Abstract
The rapid emergence of SARS-CoV-2 variants challenges vaccination strategies. Here, we collected 201 serum samples from persons with a single infection or multiple vaccine exposures, or both. We measured their neutralization titers against 15 natural variants and 7 variants with engineered spike mutations and analyzed antigenic diversity. Antigenic maps of primary infection sera showed that Omicron sublineages BA.2, BA.4/BA.5, and BA.2.12.1 are distinct from BA.1 and more similar to Beta/Gamma/Mu variants. Three mRNA COVID-19 vaccinations increased neutralization of BA.1 more than BA.4/BA.5 or BA.2.12.1. BA.1 post-vaccination infection elicited higher neutralization titers to all variants than three vaccinations alone, although with less neutralization to BA.2.12.1 and BA.4/BA.5. Those with BA.1 infection after two or three vaccinations had similar neutralization titer magnitude and antigenic recognition. Accounting for antigenic differences among variants when interpreting neutralization titers can aid the understanding of complex patterns in humoral immunity that informs the selection of future COVID-19 vaccine strains.
Keywords: antigenic cartography, SARS-CoV-2 variants, Omicron, COVID-19 vaccine, mRNA vaccine, SARS-CoV-2, spike, hybrid immunity, cartography, antigenic landscape
Graphical abstract
Wang et al. show that SARS-CoV-2 Omicron BA.1 or BA.1.1 infection after a second or third mRNA COVID-19 vaccination broadens neutralizing antibody responses to all variants, including Omicron, more than three vaccinations alone. BA.2.12.1 and BA.4/BA.5 evade neutralization more than BA.1 and BA.2 after three vaccinations or Omicron infection post-vaccination.
Introduction
COVID-19 has resulted in over 6.4 million deaths and 599 million infections worldwide.1 SARS-CoV-2 continues to circulate globally, even as population immunity increases due to infections, reinfections, and vaccination series, alone or in combination.2 Although authorized and licensed COVID-19 vaccines provide substantial protection against severe COVID-19, new and emerging SARS-CoV-2 variants continue to threaten their effectiveness. The need to develop vaccination strategies to provide the broadest and strongest immunity against emerging and future SARS-CoV-2 variants is therefore imperative.
Approved or authorized mRNA COVID-19 vaccines encode the spike protein of the first SARS-CoV-2 strain to emerge, Wuhan-Hu-1, defined as the ancestral strain. An increased reinfection risk associated with the Omicron variant compared with earlier SARS-CoV-2 variants has been observed.3 Omicron BA.1, first identified in November 2021, has led to millions of infections, including post-vaccine infections (PVIs). This has led to more recommendations for vaccine boosting. Additional variants closely related to Omicron, including BA.2 and its descendants, were detected soon afterward. These have rapidly outcompeted BA.1. For example, BA.2.12.1 and BA.4 and BA.5 (hereafter referred to as BA.4/BA.5) are now collectively the most common variants in the United States.4 , 5 , 6 Additional Omicron subvariants are also emerging, including BA.2.75 sublineages, which are spreading in various global regions.7
Vaccine formulations based on the ancestral spike antigen continue to be available for both primary series and booster vaccination schedules.8 Recent public health discussions question whether vaccinations derived from more recent strains substantially increase antibody magnitude (quantity) and breadth (recognition of many antigenically distinct variants) above boosting with the same ancestral strain, including in populations that may be unvaccinated, vaccinated, boosted, infected, reinfected, or various combinations thereof. Three doses of mRNA COVID-19 vaccines containing the ancestral strain increase immunity against a range of variants.9 , 10 , 11 , 12 , 13 However, fourth doses with the ancestral strain only transiently boost neutralization titers back to the peak observed after three doses.14 , 15 , 16 By contrast, sequential exposure to the ancestral vaccine followed by an Omicron PVI may increase neutralization titers across variants compared with vaccination with three doses alone,17 although other studies suggest protection against severe disease is similar.18
Optimal timing and composition of SARS-CoV-2 vaccines for both boosters and primary series, therefore, remain unclear. The World Health Organization (WHO) recently noted that an Omicron vaccine may provide broader protection against emerging variants in individuals who have already received two doses of ancestral vaccines. WHO recommended that unvaccinated individuals should still receive at least two doses of the ancestral-based vaccine rather than a single Omicron-based vaccine alone,19 and regulatory approvals for BA.1 antigen-containing vaccines are underway.20 More recently, the United States Food and Drug Administration (FDA) recommended that updates to COVID-19 booster vaccines include both ancestral and BA.4/BA.5 spike antigens.21 Preliminary results involving bivalent vaccines containing both the ancestral strain and Omicron BA.1 suggest that they induce similar or higher titers against BA.1 than a third dose with the ancestral strain alone.22 , 23
Antigenic diversity between Omicron variants has further complicated vaccine composition decision making. For example, a BA.1 booster may not provide sufficient protection if more recently emerged variants like BA.2.12.1 and BA.4/BA.5 further escape immunity.4 , 24 , 25 Furthermore, individuals who have been vaccinated with BNT162b2 or vaccinated and infected with BA.1 or BA.2 have lower neutralization titers against BA.2.12.1 and BA.4/BA.5 compared with BA.1 or BA.2.17 , 24 A similar observation was made with BBIBP-CorV (Sinopharm) vaccinated individuals with and without Omicron PVI.26
A challenge for informing vaccine strain selection with variant-specific antibody titers is the need to interpret antibody neutralization patterns in increasingly complex, time-varying antigenic histories derived from infection, vaccination, or both (hybrid immunity). Compounding this challenge is the need to predict humoral immunity against future variants. “Antigenic cartography” is a statistical method that geometrically interprets antibody titers, positioning variants on an “antigenic map” based on how they are neutralized by primary exposure sera.27 Compared with the direct interpretation of geometric mean titers, antigenic cartography simultaneously analyzes titers from a panel of sera and accommodates variation in individual titers when interpreting how sera recognize variant antigens. Resulting antigenic maps show the clustering of variants based on how they are recognized by distinct sera. High-quality maps perform well in cross-validation experiments by accurately predicting titers not included in making the map. Techniques that build on antigenic cartography, such as “antibody landscapes,” enable the evaluation of how neutralization titers change following antigen re-exposure compared with primary exposure. The height of an accurate antibody landscape can be used to predict titers against variants that were not used in making the landscape, as described by Fonville.28 Few antigenic maps have been made of SARS-CoV-2, likely because antigenic cartography requires the generation of large datasets of well-characterized sera from individuals with primary exposure to distinct, sequence-confirmed variants or experimentally inoculated animals.9 , 29 , 30 , 31 , 32 , 33 , 34 Although the published SARS-CoV-2 antigenic maps agree on the antigenic relationships between the ancestral strain, Delta, Beta, and Omicron BA.1 variants, the positions of BA.2, BA.2.12.1, and BA.4/BA.5 variants remain uncertain. Further, because different populations may experience different combinations of infection and vaccination exposures, more studies are needed to increase our understanding of antibody responses to SARS-CoV-2 variants.
In this study, we generated a unique dataset of neutralization titers against a large panel of SARS-CoV-2 variants, using well-characterized sera from a longitudinal cohort following primary COVID-19 cases with sequence-confirmed variant infections, and applied antigenic cartography to analyze antigenic divergences among the major SARS-CoV-2 variants. We also performed similar measurements and analyses on a separate cohort of uninfected individuals after two and three doses with an ancestral, spike-based mRNA COVID-19 vaccine (hereafter referred to as mRNA COVID-19 vaccine) to compare differences in neutralization titers across variants based on different types of antigenic exposures. We then used antibody landscapes and other related statistical metrics to quantify the gain in neutralization titer magnitude and breadth across variants following two or three mRNA COVID-19 vaccine doses and Omicron BA.1 or BA.1.1 PVIs compared with boosting with the mRNA COVID-19 vaccine alone. Together, these methods offer a useful analytical framework for quantifying and comparing the neutralization of SARS-CoV-2 variants following primary and subsequent antigen exposure. This approach along with many other considerations, including variant surveillance, and availability of candidate vaccines and clinical data, can be used by public health authorities when making final recommendations for vaccine composition.
Results
Primary infection sera display different neutralization patterns across variants
To measure the neutralization of the SARS-CoV-2 variants by sera following infections by different variants, we used serum samples from SARS-CoV-2-infected participants from the Epidemiology, Immunology, and Clinical Characteristics of Emerging Infectious Diseases with Pandemic Potential (EPICC) study (Table S1A).35 We identified serum samples collected 8–51 days post symptom onset (mean = 28 days) from 45 individuals with natural primary infections with 21 distinct variants. All individuals were unvaccinated and had sequenced, genotyped infecting viruses, as well as matched clinical and demographic data (Tables S1A and S1B). We complemented this serum set with additional serum samples from unvaccinated persons infected by variants that were underrepresented in the EPICC cohort. An additional 31 primary infection serum samples with known infecting genotypes and vaccination histories were purchased from Boca Biolistics (Pompano Beach, FL, USA, Tables S1C and S1D).34 We also included convalescent serum samples from one Beta-infected case from an unrelated FDA study protocol and six additional Beta-infected cases obtained from the HIV Vaccine Trial Network (HVTN) (see STAR Methods; Table S1A).
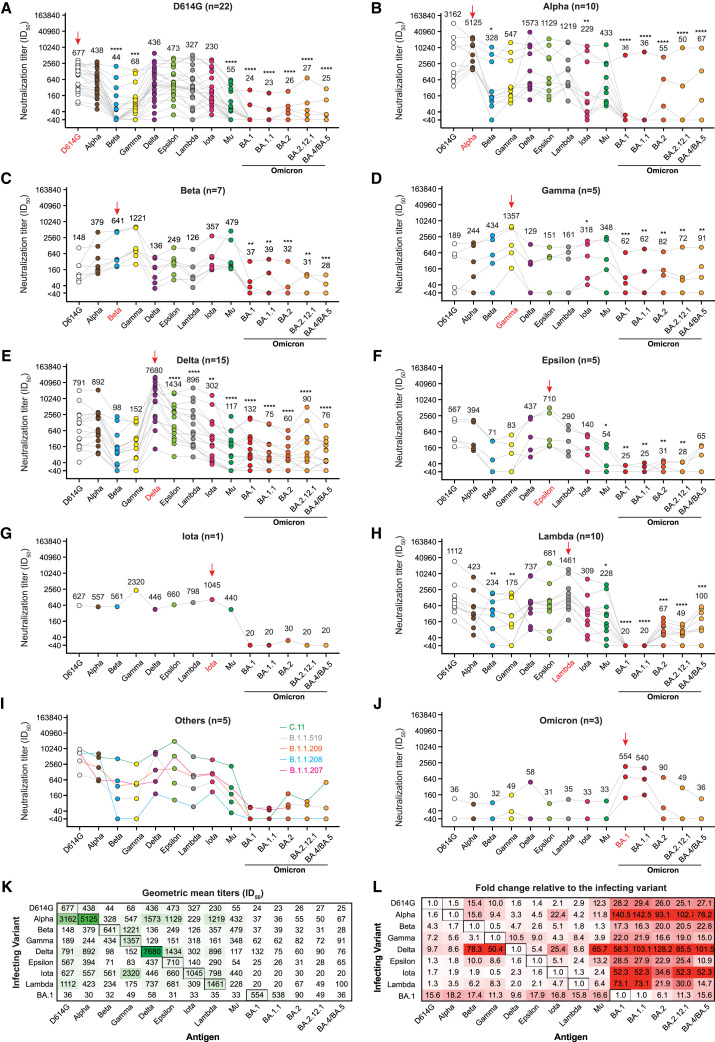
Each of the 83 serum samples was titrated for neutralization potency against a panel of 15 SARS-CoV-2 lentiviral pseudoviruses representing the major variants, including BA.1, BA.1.1, BA.2, BA.2.12.1, and BA.4/BA.5 (Table S2). Neutralization titers (50% inhibitory dilution, ID50) for sera against each variant were grouped by infecting variant and shown in Figure 1 . For each serum group, significant differences in magnitude were observed across the variant panel, although the pattern of neutralization depended on the infecting variant. Variants that temporally preceded Omicron also generally elicited lower titers against Omicron variants (Figures 1A–1I). The highest geometric mean titer (GMT) across the serum samples was generally for the infecting variant, with Alpha and Delta sera showing higher GMTs against the infecting variant compared with D614G (Figures 1B and 1E, respectively). Among the pre-Omicron infections and in agreement with previous data,36 , 37 , 38 , 39 , 40 , 41 , 42 , 43 , 44 the titers of Alpha, Delta, Epsilon, and Lambda convalescent serum samples against Beta, Gamma, and Mu variants were generally lower than against other pre-Omicron variants. Neutralization ID50 titers against the variants are shown in Figure 1K, and fold changes relative to the infecting variant are shown in Figure 1L.
