Abstract
Lysosomes receive extracellular and intracellular cholesterol and redistribute it throughout the cell. Cholesterol egress from lysosomes is critical for cholesterol homeostasis, and its failure underlies the pathogenesis of genetic disorders such as Niemann-Pick C (NPC) disease. Here we report that the BLOC one-related complex (BORC)-ARL8-homotypic fusion and protein sorting (HOPS) ensemble is required for egress of free cholesterol from lysosomes and for storage of esterified cholesterol in lipid droplets. Depletion of BORC, ARL8, or HOPS does not alter the localization of the lysosomal transmembrane cholesterol transporter NPC1 to degradative compartments but decreases the association of the luminal transporter NPC2 and increases NPC2 secretion. BORC-ARL8-HOPS depletion also increases lysosomal degradation of cation-independent (CI)-mannose 6-phosphate (M6P) receptor (MPR), which normally sorts NPC2 to the endosomal-lysosomal system and then is recycled to the trans-Golgi network. These defects likely result from impaired HOPS-dependent fusion of endosomal-lysosomal organelles and an uncharacterized function of HOPS in CI-MPR recycling. Our study demonstrates that the BORC-ARL8-HOPS ensemble is required for cholesterol egress from lysosomes by enabling CI–MPR-dependent trafficking of NPC2 to the endosomal-lysosomal system.
INTRODUCTION
Cholesterol homeostasis is essential to maintain normal functions of cells, tissues, and organs. Disruption of cholesterol homeostasis due to abnormal cholesterol levels or distribution has been linked to pathologies such as cardiovascular disease, neurodegenerative disorders and some types of cancer (Shibuya et al., 2015; Kuzu et al., 2016; Arenas et al., 2017; Huang et al., 2020). To maintain cholesterol homeostasis, cells have developed a complex machinery for the regulation of cholesterol uptake, synthesis, trafficking, and storage, which keeps cholesterol at normal levels and in the correct locations (Luo et al., 2020). Lysosomes, late endosomes, and autolysosomes are degradative organelles that play a central role in cholesterol homeostasis. Due to the similarity of their structures and functions, we henceforth refer to them collectively as “lysosomes.” Lysosomes process extracellular cholesterol contained in lipoproteins delivered to lysosomes through endocytosis and recycle intracellular cholesterol through autophagy. Lysosomal lipases produce free cholesterol by digesting lipoproteins and organelles. The free cholesterol is then redistributed into various cellular compartments. The cholesterol arriving at the endoplasmic reticulum (ER) is esterified to form cholesteryl esters and stored in lipid droplets (LDs). The level of ER cholesterol is sensed by the SREBP2 pathway that regulates cholesterol synthesis (Brown and Goldstein, 1997). Thus the trafficking of lysosomal cholesterol integrates cholesterol uptake, distribution, storage, and synthesis to control cholesterol homeostasis. In addition, recent studies have shown that lysosomal cholesterol transport regulates mTORC1 signaling (Castellano et al., 2017; Davis et al., 2021) and lysosome positioning (Willett et al., 2017), revealing new functions of lysosomal cholesterol trafficking in the regulation of cellular activities.
Cholesterol egress from lysosomes requires two Niemann-Pick C (NPC) proteins, NPC1 and NPC2 (Carstea et al., 1997; Davies et al., 2000; Naureckiene et al., 2000). Mutations in these proteins cause NPC disease characterized by accumulation of free cholesterol in lysosomes. NPC2 is a soluble lysosomal luminal protein that binds cholesterol and shuttles it to the adjacent inner surface of the lysosomal membrane. NPC1 is a transmembrane protein that takes cholesterol from NPC2 and transports it across the lysosomal membrane (Li et al., 2016; Qian et al., 2020). Additionally, other lysosomal luminal and transmembrane proteins, such as SapA, PTCH1, LIMP2, LAMP1, and LAMP2, contribute to cholesterol binding and transport (Eskelinen et al., 2004; Schneede et al., 2011; Popovic et al., 2012; Zhang et al., 2018; Meng et al., 2020). Since lysosomal proteins are key factors for cholesterol egress, correct trafficking of these proteins from their site of synthesis in the ER to lysosomes is critical for the maintenance of cholesterol homeostasis.
Newly synthesized lysosomal proteins are transported from the ER to the Golgi complex and the trans-Golgi network (TGN) where they are sorted into transport carriers for delivery to the endosomal-lysosomal system. Most lysosomal luminal proteins, including NPC2, are modified with mannose 6-phosphate (M6P) residues (Naureckiene et al., 2000), which are recognized by transmembrane M6P receptors (MPRs) at the TGN. Two such receptors, the cation-independent MPR (CI-MPR) and cation-dependent MPR (CD-MPR), have been described. MPRs and other transmembrane proteins destined for lysosomes are packaged into transport carriers and delivered to the endosomal-lysosomal system either directly (Harter and Mellman, 1992; Waguri et al., 2003; Ang et al., 2004) or indirectly after fusion with the plasma membrane and endocytosis (Lippincott-Schwartz and Fambrough, 1986; Braun et al., 1989; Janvier and Bonifacino, 2005; Chen et al., 2017). After releasing the soluble cargo proteins at the acidic pH of the endosomal-lysosomal system, MPRs are recycled back to the TGN for reuse. Failure of these trafficking pathways likely causes defects in lysosomal cholesterol transport. For example, cholesterol accumulation in endolysosomes has been found in MPR-knockdown (KD) cells (Wei et al., 2017; Willenborg et al., 2005) and cells with mutations in the TGN-tethering factor GARP, which accepts MPR carriers from endolysosomes (Wei et al., 2017).
Tethering factors are a group of proteins and protein complexes that establish physical links between transport carriers and acceptor organelles for subsequent membrane fusion. The homotypic fusion and protein sorting (HOPS) complex is a tethering factor associated with lysosomes. HOPS comprises six subunits named VPS11, VPS16, VPS18, VPS33, VPS39, and VPS41 (Solinger and Spang, 2013; Spang, 2016), two of which (VPS39 and VPS41) are specific to HOPS and the other four are shared with an early endosomal complex named CORVET (Peplowska et al., 2007). Two lysosomal small GTPases, RAB7 and ARL8, mediate HOPS association with lysosomes (Hickey et al., 2009; Stroupe et al., 2009; Khatter et al., 2015a; Jongsma et al., 2020a) and are required for HOPS-mediated membrane fusion of lysosomes with other lysosomes (homotypic fusion) (Seals et al., 2000; Stroupe et al., 2006) between lysosomes and late endosomes (Pols et al., 2013) and between lysosomes and autophagosomes (heterotypic fusion) (Jiang et al., 2014; McEwan et al., 2015; Jia et al., 2017). In previous work, we identified the lysosome-associated BLOC one-related complex (BORC), which recruits ARL8 to lysosomes (Niwa et al., 2017; Pu et al., 2015) and thus regulates the function of HOPS in lysosome-autophagosome fusion(Jia et al., 2017). BORC is composed of eight subunits named myrlysin (BORCS5), lyspersin (BORCS6), diaskedin (BORCS7), KXD1 (BORCS4), MEF2BNB (BORCS8), snapin (BORCS3; BLOC1S7), BLOS1 (BORCS1; BLOC1S1), and BLOS2 (BORCS2; BLOC1S2), the latter three of which are shared with another complex named biogenesis of lysosome-related organelles complex (BLOC1) (Pu et al., 2015). In addition to regulating HOPS-dependent lysosomal fusion, BORC mediates lysosome movement by promoting ARL8-dependent coupling of the motor proteins kinesin-1 and -3 to lysosomes (Pu et al., 2015; Guardia et al., 2016).
In this study, we show that BORC, ARL8, and HOPS are required for cholesterol egress from lysosomes. CRISPR knockout (KO) of BORC, ARL8, or HOPS increases accumulation of free cholesterol in lysosomes and decreases storage of cholesteryl esters in LDs. Mechanistic analyses show that these defects result from decreased delivery of NPC2 to lysosomes due to delayed transport from Rab7-positive endosomes to lysosomes and abnormal degradation of MPRs. We propose that the BORC-ARL8-HOPS ensemble not only enables NPC2 transport from endosomes to the lysosome but also maintains a normal level of CI-MPR probably by promoting CI-MPR recycling from endosomes.
RESULTS
BORC KO causes accumulation of free cholesterol in lysosomes
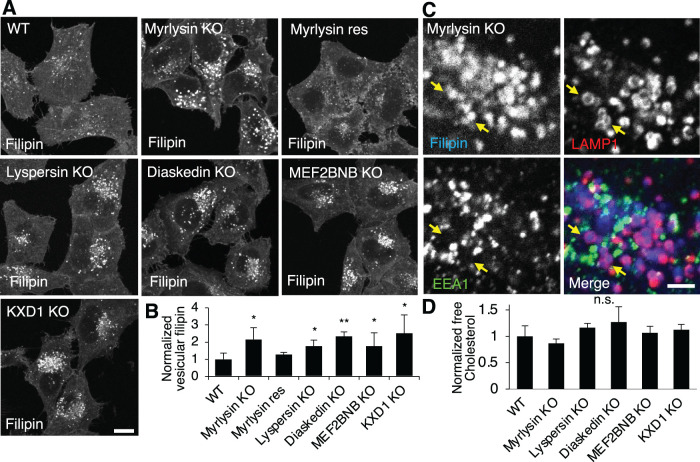
To examine whether BORC is involved in cholesterol metabolism, we used the fluorogenic compound filipin (Maxfield and Wustner, 2012) to visualize cellular free cholesterol in wildtype (WT) and BORC-KO cells. We observed that the individual deletion of genes encoding the BORC subunits myrlysin, lyspersin, diaskedin, MEF2BNB, or KXD1 by CRISPR-Cas9 (Jia et al., 2017; Pu et al., 2017; Pu et al., 2015) caused increased cholesterol accumulation in a population of cellular vesicles (Figure 1A). Quantification of the vesicular cholesterol signals revealed approximately twofold increases in BORC-subunit-KO cells compared with WT cells (Figure 1, A and B). Stable expression of the BORC subunit myrlysin in myrlysin-KO cells restored normal levels of vesicular cholesterol (Figure 1, A and B), ruling out CRISPR-Cas9 off-target effects. The cholesterol-enriched vesicles were mainly localized to the perinuclear area, consistent with the redistribution of lysosomes in BORC-KO cells (Pu et al., 2015). To determine whether these vesicles were lysosomes, we performed immunostaining for the lysosomal membrane protein LAMP1 and the early endosomal protein EEA1 after filipin staining. The results showed that the filipin-stained vesicles were positive for LAMP1 but not EEA1 (Figure 1C), indicating that cholesterol was accumulated in lysosomes. Deletion of BORC did not significantly change levels of free cholesterol in the cells, as shown by an enzymatic assay (Figure 1D), indicating that BORC controls cholesterol cellular distribution rather than total levels.
FIGURE 1:
BORC-subunit KO increases lysosomal cholesterol. (A, B) WT, BORC subunit-KO, and myrlysin-rescued (res) HeLa cells were fixed, stained with filipin, and analyzed by confocal microscopy. The intensity of vesicular filipin was quantified (n = 106, 83, 76, 46, 39, 86, and 122) and normalized to the intensity in WT cells. (C) Myrlysin-KO cells were treated as in A and costained with antibodies to EEA1 and LAMP1 for early endosomes and lysosomes, respectively. Part of a cell is shown at high magnification, and LAMP1-wrapped filipin-positive vesicles are pointed by arrows. (D) Cell lysates were extracted from the indicated cells and subjected to enzymatic measurement of free cholesterol. Bar graphs are presented as mean ± SD; p values were determined by Student’s t test. *p < 0.05, **p < 0.01, n.s., not significant (vs. WT). Scale bars, 5 μm.
