Abstract
Wilson disease (WD) is a metabolic disorder caused by inactivation of the copper-transporting ATPase 2 (ATP7B) and copper (Cu) overload in tissues. Excess Cu causes oxidative stress and pathologic changes with poorly understood mechanistic connections. In Atp7b−/− mice with established liver disease, Cu overload activates the stress-sensitive transcription factor Nrf2 (nuclear factor erythroid-derived 2-like 2). Nrf2 targets, especially sulfotransferase 1e1 (Sult1e1), are strongly induced and cause elevation of sulfated sterols, whereas oxysterols are decreased. This sterol misbalance results in inhibition of the liver X receptor (LXR) and up-regulation of LXR targets associated with inflammatory responses. Pharmacological inhibition of Sult1e1 partially reverses oxysterol misbalance and LXR inhibition. Contribution of this pathway to advanced hepatic WD was demonstrated by treating mice with an LXR agonist. Treatment decreased inflammation by reducing expression of proinflammatory molecules, diminished fibrosis by down-regulating the noncanonical transforming growth factor–β signaling pathway, and improved liver morphology and function. Thus, the identified pathway is an important driver of WD.
Oxysterol misbalance is an important driver of Wilson disease pathology downstream of elevated copper.
INTRODUCTION
Wilson disease (WD) is a potentially fatal disorder of Cu metabolism. WD is caused by inactivating mutations in the adenosine 5′-triphosphate (ATP)–driven Cu(I) transporter ATP7B, which disrupt Cu homeostasis in the liver and other organs. In hepatocytes, loss of ATP7B activity prevents Cu excretion into the bile and the delivery of a Cu cofactor to the ferroxidase, ceruloplasmin (1). Hepatic manifestations of WD range from an asymptomatic elevation of liver enzymes to cirrhosis or acute liver failure. Age and sex have modifying effects on the disease: Cirrhosis is more common in adults than pediatric patients, and acute liver failure is more common in females (2). Because of variable clinical presentations, diagnosis and treatment of WD can be challenging.
Current management of WD includes Cu chelation, zinc salts, and liver transplantation. Cu chelation is the standard-of-care therapy for WD, and it provides benefits to most patients, especially if initiated early (3). Despite proven benefits, all current therapies have limitations, including side effects, poor compliance, high cost, and potential for neurological decompensation (4, 5). Alternative and/or supplemental approaches based on a thorough understanding of WD pathogenesis are needed. Presently, the molecular mechanisms by which elevated Cu triggers hepatic fibroinflammatory disease remain unclear.
WD in humans and in animal models progresses from an asymptomatic stage, when the hepatic copper is high, but the liver morphology and function are not compromised, to advanced WD with liver inflammation, fibrosis, hepatocyte injury, and a significant functional impairment. In patients with WD and Atp7b−/− mice, the hepatic Cu content increases rapidly after birth and then remains fairly constant as the disease progresses. This observation suggests that the pathology development in WD represents a sequence of metabolic and transcriptional changes, which amplify and further modify the initial liver response to excess Cu. How this process works is unclear. Mitochondria dysfunction (6), epigenetic modifications (7), and changes in the nuclear receptor (NR) signaling (8, 9) have been described as important hallmarks of the disease, but individual contributions of these pathways to inflammation and fibrosis in WD have not been defined.
NRs are ligand-dependent transcription factors that regulate numerous cellular pathways. Defects in NR [LXR, FXR (farnesoid X receptor), and peroxisome proliferator–activated receptor α (PPARα)] signaling were identified in patients with WD and Atp7b−/− mice (10–12). The liver NR, LXR, was shown to be inhibited early in WD. At this presymptomatic stage, inhibition of LXR primarily dysregulates lipid metabolism, and activation of LXR corrects these changes and delays the development of liver injury in young Atp7b−/− mice (11). The role of LXR in advanced WD, which is characterized by liver inflammation and fibrosis, is uncertain because in advanced WD, other NRs, such as FXR, also become significantly dysregulated, and many cellular pathways are altered (8–10). The relationship between NR activity and oxidative stress, another well-known feature of advanced WD, is also unclear. To address these questions, we studied older Atp7b−/− mice with established hepatic WD. We found that the cellular response to elevated Cu involves transcriptional changes that affect enzymes involved in the production and modification of sterols. Sterol misbalance causes LXR inhibition, which contributes significantly to inflammation and fibrosis. Furthermore, we show that pharmacologic activation of LXR, even after the onset of clinical symptoms, decreases inflammation and fibrosis and improves liver function. We conclude that sterol misbalance is an important driver of pathology development downstream of elevated Cu.
RESULTS
Cu accumulation causes activation of stress-sensitive NRF2 in Atp7b−/− mice and up-regulation of NRF2 targets
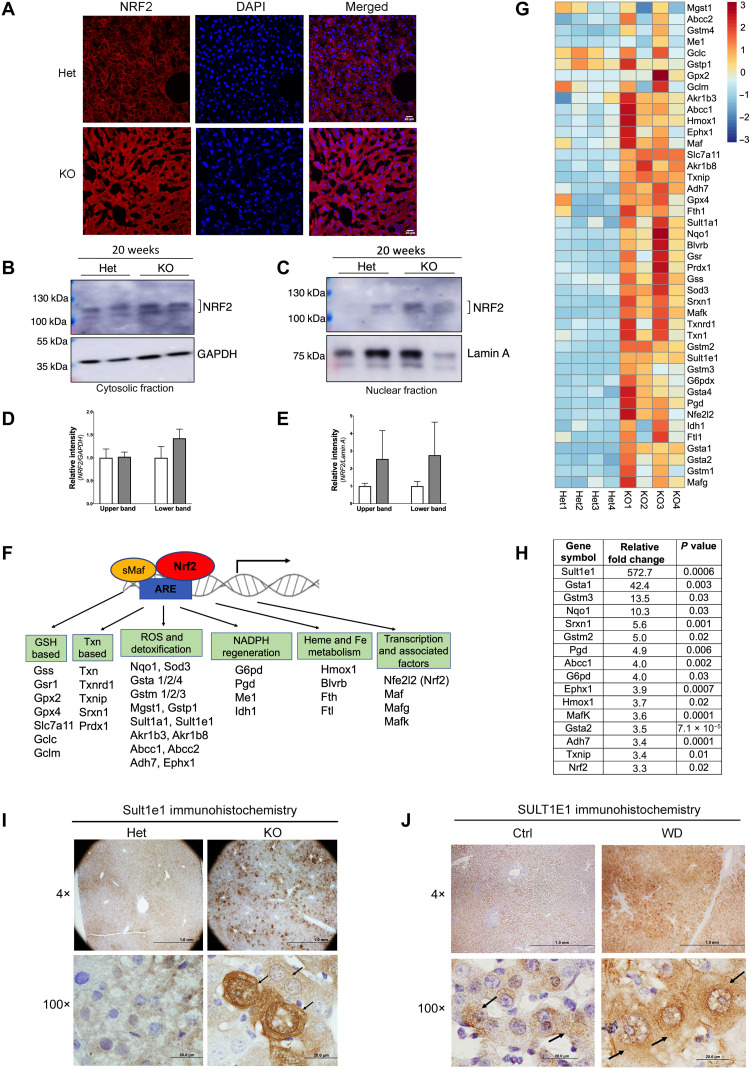
In WD, Cu accumulation in tissues decreases cells’ reductive capacity, causing oxidative stress (13). Various indicators of oxidative stress have been reported in WD [DNA damage, lipid peroxidation, and increased oxidation of glutathione (14, 15)]; however, data on protein/RNA changes that specifically define oxidative stress response in WD are very limited. To better characterize this response in Atp7b−/− mice with established liver disease (20 weeks after birth), we examined the status of the oxidative stress–sensitive transcription factor, nuclear factor erythroid-derived 2-like 2 (NRF2). Immunostaining of the liver tissue sections showed a much stronger NRF2 signal in Atp7b−/− liver compared to the age-matched controls, suggesting increased NRF2 abundance (Fig. 1A). Immunoblot analysis of liver homogenates confirmed increased NRF2 protein levels in Atp7b−/− samples (in both the cytosol and nuclear fractions) (Fig. 1, B to E).
Fig. 1. Stress-sensitive NRF2 and NRF2 targets are up-regulated in Atp7b−/− mice.
(A) Immunostaining of NRF2 (red) in tissue sections revealed stronger signal in Atp7b−/− [knockout (KO)] liver compared to control [heterozygous (Het)]. DAPI (4′,6-diamidino-2-phenylindole; blue) marks nuclei. Scale bars, 20 μm. (B and C) Western blot analysis of cellular fractions shows enrichment of elevated NRF2 protein in nuclei. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (cytosol) and lamin A (nucleus) are loading controls. (D and E) Densitometry of NRF2 signal normalized to GAPDH and lamin A, respectively. (F) Diagram showing Nrf2 along with Mafs binding to antioxidant response element (ARE) and activating expression of their target genes. (G) Heatmap analysis of Nrf2-dependent transcripts was generated using R with row scaling and clustering of transcript abundances (positive value in red denotes up-regulation, whereas negative value in blue denotes down-regulation). (H) Most significantly changed Nrf2 targets; values represent relative fold change, n = 4 male mice per group. (I) Immunohistochemistry demonstrates up-regulation of Sult1e1 protein expression in Atp7b−/− hepatocytes (indicated by arrow in the high-magnification image). Scale bars, 1 mm and 20 μm; n = 3 male mice per group. (J) Immunohistochemistry of SULT1E1 in the WD patient liver sections shows strong SULT1E1 signal in hepatocytes as compared to healthy control (indicated by arrow in the high-magnification image). Representative image of SULT1E1 staining from the control and WD patient section that we have demonstrated here. Scale bars, 1 mm and 20 μm. n = 3 per group.
