Abstract
Background & Aims
Inositol polyphosphate multikinase (IPMK), an essential enzyme for inositol phosphate metabolism, has been known to mediate major biological events such as growth. Recent studies have identified single-nucleotide polymorphisms in the IPMK gene associated with inflammatory bowel disease predisposition. Therefore, we aimed to investigate the functional significance of IPMK in gut epithelium.
Methods
We generated intestinal epithelial cell (IEC)-specific Ipmk knockout (IPMKΔIEC) mice, and assessed their vulnerability against dextran sulfate sodium–induced experimental colitis. Both bulk and single-cell RNA sequencing were performed to analyze IPMK-deficient colonic epithelial cells and colonic tuft cells.
Results
Although IPMKΔIEC mice developed normally and showed no intestinal abnormalities during homeostasis, Ipmk deletion aggravated dextran sulfate sodium–induced colitis, with higher clinical colitis scores, and increased epithelial barrier permeability. Surprisingly, Ipmk deletion led to a significant decrease in the number of tuft cells without influencing other IECs. Single-cell RNA sequencing of mouse colonic tuft cells showed 3 distinct populations of tuft cells, and further showed that a transcriptionally inactive population was expanded markedly in IPMKΔIEC mice, while neuronal-related cells were relatively decreased.
Conclusions
Cholinergic output from tuft cells is known to be critical for the restoration of intestinal architecture upon damage, supporting that tuft cell–defective IPMKΔIEC mice are more prone to colitis. Thus, intestinal epithelial IPMK is a critical regulator of colonic integrity and tissue regeneration by determining tuft cell homeostasis and affecting cholinergic output.
Keywords: Colon, IPMK, Epithelial Barrier, Tuft Cell, Single-Cell RNA-Seq, Colitis, IBD
Abbreviations used in this paper: cDNA, complementary DNA; ChAT, choline acetyltransferase; DEG, differentially expressed gene; DSS, dextran sulfate sodium; EpCAM, epithelial cell adhesion molecule; FITC, fluorescein isothiocyanate; GI, gastrointestinal; HBSS, Hank’s balanced salt solution; IBD, inflammatory bowel disease; IEC, intestinal epithelial cell; IPMK, inositol polyphosphate multikinase; IPMKf/f, inositol polyphosphate multikinase floxed; KAIST, Korea Advanced Institute of Science and Technology; mRNA, messenger RNA; mTOR, mechanistic target of rapamycin; Pou2f3, POU class 2 homeobox 3; qRT-PCR, quantitative reverse-transcription polymerase chain reaction; scRNA-seq, single-cell RNA sequencing; SNP, single-nucleotide polymorphism
Graphical abstract
Summary.
Depletion of inositol polyphosphate multikinase in mouse intestinal epithelium aggravates experimental colitis. Cholinergic tuft cells but no other intestinal cells are decreased by Ipmk deletion, revealing the role of inositol polyphosphate multikinase in maintaining gut integrity via determining tuft cell homeostasis.
The gastrointestinal (GI) tract is lined by a constantly renewing population of intestinal epithelial cells (IECs). Mammalian IECs show a multitude of physiological functions, including absorption of nutrients, secretion of hormones as well as neurotransmitters, production of antimicrobial peptides in response to infectious stimuli, and control of tissue regeneration after physical damage. The homeostatic and injury-induced regeneration of intestinal epithelium is sustained by active proliferation of crypt-based stem cells localized near the base of the crypts.1 As developing IECs progress up the crypt–villus axis, proliferation ceases, and subsequent differentiation into various cell types (eg, enterocytes, goblet, and Paneth cells) takes place.1 Specialized IECs synergistically mediate GI-specific functions in digestion, barrier, and immunity.2
Among those functionally distinctive epithelial cell types, tuft cells are not a newly discovered cell type, but their function was poorly understood until recently. The key roles of tuft cells include participation in chemosensing, type 2 immunity, as well as cholinergic signaling in the gut.3, 4, 5 Thus, tuft cells maintain epithelial integrity and are essential for protecting the host from enteric infections and inflammatory reactions.6,7 Mechanisms that regulate proliferation and differentiation of IECs, such as tuft cells, are tightly orchestrated to ensure proper maintenance of the GI tract. Still, the molecular and cellular factors that regulate intestinal homeostasis remain largely unclear.
Inflammatory bowel disease (IBD), consisting mainly of Crohn’s disease and ulcerative colitis, is a chronic inflammatory disorder of the GI tract.8 Several factors are involved in the pathogenesis of IBD, including the presence of IBD susceptibility genes, altered microbial flora, excessive innate/adaptive immunity, defective autophagy, and reduced mucosal epithelial barrier defense.9,10 IBD is associated with the dysregulation of many pathways involved in the maintenance of intestinal barrier integrity, such as proliferation, migration, differentiation, and cell death. Furthermore, excessive tissue injury and inadequate regeneration lead to an increased risk of developing diseases such as cancer.11,12
Among IBD susceptibility genes is Ipmk, encoding an enzyme with broad substrate specificity that catalyzes the production of inositol polyphosphates (eg, inositol 1,3,4,5,6-pentakisphosphate) and phosphatidylinositol 3,4,5-triphosphates.13,14 In addition to its catalytic roles, inositol polyphosphate multikinase (IPMK) regulates major signaling factors noncatalytically, including mechanistic target of rapamycin (mTOR), adenosine 5′-monophosphate–activated protein kinase, p53, and serum response factor.15 Collectively, IPMK acts as a signaling hub in mammalian cells that coordinates the activity of various signaling networks.16 Recent genome-wide association studies have shown Ipmk as a putative risk gene for IBD.17,18 Furthermore, a germ-line mutation in Ipmk was discovered in patients with familial small intestinal carcinoids, suggesting a role of IPMK in the homeostatic control of intestinal tissues.19 Accordingly, we asked whether intestinal epithelial IPMK also may play a critical role in establishing and maintaining tissue homeostasis in the gut.
In this report, we characterize the effects of conditionally ablating IPMK in IECs in the colonic response to dextran sulfate sodium (DSS)-induced colonic injury, a murine model of human IBD. Our results show that the IPMK-deficient colon is more vulnerable to experimental colitis, and IEC-specific IPMK deficiency results in significant defects in tuft cell development. In particularly, choline acetyltransferase (ChAT) expression was decreased significantly, implying that insufficient cholinergic input from tuft cells to stem cells under IPMK depletion may underlie the exacerbated colitis phenotype. Combined analyses of single-cell transcriptomics, flow cytometry, and immunofluorescence staining further suggest that IPMK deletion perturbed the composition of colonic tuft cell subtypes by increasing a transcriptionally inactive colonic tuft cell subtype and by decreasing a neuronal development-related colonic tuft cell subtype. Thus, we found that gut epithelial IPMK is a key physiological determinant for the maintenance of colonic tuft cells to promote tissue regeneration.
Results
IPMK Deficiency in Colonic Epithelium Exacerbates Experimental Colitis and Attenuates Recovery
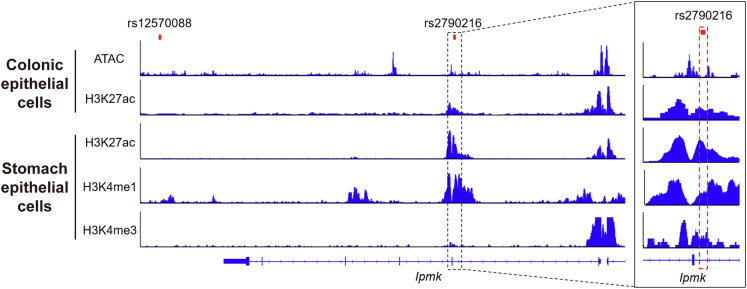
Based on the previous reports identifying several IPMK gene mutations as risk single-nucleotide polymorphisms (SNPs) for human IBD,17,18 we found that rs2790216 SNP is located at an intron region between exons 1 and 2 of the Ipmk gene,18 and rs12570088 is located approximately 12 kb downstream of the Ipmk gene17 (Figure 1). Because both variants are not present in the protein coding region, they would not change amino acid sequences and protein functions of IPMK. Interestingly, although rs12570088 is not associated with histone modification markers, rs2790216, which is close to an exon–intron junction, is associated with active enhancer markers such as H3K4me1 and H3K27ac,20, 21, 22 suggesting that rs2790216 could impact IPMK expression. Because rs2790216 is close to an exon–intron junction and is associated with H3K4me3, which usually is enriched in not only active promoters but also exon–intron junctions recruiting splicing machinery,23 rs2790216 may change the efficiency of transcript splicing that generates unstable Ipmk messenger RNA (mRNA).
Figure 1.
Gene tracks showing the genomic loci of previously reported Ipmk SNPs. Two SNPs linked to the incidence of IBD patients were associated with the Ipmk gene. SNP rs12570088 was located at approximately 12 kb downstream of the Ipmk gene,17 and rs2790216 was located at an intron region between exons 1 and 2 of the Ipmk gene.18 SNP rs2790216 was associated with active enhancer markers H3K27ac and H3K4me1, and H3K4me3, which recruits splicing machinery. ATAC, assay for transposase-accessible chromatin.
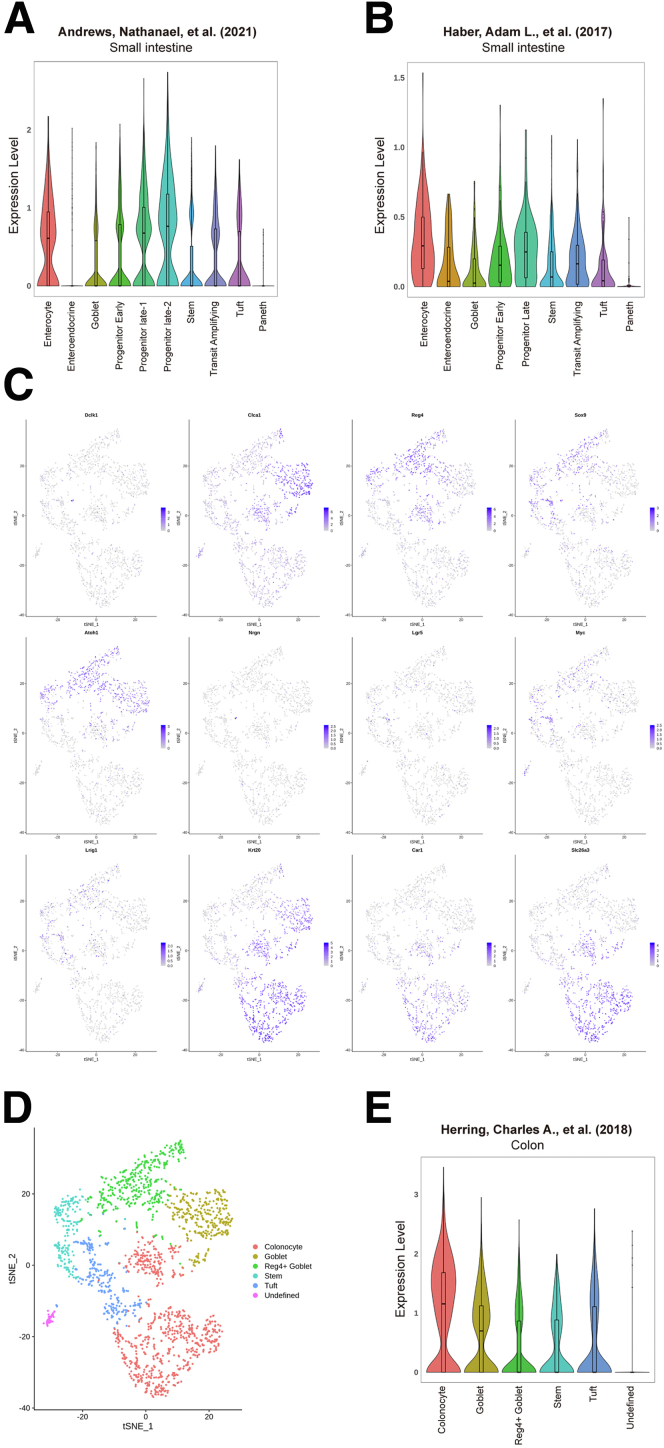
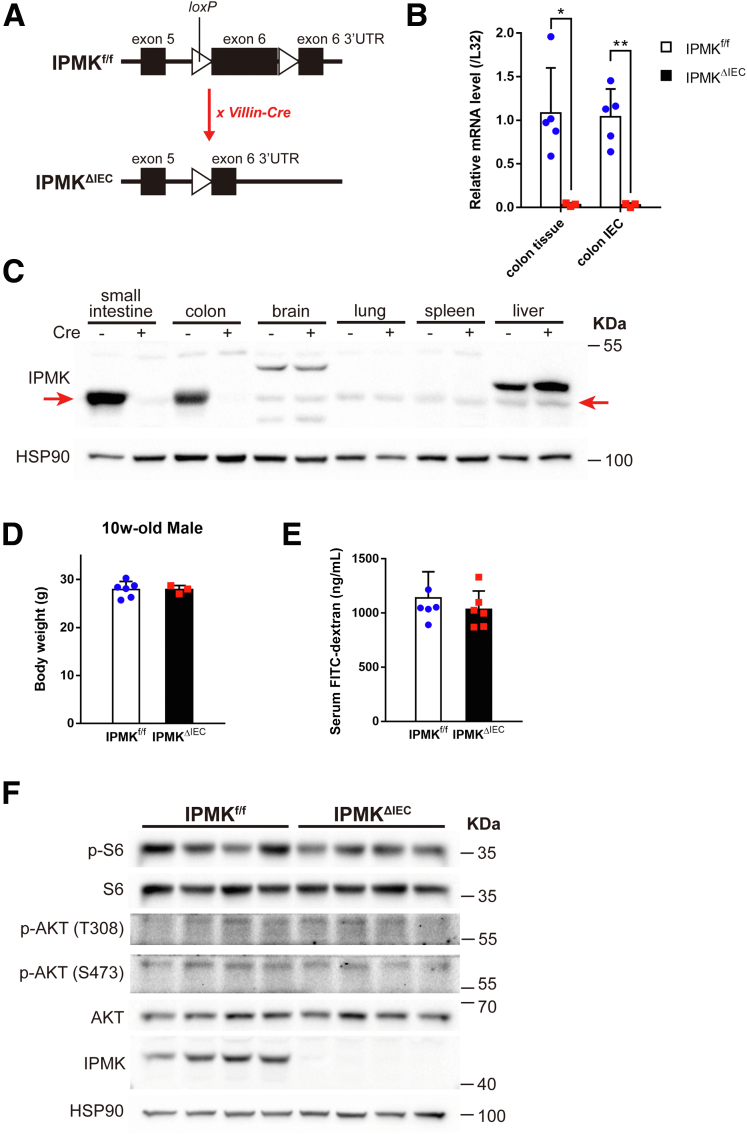
Hence, we sought to test whether IPMK plays a significant role either in intestinal homeostasis or in damaged and inflaming situations that resemble human IBD. In addition, the Human Protein Atlas database showed that IPMK is highly expressed in human colon epithelium,24 and recent single-cell RNA sequencing (scRNA-seq) projects showed that Ipmk has a pan-epithelial expression pattern in mouse intestine (Figure 2).25, 26, 27 To define the involvement of IPMK in the intestinal epithelium, an IPMKΔIEC (Ipmkf/f; Villin-Cre) mouse line was generated to achieve IEC-specific IPMK depletion (Figure 3A). Both IPMK transcript and protein were verified as successfully depleted in colon and colonic epithelia (Figure 3B and C). IPMK was unaffected in other organs.