BORC KO decreases storage of esterified cholesterol in LDs
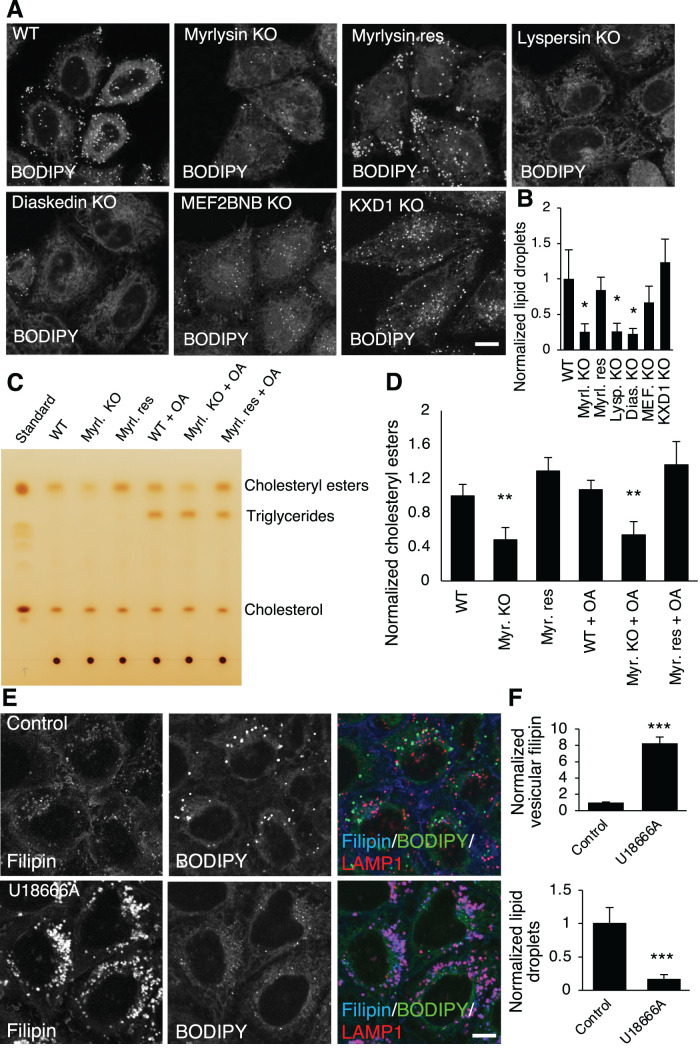
Since cells store excess cholesterol as cholesteryl esters in LDs, the above findings prompted us to ask whether BORC KO influences LDs. To address this question, we stained LDs with the neutral lipid dye BODIPY 493/503 and found decreased total intensity of LDs in some (though not all) BORC-subunit-KO cells relative to WT cells (Figure 2A). Quantification of BODIPY 493/503 intensity revealed that the total intensity of LDs decreased by ∼70% in myrlysin-, lyspersin-, or diaskedin-KO cells (Figure 2B). The decrease of LDs could be restored by re-expression of myrlysin in myrlysin-KO cells (Figure 2, A and B), similarly to the restoration of free cholesterol distribution (Figure 1, A and B). The LD decrease in MEFNB2B-KO cells was not statistically significant, and KXD1-KO cells did not show a decrease in LDs (Figure 2B). Deletion of MEF2BNB or KXD1 slightly decreased myrlysin but not the other BORC subunits (Supplemental Figure S1A). However, loss of either MEF2BNB or KXD1 dissociated ARL8b from the lysosome (Supplemental Figure S1B), similar to what happens in myrlysin-KO (Pu et al., 2015) (Supplemental Figure S1B) and other BORC subunit-KO cells (Pu et al., 2017). This result suggests that MEF2BNB and KXD1 are important for BORC function, consistent with their roles in lysosome cholesterol egress (Figure 1, A and B). We do not understand why the effects on LD levels differ between deletion of MEF2BNB or KXD1 and deletion of the other BORC subunits and will investigate this in the future.
FIGURE 2:
BORC-subunit KO decreases cellular cholesteryl esters. (A, B) WT, BORC-subunit-KO, and myrlysin-rescued HeLa cells were stained with the LD dye BODIPY493/503 (BODIPY). Live-cell imaging was performed, and BODIPY signals were quantified (n = 104, 83, 125, 114, 74, 68, and 65) and normalized to the intensity in WT cells. Myrl., myrlysin; Lysp., lyspersin; Dias., diaskedin; MEF., MEF2BNB. (C, D) Cell lysates were prepared from WT, myrlysin-KO (Myrl. KO), and myrlysin-rescued (Myrl. Res) cells treated with/without 0.3 mM OA for 3 h, and the total lipids were extracted from the lysates and subjected to TLC. Standard samples of cholesterol esters, triglycerides, and cholesterol were used to identify the bands. The intensity of cholesterol esters was quantified from three independent experiments and normalized to the intensity in WT untreated cells. (E, F) HeLa cells were treated with/without 1 μM of the NPC1 inhibitor U18666A for 16 h and fixed for staining with filipin, BODIPY, and LAMP1 antibody. The intensities of BODIPY and vesicular filipin were quantified (n = 368 and 283) and normalized to the intensities in control cells. Bar graphs are presented as mean ± SD; p values were determined by Student’s t test. *p < 0.05, **p < 0.01, ***p < 0.001 (vs. WT). Scale bars, 5 μm.
To examine if the cause for the decrease of LDs was related to the alteration of cholesterol distribution, we analyzed cellular neutral lipids using thin-layer chromatography (TLC) and iodine-vapor staining. The majority of neutral lipids in HeLa cells were cholesteryl esters, and the cholesteryl ester levels in myrlysin-KO cells were decreased by ∼50% relative to WT cells (Figure 2, C and D). The myrlysin-rescued cells exhibited cholesteryl ester levels similar to WT cells (Figure 2, C and D). Consistent with the enzymatic assay for free cholesterol (Figure 1D), the TLC results showed similar total levels of free cholesterol in all the cells. Another major type of neutral lipids found in LDs, triglycerides, were not visible in the initial assays. To boost triglyceride synthesis, we treated cells with the fatty acid oleic acid (OA) and analyzed the neutral lipid content using the same method. The addition of OA increased triglycerides, but there was no obvious difference in the triglyceride levels between WT, myrlysin-KO, and myrlysin-rescued cells (Figure 2C). In addition, OA did not change cholesteryl ester levels (Figure 2, C and D). These results indicated that the decrease of LDs in BORC-KO cells was due to the decrease of cholesteryl ester levels.
The above results suggest that cholesteryl esters are an important source for LD biogenesis in HeLa cells, and that accumulation of cholesterol in lysosomes in BORC-KO cells causes a reduction of cholesterol influx into LDs. We tested this interpretation through inhibition of the cholesterol transporter NPC1 in WT cells by using U18666A (Lu et al., 2015). As expected, U18666A caused a dramatic increase in accumulation of free cholesterol in lysosomes and a concomitant decrease in LDs (Figure 2, E and F). This observation supports the notion that interference with lysosomal cholesterol trafficking influences LD biogenesis in HeLa cells.
ARL8 regulates lysosomal cholesterol transport through the HOPS complex
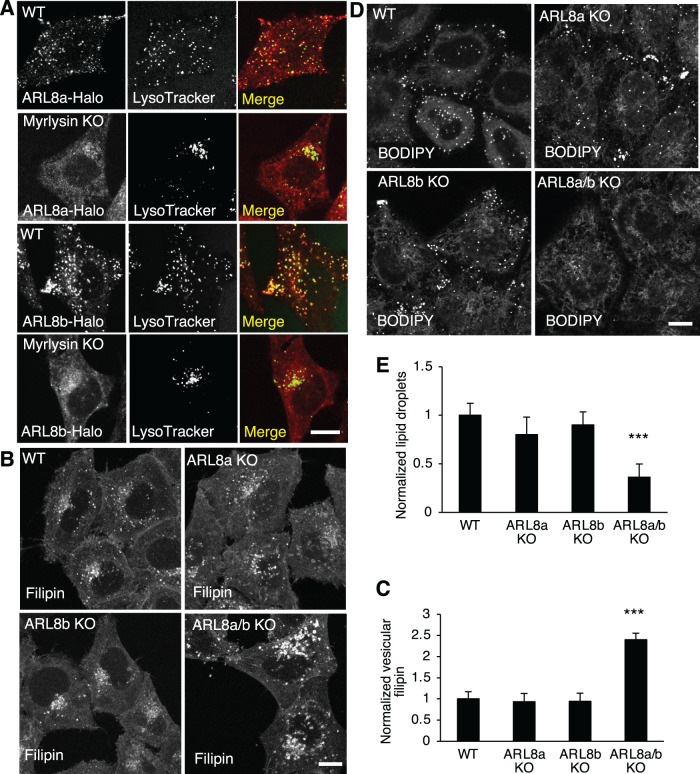
We next investigated how BORC is involved in lysosomal cholesterol trafficking. Previous studies showed that BORC recruits the small GTPase ARL8 to lysosomes, and deletion of BORC releases ARL8 from lysosomes to the cytosol. Due to the lack of suitable antibodies for immunostaining, in previous studies ARL8 was detected by the transfection of tagged constructs. To examine the localization of endogenous ARL8, we used CRISPR-Cas9 to fuse a Halo tag to the ARL8 isoforms ARL8a and ARL8b, and detected the Halo signals using Halo dyes. Consistent with the previous findings, the Halo-fused ARL8a and ARL8b localized to lysosomes in WT cells, but were mostly diffuse in myrlysin-KO cells (Figure 3A). These results confirmed that both ARL8a and ARL8b are recruited to lysosomes by BORC.
FIGURE 3:
ARL8 KO increases lysosomal cholesterol and decreases LDs. (A) Halo tag was fused to endogenous ARL8a and ARL8b isoforms by CRISPR-Cas9 in WT or myrlysin-KO HeLa cells. Halo tag and lysosomes were visualized by applying Halo dye and LysoTracker in live-cell imaging experiments, respectively. Cells as indicated were fixed and then stained with filipin (B) or stained with BODIPY (D). Fluorescence signals were observed by confocal microscopy, quantified, and normalized to the signals in WT cells. For filipin quantification, n = 105, 111, 131, and 127. For BODIPY quantification, n = 119, 88, 77, and 60. (E, F) Bar graphs are presented as mean ± SD; p values were determined using Student’s t test. ***p < 0.001 (vs. WT). Scale bars, 5 μm.
To examine whether ARL8 is involved in BORC-regulated cholesterol trafficking, we generated ARL8a-KO and ARL8a/b-double KO cells by CRISPR-Cas9, in addition to the previously generated ARL8b-KO cells (Guardia et al., 2016), and confirmed the KO by Sanger sequencing and immunoblotting (Supplemental Figure S2A). KO of either ARL8a or ARL8b did not significantly alter lysosomal cholesterol (Figure 3, B and C) or LDs (Figure 3, D and E). However, deleting both ARL8a and ARL8b increased lysosomal cholesterol (Figure 3, B and C) and decreased LDs (Figure 3, D and E), demonstrating that ARL8a and ARL8b have redundant roles in the regulation of cholesterol trafficking.
