Significance
DNA mismatch repair (MMR) prevents mutations caused by DNA-replication errors and suppresses multiple types of cancers. During MMR, the Mlh1-Pms1 complex is recruited to mispair-containing DNA and nicks the newly replicated DNA strand, targeting it for degradation and resynthesis. Here, we identified an amino acid sequence within the unstructured linker of Mlh1 required for endonuclease activity. This sequence functioned when moved within the Mlh1 linker or when moved to the Pms1 linker. These results reveal a functional role for the intrinsically disordered region, which is conserved from yeast to humans and is mutated in cancer, suggesting that it organizes the catalytically active complex even though the required sequence can be distant from the active site.
Keywords: DNA mismatch repair, intrinsically disordered protein, DNA replication, Msh2–Msh6
Abstract
Eukaryotic DNA mismatch repair (MMR) depends on recruitment of the Mlh1-Pms1 endonuclease (human MLH1-PMS2) to mispaired DNA. Both Mlh1 and Pms1 contain a long unstructured linker that connects the N- and carboxyl-terminal domains. Here, we demonstrated the Mlh1 linker contains a conserved motif (Saccharomyces cerevisiae residues 391–415) required for MMR. The Mlh1-R401A,D403A-Pms1 linker motif mutant protein was defective for MMR and endonuclease activity in vitro, even though the conserved motif could be >750 Å from the carboxyl-terminal endonuclease active site or the N-terminal adenosine triphosphate (ATP)-binding site. Peptides encoding this motif inhibited wild-type Mlh1-Pms1 endonuclease activity. The motif functioned in vivo at different sites within the Mlh1 linker and within the Pms1 linker. Motif mutations in human cancers caused a loss-of-function phenotype when modeled in S. cerevisiae. These results suggest that the Mlh1 motif promotes the PCNA-activated endonuclease activity of Mlh1-Pms1 via interactions with DNA, PCNA, RFC, or other domains of the Mlh1-Pms1 complex.
DNA mismatch repair (MMR) acts on mispairs arising from DNA-replication errors, formation of homologous recombination intermediates, and some chemically modified DNA bases (1–3). During MMR, mispair recognition by MutS homologs, primarily Msh2-Msh6 and Msh2-Msh3 in eukaryotes (4–8), is required to recruit MutL homologs to mispaired DNA, primarily Mlh1-Pms1 in eukaryotes (called MLH1-PMS2 in humans) (1–3, 9). In organisms other than Escherichia coli and related bacteria (10), the MutL homologs have an endonuclease activity that specifically nicks double-stranded DNA on strands containing pre-existing nicks (11–13). Nicking by Mlh1-Pms1 in vitro is required for Exo1-mediated repair on substrates with a nick 3′ to the mispair, as formation of a strand-specific nick 5′ to the mispair allows the 5′–3′ exonuclease activity of Exo1 to excise the mispair (11–14). The absolute requirement of this Mlh1-Pms1 nicking activity in vivo is not well understood, as both 5′ and 3′ nicks relative to mispairs are likely already present on newly synthesized DNA strands (15, 16). One proposal suggests that Mlh1-Pms1 activity maintains single-stranded discontinuities, which appear to identify the newly synthesized strand, even in the presence of the competing activities, like DNA ligation and gap filling by DNA polymerases (15, 17).
MutL homologs are comprised of an N-terminal GHKL family adenosine triphosphatase (ATPase) domain, a carboxyl-terminal dimerization domain, and a predicted unstructured linker domain that connects the folded N- and carboxyl-terminal domains (18–21). In Saccharomyces cerevisiae, the unstructured linkers of Mlh1 and Pms1 are ∼150 and 250 amino acids long, respectively (22). These linkers have a biased sequence composition with reduced hydrophobic amino acids, like the large (>50 amino acid) intrinsically disordered regions (IDRs) present in many proteins (23–25). IDRs often mediate intermolecular interactions, play functional roles, and sometimes become ordered when bound to partners (23–25).
MutL homologs, including Mlh1-Pms1, form DNA-bound rings called sliding clamps following loading by MutS homologs, ATP binding, and dimerization of the N-terminal ATPase domains; these rings rapidly diffuse along the DNA axis (26–30). The extended length of the unstructured interdomain linkers has been suggested to allow these MutL homolog clamps to migrate past protein–DNA complexes, which are normally a barrier to MutS homolog clamps, although Msh2-Msh3 clamps appear to be able to open and close on encountering a protein–DNA complex and hop over it (26–29, 31, 32). Remarkably, cleavage of the S. cerevisiae Mlh1 linker in vivo causes increased mutation rates, suggesting that intact sliding clamps are important for MMR (22). The importance of the combined lengths of the Mlh1 and Pms1 linkers in vivo is suggested by the synergistic increases in mutation rate that have been observed when combining S. cerevisiae mlh1 and pms1 mutations that shorten the linkers (26). In contrast, some linker missense mutations, which do not alter linker lengths, cause MMR defects (22, 33–35). Moreover, deletions within the S. cerevisiae Mlh1 linker tend to cause MMR defects, whereas deletions in the S. cerevisiae Pms1 linker tend not to, except for the pms1-Δ390–610 deletion that eliminates almost the entire Pms1 linker, resulting in a mutant complex that cannot be recruited by Msh2–Msh6 to mispair-containing DNA and fails to bind to DNA under low ionic strength conditions (22). Together, the data suggest that length is only one requirement for the Mlh1 and Pms1 linkers and that the Mlh1 and Pms1 linkers differ in importance for MMR.
Here, we have identified a motif in the Mlh1 linker, which spans residues 391–415, that is conserved from S. cerevisiae to humans and is required for MMR. Mutation of two of the residues in this motif, R401 and I409, to alanine caused an MMR defect, as did short deletions affecting other partially conserved residues within the motif. We found that the motif was functional when moved to different positions on the Mlh1 linker and when the distances between motif and the folded N- and carboxyl-terminal domains were altered. Moreover, moving a copy of the motif to the Pms1 subunit complemented the MMR defect caused by loss of the motif in Mlh1; in addition, swapping the Mlh1 linker with the Pms1 linker supported MMR. Mutant Mlh1-Pms1 complexes with amino acid substitutions in the conserved Mlh1 motif could not support reconstituted MMR reactions in vitro and were defective for Mlh1-Pms1 endonuclease activity but were recruited to mispair-containing DNA by Msh2-Msh6. Peptides encoding the conserved motif, but not control peptides, inhibited wild-type Mlh1-Pms1 endonuclease activity. Consistent with these observations, increased levels of Pms1-4GFP foci, which are MMR intermediates (36), were caused by mutations disrupting the conserved Mlh1 motif, similar to other mutations that reduce Mlh1-Pms1 endonuclease activity (36–38). Mutations of the motif were observed in human cancers, and these mutations disrupted MMR in vivo when modeled in the S. cerevisiae MLH1 gene. Taken together, these data are consistent with a requirement of the Mlh1 linker motif for Mlh1-Pms1 endonuclease activity in MMR, which could be due to an interaction of the motif with the DNA substrate, with the endonuclease active site, and/or with the endonuclease-activating PCNA.
Results
A Conserved Motif in the Mlh1 Linker Is Required for MMR.
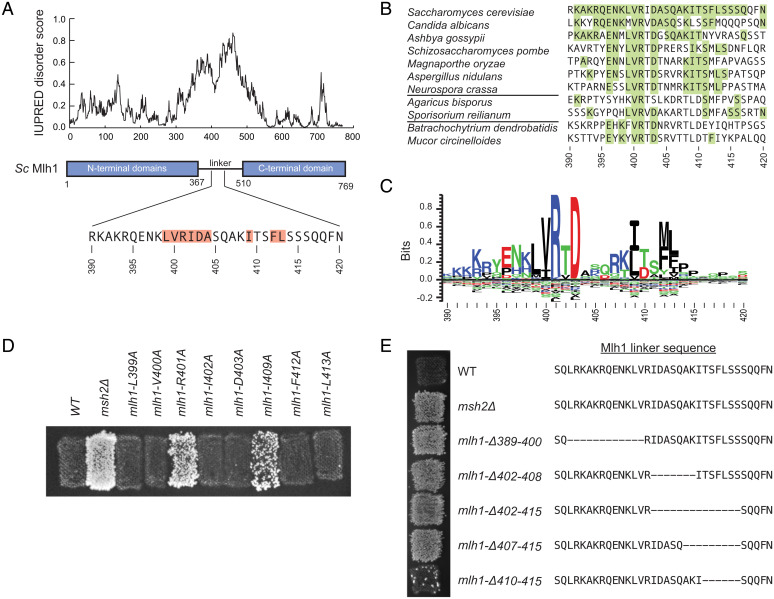
We noticed that a common region from amino acids 391–415 was affected in a series of partial deletions of the unstructured Mlh1 linker that caused MMR defects (Fig. 1A and SI Appendix, Fig. 1 (22)). Alignment of 180 fungal Mlh1 sequences identified a conserved motif roughly spanning the equivalent of S. cerevisiae residues 391–415 (Fig. 1 B and C). This motif was not located near any of the Mlh1 interfaces required for recruitment of Mlh1-Pms1 to mispaired DNA by Msh2-Msh6 or Msh2-Msh3 (9). Previously identified mutations in this region of MLH1 cause MMR defects, including mlh1-K393E,R394E, mlh1-K398E,R401E, mlh1-R401A,D430A (previously called mlh1-31), mlh1-I409N, and mlh1-Δ396–421 (33–35). Consistent with this, strains we constructed containing the mlh1-R401A,D403A and mlh1-Δ396–421 mutations at the MLH1 locus had mutation rates in the hom3-10 and lys2-10A frameshift reversion assays that were not different from those of an mlh1Δ strain based on 95% CIs (Table 1).
