ABSTRACT
Because of their extremely broad spectrum and strong biocidal power, nanoparticles of metals, especially silver (AgNPs), have been widely applied as effective antimicrobial agents against bacteria, fungi, and so on. However, the mutagenic effects of AgNPs and resistance mechanisms of target cells remain controversial. In this study, we discover that AgNPs do not speed up resistance mutation generation by accelerating genome-wide mutation rate of the target bacterium Escherichia coli. AgNPs-treated bacteria also show decreased expression in quorum sensing (QS), one of the major mechanisms leading to population-level drug resistance in microbes. Nonetheless, these nanomaterials are not immune to resistance development by bacteria. Gene expression analysis, experimental evolution in response to sublethal or bactericidal AgNPs treatments, and gene editing reveal that bacteria acquire resistance mainly through two-component regulatory systems, especially those involved in metal detoxification, osmoregulation, and energy metabolism. Although these findings imply low mutagenic risks of nanomaterial-based antimicrobial agents, they also highlight the capacity for bacteria to evolve resistance.
KEYWORDS: metallic nanoparticles, antimicrobial agents, drug resistance, experimental evolution, environmental mutagenesis
INTRODUCTION
Antibiotics, one of the greatest discoveries of modern biology, have been protecting countless humans from the infections of pathogenic microorganisms. In the early 1930s, bacterial infections killed nearly 300,000 annually, ~22% of total deaths in the United States (1). After the clinical application of antibiotics in the late 1930s, the mortality from severe infections, such as bacterial meningitis and endocarditis, declined by 60 to 75%, and life expectancy has thus increased significantly (2). However, antibiotics are also a double-edged sword for humans. With the misuse or abuse of antibiotics, morbidity and mortality resulting from infections with resistant microorganisms have increased dramatically, making antibiotic resistance, especially multidrug resistance, one of the most health-threatening events globally (3, 4). The declining efficacy of current treatments and the resultant increase in multidrug-resistant (MDR) pathogens constitute a serious worldwide public health problem (3, 5).
As an effective antimicrobial agent, silver has been widely used since ancient times, but its usage declined with the popularity of antibiotics (6–11). With the emergence of MDR microorganisms and the development of nanotechnology, silver nanoparticles (AgNPs) have drawn considerable attention due to their excellent antibiotic effects: AgNPs are exceptionally broad-spectrum antimicrobial agents which can eliminate pathogens ranging from viruses to bacteria to fungi (10, 12–16). Moreover, AgNPs can eradicate biofilms, which are the culprit of chronic infection and MDR strains (17, 18). AgNPs also have diverse action mechanisms, including destruction of the cell wall or membrane, destabilization of ribosomes or other functional proteins, interaction with DNA, and formation of free radicals (17, 19–23). Furthermore, AgNPs can be more stable and have long-lasting antimicrobial effects compared to traditional antibiotics, especially those with the corona layers, i.e., biological coating or stabilized with polyethylene glycol (PEG), polyvinyl pyrrolidone (PVP), and citrate, although their stability can also be highly dependent on their biological environments as well as their particle shape (e.g., spherical AgNPs are typically more stable than their anisotropic counterparts) (24–27). In particular, one exposure experiment of PVP-stabilized AgNPs demonstrated that there was no significant change in shape, aggregation, or dissolution and only a slight decrease in concentration even after 21 days (28). In contrast, most traditional antibiotics, such as ceftazidime and ceftriaxone, lose >10% of the initial concentration at room temperature after 2 to 3 days (29).
Because of their excellent antimicrobial properties, AgNPs are used for a variety of medical applications, such as wound dressings, dental materials, water filters, and so on (30–33). Global production of AgNPs is ~500 tons annually and will reach ~800 tons by 2025 (34, 35), occupying ~25% of total nanomaterial production (36, 37). With the development and application of silver nanotechnology, sublethal levels of AgNPs are widely found in aquatic environments; for example, the predicted concentration in water for 2020 is 225 to 1,799 ng/L (36). It is known that resistance development is a complicated genetic and evolutionary process involving the equilibrium between selection and mutations, and some mutagenic antimicrobial agents can also induce resistance mutations (38). The high stability and long-lasting antimicrobial effects of AgNPs exert more risks to the environment, and the environmental risks of AgNPs, such as microbial mutagenicity, remain to be evaluated. Moreover, bacteria are known to gain resistance to AgNPs through the regulation of osmotic pressure or transcriptional factors, efflux pumps, etc. (39–44), but how resistance generates and evolves remains unclear, and clinical strategies against AgNPs resistance are still lacking (41, 42).
Here, using the model bacterium Escherichia coli MG1655, we applied mutation accumulation (MA) experiments combined with deep whole-genome sequencing to study the mutagenic effects of AgNPs. During MA, hundreds of replicate lines from a single cell were repeatedly streaked for thousands of generations such that the strong population bottlenecks for each cell line minimized selection, allowing even highly deleterious mutations to accumulate and providing unbiased estimates for genome-wide mutation rates and their molecular spectra at single-nucleotide resolution. Differential gene expression analyses upon AgNPs treatments with sublethal or bactericidal doses were also performed to help reveal the action mechanisms of AgNPs. Finally, we did experimental evolution experiments and gene editing to study the emergence of resistance development mechanisms upon AgNPs treatment (Fig. 1).
FIG 1.
Experimental design.
RESULTS
We synthesized AgNPs by biological reduction and measured the particles’ size, shape, pH, zeta potential, and thermostability (Fig. 2; see Fig. S1A through H in the supplemental material). The minimum inhibitory concentration (MIC)—the lowest concentration preventing visible growth of bacteria—of the model strain Escherichia coli MG1655 to AgNPs were also quantified (Fig. 2C). Based on the MIC, 60 μg/mL of AgNPs were used for MA, transcriptome sequencing (RNA-seq), and experimental evolution experiments unless otherwise specified. We chose 60 μg/mL of AgNPs for a sublethal concentration as high as possible so that maximum selection pressure or significant gene expression could be achieved upon AgNPs treatment.
FIG 2.
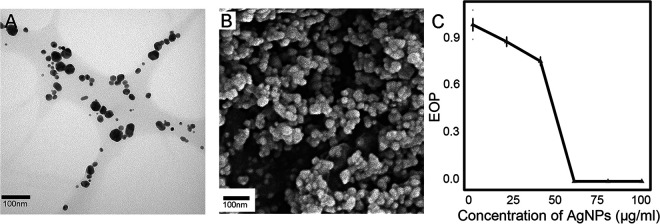
Characteristics of AgNPs in this study. (A, B) Micrographs of TEM and SEM, respectively. (C) Survival curve of E. coli under AgNPs treatment. Error bars represent standard deviation.
Characterizing AgNPs made in the lab.
Lab-made AgNPs were characterized by UV-visible (UV-vis) spectrometry, transmission electron microscopy (TEM), and scanning electron microscopy (SEM). The absorption peak of AgNPs detected by the UV-vis spectrum is at ~415 nm, which is highly consistent with previous reports (45, 46) (Fig. S1C). Because the antimicrobial potency of AgNPs is determined by size, shape, concentration, and so on (12, 47), we also measured the sizes of quasispherical AgNPs particles using the TEM micrographs in ImageJ (Fig. 2A), yielding a mean diameter of 18.21 nm (standard deviation [SD], 9.28) (Fig. S1A and B). To further evaluate the physical properties of the AgNPs nanosuspension in the culturing system, we measured the zeta potential, a critical parameter for quantifying the surface charge potential. The mean zeta potential of AgNPs is −15.27 mV (SD, 0.19), demonstrating the negative charge on the AgNPs’ surfaces as expected (Fig. S1D). We further performed the thermostability tests of the AgNPs at melted-agar temperature (60°C) by comparing the size distribution and the zeta potential of AgNPs at 25°C versus 60°C, and there was no significant difference observed; there was no significant difference in UV spectrum and zeta potential after ~24 h as well (Fig. S1B through E and G through J). The unchanged UV absorbance of the LB broth with AgNPs indicated that there was no AgNPs aggregation at least in one culturing cycle of the experiments (Fig. S1K). In addition, because pH is one of the key factors affecting mutational profiles (48), we thus measured the pH of the LB plates with and without AgNPs, finding no significant difference (Table S1; Fig. S1L, chi-square test, P ≈ 1). All of the above-described results demonstrate the regular physical properties and stability of the AgNPs in our study.
In order to determine the MIC of AgNPs in E. coli MG1655, we calculated the efficiency of plating (EOP) for the ancestor strain to estimate the survival rates under AgNPs treatments (Fig. 2C). The EOP of E. coli decreases with AgNPs concentration and drops to zero at 60 μg/mL, viz., the MIC.
AgNPs treatment dramatically changes gene expression profiles of E. coli.
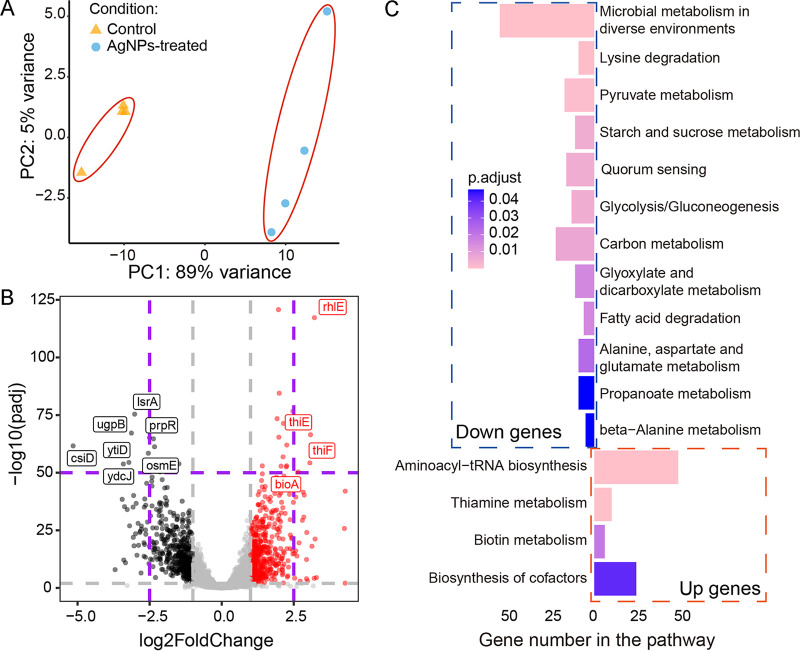
For systematically revealing the action mechanisms of AgNPs on E. coli, we applied RNA-seq on the control and the treatment and then analyzed differential gene expression (DGE) upon AgNPs treatment by using gene ontology (GO) and KEGG pathway enrichment analyses. After filtering out low-quality reads, a total of 307.85 and 314.35 million clean reads were used for the control and AgNPs treatment for downstream analysis, respectively. We then performed sample-level quality control using principal-component analysis (PCA) and heatmaps and then identified 450 upregulated and 462 downregulated genes in the AgNPs treatment compared with the control, leading to 4 and 12 KEGG pathways, respectively (Fig. 3; Fig. S2).
