Abstract
Recent methodological advances in solution NMR allow the determination of multi-state protein structures and provide insights into structurally and dynamically correlated protein sites at atomic resolution. This is demonstrated in the present work for the well-studied PDZ2 domain of protein human tyrosine phosphatase 1E for which protein allostery had been predicted. Two-state protein structures were calculated for both the free form and in complex with the RA-GEF2 peptide using the exact nuclear Overhauser effect (eNOE) method. In the apo protein, an allosteric conformational selection step comprising almost 60% of the domain was detected with an “open” ligand welcoming state and a “closed” state that obstructs the binding site by changing the distance between the β-sheet 2, α-helix 2, and sidechains of residues Lys38 and Lys72. The observed induced fit-type apo-holo structural rearrangements are in line with the previously published evolution-based analysis covering ~25% of the domain with only a partial overlap with the protein allostery of the open form. These presented structural studies highlight the presence of a dedicated highly optimized and complex dynamic interplay of the PDZ2 domain owed by the structure-dynamics landscape.
Subject terms: Solution-state NMR, Computational biophysics, Mechanism of action
In this manuscript the authors report accurate multi-state protein structures of the PDZ domain using biological NMR. By looking into protein structural states, the authors report an allosteric pathway at atomic resolution that validates previously reported low resolution findings and uncovered a structural hallmark of the allosteric ligand binding to the PDZ domain.
Introduction
An important area of structural biology is the elucidation of the molecular mechanisms behind enzyme activity, protein target recognition, and transduction of biological signals enabling various cellular pathways, that rely on protein fold and dynamics1. Correlated protein motion spanning across the scaffold is of particular interest as it gives rise to protein states and equilibrium2,3. One of the most studied conserved protein motions is ligand-induced protein rearrangement that enables to transduce the signal through a biomolecule in a specific manner known as allostery. There are two broadly accepted allosteric models including the population-shift model and the dynamic allostery model4,5. The population-shift model explains protein motion based on the protein population equilibrium between distinct protein conformations, whereas dynamic allostery relies on statistical thermodynamics that can explain dynamics-based signal transmission without a change in the average structure.
Typically for large systems including enzymes or multimeric proteins, allostery is structurally studied indirectly using X-ray crystallography of distinct states6. Despite the lack of atomic resolution, chemical shift or relaxation-based NMR studies were able to show allosteric communication within a protein fold in a solution under near-physiological conditions7–13. The ligand binding-induced chemical shift changes (for example, measured in a [15N,1H]-correlation experiment) can be used to probe both local direct ligand-induced protein conformational changes and changes far away from the ligand binding site, which are often attributed to allosteric interactions14. As chemical shifts are more sensitive to the direct interaction with the binding partner, shifts in the protein binding site usually dominate the allosterically invoked shifts.
The recent advances in biological NMR provide insights into the correlated protein sites at atomic resolution by determining multiple protein states using a plethora of NMR-based experimental restraints such as residual dipolar couplings (RDC), cross-correlated relaxation (CCR), paramagnetic relaxation enhancement (PRE), and Nuclear Overhauser Effect (NOE)15–23. In particular, the exact Nuclear Overhauser Effect (eNOE) method implemented in the eNORA2 package within CYANA yields experimental distances with a resolution of up to 0.1 Å24–28, enables the structure calculation of multiple protein states, and allows for correction of spin diffusion24,25,29,30. Furthermore, correlated protein sites can be studied using the distance and angle statistics of calculated protein conformers31–34.
One of the most studied families of allosteric molecules is PDZ domains35. These domains are crucial for protein-protein recognition and protein complex assemblies in multicellular organisms36. PDZ domains generally recognize the carboxyl-terminus of various target proteins and take part in many cellular processes including cell growth and proliferation. PDZ domains display a characteristic compact fold composed of six β-strands, two α-helices, and a unique flexible loop near the binding pocket37. The strands of the protein form an antiparallel β-sheet that serves as a platform for the binding of target molecules. Human tyrosine phosphatase 1E protein (hPTP1E) contains a PDZ2 domain and mediates a series of crucial biological processes such as protein-protein interaction38,39, signaling40, and apoptosis41. The C-terminal peptide derived from the Ras-associated guanine nucleotide exchange factor 2 (RA-GEF2) is a versatile binding partner of the PDZ2 domain of hPTP1E38. X-ray crystallography and solution NMR structures of the PDZ2 domain were solved for the free form as well as for the complex bound to the RA-GEF2 peptide42–44. The PDZ2 domain of hPTP1E binds RA-GEF2 by a β-strand addition between β-strand 2 and α-helix 2, similar to other PDZ domains45. The PDZ2 allostery was studied with various techniques including molecular dynamics46,47, evolutionary data48, and NMR relaxation of protein methyls7.
In this study, we showcase the use of the recently emerged eNOE approach to investigate multi-state structures showing correlated protein sites that may be used to infer an induced-fit ligand binding mode, and protein allostery enabled by the structural information of individual protein states at atomic resolution of both free and bound forms of the PDZ2 domain of hPTP1E.
Results and discussion
Ligand-induced dynamic changes of the PDZ2 domain
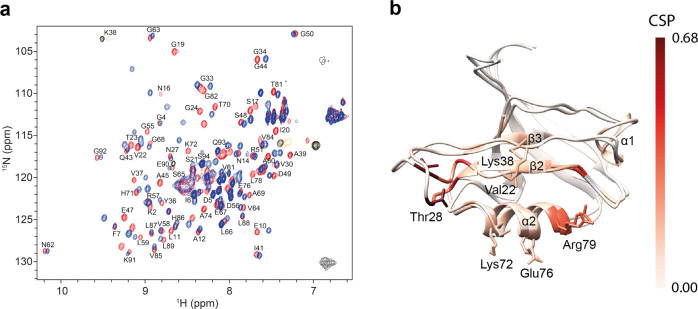
Heteronuclear 2D NMR spectroscopy was applied to gain a qualitative understanding of the binding of the PDZ2 domain of hPTP1E to the C-terminal, eight residue-long, peptide derived from the Ras-associated guanine nucleotide exchange factor 2 (RA-GEF2; Ac-ENEQVSAV-COOH) and allosteric interactions. A [1H,15N]-HSQC spectrum was acquired at 298 K for a uniformly 15N-enriched PDZ2 domain both for a free form and bound to the peptide supplied in a twofold excess. An overlay of the [1H,15N]-HSQC spectra together with the chemical shift perturbation (CSP) mapped on the protein 3D structure is shown in Fig. 1. Averaged atom-weighted chemical shift perturbations were mapped on the later calculated apo protein structure to delineate the allosteric system of the PDZ2 domain. Absolute CSP values are shown in Supplementary Fig. 1. In line with previous reports, residues of the PDZ2 binding site showed significant CSPs. Several residues far apart from the binding site also show CSP, indicating a highly dynamic scaffold7,37. Additionally, significant CSPs are observed for residues at the β-strand 2 and α-helix 2, which sandwich the ligand upon binding, with less prominent CSPs in the flexible loop Gly24-Gly34, β-strand 3, and α-helix 1. The β-strand 3 and α-helix 1 are far away from the binding site and thus have been identified as allosteric sites7.
Fig. 1. Ligand-binding induced conformational changes measured by chemical shifts.
[1H,15N]-HSQC spectra for the PDZ2 domain in apo form (red) and bound to the RA-GEF2 peptide (blue) (a). Residues of the two-state PDZ2 apo structure with the lowest CYANA target function were colored according to the mean of the chemical shift perturbation of 15N and 1H (CSPs) as indicated by the bar on the right (b).