Fig. 1.
The Mlh1 linker contains a conserved motif required for MMR. (A) IUPRED (72) long-range disorder prediction for S. cerevisiae Mlh1 (Top) indicating the position of the N- and carboxyl-terminal domains and the predicted unstructured linker as well as the location of conserved motif (Bottom). Highlighted amino acids were analyzed by alanine scanning mutagenesis. (B) Portion of an alignment of 180 fungal Mlh1 sequences with species from Ascomycota top, Basidiomycota middle, and basal fungi bottom. Highlighted residues are identical to S. cerevisiae. (C) Sequence logo for the conserved motif generated by Seq2Logo (73) derived from the 180 Mlh1 sequences. The height of the letters above the zero line corresponds to their relative presence in the alignment. (D) Patch test of strains containing alanine mutations of highly and partially conserved residues in the motif. Increased No. of papillae on CSM-Lys medium correspond to an increased lys2-10A reversion rate. The wild-type (WT) strain is MMR proficient, and the msh2Δ is completely MMR deficient. (E) Patch test of strains on CSM-Lys medium containing deletions within the conserved motif.
Table 1.
Mutation rates
| Genotype | Strain | hom3-10 reversion rate (x10−9)* | lys2-10A reversion rate (x10−9)* |
|---|---|---|---|
| Wild type | RDKY5964 | 1.77 [0.79–4.32] (1) | 5.94 [1.31–13.6] (1) |
| msh2Δ | RDKY9658 | 4200 [3390–7870] (2,373) | 136000 [124000–465000] (22,896) |
| mlh1Δ | RDKY9670 | 4170 [3300–6950] (2,356) | 142000 [110000–159000] (23,906) |
| pms1Δ | RDKY9673 | 4700 [3140–7230] (2,655) | 94300 [73300–117000] (15,875) |
| mlh1-L399A | RDKY10035 | 3.70 [3.22–4.87] (2.09) | 41.2 [31.4–54.0] (6.9) |
| mlh1-V400A | RDKY10037 | 3.19 [1.95–4.74] (1.8) | 53.1 [33.6–77.6] (8.9) |
| mlh1-R401A | RDKY10039 | 2350 [2100–4080] (1,330) | 89600 [76100–117000] (15,083) |
| mlh1-R401C | RDKY10061 | 2193 [1914–2631] (1,239) | 78228 [66900–90700] (13,170) |
| mlh1-R401H | RDKY10063 | 1288 [1100–1800] (728) | 33009 [12500–47300] (5,557) |
| mlh1-I402A | RDKY10041 | 3.18 [1.53–4.51] (1.8) | 33.9 [30.4–54] (5.7) |
| mlh1-D403A | RDKY10043 | 2.80 [1.59–4.04] (1.6) | 127 [81–145] (21) |
| mlh1-R401A,D403A | RDKY9949 | 2260 [1930–3660] (1,277) | 64000 [54300–111000] (10,774) |
| mlh1-I409A | RDKY10045 | 585 [318–928] (330) | 79800 [36800–123000] (13,432) |
| mlh1-F412A | RDKY10047 | 6.61 [4.95–9.74] (3.7) | 38.6 [28.4–46.8] (6.5) |
| mlh1-L413A | RDKY10049 | 2.60 [1.91–5.25] (1.5) | 11.3 [8.21–19.3] (1.91) |
| mlh1-Δ396–421 | RDKY9950 | 3580 [2240–3880] (2,023) | 83200 [63000–126000] (14,007) |
| mlh1-Δ391–415 | RDKY10055 | 3274 [2780–3950] (1,850) | 75323 [57200–84300] (12,680) |
| mlh1-Δ389–400 | RDKY10065 | 1570 [1370–3200] (889) | 130000 [104000–179000] (21,963) |
| mlh1-Δ402–415 | RDKY10131 | 4701 [3900–6100] (2,656) | 73800 [35700–152000] (12,424) |
| mlh1-Δ407–415 | RDKY10069 | 7010 [4610–14300] (3,958) | 200000 [165000–292000] (33,747) |
| mlh1-Δ410–415 | RDKY10109 | 5.15 [2.67–12.1] (2.9) | 166 [129–202] (28) |
| mlh1-Δ402–408 | RDKY10106 | 3259 [2500–4040] (1,841) | 220332 [206000–306000] (37,093) |
| pms1-ml | RDKY10102 | 12.9 [10.6–16.0] (7.2) | 86.0 [72.2–93.1] (14.5) |
| pms1-mlΔm | RDKY10154 | 36.1 [29.8–50.9] (20) | 320 [222–480] (54) |
| mlh1-pl | RDKY10097 | 4732 [4180–5640] (2,674) | 177675 [132000–232000] (29,912) |
| mlh1-pl pms1-ml | RDKY10144 | 41.6 [34.7–51.0] (24) | 329 [183–400] (55) |
| mlh1-pl pms1-mlΔm | RDKY10157 | 3830 [2890–4730] (2,166) | 419000 [368000–511000] (70,477) |
| mlh1-Δ391–415 pms1-ml | RDKY10141 | 17.8 [12.1–44.9] (10) | 66.5 [53.8–138] (11) |
| mlh1-Δ391–415 pms1-mlΔm | RDKY10161 | 4060 [3630–5540] (2,295) | 236000 [176000–478000] (39,745) |
*95% CIs in brackets [], fold change over wild type in parentheses ().
To further explore the motif, we made a series of chromosomal alanine-scanning mutations affecting the fully or partially conserved residues of the motif (Fig. 1D). Alanine substitutions of most of the partially conserved residues (L399, V400, F412, and L413) did not cause an increased mutation rate, consistent with previous results showing that mlh1-S415N, which alters a poorly conserved residue, did not cause an MMR defect (39) (Fig. 1D). Remarkably, the mlh1-D403A mutation, which affects one of the two most-conserved residues, did not cause a mutator phenotype. In contrast, the mlh1-R401A mutation caused a nearly complete MMR defect, and the mlh1-I409A caused a partial MMR defect (Fig. 1D and Table 1), similar to the effect of the mlh1-I409N mutation (34).
We reasoned that the lack of an effect of alanine amino acid substitutions of the partially conserved residues could be due to the fact that these residues form an interacting surface and that many of the single alanine mutations might not disrupt the interaction sufficiently to impair MMR. To test this idea, we made and tested a series of short deletions within the motif that all retained the critical R401 residue (Fig. 1E and Table 1). The mlh1-Δ389–400 mutation, which deleted partially conserved residues N-terminal to R401, caused a profound MMR defect, as did mlh1-Δ402–408, which deleted residues between R401 and I409. In addition, deletions covering I409, such as mlh1-Δ402–415 and mlh1-Δ407–415, caused a greater MMR defect than the mlh1-I409A mutation. In contrast, mlh1-Δ410–415, which only deleted residues after I409, caused a very small increase in mutation rate (Fig. 1E and Table 1). Taken together, these data indicate that Mlh1 possesses a conserved linker motif spanning S. cerevisiae residues 391–415 that is required for MMR.
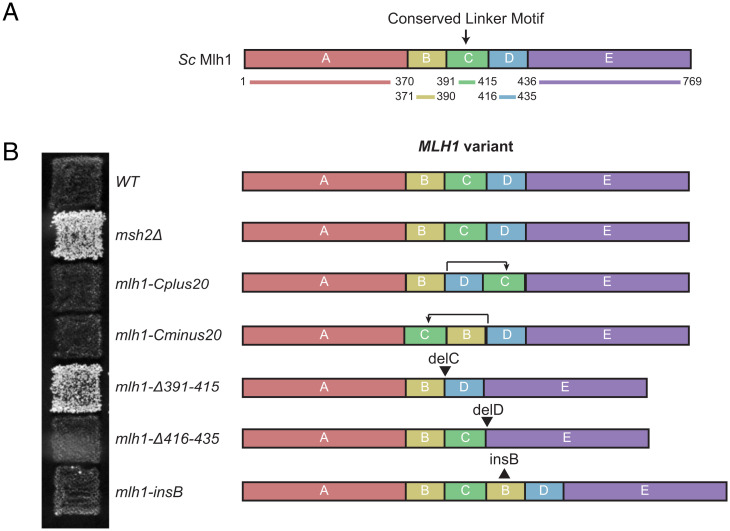
The Conserved Motif Is Functional at Different Positions within the Mlh1 Linker.