Figure 2.
IPMK shows a pan-epithelial expression pattern in mouse intestine. (A and B) Expression level of IPMK was analyzed from 2 previous scRNA-seq studies of mouse small intestine.26,27 IPMK was expressed comparably within mouse small intestinal epithelium including enterocyte, endocrine, goblet, stem, and tuft cells, except Paneth cells. (C) The data from previous scRNA-seq study of mouse colon epithelium25 were used to draw a t-SNE plot showing the mRNA level of epithelial cell type marker genes. (D) Each cell was clustered by an unsupervised method and annotated with gene expression pattern. (E) A violin plot showing the expression level of IPMK from previously clustered scRNA-seq data. IPMK expression levels also were comparable in mouse colon epithelium.
Figure 3.
IPMKΔIECmice appear normal in homeostatic conditions. (A) IPMK was specifically depleted in intestinal epithelium using Villin-Cre mice. loxP sequences were inserted in the exon 6 region of the Ipmk gene, and Cre-driven deletion of flanked sequences resulted in truncated mRNA transcripts that were degraded readily. Tissue-specific IPMK knockout was confirmed at the (B) mRNA level and (C) protein level (n = 5, 3 per genotype in panel B). Red arrows indicate the IPMK band. Data are presented as means ± SD. (D) Body weights of 10-week-old mice showed no difference between genotypes (n = 6, 3 per genotype). Data are presented as means ± SD. (E) Gut permeability did not differ significantly between genotype in a normal state. Serum FITC-dextran concentration was measured 4 hours after a 400-mg/kg dose by oral gavage (n = 6, 6 per genotype). Data are presented as means ± SD. (F) No notable difference was observed in blots of mTOR complex and AKT signaling cascade. ∗P < .05, ∗∗P < .01. HSP90, heat shock protein 90; UTR, untranslated region.
Despite IPMK SNPs being found repeatedly in IBD patients, IPMKΔIEC mice did not show a significant difference in body weight compared with Ipmk-floxed (IPMKf/f) control mice in homeostatic conditions (Figure 3D). Assessment of intestinal permeability using fluorescein isothiocyanate (FITC)-dextran also showed that IPMK-deficient intestinal epithelium has no critical defect in its barrier function (Figure 3E). To dissect the impact of IPMK depletion at the cell level, epithelial cells were isolated from mouse colonic tissues and analyzed. Contrary to previous findings in mouse embryonic fibroblasts,28 IPMK-deficient epithelial cells appeared indifferent to wild-type cells in terms of both AKT phosphorylation and mTOR complex 1 downstream signaling cascades (Figure 3F). These results collectively suggest that the deletion of IPMK in intestinal epithelium does not cause critical functional abnormalities in a normal, healthy gut.
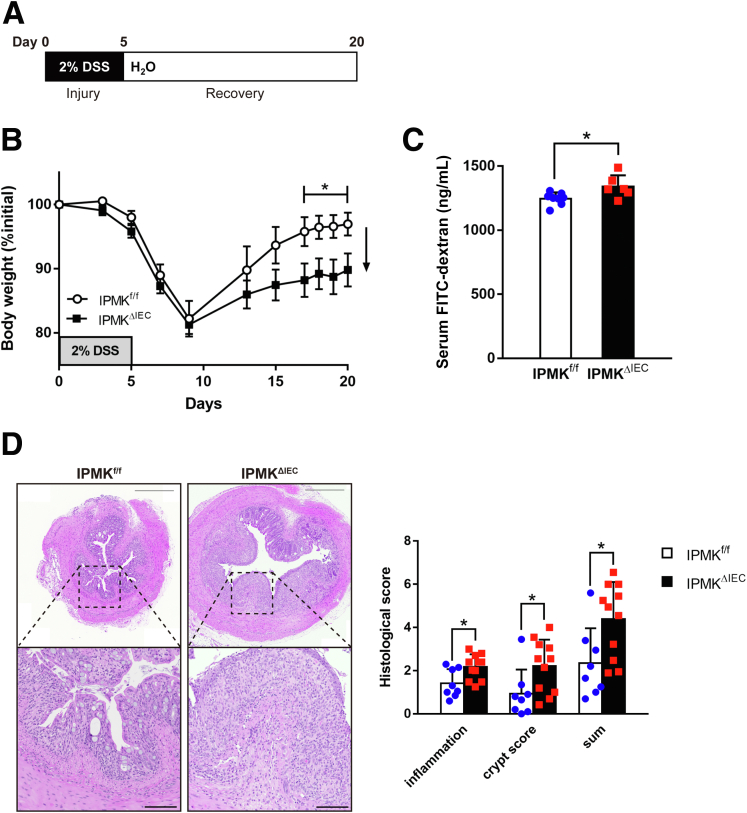
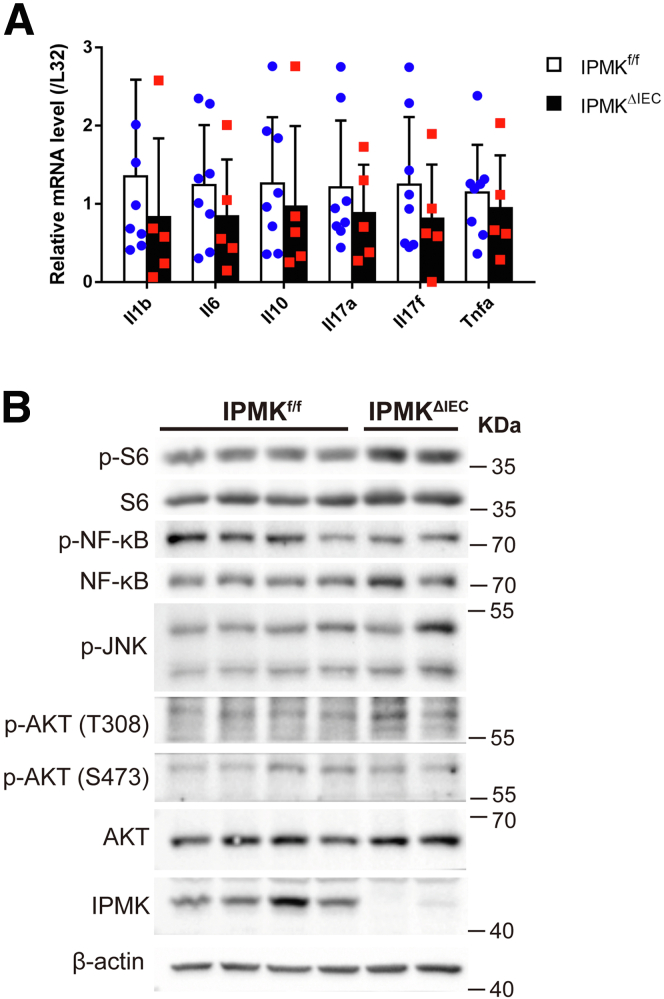
Some factors, such as Regnase-1,29 become prominent in intestinal homeostasis only when the tissue is damaged or inflamed. Therefore, we aimed to analyze IPMKΔIEC mice in pathologic conditions. A DSS-induced acute colitis model was chosen to represent human IBD (Figure 4A), and physiological features of diseased mice were assessed as in normal status. Surprisingly, IPMKΔIEC mice showed significantly attenuated recovery from colitis. Although IPMKf/f mice almost reached their initial body weight on day 17 (after 12 days post-DSS severance), IPMKΔIEC mice retained colitis-induced weight loss (Figure 4B). The gut epithelial barrier also was much more permeable in IPMKΔIEC mice, as indicated by higher FITC-dextran levels in IPMKΔIEC mice sera (Figure 4C). Histologic analyses of colonic inflammation and damage to crypt structures further confirmed that IPMK deficiency in colonic epithelium results in exacerbated colitis upon acute DSS administration (Figure 4D). This observation led us to compare the inflammation signatures of wild-type and IPMK-deficient colonic epithelium. Interestingly, expression levels of inflammatory cytokines involved in colitis were not significantly different between IPMKf/f and IPMKΔIEC mice (Figure 5A). Inflammatory signaling pathways including nuclear factor-κB were unchanged, in parallel with cytokine levels (Figure 5B).
Figure 4.
IPMK deficiency in colonic epithelium exacerbates experimental colitis and attenuates recovery. (A) Experimental scheme of a DSS-induced acute colitis model in which 2% DSS was given for 5 days and exchanged for normal water during recovery. Mice were killed on day 8 for analysis of actively inflaming tissue or on day 20 for end point analysis. (B) IEC-specific Ipmk knockout (IPMKΔIEC) mice showed attenuated recovery from colitis. Body weight was monitored at the indicated time points and calculated as the percentage of initial body weight (n = 8, 7 per genotype). Data are presented as means ± SEM. (C) FITC-dextran permeability of the colonic wall was higher in IPMKΔIEC mice on day 20. The concentration of FITC-dextran in mouse serum was measured 4 hours after 200 mg/kg was given by oral gavage (n = 8, 6 per genotype). (D) IPMK-deficient colons showed exacerbated tissue damage on day 8. Representative H&E staining images of distal colon cross-sections. Scale bars: 500 μm and 100 μm for magnified images. Histologic scoring was performed in 2 categories (inflammation and crypt score) and combined (n = 8, 11 per genotype). ∗P < .05. Data are presented as means ± SD. (A–C) Data are representative of 3 independent experiments with similar results.
Figure 5.
No significant difference in inflammatory signaling exists between IPMKΔIECand control mice. (A) Although IPMK deletion exacerbates colitis phenotypes, expression of major inflammatory cytokines in colonic epithelial cells showed no difference between genotypes (n = 8, 5 per genotype). Data are presented as means ± SD. (B) The activity of the nuclear factor-κB (NF-κB) pathway, regulating inflammation and AKT signaling, and governing cell survival, was assessed by immunoblotting. The phosphorylation level of key proteins was not different between IPMKΔIEC and IPMKf/f mice. p-AKT; p-JNK, c-Jun N-terminal kinases; p-NF-κB, nuclear factor kappa B.