NRF2 acts by relocating from the cytosol to the nucleus, where it binds to the antioxidant response element of target genes and regulates their expression (Fig. 1F). RNA sequencing of control and Atp7b−/− liver samples demonstrated a significant increase in the expression of known NRF2 targets (Fig. 1G) in Atp7b−/− mouse liver, including NRF2 itself and its partner, avian musculoaponeurotic fibrosarcoma oncogene homolog (Mafs) (Fig. 1, F and G, and table S1). The most significantly (fold change) and consistently (P value) up-regulated transcripts encode proteins with well-known roles in protection and adaptation to oxidative stress such as glutathione S-transferases Gsta1, Gstm3, Gstm2; quinone reductase Nqo1; sulfiredoxin Srxn1; phosphogluconate dehydrogenase Pgd; and others (Fig. 1H). The most changed transcript was for sulfotransferase Sult1e1 (~573-fold, P = 0.0006). The marked increase in Sult1e1 expression was verified by quantitative polymerase chain reaction (qPCR) (see in Fig. 2) and by immunostaining of Atp7b−/− mouse and human WD liver tissues, which revealed strong Sult1e1 staining in Atp7b−/− hepatocytes (Fig. 1, I and J, and fig. S1).
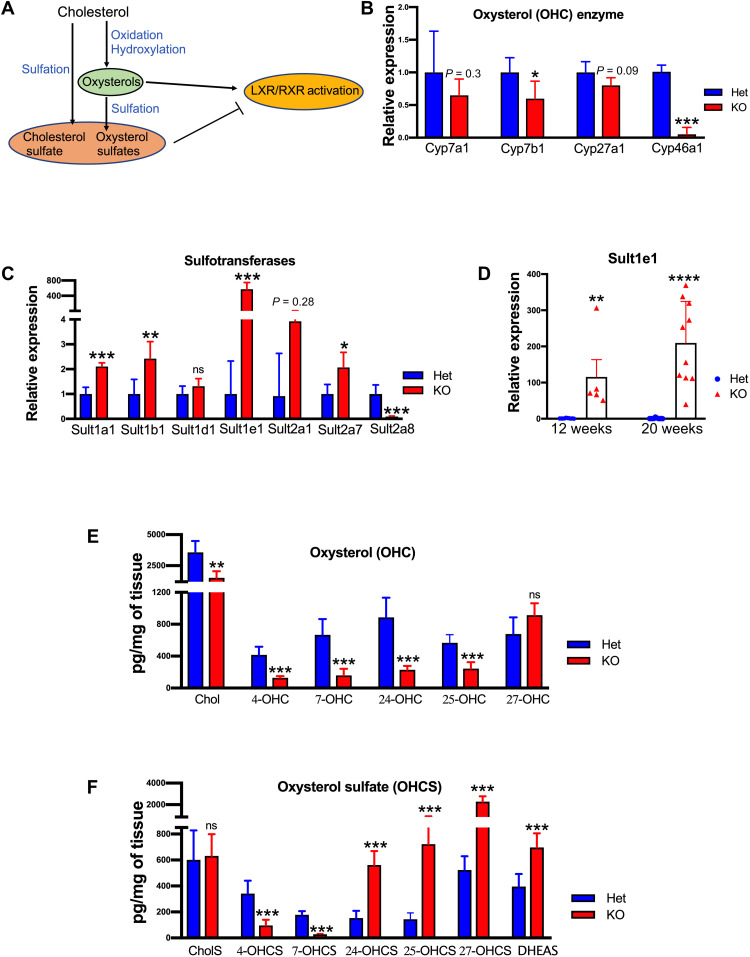
Fig. 2. Altered oxysterol synthesis (OHC) and sulfation (OHCS) result in sterol misbalance in Atp7b−/− liver.
(A) Cartoon illustrating effects of modified sterols on LXR activity. (B) Relative abundances of the transcripts for enzymes involved in OHC synthesis (Cyp450) and (C) for sulfotransferases involved in OHCS production (n = 4 male mice per group). (D) qPCR analysis of mRNA at 12 and 20 weeks shows a genotype (KO) and time-dependent up-regulation of Sult1e1 transcript (n = 5 to 10 male mice per group). (E) Liver oxysterols (4-OHC, 7-OHC, 24-OHC, 25-OHC, and Chol) and 4-OHCS and 7-OHCS were significantly reduced in KO mice. (F) Oxysterol sulfates (24-OHCS, 25-OHCS, 27-OHCS, and DHEAS) were significantly elevated in the KO liver. Values represent means ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, ****P<0.0001, n = 6 male mice per group. Blue color denotes Het, and red color denotes KO, respectively. Chol, cholesterol; 4-OHC, 4-hydroxycholesterol; 7-OHC, 7-hydroxycholesterol; 24-OHC, 24-hydroxycholesterol; 25-OHC, 25-hydroxycholesterol; 4-OHCS, 4-hydroxychole-sterol-3-sulfate; 7-OHCS, 7-hydroxycholesterol-3-sulfate; 24-OHCS, 24-hydroxy-cholesterol-3-sulfate; 25-OHCS, 25-hydroxychole-sterol-3-sulfate; 27-OHCS, 27-hydroxycholesterol-3-sulfate; DHEAS, dehydroepiandrosterone sulfate; ns, nonsignificant.
In any advanced disease, many cellular pathways are expected to be dysregulated and large number of transcripts to be altered. In Atp7b−/− liver, 3191 mRNAs were significantly changed compared to control (>2-fold change, P < 0.05). Only a subset of these transcripts represented the NRF2 targets, indicating involvement of other transcriptional regulators in the liver response to elevated Cu. Analysis of the upstream regulators using Ingenuity Pathway Analysis (IPA) pointed to IFNG, TGFB1, NR1H3 (LXRα), and TP53 as top factors responsible for downstream mRNA changes (based on the number of altered transcripts and P value; table S2). Among transcription factors, NR1H3 (LXRα) was the most significant regulator (921 targets, P = 1.06 × 10−33); the overall negative z score of LXRα-dependent changes (−0.8) pointed to partial inhibition of LXR activity.
Significant involvement of LXRα in the hepatic response to Cu overload made the strong up-regulation of sulfotransferase Sult1e1 particularly interesting. Sult1e1 catalyzes the conjugation of sulfate to several sterols including 24(S)-hydroxycholesterol (24-OHS). Oxysterols (OHC) are endogenous activators of LXR (Fig. 2A), whereas sulfated oxysterols, the products of sulfotransferase activity (OHCS), inhibit LXR (16, 17). Robust up-regulation of Sult1e1 in Atp7b−/− mice suggested that the balance of sulfated and nonsulfated oxysterols is altered in advanced liver disease, causing dysregulation of LXR.
Altered expression of enzymes involved in oxysterol synthesis and sulfation results in sterol misbalance
To test this hypothesis, we first evaluated the expression of enzymes involved in oxysterol synthesis and sulfation (Fig. 2, B to D). In Atp7b−/− liver, the relevant cytochromes were down-regulated by varying degrees compared to those in Atp7b+/− liver (Cyp7a1 by 35%, P = 0.3; Cyp7b1 by 40%, P = 0.05; Cyp27a1 by 20%, P = 0.09; and Cyp46a1 by 90%, P = 0.0009; Fig. 2B). In contrast, there was a significant up-regulation of several sulfotransferases (Fig. 2C). In addition to Sult1e1(~573-fold, P = 0.0006), the abundance of Sult1a1 transcript was increased ~2.1-fold (P = 0.0004), and that of Sult1b1 transcript was increased ~2.5-fold (P = 0.01). Sult2a8, which is involved in bile acid synthesis, was significantly down-regulated in Atp7b−/− liver (Fig. 2C). To determine whether the expression of the most up-regulated Sult1e1 changes with disease progression, the mRNA levels of Sult1e1 were analyzed by qPCR at 12 weeks and 20 weeks in Atp7b−/− liver (Fig. 2D). Sult1e1 mRNA was highly up-regulated at both time points, and the up-regulation increased with age/disease progression (Fig. 2D).
The opposite changes in the abundance of enzymes involved in oxysterol synthesis and sulfation pointed to sterol misbalance in Atp7b−/− liver. This prediction was verified by directly measuring OHC and OHCS in liver tissues. At 20 weeks after birth, the oxysterol content was significantly decreased in Atp7b−/− liver compared to controls; cholesterol (1527 versus 3568 pg/mg of tissue, P = 0.001), 4-OHC (128.6 versus 415.7 pg/mg, P = 0.0002), 7-OHC (158 versus 667 pg/mg, P < 0.0001), 24-OHC (228 versus 887 pg/mg, P < 0.0001), and 25-OHC (243 versus 566 pg/mg, P < 0.0001) were lower; 27-OHC was unchanged (Fig. 2E). In contrast, the sulfated oxysterols were significantly higher in Atp7b−/− mice compared to control: 24-OHCS (561 versus 153 pg/mg, P < 0.0001), 25-OHCS (722 versus 144 pg/mg, P < 0.0001), 27-OHCS (2280 versus 523 pg/mg, P = 0.0002), and DHEAS (dehydroepiandrosterone sulfate; 696 versus 396 pg/mg, P = 0.0002) (Fig. 2F). To determine whether the decrease in oxysterols merely reflected the decreased abundance of cholesterol, we also measured sterol levels at the earlier time point of disease progression, at 12 weeks after birth. The comparison of changes in cholesterol and OHCS at 12 and 20 weeks demonstrated that an increase in OHCS significantly outweighs changes in cholesterol and correlates with Sult1e1 up-regulation in a time-dependent manner (table S3). Thus, in Atp7b−/− mice with established WD, there was a significant decrease in the endogenous LXR agonists (OHC) with a concomitant increase in the LXR antagonists (OHCS), consistent with a predicted inhibition of LXR.
Activation of LXR corrects multiple dysregulated pathways in Atp7b−/− liver
To independently verify LXR inhibition in advanced liver disease, we compared metabolites and liver transcriptomes from control, Atp7b−/−, and Atp7b−/− mice treated with the synthetic LXR agonist T0901317. The treatment with the agonist (from 12 weeks after birth to 20 weeks after birth) was performed when clinical signs of the disease (inflammation, fibrosis, and changes in hepatocyte morphology) were already established to better mimic human WD.