Because the Mlh1 linker is predicted to be unstructured (Fig. 1A), we hypothesized that precise position of the conserved motif within the linker may not be crucial for its function. We therefore designed Mlh1 variants that shifted a 25-amino-acid region including this motif (amino acids 391–415) to a position either 20 amino acids N-terminal or 20 amino acids carboxyl-terminal of its normal position (Fig. 2A). Consistent with our hypothesis, the variants that shifted the position of the conserved motif did not cause MMR defects (Fig. 2B). During strain construction, we also isolated other variants arising due to the use of alternative homologies during recombinational repair of the Cas9-induced double-strand break at the MLH1 locus used for targeting mutations to MLH1. The mlh1-Δ391–415 allele, which deleted the conserved motif, caused a complete MMR defect, as expected (Fig. 2B). In contrast, the mlh1-Δ416–435 allele, which deleted the region immediately carboxyl-terminal to the conserved motif, did not cause an MMR defect. Additionally, a variant that inserted a duplication of amino acids 371–390 carboxyl-terminal to amino acid 415 also did not cause an MMR defect. Analysis of Mlh1 linker deletions generated here and in previous studies reveal that retention of the conserved motif, but not its precise position within the Mlh1 linker, is critical for MMR in vivo (Fig. 2B and SI Appendix, Figs. 1 and 2) (22, 26).
Fig. 2.
The conserved Mlh1 linker motif functions when moved within the Mlh1 linker. (A) Diagram of the amino acid regions of Mlh1 involved in moving a region of the Mlh1 linker containing the conserved motif (region “C”, residues 391–415). (B) Patch test of strains encoding altered Mlh1 linkers on CSM-Lys medium (Left) and diagrams of the altered Mlh1 linkers (Right).
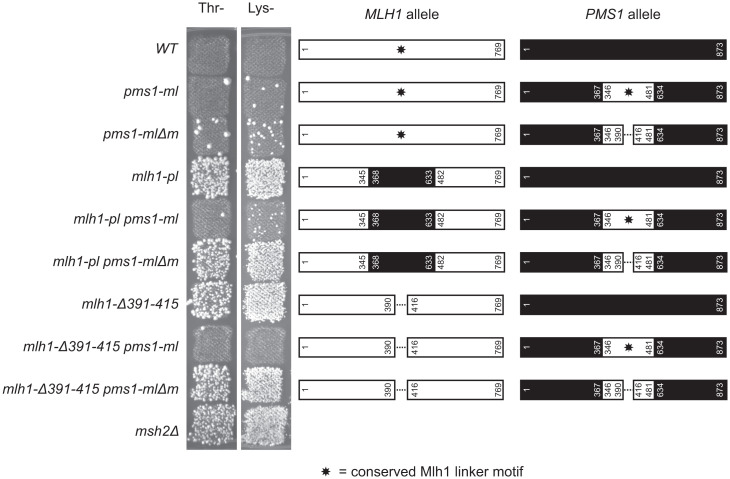
The Mlh1 Conserved Motif Is Functional When Moved to Pms1.
We next tested moving the conserved Mlh1 linker motif to Pms1. We generated the mlh1-pl mutation where the Mlh1 linker (residues 346–481) was replaced with the Pms1 linker (residues 368–633) and generated the pms1-ml mutation where the Pms1 linker was replaced with the Mlh1 linker (Fig. 3 and Table 1). The pms1-ml single mutant, in which the conserved motif was present in both Mlh1 and Pms1, was largely MMR proficient. In the pms1-ml single mutant, the length of the Pms1 linker has been shortened by 130 residues compared with the normal Pms1 linker, essentially without compromising MMR, consistent with previous observations that Pms1 linker deletions of up to 100 residues do not compromise MMR (SI Appendix, Fig. 1; (22)). In contrast and consistent with loss of the conserved motif, the mlh1-pl single mutant was MMR defective. Surprisingly, the mlh1-pl pms1-ml double mutant was largely MMR proficient. This result suggests that the conserved Mlh1 motif was functional when placed on the Pms1 subunit, although this was inconsistent with the results of a previous study that generated an MMR-defective double mutant involving Mlh1/Pms1 linker swaps with different endpoints (Mlh1 residues 335–499, Pms1 residues 364–659) (26) than those used here (Mlh1 residues 346–481, Pms1 residues 368–633). The breakpoints we used were selected by analyzing available protein structures, computational predictions of unstructured protein regions, and sequence alignments to place the breakpoints in unstructured regions outside of the structured Mlh1 and Pms1 carboxyl-terminal domains, whereas the Pms1 breakpoint at residue 659 used by others (26) appears to be located within a structured region of the Pms1 carboxyl terminus and could possibly compromise MMR (Fig. 3 and SI Appendix, Fig. 3).
Fig. 3.
The conserved Mlh1 linker motif functions when moved to the Pms1 linker. Patch test of strains on CSM-Thr and CSM-Lys medium (Left) in which the MLH1 (white boxes) and PMS1 (black boxes) genes are altered by swapping the linkers and by deletion of the conserved Mlh1 linker motif (star). Nos. indicate the position of the residues at the ends of the altered regions. The pms1-ml allele, in which the Pms1 linker is replaced by the Mlh1 linker, complements the MMR deficiency of the mlh1-pl and mlh1-Δ391–415 alleles. The pms1-mlΔm allele, in which the Pms1 linker is replaced by a version of the Mlh1 linker where residues 391–415 were deleted, does not complement MLH1 alleles lacking the motif.
To confirm this result, we tested complementation of mlh1-Δ391–415, which deletes the conserved Mlh1 motif (Fig. 2), by pms1-ml and found that the mlh1-Δ391–415 pms1-ml double mutant was largely MMR proficient (Fig. 3 and Table 1). To verify that complementation was due to the conserved Mlh1 motif, we generated a version of pms1-ml in which the conserved motif was deleted (pms1-mlΔm, deletion of the Mlh1 motif from 391 to 415). As seen for pms1-ml, the pms1-mlΔm single mutant was largely MMR proficient; however, pms1-mlΔm was unable to complement either the mlh1-pl or mlh1-Δ391–415 mutations (Fig. 3). Together, these data indicate that the conserved motif is functional when present in the interdomain linker of either of the Mlh1 or Pms1 subunits of the Mlh1-Pms1 heterodimer.
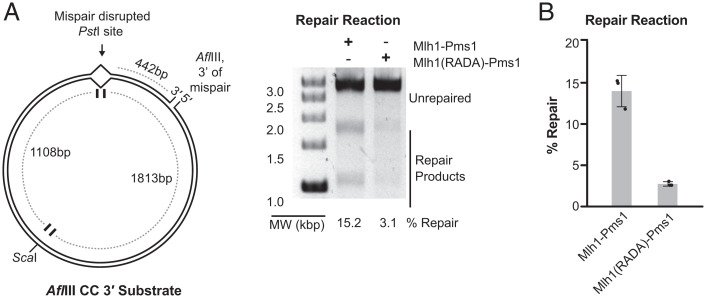
The Mlh1 Linker Motif Is Required for MMR In Vitro.
