Abstract
It is important to determine if severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections and SARS-CoV-2 mRNA vaccinations elicit different types of antibodies. Here, we characterize the magnitude and specificity of SARS-CoV-2 spike-reactive antibodies from 10 acutely infected health care workers with no prior SARS-CoV-2 exposure history and 23 participants who received SARS-CoV-2 mRNA vaccines. We found that infection and primary mRNA vaccination elicit S1- and S2-reactive antibodies, while secondary vaccination boosts mostly S1 antibodies. Using absorption assays, we found that SARS-CoV-2 infections elicit a large proportion of original antigenic sin-like antibodies that bind efficiently to the spike of common seasonal human coronaviruses but poorly to the spike of SARS-CoV-2. In converse, vaccination modestly boosts antibodies reactive to the spike of common seasonal human coronaviruses, and these antibodies cross-react more efficiently to the spike of SARS-CoV-2. Our data indicate that SARS-CoV-2 infections and mRNA vaccinations elicit fundamentally different antibody responses.
Keywords: SARS-CoV-2, antibodies, mRNA vaccines, original antigenic sin, coronavirus
Graphical abstract
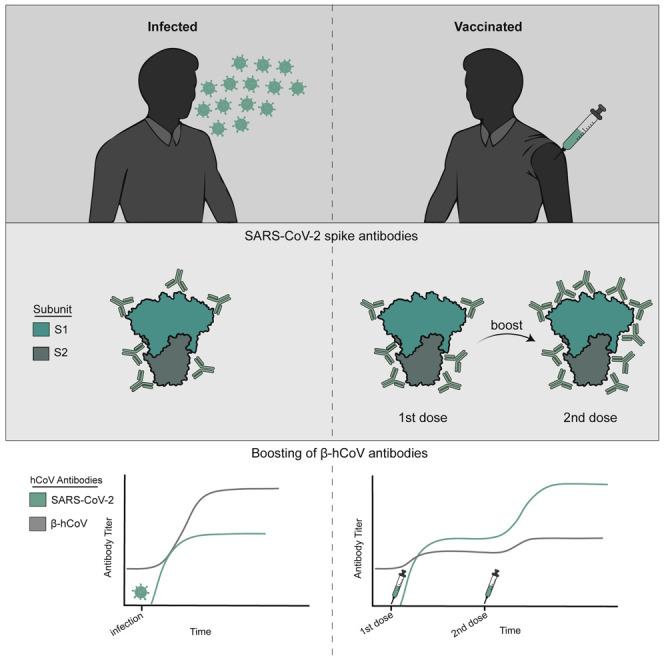
Anderson et al. show that SARS-CoV-2 infections elicit high levels of original antigenic sin-like antibodies that bind efficiently to the spike of common seasonal human coronaviruses but poorly to the spike of SARS-CoV-2. They find that SARS-CoV-2 vaccination elicits lower levels of original antigenic sin-like antibodies.
Introduction
Since late 2019 severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has spread across the world, causing a global pandemic (Carvalho et al., 2021; Zhou et al., 2020). This prompted the rapid development of several SARS-CoV-2 vaccines including two that use an mRNA-based platform (reviewed in Excler et al., 2021; Gebre et al., 2021; Golob et al., 2021). The mRNA vaccines, Pfizer BNT162b2 and Moderna mRNA-1273, employ lipid nanoparticles that encase modified mRNA encoding the spike protein of SARS-CoV-2 (Topol, 2021). SARS-CoV-2 mRNA vaccines have been found to be safe and effective at preventing severe COVID-19, hospitalizations, and death (Baden et al., 2021; Polack et al., 2020). SARS-CoV-2 mRNA vaccines elicit antibody responses that partially recognize and protect against severe disease caused by antigenically distinct SARS-CoV-2 variants (Collier et al., 2021; Doria-Rose et al., 2021; Liu et al., 2021; Wang et al., 2021a, 2021b; Widge et al., 2021; Wu et al., 2021).
Some studies suggest that prior infections with common seasonal human coronaviruses (hCoVs) impact the severity of SARS-CoV-2 infections (Gouma et al., 2021; Sagar et al., 2021). Most individuals are exposed to hCoVs early in childhood (Anderson et al., 2021; Dijkman et al., 2012; Dyrdak et al., 2021; Gaunt et al., 2010; Huang et al., 2020; Killerby et al., 2018) and then re-exposed to antigenically drifted forms of these viruses throughout life (Edridge et al., 2020; Eguia et al., 2021; Kistler and Bedford, 2021). Common hCoVs include the HKU1 and OC43 human betacoronaviruses (β-hCoVs) and 229E and NL63 human alphacoronaviruses (α-hCoVs) (Andersen et al., 2020; Boni et al., 2020; Jaimes et al., 2020; Okba et al., 2020). Studies from our group and others have shown that some individuals possessed antibodies that could bind to SARS-CoV-2 proteins prior to the COVID-19 pandemic (Anderson et al., 2021; Ng et al., 2020; Song et al., 2021). SARS-CoV-2 is a β-hCoV, and antibodies reactive to the OC43 and HKU1 β-hCoVs are boosted upon SARS-CoV-2 infections (Aguilar-Bretones et al., 2021; Anderson et al., 2021; Gouma et al., 2021; Nguyen-Contant et al., 2020; Song et al., 2021) and SARS-CoV-2 mRNA vaccinations (Amanat et al., 2021; Jackson et al., 2020; Roltgen et al., 2022). It is unknown if the recall of β-hCoV antibodies upon SARS-CoV-2 infections impacts disease outcome. A recent study suggests that the recall of OC43 β-hCoV antibodies is associated with a compromised de novo SARS-CoV-2 response in individuals with fatal COVID-19 (McNaughton et al., 2022).
The boosting of hCoV antibodies upon infection with the antigenically distinct SARS-CoV-2 is consistent with the doctrine of “original antigenic sin,” first proposed to describe influenza virus antibody responses by Thomas Francis in 1960 (Francis, 1960). We recently developed absorption assays to show that sequential heterosubtypic influenza virus infections elicit antibodies that, paradoxically, do not bind effectively to the boosting viral strain (Arevalo et al., 2020). In that study, we sequentially infected ferrets with two antigenically distinct influenza virus strains and analyzed serum samples collected after each infection. We found that many antibodies elicited by secondary influenza virus infections did not bind effectively to the secondary boosting influenza virus strain. We proposed that antigenically distinct influenza viruses engage memory B cells elicited by prior infections through multiple low-affinity interactions with thousands of B cell receptors on memory B cells. Low-affinity antibodies secreted in a soluble form through this recall response fail to bind to the antigenically distinct recall antigens because they require the level of multivalent binding that is provided on B cells.
In the current report, we used a similar absorption technique to define the specificity of β-hCoV antibodies elicited by SARS-CoV-2 infections and mRNA vaccinations. We completed a series of studies to determine if these boosted β-hCoV antibodies could cross-react to SARS-CoV-2.
Results
SARS-CoV-2 infections and vaccinations elicit antibodies against the SARS-CoV-2 spike protein
We obtained samples from individuals before and after acute SARS-CoV-2 infections (n = 10) and pre-/post-two doses of a Pfizer BNT162b2 mRNA SARS-CoV-2 vaccine (n = 23). Samples from SARS-CoV-2-infected individuals were obtained from a health care worker sero-monitoring study in which all infections were relatively mild (Gouma et al., 2021). All 10 individuals were unvaccinated and seronegative in the beginning of this study and acquired a PCR-confirmed SARS-CoV-2 infection in the spring and summer of 2020. Blood samples were collected from SARS-CoV-2-infected individuals 5–28 days (mean 17.9 days) after PCR-confirmed infections. Blood samples were collected from SARS-CoV-2 mRNA-vaccinated individuals (Goel et al., 2021a) 7–15 days post-primary immunization (mean 14.2 days), the day of or the day before the booster immunization (mean 21.2 days post-primary), and 7–12 days post-booster immunization (mean 7.5 days post-boost). Individuals in our infection and vaccination studies had similar ages, and both groups were predominantly female (Table S1).
