Summary
Widespread generation and analysis of omics data have revolutionized molecular medicine on Earth, yet its power to yield new mechanistic insights and improve occupational health during spaceflight is still to be fully realized in humans. Nevertheless, rapid technological advancements and ever-regular spaceflight programs mean that longitudinal, standardized, and cost-effective collection of human space omics data are firmly within reach. Here, we consider the practicality and scientific return of different sampling methods and omic types in the context of human spaceflight. We also appraise ethical and legal considerations pertinent to omics data derived from European astronauts and spaceflight participants (SFPs). Ultimately, we propose that a routine omics collection program in spaceflight and analog environments presents a golden opportunity. Unlocking this bright future of artificial intelligence (AI)-driven analyses and personalized medicine approaches will require further investigation into best practices, including policy design and standardization of omics data, metadata, and sampling methods.
Keywords: multiomics, European Space Agency, commercial spaceflight, personalized medicine, International Space Station, longitudinal monitoring, GDPR, astronaut ethics, biobank, artificial intelligence
Graphical abstract

The bigger picture
Omic profiling of European human subjects in space—whether they be governmental or commercial—is primed to revolutionize space medicine, as it has with medicine on Earth. Improved occupational healthcare will be important for reducing risk and increasing mission success in ambitious endeavors, including voyages to Mars.
We believe that collaborative steps should be taken today to design a standardized data resource, which will continue to be useful in the future, as data science approaches evolve. From this perspective, we introduce considerations for a routine omics program for Europeans in space. These include which data types to collect, which sampling methods to use, and at which time points. Ethical and legal considerations of European personal omics data in the context of astronauts are also discussed, with the goal of creating a policy landscape where data can be as open as possible to maximize scientific potential but as closed as necessary to protect the data subjects.
We propose that routine “omic” collection (e.g., pre-flight genomic sequencing) from European government or commercial astronauts could provide valuable data for understanding spaceflight-induced health issues. Building such a standardized data resource would help to unlock powerful data science approaches. This could improve occupational healthcare in spaceflight, including via personalized medicine approaches. We introduce considerations, including ethical and legal challenges regarding European human personal data, in the unique context of the spacefaring population and which samples to collect and when.
Introduction
In 1978, Vladimír Remek of the Czech Republic became the first human from the European Union to cross the boundary into outer space. Since then, astronauts from an array of European countries have followed suit (Figure 1A), with many of these trips occurring on board the International Space Station (ISS) since its first occupation at the turn of the millennium. Despite a steady increase in the number of European astronauts venturing beyond Earth (Figure 1B), astronauts remain a relatively scarce population. Each astronaut, therefore, presents a rare and unique opportunity to directly study the impact of spaceflight on the human body, mind, and spirit. Additionally, with the emergence of commercial spaceflight, and European citizens already flying to space via these programs (e.g., Axiom Mission 1 and Blue Origin NS-16), there is an emerging opportunity to expand the population of European human subjects who can be studied in space.1 The spaceflight environment is characterized by simultaneous exposure to stressors, including cosmic radiation, altered gravity, isolation, and confinement within a closed environment.2 Additionally, demand on the human body changes dynamically throughout different time points within a typical spaceflight mission, including pre-flight training, launch, extravehicular activity (EVA), landing, and post-flight rehabilitation.
Figure 1.
European astronaut population
The distribution and population of European astronauts who have crossed the Kármán line (as of 9/20/21).
(A) Geographically highlights the range of nationalities of European astronaut.
(B) Shows the cumulative population of European astronauts over time, based on astronauts’ first mission into outer space.
To date, studies of astronauts have shown that long-duration spaceflight induces multisystem physiological deconditioning, such as degradation of muscle and bone,3,4 and detrimental changes to the eye, known as spaceflight-associated neuro-ocular syndrome (SANS).5 However, the precise mechanisms behind these responses are yet to be fully understood, largely due to the pace and practicalities of conducting comprehensive molecular studies in astronauts. Biological changes and associated medical risks during spaceflight missions, including in the commercial spaceflight setting, have recently been reviewed elsewhere.1,2,6 The European Space Agency (ESA) and other space agencies have historically taken two complementary approaches to accelerate the understanding of molecular causes of biological changes in astronauts: translational research and space analog research. Translational research utilizes model organisms, including rodents, for molecular studies in space,7 with the aim of understanding and countering detrimental physiological changes in astronauts. Space analog research utilizes facilities on Earth, which enable one to study individual aspects of the spaceflight environment without having to conduct studies in space. In the context of human research, this can include studying inactivity via bedrest studies and isolation via use of remote research stations, such as Concordia.8
At the 2022 European Space Summit, the European Association of Space Explorers called for the development of European vehicles for transporting humans into space. Irrespective of whether or not this becomes a near- to medium-term reality, not only will ESA astronauts continue to regularly fly to the ISS but also they will fly in future exploration missions with international partners. One such example is the Artemis 2 mission to the moon, which could occur as early as 2025. The new generation of spacecraft used for these missions is likely to have highly limited cargo capacity; therefore, countermeasures, such as the Advanced Resistive Exercise Device (ARED), used on board the ISS to suppress muscle and bone loss, may not be suitable.9 Additionally, during these exploration missions, astronauts will venture beyond low Earth orbit (LEO) for the first time since the end of the Apollo program in 1972. Within LEO, the Earth’s magnetosphere provides partial protection against radiation; thus, humans venturing beyond this will experience greater exposure to solar particle event radiation and ionizing galactic cosmic radiation.2,10,11 Although there is a lack of data on the molecular effects of the beyond LEO environment, an increase in radiation-associated health risks is almost a certainty.12, 13, 14 Further considerations of future missions beyond LEO include health risks associated with unprecedentedly long-duration missions, lunar dust,15 and EVA.2 To the latter point, in the case of a Mars mission, the crew would likely need to safely perform EVA tasks in partial gravity (0.38 g) following long-duration spaceflight, without the extensive rehabilitation support available on Earth.9,16
With these new spacecraft and mission plans, it will be crucial to assess compatible countermeasures, while also gaining greater insight into the new challenges to human health posed by environments where biological and molecular adaptations remain uncharted. A key challenge for translational research from a European standpoint is that the ESA does not currently have rodent research facilities in space, and it seems unlikely that such facilities will be rapidly developed for future exploration vehicles. Thus, new approaches that do not depend on rodent research facilities should be considered by ESA. In this context, a salient opportunity for space analog research is to harmonize measures and endpoints between research in analog environments and in space, increasing fidelity. For example, the efficacy of new countermeasures could be evaluated in both environments, and the efficacy of rehabilitation following long-duration spaceflight16 could be studied and improved by testing novel rehabilitation techniques following long-duration bedrest. For these reasons, it could prove cost effective for the ESA to pursue an approach that focuses on improved capabilities for research on astronauts themselves and with increased harmonization of data and endpoint measures obtained in space and analog environments.
On Earth, medicine has undergone a revolution through the widespread generation and analysis of multiple omic data types, including genomics, transcriptomics, and proteomics. The strength of omic-based, data-driven approaches is their power to discover unanticipated effects, elucidate molecular mechanisms, and generate novel hypotheses to guide follow-up targeted studies.17,18 In scientific studies on Earth, omic-derived molecular changes are regularly correlated with performance metrics to uncover molecular drivers of physiological change.19 Omics have also become a valuable tool for clinical practice. For example, the National Health Service (NHS) in the UK (1) routinely offers all newborn babies blood spot test screening for several genomic-based disorders, including sickle cell disease, cystic fibrosis, congenital hypothyroidism, and six inherited metabolic diseases; (2) recently launched a pilot project with Genomics to evaluate incorporating polygenic risk factor data into the clinical risk assessment for cardiovascular disease; and (3) recently approved use of personalized medications for treating cystic fibrosis based on the nature of the mutation(s) present in the patient’s cystic fibrosis transmembrane conductance regulator (CFTR) encoding gene. Furthermore, the COVID-19 pandemic has highlighted how omics can rapidly be employed in combination with clinical data to determine risk of severe disease course, identify potential underlying mechanisms of susceptibility, and identify potential personalized treatments.20
Although omics have become a significant part of scientific research and standard clinical practice on Earth, for humans in space, omics are a relatively new and untapped technology. Between March 2015 and March 2016, NASA conducted the seminal Twins Study. The study monitored an astronaut during a 1-year mission in space, while the astronaut’s identical twin remained on Earth as a control. Both subjects were comprehensively profiled using multiomic measures across several time points pre-flight, in-flight, and post-flight. Spaceflight-induced changes were detected across multiple systems, with most of these alterations returning to baseline levels post-flight.21 The longitudinal profiling of the NASA Twins Study and similar profiling on the recent short-duration SpaceX Inspiration4 commercial mission demonstrate that multiomics profiling of astronauts is feasible. Accordingly, NASA has recently announced that they have elected to add omics to the standard measures program for the ISS, which ESA astronauts can opt into. NASA’s standard measures program also routinely collects select physiological measures and select biomarkers from blood and urine; commercial spaceflight missions are similarly exploring these possibilities. For example, the Translational Research Institute for Space Health (TRISH), a partner of NASA’s Human Research Program, recently created a database for commercial spaceflight health data, including crew omics data from the Inspiration4 mission.22 It is, therefore, important that the ESA also explores these new opportunities, leveraging existing expertise in European spaceflight omics23 and facilities for human subjects research. To that end, the key question is how best to develop a routine longitudinal omics program for European astronauts and/or European spaceflight participants (SFPs).
A routine omics collection program could be viewed as an initiative to develop a high-value data resource, which can be analyzed with ever-evolving big data approaches, thereby improving ESA’s artificial intelligence (AI) readiness. Access to sensitive data collected during the program would need to be controlled so that scientific research is supported, while protecting the rights of the subjects. Such a program could help to elucidate key molecular drivers behind spaceflight health issues, which could lead to improved risk quantification and approaches to spaceflight occupational health. One example of how this could be used is for the development of evidence-based recommendations on training and medical standards for spaceflight; these standards will become especially important as the spacefaring population increases in heterogeneity and begins to include humans with pre-existing conditions.1,6 Furthermore, a routine omics program could be used as a basis for personalized medicine approaches. As an example, researchers have previously found that genetic variations in one-carbon metabolism genes may contribute to susceptibility to SANS in astronauts,24 and other genomic studies have shown individual variability associated with spaceflight-relevant factors, including bone mineral density and radiation sensitivity.1 Pre-flight genomic screening of astronauts, therefore, holds potential in developing personalized medicine approaches to risk assessment and countermeasures, including pharmacogenomics and nutrigenomics.17,25,26 Pharmacogenomics aims to optimize medication therapy, with respect to the patient’s genomic data, to ensure maximum efficacy with minimal adverse effects. For example, a study of medication on board the ISS found that the metabolism of approximately a third of the drugs in the ISS repository were affected by polymorphic metabolizing enzymes.27 Through further data collection, cargo could potentially be optimized to only include the best available medication for the individual crewmembers, reducing costs and adverse drug reaction/therapy failure risk.28 Furthermore, omics could prove valuable for longitudinal health monitoring during spaceflight. For example, TRISH recently suggested that omics could be used alongside other datasets obtained via point-of-care (POC) devices to diagnose approximately one-third of NASA’s list of medical conditions that are of concern for deep-space missions.29 Finally, discoveries obtained using human-research-derived omics data could benefit health on Earth. For example, the World Health Organization has listed physical inactivity as the fourth leading risk factor for global mortality, estimated to have been responsible for 9% of global premature mortality in 2008.30 Mechanical unloading of the musculoskeletal system due to microgravity during spaceflight, or due to immobilization or reduced step count in analog environments, induces similar effects to physical inactivity, such as muscle atrophy and altered glucose handling; thus, discoveries and countermeasures developed for astronauts and/or SFPs could prove useful for terrestrial efforts against physical inactivity.
