Abstract
In this work, the network complexity should be reduced with a concomitant reduction in the number of necessary training examples. The focus thus was on the dependence of proper evaluation metrics on the number of adjustable parameters of the considered deep neural network. The used data set encompassed Hematoxylin and Eosin (H&E) colored cell images provided by various clinics. We used a deep convolutional neural network to get the relation between a model’s complexity, its concomitant set of parameters, and the size of the training sample necessary to achieve a certain classification accuracy. The complexity of the deep neural networks was reduced by pruning a certain amount of filters in the network. As expected, the unpruned neural network showed best performance. The network with the highest number of trainable parameter achieved, within the estimated standard error of the optimized cross-entropy loss, best results up to 30% pruning. Strongly pruned networks are highly viable and the classification accuracy declines quickly with decreasing number of training patterns. However, up to a pruning ratio of 40%, we found a comparable performance of pruned and unpruned deep convolutional neural networks (DCNN) and densely connected convolutional networks (DCCN).
Keywords: Cell segmentation, Pruning, Deep neural networks, Training sample size, DNN complexity
Introduction
Modern deep convolutional neural networks (DCNN) can easily encompass millions of parameters, quite a number of them being redundant or close to zero in magnitude. As such, highly complex network architectures put a heavy load to the necessary hardware as well as to computation time and require a large training sample. Especially in the medical area, data samples are often small lacking sufficient data to train complex networks, thus incurring the risk of overfitting. Moreover, it is difficult to know how many training examples are necessary for a neural network with a given number of parameters. Hence, efforts have increased recently to reduce network complexity. One way to do so is network pruning, whereby one tries to simplify the network architecture without impairing network performance. Removing weak or redundant weights speeds up learning and occasionally also improves prediction accuracy. Anyway, pruning first tries to identify the most promising network units (neurons, channels, filters) to be removed. Afterwards, the pruned network model needs further training and fine-tuning to recover the base model’s prediction performance.13 In practice, analytical proxies like the number of adjustable parameters or floating point operations (FLOPs) are commonly used to quantify pruning efficacy. Such DCNNs with reduced network complexity have several advantages to offer: Concerning server computations, less complex models reduce bandwidth usage, power consumption, and operational costs, while computations on embedded systems or edge devices guaranty privacy, low latency, and better customization.37
Work related to pruning
When considering network pruning, often heuristics are employed to identify candidate units to be removed. Generally, either data-agnostic saliency criteria are computed or data-aware techniques are considered. The former group is, for example, represented by methods considering the Hessian matrix to identify weights to be removed without harming the prediction accuracy.15 As such techniques encompass heavy computations, alternatively weights may be grouped according to some similarity measures and each group is then replaced by its prototype weight.50 If weight values, i.e. their L2-norms, represent a saliency criterion, pruning them iteratively provides another simple alternative.13 The grain of salt with such techniques is the sparse matrices they create, which don’t help speeding up inference. These drawbacks can be leveraged by applying structural pruning methods.30 The latter commonly deploy L1-norm Lasso constraints and consider channel- or filter-based pruning approaches.31,33 More generally, a Group Lasso approach58 to pruning dealt with variable selection problems at the group level. Smoothing functions were introduced to alleviate problems arising with gradient computations at the origin. Enforcing sparsity even further, the L1-norm constraint has been replaced in some studies by a L1/2-norm regularization. The latter penalty yielded better sparsity than the L1-norm regularization at similar computational costs. Chang et al.4 proposed a network pruning method based on the L1/2 penalty, which reduced incorrect pruning by increasing the sparsity of the pre-trained models. Also in Wang et al,60 the L1/2-norm constraints have been considered at the channel level, while channel importance has been evaluated by a genetic algorithm. Structured sparsity also often needs some control based on attention mechanisms as pointed out in Torfi et al,54 whose work is an extension of the work of Wen et al.61 Similarly, the work of Lin et al35 considered regularized structured filter pruning by incorporating 2 different regularizers into the objective function to fully coordinate global outputs and local filter pruning operations. Also a novel regularization-based pruning method, named IncReg, was proposed by Wang et al,57 which incrementally assigns different regularization factors to different weights based on their relative importance. Further pruning methods on the filter pruning level considered the scaling factor of batch normalization layers as salient feature65 or removed filters close to their geometric median.16 Recently structured Dirichlet filter pruning has been proposed by Adamczewski and Park.1 The authors assigned Dirichlet distributions over every channel’s convolutional layers and estimated by variational inference the parameters of the distribution. The latter allowed them to identify irrelevant filters of the architecture. As a by-product, the method provided interpretable features. Filter pruning methods also have close connections to low rank network matrix decomposition techniques.34,5 Swaminathan et al52 proposed a sparse low rank weight matrix decomposition, while considering the significance of both input and output nodes of a layer. Recent work by Yeom et al63 also explored connections between pruning and matrix decomposition methods by developing a new energy-aware pruning technique. The preserved energy corresponded to the summed singular values of the filter decomposition and filters were pruned on the basis of their energy content. Concerning data-driven pruning methods on the weight level, the average percentage of zeros of nodes19 served as saliency criterion for weight importance and network trimming. Structured pruning at the channel level was considered by He et al17 by minimizing the reconstruction error for input data. Also the entropy of channel activation was considered a suitable saliency measure for channel removal38 as well as cross entropy.3 Concerning loss functions, their derivatives can serve as cost measures for feature dropping and were deployed by Molchanov et al41 to prune network structures based on grouped feature map activation. Similarly, the magnitude of gradients has also been considered a proper saliency measure for network pruning.11
Work related to cell segmentation
A seminal recent review of deep learning methods for cellular image analysis is provided by Moen et al40 highlighting to biologists the use of deep learning techniques for biological image understanding. If image classification is intended, biological images often lack appropriate annotation with class labels, hence transfer learning deems most suitable here.44,64 Transfer learning resorts to an image classifier that has been trained on a generally huge image data set, such as ImageNet, and only re-trains the final fully connected layer with an often small image data set of interest.10,64
Changes in cell morphology may not always be captured by a labeled training data set. Here, deep learning can be used to extract feature vectors rather than labels. These features can then be clustered according to some similarity measure and appropriately classified.24,44,49 Image classification can also be employed to identify cell states. A recent study trained a classifier with images labeled with a fluorescence marker for cell differentiation. The trained classifier was then employed to identify differentiated cells directly from corresponding bright field images.48,49 Yet another study applied a deep learning classifier to fluorescence images to determine characteristic spatial patterns in the images which indicated protein localization in large data sets from yeast27,28,43 and humans.51
Another large field of image analysis, where deep learning techniques can be employed favorably, is image segmentation. Classical segmentation methods often rely on simple thresholding,23 but with moderate success only. Modern segmentation techniques come in 2 flavors: semantic segmentation and instance-based segmentation. The former partitions a cell image into semantically meaningful parts, like cytoplasm, nucleus, cytoskeleton etc. and labels all pixels belonging to the latter appropriately. Instance-based segmentation, instead, focuses on every instance of a class in any given cell image. Early examples of the latter approaches are given by the software packages U-Net9,47 and DeepCell.56 They consider instance-based segmentation as a pixel-level classification task. The trained image classifier then generates predictions about class membership pixel-wise thereby grouping pixels into categories like cell interior, cell edges, and background, for instance. A rather more classical segmentation technique is watershedding, whereby deep learning has recently been employed to learn a distance measure. This allows to build a mask encompassing those pixels which have at least a minimal distance from the background of the image.2,7 A recent application to segment single cells in images performed surprisingly well.59 Other approaches to image segmentation are based on object detection techniques, whereby the bounding box of every object is identified. Deep learning algorithms like Faster R-CNN46 and Retinanet36 combine bounding box detection with a suppression of non-maximal pixels to avoid redundant bounding box predictions.18,21,22,55 A rather different approach to image segmentation considers the segmentation problem as a vector embedding problem.6,45 Thereby, all pixels belonging to an object are assigned by a discriminative loss function to the same vector, while pixels of different objects belong to different vectors. All objects are finally identified through clustering techniques applied on the embedding space.14,39,66 Recently, most of these techniques have been generalized to be applicable to 3D data sets.12,64,53 These deep learning-based image segmentation techniques are applicable in many fields of science. They help to automatize common computer vision workflows and render possible segmentation tasks that previously deemed impossible. For example, a precise quantification of localization-based live-cell reporters became possible by way of an accurate identification of the cytoplasm in mammalian cells.42,56 A recent study explored this image segmentation method to investigate the mechanisms of cell size control during the fission of yeast cells.8 Another exciting application concerns the use of instance-based segmentation in pathology images.25,26,29,39 There interactions between tumor cells and immune cells were investigated employing spatial proteomics methods in a formalin-fixed and paraffin-embedded substrate.26
In this work, filter pruning methods are deployed to reduce network complexity and the question of the number of necessary training examples for various numbers of trainable parameters is investigated, using the example of a cell segmentation network.
Method
In this section, the used data set is described and its preprocessing is explained. Moreover, we sketch the state-of-the-art neural network applied, and provide an overview of the number of trainable parameters.
Data set
In this work, part of the data set provided by Kumar et al29 - henceforth called the Kumar data set for simplicity - was used. It encompassed Hematoxylin and Eosin stain (H & E) cell images provided by various clinics (for an example see Fig. 1). Table 1 collects all information about the various images employed in this study.
Fig. 1.
Cell image of size pixels with corresponding segmentation.62
Table 1.