FIG 3.
Differential gene expression of E. coli upon AgNPs treatment. (A) PCA plot of clustering based on gene expression. (B) Differential gene expression of AgNPs versus control treatment. Red, black, and gray dots represent significantly upregulated genes, significantly downregulated genes, and not significantly differentially expressed genes, respectively. (C) Upregulated and downregulated genes from KEGG analysis. Up genes and down genes represent the upregulated and downregulated genes under AgNPs treatment versus control.
Based on KEGG analysis, AgNPs treatment substantially slowed the overall metabolism of the cells, especially the microbial metabolism in diverse environments (54 downregulated genes, including substance modification and catabolic process, energy production and transmission, and so on) and quorum-sensing (16 downregulated genes) pathways (Fig. 3C). Among them, prpR, csiD, lsrA, ugpB, osmE, ydcJ, ytiD, prpR, and csiD are the most downregulated ones (Fig. 3B). These are mostly canonical genes involved in the transcriptional regulation of propionate catabolism, catalyzing hydroxylation of glutarate (GA), and l-2-hydroxyglutarate (L2HG). Interestingly, three genes, ydcJ, osmE, and ytiD, previously not annotated to any specific functions, might be associated with certain metabolic activities upon antimicrobial agent treatment. Further explorations of their gene functions are needed. All of the above observations suggest that AgNPs may inhibit intercellular information exchange and signal sensing of target cells and potentially reduce the formation of biofilms.
In contrast, biosynthesis of the aminoacyl-tRNA synthetases and cofactors and the metabolism of thiamine and biotin were upregulated in expression, with 164 genes involved (Fig. 3B and C), and four genes, bioA, thiF, thiE, and rhlE, having the most extreme expression elevation. The first three genes are transferases involved in the catalytic biosynthesis of thiamine and biotin, such as 7,8-diaminopelargonic acid (DAPA) and thiamine monophosphate (TMP). Thiamine and biotin are two types of vitamin B, essential for converting certain nutrients into energy. rhlE is an RNA helicase involved in ribosome assembly and improving ribosome biogenesis. Further exploration is needed to determine whether the upregulation of these biological functions reflects partial compensation for the decreased overall metabolism.
The SOS response pathway is a global response to DNA damage in which DNA repair and low-fidelity DNA polymerases are induced, leading to elevation of the genome-wide mutation rate (38). Comparing the gene expression under the AgNPs treatment with the control, we did not find significant elevation in most genes of the SOS response pathway and low-fidelity DNA polymerases (Table 1; Table S2). This shows that sublethal AgNPs treatment does not induce the SOS response, further implying that AgNPs might not damage DNA. Thus, we speculate that AgNPs do not affect bacterial genome stability and may not elevate genome-wide mutation rates. To further test this, we performed the following MA experiments, combined with deep whole-genome sequencing and mutation analyses, to assess the mutagenic effects of AgNPs at single-nucleotide resolution and whole-genome scale (38).
TABLE 1.
Information about genes with high mutation hits or in the SOS response pathwaya
| Gene name | Expression change in log2 fold change (AgNPs vs control) | P value | Change | Expt(s) | Function |
|---|---|---|---|---|---|
| narU | −1.21 | 2.46 × 10−9 | Down | MAC | Nitrate assimilation |
| alaW | 0.99 | 5.10 × 10−3 | NoDiff | MAC | Aminoacyl-tRNA synthetase |
| argG | −0.13 | 1.62 × 10−1 | NoDiff | MAC | Amino acid biosynthesis |
| pheU | 2.12 | 1.09 × 10−7 | Up | MAC | Uncharacterized protein |
| cueO | 1.4 | 2.55 × 10−7 | Up | MAA | Copper efflux oxidase |
| ynbG | 0.67 | 1.42 × 10−1 | NoDiff | MAA | Uncharacterized protein |
| isrC | 0.24 | 3.41 × 10−1 | NoDiff | MAA | Acyltransferase |
| ompR | −1.04 | 1.18 × 10−25 | Down | ARS | Osmoregulation |
| arcA | −1.19 | 3.81 × 10−32 | Down | EE | Aerobic respiration control protein |
| cusS | 0.70 | 1.15 × 10−8 | NoDiff | MAA, EE, ARS | Cu2+ or Ag+ response regulation |
| cusR | 0.61 | 6.35 × 10−6 | NoDiff | MAA, EE, ARS | Cu2+ or Ag+ response regulation |
| polB | −0.37 | 3.37 × 10−3 | NoDiff | Low-fidelity DNA polymerases | |
| dinB | −0.21 | 1.07 × 10−1 | NoDiff | Low-fidelity DNA polymerases | |
| umuD | −0.64 | 3.02 × 10−3 | NoDiff | Low-fidelity DNA polymerases | |
| umuC | −0.12 | 2.39 × 10−1 | NoDiff | Low-fidelity DNA polymerases | |
| uvrA | 0.36 | 3.08 × 10−3 | NoDiff | SOS response pathway | |
| recA | 0.24 | 2.95 × 10−2 | NoDiff | SOS response pathway | |
| uvrB | −0.68 | 3.17 × 10−5 | NoDiff | SOS response pathway | |
| uvrD | 0.06 | 5.63 × 10−1 | NoDiff | SOS response pathway |
MAC, MAA, ARS, and EE represent MA-control, MA-AgNPs, acute resistance screening, and experimental evolution, respectively. NoDiff, no difference. The cutoff P value is 0.01.
Sublethal AgNPs concentrations do not alter the genomic rate or molecular spectrum of mutations in E. coli.
In total, from a single-cell ancestor, two groups of MA lines were initiated, each with 200 replicate lines, with or without AgNPs treatment. Each of the control MA lines experienced ~1,099 cell divisions on average and ~1,112 for the AgNPs treatment, respectively. Illumina PE150 genome sequencing was done for all final lines. After filtering out cross-contaminated and low-coverage lines, 142 and 172 MA lines for the control and the treatment were used in the final analyses, respectively (Tables S3 to S10). All MA lines were sequenced at a high depth of coverage and mapping rate (Tables S3 and S7).
Despite the design of the MA experiments, sublethal AgNPs treatment might bias the mutation rate and spectrum by selecting for strong resistance mutations. We thus calculated the ratio of nonsynonymous-to-synonymous substitutions but found no significant difference from the neutral expectation in either the control (Pearson’s chi-square test, χ2 = 0.48, degrees of freedom [df] = 1, P = 0.49) or the AgNPs treatment (χ2 = 5.06 × 10−20, df = 1, P = 1.00) (Table 2; Tables S4 and S8). In addition, the Ne (effective population size, the critical parameter in population genetics, defined as the equivalent number of individuals in one idealized population) of both the control and AgNPs treatment was around 14, indicating that drift dominated selection even in the AgNPs-treated MA.
TABLE 2.
Counts and proportions of different BPSs from the control and treatmenta
| Category | Count/proportion of: |
|
|---|---|---|
| Control | Treatment | |
| Intergenic regions | 24/0.23 | 19/0.19 |
| Coding region | 81/0.77 | 79/0.81 |
| Overlap | 1/0.01 | 0/0 |
| Synonymous | 17/0.21 | 20/0.25 |
| Nonsynonymous | 63/0.78 | 59/0.75 |
| Transitions | 54/0.51 | 57/0.58 |
| A:T→G:C | 19/0.35 | 23/0.40 |
| G:C→A:T | 35/0.65 | 34/0.60 |
| Transversions | 51/0.49 | 41/0.42 |
| A:T→T:A | 13/0.25 | 14/0.34 |
| A:T→C:G | 18/0.35 | 13/0.32 |
| G:C→C:G | 5/0.10 | 2/0.05 |
| G:C→T:A | 15/0.29 | 12/0.29 |
| Insertions | 7/0.50 | 7/0.44 |
| Deletions | 7/0.50 | 9/0.56 |
Count is the total number of BPSs in the MA lines; proportion refers to the proportion of the BPSs in the category out of the total BPSs in the MA lines.
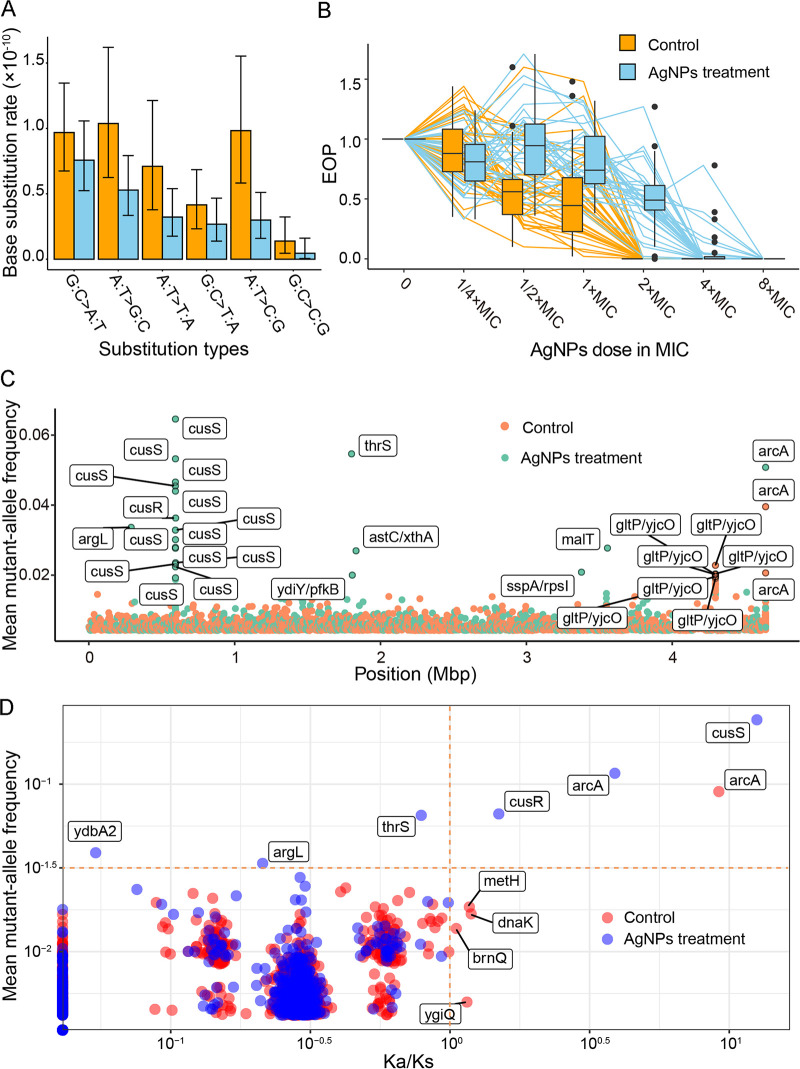
In total, 105 and 98 base pair substitutions (BPSs) were accumulated in the control and the AgNPs treatment, leading to mutation rates of 1.45 × 10−10 (95% Poisson confidence interval [CI], 1.19 × 10−10 to 1.76 × 10−10) and 1.11 × 10−10 (95% CI, 8.68 × 10−11 to 1.31 × 10−10) per nucleotide site per cell division, respectively. The mutation spectra do not differ significantly, with similar transition-to-transversion ratios (1.06 versus 1.36 in the control and AgNPs treatment, χ2 = 0.68, df = 1, P = 0.41) (Table 2; Fig. 4A). Furthermore, we also parsed out mutations at 4-fold degenerate sites, where any mutations do not change the encoded amino acids and are thus not under selection, and the mutation rates were still not significantly different between the control and the AgNPs treatment (1.32 × 10−10; 95% CI, 7.71 × 10−11 to 2.12 × 10−10, versus 1.24 × 10−10; 95% CI, 6.60 × 10−11 to 2.12 × 10−10). We also detected 14 and 16 small indels in the control and the treatment, respectively, yielding indel rates of 1.94 × 10−11 (95% CI, 1.06 × 10−11 to 3.25 × 10−11) (Table S5) and 1.81 × 10−11 (95% CI, 1.03 × 10−11 to 2.94 × 10−11) (Table S9), as well as insertion/deletion ratios of 1.00 and 0.79 (χ2 = 0, df = 1, P = 1). Thus, sublethal AgNPs concentrations do not change genome-wide mutational features of target cells.