Considerations for a European space research routine omics program
Designing an appropriate routine omics collection program for European astronauts and/or SFPs requires careful consideration of a number of factors. In this section, we discuss some of the key considerations, including collecting data with AI in mind, which sampling methods and omics data types should be collected based on practicality and perceived scientific return, and which ethical and legal challenges may need to be addressed.
AI readiness of omics data and metadata
Collecting the same standardized measures routinely from astronauts and/or SFPs across multiple missions would greatly improve the AI readiness of the generated datasets, especially when combined with accompanying standardized metadata, including spacecraft environmental measures (e.g., radiation dosimetry).7,31 One important practical consideration here will be digital storage requirements, because omics experiments generate large volumes of data. Similarly, harmonizing approaches with other space agencies will be important. In this context, there is an opportunity for the ESA to expand collaboration with the Frontier Development Lab, which it currently partners with to assist with AI approaches to Earth Observation and that is also currently partnered with NASA to assist with AI approaches across the entire Science Mission Directorate. In pursuit of AI readiness, it will be important to adhere to guidelines, such as well-implemented FAIR (findable, accessible, interoperable, and reusable) principles where possible,32,33 which could include machine-interpretable file formats, such as standardized processed omic data,18,34 alongside raw data and metadata. In order to further enhance the quantity of data, identical measures could be collected from well-designed, ground-based analog studies, to facilitate comparison and to unlock powerful machine learning (ML) techniques based on transfer learning. With the ever-increasing adoption and utility of AI-based approaches for healthcare and life sciences research,35 the importance of designing data collection and curation with AI capabilities in mind cannot be overstated. For example, it may eventually be possible to develop omic-based digital twin approaches to be used in applications, such as mission planning and countermeasure design. These approaches could model the physiological impact that mission parameters, such as duration and radiation exposure, may have on individual astronauts.36,37 Although there are a few interesting exceptions, at present, most AI approaches require large volumes of high-quality and domain-relevant training data.38, 39, 40 Notably, robust population-based analyses will be particularly challenging to achieve for humans in space, given their rarity. It is, therefore, important to initiate collection and curation of standardized astronaut and/or SFP omics data and metadata as soon as possible, thereby unlocking a future of powerful AI-based omic analyses for personalized medicine approaches and mechanistic discovery.
Omic data types
A key consideration for implementing a routine omics program will be selecting a panel of omic assays to capture from human subjects. Omics is increasingly being used as shorthand for big data approaches in biology and medicine, with numerous different omic data types and technologies now available at decreasing costs. Different omics data types and technologies offer both overlapping and unique insights into molecular changes within the body.18 Integrating multiple omic types (i.e., “multiomics”) via systems biology approaches can lead to a more holistic picture of spaceflight-associated molecular patterns.41 Initial multiomics studies during the NASA Twins mission and SpaceX Inspiration4 mission have opted for a relatively broad approach to assay selection.21 It makes sense to use findings from these missions to optimize a panel of assays for routine omics. This harmonized approach would also support comparability between data captured via a European routine omics program and these initial international studies. To further support comparability, it also seems logical to collect data types that are prevalent in large-scale terrestrial omic initiatives, such as The Cancer Genome Atlas and the European Genome-phenome Archive. The ability to easily compare astronaut data with terrestrial disease cohorts could allow for the identification of similar patterns, which could aid processes, such as countermeasure design (e.g., omic-based drug repurposing).42 While collecting the same measures over a long period of time is desirable, some flexibility is to be expected due to the continued evolution of omic technologies.
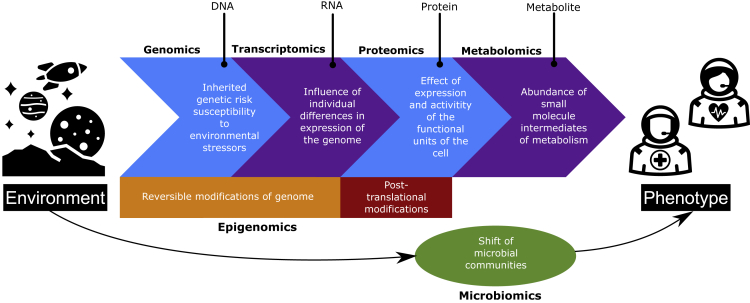
Although coverage of all potential omic types and technologies is beyond the scope of this article, there are a few clear candidates worth noting. The most immediately actionable and cost-effective measure in terms of healthcare will likely be pre-flight genomic screening,43 such as whole-genome sequencing (WGS). For example, a recent proof-of-concept study by the Mayo Clinic found that pre-emptive use of basic pharmacogenomic testing, alongside participants’ medication history, enabled pharmacists to offer medication improvement opportunities in 56% of participants.44 Some of the medications involved in the study (e.g., ondansetron, metoprolol, aripiprazole, sertraline, and phenytoin) have been documented as being part of the ISS medication kit.27 Thus, pre-flight genomic screening could potentially be used to inform medication management and personalized medicine approaches to spaceflight countermeasures.25 Similarly, microbiomic approaches, such as studying the genomic makeup of microbial communities via metagenomics, are likely to have high clinical relevance for humans in space. Metagenomic analyses of crew samples can be used to study microbiome shifts2 and potential health consequences, such as viral (e.g., herpes) reactivation and skin rashes;45,46 the addition of environmental samples to study microbial interactions between crew members and the spacecraft environment would also be useful for monitoring crew health (e.g., infectious disease management).47,48 On board the ISS, at least some coverage of metagenomics would likely fall under NASA’s microbial monitoring program. Other candidate omic data types for studying regulatory molecular changes, discovering biomarkers, and designing interventions for spaceflight-associated health risks include transcriptomics, epigenomics, proteomics, and metabolomics18 (Figure 2).
Figure 2.
Incorporating omics into astronaut studies
Overview of how candidate omic data types for studying and counteracting astronaut health risks interact with one another, the environment, and phenotype. Epigenomics is shown to span several omic data types to demonstrate that environmental exposures and activity occurring at the genomic, transcriptomic, and proteomic levels influence epigenomic status. Adapted from Yu et al.49 under CC BY 4.0 (https://creativecommons.org/licenses/by/4.0/).
Sampling time points
Due to the dynamic nature of physiological changes throughout the course of a typical spaceflight mission, it is scientifically meaningful to acquire omic measures across an array of time points. At a minimum, a sampling point just before flight and a sampling point taken shortly after the mission would prove useful for quantifying the molecular impact of the spaceflight mission. However, averaging multiple pre-flight measures would be valuable for establishing a robust molecular baseline, and multiple post-flight measures would prove useful for studying recovery and long-term health. To the latter point, long-term (i.e., “lifetime”) post-flight time points will be particularly valuable for missions beyond LEO, to study whether the human body fully recovers from spaceflight environmental factors, such as DNA damage response following the increased radiation dosage. To that point, it has recently been reported, albeit in a single pair of monozygotic twin astronauts, that onset of clonal hematopoiesis may be influenced by spaceflight.50 Clonal hematopoiesis results in lifelong changes in the blood that are associated with increased risk of cardiovascular disease and blood cancers.
The addition of in-flight measures, particularly for long-duration missions, would provide greater temporal resolution, allowing for spaceflight-induced molecular changes to be monitored over time. As a starting point, it may make sense to focus time points around expected points of significant physiological change, especially when clinically relevant. Significant physiological changes can occur rapidly, so early in-flight time points could be useful for studying their onset. For example, the first in-flight time point for the NASA Twins Study was at 14 days into the flight,21 whereas spaceflight research has indicated that significant physiological changes can occur before this point and, thus, earlier time points may be valuable. For instance, a recent study investigating anemia in astronauts collected blood and exhaled air samples at 5 ± 1 days into the 6-month flights and results already indicated significantly increased breakdown of red blood cells (hemolysis). Markers for increased hemolysis persisted throughout the in-flight time points (11 ± 1, 64 ± 5, and 157 ± 31 days), with 5 of 13 astronauts reaching clinical levels of anemia when measured 4 ± 1 days after landing.51 The potential implication that the longer the duration in space, the worse certain health risks, including anemia, suggests that in-flight time points across the full duration of spaceflight missions may be valuable. For example, other useful in-flight time points may include shortly before the end of the flight, so that this can be compared with the first post-flight time point to measure the impact of landing and initial readaptation, and shortly before and after EVA, to investigate the molecular impact of such events.
The sampling schedule also has implications for occupational health. For example, pre-flight omic measures could be used as a baseline for personalized medicine approaches. Measures taken in flight across different time points could then eventually be compared with the baseline data with ML approaches to predict health outcomes and suggest preventative interventions.37,52 Insights related to the molecular impact of mission events, such as EVA, could also be used to inform mission design, or indeed day-to-day task scheduling to manage stress responses.
While frequent sample time points are desirable, it will be important to balance the sampling schedule with regard to practical considerations, including cost, use of crew time, sample storage space, and subject recovery time (dependent on sample invasiveness). Additionally, if in-flight time points are included in the sampling schedule, it will be important to thoroughly train crew members for successful sample collection.
Accompanying functional measures
It is beneficial to routinely collect performance measures in tandem with omics, such as self-assessments (e.g., cognition, nutrition, and sleep quality), exercise outputs, and physiological measures collected from wearables and POC devices.29,53 When combined, omic and functional measures can be used to infer causal molecular-phenotypic links, which can then be used to inform and test interventions and discover novel biomarkers; if adopted, this approach would be a major advance in research capabilities, enabling widespread hypothesis-driven molecular mechanistic research to be conducted in astronauts. While outside the scope of this omic-focused article, the choice of which performance measures to collect must also be considered via an evidence-based approach, balancing scientific and clinical value with considerations regarding practicality, such as levels of invasiveness. Importantly, NASA’s standard measures program, which ESA astronauts can opt into, currently captures a curated selection of standardized functional measures from astronauts on the ISS.
Sample processing
One practical consideration regarding routine omics collection is the logistics of processing the samples. Currently, human biological samples collected on the ISS are typically preserved (e.g., via rapid freezing) and stored before being returned to Earth for sequencing and analysis. The multiomics profiling conducted for the NASA Twins Study shows that this model is feasible for longitudinal human studies on the ISS,21 yet the approach does come with some disadvantages, including sample degradation, delays in receiving results, and dependence on freezer space. Thus, while this approach with well-designed procedures for sample processing7 is likely to be appropriate for routine omics on the ISS, future commercial space stations, and short-duration missions, it may not be appropriate for all mission types. Therefore, at least for long-duration exploration missions, it may make sense to move toward an approach with reduced reliance on sample return to Earth.
One step toward enabling this in-flight autonomy is improving in-flight equipment capabilities, which has been demonstrated recently via usage of the portable Oxford Nanopore MinION sequencer on board the ISS for sequencing DNA54 and even human-derived RNA directly (Sarah Castro-Wallace, personal communication). Using this equipment, environmental swabs have been collected in-flight and then processed and sequenced on board (swab-to-sequence).47,55 As the technology becomes more compact and lightweight, autonomy of the sample processing may improve further still through robotic automation of spaceflight laboratories.56 One consideration is that in-flight generation of omics data presents challenges regarding data storage and transfer back to Earth. Importantly, downlink challenges compound with the communication delays introduced by increasing distance from Earth. In-flight computational processing of raw omics data can mitigate this issue by significantly reducing file size, and full-analysis pipelines have even been executed on-orbit, eradicating the need to downlink data to obtain results (Sarah Castro-Wallace, personal communication).
Importantly, in-flight processing of omics data has implications for occupational health in space. For example, in the context of NASA’s microbial monitoring program, in-flight processing can enable crew health to be monitored (e.g., infectious disease diagnosis) in near real time.47,55 As another use case, results obtained in-flight could eventually be inputted directly into AI models, such as clinical decision support systems (CDSSs), to inform the decisions of onboard medical officers in the event that the healthcare support teams on Earth cannot be contacted.
