Abstract
The term “SOS response” was first coined by Radman in 1974, in an intellectual effort to put together the data suggestive of a concerted gene expression program in cells undergoing DNA damage. A large amount of information about this cellular response has been collected over the following decades. In this review, we will focus on a few of the relevant aspects about the SOS response: its mechanism of control and the stressors which activate it, the diversity of regulated genes in different species, its role in mutagenesis and evolution including the development of antimicrobial resistance, and its relationship with mobile genetic elements.
Keywords: DNA damage, SOS response, mutagenesis, TLS polymerases, mobile genetic elements
The SOS response is a cellular mechanism induced by agents that threaten DNA integrity in prokaryotes that aids cell survival under stressful situations, since an unrepaired DNA damage may lead to deleterious mutations or even cell death. Cells are constantly exposed to environments that may contain DNA-damaging agents. These agents can be either a physical agent such as UV light and ionizing radiation, or a chemical compound such as alkylating and crosslinking agents. However, the threats that a cell has to face are not only external but also internal, such as the reactive oxygen species (ROS), metabolic byproducts that cause DNA damage.
Miroslav Radman used the distress signal “SOS” to define how bacterial cells sense genome instability, while studying DNA damage and replication blockages in Escherichia coli (Radman, 1974). This phenomenon triggers a pathway of physiological responses to deal with these adverse conditions, mainly DNA damage repair and/or tolerance and mutagenesis. Pathways induced by SOS include damage repair and tolerance mechanisms such as nucleotide excision repair (NER), photoreactivation, homologous recombination (HR) and translesion synthesis (TLS) (Erill et al., 2006). Despite the induction of pathways that promote DNA integrity in an error-free manner, there is also the involvement of error-prone elements in this response, responsible for improving cell survival under severe DNA damage, however exhibiting elevated mutagenesis as a consequence (Henrikus et al., 2018). SOS is subject to complex regulation controlled by the lexA and recA gene products, due to its mutagenic potential.
Fundamentals of SOS response regulation
Induction of the SOS regulon is triggered by single-stranded DNA (ssDNA) present in the cell as a consequence of replication and repair of damaged DNA (Sassanfar and Roberts, 1990). Briefly, this response is regulated by the LexA and RecA proteins in which the former plays a role as a transcriptional repressor by binding to the promoter region of genes controlled by this regulon, and the latter functions as a positive regulator of the system (Little et al., 1980; Little, 1983, 1991; Aksenov, 1999).
LexA - a self-cleaving repressor
Regulation of SOS response genes depends on transcriptional repression by the LexA protein, which binds to an operator sequence, within the promoter, known as the SOS box (Walker, 1984) and prevents RNA polymerase binding and transcription (Berg, 1988). LexA functions as a repressor in the form of a dimer consisting of two domains joined by a peptide linker: a DNA-binding domain located in the amino-terminal (NTD) and serine protease domain located in the carboxi-terminal (CTD). The CTD domain plays a role in the homodimerization of LexA (Zhang et al., 2010).
LexA repressor undergoes self-cleavage under SOS-inducing conditions (Slilaty et al., 1986). In E. coli, the enzyme has a conserved serine-lysine catalytic domain that self-cleaves its peptide bond between Ala84-Gly85 near the middle of the protein, thus losing its repressor function (Little, 1991). Structural studies in E. coli have shown that the CTD domain can be found in two different conformations: a basal cleavage-incompetent conformation and a cleavage-proficient conformation (Luo et al., 2001). In vivo, LexA self-cleavage occurs when it interacts with activated RecA protein (RecA*) (Little et al., 1980).
RecA - a DNA damage sensor
RecA protein is a key player in DNA repair, being required not only for SOS induction, but for homologous recombination and translesion synthesis as well. In the absence of ATP, RecA is found as monomers that are capable of associating with ssDNA, being able to protect DNA strand from degradation but staying in a functionally-inactive conformation (Yu and Egelman, 1992). When ATP molecules are available, the RecA-ssDNA complex is converted to the functionally-active conformation: RecA* nucleoprotein filament, a structure functioning as a co-protease responsible for inducing self-cleavage of LexA (Cox, 2007). This structure has many other functions, such as searching for homologous dsDNA to promote homologous recombination (Tsang et al., 1985). The RecA*-stimulated auto-cleavage of LexA expose previously inaccessible residues, facilitating proteolytic degradation of both fragments (Neher et al., 2003). Once LexA protein levels start to decrease, expression of SOS genes is triggered (Little and Mount, 1982).
SOS response in a nutshell
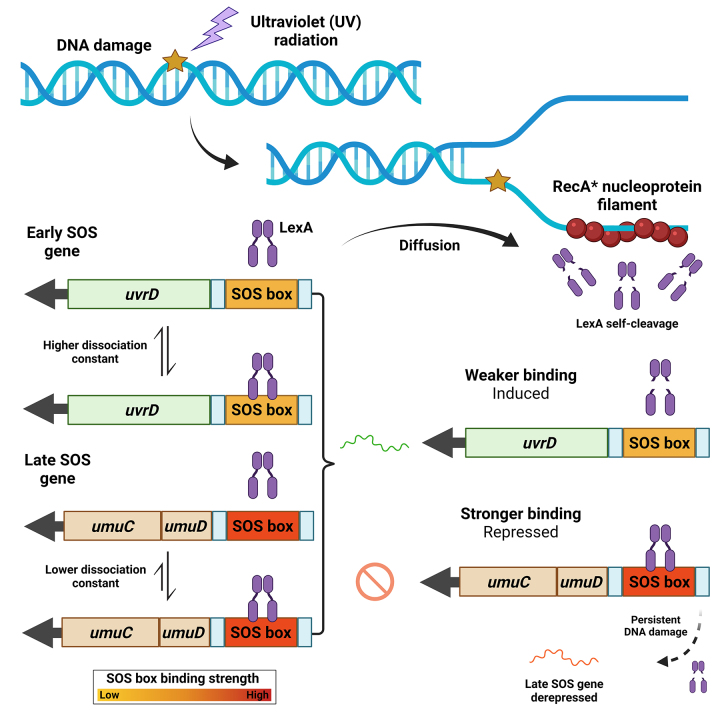
Most of the findings regarding SOS response regulation and dynamics were made using the model bacterium E. coli. Once DNA breaks or other types of damage emerge within the cell, RecA monomers readily associate with ssDNA assuming its active form (RecA*), inducing LexA self-cleavage which causes it to dissociate from SOS-regulated promoters, thereby relieving repression of the SOS regulon (Figure 1). One important aspect of SOS dynamics is that lexA itself is an SOS gene, thus generating a negative-feedback loop to re-establish repression after the induction signal is ceased. Besides that, LexA is constantly expressed during late SOS to ensure that SOS induction is interrupted once stress signal decreases and LexA degradation is not favored anymore (Walker, 1984).
Figure 1 - . Model of SOS response activation. The presence of DNA damage may block DNA replication and expose ssDNA within the cell. RecA protein associates with ssDNA, assuming its functionally-active conformation: RecA* nucleoprotein filament. This protein complex is responsible for inducing LexA self-cleavage, thus enabling transcription of SOS regulated genes. LexA repressor displays a dynamic of binding/dissociating with its target sequence and can only be cleaved once it is dissociated from DNA. Note that a stronger SOS box implies a lower dissociation constant, meaning that in this scenario, LexA is more likely to be associated with DNA and thus repressing its target. Therefore, gene expression can be modulated by SOS box strength: the weaker the operator strength, the sooner a gene will be expressed.
The dynamics of the SOS response can be manipulated by proteins that interact with RecA filament and modulate the time of induction and recovery rate of the response (Lusetti et al., 2004). The inhibitor of RecA is the RecX protein, which at low concentrations can suppress many RecA functions (Stohl et al., 2003) and blocks RecA filament polymerization (Drees et al., 2004), leading to filament dismantling (Ragone et al., 2008).
The response is orchestrated according to several variables, including the extent in which DNA was damaged and the time passed since such damage was identified (Courcelle et al., 2001; Quillardet et al., 2003), in such a way that SOS-regulated genes have different timing and levels of induction. Housekeeping and error-free repair processes comprise the initial phase of SOS, such as NER and homologous recombination. SulA protein, a division inhibitor, allows the bacterium to complete DNA repair before finalizing its cell division. Lastly, if the damage was severe and remains unrepaired, TLS-polymerases are induced leading to elevated mutagenesis but allowing replication to resolve, thus improving cell survival (Henrikus et al., 2018). It is also important to point out that RecA-mediated cleavage of LexA occurs when LexA is DNA-free but not when bound to its target DNA (Butala et al., 2011; Kovačič et al., 2013), adding more complexity to the timing of expression of SOS-regulated genes.
The dynamics of SOS genes in E. coli is also influenced by the strength of different SOS boxes (Figure 1). Usually, the consensus SOS box sequence displays a higher affinity for LexA binding since its sequence is a palindrome and optimal for LexA association: TACTG(TA)5CAGTA. Any modification within this sequence may interfere with LexA affinity for a given operator. The heterology index (HI) measures how much an SOS box differs from the consensus: the higher the HI value, the lower LexA affinity for the operator, as shown in E. coli (Lewis et al., 1994; Fernández de Henestrosa et al., 2000) and in Salmonella enterica (Mérida-Floriano et al., 2021). This process corroborates the idea that affinity of LexA is also an important factor, since lower affinity implies an earlier transcriptional derepression, consequently regulating genes that should be expressed early or late in the SOS response.
DNA damage response heterogeneity
To this date, the vast majority of studies measuring SOS induction have been using the “uniform expression model” (McCool et al., 2004). In this model, it is not clear whether the activity of a particular promoter is equally distributed across cells in a population or has a different expression for a subpopulation of cells (Kenyon and Walker, 1980; Salles and Defais, 1984). Measuring the activity of SOS regulated promoters in transcriptional fusions to reporter genes is an example that represents a population average and relies on the uniform expression model.
However, fluorescence microscopy studies to assess SOS induction at the single cell level pointed to limitations of measuring SOS induction at the population level (McCool et al., 2004; Britton et al., 2007; Jones and Uphoff, 2021). In this methodology, fluorescent proteins are fused to an SOS regulated gene and through microscopy analysis, rather than a population measurement, it is possible to determine subpopulations of cells displaying a variety of SOS induction patterns. This model is called the “two population model” and has shown how heterogeneous the induction of the SOS response within a cell population may be, allowing a much more accurate and comprehensive analysis for quantifying the SOS response.
DNA damages and other stressors leading to SOS induction
An SOS response is triggered when single-stranded DNA (ssDNA) is present in the cell, which is one of the consequences of DNA damage. Repair of double-stranded DNA (dsDNA) breaks is a fundamental aspect of genome conservation. These potentially lethal lesions frequently occur during DNA replication (Pennington and Rosenberg, 2007). The enzymes RecA and RecBCD are the initiators required for double-strand breaks (DSBs) repair and homologous recombination. The type of DNA damage determines in which state the SOS response is triggered. These two enzymes at the same time degrade and unwind DNA from DSB in vitro (Chaudhury and Smith, 1985; Anderson and Kowalczykowski, 1997).
Several stresses, including quinolone treatment, high pressure and radiation lead to SOS induction as a result of DSB (Anderson and Kowalczykowski, 1997; Anderson and Kowalczykowski, 1998). RecBCD, responsible for processing DSBs, is a molecular machinery that binds to the damaged site and initiates the unwinding of the double-helix (Singleton et al., 2004). RecB is a helicase coupled to an endonuclease domain that initially degrades the 3’-tail more efficiently than the 5’-tail. RecC splits the DNA strands to each helicase (RecB and RecD) and scans for a recombinational hotspot, known as Chi (ꭓ) site (5’ GCTGGTGG 3’), where it can bind to and prevent further degradation from the 3’ end strand (Anderson and Kowalczykowski, 1997,1998).
This event results in an ssDNA loop in the 3’ end strand to which RecA can be loaded. At the same time, RecD helicase is able to access the nuclease site more frequently, leading to a higher degradation rate of the 5’ end strand (Singleton et al., 2004). The classical DSB repair mechanism in E. coli occurs through homologous recombination (HR), which is dependent on homologous fragments from either exogenous DNA or a recently duplicated sequence after DNA replication. The nucleoprotein filament (ssDNA-RecA) from the 3’ end strand interacts with homology sequences, forming a heteroduplex structure with the intact dsDNA. Finally, DNA polymerase uses the complementary strand as a template to reconstitute double-stranded DNA and repair the DSB damage (Danilowicz et al., 2021).