Figure 1.
Neutralization antibody titers (ID50 values) against SARS-CoV-2 variant pseudoviruses for primary infection sera from individuals infected by a major SARS-CoV-2 variant
(A–J) Sera from (A) wild-type variant (D614G), (B) Alpha, (C) Beta, (D) Gamma, (E) Delta, (F) Epsilon, (G) Iota, (H) Lambda, (I) other variants, and (J) Omicron (BA.1 or BA.1.1). Each gray line corresponds to one serum sample. The red arrow denotes the infecting variant. Geometric mean neutralization titers (GMTs) are listed for each variant. Significance values for each variant are shown relative to the infecting variant.
(K) GMTs from (A)–(J) for sera from the infecting variants (rows) against all measured antigens (columns). Cells are shaded based on GMT, and serum-antigen pairs with larger titers have darker shades of green.
(L) Fold reduction in titer for each serum-antigen pair relative to the titer of the infecting variant (boxed in black). Each cell value represents the average fold change across all serum samples with the same exposure history, and darker red cells denote larger relative reductions in titer. For all neutralization assays, serum was diluted 1:40 followed by 3-fold serial dilutions. Neutralization assays were performed twice, each with an intra-assay duplicate. Neutralization curves were fitted using nonlinear dose-response regression. Titers measuring below the lowest serum dilution of 1:40 were treated as 20 for statistical analysis. Statistical analysis was performed on the paired samples using the Friedman test, followed by post hoc Dunn’s multiple comparison tests. p values for comparisons between the groups are shown, where ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, and ∗∗∗∗p ≤ 0.0001.
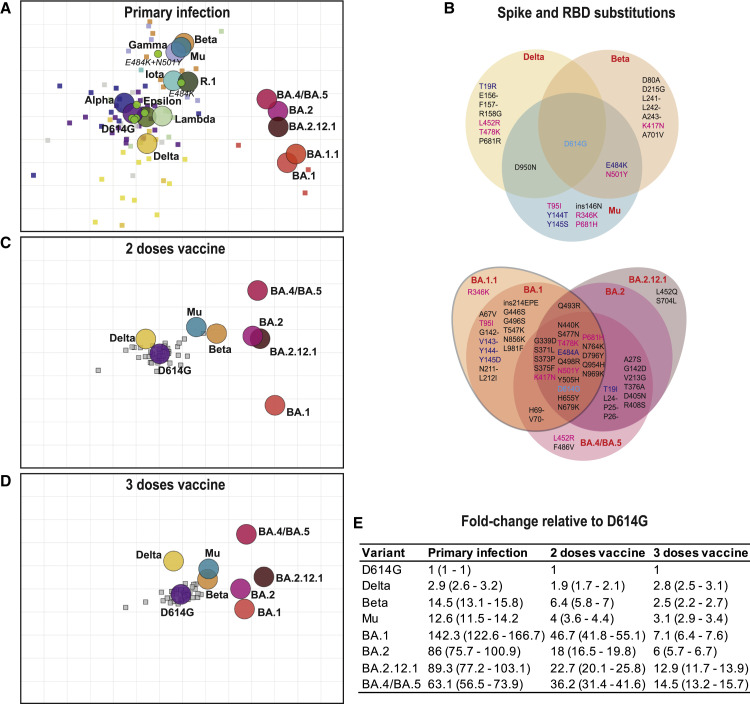
Antigenic maps using primary infection sera show Omicron variants BA.2, BA.4/BA.5, and BA.212.1 as antigenically distinct from BA.1 and more similar to the Beta/Gamma/Mu cluster
To further characterize how the primary infection sera recognize antigenic relationships among all variants, we applied antigenic cartography to an extended neutralization dataset that included neutralization titers for all 83 samples against the 15 major SARS-CoV-2 variants described above, as well as seven additional engineered spike variants with six single and double mutation in the receptor binding domain (Table S2). The cartography analysis allowed the simultaneous interpretation of 1,332 neutralization titers following primary natural infection to quantify the degree of recognition of distinct variants. Using a form of multi-dimensional scaling, the position of each strain and serum is optimized in Euclidean space such that the “antigenic distance,” a measure of antigenic recognition, between points, corresponds to the measured neutralization titer (the term antigenic distance used hereafter is always in reference to an antigenic map). The closer a serum (shown as a square on the map) is to a variant (shown as a circle on the map), the higher the titer for that serum to that variant. Overall, we find that the sera cluster near their respective infecting variants, as expected. As is standard for evaluating antigenic maps, we performed numerous validations to confirm that the maps are accurate representations of the titer data (see STAR Methods).
Consistent with previously published SARS-CoV-2 antigenic maps,9 , 29 , 30 , 31 , 32 , 33 , 34 we observed four major variant clusters (Figure 2 A). These clusters generally correspond to strains with shared amino acid changes in the spike receptor binding domain (Figure 2B), listed in parentheses below for each variant. The variants that were most similar to the ancestral strain (D614G) were Alpha (N501Y), Epsilon (L452R), and individual point mutations introduced into D614G (N501Y, L452R, T478K, R346K, and K417N). Lambda (L452Q and F490S) is only slightly further to the right of the ancestral strain, whereas Delta (L452R and T478K) is slightly below it. To the top and right of the ancestral strain are Beta and Gamma (E484K, N501Y, and K417N/T), Mu (E484K and N501Y), and D614G with both mutations E484K and N501Y. Iota (E484K), R.1 (E484K), and D614G with E484K are between the ancestral and Beta/Gamma/Mu cluster, likely because they lack the additional antigenic mutation at N501Y. Omicron BA.1 and BA.1.1 are to the right and are most distant from the ancestral variant. Both contain additional mutations in the receptor binding domain that are not observed in other variants, while BA.1.1 also contains R346K (Figure 2B). Strikingly, we found that BA.2.12.1, BA.2, and BA.4/BA.5 variants retain a large antigenic distance from the ancestral strain (89.3-, 86-, and 63.1-fold, respectively) but are shifted away from BA.1 and BA.1.1 and toward the Beta/Gamma/Mu cluster, supporting a recent observation that BA.2.12.1, BA.4, and BA.5 escape antibodies elicited by Omicron infection.45 BA.2 has numerous changes relative to BA.1 and BA.1.1 but is closely related to BA.2.12.1 and BA.4/BA.5 (Figure 2B).
Figure 2.
Antigenic maps made with neutralization titers from single-antigen exposure sera demonstrate that BA.1, BA.2, BA.2.12.1, and BA.4/BA.5 are most antigenically distinct from other major variants
(A–D) Antigenic maps were made using antigenic cartography with titers for (A) sera collected after convalescent primary infection with distinct variants and sera from uninfected individuals who received (C) two doses or (D) three doses of mRNA COVID-19 vaccines. Each grid-square side corresponds to a 2-fold dilution in the pseudovirus neutralization assay. Antigenic distance is measured in any direction on the grid. Antigens are shown as circles and labeled. Sera are shown as squares and are colored by infecting variant. (B) Substitutions in the spike and receptor binding domains for all variants used in this study.
(E) Fold difference in neutralization with 95% confidence intervals from the ancestral strain to each other variant on each map. For example, a fold difference of four corresponds to two grid squares on the antigenic map.
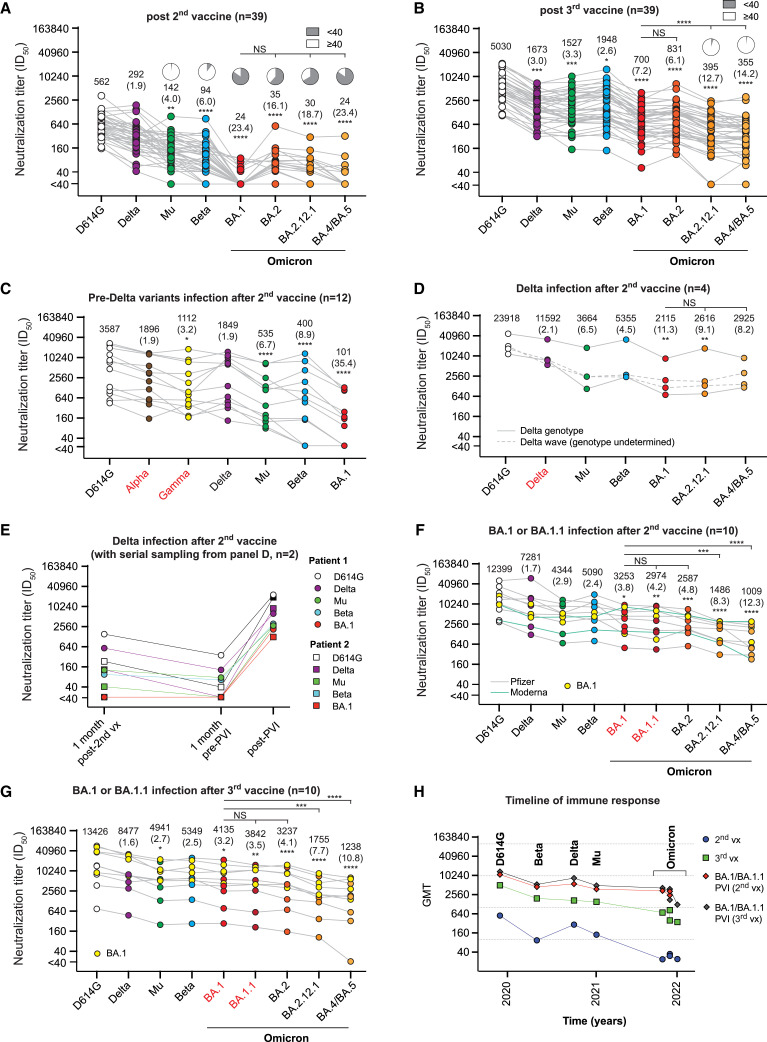
Sera after two or three doses with an mRNA COVID-19 vaccine show distinct antigenic relationships among Omicron variants
We next measured the neutralization titers for sera collected from 39 healthcare workers after two and three doses of an mRNA COVID-19 vaccine (Pfizer/BNT162b2) as part of the Prospective Assessment of SARS-CoV-2 Seroconversion (PASS) study (see STAR Methods; Table S1E). Sera were titrated against D614G, BA.1, BA.2, BA.2.12.1, and BA.4/BA.5 and Beta, Mu, and Delta. The last three were chosen because they represented the most distant clusters on the convalescent map in Figure 2A. After two doses, titers were highest for the ancestral strain and low against all Omicron variants (Figure 3 A). Both BA.1 and BA.4/BA.5 had the fewest titers above the assay cutoff (<40). By contrast, the third vaccine dose significantly boosted GMTs to all variants compared with the second vaccine dose (p < 0.0001), with BA.2 having the highest titers among the Omicron-lineage variants (GMT 831), followed by BA.1 (GMT 700), BA.2.12.1 (GMT 395), and BA.4/BA.5 (GMT 355) (Figure 3B).
Figure 3.
Neutralization titers (ID50 values) against variant pseudoviruses from post-vaccination sera with and without post-vaccination infection (PVI)
(A–G) Sera are from individuals who received (A) two doses of an mRNA COVID-19 vaccine or (B) three doses of an mRNA COVID-19 vaccine. Serum samples were obtained about 5–6 weeks following the last vaccine dose. PVI neutralization titers after 2 doses of wild-type mRNA COVID-19 vaccine in individuals infected with the (C) pre-Delta wave (Alpha or Gamma or others), (D and E) Delta, or (F and G) Omicron (BA.1 or BA.1.1) variants. Each gray line corresponds to one serum sample. GMT is listed for each variant. Significance values for each antigen are shown relative to the titer against D614G. Two of the Delta wave PVI serum samples in (D) were measured at multiple time points, shown in (E), from 1 month after the second vaccine dose and 1 month before and after PVI. (F) Shows titers from individuals with an Omicron (BA.1 or BA.1.1) PVI 2–10 months after the second vaccine, whereas (G) shows titers from individuals with an Omicron (BA.1 or BA.1.1) PVI 1–5 months after the third vaccine.
