional Biochem. 2018;56:142–151. doi: 10.1016/j.jnutbio.2018.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang Z, et al. Chlorogenic Acid Ameliorates Experimental Colitis by Promoting Growth of Akkermansia in Mice. Nutrients. 2017;9:677. doi: 10.3390/nu9070677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen M, et al. Resveratrol attenuates high-fat diet-induced non-alcoholic steatohepatitis by maintaining gut barrier integrity and inhibiting gut inflammation through regulation of the endocannabinoid system. Clin. Nutr. (Edinb., Scotl.) 2020;39:1264–1275. doi: 10.1016/j.clnu.2019.05.020. [DOI] [PubMed] [Google Scholar]
- 73.Jang YJ, Kim W-K, Han DH, Lee K, Ko G. Lactobacillus fermentum species ameliorate dextran sulfate sodium-induced colitis by regulating the immune response and altering gut microbiota. Gut microbes. 2019;10:696–711. doi: 10.1080/19490976.2019.1589281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu Y, et al. Long-term and continuous administration of Bacillus subtilis during remission effectively maintains the remission of inflammatory bowel disease by protecting intestinal integrity, regulating epithelial proliferation, and reshaping microbial structure and function. Food Funct. 2021;12:2201–2210. doi: 10.1039/D0FO02786C. [DOI] [PubMed] [Google Scholar]
- 75.Fan L, et al. B. adolescentis ameliorates chronic colitis by regulating Treg/Th2 response and gut microbiota remodeling. Gut microbes. 2021;13:1–17. doi: 10.1080/19490976.2020.1826746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chung Y, et al. A synthetic probiotic engineered for colorectal cancer therapy modulates gut microbiota. Microbiome. 2021;9:122. doi: 10.1186/s40168-021-01071-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang, T. et al. Lactobacillus coryniformis MXJ32 administration ameliorates azoxymethane/dextran sulfate sodium-induced colitis-associated colorectal cancer via reshaping intestinal microenvironment and alleviating inflammatory response. European journal of nutrition, 10.1007/s00394-021-02627-8 (2021). [DOI] [PubMed]
- 78.Wu X, Unno T, Kang S, Park S. A Korean-Style Balanced Diet Has a Potential Connection with Ruminococcaceae Enterotype and Reduction of Metabolic Syndrome Incidence in Korean Adults. Nutrients. 2021;13:495. doi: 10.3390/nu13020495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li L, et al. The effects of daily fasting hours on shaping gut microbiota in mice. BMC Microbiol. 2020;20:65. doi: 10.1186/s12866-020-01754-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zheng J, et al. Dietary inflammatory potential in relation to the gut microbiome: results from a cross-sectional study. Br. J. Nutr. 2020;124:931–942. doi: 10.1017/S0007114520001853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kong C, et al. Ketogenic diet alleviates colitis by reduction of colonic group 3 innate lymphoid cells through altering gut microbiome. Signal Transduct. Target. Ther. 2021;6:154. doi: 10.1038/s41392-021-00549-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vandeputte D, et al. Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates. Gut. 2016;65:57–62. doi: 10.1136/gutjnl-2015-309618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Asnicar F, et al. Blue poo: impact of gut transit time on the gut microbiome using a novel marker. Gut. 2021;70:1665–1674. doi: 10.1136/gutjnl-2020-323877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bian X, et al. Administration of Akkermansia muciniphila Ameliorates Dextran Sulfate Sodium-Induced Ulcerative Colitis in Mice. Front. Microbiol. 2019;10:2259. doi: 10.3389/fmicb.2019.02259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Blikslager AT, Moeser AJ, Gookin JL, Jones SL, Odle J. Restoration of barrier function in injured intestinal mucosa. Physiological Rev. 2007;87:545–564. doi: 10.1152/physrev.00012.2006. [DOI] [PubMed] [Google Scholar]
- 86.Ottman N, et al. Pili-like proteins of Akkermansia muciniphila modulate host immune responses and gut barrier function. PloS one. 2017;12:e0173004. doi: 10.1371/journal.pone.0173004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chelakkot C, et al. Akkermansia muciniphila-derived extracellular vesicles influence gut permeability through the regulation of tight junctions. Exp. Mol. Med. 2018;50:e450. doi: 10.1038/emm.2017.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang L, et al. A purified membrane protein from Akkermansia muciniphila or the pasteurised bacterium blunts colitis associated tumourigenesis by modulation of CD8+ T cells in mice. Gut. 2020;69:1988–1997. doi: 10.1136/gutjnl-2019-320105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li J, Lin S, Vanhoutte PM, Woo CW, Xu A. Akkermansia Muciniphila Protects Against Atherosclerosis by Preventing Metabolic Endotoxemia-Induced Inflammation in Apoe−/− Mice. Circulation. 2016;133:2434–2446. doi: 10.1161/CIRCULATIONAHA.115.019645. [DOI] [PubMed] [Google Scholar]