IPMK-Deficient Colon Shows Significantly Decreased Tuft Cell Number
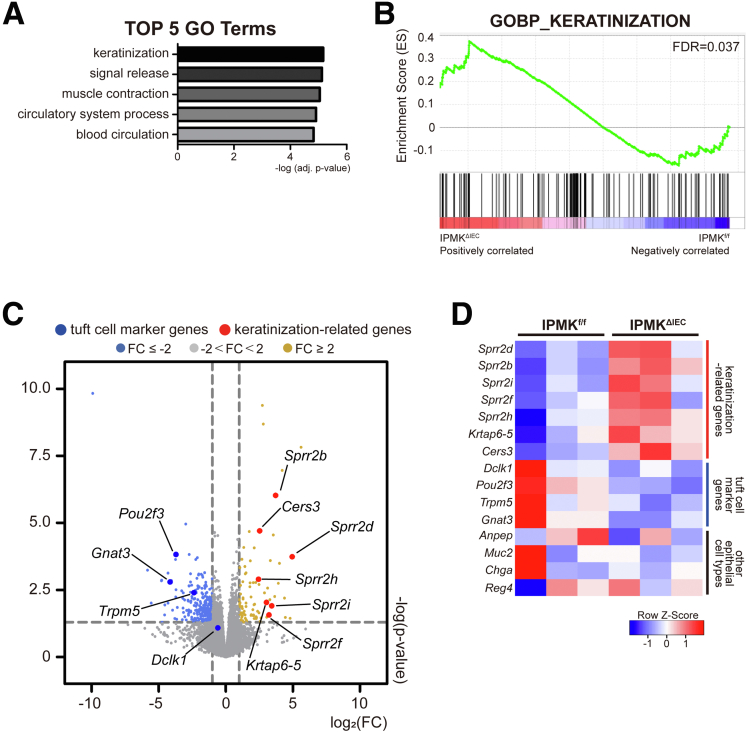
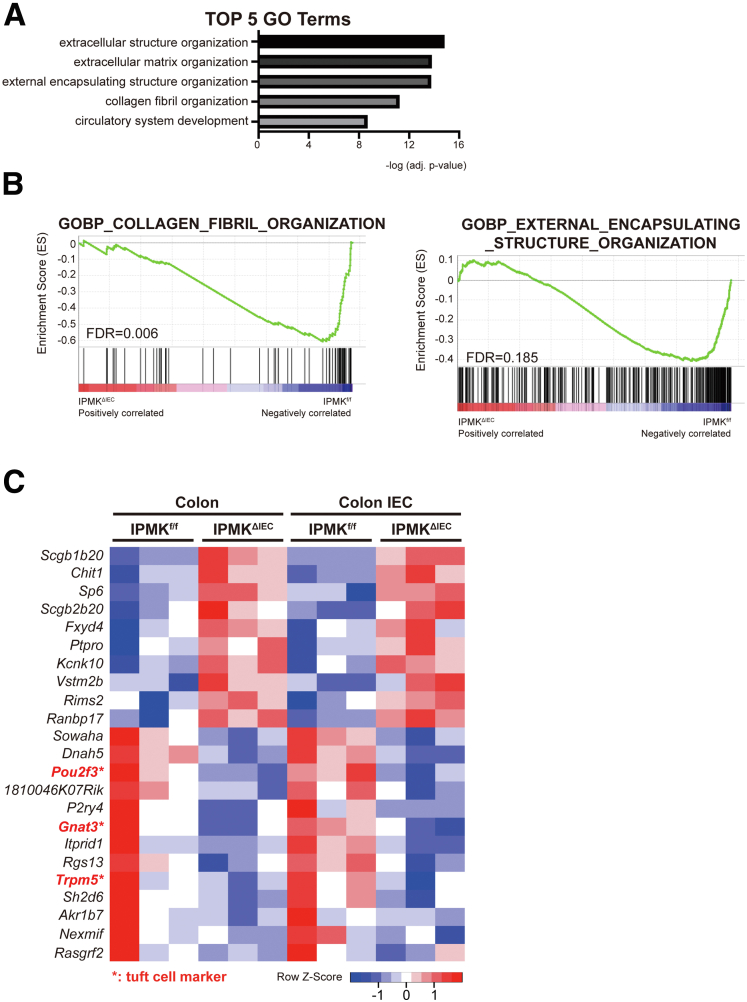
Because we did not find marked differences in inflammation in IPMK-deficient colons, transcriptomic analysis was performed to elucidate what underlies severe disease in IPMKΔIEC mice. Unexpectedly, the top-ranked category of differentially expressed genes (DEGs) was keratinization, a seemingly colitis-irrelevant biological process (Figure 6A). These genes included members of the small proline-rich protein family, such as Sprr2d and Sprr2h, and appeared to be increased dramatically in the IPMK-deficient colon (Figure 6B). Normally, these genes act as key components of keratinocyte differentiation and are strongly induced upon differentiation of stratified squamous epithelia (epidermis).30 Significant up-regulation of the Sprr gene family in the colon can be interpreted in 2 ways. First, the Sprr2h mRNA level was reported to be increased in an experimental colitis model, possibly as a compensatory action for deteriorated barrier function,31 thus higher Sprr levels may reflect functionally impaired, leaky colonic epithelia. Second, keratinocyte differentiation marker genes such as Sprr2a and Krt10 are known to be regulated by POU class 2 homeobox 3 (Pou2f3),32 a transcription factor that is expressed abundantly in the epidermis. However, in the colon, Pou2f3 is a cell fate–determining factor that is indispensable for specification of tuft cells,33 a rare and functionally distinctive population of the intestinal epithelium. We speculated that differences in Pou2f3 expression could underlie this keratinization-like transcriptomic profile. Surprisingly, Pou2f3 was down-regulated significantly in IPMKΔIEC mice along with other tuft cell markers such as Trpm5 and Gnat3 (Figure 6C and D). Other epithelial cell populations represented by Anpep (colonocytes), Muc2 (goblet cells), Chga (enteroendocrine cells), and Reg4 (deep crypt secretory cells) did not show significant variation. We speculate that some major transcriptomic changes blurred as a result of the effect of the temporal window (colon tissues were harvested on day 20 when IPMKf/f mice were recovered from DSS-induced colitis), and the heterogeneity of colonic tissues that included not only epithelial cells but also immune cells, neuronal cells, and muscle cells. Therefore, to investigate the transcriptomic profiles underlying higher disease activity of IPMK-deficient colons, actively inflaming colonic tissues were harvested earlier (on day 8) and epithelial cells were isolated further for mRNA sequencing. DEGs were again enriched for inflammation unrelated to any given process, such as extracellular organization (Figure 7A and B), and few genes overlapped with the previous analysis of colonic tissues. Although the number of common DEGs was insufficient to be categorized as valid gene ontology terms, tuft cell marker genes (Pou2f3, Gnat3, and Trpm5) were again listed (Figure 7C). Together, concurrent reduction in tuft cell–associated genes strongly implicates the alteration of tuft cell populations in the IPMK-deficient colon as a very probable cause of severe colitis pathology.
Figure 6.
Transcriptomic profile of IPMK-deficient colon reveals gene signatures associated with keratinization and tuft cell alteration. (A) The top 5 gene ontology terms were calculated using adjusted P values. (B) Enrichment set data showed significant changes of keratinization terms by gene set enrichment analysis. (C) Volcano plot of the DEGs marked with tuft cell markers and keratinization-related genes. (D) Heatmap representing selected DEGs. Genes associated with keratinization were up-regulated whereas tuft cell markers were down-regulated, and other epithelial cell type–related genes did not show significant alterations. Mice were killed on day 20 for RNA sequencing of colonic tissues (n = 3, 3 per genotype). FC, fold change; FDR, false discovery rate.
Figure 7.
Transcriptomic analysis of colonic epithelial cells at an earlier colitis phase and suggested tuft cell–related genes as major remarks of more severe colitis shown in IPMKΔIECmice. (A) The top 5 gene ontology terms were calculated using adjusted P values. (B) Enrichment set data showed significant changes of collagen fibril organization and external encapsulating structure organization by gene set enrichment analysis. (C) Heatmap representing the overlapping DEGs between colonic tissue and colonic epithelial cell mRNA sequencing. Among very few genes, tuft cell marker genes were listed as common DEGs. Mice were killed on day 8 for RNA sequencing of colonic epithelial cells (n = 3, 3 per genotype). FDR, false discovery rate.
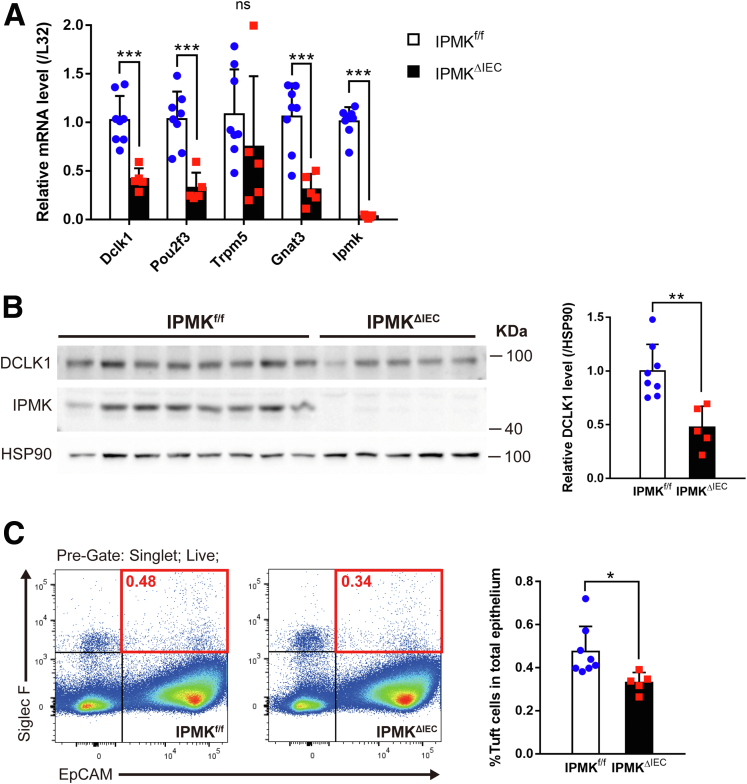
Accordingly, decreased mRNA expression of tuft cell markers was validated further through quantitative reverse-transcription polymerase chain reaction (qRT-PCR) in colonic epithelial cells. Earlier studies reported Dclk1, Pou2f3, Trpm5, and Gnat3 as tuft cell–specific genes in intestinal epithelia, and expression levels of all genes except for Trpm5 showed a significant decrease in IPMKΔIEC mice (Figure 8A). In addition, doublecortin like kinase 1 (DCLK1), a marker protein of colonic tuft cells, was down-regulated at the protein level (Figures 8B and 9A). These results emphasized the need for precise quantification of tuft cell populations in both wild-type and IPMK-deficient colitic colon tissue. Therefore, we analyzed the relative abundance of tuft cells using flow cytometry, assessing epithelial cell adhesion molecule (EpCAM)/siglecf sialic acid binding Ig-like lectin F (Siglec F) double-positive cells in isolated colonic epithelia. The percentage of tuft cells in total colonic epithelial cells was significantly lower in IPMKΔIEC mice: only approximately 70% of the control group (Figure 8C). This suggests the alteration in colonic tuft cell populations is the main cause of exacerbated colitis in IPMKΔIEC mice upon acute DSS administration.
Figure 8.
Colonic IPMK depletion leads to significantly decreased tuft cell numbers in cases of acute colitis. (A) Colonic epithelial cells from IPMKΔIEC mice showed decreased expression of tuft cell marker genes. Each mRNA was assessed by qRT-PCR and normalized to the expression of the L32 gene (n = 8, 5 per genotype). Data are presented as means ± SD. (B) DCLK1 protein level was lower in IPMK-deficient colon as shown in representative blots. Band intensities were quantified by densitometry using ImageJ (n = 8, 5 per genotype). Data are presented as means ± SD. (C) Tuft cell abundance in the colonic epithelium was quantified by flow cytometry. Epithelial cells were defined by EpCAM expression, and siglecf sialic acid binding Ig-like lectin F (Siglec F)–positive epithelial cells (red square) were considered as tuft cells (n = 8, 5 per genotype). Data are presented as means ± SD. All mice were killed on day 8. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001. Data are representative of 3 independent experiments with similar results. HSP90, heat shock protein 90.
Figure 9.
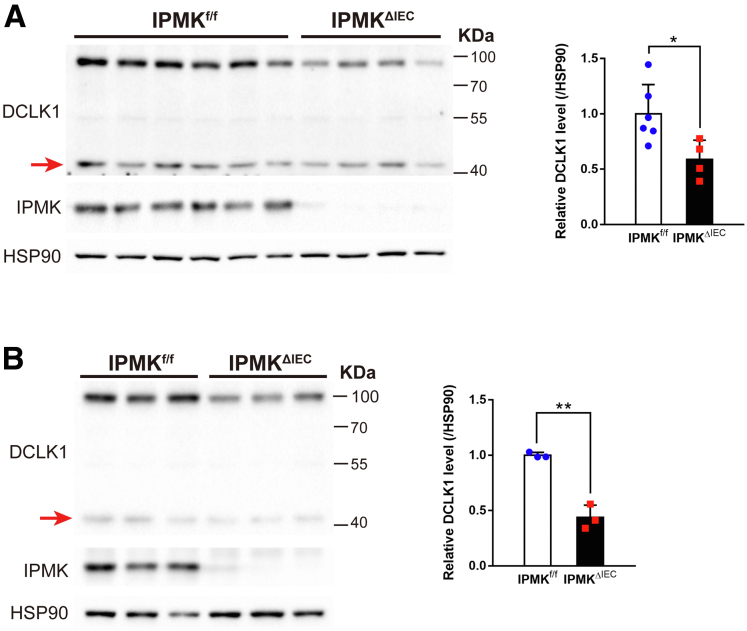
DCLK1 48-kilodalton isoforms also were down-regulated in IPMK-deficient colonic epithelial cells. Protein levels of the shorter DCLK1 isoform, with a molecular weight of 48 kilodaltons (indicated by red arrows), were decreased in colonic epithelial cells of IPMKΔIEC mice both in (A) colitic conditions (n = 6, 4 per genotype) and (B) normal status (n = 3, 3 per genotype). Band intensities were quantified by densitometry using ImageJ. Data are presented as means ± SD. ∗P < .05, ∗∗P < .01. HSP90, heat shock protein 90.
