Abstract
Gram-negative bacteria belonging to the genus Burkholderia are remarkably resistant to broad spectrum, cationic, antimicrobial peptides (AMPs). It has been proposed that this innate resistance is related to changes in the outer membrane lipopolysaccharide (OM LPS), including the constitutive, essential modification of outer membrane Lipid A phosphate groups with cationic 4-amino-4-deoxy-arabinose. This modification reduces the overall negative charge on the OM LPS which may change the OM structure and reduce the binding, accumulation, and permeation of cationic AMPs. Similarly, the Gram-negative pathogen Pseudomonas aeruginosa can quickly become resistant to many AMPs by multiple mechanisms, frequently including activation of the arn operon, which leads, transiently, to the same modification of Lipid A. We recently discovered a set of synthetically evolved AMPs that do not invoke any resistance in P. aeruginosa over multiple passages, and thus are apparently not inhibited by aminorabinosylation of Lipid A in P. aeruginosa. Here we test these resistance avoiding peptides, within a set of 18 potent AMPs, against Burkholderia thailandensis. We find that none of the AMPs tested have measurable activity against B thailandensis. Some were inactive at concentrations as high as 150 μM, despite all having sterilizing activity at ≤ 10 μM against a panel of common, human bacterial pathogens, including P. aeruginosa. We speculate that the constitutive modification of Lipid A in members of the Burkholderia genus is only part of a broader set of modifications that change the architecture of the OM to provide such remarkable levels of resistance to cationic AMPs.
INTRODUCTION
In the ongoing struggle against rising drug-resistant bacterial infections, it is imperative to identify novel antibiotic chemotypes that are less likely to induce resistance in order to stably advance infection prevention and treatment options. Resistance to conventional small molecule antibiotics can arise rapidly by selection due to their typical single site of action on a single biomolecule1. Evolved resistance genes can readily be acquired by horizontal gene transfer2–4. Furthermore, some means of resistance, such as efflux pump upregulation, can drive generic resistance to many small molecule antibiotics simultaneously. The existence of multiple active resistance pathways can lead to a state of pan drug resistance3, 5.
Cationic antimicrobial peptides (AMPs) that target microbial membranes for disruption have long been considered promising alternatives to conventional antibiotics, and many are in preclinical development6–13. There are good reasons to believe that they could be developed into effective antibiotics. In vitro, AMPs often have sterilizing microbicidal activity at low μM concentrations against many strains of bacteria8–10, 14–19 and thus are active in the same concentration range as conventional antibiotics against susceptible organisms. However, AMPs are generally also active against drug-resistant, multidrug resistant, and pan drug resistant organisms1, 20–37. In fact, resistance to conventional antibiotics is broadly associated with collateral widespread susceptibility to AMPs38.
AMPs have a unique mechanism of action, global membrane disruption39, which effectively transforms an antibiotic barrier into a targeted site of action. AMPs do not target a single site, and thus, are not able to evoke resistance through the mutation of a single target gene40, 41. Also, AMPs are not sensitive to drug-efflux pumps or other mechanisms of multi drug resistance. Nonetheless, resistance to some AMPs has been observed, and it has been experimentally selected42–45. But avoidance or delay in resistance have also been observed46–49. For example, in a direct comparison46 using the Gram-negative pathogens Acinetobacter baumannii and P. aeruginosa, it was found to be much more difficult for bacteria to evolve resistance to some AMPs than to small-molecule antibiotics under the same conditions46. Other groups, including us49, have also reported the inability to select for resistance against some AMPs46, 47, 49.
Multiple mechanisms of AMP resistance have been identified for Gram-negative bacteria50–55, but the “resistome” is small40, 41. The most commonly observed resistance mechanism is the modification of Lipid A phosphate groups with cationic moieties. Lipid A is a variable core lipid moiety that anchors the oligosaccharide component of LPS to the outer membrane (OM). Typical Lipid A molecules contain an anionic, di-phosphorylated disaccharide with 4-6 short/medium chain acyl groups. In response to exposure to sublethal concentrations of some antibiotics, Gram-negative bacteria can upregulate operons for the addition of cationic groups, including ethanolamine, glucosamine, and 4-amino-4-deoxy-arabinose (Ara4N) to the lipid A phosphates. In P. aeruginosa, for example, the addition of Ara4N to lipid A phosphates by the arn operon is a common mechanism of AMP resistance41, 51, 56, 57.
The effect of Lipid A modification on AMP activity can be rationalized by the observation that some, and probably most, AMPs kill bacteria as a result of “self-promoted uptake” 58 in which a saturating accumulation of peptide on the cell is required for lethal activity at the cytoplasmic membrane. Depending on the peptide and organism, between 1x107 and 5x108 peptide molecules must be bound to each cell for bactericidal activity58–61. The polyanionic LPS layer of Gram-negative bacteria is likely saturated with peptide under these conditions, which has led researchers to conclude that massive accumulation on the outer membrane LPS, and perhaps OM destabilization, are necessary prerequisites for the permeabilizing activity of at least some AMPs at the inner membrane55, 58, 62. It is thus reasonable that the most common mechanism of resistance to AMPs in Gram-negative bacteria is the addition of cationic groups to the anionic phosphates of the Lipid A moiety of OM LPS41, 50, 56, 63 as this modification will globally replace negative charges on LPS with positive charges, and also may change the LPS structure, reducing the OM binding potential for cationic AMPs.
However, not all AMPs are subject to the development of resistance in Gram-negative bacteria. Some AMPs are thus apparently not inhibited by the modification of Lipid A phosphates by Ara4N. For example, we recently described the synthetic molecular evolution of a family of potent, broad spectrum, hemocompatible antimicrobial peptides49 that do not invoke any resistance in multiple passages against P. aeruginosa.. We assume that this means that aminorabinosylation of lipid A cannot confer resistance to these peptides. Under the same conditions, resistance developed rapidly to conventional antibiotics49.
