Abstract
Strain variations of Helicobacter pylori have been tested by numerous methods and compared among different patient groups. The aim of this study was to investigate whether H. pylori expresses disease-specific proteins that can be detected by two-dimensional polyacrylamide gel electrophoresis (2-D PAGE). H. pylori strains isolated from duodenal ulcer, gastric cancer, and gastritis patients were analyzed. Extensive variation in spot patterns was observed between the strains, but a dendrogram analysis revealed that some strains within each disease group clustered together. Eight proteins were sequenced and found in the H. pylori genome sequence. 2-D PAGE is a useful method for studies of protein expression and for highlighting the extensive strain variation that H. pylori exhibits.
Two-dimensional polyacrylamide gel electrophoresis (2-D PAGE) is a laboratory technique with a wide variety of applications for a range of bacterial infections and diseases such as cancer. 2-D PAGE can be used to link certain gel patterns to specific diseases or modifications of specific marker proteins, and this technique can also be applied to answer molecular epidemiological questions in medical microbiology (1, 4, 7, 10, 11, 15, 16, 19, 20, 25). The studies mentioned represent some examples of the wide variety of organisms and questions concerning bacterial protein expression that may be analyzed by the 2-D gel technique. As it is now possible to obtain all of the material needed for 2-D PAGE directly from the manufacturer, including equipment, precast gels, and chemicals, it is a convenient laboratory method for the analysis of expressed bacterial proteins (8). Strain diversity among Helicobacter pylori isolates from different patient groups has been extensively studied (3, 13, 14, 21–23), and there may exist disease-specific strains (12, 17). Whether there exist “good” or “bad” strains of H. pylori is still under discussion (2). Several genetic markers of pathogenicity characteristics for different H. pylori strains have been described previously (5). Genes that are thought to be important for H. pylori pathogenesis are postulated to reside in the cag pathogenicity island. Expression of proteins from this part of the genome is now under investigation using the 2-D electrophoresis technique combined with immunoblotting (C. Lange, R. Rappuoli, and A. Covacci, Abstr. 97th Gen. Meet. Am. Soc. Microbiol., abstr. B-54, p. 38, 1997). The 2-D technique may well be used to characterize bacteria at the protein level (expression from active genes), but not at the DNA level, where both active and inactive genes may be identified. The aim of our study was to test whether H. pylori strains isolated from gastric cancer (Ca), duodenal ulcer (Du), and gastritis (Ga) patients can be grouped according to the protein patterns. We also wanted to search for disease-specific protein spots, protein marker candidates, which could be useful for H. pylori strain characterization. Risk markers for Ca and Du may be identified by this method of visualizing protein expression from different pathogenic strains of clinical isolates of H. pylori. Other issues, such as the strain variation within each group of disease outcome, as well as differences and relationships between the groups, were also addressed by this study.
Five patients with gastric adenocarcinoma (Ca), seven with Du, and four with Ga, all endoscopically and histologically verified, were included in the study. Two biopsy specimens from the antrum and two from the corpus were homogenized together and cultured on both nonselective and selective agar plates in a moist microaerobic atmosphere (10% CO2, 5% O2, 85% N2) at 37°C for 4 days. All H. pylori strains from Ca and Du patients and three of the four strains from Ga patients were positive for the cagA gene by PCR (9). Water extracts (toxin extracts) were prepared by incubation of bacteria in sterile water for 1 h followed by centrifugation at 12,250 × g for 15 min. The supernatants were removed, recentrifuged at 25,400 × g for 20 min at 4°C, and filtered through a 0.2-μm-pore-size filter. Protein concentrations were determined (18), and the samples were diluted to a final concentration of 500 μg/ml. Whole-cell extracts were prepared by sonication of cell pellets resuspended in 1 ml of water for 15 cycles of 30 s each followed by centrifugation at 16,000 × g for 5 min. The supernatants were removed, and protein concentrations were determined by a protein assay kit (Bio-Rad). Finally, 40 μl of bacterial extract was mixed with 160 μl of lysis buffer (9.9 M urea, 4% [vol/vol] Igepal CA630, 2.2% [vol/vol] ampholytes 3 to 10, 100 mM dithiothreitol, and 2% [wt/vol] 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate [CHAPS]). The protein preparations were stored at −70°C until analysis.
