Abstract
Background
High tumor mutation burden (TMB-H) could result in an increased number of neoepitopes from somatic mutations expressed by a patient’s own tumor cell which can be recognized and targeted by neighboring tumor-infiltrating lymphocytes (TILs). Deeper understanding of spatial heterogeneity and organization of tumor cells and their neighboring immune infiltrates within tumors could provide new insights into tumor progression and treatment response.
Methods
Here we first developed computational approaches using whole slide images (WSIs) to predict bladder cancer patients’ TMB status and TILs across tumor regions, and then investigate spatial heterogeneity and organization of regions harboring TMB-H tumor cells and TILs within tumors, as well as their prognostic utility. Results: In experiments using WSIs from The Cancer Genome Atlas (TCGA) bladder cancer (BLCA), our findings show that computational pathology can reliably predict patient-level TMB status and delineate spatial TMB heterogeneity and co-organization with TILs. TMB-H patients with low spatial heterogeneity enriched with high TILs show improved overall survival.
Conclusions
Computational approaches using WSIs have the potential to provide rapid and cost-effective TMB testing and TILs detection. Survival analysis illuminates potential clinical utility of spatial heterogeneity and co-organization of TMB and TILs as a prognostic biomarker in BLCA which warrants further validation in future studies.
Keywords: Tumor mutation burden prediction, Computational pathology, Bladder cancer, Survival prognosis, Spatial Heterogeneity, Tumor Immune Microenvironment
Abbreviations: TCGA, The Cancer Genome Atlas Project; TMB, Tumor Mutation Burden; TIL, Tumor-Infiltrating Lymphocyte; WSI, Whole Slide Image; BLCA, Urothelial Bladder Carcinoma
Introduction
Tumor mutation burden (TMB) is a quantitative genomic biomarker that measures the number of mutations within a tumor. High TMB (TMB-H) level has been shown to be associated with better prognosis and clinical responses to immune-checkpoint inhibitors in various cancer types such as melanoma, lung cancer, and bladder cancer.1, 2, 3, 4, 5 Higher TMB levels are correlated with higher levels of neoantigens expressed by a cancer cell, which could help the neighboring tumor-infiltrating lymphocytes (TILs) to recognize and kill them.6 Various studies including clinical trials reported that patients with TMB-H and/or high density of TILs within tumors had favorable prognosis and response to immunotherapy in many cancer types.1, 2, 3, 4, 5,7, 8, 9, 10, 11 Recent studies showed that spatial heterogeneity and composition of immune cells in the tumor microenvironment could improve our understanding of how immune environment influences patients’ prognosis and response to treatments, including immunotherapy.12, 13, 14, 15, 16 These findings might suggest that detecting regions harboring TMB-H tumor cells and TILs within the tumor microenvironment and analyzing their spatial architecture could provide new insights into the relationship between spatial TMB and TIL co-arrangement and patient’s outcome.
Tissue-based bulk DNA sequencing (e.g., whole exome sequencing (WES), targeted sequencing, etc.) and mRNA sequencing are widely used to assess patient-level TMB status and quantify TILs in tumors, respectively. However, due to the limited tissue availability, high costs, and time-consuming procedures, the clinical utility of tissue-based DNA and mRNA sequencing are limited. In addition, since these bulk DNA and mRNA sequencing approaches were not designed to take into account spatial intratumor TMB and immune heterogeneity, providing potentially biased samples may lead to inconsistent testing results.17 Although the blood-based TMB measurement (i.e., liquid biopsies) has recently become available, this approach poses similar challenges to tissue-based TMB measurements.18 The development of single-cell DNA and mRNAseq has revealed a spectrum of tumor cell and immune cell heterogeneity in the patient’s tumor, but these approaches do not provide insight into the spatial organization of tumor and immune cell architecture.11,19, 20, 21 Most recently, spatial transcriptome technologies have enabled mapping of the spatial architecture, composition. and interactions of various cell types within the tumor, but simultaneously elucidating both DNA (e.g., TMB status) and RNA-level characteristics of cells is still challenging.22, 23, 24
The use of widely available histopathological images poses a promising alternative. Routine histopathological examination is the gold-standard for diagnosis, grading, and quantification of TILs for various cancer types in the clinical setting. With recent development in deep learning, computational pathology studies have been explored to predict genetic characteristics (e.g.,mutation status, gene expression, etc.) present within tumor regions in lung,25 colon and stomach,26 and pan-cancer.27,28 Many other recent works focused on automatic segmentation and classification in pathology slides, such as automatic tumor segmentation in lung adenocarcinoma whole slide images (WSIs),29 automatic prediction of lymph node metastasis status,30 and survival prognosis based on tumoral microenvironment analysis in colorectal cancer WSIs.31 Particularly, there are also computational pathology studies on bladder cancer. For instance, convolutional neural networks (CNN) have been explored to identify different tissues such as urothelium, muscle, and stroma in urothelial carcinoma WSIs,32 and predict mutations of the FGFR3 gene from histology slides of bladder cancer.33 WSIs are being widely for TILs detection and quantification by computational analysis as well. Saltz et al. (2018) applied CNN to identify TILs in hematoxylin and eosin (H&E) stained WSIs and showed that spatial composition of TILs within tumors correlated with patient’s prognosis across cancers.34 Corredor et al. (2018) developed a set of computer-extracted variables reflecting TILs spatial distribution and co-organization with cancer cell nuclei in tissue microarrays obtained from H&E stained slides. They employed the quadratic discriminant analysis as classifier and predicted likelihood of recurrence in early-stage non-small cell lung cancer.7 Acs et al. (2019) developed algorithms to detect TILs and tested their spatial composition with cancer cells within tumors for prognosis in melanoma.35 Most recently, Abduljabbar et al. (2020) performed a study that integrated multi-region exome and RNA-sequencing (RNA-seq) data with histology image to investigate spatial tumor and immune microenvironment in lung adenocarcinoma (LUAD). The results showed that LUAD subgroup with immune cold and low neoantigen burden (i.e., low TMB) was significantly correlated with poorer disease-free survival.36 This study demonstrated that computational pathology could provide a deeper understanding of how spatial composition of tumor and immune cells within tumor microenvironment impact tumor evolution and progression.
Given these studies showing that computational approaches and deep learning algorithms utilizing morphological features present in WSIs could reliably predict characteristics of tumor and immune cells and their spatial organizations, we hypothesize that a carefully designed WSI-based computational method could accurately predict TMB status and TILs within tumors and could be used to dissect spatial heterogeneity of TMB and its co-organization with TILs across tumor regions. Specifically, we hypothesize that the comprehensive understanding of spatial co-occurrence of TILs with neighboring TMB-H or TMB-low (TMB-L) regions from pathology slides could provide a prognostic utility to identify patient subgroups with distinct survival outcome.