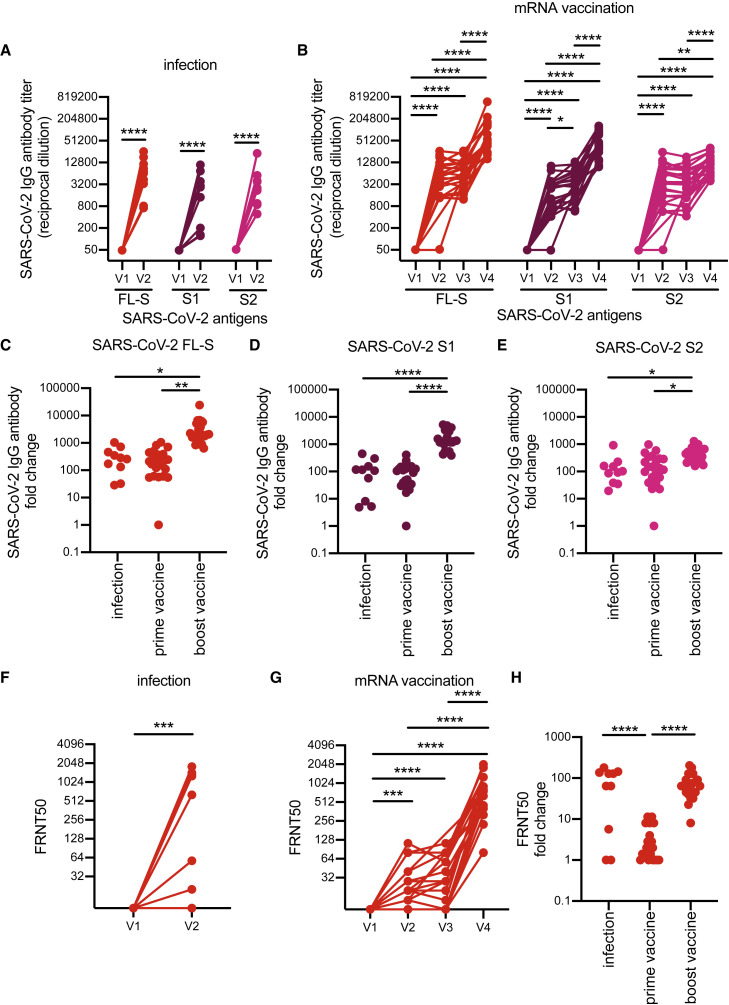
Consistent with previous studies (reviewed in Lombardi et al., 2021 and Roltgen and Boyd, 2021), antibodies against the full-length spike (FL-S) of SARS-CoV-2 increased following infection (Figure 1A) and vaccination (Figure 1B). SARS-CoV-2 FL-S antibody levels were similar following infections and primary vaccinations, and antibody levels were significantly boosted following the second dose of vaccine (Figures 1B and 1C). The SARS-CoV-2 FL-S protein consists of two domains, the S1 domain, which encompasses the receptor-binding domain (RBD) essential for cell attachment and entry (Barnes et al., 2020; Letko et al., 2020; Walls et al., 2020; Wrapp et al., 2020) and the N-terminal domain (NTD), and the S2 domain, which shares more sequence homology with hCoVs (Nguyen-Contant et al., 2020; Okba et al., 2020). Infection and primary vaccination elicited high levels of both S1 and S2 antibodies (Figures 1A and 1B), whereas secondary vaccinations boosted S1 antibodies more efficiently compared with S2 antibodies (Figures 1B, 1D, and 1E). SARS-CoV-2 neutralizing antibody levels were similar following infection and prime-boost vaccination (Figures 1F–1H).
Figure 1.
Specificity of SARS-CoV-2 antibodies induced after SARS-CoV-2 infection versus vaccination
(A and B) ELISAs were completed to quantify levels of serum antibodies binding to the SARS-CoV-2 full-length spike (FL-S) protein, the S1 domain (S1) of S, and the S2 domain (S2) of S after SARS-CoV-2 infection (A) and mRNA vaccination (B).
(C–E) We calculated fold change in antibody titers before and after seroconversion and pre-/post-prime and boost doses of a SARS-CoV-2 mRNA vaccine.
(F and G) SARS-CoV-2 pseudotype neutralization assays were completed with sera samples from SARS-CoV-2-infected individuals (F) and SARS-CoV-2 mRNA-vaccinated participants (G).
(H) Fold change in neutralization titers was calculated before and after seroconversion and pre-/post-prime and boost doses of a SARS-CoV-2 mRNA vaccine.
For (A), (B), (F), and (G), we completed paired t tests or one-way ANOVA of log2-transformed titers; ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05. For (C)–(E) and (H), we completed one-way ANOVA of titer fold changes; ∗∗∗∗p < 0.0001, ∗∗p < 0.01 ∗p < 0.05. Data are representative of two independent experiments. Neutralizing antibody titers of SARS-CoV-2 mRNA vaccinated participants have been previously reported in Goel et al. (2021a).
SARS-CoV-2 infections and vaccinations elicit antibodies against β-hCoVs
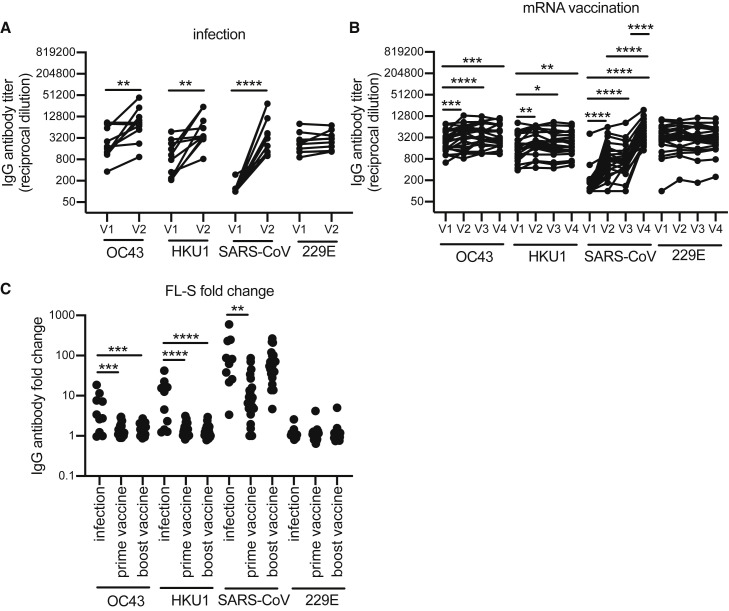
We previously found that antibodies against the FL-S of the common OC43 β-hCoV are boosted upon SARS-CoV-2 infection in hospitalized patients with severe disease (Anderson et al., 2021). Other studies have also shown that levels of antibodies against β-hCoVs including both OC43 and HKU1 are higher in COVID-19 patients compared with healthy donors (Aydillo et al., 2021; Bangaru et al., 2022; Grobben et al., 2021; McNaughton et al., 2022; Roltgen et al., 2022; Shrock et al., 2020; Song et al., 2021). Consistent with these findings, we found that antibodies reactive to the FL-S of the common OC43 and HKU1 β-hCoVs increased upon SARS-CoV-2 infections in health care workers, while antibodies to the FL-S of the common 229E α-hCoV did not (Figure 2A). Studies have shown that antibodies against common β-hCoVs increase upon SARS-CoV-2 mRNA vaccination (Amanat et al., 2021; Grobben et al., 2021; Roltgen et al., 2022). Although we found small increases in OC43 and HKU1 FL-S antibody titers following SARS-CoV-2 mRNA vaccinations (Figure 2B), the magnitude of OC43 and HKU1 FL-S antibody boosts were much lower following vaccinations compared with infections for most individuals (Figure 2C). mRNA vaccination did not increase antibodies reactive to the FL-S of the common 229E α-hCoV (Figures 2B and 2C).
Figure 2.
Antibodies to the FL-S of other betacoronaviruses are boosted upon SARS-CoV-2 infection and after vaccination to a lesser extent
(A and B) ELISAs were completed to quantify levels of serum antibodies binding to the FL-S protein of other betacoronaviruses (OC43, HKU1, SARS-CoV) and alphacoronavirus (229E) after SARS-CoV-2 infection (A) and mRNA vaccination (B). Paired t tests or one-way ANOVA of log2 transformed antibody titers; ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05. Data are representative of two independent experiments.
(C) We calculated fold change in antibody titers against spike before and after seroconversion and pre-/post-prime and boost doses of a SARS-CoV-2 mRNA vaccine. One-way ANOVA of antibody fold change; ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01.
We also completed experiments to determine how SARS-CoV-2 infections and mRNA vaccinations impact antibodies reactive to the original SARS-CoV β-hCoV, since most humans have not been exposed to this virus. There are several epitopes in both the S1 and S2 spike domains that are conserved between the original SARS-CoV and SARS-CoV-2, and others have shown that antibodies specific to SARS-CoV are boosted upon SARS-CoV-2 vaccination and infection (Aydillo et al., 2021; Roltgen et al., 2022; Walls et al., 2020). Consistent with these previous studies, antibodies reactive to the FL-S of the original SARS-CoV increased following SARS-CoV-2 infection (Figure 2A) and mRNA vaccination (Figure 2B). Antibodies reactive to the FL-S of the original SARS-CoV increased more substantially following infection relative to prime vaccination and were at similar levels following booster vaccination (Figure 2C).