All equipment and material used for 2-D PAGE and immunoblotting were obtained from Amersham Pharmacia Biotech, Uppsala, Sweden. Chemicals, except for Pharmalytes, were supplied by Sigma (St. Louis, Mo.). First-dimension runs, i.e., isoelectric focusing, were performed using Immobiline IPG strips. Water extracts were analyzed on 110-mm 4-7 linear and 180-mm 3-10 nonlinear strips, whereas whole-cell extracts were analyzed on 110-mm 3-10 linear and 180-mm 3-10 nonlinear strips. Depending on the length of the strip and the visualization method, 5 to 40 μg of protein was applied on each strip. Proteins were focused to equilibrium using the Immobiline dry strip kit and the Multiphor II electrophoresis system according to the manufacturers' instructions. Second-dimension runs, i.e., separation of proteins according to molecular weight, were performed on precast 8 to 18% or 12 to 14% gradient gels. Proteins were visualized using the PlusOne silver staining kit.
Twelve gels, four from each disease group, were digitized by scanning. The BioImage 2D analyzer 6.1 software (B.I. Systems Corporation) was used for spot matching, quantification of spot intensities, calculation of match statistics, and dendrograms. To compensate for differences in protein loading between the gels, all gels within each group were normalized to one reference gel. An intensity ratio greater than 8 or 10 and a t-test confidence interval exceeding 95% between proteins from different gels were tested. Also, to reveal unique protein spots in any of the three patient groups, the existence of equal spots in two to eight gels was assessed. By adding all 12 gels together in three composite gels, one for each patient group, dendrograms over the strain similarities and relations can be observed. Dendrograms may be derived using neighbor joining, maximum linkage, minimum linkage, or average-unweighted pair group method with averages linkage and two different calculation methods: the formula for method 1 is [(number of matched spots in AB)/(number of spots in A + number of spots in B − number of matched spots in AB)] × 100, and that for method 2 is [(2 × number of matched spots in AB)/(number of spots in A + number of spots in B)] × 100, giving the results as percentages.
Electrophoresis was performed on whole-cell extracts of one Du and one Ca strain on two separate large gels which were then used for immunoblotting (Western blots). Horizontal semidry electrophoretic transfer (0.8 mA/cm2 for 1 h) of protein spots to polyvinylidene difluoride (PVDF) membranes was performed. The membranes were air dried, rewetted in methanol, rinsed in sterile water, and blocked for 20 min in 0.5% Tween 20. Pooled sera from 10 H. pylori-positive Ca or Du patients, diluted 1:1,000 or 1:5,000 in Tris-buffered saline (TBS) with Tween (TBS-T; TBS [pH 9.0], 0.1% Tween 20), were used for a 1-h incubation. Three 10-min washes in TBS-T were performed. Secondary antibody, horseradish peroxidase-conjugated rabbit anti-human immunoglobulin G (Dakopatts, Glostrup, Denmark) at a dilution of 1:10,000, was added for 1 h of incubation. Three 10-min washes in TBS-T were performed. ECL Plus solution from the ECL Plus Western blotting detection system was added for a 5-min incubation. The blots were wrapped in Saran Wrap, placed in an X-ray film cassette with an autoradiography film (Hyperfilm ECL), and exposed for 15 s or longer. The autoradiograms were developed according to standard procedures. Immunoblotting was performed twice on each of the two different strains, first with each strain's own patient group sera, i.e., Ca sera with the Ca strain, and then with its opposite sera, i.e., Du sera with the Ca strain. Results from immunoblots with a nonlinear pH gradient were confirmed by immunoblots with a linear pH gradient. Immunoblots were analyzed by eye, as there were few high-intensity spots to be observed on the membranes.