- 90.Hansson GC, Johansson ME. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Gut microbes. 2010;1:51–54. doi: 10.4161/gmic.1.1.10470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shin J, et al. Elucidation of Akkermansia muciniphila Probiotic Traits Driven by Mucin Depletion. Front. Microbiol. 2019;10:1137. doi: 10.3389/fmicb.2019.01137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhu L, et al. Akkermansia muciniphila protects intestinal mucosa from damage caused by S. pullorum by initiating proliferation of intestinal epithelium. Vet. Res. 2020;51:34. doi: 10.1186/s13567-020-00755-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Crespo-Piazuelo D, et al. Association between the pig genome and its gut microbiota composition. Sci. Rep. 2019;9:8791. doi: 10.1038/s41598-019-45066-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hiraoka N, et al. Molecular cloning and expression of two distinct human chondroitin 4-O-sulfotransferases that belong to the HNK-1 sulfotransferase gene family. J. Biol. Chem. 2000;275:20188–20196. doi: 10.1074/jbc.M002443200. [DOI] [PubMed] [Google Scholar]
- 95.Ouwerkerk JP, Vos WMde, Belzer C. Glycobiome: bacteria and mucus at the epithelial interface. Best. Pract. Res. Clin. Gastroenterol. 2013;27:25–38. doi: 10.1016/j.bpg.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 96.Muccioli GG, et al. The endocannabinoid system links gut microbiota to adipogenesis. Mol. Syst. Biol. 2010;6:392. doi: 10.1038/msb.2010.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ansaldo E, et al. Akkermansia muciniphila induces intestinal adaptive immune responses during homeostasis. Sci. (N. Y.) 2019;364:1179–1184. doi: 10.1126/science.aaw7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kuczma MP, et al. Commensal epitopes drive differentiation of colonic Tregs. Sci. Adv. 2020;6:eaaz3186. doi: 10.1126/sciadv.aaz3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhai R, et al. Strain-Specific Anti-inflammatory Properties of Two Akkermansia muciniphila Strains on Chronic Colitis in Mice. Front. Cell. Infect. Microbiol. 2019;9:239. doi: 10.3389/fcimb.2019.00239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liu Y, et al. TLR4 regulates RORγt+ regulatory T-cell responses and susceptibility to colon inflammation through interaction with Akkermansia muciniphila. Microbiome. 2022;10:98. doi: 10.1186/s40168-022-01296-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Craft JE. Follicular helper T cells in immunity and systemic autoimmunity. Nat. Rev. Rheumatol. 2012;8:337–347. doi: 10.1038/nrrheum.2012.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lee JC, et al. Gene expression profiling of CD8+ T cells predicts prognosis in patients with Crohn disease and ulcerative colitis. J. Clin. Investig. 2011;121:4170–4179. doi: 10.1172/JCI59255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bunker JJ, et al. Innate and Adaptive Humoral Responses Coat Distinct Commensal Bacteria with Immunoglobulin A. Immunity. 2015;43:541–553. doi: 10.1016/j.immuni.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Meng X, et al. A Purified Aspartic Protease from Akkermansia Muciniphila Plays an Important Role in Degrading Muc2. Int. J. Mol. Sci. 2019;21:72. doi: 10.3390/ijms21010072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Meng X, Zhang J, Wu H, Yu D, Fang X. Akkermansia muciniphila Aspartic Protease Amuc_1434* Inhibits Human Colorectal Cancer LS174T Cell Viability via TRAIL-Mediated Apoptosis Pathway. Int. J. Mol. Sci. 2020;21:3385. doi: 10.3390/ijms21093385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gobert AP, et al. The human intestinal microbiota of constipated-predominant irritable bowel syndrome patients exhibits anti-inflammatory properties. Sci. Rep. 2016;6:39399. doi: 10.1038/srep39399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kang C-S, et al. Extracellular vesicles derived from gut microbiota, especially Akkermansia muciniphila, protect the progression of dextran sulfate sodium-induced colitis. PloS one. 2013;8:e76520. doi: 10.1371/journal.pone.0076520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bae, M. et al. Akkermansia muciniphila phospholipid induces homeostatic immune responses. Nature, 10.1038/s41586-022-04985-7 (2022). [DOI] [PMC free article] [PubMed]
- 109.Vaishnava S, et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Sci. (N. Y.) 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Alam A, et al. The microenvironment of injured murine gut elicits a local pro-restitutive microbiota. Nat. Microbiol. 2016;1:15021. doi: 10.1038/nmicrobiol.2015.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Weichselbaum L, Klein OD. The intestinal epithelial response to damage. Sci. China Life Sci. 2018;61:1205–1211. doi: 10.1007/s11427-018-9331-y. [DOI] [PubMed] [Google Scholar]