Sampling methods for spaceflight omics collection
Below, we introduce various biospecimens and sampling methods for omics data collection (Figure 3). We focus on approaches that have spaceflight heritage.
Figure 3.
Omic sampling from astronauts
The medical invasiveness of different sampling methods for spaceflight omics collection from astronauts or SFPs.
Tissue biopsy
Biopsies involve the removal of a piece of solid tissue or a sample of cells from the body. Thus, while biopsies present the gold standard for molecular profiling of human physiology, such procedures are invasive in nature, require specialized expertise, are often characterized by localized pain, and naturally carry an increased risk of infection. It is no surprise, therefore, that routine tissue biopsy sampling of astronauts in-flight is lacking, and omic studies in this sense remain constrained to those of cultured human cells flown to space—examples include transcriptomics and/or proteomics of human-induced pluripotent stem cell-derived cardiomyocytes57 and human intestinal epithelial cells.58 Given the inherent challenges of human tissue biopsy sampling in space, routine omics data collection from astronauts via this tool is most immediately likely through regular sampling programs pre- and post-flight, particularly for more readily extractable tissue types, such as skin and skeletal muscle. Indeed, in the case of the latter, muscle biopsies have previously been obtained from astronauts post- versus pre-flight to infer human skeletal muscle functional, morphological, biochemical, and, more recently, proteomic changes in space.3,59, 60, 61 With the application of various omic techniques (transcriptomics, metabolomics, proteomics, epigenomics, etc.) to biopsied skeletal muscle now being commonplace in terrestrial human physiology research, future multiomic analyses of astronaut muscle (and outwardly skin) obtained pre- and post-flight are entirely plausible. The existence of less-invasive (microbiopsy) techniques for sampling skeletal muscle62 and skin63 could make routine collection of these tissue types from astronauts during flight a realistic future prospect. In terms of a more immediate scientific return, omics collection via tissue biopsy thus lends itself as a promising tool for better understanding molecular determinants of skeletal muscle and skin responses to the spaceflight environment.
Blood
As a minimally invasive procedure, blood sampling has long been used as a primary alternative to tissue biopsies (i.e., “liquid biopsy) for molecular characterization of human health on Earth. Given the clear technical and operational advantages of blood versus tissue sampling in the spaceflight environment, blood sampling currently appears to be more viable for longitudinal monitoring of astronaut responses at the molecular level, particularly in flight. Blood samples are thus often collected from astronauts for analyses on Earth, and targeted molecular profiling of astronaut blood obtained before, during, and/or after spaceflight has subsequently been performed on numerous occasions, with particular emphasis on spaceflight-induced immune system dysregulation.64, 65, 66, 67 Additionally, comprehensive analyses of astronaut blood samples from the NASA Twins Study have yielded several new and important insights. For instance, initial multiomic (epigenomic, metabolomic, transcriptomic, and proteomic) analyses revealed wholesale gene expression changes in peripheral blood mononuclear cells and metabolite changes in plasma during spaceflight that were largely normalized upon return to Earth.21 Additional plasma proteomic profiling also uncovered patient-specific exosome protein signatures following the mission,68 with these blood samples also proving useful toward confirming human relevance of findings derived from omic analysis in model organisms.69 Other independent studies have also explored plasma metabolomic or proteomic changes in astronauts following long-term spaceflight exposure.3,70,71 Routine longitudinal multiomics of astronaut blood is thus firmly within reach. As the number of available samples across astronauts and time points continues to grow, blood omics therefore holds accelerated potential to expedite molecular understanding of human spaceflight responses in areas such as immune system dysregulation.
Urine and stool
Both normal human waste products that can be cheaply, frequently, and non-invasively sampled without specialized equipment or skill, urine, and stool (feces) naturally present themselves as strong candidate sample types for routine, longitudinal astronaut omics collection. Indeed, alongside blood, urine is a common form of liquid biopsy for terrestrial biomonitoring of humans, and can be used to screen for a variety of different (patho)physiological biomarkers (renal, cardiac, prostate, bone, etc.).72, 73, 74 However, unlike blood, urine is absent of a homeostatic mechanism and can, therefore, retain/accommodate more substances that reflect changes produced in vivo, while its low complexity lends itself to straightforward detection of low-abundance proteins.75 As such, urine may be considered as better suited for early-stage biomonitoring, particularly in terms of the kidneys.75,76 Astronaut urine omics is consequently starting to grow, with several recent studies demonstrating the multiomic (namely metabolomic and proteomic) potential of this sample type for gaining new information on human body adaptations following spaceflight.21,72,77,78 For instance, metabolomic/proteomic analyses of urine from the NASA Twins Study revealed urinary excretion of COL1A1 and COL3A1 as potential biomarkers of muscle, tendon, or bone changes during spaceflight.21 Cardiovascular-related urine proteome changes, including some associated with autonomic regulation of heart rate, have also been delineated, offering several promising urine biomarkers for screening cardiovascular responses to spaceflight.77,78
On Earth, stool has become a primary sample choice for metagenomic profiling of the gut microbiome, which plays a critical role in the internal environment and concomitantly (patho)physiological state of an individual.79, 80, 81 Recognition is now also growing for a potential role of the gut microbiome toward physiological maladaptation of astronauts during spaceflight.82 Metagenomic studies of astronaut stool have thus gained traction, demonstrating spaceflight-induced alterations in both the composition and function of an astronaut’s gut microbiome,45,83 as well as uncovering genetic and immunological evidence for transfer of environmental strains to an astronaut’s gut microbiome on board the ISS.84 Thus, astronaut stool (and outwardly other samples; see below) metagenomics can serve as a useful tool not only for guiding continual health monitoring but also mission planning and habitat design.84 Future omic studies of astronaut stool would perhaps benefit from complementary metabolomics in order to improve functional readout of gut microbiome activity.85
In closing, it appears clear that stool and urine analyses could prove practical for routine astronaut omics collection, with strong potential for rapid scientific return.
Saliva and body swabs
While a potent contributor, the human microbiome is not merely defined by the gut but rather the full complement of microbes (and their genes) that reside on or within the host. These microbes interact together alongside host genetics and the environment to contribute toward an individual’s health state.86, 87, 88, 89, 90 Wider-ranging microbiome understanding in space is thus essential for biomonitoring astronaut health and, as noted above, other factors, such as habitat design. Indeed, the ISS is a unique closed environment with rich microbial diversity91, 92, 93, 94 which, combined with possible influx and outflow of microorganisms at times of spacecraft exchange,48 may hold potential to have an impact on astronaut health. In terms of spaceflight, saliva, nose, and mouth/buccal samples present alongside stool as useful for providing insight on astronaut health and physiological conditions, whereas skin and the ear are prime sampling sites to explore interactions between an astronaut and the spaceflight environment.95 Importantly, routine, longitudinal metagenomics collection from all these locations is a highly feasible prospect in the context of spaceflight, since all are readily accessible for frequent non-invasive sampling via swab (or spit sample, in the case of saliva) at little cost or time expense and without the need for specialized skill. Demonstrating the associated scientific potential are several other recent metagenomic studies of astronauts, revealing new information on host- and site-(in)dependent microbiome responses across these various locations during spaceflight as well as microbial transmission between the astronauts and the ISS surface and vice versa.45,46,48,95,96 Thus, as complementary to stool, metagenomics collection from saliva and body swabs should serve as a strong tool for better understanding molecular determinants of astronaut health in the immediate future.
Hair
While perhaps an intriguing sample choice to note on face value, hair as a source for omics collection may represent a useful complementary source for routine biomonitoring of human physiological responses to spaceflight. Indeed, hair is readily available and easily sampled via extraction of hair follicles, requiring no specific personal expertise nor complex hardware.97,98 Moreover, hair cells can reflect host physical conditions, including human peripheral circadian clock and organismal metabolic responses to environmental changes.98 Omic studies of astronaut hair are thus beginning to emerge. Notably, a seminal study by the Japan Aerospace Exploration Agency (JAXA) conducted transcriptomics on astronaut hair follicles collected before, during, and after 6 months of spaceflight.99 Although several markers of hair growth were unsurprisingly dysregulated during flight, interestingly, hair PCDH8 was derived as a potential biomarker for astronaut neurochemical changes in the space environment.99 Terrestrial studies of human hair also span other omic lines (e.g., proteomics and metabolomics),100,101 highlighting the potential for multiomic profiling of astronaut hair samples. Nevertheless, the extent to which various physiological responses to spaceflight can be tracked by biomarkers in hair warrants a better understanding, before the true scientific potential of hair omics toward biomonitoring astronaut health can be established.
Breath
A final sample type worthy of note when it comes to astronaut omics collection is breath. Indeed, molecular analysis of exhaled breath is rapidly emerging as a useful tool on Earth for biomonitoring of an individual in health and disease (“breath-print”), both at the level of the lung as well as other vital organs via systemic circulation (e.g., the heart).102,103 Exhaled breath analysis is plausibly suited to the spaceflight environment, as samples can be acquired frequently in a non-invasive manner, with relative ease and at little expense.102 Moreover, for certain omic types, such as proteomics, exhaled breath condensate may be better suited for rapid analysis versus other forms of liquid biopsy, such as blood and urine.104 Nevertheless, omic studies of astronaut breath remain very much in their infancy. In one of the only studies to date,104 proteomic analysis of exhaled breath condensate obtained from cosmonauts before and after long-term spaceflight revealed strong enrichment for proteins involved in keratinization and pathogenic E. coli infection, offering support to the potential utility of breath analysis for non-invasive monitoring of astronaut health and respiratory tract pathology.104 On Earth, metabolomic study of human exhaled air (“breathomics”) has also proven to be highly feasible and consequently gained great traction,105 though transcriptomic studies are currently lacking, perhaps owing to poor knowledge of present mRNAs due to high fragmentation of RNA isolated from exhaled breath condensate.106 Nonetheless, sample collection advantages coupled with multiomic potential106 make exhaled breath a highly promising complementary source for molecular profiling of astronaut health, particularly with respect to the respiratory system.
Ethical and legal considerations for human space omics
In this section, the ethical challenges and legislative landscape within Europe will be considered in the context of astronaut and SFP-derived omics data. Focus is given to harmonious laws across Europe, but it should be acknowledged that national laws and the involvement of non-European law, via multinational entities (e.g., commercial spaceflight companies) and international collaborations, can introduce further complexities. Ultimately, legislation aims to find a balance between supporting scientific research and healthcare while protecting participants from harm; data should be as open as possible in order to facilitate further analysis and as protected as necessary for the participants donating data. We intend to summarize these ethical and legal frameworks to encourage productive development of policies pertaining to human space omics research, which could eventually cover biobanking and controlled utilization of omics data.
When are data personal?
In May 2018, the General Data Protection Regulation 2016/679 (GDPR) came into force in the European Union. The regulation aims to enhance individuals’ control over their data. The impact of the GDPR on omics in healthcare and research has recently been reviewed in a detailed report by the PHG Foundation.107
Article 3(1) of the GDPR states that “This Regulation applies to the processing of personal data in the context of the activities of an establishment of a controller or a processor in the Union, regardless of whether the processing takes place in the Union or not.” Therefore, a routine omics program for the ESA would most likely fall within the territorial scope of the GDPR, but importantly this is only the case if the data are deemed to be personal data.
From the definition of personal data in Article 4(1) and Recital 26 of the GDPR, it would seem apparent that omics data could be considered to be personal data if they are deemed reasonably likely to be either directly or indirectly identifiable. Hypothetically, if a database contains solely omics with all other identifiers removed, additional information would be needed to indirectly identify the natural person from whom the data are derived. This process is sometimes referred to as a linkage attack.
