ABSTRACT
The development of anti-virulence drug therapy against Acinetobacter baumannii infections would provide an alternative to traditional antibacterial therapy that are increasingly failing. Here, we demonstrate that the OmpR transcriptional regulator plays a pivotal role in the pathogenesis of diverse A. baumannii clinical strains in multiple murine and G. mellonella invertebrate infection models. We identified OmpR-regulated genes using RNA sequencing and further validated two genes whose expression can be used as robust biomarker to quantify OmpR inhibition in A. baumannii. Moreover, the determination of the structure of the OmpR DNA binding domain of A. baumannii and the development of in vitro protein-DNA binding assays enabled the identification of an OmpR small molecule inhibitor. We conclude that OmpR is a valid and unexplored target to fight A. baumannii infections and we believe that the described platform combining in silico methods, in vitro OmpR inhibitory assays and in vivo G. mellonella surrogate infection model will facilitate future drug discovery programs.
KEYWORDS: OmpR, A. baumannii, virulence, drug discovery
Introduction
The rapid development of antimicrobial resistance is an increasing serious issue and a global threat to public health [1,2]. The need for new antibacterial therapeutics is especially critical for drug-resistance Gram-negative bacteria, such as carbapenem-resistant Acinetobacter baumannii (CRAB) for which new approaches are urgently required [3]. Several European countries reported an endemic CRAB situation in 2019 [4] and the situation is also alarming in China and in U.S., [5,6]. Alternative approaches to traditional antimicrobials consist in targeting bacterial resistance or virulence mechanisms to disarm the pathogens [7–9]. The pathogenicity of A. baumannii is not fully understood but several virulence factors have been identified as part of its disease-causing ability, such as its ability to survive in adverse environmental conditions and to escape the host immune system [10,11].
Two-component systems (TCSs) play a crucial role in sensing and responding to environmental changes by adapting to diverse ecological niches, even under unfavourable conditions [12]. Typically, TCSs consist of an histidine kinase (HK) that senses specific external stimuli, and a response regulator (RR) that mediates the cellular response through differential expression of target genes. TCSs are often involved in the regulation of virulence genes expression [13], indicating that TCSs could serve as potential novel drug target in the development of anti-virulence strategies [14]. OmpR/EnvZ is a well-studied TCS in Enterobacteriaceae, where OmpR is the RR and EnvZ is the HK sensor [15–17]. Under osmolarity changes in specific cellular niches, OmpR regulates the expression of about 100 genes, including outer membrane proteins and virulence factors, such as fimbriae and pili [18,19]. In contrast, the role of OmpR is poorly explored in A. baumannii [20].
The aim of our study was to gain insight in the role of OmpR in A. baumannii pathogenesis to enable the development of OmpR inhibitors altering A. baumannii virulence. We demonstrated that OmpR is a key regulator of A. baumannii virulence using different infection models and diverse clinical strains. We further identified OmpR-regulated genes that can be used as biomarker of OmpR inhibition in whole cell assay. In addition, we solved the structure of the C-terminal domain of OmpR and developed a protein-DNA in vitro binding assay leading to the identification of an OmpR small molecule inhibitor derived from a virtual screening.
Results
OmpR is required for A. baumannii virulence in diverse mouse infection models
It has been recently suggested that OmpR plays a role in A. baumannii pathogenesis in the G. mellonella invertebrate infection model using the AB5075 strain [20]. We first evaluated if the attenuated virulence observed in G. mellonella correlates with the bacterial pathogenesis in vertebrate infection models. Neutropenic mouse thigh infection with the AB5075 wildtype strain resulted in a 2-log10 increase in thigh bacterial titre and a rapid dissemination of the infection in the blood, leading to the mortality of 1 and 3 animals at 24 and 48 hours, respectively (Figure 1(a,b)). In contrast, a 1 and 2-log10 reduction in thigh bacterial titer at 24 and 48 hours, respectively, no blood dissemination and no mortality was observed for the AB5075 ΔompR mutant (Figure 1(a,b)). These data indicate that OmpR is required for AB5075 to establish a robust infection in thigh infection model. Since A. baumannii is a threat pathogen causing sepsis, we next evaluated the role of OmpR in an immunocompetent septicaemia mouse model with the unrelated and highly virulent A. baumannii strain HUMC1 [21]. Infection with the wildtype HUMC1 strain led to 90% mortality at day one while the HUMC1 ΔompR mutant did not induce any mortality after 7 days (Figure 1(c)). Overall, ompR deletion led to strongly attenuated A. baumannii virulence protecting mice from bacteria-mediated killing and demonstrating the importance of OmpR in A. baumannii pathogenesis.
Figure 1.
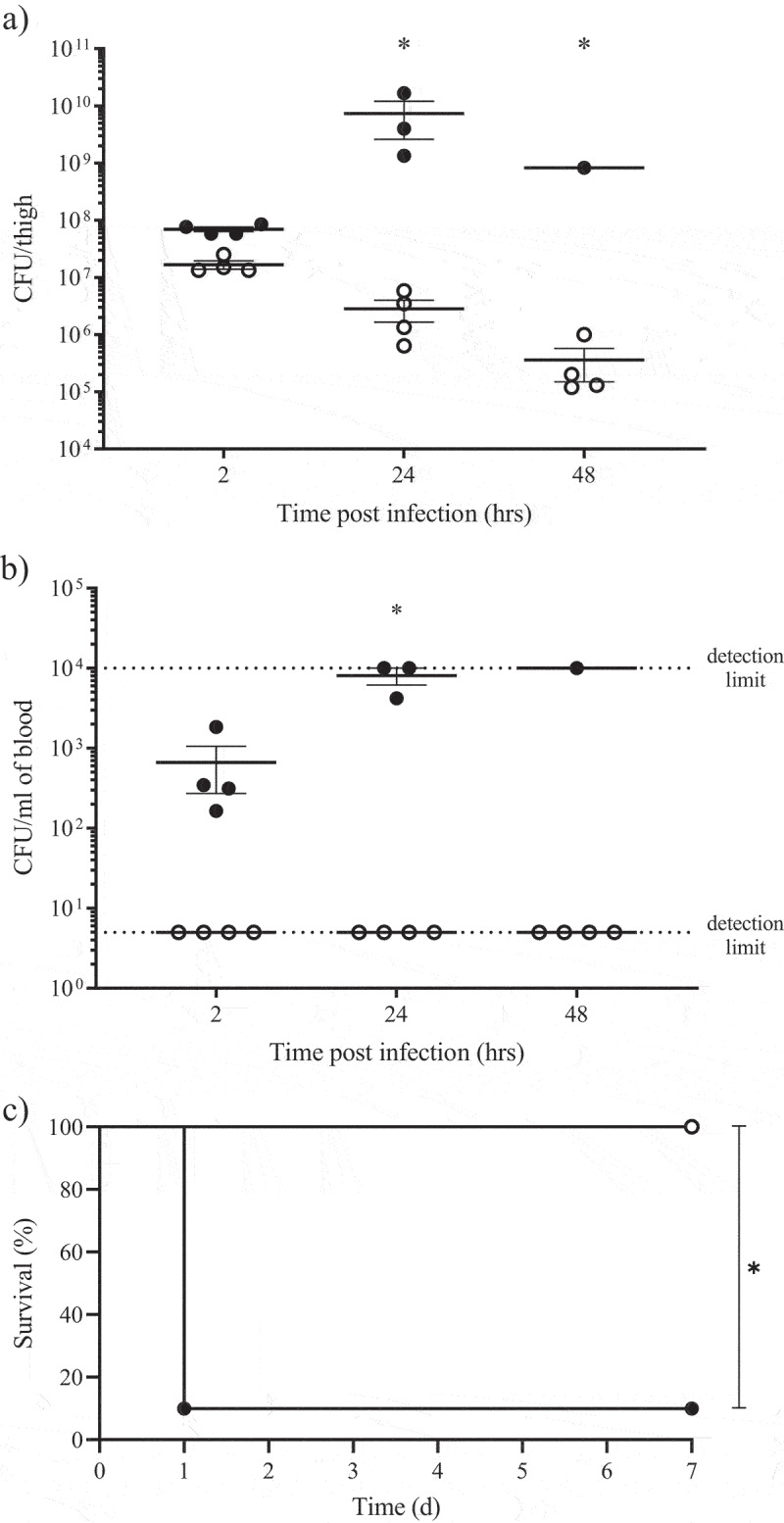
Role of OmpR in A. baumannii virulence assessed in thigh and septicaemia mouse infection models.
A) and B) Neutropenic CD-1 male mice (12 per group) were infected using thigh injection of 107 cfu of AB5075 wildtype (black) or AB5075 ΔompR mutant (white) and 4 animals per group were sacrificed at 2, 24 and 48 hours post infection for bacterial titers determination in A) thigh and B) blood. Mortality was observed in the wildtype group at 24 and 48 hours enabling bacterial titers analysis on only 3 and 1 animal, respectively. Unpaired t-test, *P-value < 0.05. C) Immunocompetent C57BL6J male mice (10 per group) were infected with intravenous injection of 5 × 107 cfu of HUMC1 wildtype (black) or HUMC1 ΔompR mutant (white) and mice survival was monitored for 7 days. Log-rank (Mantel-Cox) test, *P-value < 0.05.