Protein spots used for sequencing were localized by Coomassie blue staining on PVDF membranes according to standard procedures. Membranes were stored at −20°C until the proteins were cut out using a sterilized knife. Protein sequencing was performed on an Applied Biosystems Procise Sequencer by Edman degradation cycling on electroeluted protein samples. Nine protein spots were sequenced: eight were identified from the computer analysis, and one additional spot was identified on the immunoblot. Homologous amino acid sequences were sought in the H. pylori genome (http://www.tigr.org/tdb/mdb/hpdb/hpdb.html) and by using the BLAST program for searches in nonredundant databases such as PDB, Swissprot, GenBank CDS translations, PIR, and Spupdate (http://www.ncbi.nlm.nih.gov/BLAST).
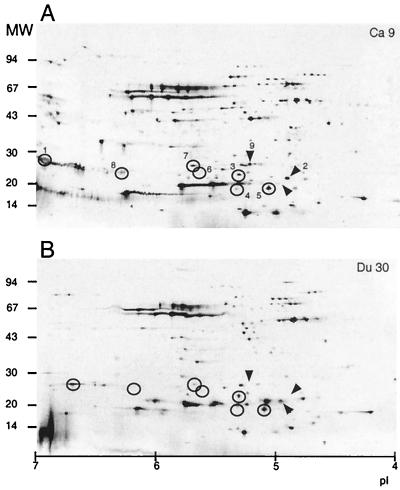
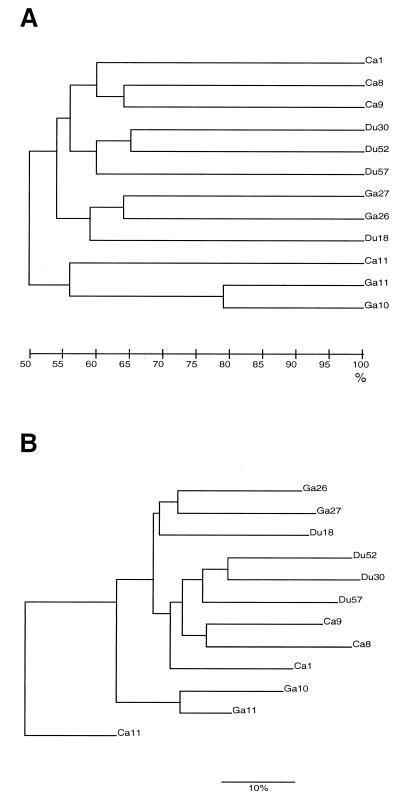
Twelve different H. pylori strains from three different patient groups were extracted and analyzed by 2-D PAGE. By eye, about 100 to 150 high-intensity spots per gel were distinguishable (Fig. 1). Many differences were evident between the strains, even between strains from the same patient group. After the initial screen, the gels were digitized and matched together. Match statistics for comparisons between the three different patient groups were determined. Again, considerable variation between the strains was evident, although the Ga strains were slightly more similar to one another (45%) than were the Du (42%) and Ca (38%) strains. In total, 46% of the spots were matched between Ca and Ga strains, 49% were matched between Ca and Du strains, and 47% were matched between Ga and Du strains. To investigate whether proteins were specific for only one type of disease, we asked the question whether certain protein spots unique for two to eight H. pylori strains were present (Table 1). The computer analysis revealed that 27 proteins were observed exclusively in four strains and that 19 proteins were found to exist in eight strains. None of the spots that matched from four strains were found within the same disease group, and none of the spots matching from eight strains were found in only two groups. Dendrograms were produced based on the match results. Slight variations were observed between the different dendrograms in the order in which the strains appeared within the groupings, but the overall impression was that strains from the same patient group clustered together independently of the calculation method. The Ca and Ga strains were more closely related than were the Du strains to either of the other two (Fig. 2).