FIG 4.
Mutation (A), resistance (B), and evolutionary patterns (C, D). (A) Mutation spectrum of MA lines from the control (orange) and AgNPs treatment (blue). (B) Survival curves of single colonies randomly chosen from experimental evolution lines. (C) Mean mutant allele frequencies of genomic sites in the control or the AgNPs-treated experimental evolution lines. The genes with the top 30 mutation frequencies are marked. Note that the symbols for identical genes (for example, cusS, arcA, and gltP/yjcO) actually represent different sites, which seemingly occur at the same site due to a genome-wide scale. (D) Mean mutant allele frequencies of genes in each group (control or AgNPs-treated) and Ka/Ks of each gene.
Rapid acquisition of resistance upon treatment with high AgNPs concentration.
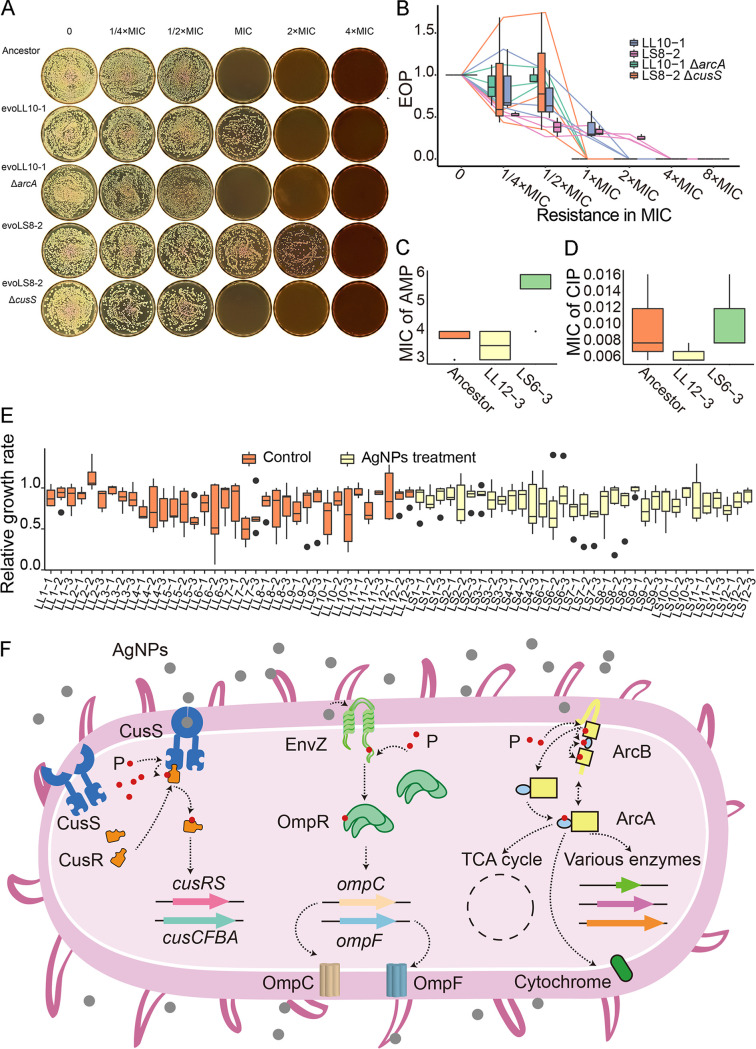
Although AgNPs do not change bacterial mutational patterns, resistance to AgNPs could arise from selection of cells carrying resistance mutations and/or other advantageous traits for clonal expansion. We then screened 75 resistant colonies, each of which was selected by treating ~1 × 105 ancestral E. coli cells at an optical density (OD) of ~1 on LB agar plates with 480 μg/mL of AgNPs (8× MIC). In total, 15 single-nucleotide polymorphism (SNP) sites were detected in three genes, cusS, cusR, and ompR (Table 1, Fig. S3A, and Table S11), belonging to two-component regulatory systems.
cusS and cusR are known to be associated with silver resistance: the Cus system is the first identified silver resistance system that can transport silver ions into the extracellular space (40, 49). ompR is one member of the two-component system EnvZ/OmpR involved in osmoregulation (50), which may decrease the infiltration of AgNPs into cells. These results demonstrate that upon acute treatment with high AgNPs concentration, populational resistance to AgNPs is mainly associated with mechanisms of silver ion resistance and/or osmoregulation.
We also iterated genes with significantly elevated mutations in the mutation accumulation lines treated with 60 μg/mL AgNPs. After the Poisson test with Bonferroni correction, five genes (cusS, cusR, cueO, ynbG, and isrC) were found to have significantly elevated mutation rates, probably selected for conferring resistance to AgNPs (Table 1; Tables S2 and S10). This confirms the above-described resistance mechanisms even at the sublethal AgNPs level.
Resistance gain of populations upon sublethal AgNPs treatment in liquid medium.
To determine whether bacteria can gain resistance at sublethal treatment over a longer time span through the above-described two-component-system resistance mutations or alternative mechanisms, with strong selection present, we initiated two groups of parallel lines cultured in liquid LB (24 in total) from a single-cell ancestor. One group of 12 lines was cultured in 100 mL LB medium with 0.1 mL transferred daily (evoLL1 to evoLL12), and the other group of 12 lines in 100 mL LB medium with 60-μg/mL AgNPs was transferred similarly (evoLS1 to evoLS12). The mean cell densities of the control and the treatment over the entire experiment were 2.75 × 109/mL and 2.82 × 109/mL (Fig. S3B), respectively (t test, P = 0.35) (density was measured by CFU every 15 transfers). The cells treated with the sublethal AgNPs concentration might take a much longer time for resistance mutations to become dominant or fixed than the above-described lethal treatment. We thus transferred each line 40 times and then plated the final evolved lines onto LB agar, randomly chose and genome sequenced three colonies from each line, and measured their MICs (Tables S12 and S13; Fig. 4B). Resistance of the final evolved lines in both the control and AgNPs treatment increased dramatically (MIC of the final control lines, 2× ancestral MIC, SD = 0; 5.5× ancestral MIC, SD = 1.94× for the AgNPs-treated final lines). Even the final evolved control lines gained resistance of 2× ancestral MIC. The resistance increase in the final evolved control was not caused by contamination of AgNPs or other lines because various precaution procedures and sufficient training of operators have been applied to avoid cross-contamination or mistakenly adding AgNPs to the control lines (see Materials and Methods for details); in addition, all 12 final AgNPs-treated experimental evolution lines accumulated cusS or cusR SNPs, while, in contrast, in the control experimental evolution lines, we did not find a single SNP in the cusS or cusR genes of any line by genome sequencing either the randomly picked colonies or the populations (Table S13; Fig. S4 and S5). For the final evolved AgNPs-treated lines, resistance ranged from about 4 to 8× the ancestral MIC. After sequencing and mutation calling of single colonies from each evolved experimental evolution line, we found that 41.67% of SNPs in the coding regions of the control lines are in arcA or arcB (6.06% in AgNPs-treated lines); 24.24% of coding-region SNPs are in cusS for the treatment (0% in control; Table S13). ArcA/ArcB is also a two-component regulatory system involved in energy generation (51).
To understand the evolution of the key genes associated with the aforementioned AgNPs resistance, we did population sequencing at ~98× and 105× depth of coverage for the control and the AgNPs treatment and analyzed mutations by using breseq (v0.35.7) at an allele frequency of >5% (Tables S14 and S15). We detected 2,000 and 1,174 variants distributed in 1,532 and 935 genes for the control and the AgNPs treatment, respectively (Tables S15 and S16). We also compared the ratio of the number of nonsynonymous substitutions per nonsynonymous site to the number of synonymous substitutions per synonymous site (Ka/Ks), which is used to evaluate selection assuming the latter is neutral (Fig. 4D; we calibrated the Ka/Ks of genes with 0 synonymous mutations with a simple regression method, shown in Fig. S3C). Positive selection appears to increase the frequency of mutations of resistance genes such as cusS, cusR, and arcA (Pearson’s product-moment correlation, r = 0.79, P = 0.21) and leads to convergent evolution of the genes in parallel lines (Fig. S4 and S5). These results confirm again the key role of the Cus system in the resistance development of AgNPs-treated bacteria. To further evaluate this hypothesis, we knocked out the gene cusS in evoLS8-2, which has a single mutation (Table S13), and found that the resistance decreased to the same level as the ancestor (Fig. 5A and B), confirming the association between the mutation and AgNPs resistance.
FIG 5.
Resistance tests and growth rates. (A, B) Tests of AgNPs resistance, given as MICs, for the ancestor, evoLL10-1, evoLL10-1 ΔarcA, evoLS8-2, and evoLS8-2 ΔcusS. (C, D) Resistance tests of strains with different genetic backgrounds, ampicillin (AMP), and ciprofloxacin (CIP). (E) Growth rates of single clones randomly chosen from the final experimental evolution lines. Growth rate measurement followed Wang et al. (112). The growth rates were assayed in the absence of AgNPs, normalized by dividing the growth rate of the ancestor. (F) Resistance mechanisms of bacteria upon AgNPs treatment, based on this and previous studies (49–51). P, phosphate group.