(H) The GMT of individual variants after vaccination with or without PVI by timeline. For all neutralization assays, the serum was diluted 1:40 followed by 3-fold serial dilutions. Neutralization assays were performed twice, each with an intra-assay duplicate. Neutralization curves were fitted using nonlinear dose-response regression. Titers measuring below the lowest serum dilution of 1:40 were treated as 20 for statistical analysis. Statistical analysis was performed on the paired samples using the Friedman test, followed by post hoc Dunn’s multiple comparison tests. p values for comparisons between the groups are shown, where ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, and ∗∗∗∗p ≤ 0.0001. NS, no significance; vx, vaccine. Pie charts indicate the percent of serum samples above the lowest tested (1:40). Numbers in parentheses indicate fold reduction in titer relative to D614G.
We also used antigenic cartography to interpret neutralization titers for sera collected after two and three vaccine doses. Titers were accurately represented as antigenic distances on antigenic maps in either one or two dimensions (Figure S1), but coordination was less accurate than for the natural infection map (Figure S2). We found that the antigenic distances between D614G and other variants (fold difference in neutralization between these variants) on the two doses vaccine antigenic map were smaller but had a similar pattern to the natural infection map (Figures 2A, 2C, and S3A), with Beta, Mu, and Delta closer (smaller fold difference) to the ancestral strain and BA.1 furthest from the ancestral strain, followed by the other Omicron variants with more intermediate antigenic distances. By contrast, sera from the same vaccinated individuals after their third vaccine dose recognized the Omicron variants differently than sera after the second vaccine dose (Figures 2D, S3B, and S3C). This is evident in the antigenic difference between the ancestral strain and BA.1 and BA.2, which dropped to 7.1- and 6-fold, respectively, whereas BA.2.12.1 and BA.4/BA.5 remained more distinct, at 12.9- and 14.5-fold difference (Figure 2E), consistent with the changes in neutralization titers against the variants after the second and third vaccine dose (Figures 3A and 3B). These changes in neutralization patterns and consequent antigenic relationships among the variants suggest that booster vaccination with the ancestral variant selectively boosts titers to some variants more than others in a way that is distinct from neutralization responses after primary infection or two vaccine doses.
Two or three doses with an mRNA COVID-19 vaccine followed by an Omicron PVI increased the neutralization of variants more than three vaccine doses alone
Although there are important differences between vaccination and infection, comparing differences in antibody neutralization of variants by individuals with Omicron PVIs to those with only three vaccine doses may be considered useful for comparing different antigen-boosting strategies. We measured neutralization titers for individuals with PVIs and compared their responses to those with only two or three vaccine doses (Tables S1A and S1E). Individuals with pre-Delta wave PVIs approximately 1 month after two doses of vaccines generally had lower titers against all variants compared with other PVI groups (p < 0.0001) but higher titers than those in the two-dose vaccine group (Figure 3C, p < 0.0001). Individuals with Delta PVIs 3–6 months after two doses of vaccines generally had the highest titers against all variants (Figure 3D). Even if the neutralization titers dropped to the background (<40) after the second vaccine, the Delta PVI boosted high neutralization titers against all variants (Figure 3E).
In agreement with a previous report,46 individuals with two vaccine doses followed by an Omicron (BA.1 or BA.1.1) PVI had high titers against previously circulating variants (GMTs 12,399–5,090) but lower titers against the early Omicron variants BA.1 and BA.2 (GMTs 3,253 and 2,587, respectively) and significantly lower titers for the later variants BA.2.12.1 and BA.4/BA.5 (GMTs 1,486 and 1,009, respectively) relative to BA.1 (p = 0.0009 and < 0.0001, respectively) and BA.2 (p = 0.0946 and 0.0028, respectively), though all but one titer was well above the assay cutoff (Figures 3F and 3G). Individuals with three vaccine doses followed by Omicron (BA.1 or BA.1.1) PVI also had high titers against BA.1 or BA.1.1 (GMTs 4135 and 3842, respectively), followed by BA.2, BA.2.12.1, and BA.4/BA.5 (GMTs 3,237, 1,755, and 1,238, respectively). Unexpectedly, titers after two or three vaccine doses followed by an Omicron PVI were not significantly different (p = 0.5013), suggesting that titers may approach a plateau with the primary vaccination series followed by an Omicron PVI. Overall, individuals who had three vaccine doses and an Omicron PVI had higher titers than those who had three vaccine doses alone (p < 0.0001).
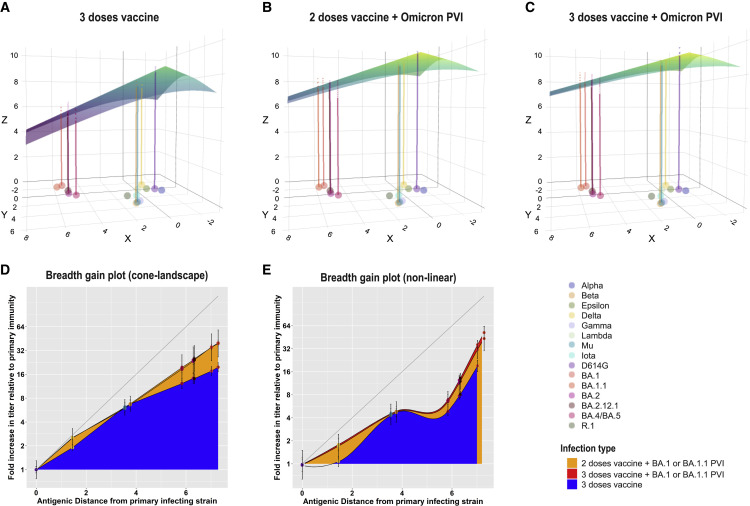
We next compared how neutralization differs across variants for sera following exposures with a single antigen versus sera following exposures with multiple antigens using antibody landscapes. Antibody landscapes can be used to evaluate both the neutralization titer magnitude and the breadth of recognition across the variants. Each serum sample has an associated three-dimensional landscape. The x and y dimensions correspond to the original two-dimensional antigenic map made with primary natural infection sera (i.e., the map in Figure 2A). In the third dimension, at each virus position on the map, the height of the landscape corresponds to the measured neutralization titer for that serum against that virus. A surface, or “landscape,” can then be fit to these data, summarizing how an individual’s neutralization titer varies as a function of the location of the infecting antigen on the two-dimensional antigenic map. Here, we use the method developed by Rössler29 that fits cone-shaped antibody landscapes, where we estimate the peak position and slope from the measured titers. Our validation analyses demonstrated that the height of the antibody landscape accurately represented titer values that were not used in making the landscape, indicating that the landscape can be used to predict titers based on their position in antigenic space (mean RMSE 1.9 versus 2.4 without information on variant antigenic relationships, Mann-Whitney, p ≤.001, further details are provided in the STAR Methods and further results in Table S3A).
We first analyzed whether infection with an antigenically distinct variant, such as BA.1 in those with prior vaccination, induced broader recognition across variants than a third dose with the ancestral strain. The slope of the landscape is a useful metric for evaluating the breadth of neuralization across measured antigens: slopes greater than 1 indicate larger fold differences between variants than after primary infection, whereas smaller slopes correspond to even neutralization across variants. We found individual antibody landscapes from those with two or three vaccine doses followed by Omicron (BA.1 or BA.1.1) PVI had landscapes with significantly more gradual slopes (means: 0.32 and 0.30, respectively), indicating relatively even neutralization titers across variants, than landscapes for individuals who received three doses of ancestral vaccine alone (mean: 0.45, Tables S3B and S3C, one-sided Mann-Whitney test, p < 0.001 and p < 0.002, respectively). Average antibody landscapes for each serum group are shown in Figures 4 A–4C. Notably, individuals with two or three doses of vaccine and Omicron PVIs had a larger gain in titer against Delta as well as Omicron variants BA.1, BA1.1, BA.2, BA.212.1, and BA.4/BA.5 than those with only three doses of vaccine (Tables S3D–S3F). Overall, as we observed in the raw neutralization titer data (Figures 3F–3H), antibody landscapes demonstrated similar breadth and magnitude of neutralization titers across variants between those with two versus three doses prior to Omicron PVI (Figures 4B and 4C). Thus, an additional boost with the ancestral-based vaccine did not appear to substantially improve antibody titer and breadth against the variants in Omicron PVI sera.
Figure 4.
Antibody landscapes and breadth gain plots show that individuals with PVIs have a large increase in both recognition and magnitude compared with those with three mRNA COVID-19 vaccine doses alone
(A–C) Antibody landscapes are shown for individuals with (A) three doses of an mRNA COVID-19 vaccine, (B) two doses of an mRNA COVID-19 vaccine followed by Omicron PVI, and (C) three doses of an mRNA COVID-19 vaccine followed by Omicron PVI. The x and y axis on each landscape correspond to the 2D antigenic map constructed from convalescent sera in Figure 2A, with colored points representing the locations of each measured antigen. The z axis in each landscape represents the interpolated log titer for all individuals with that exposure history against each antigen. The average landscape for each serum group was constructed by fitting landscapes for each individual serum sample assuming that all landscapes with the same infection history have the same slope, with the peak equal to the maximum observed titer value against any one of the measured antigens. The location of the peak titer value was fitted separately for each individual and then subsequently averaged. The colored lines represent the expected average log GMT for individuals with a particular infection history against each measured antigen. The color of the landscape, like the z axis, corresponds to the estimated log GMT across antigenic space.
(D) Breadth gain plots of the antibody landscapes in (A)–(C) for vaccinated individuals who received either a third mRNA COVID-19 vaccine dose, an Omicron PVI, or both. The x axis represents the antigenic distance from the primary convalescent sera antigenic map (Figure 2A) between the primary exposure variant and each measured antigen. Each unit on the y axis represents the gain in neutralization against a particular antigen beyond a primary infection response and is used to compare the relative gain in neutralization under different secondary exposure histories.
(E) Same as (D), but showing neutralization gain for each set of sera with the same infection history estimated directly from the titer data, with an interpolated loess fit to convey trends. Error bars represent the mean and 95% confidence intervals for each measured antigen. Shading colors denote the type of infecting variant and the number of vaccine doses received.
Omicron PVI results in the greatest breadth, although with slightly lower recognition to BA.2.12.1 and BA.4/BA.5
To further evaluate how antibody recognition of variants changed between first and subsequent exposures, we developed a method called the “breadth gain” plot (Figures 4D and 4E; Tables S3D–S3H). The x axis shows the primary antigenic map distances from Figure 2. The y axis shows the average gain in titer to each variant following secondary exposure above what was observed after primary infection. If no increase in recognition is observed against any variant, the plot shows a flat line across the x axis. Boosting that leads to breadth, i.e., similar titers across all variants, results in a straight diagonal line (see Figure S4A for further explanation). This plot aids the interpretation of whether gains in recognition are greatest against antigens that are most distant on the antigenic map. The breadth gain statistic can be directly estimated from primary and secondary titer data (if both are available) and yields similar results, but that analysis loses the comparison to antigenic distances (STAR Methods; Figure S4B; Tables S3I and S3J).
We used this method to quantify and compare the breadth of neutralization across the variants after Omicron PVIs and three doses with the ancestral vaccines. The breadth gain plots corresponding to the landscapes are shown in Figure 4D and estimated directly from the titer data in Figure 4E, with a fitted loess line as a visual guide. In Figure 4E, Omicron PVIs following two or three doses of vaccination provided significantly greater gain in recognition than three vaccine doses alone against BA.1 (one-sided Mann-Whitney: p < 0.003), BA.2 (p<0.04), as well as Delta (p ≪ 0.001, Table S3H). Compared with three vaccine doses alone, three vaccine doses and Omicron PVI induced significantly greater gain in the neutralization of BA.4/BA.5 (p < 0.03), and two and three doses and Omicron PVI induced a greater gain to BA.2.12.1 (p < 0.005). Interestingly, even for those with two and three doses of vaccine and Omicron PVI, the gain in recognition to BA.4/BA.5 and BA.212.1 was slightly lower (Figure 4E: 6.51- and 6.71-fold gain for BA.4/BA.5 and 12.2 and 11.0 for BA.212.1) than expected if breadth increased linearly across antigenic space, as represented by the landscape (Figure 4D: 12.6 and 13.5 for BA.4/BA.5 versus 17.7 and 16.1 for BA.212.1). This analysis suggests that specific epitopes were boosted more than others in a way that only partially correlated with the antigenic relationships observed after primary infection and which was not fully captured by our antibody landscapes.