IPMK Regulates Cholinergic Input to Stem Cells via Governing Tuft Cell Development
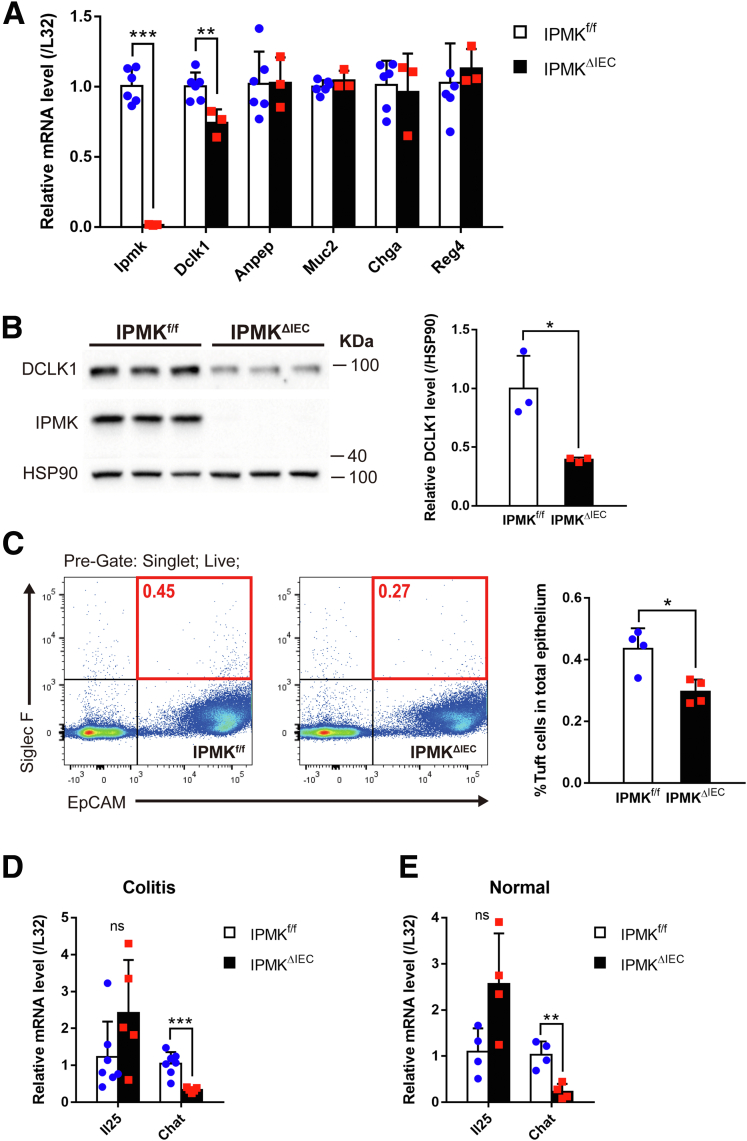
To investigate the relationship between IPMK deletion and tuft cell decrease, we first tested whether the occurrence of this phenomenon is limited to the pathologic condition. In normal, unchallenged, 10-week-old adult mice, the abundance of each colonic epithelial cell type was analyzed using marker genes. Dclk1 of tuft cells was the only gene that was decreased significantly in IPMKΔIEC mice; the other genes did not show a significant difference between groups (Figure 10A). Therefore, only tuft cells are affected specifically by the depletion of IPMK in the intestinal epithelium. The protein level of DCLK1 also was reduced in the IPMK-deficient colon at homeostasis (Figures 9B and 10B), and direct measurement of EpCAM/Siglec F double-positive cells using flow cytometry again validated the reduction of tuft cell numbers in healthy IPMKΔIEC mice (Figure 10C). Our findings collectively suggest that depletion of IPMK in colonic epithelium impaired tuft cell development before colitis progression, and these defective tuft cells subsequently result in the vulnerability of IPMKΔIEC mice upon DSS administration, without any notable abnormalities in the homeostatic condition.
Figure 10.
IPMK governs tuft cell development and regulates acetylcholine production in the colonic epithelium. (A) Basal expression of colonic epithelial cell lineage-specific marker genes was analyzed by qRT-PCR and normalized to the expression of the L32 gene. Only Dclk1, a tuft cell marker, appeared to be down-regulated (n = 6, 3 per genotype). Data are presented as means ± SD. (B) Normal colonic epithelial cells showed decreased DCLK1 protein. Band intensities of representative blots were quantified by densitometry using ImageJ (n = 3, 3 per genotype). Data are presented as means ± SD. (C) The percentage of tuft cells in the normal healthy colonic epithelium was quantified by flow cytometry and was lower in IEC-specific Ipmk knockout (IPMKΔIEC) mice. EpCAM/siglecf sialic acid binding Ig-like lectin F (Siglec F) double-positive cells (red square) were counted as tuft cells (n = 4, 4 per genotype). Data are presented as means ± SD. (D) On day 8, Chat expression was reduced in IPMK-deficient colitic epithelial cells whereas interleukin (IL)25 expression level was not altered. qRT-PCR results were normalized to the L32 gene (n = 7, 5 per genotype). Data are presented as means ± SD. (E) Chat mRNA also was down-regulated in IPMKΔIEC, mice even in normal conditions (n = 4, 4 per genotype). Data are presented as means ± SD. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001. (A–D) Data are representative of 3 independent experiments with similar results. HSP90, heat shock protein 90.
Tuft cells currently are known to be involved in intestinal biology in 2 distinctive ways: as regulators of type 2 immune response,34,35 and as paracrine supporters of intestinal stem cells.36 These functions are mediated mostly by interleukin 25 and acetylcholine, respectively, secreted by tuft cells. To elucidate how reduced tuft cells in IPMK-deficient colon worsen the DSS-induced acute colitis, the mRNA expression levels of interleukin 25 and ChAT (a key enzyme for acetylcholine synthesis in colonic tuft cells) were compared between IPMKf/f and IPMKΔIEC mice. Both in normal and pathologic states, ChAT expression appeared significantly lower in IPMK-deficient colon (Figure 10D and E), indicating that cholinergic input for intestinal stem cells is decreased.
Single-Cell Transcriptomic Analysis Reveals the Heterogeneity of Colon Tuft Cells
Recently, single-cell–level analysis of IECs showed that small intestinal tuft cells are composed of 2 populations harboring either an immune-related or a neuronal gene expression pattern.27 Because the depletion of IPMK reduced cholinergic signaling, but not immune signaling (Figure 10D and E), we speculated that the neuronal tuft cells are affected specifically by IPMK deficiency while immune-related populations remain unaffected. Therefore, scRNA-seq was performed on fluorescence-activated cell sorting (FACS)-sorted EpCAM+/Siglec F+ colonic tuft cells to analyze the transcriptomic changes of individual tuft cells in detail (Figure 11A).
Figure 11.
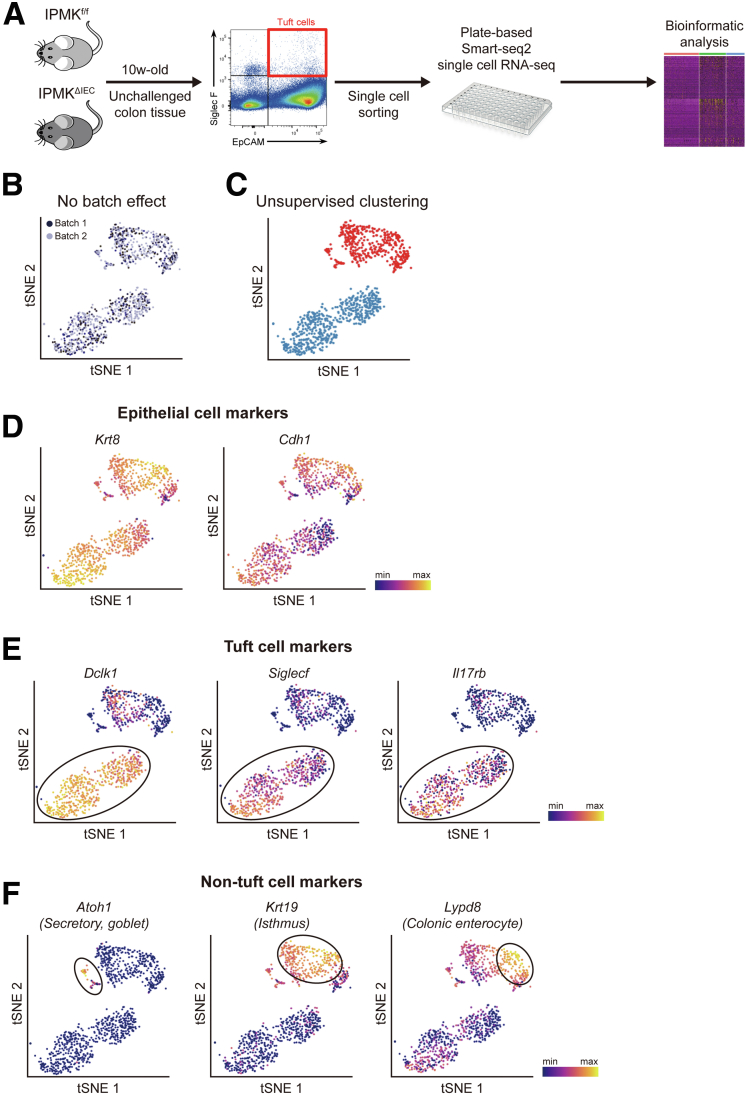
scRNA-seq analysis of tuft cells in IPMKf/fand IPMKΔIECmice. (A) An experimental scheme of fluorescence-activated cell sorting (FACS) sorting and plate-based scRNA-seq analysis. (B) A t-distributed stochastic neighbor embedding (t-SNE) plot showing that 2 independent experimental batches were overlaid well, indicating no batch effect. For each experimental batch, 2 IPMKf/f mice and 2 IPMKΔIEC mice were used. (C) Primary unsupervised hierarchical clustering analysis showed 2 clusters of cells. (D) t-SNE plots showing the expression patterns of epithelial cell marker genes (Krt8 and Cdh1), confirming the epithelial identity of analyzed cells. (E) t-SNE plots showing the expression patterns of tuft cell marker genes (Dclk1, Siglecf, and Il17rb). (F) t-SNE plots showing the expression patterns of non–tuft cell marker genes (Atoh1, secretory cells; Krt19, isthmus; and Lypd8, colonic enterocyte).
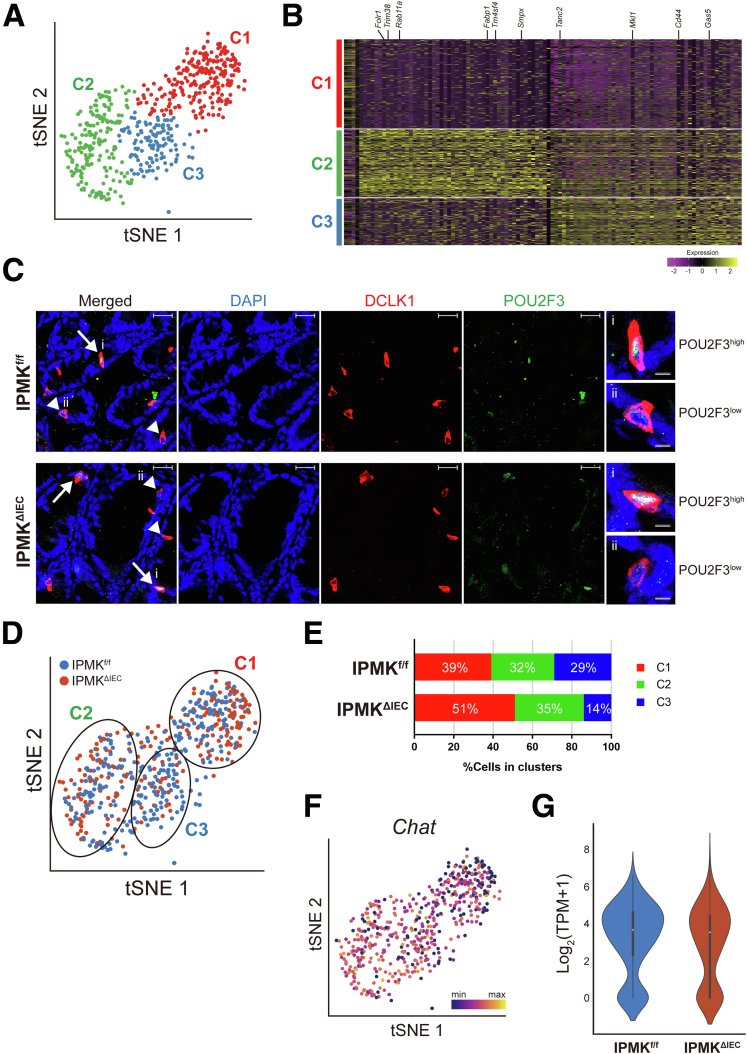
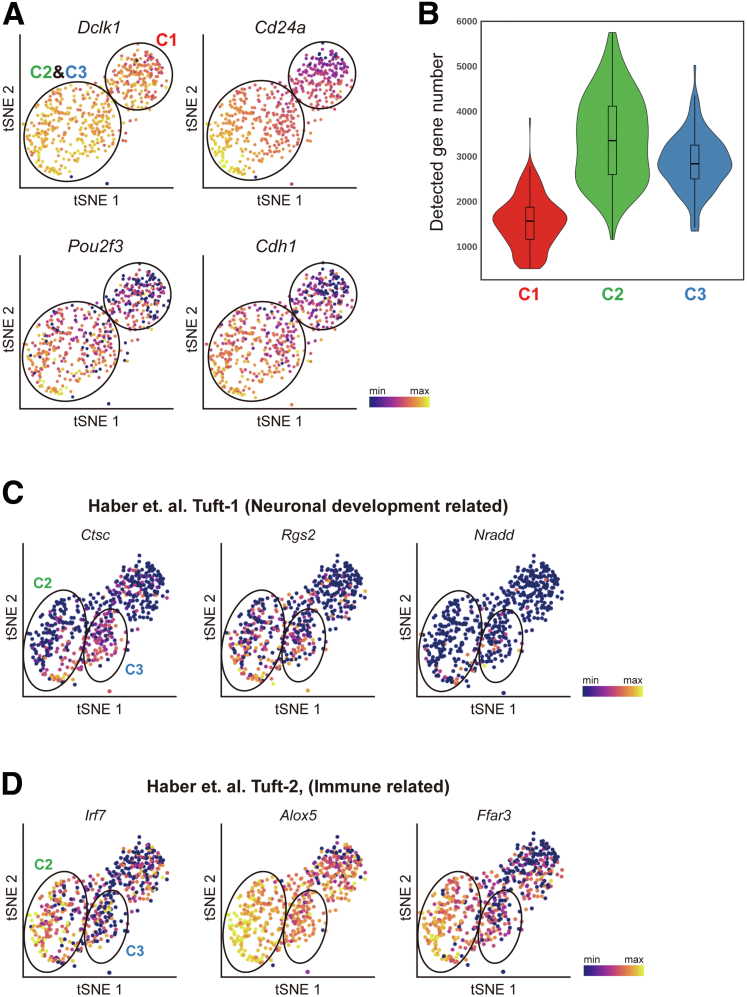
After filtering out cells with bad quality (eg, number of detected genes, <500), we analyzed 950 cells expressing epithelial markers (Krt8 and Cdh1) and found no batch effect (Figure 11B–D), indicating the high quality of our data. By analyzing the expression of known marker genes, we identified tuft cells (Dclk1+), secretory goblet cells (Atoh1+), crypt isthmus cells (Krt19+), and colonic enterocytes (Lypd8+) (Figure 11E and F). Surprisingly, in contrast to 2-clustered tuft cells of the small intestine identified previously, we first observed that colonic tuft cells are divided into 3 different populations (labeled as C1, C2, and C3) based on their transcriptional signatures (Figure 12A and B). C1 cells were distinguished from the other 2 populations in overall gene expression level. More than 2000 genes were down-regulated significantly in C1 cells, whereas only a few noncoding RNAs were discovered to be up-regulated (Supplementary Materials). The down-regulated genes included Pou2f3, which is considered to be a characteristic marker gene of tuft cells, as well as Cdh1, an epithelial cell marker gene (Figure 13A). In addition, the number of detected genes was significantly lower in C1 (Figure 13B). These results suggest that C1 tuft cells are likely to be transcriptionally inactive compared with the other colonic tuft cells. To validate the existence of C1 colonic tuft cells in vivo, we performed immunostainings of DCLK1 and POU2F3 using Swiss-roll sections of mouse colon and found that the protein level of POU2F3 clearly varied in DCLK1-positive tuft cells (Figure 12C) as in scRNA-seq, supporting the presence of C1 colonic tuft cells.