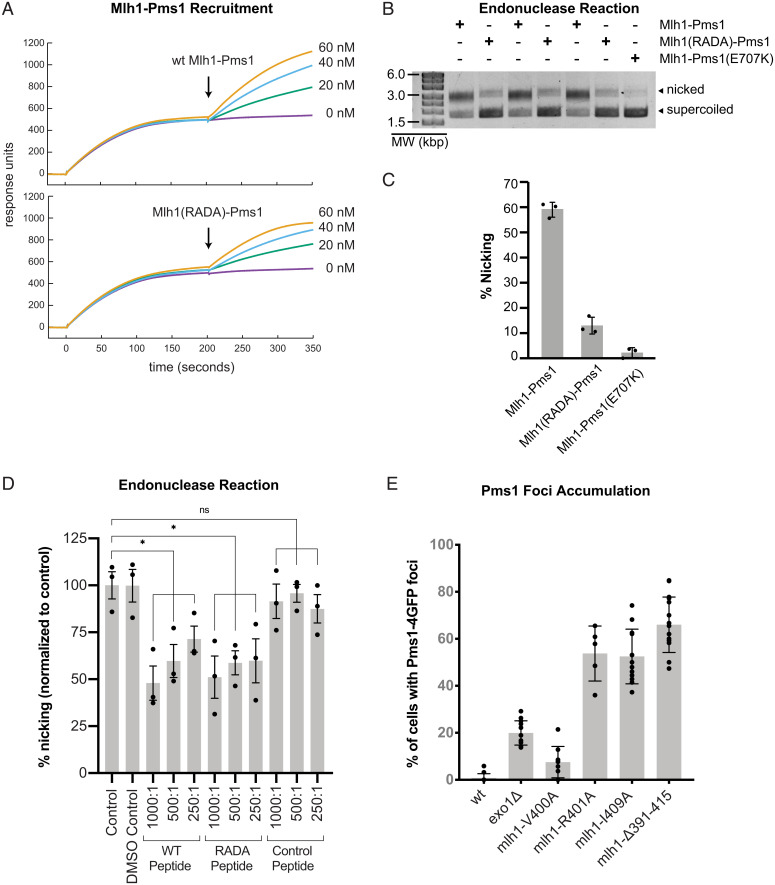
We expressed and purified the Mlh1-R401A,D403A-Pms1 (Mlh1(RADA)-Pms1) mutant complex to analyze its biochemical defects. We selected this mutant complex so that our results could be directly compared with previous studies (33), although our genetic data indicate that R401A, and not D403A, is primarily responsible for the defect in the double mutant (Table 1). Consistent with previous observations (33), we did not observe any defects in the ability of Mlh1(RADA) to interact with Pms1 during protein purification. We tested for the ability of the mutant complex to support a reconstituted Exo1- and Mlh1-Pms1-dependent MMR reaction containing a plasmid substrate with a CC mispair and a single-strand nick at the AflIII site 442 nt 3′ of the PstI-disrupting mispair and a combination of Msh2-Msh6, Mlh1-Pms1, Exo1, RFC-Δ1N, PCNA, RPA, and DNA polymerase ε (Fig. 4A). In this reaction, Exo1-mediated mispair excision on a 3′ nicked substrate is dependent upon the production of a 5′ nick by Mlh1-Pms1. Repair restores the ability of PstI to digest the plasmid such that a ScaI/PstI double digest generates diagnostic 1.1- and 1.8-kb bands. We observed ∼14% repair in the presence of wild-type Mlh1–Pms1 and only ∼2.7% repair in the presence of Mlh1(RADA)-Pms1 (Fig. 4 A and B). These MMR defects in vitro results are consistent with the MMR defects caused by mutations affecting the motif in vivo (Fig. 1 and Table 1).
Fig. 4.
Mutation of the conserved Mlh1 linker motif disrupts MMR in vitro. (A) Repair of a plasmid containing a CC mispair that disrupts the PstI site and a nick at the AflIII site 442 bp 3′ of the mispair (Left). Repair is monitored by the formation of the diagnostic 1.8 and 1.1 kbp products (Right). (B) Quantitation of the repair of the 3′ AflIII CC substrate. The bar heights show the average, and the error bars are the SD. MW, molecular weight.
The Mlh1 Linker Motif Is Required for Endonuclease Activity, but Not Recruitment by Msh2-Msh6.
In order to mediate MMR in vitro, Mlh1-Pms1 must be recruited to DNA substrates by Msh2-Msh6 (or Msh2-Msh3) and must be able to generate single-strand DNA nicks (2, 3, 17, 40). To identify the defects caused by the Mlh1(RADA)-Pms1 mutant, we first tested its ability to be recruited to an end-blocked, mispair-containing DNA by Msh2-Msh6 using a previously developed surface plasmon resonance assay (38, 41–43). In these experiments, a 236-bp DNA substrate containing a mispair was immobilized onto a streptavidin chip on one end with a 5′ biotin. The other end of the DNA substrate was blocked by binding of lac repressor, lacI, to a lac operator sequence. After immobilization and lacI end blocking, Msh2-Msh6 was loaded onto the mispaired DNA substrate in the presence of ATP, which can be observed as an increase in response units (Fig. 5A); under these conditions, Msh2-Msh6 forms mispair-dependent sliding clamps on the DNA. After 200 s, flow was switched to a solution containing Msh2-Msh6, wild-type or mutant Mlh1-Pms1, and ATP, conditions where Mlh1-Pms1 does not bind to DNA in the absence of recruitment by Msh2-Msh6 (42). With both Mlh1-Pms1 and Mlh1(RADA)-Pms1, we observed a robust increase in the response units (Fig. 5A). Thus, the Mlh1(RADA)-Pms1 mutant is recruited to the DNA substrate by Msh2-Msh6, consistent with previous observations using a gel-shift assay (33).
Fig. 5.
Mutation of the conserved Mlh1 linker motif does not alter Mlh1-Pms1 recruitment by Msh2-Msh6 but disrupts Mlh1-Pms1 endonuclease function. (A) Surface plasmon resonance shows that wild-type Mlh1-Pms1 and Mlh1(RADA)-Pms1 are recruited by Msh2-Msh6 to an immobilized end-blocked DNA containing a mispair. Msh2-Msh6 and ATP are injected at 0 s, and flow is switched to a solution containing Msh2-Msh6, Mlh1-Pms1, and ATP at 200 s, conditions where Mlh1-Pms1 does not bind to DNA in the absence of recruitment by Msh2-Msh6 (42). (B) Assay (in triplicate) for the endonuclease activity of Mlh1-Pms1 monitoring nicking of a supercoiled plasmid in the presence of PCNA, RFC-Δ1N, and ATP. (C) Quantitation of the endonuclease activity for wild-type Mlh1-Pms1, Mlh1(RADA)-Pms1, and the endonuclease-deficient Mlh1-Pms1(E707K). (D) Effect of the wild-type peptide, the RADA peptide, and the control peptide on the endonuclease activity of wild-type Mlh1-Pms1 shows inhibition increases at higher molar ratios of peptide to protein. ns, not significant. *, significant. (E) The percent of cells with Pms1-4GFP foci observed using fluorescence microscopy is shown for the wild-type strain, the exo1Δ strain, and various strains with mutations affecting the conserved Mlh1 linker. In all panels, bar heights indicate the average, error bars indicate the SD, and dots indicate the values from individual experiments.
To evaluate endonuclease activity, we tested both wild-type Mlh1-Pms1 and the Mlh1(RADA)-Pms1 mutant in an assay in which the proteins randomly nick a supercoiled DNA plasmid in the presence of both RFC-Δ1N and PCNA, where loading of Mlh1-Pms1 onto DNA by Msh2-Msh6 is not required (12, 38). In this assay, nicking of the supercoiled plasmid is monitored by altered migration of the substrate in agarose gels. We found that the Mlh1(RADA)-Pms1 mutant had ∼10% of the nicking activity of the wild-type Mlh1-Pms1 protein (Fig. 5 B and C), indicative of a defect in the nuclease activity that was roughly equivalent to the defect seen in the repair reaction. Taken together, these biochemical data indicate that the conserved Mlh1 linker motif is required for optimal Mlh1-Pms1 endonuclease activity.
Addition of a Peptide Encoding the Conserved Motif Inhibits the Mlh1-Pms1 Endonuclease Activity.
Because the conserved Mlh1 motif is in an unstructured linker, we hypothesized that a peptide encoding this motif might inhibit activity if the motif organizes the Mlh1-Pms1-DNA complex during catalysis. To test this, we synthesized a wild-type motif peptide (KAKRQENKLVRIDASQAKITSFLSS), a R401A,D403A (RADA) peptide (KAKRQENKLVaIaASQAKITSFLSS), and a control peptide generated from the linker immediately downstream of the motif (region “D” from Fig. 2, SQQFNFEGSSTKRQLSEPKV). The wild-type peptide inhibited Mlh1-Pms1 endonuclease activity in reactions where nicking of a supercoiled plasmid DNA was monitored in the presence of Mlh1-Pms1, PCNA, and RFC-1ΔN. The endonuclease activity dropped from ∼70% at a peptide-to-heterodimer molar ratio of 250:1 to ∼50% at a molar ratio of 1,000:1 (Fig. 5D). The RADA peptide showed similar levels of inhibition, whereas the control peptide did not affect the Mlh1-Pms1 endonuclease activity (Fig. 5D). Inhibition by the wild-type peptide suggests that the conserved motif helps coordinate the conformation of the endonuclease cleavage complex. Similar inhibition by the RADA peptide is consistent with a large molar excess of peptide overcoming a binding defect or with the ability of the RADA peptide to bind with normal affinity. These results suggest that the defect caused by the R401A amino acid substitution is a failure of the mutant motif to promote endonuclease activity, even after the motif is properly bound.
Mutations in the Conserved Mlh1 Linker Motif Cause Increased Levels of Pms1 Foci.