Although it is clinically relevant to predict TMB status and TILs across different cancer types, this study focuses on automatic predictions of TMB status and TILs across tumor regions in bladder cancer (BLCA), mainly due to their clinical significance in BLCA patients’ prognosis as well as integral clinical information along with WSIs provided in TCGA. In this work, we first develop and evaluate computational pipelines to predict BLCA patients’ TMB status and TILs distribution in H&E stained WSIs. We then use the tile-level (i.e., dividing a WSI into small tiles for analysis) TMB status to delineate spatial heterogeneity of TMB within WSIs. We perform a joint spatial analysis of regions harboring predicted TMB status and TILs within the tumor and use the spatial heterogeneity information to identify patient subgroups (e.g., TMB-H tumor with low spatial TMB heterogeneity enriched with high density of TILs). To the best of our knowledge, this is the first work to interrogate spatial heterogeneity and organization of TMB with TILs within tumors to evaluate its prognostic utility to stratify patients using WSIs.
Methods
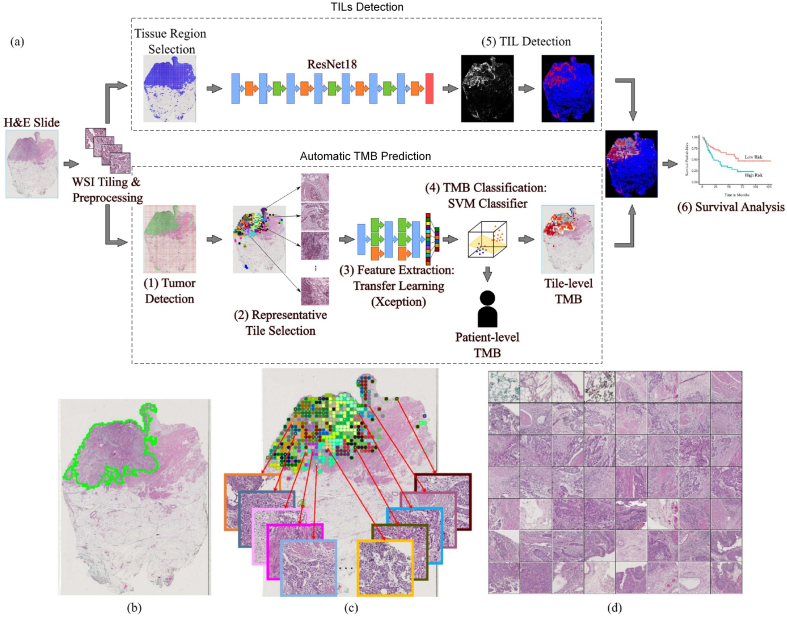
We developed a computational pipeline using WSIs to predict patient-level TMB status and delineate spatial heterogeneity of TMB present in tumors. We also trained a deep learning model to detect TILs and quantify its densities within tumor regions. The aim of our approach is to incorporate spatial TMB heterogeneity with patient-level TMB status and TIL densities to identify patient subgroups that could lead to better patient stratification. The computational analysis workflow is shown in Fig. 1(a), which includes two main modules: automatic TMB prediction and TILs detection. Table s1 lists the acronyms used in this paper.
Fig. 1.
An overview of our method to predict TMB status and TILs from WSIs. (a) An illustration of TMB and TILs prediction, and correlation with patients’ overall survival. (b) Tumor detection result (overlapped green contours). (c) Example of AP clustering on tumor tiles, where tumor tiles belonging to different clusters are indicated by different color of blocks in the image. Several representative tumor tiles indicated by arrows are zoomed-in for better viewing. (d) 56 representative tumor tiles selected by AP clustering for the slide shown in (c).
Automatic TMB prediction
Our designed patient-level TMB prediction includes the following four steps. More implementation details and parameter settings could be referred in the supplementary methods.
(1) Tumor detection: In order to focus on tumor regions for analysis, we trained a lightweight CNN (see the architecture in Fig. s1) model with only about 0.28M trainable parameters to detect tumor regions in the WSI. The utilization of the CNN model is mainly motivated by its reported superior performance in histological image classifications such as references.25,34 Given the WSI, it is first divided into non-overlapping tiles (512×512 pixels at 20× magnification). The CNN-based tumor detector then predicts each tile to be the probability of belonging to cancer regions. The prediction map corresponding to the WSI is generated by stitching predicted probabilities for all image tiles. An empirical threshold (e.g., 0.5) is applied on the prediction map to obtain tumor regions. Our quantitative evaluations showed that the designed CNN-based tumor detector could provide over 90% dice coefficient in bladder cancer detection and a superior performance than several comparative models (see Fig. s6, s7 and Table s2). Fig. 1(b) illustrates an example of cancer detection on a WSI.
(2) Representative tile selection: To improve computational efficiency in analyzing large predicted tumor regions, we selected a subset of representative tumor regions for analysis. We first divided predicted tumor regions into a set of non-overlapping tiles (128×128 pixels) at 2.5× magnification. We then characterized each tumor tile by a 42-dimensional feature vector (i.e., 40 multi-scale local binary pattern features37 and 2D location of the tumor tile). After that, affinity propagation (AP) clustering38 was applied to identify tumor regions containing tiles with similar morphological patterns. In this study, local binary patterns were used to characterize low resolution image features mainly due to its efficiency in computation, while the AP clustering was adopted mainly because it does not predefine the number of clusters. The AP clustering simultaneously identified a number of r local tumor regions and their representative tiles Rj, where 1 ≤ j ≤ r. Fig. 1(c)(d) illustrates AP clustering of tumor tiles on a WSI, where tumor tiles belonging to different clusters are indicated by different color of blocks in the image. Note that there are 56 (r = 56 for this example) representative tiles selected among 490 tumor tiles for the patient slide shown in Fig. 1(c).
(3) Feature extraction: We used transfer learning on pre-trained deep learning models to generate features for selected representative tumor tiles. First, to suppress the influence of color variations, a color deconvolution based method39 is utilized to normalize tumor tiles into a standard color appearance. Second, due to the superior performance on ImageNet classification, transfer learning on the pre-trained Xception40 model was used to extract features from selected tumor tiles. Given an input tumor tile Rj at 20× magnification (1024×1024 pixels), the transfer learning model outputs a high-level feature representation Vj which is a 2048 dimensional vector (see Fig. s4). Finally, the feature vector representing the WSI was obtained by integrating features of representative tumor tiles together, i.e., , where ρj = λj/∑j=1rλj and λj represents the number of tumor tiles belonging to the jth cluster. The feature vector is the weighted mean of features extracted from representative tiles, where each representative tile stands for the major characteristics of tumor tiles within the cluster. The features from representative tiles are integrated together to form patient-level representations as there only exist patient-level labels for the TMB status.
(4) TMB classification: We trained the Support Vector Machine (SVM) classifier based on features generated from the transfer learning model to predict patient-level TMB status. The SVM classifier has been widely-used in histological classifications due to its efficiency and efficacy, and we use it by following our previous study.41 First, principal component analysis (PCA) was used to reduce the feature dimension to prevent over-fitting. In this study, we selected the top 100 principal components which provided a superior performance in our testing. Second, feature standardization was performed on each feature component, which ensured its values have zero mean and unit variance. Finally, SVM with radial basis function (RBF) and linear kernels were trained to predict patient-level TMB status.