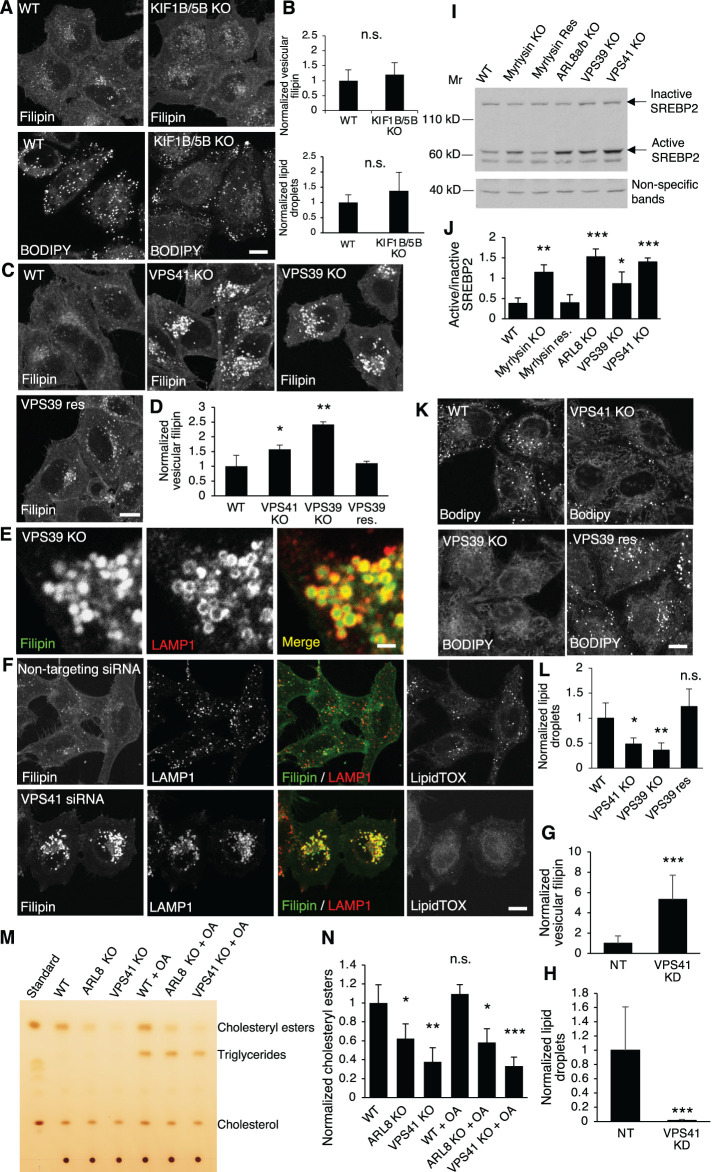
These findings prompted us to examine the downstream effectors of ARL8. Known ARL8 effectors include the motor proteins kinesin-1 and kinesin-3, which are coupled by ARL8 to lysosomes and thus mediate lysosome anterograde movement (Guardia et al., 2016; Pu et al., 2015; Rosa-Ferreira and Munro, 2011). Our previous studies showed that deleting the ubiquitous KIF5B and KIF1B isoforms of kinesin-1 and kinesin-3 clustered lysosomes in the perinuclear area (Guardia et al., 2016). However, double deletion of KIF5B and KIF1B did not change either cholesterol distribution or LD number (Figure 4, A and B), indicating that the function of ARL8 in cholesterol transport was not mediated by kinesins and, therefore, lysosome positioning or motility.
FIGURE 4:
HOPS-subunit KO increases lysosomal cholesterol and decreases LDs. (A, B) WT and KIF1B/5B-KO cells were fixed and stained with filipin or BODIPY and visualized by confocal microscopy. The signals were quantified and normalized to the signals in WT cells. For filipin quantification, n = 39 and 47. For BODIPY quantification, n = 55 and 60. Scale bar, 5 μm. (C, D) WT, VPS41-KO, VPS39-KO, and VPS39-rescued (res) cells were fixed and stained with filipin. Vesicular filipin intensity was quantified (n = 46, 35, 39, and 58) and normalized to WT cells. Scale bar, 5 μm. (E) VPS39-KO cells were fixed and costained with filipin and anti-LAMP1 antibody. Part of an enlarged cell is shown. Scale bar, 1 μm. (F-H) HeLa cells were transfected twice sequentially with a 1-d interval to deliver nontargeting siRNA or siRNA targeting VPS41 into cells, and the cells were fixed at day 5 postfirst transfection. Filipin, LAMP1 antibody, and LipidTOX staining was used to visualize free cholesterol, lysosomes, and LDs, respectively. Scale bar, 5 μm. The signals were quantified (n = 22 and 19) and normalized to the signals in the control cells. (I, J) Cell lysates were prepared from the indicated cells and subjected to immunoblotting. Quantification was from three independent experiments. (K, L) LDs were stained with BODIPY in the indicated cells, and the signals were quantified and normalized to the signals in WT cells. Scale bar, 5 μm. (M, N) Cell lysates were prepared from the indicated cells treated with/without 0.3 mM OA for 3 h, and total lipids were extracted from the lysates and subjected to TLC. Standard samples of cholesterol esters, triglycerides, and cholesterol were run alongside to identify the bands. The intensity of cholesterol esters was quantified from three independent experiments and normalized to that in WT untreated cells. Values are represented as mean ± SD; p values were determined using Student’s t test. *p < 0.05, **p < 0.01, ***p < 0.001, n.s., not significant (vs. WT).
Another effector of ARL8 is the HOPS complex, which is recruited to lysosomes by ARL8 through interaction with VPS41 (Khatter et al., 2015a). To determine the role of HOPS in lysosomal cholesterol trafficking, we knocked out the HOPS-specific subunits VPS39 and VPS41 individually by CRISPR-Cas9. Sanger sequencing and immunoblotting confirmed the deletion (Supplemental Figure S2B). We observed that deletion of either VPS39 or VPS41 significantly increased vesicular cholesterol, and the increase could be restored by stable expression of VPS39 cDNA in VPS39-KO cells (Figure 4, C and D). We then costained cells with filipin and a LAMP1 antibody to confirm that the cholesterol-enriched vesicles were lysosomes (Figure 4E). A similar observation was made in VPS41-KD cells generated by transfection of siRNA targeting VPS41 (Figure 4, F and G). These results indicated that lysosomal cholesterol egress requires BORC, ARL8, and HOPS. Moreover, KO of components of this supercomplex activated the SREBP2 pathway (Figure 4I,J), which senses ER cholesterol and regulates cholesterol synthesis and uptake. Activated SREBP2 indicated reduced ER cholesterol, which was probably caused by deficient lysosomal cholesterol egress. In addition to accumulating cholesterol in lysosomes, deletion of VPS39 or VPS41 (Figure 4, K and L) or KD of VPS41 (Figure 4, F and H) decreased LDs, consistent with the observations in the BORC-KO (Figure 2, A and B) and ARL8-KO cells (Figure 3, D and E). Furthermore, lipid analysis by TLC demonstrated significantly decreased cholesteryl ester levels in ARL8-KO and VPS41-KO cells, and the decrease was not altered by addition of OA (Figure 4, M and N). Therefore the BORC/ARL8/HOPS are required for both egress of free cholesterol from lysosomes and storage of cholesteryl esters in LDs.
BORC-ARL8-HOPS Ensemble Enables NPC2 Trafficking to Lysosomes
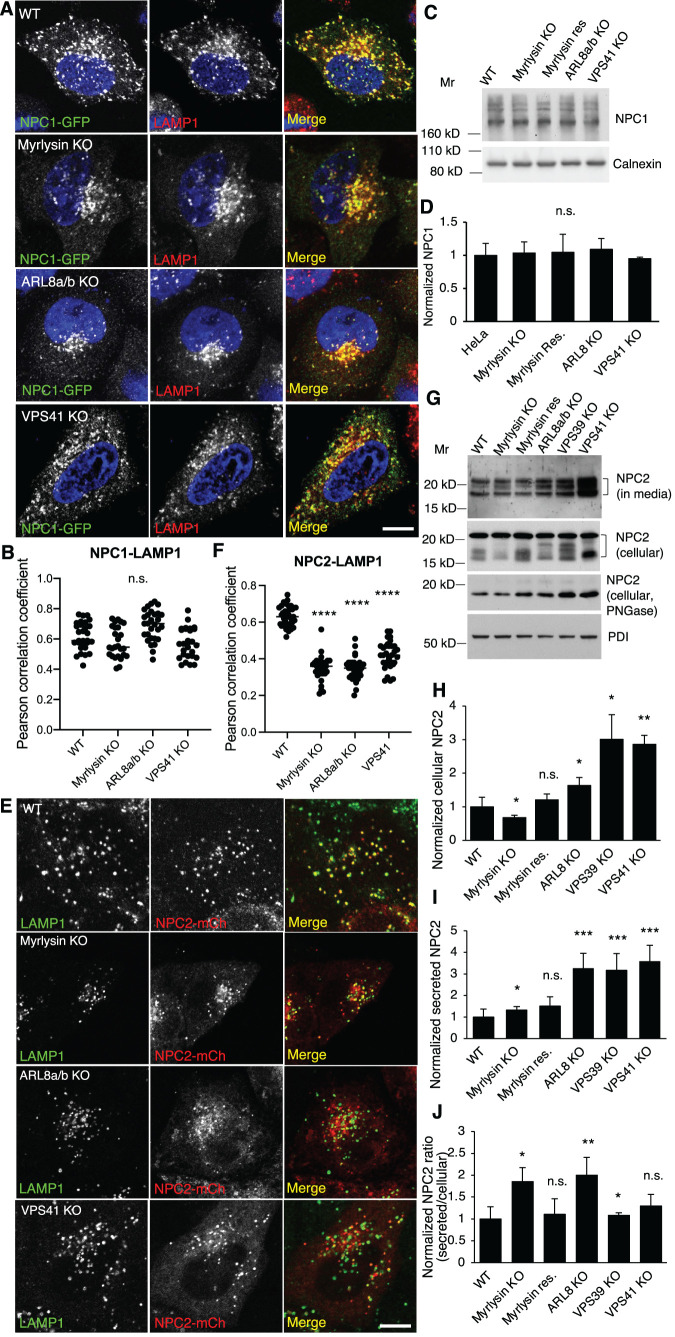
To determine how BORC, ARL8, and HOPS regulate lysosomal cholesterol egress, we examined the status of the cholesterol transport NPC proteins NPC1 and NPC2 in WT and KO cells. We found that most NPC1-GFP colocalized with LAMP1 in WT, myrlysin-, ARL8-, and VPS41-KO cells (Figure 5, A and B). Furthermore, the levels of endogenous NPC1, examined by immunoblotting, were similar in WT, myrlysin-, ARL8-, and VPS41-KO cells (Figure 5, C and D). In contrast, NPC2-mCherry (Huang et al., 2014) showed perfect colocalization with LAMP1 in WT cells, but only partial colocalization in myrlysin-, ARL8-, and VPS41-KO cells (Figure 5, E and F). We also examined NPC2-mCherry colocalization in living cells by labeling lysosomes with LysoTracker (Supplemental Figure S3A). Similar to what was observed in the fixed cells, there was less colocalization between NPC2-mCherry and LysoTracker in myrlysin-, VPS39-, and VPS41-KO cells than in WT cells. These results indicated reduced presence of NPC2 in lysosomes in myrlysin-, ARL8-, and HOPS-deficient cells, suggesting a defect in NPC2 trafficking.
