Abstract
Many genetic risk factors for Parkinson’s disease have lipid-related functions and lipid-modulating drugs such as statins may be protective against Parkinson’s disease. Moreover, the hallmark Parkinson’s disease pathological protein, α-synuclein, has lipid membrane function and pathways dysregulated in Parkinson’s disease such as the endosome–lysosome system and synaptic signalling rely heavily on lipid dynamics. Despite the potential role for lipids in Parkinson’s disease, most research to date has been protein-centric, with large-scale, untargeted serum and CSF lipidomic comparisons between genetic and idiopathic Parkinson’s disease and neurotypical controls limited. In particular, the extent to which lipid dysregulation occurs in mutation carriers of one of the most common Parkinson’s disease risk genes, LRRK2, is unclear. Further, the functional lipid pathways potentially dysregulated in idiopathic and LRRK2 mutation Parkinson’s disease are underexplored.
To better determine the extent of lipid dysregulation in Parkinson’s disease, untargeted high-performance liquid chromatography–tandem mass spectrometry was performed on serum (n = 221) and CSF (n = 88) obtained from a multi-ethnic population from the Michael J. Fox Foundation LRRK2 Clinical Cohort Consortium. The cohort consisted of controls, asymptomatic LRRK2 G2019S carriers, LRRK2 G2019S carriers with Parkinson’s disease and Parkinson’s disease patients without a LRRK2 mutation. Age and sex were adjusted for in analyses where appropriate. Approximately 1000 serum lipid species per participant were analysed.
The main serum lipids that distinguished both Parkinson’s disease patients and LRRK2 mutation carriers from controls included species of ceramide, triacylglycerol, sphingomyelin, acylcarnitine, phosphatidylcholine and lysophosphatidylethanolamine. Significant alterations in sphingolipids and glycerolipids were also reflected in Parkinson’s disease and LRRK2 mutation carrier CSF, although no correlations were observed between lipids identified in both serum and CSF. Pathway analysis of altered lipid species indicated that sphingolipid metabolism, insulin signalling and mitochondrial function were the major metabolic pathways dysregulated in Parkinson’s disease. Importantly, these pathways were also found to be dysregulated in serum samples from a second Parkinson’s disease cohort (n = 315).
Results from this study demonstrate that dysregulated lipids in Parkinson’s disease generally, and in LRRK2 mutation carriers, are from functionally and metabolically related pathways. These findings provide new insight into the extent of lipid dysfunction in Parkinson’s disease and therapeutics manipulating these pathways may be beneficial for Parkinson’s disease patients. Moreover, serum lipid profiles may be novel biomarkers for both genetic and idiopathic Parkinson’s disease.
Keywords: Parkinson’s disease, lipid, sphingolipid, LRRK2, biomarker
Galper et al. use an unbiased lipidomic approach to quantify thousands of lipids in serum and CSF from two large Parkinson’s disease cohorts. The results reveal altered lipid profiles that converge on major metabolic pathways, providing new insights into the extent of lipid dysfunction in Parkinson’s disease.
Introduction
Parkinson’s disease is a progressive neurodegenerative disorder involving the loss of dopamine-producing neurons in the brain, in conjunction with the pathological accumulation of α-synuclein protein in remaining neurons. The exact cause of Parkinson’s disease is unknown; however, genetic studies suggest a disease of complex aetiology resulting from the interplay of many genetic factors and their environmental interactions. Large-scale sequencing studies of Parkinson’s disease patients have identified several risk genes, including those with known lipid-related functions.1–10 In particular, missense mutations in GBA1, which encodes the lipid-metabolizing lysosomal hydrolase glucocerebrosidase, are the most common genetic factors that increase Parkinson’s disease risk.1,4,6,11–14 Polymorphisms in GALC (encoding galactosylceramidase) and ASAH1 (encoding acid ceramidase) and mutations in SMPD1 (encoding acid-sphingomyelinase) have also been linked to Parkinson’s disease risk, and like GBA1, encode lysosome enzymes that catabolize sphingolipids.2,3,7 Additional lipid-related Parkinson’s disease risk genes include SREBF1 (encoding sterol regulatory element binding transcription factor 1)8 that regulates sterol biosynthesis important in cell membrane maintenance15 and DGKQ (encoding diacylglycerol kinase theta)5,9,16,17 that mediates the regeneration of phosphatidylinositol from diacylglycerol important in synaptic vesicle formation.18,19 Thus, Parkinson’s disease genetics suggests that dysregulation of lipid homeostasis may contribute to the development of disease. However, the functional impact of genetic variation in lipid-associated enzymes and how this contributes to Parkinson’s disease risk remains to be determined.
Apart from their well-known roles in membrane structure, many lipids act as signalling molecules and important regulators of membrane function, allowing for the appropriate curvature and fluidity required for critical cellular processes such as the synaptic vesicle cycle, the endosome–lysosome system and phagocytosis.20 In addition, decreases in lipid substrate catabolism in the lysosome interfere with aspects of lysosomal function critical for the clearance of neurotoxic proteins, such as α-synuclein. Moreover, α-synuclein binds to lipid membranes, altering their structure and function, and lipid binding to α-synuclein monomers can result in the formation and stabilization of toxic α-synuclein oligomers.21 Therefore, alterations in lipid species due to dysregulation of lipid-metabolizing enzymes may directly promote Parkinson’s disease pathology. Moreover, meta-analyses of targeted lipid studies have indicated that higher total serum triacylglycerol and cholesterol were protective against Parkinson’s disease risk,22 or were higher in controls compared to Parkinson’s disease.23 Such results suggest widespread lipid alterations in Parkinson’s disease; however, further characterization of the collective Parkinson’s disease lipidome is required to better understand which lipids and their metabolic pathways may be involved in Parkinson’s disease.
Another established Parkinson’s disease risk gene is leucine-rich repeat kinase 2 (LRRK2).24,25 Missense mutations in LRRK2 increase the enzyme’s activity and although the biological function of LRRK2 remains to be fully elucidated, studies in cell and animal models have implicated LRRK2 in lysosomal function,26,27 driving substantial interest in the development of small molecule inhibitors of LRRK2 as potential Parkinson’s disease therapeutics.28,29 Lysosomal stress induced by LRRK2 mutations leads to increased levels of the phospholipid di-22:6 bis(monoacylglycerol)phosphate (BMP), which is excreted in the urine. Levels of BMP in the urine are regulated by LRRK2 kinase activity and thus may constitute a pharmacological biomarker in LRRK2 inhibitor clinical trials.30,31 Moreover, in differentiated human dopamine neurons and astrocyte models, LRRK2 inversely regulates glucocerebrosidase activity, implicating LRRK2 in the same sphingolipid metabolism pathways as glucocerebrosidase.32,33 Furthermore, altered ceramide metabolism has been observed in the brains of LRRK2 knockout mice.34 To date, the extent to which lipid dysregulation occurs in general in idiopathic Parkinson’s disease patients is unknown, and to be able to specifically compare lipid dysregulation in idiopathic Parkinson’s disease to Parkinson’s disease patients with LRRK2 mutations may prove highly informative.
To further determine the extent to which lipid alterations are present in Parkinson’s disease patients, a comprehensive untargeted lipidomic analysis was performed using serum and CSF from large multisite cohorts of Parkinson’s disease patients and matched controls with and without the common LRRK2 G2019S mutation. We hypothesized that altered lipid profiles would be evident in both Parkinson’s disease patients with and without the LRRK2 G2019S mutation, and that pathway analysis would identify the most critical pathways impacted by these changes in patients with Parkinson’s disease. Such findings may have implications for understanding disease pathogenesis and potential therapeutic pathways, as well as providing promising avenues for biomarker development.
Materials and methods
Patient samples and clinical data
Patient samples and matching clinical data were obtained from the MJFF LRRK2 Cohort Consortium, which is coordinated and funded by the Michael J. Fox Foundation for Parkinson’s disease research. Patient samples were collected with informed consent in accordance with the Declaration of Helsinki and the study was approved by the University of Sydney Human Research Ethics Committee (2017/076 and 2017/857). For further information on the LRRK2 cohort study, visit https://www.michaeljfox.org. For the first analysis, serum and CSF samples were obtained from the LRRK2 multi-ethnic cross-sectional cohort and consisted of controls (n = 63 serum and n = 20 CSF), LRRK2 G2019S carriers without Parkinson’s disease (n = 56 serum and n = 20 CSF), LRRK2 G2019S carriers diagnosed with Parkinson’s disease (n = 65 serum and n = 19 CSF) and Parkinson’s disease patients without a known LRRK2 mutation (n = 37 serum and n = 29 CSF). For the second analysis, serum samples were obtained from the LRRK2 Ashkenazi Jewish cohort and consisted of controls (n = 77), LRRK2 G2019S carriers without Parkinson’s disease (n = 80), LRRK2 G2019S carriers diagnosed with Parkinson’s disease (n = 79) and Parkinson’s disease patients without a known LRRK2 mutation (n = 79). During sample selection the following exclusion criteria were applied: history of repeated head injury, definite encephalitis, cerebral tumour, MPTP exposure, stroke, epilepsy, inflammatory disease of the brain and skull fractures. All participants were fasted prior to sample collection with procedures standardized for collection between 8 and 10 a.m. in a fasted state (i.e. a minimum of 8 h fasting), or if fasting was not possible then participants were to eat a low-lipid meal. The following associated clinical data were also obtained: age, sex, date of diagnosis, Movement Disorder Society Unified Parkinson’s Disease Rating Scale part three (UPDRS III), Montreal Cognitive Assessment (MoCA), Epworth Sleep Scale, Geriatric Depression Scale, University of Pennsylvania Smell Identification Test (UPSIT), Rapid Eye Movement (REM) Sleep Disorder Questionnaire, Scales for Outcomes in Parkinson’s Disease—Autonomic dysfunction (SCOPA-AUT) and the modified Schwab and England Activities of Daily Living Scale. Blood pressure, regular ibuprofen, regular aspirin and the use of other inflammatory medication were recorded and were similar across groups in the multi-ethnic cohort (the use of these medications was not available for the Ashkenazi Jewish cohort). Specific details regarding the medications or doses used were not available. Demographic and clinical data accompanying the samples are available in Supplementary Table 1 (multi-ethnic cohort) and Supplementary Table 2 (Ashkenazi Jewish cohort).
