Abstract
The fungal pathogen Histoplasma capsulatum (Hc) invades, replicates within, and destroys macrophages. To interrogate the molecular mechanisms underlying this interaction, we conducted a host-directed CRISPR-Cas9 screen and identified 361 genes that modify macrophage susceptibility to Hc infection, greatly expanding our understanding of host gene networks targeted by Hc. We identified pathways that have not been previously implicated in Hc interaction with macrophages, including the ragulator complex (involved in nutrient stress sensing), glycosylation enzymes, protein degradation machinery, mitochondrial respiration genes, solute transporters, and the ER membrane complex (EMC). The highest scoring protective hits included the complement C3a receptor (C3aR), a G-protein coupled receptor (GPCR) that recognizes the complement fragment C3a. Although it is known that complement components react with the fungal surface, leading to opsonization and release of small peptide fragments such as C3a, a role for C3aR in macrophage interactions with fungi has not been elucidated. We demonstrated that whereas C3aR is dispensable for macrophage phagocytosis of bacteria and latex beads, it is critical for optimal macrophage capture of pathogenic fungi, including Hc, the ubiquitous fungal pathogen Candida albicans, and the causative agent of Valley Fever Coccidioides posadasii. We showed that C3aR localizes to the early phagosome during Hc infection where it coordinates the formation of actin-rich membrane protrusions that promote Hc capture. We also showed that the EMC promotes surface expression of C3aR, likely explaining its identification in our screen. Taken together, our results provide new insight into host processes that affect Hc-macrophage interactions and uncover a novel and specific role for C3aR in macrophage recognition of fungi.
Author summary
Invasive fungal infections, including those caused by Histoplasma capsulatum (Hc), cause widespread disease and death in humans. Innate immune cells, such as macrophages, are important for the early detection and clearance of invading fungi, but can also act as host cells that support intracellular fungal replication. A better understanding of how macrophages recognize and ingest these fungi is critical for a complete appreciation of disease progression. To elucidate macrophage-fungal interactions, we performed a genome-scale genetic screen to identify host genes that influence macrophage survival after infection with Hc. We discovered a subset of genes that are important for macrophage capture of Hc, including a receptor, C3aR, that recognizes C3a, a cleavage product of serum complement component C3. We determined that C3aR is required for efficient capture of Hc and other fungi by macrophages, but dispensable for phagocytosis of non-fungal particles such as bacteria and latex beads. Our study uncovers a new role for C3aR in innate immune capture of pathogenic fungi such as Hc.
Introduction
The fungus Histoplasma capsulatum (Hc) is an intracellular pathogen of macrophages. Infection with Hc occurs when soil containing Hc spores or hyphal fragments is aerosolized and fungal particles are inhaled by a mammalian host [1]. In the lung, Hc invades alveolar macrophages [2,3], replicates to high intracellular levels and induces macrophage lysis [4,5]. Though many of the molecular mechanisms underpinning Hc pathogenesis are unknown, a number of Hc genes that promote immune evasion and virulence have been identified [6–10].
The initial step in Hc-macrophage interactions is phagocytosis. In general, macrophage-expressed pattern-recognition-receptors can directly bind common fungal cell-wall components [11] such as the cell-wall sugar β-glucan, which is recognized by the receptor Dectin-1 [12,13]. Engagement of phagocytosis receptors, such as Dectin-1, triggers a complex cascade of intracellular signaling events, involving small GTPase activation, membrane phospholipid remodeling, and actin cytoskeleton polymerization that allow the plasma membrane to deform and encircle the targeted particle [14,15]. Following phagocytosis, the particle is enclosed within a membrane structure termed the phagosome. Macrophage phagocytosis of Hc, unlike that of other fungi, is not dependent on β-glucan recognition by Dectin-1 [16]. Hc can prevent such recognition by shielding cell-wall β-glucan with a layer of α-glucan [17] or by secreting glucanases to prune β-glucans [18]. Instead, Hc recognition and phagocytosis is directly mediated by β2 integrin receptors [16,19] formed through dimerization of CD18 (Itgb2) with various α subunits [20], such as CD11b, a subunit of complement receptor 3 (CR3), the best studied of these receptors.
Innate immune recognition of pathogens is supported by opsonins, such as antibodies, which are recognized by Fcγ receptors expressed on macrophages, and the complement system [21]. Dozens of complement factors are secreted into biological fluids such as serum and bronchoalveolar fluid [22,23], where they react with foreign particles and facilitate their destruction and recognition by innate immune cells [24,25]. The complement cascade is triggered by three main pathways: the antibody-dependent classical pathway; the lectin pathway, through recognition of microbial sugars; and the non-specific alternative pathway, all of which culminate in the cleavage and activation of C3 [21]. Following C3 activation, C3b is covalently attached to the microbial surface, and is recognized by complement receptors (CRs) expressed on immune cells, which mediate phagocytosis [26]. C3 cleavage also releases the small peptide fragment, C3a, which is recognized by the complement C3a receptor (C3aR), a G-protein coupled receptor (GPCR) which is expressed on innate immune cells [27]. C3a acts as a chemoattractant for innate immune cells such as macrophages [28]. C3aR can also modulate the production of cytokines in response to inflammatory stimuli [29], and has been implicated in the pathogenesis of inflammatory diseases such as sepsis and allergic inflammation [30]. C5 is activated downstream of C3, leading to the release of C5a, which is also a potent chemoattractant and inflammatory modifier through its interaction with its receptor, C5aR [30]. While serum is a major source of complement, innate immune cells such as macrophages can also secrete complement components [31–33].
Ubiquitous opportunistic fungal pathogens, including Candida albicans, as well as endemic fungal pathogens such as Coccidioides immitis [34] and Hc [35], are strong activators of multiple serum complement pathways [36]. Serum enhances the phagocytosis of opportunistic fungal pathogens, and the role of C3b or inactivated C3b (iC3b) opsonization in promoting uptake of fungi due to recognition by complement receptors is well-studied [36–39]. In addition, complement plays an important role in host defense against opportunistic fungi, including Candida albicans [40] and Cryptococcus neoformans [41]. Zymosan, a cell-wall preparation of Saccharomyces cerevisiae, is well-established as a model for complement activation [42]. Additionally, complement receptors, such as CR3, can recognize other substrates, such as glucans, and promote complement-independent recognition of fungi such as Hc [16,19,26]. C5a-C5aR signaling can also promote serum-dependent neutrophil migration towards and phagocytosis of Cryptococcus neoformans [43] and promote monocyte cytokine production in response to Candida albicans infection [44]. However, the role of complement in innate immune recognition of Hc, and of C3a-C3aR signaling in macrophage interaction with fungi has not been investigated.
To characterize host genes that underlie macrophage susceptibility to infection with Hc, we took advantage of a powerful pooled host-side screening platform [45] that has been successfully employed to identify host targets of intracellular pathogens [46,47] and microbial toxins [48]. We screened a CRISPR-Cas9 knockout library in macrophage-like cells challenged with Hc, and identified genes required for macrophage susceptibility to Hc-mediated lysis. We identified a number of host pathways that affected macrophage susceptibility to Hc infection, and focused our studies on molecules that influence Hc phagocytosis. This led to the discovery that C3aR and GPCR signaling plays an important role in promoting serum-dependent phagocytosis of Hc and other fungi. In addition, our screen identified the ER membrane (EMC) complex subunit Emc1, which we discovered is critical for surface expression of C3aR but not integrin receptors. This finding suggests a role for the EMC, which facilitates folding of transmembrane helices in the ER, in the biogenesis of GPCRs in innate immune cells. Overall our findings shed light on molecular mechanisms underlying innate immune recognition of fungi, and uncover new host pathways that may be targeted by Hc to promote virulence.
Results
A large-scale pooled CRISPR screen in J774A.1 macrophage-like cells identified genes required for macrophage susceptibility to infection with Hc
To identify genes that affect macrophage sensitivity to parasitization by Hc, we conducted pooled CRISPR-Cas9 knockout screens in the J774A.1 mouse macrophage-like cell line (Fig 1). This cell line has been widely used to model macrophage interactions with pathogenic microbes, including Hc [7,9]. We demonstrated that Hc can induce lysis of J774A.1 cells in a manner dependent on the secreted effector Cbp1 (S1A Fig), which is consistent with studies in primary macrophages [7,9,10]. Screen results are listed in S1–S3 Tables.
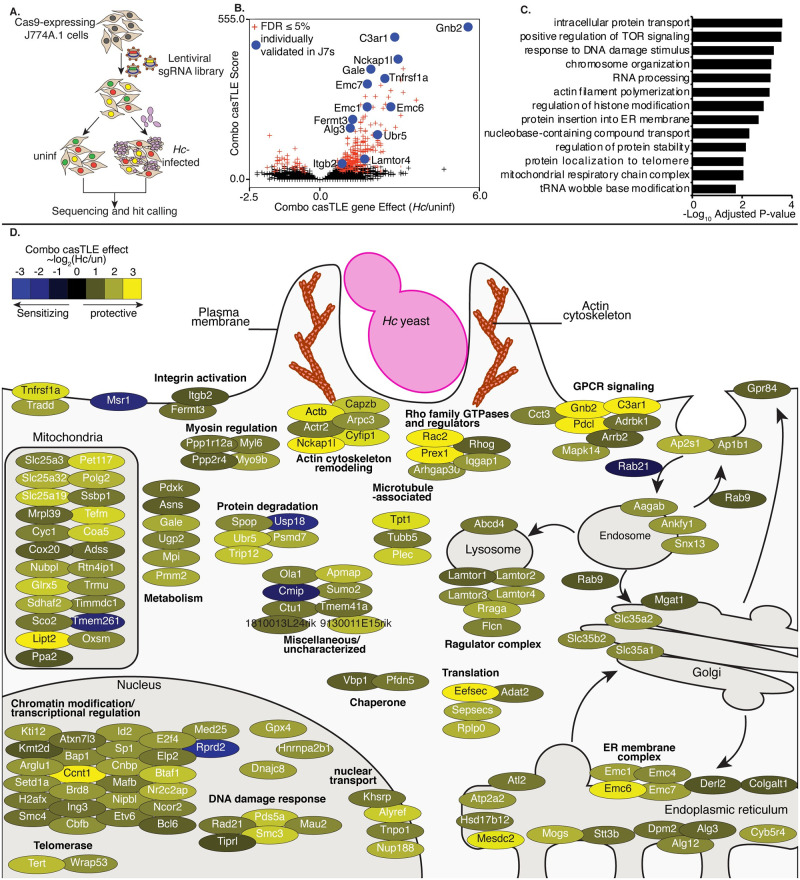
Fig 1. A pooled CRISPR screen identifies genes required for macrophage susceptibility to infection with Hc.
A. Diagram of screen approach. Cas9-expressing J774A.1 macrophage-like cells were transduced with a library of sgRNAs, challenged with Ura5-deficient Hc yeast, and subjected to 2–3 pulses of uracil treatment followed by recovery. sgRNAs amplified from Hc-infected and uninfected cells were deep-sequenced, and sequences were analyzed to identify guides that became enriched or depleted in the Hc-infected pool relative to the uninfected pool. B. Volcano plot showing the confidence score (casTLE score) versus the effect size (casTLE effect) for all genes. Genes that pass the 5% FDR cutoff are colored red, and genes individually validated in J774A.1 cells are labelled and colored in blue. C. Adjusted P-values for selected GO biological process annotations enriched in the screen hits. D. The 150 highest-scoring genes identified in the screen grouped based on their annotated function and localization in a cell, functional categories or complexes of genes are noted. Genes are colored according to their gene effect estimate, where yellow indicates enrichment in the Hc infected pool and blue indicates depletion.
To create our knockout libraries, we first generated a clonal J774A.1 cell-line with high constitutive Cas9 activity (S1B Fig). We then transduced these Cas9-expressing J774A.1 cells with pooled lentiviral sgRNAs. We used a previously designed CRISPR-Cas9 sgRNA library, which targets 23,000 protein-coding mouse genes with 10 sgRNAs/gene. The genome-wide library is split into 20 sub-libraries, each of which covers 500–1500 genes and includes 750 negative control sgRNAs [49]. We screened each sub-library separately, covering a total of 16,782 genes. These cells were infected, in duplicate, with Hc, or were left uninfected and passaged throughout the course of the screen (Fig 1A). To improve the sensitivity of our screen, we used a strain of Hc with a mutation in the URA5 gene (Hc ura5Δ) which cannot grow in media without uracil supplementation [50]. This strain does not lyse J774A.1 cells in the absence of exogenous uracil, and host cells that survive the initial round of lysis can be recovered by washing the monolayer and incubating in media without uracil supplementation (S1C–S1E Fig), thereby allowing enrichment of resistant host cells. We infected the J774A.1 pools with Hc ura5Δ and performed 2–3 rounds of uracil addition to induce Hc-mediated lysis of at least 50% of the J774A.1 cells, followed by uracil removal and recovery (see S4 Table for sub-library specific details). We pulsed the uninfected cells with uracil during passaging to match the Hc infection. The sgRNAs in the final pools were deep-sequenced to determine the enrichment of guides following challenge with Hc. We employed the Cas9 high-throughput maximum-likelihood estimator (casTLE) algorithm [51] to estimate the effect of knocking out a gene on susceptibility to Hc (caSTLE effect) based on the enrichment of guides targeting each gene in the screen compared to the enrichment of negative control sgRNAs (S1 Table). We additionally analyzed uninfected cells at the beginning and the end of passaging using the casTLE algorithm, and we were able to verify that guides targeting genes previously annotated as essential [51] dropped out of the pool during passaging as expected (S2 and S3 Tables and S1H Fig).
We identified 361 genes whose deletion modulated macrophage susceptibility to Hc infection at a 5% false-discovery rate (FDR) (Fig 1B). Confidence scores between screen replicates were moderately correlated (S1F Fig). Disruption of 322 of these genes conferred protection against Hc (combo casTLE effect >0), and disruption of 39 conferred greater susceptibility to infection (combo casTLE effect <0) (Fig 1B). We noticed that the protective hits include genes known to be required for macrophage phagocytosis, such as members of the SCAR/WAVE and ARP2/3 complexes (Fig 1C and 1D). Such regulators have been well-studied for their role in phagocytosis and chemotaxis [14,52,53]. Similarly, we identified Itgb2 (CD18), which encodes the β-subunit of CR3 that has been previously shown to facilitate recognition and phagocytosis of Hc [16,19], and Fermt3, which promotes activation of integrins [54] (Fig 1D).
Of note, we identified a number of pathways and complexes among the resistance-promoting hits that have not been previously implicated in Hc interaction with macrophages (Fig 1D), such as the ragulator complex, glycosylation enzymes, protein degradation machinery, mitochondrial respiration genes, solute transporters, and the ER membrane complex (EMC). The ragulator complex promotes nutrient stress sensing [55], and the EMC facilitates the folding of transmembrane proteins with multiple membrane-spanning regions [56,57]. The highest scoring protective hits include a group of genes (Gnb2, Pdcl, AP-1 subunits, AP-2 subunits, Arrb2) that regulate G-protein coupled receptor (GPCR) signaling and receptor trafficking following GPCR engagement (Fig 1D) [58,59]. The hit identified with the second-highest confidence score was the gene encoding the GPCR C3a receptor 1 (C3ar1/C3aR) (Fig 1D). Histograms demonstrating the enrichment of negative control sgRNAs and sgRNAs targeting Gnb2 and C3ar in the Hc-infected pool are shown in S1G Fig. We went on to investigate whether these factors play a role in macrophage phagocytosis of Hc.
Identification of genes required for phagocytosis of yeast in J774A.1 macrophage-like cells and primary macrophages
We selected 16 high-confidence hits to individually validate in J774A.1 macrophage-like cells, including two genes, SCAR/WAVE subunit Nckap1l, and Itgb2, which were expected to play a role in macrophage phagocytosis of Hc. We prioritized genes that would shed light on novel aspects of macrophage interactions with Hc and that did not appear to strongly inhibit macrophage replication. We chose the three top-performing guides, based on enrichment or depletion in the screen, for further validation.
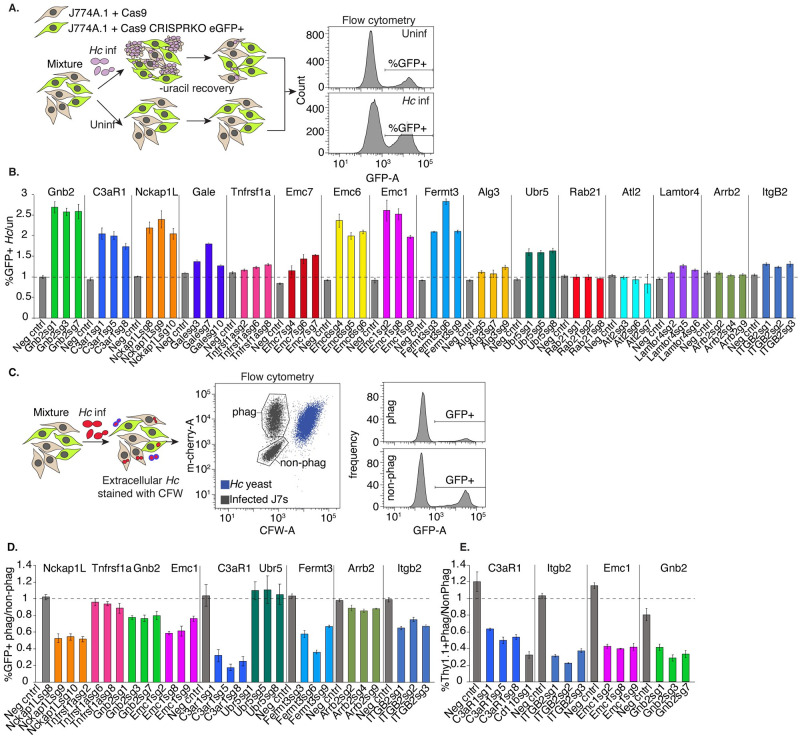
To verify susceptibility/resistance phenotypes in J774A.1 cells, we mixed GFP+, CRISPR-knockout (KO) cells with Cas9-expressing unlabeled cells, infected one pool of this mixture with Hc, and in parallel passaged the uninfected pool. Following one round of lysis and recovery, the pools were then harvested, and the proportion of GFP-expressing cells was measured by flow cytometry (Fig 2A). The ratio of GFP+ cells in the Hc-infected compared to the uninfected pool demonstrated whether targeting a specific gene conferred a fitness advantage (>1) or disadvantage (<1) to macrophages during co-culture with Hc. Of the 16 genes tested, 13 conferred a fitness advantage during Hc infection when disrupted, including Gnb2, C3ar, ER membrane complex subunits Emc1, Emc6, and Emc7, and ubiquitin ligase Ubr5 (Fig 2B). As positive controls, we included knockouts of SCAR/WAVE subunit Nckap1l, and the β-2 integrin subunit of CR3, Itgb2 (Fig 2B). The only susceptibility-promoting hit that we tested, Rab21, did not promote increased susceptibility to Hc infection when disrupted (Fig 2B).
Fig 2. Identification of genes required for phagocytosis of yeast in J774A.1 cells and primary macrophages.
A. Diagram of approach used to individually validate the role of a gene in macrophage susceptibility to Hc infection. A mixture of WT (GFP-) and CRISPRKO (GFP+) J774A.1 cells were challenged with Hc yeast in the presence of uracil, and allowed to recover. Uninfected cells from the same mixture were passaged in parallel, and the percentage of mutant cells in the Hc infected pools was compared to that of the uninfected pools via flow cytometry (n = 3 biological replicates). B. Enrichment of gene-targeting guides in the Hc infected pool relative to the control pool, compared to that of non-targeting guides. C. Diagram of approach for determining the role of a gene in phagocytosis of Hc. A mixture of WT (GFP-) and CRISPRKO (GFP+) J774A.1 cells were infected with mCherry-expressing Hc yeast. Non-internalized yeasts were excluded using calcofluor white staining. Flow cytometry was used to determine the representation of mutant cells in the phagocytic compared to the non-phagocytic populations (n = 3). D. Identification of genes required for phagocytosis of yeast in J774A.1 cells using GFP expression to measure enrichment of sgRNA-expressing cells. E. Validation of gene involvement in BMDM phagocytosis of yeast using CRISPRKO BMDMs (Thy1.1+). A mixture of transduced (Thy1.1+) and untransduced (Thy1.1-) BMDMs were similarly infected with yeast and stained with calcofluor white and a Thy1.1 antibody to determine the representation of mutants in the phagocytic and non-phagocytic populations (n = 3 biological replicates).