FIGURE 5:
Disruption of the BORC/ARL8/HOPS causes NPC2 mislocalization and increases NPC2 secretion. (A, B) WT and ARL8a/b-KO cells were transfected with a plasmid encoding NPC1-GFP. At 24 h posttransfection cells were fixed, immunostained for LAMP1, and imaged by fluorescence microscopy; 2330 cells in each group from three independent experiments were analyzed. NPC1-LAMP1 colocalization was quantified (n = 29, 26, 29, and 28) and presented as Pearson correlation coefficient. (C, D) Cell lysates were prepared from the indicated cells and subjected to SDS–PAGE and immunoblotting. Quantification was from three independent experiments. (E, F) The indicated cells were transfected with a plasmid encoding NPC2-mCherry, fixed at 24 h posttransfection, and stained with antibodies to mCherry and LAMP1. Fluorescence images were obtained by live-cell imaging with a confocal microscope. Cells in each group from three independent experiments were analyzed. NPC2-LAMP1 colocalization was quantified (n = 36, 31, 35, and 33) and presented as Pearson correlation coefficient. (G-J) Serum-free culture media were collected after 6 h incubation. Cells were recovered in complete media for 2 h and lysed to prepare cell lysates. Aliquots of cell lysates were treated with PNGase. Culture media, cell lysates, and PNGase-treated cell lysates from the indicated cells were subjected to immunoblotting. Multiple NPC2 bands were indicated with brackets. Quantification was from three independent experiments. Bar graphs are presented as mean ± SD; p values were determined by Student’s t test. *p < 0.05, **p < 0.01, ***p < 0.001, n.s., not significant (vs. WT). Scale bars, 5 μm.
We further examined the levels of endogenous NPC2 in the cells and culture media by immunoblotting. Since NPC2 is a glycosylated protein, it appears as multiple bands in immunoblotting and as a single band after treatment with the glycosidase PNGase. The results showed that cellular NPC2 increased in ARL8-KO and HOPS-KO cells and decreased in myrlysin-KO cells relative to WT cells (Figure 5, G and H). Furthermore, the amount of NPC2 secreted into the culture medium increased in myrlysin-, ARL8- and HOPS-KO cells, and normal secretion could be restored in myrlysin-rescued cells (Figure 5, G and I). Finally, the ratio of secreted to cellular NPC2 increased in myrlysin-, ARL8- and VPS39-KO cells relative to WT cells (Figure 5J). These experiments demonstrated that the efficiency of NPC2 transport to lysosomes was reduced in myrlysin-, ARL8- and HOPS-KO cells, resulting in a higher proportion of NPC2 being secreted into the culture medium. This result is consistent with the reduced colocalization of NPC2 with LAMP1 in KO cells. However, the KO cells seemed to compensate for the reduced sorting of NPC2 to lysosomes by increasing the total expression levels of the protein.
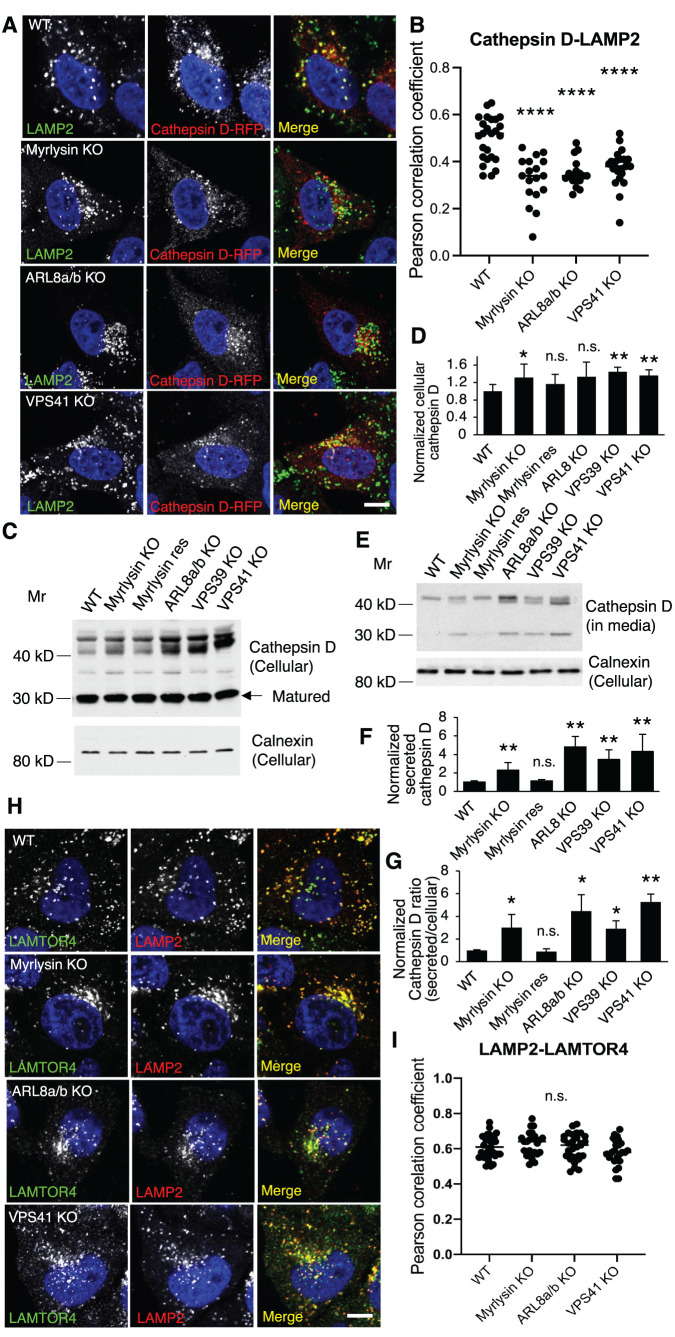
The distinct effects of the BORC/ARL8/HOPS KOs on NPC1 and NPC2 led us to speculate that lysosomal luminal proteins were more affected than the transmembrane proteins in the processes of their sorting and trafficking. To determine the generality of these effects, we examined the distribution of another lysosomal luminal protein, the acid hydrolase cathepsin D (Barrett, 1970). Cathepsin D exhibited decreased colocalization with lysosomes in myrlysin-, ARL8-, and VPS41-KO cells (Figure 6, A and B; Supplemental Figure S3B), as was the case with NPC2 (Figure 5, E and F; Supplemental Figure S3A). We also observed that the protein levels of cellular cathepsin D increased in the myrlysin-, VPS39-, and VPS41-KO cells (Figure 6, C and D), and those of secreted cathepsin D increased in myrlysin-, ARL8-, VPS39- and VPS41-KO cells (Figure 6, E and F). The ratios of secreted cathepsin D to cellular cathepsin D were significantly increased in myrlysin-, ARL8-, VPS39-, and VPS41-KO cells relative to WT cells (Figure 6G), indicating an alteration of cathepsin D trafficking. In these assays, we used the lysosome transmembrane protein LAMP1 as a lysosome marker. To determine whether the transmembrane LAMPs were normally distributed in the KO cells, we examined another lysosomal membrane protein, LAMTOR4, which is a subunit of the Ragulator complex (Bar-Peled et al., 2012; Sancak et al., 2010) and is anchored to lysosomal membrane by N-terminal myristoyl and palmitoyl groups in the LAMTOR1 subunit (Nada et al., 2009). We found that deletion of myrlysin, ARL8, or VPS41 did not change the colocalization of LAMP2 and LAMTOR4 (Figure 6, H and I), verifying the use of LAMPs as lysosomal markers. These results indicated that disruption of the BORC/ARL8/HOPS causes a trafficking defect of lysosomal luminal proteins.
FIGURE 6:
Disruption of BORC/ARL8/HOPS causes missorting of lysosomal luminal protein cathepsin D but not transmembrane proteins. (A, B) The indicated cells were transfected with plasmids encoding cathepsin D-RFP and fixed for immunostaining with antibodies to RFP and LAMP2 at 24 h posttransfection. Cells in each group were analyzed (n = 25, 18, 18, and 22) for cathepsin D-LAMP2 colocalization. The distribution of Pearson correlation coefficient was presented as Pearson correlation coefficient. (C-G) Serum-free culture media were collected after 6 h incubation. Cells were recovered in complete media for 2 h and lysed to prepare cell lysates. The culture media and cell lysates from the indicated cells were subjected to immunoblotting. Quantification was from five independent experiments. (H, I) The indicated cells were fixed for immunostaining of endogenous LAMP2 and LAMTOR4; 20–31 cells in each group from three independent experiments were analyzed. LAMP2-LAMTOR4 colocalization was quantified (n = 31, 25, 29, and 25) and presented as Pearson correlation coefficient; p values were determined by Student’s t test. *p < 0.05, **p < 0.01, ****p < 0.0001, n.s., not significant (vs. WT). Scale bars, 5 μm.
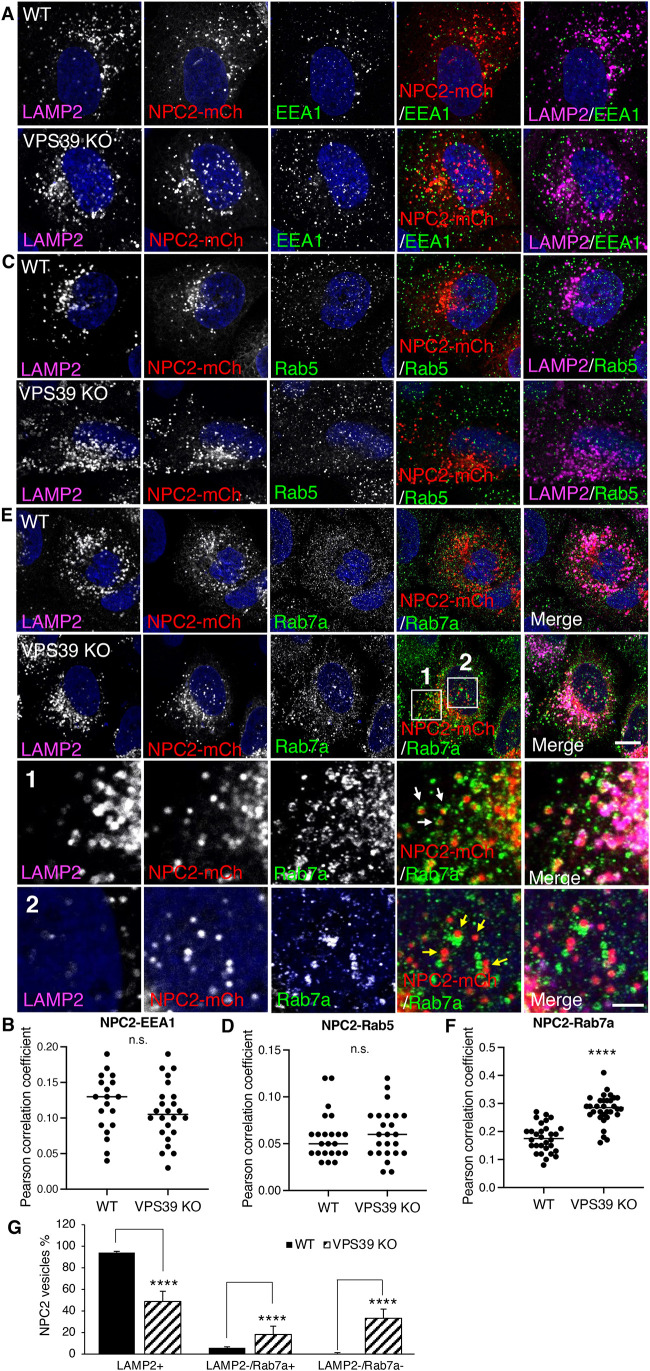
A known function of HOPS is to mediate the fusion of late endosomes and lysosomes (Pols et al., 2013). To determine whether the mislocalization of NPC2 was due to a delayed transport from endosomes to lysosomes, we examined the NPC2 localization using the endosome markers EEA1, Rab5, and Rab7a, as well as the lysosome marker LAMP2. There were very few NPC2 vesicles that colocalized with EEA1 or Rab5 early endosomes in both WT and VPS39-KO cells (Figure 7, A–D), and there was no significant difference in the colocalization of EEA1/Rab5 and LAMP2 between WT and VPS39-KO cells (Figure 7, B and D). In contrast, a small population of NPC2 vesicles that were LAMP2 negative (5.7 ± 2.3%) were colocalized with Rab7a in WT cells, and this population significantly increased in VPS39-KO cells (18.2 ± 7.7%) (Figure 7, E–G). This observation indicates that deletion of HOPS retained part of NPC2 in Rab7a endosomes, which is likely due to the deficiency of endosome-lysosome fusion. Unexpectedly, we also observed a considerable increase of LAMP2-negative NPC2 vesicles that were Rab7a negative in VPS39-KO cells (33.1 ± 8.6%) relative to WT cells (0.32 ± 0.44%). Since the mislocalized NPC2 was not in EEA1 or Rab5 endosomes (Figure 7, A–D), these LAMP2-negative and Rab7a-negative NPC2 vesicles were probably not in the endosome-lysosome system.