G. mellonella infection model can be used as surrogate of mouse model to characterize OmpR-mediated A. baumannii virulence
We implemented the G. mellonella invertebrate infection model as a more ethical and cost-effective surrogate to mouse infection models [22,23] to further evaluate the role of OmpR in A. baumannii pathogenesis using additional clinical strains. Deletion of ompR led to attenuated virulence in the four diverse clinical strains tested in G. mellonella model and protected the larvae from bacteria-mediated killing, suggesting that the role of OmpR in bacterial virulence is conserved in A. baumannii (Figure 2(a)). Importantly, chromosomal complementation of the ΔompR mutant significantly increased the virulence of A. baumannii AB5075 in the G. mellonella model, confirming that the reduced virulence of the ΔompR mutant is due to the loss of OmpR (Figure 2(b)). To further dissect the role of OmpR in A. baumannii virulence we used mutant strains expressing chromosomal OmpR variants having different activation states or DNA binding ability. Based on previous work performed on OmpR E. coli, [24] the aspartic acid at position 71 of A. baumannii OmpR, which in E. coli corresponds to the aspartic acid residue at position 55 that is phosphorylated upon EnvZ-mediated OmpR activation, was substituted by either an alanine (D71A) to abolish OmpR activation or by a glutamic acid (D71E) to induce OmpR-activated conformation. The inability of OmpR D71A and D71E mutants to be phosphorylated was confirmed in vitro (Supplementary Figure S1). In addition, the arginine at position 198 (R182 in E. coli) was substituted by a leucine (R198L) to prevent OmpR binding to DNA [25]. Infection of G. mellonella larvae with the WT and OmpR D71E chromosomal mutant led to rapid larvae killing with 50% mortality observed at 24 h that was further increasing until 72 h post infection (Figure 2(c)). In contrast, larvae infected with the ΔompR mutant and the OmpR D71A and OmpR R198L chromosomal mutants that prevent phosphorylation or DNA binding, respectively, showed reduced mortality (10–40%) at 72 h. Overall, the chromosomal mutants impaired for OmpR phosphorylation (D71A) or for DNA binding (R198L) were attenuated similarly to the ΔompR mutant, indicating that both OmpR characteristics are required for A. baumannii virulence.
Figure 2.
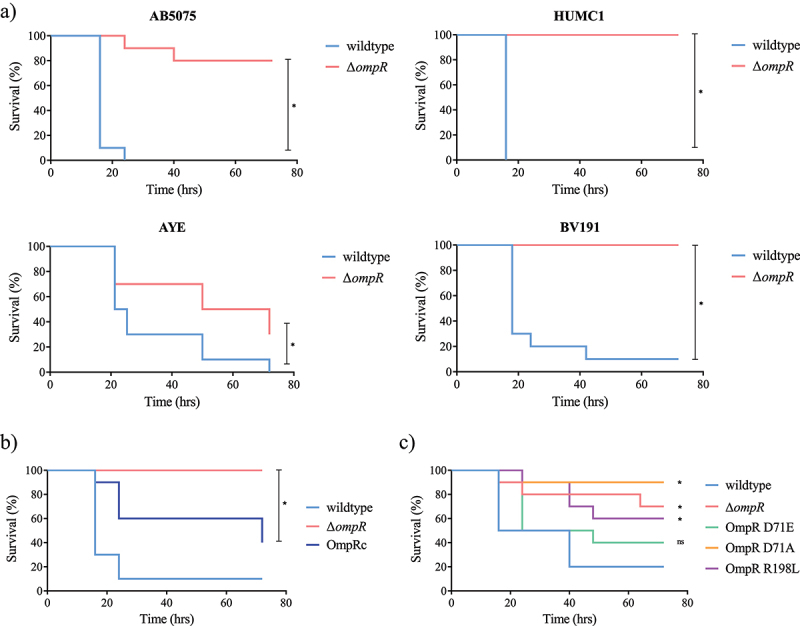
Role of OmpR in A. baumannii virulence assessed in G. mellonella infection model.
A) Groups of 10 larvae were infected with 105 cfu of AB5075, HUMC1, AYE or BV191 wildtype strains (blue) and their isogenic ΔompR mutants (red). B) Groups of 10 larvae were infected with 104 cfu of AB5075 wildtype (light blue), AB5075 ΔompR (red) and OmpR complemented (OmpRc, dark blue) strains. C) Groups of 10 larvae were infected with 104 cfu of AB5075 wildtype (light blue), AB5075 ΔompR (red), AB5075 OmpR D71E (green), AB5075 OmpR D71A (orange) and AB5075 OmpR R198L (purple) mutants. Larvae survival was monitored over 72 hours. Log-rank (Mantel-Cox) test compared to wildtype, *P-value < 0.05.
Identification of OmpR-regulated genes as biomarkers of OmpR inactivation in A. baumannii
By using different vertebrate and non-vertebrate in vivo models we have shown that OmpR is required for A. baumannii pathogenesis, suggesting that OmpR inhibition may be an alternative approach to block bacterial virulence and potentially treat A. baumannii infections. However, in vitro assays to estimate the OmpR activity are required for the identification of inhibitors. In this regard, we aimed at identifying OmpR-regulated genes that could be used as biomarkers for OmpR inhibition in A. baumannii.
A comparative RNA sequencing (RNA-seq) study was carried out with the AB5075 strain and its isogenic ∆ompR mutant, which led to the identification of 65 genes that were downregulated in ΔompR (<-2 log2 fold change, ΔompR/WT) and 71 genes that were upregulated in ΔompR (>2 log2 fold change, ΔompR/WT) (Figure 3(a) and Supplementary Table S1). The identified downregulated genes correspond to OmpR-activated candidate biomarkers while the upregulated genes correspond to OmpR-repressed candidate biomarkers. To identify OmpR biomarkers that are relevant for A. baumannii pathogenesis, we combined our RNA-seq data with the data from a transposon insertion sequencing (Tn-seq) that was conducted to identify AB5075 virulence determinant in the G. mellonella model [26]. Considering that OmpR is essential for virulence in G. mellonella, we hypothesized that genes that are downregulated in the ΔompR mutant (RNA-seq) should match with genes that are important for G. mellonella infection (underrepresented in Tn-seq). Likewise, genes overexpressed in the ΔompR mutant (RNA-seq) may match with genes that are deleterious for G. mellonella infection (overrepresented in Tn-seq). This analysis identified 19 OmpR-activated candidate genes and 12 OmpR-repressed candidate genes that may be relevant for A. baumannii pathogenesis (Figure 3(b) and Supplementary Table S2).
Figure 3.
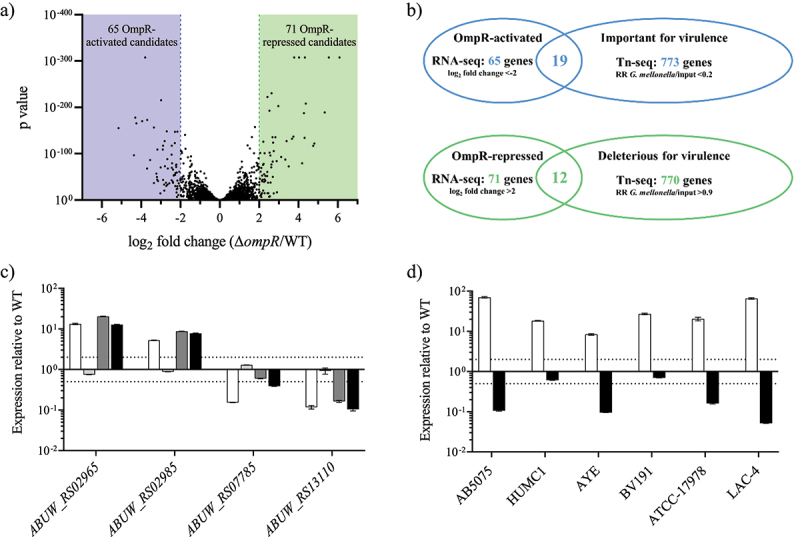
Identification and confirmation of OmpR-regulated genes.
A) RNA-seq results representation of differentially expressed genes between AB5075 wildtype and its ΔompR mutants. Sixty-five OmpR-activated candidate genes (<-2 log2 fold change, blue zone) and 71 OmpR-repressed candidate genes (>2 log2 fold change, green zone) were identified. B) The RNA-seq results were combined with a Tn-seq dataset conducted to identify A. baumannii genes that are important or deleterious for A. baumannii virulence [26]. Nineteen OmpR-activated candidate genes were important for A. baumannii virulence whereas 12 OmpR-repressed candidate genes were deleterious for A. baumannii virulence. RR: read ratio. C) The transcript levels were determined in A. baumannii AB5075 wildtype, AB5075 ΔompR (white), AB5075 OmpR D71E (light grey), AB5075 OmpR D71A (dark grey) and AB5075 OmpR R198L (black) mutants by quantitative real-time PCR and the expression level was normalized to the expression of the AB5075 wildtype strain (means ± SEM of two technical replicates). D) The transcript levels of ABUW_RS02965 (white) and ABUW_RS13110 (black) were determined in A. baumannii ΔompR mutants and normalized to the transcript levels of their respective wildtype strains (means ± SEM of two technical replicates). Horizontal dotted lines depict a 2-fold up- or downregulation.