Multi-state structure determination of the PDZ2 domain
For apo PDZ2, 1553 eNOE-derived distance restraints were extracted from a set of 3D [15N,13C]-resolved [1H,1H]-NOESY-HSQC spectra recorded at mixing times of 8, 16, 24, 32, 40, 48, and 80 ms. These distance restraints comprised 410 bidirectional distance restraints of the highest precision. In addition, 130 scalar couplings were collected, which resulted in ~17 restraints per residue, as summarized in Supplementary Table 1. Similarly, for the complex PDZ2, 1484 distance restraints from eNOEs and 65 scalar couplings that resulted in ~16 restraints per residue were collected, as summarized in Supplementary Table 2.
With the given experimental input, multi-state structure calculation with the program CYANA can be performed. CYANA simultaneously optimizes multiple structural states by minimization of the target function (TF). TF is calculated by comparing the simulated distances, which are back-calculated from all optimized states, to the experimental upper and lower limit restraints extracted from the NOE cross-peaks. Individual states are kept in proximity of each other by weak restraints, but local movements with an amplitude below 1.2 Å are allowed without penalty. These restraints are termed symmetry restraints within CYANA because they have been previously used in symmetric oligomeric protein entities and are here applied to the two states correspondingly. Spin diffusion is a major factor causing inaccuracy in deriving distances from the NOEs49. Theoretically, spin diffusion can be accounted for if all NOE cross and diagonal peaks can be measured unambiguously, which is unrealistic given a limit to NMR sensitivity50. Therefore, an alternative version of the spin diffusion correction referred to as exact NOE by Relaxation matrix Analysis (eNORA) was implemented as a part of the eNOE technique that relies on a given 3D protein structure24. So far, eNOE analysis has successfully been applied to the proteins WW domain of Pin1, full-length Pin1, GB3, cyclophilin A, ubiquitin, and a small 14-mer UUCG RNA tetraloop29,30,51–54. The elucidation of the correlated states was recently automated with the software package PDBcor, which elucidates correlated protein sites in the form of structural correlations in an unbiased and automated way from the distance statistics of individual structural entities in a multi-state structure32. It can quantify correlations in structural ensembles, uncover the protein regions that undergo synchronized rearrangements, and optimally separate conformers into states. PDBcor uses information theory and systematic clustering of protein conformers to extract mutual information between individual residues32.
In the case of the PDZ2 domain, both apo and holo multi-state protein structures were calculated following the established protocol introduced above29,51,52 using eNORA2 for the spin diffusion correction and distance extraction24,25 and CYANA for the protein structure calculation with minor modifications19. Symmetry restraints that keep structural entities in proximity of each other were relaxed in the region starting from Gly24 up to Gly34 to allow for the additional motion amplitude of the PDZ2 flexible loop. The enhanced flexibility in the loop region was shown with 15N R2/R1 NMR relaxation rates (Supplementary Fig. 2). The structure annealing algorithm was executed with 150’000 energy minimization steps for 1000 two-state conformers. A series of 1–9 state structure calculations were performed (Supplementary Fig. 3), indicating that a single-state structure does not fulfill the experimental data well, as evidenced by the high CYANA TF, which is a measure of restraint violations. However, two states appear to be sufficient to describe the experimental data well (Supplementary Figs. 3, 4, 5, 8). The relative population of the two states could be estimated with the use of the CYANA TF to be about 1:1 (Supplementary Fig. 6). A jackknife procedure, in which a certain percentage of restraints is removed randomly, followed by two-state structure calculations and correlated states analysis with the PDBcor software demonstrates that the system is well determined. About 80% of the experimental data are required for the emergence of structural correlations (Supplementary Fig. 11). The multi-state nature of the PDZ2 domain was further validated by monitoring the local sidechain dynamics, specifically rotamer averaging of the methyl groups of valines and leucines, presence of single aromatic protons HD1/HD2 and HE1/HE2 for Phe7 and Tyr36, and scalar couplings requesting rotamer averaging (Supplementary Fig. 12). The multi-state nature of the PDZ2 domain is further requested from the 15N-relaxation data showing widespread dynamics (Supplementary Figs. 12, 13).
The 20 two-state conformers with the lowest TF were selected to represent the calculated two-state structure. They satisfy the experimental restraints well, as indicated by the low CYANA target function (see Supplementary Tables 1, 2) and show favorable Ramachandran plot statistics with less than 1% of the residues in the disallowed regions (Supplementary Table 3 and Supplementary Fig. 7). In addition, the resulting structures reproduce the known PDZ2 protein fold with root mean square deviations of 1.11 Å for apo and 1.34 Å for the complex from the corresponding reported crystal structures (Supplementary Tables 1,2).
The two-state structures of the PDZ2 domain of the apo and holo forms
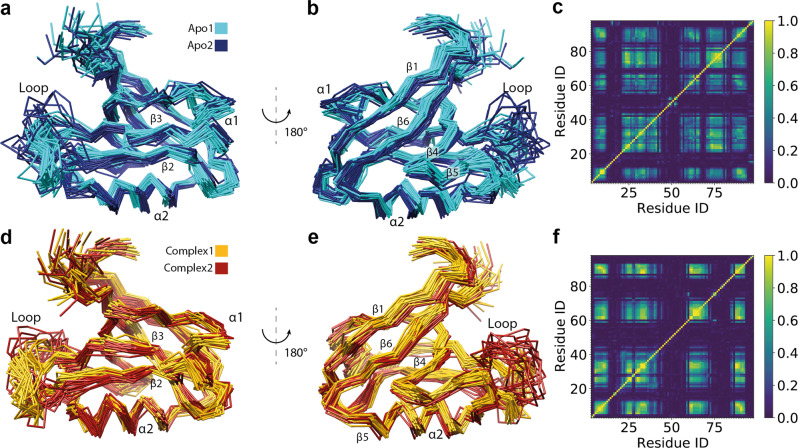
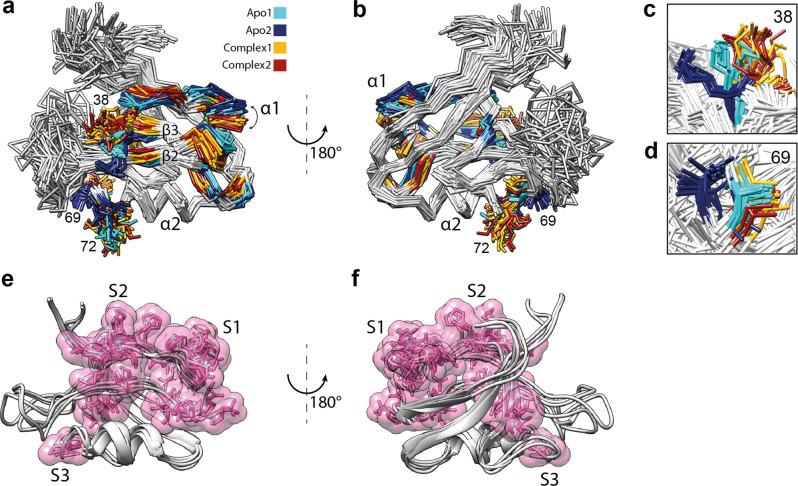
The eNOE-based two-state structures of the free PDZ2 domain (apo) and in complex with the RA-GEF2 peptide (holo) represented by 20 conformers for each state are shown in Fig. 2a, b, d, e. The two-state structures were analyzed with the PDBcor software in standard settings32 generating heatmaps (Fig. 2c, f), which were used to color the structural states in cyan/blue for the apo and yellow/red for the holo PDZ2. Each state is represented by conformers color-coded accordingly (note that for convenience, the conformer separation and coloring will be kept consistent throughout the manuscript). Inspection of the apo two-state structure yields two separated states for the β-sheet, α-helix 2, and the flexible loop Gly24-Gly34. The flexible loop adopts two very distinct conformations, one of which is relatively loose and further apart from the binding site (a dark blue state in Fig. 2a, b), while α-helix 2 is shifted along the helix axis and β-strands 2 and 3 vertically to the β-strand arrow. Hence almost 60% of the entire protein domain appears to shuffle between two states with the aforementioned secondary structures around the peptide ligand-binding site comprising β-strand 2, α-helix 2, and the loop spanning residues Gly24-Gly34 but to a smaller degree extending also into the entire β-sheet. Core residues responsible for the two-state correlations of the apo state are localized around the ligand-binding site (see also below) and the protein’s hydrophobic core (Fig. 3a, b). In Fig. 3, the 15 highest correlated residues, as determined by the correlation matrix, are highlighted as a single volume entity on the two-state apo PDZ2 structure.