Holding some similarity to rare disease cohorts,108, 109, 110 astronauts and SFPs are currently a small population of public figures. Consequently, information, including mission status, and phenotypic information, such as hair color, height, and sex, pertaining to individuals are readily available to the public. It could, therefore, be possible to link astronauts’ or SFPs’ omic data to this additional information in order to identify individuals.111 However, an important caveat is that different omics data types have different levels of identifiability when combined with phenotypic information, meaning that some will qualify as personal data while others do not.
Omics describing the unique DNA sequence of individuals, such as SNPs, are generally regarded as having the highest level of identifiability. Traits, including eye color, skin color, and sex, are considered simple to accurately predict from data sources, such as WGS.112 Thus, in the unique context of spaceflight cohorts, such data types would likely be classed as personal data. However, the identifiability of other omics types, such as transcriptomics and proteomics, is also an emerging area of research.113, 114, 115, 116 Furthermore, the generation of multiomics datasets adds an additional layer of complexity because there could be relationships between the different omic types that increase the overall likelihood of identification.117 Additionally, raw data hold increased potential for identification compared with processed data, so file formats are an important consideration for appropriate data dissemination. Finally, aggregated data, which combine datasets from a population of individuals (e.g., to produce an average value), can reduce identifiability. However, aggregated approaches can significantly reduce utility for research into individual differences associated with spaceflight-induced biological changes and, therefore, utility in personalized medicine approaches.
It is also worth mentioning that the familial element of some omic data types means that if it is deemed reasonably likely for someone to identify a family member from the astronaut or SFP’s personal omics data, then it would also be classified as that family member’s personal data, so their rights may also be brought into consideration.
Approval for data collection and processing
When it comes to navigating the ethics of conducting human research studies in Europe, there are several prominent non-legal binding instruments, including the Declaration of Helsinki, which was first formulated in 1964, with several subsequent revisions.118 These instruments have introduced key concepts that have become encoded within national laws and regulations across Europe. Two of the primary concepts are independent ethical review committees and informed consent.
First, all research collecting omics data from human subjects must gain approval from an independent ethical review committee before the research can commence. The committee consists of a panel of experts, who provide oversight for human research, ensuring that the research is conducted in agreement with internationally and locally accepted ethical guidelines and in compliance with the law. In cases of data derived from ESA astronauts flying on missions with international collaborators, approval will likely need to be granted by international ethics committees in addition to European committees. For example, the Human Research Multilateral Review Board (HRMRB) exists for overseeing human research on the ISS, with board members from the international partners.
As part of addressing the ethical review committees, investigators would be expected to demonstrate collection of informed consent from all study participants. Prior to providing such consent, participants should be clearly informed of the research purpose, experimental and data handling procedures, the potential risks, and their right to withdraw from the study at any time. As mentioned in the previous section, utility of omics is ever-increasing with the true capability and, by consequence, identifiability, of omics yet to be unlocked. This landscape of unknowns makes informed consent difficult to meaningfully define, particularly when research is not necessarily hypothesis driven, as is the case with the longitudinal omics collection program proposed in this article. In this context, broad consent has emerged in law (e.g., GDPR Recital 33) and practice as a potential means of obtaining consent for collecting omics for use in unspecified future research projects, including biobanking.119 Notably, however, in the United States, broad consent was used in the recent multiomics profiling study onboard the commercial SpaceX Inspiration4 mission, to gain consent from the crew for the archiving and future use of their samples and data for space health research.22
In order for processing to be permitted, European space omics data that are deemed to be personal data must satisfy a legal basis pursuant to Article 6 of the GDPR. In the context of space agencies or commercial companies, several of the reasons enumerated in Article 6 could prove relevant. To paraphrase, relevant bases may include (1) consent, (2) performance of a contract, and (3) public interest. However, omics personal data would also be considered special category data within the GDPR, either under the classification of genetic data or data concerning health, in Article 4 and Recitals 34 and 35.120 Article 9 states that such data types cannot be processed unless at least one of ten exceptions applies. To summarize, potentially relevant exceptions include (1) explicit consent, (2) the data subject making their data public, (3) medical purposes, and (4) archiving scientific data in the public interest. Ultimately, the restrictions on processing imposed by Articles 6 and 9 of the GDPR require careful consideration in the context of a space agency or commercial company processing astronaut or SFP personal omics data. There are many interesting angles to consider based on space agencies’ multifaceted roles as research institutions, government agencies, employers, and primary care providers for astronauts121 and the societal role of spaceflight. To the latter point, the United Nations’ Outer Space Treaty of 1967—considered to be the backbone of international space law—states that astronauts should be regarded as “envoys of mankind.” The advent of commercial spaceflight missions adds additional considerations, especially due to the variety of mission types with varying degrees of scientific value.
Ethical issues pertaining to data usage
A key ethical challenge involved in omics research is the handling of incidental findings in consideration of duty of care. Analysis of astronaut or SFP-derived omics could reveal pertinent findings, such as gene mutations associated with increased SANS risk; however, incidental findings that are not necessarily relevant in the spaceflight context, such as increased risk of late-onset Alzheimer’s disease, could also be unintentionally revealed.122 The ethical dilemma lies in the decision of whether to inform the participants, and indeed their family members, of these incidental findings, which may not be medically actionable. Ultimately, the protocol for handling incidental findings should be clearly defined when obtaining informed consent. Protocol could include giving participants the choice to opt in or out of receiving these results as well as developing appropriate support services when needed.
A further ethical challenge surrounding the processing and usage of omics data is genetic discrimination, which is when an individual receives differential treatment based on their genetic information. In Europe, genetic discrimination is typically addressed in national laws, which are influenced by regional instruments, including the 2000 Charter of Fundamental Rights of the European Union and the 1997 Convention on Human Rights and Biomedicine (also known as the Oviedo Convention).123 Chapter IV of the Oviedo Convention prohibits any form of discrimination based on genetic heritage and also prohibits the use of tests to predict genetic predisposition to diseases, unless these tests are carried out for medical or scientific purposes, together with the provision of adequate genetic counseling. Thus, this would seem to permit the use of omics to study individual susceptibility to spaceflight risks, such as radiation-induced cancers, as long as genetic counseling is provided. However, the potential role of omics-based susceptibility and predisposition in processes, such as flight assignment, nonetheless remains a contentious issue. In the United States, the 2008 Genetic Information Nondiscrimination Act (GINA) prevents NASA from making employment decisions, such as flight assignment, based on genetic data.121,122 From a European perspective, largely due to differences in national laws, the legality of using omics data in employment decisions, such as flight assignment, appears to be less clear. However, it seems likely that in flight assignment, the use of genomic susceptibility to disease, as opposed to manifestation, would be viewed as discriminatory. Additionally, ESA’s recent Parastronaut Feasibility Project could be seen to suggest a more inclusive approach to flight assignment; thus, the focus of omics technologies should probably be to reduce risk and improve health outcomes via personalized medicine approaches.25 However, some personalized medicine approaches are likely to be based on the processing of omics data deemed to be personal data under the GDPR. This could present a new challenge in operations with international partners. For example, if other space agencies such as NASA deploy personalized countermeasures but are unable to process data from ESA astronauts in compliance with the GDPR, either ESA astronauts will not be able to partake in the use of these countermeasures to improve occupational health or ESA will need to be able to deploy personalized countermeasure via an ESA program; personalized medicine approaches have recently been considered by an ESA Topical Team.25 Similarly, emergent challenges relating to GDPR compliance during processing of personal data from Europeans on commercial flights need to be considered in the context of initiatives, such as the database recently established by TRISH.22
Storing and sharing data
Because the ESA is an independent treaty organization based in Europe, sharing omics data within ESA’s network is of great importance. All 22 member states, associate members, and countries with cooperation agreements, including Canada, which sits on the Council, appear to follow the GDPR or are considered, via adequacy decisions, secure third countries due to similarities in data protection law. Thus, data can be transferred between these countries, as long as all parties first reach a consensus on issues, such as the basis for lawful processing and what constitutes personal data. While this sounds simple, reaching a consensus can be challenging because the GDPR allows scope for variation via national laws (e.g., Article 9), which may lead to key differences in interpretation and application of the GDPR.124,125 Furthermore, space biology research typically involves a great deal of international collaboration with other space agencies, posing a need for international sharing of data.
For astronaut and/or SFP omics data deemed not to be personal data, international sharing could be relatively simple. For example, the data could potentially be uploaded to a publicly accessible biobank, such as the NASA GeneLab data repository.126 For context, NASA GeneLab has emerged as the primary database for spaceflight omics data, with significant European representation in GeneLab’s Analysis Working Groups (AWGs) and experimental datasets.127 However, if the data are deemed to be personal data, transfers to countries without adequacy decisions, including the United States, become challenging and unclear at present due to the recent “Schrems II” judgment.128
Due to challenges with ensuring GDPR compliance during global sharing of sensitive healthcare and research data, alternative models for data sharing have been considered. For example, in congruence with a declaration signed by 21 European countries to transnationally share data on at least one million human genomes by the end of 2022, large-scale initiatives, such as the European Genome-Phenome Archive, are shifting toward federated approaches.129 Federated approaches can invert the traditional data-sharing paradigm of bringing the data to the analysis by instead bringing the analysis to the data. Through federated approaches, researchers can analyze data across a distributed network of databases and then combine the results, potentially avoiding the transfer of personal data across jurisdictions.130,131 This model can also be applied to the training of AI models, in a process known as federated learning. For example, a successful model for predicting dyspnea, a common side effect after radiotherapy treatment of lung cancer, was trained on sensitive clinical data from several different hospitals across Europe, without the data ever having to leave the individual hospitals.132 Federated approaches could potentially serve as an appropriate solution for enabling international access and analysis of sensitive astronaut and/or SFP healthcare or research data, while ensuring international legal compliance. Other potential approaches to ease data sharing include data sanitization methods, to convert identifiable data into non-identifiable formats.133 However, methods that modify the data should be carefully balanced against the potential loss of scientific utility.
With the specific security issues of data-sharing strategies in mind,130,134,135 data storage solutions, such as biobanks housing identifiable astronaut and/or SFP omics data, should meet high standards of security, to prevent issues such as data breaches. Chapter 4 of the GDPR requires that data controllers approach “data protection by design” and establish safeguards to protect the privacy and security of data in a manner that is proportionate to the risks involved.
Conclusions
Humanity has laid ambitious plans to venture to Mars and establish settlements on other planetary bodies, and commercial spaceflight is gaining rapid traction with the spacefaring population set to quickly increase both in terms of size and diversity. These new endeavors present new challenges for human health in space. Crucially, more data are needed, especially for environmental conditions beyond LEO, where biological and molecular adaptations remain uncharted. Recently the first multiomic profiling studies of astronauts have been performed in the NASA Twins and SpaceX Inspiration4 missions. With their feasibility for spaceflight now demonstrated, these biological big data approaches are primed to transform our understanding of health in space, as they have for health on Earth. Importantly, in addition to elucidating mechanistic understanding of spaceflight-induced physiological deconditioning, these approaches also hold strong potential to improve risk quantification and occupational health in space, such as through the design and deployment of personalized medicine approaches. Thus, routinely applying these same approaches to European human subjects, including participants in analog environments and astronauts and/or SFPs in spaceflight missions, is a golden opportunity.
In order to achieve the full potential of these big data approaches, several challenges need to be addressed in a cohesive European fashion. First, how do we best store and ensure collected data are AI ready? Second, which datasets should we collect and when? Third, how do we enable open science approaches while protecting personal data? Although we have introduced the challenges and proposed some options, it is important to achieve European consensus on these questions (Table 1). We propose forming an ESA topical team and/or holding an ESA workshop as a strategic way forward. The goal is to bring together European human omics/data science researchers and human subject researchers to address these questions in coordination with ESA and stakeholders from the commercial spaceflight sector. Importantly, this consensus will need to consider the international context of human research on the ISS, the commercial aspects of data science, and the personal nature of the data. These latter aspects could be achieved via collaboration with organizations, such as International Standards for Space Omics Processing (ISSOP), HRMRB, and TRISH.