A list of images used for pre-training, training, and testing.
| Patient ID TCGA- | Organ | Disease type | Usage |
|---|---|---|---|
| A7 − A13E − 01Z − DX1 | Breast | Breast invasive carcinoma | Pre-training |
| A7 − A13F − 01Z − DX1 | Breast | Breast invasive carcinoma | Pre-training |
| AR − A1AK − 01Z − DX1 | Breast | Breast invasive carcinoma | Training |
| E2 − A1B5 − 01Z − DX1 | Breast | Breast invasive carcinoma | Testing |
| HE − Z − DX1 | Kidney | Kidney renal papillary cell carcinoma | Training |
| B0 − Z − DX1 | Kidney | Kidney renal clear cell carcinoma | Testing |
| 38 − Z − DX1 | Liver | Lung adenocarcinoma | Training |
| 21 − Z − DX1 | Liver | Lung squamous cell carcinoma | Testing |
| G9 − Z − DX1 | Prostate | Prostate adenocarcinoma | Training |
| CH − Z − DX1 | Prostate | Prostate adenocarcinoma | Testing |
| DK − A2I6 − 01A − TS1 | Bladder | Bladder urothelial carcinoma | Training |
| G2 − A2EK − 01A − TSB | Bladder | Bladder urothelial carcinoma | Testing |
| AY − A8Y K − 01A − TS1 | Colon | Colon adenocarcinoma | Training |
| NH − A8F7 − 01A − TS1 | Colon | Colon adenocarcinoma | Testing |
| KB − A93J − 01A − TS1 | Stomach | Stomach adenocarcinoma | Training |
| RD − A8N9 − 01A − TS1 | Stomach | Stomach adenocarcinoma | Testing |
The Kumar data set consisted of an equal number of images for every tissue class. Only part of the data was used in this study, thereby assuring a balanced data set such that each tissue class was represented by one image for training and another image for testing. While selecting them, care was taken that the images came from different clinics and, if possible, represented different tumor types. This selection should render the classification task as hard as possible. Furthermore, the trained networks should be suitable for transfer learning on cell images of other clinics. All images were subdivided into overlapping image patches of size ( pixels). In total, the data set encompassed 14 654 944 such image patches. However, only Ntr = 915 934 overlapping image patches were taken of every input image and constituted the size of the training sample for an unpruned DCNN. Furthermore, the number Ntr of training samples was chosen to match the number 934 of adjustable parameters of our deep convolutional neural network (DCNN). The pixels of every image were classified as belonging either to class cell or class background, whereby the goal of the classification effort was to properly classify the center pixel of every image patch. During the training process, the total number Ntr of image patches were separated into 70% training examples and 30% validation examples using a cross-validation method. Also from every tissue, class 1 image was kept out of bag for testing and estimating standard errors. Thus, the out of bag test set consisted of 7 images, each for every tissue class, from which a corresponding number of patches was drawn for testing.
Neural network architecture
To optimize the number of training examples needed by the given neural network architecture, a deep convolutional neural network (DCNN) in analogy with Xing et al62 and a densely connected convolutional network (DCCN)20 were employed. The architecture of the networks is illustrated in Fig. 2 and Fig. 3 and the trainable parameters are given in Table 2. The DCNN network consisted of 3 convolutional layers, 3 max-pooling layers, 2 fully connected layers, and 1 softmax output layer. The input array encompassed × 3 pixels, which corresponded to a 3-channel RGB image patch of the cell images. The output layer implemented a softmax activation function. The DCCN network consisted of 4 convolutional layers, 1 max-pooling, and 3 average-pooling layers, 3 dense blocks, 1 fully connected layer, and 1 sigmoid output layer. The input array size was the same as for the DCNN. Because of the binary classification at the output, a cross-entropy loss function Lce was implemented to guide the training of both the DCNN and the DCCN.
Fig. 2.
Architecture of the DCNN for cell segmentation similar to Xing et al.62
Fig. 3.
Architecture of the DCCN for cell segmentation similar to Huang et al.20
Table 2.
Number of trainable parameters in each layer of the 2 neural network architectures.
| DCNN |
DCCN |
||
|---|---|---|---|
| Layer | Parameters | Layer | Parameters |
| conv 1 | 1225 | conv 1 | 9536 |
| conv 2 | 31 300 | DenseBlock | 335 040 |
| conv 3 | 144 080 | conv 2 | 33 280 |
| FC 1 | 738 104 | DenseBlock | 1 047 552 |
| FC 2 | 2050 | conv 3 | 132 096 |
| DenseBlock | 2 709 888 | ||
| conv 4 | 526 336 | ||
| FC | 1026 | ||
| Total number | 916 759 | Total number | 4 794 754 |
Experiments
The central question we were focusing on, was about the relation between a model’s complexity, its concomitant set of parameters, and the size of the training sample necessary to achieve a satisfactory classification accuracy. Thus, the study explored possibilities to reduce network complexity resulting in a reduced number of needed training examples. For this task, we chose a neural network with several million parameters and another with less than 1 million parameters. The selected network architectures perfectly suited this purpose, as we only wanted to achieve a classification of the central pixel of every image patch.
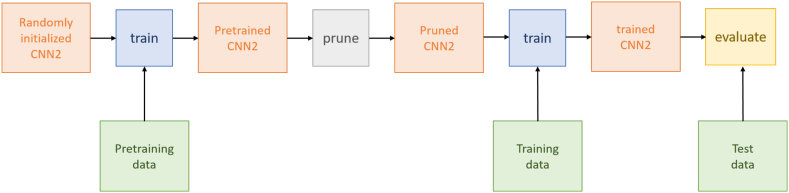
In Fig. 4, the training and evaluation process is illustrated. First, the DCNN and DCCN were implemented and randomly initialized. The networks were then pre-trained for 10 epochs employing the breast data set indicated in Table 1 of the previous section. The data set, used for pre-training, consisted of only 2 breast carcinoma cell images recorded in a clinic. These images were partitioned into 1 831 868 image patches. This set of image patches was used to train the set of 934 adjustable parameters of our network models and provided us with a pre-trained neural network that served as the basis for all further experiments. To reduce network complexity, the pre-trained neural networks were pruned by applying different pruning ratios, starting with 0% pruning and increasing to 90% pruning in steps of 10%. Thereby, pruning was done by adopting the structured L1 pruning method implemented by PyTorch.
Fig. 4.
Principal training and evaluation process.
This method is based on Li et al32 and uses the L1 norm of filters as an indicator of the importance of the filter. It is assumed that if the L1 norm of a filter is small, the importance of the filter is also low. This norm is calculated for all filters of a conv layer and then 30% of the least important filters are eliminated. This procedure is carried out one after the other for each conv layer.
All layers of the pruned networks were then further trained for 30 epochs employing various numbers of training examples Ntr(r). The latter have been computed by taking the following multiples r of the number Np of trainable parameters of the pruned DCNN architecture:
| (1) |
For the DCCN computations were performed using only r values between 0.5 and 0.001, because of the demanding compute time and the missing number of training samples. Note that this resulted in different numbers of training examples depending on the chosen pruning ratios. Additionally, all pruned neural networks were trained for further comparability with identical training sample sizes as given in Table 3. The latter have been deduced from the number Np of parameters of the unpruned DCNN or DCCN, respectively.
Table 3.
Reduction factor r and corresponding number of training patterns Ntr(r). The number of training samples with the label cell Ncell and the one with the label no cell Nnocell are also given. Remember that the number of adjustable parameters of the unpruned DCNN amounted to 934 and for the DCCN amounted to 794 754.
| DCNN | ||||||||
|---|---|---|---|---|---|---|---|---|
| r | 0.001 | 0.005 | 0.01 | 0.05 | 0.1 | 0.5 | 1 | 2 |
| Ntr(r) | 916 | 4580 | 9159 | 45 797 | 91 593 | 457 967 | 915 934 | 1 831 868 |
| Ncell | 675 | 3404 | 6816 | 34 404 | 68 827 | 344 373 | 688 780 | 1 376 401 |
| Nnocell | 241 | 1176 | 2343 | 11 393 | 22 766 | 113 594 | 227 154 | 455 467 |
| DCCN | ||||||
|---|---|---|---|---|---|---|
| r | 0.001 | 0.005 | 0.01 | 0.05 | 0.1 | 0.5 |
| Ntr(r) | 4795 | 23 974 | 47 948 | 239 738 | 479 475 | 2 397 377 |
| Ncell | 3622 | 18 045 | 36 047 | 180 505 | 360 383 | 1 800 694 |
| Nnocell | 1173 | 5929 | 11 901 | 59 233 | 119 092 | 596 683 |
After the training process, every trained network was applied to segment the test data sample. When increasing Ntr, every larger training data set contained the smaller training data set used before. Also care was taken to ensure balanced training data sets with a roughly equal number of pixels per class (cell, non-cell) in each set . To ensure that the data sets are representative, the distributions of the mutual information between all images of a data set were chosen to be similar.
Despite facing a 2-class classification problem here, we formulate the optimization problem for K classes. Hence, training was guided by a softmax cross-entropy loss function
| (2) |
where N denotes the number of pixels in input patch xi. Furthermore, yin ∈ {0.1}K denotes a 1-hot encoded ground-truth vector and represents a vector of class membership probabilities.
Evaluation metrics play an important role in assessing the outcomes of segmentation models. Hence, prediction quality was assessed deploying the following performance metrics: Dice Sørensen coefficient DSC, accuracy acc, sensitivity Se, and recall Rec, specificity Sp, and precision Pre.
In order to analyze the robustness of the training process, the unpruned DCNN was trained 7-fold with different random seed numbers and training examples, corresponding to a reduction factor r = 0.1. Finally, all test images were segmented with the trained networks. The standard deviation over these predictions was taken as a measure of the inaccuracy of the predictions for all investigated network configurations, which could not be fully exploited due to an insufficient computing capacity.
Results
Test of reproducibility
As mentioned in the last section, in order to be able to distinguish networks with different pruning configurations, hence different numbers of trainable parameters, the reproducibility of the network architecture was examined first. In 7 different training runs of an unpruned network with Ntr(r = 1.0) = 915 934 training examples, each initialized with a different random seed, the predictions of the pixel class memberships achieved with a DCNN model a standard error for the accuracy of sacc = 1.7 · 10−3 and for the cross entropy of sce = 4.3 · 10−3, respectively. These numbers were rounded to sacc = 2 · 10−3 and sce = 4 · 10−3 for convenience. For the DCCN model corresponding standard errors read sacc = 2.8 · 10 − 3 and sce = 1.3 · 10−3, respectively, which were rounded to sacc = 3 · 10−3 and sce = 1 · 10−3 for the sake of an easy comparison. These figures of merit were considered valid for all network configurations investigated in this study, as already explained above.