DNA lesions can produce chemical alteration of the base structure, modifying the coding sequence of the molecule. The best example of base damaging agent is ultraviolet radiation, which leads to photochemical reactions between neighboring bases (Sassanfar and Roberts, 1990). It was found that in E. coli the recF pathway proteins, such as RecF, RecO, and RecR, are necessary to restore replication after UV radiation-induced damage. recF, recO, and recR mutants have enhanced sensitivity to DNA damage and show delayed SOS induction. RecFOR complex proteins stabilize and strengthen the binding of RecA (Bork et al., 2001; Rangarajan et al., 2002).
Replication forks frequently stall due to physical blockages. RecA* activation following replication blockage requires RecFOR complex for processing. There are several pathways supported by genetic evidence for homologous recombination and post replication repair in E. coli and the fact that the recA gene is required in all of these pathways suggests that other genes involved in the process of repair and recombination provide activities that help RecA. It is essential for genomic integrity that accurate replication recovery occurs after DNA damage and repair (Tseng Y-C et al., 1994).
Antibiotics that do not interfere directly with DNA replication may also induce the SOS response. Penicillin and related beta-lactams interfere with peptidoglycan metabolism by disturbing the activity of penicillin-binding proteins (PBPs). Impairment of PBPs activity by beta-lactams causes the induction of the two-component signal transduction system DpiBA in E. coli. DpiA has affinity to AT rich sequences and interferes with DnaA and DnaB binding at the replication origin, leading to SOS activation (Miller et al., 2003, 2004; Cho et al., 2014).
In pathogens such as Staphylococcus aureus and Pseudomonas aeruginosa, the SOS response is involved in the mutagenesis leading to antibiotic resistance (Blázquez et al., 2006; Cirz et al., 2006, 2007; Maiques et al., 2006). SOS induction by antibiotics has many important implications, since it can increase error-prone polymerases that mediate mutagenesis and help in the spread of mobile genetic elements and pathogenicity islands (Úbeda et al., 2005), as discussed in subsequent sections.
Intracellular pH is regulated in E. coli cells by redox and proton pumps. However, a disturbance in pH regulation can lead to SOS induction (Padan et al., 1976; Padan and Schuldiner, 1986; Simmons et al., 2008). A mechanism for pH-induced expression of the SOS response is related to pH altering the structure and function of LexA (Dri and Moreau, 1994; van der Veen et al., 2010). According to Sousa et al. (2006), in the low pH of 4.0, LexA has the tendency to self-aggregate, preventing its binding to the SOS box. Further in this condition, LexA has increased affinity for non-specific DNA, meaning that SOS box is also derepressed by titrating LexA to other DNA sequences in the genome. However, operons regulated by LexA are not transcriptionally active until mild condition (pH 5.0 - 6.0) is achieved, where cell metabolism is restored and LexA operators are still predominantly free of repression. The hypothesized mechanism would explain how the SOS response can be activated in a RecA independent manner to increase bacterial survival rate after an episode of stressful low pH condition.
High pressure also leads to DNA breaks and SOS response induction. In E. coli a mechanism for high-pressure-mediated DNA break has been linked to the expression of endogenous endonucleases that promote DSB after a high-pressure stress, which consequently triggers the SOS response (Aertsen et al., 2004). During food preservation processes bacterial pathogens are often exposed to high pressure to inactivate them, and SOS induction may contribute to their survivability (Alpas et al., 2000; Aertsen and Michiels, 2005; Simmons et al., 2008).
Diversity of the SOS response among bacterial species
Escherichia coli has served as the premier model from which almost all the fundamental aspects of SOS regulation and physiology have been derived. Nevertheless, it is now clear that the SOS response displays considerable variability among phylogenetically different bacteria. This variability is observed in two key aspects: the SOS box sequence (Table 1) and the set of genes repressed by LexA.
Table 1 - . Sequence of the SOS operator (SOS box) in different bacterial species.
| Bacteria species | SOS box | Reference |
|---|---|---|
| Bacillus subtilis | CGAACN4GTTCG | Au et al., 2005 |
| Caulobacter crescentus | GTTCN7GTTC | da Rocha et al., 2008 |
| Escherichia coli | TACTG(TA)5CAGTA | Lewis et al., 1994 |
| Pseudomonas aeruginosa | CTGN2TN7CAG | Cirz et al., 2006 |
| Staphylococcus aureus | CGAACN4GTTCG | Cirz et al., 2007 |
Genes under LexA repression show variation between different bacterial species, however many cellular functions are commonly upregulated, for example genes encoding polymerases responsible for carrying out translesion synthesis, DNA repair proteins, cell division inhibitors, among others (Courcelle et al., 2001; da Rocha et al., 2008; Cirz et al., 2006, 2007).
The difference between the sequence recognized by LexA and the set of genes under its control seems to have an important role among species and how they respond to DNA damage (Erill et al., 2007). It can even be noted that it has already been described in some species, such as Pseudomonas putida and Xanthomonas axonopodis for example, the existence of two lexA regulons with independent LexA proteins and binding sequences (Yang et al., 2002; Abella et al., 2007). Furthermore, some bacteria also show SOS-independent DNA damage responses (e. g. Modell et al., 2014; Müller et al., 2018; Blanchard and Groot, 2021).
The differences in the SOS boxes makes the regulator of one species unable to exert its function in other species (Lovett Jr et al., 1994), demonstrating their evolutionary importance, and being a possible factor that led to the formation of branches in the bacterial evolutionary tree (Mazón et al., 2004), since the LexA-binding sequence is monophyletic for phyla and classes (Erill et al., 2003). P. aeruginosa have consensus SOS box almost identical to the E. coli one and both are notably unrelated to the ones present in Staphylococcus aureus and Bacillus subtillis. These latter two species share similar SOS box consensus, with the LexA homolog being called DinR the regulator in B. subtillis (Table 1) (Winterling et al., 1997). On the other hand, the model organism Caulobacter crescentus has an SOS box composed of a direct repeat, which is found in other phylogenetically related bacteria (da Rocha et al., 2008).
In B. subtillis subtilis only seven genes that are among the 33 genes under the control of LexA can be found in E. coli regulon composition. Under DNA damage, P. aeruginosa seems to upregulate the recX and recN whose gene products are recombination repair proteins, while B. subtillis upregulates uvrBA and ruvAB operons and E. coli upregulates all the genes cited above (Courcelle et al., 2003; Au et al., 2005). C. crescentus also shows upregulation in the expression of recN, uvrA and ruvCAB operon (da Rocha et al., 2008). On the other hand, S. aureus seems to downregulate the recN and ruvBA repair systems and upregulate uvrBA operon under damage induced by ciprofloxacin (Cirz et al., 2007).
The variation in the DNA damage response is illustrated by comparing the well-studied organisms E. coli and P. aeruginosa. The characterization of the SOS response in E. coli showed the derepression of 43 genes, in contrast with the 15 LexA-controlled genes in Pseudomonas aeruginosa (Courcelle et al., 2001; Cirz et al., 2006). Nevertheless, the response to DNA damage is more complex in P. aeruginosa because other regulons controlled by LexA-like repressors, with auto-cleavage promoted by activated RecA, are also induced alongside the canonical SOS response (Courcelle et al., 2001; Cirz et al., 2006). Such repressors are the PrtR protein - responsible for the repression of prtN, activator of the pyocin production (Matsui et al., 1993) and also required for expression of the type III secretion system (T3SS) through its repressive role on PtrB (Sun et al., 2014) - and the AlpR protein, which represses indirectly a self-lysis pathway promoted by the alpBCDE cluster (McFarland et al., 2015, Peña et al., 2021).
Two of the key aspects of the SOS response, cell division inhibition and translesion synthesis, show interesting variation in their main players when different bacteria are compared. Translesion synthesis and the consequent mutagenesis are mediated by error-prone DNA polymerases, mainly Pol V in E. coli (Goodman and Woodgate, 2013). Nevertheless, different bacteria use different SOS-regulated TLS pathways, as first evidenced by the characterization of DnaE2 and accessory proteins (Boshoff et al., 2003; Galhardo et al., 2005), as discussed in the next section.
To avoid DNA replication and segregation problems, the SOS response activates inhibitors of cell division. The cell division in E. coli under DNA damage stops when the product of sulA gene interacts with FtsZ and inhibits its GTPase activity (Trusca et al., 1998). FtsZ is a GTP-binding protein abundant during the early stage of cell division, responsible for polymerizing a ring structure in the middle of the bacterial cell where the future separation of cells occurs in normal conditions (De Boer et al., 1992). It has already been shown that the SulA protein also interacts with FtsZ in P. aeruginosa, however the ability to inhibit cell division per se has not yet been confirmed (Cordell et al., 2003). In SOS-inducing conditions, C. crescentus upregulates the imuA gene that shows weak, but enough homology to be confounded with sulA in a few bacterial genomic annotations, like in Pseudomonas putida, for example (Erill et al., 2006; McHenry, 2018). Yet, it is known that in C. crescentus, the filamentation caused by DNA-damage occurs through the inhibition of the final step of cell division by the interaction of a small inner membrane protein, product of sidA gene, with FtsW, one of the proteins responsible for cell constriction (Modell et al., 2011). In B. subtillis the inhibition of cell division occurs through YneA, also a membrane protein, that when expressed upon SOS-inducing conditions, promotes cell elongation (Kawai et al., 2003). However, the FtsZ ring is still polymerized, so YneA acts via protein-protein interaction with proteins, other than FtsZ, that could be part of the divisome, therefore differing in activity from SulA (Mo and Burkholder, 2010). S. aureus displays a similar mechanism where the SosA membrane protein inhibits the division septum formation causing filamentation probably through interaction with proteins responsible for a later stage of the division like in B. subtillis (Bojer et al., 2019).
Characterization of the SOS regulon of C. crescentus (da Rocha et al., 2008; Modell et al., 2011) exemplifies how the study of the SOS response in different bacterial species may reveal novel aspects of prokaryotic DNA repair and cellular defense mechanisms. Two SOS-regulated genes (mmcA and mmcB) were identified as agents protecting cells from Mitomycin C, a cross-linking agent. MmcA is probably a detoxifying enzyme, while MmcB is an endonuclease from the PD-(D/E)XK family (Lopes-Kulishev et al., 2015), also mediating resistance to cisplatin (Price et al., 2018). MmcB has been hypothesized to participate in a repair pathway also involving translesion synthesis polymerases to allow removal of interstrand crosslinks (Lopes-Kulishev et al., 2015). Another pair of SOS-regulated genes encode a toxin-antitoxin system (HigAB). The RNAse activity of the toxin HigB targets key mRNAs, therefore acting as a growth regulator after DNA damage (Kirkpatrick et al., 2016).
Besides the difference in LexA binding sites and set of regulated genes, the regulator itself can also vary between species. In the Streptococcacea family, SOS response is regulated by the HdiR repressor, a peptidase of the S24-family such as LexA, which similarly to LexA has the ability to self-cleave in the presence of ssDNA-RecA and release the transcription of an SOS regulon composed basically of error-prone polymerases (Savijoki et al., 2003). The same occurs in the Moraxellaceae family but the regulator is the UmuDab protein (Hare et al., 2014). In the phylum of Bacteroidetes, the SOS response is regulated by a new peptidase from the S24-family of phage-like repressors which, when derepressed, activates the expression of standard SOS genes (Sánchez-Osuna et al., 2021). The evolution of these peptidases with independent DNA-binding domains once again shows how heterogeneous this response can be.
Translesion synthesis, mutagenesis and bacterial evolution
One of the most intensely studied aspects of the SOS response is its influence on mutagenesis. Early studies on mutagenesis induced by ultraviolet radiation have led to the recognition that mutations are not always the result of passive replication errors caused by mutagens - on the contrary, these mutations are the result of active processing of DNA damage by the cellular machinery (reviewed by Friedberg et al., 2006). This fascinating concept has emerged from studies by Jean Weigle, in which UV irradiated λ bacteriophage was shown to have improved survival if the host cells had been pre-irradiated as well. In the same way, mutagenesis resulting from such irradiation of phages with UV light was only observed if the host cells had been pre-irradiated (Weigle, 1953). These phenomena were named respectively “Weigle reactivation” and “Weigle mutagenesis”. These observations led to the correct conclusion that mutagenesis requires an active processing of the damaged DNA by cells, which is mediated by an inducible cellular component.
This is a consequence of translesion DNA synthesis (TLS) polymerases, one of the pathways repressed by LexA and regulated by the SOS response. All living organisms are dependent on DNA polymerases for efficiently replicating their genetic material, however DNA damage causes blockage of the replisome and induction of the SOS response, a situation that can be circumvented by TLS-polymerases. These polymerases lack proofreading exonuclease activity and are error-prone, leading to incorporation of incorrect nucleotides (Goodman and Woodgate, 2013). However, their flexible active sites and additional little finger domain allow them to achieve TLS, using damaged DNA as templates and continuing replication (Boudsocq et al., 2004; Friedberg et al., 2006). Even though this process is essential for bacterial survival in adverse conditions, it could be detrimental due to the generation of deleterious mutations, making it crucial for bacteria to tightly regulate induction of TLS. On the other hand, it also may lead to bacterial evolution and diversity in virtue of mutagenesis (Galhardo et al., 2007; Goodman and Woodgate, 2013; Zhang, 2020). In fact, recent findings suggest that E. coli cells may use TLS as the first choice to deal with replication blockage, rather than error-free damage avoidance pathways, favoring the generation of genetic variability (Naiman et al., 2014).