As shown by the above-described colony and population sequencing of both the control and the AgNPs-treated experimental evolution lines, the positively selected gene arcA has high mutation hits (Fig. 4C and D). This indicates that arcA might be pleiotropic in affecting both metabolism and AgNPs resistance. The arcA mutants were possibly selected for energy generation during clonal expansion and AgNPs resistance even in the untreated lines. Taking account of the MIC increase (2× ancestral MIC; multiple precautions were applied during transfers to avoid the control lines being contaminated by AgNPs; see details in Materials and Methods) and the high frequency of arcA mutations (48%; Table S13) in the control lines, we hypothesize that ArcA mutants elevate energy generation during growth in the untreated liquid medium and also increase MIC to compensate the reduced metabolism by AgNPs. To verify this, we knocked out the mutant arcA of evoLL10-1, a subline established from a single colony of the control experimental evolution line evoLL10 (2× ancestral MIC), and found that the resistance did decrease to the ancestral level (Fig. 5A and B). Moreover, mutant lines carrying the arcA/arcB SNPs (from single colonies isolated from the final evolved control experimental evolution lines) did show elevated median resistance (0.42 versus 0.26 for lines with or without arcA/arcB mutations, respectively, estimated by the EOP at 2× ancestral MIC; Fig. S6A), though not significantly (Wilcoxon rank-sum test, P = 0.11). All of the results indicate that arcA mutations increase the resistance to AgNPs. We further compared the growth rates of the ancestor, evoLL10-1, and evoLL10-1 ΔarcA in LB broth without AgNPs (0.58 per hour, SD = 0.02; 0.53 per hour, SD = 0.02; 0.40 per hour, SD = 0.01, respectively [Fig. S6B and C]; evoLL10-1 carries only one arcA mutation, genomic position 4,639,996 G→T, compared with the ancestor and was established from a single colony of the evolved control line evoLL10 [Table S13]). We found that the growth rate of evoLL10-1 decreases to 0.92× that of the ancestor, a direct demonstration of the fitness cost of the arcA mutation (Fig. S6B). The growth rate of evoLL10-1 ΔarcA decreases even further to ~0.70× that of the ancestor. This supports the pleiotropic effects of arcA, as removing the mutant arcA not only eliminated the resistance mutation but also other functions associated with arcA, viz., the fitness costs associated with the loss of other functions overwhelm the recovered fitness by the loss of the resistance mutation.
We also observed that when the cell density is quite high (~108/mL), the ancestral cells treated even with extremely high AgNPs concentration, i.e., 960 μg/mL or 12× ancestral MIC, did not form single colonies but a lawn. We speculate that high population density might be associated with enhanced resistance, a sign of quorum sensing. To confirm this, we chose genes with mutation frequency higher than 0.09 upon AgNPs treatment, and we performed GO analysis. Surprisingly, only one pathway, quorum sensing (QS), was enriched, which included six genes (oppF, pdeR, ydcZ, sdiA, rcsA, and ftsY) functioning in DNA binding and transcription involving dozens of mutational sites (Table S15). Thus, QS of bacteria was an important response mechanism during AgNPs treatment. Even though RNA-seq shows that AgNPs reduce QS expression (Fig. 3C), QS does play an important role in population-level resistance. Future work will address the complicated issues with respect to the associations between mutations and QS.
As shown above, AgNPs are antimicrobial agents with diverse action mechanisms. An interesting question is whether resistant bacteria developed from AgNPs treatment would also gain resistance to other antibiotics. We thus chose the subline evoLS6-3 (established from a single colony of the final evolved treated line evoLS6) with the largest resistance increase (8× ancestral MIC), the final evolved control line evoLL12-3, and the ancestral line to measure resistance to ampicillin (AMP) and ciprofloxacin (CIP), which act by inhibiting cell wall synthesis and DNA replication, respectively. We did not find any significant resistance in the AgNPs-treated line (Fig. 5C and D). Interestingly, for evoLS6-3, there are only two base substitution mutations in noncoding regions detected (Table S13). Whether these mutations increase resistance by falling into regulating motifs needs further study in the future. In addition, we also randomly chose eight single colonies from each evolved control or treated line, assayed the growth rate in the absence of AgNPs, and found no significant difference (Fig. 5E). These demonstrate that AgNPs treatment does not lead to multidrug resistance in the beta-lactam and fluoroquinolone antibiotics, possibly also the case for other antimicrobial nanomaterials, further alleviating concerns about their environmental risks.
DISCUSSION
AgNPs are long-lasting antimicrobial agents, mostly because of their high metal stability compared with traditional antibiotics. While more AgNPs products are being developed and consumed, such as daily necessities and personal items, they could pose a potential threat to human health by possibly inducing or selecting resistant pathogens. In this study, we found no mutagenic effects of the silver nanoparticles, giving direct support for the low environmental risk of the application of this genuinely broad-spectrum antimicrobial agent. The ability to reduce QS is another advantage of AgNPs over traditional antibiotics. AgNPs treatment does not trigger multidrug resistance either, another hallmark of preferred antimicrobial agents. Nonetheless, E. coli can still gain AgNPs resistance, mainly through selection for mutations in multiple two-component regulatory systems (Fig. 5F). To reveal the evolutionary dynamics in higher resolution, future experiments with bacterial populations treated for longer time spans and with intermediate time points are needed; those treated with AgNPs products in clinical scenarios would be even more interesting for studying pathogen evolution or resistance mechanisms. AgNPs are also known to catalyze reactions or oxidize chemicals, such as catalyzing the reaction of borohydride reducing various dyes (52) and oxidation of benzene (53), so whether AgNPs in these scenarios indirectly affect genome stability and then elevate the emergence of drug-resistant bacteria is worth further research.
Whether AgNPs damage DNA of target bacteria, viz., mutagenic, is highly controversial. One study shows that AgNPs can interact with the phosphate groups of DNA and cause irreversible destruction (21), and Ag(I) ions can form a duplex comprising three imidazole-Ag+ imidazole base pairs in genomic DNA (54). But others argue that DNA is not affected by AgNPs treatment in bacteria or animals (20, 55, 56). In our experimental systems, we did not detect any gene expression associated with DNA damage, such as the SOS response, upon AgNPs treatment (Tables 1 and 2; Fig. 4A). Most conspicuously, AgNPs do not accelerate the genome-wide mutation rate of bacteria, contrary to the scenario with some antibiotics, such as fluoroquinolones (38, 57, 58), a sign of low environmental risk of AgNPs when released in nature. Moreover, previous studies also showed that AgNPs-treated cells would produce extra reactive oxygen species (ROS) (59, 60), but our study does not support this: in the AgNPs treatment, most genes involved in defense against oxidative stress were not upregulated (see Table S2 in the supplemental material, “Functions” column, “Defense against oxidative stress”); we did not find a significant increase in the mutation rate of oxidative damage-associated mutations either, such as G:C→T:A transversions (Fig. 4A) (61). In addition to AgNPs, more metal nanoparticles, such as gold or copper ones, have also been applied to various aspects of our lives, for example, biomedicine, biosensing, and so on (62–64), and their environmental risks could be tested with the research framework designed in this study.
Some researchers have found that cells can gain resistance to silver nanoparticles or ions through complex mechanisms related to osmotic pressure regulation; transcription regulation factors; the sil system, including silCBA, silP, silE, and silF; and/or copper-transporting efflux systems involving ompA, ompB, ompF, and ybdE (39–41, 43, 44). In our E. coli lines treated with sublethal AgNPs in liquid LB over ~40 days, these reported resistance genes do not show elevated mutation frequency or signs of positive selection (Fig. 4D; Table S15), while the point mutations in cusS, cusR, and arcA are significantly and positively selected by sublethal AgNPs treatments, similar to the results of Graves et al. (42) and Stabryla et al. (65). These results imply that appearance of resistance mutations may be time or dose dependent. To note, there is no association between SNP number and expression level for cusS/cusR genes. This could be explained by the fact that AgNPs treatment could select cells carrying SNPs in cusS/cusR genes for resistance and these SNPs could originate from spontaneous mutations caused by replication errors, escape of the DNA repair systems, oxidation, etc., without altering the expression level. thrS is the “housekeeping” gene threonyl-tRNA synthetase, which catalyzes the attachment of threonine to tRNA (66, 67). Even though its Ka/Ks is 0.79 in the AgNPs-treated experimental evolution lines, thrS has relatively high mean mutant allele frequency (0.07), and the mutant allele frequency of one thrS site (genome coordinate of 1,801,362) even reached 0.66 in the evoLS6 population (Fig. 4D; Table S15). The results suggest that thrS could be a potential AgNPs resistance gene, and synonymous mutations might still be under certain selection. thrS may speed up protein synthesis and thereby compensate for the decreased catabolism rate caused by AgNPs, i.e., keep the protein catabolism yield by increasing the protein substrates, assuming constant stoichiometry of the protein metabolism. In addition, AgNPs treatment decreases the metabolism of target cells, as inferred by our RNA-seq data set (Fig. 3), and it is also known that slowed metabolism is associated with persistence/tolerance (68). This complicates the understanding of the action mechanisms of AgNPs on bacteria, while it is consistent with previous reports of AgNPs inhibiting the formation of biofilms, which is also associated with tolerance (68–71). Interestingly, such metabolism downregulation by AgNPs treatment also exists in various organisms besides bacteria (72–74), providing ample subjects for further research on resistance, persistence, and tolerance to AgNPs.
Quorum sensing is a biological function involving series of communication circuits for regulating gene expression in response to cell density (75), and it controls several phenotypes such as swarming migration, biofilm formation, virulence, and so on (76–78). It could cause severe chronic diseases or antibiotic resistance in important pathogens, such as Serratia liquefaciens and Vibrio cholerae (78, 79). Fortunately, AgNPs, as one agent downregulating QS (80, 81), significantly reduce the expression of 16 quorum-sensing genes based on our RNA-seq data set, which represent 66.67% of all the 24 KEGG QS genes of prokaryotes (KEGG Orthology database) (Fig. 3C). This is highly consistent with previous toxicology studies which show that AgNPs decrease the expression of virulence factors involved in QS, and they inhibit the formation of biofilms (80, 82). Some antibiotics, such as daptomycin and vancomycin prophylaxis, cannot treat infections of methicillin-resistant S. aureus alone but can work when combined with silver (83). Polymyxin B rapidly penetrates the outer cell membrane, and AgNPs can synergistically enhance the permeabilization of cells (84). These have inspired antibiotic treatment plans by combining antibiotics with AgNPs, where QS and other action mechanisms facilitate efficient and effective antimicrobial therapy (84–86). Combining antibiotics with AgNPs is thus a compelling therapeutic approach that can revive ineffective antibiotics, inhibit multiresistant bacteria, and decrease resistance acquisition (87). Unavoidably, bacteria will still evolve resistance to such therapies mainly through enzymatic inactivation, modification of antibiotic target sites, reducing uptake by changes of membrane permeability or presence of porins, and drug extrusion by efflux pumps (e.g., cusS in our study) (88). To overcome these multidrug-resistant bacterial infections, it is urgent to identify more mechanisms of resistance and combine all kinds of technological approaches, such as phage therapy and CRISPRs, in addition to nanotechnology (89). Furthermore, QS is known to be associated with multiple two-component systems (90), and the exact pathways by which AgNPs downregulate QS remain unsolved. The association of the three two-component systems (CusS/CusR, ArcA/ArcB, and EnvZ/OmpR, functioning in AgNPs resistance in this study) with QS is still unknown and worth further study. AgNPs are widely used in biomedical applications, such as implanted medical devices, to prevent implant-associated infections mainly caused by biofilm-forming microorganisms (83, 91, 92). Researchers have developed various methods to prevent infections, including chemical or physical antiadhesive resurface, bactericidal materials or innate host immune molecules, and QS quenchers (92). Applying AgNPs as such topical antiseptics should have a bright future because of their long-lasting effect, low mutagenicity, and QS inhibition, as shown in this study.