Table 1.
Goals and potential topics of discussion for suggested ESA topical team/workshops
| Goal: To to explore routine omics collection by deciding which omics data to collect routinely, from which samples, and at which time points. |
|---|
| Potential topics to be addressed |
| ➔ Which omic types and technologies have the highest potential for scientific return and clinical actionability (with additional consideration to multi-omic combinations)? |
| ➔ Which sample types are the most practical to collect (i.e., cost, sample processing procedures, and invasiveness)? |
| ➔ What metadata would be the most useful and practical to standardize alongside omics collection (e.g., physiological, environmental, and lifestyle)? |
| ➔ At which time points should omics be collected? |
| ➔ How can the AI readiness of the generated datasets be maximized? |
| Goal: To help establish clear ESA policy on human omics research governance |
|---|
| Potential topics to be addressed |
| ➔ What is the potential identifiably of different omic types, in the specific context of astronauts? |
| ➔ How should ESA go about obtaining meaningful informed consent for omics research? |
| ➔ Which legal bases should be used for processing astronaut and/or SFP omics data? |
| ➔ How should ESA handle potential ethical issues, such as incidental findings and discrimination? |
| ➔ How should data storage and data sharing between ESA’s European and international network be handled? |
Acknowledgments
H.C., R.H., J.B., D.B., S.G., T.E., and N.J.S. are members of the ESA Space Omics Topical Team, funded by the ESA grant/contract 4000131202/20/NL/PG/pt “Space Omics: Towards an integrated ESA/NASA –omics database for spaceflight and ground facilities experiments” awarded to R.H., which was the main funding source for this work. H.C. is also supported by the Horizon Center for Doctoral Training at the University of Nottingham (UKRI grant no. EP/S023305/1). S.G. is supported by the Swedish Research Council VR grant 2020-04864. L.A.R. and M.M. represent the Omics Subgroup of the Japan Society for the Promotion of Science KAKENHI funding group “Living in Space” and are supported by JP15K21745, JP20H03234, and 20F20382. L.A.R. is also supported by the JSPS postdoctoral fellowship P20382. We thank Dr. Sarah Castro-Wallace, the NASA GeneLab Animal AWG, ISSOP, ESA Space Omics Topical Team, ESA Personalized Medicine Topical Team, and Global Alliance for Genomic Health (GA4GH) for useful discussions.
Author contributions
Conceptualization, H.C., L.A.R., S.G., T.E., and N.J.S.; investigation, H.C., C.R.G.W., M.J.M., T.E., N.J.S., and D.B.; writing – original draft, H.C. and C.R.G.W.; writing – review & editing, all authors; visualization, H.C. and M.J.M.; supervision, N.J.S., C.E.M., and M.M.; funding acquisition, R.H., H.C., S.G., L.A.R., and M.M.
Declarations of interests
D.B. is the CSO at yuri GmbH and co-founder of Poppy Health. M.M. is an employee of LSI Medience Corporation through the cross-appointment system of University of Tsukuba and a research advisor of Robotic Biology Institute. C.E.M. is a co-founder of Biotia and Onegevity Health.
Biography
About the author
Henry Cope is a PhD student at the United Kingdom Engineering and Physical Sciences Research Council (EPSRC)-funded Horizon Centre for Doctoral Training at the University of Nottingham, UK. The research involves investigation of data science approaches for space biology, with the goal of improving human health in space. This includes analysis of spaceflight-derived biological datasets via bioinformatics and computer vision approaches. Additionally, barriers to data science in the field of space biology are considered, including standardization of data and metadata and the ethical and legal challenges pertaining to astronaut personal data.
References
- 1.Griko Y.V., Loftus D.J., Stolc V., Peletskaya E. Private spaceflight: a new landscape for dealing with medical risk. Life Sci. Space Res. 2022;33:41–47. doi: 10.1016/j.lssr.2022.03.001. [DOI] [PubMed] [Google Scholar]
- 2.Afshinnekoo E., Scott R.T., MacKay M.J., Pariset E., Cekanaviciute E., Barker R., Gilroy S., Hassane D., Smith S.M., Zwart S.R., et al. Fundamental biological features of spaceflight: advancing the field to enable deep-space exploration. Cell. 2020;183:1162–1184. doi: 10.1016/j.cell.2020.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rittweger J., Albracht K., Flück M., Ruoss S., Brocca L., Longa E., Moriggi M., Seynnes O., Di Giulio I., Tenori L., et al. Sarcolab pilot study into skeletal muscle’s adaptation to long-term spaceflight. NPJ Microgravity. 2018;4:18. doi: 10.1038/s41526-018-0052-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gabel L., Liphardt A.M., Hulme P.A., Heer M., Zwart S.R., Sibonga J.D., Smith S.M., Boyd S.K. Pre-flight exercise and bone metabolism predict unloading-induced bone loss due to spaceflight. Br. J. Sports Med. 2022;56:196–203. doi: 10.1136/bjsports-2020-103602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee A.G., Mader T.H., Gibson C.R., Tarver W. Space flight-associated neuro-ocular syndrome. JAMA Ophthalmol. 2017;135:992. doi: 10.1001/jamaophthalmol.2017.2396. [DOI] [PubMed] [Google Scholar]
- 6.Stepanek J., Blue R.S., Parazynski S. Space medicine in the era of civilian spaceflight. N. Engl. J. Med. 2019;380:1053–1060. doi: 10.1056/nejmra1609012. [DOI] [PubMed] [Google Scholar]
- 7.Rutter L., Barker R., Bezdan D., Cope H., Costes S.V., Degoricija L., Fisch K.M., Gabitto M.I., Gebre S., Giacomello S., et al. A new era for space life science: international standards for space omics processing. Patterns. 2020;1:100148. doi: 10.1016/j.patter.2020.100148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Ombergen A., Rossiter A., Ngo-Anh T.J. “White Mars” - nearly two decades of biomedical research at the Antarctic Concordia station. Exp. Physiol. 2021;106:6–17. doi: 10.1113/ep088352. [DOI] [PubMed] [Google Scholar]
- 9.Stokes M., Evetts S., Rittweger J., Weber T., Caplan N., Danneels L., Hides J., Kuipers A., Lambrecht G., Peterson N., et al. European Space Agency; 2016. Recommendations for Future Post-mission Neuro-Musculoskeletal Reconditioning Research and Practice Post-mission Exercise (Reconditioning) Topical Team Report. [Google Scholar]
- 10.Zhang S., Wimmer-Schweingruber R.F., Yu J., Wang C., Fu Q., Zou Y., Sun Y., Wang C., Hou D., Böttcher S.I., et al. First measurements of the radiation dose on the lunar surface. Sci. Adv. 2020;6:eaaz1334. doi: 10.1126/sciadv.aaz1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeitlin C., Hassler D.M., Cucinotta F.A., Ehresmann B., Wimmer-Schweingruber R.F., Brinza D.E., Kang S., Weigle G., Böttcher S., Böhm E., et al. Measurements of Energetic Particle Radiation in Transit to Mars on the Mars Science Laboratory. Science. 2013;340 doi: 10.1126/science.1235989. [DOI] [PubMed] [Google Scholar]
- 12.Restier-Verlet J., El-Nachef L., Ferlazzo M.L., Al-Choboq J., Granzotto A., Bouchet A., Foray N. Radiation on earth or in space: what does it change? Int. J. Mol. Sci. 2021;22:3739. doi: 10.3390/ijms22073739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simonsen L.C., Slaba T.C., Guida P., Rusek A. NASA’s first ground-based Galactic Cosmic Ray Simulator: enabling a new era in space radiobiology research. PLoS Biol. 2020;18:e3000669. doi: 10.1371/journal.pbio.3000669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walsh L., Hafner L., Straube U., Ulanowski A., Fogtman A., Durante M., Weerts G., Schneider U. A bespoke health risk assessment methodology for the radiation protection of astronauts. Radiat. Environ. Biophys. 2021;60:213–231. doi: 10.1007/s00411-021-00910-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Linnarsson D., Carpenter J., Fubini B., Gerde P., Karlsson L.L., Loftus D.J., Prisk G.K., Staufer U., Tranfield E.M., van Westrenen W. Toxicity of lunar dust. Planet. Space Sci. 2012;74:57–71. doi: 10.1016/j.pss.2012.05.023. [DOI] [Google Scholar]
- 16.Petersen N., Lambrecht G., Scott J., Hirsch N., Stokes M., Mester J. Postflight reconditioning for European Astronauts - a case report of recovery after six months in space. Musculoskelet. Sci. Pract. 2017;27(Suppl 1):S23–S31. doi: 10.1016/j.msksp.2016.12.010. [DOI] [PubMed] [Google Scholar]
- 17.Schmidt M.A., Goodwin T.J. Personalized medicine in human space flight: using Omics based analyses to develop individualized countermeasures that enhance astronaut safety and performance. Metabolomics. 2013;9:1134–1156. doi: 10.1007/s11306-013-0556-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hasin Y., Seldin M., Lusis A. Multi-omics approaches to disease. Genome Biol. 2017;18:83. doi: 10.1186/s13059-017-1215-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Willis C.R.G., Gallagher I.J., Wilkinson D.J., Brook M.S., Bass J.J., Phillips B.E., Smith K., Etheridge T., Stokes T., McGlory C., et al. Transcriptomic links to muscle mass loss and declines in cumulative muscle protein synthesis during short-term disuse in healthy younger humans. FASEB. J. 2021;35:e21830. doi: 10.1096/fj.202100276rr. [DOI] [PubMed] [Google Scholar]
- 20.Kousathanas A., Pairo-Castineira E., Rawlik K., Stuckey A., Odhams C.A., Walker S., Russell C.D., Malinauskas T., Wu Y., Millar J., Shen X., Elliott K.S., Griffiths F., Oosthuyzen W., Morrice K., Keating S., Wang B., Rhodes D., Klaric L., Zechner M., Parkinson N., Siddiq A., Goddard P., Donovan S., Maslove D., Nichol A., Semple M.G., Zainy T., Maleady-Crowe F., Todd L., Salehi S., Knight J., Elgar G., Chan G., Arumugam P., Patch C., Rendon A., Bentley D., Kingsley C., Kosmicki J.A., Horowitz J.E., Baras A., Abecasis G.R., Ferreira M.A.R., Justice A., Mirshahi T., Oetjens M., Rader D.J., Ritchie M.D., Verma A., Fowler T.A., Shankar-Hari M., Summers C., Hinds C., Horby P., Ling L., McAuley D., Montgomery H., Openshaw P.J.M., Elliott P., Walsh T., Tenesa A., GenOMICC Investigators. 23andMe. Covid-19 Human Genetics Initiative. Fawkes A., Murphy L., Rowan K., Ponting C.P., Vitart V., Wilson J.F., Yang J., Bretherick A.D., Scott R.H., Hendry S.C., Moutsianas L., Law A., Caulfield M.J., Baillie J.K. Whole genome sequencing reveals host factors underlying critical Covid-19. Nature. 2022 doi: 10.1038/s41586-022-04576-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garrett-Bakelman F.E., Darshi M., Green S.J., Gur R.C., Lin L., Macias B.R., McKenna M.J., Meydan C., Mishra T., Nasrini J., et al. The NASA Twins Study: a multidimensional analysis of a year-long human spaceflight. Science. 2019;364:eaau8650. doi: 10.1126/science.aau8650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Urquieta E., Wu J., Hury J., Donoviel D. Establishment of an open biomedical database for commercial spaceflight. Nat. Med. 2022;28:611–612. doi: 10.1038/s41591-022-01761-y. [DOI] [PubMed] [Google Scholar]