Discussion
Critical public health decisions about whether and how to update current COVID-19 vaccines are challenging because of the rapid emergence of new SARS-CoV-2 variants, as well as the complex immune histories induced by past antigenic exposures from different waves of infections and vaccination campaigns. When considering vaccine antigens for new COVID-19 vaccines, the potential to elicit high neutralization titers to a broad range of variants is desirable. In this study, we measured neutralization titers against 22 SARS-CoV-2 variants in serum samples from well-characterized clinical cohorts with documented genotyped infections and vaccination histories. We then applied established and new methods for analyzing neutralization titers to quantify and interpret complex antigenic relationships among variants. We find differences in the antibody recognition of the Omicron variants after two and three vaccine doses with ancestral antigens and that Omicron PVI broadens neutralizing antibody responses across the variants similarly after two and three vaccine doses. These findings may have implications for choosing new vaccine antigens.
Moving forward, judgments for vaccine strain selection need to be made using the best available data and models, along with other considerations. Although neutralizing antibody titers against the variants provide critical information, it can be difficult to interpret complex antigenic relationships among many variants in large one-dimensional titer tables and graphs. Antigenic cartography, a method for interpreting antigenic relationships among a set of variants using antibody titers, has proven to be a useful framework for characterizing the antigenic evolution of influenza47 and dengue viruses,48 among other pathogens. Our antigenic maps of pre-Omicron variants using our primary infection sera agree with previously published antigenic maps of SARS-CoV-2,9 , 29 , 30 , 31 , 32 , 33 , 34 likely due in part to the use of similar pseudovirus assays across laboratories. We find strong clustering of the original variants with shared amino acid positions, indicating that specific amino acid changes determine antigenic phenotype. The position of BA.2 on our map agreed with an experimental animal antigenic map32 and a natural infection sera map31 but differed from another natural infection map.29 Our antigenic maps also included BA.2.12.1 and BA.4/BA.5 and showed that they have shorter antigenic distances to the Beta/Gamma/Mu cluster than BA.1 or BA.1.1, which may provide insight into related epitopes among these variants. Our work extends the literature by describing the antigenic positions of BA.1, BA.1.1, BA.2, BA.212.1, and BA.4/BA.5 along with other major variants simultaneously, enabling direct comparison within a single study.
We also evaluated the variants using well-characterized post-vaccination sera to assess how antibody titers and variant recognition after two and three vaccinations differed from primary infection. We found that three vaccinations with the ancestral variant preferentially increased neutralization to some variants more than others. Specifically, the antigenic distance from the ancestral variant to BA.1, BA.2, Beta, and Mu decreased more than the distance to BA.2.12.1, BA.4/BA.5, and Delta. Thus, although BA.1 and BA.1.1 are the most antigenically distinct variants for both primary infection and two-dose vaccine sera, the three-dose vaccine sera recognized BA.1 and BA.1.1 better than BA.4/BA.5 and BA.2.12.1. We note, however, that titers to all Omicron variants were still high after the third vaccine dose. A previous study found that additional mutations in BA.2.12.1 (L452Q) and BA.4/BA.5 (L452R, F486V, and the deletion in 69 and 70) help explain antigenic differences relative to BA.2 for three-dose vaccinee sera.4 However, that study only investigated Omicron variants and D614G using only boosted vaccinee sera. The specific mutations that explain why antigenic distances measured in sera following primary infection or two vaccine doses differ from those following three vaccine doses remain to be explained.
Importantly, the magnitude of antibody titer to the circulating variants at the time of infection likely impacts the degree of protection. We also compared the effects of the number of vaccine doses and Omicron PVIs on both neutralization titer magnitude and the breadth of neutralization across the variants. In a direct analysis of the titer data, we found that two or three doses of the ancestral vaccine followed by an Omicron PVI induced higher neutralization titers against more variants than three vaccine doses alone (p < 0.0001). This suggests that individuals who already received a third vaccine dose with the ancestral strain could still potentially broaden their immunity by receiving an Omicron vaccine. However, neutralization titers against the panel of variants were similar for those with two or three vaccine doses followed by an Omicron PVI, at least in the short-term following vaccination.
We further analyzed these data using a technique called an antibody landscape, which graphs the magnitude of titers across antigenic space as defined by the primary infection sera. This analysis can help predict titers to variants on an antigenic map that are not directly tested. Relative to individuals vaccinated with only the ancestral variant, individuals with two or three vaccine doses followed by Omicron PVIs had flatter landscapes and hence broader neutralization across all variants, including to BA.2.12.1 and BA.4/BA.5 and Delta. Because Delta is in the lower part of the antigenic map, distant from the Omicron variants, this analysis shows that re-exposure with an antigenically distinct variant may provide a stronger boost in cross-neutralizing antibodies to conserved epitopes, even against unexposed, antigenically distinct variants, than boosting with the ancestral strain. This observation may be useful in considerations for updating vaccine antigen composition and demonstrates how antigenic cartography and landscapes can be used to inform variant selection.
To evaluate whether Omicron PVI versus vaccination alone differentially boosted neutralization for certain variants and induced weaker-than-expected responses against other variants, we developed a new metric called the breadth gain plot. These analyses compare gain in antigen recognition between primary and subsequent exposure. We observed that the fold change in neutralization titers increased most the further the antigenic distance a variant was from the primary strain (in our analyses, this was always the ancestral vaccine strain). This finding is consistent with a broadening of titer responses, as illustrated in Figure 4D. Notably, however, breadth was not completely uniform across antigenic space. We find that for all vaccinated and PVI groups, the gain in titer was smaller to BA.2.12.1 and BA.4/BA.5 than expected based on their antigenic distance from the ancestral strain (Figure 4E). This finding matches our antigenic analyses showing that three vaccine doses preferentially boosted titers to BA.1 and BA.2 more than BA.2.12.1 or BA.4/BA.5. Nonetheless, the absolute magnitude of titers in both groups to BA.2.12.1 and BA.4/BA.5 was still high (GMT > 1,000), a level that has been reported to be associated with vaccine protection of 96%.49 , 50 However, a correlate of protection against BA.4/BA.5 has not yet been reported, and correlates of protection for vaccine approval purposes have not yet been established for any variant. For this reason, methods that enable the evaluation of relative differences in titer magnitude between one variant or another may be useful.
Collectively, our results point to a complex immunodominance pattern in which responses are boosted (whether by repeated ancestral vaccination or Omicron PVI) against epitopes that present to a lesser degree on BA2.12.1 and BA.4/BA.5 compared with BA.1. The uneven boosting among the variants may indicate lack of shared epitopes in some variants. Alternatively, it may indicate immune imprinting by the first antigenic exposure, as has been observed for influenza.51 For example, during the 2013–2014 flu season, the H1N1 virus infected large numbers of middle-aged adults. Subsequent analyses showed that several mutations in the virus occurred at epitopes that were targeted by antibodies in middle-aged adults that were likely elicited by prior antigenic exposures. Thus, although infection or vaccination with BA.1 could increase immunity to current Omicron variants, whether the use of BA.1 as a first Omicron antigenic exposure could affect subsequent recognition of other Omicron or future variants remains to be studied.
Overall, our results show how the antigenic co-evolution of the SARS-CoV-2 and its immune response among the host human population become more elaborate with time. We show that Omicron PVIs generally induce broader immunity than boosting with the ancestral vaccine and additional exposure to both ancestral and BA.1 antigens can increase neutralization across all variants, although to a lesser extent to BA.4/BA.5. Understanding the mechanism behind this differential boosting to distinct variants and carefully quantifying immune recognition may become increasingly important for developing vaccination strategies against future SARS-CoV-2 variants. Further, scientific insights into the evolution of antigenic diversity for SARS-COV-2 could also shed light on how other antigenically complex pathogens may have evolved.
Limitations of the study
Our study used convalescent serum samples that were collected at different time points post-COVID-19 diagnosis (2–59 days), which could have affected measured antibody magnitude and breadth to the variants. However, when we removed all samples reported as <6 days post symptom onset, it had minimal effects on the antigenic map (Figure S3D). To maximize serum coverage of variants on the antigenic map, we included ten samples that were not fully genotyped but assigned variant infections inferred by known dates of dominant circulating variants at the time of sample collection. Ideally, antibody landscapes are constructed by fitting interpolated surfaces across antigenic space as described by Fonville,28 but there are not yet enough distinct variants for this method. The emergence of future variants, titration with additional subvariants, or generation of mutant pseudoviruses that probe unoccupied areas of antigenic space may make more comprehensive antibody landscape analyses possible. There were too few individuals with PVIs with variants other than Omicron to evaluate statistical significance with landscapes and breadth gain plots. Future studies on larger numbers of individuals or samples from clinical trials will provide further information on how sequential exposure to distinct antigens covers antigenic space. Finally, although neutralization titers measured with pseudovirus neutralization assays have been correlated with protection, our study does not directly provide information on protection against variants. Further studies incorporating antibody landscapes with disease outcome data will provide further insights into how immune breadth across antigenic space is associated with clinical protection.
STAR★Methods
Key resources table
| REAGENT or RESORCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| SARS-CoV-2 pseudoviruses for variants | (Lusvarghi et al.9; Neerukonda et al.34,52), this manuscript | N/A |
| Virus spike sequence data | This manuscript | Tables S1B and S1D |
| Biological sample | ||
| Convalescent serum samples | This manuscript | Table S1A |
| Vaccinated, not boosted, serum samples | This manuscript | Table S1E |
| Vaccinated, boosted, serum samples | This manuscript | Table S1E |
| Vaccinated, not boosted, PVI, serum samples | This manuscript | Table S1A |
| Vaccinated, boosted, PVI, serum samples | This manuscript | Table S1A |
| Commercial convalescent serum samples | Boca Biolistics (Pompano Beach, FL), this manuscript | Table S1C |
| Critical commercial assays | ||
| Luciferase Assay System | Promega | Cat # E4550 |
| Experimental models: Cell lines | ||
| 293T-ACE2-TMPRSS2 | Neerukonda et al.52, BEI | Cat # NR-55293 |
| 293T/17 | ATCC | Cat # CRL-11268 |
| Recombinant DNA | ||
| pCMVΔR8.2 | VRC, USA | N/A |
| pHR’CMV-Luc | VRC, USA | N/A |
| SARS-CoV-2 variants spike plasmids | Lusvarghi et al.9; Neerukonda et al.34,52, this manuscript | Table S2 |
| Software and algorithms | ||
| Racmacs package for antigenic cartography | GitHub | https://acorg.github.io/Racmacs/ |
| R package stats | Stats-package | https://rdrr.io/r/stats/stats-package.html |
| GraphPad Prism 9 | GraphPad | https://www.graphpad.com |
| Antibody landscapes | Rössler et al.29, this manuscript | https://github.com/acorg/roessler_netzl_et_al2022 |
| Breadth Gain Plots | This manuscript | Zenodo: https://doi.org/10.5281/zenodo.7291034 |
| Deposited data | ||
| Neutralization data | This study | Zenodo: https://doi.org/10.5281/zenodo.7291034 |
| Supplementary data | This study | Zenodo: https://doi.org/10.5281/zenodo.7291034 |
Resource availability
Lead contact
Further information and reasonable requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Carol D. Weiss (carol.weiss@fda.hhs.gov).
Materials availability
All unique plasmids generated in this study are available with an MTA from the lead contact. De-identified serum samples from the PASS, EPICC, FDA, and HVTN protocols used in this study are subject to a materials transfer agreement and sample availability. The commercial serum samples can be purchased from the suppliers as listed in the key resources table.
Experimental model and subject details
Ethics
The PASS (Protocol IDCRP-126) and EPICC (Protocol IDCRP-085) studies were approved by the Uniformed Services University of the Health Sciences Institutional Review Board (IRB) in compliance with all applicable Federal regulations governing the protection of human participants. All PASS and EPICC study participants provided informed consent. The convalescent Beta sera, obtained from a traveler who had moderate-severe COVID-19 in the Republic of South Africa during the peak of the Beta (B.1.351) wave in January 2021, was obtained with informed consent and covered under the U.S. Food and Drug Administration IRB approved expedited protocol (# 2021-CBER-045). Six additional convalescent Beta serum samples were obtained from the HVTN under an approval protocol (237-EXS_Weiss_CoVPN5001), in which participants gave consent for future use of their specimens.
Gender of subjects
The genders of human subjects in this study were mixed. The influence of gender on the results of the study was not explicitly measured.
Post-vaccination sera from the PASS study
Details of the Prospective Assessment of SARS-CoV-2 Seroconversion (PASS) study protocol, including details of the inclusion/exclusion criteria, have been previously published.53 Inclusion criteria included being generally healthy, ≥ 18 years old, and employed at the Walter Reed National Military Medical Center (WRNMMC), Bethesda as a healthcare worker. Exclusion criteria included history of COVID-19, IgG seropositivity for SARS-CoV-2 (as determined by a binding antibody assay) and being severely immunocompromised at time of screening. The study was initiated in August 2020, with rolling enrollment and monthly research clinic visits to obtain serum for longitudinal SARS-CoV-2 antibody testing.