Next, we tested whether these genes play a role in macrophage phagocytosis of Hc yeast. To this end, we mixed GFP+, CRISPR-targeted cells with unlabeled, Cas9-expressing cells, infected the mixture with mCherry-expressing Hc yeast, and stained the cells with calcofluor white (CFW) to distinguish between intracellular and extracellular yeast. We used flow cytometry to measure the representation of GFP+ cells in the phagocytic compared to the non-phagocytic population (Fig 2C). As expected, targeting of Nckap1l, Fermt3, and Itgb2 led to decreased Hc phagocytosis in J774A.1 cells (Fig 2D). Additionally, we found that knockout of Emc1, Gnb2, C3ar1, and Arrb2 decreased phagocytosis of Hc (Fig 2D).
Although J774A.1 cells recapitulate many important features of primary macrophages, including phagocytosis, they also differ in characteristics such as gene expression regulation [60]. Therefore, we attempted to reproduce our findings from J774A.1 cells in bone marrow-derived macrophages (BMDMs) using CRISPR-Cas9-mediated gene disruption. We mixed CRISPR-knockout, Thy1.1+ BMDMs with WT, unlabeled BMDMs, infected the mixture with Hc yeast, and assessed phagocytosis as described above. We quantified the proportion of Thy1.1+ cells in the phagocytic compared to the non-phagocytic populations to determine whether the targeted genes promoted BMDM phagocytosis of Hc yeast. The four genes that we tested, GPCR C3ar1, integrin subunit Itgb2, ER membrane complex Emc1, and Gβ subunit Gnb2, were also required for efficient phagocytosis of Hc yeast by BMDMs (Fig 2E).
C3aR signaling plays a role in macrophage phagocytosis of fungi
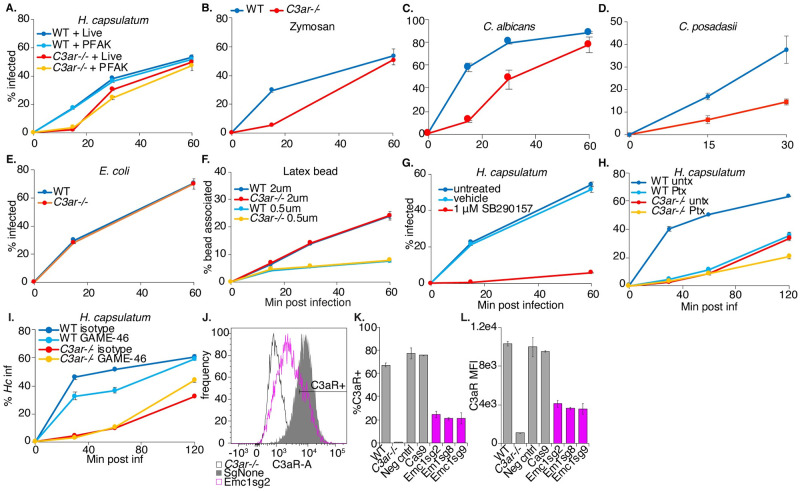
Since a role for C3aR in phagocytosis of fungi had not previously been defined, we were intrigued by the result that this receptor is required for efficient phagocytosis of Hc by J774.1 cells and BMDMs. C3aR is a GPCR that recognizes the complement C3 cleavage product, anaphylatoxin C3a, and signals through Gαi [30], which is sensitive to pertussis toxin-mediated ADP-ribosylation [61]. We further investigated the role of C3aR and GPCR signaling in macrophage phagocytosis of fungi. We generated BMDMs from C3ar-/- mice [62] and age-matched WT mice. We then infected these macrophages with several species of pathogenic fungi, including Hc yeast expressing mCherry (Fig 3A, 3B and 3G–3I), Candida albicans (Ca) yeast detected with a fluorescent antibody (Fig 3C), and Coccidioides posadasii arthroconidia labeled with FITC (Cp) (Fig 3D), and determined the extent of phagocytosis over time. We also tested phagocytosis of FITC-labeled zymosan (Fig 3B), a cell-wall extract of Saccharomyces cerevisiae. We used calcofluor white staining to distinguish between intracellular and extracellular fungi. We observed that C3aR was required for efficient phagocytosis of all three species of fungal pathogens, in addition to zymosan, suggesting an important general role for C3aR in macrophage capture and phagocytosis of fungi (Fig 3A–3D). The involvement of C3aR did not require fungal viability, as C3aR was equally important for phagocytosis of both live and killed Hc yeast, as well as zymosan (Fig 3A and 3B). The phagocytosis defect was not due to a defect in CD11b or CD18 surface expression in C3ar-/- BMDMs (S2 Fig). By 30–60 min post-infection, C3ar-/- macrophages were able to phagocytose most of the fungi we tested, especially at lower MOIs, but uptake was noticeably slower.
Fig 3. C3aR signaling plays a role in macrophage phagocytosis of fungi.
A. WT and C3ar-/- BMDMs were infected with live and PFA-killed mCherry-expressing Hc yeast (MOI2), and the phagocytosis rate was monitored over-time using flow-cytometry (n = 3 biological replicates). B. WT and C3ar-/- BMDMs were infected with FITC-labelled zymosan or mCherry-expressing Hc (MOI2) and the phagocytosis rate infected cells was monitored using flow cytometry (n = 3 biological replicates). C. BMDMs were infected with Candida albicans (Ca) (MOI3). Cells were imaged using confocal microscopy to quantify phagocytosis (n = 2 biological replicates, >350 cells/replicate). CFW staining was used to exclude extracellular Ca. D. BMDMs were infected with FITC-labelled Coccidioides posadasii (Cp) arthroconidia (MOI1), and extracellular conidia were labelled with calcofluor white. BMDM infection rates were determined using confocal microscopy (n = 3 biological replicates, 200–400 cells/rep). E. BMDMs were infected with FITC-labelled E. coli bioparticles (MOI4) and the E. coli-association with BMDMs was monitored via flow cytometry (n = 2 biological replicates). F. BMDMs were infected with 2 μm or 0.5 μm red fluorescent latex beads (MOI2), and the rate of BMDM association with the beads was measured using flow cytometry (n = 3 biological replicates). G. BMDMs were treated with a C3aR antagonist (1 μM SB290157) and infected with Hc yeast (MOI2). Phagocytosis was measured using flow cytometry (n = 3 biological replicates). H. BMDMs were pre-treated for 2 h with 1 μg/mL pertussis toxin (Ptx), which inhibits Gαi, and infected with Hc (MOI5, n = 3 biological replicates). I. BMDMs were pre-treated for 90 min with 10 μg/mL CD18 blocking antibody (GAME-46) and infected with Hc yeast (MOI5, n = 3 biological replicates) Phagocytosis was measured using flow cytometry. Emc1 is required for C3aR expression in BMDMs (J-L). J. Emc1 CRISPRKO BMDMs and control sgRNA transduced BMDMs, and C3aR levels were measured via flow cytometry following C3aR surface staining (n = 2 biological replicates). K. Histogram of C3aR levels in control and Emc1 CRISPRKO BMDMs. L. Frequency of C3aR+ cells in the indicated BMDMs. M. The mean fluorescence intensity (MFI) of the C3aR signal in the indicated BMDMs.
C3aR has been previously implicated in macrophage uptake of certain, though not all, bacterial pathogens [63,64], and in microglial phagocytosis of several substrates [65–67]. To investigate whether the requirement of C3aR for phagocytosis extends to other types of particles that can be taken up by macrophages, we measured the capture of uncoated latex beads and FITC-labelled E. coli K12 in WT and C3ar-/- BMDMs (Fig 3E and 3F). We found that C3aR was not required for uptake of E. coli (Fig 3E) or latex beads (Fig 3F), suggesting that C3aR does not play a general role in phagocytosis.
To further validate the contribution of C3aR to phagocytosis, we treated macrophages with a specific non-peptide antagonist of C3aR, SB290157 [68] five minutes before challenge with Hc (Fig 3G). We found that the C3aR antagonist was able to inhibit macrophage phagocytosis of Hc, suggesting an acute role for C3aR in macrophage phagocytosis of fungi. Treatment with the antagonist seemed to have a stronger effect on phagocytosis than C3aR deletion at 60 min post-infection, which may reflect the difference in the dynamic range of the assay due to the use of a higher MOI in the antagonist experiment. The antagonist may also have off-target effects that can inhibit phagocytosis, or macrophages from C3ar-/- mice may up-regulate other surface receptors to compensate for chronic C3aR deficiency. C3aR signaling is dependent on pertussis toxin-sensitive Gαi [30], inhibition of which interferes with macrophage phagocytosis of Zymosan particles [69]. We assessed whether Gαi inhibition by pre-treatment of macrophages with pertussis toxin (Ptx) would impact macrophage phagocytosis of Hc yeast, and whether Ptx treatment would synergize with C3aR deficiency. We found that Ptx pre-treatment inhibited macrophage phagocytosis of Hc (Fig 3H). Ptx treatment strongly phenocopies the phagocytosis defect in C3ar-/- BMDMs, and Ptx treatment modestly inhibits phagocytosis in C3ar-/- BMDMs (Fig 3H). These findings show that C3aR-dependent Gαi activation promotes phagocytosis, although Gαi activation by other receptors, and C3aR coupling to a different Gα subunit, may play a minor role in Hc phagocytosis (Fig 3H). We also investigated whether C3aR interacts with CR3 to promote phagocytosis by treating BMDMs with a CD18 blocking antibody (GAME-46) previously used to block CR3 [16] (Fig 3I). As is consistent with previous results from thioglycollate-elicited peritoneal macrophages [16], WT BMDMs treated with the CD18 inhibitor had a modest defect in phagocytosis of Hc. Treatment of C3ar-/- BMDMs with the inhibitor did not further block phagocytosis of Hc, suggesting that CR3 participates in phagocytosis downstream of C3aR (Fig 3I).
We further reasoned that Emc1 may indirectly promote phagocytosis due to its role in stabilization of proteins with multiple transmembrane helices [57], such as C3aR. To test this hypothesis, we measured C3aR surface-expression in Emc1 CRISPRKO BMDMs (Fig 3J–3L). We saw a dramatic decrease in C3aR surface expression in Emc1-targeted BMDMs compared to untransduced or control-targeted BMDMs (Fig 3J–3L), suggesting that the EMC facilitates the proper folding and biosynthesis of GPCRs, such as C3aR, in macrophages. In contrast, Emc1 CRISPRKO BMDMs did not show reduced surface expression of CD18 or CD11b (S2 Fig), verifying that the EMC is not as critical for proper folding of single-pass transmembrane proteins like integrins.
Since phagocytosis of Hc is delayed in C3ar-/- BMDMs, we expected lysis of infected BMDMs to show a corresponding delay. As expected, we found that C3ar-/- BMDMs were slightly less susceptible to Hc-mediated lysis, as measured by an established assay [9,70] (S3 Fig). Analysis of Hc colony forming units (CFUs) indicated that C3ar-/- macrophages were infected with fewer Hc yeast at the start of the experiment, and Hc yeasts did not have a major intracellular growth defect in the mutant macrophages (S3 Fig).
Since C3aR has been implicated in modulating the cytokine response to microbial infection, we measured Hc-induced cytokine release in C3ar-/- BMDMs (S4A Fig) or in C3aR-antagonist-treated thioglycollate-elicited peritoneal macrophages (Thio-Pmacs, S4B–S4D Fig). We observed moderately decreased TNFα secretion in C3ar-/- BMDMs infected with Hc at 2h post-infection, but this effect was diminished at 6h post-infection (S4A Fig). We were not able to detect IL-1β or IL-6 secretion in Hc-infected BMDMs. In Thio-Pmacs, we found that inhibition of C3aR did not affect TNFα (S4B Fig) or IL-1β secretion (S4D Fig), but slightly inhibited IL-6 secretion at 2h, but not 6h post-infection (S4C Fig). We found that C3aR may play a minor role in increasing some pro-inflammatory cytokine production at early time-points following Hc infection, and that this depends on the macrophage cell-type used. It is unclear whether the effect on cytokine release is a result of delayed phagocytosis of Hc or due to a signaling role for C3aR.
Serum C3 promotes complement opsonization, C3a release, and macrophage phagocytosis of Hc yeast
Since the canonical ligand for C3aR is C3a derived from C3 processing, we investigated whether serum represented a source of C3 that would react with Hc to generate C3a and promote phagocytosis. Macrophage infections discussed up to this point were conducted in the presence of 10% (for J774A.1 cells) or 20% (for BMDMs) heat-treated fetal bovine serum (FBS), which was not previously thought to be a robust source of complement.
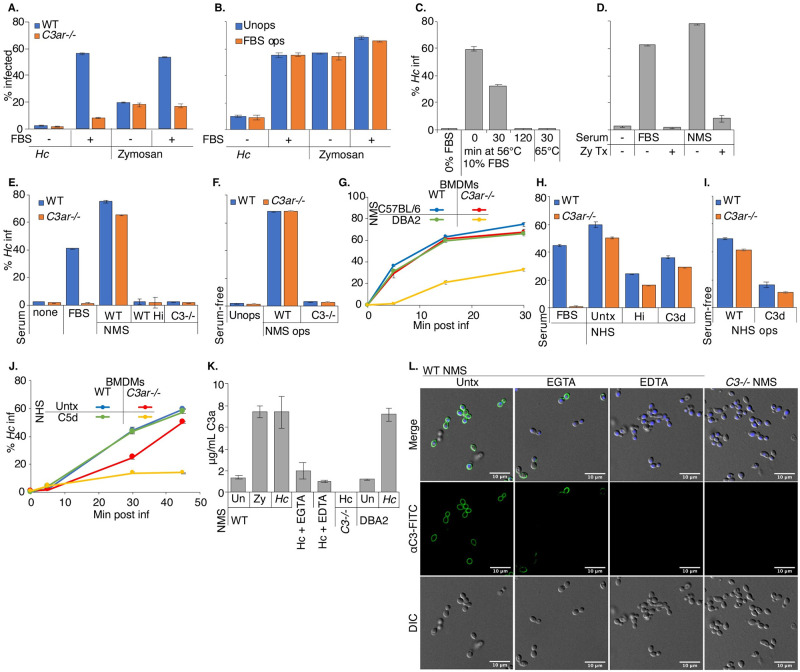
We infected WT and C3ar-/- BMDMs with mCherry+ Hc and FITC-labelled zymosan in the presence or absence of 20% heat-treated FBS, and monitored phagocytosis by flow cytometry. Surprisingly, we found that FBS promoted macrophage phagocytosis of Hc (and to a lesser extent zymosan) in a C3aR-dependent manner (Fig 4A). Even after 2h of co-culture, we did not observe efficient phagocytosis of Hc by BMDMs in serum-free media (Fig 4B). Phagocytosis of zymosan by BMDMs in serum-free media was more efficient than that of Hc, and was not dependent on C3aR (Fig 4A), as expected due to the role of Dectin 1-mediated recognition of β-glucans in non-opsonic macrophage recognition of zymosan [39]. The low level of Hc phagocytosis in serum-free media was also C3aR-independent. The ability of FBS to stimulate phagocytosis is not lot-dependent, as FBS from different lots and manufacturers promoted macrophage phagocytosis of Hc in a C3aR-dependent manner (S5 Fig). To assess whether FBS was promoting phagocytosis by opsonization of the yeast, we tested whether pre-incubation with FBS would be sufficient to stimulate phagocytosis of Hc in serum-free media (Fig 4B). We found that pre-incubation in FBS did not promote phagocytosis of Hc or zymosan (Fig 4B), suggesting either that FBS does not facilitate phagocytosis by opsonization, or that opsonization is labile. We also determined that incubating Hc with BMDM conditioned media containing FBS did not promote macrophage phagocytosis of Hc (S6B Fig), suggesting that BMDMs do not secrete a missing factor that would restore FBS-mediated opsonization of Hc.
Fig 4. Serum C3 promotes complement opsonization, C3a release, and macrophage phagocytosis of Hc yeast.
A. FBS stimulates macrophage phagocytosis of fungi in a C3aR-dependent manner. BMDMs were infected with mCherry-expressing Hc or FITC-labelled zymosan (30 min, MOI5) in the presence or absence of 20% heat-treated FBS (FBS). Phagocytosis was assessed via flow cytometry (n = 3 biological replicates). B. FBS does not promote macrophage phagocytosis of Hc via opsonization. Hc and zymosan particles were pre-incubated with 10% heat-treated FBS for 30 min at 37°C, washed, and used to infect BMDMs (2h, MOI2). Phagocytosis was measured using flow cytometry (n = 2 biological replicates). C-D. Prolonged or intense heat-treatment and zymosan treatment eliminates the phagocytosis-stimulating properties of serum. C. Macrophage phagocytosis of Hc (MOI5, 45 min, n = 3 biological replicates) was assessed in media supplemented with 10% FBS that had been subjected to heat treatment (C) at 56°C for up to 2h, at 65°C for 30 min, or that had been pre-treated with zymosan (D) (1X108 particles/mL, 60 min at 37°C). Phagocytosis was measured by flow cytometry. E. Normal mouse serum (NMS) stimulates macrophage phagocytosis of Hc in a C3-dependenent manner. BMDMs were infected with Hc yeast (MOI = 5, 60min) in serum-free media or media supplemented with 5% FBS, 5% NMS from WT mice, 5% NMS from C3-/- mice, or 5% heat-inactivated NMS (hiNMS) from WT mice and phagocytosis was measured by flow cytometry (n = 3 biological replicates). F. BMDMs in serum-free media were infected with Hc opsonized with 10% WT or C3-/- NMS. Phagocytosis was measured by flow cytometry (n = 3 biological replicates). G. C5-deficient serum promotes macrophage phagocytosis of Hc in a C3aR-dependant manner. BMDMs were infected with Hc yeast (MOI5) in media supplemented with 5% NMS from C57BL/6 mice or DBA2 (C5-deficient) mice. Phagocytosis was measured by flow cytometry (n = 2 biological replicates). H-J. Normal human serum (NHS) stimulates macrophage phagocytosis of Hc yeast. H. BMDMs were infected with Hc (MOI5, 60 min) in media supplemented with 5% untreated, heat-inactivated, or C3-depleted (C3d) NHS, and phagocytosis was monitored by flow cytometry (n = 3 biological replicates). I. Hc was opsonized with 10% untreated or C3d NHS, used to infect BMDMs in serum-free media (MOI5, 60 min), and phagocytosis was monitored by flow cytometry (n = 3 biological replicates) J. BMDMs were infected with Hc (MOI5) in media supplemented with 5% untreated or C5-depleted (C5d) NHS, and phagocytosis was monitored by flow cytometry (n = 3 biological replicates). K-L. Mouse serum promotes complement opsonization of yeast and release of C3a via multiple pathways. Hc was incubated in 10% serum from WT, C3-/-, or DBA2 mice for 30 min at 37°C. 10 mM EGTA or EDTA were added to the reactions to chelate Ca2+ or Mg2+, respectively. K. Supernatants were harvested following incubation, and mouse C3a levels were measured by ELISA (n = 3 biological replicates). L. Yeast were stained with a FITC conjugated anti-mouse C3, and imaged using confocal microscopy (representative slices are shown from 2 biological replicates).