TILs detection
In order to detect TILs, we trained and tested 144 different deep learning models by using three convolutional neural network architectures with different percentile of trainable layers, and using different parameter configurations in terms of optimizer, batch size, and learning rate (see Table s3). We make use of a public dataset34 for building TILs detector, which included 43,440 annotated image tiles. Among 144 trained TILs detectors, the best TILs detector which was trained by fine-tuning all trainable layers of Resnet18 and using Adam optimizer with the learning rate of 0.0001 and batch size of 4 provided over the 80% accuracy in distinguishing TIL and Non-TIL tiles during an independent testing (see Fig. s5(a)), which was selected to perform TILs detection. To identify TIL regions in pathology slides, the WSI was first divided into a set of non-overlapping image tiles (i.e., 112 μm×112 μm per image tile). The image tiles were then predicted as TIL tiles or non-TIL tiles by using the selected TILs detector. The WSI-level TILs detection (see the example shown in Fig. s5(b)) was then generated by stitching tile-level predictions, where tiles with prediction probabilities above 0.5 were considered as TIL regions. Based on tumor and TILs detection, we finally computed the ratio between the number of TIL pixels and the total number of tumor pixels in pathology slides, which was used as a feature variable to quantify TIL densities within tumor regions.
Results
In this section, we will first illustrate the patient cohort used in this study. We then provide quantitative evaluations on automatic patient-level TMB predictions. After that, spatial heterogeneity of TMB status is explored and correlated with BLCA patients’ OS outcomes. Finally, spatial analysis of TMB heterogeneity and TILs within tumors are explored for OS prognosis in BLCA.
Patient cohort
The dataset with digitally scanned WSIs was collected from the TCGA project through the Genomic Data Commons Portal (https://portal.gdc.cancer.gov/). The TCGA BLCA cohort consists of 386 patients (and corresponding clinical information) with 457 diagnostic H&E stained WSIs. The first diagnostic slide image (i.e., with DX1 suffix) was selected if there are multiple diagnostic slide images available for a patient. Based on the percentile of total number single nucleotide variants,1 386 TCGA BLCA patients were categorized into 3 groups: 128 low, 128 intermediate, and 130 high TMB patients. To obtain clear ground truth, patients with a so-called distinct outcome, either low or high TMB patients were used for patient-level TMB prediction evaluation. One high and four low TMB patients were excluded due to severe pen marks on slides, thus 124 low and 129 high TMB patients were used to train and test a model to predict patient-level TMB status. Based on TMB prediction and TILs detection, the whole cohort of patients with survival information was used for prognosis analysis on patients’ overall survivals.
Evaluation on patient-level TMB prediction
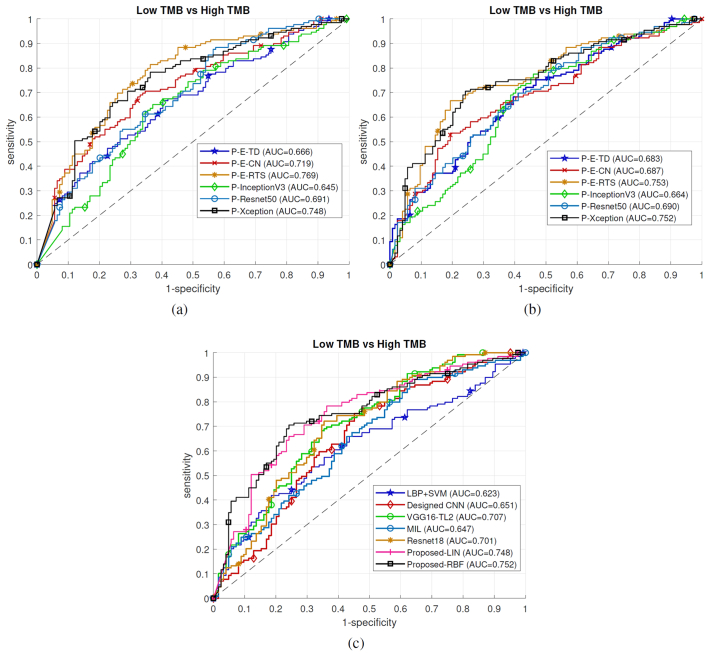
We first investigated whether the use of either tumor detection, representative tile selection, or color normalization as well as different transfer learning models could impact the performance of patient-level TMB prediction. Using TCGA BLCA dataset, we ran patient-level TMB prediction experiments by excluding tumor detection (abbreviated as P-E-TD), representative tile selection (abbreviated as P-E-RTS), or color normalization (abbreviated as P-E-CN). We also tested transfer learning on two well-known models, Inception-v3 (abbreviated as P-InceptionV3)42 and Resnet50 (abbreviated as P-Resnet50),43 in addition to Xception model (abbreviated as P-Xception), to evaluate whether different transfer learning models could impact patient-level TMB prediction performance. We trained SVM classifiers with linear or RBF kernels to predict patient-level TMB status. The leave-one-out cross validation was employed during testing different configurations. ROC curves of patient-level TMB prediction using different settings in our pipeline are shown in Figs. 2(a) and (b) using SVM with linear kernel (Linear SVM) and SVM with RBF kernel (RBF SVM), respectively (see more details in Table s4). The linear and RBF SVMs with P-Xception and P-E-RTS models achieved overall best AUROC values compared to other methods. While both approaches showed good prediction performance, the P-Xception model used the 11,164 selected representative tiles out of 125,358 tiles, which required significantly less computational time (see computational comparison example in Table s5) compared to the P-E-RTS model. This indicates that the use of AP clustering to select a set of representative tiles from a WSI increases computational efficiency without a significant loss of prediction performance. Therefore, we used the AP clustering module in our pipeline for further experiments. The patient-level TMB prediction performance using Xception model (P-Xception) is more accurate than those of Inception-v3 (P-InceptionV3) and Resnet50 (P-Resnet50), thus we used Xception model as the transfer learning algorithm for the rest of experiments.
Fig. 2.
Evaluations on TMB prediction. Ablation study of our method on TCGA BLCA TMB prediction: (a) using SVM with Linear kernel, (b) using SVM with RBF kernel. (c) Baseline comparisons of TCGA BLCA patient-level TMB predictions. Note that in (c) Proposed-LIN and Proposed-RBF represent the proposed technique using Linear SVM and RBF SVM, respectively.