Here we extend our characterization of these potent, resistance-avoiding AMPs by testing their activity against Burkholderia thailandensis, which belongs to a genus of Gram-negative bacteria (class betaproteobacteria) that is biochemically similar to Pseudomonas (class gammaproteobacteria). B. thailandensis, like most Burkholderia species, produces Lipid A molecules that are constitutively aminorabinosylated, often on both Lipid A phosphate moieties57. Here, we show that B. thailandensis is remarkably resistant to sterilization by cationic AMPs, even those against which P. aeruginosa is highly susceptible and cannot evolve resistance.
RESULTS
Synthetically evolved AMPs.
The unique resistance avoiding AMPs tested here are described elsewhere49. They are the product of three generations of synthetic molecular evolution and rational design, undertaken to identify host-compatible, broad-spectrum antimicrobial peptides17, 18, 49, 64. First, from a de novo library, we selected in parallel for members that either permeabilized bacteria-like lipid vesicles or that sterilized multiple species of bacteria simultaneously17. These peptides, exemplified by the 12-residue peptide ARVA (RRGWALRLVLAY), Fig. 1, have broad spectrum antimicrobial activity, but are potently inhibited by host cell binding, proteolysis, and serum protein binding61, 65. Next, we used ARVA as a template to design a library from which we selected potent AMPs that are not inhibited by concentrated host cells49. This screen gave rise to five “double broth sterilization”, or DBS, peptides which we have tested as both protease susceptible L-amino acid (L-DBS) peptides and as protease resistant D-amino acid (D-DBS) peptides, Fig. 1. We also tested a consensus sequence, D-CON (rrgwarrlafafgrr). Finally, a further round of rational optimization49 gave rise to our most active AMP, called D-CONGA, which stands for D-amino acid, CONsensus sequence with Glycine Absent, Fig. 1. We note that all of these evolved peptides are C-terminal amides.
Figure 1.
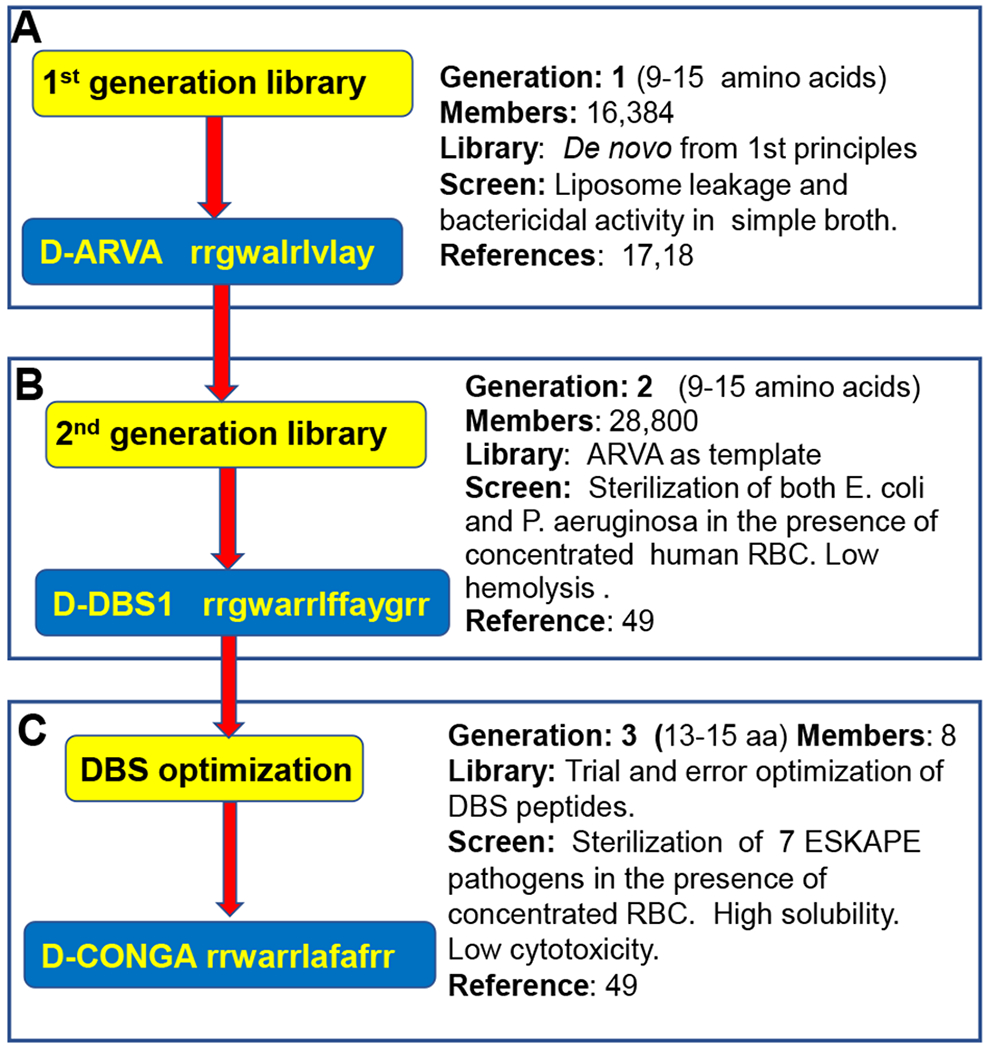
Evolution of D-CONGA. A. The 1st generation, de novo-designed library was previously screened for synthetic membrane permeabilization and for bacterial sterilization17.18. B. The 2nd generation library was designed from ARVA and screened for hemocompatible AMPs against Gram-negative pathogens49. C. The 3rd “generation” consisted of rational variants tested for broad spectrum activity against ESKAPE pathogens and low cytotoxicity. The 3rd Gen. peptide D-CONGA is the best peptide identified49.
As we previously showed, the D-DBS peptides, D-CON, and D-CONGA have excellent antimicrobial properties, suggesting that they could be developed into useful antibiotics, especially in the protection and treatment of wounds49. They have potent (MIC ≤ 10 μM) sterilizing activity against all ESKAPE pathogens (E. coli, S. aureus, K. pneumoniae, A. baumanii, P. aeruginosa, and E. faecium) which include both Gram-negative and Gram-positive bacteria. The lead peptides also have potent activity against drug-resistant bacteria in vitro and in vivo, potent activity against biofilms, in vitro and in vivo, and retain activity in the presence of concentrated host cells, tissue, and serum proteins49. Finally, they are also highly soluble, resistant to biodegradation, and have low cytotoxicity49.