FIG. 1.
2-D PAGE of two clinical isolates of H. pylori. About 10 μg of protein derived from water extracts of one Ca strain (A) and one Du strain (B) was focused on 110-mm IPG strips and separated on 8 to 18% gradient gels. Circled spots (1 and 3 to 8) exemplify proteins that were absent in certain strains or differed in intensity between strains. Spots 2 and 9 are indicated by arrows. The pI scale and the molecular weight (in thousands) are indicated at the bottom and to the left of each panel, respectively.
TABLE 1.
Number of protein spots that match on a number of gels within each disease group and on combinations of gels from the different groups
| No. of gels | % of total for disease group:
|
Total no. of spots | ||||||
|---|---|---|---|---|---|---|---|---|
| Ca | Du | Ga | CaDu | CaGa | DuGa | CaDuGa | ||
| 2 | 8 (14) | 7 (12) | 1 (2) | 19 (33) | 5 (9) | 18 (31) | 58 | |
| 3 | 0 | 3 (6) | 1 (2) | 24 (45) | 10 (19) | 9 (17) | 6 (11) | 53 |
| 4 | 0 | 0 | 0 | 9 (33) | 2 (7) | 4 (15) | 12 (44) | 27 |
| 5 | 5 (23) | 0 | 2 (9) | 15 (68) | 22 | |||
| 6 | 3 (13) | 1 (4) | 0 | 20 (83) | 24 | |||
| 7 | 1 (8) | 0 | 0 | 11 (92) | 12 | |||
| 8 | 0 | 0 | 0 | 19 (100) | 19 | |||
FIG. 2.
Two dendrograms showing the relationships among 12 H. pylori strains. Dendrogram A is produced by the method of average linkage (unweighted pair group method with averages), and B is produced by the method of neighbor joining. Both dendrograms are based on calculation method 2. At the bottom, the percentages of similarity are indicated, and to the right, there are strain identity numbers.
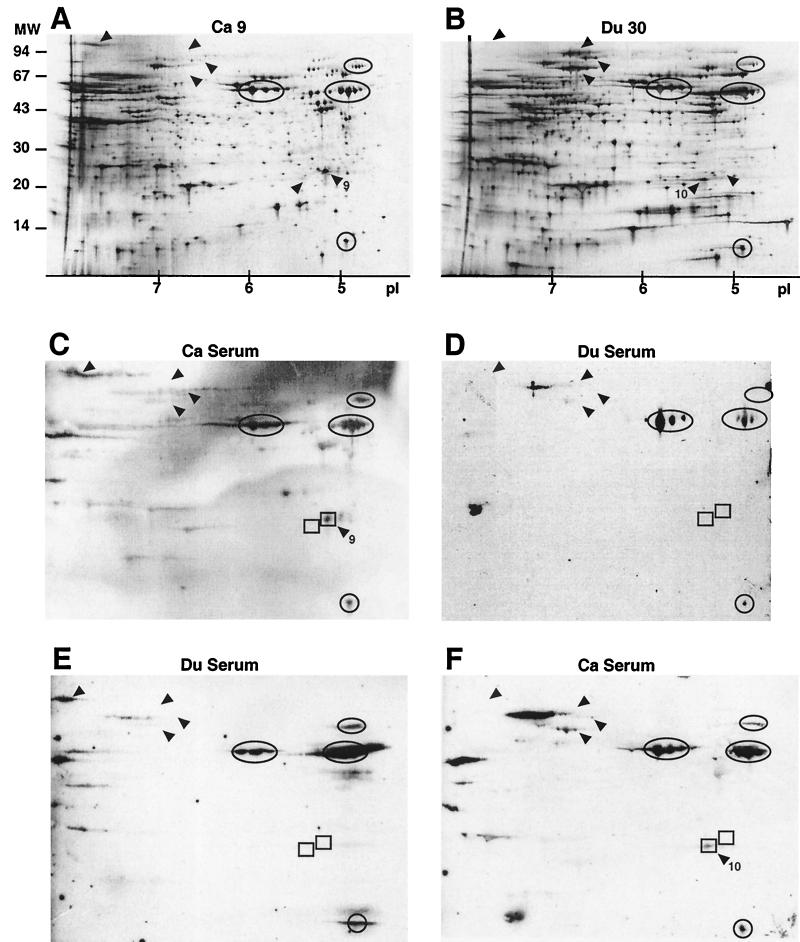
Cell extracts from one Ca and one Du strain were separated by 2-D PAGE on large gels and immunoblotted using pooled sera from 10 H. pylori-positive Ca or Du patients (Fig. 3). The immunogenic protein patterns were confirmed by small 2-D gels combined with immunoblotting. More than 600 protein spots were visualized by silver staining, and again, several differences were observed between the two strains. The immunogenic 2-D pattern of Ca 9 was similar when using sera from Ca or Du patients. One exception was spot 9, which was immunogenic with Ca serum but not with Du serum. Spot 9 was not present in the Du strain; however, a protein located close to this spot, spot 10, was immunogenic with Ca serum.