It remains a question of whether the antimicrobial effects are conferred through the silver nanoparticles and/or Ag ions. This is because the toxicology of AgNPs is highly complex, as inferred by previous studies, possibly resulting from the uncontrolled physical and chemical properties, e.g., size, shape, zeta potential, and oxidation state (93, 94). Ag ions also have a similar action mechanism to AgNPs such as interaction with thiol, induction of hydroxyl radicals, and effects on enzymes associated with the respiratory chain (95). Our study shows Ag ions’ role as one player, as two of the most significantly elevated genes, cusS and cusR, function in the transport of Cu/Ag ions, consistent with some previous reports (21, 96), while AgNPs are the main factor inhibiting QS, as shown in this and previous studies (97). Our data support that both metallic and ionic Ag are involved in the antimicrobial actions of AgNPs. Some studies have reported that the increased resistance of AgNPs originates from particle aggregations caused by flagellum-based motility (41, 65), which may be considered a shortcoming compared with Ag ions. Nevertheless, AgNPs can still inhibit the nonmotile bacteria and are effective as coated materials applied in medical devices (65). Future metal nanoparticle design needs to reduce or avoid the biologically induced aggregation of AgNPs.
In summary, all of the above observations demonstrate that AgNPs provide a compelling alternative to antibiotics and neither trigger multidrug resistance nor speed up resistance mutation generation. Moreover, AgNPs might be potentially antimutagenic, as inferred from Fig. 4A, even though there is no significant difference between the control and the AgNPs treatment due to the lack of statistical power. Despite this, strict regulations should still be applied to the production, discharge, and disposal of such metal nanomaterials because they could still efficiently select for existing resistant organisms, especially those mutated in the two-component systems mentioned above. Also, it remains a question as to whether metal nanoparticles are mutagenic to eukaryotic genomes, such as target fungal pathogens, or human consumers. A running study of ours may give one answer to this, using the fission yeast Schizosaccharomyces pombe, of which the genome is ~30% homologous to the human genome. A thorough evaluation of the mutagenicity and resistance mechanisms of various metal nanomaterials will facilitate policy-making regarding the environmental risks and usage safety of these high-tech and promising antimicrobial agents.
MATERIALS AND METHODS
Preparation and characterization of AgNPs.
To prepare the reducing solution, we followed Hebeish et al. (45). Five grams of native maize starch was dissolved in 80 mL distilled water containing 1.5 g sodium hydroxide at pH 12 and stirred by a magnetic bar at maximum speed. Until starch was dissolved completely, the silver nitrate solution (4.72 g AgNO3 dissolved in 20 mL distilled water), sterilized by filtration (Millex; catalog no. SLGP033RB), was added to the reducing solution drop by drop, and then the mixture was incubated at 70°C and stirred at maximum speed in a magnetic stirrer for 60 min. We then cooled the mixture to 25°C, added an equal volume of absolute ethanol, and rinsed the products by centrifuging at maximum speed at 25°C. We repeated the ethanol addition and centrifugation twice and air-dried the mixture.
To evaluate the stability and surface charge of AgNPs in liquid, we measured zeta potential using a size analyzer (Malvern Panalytical Zetasizer Nano ZS90). Each sample was diluted to 60 μg/mL with distilled water and measured for ~5 s three times from −100 to 100 mV. The absorption spectroscopy of reduced AgNPs (1:100 diluted by distilled water with a resistivity of 16 MΩ/cm at 25°C) was measured by a Unico UV-2100 spectrophotometer at wavelengths between 250 and 600 nm. The size and shape were measured by a Jeol transmission electron microscope (JEM-1200EX) with an acceleration voltage of 100 kV, after drying a drop of aqueous AgNPs on a carbon-coated copper TEM grid. The surface traits of nanoparticles were observed by a Carl Zeiss Merlin scanning electron microscope. We measured the pH of agar plates containing AgNPs by using a portable pH meter (Thermo Scientific Orion Star A121).
Strain, culturing, and determination of the AgNPs’ MICs for E. coli.
Escherichia coli MG1655 was from Patricia Foster’s lab, Indiana University at Bloomington. LB agar or broth (Solarbio; catalog nos. L1015 and L8291, respectively) was used for cell culturing during transferring, freezing, and DNA extraction. To determine MICs, ~1,000 cells of E. coli were incubated on LB plates with gradient concentrations of AgNPs (0, 20, 40, 60, 80, and 100 μg/mL). The efficiency of plating (EOP) was then calculated by EOP = m/N, where m is the CFU of the plate treated with AgNPs, and N is the CFU of the blank control plate without AgNPs.
MA transfers, experimental evolution, and resistance testing.
One single colony of E. coli was propagated and used as the ancestor of all of our experiments (Fig. 1). The 400 MA lines initiated were single colony transferred, and the cell divisions were estimated by CFU every 30 days. Each set of 200 lines were transferred daily on LB agar or LB agar with 60 μg/mL AgNPs at 37°C. Each MA line on LB or LB-AgNPs agar underwent 40 transfers; each transferring cycle took approximately 27.48 and 27.81 cell divisions, respectively (~1,099 and 1,112 total cell divisions for each MA line, respectively).
Acute treatment of E. coli on LB agar with 480 μg/mL AgNPs was used for selecting resistant colonies. The ancestral strain was resuscitated and serially diluted in 1× phosphate-buffered saline (PBS) buffer (10 mM). An average of 36 cells were inoculated into each of 30 test tubes and cultivated to an OD of ~1. Then, ~1 × 105 cells were plated onto LB agar containing 480 μg/mL AgNPs to select resistant bacteria. To collect mutants with diverse genotypes, we only selected one colony from one test tube and then cultured, froze, extracted DNA, and sequenced with mean depth of coverage of 78.93× (SD = 49.69) and mean mapping rate of 99.89% (SD = 0.0027). To avoid batch effect, we also performed the resistant screening at two different time points. Thirty-five and 40 resistant bacteria were selected for the first and second batches, respectively.
Twenty-four parallel populations for experimental evolution, originated from a single ancestral cell, were cultured in 100 mL LB broth in 250-mL flasks, and these included 12 control lines (evoLL1 to evoLL12) and 12 treated lines with 60 μg/mL AgNPs (evoLS1 to evoLS12). We transferred 0.1 mL of each culture into 99.9 mL of fresh medium every 24 h, and they were incubated at 37°C at 200 rpm for 40 days. The mean final cell densities of the evolved control and treatment (measured by CFU) were 2.75 × 109/mL and 2.82 × 109/mL, respectively (SDs, 0.26 × 109 and 0.20 × 109, respectively), which represent ~10 cell divisions per culturing cycle. Cross-contamination was strictly prevented by using dedicated containers, tools, operators, and so on.
Resistance tests followed Long et al. (38). Cells were cultured for 16 h to an OD at 600 nm (OD600) of ~1.5 at 37°C, and then about 1,000 cells were plated onto LB plates with AgNPs (0, 1/4×, 1/2×, 1×, 2×, 4×, and 8× MIC of AgNPs). After 24 h, the mean resistance in MIC of the three replicates for each tested line was calculated by colony counting.
Nucleic acid extraction, library construction, and sequencing.
All of the genomic DNA of E. coli from MA, mutant selection, and experimental evolution was extracted with Wizard genomic DNA purification kit (Promega, Madison, WI). The DNA libraries of E. coli were constructed using a modified protocol for TruePrep DNA library prep kit v2 for Illumina (catalog no. TD501-1; Vazyme Biotech Co., Ltd.) by following Li et al. (98), and Illumina PE150 sequencing was performed on a NovaSeq 6000 machine of Berry Genomics, Beijing.
In order to compare the differential gene expression in the control and the sublethal AgNPs treatment, colonies of E. coli were grown at the same conditions as MA: ancestor cells were streaked onto LB plates and incubated at 37°C for 16 h, and single colonies were then randomly picked and streaked onto LB plates with or without AgNPs and cultured at 37°C. Cells on each plate were then collected by rinsing with cold double-distilled water (ddH2O) and transferred into Eppendorf tubes. Four replicates were prepared for the control or the AgNPs treatment. Total RNA was then extracted by Epicentre MasterPure complete DNA and RNA purification kit (catalog no. MC85200). The concentration was quantified via a Qubit 3.0 fluorometer, and purity was measured with a microvolume spectrophotometer (Allsheng Nano-300). The cDNA library was constructed with Ribo-off rRNA depletion kit for bacteria (Vazyme; catalog no. N407-02) and VAHTS mRNA-seq V3 library prep kit for Illumina (catalog no. NR611-01), and Illumina PE150 sequencing was performed on a NovaSeq 6000 sequencer of Berry Genomics, Beijing.
Identification of mutations.
For MA lines, after trimming adaptors and filtering low-quality reads by using fastp (v0.20.0) (99), the clean reads of E. coli MA lines were mapped to the reference genome of Escherichia coli MG1655 (GenBank accession no. NC_000913.3) by using Burrows-Wheeler Aligner (v0.7.17)-MEM (100). We used the HaplotypeCaller module of Genome Analysis Toolkit (GATK v4.1.2) to call SNPs and small-scale indels using standard hard filters (101–103) and validated candidate calls by Integrative Genomics Viewer (104). The mean mutation rate (μ) was calculated by the formula , where m is the total number of mutations observed in all MA lines, n is the number of MA lines, and N and T are the total analyzed sites and number of cell divisions passed in each MA line, respectively. The standard error of the mean (SEM) of the mutation rate was calculated by the formula , where SD is the standard deviation of the line-specific mutation rates. The confidence intervals of mutation rate estimates were calculated using the Poisson cumulative distribution function approximated by the χ2 distribution. The mutations of the acutely screened resistant bacteria were called by the same method as MA lines.
For the experimental evolution, the processes of trimming and filtering raw data were the same as described above, and then the clean data were mapped to the same reference by using Bowtie2 (v2.2.5) (105). The calling of population-level mutations was performed by breseq (v0.35.7) via the –p parameter (106). Besides the three default filters, read alignments (RAs), missing coverage (MC), and new junction (NJ), we also filtered out 10 sites which were shared in all of the control and treated lines and resulted from the differences between the ancestral and reference genomes.