Previously, we have shown that Pms1-GFP foci that have a lifetime of ∼2 min can be observed using fluorescence microscopy in wild-type cells. The percentage of cells containing these foci is increased by mutations that either increase the level of mispairs or disrupt steps in MMR that are downstream of Mlh1-Pms1 recruitment, including disruption of Mlh1-Pms1 endonuclease activity (36–38, 44). We therefore tested the accumulation of Pms1 foci in mutant strains with defects in the conserved Mlh1 motif. Consistent with previous results (36–38, 44, 45), the wild-type strain had a low percentage of cells with Pms1 foci, whereas an exo1Δ mutant had an increased percentage of cells with foci (Fig. 5E). Mutations affecting the conserved motif that caused increased mutation rates, including mlh1-R401A, mlh1-I409A, and mlh1Δ391–415, also caused a substantial increase in the No. of Pms1 foci. In contrast, the mlh1-V400A mutation, which did not cause an increased mutation rate, caused levels of Pms1 foci whose distribution overlapped that of the wild-type strain (Fig. 5E). We did not quantify the intensity of the foci but observed by eye that the intensity of the foci in mlh1 mutants was similar to the foci in the exo1Δ mutant.
The Mlh1 Linker Motif Is Conserved in Humans and Is Mutated in Human Cancers.
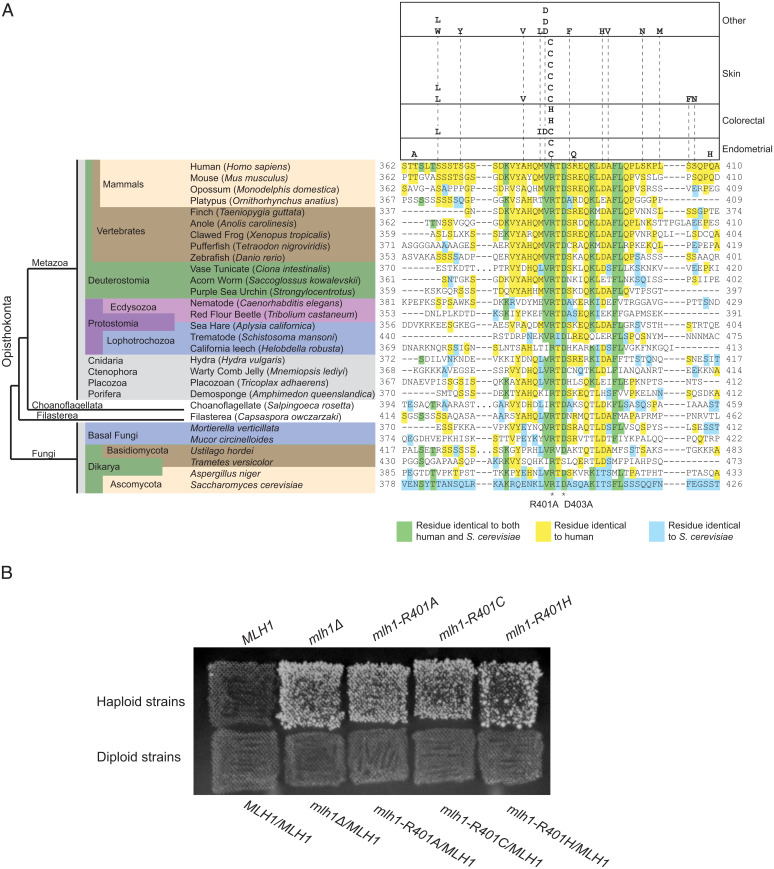
Given the extensive conservation of the Mlh1 linker motif, even in basal fungi (Fig. 1B), we investigated whether the motif was broadly conserved in opisthokonts, which includes fungi and animals. Alignment of Mlh1 sequences from selected species revealed extensive conservation of this motif, including the S. cerevisiae R401 and D403 residues (Fig. 6A). We next examined mutations in human MLH1 reported in the cBioPortal cancer data (46) and found that human equivalent of S. cerevisiae Mlh1 R401, R385, was mutated to both cysteine and histidine in multiple cancers (Fig. 6A). To test the effect of these human cancer mutations, we generated the mlh1-R401C and mlh1-R401H mutations at the chromosomal MLH1 locus and found that these mutations caused large MMR defects that were similar to those caused by mlh1-R401A (Fig. 6B). To test the dominance of these mlh1 mutations, we crossed haploid strains containing these mutations with a wild-type strain of the opposite mating type to generate diploids. All of the heterozygous diploids were MMR proficient (Fig. 6B), indicating that these cancer-associated mutations are recessive.
Fig. 6.
Mutations in the conserved Mlh1 linker motif in human cancers disrupt MMR. (A) The Mlh1 linker motif is conserved in opisthokonts from S. cerevisiae to humans. Residues highlighted in yellow are identical to the human sequence, in blue are identical to S. cerevisiae, and in green are identical to both human and S. cerevisiae. MLH1 mutations reported in cBioPortal are displayed above the alignment with the cancer types indicated at the right. (B) Engineered mutations affecting S. cerevisiae Mlh1 R401 reflecting human cancer mutations are nonfunctional for MMR as revealed by patch tests of haploid strains on CSM-Lys medium. These mutations were found to be recessive by patch tests of heterozygous diploid strains on CSM-Lys medium.
Discussion
Here, we have shown that the Mlh1 linker contains a sequence motif that is conserved from S. cerevisiae to humans and that this motif is required for MMR in vivo. This motif could be moved within the unstructured Mlh1 linker domain and onto Pms1 and remain functional. Biochemical analysis of mutant proteins demonstrated that the conserved motif was not required for Mlh1-Pms1 heterodimer formation nor for recruitment of Mlh1-Pms1 to mispaired DNAs by Msh2-Msh6 but was required for MMR in reconstituted Mlh1-Pms1-dependent MMR reactions in vitro due to a requirement for the motif in supporting optimal Mlh1-Pms1 endonuclease activity. Finally, we have also observed that human cancer mutations affecting the arginine equivalent to the conserved S. cerevisiae Mlh1 R401 also result in MMR defects when modeled in the S. cerevisiae MLH1 gene. Moreover, this region of human MLH1 has been identified in studies of protein phosphorylation, acetylation, ubiquitination, and SUMOylation (47), including the Ataxia-telangiectasia mutated (ATM)/Ataxia-telangiectasia and Rad3-related (ATR)–mediated phosphorylation of human MLH1 S406 that is promoted by BRCA1 (48), raising the possibility that some of these modifications could modulate the function of MLH1. Taken together, these results explain both the previous observations of missense mutations within the Mlh1 linker that cause MMR defects and the sensitivity of the Mlh1 linker, but not the Pms1 linker, to deletions that cause MMR defects (22, 26, 33–35).
Our ability to swap the Mlh1 and Pms1 linkers in S. cerevisiae and retain functionality is in contrast to the results of previous studies that used junction points that were closer to the N-terminal domain (11 residues for Mlh1 and four residues for Pms1) and carboxyl-terminal domains (18 residues for Mlh1 and 26 residues for Pms1) (26) than those used here. To better understand the Mlh1 and Pms1 linker regions, we used Alphafold (49) to predict the structure of the Mlh1-Pms1 carboxyl-terminal domains with attached linkers (SI Appendix, Fig. 3A). Remarkably, the resulting models for the S. cerevisiae Mlh1-Pms1 carboxyl-terminal dimer predicted additional interactions not observed in the crystal structure due to the constructs used for crystallization (20). These interactions involved an N-terminal extension of the Pms1 carboxyl terminus that was wrapped in a groove in the carboxyl terminus of Mlh1 and extension of a β sheet in the Mlh1 carboxyl terminus by a strand formed by this Pms1 extension (SI Appendix, Fig. 3A). The addition of more linker sequences to Mlh1 and Pms1 did not reveal any additional predicted interactions. To ensure that these predicted features were not specific to S. cerevisiae, we folded the carboxyl-terminal dimers of Mlh1-Pms1 homologs from an additional three fungal species and an additional three animal species. For all species, Alphafold generated predictions in which the carboxyl terminus of Mlh1 was wrapped by a similar extension of the carboxyl terminus of Pms1, despite the low sequence conservation of this region (SI Appendix, Fig. 3 B–D). This wrapping of the carboxyl terminus of Mlh1 by the Pms1 extension has several implications. First, it orients the interdomain linkers of both Mlh1 and Pms1 onto the same side of the carboxyl-terminal heterodimer (SI Appendix, Fig. 3A); in the absence of this wrapping, the linkers would be oriented to opposite faces due to the pseudo-twofold relationship between the subunits in the heterodimer (20). Second, it moves the linkers away from both the endonuclease active site and the PCNA interaction site (50) that could give rise to some steric inhibition, depending on linker conformation. Importantly, the linker swaps performed here and deletions of the Pms1 linker in previous studies performed prior the availability of Alphafold disrupt some of these interactions (SI Appendix, Fig. 3) but only cause modest MMR defects (Table 1 (22, 26)). However, the previous nonfunctional linker swaps deleted more of the Pms1 carboxyl-terminal domain and disrupted a potentially important structure, Pms1 helix B (SI Appendix, Fig. 3A); neither swap appeared to affect structural features of the Mlh1 carboxyl-terminal domain. Thus, wrapping by the Pms1 extension may promote Mlh1-Pms1 function, but it is not absolutely required for MMR in vivo.