Figure 12.
Colonic IPMK regulates the composition of colon tuft cells as shown by single cell transcriptomics. (A) A t-distributed stochastic neighbor embedding (t-SNE) plot showing 3 populations (C1, C2, and C3) of colonic tuft cells shown by unsupervised clustering analysis. (B) A heatmap showing the signature genes of each population. (C) Immunofluorescence staining confirmed the existence of DCLK1+/POU2F3high and DCLK1+/POU2F3low tuft cells in both IPMKf/f and IPMKΔIEC mice. Arrows denote POU2F3high cells (C2, C3 clusters), and arrowheads indicate POU2F3low cells (C1 cluster). Scale bars: 20 μm. Right: Magnified images of (i) POU2F3high and (ii) POU2F3low cells are shown. Scale bars: 5 μm. (D) A t-SNE plot showing the distribution of IPMKf/f (blue) and IPMKΔIEC (red) tuft cells in C1, C2, and C3 clusters. (E) A bar graph showing the proportion of C1, C2, and C3 cluster cells in IPMKf/f and IPMKΔIEC mice. (F) A t-SNE plot showing the expression level of Chat. (G) A violin plot showing the Chat expression is decreased in IPMK-deficient tuft cells compared with control cells. For scRNA-seq analysis, 2 independent batches of experiments were performed, and for each experimental batch, 2 IPMKf/f mice, and 2 IPMKΔIEC mice were used. DAPI, 4′,6-diamidino-2-phenylindole.
Figure 13.
Characterization of 3 populations of colonic tuft cells shown by scRNA-seq. (A) t-distributed stochastic neighbor embedding (t-SNE) plots showing that CD24a, Pou2f3, and Cdh1 are decreased in population C1 compared with subpopulations C2 and C3, while Dclk1 expression is comparable. (B) A violin plot showing the number of detected genes in each population, suggesting that C1 cells are transcriptionally inactive compared with C2 and C3 cells. t-SNE plots showing the expression patterns of (C) neuronal development-related genes and (D) immune-related genes, indicating that our results are consistent with a previous study.27
The gene expression patterns of C2 and C3 cells resembled those of previously reported immune-related and neuronal subtypes, respectively, of small intestinal tuft cells. Although C2 cells expressed higher levels of immune-related genes such as Irf7, Alox5, and Ffar3, neuronal-related genes (Ctsc, Rgs2, and Nradd) were highly enriched in C3 cells (Figure 13C and D), confirming the previous study.
IPMK Deficiency Alters the Proportion of Colonic Tuft Cell Populations
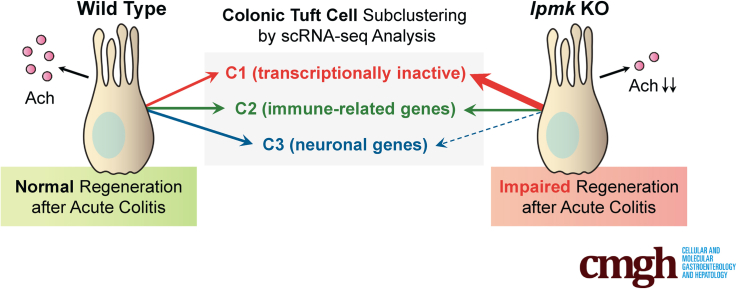
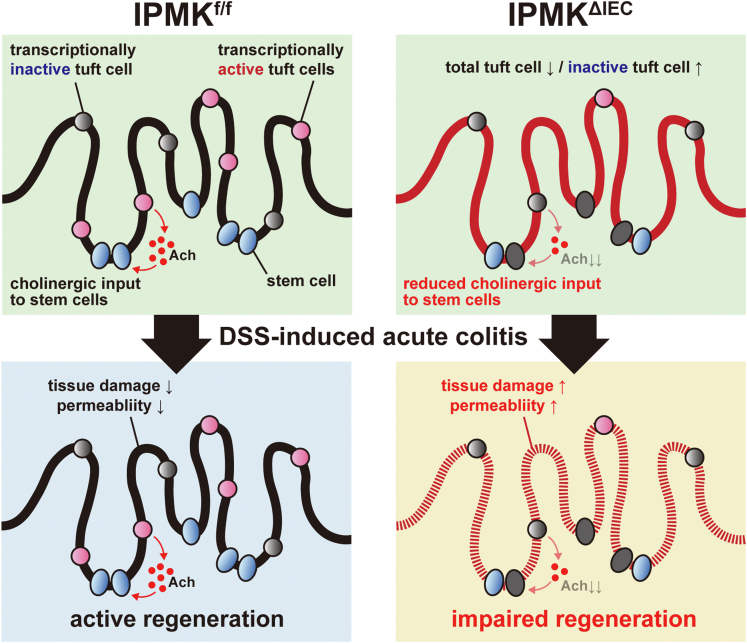
Next, to analyze the changes of cell type and gene expression induced by Ipmk deletion, we overlaid IPMKf/f cells with IPMKΔIEC cells. Although IPMK-deficient colonic epithelial cells did not form a distinct cell cluster or show dramatic gene expression changes, the clustering pattern of tuft cells differed in IPMKf/f and IPMKΔIEC animals (Figure 12D). Although the relative abundance of the immune-related C2 population was largely similar between IPMKf/f and IPMKΔIEC animals (∼30%) (Figure 12E), transcriptionally inactive C1 cells were expanded in IPMKΔIEC mice, occupying nearly half of the total tuft cell number, and the neuronal C3 population was reduced in IPMKΔIEC mice (28% → 14%) (Figure 12D and E). These results support our hypothesis that IPMK in tuft cells is important for the regeneration of colonic epithelium through a neuronal (eg, cholinergic) paracrine input. Because Chat expression was reduced in C1 (Figure 12F), the relative increase of C1 cells in IPMKΔIEC mice may lead to decreased average Chat levels of all tuft cells, and this was confirmed in IPMK-deficient tuft cells (Figure 12G). Together, these results indicate that depletion of IPMK in colonic epithelium alters the quantitative composition of tuft cell subtypes rather than modifying the characteristics of each C1/C2/C3 population. In summary, our findings show that IPMK is a critical factor for maintaining intestinal homeostasis in DSS-induced colitis through sustaining the number of transcriptionally inactive (C1) and active (C2/C3) tuft cells, thereby supporting tuft cell functions in the large intestine (Figure 14).
Figure 14.
A proposed model of IPMK actions in the gut. The number of tuft cells providing acetylcholine, which supports intestinal stem cell function, is reduced in the IPMK-deficient colon. Simultaneously, IPMK deficiency increases the relative abundance of a specific tuft cell subtype possessing decreased tuft cell–specific genes, including Chat, so that total cholinergic output from tuft cells is reduced significantly. This insufficiency in the active tuft cell population is tolerated in the normal homeostatic epithelium, and therefore IPMKΔIEC mice do not show any apparent abnormalities. However, when acute damage occurs in the colon, as in DSS-induced experimental colitis, IPMK-deficient colons show impaired regeneration, which may be owing to decreased cholinergic input to stem cells, thereby exacerbating the colitis phenotype.
Discussion
IPMK has been shown to regulate various biological events such as growth and immunity, acting either enzymatically to mediate the biosynthesis of inositol polyphosphates and phosphatidylinositol 3,4,5-trisphosphates,37,38 or noncatalytically to regulate key protein targets (eg, mTOR, p53).15 Recent evidence indicated that human IPMK SNP variants are risk factors for IBD, suggesting a key role of IPMK in the control of intestinal homeostasis.17,18 However, the specific action of IPMK in gut epithelial cells has not yet been investigated. Here, we established a physiologically important function of IPMK in maintaining gut homeostasis, showing that IPMK depletion in IECs drives the mouse colon to be more prone to DSS-induced acute colitis. Unlike our expectations from previous studies on IPMK actions in phosphatidylinositol 3-kinase (PI3K) and inflammation signaling,28,39 IPMK deletion had no impact on intestinal epithelial growth signaling (eg, PI3K-AKT-mTOR) or on nuclear factor-κB inflammatory activation. Instead, to our surprise, tuft cells, which normally promote intestinal stem cell proliferation by secreting acetylcholine, were reduced significantly in the IPMK-deficient colon. As a result, recovery of damaged tissue was attenuated, likely owing to an insufficient amount of acetylcholine. This means that IPMK normally protects intestinal epithelium from damage induced by acute inflammation, and through governing homeostasis of colonic tuft cells.
Tuft cells have been investigated intensively with regard to their role as type 2 immune response regulators, but previous research had focused mainly on small intestinal tuft cells. Small intestinal tuft cells are clearly distinguished from colonic tuft cells because succinate, a metabolite produced by helminths, triggers dramatic expansion of small intestinal tuft cells while colonic tuft cells remain unchanged.40,41 This shows that the specification mechanism of tuft cells can vary between different physiological settings. The functional significance of colonic tuft cells has been highlighted by previous studies showing that tuft cell ablation is tolerated under normal homeostatic conditions, but impaired regeneration of colonic tissues was observed upon acute damage such as irradiation7 and DSS-induced acute colitis.6,42,43 In our unbiased efforts to further dissect IPMK-mediated tuft cell dysregulation in the colon, we performed scRNA-seq analyses. In contrast to 2-clustered tuft cells of the small intestine identified previously,27 our scRNA-seq unveiled that mouse colonic tuft cells consist of 3 different populations (C1, C2, and C3). Although C2 and C3 cells appear to correspond to 2 tuft cell populations previously reported in the small intestine,27 C1 cells show unique features with extensively decreased tuft cell–specific genes. Immunohistochemistry staining further showed that tuft cell markers such as Pou2f3 were reduced among DCLK+ tuft cells in the colon, suggesting the presence of functionally inactive, degenerating tuft cells. Importantly, IPMK’s key action in the large intestine seems critical to maintain homeostatic balance among the 3 tuft cell populations because IPMKΔIEC intestine shows significantly increased C1 tuft cells, relative to C2 and C3 cells. Thus, overall reduction of Chat expression in IPMKΔIEC tuft cells suggests a diminished cholinergic signal followed by decreased tissue regeneration in colitis conditions. Notably, DEGs of colonic epithelial cell mRNA sequencing in colitis (eg, extracellular matrix organization and collagen fibril organization) (Figure 5A) strongly resembled genetic alterations in Lgr5-knockout mouse embryonic intestinal stem cells.44 This further suggests that the effect of colonic IPMK depletion eventually leads to aberrant stem cell activity and epithelial regeneration, likely through defective acetylcholine production from tuft cells.
Until now, few molecules have been identified as indispensable for the fate and development of tuft cells: Atoh1 and Pou2f3. Although Atoh1 commonly is required for differentiation of all secretory lineages,45 Pou2f3 is specific to tuft cells.33 Findings in this study suggest that IPMK is a novel factor involved in normal tuft cell development. Despite broad expression of IPMK in the gut, deletion of IPMK in Villin+ intestinal cells had a substantial impact on only the tuft cell population and not the other cell types (eg, colonocytes, goblet cells, and enteroendocrine cells), which infers that IPMK is dispensable for Atoh1-dependent cell specification processes, acting as a tuft cell–specific regulator.
Through unsupervised scRNA-seq analysis of tuft cells, we report the existence of a new tuft cell subtype (designated as C1 in our study) in the colon that shows severe down-regulation of transcripts including tuft cell markers. Colonic C1 tuft cells are speculated to be nonfunctional, degenerating cells derived from biologically active C2 and C3 tuft cells. A marked increase of C1 tuft cell populations in IPMKΔIEC colon tissues thus shows that IPMK is critical to maintaining bioactive tuft cell populations in the colon. In addition, C2 and C3 tuft cells were reduced by IPMK depletion, which suggests a possible contribution of IPMK to the developmental phase of tuft cells from intestinal stem cells. Combined with lineage tracing, further investigation will help us define how IPMK regulates tuft cell differentiation and homeostasis in the gut.
Several clinical studies have reported that reduced tuft cell numbers in the intestinal tract are associated with ileal Crohn’s disease46 and quiescent ulcerative colitis,47 suggesting that insufficient tuft cells are related to the pathology of IBDs. Of interest, IPMK-deficient colon epithelium contains reduced numbers of tuft cells in both homeostatic and DSS-induced injury conditions. Furthermore, it will be interesting to apply our discovery on C1 colonic tuft cells to examine dynamic changes in C1 tuft cells in human colonic tissues under normal and pathologic conditions to identify the clinical relevance. With an emphasis on the IPMK gene mutations as risk SNPs for human IBD, we here show a key action of IPMK in the control of the tuft cell–IBD axis, which suggests that modulating IPMK levels and activities provide insight to develop a novel therapeutic strategy to treat IBD.