FIG. 3.
2-D PAGE followed by immunoblot of one Ca and one Du strain of H. pylori. Whole-cell protein extracts were focused on 180-mm 3-10 nonlinear IPG strips, separated on 12 to 14% gradient gels, and visualized by silver staining (A and B) or transferred to PVDF membranes and blotted using sera from Ca (C and F) or Du (D and E) patients. For silver staining and blots, 20 and 40 μg of protein, respectively, were applied on each strip. Some of the highly immunogenic proteins are circled (for use as internal standards), and differences in protein pattern and/or immunogenicity are indicated by arrowheads. The rectangles (spots 9 and 10) indicate strain-specific differences. A pI scale is given for panels A and B, and markers for molecular weight (in thousands) are given for panel A.
After a search for putative strain-specific spots, eight variable proteins were subjected to N-terminal sequencing (Fig. 1; Table 2). Spots 1 and 8 were identified as alkyl hydroperoxide reductase, TsaA, which is involved in reducing organic hydroperoxides and is induced by heat shock, salt, oxidative stress, and glucose limitation. Spot 2 was identified as inorganic pyrophosphatase, an enzyme which reduces pyrophosphate, formed as the product of the many biosynthetic reactions that utilize ATP. Protein spot 3 had an unknown function. Two different protein spots with similar molecular weights but different pI values were both identified as AroD, which is involved in aromatic amino acid synthesis from chorismate, the shikimate pathway. Protein spot 6 was shown to be ScoB, an enzyme which transfers coenzyme A between various carboxylic acids. A weak, secondary sequence, which was identical to protein TsaA, was also obtained from spot 6. Spot 7 was identified as elongation factor P. Spot 9 was N-terminally blocked and could not be identified.
TABLE 2.