We selected four of those candidate marker genes (2 up- and 2 downregulated) for confirmation using qRT-PCR on the chromosomal mutants expressing different OmpR variants. The upregulated genes ABUW_RS02965 and ABUW_RS02985 and the downregulated genes ABUW_RS07785 and ABUW_RS13110 showed the expected expression pattern in the strains with active (WT and OmpR D71E) and inactive (ΔompR, OmpR D71A and OmpR R198 L) OmpR (Figure 3(c)). These results confirmed that the expression of these genes is regulated by OmpR and that both OmpR activation and DNA binding properties are important for correct regulation. Three of the confirmed OmpR-regulated genes, namely ABUW_RS02965, ABUW_RS02985 and ABUW_RS07785, encode small hypothetical proteins (<150 amino acids) with one of them (ABUW_RS02965) belonging to the RcnB protein family involved in metal ion homoeostasis [27]. The last OmpR-regulated gene (ABUW_RS13110) encodes for an amino acid efflux protein from the LysE transporter family named MatE. Interestingly, several ΔompR upregulated genes encode for small proteins similar to ABUW_RS02965 and ABUW_RS02985, and their OmpR mediated expression regulation was as well confirmed (Table S2 and Figure S2A). Finally, OmpR expression regulation of the most upregulated gene (ABUW_RS02965) was conserved in all the clinical strains tested while OmpR expression regulation of the most downregulated gene (ABUW_RS13110) was conserved in 4 of the 6 clinical strains tested (Figure 3(d)). Moreover, the expression of ABUW_RS02965 was partially restored (20- to 4-fold) while the expression of ompR and ABUW_RS13110 was fully restored in the OmpR chromosomal complemented strain, confirming that the up- and downregulation observed in the ΔompR mutant are due to the loss of OmpR (Supplementary Figure S2B). Together, our data demonstrate that the expression of the genes ABUW_RS02965 and ABUW_RS13110 is repressed and activated by OmpR, respectively, and that monitoring of their expression may serve as biomarker of OmpR inhibition in most A. baumannii strains.
Development of an OmpR DNA binding assay and identification of OmpR DNA binding sites in A. baumannii.
We aimed to develop an in vitro assay enabling the quantification of OmpR DNA binding to further expand the toolbox for OmpR inhibitor discovery. The DNA-binding motif of OmpR has been described in E. coli (Supplementary Figure S3) [18]. More specifically, the binding between OmpR and the different binding sites present upstream of the OmpR-regulated genes ompC and ompF has been extensively studied in E. coli [15,28,29]. We used this knowledge to develop a DNA binding enzyme-linked immunosorbent (D-ELISA) sandwich assay where a biotinylated oligonucleotide encoding the OmpR binding site is attached to a streptavidin coated plate and serves as a bait for the transcription factor (Supplementary Figure S3). We confirmed the functionality of the D-ELISA assay using OmpRECO and its cognate DNA binding site F1aF1b (Figure 4(a)). Furthermore, we showed that OmpRABA, which share 69% amino acid identity with OmpRECO, is also binding to the E. coli F1aF1b binding site (Figure 4(b)). Moreover, we showed that OmpRABA phosphorylation increased DNA binding whereas the OmpRABA R198 L mutant was unable to bind DNA, suggesting that the D-ELISA assay allows quantification of OmpR DNA binding (Figure 4(c)).
Figure 4.
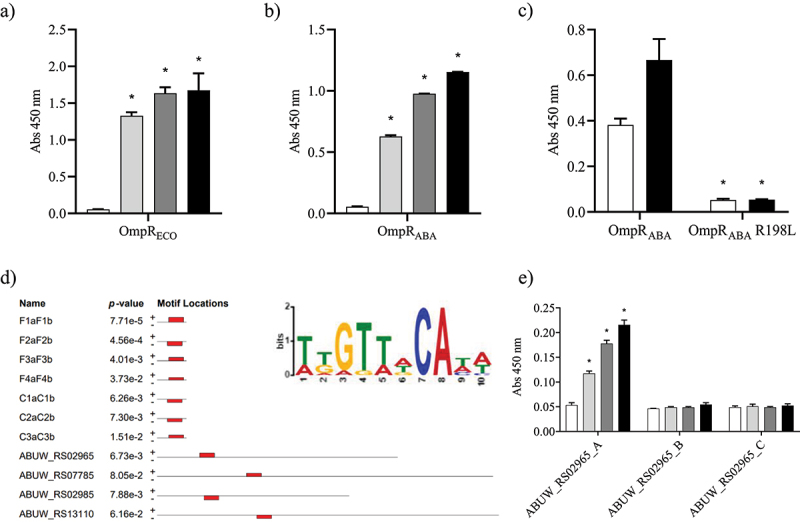
Study of OmpR protein-DNA binding using D-ELISA and in silico analysis.
OmpR binding to the F1aF1b oligonucleotide (2 µg/mL) was assessed with 3.1 µg/mL (light grey), 6.2 µg/mL (dark grey) and 12.5 µg/mL (black) of the OmpRECO (A) and OmpRABA (B) proteins. A negative control without protein was included (white). Unpaired t-test compared to control, *P-value < 0.05. C) The binding of OmpRABA and OmpRABA R198L mutant (6 µg/mL) to the F1aF1b oligonucleotide (2 µg/mL) was tested under unphosphorylated (white) and phosphorylated (black) conditions. Unpaired t-test compared to wildtype, *P-value < 0.05. D) A 10 nucleotide motif (highlighted in red) common to all known OmpRECO DNA binding sites upstream of ompF (F1aF1b, F2aF2b, F3aF3b and F4aF4b) and ompC (C1aC1b, C2aC2b and C3aC3b) was identified in the intergenic region upstream of the 4 OmpRABA regulated genes using the MEME suite [30]. The weblogo of the sequence motif is depicted in the upper right corner. E) OmpRABA binding to the ABUW_RS02965_A, B and C oligonucleotides (2 µg/mL) was assessed with 1.5 µg/mL (light grey), 3 µg/mL (dark grey) and 6 µg/mL (black) of proteins. A negative control without protein was included (white). Only the ABUW_RS02965_A encodes the identified OmpR DNA binding motif (Supplementary Figure S4). Unpaired t-test compared to control, *P-value < 0.05. The data represent the mean ± SD of at least two replicates.
As OmpRABA binds to cognate OmpRECO binding sites, we next looked whether the consensus OmpRECO DNA binding motif can be found upstream of the OmpRABA-regulated genes that we have identified previously. A 10-nucleotide motif present in all the OmpRECO DNA binding sites upstream of ompF and ompC was identified upstream of the four OmpRABA-regulated genes studied in this work (Figure 4(d) and Supplementary Figure S4). This motif corresponds to the previously identified OmpRECO DNA binding motif shifted by one nucleotide (Supplementary Figure S3). We used the intergenic region upstream of the gene ABUW_RS02965 to confirm OmpRABA binding using D-ELISA assay. Three different oligonucleotides (ABUW_RS02965_A, B and C) that span the entire intergenic region were designed and only the oligonucleotide ABUW_RS02965_A included the identified OmpR DNA binding motif (Supplementary Figure S4). OmpRABA showed dose-dependent binding only to the oligonucleotide encoding the identified OmpR DNA binding site, confirming that OmpRABA specifically binds to the identified motif (Figure 4(e)). Together, our data suggest that the expression of the four marker genes is directly regulated by OmpR in A. baumannii.
Structure of the DNA-binding domain of OmpR from A. baumannii
To enable the discovery of a small molecule inhibitor of OmpR, we solved the crystal structure of OmpRABA DNA binding domain (DBD) spanning the C-terminal residues 148 to 254 at a resolution of 2.2 Å. The analysis of the crystal structure showed that OmpRABA DBD consists of a three-stranded antiparallel β-sheet (β1, V153-F156; β2, W159-D162; β3, R166-T169) followed by three α-helices (α1, T179-E190; α2, R198-R206; α3, R215-I228) and ending with a β-hairpin (β4, I238-V241; β5, V244-F248) (Figure 5(a)). The arrangement of the β-sheets and α-helices represents the winged helix-turn-helix (wHTH) fold characteristic of DNA-binding proteins of the OmpR superfamily [31]. Notably, the OmpRABA DBD superimpose very well (root mean square deviation of 0.543 Å) with the OmpRECO DBD bound to DNA whose crystal structure has recently been determined (PDB entry: 6LXN) (Figure 5(b)) [32]. The sequence identity between OmpRABA DBD and OmpRECO DBD is 69% and all the amino acid residues of OmpRECO that are involved in the DNA binding were conserved in OmpRABA (Figure 5(c)). These observations suggest a very similar DNA-binding mode for both proteins, which is in line with OmpRABA binding to OmpRECO cognate binding site.
Figure 5.