Resistance avoidance.
We previously tested some of these peptides for the ability to induce resistance in P. aeruginosa, which is inherently resistant, or can rapidly become resistant, to many conventional antibiotics and AMPs41, 63. We tested whether P. aeruginosa could develop stable resistance to i) Four conventional antibiotics, ii) D-ARVA, the template peptide for the 2nd generation library, iii) L-DBS1 and D-DBS1, a sequence selected from the second-generation library, and iv) D-CONGA the best lead peptide evolved during the recent work. As described elsewhere49, P. aeruginosa PAO1 was grown in the presence of serially diluted antibiotic, and we selected the culture that grew at the highest concentration of antibiotic. This culture was propagated overnight in the absence of antibiotic to enable selection for stable resistance. The process was repeated for ten passages, which is equivalent to hundreds of generations. Against the four conventional antibiotics, resistance always increased (Fig. 2), and the bacteria evolved complete resistance (MIC ≥ 350 μM) to streptomycin and ceftazidime by 8 passages. Against the library template sequence D-ARVA, P. aeruginosa became resistant (MIC ≥ 67 μM) over four passages (Fig. 2). Slight changes in experimental protocols lead to resistance to D-ARVA in a single passage49. However, against the lead 2nd generation peptides L-DBS1, D-DBS1 and D-CONGA, we observed no measurable increase in resistance over ten passages. The activity of these peptides against P. aeruginosa is not sensitive to experimental details.
Figure 2.
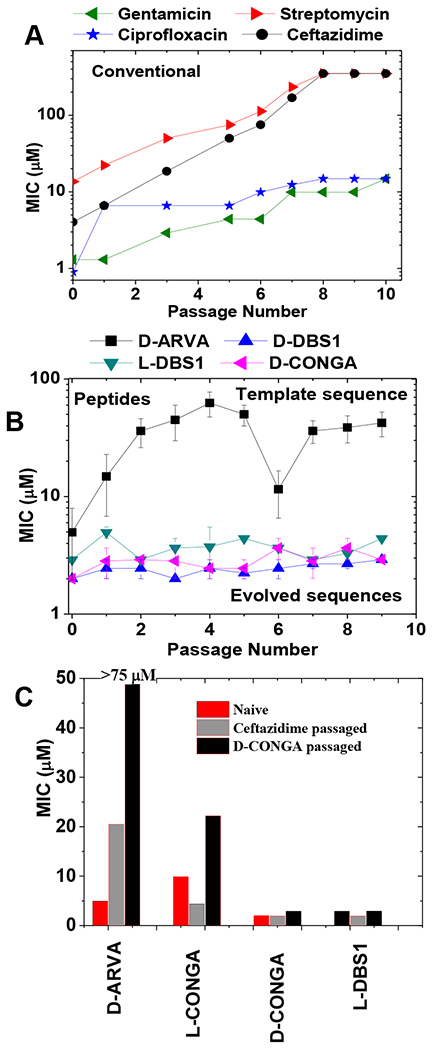
Resistance avoidance in evolved antimicrobial peptides. P. aeruginosa was treated with serially diluted conventional antibiotics or AMPs49. The culture that grew at the highest concentration was cultured overnight in the absence of antibiotic, and then the screening with antibiotic/AMP was repeated the next day. This was done for ten passages. A: The MIC values for each passage against four conventional antibiotics. B. The MIC values for each passage against four AMPs. C: Cross resistance of P. aeruginosa passaged against one antibiotic and one peptide and tested against a set of AMPs.
In Fig. 2C we show cross-resistance in P. aeruginosa after 10 passages against either the cephalosporin class antibiotic ceftazidime or the AMP D-CONGA. Passaging against D-CONGA caused no increase in resistance to D-CONGA, but it did cause resistance to D-ARVA, suggesting that the bacteria are responding to D-CONGA in a way that is ineffectual against D-CONGA, but enables resistance to D-ARVA. Even passaging against ceftazidime causes a partial increase in resistance to D-ARVA but did not increase resistance to D-CONGA. The fact that passaging against L-DBS1 did not evoke resistance to L-DBS1 or to L-CONGA indicates that protease secretion is not readily evoked as a resistance mechanism against these peptides in P. aeruginosa. Based on these data, we concluded that there is a unique, and important molecular aspect of the DBS peptides, D-CON, and D-CONGA that prevents or slows the development of resistance, compared to other AMPs. Some other AMPs have also been reported to have this property46–49. The detailed molecular mechanism of such “resistance avoidance” is unknown, however we are confident that peptides like D-CONGA that do not readily invoke resistance in P. aeruginosa, are not inhibited by aminorabinosylation of Lipid A in that species.
Most Burkholderia species are innately and highly resistant to AMPs41, 54, 55, 66. To test the correlation between resistance avoidance in P. aeruginosa and AMP resistance in Burkholderia, we measured minimum inhibitory concentrations (MIC) of D-ARVA, D-DBS peptides, D-CON, and D-CONGA against B. thailandensis. Activities are compared to Gram-negative P. aeruginosa and E. coli, and to Gram-positive S. aureus. For comparison, we also tested other AMPs from multiple sources, including indolicidin (a bovine neutrophil AMP in L- and D- forms), cecropin A (an insect AMP), melittin (a membrane lytic bee venom peptide67), MSI78/pexiganan (a synthetic analog of frog skin AMPs), WLBU2 (a synthetic AMP), and ARNY, RNNY and NATT (16 residue AMPs related to ARVA17).
We used broth dilution assays in which ~2x105 bacterial cells/ml and serially diluted peptide were mixed in nutrient broth, and the bacteria were allowed to grow overnight. We noted the lowest concentration without any growth and averaged multiple such experiments on the log scale. This assay is a stringent assay for sterilization, as overnight growth will generally allow any survivors to reach high cell density by the next day. Using susceptible bacteria, we have repeatedly shown, by plating on nutrient agar, that wells which remain clear after overnight growth do not contain any live bacteria. Thus, the MIC for an AMP is actually a minimum sterilizing concentration.