Materials and Methods
Mice
Animal protocols were performed in accordance with the guidelines and ethical policies approved by the Korea Advanced Institute of Science and Technology (KAIST) Animal Care and Use Committee. Intestinal epithelium-specific IPMK knockout mice were generated by crossing Ipmk-floxed39 and Villin-Cre mice (004586 B6.Cg-Tg[Vil1-cre]997Gum/J; The Jackson Laboratory, Bar Harbor, ME). Mice were backcrossed to C57BL/6J for at least 5 generations. Eight- to 22-week-old mice (both male and female) were used in this study. A single experimental cohort consisted of age- and sex-matched mice, age did not differ by more than 2 weeks. Mice used in this study were maintained in a specific pathogen-free facility at KAIST Laboratory Animal Resource Center.
DSS-Induced Acute Colitis Model
DSS salt (colitis grade; molecular weight, 36,000–50,000 daltons; MP Biomedicals, Irvine, CA) was given to mice as a 2% filtered solution in drinking water, ad libitum, to induce acute colitis. DSS-containing water was refreshed once every 2–3 days. After 5 days of DSS administration, mice were allowed to recover with normal drinking water. The body weight and survival of each mouse was monitored at the indicated time points. No more than 5 mice were kept in 1 cage.
FITC–Dextran Intestinal Permeability Assay
Mice were fasted (water and food) for 4 hours, and then orally gavaged with FITC-dextran (FD4; Sigma-Aldrich, Saint Louis, MO) solution in Dulbecco's phosphate buffered saline (DPBS) using a 400-mg/kg dose. For colitis-induced mice, a 200-mg/kg dose was used. After 4 hours, mice were provided with water ad libitum, and 100 μL blood was drawn from the tail. Serum was prepared by incubating at room temperature for 15 minutes and then centrifuging at 4°C, 1500 × g for 10 minutes. Serum samples (diluted in DPBS) from each mouse were loaded as technical replicates on black plates along with known concentrations of FITC-dextran standards, and fluorescence intensity was measured (excitation, 485 nm; emission, 535 nm) using a Multidetection Microplate Reader (TriStar2 LB 942; Berthold Technologies).
Tissue Fixation and Staining
Colonic tissues were harvested from mice after being killed. Fecal contents were removed by repeated flushing with prechilled phosphate-buffered saline. A distal region (∼1 cm) was cut and placed in an Eppendorf tube to make cross-sectional histology samples. Otherwise, colons were opened longitudinally along their mesenteric line and made into Swiss rolls. In both cases, tissues then were fixed in 10% neutral buffered formalin overnight at room temperature for H&E staining. Subsequent processing and H&E staining were conducted by KPNT (Cheongju, Korea), an inspection institution. Bright-field images of stained slides were obtained by Axio Scan Z1 Slide Scanner (Carl Zeiss, Oberkochen, Germany). For fluorescent immunostaining, colon Swiss rolls were fixed in 4% paraformaldehyde overnight and frozen for cryosection using Tissue-Tek O.C.T. Compound (Sakura, Torrance, CA). Heat-induced antigen retrieval was performed in sodium citrate buffer (pH 6) by autoclaving slides for 15 minutes. Primary and secondary antibodies were purchased and used as follows: DCLK1 (ab31704; Abcam, Cambridge, UK), POU2F3 (sc-293402; Santa Cruz, Dallas, TX), goat anti-mouse IgG2a, Alexa Fluor 488 (A-21131; Invitrogen, Waltham, MA), and goat anti-rabbit IgG, Alexa Fluor 594 (A-11012; Invitrogen, Waltham, MA). Nuclei were stained with 4′,6-diamidino-2-phenylindole and slides were mounted using ProLong Gold Antifade Mountant (Invitrogen, Waltham, MA). Fluorescent images were acquired on a LSM980 confocal microscope (Carl Zeiss, Oberkochen, Germany) located in KARA (KAIST Analysis Center for Research Advancement), and further processed using Zen Blue 3.4 (Carl Zeiss, Oberkochen, Germany).
Intestinal Epithelial Cell Isolation
Colonic epithelia were isolated according to the protocol of Shao et al48 with some modifications. Briefly, excised and feces-cleared mouse colons were opened along the mesenteric line, cut again into longitudinal halves, and cut into 2- to 3-mm–sized pieces. Tissue fragments were agitated gently in ice-cold Hank’s balanced salt solution (HBSS) supplemented with 2% glucose and 1 mmol/L dithiothreitol at 4°C for 10 minutes to eliminate mucus. Tissues then were moved into DPBS (Ca2+ and Mg2+-free) containing 10 mmol/L EDTA and incubated for 15 minutes at 37°C with 200 rpm shaking. After robust vortexing, colonic epithelia were released into the supernatant. Epithelium-containing supernatants were passed through a 100-μm cell strainer, and cells were collected by centrifugation at 4°C (1000 × g, 5 minutes). For flow cytometry, the epithelium releasing step was repeated 3 more times without further incubation at 37°C. Supernatants were combined and passed through a 70-μm cell strainer, collected by centrifugation, and washed with ice-cold HBSS. After centrifugation, cell pellets were resuspended in prewarmed HBSS containing 1 U/mL dispase (07913; Stemcell Technologies, Vancouver, BC, Canada) and 10 μg/mL DNase I (11284932001; Roche, Basel, Switzerland), and incubated for 10 minutes at 37°C with 200 rpm shaking to yield a single-cell suspension. After vortexing again, cells were passed through a 40-μm cell strainer and collected by centrifugation at 4°C (500 × g, 5 minutes). Cell pellets were washed with ice-cold DPBS once and counted.
Tuft Cell Quantification Using Flow Cytometry
Colonic epithelial cells were stained with Fixable Viability Dye eFluor 506 (65-0866-14; Invitrogen, Waltham, MA) in a 1:1000 ratio for dead cell exclusion. Cells were Fc-blocked using purified rat anti-mouse CD16/CD32 (clone 2.4G2, 553142; BD Pharmingen, Waltham, MA) for 10 minutes and washed with FACS buffer (Ca2+ and Mg2+-free DPBS supplemented with 0.5% bovine serum albumin and 0.3% sodium azide). For surface marker staining, phycoerythrin (PE) rat anti-mouse Siglec F (clone E50-2440, 552126; BD Pharmingen, Waltham, MA) antibody and allophycocyanin (APC) anti-mouse CD326 (EpCAM) antibody (clone G8.8, 118214; BioLegend, San Diego, CA) were used in a 1:50 and 1:100 dilution ratio, respectively. Antibody staining was performed for 30 minutes. All procedures were conducted at 4°C in the dark. Tuft cells were acquired by a BD LSRFortessa (BD Biosciences, San Diego, CA) and analyzed using FlowJo v10.7.2 (Tree Star, Ashland, OR). The percentage of tuft cells was calculated by dividing the number of EpCAM and Siglec F double-positive cells by the number of EpCAM-positive cells.
Immunoblotting
After flushing, colons were cut into appropriate sizes and mechanically homogenized in RIPA buffer (50 mmol/L Tris-HCl, pH 7.4, 150 mmol/L NaCl, 1 mmol/L EDTA, 1% NP-40, 0.25% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 2 mmol/L phenylmethylsulfonyl fluoride, 10 mmol/L sodium fluoride, 2 mmol/L sodium orthovanadate, 1 mmol/L sodium pyrophosphate, and 1× protease inhibitor cocktail) using TissueRuptor (Qiagen, Hilden, Germany). Isolated epithelial cells also were lysed in RIPA buffer. After incubation for 30 minutes at 4°C, lysates were clarified by centrifugation at 13,000 rpm for 20 minutes, and protein concentration was determined using the Pierce BCA Protein Assay Kit (23225; Thermo Scientific, Waltham, MA) according to the manufacturer’s protocol. For immunoblotting, antibodies against the following proteins were purchased from the indicated sources: DCLK1 (ab31704; Abcam); HSP90 (sc-13119; Santa Cruz Biotechnology); α-tubulin (T5168; Sigma-Aldrich); phospho-S6 (5364), S6 (2217), phospho-AKT T308 (4056), phospho-AKT S473 (9271), AKT (9272), phospho–nuclear factor-κB (3033), nuclear factor-κB (8242), phospho-c-Jun N-terminal kinase (JNK) (4668), and β-actin (4970) (Cell Signaling Technology, Danvers, MA). Anti-mouse IPMK antibody was a custom rabbit polyclonal antibody from Covance(Princeton, NJ) raised against a synthetic peptide starting with Cys followed by mouse IPMK amino acids 295 to 311 (SKAYSRHRKLYAKKHQS),28 Densitometry analysis of blots was performed using ImageJ 1.53e (National Institutes of Health, Bethesda, MD).
RNA Extraction and qRT-PCR
Total RNA was extracted from mouse tissues and epithelial cells using TRI-Reagent (TR-118; Molecular Research Center, Cincinnati, OH) or the RNeasy Lipid Tissue mini kit (74804; Qiagen) according to the manufacturer’s protocol. A maximum of 3 μg total RNA was reverse-transcribed to yield first-strand complementary DNA (cDNA) with SuperiorScript III Reverse Transcriptase (RT006L; Enzynomics, Daejeon, Korea). qPCR analyses were performed on a StepOnePlus Real-Time PCR System (Applied Biosystems, Waltham, MA) using SYBR Green (QPK-201; Toyobo, Osaka, Japan) and 20 or 25 ng cDNA. The relative expression level of each target gene was normalized to the L32 gene using the ΔΔ cycle threshold (Ct) method. Primer sequences for qRT-PCR are provided in Table 1 (all Mus musculus).
Table 1.
qRT Primer Sequences
| Forward | Reverse | |
|---|---|---|
| Ipmk | TGAAGATTGGGCGGAAGAGC | GCCATTGTGGAAAAACTTGG |
| Dclk1 | GGGTGAGAACCATCTACACCATC | CCAGCTTCTTAAAGGGCTCGAT |
| Pou2f3 | AGTGGGGATGTCGCTGATTC | TCCTCCGTCTTAATCTGTCTGTT |
| Trpm5 | CCTCCGTGCTTTTTGAACTCC | CATAGCCAAAGGTCGTTCCTC |
| Gnat3 | GAGAGCAAGGAATCAGCCAG | GTGCTTTTCCCAGATTCACC |
| Il25 | CGGAGGAGTGGCTGAAGTGGAG | ATGGGTACCTTCCTCGCCATG |
| Chat | GAGCGAATCGTTGGTATGACAA | AGGACGATGCCATCAAAAGG |
| Il1b | GCCTCGTGCTGTCGGACC | TGTCGTTGCTTGGTTCTCCTTG |
| Il6 | ATGAACAACGATGATGCACTT | TATCCAGTTTGGTAGCATCCAT |
| Il10 | CATGGGTCTTGGGAAGAGAA | AACTGGCCACAGTTTTCAGG |
| Il17a | CTCCAGAAGGCCCTCAGACTAC | GGGTCTTCATTGCGGTGG |
| Il17f | CCCATGGGATTACAACATCACTC | CACTGGGCCTCAGCGATC |
| Tnfa | ACGGCATGGATCTCAAAGAC | AGATAGCAAATCGGCTGACG |
| Anpep | CTCCTACCGGGTGATCCTGAG | GTTGCAGGTAAAGCGAACAGT |
| Chga | ACAACAGGATGGCTTTGAGG | GGTTGGTGATTGGGTATTGG |
| Reg4 | CTGGAATCCCAGGACAAAGAGTG | CTGGAGGCCTCCTCAATGTTTGC |
| Muc2 | GATGGCACCTACCTCGTTGT | GTCCTGGCACTTGTTGGAAT |
| L32 | GAAACTGGCGGAAACCCA | GGATCTGGCCCTTGAACCTT |
mRNA Sequencing and Data Processing
Library preparation, sequencing, preprocessing, genome mapping, and gene expression quantification were conducted by LAS Inc. (http://las.kr, Gimpo, Korea), as described later. Total RNA was isolated from tissue using the TRI-reagent–based method. Total RNA (500 ng) was processed for preparing a whole-transcriptome sequencing library. Enrichment of whole-transcriptome RNA by depleting ribosomal RNA was conducted to prepare the whole-transcriptome sequencing library using the MGIEasy RNA Directional Library Prep Kit (MGI, Shenzhen, China) according to the manufacturer’s instructions. After the ribosomal RNA was depleted, the remaining RNA was fragmented into small pieces using divalent cations under increased temperature. The cleaved RNA fragments were copied into first-strand cDNA using reverse transcriptase and random primers. Strand specificity was achieved in the RT directional buffer, followed by second-strand cDNA synthesis. These cDNA fragments have the addition of a single A base and subsequent ligation of the adapter. The products then were purified and enriched with PCR to create the final cDNA library. The double-stranded library was quantified using the QuantiFluor ONE dsDNA System (Promega, Madison, WI) and was equal to 330 ng in a total volume of 60 mL or less. The library was cyclized at 37°C for 60 minutes, and then digested at 37°C for 30 minutes, followed by cleanup of the circularization product. To make the DNA nanoball, the library was incubated at 30°C for 25 minutes using the DNA nanoball enzyme. Finally, the library was quantified by the QuantiFluor ssDNA System (Promega). Sequencing of the prepared DNA nanoball was conducted using the MGIseq system (MGI) with 100-bp paired-end reads.
Potentially existing sequencing adapters and raw quality bases in the raw reads were trimmed by Skewer version 0.2.2.49 The cleaned high-quality reads were mapped to the reference genome by STAR version 2.5 software.50 Because the sequencing libraries were prepared strand-specifically by using Illumina (San Diego, CA)’s strand-specific library preparation kit, the strand-specific library option, --library-type=fr-firststrand was applied in the mapping process.