FIGURE 7:
Disruption of HOPS decreases NPC2 entering endosome-lysosome system. WT and VPS39-KO cells were transfected with the plasmids encoding NPC2-mCherry and fixed for immunostaining with antibodies to LAMP2 and EEA1 (A), or Rab5 (C), or Rab7a (E) at 24 h posttransfection. Scale bars, 5 μm. Parts of a cell are shown at high magnification, and Rab7a-positive and -negative NPC2 vesicles are pointed by arrows in panels 1 and 2, respectively. Scale bar, 1 μm (E) Cells were analyzed for the colocalization between LAMP2 and EEA1 (B, n = 19 and 24), LAMP2 and Rab5 (D, n = 23 and 24), and LAMP2 and Rab7a (F, n = 30, and 32). (G) Cells in E were further analyzed for the quantities of LAMP2-positive, LAMP2-negative/Rab7a-positive, and LAMP2-negative/Rab7a-negative NPC2 vesicles in WT (n = 8) and VPS39-KO (n = 7) cells. The results were shown as the percentage of each group NPC2 vesicles out of the total number of NPC2 vesicles; p values were determined by Student’s t test. ****p < 0.0001, n.s., not significant (vs. WT).
BORC-ARL8-HOPS ensemble is required for MPR recycling
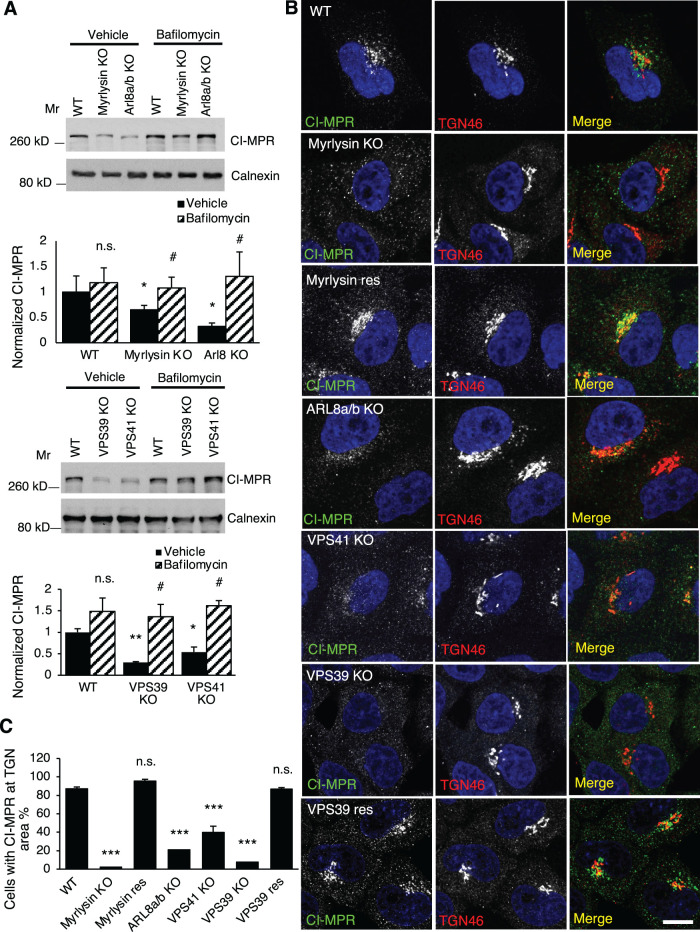
Like most lysosomal luminal proteins, NPC2 is modified with M6P groups and thus relies on MPRs for transport from the TGN to endosomes (Naureckiene et al., 2000). Deficient MPRs cause NPC2 mislocalization and secretion and cholesterol accumulation in lysosomes (Wei et al., 2017; Willenborg et al., 2005). The altered distribution of NPC2 and cholesterol in BORC/ARL8/HOPS-KO cells led us to examine the MPRs in these cells. By immunoblotting, we found that the protein levels of CI-MPR decreased by 50–70% in myrlysin-, ARL8-, VPS39-, and VPS41-KO cells (Figure 8A). Incubation with the lysosomal acidification inhibitor bafilomycin A1 restored normal CI-MPR levels, indicating that the CI-MPR was degraded in acidic compartments (Figure 8A). This treatment did not significantly increase the CI-MPR levels in WT cells probably due to the shorter period of treatment (16 h) relative to the half-life of CI-MPR (27 h) (Arighi et al., 2004), suggesting the degradation of CI-MPR in the KO cells was faster than in WT cells.
FIGURE 8:
Disruption of BORC/ARL8/HOPS increases CI-MPR lysosomal degradation. (A) WT, myrlysin-KO, myrlysin-rescued (res), VPS41-KO, VPS39-KO, and VPS39-res cells were treated with 100 nM bafilomycin A1 or the same volume of DMSO (vehicle) for 16 h and then lysed for immunoblotting. The protein levels were quantified from three independent experiments and normalized to that of WT control cells. (B, C) The indicated cells were fixed and stained with antibodies to CI-MPR and TGN46. Images were captured by confocal microscopy and analyzed. Percentages of the cells with TGN region-localized CI-MPR were quantified in each group (n = 186, 210, 77, 152, 191, 900, and 603) from three independent experiments; p values were determined by Student’s t test. */# p < 0.05, **p < 0.01, ***p < 0.001, n.s., not significant. *v.s. WT, #, v.s. vehicle. Scale bars, 5 μm.
To determine how the decreased levels of CI-MPR affect the protein cellular distribution, we performed immunostaining of the CI-MPR and the TGN protein TGN46. We observed that most of the CI-MPR was localized to a central, TGN region in WT cells (Figure 8B). The signals of this CI-MPR population, however, became much weaker or even undetectable in myrlysin-, ARL8-, VPS39-, or VPS41-KO cells. The CI-MPR levels at the TGN area were restored in myrlysin- and VPS39-rescued cells (Figure 8B). We quantified the percentage of cells that had a CI-MPR population at the TGN region. The results showed that over 90% WT cells had TGN-region CI-MPR; however, this number decreased to 2–40% in the myrlysin-, ARL8-, VPS39-, and VPS41-KO cells (Figure 8C).
We also examined the other MPR receptor, CD-MPR, by immunoblotting and did not see a significant change of the protein levels among WT, myrlysin-, ARL8-, and VPS41-KO cells (Supplemental Figure S4). Due to a lack of reagents, we could not examine the cellular distribution of CD-MPR and thus do not know whether the transport of CD-MPR was altered in BORC/ARL8/HOPS-KO cells.
These observations indicated that the depletion of BORC, ARL8, or HOPS impaired retrieval of the CI-MPR from endosomes and resulted in its degradation by lysosomal acid hydrolases. Insufficient CI-MPR likely decreases the transportation of newly synthesized lysosome luminal proteins into endosomes and as a consequence causes them to be secreted. Altogether, our results indicate that BORC/ARL8/HOPS are required for lysosomal cholesterol egress because of their role in targeting NPC2 to the lysosome by enabling the endosome-to-lysosome transport and by facilitating the recycling of CI-MPR.
DISCUSSION
Lysosomes stand at the end of endocytosis and autophagy pathways and thus serve as a distribution point for cholesterol ingested from the extracellular space as well as recycled from intracellular organelles, respectively. Lysosomal cholesterol is delivered to other organelles for membrane biogenesis and synthesis of steroid hormones, bile acids, and vitamin D. In addition, lysosomal cholesterol regulates the SREBP2 pathway that controls cholesterol synthesis and uptake. Many proteins, including the cholesterol transport proteins NPC1 and NPC2, regulate cholesterol export from lysosomes, thus contributing to cellular cholesterol homeostasis (Trinh et al., 2020). In this study, we identify BORC, ARL8, and HOPS as additional factors that are required for cholesterol egress from lysosomes. Our findings are in line with previous RNAi screens that identified the BORC/BLOC1 subunit lyspersin as a regulator of LDL uptake (Bartz et al., 2009; Kraehling et al., 2016) and another study that demonstrated a requirement of the BORC subunit BLOS1 for recycling of internalized LDL receptor to the plasma membrane (Zhang et al., 2020).
BORC and ARL8 were originally found to mediate recruitment of the kinesin-1 KIF5B and kinesin-3 KIF1B proteins to lysosomes, thus promoting anterograde transport of lysosomes along microtubule tracks (Rosa-Ferreira and Munro, 2011; Pu et al., 2015; Guardia et al., 2016). This function of BORC and ARL8 is critical for the role of lysosomes in cell migration (Pu et al., 2015; Hamalisto and Jaattela, 2016), autophagy (Jia et al., 2017), and mTOR activation (Jia and Bonifacino, 2019) but not for cholesterol egress, as shown here by the normal cholesterol content of lysosomes from KIF5B-KIF1B double KO cells (Figure 4, A and B). In contrast, lysosomal cholesterol regulates lysosome motility and positioning. For example, the lysosomal cholesterol sensor ORP1L (Johansson et al., 2007; Rocha et al., 2009) and the SREBP2-regulated gene TMEM55B (Willett et al., 2017) recruit the retrograde motor dynein-dynactin to lysosomes when lysosomal cholesterol is elevated or when its egress is blocked.