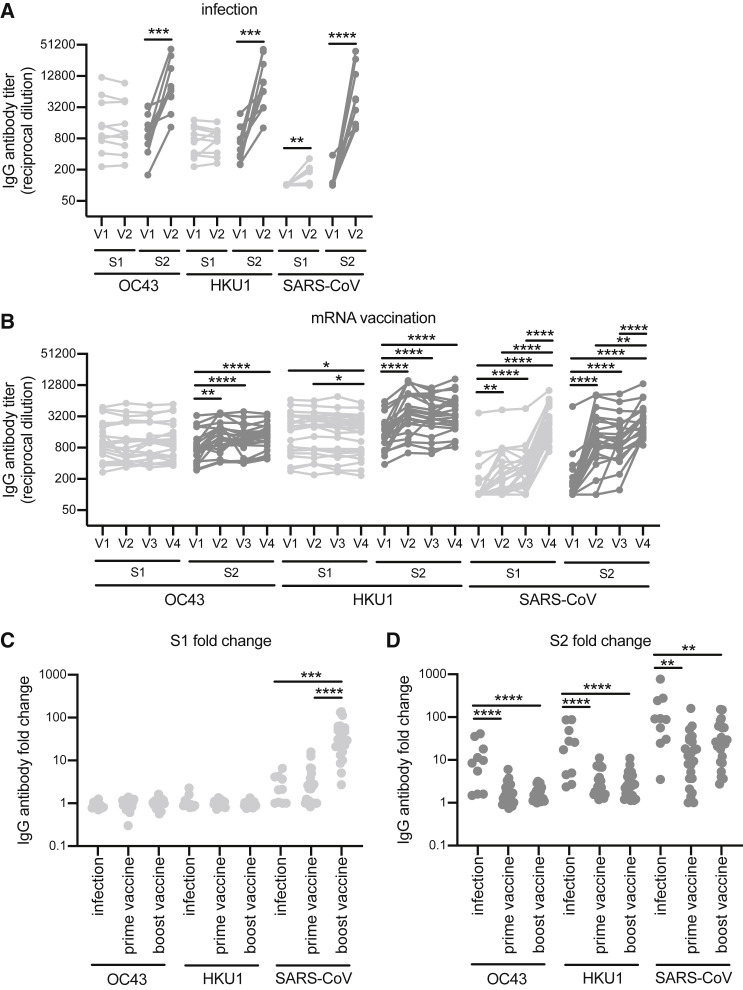
We next determined if antibodies elicited by SARS-CoV-2 infections and vaccinations targeted the S1 or S2 domains of the spike of the OC43, HKU1, and SARS-CoV β-hCoVs. Antibodies boosted by infections and vaccinations primarily targeted the S2 domain of the OC43 and HKU1 spike (Figures 3A and 3B), with infections boosting S2 responses more effectively compared with vaccinations (Figure 3D). Booster vaccinations did not affect OC43 and HKU1 S1 or S2 antibody levels (Figures 3B–3D). Antibodies elicited by infections and vaccinations targeted both the S1 and S2 domains of the original SARS-CoV (Figures 3A–3D). Antibodies elicited by infections and primary vaccinations were more biased toward the S2 domain of SARS-CoV, whereas booster vaccinations elicited more antibodies reactive to the S1 domain of SARS-CoV (Figures 3C and 3D).
Figure 3.
Antibodies reactive to the S2 domain of seasonal betacoronaviruses are boosted upon SARS-CoV-2 infection and after vaccination to a lesser extent
(A and B) ELISAs were completed to determine the levels of S1- and S2-specific antibodies against spike of OC43, HKU1, and SARS-CoV after SARS-CoV-2 infection (A) or SARS-CoV-2 mRNA vaccination (B). Paired t tests or one-way ANOVA of log2-transformed antibody titers; ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05. Data are representative of two independent experiments.
(C and D) Fold change of S1- and S2-specific antibodies was calculated before and after seroconversion and pre-/post-prime and boost mRNA vaccines. One-way ANOVA of antibody fold change; ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01.
SARS-CoV-2 infections elicit higher levels of original antigenic sin antibodies compared with SARS-CoV-2 mRNA vaccinations
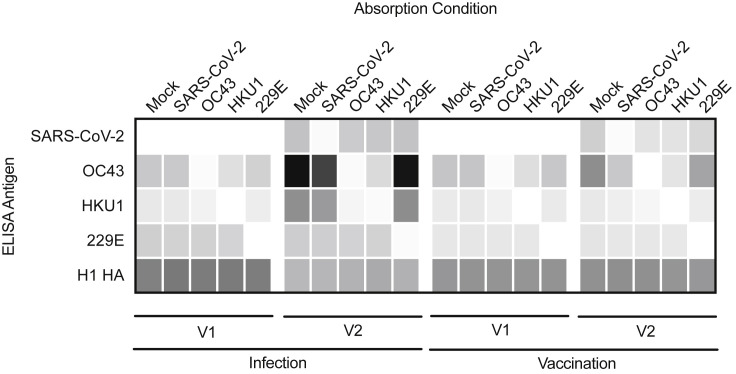
We developed a magnetic-bead-based absorption assay to determine if OC43 and HKU1 FL-S antibodies elicited by SARS-CoV-2 infection and vaccination cross-react with SARS-CoV-2 FL-S. We incubated serum samples with beads coupled with different FL-S proteins, and then we depleted bead-reactive antibodies using a magnetic column. We quantified antibody levels in serum absorbed with antigen-coupled beads to assess antibody cross-reactivity. As a control, we quantified levels of influenza virus hemagglutinin (HA) antibodies in these experiments, and we found that HA-reactive antibodies were not depleted with SARS-CoV-2, OC43, HKU1, or 229E FL-S-labeled beads (Figure 4 ).
Figure 4.
Seasonal coronavirus spike antibodies boosted by SARS-CoV-2 infection do not bind effectively to the SARS-CoV-2 spike
Sera samples from 10 SARS-CoV-2-infected health care workers and 10 SARS-CoV-2 mRNA-vaccinated participants were absorbed with SARS-CoV-2, OC43, HKU1, and 229E spike-coupled beads or mock-treated beads prior to antibody quantification by ELISA. We determined reciprocal antibody titers in samples before and after infection and pre-/post-the first dose of an mRNA vaccine for SARS-CoV-2, OC43, HKU1, 229E, and an unrelated antigen, influenza hemagglutinin H1.
SARS-CoV-2 FL-S-reactive antibodies elicited by infection were efficiently depleted with SARS-CoV-2 FL-S-coupled beads but not with OC43, HKU1, or 229E FL-S-coupled beads (Figure 4). OC43 FL-S-reactive antibodies boosted by SARS-CoV-2 infection were efficiently depleted with OC43 FL-S-coupled beads, but surprisingly, these antibodies were not efficiently depleted with SARS-CoV-2 FL-S-coupled beads (Figure 4). Similar results were obtained when we examined HKU1 antibodies. HKU1 FL-S-reactive antibodies boosted by SARS-CoV-2 infection were efficiently depleted with HKU1 FL-S-coupled beads, but these antibodies were not depleted with SARS-CoV-2 FL-S-coupled beads (Figure 4). These results are surprising since these OC43 and HKU1 FL-S-reactive antibodies were boosted upon SARS-CoV-2 infection. These data suggest that OC43 and HKU1 FL-S-reactive antibodies boosted by SARS-CoV-2 infections do not bind efficiently to SARS-CoV-2.
Similar to antibodies elicited by infection, SARS-CoV-2 FL-S-reactive antibodies elicited by vaccination were efficiently depleted with SARS-CoV-2 FL-S-coupled beads but not OC43, HKU1, or 229E FL-S-coupled beads (Figure 4). OC43 and HKU1 FL-S-reactive antibodies elicited by SARS-CoV-2 vaccination were efficiently depleted with OC43 and HKU1 FL-S beads, respectively. Interestingly, OC43 FL-S-reactive antibodies elicited by vaccination were partially depleted with SARS-CoV-2 FL-S-coupled beads (Figure 4). Following SARS-CoV-2 FL bead absorptions, vaccine-elicited OC43 FL-S-reactive antibodies were at similar levels compared with what we observed prior to SARS-CoV-2 mRNA vaccination (OC43 titer at V1 after mock absorption was 7,785; OC43 titer at V2 after mock absorption was 15,322; and OC43 titer at V2 after SARS-CoV-2 absorption was 7,326). Therefore, unlike antibodies elicited by infection, the OC43 FL-S-reactive antibodies elicited by vaccination were highly cross-reactive to SARS-CoV-2. Taken together, our data suggest that SARS-CoV-2 infection elicits OC43 and HKU1 FL-S-reactive antibodies that bind poorly to SARS-CoV-2 FL-S, while SARS-CoV-2 mRNA vaccinations elicits a relatively lower level of these types of antibodies.
