Abstract
Retinoid-binding protein7 (RBP7) is a member of the cellular retinol-binding protein (CRBP) family, which is involved in the pathogenesis of breast cancer. The study aims to illustrate the prognostic value and the potential regulatory mechanisms of RBP7 expression in breast cancer. Bioinformatics analysis with the TCGA and CPTAC databases revealed that the mRNA and protein expression levels of RBP7 in normal were higher compared to breast cancer tissues. Survival analysis displayed that the lower expression of RBP7, the worse the prognosis in ER-positive (ER+) breast cancer patients. Genomic analysis showed that low expression of RBP7 correlates with its promoter hypermethylation in breast cancer. Functional enrichment analysis demonstrated that downregulation of RBP7 expression may exert its biological influence on breast cancer through the PPAR pathway and the PI3K/AKT pathway. In summary, we identified RBP7 as a novel biomarker that is helpful for the prognosis of ER+ breast cancer patients. Promoter methylation of RBP7 is involved in its gene silencing in breast cancer, thus regulating the occurrence and development of ER+ breast cancer through the PPAR and PI3K/AKT pathways.
1. Introduction
Breast cancer is the most common cancer worldwide, accounting for 30% of female cancers [1]. Estrogen receptor-positive (ER+) breast cancer is driven by ER-mediated transcriptional activity, composing the major subtype (approximately 75%) of breast cancer [2]. Although endocrine therapy, including estrogen suppression and direct ER targeting, is widely applied in the treatment of ER+ breast cancer, acquired resistance often occurs and remains a major challenge for the treatment of ER+ patients [3]; thus, novel targets and effective therapeutic strategies for breast cancer patients are urgently needed.
Previous studies have confirmed that cellular retinol-binding protein (CRBP) family members play an important role in the pathological progression of breast cancer. CRBPs belong to the family of fatty acid-binding proteins and are required for vitamin A stability and metabolism [4]. Epigenetic silencing of CRBPs is a common event in cancers [5]. For example, Kuppumbatti et al. reported that CRBPs were underexpressed in 24% of human breast cancer [6]. Previous studies demonstrated that inhibition of the PI3K/AKT pathway by CRBPs was involved in the growth inhibition of mammary epithelial cells [7]. In breast epithelial cells, CRBP1 inhibits PI3K/AKT signalling through a retinoic acid receptor-dependent mechanism that regulates p85-p110 heterodimerization [8]. In addition, physiological retinoic acid receptor (RAR) activation is dependent on CRBP1-mediated retinol storage, and CRBP1 downregulation chronically compromises RAR activity, leading to loss of cell differentiation and tumor progression [9]. Thus, CRBPs may be used as a potential biomarker for the diagnosis and treatment of breast cancer.
Retinoid-binding protein 7 (RBP7), also named CRBP4, belongs to a clearly distinct CRBP subfamily, representing a relatively different mode of retinol binding for this protein. The RBP7 gene is located on human chromosome 1p36.22, encoding a protein of 134 amino acids in length. The size of the translated exon sequences and intron position of RBP7 is highly conserved. With a structure similar to other CRBPs, the RBP7-encoded protein binds all-trans-retinol with a lower binding affinity than other CRBPs [10]. It was reported that RBP7 regulates the occurrence and development of many diseases. For example, the PPARγ-RBP7-adiponectin pathway plays a protective role in hypertensive diseases by regulating transcriptional activity [11]. RBP7 also plays an important role in adipose tissue during adipogenesis, cold exposure, and nutritional treatment [12]. Recent studies have demonstrated that RBP7 is a strong prognostic biomarker contributing to the malignant phenotype in colon cancer [13]. However, as a novel member of the CRBP family, the clinical and prognostic significance of RBP7 in breast cancer is still unknown, and its functional role in breast cancer has never been documented.
In this study, we first analyzed the differential expression of RBP7 in breast cancer and normal tissues and evaluated the prognostic value of RBP7 by using data from the TCGA and GEO databases. Next, we determined the localization of RBP7 expression in breast cancer, the functional enrichment of its coexpressed genes, and the association between the mRNA expression and DNA methylation of RBP7. Then, we explored the association between RBP7 and multiple molecular subtypes of breast cancer and the significant KEGG pathways involved in RBP7 in ER+ breast cancer. Finally, we screened RBP7-targeting drugs from computational analysis of resistance (CARE) databases, which may provide new ideas for the treatment of breast cancer.
2. Materials and Methods
2.1. Gene Expression and Survival Analysis
The Human Protein Atlas [14, 15] (HPA; http://www.proteinatlas.org) database was used to illustrate RBP7 mRNA distribution, protein expression, and immunohistochemical maps of RBP7 in normal and breast cancer tissues. RBP7 gene expression levels in pan cancers were identified in Tumor Immune Estimation Resource [16] (TIMER; https://cistrome.shinyapps.io/timer/) and ONCOMINE [17] (http://www.oncomine.org). UALCAN [18] (http://ualcan.path.uab.edu/index.html) and the Gene Expression Omnibus [19] (GEO; https://www.ncbi.nlm.nih.gov/geo/) were used to investigate the different expression of RBP7 in normal and breast cancer tissues. To explore the prognostic role of RBP7 expression in breast cancer, Gene Expression Profiling Interactive Analysis [20] (GEPIA; gepia2.cancer-pku.cn/#index), Kaplan–Meier plotter [21] (http://kmplot.com/), and PROGgeneV2 [22] (http://www.compbio.iupui.edu/proggene) were used to determine the prognostic significance. In this study, we analyzed the prognosis of RBP7 in breast cancer with detailed hazard ratios (HRs) by setting the expression threshold at the medium or best cut-off and a log-rank pvalue less than 0.05.
2.2. Tumor Immune Single-Cell Hub (TISCH)
TISCH [23] (http://tisch.comp-genomics.org) integrates single-cell transcriptome profiles of nearly 2 million cells for 27 cancer types. In this study, we utilized the “multiple-dataset comparison” model to visualize the averaged gene expression distributed in single cells and the “Gene” module to display the heat map of the cell-type averaged expression of RBP7.
2.3. LinkedOmics
The coexpressed genes of RBP7 were screened from the TCGA BRCA (breast invasive carcinoma) cohort through the “LinkFinder” module in LinkedOmics [24] (http://www.linkedomics.org/login.php) databases, and the correlative significance was tested by the spearman correlation coefficient. The top 50 positively and negatively correlated genes are presented as heat maps. Gene Ontology biological process (GO_BP) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed with gene set enrichment analysis (GSEA) in the “LinkInterpreter” module.
2.4. RBP7 DNA Methylation Analysis
Heat maps of RBP7 in the cohort of breast cancer patients were constructed through data mining in TCGA BRCA by using the University of California Santa Cruz (UCSC) Xena [25, 26] (http://xena.ucsc.edu/). MethSurv (http://biit.cs.ut.ee/methsurv/) is used to survival analysis ground on CpG methylation patterns [27], we verified the methylation levels of probes of RBP7, and four of them with high methylation in the promoter were chosen to display the distribution of methylation under different clinical stages. SMART [28, 29] (http://www.bioinfo-zs.com/smartapp/) is to further identify the association of the mRNA expression and methylation of RBP7.
2.5. Bc-GenExMiner Online Tool and OSbrca
Based on common clinical parameters, we utilized Bc-GenExMiner (v4.7) [30] (http://bcgenex.ico.unicancer.fr) to analyze the expression data and survival curves of RBP7 in different molecular subtypes of breast cancer, including ER, PR, and HER-2 (IHC). The OSbrca [31] (http://bioinfo.henu.edu.cn/BRCA/BRCAList.jsp) was utilized to validate prognostic value of RBP7 in breast cancer.