Chemicals and materials
All solvents used were high-performance liquid chromatography grade or higher. Lipid internal standards were purchased from Avanti Polar Lipids and included phosphatidylcholine (PC, 19:0), sphingomyelin (SM, 12:0), phosphatidylethanolamine (PE, 17:0), phosphatidylglycerol (PG, 17:0), phosphatidylserine (PS, 17:0), phosphatidic acid (PA, 17:0), ceramide (Cer, d18:1/12:0), diacylglycerol (DG, 1,3 18:0 d5), cholesteryl ester (ChE, 19:0), monoacy1glycerol (MG, 17:0), triacylglycerol (TG) mix d5 (Avanti Code LM-6000), DG mix d5 (Avanti Code LM-6001), phosphatidylinositol (PI, 17:0/14:1), C12 GluCer, C12 sulphatide, C17 Cer, C17 sphingosine (So), C17 S1P, C12 C1P, D3 C20 fatty acid and C12 lactosylceramide. Lipid internal standards were prepared at 5 pmol/μl in methyl-tert butyl ether and methanol (MTBE:methanol, 1:1 v/v).
Lipid extraction
CSF and serum samples were processed in batches comprising an equal number of samples from each group due to the large cohort size. A reference sample, generated by pooling plasma from three healthy blood donors to the Australian Red Cross Blood Service, was included in duplicate in every third batch of lipid extractions to monitor batch variation. Blank samples were also included in every batch in duplicate to monitor background signal. Samples were thawed on ice and centrifuged at 14 000g for 15 min at 4°C. An aliquot of 80 µl of sample was mixed with 10 µl of the internal standard in a glass tube. Lipid extraction was based on the method by Bligh and Dyer.35 Briefly, methanol (Sigma), chloroform (Sigma) and ultrapure water (Millipore) were added sequentially and vortexed each time. Samples were then centrifuged at 1693g at 20°C for 10 min (acceleration = 9, deceleration = 5). The lower (solvent) phase was collected and transferred into a new glass tube using a glass Pasteur pipette. Chloroform was added, vortexed and centrifuged as before. The lower phase was collected and transferred into a new glass tube and dried under nitrogen gas. The dried lipid samples were reconstituted in 100 µl of isopropanol/methanol (1:1) and were stored at −80°C.
Liquid chromatography–tandem mass spectrometry
The Liquid chromatography–tandem mass spectrometry (LC-MS/MS) protocol has been previously described.36 Briefly, lipid extract (10 µl) was analysed using a Q-Exactive Plus Mass Spectrometer coupled to a U3000 UPLC system (Thermo Fisher Scientific). Chromatography was performed at 60°C on a Waters CSH C18 ulta-high-performace liquid chromatography column 2.1 × 100 mm, 1.8 µM with VanGuard guard column. Solvent A was 6:4 acetonitrile:water and Solvent B was 1:9 acetonitrile:isopropanol, both with 10 mM ammonium formate and 0.1% formic acid. Lipids were chromatographed according to the method of Castro-Perez et al.37 Briefly, a 30 min gradient running from 30 to 100% of solvent B was performed, eluting lipids in order of hydrophobicity. Column eluate was directed into the electrospray ionization (ESI) source of the mass spectrometer where a heated-ESI probe was employed. Source parameters were broadly optimized on a range of lipid standards prior to the analysis. The mass spectrometer was run in data-dependent acquisition mode. A survey scan over the mass range 200–1200 at resolution 70 K was followed by 10 data-dependent MS/MS scans on the most intense ions in the survey at 15 K resolution. Dynamic exclusion was used to improve the number of ions targeted. Samples were run in both positive and negative polarities. The mass spectrometry data acquisition and downstream analysis were performed blinded.
Lipid identification and quantification
MS data were searched against the standard LipidSearch database 4.2.23 with all common mammalian lipid classes included. Prior to LipidSearch version 4.2.23, BMP (also known as lysobisphosphatidic acid or LBPA) was not part of the searchable database within the software. To add BMP to the LipidSearch database, and consequently allow detection in samples, BMP 14:0 standard (Avanti Polar Lipids) was analysed by LC-MS/MS. As BMP and PG are isomers, PG 14:0 standard was additionally run to confirm that the data allows differentiation of BMP and PG. MS/MS scan data were sent to the LipidSearch software developers, who added BMP to the LipidSearch database. Identified lipids were normalized between samples, to correct for batch effects from the sample preparation and the LC-MS/MS analysis, using the internal standards of the same lipid category (glycerolipids, glycerophospholipids, sterol lipids, fatty acyls, sphingolipids and prenol lipids), or when not available, the average of internal standards from the corresponding ion adduct. In the event LipidSearch returned more than one of the same lipid ID, area values from each identical ID were assumed to be from the same lipid (due to software integration errors for example) or indistinguishable lipid isomers of the same class, and these were summed together.
Statistical analysis
Data analysis was performed using SPSS and R. Clinical variables across groups were assessed by one-way ANOVA with least-significant difference post hoc with the exceptions that Chi-squared was performed for comparisons of sex and Kruskal–Wallis with Mann–Whitney post hoc was performed for comparisons of medication use. Extracted lipid peak areas were square-root transformed to better meet the assumptions of normality. Peak areas that were greater than five standard deviations from the group mean were considered outliers and removed. Consequently, 2210 of 554876 data points (0.4% of the dataset) were identified as outliers. Reference samples were used to calculate the coefficient of variation between lipids identified across batches. To ensure only robustly detected lipids were included, lipid species that had >10% missing data were also excluded from analyses. The remaining missing lipid species data were considered missing at random and imputed using the group mean. Missing data for any given remaining lipid species were less than 1%, with only a few exceptions. Clinical data were not imputed. Due to software limitations and the large number of samples within the analysis, the Ashkenazi Jewish cohort required processing by LipidSearch in two batches (although it was processed by the mass spectrometer in one run). Batch was therefore included as an additional covariate in analyses of the Ashkenazi Jewish cohort. Following transformation, and removal of outliers and missing data, principal component analysis (PCA) was performed using centred and scaled data with the prcomp function in R. Following PCA, three participants—a control, an LRRK2 carrier without Parkinson’s disease and an LRRK2 carrier with Parkinson’s disease—clearly separated from the cohort majority and it was concluded that these individuals likely have an unrelated condition affecting lipids and they were therefore removed from further analysis. PCA-transformed variables were then used for linear discriminant analysis (LDA). LDA was performed using SPSS, with the within-groups covariance matrix and leave-one-out cross-validation. The assumptions for LDA (linearity, no outliers, independence, no multicollinearity, similar spread and normality) were met. Two-factor multivariate ANOVA, with age and sex included as covariates where indicated, was used to identify significantly altered principal components. To report directions of change, two-factor multivariate ANOVA was performed for individual lipid subclasses, with age and sex included as covariates and serum triacylglycerol species grouped by carbon/double bond number. Significance was accepted at P < 0.05 except when Bonferroni corrections for multiple comparisons were applied as indicated.
Pathway analysis
To understand the potential functional significance of lipids differing between Parkinson’s disease patients and/or LRRK2 G2019S carriers and controls, lipids which were significantly altered by Parkinson’s disease status or the LRRK2 mutation, as indicated in Figs 1G–J, 2E–H, 3G–J and 4E–H, were entered into an online compound and pathway database38 (https://www.genome.jp/kegg/compound/) and a list of relevant pathways were returned for each lipid. Lipids identified as differing in CSF, as indicated in Fig. 5, were also included. Pathways that contained at least two of the above lipids were reported. Glucosylceramide and galactosylceramide were entered as the search terms for monohexosylceramide (Hex1Cer) as the data obtained from LC-MS/MS do not allow these to be discriminated. Campesterol ester (CmE), dihexosyl N-acetylhexosyl ceramide (CerG2NAc1), acyl hexosyl zymosterol ester (AcHexZyE), acyl hexosyl cholesterol ester (AcHexChE), LBPA and phosphatidylethanol (PEt) were not part of the pathway database, so they were not included in the search.
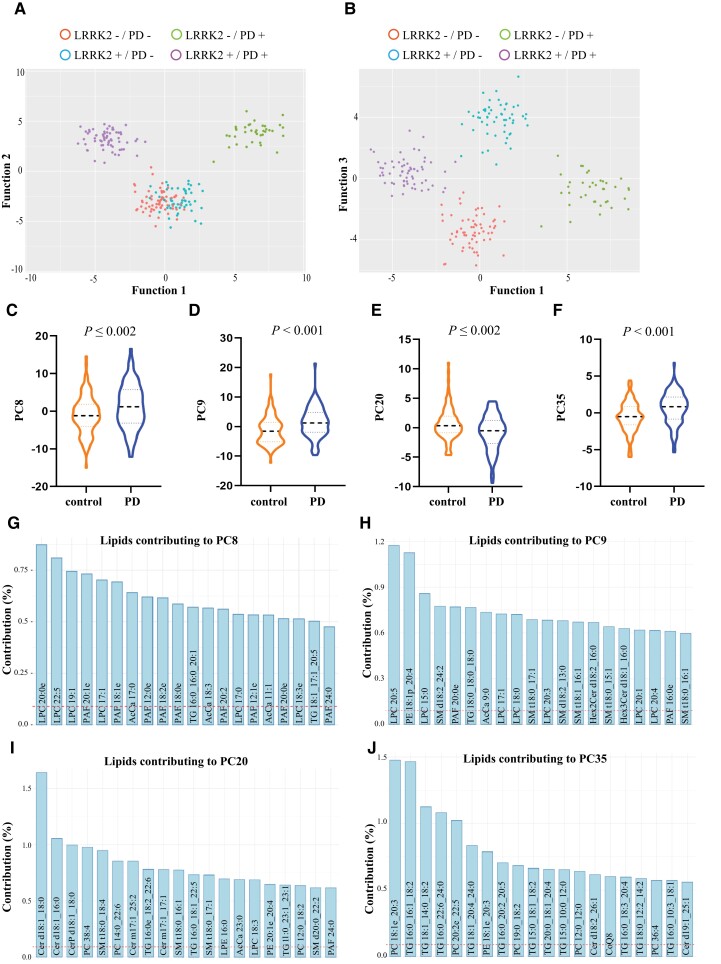
Figure 1.
Serum lipid profiles discriminate Parkinson’s disease patients from controls in a multi-ethnic cohort. Using linear discriminant analysis (LDA) on identified principal components, three canonical functions were generated that could significantly discriminate the groups. (A) A scatterplot of the first two functions clearly showed separation between the control and Parkinson’s disease groups and between the LRRK2 mutation and non-mutation Parkinson’s disease groups. (B) A scatterplot of the first and third functions shows separation of all four groups. (C) To determine which lipids may contribute to the discrimination of Parkinson’s disease patients from controls independent of LRRK2 G2019S mutation, LDA using identified principal components was performed and the results indicated control (controls plus LRRK2 carriers without Parkinson’s disease) and Parkinson’s disease (LRRK2 and non-LRRK2 Parkinson’s disease) groups were distinguishable. Multivariate analysis of variance covarying for age and sex identified four principal components, (C) PC8, (D) PC9, (E) PC20 and (F) PC35. (G–J) The top 20 lipid species that contributed to the four principal components discriminating Parkinson’s disease from controls. The dashed red line represents the expected value if the contribution of lipids were uniform. n = 221.