To compare the performance of patient-level TMB prediction with other state-of-the-art methods, we trained our designed CNN model (see Fig. s1), VGG16-TL241 and Resnet18,26 and Multiple Instance Learning based deep learning algorithm44 as baseline models. To train these deep learning models, tumor tiles of each WSI were assigned the same label (e.g., TMB high or low) as the corresponding patient-level TMB status. The final patient-level TMB prediction was obtained by averaging prediction probabilities of all tumor tiles. In addition, we also extracted local binary pattern (LBP) texture features from representative tumor tiles and made predictions using an SVM classifier with RBF kernel as the baseline. The RBF SVM was selected as it provided a slightly better performance than the linear SVM in our ablation study (see P-Xception in Figs. 2(a)(b)). Three-fold cross validation was applied to evaluate baseline deep leaning models, due to computational complexity, and the leave-one-out cross validation was used to evaluate the rest methods. Table 1 shows patient-level TMB prediction results in terms of accuracy (ACC), specificity (SPE), sensitivity (SEN) and AUROC values for our proposed method and baseline models. Fig. 2(c) shows patient-level TMB prediction performance in TCGA BLCA. Overall, the proposed pipeline provides better performance over baseline methods, which achieves from 2% to 5% improvements with respect to AUROC values. Taken together, these results indicate the efficacy of the proposed method to predict patient-level TMB status using WSIs.
Table 1.
Comparison of patient-level TMB prediction using different methods. In the table, Proposed-LIN uses SVM classifier with linear kernel, while Proposed-RBF uses SVM classifier with RBF kernel.
| Cohorts | Methods | ACC (%) | SPE (%) | SEN (%) | AUROC (95% CI) |
|---|---|---|---|---|---|
| LBP+SVM | 60.47 | 64.52 | 56.59 | 0.623 (0.550-0.689) | |
| Designed CNN | 61.66 | 62.10 | 61.24 | 0.651 (0.581-0.741) | |
| VGG16-TL241 | 65.22 | 66.94 | 63.57 | 0.707 (0.639-0.766) | |
| TCGA-BLCA | MIL44 | 58.89 | 58.87 | 58.91 | 0.647 (0.577-0.710) |
| Resnet1826 | 66.80 | 65.32 | 68.22 | 0.701 (0.638-0.765) | |
| Proposed-LIN | 69.57 | 68.55 | 70.54 | 0.748 (0.683-0.802) | |
| Proposed-RBF | 73.12 | 75.81 | 70.54 | 0.752 (0.694-0.810) |
Spatial heterogeneity of TMB status correlated with overall survival outcome in BLCA
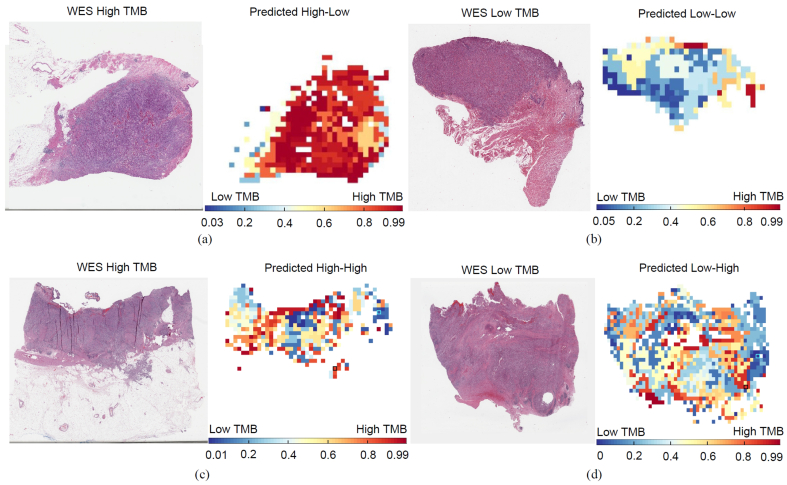
We investigated whether patient-level TMB prediction could be useful to identify patient subgroups with distinct survival outcome on the whole TCGA BLCA cohort. The TMB status of WES-based TMB high or low group was predicted by using our trained SVM with RBF kernel during the cross validation as described in above section. The TMB status for WES-based TMB intermediate group was independently predicted as TMB high or low by using our trained SVM with RBF kernel on WES-based TMB high and low groups. We grouped the whole TCGA BLCA cohort into two subgroups: predicted TMB-High vs TMB-Low, and then generated a Kaplan Meier (KM) plot of these two subgroups using overall survival (see Fig. s8(a)). While the predicted patient-level TMB-High subgroup shows a trend towards better overall survival (OS), OS difference was not significant between two subgroups using log-rank test (P=0.072). We then evaluated if the spatial heterogeneity of TMB (SH-TMB) within the patient’s tumor could help in stratifying patient into distinct survival outcome subgroups. We applied the proposed TMB prediction approach on the APC-selected representative tumor tiles. Then, the corresponding tumor regions were assigned the same TMB status as their corresponding representative tiles. To determine the SH-TMB status, we calculated the Shannon entropy45 of predicted TMB levels of tumor regions within the WSI, i.e., where Pk is the ratio between the number of the kth unique TMB prediction probability and the total number of tumor tiles within the WSI. A high entropy value indicates high SH-TMB (e.g., mixture of predicted TMB-H and low regions), while low entropy value indicates low SH-TMB within a tumor (e.g., either TMB-H or low status across most of tumor regions within WSIs). High or low entropy status was determined by using the median entropy value from all patients of TCGA BLCA cohort as the threshold (see Fig. s9(a), Table s9). Fig. 3 shows a visualization of SH-TMB heatmaps based on tile-level TMB prediction, where red and blue colors indicate predicted TMB-H and low status probability, respectively. Fig. 3(a) shows a SH-TMB heatmap of TMB-H patient based on Whole Exome Sequencing (WES) data. Our WSI-based method correctly predicted the patient-level TMB status. The entropy value based on tile-level TMB prediction indicated low SH-TMB. Specifically, the heatmap showed that most tumor regions within the WSI presented TMB-H status, while few tumor regions presented TMB low status. Similarly, Fig. 3(b) showed that our WSI-based method correctly predicted the patient as TMB low with low SH-TMB. Fig. 3(c) and (d) showed that while WSI-based patient level TMB status of these two patients agreed with WES-based TMB status, there are different mixtures of TMB-H and low status within tumor regions. Higher entropy values based on tile-level TMB status indicate higher degree of SH-TMB within WSIs.
Fig. 3.
Tile-level TMB prediction visualization. (a) Tissue-based TMB-H patient (TCGA-XF- AAN2) was predicted as patient-level TMB-H based on our WSI-based method. A heatmap of tile-level TMB prediction across tiles (i.e., tumor regions) and entropy measurement showed that most of tumor regions have TMB-H status (i.e., low SH-TMB). (b) Tissue-based TMB low patient (TCGA-XF-A9SH) was predicted as patient-level TMB low and low SH-TMB based on our WSI-based method. (c) Tissue-based TMB-H patient (TCGA-DK-A3IT) was predicted as patient-level TMB-H, while tile-level TMB prediction indicated the high SH-TMB. (d) Tissue-based TMB low patient (TCGA-FD-A3B7) was predicted as patient-level TMB low with high SH-TMB.