Varying the number of training examples
Next the unpruned DCNN was trained with various numbers of training examples. The latter were determined, given the pruning ratio δ, as Ntr(r,δ) = Ntr(r)(1 - δ). Up to a pruning ratio δ of 50% ≡ δ = 0.5 and Ntr ≥ Np, the accuracy remained practically constant with a maximal test accuracy of acc = , and a minimal cross-entropy loss of Lce ≡ H(p,q) = was achieved. Note that 99% of the maximal test accuracy could already be obtained with only 9159 training patterns, corresponding to r = 0.01 (see Table 4).
Table 4.
Cross-entropy loss Lce ≡ H(p,q), Segmentation accuracy acc, Sensitivity, Specificity, and Dice coefficient for different trained network configurations. Number of training examples: Ntr, related cross-entropy loss: Lce, test accuracy: acc. The best result per row, i.e. per reduction factor r, is highlighted as bold. Note that sce = 1 · 10−3 and sacc = 3 ⋅ 10−3 are considered to hold for all DCNN network configurations.
| r | Ntr | Sensitivity | Specificity | DiceCo | acc | Lce | Ntr | Sensitivity | Specificity | DiceCo | acc | Lce |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0% pruning | 10% pruning | |||||||||||
| 2 | 1 831 868 | 0.835 | 0.952 | 0.802 | 0.933 | 0.176 | 1 648 681 | 0.834 | 0.953 | 0.803 | 0.934 | 0.178 |
| 1 | 915 934 | 0.828 | 0.953 | 0.801 | 0.932 | 0.180 | 824 341 | 0.810 | 0.953 | 0.790 | 0.930 | 0.181 |
| 0.5 | 457 967 | 0.828 | 0.952 | 0.799 | 0.931 | 0.180 | 412 170 | 0.820 | 0.950 | 0.790 | 0.929 | 0.184 |
| 0.1 | 91 593 | 0.828 | 0.946 | 0.788 | 0.927 | 0.197 | 82 434 | 0.829 | 0.944 | 0.783 | 0.926 | 0.190 |
| 0.05 | 45 797 | 0.827 | 0.945 | 0.785 | 0.924 | 0.204 | 41 217 | 0.811 | 0.946 | 0.776 | 0.923 | 0.210 |
| 0.01 | 9159 | 0.789 | 0.946 | 0.768 | 0.921 | 0.220 | 8243 | 0.802 | 0.930 | 0.747 | 0.910 | 0.237 |
| 0.005 | 4580 | 0.744 | 0.949 | 0.743 | 0.915 | 0.241 | 4122 | 0.779 | 0.945 | 0.758 | 0.917 | 0.239 |
| 0.001 | 916 | 0.684 | 0.936 | 0.674 | 0.899 | 0.306 | 824 | 0.619 | 0.944 | 0.630 | 0.898 | 0.343 |
| 20% pruning | 30% pruning | |||||||||||
| 2 | 1 465 494 | 0.815 | 0.956 | 0.798 | 0.933 | 0.177 | 1 282 308 | 0.789 | 0.960 | 0.791 | 0.933 | 0.181 |
| 1 | 732 747 | 0.834 | 0.950 | 0.797 | 0.930 | 0.182 | 641 154 | 0.859 | 0.942 | 0.798 | 0.929 | 0.177 |
| 0.5 | 366 374 | 0.817 | 0.951 | 0.792 | 0.929 | 0.183 | 320 577 | 0.819 | 0.950 | 0.789 | 0.928 | 0.189 |
| 0.1 | 73 275 | 0.809 | 0.947 | 0.776 | 0.924 | 0.201 | 64 115 | 0.809 | 0.945 | 0.775 | 0.923 | 0.207 |
| 0.05 | 36 637 | 0.827 | 0.941 | 0.777 | 0.921 | 0.204 | 32 058 | 0.800 | 0.944 | 0.767 | 0.921 | 0.205 |
| 0.01 | 7327 | 0.825 | 0.930 | 0.765 | 0.915 | 0.231 | 6411 | 0.798 | 0.931 | 0.750 | 0.912 | 0.231 |
| 0.005 | 3664 | 0.715 | 0.951 | 0.730 | 0.913 | 0.238 | 3206 | 0.769 | 0.937 | 0.746 | 0.912 | 0.242 |
| 0.001 | 733 | 0.596 | 0.946 | 0.613 | 0.898 | 0.327 | 641 | 0.606 | 0.941 | 0.606 | 0.896 | 0.373 |
| 40% pruning | 50% pruning | |||||||||||
| 2 | 1 099 121 | 0.742 | 0.933 | 0.709 | 0.932 | 0.181 | 915 934 | 0.718 | 0.935 | 0.695 | 0.930 | 0.181 |
| 1 | 549 560 | 0.723 | 0.936 | 0.702 | 0.931 | 0.181 | 457 967 | 0.727 | 0.933 | 0.699 | 0.930 | 0.184 |
| 0.5 | 274 780 | 0.719 | 0.934 | 0.697 | 0.927 | 0.185 | 228 984 | 0.709 | 0.936 | 0.692 | 0.928 | 0.191 |
| 0.1 | 54 956 | 0.712 | 0.931 | 0.687 | 0.924 | 0.206 | 45 797 | 0.698 | 0.930 | 0.676 | 0.921 | 0.213 |
| 0.05 | 27 478 | 0.727 | 0.922 | 0.684 | 0.920 | 0.212 | 22 899 | 0.716 | 0.927 | 0.681 | 0.920 | 0.228 |
| 0.01 | 5496 | 0.661 | 0.929 | 0.653 | 0.914 | 0.237 | 4580 | 0.702 | 0.930 | 0.679 | 0.919 | 0.232 |
| 0.005 | 2748 | 0.674 | 0.930 | 0.663 | 0.915 | 0.237 | 2290 | 0.671 | 0.931 | 0.664 | 0.916 | 0.246 |
| 0.001 | 550 | 0.540 | 0.938 | 0.559 | 0.901 | 0.323 | 458 | 0.534 | 0.909 | 0.501 | 0.880 | 0.344 |
| 60% pruning | 70% pruning | |||||||||||
| 2 | 732 747 | 0.826 | 0.946 | 0.783 | 0.927 | 0.186 | 549 560 | 0.816 | 0.953 | 0.791 | 0.930 | 0.181 |
| 1 | 366 374 | 0.797 | 0.954 | 0.783 | 0.928 | 0.187 | 274 780 | 0.813 | 0.951 | 0.787 | 0.929 | 0.191 |
| 0.5 | 183 187 | 0.800 | 0.952 | 0.783 | 0.927 | 0.195 | 137 390 | 0.808 | 0.950 | 0.784 | 0.928 | 0.199 |
| 0.1 | 36 637 | 0.805 | 0.944 | 0.770 | 0.920 | 0.218 | 27 478 | 0.771 | 0.951 | 0.764 | 0.921 | 0.227 |
| 0.05 | 18 319 | 0.783 | 0.948 | 0.764 | 0.919 | 0.224 | 13 739 | 0.765 | 0.948 | 0.755 | 0.919 | 0.236 |
| 0.01 | 3664 | 0.676 | 0.950 | 0.700 | 0.907 | 0.274 | 2748 | 0.441 | 0.975 | 0.525 | 0.887 | 0.301 |
| 0.005 | 1832 | 0.476 | 0.968 | 0.539 | 0.892 | 0.359 | 1374 | 0.486 | 0.972 | 0.576 | 0.889 | 0.339 |
| 0.001 | 366 | 0.443 | 0.923 | 0.441 | 0.863 | 0.390 | 275 | 0.146 | 0.973 | 0.201 | 0.836 | 0.481 |
| 80% pruning | 90% pruning | |||||||||||
| 2 | 366 374 | 0.791 | 0.956 | 0.783 | 0.929 | 0.193 | 183 187 | 0.816 | 0.941 | 0.774 | 0.921 | 0.232 |
| 1 | 183 187 | 0.812 | 0.947 | 0.782 | 0.926 | 0.202 | 91 593 | 0.801 | 0.945 | 0.771 | 0.922 | 0.226 |
| 0.5 | 91 593 | 0.792 | 0.948 | 0.771 | 0.923 | 0.217 | 45 797 | 0.678 | 0.951 | 0.704 | 0.911 | 0.243 |
| 0.1 | 18 319 | 0.618 | 0.958 | 0.654 | 0.908 | 0.242 | 9159 | 0.653 | 0.950 | 0.685 | 0.905 | 0.266 |
| 0.05 | 9159 | 0.523 | 0.970 | 0.585 | 0.901 | 0.296 | 4580 | 0.513 | 0.962 | 0.576 | 0.893 | 0.308 |
| 0.01 | 1832 | 0.428 | 0.974 | 0.498 | 0.887 | 0.336 | 916 | 0.000 | 1.000 | 0.000 | 0.818 | 0.476 |
| 0.005 | 916 | 0.000 | 1.000 | 0.000 | 0.818 | 0.497 | 458 | 0.000 | 1.000 | 0.000 | 0.818 | 0.471 |
| 0.001 | 183 | 0.097 | 0.968 | 0.129 | 0.809 | 0.521 | 92 | 0.000 | 1.000 | 0.000 | 0.818 | 0.465 |
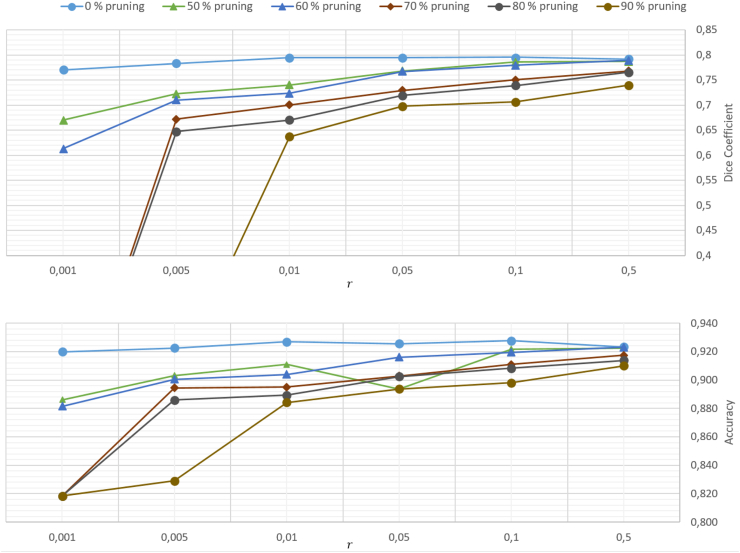
First, Fig. 5 illustrates the dependence of the test accuracies, achieved with the various DCNN pruning configurations, on the number of training examples. Corresponding result in case of the more complex DCCN are shown in Fig. 6. All networks were trained for best image segmentation by minimizing the cross-entropy loss. During training, the number of necessary training examples was varied for a given set of adjustable parameters as determined by the pruning ratio. The former was determined by the chosen network architecture via r · Np, with r denoting a factor multiplying the number Np of trainable parameters. The most complex unpruned network, either DCNN or DCCN, yielded best results among all training runs. It is remarkable to see that even a reduction of the number of training samples by a factor r = 0.001 leads to a rather modest decrease of the accuracy by roughly 5% only for both network architectures. A similar observation holds for the Dice coefficient, which decreased by 6% and 1%, respectively. With increased network pruning, however, both accuracy and Dice coefficient degrade substantially for strongly reduced training sample sizes r < 0.1. With δ = 0.9, for example, we observed a decrease of the accuracy by 11% and 9%, respectively while Dice coefficients decreased to zero for both networks. This is amazing as a reduced network complexity results in a smaller number of adjustable model parameters. Obviously, a too strong reduction of the training sample size results in a worsening of the learning ability of the resulting network. Considering the cross-entropy loss, i.e. the objective that was optimized during training, it steadily increases with decreasing reduction factor r. For the unpruned networks, cross entropy increased by 70% in case of a DCNN but only by 4% in case of a DCCN. However, the increase is especially strong for less complex networks at pruning ratios larger than 50%, where Lce more than doubles in case of the DCNN, while the increase is only 22% in case of the DCCN.