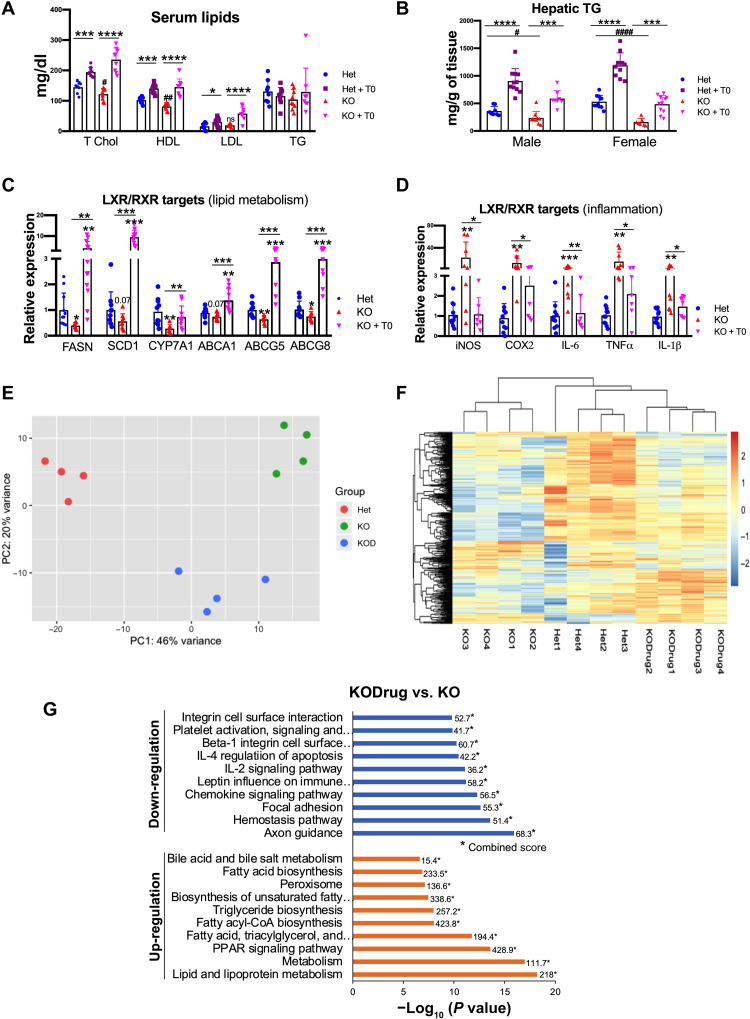
Total serum cholesterol and high-density lipoprotein were lower in Atp7b−/− mice compared to control animals and were increased by treatment with the drug T0901317 (Fig. 3A and fig. S3). Liver triglycerides (TGs) were also lower in Atp7b−/− mice, and the treatment with the agonist brought the TG levels in Atp7b−/− liver close to the healthy (untreated) control (Fig. 3B). These findings were consistent with LXR inhibition in untreated Atp7b−/− mice. The examination of known LXR-regulated transcripts (lipid metabolism and inflammation) further pointed to decreased LXR activity (Fig. 3, C and D). Compared to control, there was a significant decrease (~50 to 70%) in hepatic fatty acid synthase (FASN), stearoyl coenzyme A desaturase 1 (SCD1), and cytochrome p450 family 7A1 (CYP7A1) and ~25 to 35% decrease in ATP-binding cassette (ABC) subfamily gene expression (ABCA1, ABCG5, and ABCG8) in Atp7b−/− mouse liver. Treatment with the drug up-regulated the LXR-dependent lipid metabolism genes (Fig. 3C). Atp7b−/− mice also had a 20-fold increase in inducible nitric oxide synthase (iNOS), a 14-fold increase in tumor necrosis factor–α (TNFα), a 12.8-fold increase in cyclooxygenase-2 (Cox2), a 4.2-fold increase in interleukin-6 (IL-6), and a 4-fold increase in IL-1β expression. Treatment with the drug reduced the expression of these LXR-dependent inflammatory genes in Atp7b−/− mouse liver to levels similar to heterozygous (Het) controls (Fig. 3D).
Fig. 3. Activation of LXR corrects multiple dysregulated pathways in Atp7b−/− liver.
(A) Serum lipids [cholesterol and high-density lipoprotein (HDL)] were reduced in KO male mice and elevated in treated mice. LDL, low-density lipoprotein. (B) Hepatic TG levels were reduced in KO mice (males and females) and significantly increased in animals treated with T0901317. Values represent means ± SD. *P < 0.05, ***P < 0.001, and ****P < 0.0001 between untreated and treated mice of the same genotype; #P < 0.05, ##P < 0.01, and ####P < 0.0001 between untreated Het and KO mice, n = 8 to 11 mice per group. (C and D) Activation of LXR corrects dysregulation of targets involved in lipid metabolism (FASN, SCD1, CYP7A1, ABCA1, ABCG5, and ABCG8) and inflammation (iNOS, COX2, IL-6, TNFα, and IL-1β) in Atp7b−/− mice. Values represent means ± SD. *P < 0.05, **P < 0.01, and ***P < 0.001, n = 8 to 11 mice per group. (E) PCA plot shows that the transcriptomes of Het (red), KO (green), and drug-treated KO (KOD; blue) livers are distinctly clustered. (F) Heatmap illustrates that the significant differences in the expression profiles between KO and Het livers are decreased following treatment with the drug, making the mRNA profiles in treated liver more similar to Het. (G) EnrichR analysis demonstrates the biological pathways down- and up-regulated in KO liver in response to treatment with the drug. *Combined score. The blue color symbol denotes Het, purple denotes Het + T0, red denotes KO, and magenta denotes KO + T0, respectively. T0, T0901317.
Given that transcription factors other than LXR were also predicted to be dysregulated, we wanted to evaluate the significance of LXR inactivation to overall pathologic changes in Atp7b−/− liver. Principal components analysis (PCA) demonstrated distinct clustering of the control and knockout (KO) transcriptomes, reflecting numerous changes between these mice with established liver disease (Fig. 3E). Unsupervised clustering of differentially expressed transcripts (heatmap in Fig. 3F) confirmed significant changes in the KO samples compared to control (Het). Activation of LXR with the drug (KODrug) induced significant changes in the transcriptome compared to untreated Atp7b−/− liver, as indicated by the shift in the PCA plot (Fig. 3E). Notably, the LXR agonist affected many transcripts, and the KODrug and control liver transcriptomes became more similar (Fig. 3F). Analysis of altered pathways by IPA demonstrated that treatment of KO mice with the LXR agonist up-regulated lipid metabolism, PPAR signaling, fatty acid, TG biosynthesis, and bile acid metabolism, correcting most of the down-regulated pathways (KODrug versus KO). Treatment with the drug also down-regulated inflammatory signaling (chemokines, IL-2, and IL-4 signaling), leukocyte extravasation, focal adhesion, and integrin signaling, which were activated in untreated KO. The drug did not affect the pathways associated with DNA replication and cell cycle (Fig. 3G).
LXR agonist partly improves liver function and histology despite Cu overload
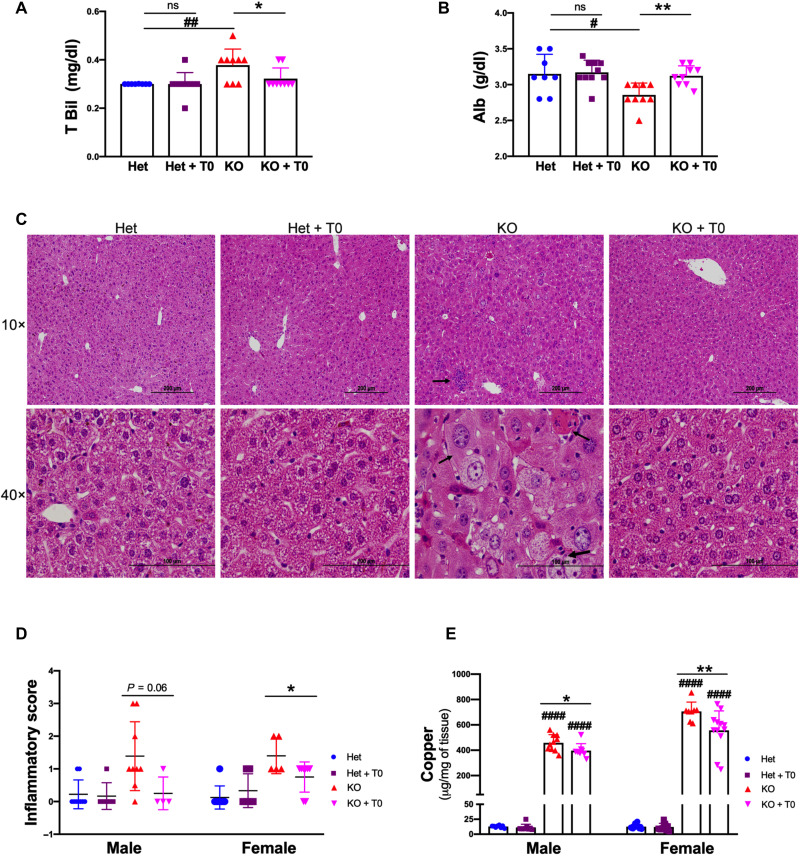
To test whether liver function was improved by LXR activation, we evaluated several parameters. Serum alanine aminotransferase (ALT) levels were higher in untreated Atp7b−/− male mice, compared to Het (mean ALT, 60 versus 24 U/liter, *P < 0.05), and were not affected by treatment with T0901317 (fig. S4A). However, serum total bilirubin showed improvement compared to untreated Atp7b−/− mice (0.32 versus 0.38 mg/dl; P < 0.05, SD = 0.02; Fig. 4A), and albumin levels also increased (3.12 versus 2.85 g/dl, *P < 0.05 and **P < 0.01) in both males (Fig. 4B) and females (fig. S4, B and C).
Fig. 4. LXR agonist partly improves liver function and histology despite Cu overload.