In Figure 3, we show the minimum inhibitory concentrations of these 18 different cationic antimicrobial peptides against B. thailandensis, and compare the results to MIC values against P. aeruginosa, E. coli, and S. aureus. The values shown are the average of 3-15 independent measurements made by broth dilution. Numbers shown in red boxes as “>conc” indicate that no sterilization was observed up to the stated maximum tested concentration. Numbers shown as “~conc” indicate that some, but not all, wells were sterilized only at the maximum concentration tested and that the MIC could be just above this maximum tested concentration.
Figure 3.
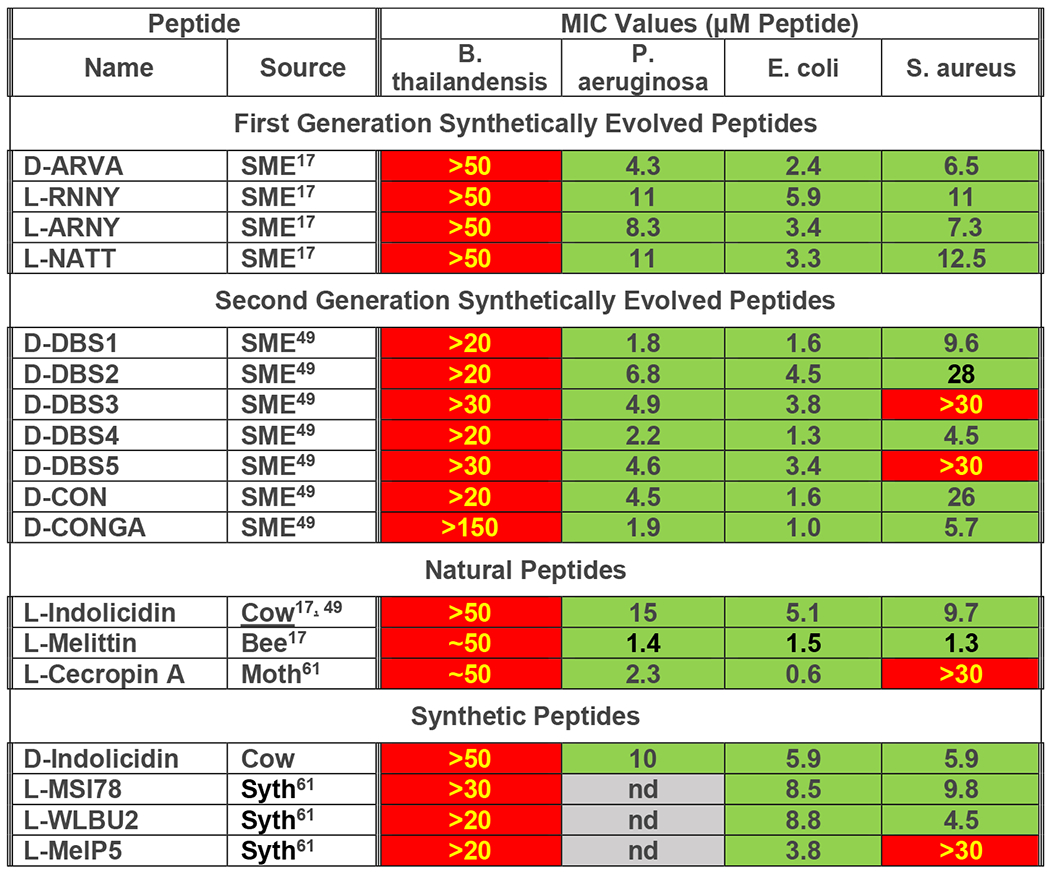
AMP activity against B. thailandensis, compared to E. coli. P. aeruginosa and S. aureus. Log averaged MIC values for 18 cationic antimicrobial peptides were measured as described49. Serially diluted peptides were added to 2x105 bacteria in growth media in 96 well plates and were incubated overnight at 37°C. After overnight incubation, optical density of each well was measured at 600 nm. Most measurements were equal either to transparent sterile media control wells or to opaque stationary phase growth control wells. In 3-15 repeat experiments the lowest sterilizing concentration was noted. The results were averaged in log space and the average was converted to concentration. Red cells with yellow text indicate little or no activity. Green cells with black text indicate measurable MIC value at ≤20 μM. nd=not determined.
Most of these peptides were studied at 20 to 50 μM concentration, but the most active one, D-CONGA, was studied up to 150 μM, as indicated in Figure 3. Remarkably, none of these potent, broad-spectrum AMPs have activity against B. thailandensis. In sharp contrast, most have sterilizing activity at less than 10 μM against most of the other microbes listed (green cells). The Gram-positive organism S. aureus is susceptible 14 of the 18 AMPs tested, which is a typical for this organism. With few exceptions, the D-DBS peptides D-CON and D-CONGA have sterilizing activity at ≤ 10 μM against all ESKAPE pathogens49.
For a few of these peptides, we also performed colony forming unit (CFU) reduction assays to measure the reduction in viable bacteria, an assay that does not depend on sterilization. For D-ARVA and D-CONGA the reduction in viable B. thailandensis was less than 10-fold (<1 log) at peptide concentrations of 50-150 μM. This can be compared to sterilization of the 2x105 susceptible P. aeruginosa/ml at 5-10 μM peptide in broth dilution experiments, demonstrating a reduction of viable P. aeruginosa by more than 4 logs at 10-20 fold lower peptide. These results show that B. thailandensis is at least 5 orders of magnitude more resistance to these evolved AMPs than P. aeruginosa.