Discussion
SARS-CoV-2 infections (Cho et al., 2021; Goel et al., 2021a; Jalkanen et al., 2021; Roltgen et al., 2022; Turner et al., 2021) and SARS-CoV-2 mRNA vaccinations (Liu et al., 2021; Widge et al., 2021; Wu et al., 2021) elicit antibody responses against the spike protein in most individuals. SARS-CoV-2 infections elicit more variable levels of antibodies, which is influenced by age and disease severity (Roltgen and Boyd, 2021; Sasson et al., 2021; Sette and Crotty, 2021; Takahashi et al., 2020). SARS-CoV-2-reactive antibodies tend to be elevated in patients with severe COVID-19 (Guthmiller et al., 2021; Kuri-Cervantes et al., 2020; Legros et al., 2021; Mathew et al., 2020; Piccoli et al., 2020; Robbiani et al., 2020; Roltgen et al., 2020; Rydyznski Moderbacher et al., 2020). Antibodies elicited by 2 doses of SARS-CoV-2 mRNA vaccines can be at similar levels as those induced by severe COVID-19 (Roltgen et al., 2022). Disease severity may also influence the breadth of SARS-CoV-2 antibody responses with more extensive epitope spreading in individuals with severe COVID-19 (Shrock et al., 2020). Studies have shown that antibodies elicited by vaccination and infections differentially recognize and neutralize SARS-CoV-2 variants (Chen et al., 2021; Cho et al., 2021; Goel et al., 2021b; Greaney et al., 2021; Planas et al., 2021; Stamatatos et al., 2021; Starr et al., 2021; Wang et al., 2021a).
We found differences in OC43 and HKU1 spike binding between antibodies elicited by SARS-CoV-2 infections versus mRNA vaccinations. SARS-CoV-2 infections elicited high levels of antibodies that bound to the S2 region of the OC43 and HKU1 spike proteins; however, our absorption assays demonstrated that these antibodies bound poorly to the SARS-CoV-2 spike. Our data are consistent with a recent study that showed that OC43-specific B cell clones that poorly recognize SARS-CoV-2 can be elicited in patients with severe COVID-19 (Aguilar-Bretones et al., 2021). We found that SARS-CoV-2 mRNA vaccinations elicited lower levels of antibodies that reacted to the S2 region of the OC43 and HKU1 spike proteins. Unlike antibodies elicited by infections, these vaccine-elicited antibodies appeared to be more cross-reactive and partially bound to both SARS-CoV-2 and common β-hCoV spike proteins. Further studies will be required to fully understand mechanisms that lead to different types of antibody responses elicited by SARS-CoV-2 infections versus vaccinations. It is possible that memory B cells elicited by prior β-hCoV infections are recalled by both SARS-CoV-2 infections and vaccinations and that long-lived germinal centers elicited by mRNA vaccinations (Lederer et al., 2022; Roltgen et al., 2022; Turner et al., 2021) are required to allow for somatic hypermutations that promote the formation of cross-reactive S2 antibodies that bind efficiently to the spike proteins of both β-hCoVs and SARS-CoV-2. Consistent with this, a recent study found that S2-specific B cells with a memory phenotype are quickly recruited following primary immunization of humans (Brewer et al., 2022).
Boosting of OC43 and HKU1 S2-reactive antibodies following SARS-CoV-2 infection is consistent with Thomas Francis’ doctrine of original antigenic sin (Francis, 1960). Francis found that antibodies elicited by influenza vaccines often bound strongly to influenza virus strains that an individual was exposed to in childhood, although it is not apparent if these recalled influenza virus antibody responses typically occur at the expense of producing de novo antibodies (as we reviewed here [Cobey and Hensley, 2017]) The functional consequences of eliciting low-affinity S2-reactive antibodies following SARS-CoV-2 infections are unclear. Our previous studies found no correlation between OC43-reactive antibody induction and disease outcome following SARS-CoV-2 infection (Anderson et al., 2021), but recent studies have suggested that the recall of OC43-reactive antibodies is associated with a compromised de novo SARS-CoV-2 response in individuals with fatal COVID-19 (McNaughton et al., 2022) and that recalled β-hCoV-specific immunoglobulin G (IgG) antibodies are unable to neutralize SARS-CoV-2 (Aguilar-Bretones et al., 2021). Further studies are required to determine how the induction of different types of hCoV and SARS-CoV-2 antibodies affect disease outcome following SARS-CoV-2 infections.
Limitations of the study
Our study has some limitations. We only analyzed samples from a relatively small number (n = 10) of individuals with mild illness at time points early after infection who were identified from an expansive sero-monitoring program (Gouma et al., 2021). The strength of this infection cohort is that we obtained serum samples before and after SARS-CoV-2 infection from seronegative individuals who had similar demographics to our cohort who received 2 doses of a SARS-CoV-2 mRNA vaccine. Future studies should further interrogate the specificity of antibodies elicited by more severe SARS-CoV-2 infections, since it has been widely reported that antibody levels are generally higher following severe COVID-19. Additional studies also should track original antigenic sin antibodies elicited by SARS-CoV-2 over longer periods of time. A recent study demonstrated severe SARS-CoV-2 infections result in an early recruitment of β-hCoV cross-reactive memory B cells followed by the accumulation of SARS-CoV-2 RBD-specific memory B cells that persist long term (Sokal et al., 2021). The duration and kinetics of recalled β-hCoV B cells after recovery is unclear and should be addressed in future studies. It is worth noting that the BNT162b2 vaccine employs a pre-fusion stabilized spike protein in which 2 prolines are substituted into the S2 region. It is possible that some of the differences that we measured in our study are due to differences in the stability of spike expressed by the vaccine relative to spike in SARS-CoV-2 virions. Finally, future studies should evaluate if “breakthrough” SARS-CoV-2 infections and additional doses of SARS-CoV-2 mRNA vaccines elicit antibodies that can bind to the spike of β-hCoVs.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat anti-human IgG-HRP | Jackson ImmunoResearch | 109-036-098; RRID: AB_2337596 |
| mAb CR3022 | Produced for this paper | N/A |
| mAb CR9114 | Produced for this paper | N/A |
| mAb 1E9F9 | Absolute Antibody | Ab01402–2.0 |
| Bacterial and virus strains | ||
| SARS-CoV-2 VSV pseudotypes | Generated for this paper | N/A |
| Biological samples | ||
| Acutely SARS-CoV-2 infected HCW serum samples | The University of Pennsylvania | Gouma et al. (2021) |
| SARS-CoV-2 mRNA vaccinated participants serum samples | The University of Pennsylvania | Goel et al. (2021a) |
| Chemicals, peptides, and recombinant proteins | ||
| SARS-CoV-2 spike protein | Produced for this paper | N/A |
| SARS-CoV-2 S1 subunit protein | ACROBiosystems | Cat. S1N-C52H3 |
| SARS-CoV-2 S2 subunit protein | ACROBiosystems | Cat. S2N-C52H5 |
| OC43 spike protein | Sino Biological | Cat. 40607-V08B |
| OC43 S1 subunit protein | Produced for this paper | N/A |
| OC43 S2 subunit protein | Produced for this paper | N/A |
| HKU1 spike protein | Sino Biological | Cat. 40606-V08B |
| HKU1 S1 subunit protein | Sino Biological | Cat. 40021-V08H |
| HKU1 S2 subunit protein | Sino Biological | Cat. 40021-V08B |
| SARS-CoV spike protein | Sino Biological | Cat. 40634-V08B |
| SARS-CoV S1 subunit protein | Sino Biological | Cat. 40150-V08B1 |
| SARS-CoV S2 subunit protein | Sino Biological | Cat. 40150-V08B3 |
| 229E spike protein | Sino Biological | Cat. 40605-V08B |
| IVR-190 H1N1 rHA | Produced for this paper | N/A |
| Experimental models: Cell lines | ||
| 293T | ATCC | Cat. CRL-3216, RRID:CVCL_0063 |
| 293F | Laboratory of Scott Hensley, University of Pennsylvania, PA | Thermo Fisher cat. R79007 |
| VeroE6/TMPRSS2 | Laboratory of Stefan Pohlman, German Primate Center, Leibniz Institute for Primate Research | Hoffmann et al. (2020) |
| Recombinant DNA | ||
| Plasmid: pCAGGS SARS-CoV-2 spike | Laboratory of Florian Krammer, Mt. Sinai, NY | Amanat et al. (2020) |
| Plasmid: pCG1 SARS-2 S | Laboratory of Stefan Pohlman, German Primate Center, Leibniz Institute for Primate Research | Hoffmann et al. (2020) |
| Plasmid: OC43 rS1 | Laboratory of Scott Hensley, University of Pennsylvania, PA | Anderson et al. (2021) |
| Plasmid: OC43 rS2 | Laboratory of Scott Hensley, University of Pennsylvania, PA | Anderson et al., (2021) |
| Plasmid: mAb CR3022 HC | Laboratory of Ian Wilson, Scripps Research Institute, CA | Yuan et al. (2020) |
| Plasmid: mAb CR3022 LC | Laboratory of Ian Wilson, Scripps Research Institute, CA | Yuan et al. (2020) |
| Plasmid: mAb CR9114 HC | Laboratory of Ian Wilson, Scripps Research Institute, CA | Dreyfus et al. (2012) |
| Plasmid: mAb CR9114 LC | Laboratory of Ian Wilson, Scripps Research Institute, CA | Dreyfus et al. (2012) |
| Software and algorithms | ||
| Prism8 | GraphPad Software | www.graphpad.com/scientific-software/prism/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Scott E. Hensley (hensley@pennmedicine.upenn.edu).