A key question raised by this work is how does this conserved interdomain linker motif promote the endonuclease activity of Mlh1-Pms1. The presence of hydrophobic residues in this otherwise unstructured region of the linker is consistent with the notion that the motif serves as some form of an interface to interact with either Mlh1, Pms1, PCNA, and/or the DNA substrate. An interaction role would explain our results demonstrating that deletion of groups of residues of the motif gives rise to an MMR defect while individual alanine substitutions of the residues in the regions covered by these deletions do not cause MMR defects. Given that defects caused by the mutant motif can be observed in an Mlh1-Pms1 endonuclease assay containing only Mlh1-Pms1, RFC, PCNA, and supercoiled DNA, it seems probable that the motif is involved in interacting with one or more of these molecules. Intriguingly, protein footprinting of Mlh1-Mlh3 (35) using a DNA-tethered Fe-ethylenediaminetetraacetic acid hydroxyl radical generating probe (FeBABE (51)) suggests that Mlh1 K393, R394, K398, and R401 are near to the DNA backbone of a bound Holliday junction. Combined with this result, the fact that the isolated motif peptide inhibits the Mlh1-Pms1 endonuclease activity further suggests that the interactions of the motif with DNA may coordinate domain motions and/or substrate positioning to convert the rapidly moving Mlh1-Pms1 clamp (26–30) to a state that is competent for catalyzing double-stranded DNA nicking.
Taken together, this work and previous studies indicate that the intrinsically disordered Mlh1 linker has at least two functions: (1) a role in forming a DNA-bound ring, which can migrate past protein blocks (26–30), and (2) a role in promoting the endonuclease activity of the carboxyl-terminal domains. The Mlh1 linker shares many of the expected features of IDRs, including high sequence variability and frequent insertions and deletions (23–25). These sequence changes are typically not expected to change IDR dynamics, which has been shown for the large subunit of RPA using NMR spectroscopy (52). Remarkably, the Mlh1 linker motif shows differences in sequence variability and in composition relative to the rest of the linker. It is highly conserved and shows a localized reduction in disorder predictions due to a short hydrophobic patch (Fig. 1), which is consistent with interaction regions in other IDRs (53–55). Potential roles for this motif in Mlh1 suggested by other IDRs include tuning DNA binding affinity (56) and facilitating enzymatic activity by displacing waters and/or contributing residues to the active site (57, 58). Ultimately understanding the motif’s precise role may require structural characterization of a Mlh1-Pms1 pre- or postcleavage complex with PCNA and DNA.
Methods
Strains.
S. cerevisiae strains were grown in YPD (1% yeast extract, 2% Bacto peptone, and 2% dextrose) or in the appropriate Complete Supplement Mixture (CSM) medium (0.67% yeast nitrogen base without amino acids, 2% dextrose, and amino acid dropout mix at the concentration recommended by the manufacturer [US Biological] at 30 °C) (36, 38, 59). All transformations with plasmids or PCR-based targeting cassettes were performed using standard lithium acetate transformation protocols (60).
All S. cerevisiae strains used for mutation-rate analysis were derived from the S288C strain RDKY5964 (MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 hom3-10 lys2::InsE-A10) (36), and all strains used for microscopy studies were derived from RDKY7588, which is RDKY5964 containing the PMS1-4GFP::kanMX6 allele (36).
Most mutant strains were constructed by introducing Cas9-generated double-stranded breaks (DSBs) in genomic DNA and simultaneously providing a homologous recombination template for repair. Cas9-generated DSBs in the region of the MLH1 gene encoding the linker were produced using pRDK2021, which was constructed from pRCC-K (61) using Gibson assembly. Cas9-generated DSBs in the region of the PMS1 gene encoding the linker were produced using pRDK2088, which was constructed by annealing the oligonucleotides 5′-atc GTG ATA GAA CCG CTT TTT CT-3′ and 5′-aac AGA AAA AGC GGT TCT ATC AC-3′ and ligating them into SapI-digested pRS425/Cas9-2xSapI (from Dr. Bruce Futcher, Stony Brook School of Medicine). Plasmids encoding the homologous recombination targets were commercially synthesized by Integrated DNA Technologies or Twist Bioscience. In all cases, the plasmid-encoded constructs included a silent mutation to disrupt the targeted Cas9 PAM site. These ∼500-bp DNA segments were amplified by PCR and transformed in combination with the appropriate Cas9 vector (pRDK2021 or pRDK2088). In some cases, the homologous recombination targets were generated by cotransforming annealed 80-bp oligonucleotides instead of PCR products. Transformants growing on selective plates (YPD+G418 for pRDK2021, CSM-Leu for pRDK2088) were then screened by PCR amplification of the genomic DNA and Sanger sequencing (RetroGen).
Mutation Rate Analysis.
Mutator phenotypes were evaluated using the hom3-10 and lys2-10A frameshift reversion assays essentially as previously described (36, 59). Qualitative analysis was done by patching colonies onto YPD plates and replica plating onto CSM-Thr and CSM-Lys synthetic dropout media for analysis of papillae growth (4, 59). Mutation rates were determined by fluctuation analysis using a minimum of two independently derived strains and 14 or more independent cultures; comparisons of mutation rates were evaluated using 95% CIs or by Mann-Whitney two-tailed tests (36, 59).
Protein Purification.
MMR proteins were purified according to standard protocols as previously described for Exo1 (62, 63), Msh2-Msh6 (54, 64), Mlh1-Pms1 (63), PCNA (63, 65), DNA polymerase ε (62, 66), RFC-Δ1N (67), and RPA (68) and were greater than 95% pure as determined by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis. Multiple protein preparations were used during the course of the experiments presented, and many of these preparations were validated in our previously published studies (13, 37, 62, 63, 69). The plasmid encoding the Mlh1-R401A,D430A mutant (pRDK2014) was generated from pRDK573 (pRS424/pGAL10-MLH1) (38) using the GeneArt Site Directed Mutagenesis Kit (Invitrogen) according to the manufacturer’s instructions.
In Vitro MMR Assays.
Reactions were performed as described (13). Three hundred ninety femtomoles of Msh2-Msh6, 390 fmol of Mlh1-Pms1, 0.38 fmol of Exo1, 290 fmol of RFC-Δ1N, 290 fmol of PCNA, 1,800 fmol of RPA, and 400 fmol of DNA pol ε were incubated with 100 ng (52 fmol) of CC mispaired substrate with a strand discontinuity at the AflIII site (3′ AflIII CC substrate) in a final volume of 10 μL containing 4 μL of proteins, 1 μL of substrate, and 5 μL of a master reaction buffer mix. The master reaction buffer mix contained 33 mM Tris (pH 7.6), 75 mM KCl, 8.3 mM MgCl2, 1 mM MnSO4, 80 μg/mL bovine serum albumin (BSA), 200 μM deoxynucleoside triphosphates, 1.66 mM glutathione, and 2.5 mM ATP. Reactions were stopped by addition of 0.42 μL of 0.5 M EDTA and 20 μL stop solution containing 0.4 mg/mL glycogen (Thermo Scientific) and 360 µg/mL Proteinase K (Sigma Aldrich) to final concentrations of 21 mM, 24 µg/mL, and 13.4 µg/mL, respectively, followed by incubation for 30 min at 55 °C. Reactions were extracted with phenol, and the DNA substrate was precipitated with ethanol, followed by digestion with PstI and ScaI. The DNA was then subjected to electrophoresis on a 0.8% agarose gel run in a buffer containing 40 mM Tris, 20 mM acetic acid, and 1 mM EDTA (pH 8.3) (TAE) (Bio-Rad) with 0.5 mg/mL ethidium bromide for 45 min at 100 V. Quantitation of the relative amounts of the different DNA species in each individual lane was performed with Alpha Imager HP software (ProteinSimple).
Surface Plasmon Resonance.
Protein–DNA and protein–protein–DNA interactions were monitored using a Biacore T100 instrument (GE Healthcare) using the conditions described previously, where Mlh1-Pms1 does not interact with DNA (5, 41, 42, 70). The DNA substrates used were 236 bp in length with biotin conjugated at one end, the lacO sequence at the other end, and a centrally located base–base mispair that was constructed as previously described (41–43). Approximately 20 ng (100 ± 5 resonance units) of DNA substrates were conjugated to streptavidin-coated Biacore SA chips (GE Healthcare), and an unmodified flow cell was used as a reference surface in each experiment. The DNA ends were blocked by flowing a buffer containing 30 nM LacI, 25 mM Tris (pH 8), 4 mM MgCl2, 110 mM NaCl, 0.01% IGEPAL CA-630 (Nonidet P-40), 2 mM dithiothreitol (DTT), and 2% glycerol over the flow cell. All experiments were performed in the same buffer and also contained 20 nM Msh2-Msh6, 40 nM Mlh1-Pms1, and 250 μM ATP. All experiments were performed at 25 °C at a flow rate of 20 μL/min, and data were collected at a frequency of 10 Hz. The data were analyzed using the BiaEvaluation v3.1 (GE Healthcare).
