Abstract
Aims
De-differentiation and activation of pro-inflammatory pathways are key transitions vascular smooth muscle cells (SMCs) make during atherogenesis. Here, we explored the upstream regulators of this ‘atherogenic transition’.
Methods and results
Genome-wide sequencing studies, including Assay for Transposase-Accessible Chromatin using sequencing and RNA-seq, were performed on cells isolated from both murine SMC-lineage-tracing models of atherosclerosis and human atherosclerotic lesions. At the bulk level, alterations in chromatin accessibility were associated with the atherogenic transitioning of lesional SMCs, especially in relation to genes that govern differentiation status and complement-dependent inflammation. Using computational biology, we observed that a transcription factor previously related to coronary artery disease, Activating transcription factor 3 (ATF3), was predicted to be an upstream regulator of genes altered during the transition. At the single-cell level, our results indicated that ATF3 is a key repressor of SMC transitioning towards the subset of cells that promote vascular inflammation by activating the complement cascade. The expression of ATF3 and complement component C3 was negatively correlated in SMCs from human atherosclerotic lesions, suggesting translational relevance. Phenome-wide association studies indicated that genetic variation that results in reduced expression of ATF3 is correlated with an increased risk for atherosclerosis, and the expression of ATF3 was significantly down-regulated in humans with advanced vascular disease.
Conclusion
Our study indicates that the plasticity of atherosclerotic SMCs may in part be explained by dynamic changes in their chromatin architecture, which in turn may contribute to their maladaptive response to inflammation-induced stress.
Keywords: Atherosclerosis, Vascular smooth muscle cell, Chromatin accessibility, Atherogenic transition
1. Introduction
Unbiased genetic efforts have confirmed that many pathways, which promote atherosclerotic cardiovascular disease (CVD) do so independently of traditional risk factors. An emerging theme from these studies is that vessel-intrinsic processes may explain much of the ‘residual risk’ that persists after lipid lowering,1 especially as they relate to the behaviour of the vascular smooth muscle cell (SMC) in the developing plaque.2 Sophisticated lineage-tracing approaches have demonstrated that mature SMCs are not terminally differentiated, and that these cells retain remarkable plasticity under periods of stress.3 Indeed, SMCs possess the capacity to undergo ‘phenotype switching’ towards a wide range of phenotypes, several of which appear to be key determinants of plaque inflammation and lesional destabilization.4,5
While the behaviour of these cells once they are present in the plaque is an area of active research, many of these studies suffer from limitations intrinsic to bulk sequencing technology (which cannot resolve differences between different SMC subpopulations) and/or were restricted to an examination of the transcriptional profile of these cells in the plaque. Also, few studies have investigated the upstream regulators driving their ‘atherogenic transition’, and their potential relationship to factors, which have been causally related to coronary artery disease (CAD) via genome-wide association studies (GWAS).
To address these limitations, we have applied a combination of whole-genome sequencing, single-cell transcriptomic profiling, and single-cell chromatin accessibility technology to lineage-traced SMCs isolated from mouse models of atherosclerosis. Identified factors were then evaluated for translational relevance, using a variety of independent clinical biobank samples. Our work highlights that disease-associated alterations in epigenetic access to CAD-related risk loci may reduce the capacity of SMCs to modulate their inflammatory stress response, and initiate their ‘atherogenic transition’ within the plaque. This study also offers a new framework for analysing changes in chromatin architecture to enhance our understanding of the epigenetic drivers of atherosclerosis.
2. Methods
Expanded methods are available in the Supplementary material online.
2.1 Experimental overview
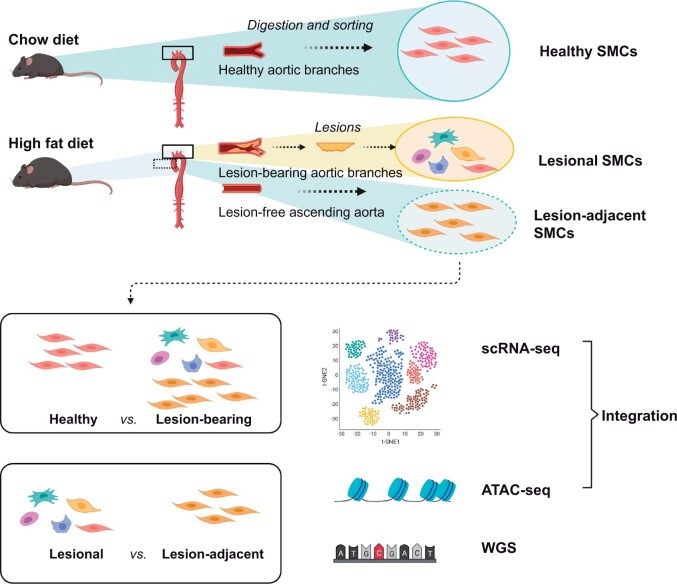
Pure collections of freshly digested and FACS-sorted SMCs were isolated from: (i) the aortae and branch vessels of chow-diet-fed animals (‘Healthy SMCs’); (ii) atherosclerotic lesions micro-dissected from high-fat-diet (HFD)-fed animals (‘Lesional SMCs’); and (iii) aortic tissue adjacent to the lesions in the HFD-fed mice (‘Lesion-adjacent SMCs’). These cells were then subjected to whole-genome sequencing, bulk and single-cell RNA sequencing (scRNA-seq) and bulk and single-cell ATAC sequencing (scATACseq), prior to integration and analysis, as described in detail below. Comparisons were made between healthy and disease (Healthy vs. Lesional SMCs), as well as cells making the ‘atherogenic transition’ (Lesion-adjacent vs. Lesional SMCs; defined in Figure 1). scRNA-seq, bulk Assay for Transposase-Accessible Chromatin with high-throughput sequencing (ATAC-seq), scATAC-seq, and bulk DNA-seq were performed as separate experiments and hence cells are derived from different animals. Except for the RNA-seq experiments, which included medial SMCs underlying the plaque, all ATAC-seq and DNA-seq experiments were performed solely on lesional cells.
Figure 1.
Overview of the experimental design. To define the genomic, epigenetic, and transcriptional profiles of SMCs during the ‘atherogenic transition’, healthy and diseased vascular SMCs were sorted from atheroprone, lineage-traced Rainbow mice by flow cytometry based on their permanent fluorescent labels. For scRNA-seq, SMCs were isolated from the ascending aorta and branch vessels of HFD-fed (Lesion-bearing) and chow-diet-fed (Healthy) animals. For whole-genome sequencing, bulk and scATACseq, pure ‘lesional SMCs’ were collected by micro-dissecting the plaque away from the medial layer; a piece of lesion-free ascending aorta adjacent to the lesion (Lesion-adjacent SMCs) was collected and used as a matched control from each HFD-fed mice. Comparisons were made between healthy and disease (Healthy vs. Lesional SMCs), as well as the atherogenic transition (Lesion-adjacent vs. Lesional SMCs). Transcriptome profiles of these SMCs served as a reference to predict what chromatin structure changes were associated with gene expression alterations.
2.2 Mouse samples
Male Myh11-CreERT2, Rosa26Rainbow/+, ApoE−/− mice (‘Rainbow’ mice) and Myh11-CreERT2, Rosa26tdTomato/tdTomato, and ApoE−/− SMC-lineage-tracing mice (‘Tomato’ mice) were generated at Stanford University as described previously.5 For both models of SMC-lineage-tracing mice, only male animals were used since the Cre allele was initially inserted into the Y chromosome [Jackson Laboratory, B6.FVB-Tg (Myh11-cre/ERT2)1Soff/J], and therefore female mice cannot be lineage traced. Animals were euthanized by cardiac perfusion of phosphate-buffered saline under deep anaesthesia by inhaling isoflurane through a nose cone (3% at a flow rate of 1 L/min). For RNA-seq, healthy control SMCs were collected from lesion-free aortic arch of chow-fed Tomato mice and Tomato+ cells were sorted by an Aria II instrument (BD Biosciences) with a 100 μm nozzle. Dead cells were excluded by SYTOXTM Blue during sorting (S34857, Thermo Fisher Scientific). For ATAC-seq and whole-genome sequencing, lesions in the aortic arch region were micro-dissected off from the medial layers of HFD-fed Rainbow mice, to minimize the inclusion of healthy cells underlying the plaque. SMCs (a combination of CFP, mOrange, and mCherry positive cells) were sorted by a Carmen (BD Biosciences) cell sorter. DRAQ5 (62251, Thermo Fisher Scientific) staining was used to ensure that only nucleated cells were included. Lesion-free segments of blood vessels in the outer curvature of the ascending aorta and the descending aorta of the same mice were also collected to isolate control SMCs.
2.3 Human samples
Human datasets utilized in this report were obtained from the Athero-Express study (scRNA-seq of carotid lesions)6 and the Stockholm-Tartu Atherosclerosis Reverse Network Engineering Task (STARNET) Study (RNA-seq data of multi-tissue, n = 672 subjects).7
2.4 Search strategy for CAD-related risk genes
We systematically searched the MEDLINE and GWAS Catalog databases from inception to December 2020, using the key words ‘cardiovascular disease’, ‘CAD’, ‘heart disease’, and ‘candidate gene’. All identified genes went through a two-step workflow where abstracts and results were first reviewed, then candidate genes, removing duplicates, were extracted. We defined a reported P-value ˂1E-05 as a CAD associated gene. All candidate genes are displayed in Supplementary material online, Table S1.
2.5 scRNA-seq and analysis
scRNA-seq of sorted SMCs from Tomato mice was performed using C1 96-well Open App IFC plates (100-8135, Fluidigm), as previously described.5 Differentially expressed genes (DEGs) identified from SCDE analysis were assessed for pathway enrichment through the web-based platform Enrichr (Bioplanet 2019, ranked by combination score) and transcription factor (TF) enrichment via CHEA3.
2.6 Whole-genome sequencing
DNA from sorted SMCs were extracted using PicoPure™ DNA extraction kit (KIT0103, Thermo Fisher Scientific) and libraries prepared using Nextera DNA Flex library preparation kit (20018705, Illumina) with Nextera DNA CD Indexes (20018708, Illumia). The raw sequencing reads were cleaned using fastp v0.20.1 and then mapped to mouse genome (GRCm38) using BWA v0.7.17. Somatic variants were called according to GATK MuTect2 Best Practices in paired mode.8
2.7 ATAC-seq
For bulk ATAC-seq, sorted SMCs were processed according to the Omni-ATAC-seq protocol.9 Differentially accessible peaks from each group were identified by MAnorm version 1.1.4 with FDR < 0.05.10 The Homer annotatePeaks tool (http://homer.salk.edu/homer/, last accessed on May 01, 2021) was used to investigate the motif enrichment of differential peaks using default parameters.