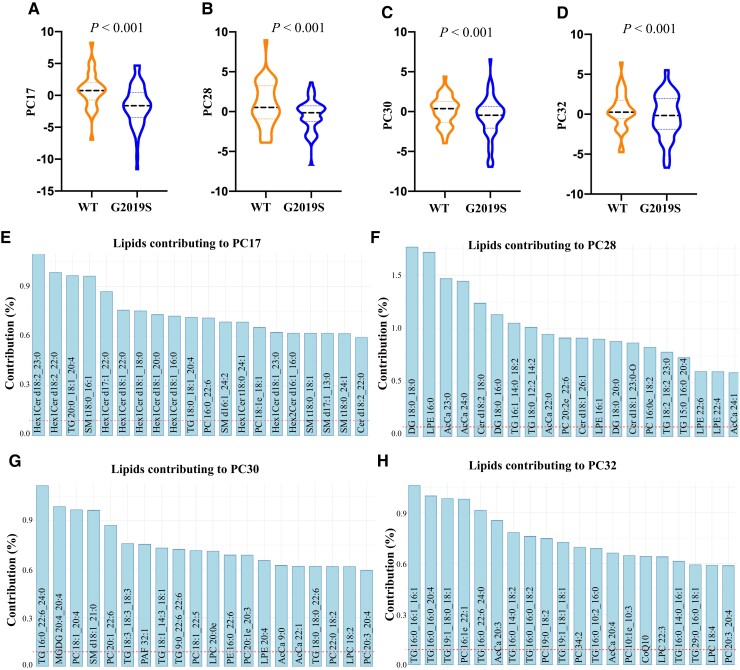
Figure 2.
Serum lipid profiles discriminate LRRK2 G2019S carriers from non-LRRK2 G2019S carriers in a multi-ethnic cohort. To determine which lipids may contribute to the discrimination of LRRK2 G2019S mutation carriers from non-LRRK2 G2019S carriers, linear discriminant analysis (LDA) was performed to identify the principal components that significantly differed between these two groups. The LRRK2 mutation group consisted of both asymptomatic carriers and manifesting Parkinson’s disease patients, while the non-LRRK2 mutation group consisted of control and idiopathic Parkinson’s disease patients. LRRK2 mutation carriers could be significantly discriminated from non-LRRK2 mutation carriers. (A–E) Multivariate ANOVA covarying for age and sex identified four principal components, (B) PC17, (C) PC28, (D) PC30 and (E) PC32, were significantly different between LRRK2 and non-LRRK2 mutation groups. (F–H) The top 20 lipid species that contributed to the four principal components discriminating LRRK2 and non-LRRK2 mutation groups. The dashed red line represents the expected value if the contribution of lipids were uniform. n = 221. WT = wild-type.
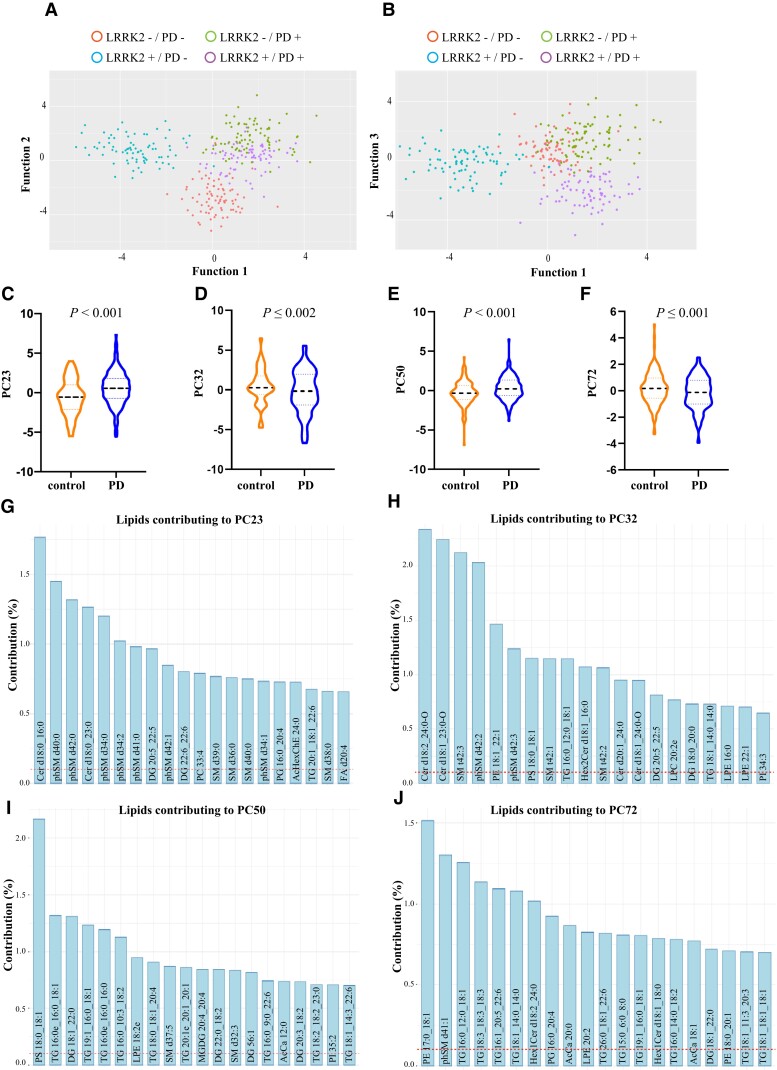
Figure 3.
Serum lipid profiles discriminate Parkinson’s disease patients from controls in the LRRK2 Ashkenazi Jewish cohort. To determine if lipid profiles could distinguish between groups in a second cohort, linear discriminant analysis (LDA) on identified principal components was performed. It was revealed that the three canonical functions generated significantly discriminated the groups. (A) A scatterplot of the first two functions clearly showed separation between the control and Parkinson’s disease groups, and between the LRRK2 mutation and non-mutation Parkinson’s disease groups. (B) A scatterplot of the first and third functions shows separation between Parkinson’s disease groups. To determine lipids that may contribute to the discrimination of Parkinson’s disease patients from controls independent of LRRK2 G2019S mutation, controls (including LRRK2 carriers without Parkinson’s disease) were compared to a Parkinson’s disease group (LRRK2 and non-LRRK2 Parkinson’s disease). (C–F) Multivariate ANOVA covarying for age and sex identified four principal components, (C) PC23, (D) PC32, (E) PC50 and (F) PC72, which remained significantly different between the two groups. (G–J) The top 20 lipid species that contributed to the top four principal components that significantly distinguished between controls and Parkinson’s disease. The dashed red line represents the expected value if the contribution of lipids were uniform. n = 315.
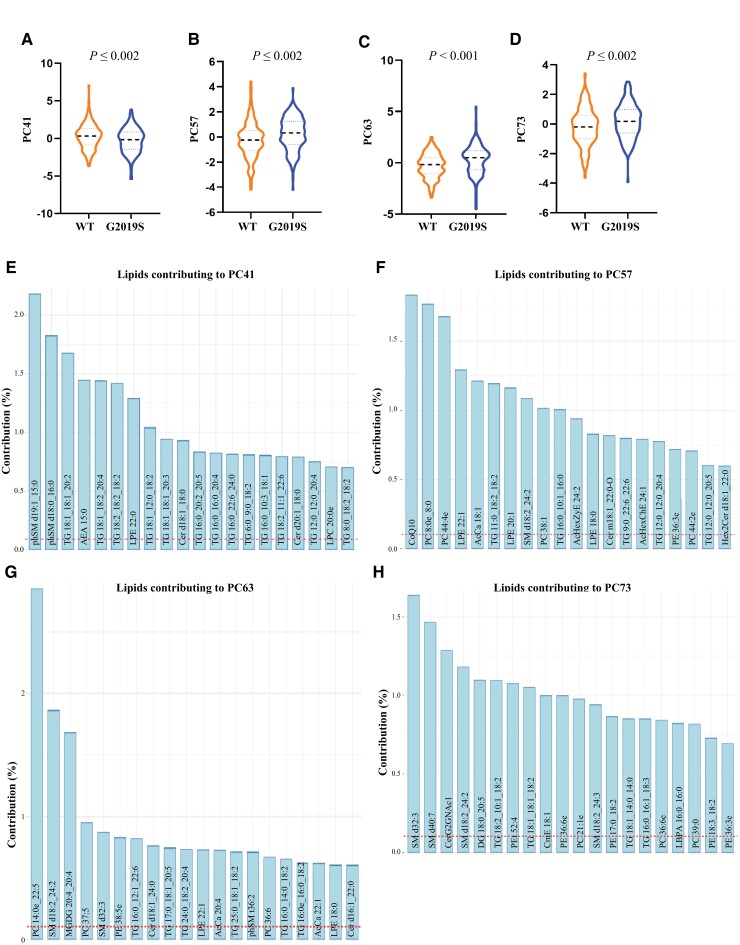
Figure 4.
Serum lipid profiles discriminate LRRK2 G2019S carriers from non-LRRK2 G2019S carriers in the LRRK2 Ashkenazi Jewish Cohort. Multivariate ANOVA covarying for age and sex identified four principal components, (A) PC41, (B) PC57, (C) PC63 and (D) PC73, that were significantly different between the LRRK2 mutation and non-mutation groups. (E–H) The top 20 lipid species that contributed to the top four components which significantly distinguished between LRRK2 mutation and non-mutation groups. The dashed red line represents the expected value if the contribution of lipids were uniform. n = 315. WT = wild-type.
Figure 5.
Sphingolipids and glycerolipids and altered in multi-ethnic LRRK2 cohort CSF. Two-factor multivariate analysis covarying for age and sex revealed a significant effect of both Parkinson’s disease and LRRK2 G2019S mutation (both P < 0.05) on CSF lipid profiles. (A–G) Post hoc analysis identified sphingolipids and glycerolipids were different between Parkinson’s disease patient and control CSF samples. (H–J) Glycerolipid and sphingolipid species were significantly different between LRRK2 G2019S mutation carriers and non-carriers. n = 88. WT = wild-type.
Data availability
The raw mass spectrometry files generated in this study have been provided to the Michael J. Fox Foundation for Parkinson’s disease research and can be requested (https://www.michaeljfox.org/news/lrrk2-cohort-consortium). Other data are available upon reasonable request from the corresponding authors and upon relevant ethical approval.