2.6. Protein–Protein Interaction (PPI) Network Analysis
PPI network analysis of RBP7 was conducted in the STRING (https://string-db.org/) database [32]. The regulatory relationships between genes were visualized via Cytoscape (ver. 3.4.0). Then, we use the starBase v3.0 [33] (http://starbase.sysu.edu.cn/index.php) to analyze the correlation between RBP7 and PIK3R3 in breast cancer.
2.7. Differentially Expressed Genes (DEGs) Analysis
We used the Limma package to screen DEGs by filtering the p.adj value of Student's t-test and the fold change (FC) and dividing the DEGs into two groups with high or low RBP7 expression. A volcano plot was generated by using the ggplot2 R software package to display the DEGs with statistical significance, i.e., p.adjust value <0.05 and absolute FC value >1. KEGG pathway analysis was performed on those DEGs by using the cluster profiler package, and the pathways with statistical significance (adjusted p < 0.05) were visualized by hierarchical clustering of a heat map [34].
2.8. Computational Analysis of Resistance (CARE)
A positive CARE score represented a high expression value, which was related to drug response in CARE [35] (http://care.dfci.harvard.edu/), and vice versa. In this study, we utilized 3 databases, i.e., Cancer Cell Line Encyclopedia (CCLE), Cancer Therapeutics Response Portal (CGP), and Genomics of Drug Sensitivity in Cancer (CTRP), to analyze the drugs targeting RBP7.
2.9. SwissDock
The PDF file of the RBP7 protein was downloaded from the RCSB Protein Data Bank (PDB) database, and the ligand and water molecules were then removed by using PyMOL software. The mol2 file of the nilotinib small molecule was downloaded from the PubChem database and converted using Open Babel software. Finally, we uploaded the two files to the SwissDock [36] (http://www.swissdock.ch) page for docking.
3. Results
3.1. Gene Expression Profiles of RBP7 in Normal and Cancer Tissues
We utilized the HPA database to analyze the mRNA and protein expression profiles of RBP7 in human normal tissues. We found that the RBP7 mRNA expression was mainly in breast and adipose tissues in normal human tissues by using the GTEx (genotype-tissue expression) database (Figure 1(a)). Consistently, the protein expression of RBP7 was highly expressed in the breast and adipose tissues (Figure 1(b)). The mRNA expression levels of RBP7 were explored by TIMER in many cancer types. Additionally, the results revealed that RBP7 mRNA expression levels were significantly lower in most cancer samples than their corresponding normal samples, including breast invasive carcinoma (BRCA), uterine corpus endometrial carcinoma (UCEC), and lung adenocarcinoma (LUAD). Besides, the data also showed that RBP7 expression was aberrantly higher in liver hepatocellular carcinoma (LIHC) and kidney renal clear cell carcinoma (KIRC) (Figure 1(c)).
Figure 1.
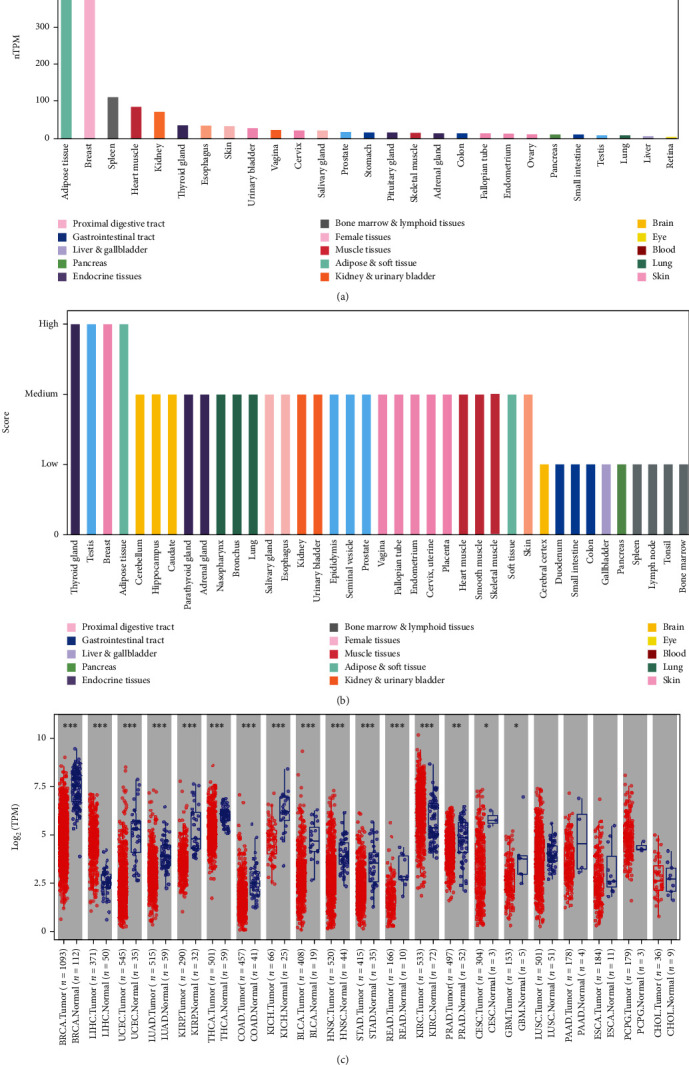
The expression of RBP7 in different human tissues and pan cancers. (a) RBP7 expression profiles in normal human tissues from the GTEx project. nTPM, normalized protein-coding transcripts per million. (b) The protein expression of RBP7 in normal human tissues. (c) The mRNA expression levels of RBP7 in different cancer types were explored by TIMER. It is presented by ranking the p value. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
To further verify the significance of RBP7 expression in cancers, the differential expression of RBP7 in tumor and normal tissues was analyzed by using ONCOMINE. We found that RBP7 was overexpressed in liver cancer and lymphoma, but decreased expression of RBP7 was found in brain and CNS cancer, breast cancer, esophageal cancer, head and neck cancer, leukemia, and ovarian cancer (Supplementary Figure 1A).
3.2. Prognostic Value of RBP7 Expression in Breast Cancer
To investigate the role of RBP7 expression in breast cancer, the UALCAN database was utilized to analyze the expression of RBP7 in 114 normal tissues and 1097 primary breast cancer tissues. The TCGA database results revealed that the mRNA expression level of RBP7 was lower in breast cancer than in normal tissues (Figure 2(a)), which was further validated by the GSE37751 dataset from the GEO database (Supplementary Figure 1B). Then, we utilized the CPTAC database to analyze the protein expression of RBP7 in breast cancer and found that the protein expression of RBP7 in breast cancer was lower than that in normal tissues (Figure 2(b)), which was consistent with the result of RBP7 expression in the TCGA database.
Figure 2.
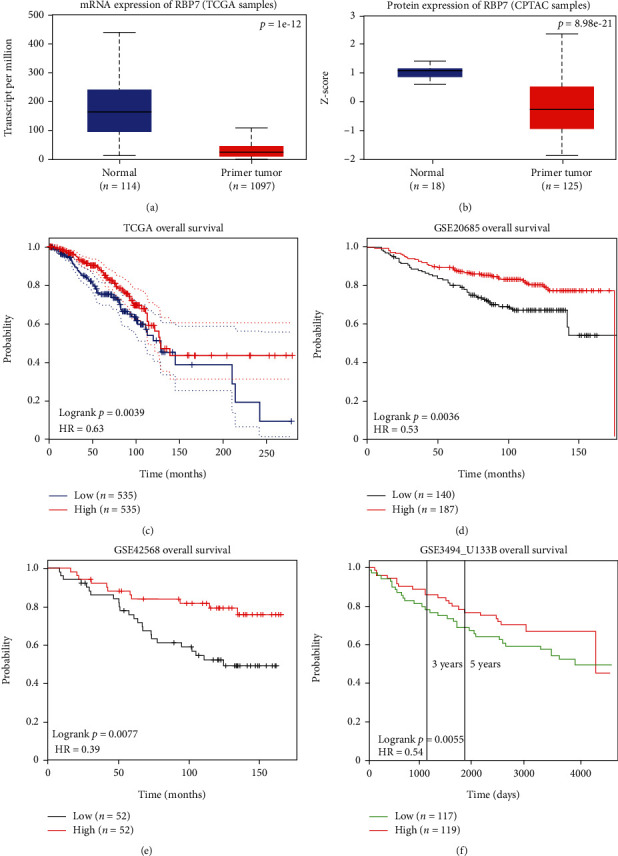
The mRNA and protein expression of RBP7 in breast cancer and the overall survival analysis with RBP7 mRNA expression. (a) Boxplot of the mRNA expression of RBP7 in normal and breast cancer tissues. (b) Boxplot of the protein expression of RBP7 in normal and breast cancer tissues. (c–f) Kaplan–Meier curve of OS based on the high and low expression of RBP7 in breast cancer patients from different databases. A log-rank p value <0.05 was considered statistically significant.