For scATACseq of mouse SMCs, sorted SMCs were captured in Chromium 10X genomics and sequenced on an Illumina NextSeq 500 (Stanford Functional Genomics Facility). Raw sequencing data were processed through 10X Genomics Cell Ranger ATAC pipeline (version 1.2.0) and were further loaded into R software using Signac v1.1.011 and ArchR v0.9.112 packages. After combination, cells were clustered with Signac and each cluster was given a name, such as Plaque 1–3.
2.8 Isolation and treatment of primary SMCs
Primary SMCs were isolated from atherosclerotic lesions and cultured as previously described.5 Purified SMCs were expanded in a 10 cm culture dish to 90% confluence when the lipofectamine RNAiMAX transfection reagent (13778150, Thermo Fisher Scientific) was used to mediate knockdown of Activating transcription factor 3 (ATF3). Cells were incubated with serum-free medium containing 75 pmol siRNA for ATF3 (AM16708, Thermo Fisher Scientific) or negative control (4390843, Thermo Fisher Scientific) for 72 h. C3 protein level in the conditioned medium was determined by ELISA (LS-F10438-1, LifeSpan Biosciences) and RNA of treated cells was extracted using miRNeasy Mini kit (217004, Qiagen) for bulk RNA-seq.
The ATF3 binding motif near the C3 promoter region, as identified by ATAC-seq in Section 2.7, was cloned into reporter constructs. The binding motif upstream of the transcription start site (TSS) (Chr17:57228039-397) was amplified and cloned into the pGL3-basic plasmid to generate a C3 pGL3-TSS construct (Promega). To determine if ATF3 has a repressive function on C3 promoter activity, mouse ATF3 plasmid (MR201634, Origene) was co-transfected with C3 PGL3-TSS. After 48 h, cells were collected and relative luciferase activity (activities of firefly to Renilla) was determined using the Dual Luciferase Reporter Assay System (E1910, Promega). Chromatin precipitation was performed using a ChIP-IT kit (53040, Active motif) with anti-ATF3 antibodies (33593, cell signalling and NBP1-85816, Novus Biologicals) according to the manufacturer’s protocol.
2.9 Statistics
Results were analysed by SPSS Statistics 20 and GraphPad Prism 5 for statistical significance between treatment groups. The normality of data for figures was determined by performing D’Agostino and Pearson omnibus normality tests. The homogeneity of variance was determined by performing an F-test. Normally distributed data of equal variance were analysed using two-tailed Student t-test or one-way ANOVA with Tukey’s post hoc. Data that did not meet either assumptions were analysed using Mann–Whitney U test. Data are presented as mean ± standard error of the mean (SEM). P-values <0.05 were considered significant.
2.10 Study approval
All animal studies were approved by the Stanford University Administrative Panel on Laboratory Animal Care (protocol 27279) and conform to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH Publication No. 85-23, revised 1996). All human studies were performed with written informed consent and with the approval of the local Ethical Committees: Institutional Review Boards at Stanford University and the University of Virginia for the scATAC-seq, Munich Vascular Biobank for PCR, and according to the UK Biobank protocol as previously described (https://www.ukbiobank.ac.uk, 1 May 2021, date last accessed). All subjects provided informed consent and the investigation conform to the principles outlined in the Declaration of Helsinki.
3. Results
3.1 Vascular SMCs develop few de novo somatic mutations during atherogenesis
Atherogenesis is now known to be driven, in part, by somatic mutations of circulating myeloid cells (a phenomenon known as ‘clonal haematopoiesis’).13 Accordingly, we hypothesized that vascular SMCs may similarly acquire de novo mutations, which could account for their observed growth and survival advantage in the vessel wall. To test this, we applied whole-genome sequencing of SMCs from healthy and diseased vessels to determine if and when mutations occur (Supplementary material online, Figure S1). While very few functional variants (which can alter the protein sequence) were observed in the healthy aorta compared to genomic DNA, we observed a modest number of somatic variants in the atherosclerotic lesions (Supplementary material online, Figure S1A, left panel), the majority of which were single nucleotide variants that led to missense mutations (Supplementary material online, Figure S1A, right panel, and Table S2). Amongst the top 20 mutated genes (allele fractions 0.6% ∼ 28.6%), Muc6, a cancer driver gene, which regulates the biosynthesis of O-glycan to maintain a protective extracellular matrix structure for epithelial cells,14 was the only gene found to be mutated in more than one animal. In the animal that carried the most mutations, we also observed unique mutations in genes that epigenetically modify chromatin structure, such as Msl115 (Supplementary material online, Figure S1B). However, most of the mutated genes were expressed at an extremely low level in plaque SMCs (based on our scRNA-seq data), and none of them overlapped with hits in previously published CAD GWAS. Hence, at least in mouse models, these rare mutations do not appear to be key drivers directly related to the development of CAD.
3.2 Vascular SMCs experience widespread alterations in chromatin accessibility during atherogenesis
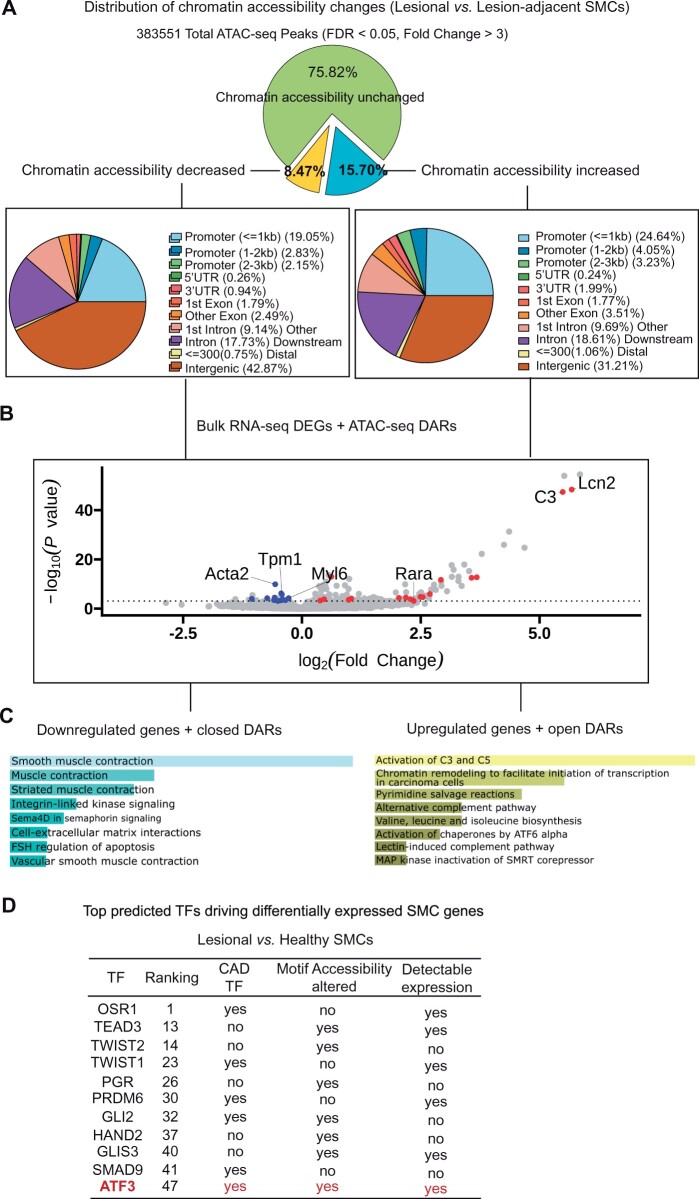
Because somatic mutations do not seem to drive the atherogenic transition of murine SMCs, we next hypothesized that epigenetic modifications may contribute to the widespread gene expression changes known to occur during lesion progression. We chose the ‘Assay for Transposase-Accessible Chromatin with high-throughput sequencing’ (ATAC-seq) to map regions of TF binding sites (TF motifs) and changes in chromatin accessibility genome-wide.16 Using bulk ATAC-seq, we observed that nearly 25% of the total accessible regions were significantly perturbed in SMCs undergoing the atherogenic transition. About 65% of these differentially accessible regions (DARs) reflect a more open chromatin structure, a quarter of which occurred in gene promoter regions (Figure 2A). To determine which of these changes might drive alterations in gene expression, we integrated RNA-seq data5 with these ATAC-seq results to determine which DEGs were altered in a concordant manner with their corresponding DARs (e.g. up-regulated DEGs for genes with more accessible DARs at the promoter region). In total, we identified 29 such genes that were dysregulated in a concordant manner during the SMC’s atherogenic transition (Supplementary material online, Table S3). These 29 genes included a marked increase in the key complement factor, C3, and significant down-regulation of classical SMC contractile genes, including Acta2 (Figure 2B). Consistent with our previous findings that genes in the complement cascade were up-regulated whereas SMC contractile proteins were down-regulated,5 pathway analyses confirmed that activation of the complement cascade and SMC contraction were the most up- and down-regulated pathways, respectively, observed for these epigenetically driven changes (BioPlanet, combination score, Figure 2C).
Figure 2.
Robust changes in chromatin accessibility predict the association of ATF3 with the ‘atherogenic transition’ of SMCs. (A) Landscape of chromatin accessibility changes illustrates the proportion and distribution of differential accessibility regions found during the SMC’s atherogenic transition. (B) Visual demonstration of DEGs that are up- (red) or down- regulated (blue) in a concordant manner with changes in chromatin accessibility at the promoter regions of SMCs. (C) Enrichr pathway enrichment analysis defines the top pathways related to the differentially expressed and epigenetically driven SMC genes identified during the atherogenic transition. (D) Features of top 50 TFs upstream of differentially expressed SMC genes during the atherogenic transition, as predicted by CHEA3 TF enrichment analysis. ATF3 is the only CAD-related TF expressed by SMCs, which also changes its chromatin accessibility at its binding sites during the transition.