Fig. 2. Presentation and correlation analysis of the calculated eNOE PDZ structures.
Two-state ensemble structures of the PDZ2 domain in the ligand-free apo form (a, b) and bound to the RA-GEF2 peptide (holo) (d, e) calculated with eNORA224,25 and CYANA software are shown in two different orientations. The two apo states are colored in cyan and dark blue, whereas the two holo states are colored in yellow and red. The secondary structures and the loop comprising residues Gly24-Gly34 are indicated. Structural correlations and optimal state separations for both protein ensembles were calculated with PDBcor in standard settings32 and shown as distance correlation matrix heatmaps for the apo (c) and holo (f) forms of the PDZ2 domain (0 (blue) means lack of any correlation and 1 (yellow) means 100% correlation between the two states in the bundle of conformers).
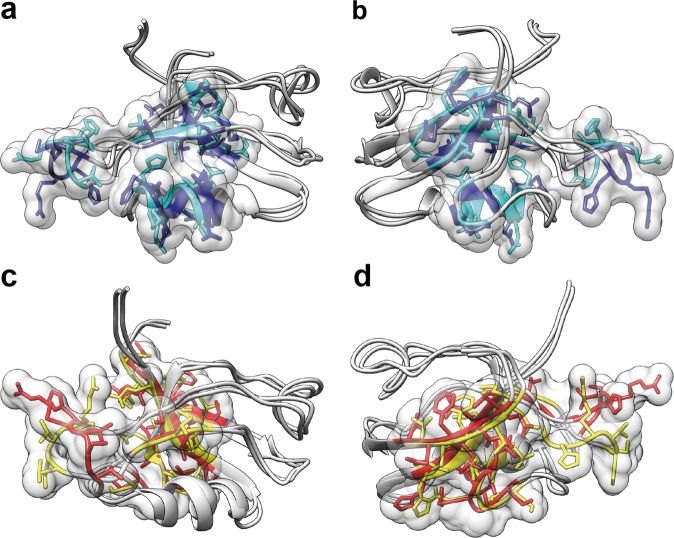
Fig. 3. Localization of the residues responsible for the correlations in apo and holo forms of the PDZ domain.
Core residues responsible for the two-state correlations of the apo state (a, b) are localized around the ligand-binding site and the protein’s hydrophobic core, whereas core residues responsible for the two-state correlations of the PDZ2 RA-GEF2 complex (c, d) are localized to the other side of the protein β-sheet. The 15 highest correlated residues, as determined by the correlation matrices shown in Fig. 2c, f are highlighted as a single volume entity on the two-state PDZ2 structures including state-dependent ribbon coloring and sidechain representation. As it is visible from the protein 3D structure, the highest correlations are concentrated in the protein binding site and all sites involved in the conformational selection mechanism shown in Figs. 4, 5.
Since dynamics in loops is believed to be inherent, one may hypothesize that the dynamics of the two states may originate from the loop comprising residues Gly24-Gly34. Due to its partial flexibility enabled by the two double glycine hinge motifs Gly23-Gly24 and Gly33-Gly34 and its location at the edge of the protein structure, the loop harbors thermal intrinsic local dynamics. Because of the conformational space that the loop covers, the relative position of helix 2 is altered between the two states via the sidechains of Thr70 and His71 and vice versa, which means that the two segments are coupled conformationally. In addition, in state 1, the covered conformational space of the C-terminal end of the loop together with Ile35 is coupled with a shift of the β-sheet via Val58/Leu59 and vice versa.
In addition to the two correlated states, the local distinct configurations of the sidechains, indicated by the Chi1 angle values shown in Supplementary Figs. 4,5, are further supported by scalar coupling measurements, shown in Supplementary Fig. 8, and state-dependent Ramachandran plots of the backbone, shown in Supplementary Figs. 9, 10.
Interestingly, the holo two-state structure also yields two separated states, including the β-sheet 2 and the flexible loop Gly24-Gly34, similar to the apo structure, but not α-helix 2 (residues 71–80) (Figs. 2c, d, 3c, d). As for the apo form, the local distinct configurations are also indicated (Supplementary Fig. 4).
Evidence for conformational selection in PDZ2 in terms of ligand binding
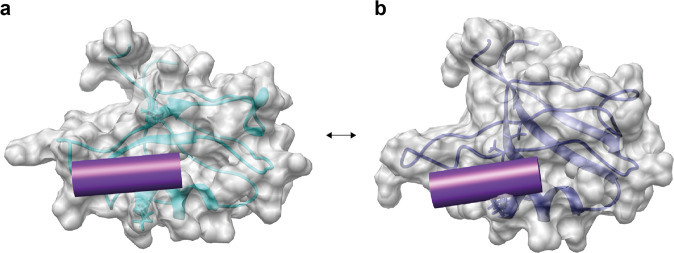
The presence of two states in the apo PDZ2 hints toward the presence of a conformational selection mechanism of ligand binding, since the binding pocket, comprised of α-helix 2 and β-strands 2, and β-strand 3 are distinct between the two states. In state 2 (blue) of the apo PDZ2 domain, the sidechains of residues Lys38 and Lys72 point directly to the binding site (Figs. 4b, 5a, c) and obstruct the binding groove. The local difference between the two states is attributed to a distinct backbone configuration for both Lys38 (Fig. 5c, with its CHI1 angle being in trans) and Lys72 with its CHI1 angle either in a trans or in gauche+ configuration, in line with scalar coupling measurements (Supplementary Fig. 8). This finding suggests that state 2 has to be a “closed” conformation, obstructing ligand-binding whereas state 1 (cyan) is the open, ligand-welcoming state that superimposes with the holo states (Figs. 4a, 5a). Furthermore, a further detailed investigation of both the apo and holo two-state structures with a focus on the sidechains of residues Ala69, Thr70, Lys72, and Lys38 close to the binding site suggests in part a conformational selection mechanism as both holo states (while distinct) are close to or are overlapping with apo state 1 (Fig. 5a, c, d). Based on the Rex contribution to the 15N-relaxation shown in Supplementary Figs. 12, 13 indicating the presence of conformational exchange dynamics in loop 2, Lys38, and Ala69 the dynamics between the two states is indicated to be in the high nanosecond to microsecond time range. Conclusively, this indicates the presence of a conformational selection model for ligand binding.