Due to the surprising finding that serum incubated at 56°C for 30 min was still able to stimulate phagocytosis, we tested whether increasing the duration or temperature of heat-treatment would affect this activity. We found that, while FBS heat-treatment at 56°C for 30 min reduced phagocytosis stimulation by 50%, treatment at 56°C for 2 h or at 65°C for 30 min nearly completely abolished the ability of FBS to stimulate phagocytosis of Hc (Fig 4C). We found that pre-treating FBS and normal mouse serum (NMS) with zymosan also eliminated their ability to stimulate phagocytosis (Fig 4D). This suggests that a consumable pathogen recognition component of serum, as opposed to a non-specific macrophage activation signal, is critical for the ability of FBS to stimulate phagocytosis. The ability of BMDMs in serum-free media to ingest zymosan (Fig 4B) and serum-opsonized Hc (Fig 4F and 4I) also suggests that BMDMs are not generally incompetent at phagocytosis in the absence of serum.
To establish a role for serum-derived C3 in macrophage recognition of Hc, we compared phagocytosis of Hc in media supplemented with no serum, FBS, or serum collected from WT or C3-/- C57BL/6 mice (WT NMS or C3-/- NMS) (Fig 4E). We found that mouse serum promoted macrophage phagocytosis of Hc in a C3-dependent manner that was sensitive to heat inactivation (Fig 4E). Surprisingly, the ability of mouse serum to stimulate phagocytosis of Hc was not strongly dependent on C3aR (Fig 4E), suggesting an additional C3aR-independent, C3-dependent mechanism of phagocytosis. Since C5 can be activated downstream of C3, leading to the release of the potent chemoattractant C5a [21], we reasoned that serum from C57BL/6 mice might stimulate phagocytosis via C5. C5a release and recognition by C5aR would then stimulate phagocytosis and compensate for C3aR-deficiency. To test this, we supplemented the media with serum from DBA2 mice, which have low levels of serum C5, but normal levels of C3 [71]. We found that C3ar-/- BMDMs were defective at phagocytosis of Hc in media supplemented with DBA2 (C5-deficient), but not C57BL/6 (C5-sufficient) serum (Fig 4G), suggesting that C5a in C57BL/6 serum acts redundantly with C3a to promote macrophage phagocytosis of Hc.
We then determined whether human serum (NHS) had a similar C3-dependent effect on BMDM recognition of Hc by supplementing media in phagocytosis assays with 5% NHS, C3, or C5 immunodepleted human serum (C3d/C5d) (Fig 4H–4J). Similar to what we observed with mouse serum-supplemented media, we found that NHS-supplemented media stimulated BMDM phagocytosis of Hc in a heat-labile and C3-dependent manner (Fig 4H). Phagocytosis of Hc with untreated NHS was also less dependent on C3aR (Fig 4H). Of note, we observed more phagocytosis of Hc in C3d NHS than in NMS from C3-/- mice, which may suggest residual C3 activity following immunodepletion or activity of other components of human serum, such as antibodies that react with Hc. We also found that, similar to our observations in mouse serum, C5 in human serum contributes to C3aR-independent recognition of Hc, since C3ar-/- BMDMs had a stronger phagocytosis defect in C5d NHS than in untreated NHS (Fig 4J).
To confirm that incubating mouse serum with Hc yeast would promote opsonization with C3, as previously described [72], and C3a release, we incubated Hc yeast with mouse serum, and measured C3a release into the supernatant by ELISA (Fig 4K) and C3 deposition on the Hc surface using immunofluorescence confocal microscopy (Fig 4L). We also found that incubating WT C57BL/6 and DBA2 serum with Hc increased C3a levels in the supernatant (Fig 4K), suggesting C3a release. We observed robust C3 staining of Hc upon incubation with WT serum, and no C3 deposition after incubation with C3-/- sera (Fig 4L). To inhibit the classical/lectin pathways, or all activation pathways, we added EGTA or EDTA, respectively, to the indicated reactions. We did not observe C3 deposition or C3a release when Mg++ was chelated with EDTA (Fig 4K and 4L). We also saw C3 deposition, although with lower efficiency and with a less uniform distribution around the yeast cell-wall, and lower levels of C3a release, in the presence of EGTA (Fig 4K and 4L). These results confirm that Hc can activate the alternative complement pathway, as previously suggested [35]. Due to the increased efficiency and uniformity of C3 deposition on yeast and the increased C3a release found in the absence of EGTA, we suggest that the classical or lectin pathways also contribute to C3 opsonization of Hc yeast. We did not find evidence of C3 deposition on the cell-surface following incubation of Hc with heat-treated FBS or BMDM conditioned media containing heat-treated FBS (S6A Fig).
To demonstrate that complement opsonization by mouse or human serum promotes macrophage phagocytosis of Hc yeast, we infected BMDMs in serum-free media with Hc opsonized by WT or C3-/- mouse serum (Fig 4F) or with untreated or C3-depleted NHS (Fig 4L). We found that opsonization with WT mouse serum, but not C3-/- serum, is sufficient to promote phagocytosis of Hc in serum-free media in a C3aR-independent manner, suggesting direct recognition of opsonized yeasts by CR3 (Fig 4F). This activity was blocked by EDTA and moderately inhibited by EGTA, suggesting contribution of both the classical/lectin and alternative pathways to phagocytosis stimulation through opsonization (S6B Fig). We similarly found that NHS-opsonized Hc was robustly recognized by BMDMs in serum-free media, and that C3d NHS-opsonized Hc were taken up less efficiently (Fig 4I). Previous work has shown C3 opsonization of Hc by human serum [72]. The increased phagocytosis of C3d NHS-opsonized Hc may be due to opsonization by Hc-reactive IgG antibodies in human serum.
Active complement C3 can also be secreted by macrophages [31–33]. We measured the release of C3 into culture supernatants by ELISA, and found that Hc infection did stimulate a modest macrophage secretion of C3 (S7A Fig). However, we did not observe a phagocytosis defect when we infected C3-/- BMDMs with Hc or zymosan in the presence of FBS (S7B Fig), suggesting that macrophage-derived C3 is not playing a major role in macrophage phagocytosis of Hc in our assay.
C3aR localizes to the early Hc-containing phagosome
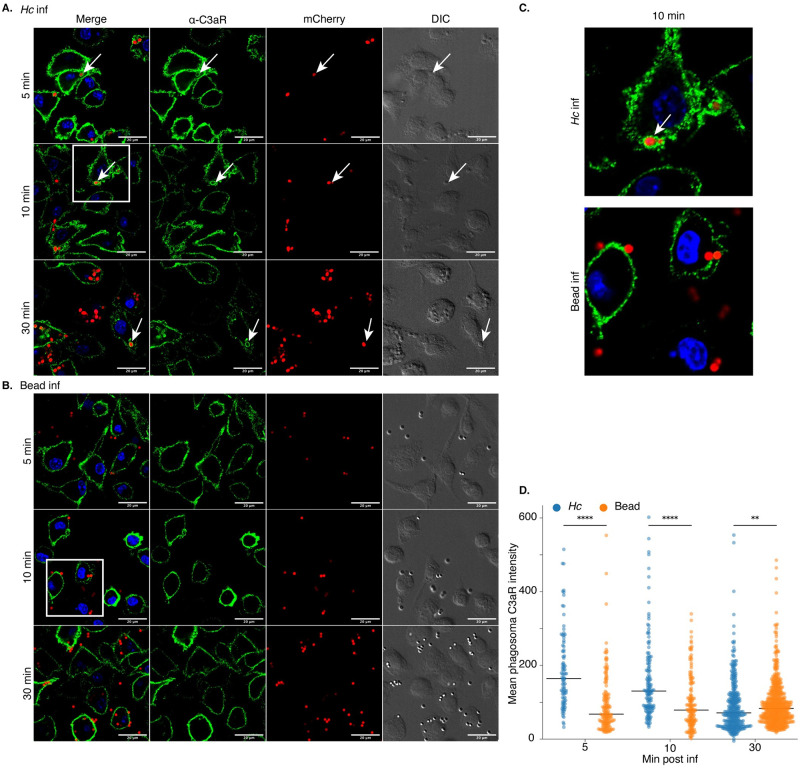
We next analyzed C3aR localization during macrophage phagocytosis of Hc and latex beads, whose uptake does not depend on C3aR. These experiments were conducted in media supplemented with 20% FBS. Localization to the Hc-containing phagosome would implicate C3aR directly in fungal capture or phagocytic cup formation, or as a passenger on the plasma membrane ruffles/filopodia involved in Hc phagocytosis. Immunofluorescence confocal microscopy confirmed that C3aR is localized at the plasma membrane in uninfected cells (S8A Fig). We observed C3aR localization to the Hc-containing phagosomes at 5- and 10-minutes post-infection, and with a lower frequency at 30 minutes post-infection (Fig 5A). Examples of C3aR-positive phagosomes are indicated by white arrows in the images. In contrast, we did not observe C3aR-positive bead-containing phagosomes at the same frequency (Fig 5B), suggesting that C3aR localizes specifically to the Hc-containing phagosome, and not to latex bead-containing phagosomes. Selected insets (indicated by a white box in Fig 5A and 5B) are enlarged to show phagosomal C3aR localization at 10 min post-infection (Fig 5C).
Fig 5. C3aR localizes to the early Hc-containing phagosome.
C3aR localizes to Hc-containing phagosomes (A) to a greater extent than latex bead-containing phagosomes (B). BMDMs were infected with the indicated particles (MOI = 5, n = 2 biological replicates per time point). Cells were then stained with a C3aR-specific antibody and imaged using optical sectioning with a confocal microscope. Representative images from a single slice are shown. C. Enlarged views of insets outlined in panels A and B by a white box. Scale bar = 20 μm. D. The mean fluorescence intensity of C3aR in the particle-containing phagosomes was quantified using ImageJ (N>91 phagosomes, **** p<0.0001, **p<0.01 by two-tailed Wilcoxon rank-sum test). The line represents the median phagosomal C3aR intensity.
To quantify C3aR localization to the Hc or bead-containing phagosome, we used ImageJ to measure the mean intensity over background of the C3aR signal surrounding the Hc or bead particle (in a 0.5 μm-thick area surrounding the particle). Example binary masks used to quantify the phagosomal C3aR intensities are shown in S8B and S8C Fig. Our analysis revealed that Hc-containing phagosomes display significantly higher C3aR enrichment than bead-containing phagosomes at 5- and 10-minutes post-infection, but to a lesser extent at 30 minutes post-infection as the phagosomes mature (Fig 5D).
C3aR promotes the formation of actin-rich protrusions that facilitate capture of Hc yeast
Since C3a is a chemoattractant for macrophages, we investigated the role of macrophage migration in the C3aR-dependent capture of Hc yeast. Although macrophages did undergo chemotaxis towards Hc in trans-well migration assays, migration was not dependent on FBS or C3aR (S9 Fig). We also were not able to rescue the phagocytosis of Hc by C3ar-/- macrophages when the likelihood of Hc-macrophage interaction was increased by centrifugation of Hc onto the monolayer, or an extended pre-incubation on ice (S10 Fig). These experiments suggest that C3aR involvement in macrophage phagocytosis of Hc is not due to its role in facilitating long-range migration of macrophages towards yeast. However, these studies do not rule out a role for C3aR-dependent control of local chemotaxis over shorter distances in macrophage capture of Hc yeast.
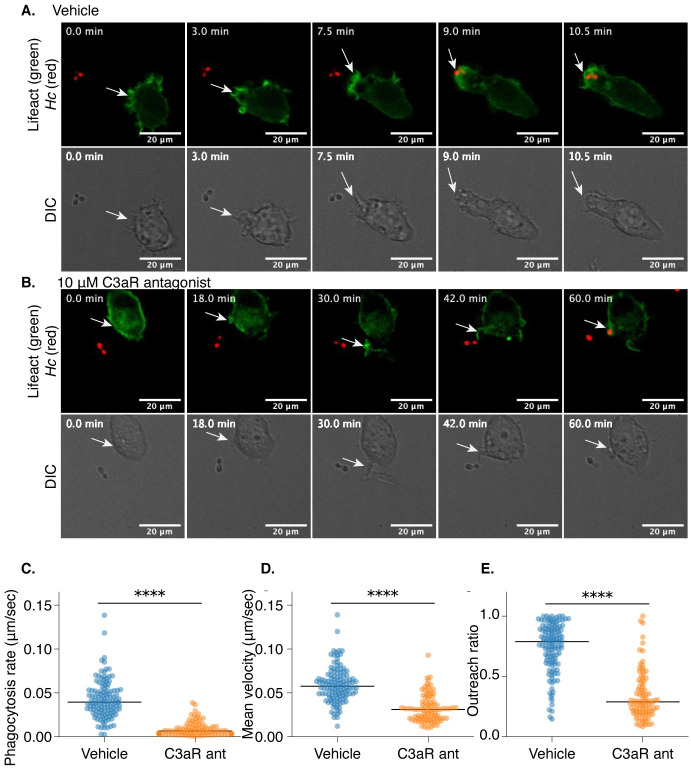
To investigate this possibility, we generated J774A.1 cells that express Lifeact-mEGFP, a probe that specifically labels F-actin [73], and performed live imaging of J774A.1 macrophages during co-culture with mCherry-labelled yeast in the presence of a C3aR antagonist (SB290157) or a vehicle control using confocal microscopy (Fig 6 and S1 and S2 Movies). These movies show macrophages extending actin-rich membrane pseudopods in the direction of nearby Hc that promote rapid Hc capture and engulfment (Fig 6A and S1 Movie). In contrast, C3aR antagonist-treated macrophages show much slower capture of Hc yeast, and fail to rapidly form such actin-rich directed membrane protrusions (Fig 6B and S2 Movie). Membrane protrusions of macrophages that eventually capture Hc yeast were tracked and analyzed [74] (Fig 6C–6E). Treatment with the C3aR antagonist significantly slowed capture of Hc yeast, as demonstrated by the lower phagocytosis rate and the lower mean velocity of the tracked protrusions (Fig 6D). Finally, the movement of the antagonist-treated membrane protrusions was less directional, as demonstrated by the lower outreach ratio of the membrane protrusions (Fig 6E).
Fig 6. C3aR promotes the formation of actin-rich protrusions that facilitate capture of Hc yeast.
J774A.1 cells were engineered to express Lifeact-mEGFP to label F-actin, co-cultured with mCherry-expressing Hc yeast, and subjected to live-cell confocal microscopy in a temperature-and-CO2 controlled chamber in media supplemented with 10% FBS. Cells were treated with a C3aR antagonist (10 μM SB290157) or a vehicle control. A. Representative images from a confocal time series (S1 Movie) showing a macrophage extending an F-actin-rich protrusion towards an mCherry expressing Hc yeast, followed by phagocytosis and formation of an actin-rich phagosome. The corresponding DIC images are shown below. B. A similar time series (S3 Movie) of macrophages treated with SB290157 showing a failure to initiate formation of a membrane protrusion and much slower capture of Hc yeast. Scale bar = 20 μm. The movement of membrane structures that successfully caputured yeast were analyzed using MtrackJ to quantify the behaviors of these structures (C-E) (n = 2 biological replicates, >50 tracks per replicate), including the phagocytosis rate, quantified as the time required for the macrophage to successfully engulf the yeast divided by the distance of the yeast to the macrophage at the start of the series (C), the mean velocity of the membrane structure closest to the yeast (D), and the outreach ratio quantified as the max displacement of the track divided by the length of the track (E) (**** p<0.0001 by two-tailed Wilcoxon rank sum test). The line represents the median measurement.
Live imaging experiments showed that C3aR facilitates the directional movement of actin-rich membrane protrusions towards Hc yeast that facilitate rapid phagocytosis. This behavior likely requires a C3a gradient that diffuses away from the Hc yeast following complement cleavage at the fungal surface, as has been proposed for anaphylatoxin-mediated microbial capture [75]. Consistent with this idea, the addition of recombinant C3a to BMDMs in the absence of a gradient was not sufficient to stimulate macrophage phagocytosis of Hc in serum-free media (S11A and S11B Fig). However, in addition to promoting local chemotaxis, C3a may also contribute to the local activation of phagocytosis receptors, such as CR3. We pre-treated BMDMs with phorbol 12-myristate 13-acetate (PMA), a PKC agonist, and assessed phagocytosis of Hc by WT and C3ar-/- BMDMs in media with and without FBS supplementation (S11C and S11D Fig). PMA activation partially rescued BMDM phagocytosis of Hc in serum-free media (S11C Fig) and in C3ar-/- BMDMs in FBS (S11D Fig). We also found that thioglycollate-elicited peritoneal macrophages (thio-Pmacs), which have been exposed to more activation signals than BMDMs, were able to take up un-opsonized Hc in serum-free media, as previously reported [16], more efficiently than BMDMs (S11E Fig). FBS did, however stimulate more efficient uptake of Hc by thio-Pmacs in a C3aR-dependent manner. These results suggest that using stimulated BMDMs or partially activated resident macrophages can partially rescue macrophage phagocytosis of Hc in serum-free media, suggesting that serum and C3aR-signaling influence the activity of other phagocytic receptors.
C3 and C3aR-deficiency do not dramatically alter mouse susceptibility to Hc infection
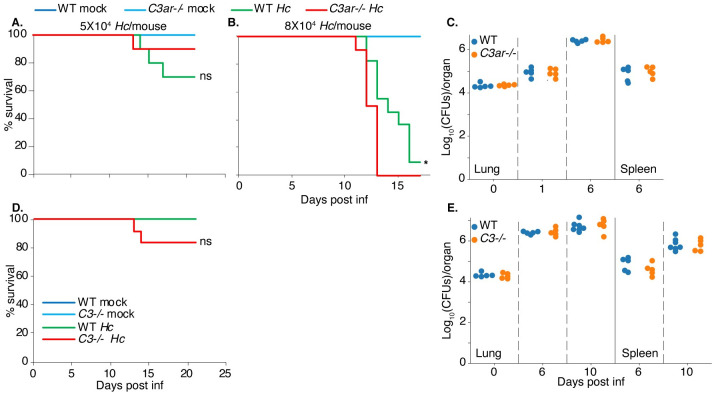
Finally, to determine whether C3aR influences disease progression in a mouse model of histoplasmosis, we compared susceptibility of WT and C3ar-/- mice to a sub-lethal or lethal dose of Hc yeast administered intranasally (Fig 7A and 7B). While C3ar-/- mice are not more susceptible to Hc infection, especially at a sub-lethal dose (Fig 7A), C3ar-/- mice succumbed to infection with a lethal dose significantly faster than the WT mice (Fig 7B). We did not find that C3ar-/- mice were more resistant to Hc challenge, suggesting that C3aR is not required for Hc to establish infection. The presence of other signals in vivo, such as C5aR, that promote macrophage phagocytosis of Hc may still allow Hc to access its replicative niche. In addition, C3aR plays other roles in the innate an adaptive immune response in vivo which may be important for the host response to fungi. We also found that C3-/- mice were not significantly more susceptible to infection with a sub-lethal dose of Hc (Fig 7D), which contrasts with the increased susceptibility of C3-/- animals to challenge with opportunistic fungi, such as Candida albicans and Cryptococcus spp [40,41,76]. We also did not observe differences in the lung or spleen fungal burden in C3ar-/- (Fig 7C) or C3-/- (Fig 7E) mice compared to WT mice following infection with a sub-lethal dose of Hc. The lung cytokine levels in C3ar-/- and C3-/- mice were also comparable to those in WT mice (S12 Fig). We noted moderately elevated levels of IL-1β (S12A Fig), TNF-α (S12C Fig), and IFN-γ (S12D Fig) in C3ar-/- lungs at 6 days post-infection. The predominantly intracellular life-cycle of Hc may render complement-mediated defenses less useful in host defense.
Fig 7. C3 and C3aR-deficiency do not dramatically alter mouse susceptibility to Hc infection.
A-B C3ar-/- mice (n≥10) and age-matched WT C57BL/6 mice (n≥10) were infected intranasally with varying doses of Hc yeast to initiate either a sub-lethal (A) or lethal (B) infection. D. C3-/- mice and age-matched WT mice were infected intranasally with a sub-lethal dose of Hc yeast. Susceptibility is illustrated by a Kaplan-Meier survival curve. ns = not significant, *p < 0.05 by logrank test. C,E. The indicated mice were infected with a sub-lethal dose of Hc. The fungal burden in lung and spleen homogenates was determined by enumeration of colony forming units (CFUs) at the indicated time points (n≥5). X-axis label for C is the same as that indicated for E.