In E. coli, three DNA polymerases are regulated by LexA: Pol II (polB), Pol IV (dinB) and Pol V (umuDC) (Courcelle et al., 2001), all of which are involved in mutagenesis to some extent (Napolitano et al., 2000). The induction of polB and dinB occurs early in the SOS response, related to the weak binding of LexA (Fernández de Henestrosa et al., 2000). These are responsible for TLS in specific DNA damages, in contrast to umuDC, considered as much more error-prone and able to bypass a more diverse set of DNA lesions, used as last resource and being strongly regulated (Sommer et al., 1993; Fernández de Henestrosa et al., 2000).
Pol II (polB) is a B-family polymerase that had its TLS function, bypass abasic lesions (Bonner et al., 1988), unveiled years after its first characterization by Knippers (1970), with low involvement in mutagenesis. UmuC and DinB are members of the Y-family of DNA polymerases, which includes many bacterial, archaeal and eukaryotic enzymes (Ohmori et al., 2001; reviewed by Jarosz et al., 2007).
Although a physiological role for DinB in DNA damage tolerance was harder to identify on the basis of phenotypes of a dinB mutant strain, it has been implicated in tolerance to some types of DNA damage, especially adducts in position N2 of guanines and alkylative lesions (Kim et al., 2001, Jarosz et al., 2006; Bjedov et al., 2007). DinB has an error rate between 10-3 and 10-5 in vitro (Tang et al., 2000; Jarosz et al., 2007) when using a non-damaged DNA as a substrate. Overexpression of dinB is heavily mutagenic to E. coli, introducing mainly -1 frameshifts at G:C runs (Kim et al., 1997), the same being observed in in vitro gap filling assays using the lacZ gene as a target (Kobayashi et al., 2002). Mutagenesis caused by overexpression of DinB occurs preferentially in the lagging strand (Kuban et al., 2005), a smaller but significant number of base substitutions are also observed.
E. coli DinB promotes TLS across adducts in the N2 position of guanine with high efficiency and accuracy (Jarosz et al., 2007). Genetic data also indicate that DinB takes place in error-free TLS in sites of endogenous alkylation damage that accumulates in repair-deficient strains (Bjedov et al., 2007). Lastly, dinB plays a major role in the process of stress-induced mutagenesis in non-growing cells (Mckenzie et al., 2001; Galhardo et al., 2009).
DinB is expressed as part of an SOS-regulated operon, which also contains the yafN-yafO toxin-antitoxin system and yafP (Singletary et al., 2009). The yafP gene encodes a putative acetyl-transferase probably involved in the metabolic transformation of genotoxic compounds (Gutierrez et al., 2011). Interestingly, umuDC is tightly repressed in SOS-uninduced cells, whereas dinB has a significant basal level of expression. In fact, about 250 molecules of DinB are present in cells, in contrast to only about 10-20 molecules of the holoenzyme of DNA Pol III, the enzyme responsible for normal replication (Fijalkowska et al., 2012). Upon SOS induction, the number of DinB molecules rises 10-fold to about 2500 molecules per cell (Kim et al., 2001). DinB expression and activity are subject to several levels of control. The dinB gene is also induced independently of the SOS response both as part of the stationary phase regulon controlled by the alternative sigma factor RpoS (Layton and Foster, 2003) and after exposure to beta-lactam antibiotics (Pérez-Capilla et al., 2005). Activity of this polymerase is modulated by a plethora of interactions, including UmuD, polyphosphate kinase (ppk), Rep helicase, RecA and the transcription elongation factor NusA (Stumpf and Foster, 2005; Godoy et al., 2007; Cohen et al., 2009; Sladewski et al., 2011).
Pol V (umuDC) is highly mutagenic, being considered the most important TLS-polymerase according to its capacity to bypass diverse forms of DNA lesions (Goodman and Woodgate, 2013). In accordance, this is the most studied TLS-polymerase, with orthologs identified in diverse prokaryotes and mobile genetic elements (Vaisman et al., 2012), such as the homologs mucAB described in plasmids (Perry and Walker, 1982) and rumAB in integrative and conjugative elements (ICEs) (Kulaeva et al., 1995). The function of UmuDC was first observed in the 70s, by Miroslav Radman and Evelyn Witkin (Radman, 1974; Sikand et al., 2021), although at that time the specific polymerase responsible for the mutagenic activity in the SOS response had not been elucidated (Sikand et al., 2021). Genetic identification of umuDC genes was first reported in a search for E. coli strains lacking UV-inducible mutagenesis (Kato and Shinoura, 1977). In the 80s umuC and umuD genes were revealed as an operon regulated by LexA and RecA (Bagg et al., 1981; Elledge and Walker, 1983; Shinagawa et al., 1983), but only in the late 90s purification and study of the mutagenic activity of UmuDC were achieved (Bruck et al., 1996; Tang et al., 1998, 1999; Reuven et al., 1999).
The complex modulation of DNA Pol V also involves the RecA protein. RecA is necessary both for the induction of the SOS response and for UmuD cleavage, in a process similar to what occurs to LexA with the involvement of RecA*. RecA* induces self-cleavage of UmuD in UmuD’, forming the complex with UmuC - UmuD’2C (Pol V) (Jiang et al., 2009). Additionally, early genetic studies have shown that RecA performs a third role in umuDC-dependent SOS mutagenesis (Blanco et al., 1982; Nohmi et al., 1988; Dutreix et al., 1989; Sweasy et al., 1990). In vitro experiments have shown that RecA bound to ssDNA is necessary for mutagenesis, with latest models suggesting that the “mutasome” complex operating in TLS is a molecular assembly of UmuD’2C-RecA-ATP (reviewed by Fujii and Fuchs 2020; Jaszczur et al., 2016; Sikand et al., 2021). RecFOR proteins also have a role in the formation of the RecA filament necessary for UmuD’2C TLS (Fujii et al., 2006). The mutagenic activity of Pol V is not only capable of incorporating incorrect nucleotides into DNA lesions, but also upstream and downstream of it (Maor-Shoshani et al., 2000; Isogawa et al., 2018; Fujii and Fuchs, 2020).
Bacteria that do not possess Pol V, approximately two thirds of the bacteria with known genomes (Sheng et al., 2021), may possess an SOS cassette consisting of imuABC (imuAB and dnaE2), responsible for TLS and mutagenic activity in stressing conditions, mainly distributed among Proteobacteria (Galhardo et al., 2005; Erill et al., 2006; McHenry, 2011). However, it is important to emphasize that genetic composition and configuration of this cassette is variable among bacterial species, some of them lacking imuA or with different genes supporting DnaE2 activity (Erill et al., 2006; Timinskas and Venclovas, 2019; Blanchard and Groot, 2021).
The relation of imuC (dnaE2) with mutagenic activity was first established in studies with Mycobacterium tuberculosis (Boshoff et al., 2003). Additionally, it was shown that dnaE2 is co-transcribed with imuA and imuB in C. crescentus and a reduced damage-induced mutagenesis activity was observed when any of these three genes were deleted (Galhardo et al., 2005). Later, a role for imuABC-like cassettes in damage-inducible mutagenesis and DNA damage tolerance was confirmed in other bacterial species (Koorits et al., 2007; Zeng et al., 2011; Blanchard and Groot, 2021; Sheng et al., 2021). In contrast to its role in TLS, involvement of ImuC in spontaneous mutagenesis in C. crescentus is minor (Valencia et al., 2020), and not enhanced by a constitutively transcribed imuABC operon (Alves et al., 2017). More recently it has been shown that non-dividing C. crescentus cells employ ImuC in DNA synthesis during gap filling of nucleotide excision repair intermediates (Joseph et al., 2021).
ImuA is a protein distantly related to SulA and RecA, ImuB is a catalytically dead Y-family polymerase, whereas ImuC (DnaE2) is a paralog of the Pol III´s alpha subunit without proofreading exonuclease activity, consequently error-prone and SOS-mutagenic (Galhardo et al., 2005; Warner et al., 2010; Timinskas et al., 2014). In M. tuberculosis, ImuC mutagenesis is also dependent on ImuA and ImuB supporting activity, ImuB being responsible for making the connection of ImuC with the β-clamp in the replication fork, making possible for ImuC to continue its function (Warner et al., 2010). Unlike SOS mutagenesis in E. coli, ImuABC activity in C. crescentus is independent of RecA, which leads to the hypothesis that ImuA may perform a similar role as the former in TLS (Alves et al., 2017). Recent results obtained in Myxococcus xanthus revealed that ImuA does not bind DNA, but interferes with RecA activity, which may indicate that this protein has a role in inhibiting competing pathways such as homologous recombination (Sheng et al., 2021).
The mutagenic activity of translesion DNA polymerases may be described as targeted (damaged DNA) or untargeted (undamaged and distant DNA sites), these events are constantly checked by DNA mismatch repair (MMR) systems, as a form of preventing misincorporations and mutations after the replication (Lewis et al., 2021). However, one of the most intriguing consequences of TLS-polymerases action is the phenomenon of antibiotic-induced mutagenesis. Antimicrobial agents of different types of action, and of regular clinical usage, are involved in the induction of the SOS response by ROS generation (Kohanski et al., 2007; Dwyer et al., 2014; Memar et al., 2018; Crane et al., 2021), consequently triggering the hypermutation phenotype and bacterial evolution that TLS polymerases may potentiate, including mutations that cause acquisition of adaptive mechanisms and resistance to antibiotics (Goodman, 2016; Memar et al., 2020). The contribution of ROS to bacterial killing by antibiotics is still under debate (Liu and Imlay, 2013; Keren et al., 2013), but it has become increasingly clear that antibiotics, at least in part through ROS generation, induce an SOS-dependent increase in mutagenesis (Pribis et al., 2019; Rodríguez-Rosado et al., 2019).
Targeting the SOS DNA repair system as a countermeasure to antibiotic resistance
The rise of antibiotic resistant bacteria poses an unprecedented concern since the discovery of penicillin (Sengupta et al., 2013). The underlying mechanism for the increasing threat is related to the large amount and misuse of antibiotics in agricultural/livestock production and therapy, where a range of sub-lethal antibiotic concentrations are released in the environment (Mann et al., 2021). Beta-lactams, quinolones and aminoglycosides are known to ultimately produce ROS in bacteria, which can directly damage proteins, DNA and cell membrane (Kohanski et al., 2007). However, while sub-therapeutic concentrations of antibiotics are not sufficient to kill bacteria, they still stimulate the SOS response by DNA damage (Kohanski et al., 2010; Thi et al., 2011). SOS increases the number of mutational events by upregulating error-prone TLS polymerases (Boshoff et al., 2003) and stimulates horizontal gene transfer (Beaber et al., 2004; Crane et al., 2018), biofilm formation (Gotoh et al., 2010) and the appearance of small colony variants, all of which have the potential to increase tolerance against antibiotics (Memar et al., 2020; Podlesek and Bertok, 2020).
It has been shown that combining antibiotics and suppression of the SOS response decreases the formation of resistant strains (e. g. Cirz et al., 2005; Thi et al., 2011; Recacha et al., 2017, Valencia et al., 2017). The most studied approaches to block the SOS response are prevention of either the activation of RecA protein or the autocatalysis of LexA cleavage. There are different alternatives to interfere with RecA activity, for example, disturbing proper filament RecA-ssDNA formation (Lee et al., 2005; Petrova et al., 2009; Nautiyal et al., 2014), or affecting the RecA ATP binding/ATPase activity that is necessary for its activation (Wigle and Singleton, 2007; Bellio et al., 2017; Ojha and Patil, 2019). Both strategies affect RecA-dependent LexA proteolysis, thus blocking the SOS response.
However, RecA has homology to a human recombinase Rad51 (Kawabata et al., 2005). This raises a concern on the usage of these compounds in combination with antibiotics. A better alternative would be to target LexA, as there are no corresponding orthologs in the human genome. A study found that phenylboronic derivatives could interfere with LexA self-cleavage by forming an acyl-enzyme intermediate with the catalytic Ser-119 (Bellio et al., 2020). Nevertheless, research on SOS inhibition directly affecting LexA is still very scarce.
Although no drug targeting the SOS machinery has been approved yet, there is no doubt that suppressing evolutionary mechanisms responsible to increase tolerance against bactericidal agents is a very promising approach to extend the shelf life of antibiotics in use today.