Fig. 4. Illustration of the conformational selection mechanism in apo form of the PDZ domain.
The apo form comprises an open ligand-welcoming (a) and a closed ligand-obstructing state (b). The surface views of the two states of the PDZ2 of the apo form, with state 1 (a) and state 2 (b) are shown with a transparent ribbon representation and the important sidechains of Lys38 and Lys72 (as sticks). The position of the RA-GEF2 peptide is visualized with a violet cylinder. The access to the binding groove is obstructed in PDZ2 apo state 2 by sidechains of residues Lys38 and Lys72 (b), which indicates the presence of a conformational selection mechanism upon ligand binding.
Fig. 5. Ligand binding-induced structural changes of the PDZ2 domain.
a, b Two views of the aligned apo and holo PDZ2 structural ensembles. All residues with distances between apo and holo averaged Cα positions of more than 1.5 Å except flexible regions are highlighted on the 3D protein structure with the color coding according to Fig. 2 with cyan and blue representing the apo form and yellow and red the holo form, respectively. Ligand-induced allosteric movement of the α-helix 1 is indicated by the arrow. Significant rearrangements in the α-helix 1 and part of the β-strand 2 facing it, the middle part of the β-sheet, and parts of the β-strand 5 and α-helix 2 in the proximity of the flexible loop validate previously reported allosteric interactions in the PDZ2 domain7,48. The conformational selection mechanism is indicated by sidechains of residues Lys38 (c), Ala69 (d), and Lys72 of the apo and holo structures. The sidechain superposition of apo state 1 (cyan) with both holo states (yellow and red) suggests a conformational selection mechanism on PDZ2 for ligand binding. e, f The three sites S1–S3 of the aforementioned correlated network. The elucidated correlated network spans from the binding site to the α-helix 1 through S1, to the Lys54 through S2, or to the Val58 through S3.
Ligand-induced conformational rearrangement of the PDZ2 domain
A conformational selection model for binding does not exclude the presence of an induced-fit mechanism as there have been several studies in this regard, including both two-step processes or simultaneous conformational selection and induced-fit mechanisms55–59. A systematic study of the conformational changes between the apo and holo PDZ2 scaffolds may provide insights into the nature of the binding mechanism. Conformational changes were first quantified in terms of the average distance between the apo and holo PDZ2 structures. For that purpose, individual conformers from both apo and holo two-state ensembles were superimposed to the first conformer of the apo ensemble using the Kabsch algorithm in UCSF Chimera60, then the Cα atom coordinates of all conformers were extracted and averaged over all apo and holo conformers to obtain mean apo and mean holo structures. Next, the distance between the Cα atom coordinates of the averaged apo and holo PDZ2 conformations was calculated for each residue. The residues for which the Cα deviate more than 1.5 Å from each other were highlighted and mapped on the apo PDZ2 structure as shown in Fig. 5. The loop comprising residues Gly24-Gly34 and the N-terminal flexible segment were excluded from the analysis due to their elevated flexibility. The largest ligand-induced backbone conformational changes are concentrated in three sites. The first site (S1) includes α-helix 1 and part of the β-strand 2 facing it. The second site (S2) includes the middle part of the β-sheet. The third site (S3) includes parts of the β-strand 5 and α-helix 2 in the proximity of the flexible loop, as summarized in Fig. 3. Ligand binding yields a shift of the α-helix 1 and a part of the β-strand 2 facing it to accommodate the ligand with a shift of the middle part of the β-sheet away from the binding site quantifiable also by the distance between the Cα atoms of residues Gly23 and Lys72, which is 7.8 ± 0.6 Å in the apo state, but 8.95 ± 0.5 Å for the PDZ2 complex state. The aforementioned structural rearrangements are coupled to the ligand-binding site, as backbone rearrangements between apo and complex PDZ2 domain are ligand-induced. Those findings correlate with one of the major findings of Lockless and Ranganathan, who showed a statistical coupling between His71 and distal residues Ala46 and Ile52 that are part of the α-helix 1 and to the findings of Fuentes et al. that indicated a coupling between residue Ile20 of the binding site and residues Ala39 and Val40 of the β-strand β37,48.
The distinct two-state structures between the apo and holo PDZ2 domain discussed indicate an induced-fit-type mechanism as previously reported by stopped-flow measurements61. In the context of multidomain proteins, the induced-fit allosteric mechanism of the PDZ2 domain may connect the PDZ2 binding site with an interdomain interface. Despite the scarce information about the exact location of the potential interdomain interface for the PDZ2 domain from hPTP1E, it is known that Cdc42 directly interacts with α-helix 1 from the Par-6 PDZ domain62 and a domain-domain interaction between the PDZ1 and PDZ2 occurs through a site located at the α-helix 1 / β-strand 1 region63. This indicates a potential involvement of the α-helix 1 in the PDZ interdomain interface, which also experiences the largest ligand-induced rearrangement according to the presented eNOE studies upon ligand binding, including a ligand-induced structural correlation with the β -sheet and the flexible loop Gly24-Gly34.
A comparison of the proposed allosteric pathways of the PDZ2 domain
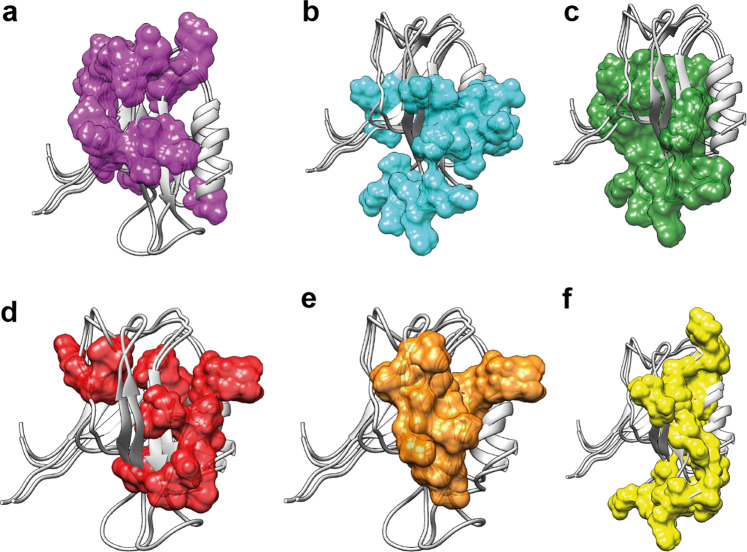
We define the allosteric pathway or network within a protein as an uninterrupted structural area composed of a set of atoms that are non-locally interconnected with each other48. The interconnection is not further defined and can be of dynamical, energetic, or structural nature. In the present study, the structural nature of the allosteric network has been investigated in the PDZ2 domain. In a comparison between the various proposed and published allosteric pathways of the PDZ2 domain under study here, the methods used need to be investigated briefly. (i) The multi-state structure determination presented shows a direct measure of a structural correlation network between states that interchange on the time scale of larger than 10 ns to micro or low milliseconds. (ii) Dynamics analysis using NMR-based relaxation measurements yields a local exchange rate between states. Similar rates for parts of the protein are then indicative of a correlated dynamical network between distinct structures. Dynamics can, in principle, be measured from ps to milliseconds. (iii) Molecular dynamics (MD) simulations are usually of a short time scale (i.e., in the ns range as exemplified with the PDZ2 domain having 16 replicas of a 2 ns MD trajectory)47. (iv) The evolutionary approach differs from the others as it does not only evaluate a single hPTP1E PDZ2 domain but a whole family of the PDZ domains. This makes this approach sensitive to a conserved allosteric interaction among all PDZ domains, which could, for example, be ligand binding. Indeed, when the various methods are applied to PDZ2 shown in Fig. 6, the allosteric pathway deduced from evolutionary data of the PDZ domain48 resembles the eNOE-based induced-fit allosteric network with the ligand-binding site and α-helix 1. In contrast, the allosteric pathways deduced from MD data46 and methyl relaxation data7 resemble rather the eNOE-based structural network of the apo PDZ2 covering the PDZ binding site with β-strands 2 and 3 and α-helix 2 (Fig. 6). Hence, in the case of the PDZ2 domain, the multi-state structure elucidation appears to cover well the structural aspect of allostery, and while this at atomic resolution, the dynamic nature must be measured kinetically or with relaxation measurements.