Discussion
We report a large-scale CRISPR-Cas9 screen conducted in macrophage-like cells challenged with Hc yeast. 361 genes emerged as high-confidence modifiers of macrophage susceptibility to Hc-mediated killing, vastly expanding our knowledge of the gene networks that underpin macrophage interaction with this important pathogen. Validation of top hits revealed an under-appreciated role for GPCR signaling through C3aR in macrophage phagocytosis of fungi. These results are particularly intriguing for Histoplasma, which is an intracellular fungal pathogen that thrives within the macrophage phagosome. Therefore, elucidating the molecular events that govern Histoplasma phagocytosis is particularly important for understanding Hc pathogenesis.
It was previously established that macrophage phagocytosis of Hc is not dependent on β-glucan recognition by Dectin-1 [16], and that Hc utilizes a number of mechanisms to minimize exposure of β-glucan on the cell surface [17,18]. In contrast, CR3 has been previously implicated in non-opsonic uptake of Hc [16,19]. Our work uncovers the important role of C3aR as a pattern recognition receptor for Hc and other fungi, potentially collaborating with CR3 to facilitate uptake of pathogenic yeasts (described in Fig 8). We also discovered that C3aR-dependent phagocytosis requires serum, and that only mouse serum that was replete with C3 could stimulate phagocytosis, suggesting that a gradient of C3a emanating from the fungal surface might be critical for the C3aR-dependent local chemotaxis and stimulation of phagocytosis, as discussed below. Since Hc cannot engage Dectin-1, there is little phagocytosis of Hc in the absence of serum. In contrast, the residual serum-independent, C3aR-independent phagocytosis of zymosan may be due to Dectin-1. Given that Hc is introduced to the host via inhalation, and since complement activity is present in the bronchoalveolar fluid [22,23], innate immune recognition of Hc likely occurs in the context of complement activation in vivo. Although we did not find that C3ar-/- mice were less susceptible to Hc infection, this does not rule out a role for C3aR in vivo, as other signals in the lung, such as C5a or C-type lectin receptors, might compensate for C3aR-deficiency to allow Hc to access its intracellular niche.
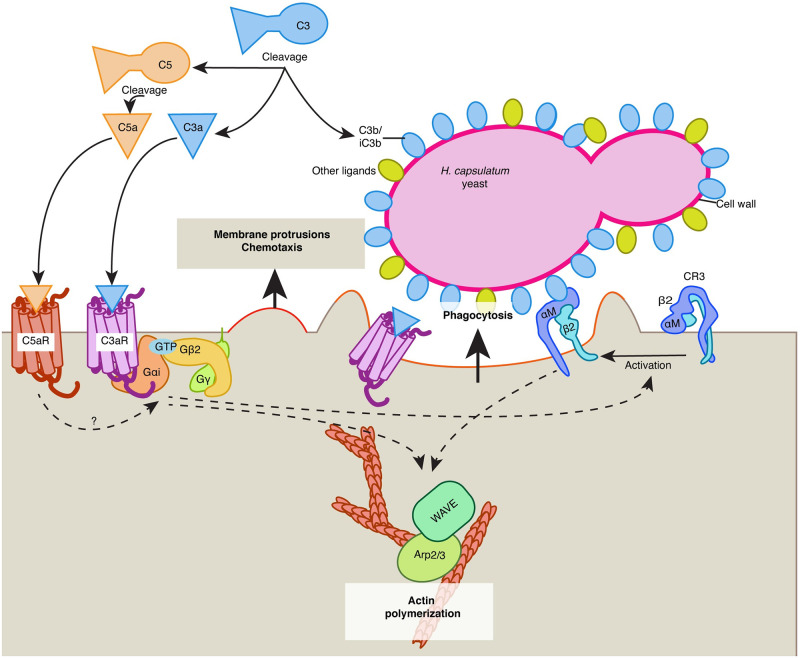
Fig 8. Model for the role of complement and C3aR in macrophage recognition of Hc yeast.
We propose the following model for the role of complement and C3aR in macrophage recognition of Hc: C3, derived from serum, reacts with the Hc cell-wall, leading to C3b/iC3b deposition on the cell-wall, and release of C3a, which diffuses away from the yeast surface leading to a concentration gradient emanating from the yeast cell-wall. C3a activates C3aR, which signals through Gαi and Gβ2 to promote the formation and directional movement of actin-rich membrane protrusions, and possibly to promote local activation or increased motility of the integrin receptor CR3. Active CR3 can then recognize C3b/iC3b or other features of the Hc cell-wall. C3aR and/or CR3 activation then coordinates actin polymerization and phagocytic cup formation by regulating the activity of actin polymerization regulators Arp2/3 and SCAR/WAVE. In the presence of C5-containing serum, the C5 convertase can similarly catalyze the cleavage of C5 at the fungal surface, leading to release of C5a and activation of C5aR, which may also drive local chemotaxis and activation of phagocytic integrins to promote phagocytosis.
The vast majority of genes identified in the screen were resistance-promoting hits, which may reflect limitations in the pooled screening approach, or the efficiency at which Hc evades macrophage defenses (in other words, it is challenging to increase macrophage sensitivity to Hc). Within these hits, we identified genes with previously described involvement in phagocytosis and Hc recognition, which validates our approach and is consistent with the requirement for Hc uptake to trigger the process of macrophage lysis. Our screen also revealed a role for GPCR signaling in Hc-host interactions. In addition to C3ar, the highest scoring protective hits included a set of genes that regulate signaling and receptor trafficking following GPCR engagement [58,59]. We validated that several of these genes, including C3ar1, Gnb2, and Arrb2, facilitate macrophage phagocytosis of Hc. While G-protein coupled receptor (GPCR) signaling is traditionally thought to play a role in chemotaxis rather than phagocytosis [77,78], several studies have implicated G-protein activity directly in cytoskeleton coordination during phagocytosis [69,79,80]. Both chemotaxis and phagocytosis depend on precise regulation of the actin cytoskeleton, and signaling often converges on the same cytoskeleton remodeling machinery [77,81]. Additionally, previous studies have shown that the mobility and activity of phagocytosis receptors is increased at the leading edge of a cell [82], and that active probing of the local environment by macrophages is critical for efficient binding of targets [83], suggesting strong coordination between chemotaxis and phagocytosis. We also identify the ER membrane complex, which facilitates the folding of transmembrane proteins with multiple membrane-spanning regions [56,57]. We show that Emc1 promotes macrophage phagocytosis of Hc, and is required for surface expression of C3aR, but not CR3 subunits. Thus, we propose that the EMC indirectly participates in phagocytosis due to its role in folding receptors such as C3aR.
Other genes and complexes identified in this screen may play important roles in Hc interaction with macrophages. To uncover the nature of their involvement will require further study. These include the ragulator complex, which activates mTORC1 upon nutrient deprivation and regulates autophagic flux that can be critical for defense against intracellular pathogens [55]. This complex also has been found in screens for phagocytosis regulators [52], and has been shown to modulate phagocytic flux in microglia [84]. Other hits may affect Hc-macrophage interactions through indirect means, or promote nutrient acquisition and intracellular replication within the phagosome. We also identified ubiquitin ligases such as Ubr5 and Trip12, which regulate histone ubiquitylation upon DNA damage [85]. Ubr5 has also been shown to down-regulate TLR signaling [86]. Validation in macrophage-like cells demonstrates that Ubr5 is required for Hc-induced lysis, but not macrophage phagocytosis of Hc, suggesting that Ubr5 promotes intracellular replication of Hc or macrophage cell death.
Since the identification of C3aR as a phagocytic receptor was intriguing, we further characterized its role in macrophage phagocytosis of Hc and other targets. While we found that C3aR was required for phagocytosis of several species of fungi, C3aR did not play a general role in phagocytosis, as C3ar-/- macrophages were not defective in uptake of E. coli or latex beads. Previous studies have demonstrated that C3aR promotes phagocytosis of damaged neurons [87,88], myelin particles [67] and protein aggregates [65]. C3aR has also been implicated in macrophage phagocytosis of uropathogenic E. coli [64], but not Pseudomonas aeruginosa [63]. Further study is needed to determine the shared characteristics of particles that require C3aR for optimal phagocytosis, such as particle size, reactivity with complement, or other biochemical properties. Nonetheless, the identification of C3aR as an important phagocytic receptor for fungi implies that it may contribute to host defense against fungal pathogens. While we do not find that C3ar-/- mice show dramatically altered susceptibility to Hc infection, more study is needed to determine whether C3aR affects host susceptibility to other fungal pathogens or modulates the immune response to fungi. Such study will be essential to determining the therapeutic benefit of targeting complement or C3aR in the treatment of invasive fungal infections.
We found that heat-treated fetal bovine serum (FBS) added to the macrophage media promoted fungal phagocytosis in a C3ar-dependent but opsonization-independent manner. This suggests that FBS promotes phagocytosis predominantly by generating C3a that activates C3aR, although the mechanism by which FBS-derived C3a might play a role independent of robust C3b opsonization is unclear. We also showed that mouse and human serum was able to stimulate macrophage phagocytosis of Hc in a C3-dependent manner, and that C3-dependent opsonization of Hc promoted macrophage phagocytosis, consistent with studies showing a role for C3b/C3bi in recognition of fungal pathogens [36]. Surprisingly, the ability of serum from C57BL/6 mice and normal human serum to promote macrophage phagocytosis of Hc was not strongly dependent on C3aR. We demonstrated that C5a-C5aR signaling may compensate for C3aR deficiency, since macrophage phagocytosis of Hc in the presence of C5 immuno-depleted human serum or serum from DBA2 mice, which are C5-deficient, showed increased C3aR dependency. This is not surprising given that C5a and C5aR have been previously implicated in innate immune recognition of fungi [43,44,89]. Future work could definitively demonstrate a role for C5aR in macrophage phagocytosis of Hc by gene deletion or pharmacological antagonism.
To investigate the role of C3aR in macrophage phagocytosis of Hc, we demonstrated that C3aR localizes to the Hc containing phagosome at early time-points during infection. Localization of C3aR to the phagosome suggests direct involvement of C3aR in Hc recognition or cytoskeleton remodeling. Alternatively, C3aR might not directly participate in phagosome formation, but reside at the plasma membrane sites that participate in Hc phagocytosis.
Finally, live imaging of actin dynamics in macrophages during Hc infection revealed that C3aR promotes the directional movement of actin-rich membrane protrusions that aid in the capture of Hc yeast. This observation is consistent with the ability of C3a to promote chemotaxis of innate immune cells including macrophages [28], and the role of G-protein signaling in activating cytoskeleton remodeling at the leading edge and the phagocytic cup [69,79,80]. We did not find strong evidence that C3aR promotes chemotaxis towards Hc yeast in trans-well assays, and we were not able to restore phagocytosis in C3ar-/- macrophages by forcing contact between macrophages and Hc, suggesting that C3aR participates in local, short-distance rather than long-distance migration during fungal phagocytosis, as has been proposed for anaphylatoxin-mediated microbial capture [75]. C3aR may also promote optimal phagocytosis by spatially coordinating receptor mobility [82] or activation [59] at the leading edge. A role for receptor activation is supported by the partial rescue of Hc phagocytosis in the absence of C3aR or in serum-free media by activating macrophages with PMA or using thioglycollate-elicited peritoneal macrophages. We postulate that a gradient of C3a diffusing away from the Hc surface is critical for this activity, as the uniform distribution of recombinant C3a alone was not sufficient to stimulate macrophage phagocytosis in the absence of serum. C3aR likely cooperates with receptors, such as CR3, that can directly recognize ligands on the Hc surface, such as C3b/iC3b, or lower-affinity ligands such as Hsp60 that require C3aR-mediated activation of phagocytic integrins for efficient recognition. Further investigation is needed to decipher the precise mechanism by which the C3a-C3aR pathway contributes to Hc recognition.
Materials and methods
Ethics statement
All mouse experiments were performed in compliance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee at the University of California San Francisco (protocol AN18753-03A). Mice were euthanized by CO2 narcosis and cervical dislocation consistent with American Veterinary Medical Association guidelines.
Strains and culture conditions
J774A.1 cells (ATCC) were cultured in Dulbecco’s modified Eagle’s medium high glucose (DMEM, UCSF media production) with 10% heat-inactivated fetal bovine serum (FBS; Corning or Atlanta), penicillin and streptomycin (pen/strep, UCSF media production). Cells were passaged by detaching with a disposable cell scraper. HEK293T cells (ATCC) were cultured in DMEM with 10% FBS and pen/strep. Platinum-E (Plat-E) retroviral packaging cells (CellBioLabs) were a gift from Jason Cyster (UCSF) and were maintained in DMEM supplemented with 10% FBS, pen/strep, glutamine, and 10mM HEPES (UCSF media production). Plat-E and HEK293T cells were passaged by detaching cells using 0.05% Trypsin-EDTA (UCSF media production). WT C57BL/6J (JAX: 000664), Rosa26-Cas9 (JAX:26179), C3ar-/- (JAX:33904), C3-/- (JAX:29661), and DBA2/J (JAX:000671) mice were obtained from Jackson Laboratories and bred in the UCSF mouse barrier facility. Bone marrow from 6-to 8-week-old female mice was isolated from femurs and tibias, and differentiated into bone marrow-derived macrophages (BMDMs) by culturing in BMM (bone marrow macrophage media) + 10mM HEPES as described previously [90]. BMM contains 10% CMG-conditioned media and 20% FBS. Mammalian cells were frozen in complete media supplemented with 10% DMSO and 50% FBS, and stored in liquid nitrogen. Thioglycollate-elicited peritoneal macrophages (Thio-Pmacs) were isolated and cultured as previously described [16,91]. Briefly, 8–12 week-old mice were injected I.P. with 3% thioglycollate broth. Four days later, mice were euthanized, peritoneal lavage was collected, and cells were cultured in RPMI supplemented with 10% FBS, pen-strep, glutamine, sodium-pyruvate, non-essential amino acids, and β-mercaptoethanol for 24h in tissue-culture conditions prior to phagocytosis or cytokine release assays. Histoplasma capsulatum (Hc) strain G217B (ATCC 26032) and G217B ura5Δ were kind gifts from William Goldman (University of North Carolina, Chapel Hill). mCherry-expressing Hc was generated as described previously [92]. The Hc cbp1 mutant strain, G217Bura5Δcbp1::T-DNA with a Ura5-containing episomal vector, was generated previously [7,9]. Hc cultures were grown on Histoplasma macrophage medium (HMM) agarose plates or in liquid HMM on an orbital shaker as previously described, and stocks generated from single colonies were frozen in HMM with 15% glycerol in single-use aliquots [93]. Mammalian cells and Hc cultures were maintained in humidified tissue-culture incubators at 37°C with 5% CO2. Hc was grown on HMM-agar plates (supplemented with 0.175 mg/mL uracil to grow Hc ura5Δ) for 1–2 weeks, and passaged in 1:25 HMM liquid culture every-other day for five days to obtain logarithmic-phase Hc yeast-cultures (OD600 = 5–7). Yeast cells were collected, resuspended in Ca++ and Mg++-free D-PBS (D-PBS), sonicated for 3 seconds on setting 2 using a Fisher Scientific Sonic Dismembrator Model 100, and counted using a hemocytometer. Hc yeast were adjusted to the appropriate concentration in D-PBS. For macrophage infections, Hc was added to the macrophage cultures, and allowed to settle onto the cells unless otherwise specified. Candida albicans (Ca) strain Sc5314 (ATCC MYA-2876) was a kind gift from Alexander Johnson (UCSF). Ca was grown on YEPD (2% peptone, 1% yeast extract, 2% glucose) agar or liquid media at 30°C. Coccidioides posadasii Silveira strain was a generous gift from Dr. Bridget Barker (Northern Arizona University). Coccidioides arthroconidia were obtained as previously described [94], by growing Coccidioides on 2xGYE (2% glucose 1% yeast extract) solid agar in flasks at 30°C for 4–6 weeks. At the time of collection, arthroconidia were dislodged with a cell scraper in PBS, filtered through miracloth to remove hyphal fragments, resuspended in PBS and stored at 4°C for up to 6 months. Arthroconidia concentration was measured by counting arthroconidia on hemocytometer.
Generation of stable J774A.1 cell-lines for CRISPRKO and live-cell imaging experiments
Gene-targeting sequences were cloned into the pMCB306 lentiguide-puro vector as previously described [95]. S5 Table lists the CRISPR targeting sequences used. The lentiviral Lifeact-monomeric eGFP-Blast vector was a kind gift from Diane Barber (UCSF). The Ef1a-Cas9-Blast lentiviral vector (pMCB393) was generated previously [49]. To generate lentivirus particles, HEK293T cells were transfected using polyethylenimine (PEI) with second-generation (sgRNA, Lifeact) or third-generation (Ef1a-Cas9-Blast) packaging plasmids and the desired transfer plasmid. Lentivirus was harvested 48-and 72-h later, and filtered through a 0.45 μm polyvinylidene fluoride (PVDF) or polyethersulfone (PES) filter (Millipore). Viruses were concentrated using the Lenti-X concentrator (Takara) according to the manufacturer’s instructions. Concentrated lentivirus (Cas9: 20X, lentiguide-puro: 1-2X, Lifeact: 5X) was added to J774A.1 cells for 12–24 h (with 8 μg/mL polybrene for Cas9), after which virus-containing media was removed and cells were grown in complete DMEM. Starting at 3 days post-transduction, cells were grown under selection with Blasticidin (2 μg/mL) or puromycin (2.5 μg/mL) for 3 days, and expanded without selection for at least 3 days or until the desired number of cells was obtained. To obtain clonal Cas9-expressing J774A.1 cells, live cells were harvested and single-cell sorted using a FACSAriaII cell-sorter into 96-well plates containing complete media supplemented with 50% sterile-filtered J774A.1 conditioned media, and expanded for 3 weeks. The Cas9 activity of the J7-Cas9 clones was determined following transduction with the lentiguide-puro-eGFP vector containing a GFP-targeting sgRNA, and measuring eGFP silencing after puromycin selection by flow cytometry. The J7-Cas9 clone with the highest eGFP-silencing activity was used to generate the pooled CRISPR libraries and individual CRISPRKO cell-lines. The efficiency of Cas9-mediated gene-targeting was assessed by PCR-amplifying the targeted locus in control and CRISPRKO cells, performing Sanger sequencing, and analyzing sequencing chromatograms using the TIDE webtool [96]. CRISPR targeting efficiency as measured by TIDE analysis for the sgRNAs used is shown in S5 Table.