Relationship between SOS response and mobile genetic elements
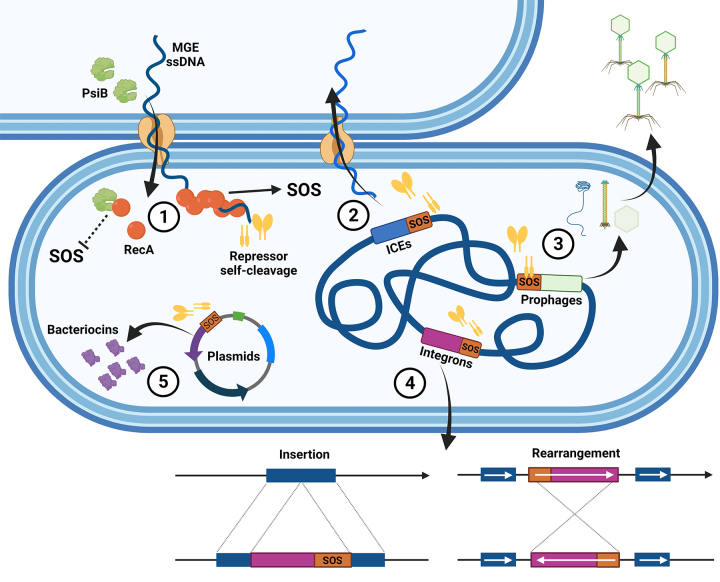
When lysogenized bacteria undergo DNA damage, bacteriophages switch to the lytic cycle, presumably to escape from an endangered host and disperse in the environment (Little, 2005). This early observation in phage biology underlies a phenomenon shared by other mobile genetic elements (MGEs), such as integrons (Guerin et al., 2009), chromosome cassettes (Liu et al., 2017), pathogenicity islands (Chittò et al., 2020) and integrative and conjugative elements (ICEs) (Beaber et al., 2004; Auchtung et al., 2005). Figure 2 depicts the relationship of the SOS response with MGEs.
Figure 2 - . Schematic representation of the involvement of SOS response with mobile genetic elements (MGE). (1) Entry of mobile elements ssDNA in the host cell induces the SOS response by the formation of RecA* filaments, however some MGE encode proteins (such as PsiB) that are able to bind free RecA, avoiding all functions of RecA including the initiation of the SOS response. (2) The SOS response regulates the transfer of integrative and conjugative elements (ICEs) from the SXT/R391 family by interacting with SetR, repressor that self-cleaves after RecA* stimulus. (3) Bacteriophages may go from lysogeny to lytic cycle after induction of the SOS response, some phages show an SOS box sequence on promoter regions, others may encode represors, like the lambda bacteriophage CI repressor that self-cleaves after RecA* stimulus. (4) Chromosomal and mobile integrons show SOS box sequences in promoter regions, with transfer and rearrangement in the chromosome after the SOS response induction. (5) The SOS response regulates expression of bacteriocins, enforcing the maintenance of plasmids in the host cell.
The SOS regulators RecA and LexA are both involved in the regulation of MGEs transfer (Fornelos et al., 2016). Several chromosomal and mobile integrons show SOS box sequences in promoter regions (Guerin et al., 2009), as well as ICEs that may possess repressors regulated by RecA (Beaber et al., 2004), consequently showing that induction of the SOS response also regulates MGEs transfer and chromosomal rearrangement during conjugation (Baharoglu et al., 2010).
It was initially observed that DNA damage was able to cause changes in the life cycle of temperate phages, from lysogeny to the lytic cycle (Little, 2005). Multiple phages are SOS-induced and regulated by LexA, others use their own RecA-controlled repressors in a similar mechanism to the self-cleavage of LexA by RecA*. The most prominent example is the λ phage that is maintained integrated in the chromosome through the CI repressor and when there is DNA damage, RecA* induces autocleavage of CI and expression of λ phage genes (Hochschild and Lewis, 2009; Fornelos et al., 2016).
ICEs from the SXT/R391 family encode the SetR repressor, from the same family of the λ-CI repressor, showing self-cleavage activity regulated by RecA in the SOS response (Beaber et al., 2004; González et al., 2019). SetR is responsible for repressing setCD, two genes involved in the transfer of ICEs (Beaber et al., 2002, 2004), and, along with LexA and CroS, also regulates the mutagenic activity of the umuDC homologs rumAB encoded in the ICE (González et al., 2019; McDonald et al., 2021). It is interesting to note that some species that often carry SXT/R391 elements such as Proteus mirabilis, are naturally devoid of chromosomal umuDC genes. This species is accordingly non-mutable by UV irradiation, but acquisition of SXT/R391 elements provides TLS and mutagenesis capacity to this bacterium. Furthermore, rumAB genes improve conjugation of the ICE to new hosts (Sato et al., 2022), demonstrating an intricate relationship of these MGEs with the SOS response.
LexA is not only able to mediate control over MGEs horizontal transfer but also over the expression of virulence factors and bacteriocins carried by MGEs (Fornelos et al., 2016). For example, clusters present in plasmids are responsible for the expression of the toxic colicin protein, which is capable of killing competing bacteria and enforcing the maintenance of the plasmids in the host through the expression of immunity proteins (Cascales et al., 2007; Budič et al., 2011; Fornelos et al., 2016). An interesting observation was also made by Kamruzzaman and Iredell (2019) that conjugative plasmids (mainly IncI and IncF) benefited from a toxin-antitoxin system (parDE I ) that is induced by stress and also elicits the SOS response, that provides antibiotic tolerance and allows the plasmid to successfully stabilize in the bacterial cell.
Diverse MGEs influence the host’s SOS response. The acquisition of new MGE is a distress event that induces the SOS response, caused by the filamentation of RecA in the ssDNA - intermediate form of transfer of MGEs - mainly in DNA with low homology to the chromosome (Baharoglu et al., 2010; Al Mamun et al., 2021). There is also development of systems to repress the SOS response so that the MGE can successfully integrate or perpetuate itself in the new host (Memar et al., 2020; Al Mamun et al., 2021). It has been shown that SOS inhibiting proteins, such as PsiB (Bagdasarian et al., 1992; Petrova et al., 2009) and SSB, are translocated through the secretion system (T4SS) together with the MGEs ssDNA, which facilitates the maintenance of these elements (Al Mamun et al., 2021). The regulation of frequency of transmission is also important for the survival of the MGE in observation that excess of horizontal transfer causes impact in the host cell (Touchon et al., 2014).
Overall, the SOS response protects cells from DNA-damaging environmental stressors and is a main player in the acquisition of antimicrobial resistance through mutagenic activity and induction of horizontal transfer of MGEs carrying these traits.
Acknowledgments
Work in the RSG lab is financially supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP, awards 2019/19435-3 and 2021/15170-5). MAL-N is beneficiary of a Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) fellowship (award number 140476/2020-2). RSO and DLHF are beneficiaries of FAPESP fellowships (award numbers 2020/12744-8 and 2020/00535-5). RRF is beneficiary of a Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) fellowship (Finance code 01).
References
- Abella M, Campoy S, Erill I, Rojo F, Barbé J. Cohabitation of two different lexA regulons in Pseudomonas putida. J Bacteriol. 2007;189:8855–8862. doi: 10.1128/JB.01213-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aertsen A, Michiels CW. Mrr instigates the SOS response after high pressure stress in Escherichia coli. Mol Microbiol. 2005;58:1381–1391. doi: 10.1111/j.1365-2958.2005.04903.x. [DOI] [PubMed] [Google Scholar]
- Aertsen A, Van Houdt R, Vanoirbeek K, Michiels CW. An SOS response induced by high pressure in Escherichia coli. J Bacteriol. 2004;186:6133–6141. doi: 10.1128/JB.186.18.6133-6141.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksenov SV. Dynamics of the inducing signal for the SOS regulatory system in Escherichia coli after ultraviolet irradiation. Math Biosci. 1999;157:269–286. doi: 10.1016/s0025-5564(98)10086-x. [DOI] [PubMed] [Google Scholar]
- Al Mamun AAM, Kishida K, Christie PJ. Protein transfer through an F Plasmid-encoded type IV secretion system suppresses the mating-induced SOS response. mBio. 2021;12:e0162921. doi: 10.1128/mBio.01629-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpas H, Kalchayanand N, Bozoglu F, Ray B. Interactions of high hydrostatic pressure, pressurization temperature and pH on death and injury of pressure-resistant and pressure-sensitive strains of foodborne pathogens. Int J Food Microbiol. 2000;60:33–42. doi: 10.1016/s0168-1605(00)00324-x. [DOI] [PubMed] [Google Scholar]
- Alves IR, Lima-Noronha MA, Silva LG, Fernández-Silva FS, Freitas ALD, Marques MV, Galhardo RS. Effect of SOS-induced levels of imuABC on spontaneous and damage-induced mutagenesis in Caulobacter crescentus. DNA Repair (Amst) 2017;59:20–26. doi: 10.1016/j.dnarep.2017.09.003. [DOI] [PubMed] [Google Scholar]
- Anderson DG, Kowalczykowski SC. The translocating RecBCD enzyme stimulates recombination by directing RecA protein onto ssDNA in a χ-regulated manner. Cell. 1997;90:77–86. doi: 10.1016/s0092-8674(00)80315-3. [DOI] [PubMed] [Google Scholar]
- Anderson DG, Kowalczykowski SC. Reconstitution of an SOS response pathway: Derepression of transcription in response to DNA breaks. Cell. 1998;95:975–979. doi: 10.1016/s0092-8674(00)81721-3. [DOI] [PubMed] [Google Scholar]
- Au N, Kuester-Schoeck E, Mandava V, Bothwell LE, Canny SP, Chachu K, Colavito SA, Fuller SN, Groban ES, Hensley LA, et al. Genetic composition of the Bacillus subtilis SOS system. J Bacteriol. 2005;187:7655–7666. doi: 10.1128/JB.187.22.7655-7666.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auchtung JM, Lee CA, Monson RE, Lehman AP, Grossman AD. Regulation of Bacillus subtilis mobile genetic element by intercellular signaling and the global DNA damage response. Proc Natl Acad Sci U S A. 2005;102:12554–12559. doi: 10.1073/pnas.0505835102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagdasarian M, Bailone A, Angulo JF, Scholz P, Bagdasarian M, Devoret R. PsiB, an anti-SOS protein, is transiently expressed by the F sex factor during its transmission to an Escherichia coli K-12 recipient. Mol Microbiol. 1992;6:885–893. doi: 10.1111/j.1365-2958.1992.tb01539.x. [DOI] [PubMed] [Google Scholar]
- Bagg A, Kenyon CJ, Walker GC. Inducibility of a gene product required for UV and chemical mutagenesis in Escherichia coli. Proc Natl Acad Sci U S A. 1981;78:5749–5753. doi: 10.1073/pnas.78.9.5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baharoglu Z, Bikard D, Mazel D. Conjugative DNA transfer induces the bacterial SOS response and promotes antibiotic resistance development through integron activation. PLoS Genet. 2010;6:e1001165. doi: 10.1371/journal.pgen.1001165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaber JW, Hochhut B, Waldor MK. Genomic and functional analyses of SXT, an integrating antibiotic resistance gene transfer element derived from Vibrio cholerae. J Bacteriol. 2002;184:4259–4269. doi: 10.1128/JB.184.15.4259-4269.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaber JW, Hochnut B, Waldor MK. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature. 2004;427:72–74. doi: 10.1038/nature02241. [DOI] [PubMed] [Google Scholar]
- Bellio P, Di Pietro L, Mancini A, Piovano M, Nicoletti M, Brisdelli F, Tondi D, Cendron L, Franceschini N, Amicosante G, et al. SOS response in bacteria: Inhibitory activity of lichen secondary metabolites against Escherichia Coli RecA protein. Phytomedicine. 2017;29:11–18. doi: 10.1016/j.phymed.2017.04.001. [DOI] [PubMed] [Google Scholar]
- Bellio P, Mancini A, Di Pietro L, Cracchiolo S, Franceschini N, Reale S, Angelis F, Perilli M, Amicosante G, Spyrakis F, et al. Inhibition of the transcriptional Withstanding drug resistance by inhibiting the bacterial mechanisms of adaptation to antimicrobials. Life Sci. 2020;241:117116. doi: 10.1016/j.lfs.2019.117116. [DOI] [PubMed] [Google Scholar]