Materials availability
All unique reagents generated in this study will be available from the lead contact upon reasonable request.
Experimental model and subject details
Samples from human Subjects
The infection cohort described in this report consists of health care workers within the University of Pennsylvania Healthcare System who were recruited into a SARS-CoV-2 sero-monitoring study that included biweekly blood draws as previously described (Gouma et al., 2021). A nasal pharyngeal (NP) swab was collected from all healthcare workers who tested positive for SARS-CoV-2 IgG and or IgM antibodies or who were experiencing COVID-like symptoms during the study period in 2020. NP swabs were PCR tested for the presence of SARS-CoV-2 viral RNA. Seroconverted health care workers with PCR-confirmed SARS-CoV-2 infection (n = 10 adults ≥18 years old) were included in this analysis. Age and sex of participants of this study are reported in Table S1. This study was approved by the Institutional Review Board of the University of Pennsylvania under protocol 842847.
The vaccination cohort described in this report consists of participants (n = 23 adults ≥18 years old) who enrolled in a study at the University of Pennsylvania that included blood draws before and after two vaccination doses with an mRNA-based COVID-19 vaccine as previously described (Goel et al., 2021a). Whole blood was collected from participants who provided proof of vaccination with Pfizer (BNT162b2) mRNA vaccines. Samples were collected at 4 timepoints: 1–2 weeks before vaccination (baseline) (visit 1; V1), 1–2 weeks post-primary immunization (visit 2; V2), the day of or day before booster immunization (visit 3; V3), and 1–2 weeks post-booster immunization (visit 4; V4). Only participants without prior SARS-CoV-2 exposure were included in this report. Plasma and PBMCs were isolated from whole blood for downstream assays. Age and sex of participants of this study are reported in Table S1. This study was approved by the Institutional Review Board of the University of Pennsylvania under protocol 844642.
All sera and plasma samples were heat-inactivated in a 56°C water bath for 1 h prior to serological testing. All samples were collected after obtaining informed consent and studies were approved by the University of Pennsylvania Institutional Review Board.
Cell lines
293F cells were from Thermo fisher (Thermo Fisher cat. R79007). 293T cells were from ATCC (ATCC cat. CRL-3216, RRID:CVCL_0063). VeroE6/TMPRSS2 cells were a gift from Stefan Pohlman (German Primate Center, Leibniz Institute for Primate Research) as described previously (Hoffmann et al., 2020). All cell lines were cultured using manufacturer’s guidelines and used as described in method details below.
Method details
Proteins for serological studies
SARS-CoV-2 full length spike (FL-S) protein was purified by Ni-NTA resin from 293F cells transfected with a plasmid that encodes the FL-S (A gift from Florian Krammer, Icahn School of Medicine at Mt. Sinai, New York City NY) (Amanat et al., 2020). S1 and S2 subunits of the SARS-CoV-2 spike were purchased from Acro Biosystems (ACROBiosystems, Newark, DE; cat. S1N-C52H3, and S2N-C52H5, respectively) and reconstituted in 200 μL Dulbecco’s phosphate buffered saline (DPBS) to a final concentration of 500 μg/mL. OC43 FL-S was also purchased (Sino Biological, Wayne PA; cat. 40588-V08B) and reconstituted in DPBS. OC43 subunit and H1N1 rHA (IVR-190) proteins were purified in our laboratory as previously described (Anderson et al., 2021; Whittle et al., 2014). Briefly, mammalian expression plasmids encoding the S1 (amino acids 15–760) or S2 (amino acids 766–1305) domains of the OC43 spike protein were cloned with an N-terminal OC43 S signal peptide, and a C-terminus encoding a Factor Xa cleavage site, a trimerization domain from T4 fibritin (FoldOn), a site-specific biotinylation sequence (AviTag), and a hexa-histidine purification tag. A mammalian expression plasmid encoded a codon-optimized H1N1 rHA (IVR-190) followed by a FoldOn trimerization domain, an AviTag, and a hexa-histidine tag at the C-terminus in place of the transmembrane domain and cytoplasmic tail of HA. 293F cells were transfected with S1, S2, or rHA encoding plasmids and proteins were purified from cell culture supernatant 6 days later with Ni-NTA resin (Qiagen, Hilden, Germany). Proteins were concentrated and buffer exchanged into PBS with Amicon centrifugal filters (Millipore, Burlington, MA) prior to quantification on a spectrophotometer (NanoDrop, Thermo Fisher Scientific, Waltham, MA). HKU1 FL-S, S1, S2, SARS-CoV FL-S, S1, S2, and 229E FL-S were purchased (Sino Biological, Wayne PA; cat. 40606-V08B, 40021-V08H, 40021-V08B, 40634-V08B, 40150-V08B1, 40150-V08B3, and 40605-V08B, respectively).
ELISAs
Antibodies reactive to SARS-CoV-2, OC43, HKU1, SARS-CoV, and 229E antigens were quantified by enzyme-linked immunosorbent assays (ELISA) as previously described (Anderson et al., 2021; Flannery et al., 2020). Absorbed sera samples were also tested for the presence of influenza virus H1 HA antibodies. In brief, ELISA plates (Thermo Fisher Scientific, Waltham, MA: cat. 14-245-153) were coated overnight at 4°C with either 2 μg/mL SARS-CoV-2, SARS-CoV, or influenza rHA antigens, 1.5 μg/mL OC43, HKU1, or 229E antigens, or Dulbecco’s phosphate buffered saline (DPBS) to control for background antibody binding. Sera was heat-inactivated in a 56°C water bath for 1 h prior to serial dilutions starting at 1:50 in dilution buffer. ELISA plates were blocked for 1 h before 50 μL of diluted sera was added and plates were incubated for 2 h on an orbital shaker. ELISA plates were washed 3 times with 1× PBS supplemented with 2% Tween (PBS-T) before the addition of goat anti-human IgG conjugated to horseradish peroxidase secondary antibody at a 1:5000 dilution (Jackson ImmunoResearch Laboratories, West Grove, PA: cat. 109-036-098). ELISA plates were developed with TMB substrate, and the reactions were stopped after 5 min by the addition of 250 mM hydrochloric acid prior to reading on a SpectraMax 190 microplate reader (Molecular Devices, San Jose, CA). Serum antibody titers were obtained from a standard curve of either serially diluted monoclonal antibody (CR3022 for SARS-CoV-2 and SARS-CoV (Yuan et al., 2020) or CR9114 for influenza virus HA (Dreyfus et al., 2012) starting at 0.5μg/mL) or serially diluted pooled serum (for OC43, HKU1, and 229E ELISAs). Standard curves were included on every plate to control for plate-to-plate variation. Antibody titers for each sample were measured in at least two technical replicates performed on separate days.
Quantification of serum SARS-CoV-2 pseudotype neutralizing antibody titers
The ability of polyclonal sera to neutralize SARS-CoV-2 was measured using a pseudo-typed VSV neutralization assay as previously described (Anderson et al., 2021; Goel et al., 2021a). Pseudotyped vesicular stomatitis virus (VSV) virions with SARS-CoV-2 Spike from wildtype D614G SARS-CoV-2 were produced through transfection (Hoffmann et al., 2020) of 293T and harvested by centrifugation prior to being aliquoted and stored at −80°C. Heat inactivated serum samples were serially diluted 2-fold and mixed with 50–200 focus forming units/well of SARS-CoV-2 VSV pseudotype virus and 600 ng/mL of 1E9F9, a mouse anti-VSV Indiana G (Absolute Antibody, Oxford, UK: cat. Ab01402–2.0). Approximately 24 h later, cells were washed, fixed with 4% paraformaldehyde, and foci were visualized and enumerated on an S6 FluoroSpot Analyzer (CTL, Shaker Heights OH). Serum SARS-CoV-2 neutralizing antibodies were measured as the greatest serum dilution at which pseudotype virus focus count was reduced in Vero E6 cells stably expressing TMPRSS2 by at least 50% (FRNT50) relative to control cells in the absence of human serum. FRNT50 titers for each sample were measured in at least two technical replicates performed on separate days. Fold-change in neutralization titers were calculated before and after infection and vaccination with an mRNA vaccine.