Fig. 6. Summary of the PDZ allosteric networks from the literature and our study.
Proposed allosteric pathways within the PDZ2 domain determined or predicted with various methods, including eNOE-based induced-fit allosteric network (a), eNOE-based conformational selection allosteric network of the free PDZ2 domain (b), eNOE-based conformational selection allosteric network of the RA-GEF2 complexed PDZ2 domain (c), allosteric network based on evolutionary coupling in the family of the PDZ domains48 (d), allosteric network based on MD simulation46 (e), and allosteric network based on methyl relaxation data7 (f).
On the multi-level allosteric mechanism of the PDZ2 domain
The multi-state structures of the apo and holo form of the PDZ2 domain indicate three levels of protein allostery (Supplementary Fig. 14). The structural correlations of the apo form comprising an extensive structural correlation network between an open ligand-welcoming and a closed ligand-obstructive state encompassing ~60% of the entire domain are in line with the presence of a conformational selection allostery mechanism of ligand binding (Fig. 5). In particular, the ligand-binding site with α-helix 2 and β-strand 2, and β-strand 3 are involved. Then, the ligand binding to the open state induces an extensive conformational change covering ~25% of the protein, including again the binding site by definition as well as most prominently α-helix 1 and β-strand 4. Interestingly this conformational change reduces the extensive allosteric network of the apo form by α-helix 2 in the complex. Moreover, while both allosteric networks comprise the binding site, they are structurally distinct also within the binding site. This can be exemplified with the two binding groove obstructing sidechains Lys38 and Lys72 (Fig. 5) in the open state 1 of apo, which overlap in the closed state 2 of apo with their orientation in the complex, while the complex shows itself again two distinct orientations. The structural change of α-helix 1 upon complex formation may be involved in the interdomain interface of the multidomain hPTP1E, but this idea remains speculative at this point.
Overall, the well-studied PDZ2 domain showcases the power of NMR with high accuracy afforded by eNOEs. This opens up the possibility to solve multiple protein states at atomic resolution under physiological conditions to study protein allostery. It elucidated a three-level allosteric network at atomic resolution and pinpoints the existence of rather extensive and sophisticated structural correlation networks of residues that, in principle, could be used for ligand binding regulation or signaling. In the context of the system of interest, the PDZ2 allosteric binding mechanism was found to be combined with the broadly accepted induced-fit and conformational selection mechanisms. In more general terms, the presented work validates also in part previously reported allosteric indicators using a genetic algorithm48, experimental data7, and MD simulation46. We expect that the presented findings of a complex structural landscape of a protein comprising correlated structural states, which interchange dynamically in the microsecond time range, are present in almost any biomolecular system awaiting to be explored.
Methods
Expression and purification of PDZ2 Domain
The DNA shuttle vector harboring the PDZ2 sequence from human tyrosine phosphatase 1E (hPTP1E) was used for bacterial expression of the PDZ2 domain. The gene of interest included an N-terminal polyhistidine tag separated by an HRV-3C protease cleavage site. Expression and purification was carried out according to the previously reported procedures with minor modifications7,64. Protein expression was performed in BL21 (DE3) Escherichia coli cells and induced with 1 mM IPTG after reaching an OD600 of 0.8. Stable isotope labeling was performed by resuspending cells in growth media supplemented with 15N-enriched ammonium chloride and 13C-enriched glucose. After overnight incubation with shaking, cells were harvested, resuspended, and lysed with a microfluidizer. The protein of interest was purified from the lysate with Ni-NTA chromatography. Then the polyhistidine tag was cleaved with HRV-3C protease and the protein of interest was further separated by passing through the Ni-NTA column. The eluted protein was concentrated at 2 mM and the buffer was exchanged for the desired buffer for NMR (150 mM sodium chloride and 50 mM phosphate buffer at pH 6.8). A peptide comprising the eight C-terminal residues from Rap Guanine Nucleotide Exchange Factor (RA-GEF2) (Ac-ENEQVSAV–COOH, BACHEM) was added to the final sample at a concentration of 2 mM (1:1) for measurements of the PDZ2 domain bound to the ligand.
NMR experiments
The NMR measurements were performed on 600, 700, and 900 MHz Bruker spectrometers equipped with a triple resonance cryoprobe at 298 K. Processing and analysis of all NMR spectra were done with NMRPipe65, Bruker Topspin 4.0, and XEASY66. The PDZ2-peptide ligand titration experiments were performed by acquiring [1H,15N]-HSQC spectra at 298 K, for a uniformly 15N-enriched PDZ2 domain both for a free form and bound to the peptide supplied in a twofold excess. The rotational correlation times τc was calculated to be 6.9 ns for the apo PDZ2 and 6.4 ns for the PDZ2 domain in complex with RA-GEF2 peptide from the 15N-relaxation experiments as described previously67. It was also optimized using a systematic screening approach with the goal of target function minimization in the structure calculations. NOE-buildups were recorded with [15N,13C]–resolved [1H,1H]–NOESY with 40 (t1,max(15N) = 14:4 ms = t1,max(13C) = 7:6 ms)* 200 (t2,max(1H) = 22:0 ms)*1024 (t3,max(1H) = 102:5 ms) complex points, an interscan delay of 0.6 s, four scans per increment and mixing times of 8, 16, 24, 32, 40, 48, and 80 ms. The data were zero-filled to 1024 points in the direct proton dimension, to 128 points in the 15N/13C–dimension and to 512 points in the indirect proton dimension, processing was done with a squared cosine window function for the proton dimensions and a cosine window function for the 15N/13C–dimension. Scalar couplings 1JHNHα were recorded as previously described in ref. 68 from a series of intensity-modulated HMQC spectra with 80(t1,max(15N) = 28.2 ms)*512(t2,max(1H) = 52.3 ms) complex points with an interscan delay of 1 s and 32 scans per increment. Scalar couplings 3JHαHβ were recorded as previously described69 from a 3D 13Cα-separated HACAHB-COSY experiment with 50(t1,max(13C) = 14.2 ms)* 54 (t2,max(1H) = 7.5 ms)* 2048 (t3,max(1H) = 204.9 ms) complex points, 16 scans per increment and 1 s of interscan delay. Scalar couplings for aromatic sidechain heavy atoms 1JNCγ and 1JCOCγ were recorded as previously described70 from intensity-modulated HSQC spectra with 200(t1,max(15N) = 150.0 ms)*512(t2,max(1H) = 51.2 ms) complex points, and interscan delay of 1.2 s and 16 scans per increment for 1JNCγ couplings and with 100(t1,max(15N) = 75.0 ms)*512(t2,max(1H) = 51.2 ms) complex points, and interscan delay of 1 s and 32 scans per increment for 1JCOCγ couplings.