GO and KEGG analyses of differential gene expression.
RNA-seq data were first filtered by fastp (v0.20.0), and the clean data were mapped to the reference and converted to SAM files by HISAT2 (v2.1.0) (107). SAMtools (v1.9) was used to convert the SAM files to BAM and to sort the BAM files, and the matrix of gene expression was generated by using featureCounts (v1.6.4) (100, 108, 109). The DGE analysis was performed with the R package DESeq2, and significance was determined by adjusted P value of >0.01 and fold change of <2 (110). GO and KEGG analyses were performed with clusterProfiler (v4.0), and the filtering parameters were pAdjustMethod of BH, pvalueCutoff of 0.05, and qvalueCutoff of 0.05.
Knockout of cusS and arcA.
We followed the Red/ET recombineering method developed by Zhang et al. (111). Briefly, we transformed the plasmids pSC101 with the Red/ET recombinase into the E. coli strains by electroporation. Then, two ~50-bp flanking sequences (primers are listed in Table S17 in the supplemental material) in the gene were selected as homology arms and then ligated to the ends of the selection marker gene (chloramphenicol resistance gene) by PCR. PCR products were introduced into the cells expressing Red/ET recombinase (expression induced by rhamnose) and recombined to knockout cusS and arcA. We also confirmed that the marker gene had no effect on AgNPs resistance.
Data availability.
All Illumina sequences were uploaded to NCBI BioProject under accession no. PRJNA551791, study no. SRP222431 in SRA.
ACKNOWLEDGMENTS
This work is supported by the National Natural Science Foundation of China (31961123002, 31872228), The Fundamental Research Funds for the Central Universities of China (202041001), The Young Taishan Scholars Program of Shandong Province (tsqn201812024), and National Institutes of Health award (R35-GM122566).
We appreciate the technical help from Vazyme Biotech Co., Ltd., Nanjing, and Wei Yang, OUC. We also thank the following undergraduate students for helping with experiments: Wenlu Zhang, An Jiang, Xia Zhou, Huilin Guo, Xiang Li, Dange Zhao, Chenchen Ruan, Xiaokai Fan, Weijian Huang, Dan Liu, Hui Li, Zehua Niu, Yuanyuan Zhang, Jiao Pan, Dapeng Ran, Xiaomeng Hu, Chengjing Zhang, and Jiyuan Hou. We also thank Xue Wang and Xiaoying Bian for kindly providing plasmids for gene knockout. All bioinformatic analyses were performed with IEMB-1 computation clusters at OUC.
Kun Wu and Hongan Long designed the study; Kun Wu, Haichao Li, Yaohai Wang, Xiao Cui, Ruobing Feng, Weizhe Chen, Yuchen Jiang, and Chao Tang performed experiments and data analyses; Kun Wu, Hongan Long, Michael Lynch, Yufei Liu, Xiaopeng Shen, and Yan Wang wrote the manuscript. All authors read and approved the manuscript before submission.
We declare that we have no competing interests.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Luepke KH, Suda KJ, Boucher H, Russo RL, Bonney MW, Hunt TD, Mohr JF. 2017. Past, present, and future of antibacterial economics: increasing bacterial resistance, limited antibiotic pipeline, and societal implications. Pharmacotherapy 37:71–84. 10.1002/phar.1868. [DOI] [PubMed] [Google Scholar]
- 2.Institute of Medicine. 2010. Antibiotic resistance: implications for global health and novel intervention strategies: workshop summary. National Academies Press, Washington, DC. [PubMed] [Google Scholar]
- 3.Walker B, Barrett S, Polasky S, Galaz V, Folke C, Engström G, Ackerman F, Arrow K, Carpenter S, Chopra K, Daily G, Ehrlich P, Hughes T, Kautsky N, Levin S, Mäler K-G, Shogren J, Vincent J, Xepapadeas T, de Zeeuw A. 2009. Looming global-scale failures and missing institutions. Science 325:1345–1346. 10.1126/science.1175325. [DOI] [PubMed] [Google Scholar]
- 4.Hofer U. 2019. The cost of antimicrobial resistance. Nat Rev Microbiol 17:3. 10.1038/s41579-018-0125-x. [DOI] [PubMed] [Google Scholar]
- 5.Done HY, Halden RU. 2015. Reconnaissance of 47 antibiotics and associated microbial risks in seafood sold in the United States. J Hazard Mater 282:10–17. 10.1016/j.jhazmat.2014.08.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klasen HJ. 2000. A historical review of the use of silver in the treatment of burns. II. Renewed interest for silver. Burns 26:131–138. 10.1016/s0305-4179(99)00116-3. [DOI] [PubMed] [Google Scholar]
- 7.Castellano JJ, Shafii SM, Ko F, Donate G, Wright TE, Mannari RJ, Payne WG, Smith DJ, Robson MC. 2007. Comparative evaluation of silver-containing antimicrobial dressings and drugs. Int Wound J 4:114–122. 10.1111/j.1742-481X.2007.00316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barillo DJ, Marx DE. 2014. Silver in medicine: a brief history BC 335 to present. Burns 40(Suppl 1):S3–S8. 10.1016/j.burns.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 9.Medici S, Peana M, Nurchi VM, Zoroddu MA. 2019. Medical uses of silver: history, myths, and scientific evidence. J Med Chem 62:5923–5943. 10.1021/acs.jmedchem.8b01439. [DOI] [PubMed] [Google Scholar]
- 10.Lansdown AB. 2006. Silver in health care: antimicrobial effects and safety in use. Curr Probl Dermatol 33:17–34. 10.1159/000093928. [DOI] [PubMed] [Google Scholar]
- 11.Sim W, Barnard RT, Blaskovich MAT, Ziora ZM. 2018. Antimicrobial silver in medicinal and consumer applications: a patent review of the past decade (2007–2017). Antibiotics (Basel) 7:93. 10.3390/antibiotics7040093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pal S, Tak YK, Song JM. 2007. Does the antibacterial activity of silver nanoparticles depend on the shape of the nanoparticle? A study of the Gram-negative bacterium Escherichia coli. Appl Environ Microbiol 73:1712–1720. 10.1128/AEM.02218-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fayaz AM, Balaji K, Girilal M, Yadav R, Kalaichelvan PT, Venketesan R. 2010. Biogenic synthesis of silver nanoparticles and their synergistic effect with antibiotics: a study against gram-positive and gram-negative bacteria. Nanomedicine (Lond) 6:103–109. 10.1016/j.nano.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 14.Xiang D, Zheng C-l, Zheng Y, Li X, Yin J, O’ Conner M, Marappan M, Miao Y, Xiang B, Duan W, Shigdar S, Zhao X. 2013. Inhibition of A/Human/Hubei/3/2005 (H3N2) influenza virus infection by silver nanoparticles in vitro and in vivo. Int J Nanomed 8:4103–4113. 10.2147/IJN.S53622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim SW, Jung JH, Lamsal K, Kim YS, Min JS, Lee YS. 2012. Antifungal effects of silver nanoparticles (AgNPs) against various plant pathogenic fungi. Mycobiology 40:53–58. 10.5941/MYCO.2012.40.1.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hajipour MJ, Fromm KM, Ashkarran AA, Jimenez de Aberasturi D, de Larramendi IR, Rojo T, Serpooshan V, Parak WJ, Mahmoudi M. 2012. Antibacterial properties of nanoparticles. Trends Biotechnol 30:499–511. 10.1016/j.tibtech.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 17.Lazar V. 2011. Quorum sensing in biofilms–how to destroy the bacterial citadels or their cohesion/power? Anaerobe 17:280–285. 10.1016/j.anaerobe.2011.03.023. [DOI] [PubMed] [Google Scholar]
- 18.Khan MR, Fromm KM, Rizvi TF, Giese B, Ahamad F, Turner RJ, Füeg M, Marsili E. 2020. Metal nanoparticle–microbe interactions: synthesis and antimicrobial effects. Part Syst Charact 37:1900419. 10.1002/ppsc.201900419. [DOI] [Google Scholar]
- 19.Yamanaka M, Hara K, Kudo J. 2005. Bactericidal actions of a silver ion solution on Escherichia coli, studied by energy-filtering transmission electron microscopy and proteomic analysis. Appl Environ Microbiol 71:7589–7593. 10.1128/AEM.71.11.7589-7593.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hwang ET, Lee JH, Chae YJ, Kim YS, Kim BC, Sang B-I, Gu MB. 2008. Analysis of the toxic mode of action of silver nanoparticles using stress-specific bioluminescent bacteria. Small 4:746–750. 10.1002/smll.200700954. [DOI] [PubMed] [Google Scholar]
- 21.Morones JR, Elechiguerra JL, Camacho A, Holt K, Kouri JB, Ramírez JT, Yacaman MJ. 2005. The bactericidal effect of silver nanoparticles. Nanotechnology 16:2346–2353. 10.1088/0957-4484/16/10/059. [DOI] [PubMed] [Google Scholar]
- 22.Eckhardt S, Brunetto PS, Gagnon J, Priebe M, Giese B, Fromm KM. 2013. Nanobio silver: its interactions with peptides and bacteria, and its uses in medicine. Chem Rev 113:4708–4754. 10.1021/cr300288v. [DOI] [PubMed] [Google Scholar]
- 23.Abram SL, Fromm KM. 2020. Handling (nano)silver as antimicrobial agent: therapeutic window, dissolution dynamics, detection methods and molecular interactions. Chemistry 26:10948–10971. 10.1002/chem.202002143. [DOI] [PubMed] [Google Scholar]
- 24.Singh P, Mijakovic I. 2022. Antibacterial effect of silver nanoparticles is stronger if the production host and the targeted pathogen are closely related. Biomedicines 10:628. 10.3390/biomedicines10030628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pinzaru I, Coricovac D, Dehelean C, Moacă E-A, Mioc M, Baderca F, Sizemore I, Brittle S, Marti D, Calina CD, Tsatsakis AM, Şoica C. 2018. Stable PEG-coated silver nanoparticles–a comprehensive toxicological profile. Food Chem Toxicol 111:546–556. 10.1016/j.fct.2017.11.051. [DOI] [PubMed] [Google Scholar]
- 26.Ronavari A, Belteky P, Boka E, Zakupszky D, Igaz N, Szerencses B, Pfeiffer I, Kónya Z, Kiricsi M. 2021. Polyvinyl-pyrrolidone-coated silver nanoparticles-the colloidal, chemical, and biological consequences of steric stabilization under biorelevant conditions. Int J Mol Sci 22:8673. 10.3390/ijms22168673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park K, Tuttle G, Sinche F, Harper SL. 2013. Stability of citrate-capped silver nanoparticles in exposure media and their effects on the development of embryonic zebrafish (Danio rerio). Arch Pharm Res 36:125–133. 10.1007/s12272-013-0005-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tejamaya M, Romer I, Merrifield RC, Lead JR. 2012. Stability of citrate, PVP, and PEG coated silver nanoparticles in ecotoxicology media. Environ Sci Technol 46:7011–7017. 10.1021/es2038596. [DOI] [PubMed] [Google Scholar]