- Berg OG. Selection of DNA binding sites by regulatory proteins: The LexA protein and the arginine repressor use different strategies for functional specificity. Nucleic Acids Res. 1988;16:5089–5105. doi: 10.1093/nar/16.11.5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjedov I, Dasgupta CN, Slade D, Le Blastier S, Selva M, Matic I. Involvement of Escherichia coli DNA polymerase IV in tolerance of cytotoxic alkylating DNA lesions in vivo. Genetics. 2007;176:1431–1440. doi: 10.1534/genetics.107.072405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard L, Groot A. Coexistence of SOS-dependent and SOS-independent regulation of DNA repair genes in radiation-resistant Deinococcus bacteria. Cells. 2021;10:924. doi: 10.3390/cells10040924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco M, Herrera G, Collado P, Rebollo JE, Botella LM. Influence of RecA protein on induced mutagenesis. Biochimie. 1982;64:633–636. doi: 10.1016/s0300-9084(82)80102-8. [DOI] [PubMed] [Google Scholar]
- Blázquez J, Gómez-Gómez J-M, Oliver A, Juan C, Kapur V, Martín S. PBP3 inhibition elicits adaptive responses in Pseudomonas aeruginosa. Mol Microbiol. 2006;62:84–99. doi: 10.1111/j.1365-2958.2006.05366.x. [DOI] [PubMed] [Google Scholar]
- Bojer MS, Wacnik K, Kjelgaard P, Gallay C, Bottomley AL, Cohn MT, Lindahl G, Frees D, Veening JW, Foster SJ, et al. SosA inhibits cell division in Staphylococcus aureus in response to DNA damage. Mol Microbiol. 2019;112:1116–1130. doi: 10.1111/mmi.14350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner CA, Randall SK, Rayssiguier C, Radman M, Eritja R, Kaplan BE, McEntee K, Goodman MF. Purification and characterization of an inducible Escherichia coli DNA polymerase capable of insertion and bypass at abasic lesions in DNA. J Biol Chem. 1988;263:18946–18952. [PubMed] [Google Scholar]
- Bork JM, Cox MM, Inman RB. The RecOR proteins modulate RecA protein function at 5′ ends of single-stranded DNA. EMBO J. 2001;20:7313–7322. doi: 10.1093/emboj/20.24.7313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boshoff HIM, Reed MB, Barry CE, Mizrahi V. DnaE2 polymerase contributes to in vivo survival and the emergence of drug resistance in Mycobacterium tuberculosis. Cell. 2003;113:183–193. doi: 10.1016/s0092-8674(03)00270-8. [DOI] [PubMed] [Google Scholar]
- Boudsocq F, Kokoska RJ, Plosky BS, Vaisman A, Ling H, Kunkel TA, Yang W, Woodgate R. Investigating the role of the little finger domain of Y-family DNA polymerases in low-fidelity synthesis and translesion replication. J Biol Chem. 2004;279:32932–32940. doi: 10.1074/jbc.M405249200. [DOI] [PubMed] [Google Scholar]
- Britton RA, Küster-Schöck E, Auchtung TA, Grossman AD. SOS induction in a subpopulation of structural maintenance of chromosome (Smc) mutant cells in Bacillus subtilis. J Bacteriol. 2007;189:4359–4366. doi: 10.1128/JB.00132-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruck I, Woodgate R, McEntee K, Goodman MF. Purification of a soluble UmuD’C complex from Escherichia coli: Cooperative binding of UmuD’C to single-stranded DNA. J Biol Chem. 1996;271:10767–10774.. doi: 10.1074/jbc.271.18.10767. [DOI] [PubMed] [Google Scholar]
- Budič M, Rijavec M, Petkovsek Z, Zgur-Bertok D. Escherichia coli bacteriocins: Antimicrobial efficacy and prevalence among isolates from patients with bacteraemia. PLoS One. 2011;6:e28769. doi: 10.1371/journal.pone.0028769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butala M, Klose D, Hodnik V, Rems A, Podlesek Z, Klare JP, Anderluh G, Busby SJW, Steinhoff HJ, Zgur-Bertok D. Interconversion between bound and free conformations of LexA orchestrates the bacterial SOS response. Nucleic Acids Res. 2011;39:6546–6557. doi: 10.1093/nar/gkr265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales E, Buchanan SK, Duché D, Kleanthous C, Lloubés R, Postle K, Riley M, Slatin S, Cavard D. Colicin biology. Microbiol Mol Biol Rev. 2007;71:158–229. doi: 10.1128/MMBR.00036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury AM, Smith GR. Role of Escherichia coli RecBC enzyme in SOS induction. Mol Gen Genet. 1985;201:525–528. doi: 10.1007/BF00331350. [DOI] [PubMed] [Google Scholar]
- Chittò M, Berger M, Klotz L, Dobrindt U. Sub-Inhibitory concentrations of SOS-Response inducing antibiotics stimulate integrase expression and excision of pathogenicity islands in uropathogenic Escherichia coli strain 536. Int J Med Microbiol. 2020;310:151361. doi: 10.1016/j.ijmm.2019.151361. [DOI] [PubMed] [Google Scholar]
- Cho H, Uehara T, Bernhardt TG. Beta-lactam antibiotics induce a lethal malfunctioning of the bacterial cell wall synthesis machinery. Cell. 2014;159:1300–1311. doi: 10.1016/j.cell.2014.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirz RT, Chin JK, Andes DR, Crécy-Lagard V, Craig WA, Romesberg FE. Inhibition of mutation and combating the evolution of antibiotic resistance. PLoS Biol. 2005;3:e176. doi: 10.1371/journal.pbio.0030176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirz RT, O’Neill BM, Hammond JA, Head SR, Romesberg FE. Defining the Pseudomonas aeruginosa SOS response and its role in the global response to the antibiotic ciprofloxacin. J Bacteriol. 2006;188:7101–7110. doi: 10.1128/JB.00807-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirz RT, Jones MB, Gingles NA, Minogue TD, Jarrahi B, Peterson SN, Romesberg FE. Complete and SOS-mediated response of Staphylococcus aureus to the antibiotic ciprofloxacin. J Bacteriol. 2007;189:531–539. doi: 10.1128/JB.01464-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SE, Godoy VG, Walker GC. Transcriptional modulator NusA interacts with translesion DNA polymerases in Escherichia coli. J Bacteriol. 2009;191:665–672. doi: 10.1128/JB.00941-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordell SC, Robinson EJH, Löwe J. Crystal structure of the SOS cell division inhibitor SulA and in complex with FtsZ. Proc Natl Acad Sci U S A. 2003;100:7889–7894. doi: 10.1073/pnas.1330742100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courcelle J, Donaldson JR, Chow K-H, Courcelle CT. DNA damage-induced replication fork regression and processing in Escherichia coli. Science. 2003;299:1064–1067. doi: 10.1126/science.1081328. [DOI] [PubMed] [Google Scholar]
- Courcelle J, Khodursky A, Peter B, Brown PO, Hanawalt PC. Comparative gene expression profiles following UV exposure in wildtype and SOS-deficient Escherichia coli. Genetics. 2001;158:41–64. doi: 10.1093/genetics/158.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox MM. Motoring along with the bacterial RecA protein. Nat Rev Mol Cell Biol. 2007;8:127–138. doi: 10.1038/nrm2099. [DOI] [PubMed] [Google Scholar]
- Crane JK, Alvarado CL, Sutton MD. Role of the SOS response in the generation of antibiotic resistance in vivo. Antimicrob Agents Chemother. 2021;65:e0001321. doi: 10.1128/AAC.00013-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane JK, Cheema MB, Olyer MA, Sutton MD. Zinc blockade of SOS response inhibits horizontal transfer of antibiotic resistance genes in enteric bacteria. Front Cell Infect Microbiol. 2018;8:410. doi: 10.3389/fcimb.2018.00410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Rocha RP, Paquola ACM, Marques MV, Menck CFM, Galhardo RS. Characterization of the SOS regulon of Caulobacter crescentus. J Bacteriol. 2008;190:1209–1218. doi: 10.1128/JB.01419-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilowicz C, Vietorisz E, Godoy-Carter V, Prévost C, Prentiss M. Influences of ssDNA-RecA filament length on the fidelity of homologous recombination. J Mol Biol. 2021;433:167143. doi: 10.1016/j.jmb.2021.167143. [DOI] [PubMed] [Google Scholar]
- De Boer P, Crossley R, Rothfield L. The essential bacterial cell-division protein FtsZ is a GTPase. Nature. 1992;359:254–256. doi: 10.1038/359254a0. [DOI] [PubMed] [Google Scholar]
- Drees JC, Lusetti SL, Chitteni-Pattu S, Inman RB, Cox MM. A RecA filament capping mechanism for RecX protein. Mol Cell. 2004;15:789–798. doi: 10.1016/j.molcel.2004.08.026. [DOI] [PubMed] [Google Scholar]
- Dri AM, Moreau PL. Control of the LexA regulon by pH: Evidence for a reversible inactivation of the LexA repressor during the growth cycle of Escherichia coli. Mol Microbiol. 1994;12:621–629. doi: 10.1111/j.1365-2958.1994.tb01049.x. [DOI] [PubMed] [Google Scholar]
- Dutreix M, Moreau PL, Bailone A, Galibert F, Battista JR, Walker GC, Devoret R. New recA mutations that dissociate the various RecA protein activities in Escherichia coli provide evidence for an additional role for RecA protein in UV mutagenesis. J Bacteriology. 1989;171:2415–2423. doi: 10.1128/jb.171.5.2415-2423.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer DJ, Belenky PA, Yang JH, MacDonald C, Martell JD, Takahashi N, Chan CTY, Lobritz MA, Braff D, Schwarz EG, et al. Antibiotics induce redox-related physiological alterations as part of their lethality. Proc Natl Acad Sci U S A. 2014;111:E2100–E2109. doi: 10.1073/pnas.1401876111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge SJ, Walker GC. Proteins required for ultraviolet light and chemical mutagenesis: Identification of the products of the umuC locus of Escherichia coli. J Mol Biol. 1983;164:175–192. doi: 10.1016/0022-2836(83)90074-8. [DOI] [PubMed] [Google Scholar]
- Erill I, Campoy S, Barbé J. Aeons of distress: An evolutionary perspective on the bacterial SOS response. FEMS Microbiol Rev. 2007;31:637–656. doi: 10.1111/j.1574-6976.2007.00082.x. [DOI] [PubMed] [Google Scholar]
- Erill I, Campoy S, Mazón G, Barbé J. Dispersal and regulation of an adaptive mutagenesis cassette in the bacteria domain. Nucleic Acids Res. 2006;34:66–77. doi: 10.1093/nar/gkj412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erill I, Escribano M, Campoy S, Barbé J. In silico analysis reveals substantial variability in the gene contents of the gamma proteobacteria LexA-regulon. Bioinformatics. 2003;19:2225–2236. doi: 10.1093/bioinformatics/btg303. [DOI] [PubMed] [Google Scholar]
- Fernández de Henestrosa AR, Ogi T, Aoyagi S, Chafin D, Hayes JJ, Ohmori H, Woodgate R. Identification of additional genes belonging to the LexA-regulon in Escherichia coli. Mol Microbiol. 2000;35:1560–1572. doi: 10.1046/j.1365-2958.2000.01826.x. [DOI] [PubMed] [Google Scholar]
- Fijalkowska IJ, Schaaper RM, Jonczyk P. DNA replication fidelity in Escherichia coli: A multi-DNA polymerase affair. FEMS Microbiol Rev. 2012;36:1105–1121. doi: 10.1111/j.1574-6976.2012.00338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornelos N, Browning DF, Butala M. The use and abuse of LexA by mobile genetic elements. Trends Microbiol. 2016;24:391–401. doi: 10.1016/j.tim.2016.02.009. [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W, Wood R, Schultz RA, Ellenberger T. DNA repair and mutagenesis. 2. ASM Press; Washington: 2006. 1129 [Google Scholar]