DISCUSSION
Members of the genus Burkholderia have high inherent resistance to conventional antibiotics and antimicrobial peptides55, 68, including resistance to the polymyxins41, 66, 69, which are approved for use in humans. Resistance in Burkholderia is important because of the potential of some species to cause serious disease in humans. These pathogenic species include the Burkholderia cepacia complex which is prevalent in the lungs cystic fibrosis patients70 and the biothreat organism Burkholderia pseudomallei, which causes meliodosis, a disease with high mortality in humans71. There are multiple mechanisms of resistance in Burkholderia55, but a dominant one that is especially effective against AMPs, is the constitutive modification of the outer membrane LPS. For example, in Burkholderia one or both of the outer membrane Lipid A phosphate moieties are modified with cationic Ara4N moieties41, 57,57, 68, 69. Some Burkholderia Lipid A variants also have one phosphate removed, with the remaining phosphate being aminoarabinosylated57. The core polysaccharide of the Burkholderia LPS can also contain Ara4N mopieties69 such that truncation of the LPS core polysaccharide has been shown to cause sensitivity to AMPs in Burkholderia72. In many other Gram-negative bacteria, stress operons can be triggered to carry out Lipid A modifications41, 51. However, in Burkholderia these modifications are constitutive68. Attempts to inactivate Ara4N modification pathways in Burkholderia69 using conditional mutants showed that both the synthesis of Ara4N and the transfer of Ara4N to Lipid A are essential to viability.
One result of these modifications is a significant reduction in the net negative charge on the OM LPS, although structural changes are also possible. These changes likely inhibit the so called “self-directed uptake” of AMPs58, a process that includes accumulation of cationic AMPs on the OM59, 61 via electrostatic interactions, including displacement of divalent cations73, and subsequent disruption of the outer membrane prior to the lethal disruption of the inner membrane. As a result of the modifications to LPS, the AMP polymyxin74 binds much more weakly to Burkholderia OM LPS than to P. aeruginosa LPS74 and does not permeabilize the OM of Burkholderia. We and others have added to the understanding of this process by measuring the accumulation of AMPs on bacteria that is necessary for killing. The killing of Gram-negative bacteria by AMPs is a saturation-dependent event in which peptides bind massively to bacterial cells with a requirement that 1x107 to 5x108 peptides are bound to each cell for activity39, 59–61. For example, about 3x108 molecules of D-ARVA are required to kill one E. coli cell, despite an MIC of only 3 μM49. Only a small portion of the total bound peptide actually interacts with the inner membrane75, which has about 2x107 lipid molecules39, to cause lethal inner membrane permeabilization.
In this work we tested the hypothesis that cationic AMPs that cannot evoke the development of resistance in P. aeruginosa through Lipid A modification will also not be inhibited by the same OM LPS modifications that are constitutive in B. thailandensis. Our results do not support the hypothesis. Instead, we verify that B. thailandensis is remarkably resistant to cationic AMPs, and we show that this innate resistance includes resistance to AMPs that are apparently not inhibited by Lipid A modification by Ara4N in P. aeruginosa.
There are testable ideas that might explain the observation that some peptides cannot invoke resistance in P. aeruginosa but simultaneously have no activity against B. thailandensis. For example, the acute aminorabinosylation of Lipid A in P. aeruginosa may be associated with a secondary increase in inherent OM permeability, or some other reduction in overall fitness. Resistance-avoiding peptides may be those that can cross the leakier OM in P. aeruginosa to access the inner membrane without first massively accumulating on the OM. On the other hand, in Burkholderia aminorabinosylation of Lipid A is both constitutive and essential. Therefore, we speculate that members of the Burkholderia genus have an array of other OM and LPS modifications which act together to stabilize the structure of the LPS. Thus, both reduced charge and increased structure could function cooperatively to reduce binding and permeation of AMPs and other antibiotics across the OM.
Funding:
Funded by NIH AI154284.
Footnotes
Conflicts of interest: Authors WCW, JG and SG have filed for protection of intellectual property rights for some of the technology described here. BJN and LM have no conflicts.
References
- 1.Arias CA; Murray BE, Antibiotic-resistant bugs in the 21st century--a clinical super-challenge. N. Engl. J. Med 2009, 360 (5), 439–443. [DOI] [PubMed] [Google Scholar]
- 2.Turner PE; Williams ES; Okeke C; Cooper VS; Duffy S; Wertz JE, Antibiotic resistance correlates with transmission in plasmid evolution. Evolution 2014, 68 (12), 3368–80. [DOI] [PubMed] [Google Scholar]
- 3.Otto M, MRSA virulence and spread. Cell Microbiol 2012, 14 (10), 1513–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith MW; Feng DF; Doolittle RF, Evolution by acquisition: The case for horizontal gene transfers. Trends Biochem. Sci 1992, 17, 489–493. [DOI] [PubMed] [Google Scholar]
- 5.Lazar V; Nagy I; Spohn R; Csorgo B; Gyorkei A; Nyerges A; Horvath B; Voros A; Busa-Fekete R; Hrtyan M; Bogos B; Mehi O; Fekete G; Szappanos B; Kegl B; Papp B; Pal C, Genome-wide analysis captures the determinants of the antibiotic cross-resistance interaction network. Nat Commun 2014, 5, 4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.White SH; Wimley WC; Selsted ME, Structure, function, and membrane integration of defensins. Cur. Opinion Struc. Biol 1995, 5 (4), 521–527. [DOI] [PubMed] [Google Scholar]