We subsequently utilized the transcriptomic sequencing data in the GEPIA database to assess the prognostic value of RBP7 in breast cancer and found that a high level of expression of RBP7 was favorable to the prognosis of breast cancer (Figure 2(c)). Kaplan–Meier survival analysis was performed with the GSE20685 (Figure 2(d)) and GSE42568 (Figure 2(e)) datasets from the GEO database to evaluate the prognostic value of RBP7 in breast cancer, which showed that lower expression of RBP7 was associated with a poorer prognosis in breast cancer. Furthermore, we used FROGgeneV2 to confirm the effect of RBP7 on the OS of breast cancer patients. The results also showed that low expression of RBP7 was significantly associated with a poor prognosis (Figure 2(f)). These results reveal that there is a significant association between RBP7 expression and breast cancer prognosis and that RBP7 serves as a protective factor in the prognosis of breast cancer.
3.3. RBP7 Expression in Different Cells from Breast Cancer Tissues
We used the TISCH database to analyze the expression of RBP7 in different cells from breast cancer tissues. It demonstrated that RBP7 is mainly expressed in endothelial and epithelial cells in TISCH database (Figures 3(a) and 3(b)). A heat map of the gene module analysis depicts the expression of RBP7 in different cell types of breast cancer datasets, in which endothelial and epithelial cells are characterized by high expression of RBP7 (Figure 3(c)). Then, we utilized the immunohistochemistry (IHC) data detected by the HPA-034749 antibody from the HPA database to determine the protein expression of RBP7 in breast cancer and normal tissues. The results showed that adipocytes were highly stained in normal breast tissues, while glandular and myoepithelial cells were mildly stained, mainly in the nucleus (Figure 3(d)). In the breast cancer samples, the expression level of RBP7 in tumor cells was ranked as weak, moderate, and strong, which was scored by the staining intensity in the pathological IHC (Figures 3(e)–3(g)).Interestingly, as a nuclear receptor, RBP7 was found to be mainly localized in the nucleus, indicating the important role of RBP7 in the regulation of gene expression in epithelial cells of breast cancer tissues.
Figure 3.
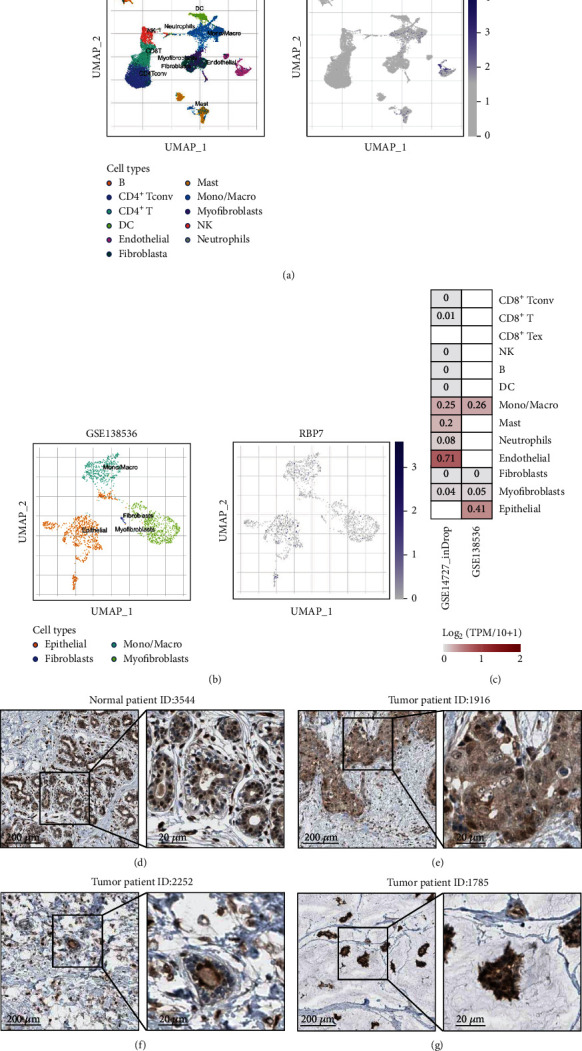
The cellular and subcellular localization of RBP7 in breast cancer. (a–b) The cellular localization of RBP7 mRNA in single breast cancer cells. The expression level is colored by marker intensity. UMAP: uniform manifold approximation and projection. (c) Heatmap showing the mRNA expression of RBP7 in different cell types. (d–g) Representative immunohistochemical staining of normal tissues (https://www.proteinatlas.org/ENSG00000162444-RBP7/tissue/breast) (d) and breast cancer with weak (e), moderate (f), or strong (g) RBP7 expression (https://www.proteinatlas.org/ENSG00000162444-RBP7/pathology/breast+cancer). Scale bars with lengths of 200 μm or 20 μm are displayed in the left and right panels, respectively.
3.4. RBP7 Coexpression Networks in Breast Cancer
To gain insight into the biological meaning of RBP7 in breast cancer, the functional module of LinkedOmics was used to examine RBP7 coexpression genes in the breast cancer cohort. The top 50 significant genes that were positively and negatively correlated with RBP7 were selected as heat maps (Supplementary Figures 2A and 2B) in which RBP7 displayed a strong positive association with the expression of FAM107A (R = 0.4767, pvalue =4.252e-01), GPIHBP1 (R = 0.4757, pvalue =8.560e-63), and FXYD1 (R = 0.4756, pvalue =9.258e-63).Remarkably, the top 50 negatively coexpressed genes had a high probability of being high-risk markers in breast cancer, of which 15 genes had significantly high HRs (pvalue<0.05). In contrast, there were no genes with high HRs (p < 0.05) in the top 50 positively coexpressed genes. These results further confirmed that RBP7 performs a protective role in the progression of breast cancer (Figure 4(a)).
Figure 4.

GO and KEGG pathway enrichment analysis of the coexpressed genes of RBP7 in breast cancer. (a) Survival maps of the top 50 positive or negative coexpressed genes of RBP7 in breast cancer. (b) GO_BP enrichment analysis of the coexpressed genes of RBP7 in breast cancer. (c) KEGG pathway analysis of the coexpressed genes of RBP7 in breast cancer.
GO term annotation of biological processes showed that RBP7 coexpressed genes mainly participate in the adrenergic signalling pathway, excretion, endothelium development, regulation of transporter activity, cell communication by electrical coupling and G protein-coupled receptor signalling pathway, with inhibition of the biological processes including double-strand break repair, cargo loading into vesicle, DNA conformation change, ncRNA transcription, and protein localization to chromosome (Figure 4(b)). KEGG pathway analysis showed that there was an enrichment in the regulation of lipolysis in adipocytes, PPAR signalling pathway, ovarian steroidogenesis, and drug metabolism (Figure 4(c)), indicating a widespread impact of RBP7 on the global transcriptome.