- Fujii S, Fuchs RP. A comprehensive view of translesion synthesis in Escherichia coli. Microbiol Mol Biol Rev. 2020;84:e00002–e00020. doi: 10.1128/MMBR.00002-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S, Isogawa A, Fuchs RP. RecFOR proteins are essential for Pol V-mediated translesion synthesis and mutagenesis. EMBO J. 2006;25:5754–5763. doi: 10.1038/sj.emboj.7601474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galhardo RS, Rocha RP, Marques MV, Menck CF. An SOS-regulated operon involved in damage-inducible mutagenesis in Caulobacter crescentus. Nucleic Acids Res. 2005;33:2603–2614. doi: 10.1093/nar/gki551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galhardo RS, Hasting PJ, Rosenberg SM. Mutation as a stress response and the regulation of evolvability. Crit Rev Biochem Mol Biol. 2007;42:399–435. doi: 10.1080/10409230701648502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galhardo RS, Do R, Yamada M, Friedberg EC, Hasting PJ, Nohmi T, Rosenberg SM. DinB upregulation is the sole role of the SOS response in stress-induced mutagenesis in Escherichia coli. Genetics. 2009;182:55–68. doi: 10.1534/genetics.109.100735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godoy VG, Jarosz DF, Simon SM, Abyzov A, Ilyin V, Walker GC. UmuD and RecA directly modulate the mutagenic potential of the Y family DNA polymerase DinB. Mol Cell. 2007;28:1058–1070. doi: 10.1016/j.molcel.2007.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González M, Huston D, McLenigan MP, McDonald JP, Garcia AM, Borden KS, Woodgate R. SetRICE391, a negative transcriptional regulator of the integrating conjugative element 391 mutagenic response. DNA Repair (Amst) 2019;73:99–109. doi: 10.1016/j.dnarep.2018.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman MF. Better living with hyper-mutation. Environ Mol Mutagen. 2016;57:421–434. doi: 10.1002/em.22023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman MF, Woodgate R. Translesion DNA polymerases. Cold Spring Harb Perspect Biol. 2013;5:a010363. doi: 10.1101/cshperspect.a010363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotoh H, Kasaraneni N, Devineni N, Dallo SF, Weitao T. SOS involvement in stress-inducible biofilm formation. Biofouling. 2010;26:603–611. doi: 10.1080/08927014.2010.501895. [DOI] [PubMed] [Google Scholar]
- Guerin E, Cambray G, Sanchez-Alberola N, Campoy S, Erill I, Da Re S, Gonzalez-Zorn B, Barbé J, Ploy MC, Mazel D. The SOS response controls integron recombination. Science. 2009;324:1034. doi: 10.1126/science.1172914. [DOI] [PubMed] [Google Scholar]
- Gutierrez A, Elez M, Clermont O, Denamur E, Matic I. Escherichia coli YafP protein modulates DNA damaging property of the nitroaromatic compounds. Nucleic Acids Res. 2011;39:4192–4201. doi: 10.1093/nar/gkr050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare JM, Ferrell JC, Witkowski TA, Grice AN. Prophage induction and differential RecA and UmuDAb transcriptome regulation in the DNA damage responses of Acinetobacter baumannii and Acinetobacter baylyi. PLoS One. 2014;9:e93861. doi: 10.1371/journal.pone.0093861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrikus SS, van Oijen AM, Robinson A. Specialized DNA polymerases in Escherichia coli: Roles within multiple pathways. Curr Genet. 2018;64:1189–1196. doi: 10.1007/s00294-018-0840-x. [DOI] [PubMed] [Google Scholar]
- Hochschild A, Lewis M. The bacteriophage lambda CI protein finds an asymmetric solution. Curr Opin Struct Biol. 2009;19:79–86. doi: 10.1016/j.sbi.2008.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isogawa A, Ong JL, Potapov V, Fuchs RP, Fujii S. Pol V-mediated translesion synthesis elicits localized untargeted mutagenesis during post-replicative gap repair. Cell Rep. 2018;24:1290–1300. doi: 10.1016/j.celrep.2018.06.120. [DOI] [PubMed] [Google Scholar]
- Jarosz DF, Beuning PJ, Cohen SE, Walker GC. Y-family DNA polymerases in Escherichia coli. Trends Microbiol. 2007;15:70–77. doi: 10.1016/j.tim.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Jarosz DF, Godoy VG, Delaney JC, Essigmann JM, Walker GC. A single amino acid governs enhanced activity of DinB DNA polymerases on damaged templates. Nature. 2006;439:225–228. doi: 10.1038/nature04318. [DOI] [PubMed] [Google Scholar]
- Jaszczur M, Bertram JG, Robinson A, Oijen AMV, Woodgate R, Cox MM, Goodman MF. Mutations for worse or better: Low-fidelity DNA synthesis by SOS DNA Polymerase V is a tightly regulated double-edged sword. Biochemistry. 2016;55:2309–2318. doi: 10.1021/acs.biochem.6b00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Q, Karata K, Woodgate R, Cox MM, Goodman MF. The active form of DNA polymerase V is UmuD’(2)C-RecA-ATP. Nature. 2009;460:359–363. doi: 10.1038/nature08178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones EC, Uphoff S. Single-molecule imaging of LexA degradation in Escherichia coli elucidates regulatory mechanisms and heterogeneity of the SOS response. Nat Microbiol. 2021;6:981–990. doi: 10.1038/s41564-021-00930-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph AM, Daw S, Sadhir I, Badrinarayanan A. Coordination between nucleotide excision repair and specialized polymerase DnaE2 action enables DNA damage survival in non-replicating bacteria. eLife. 2021;10:e67552. doi: 10.7554/eLife.67552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamruzzaman M, Iredell J. A ParDE-family toxin antitoxin system in major resistance plasmids of Enterobacteriaceae confers antibiotic and heat tolerance. Sci Rep. 2019;9:9872. doi: 10.1038/s41598-019-46318-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T, Shinoura Y. Isolation and characterization of mutants of Escherichia coli deficient in induction of mutations by ultraviolet light. Mol Gen Genet. 1977;156:121–131. doi: 10.1007/BF00283484. [DOI] [PubMed] [Google Scholar]
- Kawabata M, Kawabata T, Nishibori M. Role of recA/RAD51 family proteins in mammals. Acta Med Okayama. 2005;59:31987. doi: 10.18926/AMO/31987. [DOI] [PubMed] [Google Scholar]
- Kawai Y, Moriya S, Ogasawara N. Identification of a protein, YneA, responsible for cell division suppression during the SOS response in Bacillus subtilis. Mol Microbiol. 2003;47:1113–1122. doi: 10.1046/j.1365-2958.2003.03360.x. [DOI] [PubMed] [Google Scholar]
- Kenyon CJ, Walker GC. DNA-damaging agents stimulate gene expression at specific loci in Escherichia coli. Proc Natl Acad Sci U S A. 1980;77:2819–2823. doi: 10.1073/pnas.77.5.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren I, Wu Y, Inocencio J, Mulcahy LR, Lewis K. Killing by bactericidal antibiotics does not depend on reactive oxygen species. Science. 2013;339:1213–1216. doi: 10.1126/science.1232688. [DOI] [PubMed] [Google Scholar]
- Kim SR, Maenhaut-Michael G, Yamada M, Yamamoto Y, Matsui K, Sofuni T, Nohmi T, Ohmori H. Multiple pathways for SOS-induced mutagenesis in Escherichia coli: An overexpression of dinB/dinP results in strongly enhancing mutagenesis in the absence of any exogenous treatment to damage DNA. Proc Natl Acad Sci U S A. 1997;94:13792–13797. doi: 10.1073/pnas.94.25.13792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SR, Matsui K, Yamada M, Gruz P, Nohmi T. Roles of chromosomal and episomal dinB genes encoding DNA pol IV in targeted and untargeted mutagenesis in Escherichia coli. Mol Genet Genomics. 2001;266:207–215. doi: 10.1007/s004380100541. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick CL, Martins D, Redder P, Frandi A, Mignolet J, Chapalay JB, Chambon M, Turcatti G, Viollier PH. Growth control switch by a DNA-damage-inducible toxin-antitoxin system in Caulobacter crescentus. Nat Microbiol. 2016;1:16008. doi: 10.1038/nmicrobiol.2016.8. [DOI] [PubMed] [Google Scholar]
- Knippers R. DNA polymerase II. Nature. 1970;228:1050–1053. doi: 10.1038/2281050a0. [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Valentine MR, Pham P, O’Donnell M, Goodman MF. Fidelity of Escherichia coli DNA Polymerase IV. J Biol Chem. 2002;277:34198–34207. doi: 10.1074/jbc.M204826200. [DOI] [PubMed] [Google Scholar]
- Kohanski MA, Dwyer DJ Hayete B, Lawrence CA Collins JJ. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- Kohanski MA, DePristo MA, Collins JJ. Sub-lethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis. Mol Cell. 2010;37:311–320. doi: 10.1016/j.molcel.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koorits L, Tegova R, Tark M, Tarassova K, Tover A, Kivisaar M. Study of involvement of ImuB and DnaE2 in stationary-phase mutagenesis in Pseudomonas putida. DNA Repair (Amst) 2007;6:863–868. doi: 10.1016/j.dnarep.2007.01.010. [DOI] [PubMed] [Google Scholar]
- Kovačič L, Paulič N, Leonardi A, Hodnik V, Anderluh G, Podlesek Z, Žgur-Bertok D, Križaj I, Butala M. Structural insight into LexA-RecA* interaction. Nucleic Acids Res. 2013;41:9901–9910. doi: 10.1093/nar/gkt744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuban W, Banach-Orlowska M, Bialoskorska M, Lipowska A, Schaaper RM, Jonczyk P, Fijalkowska IJ. Mutator phenotype resulting from DNA polymerase IV overproduction in Escherichia coli: Preferential mutagenesis on the lagging strand. J Bacteriol. 2005;187:6862–6866. doi: 10.1128/JB.187.19.6862-6866.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulaeva OI, Wootton JC, Levine AS, Woodgate R. Characterization of the umu-complementing operon from R391. J Bacteriol. 1995;177:2737–2743. doi: 10.1128/jb.177.10.2737-2743.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layton JC, Foster PL. Error-prone DNA polymerase IV is controlled by the stress-response sigma factor, RpoS, in Escherichia coli. Mol Microbiol. 2003;50:549–561. doi: 10.1046/j.1365-2958.2003.03704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AM, Ross CT, Zeng B-B, Singleton SF. AA molecular target for suppression of the evolution of antibiotic resistance: Inhibition of the Escherichia coli RecA protein by N(6)-(1-naphthyl)-ADP. J Med Chem. 2005;48:5408–5411. doi: 10.1021/jm050113z. [DOI] [PubMed] [Google Scholar]
- Lewis EB, Mudipalli R, Eghbal MM, Culyba MJ. Effect of mismatch repair on the mutational footprint of the bacterial SOS mutator activity. DNA Repair (Amst) 2021;103:103130. doi: 10.1016/j.dnarep.2021.103130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis LK, Harlow GR, Gregg-Jolly LA, Mount DW. Identification of high affinity binding sites for LexA which define new DNA damage-inducible genes in Escherichia coli. J Mol Biol. 1994;241:507–523. doi: 10.1006/jmbi.1994.1528. [DOI] [PubMed] [Google Scholar]
- Little JW. The SOS regulatory system: Control of its state by the level of recA protease. J Mol Biol. 1983;167:791–808. doi: 10.1016/s0022-2836(83)80111-9. [DOI] [PubMed] [Google Scholar]
- Little JW. Mechanism of specific LexA cleavage: Autodigestion and the role of RecA coprotease. Biochimie. 1991;73:411–421. doi: 10.1016/0300-9084(91)90108-d. [DOI] [PubMed] [Google Scholar]
- Little JW. In: Phages: Their roles in bacterial pathogenesis and biotechnology. 1. Waldor MK, Friedman DI, Adhya SL, editors. ASM Press; Washington: 2005. Lysogeny, prophage induction, and lysogenic conversion; pp. 37–54. [Google Scholar]
- Little JW, Mount DW. The SOS regulatory system of Escherichia coli. Cell. 1982;29:11–22. doi: 10.1016/0092-8674(82)90085-x. [DOI] [PubMed] [Google Scholar]
- Little JW, Edmiston SH, Pacelli Z, Mount DW. Cleavage of the Escherichia coli LexA protein by the RecA protease. Proc Natl Acad Sci U S A. 1980;77:3225–3229. doi: 10.1073/pnas.77.6.3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Wu Z, Xue H, Zhao X. Antibiotics trigger initiation of SCCmec transfer by inducing SOS responses. Nucleic Acids Res. 2017;45:3944–3952. doi: 10.1093/nar/gkx153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Imlay JA. Cell death from antibiotics without the involvement of reactive oxygen species. Science. 2013;339:1210–1213. doi: 10.1126/science.1232751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes-Kulishev CO, Alves IR, Valencia EY, Pidhirnyj MI, Fernandez-Silva FS, Rodrigues TR, Guzzo CR, Galhardo RS. Functional characterization of two SOS-regulated genes involved in mitomycin C resistance in Caulobacter crescentus. DNA Repair (Amst) 2015;33:78–89. doi: 10.1016/j.dnarep.2015.06.009. [DOI] [PubMed] [Google Scholar]
- Lovett CM, Jr , O’Gara TM, Woodruff JN. Analysis of the SOS inducing signal in Bacillus subtilis using Escherichia coli LexA as a probe. J Bacteriol. 1994;176:4914–4923. doi: 10.1128/jb.176.16.4914-4923.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Pfuetzner RA, Mosimann S, Paetzel M, Frey EA, Cherney M, Kim B, Little JW, Strynadka NCJ. Crystal structure of LexA: A conformational switch for regulation of self-cleavage. Cell. 2001;106:585–594. doi: 10.1016/s0092-8674(01)00479-2. [DOI] [PubMed] [Google Scholar]