- 29.Walker SE, Iazzetta J, Law S, Biniecki K. 2010. Stability of commonly used antibiotic solutions in an elastomeric infusion device. Can J Hosp Pharm 63:212–224. 10.4212/cjhp.v63i3.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang XF, Liu ZG, Shen W, Gurunathan S. 2016. Silver nanoparticles: synthesis, characterization, properties, applications, and therapeutic approaches. Int J Mol Sci 17:1534. 10.3390/ijms17091534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chaloupka K, Malam Y, Seifalian AM. 2010. Nanosilver as a new generation of nanoproduct in biomedical applications. Trends Biotechnol 28:580–588. 10.1016/j.tibtech.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 32.Correa JM, Mori M, Sanches HL, da Cruz AD, Poiate E, Jr., Poiate IA. 2015. Silver nanoparticles in dental biomaterials. Int J Biomater 2015:485275. 10.1155/2015/485275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Das SK, Khan MM, Guha AK, Das AR, Mandal AB. 2012. Silver-nano biohybride material: synthesis, characterization and application in water purification. Bioresour Technol 124:495–499. 10.1016/j.biortech.2012.08.071. [DOI] [PubMed] [Google Scholar]
- 34.Mueller NC, Nowack B. 2008. Exposure modeling of engineered nanoparticles in the environment. Environ Sci Technol 42:4447–4453. 10.1021/es7029637. [DOI] [PubMed] [Google Scholar]
- 35.Pulit-Prociak J, Banach M. 2016. Silver nanoparticles–a material of the future…? Open Chem 14:76–91. 10.1515/chem-2016-0005. [DOI] [Google Scholar]
- 36.Massarsky A, Trudeau VL, Moon TW. 2014. Predicting the environmental impact of nanosilver. Environ Toxicol Pharmacol 38:861–873. 10.1016/j.etap.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 37.Vance ME, Kuiken T, Vejerano EP, McGinnis SP, Hochella MF, Rejeski D, Hull MS. 2015. Nanotechnology in the real world: redeveloping the nanomaterial consumer products inventory. Beilstein J Nanotechnol 6:1769–1780. 10.3762/bjnano.6.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Long H, Miller SF, Strauss C, Zhao C, Cheng L, Ye Z, Griffin K, Te R, Lee H, Chen C-C, Lynch M. 2016. Antibiotic treatment enhances the genome-wide mutation rate of target cells. Proc Natl Acad Sci USA 113:E2498–E2505. 10.1073/pnas.1601208113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Franke S, Grass G, Nies DH. 2001. The product of the ybdE gene of the Escherichia coli chromosome is involved in detoxification of silver ions. Microbiology (Reading) 147:965–972. 10.1099/00221287-147-4-965. [DOI] [PubMed] [Google Scholar]
- 40.Franke S, Grass G, Rensing C, Nies DH. 2003. Molecular analysis of the copper-transporting efflux system CusCFBA of Escherichia coli. J Bacteriol 185:3804–3812. 10.1128/JB.185.13.3804-3812.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Panáček A, Kvítek L, Smékalová M, Večeřová R, Kolář M, Röderová M, Dyčka F, Šebela M, Prucek R, Tomanec O, Zbořil R. 2018. Bacterial resistance to silver nanoparticles and how to overcome it. Nat Nanotechnol 13:65–71. 10.1038/s41565-017-0013-y. [DOI] [PubMed] [Google Scholar]
- 42.Graves JL, Tajkarimi M, Cunningham Q, Campbell A, Nonga H, Harrison SH, Barrick JE. 2015. Rapid evolution of silver nanoparticle resistance in Escherichia coli. Front Genet 6:42. 10.3389/fgene.2015.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gupta A, Matsui K, Lo JF, Silver S. 1999. Molecular basis for resistance to silver cations in Salmonella. Nat Med 5:183–188. 10.1038/5545. [DOI] [PubMed] [Google Scholar]
- 44.Silver S. 2003. Bacterial silver resistance: molecular biology and uses and misuses of silver compounds. FEMS Microbiol Rev 27:341–353. 10.1016/S0168-6445(03)00047-0. [DOI] [PubMed] [Google Scholar]
- 45.Hebeish A, El-Rafie MH, El-Sheikh MA, El-Naggar ME. 2013. Nanostructural features of silver nanoparticles powder synthesized through concurrent formation of the nanosized particles of both starch and silver. J Nanotechnol 2013:1–10. 10.1155/2013/201057. [DOI] [Google Scholar]
- 46.Abd El-Kader FH, Hakeem NA, Elashmawi IS, Menazea AA. 2015. Synthesis and characterization of PVK/AgNPs nanocomposites prepared by laser ablation. Spectrochim Acta A Mol Biomol Spectrosc 138:331–339. 10.1016/j.saa.2014.11.083. [DOI] [PubMed] [Google Scholar]
- 47.Franci G, Falanga A, Galdiero S, Palomba L, Rai M, Morelli G, Galdiero M. 2015. Silver nanoparticles as potential antibacterial agents. Molecules 20:8856–8874. 10.3390/molecules20058856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Strauss C, Long H, Patterson CE, Te R, Lynch M. 2017. Genome-wide mutation rate response to pH change in the coral reef pathogen Vibrio shilonii AK1. mBio 8:e01021-17. 10.1128/mBio.01021-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fu B, Sengupta K, Genova LA, Santiago AG, Jung W, Krzemiński Ł, Chakraborty UK, Zhang W, Chen P. 2020. Metal-induced sensor mobilization turns on affinity to activate regulator for metal detoxification in live bacteria. Proc Natl Acad Sci USA 117:13248–13255. 10.1073/pnas.1919816117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Motz M, Jung K. 2018. The role of polyproline motifs in the histidine kinase EnvZ. PLoS One 13:e0199782. 10.1371/journal.pone.0199782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brynildsen MP, Liao JC. 2009. An integrated network approach identifies the isobutanol response network of Escherichia coli. Mol Syst Biol 5:277. 10.1038/msb.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang ZJ, Liu CY, Sun LW. 2005. Catalytic properties of silver nanoparticles supported on silica spheres. J Phys Chem B 109:1730–1735. 10.1021/jp046032g. [DOI] [PubMed] [Google Scholar]
- 53.Balkis Ameen K, Rajasekar K, Rajasekharan T. 2007. Silver nanoparticles in mesoporous aerogel exhibiting selective catalytic oxidation of benzene in CO2 free air. Catal Lett 119:289–295. 10.1007/s10562-007-9233-3. [DOI] [Google Scholar]
- 54.Johannsen S, Megger N, Bohme D, Sigel RK, Muller J. 2010. Solution structure of a DNA double helix with consecutive metal-mediated base pairs. Nat Chem 2:229–234. 10.1038/nchem.512. [DOI] [PubMed] [Google Scholar]
- 55.Kim YS, Kim JS, Cho HS, Rha DS, Kim JM, Park JD, Choi BS, Lim R, Chang HK, Chung YH, Kwon IH, Jeong J, Han BS, Yu IJ. 2008. Twenty-eight-day oral toxicity, genotoxicity, and gender-related tissue distribution of silver nanoparticles in Sprague-Dawley rats. Inhal Toxicol 20:575–583. 10.1080/08958370701874663. [DOI] [PubMed] [Google Scholar]
- 56.Li Y, Bhalli JA, Ding W, Yan J, Pearce MG, Sadiq R, Cunningham CK, Jones MY, Monroe WA, Howard PC, Zhou T, Chen T. 2014. Cytotoxicity and genotoxicity assessment of silver nanoparticles in mouse. Nanotoxicology 8(Suppl 1):36–45. 10.3109/17435390.2013.855827. [DOI] [PubMed] [Google Scholar]
- 57.Song LY, Goff M, Davidian C, Mao Z, London M, Lam K, Yung M, Miller JH. 2016. Mutational consequences of ciprofloxacin in Escherichia coli. Antimicrob Agents Chemother 60:6165–6172. 10.1128/AAC.01415-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ozdemirel HO, Ulusal D, Kucukyildirim Celik S. 2021. Streptomycin and nalidixic acid elevate the spontaneous genome-wide mutation rate in Escherichia coli. Genetica 149:73–80. 10.1007/s10709-021-00114-w. [DOI] [PubMed] [Google Scholar]
- 59.Dakal TC, Kumar A, Majumdar RS, Yadav V. 2016. Mechanistic basis of antimicrobial actions of silver nanoparticles. Front Microbiol 7:1831. 10.3389/fmicb.2016.01831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Joshi N, Ngwenya BT, Butler IB, French CE. 2015. Use of bioreporters and deletion mutants reveals ionic silver and ROS to be equally important in silver nanotoxicity. J Hazard Mater 287:51–58. 10.1016/j.jhazmat.2014.12.066. [DOI] [PubMed] [Google Scholar]
- 61.Lynch M, Walsh B. 2007. The origins of genome architecture, vol 98. Sinauer Associates, Sunderland, MA. [Google Scholar]
- 62.Verma A, Uzun O, Hu Y, Hu Y, Han H-S, Watson N, Chen S, Irvine DJ, Stellacci F. 2008. Surface-structure-regulated cell-membrane penetration by monolayer-protected nanoparticles. Nat Mater 7:588–595. 10.1038/nmat2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arvizo RR, Miranda OR, Thompson MA, Pabelick CM, Bhattacharya R, Robertson JD, Rotello VM, Prakash YS, Mukherjee P. 2010. Effect of nanoparticle surface charge at the plasma membrane and beyond. Nano Lett 10:2543–2548. 10.1021/nl101140t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu Q, Zhao Y, Xu JZ, Zhu J-J. 2006. Preparation of functionalized copper nanoparticles and fabrication of a glucose sensor. Sens Actuators B Chem 114:379–386. 10.1016/j.snb.2005.06.005. [DOI] [Google Scholar]
- 65.Stabryla LM, Johnston KA, Diemler NA, Cooper VS, Millstone JE, Haig S-J, Gilbertson LM. 2021. Role of bacterial motility in differential resistance mechanisms of silver nanoparticles and silver ions. Nat Nanotechnol 16:996–1003. 10.1038/s41565-021-00929-w. [DOI] [PubMed] [Google Scholar]
- 66.Springer M, Graffe M, Butler JS, Grunberg-Manago M. 1986. Genetic definition of the translational operator of the threonine-tRNA ligase gene in Escherichia coli. Proc Natl Acad Sci USA 83:4384–4388. 10.1073/pnas.83.12.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Condon C, Putzer H, Grunberg-Manago M. 1996. Processing of the leader mRNA plays a major role in the induction of thrS expression following threonine starvation in Bacillus subtilis. Proc Natl Acad Sci USA 93:6992–6997. 10.1073/pnas.93.14.6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Balaban NQ, Helaine S, Lewis K, Ackermann M, Aldridge B, Andersson DI, Brynildsen MP, Bumann D, Camilli A, Collins JJ, Dehio C, Fortune S, Ghigo J-M, Hardt W-D, Harms A, Heinemann M, Hung DT, Jenal U, Levin BR, Michiels J, Storz G, Tan M-W, Tenson T, Van Melderen L, Zinkernagel A. 2019. Definitions and guidelines for research on antibiotic persistence. Nat Rev Microbiol 17:441–448. 10.1038/s41579-019-0196-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ciofu O, Moser C, Jensen PO, Hoiby N. 2022. Tolerance and resistance of microbial biofilms. Nat Rev Microbiol 10.1038/s41579-022-00682-4. [DOI] [PubMed] [Google Scholar]