3.5. RBP7 Methylation in Breast Cancer
To further explore the mechanism of the differential expression of RBP7 in breast cancer and normal tissues, we performed hierarchical clustering analysis of RBP7 mRNA expression related to DNA methylation by using the UCSC Cancer Genomics Browser. The results showed that RBP7 methylation mainly occurred in primary breast cancer, and the mRNA expression of RBP7 corresponding to hypermethylated samples is low (Figure 5(a)), indicating a potential correlation between mRNA expression and DNA methylation of RBP7.
Figure 5.

The methylation analysis of RBP7 in breast cancer. (a) Heat maps for the mRNA expression and DNA methylation of RBP7 in the TCGA database. (b) Visualization of methylation level and RBP7 expression in different regions of the RBP7 gene. (c) Analysis of the correlation between methylation on different methylation probes and mRNA expression of RBP7 in breast cancer.
Next, we performed methylation analysis with the Methsurv database and found that 4 methylation probes, namely, cg20413202, cg03406535, cg10796749, and cg14202757, in the promoter of RBP7 were highly methylated in breast cancer (Figure 5(b)). As routinely done, the level of methylation was represented by a beta value: a beta value ≥0.6 was considered completely methylated, a beta value ≤0.2 was considered completely unmethylated, and a beta value between 0.2 and 0.6 was considered partially methylated. According to this standard, we found that the beta values of 3 of the 4 detected methylation probes, i.e., cg20413202 (beta value = 0.947), cg10796749 (beta value = 0.798), and cg14202757 (beta value = 0.694), were higher than 0.6, indicating that almost complete methylation occurred on the promoter of RBP7 (Supplementary Figure 3A-D). Consistently, we found that most of the median beta values of different clinical stages were also above 0.6 (Supplementary Figure 3E-H), suggesting that promoter methylation leads to the inhibition of RBP7 gene transcription in breast cancer.
Subsequently, we used the SMART App to verify the Spearman correlation between gene expression and DNA methylation of the probes of RBP7 in breast cancer. The expression of RBP7 was significantly negatively correlated with the methylation probes cg03406535, cg27083689, cg18086187, cg03994053, cg27561954, cg15090005, and cg07224455. The aggregation plot for all methylation probes showed that the expression level of RBP7 was negatively correlated with the DNA methylation of RBP7 (Figure 5(c)).
3.6. Clinicopathological Association of RBP7 and Its Prognostic Value
Breast cancer is a complex disease characterized by many morphological, clinical, and molecular features. Molecular subtypes and optimal treatments for breast cancer are usually based on immunohistochemical markers such as estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2). We checked the relevance of RBP7 expression and different clinicopathological features by using the web-based tool bc-GenExMiner. We found that the expression of RBP7 was highly expressed in ER-positive breast cancer patients compared with ER-negative patients (Figure 6(a)). RBP7 expression was higher in PR-positive breast cancer patients than in PR-negative patients (Figure 6(b)), whereas RBP7 expression was higher in HER2-negative breast cancer patients than in HER2-positive patients (Figure 6(c)). To further determine the correlation of RBP7 expression and hormone receptors (HRs), we utilized DNA microarray data to perform correlation analysis of RBP7 expression with different combinations of ER and PR expression statuses, i.e., ER+/PR+, ER+/PR−, ER−/PR+, and ER−/PR−. The results showed that there were remarkable differences in RBP7 mRNA expression in some hormone receptor combinations, i.e., ER+/PR+ vs. ER−/PR−, p < 0.01, and ER+/PR+ vs. ER+/PR−, p < 0.01 (Figure 6(d)).
Figure 6.

The mRNA expression of RBP7 and the Kaplan–Meier survival curve in different molecular subtypes of breast cancer. (a–c) The mRNA expression of RBP7 in different molecular subtypes, including ER (a), PR (b), and HER2 (c), of breast cancer. (d) Bee swarm plot of RBP7 expression in breast cancer with positive or negative ER or PR expression. (e–l) Kaplan–Meier curves of breast cancer with the DNA microarray results of ER+/PR+ (e, f), ER+/PR− (g, h), ER−/PR+ (i, j), or ER−/PR− (k, l).
Furthermore, we performed survival analysis of breast cancer patients with different ER/PR combinations. Downregulated RBP7 expression was only significantly associated with poor prognosis in ER+/PR+ and ER+/PR− patients but not in ER−/PR+ and ER−/PR− breast cancer patients (Figures 6(e)–6(l)). OSbrca was used to verify the prognostic value of RBP7 in these 4 groups with the TCGA database. Consistently, we found that lower RBP7 expression was significantly correlated with poorer OS in ER+/PR+ and ER+/PR− patients (Supplementary Figure 4).
3.7. Potential Regulatory Mechanisms and Target Drugs of RBP7 in Breast Cancer
To further explore the PPI network of RBP7 in breast cancer, we used STRING to identify genes that may interact with RBP7. The interactions between RBP7 and proteins encoded by the functional genes, including SLC45A1, SLC25A33, UBE4B, TMEM201, NMNAT1, SOCS7, SOCS4, GPR157, PIK3R3, and POLR2G, are shown in Figure 7(a). Interestingly, in this interacting network, PIK3R3, a regulatory subunit of phosphatidylinositol 3-kinase (PI3K), was reported to play an important role in breast cancer. Therefore, we further analyzed the correlation between RBP7 and PIK3R3 in breast cancer and found that the expression levels of RBP7 and PIK3R3 were negatively correlated (R = −0.231, p = 6.79e − 15) (Figure 7(b)).
Figure 7.
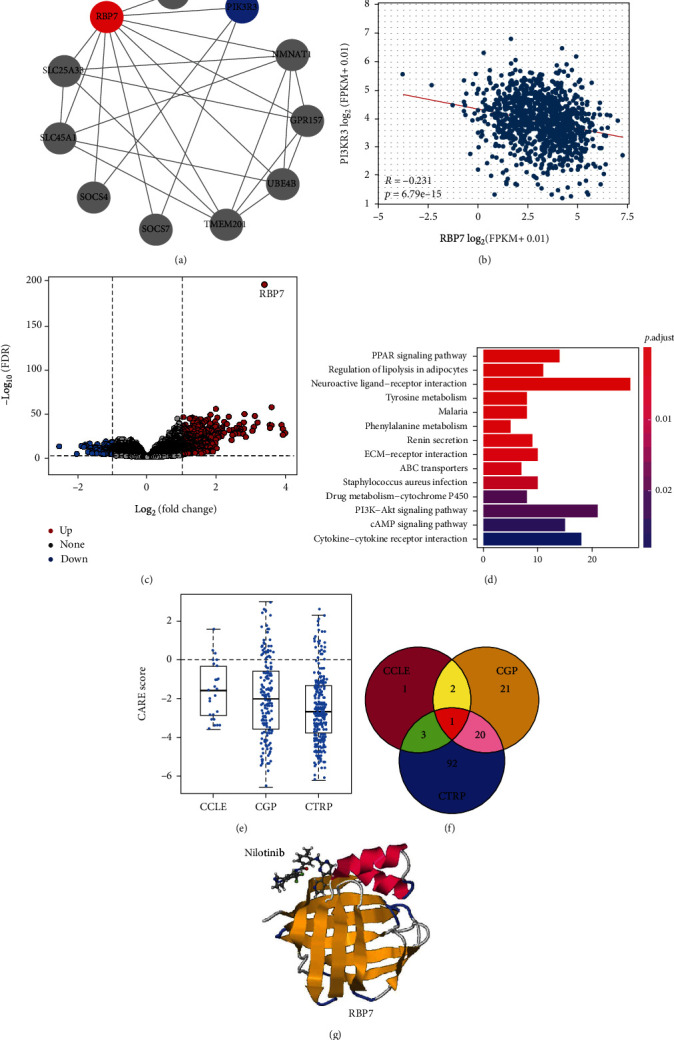
Analysis of the potential regulatory functions and RBP7-targeting drugs for breast cancer. (a) The PPI network for RBP7 and its coexpressed genes by STRING. (b) Correlation analysis of RBP7 and PIK3R3. (c) Volcano plot showing the differentially expressed genes in ER+ breast cancer with high or low expression of RBP7. (d) KEGG pathway analysis of DEGs in ER+ breast cancer with high or low expression of RBP7. (e) CARE analysis of the resistance module gene RBP7 in the CCLE, CTRP and CGP databases. (f) Venn diagram showing the overlap of the RBP7-targeting drugs in the 3 databases. (g) Prediction of the binding sites between nilotinib and RBP7.