Fig. 5.
Dice coefficient and test accuracy for the DCNN with different pruning ratios. The network configurations were trained with different data sample sizes, scaled by the factor r.
Fig. 6.
Dice coefficient and test accuracy for the DCCN with different pruning ratios. The network configurations were trained with different data sample sizes, scaled by the factor r.
Reducing network complexity by pruning
Next, the dependence of the performance metrics on network complexity was studied. With large training sample sizes, i.e. r = 1 and r = 2, respectively, in case of the DCNN network and r = 0.5 in case of the DCCN network, classification accuracy remained practically unaffected. This was expected as there the largest numbers of training examples were used. The same held true for the Dice coefficient, with the exception of 40% and 50% pruning ratios in case of the DCNN network, which unexpectedly showed a sudden drop for all numbers of training examples. Lacking any reasonable explanation, we consider the results for these 2 pruning ratios as outliers. Remarkably, up to a pruning ratio of 50% prediction accuracy remained rather high, amounting to a drop of a mere 0.2%. This is not the case for the corresponding Dice coefficient, which for the DCNN network decreased by almost 13% over the same range of network pruning (compare Table 4 but remember that the 50% values were considered outlier). More realistic might be the drop seen over the entire pruning range δ = 0 → 0.9, where the corresponding figures read 1% for the accuracy and 4% for the Dice coefficient. Remarkably, for the more complex DCCN network the corresponding decrease at r = 0.5 amounted to 1%, roughly for δ = 0 → 0.5 while it was seen to be 7% for δ = 0 → 0.9 (compare Table 5). The corresponding figures for the accuracy read 0.6% and 3%, respectively. For even larger pruning ratios, i.e. r = 0.001, both metrics degraded rapidly with decreasing training sample size and decreasing network complexity for both network architectures. Thus, the accuracy dropped by 9% for δ = 0 → 0.9 and 2% for δ = 0 → 0.5 in case of the DCNN network. For the DCCN architecture, the corresponding accuracies dropped by 11% for δ = 0 → 0.9 and 4% for δ = 0 → 0.5. This degradation was, however, especially visible for the Dice coefficient, which reduced to zero for δ = 0 → 0.9 for both architectures and yielded 25% for δ = 0 → 0.5 in case of DCNN and 13% under the same conditions in case of DCCN. Considering the cross-entropy loss, it again stood almost constant up to a pruning ratio of 50% before starting to increase with further decreasing network complexity. For instance, at r = 1 cross entropy increased in case of a DCNN by a mere 2% for δ = 0 → 0.5 but by 26% for δ = 0 → 0.9. At high reduction factors r = 0.001, the corresponding values were 12% and 52%, respectively. In case of more complex DCCN networks, we observed an increase by 3% for δ = 0 → 0.9 and by 0.3% for δ = 0 → 0.5 and r = 0.5. Again, for very parsimonious networks (r = 0.001) we, instead, observed an increase in LCE by 39% for δ = 0 → 0.9 and by 8% for δ = 0 → 0.5. The results for the cross-entropy loss Lce and the test accuracy acc for different network configurations are summarized in Table 4 and Table 5.
Table 5.
Cross-entropy loss Lce ≡ H(p,q), Segmentation accuracy acc, Sensitivity, Specificity, and Dice coefficient for different trained network configurations. Number of training examples: Ntr, related cross-entropy loss: Lce, test accuracy: acc. The best result per row, i.e. per reduction factor r, is highlighted as bold. Note that sce = 1 · 10−3 and sacc = 3 · 10−3 are considered to hold for all DCCN network configurations.
| r | Ntr | Sensitivity | Specificity | DiceCo | acc | Lce | Ntr | Sensitivity | Specificity | DiceCo | acc | Lce |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0% pruning | 50% pruning | |||||||||||
| 0.5 | 2 397 377 | 0.846 | 0.940 | 0.792 | 0.923 | 0.384 | 1 198 689 | 0.838 | 0.941 | 0.787 | 0.922 | 0.385 |
| 0.1 | 479 475 | 0.823 | 0.952 | 0.796 | 0.928 | 0.382 | 239 738 | 0.829 | 0.943 | 0.786 | 0.922 | 0.386 |
| 0.05 | 239 738 | 0.838 | 0.944 | 0.795 | 0.926 | 0.384 | 119 869 | 0.823 | 0.935 | 0.768 | 0.894 | 0.419 |
| 0.01 | 47 948 | 0.818 | 0.949 | 0.795 | 0.927 | 0.386 | 23 974 | 0.754 | 0.942 | 0.740 | 0.911 | 0.399 |
| 0.005 | 23 974 | 0.803 | 0.947 | 0.783 | 0.923 | 0.387 | 11 987 | 0.746 | 0.935 | 0.723 | 0.903 | 0.408 |
| 0.001 | 4795 | 0.771 | 0.951 | 0.770 | 0.920 | 0.396 | 2397 | 0.670 | 0.935 | 0.669 | 0.886 | 0.428 |
| 60% pruning | 70% pruning | |||||||||||
| 0.5 | 958 951 | 0.835 | 0.944 | 0.789 | 0.923 | 0.386 | 719 213 | 0.817 | 0.940 | 0.768 | 0.917 | 0.391 |
| 0.1 | 191 790 | 0.816 | 0.944 | 0.779 | 0.920 | 0.390 | 143 843 | 0.799 | 0.936 | 0.750 | 0.911 | 0.395 |
| 0.05 | 95 895 | 0.807 | 0.940 | 0.767 | 0.916 | 0.392 | 71 921 | 0.773 | 0.932 | 0.729 | 0.903 | 0.402 |
| 0.01 | 19 179 | 0.733 | 0.941 | 0.724 | 0.904 | 0.408 | 14 384 | 0.728 | 0.926 | 0.700 | 0.895 | 0.413 |
| 0.005 | 9590 | 0.715 | 0.940 | 0.710 | 0.900 | 0.415 | 7192 | 0.647 | 0.942 | 0.672 | 0.854 | 0.421 |
| 0.001 | 1918 | 0.595 | 0.939 | 0.613 | 0.882 | 0.435 | 1438 | 0.000 | 1.000 | 0.000 | 0.818 | 0.500 |
| 80% pruning | 90% pruning | |||||||||||
| 0.5 | 479 475 | 0.834 | 0.932 | 0.765 | 0.914 | 0.392 | 239 738 | 0.772 | 0.937 | 0.739 | 0.910 | 0.396 |
| 0.1 | 95 895 | 0.770 | 0.936 | 0.739 | 0.908 | 0.401 | 47 948 | 0.745 | 0.924 | 0.706 | 0.898 | 0.411 |
| 0.05 | 47 948 | 0.749 | 0.931 | 0.719 | 0.902 | 0.409 | 23 974 | 0.737 | 0.922 | 0.698 | 0.894 | 0.419 |
| 0.01 | 9590 | 0.686 | 0.927 | 0.670 | 0.889 | 0.424 | 4795 | 0.625 | 0.932 | 0.637 | 0.884 | 0.434 |
| 0.005 | 4795 | 0.651 | 0.928 | 0.647 | 0.886 | 0.428 | 2397 | 0.098 | 0.991 | 0.154 | 0.829 | 0.470 |
| 0.001 | 959 | 0.000 | 1.000 | 0.000 | 0.818 | 0.500 | 479 | 0.000 | 1.000 | 0.000 | 0.818 | 0.500 |
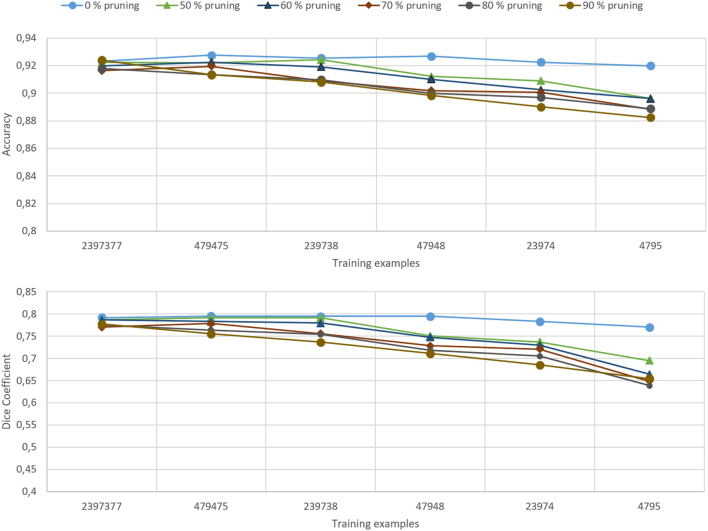
Network training with fixed training set size
In order to be able to directly compare the performance of pruned neural networks, the different network configurations were trained once again for both network architectures with identical numbers of training examples. All results are collected in Table 6 and Table 7, respectively. Note that here the number Ntr of training examples corresponds to a fraction r deduced from the number of adjustable parameters Np of the original, unpruned network. The number of predicted cells for different pruning ratios δ and number of training examples Ntr is illustrated in case of the DCNN architecture in Fig. 7 (See Fig. 8).