- 7.Yeaman MR; Yount NY, Mechanisms of antimicrobial peptide action and resistance. Pharmacol . Rev 2003, 55 (1), 27–55. [DOI] [PubMed] [Google Scholar]
- 8.Hamill P; Brown K; Jenssen H; Hancock RE, Novel anti-infectives: is host defence the answer? Curr. Opin. Biotechnol 2008, 19 (6), 628–636. [DOI] [PubMed] [Google Scholar]
- 9.Jenssen H; Hamill P; Hancock RE, Peptide antimicrobial agents. Clin. Microbiol. Rev 2006, 19 (3), 491–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marr AK; Gooderham WJ; Hancock RE, Antibacterial peptides for therapeutic use: obstacles and realistic outlook. Curr. Opin. Pharmacol 2006, 6 (5), 468–472. [DOI] [PubMed] [Google Scholar]
- 11.Zasloff M, Antimicrobial peptides of multicellular organisms. Nature 2002, 415 (6870), 389–395. [DOI] [PubMed] [Google Scholar]
- 12.Bevins CL; Zasloff M, Peptides from frog skin. Annu. Rev. Biochem 1990, 59, 395–414. [DOI] [PubMed] [Google Scholar]
- 13.Wimley WC; Hristova K, Antimicrobial Peptides: Successes, Challenges and Unanswered Questions. J. Membr. Biol 2011, 239, 27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Selsted ME; Ouellette AJ, Mammalian defensins in the antimicrobial immune response. Nat. Immunol 2005, 6 (6), 551–557. [DOI] [PubMed] [Google Scholar]
- 15.Tang YQ; Yeaman MR; Selsted ME, Antimicrobial peptides from human platelets. Infect. Immun 2002, 70 (12), 6524–6533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ouellette AJ; Selsted ME, Enteric defensins. Cur. Opin. Gastroenterology 1997, 13, 494–499. [Google Scholar]
- 17.Rathinakumar R; Wimley WC, High-throughput discovery of broad-spectrum peptide antibiotics. FASEB J 2010, 24, 3232–3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rathinakumar R; Walkenhorst WF; Wimley WC, Broad-spectrum antimicrobial peptides by rational combinatorial design and high-throughput screening: The importance of interfacial activity. J. Am. Chem. Soc 2009, 131, 7609–7617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rausch JM; Marks JR; Rathinakumar R; Wimley WC, Beta-sheet pore-forming peptides selected from a rational combinatorial library: mechanism of pore formation in lipid vesicles and activity in biological membranes. Biochemistry 2007, 46 (43), 12124–12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang Y; Wiradharma N; Xu K; Ji Z; Bi S; Li L; Yang YY; Fan W; Pollard JE; Snarr J; Chaudhary V; Jennings JD; Shaw H; Christiansen B; Wright J; Jia W; Bishop RE; Savage PB, Cationic amphiphilic alpha-helical peptides for the treatment of carbapenem-resistant Acinetobacter baumannii infection. Biomaterials 2012, 33 (34), 8841–8847. [DOI] [PubMed] [Google Scholar]
- 21.Schwab U; Gilligan P; Jaynes J; Henke D, In vitro activities of designed antimicrobial peptides against multidrug-resistant cystic fibrosis pathogens. Antimicrob. Agents Chemother 1999, 43 (6), 1435–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li W; Tailhades J; O’Brien-Simpson NM; Separovic F; Otvos L Jr.; Hossain MA; Wade JD, Proline-rich antimicrobial peptides: potential therapeutics against antibiotic-resistant bacteria. Amino. Acids 2014, 46 (10), 2287–2294. [DOI] [PubMed] [Google Scholar]
- 23.Jahnsen RD; Sandberg-Schaal A; Vissing KJ; Nielsen HM; Frimodt-Moller N; Franzyk H, Tailoring cytotoxicity of antimicrobial peptidomimetics with high activity against multidrug-resistant Escherichia coli. J Med. Chem 2014, 57 (7), 2864–2873. [DOI] [PubMed] [Google Scholar]
- 24.Moghaddam MM; Barjini KA; Ramandi MF; Amani J, Investigation of the antibacterial activity of a short cationic peptide against multidrug-resistant Klebsiella pneumoniae and Salmonella typhimurium strains and its cytotoxicity on eukaryotic cells. World J Microbiol. Biotechnol 2014, 30 (5), 1533–1540. [DOI] [PubMed] [Google Scholar]
- 25.Schlusselhuber M; Guldbech K; Sevin C; Leippe M; Petry S; Grotzinger J; Giguere S; Cauchard J, In vitro effectiveness of the antimicrobial peptide eCATH1 against antibiotic-resistant bacterial pathogens of horses. FEMS Microbiol. Lett 2014, 350 (2), 216–222. [DOI] [PubMed] [Google Scholar]
- 26.Fleming E; Heil EL; Hynicka LM, Treatment strategy for a multidrug-resistant Klebsiella UTI. Ann. Pharmacother 2014, 48 (1), 123–127. [DOI] [PubMed] [Google Scholar]
- 27.Mechkarska M; Prajeep M; Radosavljevic GD; Jovanovic IP; Al Baloushi A; Sonnevend A; Lukic ML; Conlon JM, An analog of the host-defense peptide hymenochirin-1B with potent broad-spectrum activity against multidrug-resistant bacteria and immunomodulatory properties. Peptides 2013, 50, 153–159. [DOI] [PubMed] [Google Scholar]
- 28.Deslouches B; Steckbeck JD; Craigo JK; Doi Y; Mietzner TA; Montelaro RC, Rational design of engineered cationic antimicrobial peptides consisting exclusively of arginine and tryptophan, and their activity against multidrug-resistant pathogens. Antimicrob. Agents Chemother 2013, 57 (6), 2511–2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Conlon JM; Mechkarska M; Arafat K; Attoub S; Sonnevend A, Analogues of the frog skin peptide alyteserin-2a with enhanced antimicrobial activities against Gram-negative bacteria. J Pept. Sci 2012, 18 (4), 270–275. [DOI] [PubMed] [Google Scholar]
- 30.Taira J; Kida Y; Yamaguchi H; Kuwano K; Higashimoto Y; Kodama H, Modifications on amphiphilicity and cationicity of unnatural amino acid containing peptides for the improvement of antimicrobial activity against pathogenic bacteria. J Pept. Sci 2010, 16 (11), 607–612. [DOI] [PubMed] [Google Scholar]