While a significant fraction of the DARs were found to occur in gene promoter regions, we also observed a high proportion of DARs occurring at TF motifs. In lesional SMCs, we observed 216 DARs at annotated TF motifs (Supplementary material online, Table S4). The majority of these occurred at binding sites for TFs known to control SMC differentiation and proliferation (e.g. KLF4, RUNX2, and SOX2) (Supplementary material online, Figure S2). When refining this list to only include those TFs that are both expressed by SMCs and factors, which have been previously implicated in CAD, we found TGFβ and NF-κB signalling are the top pathways implicated in the ‘atherogenic transition’ of SMCs (Supplementary material online, Figure S2). Using the computational biology tool CHEA3, ATF3 was identified as the top CAD-related TF17 predicted to drive the genes expression changes observed during the ‘atherogenic transition’ (Figure 2D, the full list of the top 50 TFs is provided in Supplementary material online, Table S5). This gene is a member of the ATF/cAMP-responsive element binding family of TFs previously shown to: (i) function as a stress response TF that represses NF-κB-dependent inflammation;18 (ii) interact with SMAD3 to enhance the TGFβ signalling pathway;19–21 and (iii) protect against atherogenesis in a murine model of disease.22
3.3 The CAD-related SMC subpopulation in the plaque is coupled to a loss of ATF3 activity
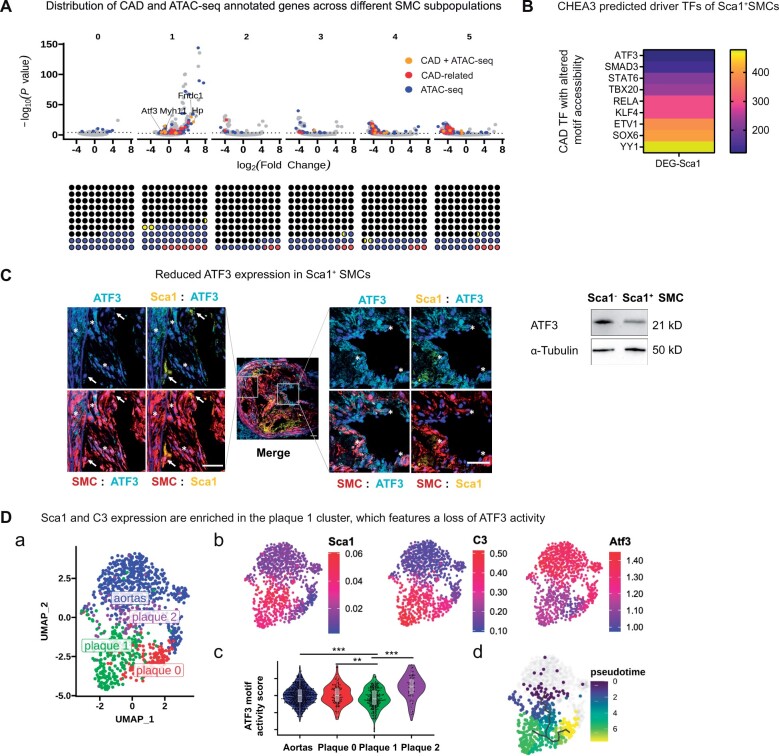
Given that the preceding studies integrated results from bulk RNA-seq and bulk ATAC-seq, the results apply to lesional SMCs as if they were a homogenous group of diseased cells. However, we and others have previously reported that vascular SMCs give rise to a vast diversity of SMC subpopulations in the developing plaque.23 Indeed, some of these subpopulations appear to be highly ‘atherogenic’, while others appear to have beneficial ‘cap-stabilizing’ properties.5,24
To further define the heterogeneity of SMC-derived cells, we previously applied scRNA-seq to lineage-traced SMCs from digested atherosclerotic plaques. This approach allowed us to identify a subpopulation of SMCs (Subset 1) that appeared particularly harmful, given that it: (i) expressed stem cell markers, such as Sca1; (ii) was hyper-proliferative relative to other cells in the plaque; and (ii) activated the pro-inflammatory complement cascade, including the key hub gene, C3. Here, we find that this group of cells (Subset 1) also harbours the highest number of dysregulated CAD-related genes, suggesting a dominant role in atherogenesis (Figure 3A, red dots). We found Subset 1 also could be the most ‘epigenetically-driven’ SMC subpopulation, given that 31.24% of its dysregulated genes had concordant chromatin accessibility changes at the promoter region during the SMC’s ‘atherogenic transition’ (Figure 4A, blue dots). One of the ‘double-hits’ (orange dots in Figure 3A, indicating a gene, which is both CAD-related and likely epigenetically driven) in SMC Subset 1 is ATF3, whose expression was significantly down-regulated in vivo (logFC = −1.643, P = 5.14E-05). Given that ATF3 is a TF whose binding site (motif) accessibility was also changed during the ‘atherogenic transition’ (Figure 2D), we used CHEA3 to investigate whether its activity could be related to the gene expression changes observed in SMC Subset 1. Using this approach, ATF3 was identified as a ‘triple-hit’ representing the top predicted CAD- associated TFs, which could be responsible for the observed changes (Figure 3B). Immunofluorescent co-staining of plaques from lineage-traced mouse models (‘Tomato’ mice) confirmed loss of this gene on lesional Sca1+ SMCs (Figure 3C), especially in Sca1+ SMCs.
Figure 3.
Loss of ATF3’s gene regulatory activity is associated with some of the atherogenic phenotypes of Sca1+ SMCs. (A) The potential atherogenic features of each SMC subpopulation (Subsets 0–5) is visualized by showing which DEGs (determined by scRNA-seq) are bona fide CAD-related risk genes (red dots, upper panel). DEGs that changed in the same direction as the chromatin accessibility at the promoter region of a given gene (determined by bulk ATAC-seq) are annotated in blue. Genes matching both criteria are shown in orange. The proportion of genes in each of the above three categories within the total list of altered genes in each population (black dots) are illustrated in the square charts (lower panels). (B) Using the list of DEGs in the Sca1+ SMC subset (Subset 1) as the input for CHEA3, the heatmap ranks TFs, which are predicted to be potential upstream regulators (yellow to purple: increasing association) of the altered genes. The illustrated TFs were selected as CAD-related TFs, which had alterations in the chromatin structure at their respective binding motifs during the atherogenic transition. (C) A set of representative pictures shows that expression of ATF3 (cyan) is low in Sca1+ (yellow) SMCs (red) in the deep intima (white arrows) and high in Sca1− SMCs in the medial and cap region (asterisks) within lineage-traced murine atherosclerotic lesions, in vivo. Scale bars: 50 μm. (D) scATAC-seq clustering identifies three subsets of lesional SMCs, based on the similarity of their chromatin landscape accessibility. The subset of SMCs within the group ‘plaque 2’ was most similar to SMCs from healthy aortas (subpanel a). ArchR gene scores predict levels of Sca1, C3, and ATF3 expression based on scATAC- seq data (red, highly expressed, subpanel b). ATF3 motif activity score was determined by chromVAR and compared among healthy and lesional SMCs. These data indicate a reduced capability of ATF3 to regulate downstream gene expression in the plaque 1 subpopulation (likelihood-ratio test against plaque 1 population, **P < 0.01, ***P < 0.001, subpanel c). Pseudotime analysis suggests the trajectory of SMCs during the atherogenic transition from healthy aortas to SMC subpopulation plaque 2. These cells then appear to give rise to the plaque 1 and 0 subgroups (subpanel d).
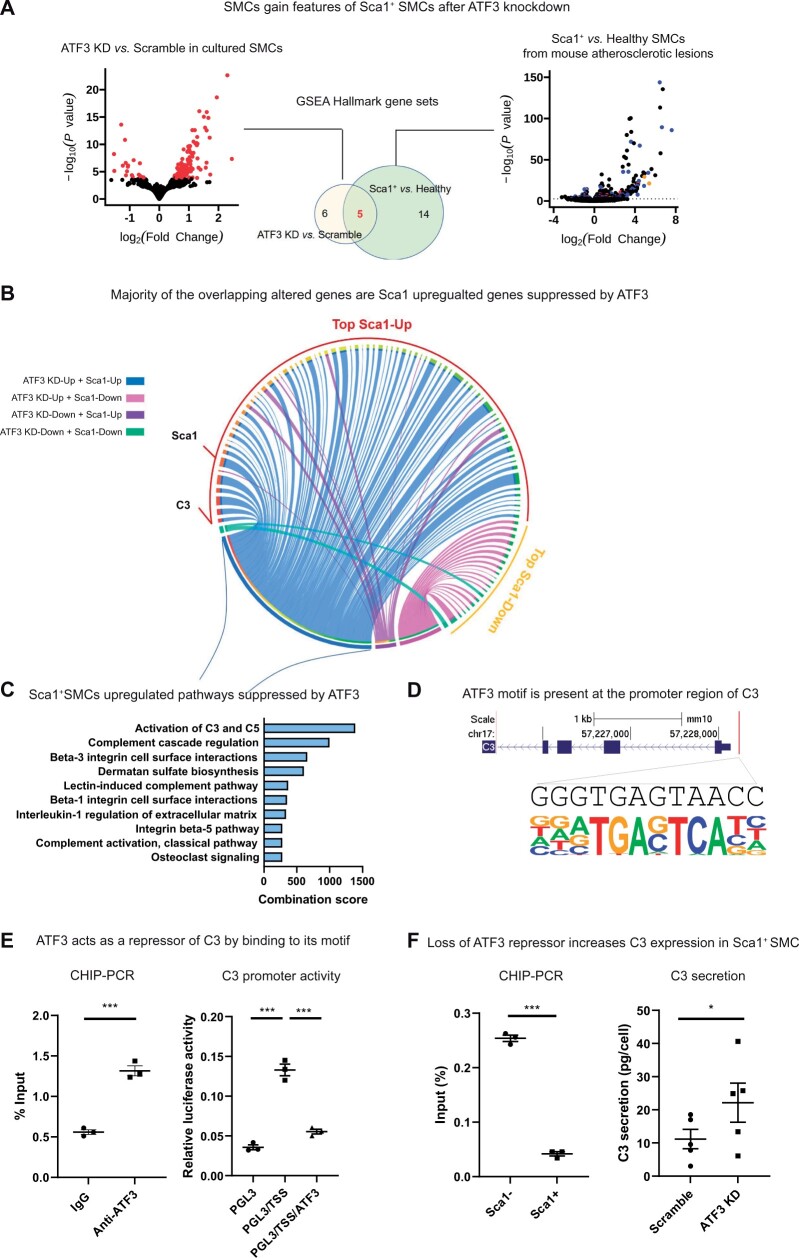
Figure 4.