Table 6.
Cross-entropy loss Lce ± 10−3 and statistical metrics (Sensitivity, Specificity, acc ± 10-3) for the segmentation prediction of different network configurations are presented. Note that here the number of training examples is the same for all tested pruning ratios. Again remember that the number of adjustable parameters of the unpruned DCNN amounted to 6 959. The rows marked yellow present results obtained with networks where the condition Ntr ≥ Np held.
| Ntr | Sensitivity | Specificity | DiceCo | acc | Lce | Sensitivity | Specificity | DiceCo | acc | Lce |
|---|---|---|---|---|---|---|---|---|---|---|
| 0% Pruning, | 10% Pruning, | |||||||||
| 1 831 868 | 0.835 | 0.952 | 0.802 | 0.933 | 0.176 | 0.828 | 0.955 | 0.804 | 0.934 | 0.178 |
| 915 934 | 0.828 | 0.953 | 0.801 | 0.932 | 0.180 | 0.821 | 0.951 | 0.792 | 0.930 | 0.180 |
| 457 967 | 0.828 | 0.952 | 0.799 | 0.931 | 0.180 | 0.824 | 0.950 | 0.792 | 0.930 | 0.180 |
| 91 593 | 0.828 | 0.946 | 0.788 | 0.927 | 0.197 | 0.803 | 0.951 | 0.780 | 0.926 | 0.193 |
| 45 797 | 0.827 | 0.945 | 0.785 | 0.924 | 0.204 | 0.800 | 0.947 | 0.773 | 0.923 | 0.211 |
| 9159 | 0.789 | 0.946 | 0.768 | 0.921 | 0.220 | 0.791 | 0.933 | 0.739 | 0.909 | 0.252 |
| 4580 | 0.744 | 0.949 | 0.743 | 0.915 | 0.241 | 0.752 | 0.950 | 0.753 | 0.918 | 0.234 |
| 916 | 0.684 | 0.936 | 0.674 | 0.899 | 0.306 | 0.743 | 0.927 | 0.704 | 0.898 | 0.305 |
| 20% Pruning, | 30% Pruning, | |||||||||
| 1 831 868 | 0.811 | 0.954 | 0.790 | 0.931 | 0.178 | 0.829 | 0.951 | 0.796 | 0.931 | 0.179 |
| 915 934 | 0.827 | 0.950 | 0.792 | 0.930 | 0.179 | 0.822 | 0.952 | 0.797 | 0.932 | 0.178 |
| 457 967 | 0.813 | 0.952 | 0.789 | 0.930 | 0.182 | 0.822 | 0.952 | 0.792 | 0.931 | 0.182 |
| 91 593 | 0.822 | 0.949 | 0.789 | 0.926 | 0.199 | 0.818 | 0.947 | 0.782 | 0.925 | 0.198 |
| 45 797 | 0.801 | 0.947 | 0.772 | 0.922 | 0.206 | 0.821 | 0.944 | 0.777 | 0.923 | 0.208 |
| 9159 | 0.795 | 0.938 | 0.756 | 0.916 | 0.224 | 0.774 | 0.946 | 0.757 | 0.918 | 0.226 |
| 4580 | 0.741 | 0.950 | 0.745 | 0.917 | 0.234 | 0.758 | 0.940 | 0.739 | 0.912 | 0.252 |
| 916 | 0.634 | 0.958 | 0.676 | 0.908 | 0.280 | 0.731 | 0.924 | 0.699 | 0.896 | 0.303 |
| 40% Pruning, | 50% Pruning, | |||||||||
| 1 831 868 | 0.825 | 0.949 | 0.789 | 0.929 | 0.179 | 0.807 | 0.954 | 0.790 | 0.931 | 0.180 |
| 915 934 | 0.838 | 0.947 | 0.793 | 0.929 | 0.176 | 0.817 | 0.951 | 0.789 | 0.929 | 0.177 |
| 457 967 | 0.817 | 0.952 | 0.791 | 0.931 | 0.185 | 0.816 | 0.952 | 0.790 | 0.930 | 0.185 |
| 91 593 | 0.819 | 0.947 | 0.783 | 0.925 | 0.203 | 0.790 | 0.953 | 0.776 | 0.926 | 0.205 |
| 45 797 | 0.834 | 0.940 | 0.778 | 0.923 | 0.209 | 0.784 | 0.949 | 0.765 | 0.922 | 0.210 |
| 9159 | 0.774 | 0.949 | 0.761 | 0.921 | 0.222 | 0.789 | 0.939 | 0.755 | 0.915 | 0.237 |
| 4580 | 0.791 | 0.930 | 0.742 | 0.909 | 0.249 | 0.748 | 0.954 | 0.757 | 0.921 | 0.228 |
| 916 | 0.660 | 0.940 | 0.667 | 0.899 | 0.321 | 0.611 | 0.944 | 0.634 | 0.896 | 0.309 |
| 60% Pruning, | 70% Pruning, | |||||||||
| 1 831 868 | 0.853 | 0.946 | 0.800 | 0.930 | 0.184 | 0.816 | 0.950 | 0.787 | 0.928 | 0.187 |
| 915 934 | 0.826 | 0.948 | 0.790 | 0.929 | 0.179 | 0.835 | 0.934 | 0.772 | 0.920 | 0.204 |
| 457 967 | 0.825 | 0.950 | 0.791 | 0.929 | 0.190 | 0.838 | 0.947 | 0.795 | 0.930 | 0.193 |
| 91 593 | 0.822 | 0.944 | 0.778 | 0.923 | 0.214 | 0.827 | 0.946 | 0.788 | 0.927 | 0.215 |
| 45 797 | 0.783 | 0.949 | 0.766 | 0.922 | 0.215 | 0.801 | 0.947 | 0.773 | 0.923 | 0.224 |
| 9159 | 0.771 | 0.942 | 0.750 | 0.915 | 0.237 | 0.654 | 0.957 | 0.687 | 0.910 | 0.258 |
| 4580 | 0.750 | 0.945 | 0.746 | 0.915 | 0.242 | 0.682 | 0.951 | 0.704 | 0.907 | 0.269 |
| 916 | 0.401 | 0.968 | 0.461 | 0.881 | 0.377 | 0.008 | 1.000 | 0.015 | 0.820 | 0.455 |
| 80% Pruning, | 90% Pruning, | |||||||||
| 1 831 868 | 0.838 | 0.949 | 0.799 | 0.930 | 0.181 | 0.839 | 0.939 | 0.780 | 0.923 | 0.207 |
| 915 934 | 0.833 | 0.950 | 0.796 | 0.930 | 0.191 | 0.831 | 0.942 | 0.780 | 0.923 | 0.218 |
| 457 967 | 0.827 | 0.947 | 0.789 | 0.928 | 0.199 | 0.831 | 0.942 | 0.780 | 0.923 | 0.224 |
| 91 593 | 0.784 | 0.951 | 0.770 | 0.925 | 0.210 | 0.831 | 0.937 | 0.775 | 0.920 | 0.231 |
| 45 797 | 0.727 | 0.955 | 0.741 | 0.920 | 0.227 | 0.795 | 0.941 | 0.761 | 0.918 | 0.239 |
| 9159 | 0.557 | 0.961 | 0.601 | 0.901 | 0.293 | 0.620 | 0.933 | 0.639 | 0.888 | 0.325 |
| 4580 | 0.563 | 0.955 | 0.602 | 0.899 | 0.270 | 0.573 | 0.952 | 0.627 | 0.894 | 0.305 |
| 916 | 0.000 | 1.000 | 0.000 | 0.818 | 0.492 | 0.000 | 1.000 | 0.000 | 0.818 | 0.482 |
Table 7.
Cross-entropy loss Lce ± 10−3 and statistical metrics (Sensitivity, Specificity, acc ± 10-3) for the segmentation prediction of different network configurations are presented. Note that here the number of training examples is the same for all tested pruning ratios. Again remember that the number of adjustable parameters of the unpruned DCCN amounted to 794 754. The rows marked yellow present results obtained with networks where the condition Ntr ≥ Np held.