Carboxyl magnetic bead absorptions
SARS-CoV-2, OC43, HKU1, and 229E FL-S antigens were coupled to carboxyl magnetic beads (RayBiotech, Peachtree Corners, GA; cat. 801-114-2) at a concentration of 35 μg antigen/100 μL magnetic beads. Mock beads were prepared by the addition of DPBS in place of antigen. Briefly, 175 μg of diluted antigen or PBS (for mock) was added to 500 μL of magnetic beads and the mixture was incubated for 2 h at 4°C with constant mixing. The unbound fractions were removed using a magnetic stand and conjugated beads were quenched by the addition of 300 μL 50 mM Tris, pH 7.4 prior to a 15-min incubation at room temperature with constant mixing. Quenching buffer was removed using a magnetic stand and the conjugated beads were washed 4 times with 300 μL wash buffer (DPBS supplemented with 0.1% BSA and 0.05% Tween-20). After the final wash, beads were resuspended in 300 μL wash buffer and were stored at 4°C prior to use in serum absorption assays.
Sera samples were absorbed with beads coupled to SARS-CoV-2, OC43, HKU1, and 229E FL-S, and mock beads. Sera samples were diluted in PBS to a final dilution of 1:25. Next, 20 μL of antigen coupled-magnetic beads or mock-treated beads were added to 100 μL of diluted sera and the mixtures were incubated for 1 h at room temperature on a plate mixer at 800rpm. Fractions containing the unabsorbed antibodies were removed using a 96-well plate magnetic stand. Unabsorbed fractions were diluted in buffer (DPBS supplemented with 1% milk and 0.1% Tween-20) prior to running in ELISA.
Quantification and statistical analysis
Statistical analyses were performed using Prism version 8 (GraphPad Software, San Diego CA). Reciprocal serum dilution antibody titers were log2 transformed for statistical analysis. ELISA antibody titers below the limit of detection (LOD) were set to a reciprocal titer equal to half the LOD. Log2 transformed antibody titers were compared with paired and unpaired t-tests, and one-way ANOVAs with Tukey’s multiple comparisons. Statistical significance was defined as a p-value <0.05.
Acknowledgments
This project has been funded in part with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contract no. 75N93021C00015. This work was supported in part by institutional funds from the University of Pennsylvania and NIH U19AI082630 (S.E.H. and E.J.W.), NIH R21AI142638 (P.B.), and NIH R01AI152236 (P.B.). E.M.A. and T.B.M. were supported by the NIH Training in Virology T32 Program (T32AI007324), and P.H. was supported by the NIH Training in Emerging Infectious Diseases T32 Program (T32AI055400). We thank J. Lurie, J. Embiid, J. Harris, and D. Blitzer for philanthropic support. The UPenn COVID Processing Unit is a unit of individuals from diverse laboratories at the University of Pennsylvania who volunteered time and effort to enable study of COVID-19 patients during the pandemic. We thank all members of the UPenn COVID- Processing Unit. We would like to thank David Anderson for assistance with the graphical abstract. The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Author contributions
Serological assays, E.M.A., S.H.L., M.A., T.E., E.G, M.J.B., T.B.M., P.H., and P.B.; data analyses, E.M.A. and S.H.L.; cohort studies and sample processing, S.G., R.R.G., M.M.P., S.A.A., D.M., D.D., D.F., A.B., J.W., J.S.M., S.D., the UPenn COVID Processing Unit, and A.R.G.; manuscript writing, E.M.A., S.H.L., and S.E.H.; supervision, I.F., D.J.R., E.J.W., P.B., and S.E.H.; funding acquisition, I.F., D.J.R, E.J.W., and S.E.H.
Declaration of interests
E.J.W. has consulting agreements with and/or is on the scientific advisory board for Merck, Elstar, Janssen, Related Sciences, Synthekine, and Surface Oncology. E.J.W. is a founder of Surface Oncology and Arsenal Biosciences. E.J.W. is an inventor on a patent (US patent number 10,370,446) submitted by Emory University that covers the use of PD-1 blockade to treat infections and cancer. S.E.H. has received consultancy fees from Sanofi Pasteur, Lumen, Novavax, and Merck for work unrelated to this report.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: September 23, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111496.
Supplemental information
Data and code availability
-
•
The published paper includes all data generated or analyzed during the study.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Aguilar-Bretones M., Westerhuis B.M., Raadsen M.P., de Bruin E., Chandler F.D., Okba N.M., Haagmans B.L., Langerak T., Endeman H., van den Akker J.P., et al. Seasonal coronavirus-specific B cells with limited SARS-CoV-2 cross-reactivity dominate the IgG response in severe COVID-19. J. Clin. Invest. 2021;131:e150613. doi: 10.1172/JCI150613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H.O., Chromikova V., McMahon M., Jiang K., Arunkumar G.A., Jurczyszak D., Polanco J., et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020;26:1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanat F., Thapa M., Lei T., Ahmed S.M.S., Adelsberg D.C., Carreño J.M., Strohmeier S., Schmitz A.J., Zafar S., Zhou J.Q., et al. SARS-CoV-2 mRNA vaccination induces functionally diverse antibodies to NTD, RBD, and S2. Cell. 2021;184:3936–3948.e10. doi: 10.1016/j.cell.2021.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson E.M., Goodwin E.C., Verma A., Arevalo C.P., Bolton M.J., Weirick M.E., Gouma S., McAllister C.M., Christensen S.R., Weaver J., et al. Seasonal human coronavirus antibodies are boosted upon SARS-CoV-2 infection but not associated with protection. Cell. 2021;184:1858–1864.e10. doi: 10.1016/j.cell.2021.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arevalo C.P., Le Sage V., Bolton M.J., Eilola T., Jones J.E., Kormuth K.A., Nturibi E., Balmaseda A., Gordon A., Lakdawala S.S., Hensley S.E. Original antigenic sin priming of influenza virus hemagglutinin stalk antibodies. Proc. Natl. Acad. Sci. USA. 2020;117:17221–17227. doi: 10.1073/pnas.1920321117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydillo T., Rombauts A., Stadlbauer D., Aslam S., Abelenda-Alonso G., Escalera A., Amanat F., Jiang K., Krammer F., Carratala J., García-Sastre A. Immunological imprinting of the antibody response in COVID-19 patients. Nat. Commun. 2021;12:3781. doi: 10.1038/s41467-021-23977-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. Overseas Ed. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangaru S., Antanasijevic A., Kose N., Sewall L.M., Jackson A.M., Suryadevara N., Zhan X., Torres J.L., Copps J., de la Peña A.T., et al. Structural mapping of antibody landscapes to human betacoronavirus spike proteins. Sci. Adv. 2022;8:eabn2911. doi: 10.1126/sciadv.abn2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes C.O., West A.P., Jr., Huey-Tubman K.E., Hoffmann M.A.G., Sharaf N.G., Hoffman P.R., Koranda N., Gristick H.B., Gaebler C., Muecksch F., et al. Structures of human antibodies bound to SARS-CoV-2 spike reveal common epitopes and recurrent features of antibodies. Cell. 2020;182:828–842.e16. doi: 10.1016/j.cell.2020.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boni M.F., Lemey P., Jiang X., Lam T.T.Y., Perry B.W., Castoe T.A., Rambaut A., Robertson D.L. Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nat. Microbiol. 2020;5:1408–1417. doi: 10.1038/s41564-020-0771-4. [DOI] [PubMed] [Google Scholar]
- Brewer R.C., Ramadoss N.S., Lahey L.J., Jahanbani S., Robinson W.H., Lanz T.V. BNT162b2 vaccine induces divergent B cell responses to SARS-CoV-2 S1 and S2. Nat. Immunol. 2022;23:33–39. doi: 10.1038/s41590-021-01088-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho T., Krammer F., Iwasaki A. The first 12 months of COVID-19: a timeline of immunological insights. Nat. Rev. Immunol. 2021;21:245–256. doi: 10.1038/s41577-021-00522-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Chen Z., Azman A.S., Sun R., Lu W., Zheng N., Zhou J., Wu Q., Deng X., Zhao Z., et al. Neutralizing antibodies against SARS-CoV-2 variants induced by natural infection or vaccination: a systematic review and pooled meta-analysis. Clin. Infect. Dis. 2021;74:734–742. doi: 10.1093/cid/ciab646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho A., Muecksch F., Schaefer-Babajew D., Wang Z., Finkin S., Gaebler C., Ramos V., Cipolla M., Mendoza P., Agudelo M., et al. Anti-SARS-CoV-2 receptor-binding domain antibody evolution after mRNA vaccination. Nature. 2021;600:517–522. doi: 10.1038/s41586-021-04060-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobey S., Hensley S.E. Immune history and influenza virus susceptibility. Curr. Opin. Virol. 2017;22:105–111. doi: 10.1016/j.coviro.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier D.A., De Marco A., Ferreira I.A.T.M., Meng B., Datir R.P., Walls A.C., Kemp S.A., Bassi J., Pinto D., Silacci-Fregni C., et al. Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nature. 2021;593:136–141. doi: 10.1038/s41586-021-03412-7. [DOI] [PubMed] [Google Scholar]