The subset of PASS uninfected vaccinee participants selected for analysis of sero-responses were those who received two doses of Pfizer/BNT162b2 vaccine by January 26, 2021, had no serological or PCR evidence of SARS-CoV-2 infection prior to two doses of vaccine, and had received a third dose of Pfizer/BNT162b2 vaccine by Nov 18, 2021 (see Table S1E). No subject included in this sub-analysis of vaccinated participants had a clinically apparent PCR-confirmed SARS-CoV-2 infection during follow-up before sera collection. Participants' serum samples were collected monthly through September of 2021, and then quarterly.
For the antibody binding assay used for screening at enrollment, serum samples were diluted 1:400 and 1:8000 and screened for immunoglobulin G (IgG) reactivity with SARS-CoV-2 spike protein and nucleocapsid protein (N), and four human coronavirus (HCoV) spike proteins using a multiplex microsphere-based immunoassay, as previously described.54
Post-infection sera from the EPICC study
The Epidemiology, Immunology, and Clinical Characteristics of Emerging Infectious Diseases with Pandemic Potential (EPICC) study is a cohort study of U.S. Military Health System (MHS) beneficiaries that includes enrollment and longitudinal follow up of those with a history of SARS-CoV-2 infection.55 Eligibility criteria for enrollment included presenting to clinical care with COVID-19-like illness and being tested for SARS-CoV-2 by polymerase chain reaction (PCR) assay (See Tables S1A and S1B). The EPICC study enrolled between March 2020 and May 2022. For this analysis derived from SARS-CoV-2 infections, EPICC enrollment occurred at eight Military Treatment Facilities (MTFs): Brooke Army Medical Center, Fort Belvoir Community Hospital, Madigan Army Medical Center, Naval Medical Center Portsmouth, Naval Medical Center San Diego, Tripler Army Medical Center, Walter Reed National Military Medical Center, and William Beaumont Army Medical Center.
Study procedures for these participants with SARS-CoV-2 infection included collection of demographic data, and completion of a clinical case report form (CRF) to characterize the acute SARS-CoV-2 infection. Biospecimen collection included serial serum samples for immune response analysis and upper respiratory specimen swabs for genotyping of SARS-CoV-2. For all enrolled participants, we also abstracted MHS-wide healthcare encounter data from the Military Health System Data Repository (MDR) to determine comorbidities. Vaccination status was ascertained by the MDR record, the CRF and self-reported questionnaire.
In addition to convalescent sera from EPICC participants, we included convalescent sera from two PASS participants with SARS-CoV-2 infection in August 2021 (during the Delta epidemic). Both participants were vaccinated with two doses of mRNA COVID-19 vaccine at the time of SARS-CoV-2 infection (Table S1A).
Post-Beta variant infection sera
Six serum samples were collection from adults following a diagnosis of COVID-19 during a period when the Beta variant was dominant (Table S1A). These infections were not genotyped but were presumed to be due to the Beta variant. One convalescent serum sample was collected under the FDA protocol (# 2021-CBER-045) from an unvaccinated adult who was diagnosed with COVID-19 by PCR during travel to the Republic of South Africa during the SARS-CoV-2 Beta infection wave in January 2021. Five additional serum samples were collected under the HVTN protocol (237-EXS_Weiss_CoVPN5001) from unvaccinated adults following a COVID-19 diagnosis during February–March 2021 in the Republic of South Africa.
Commercial post-infection sera
Convalescent sera from SARS-COV-2 infected donors were purchased from Boca Biolistics (Pompano Beach, FL). Samples were selected from the SARS-CoV-2 sequence inventory for variant infections that were not represented in the samples from the other protocols. Details about the serum donors are listed in Table S1C. Genotypes of the infecting viruses are listed in Table S1D.
Cell lines
293T/17 (ATCC CRL-11268) and 293T-ACE2-TMPRSS2 cells stably expressing human angiotensin-converting enzyme 2 (ACE2) and transmembrane serine protease 2 (TMPRSS2) (BEI Resources, Manassas, VA, USA; Cat no: NR-55293)52 were maintained at 37˚C in Dulbecco’s modified eagle medium (DMEM) supplemented with high glucose, L-glutamine, minimal essential media (MEM) non-essential amino acids, penicillin/streptomycin, HEPES, and 10% fetal bovine serum (FBS).
Plasmids
Codon-optimized, full-length open reading frames of the spike genes of SARS-CoV-2 variants in the study (Table S2) were synthesized into pVRC8400 or pcDNA3.1(+) by GenScript (Piscataway, NJ, USA). The HIV gag/pol packaging (pCMVΔR8.2) and firefly luciferase encoding transfer vector (pHR’CMV-Luc) plasmids56 , 57 were obtained from the Vaccine Research Center (VRC, National Institutes of Health, Bethesda, MD, USA).
Pseudoviruses generation
HIV-based lentiviral pseudoviruses with desired SARS-CoV-2 spike proteins were generated as previously described.52 Pseudoviruses comprising the spike glycoprotein and a firefly luciferase (FLuc) reporter gene packaged within HIV capsid were produced in 293T cells by co-transfection of 5 μg of pCMVΔR8.2, 5 μg of pHR’CMVLuc and 0.5 μg of pVRC8400 or 4 μg of pcDNA3.1(+) encoding a codon-optimized spike gene. Pseudovirus supernatants were collected approximately 48 h post transfection, filtered through a 0.45 μm low protein binding filter, and stored at -80˚C.
Method details
COVID-19 diagnosis and SARS-CoV-2 genotype
For EPICC participants, SARS-CoV-2 infection was determined by positive PCR clinical laboratory test performed at the enrolling clinical MTF site, or a follow-up upper respiratory swab collected as part of the EPICC study procedures. The specific PCR assay used at the MTF varied. The SARS-CoV-2 (2019-nCoV) CDC qPCR Probe Assay research-use-only kits (Integrated DNA Technologies, IDT, Coralville, IA) was used as the follow-up PCR assay (used for specimens collected as part of the EPICC study). This CDC qPCR assay uses two targets of the SARS-CoV-2 nucleocapsid (N) gene (N1 and N2), with an additional human RNase P gene (RP) control. We considered a positive SARS-CoV-2 infection as positive based on a cycle threshold value of less than 40 for both N1/N2 gene targets.
Whole viral genome sequencing was performed on extracted SARS-CoV-2 RNA from PCR positive specimens using a 1200bp amplicon tiling strategy.58 Amplified product was prepared for sequencing using NexteraXT library kits (Illumina Inc., San Diego, CA) and libraries were run on the Illumina NextSeq 550 sequencing platform. Genome assembly used BBMap v. 38.86 and iVar v. 1.2.2 tools. The Pango classification tool (version 4.0.6) was used for lineage classification.59 In a small minority of SARS-CoV-2 infections (Table S1B), a Pangolin lineage was unable to be ascertained and either a Nextclade clade59 was used and/or a Pangolin lineage was inferred by manual inspection of key lineage-defining amino acid substitutions. Dates of infection were also used as supplementary information to ascertain infecting genotype in such instances where spike sequence quality was lower.
Additionally, we included 10 convalescent sera with infecting genotype inferred by date of collection. Convalescent sera from seven vaccinated EPICC participants diagnosed with COVID-19 between 2/9/2021 and 4/2/2021 did not have corresponding viral sequence data to confirm the infecting genotype and were categorized as presumptive “pre-Delta” infections based on historical U.S. variant distributions (Figure 3C; Table S1A).6 The infecting genotype of two PASS participants with vaccine breakthrough infections were inferred by date of infection (late August 2021, annotated as presumptive Delta infections, Figures 3D and 3E; Table S1A). Additionally, we included in all analyses sera from a traveler who had COVID-19 in the Republic of South Africa during the peak of the Beta (B.1.351) wave in January 2021 (collected under a separate CBER protocol, 2021-CBER-045) (Table S1A) and this was annotated as a presumptive Beta infection.
Pseudovirus Neutralization assay
Neutralization assays were performed using 293T-ACE2-TMPRSS2 cells in 96-well plates as previously described.52 Pseudoviruses with titers of approximately 106 relative luminescence units per milliliter (RLU/mL) of luciferase activity were incubated with serially diluted sera for two hours at 37˚C prior to inoculation onto the plates that were pre-seeded one day earlier with 3.0 × 104 cells/well. Pseudovirus infectivity was determined 48 h post inoculation for luciferase activity by luciferase assay reagent (Promega) according to the manufacturer’s instructions. The inverse of the sera dilutions causing a 50% reduction of RLU compared to control was reported as the neutralization titer (ID50). Titers were calculated using a nonlinear regression curve fit (GraphPad Prism Software Inc., La Jolla, CA, USA). The mean titer from at least two independent experiments each with intra-assay duplicates was reported as the final titer.
Quantification and statistical analysis
Neutralization titer analysis
One-way analysis of variance (ANOVA) with Dunnett’s multiple comparisons tests (variants compared to D614G-, variants compared to BA.1), two-way ANOVA for the comparison of different groups (i.e., two-dose vaccine vs three-dose vaccine) and geometric mean titers (GMT) with 95% confidence intervals were performed using GraphPad Prism software. The p values of less than 0.05 were considered statistically significant. All neutralization titers were log2 transformed for analyses.
Antigenic cartography
We used the Racmacs package (https://acorg.github.io/Racmacs/) for antigenic cartography analyses.30 Antigenic maps are quantitative visualizations that fit antibody titers as Euclidean distances between primary infection sera and variants. Datasets with diverse variants and primary infection sera to each variant are best for making meaningful geometric interpretations, i.e., antigenic maps. Racmacs implements a modified multi-dimensional scaling approach as previously described.27 Briefly, the virus best neutralized by each serum j, is defined as b j. N ij is the neutralization titer for serum j against virus i. The antigenic distance, D ij, for serum j to each virus i is defined relative to b j: D ij =log2(b j)-log2(N ij). The map Euclidean distance d ij for each virus and serum is that which best fits the measured antigenic distance D ij in each number of dimensions. The optimal set of map coordinates for each serum and virus is identified by minimizing the stress function E=∑ije(D ij, d ij) thousands of times from random starting coordinates using a conjugate gradient optimization. For titers above the assay lower limit of quantitation, the stress function minimized is (D ij - d ij).2 For titers below the assay lower limit of quantitation, the stress function minimized is (D ij - d ij -1)2g(D ij - d ij - 1), where A unit of antigenic distance is equivalent to a two-fold dilution in neutralization titers.
We performed various quality assessments for antigenic maps including evaluation of goodness of fit and error, cross-validation, dimensionality testing (Figure S1), confidence in coordination on the map, and robustness of the map to assay error and outlier viruses and sera (Figure S2). The average absolute difference between a measured titer and its estimated map distance was only 0.50 map units, which corresponds to less than a 2-fold titer difference. Using cross-validation methods in which we made distinct antigenic maps using 90% of titers (training set) to predict the excluded 10% titers (test set), the average absolute difference between the measured titer and the predicted map distance was 1.21 map units (2.3-fold titer difference, Table S3K). In contrast, the average absolute difference between each titer and the geometric mean for the corresponding serum group and variant (as shown in Figure 1) was 1.45 map units (2.7-fold titer difference). Thus, the antigenic map is significantly more accurate at representing and predicting individual neutralization titers than can be represented by geometric mean titers alone (t-test, means of 1.21 and 1.45 antigenic units, p < 2.2e-16). We performed dimensionality testing to confirm that two dimensions were sufficient to accurately fit the data; additional goodness of fit tests and 3D maps are shown in Figure S1. Points on the antigenic map were well coordinated and robust to measurement error in the assay as well as bootstrapping of individual viruses and sera (Figure S2).