Then, we acquired ER+ breast cancer data from the TCGA database, which were quarterly ranked according to the expression level of RBP7. The high and low RBP7 expression groups were defined as the first and fourth quarters of ER+ breast cancer data, respectively, and the differential genes (|log2(FC)| > 1) between these two groups were analyzed by using the Limma software package for display as a volcano plot (Figure 7(c)).
KEGG pathway analysis was conducted to investigate the functional implications of the DEGs, and it was found that several tumor-related pathways, such as the PPAR signalling pathway, regulation of lipolysis in adipocytes, and tyrosine metabolism, were significantly enriched (Figure 7(d)). In addition, the PI3K-AKT signalling pathway, which is important in ER+ breast cancers, was also enriched (Figure 7(d)).
Furthermore, we used the CARE database to analyze the association of the molecular alteration of RBP7 with drug efficacy and found that RBP7 was negatively correlated with drug efficacy in the CCLE, CGP, and CTRP databases (Figure 7(e)). Interestingly, we found that only one drug (nilotinib) was common among the resistant drugs from these 3 databases (Figure 7(f)). Finally, we visualized the binding site of RBP7 with nilotinib by SwissDock (Figure 7(g)).
4. Discussion
RBP7 is a member of the CRBP family and is involved in retinoic acid-mediated cellular responses [37]. Previous studies demonstrated that the retinol signalling pathway might be relevant to breast cancer progression. However, the prognostic value of RBP7, a new member of the CRBP family, in breast cancer is still unclear. In this study, we utilized various databases to explore the expression, prognosis, cellular localization, coexpression network, DNA methylation, and function of RBP7 in breast cancer.
Gene expression analysis showed that RBP7 is widely expressed in various normal tissues, including thyroid, testis, breast, and adipose tissues. Notably, in breast cancer tissues, RBP7 is mainly expressed in epithelial cells with nuclear localization. Importantly, we found that both the mRNA and protein expression levels of RBP7 in breast cancer tissues were significantly lower than those in normal tissues. Survival analysis with 4 different databases showed that low expression of RBP7 was significantly associated with poor OS in breast cancer patients. To further illuminate the role of RBP7 in the progression of breast cancer, we analyzed the expression and prognosis of RBP7 in different molecular subtypes of breast cancer. The results showed that RBP7 mRNA expression in ER-positive patients was higher than that in ER-negative patients, and higher expression of RBP7 was associated with better OS and DFS in ER+ breast cancer patients, indicating that RBP7 was mainly related to the prognosis of patients with ER+ breast cancer.
Our findings were consistent with other studies on the role of RBP7 in breast cancer. For example, Kinyamu et al. used genome-wide transcriptional profiling to demonstrate that RBP7 is positively regulated by estradiol (E2) in breast cancer cells [38]; Calvo et al. reported that blockers of estrogen receptors inhibited the expression of estradiol-modulated genes, including RBP7, in the mouse mammary gland [39]. It is reasonable to propose that upregulation of RBP7 by E2 leads to a good prognosis in ER+ breast cancer. Previous studies demonstrated that the growth of human breast tumor cells is regulated by signalling pathways involving nuclear steroid thyroxine receptors [40], especially RARs, which show growth inhibitory activity against breast cancer cells both in vitro and in vivo [41].Interestingly, RAR and ER share a common coactivator, estradiol. Pemrick et al. proved that both RAR and ER have high affinity for β-estradiol by constructing chimeric RARs containing the ligand-binding domain of ER [42]. Furthermore, Fonja et al. found that anti-estrogen and Herceptin induced the expression of PDCD4, revealing that the intracellular crosstalk of RAR, ER, and Her-2 may play a role in growth inhibitory signalling in breast cancer cells [43]. In the retinoid pathway, RAR activation is associated with CRBP1-mediated retinol storage [9] [44]. RBP7, another member of the CRBP family, may also engage in crosstalk with RAR; thus, it may play an important role in mechanism regulation via RAR/ER in breast cancer. Based on the results above, we conclude that RBP7 may be a tumor suppressor gene and that high expression of RBP7 is associated with a good prognosis for ER+ breast cancer patients.
Our results demonstrated that the gene expression of RBP7 in breast cancer was significantly lower than that in normal controls. Thus, we further explored the mechanism of RBP7 downregulation in breast cancer. Bioinformatic analysis showed that there was a significant negative correlation between RBP7 mRNA expression and promoter methylation. Breast cancer is a highly complex heterogeneous disease that forms different tumor subpopulations with distinct phenotypic characteristics. The different DNA methylation patterns between cell subpopulations drive the phenotypic changes in breast cancer, which is valuable for providing novel insights into intratumor epigenetic heterogeneity [45]. Recent studies have shown that methylation of the promoter as well as intragenic and intergenic regions is involved in the modulation of tumor development and invasion [46]. The methylation of CRBPs was reported to be associated with tumor development. For example, the methylation of CRBPs is common in the esophageal mucosa of patients with esophageal squamous cell carcinoma (ESCC) in the high-risk population and tends to increase in prevalence in foci with worse pathological changes [47]. The methylation profile of CRBPs in bladder cancers is also correlated with the clinicopathological features of poor prognosis [48]. DNA hypermethylation brings about epigenetic silencing of CRBPs in human and mouse breast cancer [49]. For example, CRBP1 gene silencing was found in 60% of G2 and 66.7% of G3 carcinoma cells due to CRBP1 promoter methylation [50]. DNA methylation can occur in the whole genome, including the promoter, gene body, 3′-untranslated region (UTR) and intergenic regions, while the promoter can be further divided into TSS200, TSS1500, 5′-UTR, and the 1st exon. DNA methylation in gene promoters generally has a negative regulatory effect on gene expression, whereas methylation in intragenic regions is not always associated with gene repression [51]. We utilized the MethSurv database to identify the methylation sites of RBP7 in breast cancer and found 4 probes, namely, cg20413202, cg03406535, cg10796749, and cg14202757, located in TSS1500-N_Shore with high methylation. The analysis using SMART APP web tools showed that promoter methylation was negatively correlated with RBP7 mRNA expression. In summary, our results demonstrate that the promoter methylation of RBP7 be related to its transcriptional silencing, which may be a reasonable explanation for the gene downregulation of RBP7 in breast cancer.