ATF3 is a repressor of the complement cascade in Sca1+ SMCs. (A) GSEA found an overlap of five hallmark gene sets between genes perturbed by ATF3 knockdown in vitro and genes differentially expressed by Sca1+ SMCs in vivo. (B) A Circos plot connecting genes perturbed by ATF3 knockdown with genes that are differentially expressed by Sca1+ SMCs predicts how ATF3 regulate gene profiles in vivo. Blue and purple ribbons connect ATF3-repressed and promoted genes with genes that are up-regulated in Sca1+ SMCs, respectively. Pink and green ribbons connect ATF3- repressed and promoted genes with genes that are down-regulated in Sca1+ SMCs, respectively. (C) Pathway analysis of genes suppressed by ATF3 (blue ribbons) indicates that activation of the complement cascade is the top biological process normally repressed by ATF3 in healthy SMCs before they make the atherogenic transition into Sca1+ SMCs. (D) Predicted binding motif of ATF3 on the promoter region of C3 (Chr17: 57535101-301). (E) CHIP-PCR of primary SMCs isolated from SMC-lineage-tracing mice confirms binding of ATF3 to its predicted motif on the C3 promoter (left panel, n = 3, P = 0.0004, two-tailed Student’s t-test). Luciferase assay demonstrates that the increased C3 promoter activity in HEK293T cells transfected with its promoter region (PGL3/TSS vs. PGL3) was inhibited when ATF3 is co-transfected (PGL3/TSS/ATF3) (right panel, n = 3, one-way ANOVA with Tukey’s post hoc). (F) CHIP-PCR indicates that Sca1− SMCs have more ATF3 bound to its motif at the C3 promoter region (left panel) and when ATF3 is KD from primary SMCs, C3 secretion is increased (right panel, n = 5, P = 0.0472, two-tailed Student’s t-test). Data are presented as mean ± SEM. *P < 0.05, ***P < 0.001.
To determine if the heterogeneity of lesional SMC subpopulations was also reflected in their chromatin architecture, we next performed scATAC-seq of lineage-traced SMCs digested from atherosclerotic plaques. As shown in Figure 3D, the Signac clustering algorithm resolved three groups of lesional SMCs based on their chromatin accessibility profiles (Plaque 0–2, Figure 3D, panel a) and enrichment of TFs (Supplementary material online, Figure S3 and Table S6). Using gene scores calculated with the ArchR program, we found that SMCs in Plaque group 1 were enriched for Sca1 and C3 gene activity levels (Figure 3D, panel b) and that these cells also had the lowest ATF3 motif activity (Figure 3D, panel c). Indeed, subsequent pseudo-time analysis suggested that the ‘atherogenic transitioning’ of lesional SMCs begins with cells in Plaque group 2 (which harbours a chromatin landscape most similar to cells in lesion-free aortas), and then progresses towards the more inflammatory C3-enriched SMC population found in Plaque group 1 (Figure 3D, panel d). These changes potentially link loss of the ATF3 TF’s activity with the development of these harmful cells.
3.4 In vitro functional studies confirm an anti-inflammatory role for ATF3 on SMC physiology
While the preceding experiments associated the atherogenic transitioning of SMCs with loss of ATF3 motif activity and gene expression, those data could not establish a causal role for that gene in SMC pathophysiology. To overcome this limitation, we next knocked down (KD) ATF3 in freshly isolated and cultured vascular SMCs from our lineage-traced atherosclerotic mice (ATF3 KD vs. Scramble, log2 FC = −0.093, P = 0.015). RNA-seq of these cells confirmed that more than 40% of gene sets perturbed by the KD of ATF3 overlapped with changes observed in lesional Sca1+ SMC [gene set enrichment analysis (GSEA)] (Figure 4A and Supplementary material online, Table S7). A top overlapped hallmark gene set was Coagulation (rank first amongst Sca1+ SMCs and third amongst ATF3 KD SMCs) with genes in the complement cascade, C3, enriched in both phenotypes of SMCs. These results are summarized in a Circos Plot, which indicates how ATF3 regulates the genes, which are differentially expressed in Sca1+ SMCs, including activation of complement cascade and up-regulation of C3 when ATF3 is KD (Figure 4C, blue ribbons). Hence, we hypothesized that ATF3 is a repressor of C3. An ATF3 motif in the promoter region of C3 was found more open by bulk ATAC-seq during SMC atherogenic transition (Supplementary material online, Table S8). CHIP-PCR confirmed that ATF3 directly binds to this motif (Figure 4D) in primary SMCs isolated from mouse lesions (Figure 4E, left panel). To determine whether ATF3 is an enhancer or repressor of C3 via this motif, the C3 promoter region containing the ATF3 motif was cloned into HEK293T cells (Figure 4E, right panel, PGL3/TSS). Co-transfection of ATF3 significantly inhibited C3 promoter activity in these cells (Figure 4E, right panel, PGL3/TSS/ATF3 vs. PGL/TSS). These findings suggested that when this binding motif is occupied by ATF3 in healthy SMCs, C3 expression is repressed. Loss of ATF3’s regulatory activity in Sca1+ SMCs as shown in Figure 3 was accompanied by detachment of ATF3 from this motif (Figure 4F, left panel) and increased accessibility of this region (Supplementary material online, Table S8). Subsequent ELISA studies confirmed that knockdown of ATF3 led to a significant increase in C3 secretion into the supernatant, in vitro (Figure 4F, right panel). Given ATF3’s described role in repressing inflammation, and the overlapped phenotypes of ATF3lo cells in vitro and in the atherosclerotic plaque in vivo, we concluded that a reduction in ATF3 activity may represent a critical initial step in the transitioning of SMCs during atherogenesis.
3.5 ATF3 is implicated in clinical CVD and is inversely associated with the ‘atherogenic’ profile of human plaque SMCs
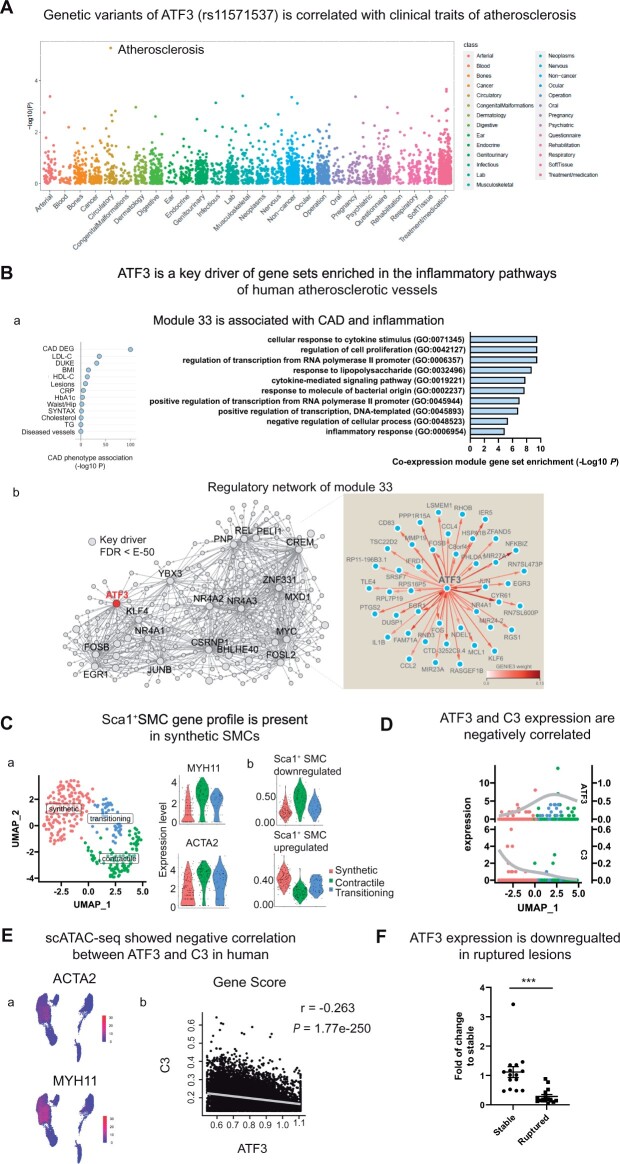
To determine the translational relevance of these findings, we next performed a series of experiments using clinical biospecimens and publicly available human biobank data. First, we conducted a phenome-wide association study (PheWAS) in 487 409 individuals from the UK Biobank to define the spectrum of clinical diagnoses associated with a commonly occurring candidate variant found within the intron of ATF3 (rs11571537; P = 8E-6). We conducted a functional annotation analysis using the quantitative scoring system embedded in the 3DSNP database,25 which revealed evidence for rs11571537 located within the TF binding site. As shown in Figure 5A, individuals who harbour this risk allele (who may have reduced expression of ATF3 in vivo) have a significantly increased risk of developing atherosclerotic disease, but no increase in other non-cardiovascular conditions (Bonferroni correction P threshold = 0.05/3100). By analysing inferred gene regulatory networks, we found that ATF3 drives the expression of genes in the STARNET co-expression module 33 (Figure 5B). In particular, these genes are: (i) predominantly co-expressed in atherosclerotic vascular tissue; (ii) enriched for CAD-DEGs; and (iii) enriched in pathways of cytokine and inflammatory responses (Figure 5B). Using 6000 SMCs isolated from human carotid plaques, we observed that while many lesional SMCs lose all detectable levels of ATF3, a modest but significant negative correlation between ATF3 and C3 expression could still be detected in atherosclerotic human vessels (correlation: −0.0563, P = 6E-04). Subsequent studies in a larger clinical dataset from the Athero-Express Study (which allowed for clustering of lesional SMCs by differentiation status according to expression of the SMC contractile genes, MYH11 and ACTA2), confirmed enrichment of genes associated with the murine Sca1+ SMCs within ‘synthetic’ human SMCs (Figure 5C). Similar to results from our mouse experiments (Figure 4B), ATF3 was anti-correlated with the up-regulated ‘Sca1+ SMC signature genes’ in the synthetic human plaque SMCs [Spearman’s rank correlation: −0.22 (Up), P = 0.01075), as graphically demonstrated in Figure 5D. We also queried sc-ATAC seq datasets from atherosclerotic human coronary artery plaques (n = 41). Similar to our murine observations, several clusters of variably differentiated SMCs could be resolved, based on their gene scores for MYH11 and ACTA2. As shown in Figure 5E, ATF3’s gene scores were inversely associated with C3, a member of the complement cascade. Finally, we measured ATF3 expression in stable and ruptured human carotid lesions from the Munich Vascular Biobank. These experiments indicated a nearly five-fold lower expression of ATF3 in ruptured lesions, suggesting a link to plaque vulnerability and clinical outcomes (Figure 5F). Together, these translational data confirm and extend observations made in lineage-traced mouse SMCs, suggesting that loss of ATF3 may also occur during the atherogenic transition of SMCs within the human plaque.
Figure 5.