Antibody landscapes
We generated antibody landscapes for all serum samples with same overall infection history (e.g. three doses of ancestral vaccine, three doses of ancestral + Omicron PVI etc.), following the general approach of Roessler and Netzel et al.,29 using code from the repository associated with that manuscript (https://github.com/acorg/roessler_netzl_et_al2022) accessed on May 22, 2022. We fitted three parameters for each landscape: the x and y coordinates of the landscape peak, and , and the landscape slope, . We first assume that all serum samples within the same serum group have the same slope. Fitted values for those parameters for each landscape are listed in Table S3D. Let represent the column basis titer, i.e. the highest titer for each serum sample j. We assume that for each serum sample, the height of the landscape at the peak is equal to column base titer of the serum. Let represent the antigenic distance between the peak and a particular measured antigen i. The predicted titer against measured antigen i for serum sample j is given by:
Let denote the observed titer for serum sample j against measured antigen i. We use the function optim() in the R package stats to minimize the square error E, which is the sum of the difference between observed and predicted titers across all measured antigens and serum samples within the serum group:
We hypothesized that re-exposure (vaccine and/or PVI) would increase neutralization breadth among variants in a way that is correlated with antigenic distances among variants. To test this hypothesis, we generated 100 bootstrap replicates in which 90% of serum-variant pairs were selected at random to be included in the training set, with the remaining 10% allocated to the test set. For each bootstrap replicate, we fit a landscape using the training set and calculated the RMSE for the 10% of the pairs in the test set comparing the measured titers with the predicted titers from the fitted landscape. As a comparison, we estimated the RMSE that would be observed if each predicted serum-variant pair were randomly assigned a variant position on the antigenic map, i.e. if we lacked information on antigenic distances. Given that there is already broad recognition across variants in our datasets, even the ‘shuffled’ landscape only had an average RMSE of 2.4 antigenic units (equivalent to 5.3-fold error) but a wide distribution of error. In contrast, the model-fitted landscapes had an error of less than 1.9 antigenic units (∼3.7 fold) indicating a better fit (see Table S3A). The distributions are significantly different (Mann-Whitney, p = < 2e-16, mean difference: 0.55 antigenic units). This result indicates that the landscapes are informative, as antigenic distances measured in the primary infection map correlated with variant recognition after multiple exposures.
We next quantify whether individuals with Omicron PVI have significantly broader landscapes (i.e. landscapes with a gentler slope) than individuals with only three doses of ancestral vaccination. To conduct this comparison and obtain confidence intervals for landscape slopes, we perform a second set of landscape fits, this time allowing each individual serum sample to have a separate slope. The results of those fits including slopes and peak locations for each landscape are shown in Table S3L.
We then calculate the mean and standard error for the landscape slope across all serum samples with the same exposure history. We obtain confidence intervals for the landscape slope by adding/subtracting twice the standard error from the mean. Those summary statistics are shown in Table S3B. We performed a one-sided Mann-Whitney test using individual slopes grouped by exposure history, with the alternate hypothesis that two or three dose Omicron PVIs have smaller slopes than three doses with the ancestral vaccine. Those results can be found in Table S3C.
Breadth gain plots
We next develop a metric, the “breadth gain”, that can be used to quantify the relative fold gain in neutralization against a particular measured antigen from sera following a secondary exposure relative to the response against that antigen that would be expected following primary exposure when both groups had the same primary exposure antigen.
The breadth gain metric is a complementary approach to the antibody landscape that avoids the assumptions made when fitting a cone-like landscape. The breadth gain against a particular measured antigen requires only the observed antibody titers from secondary exposure sera against that antigen, and a 2D antigenic map previously generated from primary convalescent sera.
Below we describe how the breadth gain can be computed from secondary exposure titer data and a primary infection antigenic map (such as the one shown in Figure 2A).
Let t(s,a) represent the log titer of serum s against each antigen a. Let k(s) represent the peak log titer for serum s across all antigens a. Let d(s,a, k(s)) denote the "drop" in log titer for serum s against antigen a with respect to the peak log titer k(s). Mathematically, this is equal to the log titer of serum s against each antigen a:
Multiple sera can have the same exposure history H. For example, these could be sera that have all had two doses of ancestral strain vaccination followed by a BA.1 breakthrough infection. Let represent the average drop in log titer against antigen a relative to the peak titer for each serum, averaging across all S sera with infection history H. We calculate the mean by averaging over all sera with the same exposure history:
Just as d(H,a) represents the average drop in log titer against antigen a relative to the peak (d(s,a, k(s))) averaged across all sera with the same exposure history, let represent the lower confidence interval bound of d(s,a, k(s)), and the upper confidence interval bound, which are obtained by adding/subtracting two times the standard error of d(s,a, k(s)) from the mean, d(H,a).
Let represent the average drop in log titer against antigen a among individuals whose exposure history is only a primary wild type infection. We estimate this quantity by assuming that it is equal to the antigenic distance between the ancestral strain and the measured antigen a on the convalescent sera map (i.e., the map Figure 2A). Mathematically:
where is the antigenic distance between antigen a and the ancestral strain in the convalescent sera antigenic map.
Let g(H,a) denote the average recognition gain for all sera with exposure history H against antigen a. This represents the difference between the average drop in log titer against antigen a for all serum samples with common exposure history H () compared to individuals whose only exposure is a primary ancestral infection .
For the 95 percent confidence interval bounds, we again have:
We then convert this average gain in recognition to units of fold-titer difference. Let f(H,a) represent the average gain in recognition from exposure history H against antigen a in terms of the fold change in the titer, where a fold change of 1 represents no gain in recognition with respect to primary ancestral infection. Mathematically:
We also convert the quantiles to the same units:
These quantiles correspond to the points and error bars in the recognition gain plot in Figure 4E and printed in Table S3G. The lines shown in Figure 4E are interpolated Loess curves connecting the measured antigens. These lines are included for visualization purposes. Loess fits were conducted using the loess() function in the R package stats with the default span setting of 0.75, and antigenic map coordinates were extracted using the R package Racmacs. All analyses for the breadth gain plots were performed using the statistical software R version 4.2.0.60
Statistical tests of gain
We use a one-sided Mann-Whitney test (the wilcox.test function in the R package stats) to determine if the individuals with exposure histories H with two or three-dose omicron PVI have significantly larger fold recognition gains against antigen a than exposure histories with only ancestral vaccination. We calculate the recognition gain separately for each secondary exposure serum sample s:
We then convert to fold units:
We then compare the distributions of the fold recognition gain f(s, a, k(s)), grouping by secondary exposure history H and measured antigen a. The results where breadth gain is estimated from the titer data is shown in Table S3H.
Empirical neutralization gain calculation
We validate the breadth gain analysis by constructing a completely empirical version of the metric that does not rely on cartography and is calculated directly from titer titer data for both primary and secondary responses. In the main methods, the average drop in log titer against antigen a among individuals whose exposure history is only a primary ancestral infection was estimated from the 2D antigenic map of convalescent primary infection sera (Figure 2A). This quantity is the only component of the breadth gain metric that utilized cartography. In this study, because we measure titers in individuals with ancestral primary infection, we can measure this quantity directly from the data. A plot of the fold neutralization gain (a completely empirical version of Figure 4E) is shown in Figure S4B, and in Table S3I. We again used a one-sided Mann-Whitney test to assess whether individuals with two or three dose Omicron PVI had larger neutralization gains against measured antigens compared to individuals with only 3 doses of wild type vaccination. Those results are shown in Table S3J.
Breadth gain plots for antibody landscapes
We also calculate the neutralization gain using the secondary exposure titers inferred by the fitted landscape (rather than using the observed secondary titers). For these calculations, we use the landscapes fitted earlier, which have shared slopes for all serum samples with the same infection history. However, the landscape corresponding to each serum sample has an individual peak location and height. The peak height is fixed to be equal to the highest observed titer for that serum sample (k(s)). The x and y-coordinates of the peak, however, are fit during the inference of the antibody landscape. Let the points (xLAND, yLAND) represent those fitted values for the location of the landscape peak. Furthermore, just as t(s,a) represents the observed log titer for serum s against antigen a, let l(s,a) represent the titer against antigen a inferred from the fitted landscape for serum s.
Based on the way landscapes are fitted, the location of the peak is allowed to be offset from the location of the observed titer peak. We adjust for this offset when calculating the neutralization gain from the landscape. For all the secondary infection serum groups in Figure 4, the serum samples had their highest titers against the ancestral strain (out of all antigens measured with those sera). We use this observation to calculate an offset term b H which represents the average drop in log titer (across all serum landscapes with the same exposure history H) between the peak titer of the fitted landscape and the predicted titer from the landscape against the ancestral strain. We can express this quantity mathematically as:
If the location of the peak of the fitted landscape was identical to the location of the peak of the observed data (the ancestral strain), the offset term would be equal to 0 since the log titer of the fitted landscape peak would be equal to the log titer against the ancestral strain.
We follow the same procedure used to generate the original breadth gain plot, except that we replace the observed titer for serum s against antigen a, (, with the estimated titer against antigen a from the fitted landscape for serum s, . As before, we calculate the drop in log titer d(s,a, k(s)) for serum s against antigen a relative to the peak titer k(s):
We then calculate the gain as before. However, here we add in the offset term:
For the points and confidence intervals in Figure 4D, we first calculate the mean and standard error of the neutralization gains for all serum samples s with the same exposure history H. For example:
We construct confidence intervals corresponding to the mean +/- 2 times the standard error, and then convert the mean and confidence interval bounds into units of fold neutralization gain. For example,
The mean and confidence intervals in units of fold neutralization gain are plotted in Figure 4D and are listed in Table S3E.
We likewise again perform one-sided Mann Whitney tests to assess whether omicron PVI have larger fold gains in neutralization (when estimated from the fitted antibody landscapes) than individuals with only three doses of ancestral vaccination. For those calculations, we convert the gain into units of fold neutralization gain and then group by secondary exposure history H and measured antigen a. For example:
Results of those tests can be found in Table S3F.
Datasets
Datasets are available at Zenodo: https://doi.org/10.5281/zenodo.7291034.
Acknowledgments
We sincerely thank members of the EPICC COVID-19 Cohort Study Group for their contributions to conducting the study and ensuring effective protocol operations. The authors also acknowledge all who contributed to the EPICC COVID-19 study: The Brooke Army Medical Center: J. Cowden, M. Darling, S. DeLeon, D. Lindholm, A. Markelz, K. Mende, S. Merritt, T. Merritt, N. Turner, and T. Wellington; the Carl R. Darnall Army Medical Center: S. Bazan, D. Hrncir, and P.K Love; the Fort Belvoir Community Hospital: N. Dimascio-Johnson, E. Ewers, K. Gallagher, C. Glinn, U. Jarral, D Jennings, D. Larson, A. Mentzos, K Reterstoff, A. Rutt, A. Silva, and C. West; the Henry M. Jackson Foundation: P. Blair, J. Chenoweth, and D. Clark; the Madigan Army Medical Center: J. Bowman, S. Chambers, C. Colombo, R. Colombo, C. Conlon, K. Everson, P. Faestel, T. Ferguson, L. Gordon, S. Grogan, S. Lis; M. Martin, C. Mount, D. Musfeldt, D. Odineal, M. Perreault, W. Robb-McGrath, R. Sainato, C. Schofield, C. Skinner, M. Stein, M. Switzer, M. Timlin, and J S. Wood; the Naval Medical Center Portsmouth: S. Banks, R. Carpenter, L. Kim, K. Kronmann, T. Lalani, T. Lee, A. Smith, R. Smith, R. Tant, and T. Warkentien; the Naval Medical Center San Diego: C. Berjohn, S. Cammarata, N. Kirkland, D. Libraty, R. Maves, and G. Utz; the Tripler Army Medical Center: C. Bradley, S. Chi, R. Flanagan, A. Fuentes, M. Jones, N. Leslie, C. Lucas, C. Madar, K. Miyasato, and C. Uyehara; the Uniformed Services University of the Health Sciences: H. Adams, B. Agan, L. Andronescu, A. Austin, C. Broder, T. Burgess, C. Byrne, K Chung, J. Davies, C. English, N. Epsi, C. Fox, M. Fritschlanski, M. Grother, A. Hadley, P. Hickey, E. Laing, C. Lanteri, J. Livezey, A. Malloy, R. Mohammed, C. Morales, L. Nevo, P. Nwachukwu, C. Olsen, E. Parmelee, S. Pollett, S. Richard, J. Rozman, J. Rusiecki, E. Samuels, M. Sanchez, A. Scher, M. Simons, A. Snow, K. Telu, D. Tribble, M. Tso, L. Ulomi, and M. Wayman; the U.S. Air Force School of Aerospace Medicine: TSgt T. Chao, R. Chapleau, M. Christian, A. Fries, C. Harrington, V. Hogan, S. Huntsberger, K. Lanter, E. Macias, J. Meyer, S. Purves, K. Reynolds, J. Rodriguez, and C. Starr; the U.S. Army Medical Research Institute of Infectious Diseases: J Kugelman; U.S. Coast Guard: J. Iskander and I. Kamara; the Womack Army Medical Center: B. Barton, D. Hostler, J. Hostler, K. Lago, C. Maldonado, and J. Mehrer, the William Beaumont Army Medical Center: T. Hunter, J. Mejia, R. Mody, J. Montes, R. Resendez, and P. Sandoval; the Walter Reed National Military Medical Center: I. Barahona, A. Baya, A. Ganesan, N. Huprikar, and B. Johnson; and the Walter Reed Army Institute of Research: S. Peel. The authors wish to also acknowledge the following individuals for their contributions to the PASS (IDCRP-126) COVID-19 study. The Henry M. Jackson Foundation: Alyssa Lindrose, Matthew Moser, Emily C. Samuels, Belinda Jackson-Thompson, Julian Davies, Luca Illinik, Mimi Sanchez, Orlando Ortega, and Edward Parmelee. NMRC-CTC (Naval Medical Research Center-Clinical Trials Center): Santina E. Maiolatesi. Christopher A. Duplessis. NMRC-CTC/General Dynamics. Kathleen F. Ramsey, Anatalio E. Reyes, Yolanda Alcorta, and Mimi A. Wong. We also thank the HVTN and CoVPN for use of samples from the CoVPN trial. Financial support: This work was supported by institutional research funds from the U.S. Food and Drug Administration (FDA), the Intramural Research Program of the National Institute of Allergy and Infectious Diseases (NIAID) of the National Institutes of Health (NIH), and awards from NIAID/NIH through an interagency agreement (AAI21013-001-00000) with Center for Biologics Evaluation and Research, FDA, as part of the U.S. Government COVID-19 response efforts, and the Defense Health Program (HU00012020067, HU00012020094, HU00012120104) and the NIAID (HU00011920111). The protocol was executed by the Infectious Disease Clinical Research Program (IDCRP), a Department of Defense (DoD) program executed by the Uniformed Services University of the Health Sciences (USU) through a cooperative agreement by the Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc. (HJF). This project has been funded in part by NIAID under an interagency agreement (Y1-AI-5072). R.S. was supported by an appointment to the NIAID Emerging Leaders in Data Science Research Participation Program. This work also used samples funded in part with federal funds from the U.S. Department of Health and Human Services, Office of the Assistant Secretary for Preparedness, and the Response, Biomedical Advanced Research & Development Authority.