- Lusetti SL, Voloshin ON, Inman RB, Camerini-Otero RD, Cox MM. The DinI protein stabilizes RecA protein filaments. J Biol Chem. 2004;279:30037–30046. doi: 10.1074/jbc.M403064200. [DOI] [PubMed] [Google Scholar]
- Maiques E, Úbeda C, Campoy S, Salvador N, Lasa Í, Novick RP, Barbé J, Penadés JR. β-lactam antibiotics induce the SOS response and horizontal transfer of virulence factors in Staphylococcus aureus. J Bacteriol. 2006;188:2726–2729. doi: 10.1128/JB.188.7.2726-2729.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann A, Nehra K, Rana JS, Dahiya T. Antibiotic resistance in agriculture: Perspectives on upcoming strategies to overcome upsurge in resistance. Curr Res Microb Sci. 2021;2:100030. doi: 10.1016/j.crmicr.2021.100030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maor-Shoshani A, Reuven NB, Tomer G, Livneh Z. Highly mutagenic replication by DNA polymerase V (UmuC) provides a mechanistic basis for SOS untargeted mutagenesis. Proc Natl Acad Sci U S A. 2000;97:565–570. doi: 10.1073/pnas.97.2.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui H, Sano Y, Ishihara H, Shinomyia T. Regulation of pyocin genes in Pseudomonas aeruginosa by positive (prtN) and negative (prtR) regulatory genes. J Bacteriol. 1993;175:1257–1263. doi: 10.1128/jb.175.5.1257-1263.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazón G, Erill I, Campoy S, Cortes P, Forano E, Barbé J. Reconstruction of the evolutionary history of the LexA-binding sequence. Microbiology (Reading) 2004;150:3783–3795. doi: 10.1099/mic.0.27315-0. [DOI] [PubMed] [Google Scholar]
- McCool JD, Long E, Petrosino JF, Sandler HA, Rosenberg SM, Sandler SJ. Measurement of SOS expression in individual Escherichia coli K-12 cells using fluorescence microscopy. Mol Microbiol. 2004;53:1343–1357. doi: 10.1111/j.1365-2958.2004.04225.x. [DOI] [PubMed] [Google Scholar]
- McDonald JP, Quiros DR, Vaisman A, Mendez AR, Reyelt J, Schmidt M, Gonzalez M, Woodgate R. CroSR391, an ortholog of the λ Cro repressor, plays a major role in suppressing polVR391-dependent mutagenesis. Mol Microbiol. 2021;116:877–889. doi: 10.1111/mmi.14777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarland KA, Dolben EL, LeRoux M, Kambara TK, Ramsey KM, Kirkpatrick RL, Mougous JD, Hogan DA, Dove SL. A self-lysis pathway that enhances the virulence of a pathogenic bacterium. Proc Natl Acad Sci U S A. 2015;112:8433–8438. doi: 10.1073/pnas.1506299112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHenry CS. Breaking the rules: Bacteria that use several DNA polymerase IIIs. EMBO Rep. 2011;12:408–414. doi: 10.1038/embor.2011.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHenry CS. In: Molecular life sciences: An encyclopedic reference. 1. Wells RD, Bond JS, Klinman J, Masters BSS, editors. Springer; New York: 2018. Many bacteria use a special mutagenic pol III in place of pol V; pp. 652–655. [Google Scholar]
- McKenzie GJ, Lee PL, Lombardo MJ, Hastings PJ, Rosenberg SM. SOS mutator DNA polymerase IV functions in adaptive mutation and not adaptive amplification. Mol Cell. 2001;7:571–579. doi: 10.1016/s1097-2765(01)00204-0. [DOI] [PubMed] [Google Scholar]
- Memar MY, Ghotaslou R, Samiei M, Adibkia K. Antimicrobial use of reactive oxygen therapy: Current insights. Infect Drug Resist. 2018;11:567–576. doi: 10.2147/IDR.S142397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memar MY, Yekani M, Celenza G, Poortahmasebi V, Naghili B, Bellio P, Baghi HB. The central role of the SOS DNA repair system in antibiotics resistance: A new target for a new infectious treatment strategy. Life Sci. 2020;262:118562. doi: 10.1016/j.lfs.2020.118562. [DOI] [PubMed] [Google Scholar]
- Mérida-Floriano A, Rowe WPM, Casadesús J. Genome-wide identification and expression analysis of SOS response genes in Salmonella enterica serovar typhimurium. Cells. 2021;10:943. doi: 10.3390/cells10040943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C, Ingmer H, Thomsen LE, Skarstad K, Cohen SN. DpiA binding to the replication origin of Escherichia coli plasmids and chromosomes destabilizes plasmid inheritance and induces the bacterial SOS response. J Bacteriol. 2003;185:6025–6031. doi: 10.1128/JB.185.20.6025-6031.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C, Thomsen LE, Gaggero C, Mosseri R, Ingmer H, Cohen SN. SOS response induction by β-lactams and bacterial defense against antibiotic lethality. Science. 2004;305:1629–1631. doi: 10.1126/science.1101630. [DOI] [PubMed] [Google Scholar]
- Mo AH, Burkholder WF. YneA, an SOS-induced inhibitor of cell division in Bacillus subtilis, is regulated posttranslationally and requires the transmembrane region for activity. J Bacteriol. 2010;192:3159–3173. doi: 10.1128/JB.00027-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modell JW, Hopkins AC, Laub MT. A DNA damage checkpoint in Caulobacter crescentus inhibits cell division through a direct interaction with FtsW. Genes Dev. 2011;25:1328–1343. doi: 10.1101/gad.2038911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modell JW, Kambara TK, Perchuk BS, Laub MT. A DNA damage-induced, SOS-independent checkpoint regulates cell division in Caulobacter crescentus. PLoS Biol. 2014;12:e1001977. doi: 10.1371/journal.pbio.1001977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller AU, Imkamp F, Weber-Ban E. The mycobacterial LexA/RecA-Independent DNA damage response is controlled by PafBC and the pup-proteasome system. Cell Rep. 2018;23:3551–3564. doi: 10.1016/j.celrep.2018.05.073. [DOI] [PubMed] [Google Scholar]
- Naiman K, Philipin G, Fuchs RP, Pagès V. Chronology in lesion tolerance gives priority to genetic variability. Proc Natl Acad Sci U S A. 2014;111:5526–5531. doi: 10.1073/pnas.1321008111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napolitano R, Janel-Bintz R, Wagner J, Fuchs RP. All three SOS-inducible DNA polymerases (pol II, pol IV and pol V) are involved in induced mutagenesis. EMBO J. 2000;19:6259–6265. doi: 10.1093/emboj/19.22.6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nautiyal A, Patil KN, Muniyappa K. Suramin is a potent and selective inhibitor of Mycobacterium tuberculosis RecA protein and the SOS response: RecA as a potential target for antibacterial drug discovery. J Antimicrob Chemother. 2014;69:1834–1843. doi: 10.1093/jac/dku080. [DOI] [PubMed] [Google Scholar]
- Neher SB, Flynn JM, Sauer RT, Baker TA. Latent ClpX-recognition signals ensure LexA destruction after DNA damage. Genes Dev. 2003;17:1084–1089. doi: 10.1101/gad.1078003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohmi T, Battista JR, Dodson LA, Walker GC. RecA-mediated cleavage activates UmuD for mutagenesis: Mechanistic relationship between transcriptional derepression and posttranslational activation. Proc Natl Acad Sci U S A. 1988;85:1816–1820. doi: 10.1073/pnas.85.6.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmori H, Friedberg EC, Fuchs RP, Goodman MF, Hanaoka F, Hinkle D, Kunkel TA, Lawrence CW, Livneh Z, Nohmi T, et al. The Y-family of DNA polymerases. Mol Cell. 2001;8:7–8. doi: 10.1016/s1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- Ojha D, Patil KN. P-coumaric acid inhibits the Listeria monocytogenes RecA protein functions and SOS response: An antimicrobial target. Biochem Biophys Res Commun. 2019;517:655–661. doi: 10.1016/j.bbrc.2019.07.093. [DOI] [PubMed] [Google Scholar]
- Padan E, Schuldiner S. Intracellular pH regulation in bacterial cells. Methods Enzymol. 1986;125:337–352. doi: 10.1016/s0076-6879(86)25029-6. [DOI] [PubMed] [Google Scholar]
- Padan E, Zilberstein D, Rottenberg H. The proton electrochemical gradient in Escherichia coli cells. Eur J Biochem. 1976;63:533–541. doi: 10.1111/j.1432-1033.1976.tb10257.x. [DOI] [PubMed] [Google Scholar]
- Peña JM, Prezioso SM, McFarland KA, Kambara TK, Ramsey KM, Deighan P, Dove SL. Control of a programmed cell death pathway in Pseudomonas aeruginosa by an antiterminator. Nat Commun. 2021;12:1702. doi: 10.1038/s41467-021-21941-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennington JM, Rosenberg SM. Spontaneous DNA breakage in single living Escherichia coli cells. Nat Genet. 2007;39:797–802. doi: 10.1038/ng2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Capilla T, Baquero M-R, Gómez-Gómez J-M, Ionel A, Martín S, Blázquez J. SOS-independent induction of dinB transcription by β-lactam-mediated inhibition of cell wall synthesis in Escherichia coli. J Bacteriol. 2005;187:1515–1518. doi: 10.1128/JB.187.4.1515-1518.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry KL, Walker GC. Identification of plasmid (pKM101) coded proteins involved in mutagenesis and UV resistance. Nature. 1982;300:278–281. doi: 10.1038/300278a0. [DOI] [PubMed] [Google Scholar]
- Petrova V, Chitteni-Pattu S, Drees JC, Inman RB, Cox MM. An SOS inhibitor that binds to free RecA protein: The PsiB protein. Mol Cell. 2009;36:121–130. doi: 10.1016/j.molcel.2009.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podlesek Z, Bertok DZ. The DNA damage inducible SOS response is a key player in the generation of bacterial persister cells and population wide tolerance. Front Microbiol. 2020;11:1785. doi: 10.3389/fmicb.2020.01785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pribis JP, García-Villada L, Zhai Y, Lewin-Epstein O, Wang AZ, Liu J, Xia J, Mei Q, Fitzgerald DM, Bos J, et al. Gamblers: An antibiotic-induced evolvable cell subpopulation differentiated by reactive-oxygen-induced general stress response. Mol Cell. 2019;74:785–800. doi: 10.1016/j.molcel.2019.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Wetmore KM, Waters RJ, Callaghan M, Ray J, Liu H, Kuehl JV, Melnyk RA, Lamson JS, Suh Y, et al. Mutant phenotypes for thousands of bacterial genes of unknown function. Nature. 2018;557:503–509. doi: 10.1038/s41586-018-0124-0. [DOI] [PubMed] [Google Scholar]
- Quillardet P, Rouffaud M-A, Bouige P. DNA array analysis of gene expression in response to UV irradiation in Escherichia coli. Res Microbiol. 2003;154:559–572. doi: 10.1016/S0923-2508(03)00149-9. [DOI] [PubMed] [Google Scholar]
- Radman M. In: Molecular and environmental aspects of mutagenesis. Prakash L, editor. Charles C. Thomas; Springfield: 1974. Phenomenology of an inducible mutagenic DNA repair pathway in Escherichia coli: SOS repair hypothesis; pp. 128–142. [Google Scholar]
- Ragone S, Maman JD, Furnham N, Pellegrini L. Structural basis for inhibition of homologous recombination by the RecX protein. EMBO J. 2008;27:2259–2269. doi: 10.1038/emboj.2008.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangarajan S, Woodgate R, Goodman MF. Replication restart in UV-irradiated Escherichia coli involving pols II, III, V, Pria, Reca and RecFOR proteins. Mol Microbiol. 2002;43:617–628. doi: 10.1046/j.1365-2958.2002.02747.x. [DOI] [PubMed] [Google Scholar]
- Recacha E, Machuca J, Díaz de Alba P, Ramos-Güelfo M, Docobo-Pérez F, Rodríguez-Beltrán J, Blázquez J, Pascual A, Rodríguez-Martínez JM. Quinolone resistance reversion by targeting the SOS response. mBio. 2017;8:e00971–e00917. doi: 10.1128/mBio.00971-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuven NB, Arad G, Maor-Shoshani A, Livneh Z. The mutagenesis protein UmuC is a DNA polymerase activated by UmuD’, RecA, and SSB and is specialized for translesion replication. J Biol Chem. 1999;274:31763–31766. doi: 10.1074/jbc.274.45.31763. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Rosado AI, Valencia EY, Rodríguez-Rojas A, Costas C, Galhardo RS, Rodríguez-Beltrán J, Blázquez J. N-acetylcysteine blocks SOS induction and mutagenesis produced by fluoroquinolones in Escherichia coli. J Antimicrob Chemother. 2019;74:2188–2196. doi: 10.1093/jac/dkz210. [DOI] [PubMed] [Google Scholar]