- 112.Lotz MM, et al. Intestinal epithelial restitution. Involvement of specific laminin isoforms and integrin laminin receptors in wound closure of a transformed model epithelium. Am. J. Pathol. 1997;150:747–760. [PMC free article] [PubMed] [Google Scholar]
- 113.Lepage M, Seltana A, Thibault M-P, Tremblay É, Beaulieu J-F. Knockdown of laminin α5 stimulates intestinal cell differentiation. Biochemical biophysical Res. Commun. 2018;495:1510–1515. doi: 10.1016/j.bbrc.2017.11.181. [DOI] [PubMed] [Google Scholar]
- 114.Coskun M, et al. Regulation of Laminin γ2 Expression by CDX2 in Colonic Epithelial Cells Is Impaired During Active Inflammation. J. Cell. Biochem. 2017;118:298–307. doi: 10.1002/jcb.25636. [DOI] [PubMed] [Google Scholar]
- 115.Thompson AI, Lees CW. Genetics of ulcerative colitis. Inflamm. bowel Dis. 2011;17:831–848. doi: 10.1002/ibd.21375. [DOI] [PubMed] [Google Scholar]
- 116.Barrett JC, et al. Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region. Nat. Genet. 2009;41:1330–1334. doi: 10.1038/ng.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pichler MJ, et al. Butyrate producing colonic Clostridiales metabolise human milk oligosaccharides and cross feed on mucin via conserved pathways. Nat. Commun. 2020;11:3285. doi: 10.1038/s41467-020-17075-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hänninen A, et al. Akkermansia muciniphila induces gut microbiota remodelling and controls islet autoimmunity in NOD mice. Gut. 2018;67:1445–1453. doi: 10.1136/gutjnl-2017-314508. [DOI] [PubMed] [Google Scholar]
- 119.Kim S, et al. Akkermansia muciniphila Prevents Fatty Liver, Decreases Serum Triglycerides, and Maintains Gut Homeostasis. Appl. Environ. Microbiol. 2020;86:e03004–e03019. doi: 10.1128/AEM.03004-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lopez-Siles M, et al. Alterations in the Abundance and Co-occurrence of Akkermansia muciniphila and Faecalibacterium prausnitzii in the Colonic Mucosa of Inflammatory Bowel Disease Subjects. Front. Cell. Infect. Microbiol. 2018;8:281. doi: 10.3389/fcimb.2018.00281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Dunn KA, et al. Early Changes in Microbial Community Structure Are Associated with Sustained Remission After Nutritional Treatment of Pediatric Crohn’s Disease. Inflamm. bowel Dis. 2016;22:2853–2862. doi: 10.1097/MIB.0000000000000956. [DOI] [PubMed] [Google Scholar]
- 122.Baxter NT, Zackular JP, Chen GY, Schloss PD. Structure of the gut microbiome following colonization with human feces determines colonic tumor burden. Microbiome. 2014;2:20. doi: 10.1186/2049-2618-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Dingemanse C, et al. Akkermansia muciniphila and Helicobacter typhlonius modulate intestinal tumor development in mice. Carcinogenesis. 2015;36:1388–1396. doi: 10.1093/carcin/bgv120. [DOI] [PubMed] [Google Scholar]
- 124.Bornet E, Westermann AJ. The ambivalent role of Bacteroides in enteric infections. Trends Microbiol. 2022;30:104–108. doi: 10.1016/j.tim.2021.11.009. [DOI] [PubMed] [Google Scholar]
- 125.Koretz RL. Response to Dr. Baldassarre. Am. J. Gastroenterol. 2018;113:1561–1562. doi: 10.1038/s41395-018-0249-7. [DOI] [PubMed] [Google Scholar]
- 126.Zhang C, Zhao L. Strain-level dissection of the contribution of the gut microbiome to human metabolic disease. Genome Med. 2016;8:41. doi: 10.1186/s13073-016-0304-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Filippis Fde, et al. Distinct Genetic and Functional Traits of Human Intestinal Prevotella copri Strains Are Associated with Different Habitual Diets. Cell host microbe. 2019;25:444–453.e3. doi: 10.1016/j.chom.2019.01.004. [DOI] [PubMed] [Google Scholar]
- 128.Karcher N, et al. Genomic diversity and ecology of human-associated Akkermansia species in the gut microbiome revealed by extensive metagenomic assembly. Genome Biol. 2021;22:209. doi: 10.1186/s13059-021-02427-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lv, Q.-B. et al. A thousand metagenome-assembled genomes of Akkermansia reveal new phylogroups and geographical and functional variations in human gut. (2020). [DOI] [PMC free article] [PubMed]
- 130.Kim S, et al. Mucin degrader Akkermansia muciniphila accelerates intestinal stem cell-mediated epithelial development. Gut microbes. 2021;13:1–20. doi: 10.1080/19490976.2021.1892441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Argüello H, et al. Early Salmonella Typhimurium infection in pigs disrupts Microbiome composition and functionality principally at the ileum mucosa. Sci. Rep. 2018;8:7788. doi: 10.1038/s41598-018-26083-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zackular JP, et al. The gut microbiome modulates colon tumorigenesis. mBio. 2013;4:e00692–13. doi: 10.1128/mBio.00692-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.