Protein sequence data for selected spots from 2-D PAGE analysis of H. pylori strains
| Spota | Spot existence | Amino acid sequence | Identification |
|---|---|---|---|
| 1 | Ca 8, 9 and 11; Du 30 and 52; Ga 27 | MLVTKLAPDFKA | Alkyl hydroperoxide reductase (TsaA), 26-kDa antigen |
| 2 | Ca 8 and 9; Ga 26 and 27 | SNLDQLEVSHDA | Inorganic pyrophosphatase (PpaA) |
| 3 | All strains | SKYTQEQIKNLV | H. pylori predicted coding region |
| 4 | Ca 8 and 9; Du 18 and 57; Ga 27 | MKILVIQNQNLX | 3-Dehydroquinase type II (AroD) |
| 5 | All strains | MKILVIQGPNL | 3-Dehydroquinase type II (AroD) |
| 6 | Ca 8 and 1; Du 18 and 30; all Ga | MREAIIKRAAKE | Succinyl coenzyme A: 3-ketoacid-coenzyme A transferase subunit B (ScoB) |
| MLVTKLAPDFKA | TsaA (weak) | ||
| 7 | All strains | AIGMSELKKGLK | Elongation factor P |
| 8 | All Ca; Du 18 and 57; all Ga | MLVTKLAPDFKA | TsaA |
| 9 | Ca 9 and 1; Du 18 | N-terminally blocked |
Spot 3 was prepared from a Du strain, Du 30, whereas the remaining spots (1, 2, and 4 to 9) were prepared from the cancer strains Ca 8 and Ca 9.
In this study, the 2-D protein patterns of H. pylori strains isolated from three patient groups with different gastric diseases were computer analyzed: (i) the relationship between and among strains within the disease groups as indicated by protein spots existing in 2 to 8 of the 12 analyzed gels, (ii) the relationships between and among strains within the disease groups as shown by dendrograms or phylogenetic trees, (iii) the existence of certain strain-specific proteins, and (iv) the intensity of the spots on the gel, i.e., level of expression of the protein in this particular strain or strains. Clinical isolates of H. pylori display a high interstrain variation at the genomic level (9, 14, 22). However, a high divergence at the genomic level does not imply functional differences between strains because of the occurrence of silent mutations, e.g., mutations in noncoding regions or in the third codon. Mutations giving rise to amino acid changes are more likely to be of functional and selective nature and may result in pI changes of the proteins. Such changes explain some, but not all, of the divergence in the protein spot patterns observed in this study. The 2-D PAGE patterns of the different H. pylori strains were highly divergent; about 47% of the observed protein spots matched among all gels. The strain variation within each group of disease outcome, as well as differences and relationships between the groups, was revealed by the dendrograms: Ca 8 and Ca 9 strains and Ga 26 and Ga 27 strains were more closely related to each other than to the other strains within the same disease group. By this, we speculate that some H. pylori strains might be more associated with a specific disease than others, giving the clustering of some, but not all, strains within each disease group. Our results differ from previous results in which dendrograms did not show any disease-specific clustering of strains (13). The dendrogram pattern suggests that there might be disease-specific proteins in each group, which would be diagnostically interesting. However, it was difficult to obtain a grouping of strains according to disease from the protein patterns, since there were few matched spots within each group. In addition, the spots that matched within a group often matched with one or more protein spots from the other two groups. Protein sequences were determined for a few putative disease-specific protein spots, protein marker candidates, which were thought to be useful for H. pylori strain characterization. None of the eight sequenced proteins showed similarities with typical virulence factors. However, it should be noted that many proteins involved in virulence in H. pylori are basic (pI > 7) and may have been overlooked in this analysis. One possible immunogenic marker was found, and the apparent molecular mass and pI of this protein were about 26 kDa and 5.2, respectively. The N-terminal sequence was blocked, and its identity remains unknown. Interestingly, a protein with similar immunogenic characteristics has been found in another study (24), but whether these proteins are the same is not known. No proteins associated with specific gastric diseases, such as Ga or Du, have yet been identified, but a protein associated with mucosa-associated lymphoid tissue lymphoma was found by conventional sodium dodecyl sulfate-PAGE (6).
In conclusion, this study has further confirmed the extensive strain variation that the bacterium H. pylori exhibits, not only in its genome but also at the protein level. Although the spot matching revealed some disease-specific clustering of strains, the 2-D PAGE technique is too laborious and expensive to use for diagnostic purposes. However, comparisons of protein pI variation might be a better choice of method for analysis of H. pylori strain heterogeneity than conventional sodium dodecyl sulfate-PAGE. If marker proteins can be found, these specific proteins may be explored further and used both for laboratory tests, which analyze disease-specific H. pylori strains, and for diagnosis of the different diseases and outcomes associated with this widespread bacterium.