To quantify the mapped reads, RSEM version 1.3.051 and default options were used. The gene annotation of the reference genome mm10 from GENCODE genome (https://www.gencodegenes.org) in GTF format was used as gene models. The DEGs between the 2 selected biological conditions were analyzed by DESeq2 version 1.26.0 software.52 To compare the expression profiles among the samples, the normalized expression values of the selected few hundred of the DEGs were clustered (unsupervised) by in-house R scripts. The scatter plots for the gene expression values and the volcano plots for the expression fold changes and P values between the 2 selected samples were drawn by in-house R scripts.
Analysis and Visualization of mRNA Sequencing Data
Gene ontology enrichment analysis (http://geneontology.org) was performed by g:Profiler version 0.2.0 and the top 5 gene ontology biological process terms were visualized by −log10–transformed adjusted P value. Gene set enrichment analysis (https://www.gsea-msigdb.org/gsea/index.jsp) was performed by gene set enrichment analysis version 4.1.0 with a normalized count and visualized with an enrichment score and calculated false discovery rate (FDR). Volcano plots were visualized by log2 transformed fold change and −log10–transformed P value. Tuft cell marker genes and keratinization-related genes were represented by specific plots. Heatmaps representing each term were visualized with row z-score calculated by log2-transformed normalized count.
Single-Cell cDNA Synthesis and Sequencing Library Generation
scRNA-seq libraries were prepared using the Smart-seq2 protocol53 with few modifications. Single-cells were sorted using a BD FACSAriaII (BD Biosciences) into 96-well PCR plates (Thermo Scientific) containing 2 μL lysis buffer (0.1% Triton X-100 [Sigma-Aldrich, Saint Louis, MO], 1 U/μL RNase Inhibitor [Enzynomics], and 0.25 μmol/L oligo-dT30VN primer) and stored at -80°C. The plates were thawed on ice and incubated for 3 minutes at 72°C to lyse the cells. Reverse-transcription (40 U/μL Maxima H minus transcriptase), template switching reaction, and PCR pre-amplification (15 cycles) were performed according to the protocol. The PCR products in each well were cleaned up using 0.5× SPRI beads (2% Sera-Mag Speed Beads [Cytiva, Marlborough, MA], 1 mol/L NaCl, 10 mmol/L Tris-HCl, pH 8.0, 1 mmol/L EDTA, 0.01% NP40, 0.05% sodium azide, and 22% w/v polyethylene glycol [PEG] 8000). To exclude empty wells and low-quality libraries, cDNA libraries were assessed by qPCR with a primer pair of glyceraldehyde-3-phosphate dehydrogenase, a housekeeping gene (forward: 5′-GTCGTGGAGTCTACTGGTGTCTTCAC-3′; reverse: 5′-GTTGTCATATTTCTCGTGGTTCACACCC-3′). Samples showing a high threshold cycle value (>35 cycles) or no PCR amplification were discarded. Fifty to 200 pg of each cDNA library were used to generate the Illumina sequencing library using EZ-Tera XT DNA library preparation kits (Enzynomics). After the final PCR amplification, samples were pooled and size-selected by SPRI beads (removing DNA bound to 0.3× beads and taking DNA bound to subsequent 0.6× beads). Pooled sequencing libraries were sequenced on an Illumina NextSeq 500 instrument using a 38-bp paired-end reads setting.
scRNA-Seq Data Analysis
Read alignment and gene quantification
Sequencing reads from scRNA-seq libraries were aligned to the mouse reference genome (version mm10 from the University of California Santa Cruz) using Bowtie 2 (version 2.4.2)54 with default parameters for paired-end reads. Expression values of each gene were quantified as transcripts per million (TPM) using RSEM (version 1.3.3).51 TPMs were transformed to log2 (TPM + 1). For downstream analyses, cells with <500 detected genes and genes expressed in <5 cells were removed.
Identification of highly variable genes, cell clustering, and t-distributed stochastic neighbor embedding (t-SNE) visualization
All analyses and visualization of data were conducted in a Python environment built on the NumPy, SciPy, Matplotlib, scikit-learn package, and Pandas libraries.55 To identify the tuft cell cluster, principal component analysis was performed on the list of highly variable genes, and the first 8 principal components were used to perform t-SNE.56 By unsupervised hierarchical clustering using Euclidean distance and Ward’s method, the tuft cell cluster was identified with higher expression of Dclk1, Siglecf, Il17rb, and Ptgs1, previously known tuft cell marker genes. To identify the subpopulations among tuft cells, principal component analysis was performed on the list of highly variable genes among tuft cells, and the first 4 principal components were used to perform t-SNE. By unsupervised hierarchical clustering using Euclidean distance and Ward’s method again, 3 different clusters were identified. Expression levels (log-transformed) of individual genes were represented by colors, as indicated in the t-SNE plots.
Differential expression test and cluster identity annotation
To identify differentially expressed genes between tuft cell population C1 and the others, normalized raw counts were applied to the DESeq2 package52 in R software, then genes with a fold-change of >2 and an FDR <0.01 were considered to be differentially expressed. Differentially expressed genes comparing each tuft cell population of C1, C2 and C3 were figured out using the FindAllMarkers function in the Seurat package57 in R software with the DESeq2 option and default parameters. The top 50 highest-averaged log2(fold-change) differentially expressed genes for C2, C3, and C4 differentially expressed genes for C1 are represented by the heatmap.
scRNA-seq data analysis from previous study
The data from Herring et al,57 2018 were downloaded from Gene Expression Omnibus as GSM2643164 and GSM2743165. Using Seurat (version 4.0.3) package in R, 2 replicates were merged, normalized, and scaled with default parameters. Principal components were analyzed with 2000 variable genes. The first 10 principal components and 0.3 resolution value for the FindClusters function in Seurat were used for unsupervised clustering.
Statistical analysis
All statistical analyses were performed using Prism 7 (GraphPad Software, San Diego, CA) and all data were presented as ±SD or SEM. Differences between groups were considered to be significant when P < .05 (Student unpaired t test, 2-tailed).
Acknowledgments
The authors thank the members of the Stem Cell and Tissue Regeneration, Dynamics of Cellular Energetics, and Metabolism Signaling Network laboratories for their assistance and advice.
Professional English proofreading was provided by https://wordvice.com on our original manuscript.
Present address of S.J.P.: ERSTEQ Co, Ltd, Daejeon, Republic of Korea.
CRediT Authorship Contributions
Seyun Kim, PhD (Conceptualization: Equal; Data curation: Equal; Formal analysis: Equal; Funding acquisition: Equal; Investigation: Equal; Methodology: Equal; Project administration: Lead; Supervision: Equal; Writing – original draft: Equal; Writing – review & editing: Equal)
Seung Eun Park (Conceptualization: Equal; Data curation: Equal; Formal analysis: Equal; Investigation: Equal; Methodology: Equal; Visualization: Equal; Writing – original draft: Equal; Writing – review & editing: Equal)
Dongeun Lee (Data curation: Equal; Formal analysis: Equal; Investigation: Equal; Methodology: Equal; Writing – review & editing: Supporting)
Jae Woong Jeong (Formal analysis: Equal; Visualization: Equal; Writing – review & editing: Supporting)
Su-Hyung Lee (Data curation: Equal; Formal analysis: Equal; Writing – review & editing: Supporting)
Seung Ju Park, PhD (Data curation: Supporting; Formal analysis: Supporting)
Jaeseung Ryu (Data curation: Supporting)
Se Kyu Oh (Methodology: Supporting)
Hanseul Yang, PhD (Data curation: Equal; Formal analysis: Equal; Funding acquisition: Equal; Investigation: Equal; Methodology: Equal; Project administration: Equal; Supervision: Equal; Writing – original draft: Supporting; Writing – review & editing: Equal)
Sungsoon Fang, PhD (Data curation: Equal; Formal analysis: Equal; Funding acquisition: Equal; Investigation: Equal; Methodology: Equal; Project administration: Equal; Supervision: Equal; Writing – review & editing: Supporting)
Data Availability Statement
The RNA sequencing data were deposited at the National Center for Biotechnology Information Sequence Read Archive. Accession number: PRJNA822083 (colon tissue RNA-seq, colitis), PRJNA823796 (colonic epithelial cell RNA-seq, colitis). Colonic tuft cell scRNA-seq data were deposited at the Gene Expression Omnibus, accession number: GSE200284.
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding Supported by a grant from the TJ Park Science Fellowship of the POSCO TJ Park Foundation (S.E.P.); National Research Foundation of Korea grants NRF-2021R1C1C100791612 and 2019R1A6A1A10073887 (H.Y.), 2021R1A2C2009749 (S.F.), 2020R1A2C3005765 (S.K.), and 2018R1A5A102426121 (H.Y. and S.K); and Korea Advanced Institute of Science and Technology KC30 grant N11210197 (S.K.). Professional English proofreading was paid by 2020R1A2C3005765 grant (S.K.).
Contributor Information
Hanseul Yang, Email: hyang@kaist.ac.kr.
Sungsoon Fang, Email: sfang@yuhs.ac.
Seyun Kim, Email: seyunkim@kaist.ac.kr.
Supplementary Material
References
- 1.Beumer J., Clevers H. Cell fate specification and differentiation in the adult mammalian intestine. Nat Rev Mol Cell Biol. 2021;22:39–53. doi: 10.1038/s41580-020-0278-0. [DOI] [PubMed] [Google Scholar]
- 2.Allaire J.M., Crowley S.M., Law H.T., Chang S.Y., Ko H.J., Vallance B.A. The intestinal epithelium: central coordinator of mucosal immunity. Trends Immunol. 2018;39:677–696. doi: 10.1016/j.it.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 3.Grencis R.K., Worthington J.J. Tuft cells: a new flavor in innate epithelial immunity. Trends Parasitol. 2016;32:583–585. doi: 10.1016/j.pt.2016.04.016. [DOI] [PubMed] [Google Scholar]
- 4.Gerbe F., Jay P. Intestinal tuft cells: epithelial sentinels linking luminal cues to the immune system. Mucosal Immunol. 2016;9:1353–1359. doi: 10.1038/mi.2016.68. [DOI] [PubMed] [Google Scholar]
- 5.Banerjee A., McKinley E.T., Von Moltke J., Coffey R.J., Lau K.S. Interpreting heterogeneity in intestinal tuft cell structure and function. J Clin Invest. 2018;128:1711–1719. doi: 10.1172/JCI120330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qu D., Weygant N., May R., Chandrakesan P., Madhoun M., Ali N., Sureban S.M., An G., Schlosser M.J., Houchen C.W. Ablation of doublecortin-like kinase 1 in the colonic epithelium exacerbates dextran sulfate sodium-induced colitis. PLoS One. 2015;10:1–14. doi: 10.1371/journal.pone.0134212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.May R., Qu D., Weygant N., Chandrakesan P., Ali N., Lightfoot S.A., Li L., Sureban S.M., Houchen C.W. Brief report: Dclk1 deletion in tuft cells results in impaired epithelial repair after radiation injury. Stem Cells. 2014;32:822–827. doi: 10.1002/stem.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baumgart D.C., Carding S.R. Inflammatory bowel disease: cause and immunobiology. Lancet. 2007;369:1627–1640. doi: 10.1016/S0140-6736(07)60750-8. [DOI] [PubMed] [Google Scholar]
- 9.Graham D.B., Xavier R.J. Pathway paradigms revealed from the genetics of inflammatory bowel disease. Nature. 2020;578:527–539. doi: 10.1038/s41586-020-2025-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ananthakrishnan A.N., Bernstein C.N., Iliopoulos D., Macpherson A., Neurath M.F., Ali R.A.R., Vavricka S.R., Fiocchi C. Environmental triggers in IBD: a review of progress and evidence. Nat Rev Gastroenterol Hepatol. 2018;15:39–49. doi: 10.1038/nrgastro.2017.136. [DOI] [PubMed] [Google Scholar]
- 11.Aardoom M.A., Joosse M.E., De Vries A.C.H., Levine A., De Ridder L. Malignancy and mortality in pediatric-onset inflammatory bowel disease: a systematic review. Inflamm Bowel Dis. 2018;24:732–741. doi: 10.1093/ibd/izx104. [DOI] [PubMed] [Google Scholar]
- 12.Nadeem M.S., Kumar V., Al-Abbasi F.A., Kamal M.A., Anwar F. Risk of colorectal cancer in inflammatory bowel diseases. Semin Cancer Biol. 2020;64:51–60. doi: 10.1016/j.semcancer.2019.05.001. [DOI] [PubMed] [Google Scholar]