With the concept that low expression of RBP7 leads to poor prognosis of ER+ breast cancer patients, we further performed KEGG pathway analysis in ER+ breast cancer and found that RBP7 exerts its biological function through crosstalk with the PPAR and PI3K/AKT signalling pathways. PPARs represent a nuclear receptor superfamily that includes PPARα, PPARβ/δ, and PPARγ. It was reported that the activation of PPARγ inhibits the cell growth of different cancers, such as colon cancer, gastric cancer, and liposarcoma [52]. For example, after cleavage by caspase-1 at Asp64, PPARγ translocates to mitochondria, leading to attenuation of medium-chain acyl-CoA dehydrogenase (MCAD) activity and inhibition of fatty acid oxidation, which brings about the accumulation of lipid droplets and differentiation of tumor-associated macrophages (TAMs), thus resulting in an ultimate suppression of tumor growth [53, 54]. Another study by Mueller et al. reported that PPARγ is highly expressed in human primary and metastatic breast cancer, and ligand activation of this receptor in breast cancer cells causes extensive lipid accumulation, which results in a reduction in the growth rate and clonogenic capacity of tumor cells [55] [56]. Intriguingly, application of the PPARγ agonist rosiglitazone in combination with the MEK inhibitor trametinib can terminally differentiate breast cancer cells that have undergone epithelial-mesenchymal transition (EMT) into adipocytes [57]. As a PPARγ target gene, RBP7 is also an upstream regulator of some other PPARγ target genes in the endothelium. PPARγ and RBP7 control the oxidative state of blood vessels by forming a transcriptional regulatory circuit (or hub) in the endothelium. Loss of RBP7 impairs this regulatory circuit, resulting in oxidative stress and dysfunction in endothelial cells [58].Cancer cells have to endure oxidative stress throughout tumorigenesis, including during initiation, matrix detachment, transmission in the circulation, and relapse after therapy [59]. In addition, endothelial injury is closely related to tumorigenesis and accompanies malignant cancer cells in almost every stage of the metastatic process [60]. Thus, impairment of the regulatory circuit between RBP7 and PPARγ increases the opportunity to promote the occurrence of cancer.
PI3K/AKT is the most frequently activated signalling pathway that promotes tumor growth [61] and progression of breast cancer [62]. Bonofiglio et al. revealed that the ERα and PPARγ pathways have an opposite effect on the regulation of the PI3K/AKT signal transduction cascade [63]. The nuclear receptor ERα has been shown to be involved in the pathophysiological process of breast cancer. Membrane-anchored ERα can activate various cytoplasmic kinases, including components of the PI3K/AKT pathway, through rapid nongenomic actions [64]. There are two signalling pathways involved in the activation of the PI3K/AKT pathway in ER+ breast cancer cells. The estrogen-dependent pathway activates the PI3K/AKT signalling process by directly binding to the p85α regulatory subunit of PI3K, thus enhancing the transcriptional activity of targeted genes in breast cancer cells [65]. For the estrogen-independent pathway, the interaction of EGFR with growth factor can directly induce ERα transcriptional activity through the PI3K/AKT signalling pathway [66]. Consequently, the downstream signalling of both pathways is activated, leading to the proliferation and survival of tumor cells. In contrast, PPARγ can inhibit the PI3K/AKT pathway by upregulating PTEN transcription in breast cancer cells [63]. As an antagonist of the PI3K/AKT pathway, PTEN plays a key role in preventing tumorigenesis [67]. Based on our results and previous studies, we hypothesize that RBP7 may regulate PTEN by targeting PPARγ, thereby suppressing the activation of the PI3K/AKT pathway. The PPI network from bioinformatics analysis revealed that RBP7 may directly interact with PIK3R3, resulting in activation of the PI3K/AKT pathway (Figure 8).
Figure 8.
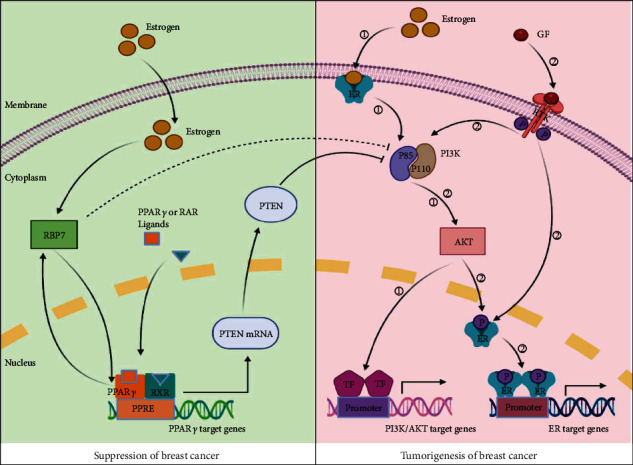
The hypothetical function of RBP7 in the regulation of the PPAR and PI3K/AKT pathways in ER+ breast cancer.
5. Conclusions
In conclusion, this study provides the first evidence that RBP7 downregulation in breast cancer is associated with promoter methylation. Furthermore, we found that RBP7 has prognostic value for ER+ breast cancer. Deep bioinformatics analysis of RBP7-related pathways reveals some vital information for the regulatory mechanism in ER+ breast cancer. However, these results need to be further validated by both in vitro and in vivo experiments. This study is helpful in providing novel approaches for clinical diagnosis and treatment.
Acknowledgments
This work has benefited from TCGA and GEO. We thank the TCGA and GEO network for its generous sharing large amounts of data. The manuscript has been presented as a preprint according to the following link “https://www.researchsquare.com/article/rs-1135383/v1” [68]. This study was supported by grants from the National Natural Science Foundation of China (No. 81971895), the Special Support Plan for Outstanding Talents of Guangdong Province (No. 2019JC05Y340), and the Guangdong Provincial Science and Technology Projects (No. 2016A020216015).
Contributor Information
Yibin Hao, Email: haoyibin450@126.com.
Yong Jiang, Email: jiang48231@163.com.
Data Availability
The original contributions presented in the study are included in the article/Supplementary material, and further inquiries can be directed to the corresponding authors.
Ethical Approval
All data of this study were public and required no ethical approval by an institutional review board or ethics committee.
Conflicts of Interest
No conflicts of interest, financial or otherwise, are declared by the authors.
Authors' Contributions
HL, QH, YH, and YJ conceived and designed research. HL and QH analyzed data. HL, JW, and ZZ interpreted results. HL, QH, and JW prepared figures. HL drafted manuscript. HHL, YH, and YJ edited and revised manuscript. HL, JW, QH, ZZ, HHL, YH, and YJ approved the final version of the manuscript.
Supplementary Materials
Supplementary Figure 1. Analysis of the mRNA expression of RBP7 in breast cancer. Supplementary Figure 2. The genes correlated with the expression of RBP7. Supplementary Figure 3. Density maps of CpG island methylation and the methylation distribution in different clinical stages of breast cancer. Supplementary Figure 4. Effect of RBP7 expression on the overall survival curves of breast cancer patients of different molecular subtypes.