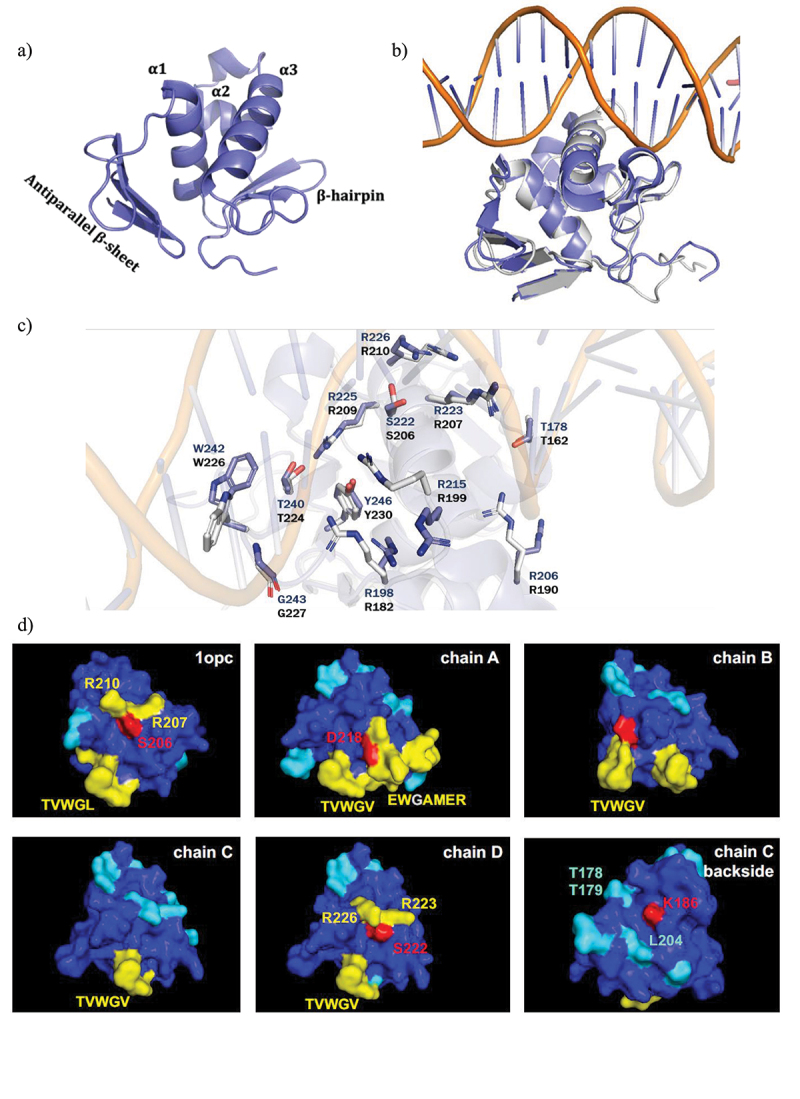
Structure of the DNA-binding domain of A. baumannii OmpR and computational hotspots analysis.
A) Overview of the crystal structure of the DNA-binding domain (DBD) of A. baumannii OmpR. B) Superposed crystal structure of OmpR DBD from A. baumannii (blue) on DNA bound structure of OmpR from E. coli (grey). C) Amino acids (blue colored for OmpRABA and grey colored for OmpRECO) that are involved in making H-bond and salt bridges with DNA are shown. D) The 3D structures of OmpRECO (1opc) and OmpRABA DBD (chain A, B, C and D) are represented and colored based on temperature scale from blue (low ligand interaction probability) to red (high ligand interaction probability). Individual residues with high predicted ligand-binding potential are highlighted. The interaction site is around the conserved residues TVWG(V/L). Hotspot 1 in OmpRABA-DBD chain A contains D218, as anchor residue for ligand binding. Hotspot 2 in OmpRABA-DBD chain D contains residues S222, R223 and R226 (S206, R207 and R210 in OmpRECO DBD). Hotspot 3 in OmpRABA-DBD chain C contains residues K186, L204, and T178/T179.
Identification of OmpR inhibitor tool compound using in silico screening approach.
We finally aimed at discovering small drug inhibitor of OmpR using the different tools developed in this study. A hotspot analysis was used to identify potential protein-ligand binding sites on the solvent-accessible surface of protein structures 1OPC and of OmpRABA DBD [33]. Three hotspots embedding highly predicted anchor residues as potential ligand binding pockets in the identification of DNA-inhibitors (Figure 5(d)) were identified. The most likely interaction site (hotspot 1) in all structures is around conserved residues T240VWG(V/L)244 in OmpRABA DBD. All structures possess this feature, most pronounced in chain A. According to RCSB-PDB annotation for 1OPC, this stretch of residues might be the RNA polymerase interaction site. The predicted residues flank a pocket that may serve as a ligand binding site. Residue D218 in chain A may serve as an anchor point. OmpRABA DBD chain D features a pronounced predicted secondary site (hotspot 2) around residues S222, R223 and R226 (S206, R207, R210 in 1OPC). The “backside” of OmpRABA DBD chain C features a third potential ligand binding pocket flanked by residues K186 (potential anchor residue), L204 and T178/T179. The structure-based pharmacophore model [34] was used for a virtual screening to search for compounds against hotspots 1 and 2 that can block OmpR DNA binding, abrogating both its repression and activation functions. More than 40 structural distinct inhibitors were retrieved as final top-scoring candidates from different commercially available chemical suppliers. We used our developed D-ELISA to evaluate the inhibitory effect of the virtual hits on OmpRABA DNA binding. We only observed a dose-dependent inhibition of OmpRABA DNA binding for the compound VSIS_039 (STL300125, Vitas-M Chemical Limited), which was identified to bind to the hotspot 2 druggable pocket of OmpR (Figure 6(a,b)). Moreover, VSIS_039 did not inhibit the binding of an unrelated transcriptional regulator (SarA from S. aureus) to its cognate DNA binding site [35], indicating that in vitro VSIS_039 mediated DNA binding inhibition is specific to OmpR. However, VSIS_039 did not affect the expression of the OmpRABA marker genes ABUW_RS02965 and ABUW_RS13110 when tested at up to 100 µM in whole cell-based assay (<2-log fold change) (Figure 5(c)). These results suggest that VSIS_039 is unable to efficiently enter in A. baumannii or is prone to rapid efflux in A. baumannii. We constructed an A. baumannii AYE mutant deleted for the three conserved and main resistance-nodulation-division (RND) efflux pumps of A. baumannii, namely adeABC, adeFGH and adeIJK. By using the ΔompR mutant of the efflux depleted strain, we confirmed that OmpR-regulation of ABUW_RS02965 and ABUW_RS13110 is not affected by the deletion of the efflux pumps (Supplementary Figure S5A), enabling to test the OmpR inhibition activity of VSIS_039 on the efflux depleted mutant. VSIS_039 demonstrated a dose dependent upregulation of ABUW_RS02965 and downregulation of ABUW_RS13110 in the AYE efflux depleted strain while it did not have antibiotic activity (MIC >200 µM, HepG2 LD50 >100 µM) (Figure 5(d)). The compound was further tested on the ΔompR mutant of the efflux depleted strain to control that the inhibitor is acting through OmpR. No effect was observed on the expression of ABUW_RS13110, suggesting that the compound is targeting OmpR, whereas the second biomarker gene (ABUW_RS02965) was upregulated as in the wildtype strain, suggesting that the compound may also have additional targets that are involved in ABUW_RS02965 regulation. Finally, using the intermediate efflux mutants, we showed that deletion of the adeABC efflux pump is not sufficient for VSIS_039 activity, which was observed in the ΔadeABC ΔadeIJK double deletion mutant and did not further increase in the ΔadeABC ΔadeIJK ΔadeFGH triple deletion mutant (Supplementary Figure S5B). Together with the D-ELISA results, the biomarker expression studies suggest that VSIS_039 alters the activity of A. baumannii OmpR through the inhibition of OmpR DNA binding and that VSIS_039 is a substrate of the A. baumannii RND efflux pumps AdeIJK, preventing its use on wildtype strains. The impact of VSIS_039 on A. baumannii virulence could not be evaluated as the AYE efflux depleted mutant was avirulent in G. mellonella model, which is consistent with efflux pumps playing an important role in A. baumannii and other Gram-negative pathogenesis [26,36–39]. Alternatively, Tipton et al. reported the role of OmpR in A. baumannii motility [20]. However, motility was not affected by the deletion of ompR in the AYE efflux depleted strain (Supplementary Figure S6), preventing the use of this phenotype to assess the activity of VSIS_039.
Figure 6.
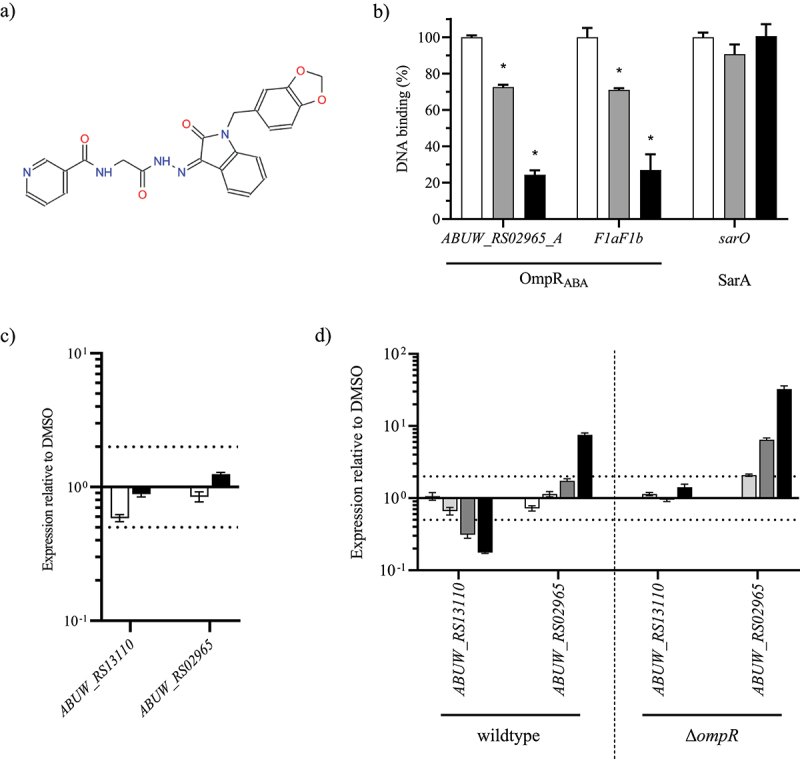
OmpR inhibitory activity of VSIS_039.