Pooled CRISPR-Cas9 screens
We used pooled mouse sgRNA sub-libraries that were generated previously [49], some of which are available on Addgene (#1000000121–1000000130). Each library covers 500–1500 genes with 10 sgRNAs/gene and includes 750 negative control sgRNAs (375 non-targeting and 375 safe-targeting sgRNAs). We performed screens on all of the sub-libraries, except for the Mouse Unique sub-libraries, which contain mouse genes that do not have known orthologues in humans. Taken together, our screens covered 16,781 mouse genes. Lentivirus was generated by transfecting HEK293T cells seeded in 15 cm dishes with sgRNA plasmids and second-generation packaging plasmid as described previously [97]. Lentivirus was harvested at 48-and 72-h post-transfection, filtered through 0.45 μm PES filters, pooled, then concentrated using the Lenti-X concentrator (Takara) according to the manufacturer’s instructions. J774A.1 cells stably expressing LentiCas9-Blast (generation described above) were incubated with 2X concentrated lentivirus for 24h at 1000X coverage in T-225 or T175 flasks for an MOI of 0.2–0.5 as determined by flow cytometry of mCherry expression at 3 days post-transduction. We then performed selection for transductants using puromycin (2.5 μg/mL) for 3 days until >90% of the cells were mCherry-positive by flow cytometry. Cells were allowed to recover from puromycin selection for three days before screening. Cells were split into two conditions, and screening was performed in duplicate. One condition was infected with Hc ura5Δ and subjected to 2–3 pulses of uracil to initiate fungal growth and macrophage lysis (see S4 Table for details specific to each sub-library). J774A.1 CRISPRKO libraries, seeded at 1000X library coverage in T-225 or T-150 flasks, were infected with Hc ura5Δ at a multiplicity of infection (MOI) of 5 yeast/macrophage. Yeast were allowed to settle onto the monolayer and incubated for 2 h. The cells were washed once with D-PBS to remove extracellular yeast, and incubated in the presence of 0.35 mg/mL uracil for 2 d until ~50% of the monolayer was cleared. Then, the monolayer was washed 3X with D-PBS to remove dead macrophages and extracellular yeast, and incubated for 2–5 days in complete media without uracil to allow the monolayer to recover. Then, uracil was re-introduced to the culture media for 1–2 d to re-initiate fungal growth and lysis. The addition and removal of uracil was performed 1–2 times depending on the speed at which the monolayer recovered. Uninfected cells were passaged in parallel every 2 d by detaching adherent cells with a cell-scraper, counting using a hemocytometer, and re-seeding into new flasks at 1000X coverage. Uninfected cells were pulsed with uracil during passaging to match the Hc infection. At the end of the screening period, cells were washed and harvested by detaching with a cell-scraper. Genomic DNA was extracted from the cells using the DNA blood midi or maxi kit according to the manufacturer’s instructions, with the inclusion of a brief centrifugation step after cell lysis to remove un-lysed Hc yeast before addition of ethanol and application to the column. Guide frequencies were quantified by PCR amplification and deep sequencing using an illumina NextSeq 500 as previously described [95].
Analysis of CRISPR-Cas9 Screens
Sub-library screens were analyzed separately using casTLE version 1.0 as previously described [51]. The results are listed in S1–S3 Tables. Briefly, the distribution of guides was compared between the uninfected and Hc-infected samples, and guide enrichments were calculated as log ratios between the infected and uninfected samples. A maximum likelihood estimator was used to estimate the effect size for each gene and the log-likelihood ratio (confidence score, or casTLE score) by comparing the distribution of the 10 gene-targeting guides to the distribution of negative control guides. An effect size of 1 roughly corresponds to one log2 fold change of the gene compared to the negative controls. P values were determined by permuting the gene-targeting guides in the screen and comparing to the distribution of negative controls using casTLE, and false discovery rate (FDR) thresholds for defining hits were calculated using the Benjamini-Hochberg procedure. We used a threshold of 5% FDR to define hits. Results from the separate sub-library screens were concatenated and visualized using JavaTreeview [98]. GO-biological process analysis was performed using Gorilla [99] using an un-ranked list of genes that passed the 5% FDR cutoff as the target list and all of the genes detected in the screen as the background list.
Competitive fitness assays in J774A.1 cells
J774A.1-Cas9 (WT) cells were mixed with CRISPRKO J774A.1-Cas9 cells harboring the lentiguide-puro vector, which drives expression of a gene-targeting sgRNA and an eGFP marker (75% WT cells, 25% CRISPRKO cells). 3.5X105 cells/well were seeded in tissue culture (TC)-treated 6-well plates. 12–24 h later, the cells were infected with Hc ura5Δ at an MOI = 5, which was incubated with the monolayer for 2 h followed by a D-PBS wash step. The cells were incubated in complete media containing 0.35 μg/mL uracil for 2 d, until lysis of >50% of the monolayer was observed. Then cells were recovered by washing 3X with D-PBS, and incubating in complete media in the absence of uracil for 2 d. Uninfected cells were detached by scraping and passaged to prevent overcrowding, and were subjected to the same washing and media conditions as the Hc-infected cells. Following the recovery period, surviving cells were harvested and stained, and GFP-expression was analyzed by flow cytometry.
Generation of CRISPR-knockout BMDMs
The pSIN MuLV sgRNA retroviral transfer plasmid (U6 guide tracer EF1a Thy1.1 P2A Neo) was a kind gift from Jason Cyster (UCSF). The sgRNA cloning site, U6 promoter, and selection marker of pSIN was replaced to match that of pMCB306 using the Gibson Assembly Cloning Kit (NEB) to generate the transfer plasmid (BAS2186) used for these studies. Gene-targeting sgRNA sequences (S5 Table) were cloned into the vector as previously described for pMCB306 [95]. To generate viral particles for expression of sgRNAs, Plat-E retroviral packaging cells were transfected with the transfer plasmid in antibiotic-free complete DMEM. Virus was harvested at 48 h and 72 h post-transfection and filtered through a 0.45μm PES filter. Bone marrow from female 6-8-week-old Rosa26-Cas9 mice was isolated and cultured for 2 d in BMM as described above. Non-adherent bone marrow cells were harvested, and 2X106 cells per well were infected with 2 mL fresh MuLV supernatant by centrifugation (2400 RPM, 2 h, RT) in 6-well non-TC-treated plates with 10 μg/mL polybrene. Viral supernatant was removed, and cells were incubated overnight in BMM. Both adherent and non-adherent bone marrow cells were infected with viral supernatant again as described above with the 72h viral harvest. 24h after the second viral spinfection, BMDMs were grown under puromycin selection (4 μg/mL) for 3 days, grown for an additional 3–5 days in BMM without puromycin, and harvested as previously described. Retroviral infection and selection were verified by Thy1.1 staining and flow cytometry. The efficiency of Cas9-mediated gene-targeting was assessed by PCR-amplifying the targeted locus in control and CRISPRKO cells, performing sanger sequencing, and analyzing sequencing chromatograms using the TIDE webtool [96]. Results are shown in S5 Table.
Competitive Hc phagocytosis assays
WT and CRISPRKO J774A.1 cells were mixed as described above (75% WT and 25% CRISPRKO), and seeded at 3X105 cells/well in tissue-culture-treated 12-well plates and incubated for 12–24 h prior to infection. Hc yeast expressing mCherry were added to the monolayers at an MOI = 2, and incubated for 1h at 37°C. Cells were then washed with ice-cold HBSS and harvested by pipetting the cells off of the well with HBSS. Similarly, Cas9-expressing BMDMs (WT) were mixed with Cas9-BMDMs transduced with a retroviral vector driving expression of a gene-targeting sgRNA (CRISPRKO) (75% WT and 25% CRISPRKO). BMDMs were added at 5X105 cells/well to non-TC-treated 12-well plates in BMM for 12–24 h, then infected with mCherry-expressing Hc for 1 h in BMM. Phagocytosis and GFP or Thy1.1 expression was measured using flow cytometry.
FITC labelling of Zymosan and Coccidioides posadasii arthroconidia
FITC-labelling was performed as described previously for Hc yeast [16]. Briefly, Zymosan A (Sigma) was sonicated for 3 seconds on setting 2 using a Fisher Scientific Sonic Dismembrator Model 100, washed with 0.05 M sodium carbonate-bicarbonate buffer, and adjusted to 2X108 particles/mL. C. posadasii arthroconidia were adjusted to 5X108 conidia/mL, and washed in a sodium carbonate-bicarbonate buffer. Fungi were incubated with in 0.05M sodium carbonate-bicarbonate buffer (pH 9.5) with 0.16mg/mL FITC (Fisher, dissolved in DMSO at 5mg/mL) for 15 min at room temperature, protected from light, then washed twice with HBSA (HBSS + 0.25% BSA). Labelled zymosan was resuspended in D-PBS, counted using a hemocytometer, and frozen in single-use aliquots at -20°C. FITC-labelled arthroconidia were resuspended in PBS and counted on a hemocytometer. FITC-labelled arthroconidia were kept at 4°C and protected from light until used in phagocytosis experiments.
Serum
FBS was purchased from Atlanta Biologicals or Corning unless otherwise indicated, and stored in 500 mL bottles at -80°C. Serum was thawed to 37°C and heat-treated by incubating in a 56°C water bath for 30 min with one inversion. Following heat-treatment, serum was aliquoted and stored at -20°C. Where indicated, serum was pre-treated with zymosan at 1X108 particles/mL for 60 min at 37°C. Zymosan was removed by centrifugation and sterile filtration (0.22 μm PVDF filter) before addition to macrophage media. To collect mouse serum, male mice were euthanized using CO2, and blood was collected by cardiac puncture. Blood was placed in a 1.5 mL Eppendorf tube and allowed to coagulate by incubation at room temperature for 1 h, and for an additional 30 min on ice. Coagulated blood was centrifuged, and the supernatant was harvested. Serum was used fresh or stored at -80°C in single-use aliquots. Untreated, C3-depleted, and C5-depleted human serum was purchased from Complement Technology, Inc.
Macrophage phagocytosis assays
BMDMs were thawed and seeded in BMM in12-well non-TC-treated plates at 5X105 cells/well (flow cytometry) or onto ethanol-sterilized glass coverslips in TC-treated 24-well plates at 2X105 cells/well (microscopy), and allowed to adhere for 12–24 h. Thioglycollate-elicited macrophages were seeded in complete media in 24-well TC-treated plates at 6X105 F4/80+ cells/well and allowed to adhere for 12-24h. Prior to infection, the cells were washed with D-PBS and fresh media was added. For some experiments, macrophages were pre-treated with the following compounds and vehicle controls: pertussis toxin (Sigma, 1 μg/mL 2 h), SB290157 (Sigma, 1 μM 5 min), anti-CD18 clone GAME-46 or isotype control (BD, 10 μg/mL 90 min), recombinant C3a (R&D systems), and phorbal 12-myristate 13-acetate (Sigma). To ensure a consistent MOI across harvests, prior to infection, a well of cells was harvested using dissociation buffer, and counted using a hemocytometer. The count was used to calculate the number of cells that had adhered to the dish to determine the number of fungal cells or particles to add to the well for the appropriate MOI. For Hc phagocytosis assays, mCherry-expressing Hc yeast was added to the macrophage monolayer. For Zymosan phagocytosis assays, FITC-labelled zymosan was sonicated and added to the monolayer. Fluorescein-conjugated E. coli bioparticles (Invitrogen) were prepared according to the manufacturer’s instructions and added to macrophage monolayers at an MOI = 4. Carboxylate-modified microspheres (2.0 μm and 0.5 μm) were sonicated and added to the macrophage monolayers at an MOI of 2. Phagocytosis of the above substrates was analyzed by flow cytometry. Logarithmic cultures of Candida albicans yeast grown in YEPD liquid media were harvested, washed 3X with D-PBS, counted, and added to macrophage monolayers on coverslips at an MOI of 3. Coverslips were washed 2X with DPBS and stained with 35 μg/mL calcofluor white for 1 min. Then, the coverslips were washed, fixed with 4% PFA at 37°C for 20 min. PFA was quenched by washing 3X with 100 mM glycine. Cells were permeabilized with 0.1% Triton-X-100 (5 min), and blocked with 1% BSA. Both intracellular and extracellular C. albicans yeast were detected by staining with a FITC-conjugated anti-C. albicans antibody (abcam, 1:1000) overnight 4°C. FITC-labelled C. posadasii arthroconidia were added to the coverslip an MOI of 1, then spun for 15 min at 550g to ensure contact between macrophages and arthroconidia. At the indicated times, the coverslips were washed and stained with 35 μg/mL CFW (2 min), washed once, fixed with 4% PFA, then washed with PBS. For C. albicans and C. posadasii experiments, Coverslips were mounted and imaged at 40X magnification. 16 fields along a grid were automatically selected. To determine the phagocytosis rate, the cell-counter plugin in FIJI was used to manually count the total number of macrophages and the number of macrophages with at least one intracellular fungus, determined by exclusion of CFW staining. The phagocytosis rate was calculated as the number of macrophages with at least one intracellular fungal particle divided by the total number of macrophages.
Serum opsonization and analysis of C3 deposition by immunofluorescence microscopy
Hc was incubated at 1X108 yeast/mL with 10% serum and the indicated chelators (10 mM EGTA or EDTA) in PBS for 30 min at 37°C. The yeast/particles were washed 2X with PBS, and co-cultured with BMDMs, or stained with a FITC-conjugated anti-mouse C3 antibody (MP Biomedicals, 1:200) for 1h at RT. Following staining, yeast/zymosan were washed 2X with PBS, and fixed with 4% PFA after spinning onto poly-L-Lysine-coated coverslips. Coverslips were washed 2X and imaged at 60X magnification to visualize mouse C3 deposition on the cell-wall.
Analysis of C3aR localization by immunofluorescence microscopy
2X105 BMDMs were seeded onto ETOH-sterilized glass coverslips in 24-well plates. Phagocytosis was synchronized by pre-incubating macrophage monolayers on ice, and centrifuging mCherry Hc yeast or fluorescent latex beads (MOI = 5) at 4°C onto the monolayers, followed by incubation at 37°C for up to 30 min. Coverslips were washed with D-PBS and fixed with 4% PFA for 20min at RT. Coverslips were blocked with PBS + 5% FBS for 1 h at RT, and stained with an anti-mouse C3aR antibody (Clone 14D4, Hycult, 1:1000) overnight at 4°C in 5% FBS. Coverslips were washed with 5% FBS and stained with AlexaFluor-488-conjugated goat anti-rat IgG (Invitrogen, 1:500) for 1 h at RT. Coverslips were imaged at 60X magnification. Optical sectioning was performed to obtain Z-stacks (0.4 μm step-size, 5 μm thickness), and 6 fields were imaged per coverslip. To quantify C3aR localization to the Hc or bead-containing phagosome, we used ImageJ to define a 0.5 μm-thick phagosome perimeter outside of the Hc or bead particle. To generate the mask, we set a threshold on the Hc (>600) or bead (>800) channels and generated a binary mask. The mask was then dilated, outlined, and dilated an additional two times. Example binary masks are shown in S8B and S8C Fig. Then, we use the 3D ROI manager [100] plugin in ImageJ to generate 3D ROI objects from the binary mask, and to quantify the mean intensity of the C3aR signal within the phagosomal volume. We subtracted the background signal, measured on phagosomes in C3ar-/- BMDMs subjected to the same staining and analysis pipeline.
Live cell-imaging and cell tracking
5X103 Lifeact-meGFP-expressing J774A.1 macrophages (generated as described in “generation of stable cell-lines”) were seeded into 96-well glass-bottom plates (Cellvis) and allowed to adhere for 12–24 h. Culture media was replaced with fresh phenol-red-free DMEM containing 10% FBS with either a vehicle control (DMSO) or 10 μM C3aR antagonist (SB290157) and incubated for 5 min. mCherry-expressing were added to the cells at an MOI of 5, and centrifuged briefly (15 sec) to facilitate contact with the macrophages. Cells were imaged every 90 sec for 45 min at 20X magnification. An Okolab stagetop incubator with temperature and humidity control was used to maintain optimal conditions (37°C and 5% CO2). Four fields were imaged per duplicate well. Actin-rich (eGFP+) membrane protrusions of macrophages that capture Hc yeast were tracked manually using the ImageJ plugin MtrackJ [101]. Tracking was started at the membrane point closest to the Hc yeast when the yeast first appeared close to the location at which it was eventually captured. The position of the yeast was used as a reference. The track was terminated when the Hc was successfully engulfed (as visualized by formation of an actin collar around the Hc yeast), or when the imaging period terminated. The phagocytosis rate is reported as the distance from the J774A.1 cell-membrane to the Hc target at the start of tracking divided by the time elapsed until the yeast was successfully engulfed. The mean velocity (mean displacement/time across tracked points) and outreach ratio of the tracks (the max displacement/net displacement) were calculated as described [74].
Confocal microscopy
For fixed imaging, coverslips were mounted onto slides using vectashield antifade mounting media, with or without DAPI (Vector labs) and sealed using nail polish. Fluorescence confocal microscopy was performed using a using a Nikon Ti-Eclipse inverted microscope with a Yokogawa spinning disk CSU-X1 and an Andor Clara CCD camera. Image analysis was performed using ImageJ.
Flow cytometry
BMDMs were washed and harvested using HBSS-based cell dissociation buffer (Thermo scientific) by incubating at 37°C for 10 min and pipetting with ice-cold HBSS. J774A.1 cells were washed with ice-cold HBSS, and harvested by spraying cells off of the well with ice-cold HBSS using a P1000 pipette. Cells were kept on ice and protected from light for subsequent steps. Cells were stained with fixable viability dye efluor450 (ebioscience; 1:1000) for competitive fitness assays and CD11b/CD18 staining, or fixable viability dye efluor780 (ebioscience; 1:500) for phagocytosis assays for 20 min. Cells were washed with FACS buffer (2% FBS and 5 mM EDTA in PBS) prior to staining with antibodies and/or Calcofluor White M2R (Sigma, 1 μg/mL) in FACS buffer for 15-20min. The following antibodies and dilutions were used: PerCP-conjugated anti-Thy1.1 (clone OX-7, biolegend, 1:100), PE, FITC, or AlexaFluor647-conjugated anti-CD11b antibody (clone M1/70, UCSF mAB core, 1:500 for BMDMs, 1:1000 for J774A.1), AlexaFluor-647-conjugated anti-F4/80 (BM8, UCSF mAB core, 1:500), and AlexaFluor-647-conjuaged anti-CD18 antibody (M18/2, Biolegend, 1:100). Cells were washed with FACS buffer. For phagocytosis and competitive fitness assays, cells were washed with D-PBS, and fixed using BD stabilizing fixative for 15min, washed with D-PBS, and kept on ice prior to data acquisition. For indirect flow cytometry measurement of C3aR expression, 5X105 BMDMs were fixed using BD stabilizing fixative (20min on ice). Cells were blocked for 20min with PBS5 (PBS+5% FBS) and stained with a C3aR antibody (Clone 14D4, Hycult, 1:500) in PBS5 for 20min on ice. Cells were washed with PBS5 and stained with APC-conjugated goat anti-rat IgG (Biolegend, 1:200) for 20 min. Cells were washed with PBS5, and resuspended in PBS for flow cytometry analysis. Flow cytometry acquisition was performed using a BD LSRII analyzer in the UCSF Parnassus Flow Core. Analysis was performed using FlowJo v. 7 or 10. Where necessary compensation was performed with single-color controls using FlowJo.
Trans-well migration assay
Cells and Hc were resuspended in migration media (DMEM with 0.5% fatty acid-free BSA, pen/strep, and 10 mM HEPES) or complete DMEM with 10% FBS. Inhibitors were added to both the well and the insert when used. 6.5 mm transwell permeable supports with 5 μm pore polycarbonate membranes (Costar) were used. Migration assays were performed in duplicate. 600 μL Hc G217B yeast was added at the indicated concentration to the well of a 24-well transwell plate. 2X105 J774A.1 cells in 100 μL media were seeded into the transwell insert, and plates were incubated at 37°C with 5% CO2 for 3 h with minimal disturbance. Media was removed from the insert, which was dipped once in D-PBS, then placed in crystal violet stain (0.5% crystal violet, 20% methanol) for 10 min at RT. Supports were rinsed with dH2O, and a Q-tip was used to gently wipe cells off of the top of the membrane, and dried at RT. Membranes were mounted on slides, and 3 fields per membrane were imaged using a Leica DM 1000 microscope at 10X magnification. Cells in each microscopic field were counted manually using the cell counter plugin in ImageJ.
Cytotoxicity assays
7.5X104 BMDMs were seeded per well of a 48-well plate and infected with Hc G217B at an MOI of 0.5 in the presence of phenol-red-free BMM. 1.875X104 J774A.1 cells were seeded per well of a 48-well plate and infected with Hc at an MOI of 5 in phenol-red-free complete DMEM. Where indicated, media was supplemented with 0.35mg/mL uracil. To recover J774A.1 cells from infection with Hc ura5Δ, cells were washed with D-PBS, and grown in complete media that did not contain uracil for 3 days. Recovered cells were re-seeded and incubated with complete media with or without uracil supplementation. At the indicated time points, the amount of lactate dehydrogenase (LDH) in the culture supernatant was measured as described previously [102]. Macrophage lysis is calculated as the percentage of total LDH from uninfected macrophages lysed in 1% Triton-X at the time of infection. Due to continued replication of macrophages during the experiment, the total LDH at later time points is greater than the total LDH from the first time point, resulting in an apparent lysis that is greater than 100%. To quantify cell depletion and recovery during infections of J774A.1 cells, we measured macrophage DNA remaining in the wells as previously described [10]. Briefly, we washed the cells with D-PBS, added ddH2O to the well to lyse the macrophages, and measured the amount of macrophage DNA in the wells using the picoGreen DsDNA reagent (Invitrogen). Fluorescence intensities were measured using the quantitative plate read option on an Mx3000P QPCR machine (Agilent).