Loss of ATF3 is correlated with activation of the complement cascade and atherosclerotic disease in humans. (A) A PheWAS study based on data from the UK biobank identified a genetic variant in ATF3 (rs11571537) that is highly correlated with atherosclerosis, but not other non-cardiovascular phenotypes. (B) In the STARNET study of RNA-seq data from n = 672 CAD patients, co-expression module 33 contains genes primarily from human atherosclerotic aorta (AOR), and is enriched for DEGs found in patients with angiographically confirmed CAD (CAD DEG). GSEA of AOR genes (Gene Ontology, Enrichr) indicated these genes are involved in the response to cytokines and inflammation (subpanel a). We found that ATF3 is a key driver (FDR<e-50, Mergeomics weigthed Key Driver Analysis) of the GENIE3 regulatory network for co-expression module 33 (subpanel b). (C) scRNA-seq data from human carotid lesions in the Athero-Express Biobank Study revealed three SMC subsets based on their transcriptome profiles and expression of contractile genes like MYH11 and ATAC2. These populations are termed: contractile, transitioning, and synthetic (subpanel a). Expression levels of the genes, which are differentially expressed in Sca1+ SMC were determined among these three subsets of human SMC (subpanel b). (D) C3 and ATF3 expression are visualized to illustrate their inverse correlation in human SMCs. (E) scATAC-seq of human coronary artery samples revealed several clusters of SMC subsets that have high expression scores of contractile genes, including MYH11 and ATAC2 (red: highly expressed, subpanel a). Gene scores of C3 and ATF3 are negatively correlated among the SMC clusters (subpanel b) by Pearson’s correlation test (r=−0.263, P = 1.77E−250). (F) PCR results from human carotid lesions in the Munich Vascular Biobank demonstrate reduced ATF3 expression in ruptured compared to stable lesions. Data are presented as mean ± SEM. ***P < 0.001, two-tailed Mann–Whitney U test (n = 15 in each group).
4. Discussion
For decades, the cornerstone of therapy for atherosclerotic conditions, such as heart attack and stroke has been based on the management of traditional risk factors.26 While the treatment of conditions, such as hypertension and diabetes has certainly improved clinical outcomes, CVD remains the world’s leading killer.27 Major advances in hypothesis-free human genetics, such as the GWAS, have allowed investigators to pursue the so-called ‘residual risk’ underlying the shortcomings of currently-available therapies.28 Surprisingly, these GWAS studies revealed that the loci most significantly associated with risk of disease appear to function independently of all known classical risk factors.29 Rather, these ‘unbiased’ studies suggest that atherosclerosis may arise due to perturbations in intrinsic vascular cell behaviour,2 especially as they relate to SMC plasticity, inflammation, and cell-fate decision-making.30–32 These discoveries build upon the ‘lipid hypothesis of atherosclerosis’33 (e.g. that high LDL is a major causal factor underlying CVD) and help explain two additional hypotheses that have recently been validated as additional drivers of atherogenesis.
The first theory that was recently confirmed is the ‘inflammatory hypothesis of atherosclerosis’.34 After decades of preclinical work, recent clinical trials convincingly reported that inflammation-suppressing therapies can reduce the risk of major adverse cardiovascular events (MACE), and do so independently of any effect on traditional risk factors.35 While more precise approaches will be necessary to avoid the sequalae of non-specific immunosuppressive therapies, it is notable that benefit has been observed with a wide range of anti-inflammatory medicines, including anti-cytokine therapies,36 pro-efferocytic therapies,37 and therapies with an unclear mechanism of action.38 These data suggest that identifying novel pathways that regulate vascular inflammation, including that related to activation of the complement cascade, could allow for the development of new translational therapies to lower the risk of developing lethal cardiovascular events.
The second theory that was recently confirmed surrounds the ‘clonal hypothesis of atherosclerosis’. Nearly 50 years ago, Earl Benditt and his son hypothesized that human atherosclerotic plaques may arise in a monoclonal fashion,39 though limitations of technology available during that era prevented additional mechanistic exploration. More recently, remarkable studies using large human genetics biobanks revealed that the clonal expansion of haematopoietic cells is independently associated with risk of MACE (a phenomenon known as ‘CHIP’ or ‘clonal haematopoiesis of indeterminate potential’),13 and sophisticated murine lineage-tracing studies confirmed that vascular SMCs expand in an oligoclonal fashion while promoting inflammation and lesion growth.5,23 While several groups have focused on the behaviour of the dominant SMC clone in the plaque, elegant studies, which applied scRNA-seq technology to lineage-traced SMCs confirmed that mature SMCs de-differentiate and actually give rise to a wide range of diverse phenotypes in the plaque, including those that resemble ‘macrophage-like’ cells24 (which are presumably deleterious) and components of the fibrous cap4 (which presumably are beneficial and prevent plaque rupture). Accordingly, understanding how the SMC ‘decides’ which phenotype to adopt, and whether it can be intervened upon to direct cells towards a more favourable phenotype, is a major area of interest to vascular biologists.40
So far, the upstream factors leading SMCs to initiate the atherogenic transition have remained elusive. Because the human phenomenon of CHIP has convincingly been shown to be driven by age-dependent somatic mutations, we hypothesized that intrinsic vessel wall cells might similarly acquire genetic variants in the developing plaque. This hypothesis is difficult to test in humans (due to the inability to definitively isolate de-differentiated SMCs from plaques), and could theoretically be challenging to test in mice (given the shorter timeline necessitated by murine atherosclerosis models). Nevertheless, we did identify a small number of SMC-specific mutations that could affect a broad spectrum of genes, including in MSL1, which is a component of histone acetyltransferase complex that modulates chromatin strucutrue.41 Similar to the key CHIP driver gene, DNMT3A, which changes the phenotype of immune cells via alterations in DNA methylation and hence chromatin structure,42 it remains possible that somatic mutations play a role in SMC phenotypes in human. However, the relative paucity of mutations documented in the ‘de-differentiated’ and pro-inflammatory murine SMCs examined in this study suggests the presence of an alternative mechanism responsible for their pathological behaviour.
For this reason, we next surveyed the chromatin architecture of lesional SMCs, and correlated disease-associated changes with DEGs in the murine atherosclerotic plaque.43 By analysing both promoter regions and TF binding sites, we found that the deleterious features of lesional SMCs, which include activation of genes in the complement cascade and down-regulation of several differentiation markers, are strongly correlated to chromatin accessibility changes observed via bulk sequencing technology. We then extended this work using single-cell RNA- and ATAC-sequencing approaches, and confirmed the marked diversity of SMCs in the plaque at both the transcriptional and epigenetic levels. When analysing the individual clusters of SMC-derived cells, we found that the Sca1+ subset harboured the highest number of dysregulated GWAS-proven, CAD-related risk genes, and also appeared to be the most ‘epigenetically-driven’ population of SMCs in the plaque.
Using a variety of informatic algorithms, we predicted that loss of accessibility to the upstream TF, ATF3, may explain how these cells adopt their harmful phenotype and pro-inflammatory status. This finding is interesting, given that ATF3 has previously been implicated in human CAD in the CARDIoGRAM Study (rs11571537, P = 7.8E-06),17 as well as several other diseases, such as diabetes and cancer, via its role in the modulation of inflammatory genes.44 ATF3 is an early responder to stress, which causes context-dependent reactions to perturbations in cellular homeostasis.44 Whole body knockout of ATF3 in mice did not manifest an obvious phenotypic change in the cardiovascular system.45 Knockout of ATF3 in ApoE−/− mice led to enlargement of atherosclerotic lesions, possibly due to increased formation of cholesterol esters facilitated by cholesterol 25-hydroxylase, which ATF3 represses when bound to its promoter.46 Because the concept of SMC ‘transdifferentiation’ towards a foam cells-like phenotype47 had not been well established at that time, a specific effect of ATF3 in SMC-derived foam cells was not observed in this study. Hepatocyte-specific depletion of ATF3 was also recently shown to strongly promote atherosclerosis.22 However, it should be noted that the regulatory effects of ATF3 may be dynamic and highly context-dependent.48 In the liver, ATF3 is immediately up-regulated in response to physiological stress, then returns to a low level within several hours.49 This pattern was reflected by our pseudotime analysis (Figure 3D), where ATF3 activity increased in the transitional SMC population (‘plaque 2’), then significantly fell in the pro-inflammatory SMC group (‘plaque 1’). We have also reanalysed a recently published scRNA-seq data from SMC-lineage-traced mice on an LDL receptor knockout background.50 Consistent with our observation, expression of ATF3 remained high in differentiated SMCs early in the disease course (after 0 and 8 weeks of HFD) but fell in the de-differentiated SMCs found within advanced lesions (16 and 26 weeks HFD). These context-dependent features of ATF3 were further exemplified in a prior study showing that it is one of the most variably expressed genes in SMCs from vascular beds of different embryological origin.23 Knockdown of ATF3 induced SMC apoptosis in vitro, while ATF3 was up-regulation in a femoral injury mouse model of restenosis.51 Taken together, these previous studies highlight how the dynamic properties of ATF3 could be beneficial or detrimental depending on the physiological condition. Because our human PheWAS and biobank studies confirmed a protective role for ATF3 in clinical outcomes, future studies should investigate whether targeting lesional ATF3 expression or chromatin accessibility is sufficient to prevent the emergence of C3-producing SMCs, and if this could ameliorate atherosclerosis in vivo.
The anti-inflammatory function of ATF3 has previously been reported in another context. In bone-marrow-derived macrophages, high-density lipoproteins induce the recruitment of ATF3 onto the promoter region of pro-inflammatory cytokines, such as interleukin-6 and tumour necrosis factor-α, whereupon it appears to suppress their expression.52 A parallel regulatory mechanism was observed in our study, where pre-bound ATF3 at the C3 promoter appeared to prevent activation of complement cascade in healthy SMCs, while this repressive activity is lost during atherogenic transition (Figure 4D–F). Hai’s group reported that ATF3 can act as its own repressor by binding to a motif immediately after the TATA box,53 potentially explaining the transient up-regulation and subsequent down-regulation of ATF3 in prior work.49 Our pseudotime analysis (Figure 3D) points to a similar negative feedback loop in SMCs, where ATF3 activity was increased in the ‘plaque 2’ subpopulation prior to its atherogenic transitioning, followed by a drop in ATF3 activity in the more diseased ‘plaque 1’.49 Additional studies on these complex molecular dynamics are needed to explore whether the loss of ATF3 function during the atherogenic transition is the result or the cause of chronic inflammatory stress.