A) Structure of the compound VSIS_039 (STL300125, Vitas-M Chemical Limited). B) OmpRABA DNA inhibition was assessed by D-ELISA using 2 µg/mL biotinylated ABUW_RS02965_A and F1aF1b oligos and 6 µg/mL of OmpRABA in the presence of DMSO (white), 10 µM (grey) and 20 µM (black) of VSIS_039 compound (mean ± SD of 2 technical replicates). As negative control, VSIS_039 was also tested on SarA and its cognate DNA binding site using 10 µg/mL of the SarA protein and 2 µg/mL of the biotinylated sar0 oligos. Unpaired t-test compared to DMSO, *P-value < 0.05. C) The transcript levels of ABUW_RS02965 and ABUW_RS13110 were determined in the A. baumannii AYE wildtype strain in the presence of VSIS_039 at 33.3 μM (white) and 100 μM (black). The transcripts levels were normalized to those of the DMSO control (mean ± SEM of 2 technical replicates). D) The transcript levels of ABUW_RS02965 and ABUW_RS13110 were determined in the A. baumannii AYE efflux depleted (ΔadeABC ΔadeFGH ΔadeIJK) wildtype and ΔompR strains in the presence of VSIS_039 at 10 μM (white), 20 μM (light grey), 40 μM (dark grey) and 80 μM (black). The transcripts levels were normalized to those of the DMSO control (mean ± SEM of 2 technical replicates). Horizontal dotted lines depict a 2-fold up- or downregulation.
Discussion
Target validation is often the first step of drug discovery and it is particularly relevant for adjuvant drugs, such as anti-virulence drugs, targeting non-essential (at least in vitro) pathways, which may be less conserved than essential pathways that are targeted by antibiotics [40–42]. In this study we demonstrated that OmpR is essential for different A. baumannii clinical isolates to establish a robust infection in thigh and septicaemia mouse models as well as in G. mellonella invertebrate model. These results are consistent with the findings of a previous study, suggesting that OmpR plays a role in AB5075 A. baumannii virulence in G. mellonella [20]. Interestingly, the results from two independent transposon mutagenesis studies on A. baumannii ATCC 17978 [43] and AB5075 [26] suggest that OmpR is important for growth in rich media while another transposon mutagenesis study on A. baumannii 307–0294 [44] identified EnvZ, the sensor kinase of the OmpR/EnvZ TCS, as an in vivo essential gene. In addition, an in silico subtractive genomic study identified EnvZ as essential for the viability of A. baumannii [45]. Together, these findings support that the OmpR/EnvZ TCS is a valid and unexploited anti-virulence drug target in A. baumannii.
The role of OmpR in A. baumannii pathogenesis echoes the important role of OmpR in the pathogenesis of adherent-invasive and uropathogenic E. coli [46,47]. OmpR-mediated adaptation to osmotic stress via differential expression of outer membrane proteins is required for uropathogenic E. coli virulence. Similarly, OmpR was shown to play an important role in A. baumannii osmotic stress response but no OmpR-regulated genes could be identified [20]. We identified 65 OmpR-downregulated candidate genes and 71 OmpR-upregulated candidate genes in A. baumannii and confirmed proper OmpR regulation of 10 of them. Interestingly, several OmpR-regulated genes encoding unknown proteins were suggested to be involved in A. baumannii virulence in G. mellonella [25]. Therefore, the mechanism of OmpR-mediated virulence in A. baumannii may be uncovered by understanding the function of those unknown proteins. However, additional work is required to first confirm the involvement of those unknow proteins in A. baumannii pathogenesis and to then uncover their role. Ultimately, the OmpR-mediated repression of ABUW_RS02965 and the OmpR-mediated activation of ABUW_RS13110 was confirmed on strains expressing different OmpR-inactivated variants, on the OmpR complemented strain and on several clinical isolates. Moreover, the identification of OmpR DNA binding site upstream of these two genes suggest that they are directly regulated by OmpR. Together, our results demonstrate that the expression of ABUW_RS02965 and ABUW_RS13110 can be used as robust biomarker of A. baumannii OmpR inhibition, both at the phosphorylation and DNA-binding levels.
The determination of the protein structure of a drug target represents an important tool for drug development [48]. The resolution of the OmpR C-terminal domain of A. baumannii enabled the identification of VSIS_039, an OmpR small molecule inhibitor that interferes with OmpR DNA binding as demonstrated by D-ELISA and mis-regulation of the OmpR biomarkers. However, VSIS_039 did not show activity in A. baumannii wildtype strains due to efflux of the drug by the AdeIJK RND efflux pumps. Cell permeability and efflux pumps are major barriers for the development of anti-infectives against Gram-negative species [49,50]. In contrast to cell based in vitro screening, the main drawback of target based in silico and in vitro screening assays is that they do not account for molecule permeation and/or efflux. Although VSIS_039 permeation into A. baumannii seems not to be a bottleneck, further optimization of VSIS_039 is required to minimize efflux and gain whole cell OmpR inhibition activity against A. baumannii.
Interestingly, the predicted binding site of VSIS_039 to OmpRABA is conserved in OmpR from Enterobacteriaceae species. The structure conservation between A. baumannii and Enterobacteriaceae OmpR proteins opens the opportunity to develop an anti-virulence drug with a broader spectrum of activity. The development of anti-virulence drugs as an alternative to antimicrobials therapeutic is in constant progression with most approaches focusing on Pseudomonas aeruginosa and Staphylococcus aureus but less commonly on A. baumannii or broad spectrum [51]. Mannoside-mediated inhibition of FimH that compromises the ability of uropathogenic and adherent-invasive E. coli to colonize and invade the bladder and gut epithelium, respectively, is a notorious anti-virulence approach with drugs in clinical development to treat urinary tract infections and Crohn’s disease [52,53]. Interestingly, the expression of FimH has been shown to be positively regulated by OmpR, further highlighting the strong potential of an OmpR inhibitor as an anti-virulence drug [54].
In conclusion, this study validated OmpR as new anti-virulence target against A. baumannii. Precision therapeutics disarming pathogens without killing them may have the potential to treat infections while limiting the development of resistance [55,56]. However, these alternative approaches come with additional challenges during drug development, with one of them being the need of alternatives to traditional MIC to measure compound efficacy [42]. Here we developed and validated biochemical, cell based and in vivo assays that, together, can be used as a drug discovery platform for OmpR inhibitor development to treat A. baumannii infections. In addition, we confirmed that inhibition of OmpR DNA binding is sufficient to block downstream virulence pathways. With this information, it will be possible to setup a bacterial OmpR-dependent reporter screening assay to screen for molecule inhibitors.
Material and methods
Strains
The A. baumannii strains used is this study are summarized in (Supplementary Table S3). Bacteria were grown in Luria Bertani (LB) broth or on LB agar, incubation was done at 37°C with shaking (220 rpm) for liquid cultures and, when required, antibiotics were added as specified.
Generation of mutants in A. baumannii clinical strains
Scarless deletions, allelic replacements and chromosomal insertion were performed using the pVT77 engineering platform and the two-step allelic exchange method previously described [40]. The lists of the engineered strains and oligonucleotides used in this study are available in (Supplementary Table S3 and Supplementary Table S4), respectively.
DNA fragments corresponding to 700-bp up- and downstream genomic regions of the gene to be deleted were amplified by PCR on the respective A. baumannii strain and cloned in the multiple cloning site (MCS) of pVT77. For the construction of the AB5075 OmpR chromosomal mutants, a 1.4-kb DNA fragment spanning ompR and centred on the codon to exchange was amplified from the A. baumannii AB5075 strain and cloned in the MCS of pVT77. Site directed PCR mutagenesis was performed on the resulting plasmids to introduce the OmpR D71A (GAC to GCC), OmpR D71E (GAC to GAG) and OmpR R198 L (CGC to CTA) mutations. For the construction of the AB5075 OmpR complemented strain, the DNA fragment containing the ompR gene and its promoter region was inserted between the ponA (ABUW_RS01420) and rrm (ABUW_RS01415) genes in the chromosome of the ΔompR mutant. The DNA fragments spanning the ponA and rrm gene were first amplified with oVT425/426 and oVT427/428 and cloned (Gibson assembly) in the MCS (XhoI/SpeI) of the pVT77 plasmid. The DNA fragment containing ompR and its promoter was amplified using oVT630/631 and cloned (Gibson assembly) between ponA and rrm (EcoRI/SpeI), finally giving the plasmid ready for ompR chromosomal insertion using allelic exchange.