- 70.Martinez-Gutierrez F, Boegli L, Agostinho A, Sánchez EM, Bach H, Ruiz F, James G. 2013. Anti-biofilm activity of silver nanoparticles against different microorganisms. Biofouling 29:651–660. 10.1080/08927014.2013.794225. [DOI] [PubMed] [Google Scholar]
- 71.Ansari MA, Khan HM, Khan AA, Cameotra SS, Saquib Q, Musarrat J. 2014. Gum arabic capped-silver nanoparticles inhibit biofilm formation by multi-drug resistant strains of Pseudomonas aeruginosa. J Basic Microbiol 54:688–699. 10.1002/jobm.201300748. [DOI] [PubMed] [Google Scholar]
- 72.Pietrzak K, Twarużek M, Czyżowska A, Kosicki R, Gutarowska B. 2015. Influence of silver nanoparticles on metabolism and toxicity of moulds. Acta Biochim Pol 62:851–857. 10.18388/abp.2015_1146. [DOI] [PubMed] [Google Scholar]
- 73.Ke M, Qu Q, Peijnenburg WJGM, Li X, Zhang M, Zhang Z, Lu T, Pan X, Qian H. 2018. Phytotoxic effects of silver nanoparticles and silver ions to Arabidopsis thaliana as revealed by analysis of molecular responses and of metabolic pathways. Sci Total Environ 644:1070–1079. 10.1016/j.scitotenv.2018.07.061. [DOI] [PubMed] [Google Scholar]
- 74.He S, Feng Y, Ni J, Sun Y, Xue L, Feng Y, Yu Y, Lin X, Yang L. 2016. Different responses of soil microbial metabolic activity to silver and iron oxide nanoparticles. Chemosphere 147:195–202. 10.1016/j.chemosphere.2015.12.055. [DOI] [PubMed] [Google Scholar]
- 75.Miller MB, Bassler BL. 2001. Quorum sensing in bacteria. Annu Rev Microbiol 55:165–199. 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- 76.Rutherford ST, Bassler BL. 2012. Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb Perspect Med 2:a012427. 10.1101/cshperspect.a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Daniels R, Vanderleyden J, Michiels J. 2004. Quorum sensing and swarming migration in bacteria. FEMS Microbiol Rev 28:261–289. 10.1016/j.femsre.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 78.Hammer BK, Bassler BL. 2003. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol Microbiol 50:101–104. 10.1046/j.1365-2958.2003.03688.x. [DOI] [PubMed] [Google Scholar]
- 79.Labbate M, Queck SY, Koh KS, Rice SA, Givskov M, Kjelleberg S. 2004. Quorum sensing-controlled biofilm development in Serratia liquefaciens MG1. J Bacteriol 186:692–698. 10.1128/JB.186.3.692-698.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shah S, Gaikwad S, Nagar S, Kulshrestha S, Vaidya V, Nawani N, Pawar S. 2019. Biofilm inhibition and anti-quorum sensing activity of phytosynthesized silver nanoparticles against the nosocomial pathogen Pseudomonas aeruginosa. Biofouling 35:34–49. 10.1080/08927014.2018.1563686. [DOI] [PubMed] [Google Scholar]
- 81.Gopalakrishnan V, Masanam E, Ramkumar VS, Baskaraligam V, Selvaraj G. 2020. Influence of N-acylhomoserine lactonase silver nanoparticles on the quorum sensing system of Helicobacter pylori: a potential strategy to combat biofilm formation. J Basic Microbiol 60:207–215. 10.1002/jobm.201900537. [DOI] [PubMed] [Google Scholar]
- 82.Qais FA, Shafiq A, Ahmad I, Husain FM, Khan RA, Hassan I. 2020. Green synthesis of silver nanoparticles using Carum copticum: assessment of its quorum sensing and biofilm inhibitory potential against gram negative bacterial pathogens. Microb Pathog 144:104172. 10.1016/j.micpath.2020.104172. [DOI] [PubMed] [Google Scholar]
- 83.Kuehl R, Brunetto PS, Woischnig A-K, Varisco M, Rajacic Z, Vosbeck J, Terracciano L, Fromm KM, Khanna N. 2016. Preventing implant-associated infections by silver coating. Antimicrob Agents Chemother 60:2467–2475. 10.1128/AAC.02934-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wan G, Ruan L, Yin Y, Yang T, Ge M, Cheng X. 2016. Effects of silver nanoparticles in combination with antibiotics on the resistant bacteria Acinetobacter baumannii. Int J Nanomedicine (Lond) 11:3789–3800. 10.2147/IJN.S104166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shahverdi AR, Fakhimi A, Shahverdi HR, Minaian S. 2007. Synthesis and effect of silver nanoparticles on the antibacterial activity of different antibiotics against Staphylococcus aureus and Escherichia coli. Nanomedicine (Lond) 3:168–171. 10.1016/j.nano.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 86.Vazquez-Muñoz R, Meza-Villezcas A, Fournier PGJ, Soria-Castro E, Juarez-Moreno K, Gallego-Hernández AL, Bogdanchikova N, Vazquez-Duhalt R, Huerta-Saquero A. 2019. Enhancement of antibiotics antimicrobial activity due to the silver nanoparticles impact on the cell membrane. PLoS One 14:e0224904. 10.1371/journal.pone.0224904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Worthington RJ, Melander C. 2013. Combination approaches to combat multidrug-resistant bacteria. Trends Biotechnol 31:177–184. 10.1016/j.tibtech.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Annunziato G. 2019. Strategies to overcome antimicrobial resistance (AMR) making use of non-essential target inhibitors: a review. Int J Mol Sci 20:5844. 10.3390/ijms20235844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lima R, Del Fiol FS, Balcao VM. 2019. Prospects for the use of new technologies to combat multidrug-resistant bacteria. Front Pharmacol 10:692. 10.3389/fphar.2019.00692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cui C, Yang C, Song S, Fu S, Sun X, Yang L, He F, Zhang L-H, Zhang Y, Deng Y. 2018. A novel two-component system modulates quorum sensing and pathogenicity in Burkholderia cenocepacia. Mol Microbiol 108:32–44. 10.1111/mmi.13915. [DOI] [PubMed] [Google Scholar]
- 91.Sendi P, Banderet F, Graber P, Zimmerli W. 2011. Periprosthetic joint infection following Staphylococcus aureus bacteremia. J Infect 63:17–22. 10.1016/j.jinf.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 92.Campoccia D, Montanaro L, Arciola CR. 2013. A review of the biomaterials technologies for infection-resistant surfaces. Biomaterials 34:8533–8554. 10.1016/j.biomaterials.2013.07.089. [DOI] [PubMed] [Google Scholar]
- 93.Kedziora A, Speruda M, Krzyzewska E, Rybka J, Lukowiak A, Bugla-Ploskonska G. 2018. Similarities and differences between silver ions and silver in nanoforms as antibacterial agents. Int J Mol Sci 19:444. 10.3390/ijms19020444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gliga AR, Skoglund S, Wallinder IO, Fadeel B, Karlsson HL. 2014. Size-dependent cytotoxicity of silver nanoparticles in human lung cells: the role of cellular uptake, agglomeration and Ag release. Part Fibre Toxicol 11:11. 10.1186/1743-8977-11-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gordon O, Vig Slenters T, Brunetto PS, Villaruz AE, Sturdevant DE, Otto M, Landmann R, Fromm KM. 2010. Silver coordination polymers for prevention of implant infection: thiol interaction, impact on respiratory chain enzymes, and hydroxyl radical induction. Antimicrob Agents Chemother 54:4208–4218. 10.1128/AAC.01830-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xiu ZM, Ma J, Alvarez PJ. 2011. Differential effect of common ligands and molecular oxygen on antimicrobial activity of silver nanoparticles versus silver ions. Environ Sci Technol 45:9003–9008. 10.1021/es201918f. [DOI] [PubMed] [Google Scholar]
- 97.Radzig MA, Koksharova OA, Khmel’ IA. 2009. Antibacterial effects of silver ions on growth of gram-negative bacteria and biofilm formation. Mol Genet Microbiol Virol 24:194–199. 10.3103/S0891416809040065. [DOI] [PubMed] [Google Scholar]
- 98.Li H, Wu K, Ruan C, Pan J, Wang Y, Long H. 2019. Cost-reduction strategies in massive genomics experiments. Mar Life Sci Technol 1:15–21. 10.1007/s42995-019-00013-2. [DOI] [Google Scholar]
- 99.Chen S, Zhou Y, Chen Y, Gu J. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34:i884–i890. 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25:1754–1760. 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43:491–498. 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A. 2013. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 43:11 10 11–11 10 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Thorvaldsdottir H, Robinson JT, Mesirov JP. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14:178–192. 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Deatherage DE, Barrick JE. 2014. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol Biol 1151:165–188. 10.1007/978-1-4939-0554-6_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37:907–915. 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 109.Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM, Li H. 2021. Twelve years of SAMtools and BCFtools. Gigascience 10:giab008. 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhang Y, Buchholz F, Muyrers JP, Stewart AF. 1998. A new logic for DNA engineering using recombination in Escherichia coli. Nat Genet 20:123–128. 10.1038/2417. [DOI] [PubMed] [Google Scholar]
- 112.Wang Y, Arenas CD, Stoebel DM, Cooper TF. 2013. Genetic background affects epistatic interactions between two beneficial mutations. Biol Lett 9:20120328. 10.1098/rsbl.2012.0328. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental methods and Fig. S1 to S6. Download aac.00628-22-s0001.pdf, PDF file, 1.6 MB (1.7MB, pdf)
Tables S1 to S17. Download aac.00628-22-s0002.xlsx, XLSX file, 1.3 MB (1.3MB, xlsx)
Data Availability Statement
All Illumina sequences were uploaded to NCBI BioProject under accession no. PRJNA551791, study no. SRP222431 in SRA.