References
- 1.Siegel R. L., Miller K. D., Jemal A. Cancer statistics, 2020. CA: a Cancer Journal for Clinicians . 2020;70(1):7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Siersbaek R., Kumar S., Carroll J. S. Signaling pathways and steroid receptors modulating estrogen receptor α function in breast cancer. Genes & Development . 2018;32(17-18):1141–1154. doi: 10.1101/gad.316646.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanker A. B., Sudhan D. R., Arteaga C. L. Overcoming endocrine resistance in breast cancer. Cancer Cell . 2020;37(4):496–513. doi: 10.1016/j.ccell.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Napoli J. L. Cellular retinoid binding-proteins, CRBP, CRABP, FABP5: effects on retinoid metabolism, function and related diseases. Pharmacology & Therapeutics . 2017;173:19–33. doi: 10.1016/j.pharmthera.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esteller M., Guo M., Moreno V., et al. Hypermethylation-associated inactivation of the cellular retinol-binding-protein 1 gene in human cancer. Cancer Research . 2002;62(20):5902–5905. [PubMed] [Google Scholar]
- 6.Kuppumbatti Y. S., Bleiweiss I. J., Mandeli J. P., Waxman S., Mira-y-Lopez R. Cellular retinol-binding protein expression and breast cancer. Journal of the National Cancer Institute . 2000;92(6):475–480. doi: 10.1093/jnci/92.6.475. [DOI] [PubMed] [Google Scholar]
- 7.Kuppumbatti Y. S., Rexer B., Nakajo S., Nakaya K., Mira-y-Lopez R. CRBP suppresses breast cancer cell survival and anchorage-independent growth. Oncogene . 2001;20(50):7413–7419. doi: 10.1038/sj.onc.1204749. [DOI] [PubMed] [Google Scholar]
- 8.Farias E. F., Marzan C., Mira-y-Lopez R. Cellular retinol-binding protein-I inhibits PI3K/Akt signaling through a retinoic acid receptor-dependent mechanism that regulates p85-p110 heterodimerization. Oncogene . 2005;24(9):1598–1606. doi: 10.1038/sj.onc.1208347. [DOI] [PubMed] [Google Scholar]
- 9.Farias E. F., Ong D. E., Ghyselinck N. B., Nakajo S., Kuppumbatti Y. S., Mira y Lopez R. Cellular retinol-binding protein I, a regulator of breast epithelial retinoic acid receptor activity, cell differentiation, and tumorigenicity. Journal of the National Cancer Institute . 2005;97(1):21–29. doi: 10.1093/jnci/dji004. [DOI] [PubMed] [Google Scholar]
- 10.Folli C., Calderone V., Ramazzina I., Zanotti G., Berni R. Ligand binding and structural analysis of a human putative cellular retinol-binding protein. The Journal of Biological Chemistry . 2002;277(44):41970–41977. doi: 10.1074/jbc.M207124200. [DOI] [PubMed] [Google Scholar]
- 11.Fang S., Sigmund C. D. PPARγ and RhoBTB1 in hypertension. Current Opinion in Nephrology and Hypertension . 2020;29(2):161–170. doi: 10.1097/MNH.0000000000000579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahn J., Kim D. H., Suh Y., Lee J. W., Lee K. Adipose-specific expression of mouse _Rbp7_ gene and its developmental and metabolic changes. Gene . 2018;670:38–45. doi: 10.1016/j.gene.2018.05.101. [DOI] [PubMed] [Google Scholar]
- 13.Elmasry M., Brandl L., Engel J., Jung A., Kirchner T., Horst D. RBP7 is a clinically prognostic biomarker and linked to tumor invasion and EMT in colon cancer. Journal of Cancer . 2019;10(20):4883–4891. doi: 10.7150/jca.35180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uhlen M., Fagerberg L., Hallstrom B. M., et al. Tissue-based map of the human proteome. Science . 2015;347(6220) doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 15.Uhlen M., Zhang C., Lee S., et al. A pathology atlas of the human cancer transcriptome. Science . 2017;357(6352) doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 16.Li T., Fan J., Wang B., et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Research . 2017;77(21):e108–e110. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rhodes D. R., Yu J., Shanker K., et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia . 2004;6(1):1–6. doi: 10.1016/S1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chandrashekar D. S., Bashel B., Balasubramanya S. A. H., et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia . 2017;19(8):649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edgar R., Domrachev M., Lash A. E. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Research . 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang Z., Li C., Kang B., Gao G., Li C., Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Research . 2017;45(W1):W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gyorffy B., Lanczky A., Eklund A. C., et al. An online survival analysis tool to rapidly assess the effect of 22, 277 genes on breast cancer prognosis using microarray data of 1, 809 patients. Breast Cancer Research and Treatment . 2010;123(3):725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 22.Goswami C. P., Nakshatri H. PROGgeneV2: enhancements on the existing database. BMC Cancer . 2014;14(1):p. 970. doi: 10.1186/1471-2407-14-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun D., Wang J., Han Y., et al. TISCH: a comprehensive web resource enabling interactive single-cell transcriptome visualization of tumor microenvironment. Nucleic Acids Research . 2021;49(D1):D1420–D1430. doi: 10.1093/nar/gkaa1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vasaikar S. V., Straub P., Wang J., Zhang B. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Research . 2018;46(D1):D956–D963. doi: 10.1093/nar/gkx1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zweig A. S., Karolchik D., Kuhn R. M., Haussler D., Kent W. J. UCSC genome browser tutorial. Genomics . 2008;92(2):75–84. doi: 10.1016/j.ygeno.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 26.Casper J., Zweig A. S., Villarreal C., et al. The UCSC genome browser database: 2018 update. Nucleic Acids Research . 2018;46(D1):D762–D769. doi: 10.1093/nar/gkx1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Modhukur V., Iljasenko T., Metsalu T., Lokk K., Laisk-Podar T., Vilo J. MethSurv: a web tool to perform multivariable survival analysis using DNA methylation data. Epigenomics . 2018;10(3):277–288. doi: 10.2217/epi-2017-0118. [DOI] [PubMed] [Google Scholar]
- 28.Li Y., Ge D., Lu C. The SMART app: an interactive web application for comprehensive DNA methylation analysis and visualization. Epigenetics & Chromatin . 2019;12(1):p. 71. doi: 10.1186/s13072-019-0316-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu W., Xu M., Wang L., et al. Integrative analysis of DNA methylation and gene expression identified cervical cancer-specific diagnostic biomarkers. Signal Transduction and Targeted Therapy . 2019;4(1):p. 55. doi: 10.1038/s41392-019-0081-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jezequel P., Gouraud W., Ben Azzouz F., et al. bc-GenExMiner 4.5: new mining module computes breast cancer differential gene expression analyses. Database: The Journal of Biological Databases and Curation . 2021;2021, article baab007 doi: 10.1093/database/baab007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan Z., Wang Q., Sun X., et al. OSbrca: a web server for breast cancer prognostic biomarker investigation with massive data from tens of cohorts. Frontiers in Oncology . 2019;9:p. 1349. doi: 10.3389/fonc.2019.01349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Szklarczyk D., Franceschini A., Kuhn M., et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Research . 2011;39(Database):D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J. H., Liu S., Zhou H., Qu L. H., Yang J. H. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Research . 2014;42(D1):D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanehisa M., Goto S., Sato Y., Furumichi M., Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Research . 2012;40(D1):D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang P., Lee W., Li X., et al. Genome-scale signatures of gene interaction from compound screens predict clinical efficacy of targeted cancer therapies. Cell Systems . 2018;6(3):343–354.e5. doi: 10.1016/j.cels.2018.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grosdidier A., Zoete V., Michielin O. Swiss Dock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Research . 2011;39:W270–W277. doi: 10.1093/nar/gkr366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choder M. Rpb4 and Rpb7: subunits of RNA polymerase II and beyond. Trends in Biochemical Sciences . 2004;29(12):674–681. doi: 10.1016/j.tibs.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 38.Kinyamu H. K., Collins J. B., Grissom S. F., Hebbar P. B., Archer T. K. Genome wide transcriptional profiling in breast cancer cells reveals distinct changes in hormone receptor target genes and chromatin modifying enzymes after proteasome inhibition. Molecular Carcinogenesis . 2008;47(11):845–885. doi: 10.1002/mc.20440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Calvo E., Luu-The V., Belleau P., Martel C., Labrie F. Specific transcriptional response of four blockers of estrogen receptors on estradiol-modulated genes in the mouse mammary gland. Breast Cancer Research and Treatment . 2012;134(2):625–647. doi: 10.1007/s10549-012-2104-7. [DOI] [PubMed] [Google Scholar]