15N-relaxation measurements and dynamics analysis
15N R1ρ, R1, and 15N{1H}-NOE relaxation data was measured at 1H fields of 600 and 900 MHz for both apo and complex PDZ2 domain. For the 15N R1 relaxation measurements, a series of interleaved experiments17 with relaxation delays of 0, 160, 320, 480, 640, 800, 960, 1120, 1280, and 1440 ms, were collected. For the 15N R1ρ relaxation measurements, the relaxation delays were 15, 20, 30, 40, 50, 60, 70, and 80 ms, under a spin-lock field of 2200 Hz. The number of complex points recorded in the direct proton dimension and the indirect 15N dimension were 1024 and 320, respectively. The interscan delay was chosen to be 2 s. The relaxation decay curves were fit using NMRPipe3 and the obtained relaxation rates, as shown in Supplementary Tables 4, 5, were analyzed using the Model-free analysis routine18 present in the software package RELAX19–21. The optimal model selected by the model-free analysis was used to determine the value of the chemical exchange rate, Rex, given in Supplementary Fig. 12 for the field frequency of 900 MHz.
Structure calculation
Single- and multi-state protein structure calculations were performed according to the previously reported procedure29,51,52 using eNORA224,25 within CYANA19. Distance extraction with eNORA2 used for spin diffusion correction an initially derived standard single-state NMR structure for apo, and in the case of the holo PDZ2, the crystal structure with PDB code 3LNY [10.2210/pdb3LNY/pdb]25,44. Lower and upper distance restraints from eNOEs, backbone, Hβ, and aromatic sidechain scalar couplings were used as inputs for the structure calculation. Calculations were done with 150’000 torsion angle dynamics steps for 1000 conformers by simulated annealing. Identical heavy atoms from multi-state conformers were kept together by a potential well with a bottom width of 1.2 Å, as previously described in ref. 71. No further refinement was done explaining at least in part the non-optimal Ramachandran statistics.
eNOE dataset for PDZ2 domain in the apo form
An exhaustive set of experimental restraints for the PDZ2 in the apo form consisted of 1143 unidirectional distance restraints, 410 bidirectional distance restraints with the highest precision of 0.1 Å and 130 scalar couplings, which results in 17 restraints per residue as summarized in Supplementary Table 1.
eNOE dataset for PDZ2 domain in holo form
For the PDZ2 domain in complex with RA-GEF2 peptide, the experimental set of restraints consisted of 995 unidirectional distance restraints, 489 bidirectional distance restraints and 130 scalar couplings, which results in 16 restraints per residue as summarized in Supplementary Table 2.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We would like to thank the Swiss National Science Foundation (SNF project 205320-182800 of R.R.), the National Science Foundation (NSF grant 1917254 for Infrastructure Innovation for Biological Research to B.V.), and the National Institutes of Health (NIH grant R01GM130694-01 to B.V.) for financial support.
Author contributions
C.N.C., H.K., and R.C. prepared the sample. B.V., H.K., A.P., and M.F. measured NMR spectra. D.A., R.R., M.M., P.K., S.K., and D.S. participated in the calculation of protein structures. D.A. analyzed both final protein structures. A.P. performed and analyzed NMR relaxation experiments. P.G., B.V., and R.R. supervised the project. D.A., P.G., B.V., and R.R. wrote the manuscript. All authors discussed the results and contributed to the final manuscript.
Peer review
Peer review information
Nature Communications thanks Andrew Lee and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Data availability
The protein structure data that support the findings of this study have been deposited in the Protein Data Bank under accession codes 3LNX [10.2210/pdb3LNX /pdb] (apo PDZ2) and 3LNY [10.2210/pdb3LNY/pdb] (PDZ2 bound to RA-GEF2 peptide). The protein structure data that was generated in this study have been deposited in the Protein Data Bank under accession codes 7QCX [10.2210/pdb7QCX/pdb] (apo PDZ2) and 7QCY [10.2210/pdb7QCY/pdb] (PDZ2 bound to RA-GEF2 peptide) and in the Biological Magnetic Resonance Bank under the accession codes 34688 [https://bmrb.io/data_library/summary/index.php?bmrbId=34688] and 34689 [https://bmrb.io/data_library/summary/index.php?bmrbId=34689].
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Beat Vögeli, Email: Beat.Vogeli@cuanschutz.edu.
Roland Riek, Email: roland.riek@phys.chem.ethz.ch.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-022-33687-x.
References
- 1.Ishima R, Torchia DA. Protein dynamics from NMR. Nat. Struct. Biol. 2000;7:740–743. doi: 10.1038/78963. [DOI] [PubMed] [Google Scholar]
- 2.Bai Y, Englander SW. Future directions in folding: the multi‐state nature of protein structure. Proteins Struct. Funct. Bioinforma. 1996;24:145–151. doi: 10.1002/(SICI)1097-0134(199602)24:2<145::AID-PROT1>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 3.Karplus M, Weaver DL. Protein-folding dynamics. Nature. 1976;260:404–406. doi: 10.1038/260404a0. [DOI] [PubMed] [Google Scholar]
- 4.Monnot C, et al. Polar residues in the transmembrane domains of the type 1 angiotensin II receptor are required for binding and coupling: reconstitution of the binding site by co-expression of two deficient mutants. J. Biol. Chem. 1996;271:1507–1513. doi: 10.1074/jbc.271.3.1507. [DOI] [PubMed] [Google Scholar]
- 5.Cooper A, Dryden D. Allostery without conformational change. Eur. Biophys. J. 1984;11:103–109. doi: 10.1007/BF00276625. [DOI] [PubMed] [Google Scholar]
- 6.Swain JF, Gierasch LM. The changing landscape of protein allostery. Curr. Opin. Struct. Biol. 2006;16:102–108. doi: 10.1016/j.sbi.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 7.Fuentes EJ, Der CJ, Lee AL. Ligand-dependent dynamics and intramolecular signaling in a PDZ domain. J. Mol. Biol. 2004;335:1105–1115. doi: 10.1016/j.jmb.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 8.Green SM, Shortle D. Patterns of nonadditivity between pairs of stability mutations in staphylococcal nuclease. Biochemistry. 1993;32:10131–10139. doi: 10.1021/bi00089a032. [DOI] [PubMed] [Google Scholar]
- 9.Frauenfelder H, McMahon BH, Austin RH, Chu K, Groves JT. The role of structure, energy landscape, dynamics, and allostery in the enzymatic function of myoglobin. Proc. Natl Acad. Sci. USA. 2001;98:2370–2374. doi: 10.1073/pnas.041614298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rod TH, Radkiewicz JL, Brooks CL. Correlated motion and the effect of distal mutations in dihydrofolate reductase. Proc. Natl Acad. Sci. USA. 2003;100:6980–6985. doi: 10.1073/pnas.1230801100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peng JW. Investigating dynamic interdomain allostery in Pin1. Biophys. Rev. 2015;7:239–249. doi: 10.1007/s12551-015-0171-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Volkman BF, Lipson D, Wemmer DE, Kern D. Two-state allosteric behavior in a single-domain signaling protein. Science. 2001;291:2429–2433. doi: 10.1126/science.291.5512.2429. [DOI] [PubMed] [Google Scholar]