- 23.Deane C.S., Borg J., Cahill T., Carnero-Diaz E., Etheridge T., Hardiman G., Leys N., Madrigal P., Manzano A., Mastroleo F., et al. Space omics research in Europe: contributions, geographical distribution and ESA member state funding schemes. iScience. 2022;25:103920. doi: 10.1016/j.isci.2022.103920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zwart S.R., Gregory J.F., Zeisel S.H., Gibson C.R., Mader T.H., Kinchen J.M., Ueland P.M., Ploutz-Snyder R., Heer M.A., Smith S.M. Genotype, B-vitamin status, and androgens affect spaceflight-induced ophthalmic changes. FASEB. J. 2016;30:141–148. doi: 10.1096/fj.15-278457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pavez Loriè E., Baatout S., Choukér A., Choukér A., Buchheim J.I., Baselet B., Dello Russo C., Wotring V., Monici M., Morbidelli L., et al. The future of personalized medicine in space: from observations to countermeasures. Front. Bioeng. Biotechnol. 2021;9:739747. doi: 10.3389/fbioe.2021.739747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lulli M., Cialdai F., Vignali L., Monici M., Luzzi S., Cicconi A., Cacchione S., Magi A., Balsamo M., Vukich M., et al. The Coenzyme Q10 as an antiapoptotic countermeasure for retinal lesions onboard the International Space Station. Front. Physiol. 2018;9 doi: 10.3389/conf.fphys.2018.26.00036. [DOI] [Google Scholar]
- 27.Stingl J.C., Welker S., Hartmann G., Damann V., Gerzer R. Where failure is not an option -personalized medicine in astronauts. PLoS One. 2015;10:e0140764. doi: 10.1371/journal.pone.0140764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sawyers L., Anderson C., Boyd M.J., Hessel V., Wotring V., Williams P.M., et al. Astropharmacy: pushing the boundaries of the pharmacists’ role for sustainable space exploration. Res. Soc. Adm. Pharm. 2022 doi: 10.1016/j.sapharm.2022.02.002. [DOI] [PubMed] [Google Scholar]
- 29.Strangman G.E., Sawyer A., Fabre K.M., Urquieta E., Hury J., III, Donoviel D., Wu J., Peterman A., Hoffman J., Donoviel D. Deep-space applications for point-of-care technologies. Curr. Opin. Biomed. Eng. 2019;11:45–50. doi: 10.1016/j.cobme.2019.08.014. [DOI] [Google Scholar]
- 30.Lee I.-M., Shiroma E.J., Lobelo F., Puska P., Blair S.N., Katzmarzyk P.T., Lancet Physical Activity Series Working Group Effect of physical inactivity on major non-communicable diseases worldwide: an analysis of burden of disease and life expectancy. Lancet. 2012;380:219–229. doi: 10.1016/s0140-6736(12)61031-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scott R.T., Grigorev K., Mackintosh G., Gebre S.G., Mason C.E., Del Alto M.E., Costes S.V. Advancing the integration of biosciences data sharing to further enable space exploration. Cell Rep. 2020;33:108441. doi: 10.1016/j.celrep.2020.108441. [DOI] [PubMed] [Google Scholar]
- 32.Wilkinson M.D., Dumontier M., Aalbersberg I.J.J., Appleton G., Axton M., Baak A., Blomberg N., Boiten J.-W., da Silva Santos L.B., Bourne P.E., et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data. 2016;3:160018. doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jacobsen A., de Miranda Azevedo R., Juty N., Batista D., Coles S., Cornet R., Courtot M., Crosas M., Dumontier M., Evelo C.T., et al. FAIR principles: interpretations and implementation considerations. Data Intell. 2020;2:10–29. doi: 10.1162/dint_r_00024. [DOI] [Google Scholar]
- 34.Overbey E.G., Saravia-Butler A.M., Zhang Z., Rathi K.S., Fogle H., da Silveira W.A., Barker R.J., Bass J.J., Beheshti A., Berrios D.C., et al. NASA GeneLab RNA-seq consensus pipeline: standardized processing of short-read RNA-seq data. iScience. 2021;24:102361. doi: 10.1016/j.isci.2021.102361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bohr A., Memarzadeh K. In: Artificial Intelligence in Healthcare. Bohr A., Memarzadeh K., editors. Academic Press; 2020. Chapter 2 - the rise of artificial intelligence in healthcare applications; pp. 25–60. [Google Scholar]
- 36.Björnsson B., Borrebaeck C., Elander N., Gasslander T., Gawel D.R., Gustafsson M., Jörnsten R., Lee E.J., Li X., Lilja S., et al. Digital twins to personalize medicine. Genome Med. 2019;12:4. doi: 10.1186/s13073-019-0701-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scott R.T., Antonsen E.L., Sanders L.M., Hastings J.J.A., Park S.-M., Mackintosh G., Reynolds R.J., Hoarfrost A.L., Sawyer A., Greene C.S., et al. Beyond low earth orbit: biomonitoring, artificial intelligence, and precision space health. arXiv. 2021 doi: 10.48550/arXiv.2112.12554. Preprint at. [DOI] [Google Scholar]
- 38.Cohain A., Divaraniya A.A., Zhu K., Scarpa J.R., Kasarskis A., Zhu J., Chang R., Dudley J.T., Schadt E.E. Exploring the reproducibility of probabilistic causal molecular network models. Pac. Symp. Biocomput. 2017;22:120–131. doi: 10.1142/9789813207813_0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mostavi M., Chiu Y.-C., Chen Y., Huang Y. CancerSiamese: one-shot learning for predicting primary and metastatic tumor types unseen during model training. BMC Bioinf. 2021;22:244. doi: 10.1186/s12859-021-04157-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma J., Fong S.H., Luo Y., Bakkenist C.J., Shen J.P., Mourragui S., Wessels L.F.A., Hafner M., Sharan R., Peng J., Ideker T. Few-shot learning creates predictive models of drug response that translate from high-throughput screens to individual patients. Nat. Can. (Que.) 2021;2:233–244. doi: 10.1038/s43018-020-00169-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.da Silveira W.A., Fazelinia H., Rosenthal S.B., Laiakis E.C., Kim M.S., Meydan C., Kidane Y., Rathi K.S., Smith S.M., Stear B., et al. Comprehensive multi-omics analysis reveals mitochondrial stress as a central biological hub for spaceflight impact. Cell. 2020;183:1185–1201.e20. doi: 10.1016/j.cell.2020.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pulley J.M., Rhoads J.P., Jerome R.N., Challa A.P., Erreger K.B., Joly M.M., Lavieri R.R., Perry K.E., Zaleski N.M., Shirey-Rice J.K., Aronoff D.M. Using what we already have: uncovering new drug repurposing strategies in existing omics data. Annu. Rev. Pharmacol. Toxicol. 2020;60:333–352. doi: 10.1146/annurev-pharmtox-010919-023537. [DOI] [PubMed] [Google Scholar]
- 43.Stark Z., Dolman L., Manolio T.A., Ozenberger B., Hill S.L., Caulfied M.J., Levy Y., Glazer D., Wilson J., Lawler M., et al. Integrating genomics into healthcare: a global responsibility. Am. J. Hum. Genet. 2019;104:13–20. doi: 10.1016/j.ajhg.2018.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matey E.T., Ragan A.K., Oyen L.J., Vitek C.R., Aoudia S.L., Ragab A.K., Fee-Schroeder K.C., Black J.L., Moyer A.M., Nicholson W.T., et al. Nine-gene pharmacogenomics profile service: the Mayo Clinic experience. Pharmacogenomics J. 2022;22:69–74. doi: 10.1038/s41397-021-00258-0. [DOI] [PubMed] [Google Scholar]
- 45.Voorhies A.A., Mark Ott C., Mehta S., Pierson D.L., Crucian B.E., Feiveson A., Oubre C.M., Torralba M., Moncera K., Zhang Y., et al. Study of the impact of long-duration space missions at the International Space Station on the astronaut microbiome. Sci. Rep. 2019;9:9911. doi: 10.1038/s41598-019-46303-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Urbaniak C., Lorenzi H., Thissen J., Jaing C., Crucian B., Sams C., Pierson D., Venkateswaran K., Mehta S. The influence of spaceflight on the astronaut salivary microbiome and the search for a microbiome biomarker for viral reactivation. Microbiome. 2020;8:56. doi: 10.1186/s40168-020-00830-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stahl-Rommel S., Jain M., Nguyen H.N., Arnold R.R., Aunon-Chancellor S.M., Sharp G.M., Castro C.L., John K.K., Juul S., Turner D.J., et al. Real-time culture-independent microbial profiling onboard the international space station using Nanopore sequencing. Genes. 2021;12:106. doi: 10.3390/genes12010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Avila-Herrera A., Thissen J., Urbaniak C., Be N.A., Smith D.J., Karouia F., Mehta S., Venkateswaran K., Jaing C. Crewmember microbiome may influence microbial composition of ISS habitable surfaces. PLoS One. 2020;15:e0231838. doi: 10.1371/journal.pone.0231838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu C.T., Chao B.N., Barajas R., Haznadar M., Maruvada P., Nicastro H.L., Ross S.A., Verma M., Rogers S., Zanetti K.A. An evaluation of the National Institutes of Health grants portfolio: identifying opportunities and challenges for multi-omics research that leverage metabolomics data. Metabolomics. 2022;18:29. doi: 10.1007/s11306-022-01878-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mencia-Trinchant N., MacKay M.J., Chin C., Afshinnekoo E., Foox J., Meydan C., Butler D., Mozsary C., Vernice N.A., Darby C., et al. Clonal hematopoiesis before, during, and after human spaceflight. Cell Rep. 2021;34:108740. doi: 10.1016/j.celrep.2021.108740. [DOI] [PubMed] [Google Scholar]
- 51.Trudel G., Shahin N., Ramsay T., Laneuville O., Louati H. Hemolysis contributes to anemia during long-duration space flight. Nat. Med. 2022;28:59–62. doi: 10.1038/s41591-021-01637-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mias G.I., Singh V.V., Rogers L.R.K., Xue S., Zheng M., Domanskyi S., Kanada M., Piermarocchi C., He J. Longitudinal saliva omics responses to immune perturbation: a case study. Sci. Rep. 2021;11:710. doi: 10.1038/s41598-020-80605-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roda A., Mirasoli M., Guardigli M., Zangheri M., Caliceti C., Calabria D., Simoni P. Advanced biosensors for monitoring astronauts’ health during long-duration space missions. Biosens. Bioelectron. 2018;111:18–26. doi: 10.1016/j.bios.2018.03.062. [DOI] [PubMed] [Google Scholar]
- 54.Castro-Wallace S.L., Chiu C.Y., John K.K., Stahl S.E., Rubins K.H., McIntyre A.B.R., Dworkin J.P., Lupisella M.L., Smith D.J., Botkin D.J., et al. Nanopore DNA sequencing and genome assembly on the international space station. Sci. Rep. 2017;7:18022. doi: 10.1038/s41598-017-18364-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burton A.S., Stahl S.E., John K.K., Jain M., Juul S., Turner D.J., Harrington E.D., Stoddart D., Paten B., Akeson M., Castro-Wallace S.L. Off earth identification of bacterial populations using 16S rDNA Nanopore sequencing. Genes. 2020;11:76. doi: 10.3390/genes11010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sanders L.M., Yang J.H., Scott R.T., Qutub A.A., Martin H.G., Berrios D.C., Hastings J.J.A., Rask J., Mackintosh G., Hoarfrost A.L., et al. Beyond low earth orbit: biological research, artificial intelligence, and self-driving labs. arXiv. 2021 doi: 10.48550/arXiv.2112.12582. Preprint at. [DOI] [Google Scholar]