(A and B) LXR agonist reduces serum total bilirubin and increases albumin in drug-treated KO male mice. (C) Representative H&E-stained liver sections from control and Atp7b−/− mice (n = 4 to 9 mice per group). Scale bars, 200 and 100 μm. Severely inflamed livers had cytomegaly (small black arrow), inflammatory infiltrates (small black arrow in 10×), and binucleate hepatocytes (big black arrow). These changes were significantly improved in treated KO mice. (D) Inflammation was scored as 0 = none, 1 = mild, 2 = moderate, and 3 = severe. Quantitation of inflammatory score in male and female, respectively. (E) T0901317 does not prevent Cu overload but reduces Cu levels partly in Atp7b−/− mice (male and female). Values represent means ± SD. *P < 0.05 and **P < 0.01 between untreated and treated mice of the same genotype; #P < 0.05, ##P < 0.01, and ####P < 0.0001 between untreated Het and KO mice, n = 8 to 11 mice per group. The blue color symbol denotes Het, purple denotes Het + T0, red denotes KO, and magenta denotes KO + T0, respectively.
Analysis of liver histology and inflammatory cytokines (Fig. 3D) further confirmed the beneficial effect of the drug. In untreated KO, hematoxylin and eosin (H&E) staining of liver sections shows enlarged hepatocytes, intracytoplasmic vacuoles, karyomegaly, multiple nuclei, and inflammatory cell infiltrates (Fig. 4C). These histological changes were notably diminished by the drug, and overall liver histology was improved. The inflammatory score was reduced by 40 to 50% in both males and females (Fig. 4D). Treatment with the drug caused a small decrease in the hepatic Cu levels compared to untreated KO animals; however, overall Cu remained high (30-fold higher in males and 46-fold higher in females compared to the drug-treated control mice; Fig. 4E). Endogenous Cu chelators, metallothioneins, also remain elevated (fig. S4D). Similarly, in the analysis of the sterol content in the tissue liver, we found no decrease in the sulfated OHCs: They were either elevated (4-OHCS, 7-OHCS, and 24-OHCS) or unchanged (cholesterol sulfate, 25-OHCS, 27-OHCS, and DHEAS) when compared to the nontreated Atp7b−/− liver. On the other hand, the treatment with LXR agonist did increase the OHC levels, bringing them closer to the normal control (fig. S2). Thus, the beneficial effects of the LXR agonist were not due to normalization of Cu levels or the full restoration of sterol balance but rather the correction of dysregulated LXR-dependent pathways.
Activation of LXR reduces fibrosis in Atp7b−/− mice primarily via a noncanonical TGFβ signaling
LXR was suggested to play an important but not yet well-defined role in liver fibrosis (18). Consequently, we examined whether the reversal of LXR inhibition would diminish fibrosis in murine WD. Inflammatory cytokines activate hepatic stellate cells (HSCs) to produce collagen 1a, leading to deposition of extracellular matrix and liver fibrosis (19). Atp7b−/− mice have an approximately fivefold increase in Col1α mRNA compared to control mice. This increase in the expression of collagen was significantly reduced by treatment with the LXR agonist (Fig. 5A). Similarly, tissue inhibitor of metalloproteinase 1 (TIMP1), another marker of inflammatory/fibrotic liver disease, was elevated in untreated Atp7b−/− mouse liver and reduced after treatment with the drug (Fig. 5A, right side).
Fig. 5. LXR agonist reduces fibrosis in Atp7b−/− mouse liver.
(A) LXR agonist reduces fibrosis. Profibrotic marker (COL1α and TIMP1) mRNA expression was significantly elevated in KO mouse liver as compared to Het. LXR agonist reduced COL1α and TIMP1 expression in KO mice compared to untreated KO (n = 8 to 11 male mice per group). (B and C) Liver sections were stained with Sirius red (collagen), a representative image of one of the Sirius red–stained liver sections is shown, and quantitation of the red stain–positive areas was performed by ImageJ (n = 4 to 5 male mice per group). Scale bars, 200 μm. (D) Activated stellate cell marker desmin (red color) expression was significantly increased in KO mice as compared to Het, which was reduced in response to drug treatment in KO, whereas F4/80 (green) macrophage marker was not significantly changed. Scale bars, 20 and 10 μm. n = 4 mice per group. (E) Significantly altered gene [log2 fold change (FC) with the type of gene] of the hepatic fibrosis signaling pathway in KO versus Het, drug-treated KO versus KO, and drug-treated KO versus Het mice is displayed in the table (n = 4 male mice per group). Values represent means ± SD. *P < 0.05 and **P < 0.01, between untreated and treated mice of the same genotype; ##P < 0.01, and ###P < 0.001 between untreated Het and KO mice. The blue color symbol denotes Het, purple denotes Het + T0, red denotes KO, and magenta denotes KO + T0, respectively.
In agreement with the changes in mRNA levels, there was significantly less collagen deposition in the drug-treated KO compared to the untreated KO. Quantification of the Sirius red–stained liver areas showed a more than 40% (P < 0.003) decrease in liver fibrosis in response to the drug (Fig. 5, B and C). These data, along with the decrease in inflammatory cytokines (Fig. 3D), suggested that the LXR agonist reverses/diminishes activation of HSC. This hypothesis was tested by immunostaining of desmin (a marker of activated HSC). Desmin is strongly up-regulated in the Atp7b−/− livers. Treatment with the drug down-regulated desmin in Atp7b−/− mice, consistent with reduced activation of HSC (Fig. 5D). Note that although treatment with the LXR agonist reversed the overexpression of many molecules associated with hepatic fibrotic signaling, some markers remained up-regulated (Fig. 5E), indicative of additional mechanisms contributing to fibrosis. The macrophage marker F4/80 was not significantly changed in Atp7b−/− livers compared to control.
The LXR agonist reverses fibrosis by changing noncanonical TGFβ signaling
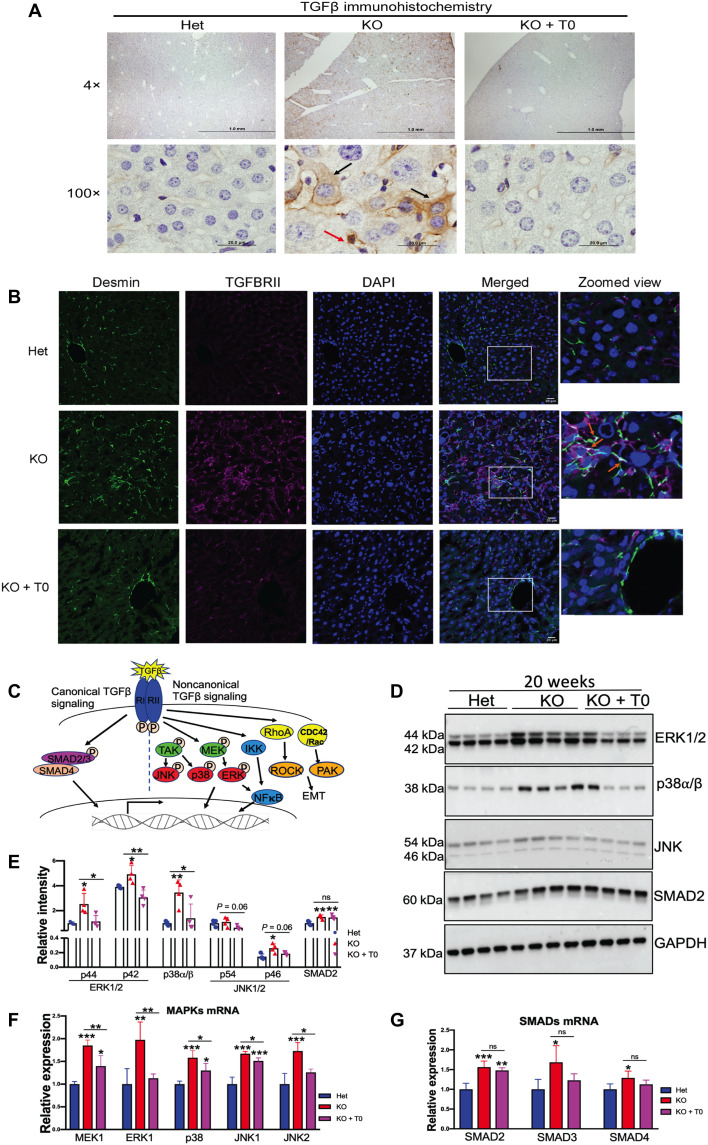
IPA analysis of transcriptomes predicted transforming growth factor–β (TGFβ) as a major upstream regulator of fibrosis (table S2). TGFβ signaling is initiated by binding of TGFβ to its cognate receptor TGFβRII. Immunohistochemistry of TGFβ revealed a positive TGFβ signal in both hepatocytes and nonparenchymal liver cells in KO mice (Fig. 6A). Transcriptome data suggested that TGFβRII was up-regulated in the KO liver, and this up-regulation was reversed by treatment with the drug (Fig. 5E). Immunostaining of activated stellate cells (identified by desmin expression) confirmed a significant increase in TGFβRII (red) expression compared to Het. Drug treatment reduced the TGFβRII signal, in agreement with the transcriptome data (Fig. 6B). To better understand how changes in TGFβRII abundance are coupled with changes in its function, we evaluated the status of downstream signaling molecules.
Fig. 6. LXR agonist reduces fibrosis in Atp7b−/− mice primarily via noncanonical TGFβ signaling.
(A) Representative immunostaining of TGFβ shows a significant increase in TGFβ signal in Atp7b−/− hepatocytes (black arrows) and nonparenchymal liver cell (red arrow). Scale bars, 20 μm. n = 3 male mice per group. (B) Activated stellate cell marker desmin (green) expression was significantly increased in KO mice and colocalized with the induced TGFβRII; zoomed view of areas marked by white quadrangle is shown on the right. Scale bars, 20 μm. (C) Diagrammatic representation of canonical and noncanonical TGFβ signaling pathways. (D) Western blots showing that MAPKs (ERK1/2, p38, and JNK1/2) and SMAD2 are elevated in KO tissues; activation of LXR reverses MAPK levels, whereas SMAD2 remains. GAPDH was used as an endogenous loading control. (E) Densitometry analysis of ERK1/2 (p44 and p42), p38, JNK (p54 and p46), and SMAD2 protein signals normalized to GAPDH. (F and G) MAPK mRNA (MEK1, ERK1, p38, JNK1, and JNK2) and SMAD (SMAD2, SMAD3, and SMAD4) mRNA expression induced in KO mouse liver compared to Het. LXR agonist reduced MAPK mRNA level; however, the drug does not affect SMAD2 to SMAD4 mRNA level in KO mice compared to untreated KO. Values represent means ± SD. *P < 0.05, **P < 0.01, and ***P < 0.001; n = 4 male mice per group. Blue color denotes Het, red denotes KO, and magenta denotes KO + T0, respectively.