- 13.Emmanouilidis L, et al. Allosteric modulation of peroxisomal membrane protein recognition by farnesylation of the peroxisomal import receptor PEX19. Nat. Commun. 2017;8:1–13. doi: 10.1038/ncomms14635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Selvaratnam R, Chowdhury S, VanSchouwen B, Melacini G. Mapping allostery through the covariance analysis of NMR chemical shifts. Proc. Natl Acad. Sci. USA. 2011;108:6133–6138. doi: 10.1073/pnas.1017311108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wüthrich K. Protein structure determination in solution by NMR spectroscopy. J. Biol. Chem. 1990;265:22059–22062. doi: 10.1016/S0021-9258(18)45665-7. [DOI] [PubMed] [Google Scholar]
- 16.Clore GM, Starich MR, Bewley CA, Cai M, Kuszewski J. Impact of residual dipolar couplings on the accuracy of NMR structures determined from a minimal number of NOE restraints. J. Am. Chem. Soc. 1999;121:6513–6514. doi: 10.1021/ja991143s. [DOI] [Google Scholar]
- 17.Iwahara J, Schwieters CD, Clore GM. Ensemble approach for NMR structure refinement against 1H paramagnetic relaxation enhancement data arising from a flexible paramagnetic group attached to a macromolecule. J. Am. Chem. Soc. 2004;126:5879–5896. doi: 10.1021/ja031580d. [DOI] [PubMed] [Google Scholar]
- 18.Kumar A. Two-dimensional nuclear Overhauser effect in biomolecules. Proc. Indian Acad. Sci. 1985;95:1–8. doi: 10.1007/BF02839713. [DOI] [Google Scholar]
- 19.Güntert P, Mumenthaler C, Wüthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 20.Güntert P, Buchner L. Combined automated NOE assignment and structure calculation with CYANA. J. Biomol. NMR. 2015;62:453–471. doi: 10.1007/s10858-015-9924-9. [DOI] [PubMed] [Google Scholar]
- 21.Güntert P. Automated NMR structure calculation with CYANA. Meth. Mol. Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 22.Vögeli B, Vugmeyster L. Distance‐independent cross‐correlated relaxation and isotropic chemical shift modulation in protein dynamics studies. ChemPhysChem. 2019;20:178–196. doi: 10.1002/cphc.201800602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fenwick RB, Schwieters CD, Vogeli B. Direct investigation of slow correlated dynamics in proteins via dipolar interactions. J. Am. Chem. Soc. 2016;138:8412–8421. doi: 10.1021/jacs.6b01447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orts J, Vögeli B, Riek R. Relaxation matrix analysis of spin diffusion for the NMR structure calculation with eNOEs. J. Chem. Theory Comput. 2012;8:3483–3492. doi: 10.1021/ct3002249. [DOI] [PubMed] [Google Scholar]
- 25.Strotz D, Orts J, Chi CN, Riek R, Vögeli B. ENORA2 exact NOE analysis program. J. Chem. Theory Comput. 2017;13:4336–4346. doi: 10.1021/acs.jctc.7b00436. [DOI] [PubMed] [Google Scholar]
- 26.Vögeli B, Olsson S, Güntert P, Riek R. The exact NOE as an alternative in ensemble structure determination. Biophys. J. 2016;110:113–126. doi: 10.1016/j.bpj.2015.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vögeli B. The nuclear Overhauser effect from a quantitative perspective. Prog. Nucl. Magn. Reson. Spectrosc. 2014;78:1–46. doi: 10.1016/j.pnmrs.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 28.Nichols PJ, et al. The exact nuclear overhauser enhancement: recent advances. Molecules. 2017;22:1176. doi: 10.3390/molecules22071176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vögeli B, Kazemi S, Güntert P, Riek R. Spatial elucidation of motion in proteins by ensemble-based structure calculation using exact NOEs. Nat. Struct. Mol. Biol. 2012;19:1053–1057. doi: 10.1038/nsmb.2355. [DOI] [PubMed] [Google Scholar]
- 30.Nichols PJ, et al. High-resolution small RNA structures from exact nuclear Overhauser enhancement measurements without additional restraints. Commun. Biol. 2018;1:61. doi: 10.1038/s42003-018-0067-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fenwick RB, et al. Weak long-range correlated motions in a surface patch of ubiquitin involved in molecular recognition. J. Am. Chem. Soc. 2011;133:10336–10339. doi: 10.1021/ja200461n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ashkinadze, D., Klukowski, P., Kadavath, H., Güntert, P. & Riek, R. PDBcor: an automated correlation extraction calculator for multi-state protein structures. Structure30, 646–652 (2022). [DOI] [PubMed]
- 33.Fenwick RB, Esteban-Martín S, Salvatella X. Understanding biomolecular motion, recognition, and allostery by use of conformational ensembles. Eur. Biophys. J. 2011;40:1339–1355. doi: 10.1007/s00249-011-0754-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fenwick RB, Orellana L, Esteban-Martín S, Orozco M, Salvatella X. Correlated motions are a fundamental property of β-sheets. Nat. Commun. 2014;5:1–9. doi: 10.1038/ncomms5070. [DOI] [PubMed] [Google Scholar]
- 35.Petit CM, Zhang J, Sapienza PJ, Fuentes EJ, Lee AL. Hidden dynamic allostery in a PDZ domain. Proc. Natl Acad. Sci. USA. 2009;106:18249–18254. doi: 10.1073/pnas.0904492106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tonikian R, et al. A specificity map for the PDZ domain family. PLoS Biol. 2008;6:e239. doi: 10.1371/journal.pbio.0060239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walma T, et al. Structure, dynamics and binding characteristics of the second PDZ domain of PTP-BL. J. Mol. Biol. 2002;316:1101–1110. doi: 10.1006/jmbi.2002.5402. [DOI] [PubMed] [Google Scholar]
- 38.Harris BZ, Lim WA. Mechanism and role of PDZ domains in signaling complex assembly. J. Cell Sci. 2001;114:3219–3231. doi: 10.1242/jcs.114.18.3219. [DOI] [PubMed] [Google Scholar]
- 39.Hung AY, Sheng M. PDZ domains: structural modules for protein complex assembly. J. Biol. Chem. 2002;277:5699–5702. doi: 10.1074/jbc.R100065200. [DOI] [PubMed] [Google Scholar]
- 40.Palmer A, et al. EphrinB phosphorylation and reverse signaling: regulation by Src kinases and PTP-BL phosphatase. Mol. Cell. 2002;9:725–737. doi: 10.1016/S1097-2765(02)00488-4. [DOI] [PubMed] [Google Scholar]
- 41.Sato T, Irie S, Kitada S, Reed JC. FAP-1: a protein tyrosine phosphatase that associates with Fas. Science. 1995;268:411–415. doi: 10.1126/science.7536343. [DOI] [PubMed] [Google Scholar]
- 42.Kozlov G, Banville D, Gehring K, Ekiel I. Solution structure of the PDZ2 domain from cytosolic human phosphatase hPTP1E complexed with a peptide reveals contribution of the β2–β3 loop to PDZ domain–ligand interactions. J. Mol. Biol. 2002;320:813–820. doi: 10.1016/S0022-2836(02)00544-2. [DOI] [PubMed] [Google Scholar]