- 40.Schneider S. M., Offterdinger M., Huber H., Grunt T. W. Involvement of nuclear steroid/thyroid/retinoid receptors and of protein kinases in the regulation of growth and of c‐erbB and retinoic acid receptor expression in MCF-7 breast cancer cells. Breast Cancer Research and Treatment . 1999;58(2):171–181. doi: 10.1023/A:1006377006816. [DOI] [PubMed] [Google Scholar]
- 41.Peng X., Maruo T., Cao Y., et al. A novel RARbeta isoform directed by a distinct promoter P 3 and mediated by retinoic acid in breast cancer cells. Cancer Research . 2004;64(24):8911–8918. doi: 10.1158/0008-5472.CAN-04-1810. [DOI] [PubMed] [Google Scholar]
- 42.Pemrick S. M., Abarzua P., Kratzeisen C., et al. Characterization of the chimeric retinoic acid receptor RARα/VDR. Leukemia . 1998;12(4):554–562. doi: 10.1038/sj.leu.2400937. [DOI] [PubMed] [Google Scholar]
- 43.Afonja O., Juste D., Das S., Matsuhashi S., Samuels H. H. Induction of PDCD4 tumor suppressor gene expression by RAR agonists, antiestrogen and HER-2/neu antagonist in breast cancer cells. Evidence for a role in apoptosis. Oncogene . 2004;23(49):8135–8145. doi: 10.1038/sj.onc.1207983. [DOI] [PubMed] [Google Scholar]
- 44.Bushue N., Wan Y. J. Retinoid pathway and cancer therapeutics. Advanced Drug Delivery Reviews . 2010;62(13):1285–1298. doi: 10.1016/j.addr.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Almendro V., Fuster G. Heterogeneity of breast cancer: etiology and clinical relevance. Clinical & Translational Oncology . 2011;13(11):767–773. doi: 10.1007/s12094-011-0731-9. [DOI] [PubMed] [Google Scholar]
- 46.Rauscher G. H., Kresovich J. K., Poulin M., et al. Exploring DNA methylation changes in promoter, intragenic, and intergenic regions as early and late events in breast cancer formation. BMC Cancer . 2015;15(1):p. 816. doi: 10.1186/s12885-015-1777-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roth M. J., Abnet C. C., Hu N., et al. P16, MGMT, RARbeta2, CLDN3, CRBP and MT1G gene methylation in esophageal squamous cell carcinoma and its precursor lesions. Oncology Reports . 2006;15(6):1591–1597. [PubMed] [Google Scholar]
- 48.Brait M., Begum S., Carvalho A. L., et al. Aberrant promoter methylation of multiple genes during pathogenesis of bladder cancer. Cancer Epidemiology, Biomarkers & Prevention . 2008;17(10):2786–2794. doi: 10.1158/1055-9965.EPI-08-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arapshian A., Bertran S., Kuppumbatti Y. S., Nakajo S., Mira-y-Lopez R. Epigenetic CRBP downregulation appears to be an evolutionarily conserved (human and mouse) and oncogene-specific phenomenon in breast cancer. Molecular Cancer . 2004;3(1):p. 13. doi: 10.1186/1476-4598-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Doldo E., Costanza G., Ferlosio A., et al. CRBP-1 expression in ovarian cancer: a potential therapeutic target. Anticancer Research . 2014;34(7):3303–3312. [PubMed] [Google Scholar]
- 51.Shenker N., Flanagan J. M. Intragenic DNA methylation: implications of this epigenetic mechanism for cancer research. British Journal of Cancer . 2012;106(2):248–253. doi: 10.1038/bjc.2011.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mirza A. Z., Althagafi I. I., Shamshad H. Role of PPAR receptor in different diseases and their ligands: physiological importance and clinical implications. European Journal of Medicinal Chemistry . 2019;166:502–513. doi: 10.1016/j.ejmech.2019.01.067. [DOI] [PubMed] [Google Scholar]
- 53.Niu Z., Shi Q., Zhang W., et al. Caspase-1 cleaves PPARγ for potentiating the pro-tumor action of TAMs. Nature Communications . 2017;8(1):p. 766. doi: 10.1038/s41467-017-00523-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fu Y., Zou T., Shen X., et al. Lipid metabolism in cancer progression and therapeutic strategies. MedComm . 2021;2(1):27–59. doi: 10.1002/mco2.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mueller E., Sarraf P., Tontonoz P., et al. Terminal differentiation of human breast cancer through PPARγ. Molecular Cell . 1998;1(3):465–470. doi: 10.1016/S1097-2765(00)80047-7. [DOI] [PubMed] [Google Scholar]
- 56.Apostoli A. J., Roche J. M., Schneider M. M., et al. Opposing roles for mammary epithelial-specific PPARγ signaling and activation during breast tumour progression. Molecular Cancer . 2015;14(1):p. 85. doi: 10.1186/s12943-015-0347-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ishay-Ronen D., Diepenbruck M., Kalathur R. K. R., et al. Gain fat–lose metastasis: converting invasive breast cancer cells into adipocytes inhibits cancer metastasis. Cancer Cell . 2019;35(1):17–32.e6. doi: 10.1016/j.ccell.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 58.Hu C., Keen H. L., Lu K. T., et al. Retinol-binding protein 7 is an endothelium-specific PPARγ cofactor mediating an antioxidant response through adiponectin. JCI Insight . 2017;2(6, article e91738) doi: 10.1172/jci.insight.91738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hayes J. D., Dinkova-Kostova A. T., Tew K. D. Oxidative stress in cancer. Cancer Cell . 2020;38(2):167–197. doi: 10.1016/j.ccell.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Blazejczyk A., Papiernik D., Porshneva K., Sadowska J., Wietrzyk J. Endothelium and cancer metastasis: perspectives for antimetastatic therapy. Pharmacological Reports . 2015;67(4):711–718. doi: 10.1016/j.pharep.2015.05.014. [DOI] [PubMed] [Google Scholar]
- 61.Miller T. W., Rexer B. N., Garrett J. T., Arteaga C. L. Mutations in the phosphatidylinositol 3-kinase pathway: role in tumor progression and therapeutic implications in breast cancer. Breast Cancer Research . 2011;13(6):p. 224. doi: 10.1186/bcr3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saal L. H., Johansson P., Holm K., et al. Poor prognosis in carcinoma is associated with a gene expression signature of aberrant PTEN tumor suppressor pathway activity. Proceedings of the National Academy of Sciences of the United States of America . 2007;104(18):7564–7569. doi: 10.1073/pnas.0702507104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bonofiglio D., Gabriele S., Aquila S., et al. Estrogen receptor alpha binds to peroxisome proliferator-activated receptor response element and negatively interferes with peroxisome proliferator-activated receptor gamma signaling in breast cancer cells. Clinical Cancer Research . 2005;11(17):6139–6147. doi: 10.1158/1078-0432.CCR-04-2453. [DOI] [PubMed] [Google Scholar]
- 64.Khatpe A. S., Adebayo A. K., Herodotou C. A., Kumar B., Nakshatri H. Nexus between PI3K/AKT and estrogen receptor signaling in breast cancer. Cancers (Basel) . 2021;13(3):p. 369. doi: 10.3390/cancers13030369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Simoncini T., Hafezi-Moghadam A., Brazil D. P., Ley K., Chin W. W., Liao J. K. Interaction of oestrogen receptor with the regulatory subunit of phosphatidylinositol-3-OH kinase. Nature . 2000;407(6803):538–541. doi: 10.1038/35035131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Miller T. W., Balko J. M., Arteaga C. L. Phosphatidylinositol 3-kinase and antiestrogen resistance in breast cancer. Journal of Clinical Oncology . 2011;29(33):4452–4461. doi: 10.1200/JCO.2010.34.4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huang J., Yan J., Zhang J., et al. SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane. Nature Communications . 2012;3(1):p. 911. doi: 10.1038/ncomms1919. [DOI] [PubMed] [Google Scholar]
- 68.Lin H., Han Q., Wang J., et al. Methylation-mediated silencing of RBP7 promotes breast cancer progression through PPAR and PI3K/AKT pathway . Research Square; 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Analysis of the mRNA expression of RBP7 in breast cancer. Supplementary Figure 2. The genes correlated with the expression of RBP7. Supplementary Figure 3. Density maps of CpG island methylation and the methylation distribution in different clinical stages of breast cancer. Supplementary Figure 4. Effect of RBP7 expression on the overall survival curves of breast cancer patients of different molecular subtypes.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary material, and further inquiries can be directed to the corresponding authors.