- 31.Fedders H; Podschun R; Leippe M, The antimicrobial peptide Ci-MAM-A24 is highly active against multidrug-resistant and anaerobic bacteria pathogenic for humans. Int. J Antimicrob. Agents 2010, 36 (3), 264–266. [DOI] [PubMed] [Google Scholar]
- 32.Park SC; Kim JY; Lee JK; Yoo S; Kim H; Seo CH; Nah JW; Hahm KS; Park Y, Synthetic diastereomeric-antimicrobial peptide: antibacterial activity against multiple drug resistant clinical isolates. Biopolymers 2011, 96 (2), 130–136. [DOI] [PubMed] [Google Scholar]
- 33.Pastewski AA; Caruso P; Parris AR; Dizon R; Kopec R; Sharma S; Mayer S; Ghitan M; Chapnick EK, Parenteral polymyxin B use in patients with multidrug-resistant gram-negative bacteremia and urinary tract infections: a retrospective case series. Ann. Pharmacother 2008, 42 (9), 1177–1187. [DOI] [PubMed] [Google Scholar]
- 34.Eley A; Ibrahim M; Kurdi SE; Conlon JM, Activities of the frog skin peptide, ascaphin-8 and its lysine-substituted analogs against clinical isolates of extended-spectrum beta-lactamase (ESBL) producing bacteria. Peptides 2008, 29 (1), 25–30. [DOI] [PubMed] [Google Scholar]
- 35.Mangoni ML; Maisetta G; Di Luca M; Gaddi LM; Esin S; Florio W; Brancatisano FL; Barra D; Campa M; Batoni G, Comparative analysis of the bactericidal activities of amphibian peptide analogues against multidrug-resistant nosocomial bacterial strains. Antimicrob. Agents Chemother 2008, 52 (1), 85–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pereira GH; Muller PR; Levin AS, Salvage treatment of pneumonia and initial treatment of tracheobronchitis caused by multidrug-resistant Gram-negative bacilli with inhaled polymyxin B. Diagn. Microbiol. Infect. Dis 2007, 58 (2), 235–240. [DOI] [PubMed] [Google Scholar]
- 37.Maisetta G; Batoni G; Esin S; Florio W; Bottai D; Favilli F; Campa M, In vitro bactericidal activity of human beta-defensin 3 against multidrug-resistant nosocomial strains. Antimicrob. Agents Chemother 2006, 50 (2), 806–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lazar V; Martins A; Spohn R; Daruka L; Grezal G; Fekete G; Szamel M; Jangir PK; Kintses B; Csorgo B; Nyerges A; Gyorkei A; Kincses A; Der A; Walter FR; Deli MA; Urban E; Hegedus Z; Olajos G; Mehi O; Balint B; Nagy I; Martinek TA; Papp B; Pal C, Antibiotic-resistant bacteria show widespread collateral sensitivity to antimicrobial peptides. Nat Microbiol 2018, 3 (6), 718–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wimley WC, Describing the mechanism of antimicrobial peptide action with the interfacial activity model. ACS Chem. Biol 2010, 5 (10), 905–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santos-Lopez A; Fritz MJ; Lombardo J; Burr AHP; Heinrich VA; Marshall CW; Cooper VS Experimental evolution to identify undescribed mechanisms of resistance to a novel cationic peptide antibiotic 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fernandez L; Alvarez-Ortega C; Wiegand I; Olivares J; Kocincova D; Lam JS; Martinez JL; Hancock RE, Characterization of the polymyxin B resistome of Pseudomonas aeruginosa. Antimicrob Agents Chemother 2013, 57 (1), 110–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peschel A; Sahl HG, The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat. Rev. Microbiol 2006, 4 (7), 529–536. [DOI] [PubMed] [Google Scholar]
- 43.Samuelsen O; Haukland HH; Jenssen H; Kramer M; Sandvik K; Ulvatne H; Vorland LH, Induced resistance to the antimicrobial peptide lactoferricin B in Staphylococcus aureus. FEBS Lett 2005, 579 (16), 3421–3426. [DOI] [PubMed] [Google Scholar]
- 44.Perron GG; Zasloff M; Bell G, Experimental evolution of resistance to an antimicrobial peptide. Proc. Biol. Sci 2006, 273 (1583), 251–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peschel A, How do bacteria resist human antimicrobial peptides? Trends Microbiol 2002, 10 (4), 179–186. [DOI] [PubMed] [Google Scholar]
- 46.Pollard JE; Snarr J; Chaudhary V; Jennings JD; Shaw H; Christiansen B; Wright J; Jia W; Bishop RE; Savage PB; Yang YY; Fan W, In vitro evaluation of the potential for resistance development to ceragenin CSA-13. J. Antimicrob. Chemother 2012, 67, 2665–2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de BA; Riool M; Cordfunke RA; Malanovic N; de BL; Koning RI; Ravensbergen E; Franken M; van der Heijde T; Boekema BK; Kwakman PHS; Kamp N; El GA; Lohner K; Zaat SAJ; Drijfhout JW; Nibbering PH, The antimicrobial peptide SAAP-148 combats drug-resistant bacteria and biofilms. Sci. Transl. Med 2018, 10 (423). [DOI] [PubMed] [Google Scholar]
- 48.Lakshmaiah Narayana J; Mishra B; Lushnikova T; Wu Q; Chhonker YS; Zhang Y; Zarena D; Salnikov ES; Dang X; Wang F; Murphy C; Foster KW; Gorantla S; Bechinger B; Murry DJ; Wang G, Two distinct amphipathic peptide antibiotics with systemic efficacy. Proc Natl Acad Sci U S A 2020, 117 (32), 19446–19454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Starr CG; Ghimire J; Guha S; Hoffmann JP; Wang Y; Sun L; Landreneau BN; Kolansky ZD; Kilanowski-Doroh IM; Sammarco MC; Morici LA; Wimley WC, Synthetic molecular evolution of host cell-compatible, antimicrobial peptides effective against drug-resistant, biofilm-forming bacteria. Proc Natl Acad Sci U S A 2020, 117 (15), 8437–8448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Joo HS; Fu CI; Otto M, Bacterial strategies of resistance to antimicrobial peptides. Philos Trans R Soc Lond B Biol Sci 2016, 371 (1695). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Muller C; Plesiat P; Jeannot K, A two-component regulatory system interconnects resistance to polymyxins, aminoglycosides, fluoroquinolones, and beta-lactams in Pseudomonas aeruginosa. Antimicrob Agents Chemother 2011, 55 (3), 1211–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Esani S; Chen T; Leung KP; Van Laar TA, Transcriptome Sequence of Antibiotic-Treated Pseudomonas aeruginosa. Microbiol Resour Announc 2019, 8 (12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morita Y; Tomida J; Kawamura Y, Responses of Pseudomonas aeruginosa to antimicrobials. Front Microbiol 2014, 4, 422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kooi C; Sokol PA, Burkholderia cenocepacia zinc metalloproteases influence resistance to antimicrobial peptides. Microbiology (Reading) 2009, 155 (Pt 9), 2818–2825. [DOI] [PubMed] [Google Scholar]