Results
Multi-ethnic LRRK2 cohort demographics and lipid identification
To determine the effect of Parkinson’s disease and the LRRK2 G2019S mutation on lipid profiles, four groups were initially analysed: controls, Parkinson’s disease patients with the LRRK2 G2019S mutation, Parkinson’s disease patients without the LRRK2 G2019S mutation and LRRK2 G2019S mutation carriers without Parkinson’s disease. Analysis of the demographic data indicated that the LRRK2 carriers without Parkinson’s disease were younger than the other groups, and that the Parkinson’s disease groups had a higher proportion of males (Supplementary Table 1). Age and sex were therefore included as covariates in analyses where appropriate. As expected, Parkinson’s disease patients showed significant symptomology on Parkinson’s disease clinical scales compared to controls, whereas the LRRK2 carriers without Parkinson’s disease were clinically indistinguishable from controls. Parkinson’s disease patients without the LRRK2 G2019S mutation had reduced olfaction and higher UPDRS III scores than LRRK2-Parkinson’s disease patients, whereas LRRK2-Parkinson’s disease patients had higher scores for depression. Using advanced mass spectrometry and the latest LipidSearch software, across the four groups, 31 lipid subclasses comprising 1118 unique lipid species were identified in serum (Supplementary Table 3). The coefficient of variance (CV) of lipids from the reference samples included across the batches of the multiethnic LRRK2 cohort was 20% ± 3%.
Phospholipid and sphingolipid metabolism are altered in Parkinson’s disease patients
To determine if serum lipid profiles could first distinguish groups of interest, PCA was employed as a dimension reduction technique. Examining the PCA eigenvalues revealed 119 principal components with a value >1 that collectively captured 95% of the variance in the dataset. The PCA-transformed variables for these 119 components were therefore extracted and used for subsequent LDA. Using LDA, three canonical functions were generated that could significantly discriminate the groups (Supplementary Table 4) with leave-one-out cross-validation indicating that 87% of cases could be correctly classified. A scatterplot of the first two functions clearly showed separation between the control and Parkinson’s disease groups, and also between the LRRK2 G2019S mutation and non-mutation Parkinson’s disease groups (Fig. 1A). Separation of all four groups was observed when the first and third functions were plotted (Fig. 1B). To identify principal components that were different between Parkinson’s disease patients and LRRK2 G2019S mutation carriers, two-factor multivariate ANOVA covarying for age and sex was employed. Significant effects were observed for Parkinson’s disease (Wilks’ lambda = 0.099, P = 3.3 × 10−20) and the LRRK2 G2019S mutation (Wilks’ lambda = 0.095, P = 8.0 × 10−21). A significant interaction between Parkinson’s disease and the LRRK2 mutation was also observed (Wilks’ lambda = 0.088, P = 2.9 × 10−23). The top four significantly different principal components for the Parkinson’s disease variable were PC8 (Fig. 1C), PC9 (Fig. 1D), PC20 (Fig. 1E) and PC35 (Fig. 1F). Age was not a significant factor in the multivariate analysis for any of these four principal components, while sex was a significant factor for PC9 (P = 0.017) and PC35 (P = 0.002). The top 20 lipid species that contributed to each of these principal components are shown in Fig. 1G–J and predominantly comprised species of ceramide, triacylglycerol, sphingomyelin, acylcarnitine, phosphatidylcholine and lysophosphatidylcholine. Pathway analysis employing the top 20 lipid species identified in Fig. 1G–J indicated that the lipids altered in Parkinson’s disease patients were from sphingolipid and phospholipid metabolism pathways, as well as oxidative phosphorylation/thermogenesis and insulin resistance pathways (Table 1).
Table 1.
Pathways altered in Parkinson’s disease patients and LRRK2 mutation carriers
| ID | Lipids | |
|---|---|---|
| Pathway altered in Parkinson’s disease | ||
| Glycerophospholipid metabolism | [map00564] | LPC, LPE, PC, PE |
| Sphingolipid metabolism | [map00600] | Cer, CerP, SM |
| Oxidative phosphorylation and thermogenesis | [map04714] | AcCa, TG |
| Glycogen production, GLUT4 translocation and glucose uptake (Insulin resistance) | [map04931] | Cer, TG |
| Sphingolipid signalling | [map04071] | Cer, SM |
| Necroptosis | [map04217] | Cer, SM |
| Retrograde endocannabinoid signalling | [map04723] | PC, PE |
| Pathways altered in LRRK2 G2019S mutation carriers | ||
| Glycerophospholipid metabolism | [map00564] | DG, LPC, LPE, PC |
| Sphingolipid metabolism | [map00600] | Cer, Hex1Cer,a SM |
| Oxidative phosphorylation and thermogenesis | [map04714] | AcCa, DG, TG |
| Glycogen production, GLUT4 translocation and glucose uptake (insulin resistance) | [map04931] | Cer, DG, TG |
| Glycerolipid metabolism | [map00561] | DG, MGDG, TG |
| Retrograde endocannabinoid signalling | [map04723] | DG, PC |
| Regulation of lipolysis in adipocytes | [map04923] | DG, TG |
| Fat digestion and absorption | [map04975] | DG, TG |
| Adipocytokine signalling pathway | [map04920] | Cer, DG |
| Necroptosis | [map04217] | Cer, SM |
| Sphingolipid signalling | [map04071] | Cer, SM |
| Neurotrophin signalling pathway | [map04722] | Cer, DG |
| Advanced glycation end products (AGE) signalling | [map04933] | Cer, DG |
Lipid subclasses of interest from the serum analyses were searched in the online KEGG compound/pathway database (Kanehisa et al.38) to identify whether lipids of interest were enriched in certain pathways. Pathways that were shared by at least two lipids are listed above. GLUT4 = Glucose transporter type 4, insulin-responsive. Glycogen production, GLUT4 translocation and glucose uptake are subpathways in an ‘insulin resistance’ pathway. Oxidative phosphorylation and thermogenesis is from ‘thermogenesis’ pathway. AcCa = acylcarnitine; LPE = lysophosphatidylethanolamine; MGDG = monogalactosyldiacylglycerol.
Glucosylceramide and galactosylceramide were entered as the search terms for monohexoyslceramide (Hex1Cer) as the data obtained from LC-MS/MS do not allow these to be discriminated. Glycerophospholipid is synonymous with phospholipid.
Phospholipid, glycerolipid and sphingolipid metabolism are altered in LRRK2 mutation carriers
The effect of the LRRK2 mutation on serum lipid profiles was also further explored. The top four significantly different principal components for the LRRK2 G2019S variable were PC17 (Fig. 2A), PC28 (Fig. 2B), PC30 (Fig. 2C) and PC32 (Fig. 2D). Age and sex were not significant factors for any of these four principal components. The top 20 lipid species that contributed to these principal components are shown in Fig. 2E–H and predominantly comprised species of hexosylceramide (glucosylceramide/galactosylceramide), acylcarnitine, phosphatidylcholine and triacylglycerol. Pathway analysis using the top 20 lipid species from Fig. 2E–H indicated that lipids affected by the LRRK2 G2019S mutation were in phospholipid, sphingolipid and glycerolipid metabolism pathways, as well as oxidative phosphorylation/thermogenesis, insulin resistance and energy/lipolysis pathways (Table 1). Thus, analysis of LRRK2 G2019S mutation carriers, which included asymptomatic carriers, showed overlap with the results of the sporadic Parkinson’s disease patients in terms of the dysregulated pathways identified. A significant interaction between LRRK2 and Parkinson’s disease status also implies a more complex scenario where specific lipid changes are also occurring in individual subgroups. The top four significantly different principal components for the interaction variable were PC6 (Supplementary Fig. 1A), PC19 (Supplementary Fig. 1B), PC38 (Supplementary Fig. 1C) and PC41 (Supplementary Fig. 1D). The top 20 lipid species that contributed to each of these principal components are shown in Supplementary Fig. 1E–H and predominantly comprised species of phosphatidylethanolamine, triacylglycerol, ceramide and phosphatidylcholine.
Phospholipid, glycerolipid and sphingolipid are altered in a second cohort of Parkinson’s patients
To determine the extent to which our findings on dysregulated lipid pathways could be replicated in a second cohort, we also obtained serum samples from the LRRK2 clinical cohort consortium Ashkenazi Jewish cohort. Analysis of the demographic data indicated that the LRRK2 G2019S carriers without Parkinson’s disease were younger than the other groups, and that the Parkinson’s disease patients without a LRRK2 G2019S mutation had a higher proportion of males (Supplementary Table 2). Age and sex were therefore again included as covariates in analyses where appropriate. Again, Parkinson’s disease patients showed significant symptomology on the clinical scales compared to controls, whereas the LRRK2 carriers without Parkinson’s disease were again clinically indistinguishable from controls. Parkinson’s disease patients without the LRRK2 G2019S mutation had reduced olfaction and higher UPDRS III and REM sleep scores than Parkinson’s disease patients with the mutation. Using the exact same mass spectrometry approaches as before, 32 lipid subclasses comprising 978 unique lipid species were identified in the second LRRK2 cohort (Supplementary Table 5). Of these 978 individual lipid species, 50% were also identified in the first cohort. The CV of lipids from the normalized reference pool samples included across the batches of the Ashkenazi Jewish cohort was 18% ± 2%. To determine if lipid profiles could distinguish groups of interest in the second cohort, PCA was again employed as a dimension reduction technique. Examining the PCA eigenvalues revealed 104 principal components with a value >1 that collectively captured 92% of the variance in the data set. The PCA-transformed variables for these 104 components were extracted and used for subsequent LDA, with three canonical functions generated that could significantly discriminate the groups (Supplementary Table 6). Leave-one-out cross-validation indicated that 81% of cases could be correctly classified. A scatterplot of the first two functions showed separation between the control and Parkinson’s disease groups, and also between the LRRK2 mutation and non-mutation Parkinson’s disease groups (Fig. 3A). There was more apparent overlap between the two Parkinson’s disease groups than observed in the previous cohort, although some separation between these groups was observed when the first and third functions were plotted (Fig. 3B). To identify principal components that were different between Parkinson’s disease patients and LRRK2 G2019S mutation carriers, two-factor multivariate ANOVA covarying for age and sex was again employed. Significant effects were observed for Parkinson’s disease (Wilks’ lambda = 0.252, P = 2.9 × 10−27) and the LRRK2 G2019S mutation (Wilks’ lambda = 0.305, P = 2.6 × 10−20). A significant interaction between Parkinson’s disease and the LRRK2 mutation was also observed (Wilks’ lambda = 0.266, P = 3.0 × 10−25). The top four significantly different principal components for the Parkinson’s disease variable were PC23 (Fig. 1C), PC32 (Fig. 1D), PC50 (Fig. 1E) and PC72 (Fig. 1F). Neither age nor sex was a significant factor for either of these two principal components. The top 20 lipid species that contributed to each of these principal components are shown in Fig. 3G–J and predominantly comprise species of ceramide, sphingomyelin and triacylglycerol. Pathway analysis again indicated that lipids altered in Parkinson’s disease patients were from phospholipid, sphingolipid and glycerolipid metabolism pathways, as well as oxidative phosphorylation/thermogenesis and insulin resistance pathways (Table 2). These data corroborated that observed in the first Parkinson’s disease cohort.