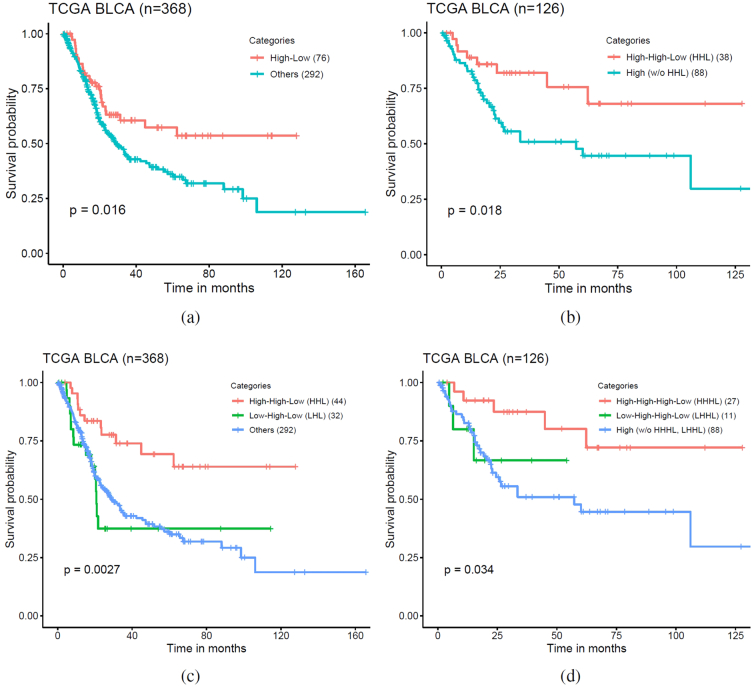
To investigate the prognostic utility of SH-TMB status, we selected patient subgroups by utilizing both patient-level TMB prediction and SH-TMB status. In experiments using TCGA BLCA cohort, we predicted patient-level TMB status for 368 patients using our proposed WSI-based method. For each patient, we assigned low or high SH-TMB status based on entropy values derived from tile-level TMB prediction. We assigned patients with predicted patient-level TMB-high and low SH-TMB into one subgroup and the rest of patients to the “Others” subgroup. Then, we generated an OS KM plot segregating by these subgroups (Fig. 4(a)), which indicates that the two subgroups have statistically significantly different OS by using log-rank test (P = 0.016). By univariate analysis using Chi-square test, the TMB subtypes correlated significantly with differences in tumor stage (P = 0.024), but not age (Age>60 vs others, P = 0.872), sex (P = 0.086), lymphovascular invasion (P = 0.064) and inflammatory infiltrate response (P = 0.428) (see Table s6). The patients in patient-level TMB-H with low spatial heterogeneity subgroup had more advanced tumor stage. The TMB subtypes did not significantly correlate with known molecular subtypes determined by Reverse Phase Protein Array (RPPA) (P = 0.761) and mRNA subtypes (P = 0.942) from TCGA BLCA study. Multivariable Cox proportional-hazard analyses of cancer stage and TMB subtypes in relation to the risk of death showed that TMB subtypes remained statistically significantly correlated with survival (see Table s7). A KM analysis based on both patient-level TMB and SH-TMB status showed that TMB subtypes with high SH-TMB status have worse OS, regardless of patient-level TMB status (see Fig. s8(b)). We further investigated whether incorporating WSI-based patient-level TMB and spatial heterogeneity with tissue-based TMB testing could improve patient stratification. While WES-based TMB-H patients tend to have better prognosis, we hypothesize that integrating WSI-based patient-level TMB status as well as spatial heterogeneity with WES-based TMB status could further improve patient stratification. To test our hypothesis, we selected 126 WES-based TMB-high patients and divided them into two subgroups: 1) WSI-based patient-level TMB-high and low SH-TMB patient subgroup (HHL) and 2) the rest of WES-based TMB-high patient subgroup (w/o HHL), respectively. Fig. 4(b) showed that WES-based TMB-H & WSI-based patient-level TMB-H and low SH-TMB patient subgroup has better OS compared to the other subgroup (log rank test P = 0.018). Taken together, these results indicate that incorporating WSI-based patient-level TMB status with SH-TMB information could lead to better patient subgroup identification with distinct OS outcome.
Fig. 4.
WSI-based patient subtypes. (a) A Kaplan-Meier (KM) plot of overall survival according to WSI-based patient-level TMB-H & low spatial TMB heterogeneity (High-Low) vs other subtypes. (b) A KM plot of overall survival for 126 WES-based TMB-H patients according to WSI-based patient-level TMB-H & low spatial TMB heterogeneity (HHL) vs other WES-based TMB-H subtypes. (c) A KM plot of overall survival according to WSI-based TILs High & patient-level TMB-H & low spatial TMB heterogeneity (HHL) vs WSI-based TILs Low & patient-level TMB-H & low spatial TMB heterogeneity (LHL) vs other subtypes. (d) A KM plot of overall survival for 126 WES-based TMB-H patients according to WSI-based TILs high & TMB-H & low spatial TMB heterogeneity (HHHL) vs WSI-based TILs Low & patient-level TMB-H & low spatial TMB heterogeneity (LHHL) vs other subtypes.
Spatial analysis of TMB heterogeneity and TILs within tumors further improved patient risk stratification in BLCA
Finally, we investigated that whether the use of spatial co-organization of predicted TMB-H and TILs within the tumor could improve prognostication. We hypothesize that a patient with most tumor regions belonging to TMB-H status (e.g., patient-level TMB-H with low spatial heterogeneity) and co-localized with high densities of TILs might have better prognosis. For instance, TMB-H patients with low spatial heterogeneity and high density of TILs (i.e., high number of both TMB-H and TILs regions within the tumor) could show better prognosis compared to patients either having low density of TILs with TMB-H or TMB low regardless of TILs status. We measured TIL densities within tumor regions for all patients of TCGA BLCA cohort and used the median TIL density score to divide patients into TIL high or low patient sub-groups (e.g., >8.12% as TIL high patient subgroup) (see Fig. s9(b)). Then we selected a subset of patients from a TIL high subgroup with the following criteria: predicted TIL High & predicted TMB High & predicted Low SH-TMB (HHL). Similarly, to investigate whether high or low level of TILs densities could be linked to patients’ prognosis, we also selected patients from a TIL low subgroup with the following criteria: predicted TIL Low & predicted TMB High & predicted Low SH-TMB (LHL). Patients belonging to the HHL subgroup tend to have most tumor regions carrying TMB-H status (i.e., a patient-level TMB-H with low SH-TMB) and higher level of TILs co-present within the patient’s tumor (ANOVA testing p<0.001) (see Fig. s10(a)). Fig. 5 shows visualization of TMB-H and TILs carrying regions within the tumors in the HHL, LHL and other subgroups. Fig. 4(c) shows a KM plot of three subgroups (e.g., the HHL subgroup vs the LHL subgroup vs other patients) and a log rank test indicates that three subgroups have statistically significant different OS (P=0.0027). The HHL subgroup showed overall best OS compared to two other subgroups. Multivariable Cox proportional-hazard analyses of cancer stage, lymphovascular invasion, mRNA-based molecular subtype, and joint TIL-TMB based patient subgroups in relation to the risk of death showed that joint TIL-TMB based patient subgroups remained statistically significantly correlated with OS (see Table s8). Interestingly, although patients in the LHL subgroup carry patient-level TMB-H with low spatial TMB-H heterogeneity, the LHL subgroup showed poorer OS compared to the HHL subgroup (HR: 3.30, 95% CI: 1.34-8.12, P<0.01). Lastly, we selected WES-based TMB-H patients and divided into three subgroups: predicted TIL High & WES-based TMB High & WSI-based predicted TMB High & predicted Low SH-TMB (HHHL) vs predicted TIL Low & WES-based TMB High & WSI-based predicted TMB High & predicted Low SH-TMB (LHHL) vs other WES-based TMB-H patients. Three subgroups from WES-based TMB-H patients have statistically different TMB-H and TILs overlapped ratio, while the HHHL subgroup has the highest TMB-H and TILs overlapped ratio among the subgroups (ANOVA testing p=0.005) (see Fig. s10(b)). Fig. 4(d) shows a KM plot of three subgroups and indicates that patients in the HHHL subgroup present better OS than other WES-based TMB-H patient subgroups (log rank test p=0.034). These results show that incorporating TILs density with patient-level and SH-TMB within the tumor based on WSIs could provide a novel prognostic biomarker to identify high or low risk patient subgroups.