These studies have several limitations that warrant consideration. In addition to difficulty extrapolating results across species due to an inability to lineage trace SMCs in humans, we did not confirm a causal relationship between ATF3 and SMC plasticity in this study. Future work should include SMC-specific knockout of ATF3 in an atheroprone multi-colour murine lineage-tracing system, to determine if this factor truly governs the ‘atherogenic transition’ of SMCs. In a recent scRNA-seq study that aimed to define the trajectory of SMCs during the atherogenic transition, Sca1+ SMCs were defined as an intermediate and multipotent group of SMCs that could differentiate into macrophage-like or fibrochondrocyte-like cells, and potentially even back towards a mature SMC phenotype.50 Formal dual lineage tracing of Sca1+ SMCs is necessary to determine whether the diverse SMC phenotypes observed in the lesion, including pro-inflammatory SMCs and SMC-derived foam cells,47 are derived from Sca1+ SMCs. Also, we do not know if other molecular mechanisms may contribute to SMC diversity in the plaque, which could be addressed with emerging simultaneous scRNA-seq/scATAC-seq platform technology. Our available scATAC-seq data and trajectory analyses suggest the involvement of multiple SMC- and CAD-related TFs networks, including those related to TCF21, SMAD3, LMOD1, and KLF4,24,31,32,54 which can be pursued with systems biology approaches to fully map the regulome of SMC diversity. For example, KLF4, a well-recognized transcription regulator of SMC phenotypic transitioning,24 has a putative binding motif on the ATF3 promoter, and may facilitate ATF3’s regulatory function.55 TCF21, a CAD GWAS TF that regulates some of the features of coronary SMCs during atherogenesis,56 also has DARs in its motif, but is not expressed in the ascending aorta, which arises from an embryological origin distinct from what gives rise to the coronary artery smooth muscle.57 The complexity of the dynamic interactions within a TF network and in different cellular contexts needs to be considered in future studies. Finally, while we observed significant overlap of changes to the chromatin architecture at loci previously implicated in CAD via GWAS studies, we do not yet definitively know the role of epigenetic changes in the heritable component of CVD, and if this should become a new translational target.
In summary, we have observed that nearly ∼25% of the chromatin landscape is altered in SMCs during atherogenesis. There appears to be marked heterogeneity in this process amongst the various subpopulations of cells, which arise from the ‘quiescent’ vascular SMC. Changes in the accessibility of certain TFs including ATF3 may constitute an early change that leads to the emergence of the inflammatory cells that drive atherosclerosis. Such changes may explain why targeting lipids alone has failed as an approach to completely limit risk of heart attack and stroke, and suggest that therapies, which suppress SMC inflammation may represent new opportunities to address vascular disease.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Authors’ contributions
Y.W., H.G., F.W., Z.Y., and N.J.L. designed research; Y.W., Z.Y., L.L., and J.Y. performed experiments on animal models and in vitro assays; Y.W., H.G., F.W., M.M., A.W.T., S.K., T.A., E.G.R., and N.J.L. analysed data. G.P., L.M., C.L.M., J.L.M.B., T.A., and E.G.R. contributed to datasets on human CVD and analytic tools; S.S.A., G.P., C.L.M., S.K., E.G.R., and J.L.M.B. provided feedback on data; and Y.W., H.G., F.W, Z.Y., A.W.T., S.K., L.M., C.L.M., S.K., M.E., X.G., and N.J.L. wrote the article.
Supplementary Material
Acknowledgements
The authors wish to acknowledge Quanyi Zhao, Max Käller and Moritz von Scheidt for critical input on the manuscript, and the Stanford Shared FACS Facility (via NIH S10 Shared Instrument Grant S10RR027431-01) for access to core facilities.
Funding
This study was supported by the National Institutes of Health (R35 HL144475 to N.J.L; R01HL125863 to J.L.M.B.; R01HL148239 and R00HL125912 to C.L.M.; 5K01HL148639-02 to E.G.R.), the American Heart Association (19EIA34770065 to N.J.L.; 18POST34030084 to Y.W.; A14SFRN20840000 to J.L.M.B.; 20POST35120545 to A.W.T.), the Swedish Research Council and Heart Lung Foundation (2018-02529 and 20170265 to J.L.M.B.), and the Fondation Leducq (‘PlaqOmics’ 18CVD02 to N.J.L., J.L.M.B., C.L.M., and G.P.).
Contributor Information
Ying Wang, Department of Surgery, Division of Vascular Surgery, Stanford University School of Medicine, 300 Pasteur drive, Stanford, CA 94305, USA; Stanford Cardiovascular Institute, Stanford University, 265 Campus Drive, Stanford, CA 94305, USA.
Hua Gao, Department of Surgery, Division of Vascular Surgery, Stanford University School of Medicine, 300 Pasteur drive, Stanford, CA 94305, USA; Stanford Cardiovascular Institute, Stanford University, 265 Campus Drive, Stanford, CA 94305, USA.
Fudi Wang, Department of Surgery, Division of Vascular Surgery, Stanford University School of Medicine, 300 Pasteur drive, Stanford, CA 94305, USA; Stanford Cardiovascular Institute, Stanford University, 265 Campus Drive, Stanford, CA 94305, USA.
Zhongde Ye, Department of Surgery, Division of Vascular Surgery, Stanford University School of Medicine, 300 Pasteur drive, Stanford, CA 94305, USA; Stanford Cardiovascular Institute, Stanford University, 265 Campus Drive, Stanford, CA 94305, USA.
Michal Mokry, Department of Cardiology and Laboratory of Clinical Chemistry, University Medical Center Utrecht, Heidelberglaan 100, Utrecht 3584 CX, the Netherlands.
Adam W Turner, Center for Public Health Genomics, Department of Public Health Sciences, University of Virginia, 1335 Lee St, Charlottesville, VA 22908, USA.
Jianqin Ye, Department of Surgery, Division of Vascular Surgery, Stanford University School of Medicine, 300 Pasteur drive, Stanford, CA 94305, USA.
Simon Koplev, Cancer Research UK Cambridge Institute, University of Cambridge, Robinson Way, Cambridge CB2 0RE, UK.
Lingfeng Luo, Department of Surgery, Division of Vascular Surgery, Stanford University School of Medicine, 300 Pasteur drive, Stanford, CA 94305, USA; Stanford Cardiovascular Institute, Stanford University, 265 Campus Drive, Stanford, CA 94305, USA.
Tom Alsaigh, Department of Surgery, Division of Vascular Surgery, Stanford University School of Medicine, 300 Pasteur drive, Stanford, CA 94305, USA; Department of Cardiovascular Medicine, Stanford University School of Medicine, 870 Quarry Road Extension, Stanford, CA 94305, USA.
Shaunak S Adkar, Department of Surgery, Division of Vascular Surgery, Stanford University School of Medicine, 300 Pasteur drive, Stanford, CA 94305, USA.
Maria Elishaev, Department of Pathology and Laboratory Medicine, Centre for Heart Lung Innvoation, University of British Columbia, 166-1081 Burrard St, Vancouver, BC V6Z 1Y6, Canada.
Xiangyu Gao, Department of Pathology and Laboratory Medicine, Centre for Heart Lung Innvoation, University of British Columbia, 166-1081 Burrard St, Vancouver, BC V6Z 1Y6, Canada.
Lars Maegdefessel, Department for Vascular and Endovascular Surgery, Klinikum rechts der Isar, Technical University Munich, and the German Center for Cardiovascular Research (DZHK partner site), Biedersteiner Str. 29, Munich 80802, Germany; Department of Internal Medicine, Center for Molecular Medicine, Karolinska Institute, Visionsgatan 18, Stockholm 171 76, Sweden.
Johan L M Björkegren, Department of Genetics and Genomic Sciences, Icahn Institute for Genomics and Multiscale Biology, Icahn School of Medicine at Mount Sinai, 1425 Madison Ave, New York, NY 10029, USA.
Gerard Pasterkamp, Department of Cardiology and Laboratory of Clinical Chemistry, University Medical Center Utrecht, Heidelberglaan 100, Utrecht 3584 CX, the Netherlands.
Clint L Miller, Center for Public Health Genomics, Department of Public Health Sciences, University of Virginia, 1335 Lee St, Charlottesville, VA 22908, USA.
Elsie G Ross, Department of Surgery, Division of Vascular Surgery, Stanford University School of Medicine, 300 Pasteur drive, Stanford, CA 94305, USA; Stanford Cardiovascular Institute, Stanford University, 265 Campus Drive, Stanford, CA 94305, USA.
Nicholas J Leeper, Department of Surgery, Division of Vascular Surgery, Stanford University School of Medicine, 300 Pasteur drive, Stanford, CA 94305, USA; Stanford Cardiovascular Institute, Stanford University, 265 Campus Drive, Stanford, CA 94305, USA.
Data availability
The raw sequencing data are available from NCBI BioProject PRJNA716327.
Translational perspective
The recent CANTOS and COLCOT trials have shown that targeting inflammatory pathways lowers the risk of major adverse cardiovascular events. However, more specific targets are needed to avoid immunosuppressive side effects. Our data identify an upstream regulator of pro-inflammatory SMCs, ATF3, which is involved in the initial atherogenic transitioning of lesional SMCs. Restoring ATF3 activity may prevent the de-differentiation of SMCs and offer a novel translational approach for the suppression of complement-dependent inflammation in atherosclerotic lesions.