Table 2.
Pathways altered in Parkinson’s disease patients and LRRK2 mutation carriers from Ashkenazi Jewish cohort
| ID | Lipids | |
|---|---|---|
| Pathway altered in Parkinson’s disease | ||
| Glycerophospholipid metabolisma | [map00564] | DG, LPE, LPC, PC, PG, PI, PS |
| Oxidative phosphorylation and thermogenesisa | [map04714] | AcCa, DG, FA, TG |
| Glycogen production, GLUT4 translocation and glucose uptake (insulin resistance)a | [map04931] | Cer, DG, FA, TG |
| Glycerolipid metabolism | [map00561] | DG, FA, MGDG, TG |
| Sphingolipid metabolisma | [map00600] | Cer, Hex1Cer, phSM, SM |
| Sphingolipid signallinga | [map04071] | Cer, DG, SM |
| Adipocytokine signalling pathway | [map04920] | Cer, DG, FA |
| Regulation of lipolysis in adipocytes | [map04923] | DG, FA, TG |
| Fat digestion and absorption | [map04975] | DG, FA, TG |
| Necroptosisa | [map04217] | Cer, SM |
| Retrograde endocannabinoid signallinga | [map04723] | DG, PC |
| Neurotrophin signalling pathway | [map04722] | Cer, DG |
| Advanced glycation end products (AGE) signalling | [map04933] | Cer, DG |
| Insulin secretion | [map04911] | DG, FA |
| Vitamin digestion and absorption | [map04977] | FA, TG |
| Cholesterol metabolism | [map04979] | FA, TG |
| Inositol phosphate metabolism | [map00562] | DG, PI |
| Phosphatidylinositol signalling system | [map04070] | DG, PI |
| Glycosylphosphatidylinositol-anchor biosynthesis | [map00563] | PE, PI |
| Pathways altered in LRRK2 G2019S mutation carriers | ||
| Glycerophospholipid metabolisma | [map00564] | DG, LPE, LPC, PC, PE |
| Sphingolipid metabolisma | [map00600] | Cer, phSM, SM |
| Oxidative phosphorylation and thermogenesisa | [map04714] | AcCa, DG, TG |
| Glycogen production, GLUT4 translocation and glucose uptake (insulin resistance)a | [map04931] | Cer, DG, TG |
| Glycerolipid metabolisma | [map00561] | DG, MGDG, TG |
| Retrograde endocannabinoid signallinga | [map04723] | DG, PC, PE |
| Sphingolipid signallinga | [map04071] | Cer, DG, SM |
| Necroptosisa | [map04217] | Cer, SM |
| Regulation of lipolysis in adipocytesa | [map04923] | DG, TG |
| Fat digestion and absorptiona | [map04975] | DG, TG |
| Adipocytokine signalling pathwaya | [map04920] | Cer, DG |
| Neurotrophin signalling pathwaya | [map04722] | Cer, DG |
| Advanced glycation end products (AGE) signallinga | [map04933] | Cer, DG |
Lipid subclasses of interest from the serum analyses were searched in the online KEGG compound/pathway database (Kanehisa et al.38) to identify whether lipids of interest were enriched in certain pathways. Pathways that were shared by at least two lipids are listed above. LPC = lysophosphatidylcholine; FA = fatty acid; phSM = sphingomyelin (phytosphingosine).
Pathway dysregulated in both the multi-ethnic and Ashkenazi Jewish serum cohorts. GLUT4 = Glucose transporter type 4, insulin-responsive. Glycogen production, GLUT4 translocation and glucose uptake are subpathways in an ‘insulin resistance’ pathway. Oxidative phosphorylation and thermogenesis is from ‘thermogenesis’ pathway. Glycerophospholipid is synonymous with phospholipid.
Phospholipid, glycerolipid and sphingolipid are altered in LRRK2 mutation carriers from the second cohort
The effect of the LRRK2 mutation on serum lipid profiles was also explored for the second cohort. The top four significantly different principal components for the LRRK2 G2019S variable were PC41 (Fig. 4A), PC57 (Fig. 4B), PC63 (Fig. 4C) and PC73 (Fig. 4D). Age and sex were not significant factors for this principal component. The top 20 lipid species that contributed to this principal component are shown in Fig. 4E–H and predominantly comprised species of triacylglycerol, phosphatidylcholine and sphingomyelin. Pathway analysis again indicated that lipids affected by the LRRK2 G2019S mutation were in phospholipid, sphingolipid and glycerolipid metabolism pathways, as well as oxidative phosphorylation/thermogenesis and insulin resistance pathways (Table 2), again showing overlap with results from the first cohort. The top four significantly different principal components for the interaction variable were PC63 (Supplementary Fig. 2A), PC73 (Supplementary Fig. 2B), PC77 (Supplementary Fig. 2C) and PC92 (Supplementary Fig. 2D). The top 20 lipid species that contributed to each of these principal components are shown in Fig. 4G and H and Supplementary Fig. 2E and F and predominantly comprised species of triacylglycerol, phosphatidylcholine and sphingomyelin.
Sphingolipids and glycerolipids are altered in CSF
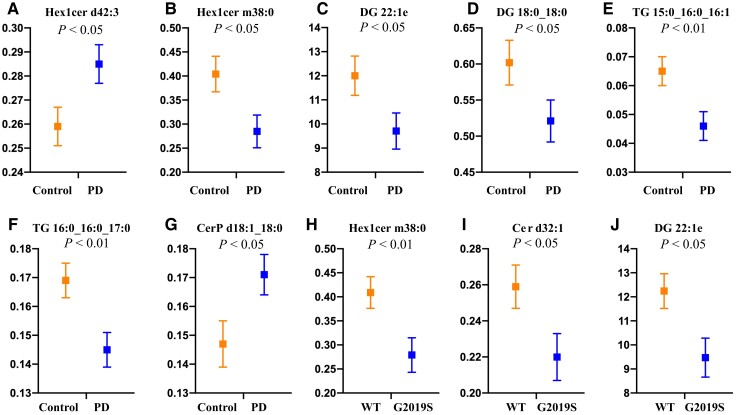
CSF from the multi-ethnic cohort (demographic data in Supplementary Table 7) underwent lipidomics as described above. Sixteen subclasses comprising 185 unique lipid species were identified in CSF (Supplementary Table 8). As the lower sample size available for CSF was not amenable to discriminant analysis, two-factor multivariate analysis was performed instead, covarying for age and sex. This analysis revealed a significant effect of both Parkinson’s disease and LRRK2 G2019S mutation (both P < 0.05) on CSF lipid profiles, whereas a significant interaction between LRRK2 and Parkinson’s disease was not observed (P = 0.23). Post hoc analysis of significantly different variables identified seven lipid species that were different between Parkinson’s disease patient and control CSF samples (Fig. 5A–G). Three lipid species were also different between LRRK2 G2109S mutation carriers, and non-carriers (Fig. 5H–J). Differentiating lipids comprised species of diacylglycerol, triacylglycerol and hexosylceramide with two species, Hex1Cer m38:0 and DG 22:1e, significantly different in both comparisons. Pathway analysis indicated that alterations in sphingolipid metabolism and insulin resistance pathways could also be observed in CSF (Supplementary Table 9). Of the multi-ethnic cohort participants that had a CSF sample analysed, 83 also had a serum sample for which lipidomics had been performed. Of the lipid species found to be significantly different in CSF, Hex1Cer d42:3, DG 18:0_18:0, ceramide phosphate (CerP) d18:1_18:0, Cer d32:1 and TG 16:0_16:0_17:0 were also detected in serum from the same individuals. However, there was no correlation between CSF and serum levels of any of these five lipid species (all P > 0.05), and unlike for CSF, univariate analysis covarying for age and sex indicated that the serum levels of these lipids were not significantly different between those with and without Parkinson’s disease (all P > 0.05).
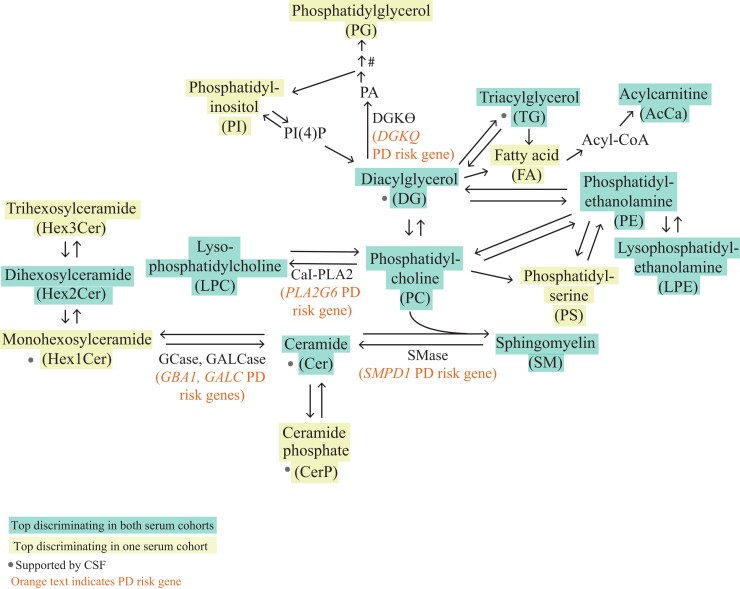
Dysregulated lipids are metabolically linked in Parkinson’s disease risk gene pathways
The top serum lipid classes contributing to the discrimination of Parkinson’s disease or LRRK2 G2019S mutation carriers as shown in Figs. 1–4 were searched in the Kyoto Encyclopedia of Genes and Genomes (KEGG) to generate a summary metabolic map. Lipids in the resulting metabolic map were highly integrated and the metabolic map included enzymes encoded by genes associated with Parkinson’s disease risk (Fig. 6). Lipids identified as significantly different in CSF by multivariate analysis, as shown in Fig. 5, also mapped to the same metabolic pathways (Fig. 6). In particular, alterations in the ceramide and triacylglycerol pathways were found in both cohorts in serum, as well as in CSF from the multi-ethnic cohort. The direction of change of significantly altered lipids from these subclasses are shown in Supplementary Table 10. Of the species significantly altered in Parkinson’s disease in Supplementary Table 10, 40% of the same lipid species were identified in serum from both cohorts, with SM d18:2_24:2 (P = 0.010 multi-ethnic cohort and P = 0.036 Ashkenazi Jewish cohort), PC 32:1e (P = 0.029 multi-ethnic cohort and P = 0.004 Ashkenazi Jewish cohort) and PC 42:6e (P = 0.005 multi-ethnic cohort and P = 0.048 Ashkenazi Jewish cohort) being significantly increased in Parkinson’s disease patient serum and Cer d18:1_24:0-O (P = 0.001 multi-ethnic cohort and P = 0.010 Ashkenazi Jewish cohort), Cer d18:2_24:0-O (P < 0.001 multi-ethnic cohort and P = 0.007 Ashkenazi Jewish cohort) and lysophosphatidylcholine (LPC) 19:1 (P = 0.002 multi-ethnic cohort and P = 0.041 Ashkenazi Jewish cohort) being significantly decreased in Parkinson’s disease patient serum. For lipid species significantly altered in G2019S mutation carriers, 38% were identified in serum from both cohorts with TG 19:1_18:1_18:2 (P = 0.041 multi-ethnic cohort and P = 0.004 Ashkenazi Jewish cohort), TG 19:1_18:2_18:2 (P = 0.019 multi-ethnic cohort and P = 0.002 Ashkenazi Jewish cohort) and TG 18:0_18:1_22:5 P = 0.020 multi-ethnic cohort and P = 0.037 Ashkenazi Jewish cohort) all being significantly decreased in G2019S mutation carriers.