TGFβ signaling is mediated through the canonical SMAD and noncanonical mitogen-activated protein kinase (MAPK) signaling pathways (Fig. 6C). SMAD2 protein levels were similarly increased (~1.5-fold) in KO and drug-treated KO livers, suggesting that the drug does not affect the components of classical SMAD pathways (Fig. 6D). By contrast, the noncanonical TGFβ (MAPKs) signaling pathway was affected by the drug. The increased abundance of extracellular signal–regulated kinase 1/2 (ERK1/2; ~2.5-fold), p38 (~3.5-fold), and c-Jun N-terminal kinase (JNK; p46, ~2-fold) was observed in KO liver compared to Het, and these changes were reversed by T0901317 treatment (Fig. 6, D and E). The changes in the transcripts were consistent with the protein data. There was more than ~1.5-fold (P < 0.05) increase in MAPK [MAPK kinase 1 (MEK1), ERK1, p38, JNK1, and JNK2] and SMAD (SMAD2-SMAD4) mRNA expression in KO mouse liver (Fig. 6, F and G). Drug treatment reduced MAPK transcript level in KO mice (Fig. 6F) but did not affect SMAD2, SMAD3, and SMAD4 levels (Fig. 6G).
Rho-like guanosine triphosphatases (GTPases) are a family of proteins that regulate TGFβ-mediated cytoskeletal organization, which works independently of classical SMAD signaling (20, 21). Atp7b−/− mice have significantly elevated Rho GTPase mRNAs (RhoA, ~1.6-fold; RhoB, ~4-fold; RhoC, ~7.4-fold; CDC42, ~1.9-fold; RAC1, ~1.7-fold, and RAC2, ~10.3-fold) (fig. S5A). Drug treatment reduces the levels of these transcripts in KO mice. To further verify changes in Rho GTPase signaling, we checked the downstream signaling kinase cascade. Rho-associated kinase (ROCK1, ~1.8-fold; ROCK2, ~2.6-fold; PAK1, ~7.5-fold; PAK2, ~1.9-fold; and PAK4, ~2.3-fold) expression was significantly elevated in KO mouse liver compared to Het mice, and drug treatment decreased their expression in KO mice compared to untreated KO mice (fig. S5B). All these data suggest that drug-dependent reduction of inflammation and fibrosis in Atp7b−/− mouse liver at 20 weeks involves the noncanonical TGFβ signaling pathway.
Inhibition of the sulfotransferase Sult1e reverses the sterol misbalance and activates LXR
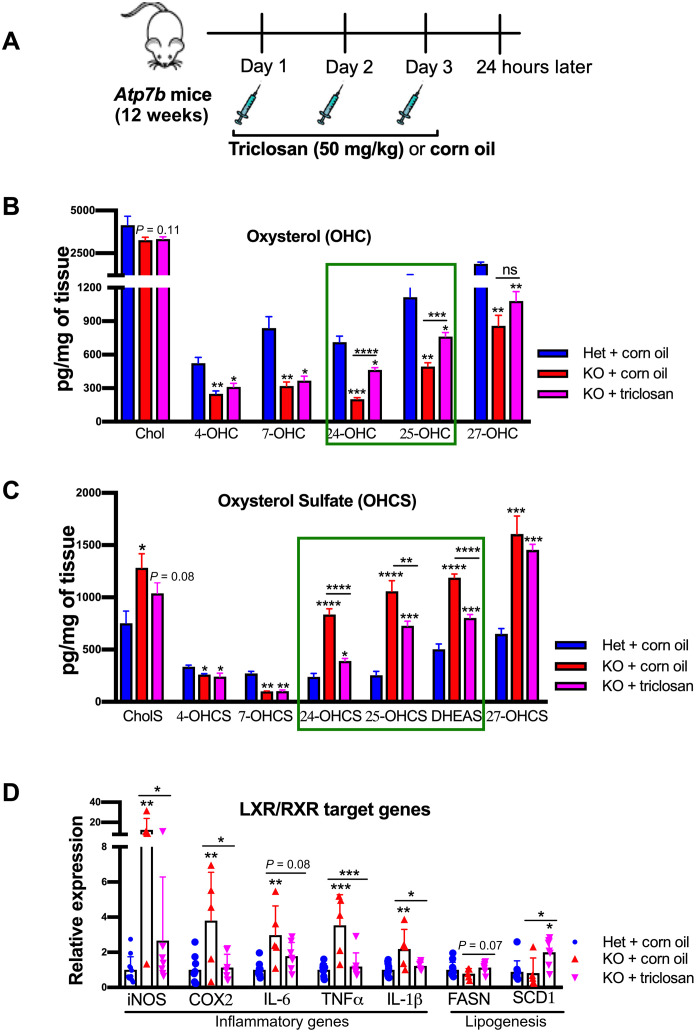
Last, we tested whether the normalization of OHC and OHCS levels would reverse the transcriptional changes caused by LXR inhibition. Toward this goal, we injected Atp7b−/− mice thrice with triclosan (Fig. 7A), an established inhibitor of sulfotransferase Sult1e1 (22), and measured the levels of oxysterols and sulfated oxysterols in the liver. As described above, oxysterol content was reduced in untreated Atp7b−/− mice as compared to control. Treatment with the Sult1e1 inhibitor significantly increased levels of 24-OHC and 25-OHC in the KO liver relative to untreated KO mice (464.8 versus 201.4 pg/mg of tissue, P < 0.0001; 762.6 versus 494.2 pg/mg of tissue, P < 0.0001, respectively). A less significant increase in 27-OHC (1080 versus 859.2 pg/mg of tissue, P = 0.10) was also detected (Fig. 7B). Concomitantly, there was a significant decrease in sulfated 24-OHCS, 25-OHCS, and DHEAS levels (391.5 versus 836.2 pg/mg of tissue, P < 0.0001; 728.7 versus 1059 pg/mg of tissue, P < 0.007; 802.2 versus 1188 pg/mg of tissue, P < 0.0001, respectively) (Fig. 7C).
Fig. 7. Inhibition of the sulfotransferase Sult1e reverses sterol misbalance and activates LXR.
(A) Cartoon illustrating the triclosan injection strategy (50 mg/kg of body weight; intraperitoneal) in 12-week-old Atp7b−/− male mice to inhibit Sult1e1. (B) Liver oxysterols (4-OHC, 7-OHC, 24-OHC, 25-OHC, and 27-OHC) were significantly reduced in KO mice, and Sult1e1 inhibition by triclosan improves 24-OHC and 25-OHC level (green box). (C) Sulfated oxysterols (24-OHCS, 25-OHCS, 27-OHCS, and DHEAS) were significantly elevated in KO mouse liver and reduced by Sult1e1 inhibition. Values represent means ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001; n = 6 male mice per group. (D) LXR inflammatory targets (iNOS, IL-1β, COX2, IL-6, and TNFα) were significantly elevated in untreated KO mice compared to Het. Treatment with triclosan down-regulates the expression of inflammatory LXR target genes and up-regulates lipogenesis (FASN and SCD1) in KO mice. Values represent means ± SD. *P < 0.05, **P < 0.01, and ***P < 0.001; n = 5 to 10 male mice per group. Blue color denotes Het + corn oil, red color denotes KO + corn oil, and magenta color denotes KO + triclosan, respectively.
An increase in LXR agonists (OHC) and a decrease in LXR antagonists (OHCS) are expected to relieve LXR inhibition and increase lipogenesis while inhibiting the inflammatory response. Similar to treatment with the LXR agonist, inhibition of Sult1e in Atp7b−/− mouse liver increased the mRNA levels for lipogenesis enzymes (FASN and SCD1) and decreased the expression of inflammatory markers (iNOS, IL-6, IL-1β, COX2, and TNFα) (Fig. 7D). These data strongly suggest that restoration of oxysterol balance reverses the effects of LXR inhibition in WD mice. To understand whether pharmacological inhibition of Sult1e1 can potentially improve fibrosis, we examined the expression of fibrotic genes. Increase in Col1α and Timp1 expression in Atp7b−/− mice was significantly reduced in the triclosan-treated KO mice (fig. S6). We also examined that FXR, an NR that regulates bile acid and lipid metabolism, also forms a heterodimer with RXR, and its expression is reduced in Atp7b−/− mice (8). However, treatment with triclosan did not restore the expression of FXR/RXR target genes (SHP and BSEP) (fig. S7).
DISCUSSION
WD is caused by Cu accumulation that triggers morphologic and functional pathologies in tissues, especially in the liver. Inactivating mutations in the Cu transporter ATP7B represent a well-established genetic cause of WD, and excess Cu is always found in WD livers. Studies in recent years have made it increasingly clear that the broad spectrum of WD phenotypes and the variable time of WD onset cannot be explained by one single factor, such as specific levels of Cu in tissues or ATP7B mutation genotype. Cu overload causes oxidative stress, mitochondrial injury and dysfunction, inhibition of NRs, and epigenetic and metabolic changes (6, 7, 14, 15). The relative contribution of these abnormalities to WD pathology is unclear and needs to be established to more accurately predict disease phenotypes and treatments. In this study, we have identified several molecular links underlying advanced hepatic WD in mice and provide evidence for the nontoxic and beneficial effects of targeting LXR after the onset of liver pathology. While the described mechanistic picture is by no means complete, our results provide strong evidence that the pathways causing oxysterol depletion and LXR inhibition should be carefully examined in human WD as potential therapeutic targets.