- 57.Wnorowski A., Sharma A., Chen H., Wu H., Shao N.-Y., Sayed N., Liu C., Countryman S., Stodieck L.S., Rubins K.H., et al. Effects of spaceflight on human induced pluripotent stem cell-derived cardiomyocyte structure and function. Stem Cell Rep. 2019;13:960–969. doi: 10.1016/j.stemcr.2019.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barrila J., Sarker S.F., Hansmeier N., Yang S., Buss K., Briones N., Park J., Davis R.R., Forsyth R.J., Ott C.M., et al. Evaluating the effect of spaceflight on the host-pathogen interaction between human intestinal epithelial cells and Salmonella Typhimurium. NPJ Microgravity. 2021;7:9. doi: 10.1038/s41526-021-00136-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Trappe S., Costill D., Gallagher P., Creer A., Peters J.R., Evans H., Riley D.A., Fitts R.H. Exercise in space: human skeletal muscle after 6 months aboard the International Space Station. J. Appl. Physiol. 2009;106:1159–1168. doi: 10.1152/japplphysiol.91578.2008. [DOI] [PubMed] [Google Scholar]
- 60.Fitts R.H., Trappe S.W., Costill D.L., Gallagher P.M., Creer A.C., Colloton P.A., Peters J.R., Romatowski J.G., Bain J.L., Riley D.A. Prolonged space flight-induced alterations in the structure and function of human skeletal muscle fibres. J. Physiol. 2010;588:3567–3592. doi: 10.1113/jphysiol.2010.188508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fitts R.H., Colloton P.A., Trappe S.W., Costill D.L., Bain J.L.W., Riley D.A. Effects of prolonged space flight on human skeletal muscle enzyme and substrate profiles. J. Appl. Physiol. 2013;115:667–679. doi: 10.1152/japplphysiol.00489.2013. [DOI] [PubMed] [Google Scholar]
- 62.Corvelyn M., De Beukelaer N., Duelen R., Deschrevel J., Van Campenhout A., Prinsen S., Gayan-Ramirez G., Maes K., Weide G., Desloovere K., et al. Muscle microbiopsy to delineate stem cell involvement in young patients: a novel approach for children with cerebral palsy. Front. Physiol. 2020;11:945. doi: 10.3389/fphys.2020.00945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Berekméri A., Tiganescu A., Alase A.A., Vital E., Stacey M., Wittmann M. Non-invasive approaches for the diagnosis of autoimmune/autoinflammatory skin diseases-A focus on psoriasis and lupus erythematosus. Front. Immunol. 2019;10:1931. doi: 10.3389/fimmu.2019.01931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Crucian B., Stowe R., Mehta S., Uchakin P., Quiriarte H., Pierson D., Sams C. Immune system dysregulation occurs during short duration spaceflight on board the space shuttle. J. Clin. Immunol. 2013;33:456–465. doi: 10.1007/s10875-012-9824-7. [DOI] [PubMed] [Google Scholar]
- 65.Crucian B., Stowe R.P., Mehta S., Quiriarte H., Pierson D., Sams C. Alterations in adaptive immunity persist during long-duration spaceflight. NPJ Microgravity. 2015;1:15013. doi: 10.1038/npjmgrav.2015.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Crucian B.E., Zwart S.R., Mehta S., Uchakin P., Quiriarte H.D., Pierson D., Sams C.F., Smith S.M. Plasma cytokine concentrations indicate that in vivo hormonal regulation of immunity is altered during long-duration spaceflight. J. Interferon Cytokine Res. 2014;34:778–786. doi: 10.1089/jir.2013.0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Buchheim J.-I., Ghislin S., Ouzren N., Albuisson E., Vanet A., Matzel S., Ponomarev S., Rykova M., Choukér A., Choukér A., Frippiat J.-P. Plasticity of the human IgM repertoire in response to long-term spaceflight. FASEB. J. 2020;34:16144–16162. doi: 10.1096/fj.202001403rr. [DOI] [PubMed] [Google Scholar]
- 68.Bezdan D., Grigorev K., Meydan C., Pelissier Vatter F.A., Cioffi M., Rao V., MacKay M., Nakahira K., Burnham P., Afshinnekoo E., et al. Cell-free DNA (cfDNA) and exosome profiling from a year-long human spaceflight reveals circulating biomarkers. iScience. 2020;23:101844. doi: 10.1016/j.isci.2020.101844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Malkani S., Chin C.R., Cekanaviciute E., Mortreux M., Okinula H., Tarbier M., Schreurs A.-S., Shirazi-Fard Y., Tahimic C.G.T., Rodriguez D.N., et al. Circulating miRNA spaceflight signature reveals targets for countermeasure development. Cell Rep. 2020;33:108448. doi: 10.1016/j.celrep.2020.108448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brzhozovskiy A.G., Kononikhin A.S., Pastushkova L.C., Kashirina D.N., Indeykina M.I., Popov I.A., Custaud M.-A., Larina I.M., Nikolaev E.N. The effects of spaceflight factors on the human plasma proteome, including both real space missions and ground-based experiments. Int. J. Mol. Sci. 2019;20:3194. doi: 10.3390/ijms20133194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pastushkova L.K., Rusanov V.B., Goncharova A.G., Nosovskiy A.M., Luchitskaya E.S., Kashirina D.N., Kononikhin A.S., Kussmaul A.R., Yakhya Y.D., Larina I.M., Nikolaev E.N. Blood plasma proteins associated with heart rate variability in cosmonauts who have completed long-duration space missions. Front. Physiol. 2021;12:760875. doi: 10.3389/fphys.2021.760875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brzhozovskiy A., Kononikhin A., Indeykina M., Pastushkova L., Popov I.A., Nikolaev E.N., Larina I.M., Brzhozovskiy A., Kononikhin A. Label-free study of cosmonaut’s urinary proteome changes after long-duration spaceflights. Eur. J. Mass Spectrom. 2017;23:225–229. doi: 10.1177/1469066717717610. [DOI] [PubMed] [Google Scholar]
- 73.Kuo T.-R., Chen C.-H. Bone biomarker for the clinical assessment of osteoporosis: recent developments and future perspectives. Biomark Res. 2017;5:18. doi: 10.1186/s40364-017-0097-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Swensen A.C., He J., Fang A.C., Ye Y., Nicora C.D., Shi T., Liu A.Y., Sigdel T.K., Sarwal M.M., Qian W.-J. A comprehensive urine proteome database generated from patients with various renal conditions and prostate cancer. Front. Med. 2021;8:548212. doi: 10.3389/fmed.2021.548212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jing J., Gao Y. Urine biomarkers in the early stages of diseases: current status and perspective. Discov. Med. 2018;25:57–65. [PubMed] [Google Scholar]
- 76.Gao Y. Urine is a better biomarker source than blood especially for kidney diseases. Adv. Exp. Med. Biol. 2015;845:3–12. doi: 10.1007/978-94-017-9523-4_1. [DOI] [PubMed] [Google Scholar]
- 77.Pastushkova L.H., Rusanov V.B., Goncharova A.G., Brzhozovskiy A.G., Kononikhin A.S., Chernikova A.G., Kashirina D.N., Nosovsky A.M., Baevsky R.M., Nikolaev E.N., Larina I.M. Urine proteome changes associated with autonomic regulation of heart rate in cosmonauts. BMC Syst. Biol. 2019;13:17. doi: 10.1186/s12918-019-0688-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pastushkova L.K., Kashirina D.N., Brzhozovskiy A.G., Kononikhin A.S., Tiys E.S., Ivanisenko V.A., Koloteva M.I., Nikolaev E.N., Larina I.M. Evaluation of cardiovascular system state by urine proteome after manned space flight. Acta Astronaut. 2019;160:594–600. doi: 10.1016/j.actaastro.2019.02.015. [DOI] [Google Scholar]
- 79.Kinross J.M., Darzi A.W., Nicholson J.K. Gut microbiome-host interactions in health and disease. Genome Med. 2011;3:14. doi: 10.1186/gm228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Valdes A.M., Walter J., Segal E., Spector T.D. Role of the gut microbiota in nutrition and health. BMJ. 2018;361:k2179. doi: 10.1136/bmj.k2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tang Q., Jin G., Wang G., Liu T., Liu X., Wang B., Cao H. Current sampling methods for gut microbiota: a call for more precise devices. Front. Cell. Infect. Microbiol. 2020;10:151. doi: 10.3389/fcimb.2020.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Turroni S., Magnani M., Kc P., Lesnik P., Vidal H., Heer M. Gut microbiome and space travelers’ health: state of the art and possible pro/prebiotic strategies for long-term space missions. Front. Physiol. 2020;11:553929. doi: 10.3389/fphys.2020.553929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu Z., Luo G., Du R., Sun W., Li J., Lan H., Chen P., Yuan X., Cao D., Li Y., et al. Effects of spaceflight on the composition and function of the human gut microbiota. Gut Microb. 2020;11:807–819. doi: 10.1080/19490976.2019.1710091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Danko D.C., Singh N., Butler D.J., Mozsary C., Jiang P., Keshavarzian A., Maienschein-Cline M., Chlipala G., Afshinnekoo E., Bezdan D., et al. Genetic and immunological evidence for microbial transfer between the international space station and an astronaut. bioRxiv. 2020 doi: 10.1101/2020.11.10.376954. Preprint at. [DOI] [Google Scholar]
- 85.Zierer J., Jackson M.A., Kastenmüller G., Mangino M., Long T., Telenti A., Mohney R.P., Small K.S., Bell J.T., Steves C.J., et al. The fecal metabolome as a functional readout of the gut microbiome. Nat. Genet. 2018;50:790–795. doi: 10.1038/s41588-018-0135-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ursell L.K., Metcalf J.L., Parfrey L.W., Knight R. Defining the human microbiome. Nutr. Rev. 2012;70(Suppl 1):S38–S44. doi: 10.1111/j.1753-4887.2012.00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Blum H.E. The human microbiome. Adv. Med. Sci. 2017;62:414–420. doi: 10.1016/j.advms.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 88.Gilbert J.A., Blaser M.J., Caporaso J.G., Jansson J.K., Lynch S.V., Knight R. Current understanding of the human microbiome. Nat. Med. 2018;24:392–400. doi: 10.1038/nm.4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Salvucci E. The human-microbiome superorganism and its modulation to restore health. Int. J. Food Sci. Nutr. 2019;70:781–795. doi: 10.1080/09637486.2019.1580682. [DOI] [PubMed] [Google Scholar]
- 90.Fan Y., Pedersen O. Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 2021;19:55–71. doi: 10.1038/s41579-020-0433-9. [DOI] [PubMed] [Google Scholar]
- 91.Be N.A., Avila-Herrera A., Allen J.E., Singh N., Checinska Sielaff A., Jaing C., Venkateswaran K. Whole metagenome profiles of particulates collected from the International Space Station. Microbiome. 2017;5:81. doi: 10.1186/s40168-017-0292-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Singh N.K., Wood J.M., Karouia F., Venkateswaran K. Succession and persistence of microbial communities and antimicrobial resistance genes associated with International Space Station environmental surfaces. Microbiome. 2018;6:204. doi: 10.1186/s40168-018-0585-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Checinska Sielaff A., Urbaniak C., Mohan G.B.M., Stepanov V.G., Tran Q., Wood J.M., Minich J., McDonald D., Mayer T., Knight R., et al. Characterization of the total and viable bacterial and fungal communities associated with the International Space Station surfaces. Microbiome. 2019;7:50. doi: 10.1186/s40168-019-0666-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Stabler R.A., Rosado H., Doyle R., Negus D., Carvil P.A., Kristjánsson J.G., Green D.A., Franco-Cendejas R., Davies C., Mogensen A., et al. Impact of the mk VI SkinSuit on skin microbiota of terrestrial volunteers and an international space station-bound astronaut. NPJ Microgravity. 2017;3:23. doi: 10.1038/s41526-017-0029-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Morrison M.D., Thissen J.B., Karouia F., Mehta S., Urbaniak C., Venkateswaran K., Smith D.J., Jaing C. Investigation of spaceflight induced changes to astronaut microbiomes. Front. Microbiol. 2021;12:659179. doi: 10.3389/fmicb.2021.659179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lee M.D., O’Rourke A., Lorenzi H., Bebout B.M., Dupont C.L., Everroad R.C. Reference-guided metagenomics reveals genome-level evidence of potential microbial transmission from the ISS environment to an astronaut’s microbiome. iScience. 2021;24:102114. doi: 10.1016/j.isci.2021.102114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Salama S.A., Serrana M., Au W.W., Au W.W. Biomonitoring using accessible human cells for exposure and health risk assessment. Mutat. Res. 1999;436:99–112. doi: 10.1016/s1383-5742(98)00021-0. [DOI] [PubMed] [Google Scholar]