Figure 6.
Implicated lipids are metabolically linked in Parkinson’s disease risk gene pathways. Pathway analysis was performed to generate a metabolic map of the main lipid classes implicated in the discrimination of Parkinson’s disease patients and LRRK2 carriers from controls. The resulting map indicated that most implicated lipids were highly integrated and were in metabolic pathways that included enzymes encoded by Parkinson’s disease risk genes (GBA1, GALC, SMPD1, PLA2G6 and DGKQ). GCase = glucocerebrosidase; GALCase = galactoceramidase; SMase = sphingomyelinase; CaI-PLA2 = 85/88 kDa calcium-independent phospholipase A2; PA = phosphatidic acid; PI(4)P = phosphatidylinositol-4-phosphate. #PG is formed from PA in a three-step reaction pathway.
Discussion
The aim of this study was to use an untargeted lipidomic approach to determine the extent to which serum and CSF lipid profiles were altered in Parkinson’s disease patients or carriers of the LRRK2 G2019S mutation and identify the main lipid pathways affected. Two large and well-characterized cohorts were employed with ∼1000 serum lipids identified for each participant. For both cohorts, the identified serum lipids could significantly discriminate Parkinson’s disease patients from matched controls, as well as carriers of the LRRK2 G2019S mutation from non-mutation carrier participants, adjusting for age and sex. Pathway analysis of Parkinson’s disease patient serum identified sphingolipid and glycerophospholipid metabolism as well as insulin resistance and oxidative phosphorylation/thermogenesis as being in the top pathways dysregulated across the two cohorts. Dysregulation of sphingolipid metabolism and insulin resistance was also observed in Parkinson’s disease and LRRK2 mutation carrier CSF samples. These data support and validate metabolomic studies that found significant alterations in species of blood phosphatidylcholine,39–44 acylcarnitine,44–49 diacylglycerol,39,40,42,50 sphingomyelin,39–42,44,51 hexosylceramide,42,52,53 ceramide,39,40,42,52,53 lysophosphatidylethanolamine,51 triacylglycerol39,40,42 and phosphatidylethanolamine39–42,54 in Parkinson’s disease patients compared to controls. Importantly, our untargeted lipidomics approach has revealed that the majority of lipids affected by Parkinson’s disease or the LRRK2 mutation were in closely linked metabolic pathways and these pathways coincided with known Parkinson’s disease risk genes encoding enzymes that regulate lipid biosynthesis and activity.
Our results also revealed that lipids significantly discriminated Parkinson’s disease patients from controls potentially providing a basis for developing objective diagnostic biomarkers for Parkinson’s disease. Such lipid changes are also potential pharmacodynamic biomarker candidates to explore in LRRK2 inhibition studies, or LRRK2 targeting clinical trials. In particular, hexosylceramide, ceramide and sphingomyelin are present in the lysosome and are known to be affected by mutations in risk genes encoding the lysosomal enzymes glucocerebrosidase, galactosylceramidase and acid-sphingomyelinase. Recent studies using animal and induced pluripotent stem cell models indicate that glucocerebrosidase may be regulated by LRRK2,32–34 consistent with the changes in these serum and CSF sphingolipids in the LRRK2 G2019S mutation carriers in this study. Another lysosomal lipid is BMP (or LBPA), which is excreted in urine in response to lysosomal stress and regulated by LRRK2.30 Although no changes in BMP were found in CSF in this study, serum BMP was involved in the discrimination of LRRK2 mutation carriers from non-carriers, and an interaction between LRRK2 and Parkinon's disease status. However, the number of BMP lipids detected was very low (and undetected in serum from the multi-ethnic cohort) and did not include the previously reported LRRK2 regulated BMP di-22:6. Therefore, further optimization of this lipid subclass from serum and CSF may be required to clarify potential changes in LRRK2 G2019S patients, or perhaps changes in BMP di-22:6 are just more easily observed in urine.
It is of interest that the lipid differences observed in Parkinson’s disease were also found in LRRK2 mutation carriers in general, indicating that changes in serum lipid species may occur early in prodromal Parkinson’s disease. Further exploration of when such lipid changes occur in asymptomatic LRRK2 mutation carriers and if further changes occur with disease onset are warranted. Of course, LRRK2 mutation carriers may develop lipid alterations independent of Parkinson’s disease, and thus an examination of lipid changes in other ‘at-risk’ cohorts, such as patients with idiopathic REM sleep behaviour disorder,55 would identify any common underlying lipid dysfunction in prodromal Parkinson’s disease. Furthermore, the finding that the same lipid pathways were identified in both idiopathic and LRRK2-associated Parkinson’s disease also agrees with clinical data indicating that these two forms of the disease are largely clinically indistinguishable.
As well as lysosomal function, the functional pathways sphingolipids are implicated in include insulin/glucose signalling (glycogen production, GLUT4 translocation and glucose uptake in the insulin resistance pathway, the advanced glycation end products - receptor for advanced glycarion end products (AGE-RAGE) signalling pathway) and those related to cell death or survival (AGE-RAGE signalling pathway, necroptosis, sphingolipid signalling and neurotrophin signalling pathway). Serum and CSF glycerolipids, which were significantly altered in Parkinson’s disease patients and LRRK2 mutation carriers in this study, are also involved in the above cell death or survival pathways and insulin/glucose signalling pathways (insulin resistance, adipocytokine signalling, insulin secretion and AGE-RAGE signalling pathway), as well as pathways relating to energy/mitochondrial ATP production (oxidative phosphorylation and thermogenesis, retrograde endocannabinoid signalling, regulation of lipolysis in adipocytes, fat digestion and absorption and adipocytokine signalling pathway). The link between lipid dysregulation in Parkinson’s disease and metabolic pathway function uncovered in our current study may be important. Meta-analyses of epidemiological studies have found that overall, there is a protective effect against Parkinson’s disease with the use of lipid-lowering statins.56–58 In addition, a recent population study with >100 000 type 2 diabetes patients found that the incidence of Parkinson’s disease was significantly lower with the use of specific diabetes drug classes [glucagon-like peptide-1 (GLP-1) receptor agonists and inhibitors of the GLP-1 degrading enzyme dipeptidyl peptidase 4 (DPP-4)].59 Although the protective mechanism against Parkinson’s disease is unclear, statins work by inhibiting cholesterol biosynthesis60 and GLP-1 and DPP-4 inhibitors are prescribed to regulate glucose levels in diabetes patients via promoting the secretion of insulin, and also appear to lower blood cholesterol. However, both statins and diabetes drugs also lower other lipids such as triacylglycerol.60–63 The unprecedented protective effect of statins and diabetic drugs have spurred considerable interest as to whether triacylglycerol and cholesterol play a role in Parkinson’s disease, with studies measuring total serum triacylglycerol and cholesterol levels as is standard practice for the cardiovascular disease field. However, the results from this have been inconsistent with two recent meta-analyses indicating that higher total serum triacylglycerol and cholesterol were protective against Parkinson’s disease risk22 or were higher levels in controls compared to Parkinson’s disease,23 while another meta-analysis published in the same year concluded total triacylglycerol and cholesterol have no effect on risk.64 These studies may suggest that more targeted therapeutics may have more consistent results.
A drawback of using total levels of lipid subclasses is that information on individual lipid species within the subclass with different saturation and chain length is lost, properties that may have critical influences on lipid functionality. A potential explanation for the inconsistencies previously reported may be that only certain lipid species within a subclass are altered, and this may be masked when total lipid levels are calculated. Our study utilized ultra-high-performance LC-MS/MS to provide a higher resolution of lipid biology to address these potential issues and provide data for future targeting of the lipid mechanisms involved. In this study, specific triacylglycerol species were altered in serum, and notably, triacylglycerol with a 16:0 acyl chain (palmitic acid), were altered in Parkinson’s disease serum and CSF. Interestingly, 16:0 fatty acid has been shown to induce insulin resistance or the blocking of insulin-stimulation of glycogen synthesis causing the accrual of diacylglycerol and ceramide,65–67 two lipids also involved in insulin signalling in the current study pathway analysis. Notably, ceramide was apparent in necroptotic and apoptotic (‘sphingolipid signalling’) pathways, suggesting the possibility that increased 16:0 fatty acids sourced from triacylglycerols cause the accumulation of ceramide, which is a known cell death pathway lipid. Indeed, 16:0 fatty acid has been shown to increase reactive oxygen species and induce mitochondrial DNA damage and apoptosis in muscle cells.65,68 Further work is required to determine the source of the altered lipids identified, and which cell or tissue types are affected and to what extent this may modulate Parkinson’s disease risk.