The cloned allelic exchange plasmids were conjugated in A. baumannii, and transconjugants were selected on LB agar plates containing 100 mg/L tellurite. The second recombination was selected on 200 mg/L 30-azido-30-deoxythymidine and the desired genetic modifications were confirmed by PCR and sequencing.
Animals
All experiments involving animals were carried out in accordance with the European directive 2010/63/UE governing animal welfare and protection, which is acknowledged by the Italian Legislative Decree no 26/2014 and according to the company policy on the care and use of laboratories animals. All the studies were revised by the Animal Welfare Body and approved by Italian Ministry of Health (authorization n. 50 and 51/2014-PR)
Male CD-1 mice (Charles River Laboratories, Italy), 6 weeks old, were allowed 5 days for acclimation after the arrival. The mice were maintained on a 12-hour light cycle with ad libitum access to rodent feed (Altromin R, A. Rieper SpA, Italy) and filtered tap water. Animals were monitored during the entire period of the studies and clinical signs were recorded.
Mouse infection models
For the neutropenic thigh infection model, male CD-1 mice of 18–20 days were made neutropenic by the administration of 150 and 100 mg kg−1 of cyclophosphamide, intraperitoneally, at 4 and 1 days before infection. Mice were briefly anaesthetized with isoflurane and infected intramuscularly in the thigh with 100 µl of 107 cfu of A. baumannii AB5075 WT and ∆ompR in exponential phase. Mice survival rate was recorded for 48 hours during the experimental phase. In addition, four animals/group were sacrificed under CO2 at 2-, 24- and 48-hours post infection and thighs were collected for cfu determination.
For the immunocompetent septicaemia infection model, C57BL6/J male mice of 7 weeks old were intravenously infected with 5 × 107 cfu of A. baumannii HUMC1 WT and ∆ompR strains in exponential phase. Mice survival rate was recorded until 7 days post infection.
Mouse infection experiments were performed at Aptuit (today Evotec), Verona, Italy.
Galleria mellonella infection model
Ten G. mellonella larvae (TruLarv™, Bio Systems Technology) per group were infected using a 10-μl injection in the right second proleg with mid-log phase (OD600 = 0.5) growing bacteria resuspended in phosphate-buffered saline (PBS) to reach the specified cfu per larva. The infected larvae were collected in a Petri dish and incubated at 37°C. The viability of the larvae was assessed twice a day up to a total of 72 hours post-infection by checking for movement. Larvae were considered dead if no movement could be observed in response to stimulus with a pipette tip.
Quantitative reverse transcription-PCR
Quantitative reverse transcription-PCR (qRT-PCR) was performed as previously described [40]. Briefly, A. baumannii isolates were grown in LB broth at 37°C to mid-log phase (OD600 = 0.5), and total RNA was extracted using a PureLink RNA minikit (Ambion) according to the manufacturer’s recommendations. When compound treatment was performed, A. baumannii isolates were grown to an OD600 of 0.3 and incubated with the specified compound concentration for 30 min before total RNA was extracted as mentioned above. qRT-PCR was performed using a GoTaq 1-Step RT-qPCR System kit (Promega) and expression levels were normalized to that of the housekeeping gene rpoD using the comparative threshold cycle (ΔΔCT) method. A detailed description of the DNA probes used for RNA detection is given in Table S4.
RNA sequencing (RNAseq)
Total RNA samples of A. baumannii AB5075 and its ΔompR mutant were prepared in triplicate following the method mentioned above for qRT-PCR. RNA libraries were prepared with an Illumina TrueSeq (including rRNA depletion) and sequenced with Illumina NextSeq (1x75-bp) at Microsynth AG (Balgach, Switzerland). The reads were mapped to A. baumannii AB5075 genome (NZ_CP008706) using TopHat (v 2.1.1), mapped reads were counted using HTSeq (v 0.6.0) and statistical analysis was performed using DESeq2 (v 1.6.3).
Protein production
A DNA fragment encoding A. baumannii OmpR full-length (OmpR-FL) (Uniprot ID: B0VPC9) without the first eighteen N-terminal residues was purchased as a codon optimized gene for E. coli expression from Genewiz. A DNA fragment encoding the C-terminal DNA-binding domain of OmpR (OmpR-DBD, amino acid 148 to 254) was amplified by PCR using the synthetic gene as a template and both OmpR-FL and OmpR-DBD DNA fragments were cloned into a modified version of the pET15 expression plasmid [57]. The DNA fragment encoding OmpR R198L was amplified by PCR from A. baumannii genomic DNA and cloned in the pET28a expression plasmid.
Bacteria harbouring the expression plasmids were grown to OD600 = 0.6 in LB medium containing the appropriate antibiotic and protein expression was induced with 1 mM IPTG at 18 °C for 18 hours for OmpR-FL and OmpR-DBD or 30°C for 6 hours for OmpR R198L. After cell lysis, the proteins were purified on a HisTrap column and the fractions containing the target proteins were pooled. The OmpR-FL and OmpR-DBD proteins were further purified on a HiLoad 16/60 Superdex 75 prep-grade column using gel filtration buffer (50 mM Tris pH 7.5, 200 mM NaCl). The OmpR R198L protein was dialysed overnight a 4°C in the dialysis buffer (20 mM Tris/HCl pH 7.5, 100 mM NaCl, 25% glycerol). Proteins were flash-frozen in liquid nitrogen and stored in aliquots at −80°C.
Crystallization of DNA binding domain of OmpR
Protein crystals were obtained using sitting-drop vapour diffusion method. Briefly, the OmpR-DBD protein was mixed in a solution consisting of 0.1 M BisTris propane (pH 8.5), 0.2 M Na2SO4 and 22% PEG 3350 and incubated at room temperature for 1 week to enable crystal growth. Diffraction data were collected on beamline PXIII at PSI, Villigen, Switzerland at 100 K. Data reduction and processing was carried out using XDS, programs from the CCP4 suite (Collaborative Computational Project 4, 1994). The crystal structure was solved via molecular replacement method using the crystal structure of DBD of OmpR from E. coli as a template (PDB code: 1OPC) [31]. Refmac was used for refinement and Coot was used for manual building of the model [58],59]. Data and refinement statistics are shown in (Supplementary Table S5). All structure figures were prepared using PyMOL (Schrödinger, LLC, New York). The structure was deposited in the protein data bank with accession code 6ZWT.
Hot-spot analysis
Hot-spot analysis and prediction of potential ligand-binding sites was performed by inSili.com LLC (inSili.com). An in-house software tool for hot-spot analysis was used for identifying potential protein-ligand binding sites on the solvent-accessible surface of protein structures 1OPC and 6ZWT (from PDB). The software captures potential protein–ligand interaction points, including protein–protein interactions. It is based on a machine-learning classifier trained to the recognition of known protein–ligand interaction sites (surface patches). Protein monomers or individual chains were analysed.
DNA binding enzyme-linked immunosorbent (D-ELISA) assay
The D-ELISA assay was used to study OmpR full length protein-DNA binding. Clear flat bottom high-binding capacity polystyrene 96-well plates (Costar™ 3590, 96-Well EIA/RIA Plates) were coated overnight at 4°C with 100 µl streptavidin (4 µg/ml) and blocked for 2 hours at room temperature using blocking buffer (10 mM Na2HPO4, 10 mM NaH2PO4, 10 mM NaCl, 0.05% of Tween 20 and 1% milk powder at pH 7). The plates were washed using the washing buffer (blocking buffer without Tween 20 and milk powder) and 100 µl of annealed biotinylated oligonucleotides diluted in blocking buffer were added to the plates for 1 hour. After washing, 100 µl of the protein diluted in blocking buffer were added and the plates were incubated for 1 hour. The plates were washed and protein-DNA binding was detected using 100 µl of monoclonal mouse antibody anti-6X His HRP (Rockland, 200-303-382) diluted to 100 ng/ml in blocking buffer for 1.5 hours followed by the addition of the TMB substrate (SouthernBiotech, 0410–01). The reaction was stopped after 30 min with 50 µl of 0.5 M H2SO4 (Sigma Aldrich, 320501) and the resulting absorbance was determined at 450 nm with the TECAN infinite F500. When specified, protein phosphorylation was done in the presence of 150 mM lithium acetyl phosphate at 30°C for 30 min. The proteins were pre-incubated (30 min) with the specified compound concentration to assess protein-DNA binding inhibition.
Phos-tag gel
The phosphorylation state of OmpR was evaluated using 13% SDS-PAGE gels supplemented with Phos-tagTM, according to the manufacturer recommendation. The gels were run at 150 V and then fixed in a solution containing 40% methanol and 10% acetic acid before staining with Coomassie Brilliant Blue (0.25%).
Swarming motility assay
Swarming motility was assessed as previously described [60]. Briefly, the inoculum was prepared by resuspending 4–6 freshly growing colonies in 100 µl LB, and 2 µl were spotted in the middle of a freshly prepared Nutrient broth agar (0.5% Eiken agar) plate. The plates were incubated in upright position at 35°C and the swarming diameter was measured after 48 hours.
Antibacterial activity and cytotoxicity assays
The antibacterial activity of VSIS_039 was determined in microbroth dilution MIC according to CLSI guidelines. Briefly, bacterial inoculum was prepared at 5 × 105 cfu/mL in cation adjusted Mueller Hinton broth (CA-MHB). Two-fold dilution series of the compound were prepared in CA-MHB at 10-fold the final concentration and 10 µl compound was mixed with 90 µl of bacterial inoculum in a 96-well assay plate. The plate was incubated at 35°C for 20 hours and the MIC was read as the lowest concentration that inhibited visual growth.