- Dijkman R., Jebbink M.F., Gaunt E., Rossen J.W.A., Templeton K.E., Kuijpers T.W., van der Hoek L. The dominance of human coronavirus OC43 and NL63 infections in infants. J. Clin. Virol. 2012;53:135–139. doi: 10.1016/j.jcv.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria-Rose N., Suthar M.S., Makowski M., O'Connell S., McDermott A.B., Flach B., Ledgerwood J.E., Mascola J.R., Graham B.S., Lin B.C., et al. Antibody persistence through 6 Months after the second dose of mRNA-1273 vaccine for covid-19. N. Engl. J. Med. 2021;384:2259–2261. doi: 10.1056/NEJMc2103916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfus C., Laursen N.S., Kwaks T., Zuijdgeest D., Khayat R., Ekiert D.C., Lee J.H., Metlagel Z., Bujny M.V., Jongeneelen M., et al. Highly conserved protective epitopes on influenza B viruses. Science. 2012;337:1343–1348. doi: 10.1126/science.1222908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyrdak R., Hodcroft E.B., Wahlund M., Neher R.A., Albert J. Interactions between seasonal human coronaviruses and implications for the SARS-CoV-2 pandemic: a retrospective study in Stockholm, Sweden, 2009-2020. J. Clin. Virol. 2021;136:104754. doi: 10.1016/j.jcv.2021.104754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edridge A.W.D., Kaczorowska J., Hoste A.C.R., Bakker M., Klein M., Loens K., Jebbink M.F., Matser A., Kinsella C.M., Rueda P., et al. Seasonal coronavirus protective immunity is short-lasting. Nat. Med. 2020;26:1691–1693. doi: 10.1038/s41591-020-1083-1. [DOI] [PubMed] [Google Scholar]
- Eguia R.T., Crawford K.H.D., Stevens-Ayers T., Kelnhofer-Millevolte L., Greninger A.L., Englund J.A., Boeckh M.J., Bloom J.D. A human coronavirus evolves antigenically to escape antibody immunity. PLoS Pathog. 2021;17:e1009453. doi: 10.1371/journal.ppat.1009453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excler J.L., Saville M., Berkley S., Kim J.H. Vaccine development for emerging infectious diseases. Nat. Med. 2021;27:591–600. doi: 10.1038/s41591-021-01301-0. [DOI] [PubMed] [Google Scholar]
- Flannery D.D., Gouma S., Dhudasia M.B., Mukhopadhyay S., Pfeifer M.R., Woodford E.C., Gerber J.S., Arevalo C.P., Bolton M.J., Weirick M.E., et al. SARS-CoV-2 seroprevalence among parturient women in Philadelphia. Sci. Immunol. 2020;5:eabd5709. doi: 10.1126/sciimmunol.abd5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis T. On the doctrine of original antigenic sin. Proc. Am. Phil. Soc. 1960;104:572–578. [Google Scholar]
- Gaunt E.R., Hardie A., Claas E.C.J., Simmonds P., Templeton K.E. Epidemiology and clinical presentations of the four human coronaviruses 229E, HKU1, NL63, and OC43 detected over 3 years using a novel multiplex real-time PCR method. J. Clin. Microbiol. 2010;48:2940–2947. doi: 10.1128/JCM.00636-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebre M.S., Brito L.A., Tostanoski L.H., Edwards D.K., Carfi A., Barouch D.H. Novel approaches for vaccine development. Cell. 2021;184:1589–1603. doi: 10.1016/j.cell.2021.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel R.R., Apostolidis S.A., Painter M.M., Mathew D., Pattekar A., Kuthuru O., Gouma S., Hicks P., Meng W., Rosenfeld A.M., et al. Distinct antibody and memory B cell responses in SARS-CoV-2 naive and recovered individuals following mRNA vaccination. Sci. Immunol. 2021;6:eabi6950. doi: 10.1126/sciimmunol.abi6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel R.R., Painter M.M., Apostolidis S.A., Mathew D., Meng W., Rosenfeld A.M., Lundgreen K.A., Reynaldi A., Khoury D.S., Pattekar A., et al. mRNA vaccines induce durable immune memory to SARS-CoV-2 and variants of concern. Science. 2021;374:abm0829. doi: 10.1126/science.abm0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golob J.L., Lugogo N., Lauring A.S., Lok A.S. SARS-CoV-2 vaccines: a triumph of science and collaboration. JCI Insight. 2021;6:149187. doi: 10.1172/jci.insight. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouma S., Weirick M.E., Bolton M.J., Arevalo C.P., Goodwin E.C., Anderson E.M., McAllister C.M., Christensen S.R., Dunbar D., Fiore D., et al. Health care worker sero-monitoring reveals complex relationships between common coronavirus antibodies and COVID-19 symptom duration. JCI Insight. 2021;6:150449. doi: 10.1172/jci.insight.150449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney A.J., Loes A.N., Gentles L.E., Crawford K.H.D., Starr T.N., Malone K.D., Chu H.Y., Bloom J.D. Antibodies elicited by mRNA-1273 vaccination bind more broadly to the receptor binding domain than do those from SARS-CoV-2 infection. Sci. Transl. Med. 2021;13:eabi9915. doi: 10.1126/scitranslmed.abi9915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grobben M., van der Straten K., Brouwer P.J., Brinkkemper M., Maisonnasse P., Dereuddre-Bosquet N., Appelman B., Lavell A.A., van Vught L.A., Burger J.A., et al. Cross-reactive antibodies after SARS-CoV-2 infection and vaccination. Elife. 2021;10:e70330. doi: 10.7554/eLife.70330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthmiller J.J., Stovicek O., Wang J., Changrob S., Li L., Halfmann P., Zheng N.Y., Utset H., Stamper C.T., Dugan H.L., et al. SARS-CoV-2 infection severity is linked to superior humoral immunity against the spike. mBio. 2021;12:e02940-20. doi: 10.1128/mBio.02940-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang A.T., Garcia-Carreras B., Hitchings M.D.T., Yang B., Katzelnick L.C., Rattigan S.M., Borgert B.A., Moreno C.A., Solomon B.D., Trimmer-Smith L., et al. A systematic review of antibody mediated immunity to coronaviruses: kinetics, correlates of protection, and association with severity. Nat. Commun. 2020;11:4704. doi: 10.1038/s41467-020-18450-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J., et al. An mRNA vaccine against SARS-CoV-2 - preliminary report. N. Engl. J. Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaimes J.A., André N.M., Chappie J.S., Millet J.K., Whittaker G.R. Phylogenetic analysis and structural modeling of SARS-CoV-2 spike protein reveals an evolutionary distinct and proteolytically sensitive activation loop. J. Mol. Biol. 2020;432:3309–3325. doi: 10.1016/j.jmb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalkanen P., Kolehmainen P., Häkkinen H.K., Huttunen M., Tähtinen P.A., Lundberg R., Maljanen S., Reinholm A., Tauriainen S., Pakkanen S.H., et al. COVID-19 mRNA vaccine induced antibody responses against three SARS-CoV-2 variants. Nat. Commun. 2021;12:3991. doi: 10.1038/s41467-021-24285-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killerby M.E., Biggs H.M., Haynes A., Dahl R.M., Mustaquim D., Gerber S.I., Watson J.T. Human coronavirus circulation in the United States 2014-2017. J. Clin. Virol. 2018;101:52–56. doi: 10.1016/j.jcv.2018.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistler K.E., Bedford T. Evidence for adaptive evolution in the receptor-binding domain of seasonal coronaviruses OC43 and 229e. Elife. 2021;10:e64509. doi: 10.7554/eLife.64509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuri-Cervantes L., Pampena M.B., Meng W., Rosenfeld A.M., Ittner C.A.G., Weisman A.R., Agyekum R.S., Mathew D., Baxter A.E., Vella L.A., et al. Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci. Immunol. 2020;5:eabd7114. doi: 10.1126/sciimmunol.abd7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederer K., Bettini E., Parvathaneni K., Painter M.M., Agarwal D., Lundgreen K.A., Weirick M., Muralidharan K., Castaño D., Goel R.R., et al. Germinal center responses to SARS-CoV-2 mRNA vaccines in healthy and immunocompromised individuals. Cell. 2022;185:1008–1024.e15. doi: 10.1016/j.cell.2022.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legros V., Denolly S., Vogrig M., Boson B., Siret E., Rigaill J., Pillet S., Grattard F., Gonzalo S., Verhoeven P., et al. A longitudinal study of SARS-CoV-2-infected patients reveals a high correlation between neutralizing antibodies and COVID-19 severity. Cell. Mol. Immunol. 