Our study was designed to provide an unbiased overview of lipid pathway dysregulation in Parkinson’s disease; however, limitations to this approach should be recognized. In particular, our lipidomic study employed relative quantification, with more precise quantification of individual lipid species requiring specific standards for each species identified. Employing a targeted approach to specifically measure individual species across the main lipid pathways identified will also allow for more precise replication studies to be performed across different cohorts, as only 50% of serum lipids identified were present in both cohorts using our untargeted lipidomics approach. However, it may be equally important that similar lipids were altered in both cohorts. For example, a long chain lipid with 20 carbons may or may not be functionally different from a lipid in the same class with 22 carbons. The functional implications of lipids with different chain lengths and saturation is an area of further development in the lipidomics field. While there are some reports of specific fatty acids such as 16:0 eliciting functional consequences, much of the functional implications of slight differences in carbon number or saturation are not completely understood, especially in the context of Parkinson’s disease. Similarly, although CSF and serum from LRRK2 mutation carriers and Parkinson’s disease patients exhibited significant glycerolipid and sphingolipid alterations, CSF was observed to have altered species (of specific chain length and double bond numbers) that were either not detected or altered in serum. This may suggest that the differences found in CSF and serum affect multiple lipids on the subclass level, or alternatively, that species variation may arise from tissue specificity of lipids. Targeted lipidomics tailored to capture even more species from each subclass may help address this. In addition, the stability of lipid changes longitudinally needs to be determined and interpretation of CSF results should be cautious based on the lower available sample size. Other limitations include that food intake can influence the levels of certain lipids and although the sample collection procedures were standardized for collection between 8 and 10 a.m. in a fasted state (i.e. a minimum of 8 h fasting) or if not possible, that participants eat a low-lipid meal, the compliance to these procedures is unknown. Moreover, body fat may correlate to certain lipids,69 and our study did not incorporate body mass index (BMI). A recent large-scale study (>500 Parkinson’s disease and >500 controls) found that BMI was not different between controls and Parkinson’s disease patients,40,70 suggesting that the lipid changes we observed in Parkinson’s disease patients are unlikely to simply reflect changes in dietary or exercise patterns. Moreover, a related measure of blood pressure medication was not significantly different between the groups in our study. Of importance, the specificity of lipid changes for Parkinson’s disease will need to be determined by comparing to similar data from other neurodegenerative disease cohorts. Although our results indicate that dysregulated lipids are in metabolic pathways regulated by the LRRK2 risk gene, other genotype–phenotype associations were not explored in this study and the extent to which mutations/polymorphisms outside of LRRK2 G2019S, including in other genes encoding lipid regulatory enzymes such as GBA1, GALC, SMPD1 and ASAH, are present in the participants is unknown. Lastly, the distinction of lipid isomers such as glucosylceramide/galactosylceramide will depend on mass spectrometry and software development, which is currently underway.
In summary, this study found that serum lipidomes were significantly altered between those with and without Parkinson’s disease and with and without the LRRK2 G2019S mutation, and that the main lipids altered were in related metabolic pathways regulated by Parkinson’s disease risk genes. These results indicate that lipids are a promising avenue of investigation for understanding the aetiology of Parkinson’s disease, as well as potential pharmacodynamic biomarkers for Parkinson’s disease. Longitudinal studies will be important to determine whether lipids are predictive of clinical onset or progression.
Supplementary Material
Acknowledgements
We kindly thank Yasuto Yokoi from Mitsui Knowledge Industry and Dr David Peake from ThermoFisher Scientific for assisting with adding BMP to the LipidSearch database. The authors acknowledge the technical assistance of Jim Matthews and Alex Shaw of the Sydney Informatics Hub, a core research facility of the University of Sydney. We acknowledge the Australian Red Cross Blood service for the provision of materials. We acknowledge the LRRK2 clinical cohort consortium (LCC) for the supply of samples and data. The investigators within the LCC contributed to the design and implementation of the LCC and/or provided data and collected biospecimens but did not participate in the analysis or writing of this report. LCC investigators include Susan Bressman (Beth Israel Medical Center, NY), Nir Giladi (Tel-Aviv Sourasky Medical Centre, Israel), Karen Marder (Columbia University Medical Centre, NY), Jose Marti Masso (Hospital Donostia, Spain), Eduardo Tolosa (Hospital Clinic de Barcelona, Spain), Jan Aasly (University Hospital, Norwegian University of Science and Technology, Norway), Daniela Berg (University of Tuebingen, Germany), Thomas Gasser (University of Tuebingen, Germany), Alexis Brice (Hospital de la Pitie-Salpetriere, France), Jean-Christopher Corval (Hospital de la Pitie-Salpetriere, France), Piu Chan (Xuanwu Hospital of Capital Medical University, China), Emily Drabant (23andMe, CA), Tatiana Foroud (Indiana University School of Medicine, IL), Faycal Hentati (National Institute of Neurology, Tunisia), Matthew Farrer (National Institute of Neurology, Tunisia), Connie Maras (Toronto Western Hospital, Canada), Anthony Lang Maras (Toronto Western Hospital, Canada) and Birgitt Schuele (The Parkinson’s Institute, CA).
Abbreviations
- BMP
bis(monoacylglycerol)phosphate
- Cer
ceramide
- DG
diacylglycerol
- FA
fatty acid
- Hex1Cer
monohexosylceramide
- LC-MS/MS
liquid chromatography–tandem mass spectrometry
- LDA
linear discriminant analysis
- PC
phosphatidylcholine
- PCA
principal component analysis
- PE
phosphatidylethanolamine
- PG
phosphatidylglycerol
- PI
phosphatidylinositol
- SM
sphingomyelin
- TG
triacylglycerol
Contributor Information
Jasmin Galper, Brain and Mind Centre and Faculty of Medicine and Health, School of Medical Sciences, University of Sydney, Camperdown, NSW 2050, Australia.
Nicholas J Dean, Faculty of Medicine and Health, Central Clinical School, University of Sydney, Camperdown, NSW 2050, Australia.
Russell Pickford, Bioanalytical Mass Spectrometry Facility, University of New South Wales, Sydney, NSW 2052, Australia.
Simon J G Lewis, Brain and Mind Centre and Faculty of Medicine and Health, School of Medical Sciences, University of Sydney, Camperdown, NSW 2050, Australia.
Glenda M Halliday, Brain and Mind Centre and Faculty of Medicine and Health, School of Medical Sciences, University of Sydney, Camperdown, NSW 2050, Australia.
Woojin S Kim, Brain and Mind Centre and Faculty of Medicine and Health, School of Medical Sciences, University of Sydney, Camperdown, NSW 2050, Australia.
Nicolas Dzamko, Brain and Mind Centre and Faculty of Medicine and Health, School of Medical Sciences, University of Sydney, Camperdown, NSW 2050, Australia.
Funding
This work was funded by the Michael J. Fox Foundation for Parkinson’s disease research and the Shake It Up Australia Foundation (#13810.01). J.G. is funded by a scholarship from Australian Rotary Health and the David Henning Memorial Foundation. G.M.H. holds an NHMRC senior leadership fellowship (#1176607). S.J.G.L. holds an NHMRC leadership fellowship (#1195830). The Dementia and Movement Disorders Laboratory is supported by ForeFront, a collaborative research group dedicated to the study of non-Alzheimer disease degenerative disorders, funded by NHMRC grants (#1037746, #1095127 and #1132524).
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Aharon-Peretz J, Rosenbaum H, Gershoni-Baruch R. Mutations in the glucocerebrosidase gene and Parkinson’s disease in Ashkenazi Jews. N Engl J Med. 2004;351(19):1972–1977. [DOI] [PubMed] [Google Scholar]
- 2. Gan-Or Z, Ozelius LJ, Bar-Shira A, et al. . The p.L302P mutation in the lysosomal enzyme gene SMPD1 is a risk factor for Parkinson disease. Neurology. 2013;80(17):1606–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chang D, Nalls MA, Hallgrímsdóttir IB, et al. . A meta-analysis of genome-wide association studies identifies 17 new Parkinson’s disease risk loci. Nat Genet. 2017;49(10):1511–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. de Carvalho Guimarães B, Valente Pereira AC, da Costa Rodrigues F, et al. . Glucocerebrosidase N370S and L444P mutations as risk factors for Parkinson’s disease in Brazilian patients. Parkinsonism Relat Disord. 2012;18(5):688–689. [DOI] [PubMed] [Google Scholar]
- 5. Simón-Sánchez J, Schulte C, Bras JM, et al. . Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat Genet. 2009;41(12):1308–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang Y, Liu L, Xiong J, et al. . Glucocerebrosidase L444P mutation confers genetic risk for Parkinson’s disease in central China. Behav Brain Funct. 2012;8:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Robak LA, Jansen IE, van Rooij J, et al. . Excessive burden of lysosomal storage disorder gene variants in Parkinson’s disease. Brain. 2017;140(12):3191–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Do CB, Tung JY, Dorfman E, et al. . Web-based genome-wide association study identifies two novel loci and a substantial genetic component for Parkinson’s disease. PLoS Genet. 2011;7(6):e1002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nalls MA, Pankratz N, Lill CM, et al. . Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat Genet. 2014;46(9):989–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nalls MA, Blauwendraat C, Vallerga CL, et al. . Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: A meta-analysis of genome-wide association studies. Lancet Neurol. 2019;18(12):1091–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Neumann J, Bras J, Deas E, et al. . Glucocerebrosidase mutations in clinical and pathologically proven Parkinson’s disease. Brain. 2009;132(Pt 7):1783–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tayebi N, Walker J, Stubblefield B, et al. . Gaucher disease with parkinsonian manifestations: Does glucocerebrosidase deficiency contribute to a vulnerability to parkinsonism? Mol Genet Metab. 2003;79(2):104–109. [DOI] [PubMed] [Google Scholar]
- 13. Gan-Or Z, Giladi N, Rozovski U, et al. . Genotype–phenotype correlations between GBA mutations and Parkinson disease risk and onset. Neurology. 2008;70(24):2277–2283. [DOI] [PubMed] [Google Scholar]
- 14. Sidransky E, Nalls MA, Aasly JO, et al. . Multicenter analysis of glucocerebrosidase mutations in Parkinson’s disease. N Engl J Med. 2009;361(17):1651–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee JH, Phelan P, Shin M, et al. . SREBP-1a-stimulated lipid synthesis is required for macrophage phagocytosis downstream of TLR4-directed mTORC1. Proc Natl Acad Sci USA. 2018;115(52):E12228–E12234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Simón-Sánchez J, van Hilten JJ, van de Warrenburg B, et al. . Genome-wide association study confirms extant PD risk loci among the Dutch. Eur J Hum Genet. 2011;19(6):655–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pankratz N, Wilk JB, Latourelle JC, et al. . Genomewide association study for susceptibility genes contributing to familial Parkinson disease. Hum Genet. 2009;124(6):593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Goldschmidt HL, Tu-Sekine B, Volk L, Anggono V, Huganir RL, Raben DM. DGKθ catalytic activity is required for efficient recycling of presynaptic vesicles at excitatory synapses. Cell Rep. 2016;14(2):200–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Puchkov D, Haucke V. Greasing the synaptic vesicle cycle by membrane lipids. Trends Cell Biol. 2013;23(10):493–503. [DOI] [PubMed] [Google Scholar]