The cytotoxicity of VSIS_039 was determined using HepG2 cell line engineered for stable expression of the human secreted embryonic alkaline phosphatase (hSEAP) [61]. HepG2-hSEAP cultured in Minimum Essential Medium Eagle supplemented with 10% foetal bovine serum were seeded at 2 × 104 cells/well in a tissue culture graded 96-well plate and incubated at 37°C (5% C02). After 24 hours, the medium was replaced by fresh medium containing 2-fold dilution of the compound. The cells were incubated for 24 hours with the compound and cell viability was assessed by hSEAP expression quantification. Briefly, heat inactivated cell culture supernatant was mixed with p-nitrophenyl phosphate at 12 mM in 2x SEAP buffer (20 mM homoarginine, 1 mM MgCl2, 21% diethanolamine, pH 9.8) and the absorbance was read at 405 nm for 15 min. Half-lethal dose (LD50) was defined as the lowest concentration that inhibited more than 50% hSEAP expression compared to untreated control.
Supplementary Material
Acknowledgement
Valentina Lucchini fellowship is supported by the Train2Target project granted from the European Union’s Horizon 2020 framework program for research and innovation (Project #721484). MN acknowledges support by the PSI-FELLOW-II-3i - International Fellowship Program. The project was partially funded by the Swiss Commission for Innovation and Technology (CTI) under the project number 18838.1 PFLS-LS. The work of Vincent Trebosc, Birgit Schellhorn, Olivia Champion, and Christian Kemmer was co-funded by the State Secretariat for Education, Research and Innovation and the European Union within the Impact2 Eurostars Project 12589. We also thank Brad Spellberg and Laurent Poirel for providing A. baumannii strains.
Funding Statement
The work was supported by the Eurostars [12589]; Horizon 2020 Framework Programme [721484]; Swiss Commission for Innovation and Technology (CTI) [18838.1 PFLS-LS].
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
All data are fully available without restriction.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/21505594.2022.2135273
References
- [1].Boucher HW, Talbot GH, Bradley JS, et al. Bad bugs, no drugs: no ESKAPE! An update from the infectious diseases society of America. Clin Infect Dis off Publ Infect Dis Soc Am. 2009;48(1):1–12. DOI: 10.1086/595011 [DOI] [PubMed] [Google Scholar]
- [2].Cassini A, Högberg LD, Plachouras D, et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European economic area in 2015: a population-level modelling analysis. Lancet Infect Dis. 2019;19(1):56–66. DOI: 10.1016/S1473-3099(18)30605-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Tacconelli E, Carrara E, Savoldi A, et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018;18(3):318–327. DOI: 10.1016/S1473-3099(17)30753-3 [DOI] [PubMed] [Google Scholar]
- [4].Lötsch F, Albiger B, Monnet DL, et al. Epidemiological situation, laboratory capacity and preparedness for carbapenem-resistant acinetobacter baumannii in Europe, 2019. Eurosurveillance. 2020;25(45):2001735. DOI: 10.2807/1560-7917.ES.2020.25.45.2001735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hu F, Zhu D, Wang F, et al. Current status and trends of antibacterial resistance in China. Clin Infect Dis off Publ Infect Dis Soc Am. 2018;67(suppl_2):S128–134 doi: 10.1093/cid/ciy657. [DOI] [PubMed] [Google Scholar]
- [6].Jernigan JA, Hatfield KM, Wolford H, et al. Multidrug-resistant bacterial infections in U.S. hospitalized patients, 2012–2017. N Engl J Med. 2020;382(14):1309–1319. DOI: 10.1056/NEJMoa1914433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Cox G, Sieron A, King AM, et al. A common platform for antibiotic dereplication and adjuvant discovery. Cell Chem Biol. 2017;24(1):98–109. DOI: 10.1016/j.chembiol.2016.11.011 [DOI] [PubMed] [Google Scholar]
- [8].Allen RC, Popat R, Diggle SP, et al. Targeting virulence: can we make evolution-proof drugs? Nat Rev Microbiol. 2014;12(4):300–308 doi: 10.1038/nrmicro3232. [DOI] [PubMed] [Google Scholar]
- [9].Clatworthy AE, Pierson E, Hung DT.. Targeting virulence: a new paradigm for antimicrobial therapy. Nat Chem Biol. 2007;3(9):541–548 doi: 10.1038/nchembio.2007.24. [DOI] [PubMed] [Google Scholar]
- [10].Morris FC, Dexter C, Kostoulias X, et al. The mechanisms of disease caused by Acinetobacter baumannii. Front Microbiol. 2019;10:10 doi: 10.3389/fmicb.2019.01601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Geisinger E, Huo W, Hernandez-Bird J, et al. Acinetobacter baumannii: envelope determinants that control drug resistance, virulence, and surface variability. Annu Rev Microbiol. 2019;73(1):481–506 doi: 10.1146/annurev-micro-020518-115714. [DOI] [PubMed] [Google Scholar]
- [12].De Silva PM, Kumar A. Signal transduction proteins in Acinetobacter baumannii: role in antibiotic resistance, virulence, and potential as drug targets. Front Microbiol. 2019;10:49 doi: 10.3389/fmicb.2019.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Beier D, Gross R. Regulation of bacterial virulence by two-component systems. Curr Opin Microbiol. 2006;9(2):143–152 doi: 10.1016/j.mib.2006.01.005. [DOI] [PubMed] [Google Scholar]
- [14].Tiwari S, Jamal SB, Hassan SS, et al. Two-component signal transduction systems of pathogenic bacteria as targets for antimicrobial therapy: an overview. Front Microbiol. 2017;8:1878 doi: 10.3389/fmicb.2017.01878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Yoshida T, Qin L, Egger LA, et al. Transcription regulation of ompF and ompC by a single transcription factor, OmpR. J Biol Chem. 2006;281(25):17114–17123 doi: 10.1074/jbc.M602112200. [DOI] [PubMed] [Google Scholar]
- [16].Barbieri CM, Wu T, Stock AM. Comprehensive analysis of OmpR phosphorylation, dimerization, and DNA binding supports a canonical model for activation. J Mol Biol. 2013;425(10):1612–1626 doi: 10.1016/j.jmb.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kenney LJ, Anand GS. EnvZ/OmpR two-component signaling: an archetype system that can function non-canonically. EcoSal Plus. 2020;9(1):9 doi: 10.1128/ecosalplus.ESP-0001-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Seo SW, Gao Y, Kim D, et al. Revealing genome-scale transcriptional regulatory landscape of OmpR highlights its expanded regulatory roles under osmotic stress in Escherichia coli K-12 MG1655. Sci Rep. 2017;7(1):2181. DOI: 10.1038/s41598-017-02110-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Shimada T, Takada H, Yamamoto K, et al. Expanded roles of two-component response regulator OmpR in Escherichia coli: genomic SELEX search for novel regulation targets. Genes Cells. 2015;20(11):915–931 doi: 10.1111/gtc.12282. [DOI] [PubMed] [Google Scholar]
- [20].Tipton KA, Rather PN. An ompR-envZ two-component system ortholog regulates phase variation, osmotic tolerance, motility, and virulence in Acinetobacter baumannii strain AB5075. J Bacteriol. 2017;199(3):e00705–16 doi: 10.1128/JB.00705-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Bruhn KW, Pantapalangkoor P, Nielsen T, et al. Host fate is rapidly determined by innate effector-microbial interactions during Acinetobacter baumannii bacteremia. J Infect Dis. 2015;211:1296–1305 doi: 10.1093/infdis/jiu593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Champion OL, Cooper IAM, James SL, et al. Galleria mellonella as an alternative infection model for yersinia pseudotuberculosis. Microbiology. 2009;155(5):1516–1522. DOI: 10.1099/mic.0.026823-0 [DOI] [PubMed] [Google Scholar]
- [23].Ramarao N, Nielsen-Leroux C, Lereclus D. The insect galleria mellonella as a powerful infection model to investigate bacterial pathogenesis. J Vis Exp JoVe. 2012;4392(70). DOI: 10.3791/4392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Rhee JE, Sheng W, Morgan LK, et al. Amino acids important for DNA recognition by the response regulator OmpR. J Biol Chem. 2008;283(13):8664–8677 doi: 10.1074/jbc.M705550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gebhardt MJ, Gallagher LA, Jacobson RK, et al. Joint transcriptional control of virulence and resistance to antibiotic and environmental stress in Acinetobacter baumannii. MBio. 2015;6:e01660–15. DOI: 10.1128/mBio.01660-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kato N, Tsuzuki M, Aiba H, et al. Gene activation by theEscherichia coli positive regulator OmpR: a mutational study of the DNA-binding domain of OmpR. Mol Gen Genet MGG. 1995;248(4):399–406 doi: 10.1007/BF02191639. [DOI] [PubMed] [Google Scholar]