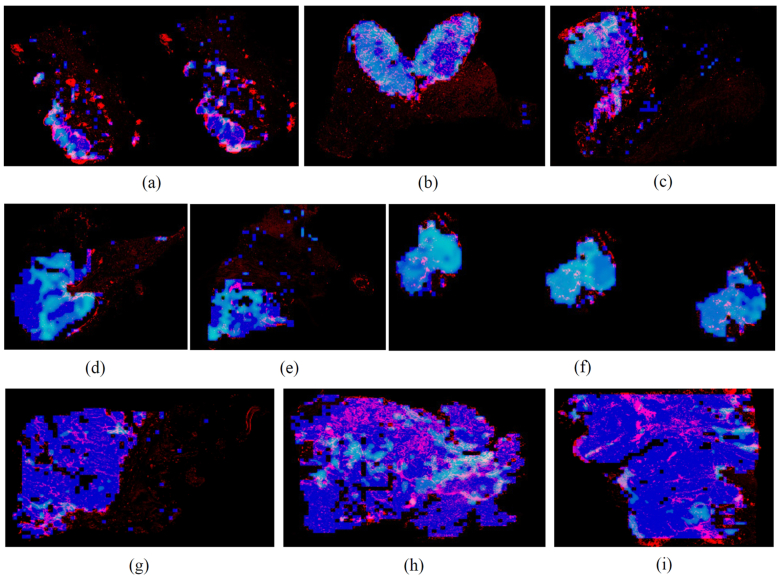
Fig. 5.
Visualization of spatial heterogeneity and organization of TMB-H and TILs within tumors. Blue color represents identified tissue regions in WSIs. Light blue (e.g., Cyan) color represents predicted TMB-H region. Red color represents predicted TILs. (a)–(c) Patients with high TILs & patient level TMB-H with low spatial TMB-H heterogeneity (HHL subtype). Most of tumor regions have been predicted as TMB-H status with high density of TILs. (d)–(f) Patients with low TILs & patient level TMB-H with low spatial TMB-H heterogeneity (LHL subtype). Most of tumor regions have been predicted as TMB-H status but with low density of TILs. (g)–(i) Patients with high TILs & TMB Low (e.g., others subtype). High TILs present within tumors with predicted TMB low status.
Discussion
Intratumor heterogeneity is one of key mechanisms driving disease progression, response and resistance to therapies.14,46 Multi-regional tissue-based sequencing from a tumor has shown spatial heterogeneity of mutational signature, mutational burden, T-cell receptor repertoire, etc.18,47, 48, 49 and its implication for treatment strategy.50 While the multi-regional tissue-based sequencing approach could provide landscape of spatial heterogeneity, it is practically challenging to generate such data, due to high costs, limited tissue availability, etc.. In this study, we present a computational pathology method to predict patient-level TMB status and investigate spatial heterogeneity of TMB within tumors. We showed that our designed method could achieve overall best performance to predict patient-level TMB status compared to other state of the art methods. We also showed that measuring and incorporating spatial heterogeneity of TMB status with patient-level TMB status based on WSIs or combined with WES-based TMB status could identify patient subgroups with distinct OS outcomes. Specifically, we found that incorporating SH-TMB information with predicted patient-level TMB status could improve patient risk stratification compared to the use of predicted patient-level TMB status alone (See Fig. s8(a) and (b)) in TCGA BLCA cohort. More specifically, patient-level TMB-H with low SH-TMB status was correlated with better OS. Visual inspection of selected tumor tiles from WSIs by our pathologist indicates that predicted TMB-H tumor tiles in patient-level TMB-H WSIs present higher densities of TILs, while they show higher grade tumors (see Table s10). This is consistent with the univariate analysis of TMB subtypes, where a higher portion of high-grade tumors belongs to patient-level TMB-H and low SH-TMB group. Although we observe an enrichment of high-grade tumors in this TMB-H subgroup, we reasoned that the higher presence of TILs within the tumors from this subgroup might lead to better prognosis. To further investigate whether higher level of TILs with SH-TMB within tumors correlates with patients’ OS, we trained end-to-end deep learning models to detect TILs and quantify TILs density within tumor regions. The predicted TILs density scores were incorporated with SH-TMB information to identify patient subgroups. The survival analysis of patient subgroups with and without high TILs presence within TMB-H tumors showed that patients carrying TMB-H status and high number of TILs within tumor regions have statistically significant better OS. It is worth to note that patient subgroup identification and survival analysis using only TILs densities information (e.g., TILs high vs low) did not show statistically significant OS difference using log rank test in TCGA BLCA (P=0.32 in Fig.s8(c)), which indicates the importance of joint spatial TILs and TMB analysis as a prognostic biomarker. Overall, our analysis demonstrated the prognostic utility of spatial TMB and TILs information based on WSIs in BLCA cohort. To the best of our knowledge, this is the first study to predict SH-TMB and investigate prognostic utility of spatial organization of TMB and TILs information for patient stratification in bladder cancer.
There are several limitations and challenges in our study. While we showed an overall better performance to predict patient-level TMB status compared with baseline methods, we have not performed further validation by using another independent dataset. This is mainly because it is practically not easy to collect patient cohorts with WSIs and corresponding sequencing data together. Larger independent cohorts from multiple institutes will be needed to validate the performance of the proposed pipeline and its generalizability. Our evaluations indicated that various deep learning-based prediction models, including end-to-end deep learning models, to predict patient-level TMB status did not show superior performance. Larger and more well-annotated WSI datasets would be needed to better train and improve the performance of deep learning-based prediction models. Our WSI-based image analysis is performed based on a tile-level not a single cell level (without distinguishing certain types of immune cells), and did not take into account specific types of spatial arrangement patterns between regions harboring TMB-H and TILs (e.g., TILs densities within local TMB-H clustered regions). For instance, the single cell level lymphocyte/immune cell detection (e.g., CD4+/CD8+/FOXP3+) and joint spatial analysis of TMB-H tumor cell and/or region and TILs and/or more advanced statistical TMB and TILs spatial modeling36 could provide higher resolution of TMB-H tumor and immune co-localization within tumor and immune microenvironment (TIME).