- 20. Xicoy H, Wieringa B, Martens GJM. The role of lipids in Parkinson’s disease. Cells. 2019;8(1):27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mazzulli JR, Xu YH, Sun Y, et al. . Gaucher disease glucocerebrosidase and α-synuclein form a bidirectional pathogenic loop in synucleinopathies. Cell. 2011;146(1):37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fu X, Wang Y, He X, Li H, Liu H, Zhang X. A systematic review and meta-analysis of serum cholesterol and triglyceride levels in patients with Parkinson’s disease. Lipids Health Dis. 2020;19(1):97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lu Y, Jin X, Zhao P. Serum lipids and the pathogenesis of Parkinson’s disease: A systematic review and meta-analysis. Int J Clin Pract. 2021;75(4):e13865. [DOI] [PubMed] [Google Scholar]
- 24. Zimprich A, Biskup S, Leitner P, et al. . Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44(4):601–607. [DOI] [PubMed] [Google Scholar]
- 25. Paisán-Ruíz C, Jain S, Evans EW, et al. . Cloning of the gene containing mutations that cause PARK8-linked Parkinson’s disease. Neuron. 2004;44(4):595–600. [DOI] [PubMed] [Google Scholar]
- 26. Madureira M, Connor-Robson N, Wade-Martins R. LRRK2: Autophagy and lysosomal activity. Front Neurosci. 2020;14:498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Roosen DA, Cookson MR. LRRK2 at the interface of autophagosomes, endosomes and lysosomes. Mol Neurodegener. 2016;11(1):73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhao Y, Dzamko N. Recent developments in LRRK2-targeted therapy for Parkinson’s disease. Drugs. 2019;79(10):1037–1051. [DOI] [PubMed] [Google Scholar]
- 29. Tolosa E, Vila M, Klein C, Rascol O. LRRK2 in Parkinson disease: Challenges of clinical trials. Nat Rev Neurol. 2020;16(2):97–107. [DOI] [PubMed] [Google Scholar]
- 30. Fuji RN, Flagella M, Baca M, et al. . Effect of selective LRRK2 kinase inhibition on nonhuman primate lung. Sci Transl Med. 2015;7(273):273ra15. [DOI] [PubMed] [Google Scholar]
- 31. Alcalay RN, Hsieh F, Tengstrand E, et al. . Higher urine bis(monoacylglycerol)phosphate levels in LRRK2 G2019S mutation carriers: Implications for therapeutic development. Mov Disord. 2020;35(1):134–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sanyal A, DeAndrade MP, Novis HS, et al. . Lysosome and inflammatory defects in GBA1-mutant astrocytes are normalized by LRRK2 inhibition. Mov Disord. 2020;35(5):760–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ysselstein D, Nguyen M, Young TJ, et al. . LRRK2 kinase activity regulates lysosomal glucocerebrosidase in neurons derived from Parkinson’s disease patients. Nat Commun. 2019;10(1):5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ferrazza R, Cogo S, Melrose H, et al. . LRRK2 deficiency impacts ceramide metabolism in brain. Biochem Biophys Res Commun. 2016;478(3):1141–1146. [DOI] [PubMed] [Google Scholar]
- 35. Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37(8):911–917. [DOI] [PubMed] [Google Scholar]
- 36. Kim WS, Jary E, Pickford R, et al. . Lipidomics analysis of behavioral variant frontotemporal dementia: A scope for biomarker development. Front Neurol. 2018;9:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Castro-Perez JM, Kamphorst J, DeGroot J, et al. . Comprehensive LC−MSE lipidomic analysis using a shotgun approach and its application to biomarker detection and identification in osteoarthritis patients. J Proteome Res. 2010;9(5):2377–2389. [DOI] [PubMed] [Google Scholar]
- 38. Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M. KEGG: Integrating viruses and cellular organisms. Nucleic Acids Res. 2021;49:D545–D551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chan RB, Perotte AJ, Zhou B, et al. . Elevated GM3 plasma concentration in idiopathic Parkinson’s disease: A lipidomic analysis. PLoS One. 2017;12(2):e0172348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang J, Zhang X, Wang L, Yang C. High performance liquid chromatography–mass spectrometry (LC-MS) based quantitative lipidomics study of ganglioside-NANA-3 plasma to establish its association with Parkinson’s disease patients. Med Sci Monit. 2017;23:5345–5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stoessel D, Schulte C, Teixeira dos Santos MC, et al. . Promising metabolite profiles in the plasma and CSF of early clinical Parkinson’s disease. Front Aging Neurosci. 2018;10:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hu L, Dong MX, Huang YL, et al. . Integrated metabolomics and proteomics analysis reveals plasma lipid metabolic disturbance in patients with Parkinson’s disease. Front Mol Neurosci. 2020;13:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yakhine-Diop SMS, Morales-García JA, Niso-Santano M, et al. . Metabolic alterations in plasma from patients with familial and idiopathic Parkinson’s disease. Aging. 2020;12(17):16690–16708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shao Y, Li T, Liu Z, et al. . Comprehensive metabolic profiling of Parkinson’s disease by liquid chromatography–mass spectrometry. Mol Neurodegener. 2021;16(1):4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Saiki S, Hatano T, Fujimaki M, et al. . Decreased long-chain acylcarnitines from insufficient β-oxidation as potential early diagnostic markers for Parkinson’s disease. Sci Rep. 2017;7(1):7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chang K-H, Cheng M-L, Tang H-Y, Huang C-Y, Wu Y-R, Chen C-M. Alternations of metabolic profile and kynurenine metabolism in the plasma of Parkinson’s disease. Mol Neurobiol. 2018;55(8):6319–6328. [DOI] [PubMed] [Google Scholar]
- 47. Schulte EC, Altmaier E, Berger HS, et al. . Alterations in lipid and inositol metabolisms in two dopaminergic disorders. PLoS One. 2016;11(1):e0147129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Burté F, Houghton D, Lowes H, et al. . Metabolic profiling of Parkinson's disease and mild cognitive impairment. Mov Disord. 2017;32(6):927–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Okuzumi A, Hatano T, Ueno S-I, et al. . Metabolomics-based identification of metabolic alterations in PARK2. Ann Clin Transl Neurol. 2019;6(3):525–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dong MX, Wei YD, Hu L. Lipid metabolic dysregulation is involved in Parkinson’s disease dementia. Metab Brain Dis. 2021;36(3):463–470. [DOI] [PubMed] [Google Scholar]
- 51. Hatano T, Saiki S, Okuzumi A, Mohney RP, Hattori N. Identification of novel biomarkers for Parkinson’s disease by metabolomic technologies. J Neurol Neurosurg Psychiatry. 2016;87(3):295–301. [DOI] [PubMed] [Google Scholar]
- 52. Mielke MM, Maetzler W, Haughey NJ, et al. . Plasma ceramide and glucosylceramide metabolism is altered in sporadic Parkinson’s disease and associated with cognitive impairment: A pilot study. PLoS One. 2013;8(9):e73094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Klatt-Schreiner K, Valek L, Kang J-S, et al. . High glucosylceramides and low anandamide contribute to sensory loss and pain in Parkinson’s disease. Mov Disord. 2020;35(10):1822–1833. [DOI] [PubMed] [Google Scholar]
- 54. Roede JR, Uppal K, Park Y, et al. . Serum metabolomics of slow vs. rapid motor progression Parkinson’s disease: A pilot study. PLoS One. 2013;8(10):e77629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Postuma RB, Aarsland D, Barone P, et al. . Identifying prodromal Parkinson’s disease: pre-motor disorders in Parkinson’s disease. Mov Disord. 2012;27(5):617–626. [DOI] [PubMed] [Google Scholar]
- 56. Sheng Z, Jia X, Kang M. Statin use and risk of Parkinson’s disease: A meta-analysis. Behav Brain Res. 2016;309:29–34. [DOI] [PubMed] [Google Scholar]
- 57. Bai S, Song Y, Huang X, et al. . Statin use and the risk of Parkinson’s disease: An updated meta-analysis. PLoS One. 2016;11(3):e0152564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Poly TN, Islam MM, Walther BA, et al. . Exploring the association between statin use and the risk of Parkinson’s disease: A meta-analysis of observational studies. Neuroepidemiology. 2017;49(3–4):142–151. [DOI] [PubMed] [Google Scholar]
- 59. Brauer R, Wei L, Ma T, et al. . Diabetes medications and risk of Parkinson’s disease: A cohort study of patients with diabetes. Brain. 2020;143(10):3067–3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Stancu C, Sima A. Statins: Mechanism of action and effects. J Cell Mol Med. 2001;5(4):378–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Schwartz EA, Koska J, Mullin MP, Syoufi I, Schwenke DC, Reaven PD. Exenatide suppresses postprandial elevations in lipids and lipoproteins in individuals with impaired glucose tolerance and recent onset type 2 diabetes mellitus. Atherosclerosis. 2010;212(1):217–222. [DOI] [PubMed] [Google Scholar]
- 62. Monami M, Lamanna C, Desideri CM, Mannucci E. DPP-4 inhibitors and lipids: Systematic review and meta-analysis. Adv Ther. 2012;29(1):14–25. [DOI] [PubMed] [Google Scholar]
- 63. Monami M, Vitale V, Ambrosio ML, et al. . Effects on lipid profile of dipeptidyl peptidase 4 inhibitors, pioglitazone, acarbose, and sulfonylureas: Meta-analysis of placebo-controlled trials. Adv Ther. 2012;29(9):736–746. [DOI] [PubMed] [Google Scholar]
- 64. Jiang Z, Xu X, Gu X, et al. . Effects of higher serum lipid levels on the risk of Parkinson’s disease: A systematic review and meta-analysis. Front Neurol. 2020;11:597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Yuzefovych L, Wilson G, Rachek L. Different effects of oleate vs. palmitate on mitochondrial function, apoptosis, and insulin signaling in L6 skeletal muscle cells: Role of oxidative stress. Am J Physiol Endocrinol Metab. 2010;299(6):E1096–E1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Chavez JA, Summers SA. Characterizing the effects of saturated fatty acids on insulin signaling and ceramide and diacylglycerol accumulation in 3T3-L1 adipocytes and C2C12 myotubes. Arch Biochem Biophys. 2003;419(2):101–109. [DOI] [PubMed] [Google Scholar]
- 67. Schmitz-Peiffer C, Craig DL, Biden TJ. Ceramide generation is sufficient to account for the inhibition of the insulin-stimulated PKB pathway in C2C12 skeletal muscle cells pretreated with palmitate. J Biol Chem. 1999;274(34):24202–24210. [DOI] [PubMed] [Google Scholar]
- 68. Rachek LI, Musiyenko SI, LeDoux SP, Wilson GL. Palmitate induced mitochondrial deoxyribonucleic acid damage and apoptosis in L6 rat skeletal muscle cells. Endocrinology. 2007;148(1):293–299. [DOI] [PubMed] [Google Scholar]
- 69. Shamai L, Lurix E, Shen M, et al. . Association of body mass index and lipid profiles: Evaluation of a broad spectrum of body mass index patients including the morbidly obese. Obes Surg. 2011;21(1):42–47. [DOI] [PubMed] [Google Scholar]
- 70. Guo X, Song W, Chen K, et al. . The serum lipid profile of Parkinson’s disease patients: a study from China. Int J Neurosci. 2015;125(11):838–844. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw mass spectrometry files generated in this study have been provided to the Michael J. Fox Foundation for Parkinson’s disease research and can be requested (https://www.michaeljfox.org/news/lrrk2-cohort-consortium). Other data are available upon reasonable request from the corresponding authors and upon relevant ethical approval.