- Salles B, Defais M. Signal of induction of recA protein in Escherichia coli. Mutat Res. 1984;131:53–59. doi: 10.1016/0167-8817(84)90011-7. [DOI] [PubMed] [Google Scholar]
- Sánchez-Osuna M, Cortés P, Lee M, Smith AT, Barbé J, Erill I. Non-canonical LexA proteins regulate the SOS response in the Bacteroidetes. Nucleic Acids Res. 2021;49:11050–11066. doi: 10.1093/nar/gkab773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassanfar M, Roberts JW. Nature of the SOS-inducing signal in Escherichia coli. J Mol Biol. 1990;212:79–96. doi: 10.1016/0022-2836(90)90306-7. [DOI] [PubMed] [Google Scholar]
- Sato JL, Fonseca DLH, Galhardo RS. rumAB genes from SXT/R391 ICEs confer UV-induced mutability to Proteus mirabilis hosts and improve conjugation after UV irradiation. DNA Repair (Amst) 2022;112:103297. doi: 10.1016/j.dnarep.2022.103297. [DOI] [PubMed] [Google Scholar]
- Savijoki K, Ingmer H, Frees D, Vogensen FK, Palva A, Varmanen P. Heat and DNA damage induction of the LexA‐like regulator HdiR from Lactococcus lactis is mediated by RecA and ClpP. Mol Microbiol. 2003;50:609–621. doi: 10.1046/j.1365-2958.2003.03713.x. [DOI] [PubMed] [Google Scholar]
- Sengupta S, Chattopadhyay MK, Grossart HP. The multifaceted roles of antibiotics and antibiotic resistance. Front Microbiol. 2013;4:47. doi: 10.3389/fmicb.2013.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng D, Wang Y, Jiang Z, Liu D, Li Y. ImuA facilitates SOS mutagenesis by inhibiting RecA-mediated activity in Myxococcus xanthus. Appl Environ Microbiol. 2021;87:e0091921. doi: 10.1128/AEM.00919-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinagawa H, Kato T, Ise T, Makino K, Nakata A. Cloning and characterization of the umu operon responsible for inducible mutagenesis in Escherichia coli. Gene. 1983;23:167–174. doi: 10.1016/0378-1119(83)90048-3. [DOI] [PubMed] [Google Scholar]
- Sikand A, Jaszczur M, Bloom LB, Woodgate R, Cox MM, Goodman MF. The SOS error-prone DNA polymerase V mutasome and β-Sliding clamp acting in concert on undamaged DNA and during translesion synthesis. Cells. 2021;10:1083. doi: 10.3390/cells10051083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons LA, Foti JJ, Cohen SE, Walker GC. The SOS regulatory network. EcoSal Plus. 2008;3:543. doi: 10.1128/ecosalplus.5.4.3. [DOI] [PubMed] [Google Scholar]
- Singletary LA, Gibson JL, Tanner EJ, McKenzie GJ, Lee PL, Gonzalez C, Rosenberg SM. An SOS-regulated type 2 toxin-antitoxin system. J Bacteriol. 2009;191:7456–7465. doi: 10.1128/JB.00963-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton MR, Dillingham MS, Gaudier M, Kowalczykowski SC, Wigley DB. Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature. 2004;432:187–193. doi: 10.1038/nature02988. [DOI] [PubMed] [Google Scholar]
- Sladewski TE, Hetrick KM, Foster PL. Escherichia coli Rep DNA helicase and error-prone DNA polymerase IV interact physically and functionally. Mol Microbiol. 2011;80:524–541. doi: 10.1111/j.1365-2958.2011.07590.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slilaty SN, Rupley JA, Little JW. Intramolecular cleavage of LexA and phage lambda repressors: Dependence of kinetics on repressor concentration, pH, temperature, and solvent. Biochemistry. 1986;25:6866–6875. doi: 10.1021/bi00370a020. [DOI] [PubMed] [Google Scholar]
- Sommer S, Bailone A, Devoret R. The appearance of the UmuD’C protein complex in Escherichia coli switches repair from homologous recombination to SOS mutagenesis. Mol Microbiol. 1993;10:963–971. doi: 10.1111/j.1365-2958.1993.tb00968.x. [DOI] [PubMed] [Google Scholar]
- Sousa FJR, Lima LMTR, Pacheco ABF, Oliveira CLP, Torriani I, Almeida DF, Foguel D, Silva JL, Mohana-Borges R. Tetramerization of the LexA repressor in solution: Implications for gene regulation of the E. coli SOS system at acidic pH. J Mol Biol. 2006;359:1059–1074. doi: 10.1016/j.jmb.2006.03.069. [DOI] [PubMed] [Google Scholar]
- Stohl EA, Brockman JP, Burkle KL, Morimatsu K, Kowalczykowski SC, Seifert HS. Escherichia coli RecX inhibits RecA recombinase and coprotease activities in vitro and in vivo. J Biol Chem. 2003;278:2278–2285. doi: 10.1074/jbc.M210496200. [DOI] [PubMed] [Google Scholar]
- Stumpf JD, Foster PL. Polyphosphate kinase regulates error-prone replication by DNA polymerase IV in Escherichia coli. Mol Microb. 2005;57:751–761. doi: 10.1111/j.1365-2958.2005.04724.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Shi J, Liu C, Jin Y, Li K, Chen R, Jin S, Wu W. PrtR homeostasis contributes to Pseudomonas aeruginosa pathogenesis and resistance against ciprofloxacin. Infect Immun. 2014;82:1638–1647. doi: 10.1128/IAI.01388-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweasy JB, Witkin EM, Sinha N, Roegner-Maniscalco V. RecA protein of Escherichia coli has a third essential role in SOS mutator activity. J Bacteriol. 1990;171:3030–3036. doi: 10.1128/jb.172.6.3030-3036.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M, Bruck I, Eritja R, Turner J, Frank EG, Woodgate R, O’Donnell M, Goodman MF. Biochemical basis of SOS mutagenesis in Escherichia coli: Reconstitution of in vitro lesion bypass dependent on the UmuD’2C mutagenic complex and RecA protein. Proc Natl Acad Sci U S A. 1998;95:9755–9760. doi: 10.1073/pnas.95.17.9755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M, Shen X, Frank EG, O’Donnell M, Woodgate R, Goodman MF. UmuD’(2)C is an error-prone DNA polymerase, Escherichia coli pol V. Proc Natl Acad Sci U S A. 1999;96:8919–8924. doi: 10.1073/pnas.96.16.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M, Pham P, Shen X, Taylor JS, O’Donnel M, Woodgate R, Goodman MF. Roles of E. coli DNA polymerases IV and V in lesion-targeted and untargeted SOS mutagenesis. Nature. 2000;404:1014–1018. doi: 10.1038/35010020. [DOI] [PubMed] [Google Scholar]
- Thi TD, López E, Rodríguez-Rojas A, Rodríguez-Beltrán J, Couce A, Guelfo JR, Castañeda-García A, Blázquez J. Effect of RecA inactivation on mutagenesis of Escherichia coli exposed to sublethal concentrations of antimicrobials. J Antimicrob Chemother. 2011;66:531–538. doi: 10.1093/jac/dkq496. [DOI] [PubMed] [Google Scholar]
- Timinskas K, Venclovas C. New insights into the structures and interactions of bacterial Y-family DNA polymerases. Nucleic Acids Res. 2019;47:4393–4405. doi: 10.1093/nar/gkz198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timinskas K, Balvociūtė M, Timinskas A, Venclovas C. Comprehensive analysis of DNA polymerase III a subunits and their homologs in bacterial genomes. Nucleic Acids Res. 2014;42:1393–1413. doi: 10.1093/nar/gkt900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touchon M, Bobay L-M, Rocha EPC. The chromosomal accommodation and domestication of mobile genetic elements. Curr Opin Microbiol. 2014;22:22–29. doi: 10.1016/j.mib.2014.09.010. [DOI] [PubMed] [Google Scholar]
- Trusca D, Scott S, Thompson C, Bramhill D. Bacterial SOS checkpoint protein SulA inhibits polymerization of purified FtsZ cell division protein. J Bacteriol. 1998;180:3946–3953. doi: 10.1128/jb.180.15.3946-3953.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang SS, Chow SA, Radding CM. Networks of DNA and recA protein are intermediates in homologous pairing. Biochemistry. 1985;24:3226–3232. doi: 10.1021/bi00334a023. [DOI] [PubMed] [Google Scholar]
- Tseng Y-C, Hung J-L, Wang T-CV. Involvement of RecF pathway recombination genes in post replication repair in UV-irradiated Escherichia coli cells. Mutat Res. 1994;315:1–9. doi: 10.1016/0921-8777(94)90021-3. [DOI] [PubMed] [Google Scholar]
- Úbeda C, Maiques E, Knecht E, Lasa Í, Novick RP, Penadés JR. Antibiotic-induced SOS response promotes horizontal dissemination of pathogenicity island-encoded virulence factors in staphylococci. Mol Microbiol. 2005;56:836–844. doi: 10.1111/j.1365-2958.2005.04584.x. [DOI] [PubMed] [Google Scholar]
- Vaisman A, McDonald JP, Woodgate R. Translesion DNA synthesis. EcoSal Plus. 2012;5:10. doi: 10.1128/ecosalplus.7.2.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia AO, Braz VS, Magalhães M, Galhardo RS. Role of error-prone DNA polymerases in spontaneous mutagenesis in Caulobacter crescentus. Genet Mol Biol. 2020;43:e20180283. doi: 10.1590/1678-4685-GMB-2018-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia EY, Esposito F, Spira B, Blázquez J, Galhardo RS. Ciprofloxacin-mediated mutagenesis is suppressed by subinhibitory concentrations of amikacin in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2017;61:e02107–e02116. doi: 10.1128/AAC.02107-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Veen S, van Schalkwijk S, Molenaar D, de Vos WM, Abee T, Wells-Bennik MHJ. The SOS response of Listeria monocytogenes is involved in stress resistance and mutagenesis. Microbiology (Reading) 2010;156:374–384. doi: 10.1099/mic.0.035196-0. [DOI] [PubMed] [Google Scholar]
- Walker GC. Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol Rev. 1984;48:60–93. doi: 10.1128/mr.48.1.60-93.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner DF, Ndwandwe DE, Abrahams GL, Kana BD, Machowski EE, Venclovas C, Mizrahi V. Essential roles for imuA’- and imuB-encoded accessory factors in DnaE2-dependent mutagenesis in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2010;107:13093–13098. doi: 10.1073/pnas.1002614107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigle JJ. Induction of mutations in a bacterial virus. Proc Natl Acad Sci U S A. 1953;39:628–636. doi: 10.1073/pnas.39.7.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigle TJ, Singleton SF. Directed molecular screening for RecA ATPase inhibitors. Bioorg Med Chem Lett. 2007;17:3249–3253. doi: 10.1016/j.bmcl.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winterling KW, Levine AS, Yasbin RE, Woodgate R. Characterization of DinR, the Bacillus subtilis SOS repressor. J Bacteriol. 1997;179:1698–1703. doi: 10.1128/jb.179.5.1698-1703.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M-K, Yang Y-C, Hsu C-H. Characterization of Xanthomonas axonopodis pv. citri LexA: Recognition of the LexA binding site. Mol Genet Genomics. 2002;268:477–487. doi: 10.1007/s00438-002-0754-6. [DOI] [PubMed] [Google Scholar]
- Yu X, Egelman EH. Structural data suggest that the active and inactive forms of the RecA filament are not simply interconvertible. J Mol Biol. 1992;227:334–346. doi: 10.1016/0022-2836(92)90702-l. [DOI] [PubMed] [Google Scholar]
- Zeng Y-H, Shen F-T, Tan C-C, Huang C-C, Young C-C. The flexibility of UV-inducible mutation in Deinococcus ficus as evidenced by the existence of the imuB-dnaE2 gene cassette and generation of superior feather degrading bacteria. Microbiol Res. 2011;167:40–47. doi: 10.1016/j.micres.2011.02.008. [DOI] [PubMed] [Google Scholar]
- Zhang APP, Pigli YZ, Rice PA. Structure of the LexA-DNA complex and implications for SOS box measurement. Nature. 2010;466:883–886. doi: 10.1038/nature09200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H. Mechanisms of mutagenesis induced by DNA lesions: Multiple factors affect mutation in translesion DNA synthesis. Crit Rev Biochem Mol Biol. 2020;55:219–251. doi: 10.1080/10409238.2020.1768205. [DOI] [PubMed] [Google Scholar]