Acknowledgments
Thanks to Tom Rohan for proofreading assistance with the manuscript. Amino acid sequence data were obtained at the Protein Analysis Center, Karolinska Institute, Stockholm, Sweden.
This project was supported by grants from the Swedish Medical Research Council (project no. 10617 and 10848) and the Nanna Svartz and Åke Wiberg Foundations.
REFERENCES
- 1.Andersen H, Birkelund S, Christiansen G, Freundt E A. Electrophoretic analysis of proteins from Mycoplasma hominis strains detected by SDS-PAGE, two-dimensional gel electrophoresis and immunoblotting. J Gen Microbiol. 1987;133:181–191. doi: 10.1099/00221287-133-1-181. [DOI] [PubMed] [Google Scholar]
- 2.Blaser M J. In a world of black and white, Helicobacter pylori is grey. Ann Intern Med. 1999;130:695–697. doi: 10.7326/0003-4819-130-8-199904200-00019. [DOI] [PubMed] [Google Scholar]
- 3.Blaser M J. Not all Helicobacter pylori strains are created equal: should all be eliminated? Lancet. 1997;349:1020–1022. doi: 10.1016/S0140-6736(96)09133-7. [DOI] [PubMed] [Google Scholar]
- 4.Cash P. The application of two-dimensional polyacrylamide gel electrophoresis to medical microbiology: molecular epidemiology of viruses and bacteria. Electrophoresis. 1991;12:592–604. doi: 10.1002/elps.1150120721. [DOI] [PubMed] [Google Scholar]
- 5.Censini S, Lang C, Crabtree J E, Ghiara P, Borodovsky M, Rappuoli R, Covacci A. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc Natl Acad Sci USA. 1996;93:14648–14653. doi: 10.1073/pnas.93.25.14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang C S, Chen L T, Yang J C, Lin J T, Chang K C, Wang J T. Isolation of a Helicobacter pylori protein, FldA, associated with mucosa-associated lymphoid tissue lymphoma of the stomach. Gastroenterology. 1999;117:82–88. doi: 10.1016/s0016-5085(99)70553-6. [DOI] [PubMed] [Google Scholar]
- 7.Coles A M, Crosby H A, Pearce J H. Analysis of the human serological response to Chlamydia trachomatis 60-kDa proteins by two-dimensional electrophoresis and immunoblotting. FEMS Microbiol Lett. 1991;65:299–304. doi: 10.1016/0378-1097(91)90231-x. [DOI] [PubMed] [Google Scholar]
- 8.Collins P J, Juhl C, Lognonné J-L. Image analysis of 2D gels: considerations and insights. Cell Mol Biol. 1994;40:77–83. [PubMed] [Google Scholar]
- 9.Enroth H, Nyrén O, Engstrand L. One stomach-one strain: does Helicobacter pylori strain variation influence disease outcome? Dig Dis Sci. 1999;44:102–107. doi: 10.1023/a:1026658301825. [DOI] [PubMed] [Google Scholar]
- 10.Exner M M, Doig P, Trust T J, Hancock R E W. Isolation and characterization of a family of porin proteins from Helicobacter pylori. Infect Immun. 1995;63:1567–1572. doi: 10.1128/iai.63.4.1567-1572.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Givskov M, Eberl L, Molin S. Responses to nutrient starvation in Pseudomonas putida KT2442: two-dimensional electrophoretic analysis of starvation- and stress-induced proteins. J Bacteriol. 1994;176:4816–4824. doi: 10.1128/jb.176.16.4816-4824.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Go M F, Chan K Y, Versalovic J, Koeuth T, Graham D Y, Lupski J R. Cluster analysis of Helicobacter pylori genomic DNA fingerprints suggests gastroduodenal disease-specific associations. Scand J Gastroenterol. 1995;30:640–646. doi: 10.3109/00365529509096306. [DOI] [PubMed] [Google Scholar]