Intracellular fungal growth assay
7.5X104 BMDMs were seeded per well of a 48-well plate and infected in triplicate with Hc at an MOI of 0.5. At the indicated time points, culture supernatants were removed and 500 μL ddH2O was added. Macrophages were osmotically and mechanically lysed by pipetting up-and-down, and plated on HMM agarose at the appropriate dilutions as described previously [7]. After incubation at 37°C with 5% CO2 for 12–14 days, colony forming units (CFUs) were enumerated. To prevent analysis of extracellular replication, CFUs were not monitored after the onset of macrophage lysis.
CBA and ELISA assays
BMDMs were seeded at 3X105 cells/well in 48-well plates (TC-treated), and infected with Hc in triplicate (MOI = 10 for CBA and MOI = 2 for C3 ELISA). Thio-Pmacs were seeded at 8X105 F4/80+ cells/well and infected with Hc in triplicate (MOI = 10). Supernatants were collected at the indicated times and either used fresh for assays or snap-frozen in LN2 and stored at -80°C. Mouse lung homogenates (see mouse infections) were thawed, and debris was pelleted by centrifugation. Macrophage supernatants were filtered using 0.2 μm glass fiber filter plates (Pall corporation) by centrifugation, and lung homogenate supernatants were filtered using spin-X cellulose acetate spin filters (Costar) by centrifugation. Tnf-α, Il-1β, Il-6, and Ifn-γ levels were measured using the appropriate mouse CBA flex set (BD) according to the manufacturer’s instructions. Data were acquired using a BD LSRII flow cytometer and analyzed using FCAP array software (BD). The colorimetric Mouse Complement C3 ELISA kit (Abcam) was used according to the manufacturer’s instructions to quantify C3 levels in macrophage culture supernatant. Mouse serum was incubated with Hc and zymosan at 10X108 particles/mL for 30 min at 37°C. The reaction was stopped by addition of 10 mM EDTA and incubation on ice. Hc and zymosan were pelleted by centrifugation, and the supernatant was filtered using Spin-X cellulose acetate filters. Supernatants were diluted 1:200. A mouse C3a ELISA pair (BD) was used as previously described [103] according to manufacturer’s instructions to measure C3a levels in the supernatants. Corning High-Bind plates were coated with 4 μg/mL capture antibody in pH 6.5 binding buffer. PBS+10% FBS was used for blocking, and PBS+10% FBS +0.05% Tween-20 was used to dilute samples, standards, and detection antibody solutions. Biotinylated C3a detection antibody was used at 6.25 ng/mL, and avidin-HRP was used at a 1:5000 dilution.
Mouse infections
C3ar-/- mice (B6.129S4(C)-C3ar1tm1Cge/BaoluJ, JAX:033904) were purchased from Jackson laboratories and bred in the UCSF mouse barrier facility. C3-/- mice (B6.129S4-C3tm1crr/J, JAX:029661) were purchased from Jackson laboratories. 8–12 week-old mice were used for infections, and age-matched WT mice (C57BL/6J, JAX:000664) were purchased from the Jackson laboratories. Hc strain G217B yeast were grown to mid-logarithmic phase (OD600 = 5–7], and washed with PBS. Yeast suspensions were sonicated for 3 seconds on setting 2 using a Fisher Scientific Sonic Dismembrator model 100, and counted using a hemacytometer. The culture was diluted in PBS to deliver the inoculum in 25 μl. Mice were infected with a sub-lethal (5X104) or lethal (8X104) dose of yeast per mouse, and were monitored daily for signs of disease, including weight loss, hunching, panting, and lack of grooming. Animals were euthanized if they exhibited 3 days of sustained weight loss greater than 25% of their maximum weight, in addition to one other symptom. To monitor fungal burden and lung cytokine levels, 5 or more mice per group were euthanized at the indicated time points. The lungs and spleens were harvested and homogenized in PBS with a protease inhibitor cocktail (Roche). Dilutions in PBS were plated on brain heart infusion (BHI) agar plates supplemented with 10% sheep’s blood (Colorado Serum Company). Plates were incubated at 30°C for 10–14 days and colony forming units (CFUs) were enumerated. The remaining homogenate was snap-frozen in liquid nitrogen, stored at -80°C, and processed for cytokine analysis (see CBA and ELISA assays).
Statistical analysis
Two-tailed t-tests were performed using Excel (Microsoft). Logrank and Wilcoxon rank-sum tests were performed using R. Details of the statistical analysis are listed in the figure legends. Error bars indicate Standard Deviation.
Supporting information
A. Characterization of Hc-mediated lysis in J774A.1 macrophage-like cells. J774A.1 cells were infected with WT Hc, or Hc with a disruption in a gene, CBP1, that is required for Hc to lyse macrophages. Lysis over time was measured using the LDH release assay. B. Validation and clonal expansion of Cas9-expressing J774A.1 cells. Cells were transduced with an Ef1a-Cas9-Blast expression vector and grown under blasticidin selection to generate a population of Cas9-expressing cells. These were subjected to single-cell sorting and clonal expansion to generate Cas9-expressing J774A.1 clones with high Cas9 activity. Cas9 activity was measured by transducing J774A.1 cells with a guide RNA vector that co-expressed EGFP with a sgRNA targeting EGFP. Cas9 activity leads to silencing of the GFP following puromycin selection. Cas9 clone 9 was chosen for the large-scale CRISPR screens due to its high-efficiency GFP silencing. C-D. Characterizing lysis and recovery from infection with uracil pulses during infection with a Ura5-deficient Hc. J774A.1 macrophages were infected with ura5 mutant Hc in the presence or absence of exogenous uracil (0.4ug/mL). Uracil-containing cells were washed and media was replaced with uracil-poor media after 2d of lysis, which allowed the macrophages to recover. Recovery was assessed using LDH release quantification to assess lysis, and the confluency of viable cells in the wells was estimated using the pico-green dsDNA assay kit following lysis of macrophages with water. E. Macrophages that had been recovered from lysis by removal of uracil from culture media were passaged for several days, and uracil was added to selected wells. Macrophage lysis over time was monitored by assessing LDH release over time to determine whether dormant yeast would be able to re-activate upon introduction of uracil. F. Reproducibility of the casTLE score across two replicates of the screens. G. Histograms comparing the distribution of negative control sgRNAs and sgRNAs targeting Gnb2 or C3ar in the Hc infected pool compared to the uninfected pool. H. Analysis of essential gene behavior during J7 library growth. Scatter plot showing the gene effect resulting from passaging of J7s, either going from the plasmid pool to the T0 pool, or the T0 pool to the uninfected pool. Genes annotated as “essential” or “non-essential” were plotted to determine whether essential genes appeared more likely to drop out of the uninfected pools.
(TIF)
BMDMs from C3ar-/- and WT mice, in addition to BMDMs expressing Cas9 and control or Emc1-targeting sgRNAs, were stained with anti-CD18 and anti-CD11b antibodies and analyzed by flow cytometry (n = 2 biological replicates). A. Representative histograms showing CD11b and CD18 levels in control, C3ar-/-, and Emc1 CRISPRKO BMDMs. The percentage of CD11b (B) and CD18 (D) positive macrophages was analyzed. The mean fluorescence intensity of CD11b (C) and CD18 (D) were also measured.
(TIF)
BMDMs were infected with Hc (MOI = 0.5), and macrophage lysis was quantified by measuring the release of lactate dehydrogenase (LDH) into the culture supernatants over-time (n = 3 biological replicates, 3 technical replicates/biorep) (A). LDH release is presented as the percentage of total LDH present in the well (supernatant and macrophage lysate) at 2 hours post-infection. At the indicated time points, macrophages were lysed using water, and lysates were spread on agar plates. Colony forming units (CFUs) were enumerated (n = 3 biological replicates, 2 technical reps/biorep) (B).
(TIF)
A. WT and C3ar-/- BMDMs were infected with Hc (MOI10), and TNFα levels in macrophage supernatants were measured using the BD Cytometric Bead Array (CBA) kit (n = 3 biological replicates). B-D. Thioglycollate-elicited peritoneal macrophages were treated with the C3aR antagonist (SB290157, 10 μM) and infected with Hc (MOI10). TNFα (B), IL-6 (C), and IL-1β (D) levels in culture supernatants were measured by CBA. *p ≤ 0.05, **p ≤ 0.01, ns = not significant by two-tailed student’s T-test.
(TIF)
(A-C) BMDMs from WT and C3ar-/- mice were infected with Hc in the presence of 20% FBS from three different lots from 2 separate suppliers. In addition, WT BMDMs differentiated in different lots of serum were treated with 10 μM of the C3aR antagonist and infected with Hc (B). Phagocytosis of Hc was measured by flow cytometry as described previously (n = 2 biological replicates).
(TIF)
BMDMs were cultured in media containing 10% FBS, and the BMDM conditioned media was harvested. Hc was incubated with macrophage conditioned media (BMDM CM), 10% FBS, or 10% normal mouse serum (NMS) with 10 mM EGTA or EDTA as indicated for 30 min 37°C. A. Incubation with conditioned media or FBS does not lead to C3 deposition on the Hc surface. C3 deposition on Hc yeast was analyzed by immunofluorescence microscopy using an anti-C3 antibody. Scale bar = 10 μm. B. Pre-incubation of yeast with conditioned media does not improve macrophage phagocytosis of Hc. Yeast were washed and used to infect BMDMs (MOI3, 30 min) in serum-free media. Phagocytosis was assessed using flow cytometry as previously described (n = 3 biological replicates).
(TIF)
A. BMDMs secrete C3 following infection with Hc. BMDMs were infected with Hc at an MOI2 for 24h, supernatants were harvested and C3 levels were quantified using a BD mouse C3 ELISA kit. B. C3-/- BMDMs are not defective in phagocytosis of Hc yeast or zymosan. WT and C3-/- BMDMs were infected with mCherry-expressing Hc or FITC-labelled zymosan, and uptake over time was measured using flow-cytometry. Extracellular yeasts were identified by Calcofluor White staining.
(TIF)
A. Uninfected WT and C3ar-/- BMDMs were stained with a C3aR-specific antibody and imaged using confocal microscopy and optical sectioning. Representative slices of 2 biological replicates are shown. The antibody specifically detects C3aR, as staining was not observed in C3ar-/- BMDMs. C3aR exhibits punctate localization near the plasma membrane in WT BMDMs. B-C. Representative slices from confocal imaging showing the region of interest mask derived from binary operations on Hc (B) or bead (C) fluorescent channel thresholding used to measure the phagosomal C3aR intensity of Hc and bead-containing phagosomes. The original and overlaid fluorescence images are shown. Scale bar = 20 μm.
(TIF)
A. Hc stimulates chemotaxis of J774A.1 macrophage-like cells. Hc yeast were seeded into multiple-well plates at varying concentrations, and WT J774A.1 cells were seeded onto transwell permeable supports with 5 μm pores. Serum-free media supplemented with 0.25% BSA was used as the diluent in both the chamber and well unless otherwise indicated. After 3 h of migration, transwells were stained with crystal violet, and non-migratory cells were wiped off of the upper side of the transwell using a Q-tip. The number of migratory cells in each condition was quantified by microscopy (n = 2 biogical replicates, 3 fields/biological replicate). B. Migration towards Hc is Gai-dependent. J774A.1 cells with or without pre-treatment with 1 μg/mL pertussis toxin (PTX) for 2 h were seeded into transwell permeable supports and migration towards 1e5 Hc/uL was quantified as described above. The number of migrating cells was quantified as described. C. The C3aR antagonist does not inhibit macrophage migration towards Hc. J774A.1 macrophages were treated with 1 μM SB290157, a C3aR antagonist, and migration towards Hc was assessed as described. D. C3aR-deficiency moderately impacts migration of J774A.1 cells towards Hc. Cas9-expressing J774A.1 macrophages transduced with non-targeting or C3aR-targeting sgRNAs were assessed for their ability to migrate towards Hc as described previously. E. Hc-dependent migration is abolished in the presence of FBS. The transwell migration assay was performed with media supplemented with BSA or 10% FBS to determine whether FBS affected the migration of macrophage-like cells towards Hc yeast.
(TIF)
A. BMDMs were infected with Hc or zymosan at an MOI = 5 for 30 min. For the control condition, particles were added to the wells and allowed to settle onto the monolayer without intervention. For the 5 min spinfection, particles were added to the cells, and the plate was spun for 5 min at 550XG at RT before transferring to a 37°C, 5% CO2 incubator. For the ice preincubation condition, BMDMs were pre-chilled for 20 min on ice, and particles were allowed to settle onto the monolayer for 1 h on ice, then were transferred to a tissue culture incubator. Phagocytosis was measured as described previously (n = 3 biological replicates).
(TIF)
BMDMs were pre-incubated in serum-free media with varying concentrations of recombinant mouse C3a (R&D systems) or with 20% FBS for 1 h, then infected with A. Hc or B. Zymosan at an MOI of 5 for 30 min. Phagocytosis was assessed by flow cytometry (n = 2 biological replicates). C-D. BMDMs In serum-free (C) or FBS-supplemented (D) media were pre-treated with PMA for 2h and infected with Hc (MOI5, 60 min). Phagocytosis was measured by flow cytometry (n = 3 biological replicates). E. Thioglycollate-elicited peritoneal macrophages in serum-free or FBS-supplemented media were treated for 5 min with a C3aR antagonist (SB290157, 10 μM) and infected with Hc. Phagocytosis was assessed by flow cytometry (n = 3 biological replicates).
(TIF)
A-D: C3ar-/- (A-D) or C3-/- (E-H) and age-matched WT mice were infected intranasally with Hc. The levels of Il-1β (A,E), Il-6 (B,F), TNF-α (C,G), and IFN-γ (D,H) in lung homogenates were measured at the indicated time points by the cytokine CBA assay. *p ≤ 0.05, ****p ≤ 0.0001, by two-tailed student’s T-test.
(TIF)
Comma-separated-values (CSV) file showing the casTLE analysis [51] results comparing the guide abundances in the Hc infected and uninfected J774A.1 pools. Each row corresponds to a gene whose targeting sgRNA sequences were identified in at least one screen replicate (count threshold = 25). The columns are shown as follows: GeneID: ENSEMBL gene ID; Symbol: EMSEMBL gene symbol; GeneInfo, Localization, Process, and Function GO-terms assigned to the gene; #Elements: the number of guides identified in either replicate (1 or 2); casTLE Effect: the magnitude and sign of the effect for one screen replicate (1 or 2) or a combination of the replicates (Combo), where a negative effect indicates guide depletion in the Hc-infected pool and a positive effect indicates guide enrichment in the Hc-infected pool compared to the uninfected pool; casTLE Score: the casTLE confidence score; Signed Combo casTLE score: the Combo casTLE confidence score with the sign of the effect; P value: indicates the p-value of the gene effect; 5% FDR: 1 indicates a gene is a screen hit under a 5% FDR cutoff, and 0 indicates a gene is not a screen hit under a 5% FDR cutoff; 10% FDR: 1 indicates a gene is a screen hit under a 10% FDR cutoff, and 0 indicates a gene is not a screen hit under a 10% FDR cutoff; Minimum Effect Estimate: Minimum effect estimate using a 95% credible interval; Maximum Effect Estimate: Maximum effect estimate using a 95% credible interval.
(CSV)
A CSV file showing the casTLE analysis results comparing the J774A.1 pool at T = 0 (time 0), or the start of the screen following sgRNA transduction and selection, and the uninfected pool that had been passaged for the duration of the screen. Row and column annotations are the same as those described above for S1 Table.
(CSV)
A CSV file showing the casTLE analysis results comparing the plasmid library pool and the uninfected J774A.1 pool that had been passaged for the duration of the screen. Row and column annotations are the same as those described above for S2 Table.
(CSV)
The sub-libraries covered in our screens are listed, with information on the number of genes covered in each screen, the rounds of Hc-mediated lysis (at least 50%) and recovery following uracil removal, the duration of the screen, the estimated selection pressure, or difference in divisions between the Hc infected and uninfected conditions assessed by cell counting. The number of sensitizing and de-sensitizing hits that pass the 5%FDR cutoff, and the replicate correlation coefficient for the confidence score is also listed.
(CSV)
Genomic DNA was harvested from CRISPR-targeted and non-targeted cells and the targeted locus was amplified by PCR (~200bp upstream and downstream of the predicted cut site), and sequenced via capillary reaction. TIDE analysis was performed on the sequence traces from control and targeted cells to estimate the percentage of alleles that had INDELs resulting from double-stranded break repair.
(CSV)
Comma-separated-values (CSV) file listing the names of the oligonucleotides used and their sequences.
(XLSX)
A representative live-cell confocal microscopy movie showing a vehicle-treated Lifeact-eGPF-expressing J774A.1 cell extending an actin-rich membrane protrusion in the direction of mCherry-expressing Hc yeast.
(MP4)
The DIC channel from the same experiment as S1 Movie.
(MP4)
A representative live-cell confocal microscopy movie showing a Lifeact-eGFP-expressing J774A.1 treated with the C3aR antagonist SB290157 (10 μM) interacting with Hc yeast and failing to initiate the formation of an actin-rich pseudopod, and as a result exhibiting delayed capture of Hc yeast.
(MP4)
The DIC channel from the same experiment as S3 Movie.
(MP4)
A ZIP archive of tables in CSV format giving counts for each sgRNA. File names indicate sub-library and experimental sample.
(ZIP)
Acknowledgments
We are grateful to Hiten Madhani, Alexander Marson, Suzanne Noble, Eric Dang, Carrie Graham, and members of the Sil, Noble, and Bassik labs for helpful discussions as we prepared this work. We appreciate the gift of cells and plasmids from Jason Cyster and Diane Barber, the gift of Candida albicans from Alexander Johnson, and Coccidioides posadasii isolates from Bridget Barker. We thank Mark Roxas for technical assistance, and Keith Walcott for BSL3 assistance. We are grateful to the the UCSF Parnassus Flow CoLab, (RRID:SCR_018206) for flow-cytometry and cell-sorting support. The PFCC is supported in part by NIH P30 DK063720 and by the NIH S10 Instrumentation Grant S10 1S10OD021822-01.
Data Availability
All data are available within the manuscript and its Supporting information files.