References
- 1. Libby P. The changing landscape of atherosclerosis. Nature 2021;592:524–533. [DOI] [PubMed] [Google Scholar]
- 2. Howson JMM, Zhao W, Barnes DR, Ho WK, Young R, Paul DS, Waite LL, Freitag DF, Fauman EB, Salfati EL, Sun BB, Eicher JD, Johnson AD, Sheu WHH, Nielsen SF, Lin WY, Surendran P, Malarstig A, Wilk JB, Tybjaerg-Hansen A, Rasmussen KL, Kamstrup PR, Deloukas P, Erdmann J, Kathiresan S, Samani NJ, Schunkert H, Watkins H, Do R, Rader DJ, Johnson JA, Hazen SL, Quyyumi AA, Spertus JA, Pepine CJ, Franceschini N, Justice A, Reiner AP, Buyske S, Hindorff LA, Carty CL, North KE, Kooperberg C, Boerwinkle E, Young K, Graff M, Peters U, Absher D, Hsiung CA, Lee WJ, Taylor KD, Chen YH, Lee IT, Guo X, Chung RH, Hung YJ, Rotter JI, Juang JJ, Quertermous T, Wang TD, Rasheed A, Frossard P, Alam DS, Majumder AAS, Di Angelantonio E, Chowdhury R, Epic CVD, Chen YI, Nordestgaard BG, Assimes TL, Danesh J, Butterworth AS, Saleheen D; CardioGramplusC4D . Fifteen new risk loci for coronary artery disease highlight arterial-wall-specific mechanisms. Nat Genet 2017;49:1113–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gomez D, Owens GK.. Smooth muscle cell phenotypic switching in atherosclerosis. Cardiovasc Res 2012;95:156–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Alencar GF, Owsiany KM, Karnewar S, Sukhavasi K, Mocci G, Nguyen AT, Williams CM, Shamsuzzaman S, Mokry M, Henderson CA, Haskins R, Baylis RA, Finn AV, McNamara CA, Zunder ER, Venkata V, Pasterkamp G, Bjorkegren J, Bekiranov S, Owens GK.. Stem cell pluripotency genes Klf4 and Oct4 regulate complex SMC phenotypic changes critical in late-stage atherosclerotic lesion pathogenesis. Circulation 2020;142:2045–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang Y, Nanda V, Direnzo D, Ye J, Xiao S, Kojima Y, Howe KL, Jarr KU, Flores AM, Tsantilas P, Tsao N, Rao A, Newman AAC, Eberhard AV, Priest JR, Ruusalepp A, Pasterkamp G, Maegdefessel L, Miller CL, Lind L, Koplev S, Bjorkegren JLM, Owens GK, Ingelsson E, Weissman IL, Leeper NJ.. Clonally expanding smooth muscle cells promote atherosclerosis by escaping efferocytosis and activating the complement cascade. Proc Natl Acad Sci USA 2020;117:15818–15826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Depuydt MAC, Prange KHM, Slenders L, Ord T, Elbersen D, Boltjes A, de Jager SCA, Asselbergs FW, de Borst GJ, Aavik E, Lonnberg T, Lutgens E, Glass CK, den Ruijter HM, Kaikkonen MU, Bot I, Slutter B, van der Laan SW, Yla-Herttuala S, Mokry M, Kuiper J, de Winther MPJ, Pasterkamp G.. Microanatomy of the human atherosclerotic plaque by single-cell transcriptomics. Circ Res 2020;127:1437–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Franzén O, Ermel R, Cohain A, Akers NK, Di Narzo A, Talukdar HA, Foroughi-Asl H, Giambartolomei C, Fullard JF, Sukhavasi K, Köks S, Gan L-M, Giannarelli C, Kovacic JC, Betsholtz C, Losic B, Michoel T, Hao K, Roussos P, Skogsberg J, Ruusalepp A, Schadt EE, Björkegren JLM.. Cardiometabolic risk loci share downstream cis- and trans-gene regulation across tissues and diseases. Science 2016;353:827–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G.. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 2013;31:213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Corces MR, Corces MR, Trevino AE, Hamilton EG, Greenside PG, Sinnott-Armstrong NA, Vesuna S, Satpathy AT, Rubin AJ, Montine KS, Wu B, Kathiria A, Cho SW, Mumbach MR, Carter AC, Kasowski M, Orloff LA, Risca VI, Kundaje A, Khavari PA, Montine TJ, Greenleaf WJ, Chang HY.. Omni-ATAC-seq: improved ATAC-seq protocol. Nat Methods 2017;14:959–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shao Z, Zhang Y, Yuan GC, Orkin SH, Waxman DJ.. MAnorm: a robust model for quantitative comparison of ChIP-Seq data sets. Genome Biol 2012;13:R16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stuart T, Srivastava A, Madad S, Lareau C, Satija R.. Single-cell chromatin analysis with Signac. Nat Methods 2021;18:1333–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Granja JM, Corces MR, Pierce SE, Bagdatli ST, Choudhry H, Chang HY, Greenleaf WJ. ArchR is a scalable software package for integrative single-cell chromatin accessibility analysis. Nat Genet 2021;53:403–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jaiswal S, Natarajan P, Silver AJ, Gibson CJ, Bick AG, Shvartz E, McConkey M, Gupta N, Gabriel S, Ardissino D, Baber U, Mehran R, Fuster V, Danesh J, Frossard P, Saleheen D, Melander O, Sukhova GK, Neuberg D, Libby P, Kathiresan S, Ebert BL.. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N Engl J Med 2017;377:111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Freire T, Bay S, von Mensdorff-Pouilly S, Osinaga E.. Molecular basis of incomplete O-glycan synthesis in MCF-7 breast cancer cells: putative role of MUC6 in Tn antigen expression. Cancer Res 2005;65:7880–7887. [DOI] [PubMed] [Google Scholar]
- 15. Wu L, Zee BM, Wang Y, Garcia BA, Dou Y.. The RING finger protein MSL2 in the MOF complex is an E3 ubiquitin ligase for H2B K34 and is involved in crosstalk with H3 K4 and K79 methylation. Mol Cell 2011;43:132–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Buenrostro JD, Wu B, Chang HY, Greenleaf WJ.. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol 2015;109:21.29.1–21.29.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. van der Harst P, Verweij N.. Identification of 64 novel genetic loci provides an expanded view on the genetic architecture of coronary artery disease. Circ Res 2018;122:433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kwon JW, Kwon HK, Shin HJ, Choi YM, Anwar MA, Choi S.. Activating transcription factor 3 represses inflammatory responses by binding to the p65 subunit of NF-kappaB. Sci Rep 2015;5:14470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mallano T, Palumbo-Zerr K, Zerr P, Ramming A, Zeller B, Beyer C, Dees C, Huang J, Hai T, Distler O, Schett G, Distler JH.. Activating transcription factor 3 regulates canonical TGFbeta signalling in systemic sclerosis. Ann Rheum Dis 2016;75:586–592. [DOI] [PubMed] [Google Scholar]
- 20. Sharma V, Dogra N, Saikia UN, Khullar M.. Transcriptional regulation of endothelial-to-mesenchymal transition in cardiac fibrosis: role of myocardin-related transcription factor A and activating transcription factor 3. Can J Physiol Pharmacol 2017;95:1263–1270. [DOI] [PubMed] [Google Scholar]
- 21. Yin X, Wolford CC, Chang YS, McConoughey SJ, Ramsey SA, Aderem A, Hai T.. ATF3, an adaptive-response gene, enhances TGF{beta} signaling and cancer-initiating cell features in breast cancer cells. J Cell Sci 2010;123:3558–3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xu Y, Li Y, Jadhav K, Pan X, Zhu Y, Hu S, Chen S, Chen L, Tang Y, Wang HH, Yang L, Wang DQ, Yin L, Zhang Y.. Hepatocyte ATF3 protects against atherosclerosis by regulating HDL and bile acid metabolism. Nat Metab 2021;3:59–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dobnikar L, Taylor AL, Chappell J, Oldach P, Harman JL, Oerton E, Dzierzak E, Bennett MR, Spivakov M, Jorgensen HF.. Disease-relevant transcriptional signatures identified in individual smooth muscle cells from healthy mouse vessels. Nat Commun 2018;9:4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shankman LS, Gomez D, Cherepanova OA, Salmon M, Alencar GF, Haskins RM, Swiatlowska P, Newman AA, Greene ES, Straub AC, Isakson B, Randolph GJ, Owens GK.. KLF4-dependent phenotypic modulation of smooth muscle cells has a key role in atherosclerotic plaque pathogenesis. Nat Med 2015;21:628–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lu Y, Quan C, Chen H, Bo X, Zhang C.. 3DSNP: a database for linking human noncoding SNPs to their three-dimensional interacting genes. Nucleic Acids Res 2017;45:D643–D649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Brown BG, Zhao XQ, Sacco DE, Albers JJ.. Lipid lowering and plaque regression. New insights into prevention of plaque disruption and clinical events in coronary disease. Circulation 1993;87:1781–1791. [DOI] [PubMed] [Google Scholar]
- 27. Virani SS, Alonso A, Benjamin EJ, Bittencourt MS, Callaway CW, Carson AP, Chamberlain AM, Chang AR, Cheng S, Delling FN, Djousse L, Elkind MSV, Ferguson JF, Fornage M, Khan SS, Kissela BM, Knutson KL, Kwan TW, Lackland DT, Lewis TT, Lichtman JH, Longenecker CT, Loop MS, Lutsey PL, Martin SS, Matsushita K, Moran AE, Mussolino ME, Perak AM, Rosamond WD, Roth GA, Sampson UKA, Satou GM, Schroeder EB, Shah SH, Shay CM, Spartano NL, Stokes A, Tirschwell DL, VanWagner LB, Tsao CW; American Heart Association Council on Epidemiology and Prevention Statistics Committee and Stroke Statistics Subcommittee . Heart disease and stroke statistics-2020 update: a report from the American Heart Association. Circulation 2020;141:e139–e596. [DOI] [PubMed] [Google Scholar]