We show that in Atp7b−/− mice, up-regulation of the stress-sensitive factor NRF2 causes increased expression of molecules involved in protection/adaptation to oxidative stress. We identified the subset of the most significantly changed transcripts, which could be considered a molecular signature of the hepatic response to oxidative stress in murine WD (Fig. 1H). Our studies also revealed strong up-regulation of sulfotransferases in Atp7b−/− mouse and human Atp7b−/− liver and led to the discovery of sterol misbalance in murine WD. Elevated Sult1e1 was also detected in Atp7b−/− mice on different genetic B6 background [see microarray supplementary table 1 in (9)]. We show that the decrease in OHCs, the endogenous activators of LXR, and increase in the sulfated OHCs, the LXR inhibitors, cause inhibition of LXR signaling. Restoration of sterol balance by lowering Sult1e1 activity diminished LXR inhibition (Fig. 7).
Unlike LXR antagonists 24-OHCS and 25-OHCS, levels of 4-OHCS and 7-OHCS are lower in Atp7b−/− mice. This is likely due to the reduced expression of Sult2a8 and the bile acid precursors 4-OHC and 7-OHC, which, in turn, may explain the previously reported (and confirmed here) dysregulation of FXR signaling (8). While our data predict the largest impact of the drug treatment on the LXR-dependent pathways, we cannot exclude a positive impact of the drug on some FXR targets (and hence added beneficial effect), since a prolonged application of T0901317 may increase intracellular concentration to levels that would be modulatory to FXR, Retimoid-related orphan receptor gamma (RORγ), and pregnane X receptor (PXR) (23). Whether this possible pleotropic effect represents the drug advantage or detriment remains to be established. At present, our data indicate that T0901317-like compounds may have significant beneficial effects for developed hepatic WD. The drug reverses many transcriptional and metabolic changes in Atp7b−/− mice. Approximately 90% of the genes, which were induced in untreated KO, were repressed in response to the drug, with the large impact on transcripts for lipid metabolism and inflammatory response (the major components of disease phenotype).
Current lifelong Cu chelation therapy improves liver function but often has notable side effects (4, 5). Treatment with LXR agonists up-regulates lipogenesis; however, there was no hepatic steatosis observed in our experiments. We did find an increase in liver weight in drug-treated Het and KO mice, which did not affect body weight (fig. S8, A and B). Increased liver weight could be due to increased TG content. In addition, PXR induces liver enlargement by activating yes-associated pathway (YAP) signaling (24). PXR and YAP1/TAZ (transcriptional coactivator wih PDZ-binding motif) levels are up-regulated in Atp7b−/− mouse liver, but the treatment with T0901317 did not alter their expression (fig. S8, C and D).
Our RNA sequencing data revealed a ~2-fold increase in suppressor of zeste homolog 12 (SUZ12) and enhancer of zeste homolog 2 (EZH2) mRNA expression in Atp7b−/− mice, and the drug treatment reduced their level to those of control (fig. S9). SUZ12, together with EZH2, forms a polycomb group repressive complex (PRC2/3) involved in transcriptional silencing by histone methylation (25). In human fibroblasts, depletion of PRC2/3 components revealed more than 1000 silenced genes belonging to TGFβ, Wnt, fibroblast growth factor, Notch, and Hedgehog signaling pathways (25). EZH2 was also identified as an important factor in TGFβ-mediated stellate cell activation and fibrosis (26). These findings suggest that LXR activation may cause transcriptional repression of TGFβ and other signaling pathways through polycomb repressive complex. Future studies should evaluate whether LXR activation causes transcriptional repression of TGFβ and other signaling pathways through the polycomb repressive complex.
The LXR agonist partially improved liver function; however, serum transaminases were not significantly changed. A possible explanation is that serum transaminases are more sensitive for acute liver injury/toxicity than chronic liver injury (27) or a longer treatment is needed. Review of histology sections revealed that LXR agonism is effective in improving pathologic changes observed in KO mouse liver at 20 weeks, although there was some individual sample variation. There was no evidence of drug-induced liver injury in the treated animals.
Liver inflammation, eventually leading to fibrosis, is commonly observed in WD. Our findings of reduced fibrotic gene expression in Atp7b−/− mice in response to the agonist agree with previous studies in LXRα/β−/− mice (28). Atp7b is expressed not only in hepatocytes but also in HSCs (www.proteinatlas.org/ENSG00000123191-ATP7B/single+cell+type/liver). In Atp7b−/− livers, both parenchymal and nonparenchymal cells accumulate Cu (29), and immunohistochemistry of TGFβ reveals increased TGFβ signal in both hepatocytes and nonparenchymal liver. At the same time, Sult1e1 was up-regulated predominantly in hepatocytes, perhaps reflecting increased Cu concentration in these cells. These data suggest that the molecular events inside different liver cells and paracrine communication between the cells may contribute to HSC activation and inflammatory response.
Despite elevated expression of TGFβ, we found no significant difference in the classical SMAD signaling pathway in response to the drug, as SMAD2 protein levels remained high in KO mice. On the other hand, the LXR agonist significantly reduced non-SMAD-associated signaling molecules (ERK1/2, p38, and JNK), which are elevated in KO livers. It was previously shown that the RAS/MAPK signaling pathway was stimulated by Cu ions (30). Recent finding demonstrated that Cu treatment induces hepatic dysfunction via activation of MAPK signaling in the mouse (31). Our data suggest that, in the liver, elevated Cu up-regulates MAPK signaling pathways in Atp7b−/− mice, and LXR agonism down-regulates MAPK signaling.
In conclusion, we identified a Cu-mediated stress-induced up-regulation of sulfotransferases and significant changes in sterol composition as the molecular basis of inhibition of the liver NR LXR. Our study also shows that inhibition of LXR signaling by sterol misbalance significantly contributes to inflammation and fibrosis in murine WD, and activation of LXR/RXR reverses liver inflammation and fibrosis via inhibition of the noncanonical TGFβ signaling pathway (Fig. 8). These findings further illustrate that dysregulation of LXR signaling contributes significantly to the pathogenesis of WD and offer a solid rationale for developing and testing NR agonists as potential novel treatment for WD.
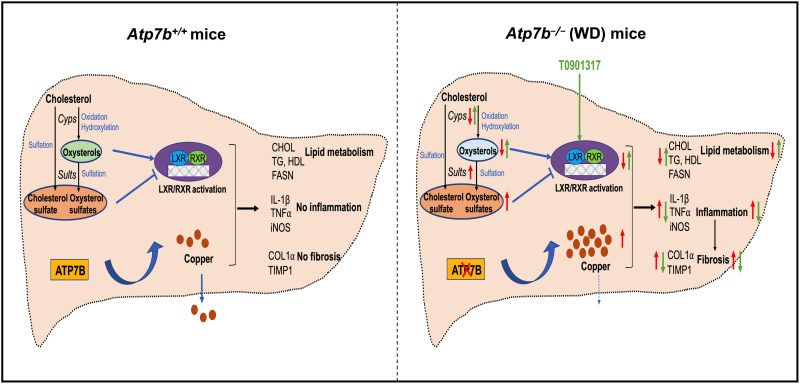
Fig. 8. Proposed model of LXR inhibition by oxysterol misbalance contributes to inflammation and fibrosis in murine WD.
Under normal conditions, functional ATP7B maintains Cu homeostasis. Inactivating mutations of ATP7B cause Cu accumulation. Cu-induced oxidative stress causes sterol misbalance by down-regulating OHC (via Cyps) enzymes and up-regulating sulfotransferases (Sult1e1, Sult1a1, and Sult1b1), which inhibit LXR in murine WD. Activation of LXR/RXR using T0901317 reverses liver inflammation and fibrosis and partially improves liver function in advanced WD. In addition, lipid functions are improved despite Cu overload. Significant decreases in inflammation and fibrosis are primarily mediated by changes in the abundance of the protein components of the noncanonical TGFβ signaling pathway. Red arrows indicate the disease state, and green arrows indicate the effect of the LXR agonist.
MATERIALS AND METHODS
Animal experiments
Animals were housed at the Johns Hopkins University, School of Medicine (JHUSOM) animal care facility, and the studies followed the National Institutes of Health guidelines. Animal protocols were approved by the Institutional Animal Care and Use Committee (protocol number M017M385). Atp7b KO (Atp7b−/−) and Het (Atp7b+/−) mice of C57BL/6x129S6/SvEv background and the drug feeding strategy were previously described (11). T0901317 (Cayman Chemical, Ann Arbor, MI) powder was mixed with Teklad 2018 powdered chow (Harlan, Madison, WI) and fed to Atp7b−/− and Atp7b+/− littermates (males and females) at 12 weeks after birth, with a dose of 50 mg/kg per day, thrice weekly (Monday, Wednesday, and Friday). Standard pellet chow was fed to the mice on days that they were not receiving the drug. Animals were weighed weekly. After 8 weeks, at 20 weeks after birth, mice were euthanized using isoflurane, blood was collected by cardiac puncture, animals were perfused with chilled phosphate-buffered saline, and tissues were harvested. Tissues were flash-frozen and stored at −80°C until required. Sections of mouse liver were immersed in RNAlater (Invitrogen) for RNA sequencing and quantitative reverse transcription PCR (qRT-PCR) and frozen at −80°C, and pieces of liver tissues were formalin and paraformaldehyde fixed overnight for paraffin and Tissue-Tek® optimum cutting temperature compound (OCT) embedding, respectively.
For the sulfotransferase 1e1 inhibition, triclosan (50 mg/kg of body weight) was injected intraperitoneally thrice (one injection each consecutive day) in Atp7b−/− male mice at 12 weeks, as previously described (22). Corn oil was used as vehicle control. Mice were euthanized 24 hours after the last injection. Sections of mouse liver were immersed in RNAlater (Invitrogen) for qRT-PCR. Tissues were flash-frozen and stored at −80°C until required.