Later studies showed that BORC and ARL8 also promote recruitment of HOPS to lysosomes, enabling their fusion with other organelles (Khatter et al., 2015b; Jia et al., 2017; Boda et al., 2019). In this study, we found that cholesterol egress from lysosomes depends not only on BORC and ARL8 but also on HOPS. Like KO of BORC subunits or ARL8 (Figures 1, A and B; 2, A and B; 3, B–E), KO of the VPS39 or VPS41 subunits of HOPS causes accumulation of free cholesterol in lysosomes and decreases both the number and the size of LDs in cells (Figure 4, C, D, K, and L). It is thus likely that BORC and ARL8 contribute to cholesterol mobilization from lysosomes through their role in promoting HOPS-dependent fusion of endosomal-lysosomal organelles. The lysosomal transmembrane protein LAMP1 and the transmembrane cholesterol-transport protein NPC1 perfectly colocalize to a population of vesicles in both WT and BORC/ARL8/HOPS-KO cells (Figure 5, A and B), indicating that accumulation of free cholesterol in those vesicles is not due to a failure of NPC1 to reach LAMP1 compartments. At least in HOPS-KO cells, these compartments are not likely normal lysosomes (Pols et al., 2013; Steel et al., 2020), although they contain several lysosomal membrane proteins and accumulate LysoTracker (Figures 4E, 5A, and 6, A and H; Supplemental Figure S2). In contrast, the luminal cholesterol transport protein NPC2 partially loses its localization to LAMP1/2 vesicles in BORC/ARL8/HOPS-KO cells relative to WT cells (Figures 5, E and F; and 7, A, C, E, and G). Moreover, a higher fraction of total NPC2 is secreted into the culture medium in BORC/ARL8/HOPS-KO cells relative to WT cells (Figure 5, G, I, and J), consistent with a reduction in the delivery efficiency of NPC2 to endosomal-lysosomal organelles. Similar observations were made for another lysosomal luminal protein, the acid hydrolase cathepsin D (Figure 6, A–G; Supplemental Figure S3B). These observations are consistent with the increased secretion of the vacuolar hydrolase carboxypeptidase Y in yeast with mutations in HOPS subunit genes (Radisky et al., 1997; Raymond et al., 1992). From these findings, we conclude that disruption of the BORC-ARL8-HOPS ensemble reduces the delivery of luminal proteins more than that of transmembrane proteins to the endosomal-lysosomal system. The partial loss of NPC2 from NPC1-containing compartments may thus be responsible for the reduced egress of free cholesterol from the endosomal-lysosomal system.
Both NPC2 and cathepsin D are M6P-modified proteins that are sorted from the TGN to endosomes by MPRs (Naureckiene et al., 2000; Qian et al., 2008). Previous studies using the RUSH system showed that the CD-MPR is exported from the TGN in a population of transport vesicles that likely delivers cargos directly to the endosomal-lysosomal system (Chen et al., 2017). In contrast, LAMP1 is exported from the TGN in a different population of tubular-vesicular carriers that travel to the plasma membrane prior to its delivery to the endosomal-lysosomal system (Chen et al., 2017). It is thus likely that BORC/ARL8/HOPS deficiency causes a greater impairment in MPR-mediated transport due to the significant loss of CI-MPR protein levels (Figure 8). In this context, NPC2 vesicles devoid of LAMP2 and Rab7a observed by immunofluorescence microscopy (Figure 7, A, C, E, and G) may correspond to the vesicles that are unable to enter endosomal compartments. By default, these vesicles may be secreted, explaining the higher fractions of NPC2 and cathepsin D that are released into the medium.
BORC/ARL8/HOPS deficiency increases the degradation of CI-MPR by acid hydrolases (Figure 8A). This could be due to the requirement of HOPS for retromer-mediated retrieval of CI-MPR from endosomes. Failed retrieval of CI-MPR, due to retromer deficiency, results in CI-MPR degradation in lysosomes (Arighi et al., 2004). HOPS and the ARL8 effector SKIP (or PLEKHM2) have been shown to recruit Rab7a GTPase-activating protein TBC1D15 to endosomes and thus maintain Rab7a dynamics (Jongsma et al., 2020b), which could be important to Rab7a-related retromer function (Rojas et al., 2008; Seaman et al., 2009; Jia et al., 2016). Considering endosomes receive CI-MPR from both the TGN- and the plasma membrane-derived vesicles, retrieval of CI-MPR from endosomes could be critical for maintaining normal CI-MPR protein levels and their distribution at the TGN area.
Unlike in cholesterol egress, deficiency of BORC and HOPS does not interfere in cathepsin activities very much (Pols et al., 2013; Pu et al., 2015; Jia et al., 2017). Although both nonlysosomal cathepsin D and NPC2 are increased, there are more cathepsin D-positive lysosomes than NPC2-positive lysosomes in HOPS-KO cells (Supplemental Figure S3C). This could be due to the existence of M6P-independent pathways that sort and transport cathepsins, which are identified in multiple cell types (Boonen et al., 2016; Canuel et al., 2008; van Meel et al., 2011), suggesting that similar mechanisms might function or be activated when needed in HeLa cells.
It is worth pointing out that, although BORC/ARL8/HOPS-KO increases the fraction of NPC2 and cathepsin D that are secreted into the culture medium, the total levels of these proteins in cells and medium, are increased (Figures 5, G–I; and 6, C–F). This could be due to increased gene transcription. NPC2 gene expression is regulated by the transcription factors PPARα (Chinetti-Gbaguidi et al., 2005) and NF-κB (Liao et al., 2018). In addition, TFEB and TFE3, which regulate lysosome biogenesis, also regulate NPC2 and cathepsin D gene expression (Sardiello et al., 2009; Palmieri et al., 2011; Settembre et al., 2012; Ballabio and Bonifacino, 2020; Carey et al., 2020). The similar levels of NPC1 in WT and BORC/ARL8/HOPS-KO cells do not support an increased lysosome biogenesis; however, TFEB/TFE3 could regulate lysosomal gene transcription to different extents. Recent studies reported that VPS41 mutations identified in patients and deletion of HOPS subunits continuously activate TFEB/TFE3 (van der Welle et al., 2021), and activation of TFEB reduces lysosomal cholesterol accumulation when NPC1 is inhibited (Contreras et al., 2020). In light of our results, it will be interesting to investigate the relationship between the BORC-ARL8-HOPS ensemble and transcription factors that regulate NPC2 expression.
EXPERIMENTAL PROCEDURES
Request a protocol through Bio-protocol.
Cell culture and transfection
HeLa cells were cultured in DMEM supplemented with 10% fetal bovine serum, 25 mM HEPES, and MycoZap Plus-CL (Lonza, Basel, Switzerland) at 37°C with 5% CO2. Plasmids (listed in the Supplemental Material) or SMARTpool siRNA (Horizon, Waterbeach, UK) were transfected using Lipofectamine 2000 and Oligofectamine, respectively, following the manufacturer’s instructions (Thermo Fisher Scientific, Waltham, MA). Cells were analyzed 24–48 h after plasmid transfection. Two-shot siRNA transfection was performed with a 1-d interval, and cells were analyzed 5 d after the first transfection.
CRISPR-Cas9 KO and KI
Genes were edited using the CRISPR-Cas9 system (Cong et al., 2013). To KO a gene, two 20-base pair (bp) targeting sequences were synthesized (Eurofins, Lancaster, PA) and introduced separately into the px458 plasmid containing the GFP sequence (Addgene, Cambridge, MA). HeLa cells were cotransfected with both plasmids, and GFP-positive cells were collected by cell sorter 48 h after transfection. Transformants were kept in normal medium for another 12 d to allow single colony formation. Genomic DNA was extracted from individual colonies, and cleavage of the target sequence was tested by PCR using a pair of primers, which produced a smaller band in KO relative to WT cells. The KO was confirmed by Sanger sequencing and immunoblotting. Alternatively, if a single target sequence was introduced to cleave genomic DNA, isolated colonies were screened by immunoblotting and confirmed by Sanger sequencing. To detect endogenous proteins, a Halo tag was inserted into the target genes to allow fused protein expression from the native promoter. Similar to the KO constructs, a 20-bp targeting sequence at the C-terminus of target gene was inserted into the px458 plasmid. A recombination template sequence was constructed into pUC57-mini plasmid (GeneScript, Piscataway, NJ), which includes ∼600 bp upstream sequence of cleavage site, 30 bp spacer, Halo tag sequence, and ∼600 bp downstream sequence with target PAM motif mutated. Both px458 and pUC57-mini were transfected into cells and sorted after 72 h for positive GFP. After culturing for an additional 10 d, the cells were sorted again to collect Halo-positive and GFP-negative cells. Single colonies were isolated and confirmed by fluorescence microscopy. To visualize Halo-fusion proteins, cells were incubated with 500 nM Janelia Fluor 594-NHS Ester (a gift from Luke Lavis, HHMI Janelia Research Campus) dissolved in complete DMEM media at 37°C with 5% CO2 for 30 min, washed twice for 10 min in total, and imaged with confocal microscope.
Guide RNA target sequences used in this study:
ARL8a KO: GTTGTGGAGCTCGTTCTTAG and GCGAGACCTTCCGGGAGCAT
VPS39 KO: GCCTCTGCAAATCGACTGTC
VPS41 KO: GTGACTCTCCCGTGGCGCCATGG and TGGGATGGCGAAAGTTGGTTTCG
ARL8a-Halo KI: CAGAAATTGCCTTTTTCAGA
ARL8b-Halo KI: TCATCATCTAAACCTGAAGC.
Free cholesterol staining and measurement
Cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) supplemented with CaCl2 and MgCl2 (PBSCM) and incubated with 25 μg/ml filipin complex (Millipore Sigma, St. Louis, MO) in PBSCM at 37°C for 30 min. When immunostaining was performed with filipin, primary antibodies and Alexa–conjugated secondary antibodies were sequentially applied to filipin-stained cells without detergent permeabilization. The total level of cellular free cholesterol was measured using a commercial kit (Cell Biolabs, San Diego, CA) and a plate reader.
Lipid analysis by TLC
Cells were collected in PBS with a cell scraper after washing twice with PBS. Total lipids were extracted by adding lipid-extraction solvent (chloroform:methanol:acetic acid = 50:50:1) of 90% vol of PBS. The organic phase and the aqueous phase were separated by a 10-min centrifugation at 21,000 × g. The organic phase was collected and loaded onto silicon-coated glass TLC plates. Free cholesterol, triglyceride mix, and cholesterol-palmitate (Millipore Sigma) were dissolved in chloroform to make 10 mg/ml stocks and loaded as lipid standards. The plates were developed in the TLC chamber filled with 150 ml developing solvent (hexane:diethyl ether:acetic acid = 80:20:1). To visualize lipid bands, the developed plates were incubated in an iodine-evaporated chamber.