Disclaimers: The contents of this publication are the sole responsibility of the author(s) and do not necessarily reflect the views, opinions, or policies of Uniformed Services University of the Health Sciences (USU); the Department of Defense (DoD); the Departments of the Army, Navy, or Air Force; the Defense Health Agency, or the Henry M. Jackson Foundation for the Advancement of Military Medicine Inc. The mention of trade names, commercial products, or organizations does not imply an endorsement by the U.S. Government. The investigators have adhered to the policies for the protection of human subjects as prescribed in 45 CFR 46. The Emerging Leaders in Data Science Research Participation Program is administered by the Oak Ridge Institute for Science and Education (ORISE) through an interagency agreement between the U.S. Department of Energy (DOE) and NIAID. ORISE is managed by Oak Ridge Associated Universities (ORAU) under DOE contract number DE-SC0014664. All opinions expressed in this paper are the author’s and do not necessarily reflect the policies and views of the FDA, NIAID, DOE, or ORAU/ORISE.
W.W., S.L., A.C.F., R.V., D.A.L., E.C.E, D.T.L., A.S., C.M.B., M.U.J., R.M., N.H., J.L., D.S., M.H.-P., M.P.S., C.C.B., D.R.T., E.D.L., E.M., L.C.K., and C.D.W. are service members or employees of the U.S. Government. This work was prepared as part of their official duties. Title 17 U.S.C. §105 provides that “Copyright protection under this title is not available for any work of the U.S. Government.” Title 17 U.S.C. §101 defines a U.S. Government work as a work prepared by a military service member or employee of the U.S. Government as part of that person’s official duties.
Author contributions
W.W., S.L., L.C.K., C.D.W., and S.D.P. conceived and designed the study and experiments. W.W., S.L., C.D.W., A.C.F., R.W., E.D.L., E.G., R.V., S.A.C., M.P.S., D.R.T., T.H.B., B.K.A., E.M., and S.D.P. acquired the data and performed the experiments. W.W., L.C.K., C.D.W., S.L., R.S., N.J.E., A.C.F., F.E., J.O.J., and S.A.R. created the detailed analysis plan and/or analyzed the data. W.W., L.C.K., C.D.W., S.L., R.S., A.C.F., S.D.P., E.M., E.D.L., and C.M.B. interpreted the findings. E.G., J.S.R., D.A.L., K.M., E.C.E., D.T.L., R.E.C., C.J.C., A.S., T.L., C.M.B., R.C.M., M.U.J., R.M., N.H., J.L., D.S., M.H.-P., G.W., A.G., M.P.S., and C.D.W. contributed resources, reagents, materials, and specimens. W.W., L.C.K., C.D.W., S.L., R.S., and S.D.P. composed the first draft of the manuscript. W.W., L.C.K., C.D.W., S.L., R.S., N.J.E., A.C.F., E.L., and E.M. provided critical revisions and edits (scientific content) to provisional drafts. All the authors reviewed and approved the final version for submission.
Declaration of interests
S.D.P., T.H.B, D.R.T., J.S.R., and M.P.S. report that the Uniformed Services University (USU) Infectious Diseases Clinical Research Program (IDCRP), a US Department of Defense institution, and the Henry M. Jackson Foundation (HJF) were funded under a Cooperative Research and Development Agreement to conduct an unrelated phase III COVID-19 monoclonal antibody immunoprophylaxis trial sponsored by AstraZeneca. The HJF, in support of the USU IDCRP, was funded by the Department of Defense Joint Program Executive Office for Chemical, Biological, Radiological, and Nuclear Defense to augment the conduct of an unrelated phase III vaccine trial sponsored by AstraZeneca. Both trials were part of the U.S. Government COVID-19 response. Neither is related to the work presented here.
Published: October 21, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.chom.2022.10.012.
Supplemental information
Data and code availability
-
•
Neutralization data have been deposited and available on Zenodo: https://doi.org/10.5281/zenodo.7291034
-
•
Additional Supplemental Items are available at Zenodo: https://doi.org/10.5281/zenodo.7291034
-
•
This study uses publicly available code, which is listed in the key resources table. Original R code used to make the figures in this manuscript are available on Zenodo: https://doi.org/10.5281/zenodo.7291034.
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request
References
- 1.World Health Organization WHO coronavirus (COVID-19) dashboard. 2020. https://covid19.who.int/
- 2.Bergeri I., Whelan M., Ware H., Subissi L., Nardone A., Lewis H.C., Li Z., Ma X., Valenciano M., Cheng B., et al. Global epidemiology of SARS-CoV-2 infection: a systematic review and meta-analysis of standardized population-based seroprevalence studies, Jan 2020–Dec 2021. Preprint at medRxiv. 2022 doi: 10.1101/2021.12.14.21267791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pulliam J.R.C., van Schalkwyk C., Govender N., von Gottberg A., Cohen C., Groome M.J., Dushoff J., Mlisana K., Moultrie H. Increased risk of SARS-CoV-2 reinfection associated with emergence of Omicron in South Africa. Science. 2022;376:eabn4947. doi: 10.1126/science.abn4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Q., Guo Y., Iketani S., Nair M.S., Li Z., Mohri H., Wang M., Yu J., Bowen A.D., Chang J.Y., et al. Antibody evasion by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4 and BA.5. Nature. 2022;608:603–608. doi: 10.1038/s41586-022-05053-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.UK Health Security Agency COVID-19 vaccine surveillance report week 24. 2022. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1083443/Vaccine-surveillance-report-week-24.pdf
- 6.Center for Disease Control COVID data tracker. 2022. https://covid.cdc.gov/covid-data-tracker/#variant-proportions
- 7.World Health Organization Tracking SARS-CoV-2 variants. 2022. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/ [PubMed]
- 8.World Health Organization WHO vaccine tracker. 2022. https://covid19.trackvaccines.org/agency/who/
- 9.Lusvarghi S., Pollett S.D., Neerukonda S.N., Wang W., Wang R., Vassell R., Epsi N.J., Fries A.C., Agan B.K., Lindholm D.A., et al. SARS-CoV-2 BA.1 variant is neutralized by vaccine booster–elicited serum but evades most convalescent serum and therapeutic antibodies. Sci. Transl. Med. 2022;14:eabn8543. doi: 10.1126/scitranslmed.abn8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Accorsi E.K., Britton A., Fleming-Dutra K.E., Smith Z.R., Shang N., Derado G., Miller J., Schrag S.J., Verani J.R. Association between 3 doses of mRNA COVID-19 vaccine and symptomatic infection caused by the SARS-CoV-2 omicron and Delta variants. JAMA. 2022;327:639–651. doi: 10.1001/jama.2022.0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ascaso-Del-Rio A., García-Pérez J., Pérez-Olmeda M., Arana-Arri E., Vergara I., Pérez-Ingidua C., Bermejo M., Castillo de la Osa M., Imaz-Ayo N., Riaño Fernández I., et al. Immune response and reactogenicity after immunization with two-doses of an experimental COVID-19 vaccine (CVnCOV) followed by a third-fourth shot with a standard mRNA vaccine (BNT162b2): RescueVacs multicenter cohort study. EClinicalmedicine. 2022;51:101542. doi: 10.1016/j.eclinm.2022.101542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia-Beltran W.F., St Denis K.J., Hoelzemer A., Lam E.C., Nitido A.D., Sheehan M.L., Berrios C., Ofoman O., Chang C.C., Hauser B.M., et al. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Cell. 2022;185 doi: 10.1016/j.cell.2021.12.033. 457–466.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gruell H., Vanshylla K., Tober-Lau P., Hillus D., Schommers P., Lehmann C., Kurth F., Sander L.E., Klein F. mRNA booster immunization elicits potent neutralizing serum activity against the SARS-CoV-2 Omicron variant. Nat. Med. 2022;28:477–480. doi: 10.1038/s41591-021-01676-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Regev-Yochay G., Gonen T., Gilboa M., Mandelboim M., Indenbaum V., Amit S., Meltzer L., Asraf K., Cohen C., Fluss R., et al. Efficacy of a fourth dose of COVID-19 mRNA vaccine against omicron. N. Engl. J. Med. 2022;386:1377–1380. doi: 10.1056/NEJMc2202542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bar-On Y.M., Goldberg Y., Mandel M., Bodenheimer O., Amir O., Freedman L., Alroy-Preis S., Ash N., Huppert A., Milo R. Protection by a fourth dose of BNT162b2 against omicron in Israel. N. Engl. J. Med. 2022;386:1712–1720. doi: 10.1056/NEJMoa2201570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Magen O., Waxman J.G., Makov-Assif M., Vered R., Dicker D., Hernán M.A., Lipsitch M., Reis B.Y., Balicer R.D., Dagan N. Fourth dose of BNT162b2 mRNA COVID-19 vaccine in a nationwide setting. N. Engl. J. Med. 2022;386:1603–1614. doi: 10.1056/NEJMoa2201688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quandt J., Muik A., Salisch N., Lui B.G., Lutz S., Krüger K., Wallisch A.K., Adams-Quack P., Bacher M., Finlayson A., et al. Omicron BA.1 breakthrough infection drives cross-variant neutralization and memory B cell formation against conserved epitopes. Sci. Immunol. 2022;7:eabq2427. doi: 10.1126/sciimmunol.abq2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gagne M., Moliva J.I., Foulds K.E., Andrew S.F., Flynn B.J., Werner A.P., Wagner D.A., Teng I.T., Lin B.C., Moore C., et al. mRNA-1273 or mRNA-Omicron boost in vaccinated macaques elicits similar B cell expansion, neutralizing responses, and protection from Omicron. Cell. 2022;185 doi: 10.1016/j.cell.2022.03.038. 1556–1571.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.World Health Organization Interim statement on the composition of current COVID-19 vaccines. 2022. https://www.who.int/news/item/17-06-2022-interim-statement-on--the-composition-of-current-COVID-19-vaccines
- 20.Medicines and Healthcare Products Regulatory Agency Regulatory approval of Spikevax bivalent Original/Omicron booster vaccine. 2022. https://www.gov.uk/government/publications/regulatory-approval-of-spikevax-bivalent-originalomicron-booster-vaccine
- 21.U.S. Food and Drug Administration Fall 2022 COVID-19 vaccine strain composition selection recommendation. 2022. https://www.fda.gov/media/159597/download
- 22.Chalkias S., Harper C., Vrbicky K., Walsh S.R., Essink B., Brosz A., McGhee N., Tomassini J.E., Chen X., Chang Y., et al. A bivalent omicron-containing booster vaccine against COVID-19. Preprint at medRxiv. 2022 doi: 10.1101/2022.06.24.22276703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pfizer Pfizer and BioNTech announce omicron-adapted COVID-19 vaccine candidates demonstrate high immune response against omicron. 2022. https://www.pfizer.com/news/press-release/press-release-detail/pfizer-and-biontech-announce-omicron-adapted-covid-19