Mlh1-Pms1 Endonuclease Assay.
Mismatch-independent endonuclease assays were performed as described previously (38). Briefly, 40 µL reactions containing 7.5 nM PCNA, 30 nM RFC-Δ1N, and 35 nM Mlh1-Pms1, 1 mM MnSO4, 20 mM Tris (pH 7.5), 0.5 mM ATP, 0.2 mg/mL BSA, 2 mM DTT, and 100 ng supercoiled pRS425 were incubated at 30 °C for 30 min. Reactions were terminated by addition of 10 µL of a stop solution containing 0.5% SDS, 70 mM EDTA, 40% glycerol, and 2.5 μg/mL proteinase K and incubated at 55 °C for 30 min. Following termination of the reaction, the samples were electrophoresed on a 0.8% agarose gel, the gel was stained with 0.5 mg/mL ethidium bromide, and the bands were quantified using an AlphaImager Gel Imaging System (ProteinSimple).
Microscopy.
Cells were grown in CSM medium to log phase and examined by live imaging essentially as previously described (36, 44), except that the images were collected with an ECHO Revolve epifluorescence microscope with an Olympus PlanApo N 60×/1.42 Oil Ph3 immersion objective. Images were visualized using Adobe Photoshop to manually score cells for the presence of GFP foci.
Alphafold Modeling.
The carboxyl-terminal regions of Mlh1 and Pms1 were simultaneously modeled as a 1:1 heterodimer using Alphafold (49) using the web interface at https://colab.research.google.com/github/sokrypton/ColabFold/blob/main/beta/AlphaFold2_advanced.ipynb. Sequences of S. cerevisiae Mlh1 and Pms1; fungal homologs from Schizosaccharomyces pombe (Ascomycota), Agaricus bisporus (Basidiomycota), and Batrachochytrium dendrobatidis (Chytridiomycota, a division of basal fungi); and animal homologs from Amphimedon queenslandica (Porifera), Caenorhabditis elegans (Protostome), and Homo sapiens (Deuterostome) were obtained from GenBank (www.ncbi.nlm.nih.gov). Sequences to be folded retained much of the interdomain linker to ensure that Pms1 residues that might wrap around Mlh1 were included. In every case, all of the models generated contained both the proper Mlh1-Pms1 dimerization interface and the wrapping of residues of Pms1 around Mlh1. Unstructured linkers N-terminal to the folded portions of the carboxyl-terminal dimers were trimmed prior to display. Images were generated using Pymol (71).
Supplementary Material
Acknowledgments
This study was supported by NIH grant R01 GM50006.
Footnotes
Reviewers: T.P., Duke University; and J.S., University at Buffalo–Downtown Campus.
Competing interest statement: Two of the authors, C.D.P. and R.D.K., are coauthors with Tom Petes, as well as an additional 45 other authors, of a review article published in 2019 (PMID 30652105) providing guidelines for cellular assays for DNA repair pathways. The production of this article did not involve experimental collaboration between C.D.P., R.D.K., and Dr. Petes. In addition, C.D.P. and R.D.K. did/do not share any research support with Dr. Petes. Other than this review article, the authors declare that they have no competing interests to report.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2212870119/-/DCSupplemental.
Data, Materials, and Software Availability
All data are included in the manuscript and/or SI Appendix.
References
- 1.Iyer R. R., Pluciennik A., Burdett V., Modrich P. L., DNA mismatch repair: Functions and mechanisms. Chem. Rev. 106, 302–323 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Reyes G. X., Schmidt T. T., Kolodner R. D., Hombauer H., New insights into the mechanism of DNA mismatch repair. Chromosoma 124, 443–462 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fishel R., Mismatch repair. J. Biol. Chem. 290, 26395–26403 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marsischky G. T., Filosi N., Kane M. F., Kolodner R., Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Dev. 10, 407–420 (1996). [DOI] [PubMed] [Google Scholar]
- 5.Srivatsan A., Bowen N., Kolodner R. D., Mispair-specific recruitment of the Mlh1-Pms1 complex identifies repair substrates of the Saccharomyces cerevisiae Msh2-Msh3 complex. J. Biol. Chem. 289, 9352–9364 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Genschel J., Littman S. J., Drummond J. T., Modrich P., Isolation of MutSbeta from human cells and comparison of the mismatch repair specificities of MutSbeta and MutSalpha. J. Biol. Chem. 273, 19895–19901 (1998). [DOI] [PubMed] [Google Scholar]
- 7.Acharya S., et al. , hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6. Proc. Natl. Acad. Sci. U.S.A. 93, 13629–13634 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dowen J. M., Putnam C. D., Kolodner R. D., Functional studies and homology modeling of Msh2-Msh3 predict that mispair recognition involves DNA bending and strand separation. Mol. Cell. Biol. 30, 3321–3328 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Groothuizen F. S., et al. , MutS/MutL crystal structure reveals that the MutS sliding clamp loads MutL onto DNA. eLife 4, e06744 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Putnam C. D., Evolution of the methyl directed mismatch repair system in Escherichia coli. DNA Repair (Amst.) 38, 32–41 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kadyrov F. A., Dzantiev L., Constantin N., Modrich P., Endonucleolytic function of MutLalpha in human mismatch repair. Cell 126, 297–308 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Kadyrov F. A., et al. , Saccharomyces cerevisiae MutLalpha is a mismatch repair endonuclease. J. Biol. Chem. 282, 37181–37190 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith C. E., et al. , Activation of Saccharomyces cerevisiae Mlh1-Pms1 endonuclease in a reconstituted mismatch repair system. J. Biol. Chem. 290, 21580–21590 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iyer R. R., et al. , The MutSalpha-proliferating cell nuclear antigen interaction in human DNA mismatch repair. J. Biol. Chem. 283, 13310–13319 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reyes G. X., et al. , Ligation of newly replicated DNA controls the timing of DNA mismatch repair. Curr. Biol. 31, 1268–1276.e6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sriramachandran A. M., et al. , Genome-wide nucleotide-resolution mapping of DNA replication patterns, single-strand breaks, and lesions by GLOE-seq. Mol. Cell 78, 975–985.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Putnam C. D., Strand discrimination in DNA mismatch repair. DNA Repair (Amst.) 105, 103161 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Groothuizen F. S., Sixma T. K., The conserved molecular machinery in DNA mismatch repair enzyme structures. DNA Repair (Amst.) 38, 14–23 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Ban C., Yang W., Crystal structure and ATPase activity of MutL: Implications for DNA repair and mutagenesis. Cell 95, 541–552 (1998). [DOI] [PubMed] [Google Scholar]
- 20.Gueneau E., et al. , Structure of the MutLα C-terminal domain reveals how Mlh1 contributes to Pms1 endonuclease site. Nat. Struct. Mol. Biol. 20, 461–468 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Guarné A., et al. , Structure of the MutL C-terminal domain: A model of intact MutL and its roles in mismatch repair. EMBO J. 23, 4134–4145 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Plys A. J., Rogacheva M. V., Greene E. C., Alani E., The unstructured linker arms of Mlh1-Pms1 are important for interactions with DNA during mismatch repair. J. Mol. Biol. 422, 192–203 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dyson H. J., Wright P. E., Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 6, 197–208 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Oldfield C. J., Dunker A. K., Intrinsically disordered proteins and intrinsically disordered protein regions. Annu. Rev. Biochem. 83, 553–584 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Wright P. E., Dyson H. J., Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 16, 18–29 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim Y., Furman C. M., Manhart C. M., Alani E., Finkelstein I. J., Intrinsically disordered regions regulate both catalytic and non-catalytic activities of the MutLα mismatch repair complex. Nucleic Acids Res. 47, 1823–1835 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gorman J., Plys A. J., Visnapuu M. L., Alani E., Greene E. C., Visualizing one-dimensional diffusion of eukaryotic DNA repair factors along a chromatin lattice. Nat. Struct. Mol. Biol. 17, 932–938 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mardenborough Y. S. N., et al. , The unstructured linker arms of MutL enable GATC site incision beyond roadblocks during initiation of DNA mismatch repair. Nucleic Acids Res. 47, 11667–11680 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu J., et al. , Cascading MutS and MutL sliding clamps control DNA diffusion to activate mismatch repair. Nature 539, 583–587 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.London J., et al. , Linker domain function predicts pathogenic MLH1 missense variants. Proc. Natl. Acad. Sci. 118, e2019215118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gorman J., et al. , Dynamic basis for one-dimensional DNA scanning by the mismatch repair complex Msh2-Msh6. Mol. Cell 28, 359–370 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown M. W., et al. , Dynamic DNA binding licenses a repair factor to bypass roadblocks in search of DNA lesions. Nat. Commun. 