Live-cell imaging
Cells were seeded onto chambered cover glasses (Cellvis, Mountain View, CA) coated with fibronectin and incubated with 200 ng/ml BODIPY 493/503 (Thermo Fisher Scientific) in complete DMEM media for LD staining or 1:10,000 diluted LysoTracker Red DND-99 (Thermo Fisher Scientific) for lysosome staining. After 30 min incubation, live-cell imaging was performed using a Zeiss LSM780 or LSM800 confocal microscope equipped with an environmental humidified chamber set at 37°C and 5% CO2 and Definite Focus. Images were acquired by using ZEN software (Zeiss, Oberkochen, Germany)
Analysis of intracellular and extracellular proteins by Western blotting
To detect secreted NPC2 and cathepsin D, cells were kept in a minimum volume of DMEM without serum for 6 h after washing 3× with the same medium. The media were collected and centrifuged at 3000 × g for 5 min to pellet any detached cells. The supernatants were mixed with SDS loading buffer and subjected to SDS–PAGE and immunoblotting. After collecting the media, cells were incubated in complete DMEM for 2 h. Cell lysates were obtained by using cold lysis buffer (300 mM NaCl, 5 mM EDTA, 50 mM Tris-HCl, pH 7.4, 1% Triton X-100, and proteinase inhibitor cocktail) after washing 3× with prechilled PBS. The cell lysates were cleared by 15-min centrifugation at 17,000 × g, 4°C, and the supernatants were collected for Western blotting. Rapid PNGase F (New England Biotech, Ipswich, MA) was used to digest glycosylated proteins following the manufacturer’s instructions.
Quantification and statistical analysis
ImageJ was used for quantifying fluorescence signals of randomly taken images from at least three independent experiments. The signal intensity of LDs and vesicular filipin was measured using the function of “Analyze particles,” and colocalization was analyzed with the plug-in “PSC Colocalization” (French et al., 2008). Western blots and TLC were quantified using the ImageJ function “Gels” from at least three independent experiments. All the bar graphs show the mean ± SD, and colocalization results include frequency distribution. Statistical significance was determined by comparing two datasets using a Student’s t test with one-tailed distribution. Probability values and number of trials are given in the figure captions and the legends where appropriate. The statistical significance is generally denoted as follows: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, and n.s., not significant.
Supplementary Material
Acknowledgments
We thank A. Doyle, B. Evans Hutzenbiler, C. Garza, and N. Chowdhury for experimental assistance and A. Doyle for her editing of the manuscript. We thank P. Lobel for the kind gifts of the NPC2 antibody and NPC2-mCherry construct, J. H. Brumell for kind gift of the ARL8b-GFP construct, and E. L. Lavis for the kind gift of the Halo dye. We thank J. Bonifacino for his suggestions, manuscript revision, and kind gift of cathepsin D-RFP construct. This research made use of the Fluorescence Microscopy and Cell Imaging Shared Resource which is partially supported by the University of New Mexico Comprehensive Cancer Center Support Grant NCI P30CA118100. This work was funded by the CoBRE Program of NIGMS, NIH (P20GM121176).
Abbreviations used:
- BORC
BLOC one-related complex
- CD-MPR
cation-dependent mannose 6-phosphate receptor
- CI-MPR
cation-independent mannose 6-phosphate receptor
- ER
endoplasmic reticulum
- HOPS
homotypic fusion and protein sorting
- KD
knockdown
- KO
knockout
- LDs
lipid droplets
- M6P
mannose 6-phosphate
- OA
oleic acid
- PBS
phosphate-buffered saline
- PBSCM
PBS supplemented with CaCl 2 and MgCl 2
- PFA
paraformaldehyde
- TGN
trans-Golgi network
- TLC
thin-layer chromatography
- WT
wildtype.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E21-11-0595-T) on June 2, 2022.
REFERENCES
- Ang AL, Taguchi T, Francis S, Folsch H, Murrells LJ, Pypaert M, Warren G, Mellman I (2004). Recycling endosomes can serve as intermediates during transport from the Golgi to the plasma membrane of MDCK cells. J Cell Biol 167, 531–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenas F, Garcia-Ruiz C, Fernandez-Checa JC (2017). Intracellular cholesterol trafficking and impact in neurodegeneration. Front Mol Neurosci 10, 382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arighi CN, Hartnell LM, Aguilar RC, Haft CR, Bonifacino JS (2004). Role of the mammalian retromer in sorting of the cation-independent mannose 6-phosphate receptor. J Cell Biol 165, 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballabio A, Bonifacino JS (2020). Lysosomes as dynamic regulators of cell and organismal homeostasis. Nat Rev Mol Cell Biol 21, 101–118. [DOI] [PubMed] [Google Scholar]
- Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM (2012). Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell 150, 1196–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett AJ (1970). Cathepsin D. Purification of isoenzymes from human and chicken liver. Biochem J 117, 601–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartz F, Kern L, Erz D, Zhu M, Gilbert D, Meinhof T, Wirkner U, Erfle H, Muckenthaler M, Pepperkok R, Runz H (2009). Identification of cholesterol-regulating genes by targeted RNAi screening. Cell Metab 10, 63–75. [DOI] [PubMed] [Google Scholar]
- Boda A, Lorincz P, Takats S, Csizmadia T, Toth S, Kovacs AL, Juhasz G (2019). Drosophila Arl8 is a general positive regulator of lysosomal fusion events. Biochim Biophys Acta Mol Cell Res 1866, 533–544. [DOI] [PubMed] [Google Scholar]
- Boonen M, Staudt C, Gilis F, Oorschot V, Klumperman J, Jadot M (2016). Cathepsin D and its newly identified transport receptor SEZ6L2 can modulate neurite outgrowth. J Cell Sci 129, 557–568. [DOI] [PubMed] [Google Scholar]
- Braun M, Waheed A, Vonfigura K (1989). Lysosomal acid-phosphatase is transported to lysosomes via the cell-surface. EMBO Journal 8, 3633–3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL (1997). The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89, 331–340. [DOI] [PubMed] [Google Scholar]
- Canuel M, Korkidakis A, Konnyu K, Morales CR (2008). Sortilin mediates the lysosomal targeting of cathepsins D and H. Biochem Biophys Res Commun 373, 292–297. [DOI] [PubMed] [Google Scholar]
- Carey KL, Paulus GLC, Wang L, Balce DR, Luo JW, Bergman P, Ferder IC, Kong L, Renaud N, Singh S, et al. (2020). TFEB transcriptional responses reveal negative feedback by BHLHE40 and BHLHE41. Cell Rep 33, 108371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carstea ED, Morris JA, Coleman KG, Loftus SK, Zhang D, Cummings C, Gu J, Rosenfeld MA, Pavan WJ, Krizman DB, et al. (1997). Niemann-Pick C1 disease gene: homology to mediators of cholesterol homeostasis. Science 277, 228–231. [DOI] [PubMed] [Google Scholar]
- Castellano BM, Thelen AM, Moldavski O, Feltes M, van der Welle RE, Mydock-McGrane L, Jiang X, van Eijkeren RJ, Davis OB, Louie SM, et al. (2017). Lysosomal cholesterol activates mTORC1 via an SLC38A9-Niemann-Pick C1 signaling complex. Science 355, 1306–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Gershlick DC, Park SY, Bonifacino JS (2017). Segregation in the Golgi complex precedes export of endolysosomal proteins in distinct transport carriers. J Cell Biol 216, 4141–4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinetti-Gbaguidi G, Rigamonti E, Helin L, Mutka AL, Lepore M, Fruchart JC, Clavey V, Ikonen E, Lestavel S, Staels B (2005). Peroxisome proliferator-activated receptor alpha controls cellular cholesterol trafficking in macrophages. J Lipid Res 46, 2717–2725. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras PS, Tapia PJ, Gonzalez-Hodar L, Peluso I, Soldati C, Napolitano G, Matarese M, Heras ML, Valls C, Martinez A, et al. (2020). c-Abl Inhibition activates TFEB and promotes cellular clearance in a lysosomal disorder. iScience 23, 101691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JP, Chen FW, Ioannou YA (2000). Transmembrane molecular pump activity of Niemann-Pick C1 protein. Science 290, 2295–2298. [DOI] [PubMed] [Google Scholar]
- Davis OB, Shin HR, Lim CY, Wu EY, Kukurugya M, Maher CF, Perera RM, Ordonez MP, Zoncu R (2021). NPC1-mTORC1 signaling couples cholesterol sensing to organelle homeostasis and is a targetable pathway in Niemann-pick type C. Developmental Cell 56, 260–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskelinen EL, Schmidt CK, Neu S, Willenborg M, Fuertes G, Salvador N, Tanaka Y, Lullmann-Rauch R, Hartmann D, Heeren J, et al. (2004). Disturbed cholesterol traffic but normal proteolytic function in LAMP-1/LAMP-2 double-deficient fibroblasts. Mol Biol Cell 15, 3132–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French AP, Mills S, Swarup R, Bennett MJ, Pridmore TP (2008). Colocalization of fluorescent markers in confocal microscope images of plant cells. Nat Protoc 3, 619–628. [DOI] [PubMed] [Google Scholar]
- Guardia CM, Farias GG, Jia R, Pu J, Bonifacino JS (2016). BORC functions upstream of Kinesins 1 and 3 to coordinate regional movement of lysosomes along different microtubule tracks. Cell Rep 17, 1950–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamalisto S, Jaattela M (2016). Lysosomes in cancer-living on the edge (of the cell). Curr Opin Cell Biol 39, 69–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harter C, Mellman I (1992). Transport of the lysosomal membrane glycoprotein lgp120 (lgp-A) to lysosomes does not require appearance on the plasma membrane. J Cell Biol 117, 311–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey CM, Stroupe C, Wickner W (2009). The Major Role of the Rab Ypt7p in Vacuole Fusion Is Supporting HOPS Membrane Association. J Biol Chem 284, 16118–16125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Song BL, Xu C (2020). Cholesterol metabolism in cancer: mechanisms and therapeutic opportunities. Nat Metab 2, 132–141. [DOI] [PubMed] [Google Scholar]