In summary, this study demonstrates the feasibility of predicting patient-level TMB status and delineating spatial heterogeneity and organization of TMB and TILs by using computational methods based on histological WSIs. Our spatial TMB and TILs analysis show that patients with more homogeneous TMB-H status across tumor regions and high density of TILs present better prognosis in bladder cancer. Joint spatial analysis of TILs and TMB within TIME for patients’ tumor provides a unique insight into how immune environment might have an influence on prognosis of patients with TMB-H status. By combining tissue-based TMB-H status with image-based TMB automatic prediction could further improve patient stratification in bladder cancer. Taken together, our work provides a new foundation of how spatial characterization of tumor (e.g., TMB-H status) and immune environment within the tumor based on WSIs could be used to improve risk stratification in bladder cancer.
Competing Interests
The authors declare that they have no competing financial interests.
Acknowledgments
This work is partially supported by Youth Fund of National Natural Science Foundation of China (NO. 82102135), the Fundamental Research Funds for the Central Universities (DUT21YG135) to HMX, and Department of Defense Translational Team Science Award (CA190578) to THH.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jpi.2022.100105.
Contributor Information
Hongming Xu, Email: mxu@dlut.edu.cn.
Jean René Clemenceau, Email: Jean.Clemenceau@mayo.edu.
Sunho Park, Email: park.sunho@mayo.edu.
Jinhwan Choi, Email: choi.jinhwan@mayo.edu.
Sung Hak Lee, Email: hakjjang@catholic.ac.kr.
Tae Hyun Hwang, Email: Hwang.taehyun@mayo.edu.
Appendix A. Supplementary data
Supplementary material 1
References
- 1.Robertson A.G., Kim J., Al-Ahmadie H., et al. Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell. 2017;171(540–56) doi: 10.1016/j.cell.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan T.A., Yarchoan M., Jaffee E., et al. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann Oncol. 2019;30:44–56. doi: 10.1093/annonc/mdy495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bandini M., Ross J.S., Raggi D., et al. Predicting the pathologic complete response after neoadjuvant pembrolizumab in muscle-invasive bladder cancer. JNCI J Natl Cancer Inst. 2021;113:48–53. doi: 10.1093/jnci/djaa076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Necchi A., Raggi D., Gallina A., et al. Updated results of PURE-01 with preliminary activity of neoadjuvant pembrolizumab in patients with muscle-invasive bladder carcinoma with variant histologies. Eur Urol. 2020;77:439–446. doi: 10.1016/j.eururo.2019.10.026. [DOI] [PubMed] [Google Scholar]
- 5.Song B.-N., Kim S.-K., Mun J.-Y., Choi Y.-D., Leem S.-H., Chu I.-S. Identification of an immunotherapy-responsive molecular subtype of bladder cancer. EBioMedicine. 2019;50:238–245. doi: 10.1016/j.ebiom.2019.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown S.D., Warren R.L., Gibb E.A., et al. Neo-antigens predicted by tumor genome meta-analysis correlate with increased patient survival. Genome Res. 2014;24:743–750. doi: 10.1101/gr.165985.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corredor G., Wang X., Zhou Y., et al. Spatial architecture and arrangement of tumor-infiltrating lymphocytes for predicting likelihood of recurrence in early-stage non–small cell lung cancer. Clin Cancer Res. 2019;25:1526–1534. doi: 10.1158/1078-0432.CCR-18-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Idos G.E., Kwok J., Bonthala N., Kysh L., Gruber S.B., Qu C. The prognostic implications of tumor infiltrating lymphocytes in colorectal cancer: a systematic review and meta-analysis. Scient Rep. 2020;10:1–14. doi: 10.1038/s41598-020-60255-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Plesca I., Tunger A., Müller L., et al. Characteristics of tumor-infiltrating lymphocytes prior to and during immune checkpoint inhibitor therapy. Front Immunol. 2020;11:364. doi: 10.3389/fimmu.2020.00364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Massi D., Rulli E., Cossa M., et al. The density and spatial tissue distribution of CD8+ and CD163+ immune cells predict response and outcome in melanoma patients receiving MAPK inhibitors. J Immunother Cancer. 2019;7:1–13. doi: 10.1186/s40425-019-0797-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oh D.Y., Kwek S.S., Raju S.S., et al. Pai C-CS, Rancan C: Intratumoral CD4+ T cells mediate anti-tumor cytotoxicity in human bladder cancer. Cell. 2020;181(1612–25) doi: 10.1016/j.cell.2020.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan Y. Spatial heterogeneity in the tumor microenvironment. Cold Spring Harb Perspect Med. 2016;6 doi: 10.1101/cshperspect.a026583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heindl A., Sestak I., Naidoo K., Cuzick J., Dowsett M., Yuan Y. Relevance of spatial heterogeneity of immune infiltration for predicting risk of recurrence after endocrine therapy of ER+ breast cancer. JNCI J Natl Cancer Inst. 2018;110:166–175. doi: 10.1093/jnci/djx137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marusyk A., Janiszewska M., Polyak K. Intratumor heterogeneity: the rosetta stone of therapy resistance. Cancer cell. 2020;37:471–484. doi: 10.1016/j.ccell.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiménez-Sánchez A., Memon D., Pourpe S., et al. Heterogeneous tumor-immune microenvironments among differentially growing metastases in an ovarian cancer patient. Cell. 2017;170(927–38) doi: 10.1016/j.cell.2017.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Binnewies M., Roberts E.W., Kersten K., et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med. 2018;24:541–550. doi: 10.1038/s41591-018-0014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jia Q., Wu W., Wang Y., et al. Local mutational diversity drives intratumoral immune heterogeneity in non-small cell lung cancer. Nat Commun. 2018;9:1–10. doi: 10.1038/s41467-018-07767-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y., Chang L., Yang Y., et al. The correlations of tumor mutational burden among single-region tissue, multi-region tissues and blood in non-small cell lung cancer. J Immunother Cancer. 2019;7:1–5. doi: 10.1186/s40425-019-0581-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Azizi E., Carr A.J., Plitas G., et al. Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell. 2018;174(1293–308) doi: 10.1016/j.cell.2018.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Z., Zhou L., Liu L., et al. Single-cell RNA sequencing highlights the role of inflammatory cancer-associated fibroblasts in bladder urothelial carcinoma. Nat Commun. 2020;11:1–12. doi: 10.1038/s41467-020-18916-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee H.W., Chung W., Lee H.-O., et al. Single-cell RNA sequencing reveals the tumor microenvironment and facilitates strategic choices to circumvent treatment failure in a chemorefractory bladder cancer patient. Genome Med. 2020;12:1–21. doi: 10.1186/s13073-020-00741-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chuah S., Chew V. High-dimensional immune-profiling in cancer: implications for immunotherapy. J Immunother Cancer. 2020;8 doi: 10.1136/jitc-2019-000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith E.A., Hodges H.C. The spatial and genomic hierarchy of tumor ecosystems revealed by single-cell technologies. Trends Cancer. 2019;5:411–425. doi: 10.1016/j.trecan.2019.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moncada R., Barkley D., Wagner F., et al. Integrating microarray-based spatial transcriptomics and single-cell RNA-seq reveals tissue architecture in pancreatic ductal adenocarcinomas. Nat Biotechnol. 2020;38:333–342. doi: 10.1038/s41587-019-0392-8. [DOI] [PubMed] [Google Scholar]