| Ntr | Sensitivity | Specificity | DiceCo | acc | Lce | Sensitivity | Specificity | DiceCo | acc | Lce |
|---|---|---|---|---|---|---|---|---|---|---|
| 0% Pruning, 934 | 50% Pruning, 341 | |||||||||
| 2 397 377 | 0.846 | 0.940 | 0.792 | 0.923 | 0.384 | 0.838 | 0.941 | 0.787 | 0.922 | 0.385 |
| 479 475 | 0.823 | 0.951 | 0.796 | 0.928 | 0.382 | 0.848 | 0.939 | 0.791 | 0.922 | 0.386 |
| 239 738 | 0.838 | 0.944 | 0.795 | 0.926 | 0.384 | 0.827 | 0.946 | 0.791 | 0.924 | 0.385 |
| 47 948 | 0.818 | 0.949 | 0.795 | 0.927 | 0.386 | 0.772 | 0.941 | 0.750 | 0.912 | 0.394 |
| 23 974 | 0.803 | 0.947 | 0.783 | 0.923 | 0.387 | 0.756 | 0.940 | 0.737 | 0.909 | 0.400 |
| 4795 | 0.771 | 0.951 | 0.770 | 0.920 | 0.396 | 0.712 | 0.935 | 0.695 | 0.896 | 0.417 |
| 60% Pruning, 374 | 70% Pruning, 780 | |||||||||
| 2 397 377 | 0.859 | 0.934 | 0.787 | 0.920 | 0.389 | 0.836 | 0.935 | 0.771 | 0.916 | 0.389 |
| 479 475 | 0.814 | 0.946 | 0.783 | 0.923 | 0.387 | 0.833 | 0.938 | 0.779 | 0.920 | 0.390 |
| 239 738 | 0.824 | 0.941 | 0.780 | 0.919 | 0.389 | 0.838 | 0.838 | 0.756 | 0.909 | 0.394 |
| 47 948 | 0.785 | 0.937 | 0.747 | 0.910 | 0.401 | 0.774 | 0.774 | 0.729 | 0.902 | 0.405 |
| 23 974 | 0.775 | 0.930 | 0.729 | 0.902 | 0.408 | 0.757 | 0.757 | 0.721 | 0.901 | 0.409 |
| 4795 | 0.638 | 0.948 | 0.665 | 0.896 | 0.421 | 0.625 | 0.625 | 0.649 | 0.888 | 0.426 |
| 80% Pruning, 187 | 90% Pruning, 593 | |||||||||
| 2 397 377 | 0.843 | 0.935 | 0.775 | 0.918 | 0.387 | 0.805 | 0.948 | 0.777 | 0.924 | 0.386 |
| 479 475 | 0.828 | 0.933 | 0.764 | 0.914 | 0.392 | 0.801 | 0.937 | 0.755 | 0.913 | 0.391 |
| 239 738 | 0.824 | 0.927 | 0.754 | 0.910 | 0.397 | 0.774 | 0.936 | 0.737 | 0.908 | 0.396 |
| 47 948 | 0.762 | 0.925 | 0.718 | 0.900 | 0.411 | 0.758 | 0.923 | 0.711 | 0.898 | 0.410 |
| 23 974 | 0.738 | 0.925 | 0.705 | 0.897 | 0.416 | 0.723 | 0.920 | 0.686 | 0.890 | 0.421 |
| 4795 | 0.622 | 0.938 | 0.639 | 0.889 | 0.426 | 0.675 | 0.921 | 0.654 | 0.882 | 0.432 |
Fig. 7.
Test accuracy and Dice coefficient for neural network configurations resulting from different pruning ratios. Each configuration was trained with an identical but variable number of training examples.
Fig. 8.
Test accuracy and Dice coefficient for neural network DCCN configurations resulting from different pruning ratios. Each configuration was trained with an identical but variable number of training examples.
Let us first consider the simpler DCNN architecture. The results showed that up to a pruning ratio of 50%, both accuracy and Dice coefficient remained rather insensitive to the actual network complexity at the maximal number of training samples Ntr = max. While the accuracy remained constant in this range, the observed reduction of the Dice coefficient amounted to 1.5%. Only for pruning ratios above 50%, both metrics declined more strongly, with the Dice coefficient responding earlier and stronger. Thus, we observed a decrease of the Dice coefficient by 3%, while the accuracy dropped by only 1% for δ = 0 → 0.9 at Ntr = max. In case of the minimal number of training samples Ntr = min, the Dice coefficient dropped to zero at δ = 0.9, however, while the accuracy was reduced by 9%. For less strongly pruned networks, i.e. δ = 0 → 0.5, the Dice coefficient dropped by 6% while the accuracy remained constant. In general, the higher the number of training examples, the higher the prediction accuracy. However, the higher the pruning rate, the lower the accuracy.
For the more complex DCCN architecture, the following observations could be made: Considering first the situation, where the number of training samples was maximal (Ntr = max) and the network pruning also reached its larges value (δ = 0.9), the Dice coefficient dropped by 5% while the accuracy only decreased by 2% and the cross-entropy objective raised by 2% as well. If, instead, the number of training samples was minimal (Ntr = min), the corresponding changes were ΔDC(Ntrmin, δ = 0.9) = 15 % , Δacc(Ntrmin, δ = 0.9) = 4 % , ΔLCE(Ntrmin, δ = 0.9) = 9%. In case of less strongly pruned DCCN networks and a maximal number of training samples we got ΔDC(Ntrmax, δ = 0.5) = 0.6 % , Δacc(Ntrmax, δ = 0.5) = 1 % , ΔLCE(Ntrmax, δ = 0.5) = 1%. If, however, the number of training patterns was minimal, the corresponding numbers read ΔDC(Ntrmin, δ = 0.5) = 10 % , Δacc(Ntrmin, δ = 0.5) = 3 % , ΔLCE(Ntrmin, δ = 0.5) = 5%. Hence, given a maximal number of training samples, strong network pruning had only a minor effect on the performance metrics, which was even less pronounced in case of less strongly pruned networks. There all changes did not exceed 1%. But the effects were much stronger, if the number of training patterns was minimal. In Fig. 9, the predictions of different networks are illustrated. The x-axis corresponds to the number of training samples and the y-axis to the pruning ratio. The test accuracy for all neural network configurations are summarized in Fig. 10.
Fig. 9.
Qualitative illustration of the predictions of different network configurations in dependence on the number of training examples and pruning ratios.
Fig. 10.
Color-coded test accuracy acc for neural network configurations resulting from different pruning rates and concomitant number of training data sample points. The accuracy is color coded according to the color bar on the right.
Analysis of computational parameters
The pruning of networks lead to a decrease in average inference time 〈tinf(δ)〉 and to a decrease in disk space V DS(see Table 8). This was verified by predicting 100 samples and taking the average of the single inference time values. For pruning ratios δ = 0.5,0.9, inference times were reduced to tinf(δ = 0.5) ≈ 0.5tinf(delta = 0) and tinf(0.9) ≈ 0.24tinf(0), respectively for both network architectures. Similarly disk space could be reduced in case of a DCNN architecture to 46 % and 9 % and for a DCCN architecture to 24% and 2%, for pruning ratios of δ = 0.5 and δ = 0.9. The experiments were performed on a GPU (GeForce GTX 1070 with total memory of 31.25 GB and 8GB RAM).
Table 8.
Compute time and disk space for different pruning ratios δ.
| δ | 0 | 0.10 | 0.20 | 0.30 | 0.40 | 0.50 | 0.60 | 0.70 | 0.80 | 0.90 |
|---|---|---|---|---|---|---|---|---|---|---|
| VDS in KB (DCNN) | 3586 | 3172 | 2822 | 2376 | 1996 | 1634 | 1282 | 947 | 623 | 315 |
| VDS in KB (DCCN) | 19 147 | 15 766 | 12 602 | 9805 | 7356 | 5058 | 3423 | 2079 | 1096 | 467 |
| 〈tinf〉 in ms (DCNN) | 7.6 | 6.1 | 5.4 | 5.9 | 5.2 | 3.9 | 3.3 | 2.8 | 2.2 | 1.8 |
| 〈tinf〉 in ms (DCCN) | 47.6 | 43.1 | 40.6 | 35.2 | 31.9 | 22.3 | 18.5 | 16.3 | 11.1 | 9.8 |
Discussion
The main goal of this work was to reduce network complexity with a concomitant reduction in the number of necessary training examples. Network optimization was driven by cross-entropy minimization, yielding a standard error as small as sce = 3 ⋅ 10−3 in case of a DCNN architecture and sce = 1 ⋅ 10−3 in case of the more complex DCCN architecture. Network predictions were evaluated employing standard statistical metrics like prediction accuracy on a test set, true-positive rate and true-negative rate. The focus thus was on the dependence of proper evaluation metrics on the number of adjustable parameters of the considered deep neural network. First, given a DCNN architecture with its concomitant number of adjustable parameters, the variance of the prediction accuracy was analyzed. With a standard error for the accuracy of sacc = 9 ⋅ 10−4, the uncertainty of the results is comparatively low. Next, the number of trainable parameters of the DCNN architecture was reduced by pruning a certain amount of weights in the network. For these parsimonious networks with reduced complexity of the weight connectivity, the training sample size was reduced by scaling down the number of training examples by a factor r, which multiplies the number of adjustable parameters of the considered DCNN. Again performance was evaluated by estimating both the standard error of the accuracy sacc and of the cross entropy sce.
The predictions of different DCNN network configurations are illustrated in Fig. 9. As expected, the unpruned DCNN neural network showed best performance. This network with the highest complexity, hence the largest number of training examples Ntr, corresponding to r = 1, achieved, within the estimated standard error of the optimized cross-entropy loss, best results, which roughly persisted up to 40% pruning. Despite the rather small standard error sacc, it appears that this observation holds across all reduction factors r, i.e. across a rather large range of training sample sizes Ntr. Remarkably, even with a pruning rate of 90%, δ = 0.9, the achieved test accuracy amounted to 98% of the accuracy of the unpruned network. Even more astonishing, if for this network the number of training examples is reduced by a factor of r = 0.01 to Ntr = 9159, the trained network (acc = 0.905) still reached 98% of the maximum achievable accuracy (acc = 0.922). Again, this accuracy was achieved on the independent test set, hence overfitting was not an issue here despite the fact that this network had 10 times more adjustable parameters than training examples.
But with further decreasing numbers of training examples, corresponding to reduction factors lower than r < 0.01, no good results could be achieved for any network configuration and number of training examples. Taken the other way round, a remarkably high accuracy remained even when the training sample size was reduced by a factor of 100, corresponding to r = 0.01 for networks with pruning ratios δ ≤ 0.5. Only then prediction accuracy degraded rapidly, especially for pruning ratios δ ≥ 0.5. The same was true for the other metric, the Dice coefficient (DC) and was also reflected in an increasing minimal cross-entropy (ce) loss. The additional metrics sensitivity (se) and specificity (sp) provided an even more detailed picture. Sensitivity measured how many of the pixels belonging to class cell were correctly predicted, while specificity measured the percentage of correctly assigned background pixels. Remarkably, the specificity or true-negative rate (TNR) was rather high and remained so across all pruning rates. On the contrary, the sensitivity or true-positive rate (TPR) was smaller and decreased further with increasing pruning rate. Furthermore, sensitivity showed a much stronger dependence on the number of training examples and decreased considerably with decreasing Ntr. Note that a high sensitivity indicates that a large number of all pixels belonging to a specific class was classified correctly. Also a high specificity indicates that a large number of pixels not belonging to a specific class were correctly classified accordingly. Fig. 10 provides a qualitative illustration of the accuracy when the number of adjustable parameters and/or the number of training examples changed. As long as the number of model parameters matched the number of training examples, accuracy was generally very high (acc > 90%). But even in case of an insufficient number of training examples, the test accuracy remained amazingly high. As this concerns the previously unseen test data set, overfitting can be safely excluded.