- [27].Blériot C, Effantin G, Lagarde F, et al. RcnB is a periplasmic protein essential for maintaining intracellular Ni and Co concentrations in Escherichia coli. J Bacteriol. 2011;193(15):3785–3793 doi: 10.1128/JB.05032-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Bergstrom LC, Qin L, Harlocker SL, et al. Hierarchical and co-operative binding of OmpR to a fusion construct containing the ompC and ompF upstream regulatory sequences of Escherichia coli. Genes Cells. 1998;3(12):777–788 doi: 10.1046/j.1365-2443.1998.00228.x. [DOI] [PubMed] [Google Scholar]
- [29].Head CG, Tardy A, Kenney LJ. Relative binding affinities of OmpR and OmpR-phosphate at the ompF and ompC regulatory sites. J Mol Biol. 1998;281(5):857–870 doi: 10.1006/jmbi.1998.1985. [DOI] [PubMed] [Google Scholar]
- [30].Bailey TL, Boden M, Buske FA, et al. MEME suite: tools for motif discovery and searching. Nucleic Acids Res. 2009;37(Web Server):W202–208. DOI: 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Martínez-Hackert E, Stock AM. The DNA-binding domain of OmpR: crystal structures of a winged helix transcription factor. Structure. 1997;5(1):109–124 doi: 10.1016/S0969-2126(97)00170-6. [DOI] [PubMed] [Google Scholar]
- [32].Sadotra S, Lou Y-C, Tang H-C, et al. Structural basis for promoter DNA recognition by the response regulator OmpR. J Struct Biol. 2021;213(1):107638 doi: 10.1016/j.jsb.2020.107638. [DOI] [PubMed] [Google Scholar]
- [33].Geppert T, Hoy B, Wessler S, et al. Context-based identification of protein-protein interfaces and “hot-spot” residues. Chem Biol. 2011;18(3):344–353 doi: 10.1016/j.chembiol.2011.01.005. [DOI] [PubMed] [Google Scholar]
- [34].Wolber G, Langer TL. LigandScout: 3-D pharmacophores derived from protein-bound ligands and their use as virtual screening filters. J Chem Inf Model. 2005;45(1):160–169 doi: 10.1021/ci049885e. [DOI] [PubMed] [Google Scholar]
- [35].Chien Y, Manna AC, Projan SJ, et al. SarA, a global regulator of virulence determinants instaphylococcus aureus, binds to a conserved motif essential for sar-dependent gene regulation. J Biol Chem. 1999;274(52):37169–37176 doi: 10.1074/jbc.274.52.37169. [DOI] [PubMed] [Google Scholar]
- [36].Colclough AL, Alav I, Whittle EE, et al. RND efflux pumps in gram-negative bacteria; regulation, structure and role in antibiotic resistance. Future Microbiol. 2020;15(2):143–157. DOI: 10.2217/fmb-2019-0235 [DOI] [PubMed] [Google Scholar]
- [37].Pérez-Varela M, Corral J, Aranda J, et al. Roles of efflux pumps from different superfamilies in the surface-associated motility and virulence of Acinetobacter baumannii ATCC 17978. Antimicrob Agents Chemother. 2019;63(3):63 doi: 10.1128/AAC.02190-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Richmond GE, Evans LP, Anderson MJ, et al. the acinetobacter baumannii two-component system AdeRS regulates genes required for multidrug efflux, biofilm formation, and virulence in a strain-specific manner. MBio. 2016;7(3). DOI: 10.1128/mBio.00430-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Yoon E-J, Chabane YN, Goussard S, et al. Contribution of resistance-nodulation-cell division efflux systems to antibiotic resistance and biofilm formation in Acinetobacter baumannii. MBio. 2015;6(2):6. DOI: 10.1128/mBio.00309-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Trebosc V, Gartenmann S, Royet K, et al. A novel genome-editing platform for drug-resistant Acinetobacter baumannii reveals an AdeR-unrelated tigecycline resistance mechanism. Antimicrob Agents Chemother. 2016;60(12):7263–7271. DOI: 10.1128/AAC.01275-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Trebosc V, Gartenmann S, Tötzl M, et al. Dissecting colistin resistance mechanisms in extensively drug-resistant Acinetobacter baumannii clinical isolates. MBio. 2019;10(4):10. DOI: 10.1128/mBio.01083-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Theuretzbacher U, Piddock LJV. Non-traditional antibacterial therapeutic options and challenges. Cell Host Microbe. 2019;26(1):61–72 doi: 10.1016/j.chom.2019.06.004. [DOI] [PubMed] [Google Scholar]
- [43].Wang N, Ozer EA, Mandel MJ, et al. Genome-wide identification of Acinetobacter baumannii genes necessary for persistence in the lung. MBio. 2014;5(3). e01163–14. DOI: 10.1128/mBio.01163-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Umland TC, Schultz LW, MacDonald U, et al. In vivo-validated essential genes identified in Acinetobacter baumannii by using human ascites overlap poorly with essential genes detected on laboratory media. MBio. 2012;3(4):3 doi: 10.1128/mBio.00113-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Kaur N, Khokhar M, Jain V, et al. Identification of druggable targets for Acinetobacter baumannii via subtractive genomics and plausible inhibitors for MurA and MurB. Appl Biochem Biotechnol. 2013;171(2):417–436 doi: 10.1007/s12010-013-0372-2. [DOI] [PubMed] [Google Scholar]
- [46].Schwan WR. Survival of uropathogenic Escherichia coli in the murine urinary tract is dependent on OmpR. Microbiology. 2009;155(6):1832–1839 doi: 10.1099/mic.0.026187-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Lucchini V, Sivignon A, Pieren M, et al. The role of OmpR in bile tolerance and pathogenesis of adherent-invasive Escherichia coli. Front Microbiol. 2021;12:684473 doi: 10.3389/fmicb.2021.684473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Singh S, Malik BK, Sharma DK. Molecular drug targets and structure based drug design: a holistic approach. Bioinformation. 2006;1(8):314–320 doi: 10.6026/97320630001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Zgurskaya HI, López CA, Gnanakaran S. Permeability barrier of gram-negative cell envelopes and approaches to bypass it. ACS Infect Dis. 2015;1(11):512–522 doi: 10.1021/acsinfecdis.5b00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Masi M, Réfregiers M, Pos KM, et al. Mechanisms of envelope permeability and antibiotic influx and efflux in gram-negative bacteria. Nat Microbiol. 2017;2(3):17001 doi: 10.1038/nmicrobiol.2017.1. [DOI] [PubMed] [Google Scholar]
- [51].Theuretzbacher U, Outterson K, Engel A, et al. The global preclinical antibacterial pipeline. Nat Rev Microbiol. 2020;18(5):275–285 doi: 10.1038/s41579-019-0288-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Mydock-McGrane LK, Hannan TJ, Janetka JW. Rational design strategies for FimH antagonists: new drugs on the horizon for urinary tract infection and crohn’s disease. Expert Opin Drug Discov. 2017;12(7):711–731 doi: 10.1080/17460441.2017.1331216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Sarshar M, Behzadi P, Ambrosi C, et al. FimH and anti-adhesive therapeutics: a disarming strategy against uropathogens. Antibiotics. 2020;9(7):397 doi: 10.3390/antibiotics9070397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Schwan WR. Regulation of fim genes in uropathogenic Escherichia coli. World J Clin Infect Dis. 2011;1(1):17–25 doi: 10.5495/wjcid.v1.i1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Spaulding CN, Klein RD, Schreiber HL, et al. Precision antimicrobial therapeutics: the path of least resistance? NPJ Biofilms Microbiomes. 2018;4(1):1–7 doi: 10.1038/s41522-018-0048-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Totsika M. Disarming pathogens: benefits and challenges of antimicrobials that target bacterial virulence instead of growth and viability. Future Med Chem. 2017;9(3):267–269 doi: 10.4155/fmc-2016-0227. [DOI] [PubMed] [Google Scholar]
- [57].Kammerer RA, Schulthess T, Landwehr R, et al. Tenascin-C hexabrachion assembly is a sequential two-step process initiated by coiled-coil α-helices. J Biol Chem. 1998;273(17):10602–10608 doi: 10.1074/jbc.273.17.10602. [DOI] [PubMed] [Google Scholar]
- [58].Emsley P, Lohkamp B, Scott W G and Cowtan K. (2010). Features and development of Coot. Acta Crystallogr D Biol CrystallogrActa Cryst DActa Cryst Sect DActa Crystallogr DActa Crystallogr Sect DActa Crystallogr Sect D Biol CrystallogrActa Crystallogr D Biol Cryst, 66(4), 486–501. 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Emsley P, Lohkamp B, Scott W G and Cowtan K. (2010). Features and development of Coot. Acta Crystallogr D Biol CrystallogrActa Cryst DActa Cryst Sect DActa Crystallogr DActa Crystallogr Sect DActa Crystallogr Sect D Biol CrystallogrActa Crystallogr D Biol Cryst, 66(4), 486–501. 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Biswas I, Machen A, Mettlach J. In Vitro motility assays for Acinetobacter species. Methods Mol Biol Clifton NJ. 2019;1946:177–187 doi: 10.1007/978-1-4939-9118-1_17. [DOI] [PubMed] [Google Scholar]
- [61].Schlatter S, Rimann M, Kelm J, et al. SAMY, a novel mammalian reporter gene derived from bacillus stearothermophilus α-amylase. Gene. 2002;282:19–31 doi: 10.1016/S0378-1119(01)00824-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are fully available without restriction.