- 25.Coudray N., Ocampo P.S., Sakellaropoulos T., et al. Classification and mutation prediction from non–small cell lung cancer histopathology images using deep learning. Nat Med. 2018;24:1559–1567. doi: 10.1038/s41591-018-0177-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kather J.N., Pearson A.T., Halama N., et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat Med. 2019;25:1054–1056. doi: 10.1038/s41591-019-0462-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu Y., Jung A.W., Torne R.V., et al. Pan-cancer computational histopathology reveals mutations, tumor composition and prognosis. Nat Cancer. 2020;1:800–810. doi: 10.1038/s43018-020-0085-8. [DOI] [PubMed] [Google Scholar]
- 28.Kather J.N., Heij L.R., Grabsch H.I., et al. Pan-cancer image-based detection of clinically actionable genetic alterations. Nat Cancer. 2020;1:789–799. doi: 10.1038/s43018-020-0087-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arlova A., Jin C., Wong-Rolle A., et al. Artificial intelligence-based tumor segmentation in mouse models of lung adenocarcinoma. J Pathol Inform. 2022;13 doi: 10.1016/j.jpi.2022.100007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brockmoeller S., Echle A., Ghaffari Laleh N., et al. Deep Learning identifies inflamed fat as a risk factor for lymph node metastasis in early colorectal cancer. J Pathol. 2022;256:269–281. doi: 10.1002/path.5831. [DOI] [PubMed] [Google Scholar]
- 31.Hacking S.M., Wu D., Alexis C., Nasim M. A novel superpixel approach to the tumoral microenvironment in colorectal cancer. J Pathol Inform. 2022;13 doi: 10.1016/j.jpi.2022.100009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wetteland R., Engan K., Eftestøl T., Kvikstad V., Janssen E.A. Treatment: a multiscale approach for whole-slide image segmentation of five tissue classes in urothelial carcinoma slides. Technol Cancer Res. 2020;19 1533033820946787. [Google Scholar]
- 33.Loeffler C.M.L., Bruechle N.O., Jung M., et al. Artificial intelligence–based detection of FGFR3 mutational status directly from routine histology in bladder cancer: a possible preselection for molecular testing? Eur Urology Focus. 2021 doi: 10.1016/j.euf.2021.04.007. [DOI] [PubMed] [Google Scholar]
- 34.Saltz J., Gupta R., Hou L., et al. Spatial organization and molecular correlation of tumor-infiltrating lymphocytes using deep learning on pathology images. Cell Rep. 2018;23(181–93) doi: 10.1016/j.celrep.2018.03.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Acs B., Ahmed F.S., Gupta S., et al. An open source automated tumor infiltrating lymphocyte algorithm for prognosis in melanoma. Nat Commun. 2019;10:1–7. doi: 10.1038/s41467-019-13043-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.AbdulJabbar K., Raza S.E.A., Rosenthal R., et al. Geospatial immune variability illuminates differential evolution of lung adenocarcinoma. Nat Med. 2020;26:1054–1062. doi: 10.1038/s41591-020-0900-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ojala T., Pietikainen M., Maenpaa T. Multiresolution gray-scale and rotation invariant texture classification with local binary patterns. IEEE Trans Pattern Anal Mach Intel. 2002;24:971–987. [Google Scholar]
- 38.Frey B.J., Dueck D. Clustering by passing messages between data points. Science. 2007;315:972–976. doi: 10.1126/science.1136800. [DOI] [PubMed] [Google Scholar]
- 39.Macenko M., Niethammer M., Marron J.S., et al. IEEE international symposium on biomedical imaging: from nano to macro. IEEE; 2009. A method for normalizing histology slides for quantitative analysis; pp. 1107–1110. [Google Scholar]
- 40.Chollet F. Proceedings of the IEEE conference on computer vision and pattern recognition. 2017. Xception: Deep learning with depthwise separable convolutions; pp. 1251–1258. [Google Scholar]
- 41.Xu H., Park S., Hwang T.H. Computerized classification of prostate cancer gleason scores from whole slide images. IEEE/ACM Trans Computat Biol Bioinform. 2020;17:1871–1882. doi: 10.1109/TCBB.2019.2941195. [DOI] [PubMed] [Google Scholar]
- 42.Szegedy C., Vanhoucke V., Ioffe S., Shlens J., Wojna Z. Proceedings of the IEEE conference on computer vision and pattern recognition. 2016. Rethinking the inception architecture for computer vision; pp. 2818–2826. [Google Scholar]
- 43.He K., Zhang X., Ren S., Sun J. Proceedings of the IEEE conference on computer vision and pattern recognition. 2016. Deep residual learning for image recognition; pp. 770–778. [Google Scholar]
- 44.Campanella G., Hanna M.G., Geneslaw L., et al. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat Med. 2019;25:1301–1309. doi: 10.1038/s41591-019-0508-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jackson H.W., Fischer J.R., Zanotelli V.R., et al. The single-cell pathology landscape of breast cancer. Nature. 2020;578:615–620. doi: 10.1038/s41586-019-1876-x. [DOI] [PubMed] [Google Scholar]
- 46.Failmezger H., Muralidhar S., Rullan A., de Andrea C.E., Sahai E., Yuan Y. Topological tumor graphs: a graph-based spatial model to infer stromal recruitment for immunosuppression in melanoma histology. Cancer Res. 2020;80:1199–1209. doi: 10.1158/0008-5472.CAN-19-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu X., Fujimoto J., Ying L., et al. Multi-region exome sequencing reveals genomic evolution from preneoplasia to lung adenocarcinoma. Nat Commun. 2019;10:1–10. doi: 10.1038/s41467-019-10877-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jamal-Hanjani M., Wilson G.A., McGranahan N., et al. Tracking the evolution of non–small-cell lung cancer. New Engl J Med. 2017;376:2109–2121. doi: 10.1056/NEJMoa1616288. [DOI] [PubMed] [Google Scholar]
- 49.Joshi K., de Massy M.R., Ismail M., et al. Spatial heterogeneity of the T cell receptor repertoire reflects the mutational landscape in lung cancer. Nat Med. 2019;25:1549–1559. doi: 10.1038/s41591-019-0592-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stanta G., Bonin S. Overview on clinical relevance of intra-tumor heterogeneity. Front Med. 2018;5:85. doi: 10.3389/fmed.2018.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1