Considering networks of various complexities, i.e. different pruning ratios δ, the latter were also trained by a fixed but variable number of training examples. The latter was deduced from the number of adjustable parameters of the unpruned network by applying the reduction factor r. If considered as a function of the number of training examples, a common observation is that as along as Np ≤ Ntr holds, all metrics evaluated remained largely insensitive to Ntr (These values are marked with a yellow background in Table 6). But if Ntr < Np is met, then some metrics like the sensitivity or the cross-entropy quickly degraded strongly with decreasing Ntr for all network configurations considered, while others like the specificity, Dice coefficient or the accuracy remained still pretty insensitive to the number of stimuli. Remember that the specificity represented the proportion of the non-cell pixels, and the sensitivity the proportion of cell pixels, that were correctly classified. The following Table 9 provides a comprehensive summary to the detailed results presented in Table 6.
Table 9.
Comprehensive summary of the results obtained with fixed numbers of training examples and a DCNN architecture.
| δ | 0 | 0.5 | 0.9 | |
|---|---|---|---|---|
| r | 0.001 | 0.01 | 0.01 | 0.01 |
| SE | 82% | 95% | 93% | 0% |
| SP | 98% | 99% | 100% | 100% |
| ACC | 96% | 99% | 99% | 89% |
| DSC | 84% | 96% | 96% | 0% |
| CE | 174% | 125% | 127% | 233% |
For example, given a pruning ratio δ = 0.9, corresponding to the least complex network architecture, and a number Ntr = 0.5Np of stimuli, the specificity (SP) was unchanged, the Dice coefficient (DSC) still achieved 97.6% and the accuracy (ACC) amounted to 99.5% of the values prevailing at Ntr > Np. At the same time, the sensitivity (SE) declined to 95% and the cross entropy (CE) increased to 115.5%. Even worse, at Ntr = 0.01Np we observed an unchanged specificity, an accuracy that still reached 88.6% of its original value, while sensitivity and Dice coefficient vanished and the cross entropy raised to 233% of its starting value. To the contrary, in case of the most complex, unpruned DCNN network, when reducing the number of stimuli by a factor of 103, we rather observed a sensitivity that reached only 82% of its starting value, a specificity of still 98% of its value, when Ntr was abundant, an accuracy of 96% of its best value, a Dice coefficient of only 84% of its starting value and a cross-entropy objective, which raised to 174% of its value with an abundant number of training examples. For an intermediate complexity with a pruning ratio of 50%, we observed an unchanged specificity, a slightly reduced sensitivity (%), a hardly reduced accuracy acc = 99%, a still respectable Dice coefficient of % but an already considerably increased cross entropy %. Still these metrics indicate that even with a considerably reduced network complexity and a rather small number of stimuli a good classification performance can be achieved.
A similar picture results in case of the more complex DCCN architecture as can be seen from Table 10, where percentage changes of all metrics are presented. The changes relate to a decrease of the number of training samples from Ntrmax to Ntrmin with all other variables kept constant. While the metrics accuracy ACC and specificity SP remain largely unchanged by a variation of the number of training samples, irrespective of the state of network pruning, sensitivity SE, Dice - Sørensen coeficient DSC and cross-entropy loss CE respond much more strongly. The changes even increase in case of parsimonious networks with a maximal pruning applied to the network weights.
Table 10.
Comprehensive summary of the percentage changes of the performance metrics obtained with fixed numbers of training examples and a DCCN architecture.
| δ | 0 | 0.5 | 0.9 |
|---|---|---|---|
| SE | 6% | 16% | 16% |
| SP | 0% | 0.4% | 2% |
| ACC | 1% | 3% | 3.4% |
| DSC | 3% | 12% | 13% |
| CE | 3.6% | 8% | 10% |
In Table 8, the inference time for the DCNN and DCCN for different pruning rates are summarized. Increasing pruning rates could decrease the inference time and the disk space. For the DCNN the inference time was reduced by 77% between no and 90% pruning, for the DCCN the reduction was 79%. Also the disc space could be reduced by 92% for the DCNN and by 98% for the DCCN.
Conclusion
This study focused on the question, how the number of training examples controlled the performance of a DCNN of given complexity, i.e. number of adjustable parameters. This is especially relevant in many biomedical problem settings, where the number of training stimuli is limited compared with the number of degrees of freedom of the deep neural network used to analyze the data. Network complexity was reduced by weight pruning, and the number of training examples Ntr was deduced from the number of degrees of freedom Np of these pruned networks. Two different scenarios where explored: First Ntr(δ) was deduced from Np(δ) of the pruned DCNN by a reduction factor r ≤ 1. Here, δ denoted the pruning ratio. Second Ntr was estimated from Np of the unpruned DCNN by the same reduction factors. While in the first scenario Np ≥ Ntr, we always considered an oversized network configuration, in the second scenario for a certain range of pruning ratios δ the condition Np(δ) ≤ Ntr held, while for larger δ the condition changed to Np(δ) ≥ Ntr. Network performance was evaluated with a set of statistical metrics which showed that up to δ ≤ 0.5, classification performance was hardly hampered, while for larger pruning ratios some statistical metrics degraded noticeably. The conclusion is that Ntr ≥ Np is not a necessary condition and that oversized networks often achieve an equally good classification performance. Finally, remember that at a pruning ratio δ = 0.5, where all evaluation metrics still achieved respectable values, average inference time was halved and the used disk space shrank to half of its size needed for the unpruned network. This may be of concern, if such evaluations have to be performed on edge devices.
Conflict of interest
None.
References
- 1.Adamczewski K., Park M. Proceedings of the 24th International Conference on Artificial Intelligence and Statistics (AISTATS) 2021, San Diego, California, USA. Vol. 130. PMLR; 2021. Dirichlet pruning for neural network compression. [Google Scholar]
- 2.Bai M., Urtasun R. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2017. Deep watershed transform for instance segmentation; pp. 5221–5229. [Google Scholar]
- 3.Bao R., Yuan X., Chen Zh., Ma R. Cross-entropy pruning for compressing convolutional neural networks. Neural Comput. 2018;30:3128–3149. doi: 10.1162/neco_a_01131. [DOI] [PubMed] [Google Scholar]
- 4.Chang J., Sha J. Prune deep neural networks with the modified l1/2 penalty. IEEE Access. 2018;7:2273–2280. [Google Scholar]
- 5.Cichocki A., Zdunek R., Amari S.-I. Nonnegative matrix and tensor factorization. IEEE Signal Process Magaz. 2008;142 doi: 10.1155/2008/852187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Brabandere B., Neven D., Van Gool L. Semantic instance segmentation with a discriminative loss function. arXiv preprint. 2017;abs/1708.02551:1–10. arXiv:1708.02551. [Google Scholar]
- 7.Duncan S., Olsson T.S.G., Hartley M., Dean C., Rosa S. A method for detecting single mrna molecules in Arabidopsis thaliana. Plant Methods. 2016;12(1):1–10. doi: 10.1186/s13007-016-0114-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Facchetti G., Knapp B., Flor-Parra I., Chang F., Howard M. Reprogramming cdr2-dependent geometry-based cell size control in fission yeast. Curr. Biol. 2019;29(2):350–358. doi: 10.1016/j.cub.2018.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falk T., Mai D., Bensch R., et al. U-net: deep learning for cell counting, detection, and morphometry. Nat. Methods. 2019;16(1):67–70. doi: 10.1038/s41592-018-0261-2. [DOI] [PubMed] [Google Scholar]
- 10.Godinez W.J., Hossain I., Lazic S.E., Davies J.W., Zhang X. A multi-scale convolutional neural network for phenotyping high-content cellular images. Bioinformatics. 2017;33(13):2010–2019. doi: 10.1093/bioinformatics/btx069. [DOI] [PubMed] [Google Scholar]
- 11.Golub M., Lemieux G., Lis M. 2019. Full deep neural network training on a pruned weight budget. [Google Scholar]
- 12.Haberl M.G., Churas C., Tindall L., et al. Cdeep3m-plug-and-play cloud based deep learning for image segmentation of light, electron and x-ray microscopy. bioRxiv. 2018:353425. doi: 10.1038/s41592-018-0106-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han S., Pool J., Tran J., Dally W. Advances in Neural Information Processing Systems. 2015. Learning both weights and connections for efficient neural network; pp. 1135–1143. [Google Scholar]
- 14.Häring M., Großhans J., Wolf F., Eule S. Automated segmentation of epithelial tissue using cycle-consistent generative adversarial networks. bioRxiv. 2018:311373. [Google Scholar]
- 15.Hassibi B., Stork D.G. Advances in Neural Information Processing Systems. 1993. Second order derivatives for network pruning: optimal brain surgeon; pp. 164–171. [Google Scholar]
- 16.He Y., Liu P., Wang Z., Hu Zh., Yang Y. The IEEE Conference on Computer Vision and Pattern Recognition (CVPR) IEEE; 2019. Filter pruning via geometric median for deep convolutional neural networks acceleration. [DOI] [Google Scholar]
- 17.He Y., Zhang X., Sun J. Proceedings of the IEEE International Conference on Computer Vision. IEEE; 2017. Channel pruning for accelerating very deep neural networks; pp. 1389–1397. [Google Scholar]
- 18.Hollandi R., Szkalisity A., Toth T., et al. A deep learning framework for nucleus segmentation using image style transfer. bioRxiv. 2019:580605. doi: 10.1016/j.cels.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu H., Peng R., Tai Y.-W., Tang Ch.-K. 2016. Network trimming: A data-driven neuron pruning approach towards efficient deep architectures. [Google Scholar]