- 43.Kozlov G, Gehring K, Ekiel I. Solution structure of the PDZ2 domain from human phosphatase hPTP1E and its interactions with C-terminal peptides from the Fas receptor. Biochemistry. 2000;39:2572–2580. doi: 10.1021/bi991913c. [DOI] [PubMed] [Google Scholar]
- 44.Zhang J, et al. Crystallographic and nuclear magnetic resonance evaluation of the impact of peptide binding to the second PDZ domain of protein tyrosine phosphatase 1E. Biochemistry. 2010;49:9280–9291. doi: 10.1021/bi101131f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Doyle DA, et al. Crystal structures of a complexed and peptide-free membrane protein–binding domain: molecular basis of peptide recognition by PDZ. Cell. 1996;85:1067–1076. doi: 10.1016/S0092-8674(00)81307-0. [DOI] [PubMed] [Google Scholar]
- 46.Gerek ZN, Ozkan SB. Change in allosteric network affects binding affinities of PDZ domains: analysis through perturbation response scanning. PLoS Comput. Biol. 2011;7:e1002154. doi: 10.1371/journal.pcbi.1002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dhulesia A, Gsponer J, Vendruscolo M. Mapping of two networks of residues that exhibit structural and dynamical changes upon binding in a PDZ domain protein. J. Am. Chem. Soc. 2008;130:8931–8939. doi: 10.1021/ja0752080. [DOI] [PubMed] [Google Scholar]
- 48.Lockless SW, Ranganathan R. Evolutionarily conserved pathways of energetic connectivity in protein families. Science. 1999;286:295–299. doi: 10.1126/science.286.5438.295. [DOI] [PubMed] [Google Scholar]
- 49.Keepers JW, James TL. A theoretical study of distance determinations from NMR. Two-dimensional nuclear Overhauser effect spectra. J. Magn. Reson. 1984;57:404–426. [Google Scholar]
- 50.Dalvit C, Fogliatto G, Stewart A, Veronesi M, Stockman B. WaterLOGSY as a method for primary NMR screening: practical aspects and range of applicability. J. Biomol. NMR. 2001;21:349–359. doi: 10.1023/A:1013302231549. [DOI] [PubMed] [Google Scholar]
- 51.Strotz D, et al. Protein allostery at atomic resolution. Angew. Chem. Int. Ed. 2020;59:22132–22139. doi: 10.1002/anie.202008734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chi CN, Strotz D, Riek R, Vögeli B. Extending the eNOE data set of large proteins by evaluation of NOEs with unresolved diagonals. J. Biomol. NMR. 2015;62:63–69. doi: 10.1007/s10858-015-9917-8. [DOI] [PubMed] [Google Scholar]
- 53.Vögeli B, et al. Exact distances and internal dynamics of perdeuterated ubiquitin from NOE buildups. J. Am. Chem. Soc. 2009;131:17215–17225. doi: 10.1021/ja905366h. [DOI] [PubMed] [Google Scholar]
- 54.Born, A. et al. Reconstruction of coupled intra-and interdomain protein motion from nuclear and electron magnetic resonance. J. Am. Chem. Soc.143, 16055–16067 (2021). [DOI] [PMC free article] [PubMed]
- 55.Wlodarski T, Zagrovic B. Conformational selection and induced fit mechanism underlie specificity in noncovalent interactions with ubiquitin. Proc. Natl Acad. Sci. USA. 2009;106:19346–19351. doi: 10.1073/pnas.0906966106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morando MA, et al. Conformational selection and induced fit mechanisms in the binding of an anticancer drug to the c-Src kinase. Sci. Rep. 2016;6:1–9. doi: 10.1038/srep24439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gianni S, Dogan J, Jemth P. Distinguishing induced fit from conformational selection. Biophysical Chem. 2014;189:33–39. doi: 10.1016/j.bpc.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 58.Trellet M, Melquiond AS, Bonvin AM. A unified conformational selection and induced fit approach to protein-peptide docking. PLoS ONE. 2013;8:e58769. doi: 10.1371/journal.pone.0058769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nussinov R, Ma B, Tsai C-J. Multiple conformational selection and induced fit events take place in allosteric propagation. Biophys. Chem. 2014;186:22–30. doi: 10.1016/j.bpc.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 61.Chi CN, et al. A sequential binding mechanism in a PDZ domain. Biochemistry. 2009;48:7089–7097. doi: 10.1021/bi900559k. [DOI] [PubMed] [Google Scholar]
- 62.Peterson FC, Penkert RR, Volkman BF, Prehoda KE. Cdc42 regulates the Par-6 PDZ domain through an allosteric CRIB-PDZ transition. Mol. Cell. 2004;13:665–676. doi: 10.1016/S1097-2765(04)00086-3. [DOI] [PubMed] [Google Scholar]
- 63.van den Berk LC, et al. An allosteric intramolecular PDZ− PDZ interaction modulates PTP-BL PDZ2 binding specificity. Biochemistry. 2007;46:13629–13637. doi: 10.1021/bi700954e. [DOI] [PubMed] [Google Scholar]
- 64.Bloem R, et al. Ligand binding studied by 2D IR spectroscopy using the azidohomoalanine label. J. Phys. Chem. B. 2012;116:13705–13712. doi: 10.1021/jp3095209. [DOI] [PubMed] [Google Scholar]
- 65.Delaglio F, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 66.Bartels C, Xia T-h, Billeter M, Güntert P, Wüthrich K. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J. Biomol. NMR. 1995;6:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- 67.Kay LE, Torchia DA, Bax A. Backbone dynamics of proteins as studied by nitrogen-15 inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 68.Kuboniwa H, Grzesiek S, Delaglio F, Bax A. Measurement of H N-H α J couplings in calcium-free calmodulin using new 2D and 3D water-flip-back methods. J. Biomol. NMR. 1994;4:871–878. doi: 10.1007/BF00398416. [DOI] [PubMed] [Google Scholar]
- 69.Grzesiek S, Kuboniwa H, Hinck AP, Bax A. Multiple-quantum line narrowing for measurement of H. alpha.-H. beta. J couplings in isotopically enriched proteins. J. Am. Chem. Soc. 1995;117:5312–5315. doi: 10.1021/ja00124a014. [DOI] [Google Scholar]
- 70.Hu J-S, Grzesiek S, Bax A. Two-dimensional NMR methods for determining χ1 angles of aromatic residues in proteins from three-bond JC'Cγ and JNCγ couplings. J. Am. Chem. Soc. 1997;119:1803–1804. doi: 10.1021/ja963625z. [DOI] [Google Scholar]
- 71.Vögeli B, Güntert P, Riek R. Multiple-state ensemble structure determination from eNOE spectroscopy. Mol. Phys. 2013;111:437–454. doi: 10.1080/00268976.2012.728257. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The protein structure data that support the findings of this study have been deposited in the Protein Data Bank under accession codes 3LNX [10.2210/pdb3LNX /pdb] (apo PDZ2) and 3LNY [10.2210/pdb3LNY/pdb] (PDZ2 bound to RA-GEF2 peptide). The protein structure data that was generated in this study have been deposited in the Protein Data Bank under accession codes 7QCX [10.2210/pdb7QCX/pdb] (apo PDZ2) and 7QCY [10.2210/pdb7QCY/pdb] (PDZ2 bound to RA-GEF2 peptide) and in the Biological Magnetic Resonance Bank under the accession codes 34688 [https://bmrb.io/data_library/summary/index.php?bmrbId=34688] and 34689 [https://bmrb.io/data_library/summary/index.php?bmrbId=34689].