- 55.Loutet SA; Valvano MA, Extreme antimicrobial peptide and polymyxin B resistance in the genus Burkholderia. Front Cell Infect Microbiol 2011, 1, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang H; Srinivas S; Xu Y; Wei W; Feng Y, Genetic and Biochemical Mechanisms for Bacterial Lipid A Modifiers Associated with Polymyxin Resistance. Trends Biochem Sci 2019. [DOI] [PubMed] [Google Scholar]
- 57.Sengyee S; Yoon SH; West TE; Ernst RK; Chantratita N, Lipopolysaccharides from Different Burkholderia Species with Different Lipid A Structures Induce Toll-Like Receptor 4 Activation and React with Melioidosis Patient Sera. Infect Immun 2019, 87 (12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Loh B; Grant C; Hancock RE, Use of the fluorescent probe 1-N-phenylnaphthylamine to study the interactions of aminoglycoside antibiotics with the outer membrane of Pseudomonas aeruginosa. Antimicrob. Agents Chemother 1984, 26 (4), 546–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Savini F; Bobone S; Roversi D; Mangoni M; Stella L, From liposomes to cells: Filling the gap between physicochemical and microbiological studies of the activity and selectivity of host-defense peptides. Peptide Science 2018, e24041. [Google Scholar]
- 60.Savini F; Luca V; Bocedi A; Massoud R; Park Y; Mangoni ML; Stella L, Cell-Density Dependence of Host-Defense Peptide Activity and Selectivity in the Presence of Host Cells. ACS Chem. Biol 2017, 12 (1), 52–56. [DOI] [PubMed] [Google Scholar]
- 61.Starr CG; He J; Wimley WC, Host Cell Interactions Are a Significant Barrier to the Clinical Utility of Peptide Antibiotics. ACS Chem. Biol 2016, 11 (12), 3391–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang Z; Choi H; Weisshaar JC, Melittin-induced Permeabilization, Re-sealing, and Re-permeabilization of E. coli Membranes. Biophys J 2017, 114 (2), 368–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moradali MF; Ghods S; Rehm BH, Pseudomonas aeruginosa Lifestyle: A Paradigm for Adaptation, Survival, and Persistence. Front Cell Infect. Microbiol 2017, 7, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rathinakumar R; Wimley WC, Biomolecular engineering by combinatorial design and high-throughput screening: small, soluble peptides that permeabilize membranes. J. Am. Chem. Soc 2008, 130 (30), 9849–9858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Starr CG; Wimley WC, Antimicrobial peptides are degraded by the cytosolic proteases of human erythrocytes. Biochim. Biophys Acta 2017, 1859 (12), 2319–2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Burtnick MN; Woods DE, Isolation of polymyxin B-susceptible mutants of Burkholderia pseudomallei and molecular characterization of genetic loci involved in polymyxin B resistance. Antimicrob Agents Chemother 1999, 43 (11), 2648–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guha S; Ferrie RP; Ghimire J; Ventura CR; Wu E; Sun L; Kim SY; Wiedman GR; Hristova K; Wimley WC, Applications and evolution of melittin, the quintessential membrane active peptide. Biochem Pharmacol 2021, 193, 114769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hamad MA; Di Lorenzo F; Molinaro A; Valvano MA, Aminoarabinose is essential for lipopolysaccharide export and intrinsic antimicrobial peptide resistance in Burkholderia cenocepacia(dagger). Mol Microbiol 2012, 85 (5), 962–74. [DOI] [PubMed] [Google Scholar]
- 69.Ortega XP; Cardona ST; Brown AR; Loutet SA; Flannagan RS; Campopiano DJ; Govan JR; Valvano MA, A putative gene cluster for aminoarabinose biosynthesis is essential for Burkholderia cenocepacia viability. J Bacteriol 2007, 189 (9), 3639–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Khanolkar RA; Clark ST; Wang PW; Hwang DM; Yau YCW; Waters VJ; Guttman DS, Ecological Succession of Polymicrobial Communities in the Cystic Fibrosis Airways. mSystems 2020, 5 (6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stone JK; DeShazer D; Brett PJ; Burtnick MN, Melioidosis: molecular aspects of pathogenesis. Expert Rev Anti Infect Ther 2014, 12 (12), 1487–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Loutet SA; Flannagan RS; Kooi C; Sokol PA; Valvano MA, A complete lipopolysaccharide inner core oligosaccharide is required for resistance of Burkholderia cenocepacia to antimicrobial peptides and bacterial survival in vivo. J Bacteriol 2006, 188 (6), 2073–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Walkenhorst WF; Sundrud JN; Laviolette JM, Additivity and synergy between an antimicrobial peptide and inhibitory ions. Biochim. Biophys Acta 2014, 1838 (9), 2234–2242. [DOI] [PubMed] [Google Scholar]
- 74.Hancock RE, Resistance mechanisms in Pseudomonas aeruginosa and other nonfermentative gram-negative bacteria. Clin. Infect. Dis 1998, 27 Suppl 1, S93–S99. [DOI] [PubMed] [Google Scholar]
- 75.Kaji T; Yano Y; Matsuzaki K, In-Cell FRET Indicates Magainin Peptide Induced Permeabilization of Bacterial Cell Membranes at Lower Peptide-to-Lipid Ratios Relevant to Liposomal Studies. ACS Infect Dis 2021, 7 (10), 2941–2945. [DOI] [PubMed] [Google Scholar]