- 20.Huang G., Liu Z., Van Der Maaten L., Weinberger K.Q. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2017. Densely connected convolutional networks; pp. 4700–4708. [Google Scholar]
- 21.Huang Z., Huang L., Gong Y., Huang C., Wang X. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. 2019. Mask scoring r-cnn; pp. 6409–6418. [Google Scholar]
- 22.Johnson J.W. Adapting mask-rcnn for automatic nucleus segmentation. arXiv preprint. 2018;abs/1805.00500:1–7. arXiv:1805.00500. [Google Scholar]
- 23.Jourdren L., Delaveau T., Marquenet E., Jacq C., Garcia M. Corsen, a new software dedicated to microscope-based 3d distance measurements: mrna–mitochondria distance, from single-cell to population analyses. RNA. 2010;16(7):1301–1307. doi: 10.1261/rna.1996810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kandaswamy C., Silva L.M., Alexandre L.A., Santos J.M. High-content analysis of breast cancer using single-cell deep transfer learning. J Biomol Screen. 2016;21(3):252–259. doi: 10.1177/1087057115623451. [DOI] [PubMed] [Google Scholar]
- 25.Keren L., Bosse M., Marquez D., et al. A structured tumor-immune microenvironment in triple negative breast cancer revealed by multiplexed ion beam imaging. Cell. 2018;174(6):1373–1387. doi: 10.1016/j.cell.2018.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khoshdeli M., Winkelmaier G., Parvin B. Fusion of encoder-decoder deep networks improves delineation of multiple nuclear phenotypes. BMC Bioinform. 2018;19(1):1–11. doi: 10.1186/s12859-018-2285-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kraus O.Z., Ba J.L., Frey B.J. Classifying and segmenting microscopy images with deep multiple instance learning. Bioinformatics. 2016;32(12):i52–i59. doi: 10.1093/bioinformatics/btw252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kraus O.Z., Grys B.T., Ba J., et al. Automated analysis of high-content microscopy data with deep learning. Mol Syst Biol. 2017;13(4):924. doi: 10.15252/msb.20177551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar N., Verma R., Sharma S., Bhargava S., Vahadane A., Sethi A. A dataset and a technique for generalized nuclear segmentation for computational pathology. IEEE Trans Med Imaging. 2017;36(7):1550–1560. doi: 10.1109/TMI.2017.2677499. [DOI] [PubMed] [Google Scholar]
- 30.Lebedev V., Lempitsky V. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016. Fast convnets using group-wise brain damage; pp. 2554–2564. [Google Scholar]
- 31.Li H., Kadav A., Durdanovic I., Samet H., Graf H.P. 2017. Pruning filters for efficient convnets. [Google Scholar]
- 32.Li H., Kadav A., Durdanovic I., Samet H., Graf H.P. Pruning filters for efficient convnets. CoRR. 2016;abs/1608.08710:1–13. abs/1608.08710. [Google Scholar]
- 33.Li R., Wang X., Quan W., Song Y., Lei L. Robust and structural sparsity auto-encoder with l21-norm minimization. Neurocomputing. 2021;425:71–81. [Google Scholar]
- 34.Li Y., Gu Sh., Van Gool L., Timofte R. Proceedings of the IEEE International Conference on Computer Vision. 2019. Learning filter basis for convolutional neural network compression; pp. 5623–5632. [Google Scholar]
- 35.Lin Sh., Ji R., Li Y., Deng Ch., Li X. Towards compact convnets via structure-sparsity regularized filter pruning. IEEE Trans Neural Netw Learn Syst. 2020;31:574–588. doi: 10.1109/TNNLS.2019.2906563. [DOI] [PubMed] [Google Scholar]
- 36.Lin T.-Y., Goyal P., Girshick R., He K., Dollár P. Proceedings of the IEEE International Conference on Computer Vision. 2017. Focal loss for dense object detection; pp. 2980–2988. [Google Scholar]
- 37.Liu J., Tripathi S., Kurup U., Shah M. Pruning algorithms to accelerate convolutional neural networks for edge applications. A survey. 2020;abs/2005.04275:1–7. [Google Scholar]
- 38.Luo J.-H., Wu J. 2017. An entropy-based pruning method for CNN compression. [Google Scholar]
- 39.Mahmood F., Borders D., Chen R.J., et al. Deep adversarial training for multi-organ nuclei segmentation in histopathology images. IEEE Trans Med Imaging. 2019;39(11):3257–3267. doi: 10.1109/TMI.2019.2927182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moen E., Bannon D., Kudo T., Graf W., Covert M., Van Valen D. Deep learning for cellular image analysis. Nat Methods. 2019;16(12):1233–1246. doi: 10.1038/s41592-019-0403-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Molchanov P., St. Tyree T., Karras T. Aila, Kautz J. 2017. Pruning convolutional neural networks for resource efficient inference. [Google Scholar]
- 42.Newby J.M., Schaefer A.M., Lee P.T., Forest M.G., Lai S.K. Convolutional neural networks automate detection for tracking of submicron-scale particles in 2d and 3d. Proc Natl Acad Scil. 2018;115(36):9026–9031. doi: 10.1073/pnas.1804420115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pärnamaa T., Parts L. Accurate classification of protein subcellular localization from high-throughput microscopy images using deep learning. G3: Genes Genomes Genet. 2017;7(5):1385–1392. doi: 10.1534/g3.116.033654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pawlowski N., Caicedo J.C., Singh S., Carpenter A.E., Storkey A. Automating morphological profiling with generic deep convolutional networks. BioRxiv. 2016:085118. [Google Scholar]
- 45.Payer C., Štern D., Neff T., Bischof H., Urschler M. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2018. Instance segmentation and tracking with cosine embeddings and recurrent hourglass networks; pp. 3–11. [Google Scholar]
- 46.Ren S., He K., Girshick R., Sun J. Faster r-cnn: towards real-time object detection with region proposal networks. IEEE Trans Pattern Anal Machine Intel. 2016;39(6):1137–1149. doi: 10.1109/TPAMI.2016.2577031. [DOI] [PubMed] [Google Scholar]
- 47.Ronneberger O., Fischer P., Brox T. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2015. U-net: convolutional networks for biomedical image segmentation; pp. 234–241. [Google Scholar]
- 48.Simm J., Klambauer G., Arany A., et al. Repurposing high-throughput image assays enables biological activity prediction for drug discovery. Cell Chem Biol. 2018;25(5):611–618. doi: 10.1016/j.chembiol.2018.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sommer C., Hoefler R., Samwer M., Gerlich D.W. A deep learning and novelty detection framework for rapid phenotyping in high-content screening. Mol Biol Cell. 2017;28(23):3428–3436. doi: 10.1091/mbc.E17-05-0333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Srinivas S., Babu R.V. Procedings of the British Machine Vision Conference 2015. British Machine Vision Association; 2015. Data-free parameter pruning for deep neural networks; pp. 31.1–31.12. [Google Scholar]
- 51.Sullivan D.P., Winsnes C.F., Åkesson L., et al. Deep learning is combined with massive-scale citizen science to improve large-scale image classification. Nat Biotechnol. 2018;36(9):820–828. doi: 10.1038/nbt.4225. [DOI] [PubMed] [Google Scholar]
- 52.Swaminathan S., Garg D., Kannan R., Andres F. Sparse low rank factorization for deep neural network compression. Neurocomputing. 2020;398:185–196. [Google Scholar]
- 53.Tokuoka Y., Yamada T.G., Hiroi N.F., Kobayashi T.J., Yamagata K., Funahashi A. Convolutional neural network-based instance segmentation algorithm to acquire quantitative criteria of early mouse development. BioRxiv. 2018:324186. doi: 10.1038/s41540-020-00152-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Torfi A., Shirvani R.A., Soleymani S., Nasrabadi N.M. 2018. Attention-based guided structured sparsity of deep neural networks. [Google Scholar]
- 55.Tsai H.-F., Gajda J., Sloan T.F.W., Rares A., Shen A.Q. Usiigaci: instance-aware cell tracking in stain-free phase contrast microscopy enabled by machine learning. SoftwareX. 2019;9:230–237. [Google Scholar]
- 56.Van Valen D.A., Kudo T., Lane K.M., et al. Deep learning automates the quantitative analysis of individual cells in live-cell imaging experiments. PLoS Computat. Biol. 2016;12(11):e1005177. doi: 10.1371/journal.pcbi.1005177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang H., Hu X., Zhang Q., Wang Y., Yu L., Hu H. Structured pruning for efficient convolutional neural networks via incremental regularization. IEEE J Select Topics Signal Process. 2020;14(4):775–788. [Google Scholar]
- 58.Wang J., Xu Ch., Yang X., Zurada J.M. A novel pruning algorithm for smoothing feedforward neural networks based on group lasso method. IEEE Trans Neural Netw Learn Syst. 2018;29:2012–2024. doi: 10.1109/TNNLS.2017.2748585. [DOI] [PubMed] [Google Scholar]
- 59.Wang W., Taft D.A., Chen Y.-J., Zhang J., Wallace C.T., Min X., Watkins S.C., Xing J., editors. Learn to segment single cells with deep distance estimator and deep cell detectorComput Biol Med. 2019;108:133–141. doi: 10.1016/j.compbiomed.2019.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Zh., Li F., Shi G., Xie X., Wang F. Network pruning using sparse learning and genetic algorithm. Neurocomputing. 2020;404:247–256. [Google Scholar]
- 61.Wen W., Wu Ch., Wang Y., Chen Y., Li H. 30th Conference on Neural Information Processing Systems (NIPS 2016), Barcelona, Spain. 2016. Learning structured sparsity in deep neural networks; pp. 1–9. [Google Scholar]
- 62.Xing F., Xie Y., Yang L. An automatic learning-based framework for robust nucleus segmentation. IEEE Trans Med Imaging. 2015;35(2):550–566. doi: 10.1109/TMI.2015.2481436. [DOI] [PubMed] [Google Scholar]
- 63.Yeom S.-K., Shim K.-H., Hwang J.-H. 2021. Toward compact deep neural networks via energy-aware pruning. [Google Scholar]
- 64.Zhang S., Zhou J., Hailin H., et al. A deep learning framework for modeling structural features of rna-binding protein targets. Nucleic Acids Res. 2016;44(4):e32. doi: 10.1093/nar/gkv1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao Ch., Ni B., Zhang J., Zhao Q. The IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2019. Variational convolutional neural network pruning. [Google Scholar]
- 66.Zhu J.-Y., Park T., Isola P., Efros A.A. Proceedings of the IEEE International Conference on Computer Vision. 2017. Unpaired image-to-image translation using cycle-consistent adversarial networks; pp. 2223–2232. [Google Scholar]