- 98.Terada M., Seki M., Higashibata A., Yamada S., Takahashi R., Majima H.J., Yamazaki T., Watanabe-Asaka T., Niihori M., Mukai C., Ishioka N. Genetic analysis of the human hair roots as a tool for spaceflight experiments. Adv. Biosci. Biotechnol. 2013;04:75–88. doi: 10.4236/abb.2013.410a3009. [DOI] [Google Scholar]
- 99.Terada M., Seki M., Takahashi R., Yamada S., Higashibata A., Majima H.J., Sudoh M., Mukai C., Ishioka N. Effects of a closed space environment on gene expression in hair follicles of astronauts in the international space station. PLoS One. 2016;11:e0150801. doi: 10.1371/journal.pone.0150801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Adav S.S., Subbaiaih R.S., Kerk S.K., Lee A.Y., Lai H.Y., Ng K.W., Sze S.K., Schmidtchen A. Studies on the proteome of human hair - identification of histones and deamidated keratins. Sci. Rep. 2018;8:1599. doi: 10.1038/s41598-018-20041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chen Y., Guo J., Xing S., Yu H., Huan T. Global-scale metabolomic profiling of human hair for simultaneous monitoring of endogenous metabolome, short- and long-term exposome. Front. Chem. 2021;9:674265. doi: 10.3389/fchem.2021.674265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cikach F.S., Jr., Dweik R.A. Cardiovascular biomarkers in exhaled breath. Prog. Cardiovasc. Dis. 2012;55:34–43. doi: 10.1016/j.pcad.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ibrahim W., Carr L., Cordell R., Wilde M.J., Salman D., Monks P.S., Thomas P., Brightling C.E., Siddiqui S., Greening N.J. Breathomics for the clinician: the use of volatile organic compounds in respiratory diseases. Thorax. 2021;76:514–521. doi: 10.1136/thoraxjnl-2020-215667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kononikhin A.S., Brzhozovskiy A.G., Ryabokon A.M., Fedorchenko K., Zakharova N.V., Spasskii A.I., Popov I.A., Ilyin V.K., Solovyova Z.O., Pastushkova L.K., et al. Proteome profiling of the exhaled breath condensate after long-term spaceflights. Int. J. Mol. Sci. 2019;20:4518. doi: 10.3390/ijms20184518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hauschild A.-C., Frisch T., Baumbach J.I., Baumbach J. Carotta: revealing hidden confounder markers in metabolic breath profiles. Metabolites. 2015;5:344–363. doi: 10.3390/metabo5020344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Campanella A., De Summa S., Tommasi S. Exhaled breath condensate biomarkers for lung cancer. J. Breath Res. 2019;13:044002. doi: 10.1088/1752-7163/ab2f9f. [DOI] [PubMed] [Google Scholar]
- 107.Mitchell C., Ordish J., Johnson E., Brigden T., Hall A. 2020. The GDPR and Genomic Data—The Impact of the GDPR and DPA 2018 on Genomic Healthcare and Research. [Google Scholar]
- 108.Haendel M., Vasilevsky N., Unni D., Bologa C., Harris N., Rehm H., Hamosh A., Baynam G., Groza T., McMurry J., et al. How many rare diseases are there? Nat. Rev. Drug Discov. 2020;19:77–78. doi: 10.1038/d41573-019-00180-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nguengang Wakap S., Lambert D.M., Olry A., Rodwell C., Gueydan C., Lanneau V., Murphy D., Le Cam Y., Rath A. Estimating cumulative point prevalence of rare diseases: analysis of the Orphanet database. Eur. J. Hum. Genet. 2020;28:165–173. doi: 10.1038/s41431-019-0508-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nguyen M.T., Goldblatt J., Isasi R., Jagut M., Jonker A.H., Kaufmann P., Ouillade L., Molnar-Gabor F., Shabani M., Sid E., et al. Model consent clauses for rare disease research. BMC Med. Ethics. 2019;20:55. doi: 10.1186/s12910-019-0390-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shabani M., Marelli L. Re-identifiability of genomic data and the GDPR: assessing the re-identifiability of genomic data in light of the EU General Data Protection Regulation. EMBO Rep. 2019;20:e48316. doi: 10.15252/embr.201948316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lippert C., Sabatini R., Maher M.C., Kang E.Y., Lee S., Arikan O., Harley A., Bernal A., Garst P., Lavrenko V., et al. Identification of individuals by trait prediction using whole-genome sequencing data. Proc. Natl. Acad. Sci. USA. 2017;114:10166–10171. doi: 10.1073/pnas.1711125114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Harmanci A., Gerstein M. Quantification of private information leakage from phenotype-genotype data: linking attacks. Nat. Methods. 2016;13:251–256. doi: 10.1038/nmeth.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gürsoy G., Li T., Liu S., Ni E., Brannon C.M., Gerstein M.B. Functional genomics data: privacy risk assessment and technological mitigation. Nat. Rev. Genet. 2021;23:245–258. doi: 10.1038/s41576-021-00428-7. [DOI] [PubMed] [Google Scholar]
- 115.Elhaik E., Ahsanuddin S., Robinson J.M., Foster E.M., Mason C.E. The impact of cross-kingdom molecular forensics on genetic privacy. Microbiome. 2021;9:114. doi: 10.1186/s40168-021-01076-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bandeira N., Deutsch E.W., Kohlbacher O., Martens L., Vizcaíno J.A. Data management of sensitive human proteomics data: current practices, recommendations, and perspectives for the future. Mol. Cell. Proteomics. 2021;20:100071. doi: 10.1016/j.mcpro.2021.100071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dupras C., Bunnik E.M. Toward a framework for assessing privacy risks in multi-omic research and databases. Am. J. Bioeth. 2021;21:46–64. doi: 10.1080/15265161.2020.1863516. [DOI] [PubMed] [Google Scholar]
- 118.World Medical Association World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA. 2013;310:2191–2194. doi: 10.1001/jama.2013.281053. [DOI] [PubMed] [Google Scholar]
- 119.Hallinan D. Broad consent under the GDPR: an optimistic perspective on a bright future. Life Sci Soc Policy. 2020;16:1. doi: 10.1186/s40504-019-0096-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Shabani M., Borry P. Rules for processing genetic data for research purposes in view of the new EU General Data Protection Regulation. Eur. J. Hum. Genet. 2018;26:149–156. doi: 10.1038/s41431-017-0045-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Reed R.D., Antonsen E.L. Should NASA collect astronauts’ genetic information for occupational surveillance and research? AMA J Ethics. 2018;20:E849–E856. doi: 10.1001/amajethics.2018.849. [DOI] [PubMed] [Google Scholar]
- 122.Antonsen E.L., Reed R.D. Policy considerations for precision medicine in human spaceflight. Hous. J. Health L. & Pol’y. 2019;19:1. [Google Scholar]
- 123.Joly Y., Dupras C., Pinkesz M., Tovino S.A., Rothstein M.A. Looking beyond GINA: policy approaches to address genetic discrimination. Annu. Rev. Genom. Hum. Genet. 2020;21:491–507. doi: 10.1146/annurev-genom-111119-011436. [DOI] [PubMed] [Google Scholar]
- 124.Molnár-Gábor F., Korbel J.O. Genomic data sharing in Europe is stumbling-Could a code of conduct prevent its fall? EMBO Mol. Med. 2020;12:e11421. doi: 10.15252/emmm.201911421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Molnár-Gábor F., Sellner J., Pagil S., Slokenberga S., Tzortzatou-Nanopoulou O., Nyström K. Harmonization after the GDPR? Divergences in the rules for genetic and health data sharing in four member states and ways to overcome them by EU measures: insights from Germany, Greece, Latvia and Sweden. Semin. Cancer Biol. 2021 doi: 10.1016/j.semcancer.2021.12.001. [DOI] [PubMed] [Google Scholar]
- 126.Berrios D.C., Galazka J., Grigorev K., Gebre S., Costes S.V. NASA GeneLab: interfaces for the exploration of space omics data. Nucleic Acids Res. 2021;49:D1515–D1522. doi: 10.1093/nar/gkaa887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Madrigal P., Gabel A., Villacampa A., Manzano A., Deane C.S., Bezdan D., Carnero-Diaz E., Medina F.J., Hardiman G., Grosse I., et al. Revamping space-omics in Europe. Cell Syst. 2020;11:555–556. doi: 10.1016/j.cels.2020.10.006. [DOI] [PubMed] [Google Scholar]
- 128.Hallinan D., Bernier A., Cambon-Thomsen A., Crawley F.P., Dimitrova D., Medeiros C.B., Nilsonne G., Parker S., Pickering B., Rennes S. International transfers of personal data for health research following Schrems II: a problem in need of a solution. Eur. J. Hum. Genet. 2021;29:1502–1509. doi: 10.1038/s41431-021-00893-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Saunders G., Baudis M., Becker R., Beltran S., Béroud C., Birney E., Brooksbank C., Brunak S., Van den Bulcke M., Drysdale R., et al. Leveraging European infrastructures to access 1 million human genomes by 2022. Nat. Rev. Genet. 2019;20:693–701. doi: 10.1038/s41576-019-0156-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Thorogood A., Rehm H.L., Goodhand P., Page A.J.H., Joly Y., Baudis M., Rambla J., Navarro A., Nyronen T.H., Linden M., et al. International federation of genomic medicine databases using GA4GH standards. Cell Genom. 2021;1:100032. doi: 10.1016/j.xgen.2021.100032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Rehm H.L., Page A.J.H., Smith L., Adams J.B., Alterovitz G., Babb L.J., Barkley M.P., Baudis M., Beauvais M.J.S., Beck T., et al. GA4GH: international policies and standards for data sharing across genomic research and healthcare. Cell Genom. 2021;1:100029. doi: 10.1016/j.xgen.2021.100029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Jochems A., Deist T.M., van Soest J., Eble M., Bulens P., Coucke P., Dries W., Lambin P., Dekker A. Distributed learning: developing a predictive model based on data from multiple hospitals without data leaving the hospital - a real life proof of concept. Radiother. Oncol. 2016;121:459–467. doi: 10.1016/j.radonc.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 133.Gürsoy G., Emani P., Brannon C.M., Jolanki O.A., Harmanci A., Strattan J.S., Cherry J.M., Miranker A.D., Gerstein M. Data sanitization to reduce private information leakage from functional genomics. Cell. 2020;183:905–917.e16. doi: 10.1016/j.cell.2020.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Rieke N., Hancox J., Li W., Milletarì F., Roth H.R., Albarqouni S., Bakas S., Galtier M.N., Landman B.A., Maier-Hein K., et al. The future of digital health with federated learning. NPJ Digit Med. 2020;3:119. doi: 10.1038/s41746-020-00323-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Truong N., Sun K., Wang S., Guitton F., Guo Y. Privacy preservation in federated learning: an insightful survey from the GDPR perspective. Comput. Secur. 2021;110:102402. doi: 10.1016/j.cose.2021.102402. [DOI] [Google Scholar]