- 13.Hazell S L, Andrews R H, Mitchell H M, Daskalopoulous G. Genetic relationship among isolates of Helicobacter pylori: evidence for the existence of a Helicobacter pylori species-complex. FEMS Microbiol Lett. 1997;150:27–32. doi: 10.1111/j.1574-6968.1997.tb10345.x. [DOI] [PubMed] [Google Scholar]
- 14.Jiang Q, Hiratsuka K, Taylor D E. Variability of gene order in different Helicobacter pylori strains contributes to genome diversity. Mol Microbiol. 1996;20:833–842. doi: 10.1111/j.1365-2958.1996.tb02521.x. [DOI] [PubMed] [Google Scholar]
- 15.Kahn P. From genome to proteome: looking at a cell's proteins. Science. 1995;270:369–370. doi: 10.1126/science.270.5235.369. [DOI] [PubMed] [Google Scholar]
- 16.Kovarova H, Stulik J, Macela A, Lefkovits I, Skrabkova Z. Using two-dimensional gel electrophoresis to study immune response against intracellular bacterial infection. Electrophoresis. 1992;13:741–742. doi: 10.1002/elps.11501301160. [DOI] [PubMed] [Google Scholar]
- 17.Kwon D-H, El-Zaatari F A K, Woo J-S, Perng C-L, Graham D Y, Go M F. REP-PCR fragments as biomarkers for differentiating gastroduodenal disease-specific Helicobacter pylori strains. Dig Dis Sci. 1998;43:980–987. doi: 10.1023/a:1018818431828. [DOI] [PubMed] [Google Scholar]
- 18.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 19.Luft B, Jiang W, Munoz P, Dattwyler R J, Gorevic P D. Biochemical and immunological characterization of the surface proteins of Borrelia burgdorferi. Infect Immun. 1989;57:3637–3645. doi: 10.1128/iai.57.11.3637-3645.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McAtee C P, Fry K E, Berg D E. Identification of potential diagnostic and vaccine candidates of Helicobacter pylori by “proteome” technologies. Helicobacter. 1998;3:163–169. [PubMed] [Google Scholar]
- 21.Salaün L, Audibert C, Le Lay G, Burucoa C, Fauchère J-L, Picard B. Panmictic structure of Helicobacter pylori demonstrated by the comparative study of six genetic markers. FEMS Microbiol Lett. 1998;161:231–239. doi: 10.1111/j.1574-6968.1998.tb12953.x. [DOI] [PubMed] [Google Scholar]
- 22.Taylor D E, Eaton M, Chang N, Salama S M. Construction of a Helicobacter pylori genome map and demonstration of diversity at the genome level. J Bacteriol. 1992;174:6800–6806. doi: 10.1128/jb.174.21.6800-6806.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van der Ende A, Rauws E A J, Feller M, Mulder C J J, Tytgat G N, Dankert J. Heterogenous Helicobacter pylori isolates from members of a family with a history of peptic ulcer disease. Gastroenterology. 1996;111:638–647. doi: 10.1053/gast.1996.v111.pm8780568. [DOI] [PubMed] [Google Scholar]
- 24.Wang J-T, Chang C S, Lee C Z, Yang J C, Lin J T, Wang T H. Antibody to a Helicobacter pylori species specific antigen in patients with adenocarcinoma of the stomach. Biochem Biophys Res Commun. 1998;244:360–363. doi: 10.1006/bbrc.1998.8271. [DOI] [PubMed] [Google Scholar]
- 25.Zaiger Abshire K, Neidhardt F. Analysis of proteins synthesized by Salmonella typhimurium during growth within a host macrophage. J Bacteriol. 1993;175:3734–3743. doi: 10.1128/jb.175.12.3734-3743.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]