Funding Statement
This work was supported by a National Science Foundation Graduate Research Fellowship to AC, R01AI136735 and 2R37AI066224 to AS, and the NIH Director’s New Innovator Award Program 1DP2HD084069-01 to MCB. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Pfaller MA, Diekema DJ. Epidemiology of invasive mycoses in North America. Critical reviews in microbiology. 2010;36(1):1–53. doi: 10.3109/10408410903241444 [DOI] [PubMed] [Google Scholar]
- 2.Deepe GS Jr., Gibbons RS, Smulian AG. Histoplasma capsulatum manifests preferential invasion of phagocytic subpopulations in murine lungs. J Leukoc Biol. 2008;84(3):669–78. doi: 10.1189/jlb.0308154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones GS, Sepúlveda VE, Goldman WE. Biodiverse Histoplasma Species Elicit Distinct Patterns of Pulmonary Inflammation following Sublethal Infection. mSphere. 2020;5(4). doi: 10.1128/mSphere.00742-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horwath MC, Fecher RA, Deepe GS Jr. Histoplasma capsulatum, lung infection and immunity. Future microbiology. 2015;10(6):967–75. doi: 10.2217/fmb.15.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garfoot AL, Rappleye CA. Histoplasma capsulatum surmounts obstacles to intracellular pathogenesis. The FEBS journal. 2016;283(4):619–33. doi: 10.1111/febs.13389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ray SC, Rappleye CA. Flying under the radar: Histoplasma capsulatum avoidance of innate immune recognition. Semin Cell Dev Biol. 2019;89:91–8. doi: 10.1016/j.semcdb.2018.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.English BC, Van Prooyen N, Ord T, Ord T, Sil A. The transcription factor CHOP, an effector of the integrated stress response, is required for host sensitivity to the fungal intracellular pathogen Histoplasma capsulatum. PLoS pathogens. 2017;13(9):e1006589. doi: 10.1371/journal.ppat.1006589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woods JP. Revisiting old friends: Developments in understanding Histoplasma capsulatum pathogenesis. Journal of Microbiology. 2016;54(3):265–76. doi: 10.1007/s12275-016-6044-5 [DOI] [PubMed] [Google Scholar]
- 9.Isaac DT, Berkes CA, English BC, Hocking Murray D, Lee YN, Coady A, et al. Macrophage cell death and transcriptional response are actively triggered by the fungal virulence factor Cbp1 during H. capsulatum infection. Mol Microbiol. 2015;98(5):910–29. doi: 10.1111/mmi.13168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sebghati TS, Engle JT, Goldman WE. Intracellular parasitism by Histoplasma capsulatum: fungal virulence and calcium dependence. Science (New York, NY). 2000;290(5495):1368–72. doi: 10.1126/science.290.5495.1368 [DOI] [PubMed] [Google Scholar]
- 11.Erwig LP, Gow NA. Interactions of fungal pathogens with phagocytes. Nature reviews Microbiology. 2016;14(3):163–76. doi: 10.1038/nrmicro.2015.21 [DOI] [PubMed] [Google Scholar]
- 12.Brown GD, Taylor PR, Reid DM, Willment JA, Williams DL, Martinez-Pomares L, et al. Dectin-1 is a major beta-glucan receptor on macrophages. The Journal of experimental medicine. 2002;196(3):407–12. doi: 10.1084/jem.20020470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goodridge HS, Reyes CN, Becker CA, Katsumoto TR, Ma J, Wolf AJ, et al. Activation of the innate immune receptor Dectin-1 upon formation of a ’phagocytic synapse’. Nature. 2011;472(7344):471–5. doi: 10.1038/nature10071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flannagan RS, Jaumouillé V, Grinstein S. The cell biology of phagocytosis. Annu Rev Pathol. 2012;7:61–98. doi: 10.1146/annurev-pathol-011811-132445 [DOI] [PubMed] [Google Scholar]
- 15.Freeman SA, Grinstein S. Phagocytosis: receptors, signal integration, and the cytoskeleton. Immunological reviews. 2014;262(1):193–215. doi: 10.1111/imr.12212 [DOI] [PubMed] [Google Scholar]
- 16.Lin JS, Huang JH, Hung LY, Wu SY, Wu-Hsieh BA. Distinct roles of complement receptor 3, Dectin-1, and sialic acids in murine macrophage interaction with Histoplasma yeast. J Leukoc Biol. 2010;88(1):95–106. doi: 10.1189/jlb.1109717 [DOI] [PubMed] [Google Scholar]
- 17.Rappleye CA, Eissenberg LG, Goldman WE. Histoplasma capsulatum alpha-(1,3)-glucan blocks innate immune recognition by the beta-glucan receptor. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(4):1366–70. doi: 10.1073/pnas.0609848104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garfoot AL, Shen Q, Wuthrich M, Klein BS, Rappleye CA. The Eng1 beta-Glucanase Enhances Histoplasma Virulence by Reducing beta-Glucan Exposure. mBio. 2016;7(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bullock WE, Wright SD. Role of the adherence-promoting receptors, CR3, LFA1, and P150,95, in binding of Histoplasma capsulatum by human macrophages. Journal of Experimental Medicine. 1987;165(1):195–210. doi: 10.1084/jem.165.1.195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan SM. The leucocyte β2 (CD18) integrins: the structure, functional regulation and signalling properties. Biosci Rep. 2012;32(3):241–69. doi: 10.1042/BSR20110101 [DOI] [PubMed] [Google Scholar]
- 21.Noris M, Remuzzi G. Overview of complement activation and regulation. Semin Nephrol. 2013;33(6):479–92. doi: 10.1016/j.semnephrol.2013.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong SSW, Daniel I, Gangneux JP, Jayapal JM, Guegan H, Dellière S, et al. Differential Interactions of Serum and Bronchoalveolar Lavage Fluid Complement Proteins with Conidia of Airborne Fungal Pathogen Aspergillus fumigatus. Infect Immun. 2020;88(9). doi: 10.1128/IAI.00212-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bolger MS, Ross DS, Jiang H, Frank MM, Ghio AJ, Schwartz DA, et al. Complement levels and activity in the normal and LPS-injured lung. Am J Physiol Lung Cell Mol Physiol. 2007;292(3):L748–59. doi: 10.1152/ajplung.00127.2006 [DOI] [PubMed] [Google Scholar]
- 24.Lambris JD, Ricklin D, Geisbrecht BV. Complement evasion by human pathogens. Nature reviews Microbiology. 2008;6(2):132–42. doi: 10.1038/nrmicro1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee CY, Herant M, Heinrich V. Target-specific mechanics of phagocytosis: protrusive neutrophil response to zymosan differs from the uptake of antibody-tagged pathogens. Journal of cell science. 2011;124(Pt 7):1106–14. doi: 10.1242/jcs.078592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Torres-Gomez A, Cabañas C, Lafuente EM. Phagocytic Integrins: Activation and Signaling. Front Immunol. 2020;11:738. doi: 10.3389/fimmu.2020.00738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quell KM, Karsten CM, Kordowski A, Almeida LN, Briukhovetska D, Wiese AV, et al. Monitoring C3aR Expression Using a Floxed tdTomato-C3aR Reporter Knock-in Mouse. Journal of immunology (Baltimore, Md: 1950). 2017;199(2):688–706. doi: 10.4049/jimmunol.1700318 [DOI] [PubMed] [Google Scholar]
- 28.Zwirner J, Werfel T, Wilken HC, Theile E, Gotze O. Anaphylatoxin C3a but not C3a(desArg) is a chemotaxin for the mouse macrophage cell line J774. European journal of immunology. 1998;28(5):1570–7. doi: [DOI] [PubMed] [Google Scholar]
- 29.Coulthard LG, Woodruff TM. Is the complement activation product C3a a proinflammatory molecule? Re-evaluating the evidence and the myth. Journal of immunology (Baltimore, Md: 1950). 2015;194(8):3542–8. doi: 10.4049/jimmunol.1403068 [DOI] [PubMed] [Google Scholar]
- 30.Klos A, Tenner AJ, Johswich KO, Ager RR, Reis ES, Köhl J. The role of the anaphylatoxins in health and disease. Mol Immunol. 2009;46(14):2753–66. doi: 10.1016/j.molimm.2009.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ezekowitz RA, Sim RB, Hill M, Gordon S. Local opsonization by secreted macrophage complement components. Role of receptors for complement in uptake of zymosan. The Journal of experimental medicine. 1984;159(1):244–60. doi: 10.1084/jem.159.1.244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goodrum KJ. Complement component C3 secretion by mouse macrophage-like cell lines. J Leukoc Biol. 1987;41(4):295–301. doi: 10.1002/jlb.41.4.295 [DOI] [PubMed] [Google Scholar]
- 33.Luo C, Chen M, Madden A, Xu H. Expression of complement components and regulators by different subtypes of bone marrow-derived macrophages. Inflammation. 2012;35(4):1448–61. doi: 10.1007/s10753-012-9458-1 [DOI] [PubMed] [Google Scholar]
- 34.Galgiani JN, Yam P, Petz LD, Williams PL, Stevens DA. Complement activation by Coccidioides immitis: in vitro and clinical studies. Infect Immun. 1980;28(3):944–9. doi: 10.1128/iai.28.3.944-949.1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ratnoff WD, Pepple JM, Winkelstein JA. Activation of the alternative complement pathway by Histoplasma capsulatum. Infect Immun. 1980;30(1):147–9. doi: 10.1128/iai.30.1.147-149.1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kozel TR. Activation of the complement system by pathogenic fungi. Clinical microbiology reviews. 1996;9(1):34–46. doi: 10.1128/CMR.9.1.34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zaragoza O, Taborda CP, Casadevall A. The efficacy of complement-mediated phagocytosis of Cryptococcus neoformans is dependent on the location of C3 in the polysaccharide capsule and involves both direct and indirect C3-mediated interactions. European journal of immunology. 2003;33(7):1957–67. doi: 10.1002/eji.200323848 [DOI] [PubMed] [Google Scholar]
- 38.Harpf V, Rambach G, Würzner R, Lass-Flörl C, Speth C. Candida and Complement: New Aspects in an Old Battle. Front Immunol. 2020;11:1471. doi: 10.3389/fimmu.2020.01471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taylor PR, Tsoni SV, Willment JA, Dennehy KM, Rosas M, Findon H, et al. Dectin-1 is required for beta-glucan recognition and control of fungal infection. Nature immunology. 2007;8(1):31–8. doi: 10.1038/ni1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsoni SV, Kerrigan AM, Marakalala MJ, Srinivasan N, Duffield M, Taylor PR, et al. Complement C3 plays an essential role in the control of opportunistic fungal infections. Infect Immun. 2009;77(9):3679–85. doi: 10.1128/IAI.00233-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shapiro S, Beenhouwer DO, Feldmesser M, Taborda C, Carroll MC, Casadevall A, et al. Immunoglobulin G monoclonal antibodies to Cryptococcus neoformans protect mice deficient in complement component C3. Infect Immun. 2002;70(5):2598–604. doi: 10.1128/IAI.70.5.2598-2604.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pillemer L, Blum L, Lepow IH, Ross OA, Todd EW, Wardlaw AC. The properdin system and immunity. I. Demonstration and isolation of a new serum protein, properdin, and its role in immune phenomena. Science (New York, NY). 1954;120(3112):279–85. [DOI] [PubMed] [Google Scholar]
- 43.Sun D, Zhang M, Liu G, Wu H, Zhu X, Zhou H, et al. Real-Time Imaging of Interactions of Neutrophils with Cryptococcus neoformans Demonstrates a Crucial Role of Complement C5a-C5aR Signaling. Infect Immun. 2016;84(1):216–29. doi: 10.1128/IAI.01197-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng SC, Sprong T, Joosten LA, van der Meer JW, Kullberg BJ, Hube B, et al. Complement plays a central role in Candida albicans-induced cytokine production by human PBMCs. European journal of immunology. 2012;42(4):993–1004. doi: 10.1002/eji.201142057 [DOI] [PubMed] [Google Scholar]
- 45.Kampmann M, Bassik MC, Weissman JS. Functional genomics platform for pooled screening and generation of mammalian genetic interaction maps. Nature Protocols. 2014;9(8):1825–47. doi: 10.1038/nprot.2014.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jeng EE, Bhadkamkar V, Ibe NU, Gause H, Jiang L, Chan J, et al. Systematic Identification of Host Cell Regulators of Legionella pneumophila Pathogenesis Using a Genome-wide CRISPR Screen. Cell Host Microbe. 2019;26(4):551–63.e6. doi: 10.1016/j.chom.2019.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park RJ, Wang T, Koundakjian D, Hultquist JF, Lamothe-Molina P, Monel B, et al. A genome-wide CRISPR screen identifies a restricted set of HIV host dependency factors. Nat Genet. 2017;49(2):193–203. doi: 10.1038/ng.3741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Napier BA, Brubaker SW, Sweeney TE, Monette P, Rothmeier GH, Gertsvolf NA, et al. Complement pathway amplifies caspase-11-dependent cell death and endotoxin-induced sepsis severity. The Journal of experimental medicine. 2016;213(11):2365–82. doi: 10.1084/jem.20160027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morgens DW, Wainberg M, Boyle EA, Ursu O, Araya CL, Tsui CK, et al. Genome-scale measurement of off-target activity using Cas9 toxicity in high-throughput screens. Nature communications. 2017;8:15178. doi: 10.1038/ncomms15178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Worsham PL, Goldman WE. Selection and characterization of ura5 mutants of Histoplasma capsulatum. Molecular & general genetics: MGG. 1988;214(2):348–52. doi: 10.1007/BF00337734 [DOI] [PubMed] [Google Scholar]
- 51.Morgens DW, Deans RM, Li A, Bassik MC. Systematic comparison of CRISPR/Cas9 and RNAi screens for essential genes. Nature biotechnology. 2016;34(6):634–6. doi: 10.1038/nbt.3567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haney MS, Bohlen CJ, Morgens DW, Ousey JA, Barkal AA, Tsui CK, et al. Identification of phagocytosis regulators using magnetic genome-wide CRISPR screens. Nat Genet. 2018;50(12):1716–27. doi: 10.1038/s41588-018-0254-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Devreotes P, Horwitz AR. Signaling networks that regulate cell migration. Cold Spring Harb Perspect Biol. 2015;7(8):a005959. doi: 10.1101/cshperspect.a005959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moser M, Nieswandt B, Ussar S, Pozgajova M, Fässler R. Kindlin-3 is essential for integrin activation and platelet aggregation. Nat Med. 2008;14(3):325–30. doi: 10.1038/nm1722 [DOI] [PubMed] [Google Scholar]
- 55.Jewell JL, Russell RC, Guan KL. Amino acid signalling upstream of mTOR. Nature reviews Molecular cell biology. 2013;14(3):133–9. doi: 10.1038/nrm3522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chitwood PJ, Hegde RS. The Role of EMC during Membrane Protein Biogenesis. Trends Cell Biol. 2019. doi: 10.1016/j.tcb.2019.01.007 [DOI] [PubMed] [Google Scholar]
- 57.Shurtleff MJ, Itzhak DN, Hussmann JA, Schirle Oakdale NT, Costa EA, Jonikas M, et al. The ER membrane protein complex interacts cotranslationally to enable biogenesis of multipass membrane proteins. Elife. 2018;7. doi: 10.7554/eLife.37018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Irannejad R, von Zastrow M. GPCR signaling along the endocytic pathway. Current opinion in cell biology. 2014;27:109–16. doi: 10.1016/j.ceb.2013.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Block H, Stadtmann A, Riad D, Rossaint J, Sohlbach C, Germena G, et al. Gnb isoforms control a signaling pathway comprising Rac1, Plcβ2, and Plcβ3 leading to LFA-1 activation and neutrophil arrest in vivo. Blood. 2016;127(3):314–24. doi: 10.1182/blood-2015-06-651034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Andreu N, Phelan J, de Sessions PF, Cliff JM, Clark TG, Hibberd ML. Primary macrophages and J774 cells respond differently to infection with Mycobacterium tuberculosis. Scientific reports. 2017;7:42225. doi: 10.1038/srep42225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Haas PJ, van Strijp J. Anaphylatoxins: their role in bacterial infection and inflammation. Immunologic research. 2007;37(3):161–75. doi: 10.1007/BF02697367 [DOI] [PubMed] [Google Scholar]
- 62.Humbles AA, Lu B, Nilsson CA, Lilly C, Israel E, Fujiwara Y, et al. A role for the C3a anaphylatoxin receptor in the effector phase of asthma. Nature. 2000;406(6799):998–1001. doi: 10.1038/35023175 [DOI] [PubMed] [Google Scholar]
- 63.Mueller-Ortiz SL, Hollmann TJ, Haviland DL, Wetsel RA. Ablation of the complement C3a anaphylatoxin receptor causes enhanced killing of Pseudomonas aeruginosa in a mouse model of pneumonia. Am J Physiol Lung Cell Mol Physiol. 2006;291(2):L157–65. doi: 10.1152/ajplung.00358.2005 [DOI] [PubMed] [Google Scholar]
- 64.Wu KY, Zhang T, Zhao GX, Ma N, Zhao SJ, Wang N, et al. The C3a/C3aR axis mediates anti-inflammatory activity and protects against uropathogenic E coli-induced kidney injury in mice. Kidney Int. 2019;96(3):612–27. doi: 10.1016/j.kint.2019.03.005 [DOI] [PubMed] [Google Scholar]
- 65.Lian H, Litvinchuk A, Chiang AC, Aithmitti N, Jankowsky JL, Zheng H. Astrocyte-Microglia Cross Talk through Complement Activation Modulates Amyloid Pathology in Mouse Models of Alzheimer’s Disease. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2016;36(2):577–89. doi: 10.1523/JNEUROSCI.2117-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Litvinchuk A, Wan YW, Swartzlander DB, Chen F, Cole A, Propson NE, et al. Complement C3aR Inactivation Attenuates Tau Pathology and Reverses an Immune Network Deregulated in Tauopathy Models and Alzheimer’s Disease. Neuron. 2018;100(6):1337–53.e5. doi: 10.1016/j.neuron.2018.10.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang LY, Pan J, Mamtilahun M, Zhu Y, Wang L, Venkatesh A, et al. Microglia exacerbate white matter injury via complement C3/C3aR pathway after hypoperfusion. Theranostics. 2020;10(1):74–90. doi: 10.7150/thno.35841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ames RS, Lee D, Foley JJ, Jurewicz AJ, Tornetta MA, Bautsch W, et al. Identification of a selective nonpeptide antagonist of the anaphylatoxin C3a receptor that demonstrates antiinflammatory activity in animal models. Journal of immunology (Baltimore, Md: 1950). 2001;166(10):6341–8. doi: 10.4049/jimmunol.166.10.6341 [DOI] [PubMed] [Google Scholar]
- 69.Huang NN, Becker S, Boularan C, Kamenyeva O, Vural A, Hwang IY, et al. Canonical and noncanonical g-protein signaling helps coordinate actin dynamics to promote macrophage phagocytosis of zymosan. Molecular and cellular biology. 2014;34(22):4186–99. doi: 10.1128/MCB.00325-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Decker T, Lohmann-Matthes ML. A quick and simple method for the quantitation of lactate dehydrogenase release in measurements of cellular cytotoxicity and tumor necrosis factor (TNF) activity. J Immunol Methods. 1988;115(1):61–9. doi: 10.1016/0022-1759(88)90310-9 [DOI] [PubMed] [Google Scholar]
- 71.Nilsson UR, Müller-Eberhard HJ. Deficiency of the fifth component of complement in mice with an inherited complement defect. The Journal of experimental medicine. 1967;125(1):1–16. doi: 10.1084/jem.125.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang MX, Klein B. Activation, binding, and processing of complement component 3 (C3) by Blastomyces dermatitidis. Infect Immun. 1997;65(5):1849–55. doi: 10.1128/iai.65.5.1849-1855.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, et al. Lifeact: a versatile marker to visualize F-actin. Nature methods. 2008;5(7):605–7. doi: 10.1038/nmeth.1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Svensson CM, Medyukhina A, Belyaev I, Al-Zaben N, Figge MT. Untangling cell tracks: Quantifying cell migration by time lapse image data analysis. Cytometry Part A: the journal of the International Society for Analytical Cytology. 2018;93(3):357–70. doi: 10.1002/cyto.a.23249 [DOI] [PubMed] [Google Scholar]
- 75.Heinrich V, Simpson WD 3rd, Francis EA. Analytical Prediction of the Spatiotemporal Distribution of Chemoattractants around Their Source: Theory and Application to Complement-Mediated Chemotaxis. Front Immunol. 2017;8:578. doi: 10.3389/fimmu.2017.00578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mershon KL, Vasuthasawat A, Lawson GW, Morrison SL, Beenhouwer DO. Role of complement in protection against Cryptococcus gattii infection. Infect Immun. 2009;77(3):1061–70. doi: 10.1128/IAI.01119-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rougerie P, Miskolci V, Cox D. Generation of membrane structures during phagocytosis and chemotaxis of macrophages: role and regulation of the actin cytoskeleton. Immunological reviews. 2013;256(1):222–39. doi: 10.1111/imr.12118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Van Haastert PJ, Devreotes PN. Chemotaxis: signalling the way forward. Nature reviews Molecular cell biology. 2004;5(8):626–34. doi: 10.1038/nrm1435 [DOI] [PubMed] [Google Scholar]