- 24.Hachmann N.P., Miller J., Collier A.Y., Ventura J.D., Yu J., Rowe M., Bondzie E.A., Powers O., Surve N., Hall K., Barouch D.H. Neutralization escape by SARS-CoV-2 omicron subvariants BA.2.12.1, BA.4, and BA.5. N. Engl. J. Med. 2022;387:86–88. doi: 10.1056/NEJMc2206576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qu P., Faraone J., Evans J.P., Zou X., Zheng Y.M., Carlin C., Bednash J.S., Lozanski G., Mallampalli R.K., Saif L.J., et al. Neutralization of the SARS-CoV-2 omicron BA.4/5 and BA.2.12.1 Subvariants. N. Engl. J. Med. 2022;386:2526–2528. doi: 10.1056/NEJMc2206725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yao L., Zhu K.L., Jiang X.L., Wang X.J., Zhan B.D., Gao H.X., Geng X.Y., Duan L.J., Dai E.H., Ma M.J. Omicron subvariants escape antibodies elicited by vaccination and BA.2.2 infection. Lancet Infect. Dis. 2022;22:1116–1117. doi: 10.1016/S1473-3099(22)00410-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith D.J., Lapedes A.S., de Jong J.C., Bestebroer T.M., Rimmelzwaan G.F., Osterhaus A.D., Fouchier R.A. Mapping the antigenic and genetic evolution of influenza virus. Science. 2004;305:371–376. doi: 10.1126/science.1097211. [DOI] [PubMed] [Google Scholar]
- 28.Fonville J.M., Wilks S.H., James S.L., Fox A., Ventresca M., Aban M., Xue L., Jones T.C., Le N.M.H., Pham Q.T., et al. Antibody landscapes after influenza virus infection or vaccination. Science. 2014;346:996–1000. doi: 10.1126/science.1256427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rössler A., Netzl A., Knabl L., Schäfer H., Wilks S.H., Bante D., Falkensammer B., Borena W., von Laer D., Smith D., et al. BA.2 omicron differs immunologically from both BA.1 omicron and pre-omicron variants. Preprint at medRxiv. 2022 doi: 10.1101/2022.05.10.22274906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilks S.H., Mühlemann B., Shen X., Türeli S., LeGresley E.B., Netzl A., Caniza M.A., Chacaltana-Huarcaya J.N., Daniell X., Datto M.B., et al. Mapping SARS-CoV-2 antigenic relationships and serological responses. Preprint at bioRxiv. 2022 doi: 10.1101/2022.01.28.477987. [DOI] [PubMed] [Google Scholar]
- 31.van der Straten K., Guerra D., van Gils M.J., Bontjer I., Caniels T.G., van Willigen H.D.G., Wynberg E., Poniman M., Burger J.A., Bouhuijs J.H., et al. Antigenic cartography using sera from sequence-confirmed SARS-CoV-2 variants of concern infections reveals antigenic divergence of Omicron. Immunity. 2022;55 doi: 10.1016/j.immuni.2022.07.018. 1725–1731.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mykytyn A.Z., Rissmann M., Kok A., Rosu M.E., Schipper D., Breugem T.I., van den Doel P.B., Chandler F., Bestebroer T., de Wit M., et al. Omicron BA.1 and BA.2 are antigenically distinct SARS-CoV-2 variants. Preprint at bioRxiv. 2022 doi: 10.1101/2022.02.23.481644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amanat F., Strohmeier S., Meade P.S., Dambrauskas N., Mühlemann B., Smith D.J., Vigdorovich V., Sather D.N., Coughlan L., Krammer F. Vaccination with SARS-CoV-2 variants of concern protects mice from challenge with wild-type virus. PLoS Biol. 2021;19 doi: 10.1371/journal.pbio.3001384. e3001384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neerukonda S.N., Vassell R., Lusvarghi S., Wang R., Echegaray F., Bentley L., Eakin A.E., Erlandson K.J., Katzelnick L.C., Weiss C.D., et al. SARS-CoV-2 Delta variant displays moderate resistance to neutralizing antibodies and spike protein properties of higher soluble ACE2 sensitivity, enhanced cleavage and fusogenic activity. Viruses. 2021;13:2485. doi: 10.3390/v13122485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Epsi N.J., Richard S.A., Lindholm D.A., Mende K., Ganesan A., Huprikar N., Lalani T., Fries A.C., Maves R.C., Colombo R.E., et al. Understanding “hybrid immunity”: comparison and predictors of humoral immune responses to SARS-CoV-2 infection and COVID-19 vaccines. Clin. Infect. Dis. 2022 doi: 10.1093/cid/ciac392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wibmer C.K., Ayres F., Hermanus T., Madzivhandila M., Kgagudi P., Oosthuysen B., Lambson B.E., de Oliveira T., Vermeulen M., van der Berg K., et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat. Med. 2021;27:622–625. doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- 37.Guo H., Fan Q., Song S., Shen S., Zhou B., Wang H., Cheng L., Ge X., Ju B., Zhang Z. Increased resistance of SARS-CoV-2 Lambda variant to antibody neutralization. J. Clin. Virol. 2022;150–151:105162. doi: 10.1016/j.jcv.2022.105162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang P., Casner R.G., Nair M.S., Wang M., Yu J., Cerutti G., Liu L., Kwong P.D., Huang Y., Shapiro L., Ho D.D. Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization. Cell Host Microbe. 2021;29 doi: 10.1016/j.chom.2021.04.007. 747–751.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uriu K., Kimura I., Shirakawa K., Takaori-Kondo A., Nakada T.A., Kaneda A., Nakagawa S., Sato K., Genotype to Phenotype Japan (G2P-Japan) Consortium Neutralization of the SARS-CoV-2 mu variant by convalescent and vaccine serum. N. Engl. J. Med. 2021;385:2397–2399. doi: 10.1056/NEJMc2114706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ho D., Wang P., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P., et al. Increased resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7 to antibody neutralization. Preprint at Res Sq. 2021 doi: 10.21203/rs.3.rs-155394/v1. [DOI] [Google Scholar]
- 41.Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 42.Collier D.A., De Marco A., Ferreira I.A.T.M., Meng B., Datir R.P., Walls A.C., Kemp S.A., Bassi J., Pinto D., Silacci-Fregni C., et al. Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nature. 2021;593:136–141. doi: 10.1038/s41586-021-03412-7. [DOI] [PubMed] [Google Scholar]
- 43.Aleem A., Akbar Samad A.B., Slenker A.K. StatPearls. StatPearls Publishing; 2022. Emerging variants of SARS-CoV-2 and novel therapeutics against coronavirus (COVID-19) [PubMed] [Google Scholar]
- 44.Mlcochova P., Kemp S.A., Dhar M.S., Papa G., Meng B., Ferreira I.A.T.M., Datir R., Collier D.A., Albecka A., Singh S., et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature. 2021;599:114–119. doi: 10.1038/s41586-021-03944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cao Y., Yisimayi A., Jian F., Song W., Xiao T., Wang L., Du S., Wang J., Li Q., Chen X., et al. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature. 2022;608:593–602. doi: 10.1038/s41586-022-04980-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Richardson S.I., Madzorera V.S., Spencer H., Manamela N.P., van der Mescht M.A., Lambson B.E., Oosthuysen B., Ayres F., Makhado Z., Moyo-Gwete T., et al. SARS-CoV-2 Omicron triggers cross-reactive neutralization and Fc effector functions in previously vaccinated, but not unvaccinated, individuals. Cell Host Microbe. 2022;30 doi: 10.1016/j.chom.2022.03.029. 880–886.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Russell C.A., Jones T.C., Barr I.G., Cox N.J., Garten R.J., Gregory V., Gust I.D., Hampson A.W., Hay A.J., Hurt A.C., et al. The global circulation of seasonal influenza A (H3N2) viruses. Science. 2008;320:340–346. doi: 10.1126/science.1154137. [DOI] [PubMed] [Google Scholar]
- 48.Katzelnick L.C., Coello Escoto A., Huang A.T., Garcia-Carreras B., Chowdhury N., Maljkovic Berry I., Chavez C., Buchy P., Duong V., Dussart P., et al. Antigenic evolution of dengue viruses over 20 years. Science. 2021;374:999–1004. doi: 10.1126/science.abk0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gilbert P.B., Montefiori D.C., McDermott A.B., Fong Y., Benkeser D., Deng W., Zhou H., Houchens C.R., Martins K., Jayashankar L., et al. Immune correlates analysis of the mRNA-1273 COVID-19 vaccine efficacy clinical trial. Science. 2022;375:43–50. doi: 10.1126/science.abm3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khoury D.S., Schlub T.E., Cromer D., Steain M., Fong Y., Gilbert P.B., Subbarao K., Triccas J.A., Kent S.J., Davenport M.P. Correlates of protection, thresholds of protection, and immunobridging in SARS-CoV-2 infection. Preprint at medRxiv. 2022 doi: 10.1101/2022.06.05.22275943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Linderman S.L., Chambers B.S., Zost S.J., Parkhouse K., Li Y., Herrmann C., Ellebedy A.H., Carter D.M., Andrews S.F., Zheng N.Y., et al. Potential antigenic explanation for atypical H1N1 infections among middle-aged adults during the 2013–2014 influenza season. Proc. Natl. Acad. Sci. USA. 2014;111:15798–15803. doi: 10.1073/pnas.1409171111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Neerukonda S.N., Vassell R., Herrup R., Liu S., Wang T., Takeda K., Yang Y., Lin T.L., Wang W., Weiss C.D. Establishment of a well-characterized SARS-CoV-2 lentiviral Pseudovirus neutralization assay using 293T cells with stable expression of ACE2 and TMPRSS2. PLoS One. 2021;16 doi: 10.1371/journal.pone.0248348. e0248348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jackson-Thompson B.M., Goguet E., Laing E.D., Olsen C.H., Pollett S., Hollis-Perry K.M., Maiolatesi S.E., Illinik L., Ramsey K.F., Reyes A.E., et al. Prospective assessment of SARS-CoV-2 seroconversion (PASS) study: an observational cohort study of SARS-CoV-2 infection and vaccination in healthcare workers. BMC Infect. Dis. 2021;21:544. doi: 10.1186/s12879-021-06233-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Laing E.D., Sterling S.L., Richard S.A., Epsi N.J., Coggins S., Samuels E.C., Phogat S., Yan L., Moreno N., Coles C.L., et al. Antigen-based multiplex strategies to discriminate SARS-CoV-2 natural and vaccine induced immunity from seasonal human coronavirus humoral responses. Preprint at medRxiv. 2021 doi: 10.1101/2021.02.10.21251518. [DOI] [Google Scholar]
- 55.Richard S.A., Pollett S.D., Lanteri C.A., Millar E.V., Fries A.C., Maves R.C., Utz G.C., Lalani T., Smith A., Mody R.M., et al. COVID-19 outcomes among US Military Health System beneficiaries include complications across multiple organ systems and substantial functional impairment. Open Forum Infect. Dis. 2021;8:ofab556. doi: 10.1093/ofid/ofab556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zufferey R., Nagy D., Mandel R.J., Naldini L., Trono D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotechnol. 1997;15:871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]
- 57.Naldini L., Blömer U., Gallay P., Ory D., Mulligan R., Gage F.H., Verma I.M., Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 58.Freed N.E., Vlková M., Faisal M.B., Silander O.K. Rapid and inexpensive whole-genome sequencing of SARS-CoV-2 using 1200 bp tiled amplicons and Oxford nanopore rapid barcoding. Biol. Methods Protoc. 2020;5:bpaa014. doi: 10.1093/biomethods/bpaa014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aksamentov I., Roemer C., Hodcroft E.B., Neher R.A. Nextclade: clade assignment, mutation calling and quality control for viral genomes. J. Open Source Software. 2021;6:3773. doi: 10.21105/joss.03773. [DOI] [Google Scholar]
- 60.R Core Team R: a language and environment for statistical computing. R Foundation for Statistical Computing. 2022 https://www.R-project.org/ [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Neutralization data have been deposited and available on Zenodo: https://doi.org/10.5281/zenodo.7291034
-
•
Additional Supplemental Items are available at Zenodo: https://doi.org/10.5281/zenodo.7291034
-
•
This study uses publicly available code, which is listed in the key resources table. Original R code used to make the figures in this manuscript are available on Zenodo: https://doi.org/10.5281/zenodo.7291034.
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request