Human liver sections
Human liver sections for immunostaining were obtained from the Johns Hopkins Hospital Pathology Archives under Institutional Review Board protocol no. 228609. The WD sections were from liver biopsies done for clinical reasons. The normal liver sections were obtained from the caudate lobe of donor livers during liver transplants.
Western blotting
Liver tissue (~50 mg) was homogenized with 500 μl of radioimmunoprecipitation assay (RIPA) buffer (Sigma-Aldrich), 1× protease, and phosphatase inhibitors (Roche). Protein concentration was estimated by bicinchoninic acid (BCA) using bovine serum albumin standard. Protein lysates (80 to 100 μg) were separated on 10% SDS–polyacrylamide gel electrophoresis gel (Bio-Rad) and transferred using a Trans-Blot turbo system (Bio-Rad). After blocking with 5% fat-free milk for 1 hour, membranes were incubated with primary antibodies overnight at 4°C on a shaker, followed by a secondary antibody for 1 hour at room temperature. For glyceraldehyde-3-phosphate dehydrogenase (GAPDH), membranes were directly incubated with GAPDH–horseradish peroxidase antibody for 1 hour and visualized using Amersham Imager 600 (GE Healthcare). Antibodies’ source and dilutions are given in table S4.
RNA isolation and qRT-PCR
Total liver RNA was isolated from 20 to 30 mg of liver tissue using the QIAshredder and RNeasy Mini Kit (QIAGEN) following the manufacturers’ protocol. First-strand complementary DNA (cDNA) was synthesized from 1 μg of RNA (quantified by IMPLEN NanoPhotometer) using the High Capacity RNA-to-cDNA Kit (Applied Biosystems). PCR reactions were performed with 2× PowerUp SYBR Green Master Mix and 5 μl of cDNA (1:50 diluted) on the QuantStudio6 Flex system (Applied Biosystems) using primers described in table S5. The transcript abundances were determined using a relative quantitative Ct method (32), and Rpl19 was used as an endogenous control.
RNA sequencing and data analysis
RNA was isolated using the RNeasy Mini Kit (QIAGEN). RNA concentration and quality were determined by the Ribogreen method (Victor X2 fluorimetry) and Agilent bioanalyzer, respectively. RNA samples (500 ng) with RNA integrity values of 8 or higher were used for library preparation (TrueSeq Stranded Total RNA library). Libraries were generated, and sequencing was performed by Psomagen Inc. (Rockville, MD), using Illumina NovaSeq6000 (150 base pairs, paired-end). Read quality was checked with FastQC v0.11.7 and trimmed with trimmometric 0.38. Trimmed reads were mapped to reference genome UCSC mm10 using HISAT2v 2.1.0 (~50 million mapped reads per sample). The transcripts were assembled by StringTie v.2.1.3b with aligned reads. Expression profiles were represented as read count and FPKM (fragments per kilobase of transcript per million mapped reads) values. Data are available at the Gene Expression Omnibus database under accession number GSE174015. Analysis of differences in transcript abundance and data visualization were done using the R package (DSeq2) and IPA. These analyses included evaluation of associations between significantly altered genes (more than 1.5-fold change, P < 0.05) and metabolic and signaling pathways. The significance of associations was calculated using the Z score (+2 or −2), P value (0.05), or the combined score. Transcription factor–binding sites enriched within regulated regions of the significantly changed gene (up- and down-regulated) were identified using EnrichR (33).
Sterol measurement (liquid chromatography–tandem mass spectrometry)
Oxysterols and sterol sulfates were extracted from liver tissue homogenates (25 mg/10× of ddH2O) using a modified Folch extraction as described earlier (34, 35). Cholesterol sulfate-D7, chenodeoxycholic acid-D5, and 25-hydroxycholesterol-D6 internal standards (each 200 ng/ml) (CDN Isotopes, Quebec, Canada and Avanti Polar Lipids, AL, USA) were spiked in the Folch extraction solvent (methanol:dichloromethane, 2:1) containing butylated hydroxytoluene (50 μg/ml) before the analyte extraction. The organic layer containing the sterol derivatives was dried under a nitrogen evaporator (Organomation, MA, USA) and resuspended in 100 μl of methanol containing 0.2% formic acid (FA). Chromatographic separations of sterol derivatives from the extracts were achieved on pentafluorophenyl column (pursuit PFP, 150 mm by 2 mm, 3 μm in particle size, Agilent Technologies, CA, USA) by Shimadzu ultrafast liquid chromatography (Shimadzu, Kyoto, Japan). Binary mobile phase gradient using methanol (Eluent-A) and deionized water (ddH2O-Eluent-B), both containing 0.1% FA, was used to elute the compounds from the column within a 14-min analytical run. The column was operated at 40°C with a constant mobile phase flow rate of 250 μl/min. Eluted compounds were introduced into a Quadrupole ion trap mass spectrometer (API4000 QTRAP LC-MS/MS, AB Sciex, ON, Canada), where individual oxysterols and sterol sulfates were detected by multiple reaction monitoring mode (MRM) and pseudo-MRM mode. Cholesterol and oxysterols were analyzed in electrospray ionization (ESI)–positive mode, and sterol sulfates were analyzed in ESI-negative mode. Cholesterol and oxysterols produced [M + H-H2O] + prominent ions in ESI-positive mode, whereas sterol sulfates produced [M − H] ions in ESI-negative mode. Sterol (hydroxy and sulfo cocktail) standards (Cayman Chemicals, MI, USA and Avanti Polar Lipids, AL, USA) were used to construct eight-point (0.1 to 1000 ng/ml) calibration curves by plotting the area under the curve (AUC) response to the standard concentrations. These calibration curves were used for the measured AUC for analytes in the extracted samples to calculate measured quantities present in each sample. The AUC ratios were normalized to the wet weight of the tissue and reported as nanograms of analyte per milligram of tissue. Analyte values were multiplied by 1000 to report as picograms per milligram of tissue. Instrument control and quantification were performed using Analyst 1.4.2 and MultiQuant software (AB Sciex, Thornhill, ON, Canada). The mobile phase gradient program and MRM transitions of each analyte are provided in the Supplementary Materials (tables S6 and S7).
Immunofluorescence
Liver pieces were fixed (4% paraformaldehyde), cryosectioned (7 to 10 μm), and stained with anti-NRF2, anti-F4/80, anti-desmin, and anti-TGFβRII antibodies as detailed in the table S4. Images were acquired by using an LSM800 confocal laser microscope. TIFF files were imported into Adobe Photoshop, and the entire field was enhanced and sharpened by using the levels command.
Immunohistochemistry
Liver tissue pieces were fixed (10% formalin), processed, embedded (paraffin), and cut at the Johns Hopkins Pathology Core. Sections underwent antigen retrieval [GTX 30396; citrate buffer (pH 6.0)], blocking (normal goat serum), and staining with anti-SULT1E1 (sc-376009; 1:250 for mouse and 1:100 for human WD section) and anti-TGFβ (sc-130348; 1:250) overnight at 4°C, followed by biotinylated secondary antibody (BA9200; 1:1000) incubation for 1 hour at room temperature and developed using ABC reagent (PK-4000) and 3, 3′-diaminobenzidine (DAB) (BD-550880). Sections were counterstained with hematoxylin (14166S). Images were acquired using an Olympus light color microscope.
Histology (H&E) and Picro-Sirius red staining
Formalin-fixed liver tissues were cut and stained with H&E at the Johns Hopkins Pathology Core. The histology images were collected using an Olympus light color microscope station.
To detect and quantify fibrotic changes, sections were stained for collagen for 1 hour using Picro-Sirius red solution (0.1% Direct Red 80; Sigma-Aldrich, #365548) in 1.3% saturated picric acid (Sigma-Aldrich, #P6744) after 8 min of hematoxylin nuclei stain. Slides were washed with 0.5% acetic acid twice before dehydration and mounting. Images were acquired using an Olympus light color microscope. Sirius red positivity was quantitated using ImageJ.
Serum biochemical analysis
Following cardiac puncture, whole blood was collected in amber Microtainer tubes (Beckton Dickinson, Franklin Lakes, NJ). The serum was separated after centrifugation at 2400g for 10 min. Serum liver function tests and lipid panels were performed in the Phenotyping and Pathology Core Laboratory of JHUSOM.
Hepatic TG measurements
The Infinity TG assay kit (Thermo Fisher Scientific) was used to measure hepatic TG content. Liver tissue (30 to 40 mg) was homogenized in 500 μl of RIPA lysis buffer (Sigma-Aldrich) with 1× protease and phosphatase inhibitors (Roche), and samples were assayed in duplicate. Absorbance was measured at 540 nm using a FLUOstar Omega (BMG LabTech) plate reader. TG concentrations were calculated using a standard curve and then normalized to the mass of liver tissue.
Hepatic copper measurements
Hepatic Cu levels were measured by a PerkinElmer Atomic Absorption Spectrometer PinAAcle 900 T. Briefly, 50 to 60 mg of the liver were digested in 200 μl of HNO3 at 65°C for 2 hours. After digestion, the samples were cooled, 300 μl of HPLC-grade H2O was added, and samples were stored at 4°C. Before measurement, the sample was diluted with HPLC-grade H2O to decrease the HNO3 concentration to less than 2%. Samples were measured in triplicates, and Cu concentration was calculated using Cu standards and the dilution factor and then normalized to tissue weight.
Statistics
Prism 8 (GraphPad) was used for statistical analysis. As indicated in figure legends, one-way analysis of variance (ANOVA) with Tukey’s post hoc test or an unpaired t test was used. P < 0.05 was considered significant.
Acknowledgments
We thank N.J.H. for access and assistance for sterol analysis. We thank Psomagen Inc., Rockville, MD, for performing RNA sequencing and basic analysis. We thank B. Devenney for assisting in mice handling and triclosan injection. We also thank the Phenocore, Phenotyping, and Pathology Core Laboratory of JHUSOM for running serum biochemistry, Johns Hopkins Pathology Core for sectioning and H&E staining, and C. C. Talbot Jr. for help with the statistical analysis of data.
Funding: This work was supported by the NIH grant RO1-DK117396 to S.L. and J.P.H.