- 79.Peracino B, Borleis J, Jin T, Westphal M, Schwartz JM, Wu L, et al. G protein beta subunit-null mutants are impaired in phagocytosis and chemotaxis due to inappropriate regulation of the actin cytoskeleton. The Journal of cell biology. 1998;141(7):1529–37. doi: 10.1083/jcb.141.7.1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pan M, Neilson MP, Grunfeld AM, Cruz P, Wen X, Insall RH, et al. A G-protein-coupled chemoattractant receptor recognizes lipopolysaccharide for bacterial phagocytosis. PLoS Biol. 2018;16(5):e2005754. doi: 10.1371/journal.pbio.2005754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Heinrich V, Lee CY. Blurred line between chemotactic chase and phagocytic consumption: an immunophysical single-cell perspective. Journal of cell science. 2011;124(Pt 18):3041–51. doi: 10.1242/jcs.086413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Freeman SA, Vega A, Riedl M, Collins RF, Ostrowski PP, Woods EC, et al. Transmembrane Pickets Connect Cyto- and Pericellular Skeletons Forming Barriers to Receptor Engagement. Cell. 2018;172(1–2):305–17.e10. doi: 10.1016/j.cell.2017.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Flannagan RS, Harrison RE, Yip CM, Jaqaman K, Grinstein S. Dynamic macrophage "probing" is required for the efficient capture of phagocytic targets. The Journal of cell biology. 2010;191(6):1205–18. doi: 10.1083/jcb.201007056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shen K, Sidik H, Talbot WS. The Rag-Ragulator Complex Regulates Lysosome Function and Phagocytic Flux in Microglia. Cell Rep. 2016;14(3):547–59. doi: 10.1016/j.celrep.2015.12.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gudjonsson T, Altmeyer M, Savic V, Toledo L, Dinant C, Grøfte M, et al. TRIP12 and UBR5 suppress spreading of chromatin ubiquitylation at damaged chromosomes. Cell. 2012;150(4):697–709. doi: 10.1016/j.cell.2012.06.039 [DOI] [PubMed] [Google Scholar]
- 86.Cho JH, Kim SA, Seo YS, Park SG, Park BC, Kim JH, et al. The p90 ribosomal S6 kinase-UBR5 pathway controls Toll-like receptor signaling via miRNA-induced translational inhibition of tumor necrosis factor receptor-associated factor 3. The Journal of biological chemistry. 2017;292(28):11804–14. doi: 10.1074/jbc.M117.785170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Surugiu R, Catalin B, Dumbrava D, Gresita A, Olaru DG, Hermann DM, et al. Intracortical Administration of the Complement C3 Receptor Antagonist Trifluoroacetate Modulates Microglia Reaction after Brain Injury. Neural Plast. 2019;2019:1071036. doi: 10.1155/2019/1071036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vasek MJ, Garber C, Dorsey D, Durrant DM, Bollman B, Soung A, et al. A complement-microglial axis drives synapse loss during virus-induced memory impairment. Nature. 2016;534(7608):538–43. doi: 10.1038/nature18283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sun D, Sun P, Li H, Zhang M, Liu G, Strickland AB, et al. Fungal dissemination is limited by liver macrophage filtration of the blood. Nature communications. 2019;10(1):4566. doi: 10.1038/s41467-019-12381-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hwang LH, Mayfield JA, Rine J, Sil A. Histoplasma requires SID1, a member of an iron-regulated siderophore gene cluster, for host colonization. PLoS pathogens. 2008;4(4):e1000044. doi: 10.1371/journal.ppat.1000044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Huang JH, Lin CY, Wu SY, Chen WY, Chu CL, Brown GD, et al. CR3 and Dectin-1 Collaborate in Macrophage Cytokine Response through Association on Lipid Rafts and Activation of Syk-JNK-AP-1 Pathway. PLoS pathogens. 2015;11(7):e1004985. doi: 10.1371/journal.ppat.1004985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Van Prooyen N, Henderson CA, Hocking Murray D, Sil A. CD103+ Conventional Dendritic Cells Are Critical for TLR7/9-Dependent Host Defense against Histoplasma capsulatum, an Endemic Fungal Pathogen of Humans. PLoS pathogens. 2016;12(7):e1005749. doi: 10.1371/journal.ppat.1005749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Worsham PL, Goldman WE. Quantitative plating of Histoplasma capsulatum without addition of conditioned medium or siderophores. Journal of medical and veterinary mycology: bi-monthly publication of the International Society for Human and Animal Mycology. 1988;26(3):137–43. [PubMed] [Google Scholar]
- 94.Mead HL, Van Dyke MCC, Barker BM. Proper Care and Feeding of Coccidioides: A Laboratorian’s Guide to Cultivating the Dimorphic Stages of C. immitis and C. posadasii. Curr Protoc Microbiol. 2020;58(1):e113. doi: 10.1002/cpmc.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Deans RM, Morgens DW, Okesli A, Pillay S, Horlbeck MA, Kampmann M, et al. Parallel shRNA and CRISPR-Cas9 screens enable antiviral drug target identification. Nat Chem Biol. 2016;12(5):361–6. doi: 10.1038/nchembio.2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Brinkman EK, Chen T, Amendola M, van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic acids research. 2014;42(22):e168. doi: 10.1093/nar/gku936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bassik MC, Kampmann M, Lebbink RJ, Wang SY, Hein MY, Poser I, et al. A Systematic Mammalian Genetic Interaction Map Reveals Pathways Underlying Ricin Susceptibility. Cell. 2013;152(4):909–22. doi: 10.1016/j.cell.2013.01.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Saldanha AJ. Java Treeview—extensible visualization of microarray data. Bioinformatics. 2004;20(17):3246–8. doi: 10.1093/bioinformatics/bth349 [DOI] [PubMed] [Google Scholar]
- 99.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ollion J, Cochennec J, Loll F, Escudé C, Boudier T. TANGO: a generic tool for high-throughput 3D image analysis for studying nuclear organization. Bioinformatics. 2013;29(14):1840–1. doi: 10.1093/bioinformatics/btt276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Meijering E, Dzyubachyk O, Smal I. Methods for cell and particle tracking. Methods Enzymol. 2012;504:183–200. doi: 10.1016/B978-0-12-391857-4.00009-4 [DOI] [PubMed] [Google Scholar]
- 102.Isaac DT, Coady A, Van Prooyen N, Sil A. The 3-Hydroxy-Methylglutaryl Coenzyme A Lyase HCL1 Is Required for Macrophage Colonization by Human Fungal Pathogen Histoplasma capsulatum. Infection and Immunity. 2013;81(2):411–20. doi: 10.1128/IAI.00833-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Roy RM, Paes HC, Nanjappa SG, Sorkness R, Gasper D, Sterkel A, et al. Complement component 3C3 and C3a receptor are required in chitin-dependent allergic sensitization to Aspergillus fumigatus but dispensable in chitin-induced innate allergic inflammation. mBio. 2013;4(2). doi: 10.1128/mBio.00162-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A. Characterization of Hc-mediated lysis in J774A.1 macrophage-like cells. J774A.1 cells were infected with WT Hc, or Hc with a disruption in a gene, CBP1, that is required for Hc to lyse macrophages. Lysis over time was measured using the LDH release assay. B. Validation and clonal expansion of Cas9-expressing J774A.1 cells. Cells were transduced with an Ef1a-Cas9-Blast expression vector and grown under blasticidin selection to generate a population of Cas9-expressing cells. These were subjected to single-cell sorting and clonal expansion to generate Cas9-expressing J774A.1 clones with high Cas9 activity. Cas9 activity was measured by transducing J774A.1 cells with a guide RNA vector that co-expressed EGFP with a sgRNA targeting EGFP. Cas9 activity leads to silencing of the GFP following puromycin selection. Cas9 clone 9 was chosen for the large-scale CRISPR screens due to its high-efficiency GFP silencing. C-D. Characterizing lysis and recovery from infection with uracil pulses during infection with a Ura5-deficient Hc. J774A.1 macrophages were infected with ura5 mutant Hc in the presence or absence of exogenous uracil (0.4ug/mL). Uracil-containing cells were washed and media was replaced with uracil-poor media after 2d of lysis, which allowed the macrophages to recover. Recovery was assessed using LDH release quantification to assess lysis, and the confluency of viable cells in the wells was estimated using the pico-green dsDNA assay kit following lysis of macrophages with water. E. Macrophages that had been recovered from lysis by removal of uracil from culture media were passaged for several days, and uracil was added to selected wells. Macrophage lysis over time was monitored by assessing LDH release over time to determine whether dormant yeast would be able to re-activate upon introduction of uracil. F. Reproducibility of the casTLE score across two replicates of the screens. G. Histograms comparing the distribution of negative control sgRNAs and sgRNAs targeting Gnb2 or C3ar in the Hc infected pool compared to the uninfected pool. H. Analysis of essential gene behavior during J7 library growth. Scatter plot showing the gene effect resulting from passaging of J7s, either going from the plasmid pool to the T0 pool, or the T0 pool to the uninfected pool. Genes annotated as “essential” or “non-essential” were plotted to determine whether essential genes appeared more likely to drop out of the uninfected pools.
(TIF)
BMDMs from C3ar-/- and WT mice, in addition to BMDMs expressing Cas9 and control or Emc1-targeting sgRNAs, were stained with anti-CD18 and anti-CD11b antibodies and analyzed by flow cytometry (n = 2 biological replicates). A. Representative histograms showing CD11b and CD18 levels in control, C3ar-/-, and Emc1 CRISPRKO BMDMs. The percentage of CD11b (B) and CD18 (D) positive macrophages was analyzed. The mean fluorescence intensity of CD11b (C) and CD18 (D) were also measured.
(TIF)
BMDMs were infected with Hc (MOI = 0.5), and macrophage lysis was quantified by measuring the release of lactate dehydrogenase (LDH) into the culture supernatants over-time (n = 3 biological replicates, 3 technical replicates/biorep) (A). LDH release is presented as the percentage of total LDH present in the well (supernatant and macrophage lysate) at 2 hours post-infection. At the indicated time points, macrophages were lysed using water, and lysates were spread on agar plates. Colony forming units (CFUs) were enumerated (n = 3 biological replicates, 2 technical reps/biorep) (B).
(TIF)
A. WT and C3ar-/- BMDMs were infected with Hc (MOI10), and TNFα levels in macrophage supernatants were measured using the BD Cytometric Bead Array (CBA) kit (n = 3 biological replicates). B-D. Thioglycollate-elicited peritoneal macrophages were treated with the C3aR antagonist (SB290157, 10 μM) and infected with Hc (MOI10). TNFα (B), IL-6 (C), and IL-1β (D) levels in culture supernatants were measured by CBA. *p ≤ 0.05, **p ≤ 0.01, ns = not significant by two-tailed student’s T-test.
(TIF)
(A-C) BMDMs from WT and C3ar-/- mice were infected with Hc in the presence of 20% FBS from three different lots from 2 separate suppliers. In addition, WT BMDMs differentiated in different lots of serum were treated with 10 μM of the C3aR antagonist and infected with Hc (B). Phagocytosis of Hc was measured by flow cytometry as described previously (n = 2 biological replicates).
(TIF)
BMDMs were cultured in media containing 10% FBS, and the BMDM conditioned media was harvested. Hc was incubated with macrophage conditioned media (BMDM CM), 10% FBS, or 10% normal mouse serum (NMS) with 10 mM EGTA or EDTA as indicated for 30 min 37°C. A. Incubation with conditioned media or FBS does not lead to C3 deposition on the Hc surface. C3 deposition on Hc yeast was analyzed by immunofluorescence microscopy using an anti-C3 antibody. Scale bar = 10 μm. B. Pre-incubation of yeast with conditioned media does not improve macrophage phagocytosis of Hc. Yeast were washed and used to infect BMDMs (MOI3, 30 min) in serum-free media. Phagocytosis was assessed using flow cytometry as previously described (n = 3 biological replicates).
(TIF)
A. BMDMs secrete C3 following infection with Hc. BMDMs were infected with Hc at an MOI2 for 24h, supernatants were harvested and C3 levels were quantified using a BD mouse C3 ELISA kit. B. C3-/- BMDMs are not defective in phagocytosis of Hc yeast or zymosan. WT and C3-/- BMDMs were infected with mCherry-expressing Hc or FITC-labelled zymosan, and uptake over time was measured using flow-cytometry. Extracellular yeasts were identified by Calcofluor White staining.
(TIF)
A. Uninfected WT and C3ar-/- BMDMs were stained with a C3aR-specific antibody and imaged using confocal microscopy and optical sectioning. Representative slices of 2 biological replicates are shown. The antibody specifically detects C3aR, as staining was not observed in C3ar-/- BMDMs. C3aR exhibits punctate localization near the plasma membrane in WT BMDMs. B-C. Representative slices from confocal imaging showing the region of interest mask derived from binary operations on Hc (B) or bead (C) fluorescent channel thresholding used to measure the phagosomal C3aR intensity of Hc and bead-containing phagosomes. The original and overlaid fluorescence images are shown. Scale bar = 20 μm.
(TIF)
A. Hc stimulates chemotaxis of J774A.1 macrophage-like cells. Hc yeast were seeded into multiple-well plates at varying concentrations, and WT J774A.1 cells were seeded onto transwell permeable supports with 5 μm pores. Serum-free media supplemented with 0.25% BSA was used as the diluent in both the chamber and well unless otherwise indicated. After 3 h of migration, transwells were stained with crystal violet, and non-migratory cells were wiped off of the upper side of the transwell using a Q-tip. The number of migratory cells in each condition was quantified by microscopy (n = 2 biogical replicates, 3 fields/biological replicate). B. Migration towards Hc is Gai-dependent. J774A.1 cells with or without pre-treatment with 1 μg/mL pertussis toxin (PTX) for 2 h were seeded into transwell permeable supports and migration towards 1e5 Hc/uL was quantified as described above. The number of migrating cells was quantified as described. C. The C3aR antagonist does not inhibit macrophage migration towards Hc. J774A.1 macrophages were treated with 1 μM SB290157, a C3aR antagonist, and migration towards Hc was assessed as described. D. C3aR-deficiency moderately impacts migration of J774A.1 cells towards Hc. Cas9-expressing J774A.1 macrophages transduced with non-targeting or C3aR-targeting sgRNAs were assessed for their ability to migrate towards Hc as described previously. E. Hc-dependent migration is abolished in the presence of FBS. The transwell migration assay was performed with media supplemented with BSA or 10% FBS to determine whether FBS affected the migration of macrophage-like cells towards Hc yeast.
(TIF)
A. BMDMs were infected with Hc or zymosan at an MOI = 5 for 30 min. For the control condition, particles were added to the wells and allowed to settle onto the monolayer without intervention. For the 5 min spinfection, particles were added to the cells, and the plate was spun for 5 min at 550XG at RT before transferring to a 37°C, 5% CO2 incubator. For the ice preincubation condition, BMDMs were pre-chilled for 20 min on ice, and particles were allowed to settle onto the monolayer for 1 h on ice, then were transferred to a tissue culture incubator. Phagocytosis was measured as described previously (n = 3 biological replicates).
(TIF)
BMDMs were pre-incubated in serum-free media with varying concentrations of recombinant mouse C3a (R&D systems) or with 20% FBS for 1 h, then infected with A. Hc or B. Zymosan at an MOI of 5 for 30 min. Phagocytosis was assessed by flow cytometry (n = 2 biological replicates). C-D. BMDMs In serum-free (C) or FBS-supplemented (D) media were pre-treated with PMA for 2h and infected with Hc (MOI5, 60 min). Phagocytosis was measured by flow cytometry (n = 3 biological replicates). E. Thioglycollate-elicited peritoneal macrophages in serum-free or FBS-supplemented media were treated for 5 min with a C3aR antagonist (SB290157, 10 μM) and infected with Hc. Phagocytosis was assessed by flow cytometry (n = 3 biological replicates).
(TIF)
A-D: C3ar-/- (A-D) or C3-/- (E-H) and age-matched WT mice were infected intranasally with Hc. The levels of Il-1β (A,E), Il-6 (B,F), TNF-α (C,G), and IFN-γ (D,H) in lung homogenates were measured at the indicated time points by the cytokine CBA assay. *p ≤ 0.05, ****p ≤ 0.0001, by two-tailed student’s T-test.
(TIF)
Comma-separated-values (CSV) file showing the casTLE analysis [51] results comparing the guide abundances in the Hc infected and uninfected J774A.1 pools. Each row corresponds to a gene whose targeting sgRNA sequences were identified in at least one screen replicate (count threshold = 25). The columns are shown as follows: GeneID: ENSEMBL gene ID; Symbol: EMSEMBL gene symbol; GeneInfo, Localization, Process, and Function GO-terms assigned to the gene; #Elements: the number of guides identified in either replicate (1 or 2); casTLE Effect: the magnitude and sign of the effect for one screen replicate (1 or 2) or a combination of the replicates (Combo), where a negative effect indicates guide depletion in the Hc-infected pool and a positive effect indicates guide enrichment in the Hc-infected pool compared to the uninfected pool; casTLE Score: the casTLE confidence score; Signed Combo casTLE score: the Combo casTLE confidence score with the sign of the effect; P value: indicates the p-value of the gene effect; 5% FDR: 1 indicates a gene is a screen hit under a 5% FDR cutoff, and 0 indicates a gene is not a screen hit under a 5% FDR cutoff; 10% FDR: 1 indicates a gene is a screen hit under a 10% FDR cutoff, and 0 indicates a gene is not a screen hit under a 10% FDR cutoff; Minimum Effect Estimate: Minimum effect estimate using a 95% credible interval; Maximum Effect Estimate: Maximum effect estimate using a 95% credible interval.
(CSV)
A CSV file showing the casTLE analysis results comparing the J774A.1 pool at T = 0 (time 0), or the start of the screen following sgRNA transduction and selection, and the uninfected pool that had been passaged for the duration of the screen. Row and column annotations are the same as those described above for S1 Table.
(CSV)
A CSV file showing the casTLE analysis results comparing the plasmid library pool and the uninfected J774A.1 pool that had been passaged for the duration of the screen. Row and column annotations are the same as those described above for S2 Table.
(CSV)
The sub-libraries covered in our screens are listed, with information on the number of genes covered in each screen, the rounds of Hc-mediated lysis (at least 50%) and recovery following uracil removal, the duration of the screen, the estimated selection pressure, or difference in divisions between the Hc infected and uninfected conditions assessed by cell counting. The number of sensitizing and de-sensitizing hits that pass the 5%FDR cutoff, and the replicate correlation coefficient for the confidence score is also listed.
(CSV)
Genomic DNA was harvested from CRISPR-targeted and non-targeted cells and the targeted locus was amplified by PCR (~200bp upstream and downstream of the predicted cut site), and sequenced via capillary reaction. TIDE analysis was performed on the sequence traces from control and targeted cells to estimate the percentage of alleles that had INDELs resulting from double-stranded break repair.
(CSV)
Comma-separated-values (CSV) file listing the names of the oligonucleotides used and their sequences.
(XLSX)
A representative live-cell confocal microscopy movie showing a vehicle-treated Lifeact-eGPF-expressing J774A.1 cell extending an actin-rich membrane protrusion in the direction of mCherry-expressing Hc yeast.
(MP4)
The DIC channel from the same experiment as S1 Movie.
(MP4)
A representative live-cell confocal microscopy movie showing a Lifeact-eGFP-expressing J774A.1 treated with the C3aR antagonist SB290157 (10 μM) interacting with Hc yeast and failing to initiate the formation of an actin-rich pseudopod, and as a result exhibiting delayed capture of Hc yeast.
(MP4)
The DIC channel from the same experiment as S3 Movie.
(MP4)
A ZIP archive of tables in CSV format giving counts for each sgRNA. File names indicate sub-library and experimental sample.
(ZIP)
Data Availability Statement
All data are available within the manuscript and its Supporting information files.