- Huang L, Pike D, Sleat DE, Nanda V, Lobel P (2014). Potential Pitfalls and Solutions for Use of Fluorescent Fusion Proteins to Study the Lysosome. Plos One 9, e88893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janvier K, Bonifacino JS (2005). Role of the endocytic machinery in the sorting of lysosome-associated membrane proteins. Mol Biol Cell 16, 4231–4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia D, Zhang JS, Li F, Wang J, Deng Z, White MA, Osborne DG, Phillips-Krawczak C, Gomez TS, Li H, et al. (2016). Structural and mechanistic insights into regulation of the retromer coat by TBC1d5. Nat Commun 7, 13305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia R, Bonifacino JS (2019). Lysosome Positioning Influences mTORC2 and AKT Signaling. Mol Cell 75, 26–38.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia R, Guardia CM, Pu J, Chen Y, Bonifacino JS (2017). BORC coordinates encounter and fusion of lysosomes with autophagosomes. Autophagy 13, 1648–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P, Nishimura T, Sakamaki Y, Itakura E, Hatta T, Natsume T, Mizushima N (2014). The HOPS complex mediates autophagosome-lysosome fusion through interaction with syntaxin 17. Mol Biol Cell 25, 1327–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M, Rocha N, Zwart W, Jordens I, Janssen L, Kuijl C, Olkkonen VM, Neefjes J (2007). Activation of endosomal dynein motors by stepwise assembly of Rab7-RILP-p150Glued, ORP1L, and the receptor betalll spectrin. J Cell Biol 176, 459–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongsma M, Bakker J, Cabukusta B, Liv N, Elsland DV, Fermie J, Akkermans J, Kuijl C, Janssen L, Hoogzaad D, et al. (2020a). SKIP-HOPS recruits TBC 1D15 for a Rab7-to-Arl8b identity switch to control late endosome transport. EMBO J 39, e102301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongsma ML, Bakker J, Cabukusta B, Liv N, van Elsland D, Fermie J, Akkermans JL, Kuijl C, van der Zanden SY, Janssen L, et al. (2020b). SKIP-HOPS recruits TBC1D15 for a Rab7-to-Arl8b identity switch to control late endosome transport. EMBO J 39, e102301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatter D, Raina VB, Dwivedi D, Sindhwani A, Bahl S, Sharma M (2015a). The small GTPase Arl8b regulates assembly of the mammalian HOPS complex on lysosomes. J Cell Sci 128, 1746–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatter D, Sindhwani A, Sharma M (2015b). Arf-like GTPase Arl8: Moving from the periphery to the center of lysosomal biology. Cell Logist 5, e1086501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraehling JR, Chidlow JH, Rajagopal C, Sugiyama MG, Fowler JW, Lee MY, Zhang X, Ramirez CM, Park EJ, Tao B, et al. (2016). Genome-wide RNAi screen reveals ALK1 mediates LDL uptake and transcytosis in endothelial cells. Nat Commun 7, 13516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzu OF, Noory MA, Robertson GP (2016). The Role of Cholesterol in Cancer. Cancer Res 76, 2063–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Saha P, Li J, Blobel G, Pfeffer SR (2016). Clues to the mechanism of cholesterol transfer from the structure of NPC1 middle lumenal domain bound to NPC2. Proc Natl Acad Sci USA 113, 10079–10084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Wei J, Wang J, Shi X, Luo J, Song BL (2018). The noncanonical NF-kappaB pathway promotes NPC2 expression and regulates intracellular cholesterol trafficking. Sci China Life Sci 61, 1222–1232. [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Fambrough DM (1986). Lysosomal membrane dynamics: structure and interorganellar movement of a major lysosomal membrane glycoprotein. J Cell Biol 102, 1593–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F, Liang Q, Abi-Mosleh L, Das A, De Brabander JK, Goldstein JL, Brown MS (2015). Identification of NPC1 as the target of U18666A, an inhibitor of lysosomal cholesterol export and Ebola infection. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Yang H, Song BL (2020). Mechanisms and regulation of cholesterol homeostasis. Nat Rev Mol Cell Biol 21, 225–245. [DOI] [PubMed] [Google Scholar]
- Maxfield FR, Wustner D (2012). Analysis of cholesterol trafficking with fluorescent probes. Methods Cell Biol 108, 367–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwan DG, Popovic D, Gubas A, Terawaki S, Suzuki H, Stadel D, Coxon FP, Miranda de Stegmann D, Bhogaraju S, Maddi K, et al. (2015). PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins. Mol Cell 57, 39–54. [DOI] [PubMed] [Google Scholar]
- Meng Y, Heybrock S, Neculai D, Saftig P (2020). Cholesterol Handling in Lysosomes and Beyond. Trends Cell Biol 30, 452–466. [DOI] [PubMed] [Google Scholar]
- Nada S, Hondo A, Kasai A, Koike M, Saito K, Uchiyama Y, Okada M (2009). The novel lipid raft adaptor p18 controls endosome dynamics by anchoring the MEK-ERK pathway to late endosomes. EMBO J 28, 477–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naureckiene S, Sleat DE, Lackland H, Fensom A, Vanier MT, Wattiaux R, Jadot M, Lobel P (2000). Identification of HE1 as the second gene of Niemann-Pick C disease. Science 290, 2298–2301. [DOI] [PubMed] [Google Scholar]
- Niwa S, Tao L, Lu SY, Liew GM, Feng W, Nachury MV, Shen K (2017). BORC regulates the axonal transport of synaptic vesicle precursors by activating ARL-8. Curr Biol 27, 2569–2578.e2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri M, Impey S, Kang H, di Ronza A, Pelz C, Sardiello M, Ballabio A (2011). Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum Mol Genet 20, 3852–3866. [DOI] [PubMed] [Google Scholar]
- Peplowska K, Markgraf DF, Ostrowicz CW, Bange G, Ungermann C (2007). The CORVET tethering complex interacts with the yeast Rab5 homolog Vps21 and is involved in endo-lysosomal biogenesis. Dev Cell 12, 739–750. [DOI] [PubMed] [Google Scholar]
- Pols MS, ten Brink C, Gosavi P, Oorschot V, Klumperman J (2013). The HOPS proteins hVps41 and hVps39 are required for homotypic and heterotypic late endosome fusion. Traffic 14, 219–232. [DOI] [PubMed] [Google Scholar]
- Popovic K, Holyoake J, Pomes R, Prive GG (2012). Structure of saposin A lipoprotein discs. Proc Natl Acad Sci USA 109, 2908–2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu J, Keren-Kaplan T, Bonifacino JS (2017). A Ragulator-BORC interaction controls lysosome positioning in response to amino acid availability. J Cell Biol 216, 4183–4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu J, Schindler C, Jia R, Jarnik M, Backlund P, Bonifacino JS (2015). BORC, a multisubunit complex that regulates lysosome positioning. Dev Cell 33, 176–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian H, Wu X, Du X, Yao X, Zhao X, Lee J, Yang H, Yan N (2020). Structural basis of low-pH-dependent lysosomal cholesterol egress by NPC1 and NPC2. Cell 182, 98–111.e118. [DOI] [PubMed] [Google Scholar]
- Qian M, Sleat DE, Zheng H, Moore D, Lobel P (2008). Proteomics analysis of serum from mutant mice reveals lysosomal proteins selectively transported by each of the two mannose 6-phosphate receptors. Mol Cell Proteomics 7, 58–70. [DOI] [PubMed] [Google Scholar]
- Radisky DC, Snyder WB, Emr SD, Kaplan J (1997). Characterization of VPS41, a gene required for vacuolar trafficking and high-affinity iron transport in yeast. Proc Natl Acad Sci USA 94, 5662–5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond CK, Howald-Stevenson I, Vater CA, Stevens TH (1992). Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol Biol Cell 3, 1389–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha N, Kuijl C, van der Kant R, Janssen L, Houben D, Janssen H, Zwart W, Neefjes J (2009). Cholesterol sensor ORP1L contacts the ER protein VAP to control Rab7-RILP-p150 Glued and late endosome positioning. J Cell Biol 185, 1209–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas R, van Vlijmen T, Mardones GA, Prabhu Y, Rojas AL, Mohammed S, Heck AJ, Raposo G, van der Sluijs P, Bonifacino JS (2008). Regulation of retromer recruitment to endosomes by sequential action of Rab5 and Rab7. J Cell Biol 183, 513–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa-Ferreira C, Munro S (2011). Arl8 and SKIP act together to link lysosomes to kinesin-1. Dev Cell 21, 1171–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM (2010). Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141, 290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, et al. (2009). A gene network regulating lysosomal biogenesis and function. Science 325, 473–477. [DOI] [PubMed] [Google Scholar]
- Schneede A, Schmidt CK, Holtta-Vuori M, Heeren J, Willenborg M, Blanz J, Domanskyy M, Breiden B, Brodesser S, Landgrebe J, et al. (2011). Role for LAMP-2 in endosomal cholesterol transport. J Cell Mol Med 15, 280–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seals DF, Eitzen G, Margolis N, Wickner WT, Price A (2000). A Ypt/Rab effector complex containing the Sec1 homolog Vps33p is required for homotypic vacuole fusion. Proc Natl Acad Sci USA 97, 9402–9407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MN, Harbour ME, Tattersall D, Read E, Bright N (2009). Membrane recruitment of the cargo-selective retromer subcomplex is catalysed by the small GTPase Rab7 and inhibited by the Rab-GAP TBC1D5. J Cell Sci 122, 2371–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, et al. (2012). A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J 31, 1095–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya Y, Chang CCY, Chang TY (2015). ACAT1/SOAT1 as a therapeutic target for Alzheimer’s disease. Future Med. Chem. 7, 2451–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solinger JA, Spang A (2013). Tethering complexes in the endocytic pathway: CORVET and HOPS. FEBS J 280, 2743–2757. [DOI] [PubMed] [Google Scholar]
- Spang A (2016). Membrane tethering complexes in the endosomal system. Front Cell Dev Biol 4, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steel D, Zech M, Zhao C, Barwick KES, Burke D, Demailly D, Kumar KR, Zorzi G, Nardocci N, Kaiyrzhanov R, et al. (2020). Loss-of-function variants in HOPS complex genes VPS16 and VPS41 cause early onset dystonia associated with lysosomal abnormalities. Ann Neurol 88, 867–877. [DOI] [PubMed] [Google Scholar]
- Stroupe C, Collins KM, Fratti RA, Wickner W (2006). Purification of active HOPS complex reveals its affinities for phosphoinositides and the SNARE Vam7p. EMBO J 25, 1579–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroupe C, Hickey CM, Mima J, Burfeind AS, Wickner W (2009). Minimal membrane docking requirements revealed by reconstitution of Rab GTPase-dependent membrane fusion from purified components. Proc Natl Acad Sci USA 106, 17626–17633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh MN, Brown MS, Goldstein JL, Han J, Vale G, McDonald JG, Seemann J, Mendell JT, Lu F (2020). Last step in the path of LDL cholesterol from lysosome to plasma membrane to ER is governed by phosphatidylserine. Proc Natl Acad Sci USA 117, 18521–18529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Welle REN, Jobling R, Burns C, Sanza P, van der Beek JA, Fasano A, Chen L, Zwartkruis FJ, Zwakenberg S, Griffin EF, et al. (2021). Neurodegenerative VPS41 variants inhibit HOPS function and mTORC1-dependent TFEB/TFE3 regulation. EMBO Mol Med 13, e13258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meel E, Boonen M, Zhao H, Oorschot V, Ross FP, Kornfeld S, Klumperman J (2011). Disruption of the Man-6-P targeting pathway in mice impairs osteoclast secretory lysosome biogenesis. Traffic 12, 912–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waguri S, Dewitte F, Le Borgne R, Rouille Y, Uchiyama Y, Dubremetz JF, Hoflack B (2003). Visualization of TGN to endosome trafficking through fluorescently labeled MPR and AP-1 in living cells. Mol Biol Cell 14, 142–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J, Zhang YY, Luo J, Wang JQ, Zhou YX, Miao HH, Shi XJ, Qu YX, Xu J, Li BL, Song BL (2017). The GARP complex is involved in intracellular cholesterol transport via targeting NPC2 to lysosomes. Cell Rep 19, 2823–2835. [DOI] [PubMed] [Google Scholar]
- Willenborg M, Schmidt CK, Braun P, Landgrebe J, von Figura K, Saftig P, Eskelinen EL (2005). Mannose 6-phosphate receptors, Niemann-Pick C2 protein, and lysosomal cholesterol accumulation. J Lipid Res 46, 2559–2569. [DOI] [PubMed] [Google Scholar]
- Willett R, Martina JA, Zewe JP, Wills R, Hammond GRV, Puertollano R (2017). TFEB regulates lysosomal positioning by modulating TMEM55B expression and JIP4 recruitment to lysosomes. Nat Commun 8, 1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Bulkley DP, Xin Y, Roberts KJ, Asarnow DE, Sharma A, Myers BR, Cho W, Cheng Y, Beachy PA (2018). Structural basis for cholesterol transport-like activity of the hedgehog receptor patched. Cell 175, 1352–1364.e1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Hao C, Shui G, Li W (2020). BLOS1 mediates kinesin switch during endosomal recycling of LDL receptor. Elife 9, e58069. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.