Author contributions: S.D., S.L., and J.P.H. designed the research. S.D., A.M., P.D., Y.D., and R.H. performed the research. S.D., A.G., S.M., P.D., K.L.G., N.J.H., S.L., and J.P.H. analyzed the data. S.D., S.L., and J.P.H. wrote the paper.
Competing interests: Patent: Targeting liver NRs as a treatment for WD; inventors: J.P.H and S.L. (PCT/US2015/054135 filed 10/7/2015). The authors declare that they have no other competing interests.
Data and materials availability: Data are available at the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE174015. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S9
Tables S1 to S7
REFERENCES AND NOTES
- 1.Gross J. B. Jr., Myers B. M., Kost L. J., Kuntz S. M., LaRusso N. F., Biliary copper excretion by hepatocyte lysosomes in the rat. Major excretory pathway in experimental copper overload. J. Clin. Invest. 83, 30–39 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boga S., Ala A., Schilsky M. L., Hepatic features of Wilson disease. Handb. Clin. Neurol. 142, 91–99 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Członkowska A., Litwin T., Karlinski M., Dziezyc K., Chabik G., Czerska M., D-penicillamine versus zinc sulfate as first-line therapy for Wilson’s disease. Eur. J. Neurol. 4, 599–606 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Merle U., Schaefer M., Ferenci P., Stremmel W., Clinical presentation, diagnosis and long-term outcome of Wilson’s disease: A cohort study. Gut 56, 115–120 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schilsky M. L., Roberts E. A., Hahn S., Askari F., Costly choices for treating Wilson’s disease. Hepatology 61, 1106–1108 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Zischka H., Einer C., Mitochondrial copper homeostasis and its derailment in Wilson disease. Int. J. Biochem. Cell Biol. 102, 71–75 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Mordaunt C. E., Kieffer D. A., Shibata N. M., Czlonkowska A., Litwin T., Weiss K., Zhu Y., Bowlus C. L., Sarkar S., Cooper S., Wan Y. Y., Ali M. R., LaSalle J. M., Medici V., Epigenomic signatures in liver and blood of Wilson disease patients include hypermethylation of liver-specific enhancers. Epigenetics Chromatin 12, 10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wooton-Kee C. R., Jain A. K., Wagner M., Grusak M. A., Finegold M. J., Lutsenko S., Moore D. D., Elevated copper impairs hepatic nuclear receptor function in Wilson’s disease. J. Clin. Invest. 125, 3449–3460 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wooton-Kee C. R., Robertson M., Zhou Y., Dong B., Sun Z., Kim K. H., Liu H., Xu Y., Putluri N., Saha P., Coarfa C., Moore D. D., Nuotio-Antar A. M., Metabolic dysregulation in the Atp7b−/− Wilson’s disease mouse model. Proc. Natl. Acad. Sci. U.S.A. 117, 2076–2083 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilmarth P. A., Short K. K., Fiehn O., Lutsenko S., David L. L., Burkhead J. L., A systems approach implicates nuclear receptor targeting in the Atp7b(−/−) mouse model of Wilson’s disease. Metallomics 4, 660–668 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamilton J. P., Koganti L., Muchenditsi A., Pendyala V. S., Huso D., Hankin J., Murphy R. C., Huster D., Merle U., Mangels C., Yang N., Potter J. J., Mezey E., Lutsenko S., Activation of liver X receptor/retinoid X receptor pathway ameliorates liver disease in Atp7B−/− (Wilson disease) mice. Hepatology 63, 1828–1841 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagasaka H., Milda T., Inui A., Inoue I., Tsukahara H., Komatsu H., Hiejima E., Fujisawa T., Yorifuji T., Hiranao K., Okajima H., Inomata Y., Fatty liver and antioxidant enzyme activities along with peroxisome proliferator-activated receptors γ and α expressions in the liver of Wilson’s disease. Mol. Genet. Metab. 107, 542–547 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Buiakova O. I., Xu J., Lutsenko S., Zeitlin S., Das K., Ross B. M., Mekios C., Scheinberg I. H., Gilliam T. C., Null mutation of the murine ATP7B (Wilson disease) gene results in intracellular copper accumulation and late-onset hepatic nodular transformation. Hum. Mol. Genet. 8, 1665–1671 (1999). [DOI] [PubMed] [Google Scholar]
- 14.Nagasaka H., Inoue I., Inui A., Komatsu H., Sogo T., Murayama K., Murakami T., Yorifuji T., Asayama K., Katayama S., Uemoto S., Kobayashi K., Takayanagi M., Fujisawa T., Tsukahara H., Relationship between oxidative stress and antioxidant systems in the liver of patients with Wilson disease: Hepatic manifestation in Wilson disease as a consequence of augmented oxidative stress. Pediatr. Res. 60, 472–477 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Kalita J., Kumar V., Ranjan A., Misra U. K., Role of oxidative stress in the worsening of neurologic Wilson disease following chelating therapy. Neuromolecular Med. 17, 364–372 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Bai Q., Zhang X., Xu L., Kakiyama G., Heuman D., Sanyal A., Pandak W. M., Yin L., Xie W., Ren S., Oxysterol sulfation by cytosolic sulfotransferase suppresses liver X receptor/sterol regulatory element binding protein-1c signaling pathway and reduces serum and hepatic lipids in mouse models of nonalcoholic fatty liver disease. Metabolism 61, 836–845 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ren S., Li X., Rodriguez-Agudo D., Gil G., Hylemon P., Pandak W. M., Sulfated oxysterol, 25HC3S, is a potent regulator of lipid metabolism in human hepatocytes. Biochem. Biophys. Res. Commun. 360, 802–808 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Königshofer P., Brusilovskaya K., Petrenko O., Hofer B. S., Schwabl P., Trauner M., Reiberger T., Nuclear receptors in liver fibrosis. Biochim. Biophys. Acta Mol. Basis Dis. 1867, 166235 (2021). [DOI] [PubMed] [Google Scholar]
- 19.Friedman S. L., Mechanisms of hepatic fibrogenesis. Gastroenterology 134, 1655–1669 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y. E., Non-Smad signaling pathways of the TGF-β family. Cold Spring Harb. Perspect. Biol. 9, a022129 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hervé J. C., Bourmeyster N., Rho GTPases at the crossroad of signaling networks in mammals. Small GTPases 6, 43–48 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barbosa A. C. S., Zhou D., Xie Y., Choi Y., Tung H., Chen X., Xu M., Gibbs R. B., Poloyac S. M., Liu S., Yu Y., Luo J., Liu Y., Xie W., Inhibition of estrogen sulfotransferase (SULT1E1/EST) ameliorates ischemic acute kidney injury in mice. J. Am. Soc. Nephrol. 31, 1496–1508 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toporova L., Grimaldi M., Boulahtouf A., Balaguer P., Assessing the selectivity of FXR, LXRs, CAR, and RORγ pharmaceutical ligands with reporter cell lines. Front. Pharmacol. 11, 1122 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang Y., Feng D., Ma X., Fan S., Gao Y., Fu K., Wang Y., Sun J., Yao X., Liu C., Zhang H., Xu L., Liu A., Gonzalenz F. J., Yang Y., Gao B., Huang M., Bi H., Pregnane X receptor regulates liver size and liver cell fate by yes-associated protein activation in mice. Hepatology 69, 343–358 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bracken A. P., Dietrich N., Pasini D., Hansen K. H., Helin K., Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 20, 1123–1136 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin-Mateos R., Assuncao T. M. D., Arab J. P., Jalan-Sakrikar N., Yaqoob U., Greuter T., Verma V. K., Mathison A. J., Cao S., Lomberk G., Mathurin P., Urrutia R., Huebert R. C., Shaw V. H., Enhancer of zeste homologue 2 inhibition attenuates TGF-β dependent hepatic stellate cell activation and liver fibrosis. Cell. Mol. Gastroenterol. Hepatol. 7, 197–209 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giannini E. G., Testa R., Savarino V., Liver enzyme alteration: A guide for clinicians. CMAJ 172, 367–379 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beaven S. W., Wroblewski K., Wang J., Hong C., Bensinger S., Tsukamoto H., Tontonoz P., Liver X receptor signaling is a determinant of stellate cell activation and susceptibility to fibrotic liver disease. Gastroenterology 140, 1052–1062 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muchenditsi A., Yang H., Hamilton J. P., Koganti L., Housseau F., Aronov L., Fan H., Pierson H., Bhattacharjee A., Murphy R., Sears C., Potter J., Wooton-Kee C. R., Lutsenko S., Targeted inactivation of copper transporter Atp7b in hepatocytes causes liver steatosis and obesity in mice. Am. J. Physiol. Gastrointest. Liver Physiol. 313, G39–G49 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turski M. L., Brady D. C., Kim H. J., Kim B., Nose Y., Counter C. M., Winge D. R., Thiele D. J., A novel role for copper in Ras/mitogen-activated protein kinase signaling. Mol. Cell. Biol. 32, 1284–1295 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu H., Guo H., Deng H., Cui H., Fang J., Zuo Z., Deng J., Li Y., Wang X., Zhao L., Copper induces hepatic inflammatory responses by activation of MAPKs and NF-κB signalling pathways in the mouse. Ecotoxicol. Environ. Saf. 201, 110806 (2020). [DOI] [PubMed] [Google Scholar]
- 32.Schmittgen T. D., Livak K. J., Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Kuleshov M. V., Jones M. R., Rouillard A. D., Fernandenz N. F., Duan Q., Wang Z., Koplev S., Jenkins S. L., Jagodnik K. M., Lachmann A., McDermott M. G., Monteiro C. D., Gundersen G. W., Ma’ayan A., Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Folch J., Lees M., Stanley G. H. S., A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226, 497–509 (1957). [PubMed] [Google Scholar]
- 35.Reis A., Rudnitskaya A., Blackburn G. J., Fauzi N. M., Pitt A. R., Spickett C. M., A comparison of five lipid extraction solvent systems for lipidomic studies of human LDL. J. Lipid Res. 54, 1812–1824 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S9
Tables S1 to S7