2021;18:318–327. doi: 10.1038/s41423-020-00588-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Liu J., Xia H., Zhang X., Fontes-Garfias C.R., Swanson K.A., Cai H., Sarkar R., Chen W., Cutler M., et al. Neutralizing activity of BNT162b2-elicited serum. N. Engl. J. Med. 2021;384:1466–1468. doi: 10.1056/NEJMc2102017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi A., Bozzi G., Ungaro R., Villa S., Castelli V., Mangioni D., Muscatello A., Gori A., Bandera A. Mini review immunological consequences of immunization with COVID-19 mRNA vaccines: preliminary results. Front. Immunol. 2021;12:657711. doi: 10.3389/fimmu.2021.657711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew D., Giles J.R., Baxter A.E., Oldridge D.A., Greenplate A.R., Wu J.E., Alanio C., Kuri-Cervantes L., Pampena M.B., D'Andrea K., et al. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020;369:eabc8511. doi: 10.1126/science.abc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNaughton A.L., Paton R.S., Edmans M., Youngs J., Wellens J., Phalora P., Fyfe A., Belij-Rammerstorfer S., Bolton J.S., Ball J., et al. Fatal COVID-19 outcomes are associated with an antibody response targeting epitopes shared with endemic coronaviruses. JCI Insight. 2022;7:e156372. doi: 10.1172/jci.insight.156372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng K.W., Faulkner N., Cornish G.H., Rosa A., Harvey R., Hussain S., Ulferts R., Earl C., Wrobel A.G., Benton D.J., et al. Preexisting and de novo humoral immunity to SARS-CoV-2 in humans. Science. 2020;370:1339–1343. doi: 10.1126/science.abe1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Contant P., Embong A.K., Kanagaiah P., Chaves F.A., Yang H., Branche A.R., Topham D.J., Sangster M.Y. S protein-reactive IgG and memory B cell production after human SARS-CoV-2 infection includes broad reactivity to the S2 subunit. mBio. 2020;11:e01991-20. doi: 10.1128/mBio.01991-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okba N.M.A., Müller M.A., Li W., Wang C., GeurtsvanKessel C.H., Corman V.M., Lamers M.M., Sikkema R.S., de Bruin E., Chandler F.D., et al. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease patients. Emerg. Infect. Dis. 2020;26:1478–1488. doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccoli L., Park Y.J., Tortorici M.A., Czudnochowski N., Walls A.C., Beltramello M., Silacci-Fregni C., Pinto D., Rosen L.E., Bowen J.E., et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183:1024–1042.e21. doi: 10.1016/j.cell.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., Planchais C., Porrot F., Robillard N., Puech J., et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596:276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., et al. Safety and efficacy of the BNT162b2 mRNA covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röltgen K., Boyd S.D. Antibody and B cell responses to SARS-CoV-2 infection and vaccination. Cell Host Microbe. 2021;29:1063–1075. doi: 10.1016/j.chom.2021.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röltgen K., Nielsen S.C.A., Silva O., Younes S.F., Zaslavsky M., Costales C., Yang F., Wirz O.F., Solis D., Hoh R.A., et al. Immune imprinting, breadth of variant recognition, and germinal center response in human SARS-CoV-2 infection and vaccination. Cell. 2022;185:1025–1040.e14. doi: 10.1016/j.cell.2022.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röltgen K., Powell A.E., Wirz O.F., Stevens B.A., Hogan C.A., Najeeb J., Hunter M., Wang H., Sahoo M.K., Huang C., et al. Defining the features and duration of antibody responses to SARS-CoV-2 infection associated with disease severity and outcome. Sci. Immunol. 2020;5:eabe0240. doi: 10.1126/sciimmunol.abe0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydyznski Moderbacher C., Ramirez S.I., Dan J.M., Grifoni A., Hastie K.M., Weiskopf D., Belanger S., Abbott R.K., Kim C., Choi J., et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell. 2020;183:996–1012.e19. doi: 10.1016/j.cell.2020.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagar M., Reifler K., Rossi M., Miller N.S., Sinha P., White L.F., Mizgerd J.P. Recent endemic coronavirus infection is associated with less-severe COVID-19. J. Clin. Invest. 2021;131:143380. doi: 10.1172/JCI143380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasson J.M., Campo J.J., Carpenter R.M., Young M.K., Randall A.Z., Trappl-Kimmons K., Oberai A., Hung C., Edgar J., Teng A.A., et al. Diverse humoral immune responses in younger and older adult COVID-19 patients. mBio. 2021;12:e0122921. doi: 10.1128/mBio.01229-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette A., Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021;184:861–880. doi: 10.1016/j.cell.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrock E., Fujimura E., Kula T., Timms R.T., Lee I.H., Leng Y., Robinson M.L., Sie B.M., Li M.Z., Chen Y., et al. Viral epitope profiling of COVID-19 patients reveals cross-reactivity and correlates of severity. Science. 2020;370:eabd4250. doi: 10.1126/science.abd4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokal A., Chappert P., Barba-Spaeth G., Roeser A., Fourati S., Azzaoui I., Vandenberghe A., Fernandez I., Meola A., Bouvier-Alias M., et al. Maturation and persistence of the anti-SARS-CoV-2 memory B cell response. Cell. 2021;184:1201–1213.e14. doi: 10.1016/j.cell.2021.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song G., He W.T., Callaghan S., Anzanello F., Huang D., Ricketts J., Torres J.L., Beutler N., Peng L., Vargas S., et al. Cross-reactive serum and memory B-cell responses to spike protein in SARS-CoV-2 and endemic coronavirus infection. Nat. Commun. 2021;12:2938. doi: 10.1038/s41467-021-23074-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatatos L., Czartoski J., Wan Y.H., Homad L.J., Rubin V., Glantz H., Neradilek M., Seydoux E., Jennewein M.F., MacCamy A.J., et al. mRNA vaccination boosts cross-variant neutralizing antibodies elicited by SARS-CoV-2 infection. Science. 2021;372:1413–1418. doi: 10.1126/science.abg9175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.N., Czudnochowski N., Liu Z., Zatta F., Park Y.J., Addetia A., Pinto D., Beltramello M., Hernandez P., Greaney A.J., et al. SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape. Nature. 2021;597:97–102. doi: 10.1038/s41586-021-03807-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T., Ellingson M.K., Wong P., Israelow B., Lucas C., Klein J., Silva J., Mao T., Oh J.E., Tokuyama M., et al. Sex differences in immune responses that underlie COVID-19 disease outcomes. Nature. 2020;588:315–320. doi: 10.1038/s41586-020-2700-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topol E.J. Messenger RNA vaccines against SARS-CoV-2. Cell. 2021;184:1401. doi: 10.1016/j.cell.2020.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner J.S., O'Halloran J.A., Kalaidina E., Kim W., Schmitz A.J., Zhou J.Q., Lei T., Thapa M., Chen R.E., Case J.B., et al. SARS-CoV-2 mRNA vaccines induce persistent human germinal centre responses. Nature. 2021;596:109–113. doi: 10.1038/s41586-021-03738-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Muecksch F., Schaefer-Babajew D., Finkin S., Viant C., Gaebler C., Hoffmann H.H., Barnes C.O., Cipolla M., Ramos V., et al. Naturally enhanced neutralizing breadth against SARS-CoV-2 one year after infection. Nature. 2021;595:426–431. doi: 10.1038/s41586-021-03696-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C.O., Finkin S., Schaefer-Babajew D., Cipolla M., Gaebler C., Lieberman J.A., et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592:616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle J.R.R., Wheatley A.K., Wu L., Lingwood D., Kanekiyo M., Ma S.S., Narpala S.R., Yassine H.M., Frank G.M., Yewdell J.W., et al. Flow cytometry reveals that H5N1 vaccination elicits cross-reactive stem-directed antibodies from multiple Ig heavy-chain lineages. J. Virol. 2014;88:4047–4057. doi: 10.1128/JVI.03422-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widge A.T., Rouphael N.G., Jackson L.A., Anderson E.J., Roberts P.C., Makhene M., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., et al. Durability of responses after SARS-CoV-2 mRNA-1273 vaccination. N. Engl. J. Med. 2021;384:80–82. doi: 10.1056/NEJMc2032195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Werner A.P., Koch M., Choi A., Narayanan E., Stewart-Jones G.B.E., Colpitts T., Bennett H., Boyoglu-Barnum S., Shi W., et al. Serum neutralizing activity elicited by mRNA-1273 vaccine. N. Engl. J. Med. 2021;384:1468–1470. doi: 10.1056/NEJMc2102179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Wu N.C., Zhu X., Lee C.C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The published paper includes all data generated or analyzed during the study.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.