- 13.Saiardi A., Nagata E., Luo H.R., Sawa A., Luo X., Snowman A.M., Snyder S.H. Mammalian inositol polyphosphate multikinase synthesizes inositol 1,4,5-trisphosphate and an inositol pyrophosphate. Proc Natl Acad Sci U S A. 2001;98:2306–2311. doi: 10.1073/pnas.041614598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Resnick A.C., Snowman A.M., Kang B.M., Hurt K.J., Snyder S.H., Saiardi A. Inositol polyphosphate multikinase is a nuclear PI3-kinase with transcriptional regulatory activity. Proc Natl Acad Sci U S A. 2005;102:12783–12788. doi: 10.1073/pnas.0506184102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee B., Park S.J., Hong S., Kim K., Kim S. Inositol polyphosphate multikinase signaling: multifaceted functions in health and disease. Mol Cells. 2021;44:187–194. doi: 10.14348/molcells.2021.0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim E., Ahn H., Kim M.G., Lee H., Kim S. The expanding significance of inositol polyphosphate multikinase as a signaling hub. Mol Cells. 2017;40:315–321. doi: 10.14348/molcells.2017.0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yokoyama J.S., Wang Y., Schork A.J., Thompson W.K., Karch C.M., Cruchaga C., McEvoy L.K., Witoelar A., Chen C.H., Holland D., Brewer J.B., Franke A., Dillon W.P., Wilson D.M., Mukherjee P., Hess C.P., Miller Z., Bonham L.W., Shen J., Rabinovici G.D., Rosen H.J., Miller B.L., Hyman B.T., Schellenberg G.D., Karlsen T.H., Andreassen O.A., Dale A.M., Desikan R.S. Association between genetic traits for immune-mediated diseases and Alzheimer disease. JAMA Neurol. 2016;73:691–697. doi: 10.1001/jamaneurol.2016.0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Donnell S., Borowski K., Espin-Garcia O., Milgrom R., Kabakchiev B., Stempak J., Panikkath D., Eksteen B., Xu W., Steinhart A.H., Kaplan G.G., McGovern D.P.B., Silverberg M.S. The unsolved link of genetic markers and Crohn’s disease progression: a North American cohort experience. Inflamm Bowel Dis. 2019;25:1541–1549. doi: 10.1093/ibd/izz016. [DOI] [PubMed] [Google Scholar]
- 19.Sei Y., Zhao X., Forbes J., Szymczak S., Li Q., Trivedi A., Voellinger M., Joy G., Feng J., Whatley M., Jones M.P.S., Harper U.L., Marx S.J., Venkatesan A.M., Chandrasekharappa S.C., Raffeld M., Quezado M.M., Louie A., Chen C.C., Lim R.M., Agarwala R., Schäffer A.A., Hughes M.S., Bailey-Wilson J.E., Wank S.A. A hereditary form of small intestinal carcinoid associated with a germline mutation in inositol polyphosphate multikinase. Gastroenterology. 2015;149:67–78. doi: 10.1053/j.gastro.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okabe A., Huang K.K., Matsusaka K., Fukuyo M., Xing M., Ong X., Hoshii T., Usui G., Seki M., Mano Y., Rahmutulla B., Kanda T., Suzuki T., Rha S.Y., Ushiku T., Fukayama M., Tan P., Kaneda A. Cross-species chromatin interactions drive transcriptional rewiring in Epstein–Barr virus–positive gastric adenocarcinoma. Nat Genet. 2020;52:919–930. doi: 10.1038/s41588-020-0665-7. [DOI] [PubMed] [Google Scholar]
- 21.Yin S., Ray G., Kerschner J.L., Hao S., Perez A., Drumm M.L., Browne J.A., Leir S.H., Longworth M., Harris A. Functional genomics analysis of human colon organoids identifies key transcription factors. Physiol Genomics. 2020;52:234–244. doi: 10.1152/physiolgenomics.00113.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Indukuri R., Jafferali M.H., Song D., Damdimopoulos A., Hases L., Zhao C., Archer A., Williams C. Genome-wide estrogen receptor β chromatin binding in human colon cancer cells reveals its tumor suppressor activity. Int J Cancer. 2021;149:692–706. doi: 10.1002/ijc.33573. [DOI] [PubMed] [Google Scholar]
- 23.Sims R.J., Millhouse S., Chen C.F., Lewis B.A., Erdjument-Bromage H., Tempst P., Manley J.L., Reinberg D. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell. 2007;28:665–676. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A., Olsson I.M., Edlund K., Lundberg E., Navani S., Szigyarto C.A.K., Odeberg J., Djureinovic D., Takanen J.O., Hober S., Alm T., Edqvist P.H., Berling H., Tegel H., Mulder J., Rockberg J., Nilsson P., Schwenk J.M., Hamsten M., Von Feilitzen K., Forsberg M., Persson L., Johansson F., Zwahlen M., Von Heijne G., Nielsen J., Pontén F. Tissue-based map of the human proteome. Science. 2015;347 doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 25.Herring C.A., Banerjee A., McKinley E.T., Simmons A.J., Ping J., Roland J.T., Franklin J.L., Liu Q., Gerdes M.J., Coffey R.J., Lau K.S. Unsupervised trajectory analysis of single-cell RNA-Seq and imaging data reveals alternative tuft cell origins in the gut. Cell Syst. 2018;6:37–51.e9. doi: 10.1016/j.cels.2017.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andrews N., Serviss J.T., Geyer N., Andersson A.B., Dzwonkowska E., Šutevski I., Heijboer R., Baryawno N., Gerling M., Enge M. An unsupervised method for physical cell interaction profiling of complex tissues. Nat Methods. 2021;18:912–920. doi: 10.1038/s41592-021-01196-2. [DOI] [PubMed] [Google Scholar]
- 27.Haber A.L., Biton M., Rogel N., Herbst R.H., Shekhar K., Smillie C., Burgin G., Delorey T.M., Howitt M.R., Katz Y., Tirosh I., Beyaz S., Dionne D., Zhang M., Raychowdhury R., Garrett W.S., Rozenblatt-Rosen O., Shi H.N., Yilmaz O., Xavier R.J., Regev A. A single-cell survey of the small intestinal epithelium. Nature. 2017;551:333–339. doi: 10.1038/nature24489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maag D., Maxwell M.J., Hardesty D.A., Boucher K.L., Choudhari N., Hanno A.G., Ma J.F., Snowman A.S., Pietropaoli J.W., Xu R., Storm P.B., Saiardi A., Snyder S.H., Resnick A.C. Inositol polyphosphate multikinase is a physiologic PI3-kinase that activates Akt/PKB. Proc Natl Acad Sci U S A. 2011;108:1391–1396. doi: 10.1073/pnas.1017831108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagahama Y., Shimoda M., Mao G., Singh S.K., Kozakai Y., Sun X., Motooka D., Nakamura S., Tanaka H., Satoh T., Maeda K., Akira S. Regnase-1 controls colon epithelial regeneration via regulation of mTOR and purine metabolism. Proc Natl Acad Sci U S A. 2018;115:11036–11041. doi: 10.1073/pnas.1809575115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tesfaigzi J., Carlson D.M. Expression, regulation, and function of the SPR family of proteins: a review. Cell Biochem Biophys. 1999;30:243–265. doi: 10.1007/BF02738069. [DOI] [PubMed] [Google Scholar]
- 31.Fang K., Zhang S., Glawe J., Grisham M.B., Kevil C.G. Temporal genome expression profile analysis during t-cell-mediated colitis: Identification of novel targets and pathways. Inflamm Bowel Dis. 2012;18:1411–1423. doi: 10.1002/ibd.22842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma G.W., Chu Y.K., Yang H., Yan X.H., Rong E.G., Li H., Wang N. Functional analysis of sheep POU2F3 isoforms. Biochem Genet. 2020;58:335–347. doi: 10.1007/s10528-019-09945-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamashita J., Ohmoto M., Yamaguchi T., Matsumoto I., Hirota J. Skn-1a/Pou2f3 functions as a master regulator to generate Trpm5-expressing chemosensory cells in mice. PLoS One. 2017;12:1–14. doi: 10.1371/journal.pone.0189340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Howitt M.R., Lavoie S., Michaud M., Blum A.M., Tran S.V., Weinstock J.V., Gallini C.A., Redding K., Margolskee R.F., Osborne L.C., Artis D., Garrett W.S. Tuft cells, taste-chemosensory cells, orchestrate parasite type 2 immunity in the gut. Science. 2016;351:1329–1333. doi: 10.1126/science.aaf1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gerbe F., Sidot E., Smyth D.J., Ohmoto M., Matsumoto I., Dardalhon V., Cesses P., Garnier L., Pouzolles M., Brulin B., Bruschi M., Harcus Y., Zimmermann V.S., Taylor N., Maizels R.M., Jay P. Intestinal epithelial tuft cells initiate type 2 mucosal immunity to helminth parasites. Nature. 2016;529:226–230. doi: 10.1038/nature16527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Middelhoff M., Westphalen C.B., Hayakawa Y., Yan K.S., Gershon M.D., Wang T.C., Quante M. Dclk1-expressing tuft cells: critical modulators of the intestinal niche? Am J Physiol Gastrointest Liver Physiol. 2017;313:G285–G299. doi: 10.1152/ajpgi.00073.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saiardi A., Erdjument-Bromage H., Snowman A.M., Tempst P., Snyder S.H. Synthesis of diphosphoinositol pentakisphosphate by a newly identified family of higher inositol polyphosphate kinases. Curr Biol. 1999;9:1323–1326. doi: 10.1016/s0960-9822(00)80055-x. [DOI] [PubMed] [Google Scholar]
- 38.Anutosh Chakraborty, Kim S., Snyder S.H. Inositol pyrophosphates as mammalian cell signals. Sci Signal. 2011;4:re1. doi: 10.1126/scisignal.2001958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim E., Beon J., Lee S., Park S.J., Ahn H., Kim M.G., Park J.E., Kim W., Yuk J.-M., Kang S.-J., Lee S.-H., Jo E.-K., Seong R.H., Kim S. Inositol polyphosphate multikinase promotes Toll-like receptor–induced inflammation by stabilizing TRAF6. Sci Adv. 2017;3 doi: 10.1126/sciadv.1602296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nadjsombati M.S., McGinty J.W., Lyons-Cohen M.R., Jaffe J.B., DiPeso L., Schneider C., Miller C.N., Pollack J.L., Nagana Gowda G.A., Fontana M.F., Erle D.J., Anderson M.S., Locksley R.M., Raftery D., von Moltke J. Detection of succinate by intestinal tuft cells triggers a type 2 innate immune circuit. Immunity. 2018;49:33–41.e7. doi: 10.1016/j.immuni.2018.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lei W., Ren W., Ohmoto M., Urban J.F., Matsumoto I., Margolskee R.F., Jiang P. Activation of intestinal tuft cell-expressed sucnr1 triggers type 2 immunity in the mouse small intestine. Proc Natl Acad Sci U S A. 2018;115:5552–5557. doi: 10.1073/pnas.1720758115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Westphalen C.B., Asfaha S., Hayakawa Y., Takemoto Y., Lukin D., Nuber A., Brandtner A., Setlik W., Remotti H., Muley A., Chen X., May R., Houchen C., Fox J., Gershon M., Quante M., Wang T. Long-lived intestinal tuft cells serve as colon cancer–initiating cells. J Clin Invest. 2014;124:1283–1295. doi: 10.1172/JCI73434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hayakawa Y., Sakitani K., Konishi M., Asfaha S., Niikura R., Tomita H., Renz B.W., Tailor Y., Macchini M., Middelhoff M., Jiang Z., Tanaka T., Dubeykovskaya Z.A., Kim W., Chen X., Urbanska A.M., Nagar K., Westphalen C.B., Quante M., Lin C.S., Gershon M.D., Hara A., Zhao C.M., Chen D., Worthley D.L., Koike K., Wang T.C. Nerve growth factor promotes gastric tumorigenesis through aberrant cholinergic signaling. Cancer Cell. 2017;31:21–34. doi: 10.1016/j.ccell.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fernandez Vallone V., Leprovots M., Ribatallada-Soriano D., Gerbier R., Lefort A., Libert F., Vassart G., Garcia M. LGR5 controls extracellular matrix production by stem cells in the developing intestine. EMBO. 2020;21 doi: 10.15252/embr.201949224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gerbe F., Van Es J.H., Makrini L., Brulin B., Mellitzer G., Robine S., Romagnolo B., Shroyer N.F., Bourgaux J.F., Pignodel C., Clevers H., Jay P. Distinct ATOH1 and Neurog3 requirements define tuft cells as a new secretory cell type in the intestinal epithelium. J Cell Biol. 2011;192:767–780. doi: 10.1083/jcb.201010127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Banerjee A., Herring C.A., Chen B., Kim H., Simmons A.J., Southard-Smith A.N., Allaman M.M., White J.R., Macedonia M.C., Mckinley E.T., Ramirez-Solano M.A., Scoville E.A., Liu Q., Wilson K.T., Coffey R.J., Washington M.K., Goettel J.A., Lau K.S. Succinate produced by intestinal microbes promotes specification of tuft cells to suppress ileal inflammation. Gastroenterology. 2020;159:2101–2115.e5. doi: 10.1053/j.gastro.2020.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kjærgaard S., Jensen T.S.R., Feddersen U.R., Bindslev N., Grunddal K.V., Poulsen S.S., Rasmussen H.B., Budtz-Jørgensen E., Berner-Hansen M. Decreased number of colonic tuft cells in quiescent ulcerative colitis patients. Eur J Gastroenterol Hepatol. 2021;25:817–824. doi: 10.1097/MEG.0000000000001959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shao L., Oshima S., Duong B., Advincula R., Barrera J., Malynn B.A., Ma A. A20 restricts Wnt signaling in intestinal epithelial cells and suppresses colon carcinogenesis. PLoS One. 2013;8:1–7. doi: 10.1371/journal.pone.0062223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang H., Lei R., Ding S.W., Zhu S. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics. 2014;15:1–12. doi: 10.1186/1471-2105-15-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:1–21. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Picelli S., Björklund Å.K., Faridani O.R., Sagasser S., Winberg G., Sandberg R. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods. 2013;10:1096–1100. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- 54.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pedregosa F., Grisel O., Weiss R., Passos A., Brucher M., Varoquax G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V., Brucher M. Scikit-learn: machine learning in Python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- 56.Laurens van der Maaten G.H. Visualizing data using t-SNE Laurens. J Mach Learn Res. 2008;9:2579–2605. [Google Scholar]
- 57.Hao Y., Hao S., Andersen-Nissen E., Mauck W.M., Zheng S., Butler A., Lee M.J., Wilk A.J., Darby C., Zager M., Hoffman P., Stoeckius M., Papalexi E., Mimitou E.P., Jain J., Srivastava A., Stuart T., Fleming L.M., Yeung B., Rogers A.J., McElrath J.M., Blish C.A., Gottardo R., Smibert P., Satija R. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–3587.e29. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA sequencing data were deposited at the National Center for Biotechnology Information Sequence Read Archive. Accession number: PRJNA822083 (colon tissue RNA-seq, colitis), PRJNA823796 (colonic epithelial cell RNA-seq, colitis). Colonic tuft cell scRNA-seq data were deposited at the Gene Expression Omnibus, accession number: GSE200284.