- 28. Nikpay M, Goel A, Won HH, Hall LM, Willenborg C, Kanoni S, Saleheen D, Kyriakou T, Nelson CP, Hopewell JC, Webb TR, Zeng L, Dehghan A, Alver M, Armasu SM, Auro K, Bjonnes A, Chasman DI, Chen S, Ford I, Franceschini N, Gieger C, Grace C, Gustafsson S, Huang J, Hwang SJ, Kim YK, Kleber ME, Lau KW, Lu X, Lu Y, Lyytikainen LP, Mihailov E, Morrison AC, Pervjakova N, Qu L, Rose LM, Salfati E, Saxena R, Scholz M, Smith AV, Tikkanen E, Uitterlinden A, Yang X, Zhang W, Zhao W, de Andrade M, de Vries PS, van Zuydam NR, Anand SS, Bertram L, Beutner F, Dedoussis G, Frossard P, Gauguier D, Goodall AH, Gottesman O, Haber M, Han BG, Huang J, Jalilzadeh S, Kessler T, Konig IR, Lannfelt L, Lieb W, Lind L, Lindgren CM, Lokki ML, Magnusson PK, Mallick NH, Mehra N, Meitinger T, Memon FU, Morris AP, Nieminen MS, Pedersen NL, Peters A, Rallidis LS, Rasheed A, Samuel M, Shah SH, Sinisalo J, Stirrups KE, Trompet S, Wang L, Zaman KS, Ardissino D, Boerwinkle E, Borecki IB, Bottinger EP, Buring JE, Chambers JC, Collins R, Cupples LA, Danesh J, Demuth I, Elosua R, Epstein SE, Esko T, Feitosa MF, Franco OH, Franzosi MG, Granger CB, Gu D, Gudnason V, Hall AS, Hamsten A, Harris TB, Hazen SL, Hengstenberg C, Hofman A, Ingelsson E, Iribarren C, Jukema JW, Karhunen PJ, Kim BJ, Kooner JS, Kullo IJ, Lehtimaki T, Loos RJF, Melander O, Metspalu A, Marz W, Palmer CN, Perola M, Quertermous T, Rader DJ, Ridker PM, Ripatti S, Roberts R, Salomaa V, Sanghera DK, Schwartz SM, Seedorf U, Stewart AF, Stott DJ, Thiery J, Zalloua PA, O'Donnell CJ, Reilly MP, Assimes TL, Thompson JR, Erdmann J, Clarke R, Watkins H, Kathiresan S, McPherson R, Deloukas P, Schunkert H, Samani NJ, Farrall M.. A comprehensive 1,000 genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet 2015;47:1121–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. O'Donnell CJ, Kavousi M, Smith AV, Kardia SL, Feitosa MF, Hwang SJ, Sun YV, Province MA, Aspelund T, Dehghan A, Hoffmann U, Bielak LF, Zhang Q, Eiriksdottir G, van Duijn CM, Fox CS, de Andrade M, Kraja AT, Sigurdsson S, Elias-Smale SE, Murabito JM, Launer LJ, van der Lugt A, Kathiresan S, Consortium CA, Krestin GP, Herrington DM, Howard TD, Liu Y, Post W, Mitchell BD, O'Connell JR, Shen H, Shuldiner AR, Altshuler D, Elosua R, Salomaa V, Schwartz SM, Siscovick DS, Voight BF, Bis JC, Glazer NL, Psaty BM, Boerwinkle E, Heiss G, Blankenberg S, Zeller T, Wild PS, Schnabel RB, Schillert A, Ziegler A, Munzel TF, White CC, Rotter JI, Nalls M, Oudkerk M, Johnson AD, Newman AB, Uitterlinden AG, Massaro JM, Cunningham J, Harris TB, Hofman A, Peyser PA, Borecki IB, Cupples LA, Gudnason V, Witteman JC; CARDIoGRAM Consortium . Genome-wide association study for coronary artery calcification with follow-up in myocardial infarction. Circulation 2011;124:2855–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McPherson R, Davies RW.. Inflammation and coronary artery disease: insights from genetic studies. Can J Cardiol 2012;28:662–666. [DOI] [PubMed] [Google Scholar]
- 31. Nanda V, Wang T, Pjanic M, Liu B, Nguyen T, Matic LP, Hedin U, Koplev S, Ma L, Franzen O, Ruusalepp A, Schadt EE, Bjorkegren JLM, Montgomery SB, Snyder MP, Quertermous T, Leeper NJ, Miller CL.. Functional regulatory mechanism of smooth muscle cell-restricted LMOD1 coronary artery disease locus. PLoS Genet 2018;14:e1007755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim JB, Pjanic M, Nguyen T, Miller CL, Iyer D, Liu B, Wang T, Sazonova O, Carcamo-Orive I, Matic LP, Maegdefessel L, Hedin U, Quertermous T.. TCF21 and the environmental sensor aryl-hydrocarbon receptor cooperate to activate a pro-inflammatory gene expression program in coronary artery smooth muscle cells. PLoS Genet 2017;13:e1006750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Goldstein JL, Brown MS.. Atherosclerosis: the low-density lipoprotein receptor hypothesis. Metabolism 1977;26:1257–1275. [DOI] [PubMed] [Google Scholar]
- 34. Ross R. Atherosclerosis–an inflammatory disease. N Engl J Med 1999;340:115–126. [DOI] [PubMed] [Google Scholar]
- 35. Ridker PM, Everett BM, Thuren T, MacFadyen JG, Chang WH, Ballantyne C, Fonseca F, Nicolau J, Koenig W, Anker SD, Kastelein JJP, Cornel JH, Pais P, Pella D, Genest J, Cifkova R, Lorenzatti A, Forster T, Kobalava Z, Vida-Simiti L, Flather M, Shimokawa H, Ogawa H, Dellborg M, Rossi PRF, Troquay RPT, Libby P, Glynn RJ, Group CT; CANTOS Trial Group . Antiinflammatory therapy with canakinumab for atherosclerotic disease. N Engl J Med 2017;377:1119–1131. [DOI] [PubMed] [Google Scholar]
- 36. Ridker PM, MacFadyen JG, Thuren T, Libby P.. Residual inflammatory risk associated with interleukin-18 and interleukin-6 after successful interleukin-1beta inhibition with canakinumab: further rationale for the development of targeted anti-cytokine therapies for the treatment of atherothrombosis. Eur Heart J 2020;41:2153–2163. [DOI] [PubMed] [Google Scholar]
- 37. Jarr KU, Nakamoto R, Doan BH, Kojima Y, Weissman IL, Advani RH, Iagaru A, Leeper NJ.. Effect of CD47 blockade on vascular inflammation. N Engl J Med 2021;384:382–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Samuel M, Tardif JC, Khairy P, Roubille F, Waters Dd Gregoire JC, Pinto FJ, Maggioni AP, Diaz R, Berry C, Koenig W, Ostadal P, Lopez-Sendon J, Gamra H, Kiwan GS, Dube MP, Provencher M, Orfanos A, Blondeau L, Kouz S, L'Allier PL, Ibrahim R, Bouabdallaoui N, Mitchell D, Guertin MC, Lelorier J.. Cost-effectiveness of low-dose colchicine after myocardial infarction in the Colchicine Cardiovascular Outcomes Trial (COLCOT). Eur Heart J Qual Care Clin Outcomes 2020;7:486–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Benditt EP, Benditt JM.. Evidence for a monoclonal origin of human atherosclerotic plaques. Proc Natl Acad Sci USA 1973;70:1753–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. DiRenzo D, Owens GK, Leeper NJ.. "Attack of the Clones": commonalities between cancer and atherosclerosis. Circ Res 2017;120:624–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Smith ER, Cayrou C, Huang R, Lane WS, Cote J, Lucchesi JC.. A human protein complex homologous to the Drosophila MSL complex is responsible for the majority of histone H4 acetylation at lysine 16. Mol Cell Biol 2005;25:9175–9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Abplanalp WT, Cremer S, John D, Hoffmann J, Schuhmacher B, Merten M, Rieger MA, Vasa-Nicotera M, Zeiher AM, Dimmeler S.. Clonal hematopoiesis-driver DNMT3A mutations alter immune cells in heart failure. Circ Res 2021;128:216–228. [DOI] [PubMed] [Google Scholar]
- 43. Zhao Y, Zheng D, Cvekl A.. Profiling of chromatin accessibility and identification of general cis-regulatory mechanisms that control two ocular lens differentiation pathways. Epigenetics Chromatin 2019;12:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hai T, Wolford CC, Chang YS.. ATF3, a hub of the cellular adaptive-response network, in the pathogenesis of diseases: is modulation of inflammation a unifying component? Gene Expr 2010;15:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Soraya AS, Tali H, Rona S, Tom F, Roy K, Ami A.. ATF3 expression in cardiomyocytes and myofibroblasts following transverse aortic constriction displays distinct phenotypes. Int J Cardiol Heart Vasc 2021;32:100706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gold ES, Ramsey SA, Sartain MJ, Selinummi J, Podolsky I, Rodriguez DJ, Moritz RL, Aderem A.. ATF3 protects against atherosclerosis by suppressing 25-hydroxycholesterol-induced lipid body formation. J Exp Med 2012;209:807–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang Y, Dubland JA, Allahverdian S, Asonye E, Sahin B, Jaw JE, Sin DD, Seidman MA, Leeper NJ, Francis GA.. Smooth muscle cells contribute the majority of foam cells in ApoE (Apolipoprotein E)-deficient mouse atherosclerosis. Arterioscler Thromb Vasc Biol 2019;39:876–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ku HC, Cheng CF.. Master regulator activating transcription factor 3 (ATF3) in metabolic homeostasis and cancer. Front Endocrinol (Lausanne) 2020;11:556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chen BP, Wolfgang CD, Hai T.. Analysis of ATF3, a transcription factor induced by physiological stresses and modulated by gadd153/Chop10. Mol Cell Biol 1996;16:1157–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pan H, Xue C, Auerbach BJ, Fan J, Bashore AC, Cui J, Yang DY, Trignano SB, Liu W, Shi J, Ihuegbu CO, Bush EC, Worley J, Vlahos L, Laise P, Solomon RA, Connolly ES, Califano A, Sims PA, Zhang H, Li M, Reilly MP.. Single-cell genomics reveals a novel cell state during smooth muscle cell phenotypic switching and potential therapeutic targets for atherosclerosis in mouse and human. Circulation 2020;142:2060–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lv D, Meng D, Zou FF, Fan L, Zhang P, Yu Y, Fang J.. Activating transcription factor 3 regulates survivability and migration of vascular smooth muscle cells. IUBMB Life 2011;63:62–69. [DOI] [PubMed] [Google Scholar]
- 52. De Nardo D, Labzin LI, Kono H, Seki R, Schmidt SV, Beyer M, Xu D, Zimmer S, Lahrmann C, Schildberg FA, Vogelhuber J, Kraut M, Ulas T, Kerksiek A, Krebs W, Bode N, Grebe A, Fitzgerald ML, Hernandez NJ, Williams BR, Knolle P, Kneilling M, Rocken M, Lutjohann D, Wright SD, Schultze JL, Latz E.. High-density lipoprotein mediates anti-inflammatory reprogramming of macrophages via the transcriptional regulator ATF3. Nat Immunol 2014;15:152–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wolfgang CD, Liang G, Okamoto Y, Allen AE, Hai T.. Transcriptional autorepression of the stress-inducible gene ATF3. J Biol Chem 2000;275:16865–16870. [DOI] [PubMed] [Google Scholar]
- 54. Shi X, DiRenzo D, Guo LW, Franco SR, Wang B, Seedial S, Kent KC.. TGF-beta/Smad3 stimulates stem cell/developmental gene expression and vascular smooth muscle cell de-differentiation. PLoS One 2014;9:e93995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Whitlock NC, Bahn JH, Lee SH, Eling TE, Baek SJ.. Resveratrol-induced apoptosis is mediated by early growth response-1, Kruppel-like factor 4, and activating transcription factor 3. Cancer Prev Res (Phila) 2011;4:116–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wirka RC, Wagh D, Paik DT, Pjanic M, Nguyen T, Miller CL, Kundu R, Nagao M, Coller J, Koyano TK, Fong R, Woo YJ, Liu B, Montgomery SB, Wu JC, Zhu K, Chang R, Alamprese M, Tallquist MD, Kim JB, Quertermous T.. Atheroprotective roles of smooth muscle cell phenotypic modulation and the TCF21 disease gene as revealed by single-cell analysis. Nat Med 2019;25:1280–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nurnberg ST, Cheng K, Raiesdana A, Kundu R, Miller CL, Kim JB, Arora K, Carcamo-Oribe I, Xiong Y, Tellakula N, Nanda V, Murthy N, Boisvert WA, Hedin U, Perisic L, Aldi S, Maegdefessel L, Pjanic M, Owens GK, Tallquist MD, Quertermous T.. Coronary artery disease associated transcription factor TCF21 regulates smooth muscle precursor cells that contribute to the fibrous cap. PLoS Genet 2015;11:e1005155. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequencing data are available from NCBI BioProject PRJNA716327.
Translational perspective
The recent CANTOS and COLCOT trials have shown that targeting inflammatory pathways lowers the risk of major adverse cardiovascular events. However, more specific targets are needed to avoid immunosuppressive side effects. Our data identify an upstream regulator of pro-inflammatory SMCs, ATF3, which is involved in the initial atherogenic transitioning of lesional SMCs. Restoring ATF3 activity may prevent the de-differentiation of SMCs and offer a novel translational approach for the suppression of complement-dependent inflammation in atherosclerotic lesions.