7, 10607 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Argueso J. L., et al. , Systematic mutagenesis of the Saccharomyces cerevisiae MLH1 gene reveals distinct roles for Mlh1p in meiotic crossing over and in vegetative and meiotic mismatch repair. Mol. Cell. Biol. 23, 873–886 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sia E. A., Dominska M., Stefanovic L., Petes T. D., Isolation and characterization of point mutations in mismatch repair genes that destabilize microsatellites in yeast. Mol. Cell. Biol. 21, 8157–8167 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Claeys Bouuaert C., Keeney S., Distinct DNA-binding surfaces in the ATPase and linker domains of MutLγ determine its substrate specificities and exert separable functions in meiotic recombination and mismatch repair. PLoS Genet. 13, e1006722 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hombauer H., Campbell C. S., Smith C. E., Desai A., Kolodner R. D., Visualization of eukaryotic DNA mismatch repair reveals distinct recognition and repair intermediates. Cell 147, 1040–1053 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goellner E. M., et al. , PCNA and Msh2-Msh6 activate an Mlh1-Pms1 endonuclease pathway required for Exo1-independent mismatch repair. Mol. Cell 55, 291–304 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith C. E., et al. , Dominant mutations in S. cerevisiae PMS1 identify the Mlh1-Pms1 endonuclease active site and an exonuclease 1-independent mismatch repair pathway. PLoS Genet. 9, e1003869 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wanat J. J., Singh N., Alani E., The effect of genetic background on the function of Saccharomyces cerevisiae mlh1 alleles that correspond to HNPCC missense mutations. Hum. Mol. Genet. 16, 445–452 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Li G. M., Mechanisms and functions of DNA mismatch repair. Cell Res. 18, 85–98 (2008). [DOI] [PubMed] [Google Scholar]
- 41.Hargreaves V. V., Shell S. S., Mazur D. J., Hess M. T., Kolodner R. D., Interaction between the Msh2 and Msh6 nucleotide-binding sites in the Saccharomyces cerevisiae Msh2-Msh6 complex. J. Biol. Chem. 285, 9301–9310 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mendillo M. L., Mazur D. J., Kolodner R. D., Analysis of the interaction between the Saccharomyces cerevisiae MSH2-MSH6 and MLH1-PMS1 complexes with DNA using a reversible DNA end-blocking system. J. Biol. Chem. 280, 22245–22257 (2005). [DOI] [PubMed] [Google Scholar]
- 43.Shell S. S., Putnam C. D., Kolodner R. D., Chimeric Saccharomyces cerevisiae Msh6 protein with an Msh3 mispair-binding domain combines properties of both proteins. Proc. Natl. Acad. Sci. U.S.A. 104, 10956–10961 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Campbell C. S., et al. , Mlh2 is an accessory factor for DNA mismatch repair in Saccharomyces cerevisiae. PLoS Genet. 10, e1004327 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Calil F. A., et al. , Rad27 and Exo1 function in different excision pathways for mismatch repair in Saccharomyces cerevisiae. Nat. Commun. 12, 5568 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao J., et al. , Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hornbeck P. V., et al. , PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 43, D512–D520 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Romeo F., et al. , BRCA1 is required for hMLH1 stabilization following doxorubicin-induced DNA damage. Int. J. Biochem. Cell Biol. 43, 1754–1763 (2011). [DOI] [PubMed] [Google Scholar]
- 49.Jumper J., et al. , Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Genschel J., et al. , Interaction of proliferating cell nuclear antigen with PMS2 is required for MutLα activation and function in mismatch repair. Proc. Natl. Acad. Sci. U.S.A. 114, 4930–4935 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miller G., Hahn S., A DNA-tethered cleavage probe reveals the path for promoter DNA in the yeast preinitiation complex. Nat. Struct. Mol. Biol. 13, 603–610 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Daughdrill G. W., Narayanaswami P., Gilmore S. H., Belczyk A., Brown C. J., Dynamic behavior of an intrinsically unstructured linker domain is conserved in the face of negligible amino acid sequence conservation. J. Mol. Evol. 65, 277–288 (2007). [DOI] [PubMed] [Google Scholar]
- 53.Cheng Y., et al. , Mining alpha-helix-forming molecular recognition features with cross species sequence alignments. Biochemistry 46, 13468–13477 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shell S. S., Putnam C. D., Kolodner R. D., The N terminus of Saccharomyces cerevisiae Msh6 is an unstructured tether to PCNA. Mol. Cell 26, 565–578 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vacic V., et al. , Characterization of molecular recognition features, MoRFs, and their binding partners. J. Proteome Res. 6, 2351–2366 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu Y., Matthews K. S., Bondos S. E., Multiple intrinsically disordered sequences alter DNA binding by the homeodomain of the Drosophila hox protein ultrabithorax. J. Biol. Chem. 283, 20874–20887 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McElheny D., Schnell J. R., Lansing J. C., Dyson H. J., Wright P. E., Defining the role of active-site loop fluctuations in dihydrofolate reductase catalysis. Proc. Natl. Acad. Sci. U.S.A. 102, 5032–5037 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fersht A. R., Knill-Jones J. W., Bedouelle H., Winter G., Reconstruction by site-directed mutagenesis of the transition state for the activation of tyrosine by the tyrosyl-tRNA synthetase: A mobile loop envelopes the transition state in an induced-fit mechanism. Biochemistry 27, 1581–1587 (1988). [DOI] [PubMed] [Google Scholar]
- 59.Amin N. S., Nguyen M. N., Oh S., Kolodner R. D., exo1-Dependent mutator mutations: Model system for studying functional interactions in mismatch repair. Mol. Cell. Biol. 21, 5142–5155 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sherman F., Fink G. R., Hicks J. B., Methods in Yeast Genetics (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, 1986). [Google Scholar]
- 61.Generoso W. C., Gottardi M., Oreb M., Boles E., Simplified CRISPR-Cas genome editing for Saccharomyces cerevisiae. J. Microbiol. Methods 127, 203–205 (2016). [DOI] [PubMed] [Google Scholar]
- 62.Bowen N., Kolodner R. D., Reconstitution of Saccharomyces cerevisiae DNA polymerase ε-dependent mismatch repair with purified proteins. Proc. Natl. Acad. Sci. U.S.A. 114, 3607–3612 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bowen N., et al. , Reconstitution of long and short patch mismatch repair reactions using Saccharomyces cerevisiae proteins. Proc. Natl. Acad. Sci. U.S.A. 110, 18472–18477 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Antony E., Hingorani M. M., Mismatch recognition-coupled stabilization of Msh2-Msh6 in an ATP-bound state at the initiation of DNA repair. Biochemistry 42, 7682–7693 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fien K., Stillman B., Identification of replication factor C from Saccharomyces cerevisiae: A component of the leading-strand DNA replication complex. Mol. Cell. Biol. 12, 155–163 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Georgescu R. E., et al. , Mechanism of asymmetric polymerase assembly at the eukaryotic replication fork. Nat. Struct. Mol. Biol. 21, 664–670 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gomes X. V., Gary S. L., Burgers P. M., Overproduction in Escherichia coli and characterization of yeast replication factor C lacking the ligase homology domain. J. Biol. Chem. 275, 14541–14549 (2000). [DOI] [PubMed] [Google Scholar]
- 68.Nakagawa T., Flores-Rozas H., Kolodner R. D., The MER3 helicase involved in meiotic crossing over is stimulated by single-stranded DNA-binding proteins and unwinds DNA in the 3′ to 5′ direction. J. Biol. Chem. 276, 31487–31493 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Goellner E. M., et al. , Identification of Exo1-Msh2 interaction motifs in DNA mismatch repair and new Msh2-binding partners. Nat. Struct. Mol. Biol. 25, 650–659 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.W. J. Graham, 5th, Putnam C. D., Kolodner R. D., The properties of Msh2-Msh6 ATP binding mutants suggest a signal amplification mechanism in DNA mismatch repair. J. Biol. Chem. 293, 18055–18070 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schrodinger L. (2015) The PyMOL molecular graphics system. https://pymol.org/2/.
- 72.Erdős G., Pajkos M., Dosztányi Z., IUPred3: Prediction of protein disorder enhanced with unambiguous experimental annotation and visualization of evolutionary conservation. Nucleic Acids Res. 49, W297–W303 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thomsen M. C., Nielsen M., Seq2Logo: A method for construction and visualization of amino acid binding motifs and sequence profiles including sequence weighting, pseudo counts and two-sided representation of amino acid enrichment and depletion. Nucleic Acids Res. 40, W281–W287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included in the manuscript and/or SI Appendix.